- Department of Physics and Astronomy, California State University, Northridge, CA, United States
Antibiotic resistance is a growing global problem that requires innovative therapeutic approaches and strategies for administering antibiotics. One promising approach is combination therapy, in which two or more drugs are combined to combat an infection. Along this line, the combination of antimicrobial peptides (AMPs) with conventional antibiotics has gained attention mainly due to the complementary mechanisms of action of AMPs and conventional antibiotics. In this article, we review both in vitro and in vivo studies that explore the synergy between AMPs and antibiotics. We highlight several mechanisms through which synergy is observed in in vitro experiments, including increasing membrane permeability, disrupting biofilms, directly potentiating antibiotic efficacy, and inhibiting resistance development. Moreover, in vivo studies reveal additional mechanisms such as enhanced/modulated immune responses, reduced inflammation, and improved tissue regeneration. Together, the current literature demonstrates that AMP-antibiotic combinations can substantially enhance efficacy of antibiotic therapies, including therapies against resistant bacteria, which represents a valuable enhancement to current antimicrobial strategies.
1 Introduction
Antimicrobial resistance represents a major global health challenge, threatening public health worldwide (World Health Organization et al., 2015; WHO, 2020, 2023). The emergence of multidrug-resistant bacteria has made many traditional antibiotics ineffective, limiting treatment options for infectious diseases (Rossolini et al., 2014; CDC, 2019). Innovative approaches that both enhance the effectiveness of existing antibiotics and prevent the development of resistance are crucial for addressing this growing crisis. Among these, synergistic therapeutic strategies that combine antibiotics have shown promise in combating drug-resistant infections (Lehár et al., 2009; Cokol et al., 2011; Tamma et al., 2012; León-Buitimea et al., 2020; Zhu et al., 2022).
In this context, the use of antimicrobial peptides (AMPs) in combination therapies has attracted significant attention. AMPs are natural defense molecules found in most multicellular organisms, including humans, that exhibit a broad spectrum of antimicrobial activity. Their unique “physics-based” mechanisms of action, which involve intricate electrostatic and hydrophobic interactions with lipid membrane structures, allow them to penetrate cell membranes and disrupt their integrity (Zasloff, 2002; Brogden, 2005; Gordon et al., 2005; Hancock et al., 2012; LaRock and Nizet, 2015). AMPs bind onto the membrane of their target cells and form pores across the membrane–a mechanism that is markedly different from conventional antibiotics that target specific bacterial enzymes, proteins, or pathways related to growth and proliferation (Matsuzaki et al., 1995, 1998; Epand and Vogel, 1999; Shai, 1999; Heller et al., 2000; Huang, 2000; Yang et al., 2001; Taheri-Araghi and Ha, 2007, 2010; Gualerzi et al., 2013). AMPs are known for their rapid bactericidal activity, low likelihood of resistance development, immunomodulatory properties, and potential for enhancing wound healing (Ganz, 2003; Jenssen et al., 2006; Hancock et al., 2012; Mookherjee et al., 2020).
The distinct pore-forming mechanism of AMPs made them promising candidates for combination therapies with conventional antibiotics. The outcome of combination therapy depends on various factors, including the choice of antibiotics, the target pathogen, and the environmental conditions (Greco, 1995; Chou, 2006; Cokol et al., 2011; Imamovic and Sommer, 2013; Yu et al., 2016; Grassi et al., 2017). Over recent decades, numerous in vivo, in vitro, and preclinical studies have explored combinations of AMPs with conventional antibiotics against various pathogens. While most published studies report synergistic effects that enhance antimicrobial activity and potentially mitigate resistancecite (Paula Jorge and Pereira, 2012; Yu et al., 2016; Grassi et al., 2017; Mhlongo et al., 2023), instances where combinations fail to exhibit synergy were also reported (He et al., 2015).
The current body of literature on AMP combination therapies, although still evolving, provides valuable insights into the mechanisms driving their synergistic action. Further research is needed to evaluate their long-term efficacy and toxicity in clinical settings.
This review article examines the existing literature on the use of AMPs in combination with traditional antibiotics. It explores the mechanisms of synergy, reviews empirical evidence from in vitro and in vivo studies, and discusses strategies to optimize these combinations. We also address the challenges and future directions in the development of these combination therapies to combat the growing threat of antimicrobial resistance.
2 Synergistic therapeutic approaches: AMPs and conventional antibiotics
The investigation of combination therapies include both AMP-AMP and AMP-conventional antibiotic combinations (Westerhoff et al., 1995; Mittal et al., 2016; León-Buitimea et al., 2020; Zhu et al., 2022; Mhlongo et al., 2023). Researchers have employed a variety of experimental techniques to assess the efficacy of these combinations, which can be broadly categorized into in vitro, in vivo, and preclinical studies (Paula Jorge and Pereira, 2012; Grassi et al., 2017). Various methodologies were used in the experimental approaches, which reflect the diverse expertise of research groups in this field, ranging from physics, chemistry, biochemistry, and biology to medical disciplines.
A significant portion of the literature focuses on in vitro experiments (Grassi et al., 2017; Mhlongo et al., 2023), in many cases targeting antimicrobial-resistant bacteria (Graham and Coote, 2007; Iwasaki et al., 2007; Desbois and Coote, 2011; Almaaytah et al., 2019; Morroni et al., 2019; Shang et al., 2019; Wongkaewkhiaw et al., 2019). This focus highlights a collective hope that combination therapies can address drug resistance challenges (Rossolini et al., 2014; CDC, 2019; WHO, 2023). While the knowledge on the outcomes of specific drug combinations are crucial, the broader objective is to find the underlying mechanisms that govern these interactions.
The effectiveness of AMP-antibiotic combinations is influenced by several factors besides the choice of specific AMP and antibiotic, including their concentrations and dosing regimens, the target organism, the presence and type of resistance mechanisms, and the local microenvironment (Maisetta et al., 2009; León-Buitimea et al., 2020; Zhu et al., 2022). A comprehensive understanding of these factors is essential for optimizing combination therapies and advancing the development of novel therapeutic strategies (Gordon et al., 2005; Jenssen et al., 2006; Mittal et al., 2016; Nešuta et al., 2016). Various mechanisms have been proposed to explain the synergy between AMPs and antibiotics (Grassi et al., 2017; Mhlongo et al., 2023). Detailed understanding of these mechanisms is crucial for the optimization and successful application of AMP-antibiotic combinations in combating bacterial infections. This understanding will not only facilitate the development of effective therapeutic strategies but also contribute to the broader goal of overcoming the challenges posed by antibiotic resistance (Brogden, 2005; Straus and Hancock, 2006; Hancock et al., 2012).
To enhance the antimicrobial activity of AMPs, several optimization strategies have been tested, such as peptide modification, the creation of synthetic analogs, and formulation techniques aimed at improving stability and bioavailability (Khara et al., 2015; Mittal et al., 2016; Nešuta et al., 2016). These efforts proved promising in improving efficacy of AMPs and broadening their clinical applications (Zhu et al., 2022). However, there is a need for further research to understand the long-term efficacy, toxicity, and pharmacokinetics of these therapeutic combinations (WHO, 2020, 2023).
3 In vitro studies on the efficacy of synergistic AMP combinations
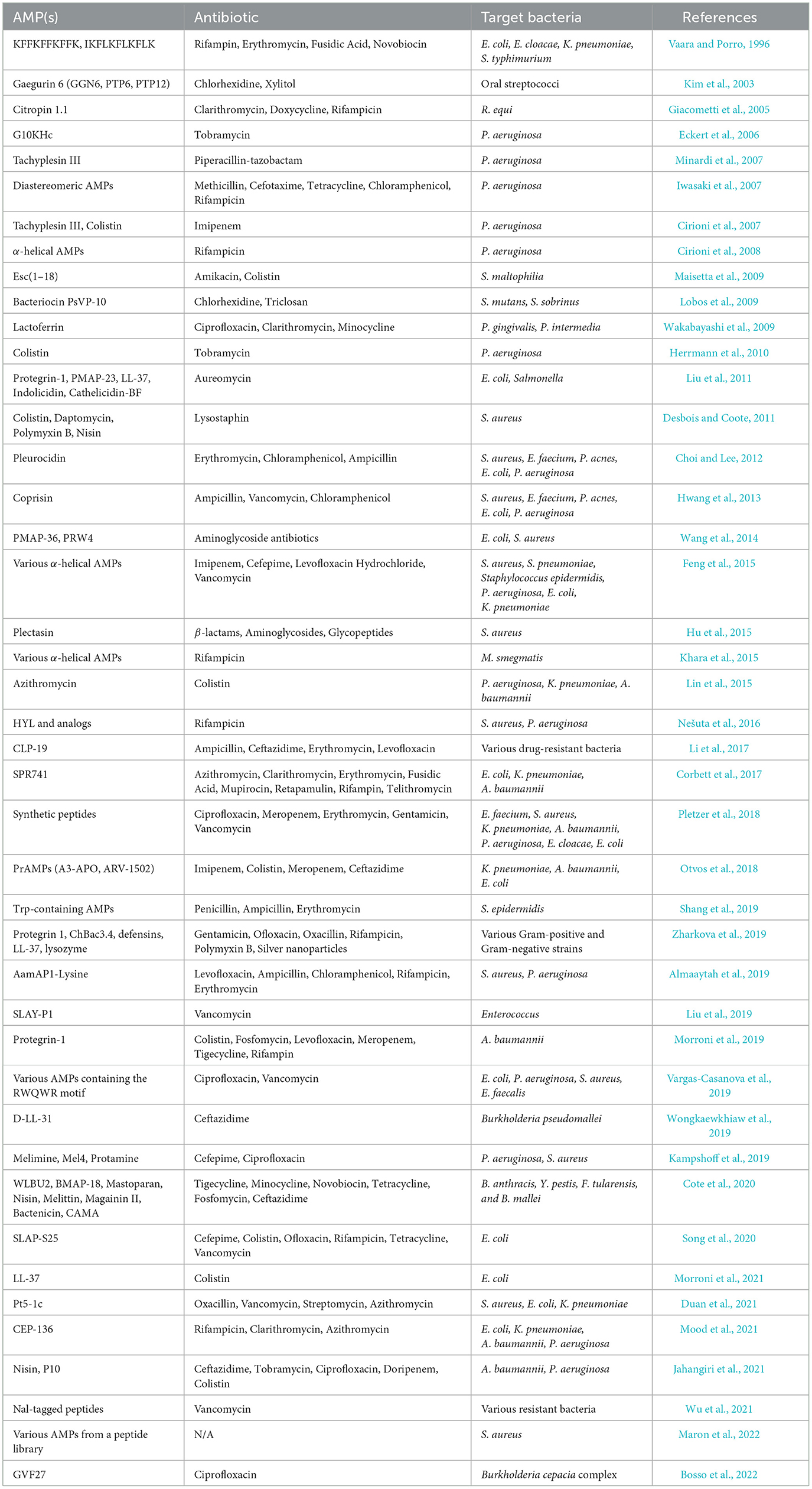
Numerous in vitro studies have demonstrated the potential of combination therapy using AMPs and conventional antibiotics. In vitro experiments provide a controlled environment to investigate the actions of AMP-antibiotic combinations and may reveal the underlying mechanisms, including effectiveness against antibiotic-resistant strains. Table 1 presents a comprehensive list of in vitro studies, detailing the specific AMPs, antibiotics, and organisms tested. From these references, four main categories of mechanisms by which AMPs enhance the efficacy of antibiotics have been identified. These mechanisms include increased membrane permeability, disruption of biofilms, direct enhancement of antibiotic efficacy, and inhibition of resistance mechanisms. These categories are schematically depicted in Figure 1 and are briefly discussed in this section to provide insights into the synergistic actions that can potentially overcome antibiotic resistance in clinical settings.
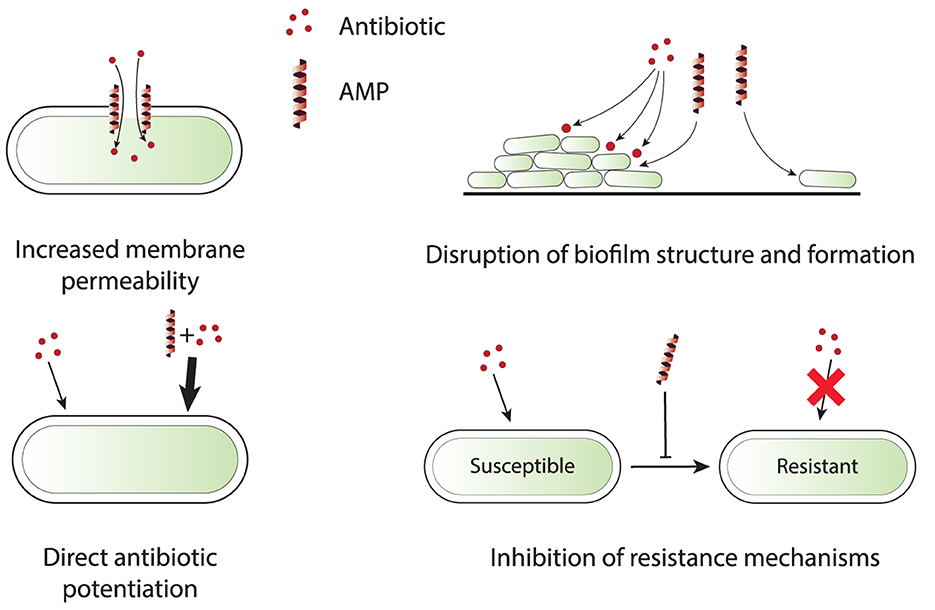
Figure 1. Schematic representation of four primary synergy mechanisms that are observed in in vitro experiments for AMP-antibiotic combinations. Increased membrane permeability: AMPs disrupt bacterial membrane integrity, allowing antibiotics to penetrate more efficiently, especially those targeting intracellular processes. Disruption of biofilm structure and formation: AMPs break down the biofilm matrix or inhibit its formation, exposing bacteria to antibiotics that can then effectively clear infections. Direct antibiotic potentiation: AMPs directly enhance the potency of antibiotics by modifying bacterial metabolic processes and increasing antibiotic sensitivity. Inhibition of resistance mechanisms: AMPs interfere with bacterial resistance mechanisms, such as efflux pumps and resistance gene expression, reducing the likelihood of resistance development and improving antibiotic efficacy.
3.1 Increased membrane permeability
AMPs compromise bacterial membrane integrity, thus enhancing the penetration and efficacy of antibiotics. Specifically, AMPs enable antibiotics that target intracellular processes to reach their sites of action more efficiently. This action is crucial against bacteria equipped with efflux pumps that actively expel antibiotic molecules. For instance, the synthetic peptide β-Ala-modified analogs of anoplin demonstrate significant membrane disruption, followed by enhanced antimicrobial potency and synergistic effects with conventional antibiotics against drug-resistant Pseudomonas aeruginosa, without prompting resistance development (Zhong et al., 2020).
Similarly, the antimicrobial peptide LL-37, when combined with colistin, showed strong synergy, drastically reducing the minimum inhibitory concentrations (MIC) against multidrug-resistant Escherichia coli. This highlights LL-37's role in membrane permeabilization and efflux pump circumvention (Morroni et al., 2021). In another study, pleurocidin was found to enhance antibiotic effectiveness by inducing hydroxyl radical formation, which contributes to membrane damage, as well as by disrupting bacterial cytoplasmic membranes, thereby promoting antibiotic entry (Choi and Lee, 2012).
Cathelicidin peptides also demonstrated such mechanisms by disrupting bacterial cell membranes and enhancing the bactericidal activity of co-administered antibiotics such as aureomycin against enteric pathogens (Liu et al., 2011). In similar synergistic interactions, the peptide coprisin not only exhibited intrinsic antimicrobial properties but also enhanced the activity of conventional antibiotics by facilitating their access to internal bacterial targets, crucial for combating biofilm-forming bacteria (Hwang et al., 2013).
The combined use of peptides PMAP-36 and PRW4 with aminoglycosides also showcased the role of membrane permeabilization, where these peptides enhance the intracellular delivery of antibiotics, contributing to a synergistic antibacterial effect (Wang et al., 2014).
These findings collectively highlight the critical role of AMPs in disrupting bacterial membranes, which not only enhances the efficacy of antibiotics against resistant strains but also broadens the therapeutic window of existing antimicrobial agents. This provides a robust strategy against infections that are difficult to treat with traditional antibiotics alone.
3.2 Disruption of biofilms
AMPs have demonstrated an ability not only for disrupting the biofilm matrix but also for inhibiting the formation of biofilms in the first place. As such, AMPs can facilitate the action of antibiotics by exposing otherwise protected bacteria within a biofilm structure, thus enhancing their susceptibility to treatments. This mechanism is particularly important in addressing persistent infections.
The peptide Cecropin A has been shown to disrupt uropathogenic E. coli biofilms, enhancing the efficacy of the antibiotic nalidixic acid, and leading to a synergistic clearance of infections without inducing resistance, a vital aspect in treating recurrent infections (Kalsy et al., 2020). Similarly, the peptide GVF27 targets biofilms formed by Burkholderia cepacia complex, known for its robust antibiotic resistance, enhancing the effects of traditional antibiotics like ciprofloxacin (Bosso et al., 2022).
In another study involving both in vitro and in vivo experiments with murine subcutaneous abscess model, the combination of synthetic peptides with meropenem and erythromycin significantly reduced infection sizes caused by ESKAPE pathogens (E. faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter spp), which are notorious for their biofilm-forming capabilities and multidrug resistance (Pletzer et al., 2018).
Additionally, peptide Pt5-1c, when combined with vancomycin and streptomycin, has shown to not only disrupt biofilms but also restore antibiotic sensitivity in multidrug-resistant strains, offering a dual advantage (Duan et al., 2021). The LL-37 peptide has also been reported to significantly reduce biofilm formation and, in combination with colistin, shows enhanced bactericidal activity against multidrug-resistant bacteria (Morroni et al., 2021).
The AMP-antibiotic synergy against biofilms have also been observed citropin 1.1, which, when combined with rifampicin, shows enhanced activity against Rhodococcus equi biofilms (Giacometti et al., 2005). Furthermore, combinations of early generation antibiotics with AMPs have been effective against biothreat agents like Burkholderia mallei and Yersinia pestis, indicating the potential of these combinations in both clinical applications (Cote et al., 2020).
3.3 Direct antibiotic potentiation
AMPs can also directly augment the potency of conventional antibiotics through mechanisms that modify bacterial metabolic processes and increase antibiotic sensitivity. For instance, the peptide CLP-19 was reported to synergistically enhance the effects of both bactericidal and bacteriostatic agents. It significantly reduces the adverse effects associated with antibiotic-induced endotoxin release, which is especially important in severe infections (Li et al., 2017).
Additionally, studies have shown that some AMP SPR741 can potentiate antibiotics by circumventing bacterial resistance mechanisms, such as efflux pumps in E. coli, thus enabling higher intracellular concentrations of antibiotics (Corbett et al., 2017).
In another example, SLAP-S25, a peptide incorporating non-natural amino acids, has a minimal antibacterial effect on its own but substantially increases the efficacy of a broad range of antibiotics against multidrug-resistant pathogens. This enhancement is due to its ability to act alongside antibiotics in overcoming bacterial defenses (Song et al., 2020).
Antibiotic azithromycin was shown to have enhance activity when used in combination with AMPs. Although traditionally not recommended for multidrug-resistant Gram-negative infections, azithromycin showed significant bactericidal activity when combined with colistin, pointing to potential against difficult-to-treat infections (Lin et al., 2015).
The combination of AMPs and conventional antibiotics not only holds promise in vitro but has also proven effective in vivo, significantly reducing infection levels and suggesting a viable strategy for enhancing the efficacy of existing antibiotics. This approach may help mitigate the development of antibiotic resistance and improve treatment outcomes for infections caused by resistant bacteria (Kampshoff et al., 2019).
3.4 Inhibition of resistance mechanisms
Some studies have reported the role of AMPs in inhibiting antibiotic resistance through various mechanisms, which is crucial for managing drug-resistant infections (Maron et al., 2022). This offers mechanisms that traditional antibiotics cannot exploit. For instance, proline-rich AMP A3-APO, in combination with colistin, was shown to significantly improve treatment efficacy and hindered resistance in treated pathogens (Otvos et al., 2018). This synergy is also seen in other combinations, where AMPs and antibiotics together achieve a greater antimicrobial effect and reduce the likelihood of resistance development (Zharkova et al., 2019).
Further, the AMP Cecropin A disrupts uropathogenic E. coli biofilms and inhibits efflux pump activity, the two mechanisms that induce bacterial resistance. Cecropin A was shown to slow the emergence of resistance while clearing infections (Kalsy et al., 2020). Similarly, β-Ala modified peptides like Ano-1β and Ano-8β exhibit strong membrane disruption and are noted for their low propensity for resistance development compared to standard antibiotics (Zhong et al., 2020).
Moreover, novel AMPs like CSM5-K5 not only exhibit potent bactericidal activity but also restore antibiotic sensitivity in previously resistant strains. Such findings are crucial, as they show that AMPs can reverse resistance trends-a significant advantage in the current era of high antibiotic failure rates (Thappeta et al., 2020).
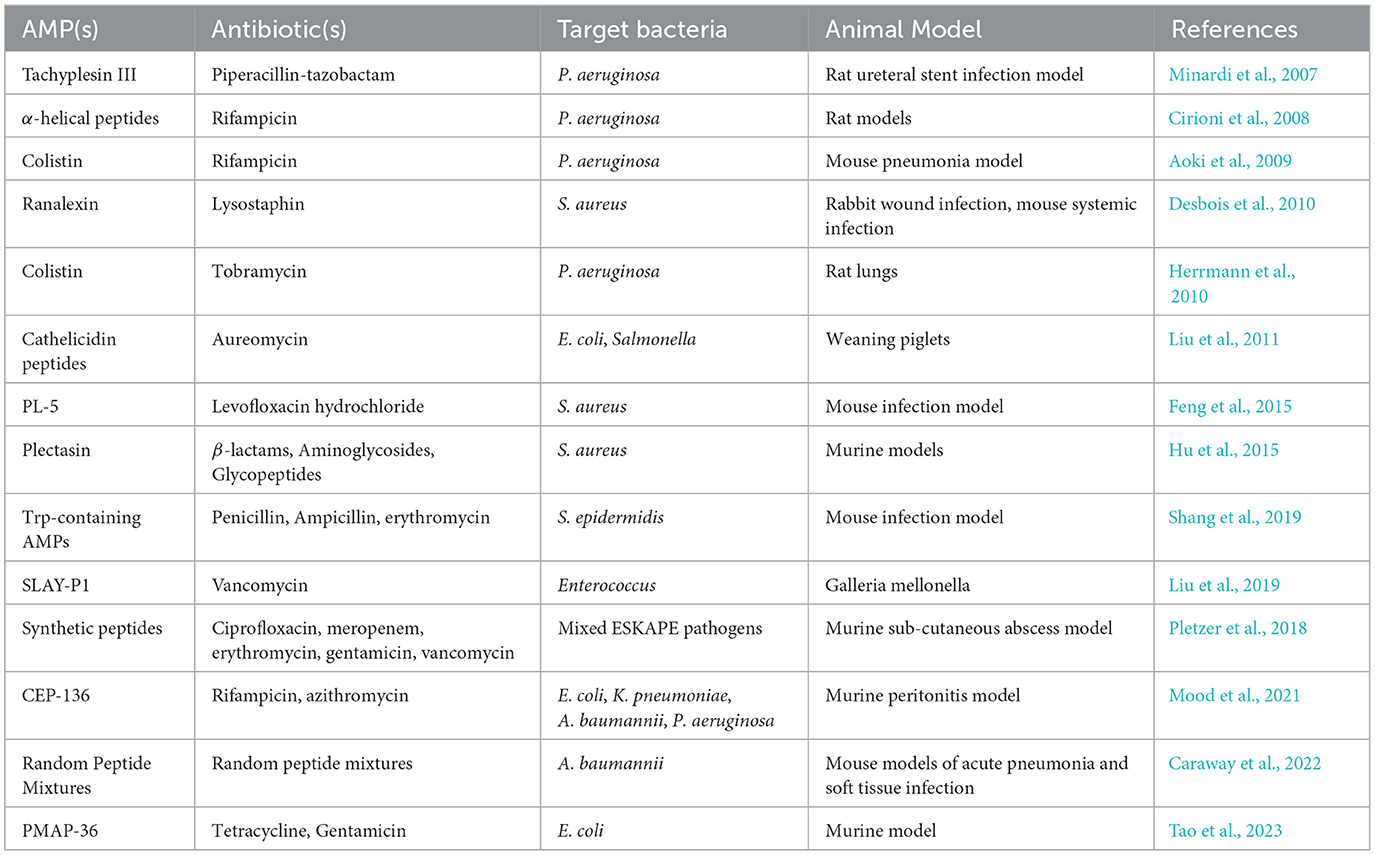
4 In vivo studies on the efficacy of synergistic AMP combinations
The translation of in vitro synergistic effects of AMP-antibiotic combinations to in vivo models represents a crucial step in the development of effective therapeutic strategies against infectious diseases. In the complex biological systems of living organisms experimented with in in vivo experiments, certain synergistic mechanisms not identifiable in in vitro studies come to light, providing additional insight into how these combinations might be optimized for clinical use. Table 2 presents a list of in vivo studies with information on the AMP-antibiotic combinations that were tested, as well as target bacteria and the animal model. Figure 2 highlight four mechanisms identified in in vivo experiments which are related to the host organism, thus not available to study in in vitro settings. In this section, we briefly discuss these mechanisms and their implications for enhancing the effectiveness of AMP-antibiotic therapies in clinical applications.
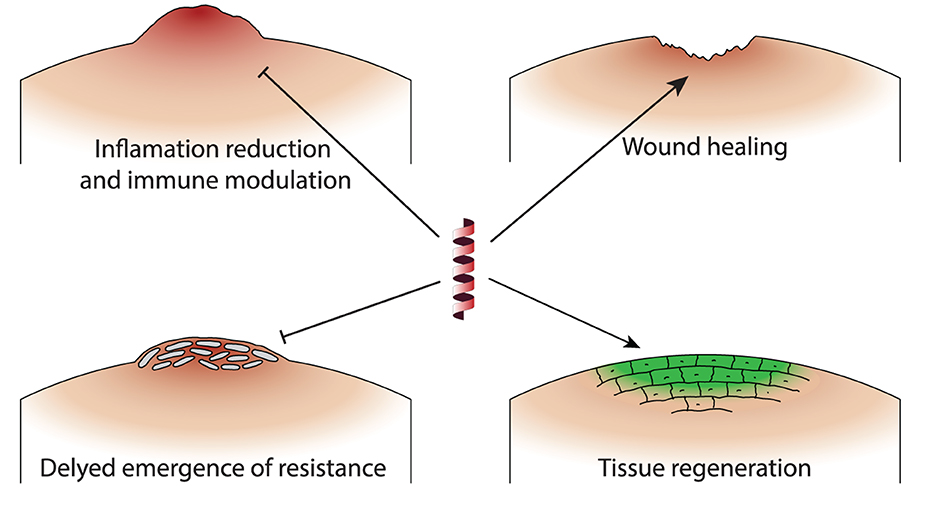
Figure 2. Schematic representation of the primary mechanisms by which AMP-antibiotic combinations demonstrate unique effects in in vivo studies. Inflammation reduction and immune modulation: AMPs enhance immune responses and reduce inflammation, promoting monocyte/macrophage migration to infection sites. Enhancing wound healing: Certain combinations prevent biofilm formation and support tissue regeneration, beneficial for infections with indwelling medical devices. Delayed emergence of resistance: AMPs sustain antibiotic efficacy and delay the development of resistant bacterial strains, extending the usefulness of antibiotics. Tissue regeneration: AMP-antibiotic combinations reduce MRSA burden in wound and systemic infections, demonstrating enhanced tissue regenerative properties.
4.1 Inflammation reduction and immune modulation
In in vivo settings, AMPs have been observed to enhance immune responses, a mechanism not tested in in vitro experiments. For example, the combination of PMAP-36 and tetracycline not only reduced bacterial load but also promoted the migration of monocytes/macrophages to the site of infection, significantly increasing survival in murine models (Tao et al., 2023).
Another unique in vivo mechanism involves the reduction of inflammation and cytokine production. Certain AMPs, when combined with antibiotics, have demonstrated a significant decrease in inflammatory markers and cytokine levels, contributing to better clinical outcomes. This effect is particularly beneficial in treating infections where inflammation exacerbates the disease process, such as in cystic fibrosis or sepsis (Aoki et al., 2009; Herrmann et al., 2010).
4.2 Delayed emergence of resistance
An important advantage of using AMPs in combination with antibiotics is the delayed emergence of bacterial resistance. In vivo studies demonstrate that AMPs can sustain antibiotic efficacy and reduce the evolution of resistant strains, providing a sustainable approach to managing bacterial infections. For instance, PMAP-36 has been shown to delay the development of resistance to tetracycline in porcine extraintestinal pathogenic E. coli, highlighting its potential as part of a combination therapy to extend the usefulness of existing antibiotics (Hu et al., 2015; Tao et al., 2023).
4.3 Enhancing wound healing and tissue regeneration
Beyond their antimicrobial action, some AMP-antibiotic combinations have been shown to possess wound healing and tissue-regenerative properties. Minardi et al. (2007) demonstrated that Tachyplesin III, when combined with piperacillin-tazobactam, prevented P. aeruginosa biofilm formation on ureteral stents in a rat model, highlighting the potential for preventing infections in patients with indwelling medical devices. Desbois et al. (2010) explored the synergistic effect of ranalexin with lysostaphin in wound and systemic infections, showing significant reduction in MRSA burden in both rabbit and mouse models.
5 Optimization strategies for enhancing synergy between AMPs and conventional antibiotics
Several methods have been used to optimize the efficacy of AMPs and their synergistic action with antibiotics. Optimization is essential for enhancing the collective efficacy of drug combinations, reducing the potential for resistance development, and minimizing toxicity. Here, we outline some of the strategies utilized to enhance the efficacy of AMP-antibiotic combinations.
5.1 AMP modification for enhanced stability and specificity
Modifying the structure of AMPs is a fundamental approach to enhance their utility in combination therapies. Strategies such as amino acid substitution, cyclization, and the incorporation of non-natural amino acids are employed to improve stability, membrane penetration, and specificity. Such modifications are designed to optimize the interaction of AMPs with bacterial membranes, thereby enhancing their antimicrobial effectiveness while reducing potential cytotoxic effects on mammalian cells (Giacometti et al., 2005; Taheri-Araghi and Ha, 2007; Choi and Lee, 2012; Khara et al., 2015; Mittal et al., 2016; Nešuta et al., 2016).
5.2 Tailored combination therapy based on pathogen profile
Tailoring therapy to the specific pathogens and their resistance mechanisms can significantly enhance the efficacy of AMP-antibiotic combinations. This precision medicine approach involves selecting AMPs and antibiotics that complement each other's mechanisms of action and are effective against the resistance profiles of the target bacteria. Consideration of the local microenvironment at the infection site is crucial to this strategy. Although still in its infancy, such targeted approaches hold great promise for improving therapeutic outcomes (Yu et al., 2016; Zhu et al., 2022).
5.3 Utilization of high-throughput screening and computational models
To further refine optimization strategies, high-throughput screening methods and computational modeling serve as essential tools. These techniques allow for the rapid identification of effective AMP-antibiotic pairs by predicting potential synergies based on the properties of the drugs and the characteristics of the target bacteria. This can significantly accelerate the development of effective combination therapies (Maisetta et al., 2009; Paula Jorge and Pereira, 2012; Mookherjee et al., 2020).
5.4 Bioinformatics tools and AMP databases
The use of bioinformatics tools and comprehensive databases that catalog information on AMPs supports the discovery and design of novel AMPs. These resources are invaluable for researchers seeking to develop new synergistic combinations that can be effectively integrated into clinical practice (Maisetta et al., 2009; Paula Jorge and Pereira, 2012).
These strategies represent a multifaceted approach to enhancing the synergy between AMPs and conventional antibiotics. Each method contributes to a deeper understanding and more effective application of these combinations in the battle against resistant infections. As research progresses, these optimization strategies will likely evolve, offering new avenues for combating antimicrobial resistance and optimizing therapeutic strategies.
6 Future directions
Research into synergistic therapeutic approaches involving AMPs and conventional antibiotics has made significant progress in recent years, which presents a promising strategy against infections, including resistant bacteria. Both laboratory (in vitro) and live subject (in vivo) studies have demonstrated the efficacy of AMP-antibiotic combinations against a wide range of pathogens, including drug-resistant strains and infections associated with biofilms (Tables 1, 2 and references therein). These studies have also demonstrated a range of mechanistic models that lead to the synergistic action of that enhance antimicrobial effectiveness, suppress the emergence of resistance, and even promote wound healing and tissue regeneration (Sections 3 and 4).
However, despite these advances, there are several challenges and limitations to the future development of AMP-antibiotic combination therapies. Firstly, while in vitro assays offer valuable insights into the benefits of AMP-antibiotic synergy, translating these findings into in vivo models and further in clinical settings is highly complex. More extensive preclinical assessments and clinical trials are needed to confirm the safety, efficacy, and pharmacokinetics of these combination therapies in human subjects (Seyhan, 2019; Dijksteel et al., 2021; Talapko et al., 2022).
Furthermore, substantially more detailed understanding of the mechanisms driving synergistic interactions is necessary to inform the design of effective combination therapies for treating infectious diseases. Precise mechanistic knowledge would predict the expected outcomes of combination therapy. Among various methodologies, high-throughput screening and computational models can expedite the identification of optimal AMP-antibiotic pairs and forecast synergistic effects. This knowledge would enable us to refine optimization strategies for enhancing synergy and mitigating resistance development. Peptide engineering, dosage optimization, and tailored delivery mechanisms for different pathogens and resistance profiles hold great potential but require further refinement and validation (Tan et al., 2021; Zhang and Yang, 2022).
Addressing biofilm-related infections remains paramount. While AMP-antibiotic combinations have shown promise in combating biofilms, further research is needed to confirm the efficacy of AMPs against biofilms, as some studies have highlighted limitations in their effectiveness (Taheri-Araghi and Guerbidjian, 2020). Similarly, efforts to overcome drug resistance and expand the use of AMP-antibiotic combinations are ongoing. Besides studies reporting the lack of resistant development against AMPs, there is also evidence that microorganisms have developed resistance to certain AMPs (Perron et al., 2006).
In conclusion, research on AMP-antibiotic combinations offers hope for combating antimicrobial resistance. However, addressing remaining challenges and guiding future research will be essential to fully realize their potential in treating infections. Collaborative efforts and innovation are key to revolutionizing infectious disease management and reducing antimicrobial resistance worldwide.
Author contributions
ST-A: Investigation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a Research, Scholarship, and Creative Activity (RSCA) grant from the California State University Office of the Chancellor, through the Office of Research and Sponsored Projects at California State University, Northridge (CSUN).
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Almaaytah, A., Abualhaijaa, A., and Alqudah, O. (2019). The evaluation of the synergistic antimicrobial and antibiofilm activity of AamAP1-Lysine with conventional antibiotics against representative resistant strains of both Gram-positive and Gram-negative bacteria. Infect. Drug Resist. 12, 1371–1380. doi: 10.2147/IDR.S204626
Aoki, N., Tateda, K., Kikuchi, Y., Kimura, S., Miyazaki, C., Ishii, Y., et al. (2009). Efficacy of colistin combination therapy in a mouse model of pneumonia caused by multidrug-resistant Pseudomonas aeruginosa. J. Antimicrob. Chemother. 63, 534–542. doi: 10.1093/jac/dkn530
Bosso, A., Gaglione, R., Di Girolamo, R., Veldhuizen, E. J., García-Vello, P., Fusco, S., et al. (2022). Human cryptic host defence peptide gvf27 exhibits anti-infective properties against biofilm forming members of the burkholderia cepacia complex. Pharmaceuticals 15:260. doi: 10.3390/ph15020260
Brogden, K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250. doi: 10.1038/nrmicro1098
Caraway, H. E., Lau, J. Z., Maron, B., Oh, M. W., Belo, Y., Brill, A., et al. (2022). Antimicrobial random peptide mixtures eradicate Acinetobacter baumannii biofilms and inhibit mouse models of infection. Antibiotics 11:413. doi: 10.3390/antibiotics11030413
CDC (2019). Antibiotic resistance threats in the United States, 2019. Atlanta, GA: US Department of Health and Human Services, Centres for Disease Control and Prevention.
Choi, H., and Lee, D. G. (2012). Antimicrobial peptide pleurocidin synergizes with antibiotics through hydroxyl radical formation and membrane damage, and exerts antibiofilm activity. Biochim. Biophys. Acta-Gen. Subj. 1820, 1831–1838. doi: 10.1016/j.bbagen.2012.08.012
Chou, T.-C. (2006). Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 58, 621–681. doi: 10.1124/pr.58.3.10
Cirioni, O., Ghiselli, R., Silvestri, C., Kamysz, W., Orlando, F., Mocchegiani, F., et al. (2007). Efficacy of tachyplesin iii, colistin, and imipenem against a multiresistant Pseudomonas aeruginosa strain. Antimicrob. Agents Chemother. 51, 2005–2010. doi: 10.1128/AAC.01576-06
Cirioni, O., Silvestri, C., Ghiselli, R., Orlando, F., Riva, A., Mocchegiani, F., et al. (2008). Protective effects of the combination of α-helical antimicrobial peptides and rifampicin in three rat models of Pseudomonas aeruginosa infection. J. Antimicrob. Chemother. 62, 1332–1338. doi: 10.1093/jac/dkn393
Cokol, M., Chua, H. N., Tasan, M., Mutlu, B., Weinstein, Z. B., Suzuki, Y., et al. (2011). Systematic exploration of synergistic drug pairs. Mol. Syst. Biol. 7:544. doi: 10.1038/msb.2011.71
Corbett, D., Wise, A., Langley, T., Skinner, K., Trimby, E., Birchall, S., et al. (2017). Potentiation of antibiotic activity by a novel cationic peptide: potency and spectrum of activity of spr741. Antimicrob. Agents Chemother. 61, 10–1128. doi: 10.1128/AAC.00200-17
Cote, C. K., Blanco, I. I., Hunter, M., Shoe, J. L., Klimko, C. P., Panchal, R. G., et al. (2020). Combinations of early generation antibiotics and antimicrobial peptides are effective against a broad spectrum of bacterial biothreat agents. Microb. Pathog. 142:104050. doi: 10.1016/j.micpath.2020.104050
Desbois, A. P., and Coote, P. J. (2011). Bactericidal synergy of lysostaphin in combination with antimicrobial peptides. Eur. J. Clin. Microbiol. Infect. Dis. 30, 1015–1021. doi: 10.1007/s10096-011-1188-z
Desbois, A. P., Gemmell, C. G., and Coote, P. J. (2010). In vivo efficacy of the antimicrobial peptide ranalexin in combination with the endopeptidase lysostaphin against wound and systemic meticillin-resistant Staphylococcus aureus (MRSA) infections. Int. J. Antimicrob. Agents 35, 559–565. doi: 10.1016/j.ijantimicag.2010.01.016
Dijksteel, G. S., Ulrich, M. M. W., Middelkoop, E., and Boekema, B. K. H. L. (2021). Review: lessons learned from clinical trials using antimicrobial peptides (AMPS). Front. Microbiol. 12:616979. doi: 10.3389/fmicb.2021.616979
Duan, H., Zhang, X., Li, Z., Yuan, J., Shen, F., Zhang, S., et al. (2021). Synergistic effect and antibiofilm activity of an antimicrobial peptide with traditional antibiotics against multi-drug resistant bacteria. Microb. Pathog. 158:105056. doi: 10.1016/j.micpath.2021.105056
Eckert, R., Brady, K. M., Greenberg, E. P., Qi, F., Yarbrough, D. K., He, J., et al. (2006). Enhancement of antimicrobial activity against Pseudomonas aeruginosa by coadministration of g10khc and tobramycin. Antimicrob. Agents Chemother. 50, 3833–3838. doi: 10.1128/AAC.00509-06
Epand, R. M., and Vogel, H. J. (1999). Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta, 1462, 11–28. doi: 10.1016/S0005-2736(99)00198-4
Feng, Q., Huang, Y., Chen, M., Li, G., and Chen, Y. (2015). Functional synergy of α-helical antimicrobial peptides and traditional antibiotics against gram-negative and gram-positive bacteria in vitro and in vivo. Eur. J. Clin. Microbiol. Infect. Dis. 34, 197–204. doi: 10.1007/s10096-014-2219-3
Ganz, T. (2003). Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3, 710–720. doi: 10.1038/nri1180
Giacometti, A., Cirioni, O., Kamysz, W., Silvestri, C., Del Prete, M. S., Licci, A., et al. (2005). In vitro activity of citropin 1.1 alone and in combination with clinically used antimicrobial agents against rhodococcus equi. J. Antimicrob. Chemother. 56, 410–412. doi: 10.1093/jac/dki236
Gordon, Y. J., Romanowski, E. G., and McDermott, A. M. (2005). A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 30, 505–515. doi: 10.1080/02713680590968637
Graham, S., and Coote, P. J. (2007). Potent, synergistic inhibition of Staphylococcus aureus upon exposure to a combination of the endopeptidase lysostaphin and the cationic peptide ranalexin. J. Antimicrob. Chemother. 59, 759–762. doi: 10.1093/jac/dkl539
Grassi, L., Maisetta, G., Esin, S., and Batoni, G. (2017). Combination strategies to enhance the efficacy of antimicrobial peptides against bacterial biofilms. Front. Microbiol. 8:2409. doi: 10.3389/fmicb.2017.02409
Greco, W. R. (1995). The search for synergy: a critical review from a response surface perspective. Pharmacol. Rev. 47, 331–385.
Gualerzi, C. O., Brandi, L., Fabbretti, A., and Pon, C. L. (2013). Antibiotics: targets, mechanisms and resistance. Hoboken, NJ: John Wiley & Sons. doi: 10.1002/9783527659685
Hancock, R. E., Nijnik, A., and Philpott, D. J. (2012). Modulating immunity as a therapy for bacterial infections. Nat. Rev. Microbiol. 10, 243–254. doi: 10.1038/nrmicro2745
He, J., Starr, C. G., and Wimley, W. C. (2015). A lack of synergy between membrane-permeabilizing cationic antimicrobial peptides and conventional antibiotics. Biochim. Biophys. Acta 1848, 8–15. doi: 10.1016/j.bbamem.2014.09.010
Heller, W. T., Waring, A. J., Lehrer, R. I., Harroun, T. A., Weiss, T. M., Yang, L., et al. (2000). Membrane thinning effect of the beta-sheet antimicrobial protegrin. Biochemistry 39, 139–145. doi: 10.1021/bi991892m
Herrmann, G., Yang, L., Wu, H., Song, Z., Wang, H., Høiby, N., et al. (2010). Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J. Infect. Dis. 202, 1585–1592. doi: 10.1086/656788
Hu, Y., Liu, A., Vaudrey, J., Vaiciunaite, B., Moigboi, C., McTavish, S. M., et al. (2015). Combinations of β-lactam or aminoglycoside antibiotics with plectasin are synergistic against methicillin-sensitive and methicillin-resistant Staphylococcus aureus. PLoS ONE 10:e0117664. doi: 10.1371/journal.pone.0117664
Huang, H. W. (2000). Action of antimicrobial peptides: two-state model. Biochemistry 39, 8347–8352. doi: 10.1021/bi000946l
Hwang, I.-s., Hwang, J.-S., Hwang, J. H., Choi, H., Lee, E., Kim, Y., and Lee, D. G. (2013). Synergistic effect and antibiofilm activity between the antimicrobial peptide coprisin and conventional antibiotics against opportunistic bacteria. Curr. Microbiol. 66, 56–60. doi: 10.1007/s00284-012-0239-8
Imamovic, L., and Sommer, M. O. (2013). Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 5:204ra132. doi: 10.1126/scitranslmed.3006609
Iwasaki, T., Saido-Sakanaka, H., Asaoka, A., Taylor, D., Ishibashi, J., Yamakawa, M., et al. (2007). In vitro activity of diastereomeric antimicrobial peptides alone and in combination with antibiotics against methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa. J. Insect Biotechnol. Sericol. 76, 25–29.
Jahangiri, A., Neshani, A., Mirhosseini, S. A., Ghazvini, K., Zare, H., Sedighian, H., et al. (2021). Synergistic effect of two antimicrobial peptides, nisin and p10 with conventional antibiotics against extensively drug-resistant Acinetobacter baumannii and colistin-resistant Pseudomonas aeruginosa isolates. Microb. Pathog. 150:104700. doi: 10.1016/j.micpath.2020.104700
Jenssen, H., Hamill, P., and Hancock, R. E. W. (2006). Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491–511. doi: 10.1128/CMR.00056-05
Kalsy, M., Tonk, M., Hardt, M., Dobrindt, U., Zdybicka-Barabas, A., Cytrynska, M., et al. (2020). The insect antimicrobial peptide cecropin a disrupts uropathogenic Escherichia coli biofilms. Npj Biofilms Microbiomes 6:6. doi: 10.1038/s41522-020-0116-3
Kampshoff, F., Willcox, M. D., and Dutta, D. (2019). A pilot study of the synergy between two antimicrobial peptides and two common antibiotics. Antibiotics 8:60. doi: 10.3390/antibiotics8020060
Khara, J. S., Lim, F. K., Wang, Y., Ke, X.-Y., Voo, Z. X., Yang, Y. Y., et al. (2015). Designing α-helical peptides with enhanced synergism and selectivity against mycobacterium smegmatis: discerning the role of hydrophobicity and helicity. Acta Biomater. 28, 99–108. doi: 10.1016/j.actbio.2015.09.015
Kim, S. S., Kim, S., Kim, E., Hyun, B., Kim, K.-K., Lee, B. J., et al. (2003). Synergistic inhibitory effect of cationic peptides and antimicrobial agents on the growth of oral streptococci. Caries Res. 37, 425–430. doi: 10.1159/000073394
LaRock, C. N., and Nizet, V. (2015). Cationic antimicrobial peptide resistance mechanisms of streptococcal pathogens. Biochim. Biophys. Acta 1848, 3047–3054. doi: 10.1016/j.bbamem.2015.02.010
Lehár, J., Krueger, A. S., Avery, W., Heilbut, A. M., Johansen, L. M., Price, E. R., et al. (2009). Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol. 27, 659–666. doi: 10.1038/nbt.1549
León-Buitimea, A., Garza-Cárdenas, C. R., Garza-Cervantes, J. A., Lerma-Escalera, J. A., and Morones-Ramírez, J. R. (2020). The demand for new antibiotics: antimicrobial peptides, nanoparticles, and combinatorial therapies as future strategies in antibacterial agent design. Front. Microbiol. 11:1669. doi: 10.3389/fmicb.2020.01669
Li, D., Yang, Y., Tian, Z., Lv, J., Sun, F., Wang, Q., et al. (2017). Synergistic antibiotic effect of looped antimicrobial peptide clp-19 with bactericidal and bacteriostatic agents. Oncotarget 8:55958. doi: 10.18632/oncotarget.18124
Lin, L., Nonejuie, P., Munguia, J., Hollands, A., Olson, J., Dam, Q., et al. (2015). Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant gram-negative bacterial pathogens. EBioMedicine 2, 690–698. doi: 10.1016/j.ebiom.2015.05.021
Liu, Y., Jia, Y., Yang, K., Li, R., Xiao, X., Wang, Z., et al. (2019). Antagonizing vancomycin resistance in Enterococcus by surface localized antimicrobial display-derived peptides. ACS Infect. Dis. 6, 761–767. doi: 10.1021/acsinfecdis.9b00164
Liu, Y., Luan, C., Xia, X., An, S., and Wang, Y. (2011). Antibacterial activity, cytotoxicity and mechanisms of action of cathelicidin peptides against enteric pathogens in weaning piglets. Int. J. Pept. Res. Ther. 17, 175–184. doi: 10.1007/s10989-011-9255-y
Lobos, O., Padilla, A., and Padilla, C. (2009). In vitro antimicrobial effect of bacteriocin psvp-10 in combination with chlorhexidine and triclosan against Streptococcus mutans and Streptococcus sobrinus strains. Arch. Oral Biol. 54, 230–234. doi: 10.1016/j.archoralbio.2008.11.007
Maisetta, G., Mangoni, M. L., Esin, S., Pichierri, G., Capria, A. L., Brancatisano, F. L., et al. (2009). In vitro bactericidal activity of the n-terminal fragment of the frog peptide esculentin-1b (esc 1-18) in combination with conventional antibiotics against Stenotrophomonas maltophilia. Peptides 30, 1622–1626. doi: 10.1016/j.peptides.2009.06.004
Maron, B., Rolff, J., Friedman, J., and Hayouka, Z. (2022). Antimicrobial peptide combination can hinder resistance evolution. Microbiol. Spectr. 10, e00973-22. doi: 10.1128/spectrum.00973-22
Matsuzaki, K., Sugishita, K., Fujii, N., and Miyajima, K. (1995). Molecular basis for membrane selectivity of an antimicrobial peptide, magainin 2. Biochemistry 34, 3423–3429. doi: 10.1021/bi00010a034
Matsuzaki, K., Sugishita, K., Ishibe, N., Ueha, M., Nakata, S., Miyajima, K., et al. (1998). Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry 37, 11856–11863. doi: 10.1021/bi980539y
Mhlongo, J. T., Waddad, A. Y., Albericio, F., and de la Torre, B. G. (2023). Antimicrobial peptide synergies for fighting infectious diseases. Adv. Sci. 10:2300472. doi: 10.1002/advs.202300472
Minardi, D., Ghiselli, R., Cirioni, O., Giacometti, A., Kamysz, W., Orlando, F., et al. (2007). The antimicrobial peptide tachyplesin iii coated alone and in combination with intraperitoneal piperacillin-tazobactam prevents ureteral stent Pseudomonas infection in a rat subcutaneous pouch model. Peptides 28, 2293–2298. doi: 10.1016/j.peptides.2007.10.001
Mittal, S., Maurya, I. K., Kaur, S., Swami, A., Jain, R., Wangoo, N., et al. (2016). Insights into mechanistic and synergistic aspects of novel synthetic short cationic antibacterial peptides. ChemistrySelect 1, 5510–5516. doi: 10.1002/slct.201600947
Mood, E. H., Goltermann, L., Brolin, C., Cavaco, L. M., Nejad, A. J., Yavari, N., et al. (2021). Antibiotic potentiation in multidrug-resistant gram-negative pathogenic bacteria by a synthetic peptidomimetic. ACS Infect. Dis. 7, 2152–2163. doi: 10.1021/acsinfecdis.1c00147
Mookherjee, N., Anderson, M. A., Haagsman, H. P., and Davidson, D. J. (2020). Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug Discov. 19, 311–332. doi: 10.1038/s41573-019-0058-8
Morroni, G., Sante, L. D., Simonetti, O., Brescini, L., Kamysz, W., Kamysz, E., et al. (2021). Synergistic effect of antimicrobial peptide ll-37 and colistin combination against multidrug-resistant Escherichia coli isolates. Future Microbiol. 16, 221–227. doi: 10.2217/fmb-2020-0204
Morroni, G., Simonetti, O., Brenciani, A., Brescini, L., Kamysz, W., Kamysz, E., et al. (2019). In vitro activity of protegrin-1, alone and in combination with clinically useful antibiotics, against Acinetobacter baumannii strains isolated from surgical wounds. Med. Microbiol. Immunol. 208, 877–883. doi: 10.1007/s00430-019-00624-7
Nešuta, O., Hexnerová, R., Budesinsky, M., Slaninová, J., Bednárová, L., Hadravová, R., et al. (2016). Antimicrobial peptide from the wild bee hylaeus signatus venom and its analogues: structure-activity study and synergistic effect with antibiotics. J. Nat. Prod. 79, 1073–1083. doi: 10.1021/acs.jnatprod.5b01129
Otvos Jr, L., Ostorhazi, E., Szabo, D., Zumbrun, S. D., Miller, L. L., Halasohoris, S. A., et al. (2018). Synergy between proline-rich antimicrobial peptides and small molecule antibiotics against selected gram-negative pathogens in vitro and in vivo. Front. Chem. 6:309. doi: 10.3389/fchem.2018.00309
Paula Jorge, A. L., and Pereira, M. O. (2012). New trends in peptide-based anti-biofilm strategies: a review of recent achievements and bioinformatic approaches. Biofouling 28, 1033–1061. doi: 10.1080/08927014.2012.728210
Perron, G. G., Zasloff, M., and Bell, G. (2006). Experimental evolution of resistance to an antimicrobial peptide. Proc. R. Soc. B Biol. Sci. 273, 251–256. doi: 10.1098/rspb.2005.3301
Pletzer, D., Mansour, S. C., and Hancock, R. E. (2018). Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant eskape pathogens. PLoS Pathog. 1:e1007084. doi: 10.1371/journal.ppat.1007084
Rossolini, G. M., Arena, F., Pecile, P., and Pollini, S. (2014). Update on the antibiotic resistance crisis. Curr. Opin. Pharmacol. 18, 56–60. doi: 10.1016/j.coph.2014.09.006
Seyhan, A. A. (2019). Lost in translation: the valley of death across preclinical and clinical divide-identification of problems and overcoming obstacles. Transl. Med. Commun. 4, 1–19. doi: 10.1186/s41231-019-0050-7
Shai, Y. (1999). Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1462, 55–70. doi: 10.1016/S0005-2736(99)00200-X
Shang, D., Liu, Y., Jiang, F., Ji, F., Wang, H., Han, X., et al. (2019). Synergistic antibacterial activity of designed trp-containing antibacterial peptides in combination with antibiotics against multidrug-resistant Staphylococcus epidermidis. Front. Microbiol. 10:2719. doi: 10.3389/fmicb.2019.02719
Song, M., Liu, Y., Huang, X., Ding, S., Wang, Y., Shen, J., et al. (2020). A broad-spectrum antibiotic adjuvant reverses multidrug-resistant gram-negative pathogens. Nat. Microbiol. 5, 1040–1050. doi: 10.1038/s41564-020-0723-z
Straus, S. K., and Hancock, R. E. (2006). Mode of action of the new antibiotic for gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta 1758, 1215–1223. doi: 10.1016/j.bbamem.2006.02.009
Taheri-Araghi, S., and Guerbidjian, O. (2020). A layer of dead cells at the periphery protects biofilms from antimicrobial peptides. Biophys. J. 118:382a. doi: 10.1016/j.bpj.2019.11.2180
Taheri-Araghi, S., and Ha, B.-Y. (2007). Physical basis for membrane-charge selectivity of cationic antimicrobial peptides. Phys. Rev. Lett. 98:168101. doi: 10.1103/PhysRevLett.98.168101
Taheri-Araghi, S., and Ha, B.-Y. (2010). Cationic antimicrobial peptides: a physical basis for their selective membrane-disrupting activity. Soft Matter 6, 1933–1938. doi: 10.1039/b922985j
Talapko, J., Meštrović, T., Juzbašić, M., Tomas, M., Erić, S., Horvat Aleksijević, L., et al. (2022). Antimicrobial peptides–mechanisms of action, antimicrobial effects and clinical applications. Antibiotics 11:1417. doi: 10.3390/antibiotics11101417
Tamma, P. D., Cosgrove, S. E., and Maragakis, L. L. (2012). Combination therapy for treatment of infections with gram-negative bacteria. Clin. Microbiol. Rev. 25, 450–470. doi: 10.1128/CMR.05041-11
Tan, P., Fu, H., and Ma, X. (2021). Design, optimization, and nanotechnology of antimicrobial peptides: from exploration to applications. Nano Today 39:101229. doi: 10.1016/j.nantod.2021.101229
Tao, Q., Lu, Y., Liu, Q., Chen, R., Xu, Y., Li, G., et al. (2023). Antibacterial activity of antimicrobial peptide pmap-36 combined with tetracycline against porcine extraintestinal pathogenic Escherichia coli in vitro and in vivo. PPR:PPR717413. doi: 10.21203/rs.3.rs-3282565/v1
Thappeta, K. R., Vikhe, Y. S., Yong, A. M., Chan-Park, M. B., and Kline, K. A. (2020). Combined efficacy of an antimicrobial cationic peptide polymer with conventional antibiotics to combat multidrug-resistant pathogens. ACS Infect. Dis. 6, 1228–1237. doi: 10.1021/acsinfecdis.0c00016
Vaara, M., and Porro, M. (1996). Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 40, 1801–1805. doi: 10.1128/AAC.40.8.1801
Vargas-Casanova, Y., Rodríguez-Mayor, A. V., Cardenas, K. J., Leal-Castro, A. L., Muñoz-Molina, L. C., Fierro-Medina, R., et al. (2019). Synergistic bactericide and antibiotic effects of dimeric, tetrameric, or palindromic peptides containing the rwqwr motif against gram-positive and gram-negative strains. RSC Adv. 9, 7239–7245. doi: 10.1039/C9RA00708C
Wakabayashi, H., Yamauchi, K., Kobayashi, T., Yaeshima, T., Iwatsuki, K., Yoshie, H., et al. (2009). Inhibitory effects of lactoferrin on growth and biofilm formation of porphyromonas gingivalis and prevotella intermedia. Antimicrob. Agents Chemother. 53, 3308–3316. doi: 10.1128/AAC.01688-08
Wang, Z., Zhang, L., Wang, J., Wei, D., Shi, B., Shan, A., et al. (2014). Synergistic interaction of pmap-36 and prw4 with aminoglycoside antibiotics and their antibacterial mechanism. World J. Microbiol. Biotechnol. 30, 3121–3128. doi: 10.1007/s11274-014-1739-4
Westerhoff, H. V., Zasloff, M., Rosner, J. L., Hendler, R. W., Waal, D. E., Gomes, A., et al. (1995). Functional synergism of the magainins pgla and magainin-2 in Escherichia coli, tumor cells and liposomes. Eur. J. Biochem. 228, 257–264. doi: 10.1111/j.1432-1033.1995.00257.x
WHO (2023). No time to wait: Securing the future from drug-resistant infections–report to the secretary-general of the United Nations. Geneva: WHO.
Wongkaewkhiaw, S., Taweechaisupapong, S., Anutrakunchai, C., Nazmi, K., Bolscher, J. G., Wongratanacheewin, S., et al. (2019). D-ll-31 in combination with ceftazidime synergistically enhances bactericidal activity and biofilm destruction in Burkholderia pseudomallei. Biofouling 35, 573–584. doi: 10.1080/08927014.2019.1632835
World Health Organization Food and Agriculture Organization of the United Nations, and World Organization for Animal Health. (2015). Global action plan on antimicrobial resistance. Geneva: WHO.
Wu, C.-L., Peng, K.-L., Yip, B.-S., Chih, Y.-H., and Cheng, J.-W. (2021). Boosting synergistic effects of short antimicrobial peptides with conventional antibiotics against resistant bacteria. Front. Microbiol. 12:747760. doi: 10.3389/fmicb.2021.747760
Yang, L., Harroun, T. A, Weiss, T. M, Ding, L., and Huang, H. W (2001). Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 81, 1475–1485. doi: 10.1016/S0006-3495(01)75802-X
Yu, G., Baeder, D. Y., Regoes, R. R., and Rolff, J. (2016). Combination effects of antimicrobial peptides. Antimicrob. Agents Chemother. 60, 1717–1724. doi: 10.1128/AAC.02434-15
Zasloff, M. (2002). Antimicrobial peptides of multicellular organisms. Nature 415, 389–395. doi: 10.1038/415389a
Zhang, C., and Yang, M. (2022). Antimicrobial peptides: from design to clinical application. Antibiotics 11:349. doi: 10.3390/antibiotics11030349
Zharkova, M. S., Orlov, D. S., Golubeva, O. Y., Chakchir, O. B., Eliseev, I. E., Grinchuk, T. M., et al. (2019). Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics–a novel way to combat antibiotic resistance? Front. Cell. Infect. Microbiol. 9:128. doi: 10.3389/fcimb.2019.00128
Zhong, C., Zhu, Y., Zhu, N., Liu, T., Gou, S., Zhang, F., et al. (2020). Synthesis and anti-pseudomonal activity of new β-ala modified analogues of the antimicrobial peptide anoplin. Int. J. Med. Microbiol. 310:151433. doi: 10.1016/j.ijmm.2020.151433
Keywords: antimicrobial resistance, antimicrobial peptides, combination therapy, antibiotic synergy, drug combination
Citation: Taheri-Araghi S (2024) Synergistic action of antimicrobial peptides and antibiotics: current understanding and future directions. Front. Microbiol. 15:1390765. doi: 10.3389/fmicb.2024.1390765
Received: 23 February 2024; Accepted: 05 July 2024;
Published: 31 July 2024.
Edited by:
Corina Ciobanasu, Alexandru Ioan Cuza University, RomaniaReviewed by:
Axel Hollmann, National Scientific and Technical Research Council (CONICET), ArgentinaJianhua Wang, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2024 Taheri-Araghi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sattar Taheri-Araghi, c2F0dGFyLnRhaGVyaUBjc3VuLmVkdQ==
 Sattar Taheri-Araghi
Sattar Taheri-Araghi