- 1Key Laboratory of Marine Biogenetic Resources, Third Institute of Oceanography, Ministry of Natural Resources, Xiamen, China
- 2School of Marine Sciences, China University of Geosciences, Beijing, China
- 3Faculty of Marine Biology, Xiamen Ocean Vocational College, Xiamen, China
Introduction: Hydrothermal vents, rich in heavy metals, provided a unique niche for heavy metal resistant microbes. However, knowledge about copper resistant microbes in deep sea hydrothermal vents is still limited.
Methods: The copper-resistant bacteria were isolated from deep-sea hydrothermal vent samples and conducted thorough physical, phylogenetic, and genomic analyses to elucidate their copper resistance capability and related genes.
Results: Twelve highly copper-resistant bacteria (up to 6-10 mM) were isolated from deep sea hydrothermal fields They were affiliated with the Pseudoalteromonas (4), Marinobacter (3), Halomonas (2), Psychrobacter (1), and Pseudomonas (1) genus in the α-Proteobacteria, and the Sphingomonas (1) genus in the β-Proteobacteria. The presence of copper in the medium obviously induced the amount of polysaccharides and proteins in the crude extracellular polymeric substances (EPS) produced by Halomonas sp. CuT 3-1, Pseudoalteromonas sp. CuT 4-3 and Marinobacter metalliresistant CuT 6, which could absorb 40 to 50 mg•g−1 copper. We further described a novel species, Marinobacter metalliresistant sp. nov. CuT 6T, which exhibited a higher copper resistance and encoded more heavy metal resistance-related genes than other Marinobacter species.
Discussion: It revealed that the copper resistance capability exhibited by these strains in hydrothermal fields is likely attributed to the production of exopolymeric substances, such as polysaccharides and proteins, as well as active transport or efflux mechanisms for heavy metals.
1 Introduction
Deep-sea hydrothermal fields are a source of heavy metals such as Cu, Zn, V, and U (Hu et al., 2022). For example, hydrothermal fluids continued to contribute a significant proportion (5–14%) of the Earth’s oceanic copper inventory in the form of organically bound complexes, including metallic mineral deposits (Zierenberg et al., 2000; Sander and Koschinsky, 2011; Stüeken, 2020). The content of copper in deep-sea hydrothermal vents has been documented to reach notably high levels, 141.15–551.58 mg/kg in the hydrothermal sediments (Zhu and Zhou, 2021) and 9.6–228.0 g/kg in the hydrothermal sulfides (Bonatti et al., 1979; Dekov et al., 2010; Sander and Koschinsky, 2011; Sun et al., 2011; Liao et al., 2018). These values are notably higher than the average crustal abundance of copper (63 mg/kg), highlighting the remarkable potential of these vents for copper-resistant bacteria.
Heavy metal resistance and acquisition are important adaptive strategies in bacteria in terrestrial and aquatic environments, but their distribution and mechanisms were unexplored (Mathivanan et al., 2021). Most metallophiles were found in the metal processing industries or dumping sites, not natural habitats such as deep-sea or terrestrial hydrothermal sources (Dopson and Holmes, 2014; Kanekar and Kanekar, 2022). The reported neutrophilic copper-resistant microorganisms predominantly belong to diverse bacterial phyla, including α-proteobacteria (Methylobacterium sp.), β-proteobacteria (Alcaligenes sp.), γ-proteobacteria (Pseudomonas sp., Salmonella sp., and Acinetobacter sp.), Firmicutes (Bacillus sp. and Streptococcus sp.), and fungi (Paecilomyces lilacinus, Aspergillus terreus, and Cladosporium sp.). Furthermore, some mineral-oxidizing hyperthermophilic archaea such as Acidianus brierleyi, Sulfolobus metallicus, and Metallosphaera sedula, which were from terrestrial hot springs, exhibited remarkable tolerance to elevated copper concentrations (Harrison, 2016). Among them, Bacillus sp. recorded the highest copper-resistant ability up to 5.5 mM (Kunito et al., 1997). However, little attention has been paid to heavy metal-related microorganisms in deep-sea hydrothermal fields before. The bacterial strain Alcanivorax sp. VBW004 from a shallow hydrothermal vent has demonstrated remarkable copper resistance capabilities, tolerating concentrations up to 2.4 mM. This is attributed to the presence of copper resistance genes, such as copB (Ramasamy et al., 2020). Vetriani et al. reported these bacteria including Alcanivorax, Bacillus, Pseudomonas, Marinobacter, Pseudoalteromonas, and Halomonas had mercury resistance ability and an ecological role in mercury detoxification in the vent environment (Vetriani et al., 2005). Microorganisms inhabiting hydrothermal vents exhibit a tolerance to heavy metals, yet most of them are unable to thrive in environments with elevated concentrations. For example, Nitratiruptor sp. SB155-2 (belonging to phylum Campylobacterota) from deep-sea hydrothermal vents could only grow under Cu (II) stress below 0.1 mM copper through high-affinity efflux systems (Ares et al., 2022). In this study, we first isolated several high copper-resistant bacteria from deep-sea hydrothermal fields in the South West Indian Ocean ridge. These strains could tolerate concentrations of up to 6–10 mM Cu (II) and could grow under the stress of higher than 2 mM Cu (II). Their potential resistant mechanism based on their genome analysis was discussed to understand their adaptation in deep-sea hydrothermal vents.
2 Materials and methods
2.1 Sample collection
The samples were collected using a TV grab sampler at several sites at a depth of 1,453–2,653 m in hydrothermal fields of the southwest Indian Ocean in February 2020 during the cruise of DaYang Hao (Table S1). The sediments, sulfides, and oxides were fractionated on a clean bench and subsample was further used to inoculate (1/10, w/v) a sterile liquid enrichment media at 25°C on board. The sulfides were composed primarily of hydrothermal chimney fragments, predominantly consisting of pyrite (FeS₂), chalcopyrite (CuFeS₂), and sphalerite (ZnS), among others. The oxide samples were dominated by oxides/hydroxides of Fe, Mn, and Si. Furthermore, the sediments were composed of calcareous ooze, with traces of sulfides, exhibiting a significant proportion of calcium carbonate, reaching up to 50% in volume.
2.2 Enrichment and isolation of copper-resistant bacteria
Enrichment of copper-resistant bacteria was conducted in 100 mL of marine broth (MB) with 6 mM Cu (II) as copper sulfate (CuSO4•5H2O) in 250 mL Erlenmeyer flasks. Per 1,000 mL of MB (Becton, Dickinson) contained 5.0 g peptone, 1.0 g yeast extract, 0.1 g ferric citrate, 19.45 g NaCl, 5.9 g MgCl2, 1.8 g CaCl2, 0.55 g KCl, 0.16 g NaHCO3, 34.0 mg SrCl2, 22.0 mg H3BO3, 0.08 g KBr, 4.0 mg Na2SiO3, 2.4 mg NaF, 1.6 mg NH4NO3, 8.0 mg Na2HPO4, 3.24 g MgSO4, and the solution was adjusted to pH 7.0. The enrichment culture was incubated at 28°C for 3–5 days on board and stored with glycerol at −20°C. In the lab, the subculture was incubated at 28°C for 24 h, with shaking (150 rpm). Cu (II)-resistant bacterial strains were purified by repeatedly streaking them on marine broth agar (MA) plates containing 6 mM Cu (II). These isolates were deposited in the Marine Culture Collection of China, MCCC1 (Table 1).
2.3 Identification and phylogenetic analysis of isolates
DNA of each isolate was extracted by the Genomic DNA extraction kit (SBS Genetech Co., Ltd., Shanghai). Two universal primers corresponding to E. coli positions 27F (5’-AGATTTGATCMTG GCTCAG-3′) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3′) were used for PCR amplification of the 16S ribosomal RNA of the isolates, and sequences information were obtained through Sanger sequencing. EzBioCloud2 was used for homology searches on 16 s rDNA genes (Yoon et al., 2017a). Phylogenetic analysis was performed using MEGA X (64-bit) (Larkin et al., 2007) after multiple alignments of the sequence data with Clustal W. Distances were calculated using distance options according to Kimura’s two-parameter model and clustering was performed using the neighbor-joining (NJ) methods. Sequences of related taxa were obtained from the GenBank database and supported with bootstrap values based on 1,000 replications.
2.4 Detection and identification of EPS production
To identify potential EPS production by bacterial isolates, the Congo red agar (CRA) plate test was utilized, which contained 5.0 g/L peptone, 1.0 g/L yeast extract, 30.0 g/L NaCl, and 15.0 g/L agar, with pH adjusted to 7.0. After autoclaving (121°C, 15 min), 10.0 g/L of glucose and 0.8 g/L of Congo red indicator were added (Maalej et al., 2015). The purified strains were inoculated onto a CRA plate and incubated at 28°C for 48 h. Positive colonies were observed to turn red to black.
The selected strains for EPS production were inoculated into MB medium containing 1 mM copper (250 mg/L) and incubated at 28°C for 72 h in a shaker at 150 rpm. The MB medium without copper served as a control. The cells were centrifuged at 8000 rpm for 10 min, and the supernatant was mixed with two volumes of cold 95% ethanol, followed by overnight incubation at 4°C. Most EPS was precipitated by centrifugation at 8000 rpm for 20 min at 4°C. It was then washed twice with cold 95% ethanol for purification dissolved in 10 mL of ddH2O and filtered through a 0.22-μm sterile filter (Whatman Uniflo, United States). The polysaccharide in EPS extract was determined using the phenol–sulfuric acid method with glucose as the standard (Dubois et al., 1951). The proteins in EPS extract were detected using the BCA Protein Assay Kit (Thermo Scientific, USA) with bovine serum albumin as the standard (Smith et al., 1985). All the optical density measurements were detected using a multimode microplate reader (Thermo Scientific, Model: Varioskan LUX, United States).
2.5 Determination of the cu (II) resistance ability and biosorption capacity of isolates
The resistance of the isolates to Cu was tested by determining their growth on solid and liquid MB medium with various concentrations of CuSO4 (0, 0.4, 0.8, 1.2, 1.6, 2, 4, 6, 7, 8, 9, 10 mM) at 28°C for 5 days in triplicate. Use the formula (Ri = ∣(V0-Vi)/V0∣) to determine the effect of copper concentration on growth rates. In this formula, Ri represents the relative change in growth rate at an i mM copper concentration. V denotes the growth rate of the strains during the exponential growth phase, which is observed from 8 to 16 h in this study. V0 is the exponential growth rate at 0 mM copper concentration, serving as a baseline control. Vi is the exponential growth rate at the i mM copper concentration. Use statistical software SPSS to perform an analysis of variance (ANOVA) to assess the statistical significance of the growth rate differences under different copper concentrations among strains.
The tolerance of the bacteria against various concentrations of heavy metals was tested. To evaluate the sorption capability, isolates were cultured in triplicate in 100 mL of MB medium containing 1 mM Cu (250 mg/L) in a 250 mL flask at 150 rpm for 24 h at 28°C. The supernatants of cultures were sampled every 24 h, filtered, and then diluted with 10% HNO3 for the estimation of copper in the medium. To estimate the biosorption capacity of EPS, the extracted EPS was dissolved to a final concentration of 1 g/L, followed by the addition of different concentrations of CuSO4 solution ranging from 0 to 400 mg/L and after 1-h incubation then filter diluted with 10% HNO3. Copper ion concentrations were estimated using an atomic adsorption spectrometer (Srivastava and Majumder, 2008) and measured using an inductively coupled plasma optical emission spectrometer (ICP-OES, PerkinElmer, Model: Optima 7,300 V, United States). Using an uninoculated medium without CuSO4 standard solution served as a control.
2.6 Characterization of Marinobacter metalliresistant CuT 6
2.6.1 Physiological characterization of Marinobacter metalliresistant CuT 6
The determination of the temperature range for growth was tested over the range of 4–45°C (i.e., 4, 10, 15, 20, 28, 32, 37, 40, 45, 50, and 55). The pH range for growth was tested from initial pH 4.0 to initial pH 10.0, at 28°C, with MES buffer (pH 4.0–6.0), PIPES buffer (pH 7.0–8.0), HEPES buffer (pH 8.0–9.0), and AMPSO buffer (pH 9.0–10.0). Salt tolerance was tested at 28°C in the MB medium prepared with various concentrations of NaCl (0–20% w/v, 1% intervals), which substitute for sea salts. The carbon substrate utilization patterns of the isolate were investigated using the BIOLOG Gen III microPlate in triplicates under optimal growth conditions. The enzymatic activities were assessed using API ZYM strips (bioMérieux). Membrane polar lipids for the PLFA analysis were extracted from freeze-dried biomass using acidic methanol, and their fatty acid composition was examined using GC–MS as previously described by Zhilina et al. (1997). The total phospholipids were identified by two-dimensional TLC as described by Sorokin et al. (2012). Respiratory lipoquinones were analyzed in cold acetone extract first by TLC (Collins, 1985) and then a major UV-absorbing band was eluted and subjected to tandem mass spectrometry (Thermo Fisher Scientific, Model: LCG Advantage Max, United States) with chemical ionization at atmospheric pressure. The quinones were identified by ionic mass by HPLC-MS. Morphological characteristics of cells of the novel isolate were determined by using light microscopy (Olympus, Model: CX21, Japan) and transmission electron microscopy (JEOL, Model: JEM-1230 and JEM2100, Japan).
2.6.2 Genome analysis of Marinobacter metalliresistant CuT 6
The whole-genome sequencing was conducted by Gene Denovo Biotechnology Co., Ltd. (Guangzhou, China) using Illumina NovaSeq 6,000 (Illumina, San Diego, CA, USA) and PacBio Sequel II system (PacBio, Menlo Park, CA, United States) according to the manufacturer’s suggested protocols. Fastp version 0.20.0 and Pilon version 1.23 were used to control and correct the data (Walker et al., 2014; Chen et al., 2018). The complete genome was assembled using Falcon version 0.3.0(Chin et al., 2016). The obtained genome sequences were annotated using the NCBI Prokaryotic Genome Annotation Pipeline and deposited at DDBJ/ENA/GenBank. The tRNA genes were predicted using tRNAscan-SE version 1.3.1 (Lowe and Chan, 2016), and the rRNA genes using RNAmmer version 1.2 (Lagesen et al., 2007). The content of the genomic G + C was determined by the complete genome sequence.
2.6.3 Phylogenetic analyses of Marinobacter metalliresistant CuT 6
The average nucleotide identity (ANI) of the genome was calculated with the algorithm of OrthoANIu using the EzBioCloud web service3 (Yoon et al., 2017b). The digital DNA–DNA hybridization (diDDH) estimate values were analyzed using the Genome-to-Genome Distance Calculator4 (Meier-Kolthoff et al., 2022). UBCG 3.0 analysis (Kim et al., 2021) based on a set of 92 single-copy core genes was performed to investigate the relationships among strain CuT 6 and the related species of Marinobacter (n = 22). The UBCG utilizes a collection of external tools to derive a phylogenomic tree from a range of genomic sequences. These tools include Prodigal (Hyatt et al., 2010), Hmmsearch,5 Mafft (Katoh and Standley, 2013), RaxML (Stamatakis, 2014), and FastTree (Price et al., 2010).
2.7 Data availability
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequence of isolates are shown in Table 1. The whole-genome project for M. metalliresistant CuT 6 has been deposited in GenBank under the accession number ASM3809864v1 and is submitted in the NCBI under BioSample number SAMN29498721.
3 Results and discussions
3.1 Isolation and identification of copper-resistant bacteria
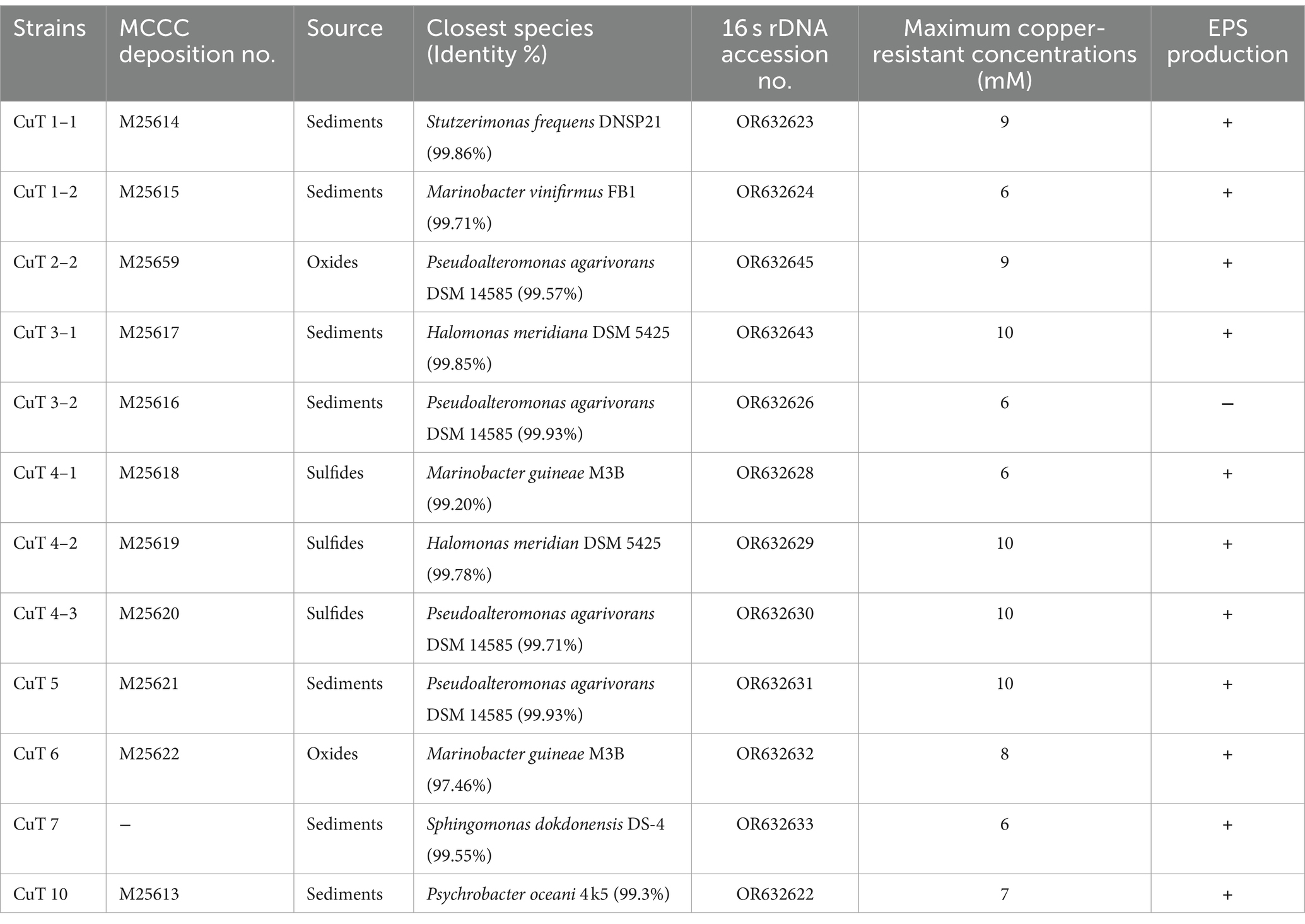
Twelve copper-resistant bacteria were isolated from hydrothermal fields, of which 11 were assigned to the γ-Proteobacteria and one to the α-Proteobacteria as determined by their 16S rDNA sequences. They were affiliated with the Pseudoalteromonas (4), Marinobacter (3), Halomonas (2), Psychrobacter (1), Pseudomonas (1) genus in the γ-Proteobacteria group, and the Sphingomonas (1) genera in the α-Proteobacteria group (Figure 1; Table 1). All the isolates had growth temperatures between 4 and 37°C, with the optimal growth temperature being higher than 15°C, specifically approximately 28°C. As all the strains had growth at 4°C, they could be assigned as psychrotrophs. All 12 strains were Gram-negative, neutrophilic, and aerobic.
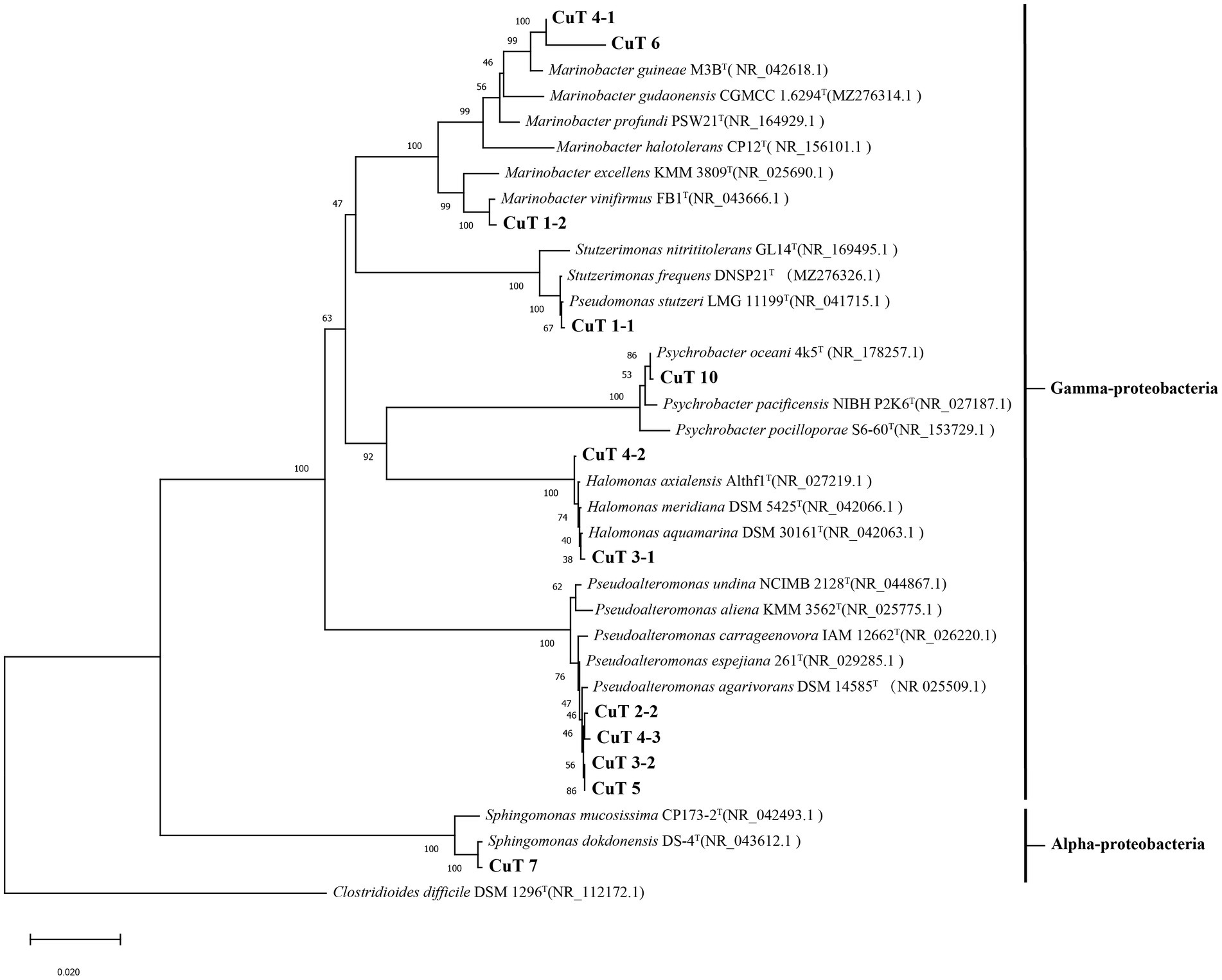
Figure 1. Phylogenetic tree reconstructed based on the small-subunit (16S) rRNA gene (1,399 bp) of isolates by the neighbor-joining method showed their positions and bootstrap values (out of 1,000 replicates).
Among these genera, Pseudomonas and Halomonas strains have often been found heavy metal resistant ability and used to sequester heavy metals such as Cd (II), Cu (II), Cr (VI), and Ni (II) from aqueous solution (Gabr et al., 2008; Fang et al., 2011; Ozturk et al., 2012; Govarthanan et al., 2015; Mădălina et al., 2016; Chug et al., 2021). Strain CuT 1–1 closest to Pseudomonas stutzeri ATCC 17588 was isolated from sediments and tolerated with 9 mM copper. Two Halomonas strains CuT 3–1 and CuT 4–2 showed extra high copper-resistant capacity of up to 10 mM, which are close to extremely halotolerant bacteria Halomonas meridiana DSM 5425 isolated from Antarctic saline lake (Kaye and Baross, 2000; Jonathan et al., 2011).
Pseudoalteromonas, Marinobacter, and Psychrobacter are broadly distributed in marine environments, including hydrothermal vents, subseafloor, and deep-sea sediments (Holmstrom and Kjelleberg, 1999). However, little reported their metal-resistant ability. For example, the EPS-producing Pseudoalteromonas sp. MER144 from Antarctic seawater showed metal adsorption capacity (Caruso et al., 2017). In this study, three strains showed 99.57–99.93% similarity with marine Pseudoalteromonas agarivorans DSM 14585 and had high copper-resistant ability 9–10 mM. Another Pseudoalteromonas sp. strain CuT 3–2 showed 99.01% 16 s rDNA similarity with Pseudoalteromonas carrageenovora IAM12662. Three Marinobacter strains CuT 1–2, CuT 4–1, and CuT 6 were close to M. vinifirmus FB1T (99.93% 16 s rDNA similarity) and M. guineae M3BT (99.21, 96.88% 16 s rDNA similarity). Strain CuT 6 was identified as a novel species and further characterized as follows. Strain CuT 10 had 98.89% 16 s rDNA similarity with Psychrobacter oceani 4k5T, which was isolated from marine sediment.
Sphingomonas genus in the α-Proteobacteria group was frequently isolated from soil and encoded heavy metal resistance-related genes such as multicopper oxidase and the czc efflux system (Altimira et al., 2012; Kim et al., 2020). Those strains possess multifaceted functions ranging from remediation of environmental contaminations to promoting plant growth (Asaf et al., 2020). In this study, strain CuT 7 closest to Sphingomonas dokdonensis DS-4 was isolated. Sphingomonas dokdonensis DS-4 was isolated from soil, but no metal-resistant ability was found previously (Yoon et al., 2006).
Diverse and adaptable heterotrophic α- and γ-proteobacteria have consistently been detected and repeatedly isolated from different venting areas throughout a wide range of environmental conditions in hydrothermal plumes, sulfide structures, and associated sediments, demonstrating their prevalence and dominance in the deep-sea hydrothermal field (Edwards et al., 2003; Kato et al., 2013; Meier et al., 2016; He et al., 2022). In the sample area (Southwest Indian Ocean Ridge), metagenomic analysis also showed that α- and γ-proteobacteria were abundant in two sulfides samples, accounting for 7.79–24.54% and 20.6–61.0% (Zhong et al., 2022). Our findings implied that their adaptability might encompass their remarkable resistance ability to heavy metals.
3.2 Copper resistance profile and adsorption capacity by isolates
In this study, all 12 isolates showed highly copper-resistant activities up to 6 to 10 mM in the liquid MB medium with various concentrations of CuSO4 (Table 1; Supplementary Figure S1). Four strains, including Halomonas sp. CuT 3–1 and CuT 4–2, Pseudoalteromonas sp. CuT 4–3 and CuT 5, could tolerate high concentrations of Cu (II) up to 10 mM. Among the three isolated Marinobacter strains (CuT 1–2, CuT 4–1, and CuT 6), strain CuT 6 exhibited an exceptional capacity to withstand copper concentrations reaching as high as 8 mM. These three distinct genera, including Halomonas sp. CuT 3–1, Pseudoalteromonas sp. CuT 4–3, and Marinobacter metalliresistant CuT 6, were chosen as representatives for the analysis of their copper-resistant abilities, as they exhibited high levels of copper resistance.
The effects of copper with treatment concentrations ranging from 0 to 10 mM on the bacterial growth curve and the maximum bacterial growth number are shown in Supplementary Figure S1 and Figure 2. The growth of Halomonas sp. CuT 3–1 was inhibited in the presence of higher than 2 mM copper (Supplementary Figure S1A). Furthermore, it could have growth with OD600 (optical density at 600 nm) of 0.09 in the 18 h culture with 10 mM Cu (II) (Supplementary Figure S1A). There was almost no effect on the highest cell growth number of Pseudoalteromonas sp. CuT 4–3 under 2.0 mM Cu (II) and M. metalliresistant CuT 6 under 1.6 mM Cu (II) (Figure 2). Pseudoalteromonas sp. CuT 4–3 could grow up to OD600 = 0.3 at 6 mM Cu (II) (Supplementary Figure S1B). M. metalliresistant CuT 6 showed the highest growth in MB with 0.4–0.8 mM Cu (II) and reached an optical density (OD600) of 0.25 with 8 mM Cu (II). Previous reports showed that most copper-resistant bacteria tolerate 0.5–3.5 mM of Cu (II) (Altimira et al., 2012). Bacillus sp. showed the highest copper-resistant ability up to 5.5 mM (Kunito et al., 1997). Some copper-resistant bacteria could decrease copper (<1.5 mM) for the remediation of metal-contaminated waters or soils (Kugler et al., 2022). The isolates in this study displayed high resistance to Cu (II) up to 6–10 mM, which indicated their highly heavy metal resistant ability might contribute to their adaptation in the hydrothermal fields. Besides, the genus Marinobacter is very broadly distributed in marine and terrestrial environments (Cooper et al., 2022). However, only one report demonstrated that Marinobacter adhaerens HP15 had zinc resistance by two CzcCBA efflux pumps and helped it to colonize aggregates (Stahl et al., 2015; Will et al., 2020). In this study, typical heavy metals, including Cu (II), Cr (VI), Cd (II), Co (II), Zn (II), and Hg (II), were further determined to study the heavy metal-resistant abilities of 30 Marinobacter species (28 type species and 2 new isolates from hydrothermal vents) (Supplementary Figure S2). Most of them exhibited certain copper resistance, with a minimum tolerance of 2 mM and a maximum tolerance of 6 mM Cu (II). Among them, strain CuT 6 showed the highest copper resistance capacity up to 8 mM and could tolerate 2 mM Co (II), 3 mM Zn (II), and 0.3 mM Hg (II) (Figure 3; Supplementary Figure S2). Among their phylogenetically closest relatives, M. guineae strain M3BT and M. profundi strain PSW21T demonstrated the highest copper resistance at a concentration of only 2 mM, which is different from the growth rate changes observed in CuT 6 when CuT 6 was exposed to copper. The differences between strains were especially obvious at concentrations of 4, 6, and 8 mM. (p < 0.05, Table. S6). Besides, each of Marinobacter spp. displayed resistance to at least one type of heavy metal. Among them, Marinobacter fuscus NH169-3T showed metal-resistant ability to all the tested heavy metals.
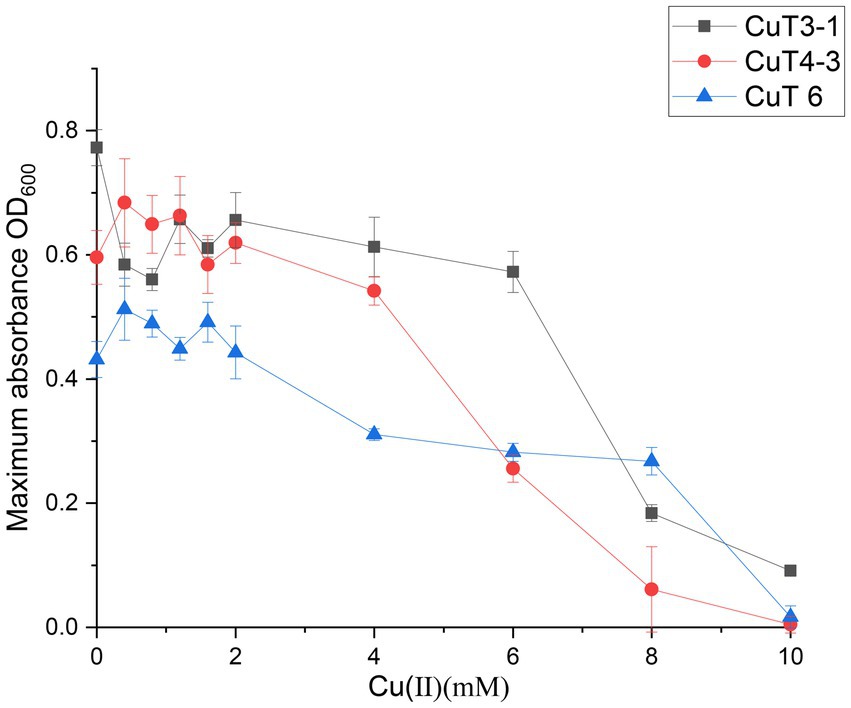
Figure 2. Highest growth number of highly copper-resistant strains, including Halomonas sp. CuT 3–1, Pseudoalteromonas sp. CuT 4–3 and Marinobacter metalliresistant CuT 6, were grown in the MB medium supplemented with the concentration of CuSO4 at 28°C for 85 h.
Figure 3. Growth curve of bacterium Marinobacter metalliresistant CuT 6 (C), in comparison to its phylogenetically closest relatives, M. guineae M3BT (A) and M. profundi PSW21T (B). Strains were grown at 28°C for 20 h in MB medium supplemented with 0–10 mM CuSO4. Growth was observed using turbidity (OD600).
3.3 Biosorption of copper by produced EPS of isolates
Except for strain CuT 3–2, 11 out of 12 isolates exhibited positive EPS production ability, forming red or black colonies on the CRA plates compared with colonies on MB plates (Supplementary Figure S3). The EPS was subsequently purified from a 24-h liquid culture of Halomonas sp. CuT 3–1, Pseudoalteromonas sp. CuT 4–3, and M. metalliresistant CuT 6 as described methods (Maalej et al., 2015).
Compared to the absence of Cu (II) in the medium, the addition of 1 mM Cu (II) significantly induced the production of EPS by these three isolates, which were mainly composed of proteins and sugars (Supplementary Figure S4). The increase in the concentration of EPS from 379.65 mg/L to 1013.00 mg/L in the culture of Halomonas sp. strain CuT 3–1 with copper stimulation. The polysaccharide and the protein contents in the EPS of CuT 3–1 increased by 3.64-fold [from 21.64 mg/L (5.70%) to 100.49 mg/L (9.92%)] and 2.55-fold [from 358.04 mg/L (94.30%) to 912.06 mg/L (90.08%)] with copper stimulation. Contents of EPS of Pseudoalteromonas sp. strain CuT 4–3 consistently increased from 366.70 mg/L to 1057.92 mg/L as Cu increased. The polysaccharide content of strain CuT 4–3 increased from 33.37 mg/L (9.10%) to 66.12 mg/L (6.25%), and the protein part increased from 333.44 mg/L (90.10%) to 992.00 mg/L (93.75%). The total EPS content of strain CuT 6 was lower than those of strain CuT 3–1 and strain CuT 4–3. The EPS content of strain CuT 6 increased a little from 214.35 mg/L to 447.17 mg/L with copper stimulation. The copper ion addition also induced an increase in polysaccharide content [from 50.48 mg/L (23.55%) to 82.19 mg/L (18.38%)] and protein content [from 214.31 mg/L (76.45%) to 364.96 mg/L (81.62%)] of strain CuT 6. In addition, we examined the composition and contents of EPS produced by strain CuT 6 and its closely related type strains under conditions with and without copper. The results indicated that strain CuT 6 produced different components and contents of EPS, and it responded more sensitively to copper stimulation compared to the other five Marinobacter strains (Supplementary Figure S10).
The copper adsorption capacity of the culture medium of copper-tolerant bacteria during growth at 250 mg/L of CuSO4 was monitored (Supplementary Figure S5). The binding ability of EPS produced by these bacteria at different concentrations of 0–400 mg/L of CuSO4 was also determined, as shown in Figure 4. With the addition of CuSO4, the copper ion content in the mediums was reduced by copper-resistant bacterial strains. After 24 h, strain CuT 3–1 reduced 75.38 mg/L of CuSO4. After 48-h cultivation, strain CuT 4–3 and strain CuT 6 reduced 74.60 mg/L and 79.21 mg/L of CuSO4, respectively. The binding of metal by EPS increased linearly with the content of Cu (II). The EPS of M. metalliresistant CuT 6 was found to have the highest binding ability up to 52.24 mg CuSO4/g EPS. The maximum amount of Cu (II) bound to the EPS of strain CuT 4–3 and CuT 3–1 was 37.78 and 35.92 mg CuSO4/g EPS. These three copper-tolerant bacterial strains all exhibited abilities to reduce CuSO4 concentrations in the medium. However, their EPS adsorption capacities varied significantly. The EPS produced by strain CuT 6 had a higher capacity to adsorb CuSO4 than those produced by the other two strains. The results indicated that the copper ions could be adsorbed by the cells themselves and by the secreted EPS. The findings revealed that upon exposure to copper, the production of EPSs by copper-resistant bacteria increased, along with a fluctuating ratio of polysaccharides and proteins within the EPS matrix. Notably, the EPS surrounding the cells displayed remarkable adsorption capabilities for heavy metals, which might contribute to bacterial adaptation in the harsh environments of deep-sea hydrothermal vents.
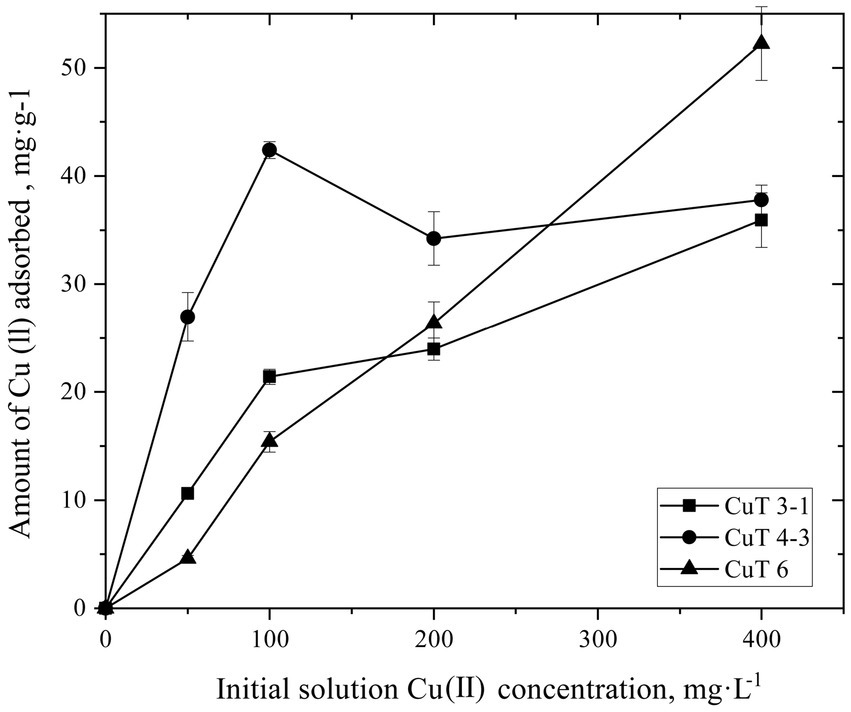
Figure 4. Biosorption of copper by the crude exopolysaccharide (EPS) produced by Halomonas sp. CuT 3–1, Pseudoalteromonas sp. CuT 4–3 and Marinobacter metalliresistant CuT 6 over a range of metal concentration (0–400 mg/L of Cu (II)).
3.4 Characterization of Marinobacter metalliresistant CuT 6
3.4.1 Phylogenetic and genomic analysis of Marinobacter metalliresistant CuT 6
The maximum-likelihood phylogenetic tree based on the 16S rRNA gene and the genome-based phylogenomic tree by using the up-to-date bacterial core gene sets (92 genes) were established to elucidate the phylogenetic position of strain CuT 6 in the phylum of Proteobacteria. It showed that strain CuT 6 belonged to the genus Marinobacter and was closest to M. guineae and M. profundi (Figure 1; Supplementary Figure S6). Phylogenetic analysis of the 16S rRNA gene sequences showed that strain CuT 6’s closest relative was M. guineae M3BT, with which it shared 99.1% similarity. The ANI values of strain CuT 6 to M. guineae M3BT, M. profundi PSW21T, and M. lipolyticus SM19T were 88.3, 76.91, 77.3%, respectively, which are below the standard ANI criterion for species identity (95.0–96.0%) (Kim et al., 2014). The diDDH estimate values between strain CuT 6 and M. guineae M3BT (Montes et al., 2008), M. lipolyticus SM19T, and M. gudaonensis CGMCC 1.6294T were 43.2 ± 6.5%, 21.7 ± 0.9%, and 30.2 ± 6.6% (Table S5), respectively. These values were far below the standard criterion (70%) for the delineation of prokaryotic species (Stackebrandt and Goebel, 1994). These results confirm that strain CuT 6 represents a novel species of the genus Marinobacter.
The genome of strain CuT 6 consists of a single circular 4,411,896 bp chromosome without an extrachromosomal element. A total of 1,949 genes and 1,897 protein-coding sequences were predicted. Nine rRNA loci and 50 tRNA genes were identified. The G + C content of the genomic DNA of strain CuT 6 was 57.60%, calculated in silico using the complete genome sequence of strain CuT 6. The genome characteristics of CuT 6 and its relatives are listed in Table S4.
3.4.2 Physiological characterization of Marinobacter metalliresistant CuT 6
Cells of strain CuT 6 are rod-shaped, measuring 0.6 μm wide and 2.0–3.0 μm long. They are Gram-negative and non-spore-forming (Supplementary Figure S7). Cells are motile with the polar flagella. Growth was observed between 4°C and 45°C, and the optimal temperature for growth was 28°C. Growth was observed from 0 to 15% (w/v) NaCl. The optimal salt concentration was 3–4% for NaCl. The novel isolate is obligately chemoorganoheterotrophic, and its growth is observed under aerobic or anaerobic conditions. Strain CuT 6 was found to be able to utilize carbohydrates (including dextrin gentiobiose L-fucose, D-fucose, D-mannitol, glycerol, and D-galacturonic acid), organic acids (L-lactic acid, α-ketoglutaric acid L-malic acid, D-malic acid, β-hydroxybutyric acid, acetoacetic acid, propanoic acid, and trisodium citrate), and amino acids (including L-alanine, L-glutamic acid, and L-Histidine), but not L-arabinose, D-mannose, maltose, potassium gluconate, adipic acid, or capric acid. Nitrate and nitrite are reduced. Tween 40 is hydrolysed, but lecithin, casein and starch are not. Using the API 20NE test, it was found that D-glucose fermentation is positive, but gelatin hydrolysis, indole production, glucose, arginine dihydrolase, and urease are negative, which was different from M. guineae M3BT. Strain CuT 6 could produce alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14) leucine arylamidase, valine arylamidase, cystine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase and N-acetyl-b-glucosaminidase, but not α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase, and α-fucosidase (API ZYM). The main fatty acids are summed feature 3 (C16:1 ω7c and/or iso-C16:1 ω6c), C16:1 ω9c, C16:0, and C18:1 ω9c. The major respiratory quinone of strain CuT 6 was identified as ubiquinone 9 (Q-9 and Q-9(H2)). The polar lipids of strain CuT 6 were identified as phosphatidylethanolamine (PE), phosphatidylglycerol (PG), diphosphatidylglycerol (GL), and aminolipid (AL), as shown in Supplementary Figure S8. Differences in physiological, biochemical, and chemotaxonomic characteristics between strain CuT6 and its relatives are given in Table 2.
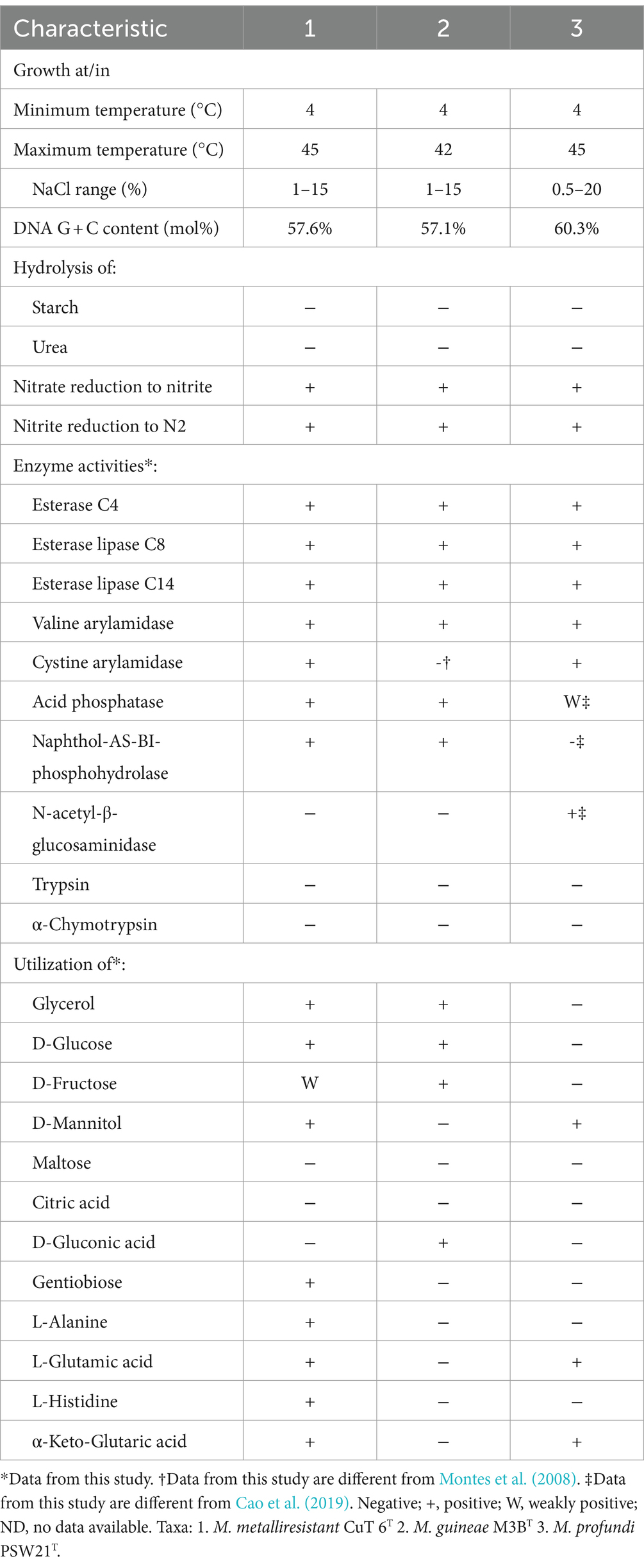
Table 2. Comparison of the phenotypic characteristics of strain CuT 6 and their closely related reference strains.
3.4.3 Possible mechanisms of copper resistance in Marinobacter metalliresistant CuT 6
Microbial copper resistance systems span copper efflux (cop, pco, cus, cue, and other extrachromosomal efflux systems), copper sequestration (cusF and siderophores), and copper oxidation (mixed copper oxidases and superoxide dismutase) (Rademacher and Masepohl, 2012). CopA is considered to be a marker gene for copper-tolerant microorganisms, which mainly mediates copper ion efflux reactions in microorganisms. CopA is divided into two groups encoding for multicopper oxidase and P-type ATPase (Li et al., 2015). In addition, the Mer family genes are not only involved in enhancing microbial mercury tolerance but are also thought to act as regulatory factors for genes conferring copper resistance, such as MerR (Li et al., 2014). Other multiple heavy metal resistance genes include the cobalt–zinc–cadmium efflux system (czcABCD), arsenic (arsABCR), and cation efflux genetic system (cusAB) (Silver and Phung, 1996).
We searched the copper resistance-related genes in the genus Marinobacter genomes including strain CuT 6 and other type strains (Figure 5; Table S2). Based on an analysis of the genomic copper resistance genes, it was discovered that strain CuT 6 uniquely encoded four copies of the copA gene, encompassing two multicopper oxidases and two ATPases, as well as a single copB gene within its genome (Supplementary Figure S9). It also encoded the copper resistance protein PcoA (NLK58_09295), which is required for the copper-inducible expression of copper resistance. This is consistent with its high copper-resistant ability. In addition, at least one Mer operon (NLK58_12425-NLK58_12440), including mercuric reductase (merA), was harbored by strain CuT 6. Additionally, strain CuT6 contains a partial cation efflux genetic system (cusAB) and cobalt–zinc–cadmium efflux system (czcAR), such as two cusA (NLK58_09210 and NLK58_12420) and two czcA (NLK58_09270 and NLK58_16520) genes (Table S2). These efflux genes within strain CuT 6, including copA (ATPases), copB, cusAB, czcA, and MerPT, potentially play a key role in the heavy metal transport capabilities. The enzymes copA (multicopper oxidase) and pcoA are responsible for oxidizing copper. In the M. metalliresistant CuT 6 genome, we have also identified some potential EPS synthesis-related genes and gene clusters, mainly composed of glycosyltransferases, forming functional structures for polysaccharide synthesis (Table S3).
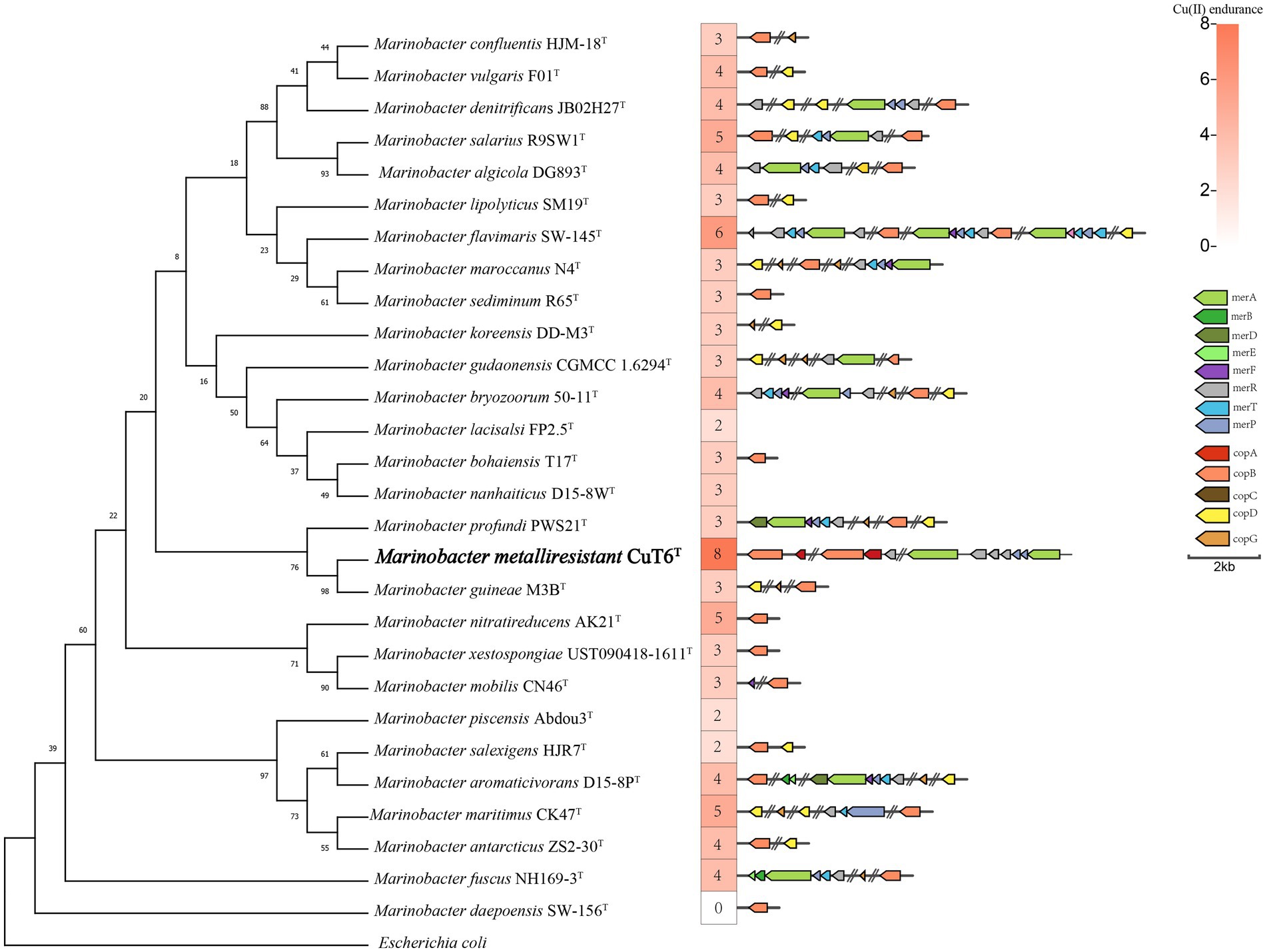
Figure 5. Maximum-likelihood phylogenetic tree reconstructed based on the 16S rRNA gene sequences of strain CuT 6 and representative members in the genus Marinobacter, with their copper resistant ability and related genes. Escherichia coli (NR_024570.1) was selected as an outgroup. Bootstrap values are given on the nodes of the tree. Branch node values below 50 are not shown here. There were a total of 1,249 positions in the final dataset. Bar, 0.01 substitutions per nucleotide position.
As reported in the genome of Pseudoalteromonas sp. CuT 4–3 by Ji et al. (2024), our study by genomic analysis concluded that multiple heavy metal resistance genes as active transport or efflux mechanisms and EPS biosynthesis for the absorption of these copper-resistant bacteria were responsible for heavy metal detoxification. These genes included copper-resistant genes (copABCD and pcoAB), cobalt–zinc–cadmium efflux system protein (czcABCD), arsenic (arsABCR), and cation efflux genetic system (cusAB), as well as proteins involved in polysaccharide syntheses such as the epsG family, cellulose biosynthesis bcs operons, Wzx flippase, Wzy polymerase, and polysaccharide copolymerase (Wzz) proteins.
3.4.4 Description of Marinobacter metalliresistant sp.nov
The cells are motile, round-ended rods (2.0–3.0 μm in length, 0.6 μm in width) with flagellum. Endospores are never observed. Growth is observed at salinities from 0 to 15% (optimum 3–4%), from pH 5 to 8 (optimum 7), and at temperatures between 4 and 45°C (optimum 28°C). Doubling time is 56 min under optimal growth conditions. Aerobic or facultative anaerobe. Uses complex organic compounds and organic acids including yeast extract, peptone, tryptone, casein, casamino acids, citrate, lactate, acetate, fumarate, propanoate and pyruvate. Does not reduce sulfite, sulfate, thiosulfate or nitrate. The type strain, CuT 6T (=MCCC M25622T = KCTC 92387T), was isolated from a hydrothermal sulfide sample collected at a depth of 2,901 m in a hydrothermal area of the southwest Indian Ocean (50.75 E, 40.04 S). The DNA G + C content of the type strain is 57.6 mol%.
4 Conclusion
Twelve highly copper-resistant bacteria were isolated from hydrothermal fields, which tolerate copper concentrations of up to 6–10 mM. They were affiliated with α-Proteobacteria and γ-Proteobacteria, including Sphingomonas (1), Pseudoalteromonas (4), Marinobacter (3), Halomonas (2), Psychrobacter (1), and Pseudomonas (1). The EPS content of strains was induced by copper ion, which could absorb 40 to 50 mg•g−1 copper. By physiologic and genomic analysis, we concluded that the copper resistance mechanisms, mainly the production of exopolymeric substances, and copper resistance genes as active transport or efflux mechanisms, of these strains in deep-sea hydrothermal fields. A novel species, Marinobacter metalliresistant sp. nov. CuT 6T, was identified.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
TY: Data curation, Investigation, Writing – original draft. MQ: Investigation, Writing – review & editing, Data curation. ZS: Writing – review & editing, Supervision. YZ: Writing – review & editing, Investigation. XZ: Data curation, Funding acquisition, Investigation, Project administration, Writing – original draft, Writing – review & editing, Supervision.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key R&D Program of China (No. 2021YFF0501304) and the National Natural Science Foundation of China (No.91951201) and Scientific Research Foundation of Third Institute of Oceanography, MNR (No. 2019021).
Acknowledgments
The authors are very grateful to the whole team of the oceanic cruises of COMRA DY65 via the R/V ‘Dayang hao’ for helping them collect the samples from the deep-sea hydrothermal vent fields.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1390451/full#supplementary-material
Footnotes
2. ^https://www.ezbiocloud.net/
3. ^https://www.ezbiocloud.net/tools/ani, accessed on 1 May 2023.
4. ^GGDC 3.0, https://ggdc.dsmz.de/, accessed on 7 May 2023.
References
Altimira, F., Yáñez, C., Bravo, G., González, M., Rojas, L. A., and Seeger, M. (2012). Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of Central Chile. BMC Microbiol. 12:193. doi: 10.1186/1471-2180-12-193
Ares, Á., Sakai, S., Sasaki, T., Mitarai, S., and Nunoura, T. (2022). Sequestration and efflux largely account for cadmium and copper resistance in the deep sea epsilon proteobacterium, Nitratiruptor sp. SB155-2. Environ. Microbiol. 24, 6144–6163. doi: 10.1111/1462-2920.16255
Asaf, S., Numan, M., Khan, A. L., and Al-Harrasi, A. (2020). Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 40, 138–152. doi: 10.1080/07388551.2019.1709793
Bonatti, E., Kolla, V., Moore, W. S., and Stern, C. (1979). Metallogenesis in marginal basins: fe-rich basal deposits from the philippine sea. Mar. Geol. 32, 21–37. doi: 10.1016/0025-3227(79)90144-0
Cao, J., Liu, P., Liu, R., Su, H., Wei, Y., and Liu, R. (2019). Marinobacter profundi sp. nov., a slightly halophilic bacterium isolated from a deep-sea sediment sample of the New Britain trench. Antonie Van Leeuwenhoek 112, 425–434. doi: 10.1007/s10482-018-1176-8
Caruso, C., Rizzo, C., Mangano, S., Poli, A., Donato, P. D., Nicolaus, B., et al. (2017). Extracellular polymeric substances with metal adsorption capacity produced by Pseudoalteromonas sp. MER144 from Antarctic seawater. Environ. Sci. Pollut. Res. Int. 25, 4667–4677. doi: 10.1007/s11356-017-0851-z
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (Oxford, England) 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chin, C. S., Peluso, P., Sedlazeck, F. J., Nattestad, M., Concepcion, G. T., Clum, A., et al. (2016). Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054. doi: 10.1038/nmeth.4035
Chug, R., Mathur, S., Kothari, S. L., and Harish, G. V. S. (2021). Maximizing EPS production from Pseudomonas aeruginosa and its application in Cr and Ni sequestration. Biochem. Biophys. Rep. 26:100972. doi: 10.1016/j.bbrep.2021.100972
Collins, M. D. (1985). Analysis of isoprenoid quinones. Methods Microbiol. 18, 329–366. doi: 10.1016/S0580-9517(08)70480-X
Cooper, Z. S., Rapp, J. Z., Shoemaker, A. M. D., Anderson, R. E., Zhong, Z. P., and Deming, J. W. (2022). Evolutionary divergence of Marinobacter strains in cryopeg brines as revealed by pangenomics. Front. Microbiol. 13:879116. doi: 10.3389/fmicb.2022.879116
Dekov, V. M., Cuadros, J., Kamenov, G. D., Weiss, D., Arnold, T., Basak, C., et al. (2010). Metalliferous sediments from the h.m.s. challenger voyage (1872-1876). Geochim. Cosmochim. Acta 74, 5019–5038. doi: 10.1016/j.gca.2010.06.001
Dopson, M., and Holmes, D. S. (2014). Metal resistance in acidophilic microorganisms and its significance for biotechnologies. Appl. Microbiol. Biotechnol. 98, 8133–8144. doi: 10.1007/s00253-014-5982-2
Dubois, M., Gilles, K., Hamilton, J. K., Rebers, P. A., and Smith, F. (1951). A colorimetric method for the determination of sugars. Nature 168:167. doi: 10.1038/168167a0
Edwards, K. J., Rogers, D. R., Wirsen, C. O., and McCollom, T. (2003). Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic α-and γ-Proteobacteria from the deep sea. Appl. Environ. Microbiol. 69, 2906–2913. doi: 10.1128/AEM.69.5.2906-2913
Fang, L., Wei, X., Cai, P., Huang, Q., Chen, H., Liang, W., et al. (2011). Role of extracellular polymeric substances in cu(II) adsorption on Bacillus subtilis and Pseudomonas putida. Bioresour. Technol. 102, 1137–1141. doi: 10.1016/j.biortech.2010.09.006
Gabr, R. M., Hassan, S. H. A., and Shoreit, A. A. M. (2008). Biosorption of lead and nickel by living and non-living cells of Pseudomonas aeruginosa asu 6a. Int. Biodeterior. Biodegradation 62, 195–203. doi: 10.1016/j.ibiod.2008.01.008
Govarthanan, M., Shim, J., Kim, S. A., Kamala-Kannan, S., and Oh, B. T. (2015). Isolation and characterization of multi-metal-resistant Halomonas sp. MG from Tamil Nadu magnesite ore soil in India. Curr. Microbiol. 71, 618–623. doi: 10.1007/s00284-015-0897-4
Harrison, S. T. L. (2016). “Biotechnologies that utilize acidophiles” in Acidophiles. Life in extremely acidic environments. eds. R. J. Quatrini and D. B. Johnson (Norfolk: Caister Academic Press), 265–283.
He, Y., Zeng, X., Xu, F., and Shao, Z. (2022). Diversity of mixotrophic neutrophilic thiosulfate-and iron-oxidizing bacteria from deep-sea hydrothermal vents. Microorganisms 11:100. doi: 10.3390/microorganisms11010100
Holmstrom, C., and Kjelleberg, S. (1999). Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 30, 285–293. doi: 10.1111/j.1574-6941.1999.tb00656.x
Hu, S., Tao, C., Liao, S., Zhu, C., and Qiu, Z. (2022). Transformation of minerals and mobility of heavy metals during oxidative weathering of seafloor massive sulfide and their environmental significance. Sci. Total Environ. 5:153091:819. doi: 10.1016/j.scitotenv.2022.153091
Hyatt, D., Chen, G. L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Ji, B., Yu, T., and Zeng, X. (2024). Complete genome analysis of copper resistant bacteria Pseudoalteromonas sp. CuT 4-3 isolated from a deep-sea hydrothermal vent. Mar. Genomics 75:101106. doi: 10.1016/j.margen.2024.101106
Jonathan, Z., Kaye, J. B. S., Edwards, K. J., and Baross, J. A. (2011). Halomonas and Marinobacter ecotypes from hydrothermal vent, subseafloor and deep-sea environments. FEMS Microbiol. Ecol. 75, 123–133. doi: 10.1111/j.1574-6941.2010.00984.x
Kanekar, P. P., and Kanekar, S. P. (2022). Metallophilic, metal-resistant, and metal-tolerant microorganisms. Singapore: Springer.
Kato, S., Nakawake, M., Kita, J., Yamanaka, T., Utsumi, M., Okamura, K., et al. (2013). Characteristics of microbial communities in crustal fluids in a deep-sea hydrothermal field of the Suiyo seamount. Front. Microbiol. 4:85. doi: 10.3389/fmicb.2013.00085
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kaye, J. Z., and Baross, J. A. (2000). High incidence of halotolerant bacteria in Pacific hydrothermal-vent and pelagic environments. FEMS Microbiol. Ecol. 32, 249–260. doi: 10.1111/j.1574-6941.2000.tb00718.x
Kim, J., Na, S.-I., Kim, D., and Chun, J. (2021). UBCG2: up-to-date bacterial core genes and pipeline for phylogenomic analysis. J. Microbiol. 59, 609–615. doi: 10.1007/s12275-021-1231-4
Kim, M., Oh, H.-S., Park, S.-C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Kim, Y. J., Park, J. Y., Balusamy, S. R., Huo, Y., Nong, L. K., and Thi Le, H. (2020). Comprehensive genome analysis on the novel species Sphingomonas panacis DCY99T reveals insights into iron tolerance of ginseng. Int. J. Mol. Sci. 21:2019. doi: 10.3390/ijms21062019
Kugler, A., Brigmon, R. L., Friedman, A., Coutelot, F. M., Polson, S. W., Seaman, J. C., et al. (2022). Bioremediation of copper in sediments from a constructed wetland ex situ with the novel bacterium Cupriavidus basilensis SRS. Sci. Rep. 12:17615. doi: 10.1038/s41598-022-20930-0
Kunito, T., Nagaoka, K., Tada, N., Saeki, K., Senoo, K., Oyaizu, H., et al. (1997). Characterization of cu-resistant bacterial communities in cu-contaminated soils. Soil Sci. Plant Nutr. 43, 709–717. doi: 10.1080/00380768.1997.10414795
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. NAR. 35, 3100–3108. doi: 10.1093/nar/gkm160
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, Z., Ma, Z., Hao, X., Rensing, C., and Wei, G. (2014). Genes conferring copper resistance in Sinorhizobium meliloti CCNWSX0020 also promote the growth of Medicago lupulina in copper-contaminated soil. Appl. Environ. Microbiol. 80, 1961–1971. doi: 10.1128/AEM.03381-13
Li, X., Zhu, Y. G., Shaban, B., Bruxner, T. J. C., Bond, P. L., and Huang, L. (2015). Assessing the genetic diversity of cu resistance in mine tailings through high-throughput recovery of full-length copA genes. Sci. Rep. 5:13258. doi: 10.1038/srep13258
Liao, S. L., Tao, C. H., Li, H. M., Fernando, J. A. S., Liang, J., and Yang, W. F. (2018). Bulk geochemistry, sulfur isotope characteristics of the Yuhuang-1 hydrothermal field on the ultraslow-spreading southwest Indian ridge. Ore Geol. Rev. 96, 13–27. doi: 10.1016/j.oregeorev.2018.04.007
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Maalej, H., Hmidet, N., Boisset, C., Buon, L., Heyraud, A., and Nasri, M. (2015). Optimization of exopolysaccharide production from Pseudomonas stutzeri AS22 and examination of its metal-binding abilities. J. Appl. Microbiol. 118, 356–367. doi: 10.1111/jam.12688
Mădălina, V. D., Laszlo, B., Leonard, B. H., and Aharon, O. (2016). Heavy metal resistance in halophilic Bacteria and Archaea. FEMS Microbiol. Lett. 363:fnw 146. doi: 10.1093/femsle/fnw146
Mathivanan, K., Chandirika, J. U., Vinothkanna, A., Yin, H., Liu, X., and Meng, D. (2021). Bacterial adaptive strategies to cope with metal toxicity in the contaminated environment-a review. Ecotoxicol. Environ. Saf. 226:112863. doi: 10.1016/j.ecoenv.2021.112863
Meier, D. V., Bach, W., Girguis, P. R., Gruber-Vodicka, H. R., Reeves, E. P., Richter, M., et al. (2016). Heterotrophic Proteobacteria in the vicinity of diffuse hydrothermal venting. Environ. Microbiol. 18, 4348–4368. doi: 10.1111/1462-2920.13304
Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L., and Göker, M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 50, D801–D807. doi: 10.1093/nar/gkab902
Montes, M. J., Bozal, N., and Mercadé, E. (2008). Marinobacter guineae sp. nov., a novel moderately halophilic bacterium from an Antarctic environment. Int. J. Syst. Evol. 58, 1346–1349. doi: 10.1099/ijs.0.65298-0
Ozturk, S., Kaya, T., Aslim, B., and Tan, S. (2012). Removal and reduction of chromium by Pseudomonas spp. and their correlation to rhamnolipid production. J. Hazard. Mater. 231-232, 64–69. doi: 10.1016/j.jhazmat.2012.06.038
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Rademacher, C., and Masepohl, B. (2012). Copper-responsive gene regulation in bacteria. Microbiology 158, 2451–2464. doi: 10.1099/mic.0.058487-0
Ramasamy, K. P., Rajasabapathy, R., Lips, I., Mohandass, C., and James, R. A. (2020). Genomic features and copper biosorption potential of a new Alcanivorax sp. VBW004 isolated from the shallow hydrothermal vent (Azores, Portugal). Genomics 112, 3268–3273. doi: 10.1016/j.ygeno.2020.06.015
Sander, S. G., and Koschinsky, A. (2011). Metal flux from hydrothermal vents increased by organic complexation. Nat. Geosci. 4, 145–150. doi: 10.1038/ngeo1088
Silver, S., and Phung, L. T. (1996). Bacterial heavy metal resistance: new surprises. Ann. Rev. Microbiol. 50, 753–789. doi: 10.1146/annurev.micro.50.1.753
Smith, P. K., Krohn, R. I., Hermanson, G. T., Mallia, A. K., Gartner, F. H., Provenzano, M. D., et al. (1985). Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85. doi: 10.1016/0003-2697(85)90442-7
Sorokin, D. Y., Tourova, T. P., Sukhacheva, M. V., and Muyzer, G. (2012). Desulfuribacillus alkaliarsenatis gen. Nov. sp. nov., a deep-lineage, obligately anaerobic, dissimilatory sulfur and arsenate-reducing, haloalkaliphilic representative of the order Bacillales from soda lakes. Extremophiles 16, 597–605. doi: 10.1007/s00792-012-0459-7
Srivastava, N. K., and Majumder, C. B. (2008). Novel biofiltration methods for the treatment of heavy metals from industrial wastewater. J. Hazard. Mater. 151, 1–8. doi: 10.1016/j.jhazmat.2007.09.101
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Stahl, A., Pletzer, D., Mehmood, A., and Ullrich, M. S. (2015). Marinobacter adhaerens HP15 harbors two CzcCBA efflux pumps involved in zinc detoxification. Antonie van Leeuwenhoek. 108, 649–58. doi: 10.1007/s10482-015-0520-5
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stüeken, E. E. (2020). Hydrothermal vents and organic ligands sustained the Precambrian copper budget. Geochem. Perspect. Lett. 16, 12–16. doi: 10.7185/geochemlet.2037
Sun, Z., Zhou, H., Yang, Q., Sun, Z., Bao, S., and Yao, H. (2011). Hydrothermal Fe-Si-Mn oxide deposits from the central and south valu fa ridge, lau basin. Appl. Geochem. 26, 1192–1204. doi: 10.1016/j.apgeochem.2011.04.008
Vetriani, C., Chew, Y. S., Miller, S. M., Yagi, J., Coombs, J., Lutz, R. A., et al. (2005). Mercury adaptation among bacteria from a deep-sea hydrothermal vent. Appl. Environ. Microbiol. 71, 220–226. doi: 10.1128/AEM.71.1.220-226.2005
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Will, V., Stahl, A., and Ullrich,. (2020). Heavy metal resistance in Marinobacter adhaerens HP15 supports colonization of transparent exopolymer particles during its interaction with diatoms. Mar. Ecol. Prog. Ser. doi: 10.3354/meps13558
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017a). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017b). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Yoon, J. H., Lee, M. H., Kang, S. J., Lee, S. Y., and Oh, T. K. (2006). Sphingomonas dokdonensis sp. nov., isolated from soil. Int. J. Syst. Evol. 56, 2165–2169. doi: 10.1099/IJS.0.64114-0
Zhilina, T. N., Zavarzin, Z. G., Rainey, F. A., Pikuta, E. N., Osipov, G. A., and Kostrikina, N. A. (1997). Desulfonatronovibrio hydrogenovorans gen. Nov., sp. nov., an alkaliphilic, sulfate-reducing bacterium. Int. J. Syst. Bacteriol. 47, 144–149. doi: 10.1099/00207713-47-1-144
Zhong, Y. W., Zhou, P., Cheng, H., Zhou, Y. D., Pan, J., Xu, L., et al. (2022). Metagenomic features characterized with microbial Iron Oxidoreduction and mineral interaction in southwest Indian ridge. Microbiol. Spectr. 10:e0061422. doi: 10.1128/spectrum.00614-22
Zhu, Q. K., and Zhou, H. Y. (2021). Reviews and prospects of submarine metalliferous sediments of hydrothermal origin. Mar. Sci. 45, 69–80. doi: 10.11759/hykx20191221001
Keywords: copper-resistant bacteria, deep-sea hydrothermal fields, Indian Ocean, extracellular polymeric substances, Marinobacter metalliresistant sp. nov
Citation: Yu T, Qin M, Shao Z, Zhao Y and Zeng X (2024) Isolation of highly copper-resistant bacteria from deep-sea hydrothermal fields and description of a novel species Marinobacter metalliresistant sp. nov. Front. Microbiol. 15:1390451. doi: 10.3389/fmicb.2024.1390451
Edited by:
L. F. Wu, Center National de la Recherche Scientifique, FranceReviewed by:
Juan Castro-Severyn, Catholic University of the North, ChileJeffrey Henderson, Washington University in St. Louis, United States
Copyright © 2024 Yu, Qin, Shao, Zhao and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Zeng, emVuZ3hpYW5nQHRpby5vcmcuY24=
†These authors have contributed equally to this work
 Tong Yu
Tong Yu Meng Qin
Meng Qin Zongze Shao
Zongze Shao Yuemei Zhao1
Yuemei Zhao1 Xiang Zeng
Xiang Zeng