
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 June 2024
Sec. Microbe and Virus Interactions with Plants
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1390422
 Zhiting Li1,2†
Zhiting Li1,2† Wanzhen Feng1,2†
Wanzhen Feng1,2† Zaobing Zhu3†
Zaobing Zhu3† Shengdan Lu1,2
Shengdan Lu1,2 Mingze Lin1,2
Mingze Lin1,2 Jiali Dong4
Jiali Dong4 Zhixin Wang1,5
Zhixin Wang1,5 Fuxiu Liu5*
Fuxiu Liu5* Qinghe Chen1,2*
Qinghe Chen1,2*Phytophthora sojae is a devastating plant pathogen that causes soybean Phytophthora root rot worldwide. Early on-site and accurate detection of the causal pathogen is critical for successful management. In this study, we have developed a novel and specific one-pot RPA/PCR-CRISPR/Cas12 assay for on-site detection (Cas-OPRAD) of Phytophthora root rot (P. sojae). Compared to the traditional RPA/PCR detection methods, the Cas-OPRAD assay has significant detection performance. The Cas-OPRAD platform has excellent specificity to distinguish 33 P. sojae from closely related oomycetes or fungal species. The PCR-Cas12a assay had a consistent detection limit of 100 pg. μL−1, while the RPA-Cas12a assay achieved a detection limit of 10 pg. μL−1. Furthermore, the Cas-OPRAD assay was equipped with a lateral flow assay for on-site diagnosis and enabled the visual detection of P. sojae on the infected field soybean samples. This assay provides a simple, efficient, rapid (<1 h), and visual detection platform for diagnosing Phytophthora root rot based on the one-pot CRISPR/Cas12a assay. Our work provides important methods for early and accurate on-site detection of Phytophthora root rot in the field or customs fields.
Phytophthora sojae is a major oomycete pathogen that causes damping-off of seedlings, and root and stem rot of soybean plants in soybean-producing regions worldwide (Erwin and Ribeiro, 1996; Tyler, 2007). Phytophthora sojae can infect soybean at all growth stages and yield losses can reach 100% in soybean fields when susceptible varieties are planted (Wang et al., 2006; Dorrance, 2018). Phytophthora sojae is listed as a plant quarantine pathogen in China due to its high risk to agricultural and economic security (Wang et al., 2006; Dai Y. et al., 2019). With the increasing global transport of soybeans, P. sojae presents a serious threat to soybean production worldwide. Phytophthora root rot is difficult to diagnose due to the symptoms resembling those caused by other pathogens such as Fusarium, Pythium, or Rhizoctonia species. Therefore, the development of a new technique for the detection of the pathogen will be essential to prevent transmission of P. sojae into disease-free soybean growing regions.
Various methods are available for detecting P. sojae infection, isolation and cultivation, leaf disc baiting, and molecular detection. Isolation-based methods offer incontrovertible evidence of P. sojae infection (Su and Shen, 1993; Chen et al., 2004), while the leaf-disc baiting assay is less complicated and more effective than isolating the pathogen from plant tissues (Jinhuo and Anderson, 1998; Dai T. et al., 2019; Matthiesen et al., 2021). Molecular approaches, such as conventional polymerase chain reaction (PCR) and real-time PCR (qPCR), are well-established and considered powerful diagnosis tools (Bienapfl et al., 2011; Haudenshield et al., 2017; Xiong et al., 2019). However, culturing and baiting detection methods are time-consuming and relatively insensitive. Furthermore, conventional PCR or real-time PCR require well-equipped laboratories and well-trained personnel, hindering their application in resource-limited environments or on-site detection. In contrast, various isothermal nucleic acid amplification techniques, such as loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA), allow nucleic acid amplification at a single temperature, making them suitable for field and on-site diagnosis in low-resource areas (Dai et al., 2012; Rojas et al., 2017). However, their practical application remains challenging due to the issues of false positives caused by non-specific amplification.
The clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated (Cas) system has recently been applied for molecular detection (Gootenberg et al., 2017; Chen et al., 2018; Li et al., 2018; Wheatley and Yang, 2021; Eisenstein, 2022). Several CRISPR/Cas-based detection platforms, such as DETECTR (Chen et al., 2018), HOLMES (Li et al., 2018), and SHERLOCK (Gootenberg et al., 2017; Kellner et al., 2019), have been developed as point-of-care diagnostic tools for highly specific and sensitive detection of the pathogen. CRISPR-based molecular detection offers several advantages, including rapid detection times, high specificity, and sensitivity comparable to or exceeding traditional methods like PCR (Islam and Kasfy, 2023; Kasfy et al., 2024). Additionally, these methods are relatively simple and can be adapted for use in various settings, including point-of-care diagnostics and field surveillance (Li et al., 2019; Islam and Kasfy, 2023). Most recently, Cas12a-based detection methods have been used to diagnose plant pathogenic fungi (Magnaporthe oryzae), bacterial pathogens (Candidatus Liberibacter asiaticus), and viruses (PVX, PVY, TMV, ASPV, RSV) (Aman et al., 2020; Jiao et al., 2021; Kang et al., 2021; Wheatley and Yang, 2021; Sánchez et al., 2022; Zhu et al., 2022). However, these platforms currently employ step-by-step nucleic acid preamplification and Cas12a cleavage steps, which could potentially increase the risk of carryover contamination between processes. This is particularly concerning for specific detection, as the extra processes of tube opening during preamplification can introduce non-specific amplification and cross-contamination (Wang SY et al., 2021; Guo et al., 2023). To address these challenges, there is a need to develop one-pot CRISPR/Cas-based assays for plant pathogen detection. Such assays would integrate the preamplification and cleavage steps into a single reaction, reducing the risk of contamination and improving specificity.
In this study, we present a novel one-pot RPA/PCR CRISPR/Cas12a assay for the simple, rapid, specific, and on-site visual detection of P. sojae, combining either PCR or RPA with CRISPR/Cas12a. Following amplification, an optimized Cas12a solution was added to the reaction mixture to initiate cleavage. The results were visualized by naked-eye observation under blue or UV light. To facilitate in-field detection in resource-limited settings, we further combined the CRISPR/Cas12a assay with lateral flow strips. Except for the analytical sensitivity and specificity, the feasibility of this CRISPR/Cas12a assay was evaluated using inoculated soybeans samples and field samples. The Cas-OPRAD assay is a simple, specific, sensitive, and visual method for P. sojae detection, which has the potential to be widely applied in the management of plant pathogens.
A total of 33 P. sojae isolates from different geographic areas were tested in this study, as well as 37 isolates from 17 different oomycetes (including 34 isolates of 14 species representing 10 clades of Phytophthora), and 16 other fungi and bacteria isolated from soybean and other hosts. The origin, host affiliation, and number of tested isolates are listed in Table 1. All isolates were routinely maintained in liquid nitrogen at the School of Breeding and Multiplication, Hainan University, China.
Genomic DNA was extracted from mycelial cultures using a DNAsecure Plant kit (Tiangen, Beijing, China) as described previously (Wu et al., 2017). The purified genomic DNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific), and stored as aliquots at a concentration of 100 ng μL−1 in sterile distilled water at −20°C.
All the primers, DNA fragments (including CRISPR RNA [crRNA], and ssDNA probe) were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). REnGen® Lba Cas12a and NEBufferTM were purchased from New England BioLabs Ltd. (Beijing, China). High-Affinity HotStart Taq (ET108-1) and 2 × PCR SuperMix were purchased from Tiangen Biotech (Beijing, China), while RPA Kits were obtained from AMP-Future Biothech Co., Ltd. (Changzhou, China). The lateral flow strips were purchased from Tiosbio Biotech Co. Ltd. (Beijing China). Nuclease-free H2O was obtained from Sangon Biotech (Shanghai, China). Genomic DNA Kits were purchased from Tiangen Biotech (Beijing, China).
To identify the conserved, single-copy, candidate detection targets for P. sojae specific PCR or RPA assays, we retrieved the annotated genomic sequence of P. sojae.1 All the gene sequences of P. sojae were used to perform BLAST searches against the publicly available genomic sequences of Phytophthora and related taxa (e.g., Pythium, and Phytopythium species) in the National Center for Biotechnology Information (NCBI) database. A total of 1,187 P. sojae genes did not share any sequence similarity with genomic sequences of any other species. Three loci (PHYSODRAFT_299276, PHYSODRAFT_255386, and PHYSODRAFT_531894) were randomly selected as detection targets to test the specificity using a collection of Phytophthora species (Table 1) in conventional PCR. Ultimately, a single-copy gene (PHYSODRAFT_299276) was selected as the specific target for P. sojae detection.
The target sequence of P. sojae (PHYSODRAFT_299276) was used to design the crRNAs and primers for the CRISPR/Cas assay. RPA primers were designed and analyzed using Primer 5 and Primer-Explore V5 software.2 The crRNA with 20–24 bp target-dependent sequence following PAM (5’-TTTN-3′) sit was designed using EuPaGDT3 and an RNA scaffold to assist in protein binding. RPA primers and crRNA were synthesized by Sangon Biotech (Shanghai, China). The sequences of all primers are listed in Supplementary Table S1.
Conventional PCR assays were performed in 10 μL reactions. Each reaction included 5 μL of 2 × Taq PCR Master Mix (Tiangen Biotech Co., Ltd., Beijing, China), 0.5 μL of each forward and reverse primer (10 μM), 1 μL of template DNA (100 ng μL−1), and 3.0 μL of nuclease-free H2O. Thermal cycling was carried out using PTC-200 Thermo Cycler (MJ Research, Watertown, MA, United States) with the following conditions: 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 58°C for 45 s, and 72°C for 30 s, and a final extension at 72°C for 10 min. The PCR amplification products were separated by electrophoresis on 2.0% agarose gel, stained with ethidium bromide, and visualized under UV light. Each experiment was performed in triplicate.
RPA assay was performed using the AMP-Future Biotech Co. Ltd. (Changzhou, China) according to the manufacturer’s instructions. Briefly, each 10 μL reaction contained 5.9 μL of A buffer, 0.5 μL of B buffer, 0.4 μL of each forward and reverse primers (10 μM), 1 μL of DNA (100 ng μL−1), and 0.8 μL of nuclease-free H2O. The mixture was incubated in a conventional water bath at 38°C for 15 min for RPA reaction.
The efficiency of the CRISPR/Cas12a cleavage system is directly influenced by the concentration of crRNA and Cas12a and affects both trans-cleavage efficiency and fluorescence intensity. To determine the optimal CRISPR/Cas12a reaction conditions, a range of Cas12a and crRNA concentrations (50 nM ~ 300 nM), Cas12a-mediated cleavage temperature (37 ~ 42°C), RPA reaction time (5 min ~ 30 min), and trehalose concentrations (0.006 mM ~ 6 mM) were tested. Nuclease-free water was used as a negative control. The CRISPR/Cas12a reaction products were analyzed by naked eye observation or by detecting the maximal fluorescence signal value to determine the optimal CRISPR/Cas reaction conditions. All experiments were repeated three times.
The CRISPR/Cas12a reaction was performed in a 5 μL reaction mixture containing 0.75 μL crRNA (4 μM), 1.5 μL Cas12a (2 μM), 0.75 μL ssDNA FQ reporter (100 nM), and 1.5 μL of 10× NEB Buffer r2.1, 0.5 μL of nuclease-free H2O, along with 10 μL of RPA products (or 0.5 μL of 6 mM trehalose and 10 μL of PCR products). In the PCR-CRISPR/Cas12a assay, 6 mM trehalose was added to the reaction solution to protect the activity of the Cas12a protein. The reactions were incubated at 38°C for 10 min and then placed on an ultraviolet (UV) or blue light transilluminator for naked-eye detection.
The one-pot CRISPR/Cas12a detection combines RPA/PCR reaction with Cas12a cleavage in a single tube reaction. To perform the assay, 10 μL of the RPA/PCR assay containing targets was added to the bottom of the tube, and 5 μL of CRISPR/Cas12a reaction solution was initially added inside the inner wall of the tube lid. After incubating the RPA/PCR reaction for 10–60 min, the CRISPR/Cas12a solution was swung into the RPA reaction solution by hand on-site, and the mixture was incubated for an additional 15 min. The reaction tubes were then placed on a blue or UV light transilluminator for naked eye detection (Figure 1).
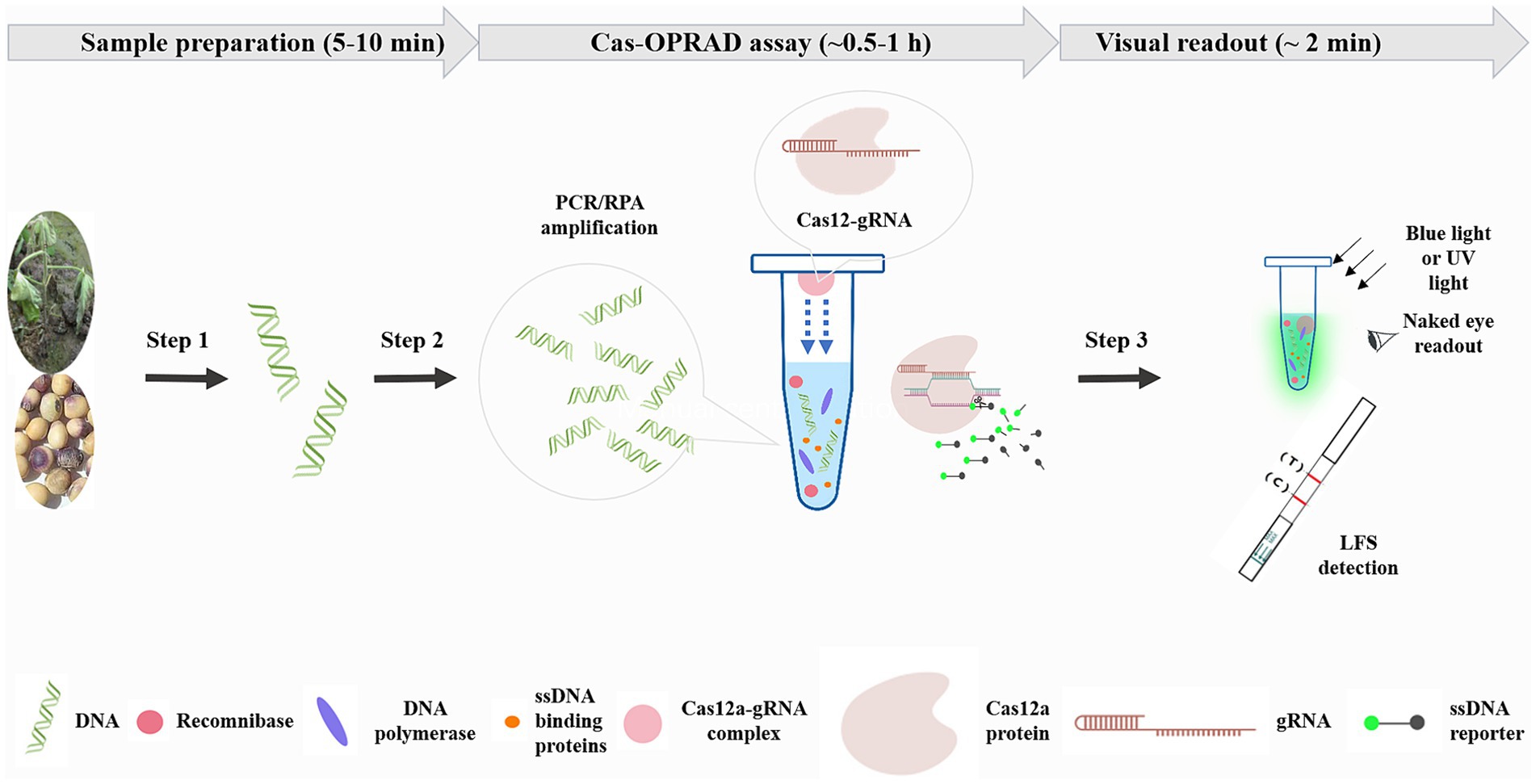
Figure 1. Schematic of Cas-OPRAD workflow for rapid sensitivity, and visual detection of P. sojae. Step 1: sample preparation. Step 2: Cas-OPRAD assay. Step 3: visual readout.
The structure of the lateral flow strip (LFS) incorporated an absorbent pad, a conjugate pad, a nitrocellulose (NC) membrane, and a sample pad on a plastic adhesive backing card. The capture reagents streptavidin and anti-IgG antibody were dispensed onto the NC membrane, and each band was separated by 5 mm. When there is no target in the system, the conjugate was captured by the streptavidin through biotin, the control line became red because of the aggregation of the colloidal gold in this area, while the test line without the flow-through of the conjugate had no color change. However, if there are targets in the system, the ssDNA reporter is cleavaged by CRISPR/Cas12a, and FAM and biotin molecules are separated. After the integration of ssDNA-biotin captured by streptavidin at the C line, the remaining conjugate of FAM-ssDNA and anti-FAM antibody would flow into the test line and be captured by anti-lgG antibody. The lateral flow detection was performed according to the Tiosbio protocol (Tiosbio Biotech Co. Ltd., Beijing China). Briefly, 85 μL Tiosbio assay buffer was added to 15 μL of the RPA-CRISPR/Cas12a reaction and incubated for 5 min. The lateral flow test strip was then inserted into the reaction mixture for 5 ~ 10 min to allow the reaction to occur. A positive test result was indicated by the appearance of both the control and test lines, while a negative result showed only the control line. All lateral flow assays were performed in triplicate.
To test the specificity of the one-pot RPA/PCR CRISPR/Cas12a assays, a total of 33 P. sojae isolates from different geographic origins, 37 isolates from 17 different oomycetes species, and 16 other fungi and bacteria were used (Table 1). To determine the sensitivity of the CRISPR/Cas12a assay, 10-fold serial dilutions of P. sojae purified gDNA ranging from 100 to 0.001 ng μL−1 were used as DNA templates. Nuclease-free H2O was used as the negative control. All assays were repeated in triplicate for each concentration of the gDNA template.
To assess the practical application of the one-pot CRISPR/Cas12a detection for P. sojae, soybean seedlings were inoculated with P. sojae as described previously (Zhao et al., 2015). The DNA was extracted and evaluated using the developed RPA/PCR-CRISPR/Cas12a assays. Each experiment was repeated three times. In addition, a total of 20 soybean plants collected from different fields in Fujian and Hainan provinces were assayed using the RPA/PCR-CRISPR/Cas12a assays. The DNA was extracted from infected soybeans as described previously (Chen et al., 2013), and the RPA/PCR-CRISPR/Cas12a assay was performed as described above. Purified DNA of P. sojae was used as the positive control and healthy soybeans were used as negative control. The P. sojae infection was confirmed using the conventional isolation culture method for the identification of the pathogens based on morphological characteristics. Each experiment was repeated three times.
A schematic description of the Cas-OPRAD assay and the approximate time of each step are shown in Figure 1. The shortest time of Cas-OPRAD assay can reach about 0.5–1 h, including three primary steps: sample preparation, Cas-OPRAD assay, and visual readout. In the first step, the sample was prepared by using a rapid nucleic acids extraction method, and then the RPA or PCR technique ensured the efficient production of DNA amplification from the sample. In the first step, the sample was prepared by using a rapid nucleic acids extraction kit, the RPA or PCR technique was employed to enrich the target DNA fragments, which ensures the efficient production of DNA amplification from the sample. We utilized the principle of liquid surface tension and spatial isolation to separate the RPA/PCR reaction from sgRNA-Cas12 in a one-pot reaction. Specifically, the sgRNA-Cas12 complex was adsorption suspended on the top cover of the reaction tube, while the RPA or PCR reaction was located at the bottom of the tube. After the nucleic amplification reaction was completed, a simple centrifugation or manual shaking was used to mix the two components for reaction, this design avoids cross-contamination of various components in complex systems. For amplification product identification, the gRNA was specially designed to target RPA/PCR products, and the amplification products were recognized and cleaved, especially by the sgRNA and Cas12a complex. Furthermore, the Cas12a-sgRNA-target ternary complex activated the trans-cleavage activity of Cas12a, resulting in ultrafast cleavage of the ssDNA fluorescent probe and consequent fluorescent signal generation. The fluorescent signal generated from trans-cleavage was monitored by a fluorescence detector or LFS device.
The specificity of Cas OPRAD was carried out in three steps. Firstly, we determine sequence specificity through comparative genomic analysis, 1,187 candidate genes were identified from P. sojae. Pair-wise gene sequence alignments were then performed, and three P. sojaespecific genes were selected as the detection target candidates: PHYSODRAFT_299276, PHYSODRAFT_255386, and PHYSODRAFT_531894. Then, the primers were designed for these genes, and a conventional PCR assay was used to evaluate the specificity of the primer pairs. Primers So-F/So-R (Supplementary Table S1), designed from PHYSODRAFT_299276, successfully distinguished P. sojae from other plant pathogens by amplifying a 1,310 bp target band. In contrast, PHYSODRAFT_255386 and PHYSODRAFT_531894 amplified non-target bands from the gDNA of P. sojae (data not shown). These results suggest that the primers designed from PHYSODRAFT_299276 had good specificity for P. sojae detection. Therefore, a probe for PHYSODRAFT_299276 was used as the detection target.
To verify the feasibility of our scheme, we first optimized the concentration of crRNA and the ratio of Cas12a to crRNA in the CRISPR/Cas12a assay, as the crRNA concentration plays a crucial role in CRISPR/Cas12a reaction. The optimal concentration of crRNA and the optimal ratio of Cas12a to crRNAs were determined to be 200 nM and 1:1, respectively (Supplementary Figure S1A). Under the optimized conditions of crRNA concentration and the ratio of Cas12a to crRNAs, we further optimized CRISPR/Cas12a reaction time and temperature. The ideal conditions for the cleavage were determined to be 38°C for 10 min (Supplementary Figures S1B,C).
To maintain the activity of Cas12a in one pot, according previous report by Saisawang et al. (2023), we adjusted the temperature of the heat cap in the PCR-CRISPR/Cas12a reaction to 50°C and trehalose was added into CRISPR/Cas reaction to help enzyme stabilization in target cleavage. The optimized concentration of trehalose for PCR-CRISPR/Cas12a was 0.6 mM (Supplementary Figure S1D). Therefore, subsequent PCR-CRISPR/Cas12a assays were carried out under these conditions. We tested eight reaction systems (reactions # 1–8) for PCR-CRISPR/Cas12a (Supplementary Figure S2) and six reactions (reactions # 1–6) for RPA-CRISPR/Cas12a, respectively (Figure 2). The generation of fluorescence comes from Cas12 cleavage FQ ssDNA probe molecules, after incubation at 38°C for 15 min and 5 ~ 10 min of CRISPR/Cas detection, only reaction #1, containing Cas12a, crRNAs, target DNA, ssDNA-FQ, and RPA or PCR reaction mixture, generated a naked-eye visual result that was visible under blue or UV light (Figure 2; Supplementary Figure S2). To further confirm the specificity and reaction time of the fluorescence signal, the one-pot RPA-CRISPR/Cas12a assay was analyzed on an AriaMx quantitative PCR instrument. These fluorescence value results demonstrate that the CRISPR/Cas12a assay provides a high fluorescence intensity value (>30,000 AU) within 15 min, which indicates that Cas-OPRAD can be offered a rapid, specific, and simple method for detecting P. sojae (Figure 2B).
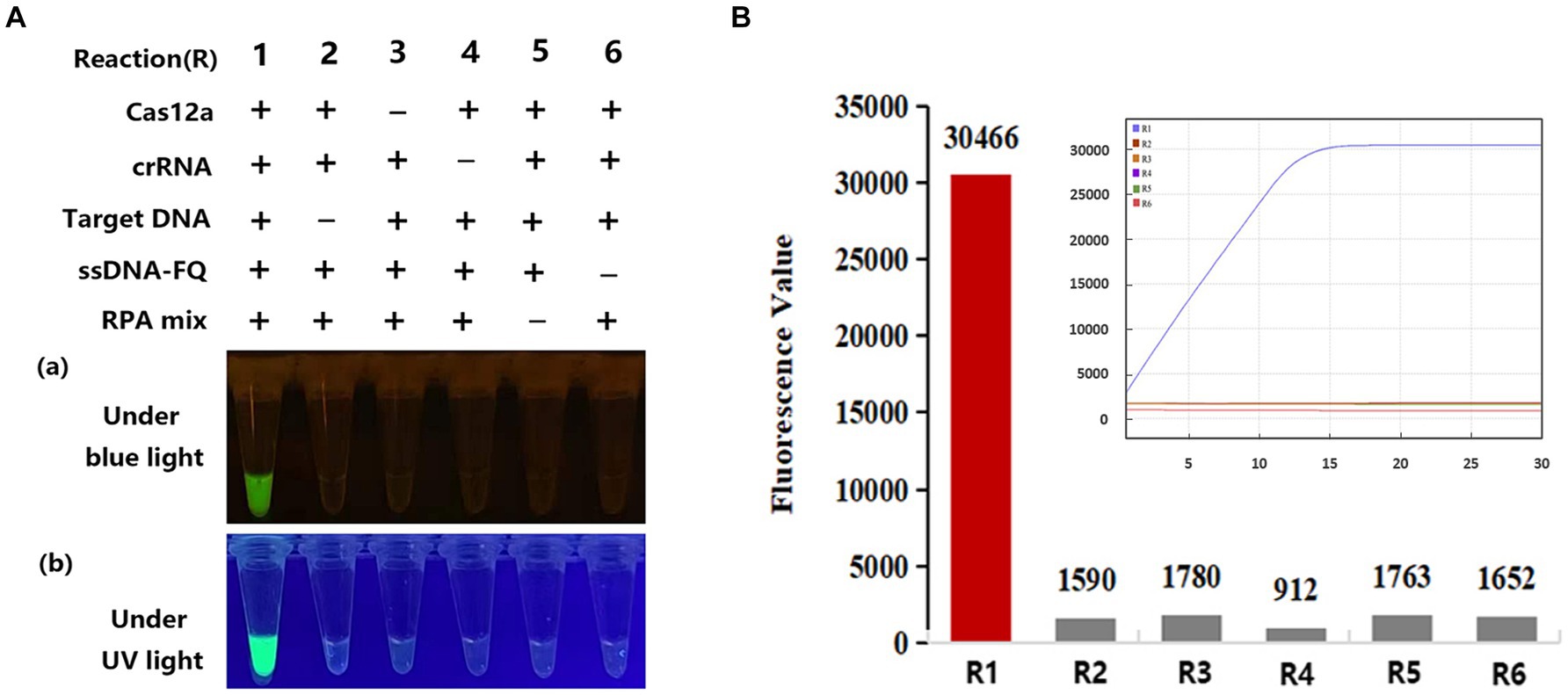
Figure 2. Evaluation of Cas-OPRAD reactions with various components. (A) RPA-CRISPR/Cas12a reaction. (a) Visualization under blue light; (b) visualization under UV light; (B) Fluorescent readout with quantitative PCR instrument. Each experiment was repeated three times with similar results.
To validate the specificity of Cas OPRAD detection, DNA templates were extracted from 33 P. sojae isolates and 53 isolates of other non-P. sojae were evaluated (Table 1). After the Cas-OPRAD reaction, only P. sojae showed a positive reaction, while no color change was observed for the other pathogens, including other Phytophthora spp. and other fungal and bacteria isolates from soybeans (Figures 3A,B; Supplementary Figures S3A,B). Notably, there was no cross-reaction with closely related species, such as P. melonis, P. cinnamomi, and P. vignae, which belong to the same clad 7b as P. sojae. These results demonstrate that the Cas-OPRAD assay developed in this study is highly specific for P. sojae.
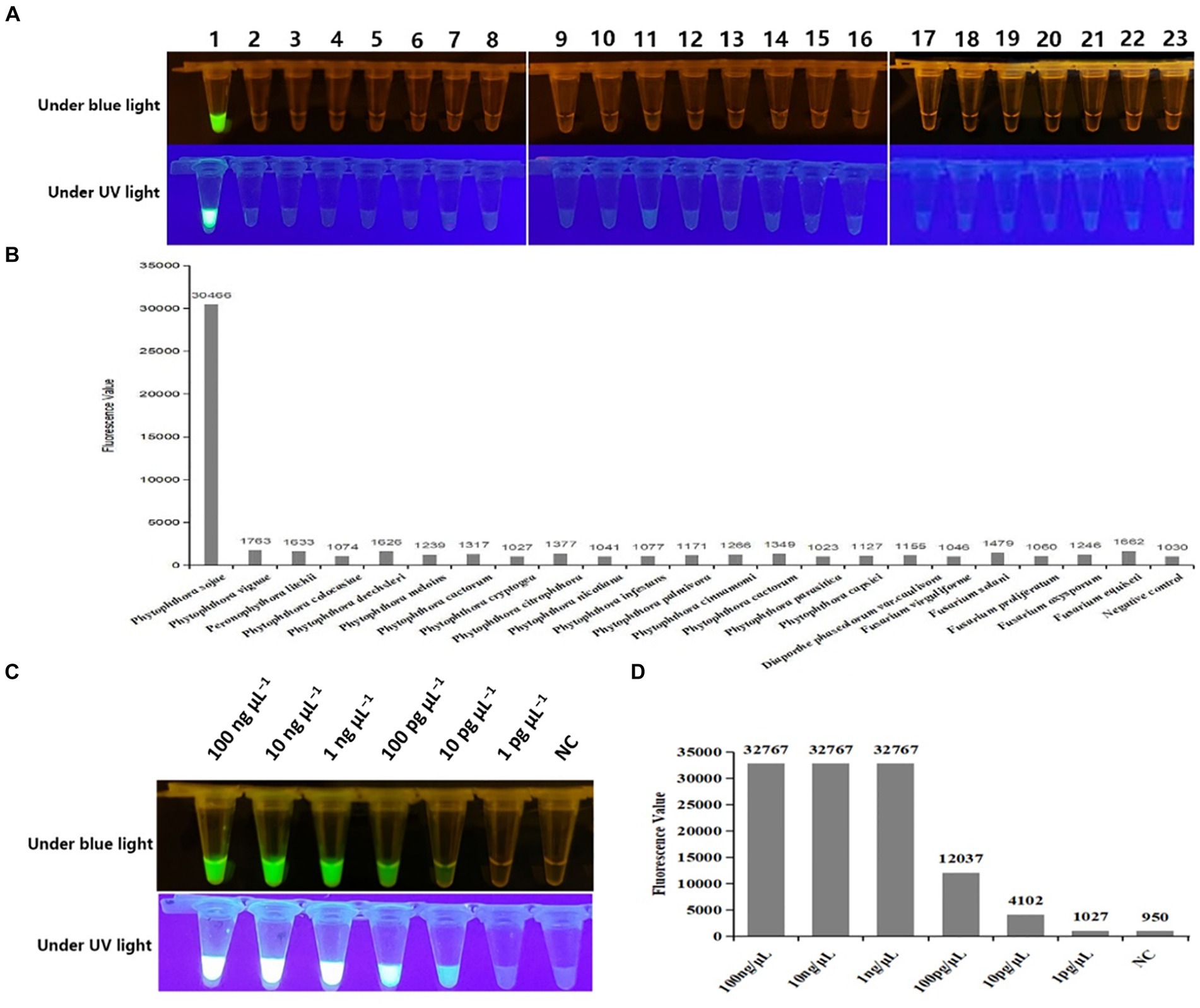
Figure 3. Specificity and sensitivity test of the Cas-OPRAD assay for detection of P. sojae. (A) Specificity visualization assay under blue and UV light; (B) Fluorescent readout detection for specificity assay of P. sojae; Lane 1: Phytophthora sojae; 2: Phytophthora vignae; 3: Phytophthora infestans; 4: Phytophthora meloins; 5: Phytophthora cryptogea; 6: Phytophthora parasitica; 7: Phytophthora drechsleri; 8: Phytophthora capsici; 9: Phytophthora cactorum; 10: Phytophthora cinnamomi; 11: Phytophthora citrophthora; 12: Phytophthora colocasiae; 13: Phytophthora cactorum; 14: Phytophthora nicotiana; 15: Peronophythora litchii; 16: Phytophthora palmivora; 17: Diaporthe phaseolorum var.caulivora; 18: Fusarium virguliforme; 19: Fusarium equiseti; 20: Fusarium solani; 21: Fusarium proliferatum; 22: Fusarium oxysporum; 23: Negative control. (C) Sensitivity visualization assay under blue and UV light; (D) Fluorescent readout detection of P. sojae.
To further evaluate the sensitivity of the CRISPR/Cas12a assay, 10-fold serially diluted DNA templates of P. sojae were used. As shown in Supplementary Figures S4A,B the detection limit for the PCR-CRISPR/Cas12a was 100 pg. μL−1, while the sensitivity of the RPA-CRISPR/Cas12a assay reached 10 pg. μL−1 in both fluorescence detection (Figures 3C,D). Notably, the sensitivity for Cas-OPRAD was found to be at least 10 times higher than that of the PCR-CRISPR/Cas12a method. These results suggest that the RPA-CRISPR/Cas12a-based assay is more sensitive for early detection of the soybean pathogen P. sojae.
The Cas-OPRAD assay is performed at an isothermal temperature, making it potentially suitable for on-site detection. In addition to fluorescence detection, we also utilized lateral flow strips (LFS) for a visual CRISPR/Cas12a assay that can facilitate on-site detection of P. sojae. The principle of the test strip is shown in Figure 4A, the structure of LFS incorporated an absorbent pad, a conjugate pad, a nitrocellulose (NC) membrane, and a sample pad on a plastic adhesive backing card. The capture reagents streptavidin and anti-IgG antibody were dispensed onto the NC membrane, and each band was separated by 5 mm. When there is no target in the system, the conjugate was captured by the streptavidin through biotin, the control line became red because of the aggregation of the colloidal gold in this area, while the test line without the flow-through of the conjugate had no color change. However, if there are targets in the system, the ssDNA reporter is cleavaged by CRISPR/Cas12a, and FAM and biotin molecules are separated. After the integration of ssDNA-biotin captured by streptavidin at the C line, the remaining conjugate of FAM-ssDNA and anti-FAM antibody would flow into the test line and be captured by anti-lgG antibody. As expected, bands appeared on both the test line and control line of the strip in a positive reaction, while only a band appeared on the control line in a negative reaction. Therefore, the lateral flow assay format provides a rapid, simple, instrument-free, on-site field detection method for P. sojae.
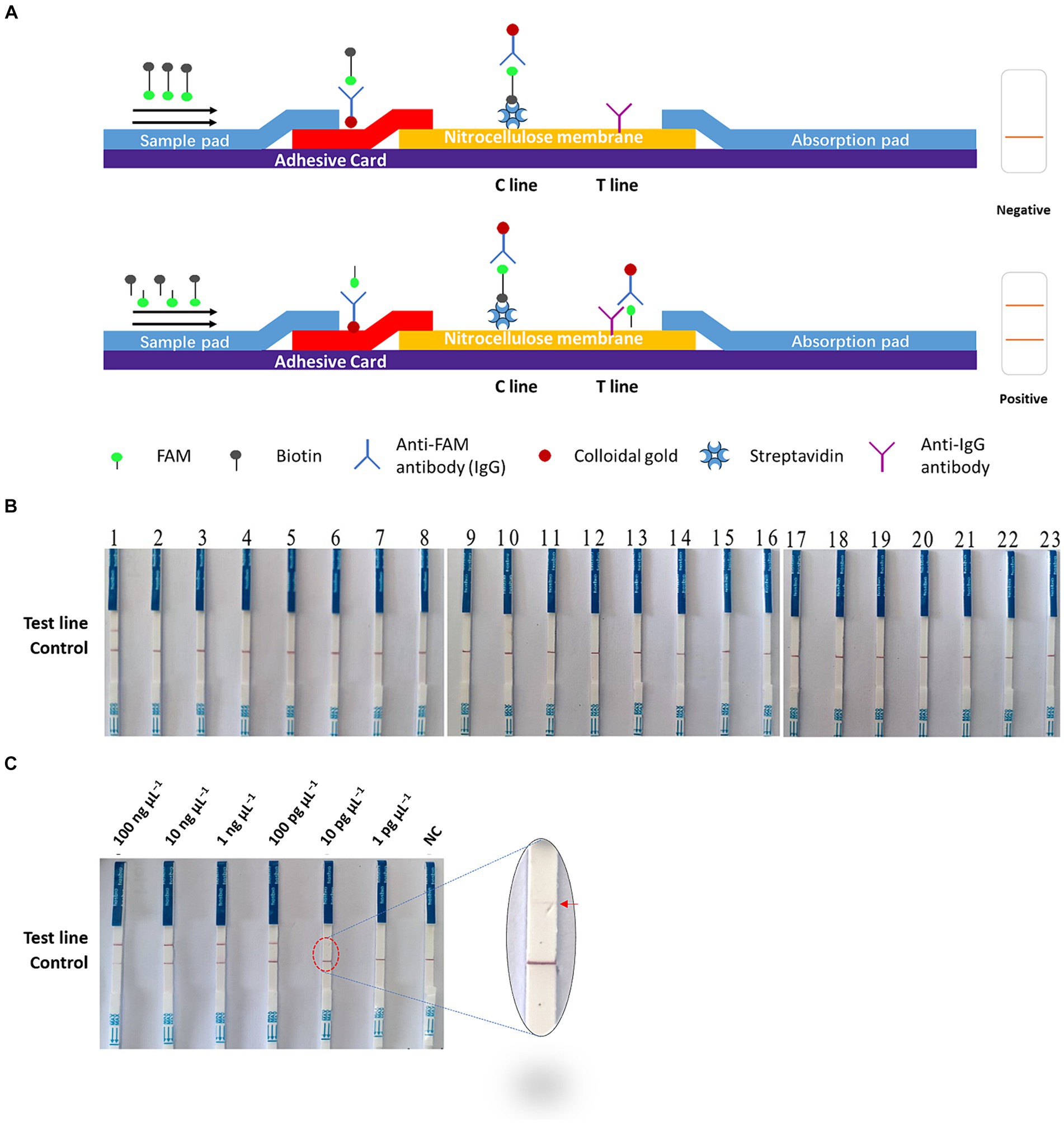
Figure 4. On-site Cas-OPRAD assays for detection of P. sojae. (A) Structure and design of the LFS. (B) Lateral-flow readout detection of P. sojae. Lane 1: Phytophthora sojae; 2: Phytophthora vignae; 3: Phytophthora infestans; 4: Phytophthora meloins; 5: Phytophthora cryptogea; 6: Phytophthora parasitica; 7: Phytophthora drechsleri; 8: Phytophthora capsici; 9: Phytophthora cactorum; 10: Phytophthora cinnamomi; 11: Phytophthora citrophthora; 12: Phytophthora colocasiae; 13: Phytophthora cactorum; 14: Phytophthora nicotiana; 15: Peronophythora litchii; 16: Phytophthora palmivora; 17: Diaporthe phaseolorum. (C) The sensitivity assay of lateral flow strips readout detection for P. sojae. Lane 1:100 ng μL−1; 2: 10 ng μL−1; 3: 1 ng μL−1; 4: 100 pg μL−1; 5: 10 pg μL−1; 6: 1 pg μL−1; and 7: NC (negative control).
The specificity and sensitivity of the Cas-OPRAD LFS platform were also evaluated for the detection of P. sojae. The specificity test results were visualized using LFS tests (Figure 4B). In the LF-OPRAD assay, red bands at the T line were only observed for the Cas-OPRAD products of the P. sojae DNA sample, and no red band at the T line was observed for reactions with other pathogens. These results indicate that the developed Cas-OPRAD platform for the P. sojae pathogens was highly specific. In sensitivity tests, the results from LFS indicated are same sensitivity as for the Cas-OPRAD assays. For example, a red band at the T line was observed for reactions at all P. sojae DNA dilutions except for 1 pg. μL−1 and NC (Figure 4C), indicating that the sensitivity of Cas-OPRAD was 10 pg. μL−1, these results indicate that the on-site platform could be used for P. sojae pathogen detection without dedicated professional equipment.
To verify the validity and feasibility of the Cas-OPRAD for diagnosing soybean samples, the DNA was extracted from three regions (I: roots and underground stems. II: cotyledons and stem above the part of ground. III: epicotyl and first leaf) of inoculated soybean seedlings (Figure 5A). As shown in Figures 5B–D, positive reactions were observed in regions I and II, but not in region III or healthy soybeans using Cas-OPARD or PCR-CRISPR/Cas12a, the result indicates that under artificial inoculation, the P. sojae infection in the roots of soybeans is higher than that in the leaves. Therefore, in field sample collection, it is necessary to prioritize collecting regions I or II for Cas-OPARD analysis.
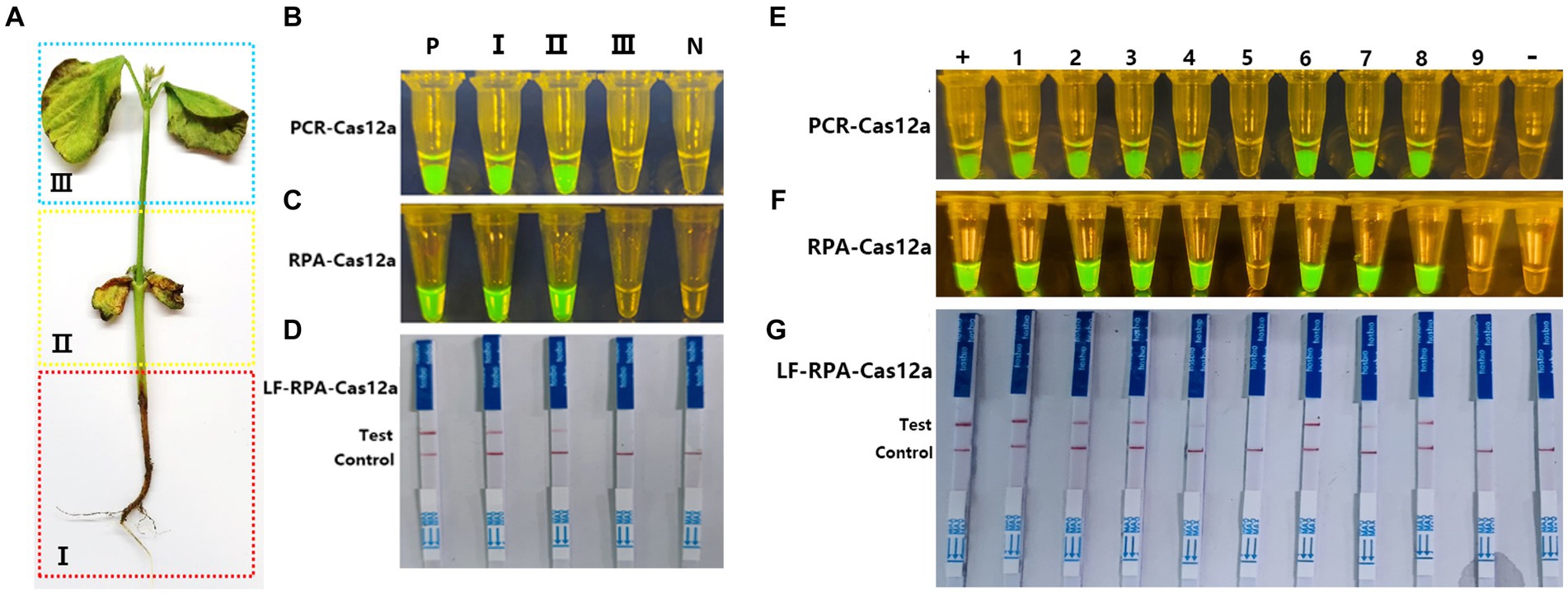
Figure 5. Application of Cas-OPRAD assays for detection of P. sojae in filed samples. (A) Artificial division of different areas of the soybean represents the varying stages of infection; (B) Blue and UV light visualization assay of one-pot PCR-Cas12a in inoculated soybean seedlings; (C) Blue and UV light visualization assay of one-pot RPA-Cas12a in inoculated soybean seedlings; (D) One-pot RPA-Cas12a LFS assay in filed soybean sample; +: Positive control; −: negative control; Lane1: Region I; Lane 2: Region II; Lane3: Region III. (E) one-pot PCR-Cas12a LFS assay; (F) one-pot RPA-Cas12a LFS assay; (G) RPA-Cas12a LFS assay in filed soybean sample. +, Positive control; −, negative control; Lane1-8: naturally infected soybean root samples; 9: healthy soybean sample.
According to the results above, a total of 20 field soybean samples collected from natural fields of Fujian and Haina province, China, were used for Cas-OPARD assay or PCR-CRISPR/Cas12a with both fluorescence and lateral flow readouts. Of the 20 collected samples, 16 were positive and four were negative using both the Cas-OPARD, the results obtained by using the PCR-CRISPR/Cas12a method are consistent with those obtained by Cas-OPRAD (Figures 5E,F). The detection results showed 100% consistency between the visual detection under blue or UV light and the lateral flow strip readout (Figures 5F,G). These results demonstrate the high detection capabilities of the CRISPR/Cas12a assay for P. sojae.
The identification of a specific target gene is critical for the development of accurate and sensitive molecular diagnosis methods. Although several genes, such as the internal transcribed spacer (ITS) region, β-tubulin (TUB) loci, elongation factor 1α, and ras-related protein Ypt1 (YPT), have been used to design primers for detecting Phytophthora (Kroon et al., 2004; Blair et al., 2008). However, molecular detection based on these target genes may pose false positives due to high homology (Kunadiya et al., 2017). Therefore, the combination of comparative genomics and bioinformatics has been applied to identify specific target genes for various plant pathogen species, including Pectobacterium carotovorum (Ahmed et al., 2018), P. infestans (Kong et al., 2020), P. cinnamomi (Chen et al., 2022), Magnaporthe oryzae Triticum (MoT) (Kang et al., 2021), and Fusarium circinatum (Xu et al., 2023). In this study, we performed a comparative genomic analysis based on public genomic sequence data of several oomycetes species to identify a candidate detection target gene that is specific to P. sojae. We identified a single-copy gene (PHYSODRAFT_299276) and developed RPA/PCR CRISPR/Cas12 assays to verify its specificity using 33 P. sojae isolates and 53 isolates of other non-P. sojae species. Moreover, this assay did not show cross-reaction with closely related species clade 7b, P. melonis, P. cinnamomi, and P. vignae. RPA/PCR CRISPR/Cas12 assay that showed 100% inclusivity and 100% exclusivity when tested among closely related species. Our results suggest that this assay is highly specific for P. sojae detection.
One-pot CRISPR/Cas coupled with nucleic acid amplification detection streamlines genetic analysis by integrating multiple steps into a single reaction. This synergy enhances efficiency, reduces time, and minimizes contamination risks, crucial for applications like pathogen detection, genetic screening, and diagnostics. By combining CRISPR’s precision gene detection with amplification’s sensitivity, it enables rapid and accurate identification of target sequences, revolutionizing research, and medical diagnostics (Wang R et al., 2021; Hu et al., 2022). The simplicity and versatility of this approach hold immense potential for advancing various fields, from healthcare to agriculture and beyond, promising faster, more accessible, and reliable genetic analysis. To reduce the risks of cross-contamination and improve the convenience of on-site detection of P. sojae, we developed a one-pot CRISPR/Cas12a OPRAD assay. In the One-pot design, we utilized the principle of liquid surface tension and spatial isolation to separate the RPA/PCR reaction from the sgRNA-Cas12 a one-pot reaction. This integrated strategy initially provided a new idea for on-site diagnosis and epidemic control of COVID-19 (Ding et al., 2020). Additionally, to maintain the activity of Cas12a in one pot PCR-CRISPR/Cas12, we adjusted the temperature to 50°C of the heat cap and optimized 0.6 mM trehalose to maintain the stability in one-pot PCR-CRISPR/Cas12a reaction. As a nonreducing disaccharide, trehalose can provide proteins with the most stability in a one-pot reaction, as they form hydrogen bonds with the protein to minimize damage from desorbing the hydration shell (Lin et al., 2022; Ghouneimy et al., 2023). CRISPR/Cas12a reagents were pre-loaded into the inner wall of the tube lid. Following RPA/PCR amplification, the reagents were mixed into the reaction solution by centrifugation, avoiding potential aerosol contamination from re-opening the tube lid post-amplification. This assay minimizes potential cross-contamination issues by avoiding the need to reopen the reaction tube lid after the RPA reaction.
Isothermal amplification techniques have emerged as powerful alternatives to PCR (Polymerase Chain Reaction) due to their simplicity, rapidity, and applicability in various settings. While PCR relies on thermal cycling to amplify DNA, isothermal amplification methods operate at a constant temperature, making them more accessible for point-of-care diagnostics and field applications (Qi et al., 2018). RPA employs recombinase proteins to facilitate strand exchange, enabling DNA amplification at a constant temperature (usually 37–42°C). RPA reactions are highly specific and can be completed within 20–30 min, making them suitable for rapid diagnostics in resource-limited settings (Piepenburg et al., 2006; Guo et al., 2023). These isothermal techniques offer several advantages over PCR, including faster turnaround times, simplified instrumentation requirements, and tolerance to inhibitors present in crude samples. They have been extensively applied in fields such as infectious disease diagnosis, food safety monitoring, and environmental surveillance. As isothermal amplification methods continue to evolve, they hold immense promise for decentralized molecular testing and point-of-care applications (Sánchez et al., 2022; Kasfy et al., 2024). Although our results show that both amplification methods can be combined with one-pot CRISPR/Cas12 detection, however, the one-pot RPA shows higher sensitivity (about 10 times) since RPA has a higher amplification efficiency in the one-pot-CRISPR/Cas12 reaction, indicating that Cas-OPRAD based on RPA-CRISPR/Cas is more practical for detection due to lower reaction temperature, shorter detection time, and is easier to operate in the field. Moreover, the amplification temperature of the RPA reaction allows it to be paired with CRISPR/Cas12a cleavage in a one-pot reaction. This approach simplifies the assay by using a single temperature and a single tube for the amplification reaction, making it amenable to on-site detection without the need for highly equipped laboratories or well-trained personnel.
On-site detection is pivotal for swift response, cost-efficiency, and preventive action in various fields. It enables immediate identification of substances, pathogens, and anomalies, saving time and resources by eliminating the need for off-site analysis (Hu et al., 2017; Bai et al., 2019; Zhu et al., 2022). The established Cas-OPRAD has powerful field sample processing capabilities, as illustrated in Scheme 1. The one-pot CRISPR/Cas12a-based detection platform works with four processes that integrate (i) Sample preparation (ii) PCR or RPA amplification of the target DNA, sequence-specific recognition, and trans-cleavage by Cas12a/crRNA (Cas-OPRAD assay), and (iii) visual readout of results. This streamlined approach provides a powerful tool for on-site detection of this destructive soybean pathogen, which can straightforward visual interpretation of results by the naked eye without requiring specialized equipment.
In this study, we developed a one-pot RPA/PCR-CRISPR/Cas12a detection platform for sensitive and specific detection of the soybean pathogenic oomycete P. sojae, named Cas-OPRAD. The advantages of Cas-OPRAD are shown in Table 2 compared with previously reported methods. The Cas-OPRAD assay has the advantages of sensitivity (<10 pg./μL), due to the RPA amplification and trans cleavage of activated Cas12a protein. The assay demonstrates exceptional analytical specificity without cross-reactivity against closely related species with specific RPA primers and CRISPR sgRNA design. In addition, to achieve on-site detection, we utilized the principle of liquid surface tension and spatial isolation to separate the RPA/PCR reaction from sgRNA-Cas12 in a one-pot reaction. We provide a straightforward visual readout for the soybean pathogen P. sojae via both fluorescent and lateral flow detection modalities, which extends the application of Cas-OPRAD to point-of-care self-diagnoses. The whole test procedure can be performed in the field with simple operator training, using a smart thermos cup (Zhu et al., 2022), and visual inspection using a LFS device. In field sample tests, the time required for the whole process can be shortened to 40 min for all 20 field soybean samples, considerably shorter than conventional PCR or RPA-CRISPR/Cas assay. By coupling target amplification by RPA with Cas12a-mediated cleavage in a streamlined single-reaction format, this platform enables on-site application for early and precise identification of P. sojae outbreaks to empower effective disease management. Overall, this integrated isothermal CRISPR-based diagnostic tool has tremendous potential to aid in monitoring and control of destructive soybean diseases attributed to P. sojae infection.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
ZL: Writing – review & editing, Writing – original draft, Validation, Methodology, Formal analysis, Data curation. WF: Writing – review & editing, Writing – original draft, Validation, Methodology, Formal analysis, Data curation. ZZ: Writing – review & editing, Validation, Methodology, Formal analysis, Data curation. SL: Writing – review & editing, Methodology, Data curation. ML: Writing – review & editing, Methodology, Data curation. JD: Writing – review & editing, Methodology, Data curation. ZW: Writing – review & editing, Methodology, Data curation. FL: Writing – review & editing, Supervision, Investigation, Funding acquisition, Data curation, Conceptualization. QC: Writing – review & editing, Writing – original draft, Supervision, Project administration, Investigation, Funding acquisition, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the Hainan Province Science and Technology Special Fund (ZDYF2022XDNY241), the National Key Research and Development Program of China (2023YFD1401005), and the Scientific Research Foundation of Hainan University [KYQD(ZR)-20080].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1390422/full#supplementary-material
Ahmed, F. A., Larrea-Sarmiento, A., Alvarez, A. M., and Arif, M. (2018). Genome-informed diagnostics for specific and rapid detection of Pectobacterium species using recombinase polymerase amplification coupled with a lateral flow device. Sci. Rep. 8:15972. doi: 10.1038/s41598-018-34275-0
Aman, R., Mahas, A., Marsic, T., Hassan, N., and Mahfouz, M. M. (2020). Efficient, rapid, and sensitive detection of plant RNA viruses with one-pot RT-RPA-CRISPR/Cas12a assay. Front. Microbiol. 11:610872. doi: 10.3389/fmicb.2020.610872
Bai, J., Lin, H., Li, H., Zhou, Y., Liu, J., Zhong, G., et al. (2019). Cas12a-based on-site and rapid nucleic acid detection of African swine fever. Front. Microbiol. 10:2830. doi: 10.3389/fmicb.2019.02830
Bienapfl, J. C., Malvick, D. K., and Percich, J. A. (2011). Specific molecular detection of Phytophthora sojae using conventional and real-time PCR. Fungal Biol. 115, 733–740. doi: 10.1016/j.funbio.2011.05.007
Blair, J. E., Coffey, M. D., Park, S.-Y., Geiser, D. M., and Kang, S. (2008). A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet. Biol. 45, 266–277. doi: 10.1016/j.fgb.2007.10.010
Chen, Z., Jiao, B., Zhou, J., He, H., and Dai, T. (2022). Rapid detection of Phytophthora cinnamomi based on a new target gene Pcinn13739. Front. Cell. Infect. Microbiol. 12:923700. doi: 10.3389/fcimb.2022.923700
Chen, Q., Li, B., Liu, P., Lan, C., Zhan, Z., and Weng, Q. (2013). Development and evaluation of specific PCR and LAMP assays for the rapid detection of Phytophthora melonis. Eur. J. Plant Pathol. 137, 597–607. doi: 10.1007/s10658-013-0273-9
Chen, J. S., Ma, E., Harrington, L. B., Da Costa, M., Tian, X., Palefsky, J. M., et al. (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439. doi: 10.1126/science.aar6245
Chen, Q., Weng, Q., Wang, Y., and Zheng, X. (2004). Identification and sequencing of ribosomal DNA-ITS of Phytophthora sojae in Fujian. Acta Phytopathol. Sin. 34, 112–116. doi: 10.13926/j.cnki.apps.2004.02.003
Dai, T.-T., Lu, C.-C., Lu, J., Dong, S., Ye, W., Wang, Y., et al. (2012). Development of a loop-mediated isothermal amplification assay for detection of Phytophthora sojae. FEMS Microbiol. Lett. 334, 27–34. doi: 10.1111/j.1574-6968.2012.02619.x
Dai, Y., Somoza, R. A., Wang, L., Welter, J. F., Li, Y., Caplan, A. I., et al. (2019). Exploring the trans-cleavage activity of CRISPR-Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew. Chem. 131, 17560–17566. doi: 10.1002/ange.201910772
Dai, T., Yang, X., Hu, T., Jiao, B., Xu, Y., Zheng, X., et al. (2019). Comparative evaluation of a novel recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) assay, LAMP, conventional PCR, and leaf-disc baiting methods for detection of Phytophthora sojae. Front. Microbiol. 10:1884. doi: 10.3389/fmicb.2019.01884
Ding, X., Yin, K., Li, Z., Lalla, R. V., Ballesteros, E., Sfeir, M. M., et al. (2020). Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 11:4711. doi: 10.1038/s41467-020-18575-6
Dorrance, A. E. (2018). Management of Phytophthora sojae of soybean: a review and future perspectives. Can. J. Plant Pathol. 40, 210–219. doi: 10.1080/07060661.2018.1445127
Eisenstein, M. (2022). Seven technologies to watch in 2022. Nature 601, 658–661. doi: 10.1038/d41586-022-00163-x
Erwin, D. C., and Ribeiro, O. K. (1996). Phytophthora Diseases Worldwide. St. Paul, MN, USA: The American Phytopathological Society.
Ghouneimy, A., Mahas, A., Marsic, T., Aman, R., and Mahfouz, M. (2023). CRISPR-based diagnostics: challenges and potential solutions toward point-of-care applications. ACS Synth. Biol. 12, 1–16. doi: 10.1021/acssynbio.2c00496
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442. doi: 10.1126/science.aam9321
Guo, Y., Xia, H., Dai, T., and Liu, T. (2023). RPA-CRISPR/Cas12a mediated isothermal amplification for visual detection of Phytophthora sojae. Front. Cell. Infect. Microbiol. 13:1208837. doi: 10.3389/fcimb.2023.1208837
Haudenshield, J. S., Song, J. Y., and Hartman, G. L. (2017). A novel, multiplexed, probe-based quantitative PCR assay for the soybean root- and stem-rot pathogen, Phytophthora sojae, utilizes its transposable element. PLoS One 12:e0176567. doi: 10.1371/journal.pone.0176567
Hu, X. R., Dai, D. J., Wang, H. D., and Zhang, C. Q. (2017). Rapid on-site evaluation of the development of resistance to quinone outside inhibitors in Botrytis cinerea. Sci. Rep. 7:13861. doi: 10.1038/s41598-017-13317-z
Hu, F., Liu, Y., Zhao, S., Zhang, Z., Li, X., Peng, N., et al. (2022). A one-pot CRISPR/Cas13a-based contamination-free biosensor for low-cost and rapid nucleic acid diagnostics. Biosens. Bioelectron. 202:113994. doi: 10.1016/j.bios.2022.113994
Islam, T., and Kasfy, S. H. (2023). CRISPR-based point-of-care plant disease diagnostics. Trends Biotechnol. 41, 144–146. doi: 10.1016/j.tibtech.2022.10.002
Jiao, J., Kong, K., Han, J., Song, S., Bai, T., Song, C., et al. (2021). Field detection of multiple RNA viruses/viroids in apple using a CRISPR/Cas12a-based visual assay. Plant Biotechnol. J. 19, 394–405. doi: 10.1111/pbi.13474
Jinhuo, P., and Anderson, T. (1998). Detection of Phytophthora sojae from soil with soybean leaf-disc baiting. Plant Quarantine 5, 1998–2004.
Kang, H., Peng, Y., Hua, K., Deng, Y., Bellizzi, M., Gupta, D. R., et al. (2021). Rapid detection of wheat blast pathogen Magnaporthe oryzae Triticum Pathotype using genome-specific primers and Cas12a-mediated technology. Engineering 7, 1326–1335. doi: 10.1016/j.eng.2020.07.016
Kasfy, S. H., Hia, F. T., and Islam, T. (2024). Do CRISPR-based disease diagnosis methods qualify as point-of-care diagnostics for plant diseases? Nucleus 67, 11–24. doi: 10.1007/s13237-024-00489-z
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O., and Zhang, F. (2019). SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 14, 2986–3012. doi: 10.1038/s41596-019-0210-2
Kong, L., Wang, H., Wang, S., Xu, P., Zhang, R., Dong, S., et al. (2020). Rapid detection of potato late blight using a loop-mediated isothermal amplification assay. J. Integr. Agric. 19, 1274–1282. doi: 10.1016/S2095-3119(19)62816-9
Kroon, L. P. N. M., Bakker, F. T., van den Bosch, G. B. M., Bonants, P. J. M., and Flier, W. G. (2004). Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genet. Biol. 41, 766–782. doi: 10.1016/j.fgb.2004.03.007
Kunadiya, M., White, D., Dunstan, W. A., Hardy, G. E. S. J., Andjic, V., and Burgess, T. I. (2017). Pathways to false-positive diagnoses using molecular genetic detection methods; Phytophthora cinnamomi a case study. FEMS Microbiol. Lett. 364:fnx0009. doi: 10.1093/femsle/fnx0009
Li, S.-Y., Cheng, Q.-X., Wang, J.-M., Li, X.-Y., Zhang, Z.-L., Gao, S., et al. (2018). CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov 4:20. doi: 10.1038/s41421-018-0028-z
Li, Y., Li, S., Wang, J., and Liu, G. (2019). CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 37, 730–743. doi: 10.1016/j.tibtech.2018.12.005
Lin, M., Yue, H., Tian, T., Xiong, E., Zhu, D., Jiang, Y., et al. (2022). Glycerol additive boosts 100-fold sensitivity enhancement for one-pot RPA-CRISPR/Cas12a assay. Anal. Chem. 94, 8277–8284. doi: 10.1021/acs.analchem.2c00616
Matthiesen, R. L., Schmidt, C., and Robertson, A. E. (2021). Comparison of baiting techniques for recovering Phytophthora sojae from soybean fields in Iowa. Plant Health Progress 22, 316–322. doi: 10.1094/PHP-02-21-0040-FI
Piepenburg, O., Williams, C. H., Stemple, D. L., and Armes, N. A. (2006). DNA detection using recombination proteins. PLoS Biol. 4:e204. doi: 10.1371/journal.pbio.0040204
Qi, H., Yue, S., Bi, S., Ding, C., and Song, W. (2018). Isothermal exponential amplification techniques: from basic principles to applications in electrochemical biosensors. Biosens. Bioelectron. 110, 207–217. doi: 10.1016/j.bios.2018.03.065
Rojas, J. A., Miles, T. D., Coffey, M. D., Martin, F. N., and Chilvers, M. I. (2017). Development and application of qPCR and RPA genus- and species-specific detection of Phytophthora sojae and P. sansomeana root rot pathogens of soybean. Plant Dis. 101, 1171–1181. doi: 10.1094/PDIS-09-16-1225-RE
Saisawang, C., Naksith, P., Sakdee, S., Ketterman, A. J., Tuntithavornwat, S., Nimsamer, P., et al. (2023). Optimal stabilization for long-term storage of nucleic acid-based CRISPR/Cas12a assay for SARS-CoV-2 detection. Karbala Int. J. Mod. Sci. 9:3288. doi: 10.33640/2405-609X.3288
Sánchez, E., Ali, Z., Islam, T., and Mahfouz, M. (2022). A CRISPR-based lateral flow assay for plant genotyping and pathogen diagnostics. Plant Biotechnol. J. 20, 2418–2429. doi: 10.1111/pbi.13924
Su, Y., and Shen, C. (1993). The discovery and biological characteristic studies of Phytophthora megasperma f. sp. glycinea on soyabean in China. Acta Phytopathol. Sin. 23, 341–347.
Sun, X., Lei, R., Zhang, H., Chen, W., Jia, Q., Guo, X., et al. (2024). Rapid and sensitive detection of two fungal pathogens in soybeans using the recombinase polymerase amplification/CRISPR-Cas12a method for potential on-site disease diagnosis. Pest Manag. Sci. 80, 1168–1181. doi: 10.1002/ps.7847
Tyler, B. M. (2007). Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
Wang, S.-Y., Du, Y.-C., Wang, D.-X., Ma, J.-Y., Tang, A.-N., and Kong, D.-M. (2021). Signal amplification and output of CRISPR/Cas-based biosensing systems: a review. Anal. Chim. Acta 1185:338882. doi: 10.1016/j.aca.2021.338882
Wang, R., Qian, C., Pang, Y., Li, M., Yang, Y., Ma, H., et al. (2021). opvCRISPR: one-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 172:112766. doi: 10.1016/j.bios.2020.112766
Wang, Y., Zhang, W., Wang, Y., and Zheng, X. (2006). Rapid and sensitive detection of Phytophthora sojae in soil and infected soybeans by species-specific polymerase chain reaction assays. Phytopathology 96, 1315–1321. doi: 10.1094/PHYTO-96-1315
Wheatley, M. S., and Yang, Y. (2021). Versatile applications of the CRISPR/Cas toolkit in plant pathology and disease management. Phytopathology 111, 1080–1090. doi: 10.1094/PHYTO-08-20-0322-IA
Wu, M., Li, B., Liu, P., Weng, Q., Zhan, J., and Chen, Q. (2017). Genetic analysis of Phytophthora sojae populations in Fujian, China. Plant Pathol. 66, 1182–1190. doi: 10.1111/ppa.12666
Xiong, Q., Xu, J., Zheng, X., Zhu, Y., Zhang, C., Wang, X., et al. (2019). Development of seven novel specific SCAR markers for rapid identification of Phytophthora sojae: the cause of root-and stem-rot disease of soybean. Eur. J. Plant Pathol. 153, 517–531. doi: 10.1007/s10658-018-1579-4
Xu, J., Yang, X., Wu, C., Chen, Z., and Dai, T. (2023). Recombinase polymerase amplification–lateral flow dipstick assay for rapid detection ofFusarium circinatumBased on a newly identified unique target gene. Plant Dis. 107, 1067–1074. doi: 10.1094/PDIS-04-22-0864-RE
Zhao, W., Wang, T., and Qi, R. (2015). Ypt1 gene-based detection of Phytophthora sojae in a loop-mediated isothermal amplification assay. J. Plant Dis. Prot. 122, 66–73. doi: 10.1007/BF03356533
Keywords: Phytophthora sojae, one-pot RPA/PCR, recombinase polymerase amplification, CRISPR/Cas12a, on-site detection, visual detection
Citation: Li Z, Feng W, Zhu Z, Lu S, Lin M, Dong J, Wang Z, Liu F and Chen Q (2024) Cas-OPRAD: a one-pot RPA/PCR CRISPR/Cas12 assay for on-site Phytophthora root rot detection. Front. Microbiol. 15:1390422. doi: 10.3389/fmicb.2024.1390422
Received: 23 February 2024; Accepted: 21 May 2024;
Published: 05 June 2024.
Edited by:
Tofazzal Islam, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Yogesh K. Ahlawat, University of Florida, United StatesCopyright © 2024 Li, Feng, Zhu, Lu, Lin, Dong, Wang, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinghe Chen, cWhjaGVuQGhhaW5hbnUuZWR1LmNu; Fuxiu Liu, ZnV4aXVsaXVAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.