- 1Department of Biotechnology and Life Sciences, Faculty of Biotechnology and Life Sciences, Sojo University, Kumamoto, Japan
- 2Division of Infection and Host Defense, Tohoku Medical and Pharmaceutical University, Sendai, Japan
The cell surface of Cryptococcus neoformans is covered by a thick capsular polysaccharide. The capsule is the most important virulence factor of C. neoformans; however, the complete mechanism of its biosynthesis is unknown. The capsule is composed of glucuronoxylomannan (GXM) and glucuronoxylomannogalactan (GXMGal). As GXM is the most abundant component of the capsule, many studies have focused on GXM biosynthesis. However, although GXMGal has an important role in virulence, studies on its biosynthesis are scarce. Herein, we have identified a GT31 family β-(1 → 3)-galactosyltransferase Ggt2, which is involved in the biosynthesis of the galactomannan side chain of GXMGal. Comparative analysis of GXMGal produced by a ggt2 disruption strain revealed that Ggt2 is a glycosyltransferase that catalyzes the initial reaction in the synthesis of the galactomannan side chain of GXMGal. The ggt2 disruption strain showed a temperature-sensitive phenotype at 37°C, indicating that the galactomannan side chain of GXMGal is important for high-temperature stress tolerance in C. neoformans. Our findings provide insights into complex capsule biosynthesis in C. neoformans.
1 Introduction
Cryptococcus neoformans, a basidiomycete yeast, is the primary pathogen responsible for cryptococcosis, a globally prevalent disease that affects immunocompromised individuals, specifically those infected with HIV (Maziarz and Perfect, 2016; Altamirano et al., 2020). Upon pulmonary infection, C. neoformans disseminates to the central nervous system and causes severe meningoencephalitis with a high mortality rate.
Capsular polysaccharides, specifically glucuronoxylomannan (GXM), glucuronoxylomannogalactan (GXMGal), and a small amount of mannoproteins, are the key virulence factors of C. neoformans (Bose et al., 2003). Several acapsular mutants of C. neoformans have been shown to nearly completely lack pathogenicity in mice models, suggesting that enzymes involved in capsule biosynthesis are targets for developing antifungal drugs against cryptococcosis (O’Meara and Alspaugh, 2012; Casadevall et al., 2019). The detailed structures of GXM and GXMGal have been determined. GXM is a larger polysaccharide (MW: 1,000,000–7,000,000) consisting of α-mannan core chains of α-(1 → 3)-linked mannose (Man) and β-linked glucuronic acid (GlcA) and xylose (Xyl) (Cherniak et al., 1998). GXMGal is a relatively smaller polysaccharide (MW: ~100,000), consisting of α-galactan core chains of α-(1 → 6)-linked galactose (Gal) with galactomannan side chains (Vaishnav et al., 1998). Variable numbers of Xyl and GlcA residues are attached to the galactomannan side chain (Heiss et al., 2009). Galactofuranose (Galf) is also found attached to the α-galactan backbone (Heiss et al., 2013). Man residues in GXM and GXMGal are O-acetylated; in GXMGal, 80% of the Man residues in the galactomannan side chain are acetylated, influencing the immunomodulatory and immunogenic properties of this polysaccharide (Previato et al., 2017).
GXM is present in ~90% of the capsules, whereas GXMGal is present in only 7–10% of the capsules (Doering, 2009). Thus, although there is a large difference in the amounts of GXM and GXMGal, mutants lacking either polysaccharide lose virulence (Chang and Kwon-Chung, 1994; Moyrand et al., 2007; Kumar et al., 2011). The role of UDP-Gal is crucial, which is evident as disruptants of the genes UGE1, encoding UDP-glucose 4-epimerase, and UGT1, encoding a Golgi-localized UDP-Gal transporter, are avirulent, likely due to a deficiency in GXMGal (Moyrand et al., 2008; Li et al., 2017). This indicates that galactosyltransferases involved in Golgi-localized GXMGal biosynthesis are targets for antifungal drug development. However, GXMGal biosynthesis-related glycosyltransferases are limited to only two species, β-(1 → 2)-xyltransferases Cxt1 and Cxt2 (Klutts et al., 2007; Klutts and Doering, 2008; Reilly et al., 2009; Wang et al., 2018). The putative α-Gal β-(1 → 3)-Gal transferase Pvg3 in Schizosaccharomyces pombe, which belongs to the GT31 family, highlights the diversity of glycosyltransferases (Andreishcheva et al., 2004; Fukunaga et al., 2023). In C. neoformans, the genome encodes three GT31 family glycosyltransferases, one of which, Ggt1, functions as an α-Man β-(1 → 6)-Gal transferase in GIPC biosynthesis (Wohlschlager et al., 2013). However, the functions of other presumptive glycosyltransferases of the GT31 family in the C. neoformans genome are unclear.
Herein, we report that a putative glycosyltransferase belonging to the GT31 family in C. neoformans is involved in GXMGal biosynthesis. Using the cap59 disruption strain (a GXM-deficient strain of C. neoformans) as the parental strain, we constructed disruption strains of GT31 family glycosyltransferases (ggt1, ggt2, and ggt3). A nuclear magnetic resonance and methylation gas chromatography–mass spectrometry analysis of the structure of GXMGal produced by the cap59 and ggt2 double-disruptant strain revealed that the galactomannan side chain was reduced or almost completely lost. The ggt2 disruption strain exhibited a temperature-sensitive (Ts) phenotype at 37°C. These results indicate that, in C. neoformans, the galactomannan side chain of GXMGal has an important role in high-temperature stress tolerance.
2 Materials and methods
2.1 Strains and medium
The C. neoformans strains used in this study are listed in Supplementary Table S1. The C. neoformans var. grubii H99 strain was obtained from the Fungal Genetics Stock Center (Kansas City, USA).
The strains were cultured on YPD medium (2% w/v glucose, 2% w/v peptone, and 1% w/v yeast extract). To produce GXMGal, the strains were cultured in 10% Sabouraud liquid medium [0.4% w/v glucose, 0.1% w/v peptone, 0.1% w/v tryptone, and 50 mM 3-morpholinopropanesulfonic acid (pH 7.3)].
2.2 Construction of the cap59 disruption strain
CAP59 (CNAG_00721) was disrupted in C. neoformans H99 by inserting NEO using the CRISPR/Cas9 system (Huang et al., 2022). A gene replacement cassette encompassing a 50-bp homology arm at the 5′ end and a 50-bp homology arm at the 3′ end of CAP59 was amplified by recombinant PCR using pNEO_6xHA (So et al., 2017) as a template and the cap59-del-F–cap59-del-R primer pair (Supplementary Table S2). The Cas9 expression cassette was amplified by PCR using pBHM2403 as a template and the M13-F–M13-R primer pair. The sgRNA expression cassette was amplified in two PCR steps. An sgRNA scaffold containing a 20-bp target sequence and U6 promoter and an sgRNA scaffold containing a 20-bp target sequence and U6 terminator was amplified by PCR using pBHM2329 as a template and the M13-F–cap59-gRNA-R1 and cap59-gRNA-F2–M13-R primer pairs. The PCR fragments were combined by fusion PCR using the U6-F–U6-R primer pair. All PCR fragments were introduced into C. neoformans by electroporation using Gene Pulser II (Bio-Rad, Hercules, CA), yielding cap59Δ strain. Transformants were selected using YPD agar plates supplemented with 200 μg/mL G418. The introduction of NEO into each locus was confirmed by PCR using the cap59-comf-F–cap59-comf-R primer pair (Supplementary Figure S1).
2.3 Construction of the ggt1, ggt2, and ggt3 disruption strains
GGT1 (CNAG_01385), GGT2 (CNAG_01050), and GGT3 (CNAG_06918) were disrupted in C. neoformans H99 and cap59Δ strains by inserting HYG. Gene replacement cassettes encompassing a 50-bp homology arm at the 5′ end and a 50-bp homology arm at the 3′ end of GGT1, GGT2, and GGT3 were amplified by recombinant PCR using pHYG_GFP (So et al., 2017) as a template and the xxxx-del-F–xxxx-del-R (where “xxxx” indicates GGT1, GGT2, or GGT3) primer pair (Supplementary Table S2). An sgRNA scaffold containing a 20-bp target sequence and U6 promoter and an sgRNA scaffold containing a 20-bp target sequence and U6 terminator were amplified by PCR using pBHM2329 as a template and the M13-F–xxxx-gRNA-R1 and xxxx-gRNA-F2–M13-R primer pairs. All PCR fragments were introduced into C. neoformans H99 and cap59Δ by electroporation using Gene Pulser II (Bio-Rad, Hercules, CA), yielding ggt1Δ, ggt2Δ, ggt3Δ, cap59Δggt1Δ, cap59Δggt2Δ, and cap59Δggt3Δ strains. Moreover, transformants were selected on YPD agar plates supplemented with 200 μg/mL hygromycin B. The introduction of HYG into each locus was confirmed by PCR using the xxxx-comf-F–xxxx-comf-R primer pair (Supplementary Figure S1).
2.4 Construction of the cap59, ggt2, and ggt3 triple disruption strain
GGT3 was disrupted in C. neoformans cap59Δggt2Δ strain by inserting NAT. A gene replacement cassette encompassing a 50-bp homology arm at the 5′ end and a 50-bp homology arm at the 3′ end of GGT3 was amplified by recombinant PCR using pNAT_mCherry (So et al., 2017) as a template and the ggt3-del-F–ggt3-del-R primer pair (Supplementary Table S2). Transformants were selected on YPD agar plates supplemented with 100 μg/mL nourseothricin sulfate. The introduction of NAT into each locus was confirmed by PCR using the ggt3-comf-F–ggt3-comf-R primer pair (Supplementary Figure S2).
2.5 Complementation of the ggt2 disruption strain with wild-type GGT2
For complementation, analysis of GGT2 using a gene replacement cassette, encompassing a homology arm at the 5′ end of GGT2, wild-type GGT2 containing 3′-UTR, hygromycin B resistance gene (hph), and a homology arm at the 3′ end of GGT2, was constructed by recombinant PCR using H99 genomic DNA; pNAT_mCherry as a template; and the ggt2-comp-1–ggt2-comp-2, ggt2-comp-3–ggt2-comp-4, and ggt2-comp-5–ggt2-comp-6 primer pairs. The resultant DNA fragment was amplified with the ggt2-comp-1–ggt2-comp-6 primer pair. An sgRNA scaffold containing a 20-bp target sequence and U6 promoter and an sgRNA scaffold containing a 20-bp target sequence and U6 terminator were amplified by PCR using pBHM2329 as a template and the M13-F–HYG-gRNA-R1 and HYG-gRNA-F2–M13-R primer pairs. All PCR fragments were introduced into C. neoformans ggt2Δ and cap59Δggt2Δ by electroporation. Transformants were selected on YPD agar plates supplemented with 100 μg/mL nourseothricin sulfate. The introduction of NAT into each locus was confirmed by PCR using the ggt3-comp-comf-F–ggt3-comp-comf-R primer pair (Supplementary Figure S1).
2.6 Measurement of capsule size
C. neoformans strains were cultured in 3 mL of YPD liquid medium at 30°C for 24 h. The cells were then collected by centrifugation and washed three times with sterile phosphate-buffered saline (PBS), suspended in 2 mL of 10% Sabouraud liquid medium, and incubated at 30°C for 24 h to induce capsule production. The culture medium was diluted with PBS mixed at a ratio of 1:1 with India ink (Syogeikuretake Shikon BB1-18; Kuretake Co., Ltd. Nara, Japan) and incubated for 15 min. Images of stained cells were acquired using a microscope equipped with a digital camera. The diameters of the cells and capsules were measured immediately (50 cells), and the average diameter was calculated.
2.7 Preparation of the GXMGal fraction
Purification of GXMGal was performed as described (Rocha et al., 2015). Briefly, cap59Δ strains were cultivated in 1 L of 10% Sabouraud medium at 30°C with shaking (160 rpm) for 5 days. The culture supernatant was collected by centrifugation, mixed with an equal volume of phenol:chloroform, and centrifuged. The collected supernatant was dialyzed overnight at 4°C using a Visking Tube (Nihon Medical Science, Inc. Japan). The polysaccharides were powdered by lyophilization. GXMGal powder was dissolved in 3% cetyltrimethylammonium bromide solution in 1% borate at pH 9.5. The GM fraction was collected as precipitate, washed with 75% ethanol, dialyzed with water, and lyophilized.
2.8 Methylation GC–MS and nuclear magnetic resonance spectroscopy
Glycosidic linkages were analyzed as previously described (Klutts and Doering, 2008; Katafuchi et al., 2017). Briefly, GXMGal was separately dissolved in dimethyl sulfoxide, followed by NaOH addition. After stirring for 3 h, methyl iodide was added, and the suspension was stirred for 24 h. The methylated products were extracted in chloroform and washed using dH2O. Then, the methylated samples were hydrolyzed using 2 M trifluoroacetic acid, reduced, and acetylated. The partially methylated alditol acetates were analyzed by GC–MS using a capillary column (30 m × 0.25 mm; DB-5, Agilent, CA) with helium as the carrier gas at a gradient temperature program of 210°C–260°C at 5°C/min. The GC–MS analyses were performed using a JMS-K9 mass spectrometer (JEOL, Tokyo, Japan). NMR experiments were performed as previously described (Klutts and Doering, 2008; Katafuchi et al., 2017). The NMR spectra were recorded using a JNM-LA600 spectrometer (JEOL) at 45°C. Proton and carbon chemical shifts were referenced relative to internal acetone at δ 2.225 and 31.07 ppm, respectively.
3 Results
3.1 Identification of candidate galactosyltransferases involved in GXMGal biosynthesis in C. neoformans
First, we searched the C. neoformans H99 genome for candidate genes encoding enzymes that transfer the β-galactosyl residue to the hydroxyl group at position 3 of the α-galactosyl residue in GXMGal. S. pombe Pvg3 is a glycosyltransferase that exhibits similar enzymatic activity. Therefore, we selected candidate genes by PSI-BLAST search using Pvg3 as a query. Two homologous proteins were selected (CNAG_01050 and CNAG_01385). CNAG_01385 is Ggt1––an α-mannoside β-(1 → 6)-galactosyltransferase involved in GIPC biosynthesis in C. neoformans. Then, a BLASTp search was performed using CNAG_01050 as a query to select candidate genes. CNAG_06918 was selected in addition to CNAG_01385. Therefore, we named CNAG_01050 and CNAG_06918 as Ggt2 and Ggt3, respectively, the two α-galactoside β-(1 → 3)-galactosyltransferase candidates. Ggt2 and Ggt3 are members of the GT31 family. Ggt2 has an amino acid sequence homology of 23% (in the 434–645 amino acid region of Ggt1) and 33% (in the 122–320 amino acid region of Ggt3) with Ggt1 and Ggt3, respectively (Figure 1). Ggt1 does not have a transmembrane domain, but analysis using DeepLoc 2.0 (Thumuluri et al., 2022) predicted a transmembrane region at 338–360 aa. Ggt2 and Ggt3 were predicted to be Golgi-localized type II membrane proteins with one transmembrane region at N-terminal 35–57 and 21–43 aa, respectively, according to DeepLoc 2.0 (Figure 1).
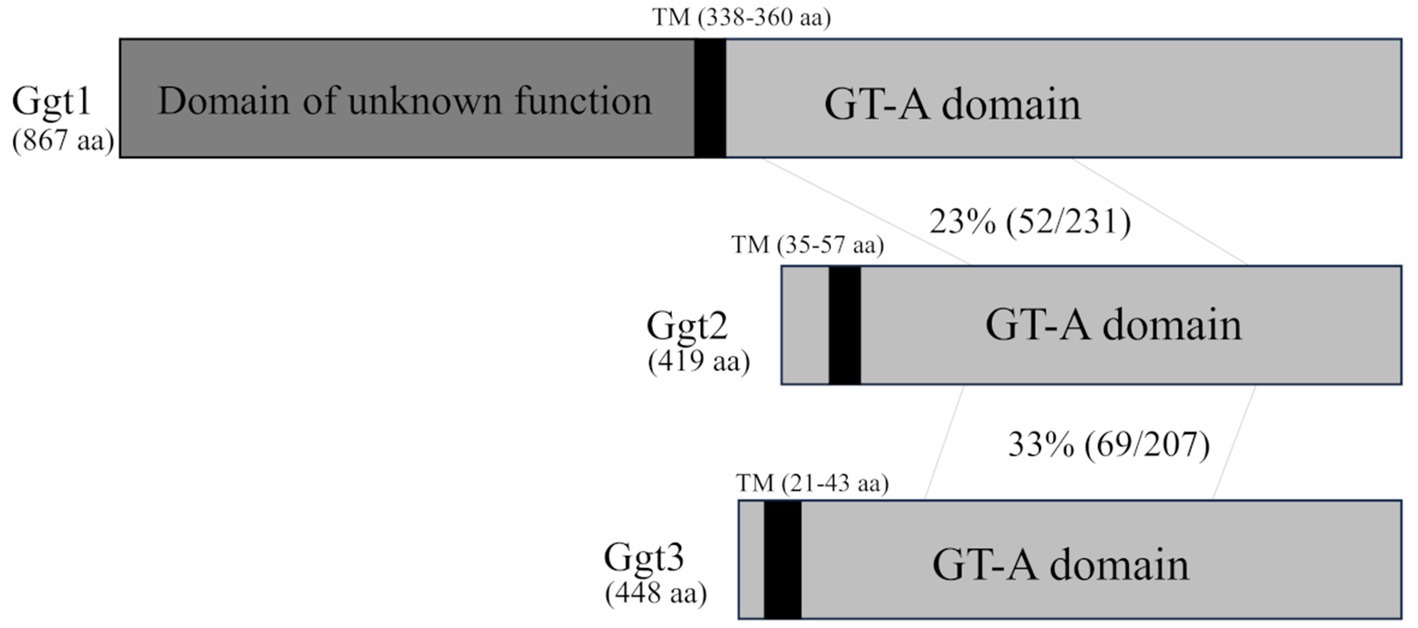
Figure 1. Schematic of the Ggt1, Ggt2, and Ggt3 proteins. The vertical black bars indicate transmembrane (TM) domains of Ggt1 (338–360 aa), Ggt2 (35–57 aa), and Ggt3 (21–43 aa). The gray bars indicate GT-A fold domains of Ggt1, Ggt2, and Ggt3. The dark gray bar indicates an unknown domain of Ggt1. The identity of the amino acid sequences of Ggt1 and Ggt2 and Ggt2 and Ggt3 in the GT-A fold domain are indicated.
3.2 1H-NMR analysis of GXMGal from GGT mutants
To investigate the effect of loss of presumptive GT31 family glycosyltransferases on GXMGal biosynthesis, we constructed GGT1, GGT2, and GGT3 triple disruptants and GGT2 and GGT3 double disruptants using cap59Δ as parental strain. All strains were cultured in 10% Sabouraud medium for 5 days, and GXMGal was purified from the culture supernatant. Purified GXMGal was analyzed by 1H-NMR (Figure 2). Chemical shifts at ~5.22, 5.16, and 4.99 ppm in cap59Δ, cap59Δggt1Δ, and cap59Δggt3Δ strains indicated -(1 → 3)-Manα-, terminal Manα, and -(1 → 3)-Manα-(2 ← 1)-galactomannan side chain of GXMGal, respectively (Klutts and Doering, 2008). In cap59Δggt2Δ and cap59Δggt2Δggt3Δ, chemical shifts appeared at 4.98 ppm, indicating the presence of the -(1 → 6)-Galα- side chain. These results indicate that loss of GGT2 results in a reduction or loss of the galactomannan side chain of GXMGal.
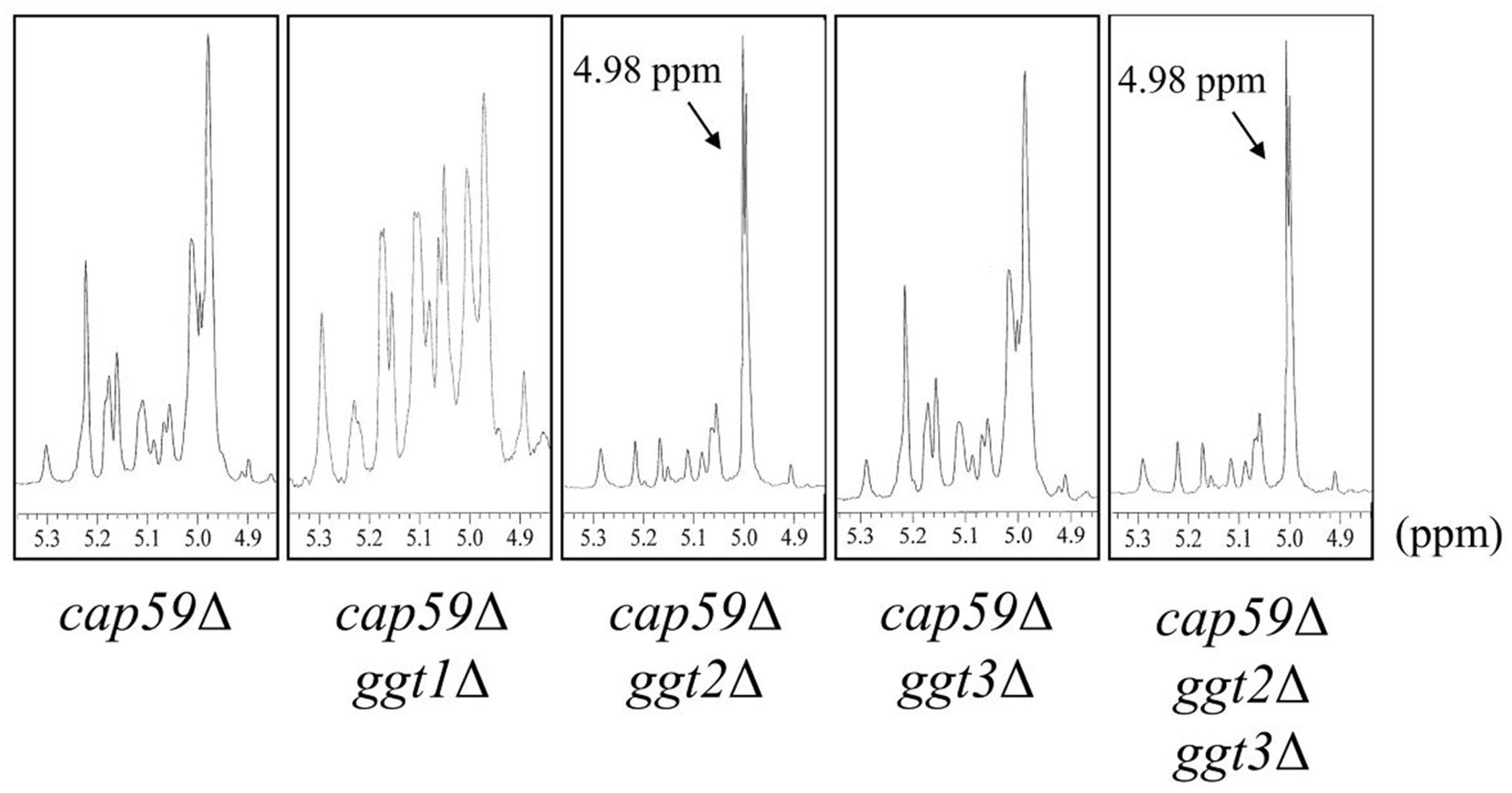
Figure 2. 1H-NMR analysis of GXMGal from cap59Δ, cap59Δ ggt1Δ, cap59Δ ggt2Δ, cap59Δ ggt3Δ, and cap59Δ ggt2Δ ggt3Δ strains. 1H-NMR signals at 5.22, 5.16, and 4.99 ppm in cap59Δ, cap59Δ ggt1Δ, and cap59Δ ggt3Δ are derived from H-1 at position C-1 of the underlined Man residues in -(1 → 3)-Manα- and terminal Manα and the -(1 → 3)- Manα-(2 ← 1)- side chain of GXMGal. 1H-NMR signals at 4.98 ppm in cap59Δ ggt2Δ and cap59Δ ggt2Δ ggt3Δ are derived from H-1 at position C-1 of the underlined Gal residues in -(1 → 6)-Galα-. The proton chemical shifts were referenced relative to internal acetone at δ 2.225 ppm.
3.3 Phenotypic analysis of GGT mutants
To clarify the physiological role of GGT in C. neoformans cells, ggt1, ggt2, and ggt3 single-disruptant strains were constructed using H99 as the parental strain. As uge1Δ and ugt1Δ cannot supply UDP-Gal to the Golgi, they are deficient in GXMGal and GIPC biosynthesis and exhibit a Ts phenotype at 37°C (Moyrand et al., 2008; Li et al., 2017). ggt1Δ lacks GIPC and exhibits a Ts phenotype at 37°C. Therefore, we observed growth of GGT disruptants at 37°C (Figure 3). At 30°C, ggt1Δ showed slightly delayed growth compared with the wild-type; moreover, ggt2Δ and ggt3Δ showed similar growth compared with the H99 strain. By contrast, at 37°C, ggt1Δ and ggt2Δ showed dramatically delayed growth, indicating a Ts phenotype. The growth of ggt3Δ and H99 was similar. These results indicate that the glycan structure synthesized by Ggt2 is important for high-temperature stress tolerance in C. neoformans.
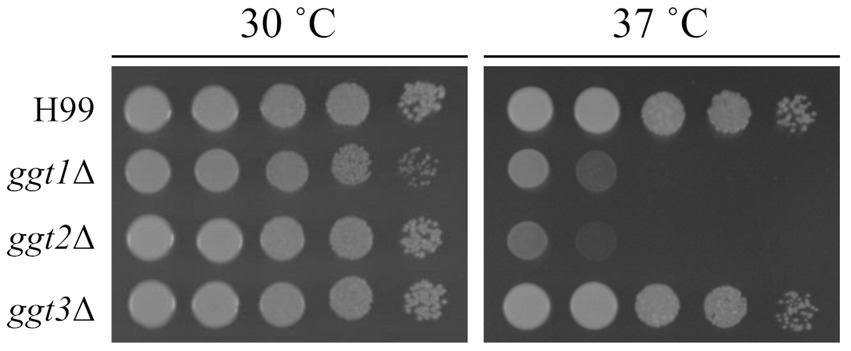
Figure 3. Phenotype of ggt1, ggt2, and ggt3 single disruptants. Colony morphology of H99, ggt1Δ, ggt2Δ, and ggt3Δ cultured on YPD agar at 30°C and 37°C for 3 days, respectively. The agar medium was inoculated with 10-fold serial dilutions of cells adjusted to 106 cells.
3.4 Drug resistance of ggt2 disruptant strain
We examined the growth of ggt2Δ on media containing various drugs. ugt1 disruptant strains show sensitivity to NaCl, Congo red, H2O2, and sodium dodecyl sulfate (SDS) (Li et al., 2017). Therefore, we tested the sensitivity of ggt2Δ to these drugs (Figure 4). ggt2Δ was not significantly sensitive to any of the drugs. Conversely, the Ts phenotype of ggt2Δ was completely rescued by complementation with wild-type GGT2 and slightly rescued by 1 M sorbitol, which exerts high osmotic pressure. These results indicate that the galactomannan side chain of GXMGal biosynthesized by Ggt2 is important for high-temperature stress tolerance in C. neoformans.
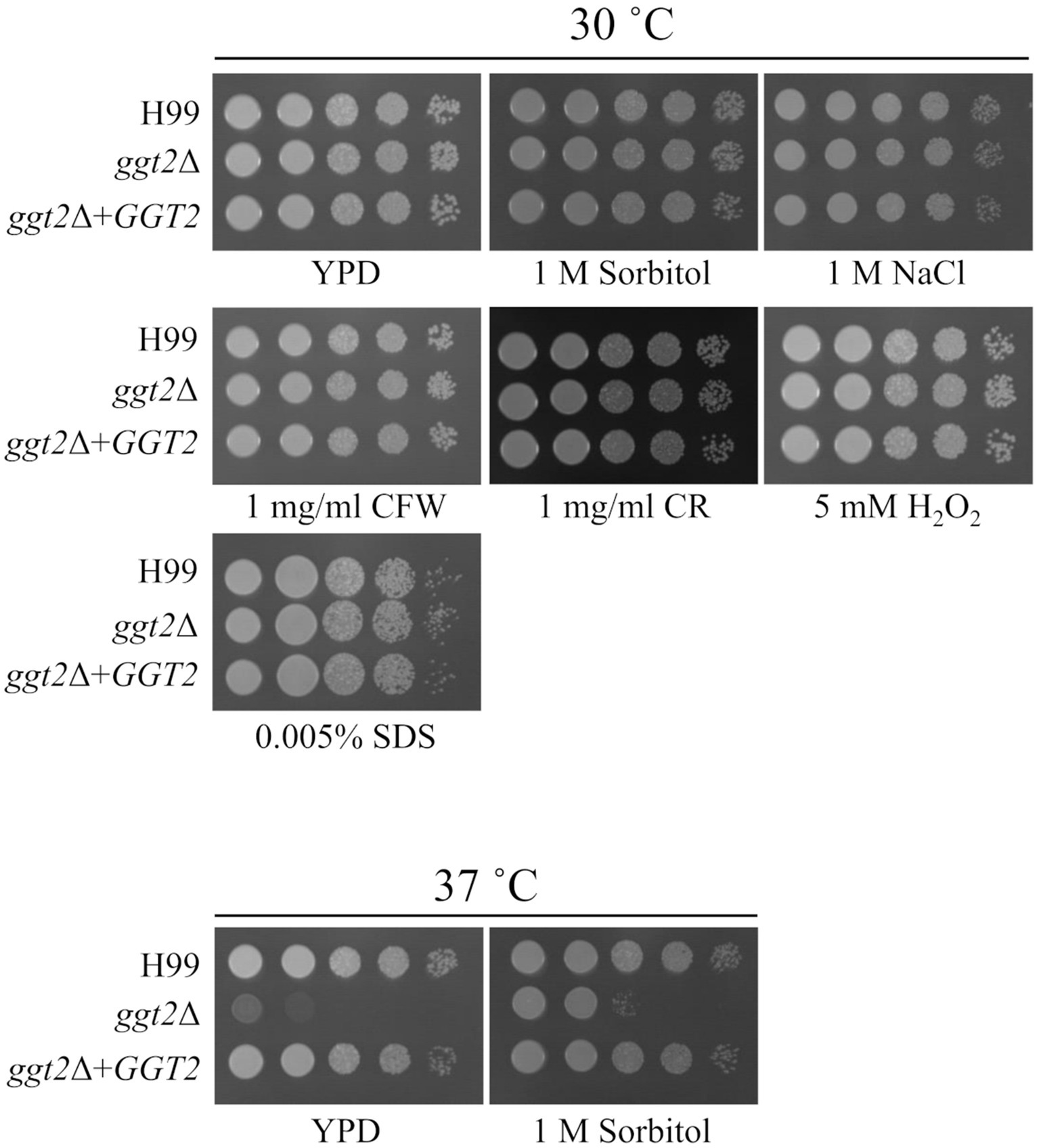
Figure 4. Drug sensitivity of the ggt2 disruptant. Colony morphology of H99, ggt2Δ, and ggt2Δ + GGT2 on YPD agar supplemented with or without 1 M sorbitol, 1 M NaCl, 1 mg/mL calcofluor white (CFW), 1 mg/mL Congo red (CR), 5 mM H2O2, and 0.005% sodium dodecyl sulfate (SDS) at 30°C and 37°C for 3 days. The agar medium was inoculated with 10-fold serial dilutions of cells adjusted to 106 cells.
3.5 Capsule productivity of ggt2 disruptant strain
To further analyze the ggt2Δ phenotype, the capsule structure was stained with India ink for microscopic observation (Supplementary Figure S3). Quantification of cell and capsule sizes revealed no significant differences between ggt2Δ and the wild-type strain. These findings indicated that Ggt2 absence did not affect GXM production.
3.6 Role of Ggt2 in GXMGal biosynthesis
To examine the role of Ggt2 in GXMGal biosynthesis, we complemented wild-type GGT2 in cap59Δggt2Δ and analyzed the structure of GXMGal. The structure of GXMGal produced by each strain was analyzed by 1H-NMR (Figure 5). Sharp doublet peaks at 4.98 ppm of the α-galactan backbone seen in cap59Δggt2Δ disappeared due to GGT2 complementation, and the Man-derived chemical shift seen in wild-type GXMGal reappeared.
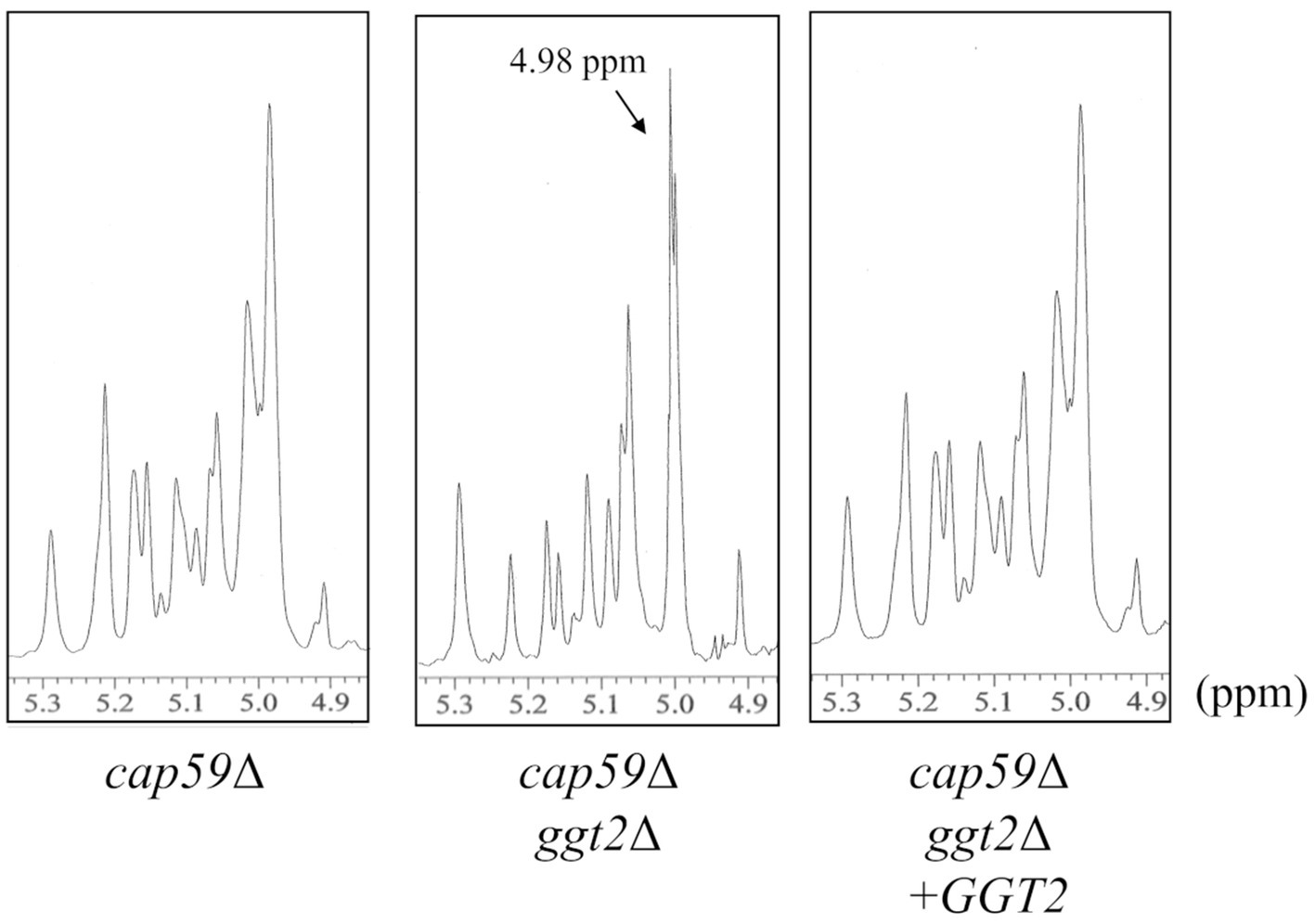
Figure 5. 1H-NMR analysis of GXMGal from cap59Δ, cap59Δ ggt2Δ, and cap59Δ ggt2Δ + GGT2 strains. 1H-NMR signals at 5.22, 5.16, and 4.99 ppm in cap59Δ and cap59Δ ggt2Δ + GGT2 are derived from H-1 at position C-1 of the underlined Man residues in -(1 → 3)-Manα- and terminal Manα and the -(1 → 3)- Manα-(2 ← 1)- side chain of GXMGal. 1H-NMR signal at 4.98 ppm in cap59Δggt2Δ is derived from H-1 at position C-1 of the underlined Gal residues in -(1 → 6)-Galα-. The proton chemical shifts were referenced relative to internal acetone at δ 2.225 ppm.
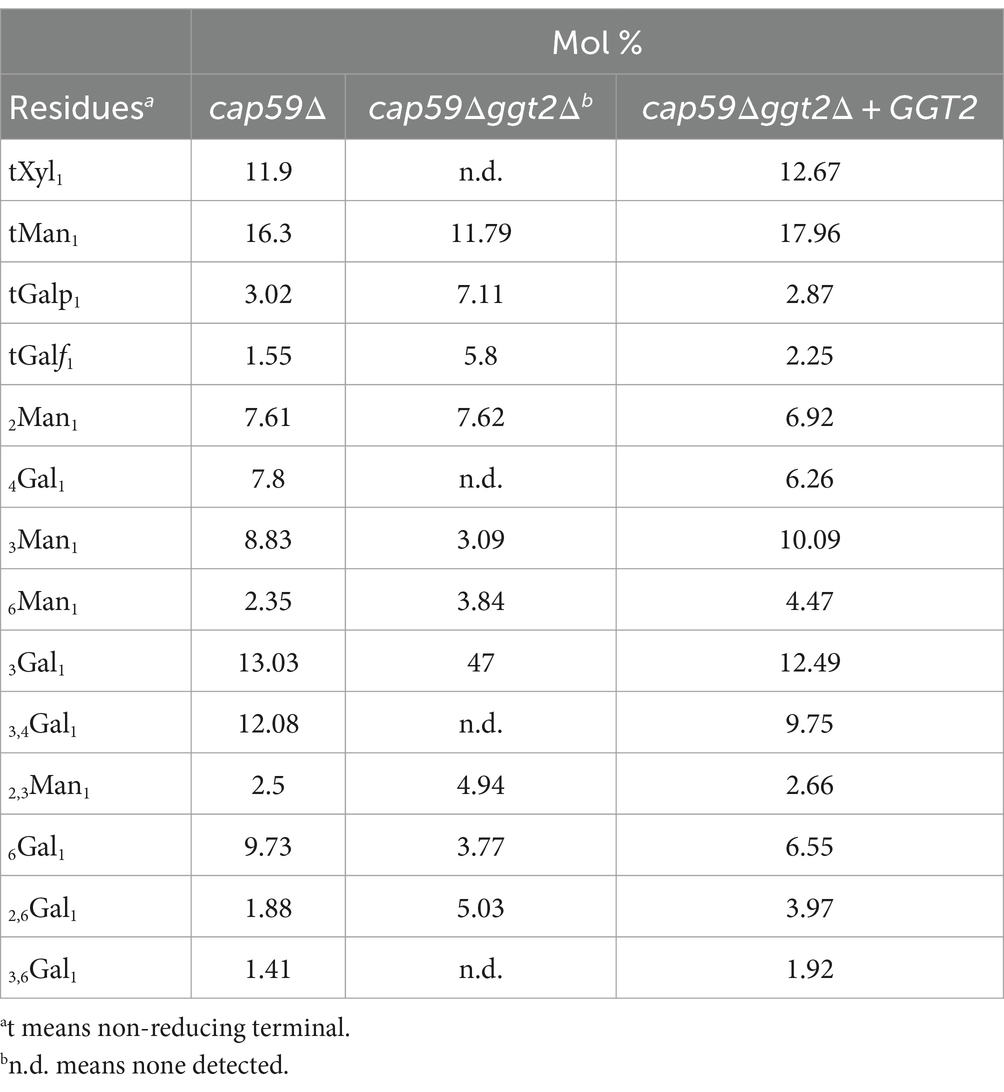
Methylation GC–MS analysis was used to investigate constituent sugars and the linkage mode of GXMGal produced by each strain (Table 1). In GXMGal produced by cap59Δggt2Δ, terminal Xyl (tXyl1) residues were reduced to below the detection limit. Additionally, 4-substituted Gal (4Gal1) and 3,4-substituted Gal (3,4Gal1) (indicating β-Gal residues in the galactomannan side chain) and 3,6-substituted Gal (3,6Gal1) (indicating α-Gal residues attached to galactomannan side chains) were reduced to below detection limit. The ratio of 2,6-substituted Gal (indicating α-Gal with Galf side chain) and terminal Gal (tGal1) and Galf (tGalf1) increased. By contrast, the ratio of 3-substituted Gal (3Gal1) and 6-substituted Man (6Gal1), which are not present in GXMGal, increased drastically. Terminal Man (tMan1), 3-substituted Man (3Man1), and 2,3-substituted Man (2,3Man1) decreased but did not disappear.
We analyzed the structure of GXMGal in detail by 13C-NMR (Figure 6). In GXMGal from cap59Δ, we detected a broad chemical shift at 100.6–105.1 ppm, possibly derived from position C-1 of Manα-(1 → 3)-, 2,3-O-substituted-Manα, Xylβ-(1 → 2)-, and Xylβ-(1 → 3)- in the galactomannan side chain (Klutts and Doering, 2008). These structures were suggested to be lost with the loss of GGT2. The chemical shifts at 78.5 and 77.1 ppm, which originate from position C-4 of 4-substituted Galβ and 3,4-substituted Galβ in wild-type GXMGal, disappeared in ggt2Δ. The chemical shift at 80.9 ppm originating from position C-3 of 3,6-substituted Galα disappeared in ggt2Δ. These results strongly support our hypothesis that Ggt2 catalyzes β-(1 → 3)-Gal transfer to α-galactoside––the initial reaction in the biosynthesis of the galactomannan side chain of GXMGal.
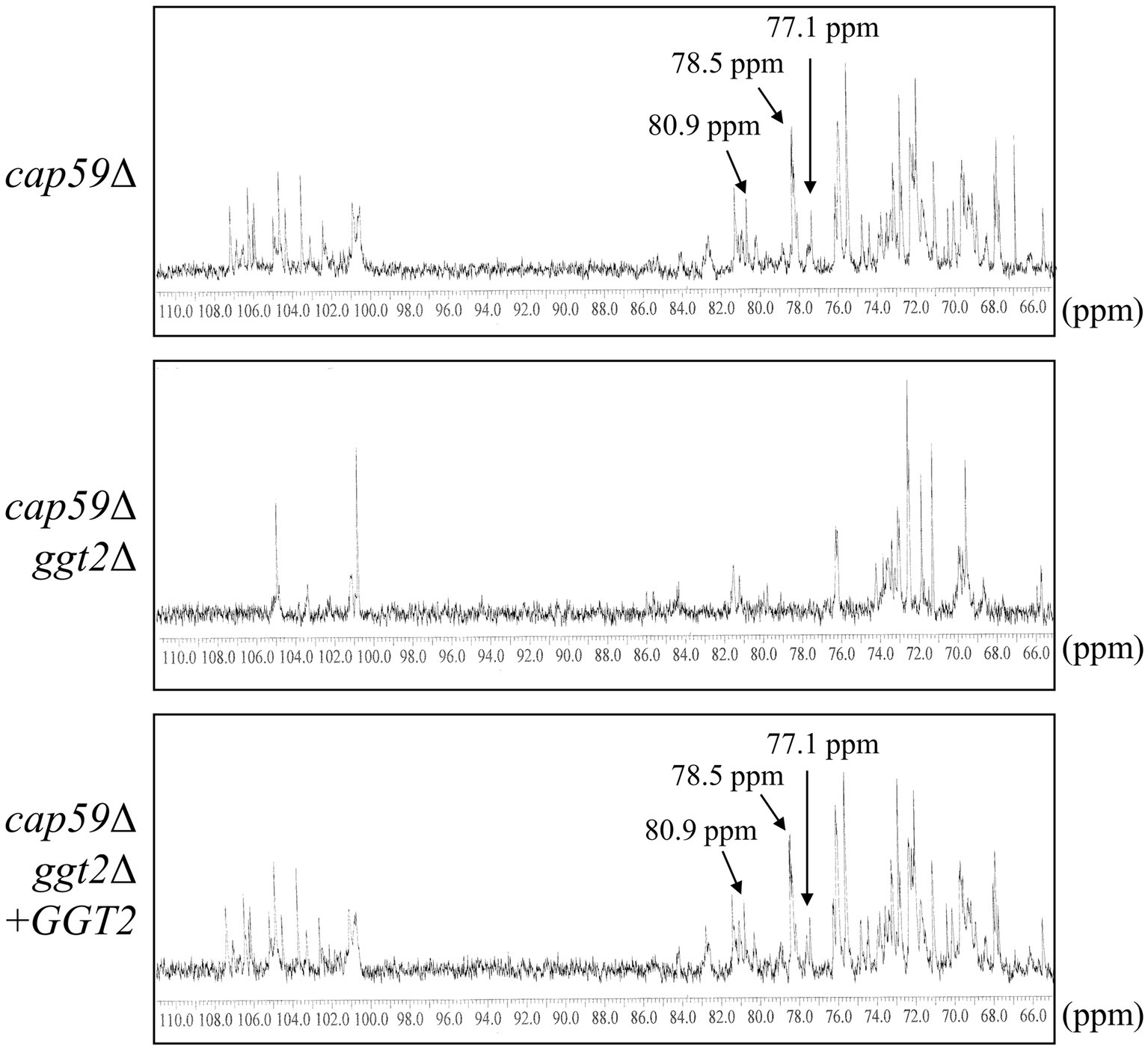
Figure 6. 13C-NMR analysis of GXMGal from cap59Δ, cap59Δ ggt2Δ, and cap59Δ ggt2Δ + GGT2 strains. Broad chemical signals at 100.6–105.1 ppm in cap59Δ and cap59Δ ggt2Δ + GGT2 are derived from position C-1 of Manα-(1 → 3)-, 2,3-O-substituted-Manα, Xylβ-(1 → 2)-, and Xylβ-(1 → 3)- in the galactomannan side chain. 13C-NMR signals at 78.5 and 77.1 ppm in cap59Δ and cap59Δ ggt2Δ + GGT2 are derived from position C-4 of Gal residues in 4-substituted Galβ and the 3,4-substituted Galβ side chain of GXMGal. The 13C-NMR signal at 80.9 ppm in cap59Δ and cap59Δ ggt2Δ + GGT2 is derived from position C-3 of Gal residues in 3,6-substituted Galα. The carbon chemical shifts were referenced relative to internal acetone at δ 31.07 ppm.
3.7 Phylogenetic analysis of Pvg3, Ggt1, Ggt2, and Ggt3 family proteins belonging to the GT31 family
Sequences of Pvg3, Ggt1, Ggt2, and Ggt3 family proteins were used to construct an evolutionary phylogenetic tree (Figure 7). The data set for analysis was obtained from FungiDB using the amino acid sequences of S. pombe Pvg3 and C. neoformans Ggt1, Ggt2, and Ggt3 as search queries.1 The protein sequences were clearly divided into the Pvg3, Ggt1, Ggt2, and Ggt3 clades (Figure 7). Ggt1 was widely distributed in Basidiomycota, including Ustilago, Coprinopsis, Schizophyllum, and Cryptococcus. Ggt2 was distributed in Pucciniomycetes and Tremellomycetes in Basidiomycota. Furthermore, Ggt3 was distributed only in Tremellomycetes in Basidiomycota.
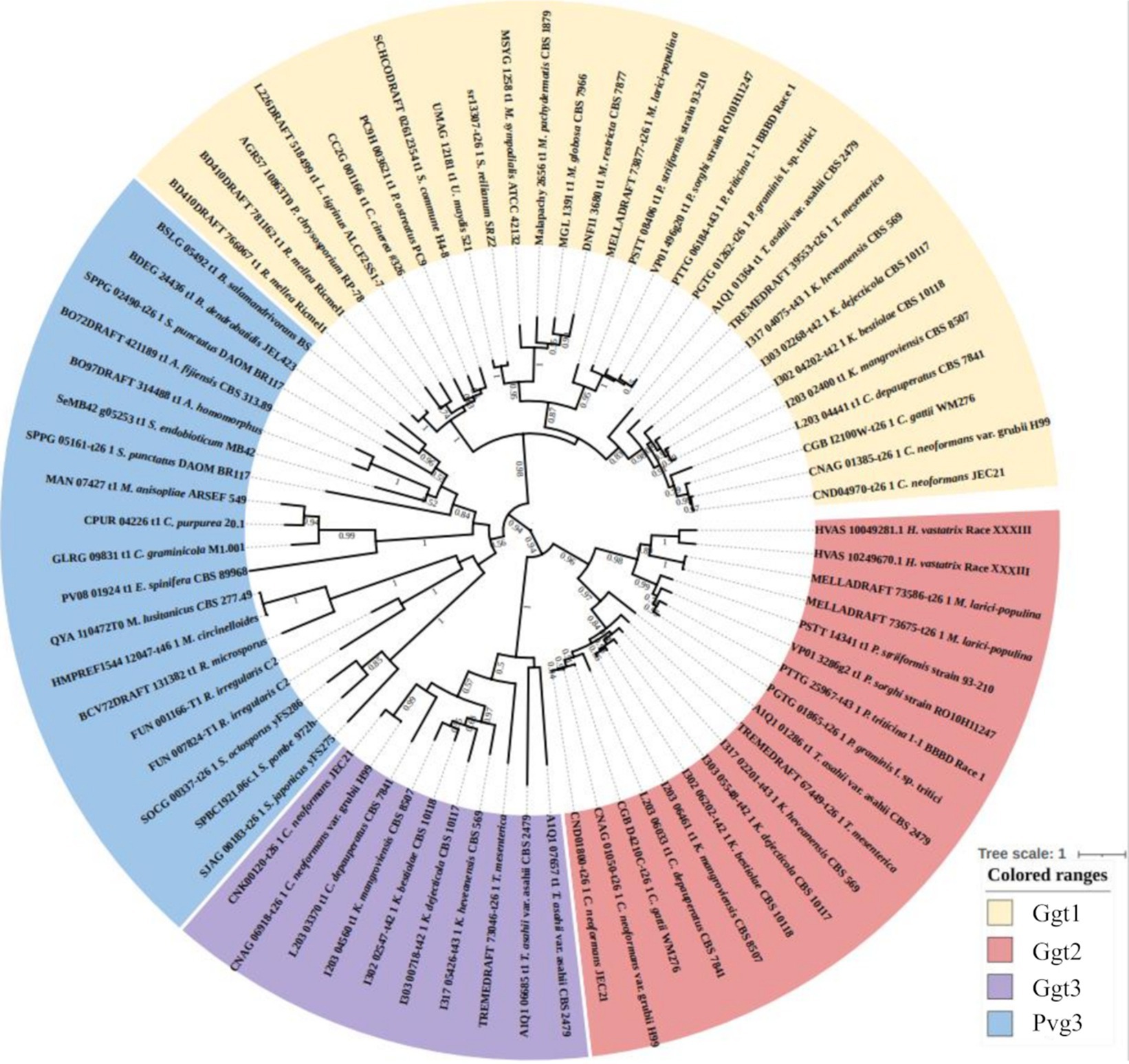
Figure 7. Phylogenetic analysis of Ggt1, Ggt2, and Ggt3 homologs in basidiomycetes and Pvg3 homologs in basidiomycetes and fission yeasts. Protein sequences were downloaded from FungiDB. The phylogenetic tree was drawn using iTOL. Alignment and phylogenetic tree inference were performed using MAFFT and RAxML, respectively, included in ETE v3.
4 Discussion
This study aimed to identify glycosyltransferases involved in the biosynthesis of C. neoformans capsules. We identified a putative β-(1 → 3)-Gal transferase belonging to the GT31 family that plays a role in GXMGal biosynthesis (Figure 8). Ggt1 is conserved in a wide range of species in the phylum Basidiomycota, whereas Ggt2 is only conserved in certain basidiomycete yeasts, such as Pucciniomycetes and Tremellomycetes (Figure 7). Multiple alignments of Ggt2 homologs revealed that amino acids in the GT-A domain are highly conserved (Supplementary Figure S4). Therefore, Ggt2 homologs may be responsible for synthesizing important glycan structures, including GXMGal. The enzyme involved in capsule biosynthesis has attracted attention as a novel antifungal drug target due to its contribution to virulence (Almeida et al., 2015). Many putative glycosyltransferases, involved in GXM biosynthesis, have been identified as CAP genes, because they can be easily screened based on phenotypes, such as India ink-negative staining (Fromtling et al., 1982). However, identification of glycosyltransferases involved in capsule biosynthesis is challenging, and only a few glycosyltransferases have been identified: α-(1 → 3)-Man transferase Cmt1 (Doering, 1999; Sommer et al., 2003) is involved in GXM biosynthesis and β-(1 → 2)-Xyl transferases Cxt1 and Cxt2 are involved in GXMGal biosynthesis (Klutts et al., 2007; Klutts and Doering, 2008; Reilly et al., 2009; Wang et al., 2018). We believe that our discovery of Ggt2 will contribute to studies on capsule biosynthesis in C. neoformans.
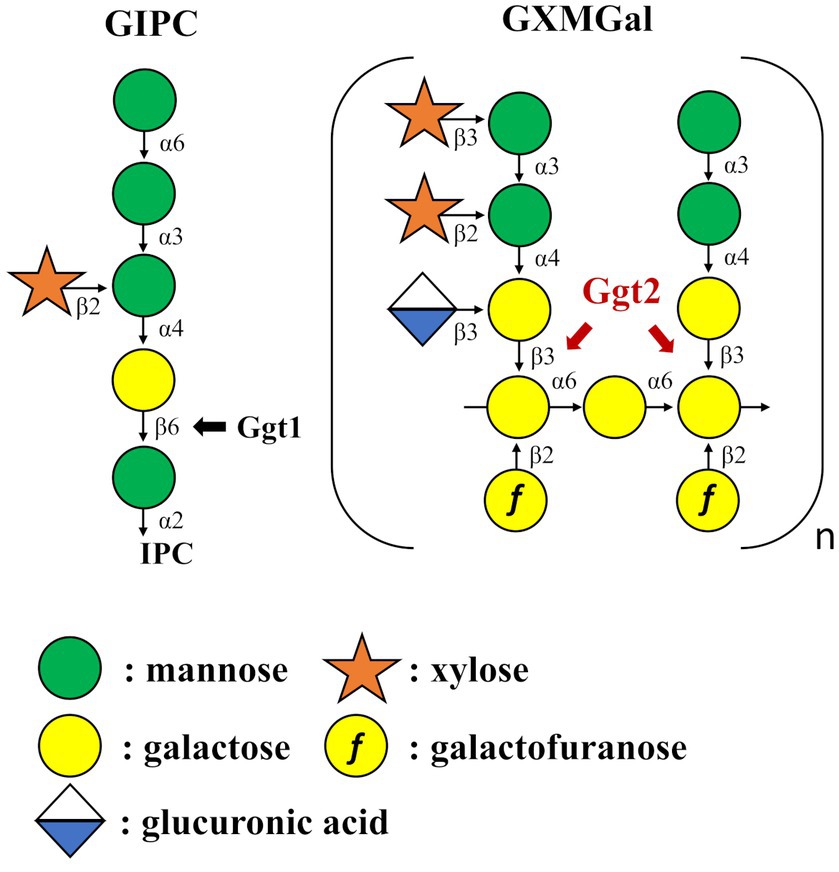
Figure 8. Summary model of GIPC and GXMGal biosynthesis in Cryptococcus neoformans. Ggt1 represents GIPC α-mannoside β-(1 → 6)-galactosyltransferase. Ggt2 represent α-galactoside β-(1 → 3)-galactosyltransferase.
Methylation GC–MS and NMR analyses revealed the detailed role of Ggt2 in GXMGal biosynthesis (Table 1; Figure 6). The loss or severe reduction of the galactomannan side chain of GXMGal in the ggt2 disruptant strain indicates that Ggt2 is the only α-Gal β-(1 → 3)-Gal transferase involved in GXMGal biosynthesis (Figure 2) because Ggt1 and Ggt2 accept α-mannoside and α-galactoside as receptor substrates, respectively. This is a logical result because the structure of the sugar chains involved in their biosynthesis suggests that Ggt1 and Ggt2 are likely to use α-mannoside and α-galactoside as their acceptor substrates, respectively. The GXMGal structure between the ggt3Δ and wild-type strains was not notably different and did not exhibit a Ts phenotype, suggesting that Ggt3 is not involved in GXMGal or GIPC biosynthesis (Figures 3, 4). The function of Ggt3 must be analyzed in detail. Interestingly, methylation GC–MS analysis detected methyl-esterified sugars that may have originated from glycans other than GXM and GXMGal. These indicate the presence of unknown glycan structures, such as N- or O-glycans or glycolipids. Considering how GXMGal was first discovered in the culture supernatant of a mutant strain lacking GXM (Cherniak et al., 1982), C. neoformans may possess unknown glycan structures. Thus, the detailed structures of these glycans should be clarified in the future.
Phenotypic analysis of gene disruptant strains revealed the physiological functions of Ggt2. ggt2Δ, like ggt1Δ, exhibited a Ts phenotype at 37°C, indicating that the galactomannan side chain of GXMGal, like GIPC, is important for high-temperature stress tolerance in C. neoformans. Notably, GXMGal is important for high-temperature stress tolerance in C. neoformans, although it is less abundant than GXM. In C. neoformans, Saccharomyces cerevisiae, and Aspergillus fumigatus, O-glycan deficiency leads to reduced high-temperature stress tolerance and disruption of cell wall integrity (Wagener et al., 2008; Kadooka et al., 2022; Thak et al., 2022). However, ggt2Δ was insensitive to Congo red or calcofluor white, which are inhibitors of cell wall synthesis, suggesting that loss of galactomannan side chains in GXMGal does not affect cell wall integrity in C. neoformans. Additionally, ugt1Δ is sensitive to Congo red, NaCl, and SDS (Li et al., 2017), but ggt2Δ is not, suggesting that the phenotype of ugt1Δ is not due to the loss of galactomannan side chains of GXMGal but rather due to a loss of GIPC. Consistently, GXM-deficient mutants were sensitive to NaCl and SDS (Li et al., 2018a,b). The phenotypic differences in polysaccharide-deficient mutants are interesting and should be analyzed further.
We used a bacterial heterologous expression system to generate recombinant Ggt2 and measured its β-Gal transfer activity to 4-methylumbelliferylated α-Gal. However, we could not detect glycosyltransfer activity (data not shown). This may be characteristic of the substrate specificity of Ggt2. The galactomannan side chain of GXMGal is added in succession, probably because Ggt2 recognizes di- or trisaccharide α-galacto-oligosaccharides as substrates and can only transfer β-Gal to a certain location. Another hypothesis is that Ggt2 does not exhibit glycosyltransferase activity by itself. For example, S. pombe Pvg3, which belongs to the GT31 family, is not active alone but exhibits glycosyltransferase activity by forming a complex with several proteins (Fukunaga et al., 2023). The enzymatic features of Ggt2 may be essential for the formation of the unique GXMGal structure and should be studied in more detail.
In conclusion, we have partially identified the mechanism of GXMGal biosynthesis in C. neoformans. Our findings will contribute substantially to understanding the structure and biosynthesis of the fungal cell wall and developing anticryptococcal agents.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
CK: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration. YT: Writing – review & editing, Data curation, Methodology, Validation. DH: Writing – review & editing, Data curation, Methodology, Validation. TO: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported in part by Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (JSPS KAKENHI) (21 K05373 to TO and 22 K06600 to YT) and Grant-in-Aid for Early-Career Scientists from the Japan Society for the Promotion of Science (JSPS KAKENHI) (22 K14817 to CK and 20 K15997 to YT). Additionally, a part of this study was supported by a Grant-in-Aid from the Institute for Fermentation, Osaka (IFO)(https://www.ifo.or.jp/) to CK (grant number Y-2023-2-030), TO (grant number G-2023-2-068), and a 2022 Young Investigator Research Grant from the Noda Institute for Scientific Research (to CK). Furthermore, a part of this study was supported by a Grant-in-Aid from the Amano Enzyme Foundation (https://www.amano-enzyme.co.jp/corporate/foundation/index.html) to TO.
Acknowledgments
We thank Ayumi Izuno (Sojo University) for assisting with some experiments. The strains were procured from the Fungal Genetics Stock Center (Kansas City, MO). The CRISPR/Cas9 plasmid and the plasmid carrying the drug resistance marker were purchased from Addgene. We thank Hiten D Madhani (University of California) and Yong-Sun Bahn (Yonsei University) for their donation of plasmids.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1390371/full#supplementary-material
SUPPLEMENTARY FIGURE S1
Construction of Cryptococcus neoformans gene disruption strains. (A) Chromosomal maps of cap59, ggt1, ggt2, and ggt3 disruptant strains. The positions of primers are indicated by arrows. (B) Confirmation of correct recombination of each strain using PCR analysis. Electrophoretic analysis of products amplified by PCR using the xxxX-comf-F–xxxX-comf-R primer pair. M, DNA size markers (Gene Ladder Wide 2; Nippon Gene, Tokyo, Japan).
SUPPLEMENTARY FIGURE S2
Construction of GGT2 complementation strains. (A) Chromosomal map of ggt2 disruptant strain. The positions of primers are indicated by arrows. (B) Confirmation of correct recombination of each strain using PCR analysis. Electrophoretic analysis of products amplified by PCR using the ggt2-comp-comf-F–ggt2-comp-comf-R primer pair. M, DNA size markers (Gene 493 Ladder Wide 2; Nippon Gene, Tokyo, Japan).
SUPPLEMENTARY FIGURE S3
Visualization of capsular structure. (A) Capsule imaging of H99, ggt2Δ, ggt2Δ+GGT2, and cap59Δ. C. neoformans strains were pre-cultured in YPD liquid medium at 30°C for 24 h, transferred to 10% Sabouraud liquid medium and further cultured at 30°C for 24 h. Finally, the cellswere stained with India ink and observed microscopically. Scale bars: 10 μm. (B) Capsule size quantification. The average diameter was calculated for the cells and capsules of H99, ggt2Δ, ggt2Δ+GGT2, and cap59Δ.
SUPPLEMENTARY FIGURE S4
Multiple sequence alignment of Ggt2 homologs. Multiple sequence alignment was performed using MAFFT v6.864 in GenomeNet Bioinformatics Tools and displayed in ESPript 3.0
Footnotes
References
Almeida, F., Wolf, J. M., and Casadevall, A. (2015). Virulence-associated enzymes of Cryptococcus neoformans. Eukaryot. Cell 14, 1173–1185. doi: 10.1128/EC.00103-15
Altamirano, S., Jackson, K. M., and Nielsen, K. (2020). The interplay of phenotype and genotype in Cryptococcus neoformans disease. Biosci. Rep. 40:BSR20190337. doi: 10.1042/BSR20190337
Andreishcheva, E. N., Kunkel, J. P., Gemmill, T. R., and Trimble, R. B. (2004). Five genes involved in biosynthesis of the pyruvylated Galbeta1,3-epitope in Schizosaccharomyces pombe N-linked glycans. J. Biol. Chem. 279, 35644–35655. doi: 10.1074/jbc.M403574200
Bose, I., Reese, A. J., Ory, J. J., Janbon, G., and Doering, T. L. (2003). A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2, 655–663. doi: 10.1128/EC.2.4.655-663.2003
Casadevall, A., Coelho, C., Cordero, R. J. B., Dragotakes, Q., Jung, E., Vij, R., et al. (2019). The capsule of Cryptococcus neoformans. Virulence 10, 822–831. doi: 10.1080/21505594.2018.1431087
Chang, Y. C., and Kwon-Chung, K. J. (1994). Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14, 4912–4919. doi: 10.1128/mcb.14.7.4912-4919.1994
Cherniak, R., Reiss, E., and Turner, S. H. (1982). A galactoxylomannan antigen of Cryptococcus neoformans serotype a. Carbohydr. Res. 103, 239–250. doi: 10.1016/S0008-6215(00)80686-2
Cherniak, R., Valafar, H., Morris, L. C., and Valafar, F. (1998). Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin. Diagn. Lab. Immunol. 5, 146–159. doi: 10.1128/CDLI.5.2.146-159.1998
Doering, T. L. (1999). A unique alpha-1,3 mannosyltransferase of the pathogenic fungus Cryptococcus neoformans. J. Bacteriol. 181, 5482–5488. doi: 10.1128/JB.181.17.5482-5488.1999
Doering, T. L. (2009). How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Ann. Rev. Microbiol. 63, 223–247. doi: 10.1146/annurev.micro.62.081307.162753
Fromtling, R. A., Shadomy, H. J., and Jacobson, E. S. (1982). Decreased virulence in stable, acapsular mutants of Cryptococcus neoformans. Mycopathologia 79, 23–29. doi: 10.1007/BF00636177
Fukunaga, T., Watanabe, M., Nakamichi, Y., Morita, T., Higuchi, Y., Maekawa, H., et al. (2023). Mechanistic insights into Schizosaccharomyces pombe GT-A family protein Pvg3 in the biosynthesis of pyruvylated β1,3-galactose of N-linked oligosaccharides. J. Biosci. Bioeng. 135, 423–432. doi: 10.1016/j.jbiosc.2023.03.002
Heiss, C., Klutts, J. S., Wang, Z., Doering, T. L., and Azadi, P. (2009). The structure of Cryptococcus neoformans galactoxylomannan contains beta-D-glucuronic acid. Carbohydr. Res. 344, 915–920. doi: 10.1016/j.carres.2009.03.003
Heiss, C., Skowyra, M. L., Liu, H., Klutts, J. S., Wang, Z., Williams, M., et al. (2013). Unusual galactofuranose modification of a capsule polysaccharide in the pathogenic yeast Cryptococcus neoformans. J. Biol. Chem. 288, 10994–11003. doi: 10.1074/jbc.M112.441998
Huang, M. Y., Joshi, M. B., Boucher, M. J., Lee, S., Loza, L. C., Gaylord, E. A., et al. (2022). Short homology-directed repair using optimized Cas9 in the pathogen Cryptococcus neoformans enables rapid gene deletion and tagging. Genetics 220:iyab180. doi: 10.1093/genetics/iyab180
Kadooka, C., Hira, D., Tanaka, Y., Chihara, Y., Goto, M., and Oka, T. (2022). Mnt1, an α-(1 → 2)-mannosyltransferase responsible for the elongation of N-glycans and O-glycans in Aspergillus fumigatus. Glycobiology 32, 1137–1152. doi: 10.1093/glycob/cwac049
Katafuchi, Y., Li, Q., Tanaka, Y., Shinozuka, S., Kawamitsu, Y., Izumi, M., et al. (2017). GfsA is a β1,5-galactofuranosyltransferase involved in the biosynthesis of the galactofuran side chain of fungal-type galactomannan in Aspergillus fumigatus. Glycobiology 27, 568–581. doi: 10.1093/glycob/cwx028
Klutts, J. S., and Doering, T. L. (2008). Cryptococcal xylosyltransferase 1 (Cxt1p) from Cryptococcus neoformans plays a direct role in the synthesis of capsule polysaccharides. J. Biol. Chem. 283, 14327–14334. doi: 10.1074/jbc.M708927200
Klutts, J. S., Levery, S. B., and Doering, T. L. (2007). A beta-1,2-xylosyltransferase from Cryptococcus neoformans defines a new family of glycosyltransferases. J. Biol. Chem. 282, 17890–17899. doi: 10.1074/jbc.M701941200
Kumar, P., Yang, M., Haynes, B. C., Skowyra, M. L., and Doering, T. L. (2011). Emerging themes in cryptococcal capsule synthesis. Curr. Opin. Struct. Biol. 21, 597–602. doi: 10.1016/j.sbi.2011.08.006
Li, L. X., Ashikov, A., Liu, H., Griffith, C. L., Bakker, H., and Doering, T. L. (2017). Cryptococcus neoformans UGT1 encodes a UDP-galactose/UDP-GalNAc transporter. Glycobiology 27, 87–98. doi: 10.1093/glycob/cww078
Li, L. X., Rautengarten, C., Heazlewood, J. L., and Doering, T. L. (2018a). Xylose donor transport is critical for fungal virulence. PLoS Pathog. 14:e1006765. doi: 10.1371/journal.ppat.1006765
Li, L. X., Rautengarten, C., Heazlewood, J. L., and Doering, T. L. (2018b). UDP-glucuronic acid transport is required for virulence of Cryptococcus neoformans. MBio 9, e02319–e02317. doi: 10.1128/mBio.02319-17
Maziarz, E. K., and Perfect, J. R. (2016). Cryptococcosis. Infect. Dis. Clin. N. Am. 30, 179–206. doi: 10.1016/j.idc.2015.10.006
Moyrand, F., Fontaine, T., and Janbon, G. (2007). Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol. Microbiol. 64, 771–781. doi: 10.1111/j.1365-2958.2007.05695.x
Moyrand, F., Lafontaine, I., Fontaine, T., and Janbon, G. (2008). UGE1 and UGE2 regulate the UDP-glucose/UDP-galactose equilibrium in Cryptococcus neoformans. Eukaryot. Cell 7, 2069–2077. doi: 10.1128/EC.00189-08
O’Meara, T. R., and Alspaugh, J. A. (2012). The Cryptococcus neoformans capsule: a sword and a shield. Clin. Microbiol. Rev. 25, 387–408. doi: 10.1128/CMR.00001-12
Previato, J. O., Vinogradov, E., Maes, E., Fonseca, L. M., Guerardel, Y., Oliveira, P. A. V., et al. (2017). Distribution of the O-acetyl groups and β-galactofuranose units in galactoxylomannans of the opportunistic fungus Cryptococcus neoformans. Glycobiology 27, 582–592. doi: 10.1093/glycob/cww127
Reilly, M. C., Levery, S. B., Castle, S. A., Klutts, J. S., and Doering, T. L. (2009). A novel xylosylphosphotransferase activity discovered in Cryptococcus neoformans. J. Biol. Chem. 284, 36118–36127. doi: 10.1074/jbc.M109.056226
Rocha, J. D., Nascimento, M. T., Decote-Ricardo, D., Côrte-Real, S., Morrot, A., Heise, N., et al. (2015). Capsular polysaccharides from Cryptococcus neoformans modulate production of neutrophil extracellular traps (NETs) by human neutrophils. Sci. Rep. 5:8008. doi: 10.1038/srep08008
So, Y. S., Yang, D. H., Jung, K. W., Huh, W. K., and Bahn, Y. S. (2017). Molecular characterization of adenylyl cyclase complex proteins using versatile protein-tagging plasmid systems in Cryptococcus neoformans. J. Microbiol. Biotechnol. 27, 357–364. doi: 10.4014/jmb.1609.09036
Sommer, U., Liu, H., and Doering, T. L. (2003). An alpha-1,3-mannosyltransferase of Cryptococcus neoformans. J. Biol. Chem. 278, 47724–47730. doi: 10.1074/jbc.M307223200
Thak, E. J., Son, Y. J., Lee, D. J., Kim, H., Kim, J. H., Lee, S. B., et al. (2022). Extension of O-linked mannosylation in the Golgi apparatus is critical for cell wall integrity signaling and interaction with host cells in Cryptococcus neoformans pathogenesis. MBio 13:e0211222. doi: 10.1128/mbio.02112-22
Thumuluri, V., Almagro Armenteros, J. J., Johansen, A. R., Nielsen, H., and Winther, O. (2022). DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 50, W228–W234. doi: 10.1093/nar/gkac278
Vaishnav, V. V., Bacon, B. E., O’Neill, M., and Cherniak, R. (1998). Structural characterization of the galactoxylomannan of Cryptococcus neoformans Cap67. Carbohydr. Res. 306, 315–330. doi: 10.1016/s0008-6215(97)10058-1
Wagener, J., Echtenacher, B., Rohde, M., Kotz, A., Krappmann, S., Heesemann, J., et al. (2008). The putative alpha-1,2-mannosyltransferase AfMnt1 of the opportunistic fungal pathogen Aspergillus fumigatus is required for cell wall stability and full virulence. Eukaryot. Cell 7, 1661–1673. doi: 10.1128/EC.00221-08
Wang, Z. A., Li, L. X., and Doering, T. L. (2018). Unraveling synthesis of the cryptococcal cell wall and capsule. Glycobiology 28, 719–730. doi: 10.1093/glycob/cwy030
Wohlschlager, T., Buser, R., Skowyra, M. L., Haynes, B. C., Henrissat, B., Doering, T. L., et al. (2013). Identification of the galactosyltransferase of Cryptococcus neoformans involved in the biosynthesis of basidiomycete-type glycosylinositolphosphoceramide. Glycobiology 23, 1210–1219. doi: 10.1093/glycob/cwt057
Keywords: Cryptococcus neoformans , galactosyltransferase, glucuronoxylomannogalactan, polysaccharide, virulence factor
Citation: Kadooka C, Tanaka Y, Hira D and Oka T (2024) Identification of a putative α-galactoside β-(1 → 3)-galactosyltransferase involved in the biosynthesis of galactomannan side chain of glucuronoxylomannogalactan in Cryptococcus neoformans. Front. Microbiol. 15:1390371. doi: 10.3389/fmicb.2024.1390371
Edited by:
Allan J. Guimarães, Fluminense Federal University, BrazilReviewed by:
Yong Fu, Washington University in St. Louis, United StatesAngie Gelli, University of California, Davis, United States
Copyright © 2024 Kadooka, Tanaka, Hira and Oka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chihiro Kadooka, a2Fkb29rYUBiaW8uc29qby11LmFjLmpw; Takuji Oka, b2thQGJpby5zb2pvLXUuYWMuanA=
 Chihiro Kadooka
Chihiro Kadooka Yutaka Tanaka
Yutaka Tanaka Daisuke Hira
Daisuke Hira Takuji Oka
Takuji Oka