
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 May 2024
Sec. Virology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1390328
Porcine viral diarrhea is caused by many pathogens and can result in watery diarrhea, dehydration and death. Various detection methods, such as polymerase chain reaction (PCR) and real-time quantitative PCR (qPCR), have been widely used for molecular diagnosis. We developed a triplex real-time quantitative reverse transcription PCR (qRT-PCR) for the simultaneous detection of three RNA viruses potentially associated with porcine viral diarrhea: porcine epidemic diarrhea virus (PEDV), porcine transmissible gastroenteritis virus (TGEV), and porcine rotavirus A (PoRVA). The triplex qRT-PCR had R2 values of 0.999 for the standard curves of PEDV, TGEV and PoRVA. Importantly, the limits of detection for PEDV, TGEV and PoRVA were 10 copies/μL. The specificity test showed that the triplex qRT-PCR detected these three pathogens specifically, without cross-reaction with other pathogens. In addition, the approach had good repeatability and reproducibility, with intra-and inter-assay coefficients of variation <1%. Finally, this approach was evaluated for its practicality in the field using 256 anal swab samples. The positive rates of PEDV, TGEV and PoRVA were 2.73% (7/256), 3.91% (10/256) and 19.14% (49/256), respectively. The co-infection rate of two or more pathogens was 2.73% (7/256). The new triplex qRT-PCR was compared with the triplex RT-PCR recommended by the Chinese national standard (GB/T 36871-2018) and showed 100% agreement for PEDV and TGEV and 95.70% for PoRVA. Therefore, the triplex qRT-PCR provided an accurate and sensitive method for identifying three potential RNA viruses for porcine viral diarrhea that could be applied to diagnosis, surveillance and epidemiological investigation.
The main pathogens causing diarrhea in piglets are porcine epidemic diarrhea virus (PEDV), porcine transmissible gastroenteritis virus (TGEV) and porcine rotavirus A (PoRVA) (Zhang et al., 2013; Monteagudo et al., 2022). Co-infection by these viruses is common in swine and poses a serious challenge for diarrhea control in swine farms (Zhang et al., 2019; Shi et al., 2021). PEDV is an enveloped, single-stranded, positive-sense RNA virus belonging to the genus Coronavirus in the family Coronaviridae. In 1978, researchers first isolated PEDV from the intestinal contents of pigs in the UK (Pensaert and de Bouck, 1978). PEDV has spread globally since then, causing watery diarrhea, vomiting, dehydration and death in pigs, resulting in severe economic losses for the swine industry (Wang et al., 2016). The PEDV genome is approximately 28 kb long and comprises seven open reading frames (ORFs). The M gene (ORF5) encodes the membrane protein M and has a relatively conserved sequence, which makes it a suitable molecular detection target for PEDV diagnosis (Kocherhans et al., 2001; Yang et al., 2014; Rasmussen et al., 2018). TGEV is also an RNA virus that belongs to the Coronavirus genus and Coronaviridae family. It was the first coronavirus identified in pigs and is responsible for porcine viral diarrhea. The genome of TGEV is approximately 28.6 kb in length and comprises nine major ORFs. The N gene (ORF6) encodes the capsid protein N and is relatively conserved in the TGEV genome (Eleouet et al., 1995; Yount et al., 2000). Another porcine enteric virus, PoRVA, is a non-enveloped double-stranded RNA virus belonging to the genus Rotavirus and family Reoviridae. It is one of the major pathogens responsible for severe diarrhea in piglets worldwide (Vlasova et al., 2017; Luo et al., 2023). The PoRVA genome is approximately 18.5 kb and has 11 double-stranded RNA segments. NSP3 is a relatively conserved gene that plays a key role in viral replication and transcription, and is a common target gene for detecting PoRVA infection (Ghosh and Kobayashi, 2011).
Porcine viral diarrhea caused by these three enteric viruses poses serious health and economic threats to pig farming in China. It is also a challenge to the microbiological quality control of specific pathogen-free pigs for scientific research. To cope with the challenges of PEDV, TGEV and PoRVA, 34 standards have been publicly released in China so far, including 3 national standards, 6 agricultural industry and entry-exit inspection and quarantine industry standards, and 25 provincial local standards. These standards specify the detection methods for the three pathogens, such as reverse transcription polymerase chain reaction (RT-PCR), nested RT-PCR, single real-time quantitative RT-PCR (qRT-PCR) and duplex qRT-PCR. Among the 34 standards, a triplex RT-PCR technique was established only in the Chinese national standard (GB/T 36871-2018, 2018) for simultaneous detection and diagnosis of PEDV, TGEV and PoRVA. In addition, the duplex RT-PCR and duplex qRT-PCR techniques for differential diagnosis of dual infections by porcine viral diarrhea viruses were developed in some Chinese provincial local standards, e.g., Zhejiang provincial local standard (DB33/T 2254-2020, 2020) and Anhui provincial local standard (DB34/T 2795-2016, 2016). Porcine viral diarrhea is also severe worldwide, which has led to development of various pathogen detection techniques, such as triplex RT-PCR, nested RT-PCR and qRT-PCR (Huang et al., 2019; Chen et al., 2023). The accurate and rapid molecular diagnosis is essential for the prevention and control of the diseases caused by PEDV, TGEV and PoRVA. Therefore, it is necessary to establish a detection method with high specificity, sensitivity and efficiency.
Real-time qPCR is an accurate, sensitive, and rapid method for detecting and quantifying target genomes. Compared to conventional single qPCR, multiplex qPCR can simultaneously detect multiple target genes in a single reaction, showing many advantages such as high efficiency, throughput, and cost effective (Mackay, 2004; Yang et al., 2022). Advances in molecular biology techniques have led to widespread use of multiplex qPCR in clinical detection (Wang et al., 2020). In this study, we designed primers and probes based on the conserved fragments of PEDV M gene, TGEV N gene and PoRVA NSP3 gene, and successfully developed a triplex qRT-PCR based on TaqMan probes. This method was highly sensitive and specific and did not cross-react with the genomes of other swine pathogens. It could be used for diagnosis, epidemiological investigation and microbiological quality control of specific pathogen-free pigs.
The genomes (DNA or RNA) of PEDV, TGEV, PoRVA, pseudorabies virus (PRV), porcine circovirus type 2 (PCV2), porcine parvovirus (PPV), porcine deltacoronavirus (PDCoV), Seneca virus A (SVA), Toxoplasma gondii, Leptospira interrogans, Mycoplasma hyopneumoniae, Mycoplasma hyorhinis, Haemophilus parasuis, Streptococcus suis, Pasteurella multocida and Actinobacillus pleuropneumoniae were preserved by the State Key Laboratory for Animal Disease Control and Prevention of China or Heilongjiang Provincial Key Laboratory of Laboratory Animal and Comparative Medicine. In addition, 256 anal swab samples from pigs with or without clinical diarrhea were obtained from Animal Health Testing Center of Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences. The Institutional Review Board of Harbin Veterinary Research Institute or the Regulations on the Administration of Laboratory Animals of China did not require the study to be reviewed or approved by an ethics committee because the samples were collected from animals that were already dead or euthanized for other purposes, and no additional harm or intervention was imposed on the animals.
To ensure the detection performance of the primers and probes used in the triplex qRT-PCR, M gene of PEDV genome, N gene of TGEV genome and NSP3 gene of PoRVA genome were selected as the detection targets, based on their relative conservation. We obtained 86 PEDV M gene sequences, 46 TGEV N gene sequences and 20 PoRVA NSP3 gene sequences from GenBank database. We used MegAlign to align them and determine the most conserved regions of each target gene. Using Primer Express 3.0.1, we designed primers and probes for the three viruses with the following conditions: primer length 18–30 bp, primer melting temperature (Tm) 58–62°C, primer GC content 40–60%, product Tm 70–90°C and product size 70–150 bp. We ensured that the probe Tm value was higher than that of the primers. The specificity of the primers and probes was verified using the BLAST tool provided by the National Center for Biotechnology Information. For triplex detection, the probes for the three viral genes were labeled with different 5′-reporting dyes: Victoria Blue (VIC), Cyanine 5 (Cy5) and Fluorescein (FAM) and corresponding 3′-quenchers: Black Hole Quencher 1 (BHQ1), Black Hole Quencher 2 (BHQ2) and Minor Groove Binder (MGB). The triplex RT-PCR recommended by the Chinese national standard (GB/T 36871-2018, 2018) was used to verify the accuracy of the results for clinical samples. The details of the primers and probes are provided in Table 1. Figure 1 shows the locations of the triplex qRT-PCR primers and probes for PEDV, TGEV and PoRVA in different reference strains.
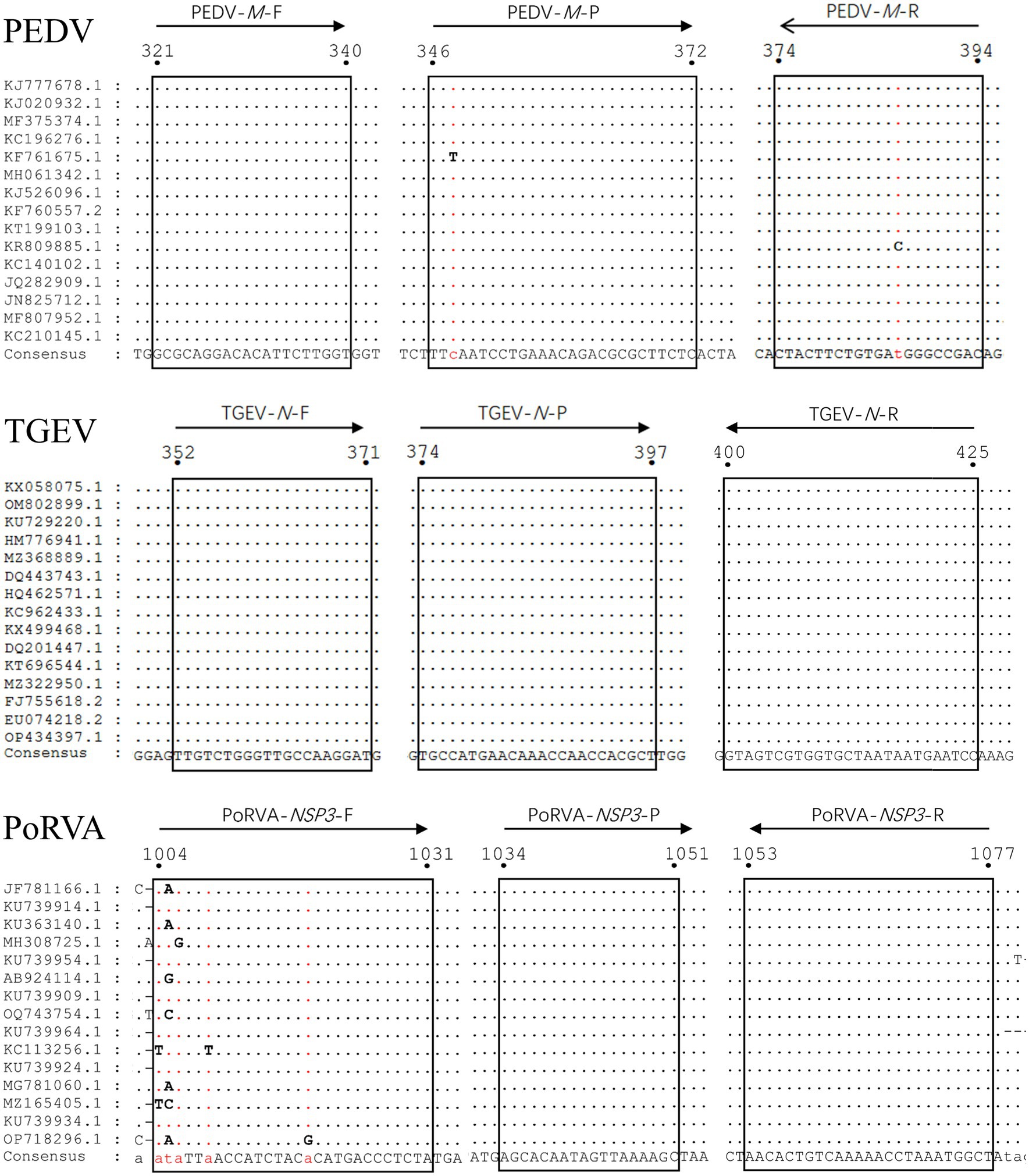
Figure 1. Alignment of sequences of reference viral strains collected from GenBank database. The locations of the primers/probes specific for PEDV M gene, TGEV N gene and PoRVA NSP3 gene are shown. The positions of the partial nucleotide fragments are indicated by numbers.
A synthetic gene fragment (PEDV-M-TGEV-N-PoRVA-NSP3) containing partial sequences of PEDV M gene, TGEV N gene and PoRVA NSP3 gene was constructed in Sangon Biotech Co., Ltd. (Shanghai, China). This fragment was inserted into the pUC57 cloning vector, forming a standard plasmid (pUC57-PEDV M & TGEV N & PoRVA NSP3) for subsequent detection (Figure 2). The sequence of the synthetic gene fragment is shown in Supplementary Table S1. The plasmids were quantified by ultraviolet absorbance at 260 and 280 nm wavelengths using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and their copy number was calculated based on the size of the standard plasmid template using the following formula: copies/μL = (A260 (ng/μL) × 10−9 × 6.02 × 1023)/(DNA length × 650). The standard plasmids were serially diluted 10-fold to a concentration gradient of 108–100 copies/μL with EASY Dilution (TaKaRa, China, Dalian).
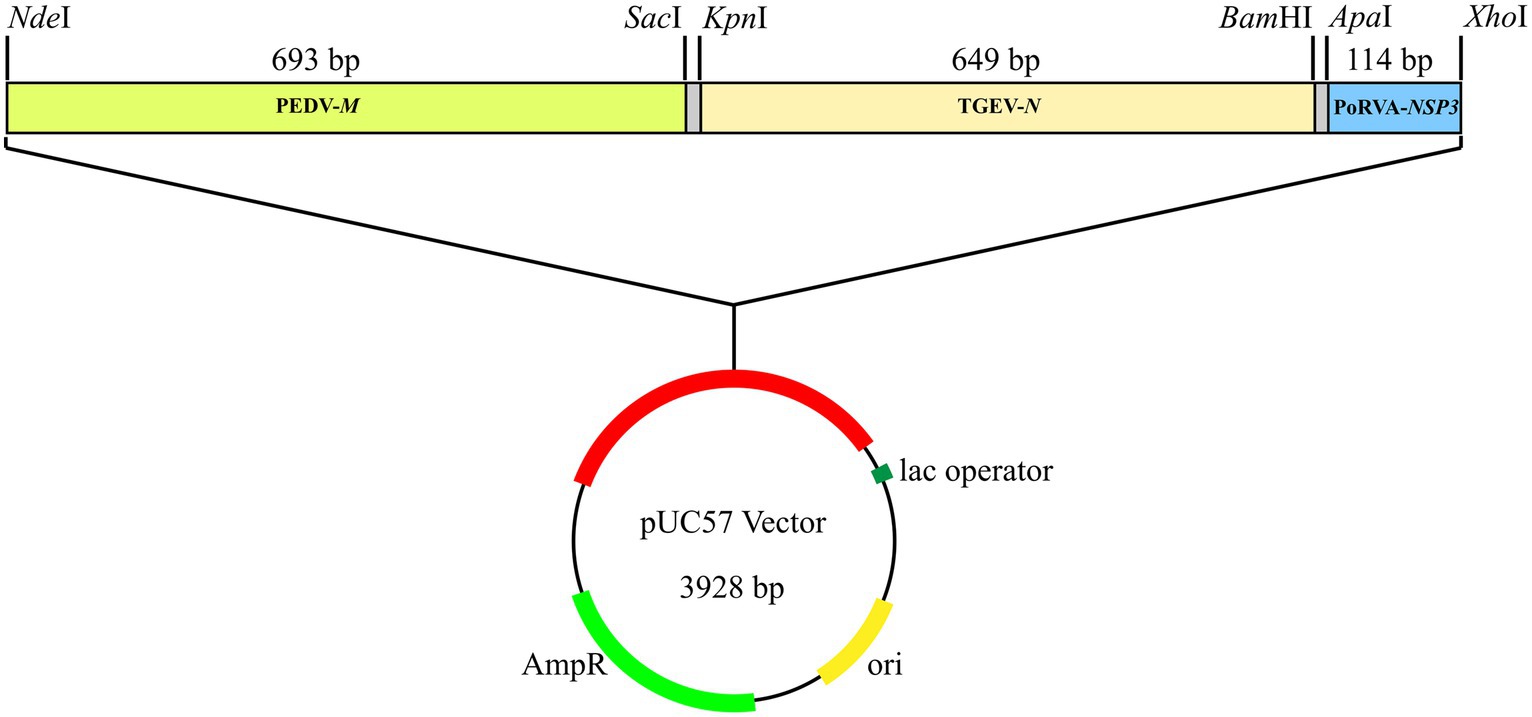
Figure 2. Standard plasmid containing three conserved gene fragments: PEDV M (693 bp), TGEV N (649 bp) and PoRVA NSP3 (114 bp). Each fragment had specific restriction enzyme cutting sites at both ends: NdeI and SacI for PEDV M; KpnI and BamHI for TGEV N; and ApaI and XhoI for PoRVA NSP3.
The unoptimized triplex qRT-PCR consisted of 10 μL 2× One Step U+ Mix (Vazyme, China, Nanjing), 1 μL One Step U+ Enzyme Mix (Vazyme), 0.4 μL 50× ROX Reference Dye II, 0.4 μL each primer (final concentration of 200 nM), 0.2 μL each probe (final concentration of 100 nM), 4 μL template and 1.6 μL RNase-free water in a total volume of 20 μL. The amplification was performed on an ABI QuantStudio5 real-time PCR system (Thermo Fisher Scientific) with the following program: 55°C for 15 min, 95°C for 30 s; 40 cycles of 95°C for 10 s and 60°C for 30 s. The fluorescence signal was automatically collected at the end of each cycle. To optimize the reaction system, we explored different primer volumes (10 μM) and probe volumes (10 μM). A range of primer volumes (0.3–1.2 μL) was assessed to achieve final concentrations spanning 150–600 nM. Additionally, probe volumes were varied from 0.1 to 0.6 μL, covering a concentration range of 50–300 nM. Recombinant plasmids (107 copies/μL) served as the detection template for optimization. Finally, the fluorescence intensity and cycle threshold (Ct) values of each primer and probe concentration were compared to determine the optimal volumes. The annealing temperature was also optimized by setting six gradients from 56 to 61°C and comparing the fluorescence intensity and Ct values of each gradient.
On the basis of the optimized reaction and protocol, three replicates of plasmid samples with serial dilutions from 108 to 10 copies/μL were detected using the triplex qRT-PCR and subjected to linear regression between Ct values and the logarithm of plasmid copy numbers. Eight-point standard curves were established for PEDV, TGEV and PoRVA, including negative controls.
To evaluate the specificity of the primer and probe sets, genomes (DNA or RNA) of PEDV, TGEV, PoRVA, PRV, PCV2, PPV, PDCoV, SVA, T. gondii, L. interrogans, M. hyopneumoniae, M. hyorhinis, H. parasuis, S. suis, P. multocida and A. pleuropneumoniae were tested using the triplex qRT-PCR. All nucleic acid samples were stored previously in our laboratory.
For sensitivity assessment, standard plasmids were serially diluted 10-fold to final concentrations ranging from 108 to 1 copies/μL. These dilutions were used as templates to determine the limit of detection (LoD) for triplex qRT-PCR and each reaction was repeated three times in a single test. We tested the standard plasmids at 100, 10 and 1 copies/μL 20 times to ensure the LoD accuracy. We set the LoD as the lowest concentration of standard plasmids that gave positive results in 85% of the replicates and marked it on the amplification curves. The threshold was set in the middle of the exponential amplification phase in the logarithmic view. A positive test result was defined as an exponential fluorescence curve that crossed the threshold within 35 cycles [(Ct) <35]. According to this definition, we calculated the positive rates at 100, 10 and 1 copies/μL of standard plasmids.
The repeatability (intra-assay precision) and reproducibility (inter-assay precision) of the developed triplex qRT-PCR were determined using standard plasmids at three different concentrations (106, 104 and 100 copies/μL). We analyzed each dilution in triplicate on the same day for intra-assay variability and in six independent experiments by two different operators on different days for inter-assay variability. The coefficient of variation (CV) of the Ct values of the samples at different concentrations was calculated in both intra-assay and inter-assay tests to estimate the repeatability and reproducibility.
Viral RNA was extracted from 256 anal swab samples using AxyPrep Body Fluid Viral DNA/RNA Miniprep Kit (Corning Life Sciences, China, Wujiang). The RNA samples were tested in triplicate by the optimized triplex qRT-PCR. Subsequently, the sample RNA was reverse transcribed into cDNA using PrimeScript™ RT Master Mix (Perfect Real Time) (TaKaRa) and detected by the triplex RT-PCR recommended by the Chinese national standard (GB/T 36871-2018, 2018), to validate the clinical performance of the developed triplex qRT-PCR assay. For the triplex RT-PCR, the reaction mixture (25 μL) contained 12.5 μL 2× Taq PCR Star Mix (Genstar, China, Beijing), 0.2 μL PEDV primers (final concentration of 80 nM), 0.4 μL TGEV primers (final concentration of 160 nM), 1 μL PoRVA primers (final concentration of 400 nM), 4 μL cDNA template and 5.3 μL RNase-free water. We performed the triplex RT-PCR with the following parameters: pre-denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 1 min, and final extension at 72°C for 10 min.
A plasmid with three conserved gene fragments of PEDV M (693 bp), TGEV N (649 bp) and PoRVA NSP3 (114 bp) was constructed, all containing their respective qRT-PCR amplicons. The plasmid was used as the standard for subsequent detection.
Using the standard plasmid pUC57-PEDV M & TGEV N & PoRVA NSP3 with the target fragments as the template, we optimized the reaction conditions of the triplex qRT-PCR. Orthogonal experiments determined the optimal annealing temperature and concentrations of primers and probes. For TGEV and PoRVA, the optimal volumes of probes and primers were 0.4 and 0.7 μL, respectively. For PEDV, both were 0.3 μL (Figure 3). The confirmed reaction system was listed in Table 2. The optimal annealing temperature was 60°C, which yielded the highest amplification efficiency (Figure 4).
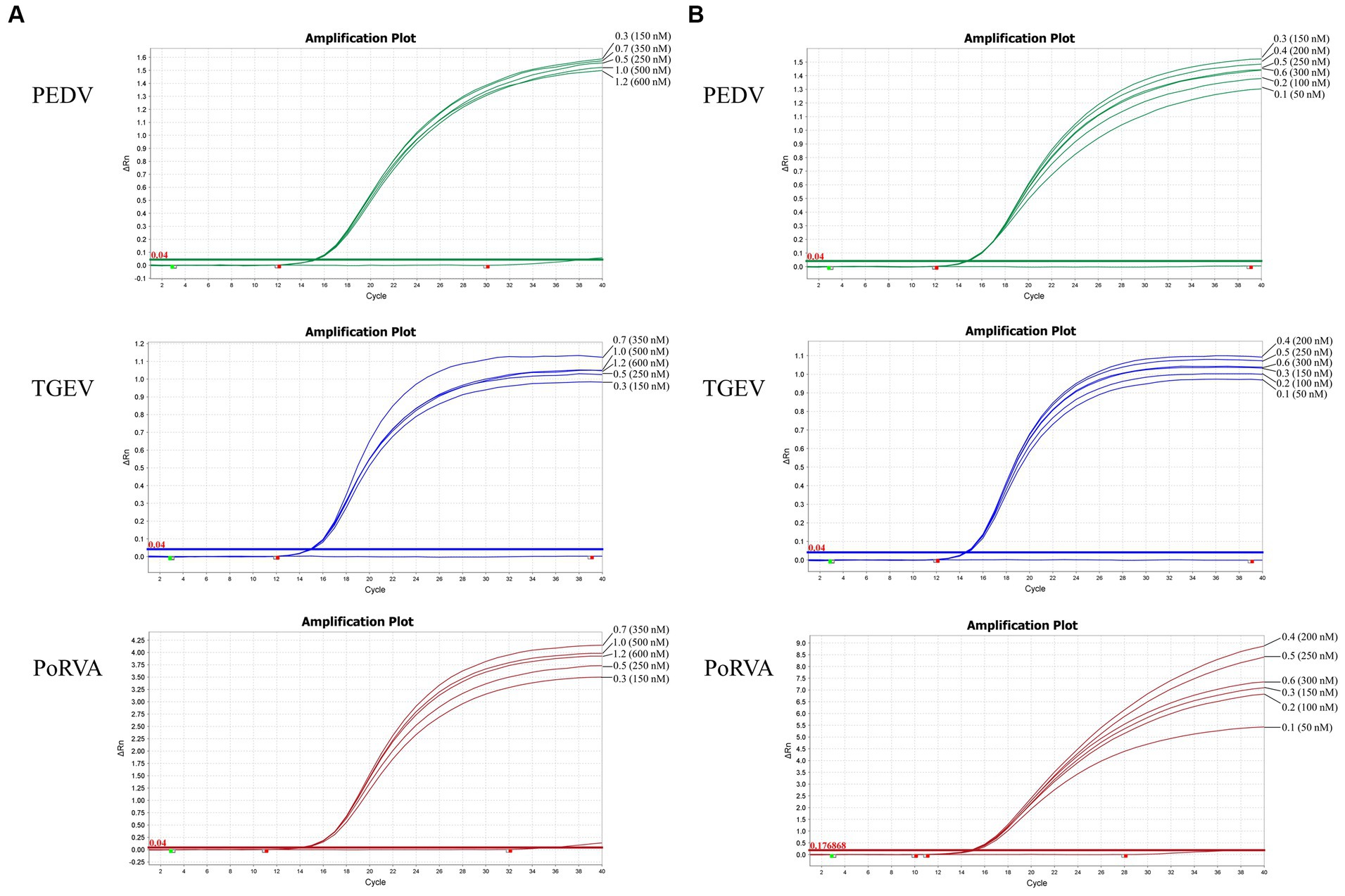
Figure 3. Optimization of the triplex qRT-PCR assay. (A) Optimization of primer volumes and the final concentrations in the reaction. The optimal volumes of forward and reverse primers were 0.7 μL for TGEV (final concentration: 350 nM) and PoRVA (final concentration: 350 nM), and 0.3 μL for PEDV (final concentration: 150 nM). (B) Optimization of probe volumes and the final concentrations in the reaction. The optimal volumes of probes were 0.4 μL for TGEV (final concentration: 200 nM) and PoRVA (final concentration: 200 nM), and 0.3 μL for PEDV (final concentration: 150 nM).
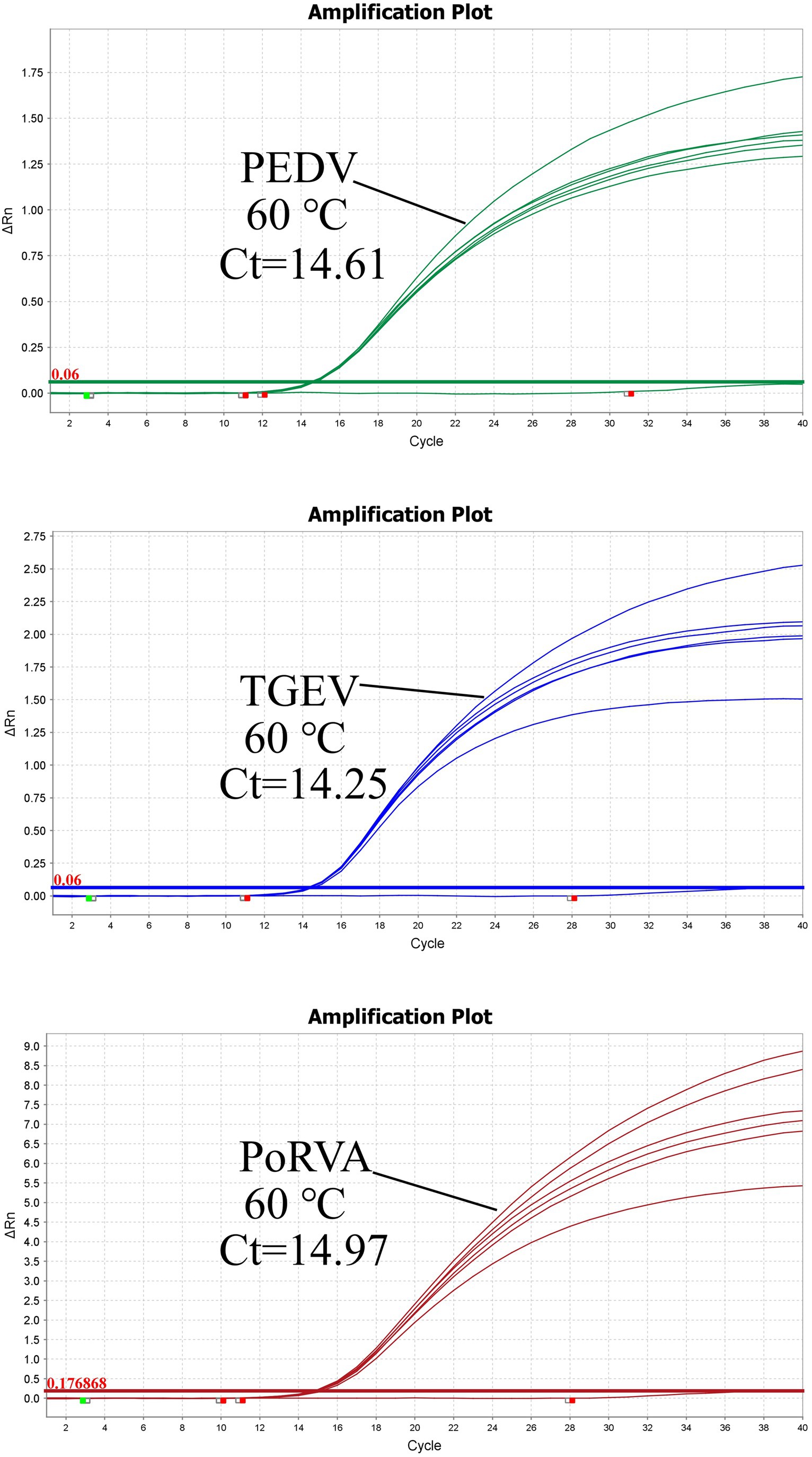
Figure 4. Triplex qRT-PCR amplification curves of different annealing temperatures. The optimal annealing temperature for triplex qRT-PCR was determined by measuring the amplification efficiency of the reaction at different Tm values. The highest amplification efficiency was achieved at 60°C.
The standard plasmid was diluted in a 10-fold series and eight standard samples (108–10 copies/μL) were selected as templates to establish the standard curve of the triplex qRT-PCR. Figure 5 shows the correlation coefficients (R2), equation slopes and amplification efficiencies (E) for each virus: PEDV, 0.999, −3.272 and 102.118%; TGEV, 0.999, −3.294 and 101.179%; and PoRVA, 0.999, −3.22 and 104.441%. The initial template and Ct value had a good linear relationship, as indicated by R2 and E.
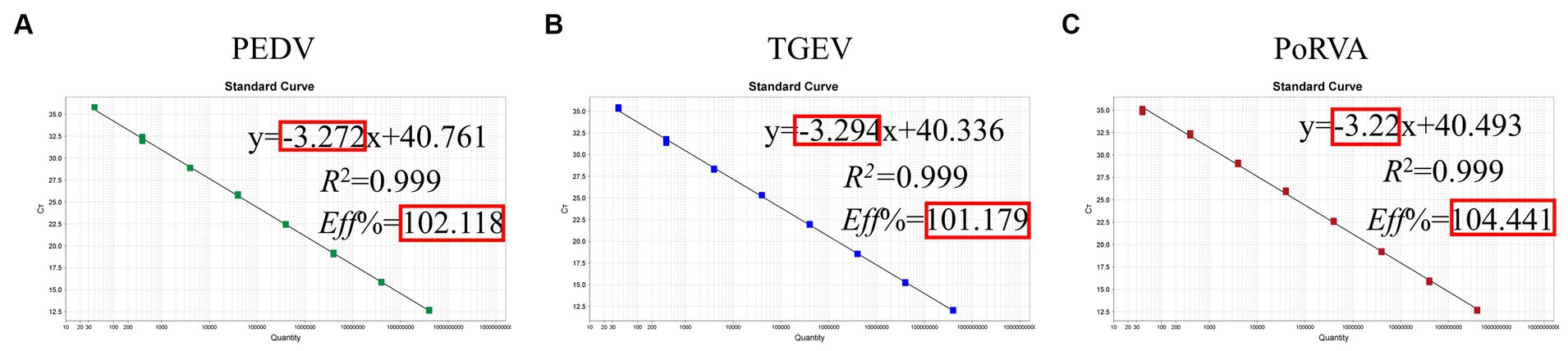
Figure 5. Standard curve for the triplex qRT-PCR assay. (A) Standard curve for PEDV M gene. (B) Standard curve for TGEV N gene. (C) Standard curve for PoRVA NSP3 gene.
Genomic DNA or RNA of 16 porcine pathogens (PEDV, TGEV, PoRVA, PRV, PCV2, PPV, PDCoV, SVA, T. gondii, L. interrogans, M. hyopneumoniae, M. hyorhinis, H. parasuis, S. suis, P. multocida and A. pleuropneumoniae) was used as a template for the triplex qRT-PCR. Amplification curves were obtained for PEDV, TGEV and PoRVA but not for the other porcine pathogens (Figure 6). Therefore, the triplex qRT-PCR assay was specific for detection of PEDV, TGEV and PoRVA, and had no cross-reaction with other porcine pathogens.
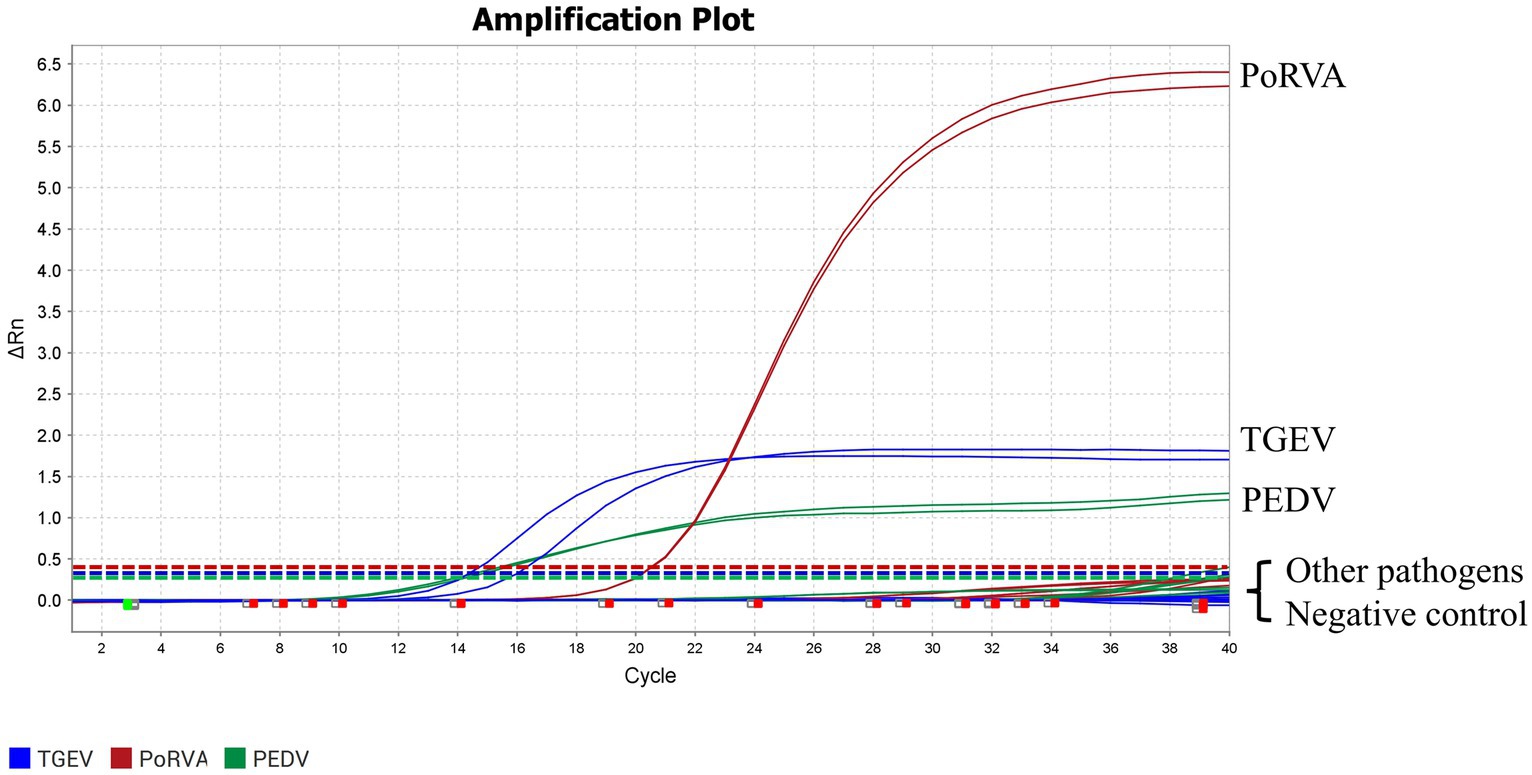
Figure 6. Specific amplification curves for the triplex qRT-PCR assay. Three fluorescent signals were monitored by triplex qRT-PCR. RNA of PEDV, TGEV and PoRVA was used as a positive control. No fluorescent signal was observed when genomes of other porcine pathogens were used as templates. The graph type was set in linear phase to simultaneously display the three different fluorescent signals (VIC, Cy5 and FAM) with distinct signal intensities. Other pathogens included PRV, PCV2, PPV, PDCoV, SVA, T. gondii, L. interrogans, M. hyopneumoniae, M. hyorhinis, H. parasuis, S. suis, P. multocida and A. pleuropneumoniae.
Different concentrations of standard plasmids were used as templates for the triplex qRT-PCR. Table 3 shows that a plasmid concentration of 100 copies/μL resulted in 100% positive detection rates for PEDV, TGEV and PoRVA. At 10 copies/μL, the positive detection rates were 100, 90 and 95% for PEDV, TGEV and PoRVA, respectively. At 1 copy/μL, the positive detection rates were 75, 5% and 0 for PEDV, TGEV and PoRVA, respectively. The LoD was defined as the lowest standard plasmid concentration with positive results in 85% of 20 replicates. Therefore, the triplex qRT-PCR showed high sensitivity, with a LoD of 10 copies/μL for PEDV, TGEV and PoRVA (Figure 7).
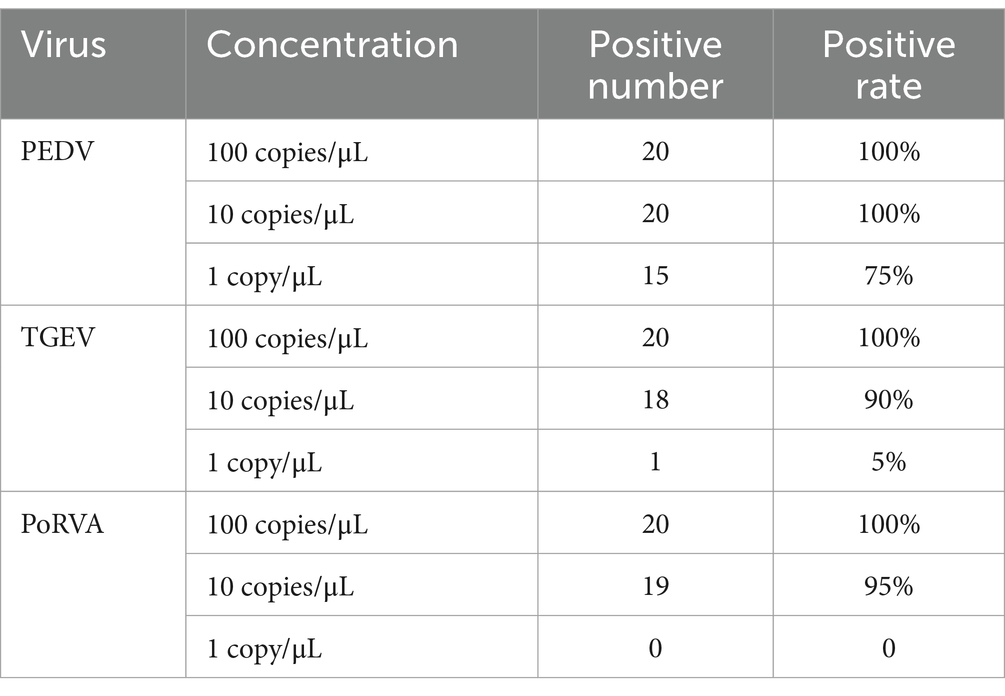
Table 3. Positive detection rate of 100 copies, 10 copies and 1 copy standard plasmids for 20 times.
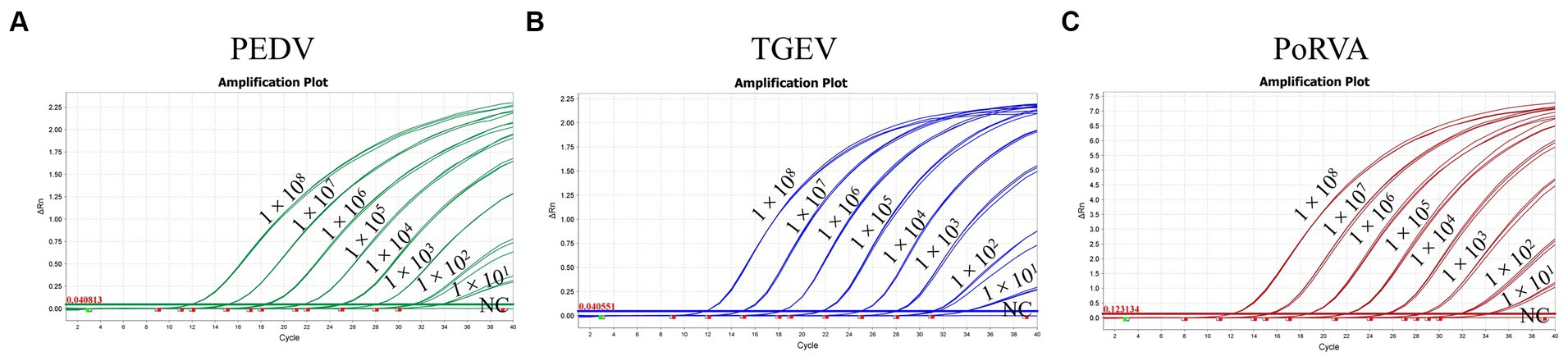
Figure 7. The sensitivity of the triplex qRT-PCR assay. (A) Sensitivity for PEDV M gene. (B) Sensitivity for TGEV N gene. (C) Sensitivity for PoRVA NSP3 gene.
Three concentrations of standard plasmids, 106, 104 and 100 copies/μL, were used to assess the repeatability and reproducibility of the triplex qRT-PCR. The intra-and inter-assay CVs were 0.08–0.79% and 0.37–0.83%, respectively (Table 4), which indicated good repeatability and reproducibility.
A total of 256 porcine anal swab samples were tested using the triplex qRT-PCR. The positive rates for PEDV, TGEV and PoRVA were 2.73% (7/256), 3.91% (10/256) and 19.14% (49/256), respectively. To verify the accuracy of the method, the clinical samples were also tested by the triplex RT-PCR recommended by the Chinese national standard (GB/T 36871-2018, 2018). The triplex RT-PCR results showed that the positive rates for PEDV, TGEV and PoRVA were 2.73% (7/256), 3.91% (10/256) and 14.84% (38/256), respectively. Both methods detected seven samples co-infected with PEDV and PoRVA. The new triplex qRT-PCR had 100% (PEDV), 100% (TGEV) and 95.70% (PoRVA) agreement with the triplex RT-PCR, indicating that the new approach was accurate, reliable and more sensitive (Table 5).
PEDV, TGEV and PoRVA are porcine enteric RNA viruses that cause porcine viral diarrhea (Chen et al., 2010; Zhao et al., 2016; Zhang et al., 2017). Co-infections with various combinations and all three viruses are common in swine herds worldwide (Song et al., 2006; Liu et al., 2019; El-Tholoth et al., 2021). These co-infections severely compromise the herd immunity and result in an increased risk of secondary infections, higher piglet mortality and significant economic losses, and they are a major concern for the swine industry (Jung et al., 2008; Mesonero-Escuredo et al., 2018; Song et al., 2022).
Currently, the commonly used molecular detection techniques for PEDV, TGEV and PoRVA include RT-PCR, nested RT-PCR, qRT-PCR, reverse transcription loop-mediated isothermal amplification, reverse transcription recombinase-aided amplification, and CRISPR-Cas nucleic acid detection (Marthaler et al., 2014; Areekit et al., 2022; Wu et al., 2022; Lazov et al., 2023; Xia et al., 2024). RT-PCR and nested RT-PCR are not capable of quantitative analysis, and their operation is cumbersome, time-consuming and less sensitive. Isothermal amplification techniques, including reverse transcription loop-mediated isothermal amplification and reverse transcription recombinase-aided amplification, are prone to false-positive results. CRISPR-Cas nucleic acid detection is expensive and not suitable for large-scale detection, and its multiplex technology is not yet matured. qRT-PCR is a highly specific and sensitive method for quantifying trace amounts of RNA in samples and is the most practical technique for the detection of viral RNA. It displays the results as fluorescent signals, which are easy to interpret. In particular, the multiplex qRT-PCR technique can detect multiple target genes of various pathogens in a single-tube reaction, using specific primers and probes with different fluorescent labels. Researchers have established some multiplex qRT-PCR detection methods for pathogens related to porcine viral diarrhea (Han et al., 2019; Huang et al., 2019; Jia et al., 2019). The detection method using SYBR Green fluorescent dye requires validation of product specificity through melting curve analysis. However, in practical applications, issues such as false-positive signals, dye redistribution, and low sensitivity can affect the reliability of detection results. However, the one-step TaqMan probe-based multiplex qRT-PCR method allows simultaneous detection of various RNA viruses without prior reverse transcription. In practical applications, it is easier to operate and has strong practicality for daily monitoring of pig diseases.
In this study, we designed three pairs of virus-gene-specific primers and corresponding probes for one-step triplex qRT-PCR, which can simultaneously detect PEDV, TGEV and PoRVA in one tube. We inserted three gene fragments into the same vector to generate a standard plasmid containing three gene targets for the triplex qRT-PCR, rather than using a mixture of three standard plasmids. This approach reduced the preparation cost of standard plasmids and minimized the systematic errors from adding three different plasmids. The sensitivity test revealed a LoD of 10 copies/μL for each pathogen. A strong linear correlation between Ct values and standard copy numbers was demonstrated by the standard curve plots. The primer and probe sequences used in the detection method were highly specific, and the fluorescent dyes VIC, Cy5 and FAM did not interfere with each other. Thus, PEDV, TGEV and PoRVA were accurately detected without cross-reaction with other swine pathogens (PRV, PCV2, PPV, PDCoV, SVA, T. gondii, L. interrogans, M. hyopneumoniae, M. hyorhinis, H. parasuis, S. suis, P. multocida and A. pleuropneumoniae). Furthermore, we tested 256 porcine anal swab samples with the developed triplex qRT-PCR method to verify its practicality and usefulness in clinical samples. The results indicated that PEDV, TGEV and PoRVA were detected in 7 (2.73%), 10 (3.91%) and 49 (19.14%) samples, respectively. This suggested that PEDV, TGEV and PoRVA persisted in pig herds. Co-infection with two or more of PEDV, TGEV and PoRVA was also common, which can worsen immunosuppression and inflammation, increase the risk of secondary infection by other pathogens, and exacerbate these diseases. This was supported by the detection of seven samples that were co-infected with PEDV and PoRVA in 256 clinical samples. We also tested 256 samples with the triplex RT-PCR detection method recommended by the Chinese national standard (GB/T 36871-2018, 2018). The results showed that consistency rates of 100% (PEDV), 100% (TGEV) and 95.70% (PoRVA) between the two methods. The sensitivity of the triplex qRT-PCR was significantly higher than that of triplex RT-PCR.
We developed a triplex qRT-PCR for simultaneous and differential detection of PEDV, TGEV and PoRVA. This new method is cost-effective, efficient and user-friendly. It can obtain results within an hour regardless of the number of samples to be tested or diagnosed, whether for routine screening or temporary diagnosis of these three pathogens in pig herds. It provides a reliable detection technique for accurate diagnosis, epidemiological investigation and microbiological quality control of laboratory pigs.
The sequence presented in the study is showed in Supplementary Table S1, further inquiries can be directed to the corresponding authors.
The manuscript presents research on animals that do not require ethical approval for their study.
TL: Investigation, Methodology, Writing – original draft. KL: Methodology, Validation, Writing – original draft. CL: Investigation, Project administration, Writing – original draft. CX: Project administration, Supervision, Writing – original draft. CG: Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program (2021YFF0703000), Pilot Technology Project of National Pig Technology Innovation Center (NCTIP-XD1C09), Special Funds for Basic Scientific Research Operations of Central Public Welfare Scientific Research Institutions (1610302022018).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1390328/full#supplementary-material
Anhui provincial local standard DB34/T 2795-2016 . (2016). Duplex RT-PCR assay for detection of porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus. Available at:. (in Chinese)https://std.samr.gov.cn/search/std?q=DB34%2FT%202795-2016
Areekit, S., Tangjitrungrot, P., Khuchareontaworn, S., Rattanathanawan, K., Jaratsing, P., Yasawong, M., et al. (2022). Development of duplex LAMP technique for detection of porcine epidemic diarrhea virus (PEDV) and porcine circovirus type 2 (PCV 2). Curr. Issues Mol. Biol. 44, 5427–5439. doi: 10.3390/cimb44110368
Chen, Q., Li, J., Fang, X., and Xiong, W. (2010). Detection of swine transmissible gastroenteritis coronavirus using loop-mediated isothermal amplification. Virol. J. 7:206. doi: 10.1186/1743-422X-7-206
Chen, J., Liu, R., Liu, H., Chen, J., Li, X., Zhang, J., et al. (2023). Development of a multiplex quantitative PCR for detecting porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine deltacoronavirus simultaneously in China. Vet. Sci. 10:402. doi: 10.3390/vetsci10060402
Chinese national standard GB/T 36871-2018 . (2018). Multiplex RT-PCR to detect porcine transmissible gastroenteritis virus, porcine epidemic diarrhea virus and porcine rotavirus. Available at:. (in Chinese)https://std.samr.gov.cn/search/std?q=GB%2FT%2036871-2018
Eleouet, J. F., Rasschaert, D., Lambert, P., Levy, L., Vende, P., and Laude, H. (1995). Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology 206, 817–822. doi: 10.1006/viro.1995.1004
El-Tholoth, M., Bai, H., Mauk, M. G., Saif, L., and Bau, H. H. (2021). A portable, 3D printed, microfluidic device for multiplexed, real time, molecular detection of the porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine deltacoronavirus at the point of need. Lab Chip 21, 1118–1130. doi: 10.1039/d0lc01229g
Ghosh, S., and Kobayashi, N. (2011). Whole-genomic analysis of rotavirus strains: current status and future prospects. Future Microbiol. 6, 1049–1065. doi: 10.2217/fmb.11.90
Han, H., Zheng, H., Zhao, Y., Tian, R., Xu, P., Hou, H., et al. (2019). Development of a SYBR green I-based duplex real-time fluorescence quantitative PCR assay for the simultaneous detection of porcine epidemic diarrhea virus and porcine circovirus 3. Mol. Cell. Probes 44, 44–50. doi: 10.1016/j.mcp.2019.02.002
Huang, X., Chen, J., Yao, G., Guo, Q., Wang, J., and Liu, G. (2019). A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl. Microbiol. Biotechnol. 103, 4943–4952. doi: 10.1007/s00253-019-09835-7
Jia, S., Feng, B., Wang, Z., Ma, Y., Gao, X., Jiang, Y., et al. (2019). Dual priming oligonucleotide (DPO)-based real-time RT-PCR assay for accurate differentiation of four major viruses causing porcine viral diarrhea. Mol. Cell. Probes 47:101435. doi: 10.1016/j.mcp.2019.101435
Jung, K., Kang, B. K., Lee, C. S., and Song, D. S. (2008). Impact of porcine group A rotavirus co-infection on porcine epidemic diarrhea virus pathogenicity in piglets. Res. Vet. Sci. 84, 502–506. doi: 10.1016/j.rvsc.2007.07.004
Kocherhans, R., Bridgen, A., Ackermann, M., and Tobler, K. (2001). Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes 23, 137–144. doi: 10.1023/A:1011831902219
Lazov, C. M., Papetti, A., Belsham, G. J., Bøtner, A., Rasmussen, T. B., and Boniotti, M. B. (2023). Multiplex real-time RT-PCR assays for detection and differentiation of porcine enteric coronaviruses. Pathogens 12:1040. doi: 10.3390/pathogens12081040
Liu, G., Jiang, Y., Opriessnig, T., Gu, K., Zhang, H., and Yang, Z. (2019). Detection and differentiation of five diarrhea related pig viruses utilizing a multiplex PCR assay. J. Virol. Methods 263, 32–37. doi: 10.1016/j.jviromet.2018.10.009
Luo, S., Chen, X., Yan, G., Chen, S., Pan, J., Zeng, M., et al. (2023). Emergence of human-porcine reassortment G9P [19] porcine rotavirus A strain in Guangdong province, China. Front. Vet. Sci. 9:1111919. doi: 10.3389/fvets.2022.1111919
Mackay, I. M. (2004). Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 10, 190–212. doi: 10.1111/j.1198-743x.2004.00722.x
Marthaler, D., Homwong, N., Rossow, K., Culhane, M., Goyal, S., Collins, J., et al. (2014). Rapid detection and high occurrence of porcine rotavirus A, B, and C by RT-qPCR in diagnostic samples. J. Virol. Methods 209, 30–34. doi: 10.1016/j.jviromet.2014.08.018
Mesonero-Escuredo, S., Strutzberg-Minder, K., Casanovas, C., and Segalés, J. (2018). Viral and bacterial investigations on the aetiology of recurrent pig neonatal diarrhoea cases in Spain. Porcine Health Manag. 4:5. doi: 10.1186/s40813-018-0083-8
Monteagudo, L. V., Benito, A. A., Lázaro-Gaspar, S., Arnal, J. L., Martin-Jurado, D., Menjon, R., et al. (2022). Occurrence of rotavirus A genotypes and other enteric pathogens in diarrheic suckling piglets from Spanish swine farms. Animals 12:251. doi: 10.3390/ani12030251
Pensaert, M. B., and de Bouck, P. (1978). A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 58, 243–247. doi: 10.1007/BF01317606
Rasmussen, T. B., Boniotti, M. B., Papetti, A., Grasland, B., Frossard, J., Dastjerdi, A., et al. (2018). Full-length genome sequences of porcine epidemic diarrhoea virus strain CV777; use of NGS to analyse genomic and sub-genomic RNAs. PLoS One 13:e0193682. doi: 10.1371/journal.pone.0193682
Shi, Y., Li, B., Tao, J., Cheng, J., and Liu, H. (2021). The complex co-infections of multiple porcine diarrhea viruses in local area based on the luminex xTAG multiplex detection method. Front. Vet. Sci. 8:602866. doi: 10.3389/fvets.2021.602866
Song, L., Chen, J., Hao, P., Jiang, Y., Xu, W., Li, L., et al. (2022). Differential transcriptomics analysis of IPEC-J2 cells single or coinfected with porcine epidemic diarrhea virus and transmissible gastroenteritis virus. Front. Immunol. 13:844657. doi: 10.3389/fimmu.2022.844657
Song, D. S., Kang, B. K., Oh, J. S., Ha, G. W., Yang, J. S., Moon, H. J., et al. (2006). Multiplex reverse transcription-PCR for rapid differential detection of porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine group A rotavirus. J. Vet. Diagn. Invest. 18, 278–281. doi: 10.1177/104063870601800309
Vlasova, A. N., Amimo, J. O., and Saif, L. J. (2017). Porcine rotaviruses: epidemiology, immune responses and control strategies. Viruses 9:48. doi: 10.3390/v9030048
Wang, Y., Das, A., Zheng, W., Porter, E., Xu, L., Noll, L., et al. (2020). Development and evaluation of multiplex real-time RT-PCR assays for the detection and differentiation of foot-and-mouth disease virus and Seneca Valley virus 1. Transbound. Emerg. Dis. 67, 604–616. doi: 10.1111/tbed.13373
Wang, D., Fang, L., and Xiao, S. (2016). Porcine epidemic diarrhea in China. Virus Res. 226, 7–13. doi: 10.1016/j.virusres.2016.05.026
Wu, X., Liu, Y., Gao, L., Yan, Z., Zhao, Q., Chen, F., et al. (2022). Development and application of a reverse-transcription recombinase-aided amplification assay for porcine epidemic diarrhea virus. Viruses 14:591. doi: 10.3390/v14030591
Xia, Y., Li, Y., He, Y., Wang, X., Qiu, W., Diao, X., et al. (2024). Development of a CRISPR-Cas12a based assay for the detection of swine enteric coronaviruses in pig herds in China. Adv. Biotechnol. 2:7. doi: 10.1007/s44307-024-00015-x
Yang, D., Ge, F., Ju, H., Wang, J., Liu, J., Ning, K., et al. (2014). Whole-genome analysis of porcine epidemic diarrhea virus (PEDV) from eastern China. Arch. Virol. 159, 2777–2785. doi: 10.1007/s00705-014-2102-7
Yang, J., Li, D., Wang, J., Zhang, R., and Li, J. (2022). Design, optimization, and application of multiplex rRT-PCR in the detection of respiratory viruses. Crit. Rev. Clin. Lab. Sci. 59, 555–572. doi: 10.1080/10408363.2022.2072467
Yount, B., Curtis, K. M., and Baric, R. S. (2000). Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74, 10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000
Zhang, Q., Hu, R., Tang, X., Wu, C., He, Q., Zhao, Z., et al. (2013). Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch. Virol. 158, 1631–1636. doi: 10.1007/s00705-013-1659-x
Zhang, Q., Liu, X., Fang, Y., Zhou, P., Wang, Y., and Zhang, Y. (2017). Detection and phylogenetic analyses of spike genes in porcine epidemic diarrhea virus strains circulating in China in 2016–2017. Virol. J. 14:194. doi: 10.1186/s12985-017-0860-z
Zhang, F., Luo, S., Gu, J., Li, Z., Li, K., Yuan, W., et al. (2019). Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet. Res. 15:470. doi: 10.1186/s12917-019-2212-2
Zhao, Z., Yang, Z., Lin, W., Wang, W., Yang, J., Jin, W., et al. (2016). The rate of co-infection for piglet diarrhea viruses in China and the genetic characterization of porcine epidemic diarrhea virus and porcine kobuvirus. Acta Virol. 60, 55–61. doi: 10.4149/av_2016_01_55
Zhejiang provincial local standard DB33/T 2254-2020 . (2020). Method of duplex fluorescence quantitative RT-PCR for the detection of porcine epidemic diarrhea virus and transmissible gastroenteritis virus. Available at:. (in Chinese)https://std.samr.gov.cn/search/std?q=DB33%2FT%202254-2020
Keywords: porcine epidemic diarrhea virus, porcine transmissible gastroenteritis virus, porcine rotavirus A, porcine enteric viruses, triplex real-time qRT-PCR
Citation: Luo T, Li K, Li C, Xia C and Gao C (2024) Development of a triplex quantitative reverse transcription-polymerase chain reaction for the detection of porcine epidemic diarrhea virus, porcine transmissible gastroenteritis virus, and porcine rotavirus A. Front. Microbiol. 15:1390328. doi: 10.3389/fmicb.2024.1390328
Received: 23 February 2024; Accepted: 17 April 2024;
Published: 10 May 2024.
Edited by:
Peirong Jiao, South China Agricultural University, ChinaReviewed by:
Mengmeng Zhao, Foshan University, ChinaCopyright © 2024 Luo, Li, Li, Xia and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changyou Xia, eGlhY2hhbmd5b3VAY2Fhcy5jbg==; Caixia Gao, Z2FvY2FpeGlhQGNhYXMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.