
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 03 June 2024
Sec. Food Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1390030
Introduction: Aspergillus cristatus is a homothallic fungus that is used in the natural fermentation process of Chinese Fuzhuan tea and has been linked to the production of bioactive components. However, not much is known about the variations present in the fungus. To understand the variation of the dominant microorganism, A. cristatus, within dark tea, the present study investigated the genetic and morphological diversity of 70 A. cristatus collected across six provinces of China.
Methods: Expressed sequence tags-simple sequence repeats (EST-SSR) loci for A. cristatus were identified and corresponding primers were developed. Subsequently, 15 specimens were selected for PCR amplification.
Results: The phylogenetic tree obtained revealed four distinct clusters with a genetic similarity coefficient of 0.983, corresponding to previously identified morphological groups. Five strains (A1, A11, B1, D1, and JH1805) with considerable differences in EST-SSR results were selected for further physiological variation investigation. Microstructural examinations revealed no apparent differentiation among the representative strains. However, colony morphology under a range of culture media varied substantially between strains, as did the extracellular enzymatic activity (cellulase, pectinase, protease, and polyphenol oxidase); the data indicate that there are differences in physiological metabolic capacity among A. cristatus strains.
Discussion: Notably, JH1805, B1, and A11 exhibited higher enzymatic activity, indicating their potential application in the production of genetically improved strains. The findings provide valuable insights into species identification, genetic diversity determination, and marker-assisted breeding strategies for A. cristatus.
Aspergillus cristatus, known as the “Golden flower fungus,” is the sexual type of Eurotium cristatum (Xiao et al., 2020; Efimenko et al., 2021), and it is the predominant microorganism in Fuzhuan tea, which is a notable dark tea variant produced through the fermentation of Camellia sinensis (An et al., 2021; Zhang et al., 2022a). The fungus ferments tea leaves and secretes enzymes that catalyze the oxidation, polymerization, transformation, and degradation of cellulose, pectin, fat, and polyphenols. The enzymatic activity reduces undesirable bitterness, while contributing to the distinctive color, aroma, and taste of dark tea (Xiao et al., 2021; Du et al., 2022; Xiao et al., 2022). Traditionally, A. cristatus was used for the processing of Fuzhuan tea. However, advances in production technology in recent years have enabled the development of different types of dark tea products, such as “loose dark” (Yao et al., 2017; Chen et al., 2023), “instant dark” (An et al., 2021; Chen et al., 2021), and “Pu′er” teas (Jiang et al., 2018) using A. cristatus, which effectively improved the quality of the tea. Additionally, A. cristatus fermentation yields a range of bioactive components, including monacolin K, which regulates blood lipids (Lu et al., 2022a), antibacterial benzaldehyde derivatives (Shi et al., 2019), amide derivatives (Wen et al., 2022), flavonoids with hypoglycemic effects (Zhao et al., 2023a), and fungal polysaccharides that modulate intestinal flora composition (Lu et al., 2022b). The health benefits of dark tea have made it highly desirable among consumers. In 2022, China’s dark tea output reached 426,300 tons, an increase of approximately 148% compared with 172,000 tons in 2012. Recently, researchers have identified the potential use of A. cristatus to improving the quality of other plant materials such as mulberry leaves (Yang et al., 2023), Ginkgo biloba seeds (Zou et al., 2022), turmeric (Xiang et al., 2020), and Siraitia grosvenorii (Yin et al., 2023a). Beyond catering to the health needs of contemporary consumers, A. cristatus-fermented foods also offer opportunities for deep processing and diversified utilization of agricultural and forestry products.
As a crucial microbial resource, A. cristatus has been extensively investigated with regard to fermentation technology, metabolites, and healthcare effects; however, its genetic diversity remains poorly understood. Because dark tea production is widely distributed in China, variations in geography and anthropogenic factors likely contribute to A. cristatus evolution, as some researchers have discovered that A. cristatus isolated from different dark teas does not form complete homogenous populations regarding their physical characteristics. Furthermore, previous studies have provided supporting evidence that confirms the presence of diversity within A. cristatus. Li et al. (2023a) conducted a study on 20 wild strains of A. cristatus, which exhibited diverse morphological traits, and they determined that only six were capable of lovastatin (a lipid-lowering drug) biosynthesis. Moreover, the observed yields metabolized varied substantially. Wang et al. (2019) compared the internal transcribed spacer (ITS) sequence differences of 33 A. cristatus and found that 24 were different from that of the standard strain to varying degrees (>99%); in addition, different strains had certain variations in their morphology and growth rate. Shi et al. (2019) analyzed benzaldehyde derivatives from eight A. cristatus using the high-performance liquid chromatography, revealing substantial variations in both the composition and yield of metabolic benzaldehyde derivatives among the different strains. Furthermore, Wang et al. (2022a) determined that the ITS sequences of 18 A. cristatus strains isolated from Fuzhuan tea demonstrated high similarity (with one exception showing 98.46% similarity to corresponding reference strains, and the rest being ≥99%), with notable differences among the strains in terms of tolerance to acidic environments, concentrations of bile salts, capacity to inhibit intestinal pathogenic bacteria, as well as hydrophobicity. Because the differences influence dark tea quality, the fermentation processes must be controlled to improve the product. Such a level of control requires the elucidation of A. cristatus genetic diversity and strain variations. However, existing studies have some limitations. Firstly, the physiological characteristics of fungi are influenced by cultural factors, leading to inconsistent gene expression and unstable morphological identification results. Secondly, the molecular marker sequences used (such as ITS, 18S rDNA, etc.) are relatively conservative and provide less phylogenetic information, making it difficult to distinguish intraspecific units. Therefore, there is still a need for more accurate and high-resolution molecular markers to establish a basis for identifying the genetic diversity of A. cristatus. Expressed sequence tags-simple sequence repeats (EST-SSR) are localized within the transcriptional regions of the genome and potentially play roles similar to those of functional genes, making such markers ideal for evaluating intraspecific genetic diversity (Lu et al., 2021). Moreover, EST-SSR technology has been employed extensively in microbial population genetics, germplasm resource evaluation, and taxonomic studies (Zhang et al., 2022c; Darshan et al., 2023; Wang et al., 2023a).
In the present study, A. cristatus strains from different regions of China were collected to analyze their morphology, enzyme activity, and microsatellite-based genetic diversity. The aim of the study was to clarify the A. cristatus strains and identify candidates suitable for improving dark-tea production.
Sixty-nine A. cristatus strains from dark tea samples were collected across various regions of China (Hunan, Hubei, Zhejiang, Shaanxi, Guangxi, and Guizhou Province) and isolated using a gradient dilution (Figures 1A,B). Three standard strains provided by Hunan City University (A. cristatus JH1805, A. pseudoglaucus HL1801, and A. chevalieri XW1803) were also included as controls for the isolated strains to effectively analyze the variation of wild-type strains. The sources of the experimental strains are listed in Supplementary Table S1.
Potato dextrose agar (PDA), modified PDA, 20% Czapek-Dox medium (20% CDA), 60% CDA, czapek yeast extract agar (CYA), potato glucose liquid medium (PDL), and dark tea liquid medium (DTL) were used as culture media. Among them, PDA, modified PDA, 20% CDA, and 60% CDA were used for morphological observations. Whereas PDL and DTL were used for the cultivation of mycelium for DNA extraction and detection of extracellular enzyme activity of the strains, respectively. The formulas for the different culture media are shown in Supplementary Table S2.
Strains were inoculated onto PDA plates and cultured at 28°C for 6 d (Figure 1C). Their characteristics were observed and recorded (for methodology, see Supplementary Figure S1). The assessment and clustering of the morphological characteristics of the colony were performed according to the method described by Yang and Song (2014). The average colony size was determined based on the crossover method. Edge shape was assessed based on the orderliness of colony edges, pigment secretion was evaluated based on the sizes and depths of melanin-containing areas in the colony center; surface characteristics were evaluated to determine colony smoothness; cluster analysis was determined using SPSS AU (Xu et al., 2023a) based on four physiological traits of the tested strains.
The RNA was extracted from sexual and asexual mycelia of A. cristatus JH1805 cultured on PDA medium for 6 d (Hu et al., 2023). The extracted RNA was then used for library construction and sequencing using the methods described by Hu et al. (2023).
MISA software (Liu et al., 2023a; https://webblast.ipk-gatersleben.de/misa/) was used to identify SSR loci within the A. cristatus transcriptome (Figure 1D). The criteria were as follows: single nucleotide repeats >10; dinucleotide repeats >5; trinucleotide repeats >4; and tetranucleotide, pentanucleotide, and hexanucleotide repeats >3.
Primers for SSR-containing sequences ≥12 base pairs (bp) were designed in Primer Premier 5.0. The design criteria were as follows: (1) flanking sequences ≥50 bp; (2) annealing temperature (Tm) of 58–65°C, and a ≤2°C difference between the Tm of positive and negative primers; (3) expected amplicon size, 80–250 bp; (4) primer length, 18–28 bp; and (5) GC content, 40–60%. Hairpins, dimers, false priming, and cross-dimer formation were avoided as much as possible during the primer design. Primer verification was done using BLAST on the unigene library, which resulted in 30 pairs selected for subsequent primer synthesis (Supplementary Table S3).
Representative strains, such as JH1805, A1, A11, A13, A12, A27, A30, B1, B8, C5, C6, D1, E1, E13, and F3 were selected from different provinces as test samples for DNA extraction (Alcaide et al., 2019; Xu et al., 2023b), and A. chevalieri XW1803 was used as the control. The 16 strains were cultured at 28°C in PDL liquid medium for 6 d, then centrifuged at 2550 × g for 5 min. Subsequently, mycelia were collected for DNA extraction.
Extracted DNA was amplified with the 30 EST-SSR primer pairs. The reaction mix comprised 34 μL T3 Super PCR Mix, 2 μL each of upstream and downstream primer, and 2 μL of the DNA template. The parameter settings for PCR amplification of EST-SSR sequences are shown in Supplementary Table S4. Subsequently, PCR products were electrophoresed for 120 min on a 10% polyacrylamide gel at 100 V. Bands were visualized using an EB-stained gel imaging system (Vidya et al., 2021).
To assess the suitability (presence) of SSR markers, their degree of polymorphism (based on length) was used (Zhao et al., 2023b). High, intermediate, and low polymorphism is typically defined as SSR length ≥ 20 bp, 12–19 bp, and <12 bp, respectively (Zhang et al., 2022b).
The analysis of EST-SSR genetic diversity was conducted as follows: the images captured by the gel imaging system in step 2.2.3 were analyzed, and each pair of EST-SSR primer corresponds to a locus. If a polymorphic band is observed, it is considered an allelic variation. The electrophoretic bands were analyzed statistically using the “0,1” system, where no bands were recorded as “0” and bands were recorded as “1.” The statistical results were summarized in an MS Excel table to establish the original database (Li et al., 2023b), which was used for the comparison and clustering analysis of EST-SSR amplified bands. The genetic similarity coefficients among 16 strains were analyzed using NTSYS-pc 2.2, and unweighted pair group method with arithmetic mean (UPGMA) hierarchical clustering analysis was performed based on the similarity coefficient to construct a system tree. The positions of different strains on the system tree can be used to evaluate their EST-SSR genetic diversity (Figure 1E) (Chen et al., 2022). In addition, the conjugate values and matrix correlation coefficients of the UPGMA clustering results were calculated using NTSYS-pc 2.2 to evaluate the clustering results (Yildiz et al., 2021).
The 15 A. cristatus phenotypic traits obtained from the PDA culture were standardized in NTSYS-pc 2.2 to obtain Euclidean distance and genetic distance matrices of EST-SSR markers. The two matrices were subjected to a Mantel correlation analysis (Yang et al., 2022).
According to the results of EST-SSR genetic diversity analysis, A. cristatus A1, A11, B1, and D1 were selected as representative strains from different branches of the clustering tree (Figure 1F), which was then compared with the standard strain A. cristatus JH1805, A. pseudoglaucus HL1801, and A. chevalieri XW1803.
Using the three-point method, strains were inoculated onto plates containing PDA, modified PDA, 20% CDA, 60% CDA, and CYA (Figure 1G). Since culture temperature and osmotic pressure affect the reproductive pattern of A. cristatus (Tan et al., 2018; Shao et al., 2022), to observe the different reproductive structures of A. cristatus, all plates were incubated at 28°C except for 60% CDA, which was incubated at 32°C. After culturing for 6 d, colony morphology was recorded by photographing.
Sexual and asexual mycelia cultured in 60% CDA medium for 6 d were selected and prepared as fixed specimens (Figure 1H). Mycelium structure was observed under a scanning electron microscope (SEM). Different regions from the electron microscope images were selected for length and diameter measurements in ImageJ Software (Wainaina and Taherzadeh, 2023). Twenty samples per structure were measured randomly.
Spore suspensions (1.0 mL) for each strain were pipetted into a dark tea liquid medium and then cultivated at 28°C (Figure 1I). After fermentation for 3–8 d, suspensions were centrifuged at 8263 × g for 10 min to obtain the supernatant crude enzyme solution. The enzyme activity per unit volume was calculated by adding the volume of the sample (U/mL).
Cellulase activity was measured using the dinitrosalicylic acid (DNS) method (Sudeep et al., 2020): 0.5 mL of crude enzyme solution was added to 2.0 mL of 1.0% CMC-Na solution, and the solution was placed in a water bath at 40°C for 30 min. Subsequently, 2.0 mL of DNS solution was added to terminate the reaction immediately. The solution was then heated in boiling water for 5 min, after which it was removed and cooled. The volume was maintained at 20 mL. The optical density value was measured at 540 nm using an ultraviolet spectrophotometer, with glucose as the standard curve. One unit of cellulase activity was defined as the amount of enzyme required to produce glucose (1 μg/min) under the above reaction conditions.
During the determination of pectinase activity, 1% CMC-Na solution was replaced by 1% pectin solution, the other methods were similar to those used for cellulase determination. One unit of pectinase activity was defined as the amount of enzyme required to produce glucose (1 μg/min) under the above reaction conditions.
The protease activity was determined using the Folin–Ciocalteu method (Sharma et al., 2021): 1 mL of crude enzyme solution was added to 1 mL of casein. Both the crude enzyme solution and casein substrate were pre-incubated at 40°C in a water bath for 3 min before mixing. Once combined, the reaction mixture was incubated further for 10 min at 40°C in a water bath. Afterward, 2 mL of 0.4 mol/L trichloroacetic acid solution, which is used to inhibit the enzyme, was removed and allowed to sit for 10 min at 20°C and then the solution was centrifuged at 1632 × g for 5 min. After centrifugation, 1 mL of supernatant was added to 5 mL of 0.4 mol/L sodium carbonate solution and then 1 mL of Folin reagent was added to the solution. This was then shaken well and kept in a 40°C water bath for 20 min. An ultraviolet spectrophotometer was used to measure the optical density of the solution at 660 nm, with tyrosine used to plot the standard curve. One unit of protease activity was defined as the amount of enzyme required to hydrolyze casein to produce 1 μg/min tyrosine under the above reaction conditions.
Polyphenol oxidase activity was determined using the catechol method (González-Ceballos et al., 2023): 1.0 mL of crude enzyme solution was added to 3 mL of reaction mixture (citrate buffer: 0.1% proline: 1%, catechol = 10:2:3). The solution was then kept in a thermostatic water bath at 37°C for 10 min, and 3 mL of trichloroacetic acid (1 mL) was added immediately to terminate the reaction. The optical density was measured at 460 nm using an ultraviolet spectrophotometer, and the catechol in the blank control group was replaced by 0.05 mol/L PBS Buffer Solution. One unit of polyphenol oxidase activity was defined as the amount of enzyme required to increase the absorbance by 1 within 1 min under the above reaction conditions.
To ensure the reliability of the test results, all enzymatic activity test experiments were repeated three times. DPS 15.10 (Zheng et al., 2023) statistical software was used to analyze the variance in the data. p < 0.05 indicated a significant difference and the test results were expressed as mean ± standard error.
The 70 A. cristatus strains, including the standard JH1805, exhibited several common morphological features (Figure 2). First, the mycelial structure was relatively dense and firmly attached to the medium; colony edges were relatively shallow, mostly light yellow to yellow, while the melanin-dyed central area was orange yellow to brown. The overall colony structure was presented as concentric rings of 2–3 colors. The sporogenous structure was mainly closed ascocarp, and small amounts of gray-green conidia heads were present along the edges of several colonies. Finally, the medium around the colony formed a black-brown halo from pigment diffusion.
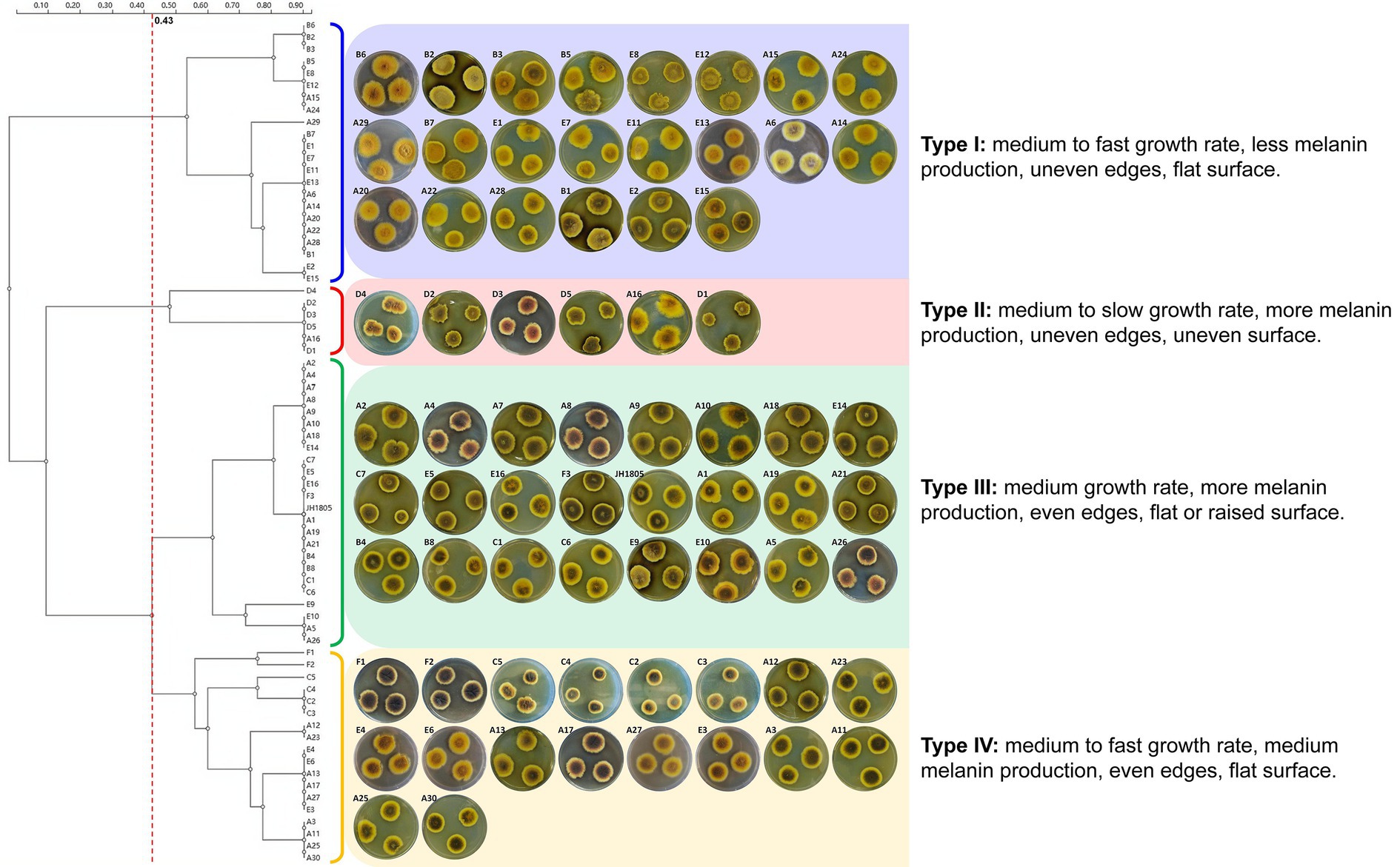
Figure 2. Colony characteristics and morphological clustering of 70 A. cristatus cultured in PDA medium (28°C, 6 d).
The 70 strains varied in growth rate, melanin secretion, colony edge shape, and colony surface characteristics. A significant disparity in growth rate was reflected on day 6, when A2, B1, and E2 colony diameters exceeded 30 mm, whereas A30, D1, and C4 were 18–26 mm in diameter. The A11, A12, and F1 colonies that had high pigment secretion were dyed black-brown to brown, and the dark area accounted for approximately 3/4 of the diameter of the mycelia. In contrast, the dyed area of A1 and C6 were less than half the colony diameter, and the A14, A22, and E7 strains only secreted a small amount of pigment. In terms of colony edge shape, JH1805, A11, E5, and E15 were all relatively uniform, whereas other strains were uneven, as they were either serrated (A9, A12), or petal-shaped (A2, A4). Surface characteristics also differed across strains. Most colonies were flat, with the center only slightly thicker than the edge, although a few strains exhibited notable uplift (A21, C2) or folds (A17, F1). Reverse-side colony color and conidia number also differed between strains.
After investigating growth rate, pigment secretion, colony edge shape, and surface characteristics of the 70 A. cristatus cultured on PDA plates (Supplementary Table S5), a clustering analysis was performed (Figure 2). The results indicated that, at a 0.430 threshold, the tested isolates can be divided into four groups.
To some extent, the tested A. cristatus strains were clustered based on geographical distribution (Figure 3). For example, the five A. cristatus strains from Zhejiang were clustered in type II, whereas Hubei strains were mainly type I. Type I strains were mainly distributed in the northern dark tea-producing regions, while types III and IV were mainly from the south. Thus, a north–south distribution was observed overall, but no consistent pattern between the distribution type and geographical region was observed.
The 10,542 transcriptome unigene sequences (totaling 14,972.05 kb in length) yielded 2,183 SSR loci (Table 1). On average, one SSR locus was present for every 6.86 kb sequence. Overall, the SSR frequency was 20.71%, with 358 unigenes containing two or more SSR loci.
Based on Figure 4, out of 2,183 A. cristatus SSR sequences, there are a total of 194 highly polymorphism SSR sequences (8.89%) with a length ≥ 20 bp, 1792 moderately polymorphism SSR sequences (82.09%) with a length between 12–19 bp, and 197 lowly polymorphism SSR sequences (9.02%) with a length < 12 bp. In total, 90.98% of the SSR sequences exhibit moderate to high levels of polymorphism.
Most of the 30 EST-SSR primers successfully amplified the templates from A. cristatus (JH1805, A1, A11, A12, A13, A27, A30, B8, C5, C6, D1, E1, E13, B1 and F3) and A. chevalieri XW1803 (Supplementary Figure S2).
Cluster analysis with UPGMA and phylogenetic analyses (Figure 5) revealed that A. chevalieri formed a distinct branch, as expected of the outgroup, and exhibited a genetic similarity of 0.775 with A. cristatus. The value was significantly lower than the genetic similarity between different A. cristatus strains (0.965–0.983). Therefore, the selected primers effectively discriminated between intra- and interspecies units. The 15 A. cristatus strains were divided into five branches with a genetic similarity coefficient of 0.983: the first branch included A1, C5, A13, A27, C6, B8, A12, JH1805, F3, and A30; the second branch comprised E1 and E13 from Shaanxi Province; finally, B1, D1 and A11 each formed a branch.

Figure 5. Genetic diversity clustering analysis of 16 fungi was conducted using 30 EST-SSR molecular markers.
A Mantel correlation analysis was conducted on the Euclidean distance matrix of phenotypic traits and the genetic distance matrix of EST-SSR markers for the 15 A. cristatus strains (Supplementary Figure S3). Strain morphology and EST-SSR genetic diversity were significantly correlated (r = 0.515, p = 0.9959).
All seven tested strains grew successfully on the five cultured media (PDA, modified PDA, 20% CDA, 60% CDA, and CYA), forming colonies with different morphologies (Figure 6A). In general, the growth rate of the seven fungi strains was higher on the modified PDA and 60% CDA media, and lower on the PDA and 20% CDA media, the growth rate of the colony was positively correlated with the concentrations of nutrients, such as carbon source. Moreover, the fungal colonies from the seven strains predominantly displayed ascocarps (structure related to sexual reproduction) grown on the PDA, modified PDA and CYA medium. Conversely, asexual reproduction (conidia head) structures were rarely observed, whereas asexual reproduction was predominantly observed in the colonies grown on the 60% CDA medium. Notably, although the growth rate and pigment secretion of the five strains of A. cristatus were affected greatly by the type of medium, the strains showed similar physiological characteristics on different media: for example, B1 grew faster and produced fewer pigments on all the media; A11 grew at a moderate rate and produced more pigments, exhibiting stability of certain physiological characteristics in different strains.
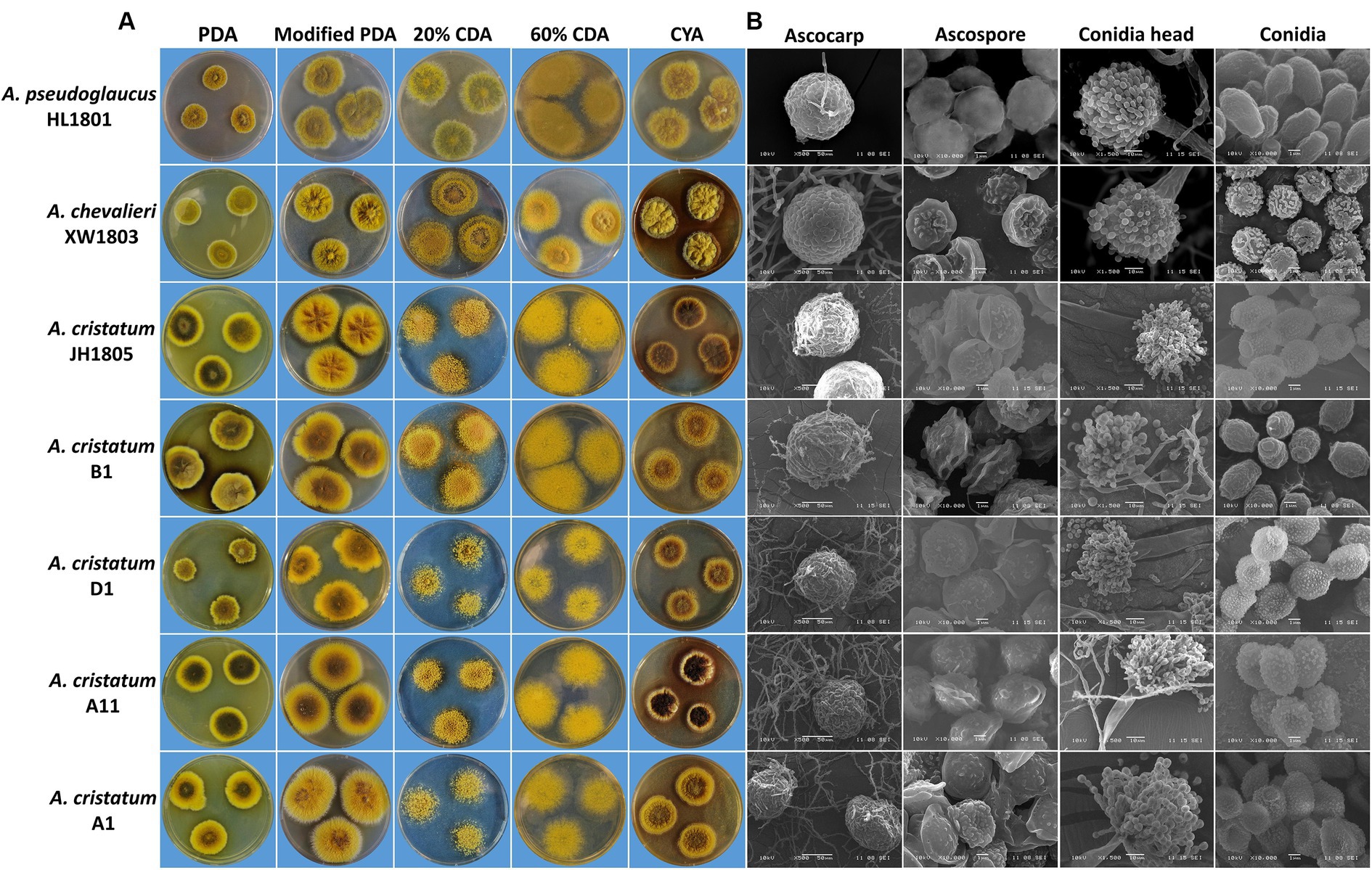
Figure 6. Colony morphology (A) and microstructural characteristics under scanning electron microscopy (B) of seven A. cristatus strains.
Strain structural SEM images of seven tested strains were quantified in ImageJ (Figure 6B) and analyzed (Table 2). Sexual A. cristatus has a spherical ascocarp (diameter: 50.0–180.0 μm) surrounded by numerous mycelia. As growth progressed, the ascocarp wall became transparent and eventually ruptured, releasing ascospores upon maturation. Ascospores were 3.4–4.7 μm × 4.4–6.4 μm in size and presented a double-convex shape. Their surfaces were rough, with two prominent corona protrusions at the equator and grooves between the protrusions.
Asexual A. cristatus possessed a conidial head of 35.0–80.0 μm in length. At the immature stage, the conidial head exhibited multiple conidial chains, each containing approximately three to seven conidia, which increased in a time-dependent manner. The mature conidia detached from conidial chains and were dispersed within the matrix. The most mature conidia were observed to be ellipsoidal, with small spiny protrusions on the outer wall and 3.2–3.8 μm × 4.2–5.2 μm in size.
The related A. pseudoglaucus is mainly distinguished from A. cristatus by a smooth ascospore surface, less obvious coronal protrusions at the equator, and shallower grooves. In A. chevalieri, ascospores were flat, with a depression in the middle of the bearing-like equator, while conidia were thicker and shorter.
In all seven strains, cellulase activity increased rapidly during fermentation, peaking on days 6 and 7 and then leveling off (Figure 7A). Cellulase activity was the highest among A. cristatus B1 and A11 (1.779 U/mL and 1.786 U/mL on day 7, respectively). Strong cellulase activity was also observed in A. chevalieri XW1803, with a value of 1.607 U/mL on day 6. At the peak of cellulase activity (7 d), the cellulase activity values of each strain were compared in pairs, and there were no significant differences among XW1803, JH1805, B1, A11, and A1 (p > 0.05); there were no significant differences among XW1803, JH1805, D1, and A1 (p > 0.05); and there were significant differences among the other strains (p < 0.05). Among the 5 A. cristatus, D1 had the lowest cellulase activity.
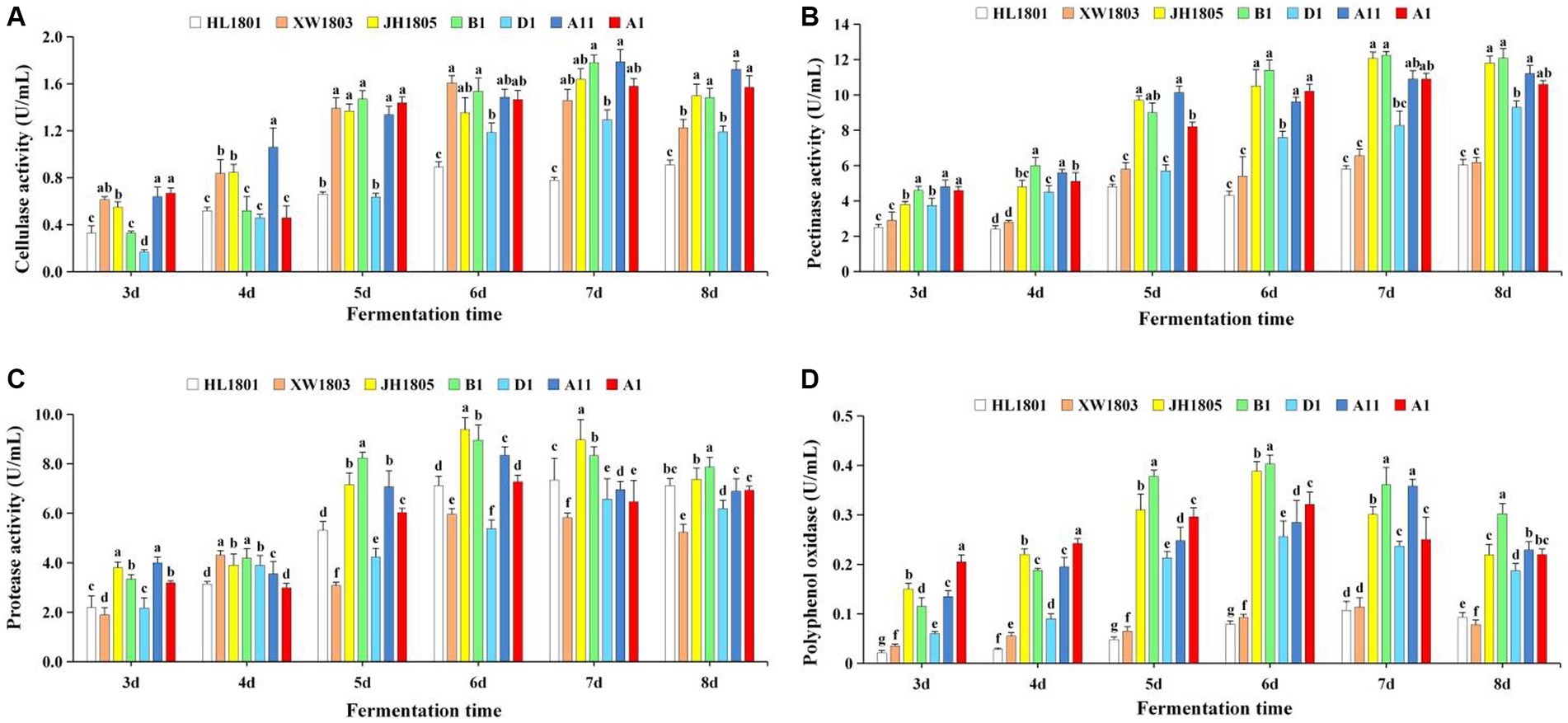
Figure 7. Extracellular enzyme activity in a fermentation broth of seven A. cristatus strains [(A) Cellulase activity, (B) Pectinase activity, (C) Protease activity, (D) Polyphenol oxidase]. Each column of data for the same fermentation period is marked with a different lowercase letter to indicate significant differences (p < 0.05).
The pectinase activity of the seven test strains is shown in Figure 7B. Activity was the highest in JH1805 and B1, peaking on day 7 at 12.071 U/mL and 12.242 U/mL, respectively. In contrast, pectinase activity was low for A. pseudoglaucus HL1801 and A. chevalieri XW1803, peaking on day 8 (6.029 U/mL) and 7 (6.547 U/mL), respectively. On the 8th day of fermentation, the pectinase activity among the strains was varied. Notably, there were no significant differences observed between strains JH1805, B1, and A11, and A1, as well as between A1 and D1, and between HL1801 and XW1803 (p > 0.05). However, the remaining strains exhibited statistically significant differences (p < 0.05), demonstrating their diverse methods of hydrolyzing pectin. Notably, most strains maintained high pectinase activity during late fermentation.
Protease activity in all seven strains increased initially before decreasing (Figure 7C). Notably, A. cristatus JH1805 and B1 demonstrated robust protease activity, reaching 9.382 U/mL and 8.951 U/mL on day 6, respectively. In contrast, A. chevalieri XW1803 and A. cristatus D1 had low protease activity, peaking at 5.960 U/mL and 6.568 U/mL on fermentation days 6 and 7, respectively. On the 6th day of the peak period of protease activity, each strain displayed a normal distribution, with significant differences between different strains (p < 0.05).
Polyphenol oxidase activity was robust on days 6 and 7 of fermentation, before decreasing (Figure 7D). Activity was particularly high on day 6 in A. cristatus B1 and JH1805, peaking at 0.403 U/mL and 0.388 U/mL, respectively. Activity was lower in A. pseudoglaucus HL1801 and A. chevalieri XW1803, with peak values being 0.107 and 0.114 U/mL, respectively, on day 7. On the 7th day of fermentation, there was no significant difference in polyphenol oxidase activity between B1 and A11, D1 and A1, and HL1801 and XW1803. However, there were significant differences between the other remaining strains (p < 0.05).
In the present study, the morphological variations of 70 A. cristatus strains (representing six provinces in China) were identified, and they were classified into four distinct groups. A distinct north–south pattern in strain type distribution was observed; however, a full alignment between the distribution and specific geographical regions was absent. The lack of a strong correlation may be attributed to a geographical separation between raw material production and processing for dark tea. For example, in the southern region of Shaanxi Province, the cultivation of A. cristatus is limited and the raw materials used for processing dark tea are mainly imported from other provinces (Xiao et al., 2018). Similarly, shortages in locally produced raw tea leaves necessitate the import of these raw leaves from other provinces to maintain the production of processed products. The process of using blended raw materials to produce dark tea is common in the industry (Fu et al., 2016), facilitating the dissemination of A. cristatus, and thereby promotes genetic exchange among microbial populations across varying provinces. Additionally, China’s current dark tea production approach is less dependent on artificial fermentation agents; instead, China relies on the natural inoculation of A. cristatus to drive the fermentation process (Wu et al., 2023). The use of such a natural method reportedly contributes to the genetic diversity present in the strains involved (Albertin et al., 2014; Steensels et al., 2014).
In the present study, 10,542 Unigenes of A. cristatus transcriptome sequences were retrieved, and a total of 2,183 EST-SSR loci were found, with an SSR frequency of 20.71%. Among the EST-SSR sequences in A. cristatus, 194 (8.89% of all SSRs) had high polymorphism, and 1,792 (82.09%) were moderately polymorphic. The phylogenetic tree constructed from PCR data showed that A. chevalieri clustered separately as an outgroup, with far lower genetic similarity (0.775) than coefficients within A. cristatus (0.965–0.983). Furthermore, clustering the strains based on genetic similarity resulted in a strong correlation with morphology-based groups. The results clearly demonstrate that the selected EST-SSR primers effectively identified intra- and inter-species units among the tested strains. Although the clustering tree topology of EST-SSR markers and morphological markers were not completely consistent, the traits of species are usually determined by multiple genes, and the detected EST-SSR loci are not necessarily associated with the traits. The integration of EST-SSR markers and morphological markers enhances the elucidation of the genetic diversity present within a species (Yin et al., 2023b).
Morphological differences between Aspergillus species (Diba et al., 2007) can be distinguished reliably under different culture media. The results of culturing experiments revealed substantial differences in growth rate, colony shape, and surface color across the five representative A. cristatus strains, reflecting physiological differentiation. When compared with the related A. pseudoglaucus and A. chevalieri, A. cristatus morphology was similar to the former on modified PDA and CYA, as well as the latter on modified PDA and 20% CDA. Such similarities often cause confusion of the three fungal species. Changes in media induce morphological differences in A. cristatus and related species due to variation in osmotic pressure (Huang et al., 2017; Tan et al., 2018; Hu et al., 2021). In particular, elevated osmotic pressure shifted reproductive mode from sexual to asexual (Ge et al., 2016). The asexual colonies cultured on 60% CDA were different across the three species. Therefore, hypertonic media-induced variation in asexual colonies is an effective means of identifying A. cristatus and related species under conventional culture conditions.
Spore characteristics are typically the primary basis for morphological identification of fungi (Simões et al., 2013; van den Brule et al., 2020). Electron microscopy revealed that A. cristatus strains had substantially different spores from their relatives. However, ascospores and conidia were highly similar among the five strains, consistent with the findings of previous studies on A. cristatus (Ren et al., 2017; Liu et al., 2018; Rui et al., 2019). Slight variations in spore size across the five A. cristatus strains (differences in average ascospore and conidia lengths were ≤0.2 μm and ≤0.3 μm, respectively) were observed; however, the fluctuations could be attributed to inconsistencies in maturity. Overall, the microstructure of the five A. cristatus strains did not differ considerably.
The extracellular enzyme activity of dominant microorganisms is a key factor promoting fermentation, among which cellulase, pectinase, protease, and polyphenol oxidase are important to the formation of dark tea quality. In tea leaves, cellulase hydrolyzes cellulose to smaller sugar molecules (Bhati et al., 2021), softening leaf stalk tissue and facilitating cell-wall breakdown to enhance water content (Chandini et al., 2011; Xu et al., 2022; Baruah et al., 2023). Pectinase hydrolyzes pectin to generate small water-soluble molecules (de Souza and Kawaguti, 2021), promoting the release of soluble sugars, polysaccharides, and other substances from leaf stalk cells. Such compounds contribute to the flavor profile and clarify tea infusions (Liang et al., 2021; Wang et al., 2023b). Proteases hydrolyze peptide bonds in proteins to improve protein utilization in tea. The generated amino acids contribute components that improve tea aroma and flavor (Yu and Yang, 2020). During tea processing, polyphenol oxidase facilitates the production of pigments, such as theaflavins and thearubigins (Zheng et al., 2016; Wang et al., 2022b; Parveen et al., 2023). This reaction lowers the bitterness of the tea soup, while deepening its color intensity. Furthermore, polyphenol oxidase is critical to tea aroma through the formation of aromatic compounds (e.g., terpenes and vanillin) (Liu et al., 2023b). Notably, the seven strains exhibited distinct activity patterns for all four extracellular enzymes, reflecting variations in metabolic rate and fermentation performance. In addition, the key factor for the formation of dark tea quality is the transformation of its chemical components catalyzed by microbial enzymes. Highly active microbial extracellular enzymes can promote the transformation of its contents, thus shortening the fermentation time and improving the quality of products (Lin et al., 2021). In the present study, three A. cristatus strains JH1805, B1, and A11 had stronger extracellular enzyme activities, and they are excellent candidates for use in breeding programs aimed at enhancing dark tea fermentation.
Genetic diversity is the basis of functional diversity and a prerequisite for the development of functional microorganisms. A. cristatus is not only a key microorganism for dark tea fermentation, but is also used widely in the development of health products and fungal active substances. However, there is currently less information on the genetic diversity of this strain, which limits the development of its genetic resources and quality control of dark tea. In the present study, a set of EST-SSR markers was developed for the evaluation of genetic resources of A. cristatus using high-throughput sequencing technology. The genetic diversity of different A. cristatus genetic resources from different sources were analyzed based on EST-SSR markers and phenotypic characteristics, demonstrating the differences in genetic diversity between geographic and temporal locations and providing key molecular marker resources for identification, genetic variation determination, and other A. cristatus studies. At the same time, EST-SSR markers originate from transcriptome sequences, reflecting the coding part of genes, and may directly identify the alleles that determine important phenotypes or physiological traits (Yang et al., 2020), providing convenience for A. cristatus breeding. Although the application of EST-SSR markers in microorganisms is relatively late, it has the advantages of high polymorphism, good repeatability, codominant inheritance etc. (Zhou et al., 2023), and has great application prospects in the development of A. cristatus and other functional microorganisms. Notably, a key limitation of the present study is that only an initial evaluation of the genetic diversity of A. cristatus has been completed. Subsequent work can be based on the research results to select core germplasm and further study the physiological characteristics, metabolite activity, and their correlations with molecular markers of the strains, to improve the EST-SSR molecular marker database. In addition, additional A. cristatus resources should be collected to enlarge the source range of germplasm resources and facilitate scientific management and exploitation of A. cristatus resources.
This study investigated A. cristatus genetic diversity and phylogeny through morphology, microstructure, enzyme activity, and EST-SSR marker analysis. The authors confirmed the presence of distinct populations in A. cristatus, identifiable through physiological differentiation and genetic diversity. We further demonstrated the utility of EST-SSR markers as effective tools for strain discrimination. In conclusion, the results have provided a scientific foundation for species identification, genetic diversity analysis, and marker-assisted breeding programs targeting A. cristatus.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA827193.
ZYH: Writing – original draft, Investigation, Methodology, Writing – review & editing. SQL: Methodology, Writing – review & editing. XHZ: Data curation, Software, Writing – review & editing. ZJL: Data curation, Software, Writing – review & editing. TTL: Investigation, Resources, Writing – review & editing. SLY: Funding acquisition, Writing – review & editing. XYZ: Funding acquisition, Writing – review & editing. ZGX: Methodology, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Hunan Province (2023JJ50338, 2022JJ50285) and Research Foundation of Education Bureau of Hunan Province (22B0785, 23B0748).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1390030/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Methods for observation of A. cristatus colony morphology. (A) Colony size. (B) Ability to secrete pigment. (C) Colony edge characteristics, and (D) Colony surface characteristics.
SUPPLEMENTARY FIGURE S2 | Results were obtained from amplification of 30 EST-SSR primers using 16 fungi DNA samples (Lane 1–15: A. cristatus A1, B1, C5, D1, A13, A27, C6, E1, A30, F3, JH1805, A12, B8, E13, A11; lane 16: A. chevalieri XW1803).
SUPPLEMENTARY FIGURE S3 | Correlation of strain morphology with EST-SSR genetic diversity.
SUPPLEMENTARY TABLE S1 | Source information of experimental strains.
SUPPLEMENTARY TABLE S2 | Different culture medium formulations.
SUPPLEMENTARY TABLE S3 | 30 EST-SSR primers information of A. cristatus.
SUPPLEMENTARY TABLE S4 | Parameter setting for PCR amplification of EST-SSR sequences.
SUPPLEMENTARY TABLE S5 | Colony characteristics of 70 A. cristatus on PDA plate (28 °C, 6 d).
Albertin, W., Chasseriaud, L., Comte, G., Panfili, A., Delcamp, A., Salin, F., et al. (2014). Winemaking and bioprocesses strongly shaped the genetic diversity of the ubiquitous yeast Torulaspora delbrueckii. PLoS One 9:e94246. doi: 10.1371/journal.pone.0094246
Alcaide, F., Solla, A., Mattioni, C., Castellana, S., and Martín, M. Á. (2019). Adaptive diversity and drought tolerance in Castanea sativa assessed through EST-SSR genic markers. Forestry 92, 287–296. doi: 10.1093/forestry/cpz007
An, T., Chen, M., Zu, Z., Chen, Q., Lu, H., Yue, P., et al. (2021). Untargeted and targeted metabolomics reveal changes in the chemical constituents of instant dark tea during liquid-state fermentation by Eurotium cristatum. Food Res. Int. 148:110623. doi: 10.1016/j.foodres.2021.110623
Baruah, K. N., Singha, S., Mukherjee, P., and Uppaluri, R. V. (2023). Optimization of the enzymatic extraction of catechins from Assam tea leaves. Biomass Conv. Bioref. 2023:04684. doi: 10.1007/s13399-023-04684-x
Bhati, N., Shreya,, and Sharma, A. K. (2021). Cost-effective cellulase production, improvement strategies, and future challenges. J. Food Process Eng. 44:e13623. doi: 10.1111/jfpe.13623
Chandini, S. K., Rao, L. J., Gowthaman, M. K., Haware, D. J., and Subramanian, R. (2011). Enzymatic treatment to improve the quality of black tea extracts. Food Chem. 127, 1039–1045. doi: 10.1016/j.foodchem.2011.01.078
Chen, J., Cui, H., Huang, H., Wei, S., Liu, Y., Yu, H., et al. (2022). EST-SSR markers’ development based on RNA-sequencing and their application in population genetic structure and diversity analysis of Eleusine indica in China. Curr. Issues Mol. Biol. 45, 141–150. doi: 10.3390/cimb45010011
Chen, Q., Zhang, M., Chen, M., Li, M., Zhang, H., Song, P., et al. (2021). Influence of Eurotium cristatum and aspergillus Niger individual and collaborative inoculation on volatile profile in liquid-state fermentation of instant dark teas. Food Chem. 350:129234. doi: 10.1016/j.foodchem.2021.129234
Chen, M., Zu, Z., Shen, S., An, T., Zhang, H., Lu, H., et al. (2023). Dynamic changes in the metabolite profile and taste characteristics of loose-leaf dark tea during solid-state fermentation by Eurotium cristatum. LWT-Food Sci. Technol. 176:114528. doi: 10.1016/j.lwt.2023.114528
Darshan, K., Aggarwal, R., Bashyal, B. M., Singh, J., Saharan, M. S., Gurjar, M. S., et al. (2023). Characterization and development of transcriptome-derived novel EST-SSR markers to assess genetic diversity in Chaetomium globosum. 3Biotech 13:379. doi: 10.1007/s13205-023-03794-7
de Souza, T. S., and Kawaguti, H. Y. (2021). Cellulases, hemicellulases, and pectinases: applications in the food and beverage industry. Food Bioprocess Tech. 14, 1446–1477. doi: 10.1007/s11947-021-02678-z
Diba, K., Kordbacheh, P., Mirhendi, S. H., Rezaie, S., and Mahmoudi, M. (2007). Identification of aspergillus species using morphological characteristics. Pak. J. Med. Sci. 23, 867–872. Available at: https://www.pjms.com.pk/issues/octdec207/article/article9.html
Du, Y., Yang, W., Yang, C., and Yang, X. (2022). A comprehensive review on microbiome, aromas and flavors, chemical composition, nutrition and future prospects of Fuzhuan brick tea. Trends Food Sci. Tech. 119, 452–466. doi: 10.1016/j.tifs.2021.12.024
Efimenko, T. A., Shanenko, E. F., Mukhamedzhanova, T. G., Efremenkova, O. V., Nikolayev, Y. A., Bilanenko, E. N., et al. (2021). Eurotium cristatum postfermentation of fireweed and apple tree leaf herbal teas. Int. J. Food Sci. 2021:6691428. doi: 10.1155/2021/6691428
Fu, J., Lv, H., and Chen, F. (2016). Diversity and variation of bacterial community revealed by MiSeq sequencing in Chinese dark teas. PLoS One 11:e0162719. doi: 10.1371/journal.pone.0162719
Ge, Y., Wang, Y., Liu, Y., Tan, Y., Ren, X., Zhang, X., et al. (2016). Comparative genomic and transcriptomic analyses of the Fuzhuan brick tea-fermentation fungus aspergillus cristatus. BMC Genomics 17:428. doi: 10.1186/s12864-016-2637-y
González-Ceballos, L., Guirado-Moreno, J. C., Utzeri, G., García, J. M., Fernández-Muino, M. A., Osés, S. M., et al. (2023). Straightforward purification method for the determination of the activity of glucose oxidase and catalase in honey by extracting polyphenols with a film-shaped polymer. Food Chem. 405:134789. doi: 10.1016/j.foodchem.2022.134789
Hu, Z. Y., Liu, S. Q., Xu, Z. G., and Dong, M. (2023). Comparative transcriptomic analysis of different reproductive types of Eurotium cristatum. GAB 42, 400–417. doi: 10.13417/j.gab.042.000400
Hu, Z. Y., Liu, S. C., Xu, Z. G., Liu, S. Q., Li, T. T., Yu, S. L., et al. (2021). Comparison of aspergillus chevalieri and related species in dark tea at different aspects: morphology, enzyme activity and mitochondrial genome. J. Food Process Pres. 45:e15903. doi: 10.1111/jfpp.15903
Huang, Q., Cao, Y., Liu, Z., Tan, Y., and Liu, Y. (2017). Efficient gene replacements in ku70 disruption strain of aspergillus chevalieri var. intermedius. Biotechnol. Biotec. EQ 31, 16–22. doi: 10.1080/13102818.2016.1251828
Jiang, C., Zeng, Z., Huang, Y., and Zhang, X. (2018). Chemical compositions of Pu'er tea fermented by Eurotium Cristatum and their lipid-lowering activity. LWT-Food Sci. Technol. 98, 204–211. doi: 10.1016/j.lwt.2018.08.007
Li, T., Liu, Z., Li, J., Zheng, Y., Liu, Z., and Ling, P. (2023b). Lovastatin production by wild Eurotium cristatum isolated from fuzhuan brick tea produced using forest resources in Auhua. Forests 14:1409. doi: 10.3390/f14071409
Li, J., Zhang, C., Chen, S., Jiang, K., Guan, H., and Liu, W. (2023a). Characterization and application of EST-SSR markers developed from transcriptome sequences in Elymus breviaristatus (Poaceae: Triticeae). Genes 14:302. doi: 10.3390/genes14020302
Liang, S., Granato, D., Zou, C., Gao, Y., Zhu, Y., Zhang, L., et al. (2021). Processing technologies for manufacturing tea beverages: from traditional to advanced hybrid processes. Trends Food Sci. Tech. 118, 431–446. doi: 10.1016/j.tifs.2021.10.016
Lin, F. J., Wei, X. L., Liu, H. Y., Li, H., Xia, Y., Wu, D. T., et al. (2021). State-of-the-art review of dark tea: from chemistry to health benefits. Trends Food Sci. Tech. 109, 126–138. doi: 10.1016/j.tifs.2021.01.030
Liu, K., Chen, Q., Luo, H., Li, R., Chen, L., Jiang, B., et al. (2023a). An in vitro catalysis of tea polyphenols by polyphenol oxidase. Molecules 28:1722. doi: 10.3390/molecules28041722
Liu, S., Qiao, Z., Zeng, H., Li, Y., Cai, N., Liu, X., et al. (2023b). Analysis on SSR loci in transcriptome and development of EST-SSR molecular markers in Lonicera macranthoides. MPB 14, 1–8. doi: 10.5376/mpb.2023.14.0001
Liu, H., Sang, S., Wang, H., Ren, X., Tan, Y., Chen, W., et al. (2018). Comparative proteomic analysis reveals the regulatory network of the veA gene during asexual and sexual spore development of aspergillus cristatus. Biosci. Rep. 38:BSR20180067. doi: 10.1042/BSR20180067
Lu, X., Jing, Y., Li, Y., Zhang, N., and Cao, Y. (2022a). Eurotium cristatum produced β-hydroxy acid metabolite of monacolin K and improved bioactive compound contents as well as functional properties in fermented wheat bran. LWT-Food Sci. Technol. 158:113088. doi: 10.1016/j.lwt.2022.113088
Lu, X., Jing, Y., Zhang, N., and Cao, Y. (2022b). Eurotium cristatum, a probiotic fungus from Fuzhuan brick tea, and its polysaccharides ameliorated DSS-induced ulcerative colitis in mice by modulating the gut microbiota. J. Agric. Food Chem. 70, 2957–2967. doi: 10.1021/acs.jafc.1c08301
Lu, J., Zhang, Y., Diao, X., Yu, K., Dai, X., Qu, P., et al. (2021). Evaluation of genetic diversity and population structure of Fragaria nilgerrensis using EST-SSR markers. Gene 796-797:145791. doi: 10.1016/j.gene.2021.145791
Parveen, A., Qin, C. Y., Zhou, F., Lai, G., Long, P., Zhu, M., et al. (2023). The chemistry, sensory properties and health benefits of aroma compounds of black tea produced by camellia sinensis and camellia assamica. Horticulturae. 9:1253. doi: 10.3390/horticulturae9121253
Ren, C. G., Tan, Y. M., Ren, X. X., Liu, Y. X., and Liu, Z. Y. (2017). Metabolomics reveals changes in metabolite concentrations and correlations during sexual development of Eurotium cristatum (synonym: aspergillus cristatus). Mycosphere 8, 1626–1639. doi: 10.5943/mycosphere/8/10/3
Rui, Y., Wan, P., Chen, G., Xie, M., Sun, Y., Zeng, X., et al. (2019). Analysis of bacterial and fungal communities by Illumina MiSeq platforms and characterization of aspergillus cristatus in Fuzhuan brick tea. LWT-Food Sci. Technol. 110, 168–174. doi: 10.1016/j.lwt.2019.04.092
Shao, L., Tan, Y., Song, S., Wang, Y., Liu, Y., Huang, Y., et al. (2022). Achog1 is required for the asexual sporulation, stress responses and pigmentation of aspergillus cristatus. Front. Microbiol. 13:1003244. doi: 10.3389/fmicb.2022.1003244
Sharma, S., Kumar, S., Kaur, R., and Kaur, R. (2021). Multipotential alkaline protease from a novel Pyxidicoccus sp. 252: ecofriendly replacement to various chemical processes. Front. Microbiol. 12:722719. doi: 10.3389/fmicb.2021.722719
Shi, J., Liu, J., Kang, D., Huang, Y., Kong, W., Xiang, Y., et al. (2019). Isolation and characterization of benzaldehyde derivatives with anti-inflammatory activities from Eurotium cristatum, the dominant fungi species in fuzhuan brick tea. ACS Omega 4, 6630–6636. doi: 10.1021/acsomega.9b00593
Simões, M. F., Santos, C., and Lima, N. (2013). Structural diversity of aspergillus (section Nigri) spores. Microsc. Microanal. 19, 1151–1158. doi: 10.1017/S1431927613001712
Steensels, J., Snoek, T., Meersman, E., Nicolino, M. P., Voordeckers, K., and Verstrepen, K. J. (2014). Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol. Rev. 38, 947–995. doi: 10.1111/1574-6976.12073
Sudeep, K. C., Upadhyaya, J., Joshi, D. R., Lekhak, B., Kumar Chaudhary, D., Raj Pant, B., et al. (2020). Production, characterization, and industrial application of pectinase enzyme isolated from fungal strains. Fermentation 6:59. doi: 10.3390/fermentation6020059
Tan, Y., Wang, H., Wang, Y., Ge, Y., Ren, X., Ren, C., et al. (2018). The role of the veA gene in adjusting developmental balance and environmental stress response in aspergillus cristatus. Fungal Biol 122, 952–964. doi: 10.1016/j.funbio.2018.05.010
van den Brule, T., Lee, C. L. S., Houbraken, J., Haas, P. J., Wösten, H., and Dijksterhuis, J. (2020). Conidial heat resistance of various strains of the food spoilage fungus Paecilomyces variotii correlates with mean spore size, spore shape and size distribution. Food Res. Int. 137:109514. doi: 10.1016/j.foodres.2020.109514
Vidya, V., Prasath, D., Snigdha, M., Gobu, R., Sona, C., and Maiti, C. S. (2021). Development of EST-SSR markers based on transcriptome and its validation in ginger (Zingiber officinale Rosc.). PLoS One 16:e0259146. doi: 10.1371/journal.pone.0259146
Wainaina, S., and Taherzadeh, M. J. (2023). Automation and artificial intelligence in filamentous fungi-based bioprocesses: a review. Bioresour. Technol. 369:128421. doi: 10.1016/j.biortech.2022.128421
Wang, X., Cui, Y., Sang, C., Wang, B., Yuan, Y., Liu, L., et al. (2022a). Fungi with potential probiotic properties isolated from Fuzhuan brick tea. Food Sci. Hum. Well. 11, 686–696. doi: 10.1016/j.fshw.2021.12.026
Wang, D., Lin, F., Qin, Z., Luo, R., Li, T., and Zou, W. (2023a). Enzymes in ready-to-drink tea and coffee products. AP 2023, 105–124. doi: 10.1016/B978-0-323-85683-6.00005-3
Wang, H., Shen, S., Wang, J., Jiang, Y., Li, J., Yang, Y., et al. (2022b). Novel insight into the effect of fermentation time on quality of Yunnan congou black tea. LWT-Food Sci. Technol. 155:112939. doi: 10.1016/j.lwt.2021.112939
Wang, F., Tang, T., Mao, T., Duan, Y., Guo, X., and You, J. (2023b). Development of EST-SSR primers and genetic diversity analysis of the southern blight pathogen Sclerotium rolfsii using transcriptome data. Front. Microbiol. 14:1152865. doi: 10.3389/fmicb.2023.1152865
Wang, X., Zhang, Y., Ren, T., Zhao, Q., Yue, T., and Yuan, Y. (2019). Isolation and identification of Eurotium cristatum from Fuzhuan tea and its application in liquid-state fermentation. Food Sci. 40, 172–178. doi: 10.7506/spkx1002-6630-20180820-207
Wen, Z., Mingyu, W., Jingguan, Y., and Li, N. (2022). Fermentation optimization of culture medium and isolation and identification of antimicrobial active substances from Eurotium cristatum. JCIFST 22, 345–356. doi: 10.16429/j.1009-7848.2022.02.037
Wu, H., Zhao, H., Ding, J., Wang, Y., Hou, J., and Yang, L. (2023). Metabolites and microbial characteristics of Fu brick tea after natural fermentation. LWT-Food Sci. Technol. 181:114775. doi: 10.1016/j.lwt.2023.114775
Xiang, X., Song, C., Shi, Q., Tian, J., Chen, C., Huang, J., et al. (2020). A novel predict-verify strategy for targeted metabolomics: comparison of the curcuminoids between crude and fermented turmeric. Food Chem. 331:127281. doi: 10.1016/j.foodchem.2020.127281
Xiao, Y., He, C., Chen, Y., Ho, C. T., Wu, X., Huang, Y., et al. (2022). UPLC–QQQ–MS/MS-based widely targeted metabolomic analysis reveals the effect of solid-state fermentation with Eurotium cristatum on the dynamic changes in the metabolite profile of dark tea. Food Chem. 378:131999. doi: 10.1016/j.foodchem.2021.131999
Xiao, Z., Huang, X., Zang, Z., and Yang, H. (2018). Spatio-temporal variation and the driving forces of tea production in China over the last 30 years. J. Geogr. Sci. 28, 275–290. doi: 10.1007/s11442-018-1472-2
Xiao, Y., Li, M., Liu, Y., Xu, S., Zhong, K., Wu, Y., et al. (2021). The effect of Eurotium cristatum (MF800948) fermentation on the quality of autumn green tea. Food Chem. 358:129848. doi: 10.1016/j.foodchem.2021.129848
Xiao, Y., Zhong, K., Bai, J. R., Wu, Y. P., and Gao, H. (2020). Insight into effects of isolated Eurotium cristatum from Pingwu Fuzhuan brick tea on the fermentation process and quality characteristics of Fuzhuan brick tea. J. Sci. Food Agr. 100, 3598–3607. doi: 10.1002/jsfa.10353
Xu, J., Hussain, M., Su, W., Yao, Q., Yang, G., Zhong, Y., et al. (2022). Effects of novel cellulase (Cel 906) and probiotic yeast fermentation on antioxidant and anti-inflammatory activities of vine tea (Ampelopsis grossedentata). Front. Bioeng. Biotech. 10:1006316. doi: 10.3389/fbioe.2022.1006316
Xu, L., Li, P., Su, J., Wang, D., Kuang, Y., Ye, Z., et al. (2023b). EST-SSR development and genetic diversity in the medicinal plant Pseudostellaria heterophylla (Miq.) pax. J. Appl. Res. Med. Aroma. 33:100450. doi: 10.1016/j.jarmap.2022.100450
Xu, C., Ma, Y., Zhang, H., Li, X., and Chen, Q. (2023a). The impact of environmental factors on the spatiotemporal heterogeneity of phytoplankton community structure and biodiversity in the Qiongzhou strait. Water 15:3792. doi: 10.3390/w15213792
Yang, W., Bai, Z., Wang, F., Zou, M., Wang, X., Xie, J., et al. (2022). Analysis of the genetic diversity and population structure of Monochasma savatieri Franch. Ex maxim using novel EST-SSR markers. BMC Genomics 23:597. doi: 10.1186/s12864-022-08832-x
Yang, S., Fan, L., Tan, P., Lei, W., Liang, J., and Gao, Z. (2023). Effects of Eurotium cristatum on chemical constituents and α-glucosidase activity of mulberry leaf tea. Food Biosci. 53:102557. doi: 10.1016/j.fbio.2023.102557
Yang, Y., He, R., Zheng, J., Hu, Z., Wu, J., and Leng, P. (2020). Development of EST-SSR markers and association mapping with floral traits in Syringa oblata. BMC Plant Biol. 20:436. doi: 10.1186/s12870-020-02652-5
Yang, M., and Song, R. (2014). Colony characteristics and genetic diversity of Cytospora chrysosperma strains from ten provinces in China. Scientia Silvae Sinicae 50, 160–166. doi: 10.11707/j.1001-7488.20140621
Yao, Y., Wu, M., Huang, Y., Li, C., Pan, X., Zhu, W., et al. (2017). Appropriately raising fermentation temperature beneficial to the increase of antioxidant activity and gallic acid content in Eurotium cristatum-fermented loose tea. LWT-Food Sci. Technol. 82, 248–254. doi: 10.1016/j.lwt.2017.04.032
Yildiz, E., Pinar, H., Uzun, A., Yaman, M., Sumbul, A., and Ercisli, S. (2021). Identification of genetic diversity among Juglans regia L. genotypes using molecular, morphological, and fatty acid data. Genet. Resour. Crop. Ev. 68, 1425–1437. doi: 10.1007/s10722-020-01072-6
Yin, J., Liu, X., Peng, F., Wang, Q., Xiao, Y., and Liu, S. (2023a). Metabolite profiling, antioxidant and anti-aging activities of Siraitia grosvenorii pomace processed by solid-state fermentation with Eurotium cristatum. Process Biochem. 133, 109–120. doi: 10.1016/j.procbio.2023.08.016
Yin, J., Zhao, H., Wu, X., Li, Y., Chen, H., and Han, R. (2023b). SSR marker based analysis for identification and of genetic diversity of non-heading Chinese cabbage varieties. Front. Plant Sci. 14:1112748. doi: 10.3389/fpls.2023.1112748
Yu, Z., and Yang, Z. (2020). Understanding different regulatory mechanisms of proteinaceous and non-proteinaceous amino acid formation in tea (Camellia sinensis) provides new insights into the safe and effective alteration of tea flavor and function. Crit. Rev. Food Sci. 60, 844–858. doi: 10.1080/10408398.2018.1552245
Zhang, Y., Liu, X., Li, Y., Liu, X., Ma, H., Qu, S., et al. (2022a). Basic characteristics of flower transcriptome data and derived novel EST-SSR markers of Luculia yunnanensis, an endangered species endemic to Yunnan, southwestern China. Plants 11:1204. doi: 10.3390/plants11091204
Zhang, B., Ren, D., Zhao, A., Cheng, Y., Liu, Y., Zhao, Y., et al. (2022b). Eurotium cristatum reduces obesity by alleviating gut microbiota dysbiosis and modulating lipid and energy metabolism. J. Sci. Food Agr. 102, 7039–7051. doi: 10.1002/jsfa.12065
Zhang, Y., Wu, X., Wang, W., Xu, Y., Sun, H., Cao, Y., et al. (2022c). Virulence characteristics of Blumeria graminis f. sp. tritici and its genetic diversity by EST-SSR analyses. PeerJ 10:e14118. doi: 10.7717/peerj.14118
Zhao, M., Shu, G., Hu, Y., Cao, G., and Wang, Y. (2023a). Pattern and variation in simple sequence repeat (SSR) at different genomic regions and its implications to maize evolution and breeding. BMC Genomics 24:136. doi: 10.1186/s12864-023-09156-0
Zhao, Q., Yan, X., Yue, Y., Yue, T., and Yuan, Y. (2023b). Improved flavonoid content in mulberry leaves by solid-state fermentation: metabolic profile, activity, and mechanism. Innov. Food Sci. Emerg. 84:103308. doi: 10.1016/j.ifset.2023.103308
Zheng, H., Huang, W., Li, X., Huang, H., Yuan, Q., Liu, R., et al. (2023). CRISPR/Cas9-mediated BoaAOP2s editing alters aliphatic glucosinolate side-chain metabolic flux and increases the glucoraphanin content in Chinese kale. Food Res. Int. 170:112995. doi: 10.1016/j.foodres.2023.112995
Zheng, X. Q., Li, Q. S., Xiang, L. P., and Liang, Y. R. (2016). Recent advances in volatiles of teas. Molecules 21:338. doi: 10.3390/molecules21030338
Zhou, Y., Ye, Y., Zhu, G., Xu, Y., Tan, J., and Liu, J. (2023). Diversity, classification, and EST-SSR-based association analysis of caladium ornamental traits. Physiol. Plantarum 175:e13841. doi: 10.1111/ppl.13841
Keywords: Aspergillus cristatus, molecular marker, cluster analysis, polymorphism, microbial resources
Citation: Hu Z, Liu S, Zhou X, Liu Z, Li T, Yu S, Zhang X and Xu Z (2024) Morphological variation and expressed sequence tags-simple sequence repeats-based genetic diversity of Aspergillus cristatus in Chinese dark tea. Front. Microbiol. 15:1390030. doi: 10.3389/fmicb.2024.1390030
Received: 22 February 2024; Accepted: 22 May 2024;
Published: 03 June 2024.
Edited by:
Haifeng Zhao, South China University of Technology, ChinaReviewed by:
Kunyi Liu, Yibin Vocational and Technical College, ChinaCopyright © 2024 Hu, Liu, Zhou, Liu, Li, Yu, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenggang Xu, eHV6aGVuZ2dhbmdAbndhZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.