
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 July 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1388651
 Na Liu1†
Na Liu1† Biao Tang2†
Biao Tang2† Hui Wang1
Hui Wang1 Xiangyang Chen3
Xiangyang Chen3 Peipei Wen1
Peipei Wen1 Zhaorui Wang1
Zhaorui Wang1 Xu Chen1
Xu Chen1 Xiaobing Guo4
Xiaobing Guo4 Jianjun Gou4*
Jianjun Gou4* Yinsen Song1*
Yinsen Song1*Objectives: To investigate the genetic characteristics and transmission mechanism of the NDM-1-, IMP-4-, and SHV-12-producing multidrug-resistant (MDR) clinical isolate, Citrobacter freundii BC73.
Methods: C. freundii BC73 was isolated from a urine specimen of a urological patient diagnosed with bladder cancer at a Chinese teaching hospital. Antimicrobial susceptibility testing was carried out using DL-120E susceptibility cards and DL-96A system. Whole genome sequencing (WGS) of the isolate was performed using the Illumina and Oxford Nanopore platforms to analyze the genetic context of drug resistance genes and plasmid characteristics. The phylogenetic tree was constructed and visualized by KSNP3.0 software and iTOL5.0 online database.
Results: C. freundii isolate BC73 co-carrying blaNDM-1, blaIMP-4 and blaSHV-12 were multidrug-resistant. blaNDM-1 and blaIMP-4 were located on a novel IncFIB-like plasmid, pCFBC1, and an IncN-IncU hybrid plasmid, pCFBC2, respectively. The transferability of blaNDM-1 and blaIMP-4 from C. freundii BC73 to E. coli J53 was successfully demonstrated. The genetic context of the blaNDM-1 and blaIMP-4 genes were ISCR27-groEL-∆groES-cutA-dsbD-trpF-bleMBL-blaNDM-1-∆ISAba125-IS3000 and intI1-blaIMP-4-Kl.pn.13-mobC-IS6100, respectively. Additionally, two extensive transposition units (MGE1 in pCFBC1, MGE2 in pCFBC2) were identified and numerous antimicrobial resistance genes were discovered on it.
Conclusion: To our knowledge, our study represents the first characterization of a ST22 C. freundii isolate co-harboring blaNDM-1, blaIMP-4, and blaSHV-12, obtained from a urine sample. The dissemination of this MDR isolate should be of close concern in future clinical surveillance.
Infections due to carbapenemase-producing Enterobacteriaceae (CPE) remain pose a major threat to the public health (Nordmann et al., 2011; Tang et al., 2023). In particular, the co-production of two or three carbapenemases in a single bacterial isolate has become increasingly prevalent over the past 5 years, and resistance has shown an increase compared to the presence of a single gene. Such as in the study by Biez et al., the MICs of imipenem, meropenem and ertapenem in blaNDM-1-E. coli J53 or blaOXA-48-E. coli J53 transconjugants (Tc) or blaVIM-1-E. coli TOP10 transformant (Tf) were significantly lower than the original strain NDM-1-, VIM-1- and OXA-48-producing C. freundii 255A1. In another report, the MIC of meropenem in the original strain 112,298 was the same as the highest MIC in the transformants (112298-KPC-TOP10 and 112,298-NDM-TOP10) (Feng et al., 2015; Biez et al., 2022). We should beware of the emergence of such strains. The blaNDM-1 and blaIMP-4 genes, both encoding metallo-beta-lactamases (MBLs) with high carbapenemase activity, enable them to hydrolyze nearly all β-lactams including carbapenems. In recent years, they have been frequently detected in a diverse array of gram-negative bacteria, leading to the occurrence of numerous serious outbreaks (Yong et al., 2009; Dolejska et al., 2016; Xiong et al., 2016; Matsumura et al., 2017; Guducuoglu et al., 2018; Roberts et al., 2020). The simultaneous presence of these two resistance genes in a single strain may result in the emergence of highly drug-resistant variants, presenting a significant challenge for the treatment of infections.
Citrobacter freundii, a member of Enterobacteriaceae family and widely existed in water, soil, and the intestines of both animals and humans, has been identified as an opportunistic pathogen responsible for various infections including urinary, gastrointestinal, respiratory, peritoneal and bloodstream infections (Bodey, 2005). Unfortunately, the indiscriminate use of carbapenems has led to an escalating acquired resistance to antibiotics in C. freundii in recent years. So far, carbapenemases such as KPC-2-, NDM-1-, IMP-4-, OXA-48- and VIM-1-type have been reported in C. freundii, with affected regions including China, India, Spain, France and Italy (Yong et al., 2009; Gaibani et al., 2013; Feng et al., 2015; Lalaoui et al., 2019; Biez et al., 2022). However, the coexistence of NDM-1 and IMP-4 in single C. freundii isolate, along with its characteristics of transmission and resistance, has been rarely documented.
In this study, we identified a ST22 isolate of C. freundii, named BC73, which is the first reported case of co-carrying blaNDM-1, blaIMP-4 and blaSHV-12 from urine. Upon comprehensive investigation, we discovered that blaNDM-1 and blaIMP-4 were carried by a novel MDR plasmid and an IncN-IncU hybrid plasmid, respectively. Additionally, two extensive transposition units (MGE1 in pCFBC1, MGE2 in pCFBC2) harboring multiple resistance genes were identified, which were a potential contribution to the dissemination of multiple drug resistance.
A urine specimen was obtained from a hospital patient undergoing examination at the Fifth Clinical Medical College of Henan University of Chinese Medicine (FCMC-HUCM), Zhengzhou, China, in December 2021. The sample were cultured on MacConkey agar (OXOID, Hampshire, United Kingdom) plates supplemented with 2 mg/L meropenem and incubated at 37°C for 18–24 h. Species identification was conducted using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (Bruker, Bremen, Germany) and 16S rRNA gene sequencing. In vitro susceptibility test was performed using DL-120E susceptibility cards and DL-96A system (Zhuhai Deere Biological Engineering Co., LTD), which included 25 antibacterial agents as listed in Table 1. The interpretation of results followed the guidelines of the Clinical Laboratory Standards Institute (CLSI 2021; Humphries et al., 2021), with the exceptions of tigecycline and colistin, for which clinical breakpoints were determined according to the U.S. Food and Drug Administration (FDA) (Marchaim et al., 2014) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2022) guidelines1, respectively. Ultimately, C. freundii BC73 was confirmed and its details were presented in the subsequent results.
The blaNDM/IMP-carrying plasmids were visualized through PFGE/S1 nuclease analysis, followed by southern hybridization, utilizing digoxigenin-labeled blaNDM-1 and blaIMP-4-specific probes. The conjugation transfer of plasmids was executed by co-culturing with the recipient E. coli J53 at a 1:10 donor-to-recipient ratio, maintained at 25°C (Gou et al., 2020). Transconjugants were selectively cultivated on Mueller-Hinton medium supplemented with sodium azide (150 mg/L) and meropenem (2 mg/L). The confirmation of the selected transconjugants were carried out through PCR experiments.
Total DNA was extracted utilizing the Tiangen Genomic DNA Extraction Kit (Tiangen, Beijing, China) and subsequently sequenced using the Illumina HiSeq 4,000-PE150 (Illumina, San Diego, United States) and Oxford Nanopore GridION (Nanopore, Oxford, United Kingdom) platforms. De novo assembly of both short reads and long reads was conducted using Unicycler v0.4.8 (Wick et al., 2017), and the genomic sequences were annotated through the NCBI prokaryotic genome annotation pipeline. To identify sequence types (ST) and antimicrobial resistance genes, PubMLST2 and ResFinder 4.53 were employed. Replicon types of plasmid were performed by PlasmidFinder 2.1.4 The conjugation transfer modules in plasmids were predicted by oriTfinder5 and ICEfinder.6 Virulence factors (VFs) and mobile genetic elements (MGEs) were identified using VRprofile 2.7 CRISPR arrays were conducted by CRISPR Finder.8 Additionally, sequence comparisons were executed using BLAST.9 Comparative maps of the gene environment surrounding blaNDM-1 and blaIMP-4 genes were generated by Easyfig (Sullivan et al., 2011) and the BLAST Ring Image Generator (BRIG) (Alikhan et al., 2011) tool.
Genome sequences of 45 available C. freundii isolates were downloaded from the NCBI10 database, with C. freundii B38 (GCA_001702455.1) selected as the reference genome for comparative analysis. Subsequently, C. freundii BC73 and other C. freundii genomes were analyzed based on core genomic single nucleotide polymorphisms (SNPs) using KSNP3.0 (Gardner et al., 2015). Finally, a maximum likelihood tree was generated and visualized by iTOL5.0 (Letunic and Bork, 2021).
The complete genome sequence of C. freundii BC73 has been submitted to GenBank and assigned the accession numbers CP117475-CP117478.
Carbapenem-resistant C. freundii BC73 was isolated from a urine specimen of the patient who was hospitalized for urinary tract infection. The patient had a history of bladder cancer and had undergone total cystotomy and abdominal fistula drainage three months prior. Subsequent PCR and sequencing confirmed that the isolate was C. freundii carrying both blaNDM-1 and blaIMP-4 (Supplement 1).
The antimicrobial susceptibility results were presented in Table 1 and the image of the inhibition zones was deposited in Figure 1. Both C. freundii BC73 and transconjugant BC73-J53 exhibited resistance to all β-lactams, chloramphenicol, minocycline, azithromycin, and gentamicin antibiotics tested while they were sensitive to nitrofurantoin, tigecycline, polymyxin B, and amikacin. In addition, when C. freundii BC73 was resistant to levofloxacin and trimethoprim/sulfamethoxazole, transconjugant BC73-J53 was sensitive to it. Interestingly, the resistance of transconjugant BC73-J53 to Imipenem and cefepime was greater than that of C. freundii BC73 to it. The recipient E. coli J53 was susceptible to all antibacterial agents tested.
The isolate BC73 was identified as ST22. The genome was a single chromosome spanning 5,160,079 bp, exhibiting an average G + C content of 51.6%. Additionally, three plasmids (pCFBC1, pCFBC2, pCFBC3) were identified. The chromosome possessed 4,973 coding genes, 29 ISs, and the virulence genes csgABC, rcsA, wbtL, misL, and galE. Notably, anti-microbial resistance (AMR) genes blaCMY-48, aadA1 and dfrA1 were located on chromosome while blaNDM-1 on pCFBC1 and blaIMP-4 on pCFBC2 (Table 2).
C. freundii BC73 carried a ~131 kb plasmid harboring blaNDM-1 gene and a ~68 kb plasmid encoding blaIMP-4 gene (Figure 2). pCFBC2 was successfully transferred to E. coli J53 from C. freundii BC73 by conjugative assays and the conjugative efficiency was (1.11 ± 0.29) × 10−3 (Figure 3). pCFBC1 was a novel nonseparable plasmid, designated as IncFIB-like plasmid, with 130,842 bp in length and an average GC content of 53.1% (Table 2). A collection of replication initiation and stability proteins (repB, parAB), transcriptional regulators (acrR, deoR, frmBR, uidABC, uxuAR, ampR, lacI) formed the backbone of pCFBC1. Furthermore, four mobile genetic elements (MGEs) including MGE1, MGE2, MGE3 and MGE4 were found in this plasmid. In these MGEs, a lot of transposition units comprising ISs and antimicrobial resistance genes such as IS26-aac(3)-IId module, IS6100-mph(A)-mrx-mphR module, chrA-IS5075-sul1 module, ISCR1-sul1-qacE∆1-arr-3-catB3-IS1 module, and IS26-based module (IS26-blaSHV-12-IS26-tet(D)-IS26-catA2-IS26-ISVsa3-sul2-IS5075-∆Tn3-IS26-insB-IS26) were found. In comparison with selected plasmids, pCFBC1 exhibited 100.00 and 99.99% nucleotide identity with DY2010 plasmid 1 (CP086288) and pCFR17_1 (CP035277), respectively (Figure 4A). On the other hand, pCFBC2 emerged as an IncN and IncU hybrid plasmid, with 68,426 bp in length and an average GC content of 51.4% (Table 2). It featured two repB, mobC, frmBR, stbABC and ardABKR genes essential for replication and maintenance. In addition, the complete system for conjugation transfer including traKN, kikA, oriT, relaxase, the type IV coupling proteins (T4CP) and the type IV secretion system (T4SS) (virB1-11) was found. Two variable regions (VR1 and VR2) including a blaIMP-4 associated In823 and an extensive transposition unit (MGE2 in pCFBC2) were identified. These two regions harbored antimicrobial resistance genes including blaIMP-4, aac(6′)-Ib3, qacE∆1, qnrS1, and arr-3. pCFBC2 exhibited 99.97% nucleotide identity with pCA71-IMP (CP064181) and pIMP-HK1500 (KT989599), as detailed in Figure 4B.
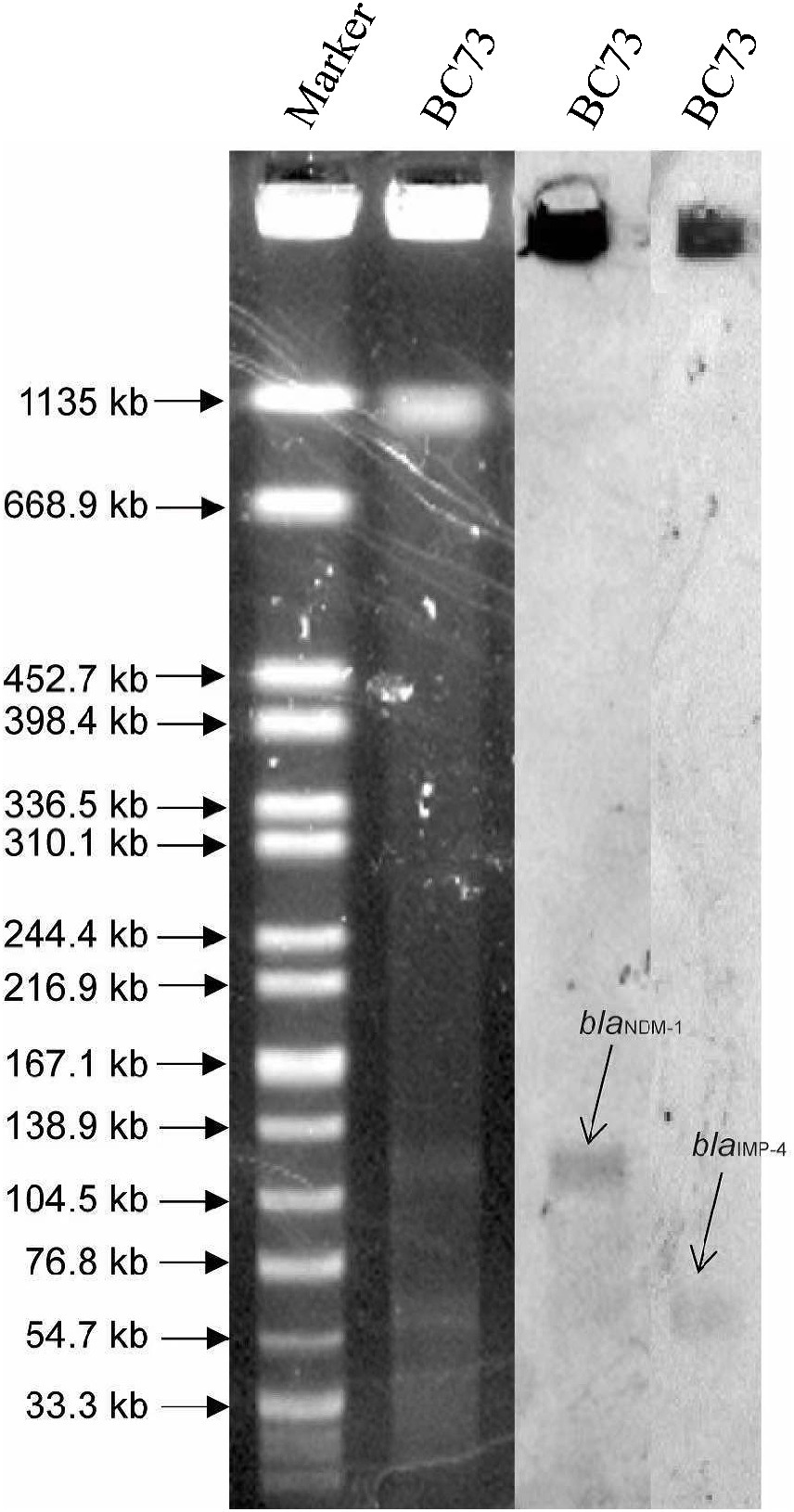
Figure 2. The identification of plasmids size using S1-PFGE (left) and southern blot and hybridization (right). pCFBC1 plasmid was between 104.5 kb and 138.9 kb, which was positive for a probe against blaNDM-1. pCFBC2 plasmid was between 54.7 kb and 76.8 kb, which was positive for a probe against blaIMP-4.
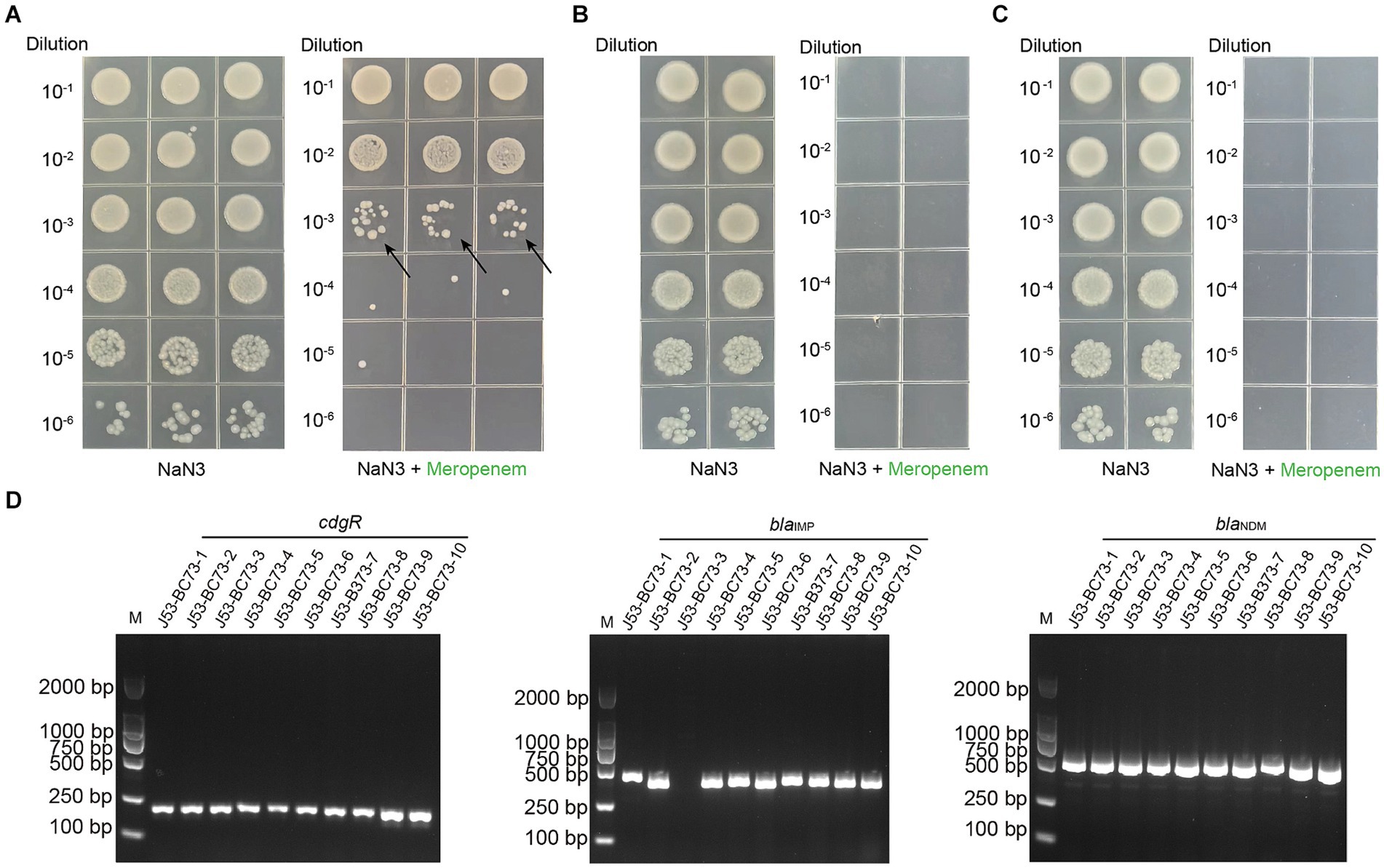
Figure 3. Conjugation transfer test and PCR verification of transconjugants of C. freundii strain BC73 with sodium azide-resistant E. coli strain J53 served as the recipient strain. (A) Conjugation transfer test of C. freundii strain BC73. From left to right are the screening results under the selective pressure of sodium azide and meropenem. The arrow represents the transconjugants. (B&C) Testing the transformation of the strain BC73’s genome DNA into the natural state of E. coli J53. One microliter of DNA at a concentration of 100 (B) & 1 (C) micrograms per milliliter was mixed with the BC73 strain and two repetitions were made. (D) PCR verification of transconjugants with sodium azide and meropenem. cdgR gene is a specific primer for E. coli.

Figure 4. Plasmid comparison of pCFBC1 and pCFBC2 plasmids. (A) Comparison of the pCFBC1 with DY2010 plasmid 1 (CP086288) and pCFR17_1 (CP035277). (B) Comparison of the pCFBC2 with pCA71-IMP (CP064181), pIMP-CF-127 (CP068026), pIMP-FJ1503 (KU051710) and pIMP-HK1500 (KT989599).
In pCFBC1, the blaNDM-1 gene resided within the composite structure of ∆Tn3000 and ∆Tn125 (ISCR27-groEL -∆groES-cutA-dsbD-trpF-bleMBL-blaNDM-1-∆ISAba125-IS3000). Upstream of this structure was MGE1 (IS26-based), comprised of a transposon from the Tn3 family, insertion sequences IS26, IS5075 and ISVsa3 and antimicrobial resistance genes (blaSHV-12, tet(D), catA2, sul2). Sequence comparisons revealed minor variations in the immediate genetic context of blaNDM-1 among the three plasmids (pCFBC1, pZY-NDM1, pNDM-Cf7308) (Figure 5A). In VR1 of pCFBC-2 (Figure 5B), the class 1 integron In823 carrying the resistance gene cassette blaIMP-4, the group II intron Kl.pn.13 and a mobilization protein (mobC) was inserted between the EcorII and uvp1 gene. And IS6100 was inserted downstream of the integron. In VR2 of pCFBC-2 (Figure 5B), the fipA gene was interrupted by the insertion of a ~17 kb region (MGE2) including Tn6292, splitting it into two fragments (fipA∆1 and fipA∆2). Tn6292 consisted of relics of Tn6292 tnp genes (tnpA and tnpR), insertion sequences (ISKpn19, IS2-IS26), and the qnrS1 gene. Interestingly, tnpA was disrupted and divided into two parts (tnpA∆1 and tnpA∆2) by ISKpn19 and a ~13 kb complex sequence, which was flanked by mobile elements tnpR-IS5075-∆Tn3-IS26 and a class 1 integron structure: intI1-aac(6′)-Ib3-arr-3-qacE∆1, lacking a common sul1 gene. In the middle, a replication initiation protein repB and two toxin-antitoxin proteins (higA, yafQ) were identified. A linear comparison of the blaIMP-4 genetic background among these plasmids displayed several differences: (1) the integrase gene intI1 immediately upstream of blaIMP-4 was complete in p11219-IMP (MF344561) and pCFBC-2, but interrupted by IS26 in pCA71-IMP, pIMP-CF-15-127 (CP068026), pIMP-FJ1503 (KU051710), and pIMP-HK1500. (2) The group II intron Kl.pn.13 immediately downstream of blaIMP-4 was disturbed by ISSen4 only in pIMP-HK1500. (3) A common 3′-conserved segment (qacE∆1-sul1) of class 1 integron In823 in p11219-IMP was absent in others plasmid and IS6100 was inserted at the far-end downstream of blaIMP-4. (4) There was no IS26 upstream of blaIMP-4 only in p11219-IMP and pCFBC-2 (VR1 of Figure 5B). Additionally, due to the fragmentation and rearrangement of genetic content, different variants of Tn6292 were generated in four plasmids, but the most significant variation was found in pCFBC-2, reflecting in a complex sequence inserted above-mentioned (VR2 of Figure 5B).
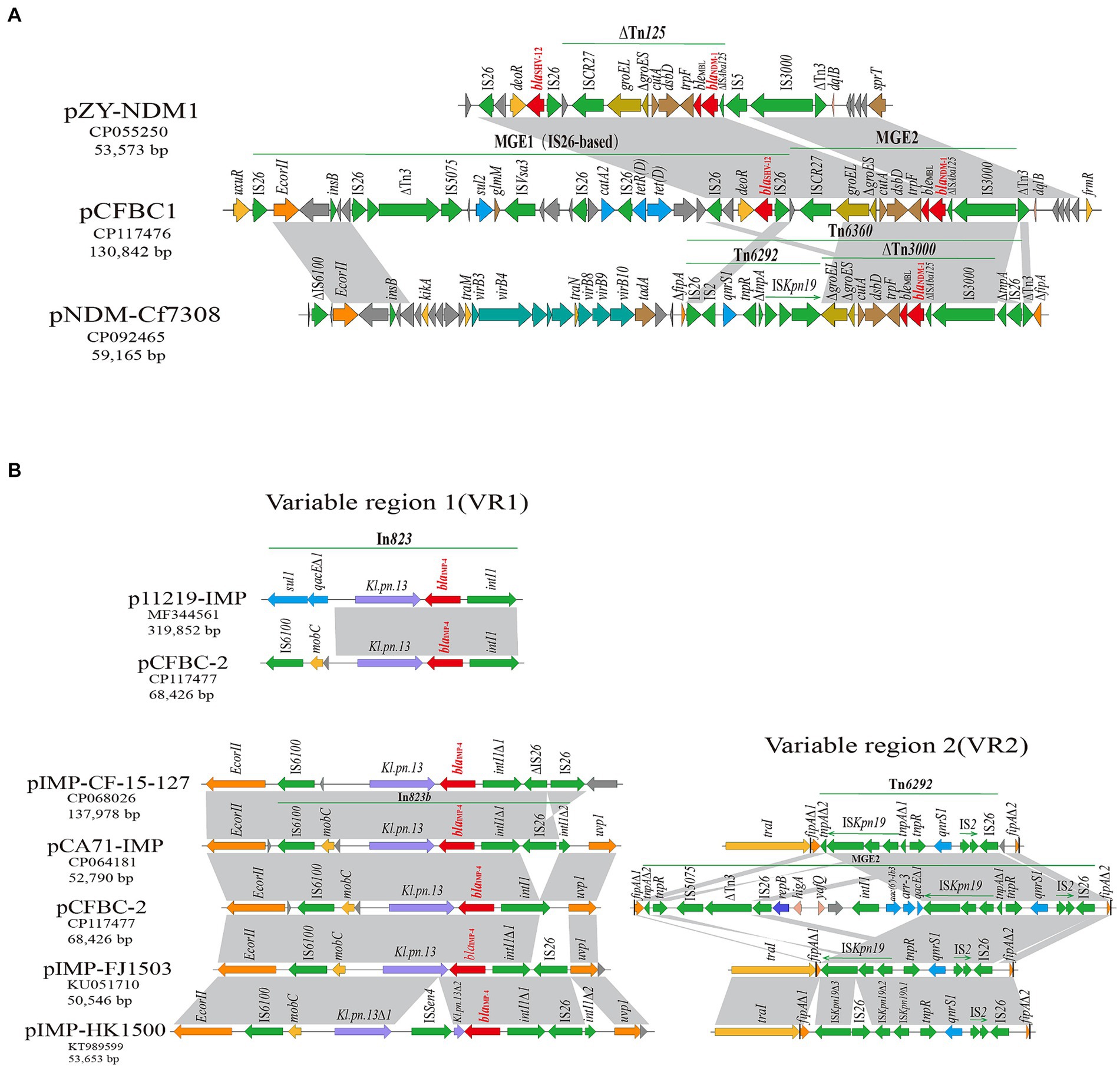
Figure 5. Gene-environment comparison of blaNDM-1 and blaIMP-4. (A) Genetic environment of blaNDM-1 on pCFBC1 and related plasmids. The regions with highly similar sequences were marked by light gray. The blaNDM-1 genes were shown by red; mobile elements were drawn by green. (B) In VR1, genetic context of blaIMP-4 on pCFBC2 and related plasmids. The blaIMP-4 genes were shown by red. In VR2, the genetic feature of Tn6292-associated area on pCFBC2 and related plasmids.
The diverse STs of 46 C. freundii isolates were displayed in Figure 6, which mainly included 22, 98, 116, 64, 396. These isolates, obtained from various specimen types including rectal swab, urine, stool, blood, abscess, nose throat swab, drainage liquid, lavabo, toilette, wastewater, soil, grass, sediment around river and food, were sourced from different hosts (homo sapiens, environment, food) across multiple countries including China, Germany, France, Spain, Switzerland, United States, Czech Republic, and Viet Nam spanning the period from 1998 to the present. The majority of C. freundii isolates carried resistance determinants such as β-lactams (blaNDM, blaKPC, blaVIM, blaIMP, blaOXA, blaSHV, blaTEM, blaCMY), aminoglycosides (aac(6′)-Ib-cr, aac(3)-IId, aac(6′)-Ib3, aadA1, aadA2), and folate pathway antagonists (sul1, sul2). Furthermore, C. freundii BC73 exhibited clustering with C. freundii MEI002, C. freundii CAV1321, C. freundii 064C1, C. freundii CF8_ST22, C. freundii IDR1800045912-01-00, C. freundii P7699, C. freundii MH17-012 N, and C. freundii DY2007. Notably, C. freundii DY2007, encoded blaNDM-5 and blaOXA-1 genes, was isolated from a blood specimen in China in 2020 and demonstrated the closest relationship to C. freundii BC73.
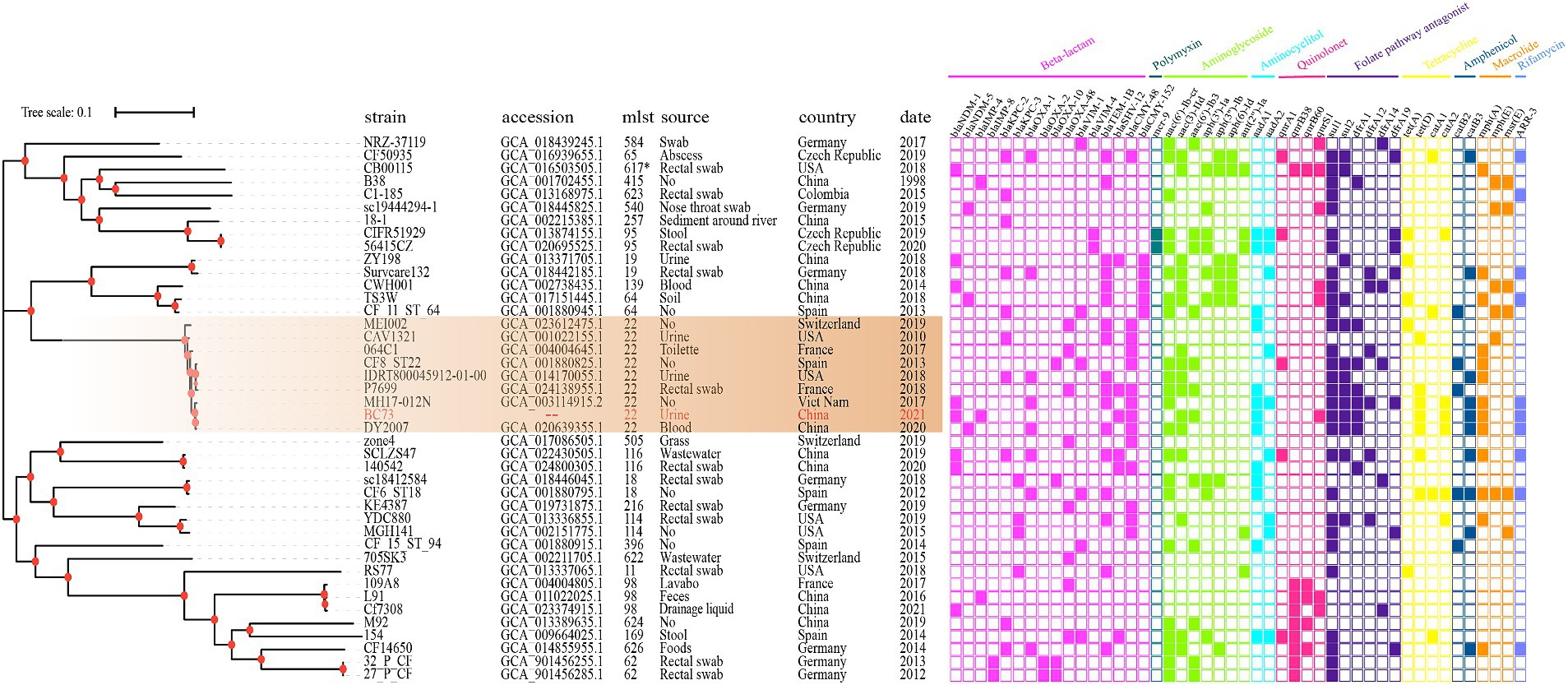
Figure 6. The phylogenetic tree composed of 46 C. freundii genome generated by KSNP3.0, where C. freundii B38 was used as the standard strain. Different color of the right panel indicated different positive antibiotic resistance genes of the corresponding strains.
Citrobacter freundii resistant to carbapenems has been gradually observed in patients with hospital-acquired infections (Hammerum et al., 2016; Zhang et al., 2023). However, studies on its transmission mechanisms and resistance characteristics, especially in case involving C. freundii carrying multiple carbapenemase genes, were scarce. In the routine collection of CRE strains, we obtained the C. freundii isolate BC73 co-carrying the carbapenemase genes blaNDM-1, blaIMP-4, and ESBLs gene blaSHV-12. It was isolated from the urine of a 64-year-old patient with urinary tract infections resulting from total cystotomy and abdominal fistula drainage. Carbapenems resistant Citrobacter freundii isolates, especially multidrug-resistant strains, were gradually being found in urinary tract infections, which may cause the extension of infection and the increase of patients’ suffering (Prussing et al., 2020; Zhang et al., 2021; Ye et al., 2023). The AST results presented in Table 1 showed that C. freundii BC73 and transconjugant BC73-J53 were resistance to all β-lactams, chloramphenicol, minocycline, azithromycin, and gentamicin antibiotics tested. Conversely, they were susceptible to nitrofurantoin, tigecycline, polymyxin B, and amikacin. The resistant phenotype of C. freundii BC73 was consistent with resistant genotype of it, implying resistance determinants were likely responsible for multiple drug resistance.
Recently, IncX3 plasmids carrying blaNDM have commonly been identified in different species of the Enterobacteriaceae (Ye et al., 2023), implying their significant role in blaNDM transfer. Furthermore, a study from Zhang et al. showed a high transfer frequency were observed in pZY-NDM1, a IncX3 plasmid harboring blaNDM-1 gene (Zhang et al., 2021). Interestingly, we identified a nonseparable plasmid, pCFBC-1, co-carrying blaNDM-1, blaSHV-12 and blaDHA-1. In this plasmid, we found three noteworthy features: (1) pCFBC-1 (~131 kb) was a novel large plasmid carrying blaNDM-1 gene. Firstly, compared to the most homologous plasmids in Figure 4A, we speculated that pCFBC1 underwent a genetic recombination and a conserved structure (groEL-∆groES-cutA-dsbD-trpF-bleMBL-blaNDM-1-∆ISAba125-IS3000) was recombined into pCFBC1. To our knowledge, this was the first time that the blaNDM-1 gene was present in this type plasmid and the resistance of it may be increased. Second, the most homologous plasmids of pCFBC1 have not been systematically analyzed. Based on the above, we believed that pCFBC1 was a novel plasmid. (2) After comparison with the PlasmidFinder database, pCFBC1 was unable to obtain the replicon type of the plasmid and was defined as a nonseparable plasmid. According to the replicon type of the plasmid with higher homology to pCFBC1, we named it InFIB-like plasmid. (3) Lots of transposition units (ISs + resistant determinants) were discovered in MGEs of pCFBC-1 such as IS26-aac(3)-IId module (aminoglycosides resistance), IS6100-mph(A)-mrx-mphR module (macrolides resistance), chrA-IS5075-sul1 module (chromates and folate pathway antagonists resistance), ISCR1-sul1-qacE∆1-arr-3-catB3-IS1 module (folate pathway antagonists resistance, quaternary ammonium compounds, rifamycin and phenicol resistance) and IS26-based module (IS26-blaSHV-12-IS26-tet(D)-IS26-catA2-IS26-ISVsa3-sul2-IS5075-∆Tn3-IS26-insB-IS26/β-lactams, tetracyclines, phenicol and folate pathway antagonists resistance) (Figure 4A). The presence of IS26 was highlighted as a potential contributor to the dissemination of resistance genes (Jia et al., 2022; Li et al., 2022). Wang et al. ever emphasized that genes encoding resistance could be recruited into a variable genetic locus flanked by IS elements and transposons, facilitating their common transfer in Enterobacteriaceae (Wang et al., 2017a). Additionally, three reports demonstrated (Toleman et al., 2006; Liu et al., 2015; Li et al., 2020) that ISCR1 may contribute to the mobilization of blaNDM-1 through rolling-circle transposition, manifesting the potential of ISCR1 in transferring resistance genes. At present, while blaIMP-4 has been found in various plasmid types (N, HI2, L/M and A/C), the IncN type, known for its broad-host-range and self-conjugative properties, remains predominant for the spread of blaIMP-4 in China (Lai et al., 2017; Wang et al., 2017b; Liu et al., 2021). In our study, we identified a IncN-IncU hybrid plasmid, pCFBC-2, carrying blaIMP-4 and a class 1 integron In823. The elements related to conjugation transfer such as traKN, kikA, oriT, relaxase, the type IV coupling proteins (T4CP) and the type IV secretion system (T4SS) (virB1-11) were revealed in pCFBC2. Furthermore, an extensive transposition unit (MGE2 in pCFBC2) were described. Compared to the most homologous IncN plasmids in Figure 4B, pCFBC2 not only possessed both IncN and IncU replicon types, but also added Tn3 family, aac(6′)-Ib3 and arr-3 on it, making its host range wider and mobility more flexible.
Horizontal gene transfer (HGT) plays a crucial role in the dissemination of bacterial resistance. The primary vehicle of HGT included plasmids, transposons (Tn), insertion sequences (IS) and integrons (In), which possessed the capability to capture and recombine genes associated with antibiotic resistance, heavy metal resistance and virulence, disseminating them with mobile characteristics (Qiao et al., 2023). Our investigation revealed that C. freundii BC73 successfully transferred blaNDM-1 and blaIMP-4, along with a carbapenem non-susceptible phenotype, to the recipient E. coli J53. This confirmed the natural horizontal gene transfer characteristic across species for blaNDM-1 and blaIMP-4. From the gene point of view, pCFBC2 had the complete conjugation transfer system, which further verified its autonomous conjugation transfer ability. According to our experimental validation in Figure 3, although pCFBC1 lacked elements related to conjugation transfer except part of relaxase, blaNDM-1 gene in it can be transferred to the recipient E. coli J53, suggesting that pCFBC1 may have been transferred to the recipient E. coli J53 with the help of pCFBC2. In pCFBC-1, blaNDM-1 was located in a conserved structure: ISCR27-groEL-∆groES-cutA-dsbD-trpF-bleMBL-blaNDM-1-∆ISAba125-IS3000, resembling the structures found in pZY-NDM1 and pNDM-Cf7308. This structure was a combination of Tn3000 and Tn125 remnants. Previous studies have described the prototype structures of Tn3000 and Tn125 associated with blaNDM-1, as vital vehicles for its dissemination, namely IS3000-groEL-groES-cutA-dsbD-trpF-bleMBL-blaNDM-1-ISAba125-IS3000 and ISAba125-ISCR27-groEL-∆groES-cutA-dsbD-trpF-bleMBL-blaNDM-1-ISAba125, respectively (Poirel et al., 2012; Campos et al., 2015). Compared with traditional Tn3000 and Tn125, one copy of IS3000 and ISAba125 was absent and another copy of ISAba125 was incomplete in pCFBC-1 (Figure 5A). The genetic background of blaIMP-4 was intI1-blaIMP-4-Kl.pn.13-mobC-IS6100 (MGE1 of pCFBC2). Compared with the genetic context of blaIMP-4 in other plasmids (VR1 of Figure 5B), we observed its subtle changes in pCFBC-2, suggesting the occurrence of gene recombination. Class 1 integrons should be responsible for the transfer of blaIMP gene. Thus far, blaIMP-4-associated class 1 integrons, including In809, 823, 823b, 1,377, 1,456, 1,460, and 1,589, has been reported in Enterobacteriaceae (Lee et al., 2017; Matsumura et al., 2017; Dolejska et al., 2018; Liang et al., 2018; Liu et al., 2021; Zhao et al., 2021). The distinction between In823 and In823b depends on the integrality of the intI1 gene. Furthermore, we identified a large MGE2 situated between two fragments (fipA∆1 and fipA∆2) in pCFBC2. In addition to containing the common Tn6292 (IS2-IS26-qnrS1, ISKpn19 and tnp genes) (VR2 of Figure 5B), one repB gene, two toxin-antitoxin proteins (higA and yafQ), mobile elements (IS5075-∆Tn3-IS26) and a class 1 integron carrying gene cassettes aac(6′)-Ib3 and arr-3 were assembled on it. The interruption of the fipA gene has been reported could promote the accumulation of plasmids in diverse hosts and facilitate the aggregation of mobile elements (Yang et al., 2018), which was a beneficial explanation for the formation of this MGE2.
Phylogenetic analysis (Figure 6) was conducted to unveil evolutionary characteristics and homology of C. freundii. The results revealed that C. freundii BC73 clustered with C. freundii MEI002, C. freundii CAV1321, C. freundii 064C1, C. freundii CF8_ST22, C. freundii IDR1800045912-01-00, C. freundii P7699, C. freundii MH17-012 N and C. freundii DY2007. Interestingly, these isolates all belonged to the ST22 C. freundii strain and were distributed in different countries over the span of a decade, which suggested that the ST22 C. freundii strains have disseminated globally and they may be highly clonal. Notably, C. freundii BC73 co-carrying blaNDM-1 and blaIMP-4 and C. freundii DY2007 harboring blaNDM-5, isolated from dongyang, China in 2020 (Ye et al., 2023), were found to be the most closely related isolates. Previous report has described that the differences between blaNDM-1 and blaNDM-5 were represented by mutations at only two specific sites (88, 154) (Sun et al., 2019). Based on above findings, we proposed a bold hypothesis that C. freundii BC73 likely evolved from DY2007 through vertical propagation.
Since the initial discovery of NDM-1 and IMP-4 in Enterobacteriaceae, these carbapenemases have rapidly disseminated worldwide (Chu et al., 2001; Yong et al., 2009). In recent years, NDM-1 or IMP-4 producing C. freundii has frequently identified in the clinical setting, which further aggravated the concerns for public health (Wu et al., 2016; Liu et al., 2021). However, C. freundii with the coexistence of NDM-1 and IMP-4 has been rarely reported. To our knowledge, only one such isolate, named wang9, has been reported in China. The C. freundii wang9 isolate belonged to ST415, and the genes blaNDM-1 and blaIMP-4 were located on a conjugative IncHI1B plasmid, pwang9-1. The blaNDM-1 gene was located on the transposon TnAS3 (IS91-sul-ISAba14-aph (3′)-VI-IS30-blaNDM-1-bleMBL-trpF-dsbD-IS91) while the blaIMP-4 gene was carried by integron In1337 (intI1-blaIMP-4-∆Kl.pn.13-qacG2-aac(6′)-Ib4-∆catB3) (Qiao et al., 2023). However, the above characteristics were markedly distinct in our isolate. The more attention should be paid on further monitoring and genetic analysis of NDM-1 and IMP-4-producing C. freundii isolates and flexible transposition units for better understanding of multiple drug resistance transfer.
In this study, we identified and characterized the genome of C. freundii BC73 co-carrying blaNDM-1, blaIMP-4 and blaSHV-12 from an inpatient with urinary tract infection after bladder cancer surgery. The blaNDM-1 and blaIMP-4 genes were located in a novel MDR plasmid and an IncN-IncU hybrid plasmid (pCFBC1, pCFBC2), respectively. In addition, multiple transposition units (ISs + resistant determinants), especially including two extensive transposition units (MGE1 in pCFBC1, MGE2 in pCFBC2), were found on it. The dissemination of NDM-1 and IMP-4-producing C. freundii isolates and ISs + resistant determinants should be of close concern in future clinical surveillance.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The ethical protocol was approved by the Ethics Committee of The Fifth Clinical Medical College of Henan University of Chinese Medicine (Zhengzhou People’s Hospital).
NL: Data curation, Funding acquisition, Resources, Software, Visualization, Writing – original draft, Writing – review & editing. BT: Data curation, Funding acquisition, Software, Supervision, Writing – review & editing. HW: Software, Writing – review & editing. XiC: Software, Writing – review & editing. PW: Data curation, Resources, Writing – review & editing. ZW: Data curation, Writing – review & editing. XuC: Data curation, Writing – review & editing. XG: Supervision, Writing – review & editing. JG: Supervision, Writing – review & editing. YS: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by joint construction project of Henan Province medical science and technology research plan (LHGJ20220793), the famous doctor support project of Zhengzhou (2021ZW-41), the collaborative innovation project of Zhengzhou (2023XTCX052), the Zhejiang Provincial Natural Science Foundation of China (LY23C180001), the ‘Leading Goose’R&D program of Zhejiang Province (2023C03045) and the program of Zhejiang agriculture and rural affairs (2023SN|F058).
We are grateful to the reviewers who helped to improve this paper and all members of our team.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1388651/full#supplementary-material
1. ^https://www.eucast.org/clinical_breakpoints/
3. ^https://cge.food.dtu.dk/services/ResFinder/
4. ^https://cge.food.dtu.dk/services/PlasmidFinder/
5. ^https://tool-mml.sjtu.edu.cn/oriTfinder/oriTfinder.html
6. ^https://bioinfo-mml.sjtu.edu.cn/ICEfinder/ICEfinder.html
7. ^https://tool2-mml.sjtu.edu.cn/VRprofile/VRprofile.php
8. ^http://crispr.i2bc.paris-saclay.fr/
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Biez, L., Bonnin, R. A., Naas, T., and Dortet, L. (2022). Characterization of VIM-1-, NDM-1- and OXA-48-producing Citrobacter freundii in France. J. Antimicrob. Chemother. 77, 1200–1202. doi: 10.1093/jac/dkac005
Bodey, G. P. (2005). Managing infections in the immunocompromised patient. Clin. Infect. Dis. 40:S239. doi: 10.1086/427328
Campos, J. C., Da Silva, M. J., Dos Santos, P. R., Barros, E. M., Pereira Mde, O., Seco, B. M., et al. (2015). Characterization of Tn3000, a transposon responsible for blaNDM-1 dissemination among Enterobacteriaceae in Brazil, Nepal, Morocco, and India. Antimicrob. Agents Chemother. 59, 7387–7395. doi: 10.1128/AAC.01458-15
Chu, Y. W., Afzal-Shah, M., Houang, E. T., Palepou, M. I., Lyon, D. J., Woodford, N., et al. (2001). IMP-4, a novel metallo-beta-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45, 710–714. doi: 10.1128/AAC.45.3.710-714.2001
Dolejska, M., Masarikova, M., Dobiasova, H., Jamborova, I., Karpiskova, R., Havlicek, M., et al. (2016). High prevalence of Salmonella and IMP-4-producing enterobacteriaceae in the silver gull on five islands, Australia. J. Antimicrob. Chemother. 71, 63–70. doi: 10.1093/jac/dkv306
Dolejska, M., Papagiannitsis, C. C., Medvecky, M., Davidova-Gerzova, L., and Valcek, A. (2018). Characterization of the complete nucleotide sequences of IMP-4-encoding plasmids, belonging to diverse Inc families, recovered from Enterobacteriaceae isolates of wildlife origin. Antimicrob. Agents Chemother. 62:e02434–17. doi: 10.1128/AAC.02434-17
Feng, J., Qiu, Y., Yin, Z., Chen, W., Yang, H., Yang, W., et al. (2015). Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J. Antimicrob. Chemother. 70, 2987–2991. doi: 10.1093/jac/dkv232
Gaibani, P., Ambretti, S., Farruggia, P., Bua, G., Berlingeri, A., Tamburini, M. V., et al. (2013). Outbreak of Citrobacter freundii carrying VIM-1 in an Italian hospital, identified during the carbapenemases screening actions, June 2012. Int. J. Infect. Dis. 17, e714–e717. doi: 10.1016/j.ijid.2013.02.007
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Gou, J. J., Liu, N., Guo, L. H., Xu, H., Lv, T., Yu, X., et al. (2020). Carbapenem-resistant Enterobacter hormaechei ST1103 with IMP-26 Carbapenemase and ESBL gene Bla (SHV-178). Infect Drug Resist 13, 597–605. doi: 10.2147/IDR.S232514
Guducuoglu, H., Gursoy, N. C., Yakupogullari, Y., Parlak, M., Karasin, G., Sunnetcioglu, M., et al. (2018). Hospital outbreak of a Colistin-resistant, NDM-1- and OXA-48-producing Klebsiella pneumoniae: high mortality from Pandrug resistance. Microb. Drug Resist. 24, 966–972. doi: 10.1089/mdr.2017.0173
Hammerum, A. M., Hansen, F., Nielsen, H. L., Jakobsen, L., Stegger, M., Andersen, P. S., et al. (2016). Use of WGS data for investigation of a long-term NDM-1-producing Citrobacter freundii outbreak and secondary in vivo spread of blaNDM-1 to Escherichia coli, Klebsiella pneumoniae and Klebsiella oxytoca. J. Antimicrob. Chemother. 71, 3117–3124. doi: 10.1093/jac/dkw289
Humphries, R., Bobenchik, A. M., Hindler, J. A., and Schuetz, A. N. (2021). Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 59:e0021321. doi: 10.1128/JCM.00213-21
Jia, X., Jia, P., Zhu, Y., Yu, W., Li, X., Xi, J., et al. (2022). Coexistence of Bla (NDM-1) and Bla (IMP-4) in one novel hybrid plasmid confers transferable Carbapenem resistance in an ST20-K28 Klebsiella pneumoniae. Front. Microbiol. 13:891807. doi: 10.3389/fmicb.2022.891807
Lai, K., Ma, Y., Guo, L., An, J., Ye, L., and Yang, J. (2017). Molecular characterization of clinical IMP-producing Klebsiella pneumoniae isolates from a Chinese tertiary hospital. Ann. Clin. Microbiol. Antimicrob. 16:42. doi: 10.1186/s12941-017-0218-9
Lalaoui, R., Djukovic, A., Bakour, S., Hadjadj, L., Sanz, J., Salavert, M., et al. (2019). Genomic characterization of Citrobacter freundii strains coproducing OXA-48 and VIM-1 carbapenemase enzymes isolated in leukemic patient in Spain. Antimicrob. Resist. Infect. Control 8:167. doi: 10.1186/s13756-019-0630-3
Lee, J. H., Bae, I. K., Lee, C. H., and Jeong, S. (2017). Molecular characteristics of first IMP-4-producing Enterobacter cloacae sequence type 74 and 194 in Korea. Front. Microbiol. 8:2343. doi: 10.3389/fmicb.2017.02343
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–w296. doi: 10.1093/nar/gkab301
Li, Y., Fang, C., Qiu, Y., Dai, X., and Zhang, L. (2022). Genomic characterization of a carbapenem-resistant Citrobacter freundii cocarrying Bla(KPC-2) and Bla(NDM-1). J Glob Antimicrob Resist 29, 289–292. doi: 10.1016/j.jgar.2022.04.014
Li, Z., Lin, Y., Lu, L., Wang, K., Yang, L., Li, P., et al. (2020). Genetic characterisation of a complex class 1 integron in an NDM-1-producing Citrobacter freundii ST396 clinical strain isolated from a urine sample. J Glob Antimicrob Resist 23, 64–66. doi: 10.1016/j.jgar.2020.08.002
Liang, Q., Jiang, X., Hu, L., Yin, Z., Gao, B., Zhao, Y., et al. (2018). Sequencing and genomic diversity analysis of IncHI5 plasmids. Front. Microbiol. 9:3318. doi: 10.3389/fmicb.2018.03318
Liu, W., Dong, H., Yan, T., Liu, X., Cheng, J., Liu, C., et al. (2021). Molecular characterization of Bla (IMP) (−) (4) -carrying Enterobacterales in Henan Province of China. Front. Microbiol. 12:626160. doi: 10.3389/fmicb.2021.626160
Liu, Y., Wan, L. G., Deng, Q., Cao, X. W., Yu, Y., and Xu, Q. F. (2015). First description of NDM-1-, KPC-2-, VIM-2- and IMP-4-producing Klebsiella pneumoniae strains in a single Chinese teaching hospital. Epidemiol. Infect. 143, 376–384. doi: 10.1017/S0950268814000995
Marchaim, D., Pogue, J. M., Tzuman, O., Hayakawa, K., Lephart, P. R., Salimnia, H., et al. (2014). Major variation in MICs of tigecycline in gram-negative bacilli as a function of testing method. J. Clin. Microbiol. 52, 1617–1621. doi: 10.1128/JCM.00001-14
Matsumura, Y., Peirano, G., Motyl, M. R., Adams, M. D., Chen, L., Kreiswirth, B., et al. (2017). Global molecular epidemiology of IMP-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 61:e16. doi: 10.1128/AAC.02729-16
Nordmann, P., Naas, T., and Poirel, L. (2011). Global spread of carbapenemase-producing enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798. doi: 10.3201/eid1710.110655
Poirel, L., Bonnin, R. A., Boulanger, A., Schrenzel, J., Kaase, M., and Nordmann, P. (2012). Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1087–1089. doi: 10.1128/AAC.05620-11
Prussing, C., Snavely, E. A., Singh, N., Lapierre, P., Lasek-Nesselquist, E., Mitchell, K., et al. (2020). Nanopore MinION sequencing reveals possible transfer of Bla (KPC-2) plasmid across bacterial species in two healthcare facilities. Front. Microbiol. 11:2007. doi: 10.3389/fmicb.2020.02007
Qiao, J., Chen, Y., Ge, H., Xu, H., Guo, X., Liu, R., et al. (2023). Coexistence of Bla(IMP-4), Bla(NDM-1) and Bla(OXA-1) in Bla(KPC-2)-producing Citrobacter freundii of clinical origin in China. Front. Microbiol. 14:1074612. doi: 10.3389/fmicb.2023.1074612
Roberts, L. W., Catchpoole, E., Jennison, A. V., Bergh, H., Hume, A., Heney, C., et al. (2020). Genomic analysis of carbapenemase-producing Enterobacteriaceae in Queensland reveals widespread transmission of Bla(IMP-4) on an IncHI2 plasmid. Microb Genom 6:e321. doi: 10.1099/mgen.0.000321
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, P., Xia, W., Liu, G., Huang, X., Tang, C., Liu, C., et al. (2019). Characterization of Bla (NDM-5)-positive Escherichia coli prevalent in a university hospital in eastern China. Infect Drug Resist 12, 3029–3038. doi: 10.2147/IDR.S225546
Tang, B., Guan, C., Lin, H., Liu, C., Yang, H., Zhao, G., et al. (2023). Emergence of co-existence of mcr-1 and blaNDM-5 in Escherichia fergusonii. Int J Antimicrob Agents. 61:106742. doi: 10.1016/j.ijantimicag.2023.106742
Toleman, M. A., Bennett, P. M., and Walsh, T. R. (2006). ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70, 296–316. doi: 10.1128/MMBR.00048-05
Wang, Y., Lo, W. U., Lai, R. W., Tse, C. W., Lee, R. A., Luk, W. K., et al. (2017b). IncN ST7 epidemic plasmid carrying blaIMP-4 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. J. Antimicrob. Chemother. 72, 99–103. doi: 10.1093/jac/dkw353
Wang, J., Yuan, M., Chen, H., Chen, X., Jia, Y., Zhu, X., et al. (2017a). First report of Klebsiella oxytoca strain simultaneously producing NDM-1, IMP-4, and KPC-2 Carbapenemases. Antimicrob. Agents Chemother. 61:e17. doi: 10.1128/AAC.00877-17
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, W., Espedido, B., Feng, Y., and Zong, Z. (2016). Citrobacter freundii carrying blaKPC-2 and blaNDM-1: characterization by whole genome sequencing. Sci. Rep. 6:30670. doi: 10.1038/srep30670
Xiong, J., Déraspe, M., Iqbal, N., Ma, J., Jamieson, F. B., Wasserscheid, J., et al. (2016). Genome and plasmid analysis of blaIMP-4-carrying Citrobacter freundii B38. Antimicrob. Agents Chemother. 60, 6719–6725. doi: 10.1128/AAC.00588-16
Yang, L., Li, P., Liang, B., Hu, X., Li, J., Xie, J., et al. (2018). Multidrug-resistant Citrobacter freundii ST139 co-producing NDM-1 and CMY-152 from China. Sci. Rep. 8:10653. doi: 10.1038/s41598-018-28879-9
Ye, J., Jin, L., Li, Y., Xu, H., Lin, Y., Zhou, T., et al. (2023). Complete-genome sequencing and comparative genomic characterization of Bla(NDM-5) carrying Citrobacter freundii isolates from a patient with multiple infections. BMC Genomics 24:506. doi: 10.1186/s12864-023-09579-9
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, Bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Zhang, F., Li, Z., Liu, X., Hu, Y., Zhao, J., Zhang, Y., et al. (2023). Carbapenem-resistant Citrobacter freundii harboring Bla(KPC-2) and Bla(NDM-1): a study on their transferability and potential dissemination via generating a transferrable hybrid plasmid mediated by IS6100. Front. Microbiol. 14:1239538. doi: 10.3389/fmicb.2023.1239538
Zhang, T., Lin, Y., Li, P., Li, Z., Liu, X., Li, J., et al. (2021). Characterization of plasmid co-harboring NDM-1 and SHV-12 from a multidrug-resistant Citrobacter freundii strain ZT01-0079 in China. Infect Drug Resist 14, 947–952. doi: 10.2147/IDR.S301736
Keywords: blaNDM-1, blaIMP-4, Citrobacter freundii, MDR, genomics
Citation: Liu N, Tang B, Wang H, Chen X, Wen P, Wang Z, Chen X, Guo X, Gou J and Song Y (2024) Coexistence of a novel NDM-1-encoding MDR plasmid and an IMP-4-encoding IncN-IncU hybrid plasmid in a clinical isolate of Citrobacter freundii BC73. Front. Microbiol. 15:1388651. doi: 10.3389/fmicb.2024.1388651
Received: 20 February 2024; Accepted: 28 June 2024;
Published: 10 July 2024.
Edited by:
Zhigang Qiu, Tianjin Institute of Environmental and Operational Medicine, ChinaReviewed by:
Xiaolong Wang, Nanjing University, ChinaCopyright © 2024 Liu, Tang, Wang, Chen, Wen, Wang, Chen, Guo, Gou and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinsen Song, c29uZ3lzQGhhY3RjbS5lZHUuY24=; Jianjun Gou, amlhbmp1bmdAenp1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.