- 1Laser Research Center of Dentistry, Dentistry Research Institute, Tehran University of Medical Sciences, Tehran, Iran
- 2Experimental Medicine Research Center, Tehran University of Medical Sciences, Tehran, Iran
Persistent infection caused by biofilm is an urgent in medicine that should be tackled by new alternative strategies. Low efficiency of classical treatments and antibiotic resistance are the main concerns of the persistent infection due to biofilm formation which increases the risk of morbidity and mortality. The gene expression patterns in biofilm cells differed from those in planktonic cells. One of the promising approaches against biofilms is nanoparticle (NP)-based therapy in which NPs with multiple mechanisms hinder the resistance of bacterial cells in planktonic or biofilm forms. For instance, NPs such as silver (Ag), zinc oxide (ZnO), titanium dioxide (TiO2), copper oxide (Cu), and iron oxide (Fe3O4) through the different strategies interfere with gene expression of bacteria associated with biofilm. The NPs can penetrate into the biofilm structure and affect the expression of efflux pump, quorum-sensing, and adhesion-related genes, which lead to inhibit the biofilm formation or development. Therefore, understanding and targeting of the genes and molecular basis of bacterial biofilm by NPs point to therapeutic targets that make possible control of biofilm infections. In parallel, the possible impact of NPs on the environment and their cytotoxicity should be avoided through controlled exposure and safety assessments. This study focuses on the biofilm-related genes that are potential targets for the inhibition of bacterial biofilms with highly effective NPs, especially metal or metal oxide NPs.
1 Introduction
Approximately 40 to 80% of bacteria possess the ability to create biofilms under harsh environmental conditions. This phenomenon results in a major global concern for human health, as it greatly contributes to the development of persistent infections (Muhammad et al., 2020). Infections caused by biofilms are highly resistant to antibiotics and host immune cells compared to planktonic cells. Unlike planktonic bacteria, biofilms are resistant to conventional cleaning, washing, heating, and disinfection procedures (Sharma et al., 2019). The main reasons for antibiotic resistance of biofilms are the restriction of antibiotic entry to the biofilm structure, slow growth and metabolism, and resistance genes. Biofilm formation on medical implants such as sutures, catheters, heart valves, joint prostheses, and dental implants leads to chronic infections such as wound infections, urinary tract infections, lung infections, and chronic osteomyelitis. Infections caused by implanted medical devices account for about 60–70% of all nosocomial infections. The only way to eliminate biofilms from medical implants is the implant removal which is an expensive and problematic treatment for patients (Cangui-Panchi et al., 2022). Biofilms are homogeneous or heterogeneous microbial communities of microorganisms that live on abiotic or biotic surfaces and consist of cells, extracellular matrix (ECM), and other polymeric substances. EMC is a biopolymer matrix consisting of exopolysaccharides (EPS), extracellular DNA, proteins, and amyloidogenic proteins (Gupta et al., 2016).
There are important genes associated with bacteria that grow in biofilm, including rhlI-rhlR, rhlAB, pqsR-pqsA, lasR, lecA, and pel A in Pseudomonas aeruginosa (Xu et al., 2018; Abdelraheem et al., 2020), while icaADBC, eno (laminin), ebps (elastin), fib (fibrinogen-binding protein) are necessary for biofilm formation in Staphylococcus aureus (Kot et al., 2018). The gtfs genes are essential for the adhesion of Streptococcus mutans. In addition, the genes gbps, smu630, relA, and comDE are involved in bacterial adhesion and biofilm formation (You, 2019). Type I fimbriae encoded by the gene cluster fimABCDEFGH are involved in the attachment of Escherichia coli cells (Ren et al., 2004).
Some of the important strategies being developed against biofilms are quorum sensing inhibitors (QSIs), bacteriophages, enzymes, surfactants, nanoparticles (NPs), antimicrobial photodynamic therapy, ethnopharmacology, and diguanylate cyclase inhibitors (Qayyum and Khan, 2016). Although numerous antimicrobial drugs are commercially available, they often lack efficacy against multidrug-resistant (MDR) microorganisms, posing a major challenge to healthcare teams (Ahmed et al., 2021). The emergence of antimicrobial resistance has increased the need for the development of alternate antimicrobial agents (Uddin et al., 2021).
Nanotechnology is a promising tool for the management of MDR strains and bacterial biofilms. In recent years, the applications of NPs in the medical field have increased for the treatment of infectious diseases. NPs possess different physicochemical properties than bulk forms due to their unique size and structure at the nanoscale. In the size range of 1 nm to 100 nm, NPs exhibit enhanced surface-to-volume ratio, increased reactivity, altered electronic properties, etc., depending on size (Baig et al., 2021). The ability of NPs to inhibit the proliferation of microbial cells makes them the most widely used drug delivery system to combat pathogenic organisms (Lahiri et al., 2021).
The unique properties of metal NPs have led to their great interest as antimicrobial agents. The antimicrobial properties of various metal NPs, including silver (Ag), zinc oxide (ZnO), titanium dioxide (TiO2), copper (Cu), iron oxide (Fe3O4), selenium (Se), and gold (Au) have been extensively investigated (Hemeg, 2017). Previous research suggests that NPs have different mechanisms of action than antibiotics, which may improve their ability to combat MDR bacteria. Antibacterial activity studies have shown that NPs can enter the cell and attach to cell receptors, leading to intracellular disintegration and consequent cell death, and inhibit essential metabolic enzymes, thereby disrupting bacterial cell reproduction and respiration. NPs have been developed that interfere with signal-based biofilm formation to prevent biofilm-associated infections (Gupta et al., 2017; Wang et al., 2017). The review describes the efficacy of highly potent NPs, especially metal or metal oxide NPs, as potent anti-biofilm therapeutics with particular reference to their ability to inhibit bacterial biofilm-related genes.
2 Biofilm formation process
Biofilm formation is a survival strategy of microorganisms in which microbial cells adapt to their environment and adopt a multicellular lifestyle in which bacterial cells are immobilized within their EPS matrix. The bacteria within this matrix are protected from antibacterial compounds and are up to 1,000 times more resistant to antibiotics (Uneputty et al., 2022). Biofilm formation begins with reversible attachment of free-floating bacterial cells to a surface. This is followed by irreversible attachment, which is facilitated by bacterial adhesion structures and short-range forces. The process of EPS production drives reversible attachment to the surface. They then develop into a well-organized structure encased in an EPS matrix. Ultimately, the bacterial cells are able to escape and access new niches (Muhammad et al., 2020). The stages of biofilm life cycle are depicted in Figure 1. Five key stages of biofilm formation are presented below.
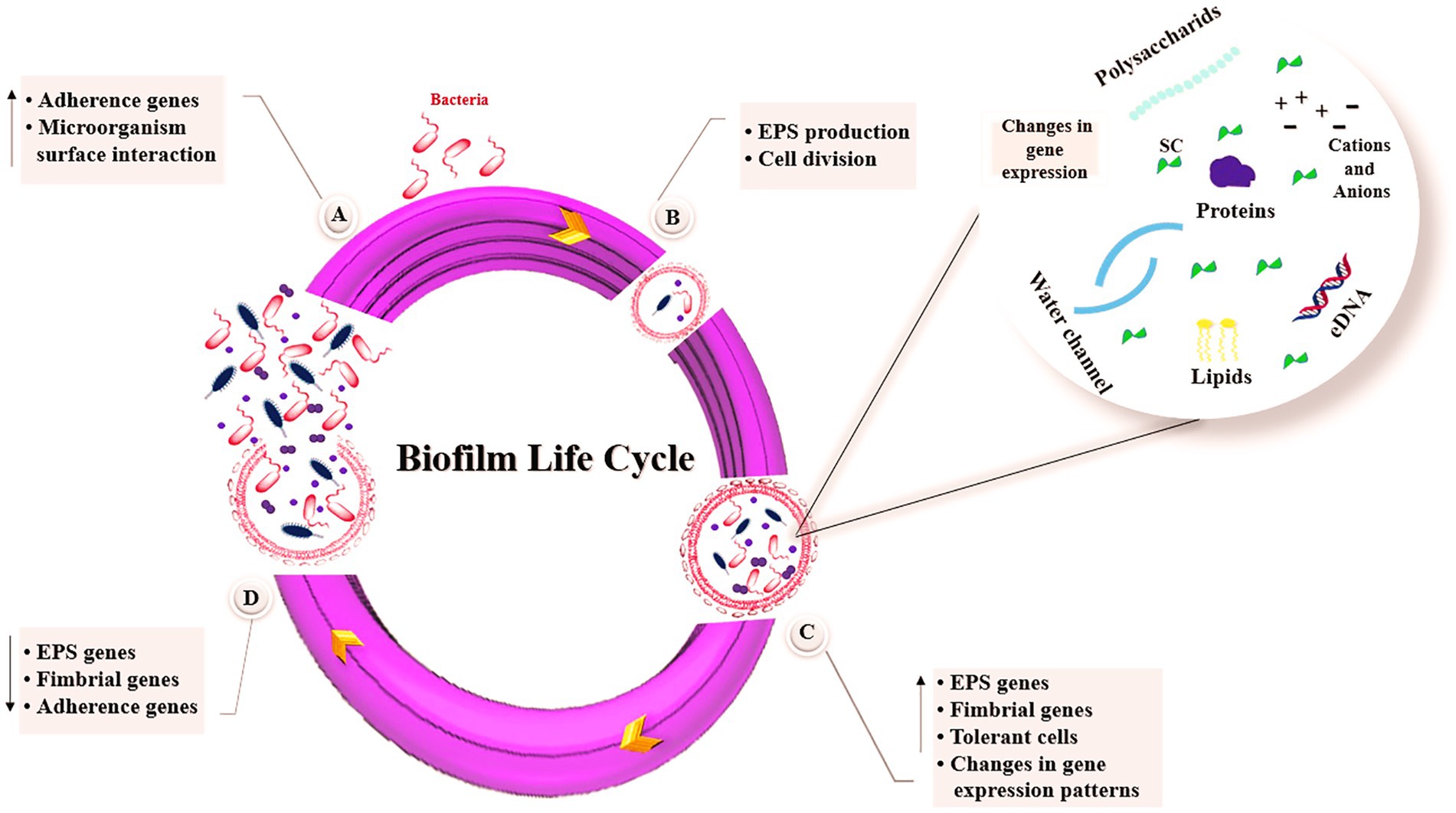
Figure 1. Stages of biofilm formation. (A) Bacteria attachment, (B) Microcolony formation, (C) Biofilm maturation, (D) Biofilm dispersal. SC, signaling compounds; eDNA, extracellular DNA; EPS, exopolysaccharides.
2.1 Bacterial initial attachment and irreversible adhesion
Bacterial attachment to surfaces favors in biofilm formation in initial step. Initially, the bacterium gets close enough to allow initial attachment, and the forces involved in this initial attachment are van der Waals forces, electrostatic and hydrophobic interactions. During this initial contact, the bacteria still exhibit Brownian motion and can be easily removed by the flow rate of the liquid. Subsequently, an irreversible step occurs, as the bacteria attach to the surface by producing EPS and or filamentous appendages, such as pili or flagella and non-fimbrial adhesions. At the end of this phase, much stronger physical or chemical forces are needed to remove the bacteria attached to the surface (Palmer et al., 2007). The size of the contact area between bacterial cell and material surface, the total of the repulsive or attractive forces between two surfaces, the hydrophobicity/hydrophilicity, roughness and surface charge, the physicochemical and topographic properties of a substrate surface can all influence the attachment of bacteria (Renner and Weibel, 2011). In S. aureus, atlE gene expression mediates bacterial adhesion and secretes autolysin. Strains lacking the atlE gene exhibit a marked reduction in adhesion capacity. The expression of fbe and sap genes leads to synthesis of fiber protein binding protein, which mediates the attachment of S. aureus and fibrin. During the accumulation phase, iapABD gene is expressed to synthesize the ECM and eventually form pileus-like multilayer structure in mature biofilms (Zhao and Ashraf, 2015).
2.2 EPS synthesis
Irreversible adhesion is promoted by the synthesis of EPS, which is controlled by the QS of the bacterial cells. Bacteria produce EPS, which is an important element of biofilm ECM. EPS can mediate bacterial adhesion–cohesion via hydrophobic and ion-bridging forces. Overall, EPS directly mediates adherence to microbial cell surfaces, cell-to-cell adhesion, biofilm structure, biofilm formation, water retention, cell–cell signaling, cell protection, nutrient trapping capability, and genetic adaptation. In addition, the cyclic messenger di-GMP (c-di-GMP) is considered to be one of the triggers for the switching of reversible to irreversible attachment via EPS and cell surface structures. EPS are mainly enzymes and structural proteins, DNAs, polysaccharides, phospholipids, and glycoproteins (Czaczyk and Myszka, 2007). Polysaccharides are a key fraction of the EPS matrix and are necessary during the biofilm development process and growth of the bacteria (Flemming, 2016).
2.3 Microcolony formation
Microbial cells begin to multiply and divide after they connect to a biotic or abiotic surface and this attachment becomes stable. This process is started by specific chemical signaling within the EPS. The next step in this process is the formation of microcolonies. The different micro-communities that make up the bacterial colonies in a biofilm usually coordinate with each other in different ways. The exchange of substrate, the distribution of important metabolic products, and the excretion of metabolic end products all depend on this coordination (Gestel et al., 2015).
2.4 Biofilm maturation
The EPS-embedded bacteria conduct to microcolonies formation and biofilms maturation. The EPS then acts as a biological ‘glue’ between the embedded bacterial cells through changes in gene expression. Through matrix formation, nutrients are transported to the cell communities and unwanted products are removed through the formation of water-filled channels. Often a mushroom-shaped multicellular structure of microcolonies is displayed. During the maturation process, inhibition of the production of surface structures restricts motility in microcolonies and the pattern of gene expression differs significantly from that of planktonic cells (Muhammad et al., 2020).
2.5 Biofilm dispersal
The dispersal process is a tactic used by bacterial cells to breakdown biofilms and begin a new biofilm life-cycle. The complicated process of dispersal is controlled by effectors, signal transduction pathways, and environmental signals. Genes responsible for the EPS generation, initial attachment, and fimbriae synthesis are often reduced during dispersal, but genes related to EPS degradation and flagella synthesis are typically enhanced. Inhibition of c-di-GMP signaling pathways is another effective method of biofilm dispersion. Furthermore, environmental factors (i.e., temperature, nutrients, and pH) increase in glucose supply, and lack of oxygen can contribute to biofilm dispersal (Toyofuku et al., 2016).
3 QS mechanism
QS is a chemical communication mechanism that correlates with population density and is used by bacteria to regulate the production of virulence factors and biofilm formation. The elimination of QS is a new approach to combat their pathogenicity (Colino et al., 2018). They implement QS by secreting small extracellular signaling molecules that act as autoinducers (AI) to start genetic programming. Three main QS systems can be distinguished: the acyl-homoserine lactone (AHL) QS system in Gram-negative bacteria, the autoinducing peptide (AIP) QS system in Gram positive bacteria, and the AI-2 QS system in both Gram-negative and Gram-positive bacteria. QS is associated with the control of the swarming behavior of bacteria and the development of biofilm architecture (Sadekuzzaman et al., 2015).
Quorum quenching (QQ) refers to any strategy that interferes with proper microbial QS signaling. This can occur in two ways: inhibition and degradation of AI and disruption of its interaction with the receptor. These compounds have been characterized as QQ, which interfere with bacterial communication, reducing virulence without affecting growth. Depending on the type of regulation (i.e., whether QS induces or suppresses virulence), agents must either inhibit or stimulate QS-regulated gene expression (Colino et al., 2018). QQ enzymes have frequently been found to inhibit QS by targeting AHLs (Hong et al., 2012).
4 Efflux pumps
Efflux pumps are membrane proteins that are involved in the development of antibiotic resistance and the export of molecules such as QS signals, drugs, detergents, and heavy metals outside of the cell. Efflux pump genes are present in bacterial chromosomes and mobile genetic elements such as plasmids (Webber and Piddock, 2003). At least four different roles for efflux pumps in biofilm formation are possible. Efflux of EPSs and/or QS and QQ molecules to facilitate biofilm matrix formation and regulate QS, respectively; they could also indirectly regulate genes involved in biofilm formation; efflux of harmful molecules, such as antibiotics and metabolic intermediates; and influence aggregation by inhibiting adhesion to surfaces and other cells (Alav et al., 2018). In E. coli biofilm development, the efflux genes araJ, ddpD, emrK, gltK, ycbO, and yhdX increased during biofilm growth (May et al., 2009). Sánchez et al. reported that P. aeruginosa nalB and nfxB mutants, which overexpress MexAB-OprM and MexCD-OprJ efflux systems, respectively, did not show any defects in biofilm formation and, in fact, the nalB mutant strains were found to exhibit significantly denser biofilm formation when compared to the wild-type (Sánchez et al., 2002). One possibility is that the overexpression of multidrug efflux pump MexEF-OprN reduces the intracellular concentration of QS signals exhibited impaired biofilm formation (Alav et al., 2018). Holling et al. reported that disruption of the bcr efflux gene in Proteus mirabilis led to reduced biofilm formation. Furthermore, the bcr mutant exhibited deficiencies in both swarming and swimming motility (Holling et al., 2014).
5 Intercellular adhesion operon
The intercellular adhesion (ica) operon, which includes the genes icaA, icaD, icaB, and icaC, encodes biosynthetic enzymes involved in the synthesis of polysaccharide intercellular adhesin (PIA). EPS synthesis is closely linked to the icaA and icaD genes. Similarly, icaB and icaC contribute to the production of poly-N-acetylglucosamine polymer by transporting it to the bacterial cell surface and deacetylating exopolysaccharide molecules. The N-acetylglucosaminyltransferase acts on the substrate UDP-N-acetylglucosamine with the help of icaD and forms an EPS. The reaction product is then transported across the cytoplasmic membrane by icaC. Deacetylation of PIA by icaB in turn enables PIA to adhere to living and non-living surfaces and facilitates biofilm formation. Although the PIA adhesin plays a key role in the process of biofilm synthesis, it is not essential for biofilm production. There are biofilm-forming strains that do not contain ica operon genes (Swolana et al., 2022).
6 Strategies for prevention of biofilm formation
Biofilm formation is a complicated process that can be targeted by anti-biofilm agents to prevent the various stages of biofilm development (Mahamuni-Badiger et al., 2020). Several genes are involved in biofilm formation, so actions can be taken at the genetic level, such as interfering with second messenger signals and the two-component system (TCS), to modulate EPS metabolism (Karatan and Watnick, 2009). C-di-AMP and c-di-GMP are conserved second messenger signals that are crucial for important functions of virulence, such as biofilm formation. They control various EPS-producing exoenzymes, polysaccharides, and adhesins. It is therefore possible to target them to disrupt EPS (Jiang et al., 2020). Antisense RNA is complementary to messenger RNA and its duplex can interfere with the translation of specific proteins (Yoshioka et al., 2019). Antisense RNA could manipulate the expression of target genes and control the biofilm (Wang and Kuramitsu, 2005). Bacteria have an adaptive immune defense mechanism known as the CRISPR/Cas system (clustered regularly interspaced short palindromic repeats). Biofilm formation, EPS synthesis, interspecific competitiveness, and other aspects of bacterial virulence are influenced by the CRISPR/Cas system (Arora, 2024). The CRISPR/Cas9 system targeting sdiA has been reported to have effects on cell adhesion and biofilm formation of Salmonella enterica (Askoura et al., 2021). Furthermore, the CRISPR/Cas-HDR approach was used to prevent biofilm formation of E. coli by knockout genes involved in adhesion (fimH/bolA) and QS (luxS; Alshammari et al., 2023). On the other hand, the expression of wcaF, which is involved in colanic acid synthesis and has been identified as a critical component of the E. coli biofilm, was controlled using the CRISPRi/dCas9 system (Zhang and Poh, 2018).
Biofilm formation can be prevented by avoiding the initial attachment of planktonic cells to surfaces by remodeling the surface or treating the cells to block cell attachment. Preformed biofilms can be removed by detachment, erosion, and dispersal (Mahamuni-Badiger et al., 2020). Furthermore, anti-biofilm agents can act in different ways by inhibiting or altering QS signaling pathways, membrane permeabilization, biofilm assembly, lipopolysaccharide assembly, and enzymatic dispersion of EPS (Roy et al., 2018; Asma et al., 2022).
7 NPs as inhibitors of biofilm formation
Nanotechnology approaches have great potential to offer effective and practical solutions for biofilm management and prevention, as they deal with the engineering of materials at the atomic and molecular level that have high surface-to-volume ratios and high activation energy of atoms (Mohanta et al., 2023). It is a good platform to generate effective antimicrobial agents by optimizing the physicochemical properties of metals. Metal oxides are toxic and harmful to natural sources, but at the nanoscale they change their physicochemical properties. The ability of NPs to penetrate biofilms also inhibits biofilm formation (Mohanta et al., 2023). Three stages can be distinguished in the interaction between NPs and biofilm: NP transport in the vicinity of the biofilm, NP attachment to the biofilm surface, and NP migration in biofilms. Many factors, including the environment, EPM, and the physico-chemical properties of the NPs, influence how the individual phases take place (Shkodenko et al., 2020).
Some NPs themselves have the ability to demonstrate anti-biofilm activities because they contain antibacterial components such metal oxides and cationic surfactants (Sadekuzzaman et al., 2015). Their large surface area to mass ratio, high reactivity, and surface functionalization have given them special qualities that enable effective biofilm eradication. NPs act as anti-QS by interfering with the mechanism of bacterial cell–cell communication or inhibiting the QS signaling system. This is able to prevent production of molecule-receptor complex and the formation of various signaling molecules. The abilities of metallic NPs to exert QQ activity have been particularly noted (Lahiri et al., 2021). As given in Figure 2, NPs can exert their antibacterial activity through different mechanisms, such as cell wall disruption, interactions with DNA and/or proteins, inhibition of biofilm development, or generation of reactive oxygen species (ROS; Wang et al., 2017).
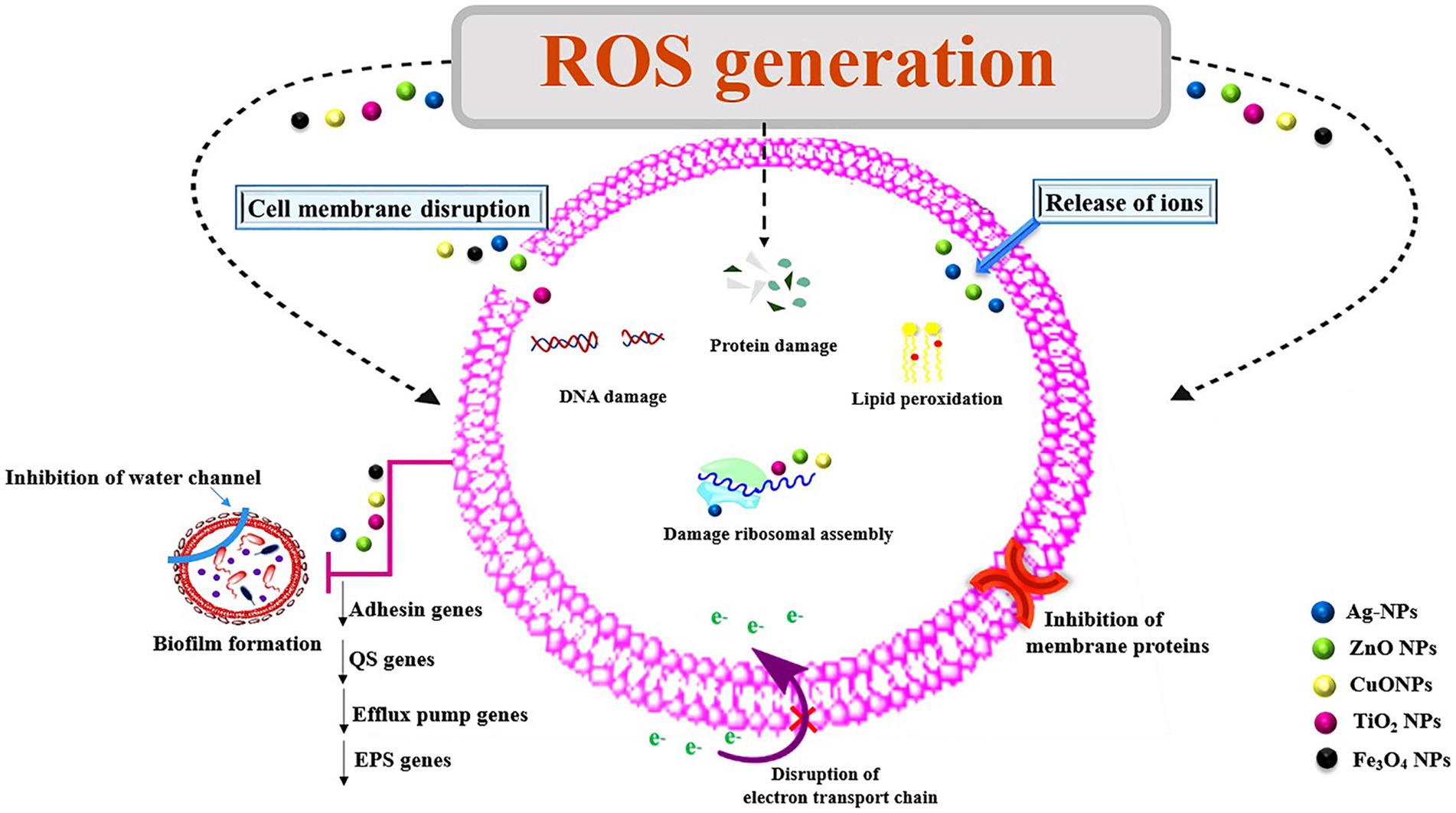
Figure 2. Mechanisms of action of nanoparticles for their antimicrobial properties. ROS, reactive oxygen species; NPs, nanoparticles; Ag, silver; ZnO, zinc oxide; TiO2, titanium dioxide; Cu, copper oxide; Fe3O4, iron oxide; EPS, exopolysaccharides; QS, quorum sensing.
7.1 Ag NPs
Ag NPs are widely used in healthcare and biomedicine, food storage, and environmental applications. Ag NPs are remarkable for their unique physicochemical features such as excellent catalytic activity and stability, high conductivity, and significant anti-inflammatory and antibacterial activities (Lee and Jun, 2019). Interference with cell-membrane function and intracellular ROS generation have been the key pathways for antibacterial activity (Liao et al., 2019).
Ag NPs probably destabilize the cell membrane and produce Ag ions that react with membrane proteins. These NPs have the ability to pass through bacterial cell walls and/or membranes, attach to bacterial DNA, and obstruct DNA replication. Furthermore, they have the ability to disrupt ribosomal function to translate mRNA into protein forms, thereby activating cytochrome B proteins (Yan et al., 2018). The antibacterial mechanism of Ag NPs works by inhibiting O2 metabolism, eventually killing microorganisms (Shahverdi et al., 2007). Furthermore, Ag NPs can efficiently diminish bacterial biofilm biomass (Montazeri et al., 2020).
The anti-biofilm activity and downregulation of bacterial biofilm-related genes of Ag NPs against a variety of bacteria has been reported in several research papers (Table 1). Ag NPs have the ability to block the synthesis of bacterial EPS and then biofilm. The ability of biofilm inhibition by Ag NPs may be due to the existence of water channels throughout the biofilm (Kalishwaralal et al., 2010; Montazeri et al., 2020; Pourmbarak Mahnaie and Mahmoudi, 2020). They can enter biofilms and inhibit biofilm development by suppressing gene expression. These ions are internalized and prevent the penetration of amines, thiols, or carboxylates. Ag NPs tend to agglomerate, which negatively affects their antimicrobial efficacy. Therefore, their surface must be functionalized before application (Le Ouay and Stellacci, 2015).
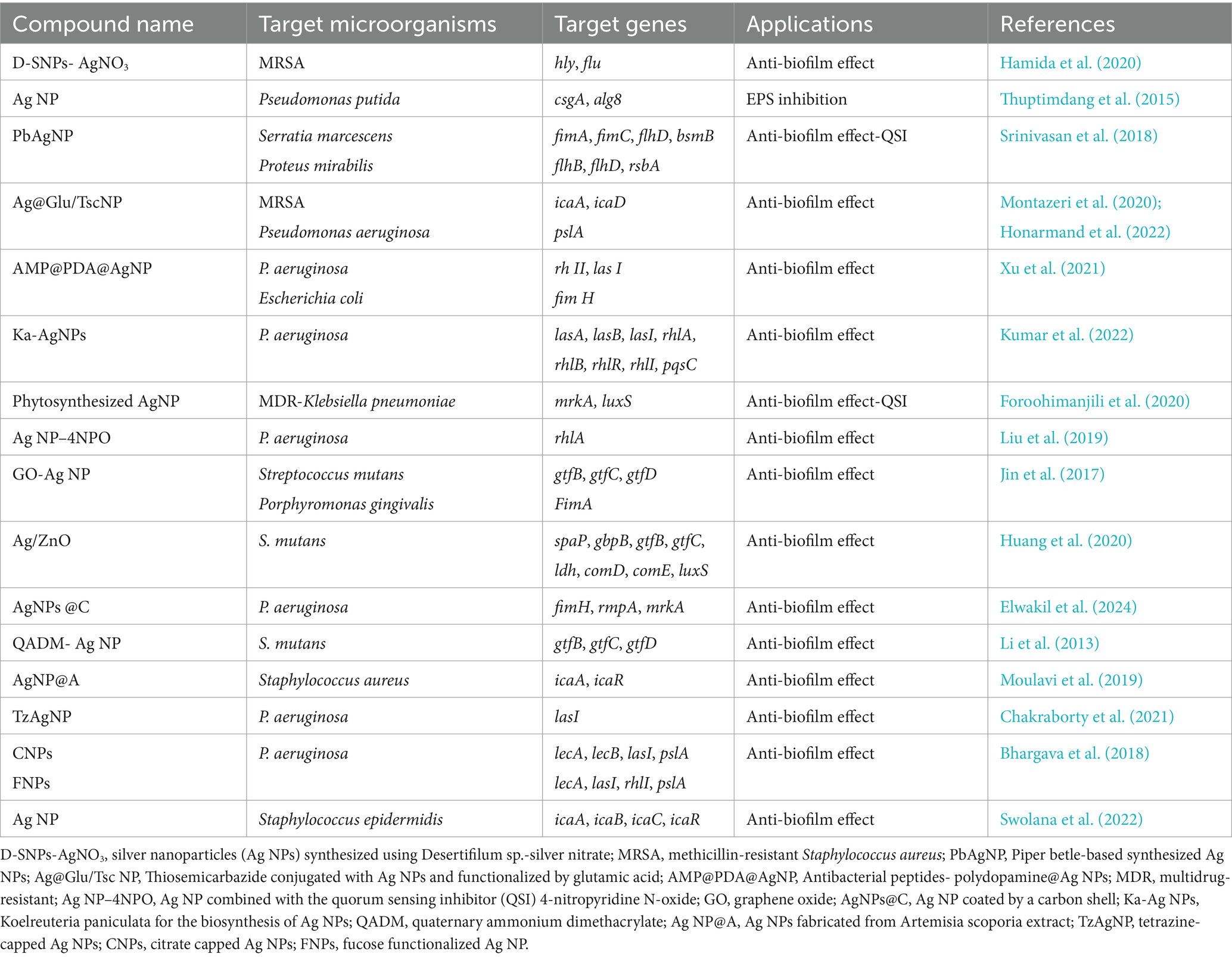
Table 1. Summary of the effect of silver nanoparticles and their related products on the downregulation of bacterial biofilm-related genes.
Ag has a strong inhibitory effect on the strong biofilms produced by Acinetobacter baumannii. In presence of Ag NPs, the expression of major adhesion virulence factors of A. baumannii such as group 2 capsule synthesis (kpsMII) and afa/draBC adhesin genes decreased by 4.6-folds and 3.4-folds, respectively. Furthermore, in comparison to the control, the transcription of biofilm-related genes (bap, OmpA, and csuA/B) was drastically reduced by 4.5-, 3.1-, and 3.1-folds, respectively (Hetta et al., 2021).
Miola et al. (2015) assert that Ag NPs mixed with poly methyl methacrylate (PMMA)-based bone cement significantly reduced biofilm formation. The main mechanism of this Ag NP–PMMA is inhibition of bacterial colonization. Ag hydroxyapatite nanocomposite (Ag/HA-NC) can obviously inhibit S. aureus biofilm formation. Bacterial adhesion and biofilm formation on the Ag/HA-NC coating were significantly lower than on the HA coating. In addition, the expression of atlE, fbe, sap, and iapB genes, all involved in initial attachment of S. aureus was inhibited with Ag/HA-NC. Therefore, it can be concluded that the antibacterial effect of Ag/HA-NC is achieved due to the release of Ag NPs, which supports the clinical use of Ag/HA-NC (Zhao and Ashraf, 2015).
Silencing of QS is a novel approach to combat the pathogenicity of P. aeruginosa. Mycofabricated Ag NPs with metabolites of the soil fungus Rhizopus arrhizus BRS-07 were able to decrease LasIR-RhlIR levels, inhibit biofilm formation, and significantly reduce the expression of QS-regulated genes (lasI, lasR, rhlI, rhlR; Singh et al., 2015). In addition, Ag NPs stabilized with glutathione (GSH-Ag NPs) at a concentration of ½ MIC was found to have anti-biofilm activity in P. aeruginosa by reducing the expression of lasR and lasI genes (Pourmbarak Mahnaie and Mahmoudi, 2020). One study found that in resistant Klebsiella pneumoniae strains, the expression of the efflux pump gene OxqAB was decreased by both commercial and biosynthesized Ag NPs (Dolatabadi et al., 2021). What is more, Ag NPs were potential NPs that showed excellent reduction of biofilm-related genes (fimH, rmpA, and mrkA) in K. pneumoniae (Mousavi et al., 2023).
Wang et al., developed the Ag NP-functionalized titanium implant surface. The Ag NP-doped surface reduced the expression of icaA gene for Staphylococcus epidermidis and fnbA and fnbB genes for methicillin-resistant S. aureus to decrease the production of intercellular polysaccharide adhesin and fibronectin-binding proteins, thereby reducing bacterial adhesion and biofilm formation (Wang et al., 2016). Although Ag NPs have been extensively studied for antibacterial applications, their production and use are subject to many limitations, including the inability to control size and shape, aggregation, colloidal stability, and potential toxicity at high doses (Jena et al., 2012; Zhang et al., 2019).
7.2 ZnO NPs
ZnO NPs are widely known for their antibacterial and anti-biofilm activity against a wide range of bacteria such as P. aeruginosa, Streptococcus pneumoniae, Listeria monocytogens, Salmonella enteritidis, and E. coli with low toxicity to human cells (Mahamuni-Badiger et al., 2020). Thanks to their potent antibacterial action, these NPs can reduce microbial adhesion, proliferation, and biofilm growth (Campoccia et al., 2013). However, their antibacterial activity is influenced by various factors such as UV illumination, size, shape, concentration, surface modifications, and surface defects (Mahamuni-Badiger et al., 2020).
Recently, ZnO NPs have gained even more attention as they possess the highest toxicity against drug resistant microorganisms. ZnO NPs exhibit a different type of antimicrobial action compared to other NPs. They first destroy the bacterial cell wall, then enter the cell, and finally accumulate in the cell membrane, leading to death (Li et al., 2012). These NPs can alter the microenvironment near the bacteria and liberate ROS species and Zn2+ ions cause damaging lipids, proteins, carbohydrates, and DNA by oxidative stress, lipid peroxidation, and disrupting vital cellular functions (Sondi and Salopek-Sondi, 2004). Among NPs, they are strongly preferred because ZnO is recognized as a safe material by the US Food and Drug Administration (De Romana et al., 2002). ZnO meets the requirements for an ideal anti-biofilm agent (Hou et al., 2018).
QS inhibitory effect of ZnO NPs has been demonstrated by reducing the production of P. aeruginosa virulence factors such as rhamnolipids, pyocyanin, pyoverdin, hemolysins, elastase, and proteases. In addition, the inhibitory effect of ZnO NPs on the QS regulatory genes lasI, lasR, rhlI, rhlR, pqsA, and pqsR, which control the secretion of virulence factors, was confirmed (Saleh et al., 2019).
Further, norA, norB, norC, and tet38 are important efflux pump genes that contribute to the MDR phenotype in S. aureus. The simultaneous use of ZnO@glutamic acid–thiosemicarbazide NP at sub-MIC concentrations in combination with ciprofloxacin decreased the expression of norA, norB, norC, and tet38 by 5.4-, 3.8-, 2.1-, and 3.4-fold, respectively, compared to ciprofloxacin alone (Nejabatdoust et al., 2019).
A previous study showed that ZnO/zeolite NC at concentrations below the MIC in combination with chitosan reduced the expression of gtfB, gtfC, and ftf genes in S. mutans by 4.16-, 4.40-, and 2.88-fold, respectively (Afrasiabi et al., 2021). Along these lines, inhibition of the esp gene five-fold in Enterococcus faecalis decreased the ability to form biofilm (Partoazar et al., 2019). According to Khan et al. (2016), the expression level of the gtfB gene decreased 0.93-fold when Streptococcus mitis was grown with 30 μg/mL ZnO NP. Abdelghafar et al. also found a significant decrease in icaA and sarA genes after treatment of S. aureus with ZnO NPs (Abdelghafar et al., 2022). Similarly, sub-MIC of ZnO NPs had a significant effect on the biofilm formation rate of S. aureus and the gene expression of ica A, ica D, and fnb A (Abdelraheem et al., 2021).
7.3 TiO2 NPs
TiO2 NPs have been found as antimicrobial agents against various microorganisms that exert anti-QS activity. TiO2 NPs exhibited improved antibacterial and anti-biofilm activity in comparison to bulk form. In addition, it reduces expression of the efflux pump genes (MexY, MexB, MexA) and QS-regulated genes (lasR, lasI, rhll, rhlR, pqsA, pqsR) of P. aeruginosa. TiO2 NPs can increase the effectiveness of conventional antibiotics (Ahmed et al., 2021). The diminutive size, large surface area, and their ability to penetrate the cell wall are the factors determining the antimicrobial activities (Saranya et al., 2018). Moreover, exposure to UV light leads to the generation of ROS, which may enhance the antibacterial activity of TiO2 NPs (de Dicastillo et al., 2019). Several studies reported the antibacterial and antifungal activities of TiO2 NPs (Anupong et al., 2023; Younis et al., 2023). Abdulazeem et al. found that biofilm growth is completely reduced in A. baumannii, P. aeruginosa, Proteus vulgaris, and Serratia marcescens as TiO2 NPs target sulfhydryl groups in the cell membrane to form S–TiO2 bond. This reaction inhibits the electron transport chain and enzymes needed for biofilm formation (Abdulazeem et al., 2019). However, Abdel-Fatah et al. stated that TiO2 NPs had no bactericidal effect against S. aureus, E. coli, and Bacillus subtilis (Abdel-Fatah et al., 2016). These variations among different results may reflect differences in concentration, zeta potential, particle shape, size, and the examined pathogen (Skandalis et al., 2017).
The overexpression of efflux pump genes was dramatic in non-treated P. aeruginosa, especially the MexY gene. TiO2 NPs significantly reduce the expression of efflux pump genes with its efflux pump inhibitor activity (Ahmed et al., 2021). The mxdABCD complex is a new set of genes that are important essential for biofilm formation and growth and can be influenced by TiO2 NPs. Of these, mxdA gene encoding diguanylate cyclase and mxdB gene encoding glycosyl transferase, both of which confer to cell attachment and EPS synthesis. These genes are required for the three-dimensional of biofilm architecture (Maurer-Jones et al., 2013). In Liu et al. (2015) reported that TiO2 NPs on the surface and inside titania nanotubes reduced gtfB, gtfC, and gtfD genes of S. mutans, which improves the efficacy of orthopedic and dental implants. In a recent research, the antibacterial activity of TiO2 NPs and Ganoderma extract against P. aeruginosa and methicillin-resistant Staphylococcus aureus (MRSA) was investigated. As a result, the algD gene was reduced when TiO2 NPs were used alone or in combination with Ganoderma extract. None of them affected the iacA gene (Marzhoseyni et al., 2023). However, DNA damage induced by TiO2 NPs limits the efficacy of them (Chen et al., 2014).
7.4 Cu NPs
Cu NPs have proven to be efficient antibacterial agents due to their low cost, ease of mixing with polarized liquids such as water, and their relatively stable physical and chemical properties (LewisOscar et al., 2015). Graphene oxide-Cu NC has been shown to alter biofilm architecture, inhibit EPS formation and distribution, and dysregulate the expression of EPS-related genes (Mao et al., 2021). The gtf gene family, a glucosyltransferase (Gtf) encoding gene in S. mutans, can synthesize EPSs directly from sucrose. GTF-produced glucan maintains the integrity of the matrix structure. In exposure to graphene oxide-Cu NCs, the expression of gtfB, gtfC, and gbpB decreased, while the expression of the rnc gene increased. In addition, the expression of vicRKX was suppressed by rnc genes. Cell wall homeostasis and expression of gtfB/C genes can be enhanced by the VicRK two-component system. The group treated with graphene oxide and Cu NCs showed a significant increase in rnc expression, which is consistent with the negative effect of the rnc gene (Mao et al., 2021). Further, it was also reported that Cu/graphitic carbon nitride NCs downregulated icaA gene of S. aureus to reduce biofilm formation (Rashki et al., 2023b).
Cu NPs lowered the biofilm of P. aeruginosa by reducing the ppyR gene. In addition to this, the presence of Cu NPs led to an increase in the expression of the biofilm dispersion locus gene (bdlA). Also, Cu NPs caused a decrease in the rsaL gene as well as the mexA and mex B efflux genes (Singh et al., 2019). Aziz et al. found that the virulence gene-related to adhesion (fimH, papC) of Klebsiella oxytoca was reduced 8.5- and 9.0-fold by using Cu cobalt oxide NPs (Aziz et al., 2023).
The high toxicity of Cu NPs causes oxidative lesions, which limits the effectiveness of these NPs (Naz et al., 2020). Quercetin is a natural substance with anti-QS potential that has anti-biofilm properties. However, it has some disadvantages, including low water solubility and bioavailability issues. Quercetin/Cu NPs have been found to prevent biofilm formation by disrupting the integrity of the cell membrane and processes involved in QS, and by reducing the expression of lasI, lasR, rhlI, and rhIR genes in P. aeruginosa and agrA and icaA in S. aureus, and are non-toxic to mouse fibroblast cells (Cheng et al., 2024).
7.5 Fe3O4 NPs
Fe3O4 NPs have attracted attention due to their unique properties, which include larger surface area, superparamagnetic nature, surface-to-volume ratio, and easy separation process. These NPs have antibacterial and anti-biofilm properties via various mechanisms such as intracellular ROS generation, electrostatic attraction with the cell membrane and its proteins, leading to physical destruction and eventual death of the microbes. In addition, there are a number of advantages, including low toxicity, biocompatibility for medical applications and environmental friendliness (Das et al., 2021).
The clinical management of antibiotic-resistant S. aureus and E. coli raises a number of public health issues. Furthermore, increasingly serious antimicrobial resistance and biofilm formation pose a barrier to conventional therapy. Rhamnolipid (RHL)-coated Fe3O4 NPs- p-coumaric acid (p-CoA) and gallic acid (GA)-polymer common coatings (PVA; RHL-Fe3O4@PVA@p-CoA/GA) can strangely inhibit bacterial growth and biofilm formation via downregulating icaABCD and csgBAC operons, which encode the production of slime layer (icaA and icaD) and the production of curli fimbriae (csgA, csgD, and crl) in S. aureus and E. coli, respectively (Sharaf et al., 2022). Analysis of fimA and csgA genes for biofilm formation of E. coli confirmed the ability of clay halloysite nanotubules with tannic acid and Fe3O4 NPs to prevent bacterial adhesion and biofilm formation (Bu et al., 2024). A summary of the effect of the other NPs on the downregulation of bacterial biofilm-related genes can be found in Table 2.
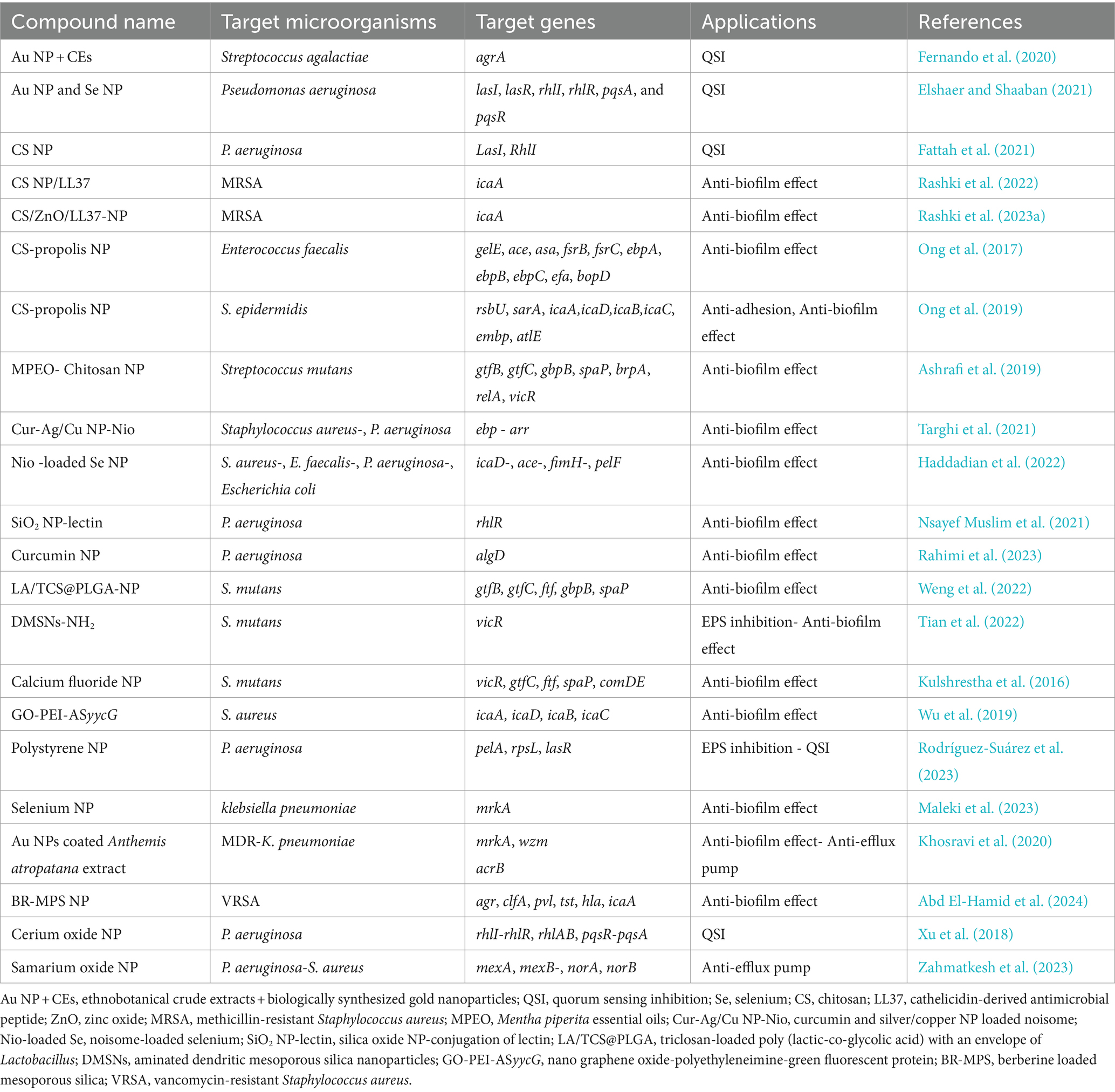
Table 2. Summary of the effect of the other nanoparticles on the downregulation of bacterial biofilm-related genes.
Although metal NPs have been shown to be toxic in human cells, efforts are being focused on reducing their toxicity through various strategies, e.g., by producing composites with clays, doping with polymers and proteins, etc. (Odatsu et al., 2020). The limited use of NPs in clinical applications is due to the incomplete understanding of their side effects, and further studies are needed to utilize them more effectively. The routine use of NPs in the fight against bacterial infections will be possible once the toxicity of NPs is clarified by detailed in vivo and clinical studies. The correct determination of the threshold dose and exposure time is essential for the safe administration of NPs (Ozdal and Gurkok, 2022).
8 Conclusion
Reviewing the studies revealed that NPs target the gene expression and biomolecular of bacterial biofilm, resulting in destroying or hindering biofilm formation. NPs based on Ag NPs, ZnO NPs, TiO2 NPs, and Cu NPs are highly effective against biofilms of important pathogens due to their ability to inhibit lasI, lasR, rhlI, rhlR of P. aeruginosa, gtfB, gtfC of S. mutans, and icaA of S. aureus. Fe3O4 NPs reduced biofilm formation of S. aureus by inhibiting icaA/D expression. The versatility of NPs in combat with bacterial biofilm causes cells not to have enough opportunity to bypass or trigger the resistance mechanisms that are affected under gene regulation. For instance, NPs directly or via their derivatives like ROS or released ions, inhibit genes of virulence factors in bacteria. This phenomenon hinders the main factors that are necessary for the development and persistency of the biofilm. The bacterial genes associated with the flagella synthesis and other adhesion factors, penetration or internalization of vital elements, QS regulation, efflux pump, oxidative stress, etc. can be suppressed during exposure with the kind of NPs. Even though the antibacterial activity of NPs has been extensively studied, the mechanisms of action of them on biofilm regulatory genes are still poorly understood and controversial. So far, investigation of the mechanisms of action of NPs on the biofilm cells has developed targeted therapy and increased antibacterial efficacy. On the other hand, potential mutagenicity of the NPs may produce new dangerous antibiotic-resistant strains. The evolution of bacteria can be accelerated by NPs especially under co-culture in biofilm conditions. The use of NPs on an industrial scale can also have serious consequences for the environment which address specific issues to advance in research. As there is a necessity to develop new approaches to combat bacterial biofilms, new information about the efficacy of NPs against biofilms, the mechanisms of action on cells, mutagenicity, and genotoxicity will be also crucial.
Author contributions
SA: Conceptualization, Writing – original draft, Writing – review & editing. AP: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abd El-Hamid, M. I., Ibrahim, D., Elazab, S. T., Gad, W. M., Shalaby, M., El-Neshwy, W. M., et al. (2024). Tackling strong biofilm and multi-virulent vancomycin-resistant Staphylococcus aureus via natural alkaloid-based porous nanoparticles: perspective towards near future eradication. Front. Cell. Infect. Microbiol. 13:1287426. doi: 10.3389/fcimb.2023.1287426
Abdel-Fatah, W. I., Gobara, M. M., Mustafa, S. F., Ali, G. W., and Guirguis, O. W. (2016). Role of silver nanoparticles in imparting antimicrobial activity of titanium dioxide. Mater. Lett. 179, 190–193. doi: 10.1016/j.matlet.2016.05.063
Abdelghafar, A., Yousef, N., and Askoura, M. (2022). Zinc oxide nanoparticles reduce biofilm formation, synergize antibiotics action and attenuate Staphylococcus aureus virulence in host; an important message to clinicians. BMC Microbiol. 22:244. doi: 10.1186/s12866-022-02658-z
Abdelraheem, W. M., Abdelkader, A. E., Mohamed, E. S., and Mohammed, M. S. (2020). Detection of biofilm formation and assessment of biofilm genes expression in different Pseudomonas aeruginosa clinical isolates. Meta Gene 23:100646. doi: 10.1016/j.mgene.2020.100646
Abdelraheem, W. M., Khairy, R. M., Zaki, A. I., and Zaki, S. H. (2021). Effect of ZnO nanoparticles on methicillin, vancomycin, linezolid resistance and biofilm formation in Staphylococcus aureus isolates. Ann. Clin. Microbiol. Antimicrob. 20:54. doi: 10.1186/s12941-021-00459-2
Abdulazeem, L., Al-Amiedi, B., Alrubaei, H. A., and Al-Mawlah, Y. H. (2019). Titanium dioxide nanoparticles as antibacterial agents against some pathogenic bacteria. Drug Invent. Today 12, 963–967.
Afrasiabi, S., Bahador, A., and Partoazar, A. (2021). Combinatorial therapy of chitosan hydrogel-based zinc oxide nanocomposite attenuates the virulence of Streptococcus mutans. BMC Microbiol. 21:62. doi: 10.1186/s12866-021-02128-y
Ahmed, F. Y., Aly, U. F., Abd El-Baky, R. M., and Waly, N. G. (2021). Effect of titanium dioxide nanoparticles on the expression of efflux pump and quorum-sensing genes in Mdr Pseudomonas aeruginosa isolates. Antibiotics 10:625. doi: 10.3390/antibiotics10060625
Alav, I., Sutton, J. M., and Rahman, K. M. (2018). Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 73, 2003–2020. doi: 10.1093/jac/dky042
Alshammari, M., Ahmad, A., Alkhulaifi, M., Al Farraj, D., Alsudir, S., Alarawi, M., et al. (2023). Reduction of biofilm formation of Escherichia coli by targeting quorum sensing and adhesion genes using the Crispr/Cas9-Hdr approach, and its clinical application on urinary catheter. J. Infect. Public Health 16, 1174–1183. doi: 10.1016/j.jiph.2023.05.026
Anupong, W., On-Uma, R., Jutamas, K., Salmen, S. H., Alharbi, S. A., Joshi, D., et al. (2023). Antibacterial, antifungal, antidiabetic, and antioxidant activities potential of Coleus aromaticus synthesized titanium dioxide nanoparticles. Environ. Res. 216:114714. doi: 10.1016/j.envres.2022.114714
Arora, A. (2024). Crispri-mediated gene silencing in biofilm cycle and quorum sensing. Gene Editing in Plants: Crispr-Cas and Its Applications, 139–178. doi: 10.1007/978-981-99-8529-6_6
Ashrafi, B., Rashidipour, M., Marzban, A., Soroush, S., Azadpour, M., Delfani, S., et al. (2019). Mentha piperita essential oils loaded in a chitosan nanogel with inhibitory effect on biofilm formation against S. mutans on the dental surface. Carbohydr. Polym. 212, 142–149. doi: 10.1016/j.carbpol.2019.02.018
Askoura, M., Almalki, A. J., Lila, A. S. A., Almansour, K., Alshammari, F., Khafagy, E.-S., et al. (2021). Alteration of Salmonella enterica virulence and host pathogenesis through targeting sdiA by using the Crispr-Cas9 system. Microorganisms 9:2564. doi: 10.3390/microorganisms9122564
Asma, S. T., Imre, K., Morar, A., Herman, V., Acaroz, U., Mukhtar, H., et al. (2022). An overview of biofilm formation–combating strategies and mechanisms of action of antibiofilm agents. Lifestyles 12:1110. doi: 10.3390/life12081110
Aziz, S. N., Al-Kadmy, I. M., Rheima, A. M., Al-Sallami, K. J., Abd Ellah, N. H., El-Saber Batiha, G., et al. (2023). Binary CuO\CoO nanoparticles inhibit biofilm formation and reduce the expression of papC and fimH genes in multidrug-resistant Klebsiella oxytoca. Mol. Biol. Rep. 50, 5969–5976. doi: 10.1007/s11033-023-08447-9
Baig, N., Kammakakam, I., and Falath, W. (2021). Nanomaterials: a review of synthesis methods, properties, recent progress, and challenges. Mater. Adv 2, 1821–1871. doi: 10.1039/D0MA00807A
Bhargava, A., Pareek, V., Roy Choudhury, S., Panwar, J., and Karmakar, S. (2018). Superior bactericidal efficacy of fucose-functionalized silver nanoparticles against Pseudomonas aeruginosa Pao1 and prevention of its colonization on urinary catheters. Acs Appl Bio Mater 10, 29325–29337. doi: 10.1021/acsami.8b09475
Bu, K.-B., Kim, M., Sung, J.-S., and Kadam, A. A. (2024). Halloysite nanotubes decorated with Fe3O4 nanoparticles and tannic acid for effective inhibition of E. coli biofilm. Acs Appl Nano Mater 7, 1, 313–322. doi: 10.1021/acsanm.3c04518
Campoccia, D., Montanaro, L., and Arciola, C. R. (2013). A review of the biomaterials technologies for infection-resistant surfaces. Biomaterials 34, 8533–8554. doi: 10.1016/j.biomaterials.2013.07.089
Cangui-Panchi, S. P., Ñacato-Toapanta, A. L., Enríquez-Martínez, L. J., Reyes, J., Garzon-Chavez, D., and Machado, A. (2022). Biofilm-forming microorganisms causing hospital-acquired infections from intravenous catheter: a systematic review. Curr Res Microb Sci. 3:100175. doi: 10.1016/j.crmicr.2022.100175
Chakraborty, P., Paul, P., Kumari, M., Bhattacharjee, S., Singh, M., Maiti, D., et al. (2021). Attenuation of Pseudomonas aeruginosa biofilm by thymoquinone: an individual and combinatorial study with tetrazine-capped silver nanoparticles and tryptophan. Folia Microbiol. 66, 255–271. doi: 10.1007/s12223-020-00841-1
Chen, Z., Wang, Y., Ba, T., Li, Y., Pu, J., Chen, T., et al. (2014). Genotoxic evaluation of titanium dioxide nanoparticles in vivo and in vitro. Toxicol. Lett. 226, 314–319. doi: 10.1016/j.toxlet.2014.02.020
Cheng, J., Zhang, H., Lu, K., Zou, Y., Jia, D., Yang, H., et al. (2024). Bi-functional quercetin/copper nanoparticles integrating bactericidal and anti-quorum sensing properties for preventing the formation of biofilms. Biomater. Sci. 12, 1788–1800. doi: 10.1039/D4BM00034J
Colino, C. I., Millán, C. G., and Lanao, J. M. (2018). Nanoparticles for signaling in biodiagnosis and treatment of infectious diseases. Int. J. Mol. Sci. 19:1627. doi: 10.3390/ijms19061627
Czaczyk, K., and Myszka, K. (2007). Biosynthesis of extracellular polymeric substances (eps) and its role in microbial biofilm formation. Pol. J. Environ. Stud. 16, 799–806.
Das, P., Ghosh, S., and Nayak, B. (2021). Phyto-fabricated nanoparticles and their anti-biofilm activity: Progress and current status. Front Nanotechnol 3:739286. doi: 10.3389/fnano.2021.739286
De Dicastillo, C. L., Patiño, C., Galotto, M. J., Vásquez-Martínez, Y., Torrent, C., Alburquenque, D., et al. (2019). Novel hollow titanium dioxide nanospheres with antimicrobial activity against resistant bacteria. Beilstein J Nanotechnol 10, 1716–1725. doi: 10.3762/bjnano.10.167
De Romana, D. L., Brown, K., and Guinard, J. X. (2002). Sensory trial to assess the acceptability of zinc fortificants added to iron-fortified wheat products. J. Food Sci. 67, 461–465. doi: 10.1111/j.1365-2621.2002.tb11429.x
Dolatabadi, A., Noorbazargan, H., Khayam, N., Moulavi, P., Zamani, N., Asghari Lalami, Z., et al. (2021). Ecofriendly biomolecule-capped Bifidobacterium bifidum-manufactured silver nanoparticles and efflux pump genes expression alteration in Klebsiella pneumoniae. Microb. Drug Resist. 27, 247–257. doi: 10.1089/mdr.2019.0366
Elshaer, S. L., and Shaaban, M. I. (2021). Inhibition of quorum sensing and virulence factors of Pseudomonas aeruginosa by biologically synthesized gold and selenium nanoparticles. Antibiotics 10:1461. doi: 10.3390/antibiotics10121461
Elwakil, B. H., Eldrieny, A. M., Almotairy, A. R. Z., and El-Khatib, M. (2024). Potent biological activity of newly fabricated silver nanoparticles coated by a carbon shell synthesized by electrical arc. Sci. Rep. 14, 1–12. doi: 10.1038/s41598-024-54648-y
Fattah, R. A. F. A., Mohamed, T. A. H., and Elsayed, M. S. (2021). Effect of chitosan nanoparticles on quorum sensing-controlled virulence factors and expression of LasI and RhlI genes among Pseudomonas aeruginosa clinical isolates. Aims microbiol 7, 415–430. doi: 10.3934/microbiol.2021025
Fernando, S. I. D., Judan Cruz, K. G., and Watanabe, K. (2020). Quorum sensing-linked agrA expression by ethno-synthesized gold nanoparticles in Tilapia Streptococcus agalactiae biofilm formation. BioNanoScience 10, 696–704. doi: 10.1007/s12668-020-00758-6
Foroohimanjili, F., Mirzaie, A., Hamdi, S. M. M., Noorbazargan, H., Hedayati Ch, M., Dolatabadi, A., et al. (2020). Antibacterial, antibiofilm, and antiquorum sensing activities of phytosynthesized silver nanoparticles fabricated from Mespilus germanica extract against multidrug resistance of Klebsiella pneumoniae clinical strains. J. Basic Microbiol. 60, 216–230. doi: 10.1002/jobm.201900511
Gestel, J. V., Vlamakis, H., and Kolter, R. (2015). Division of labor in biofilms: the ecology of cell differentiation. Microbiol Spectr 3:MB-0002-2014. doi: 10.1128/microbiolspec.MB-0002-2014
Gupta, P., Sarkar, S., Das, B., Bhattacharjee, S., and Tribedi, P. (2016). Biofilm, pathogenesis and prevention—a journey to break the wall: a review. Arch. Microbiol. 198, 1–15. doi: 10.1007/s00203-015-1148-6
Gupta, D., Singh, A., and Khan, A. U. (2017). Nanoparticles as efflux pump and biofilm inhibitor to rejuvenate bactericidal effect of conventional antibiotics. Nanoscale Res. Lett. 12:454. doi: 10.1186/s11671-017-2222-6
Haddadian, A., Robattorki, F. F., Dibah, H., Soheili, A., Ghanbarzadeh, E., Sartipnia, N., et al. (2022). Niosomes-loaded selenium nanoparticles as a new approach for enhanced antibacterial, anti-biofilm, and anticancer activities. Sci. Rep. 12:21938. doi: 10.1038/s41598-022-26400-x
Hamida, R. S., Ali, M. A., Goda, D. A., Khalil, M. I., and Al-Zaban, M. I. (2020). Novel biogenic silver nanoparticle-induced reactive oxygen species inhibit the biofilm formation and virulence activities of methicillin-resistant Staphylococcus aureus (Mrsa) strain. Front. Bioeng. Biotechnol. 8:433. doi: 10.3389/fbioe.2020.00433
Hemeg, H. A. (2017). Nanomaterials for alternative antibacterial therapy. Int. J. Nanomedicine 12, 8211–8225. doi: 10.2147/IJN.S132163
Hetta, H. F., Al-Kadmy, I. M., Khazaal, S. S., Abbas, S., Suhail, A., El-Mokhtar, M. A., et al. (2021). Antibiofilm and antivirulence potential of silver nanoparticles against multidrug-resistant Acinetobacter baumannii. Sci. Rep. 11:10751. doi: 10.1038/s41598-021-90208-4
Holling, N., Lednor, D., Tsang, S., Bissell, A., Campbell, L., Nzakizwanayo, J., et al. (2014). Elucidating the genetic basis of crystalline biofilm formation in Proteus mirabilis. Infect. Immun. 82, 1616–1626. doi: 10.1128/IAI.01652-13
Honarmand, T., Sharif, A. P., Salehzadeh, A., Jalali, A., and Nikokar, I. (2022). Does conjugation of silver nanoparticles with thiosemicarbazide increase their antibacterial properties? Microb. Drug Resist. 28, 293–305. doi: 10.1089/mdr.2020.0557
Hong, K.-W., Koh, C.-L., Sam, C.-K., Yin, W.-F., and Chan, K.-G. (2012). Quorum quenching revisited—from signal decays to signalling confusion. Sensors 12, 4661–4696. doi: 10.3390/s120404661
Hou, J., Wu, Y., Li, X., Wei, B., Li, S., and Wang, X. (2018). Toxic effects of different types of zinc oxide nanoparticles on algae, plants, invertebrates, vertebrates and microorganisms. Chemosphere 193, 852–860. doi: 10.1016/j.chemosphere.2017.11.077
Huang, Q., Wang, S., Sun, Y., Shi, C., Yang, H., and Lu, Z. (2020). Effects of ag/ZnO nanocomposite at sub-minimum inhibitory concentrations on virulence factors of Streptococcus mutans. Arch. Oral Biol. 111:104640. doi: 10.1016/j.archoralbio.2019.104640
Jena, P., Mohanty, S., Mallick, R., Jacob, B., and Sonawane, A. (2012). Toxicity and antibacterial assessment of chitosancoated silver nanoparticles on human pathogens and macrophage cells. Int. J. Nanomedicine 7, 1805–1818. doi: 10.2147/IJN.S28077
Jiang, Y., Geng, M., and Bai, L. (2020). Targeting biofilms therapy: current research strategies and development hurdles. Microorganisms 8:1222. doi: 10.3390/microorganisms8081222
Jin, J., Zhang, L., Shi, M., Zhang, Y., and Wang, Q. (2017). Ti-go-ag nanocomposite: the effect of content level on the antimicrobial activity and cytotoxicity. Int. J. Nanomedicine 12, 4209–4224. doi: 10.2147/IJN.S134843
Kalishwaralal, K., Barathmanikanth, S., Pandian, S. R. K., Deepak, V., and Gurunathan, S. (2010). Silver nanoparticles impede the biofilm formation by Pseudomonas aeruginosa and Staphylococcus epidermidis. Colloids Surf. B: Biointerfaces 79, 340–344. doi: 10.1016/j.colsurfb.2010.04.014
Karatan, E., and Watnick, P. (2009). Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347. doi: 10.1128/MMBR.00041-08
Khan, S. T., Ahmad, J., Ahamed, M., Musarrat, J., and Al-Khedhairy, A. A. (2016). Zinc oxide and titanium dioxide nanoparticles induce oxidative stress, inhibit growth, and attenuate biofilm formation activity of Streptococcus mitis. J. Biol. Inorg. Chem. 21, 295–303. doi: 10.1007/s00775-016-1339-x
Khosravi, M., Mirzaie, A., Kashtali, A. B., and Noorbazargan, H. (2020). Antibacterial, anti-efflux, anti-biofilm, anti-slime (exopolysaccharide) production and urease inhibitory efficacies of novel synthesized gold nanoparticles coated Anthemis atropatana extract against multidrug-resistant Klebsiella pneumoniae strains. Arch. Microbiol. 202, 2105–2115. doi: 10.1007/s00203-020-01930-y
Kot, B., Sytykiewicz, H., and Sprawka, I. (2018). Expression of the biofilm-associated genes in methicillin-resistant Staphylococcus aureus in biofilm and planktonic conditions. Int. J. Mol. Sci. 19:3487. doi: 10.3390/ijms19113487
Kulshrestha, S., Khan, S., Hasan, S., Khan, M. E., Misba, L., and Khan, A. U. (2016). Calcium fluoride nanoparticles induced suppression of Streptococcus mutans biofilm: an in vitro and in vivo approach. Appl. Microbiol. Biotechnol. 100, 1901–1914. doi: 10.1007/s00253-015-7154-4
Kumar, S., Paliya, B. S., and Singh, B. N. (2022). Superior inhibition of virulence and biofilm formation of Pseudomonas aeruginosa Pao1 by phyto-synthesized silver nanoparticles through anti-quorum sensing activity. Microb. Pathog. 170:105678. doi: 10.1016/j.micpath.2022.105678
Lahiri, D., Nag, M., Sheikh, H. I., Sarkar, T., Edinur, H. A., Pati, S., et al. (2021). Microbiologically-synthesized nanoparticles and their role in silencing the biofilm signaling cascade. Front. Microbiol. 12:636588. doi: 10.3389/fmicb.2021.636588
Le Ouay, B., and Stellacci, F. (2015). Antibacterial activity of silver nanoparticles: a surface science insight. Nano Today 10, 339–354. doi: 10.1016/j.nantod.2015.04.002
Lee, S. H., and Jun, B.-H. (2019). Silver nanoparticles: synthesis and application for nanomedicine. Int. J. Mol. Sci. 20:865. doi: 10.3390/ijms20040865
Lewisoscar, F., Mubarakali, D., Nithya, C., Priyanka, R., Gopinath, V., Alharbi, N. S., et al. (2015). One pot synthesis and anti-biofilm potential of copper nanoparticles (Cunps) against clinical strains of Pseudomonas aeruginosa. Biofouling 31, 379–391. doi: 10.1080/08927014.2015.1048686
Li, C.-H., Shen, C.-C., Cheng, Y.-W., Huang, S.-H., Wu, C.-C., Kao, C.-C., et al. (2012). Organ biodistribution, clearance, and genotoxicity of orally administered zinc oxide nanoparticles in mice. Nanotoxicology 6, 746–756. doi: 10.3109/17435390.2011.620717
Li, F., Weir, M. D., Chen, J., and Xu, H. H. (2013). Comparison of quaternary ammonium-containing with nano-silver-containing adhesive in antibacterial properties and cytotoxicity. Dent. Mater. 29, 450–461. doi: 10.1016/j.dental.2013.01.012
Liao, C., Li, Y., and Tjong, S. C. (2019). Bactericidal and cytotoxic properties of silver nanoparticles. Int. J. Mol. Sci. 20:449. doi: 10.3390/ijms20020449
Liu, L., Li, J.-H., Zi, S.-F., Liu, F.-R., Deng, C., Ao, X., et al. (2019). Agnp combined with quorum sensing inhibitor increased the antibiofilm effect on Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 103, 6195–6204. doi: 10.1007/s00253-019-09905-w
Liu, W., Su, P., Chen, S., Wang, N., Wang, J., Liu, Y., et al. (2015). Antibacterial and osteogenic stem cell differentiation properties of photoinduced TiO2 nanoparticle-decorated TiO2 nanotubes. Nanomedicine 10, 713–723. doi: 10.2217/nnm.14.183
Mahamuni-Badiger, P. P., Patil, P. M., Badiger, M. V., Patel, P. R., Thorat-Gadgil, B. S., Pandit, A., et al. (2020). Biofilm formation to inhibition: role of zinc oxide-based nanoparticles. Mater. Sci. Eng. C Mater. Biol. Appl. 108:110319. doi: 10.1016/j.msec.2019.110319
Maleki, A. R., Tabatabaei, R. R., Aminian, F., Ranjbar, S., Ashrafi, F., and Ranjbar, R. (2023). Antibacterial and antibiofilm effects of green synthesized selenium nanoparticles on clinical Klebsiella pneumoniae isolates. J. Basic Microbiol. 63, 1373–1382. doi: 10.1002/jobm.202300332
Mao, M., Zhang, W., Huang, Z., Huang, J., Wang, J., Li, W., et al. (2021). Graphene oxide-copper nanocomposites suppress cariogenic Streptococcus mutans biofilm formation. Int. J. Nanomedicine 16, 7727–7739. doi: 10.2147/IJN.S303521
Marzhoseyni, Z., Rashki, S., and Nazari-Alam, A. (2023). Evaluation of the inhibitory effects of TiO2 nanoparticle and Ganoderma lucidum extract against biofilm-producing bacteria isolated from clinical samples. Arch. Microbiol. 205:59. doi: 10.1007/s00203-023-03403-4
Maurer-Jones, M. A., Gunsolus, I. L., Meyer, B. M., Christenson, C. J., and Haynes, C. L. (2013). Impact of TiO2 nanoparticles on growth, biofilm formation, and flavin secretion in Shewanella oneidensis. Anal. Chem. 85, 5810–5818. doi: 10.1021/ac400486u
May, T., Ito, A., and Okabe, S. (2009). Induction of multidrug resistance mechanism in Escherichia coli biofilms by interplay between tetracycline and ampicillin resistance genes. Antimicrob. Agents Chemother. 53, 4628–4639. doi: 10.1128/AAC.00454-09
Miola, M., Fucale, G., Maina, G., and Verne, E. (2015). Antibacterial and bioactive composite bone cements containing surface silver-doped glass particles. Biomed. Mater. 10:055014. doi: 10.1088/1748-6041/10/5/055014
Mohanta, Y. K., Chakrabartty, I., Mishra, A. K., Chopra, H., Mahanta, S., Avula, S. K., et al. (2023). Nanotechnology in combating biofilm: a smart and promising therapeutic strategy. Front. Microbiol. 13:1028086. doi: 10.3389/fmicb.2022.1028086
Montazeri, A., Salehzadeh, A., and Zamani, H. (2020). Effect of silver nanoparticles conjugated to thiosemicarbazide on biofilm formation and expression of intercellular adhesion molecule genes, icaad, in Staphylococcus aureus. Folia Microbiol. 65, 153–160. doi: 10.1007/s12223-019-00715-1
Moulavi, P., Noorbazargan, H., Dolatabadi, A., Foroohimanjili, F., Tavakoli, Z., Mirzazadeh, S., et al. (2019). Antibiofilm effect of green engineered silver nanoparticles fabricated from Artemisia scoporia extract on the expression of icaA and icaR genes against multidrug-resistant Staphylococcus aureus. J. Basic Microbiol. 59, 701–712. doi: 10.1002/jobm.201900096
Mousavi, S. M., Mousavi, S. M., Moeinizadeh, M., Aghajanidelavar, M., Rajabi, S., and Mirshekar, M. (2023). Evaluation of biosynthesized silver nanoparticles effects on expression levels of virulence and biofilm-related genes of multidrug-resistant Klebsiella pneumoniae isolates. J. Basic Microbiol. 63, 632–645. doi: 10.1002/jobm.202200612
Muhammad, M. H., Idris, A. L., Fan, X., Guo, Y., Yu, Y., Jin, X., et al. (2020). Beyond risk: bacterial biofilms and their regulating approaches. Front. Microbiol. 11:928. doi: 10.3389/fmicb.2020.00928
Naz, S., Gul, A., and Zia, M. (2020). Toxicity of copper oxide nanoparticles: a review study. IET Nanobiotechnol. 14, 1–13. doi: 10.1049/iet-nbt.2019.0176
Nejabatdoust, A., Zamani, H., and Salehzadeh, A. (2019). Functionalization of ZnO nanoparticles by glutamic acid and conjugation with thiosemicarbazide alters expression of efflux pump genes in multiple drug-resistant Staphylococcus aureus strains. Microb. Drug Resist. 25, 966–974. doi: 10.1089/mdr.2018.0304
Nsayef Muslim, S., Mohammed Ali, A. N., and Auda, I. G. (2021). Anti-biofilm and anti-virulence effects of silica oxide nanoparticle–conjugation of lectin purified from Pseudomonas aeruginosa. IET Nanobiotechnol. 15, 318–328. doi: 10.1049/nbt2.12022
Odatsu, T., Kuroshima, S., Sato, M., Takase, K., Valanezhad, A., Naito, M., et al. (2020). Antibacterial properties of nano-ag coating on healing abutment: an in vitro and clinical study. Antibiotics 9:347. doi: 10.3390/antibiotics9060347
Ong, T. H., Chitra, E., Ramamurthy, S., Ling, C. C. S., Ambu, S. P., and Davamani, F. (2019). Cationic chitosan-propolis nanoparticles alter the zeta potential of S. epidermidis, inhibit biofilm formation by modulating gene expression and exhibit synergism with antibiotics. PLoS One 14:e0213079. doi: 10.1371/journal.pone.0213079
Ong, T. H., Chitra, E., Ramamurthy, S., Siddalingam, R. P., Yuen, K. H., Ambu, S. P., et al. (2017). Chitosan-propolis nanoparticle formulation demonstrates anti-bacterial activity against Enterococcus faecalis biofilms. PLoS One 12:e0174888. doi: 10.1371/journal.pone.0174888
Ozdal, M., and Gurkok, S. (2022). Recent advances in nanoparticles as antibacterial agent. Admet Dmpk 10, 115–129. doi: 10.5599/admet.1172
Palmer, J., Flint, S., and Brooks, J. (2007). Bacterial cell attachment, the beginning of a biofilm. J. Ind. Microbiol. Biotechnol. 34, 577–588. doi: 10.1007/s10295-007-0234-4
Partoazar, A., Talaei, N., Bahador, A., Pourhajibagher, M., Dehpour, S., Sadati, M., et al. (2019). Antibiofilm activity of natural zeolite supported NanoZnO: inhibition of Esp gene expression of Enterococcus faecalis. Nanomedicine 14, 675–687. doi: 10.2217/nnm-2018-0173
Pourmbarak Mahnaie, M., and Mahmoudi, H. (2020). Effect of glutathione-stabilized silver nanoparticles on expression of las I and las R of the genes in Pseudomonas aeruginosa strains. Eur. J. Med. Res. 25:17. doi: 10.1186/s40001-020-00415-4
Qayyum, S., and Khan, A. U. (2016). Nanoparticles vs. biofilms: a battle against another paradigm of antibiotic resistance. Med. Chem. Commun. 7, 1479–1498. doi: 10.1039/C6MD00124F
Rahimi, M., Piroozmand, A., Shayestehpour, M., Salamat, S., Peik Falak, F., Shakerimoghaddam, A., et al. (2023). Effect of curcumin nanoparticles and alcoholic extract of Falcaria vulgaris on the growth rate, biofilm, and gene expression in Pseudomonas aeruginosa isolated from burn wound infection. Mol. Biol. Rep. 50, 6681–6690. doi: 10.1007/s11033-023-08559-2
Rashki, S., Dawi, E. A., Zilaei, M. R., Safardoust-Hojaghan, H., Ghanbari, M., Ryadh, A., et al. (2023a). ZnO/chitosan nanocomposites as a new approach for delivery Ll37 and evaluation of the inhibitory effects against biofilm-producing methicillin-resistant Staphylococcus aureus isolated from clinical samples. Int. J. Biol. Macromol. 253:127583. doi: 10.1016/j.ijbiomac.2023.127583
Rashki, S., Ghanbari, M., Khudhair, Z. H., Marzhoseyni, Z., Bameri, Z., Afsharikhah, S., et al. (2023b). Evaluate the effect of graphitic carbon nitride nanosheets decorated with copper nanoparticles on biofilm and icaA gene expression in Staphylococcus aureus isolated from clinical samples. Arab. J. Chem. 16:104882. doi: 10.1016/j.arabjc.2023.104882
Rashki, S., Safardoust-Hojaghan, H., Mirzaei, H., Abdulsahib, W. K., Mahdi, M. A., Salavati-Niasari, M., et al. (2022). Delivery Ll37 by chitosan nanoparticles for enhanced antibacterial and antibiofilm efficacy. Carbohydr. Polym. 291:119634. doi: 10.1016/j.carbpol.2022.119634
Ren, D., Bedzyk, L., Thomas, S., Ye, R., and Wood, T. K. (2004). Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64, 515–524. doi: 10.1007/s00253-003-1517-y
Renner, L. D., and Weibel, D. B. (2011). Physicochemical regulation of biofilm formation. MRS Bull. 36, 347–355. doi: 10.1557/mrs.2011.65
Rodríguez-Suárez, J. M., Gershenson, A., Onuh, T. U., and Butler, C. S. (2023). The heterogeneous diffusion of polystyrene nanoparticles and the effect on the expression of quorum-sensing genes and eps production as a function of particle charge and biofilm age. Environ. Sci. Nano 10, 2551–2565. doi: 10.1039/D3EN00219E
Roy, R., Tiwari, M., Donelli, G., and Tiwari, V. (2018). Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence 9, 522–554. doi: 10.1080/21505594.2017.1313372
Sadekuzzaman, M., Yang, S., Mizan, M., and Ha, S. (2015). Current and recent advanced strategies for combating biofilms. Compr. Rev. Food Sci. Food Saf. 14, 491–509. doi: 10.1111/1541-4337.12144
Saleh, M. M., Sadeq, R.'. A., Abdel Latif, H. K., Abbas, H. A., and Askoura, M. (2019). Zinc oxide nanoparticles inhibits quorum sensing and virulence in Pseudomonas aeruginosa. Afr. Health Sci. 19, 2043–2055. doi: 10.4314/ahs.v19i2.28
Sánchez, P., Linares, J. F., Ruiz-Díez, B., Campanario, E., Navas, A., Baquero, F., et al. (2002). Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 50, 657–664. doi: 10.1093/jac/dkf185
Saranya, K. S., Vellora Thekkae Padil, V., Senan, C., Pilankatta, R., Saranya, K., George, B., et al. (2018). Green synthesis of high temperature stable anatase titanium dioxide nanoparticles using gum kondagogu: characterization and solar driven photocatalytic degradation of organic dye. Nano 8:1002. doi: 10.3390/nano8121002
Shahverdi, A. R., Fakhimi, A., Shahverdi, H. R., and Minaian, S. (2007). Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomedicine 3, 168–171. doi: 10.1016/j.nano.2007.02.001
Sharaf, M., Sewid, A. H., Hamouda, H., Elharrif, M. G., El-Demerdash, A. S., Alharthi, A., et al. (2022). Rhamnolipid-coated iron oxide nanoparticles as a novel multitarget candidate against major foodborne E. coli serotypes and methicillin-resistant S. aureus. Microbiol Spectr 10, e00250–e00222. doi: 10.1128/spectrum.00250-22
Sharma, D., Misba, L., and Khan, A. U. (2019). Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 8:76. doi: 10.1186/s13756-019-0533-3
Shkodenko, L., Kassirov, I., and Koshel, E. (2020). Metal oxide nanoparticles against bacterial biofilms: perspectives and limitations. Microorganisms 8:1545. doi: 10.3390/microorganisms8101545
Singh, N., Paknikar, K. M., and Rajwade, J. (2019). Gene expression is influenced due to ‘nano’and ‘ionic’copper in pre-formed Pseudomonas aeruginosa biofilms. Environ. Res. 175, 367–375. doi: 10.1016/j.envres.2019.05.034
Singh, B. R., Singh, B. N., Singh, A., Khan, W., Naqvi, A. H., and Singh, H. B. (2015). Mycofabricated biosilver nanoparticles interrupt Pseudomonas aeruginosa quorum sensing systems. Sci. Rep. 5:13719. doi: 10.1038/srep13719
Skandalis, N., Dimopoulou, A., Georgopoulou, A., Gallios, N., Papadopoulos, D., Tsipas, D., et al. (2017). The effect of silver nanoparticles size, produced using plant extract from Arbutus unedo, on their antibacterial efficacy. Nano 7:178. doi: 10.3390/nano7070178
Sondi, I., and Salopek-Sondi, B. (2004). Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for gram-negative bacteria. J. Colloid Interface Sci. 275, 177–182. doi: 10.1016/j.jcis.2004.02.012
Srinivasan, R., Vigneshwari, L., Rajavel, T., Durgadevi, R., Kannappan, A., Balamurugan, K., et al. (2018). Biogenic synthesis of silver nanoparticles using Piper betle aqueous extract and evaluation of its anti-quorum sensing and antibiofilm potential against uropathogens with cytotoxic effects: an in vitro and in vivo approach. Environ. Sci. Pollut. Res. Int. 25, 10538–10554. doi: 10.1007/s11356-017-1049-0
Swolana, D., Kępa, M., Kruszniewska-Rajs, C., and Wojtyczka, R. D. (2022). Antibiofilm effect of silver nanoparticles in changing the biofilm-related gene expression of Staphylococcus epidermidis. Int. J. Mol. Sci. 23:9257. doi: 10.3390/ijms23169257
Targhi, A. A., Moammeri, A., Jamshidifar, E., Abbaspour, K., Sadeghi, S., Lamakani, L., et al. (2021). Synergistic effect of curcumin-cu and curcumin-ag nanoparticle loaded niosome: enhanced antibacterial and anti-biofilm activities. Bioorg. Chem. 115:105116. doi: 10.1016/j.bioorg.2021.105116
Thuptimdang, P., Limpiyakorn, T., Mcevoy, J., Pruß, B. M., and Khan, E. (2015). Effect of silver nanoparticles on Pseudomonas putida biofilms at different stages of maturity. J. Hazard. Mater. 290, 127–133. doi: 10.1016/j.jhazmat.2015.02.073
Tian, Y., Zhang, Y., Zhang, M., Chen, X., Lei, L., and Hu, T. (2022). Antisense vicR-loaded dendritic mesoporous silica nanoparticles regulate the biofilm organization and cariogenicity of Streptococcus mutans. Int. J. Nanomedicine 17, 1255–1272. doi: 10.2147/IJN.S334785
Toyofuku, M., Inaba, T., Kiyokawa, T., Obana, N., Yawata, Y., and Nomura, N. (2016). Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 80, 7–12. doi: 10.1080/09168451.2015.1058701
Uddin, T. M., Chakraborty, A. J., Khusro, A., Zidan, B. R. M., Mitra, S., Emran, T. B., et al. (2021). Antibiotic resistance in microbes: history, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 14, 1750–1766. doi: 10.1016/j.jiph.2021.10.020
Uneputty, A., Dávila-Lezama, A., Garibo, D., Oknianska, A., Bogdanchikova, N., Hernández-Sánchez, J., et al. (2022). Strategies applied to modify structured and smooth surfaces: a step closer to reduce bacterial adhesion and biofilm formation. Colloid Interface Sci Commun 46:100560. doi: 10.1016/j.colcom.2021.100560
Wang, L., Hu, C., and Shao, L. (2017). The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. J. Nanomedicine 12, 1227–1249. doi: 10.2147/IJN.S121956
Wang, B., and Kuramitsu, H. K. (2005). Inducible antisense Rna expression in the characterization of gene functions in Streptococcus mutans. Infect. Immun. 73, 3568–3576. doi: 10.1128/IAI.73.6.3568-3576.2005
Wang, J., Li, J., Guo, G., Wang, Q., Tang, J., Zhao, Y., et al. (2016). Silver-nanoparticles-modified biomaterial surface resistant to staphylococcus: new insight into the antimicrobial action of silver. Sci. Rep. 6:32699. doi: 10.1038/srep32699
Webber, M., and Piddock, L. (2003). The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 51, 9–11. doi: 10.1093/jac/dkg050
Weng, L., Wu, L., Guo, R., Ye, J., Liang, W., Wu, W., et al. (2022). Lactobacillus cell envelope-coated nanoparticles for antibiotic delivery against cariogenic biofilm and dental caries. J Nanobiotechnology 20:356. doi: 10.1186/s12951-022-01563-x
Wu, S., Liu, Y., Zhang, H., and Lei, L. (2019). Nano-graphene oxide improved the antibacterial property of antisense yycG Rna on Staphylococcus aureus. J. Orthop. Surg. Res. 14:305. doi: 10.1186/s13018-019-1356-x
Xu, J., Li, Y., Wang, H., Zhu, M., Feng, W., and Liang, G. (2021). Enhanced antibacterial and anti-biofilm activities of antimicrobial peptides modified silver nanoparticles. Int. J. Nanomedicine 16, 4831–4846. doi: 10.2147/IJN.S315839
Xu, Y., Wang, C., Hou, J., Wang, P., You, G., and Miao, L. (2018). Mechanistic understanding of cerium oxide nanoparticle-mediated biofilm formation in Pseudomonas aeruginosa. Environ. Sci. Pollut. Res. Int. 25, 34765–34776. doi: 10.1007/s11356-018-3418-8
Yan, X., He, B., Liu, L., Qu, G., Shi, J., Hu, L., et al. (2018). Antibacterial mechanism of silver nanoparticles in Pseudomonas aeruginosa: proteomics approach. Metallomics 10, 557–564. doi: 10.1039/C7MT00328E
Yoshioka, K., Kunieda, T., Asami, Y., Guo, H., Miyata, H., Yoshida-Tanaka, K., et al. (2019). Highly efficient silencing of microrna by heteroduplex oligonucleotides. Nucleic Acids Res. 47, 7321–7332. doi: 10.1093/nar/gkz492
You, Y.-O. (2019). Virulence genes of Streptococcus mutans and dental caries. Int. J. Oral Biol. 44, 31–36. doi: 10.11620/IJOB.2019.44.2.31
Younis, A. B., Haddad, Y., Kosaristanova, L., and Smerkova, K. (2023). Titanium dioxide nanoparticles: Recent progress in antimicrobial applications. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 15:e1860. doi: 10.1002/wnan.1860
Zahmatkesh, H., Mirpour, M., Zamani, H., and Rasti, B. (2023). Effect of samarium oxide nanoparticles fabricated by curcumin on efflux pump and virulence genes expression in Mdr Pseudomonas aeruginosa and Staphylococcus aureus. J. Clust. Sci. 34, 1227–1235. doi: 10.1007/s10876-022-02274-x
Zhang, J., and Poh, C. L. (2018). Regulating exopolysaccharide gene wcaF allows control of Escherichia coli biofilm formation. Sci. Rep. 8:13127. doi: 10.1038/s41598-018-31161-7
Zhang, Y., Sun, P., Zhang, L., Wang, Z., Wang, F., Dong, K., et al. (2019). Silver-infused porphyrinic metal–organic framework: surface-adaptive, on-demand nanoplatform for synergistic bacteria killing and wound disinfection. Adv. Funct. Mater. 29:1808594. doi: 10.1002/adfm.201808594
Keywords: antibiotic resistance, bacterial pathogens, biofilms, efflux pumps, gene expression, quorum quenching, quorum-sensing, nanoparticles
Citation: Afrasiabi S and Partoazar A (2024) Targeting bacterial biofilm-related genes with nanoparticle-based strategies. Front. Microbiol. 15:1387114. doi: 10.3389/fmicb.2024.1387114
Edited by:
Santi M. Mandal, Indian Institute of Technology Kharagpur, IndiaReviewed by:
Richa Priyadarshini, Shiv Nadar University, IndiaMurat Ozdal, Atatürk University, Türkiye
Sandhimita Mondal, Techno India University, India
Copyright © 2024 Afrasiabi and Partoazar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alireza Partoazar, cGFydG9hemFyQHR1bXMuYWMuaXI=
 Shima Afrasiabi
Shima Afrasiabi Alireza Partoazar2*
Alireza Partoazar2*