
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 05 June 2024
Sec. Microbiotechnology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1386855
Cordyceps militaris, also called as bei-chong-cao, is an insect-pathogenic fungus from the Ascomycota phylum and the Clavicipitaceae family. It is a valuable filamentous fungus with medicinal and edible properties that has been utilized in traditional Chinese medicine (TCM) and as a nutritious food. Cordycepin is the bioactive compound firstly isolated from C. militaris and has a variety of nutraceutical and health-promoting properties, making it widely employed in nutraceutical and pharmaceutical fields. Due to the low composition and paucity of wild resources, its availability from natural sources is limited. With the elucidation of the cordycepin biosynthetic pathway and the advent of synthetic biology, a green cordycepin biosynthesis in Saccharomyces cerevisiae and Metarhizium robertsii has been developed, indicating a potential sustainable production method of cordycepin. Given that, this review primarily focused on the metabolic engineering and heterologous biosynthesis strategies of cordycepin.
Cordyceps militaris, belonging to the genus Cordyceps of Ascomycota phylum and the Clavicipitaceae family, is among the most important “traditional medicines” with a long history of medicinal use in China. This highly valued medicinal fungus has various clinical components, including cordycepin, carotenoid, D-mannitol, polysaccharides, ergosterol and mannitol, which have a wide range of pharmacological benefits, such as anticancer, antioxidant, antibacterial, and immunomodulatory effects. Among them, cordycepin, or 3′-deoxyadenosine, is a derivative of the nucleoside adenosine. Cordycepin was initially isolated from C. militaris by Cunningham et al. (1950) and is the main bioactivity ingredient in this fungus. In reality, except for C. militaris, cordycepin can also be produced by other cordyceps genus, such as Cordyceps kyushensis Kob and Cordyceps cicadae.
Cordycepin is recognized as a bioactive metabolite with therapeutic potentials, exhibiting an extensive range of biology activities, such as anti-cancer, anti-diabetic, anti-hyperlipidemia, anti-inflammatory, anti-photoaging, anti-microbial, anti- platelet aggregation, hypolipidemic, analgesic and immunomodulatory (Sharma et al., 2022). Recent study showed that cordycepin had a strong binding affinity with SARS-CoV-2 spike protein (interaction score: −145.3) and main proteases (M pro, interaction score: −180.5). It was also confirmed to have inhibitory potential against the COVID-19 polymerase enzyme (RdRp) (Verma, 2022), implying cordycepin has the potential for the treatment of COVID-19.
Nevertheless, due to its trace amount in natural sources, the production of cordycepin remains a challenge and is still far from the market demand. As a result, the global market prices of cordycepin gradually rose, approximately $440,000/kg in 2022 (Yang et al., 2020). In recent decades, considerable efforts were focused on the improvement of cordycepin productivity though chemical synthetic and biosynthesis methods. e.g., Masuda et al. (2014) have found that after 45 days (ds) of cultivation, the highest synthesis of cordycepin in the C. militaris mutant G81-3 can reach 14.3 g/L. The late-breaking study enclosed a single gene cluster containing four genes (Cns1-4) capable of synthesizing cordycepin and pentostatin (PTN). Cordycepin was mainly synthesized by the dephosphorylation of adenosine or its 2′, 3′ cyclic monophosphate (2′,3′-cAMP) to adenosine 3′- monophosphate (3′-AMP) catalyzed by Cns2, which was followed by oxidoreduction reactions by Cns1 (Xia et al., 2017). Based on the main biosynthetic pathways of cordycepin, synthetic biology approaches were gradually investigated to introduce these key enzyme genes into the expression system of various hosts to increase the yield and titer of cordycepin. The following review focused on the biosynthesis and metabolic engineering strategies, including microbial fermentation, mutation breeding, modification of pathways enzymes, genome-scale metabolic model (GEMs), etc.
Cordycepin was mainly obtained from the fruiting bodies of C. militaris, with the limited concentration of 2–3 mg/kg (Yang et al., 2020). Relying solely on fermentation may not be sufficient to meet the significantly increased market demands for cordycepin. Mutation breeding is a technique that induces genetic mutations in organisms to create new traits. C. militaris mutants with increased cordycepin yield was obtained using mutation breading (Figure 1 and Table 1).
Through a multifunctional plasma mutation system (MPMS), the high cordycepin-yielding mutant strain GYS60 yielded about 7.883 mg/mL cordycepin, over 20 times higher than that in the wild strain C. militaris G81-3 (Zhang et al., 2020). By exposing to a proton beam irradiation, 14.3 g/L of cordycepin was yielded without crystallization via the former method (Masuda et al., 2014). Wang L. et al. (2017) and Wang Y. et al. (2017) obtained xanthine-guanine double auxotrophic mutants through ultrasonic mutagenesis in conjunction with diethyl sulfate treatment, improving the yield of cordycepin by 40% to 1.76 g/L on 20 ds after batch fermentation. The potential causes may be the obstruction of xanthine and guanine synthesis and metabolism, which resulted in an augmented metabolic flux directed towards the precursor substances adenine or adenosine (Wang L. et al., 2017; Wang Y. et al., 2017). Genome shuffling is a powerful technology that can rapidly improve the microbial phenotypes without revealing the genetic background. This method has been successfully used to improve tyrosine production in Streptomyces fradiae. Six cordycepin-improved C. militaris strains were screened using UV and HNO2 mutagenesis. The maximum cordycepin output was 978.25 μg/g, 9.62-fold increase over the ancestor strain (Wang L. et al., 2017; Wang Y. et al., 2017). Besides, a great number of strains with a high yield of target metabolites were created through rounds of protoplast fusion. For example, the cordycepin content of C. militaris mycelium generated using protoplast fusion technique was enhanced to 1.73 times of that in the parent strains (Zhou and Bian, 2007). Using UV mutagenesis on C. militaris protoplasts, cordycepin content increased to 4.51 mg/g, 1.74 times that of the parent strain (Liu T. et al., 2018; Liu H. et al., 2018). Regardless of the controversial issues surrounding genetically modified organisms or food, mating of parental strains remains a highly effective approach for enhancing cordycepin production. Kang et al. (2017) used spores hybridization to obtain the C. militaris KSP8 with a high cordycepin yield of about 6.63 mg/g and an increase of nearly 35% compared to parent strains.
Overall, mutation breeding has widely used in conjunction with other techniques such as genetic engineering and optimal fermentation conditions to improve the cordycepin yield and fulfill rising market demand for this valuable compound.
Cordycepin is naturally produced in C. militaris, Aspergillus nidulans, C. kyushuensis and C. cicadae (Kaczka et al., 1964; Liu T. et al., 2018; Liu H. et al., 2018; Zhao et al., 2019; Wang K. et al., 2022; Wang L. et al., 2022). In most previous studies, cordycepin was enhanced by solid state fermentation, submerged fermentation, and surface/static fermentation. Changes in the culture medium composition have obviously effect on the growth and quality of Cordyceps (Sung et al., 2010).Rice media, produced the maximum cordycepin (814.60 mg/g) followed by oat and wheat media (638.85 and 565.20 mg/g, respectively) (Adnan et al., 2017). To better understand the causes, the effect of fermented factors on cordycepin synthesis, including nutrients in the basic medium, physics conditions (light, temperature) and various exogenous substances (nitrogen sources, carbon sources, precursor of cordycepin synthesis, plant growth hormone, salt, vegetable oil) (Zhao et al., 2020), were systematically investigated and optimized (Figure 1 and Table 1).
Carbon sources are directly associated with the biosynthesis of several metabolites in fungal cells (Ruiz et al., 2010). Hexose, a six-carbon (C6) sugar, such as glucose, has been shown to preferred carbon source for cordycepin production due to the relationship between high sugar intake and cordycepin accumulation. The maximum cordycepin production (about 245.7 mg/L) was produced in the media containing 40 g glucose/L after 18 ds of culture (Mao et al., 2005). Another study found that the highest titer of cordycepin was produced in the medium broth with xylose compared to sucrose and glucose. Whereas, prolonged cultivation was required to achieve the extracellular cordycepin titer in xylose. Transcriptomic analysis revealed that the assimilated xylose might be directed towards cordycepin biosynthetic pathway rather than central carbon metabolism (Raethong et al., 2018).
Apart from the carbon source, nitrogen supply also aids in increasing cordycepin synthesis. Through the optimization of the feeding time and amount of NH4+, cordycepin concentration achieved to 420.5 ± 15.1 mg/L, and 70% higher compared to batch cultivation methods (Mao and Zhong, 2006). Besides, animal-free nitrogen source (vegetable seed extract), was also investigated. The addition of 80 g/L soybean extract powder (SBEP) increased the cordycepin production up to 2.52 g/L, which was greater than the control (peptone). Quantitative polymerase chain reaction (qPCR) result revealed that 80 g/L SBEP significantly increased the expression of genes linked with the carbon metabolic pathway, amino acid metabolism, and two crucial genes involved in the cordycepin biosynthesis (Cns1 and NT5E) compared to peptone-supplemented culture (Sripilai et al., 2023).
Balance carbon and nitrogen metabolism is essential for metabolic system. The ratio of carbon and nitrogen in the media are also considered to be crucial for the cordycepin production. For examples, the maximal cordycepin production obtained was 345.4 mg/L with 42.0 g/L glucose and 15.8 g/L peptone (Mao et al., 2005). Adenosine and cordycepin content rose with a C/N ratio of 1:1.5, decreased with a C/N ratio of 1:3 (Shih et al., 2007). Cordycepin production increased to 2.64 g/L at a working volume of 147.5 mL, an inoculum size of 8.8% v/v, and a cultivation time of 40.0 ds with an 8:1 C/N ratio and 80 g/L SBEP supplementation (Raethong et al., 2020).
Biotic elicitors are crude extracts and purified products derived from fungal, plant and other sources (Halder et al., 2019). Adding biotic elicitors is an effective strategy to enhance cordycepin accumulation in some fungi. Naphthalene acetic acid (NAA) and vitamin are plant growth regulators with physiological effects with similar to plant hormone auxin (IAA) in terms of regulating plant growth and development. The addition of NAA to the solid medium of C. militaris significantly enhanced cordycepin content in the fruit-body. The addition of 10 mg/L Vitamin B1 (VB1) resulted in the highest yield of cordycepin, about 1.16 g/L after 35 ds of culture, approximately 95.8% higher than that control group (Kang et al., 2014). Through genes expressions and metabolome analysis, cordycepin synthesis might be associated with the metabolic pathways, including purine metabolism, TCA cycle, pentose phosphate pathway, alanine, aspartate, and glutamate metabolism, histidine metabolism and ABC transporter pathway (Wang et al., 2023). Besides, 5 biotic elicitors in different concentrations (0.05, 0.5, 1, and 2 mg/mL), including chitosan (CHT), 2,4-dichlorophenoxyacetic acid (2,4-D), methyl jasmonate (MeJA), gibberellic acid (GA), and triacontanol (TRIA), were firstly introduced to enhance the production of 10 major components in the fruit-body of C. militaris, showing that the addition of 1 mg/L of chitosan resulted in a 0.34-fold increase in cordycepin production compared to control (Lin et al., 2020). It was demonstrated that the application of biotic elicitors is an effective strategy to enhance the production of bioactivity components and the effect of biotic elicitors depended on their types and concentrations.
Trace metals have been reported to have a significant effect on the growth of C. militaris. For instances, 0.1 mM Mn2+ stimulated cell growth and promote the synthesis of Ns, such as uridine, guanosine, and adenosine (Gu et al., 2007). The yield of cordycepin reached to 596.59 mg/L after initially adding 1 g/L FeSO4·7H2O, roughly 70% higher than control, which might be related to the rapidly decreased consumption of inosine 5′-monophosphate (IMP), a putative cordycepin precursor (Fan et al., 2012). Besides, the addition of zinc (Zn) resulted in cordycepin reaching 1.55.g/L after 15 ds of cultivation at 25°C, 6.88 times higher than the control (225.66 mg/L). It suggested that Zn was involved in global transcriptional control and influenced cordycepin biosynthesis (Zhao et al., 2020). These findings underlined the importance of trace metals in modulating cellular processes and enzyme activities in C. militaris, enhancing their roles in the fungus’s growth and metabolic functions.
In engineering process, physical parameters such as pH, dissolved oxygen, light, temperature and biotic elicitors were extensively exploited to improve cordycepin yield. Adnan et al. (2017) found that the optimal conditions for maximum cordycepin production were 25°C, pH5.5 and 21 ds of culture. C. militaris liquid culture was exposed to light-emitting diode (LED) blue light (440-450 nm) for 16 h per day, significantly increasing cordycepin accumulation (Lin et al., 2018).
Cordycepin has a complex metabolic pathway and synthetic regulation that has yet to be fully disclosed. Kredich and Guarino (1961) firstly proposed adenosine as a potential precursor of cordycepin biosynthesis in C. militaris. Lennon and Suhadolnik (1976) hypothesized that the mechanism for cordycepin synthesis might be similar to that of 2′-dA. The biosynthesis of cordycepin was investigated using [U-14C] adenosine and [3-3H] ribose. It was proposed that cordycepin can be produced by converting adenosine to 3′-deoxyadenosin (dA) without hydrolyzing the N-riboside bond, similar to 2′-dA synthesis (Yang et al., 2020). Lin et al. (2016) observed a significant up-regulation of 5′-nucleotidase (5’-NT) expression in Ophiocordyceps sinensis through qPCR analysis, indicating the potential regulatory role for 5’-NT in cordycepin production.
Based on transcriptomics analysis, Xiang proposed that adenosine kinase (ADK), adenylate kinase (ADEK) and 5′-NT, which are involved in phosphorylation and dephosphorylation in the adenosine metabolism pathway, may also be involved in phosphorylation and dephosphorylation in the cordycepin biosynthesis pathway and additional subunits also be involved in the reduction mechanism of the cordycepin synthesis (Xiang et al., 2014). Ribonucleotide reductases small subunit (RNRM) expression significantly increased in C. militaris with the high yield of cordycepin compared to that in wild (Liu G. et al., 2020; Liu W. Y. et al., 2020). The proposed pathway of cordycepin production in C. militaris is described as: Adenylate kinase (AK) converts AMP to adenosine diphosphate (ADP), which is then catalyzed by ribonucleotide reductase (nrdJ) to 3′-deoxyadenosine 5′-diphosphate (3’-dADP). Next, AK converts 3’-dADP to 3′-deoxyadenosine 5′-phosphate (3’-dAMP), which is further converted to cordycepin by 5′-nucleotidase. Additionally, the involvement of 3′, 5′-cyclic-nucleotide phosphodiesterase (PDEs) was proposed to be involved in the synthesis of cordycepin in Paecilomyces hepialid through the differential gene expression analysis (Pang et al., 2018). The addition of 5 mg/L rotenone significantly increased cordycepin production by 316.09%, along with mycelial growth inhibition and cell wall destruction. The integrated analysis of the two omics confirmed that rotenone altered the nucleosides metabolism pathway toward adenosine and activated the genes responsible for producing cordycepin (cns1-3) from adenosine (Ma et al., 2023). The L-alanine addition to C. militaris fermentation increased cordycepin yield by up-regulating the transcriptome level of genes involved in the bio-pathways of energy generation and amino acid conversion. And two Zn2Cys6-type transcription factors, CmTf1 and CmTf2, were also found to be highly up-regulated after the addition of L-alanine, probably participating into cordycepin metabolism (Chen et al., 2020).
Metabolomics studies showed that many metabolic pathways have been changed in the high-yielding cordycepin strains, e.g., citric acid cycle, purine metabolism, and pyrimidine metabolism (Ma et al., 2023). Changes in polysaccharide and protein synthesis metabolic poly-pathways, such as galactose metabolism and amino acid metabolism, would boost cordycepin′s production (Yu et al., 2023). Besides, the global metabolic response of C. militaris strain TBRC6039 in cordycepin synthesis on diverse carbon sources, namely sucrose, glucose and xylose, were investigated using transcriptome analysis and the genome-scale network-driven analysis. A gene (CL1730.Contig2) encoding ATP phosphoribosyltransferase (ATPPRT) was found to be highly expressed in xylose containing conditions, suggesting that ATPPRT might function in the biosynthesis of cordycepin (Raethong et al., 2018). Xia et al. (2017) also found that ATPPRT might be associated with PTN biosynthesis when xylose and phosphoribosyl pyrophosphate (PRPP) and adenosine were used as substrates.
Xia et al. (2017) found that C. militaris possesses a newly evolved gene cluster of Cns1-Cns4, which mediate the dual biosynthesis of cordycepin and another adenosine analog, PTN, gene locations and protein structural domains are shown in Figure 2. Cordycepin biosynthesis starts from adenosine and proceeds through the stepwise reactions of phosphorylation, dephosphorylation, and reduction catalyzed by Cns1/Cns2 complex. Cns1 and Cns2 interact tightly with each other and that one enzyme cannot function alone. Cns1 and Cns2 are required for cordycepin biosynthesis, while the Cns3 gene is involved in biosynthesis of PTN. The Cns1/Cns2 complex catalyzes cordycepin biosynthesis in parallel with PTN biosynthesis. This dual production is analogous to the bacterial “protector-protégé strategy” of purine metabolism, in which the protected cordycepin can be deaminated to 3′-deoxyinosine once it reaches a self-toxic level in fungal cells. Cns4 regulates the cell’s outpumping of PTN. So, the biosynthesis of cordycepin is coupled with PTN production by a gene cluster and the coupling is important for metabolic regulation where PTN safeguards cordycepin from deamination by inhibiting adenosine deaminase (ADA) activity (Xia et al., 2017). The requirement of multienzyme complexes for purine biosynthesis has long been known. Based on the proposed biosynthesis principles, C. militaris was engineered to produce higher levels of cordycepin and PTN. Besides, ADA is widely distributed in mammalian blood and tissues, which caused that cordycepin is quickly metabolized and converted into the inactive metabolite upon entering the body. Guan constructed a non-competitive inhibition to screen ADA inhibitors against the degradation of cordycepin via UHPLC–MS/MS. Oleanolic acid and ursolic acid from Ligustri Lucidi Fructus were chosen as ADA inhibitors with half maximal inhibitory concentration of 21.82 ± 0.39 and 18.41 ± 0.14 μM, respectively (Guan et al., 2020). Another research discovered that emodin inhibits ADA, as well as a high cytotoxic activity against K562 leukemia cells (Zhang et al., 2019). The proposed biosynthetic pathway of cordycepin in C. militaris is described as:using adenosine as the starting material, the nucleoside / nucleotide kinase domain of Cns 3 catalyses the hydroxyl phosphorylation of adenosine at 3’-OH site to yield adenosine-3′-monophosphate (3’-AMP). Meanwhile, 3’-AMP could also be produced from 2′, 3′-cyclic monophosphate (2′,3’-cAMP) by an unidentified enzyme. Cns2 then phosphohydrolates 3′-AMP to 2′-carbonyl-3′-deoxyadenosine (2’-C-3′-dA). 2’-C-3′-dA is then converted to cordycepin by oxidoreduction reaction via Cns1.
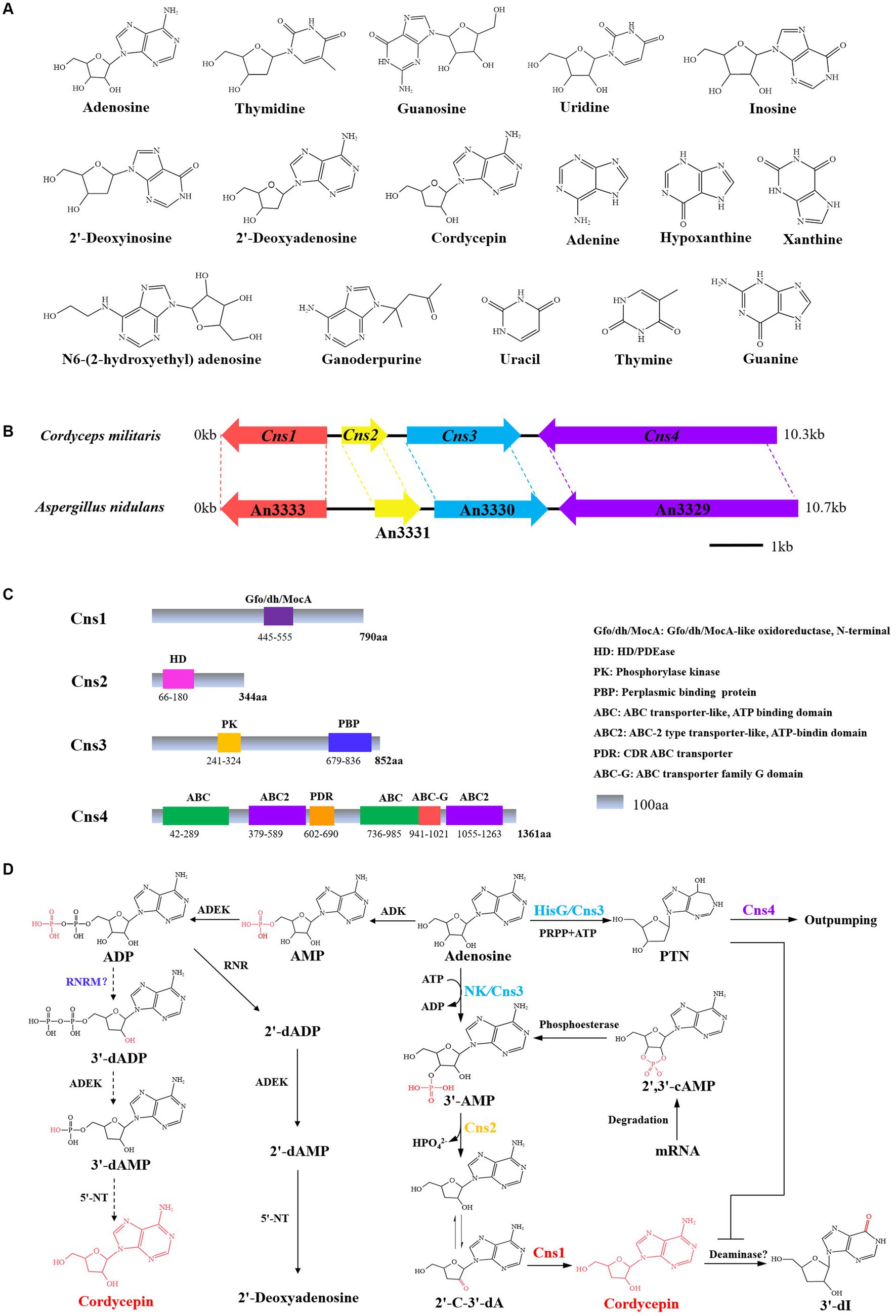
Figure 2. Cordycepin biosynthetic gene cluster and biosynthesis pathway. (A) Chemical structure formula of Nt/Ns. (B) The gene cluster for synthesize cordycepin in C. militaris and A. nidulans. (C) Schematic structure of the proteins Cns-4. (D) Biosynthetic pathway of cordycepin. PRPP, phosphoribosyl pyrophosphate; 2′-C-3′-dA, 2′-carbonyl-3′-deoxyadenosine; NK, an N-terminal nucleoside kinase; HisG, a C-terminal HisG family of ATP phosphoribosyltransferases; 3′-dI, 3′-deoxyinosine.
In addition to C. militaris, the orthologous genes also present in the genome of A. nidulans and C. kyusyuensis. It was suggested that the both species can synthesize cordycepin. Through the high-throughput analysis of the transcriptome and proteome of C. kyushuensis Kobayasi, the gene cluster ck1-ck4 was found to enhance the production of cordycepin and the biosynthesis pathways of cordycepin-PTN coupling was also confirmed (Zhao et al., 2019). United States FDA approved clinical trials of cordycepin combined with PTN for the treatment of terminal deoxynucleotide-positive leukemia (Foss, 2000). Furthermore, the modification of epigenetic status by valproic acid (VPA) significantly enhanced cordycepin production by altering key gene regulatory in C. militaris. The production of cordycepin peaked at 2,835.32 ± 34.35 mg/L in 400 mL working volume and increased by 41.187% after 50 μM VPA treatment, compared to the basal media. The explanation could be related to up-regulation of 5’-NT, ADEK, phosphoribosyltransferase (PRTase) and Cns1-Cns4 in C. militaris (Kunhorm et al., 2022). The proposed cordycepin biosynthetic pathways were sketched in Figure 2.
Traditional mutation breeding is untargeted, laborious and time-consuming. Based on the elucidation of cordycepin biosynthesis pathway, genetic engineering of the Cordyceps strains has a great potential in cordycepin synthesis. Genome/gene editing technologies methods were based on homologous recombination (HR), split-marker approach, Agrobactirium tumfacience mediated-transformant (ATMT)/protoplast-polyethylene glycol (PEG)-mediated transformation (PMT) random/targeted gene mutagenesis and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) (Chen et al., 2018; Lou et al., 2018; Liu et al., 2023). The commonly used transformation methods are summarized in Table 2.
ATMT has been widely and effectively used for transformation, random or targeted gene insertion in several filamentous fungi (Zheng et al., 2011). Compared to other transformation techniques, ATMT allows the flexibility with respect to choose of protoplasts, mycelium, conidia and other cell types as recipient for transformation (Sun et al., 2018). To obtain insertional mutagenesis in C. militaris, the transformants was successfully obtained by co-cultivation of A. tumefaciens AGL-1 carrying vector pATMT1 with the conidia of JM strain, with efficiency in the range of 30–600 transformants/1 × 105 conidia and 55–80% of transformants harbored a single-copy (Zheng et al., 2011). The transformation efficiency can be optimized as ratio of bacteria to conidia, pH of co-culture media and co-culture duration.
PEG, an important method for studying gene function in filamentous fungi, has been successfully applied to C. militaris for transformation (Lou et al., 2019). The yield of protoplasts was significantly affected by the enzyme system used to digest the hyphae. In one study, the 4-day-old mononuclear C. militaris mycelia were incubated in an enzymolysis solution (pH 6.5) composing of 1.00% lyticase, 1.00% lywallzyme, and 0.8 M KCl at 32°C for 3.0 h, achieving the highest protoplast yield of 2.25 × 107 protoplasts/g fresh weight. 10 μg of the linear DNA fragment and 100 μL of protoplasts were mixed, obtaining 87 PCR-positive mutants (Lou et al., 2019). Besides, the applications of remnerative fungi A. cinnamomea and C. militaris were expanded to promote health and well-being through the protoplasts fusion that can be developed as innovative materials for uses. The optimal digestion condition for the preparation of C. militaris protoplasts was 2.0% lywallzyme, 1.0% cellulase, 0.5 M KCl, temperature as 32°C and time as 2.5 h, achieving the highest yield of 1.10 × 108 protoplasts/mL. Taken together, the primary factors influencing protoplast transformation include the enzyme digestion conditions (lytic enzymes, the concentration of the enzyme mixture, the concentration of KCl, digestion temperature, duration and pH), protoplasts regeneration, concentrations of PEG4000 and plasmid DNA.
However, random and HR-based targeted mutagenesis techniques were insufficient and too complicated to meet the requirement for precise, highly efficient and repeatable editing. The breakthrough discovery of Cns gene cluster integrated with targeted genome editing techniques, such as gene knockout and knockin. CRISPR technology is an innovative and highly efficient genome-editing tool capable of gene editing (Alamillo et al., 2023), presenting a promising approach to enhance cordycepin production and related compounds. CRISPR Type II is the most widely used, comprising of two key components: the CRISPR-Cas9 derived from Streptococcus pyogenes and a single-guide RNA (sgRNA) that combines precrRNA and tracrRNA (Mir et al., 2018). CRISPR-Cas9 system has been applied in filamentous fungi, such as Aspergillus (Nødvig et al., 2015; Vanegas et al., 2019), Trichoderma reesei (Liu et al., 2015), and Neurospora crassa (Matsu-ura et al., 2015), whereas its application in Cordyceps genus poses distinct challenges. Cordyceps genus undergo a complex two-stage growth process and face high rates of fruit-body gemmated degeneration. Above factors contribute to additional difficulties in implementing the CRISPR system in these species (Zhang et al., 2023). In C. militaris, the pyrG gene was successfully edited using CRISPR/Cas9 system, with the efficiency of 11.76%. The editing efficiency of Cmura5 reached 100% when an optimized ribonucleoprotein (RNP)-based method was employed, indicating that CRISPR-Cas9-based gene editing techniques can be applied in this species. One study used a codon-optimized Cas9, along with a newly reported promoter (Pcmlsm3) and terminator (Tcmura3), to enhance Cas9 expression (Chen et al., 2018). Furthermore, a CRISPR/Cas9 system based on AMA1 sequence was also successfully constructed to edit multiple genes in C. militaris, with efficiencies of 55.1 and 89% for Cmwc-1 and Cmvvd, respectively. Using this system, a single RNA was edited with an efficiency of 73.9%, while, double genes (Cmwc-1 and Cmvvd) were edited simultaneously with an efficiency of 10% (Meng et al., 2022). CRISPR/Cas9 system can be applied to edit the biosynthetic pathway of cordycepin, the supply and use of adenosine, and PRPP utilization, increasing cordycepin production and facilitating the rapid development of the edible and medicinal fungi industry.
Introducing microbes by virtue of synthetic biology holds a great potentially strategies for cordycepin synthesis. Cordycepin heterologous biosynthesis in Saccharomyces cerevisiae, Yarrowia lipolytica, and Komagataella phaffii uses an enzyme complex of Cns1 and Cns2 and 3′-AMP as the substrate. Cordycepin was enhanced through metabolic engineering strategies, i.e., promoter, protein, adenosine triphosphate, and precursor engineering (Song et al., 2023). S. cerevisiae S288c was successfully employed to synthesize cordycepin by introducing ScCNS1 and ScCNS2 cloned from C. millitaris L5111, and the titer reached 137. 27 mg/L after fermentation optimization (Huo et al., 2021). The emerging non-conventional yeasts, including K. phaffii, Y. lipolytica, Kluyveromyces lactis, Kluyveromyces marxianus, Hansenula polymorpha and Rhodotorula toruloides, have received a lot of attention these years and are being considered as alternatives for biorefinery platforms due to their inherent qualities, ranging from a broad spectrum of carbon-source usage to high environmental tolerance (Patra et al., 2021). Y. lipolytica, an FDA-approved non-conventional yeast, is regarded as a possible platform for manufacturing non-native compounds due to its unique properties (Wang K. et al., 2022; Wang L. et al., 2022). Y. lipolytica has gained particular interest as chassis strains in industry due to the ability of the biosynthesis of value-added molecules by utilizing low-cost bioresources. Moreover, the 3′-AMP biosynthetic pathway naturally present in Y. lipolytica, which is critical to the synthesis of cordycepin. One study showed that the engineered Y. lipolytica produce cordycepin as high as 3.26 g/L from glucose with the metabolic engineering strategies, including the optimizing enzyme fusion and the integration sites, the precursor supply, purine biosynthetic pathway optimization through modular engineering, and using glucose and molasses as the carbon source (Duan et al., 2022). This study shows that the exiting potential of non-conventional yeasts in producing cordycepin and other Ns antibotics using artificial microbial cell factories. The maximum cordycepin productivity of 656.27 mg/L/d (72 h) and cordycepin titer of about 2.29 g/L (120 h) was produced from agro-industrial waste by Y. lipolytica in the optimized media. This study lays the groundwork for effective cordycepin production from agro-industrial waste (Duan et al., 2023). In recent study, the production titer and yields of cordycepin in the engineered Y. lipolytica YlCor-18 under fed-batch fermentation were enhanced to 4.36 g/L and 213.85 mg/g after 168 h of culture (Song et al., 2023). Based on above studies, Y. lipolytica was considered to be a cell factory for cordycepin production and also serve as the model for the green biomanufacturing of other nucleoside antibiotics employing artificial cell factories. Besides, K. phaffii has the intriguing ability to efficiently digest methanol as a sole source of carbon and energy. It was engineered to biosynthesize cordycepin from methanol, which could be converted from CO2. By implementing the fermentation optimization, cordycepin content in broth reached 2.68 g/L within 168 h. This corresponded to a productivity rate of about 15.95 mg/L/h (Tan et al., 2023). The production of cordycepin in strains through heterologous biosynthesis is shown in Table 3. Besides, Xia et al. (2017) successfully expressed Cns2 in E.coli, but failed to express Cns1. In vitro attempts to validate the feasibility of Cns2 directly converting 3’-AMP to 2’-C-3′-dA were unsuccessful, validating that Cns2 functions in the presence of Cns1 (Xia et al., 2017). Various certain drawbacks, such as biased codon usage, protein solubility, mRNA stability, and secondary structure, may hinder the E. coli system from being employed efficiently to overexpress heterologous genes (Gu et al., 2010; Pellizza et al., 2018).To address the potential challenges, some engineering strategies will be adopted across several sizes, such as replacing rare codons with more favorable major codons and periplasmic secretion of heterologous proteins, which provides several benefits, including proper folding, solubility, ease of purification, and enhanced protein output (Terpe, 2006).
Therefore, cordycepin yield will be enhanced through editing the pathways of cordycepin production in C. militaris and, on the other hand, cordycepin can be also heterologous biosynthesized by introducing the cordycepin biosynthetic pathway into engineered yeast and E. coli strains (Figure 3). Metabolic engineering strategies and the fermentation conditions will be combined to enhance the synthetic capability of engineered strains.
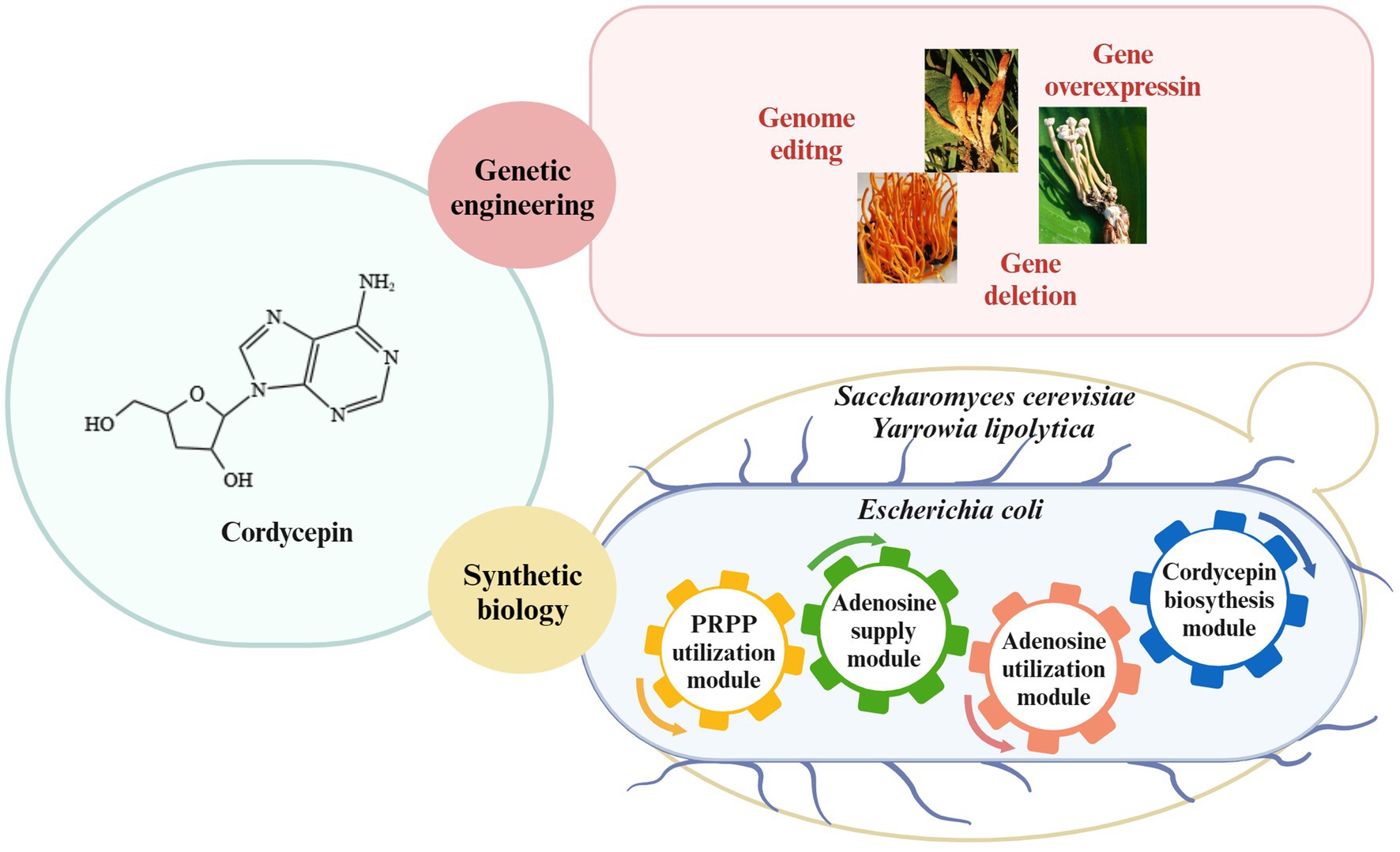
Figure 3. Strategies for cordycepin biosynthesis. Enhancement of cordycepin content in Cordyceps spp. through genetic engineering; synthesis of cordycepin in E. coli, S. cerevisiae and Y. lipolytica through synthetic biology.
ivME is the process of assembling many purified or partially purified enzymes along with nonenzyme components in accordance with the designed pathways to convert certain substrates into target compounds in vitro (Wei et al., 2020). ivME offers advantages such as precise reaction control and increased product yield, eliminating the need for complex cell metabolism engineering and route development. ivME has been used to investigate functional enzymes and metabolites, such as camalexin, mevalonate and isobutanol (Taniguchi et al., 2017). In 2019, a new in vitro approach for cordycepin synthesis was devised and announced as a patent for innovation [CN109628528A] by adding cordycepin synthesis-related enzymes from C. militaris and substrates. However, ivME encounters the problem of enzyme inactivation, the complexity of the experimental techniques, and adenosine substrate inefficiency. This system is challenging to scale up for biomanufacturing. So a combined in vivo/in vitro metabolic engineering strategy was proposed to accelerate the potential for increased cordycepin yield.
The Cordyceps species, as the important sources of biochemical molecules and clinic drugs, has gained a considerable significance in several biotechnology, industrial and clinical applications. Among them, C. militaris was a promising candidate for the industrial applications of cordycepin production. Currently, most studies have focused on the fermentation strategies to increase cordycepin yield. Moreover, the integrated application of multi-omics, systems biology and synthetic biology techniques on Cordyceps allowed a much deeper understanding of the biology of Cordyceps and cordycepin synthesis, opening new and exciting approaches for exploring the biological and anabolic characteristics of cordycepin. Till now, some effective systems as useful tools of insertional mutagenesis in C. militaris have been successfully developed and utilized, including ATMT, PMT, particle bombardment (PB), split marker approach and CRISPR/Cas9. In particular, CRISPR/Cas9 has been successfully applied to precisely edit certain key genes in C. militaris, which will be used to edit the pathways of cordycepin production. It was reported that Agaricus bisporus with a CRISPR/Cas9-edited genome escaped the supervision of the US Department of Agriculture in 2016. CRISPR-based genome engineering technique led to a new era of the genome reconstruction of C. militaris, accelerating industrialization of mushroom.
Moreover, with the broad application of synthetic biology and the elucidation of cordycepin biosynthetic pathway, cordycepin can be heterologously biosynthesized in engineered yeast strains. Various system metabolic engineering methodologies, such as enzyme and precursor engineering, modular engineering and fermentation condition optimization, will be applied to enhance the synthetic potential of engineered strains. Besides, ivME is an alternative strategy that can overcome the limitations of the cell membrane and cell growth while displaying several advantages in terms of flexibility and control. ivME has been developed as an effective technique for synthesising functional enzymes and valuable chemicals, potentially leading to a high yield of cordycepin.
Therefore, the advancement of gene editing method, synthetic biology and in vivo / in vitro metabolic engineering would improve cordycepin synthesis and facilitate commercial-scale production.
XT: Conceptualization, Writing – original draft, Writing – review & editing, funding acquisition, supervision. JG: Writing – review & editing, supervision. TP: Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by Natural Sciences Foundation of China Science (82003900).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adnan, M., Ashraf, S. A., Khan, S., Alshammari, E., and Awadelkareem, A. M. (2017). Effect of pH, temperature and incubation time on cordycepin production from Cordyceps militaris using solid-state fermentation on various substrates. Cyta J. Food 15, 617–621. doi: 10.1080/19476337.2017.1325406
Alamillo, J. M., López, C. M., Martínez Rivas, F. J., Torralbo, F., Bulut, M., and Alseekh, S. (2023). Clustered regularly interspaced short palindromic repeats/Crispr-associated protein and hairy roots: a perfect match for gene functional analysis and crop improvement. Curr. Opin. Biotechnol. 79:102876. doi: 10.1016/j.copbio.2022.102876
Chen, B. X., Wei, T., Xue, L. N., Zheng, Q. W., Ye, Z. W., Zou, Y., et al. (2020). Transcriptome analysis reveals the flexibility of Cordycepin network in Cordyceps militaris activated by L-alanine addition. Front. Microbiol. 11:577. doi: 10.3389/fmicb.2020.00577
Chen, B. X., Wei, T., Ye, Z. W., Yun, F., Kang, L. Z., Tang, H. B., et al. (2018). Efficient Crispr-Cas9 gene disruption system in edible-medicinal mushroom Cordyceps militaris. Front. Microbiol. 9:1157. doi: 10.3389/fmicb.2018.01157
Cunningham, K. G., Manson, W., Spring, F. S., and Hutchinson, S. A. (1950). Cordycepin, a metabolic product isolated from cultures of Cordyceps militaris (Linn.) link. Nature 166:949. doi: 10.1038/166949a0
Duan, X.-Y., Liu, H.-H., Song, L.-P., Wang, C., Yang, H., Lu, X. Y., et al. (2023). Efficient production of cordycepin by engineered Yarrowia lipolytica from agro-industrial residues. Bioresour. Technol. 377:128964. doi: 10.1016/j.biortech.2023.128964
Duan, X. Y., Tian, Y., Song, Z. Q., Song, L. P., Lin, W. B., Wang, C., et al. (2022). High-level de novo biosynthesis of cordycepin by systems metabolic engineering in Yarrowia lipolytica. Bioresour. Technol. 363:127862. doi: 10.1016/j.biortech.2022.127862
Fan, D.-D., Wang, W., and Zhong, J.-J. (2012). Enhancement of cordycepin production in submerged cultures of Cordyceps militaris by addition of ferrous sulfate. Biochem. Eng. J. 60, 30–35. doi: 10.1016/j.bej.2011.09.014
Gu, Y.-X., Wang, Z.-S., Li, S.-X., and Yuan, Q. S. (2007). Effect of multiple factors on accumulation of nucleosides and bases in Cordyceps militaris. Food Chem. 102, 1304–1309. doi: 10.1016/j.foodchem.2006.07.018
Gu, W., Zhou, T., and Wilke, C. O. (2010). A universal trend of reduced mrna stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput. Biol. 6:e1000664. doi: 10.1371/journal.pcbi.1000664
Guan, H., Qi, S., Liu, W., Ma, C., and Wang, C. (2020). A rapid assay to screen adenosine deaminase inhibitors from ligustri lucidi fructus against metabolism of cordycepin utilizing ultra-high-performance liquid chromatography-tandem mass spectrometry. Biomed. Chromatogr. 34:e4779. doi: 10.1002/bmc.4779
Halder, M., Sarkar, S., and Jha, S. (2019). Elicitation: a biotechnological tool for enhanced production of secondary metabolites in hairy root cultures. Eng. Life Sci. 19, 880–895. doi: 10.1002/elsc.201900058
Huo, C., Li, H., Li, Q., Wang, J., Li, C., and Wang, L. (2021). Construction and optimization of cordycepin-producing Saccharomyces cerevisiae. Sheng Wu Gong Cheng Xue Bao 37, 3334–3347. doi: 10.13345/j.cjb.200738
Kaczka, E. A., Dulaney, E. L., Gitterman, C. O., Boyd Woodruff, H., and Folkers, K. (1964). Isolation and inhibitory effects of kb cell cultures of 3′-deoxyandenosine from Aspergillus nidulans (Eidam) Wint. Biochem. Biophys. Res. Commun. 14, 452–455. doi: 10.1016/0006-291X(64)90085-3
Kang, N., Lee, H. H., Park, I., and Seo, Y. S. (2017). Development of high Cordycepin-producing Cordyceps militaris strains. Mycobiology 45, 31–38. doi: 10.5941/MYCO.2017.45.1.31
Kang, C., Wen, T. C., Kang, J. C., Meng, Z. B., Li, G. R., and Hyde, K. D. (2014). Optimization of large-scale culture conditions for the production of cordycepin with Cordyceps militaris by liquid static culture. Sci. World J. 2014:510627, 1–15. doi: 10.1155/2014/510627
Kredich, N. M., and Guarino, A. J. (1961). Studies on the biosynthesis of cordycepin. Biochim. Biophys. Acta 47, 529–534. doi: 10.1016/0006-3002(61)90546-7
Kunhorm, P., Chueaphromsri, P., Chaicharoenaudomrung, N., and Noisa, P. (2022). Enhancement of cordycepin production from Cordyceps militaris culture by epigenetic modification. Biotechnol. Lett. 44, 581–593. doi: 10.1007/s10529-022-03241-2
Lennon, M. B., and Suhadolnik, R. J. (1976). Biosynthesis of 3′-deoxyadenosine by Cordyceps militaris. Mechanism of reduction. Biochim. Biophys. Acta 425, 532–536. doi: 10.1016/0005-2787(76)90017-4
Lin, L. T., Lai, Y. J., Wu, S. C., Hsu, W. H., and Tai, C. J. (2018). Optimal conditions for cordycepin production in surface liquid-cultured Cordyceps militaris treated with porcine liver extracts for suppression of oral cancer. J. Food Drug Anal. 26, 135–144. doi: 10.1016/j.jfda.2016.11.021
Lin, S., Liu, Z. Q., Xue, Y. P., Baker, P. J., Wu, H., Xu, F., et al. (2016). Biosynthetic pathway analysis for improving the cordycepin and cordycepic acid production in Hirsutella sinensis. Appl. Biochem. Biotechnol. 179, 633–649. doi: 10.1007/s12010-016-2020-0
Lin, L. Z., Wei, T., Yin, L., Zou, Y., Bai, W. F., Ye, Z. W., et al. (2020). An efficient strategy for enhancement of bioactive compounds in the fruit body of Caterpillar medicinal mushroom, Cordyceps militaris (Ascomycetes), by spraying biotic elicitors. Int. J. Med. Mushrooms 22, 1161–1170. doi: 10.1615/IntJMedMushrooms.2020037155
Liu, G., Cao, L., Rao, Z., Qiu, X., and Han, R. (2020). Identification of the genes involved in growth characters of medicinal fungus Ophiocordyceps sinensis based on Agrobacterium tumefaciens-mediated transformation. Appl. Microbiol. Biotechnol. 104, 2663–2674. doi: 10.1007/s00253-020-10417-1
Liu, R., Chen, L., Jiang, Y., Zhou, Z., and Zou, G. (2015). Efficient genome editing in filamentous fungus Trichoderma reesei using the Crispr/Cas9 system. Cell Discov. 1:15007. doi: 10.1038/celldisc.2015.7
Liu, T., Liu, Z., Yao, X., Huang, Y., Qu, Q., Shi, X., et al. (2018). Identification of cordycepin biosynthesis-related genes through de novo transcriptome assembly and analysis in Cordyceps cicadae. R. Soc. Open Sci. 5:181247. doi: 10.1098/rsos.181247
Liu, H., Ma, H., Mao, L. R., Li, Y., Zhang, T., Bao, J., et al. (2018). Breeding of high yield Cordyceps militaris mutant strains by protoplast mutagenesis. Sci. Sericult. 44, 450–457. doi: 10.13441/j.cnki.cykx.2018.03.014
Liu, Q., Meng, G., Wang, M., Li, X., Liu, M., Wang, F., et al. (2023). Safe-Harbor-targeted Crispr/Cas9 system and Cmhyd1 overexpression enhances disease resistance in Cordyceps militaris. J. Agric. Food Chem. 71, 15249–15260. doi: 10.1021/acs.jafc.3c05131
Liu, W. Y., Wang, T. T., Chen, S. X., Zhan, Y. N., Li, M. C., and Sun, L. Y. (2020). Transcriptional regulation of cordycepin by a ribonucleotide reductase gene. Modern Food Sci. Technol. 36, 41–46. doi: 10.13982/j.mfst.1673-9078.2020.9.0276
Lou, H.-W., Ye, Z.-W., Yu, Y.-H., Lin, J. F., Guo, L. Q., Chen, B. X., et al. (2019). The efficient genetic transformation of Cordyceps militaris by using mononuclear protoplasts. Sci. Hortic. 243, 307–313. doi: 10.1016/j.scienta.2018.08.043
Lou, H., Ye, Z., Yun, F., Lin, J., Guo, L., Chen, B., et al. (2018). Targeted gene deletion in Cordyceps militaris using the Split-marker approach. Mol. Biotechnol. 60, 380–385. doi: 10.1007/s12033-018-0080-9
Ma, Y. C., Huang, P., Wang, X. L., and Liu, G. Q. (2023). Multi-omics analysis unravels positive effect of rotenone on the cordycepin biosynthesis in submerged fermentation of Cordyceps militaris. Bioresour. Technol. 373:128705. doi: 10.1016/j.biortech.2023.128705
Mao, X.-B., Eksriwong, T., Chauvatcharin, S., and Zhong, J. J. (2005). Optimization of carbon source and carbon/nitrogen ratio for cordycepin production by submerged cultivation of medicinal mushroom Cordyceps militaris. Process Biochem. 40, 1667–1672. doi: 10.1016/j.procbio.2004.06.046
Mao, X. B., and Zhong, J. J. (2004). Hyperproduction of cordycepin by two-stage dissolved oxygen control in submerged cultivation of medicinal mushroom Cordyceps militaris in bioreactors. Biotechnol. Prog. 20, 1408–1413. doi: 10.1021/bp049765r
Mao, X.-B., and Zhong, J.-J. (2006). Significant effect of Nh4+ on cordycepin production by submerged cultivation of medicinal mushroom Cordyceps militaris. Enzym. Microb. Technol. 38, 343–350. doi: 10.1016/j.enzmictec.2004.10.010
Masuda, M., Das, S. K., Hatashita, M., Fujihara, S., and Sakurai, A. (2014). Efficient production of cordycepin by the Cordyceps militaris mutant G81-3 for practical use. Process Biochem. 49, 181–187. doi: 10.1016/j.procbio.2013.10.017
Matsu-ura, T., Baek, M., Kwon, J., and Hong, C. (2015). Efficient gene editing in neurospora crassa with Crispr technology. Fungal Biol. Biotechnol. 2:4. doi: 10.1186/s40694-015-0015-1
Meng, G., Wang, X., Liu, M., Wang, F., Liu, Q., and Dong, C. (2022). Efficient Crispr/Cas9 system based on autonomously replicating plasmid with an Ama1 sequence and precisely targeted gene deletion in the edible fungus, Cordyceps militaris. Microb. Biotechnol. 15, 2594–2606. doi: 10.1111/1751-7915.14107
Mir, A., Edraki, A., Lee, J., and Sontheimer, E. J. (2018). Type ii-C Crispr-Cas9 biology, mechanism, and application. ACS Chem. Biol. 13, 357–365. doi: 10.1021/acschembio.7b00855
Nødvig, C. S., Nielsen, J. B., Kogle, M. E., and Mortensen, U. H. (2015). A Crispr-Cas9 system for genetic engineering of filamentous Fungi [J]. PLoS One 10:e0133085. doi: 10.1371/journal.pone.0133085
Pang, F., Wang, L., Jin, Y., Guo, L., Song, L., Liu, G., et al. (2018). Transcriptome analysis of Paecilomyces hepiali at different growth stages and culture additives to reveal putative genes in cordycepin biosynthesis. Genomics 110, 162–170. doi: 10.1016/j.ygeno.2017.09.008
Patra, P., das, M., Kundu, P., and Ghosh, A. (2021). Recent advances in systems and synthetic biology approaches for developing novel cell-factories in non-conventional yeasts. Biotechnol. Adv. 47:107695. doi: 10.1016/j.biotechadv.2021.107695
Pellizza, L., Smal, C., Rodrigo, G., and Arán, M. (2018). Codon usage clusters correlation: towards protein solubility prediction in heterologous expression systems in E. coli. Sci. Rep. 8:10618. doi: 10.1038/s41598-018-29035-z
Raethong, N., Laoteng, K., and Vongsangnak, W. (2018). Uncovering global metabolic response to cordycepin production in Cordyceps militaris through transcriptome and genome-scale network-driven analysis. Sci. Rep. 8:9250. doi: 10.1038/s41598-018-27534-7
Raethong, N., Wang, H., Nielsen, J., and Vongsangnak, W. (2020). Optimizing cultivation of Cordyceps militaris for fast growth and cordycepin overproduction using rational design of synthetic media. Comput. Struct. Biotechnol. J. 18, 1–8. doi: 10.1016/j.csbj.2019.11.003
Ruiz, B., Chávez, A., Forero, A., García-Huante, Y., Romero, A., Sánchez, M., et al. (2010). Production of microbial secondary metabolites: regulation by the carbon source. Crit. Rev. Microbiol. 36, 146–167. doi: 10.3109/10408410903489576
Sharma, S., Madaan, K., and Kaur, R. (2022). Cordycepin as a metabolite with pharmacological potential: a review. Int. J. Med. Mushrooms 24, 1–20. doi: 10.1615/IntJMedMushrooms.2022044442
Shih, I.-L., Tsai, K.-L., and Hsieh, C. (2007). Effects of culture conditions on the mycelial growth and bioactive metabolite production in submerged culture of Cordyceps militaris. Biochem. Eng. J. 33, 193–201. doi: 10.1016/j.bej.2006.10.019
Song, Z., Lin, W., Duan, X., Song, L., Wang, C., Yang, H., et al. (2023). Increased cordycepin production in Yarrowia lipolytica using combinatorial metabolic engineering strategies. ACS Synth. Biol. 12, 780–787. doi: 10.1021/acssynbio.2c00570
Sripilai, K., Chaicharoenaudomrung, N., Phonchai, R., Chueaphromsri, P., Kunhorm, P., and Noisa, P. (2023). Development of an animal-free nitrogen source for the liquid surface culture of Cordyceps militaris. Lett. Appl. Microbiol. 76:ovad053. doi: 10.1093/lambio/ovad053
Sun, J., Xu, R., Xiao, S., Lu, Y., Zhang, Q., and Xue, C. (2018). Agrobacterium tumefaciens-mediated transformation as an efficient tool for insertional mutagenesis of Kabatiella zeae. J. Microbiol. Methods 149, 96–100. doi: 10.1016/j.mimet.2018.05.004
Sung, G. H., Shrestha, B., Han, S. K., Kim, S. Y., and Sung, J. M. (2010). Growth and cultural characteristics of Cordyceps cardinalis collected from Korea. Mycobiology 38, 274–281. doi: 10.4489/MYCO.2010.38.4.274
Tan, H., Wang, L., Wang, H., Cheng, Y., Li, X., Wan, H., et al. (2023). Engineering Komagataella phaffii to biosynthesize cordycepin from methanol which drives global metabolic alterations at the transcription level. Synthetic Syst. Biotechnol. 8, 242–252. doi: 10.1016/j.synbio.2023.03.003
Taniguchi, H., Okano, K., and Honda, K. (2017). Modules for in vitro metabolic engineering: pathway assembly for bio-based production of value-added chemicals. Synthetic Syst. Biotechnol. 2, 65–74. doi: 10.1016/j.synbio.2017.06.002
Terpe, K. (2006). Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. 72, 211–222. doi: 10.1007/s00253-006-0465-8
Vanegas, K. G., Jarczynska, Z. D., Strucko, T., and Mortensen, U. H. (2019). Cpf1 enables fast and efficient genome editing in aspergilli. Fungal Biol. Biotechnol 6:6. doi: 10.1186/s40694-019-0069-6
Verma, A. K. (2022). Cordycepin: a bioactive metabolite of Cordyceps militaris and polyadenylation inhibitor with therapeutic potential against Covid-19. J. Biomol. Struct. Dyn. 40, 3745–3752. doi: 10.1080/07391102.2020.1850352
Wang, X., Li, Y., Li, X., Sun, L., Feng, Y., Sa, F., et al. (2023). Transcriptome and metabolome profiling unveils the mechanisms of naphthalene acetic acid in promoting cordycepin synthesis in Cordyceps militaris. Front. Nutr. 10:1104446. doi: 10.3389/fnut.2023.1104446
Wang, L., Liu, W. J., Cao, Z. P., Tian, J., Jin, X. Z., and Zhou, G. Q. (2017). Screening of a double auxotrophic Cordyceps militaris mutant and its high fermentation yield of cordycepin. Modern Food Sci. Technol. 33, 54–59. doi: 10.13982/j.mfst.1673-9078.2017.3.009
Wang, K., Shi, T.-Q., Lin, L., Wei, P., Ledesma-Amaro, R., Ji, X. J., et al. (2022). Advances in synthetic biology tools paving the way for the biomanufacturing of unusual fatty acids using the Yarrowia lipolytica chassis. Biotechnol. Adv. 59:107984. doi: 10.1016/j.biotechadv.2022.107984
Wang, L., Yan, H., Zeng, B., and Hu, Z. (2022). Research progress on cordycepin synthesis and methods for enhancement of cordycepin production in Cordyceps militaris. Bioengineering 9:69. doi: 10.3390/bioengineering9020069
Wang, Y., Zhang, G., Zhao, X., and Ling, J. (2017). Genome shuffling improved the nucleosides production in Cordyceps kyushuensis. J. Biotechnol. 260, 42–47. doi: 10.1016/j.jbiotec.2017.08.021
Wei, X., Meng, D., and You, C. (2020). “Chapter 8 - in vitro metabolic engineering: current status and recent progress” in Systems and synthetic metabolic engineering. eds. L. Liu, G. Du, and Y. Liu (Amsterdam: Academic Press), 183–206.
Wongsa, B., Raethong, N., Chumnanpuen, P., Wong-ekkabut, J., Laoteng, K., and Vongsangnak, W. (2020). Alternative metabolic routes in channeling xylose to cordycepin production of Cordyceps militaris identified by comparative transcriptome analysis. Genomics 112, 629–636. doi: 10.1016/j.ygeno.2019.04.015
Xia, Y., Luo, F., Shang, Y., Chen, P., Lu, Y., and Wang, C. (2017). Fungal cordycepin biosynthesis is coupled with the production of the safeguard molecule pentostatin. Cell Chem. Biol. 24, 1479–1489.e4. doi: 10.1016/j.chembiol.2017.09.001
Xiang, L., Li, Y., Zhu, Y., Luo, H., Li, C., Xu, X., et al. (2014). Transcriptome analysis of the Ophiocordyceps sinensis fruiting body reveals putative genes involved in fruiting body development and cordycepin biosynthesis. Genomics 103, 154–159. doi: 10.1016/j.ygeno.2014.01.002
Yang, T., Guo, M., Yang, H., Guo, S., and Dong, C. (2016). The blue-light receptor Cmwc-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl. Microbiol. Biotechnol. 100, 743–755. doi: 10.1007/s00253-015-7047-6
Yang, L., Li, G., Chai, Z., Gong, Q., and Guo, J. (2020). Synthesis of cordycepin: current scenario and future perspectives. Fungal Genet. Biol. 143:103431. doi: 10.1016/j.fgb.2020.103431
Yu, J., Sun, M., Wang, X., Qi, D., and Han, C. (2023). Poly-pathways metabolomics for high-yielding cordycepin of Cordyceps militaris [J]. Biomed. Chromatogr. 37:e5551. doi: 10.1002/bmc.5551
Zhang, Y., Chen, S., Yang, L., and Zhang, Q. (2023). Application progress of Crispr/Cas9 genome-editing technology in edible fungi. Front. Microbiol. 14:1169884. doi: 10.3389/fmicb.2023.1169884
Zhang, H., Deng, L., Zhang, Z., Guan, Y., Li, B., Yang, J., et al. (2020). Enhanced Cordycepin production in Caterpillar medicinal mushroom, Cordyceps militaris (Ascomycetes), mutated by a multifunctional plasma mutagenesis system. Int. J. Med. Mushrooms 22, 1147–1159. doi: 10.1615/IntJMedMushrooms.2020037153
Zhang, X. G., Ma, G. Y., Kou, F., Liu, W. J., Sun, Q. Y., Guo, G. J., et al. (2019). Reynoutria Japonica from traditional Chinese medicine: a source of competitive adenosine deaminase inhibitors for anticancer. Comb. Chem. High Throughput Screen 22, 113–122. doi: 10.2174/1386207322666190415100618
Zhao, X., Li, Q., Liu, W., Guan, H., Li, C., Wang, J., et al. (2020). Advances in biosynthesis of cordycepin from Cordyceps militaris. Sheng Wu Gong Cheng Xue Bao 36, 1293–1304. doi: 10.13345/j.cjb.190500
Zhao, X., Zhang, G., Li, C., and Ling, J. (2019). Cordycepin and pentostatin biosynthesis gene identified through transcriptome and proteomics analysis of Cordyceps kyushuensis kob. Microbiol. Res. 218, 12–21. doi: 10.1016/j.micres.2018.09.005
Zheng, Z., Huang, C., Cao, L., Xie, C., and Han, R. (2011). Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in medicinal fungus Cordyceps militaris. Fungal Biol. 115, 265–274. doi: 10.1016/j.funbio.2010.12.011
Keywords: Cordyceps militaris, cordycepin, microbial fermentation, biosynthesis, metabolic engineering
Citation: Peng T, Guo J and Tong X (2024) Advances in biosynthesis and metabolic engineering strategies of cordycepin. Front. Microbiol. 15:1386855. doi: 10.3389/fmicb.2024.1386855
Received: 16 February 2024; Accepted: 26 April 2024;
Published: 05 June 2024.
Edited by:
Yechun Wang, Trait Biosciences, United StatesReviewed by:
Jun-Wei Xu, Kunming University of Science and Technology, ChinaCopyright © 2024 Peng, Guo and Tong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinlin Guo, Z3VvNTk2QGNkdXRjbS5lZHUuY24=; Xinxin Tong, dG9uZ3h4QGNkdXRjbS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.