- 1Microbial Biotechnology Research Center, Iran University of Medical Sciences, Tehran, Iran
- 2Department of Microbiology, School of Medicine, Iran University of Medical Sciences, Tehran, Iran
- 3Vice Chancellery of Education and Research, Torbat Heydariyeh University of Medical Sciences, Torbat Heydariyeh, Iran
- 4Molecular and Medicine research center, Khomein University of Medical Sciences, Khomein, Iran
- 5Infectious Diseases Research Center (IDRC), Arak University of Medical Sciences, Arak, Iran
Background: Colistin is used as a last resort for managing infections caused by multidrug-resistant bacteria. However, the high emergence of colistin-resistant strains has restricted the clinical use of this antibiotic in the clinical setting. In the present study, we evaluated the global prevalence of the mutation in the mgrB gene, one of the most important mechanisms of colistin resistance in Klebsiella pneumoniae.
Methods: Several databases, including Scopus, Medline (via PubMed), and Web of Science, were searched (until August 2023) to identify those studies that address the mgrB mutation in clinical isolates of K. pneumoniae. Using Stata software, the pooled prevalence of mgrB mutation and subgroup analyses for the year of publication, country, continent, mgrB mutation types, and detection methods of mgrB mutation were analyzed.
Results: Out of the 115 studies included in the analysis, the prevalence of mgrB mutations in colistin-resistant K. pneumoniae isolates was estimated at 65% of isolates, and mgrB variations with insertional inactivation had the highest prevalence among the five investigated mutations with 69%. The year subgroup analysis indicated an increase in mutated mgrB from 46% in 2014 to 61% in 2022. Europe had the highest prevalence of mutated mgrB at 73%, while Africa had the lowest at 54%.
Conclusion: Mutations in the mgrB gene are reported as one of the most common mechanisms of colistin resistance in K. pneumoniae, and the results of the present study showed that 65% of the reported colistin-resistant K. pneumoniae had a mutation in this gene.
1 Introduction
The increasing prevalence of infections due to multidrug-resistant (MDR) bacteria is a major public health concern, and the emergence of antimicrobial resistance has created a difficult challenge for treating a wide variety of infectious diseases (Dadashi et al., 2022). Today, colistin is considered one of the last remaining options for physicians in the fight against MDR and pan-drug-resistant (PDR) bacteria (Moubareck et al., 2018; Menekşe et al., 2019; Moghadam et al., 2022). Colistin, or polymixin E, is a cationic antibiotic and belongs to the polymixin antibiotic class that has that have activity against most Gram-negative bacteria. In the past, colistin had limited use in medicine because of its toxicity, especially nephrotoxicity, but in recent years, due to the increasing rate of MDR bacteria, especially carbapenemase-producing strains, the application of colistin has become more common (Caniaux et al., 2017; Poirel et al., 2017).
However, the high prevalence of colistin-resistant (ColR) strains has restricted the clinical use of colistin. Moreover, a worrying 25–71% mortality rate is reported for colistin-resistant infections (Moubareck et al., 2018; Menekşe et al., 2019; Moghadam et al., 2022).
Enterobacteriaceae cause a wide range of infections in humans. They are capable of acquiring resistance to many antibiotics through horizontal gene transfer (Hasani et al., 2017; Dadashi et al., 2022). Among the bacteria in this family, K. pneumoniae is the most common species that has developed resistance to colistin. Colistin resistance in K. pneumoniae has been reported worldwide in Asia, Europe, North America, South America, and Africa (Ah et al., 2014; Giamarellou, 2016).
Furthermore, resistance to colistin is mainly mediated through chromosomes or horizontal gene transfer. For the first time, the plasmid-borne mcr-1 gene was reported from China, and to date, 10 different types of mcr genes have been reported (Liu et al., 2016; Caniaux et al., 2017; Aris et al., 2020; Hussein et al., 2021). Additionally, chromosomal gene mutations such as pmrA/pmrB, crrA/crrB, and phoP/phoQ, as well as variations in mgrB, are believed to be significant factors in the development of colistin resistance in K. pneumoniae (Cannatelli et al., 2014; Poirel et al., 2017).
The PmrAB and PhoPQ two-component systems are associated with bacterial survival and are usually activated when macrophages attack bacteria. The Pmr system consists of genes and operons involved in adding phosphoethanolamine and 4-amino-4-deoxy-L-arabinose to lipopolysaccharide (LPS; Gunn, 2008; Poirel et al., 2017).
To this end, the inactivation of mgrB causes a negative feedback regulator of the PhoQ-PhoP signaling system, which leads to the acquisition of colistin resistance in K. pneumoniae. This phenomenon ultimately activated the Pmr system, causing modification and reduced affinity of the LPS, which is the colistin target (Cannatelli et al., 2013; Khoshbayan et al., 2021). Collectively, mgrB variation is reported as one of the most common resistance mechanisms among ColR K. pneumoniae isolates (Aghapour et al., 2019). However, there is no exact report on its prevalence among clinical isolates of K. pneumoniae. Therefore, this study aims to investigate the global prevalence of the mutation in the mgrB among clinical isolates of ColR K. pneumoniae.
2 Methods
2.1 Search strategy
A comprehensive and systematic search was conducted for relevant articles by two authors (AKH and NB) until August 2023 in the electronic databases, including Medline (via PubMed), Scopus, and Web of Science. The following search keywords were obtained from the National Library of Medicine’s medical subject heading (MeSH) terms, titles, or abstracts with the help of Boolean operators (and/or) including “Klebsiella pneumoniae” AND “mgrB” with their Mesh terms. The present study was conducted according to the Preferred Reporting Items of the Systematic Review and Meta-Analysis (PRISMA) guidelines.
2.2 Selection criteria and data extraction
Two authors (AKH and NB) worked independently to review the titles, abstracts, and full texts of all retrieved studies, and they excluded irrelevant articles (review articles, case reports, short communication, letters to the editor, brief reports, conference abstracts, and studies with ambiguous results). The search was limited to articles published in English that reported the prevalence of the mgrB in clinical isolates of ColR K. pneumoniae. Disagreements among authors were resolved through discussion and consensus. The information extracted from each of the included articles is as follows: first author name, publication year, country, continent, the total number of K. pneumoniae isolates, number of ColR isolates, number of ColR isolates carrying the mutated mgrB, the mgrB mutation types, and method used for detection of mgrB mutation.
2.3 Quality assessment
An adapted version of the Joanna Briggs Institute (JBI) checklist was used to independently assess study quality by two review authors (ZE and NN; Moola et al., 2017).
2.4 Statistical analysis
A meta-analysis was performed using Stata software v. 17, and a random-effects model estimated the pooled prevalence of the mutated mgrB in ColR K. pneumoniae isolates and the prevalence of five types of mgrB mutation (insertional inactivation, substitution, nonsense mutation, complete and partial deletion) with 95% confidence intervals (95% CI). A Freeman-Tukey double arcsine transformation was performed using the metaprop command of Stata software to estimate the weighted pooled fractions. The I2 value was used to examine statistical heterogeneity between studies. In this regard, I2 ≤ 25% was considered low homogeneity, 25% < I2 ≤ 75% shows moderate heterogeneity, and I2 > 75% indicates high heterogeneity. Potential publication bias was checked using funnel plots and Begg tests. Subgroup analyses were performed for the year of publication, country, continent, and methods used to detect mgrB variations.
3 Results
3.1 Search results
A total of 769 studies were identified in the three electronic databases up to August 2023, and 592 articles were included after duplicate removal. 258 studies after an initial screening of the title and abstract, were eligible for further analysis, of which 115 were included in the final analysis (Supplementary 2, Figure 1).
3.2 Meta-analysis results
In the 115 studies, 2,652 ColR K. pneumoniae and 1,448 ColR isolates with a change in mgrB were found (Table 1). The pooled prevalence of mgrB variations in ColR K. pneumoniae isolates was detected in 65% of isolates (95% CI: 56–72%; I 2 = 91.67%; p < 0.001; Supplementary File 3). The results of Begg’s test (p = 0.4202) showed no publication bias in our study. Noteworthy, the result of publication bias was shown in the funnel plot (Supplementary 2, Figure 2). The year subgroup analysis indicated an increase in mutated mgrB from 46% (95% CI: 27–65%) in 2014 to 61% (95% CI: 43–78%) in 2022. However, in 2023, the results showed a decrease in the rate of mutation to 39% (95% CI: 5–80%), which could be due to the small number of studies compared to 2022 (p = 0.259; Supplementary 2, Figure 3). A subgroup meta-analysis of continents also showed that Europe had the highest rate of mutated mgrB (73%; 95% CI: 63–82%), while Africa had the lowest rate (54%; 95% CI: 9–96%; p = 0.445; Supplementary 2, Figures 4, 5). Among the countries analyzed, Tunisia (95% CI: 97–100%) and Israel (95% CI: 80–100%) with 100% had the highest prevalence of mutated mgrB, while Spain with 8% (95% CI: 0–33%) showed the lowest (p < 0.001; Supplementary 2, Figure 6). Subgroup meta-analysis based on the detection method of mutated mgrB revealed 59% (95% CI: 49–69%) for the polymerase chain reaction (PCR) method and 71% (95% CI: 57–84%) for the whole genome sequencing (WGS) method (p = 0. 219; Supplementary 2, Figure 7). The pooled prevalence of mgrB variations with insertional inactivation in the total number of mgrB variations of ColR K. pneumoniae isolates was 69% (95% CI: 56–72%; I2 = 79.37%; p < 0.001; Supplementary 2, Figure 8). The results of the subgroup meta-analysis showed the only significant difference in the subgroup of countries. Spain had the highest mutation rate with 100% (95% CI: 57–100%) and Serbia had the lowest mutation rate with 0.0% (95% CI: 0–4%), (p < 0.001; Supplementary 4, Figure 3). The pooled prevalence of mgrB variations with substitution in the total number of mgrB variations of ColR K. pneumoniae isolates was 36% (95% CI: 25–48%; I2 = 87.31%; p < 0.001; Supplementary 2, Figure 9). The results of the subgroup meta-analysis showed an increase in the substitution mutation from 18% (95% CI: 8–30%) in 2014 to 50% (95% CI: 19–81%) in 2022 (p < 0.001; Supplementary 4, Figure 5). The highest prevalence of substitution mutation was observed in Brazil at 73% (95% CI: 4–100%), while Taiwan and Greece had the lowest rates with 11% each (95% CI: 2–24% and 6–18%, respectively; p = 0.003; Supplementary 4, Figure 7). Moreover, the subgroup meta-analysis based on the diagnostic method revealed that WGS detected the mutations in 60% of cases (95% CI: 39–80%), while PCR detected mutations in 16% of cases (95% CI: 10–24%; p < 0.001; Supplementary 4, Figure 8). The pooled prevalence of mgrB variations with nonsense mutations in the total number of mgrB variations of ColR K. pneumoniae isolates was 30% (95% CI: 19–42%; I2 = 88.63%; p < 0.001; Supplementary 2, Figure 10). The results of the subgroup meta-analysis showed an increase in nonsense mutations from 18% (95% CI: 9–29%) in 2014 to 100% (95% CI: 100–100%) in 2023 (p < 0.001; Supplementary 4, Figure 9). In addition, Asia had the highest rate of nonsense mutation with 36% (95% CI: 19–55%), while South America had the lowest rate with only 7% (95% CI: 1–17%; p < 0.001; Supplementary 4, Figure 10). Of the countries studied, Iran had the highest prevalence of nonsense mutation, which was 69% (95% CI: 49–87%). On the other hand, Brazil and Serbia had the lowest rate of this mutation, which was 8% (95% CI: 1–18%) and 8% (95% CI: 0–22%), respectively (p < 0.001; Supplementary 4, Figure 11). The pooled prevalence of mgrB variations with complete deletion in the total number of mgrB variations of ColR K. pneumoniae isolates was 19% (95% CI: 11–28%; I2 = 56.99%; p < 0.001; Supplementary 2, Figure 11). The results of the subgroup meta-analysis showed an increase in complete deletion in mgrB from 9% (95% CI: 1–21%) in 2014 to 30% (95% CI: 13–49%) in 2022 (p = 0.002; Supplementary 4, Figure 13). Furthermore, the pooled prevalence of mgrB variations with partial deletion in the total number of mgrB variations of ColR K. pneumoniae isolates was 14% (95% CI: 6–22%; I2 = 69.78%; p < 0.001; Supplementary 2, Figure 12). Among the countries investigated, Brazil had the highest prevalence of partial deletion in mgrB with 52% (95% CI: 9–94%), while Taiwan had the lowest rate of this mutation with 6% (95% CI: 1–14%; p = 0.003; Supplementary 4, Figure 18).
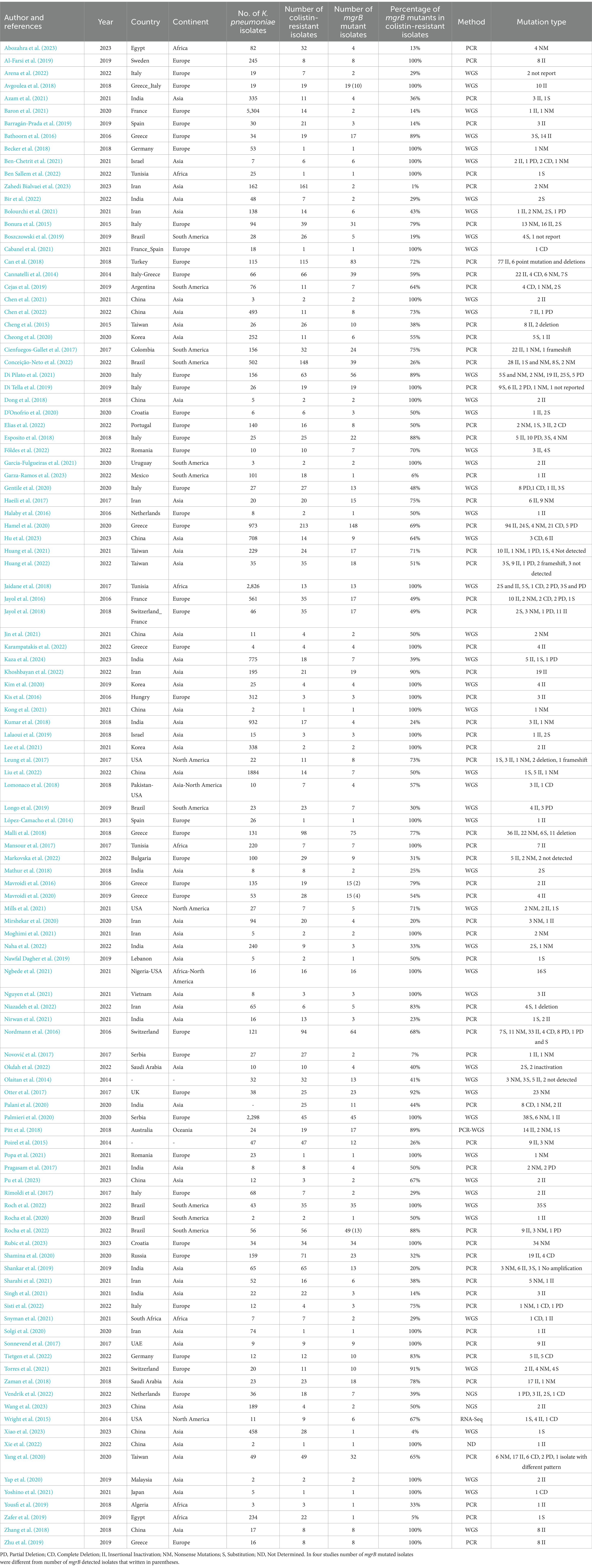
Table 1. Characteristics of included studies that reported resistance to colistin by mgrB mutation in the present meta-analysis.
4 Discussion
In recent years, the effectiveness of antibiotics against MDR pathogens has decreased, leaving colistin as the last available option (Lim et al., 2010). Numerous mechanisms in Gram-negative bacteria result in changes to the outer membrane, which are the main causes of colistin resistance (Li et al., 2006). As mentioned, mgrB inactivation leads to dysregulation of the PhoQ-PhoP signaling system, eventually leading to LPS modification (Cannatelli et al., 2013).
A recent study declared that MgrB alteration could create a fitness cost in K. pneumoniae related to the bacteria’s environmental survival. This phenomenon could pose a silent threat to hospital transmission, as the physical changes resulting from the mgrB mutation seem to cause resistance to disinfectants.
Furthermore, during a two-year period, Xie et al. isolated one colistin-susceptible isolate and one mgrB-mutated ColR isolate from a patient. The ColR isolate exhibits an increased growth rate, but the colistin-susceptible isolate showed significantly decreased growth during a three-hour period, indicating that colistin resistance might result in resistance to human serum (Xie et al., 2022; Yap et al., 2022). Furthermore, the results of a recently published study showed that mutation of mgrB led to resistance to the Galleria mellonella antimicrobial peptides, and in both in vivo and in vitro experiments, it stimulated little activation of inflammatory responses. This phenomenon could be related to the increased virulence associated with this mutation, as many studies have shown the importance of an inflammatory response for K. pneumoniae clearance (Kidd et al., 2017). Interestingly, another study demonstrated that MgrB-dependent ColR K. pneumoniae isolates exhibit increased survival outside the host, leading to enhanced host-to-host transmission (Bray et al., 2022). Therefore, physicians and researchers must appreciate the importance of mgrB mutant isolates for cautious consideration of colistin utilization in K. pneumoniae infections. The significant rise in ColR isolates observed in recent years is related to the rapidly increasing use of colistin in hospital settings, which eventually accelerates the selection pressure for resistance (Wang et al., 2017; Liu and Liu, 2018). Nevertheless, the precise prevalence of mgrB variations was not reported in the recently published studies, therefore, the current study investigates the prevalence of mutated mgrB among the clinical isolates of ColR K. pneumoniae worldwide.
According to our analysis, 65% of all the ColR K. pneumoniae isolates carried mutated mgrB. Furthermore, the prevalence of the mgrB mutation has steadily increased from 46% in 2014 to 61% in 2022, which is a 15% increase. Similarly, a recent study demonstrates an increase in ColR from 4.8% in 2013–2018 to 8.2% in 2019–2021 in Iran (Narimisa et al., 2022). Moreover, the annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) declared that ColR K. pneumoniae has reached a high level of more than 20% in Italy and Greece (Prevention ECfD, Control, 2017; Liu and Liu, 2018). The increasing global use of colistin could lead to an enhanced increase in resistance to the antibiotic, as shown by our analysis of a 15% increase. This phenomenon highlights the urgent need to evaluate the strategies of antimicrobial resistance management internationally (Yusof et al., 2022).
Our results showed that Europe showed the highest rate of mutated mgrB among the continents with 73%, and Africa had the lowest prevalence, with 54%. In 2012, Jaidane et al. demonstrated the emergence of colistin resistance in Tunisia and showed the critical role of MgrB in ColR K. pneumoniae isolates (Jaidane et al., 2018). Furthermore, of the 47 ColR K. pneumoniae isolates in Thailand, mutated mgrB was the leading cause of ColR, which was observed among 43 (91.5%) isolates (Shein et al., 2022). Moreover, a recently published study declared that the most common resistance mechanism among ColR K. pneumoniae isolates in the Middle East is mutations and insertion sequence transpositions in the mgrB (Aris et al., 2020). Moreover, a recent study investigating the prevalence of mutated ColR K. pneumoniae reported that four countries in the Middle East had a high prevalence (>50%) of mutated ColR K. pneumoniae (Saudi Arabia, Qatar, Tunisia, and Iran; Yusof et al., 2022). We observed various mutations in the mgrB locus and categorized them into five groups: insertional inactivation, substitution, nonsense mutation, complete deletion, and partial deletion To view the details, you can refer to the Supplementary Excel file. The prevalence of substitution and complete deletion increased from 2014 to 2022 from 18 to 50% and 9 to 30%, respectively. Additionally, the prevalence of nonsense mutations has increased from 18% in 2014 to 100% in 2023. Insertional inactivation had the highest pooled prevalence among the mgrB variations, at 69%. These small mobile genetic elements are found in the genomes of most bacteria and pose a severe danger to gene structure and expression (Consuegra et al., 2021).
The insertion of IS elements leads to the inactivation or truncation of mgrB, resulting in the malfunction of MgrB (Yang et al., 2020). On many occasions, IS elements are carried by Inc. plasmid groups, and some studies indicate that these plasmids may also carry other resistance genes, like carbapenemase (Fordham et al., 2022). The presence of multidrug-resistant IS-carrying plasmids is a significant concern. The emergence of antimicrobial resistance can lead to colistin therapy, which can mobilize IS elements and potentially create extensively drug-resistant (XDR) or PDR isolates (Fordham et al., 2022). Therefore, monitoring the mutations caused by IS elements in K. pneumoniae is crucial to prevent the worldwide spread of colistin resistance (Yang et al., 2020; Yusof et al., 2022).
Generally, in the analysis of detection methods, it was found that both PCR and WGS methods were equally effective in detecting mutations, with no clear superiority of one over the other. However, WGS was more effective in detecting substitution mutations in 60% of cases, while PCR was effective only in 16%. Therefore, WGS can be considered to be the ideal method for detecting this specific mutation. In combination with Sanger sequencing, PCR has been traditionally used as the gold standard for mutation detection for many years due to its high specificity and low rate of false positives. Although this method has some limitations, such as low sensitivity, it is also time-consuming because of the need for manual analysis of sequencing chromatograms (Gao et al., 2016). Despite these limitations, due to its accessibility and low cost, PCR is still a reasonable and affordable method, especially in developing countries.
5 Limitations
Our study has certain limitations. Because only one study was conducted on the Oceania continent, we could not compare the prevalence of the mgrB mutation in ColR K. pneumoniae with other continents. We did not investigate the sequence type (ST) of resistant isolates because some studies did not report or determine the ST type. In addition, the heterogeneity among studies was relatively high; therefore, subgroup analysis was used to find and reduce the source of heterogeneity.
6 Conclusion
Given the high importance and rise in the global prevalence of ColR K. pneumoniae isolates, it is vital to know the underlying mechanisms related to colistin resistance. The results of the present study showed that 65% of the ColR K. pneumoniae had variation in this gene. Collectively, these findings emphasize the importance of regular monitoring of ColR isolates in clinical settings to stop the spread of ColR isolates. Additionally, adopting innovative screening techniques, practicing antibiotic stewardship, lowering the usage of antibiotics in agriculture, and emphasizing the urgent need to design an organized plan to measure the colistin resistance level are effective strategies to combat antibiotic resistance. In this concept, the exact detection of mechanisms that lead to the mutation in mgrB could significantly decrease the extension of ColR K. pneumoniae. However, more confirmatory studies are needed to advance our knowledge in this field.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
AK: Investigation, Writing – original draft, Writing – review & editing. NN: Writing – original draft, Writing – review & editing. ZE: Writing – review & editing. NB: Writing – review & editing. SR: Writing – review & editing. AS: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We would like to thank Mahmoud Yousefifard from the Physiology Research Center, Iran University of Medical Sciences, for supporting us during this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1386478/full#supplementary-material
References
Abozahra, R., Gaballah, A., and Abdelhamid, S. M. (2023). Prevalence of the colistin resistance gene MCR-1 in colistin-resistant Klebsiella pneumoniae in Egypt. AIMS microbiology. 9, 177–194. doi: 10.3934/microbiol.2023011
Aghapour, Z., Gholizadeh, P., Ganbarov, K., Bialvaei, A. Z., Mahmood, S. S., Tanomand, A., et al. (2019). Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infection and drug resistance. 12, 965–975. doi: 10.2147/IDR.S199844
Ah, Y.-M., Kim, A.-J., and Lee, J.-Y. (2014). Colistin resistance in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 44, 8–15. doi: 10.1016/j.ijantimicag.2014.02.016
Al-Farsi, H. M., Al-Adwani, S., Ahmed, S., Vogt, C., Ambikan, A. T., Leber, A., et al. (2019). Effects of the antimicrobial peptide LL-37 and innate effector mechanisms in colistin-resistant Klebsiella pneumoniae with mgrB insertions. Front. Microbiol. 10:2632. doi: 10.3389/fmicb.2019.02632
Arena, F., Menchinelli, G., Di Pilato, V., Torelli, R., Antonelli, A., Henrici De Angelis, L., et al. (2022). Resistance and virulence features of hypermucoviscous Klebsiella pneumoniae from bloodstream infections: results of a nationwide Italian surveillance study. Front. Microbiol. 13:983294. doi: 10.3389/fmicb.2022.983294
Aris, P., Robatjazi, S., Nikkhahi, F., and Marashi, S. M. A. (2020). Molecular mechanisms and prevalence of colistin resistance of Klebsiella pneumoniae in the Middle East region: a review over the last 5 years. J. Global Antimicrobial Resistance. 22, 625–630. doi: 10.1016/j.jgar.2020.06.009
Avgoulea, K., Di Pilato, V., Zarkotou, O., Sennati, S., Politi, L., Cannatelli, A., et al. (2018). Characterization of extensively drug-resistant or pandrug-resistant sequence type 147 and 101 OXA-48-producing Klebsiella pneumoniae causing bloodstream infections in patients in an intensive care unit. Antimicrob. Agents Chemother. 62, 02457–02417. doi: 10.1128/AAC.02457-17
Azam, M., Gaind, R., Yadav, G., Sharma, A., Upmanyu, K., Jain, M., et al. (2021). Colistin resistance among multiple sequence types of Klebsiella pneumoniae is associated with diverse resistance mechanisms: a report from India. Front. Microbiol. 12:609840. doi: 10.3389/fmicb.2021.609840
Baron, S. A., Cassir, N., Hamel, M., Hadjadj, L., Saidani, N., Dubourg, G., et al. (2021). Risk factors for acquisition of colistin-resistant Klebsiella pneumoniae and expansion of a colistin-resistant ST307 epidemic clone in hospitals in Marseille, France, 2014 to 2017. Eur. Secur. 26:2000022. doi: 10.2807/1560-7917.ES.2021.26.21.2000022
Barragán-Prada, H., Ruiz-Hueso, P., Tedim, A. P., González-Candelas, F., Galán, J. C., Cantón, R., et al. (2019). Emergence and dissemination of colistin-resistant Klebsiella pneumoniae isolates expressing OXA-48 plus CTX-M-15 in patients not previously treated with colistin in a Spanish university hospital. Diagn. Microbiol. Infect. Dis. 93, 147–153. doi: 10.1016/j.diagmicrobio.2018.08.014
Bathoorn, E., Tsioutis, C., da Silva, V. J., Scoulica, E., Ioannidou, E., Zhou, K., et al. (2016). Emergence of pan-resistance in KPC-2 carbapenemase-producing Klebsiella pneumoniae in Crete, Greece: a close call. J. Antimicrob. Chemother. 71, 1207–1212. doi: 10.1093/jac/dkv467
Becker, L., Fuchs, S., Pfeifer, Y., Semmler, T., Eckmanns, T., Korr, G., et al. (2018). Whole genome sequence analysis of CTX-M-15 producing Klebsiella isolates allowed dissecting a polyclonal outbreak scenario. Front. Microbiol. 9:322. doi: 10.3389/fmicb.2018.00322
Ben Sallem, R., Laribi, B., Arfaoui, A., Ben Khelifa Melki, S., Ouzari, H., Ben Slama, K., et al. (2022). Co-occurrence of genes encoding carbapenemase, ESBL, pAmpC and non-β-lactam resistance among Klebsiella pneumonia and E. coli clinical isolates in Tunisia. Lett. Appl. Microbiol. 74, 729–740. doi: 10.1111/lam.13658
Ben-Chetrit, E., Mc Gann, P., Maybank, R., Stam, J., Assous, M. V., and Katz, D. E. (2021). Colistin-resistant Klebsiella pneumoniae bloodstream infection: old drug, bad bug. Arch. Microbiol. 203, 2999–3006. doi: 10.1007/s00203-021-02289-4
Bir, R., Gautam, H., Arif, N., Chakravarti, P., Verma, J., Banerjee, S., et al. (2022). Analysis of colistin resistance in carbapenem-resistant Enterobacterales and XDR Klebsiella pneumoniae. Therapeutic Advan. Infectious Dis. 9:204993612210806. doi: 10.1177/20499361221080650
Bolourchi, N., Shahcheraghi, F., Giske, C. G., Nematzadeh, S., Noori Goodarzi, N., Solgi, H., et al. (2021). Comparative genome analysis of colistin-resistant OXA-48-producing Klebsiella pneumoniae clinical strains isolated from two Iranian hospitals. Ann. Clin. Microbiol. Antimicrob. 20, 1–11. doi: 10.1186/s12941-021-00479-y
Bonura, C., Giuffrè, M., Aleo, A., Fasciana, T., Di Bernardo, F., Stampone, T., et al. (2015). An update of the evolving epidemic of Bla KPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: emergence of multiple non-ST258 clones. PLoS One 10:e0132936. doi: 10.1371/journal.pone.0132936
Boszczowski, I., Salomão, M. C., Moura, M. L., Freire, M. P., Guimarães, T., Cury, A. P., et al. (2019). Multidrug-resistant Klebsiella pneumoniae: genetic diversity, mechanisms of resistance to polymyxins and clinical outcomes in a tertiary teaching hospital in Brazil. Rev. Inst. Med. Trop. São Paulo 61:e29. doi: 10.1590/s1678-9946201961029
Bray, A. S., Smith, R. D., Hudson, A. W., Hernandez, G. E., Young, T. M., George, H. E., et al. (2022). MgrB-dependent colistin resistance in Klebsiella pneumoniae is associated with an increase in host-to-host transmission. MBio 13, e03595–e03521. doi: 10.1128/mbio.03595-21
Cabanel, N., Rosinski-Chupin, I., Chiarelli, A., Botin, T., Tato, M., Canton, R., et al. (2021). Evolution of VIM-1-producing Klebsiella pneumoniae isolates from a hospital outbreak reveals the genetic bases of the loss of the urease-positive identification character. Msystems. 6, e00244–e00221. doi: 10.1128/mSystems.00244-21
Can, F., Menekse, S., Ispir, P., Atac, N., Albayrak, O., Demir, T., et al. (2018). Impact of the ST101 clone on fatality among patients with colistin-resistant Klebsiella pneumoniae infection. J. Antimicrob. Chemother. 73, 1235–1241. doi: 10.1093/jac/dkx532
Caniaux, I., Van Belkum, A., Zambardi, G., Poirel, L., and Gros, M. (2017). MCR: modern colistin resistance. Eur. J. Clin. Microbiol. Infect. Dis. 36, 415–420. doi: 10.1007/s10096-016-2846-y
Cannatelli, A., D'Andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13
Cannatelli, A., Giani, T., D'Andrea, M. M., Pilato, V. D., Arena, F., Conte, V., et al. (2014). MgrB inactivation is a common mechanism of Colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703. doi: 10.1128/AAC.03110-14
Cejas, D., Elena, A., Nuñez, D. G., Platero, P. S., De Paulis, A., Magariños, F., et al. (2019). Changing epidemiology of KPC-producing Klebsiella pneumoniae in Argentina: emergence of hypermucoviscous ST25 and high-risk clone ST307. J. global antimicrobial resistance. 18, 238–242. doi: 10.1016/j.jgar.2019.06.005
Chen, X., Li, P., Sun, Z., Xu, X., Jiang, J., and Su, J. (2022). Insertion sequence mediating mrgB disruption is the major mechanism of polymyxin resistance in carbapenem-resistant Klebsiella pneumoniae isolates from China. J. Global Antimicrobial Resistance. 30, 357–362. doi: 10.1016/j.jgar.2022.07.002
Chen, J., Zeng, Y., Zhang, R., and Cai, J. (2021). In vivo emergence of colistin and tigecycline resistance in carbapenem-resistant hypervirulent Klebsiella pneumoniae during antibiotics treatment. Front. Microbiol. 12:702956. doi: 10.3389/fmicb.2021.702956
Cheng, Y.-H., Lin, T.-L., Pan, Y.-J., Wang, Y.-P., Lin, Y.-T., and Wang, J.-T. (2015). Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob. Agents Chemother. 59, 2909–2913. doi: 10.1128/AAC.04763-14
Cheong, H. S., Kim, S. Y., Seo, J., Wi, Y. M., Peck, K. R., and Ko, K. S. (2020). Colistin resistance and extensive genetic variations in PmrAB and PhoPQ in Klebsiella pneumoniae isolates from South Korea. Curr. Microbiol. 77, 2307–2311. doi: 10.1007/s00284-020-02074-4
Cienfuegos-Gallet, A. V., Chen, L., Kreiswirth, B. N., and Jiménez, J. N. (2017). Colistin resistance in carbapenem-resistant Klebsiella pneumoniae mediated by chromosomal integration of plasmid DNA. Antimicrob. Agents Chemother. 61, e00404–e00417. doi: 10.1128/AAC.00404-17
Conceição-Neto, O. C., da Costa, B. S., Pontes, L. S., Silveira, M. C., Justo-da-Silva, L. H., de Oliveira Santos, I. C., et al. (2022). Polymyxin resistance in clinical isolates of K. pneumoniae in Brazil: update on molecular mechanisms, clonal dissemination and relationship with KPC-producing strains. Front. Cell. Infect. Microbiol. 12:898125. doi: 10.3389/fcimb.2022.898125
Consuegra, J., Gaffé, J., Lenski, R. E., Hindré, T., Barrick, J. E., Tenaillon, O., et al. (2021). Insertion-sequence-mediated mutations both promote and constrain evolvability during a long-term experiment with bacteria. Nat. Commun. 12:980. doi: 10.1038/s41467-021-21210-7
D’Onofrio, V., Conzemius, R., Varda-Brkić, D., Bogdan, M., Grisold, A., Gyssens, I. C., et al. (2020). Epidemiology of colistin-resistant, carbapenemase-producing Enterobacteriaceae and Acinetobacter baumannii in Croatia. Infect. Genet. Evol. 81:104263. doi: 10.1016/j.meegid.2020.104263
Dadashi, M., Sameni, F., Bostanshirin, N., Yaslianifard, S., Khosravi-Dehaghi, N., Nasiri, M. J., et al. (2022). Global prevalence and molecular epidemiology of mcr-mediated colistin resistance in Escherichia coli clinical isolates: a systematic review. J. Global Antimicrobial Resistance. 29, 444–461. doi: 10.1016/j.jgar.2021.10.022
Di Pilato, V., Errico, G., Monaco, M., Giani, T., Del Grosso, M., Antonelli, A., et al. (2021). The changing epidemiology of carbapenemase-producing Klebsiella pneumoniae in Italy: toward polyclonal evolution with emergence of high-risk lineages. J. Antimicrob. Chemother. 76, 355–361. doi: 10.1093/jac/dkaa431
Di Tella, D., Tamburro, M., Guerrizio, G., Fanelli, I., Sammarco, M. L., and Ripabelli, G. (2019). Molecular epidemiological insights into colistin-resistant and carbapenemases-producing clinical Klebsiella pneumoniae isolates. Infection and Drug Resistance. 12, 3783–3795. doi: 10.2147/IDR.S226416
Dong, N., Yang, X., Zhang, R., Chan, E. W.-C., and Chen, S. (2018). Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerging microbes & infections. 7, 1–8. doi: 10.1038/s41426-018-0146-6
Elias, R., Spadar, A., Phelan, J., Melo-Cristino, J., Lito, L., Pinto, M., et al. (2022). A phylogenomic approach for the analysis of colistin resistance-associated genes in Klebsiella pneumoniae, its mutational diversity and implications for phenotypic resistance. Int. J. Antimicrob. Agents 59:106581. doi: 10.1016/j.ijantimicag.2022.106581
Esposito, E. P., Cervoni, M., Bernardo, M., Crivaro, V., Cuccurullo, S., Imperi, F., et al. (2018). Molecular epidemiology and virulence profiles of Colistin-resistant Klebsiella pneumoniae blood isolates from the hospital agency “Ospedale Dei Colli,” Naples, Italy. Front. Microbiol. 9:1463. doi: 10.3389/fmicb.2018.01463
Főldes, A., Oprea, M., Székely, E., Usein, C.-R., and Dobreanu, M. (2022). Characterization of Carbapenemase-producing Klebsiella pneumoniae isolates from two Romanian hospitals co-presenting resistance and Heteroresistance to Colistin. Antibiotics. 11:1171. doi: 10.3390/antibiotics11091171
Fordham, S. M. E., Mantzouratou, A., and Sheridan, E. (2022). Prevalence of insertion sequence elements in plasmids relating to mgrB gene disruption causing colistin resistance in Klebsiella pneumoniae. Microbiology 11:e1262. doi: 10.1002/mbo3.1262
Gao, J., Wu, H., Wang, L., Zhang, H., Duan, H., Lu, J., et al. (2016). Validation of targeted next-generation sequencing for RAS mutation detection in FFPE colorectal cancer tissues: comparison with sanger sequencing and ARMS-scorpion real-time PCR. BMJ Open 6:e009532. doi: 10.1136/bmjopen-2015-009532
Garcia-Fulgueiras, V., Magallanes, C., Reyes, V., Cayota, C., Galiana, A., Vieytes, M., et al. (2021). In vivo high plasticity of multi-drug resistant ST258 Klebsiella pneumoniae. Microb. Drug Resist. 27, 1126–1130. doi: 10.1089/mdr.2020.0310
Garza-Ramos, U., Silva-Sánchez, J., López-Jácome, L. E., Hernández-Durán, M., Colín-Castro, C. A., Sánchez-Pérez, A., et al. (2023). Carbapenemase-encoding genes and Colistin resistance in gram-negative Bacteria during the COVID-19 pandemic in Mexico: results from the Invifar network. Microb. Drug Resist. 29, 239–248. doi: 10.1089/mdr.2022.0226
Gentile, B., Grottola, A., Orlando, G., Fregni Serpini, G., Venturelli, C., Meschiari, M., et al. (2020). A retrospective whole-genome sequencing analysis of carbapenem and colistin-resistant Klebsiella pneumoniae nosocomial strains isolated during an MDR surveillance program. Antibiotics. 9:246. doi: 10.3390/antibiotics9050246
Giamarellou, H. (2016). Epidemiology of infections caused by polymyxin-resistant pathogens. Int. J. Antimicrob. Agents 48, 614–621. doi: 10.1016/j.ijantimicag.2016.09.025
Gunn, J. S. (2008). The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16, 284–290. doi: 10.1016/j.tim.2008.03.007
Haeili, M., Javani, A., Moradi, J., Jafari, Z., Feizabadi, M. M., and Babaei, E. (2017). MgrB alterations mediate colistin resistance in Klebsiella pneumoniae isolates from Iran. Front. Microbiol. 8:2470. doi: 10.3389/fmicb.2017.02470
Halaby, T., Kucukkose, E., Janssen, A. B., Rogers, M. R., Doorduijn, D. J., van der Zanden, A. G., et al. (2016). Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob. Agents Chemother. 60, 6837–6843. doi: 10.1128/AAC.01344-16
Hamel, M., Chatzipanagiotou, S., Hadjadj, L., Petinaki, E., Papagianni, S., Charalampaki, N., et al. (2020). Inactivation of mgrB gene regulator and resistance to colistin is becoming endemic in carbapenem-resistant Klebsiella pneumoniae in Greece: a nationwide study from 2014 to 2017. Int. J. Antimicrob. Agents 55:105930. doi: 10.1016/j.ijantimicag.2020.105930
Hasani, A., Purmohammad, A., Rezaee, M. A., Hasani, A., and Dadashi, M. (2017). Integron-mediated multidrug and quinolone resistance in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Archives of pediatric. Infect. Dis. Ther. 5:e36616. doi: 10.5812/pedinfect.36616
Hu, H., Shi, Q., Zhang, P., Quan, J., Han, X., Zhao, D., et al. (2023). Prevalence and molecular characteristics of colistin-resistant isolates among clinically isolated carbapenem-resistant Klebsiella pneumoniae in China. Int. J. Antimicrob. Agents 62:106873. doi: 10.1016/j.ijantimicag.2023.106873
Huang, P.-H., Chen, W.-Y., Chou, S.-H., Wang, F.-D., and Lin, Y.-T. (2022). Risk factors for the development of colistin resistance during colistin treatment of carbapenem-resistant Klebsiella pneumoniae infections. Microbiology spectrum. 10, e00381–e00322. doi: 10.1128/spectrum.00381-22
Huang, P.-H., Cheng, Y.-H., Chen, W.-Y., Juan, C.-H., Chou, S.-H., Wang, J.-T., et al. (2021). Risk factors and mechanisms of in vivo emergence of colistin resistance in carbapenem-resistant Klebsiella pneumoniae. Int. J. Antimicrob. Agents 57:106342. doi: 10.1016/j.ijantimicag.2021.106342
Hussein, N. H., Al-Kadmy, I. M. S., Taha, B. M., and Hussein, J. D. (2021). Mobilized colistin resistance (mcr) genes from 1 to 10: a comprehensive review. Mol. Biol. Rep. 48, 2897–2907. doi: 10.1007/s11033-021-06307-y
Jaidane, N., Bonnin, R. A., Mansour, W., Girlich, D., Creton, E., Cotellon, G., et al. (2018). Genomic insights into colistin-resistant Klebsiella pneumoniae from a Tunisian teaching hospital. Antimicrob. Agents Chemother. 62, e01601–e01617. doi: 10.1128/AAC.01601-17
Jayol, A., Nordmann, P., Lehours, P., Poirel, L., and Dubois, V. (2018). Comparison of methods for detection of plasmid-mediated and chromosomally encoded colistin resistance in Enterobacteriaceae. Clin. Microbiol. Infect. 24, 175–179. doi: 10.1016/j.cmi.2017.06.002
Jayol, A., Poirel, L., Dortet, L., and Nordmann, P. (2016). National survey of colistin resistance among carbapenemase-producing Enterobacteriaceae and outbreak caused by colistin-resistant OXA-48-producing Klebsiella pneumoniae, France, 2014. Eur. Secur. 21:30339. doi: 10.2807/1560-7917.ES.2016.21.37.30339
Jin, X., Chen, Q., Shen, F., Jiang, Y., Wu, X., Hua, X., et al. (2021). Resistance evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11 during treatment with tigecycline and polymyxin. Emerging microbes & infections. 10, 1129–1136. doi: 10.1080/22221751.2021.1937327
Karampatakis, T., Zarras, C., Pappa, S., Vagdatli, E., Iosifidis, E., Roilides, E., et al. (2022). Emergence of ST39 carbapenem-resistant Klebsiella pneumoniae producing VIM-1 and KPC-2. Microb. Pathog. 162:105373. doi: 10.1016/j.micpath.2021.105373
Kaza, P., Britto, X. B., Mahindroo, J., Singh, N., Baker, S., Nguyen, T. N. T., et al. (2024). Extensively-drug resistant (XDR) Klebsiella pneumoniae associated with complicated urinary tract infection in northern India. Jpn. J. Infect. Dis. 77, 7–15. doi: 10.7883/yoken.JJID.2023.009
Khoshbayan, A., Shariati, A., Razavi, S., Baseri, Z., Ghodousi, A., and Darban-Sarokhalil, D. (2022). Mutation in mgrB is the major colistin resistance mechanism in Klebsiella pneumoniae clinical isolates in Tehran, Iran. Acta Microbiol. Immunol. Hung. 69, 61–67. doi: 10.1556/030.2022.01679
Khoshbayan, A., Shariati, A., Shahmoradi, S., Baseri, Z., Mozafari, H., and Darban-Sarokhalil, D. (2021). Prevalence and molecular mechanisms of colistin resistance in Acinetobacter baumannii clinical isolates in Tehran, Iran. Acta Microbiol. Immunol. Hung. 68, 262–266. doi: 10.1556/030.2021.01420
Kidd, T. J., Mills, G., Sá-Pessoa, J., Dumigan, A., Frank, C. G., Insua, J. L., et al. (2017). A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol. Med. 9, 430–447. doi: 10.15252/emmm.201607336
Kim, Y.-J., Kim, S., Kim, J., and Bae, S. (2020). Tracking short-term changes in the genetic diversity and antimicrobial resistance of OXA-232-producing Klebsiella pneumoniae ST14 in clinical settings. Clin. Microbiol. Infect. 26, 78–86. doi: 10.1016/j.cmi.2019.05.008
Kis, Z., Tóth, Á., Jánvári, L., and Damjanova, I. (2016). Countrywide dissemination of a DHA-1-type plasmid-mediated AmpC β-lactamase-producing Klebsiella pneumoniae ST11 international high-risk clone in Hungary, 2009–2013. J. Med. Microbiol. 65, 1020–1027. doi: 10.1099/jmm.0.000302
Kong, Y., Li, C., Chen, H., Zheng, W., Sun, Q., Xie, X., et al. (2021). In vivo emergence of colistin resistance in carbapenem-resistant Klebsiella pneumoniae mediated by premature termination of the mgrB gene regulator. Front. Microbiol. 12:656610. doi: 10.3389/fmicb.2021.656610
Kumar, A., Biswas, L., Omgy, N., Mohan, K., Vinod, V., Sajeev, A., et al. (2018). Colistin resistance due to insertional inactivation of the mgrB in Klebsiella pneumoniae of clinical origin: first report from India. Spanish J. chemotherapy 31, 406–410
Lalaoui, R., Bakour, S., Livnat, K., Assous, M. V., Diene, S. M., and Rolain, J.-M. (2019). Spread of carbapenem and colistin-resistant Klebsiella pneumoniae ST512 clinical isolates in Israel: a cause for vigilance. Microb. Drug Resist. 25, 63–71. doi: 10.1089/mdr.2018.0014
Lee, T. H., Cho, M., Lee, J., Hwang, J.-H., Lee, C.-S., and Chung, K. M. (2021). Molecular characterization of Carbapenem-resistant, Colistin-resistant Klebsiella pneumoniae isolates from a tertiary Hospital in Jeonbuk, Korea. J. Bacteriol. Virol. 51, 120–127. doi: 10.4167/jbv.2021.51.3.120
Leung, L. M., Cooper, V. S., Rasko, D. A., Guo, Q., Pacey, M. P., McElheny, C. L., et al. (2017). Structural modification of LPS in colistin-resistant, KPC-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 72, 3035–3042. doi: 10.1093/jac/dkx234
Li, J., Nation, R. L., Turnidge, J. D., Milne, R. W., Coulthard, K., Rayner, C. R., et al. (2006). Colistin: the re-emerging antibiotic for multidrug-resistant gram-negative bacterial infections. Lancet Infect. Dis. 6, 589–601. doi: 10.1016/S1473-3099(06)70580-1
Lim, L. M., Ly, N., Anderson, D., Yang, J. C., Macander, L., Jarkowski, A. III, et al. (2010). Resurgence of colistin: a review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy: J. Human Pharmacol. Drug Therapy. 30, 1279–1291. doi: 10.1592/phco.30.12.1279
Liu, Y., and Liu, J.-H. (2018). Monitoring Colistin resistance in food animals, an urgent threat. Expert Rev. Anti-Infect. Ther. 16, 443–446. doi: 10.1080/14787210.2018.1481749
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Liu, X., Wu, Y., Zhu, Y., Jia, P., Li, X., Jia, X., et al. (2022). Emergence of colistin-resistant hypervirulent Klebsiella pneumoniae (CoR-HvKp) in China. Emerging Microbes & Infections. 11, 648–661. doi: 10.1080/22221751.2022.2036078
Lomonaco, S., Crawford, M. A., Lascols, C., Timme, R. E., Anderson, K., Hodge, D. R., et al. (2018). Resistome of carbapenem-and colistin-resistant Klebsiella pneumoniae clinical isolates. PLoS One 13:e0198526. doi: 10.1371/journal.pone.0198526
Longo, L. G., de Sousa, V. S., Kraychete, G. B., Justo-da-Silva, L. H., Rocha, J. A., Superti, S. V., et al. (2019). Colistin resistance emerges in pandrug-resistant Klebsiella pneumoniae epidemic clones in Rio de Janeiro. Brazil. Int. J. Antimicrobial Agents. 54, 579–586. doi: 10.1016/j.ijantimicag.2019.08.017
López-Camacho, E., Gómez-Gil, R., Tobes, R., Manrique, M., Lorenzo, M., Galván, B., et al. (2014). Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumoniae outbreak including carbapenem and colistin resistance. J. Antimicrob. Chemother. 69, 632–636. doi: 10.1093/jac/dkt419
Malli, E., Florou, Z., Tsilipounidaki, K., Voulgaridi, I., Stefos, A., Xitsas, S., et al. (2018). Evaluation of rapid polymyxin NP test to detect colistin-resistant Klebsiella pneumoniae isolated in a tertiary Greek hospital. J. Microbiol. Methods 153, 35–39. doi: 10.1016/j.mimet.2018.08.010
Mansour, W., Haenni, M., Saras, E., Grami, R., Mani, Y., Ben Haj Khalifa, A., et al. (2017). Outbreak of colistin-resistant carbapenemase-producing Klebsiella pneumoniae in Tunisia. J Glob Antimicrob Resist. 10, 88–94. doi: 10.1016/j.jgar.2017.03.017
Markovska, R., Marteva-Proevska, Y., Velinov, T., Pavlov, I., Kaneva, R., and Boyanova, L. (2022). Detection of different colistin resistance mechanisms among multidrug resistant Klebsiella pneumoniae isolates in Bulgaria. Acta Microbiol. Immunol. Hung. 69, 220–227. doi: 10.1556/030.2022.01746
Mathur, P., Veeraraghavan, B., Devanga Ragupathi, N. K., Inbanathan, F. Y., Khurana, S., Bhardwaj, N., et al. (2018). Multiple mutations in lipid-a modification pathway & novel fosA variants in colistin-resistant Klebsiella pneumoniae. Future Sci. OA 4:FSO319. doi: 10.4155/fsoa-2018-0011
Mavroidi, A., Katsiari, M., Likousi, S., Palla, E., Roussou, Z., Nikolaou, C., et al. (2016). Characterization of ST258 Colistin-resistant, blaKPC-producing Klebsiella pneumoniae in a Greek hospital. Microbial drug resistance 22, 392–398. doi: 10.1089/mdr.2015.0282
Mavroidi, A., Katsiari, M., Likousi, S., Palla, E., Roussou, Z., Nikolaou, C., et al. (2020). Changing characteristics and in vitro susceptibility to ceftazidime/avibactam of bloodstream extensively drug-resistant Klebsiella pneumoniae from a Greek intensive care unit. Microb. Drug Resist. 26, 28–37. doi: 10.1089/mdr.2019.0090
Menekşe, Ş., Çağ, Y., Işık, M. E., Şahin, S., Hacıseyitoğlu, D., Can, F., et al. (2019). The effect of colistin resistance and other predictors on fatality among patients with bloodstream infections due to Klebsiella pneumoniae in an OXA-48 dominant region. Int. J. Infect. Dis. 86, 208–211. doi: 10.1016/j.ijid.2019.06.008
Mills, J. P., Rojas, L. J., Marshall, S. H., Rudin, S. D., Hujer, A. M., Nayak, L., et al. (2021). Risk factors for and mechanisms of COlistin resistance among Enterobacterales: getting at the CORE of the issue. Open Forum Infect. Dis. 8:ofab145. doi: 10.1093/ofid/ofab145
Mirshekar, M., Darbandi, A., Ghanavati, R., Shivaee, A., and Masjedian, F. (2020). Analysis of mgrB gene mutations in colistin-resistant Klebsiella pneumoniae in Tehran, Iran. Gene Reports. 21:100864. doi: 10.1016/j.genrep.2020.100864
Moghadam, M. T., Mojtahedi, A., Moghaddam, M. M., Fasihi-Ramandi, M., and Mirnejad, R. (2022). Rescuing humanity by antimicrobial peptides against colistin-resistant bacteria. Appl. Microbiol. Biotechnol. 106, 3879–3893. doi: 10.1007/s00253-022-11940-z
Moghimi, M., Haeili, M., and Mohajjel, S. H. (2021). Characterization of Tigecycline resistance among Tigecycline non-susceptible Klebsiella pneumoniae isolates from humans, food-producing animals, and in vitro selection assay. Front. Microbiol. 12:702006. doi: 10.3389/fmicb.2021.702006
Moola, S., Munn, Z., Tufanaru, C., Aromataris, E., Sears, K., Sfetcu, R., et al. (2017). “Systematic reviews of etiology and risk” in Joanna briggs institute reviewer's manual, vol. 5 (Adelaide, Australia: The Joanna Briggs Institute).
Moubareck, C. A., Mouftah, S. F., Pál, T., Ghazawi, A., Halat, D. H., Nabi, A., et al. (2018). Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int. J. Antimicrob. Agents 52, 90–95. doi: 10.1016/j.ijantimicag.2018.03.003
Naha, S., Sands, K., Mukherjee, S., Dutta, S., and Basu, S. (2022). A 12 year experience of colistin resistance in Klebsiella pneumoniae causing neonatal sepsis: two-component systems, efflux pumps, lipopolysaccharide modification and comparative phylogenomics. J. Antimicrob. Chemother. 77, 1586–1591. doi: 10.1093/jac/dkac083
Narimisa, N., Goodarzi, F., and Bavari, S. (2022). Prevalence of colistin resistance of Klebsiella pneumoniae isolates in Iran: a systematic review and meta-analysis. Ann. Clin. Microbiol. Antimicrob. 21, 1–9. doi: 10.1186/s12941-022-00520-8
Nawfal Dagher, T., Azar, E., Al-Bayssari, C., Chamieh, A. S., and Rolain, J. M. (2019). First detection of Colistin-resistant Klebsiella pneumoniae in association with NDM-5 Carbapenemase isolated from clinical Lebanese patients. Microbial drug resistance 25, 925–930. doi: 10.1089/mdr.2018.0383
Ngbede, E. O., Adekanmbi, F., Poudel, A., Kalalah, A., Kelly, P., Yang, Y., et al. (2021). Concurrent resistance to Carbapenem and Colistin among Enterobacteriaceae recovered from human and animal sources in Nigeria is associated with multiple genetic mechanisms. Front. Microbiol. 12:740348. doi: 10.3389/fmicb.2021.740348
Nguyen, T. N. T., Nguyen, P. L. N., Le, N. T. Q., Nguyen, L. P. H., Duong, T. B., Ho, N. D. T., et al. (2021). Emerging carbapenem-resistant Klebsiella pneumoniae sequence type 16 causing multiple outbreaks in a tertiary hospital in southern Vietnam. Microbial genomics. 7:000159. doi: 10.1099/mgen.0.000519
Niazadeh, M., Nikkhahi, F., Robatjazi, S., Javadi, A., Farzam, S. A., Babaei, S., et al. (2022). Evaluation of mechanisms of colistin resistance in Klebsiella pneumoniae strains isolated from patients with urinary tract infection in ICU. Iranian J. Microbiol. 14, 31–37. doi: 10.18502/ijm.v14i1.8798
Nirwan, P. K., Chatterjee, N., Panwar, R., Dudeja, M., and Jaggi, N. (2021). Mutations in two component system (PhoPQ and PmrAB) in colistin resistant Klebsiella pneumoniae from north Indian tertiary care hospital. J. Antibiot. 74, 450–457. doi: 10.1038/s41429-021-00417-2
Nordmann, P., Jayol, A., and Poirel, L. (2016). Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg. Infect. Dis. 22, 1038–1043. doi: 10.3201/eid2206.151840
Novović, K., Trudić, A., Brkić, S., Vasiljević, Z., Kojić, M., Medić, D., et al. (2017). Molecular epidemiology of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae in Serbia from 2013 to 2016. Antimicrob. Agents Chemother. 61, e02550–e02516. doi: 10.1128/AAC.02550-16
Okdah, L., AlDosary, M. S., AlMazyed, A., Alkhurayb, H. M., Almossallam, M., Al Obaisi, Y. S., et al. (2022). Genomic characterization of Colistin-resistant isolates from the king Fahad Medical City, Kingdom of Saudi Arabia. Antibiotics. 11:1597. doi: 10.3390/antibiotics11111597
Olaitan, A. O., Diene, S. M., Kempf, M., Berrazeg, M., Bakour, S., Gupta, S. K., et al. (2014). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44, 500–507. doi: 10.1016/j.ijantimicag.2014.07.020
Otter, J. A., Doumith, M., Davies, F., Mookerjee, S., Dyakova, E., Gilchrist, M., et al. (2017). Emergence and clonal spread of colistin resistance due to multiple mutational mechanisms in carbapenemase-producing Klebsiella pneumoniae in London. Sci. Rep. 7, 1–8. doi: 10.1038/s41598-017-12637-4
Palani, G. S., Ghafur, A., Krishnan, P., Rayvathy, B., and Thirunarayan, M. (2020). Intestinal carriage of colistin resistant Enterobacteriaceae in hospitalized patients from an Indian center. Diagn. Microbiol. Infect. Dis. 97:114998. doi: 10.1016/j.diagmicrobio.2020.114998
Palmieri, M., D’Andrea, M. M., Pelegrin, A. C., Mirande, C., Brkic, S., Cirkovic, I., et al. (2020). Genomic epidemiology of carbapenem-and colistin-resistant Klebsiella pneumoniae isolates from Serbia: predominance of ST101 strains carrying a novel OXA-48 plasmid. Front. Microbiol. 11:294. doi: 10.3389/fmicb.2020.00294
Pitt, M. E., Elliott, A. G., Cao, M. D., Ganesamoorthy, D., Karaiskos, I., Giamarellou, H., et al. (2018). Multifactorial chromosomal variants regulate polymyxin resistance in extensively drug-resistant Klebsiella pneumoniae. Microbial genomics. 4:e000158. doi: 10.1099/mgen.0.000158
Poirel, L., Jayol, A., Bontron, S., Villegas, M.-V., Ozdamar, M., Türkoglu, S., et al. (2015). The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 70, 75–80. doi: 10.1093/jac/dku323
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Popa, L. I., Gheorghe, I., Barbu, I. C., Surleac, M., Paraschiv, S., Măruţescu, L., et al. (2021). Multidrug resistant Klebsiella pneumoniae ST101 clone survival chain from inpatients to hospital effluent after chlorine treatment. Front. Microbiol. 11:610296. doi: 10.3389/fmicb.2020.610296
Pragasam, A. K., Shankar, C., Veeraraghavan, B., Biswas, I., Nabarro, L. E., Inbanathan, F. Y., et al. (2017). Molecular mechanisms of colistin resistance in Klebsiella pneumoniae causing bacteremia from India—a first report. Front. Microbiol. 7:2135. doi: 10.3389/fmicb.2016.02135
Prevention ECfD, Control. European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2015. Available at: https://ecdceuropaeu/en/publications-data/antimicrobial-resistance-surveillance-europe-2015, ECDC Stockholm; (2017).
Pu, D., Zhao, J., Lu, B., Zhang, Y., Wu, Y., Li, Z., et al. (2023). Within-host resistance evolution of a fatal ST11 hypervirulent carbapenem-resistant Klebsiella pneumoniae. Int. J. Antimicrob. Agents 61:106747. doi: 10.1016/j.ijantimicag.2023.106747
Rimoldi, S. G., Gentile, B., Pagani, C., Di Gregorio, A., Anselmo, A., Palozzi, A. M., et al. (2017). Whole genome sequencing for the molecular characterization of carbapenem-resistant Klebsiella pneumoniae strains isolated at the Italian ASST Fatebenefratelli Sacco Hospital, 2012–2014. BMC Infect. Dis. 17, 1–11. doi: 10.1186/s12879-017-2760-7
Roch, M., Martins, W. M. B. S., Sierra, R., Gales, A. C., and Andrey, D. O. (2022). Characterization of amino acid substitution W20S in MgrB involved in Polymyxin resistance in Klebsiella pneumoniae. Microbiology Spectrum. 10, e01766–e01721. doi: 10.1128/spectrum.01766-21
Rocha, V. F. D., Barbosa, M. S., Leal, H. F., Silva, G. E. O., Monteiro, A. S. S., Azevedo, J., et al. (2022). Prolonged outbreak of carbapenem and colistin-resistant klebsiella pneumoniae at a large tertiary hospital in Brazil. Front. Microbiol. 13:513. doi: 10.3389/fmicb.2022.831770
Rocha, I. V., dos Santos Silva, N., das Neves Andrade, C. A., de Lacerda Vidal, C. F., Leal, N. C., and Xavier, D. E. (2020). Diverse and emerging molecular mechanisms award polymyxins resistance to Enterobacteriaceae clinical isolates from a tertiary hospital of Recife, Brazil. Infect. Genet. Evol. 85:104584. doi: 10.1016/j.meegid.2020.104584
Rubic, Z., Jelic, M., Soprek, S., Tarabene, M., Ujevic, J., Goic-Barisic, I., et al. (2023). Molecular characterization of colistin resistance genes in a high-risk ST101/KPC-2 clone of Klebsiella pneumoniae in a University Hospital of Split, Croatia. Int. Microbiol. 26, 631–637. doi: 10.1007/s10123-023-00327-3
Shamina, O., Kryzhanovskaya, O., Lazareva, A., Alyabieva, N., Polikarpova, S., Karaseva, O., et al. (2020). Emergence of a ST307 clone carrying a novel insertion element MITEKpn1 in the mgrB gene among carbapenem-resistant Klebsiella pneumoniae from Moscow, Russia. Int. J. Antimicrob. Agents 55:105850. doi: 10.1016/j.ijantimicag.2019.11.007
Shankar, C., Venkatesan, M., Rajan, R., Mani, D., Lal, B., Prakash, J. A. J., et al. (2019). Molecular characterization of colistin-resistant Klebsiella pneumoniae & its clonal relationship among Indian isolates. Indian J. Med. Res. 149, 199–207. doi: 10.4103/ijmr.IJMR_2087_17
Sharahi, J. Y., Hashemi, A., Ardebili, A., and Davoudabadi, S. (2021). Molecular characteristics of antibiotic-resistant Escherichia coli and Klebsiella pneumoniae strains isolated from hospitalized patients in Tehran, Iran. ACMA 20, 1–14. doi: 10.1186/s12941-021-00437-8
Shein, A. M. S., Wannigama, D. L., Higgins, P. G., Hurst, C., Abe, S., Hongsing, P., et al. (2022). High prevalence of mgrB-mediated colistin resistance among carbapenem-resistant Klebsiella pneumoniae is associated with biofilm formation, and can be overcome by colistin-EDTA combination therapy. Sci. Rep. 12:12939. doi: 10.1038/s41598-022-17083-5
Singh, S., Pathak, A., Rahman, M., Singh, A., Nag, S., Sahu, C., et al. (2021). Genetic characterisation of colistin resistant Klebsiella pneumoniae clinical isolates from North India. Front. Cell. Infect. Microbiol. 11:666030. doi: 10.3389/fcimb.2021.666030
Sisti, S., Diotti, R. A., Caputo, V., Libera, M., Ferrarese, R., Carletti, S., et al. (2022). Identification of a novel mutation involved in colistin resistance in Klebsiella pneumoniae through next-generation sequencing (NGS) based approaches. New Microbiol. 45, 199–209
Snyman, Y., Whitelaw, A. C., Reuter, S., Maloba, M. R. B., and Newton-Foot, M. (2021). Colistin resistance mechanisms in clinical Escherichia coli and Klebsiella spp. isolates from the Western cape of South Africa. Microb. Drug Resist. 27, 1249–1258. doi: 10.1089/mdr.2020.0479
Solgi, H., Shahcheraghi, F., Bolourchi, N., and Ahmadi, A. (2020). Molecular characterization of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae ST11 harbouring blaNDM-1 and blaOXA-48 carbapenemases in Iran. Microb. Pathog. 149:104507. doi: 10.1016/j.micpath.2020.104507
Sonnevend, Á., Ghazawi, A., Hashmey, R., Haidermota, A., Girgis, S., Alfaresi, M., et al. (2017). Multihospital occurrence of pan-resistant Klebsiella pneumoniae sequence type 147 with an IS Ecp1-directed Bla OXA-181 insertion in the mgrB gene in the United Arab Emirates. Antimicrob. Agents Chemother. 61, e00418–e00417. doi: 10.1128/AAC.00418-17
Tietgen, M., Sedlaczek, L., Higgins, P. G., Kaspar, H., Ewers, C., and Göttig, S. (2022). Colistin resistance mechanisms in human and veterinary Klebsiella pneumoniae isolates. Antibiotics. 11:1672. doi: 10.3390/antibiotics11111672
Torres, D. A., Seth-Smith, H. M., Joosse, N., Lang, C., Dubuis, O., Nüesch-Inderbinen, M., et al. (2021). Colistin resistance in gram-negative bacteria analysed by five phenotypic assays and inference of the underlying genomic mechanisms. BMC Microbiol. 21, 1–12. doi: 10.1186/s12866-021-02388-8
Vendrik, K. E., de Haan, A., Witteveen, S., Hendrickx, A. P., Landman, F., Notermans, D. W., et al. (2022). A prospective matched case-control study on the genomic epidemiology of colistin-resistant Enterobacterales from Dutch patients. Commun. Med. 2:55. doi: 10.1038/s43856-022-00115-6
Wang, Y., Tian, G.-B., Zhang, R., Shen, Y., Tyrrell, J. M., Huang, X., et al. (2017). Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 17, 390–399. doi: 10.1016/S1473-3099(16)30527-8
Wang, S., Wang, L., Jin, J., Li, G., Shao, H., Song, Y., et al. (2023). Genomic epidemiology and characterization of Carbapenem-resistant Klebsiella pneumoniae in ICU inpatients in Henan Province, China: a multicenter cross-sectional study. Microbiology Spectrum. 11, e04197–e04122. doi: 10.1128/spectrum.04197-22
Wright, M. S., Suzuki, Y., Jones, M. B., Marshall, S. H., Rudin, S. D., van Duin, D., et al. (2015). Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 59, 536–543. doi: 10.1128/AAC.04037-14
Xiao, C., Li, X., Huang, L., Cao, H., Han, L., Ni, Y., et al. (2023). Prevalence and molecular characteristics of polymyxin-resistant Enterobacterales in a Chinese tertiary teaching hospital. Front. Cell. Infect. Microbiol. 13:418. doi: 10.3389/fcimb.2023.1118122
Xie, M., Chen, K., Dong, N., Xu, Q., Chan, E. W.-C., Zhang, R., et al. (2022). Phenotypic changes associated with in vivo evolution of Colistin resistance in ST11 Carbapenem-resistant Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 12:841748. doi: 10.3389/fcimb.2022.841748
Yang, T.-Y., Wang, S.-F., Lin, J.-E., Griffith, B. T. S., Lian, S.-H., Hong, Z.-D., et al. (2020). Contributions of insertion sequences conferring colistin resistance in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 55:105894. doi: 10.1016/j.ijantimicag.2020.105894
Yap, P. S. X., Ahmad Kamar, A., Chong, C. W., Ngoi, S. T., and Teh, C. S. J. (2020). Genomic insights into two colistin-resistant Klebsiella pneumoniae strains isolated from the stool of preterm neonate during the first week of life. Microb. Drug Resist. 26, 190–203. doi: 10.1089/mdr.2019.0199
Yap, P. S.-X., Cheng, W.-H., Chang, S.-K., Lim, S.-H. E., and Lai, K.-S. (2022). MgrB mutations and altered cell permeability in Colistin resistance in Klebsiella pneumoniae. Cells 11:2995. doi: 10.3390/cells11192995
Yoshino, M., Aihara, M., Gotoh, Y., Akimoto, M., Tatsuhara, W., Kiyosuke, M., et al. (2021). Stepwise evolution of a Klebsiella pneumoniae clone within a host leading to increased multidrug resistance. Msphere. 6, e00734–e00721. doi: 10.1128/mSphere.00734-21
Yousfi, H., Hadjadj, L., Dandachi, I., Lalaoui, R., Merah, A., Amoura, K., et al. (2019). Colistin-and carbapenem-resistant Klebsiella pneumoniae clinical_isolates: Algeria. Microb. Drug Resist. 25, 258–263. doi: 10.1089/mdr.2018.0147
Yusof, N. Y., Norazzman, N. I. I., Hakim, S. N.’. W. A., Azlan, M. M., Anthony, A. A., Mustafa, F. H., et al. (2022). Prevalence of mutated Colistin-resistant Klebsiella pneumoniae: a systematic review and Meta-analysis. Tropical Med. Infectious Dis. 7:414. doi: 10.3390/tropicalmed7120414
Zafer, M. M., El-Mahallawy, H. A., Abdulhak, A., Amin, M. A., Al-Agamy, M. H., and Radwan, H. H. (2019). Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann. Clin. Microbiol. Antimicrob. 18, 1–8. doi: 10.1186/s12941-019-0339-4
Zahedi Bialvaei, A., Eslami, P., Ganji, L., Dolatyar Dehkharghani, A., Asgari, F., Koupahi, H., et al. (2023). Prevalence and epidemiological investigation of mgrB-dependent colistin resistance in extensively drug resistant Klebsiella pneumoniae in Iran. Sci. Rep. 13:10680. doi: 10.1038/s41598-023-37845-z
Zaman, T. U., Albladi, M., Siddique, M. I., Aljohani, S. M., and Balkhy, H. H. (2018). Insertion element mediated mgrB disruption and presence of ISKpn28 in colistin-resistant Klebsiella pneumoniae isolates from Saudi Arabia. Infection and Drug Resistance. 11, 1183–1187. doi: 10.2147/IDR.S161146
Zhang, R., Dong, N., Huang, Y., Zhou, H., Xie, M., Chan, E. W.-C., et al. (2018). Evolution of tigecycline-and colistin-resistant CRKP (carbapenem-resistant Klebsiella pneumoniae) in vivo and its persistence in the GI tract. Emerging microbes & infections. 7, 1–11. doi: 10.1038/s41426-018-0129-7
Zhu, Y., Galani, I., Karaiskos, I., Lu, J., Aye, S. M., Huang, J., et al. (2019). Multifaceted mechanisms of colistin resistance revealed by genomic analysis of multidrug-resistant Klebsiella pneumoniae isolates from individual patients before and after colistin treatment. J. Inf. Secur. 79, 312–321. doi: 10.1016/j.jinf.2019.07.009
Keywords: colistin, mgrB, Klebsiella pneumoniae, colistin-resistant, global prevalence
Citation: Khoshbayan A, Narimisa N, Elahi Z, Bostanghadiri N, Razavi S and Shariati A (2024) Global prevalence of mutation in the mgrB gene among clinical isolates of colistin-resistant Klebsiella pneumoniae: a systematic review and meta-analysis. Front. Microbiol. 15:1386478. doi: 10.3389/fmicb.2024.1386478
Edited by:
Giovanni Gherardi, Campus Bio-Medico University, ItalyReviewed by:
Ramesh N., Vellore Institute of Technology, IndiaArta Karruli, University Medical Center Mother Teresa (QSUT), Albania
Copyright © 2024 Khoshbayan, Narimisa, Elahi, Bostanghadiri, Razavi and Shariati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aref Shariati, YXJlZnNoYXJpYXRpMDExMUBzYm11LmFjLmly; YXJlZnNoYXJpYXRpMDExMUBnbWFpbC5jb20=
†These authors have contributed equally to this work
 Amin Khoshbayan
Amin Khoshbayan Negar Narimisa
Negar Narimisa Zahra Elahi2,3
Zahra Elahi2,3 Narjess Bostanghadiri
Narjess Bostanghadiri Aref Shariati
Aref Shariati