
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 22 March 2024
Sec. Microbe and Virus Interactions with Plants
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1386417
This article is part of the Research TopicAdvances in Beneficial and Pathogenic Plant-Microbe Interactions in Cereal CropsView all 7 articles
Black scurf caused by Rhizoctonia solani severely affects potato production. Through amplification of V3-V4 and ITS1-5f variable regions of 16S and internal transcribed spacer (ITS) rRNA, the study was based on the location (Kunming, Qujing, and Zhaotong), plant components (rhizosphere soil and roots), and sample types (healthy and diseased) to assess the diversity of bacterial and fungal communities. We found plant components significantly influence microbial diversity, with rhizosphere soil being more diverse than roots, and the microbial community in the root is mainly derived from the rhizosphere soil. Moreover, the rhizosphere soil and roots of healthy potato plants exhibit greater microbial diversity compared to those of potato plants infected by Rhizoctonia solani. Bacterial phyla Actinobacteriota and Acidobacteriota were enriched in rhizosphere soil compared to that of roots, whereas Proteobacteria and Cyanobacteria showed the opposite trend. Fungal phylum Ascomycota was found in low relative abundance in rhizosphere soil than in roots, whereas Basidiomycota showed the opposite trend. Bacterial genera including Streptomyces, Lysobacter, Bacillus, Pseudomonas, Ensifer, Enterobacter, and the Rhizobium group (Allorhizobium, Neorhizobium, Pararhizobium, Rhizobium), along with fungal genera such as Aspergillus, Penicillium, Purpureocillium, and Gibberella moniliformis, have the potential ability of plant growth promotion and disease resistance. However, most fungal species and some bacterial species are pathogenic to potato and could provide a conducive environment for black scurf infection. Interaction within the bacterial network increased in healthy plants, contrasting with the trend in the fungal network. Our findings indicate that R. solani significantly alters potato plant microbial diversity, underscoring the complexity and potential interactions between bacterial and fungal communities for promoting potato plant health and resistance against black scurf.
The soilborne pathogenic fungus Rhizoctonia solani Kühn, also known as Thanatephorus cucumeris, is a basidiomycete with a large host range and different anastomosis groups (AGs). The pathogen can cause diseases in plant families including Solanaceae, Poaceae, Amaranthaceae, Fabaceae, Brassicaceae, Rubiaceae, Araceae, Malvaceae, Moraceae, and Linaceae (Mayo et al., 2015; Verwaaijen et al., 2017; Ajayi-Oyetunde and Bradley, 2018). It is divided into 14 AGs based on its special genetic and biological characteristics, including AG-1 to AG-13 and a bridging isolate AG-BI (Parissa and Tarighi, 2011), among which AG-1 to AG-13 were reported to cause Rhizoctonia disease on potato (Balali et al., 1995; Truter and Wehner, 2004; Yanar et al., 2005; Woodhall et al., 2008; Yang et al., 2015; Gush et al., 2019; Murdock et al., 2019; Yang et al., 2019; Lopez Corrales et al., 2023). It is widely known that R. solani AG-3 PT is notably aggressive, primarily causing stem canker or black scurf on potatoes (Balali et al., 1995; Woodhall et al., 2008, 2013; Fiers et al., 2011; Yang et al., 2015).
Rhizoctonia solani AG-3 PT infects all parts of the potato plant, including sprout, tubers, stems, stolons, roots, and leaves (Atkinson et al., 2010). Infected plants exhibit a range of symptoms such as sprout canker formation (Figure 1A; Gutierrez et al., 1997); sunken, brown, necrotic lesions on stems, stolons, and roots and sclerotia development on tubers (Figures 1B,C; Lehtonen et al., 2009); leaves turning red and upward curling (Figure 1D; Tsror, 2010; Malik et al., 2014); an absence of tuber formation or the growth of small tubers (Figure 1E); aerial tubers formation on stems (Figure 1F; Beukema and van der Zang, 1990); and a grayish-white, felt-like mycelium mat emerging at the base of stems and on the plant parts that are in contact with soil (Figure 1G; Banville and Carling, 2001). All of these symptoms may appear on infected potato plants either separately or in combination (Zheng et al., 2014; Muzhinji et al., 2018). Currently, Black scurf of potato disease occurs in all potato-growing regions around the world, including Yunnan Province, China, leading to marketable yield losses of up to 30% (Banville, 1989; Samsatly et al., 2020; Zrenner et al., 2021). Yunnan Province is located in southwest China, with unique geographical and climatic characteristics (Li et al., 2022). Through an investigation of the occurrence of Rhizoctonia diseases in the main potato production areas of Yunnan Province, it was found that the principal production regions, such as the Diqing Tibetan Autonomous Prefecture, Lijiang, and Dali Bai Autonomous Prefecture in Northwestern Yunnan, as well as Zhaotong, Qujing, and Kunming in Northeastern Yunnan, all experienced outbreaks of potato black scurf disease. The incidence rate in fields with severe infections can reach up to 70% to 80%. Particularly in Kunming, Qujing, and Zhaotong, the highest incidence rates were observed at 36.00%, 29.75%, and 29.01%, respectively, followed by Lijiang, Diqing, and Dali, with rates of 27.28%, 20.91%, and 12.36% (Wang et al., 2014). The widespread occurrence of this disease has significantly impacted the yield and quality of local potatoes. Locations play an important role on the occurrence of soilborne diseases (Cai et al., 2021).
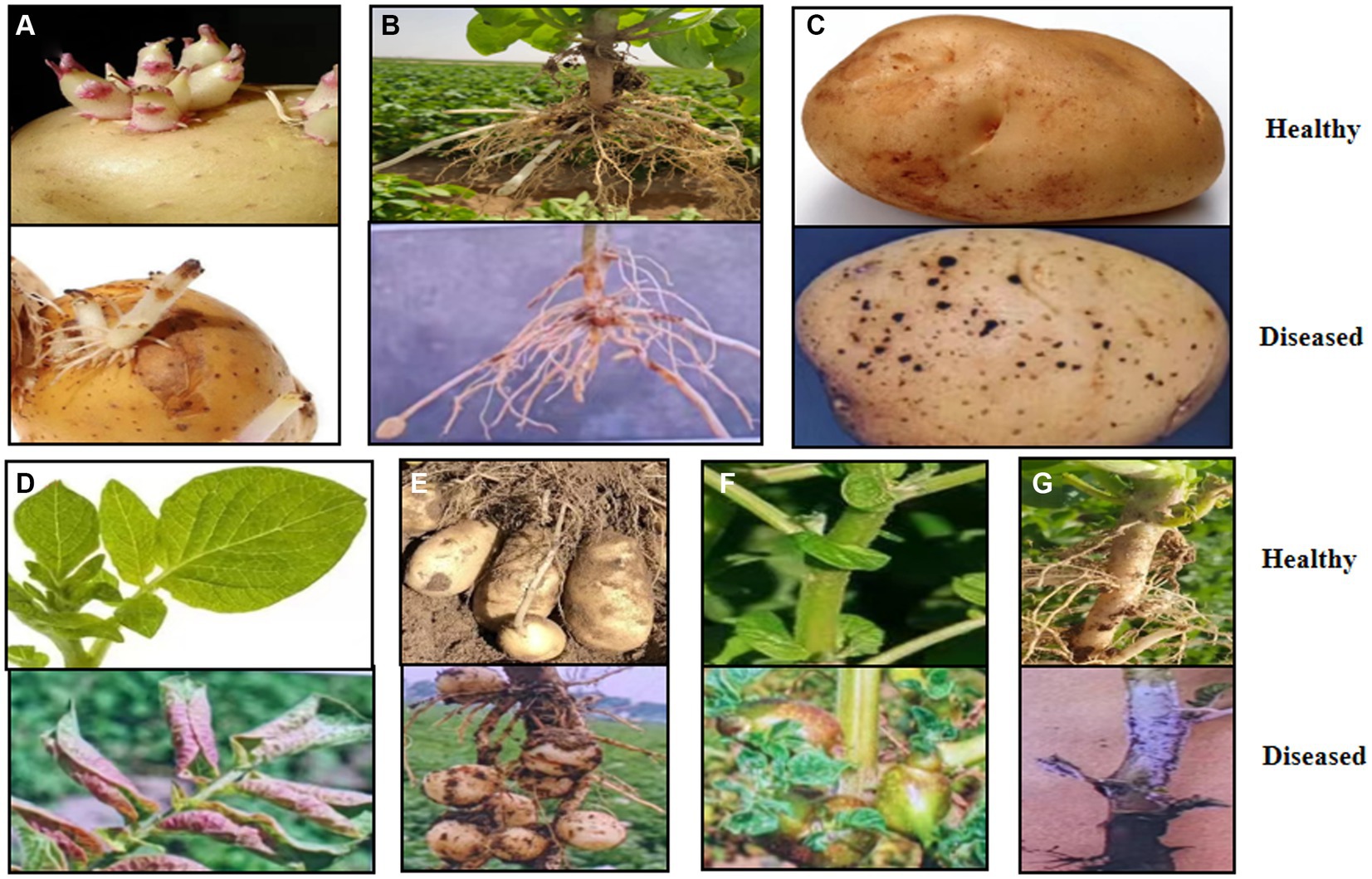
Figure 1. Disease potato plant with typical black scurf symptoms. (A) Causing sprout nipping and cankers. (B) Sunken brown necrotic lesions formation on stems, stolons, and roots. (C) Sclerotia formation on tubers. (D) Reddening and inward curling of leaves. (E) No tuber formation or small tubers. (F) Aerial potato formation on stems. (G) White fruiting bodies forming at stem base.
It is important to note that R. solani AG3 PT predominantly colonizes the belowground parts of potato, including roots (Schreiter et al., 2018). It is a seed- and soil-borne pathogen, which survives through sclerotia and mycelia in infected seeds or soil in tropical environments. Exhibiting facultative parasitism, it can survive in soil residues as a saprophyte without a specific host (Akber et al., 2022). Infected soil is a primary inoculum source, with the pathogen emerging in the rhizosphere soil from roots (Senapati et al., 2022). The spread of the pathogen occurs through mechanisms such as sclerotia dispersal by rain, contaminated soil particles, and mycelial networks connecting plants, as well as via seeds that are already infected (Schreiter et al., 2018). While the teleomorph stage allows airborne basidiospore transmission but is less common in the fields. The sclerotia, the asexual stage, can remain viable in soil for years (Kouzai et al., 2020).
The rhizosphere, the root-surrounding zone, is among Earth’s most intricate ecosystems (Wierzbicka-Woś et al., 2019). Plant disease resistance and growth largely depend on rhizosphere microbial diversity (Mendes et al., 2013; Dong et al., 2019). Plant genetics and soil type play key roles in forming a beneficial rhizosphere microbiome (Mendes et al., 2013; Dong et al., 2019). Recent scientific progress and advanced sequencing technologies have simplified studying plant-microbe interactions (Govindasamy et al., 2014; Mhlongo et al., 2018). Nonetheless, further research is needed to fully understand these interactions and their mechanisms (Bulgarelli et al., 2013).
Currently, biological control involving specific endophytes and rhizosphere microbiome like Aspergillus, Bacillus, Pseudomonas, Streptomyces, and Lysobacter is a viable method for managing various soilborne diseases and this approach works through mechanisms such as antagonism, altering rhizospheric microbial diversity, and metabolite production (Ma et al., 2018; Schreiter et al., 2018; Wu et al., 2020; Zhang J. et al., 2020; Asaturova et al., 2021; Wei et al., 2021; Abdelaziz et al., 2023; Rashad et al., 2023). In recent years, there have been successive reports on the biological control of potato black scurf. Ikeda et al. (2012) found that Pythium oligandrum effectively reduces potato black scurf through mechanisms like hyperparasitism and triggering the plant’s resistance. Furthermore, Tsror et al. (2001) observed that in organic potato cultivation, applying Trichoderma harzianum, nonpathogenic Rhizoctonia (np-R), and cattle manure compost amendment (CMC-H) directly into the soil furrows effectively reduced the occurrence of black scurf. Additionally, studies have shown that strains StS3 and StT2 of Pseudomonas spp. are promising as biocontrol agents. They not only enhance plant growth but also effectively diminish the occurrence of black scurf in potatoes (Tariq et al., 2010). The challenge in effectively controlling this disease stems from its ability to infect a wide variety of hosts, its complex species characteristics, extensive geographical spread, and its resilience (Mayo et al., 2015; Verwaaijen et al., 2017; Ajayi-Oyetunde and Bradley, 2018). Currently, there is no comprehensive solution to fully manage this widespread and stubborn disease.
Potato is a major staple food and the fourth largest crop grown worldwide (Liu et al., 2016). It serves not only as a food source, consumable as a vegetable or processed into snack foods, but also plays a vital role as an industrial raw material (Kowalczewski et al., 2019). The primary regions for potato cultivation in China include areas like Inner Mongolia, Gansu, Guizhou, and Yunnan, which provide ideal natural conditions for its growth. However, R. solani annually causes substantial decreases in both the yield and quality of potato crops. Addressing this challenge necessitates a comprehensive understanding of both the microbiome’s population dynamics and its distribution within potato plants, to effectively control the black scurf disease. Therefore, the present study aims to explore the core microbiota (bacteria and fungi) associated with different locations (Kunming, Qujing, and Zhaotong), plant components (rhizosphere soil and roots), and sample types (healthy and diseased). This study posits that exploring the natural potato microbiome can aid in developing strategies to reduce the prevalence of black scurf disease in potatoes.
Rhizosphere soil and root samples of healthy and diseased potato plants were collected from three various locations: Kunming (24.9195°N, 102.4785°E), Qujing (25.6742°N, 104.2550°E), and Zhaotong (28.6299°N, 104.4160°E) in Yunnan Province, China, in September 2021 (Figure 2). Over the past 4 years, these agricultural fields have been consistently used for potato cultivation, employing conventional management practices without the use of any probiotic microorganisms. During the collection process, the top layer of soil, measuring 4–5 cm, was removed, and the potato plants were then carefully uprooted (three plants per field from three different fields for both healthy and diseased plants). The bulk soil was separated by gently shaking the roots, and the finer soil particles adhering to the roots were retained as samples of rhizosphere soil, along with fibrous root samples. In total, 12 composite samples were acquired (three replicates per sample) from these locations in Yunnan. These samples were immediately placed in polythene bags and stored in an icebox for transport. Upon arrival at the laboratory, they were preserved at −80°C for subsequent analysis.
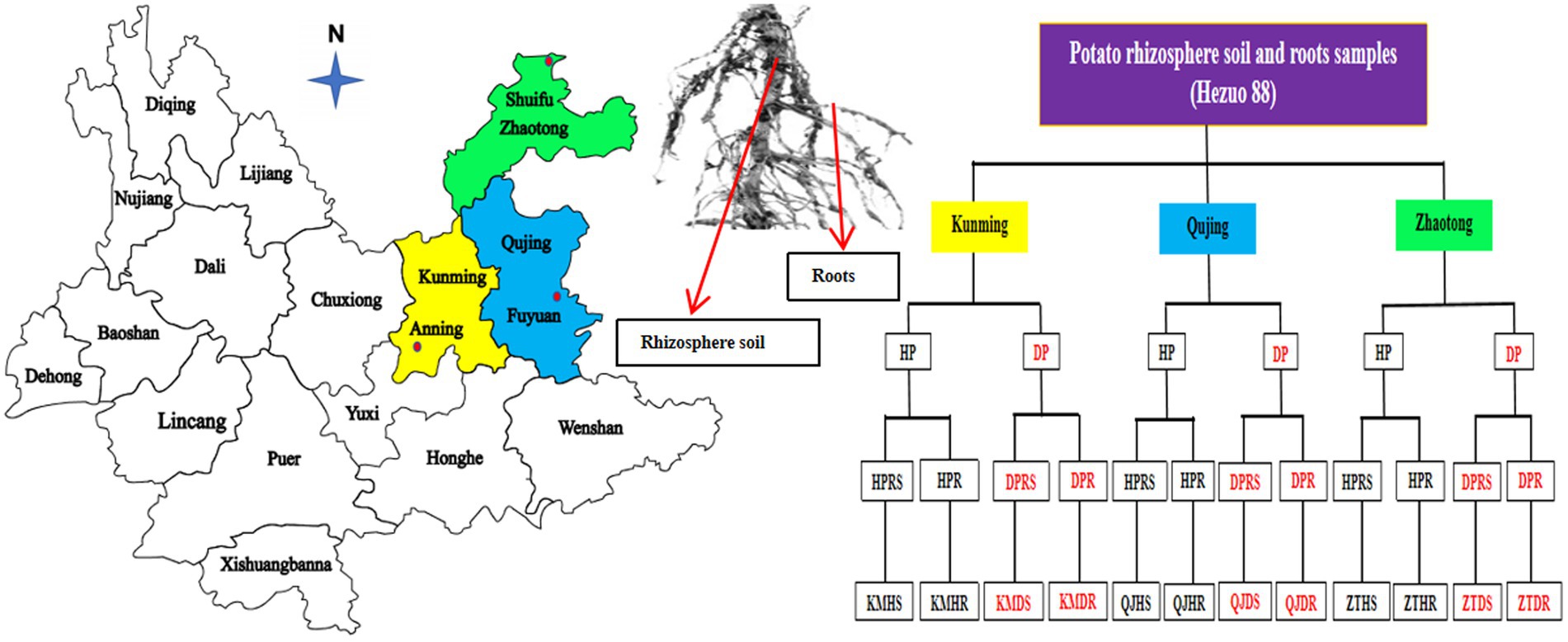
Figure 2. Overview of sampling strategy for both healthy and black scurf-infected diseased Hezuo 88 potato plants. We gathered samples of rhizosphere soil and roots from three distinct sites in Yunnan Province: Kunming (Anning), Qujing (Fuyuan), and Zhaotong (Shuifu). Here: Kunming (KM), Qujing (QJ), Zhaotong (ZT), Healthy plants (HP), Diseased plants (DP), Healthy plants rhizosphere soil (HPRS), Healthy plants roots (HPR), Diseased plants rhizosphere soil (DPRS), Diseased plants roots (DHR), Kunming healthy plants rhizosphere soil (KMHS), Kunming healthy plants roots (KMHR), Kunming diseased plants rhizosphere soil (KMDS), Kunming diseased plants roots (KMDR), Qujing healthy plants rhizosphere soil (QJHS), Qujing healthy plants roots (QJHR), Qujing diseased plants rhizosphere soil (QJDS), Qujing diseased plants roots (QJDR), Zhaotong healthy plants rhizosphere soil (ZTHS), Zhaotong healthy plants roots (ZTHR), Zhaotong diseased plants rhizosphere soil (ZTDS), Zhaotong diseased plants roots (ZTDR).
Genomic DNA was isolated from each sample using the Soil and Plant DNA Extraction Kit (Zymo Research Corp., Irvine, CA, United States). The process involved extracting DNA from 0.5 grams of soil and 1 gram of roots per sample, adhering to the kit’s provided guidelines. The DNA’s purity was then assessed using a NanoDrop spectrophotometer (ND2000, Thermo Scientific, Madison, WI, United States), ensuring an optical density (OD) ratio of 260/280 nm between 1.7 and 1.9. The isolated DNA was subsequently stored at −20°C for later analysis. For investigating the bacterial and fungal diversity, the V3-V4 and ITS1-5F regions of the 16S and internal transcribed spacer (ITS) rRNA genes were amplified. This was done using two sets of universal primers: 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′) for bacteria, and 1743F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and 2043R (5′-GCTGCGTTCTTCATCGATGC-3′) for fungi (Zhang J. et al., 2020).
The construction of the amplicon library followed the protocols for 16S and ITS Metagenomic Sequencing Library preparation, utilizing the Nextera XT Index Kit (Illumina Inc. Madison, WI, United States). To evaluate the quality of the amplicons, gel electrophoresis was employed. The purification of the amplicon library was conducted using 1X AMPure XP beads, with further assessment on an Agilent DNA1000 chip via a Bioanalyzer2100. Quantification was performed using the Qubit Fluorometer 2.0 and a Qubit dsDNA assay kit (Life Technologies, Cat. No. Q328520; Gao et al., 2018). Equal amounts of these purified amplicons were combined for further sequencing analysis. The sequencing was carried out using the Illumina MiSeq platform at Novogene Bioinformatics Technology Co. Ltd., based in Beijing, China.
The initial sequencing data were gathered in FASTQ format, and the Trimmomatic software was employed to eliminate low-quality reads (those with a score below 20) and to arrange the reads into paired ends (Bolger et al., 2014). For the assembly of these paired-end reads, FLASH software was utilized, setting parameters for a minimum and maximum overlap of 10 and 200 bp, respectively, with a maximum mismatch rate of 20%. The UCHIME software was then applied for the exclusion of chimeric sequences, facilitating the generation of clean reads (Edgar et al., 2011).
The UPARSE pipeline was utilized to process the clean reads, leading to the formation of operational taxonomic units (OTUs) at a similarity threshold of 97% or higher (Edgar, 2013). Taxonomic classification of each representative read and OTU was carried out using the ribosomal database project (RDP) classifier within the SILVA database for bacterial species (with a confidence level of 70%) and the UNITE database for fungal species (Ikeda et al., 2012; Kõljalg et al., 2013). The analysis of OTUs included their relative abundance at both the genus and phylum levels. Furthermore, alpha and beta diversity indices were computed to assess species richness and evenness. To identify common and unique OTUs across different variables, such as sample types, plant components, and locations, a Venn diagram was employed.
Statistical analysis of the data was carried out using the t-test method, setting the significance level at p < 0.05. IBM SPSS software, version 20.0 (SPSS Inc., Chicago, IL, United States), was the tool of choice for all statistical computations. The QIIME software, version 1.9.1, was employed to determine various indices, including observed OTUs, Chao1, Shannon, and the abundance-based coverage estimator (ACE). For the beta diversity assessment of both bacterial and fungal communities, the Bray–Curtis dissimilarity index was calculated and then applied in principal coordinate analysis (PCoA) using QIIME. The creation of relative abundance bar plots and heatmaps at the genera level, as well as bar plots at the species level, was achieved through R scripts in R software (version 2.15.3; Dong et al., 2018). Co-occurrence network analysis at the genera level for OTUs was conducted using the SPARCC method in R, adhering to criteria of p < 0.05 and a correlation coefficient greater than 0.3. Network characteristics were computed and visualized utilizing Gephi version 0.9.2. For the processing and illustration of all figures, Adobe Illustrator CC 2019, provided by Adobe Systems Inc. in San Francisco, CA, United States, was used.
This study delved into the composition of bacterial and fungal communities in various segments of the potato plant, including the rhizosphere soil and roots, across different regions (Kunming, Qujing, and Zhaotong), and between contrasting types of samples (healthy vs. diseased). Supplementary Table 1 displays information on the number of raw and clean reads, along with quality control metrics (Q20% and Q30%), derived from amplifying the 16S (V3-V4) and ITS (1-5f) rRNA sequences of bacteria and fungi, respectively. Post quality assurance and the removal of chimeric sequences, the average yield was 70,327 bacterial and 77,241 fungal clean reads for each sample, with an average sequence length of 415 bps for bacteria and 265 bps for fungi, as obtained through Illumina sequencing (Supplementary Table 1). Furthermore, rarefaction curves constructed from the OTUs indicated comprehensive sampling coverage for all samples in both the bacterial and fungal communities (Supplementary Figure 1).
The impact of various factors, including geographical locations (Kunming, Qujing, and Zhaotong), parts of the plant (rhizosphere soil and roots), and the nature of the samples (healthy vs. diseased), on the composition of bacterial and fungal communities was examined. The Bray–Curtis dissimilarity metric was employed to assess beta diversity, which reflects variations in the structure of bacterial and fungal communities, across all 12 composite samples (Figure 3). Among these factors, the components of the plant, namely rhizosphere soil and roots, were found to have a significant effect on the composition of both bacterial and fungal communities. The distinct separation of samples along one axis for the bacterial community composition suggests a more pronounced influence of this variable on the structure of bacterial communities compared to fungal ones. The types of samples and their locations exhibited a minimal impact on both bacterial and fungal communities. A different trend was noted in the principal coordinate analysis (PCoA), revealing differences of 40.44% and 40.46% in the composition of bacterial and fungal communities, respectively. To further evaluate beta diversity, distance heatmap graphs were created for all 12 samples using both Weighted UniFrac (which considers taxa abundances) and Unweighted UniFrac (sensitive to less common taxa), providing insight into the diversity of bacterial and fungal communities (Supplementary Figure 2).
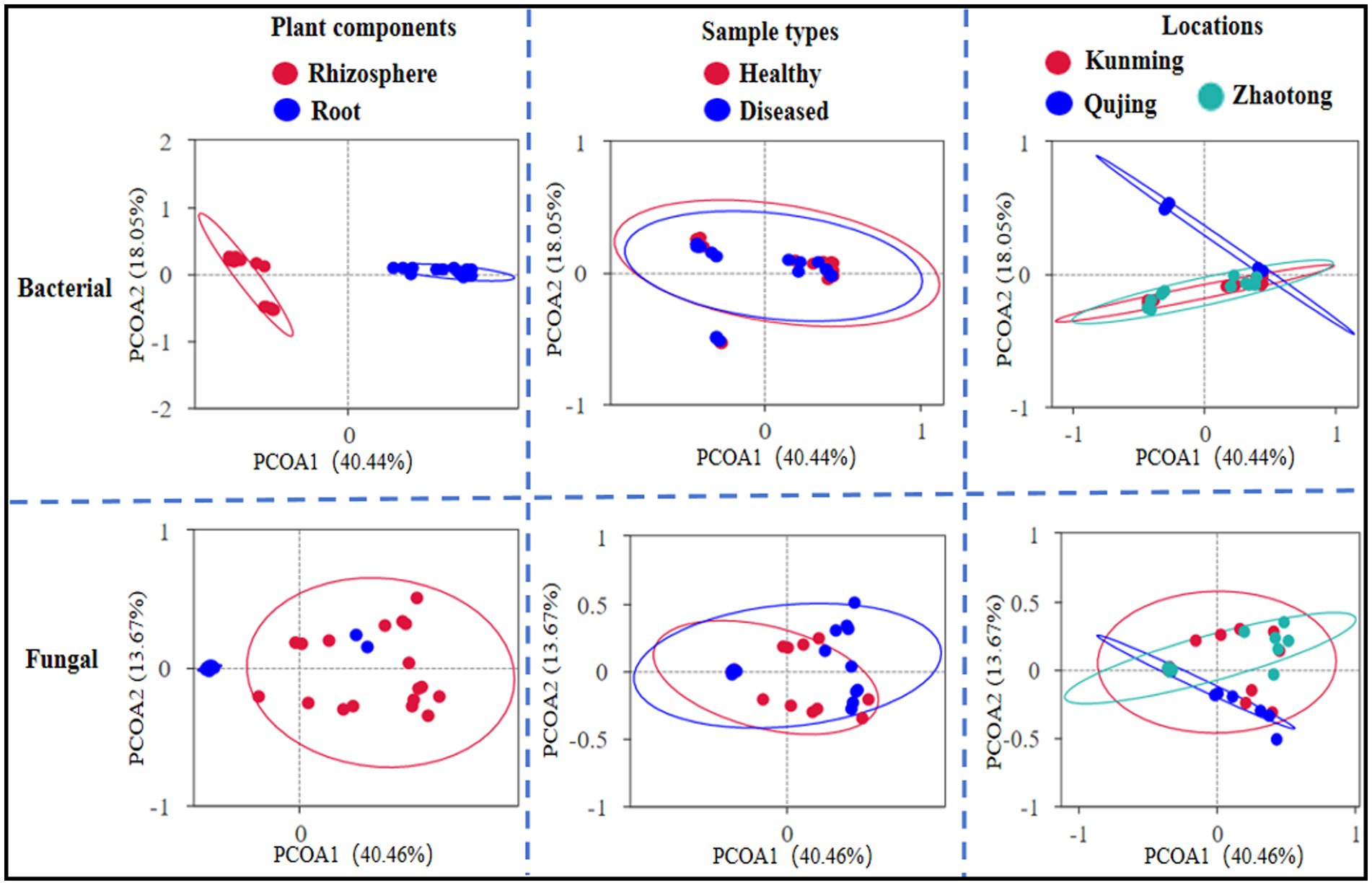
Figure 3. Principal coordinate analysis (PCoA) utilized Bray–Curtis dissimilarity metrics to exhibit the beta diversity analysis across all 12 combined samples (with each sample having three replicates) of potato plants, considering three different variables.
Figure 4 presents the alpha diversity indices, including the Observed species, Shannon, Chao 1, and Pielou_e at cutoff levels of 3%. Within the different plant parts (rhizosphere soil and roots), it was observed that the alpha diversity indices, namely Observed species, Shannon, Chao 1, and Pielou_e, were significantly greater in rhizosphere soil compared to the roots for both bacterial and fungal communities. This suggests a notably higher count of bacteria and fungi in the rhizosphere soil than in the roots. Regarding the types of samples (healthy and diseased), the alpha diversity indices for bacterial and fungal communities were found to be higher in healthy samples as opposed to diseased ones. The locations have little impact on alpha diversity indices of bacterial and fungal communities.
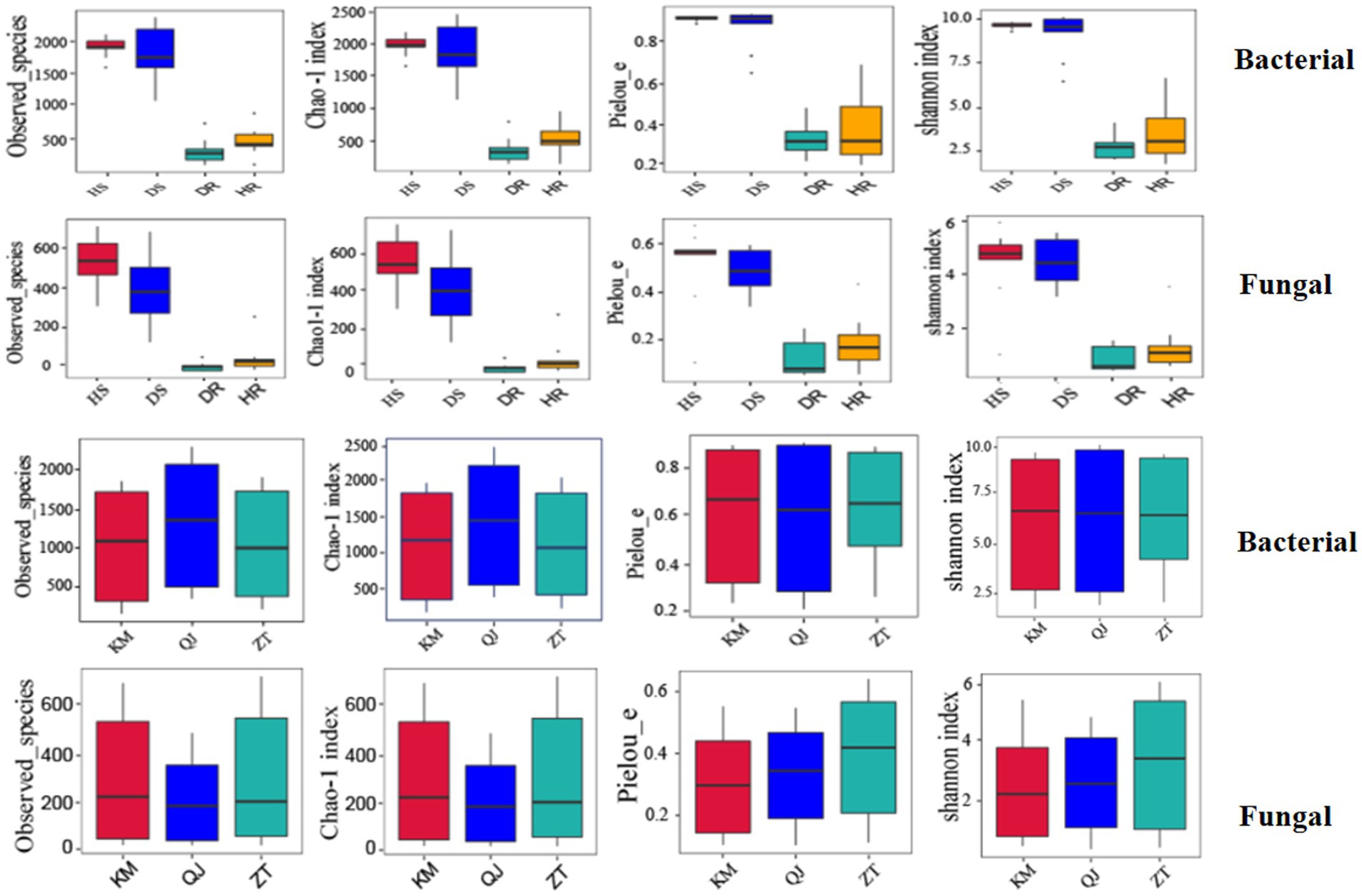
Figure 4. Boxplot of bacterial (top) and fungal (bottom) showing alpha diversity indexes of potato plants under three variables (locations, sample types, and plant components). HS, healthy rhizosphere soil; DS, diseased rhizosphere soil; HR, healthy roots; DR, diseased roots; KM, Kunming; QJ, Qujing; ZT, Zhaotong.
The number of operational taxonomic units (OTUs) was observed to be higher in rhizosphere soil than in root samples. Rhizosphere soil samples displayed greater diversity and richness in both bacterial and fungal OTUs compared to root samples. Furthermore, a higher diversity of bacterial communities was noted in healthy plant samples as opposed to diseased ones (Figure 5). In terms of specific bacterial OTUs, rhizosphere soil contained a substantially larger count (13,579) than root tissue (2,858), with 1,511 OTUs shared between them. Conversely, root tissues fewer specific fungal OTUs (481) compared to those in rhizosphere soil (3,704), with 173 OTUs common to both. When comparing healthy and diseased plant samples, unique bacterial OTUs numbered 6,988 and 6,672, respectively, with 4,288 common OTUs between them. For fungal OTUs, healthy samples had 1,859 unique OTUs, surpassing the 1,521 in diseased samples, with 978 OTUs shared. Further analysis of OTUs from different locations (Kunming, Qujing, and Zhaotong) showed that a total of 17,948 and 4,358 specific OTUs were recovered for both bacterial and fungal communities, respectively, and 713 (bacterial) and 144 (fungal) OTUs were found as common OTUs.
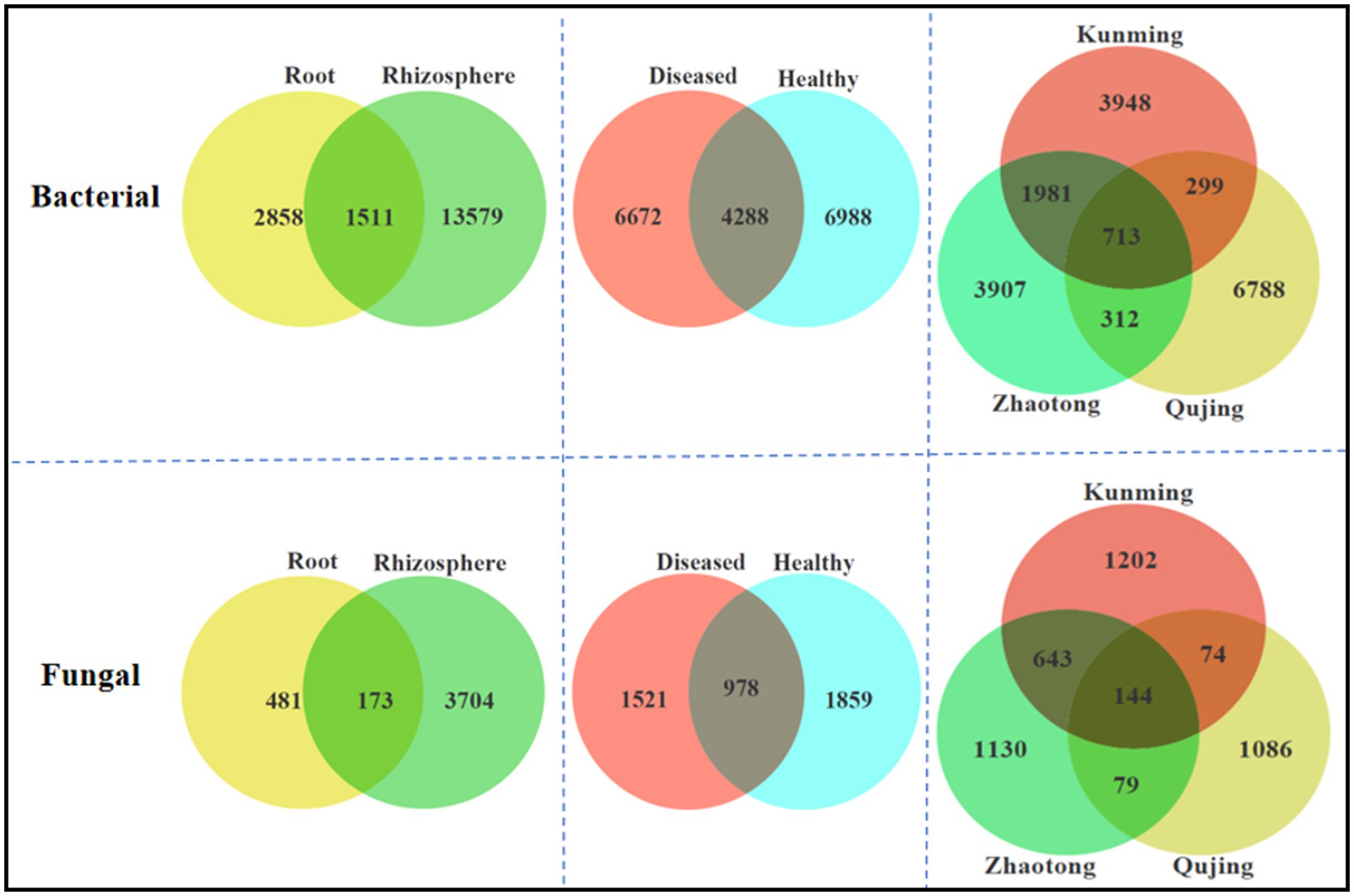
Figure 5. Distribution of bacterial (top) and fungal (bottom) operational taxonomic units in three variables, i.e., plant components (rhizosphere soil and roots), sample types (healthy and diseased), and locations (Kunming, Qujing, and Zhaotong).
Figure 6 and Supplementary Table 2 display the top 10 bacterial and fungal phyla whose relative abundance exceeds 1%. The predominant bacterial phyla in all samples of rhizosphere soil and roots, with a relative abundance over 1%, include Proteobacteria, Cyanobacteria, Firmicutes, Actinobacteriota, Chloroflexi, Acidobacteriota, Gemmatimonadota, Bacteroidota, Myxococcota, Crenarchaeota, and Others (Figure 6A). Similarly, the leading fungal phyla in these samples, with a relative abundance above 1%, are Ascomycota, Basidiomycota, Mortierellomycota, Mucoromycota, Rozellomycota, Fungi_phy_Incertae_sedis, Chytridiomycota, Aphelidiomycota, Basidiobolomycota, Olpidiomycota, and Others (Figure 6B). Among the three factors (plant components, sample types, and locations), the plant components, namely rhizosphere soil and roots, markedly affect the composition of bacterial and fungal communities. In rhizosphere soil, a high relative abundance of the phyla Actinobacteriota (19.64% average) and Acidobacteriota (10.35% average) was observed. Conversely, in root samples, Proteobacteria (46.54% average) and Cyanobacteria (45.67% average) were more abundantly present (Figure 6A). For fungal phyla, Ascomycota showed a lower relative abundance in rhizosphere soil (64.84% average) compared to roots (87.22% average). On the other hand, the phylum Basidiomycota demonstrated a higher relative abundance in rhizosphere soil (15.33% average) compared to roots (0.48% average; Figure 6B).
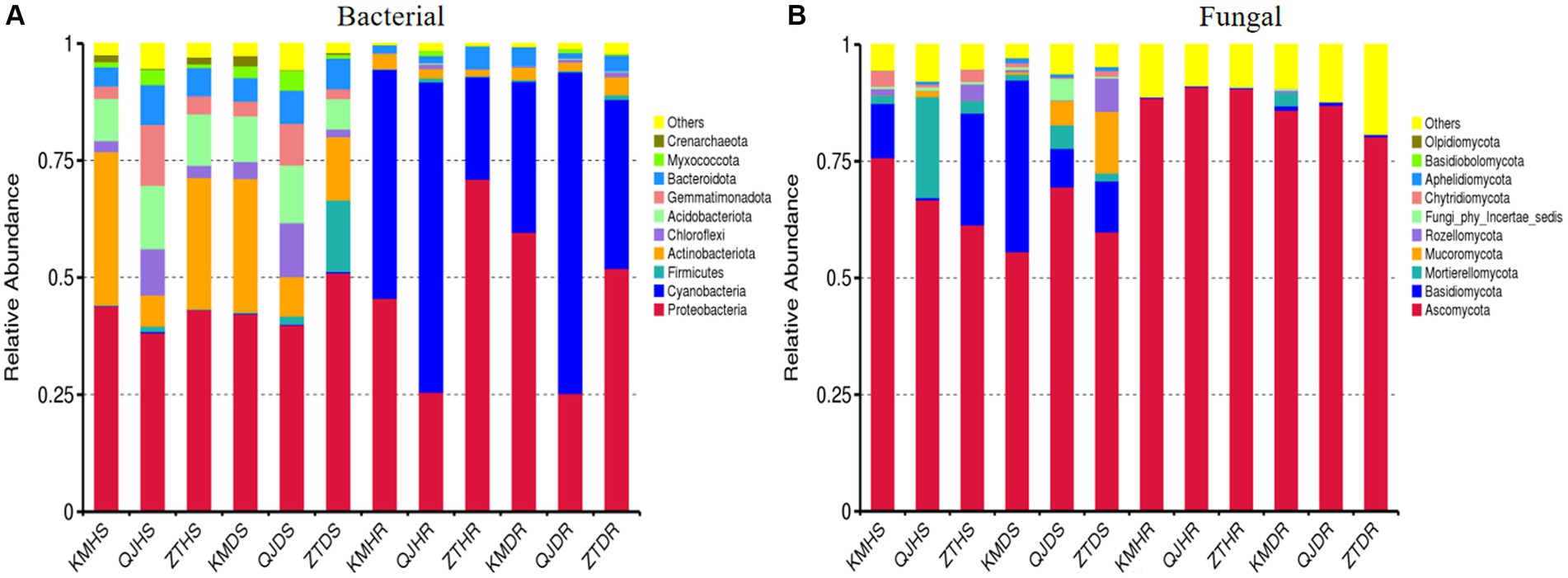
Figure 6. Relative abundance bar plots at phylum level based on the species annotation results in 12 composite samples (average of three replicates per sample) of potato plants under three variables. (A) Relative abundance at the phylum level in bacterial communities and (B) relative abundance at the phylum level in fungal communities. KM, Kunming; QJ, Qujing; ZT, Zhaotong; HS, healthy rhizosphere soil; DS, diseased rhizosphere soil; HR, healthy roots; DR, diseased roots.
In our study, we assessed the top 35 bacterial and fungal genera from the previously identified leading 10 phyla. Based on their relative abundance across all samples, these 35 genera were chosen for a heatmap analysis to ascertain their prevalence in varying sample types (healthy and diseased). The heatmaps, illustrating the relative abundance of these top 35 bacterial and fungal genera, are categorized and compared across three different factors: locations, plant components, and sample types, as presented in Figure 7. In healthy rhizosphere soil (HS), genera Nocardioides, Streptomyces, Pontibacter, Kribbella, Lysobacter, Bacillus, and Pseudomonas showed high abundance. In diseased rhizosphere soil (DS), genera Aquicella, Hydrogenophaga, Trichococcus, and Acidovorax were more prevalent. In healthy roots (HR), genera including Enterobacter, Actinoplanes, Chryseobacterium, Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Dyadobacter, and Brevundimonas were dominant, while in diseased roots (DR), Sphingobium, Burkholderia-Caballeronia-Paraburkholderia, and Ensifer were notably abundant. Notably, Ralstonia genus exhibited high abundance in both diseased rhizosphere soil and root samples (Figure 7A). Similarly, for fungi, genera like Paramyrothecium, Rhizophlyctis, Humicola, Plectosphaerella, Penicillium, and Aspergillus in HS; Alternaria, Colletotrichum, Mucor, Fusicolla, Varicosporellopsis, Scutellinia, Volutella, and Fusarium in DS; Pestalotiopsis, and Purpureocillium in HR; Meyerozyma, Bisifusarium, Ceratobasidium, and Gibberella moniliformis in DR, were highly abundant. The genus Thanatephorus, however, was particularly abundant in diseased rhizosphere soil and root samples (Figure 7B).
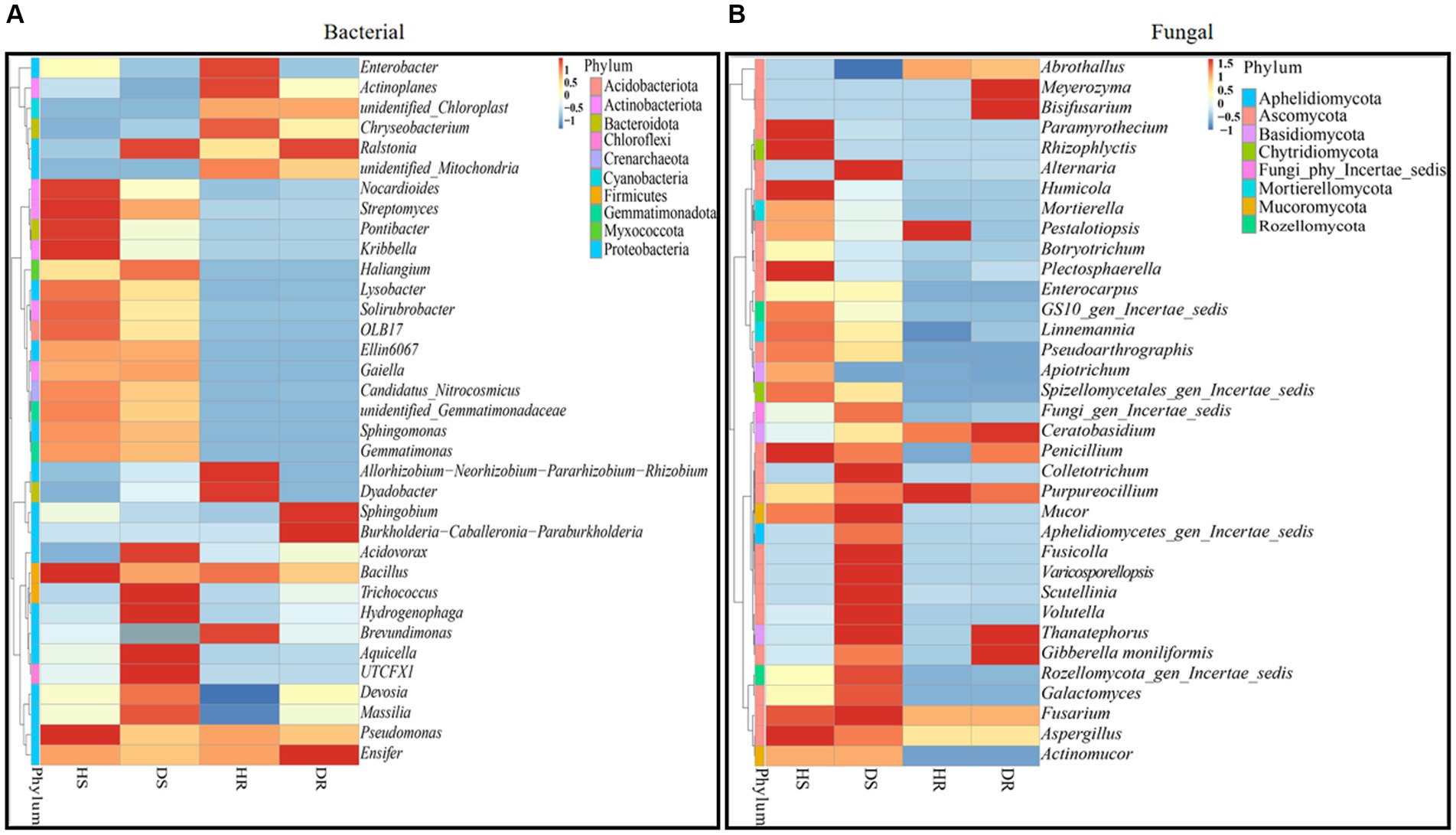
Figure 7. Relative abundance heatmaps at the genus level for top 35 bacterial (A) and fungal (B) genera in group-wise comparison under three variables. HS, healthy rhizosphere soil; DS, diseased rhizosphere soil; HR, healthy roots; DS, diseased roots.
Our analysis focused on the relative abundance of the leading 10 bacterial and fungal species within the top 35 genera. Bar plots depicting the relative abundance of these top 10 bacteria and fungi, categorized according to three criteria (locations, plant components, and sample types), are illustrated in Figure 8. In diseased roots and rhizosphere soil, the bacterial wilt pathogen Ralstonia solanacearum was notably prevalent. High abundances of Bacillus sp., Pseudomonas sp., Streptomyces flaveolus, and Lysobacter_daejeonensis were observed in healthy soil (HS). Furthermore, Ensifer adhaerens showed a high prevalence in diseased roots (DR; Figure 8A). In the context of fungi, species like Aspergillus sp. and Penicillium sp. were predominantly found in HS. Additionally, species such as Alternaria alternata, Thanatephorus cucumeris, Colletotrichum gloeosporioides, Ceratobasidium endornavirus, and Gibberella moniliformis were particularly abundant in diseased rhizosphere soil and root samples (Figure 8B).
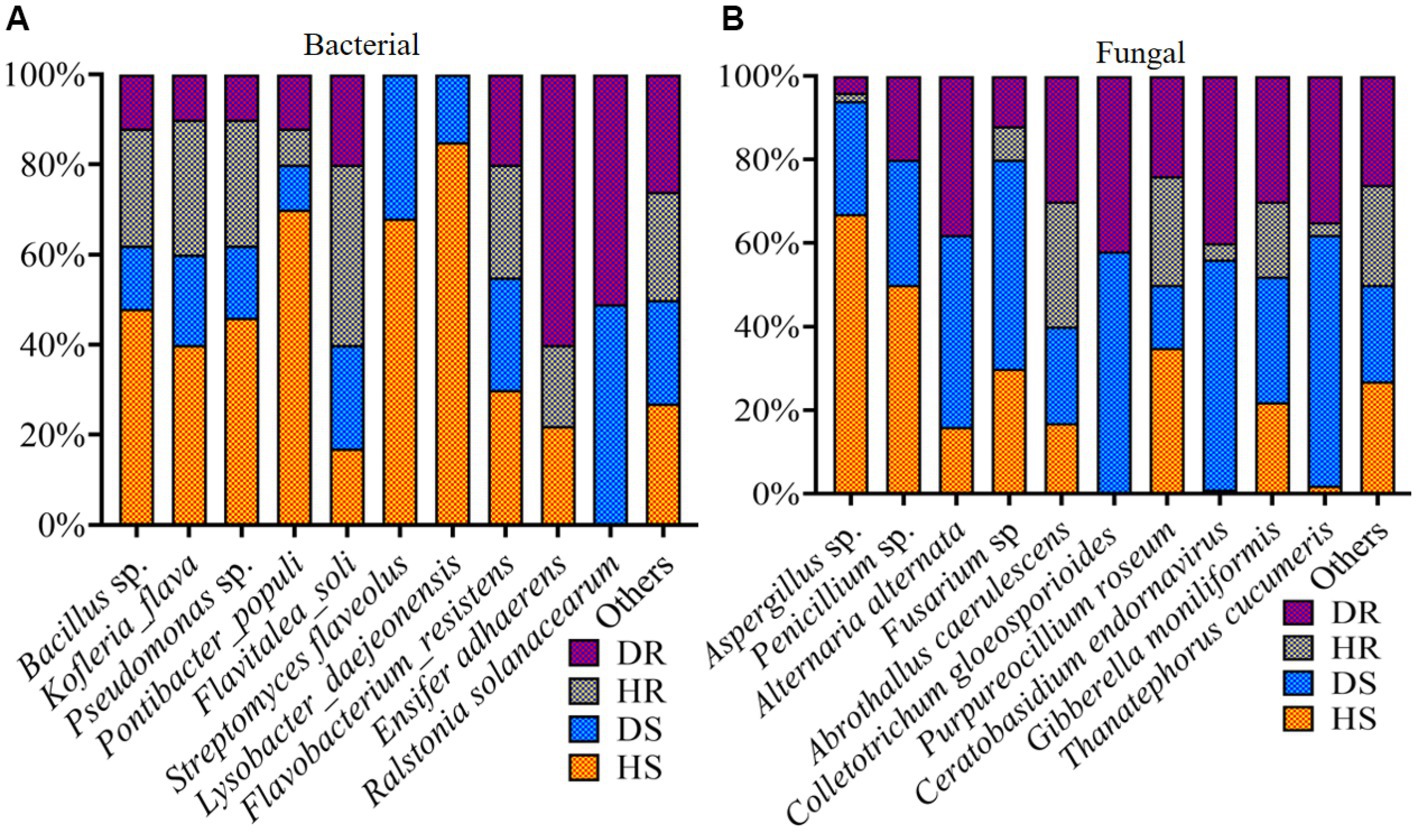
Figure 8. Relative abundance bar plots for top 10 bacterial (A) and fungal (B) species in group-wise comparison under three variables. HS, healthy rhizosphere soil; DS, diseased rhizosphere soil; HR, healthy roots; DS, diseased roots.
For each healthy and diseased sample, a microbial co-occurrence network at the genus level was developed, encompassing both bacterial and fungal Operational Taxonomic Units (OTUs) and relating to different plant components (rhizosphere soil and roots; Figure 9). Analysis revealed that in the bacterial communities, metrics such as the average degree, the total number of nodes, and the count of edges were more pronounced in healthy samples compared to those in diseased samples. Conversely, in the fungal communities, these same metrics—average degree, number of nodes, and number of edges—were elevated in diseased samples relative to healthy ones. This pattern suggests an enhanced level of interaction and connectivity within the bacterial microbial network in healthy samples, whereas the fungal microbial network exhibited a reverse trend in its connectivity.
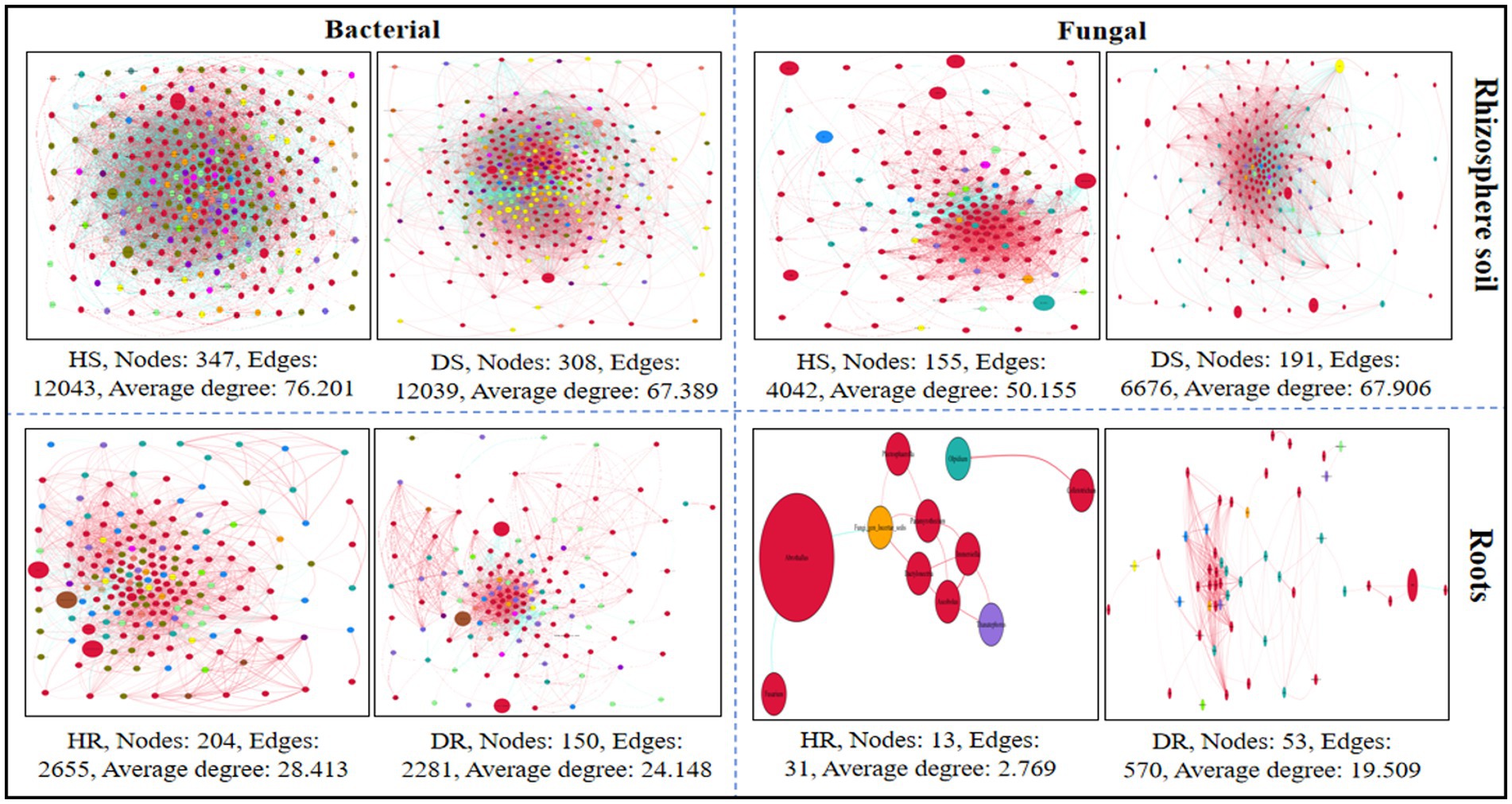
Figure 9. Co-occurrence network analysis of bacterial and fungal communities at genus level associated with healthy and diseased samples. Plant components (rhizosphere soil and roots). Here, HS; healthy soil, DS; diseased soil, HR; healthy roots, DR; diseased roots.
The potato’s ability to adapt to various climates has made it crucial for both economy and global food security (Xia et al., 2017; Ajayi-Oyetunde and Bradley, 2018; Iradukunda et al., 2022; Shuang et al., 2022). However, its continuous cultivation has led to an increase in black scurf disease, caused by Rhizoctonia solani, affecting potato yields worldwide (Lankau et al., 2020; Zhang X.-Y. et al., 2020; Hiltunen et al., 2021; Kankam et al., 2021; Kiptoo et al., 2021; Qin et al., 2022; Yang et al., 2022). To combat this, strategies such as resistance breeding, crop rotation, adjusting sowing dates and methods, chemical control, and, more recently, the use of disease-suppressive biocontrol agents have been employed (Kiptoo et al., 2021; Zrenner et al., 2021; Munir et al., 2022; Ahmed et al., 2022b). Additionally, the health of soil and microbial diversity in the rhizosphere play vital roles in controlling soilborne diseases and enhancing plant health (Köberl et al., 2013; Dong et al., 2018; Hao and Ashley, 2021).
Repeated cultivation of the same or closely related crops in the same soil, known as monoculture, leads to soil sickness, reducing crop yield and quality (Zheng et al., 2014). The microbiome in the rhizosphere serves as the initial defense mechanism against infections from soilborne pathogens and various forms of abiotic stress (Bulgarelli et al., 2013; Mendes et al., 2013). However, there is limited understanding of the microbial communities in the rhizosphere soil and roots of healthy and black scurf-infected potato plants across various geographical locations. In our study, we conducted a thorough examination of bacterial and fungal communities in different locations, sample types, and plant components by amplifying the V3-V4 and ITS1-5f variable regions of the 16S and internal transcribed spacer (ITS) rRNA genes. Our findings reinforce the notion that distinct plant components markedly influence the bacterial and fungal community structures, regardless of the sample types and geographical locations.
Wang et al. (2022) studied the impact of Alternaria solani infection on the microbial communities and multifunctionality in healthy vs. infected potato rhizosphere soils to explore the relationship between soil microbes, functionality, and pathogens. Despite varied observations on how rhizospheric and endophytic bacteria influence plant growth and health, no specific assembly pattern was noted (Lundberg et al., 2012; Lebeis et al., 2015). The rhizosphere is highlighted as a crucial habitat for microbial colonization into plant components, especially bacterial communities (Bulgarelli et al., 2013; Kaushal et al., 2020). In this research, we gathered samples from areas where potatoes have been cultivated consecutively for 4 years, identifying soil, diseased potatoes, and mechanical equipment as key vectors for spreading the black scurf pathogen.
Our study showed that the rhizosphere soil significantly affects microbial diversity, hosting more bacterial and fungal populations than the roots, with most root microbes also present in the rhizosphere across different locations. This suggests root microbes likely originate from the rhizosphere soil, supported by findings that host plants select specific microbial communities for their roots from the rhizosphere (Lundberg et al., 2012). Moreover, we observed also healthy potato plants exhibited richer microbial communities than those infected with black scurf. This may be due to disease stress impacting carbon availability and microbial growth (Kaushal et al., 2020), aligning with research showing healthier soils have more diverse microbes (Fu et al., 2017; Wu et al., 2017). However, some studies report higher microbial diversity in diseased soils (Wu et al., 2021; Zhang et al., 2022), potentially due to factors like soil type, nutrient levels, crop rotation practices, local climate, host plant species, and disease progression (Yuan et al., 2020). Potato plant resistance and severity of R. solani in different plant components also contribute to the prevalence of microbial communities. Bacterial and fungal OTU count was recorded maximum in all locations and plant components in rhizosphere soil samples than in the root samples.
Host plant’s environment can either support or hinder the colonization of specific bacterial and fungal groups in various plant components. Thus, bacterial and fungal communities in these specific plant components are either boosted or exhausted (Kaushal et al., 2020). In our study, we found stable core bacterial and fungal communities across all samples, with notable differences in their distribution among plant components. Specifically, Actinobacteriota and Acidobacteriota were more common in the rhizosphere soil, while Proteobacteria and Cyanobacteria were more prevalent in the roots. For fungi, Ascomycota was less abundant in the rhizosphere compared to the roots, whereas Basidiomycota showed the opposite pattern. Results showed that the black scurf pathogen significantly impacts these microbial communities.
A substantial number of varied bacteria and fungi are known to boost plant growth, reduce the occurrence of diseases, and provide other advantageous biological functions for plants (Santoyo et al., 2016; Gouda et al., 2018). Our research identifies core microbial communities that promote plant growth, enhance resistance, and lower disease rates by producing antibiotics, volatile compounds, secondary metabolites, and fixing nitrogen. Notably abundant bacterial genera include Streptomyces, Lysobacter, Bacillus, Pseudomonas, Ensifer, Enterobacter, and the Rhizobium group (Allorhizobium, Neorhizobium, Pararhizobium, Rhizobium), which are crucial for plant growth, nutrient acquisition, and suppressing diseases. Among fungi, Gibberella, Aspergillus, Penicillium sp., and Purpureocillium act as growth enhancers and biocontrol agents, while others may be saprophytic or pathogenic to crops.
The black scurf pathogen, R. solani (also known as Thanatephorus cucumeris), was found in high concentrations in the rhizosphere soil and roots of diseased plants, posing a significant threat to agriculture by infecting plant roots and disrupting symbiotic relationships with beneficial soil microbes. R. solani competes for nutrients and may even release toxins to suppress beneficial microbes like Streptomyces, Lysobacter, Bacillus, Pseudomonas, Ensifer, Gibberella, Aspergillus, Penicillium sp., and Purpureocillium, reducing their population and negatively affecting the soil’s microbial community and plant health. Additionally, pathogenic bacteria and fungi such as Ralstonia solanacearum, Alternaria alternata, Thanatephorus cucumeris, Colletotrichum gloeosporioides, and Ceratobasidium endornavirus were also present in high relative abundance in diseased rhizosphere soil and roots, indicating that the health of potato plants is closely linked to these microbial communities, with pathogenic ones contributing to the spread of R. solani and black scurf disease.
Microbial co-occurrence networks show direct and indirect relationships among microbes, highlighting their roles and ecological niches. Our findings indicate that R. solani has a more pronounced effect on the structure of bacterial communities than on fungal communities. This aligns with other research, such as that by Ahmed et al. (2022a), showing Ralstonia solanacearum’s significant impact on bacterial structures over fungal ones (Ahmed et al., 2022a). Healthy potato plants showed fewer negative bacterial interactions than diseased ones, echoing the findings of Wang et al. (2021) and Wu et al. (2021). Stronger and more resilient inter-microbial relationships, less vulnerable to environmental shifts, and more effective in suppressing soil-borne diseases (Tao et al., 2018). The heightened competition among microbial taxa in diseased conditions may result from an increase in potential pathogenic fungal and bacterial taxa in the rhizospheres of diseased potatoes. Simultaneously, the plant roots may recruit beneficial microbes to counteract pathogenic fungi (Zhang et al., 2022). Key species identified, such as Streptomyces, Lysobacter, Bacillus, Pseudomonas, Ensifer, Enterobacter, the Rhizobium group (Allorhizobium, Neorhizobium, Pararhizobium, Rhizobium), Aspergillus, Penicillium, Purpureocillium, and Gibberella moniliformis, are recognized for enhancing plant growth and disease resistance (Ma et al., 2018; Schreiter et al., 2018; Wu et al., 2020; Zhang J. et al., 2020; Asaturova et al., 2021; Wei et al., 2021; Abdelaziz et al., 2023; Rashad et al., 2023).
Our study concludes that potato plants host diverse bacterial and fungal communities, with their composition significantly varying across different plant components. The rhizosphere soil exhibited richer microbial diversity than the roots, and healthy plants showed greater microbial diversity than diseased ones. We found that certain microbes in the potato microbiome contribute to growth and disease resistance, identifying pathogenic communities in diseased plants linked to potato black scurf disease. Future research should focus on the microbiomes of black scurf-resistant potato varieties, exploring potential for disease control through microbiome manipulation to improve yield and quality.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
YY: Conceptualization, Data curation, Investigation, Methodology, Software, Supervision, Writing – original draft, Writing – review & editing. JH: Data curation, Investigation, Writing – review & editing. XW: Investigation, Writing – review & editing. KH: Investigation, Writing – review & editing. CL: Conceptualization, Formal analysis, Methodology, Writing – review & editing. GY: Conceptualization, Formal Analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This project was financially supported by the National Key R&D Program of China (2023YFD1400800), the National Natural Scientific Foundation of China (31960525, 31160352, and 31360423), and the National Natural Scientific Foundation of Yunnan (202207AA11000).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1386417/full#supplementary-material
Abdelaziz, A. M., El-Wakil, D. A., Hashem, A. H., Al-Askar, A. A., AbdElgawad, H., and Attia, M. S. (2023). Efficient role of endophytic aspergillus terreus in biocontrol of rhizoctonia solani causing damping-off disease of Phaseolus vulgaris and Vicia faba. Microorganisms 11:1487. doi: 10.3390/microorganisms11061487
Ahmed, W., Dai, Z., Liu, Q., Munir, S., Yang, J., Karunarathna, S. C., et al. (2022a). Microbial cross-talk: dissecting the Core microbiota associated with flue-cured tobacco (Nicotiana tabacum) plants under healthy and diseased state. Front. Microbiol. 13:845310. doi: 10.3389/fmicb.2022.845310
Ahmed, W., Yang, J., Tan, Y., Munir, S., Liu, Q., Zhang, J., et al. (2022b). Ralstonia solanacearum, a deadly pathogen: revisiting the bacterial wilt biocontrol practices in tobacco and other Solanaceae. Rhizosphere 21:100479. doi: 10.1016/j.rhisph.2022.100479
Ajayi-Oyetunde, O. O., and Bradley, C. A. (2018). Rhizoctonia solani: taxonomy, population biology and management of rhizoctonia seedling disease of soybean. Plant Pathol. 67, 3–17. doi: 10.1111/ppa.12733
Akber, M. A., Mubeen, M., Sohail, M. A., Khan, S. W., Solanki, M. K., Khalid, R., et al. (2022). Global distribution, traditional and modern detection, diagnostic, and management approaches of rhizoctonia solani associated with legume crops. Front. Microbiol. 13:1091288. doi: 10.3389/fmicb.2022.1091288
Asaturova, A., Shternshis, M., Tsvetkova, V., Shpatova, T., Maslennikova, V., Zhevnova, N., et al. (2021). Biological control of important fungal diseases of potato and raspberry by two Bacillus velezensis strains. PeerJ 9:e11578. doi: 10.7717/peerj.11578
Atkinson, D., Thornton, M. K., and Miller, J. S. (2010). Development of rhizoctonia solani on stems, Stolons and tubers of potatoes I. Effect of inoculum source. Am. J. Potato Res. 87, 374–381. doi: 10.1007/s12230-010-9143-6
Balali, G., Neate, S., Scott, E., Whisson, D., and Wicks, T. (1995). Anastomosis group and pathogenicity of isolates of rhizoctonia solani from potato crops in South Australia. Plant Pathol. 44, 1050–1057. doi: 10.1111/j.1365-3059.1995.tb02664.x
Banville, G. J. (1989). Yield losses and damage to potato plants caused by rhizoctonia solani kuhn. Am Potato J 66, 821–834. doi: 10.1007/BF02853963
Banville, G. B., and Carling, D. E. (2001). “Rhizoctonia canker and black scurf,” In Compendium of Potato Diseases. Eds. W. R. Stevenson, R. Loria, G. Franc, and D. P. Weingartner St Paul: APS Press, 36–37.
Beukema, HP, and van der Zang, DE (1990). Introduction to Potato Production. Wageningen: Centre for Agriculture Publishing and Documentation.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Loren, V., van Themaat, E., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Cai, Q., Zhou, G., Ahmed, W., Cao, Y., Zhao, M., Li, Z., et al. (2021). Study on the relationship between bacterial wilt and rhizospheric microbial diversity of fluecured tobacco cultivars. Eur. J. Plant Pathol. 160, 265–276. doi: 10.1007/s10658-021-02237-4
Dong, C.-J., Wang, L.-L., Li, Q., and Shang, Q.-M. (2019). Bacterial communities in the rhizosphere, phyllosphere and endosphere of tomato plants. PLoS One 14:e0223847. doi: 10.1371/journal.pone.0223847
Dong, L., Xu, J., Zhang, L., Cheng, R., Wei, G., Su, H., et al. (2018). Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta Pharm. Sin. B 8, 272–282. doi: 10.1016/j.apsb.2017.12.011
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., and Haas, B. J., Clemente, J. C., and Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fiers, M., Edel-Hermann, V., Heraud, C., Gautheron, N., Chatot, C., Hingrat, Y., et al. (2011). Genetic diversity of rhizoctonia solani associated with potato tubers in France. Mycologia 103, 1230–1244. doi: 10.3852/10-231
Fu, L., Penton, C., Ruan, Y., Shen, Z., Xue, C., Rong, L., et al. (2017). Inducing the rhizosphere microbiome by biofertilizer application to suppress banana fusarium wilt disease. Soil Biol. Biochem. 104, 39–48. doi: 10.1016/j.soilbio.2016.10.008
Gao, H., Hua, C., and Tong, M. (2018). Impact of Dinophysis acuminata feeding Mesodinium rubrum on nutrient dynamics and bacterial composition in a microcosm. Toxins 10:443. doi: 10.3390/toxins10110443
Gouda, S., Kerry, R. G., Das, G., Paramithiotis, S., Shin, H.-S., and Patra, J. K. (2018). Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 206, 131–140. doi: 10.1016/j.micres.2017.08.016
Govindasamy, V., Franco, C., and Vadakattu, G. (2014). “Endophytic actinobacteria: diversity and ecology” in Advances in endophytic research, Eds. V. C. Verma and A. C. Gange. (New Delhi: Springer). 27–59.
Gush, S.-L., Muzhinji, N., Truter, M., and Waals, J. E. (2019). First report of rhizoctonia solani AG 2-2IIIB causing elephant Hide on potato tubers in South Africa. Plant Dis. 103:1776. doi: 10.1094/PDIS-10-18-1850-PDN
Gutierrez, W. A., Shew, H. D., and Melton, T. A. (1997). Sources of inoculum and Management for Rhizoctonia solani damping-off on tobacco transplants under greenhouse conditions. Plant Dis. 81, 604–606. doi: 10.1094/PDIS.1997.81.6.604
Hao, J., and Ashley, K. (2021). Irreplaceable role of amendment-based strategies to enhance soil health and disease suppression in potato production. Microorganisms 9:1660. doi: 10.3390/microorganisms9081660
Hiltunen, L., Tarvainen, O., Kelloniemi, J., Tanskanen, J., Karhu, J., and Valkonen, J. P. T. (2021). Soil bacterial community in potato tuberosphere following repeated applications of a common scab suppressive antagonist. Appl. Soil Ecol. 167:104096. doi: 10.1016/j.apsoil.2021.104096
Ikeda, S., Shimizu, A., Shimizu, M., Takahashi, H., and Takenaka, S. (2012). Biocontrol of black scurf on potato by seed tuber treatment with Pythium oligandrum. Biol Control 60, 297–304. doi: 10.1016/j.biocontrol.2011.10.016
Iradukunda, L., Wang, Y.-P., Nkurikiyimfura, O., Wang, T., Yang, L.-N., and Zhan, J. (2022). Establishment and application of a multiplex PCR assay for the rapid detection of rhizoctonia solani anastomosis group (AG)-3PT, the pathogen causing potato black scurf and stem canker. Pathogens 11:627. doi: 10.3390/pathogens11060627
Kankam, F., Larbi-Koranteng, S., and Adomako, J. (2021). Rhizoctonia disease of potato: epidemiology, toxin types and management. Egypt J Phytopathol 49, 197–209. doi: 10.21608/ejp.2021.72057.1028
Kaushal, M., Swennen, R., and Mahuku, G. (2020). Unlocking the microbiome communities of Banana (Musa spp.) under disease stressed (fusarium wilt) and non-stressed conditions. Microorganisms 8:443. doi: 10.3390/microorganisms8030443
Kiptoo, J., Abbas, A., Bhatti, A., Muhammad Usman, H., Shad, M., Umer, M., et al. (2021). Rhizoctonia solani of potato and its management: a review. Plant Prot. 5, 157–169. doi: 10.33804/pp.005.03.3925
Köberl, M., Schmidt, R., Ramadan, E. M., Bauer, R., and Berg, G. (2013). The microbiome of medicinal plants: diversity and importance for plant growth, quality and health. Front. Microbiol. 4:400. doi: 10.3389/fmicb.2013.00400
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Kouzai, Y., Shimizu, M., Inoue, K., Uehara-Yamaguchi, Y., Takahagi, K., Nakayama, R., et al. (2020). BdWRKY38 is required for the incompatible interaction of Brachypodium distachyon with the necrotrophic fungus Rhizoctonia solani. Plant J. 104, 995–1008. doi: 10.1111/tpj.14976
Kowalczewski, P. Ł., Olejnik, A., Białas, W., Rybicka, I., Zielińska-Dawidziak, M., Siger, A., et al. (2019). The nutritional value and biological activity of concentrated protein fraction of potato juice. Nutrients 11:1523. doi: 10.3390/nu11071523
Lankau, E. W., Xue, D., Christensen, R., Gevens, A. J., and Lankau, R. A. (2020). Management and soil conditions influence common scab severity on potato tubers via indirect effects on soil microbial communities. Phytopathology 110, 1049–1055. doi: 10.1094/PHYTO-06-19-0223-R
Lebeis, S., Herrera Paredes, S., Lundberg, D., Breakfield, N., Gehring, J., McDonald, M., et al. (2015). PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. doi: 10.1126/science.aaa8764
Lehtonen, M. J., Wilson, P. S., Ahvenniemi, P., Valkonen, J. P. T., Vanhatalo, A., Alakukku, L., et al. (2009). Formation of canker lesions on stems and black scurf on tubers in experimentally inoculated potato plants by isolates of AG2-1, AG3 and AG5 of rhizoctonia solani: a pilot study and literature review. Agric. Food Sci. 18, 223–233. doi: 10.2137/145960609790059415
Li, Y.-X., Yang, Y.-T., Ma, L., Liu, J.-P., An, Q.-C., Zhang, C.-Y., et al. (2022). Comparative analyses of antibiotic resistance genes in jejunum microbiota of pigs in different areas. Front. Cell. Infect. Microbiol. 12:887428. doi: 10.3389/fcimb.2022.887428
Liu, Y., Lin-Wang, K., Espley, R. V., Wang, L., Yang, H., Yu, B., et al. (2016). Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 67, 2159–2176. doi: 10.1093/jxb/erw014
Lopez Corrales, R., Michereff, S., García-Estrada, R., León-Félix, J., Correia, K., Rabago Zavala, K., et al. (2023). First confirmed report of rhizoctonia solani AG-7 causing potato stem canker in Mexico. Plant Dis. 107:2260. doi: 10.1094/PDIS-11-22-2661-PDN
Lundberg, D., Lebeis, S., Herrera Paredes, S., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Ma, L., Zhang, H.-Y., Zhou, X.-K., Yang, C.-G., Zheng, S.-C., Duo, J.-L., et al. (2018). Biological control tobacco bacterial wilt and black shank and root colonization by bio-organic fertilizer containing bacterium Pseudomonas aeruginosa NXHG29. Appl. Soil Ecol. 129, 136–144. doi: 10.1016/j.apsoil.2018.05.011
Malik, O., Chohan, S., and Naqvi, S. A. H. (2014). Occurrence of black scurf disease of potato in Multan (Punjab) Alongwith its in vitro chemical and biotic elicitor mediated management. J. Agric. Sci. 6:134. doi: 10.5539/jas.v6n9p134
Mayo, S., Gutiérrez, S., Malmierca, M. G., Lorenzana, A., Campelo, M. P., Hermosa, R., et al. (2015). Influence of rhizoctonia solani and Trichoderma spp. in growth of bean (Phaseolus vulgaris L.) and in the induction of plant defense-related genes. Front. Plant Sci. 6:685. doi: 10.3389/fpls.2015.00685
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Mhlongo, M. I., Piater, L. A., Madala, N. E., Labuschagne, N., and Dubery, I. A. (2018). The chemistry of plant-microbe interactions in the rhizosphere and the potential for metabolomics to reveal Signaling related to Defense priming and induced systemic resistance. Front. Plant Sci. 9:112. doi: 10.3389/fpls.2018.00112
Munir, S., Ahmed, A., Li, Y., He, P., Singh, B. K., He, P., et al. (2022). The hidden treasures of citrus: finding Huanglongbing cure where it was lost. Crit. Rev. Biotechnol. 42, 1–16. doi: 10.1080/07388551.2021.1942780
Murdock, M., Woodhall, J., Maggard, S., Keith, S., Harrington, M., Oropeza, L., et al. (2019). First report of rhizoctonia solani AG 2-1 causing stem canker of potato (Solanum tuberosum) in Idaho. Plant Dis. 103:2130. doi: 10.1094/PDIS-02-19-0364-PDN
Muzhinji, N., Woodhall, J. W., Truter, M., and van der Waals, J. E. (2018). Variation in fungicide sensitivity among rhizoctonia isolates recovered from potatoes in South Africa. Plant Dis. 102, 1520–1526. doi: 10.1094/PDIS-09-17-1470-RE
Parissa, T., and Tarighi, S. (2011). Cytomolecular aspects of rice sheath blight caused by rhizoctonia solani. Eur. J. Plant Pathol. 129, 511–528. doi: 10.1007/s10658-010-9725-7
Qin, J., Bian, C., Duan, S., Wang, W., Li, G., and Jin, L. (2022). Effects of different rotation cropping systems on potato yield, rhizosphere microbial community and soil biochemical properties. Front. Plant Sci. 13:999730. doi: 10.3389/fpls.2022.999730
Rashad, Y. M., Al Tami, M. S., and Abdalla, S. A. (2023). Eliciting transcriptomic and antioxidant defensive responses against rhizoctonia root rot of sorghum using the endophyte aspergillus oryzae YRA3. Sci. Rep. 13:19823. doi: 10.1038/s41598-023-46696-7
Samsatly, J., Bayen, S., and Jabaji, S. H. (2020). Vitamin B6 is under a tight balance during disease development by rhizoctonia solani on different cultivars of potato and on Arabidopsis thaliana mutants. Front. Plant Sci. 11:875. doi: 10.3389/fpls.2020.00875
Santoyo, G., Moreno-Hagelsieb, G., Orozco-Mosqueda, M. D. C., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Schreiter, S., Babin, D., Smalla, K., and Grosch, R. (2018). Rhizosphere competence and biocontrol effect of pseudomonas sp. RU47 independent from plant species and soil type at the field scale. Front. Microbiol. 9:97. doi: 10.3389/fmicb.2018.00097
Senapati, M., Tiwari, A., Sharma, N., Chandra, P., Bashyal, B. M., Ellur, R. K., et al. (2022). Rhizoctonia solani Kühn pathophysiology: status and prospects of sheath blight disease Management in Rice. Front. Plant Sci. 13:881116. doi: 10.3389/fpls.2022.881116
Shuang, M., Wang, Y., Teng, W., and Jin, G. (2022). Isolation and identification of an endophytic bacteria bacillus sp. K-9 exhibiting biocontrol activity against potato common scab. Arch. Microbiol. 204:483. doi: 10.1007/s00203-022-02989-5
Tao, J., Meng, D., Qin, C., Liu, X., Liang, Y., Xiao, Y., et al. (2018). Integrated network analysis reveals the importance of microbial interactions for maize growth. Appl. Microbiol. Biotechnol. 102, 3805–3818. doi: 10.1007/s00253-018-8837-4
Tariq, M., Yasmin, S., and Hafeez, F. Y. (2010). Biological control of potato black scurf by rhizosphere associated bacteria. Braz. J. Microbiol. 41, 439–451. doi: 10.1590/S1517-838220100002000026
Truter, M., and Wehner, F. (2004). Anastomosis grouping of rhizoctonia solani associated with black scurf and stem canker of potato in South Africa. Plant Dis 88:83. doi: 10.1094/PDIS.2004.88.1.83B
Tsror, L. (2010). Biology, epidemiology and Management of Rhizoctonia solani on potato. J. Phytopathol. 158, 649–658. doi: 10.1111/j.1439-0434.2010.01671.x
Tsror, L., Barak, R., and Sneh, B. (2001). Biological control of black scurf on potato under organic management. Crop Prot. 20, 145–150. doi: 10.1016/S0261-2194(00)00124-1
Verwaaijen, B., Wibberg, D., Kröber, M., Winkler, A., Zrenner, R., Bednarz, H., et al. (2017). The rhizoctonia solani AG1-IB (isolate 7/3/14) transcriptome during interaction with the host plant lettuce (Lactuca sativa L.). PLoS One 12:e0177278. doi: 10.1371/journal.pone.0177278
Wang, M., Han, Y., Xu, Z., Wang, S., Jiang, M., and Wang, G. (2021). Hummock-hollow microtopography affects soil enzyme activity by creating environmental heterogeneity in the sedge-dominated peatlands of the Changbai Mountains, China. Ecol. Indic. 121:107187. doi: 10.1016/j.ecolind.2020.107187
Wang, D.-Y., Liu, X., Yang, Y.-L., Feng, R., Cao, Y.-S., Zheng, X., et al. (2014). Control experiment and occurrence of potato black scurf disease in Yunnan Province. Chin Potato J 72, 225–227. doi: 10.1086/677128
Wang, F., Wang, F., Zhang, H., Qin, F., Xiang, W., Wu, C., et al. (2022). Deciphering differences in microbial community composition and multifunctionality between healthy and Alternaria solani-infected potato rhizosphere soils. Plant and Soil 484, 347–362. doi: 10.1007/s11104-022-05797-4
Wei, L., Yang, J., Ahmed, W., Xiong, X., Liu, Q., Huang, Q., et al. (2021). Unraveling the association between metabolic changes in inter-genus and intra-genus bacteria to mitigate clubroot disease of Chinese cabbage. Agronomy 11:2424. doi: 10.3390/agronomy11122424
Wierzbicka-Woś, A., Henneberger, R., Batista-García, R. A., Martínez-Ávila, L., Jackson, S. A., Kennedy, J., et al. (2019). Biochemical characterization of a novel monospecific Endo-β-1,4-Glucanase belonging to GH family 5 from a rhizosphere metagenomic library. Front. Microbiol. 10:1342. doi: 10.3389/fmicb.2019.01342
Woodhall, J., Adams, I., Peters, J., Harper, G., and Boonham, N. (2013). A new quantitative real-time PCR assay for rhizoctonia solani AG3-PT and the detection of AGs of rhizoctonia solani associated with potato in soil and tuber samples in Great Britain. Eur. J. Plant Pathol. 136, 273–280. doi: 10.1007/s10658-012-0161-8
Woodhall, J., Lees, A., Edwards, S., and Jenkinson, P. (2008). Infection of potato by rhizoctonia solani: effect of anastomosis group. Plant Pathol. 57, 897–905. doi: 10.1111/j.1365-3059.2008.01889.x
Wu, X., Hongli, L., Wang, Y., and Zhang, X. (2020). Effects of bio-organic fertiliser fortified by Bacillus cereus QJ-1 on tobacco bacterial wilt control and soil quality improvement. Biocontrol Sci. Tech. 30, 351–369. doi: 10.1080/09583157.2020.1711870
Wu, J., Jiao, Z., Zhou, J., Guo, F., Ding, Z., and Qiu, Z. (2017). Analysis of bacterial communities in rhizosphere soil of continuously cropped healthy and diseased konjac. World J. Microbiol. Biotechnol. 33:134. doi: 10.1007/s11274-017-2287-5
Wu, C., Wang, F., Ge, A., Zhang, H., Chen, G., Deng, Y., et al. (2021). Enrichment of microbial taxa after the onset of wheat yellow mosaic disease. Agric. Ecosyst. Environ. 322:107651. doi: 10.1016/j.agee.2021.107651
Xia, Y., Fei, B., He, J., Zhou, M., Zhang, D., Pan, L., et al. (2017). Transcriptome analysis reveals the host selection fitness mechanisms of the rhizoctonia solani AG1IA pathogen. Sci. Rep. 7:10120. doi: 10.1038/s41598-017-10804-1
Yanar, Y., Yilmaz, G., Cesmeli, I., and Coskun, S. (2005). Characterization of rhizoctonia solani isolates from potatoes in Turkey and screening potato cultivars for resistance to AG-3 isolates. Phytoparasitica 33, 370–376. doi: 10.1007/BF02981304
Yang, S., Kong, Y., Fanxiang, M., Zhang, J., Wang, L., Hu, L., et al. (2019). First report of rhizoctonia solani AG-1-IB infecting potato with stem canker in Heilongjiang Province, China. Plant Dis. 103:2126. doi: 10.1094/PDIS-02-19-0390-PDN
Yang, S., Kong, Y., Guo, M., Dong, X., Min, F., Wei, Q., et al. (2022). First report of black scurf caused by rhizoctonia solani AG 2-2IV on potato tubers in Heilongjiang Province, China. Plant Dis. 106:2996. doi: 10.1094/PDIS-12-21-2801-PDN
Yang, Y., Zhao, C., Guo, Z., and Wu, X. (2015). Anastomosis group and pathogenicity of rhizoctonia solani associated with stem canker and black scurf of potato in China. Eur. J. Plant Pathol. 143, 99–111. doi: 10.1007/s10658-015-0668-x
Yuan, J., Wen, T., Zhang, H., Zhao, M., Penton, C. R., Thomashow, L. S., et al. (2020). Predicting disease occurrence with high accuracy based on soil macroecological patterns of fusarium wilt. ISME J. 14, 2936–2950. doi: 10.1038/s41396-020-0720-5
Zhang, X.-Y., Li, C., Hao, J.-J., Li, Y.-C., Li, D.-Z., Zhang, D.-M., et al. (2020). A novel streptomyces sp. strain PBSH9 for controlling potato common scab caused by Streptomyces galilaeus. Plant Dis. 104, 1986–1993. doi: 10.1094/PDIS-07-19-1469-RE
Zhang, J., Wei, L., Yang, J., Ahmed, W., Wang, Y., Fu, L., et al. (2020). Probiotic consortia: reshaping the Rhizospheric microbiome and its role in suppressing root-rot disease of panax notoginseng. Front. Microbiol. 11:701. doi: 10.3389/fmicb.2020.00701
Zhang, H., Wu, C., Wang, F., Wang, H., Chen, G., Cheng, Y., et al. (2022). Wheat yellow mosaic enhances bacterial deterministic processes in a plant-soil system. Sci. Total Environ. 812:151430. doi: 10.1016/j.scitotenv.2021.151430
Zheng, L., Zhang, M., Chen, Q., Zhu, M., and Zhou, E. (2014). A novel mycovirus closely related to viruses in the genus Alphapartitivirus confers hypovirulence in the phytopathogenic fungus rhizoctonia solani. Virology 456-457, 220–226. doi: 10.1016/j.virol.2014.03.029
Keywords: disease resistance, microbial diversity, plant components, potato black scurf, soil-borne diseases
Citation: Yang Y, Hu J, Wei X, Huang K, Li C and Yang G (2024) Deciphering core microbiota in rhizosphere soil and roots of healthy and Rhizoctonia solani-infected potato plants from various locations. Front. Microbiol. 15:1386417. doi: 10.3389/fmicb.2024.1386417
Received: 15 February 2024; Accepted: 11 March 2024;
Published: 22 March 2024.
Edited by:
Febri Doni, Padjadjaran University, IndonesiaReviewed by:
Mia Miranti, Padjadjaran University, IndonesiaCopyright © 2024 Yang, Hu, Wei, Huang, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Genhua Yang, Z2h5YW5nMjAwM0AxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.