
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 May 2024
Sec. Food Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1383959
 Marzie Mahdizade Ari1
Marzie Mahdizade Ari1 Shiva Mirkalantari1
Shiva Mirkalantari1 Davood Darban-Sarokhalil1
Davood Darban-Sarokhalil1 Atieh Darbandi1
Atieh Darbandi1 Shabnam Razavi1,2*
Shabnam Razavi1,2* Malihe Talebi1*
Malihe Talebi1*Background: The use of probiotics is emerging as an innovative approach to managing oral health issues and mediating the immune system. The current study assessed the in vitro impacts of non-orally isolated probiotics on periodontitis and tooth decay pathogens.
Methods: Briefly, the persistence of probiotics in exposure to oral cavity enzymes, hydrogen peroxide, and saliva samples was examined. It was also investigated the biofilm formation and aggregation ability of probiotics, the adherence of probiotics in human gingival fibroblast cell (HGFC) lines and molar teeth samples, and the potential of probiotics to co-aggregate with oral pathogens. Additionally, the current study evaluated the effects of live probiotics on virulence gene expression, biofilm production of main oral pathogens, and changes in inflammation markers.
Results: The probiotics remained alive when exposed to enzymes in the oral cavity, hydrogen peroxide, and saliva at baseline, 1, 3, and 5 h after incubation at 37°C (p-value <0.05). Probiotics demonstrated to produce biofilm and aggregation, as well as adherence to HGFCs and maxillary molars (p-value >0.05). They showed significant co-aggregation with oral pathogens, which were recorded as 65.57% for B. bifidum 1001 with S. mutans, 50.06% for B. bifidum 1005 with P. gingivalis, 35.6% for L. plantarum 156 with F. nucleatum, and 18.7% for B. longum 1044 with A. actinomycetemcomitans after 8 h of incubation. A balance between pro-inflammatory and anti-inflammatory cytokines, along with inhibition of biofilm formation and changes in virulence gene transcripts, were observed. However, most of these changes were not statistically significant (p-value >0.05).
Conclusion: This study demonstrated the direct link between adhesiveness, aggregation, and biofilm formation with probiotic antibacterial activity. In addition to the careful selection of suitable probiotic strains, the concentration and origin of probiotic isolates should be considered.
Probiotics, when administered in adequate quantities, are live microorganisms that significantly impact human health (Saraf et al., 2010). These beneficial bacteria need to withstand the harsh environment of the gastrointestinal tract. Probiotics directly fight pathogens by producing antibacterial compounds such as bacteriocins, and various metabolites like organic acids (Ibarburu et al., 2015). Probiotics are widely utilized in food, pharmaceutical, chemical industries as well as cosmetic. Several types of microorganisms are introduced as probiotics, such as Lactobacillus acidophilus (L. acidophilus), Lactiplantibacillus plantarum (L. plantarum), Lacticaseibacillus casei (L. casei), Lactobacillus delbrueckii (L. delbrueckii), Lacticaseibacillus rhamnosus (L. rhamnosus), Limosilactobacillus fermentum (L. fermentum), Limosilactobacillus reuteri (L. reuteri), and Lacticaseibacillus paracasei (L. paracasei), Bifidobacterium bifidum (B. bifidum), Bifidobacterium breve (B. breve), Bifidobacterium longum (B. longum), Bifidobacterium lactis (B. lactis), and Bifidobacterium animalis (B. animalis). Streptococcus spp.; Enterococcus; Saccharomyces; Pediococcus; and Leuconostoc are also recognized for their probiotic properties (Padmavathi et al., 2018). Probiotics have been found to maintain the stability and diversity of oral biofilms by interacting with the oral microbial population, potentially alleviating symptoms of metabolic disorders, cancer, allergic reactions, and autoimmune disorders (Menon, 2016). The precise mechanism of action of probiotics in the oral cavity is not fully understood, and it is still under investigation. Studies suggested that the primary function of probiotics involves competing for attachment sites, producing of metabolites against pathogens, and regulating immune responses. In particular, L. casei strain Shirota and L. reuteri have been shown to effectively hinder the growth and biofilm formation of Streptococcus mutans (S. mutans) and Porphyromonas gingivalis (P. gingivalis) (Widyarman et al., 2019). Moreover, the levels of pro-inflammatory cytokines like interleukin-1 beta (IL-1β), tumor necrosis factor alpha (TNF-α), and interleukin-8 (IL-8) decreased in the gingival crevicular fluid (GCF) following chewing gum containing L. reuteri (Menon, 2016). Periodontitis is a severe infectious disease that impacts the tooth-supporting tissues and is characterized by bleeding and redness of the gums. Apart from the enrollment of host factors in periodontitis, the main reason behind periodontitis is attributed to a combination of virulence factors present in pathogens responsible for colonization, tissue destruction, and biofilm formation. Among all the factors mentioned earlier, this stands out as the primary and crucial element in the colonization. Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans), P. gingivalis, and Fusobacterium nucleatum (F. nucleatum) are the etiological causative agents of periodontitis (Sujeetha et al., 2019). Tooth decay, another frequent oral health issue, occurs when a sticky mass known as dental plaque or biofilm develops on the surface of the teeth. During this procedure, certain species of Streptococcus, including S. mutans and S. sobrinus, play a role in creating substances like glucan and fructan (Lee and Kim, 2014), and increasing biofilm-producing proteins to enhance bacterial biofilm formation ability (Matsumoto-Nakano, 2018). Periodontal disease occurs when the balance of microorganisms in the oral cavity is disturbed by the excessive growth of harmful periopathogens and the formation of biofilm. This process ultimately causes the development of periodontal pockets, damage to the surrounding tissues, and deterioration of the bone supporting the teeth (Cugini et al., 2021). It has been suggested that probiotics have the ability to treat and prevent periodontitis and dental caries. To select the most effective probiotic for combating oral pathogens, we conducted an in vitro study to explore the effects of probiotics isolated from milk and feces on the expression of minor fimbriae (Mfa1) and arginine-gingipain (Rgp) genes in P. gingivalis, rough-colony protein A (RcpA) in A. actinomycetemcomitans, glucosyltransferase B (GtfB) in S. mutans, and fibroblast activation protein-2 (Fap2) in F. nucleatum; the formation of biofilms (microbial plaque) by oral pathogens; as well as the inflammation in the oral cavity, which characterized by inflammatory cytokines like IL-8, and anti-inflammatory cytokines like IL-10. Before that, it is important to address the effectiveness and viability of probiotics in the rough condition of the oral cavity.
The effectiveness of probiotics in the oral cavity was assessed when exposed to various enzymes, hydrogen peroxide, and saliva. The ability of probiotics to attach to human gingival fibroblast cells (HGFCs) and maxillary teeth was also analyzed. In the following section, the effect of each strain of Bifidobacterium and Lactobacillus spp., a combination of five Lactobacillus strains, a combination of five Bifidobacterium strains, and a combination of the most effective Lactobacillus and Bifidobacterium strains were analyzed on the expression of the main virulence genes in A. actinomycetemcomitans, P. gingivalis, F. nucleatum, and S. mutans. Analysis of the gene expression of IL-8 and IL-10 cytokines was also conducted on the supernatant of HGFC. Except for the competition and biofilm formation assays, which were carried out in duplicate, all other experiments were performed in triplicate.
Five Lactobacillus strains were isolated from the stool and five Bifidobacterium strains were taken from breast milk and infant feces. These strains were chosen for investigation in the current study, as outlined in Table 1. The study included Lactiplantibacillus plantarum (L. plantarum 42 and L. plantarum 156), Levilactobacillus brevis (L. brevis 205), Limosilactobacillus reuteri (L. reuteri 100), and Lacticaseibacillus rhamnosus (L. rhamnosus 195) (Rohani et al., 2015). Additionally, Bifidobacterium bifidum (Bifidobacterium bifidum 1001 and Bifidobacterium bifidum 1005), Bifidobacterium breve (Bifidobacterium breve 1015 and Bifidobacterium breve 1063), and Bifidobacterium longum (Bifidobacterium longum 1044) were also studied (Eshaghi et al., 2017). The current study involved the use of four pathogens, including A. actinomycetemcomitans JP2 genotype strain HK1651 and S. mutans purchased from the microbial collection bank of Shahid Beheshti University of Medical Sciences, faculty of dentistry, as well as P. gingivalis ATCC 33277 and F. nucleatum ATCC 27725 from Tehran University of Medical Sciences (kindly donated by Dr. Douraghi).
Probiotics were grown on Man-Rogosa-Sharpe (MRS) agar (Merck, Darmstadt, Germany) and incubated at 37°C for 48–72 h under microaerophilic conditions. A. actinomycetemcomitans and P. gingivalis were cultivated on brain heart infusion (BHI) agar (Merck, Darmstadt, Germany) supplemented with 0.5% defibrinated sheep blood and incubated at 37°C for 48–72 h under anaerobic conditions. F. nucleatum was grown on Brucella agar based medium (Condalab, Spain) supplemented with 5% horse serum (Baharafshan, Iran) and 10% defibrinated sheep blood (Baharafshan, Iran) and incubated at 37°C for 48–72 h in anaerobic conditions. S. mutans was cultured on blood agar (Merck, Darmstadt, Germany) and incubated at 37°C for 24 h under aerobic conditions. In all experiments, to achieve an optical density (OD) of 0.08–0.13 equivalent to 1.5 × 108 colony forming units (CFUs)/mL, bacterial suspensions were first prepared by transferring fresh colonies of probiotics and pathogens grown on agar plates to MRS and BHI broth, respectively, and then incubated at 37°C under microaerophilic conditions.
Suspensions of Lactobacillus and Bifidobacterium strains were prepared at a concentration of 1.5 × 108 CFU/mL, exposed to 0.4 mM H2O2 (30% w/v) (Mojallali, Iran), and then incubated at 37°C for 18 h in the presence of 5% CO2 under microaerophilic conditions. Survival of probiotics was evaluated at different time intervals at baseline, 1, 3, and 5 h of incubation using a colony count assay. Following incubation, each treatment was subjected to serial dilution and spot culture on MRS agar to determine the survived strains, and then incubated at 37°C for 24 h. After 24 h, colonies growing on MRS agar that belonged to Lactobacillus or Bifidobacterium spp. were counted (Sun et al., 2013).
Probiotics at a concentration of 1.5 × 108 CFU/mL were treated with PBS (phosphate-buffered saline) (Bio-IDEA, Iran) containing α-amylase (220 IU/mg), lipase (700 IU/mg), lysozyme (22 IU/mg), and proteinase K (1 mg/mL) (Mojallali, Iran) (Yang et al., 2021). NaOH was introduced to enhance the effectiveness of α-amylase and lipase by adjusting the pH level to 6.5. A colony count assay was performed to evaluate the survival rate of the treated probiotics compared to the control group (probiotics without enzymes) at baseline, 1, 3, and 5 h after incubation at 37°C.
Briefly, 1.5 h after eating, drinking, or tooth brushing, 3 mL of unstimulated saliva from healthy volunteers was collected in sterile tubes (MAXWELL, China), and immediately placed on an ice pack. The collected saliva sample was centrifuged (Beckman, United States) at 10,000 rpm and 4°C for 10 min (min) to obtain a homogenous sample. Subsequently, 1 mL of the supernatant was carefully transferred into an Eppendorf tube. Then, 10 μL of supernatant was placed on blood agar and kept for incubation in both aerobic and anaerobic conditions. In order to obtain germ-free saliva and prevent any interference with the probiotic treatment process, the supernatant was pasteurized by heating at 65°C for 30 min. Following centrifugation, the supernatant was transferred to a new tube, and kept at −70°C. Before that, 10 μL of pasteurized supernatant was cultured on blood agar, and incubated in aerobic and anaerobic conditions to assess for any potential contamination.
A fresh overnight culture of probiotics was prepared at 1.5 × 108 CFU/mL. Then, 200 μL of probiotic suspension was mixed with 1.8 mL of pasteurized saliva and incubated at 37°C for 24 h. Serial dilution was carried out before and after the incubation period. For the control group, the same procedures were followed, while 0.9% NaCl was used instead of saliva. All experiments were repeated twice.
Intact maxillary molars were provided by healthy women who have been replaced with gold teeth for cosmetic reasons. In order to investigate the adherence ability of probiotics to maxillary molars, suspensions of probiotics at 1.5 × 108 CFU/mL were prepared. Then, 1.5 mL of the prepared suspension was placed into a 2-mL sterile Eppendorf tube containing a sterilized maxillary molar sample, and incubated at 37°C for 48 h under microaerophilic conditions. After incubation, the maxillary molar was rinsed three times with either normal saline or PBS to eliminate unattached bacteria. The maxillary molar was gently washed with PBS and transferred into a 2-mL sterile Eppendorf tube containing 1.5 mL of PBS. It could be either sonicated for 15 s at a frequency of 30 kHz and an output power of 7 W or centrifuged at 5,000 rpm and 4°C for 3–5 min to remove unbound and residual periopathogens (Pourhajibagher et al., 2020). Finally, 10 μL of Eppendorf content containing maxillary molar was placed on MRS agar. After incubation at 37°C for 72 h under microaerophilic conditions, the CFU/mL of the probiotic strain adhered to the maxillary molar was calculated based on the method described by Miles et al. (1938).
The biofilm formation ability of probiotics was evaluated according to Mirzaei et al. (2022) using a microtiter plate assay. Briefly, the suspension of fresh probiotics was cultured in MRS broth supplemented with 2.5% glucose and incubated at 37°C for 12–36 h with 5% CO2. Then standard suspensions (1.5 × 108 CFU/mL) of probiotics were prepared, and 200 μL of suspension containing probiotic-MRS broth was added to a 96-well microplate (NEST, China) and incubated in an incubator with 5% CO2 at 37°C for 36 h. After incubation, the suspension was gently removed from the well, and washed three times with PBS and air-dried for 30 min. Then, 200 μL of methanol (Mojallali, Iran) was added to each well to stabilize the biofilm. After 15 min, the solution was aspirated, and the plate was air-dried at room temperature. The wells were stained with 200 μL of crystal violet (0.05%) for 5 min, and the solution was aspirated, and the wells were washed three times with PBS and left to dry naturally at room temperature for 30 min. Finally, 200 μL of ethanol (95%) (Mojallali, Iran) was added to each well and incubated at 37°C for 30 min in a shaker incubator (IKA, Germany). The content of each well was transferred to the corresponding well in another microplate, and the absorbance was measured at 492 nm using a microplate reader (BioTek, United States). All experiments were performed in triplicate. The classification of probiotic isolates based on their optical absorbance (OD) is showed in Table 2.
Aggregation assay was performed according to Del Re et al. (2000). Certain probiotics with the ability to form aggregates, can effectively inhibit the formation of dental plaque by biofilm-producing bacteria. Briefly, probiotic isolates were incubated overnight in MRS broth medium in an incubator with 5% CO2 at 37°C. After washing the cells with phosphate buffered saline (PBS), the probiotic suspension was prepared with PBS to achieve a concentration of 1.5 × 108 CFU/mL. The ability of the current probiotics to induce aggregation was investigated by measuring the decrease in optical absorbance of bacterial suspensions at 600 nm due to aggregation and diffusion. The aggregation capacity (AC) was determined using the following formula, where ODt and OD0 are the optical absorbance at the end (8 h) and initial times, respectively (Bosch et al., 2012).
The methods employed for co-aggregation and aggregation assays were identical. Briefly, probiotic isolates were incubated overnight in MRS broth in an incubator with 5% CO2 at 37°C. After washing the cells with phosphate buffered saline (PBS), the probiotic suspension was prepared with PBS to achieve a concentration of 1.5 × 108 CFU/mL. The capability of the probiotics to co-aggregate with pathogens was investigated by combining equal quantities of probiotics and pathogens. After 8 h, the turbidity of the mixture was examined and compared to that of individual suspensions of each pathogen and each probiotic as the control groups (MacDonald et al., 2021). The co-aggregation capacity was reported as the percentage reduction in optical absorbance and calculated according to Handley et al. (1987) using the following formula: Where X and y indicate each of the two strains in the control tubes, (x + y) represents the mixture, and A represents absorbance at a wavelength of 600 nm.
The HGFC cells with the NCBI code of C165 were obtained from the Cell Bank Department of the Pasteur Institute of Iran. The HGFC were grown in DMEM (Dulbecco’s Modified Eagle Medium) (Biosera, France) supplemented with 15% heated-inactivated fetal bovine serum (FBS) (Biosera, France) and 2% glutamine (Bio-IDEA, Iran). Then, the cells were incubated in a 5% CO2 incubator at 37°C. HGF cells were seeded in 6-and 12-well plates (SPL, Korea) in order to conduct adhesion and competition assays, respectively.
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, a tetrazole) assay was used to determine the potential inhibitory effects of probiotics on HGFCs. MTT determines the survival rate of cells based on cell metabolic activity after treatment with drugs or agents. A living cell has a mitochondrial dehydrogenase enzyme which converts tetrazolium into formazan, and the color of cell changes to purple following the cleavage of tetrazolium to into formazan. Finally, color intensity is determined based on optical absorbance. Briefly, the cells were seeded at a concentration of 2.5 × 104 cells per well in 96 well plates. At a confluency of 70–90%, cells were treated with 250 μL of probiotics and incubated in an incubator with 5% CO2 at 37°C for 1, 3, and 24 h. To reduce MTT into formazan, 100 μL of MTT (Sigma, United States) was added to each well containing cells and probiotics and incubated in an incubator with 5% CO2 at 37°C. After incubation, 100 μL of DMSO lysing solution was added to each well to lyse the insoluble formazan crystals and release the formazan. The contents of the wells were transferred to a sterile 96-well microtiter plate, and the optical absorbance (optical density, OD) was read at 490 nm using a microplate reader (BioTek, United States). To determine the percentage survival rate of HGFC, the OD of the treated, untreated, and blank wells was measured using the following formula:
(AB: absorbance of blank well, AC: absorbance of control, and AT: absorbance of treated well)
The adhesion potential of bacteria to surfaces is studied using qualitative and quantitative approaches. In the qualitative method, the number of attached bacteria is determined using the gram staining technique with a light microscope, and the findings are reported as a percentage. In the quantitative method, the bacteria attached to the well are transferred to a plate containing agar medium. Following the incubation, the number of bacteria in the plate is determined using serial dilution and colony count methods. This study examined the binding ability of probiotics to HGFCs through a qualitative adhesion assay. Briefly, 6-well plates with 70–90% confluency of HGFCs were prepared. After 24 h of incubation, the plate wells were washed three times with PBS, and then 2 mL of fresh high-glucose DMEM medium enriched with 10% FBS was added to each well and incubated at 37°C for 30 min in the presence of 5% CO2. A probiotic suspension at a concentration of 1.5 × 108 CFU/mL was prepared using DMEM and transferred to the wells containing 2 mL of HGFCs. The experiment was performed in triplicate, and HGFCs that were not exposed to probiotics were used as a negative control. After 120 min of incubation at 37°C with 5% CO2, the wells were slowly aspirated and washed three times with PBS. Then, the gram staining method was performed, and adherent probiotics were counted with a light microscope (X100) in 20 microscopic fields (Zhang and Duan, 2022). Following counting visible probiotics on HGFC surfaces, the adhesion index was calculated by the formula “number of adherent bacteria/number of cells × 100%,” probiotic strains were classified (D'Alessandro et al., 2021).
In this research, a competition assay was conducted to assess the ability of Lactobacillus and Bifidobacterium to reduce the virulence of F. nucleatum, P. gingivalis, A. actinomycetemcomitans, and S. mutans. To carry out this experiment, 12-well plates containing HGFCs with a confluency of 70–90% were prepared. The wells were rinsed with PBS three times, and, 2 mL of fresh DMEM medium with 10% FBS was added to each well. The plates were then placed in an incubator at 37°C with 5% CO2 for 30 min. Bacterial suspensions with a concentration of 1.5 × 108 CFU/mL were prepared from fresh cultures of Lactobacillus, Bifidobacterium, and pathogens using PBS or DMEM, and introduced to the wells containing 2 mL of HGF cells. Wells with and without bacteria were considered as positive and negative controls, respectively. All analyses were performed in duplicate.
The intervention groups were divided into two main groups; including the treatment and prevention groups. To prepare the treatment group, HGFCs were first exposed to the pathogen, then probiotics were added separately. Conversely, in the prevention group, HGFCs were first treated with probiotics before the pathogen was introduced. Briefly, 200 μL of the probiotics at 1.5 × 108 CFU/mL were added to the wells containing HGFCs and incubated for 60 min, then 200 μL of each pathogen (F. nucleatum, P. gingivalis, A. actinomycetemcomitans, and S. mutans) in equal volume was added separately and incubated anaerobically for 60 min at 37°C to evaluate changes in virulence gene expression of pathogens in the presence of probiotics (Haukioja et al., 2008). Similar to the prevention group, 200 μL of the pathogens at a concentration of 1.5 × 108 CFU/mL were added to the wells containing HGFCs and allowed to bind for 60 min of anaerobically incubated at 37°C, then 200 μL of each probiotic with the same concentration was added and incubated for another 60 min (Haukioja et al., 2008).
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to investigate the expression of Mfa1 and Rgp genes in P. gingivalis, RcpA in A. actinomycetemcomitans, Fap2 in F. nucleatum, and GtfB in S. mutans following exposure to single probiotics, a cocktail of five Lactobacillus species, a cocktail of five Bifidobacterium species, and cocktail of Lactobacillus plus Bifidobacterium. REX solution (YTA, Iran) was used to extract bacterial RNA from HGFCs according to the manufacturer’s instructions. Following the extraction of RNA, the purity and quality of the RNA were assessed using a NanoDrop spectrophotometer (Beckman, United States) and agarose gel electrophoresis (Fanavaranakhtarian, Iran), respectively. Genomic DNA was eliminated by RNase free DNase I treatment (Thermo Fisher Scientific, United States), and cDNA was synthesized using cDNA Synthesis kit (Favorgen Biotech, Austria). qRT-PCR was performed using SYBR Green qPCR Master Mix (YTA, Iran) in a final volume of 20 μL. The primers used in this study, were designed and verified by NCBI primer BLAST (Table 3). qRT-PCR thermal cycling conditions were specific for each gene but usually considered as: 95°C for 3 min (initial denaturation), followed by 40 cycles of 95°C for 15 s (denaturation), 54°C for 20 s (annealing), and 72°C for 30 s (extension) conducted on a real-time PCR cycler (Rotor-Gene, Germany). Gene expression levels were calculated according to the 2-ΔΔCT method.
The HGFC were cultured in DMEM supplemented with 10% FBS (without antibiotics) and seeded in 24-well plates after reaching 70–90% confluency. To induce inflammatory and anti-inflammatory cytokines in the HGF cell line, HGFC were pre-treated with P. gingivalis at a MOI of 25:1 and an incubated for 8 h. The probiotics were then added to the pre-treated wells at an equal MOI and incubated in incubator with 5% CO2 at 37°C. The wells were washed with PBS to remove any cells that were not bound (MacDonald et al., 2021). The collected supernatants were centrifuged at 2,000 rpm for 5 min. Then, the resulting supernatant was used for the analysis of cytokines. Changes in IL-8 and IL-10 gene expression in HGFC supernatant were evaluated using qRT-PCR. The RNA of the infected cells was extracted using REX solution (YTA, Iran), and cDNA synthesis was done by a cDNA synthesis kit (Favorgen Biotech, Austria). Gene expression levels were calculated according to the 2-ΔΔCT method.
ΔΔCT = ΔCT test sample – ΔCT calibrator sample = 2-ΔΔCT
F. nucleatum, P. gingivalis, A. actinomycetemcomitans, and S. mutans, along with probiotic strains, were prepared in a solution containing 1.5 × 108 CFU/mL. The equal volumes (200 μL) of the probiotic and the pathogen were cultured in TSB (tryptic soy broth) (Merck, Darmstadt, Germany) with 1% sucrose, then transferred to a 96-well plate and incubated in an incubator with 5% CO2 at 37°C. The control group was considered for each pathogen and probiotic. Following the incubation, the suspension was carefully removed from the wells, and the wells were rinsed three times with PBS, before being left to dry for 30 min. Then, 200 μL of methanol was added to each well to stabilize the biofilm. After 15 min, the solution was aspirated, and the plate was air-dried at room temperature. The wells were stained with 200 μL of crystal violet (0.05%) and incubate for 5 min. Then, the solution was aspirated, and the wells were washed three times with PBS and left to air dry for 30 min at room temperature. Finally, 200 μL of ethanol (95%) was added to each well, and left to incubated at 37°C for 30 min in a shaker incubator. The content of each well was transferred to its equivalent well in the same microplate, and the absorbance was measured at 570 nm using a microplate reader.
GraphPad software version 9 was used for statistical analysis of the data. Paired-sample t-test was used to show significant differences between the groups receiving probiotics and the control group. Dunnet’s two-sided t-test was used to investigate the effect of probiotics on gene expression and biofilm formation of periodontal pathogens in the groups receiving probiotics either as prevention or as treatment, which allows multiple comparisons. The statistical significance level of the data was considered to be less than 0.05 (<0.05).
Lactobacillus and Bifidobacterium strains remained alive at baseline and after being exposed to 0.4 mM H2O2 for 1, 3, and 5 h (Figure 1). However, L. reuteri 100, L. brevis 205, B. breve 1,015, and B. breve 1,063 were more resistant to H2O2, all strains survived notably up to 5 h after exposure to H2O2.
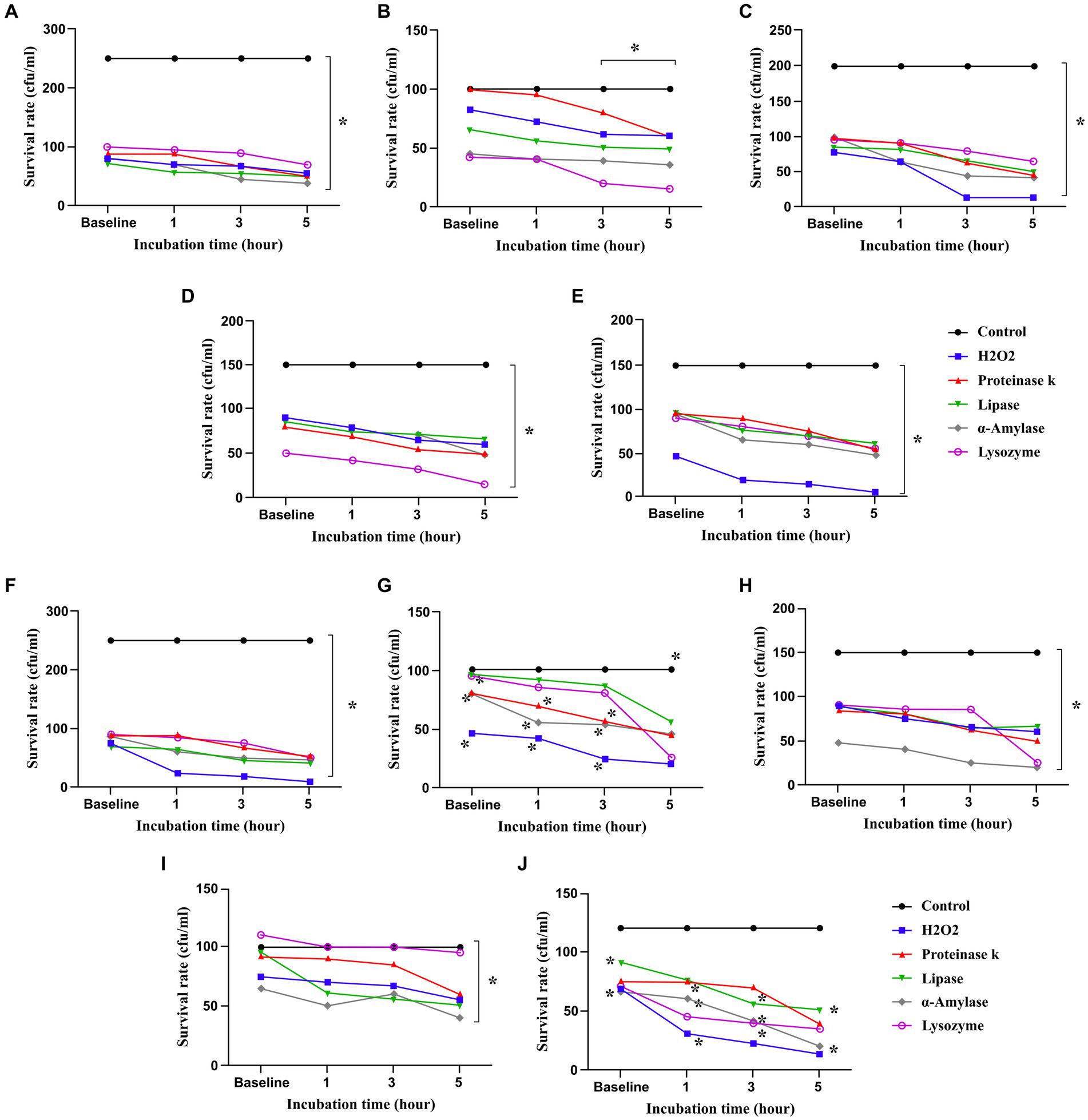
Figure 1. Probiotics survived in presence of hydrogen peroxide (H2O2), proteinase K, lipase, α-amylase and lysozyme. All data describe as mean ± SEM. *p < 0.0001. (A) L. plantarum 42, (B) L. reuteri 100, (C) L. rhamnosus 195, (D) L. brevis 205, (E) L. plantarum 156, (F) B. bifidum 1001, (G) B. bifidum 1005, (H) B. breve 1015, (I) B. breve 1063, (J) B. longum 1044.
According to the results shown in Figure 1, L. rhamnosus 195, L. plantarum 156, B. bifidum 1005, B. breve 1015, B. longum 1044, and B. breve 1063 showed great tolerance among other strains when exposed to lysozyme (22 IU/mg). The findings demonstrated a consistent outcome when using proteinase-treated probiotics, suggesting the durability of all strains following exposure for 1, 3, and 5 h. Indeed, L. reuteri 100, L. plantarum 156, B. bifidum 1005, B. breve 1015, B. breve 1063, and B. longum 1044 exhibited the greatest resistance to proteinase. In addition, after being exposed to α-amylase (220 IU/mg), all the probiotic strains, particularly L. reuteri 100, L. rhamnosus 195, L. brevis 205, L. plantarum 156, B. bifidum 1005, B. breve 1015, and B. breve 1063, remained viable for 5 h after the exposure. The survival rates of the probiotic strains against lipase (700 IU/mg) were comparable to those observed with other enzymes, however, the differences were not statistically significant. All strains survived up to 5 h after treatment with lysozyme, proteinase, α-amylase, and lipase (Figure 1).
Although the number of probiotic strains exposed to the saliva and teeth after 24 h was significantly decreased compared to the control group, as shown in Table 4, probiotics displayed tolerance to saliva and showed weak adherence to the teeth (p > 0.05).
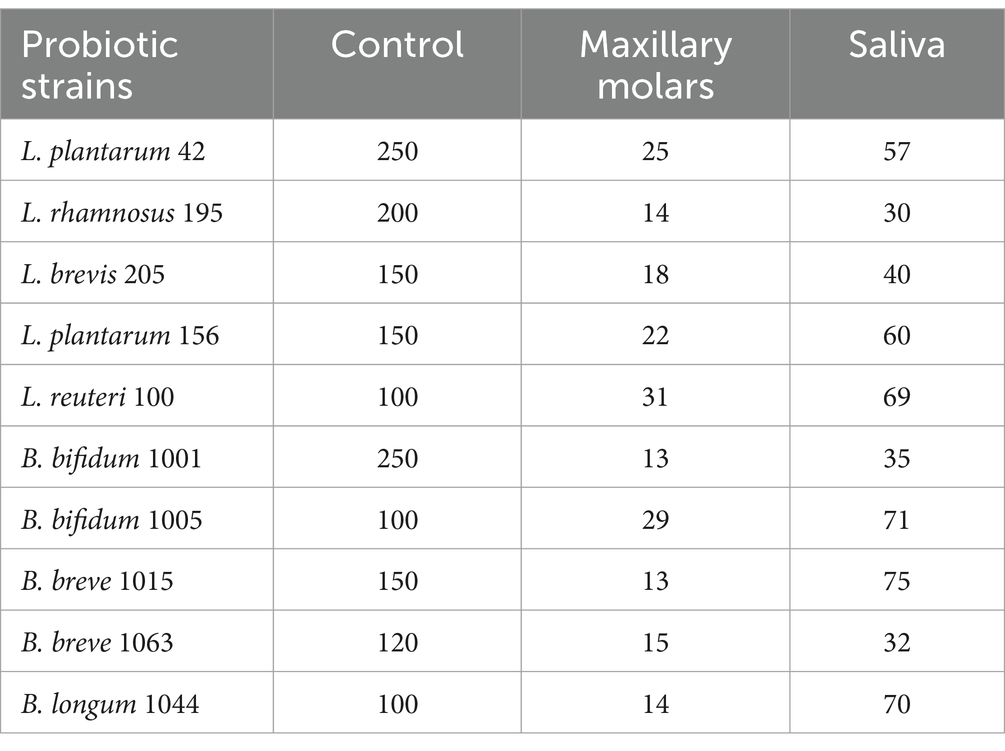
Table 4. Log 10 CFU/mL of probiotics after 24 h treatment on maxillary molars and exposure to saliva.
All probiotics were able to form biofilm. In Table 5, the probiotic isolates are classified based on biofilm production ability as strong, medium, weak, and non-biofilm producers. None of the probiotics showed strong abilities in producing biofilms, the maximum and minimum levels of biofilm production by probiotics were measured at 0.144 and 0.112, respectively. There was no significant difference among probiotics in biofilm production (p > 0.05).
All probiotics showed moderate to weak auto-aggregation ability (Table 6); however, no significant difference was observed in aggregation formation between probiotics (p > 0.05). The present study examined the co-aggregation of probiotics with oral pathogens, and measured the reduction in optical absorbance of bacterial tubes after 8 h. The findings revealed that all probiotics were able to co-aggregate with S. mutans, P. gingivalis, F. nucleatum, and A. actinomycetemcomitans. Strong co-aggregation was observed between B. bifidum 1001, L. plantarum 156, L. reuteri 100, and L. brevis 205 with S. mutans; B. bifidum 1005 and L. reuteri 100 with P. gingivalis; L. plantarum 156 and L. plantarum 42 with F. nucleatum; and B. longum 1044 and L. plantarum 156 with A. actinomycetemcomitans (Table 6).
The MTT viability assay results showed that none of the probiotics had toxicity on HGFC when administered at a concentration of 108 CFU/mL for 1, 3, and 24 h (Figure 2). This concentration was used in subsequent cell-related experiments. Notably, the negative control in this experiment was considered HGFC without treatment by any probiotics.
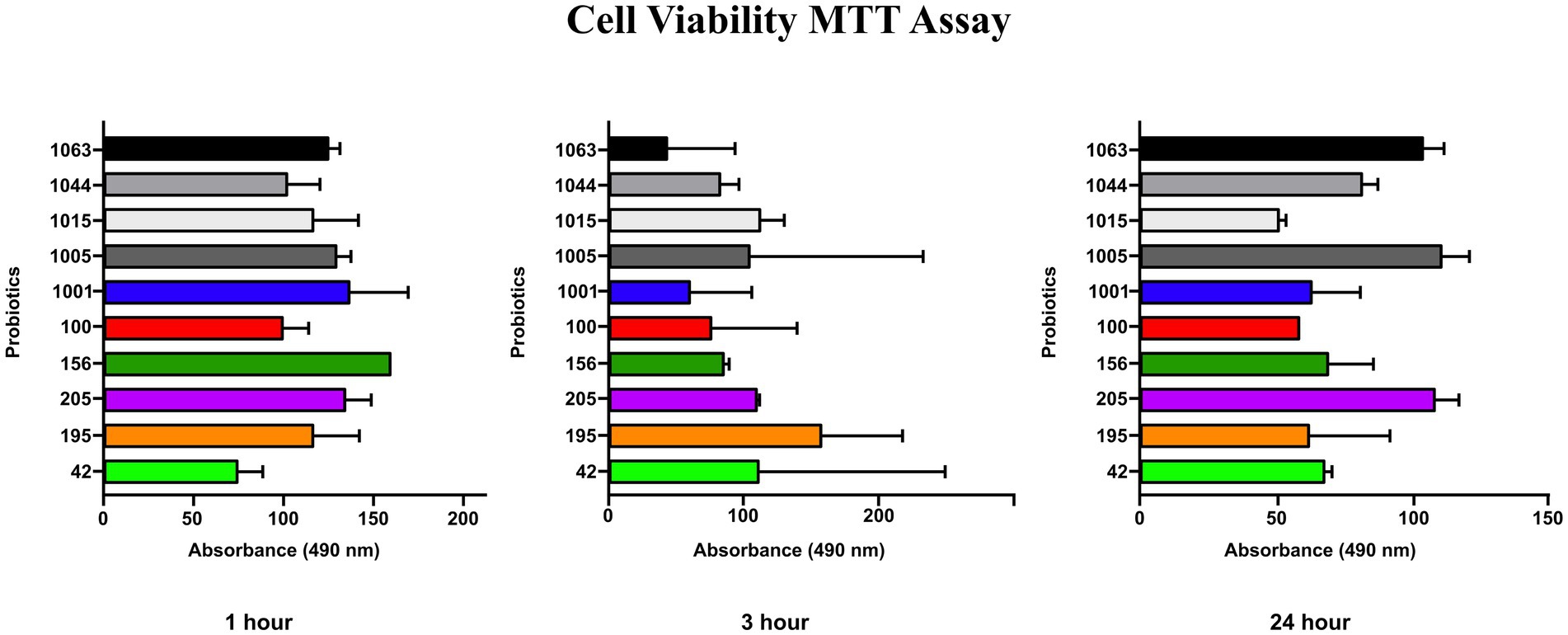
Figure 2. MTT assay (cell viability %) of probiotics after 1, 3 and 24 h for HGF cell line. No significant influence on gingival fibroblast cell viability was observed.
In vitro binding of probiotics to HGFC was determined after 2 h of incubation. The findings showed that all probiotics possessed binding ability to HGFC (Table 7). As indicated in Table 7, the comparison of probiotics showed that L. reuteri 100 and B. bifidum 1,005 exhibited the highest cell attachment capability, with an average of 23.8 and 20.5 adherent bacteria per HGF cell, respectively. The present study results indicated that eight strains were found to be adhesive, while the remaining strains did not show adhesive properties.
The qRT-PCR was used to investigate the effect of probiotics on the expression of virulence genes of oral pathogens (Figure 3). Most of the living probiotics and cocktail of Bifidobacterium plus Lactobacillus in the present study changed the expression of Mfa1 and RgpA, which was not statistically significant (p > 0.05). L. plantarum 42 and B. longum 1044 in the prevention group, B. breve 1015 and a cocktail of Lactobacillus in the treatment group, and a cocktail of Bifidobacterium plus Lactobacillus in both groups were effective in reducing RcpA in A. actinomycetemcomitans (p > 0.05). L. brevis 205, B. longum 1044, B. breve 1015, and a cocktail of Bifidobacterium plus Lactobacillus in both groups, L. plantarum 156 in the treatment group, and B. bifidum 1001 in the prevention group were effective in reducing GtfB in S. mutans (p > 0.05). L. reuteri 100, B. bifidum 1001, and a cocktail of Bifidobacterium plus Lactobacillus in both groups, B. bifidum 1005 and L. plantarum 42 in the prevention group, and L. rhamnosus 195 in the treatment group could reduce FapA in F. nucleatum (p > 0.05). Using Dunnett’s multiple comparison test, no statistically significant difference was found in the virulence gene expression of oral pathogens between the prevention and treatment groups (p = 0.9).
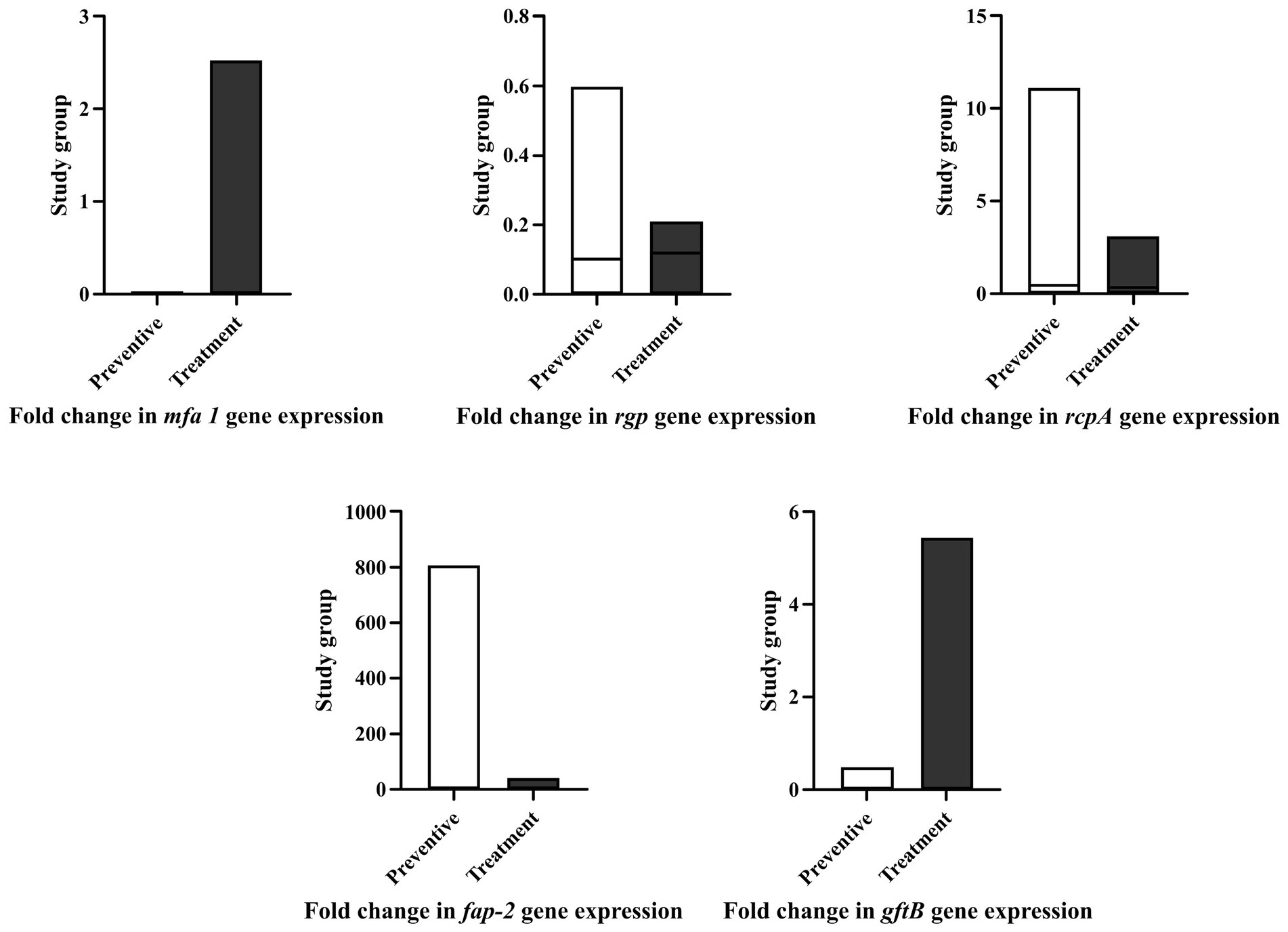
Figure 3. Most of single and live probiotics as well as cocktails of Bifidobacterium plus Lactobacillus in preventive and treatment group reduced the fold change of virulence gene expression in P. gingivalis, A. actinomycetemcomitans, F. nucleatum, and S. mutans. None of gene expression changes were statistically significant. All data represent as mean ± SEM.
By cell culture analysis, single probiotics including L. plantarum 42, B. breve 1015, and B. bifidum 1001 were more effective in increasing IL-10, and B. bifidum 1005 was effective in decreasing 1 L-8 in both intervention groups. The present research showed that a cocktail of Bifidobacterium and Lactobacillus strains was the most effective in decreasing IL-8 and enhancing the secretion of IL-10 by HGFCs in both prevention and treatment groups. However, none of these changes were statistically significant (p > 0.05) (Figure 4).
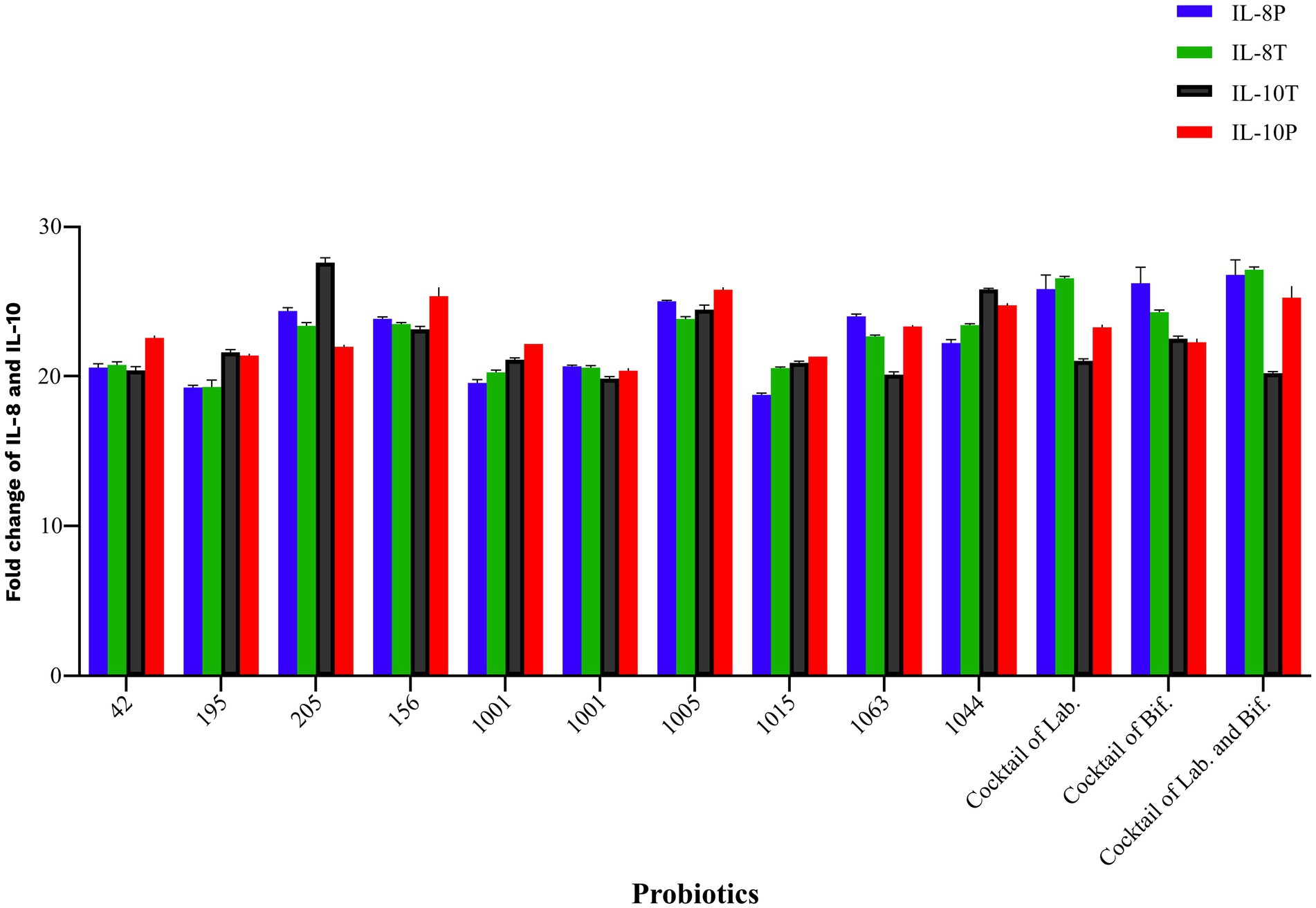
Figure 4. Probiotics modulate the inflammatory and non-inflammatory gene expression following challenging with P. gingivalis in HGF cell line. All data indicate the mean ± SEM. None of these changes were statistically significant. (P), preventive; (T), treatment; (Lab), Lactobacillus; (Bif), Bifidobacterium.
L. plantarum 42 and B. breve 1,015 in the prevention group and L. rhamnosus 195, L. plantarum 156, and B. longum 1,044 in the treatment group significantly reduced the biofilm production ability of A. actinomycetemcomitans (p < 0.05), compared to the control group that did not receive any treatment. Furthermore, intra-group comparison showed no difference between the probiotics in the prevention and treatment groups in reducing A. actinomycetemcomitans biofilm production (p > 0.05). All probiotics in the prevention group were able to significantly reduce the biofilm production ability of P. gingivalis in comparison to the treatment group (p = 0.01). However, L. reuteri 100, B. bifidum 1001, B. bifidum 1005, B. breve 1015, and B. breve 1063 in the treatment group were able to reduce P. gingivalis biofilm formation, but it was not significant (p > 0.05). When comparing the effects of probiotics on F. nucleatum biofilm formation, it was observed that L. plantarum 42, L. reuteri 100, B. bifidum 1001, and B. longum 1044 in the prevention group, as well as L. plantarum 42, L. brevis 205, L. plantarum 156, B. bifidum 1001, B. bifidum 1005, B. breve 1015, and B. longum 1044 in the treatment group, reduced F. nucleatum biofilm formation in compare to the control group (p > 0.05).
The group of S. mutans that received probiotics for prevention showed a significant decrease in biofilm production compared to the control group (p = 0.04). In contrast, the difference in biofilm formation was not significant between the treated and untreated S. mutans groups (p = 0.05). No significant difference was observed following an intra-group comparison between either S. mutans isolates receiving probiotics as a prevention or treatment group (p > 0.05). Notably, L. plantarum 156 and B. bifidum 1001 in the prevention group and L. plantarum 42 and B. longum 1044 in the treatment group resulted in the highest reduction in S. mutans biofilm formation. All results are shown in Figure 5.
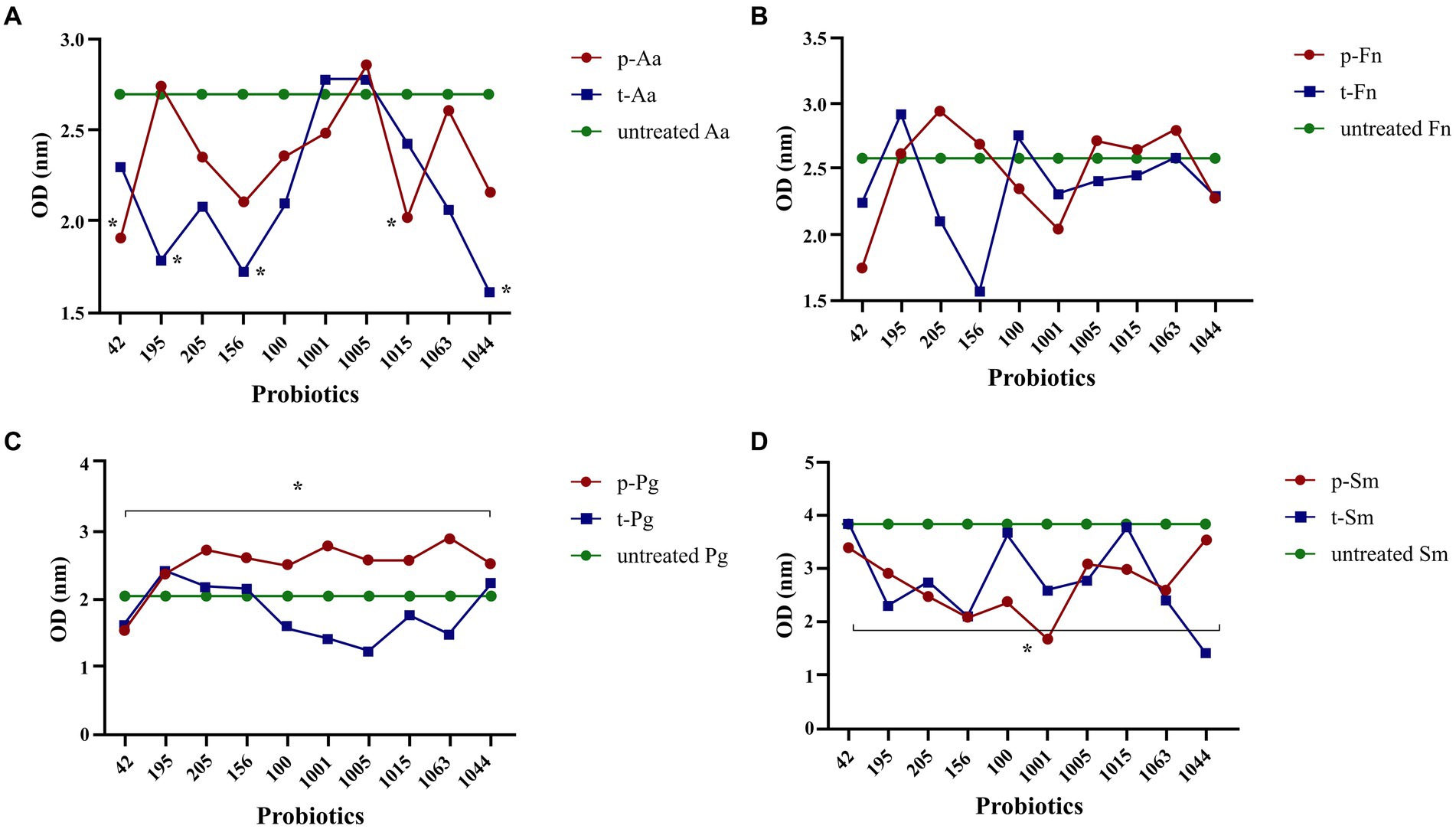
Figure 5. Probiotics reduced biofilm formation by oral pathogens, including A. actinomycetemcomitans (A), F. nucleatum (B), P. gingivalis (C), and S. mutans (D). All data indicate the mean ± SEM. *p < 0.05. Statistically significant decrease in the biofilm production in A. actinomycetemcomitans was observed in both treatment and preventive groups. In all preventive group, probiotics dereased the biofilm formation in S. mutans and P. gingivalis, while the reduction was not significant for the treatment group. Biofilm formation reduction for F. nucleatum was not significant in preventive and treatment groups.
Antibiotics and mechanical removal of dental plaque are commonly employed to reduce the biofilm and periodontal pockets in dental plaque-related diseases. There is concern regarding the diminishing effectiveness of antibiotics due to the emergence of antibiotic resistance and potential side effects (Homayouni Rad et al., 2023), however, antibiotics are still used in treatment. Herbal-based medicines with antimicrobial properties like Aloe barbadensis Miller, Trifolium pratense, and Medicago sativa (Palombo, 2011), as well as enzyme inhibitors like protease inhibitors against P. gingivalis (Hosn et al., 2015), and microbial therapy like probiotics (Saraf et al., 2010), are innovative therapeutic strategies which have been developed to combat oral diseases. Medicinal plants are known for their ability to provide antioxidants and anti-inflammatory benefits, with minimal adverse effects and the rarely emergence of resistance strains (Pasupuleti et al., 2023). Designing inhibitors targeting cysteine proteases of P. gingivalis would be beneficial in the management of periodontitis by preventing tissue destruction (Hosn et al., 2015).
The interest in utilizing probiotics in dentistry has increased. As probiotics target the pathogens while maintaining a balance within the oral microbiota, antibiotics disrupt normal flora besides eliminating pathogenic bacteria. According to in vitro studies, probiotics prevent the growth of cariogenic bacteria like S. mutans and periodontal pathogens such as P. gingivalis, A. actinomycetemcomitans, Prevotella intermedia, and F. nucleatum by colonizing the oral cavity, forming biofilm, and reducing the acidity level, which helps to maintain hemostasis and manage the immune system (Haukioja, 2010; Homayouni Rad et al., 2023; Shirbhate et al., 2023). Furthermore, clinical trials have shown a significant decrease in probing pocket depth (PPD), clinical attachment loss (CAL), bleeding on probing (BOP), plaque index (PI), and gingival index (GI) in the probiotic group (Dhaliwal et al., 2017; Costacurta et al., 2018; Kumar et al., 2021; Hardan et al., 2022). An ideal probiotic for controlling oral diseases should effectively colonize oral surfaces, be resistant to oral flora, have the ability to form biofilm, not produce foul-smelling compounds, lack resistance to antibiotics and toxicity, establish homeostasis in the oral cavity and regulate immune responses, and not promote caries (Bosch et al., 2012; Wen et al., 2022).
The main finding of this study is the durability of probiotics isolated from breast milk and infant feces in exposure to various enzymes as well as their binding ability to gingival fibroblast cells. To determine the most resistant probiotic in the oral cavity, the concentrations of protease, lipase, and α-amylase enzymes, tested in this study were adjusted near their natural concentrations in saliva, and the results showed all probiotics were stable in the oral cavity and saliva. Probiotics that possess the capability to produce biofilm, can establish long-term colonization in the gut, effectively limiting the growth of harmful bacteria. All probiotics demonstrated weak biofilm formation and aggregation. The development of biofilm and aggregation by probiotics is primarily influenced by their ability to adhere to surfaces, and all probiotics examined in this study showed the ability to attach to maxillary molars and gingival fibroblast cells (D'Alessandro et al., 2021). Nevertheless, probiotics showed weak adhesion capabilities, Lactobacillus spp. demonstrated great adhesiveness compared to other probiotic strains. This supports the idea that probiotics originated from fecal samples are the same as to those found in the mouth (Rohani et al., 2015). In order to establish long-term colonization of probiotics in the oral cavity, a strong and permanent adherence is required to colonization, however, probiotics remain for a short period in the oral cavity, and most of them establish a reversible adherence with cells that can be easily detached (Van Holm et al., 2023).
Biofilm serves as a niche for protecting microorganisms (Jayathilake et al., 2017). This study revealed, S. mutans exhibited a robust ability to form biofilms, although P. gingivalis showed weaker biofilm formation capability. Targeting biofilm formation in biofilm-producing pathogens is the most important objective in the management of caries and periodontal disease. An efficient probiotic hinders the growth of pathogens in dental plaque through biofilm formation and co-aggregation with pathogens. Our probiotics reduced the biofilm formation of A. actinomycetemcomitans, P. gingivalis, F. nucleatum, and S. mutans in the prevention and treatment groups. A significant decrease in biofilm formation of S. mutans was observed, which was confirmed by a significant reduction in the GtfB transcript. This is in accordance with observations made by Ahmed et al. (2014) and Hasan et al. (2015) that demonstrated the reduction in Gft enzyme expression plays an important role in preventing biofilm formation and developing caries in L. rhamnosus. These claims highlight the potential of Lactobacillus spp. as a promising alternative to antibiotics due to their anti-biofilm activity (Ahmed et al., 2014), and suppress the expression of GtfB, GtfC, and GtfD in S. mutans (Hasan et al., 2015). Although the main mechanism of biofilm inhibition by probiotics is still not fully understood, competing for nutrients, disrupting the attachment of pathogens, and producing antimicrobial peptides have been described as potential mechanisms (Barzegari et al., 2020). Some probiotics, like L. plantarum, reduce the biofilm mass by positively-charged D-Alanine in lipoteichoic acid (LTA). LTA has an inhibitory effect on the production of extracellular polymeric substances (EPS) (Ahn et al., 2018). The cationic amphiphilic structure of LTA also prevents aggregation by binding biofilm-associated genes to bacterial DNA (Anunthawan et al., 2015). In addition to LTA, probiotics such as L. fermentum can limit the biofilm production in S. mutans by using antibiofilm substances like biosurfactants (Tahmourespour et al., 2011). It is similar to the suppression of biofilm formation in Actinomyces naeslundii and Staphylococcus aureus through rhamnolipid and lipopeptide production by Burkholderia thailandensis and Bacillus subtilis, respectively (Rivardo et al., 2009; Elshikh et al., 2017). Interestingly, probiotics prevent the production of biofilm by other pathogenic bacteria due to their biofilm formation ability. According to the Ramos et al. study (Ramos et al., 2012), the supernatant of L. plantarum reduced the formation of P. aeruginosa biofilm without changing its matrix composition. By viability assay, they showed lactic acid solution prepared from L. plantarum not only has bacteriostatic and bactericidal properties on biofilm and planktonic forms of P. aeruginosa, but also suppresses the expression of virulence factors which are regulated by quorum sensing (QS), like elastase, rhamnolipid, and pyocyanin (Durant et al., 2000; Ramos et al., 2012). Differences in bacterial source, LTA structure, and probiotic strain led to controversies in biofilm inhibition results (Ryu et al., 2008).
Studies have shown that probiotics exert antimicrobial properties through the production of enzymes, acids, bacteriocins, and hydrogen peroxide (Duncker et al., 2011). In addition to preventing biofilm formation, the present study also showed L. plantarum 42, L. reuteri 100, and B. bifidum 1001 have inhibitory impacts on gene transcripts of RcpA, Mfa1, RgpA, Fap2, and GtfB in treated A. actinomycetemcomitans, P. gingivalis, F. nucleatum, and S. mutans compared to the untreated control group and other probiotics. The aforementioned genes are involved in biofilm formation (Smith et al., 2016), colonization, auto-aggregation, and the interaction of pathogens with the surrounding (Zijnge et al., 2012).
Probiotics also regulate host-related factors and modulate inflammation in the oral cavity. In this study, probiotics were effective in decreasing IL-8 and increasing IL-10 in HGFCs in both prevention and treatment groups. This is similar to the Kaci et al. (2011, 2014) findings which showed S. salivarius reduced TNF-α, IL-1, and IL-8 in stimulated intestinal epithelial cells and HT-29 by a small heat-resistant protein produced by some probiotics. Probiotics also modulate inflammation by producing short-chain fatty acids (SCFA) as postbiotic compounds like propionate and butyrate. Postbiotics produced by probiotics not only suppress the NF-κB pathway and interfere with suppression of Treg function (Vinolo et al., 2011), but also stimulate the production of anti-inflammatory cytokines and Treg differentiation (Kespohl et al., 2017).
The efficacy of our probiotics did not meet significant results as expected, which was similar to the Bosch et al. (2012) study. Lack of significant impressiveness by probiotics arises from their limited potential in biofilm formation, aggregation, and adhesion to HGFCs, which determine the eligibility of a candidate probiotic for use in the treatment of oral diseases (Sorroche et al., 2012). Moreover, the pathogen-specific inhibitory effect of probiotics and the presence of thick peptidoglycan in Gram-positive bacteria (Yang et al., 2021), should be considered which require a higher dosage of probiotics and a cell-free culture supernatant to significantly prevent the growth of harmful bacteria (Ding et al., 2021). In this context, Yang et al. (2021) showed that cell-free culture supernatant of L. reuteri had greater antimicrobial activity than live strains, due to the production of secondary bioactive metabolites (Ren et al., 2018), and the reduction of intracellular ATP required for the growth of oral pathogens. This evidences encourages us to prioritize the use of cell-free culture supernatants in future studies. In the present study, L. reuteri represents great function in tolerating oral conditions, binding to gingival fibroblast cells, and reducing biofilm formation, virulence gene expression, and inflammatory responses. The beneficial effect of L. reuteri and its survival in rough conditions were already proven and attributed to the production of inulin-type fructansucrase (Duncker et al., 2011), reutericyclin, reuteran, and reuterin (Sun et al., 2022).
To determine whether the origin of the probiotic isolation involved in lack of significant results or not, the functional equivalences and differences of oral and fecal probiotics should be addressed. Shimabukuro et al., in an animal model, evaluated the management of periodontitis by B. breve and B. bifidum, which were extracted from fecal samples. They showed complete clearance of P. gingivalis from oral biofilms and improvement in alveolar bone destruction by B. bifidum (Shimabukuro et al., 2021). The study evaluated the adherence ability and the modulatory effect of the immune system of L. salivarius AR809 extracted from the oral cavity of a healthy individual (Jia et al., 2019). The efficient adherence of oral isolated probiotics to pharyngeal epithelial FaDu cells and modulating the host’s immune response by enhancing IL-10 production and reducing the expression of TNF-α and IL-1B were observed. Despite being less studied about the effect of probiotics on dental caries disease and the need for further comprehensive studies, recent studies have shown probiotics isolated from oral and fecal sources to be promising candidates for clinical application (Shokryazdan et al., 2017).
The main challenges in the widespread application of probiotics for oral treatment are the introduction of single probiotic strains and the limited global access to probiotics (Spacova et al., 2020). Another important consideration in probiotic use is patient acceptance. However, following the use of mouthwash-containing probiotics, patient compliance was evaluated which more than 95% of volunteers were satisfied (Nisha et al., 2023). The effectiveness of probiotics also depends on the delivery method in terms of fluctuations of temperature, oxidation, pH level, and resistance to decomposition by enzymes. Various methods have been introduced to overcome delivery limitations, among them oral delivery is more common due to patient compliance, cost-effectiveness, and easy application for prevention and treatment purposes (Baral et al., 2021). Oral delivery of probiotics is achieved by dietary supplements, oral suspensions, lozenges, mouthwash, granules, capsules, oral films, and tablets for the treatment of oral diseases (Lee et al., 2017; Nie et al., 2023). Mouthwash-containing probiotics contribute to reducing plaque formation and compete with pathogens for adhesion sites, thereby regulating plaque ecology in normal flora (Meurman and Stamatova, 2007). Chewing gums are another common formulation of probiotics for oral diseases which are designed by probiotics (Ribeiro et al., 2020). Probiotic based gums successfully prevent S. mutans growth, and manage periodontal diseases (Krasse et al., 2006). The simple way to administer of probiotics is through dietary supplements in the form of granules to achieve a gradual and slow release of probiotics (Shirbhate et al., 2023).
Despite the safety history of probiotics for clinical application, a rarely occurring bacteremia in high-risk individuals was reported following consumption of probiotics (Doron and Snydman, 2015). Therefore, it is essential to properly examine the characteristics of microbial therapy with probiotics. The general safety of probiotics is described as their stability against rough conditions like gastric juice and enzymes, and their the binding ability, while functional safety is determined by having an antagonistic ability against pathogens, modulating immune responses, and selective activity in stimulating or suppressing the growth of certain bacteria in the oral cavity (Shirbhate et al., 2023). In addition to the survival of probiotics in the oral cavity, we have previously confirmed that present probiotics are resistant at pH = 2 and 0.4% bile salt, do not harbor any antibiotic resistance genes, or induce hemolysis (Rohani et al., 2015; Eshaghi et al., 2017). Also, the safety of present probiotics based on their mutagenicity, genotoxicity, and possible adverse effects was shown in the Darbandi et al. (2022) study, in which no mutations, genotoxicity in cell line examination, or adverse effects in animal models like mortality, abnormality, and weight change were observed in any dose range of 2000, 1,000, and 500 mg/kg. The examination of probiotics to produce lactic acid is important. Bosch et al. (2012) excluded 7 out of 46 probiotic candidates due to lactic acid production and the risk of dental caries.
The long-term effect of probiotics in the oral cavity is not well-known, but the short-term effects of probiotics showed they have significant anti-caries impacts, and significant changes in the alpha diversity of the oral microbiome (Dassi et al., 2018). Hradicka et al. (2023) studied the safety concerns of long-term probiotics in people who have taken probiotics daily for more than 10 years. They showed the long-term administration of probiotics does not have a significant effect on health, and causes gut microbiota alteration, and a significant increase in serum biochemical parameters, lipid metabolism, and inflammatory response. Meanwhile, the short-term administration of probiotics has significant effects on strengthening the gut-liver axis pathway and stimulating the innate immune response. This claim emphasizes the importance of the duration of probiotic consumption in the outcome of the clinical application.
For future prospective research, evaluation of the safety of probiotics, the composition of the microbiota before and after the administration of probiotics, the optimal dosage, the duration of administration, and determining whether preventive or treatment purposes were recommended to be considered. Despite the various animal model studies that have been conducted to investigate caries-related disease, rats are not an ideal representation of humans. Therefore, probiotics should be studied in all human groups, especially high-risk populations such as immunocompromised individuals and patients with severe underlying diseases. Also, it is important to consider the characteristics of the participants based on gender and lifestyle habits (smoking, drugs, alcohol, and lifestyle) in a long-term clinical trial.
The current study did not investigate the production of lactic acid, unpleasant volatile compounds, hydrogen sulfide, and soluble glucan by probiotics, as well as the scanning electron microscope (SEM) analysis that could provide more insight into microbial communities. These experiments will be examined in upcoming studies. In this study, different combinations of probiotics in cocktails were used to determine their efficacy against oral pathogens. The key distinction of the current study from similar studies lies in the source of probiotics utilized. While many studies have focused on the effects of commercially available oral probiotic tablets or isolated strains, the present study employed probiotics derived from feces and breast milk, which has shed light on their potential for oral treatment. Further studies are needed to examine their protective role in the mouth and confidently introduce all or some of them as potential candidates for the treatment of dental caries disease.
According to the specific criteria of selection for oral application of probiotics, the functional capabilities of present probiotics showed they had the ability to survive in the oral cavity, form biofilm and aggregation with oral pathogens, modulate immune responses, prevent the growth of pathogens, and adhere to the HGFC and maxillary molar. However, some of these effects were not statistically significant. Moreover, no antibiotic resistance, mutagenicity, toxicity, or genotoxicity were reported from the aforementioned probiotics. Our results also confirm the direct relationship between biofilm production, aggregation and co-agglutination ability, which significantly depends on the adherence ability of bacteria. Our finding discovered L. reuteri as a promising probiotic candidate which effectively reduces the expression of virulence genes, inhibits biofilm formation, remains stable in oral cavity conditions, and binds to gingival fibroblast cells. For future study, we suggest using the cell-free culture supernatant of L. reuteri to treat or prevent caries and periodontitis.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by Ethics Committee of the Iran University of Medical Sciences (IR.IUMS.FMD.REC.1400.544). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
MM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. SM: Formal analysis, Writing – review & editing. DD-S: Data curation, Writing – review & editing. AD: Formal analysis, Writing – review & editing. SR: Supervision, Writing – review & editing. MT: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by Iran University of Medical Sciences (grant number 21734).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmed, A., Dachang, W., Lei, Z., Jianjun, L., Juanjuan, Q., and Yi, X. (2014). Effect of Lactobacillus species on Streptococcus mutans biofilm formation. Pak. J. Pharm. Sci. 27, 1523–1528
Ahn, K. B., Baik, J. E., Park, O. J., Yun, C. H., and Han, S. H. (2018). Lactobacillus plantarum lipoteichoic acid inhibits biofilm formation of Streptococcus mutans. PLoS One 13:e0192694. doi: 10.1371/journal.pone.0192694
Anunthawan, T., de la Fuente-Núñez, C., Hancock, R. E. W., and Klaynongsruang, S. (2015). Cationic amphipathic peptides KT2 and RT2 are taken up into bacterial cells and kill planktonic and biofilm bacteria. Biochim. Biophys. Acta 1848, 1352–1358. doi: 10.1016/j.bbamem.2015.02.021
Baral, K. C., Bajracharya, R., Lee, S. H., and Han, H. K. (2021). Advancements in the pharmaceutical applications of probiotics: dosage forms and formulation technology. Int. J. Nanomedicine 16, 7535–7556. doi: 10.2147/IJN.S337427
Barzegari, A., Kheyrolahzadeh, K., Hosseiniyan Khatibi, S. M., Sharifi, S., Memar, M. Y., and Zununi Vahed, S. (2020). The Battle of probiotics and their derivatives against biofilms. Infect Drug Resist 13, 659–672. doi: 10.2147/IDR.S232982
Bosch, M., Nart, J., Audivert, S., Bonachera, M. A., Alemany, A. S., Fuentes, M. C., et al. (2012). Isolation and characterization of probiotic strains for improving oral health. Arch. Oral Biol. 57, 539–549. doi: 10.1016/j.archoralbio.2011.10.006
Costacurta, M., Sicure, L., Margiotta, S., Ingrasciotta, L., and Docimo, R. (2018). Clinical effects of Lactobacillus reuteri probiotic in treatment of chronic periodontitis. A randomized, controlled trial. Oral Implantol. 11, 191–198,
Cugini, C., Ramasubbu, N., Tsiagbe, V. K., and Fine, D. H. (2021). Dysbiosis from a microbial and host perspective relative to oral health and disease. Front. Microbiol. 12:617485. doi: 10.3389/fmicb.2021.617485
D'Alessandro, M., Parolin, C., Bukvicki, D., Siroli, L., Vitali, B., De Angelis, M., et al. (2021). Probiotic and metabolic characterization of vaginal lactobacilli for a potential use in functional foods. Microorganisms 9:833. doi: 10.3390/microorganisms9040833
Darbandi, A., Mirkalantari, S., Golmoradi Zadeh, R., Esghaei, M., Talebi, M., and Kakanj, M. (2022). Safety evaluation of mutagenicity, genotoxicity, and cytotoxicity of Lactobacillus spp. isolates as probiotic candidates. J. Clin. Lab. Anal. 36:e24481. doi: 10.1002/jcla.24481
Dassi, E., Ferretti, P., Covello, G., HTM-CMB-2015Speccher, A., Migazzi, A., et al. (2018). The short-term impact of probiotic consumption on the oral cavity microbiome. Sci. Rep. 8:10476. doi: 10.1038/s41598-018-28491-x
Del Re, B., Sgorbati, B., Miglioli, M., and Palenzona, D.. (2000). Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 31, 438–442. doi: 10.1046/j.1365-2672.2000.00845.x
Dhaliwal, P. K., Grover, V., Malhotra, R., and Kapoor, A. (2017). Clinical and microbiological investigation of the effects of probiotics combined with scaling and root planning in the Management of Chronic Periodontitis: a randomized controlled study. J Int Acad Periodontol 19, 101–108
Ding, Q., Sun, X., Cao, S., Zhao, C., Wang, Y., and Wang, X. (2021). Heat-killed Lactobacillus acidophilus mediates Fusobacterium nucleatum induced pro-inflammatory responses in epithelial cells. FEMS Microbiol. Lett. 368:fnaa160. doi: 10.1093/femsle/fnaa160
Doron, S., and Snydman, D. R. (2015). Risk and safety of probiotics. Clin. Infect. Dis. 60, S129–S134. doi: 10.1093/cid/civ085
Duncker, S. C., Kamiya, T., Wang, L., Yang, P., and Bienenstock, J. (2011). Probiotic Lactobacillus reuteri alleviates the response to gastric distension in rats. J. Nutr. 141, 1813–1818. doi: 10.3945/jn.110.136689
Durant, J. A., Corrier, D. E., and Ricke, S. C. (2000). Short-chain volatile fatty acids modulate the expression of the hilA and invF genes of Salmonella typhimurium. J. Food Prot. 63, 573–578. doi: 10.4315/0362-028X-63.5.573
Elshikh, M., Funston, S., Chebbi, A., Ahmed, S., Marchant, R., and Banat, I. M. (2017). Rhamnolipids from non-pathogenic Burkholderia thailandensis E264: physicochemical characterization, antimicrobial and antibiofilm efficacy against oral hygiene related pathogens. New Biotechnol. 36, 26–36. doi: 10.1016/j.nbt.2016.12.009
Eshaghi, M., Bibalan, M. H., Rohani, M., Esghaei, M., Douraghi, M., Talebi, M., et al. (2017). Bifidobacterium obtained from mother’s milk and their infant stool; a comparative genotyping and antibacterial analysis. Microb. Pathog. 111, 94–98. doi: 10.1016/j.micpath.2017.08.014
Handley, P. S., Harty, D. W. S., Wyatt, J. E., Brown, C. R., Doran, J. P., and Gibbs, A. C. C. (1987). A comparison of the adhesion, coaggregation and cell-surface hydrophobicity properties of fibrillar and fimbriate strains of Streptococcus salivarius. J. Gen. Microbiol. 133, 3207–3217. doi: 10.1099/00221287-133-11-3207
Hardan, L., Bourgi, R., Cuevas-Suárez, C. E., Flores-Rodríguez, M., Omaña-Covarrubias, A., Nicastro, M., et al. (2022). The use of probiotics as adjuvant therapy of periodontal treatment: a systematic review and Meta-analysis of clinical trials. Pharmaceutics 14:1017. doi: 10.3390/pharmaceutics14051017
Hasan, S., Danishuddin, M., and Khan, A. U. (2015). Inhibitory effect of Zingiber officinale towards Streptococcus mutans virulence and caries development: in vitro and in vivo studies. BMC Microbiol. 15:1. doi: 10.1186/s12866-014-0320-5
Haukioja, A., Loimaranta, V., and Tenovuo, J. (2008). Probiotic bacteria affect the composition of salivary pellicle and streptococcal adhesion in vitro. Oral Microbiol. Immunol. 23, 336–343. doi: 10.1111/j.1399-302X.2008.00435.x
Homayouni Rad, A., Pourjafar, H., and Mirzakhani, E. (2023). A comprehensive review of the application of probiotics and postbiotics in oral health. Front. Cell. Infect. Microbiol. 13:1120995. doi: 10.3389/fcimb.2023.1120995
Hosn, K. N., Jefferson, M. M., Leding, C., Shokouh-Amiri, S., and Thomas, E. L. (2015). Inhibitors of bacterial protease enzymes for periodontal therapy. Clin Exp Dent Res 1, 18–25. doi: 10.1002/cre2.4
Hradicka, P., Adamkova, P., Lenhardt, L., Gancarcikova, S., Iannaccone, S. F., and Demeckova, V. (2023). Addressing safety concerns of long-term probiotic use: in vivo evidence from a rat model. J. Funct. Foods 104:105521. doi: 10.1016/j.jff.2023.105521
Ibarburu, I., Puertas, A. I., Berregi, I., Rodríguez-Carvajal, M. A., Prieto, A., and Dueñas, M. T. (2015). Production and partial characterization of exopolysaccharides produced by two Lactobacillus suebicus strains isolated from cider. Int. J. Food Microbiol. 214, 54–62. doi: 10.1016/j.ijfoodmicro.2015.07.012
Jayathilake, P. G., Jana, S., Rushton, S., Swailes, D., Bridgens, B., Curtis, T., et al. (2017). Extracellular polymeric substance production and aggregated bacteria colonization influence the competition of microbes in biofilms. Front. Microbiol. 8:1865. doi: 10.3389/fmicb.2017.01865
Jia, G. C., Che, N., Xia, Y. J., Lai, P. F. H., Xiong, Z. Q., Wang, G. Q., et al. (2019). Adhesion to pharyngeal epithelium and modulation of immune response: Lactobacillus salivarius AR809, a potential probiotic strain isolated from the human oral cavity. J. Dairy Sci. 102, 6738–6749. doi: 10.3168/jds.2018-16117
Kaci, G., Goudercourt, D., Dennin, V., Pot, B., Doré, J., Ehrlich, S. D., et al. (2014). Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl. Environ. Microbiol. 80, 928–934. doi: 10.1128/AEM.03133-13
Kaci, G., Lakhdari, O., Doré, J., Ehrlich, S. D., Renault, P., Blottière, H. M., et al. (2011). Inhibition of the NF-kappaB pathway in human intestinal epithelial cells by commensal Streptococcus salivarius. Appl. Environ. Microbiol. 77, 4681–4684. doi: 10.1128/AEM.03021-10
Kespohl, M., Vachharajani, N., Luu, M., Harb, H., Pautz, S., Wolff, S., et al. (2017). The microbial metabolite butyrate induces expression of Th1-associated factors in CD4(+) T cells. Front. Immunol. 8:1036. doi: 10.3389/fimmu.2017.01036
Krasse, P., Carlsson, B., Dahl, C., Paulsson, A., Nilsson, A., and Sinkiewicz, G. (2006). Decreased gum bleeding and reduced gingivitis by the probiotic Lactobacillus reuteri. Swed. Dent. J. 30, 55–60
Kumar, V., Singhal, R., Rastogi, P., Lal, N., Pandey, S., and Mahdi, A. A. (2021). Localized probiotic-guided pocket recolonization in the treatment of chronic periodontitis: a randomized controlled clinical trial. J Periodontal Implant Sci 51, 199–212. doi: 10.5051/jpis.2004140207
Lee, S. H., and Kim, Y. J. (2014). A comparative study of the effect of probiotics on cariogenic biofilm model for preventing dental caries. Arch. Microbiol. 196, 601–609. doi: 10.1007/s00203-014-0998-7
Lee, Y., Kim, K., Kim, M., Choi, D. H., and Jeong, S. H. (2017). Orally disintegrating films focusing on formulation, manufacturing process, and characterization. J. Pharm. Investig. 47, 183–201. doi: 10.1007/s40005-017-0311-2
MacDonald, K. W., Chanyi, R. M., Macklaim, J. M., Cadieux, P. A., Reid, G., and Burton, J. P. (2021). Streptococcus salivarius inhibits immune activation by periodontal disease pathogens. BMC Oral Health 21:245. doi: 10.1186/s12903-021-01606-z
Matsumoto-Nakano, M. (2018). Role of Streptococcus mutans surface proteins for biofilm formation. Japanese Dent. Sci. Rev. 54, 22–29. doi: 10.1016/j.jdsr.2017.08.002
Menon, A. (2016). Implications of probiotics on oral health: past-to-present. J. Dent. Res. Rev. 3:36. doi: 10.4103/2348-2915.180118
Meurman, J. H., and Stamatova, I. (2007). Probiotics: contributions to oral health. Oral Dis. 13, 443–451. doi: 10.1111/j.1601-0825.2007.01386.x
Miles, A. A., Misra, S. S., and Irwin, J. O. (1938). The estimation of the bactericidal power of the blood. J Hyg 38, 732–749. doi: 10.1017/S002217240001158X
Mirzaei, R., Alikhani, M. Y., Arciola, C. R., Sedighi, I., Yousefimashouf, R., and Bagheri, K. P. (2022). Prevention, inhibition, and degradation effects of melittin alone and in combination with vancomycin and rifampin against strong biofilm producer strains of methicillin-resistant Staphylococcus epidermidis. Biomed. Pharmacother. 147:112670. doi: 10.1016/j.biopha.2022.112670
Nie, Q., Wan, X., Tao, H., Yang, Q., Zhao, X., Liu, H., et al. (2023). Multi-function screening of probiotics to improve oral health and evaluating their efficacy in a rat periodontitis model. Front. Cell. Infect. Microbiol. 13:1261189. doi: 10.3389/fcimb.2023.1261189
Nisha, D. S., Sebastian, B., Kalappurakkal, R., and Kirubakaran, R. (2023). Efficacy of aloe vera and probiotic mouthwashes vs. fluoride mouthwash on Streptococcus mutans in plaque around brackets of orthodontic patients: a randomized clinical trial. Angle Orthod. 93, 538–544. doi: 10.2319/082222-595.1
Padmavathi, T., Bhargavi, R., Priyanka, P. R., Niranjan, N. R., and Pavitra, P. V. (2018). Screening of potential probiotic lactic acid bacteria and production of amylase and its partial purification. J. Genet. Eng. Biotechnol. 16, 357–362. doi: 10.1016/j.jgeb.2018.03.005
Palombo, E. A. (2011). Traditional medicinal plant extracts and natural products with activity against oral bacteria: potential application in the prevention and treatment of oral diseases. Evid. Based Complement. Alternat. Med. 2011:680354. doi: 10.1093/ecam/nep067
Pasupuleti, M. K., Nagate, R. R., Alqahtani, S. M., Penmetsa, G. S., Gottumukkala, S. N. V. S., and Ramesh, K. S. V. (2023). Role of medicinal herbs in periodontal therapy: a systematic review. J. Int. Soc. Prev. Community Dent. 13, 9–16. doi: 10.4103/jispcd.JISPCD_210_22
Pourhajibagher, M., Rokn, A., Barikani, H., and Bahador, A. (2020). Photo-sonodynamic antimicrobial chemotherapy via chitosan nanoparticles-indocyanine green against polymicrobial periopathogenic biofilms: ex vivo study on dental implants. Photodiagn. Photodyn. Ther. 31:101834. doi: 10.1016/j.pdpdt.2020.101834
Ramos, A. N., Sesto Cabral, M. E., Noseda, D., Bosch, A., Yantorno, O. M., and Valdez, J. C. (2012). Antipathogenic properties of Lactobacillus plantarum on Pseudomonas aeruginosa: the potential use of its supernatants in the treatment of infected chronic wounds. Wound Repair Regen. 20, 552–562. doi: 10.1111/j.1524-475X.2012.00798.x
Ren, D., Zhu, J., Gong, S., Liu, H., and Yu, H. (2018). Antimicrobial characteristics of lactic acid Bacteria isolated from homemade fermented foods. Biomed. Res. Int. 2018:5416725. doi: 10.1155/2018/5416725
Ribeiro, F., Junqueira, J. C., dos Santos, J. D., de Barros, P. P., Rossoni, R. D., Shukla, S., et al. (2020). Development of probiotic formulations for Oral candidiasis prevention: Gellan gum as a carrier to deliver Lactobacillus paracasei 28.4. Antimicrob. Agents Chemother. 64:64. doi: 10.1128/AAC.02323-19
Rivardo, F., Turner, R. J., Allegrone, G., Ceri, H., and Martinotti, M. G. (2009). Anti-adhesion activity of two biosurfactants produced by Bacillus spp. prevents biofilm formation of human bacterial pathogens. Appl. Microbiol. Biotechnol. 83, 541–553. doi: 10.1007/s00253-009-1987-7
Rohani, M., Noohi, N., Talebi, M., Katouli, M., and Pourshafie, M. R. (2015). Highly heterogeneous probiotic Lactobacillus species in healthy Iranians with low functional activities. PLoS One 10:e0144467. doi: 10.1371/journal.pone.0144467
Ryu, Y., Baik, J. E., Yang, J. S., Kang, S. S., Im, J., Yun, C. H, et al. (2008). Differential immunostimulatory effects of gram-positive bacteria due to their lipoteichoic acids. Int. Immunopharmacol. 9, 127–133. doi: 10.1016/j.intimp.2008.10.014
Saraf, K., Shashikanth, M. C., Priy, T., Sultana, N., and Chaitanya, N. C. (2010). Probiotics--do they have a role in medicine and dentistry? J. Assoc. Physicians India 58, 488-90–495-6,
Shimabukuro, N., Cataruci, A. C. S., Ishikawa, K. H., Oliveira, B. E., Kawamoto, D., Ando-Suguimoto, E. S., et al. (2021). Bifidobacterium strains present distinct effects on the control of alveolar bone loss in a periodontitis experimental model. Front. Pharmacol. 12:713595. doi: 10.3389/fphar.2021.713595
Shirbhate, U., Bajaj, P., Chandak, M., Jaiswal, P., Sarangi, S., Suchak, D., et al. (2023). Clinical implications of probiotics in oral and periodontal health: a comprehensive review. Cureus 15:e51177. doi: 10.7759/cureus.51177
Shokryazdan, P., Faseleh Jahromi, M., Liang, J. B., and Ho, Y. W. (2017). Probiotics: from isolation to application. J. Am. Coll. Nutr. 36, 666–676. doi: 10.1080/07315724.2017.1337529
Smith, K. P., Ruiz, T., and Mintz, K. P. (2016). Inner-membrane protein MorC is involved in fimbriae production and biofilm formation in Aggregatibacter actinomycetemcomitans. Microbiology 162, 513–525. doi: 10.1099/mic.0.000246
Sorroche, F. G., Spesia, M. B., Zorreguieta, Á., and Giordano, W. (2012). A positive correlation between bacterial autoaggregation and biofilm formation in native Sinorhizobium meliloti isolates from Argentina. Appl. Environ. Microbiol. 78, 4092–4101. doi: 10.1128/AEM.07826-11
Spacova, I., Dodiya, H. B., Happel, A. U., Strain, C., Vandenheuvel, D., Wang, X., et al. (2020). Future of probiotics and prebiotics and the implications for early career researchers. Front. Microbiol. 11:1400. doi: 10.3389/fmicb.2020.01400
Sujeetha, M., Logaranjani, A., Rajapriya, M. J., and Rajaram, V. (2019). Host bacterial interactions in periodontal disease-an overview. International Journal of Current Research and Review 11, 01–06. doi: 10.31782/IJCRR.2019.11101
Sun, J., Hu, X. L., le, G. W., and Shi, Y. H. (2013). Inhibition of Fe-induced colon oxidative stress by lactobacilli in mice. World J. Microbiol. Biotechnol. 29, 209–216. doi: 10.1007/s11274-012-1172-5
Sun, M. C., Hu, Z. Y., Li, D. D., Chen, Y. X., Xi, J. H., and Zhao, C. H. (2022). Application of the Reuterin system as food preservative or health-promoting agent: a critical review. Food Secur. 11:4000. doi: 10.3390/foods11244000
Tahmourespour, A., Salehi, R., Kermanshahi, R. K., and Eslami, G. (2011). The anti-biofouling effect of Lactobacillus fermentum-derived biosurfactant against Streptococcus mutans. Biofouling 27, 385–392. doi: 10.1080/08927014.2011.575458
Van Holm, W., Lauwens, K., De Wever, P., Schuermans, A., Zayed, N., Pamuk, F., et al. (2023). Probiotics for oral health: do they deliver what they promise? Front. Microbiol. 14:1219692. doi: 10.3389/fmicb.2023.1219692
Vinolo, M. A., Rodrigues, H. G., Hatanaka, E., Sato F. T., Sampaio, S. C., and Curi, R., H. G., Hatanaka, E., Sato, F. T., Sampaio, S. C., and Curi, R. (2011). Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J. Nutr. Biochem. 22, 849–855. doi: 10.1016/j.jnutbio.2010.07.009
Wen, Z. T., Huang, X., Ellepola, K., Liao, S., and Li, Y. (2022). Lactobacilli and human dental caries: more than mechanical retention. Microbiology 168, 1–11. doi: 10.1099/mic.0.001196
Widyarman, A., Bachtiar, E. W., Bachtiar, B. M., and Seneviratne, C. J. (2019). Inhibitory effect of probiotic lactobacilli against Streptococcus mutans and Porphyromonas gingivalis biofilms. Sci. Dent. J. 3:50. doi: 10.4103/SDJ.SDJ_8_19
Yang, K. M., Kim, J. S., Kim, H. S., Kim, Y. Y., Oh, J. K., Jung, H. W., et al. (2021). Lactobacillus reuteri AN417 cell-free culture supernatant as a novel antibacterial agent targeting oral pathogenic bacteria. Sci. Rep. 11:1631. doi: 10.1038/s41598-020-80921-x
Zhang, J., and Duan, Z. (2022). Identification of a new probiotic strain, Lactiplantibacillus plantarum VHProbi(®) V38, and its use as an oral health agent. Front. Microbiol. 13:1000309. doi: 10.3389/fmicb.2022.1000309
Keywords: probiotics, Lactobacillus, Bifidobacterium, oral health, dental caries, periodontitis, inflammation
Citation: Mahdizade Ari M, Mirkalantari S, Darban-Sarokhalil D, Darbandi A, Razavi S and Talebi M (2024) Investigating the antimicrobial and anti-inflammatory effects of Lactobacillus and Bifidobacterium spp. on cariogenic and periodontitis pathogens. Front. Microbiol. 15:1383959. doi: 10.3389/fmicb.2024.1383959
Received: 08 February 2024; Accepted: 03 May 2024;
Published: 30 May 2024.
Edited by:
Valério Monteiro-Neto, Federal University of Maranhão, BrazilReviewed by:
Karin Hitomi Ishikawa, University of São Paulo, BrazilCopyright © 2024 Mahdizade Ari, Mirkalantari, Darban-Sarokhalil, Darbandi, Razavi and Talebi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Malihe Talebi, VGFsZWJpLm1AaXVtcy5hYy5pcg==; Shabnam Razavi, cmF6YXZpLnNoQGl1bXMuYWMuaXI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.