
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 23 May 2024
Sec. Microbial Physiology and Metabolism
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1383923
The Epichloë genus represents a significant group of above-ground endophytes extensively researched for their potential applications in agriculture and ecology. Additionally, Epichloë species synthesize bioactive alkaloids, which generally cause health problems in livestock and have detrimental effects on the performance of insect herbivores. Psathyrostachys lanuginosa serves as a valuable forage grass for livestock owing to its high nutritional value and resilience in adverse environmental conditions. Nevertheless, to date, no reports have documented Epichloë as endophytes of P. lanuginosa. In this study, four strains (PF5, PF9, QG2, and QG4) were isolated and identified through morphological, molecular, and phylogenetic analyses as endophytes of P. lanuginosa. Morphological analysis indicated colony characteristics and conidia features consistent with symbiotic Epichloë, with no significant differences observed in growth rates or conidia dimensions among the four strains. Phylogenetic analysis confirmed all strains as E. bromicola. Additionally, alkaloid biosynthetic genes were detected, revealing differences in the potential synthesis of peramine and indole diterpenoid alkaloids among strains from different geographic origins. However, all four E. bromicola strains exhibited similar potential for synthesizing ergot alkaloids, but not loline alkaloids. Overall, this study identified P. lanuginosa as a novel host for E. bromicola and provided insights into the alkaloid profiles of these strains, laying a solid foundation for the scientific and rational utilization of Epichloë resources.
The interaction between plants and microorganisms is common in nature and plays a vital role in plant ecology and agriculture (Shalev et al., 2022). The endophytic genus Epichloë consists of above-ground filamentous fungal endophytes known for their host specificity. Epichloë primarily infects above-ground plant parts, such as seeds, stems, and sheaths, while it does not grow in the roots (Christensen et al., 2008). The grass family Poaceae is the sole documented host of Epichloë to date. Host grasses infected with Epichloë exhibit no discernible disease symptoms and serve as habitats for the endophyte’s life cycle (Siegel et al., 1987). Epichloë relies on host grasses for nutrients, while in turn, it contributes to the host’s resilience against external stressors. This mutualistic relationship between Epichloë and host grasses underscores its ecological significance. Research on Epichloë species gained scholarly attention in the late 1970s, particularly following Bacon et al.’s (1977) findings on the production of alkaloids by Epichloë, which could induce toxic reactions in herbivorous livestock. Subsequent research deepened our understanding of the relationship between Epichloë and its host grasses, revealing the diverse effects of Epichloë on its hosts and ecological roles. Furthermore, Epichloë species have emerged as important agricultural microbial resources (O'Keeffe et al., 2022). Epichloë species employ various modes of transmission, including vertical and horizontal transmission. Vertical transmission occurs asexually through the seeds of the mother plant lineage, while horizontal transmission occurs sexually via ascospores. Epichloë species transmitted horizontally may exhibit antagonistic characteristics with their hosts, leading to “choke” or “cattail” disease (White, 1997). Some Epichloë species can be facultatively transmitted, utilizing both ascospores and seeds for transmission (Tintjer et al., 2008; Gundel et al., 2017). However, vertical transmission remains the predominant mode reported for Epichloë species (Chen et al., 2015).
The production of alkaloids by symbiotic Epichloë is a significant area of research within the field of endophytic fungi. Alkaloids play a crucial role in deterring herbivorous animals from consuming host grasses. Epichloë species can synthesize four types of alkaloids: peramine, ergot alkaloids, indole-diterpenes, and lolines (Bush et al., 1997). These alkaloids exhibit varying degrees of toxicity to insects and livestock, with ergot alkaloids and indole-diterpenes being toxic to both, while peramine and lolines are toxic to insects but safe for livestock (Schardl et al., 2013; Roberts and Lindow, 2014; Guerre, 2015; Philippe, 2016). Utilizing Epichloë for the improvement of plant germplasm has gained traction in recent years. Researchers have identified novel strains of Epichloë that impart robust insect resistance without causing harm to livestock. These strains have been introduced into various grasses or crops through artificial inoculation, thereby directly or indirectly assisting host grasses by producing alkaloids and influencing interspecific and intraspecific competition (Clay and Holah, 1999; Hare, 2011; Li et al., 2014). Vertical transmission is essential for maintaining genetic stability and is a prerequisite for plant breeding (Becker and Leon, 1988). However, challenges such as host specificity limit the success of artificial inoculation with Epichloë species (Becker et al., 2018). Psathyrostachys, a small genus within the family Poaceae, commonly known as the grass family, was described by Nevski (1934). It includes several species of perennial grasses native to Asia, particularly China and Mongolia. These grasses are valued for their forage qualities and ability to withstand harsh environmental conditions, making them important resources for livestock grazing and soil stabilization in arid and semi-arid regions. So far, eight recognized species of Psathyrostachys, with two subspecies, have been identified, six of which were previously classified under the genera Hordeum and Elymus (Baden et al., 1989; Baden, 1991). Among the species, P. lanuginosa has garnered attention due to its valuable biological characteristics, including early maturity, high quality, and stress resistance (Kang et al., 2011). As aforementioned, Epichloë species have been identified as endophytic fungi of various grass plants, particularly those belonging to the family Poaceae. However, to date, Epichloë species have not been detected in P. lanuginosa. In the present study, we isolated and identified Epichloë strains as endophytes of P. lanuginosa using morphological keys and phylogenetic analysis. Furthermore, we conducted alkaloid profiling of four strains of Epichloë. This research contributes to our understanding of Epichloë species from a new host, P. lanuginosa, and expands our knowledge of the host diversity of Epichloë species.
Plant samples of P. lanuginosa were collected from Yulin, Shaanxi Province, China (N37°32′04″, E108°52′06″; August 2021) and Lanzhou, Gansu Province, China (N36°07′10″, E103°42′05″; August 2021). Following collection, the plant samples were promptly transported to the laboratory for microscopic assessment of Epichloë species infection by staining the plant stalks with aniline blue (Li, 2005). Roughly 50% of the seeds obtained from the plant samples were allocated for propagation purposes, while the remaining were designated for the isolation of Epichloë species. To achieve endophyte isolation, the seeds were subjected to surface sterilization using 70% ethanol for 3 min, followed by treatment with a 5% sodium hypochlorite solution for an equivalent duration. The sterilized seeds were subsequently subjected to a triple wash with sterile water, and their surface moisture was removed by blotting with sterile filter paper. These sterilized seeds were then introduced into PDA media, which was supplemented with 100 μg mL−1 of ampicillin and 50 μg mL−1 of streptomycin sulfate. Finally, the PDA plates were wrapped with sealing film and incubated in darkness at a temperature of 22°C. Throughout this period, contaminated seeds were removed, and uncontaminated seeds were monitored until the emergence of endophytic fungi. Finally, four strains: PF5, PF9, QG2, and QG4 were obtained. Among them, strains PF5 and PF9 were isolated from the Psathyrostachys grown in Lanzhou, while strains QG2 and QG4 were isolated from the Psathyrostachys grown in Yulin.
The morphological examination of endophytes was conducted on PDA plates. Using a sterile puncher, 0.4 cm diameter mycelial plugs were taken from a 30-day-old colony and placed in the center of the PDA medium. The plates were then sealed with sealing film and cultured at 22°C in the dark for 32 days. After the incubation period, colony morphology was observed, recorded, and photographed, and a comparison was made to determine if there were any differences in colony morphology between the different strains. Similarly, the growth rate of the strains was measured using 0.4 cm diameter mycelia plug taken from the 30-day-old colonies. Each strain was tested on six replicate plates, which were then placed in the center of the PDA medium and cultured under dark conditions at both 22°C and 25°C, respectively. Weekly measurements of colony diameter were conducted for 8 weeks using the ‘crossing method’. The PDA medium was also used for observing and measuring conidia and the length of conidiogenous cells. After all the strains were cultured for 2 weeks, sterile coverslips were inserted into the PDA medium at a 45° angle, and the PDA plates were sealed with sealing film. Culture continued until the mycelia of strains grew to the surface of the coverslips. The coverslips were then removed, placed on a glass slide with a drop of toluidine blue solution, and examined using an automated upright fluorescence microscope (Olympus, BX63). Measurements of 50 conidia and 30 conidiogenous cells of each isolate were taken, including their width and length.
Purified strains were cultured on PDA medium for 2 weeks, and the mycelium was collected into 2 mL tubes by gently scraping the surface of PDA plates with a sterile glass rod. The total DNA of the endophytic strains was extracted following the manufacturer’s instructions using a fungal DNA extraction kit (Omega, Beijing, China). After extraction, the DNA was stored at -20°C until further use. Species identification of the fungal strains was conducted by direct sequencing of the housekeeping genes tefA and tubB using the extracted DNA with the highest concentration. The primer sets tef1-exon 5u-1 (GGCAGCGATAATCAGGATAG) and tef1-exon 1d-1 (GGGTAAGGACGAAAAGACTCA) were employed for tefA (Moon et al., 2002), while tub2-exon 4u-2 (GTTTCGTCCGAGTTCTCGAC) and tub2-exon 1d-1 (GAGAAAATGCGTGAGATTGT) were used for tubB (Moon et al., 2007). PCR reactions were performed in 25 μL volumes, consisting of 12.5 μL 2× SanTaq PCR Master Mix, 9.5 μL ddH2O, 1 μL DNA (40 ng μL−1), and 1 μL each of the forward and reverse primers (10 μM). The PCR protocol included an initial denaturation step at 94°C for 5 min, followed by 34 cycles of denaturation at 94°C for 30 s, annealing at 55°C (tefA) or 45°C (tubB) for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 10 min, with a hold at 4°C. Sequencing of all PCR products was conducted by Shanghai Sangon Biology Engineering Technology and Service Co., Ltd. The obtained sequences were compared against published nucleotide sequences using Blast on the NCBI website to preliminarily determine their classification within the Epichloë genus. Subsequently, the sequences were aligned with other Epichloë species using MAFFT software (v. 7.505) (Dereeper et al., 2008), and poorly aligned regions were removed with Gblocks v. 0.91b (Katoh and Standley, 2013). Substitutional saturation of the sequences was assessed using DAMBE software (Xia, 2017), maximum-likelihood phylogenetic trees (ML) with substitution model TNe + G4 were constructed using IQ-tree software (v. 2.2.0) (Minh et al., 2020), with a bootstrap value of 1,000. The Epichloë species names, strain names, hosts, and accession numbers used for the construction of phylogenetic trees are listed in Table 1.
PCR analysis was conducted to assess the presence of 35 genes associated with the biosynthesis of four major groups of alkaloids in all endophytic strains. Among the 35 genes, one gene is involved in peramine biosynthesis, 14 genes are involved in ergot alkaloid biosynthesis, 11 genes are involved in indole-diterpene biosynthesis, and 11 genes are involved in loline alkaloid biosynthesis. Additionally, the mating-type genes of the strains were also determined using PCR. Details of the 46 pairs of primers used are provided in Supplementary Table S1. PCR amplification was conducted in 25 μL reaction volumes. The protocol included an initial pre-denaturation step at 94°C for 1 min, followed by 30 cycles of denaturation at 94°C for 15 s, annealing at 56°C for 30 s, and extension at 72°C for 1 min. A final extension step at 72°C for 10 min was performed, followed by holding at 4°C. Subsequently, PCR products were analyzed by 1.5% agarose gel electrophoresis to determine the presence of the target genes in the strains.
A total of four endophytic fungal strains were recovered from surface-sterilized P. lanuginosa samples infected by Epichloë. Strains PF5 and PF9 were obtained from P. lanuginosa in Lanzhou, while strains QG2 and QG4 originated from P. lanuginosa in Yulin. Although all strains exhibited typical traits of Epichloë endophytic fungi, slight variations were observed. Overall, the colonies of the four endophytic fungi appeared white on the front with sparse outer aerial hyphae. The central region of the colonies was yellowish, gradually fading toward the edges. However, the colony edge of isolate QG2 was irregular compared to the other three strains, isolate PF5 had a slightly tougher colony texture, and isolate QG4 displayed an obvious growth circle (Figure 1). The colonies exhibited moderate growth rates on PDA at 22°C/25°C, reaching diameters of 11.29–15.97/16.96–19.89 mm (14 days), 20.65–29.40/31.26–44.98 mm (28 days), 26.45–43.31/51.75–61.78 mm (42 days), and 30.74–54.26/54.99–65.15 mm (56 days). Conidia shapes were predominantly oval and asymmetrical, with an average size of 3.5–3.9 × 1.8–1.9 μm, and an average length of the conidiogenous cell of 10.2–11.3 μm (Table 2). There were no significant differences observed in growth rate, length of conidiogenous cells, and conidia size among the four strains studied. The morphological characteristics of the other E. bromicola strains listed in Table 2 included a growth rate ranging from 0.88 to 1.29 mm day−1, length of conidiogenous cells ranging from 8 to 29 μm, length of conidia ranging from 3.7 to 5.3 μm, and width of conidia ranging from 1.8 to 3.5 μm (Table 2). When compared to previously reported E. bromicola endophytes, the morphological features (growth rate, length of conidiogenous cells, and conidia size) of the four strains examined in this study were slightly smaller but still fell within the normal range.
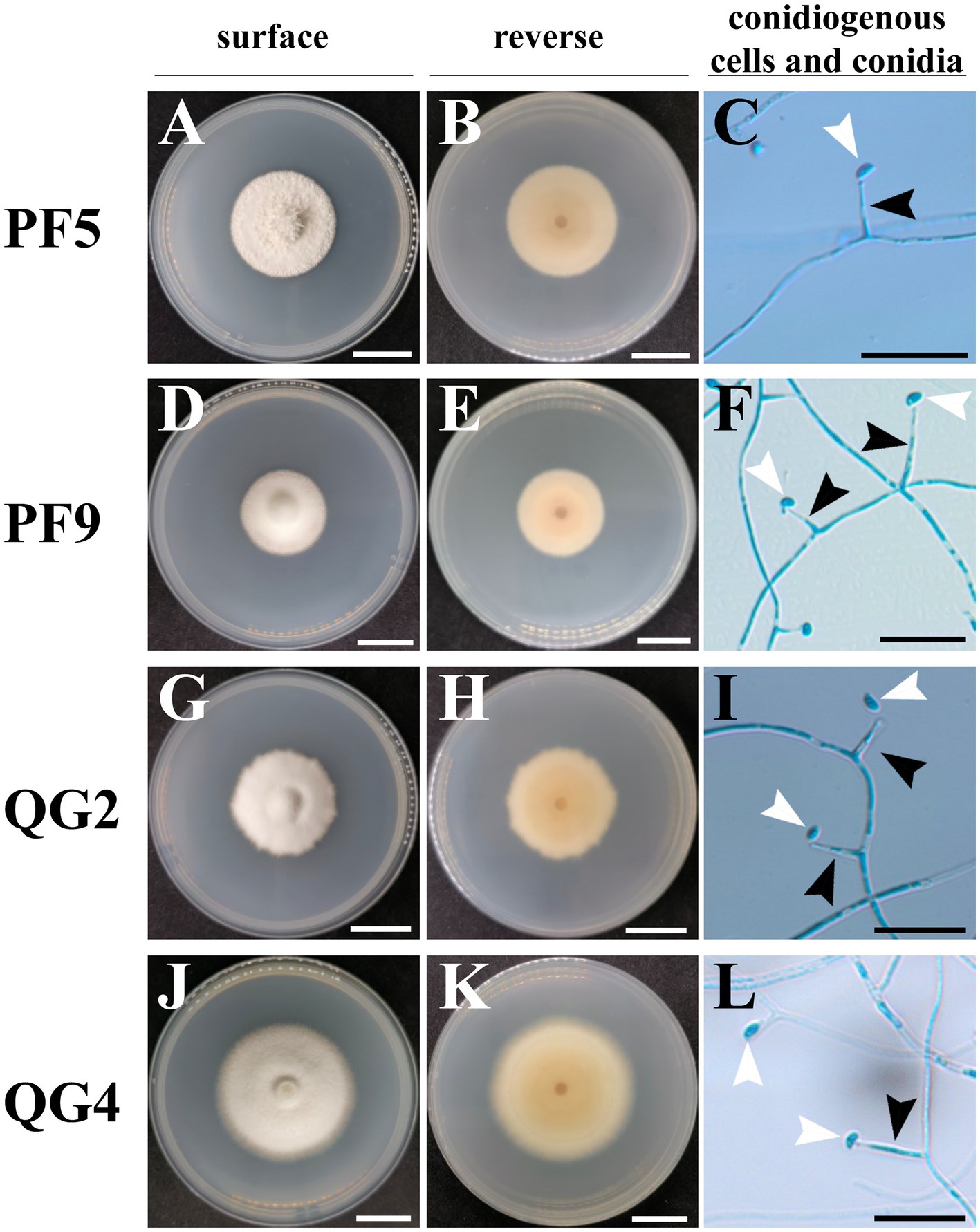
Figure 1. Colony morphology, conidiogenous cells, and conidia of Epichloë strains from Psathyrostachys lanuginosa. The colony is from cultures grown on PDA at 22°C for 32 days. (A,D,G,J) The surface view of colonies of strains PF5, PF9, QG2, and QG4; (B,E,H,K) The reverse view of colonies of strains PF5, PF9, QG2, and QG4; (C,F,I,L) The micrographs of toluidine blue-stained conidiogenous cells (black arrow) and conidia (white arrow) of strains PF5, PF9, QG2, and QG4; white scale bars = 2 cm, black scale bars = 20 μm.
The amplified PCR products yielded single peaks in the sequencing results, indicating that the four strains belong to non-hybrid species. This classification was further reinforced by the construction of maximum likelihood phylogenetic trees using tefA and tubB gene sequences. Specifically, strains PF5 and PF9 exhibited taxonomic congruence with strains QG2 and QG4. Phylogenetic analysis, employing 42 tefA gene sequences, revealed that all four strains formed a distinct clade with E. bromicola, supported by a bootstrap value of 96% (Figure 2). Similarly, in the phylogenetic analysis based on 43 tubB gene sequences, all strains grouped together with E. bromicola, with a bootstrap value of 98% (Figure 3). Thus, our investigation confirms the identification of the endophytic strains infecting P. lanuginosa as E. bromicola.
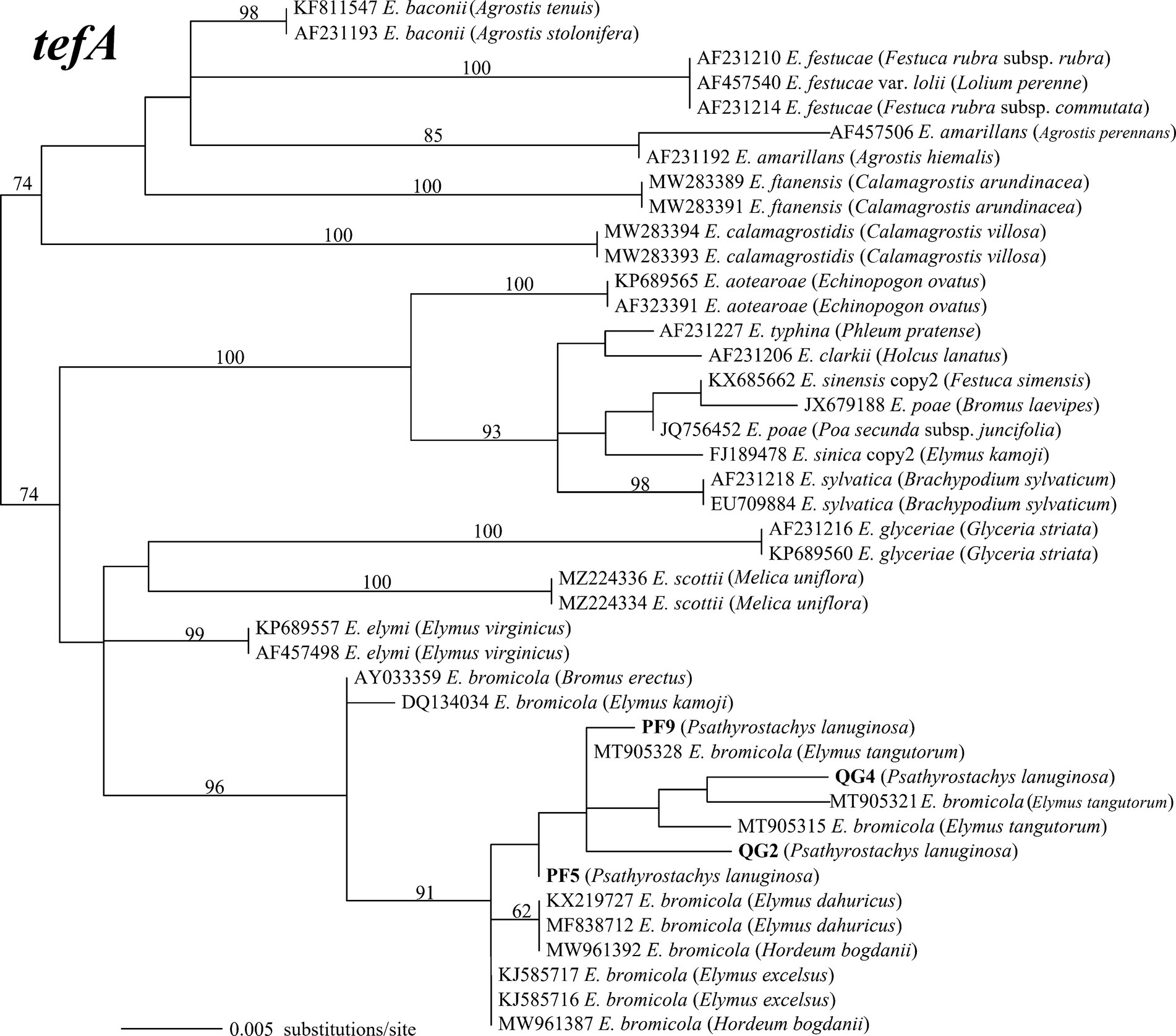
Figure 2. The molecular phylogeny derived from maximum likelihood (substitution model TNe + G4) analysis of the tefA gene from representative Epichloë species from Psathyrostachys lanuginosa. Numbers above the branches are bootstrap support percentages assessed with 1,000 replications.
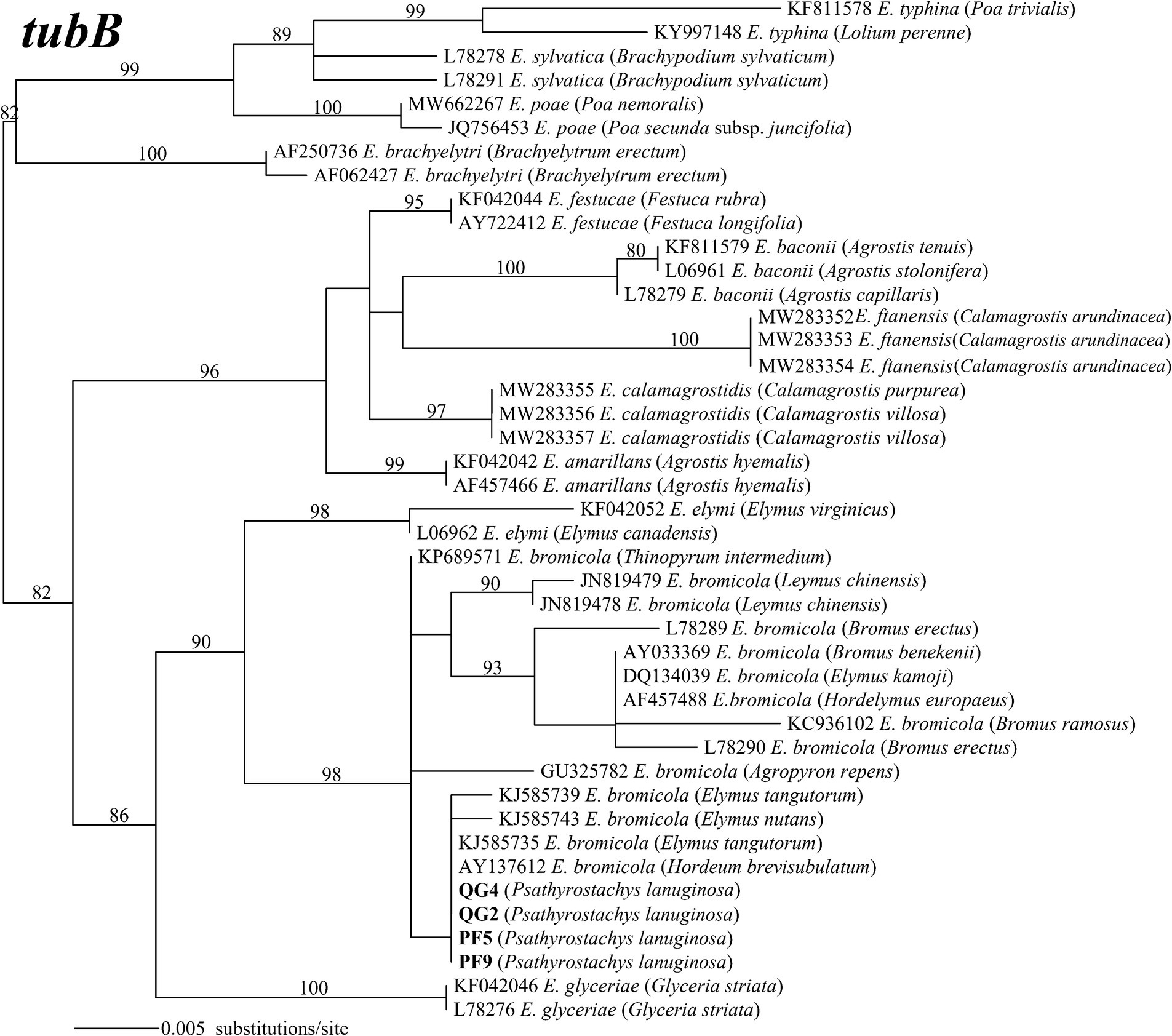
Figure 3. The molecular phylogeny derived from maximum likelihood (substitution model K2P + G4) analysis of the tubB gene from representative Epichloë species from the Psathyrostachys lanuginosa. Numbers above the branches are bootstrap support percentages assessed with 1,000 replications.
All four strains exhibited differences in alkaloid synthesis genes and mating-type genes, reflecting variations in alkaloid production among the four strains as shown in Table 3. While the synthesis genes for ergot and loline alkaloids were consistent across all strains, disparities were observed in the synthesis genes of peramine and indole-diterpene alkaloids (Table 3). Notably, all four strains exclusively contained the lolC gene within the genes responsible for loline alkaloid synthesis, suggesting a potential deficiency in synthesizing loline alkaloids. Among the 14 genes implicated in ergot alkaloid synthesis, all four strains possessed genes dmaW, easF, easE, easC, easD, easA, easG, cloA, lpsA, lpsB, and easH, while lacking genes lpsC, easO, and easP, which encode enzymes for a separate branch to ergonovine, lysergic acid alpha-hydroxylamide, and to ergine. Consequently, the four strains demonstrated potential for synthesizing chanolavine I (CC), D-lysergic acid, and ergovaline (ERV), albeit lacking the potential for synthesizing ergonovine (EN) or lysergic acid α-hydroxyethylamide (LAH). Within the eight domain structures of the peramine synthetase-encoding gene, ppzA (formly perA), all four strains harbored six domains, including ppzA–A1, ppzA–T1, ppzA–C, ppzA–A2, ppzA–M, and ppzA–T2. However, strains PF5 and PF9 possessed the ppzA–R domain (representing allele ppzA-1), while strains QG2 and QG4 harbored the ppzA–ΔR domain (representing allele ppzA-2) in reverse. ppzA-∆R means ppzA from which the R-domain was deleted, the implication of the deletion is that the final enzymatic step from the diketopiperazine to peramine is missing in the ∆R versions, such that pyrrolopyrazine-1,4-diones are produced instead of peramine. Recent studies have indicated its capacity to encode different metabolites and confer protective effects on the host (Berry et al., 2019). Regarding the 11 genes within the IDT/LTM clusters, strains PF5 and PF9 contained nine of them, including idtG, idtB, idtM, idtC, idtS, idtP, idtO, idtF, and idtK, suggesting their potential to synthesize paspaline, terpendole I, paxilline (PAX), and terpendole K (TDK), but not lolitrem B (LTM) theoretically. Conversely, strains QG2 and QG4 only possessed three genes (idtM, idtS, and idtK) related to indole-diterpene synthesis, probably rendering them incapable of synthesizing any type of indole-diterpene alkaloids due to the absence of the pivotal gene idtG in the IDT/LTM cluster.
In this study, we identified P. lanuginosa as a previously unreported host of Epichloë species, Four endophytic fungal strains of Epichloë were isolated from P. lanuginose from two distinct geographical locations, i.e., Yulin, Shaanxi Province and Lanzhou, Gansu Province, China. Morphological and phylogenetic analyses based on tefA and tubB sequences confirmed the taxonomic status of these four strains as E. bromicola. Furthermore, we elucidated the presence of alkaloid synthesis genes within these four E. bromicola strains. Epichloë bromicola exhibits a broad host range within the Poaceae family. Previous studies have identified this endophytic fungus in various grass species, including Hordeum (Iannone et al., 2015; Yi et al., 2018; Chen et al., 2019), Leymus (Zhu et al., 2013), Elymus (Li et al., 2006; Song and Nan, 2015; Shi et al., 2017), Bromus (Leuchtmann and Schardl, 1998; Groppe et al., 1999), and Agropyron (Lembicz et al., 2010). In a study by Shi et al. (2017), E. bromicola isolated from E. dahuricus revealed that all but one isolate out of 10 belonged to mating type A (MTA). Phylogenetic analysis of seven strains using tefA and tubB showed that six grouped together, while the seventh, the only mating type B (MTB) strain, grouped with those from E. kamoji, known to be sexual (Li et al., 2006). Li et al. (2006) conducted a study where they isolated eight strains of E. bromicola from E. kamoji native to China. Among these strains, two were classified as MTA, while the remaining six were categorized as MTB. Similarly, Yi et al. (2018) analyzed E. bromicola from six different seed accessions, all of which were MTA. Furthermore, Chen et al. (2019) found that three E. bromicola isolates that were symbiotic with H. brevisubulatum, and all were MTA. These studies collectively suggest that mating type diversity is extremely low in E. dahuricus, H. bogdanii, and H. brevisubulata. In the current research, we found two MTA and two MTB isolates from P. lanuginosa. This discovery suggests the presence of a sexual population in P. lanuginosa. However, stromata were not observed on this host under natural conditions. This novel endophyte-grass combination raises questions about the widespread occurrence of this association and warrants further investigation. In some species with sexual Epichloë, stromata rarely form, and even if sexual reproduction is infrequent, it may still play a significant role in endophyte diversification.
Significant variation in alkaloid biosynthetic potential among E. bromicola isolates from different or the same hosts has been observed in previous studies (Shi et al., 2017; Chen et al., 2019). This phenomenon was further supported in our study. We investigated the alkaloid biosynthesis gene profiles of four E. bromicola strains (PF5, PF9, QG2, QG4). All strains lacked genes necessary for loline alkaloid synthesis but possessed the potential to produce ergot and peramine alkaloids. Genetic polymorphisms within the ppzA gene results in differential peramine vs. pyrrolopyrazine-1,4-diones production among the strains (Berry et al., 2019). Furthermore, PF5 and PF9 harbored genes potentially involved in indole-diterpene alkaloid biosynthesis, absent in QG2 and QG4 strains. The QG2 and QG4 strains lacked idtG required for paspaline production, suggesting limitations in synthesizing any type of indole-diterpene alkaloids. Similar observations were reported for E. bromicola isolated from Elymus dahuricus, highlighting remarkable intraspecific diversity within E. bromicola regarding its alkaloid biosynthetic potential (Shi et al., 2017). This diversity appears to be influenced by both host plant species and genetic polymorphisms within the fungal population.
The genus Psathyrostachys, a perennial member of the Triticeae tribe, has primarily been investigated in the context of agricultural applications. Unlike common wheat (Triticum aestivum) with its A, B, and D genomes, or other Triticeae members with I, H, R, St, P, E, and W genomes, the entire Psathyrostachys genus possesses a distinct Ns genome (Hsiao et al., 1986). This unique genetic makeup offers a valuable resource for the improvement of common wheat due to the presence of beneficial traits and genes (Cao et al., 2008; Ma et al., 2016). For example, P. huashanica, is an endemic species found in China’s Qinling Mountains, exemplifies the potential of this genus. This species exhibits cold, drought, and barren tolerance, early maturity, high grain quality, and resistance to stripe rust, take-all, and scab (Shu et al., 1991; Du et al., 2014; Ma et al., 2016). These characteristics position it as a significant source of novel genetic diversity within Triticeae. Furthermore, distant hybridization techniques have enabled the successful transfer of superior high-molecular-weight (HMW) gliadin genes from the Psathyrostachys Ns genome into common wheat (Zhao et al., 2010). These findings underline the agricultural importance of Psathyrostachys, independent of its potential as a microbial resource. The present study unveils a novel symbiotic association between P. lanuginosa and E. bromicola, a finding with significant implications. While Epichloë symbioses typically confer benefits to host grasses, no prior reports documented such interactions within Psathyrostachys (Song et al., 2015). The combination of Psathyrostachys, known for its exceptional traits, with an symbiotic Epichloë raises the possibility of further enhanced performance, considering the well-documented benefits provided by Epichloë symbioses in other grasses (Song et al., 2015). However, the absence of previous research on Psathyrostachys-Epichloë interactions and the exclusion of growth and stress resistance evaluations in this study necessitate further investigation.
The study of Epichloë species in grasses has emerged as a significant discipline in research history. Our understanding of Epichloë species has evolved considerably over time, transitioning from early incidences of livestock poisoning to contemporary insights gained from diverse perspectives (Bacon et al., 1977). As our comprehension of Epichloë species continues to advance, it also presents an increasing array of challenges. The utilization of Epichloë species in plant breeding has been progressively adopted due to their host stress resistance traits and the detectability and facile screening of alkaloids synthesized by these endophytes. One strategy involves the inoculation of Epichloë species that do not synthesize harmful alkaloids into other grass species, thereby generating novel germplasm with desirable attributes (White et al., 2019). However, practical outcomes are often suboptimal. Firstly, the success rate of Epichloë species inoculation is limited by their host specificity. This is influenced by factors such as inoculation technique, plant genotype, and Epichloë strains, necessitating ongoing optimization and adjustment of inoculation methods (Becker et al., 2018). Secondly, some materials successfully inoculated with Epichloë species may exhibit poor performance, such as severe stunting (Simpson et al., 2014). Nonetheless, it’s noteworthy that while some materials artificially inoculated with Epichloë have demonstrated successful performance, there exists a potential barrier to transmission across generations due to incompatibility between certain Epichloë species and specific grasses. Although the underlying mechanisms are not fully understood, this underscores the importance of exploring the compatibility of Epichloë species with different grasses in future endeavors. Importantly, numerous successful cases demonstrate the utility of Epichloë species in plant germplasm innovation. For instance, researchers have utilized renowned Epichloë strains AR1, AR37, and NEA2 to cultivate numerous commercially viable grass cultivars, accounting for more than 70% of proprietary seed sales in New Zealand a decade ago (Caradus et al., 2013). Experimental manipulation of Epichloë species through fungal culture and inoculation suggests that the degree of genetic similarity between native and novel host plants positively correlates with the likelihood of establishing a mutually beneficial symbiotic relationship (Simpson and Mace, 2012). To date, no reports have been found of Epichloë species infecting cereal crops naturally. Given the significance of cereal crops, there is a growing interest in exploring Epichloë species as a means to expedite the cultivation of novel cereal crop varieties with exceptional traits (Kang et al., 2011). Several researchers have investigated the artificial inoculation of E. bromicola strains in cultivated barley, reporting notable successes such as enhancements in aboveground biomass, seed yield per plant, and growth period advancements. The E. bromicola strain utilized in these studies was isolated from wild barley and exhibited close genetic relatedness to cultivated barley (Li et al., 2021). Therefore, it is imperative to explore Epichloë species in wild-related species of cultivated plants. The Epichloë strains analyzed in this research were isolated from Psathyrostachys, a taxonomically related species to wheat. However, further experimentation is required to evaluate their alkaloid-producing capabilities in plants for potential application in artificial inoculation studies, which will be the primary focus of our forthcoming research.
In conclusion, we have identified P. lanuginosa as a previously unreported host of four endophytic fungal strains of Epichloë from two distinct geographical locations in China. Morphological and phylogenetic analyses confirmed the taxonomic status of these strains as E. bromicola, elucidating the presence of alkaloid synthesis genes within them. Epichloë bromicola exhibits a broad host range within the Poaceae family, with significant variation observed in alkaloid biosynthetic potential across different host species. Our investigation into the alkaloid biosynthesis gene profiles of four E. bromicola strains revealed variations in the presence of genes associated with alkaloid synthesis, suggesting intraspecific diversity influenced by both host plant species and genetic polymorphisms within the fungal population. Additionally, we underscore the agricultural significance of Psathyrostachys genus, highlighting its potential for genetic improvement of common wheat and its exceptional traits. The revelation of a novel symbiotic association between P. lanuginosa and E. bromicola prompts further exploration into the potential benefits of this interaction, emphasizing the need for future research to elucidate its implications for host grass performance. While challenges remain in optimizing Epichloë species inoculation and understanding the mechanisms underlying host compatibility, successful applications in plant breeding underscore the utility of these endophytes in generating novel germplasm with desirable attributes. As we continue to explore the diversity and applications of Epichloë species, further investigations into their interactions with different grass species and their potential for enhancing cereal crop varieties are warranted, with a particular focus on evaluating the alkaloid-producing capabilities of Epichloë strains isolated from wild-related species of cultivated plants. This comprehensive approach will advance our understanding of Epichloë biology and its agricultural applications, paving the way for the development of improved crop varieties with enhanced resilience and productivity.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
TC: Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. TW: Methodology, Software, Writing – review & editing. MD: Investigation, Writing – review & editing. KM: Formal analysis, Investigation, Writing – review & editing. CL: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. GB: Funding acquisition, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Science Foundation of China (U21A20239 and 32001396), the Open Project of State Key Laboratory of Plateau Ecology and Agriculture, Qinghai University (2022-KF-02), the Science and Technology Planning Project of Gansu Province, China (Natural Science Foundation) (22JR5RA458), and Gansu Provincial Science and Technology Major Projects (22ZD6NA007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1383923/full#supplementary-material
Bacon, C. W., Porter, J. K., Robbins, J. D., and Luttrell, E. S. (1977). Epichloë typhina from toxic tall fescue grasses. Appl. Environ. Microbiol. 34, 576–581. doi: 10.1128/aem.34.5.576-581.1977
Baden, C. (1991). A taxonomic revision of Psathyrostachys (Poaceae). Nord. J. Bot. 11, 3–26. doi: 10.1111/j.1756-1051.1991.tb01790.x
Baden, C., Vonbothmer, R., Flink, J., and Jacobsen, N. (1989). Intergeneric hybridization between Psathyrostachys and Hordeum. Nord. J. Bot. 9, 333–342. doi: 10.1111/j.1756-1051.1989.tb01008.x
Becker, Y., Green, K. A., Scott, B., and Becker, M. (2018). Artificial inoculation of Epichloë festucae into Lolium perenne, and visualization of endophytic and epiphyllous fungal growth. Bioprotocol. 8:2990. doi: 10.21769/BioProtoc.2990
Becker, H., and Leon, J. (1988). Stability analysis in plant breeding. Plant Breed. 101, 1–23. doi: 10.1111/j.1439-0523.1988.tb00261.x
Berry, D., Mace, W., Grage, K., Wesche, F., Gore, S., Schardl, C. L., et al. (2019). Efficient nonenzymatic cyclization and domain shuffling drive pyrrolopyrazine diversity from truncated variants of a fungal NRPS. PNAS 116, 25614–25623. doi: 10.1073/pnas.1913080116
Brem, D., and Leuchtmann, A. (2003). Molecular evidence for host-adapted races of the fungal endophyte Epichloë bromicola after presumed host shifts. Evolution 57, 37–51. doi: 10.1554/0014-3820(2003)057[0037,MEFHAR]2.0.CO;2
Bush, L. P., Wilkinson, H. H., and Schardl, C. L. (1997). Bioprotective alkaloids of grass-fungal endophyte symbioses. Plant Physiol. 114, 1–7. doi: 10.1104/pp.114.1.1
Cao, Z. J., Deng, Z. Y., Wang, M. N., Wang, X. P., Jing, J. X., Zhang, X. Q., et al. (2008). Inheritance and molecular mapping of an alien stripe-rust resistance gene from a wheat-Psathyrostachys huashanica translocation line. Plant Sci. 174, 544–549. doi: 10.1016/j.plantsci.2008.02.007
Caradus, J., Lovatt, S., and Belgrave, B. (2013). Adoption of forage technologies. In: Proceedings of the New Zealand Grassland Association. 39–44.
Chen, L., Li, X. Z., Li, C. J., Swoboda, G. A., Young, C. A., Sugawara, K., et al. (2015). Two distinct Epichloë species symbiotic with Achnatherum inebrians, drunken horse grass. Mycologia 107, 863–873. doi: 10.3852/15-019
Chen, T. X., Simpson, W. R., Song, Q. Y., Chen, S. H., Li, C. J., and Ahmad, R. Z. (2019). Identification of Epichloë endophytes associated with wild barley (Hordeum brevisubulatum) and characterisation of their alkaloid biosynthesis. N. Z. J. Agric. Res. 62, 131–149. doi: 10.1080/00288233.2018.1461658
Christensen, M. J., Bennett, R. J., Ansari, H. A., Koga, H., Johnson, R. D., Bryan, G. T., et al. (2008). Epichloë endophytes grow by intercalary hyphal extension in elongating grass leaves. Fungal Genet. Biol. 45, 84–93. doi: 10.1016/j.fgb.2007.07.013
Clay, K., and Holah, J. (1999). Fungal endophyte symbiosis and plant diversity in successional fields. Science 285, 1742–1744. doi: 10.1126/science.285.5434.1742
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Du, W. L., Wang, J., Lu, M., Sun, S. G., Chen, X. H., Zhao, J. X., et al. (2014). Characterization of a wheat-Psathyrostachys huashanica Keng 4Ns disomic addition line for enhanced tiller numbers and stripe rust resistance. Planta 239, 97–105. doi: 10.1007/s00425-013-1957-2
Groppe, K., Steinger, T., Sanders, I., Schmid, B., Wiemken, A., and Boller, T. (1999). Interaction between the endophytic fungus Epichloë bromicola and the grass Bromus erectus: effects of endophyte infection, fungal concentration and environment on grass growth and flowering. Mol. Ecol. 8, 1827–1835. doi: 10.1046/j.1365-294x.1999.00772.x
Guerre, P. (2015). Ergot alkaloids produced by endophytic fungi of the genus Epichloë. Toxins. 7, 773–790. doi: 10.3390/toxins7030773
Gundel, P. E., Rudgers, J. A., and Whitney, K. D. (2017). Vertically transmitted symbionts as mechanisms of transgenerational effects. Am. J. Bot. 104, 787–792. doi: 10.3732/ajb.1700036
Hare, J. D. (2011). Ecological role of volatiles produced by plants in response to damage by herbivorous insects. Annu. Rev. Entomol. 56, 161–180. doi: 10.1146/annurev-ento-120709-144753
Hsiao, C., Wang, R. R. C., and Dewey, D. R. (1986). Karyotype analysis and genome relationships of 22 diploid species in the tribe triticeae. Can. J. Genet. Cytol. 28, 109–120. doi: 10.1139/g86-015
Iannone, L. J., Irisarri, J. G. N., Mc Cargo, P. D., Perez, L. I., and Gundel, P. E. (2015). Occurrence of Epichloë fungal endophytes in the sheep-preferred grass Hordeum comosum from Patagonia. J. Arid Environ. 115, 19–26. doi: 10.1016/j.jaridenv.2014.12.008
Kang, H. Y., Wang, Y., Fedak, G., Cao, W. G., Zhang, H. Q., Fan, X., et al. (2011). Introgression of chromosome 3ns from Psathyrostachys huashanica into wheat specifying resistance to stripe rust. PLoS One 6:e21802. doi: 10.1371/journal.pone.0021802
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Lembicz, M., Górzynska, K., and Leuchtmann, A. (2010). Choke disease caused by Epichloë bromicola in the grass Agropyron repens in Poland. Plant Dis. 94:1372. doi: 10.1094/pdis-12-09-0810
Leuchtmann, A., and Oberhofer, M. (2013). The Epichloë endophytes associated with the woodland grass Hordelymus europaeus including four new taxa. Mycologia 105, 1315–1324. doi: 10.3852/12-400
Leuchtmann, A., and Schardl, C. L. (1998). Mating compatibility and phylogenetic relationships among two new species of Epichloë and other congeneric European species. Mycol. Res. 102, 1169–1182. doi: 10.1017/s0953756298006236
Li, C. J. (2005). Biological and ecological characteristics of Achnatherum inebrians/Neotyphodium endophyte symbiont. Lanzhou: Lanzhou University.
Li, T., Blande, J. D., Gundel, P. E., Helander, M., and Saikkonen, K. (2014). Epichloë endophytes alter inducible indirect defences in host grasses. PLoS One 9:e101331. doi: 10.1371/journal.pone.0101331
Li, W., Ji, Y. L., Yu, H. S., and Wang, Z. W. (2006). A new species of Epichloë symbiotic with Chinese grasses. Mycologia 98, 560–570. doi: 10.3852/mycologia.98.4.560
Li, C. J., Wang, Z. F., Chen, T. X., and Nan, Z. B. (2021). Creation of novel barley germplasm using an Epichloë endophyte. Chin. Sci. Bull. 66, 2608–2617. doi: 10.1360/TB-2020-1587
Ma, D. F., Fang, Z. W., Yin, J. L., Chao, K. X., Jing, J. X., Li, Q., et al. (2016). Molecular mapping of stripe rust resistance gene YrHu derived from Psathyrostachys huashanica. Mol. Breed. 36:64. doi: 10.1007/s11032-016-0487-6
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Moon, C. D., Guillaumin, J. J., Ravel, C., Li, C. J., Craven, K. D., and Schardl, C. L. (2007). New Neotyphodium endophyte species from the grass tribes Stipeae and Meliceae. Mycologia 99, 895–905. doi: 10.3852/mycologia.99.6.895
Moon, C. D., Miles, C. O., Järlfors, U., and Schardl, C. L. (2002). The evolutionary origins of three new Neotyphodium endophyte species from grasses indigenous to the southern hemisphere. Mycologia 94, 694–711. doi: 10.1080/15572536.2003.11833197
O'Keeffe, K. R., Wheeler, B. T., and Mitchell, C. E. (2022). A microbial mutualist within host individuals increases parasite transmission between host individuals: evidence from a field mesocosm experiment. Front. Microbiol. 13:824211. doi: 10.3389/fmicb.2022.824211
Philippe, G. (2016). Lolitrem B and indole diterpene alkaloids produced by endophytic fungi of the genus Epichloë and their toxic effects in livestock. Toxins. 8:47. doi: 10.3390/toxins8020047
Roberts, E., and Lindow, S. (2014). Loline alkaloid production by fungal endophytes of fescue species select for particular epiphytic bacterial microflora. ISME J. 8, 359–368. doi: 10.1038/ismej.2013.170
Schardl, C. L., Florea, S., Pan, J., Nagabhyru, P., Bec, S., and Calie, P. J. (2013). The Epichloë: alkaloid diversity and roles in symbiosis with grasses. Curr. Opin. Plant Biol. 16, 480–488. doi: 10.1016/j.pbi.2013.06.012
Shalev, O., Karasov, T. L., Lundberg, D. S., Ashkenazy, H., Ayutthaya, P. P. N., and Weigel, D. (2022). Commensal Pseudomonas strains facilitate protective response against pathogens in the host plant. Nat Ecol Evol. 6, 383–396. doi: 10.1038/s41559-022-01673-7
Shi, C., An, S. Z., Yao, Z. P., Young, C. A., Panaccione, D. G., Lee, S. T., et al. (2017). Toxin-producing Epichloë bromicola strains symbiotic with the forage grass Elymus dahuricus in China. Mycologia 109, 847–859. doi: 10.1080/00275514.2018.1426941
Shu, Y. C., An, J. Z., and Jie, F. (1991). The hybridization between Triticum aestivum and Psathyrostachys huashanica. J. Genet. Genomics 18, 508–512,
Siegel, M. R., Jarlfors, U., Latch, G. C. M., and Johnson, M. C. (1987). Ultrastructure of Aremonium-coenophialum, Aremonium-lolii, and Epichloë-typhina endophytes in host and nonhost Festuca and Lolium species of grasses. Can. J. Bot. 65, 2357–2367. doi: 10.1139/b87-320
Simpson, W. R., Faville, M. J., Moraga, R. A., Williams, W. M., McManus, M. T., and Johnson, R. D. (2014). Epichloë fungal endophytes and the formation of synthetic symbioses in Hordeeae (=Triticeae) grasses. J. Syst. Evol. 52, 794–806. doi: 10.1111/jse.12107
Simpson, W., and Mace, W. (2012). Novel associations between Epichloë endophytes and grasses: possibilities and outcomes[C]// Epichloë, endophytes of cool season grasses: implications, utilization and biology. In: Proceedings of the 7th International Symposium on Fungal Endophytes of Grasses, Lexington, Kentucky, USA, 28 June to 1 July 2010. Samuel Roberts Noble Foundation. 35–39.
Song, M. L., Chai, Q., Li, X. Z., Yao, X., Li, C. J., Christensen, M. J., et al. (2015). An asexual Epichloë endophyte modifies the nutrient stoichiometry of wild barley (Hordeum brevisubulatum) under salt stress. Plant and Soil 387, 153–165. doi: 10.1007/s11104-014-2289-0
Song, H., and Nan, Z. B. (2015). Origin, divergence, and phylogeny of asexual Epichloë endophyte in Elymus species from Western China. PLoS One 10:e0127096. doi: 10.1371/journal.pone.0127096
Tintjer, T., Leuchtmann, A., and Clay, K. (2008). Variation in horizontal and vertical transmission of the endophyte Epichloë elymi infecting the grass Elymus hystrix. New Phytol. 179, 236–246. doi: 10.1111/j.1469-8137.2008.02441.x
White, J. F. (1997). Perithecial ontogeny in the fungal genus Epichloë: An examination of the Clavicipitalean centrum. Am. J. Bot. 84, 170–178. doi: 10.2307/2446078
White, J. F., Kingsley, K. L., Zhang, Q. W., Verma, R., Obi, N., Dvinskikh, S., et al. (2019). Review: endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 75, 2558–2565. doi: 10.1002/ps.5527
Xia, X. H. (2017). DAMBE6: new tools for microbial genomics, phylogenetics, and molecular evolution. J. Hered. 108, 431–437. doi: 10.1093/jhered/esx033
Yi, M., Hendricks, W. Q., Kaste, J., Charlton, N. D., Nagabhyru, P., Panaccione, D. G., et al. (2018). Molecular identification and characterization of endophytes from uncultivated barley. Mycologia 110, 453–472. doi: 10.1080/00275514.2018.1464818
Zhao, J. X., Ji, W. Q., Wu, J., Chen, X. H., Cheng, X. N., Wang, J. W., et al. (2010). Development and identification of a wheat-Psathyrostachys huashanica addition line carrying HMW-GS, LMW-GS and gliadin genes. Genet. Resour. Crop. Evol. 57, 387–394. doi: 10.1007/s10722-009-9477-4
Keywords: endophyte, Psathyrostachys lanuginosa, Epichloë bromicola, alkaloids, vertical transmission
Citation: Chen T, Wang T, Du M, Malik K, Li C and Bao G (2024) Discovery of Epichloë as novel endophytes of Psathyrostachys lanuginosa in China and their alkaloid profiling. Front. Microbiol. 15:1383923. doi: 10.3389/fmicb.2024.1383923
Received: 08 February 2024; Accepted: 13 May 2024;
Published: 23 May 2024.
Edited by:
David Romero, National Autonomous University of Mexico, MexicoReviewed by:
Irene Castano, Instituto Potosino de Investigación Científica y Tecnológica (IPICYT), MexicoCopyright © 2024 Chen, Wang, Du, Malik, Li and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taixiang Chen, Y2hlbnR4QGx6dS5lZHUuY24=; Gensheng Bao, YmFvZ2Vuc2hlbmcyMDA4QGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.