
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 22 May 2024
Sec. Microbial Symbioses
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1383854
This article is part of the Research TopicPlant Microbiome: Interactions, Mechanisms of Action, and Applications, Volume IIIView all 22 articles
 Matsobane Tlou1*
Matsobane Tlou1* Benedict Ndou1
Benedict Ndou1 Nokufa Mabona1
Nokufa Mabona1 Adivhaho Khwathisi2
Adivhaho Khwathisi2 Collins Ateba3
Collins Ateba3 Ntakadzeni Madala2
Ntakadzeni Madala2 Mahloro Hope Serepa-Dlamini4
Mahloro Hope Serepa-Dlamini4Elephantorrhiza elephantina, a wild plant in southern Africa, is utilized in traditional medicine for various ailments, leading to its endangerment and listing on the Red List of South African Plants. To date, there have been no reports on bacterial endophytes from this plant, their classes of secondary metabolites, and potential medicinal properties. This study presents (i) taxonomic characterization of bacterial endophytes in leaf and root tissues using 16S rRNA, (ii) bacterial isolation, morphological, and phylogenetic characterization, (iii) bacterial growth, metabolite extraction, and LC–MS-based metabolite fingerprinting, and (iv) antimicrobial testing of bacterial crude extracts. Next-generation sequencing yielded 693 and 2,459 DNA read counts for the rhizomes and leaves, respectively, detecting phyla including Proteobacteria, Bacteroidota, Gemmatimonadota, Actinobacteriota, Verrucomicrobiota, Dependentiae, Firmicutes, and Armatimonodata. At the genus level, Novosphingobium, Mesorhizobium, Methylobacterium, and Ralstonia were the most dominant in both leaves and rhizomes. From root tissues, four bacterial isolates were selected, and 16S rRNA-based phylogenetic characterization identified two closely related Pseudomonas sp. (strain BNWU4 and 5), Microbacterium oxydans BNWU2, and Stenotrophomonas maltophilia BNWU1. The ethyl acetate:chloroform (1:1 v/v) organic extract from each isolate exhibited antimicrobial activity against all selected bacterial pathogens. Strain BNWU5 displayed the highest activity, with minimum inhibitory concentrations ranging from 62.5 μg/mL to 250 μg/mL against diarrhoeagenic Escherichia coli, Escherichia coli O157:H7, Salmonella enterica, antibiotic-resistant Vibrio cholerae, Staphylococcus aureus, Bacillus cereus, and Enterococcus durans. LC–MS analysis of the crude extract revealed common antimicrobial metabolites produced by all isolates, including Phenoxomethylpenicilloyl (penicilloyl V), cis-11-Eicosenamide, 3-Hydroxy-3-phenacyloxindole, and 9-Octadecenamide.
The plant microbiome comprises a group of commensal microbes that are classified into epiphytic, phyllospheric, rhizospheric, and endophytic based on their association with the plant host. The microbiome is influenced by complex interactions between the hosts and environmental factors such as salinity, soil-type, −structure, −moisture, −organic matter and root exudates (Fierer, 2017). The endophytes are communities of microorganisms, including bacteria, fungus, or actinomycetes that spend all or part of their life cycles inside the tissues of a plant while often presenting no disease symptoms (Arora and Ramawat, 2017). They are ubiquitous in nature and exhibit complex interactions with their hosts, which may involve mutualism, antagonism and rarely parasitism (Nair and Padmavathy, 2014; Gouda et al., 2016).
Endophytic microbes exhibit two distinct categories: obligate, characterized by their uncultivability and reliance solely on the host for survival and growth; and facultative survivors, which are culturable and can operate independently from the host. Despite the extensive history on microbial culture and enumeration, the uncultivability of obligate endophytes presents a bottleneck in the identification and measurement of endophyte diversity and community structure in plants (Gaiero et al., 2013). The paradigm that only 1% of microbes for a given environment is culturable has had a profound impact on our understanding of microbial ecology and is still a major motivation for using molecular tools for studying microbial communities (Martiny, 2019). However, there has been substantial progress in cultivation techniques which suggest that the paradigm may have shifted (Martiny, 2019).
Medicinal plants are wild plant species that have been used for centuries in traditional or herbal medicine for the treatment of various diseases. According to the International Union for Conservation of Nature and the World Wildlife Fund, over 15,000 flowering medicinal plants face the threat of extinction, primarily because of overharvesting and habitat destruction driven by the growing global population and increased demand for plant-based resources (Chen et al., 2016). Elephantorrhiza elephantina is a member of a small and purely African genus represented by nine plant species on the continent (Glen, 2003). It is an important plant resource in southern Africa, where the tuberous rhizome it is used as food and medicine by the Indigenous people (Coates and Coates, 2002). The underground rhizomes are one of the primary herbal medicines in southern Africa and consequently sold in various traditional medicine markets (Williams et al., 2001; Mander et al., 2006). The ethnomedicinal uses for this plant include the treatment of ailments such as diarrhea, diabetes, chest complaints, heart conditions, hypertension, syphilis, infertility in women, bladder problems, skin problems, waist pain in infants, fever, and hemorrhoids (Semenya and Potgieter, 2013; Lall and Kishore, 2014; Maroyi, 2017). Due to the unsustainable use of E. elephantina rhizomes driven by the demand, the plant is considered endangered and has been included on the Red List of South African Plants by the South African National Biodiversity Institute (Williams, 2008).
Some of the ethnomedical properties associated with E. elephantina have been corroborated by the pharmacological activities reported for this plant, i.e., antifungal (Mukanganyama et al., 2011), antibacterial (Mpofu S. et al., 2014), anthelmintic (Maphosa et al., 2010), antiplasmodial (Clarkson et al., 2004), anti-rickettsial (Naidoo et al., 2005), antioxidant, (Mpofu S. J. et al., 2014) and anti-inflammatory activities (Maphosa et al., 2009). Furthermore, E. elephantina is known to produce a wide array of secondary metabolites/bioactive metabolites including anthocyanidins, anthraquinones, esters, phenolic compounds, flavonoids, glycosides, phytosterols, saponins, tannins, and triterpenoids (Msimanga et al., 2013; Maroyi, 2017), which can be linked to the reported pharmacological activities. However, in 2015, Ludwig-Müller highlighted the need to re-evaluate the well-known plant production systems based on the findings that interesting metabolites could be produced by endophytes instead of the host. The anticancer drug, taxol is a prominent example of plant-derived metabolite synthesized by fungal endophyte, Taxomyces andreanae which was isolated from the plant, Taxus brevifolia (Strobel et al., 1993). The search for taxane producing endophytes was prompted by the high demand for the drug and consequently, the search for alternative sources to the plant material (Ludwig-Müller, 2015). The production of plant-derived metabolites has also been reported for endophytic bacterial isolates (Trapp et al., 2015; Photolo et al., 2020) and this, further underscores a report by Chatterjee et al. (2019), which highlighted that the biologically active compounds are frequently not the plant’s own metabolic products but are instead produced by the endophytes.
To date, there have been no reports on the plant microbiome of E. elephantina. Consequently, our understanding of the nature of endophytes that inhabit these plants and the interactions between the host plant and various classes of microbes remains limited. Furthermore, it is deducible from literature that, the high demand for the rhizome implies that the plant is often uprooted and destroyed during rhizome harvesting, hence the endangerment of the plant species. This study hypothesized that, akin to all plants, the internal tissues of E. elephantina harbor various forms of endophytes, including bacteria. The secondary metabolites produced by the endophytic bacteria are also likely to influence the diverse pharmacological activities associated with the plant and therefore, the bacteria can be explored as possible alternative sources for the desired biological activity. Moreover, the characteristic rapid growth on relatively inexpensive culture media coupled to the ease of genetic manipulation in vitro indicates that bacterial endophytes can be useful as easily accessible and sustainable producers of natural products (Gouda et al., 2016). Therefore, in this study we report on 16S rRNA-based screening of the bacterial metagenome of the leaf and rhizome tissue using next-generation sequencing and the relative taxonomic classification of bacterial populations from both tissues. The study also reports on the culture-based isolation of bacterial endophytes from the rhizome, phylogenetic classification, the production of bioactive metabolites and antimicrobial activity assays.
Healthy disease-free plant samples (roots and leaves) were collected (Date: 22/03/2022) from E. elephantina, in Mahikeng (25°57′ 27,5”S 25°26′30,6″E), North West province, South Africa. The identification of the plant and sample collection was performed under the guidance of Dr. Madeleen Struwig (S.D. Phalatse Herbarium, North-West University, Mafikeng Campus). The plant samples were thoroughly washed with tap water to remove soil, cut into small segments, treated with Tween 80 for 10 min (min) with vigorous shaking and then rinsed with distilled water. The plant samples were further immersed in 70% ethanol for 1 min and then treated with 1% NaOCl for 10 min. The samples were then rinsed five times with sterile distilled water and the final wash was plated on Luria Bertani (LB) agar plates (g/L; peptone 10, yeast extract 5, sodium chloride 10, and agar 15, pH 7.0) as a control.
The surface sterilized leaf and rhizome samples were homogenized in Phosphate-Buffered Saline (PBS) and 10 mL of each sample sent to Inqaba Biotechnical Industries (Pretoria, South Africa) a commercial service provider, for bacterial metagenome analysis. Briefly, bacterial genomic DNA was extracted from the plant samples using the Quick-DNA™ Fungal/Bacterial Miniprep Kit. The extracted DNA samples were used as a template in a PCR using a universal primer pair 27F (5′-AGAGTTTGATCTGGCTCAG-3′) and 1492R (5′-AAGGAGGTGWTCCARCC-3′) targeting the V1 -V9 region of the bacterial 16S rRNA gene (Huse et al., 2008). The following conditions were used for polymerase chain reaction (PCR): 1X initiation cycle at 92°C for 2 min, 30X denaturation cycles at 92°C for 30 s, 30X primer annealing cycles at 52°C for 30 s, 30X extension cycles at 72°C for 2 min, and a 1X elongation cycle at 72°C for 2 min. The resulting amplicons were barcoded with Pacbio M13 barcodes for multiplexing through limited cycle PCR.1 The resulting barcoded amplicons were quantified and pooled with equimolar and then AMPure PB bead-based purification step was performed. The PacBio SMRTbell library was prepared from the pooled amplicons following the manufacture protocol. Sequencing primer annealing and polymerase binding were done following SMRTlink Link software protocol to prepare the library for sequencing on PacBio Sequel IIe system. Raw subreads were processed through the SMRTlink (v10.2) Circular Consensus Sequences (CCS) algorithm to produce highly accurate reads (>QV40). These highly accurate reads were processed through DADA22 and qiime23 for quality control assessment and taxonomic classification, respectively.
The bacterial endophytes were isolated from the plant samples using a method described by Jasim et al. (2014). Briefly, the outer surface of the plant material was trimmed off and the sample homogenized in PBS (g/L; sodium chloride 8, potassium chloride 0.2, disodium hydrogen phosphate 1.44 and potassium dihydrogen phosphate 0.24, pH 7.4). The samples were serially diluted up to 10−3 and 0.1 mL of the dilution plated on LB agar (g/L; peptone 10, yeast extract 5, sodium chloride 10, and agar 15, pH 7.0) and incubated at 30°C for a few days. The plates were observed daily for bacterial colony growth followed by isolation and sub-culturing in LB agar until pure colonies were obtained. Glycerol stocks (50%, glycerol diluted in sterile LB broth) of each bacterial isolate were prepared and stored at −80°C for future use. Afterwards, the pure colonies were subjected to Gram staining as described by Collins et al. (2004) to establish morphological characteristics such as shape and Gram stain reaction. Gram stain slides were observed using an OLYMPUS CH20BIMF200 compound bright-field microscope (Lasec® Group, Cape town, South Africa) with 100x magnification.
Genomic DNA was extracted from an overnight starter culture using Quick-DNA™ Fungal/Bacterial Miniprep Kit following the manufacturer’s protocol. The extracted genomic DNA was then quantified using a NanoDrop™ ND-2000 UV–vis spectrophotometer (Thermo Fisher Scientific Inc., Waltham, USA). The 16S rRNA gene for each isolate was amplified according to a method described by Hongoh et al. (2003). The PCR was performed using the universal primers 27F (5′-AGAGTTTGATCTGGCTCAG-3′) and 1492R (5′-AAGGAGGTGWTCCARCC-3′) which were purchased from Inqaba Biotechnological Industries (Pretoria, South Africa). PCR was in 25 μL total volumes using the following conditions: 1X initiation cycle at 92°C for 2 min, 30X denaturation cycles at 92°C for 30 s, 30X primer annealing cycles at 52°C for 30 s, 30X extension cycles at 72°C for 2 min, and a 1X elongation cycle at 72°C for 2 min followed by termination at 4°C. The PCR products were analyzed on a 1% agarose gel by electrophoresis at a constant 100 V and 200 mA. The positive products were then excised from the gel and purified u + sing the GeneJet gel extraction kit. The purified PCR products were sequenced at Inqaba Biotechnical Industries (Pretoria, South Africa).
The CLC Bio Main Workbench was used to assemble the forward and reverse sequencing reads to form a consensus sequence for each isolate. This was followed by a Basic Local Alignment Search Tool (BLAST) search on the National Center for Biotechnology Information (NCBI) GenBank nucleotide sequence database4 to determine if a sequence in the database matches the query sequence (Altschul et al., 1997). Sequences of species closely matching the BLAST query to each sample along with their closely related taxa were obtained for the phylogenetic analysis. The sequences were aligned using the multiple sequences alignment tool, Clustal X 2.1 version (Thompson et al., 1994). The phylogenetic and molecular evolutionary analysis was conducted with MEGA X using maximum likelihood method with 1,000 bootstrap replicates (Kumar et al., 2018).
A starter culture was prepared by inoculating a single colony of the bacterial sample into 5 mL of nutrient broth (NB) and incubating it overnight at 37°C with continuous agitation at 120 rpm. Subsequently, 1 mL of the starter culture was transferred into 50 mL of LB broth (g/L; tryptone 10, yeast extract 5, and sodium chloride 10, pH 7.0) and cultivated at 37°C with continuous agitation at 120 rpm. Optical density readings were recorded in triplicate every 2 h (h) over a 48 h period using the ONDA (UV/VIS) spectrophotometer (Lasec® Group, Cape town, South Africa) at a wavelength of 600 nm.
The extraction of endophytic bacterial secondary metabolites was carried out using the method previously described in Balachandran et al. (2012) with minor modifications. Briefly, LB broth (1 L) was prepared in 2 L Erlenmeyer flasks and autoclaved at 121°C for 20 min. Each of the 2 L flasks was inoculated with the isolates and incubated at 37°C for 7 days agitating at 120 rpm. The cells were harvested, centrifuged at 10000 rpm for 15 min for biomass removal and equal volumes of ethyl acetate and chloroform (1,1 v/v) were added to the supernatant followed by vigorous agitation. The organic solvent layer was collected in a boiling flask, and it was further concentrated using a vacuum rotary evaporator (Lasec® Group, Cape town, South Africa) at 40°C. The crude extracts were transferred to 5 mL sterile vials and left to dry at room temperature ± 25°C.
Minimum inhibitory concentration (MIC) studies of the crude extracts from the isolates were conducted following the method described by Andrews (Photolo et al., 2020) with minor modifications. Briefly, stock solutions were prepared by dissolving 1 mg of the extract in 1 mL of dimethyl sulfoxide (DMSO) to a final concentration of 1 mg/mL. These stock solutions were then serially diluted to concentrations of 500, 250, 125, 62.5, 31.25, 15.625, 7.813, 3.906, and 1.953 μg/mL using tryptic soy broth (TSB) (g/L; casein peptone (pancreatic) 17, dipotassium hydrogen phosphate 2.5, glucose 2.5, sodium chloride 5, and soya peptone (papain digest) 3). The pathogenic test strains used included Gram-negative bacteria diarrhoeagenic Escherichia coli DEC-NWU (GenBank Ascension Number: CP121294), an environmental Escherichia coli O157:H7 strain (GenBank Ascension Number: JAVLRS000000000), S. enterica DSS_NWU (GenBank Ascension Number: CP123007), and antibiotic resistant Vibrio cholerae (GenBank Ascension Number: CP122254) and Gram-positive bacteria Staphylococcus aureus (ATCC 26923), Bacillus cereus. (ATCC 10876), and Enterococcus durans NWUTAL1 (GenBank Ascension Number: VMRQ00000000) (Foka et al., 2020). Using McFarland 0.5 standard, 50 μL of each pathogen was inoculated in 15 mL of TSB and incubated at 37°C for 24 h. In a 96 well microtiter plate, 100 μL of the pathogenic test strains were added horizontally and 100 μL of the different dilutions of the crude extracts were added vertically starting from the highest to the lowest concentrations. This was done following aseptic techniques. Similarly, this was done with the positive controls of ampicillin and streptomycin and for the negative control, DMSO was used. The microtiter plate was sealed with parafilm and incubated at 37°C for 16–20 h. After incubation, 10 μL of 4 mg/mL iodonitrotetrazolium chloride was added to each well. The MIC was recorded as the lowest concentration with clear wells, which indicated the absence of microbial growth. The antimicrobial experiments were performed in triplicates (n = 3).
The crude extracts were analyzed on a liquid chromatography–quadrupole time-of-flight tandem MS instrument (LCMS-9030 qTOF) (Shimadzu Corporation, Kyoto, Japan) using the method previously described in Ramabulana et al. (2021) with minor modifications. The chromatographic separation was performed on a Shim-pack Velox C18 column (100 mm × 2.1 mm with particle size of 2.7 μm) (Shimadzu Corporation, Kyoto, Japan) at 55°C. For all samples, an injection volume of 3 μL was used and run using a binary mobile phase gradient which consisted of solvent A: 0.1% formic acid in Milli-Q water and solvent B: Methanol with 0.1% formic acid. The flow rate was set to 0.3 mL/min throughout the set 20 min gradient with the following separation conditions: 5% B maintained for 3 min, 5–40% B over 3–5 min, 40–95% B over 5–12 min, from 12–16 min the conditions were maintained at 95% B, then the gradient was changed to 5% B between 16–18 min and maintained at 5% B for 2 min. The gradient was returned to initial conditions between 18–20 min which was followed by a 3-min column re-equilibration time. The chromatographic effluents were further analysed utilizing the qTOF high-definition mass spectrometer set to acquire negative electrospray ionization data. The subsequent parameters were set as: interface voltage of 4.0 kV, interface temperature of 300°C, nebulization, and dry gas flow 3 L/min, heat block temperature of 400°C, DL temperature of 280°C, detector voltage of 1.8 kV and the flight tube temperature at 42°C. Sodium iodide (NaI) was used as a calibration solution to monitor high mass accuracy. MS1 and MS2 (through data dependent acquisition) were generated simultaneously for all ions with an m/z range between 100–1,000 surpassing an intensity threshold of 5,000. Fragmentation experiments were performed using argon as a collision gas at a collision energy of 30 eV with a spread of 5 eV. Quality control (QC), pooled samples were used to condition the LC–MS system and for non-linear signal correction. The QC samples were injected at the beginning and end of the batch to ensure system equilibration. Furthermore, sample acquisition was randomized, and the QC sample analyzed every 10 injections to monitor and correct changes in the instrument response.
The SIRIUS software was used for the identification of metabolites (Dührkop et al., 2015). The SIRIUS software (version 4.9.12), was downloaded from Lehrstuhl Bioinformatik Jena website.5 Raw data obtained from the Shimadzu LCMS-9030 qTOF was converted to an open-source format (.mzML) prior to being uploaded to the SIRIUS software. To compute the molecular formulas, instrument type was set as qTOF, mass accuracy was set as 10 ppm and possible ionization was selected as [M–H]+. C, H, N, and O were selected for element searches, and the number of candidates was set to 10. Structure elucidation by CSI: FingerID was set to search in all databases and [M–H]+ as the only adduct (Dührkop et al., 2015). Canopus Class Prediction was also abled (Hoffmann et al., 2021).
The data was analyzed using analysis of variance (ANOVA). The antimicrobial assay data were described as mean ± standard deviations (SD). This analysis was done using GraphPad prism 9 (Belayneh et al., 2022) and p values less than 0.05 were statistically different.
The NGS data analysis of the bacterial metagenome in E. elephantina yielded 693.0 read counts for the rhizome, revealing a diverse composition with 7 phyla, 11 classes, 18 orders, 21 families, and 23 genera (data not shown). Similarly, the leaves exhibited a total of 2,459 read counts, reflecting a rich biodiversity comprising 8 phyla, 8 classes, 12 orders, 17 families, and 19 genera (data not shown). The NGS data obtained for leaves and rhizome were deposited to the NCBI BioSample database and, respectively, assigned BioSample accessions SAMN40452947 and SAMN40452948.
At the phylum level, the bacterial composition in both leaves and rhizome of E. elephantina encompassed Proteobacteria, Bacteroidota, Gemmatimonadota, Actinobacteriota, Verrucomicrobiota, Dependentiae, Firmicutes, and Armatimonodata, with Proteobacteria predominating in both plant parts at 90 and 78% in the rhizome and leaves, respectively. Additionally, 17.6 and 4.51% of unidentified bacteria were noted in the rhizome and leaves, respectively. Notably, Verrucomicrobiota and Dependentiae phyla were exclusive to the leaves, while Armatimonadota phylum was solely detected in the rhizome (Figure 1A). Among the genera, Novosphingobium, Mesorhizobium, Methylobacterium, and Ralstonia, all belonging to the Proteobacteria phylum, dominated the microbiota in both leaves and rhizome. At the genus level, the rhizome exhibited greater bacterial diversity compared to the leaves (Figure 1B).
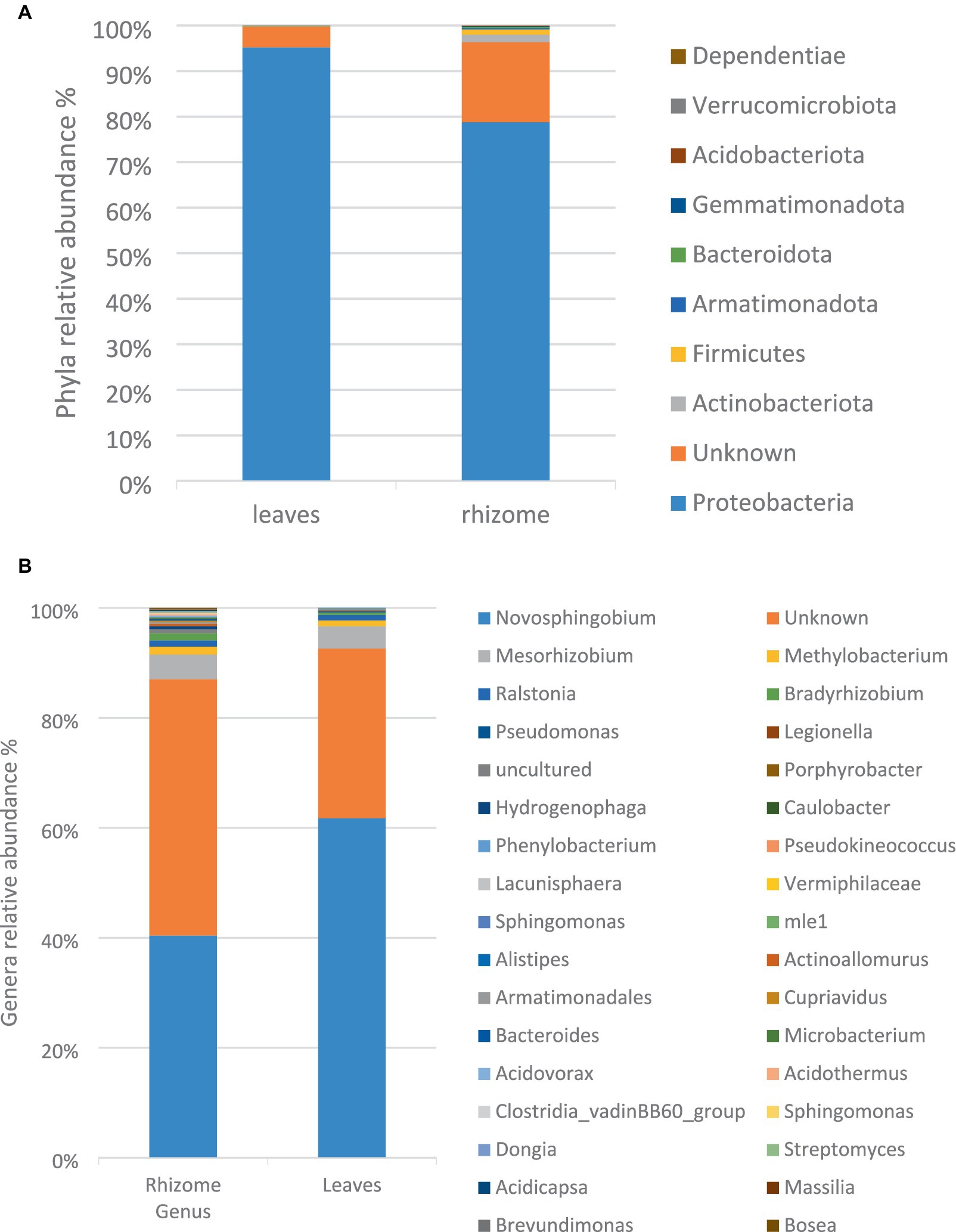
Figure 1. The relative taxonomic abundance of bacterial phyla (A) and genus (B) in leaves and rhizome samples based on analysis of full-length 16S gene amplicons.
The bacterial isolation experiment yielded 10 pure colonies designated as EER-1 to EER-10. To molecularly identify these bacterial isolates, the 16S rRNA gene was sequenced using universal primers 27F and 1492R and sequencing data revealed that EER-1, EER6, EER-9 and 10 exhibited a maximum identity of 99% or greater to specific reference strains, namely Stenotrophomonas maltophilia, Microbacterium oxydans, Pseudomonas chengduensis, and Pseudomonas alcaligenes, respectively (Table 1). Notably, EER-2 to EER-5 and EER-7 and 8 were either identified as mixed cultures or re-isolates and, consequently, were excluded from further consideration. The obtained 16S rRNA sequences were submitted to the National Center for Biotechnology Information (NCBI) and were assigned unique accession numbers, as detailed in Table 1.
The phylogenetic analysis of the 16S rRNA sequence of the isolates along with the sequences retrieved from the NCBI was carried out with MEGA X using the maximum likelihood method with 1,000 bootstrap replicates. The results showed that all the endophytic bacterial isolates grouped with various closely related bacterial species. Each genus was analyzed in separate phylogenetic trees (Figure 2) and it was observed that, Stenotrophomonas sp. BNWU1 (Figure 2A) has a monophyletic relationship with Stenotrophomonas maltophilia strain JM11, S. pavanii strain SCSB and S. geniculate strain WK16 supported by 48% bootstrap value. It also formed a separate clade indicating it could be different from the other 3 Stenotrophomonas species. Microbacterium sp. BNWU2 (Figure 2B) had a paraphyletic relationship with other Microbacterium species and formed a separate clade. On the other hand, Pseudomonas sp. BNWU4 and BNWU5 both formed separate clades, indicating they are different species, and both formed paraphyletic relationship with other Pseudomonas species (Figure 2C).
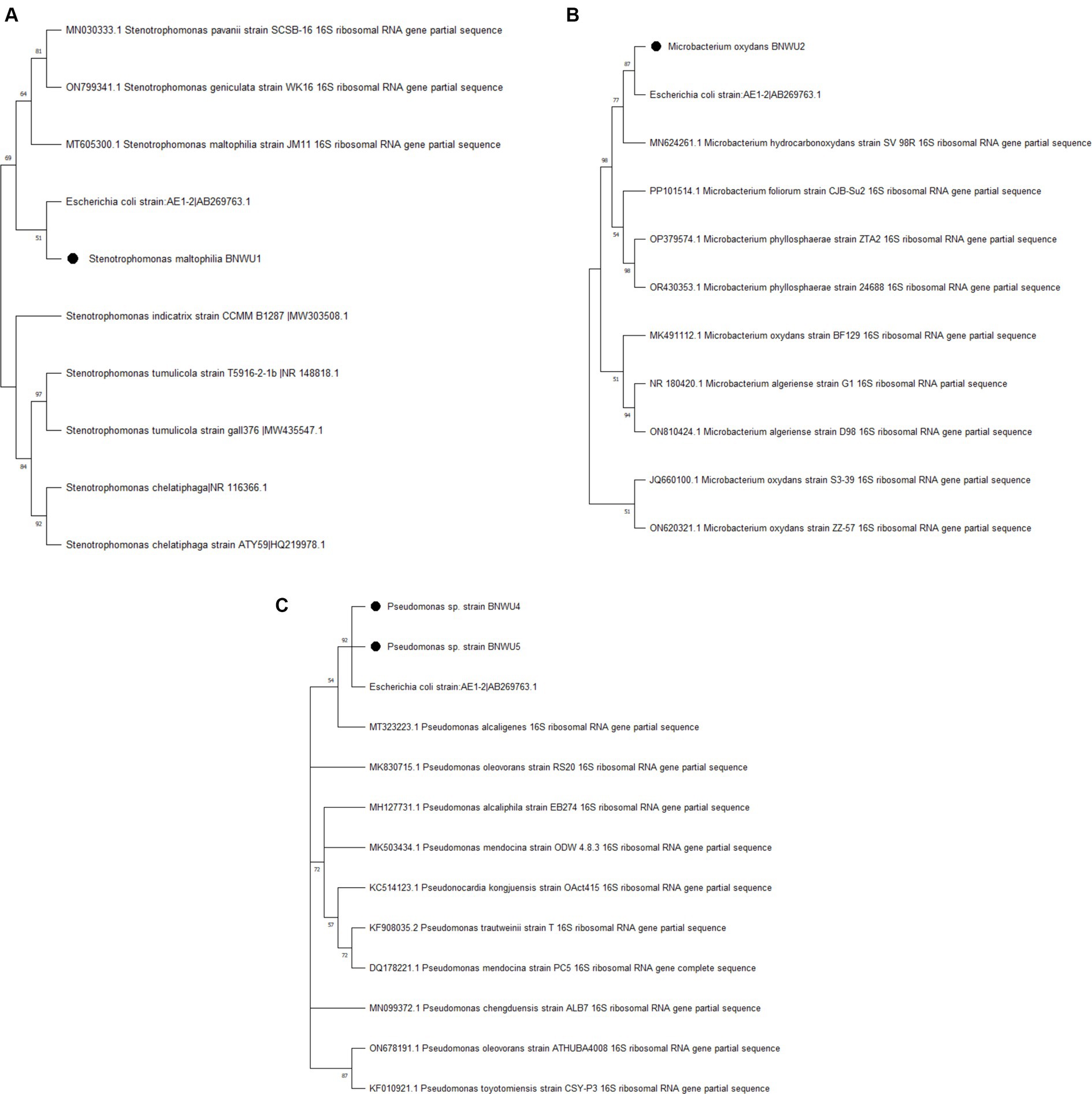
Figure 2. Maximum likelihood phylogenetic tree based on 16S rRNA gene sequence for BNWU1 (A), BNWU2 (B), and BNWU 4 and 5 (C) versus selected bacterial species from GenBank. Numbers above or below the nodes indicate bootstrap values generated after 1,000 replications.
Gram staining was employed to assess the morphological characteristics and Gram stain reactions of the bacterial isolates. The results revealed that all isolates exhibited a rod-shaped or bacilli morphology and displayed a Gram-negative staining pattern, except for BNWU2, which exhibited a Gram-positive staining reaction (data not shown).
The bacterial isolates were cultured in LB broth for a duration of 48 h to track the growth dynamics over time. Notably, uniform growth profiles were observed for the Pseudomonas isolates (BNWU4 and 5). Additionally, it was observed that the Pseudomonas isolates entered the stationary phase of growth within the initial 12 h post-inoculation, while BNWU1 and BNWU2 reached this phase after 24 h (data not shown). The stationary growth phase was selected for the extraction of secondary metabolites because various biological activities, are typically produced during this phase in a fermentation medium after the completion of microbial growth (Seyedsayamdost, 2019).
The chloroform: ethyl acetate extracts from all isolates were tested for antibacterial activity against seven pathogenic strains: Gram-negative bacteria Escherichia coli (ATCC25922), E. coli 0.157:H7 (Salmonella sp., and Vibrio cholerae); Gram-positive bacteria Staphylococcus aureus, Bacillus sp., and Enterococcus durans. The MIC for the crude extracts for all isolates ranged from 62.5 μg/mL to 250 μg/mL. with BNWU5 recording the lowest MIC values/high antimicrobial activity when compared to other isolates (Table 2). When the endophytic bacteria’s crude extracts were tested against Salmonella sp., the endophytic bacterial crude extracts showed lower MIC values (250 μg/mL) than the positive controls (ampicillin) (Table 2). The lowest MIC value was 62.5 μg/mL which was observed for Pseudomonas sp. Strain BNWU5 against pathogenic strain Bacillus sp. (Table 2).
To unravel the compounds contributing to the observed antimicrobial activity or antioxidant properties, LC-qTOFMS analysis was conducted. The identification of these compounds was based on their retention times (RT), parent masses, and molecular formulas, as outlined in Table 3 and Supplementary Figures S1A–D. The LC-qTOFMS analysis revealed a diverse array of bioactive compounds within the bacterial endophyte extracts. This included a range of fatty acids, alkaloids, flavonoids, peptides, and various phenolic compounds. The presence of these chemical classes is noteworthy, as they have established associations with significant antimicrobial and antioxidant properties, aligning with the observed effects of the bacterial endophyte extracts. The comprehensive identification of these compounds paves the way for a deeper understanding of the molecular basis of the observed bioactivity and opens avenues for further exploration of these endophytic bacterial extracts for potential therapeutic applications. Refer to Table 3 for detailed information on the identified compounds.
This study provides initial insights into the endophytic bacterial community residing in the leaves and roots of the medicinal plant E. elephantina. The examination of leaves and rhizome microbiome revealed a diverse composition of phyla, namely Proteobacteria, Bacteroidota, Gemmatimonadota, Actinobacteriota, Verrucomicrobiota, Dependentiae, Firmicutes, and Armatimonodata, with Proteobacteria being the predominant phylum in both tissues. The prevalence of this phylum in the plant samples is unsurprising and is aligned with observations in various environmental samples (Spain et al., 2009; Tkacz et al., 2018). Notably, some bacteria identified in the study were novel, showing no homologies to previously studied bacterial phyla in the databases. These findings lay the groundwork for future research focused on the isolation, characterization, and application of metabolite-producing endophytic bacteria from E. elephantina.
Given the notably higher bacterial diversity found in the rhizome compared to the leaves, the rhizome was selected for the isolation of bacteria. Subsequently, four isolates were selected and tentatively identified as, Stenotrophomonas sp. BNWU1, Microbacterium sp. BNWU2, and two Pseudomonas sp. BNWU4 and BNWU5 based on the 16S rRNA blast search results. The phylogenetic characterization revealed that both BNWU2, 4 and 5 are characterized by a paraphyletic relationship to closely related species while, BNWU1 shows a monophyletic relationship with other Microbacterium species. Therefore, in our study the ancestry for most of the isolates has not been unambiguously established and it represents a failure of resolution in the complete identification of the two isolates (Hoelzer and Meinick, 1994; Lewis et al., 2005). This could be due to thefact that the molecular marker, 16S rRNA is not infallible and there are reports to this effect (Boudewijns et al., 2006). Therefore, techniques such as the multilocus sequence analysis (MLSA) and whole genome sequencing can be very useful in the resolution of the identity of the isolates as demonstrated by Maela and Serepa-Dlamini (2023) and Diale et al. (2021).
All isolates can be categorized within the phylum Proteobacteria, except for Microbacterium BNWU2, which belongs to Actinobacteriota. Additionally, the NGS data accounted for genera Pseudomonas and Microbacterium indicating that the screening complemented the bacterial isolation method. However, Strenotrophomonas was not detected, and this may be due to the inherent limitation associated with using a single molecular marker in the resolution of closely related species (Poretsky et al., 2014) and the fact that samples were only collected once in this study. This is a first report of its kind albeit the lower diversity of bacterial isolates which may be due to the influence of culture conditions such as media and temperature (Ifeanyi et al., 2014). Moreover, it is widely acknowledged that metagenome analysis faces a limitation in discriminating between dead and viable cells, as highlighted by Tan et al. (2015). In the specific context of this study, it is crucial to recognize that the identification of bacteria through the screening technique does not always correlate with their cultivability. Nevertheless, employing various microbiological culture media, as suggested by Chauhan et al. (2020), holds the potential to enhance the diversity of bacterial isolates.
In this study, all the isolates were recorded to reach the stationary growth phase post incubation for 24-h. The decision to focus on this growth phase for secondary metabolite extraction was informed by the well-established tendency of bacteria during the stationary phase to produce secondary metabolites, including antibiotics and siderophores (Ruiz et al., 2010). Our analysis revealed a similar growth profile for the Pseudomonas species (BNWU4 and 5) when cultured in LB broth. This similarity suggests a genus-specific phenotype, reinforcing the notion of a close relationship between these isolates.
The antibacterial activity for the crude extracts obtained from all the bacterial isolates was determined against seven pathogenic strains: Escherichia coli, E. coli 0.157, Salmonella sp., Vibrio cholera, Staphylococcus aureus (ATCC26923), Bacillus sp., and Enterococcus durans. The MIC varied depending on the test strain used, the extracts showed interesting results as they were effective against all the test strains at MIC lower than 1 mg/mL. Crude extracts exhibiting a minimum inhibitory concentration of 1 mg/mL or lower are considered significantly active (Van Vuuren, 2008). This shows that endophytic bacteria from E. elephantina have great potential for the development of compounds containing bioactivities against human pathogenic microorganisms as shown in Table 3. The obtained results agree with other previous studies, that have proven endophytic bacteria are potential sources of novel bioactive compounds with antimicrobial activity (Makuwa and Serepa-Dlamini, 2021). Furthermore, the alkaloids, flavonoids, and phenolic compounds that were detected in this study have been previously reported to possess antimicrobial activities and antioxidant properties both in vitro and in vivo studies (Martins et al., 2016; Yin et al., 2016).
Bioactive compounds including cinchophen, 9-octadecenamide, tyrosylproline, beauvericin, and phenoxomethylpenicilloyl (penicilloyl V) (Table 3) which are known for antimicrobial activities were detected in this study. Beauvericin which was only detected in Pseudomonas sp. Strain BNWU5 and belongs to the enniatins antibiotic family, characterized by a cyclic hexadepsipeptide and the alternate presence of three d-hydroxy-isovaleryl and three N-methyl-phenylalanyl groups (Hamill et al., 1969). Caloni et al. (2020) reported that beauvericin has many biological activities such as antibacterial, antiviral, antifungal, antiparasitic, insecticidal and anticarcinogenic activities, and this may explain the high antibacterial activity of isolate BNWU5. Interestingly, the crude extracts from all the isolates tested positive for phenoxomethylpenicilloyl that is currently used as a prescription antibiotic for certain bacterial infections, such as streptococcal and pneumococcal upper respiratory tract infections, and the prevention of rheumatic fever and chorea (Rawson et al., 2021). Epicatechin-(4beta- > 8)-4’-O-methylgallocatechin a flavonoid that is known for wound healing (Schmidt et al., 2011; Ramalingam et al., 2022) activity was also identified in all the four isolates. Interestingly, similar/related compounds include epigallocatechin gallate, epicatechin gallate, and epicatechin have been reported from E. elephantina (Maroyi, 2017). Therefore, this supports the reports on endophytes that produce similar bioactive compounds as their host plant and therefore, can be further explored as alternative sources for the sought-after bioactivities (Deshmukh et al., 2020; Agrawal et al., 2022).
The robust antimicrobial properties observed in this study may also be attributed to the presence of various flavonoids, alkaloids, and phenolic compounds in the crude extracts, as revealed by our LC/MS findings (Table 3). Flavonoids and phenolic compounds are recognized for their ideal structural chemistry conducive to free radical scavenging activity (Kannan et al., 2016). Among the identified compounds, dibutyl phthalate, a phenolic compound, stood out. This compound has been reported to possess a spectrum of activities, including antifungal, antibacterial, antiviral, and antioxidant properties (Khatiwora et al., 2012; Shobi and Viswanathan, 2018). The antibacterial activity of dibutyl phthalate, as reported by Shobi and Viswanathan (2018), further supports its potential therapeutic applications. The previously mentioned studies on dibutyl phthalate focus on the identification and isolation of this metabolite from medicinal plant species. The recognition of this metabolite in the crude extract derived from the bacterial isolates in our study lends support to the idea that these compounds may originate from endophytic organisms rather than the metabolic processes of the host plant, as proposed by Ludwig-Müller (2015). Furthermore, numerous reports highlight instances where endophytes produce molecules initially attributed to plants. This underscores the growing understanding of endophytes as promising alternative sources for pharmaceutically active metabolites. Additionally, alkaloids and flavonoids, such as octocrylene, cyclo (histidyl-leucyl), avobenzone, indoleacrylic acid, and L-Abrine, were identified in our study. These compounds are known for their significant antimicrobial and antioxidant properties. Notably, avobenzone and octocrylene, recognized antioxidant agents, are commonly used in skin care products and cosmetics (Jesus et al., 2023). Indoleacrylic acid, identified in this study, emerges as a promising compound in drug discovery due to its multifaceted properties, including antibacterial, antioxidant, anti-inflammatory, and antidiabetic activities (Wlodarska et al., 2017; Liu et al., 2023). These findings underscore the diverse array of bioactive compounds present in the extracts from endophytic bacterial isolates, suggesting their potential as valuable sources for the development of antimicrobial drugs and antioxidant supplements.
Another exciting advancement in this study is the remarkable similarity observed in the LC–MS chromatograms (Supplementary Figures S1A–D) and chemical profiles of the secondary metabolites (Table 3) among the bacterial isolates. Several factors could account for this phenomenon: firstly, methodological nuances such as extraction and analytical techniques (Olivon et al., 2017). Secondly, the conservation of biosynthetic pathways, as demonstrated by Cimermancic et al. (2014). Thirdly, the possibility of horizontal gene transfer, wherein different bacterial strains acquire genetic material enabling the production of similar secondary metabolites, as discussed by Smillie et al. (2011). Lastly, the influence of common environmental factors cannot be overlooked. Bacteria residing in similar ecological niches or exposed to comparable environmental conditions may exhibit parallel adaptation strategies, leading to the production of comparable secondary metabolites, as suggested by Blin et al. (2013).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, OQ859587, OQ859588, OQ859589, and OQ859590.
MT: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Methodology, Funding acquisition, Formal analysis, Data curation, Conceptualization. BN: Writing – review & editing, Software, Methodology, Investigation, Data curation. NoM: Writing – review & editing, Software, Methodology, Investigation. AK: Writing – review & editing, Software, Methodology, Formal analysis. CA: Writing – review & editing, Resources, Methodology. NtM: Writing – review & editing, Software, Resources, Methodology, Investigation. MS-D: Writing – review & editing, Validation, Software, Methodology, Formal analysis.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. NRF Thuthuka (Grant no. TTK210216586709).
The authors would like to thank the S.D. Phalatse Herbarium, North-West University, (Mafikeng Campus) for assistance with the collection of the plant sample and Faculty of Natural Sciences and Agriculture (North-West University) for providing funding for research consumables and article processing charges (APC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1383854/full#supplementary-material
2. ^https://benjjneb.github.io/dada2/index.html
3. ^https://docs.qiime2.org/2021.11/
Agrawal, S., Samanta, S., and Deshmukh, S. K. (2022). The antidiabetic potential of endophytic fungi: future prospects as therapeutic agents. Biotechnol. Appl. Biochem. 69, 1159–1165. doi: 10.1002/bab.2192
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Arora, J., and Ramawat, K. G. (2017). An introduction to endophytes. Endophytes 15, 1–23. doi: 10.1007/978-3-319-66541-2_1
Balachandran, C., Duraipandiyan, V., and Ignacimuthu, S. (2012). Cytotoxic (A549) and antimicrobial effects of Methylobacterium sp. isolate (ERI-135) from Nilgiris forest soil, India. Asian Pac. J. Trop. Biomed. 2, 712–716. doi: 10.1016/S2221-1691(12)60215-9
Barupal, D. K., and Fiehn, O. (2019). Generating the blood exposome database using a comprehensive text mining and database fusion approach. Environ. Health Perspect. 127:097008. doi: 10.1289/EHP4713
Belayneh, T., Gebremichael, S., Chinchkar, A. V., Berhanu, T., Singh, A., and Upadhyay, A. (2022). Comparative study on chemical composition and antioxidant properties (GraphPad prism approach) of wild Ethiopian Z. Spina-christi and Indian Z. Jujube fruit species. Food Anal. Methods 15, 2224–2237. doi: 10.1007/s12161-022-02274-7
Blin, K., Medema, M. H., Kazempour, D., Fischbach, M. A., Breitling, R., Takano, E., et al. (2013). antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 41, W204–W212. doi: 10.1093/nar/gkt449
Boudewijns, M., Bakkers, J. M., Sturm, P. D., and Melchers, W. J. (2006). 16S rRNA gene sequencing and the routine clinical microbiology laboratory: a perfect marriage? J. Clin. Microbiol. 44, 3469–3470. doi: 10.1128/JCM.01017-06
Caloni, F., Fossati, P., Anadón, A., and Bertero, A. (2020). Beauvericin: the beauty and the beast. Environ. Toxicol. Pharmacol. 75:103349. doi: 10.1016/j.etap.2020.103349
Chatterjee, S., Ghosh, R., and MandalID, N. C. (2019). Production of bioactive compounds with bactericidal and antioxidant potential by endophytic fungus Alternaria alternata AE1 isolated from Azadirachta indica a. Juss. PLoS One 14:436. doi: 10.1371/journal.pone.0214744
Chauhan, A., and Jindal, T. (2020). Microbiological culture media: types, role and composition. In: Microbiological Methods for Environment, Food and Pharmaceutical Analysis. Springer, Cham.
Chen, S. L., Yu, H., Luo, H. M., Wu, Q., Li, C. F., and Steinmetz, A. (2016). Conservation and sustainable use of medicinal plants: problems, progress, and prospects. Chin. Med. 11, 1–10. doi: 10.1186/s13020-016-0108-7
Cimermancic, P., Medema, M. H., Claesen, J., Kurita, K., Brown, L. C. W., Mavrommatis, K., et al. (2014). Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421. doi: 10.1016/j.cell.2014.06.034
Clarkson, C., Maharaj, V. J., Crouch, N. R., Grace, O. M., Pillay, P., Matsabisa, M. G., et al. (2004). In vitro antiplasmodial activity of medicinal plants native to or naturalised in South Africa. J. Ethnopharmacol. 92, 177–191. doi: 10.1016/j.jep.2004.02.011
Collins, M. D., Falsen, E., Brownlee, K., and Lawson, P. A. (2004). Helcococcus sueciensis sp. nov., isolated from a human wound. Int. J. Syst. Evol. Microbiol. 54, 1557–1560. doi: 10.1099/ijs.0.63077-0
Deshmukh, S. K., Agrawal, S., Prakash, V., Gupta, M. K., and Reddy, M. S. (2020). Anti-infectives from mangrove endophytic fungi. S. Afr. J. Bot. 134, 237–263. doi: 10.1016/j.sajb.2020.01.006
Diale, M. O., Kayitesi, E., and Serepa-Dlamini, M. H. (2021). Genome in silico and in vitro analysis of the probiotic properties of a bacterial endophyte, Bacillus paranthracis strain MHSD3. Front. Genet. 12:672149. doi: 10.3389/fgene.2021.672149
dos Reis, C. M., da Rosa, B. V., da Rosa, G. P., do Carmo, G., Morandini, L. M. B., Ugalde, G. A., et al. (2019). Antifungal and antibacterial activity of extracts produced from Diaporthe schini. J. Biotechnol. 294, 30–37. doi: 10.1016/j.jbiotec.2019.01.022
Dührkop, K., Shen, H., Meusel, M., Rousu, J., and Böcker, S. (2015). Searching molecular structure databases with tandem mass spectra using CSI: FingerID. Proc. Natl. Acad. Sci. 112, 12580–12585. doi: 10.1073/pnas.1509788112
Ezelarab, H. A., Hassan, H. A., Abuo-Rahma, G. E. D. A., and Abbas, S. H. (2023). Design, synthesis, and biological investigation of quinoline/ciprofloxacin hybrids as antimicrobial and anti-proliferative agents. J. Iran. Chem. Soc. 20, 683–700. doi: 10.1007/s13738-022-02704-7
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Foka, F. E. T., Mienie, C., Bezuidenhout, C. C., and Ateba, C. N. (2020). Complete genomic analysis of VRE from a cattle feedlot: focus on 2 antibiotic resistances. Front. Microbiol. 11:571958. doi: 10.3389/fmicb.2020.571958
Furukawa, T., Akutagawa, T., Funatani, H., Uchida, T., Hotta, Y., Niwa, M., et al. (2012). Cyclic dipeptides exhibit potency for scavenging radicals. Bioorg. Med. Chem. 20, 2002–2009. doi: 10.1016/j.bmc.2012.01.050
Gamit, H. A., Naik, H., Chandarana, K. A., Chandwani, S., and Amaresan, N. (2023). Secondary metabolites from methylotrophic bacteria: their role in improving plant growth under a stressed environment. Environ. Sci. Pollut. Res. 30, 28563–28574. doi: 10.1007/s11356-023-25505-8
Gaiero, J. R., McCall, C. A., Thompson, K. A., Day, N. J., Best, A. S., and Dunfield, K. E. (2013). Inside the root microbiome: bacterial root endophytes and plant growth promotion. American journal of botany, 100, 1738–1750. doi: 10.3732/ajb.1200572
Gholap, A. D., Sayyad, S. F., Hatvate, N. T., Dhumal, V. V., Pardeshi, S. R., Chavda, V. P., et al. (2023). Drug delivery strategies for Avobenzone: a case study of Photostabilization. Pharmaceutics 15:1008. doi: 10.3390/pharmaceutics15031008
Glen, H. F. (2003). “Elephantorrhiza” in Plants of southern Africa: An annotated checklist. eds. G. Germishuizen and N. L. Meyer, vol. 14 (Pretoria: National Botanical Institute), 508.
Gouda, S., Das, G., Sen, S. K., Shin, H. S., and Patra, J. K. (2016). Endophytes: a treasure house of bioactive compounds of medicinal importance. Front. Microbiol. 7:1538. doi: 10.3389/fmicb.2016.01538
Hamill, R. L., Higgens, C. E., Boaz, H. E., and Gorman, M. (1969). The structure op beauvericin, a new depsipeptide antibiotic toxic to Artemia salina. Tetrahedron Lett. 10, 4255–4258. doi: 10.1016/S0040-4039(01)88668-8
Hoelzer, G. A., and Meinick, D. J. (1994). Patterns of speciation and limits to phylogenetic resolution. Trends Ecol. Evol. 9, 104–107. doi: 10.1016/0169-5347(94)90207-0
Hoffmann, M. A., Nothias, L. F., Ludwig, M., Fleischauer, M., Gentry, E. C., Witting, M., et al. (2021). Assigning confidence to structural annotations from mass spectra with COSMIC. BioRxiv, 2021–2003. doi: 10.1101/2021.03.18.435634
Hongoh, Y., Yuzawa, H., Ohkuma, M., and Kudo, T. (2003). Evaluation of primers and PCR conditions for the analysis of 16S rRNA genes from a natural environment. FEMS Microbiol. Lett. 221, 299–304. doi: 10.1016/S0378-1097(03)00218-0
Huse, S. M., Dethlefsen, L., Huber, J. A., Welch, D. M., Relman, D. A., and Sogin, M. L. (2008). Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. doi: 10.1371/journal.pgen.1000255
Ifeanyi, V. O., Nwosu, S. C., Okafor, J. O., Onnegbu, C. P., and Nwabunnia, E. (2014). Comparative studies on five culture media for bacterial isolation. Afr. J. Microbiol. Res. 8, 3330–3334. doi: 10.5897/AJMR12.1604
Jasim, B., Joseph, A. A., John, C. J., Mathew, J., and Radhakrishnan, E. K. (2014). Isolation and characterization of plant growth promoting endophytic bacteria from the rhizome of Zingiber officinale. Three biotech 4, 197–204. doi: 10.1007/s13205-013-0143-3
Jesus, A., Mota, S., Torres, A., Cruz, M. T., Sousa, E., Almeida, I. F., et al. (2023). Antioxidants in sunscreens: which and what for? Antioxidants, 12, 138. doi: 10.3390/antiox12010138
Kannan, M., Kumar, T. S., and Rao, M. V. (2016). Antidiabetic and antioxidant properties of Waltheria indica L., an ethnomedicinal plant. Int. J. Pharma Res. Health Sci. 4, 1376–1384. doi: 10.21276/ijprhs.2016.05.07
Khatiwora, E., Adsul, V. B., Kulkarni, M., Deshpande, N. R., and Kashalkar, R. V. (2012). Antibacterial activity of Dibutyl phthalate: a secondary metabolite isolated from Ipomoea carnea stem. J. Pharm. Res. 5, 150–152.
Kumar, S, Stecher, G, Li, M, Knyaz, C, and Tamura, K (2018).Mega, X: molecular evolutionary genetics analysis across computing platforms;. Molecular Biology and Evolution, 35, 1547–1549. doi: 10.1093/molbev/msy096
Lall, N., and Kishore, N. (2014). Are plants used for skin care in South Africa fully explored? J. Ethnopharmacol. 153, 61–84. doi: 10.1016/j.jep.2014.02.021
Laskar, S., Espino, O., and Bandyopadhyay, D. (2019). Isolation, solid-state structure determination, in silico and in vitro anticancer evaluation of an indole amino acid alkaloid L-Abrine. Curr. Cancer Drug Targets 19, 707–715. doi: 10.2174/1568009619666190111111937
Lewis, P. O., Holder, M. T., and Holsinger, K. E. (2005). Polytomies and Bayesian phylogenetic inference. Syst. Biol. 54, 241–253. doi: 10.1080/10635150590924208
Li, C., Ji, C., and Tang, B. (2018). Purification, characterisation and biological activity of melanin from Streptomyces sp. FEMS Microbiol. Lett. 365:fny077. doi: 10.1093/femsle/fny077
Liu, D., Zhang, S., Li, S., Zhang, Q., Cai, Y., Li, P., et al. (2023). Indoleacrylic acid produced by Parabacteroides distasonis alleviates type 2 diabetes via activation of AhR to repair intestinal barrier. BMC Biol. 21:90. doi: 10.1186/s12915-023-01578-2
Ludwig-Müller, J. (2015). Plants and endophytes: equal partners in secondary metabolite production? Biotechnol. Lett. 37, 1325–1334. doi: 10.1007/s10529-015-1814-4
Maela, M. P., and Serepa-Dlamini, M. H. (2023). Genome sequence and characterisation of Peribacillus sp. strain AS_2, a bacterial endophyte isolated from Alectra sessiliflora. Microbiol. Res. 15, 50–65. doi: 10.3390/microbiolres15010004
Makuwa, S. C., and Serepa-Dlamini, M. H. (2021). The antibacterial activity of crude extracts of secondary metabolites from bacterial endophytes associated with Dicoma anomala. Int. J. Microbiol. 2021, 1–12. doi: 10.1155/2021/8812043
Mander, M., Diederichs, N., and Steytler, N. (2006). “Marketing of medicinal plants and products” in Commercialising Medicinal Plants: A Southern African Guide. ed. N. Diederichs (Stellenbosch: African Sun Media) 77, 812–829.
Maphosa, V., Masika, P. J., Bizimenyera, E. S., and Eloff, J. N. (2010). In-vitro anthelminthic activity of crude aqueous extracts of Aloe ferox, Leonotis leonurus and Elephantorrhiza elephantina against Haemonchus contortus. Trop. Anim. Health Prod. 42, 301–307. doi: 10.1007/s11250-009-9421-9
Maphosa, V., Masika, P. J., and Moyo, B. (2009). Investigation of the anti-inflammatory and antinociceptive activities of Elephantorrhiza elephantina (Burch.) Skeels root extract in male rats. Afr. J. Biotechnol. 8.
Maroyi, A. (2017). Phytochemical and ethnopharmacological review of Elephantorrhiza goetzei (harms) harms. Asian Pac J Trop Med 10, 107–113. doi: 10.1016/j.apjtm.2017.01.014
Martins, N., Barros, L., and Ferreira, I. C. (2016). In vivo antioxidant activity of phenolic compounds: facts and gaps. Trends Food Sci. Technol. 48, 1–12. doi: 10.1016/j.tifs.2015.11.008
Martiny, A. C. (2019). High proportions of bacteria are culturable across major biomes. ISME J. 13, 2125–2128. doi: 10.1038/s41396-019-0410-3
Mpofu, S. J., Msagati, T. A., and Krause, R. W. (2014). Cytotoxicity, phytochemical analysis and antioxidant activity of crude extracts from rhizomes of Elephantorrhiza elephantina and Pentanisia prunelloides. AJTCAM 11, 34–52. doi: 10.4314/ajtcam.v11i1.6
Mpofu, S., Ndinteh, D. T., Van Vuuren, S. F., Olivier, D. K., and Krause, R. W. M. (2014). Interactive efficacies of Elephantorrhiza elephantina and Pentanisia prunelloides extracts and isolated compounds against gastrointestinal bacteria. S. Afr. J. Bot. 94, 224–230. doi: 10.1016/j.sajb.2014.07.002
Msimanga, H. Z., Fenstermacher, J., Levitz, A., Najimudeen, I., Phillips, C., and Wysocki, E. M. (2013). Identification of compounds in hexane extracts of Elephantorrhiza elephantina and their comparison with selected over the counter products. J. Med. Plants Res. 7, 198–208. doi: 10.5897/JMPR12.1212
Mukanganyama, S., Ntumy, A. N., Maher, F., Muzila, M., and Andrae-Marobela, K. (2011). Screening for anti-infective properties of selected medicinal plants from Botswana. African J. Plant Sci. Biotechnol. 5.
Naidoo, V., Zweygarth, E., Eloff, J. N., and Swan, G. E. (2005). Identification of anti-babesial activity for four ethnoveterinary plants in vitro. Vet. Parasitol. 130, 9–13. doi: 10.1016/j.vetpar.2005.03.001
Nair, D. N., and Padmavathy, S. (2014). Impact of endophytic microorganisms on plants, environment and humans. Sci. World J. 2014, 1–11. doi: 10.1155/2014/250693
Olivon, F., Allard, P. M., Koval, A., Righi, D., Genta-Jouve, G., Neyts, J., et al. (2017). Bioactive natural products prioritization using massive multi-informational molecular networks. ACS Chem. Biol. 12, 2644–2651. doi: 10.1021/acschembio.7b00413
Paudel, B., Maharjan, R., Rajbhandari, P., Aryal, N., Aziz, S., Bhattarai, K., et al. (2021). Maculosin, a non-toxic antioxidant compound isolated from Streptomyces sp. KTM18. Pharm. Biol. 59, 931–934. doi: 10.1080/13880209.2021.1946091
Photolo, M. M., Mavumengwana, V., Sitole, L., and Tlou, M. G. (2020). Antimicrobial and antioxidant properties of a bacterial endophyte, Methylobacterium radiotolerans MAMP 4754, isolated from Combretum erythrophyllum seeds. Int. J. Microbiol. 2020, 1–11. doi: 10.1155/2020/9483670
Poretsky, R., Rodriguez-R, L. M., Luo, C., Tsementzi, D., and Konstantinidis, K. T. (2014). Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One 9:e93827. doi: 10.1371/journal.pone.0093827
Qi, D., Zou, L., Zhou, D., Zhang, M., Wei, Y., Li, K., et al. (2022). Biocontrol potential and antifungal mechanism of a novel Streptomyces sichuanensis against fusarium oxysporum f. sp. cubense tropical race 4 in vitro and in vivo. Appl. Microbiol. Biotechnol. 106, 1633–1649. doi: 10.1007/s00253-022-11788-3
Ramabulana, A. T., Petras, D., Madala, N. E., and Tugizimana, F. (2021). Metabolomics and molecular networking to characterize the chemical space of four Momordica plant species. Meta 11:763. doi: 10.3390/metabo11110763
Ramalingam, S., Chandrasekar, M. J., and Nanjan, M. J. (2022). Plant-based natural products for wound healing: a critical review. Curr. Drug Res. Rev. Formerly 14, 37–60. doi: 10.2174/2589977513666211005095613
Rawson, T. M., Wilson, R. C., Moore, L. S., Macgowan, A. P., Lovering, A. M., Bayliss, M., et al. (2021). “Exploring the pharmacokinetics of phenoxymethylpenicillin (penicillin-v) in adults: a healthy volunteer study” in Open forum infectious diseases, vol. 8 (New York: Oxford University Press), ofab573.
Ruiz, B., Chávez, A., Forero, A., García-Huante, Y., Romero, A., Sánchez, M., et al. (2010). Production of microbial secondary metabolites: regulation by the carbon source. Crit. Rev. Microbiol. 36, 146–167. doi: 10.3109/10408410903489576
Saboon, B., Bibi, Y., Ayoubi, S. A., Afzal, T., Gilani, S., Malik, K., et al. (2023). Antioxidant-activity-guided purification and separation of Octocrylene from Saussurea heteromalla. Separations 10:107. doi: 10.3390/separations10020107
Schmidt, C. A., Murillo, R., Heinzmann, B., Laufer, S., Wray, V., and Merfort, I. (2011). Structural and conformational analysis of proanthocyanidins from Parapiptadenia rigida and their wound-healing properties. J. Nat. Prod. 74, 1427–1436. doi: 10.1021/np200158g
Semenya, S. S., and Potgieter, M. J. (2013). Ethnobotanical survey of medicinal plants used by Bapedi traditional healers to treat erectile dysfunction in the Limpopo Province, South Africa. J. Med. Plants Res. 7, 349–357. doi: 10.5897/JMPR12.499
Seyedsayamdost, M. R. (2019). Toward a global picture of bacterial secondary metabolism. J. Ind. Microbiol. Biotechnol. 46, 301–311. doi: 10.1007/s10295-019-02136-y
Shobi, T., and Viswanathan, M. (2018). Antibacterial activity of di-butyl phthalate isolated from Begonia malabarica. J. Appl. Biotechnol. Bioeng. 5, 97–100. doi: 10.15406/jabb.2018.05.00123
Smillie, C. S., Smith, M. B., Friedman, J., Cordero, O. X., David, L. A., and Alm, E. J. (2011). Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244. doi: 10.1038/nature10571
Spain, A. M., Krumholz, L. R., and Elshahed, M. S. (2009). Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 3, 992–1000. doi: 10.1038/ismej.2009.43
Strobel, G., Stierle, A., Stierle, D., and Hess, W. M. (1993). Taxomyces andreanae, a proposed new taxon for a bulbilliferous hyphomycete associated with Pacific yew (Taxus brevifolia). Mycotaxon 47, 71–80.
Tan, B., Ng, C., Nshimyimana, J. P., Loh, L. L., Gin, K. Y. H., and Thompson, J. R. (2015). Next-generation sequencing (NGS) for assessment of microbial water quality: current progress, challenges, and future opportunities. Front. Microbiol. 6:1027. doi: 10.3389/fmicb.2015.01027
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tkacz, A., Hortala, M., and Poole, P. S. (2018). Absolute quantitation of microbiota abundance in environmental samples. Microbiome 6, 1–13. doi: 10.1186/s40168-018-0491-7
Trapp, M. A., Kai, M., Mithöfer, A., and Rodrigues-Filho, E. (2015). Antibiotic oxylipins from Alternanthera brasiliana and its endophytic bacteria. Phytochemistry 110, 72–82. doi: 10.1016/j.phytochem.2014.11.005
Van Vuuren, S. F. (2008). Antimicrobial activity of South African medicinal plants. Journal of ethnopharmacology, 119, 462–472. doi: 10.1016/j.jep.2008.05.038
Wattana-Amorn, P., Charoenwongsa, W., Williams, C., Crump, M. P., and Apichaisataienchote, B. (2016). Antibacterial activity of cyclo (L-pro-L-Tyr) and cyclo (D-pro-L-Tyr) from Streptomyces sp. strain 22-4 against phytopathogenic bacteria. Nat. Prod. Res. 30, 1980–1983. doi: 10.1080/14786419.2015.1095747
WHOStuart, M. C., Kouimtzi, M., and Hill, S. (2009). WHO model formulary 2008. Geneva: World Health Organization.
Williams, V. L. (2008). Elephantorrhiza elephantina (Burch.) Skeels. National Assessment: Red list of South African plants version 2020.1.
Williams, V. L., Balkwill, K., and Witkowski, E. T. F. (2001). A lexicon of plants traded in the Witwatersrand umuthi shops, South Africa. Bothalia 31, 71–98. doi: 10.4102/abc.v31i1.508
Wlodarska, M., Luo, C., Kolde, R., d’Hennezel, E., Annand, J. W., Heim, C. E., et al. (2017). Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe 22, 25–37.e6. doi: 10.1016/j.chom.2017.06.007
Wu, Q., Patocka, J., Nepovimova, E., and Kuca, K. (2018). A review on the synthesis and bioactivity aspects of beauvericin, a fusarium mycotoxin. Front. Pharmacol. 9:1338. doi: 10.3389/fphar.2018.01338
Xu, Y., Zhan, J., Wijeratne, E. K., Burns, A. M., Gunatilaka, A. L., and Molnár, I. (2007). Cytotoxic and antihaptotactic beauvericin analogues from precursor-directed biosynthesis with the insect pathogen Beauveria bassiana ATCC 7159. J. Nat. Prod. 70, 1467–1471. doi: 10.1021/np070262f
Yang, Y., and McClements, D. J. (2013). Vitamin E bioaccessibility: influence of carrier oil type on digestion and release of emulsified α-tocopherol acetate. Food Chem. 141, 473–481. doi: 10.1016/j.foodchem.2013.03.033
Yin, T. P., Cai, L., Xing, Y., Yu, J., Li, X. J., Mei, R. F., et al. (2016). Alkaloids with antioxidant activities from Aconitum handelianum. J. Asian Nat. Prod. Res. 18, 603–610. doi: 10.1080/10286020.2015.1114473
Keywords: Elephantorrhiza elephantina , next-generation sequencing, taxonomy, bacterial endophytes, phylogeny, secondary metabolites, antimicrobial activity
Citation: Tlou M, Ndou B, Mabona N, Khwathisi A, Ateba C, Madala N and Serepa-Dlamini MH (2024) Next generation sequencing-aided screening, isolation, molecular identification, and antimicrobial potential for bacterial endophytes from the medicinal plant, Elephantorrhiza elephantina. Front. Microbiol. 15:1383854. doi: 10.3389/fmicb.2024.1383854
Received: 08 February 2024; Accepted: 06 May 2024;
Published: 22 May 2024.
Edited by:
George Newcombe, University of Idaho, United StatesReviewed by:
Shivankar Agrawal, Indian Council of Medical Research (ICMR), IndiaCopyright © 2024 Tlou, Ndou, Mabona, Khwathisi, Ateba, Madala and Serepa-Dlamini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matsobane Tlou, TWF0c29iYW5lLlRsb3VAbnd1LmFjLnph; bWF0dGd0bG91QGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.