- Department of Civil and Environmental Engineering, School of Engineering and Applied Science, The George Washington University, Washington, DC, United States
Free-living amoebae (FLA) are prevalent in nature and man-made environments, and they can survive in harsh conditions by forming cysts. Studies have discovered that some FLA species are able to show pathogenicity to human health, leading to severe infections of central nervous systems, eyes, etc. with an extremely low rate of recovery. Therefore, it is imperative to establish a surveillance framework for FLA in environmental habitats. While many studies investigated the risks of independent FLA, interactions between FLA and surrounding microorganisms determined microbial communities in ecosystems and further largely influenced public health. Here we systematically discussed the interactions between FLA and different types of microorganisms and corresponding influences on behaviors and health risks of FLA in the environment. Specifically, bacteria, viruses, and eukaryotes can interact with FLA and cause either enhanced or inhibited effects on FLA infectivity, along with microorganism community changes. Therefore, considering the co-existence of FLA and other microorganisms in the environment is of great importance for reducing environmental health risks.
1 Introduction
Free-living amoebae (FLA) are a type of eukaryotic organisms that can be widely present in different environments. Unlike prokaryotic unicellular organisms such as bacteria and archaea, FLA have more complicated cell structures. Compared with bacteria or viruses, this type of microorganisms has nucleus packing their DNA and abundant organelles. These features allow them to differ from bacteria or viruses in interaction behavior and mechanism with other microorganisms in the environment. Compared to parasitic amoebae, FLA can survive and reproduce independently without hosts. They inhabit a diverse range of habitats, such as water, soil, air, and even extreme environments (Rivera et al., 1987; Visvesvara and Stehr-Green, 1990). The two main life stages of FLA are the trophozoite (active, feeding stage) and the cyst (dormant, resistant stage). Since FLA reproduce under favorable conditions and differentiate into dormant forms under unfavorable conditions, FLA can persist in different harsh conditions without perishing (De Jonckheere, 1991; Khan et al., 2015). However, this may not always happen when they are treated in some extreme cases (e.g., boiling). In addition, FLA are also prevalent in man-made environments such as swimming pools, sewage, cooling water, eyewash solutions, contact lenses, dialysis units, and dental treatment units (Mergeryan, 1991; Water, 2010). To date, FLA include four genera known to contain pathogenic species: Acanthamoeba, Balamuthia, Naegleria, and Sappinia (Schuster and Visvesvara, 2004). The T4 genotype of Acanthamoeba is implicated in various severe human pathogenic diseases, including granulomatous amebic encephalitis (GAE), a fatal infection of the central nervous system (Otero-Ruiz et al., 2022). Acanthamoeba keratitis (AK), primarily affecting long-term contact lens wearers, has been reported in over 3,000 patients worldwide so far (Stehr-Green et al., 1989; Juarez et al., 2018). Moreover, Balamuthia mandrillaris and Sappinia pedata can also cause amebic encephalitis, referred to as Balamuthia amebic encephalitis (BAE) and Sappinia amebic encephalitis (SAE), respectively. Among the ~100 reported cases of BAE, there have been only three known survivors (Qvarnstrom et al., 2013; Luca, 2022). Likewise, Naegleria fowleri can cause primary amebic meningoencephalitis (PAM), a disease with a notably high fatality rate, typically contracted through exposure to contaminated water sources (Capewell et al., 2015).
Given the considerable pathogenic threats posed by FLA to human health, it is important to comprehensively understand how FLA behave in real environments and to mitigate the associated public health risks. Achieving this goal requires a deeper understanding of FLA within intricate ecosystems. While extensive research has explored amoeba growth, reproduction, mutation, and pathogenicity with human infection, the relationship between FLA and microbial communities has received limited attention.
Studies have highlighted that interactions between FLA and other microorganisms can also facilitate the transmission of diseases caused by other types of microorganisms. FLA play an important role in the disposal of organisms by acting as a “Trojan horse” that serves as a vector for the colonization of new habitats or hosts (Vaerewijck et al., 2014). FLA typically use bacteria and other microorganisms as food sources, but some prey species can escape predation and survive within the host or even proliferate within its cytoplasm or nucleus, which is called amoeba-resistant microorganisms (ARM) (Rayamajhee et al., 2021). Some eukaryotes even reversely cleave FLA as a host to obtain their nutrients (Steenbergen et al., 2003). Hence, a comprehensive review of the interplay between FLA and environmental microorganisms is imperative for future studies in microbial risk management.
Therefore, here we aim to (1) systematically review the various common types of microbial groups that can interact with FLA; (2) understand the different patterns and mechanisms of interactions between these microorganisms and FLA, so as to understand the neighboring impact to FLA in complex ecological environments. This review will provide crucial information for public health forecasting.
2 Interactions between FLA and bacteria
2.1 The impact of FLA on bacterial growth and community change
FLA influence the survival, growth, and community of bacteria in water through predation. Studies have shown that FLA can discern and selectively target certain bacteria as food sources while avoiding pathogenic ones (Bornier et al., 2021). This preferential hunting behavior significantly shapes the co-existing bacterial community. Acanthamoeba castellanii displays a marked preference for consuming bacteria from the Betaproteobacteria class and Firmicutes phylum in soil systems, leading to a notable decrease in these dominant bacterial taxa and a concurrent increase in other grazing-tolerant bacteria (Rosenberg et al., 2009). Moreover, this influence on bacterial communities can also pose a threat to public health. In engineered water systems, it has been shown that FLA have a preference for consuming bacteria from the environment, with an order of Escherichia coli strains and then Legionella pneumophila. However, L. pneumophila are not indeed “consumed,” since they can use FLA as a Trojan horse and replicate within them (Shaheen and Ashbolt, 2021). This will also lead to a change in population ratios for microorganisms in water. Prey bacteria for FLA will decrease first with an increase of FLA, which will later facilitate the growth and replication of other pathogenic bacteria like L. pneumophila (Shaheen and Ashbolt, 2021). Therefore, FLA have a significant impact on controlling bacterial populations and altering community composition.
Some bacteria show resistance to FLA predation and their growth can be even supported by the existence of FLA. These bacteria are known as amoeba-resistant bacteria (ARB). Typically, ARB can survive and be protected within FLA, and the detection of ARB correlates with FLA abundance in environments (Delafont et al., 2016; Malinowski et al., 2022). For example, Legionella spp. have been reported to strongly associate with FLA in potable water distribution systems from 10 hospitals in southwest France, with this relationship being partly temperature-dependent (Lasheras et al., 2006). A previous study also revealed a statistically significant co-occurrence of Legionella spp. and FLA, with all Legionella-positive samples tested positive for FLA (Nisar et al., 2022). Gene expression may also be altered for L. pneumophila during the interaction with FLA (Buse et al., 2015). The study showed that interactions between three strains of L. pneumophila and FLA hosts were promoted after gray water exposure, with an inhibition of amoebal encystment and an alteration of bacterial gene expression. In addition, biofilms can greatly impact the interactions between Legionella and FLA. The densities of Legionella spp. positively correlated with Vermamoeba vermiformis densities (r = 0.83, p < 0.028) in biofilms within garden hoses (Thomas et al., 2014). It was reported that FLA, including Willaertia magna and Acanthamoeba polyphaga, showed long-term interactions with L. pneumophila in natural biofilms (Shaheen et al., 2019). A nutrient recycling within the system of FLA and culturable Legionella can lead to a balance in concentrations of FLA and Legionella at room temperature, especially after a 85-week's co-culturing. And this can allow the detection of Legionella even after a quite long period.
Mycobacterium is another significant group of ARB. The growth of Mycobacterium avium was markedly enhanced in the presence of A. castellanii. Notably, these bacteria also exhibited increased virulence when infecting beige mice after growth within amoebae (Cirillo et al., 1997). A strong association between FLA and nontuberculous mycobacteria has also been observed in drinking water distribution systems (Delafont et al., 2014). Specifically, Acanthamoeba, Vermamoeba, Echinamoeba, and Protacanthamoeba have been shown to carry and support the replication of mycobacteria, including Mycobacterium llatzerense and Mycobacterium chelonae. The mechanism for M. avium surviving intracellularly in amoeba has also been proposed. Upon Acanthamoeba infection, M. avium rapidly enters individual vacuoles that subsequently merge into a single large vacuole. By inhibiting lysosomal fusion akin to macrophages, M. avium can survive and replicate within amoeba cells. M. avium replicates at temperatures as low as 24°C and that its growth and interaction with FLA can enhance virulence (Rubenina et al., 2017). Some Pseudomonas can also be supported by FLA. It was shown that viable but non-culturable (VBNC) Pseudomonas aeruginosa could be even resuscitated to an active form within 2 h when associated with A. polyphaga (Dey et al., 2019).
Finally, FLA can also facilitate the resistance of bacterial pathogens toward disinfectants in drinking water. It was revealed that the resistance of M. avium to water disinfectant monochloramine was largely enhanced when bacteria were co-cultured with Acanthamoeba (Berry et al., 2010). Moreover, the effectiveness of chlorine and heat treatments to Legionella spp. strains were observed to be reduced when Legionella was associated with Acanthamoeba (Cervero-Aragó et al., 2015). However, the supporting effect is not the only way FLA influence co-existing bacterial species. It was reported that an inhibition effect was surprisingly observed for Willaertia magna c2c that the growth of one strain of L. pneumophila was inhibited, while other similar strains did not show this effect (Dey et al., 2009). Therefore, since many pathogenic bacteria can be supported when associated with FLA, FLA can thus be considered as a shelter for bacteria, and can further increase the health risks caused by associated bacterial pathogens.
Overall, FLA can influence the occurrence and abundance of bacteria by acting as a predator and a harbor (Figure 1). It is particularly important to note that the presence of FLA in engineered water systems can substantially increase the occurrence of opportunistic bacterial pathogens, including Legionella and Mycobacterium, posing risks to public health. Thus, establishing more robust monitoring and detection methods for FLA in water distribution systems is crucial.
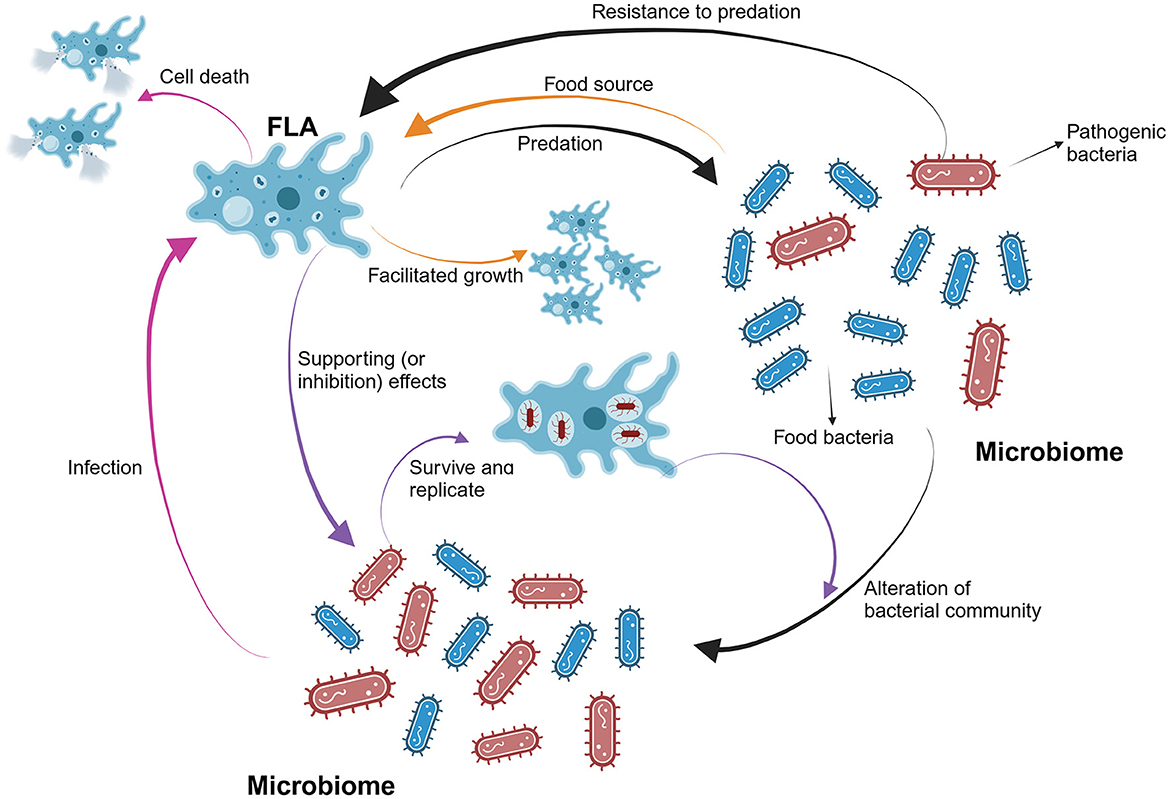
Figure 1. The overall interactions between FLA and bacteria in water systems. FLA can selectively consume food bacteria and thereby alter the populations of different bacteria species in the bacterial community. The abundance of FLA will increase first when sufficient food bacteria are available. Also, pathogenic bacteria can resist the predation and infect FLA. Serving as a harbor for pathogenic bacteria, FLA will facilitate the growth and replications of them. At this point, the abundance of FLA will decrease due to the induced death caused by the pathogenic bacteria.
2.2 The impact of bacteria on FLA survival
The growth of FLA could be supported by co-culturing with bacteria. It has been shown that the external bacteria enhance Acanthamoeba growth and binding to contact lenses, as well as the survival of amoebae in biocidal lens care solutions (Bottone et al., 1992). However, this is not always the case. It is reported that at low densities, gram-negative bacteria in PBS supported A. castellanii growth. However, at amoebae to bacteria ratios exceeding 1:10,000, gram-negative bacteria, including E. coli, became inhibitory to the amoebae (Wang and Ahearn, 1997).
Certain bacterial species, especially the pathogenic ones, can alter the life cycles of FLA. L. pneumophila was reported to induce the death of FLA after replication within them (Dey et al., 2009). Different L. pneumophila strains have been shown to inhibit the growth of amoebae of A. castellanii, Hartmannella vermiformis, and W. magna, although susceptibility varies among different amoebae. The bacteria of Burkholderia cepacian was observed to slightly reduce the viability and cyst formation of A. polyphaga in co-cultures (Anacarso et al., 2010). The bacteria of Pseudomonas strains demonstrated a clear type of endocytosis inside amoebae, with Pseudomonas fluorescens causing lysis of the host, whereas P. aeruginosa did not (Anacarso et al., 2010). FLA were found to show dependence on their endosymbionts. It was reported that symbiotic bacteria could provide essential cellular functions to amoebae, compensating for genetic defects in housekeeping genes caused by the bacteria themselves (Choi et al., 1997). In summary, some pathogenic bacteria cause significant damage to FLA cells during their intracellular replication, leading to a decrease in the viability and abundance of FLA communities.
3 Interactions between FLA and viruses
3.1 FLA as hosts for giant viruses
The discovery of giant viruses has highlighted a new dimension of complexity in the study of amoebae (Balczun and Scheid, 2017). The structure of giant viruses is far more complex than ordinary viruses. Most giant viruses interact with host FLA through infecting their trophozoites. Initiating the amoeboid phagocytosis process is necessary for these viruses to start their replication cycle and facilitate the formation of their progeny (Raoult and Boyer, 2010). This enhanced comprehension of giant viruses' dynamics with amoebae paves the way for an in-depth examination of particular instances, exemplified by mimiviruses, which redefine conventional notions of viral morphology and activity while underscoring their complex symbiosis with amebic hosts. Here, we illustrated the interactions between typical giant viruses and their FLA hosts in Figure 2.
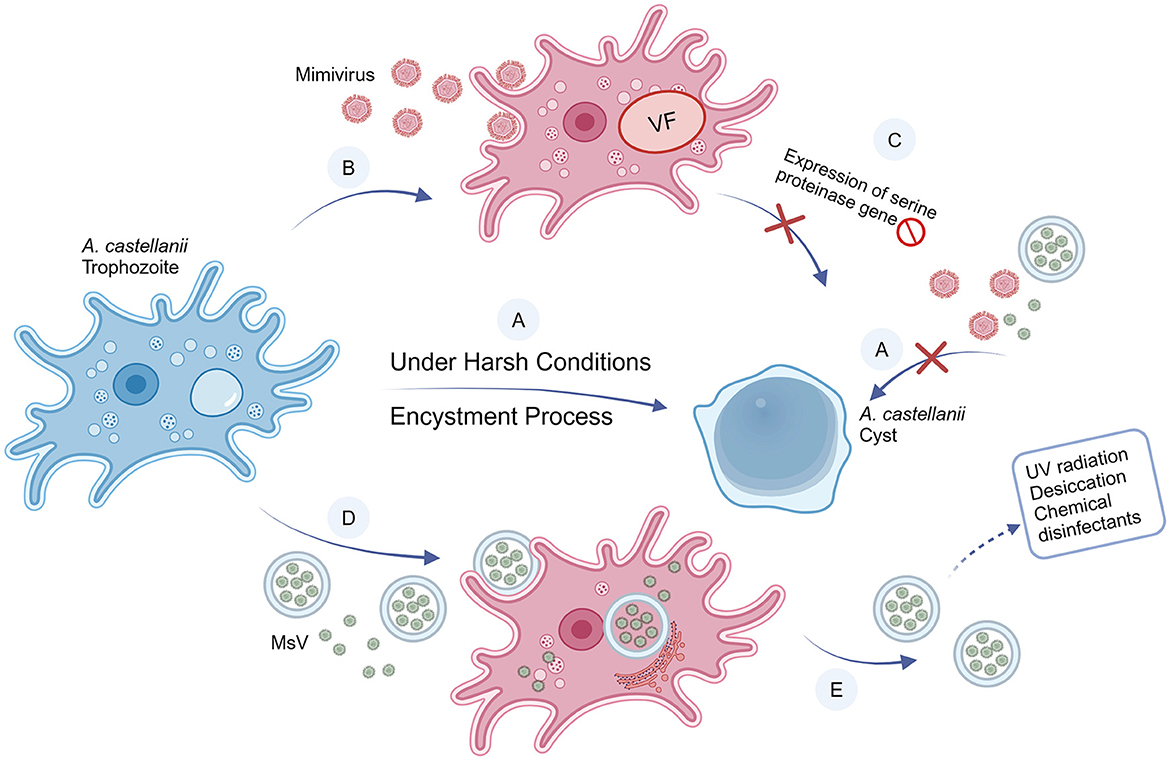
Figure 2. Interactions between A. castellanii and mimivirus and MsV. (A) Under harsh conditions (i.e., extreme temperatures, hunger, unfavorable pH, etc), A. castellanii develops a coping strategy by differentiating from trophozoites (vegetative form) into cysts (resistant form), a process called encystment. Once triggered encystment, the giant viruses are unable to infect cysts. (B) Mimiviruses can infect and replicate in A. castellanii trophozoites. (C) When A. castellanii is infected, the mimivirus blocks the expression of the serine proteinase gene, preventing the encystment process, thereby the cyst cannot form. (D) Compared with a single MsV particle, the size of vesicles containing MsV particles is large enough to trigger phagocytosis of A. castellanii. Particles can be released into the trophozoite as individual particles or vesicles, thereby initiating infection. (E) Compared with free particles, vesicles can increase infectivity and improve resistance to the external environment.
Mimiviruses, distinguishable by light microscope due to their coccobacillus-like structure, constitute a unique group of substantial viruses. The Acanthamoeba polyphaga mimivirus (APMV), colloquially referred to as mimivirus, was first isolated from the cytoplasm of amoebae (Suzan-Monti et al., 2007), marking its first recognition as a viral entity. Subsequent research identified mimiviruses as having a circular, double-stranded DNA genome of more than 1 million base pairs (Raoult et al., 2004). Beyond their large genome size, mimiviruses are unique for their departure from the typical viral reliance on host cell machinery for protein translation, showcasing greater complexity and autonomy. Simply, mimiviruses can replicate in amoebae rather than being digested. Unlike most other viruses, mimiviruses actively transcribe their genome while proliferating in its FLA hosts, suggesting an active utilization of their genetic information, instead of passive replication (Suzan-Monti et al., 2007; Colson et al., 2017).
Another giant virus, Marseillevirus (MsV), was also isolated from Acanthamoeba. MsV shares a very similar icosahedral structure with mimivirus (Boyer et al., 2009), but is much smaller (<500 nm). A free MsV would not enter FLA through phagocytosis, because the phagocytosis is triggered when FLA recognize particles >500 nm (Weisman and Korn, 1967). Instead, MsV can enter FLA through enormous vesicles derived from the endoplasmic reticulum (ER). It was found that these vesicles consisted of various numbers of membranes. These vesicles may contain at least 10 virus particles. When the vesicles are single-layered, they can merge with the FLA cell membrane, and release the viruses directly into the cytoplasm. In multi-layered vesicles, a similar fusion process occurs, but the inner membrane remains intact, containing a cluster of viral particles. This novel entry mechanism of MsV via giant vesicles represents an innovative mode of large DNA virus invasion into host cells (Arantes et al., 2016). Giant vesicles could confer resistance to MsV against environmental factors, boost the replicative success of MsV within its natural hosts, and facilitate MsV transmission in the environment. This phagocytosis process provides an avenue for small-sized viruses, which will otherwise go unnoticed by FLA, to invade host cells.
While mimiviruses and MsV grow and thrive in specific FLA, there are more complex giant viruses infecting a wide range of FLA. Tupanvirus, known as one of the most complex giant viruses, was found to infect Acanthamoeba castellanii, Acanthamoeba polyphaga, Acanthamoeba griffini, and Vermamoeba vermiformis (Abrahão et al., 2018). Tupanvirus can manipulate the host cells in a peculiar way which is known as the Tupanvirus-induced cytopathic effect (CPE) to continually provide a site for their progeny to grow and proliferate. Tupanvirus encodes a mannose-specific lectin (MBP) that activates cellular and viral mannose receptor genes, enhancing FLA cell attachment (Barre et al., 1997). Tupanvirus-infected FLA cells mix with uninfected cells to form large bundles of host cells, boosting the likelihood that viral progeny finds additional host cells (Oliveira et al., 2019).
3.2 Emerging interactions with viruses
In addition to giant viruses, several other viruses were also found to interact with FLA. Reovirus, for example, can be internalized into and enriched in the nucleus of trophozoites within FLA (Silverstein et al., 1976). Accumulation of Reovirus particles in the nucleus after phagocytosis does not impede the normal growth of trophozoites. Instead, Reovirus can persist as aggregates. This peculiar symbiotic relationship allows Reovirus to successfully find a harbor suitable for their own survival, thus easily avoiding the threat of external disinfection (Folkins et al., 2020). Similarly, human adenovirus type 5 (HAdV5), Norovirus, and others can also be internalized by FLA to avoid physical and chemical disinfection processes (Hsueh and Gibson, 2015; Verani et al., 2016).
While FLA were shown to harbor non-enveloped viruses, the interactions between enveloped viruses and FLA were rarely studied. To explore the possible interactions between FLA and enveloped severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), surrogate experiments using the enveloped bacteriophage Phi6 were conducted to investigate the vectorial capacity of V. vermiformis. Phi6's induction of apoptosis and impact on amoeboid mitochondria, facilitating trophoblast persistence, might suggest a mechanism for SARS-CoV-2's interaction with FLA (Dey et al., 2022).
Overall, understanding how giant viruses interact with FLA hosts, has revealed the viral complexity and diversity. The intricate mechanisms of infection and replication challenge the conventional paradigms of viral-host dynamics. The observation of cytopathic effects signifies an advanced level of host-virus interaction. Furthermore, understanding the dynamics of these viral entities can shed light on the mechanisms of pathogen persistence and transmission, offering insights into the risks they may pose to human health, especially in the context of waterborne and zoonotic diseases.
4 Interactions between FLA, fungi, and protozoans
FLA can also interact with numerous eukaryotes. One such group, referred to as amoebophagous fungi, can function either as predators or parasites of their prey, the FLA (Corsaro et al., 2018; Scheid, 2018). In particular, dimorphic fungi, which existed in the form of mold or yeast, were found to kill FLA and use them as a source of nourishment through cell lysis. For example, Cryptococcus neoformans, a human pathogenic fungus, exhibits sustained growth when co-cultivated with amoebae over an extended period of time. In detail, Dictyostelium discoideum, one amoeba host, engulfed C. neoformans cells, and the presence of budding fungal cells inside D. discoideum suggested that C. neoformans cells replicated intracellularly (Steenbergen et al., 2003). Nevertheless, those fungi can not stay extensively within FLA. Instead, they can break down the FLA cells and grow extracellularly (Steenbergen et al., 2003). This process involves killing and attacking the host cells from inside and then extracting the required nutrients from the remains (Steenbergen et al., 2004). Additionally, it is important to point out that FLA do not only rely on phagocytosis to engage with dimorphic fungi. Blastomyces dermatitidis primarily utilizes proteins on the membrane surface to adhere to the FLA cell membrane surface, resulting in cytotoxic effects on the FLA and ultimately causing its demise (Steenbergen et al., 2004). In addition, it was found that the fungi increased their virulence against the great wax borer which was used as an infection model for the pathogenic dimorphic fungi, through the transmission of A. castellanii (Thomaz et al., 2013). Therefore, the complex and predatory interactions between FLA and dimorphic fungi significantly affect ecosystems and human health, potentially enhancing fungal virulence and influencing infection spread.
In addition to fungi, FLA can interact with other protozoan parasites. Specifically, certain FLA, such as Acanthamoeba and Naegleria, may serve as potential hosts for oocysts of Toxoplasma gondii. The T. gondii oocysts taken up by A. castellanii were found to maintain their capacity to cause an infection in mice (Winiecka-Krusnell et al., 2009). The same study also suggested that T. gondii oocysts might block lysosomal binding thus allowing themselves to escape digestion.
In conclusion, the predatory behavior of dimorphic fungi toward FLA and the ability of FLA to potentially harbor and interact with protozoan parasites highlight possible pathways for disease transmission and the enhancement of pathogen virulence. The multifaceted interactions between FLA and a range of eukaryotic organisms underscore a critical microbial ecology with direct implications for human health and environmental stability.
5 Discussion
FLA play a significant role in microbial ecosystems within natural and engineered water environments, where their interactions with other microorganisms profoundly influence community dynamics and health outcomes. This review provides an overview of documented interactions between FLA and various microorganisms across taxonomic divisions. For bacteria and some fungi, FLA exhibit predatory behavior toward them, which serve as a food source, while parasitism represents another mode of interaction with certain bacteria and fungi. Current research has demonstrated FLA serve dual roles for pathogenic bacteria by providing a protective environment and facilitating their competitive adaptation (Rubenina et al., 2017), yet significant knowledge gaps remain. Despite studies have reported how the infection of FLA by bacteria like L. pneumophila or M. avium occurs, the mechanism of surviving and evolving within FLA for many other bacteria still largely remains unclear. In particular, it remains uncertain whether FLA can provide a unique niche to select certain mutations or induce genetic drift and transfer in bacterial populations. The future study on the evolution and transmission of bacterial pathogens within FLA is needed. While exogenous bacterial interactions with FLA have been extensively studied, exploring endogenous bacterial environments within FLA presents an intriguing avenue for investigation. These environments create specific micro-ecosystems where various ARB evolve within FLA cells. The evolution may be associated with horizontal gene transfers within FLA, and it is also worth investigating whether ARB can acquire antibiotic resistance genes (ARG) from other bacteria. Various methods can be utilized for exploring the bacteria within FLA. In particular, metagenomics may help in determining the ARG they contain, as well as the evolution of wide-ranged pathogenic bacteria, due to its broad applicability for unknown microbes. Additionally, FLA's mobility may facilitate the transport of pathogenic bacteria in the environment. Despite the relevance to public health risks, its influence on bacterial fate and transport remains understudied. Therefore, future research efforts could prioritize the study of ARB evolution and transmission within FLA.
It is crucial to conduct further studies on how human viruses coexist with FLA in the environment, but there are gaps in our current understanding of the interactions between viruses and FLA. Focusing on key factors such as environmental conditions to streamline variables, and ensuring consistent experimental setups to minimize heterogeneity, such research will deepen our knowledge of these interactions and help develop targeted strategies for mitigating virus-related risks. Meanwhile, the mechanism of gaining resistance for virus harboring in FLA is largely neglected. For instance, giant viruses have the ability to reproduce within FLA and simultaneously modify the host's internal environment to support their propagation. This necessitates further investigation into molecular mechanisms, such as whether giant viruses utilize a ribosome-independent translation system. Such a system would allow them to synthesize proteins more efficiently without competing for the host's ribosomes, potentially explaining their enhanced replication within FLA. Regarding the fungal aspect, there is currently very little research on the targeted application of amoebophagous fungi as biocontrol tools in environments where abundant pathogenic FLA exists (Scheid, 2018). However, this type of research is crucial, especially for the application of such fungi with no impact on human health.
Prior research has established a solid base of data and valuable insights into the interactions between FLA and other microorganisms. Future efforts must address the research gaps resulting from the limited diversity and mechanism study of microorganisms examined.
Author contributions
SF: Writing – review & editing, Writing – original draft. YS: Writing – review & editing, Writing – original draft. LQ: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrahão, J., Silva, L., Silva, L. S., Khalil, J. Y. B., Rodrigues, R., Arantes, T., et al. (2018). Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 9:749. doi: 10.1038/s41467-018-03168-1
Anacarso, I., Guerrieri, E., Bondi, M., de Niederhäusern, S., Iseppi, R., Sabia, C., et al. (2010). Influence of Legionella pneumophila and other water bacteria on the survival and growth of Acanthamoeba polyphaga. Arch. Microbiol. 192, 877–882. doi: 10.1007/s00203-010-0618-0
Arantes, T. S., Rodrigues, R. A. L., dos Santos Silva, L. K., Oliveira, G. P., De Souza, H. L., Khalil, J. Y., et al. (2016). The large Marseillevirus explores different entry pathways by forming giant infectious vesicles. J. Virol. 90, 5246–5255. doi: 10.1128/JVI.00177-16
Balczun, C., and Scheid, P. L. (2017). Free-living amoebae as hosts for and vectors of intracellular microorganisms with public health significance. Viruses 9:65. doi: 10.3390/v9040065
Barre, A., Van Damme, E. J., Peumans, W. J., and Rougé, P. (1997). Curculin, a sweet-tasting and taste-modifying protein, is a non-functional mannose-binding lectin. Plant Mol. Biol. 33, 691–698. doi: 10.1023/A:1005704616565
Berry, D., Horn, M., Xi, C., and Raskin, L. (2010). Mycobacterium avium infections of Acanthamoeba strains: host strain variability, grazing-acquired infections, and altered dynamics of inactivation with monochloramine. Appl. Environ. Microbiol. 76, 6685–6688. doi: 10.1128/AEM.00644-10
Bornier, F., Zas, E., Potheret, D., Laaberki, M.-H., Coupat-Goutaland, B., Charpentier, X., et al. (2021). Environmental free-living amoebae can predate on diverse antibiotic-resistant human pathogens. Appl. Environ. Microbiol. 87, e00747–00721. doi: 10.1128/AEM.00747-21
Bottone, E. J., Madayag, R. M., and Qureshi, M. N. (1992). Acanthamoeba keratitis: synergy between amebic and bacterial cocontaminants in contact lens care systems as a prelude to infection. J. Clin. Microbiol. 30, 2447–2450. doi: 10.1128/jcm.30.9.2447-2450.1992
Boyer, M., Yutin, N., Pagnier, I., Barrassi, L., Fournous, G., Espinosa, L., et al. (2009). Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Nat. Acad. Sci. 106, 21848–21853. doi: 10.1073/pnas.0911354106
Buse, H. Y., Lu, J., and Ashbolt, N. J. (2015). Exposure to synthetic gray water inhibits amoeba encystation and alters expression of Legionella pneumophila virulence genes. Appl. Environ. Microbiol. 81, 630–639. doi: 10.1128/AEM.03394-14
Capewell, L. G., Harris, A. M., Yoder, J. S., Cope, J. R., Eddy, B. A., Roy, S. L., et al. (2015). Diagnosis, clinical course, and treatment of primary amoebic meningoencephalitis in the United States, 1937–2013. J. Pediatric Infect. Dis. Soc. 4, e68–e75. doi: 10.1093/jpids/piu103
Cervero-Aragó, S., Rodríguez-Martínez, S., Puertas-Bennasar, A., and Araujo, R. M. (2015). Effect of common drinking water disinfectants, chlorine and heat, on free Legionella and amoebae-associated Legionella. PLoS ONE 10:e0134726. doi: 10.1371/journal.pone.0134726
Choi, J. Y., Lee, T. W., Jeon, K. W., and Ahn, T. I. (1997). Evidence for symbiont-induced alteration of a host's gene expression: irreversible loss of SAM synthetase from Amoeba proteus. J. Eukaryot. Microbiol. 44, 412–419. doi: 10.1111/j.1550-7408.1997.tb05717.x
Cirillo, J. D., Falkow, S., Tompkins, L. S., and Bermudez, L. E. (1997). Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65, 3759–3767. doi: 10.1128/iai.65.9.3759-3767.1997
Colson, P., La Scola, B., Levasseur, A., Caetano-Anollés, G., and Raoult, D. (2017). Mimivirus: leading the way in the discovery of giant viruses of amoebae. Nat. Rev. Microbiol. 15, 243–254. doi: 10.1038/nrmicro.2016.197
Corsaro, D., Köhsler, M., Wylezich, C., Venditti, D., Walochnik, J., Michel, R., et al. (2018). New insights from molecular phylogenetics of amoebophagous fungi (Zoopagomycota, Zoopagales). Parasitol. Res. 117, 157–167. doi: 10.1007/s00436-017-5685-6
De Jonckheere, J. F. (1991). Ecology of acanthamoeba. Rev. Infect. Dis. 13(Supplement_5), S385–S387. doi: 10.1093/clind/13.Supplement_5.S385
Delafont, V., Bouchon, D., Héchard, Y., and Moulin, L. (2016). Environmental factors shaping cultured free-living amoebae and their associated bacterial community within drinking water network. Water Res. 100, 382–392. doi: 10.1016/j.watres.2016.05.044
Delafont, V., Mougari, F., Cambau, E., Joyeux, M., Bouchon, D., Héchard, Y., et al. (2014). First evidence of amoebae–mycobacteria association in drinking water network. Environ. Sci. Technol. 48, 11872–11882. doi: 10.1021/es5036255
Dey, R., Bodennec, J., Mameri, M. O., and Pernin, P. (2009). Free-living freshwater amoebae differ in their susceptibility to the pathogenic bacterium Legionella pneumophila. FEMS Microbiol. Lett. 290, 10–17. doi: 10.1111/j.1574-6968.2008.01387.x
Dey, R., Dlusskaya, E., and Ashbolt, N. J. (2022). SARS-CoV-2 surrogate (Phi6) environmental persistence within free-living amoebae. J. Water Health 20, 83–91. doi: 10.2166/wh.2021.167
Dey, R., Rieger, A. M., Stephens, C., and Ashbolt, N. J. (2019). Interactions of Pseudomonas aeruginosa with Acanthamoeba polyphaga observed by imaging flow cytometry. Cytometry Part A 95, 555–564. doi: 10.1002/cyto.a.23768
Folkins, M. A., Dey, R., and Ashbolt, N. J. (2020). Interactions between human reovirus and free-living amoebae: implications for enteric virus disinfection and aquatic persistence. Environ. Sci. Technol. 54, 10201–10206. doi: 10.1021/acs.est.0c02896
Hsueh, T.-Y., and Gibson, K. E. (2015). Interactions between human norovirus surrogates and Acanthamoeba spp. Appl. Environ. Microbiol. 81, 4005–4013. doi: 10.1128/AEM.00649-15
Juarez, M. M., Tartara, L. I., Cid, A. G., Real, J. P., Bermudez, J. M., Rajal, V. B., et al. (2018). Acanthamoeba in the eye, can the parasite hide even more? Latest developments on the disease. Contact Lens Anterior Eye 41, 245–251. doi: 10.1016/j.clae.2017.12.017
Khan, N. A., Baqir, H., and Siddiqui, R. B. (2015). The immortal amoeba: a useful model to study cellular differentiation processes? Pathog. Glob. Health 109, 305–306. doi: 10.1080/20477724.2015.1103504
Lasheras, A., Boulestreau, H., Rogues, A.-M., Ohayon-Courtes, C., Labadie, J.-C., Gachie, J.-P., et al. (2006). Influence of amoebae and physical and chemical characteristics of water on presence and proliferation of Legionella species in hospital water systems. Am. J. Infect. Control 34, 520–525. doi: 10.1016/j.ajic.2006.03.007
Luca, I. (2022). “Fungi and oomycetes–allies in eliminating environmental pathogens,” in Animal Welfare-New Insights, eds. S.-W. Hung, C.-C. Chen, C.-L. Lu, T.-T. Kao, and R. Payan-Carreira (London: IntechOpen). doi: 10.5772/intechopen.106498
Malinowski, N., Domingos, S., Wylie, J., Morgan, M. J., Metcalfe, S., Walsh, T., et al. (2022). Free-living amoeba and associated pathogenic bacteria in well-chlorinated drinking water storage tanks. ACS ES&T Water 2, 1511–1520. doi: 10.1021/acsestwater.1c00428
Mergeryan, H. (1991). The prevalence of Acanthamoeba in the human environment. Rev. Infect. Dis. 13(Supplement_5), S390–S391. doi: 10.1093/clind/13.Supplement_5.S390
Nisar, M. A., Ross, K. E., Brown, M. H., Bentham, R., Hinds, J., Whiley, H., et al. (2022). Molecular screening and characterization of Legionella pneumophila associated free-living amoebae in domestic and hospital water systems. Water Res. 226:119238. doi: 10.1016/j.watres.2022.119238
Oliveira, G., Silva, L., Leão, T., Mougari, S., da Fonseca, F. G., Kroon, E. G., et al. (2019). Tupanvirus-infected amoebas are induced to aggregate with uninfected cells promoting viral dissemination. Sci. Rep. 9:183. doi: 10.1038/s41598-018-36552-4
Otero-Ruiz, A., Gonzalez-Zuñiga, L. D., Rodriguez-Anaya, L. Z., Lares-Jiménez, L. F., Gonzalez-Galaviz, J. R., Lares-Villa, F., et al. (2022). Distribution and current state of molecular genetic characterization in pathogenic free-living amoebae. Pathogens 11:1199. doi: 10.3390/pathogens11101199
Qvarnstrom, Y., Nerad, T. A., and Visvesvara, G. S. (2013). Characterization of a new pathogenic Acanthamoeba species, A. byersi n. sp., isolated from a human with fatal amoebic encephalitis. J. Eukaryot. Microbiol. 60, 626–633. doi: 10.1111/jeu.12069
Raoult, D., Audic, S., Robert, C., Abergel, C., Renesto, P., Ogata, H., et al. (2004). The 1.2-megabase genome sequence of Mimivirus. Science 306, 1344–1350. doi: 10.1126/science.1101485
Raoult, D., and Boyer, M. (2010). Amoebae as genitors and reservoirs of giant viruses. Intervirology 53, 321–329. doi: 10.1159/000312917
Rayamajhee, B., Subedi, D., Peguda, H. K., Willcox, M. D., Henriquez, F. L., Carnt, N., et al. (2021). A systematic review of intracellular microorganisms within Acanthamoeba to understand potential impact for infection. Pathogens 10:225. doi: 10.3390/pathogens10020225
Rivera, F., Roy-Ocotla, G., Rosas, I., Ramirez, E., Bonilla, P., Lares, F., et al. (1987). Amoebae isolated from the atmosphere of Mexico City and environs. Environ. Res. 42, 149–154. doi: 10.1016/S0013-9351(87)80016-6
Rosenberg, K., Bertaux, J., Krome, K., Hartmann, A., Scheu, S., Bonkowski, M., et al. (2009). Soil amoebae rapidly change bacterial community composition in the rhizosphere of Arabidopsis thaliana. ISME J. 3, 675–684. doi: 10.1038/ismej.2009.11
Rubenina, I., Kirjušina, M., Bērzinš, A., Valcina, O., and Jahundoviča, I. (2017). Relationships between free-living amoeba and their intracellular bacteria. Proc. Latv. Acad. Sci. B: Nat. Exact Appl. Sci. 71. doi: 10.1515/prolas-2017-0044
Scheid, P. L. (2018). Amoebophagous fungi as predators and parasites of potentially pathogenic free-living amoebae. Open Parasitol. J. 6. doi: 10.2174/1874421401806010075
Schuster, F. L., and Visvesvara, G. S. (2004). Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int. J. Parasitol. 34, 1001–1027. doi: 10.1016/j.ijpara.2004.06.004
Shaheen, M., and Ashbolt, N. J. (2021). Differential bacterial predation by free-living amoebae may result in blooms of Legionella in drinking water systems. Microorganisms 9:174. doi: 10.3390/microorganisms9010174
Shaheen, M., Scott, C., and Ashbolt, N. J. (2019). Long-term persistence of infectious Legionella with free-living amoebae in drinking water biofilms. Int. J. Hyg. Environ. Health 222, 678–686. doi: 10.1016/j.ijheh.2019.04.007
Silverstein, S. C., Christman, J. K., and Acs, G. (1976). The reovirus replicative cycle. Annu. Rev. Biochem. 45, 375–408. doi: 10.1146/annurev.bi.45.070176.002111
Steenbergen, J. N., Nosanchuk, J. D., Malliaris, S. D., and Casadevall, A. (2003). Cryptococcus neoformans virulence is enhanced after growth in the genetically malleable host Dictyostelium discoideum. Infect. Immun. 71, 4862–4872. doi: 10.1128/IAI.71.9.4862-4872.2003
Steenbergen, J. N., Nosanchuk, J. D., Malliaris, S. D., and Casadevall, A. (2004). Interaction of Blastomyces dermatitidis, Sporothrix schenckii, and Histoplasma capsulatum with Acanthamoeba castellanii. Infect. Immun. 72, 3478–3488. doi: 10.1128/IAI.72.6.3478-3488.2004
Stehr-Green, J. K., Bailey, T. M., and Visvesvara, G. S. (1989). The epidemiology of Acanthamoeba keratitis in the United States. Am. J. Ophthalmol. 107, 331–336. doi: 10.1016/0002-9394(89)90654-5
Suzan-Monti, M., Scola, B. L., Barrassi, L., Espinosa, L., and Raoult, D. (2007). Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS ONE 2:e328. doi: 10.1371/journal.pone.0000328
Thomas, J. M., Thomas, T., Stuetz, R. M., and Ashbolt, N. J. (2014). Your garden hose: a potential health risk due to Legionella spp. growth facilitated by free-living amoebae. Environ. Sci. Technol. 48, 10456–10464. doi: 10.1021/es502652n
Thomaz, L., García-Rodas, R., Guimarães, A. J., Taborda, C. P., Zaragoza, O., Nosanchuk, J. D., et al. (2013). Galleria mellonella as a model host to study Paracoccidioides lutzii and Histoplasma capsulatum. Virulence 4, 139–146. doi: 10.4161/viru.23047
Vaerewijck, M. J., Baré, J., Lambrecht, E., Sabbe, K., and Houf, K. (2014). Interactions of foodborne pathogens with free-living protozoa: potential consequences for food safety. Compr. Rev. Food Sci. Food Saf. 13, 924–944. doi: 10.1111/1541-4337.12100
Verani, M., Di Giuseppe, G., Tammaro, C., and Carducci, A. (2016). Investigating the role of Acanthamoeba polyphaga in protecting Human Adenovirus from water disinfection treatment. Eur. J. Protistol. 54, 11–18. doi: 10.1016/j.ejop.2016.02.006
Visvesvara, G. S., and Stehr-Green, J. K. (1990). Epidemiology of free-living ameba infections 1. J. Protozool. 37, 25s−33s. doi: 10.1111/j.1550-7408.1990.tb01142.x
Wang, X., and Ahearn, D. G. (1997). Effect of bacteria on survival and growth of Acanthamoeba castellanii. Curr. Microbiol. 34, 212–215. doi: 10.1007/s002849900170
Water, I. D. U. (2010). Free-living amoebae (FLA). Parasite 17, 67–70. doi: 10.1051/parasite/2010171067
Weisman, R. A., and Korn, E. D. (1967). Phagocytosis of latex beads by Acanthamoeba. I. Biochemical properties. Biochemistry 6, 485–497. doi: 10.1021/bi00854a017
Keywords: free-living amoebae, FLA, bacteria, viruses, microorganisms, eukaryotes
Citation: Fan S, Shen Y and Qian L (2024) Social life of free-living amoebae in aquatic environment— comprehensive insights into interactions of free-living amoebae with neighboring microorganisms. Front. Microbiol. 15:1382075. doi: 10.3389/fmicb.2024.1382075
Received: 05 February 2024; Accepted: 03 June 2024;
Published: 19 June 2024.
Edited by:
Geoffrey James Puzon, CSIRO Land and Water, AustraliaReviewed by:
Caitlin Proctor, Purdue University, United StatesCopyright © 2024 Fan, Shen and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Qian, bGkucWlhbkBnd3UuZWR1
 Shi Fan
Shi Fan Yun Shen
Yun Shen Li Qian
Li Qian