
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 April 2024
Sec. Terrestrial Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1377721
Subsurface karst caves provide unique opportunities to study the deep biosphere, shedding light on microbial contribution to elemental cycling. Although ammonia oxidation driven by both ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) is well explored in soil and marine environments, our understanding in the subsurface biosphere still remained limited to date. To address this gap, weathered rock and sediment samples were collected from the Xincuntun Cave in Guilin City, an alkaline karst cave, and subjected to high-throughput sequencing and quantification of bacterial and archaeal amoA, along with determination of the potential nitrification rates (PNR). Results revealed that AOA dominated in ammonia oxidation, contributing 48–100% to the PNR, and AOA amoA gene copies outnumbered AOB by 2 to 6 orders. Nitrososphaera dominated in AOA communities, while Nitrosopira dominated AOB communities. AOA demonstrated significantly larger niche breadth than AOB. The development of AOA communities was influenced by deterministic processes (50.71%), while AOB communities were predominantly influenced by stochastic processes. TOC, NH4+, and Cl− played crucial roles in shaping the compositions of ammonia oxidizers at the OTU level. Cross-domain co-occurrence networks highlighted the dominance of AOA nodes in the networks and positive associations between AOA and AOB, especially in the inner zone, suggesting collaborative effort to thrive in extreme environments. Their high gene copies, dominance in the interaction with ammonia oxidizing bacteria, expansive niche breadth and substantial contribution to PNR collectively confirmed that AOA better adapted to alkaline, oligotrophic karst caves environments, and thus play a fundamental role in nitrogen cycling in subsurface biosphere.
Earth’s subsurface environments are isolated from phototrophic energy sources, which are characterized by oligotrophic condition and limitation in electron donors or electron acceptors (Jones et al., 2018). Microorganisms living in such environments are highly dependent on the oxidation of limited inorganic matter for energy (Zhang et al., 2018; Dong et al., 2020; Jones and Northup, 2021). Nevertheless, a large number of microbial cells is estimated to be 2 × 1029–6 × 1029 in the terrestrial subsurface biosphere excluding those in soils (Magnabosco et al., 2018). The subsurface biosphere serves as a fascinating place to decipher microbial dark matter and offer new knowledge about life, particularly in examining minimum energetic requirements and adaptations to oligotrophic environments.
Karst caves are known as subsurface extreme ecosystems with twilight or dark conditions, nutrient deprivation and isolation from the surficial environments, which harbor numerous chemoautotrophic microorganisms (Marques et al., 2019). Currently, progresses have been made on microbial communities, niche differentiation, community assembly and their correlation with environmental variables (Yun et al., 2016; Zhao et al., 2017; Cheng et al., 2019, 2021, 2022; Cao et al., 2021). Bacteria, microalgae, and fungi are found to be involved in the elemental cycles of carbon, nitrogen, sulfur, manganese, and iron, as well as in the dissolution and precipitation of limestone (Grobbelaar, 2000; Mulec, 2008; Sebela et al., 2015; Jones and Northup, 2021). Moreover, microbial functional groups such as ammonia-oxidizers are also been investigated and ammonia-oxidizing archaea (AOA) contribute more to ammonia oxidation than their bacterial counterparts as indicated by clone libraries in cave sediments (Zhao et al., 2017). Nevertheless, the limitation of the first-generation sequencing technique may not reveal the nitrifying communities comprehensively. Moreover, the adaption and ubiquitous occurrence of ammonia oxidizers in different cave habitats such as sediments and weathered rocks and how AOA and AOB interact with each other remain largely unexplored in the subsurface biosphere.
Nitrification is one of the fundamental biogeochemical processes mediated by microorganisms in natural environments, which has been well-reported in ecosystems such as soils, wetlands, farmland, estuary, and marine environments (He et al., 2007; Beman et al., 2012; Veresoglou et al., 2012; Phillips et al., 2015; Chen et al., 2019; Luvizott et al., 2019; Wei et al., 2023). The oxidation of ammonia (NH3) to nitrite (NO2−) is the first and rate-limiting step in nitrification, catalyzed by ammonia monooxygenase enzymes encoded by the amoA gene of AOA and AOB (Daims et al., 2015). Therefore, the amoA gene is widely used to explore ammonia-oxidizing communities in natural environments (Heiss et al., 2022). AOA and AOB are widely distributed in natural ecosystems, and they dominate ammonia-oxidizing communities under different conditions. Usually, AOA dominate in strong acidic environments with low ammonia concentration (Zhang et al., 2012; Kozlowski et al., 2016), whereas AOB is dominant in nitrogen rich environments (Di et al., 2009) due to their different substrate affinity (Martens-Habbena et al., 2009). Nevertheless, recent study showed that not all AOA possess such higher substrate affinity than AOB. In fact, the substrate affinity of ammonia-oxidizers correlated with their cell surface area to volume ratio (Jung et al., 2022). These new information on enzyme kinetic may indicate more complexity about niche differentiation between AOA and AOB. In addition, oxygen concentration, light condition, temperature, metal and organic compound also contribute to their distinct ecological niche differentiation in natural environments (Liu et al., 2015; Ouyang et al., 2017; Cheng et al., 2019; Khanom et al., 2021). The isolation and relative stable conditions create specific microhabitats within caves, especially loose sediments on the ground and weathered rocks of the cave passage (Cheng et al., 2023; Liu X. Y. et al., 2023), providing excellent conditions for the exploration of the niche differences of ammonia-oxidizing microorganisms in oligotrophic environments. However, the distribution of AOA and AOB and the ecological processes responsible for their development in these unique microhabitats remain poorly understood.
The theory of microbial assembly based on ecological niches is one of the universal tools to study microbial communities (Stegen et al., 2012; Ferrenberg et al., 2016; Yuan et al., 2019; Zhao et al., 2019a; Yu et al., 2021; Wang et al., 2024). Deterministic processes are the selection and filtering of species by ecological choices imposed by biotic and abiotic factors, while stochastic processes are the role of unpredictable interventions such as births and deaths on microbial communities (Zhou and Ning, 2017; Zhang et al., 2022; Fang W. K. et al., 2023). Ecological guilds exhibit different community structures due to different responses to environmental selection (Zhao et al., 2019b). Studies have clearly demonstrated the strong niche specificity of bacterial communities in loose sediments and those living on weathered rocks (Cao et al., 2021; Liu X. Y. et al., 2023) as well as for methanotrophs (Cheng et al., 2022), which may be also true in microbial functional groups involved in nitrification. If it is the case, what ecological processes contribute to the differences in ammonia-oxidizing communities in different niches in karst caves?
To fill these knowledge gaps, we collected loose sediment and weathered rock samples from the Xincuntun Cave in Guilin city, along the cave passage and subjected to amplicon high-throughput sequencing and quantification of the amoA gene and PNR (potential nitrification rate) measurement. Our aims are to investigate: (i) composition and niche differentiation of ammonia-oxidizing microbial communities; (ii) potential role of the environmental factors in ecological niche differentiation; (iii) the contribution of AOA and AOB to nitrification and their adaption to subsurface caves.
The Xincuntun (XCT) Cave is a pristine karst cave without any tourists, located in Yongfu County, Guilin city, Guangxi Province (24°58′38.5″N, 109°44′15.7″E). It has a subtropical monsoon climate with an annual average temperature of 18.8°C and an annual average rainfall of 1,950 mm, which is mainly concentrated in March to August. The XCT Cave consists of two branches with a total length of 386 m. We sampled the left branch in this study, which is 100 m long, 2–7 m high, and about 3 m wide.
The weathered rock (W) and loose sediment (S) samples were collected from the cave with an interval of 20 m. The first two sampling sites were located in the entrance nearby zone with weak light (designated as ENZ), whereas the other three sampling sites located in the dark inner zones (IZ). Surface sediment samples were collected by five-point sampling method with a depth <5 cm with sterilized shovels and weathered rock samples were gently scraped from the weathered cave wall. All samples were stored in 50 mL sterile centrifuge tubes and kept on ice. They were immediately stored in freezer upon arrival in the hotel (−20°C). All utensils used for sampling were sterilized beforehand, and sterile gloves and masks were worn throughout the sampling to avoid contamination. Samples were transported back to the laboratory on ice in an insulated box and stored at −80°C for future use.
Samples were freeze-dried for 24 h using a freeze-dryer (ALPHA 1-2LD, Christ, Germany). 10 g of ground sample was mixed with ultrapure water in a ratio of 1:5 (w/v), shaken for 5 min (Vortex-Genie®2, QIAGEN, Germany) and centrifuged at 6,800 × g (TGL-16A, Changsha) for 15 min. The supernatant was filtered through a 0.22 μm membrane, acidified with 3 M HCl for cation except NH4+ measurement using ICS-600 (Thermo, United States). The ammonium of the filtrate without acidification was measured by the salicylic acid assay (Kandeler and Gerber, 1988), and the filtrate without acidification was determined for anion with ICP-OES (iCAP 7,600+) ion chromatography. pH was determined using a multi-parameter water quality tester (HACH, Loveland, CO, United States), and TOC was determined using an elemental analyzer (Vario MACRO cube, Elementar, Germany). The amount of un-ionized NH3 based on pH was calculated using the following formula (Emerson et al., 1975):
Where pKa is the dissociation constant of NH3 + H+/NH4+ pair in solution.
Total DNA was extracted from 0.5 g of freeze-dried weathered rocks and sediments using the FastDNA® SPIN kits for soil (MP Biomedicals, United States). The concentration and quality of the extracted DNA were determined using a Nanodrop 2000 (ND2000, Thermo Fisher Scientific) spectrophotometer for subsequent experiments. AOA was amplified using the primer set of Arch-amoA26F/Arch-amoA417R (Park et al., 2008), and the primer set of amoA1F/amoA2R was used for AOB (Weiner and Maizels, 1999). Paired-end sequencing of the amoA functional genes of AOA and AOB was performed on the Illumina Miseq PE300 platform at Shanghai Personal Biotechnology, Co., Ltd., Shanghai, China. Quantification of the amoA genes of AOA and AOB was performed using the primer sets of Arch-amoAF/Arch-amoAR and amoA1F/amoA2R, respectively, with the systems and reactions as described previously (Rotthauwe et al., 1997; Francis et al., 2005; Gao et al., 2016). The R2 values of the standard curves were 0.95 or higher in this study. The abundance of each gene was normalized to the number of qPCR-derived gene copies per gram of dry weight sample. All raw sequence reads were deposited in National Omics Data Encyclopedia (NODE) with the project numbers OER444534 for AOA and OER445462 for AOB.
Primer fragments were excised with cutadapt plugin, sequences were spliced with the fastq_mergepairs module of Vsearch, and quality control was performed with the fastq_filter module. Repetitive sequences and chimeras were removed with the derep_fulllength module and uchime_denovo module, respectively. A perl1 script was run to filter chimeras from the sequence set after quality control. High quality sequences were clustered at 85% similarity (Pester et al., 2012). Species annotation was performed using the National Center for Biotechnology Information (NCBI) database.2 The sequence numbers of amoA genes were, respectively, resampled to 58,405 (for AOA) and 39,470 (for AOB) reads to avoid the influence of sequencing depth on microbial diversity.
Alpha diversity indices (Chao 1, Shannon, Simpson), and principal coordinate analysis (PCoA) were calculated via the vegan and ggplot packages in R (Dixon, 2003). The permutational multivariate analysis of ANOVA (PERMANOVA) was conducted based on Bray–Curtis dissimilarity via the vegan package in R (Rui et al., 2015). Redundancy analysis (RDA) of environmental factors and relative abundance of dominant OTUs were conducted with Canoco 5, and co-occurrence network of ammonia-oxidizing microorganisms was analyzed with Hmisc package and were visualized with the Fruchterman-Reingold layout in Gephi (Bastian et al., 2009). Keystone taxa was characterized using betweenness centrality values (Jiao et al., 2016; Xiang et al., 2017). Community construction was based on iCAMP in the R package implemented in the Galaxy platform online website3 (Ning et al., 2020). The Pearson test was used in the correlation between βNTI and environmental factors for those that conformed to a normal distribution and the Spearman test for those that did not. The niche breadth index was calculated using the spaa package in R. The phylogenetic tree was constructed by selecting the dominant OTUs with >10% abundance and using the National Center for Biotechnology Information (NCBI) GenBank database for BLAST comparison to obtain highly homologous sequences (> 97%) (Liu H. Y. et al., 2023). Phylogenetic analysis was performed using the maximum likelihood method using MEGA 11 software (Ren et al., 2023).
PNR measurements were conducted in triplicates with two experimental groups. The air-dried sample of 5 g was transferred into a 50 mL corning tube and 20 mL of phosphate buffer was added. 1 mM (NH4)2SO4 was added into the centrifuge tubes to serve as the substrate for ammonia oxidation (Zhao et al., 2017). The control groups were treated with 800 μg/mL kanamycin to inhibit the growth of AOB (Taylor et al., 2010). Subsequently samples were incubated at 19°C (close to the mean annual temperature in XCT Cave) for 24 h in the dark at rest followed by the addition of 1 mM KClO3 to stop nitrite oxidation (He et al., 2007; Tourna et al., 2011; Zhao et al., 2017). Nitrite concentration was measured with sulfonamide colorimetric method with a 1:4 ratio of sulfonamide to nitrite (Yu et al., 2007). Variation of nitrite was used to calculate the potential nitrification rate based on the formula of:
The definition of abbreviation in the formula are showed as follow:
PNRAOA: the potential nitrification rate by AOA; CkanamycinNO2−: NO2− concentration in the system with the addition of kanamycin; Cno-canamycinNO2−: NO2− concentration in the system without the addition of kanamycin; PNRAOB: potential nitrification rate of AOB.
The NH4+ concentration was higher in the sediment samples than those in weathered rock. Toward the interior of the cave, NH4+ concentration decreased, and NH3 increased (Table 1). The highest NH4+ concentration (23.43 ± 4.54 mg/kg) was observed in the sediment collected in entrance nearby zone (ENZ-S), and the lowest NH4+ concentration (10.40 ± 4.15 mg/kg) was observed in the weathered rock collected in the dark inner zone (IZ-W). The variation of NH3 ranged from 88.04 ± 22.46 to 151.53 ± 47.29 mg/kg (Table 1). All samples were weakly alkaline and the pH value (8.03 ± 0.47 and 8.38 ± 0.40) of IZ samples was higher than those of ENZ samples (7.69 ± 0.47 and 7.81 ± 0.18) (Table 1). Temperature in inner zone was higher (16.30 ± 0.92°C) than that in ENZ (10.00 ± 0.14°C).
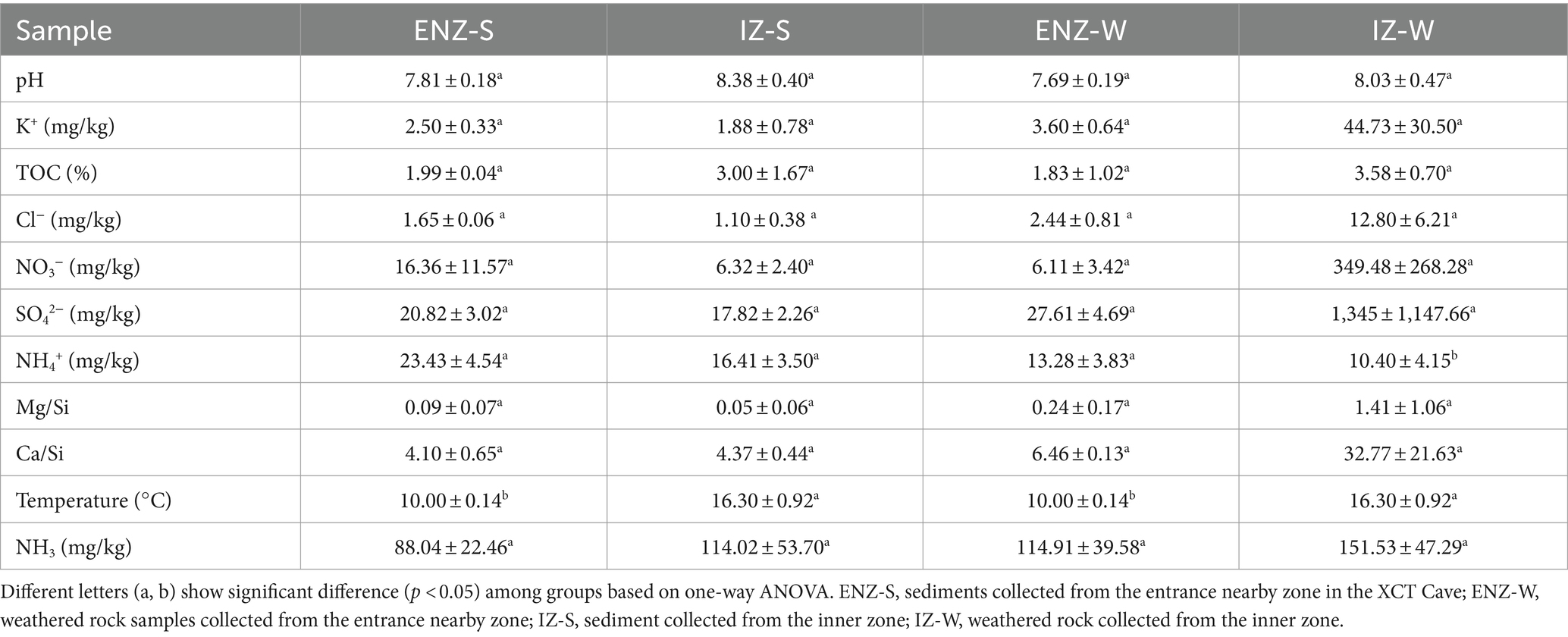
Table 1. Physicochemical parameters of weathered rocks and sediments within the XCT Cave, Guilin City, Guangxi Province.
Alpha diversity of ammonia-oxidizers was significantly different among different niches. Overall AOA communities showed higher alpha diversity than those of AOB as indicated by Chao 1 index, Simpson index and Shannon index (Figure 1). The AOA alpha diversity in inner zone was higher than that that in the entrance nearby zone, and the alpha diversity of the weathered rock was higher than that of the sediments (Figure 1A). In contrast, AOB showed a higher alpha index in IZ than those in ENZ, whereas the significant difference was only found in the rock samples in ENZ and sediments in IZ (Figure 1B). PCoA analysis showed significant differences in AOA (p = 0.002) and AOB (p = 0.001) communities between different niches such as sediments versus weathered rocks and samples in ENZ versus those in IZ (Figures 1C,D).
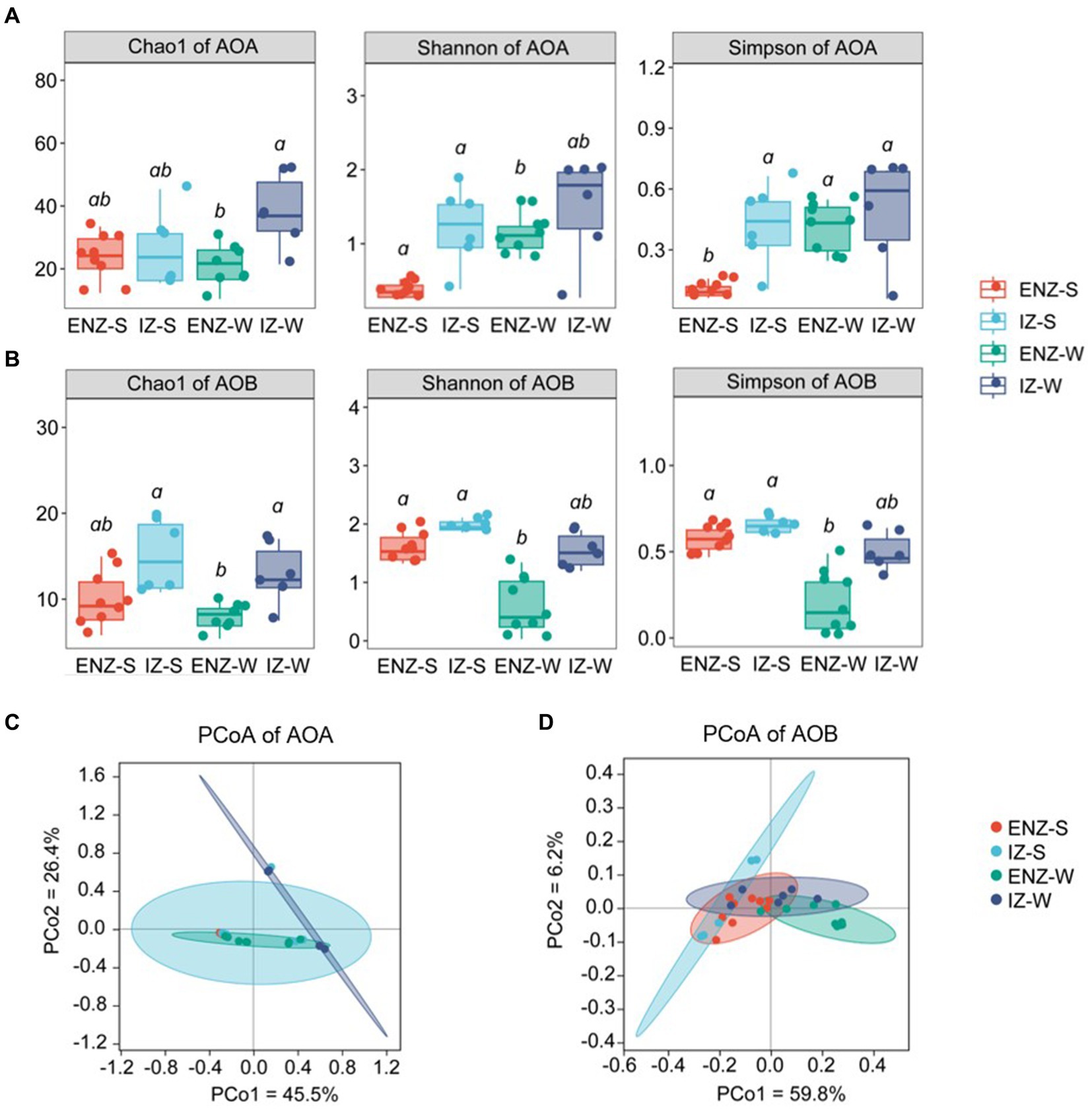
Figure 1. Alpha diversity of ammonia-oxidizing archaea (A) and ammonia-oxidizing bacteria (B) within the XCT Cave, Guilin City. Different letters (a, b) above the box show significant difference (p < 0.05) among groups based on one-way ANOVA. Panels (C,D) revealed beta diversity based on the Bray-Curtis distance based on amoA gene sequences of ammonia-oxidizing archaea and ammonia-oxidizing bacteria as indicated by PCoA plots, respectively. ENZ-S, sediments collected from the entrance nearby zone; ENZ-W, weathered rock samples collected from the entrance nearby zone; IZ-S, sediment from the interior zone; IZ-W, weathered rock from the interior zone.
A total of 200 AOA OTUs was recovered from the cave samples and OTU1 was numerically dominant in all habitats (Figure 2A). All the AOA OTUs were affiliated with Thaumarchaeota, which was furthered divided into two orders (Nitrosopumilales and Nitrososphaeria) and three genera (Nitrososphaera, Candidatus Nitrosocosmicus, and Nitrosopumilus). AOTU1 dominated AOA communities with the highest relative abundance of 94.86%, 53.17%, and 61.16% in ENZ-S, IZ-S and ENZ-W, respectively, while AOTU2 dominated IZ-W with the relative abundance of 43.17% (Figure 2A). At the genus level, Nitrososphaera was dominant in samples from ENZ-S, IZ-S and ENZ-W, accounting for 95.70%, 61.28%, and 68.18%, respectively. However, an unclassified genus of Nitrosophaeraceae had the highest relative abundance (45.30%) in IZ-W, followed by the Nitrosopumilus with a relative abundance of 37.54% (Figure 2C).
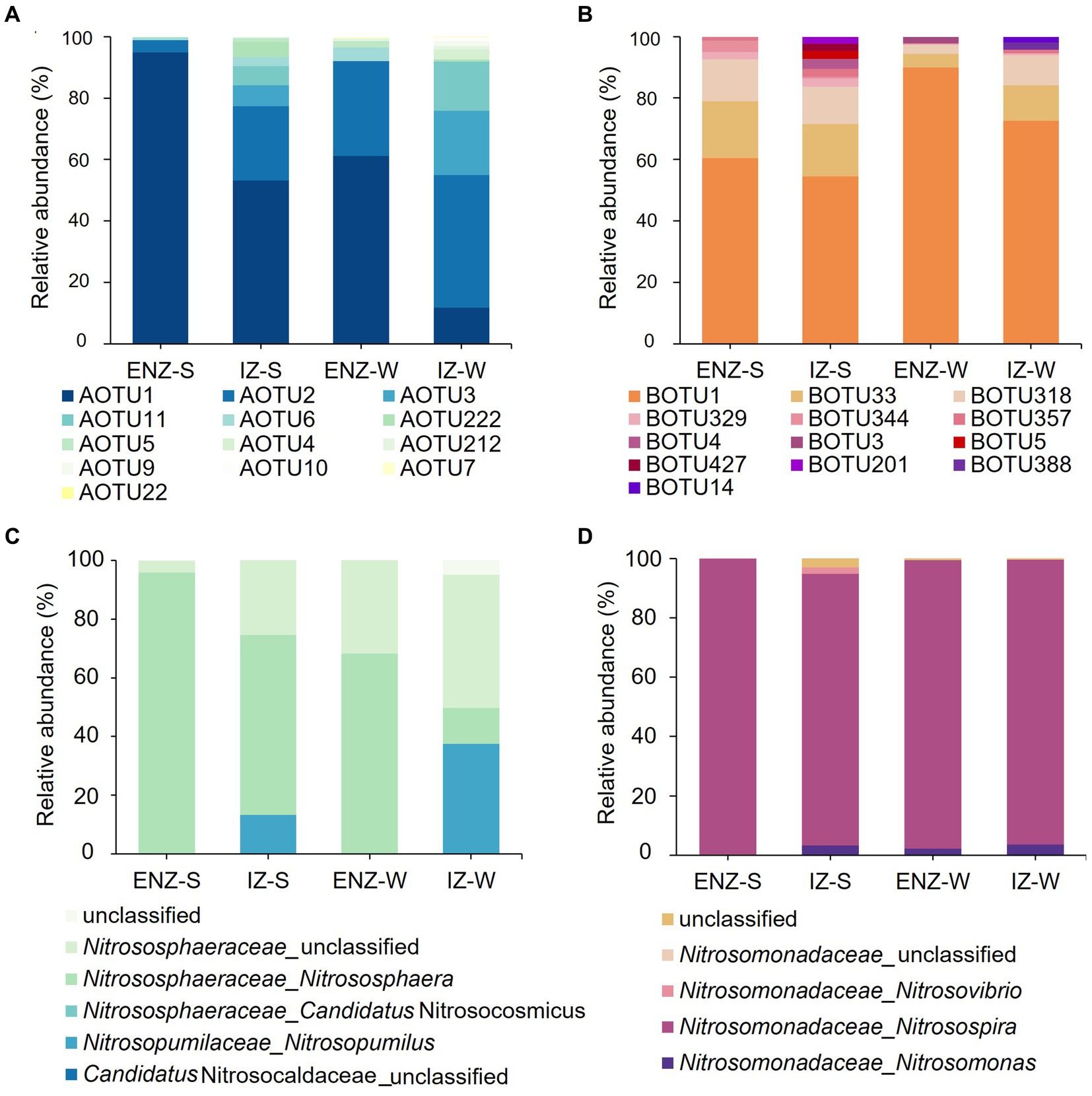
Figure 2. Compositions of ammonia oxidizing archaea and bacteria within the XCT Cave. The relative abundance of ammonia oxidizers at the OTU level (top 13) (A) and the genus level (C). The compositions of ammonia-oxidizing bacteria at the OTU level (top13) (B) and the genus level (D) in the XCT Cave. Abbreviations of ENZ-S, ENZ-W, IZ-S ang IZ-W are the same as those in Figure 1.
In total 35 AOB OTUs were assigned with three genera, Nitrosomonas, Nitrosospira, and Nitrosovibrio, which belonged to the orders of Nitrosomonadales and Nitrosomonadaceae, affiliated to Betaproteobacteria class. BOTU1 dominated in AOB communities with the relative abundance of 60.47%, 54.42%, 90.00%, and 72.54% in ENZ-S, IZ-S, ENZ-W and IZ-W, respectively (Figure 2B). Whereas, at the genus level, Nitrosospira dominated in all samples with 99.91%, 91.57%, 97.24%, and 95.94% in ENZ-S, IZ-S, ENZ-W, and IZ-W, respectively (Figure 2D).
Phylogenetically all the dominant AOA OTUs (AOTU) in the XCT Cave belonged to Group I.1b (Figure 3A) and the AOB OTUs (BOTU) belonged to Cluster D and Cluster C (Figure 3B).
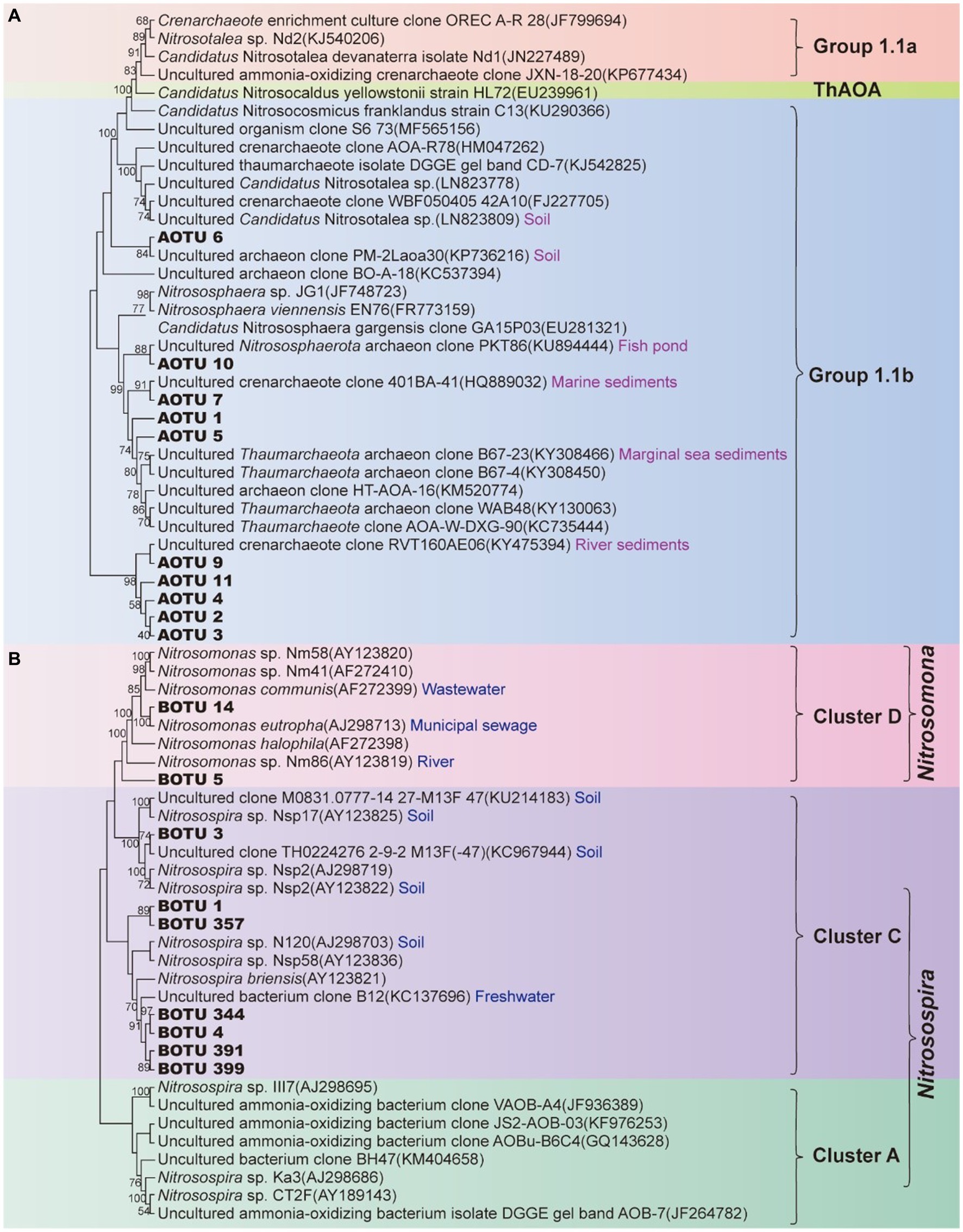
Figure 3. Phylogenetic tree of ammonia-oxidizing archaea (A) and ammonia-oxidizing bacteria (B) based on amoA gene sequences. Branching patterns in the maximum likelihood tree were expressed using the respective bootstrap values (1,000 iterations). OTU in bolds are from this study. AOTU, archaeal OTU; BOTU, bacterial OTU.
The AOA outnumbered AOB by 2 to 6 orders magnitude as confirmed by the quantification of amoA gene. In samples close to the cave entrance, the absolute abundance of AOA amoA gene ranged from 3.02 × 1010 to 6.25 × 1010 copies/g in weathered rock, whereas AOB was ranging from below detection to 4.87 × 107 copies/g (Figure 4A). In sediments samples, AOA varied from 5.05 × 109 to 6.64 × 1010 copies/g, whereas the AOB ranged from 1.76 × 105 to 7.48 × 106 copies/g (Figure 4A). In the inner zone, AOA had 1.73 × 1010 to 5.61 × 1010 copies/g in weathered rock, while AOB ranged from 2.48 × 104 to 2.51 × 108 copies/g sample. In sediments samples, AOA had 4.22 × 109 to 5.62 × 109 copies/g, and AOB varied from 1.78 × 104 to 6.85 × 106 copies/g (Figure 4A).
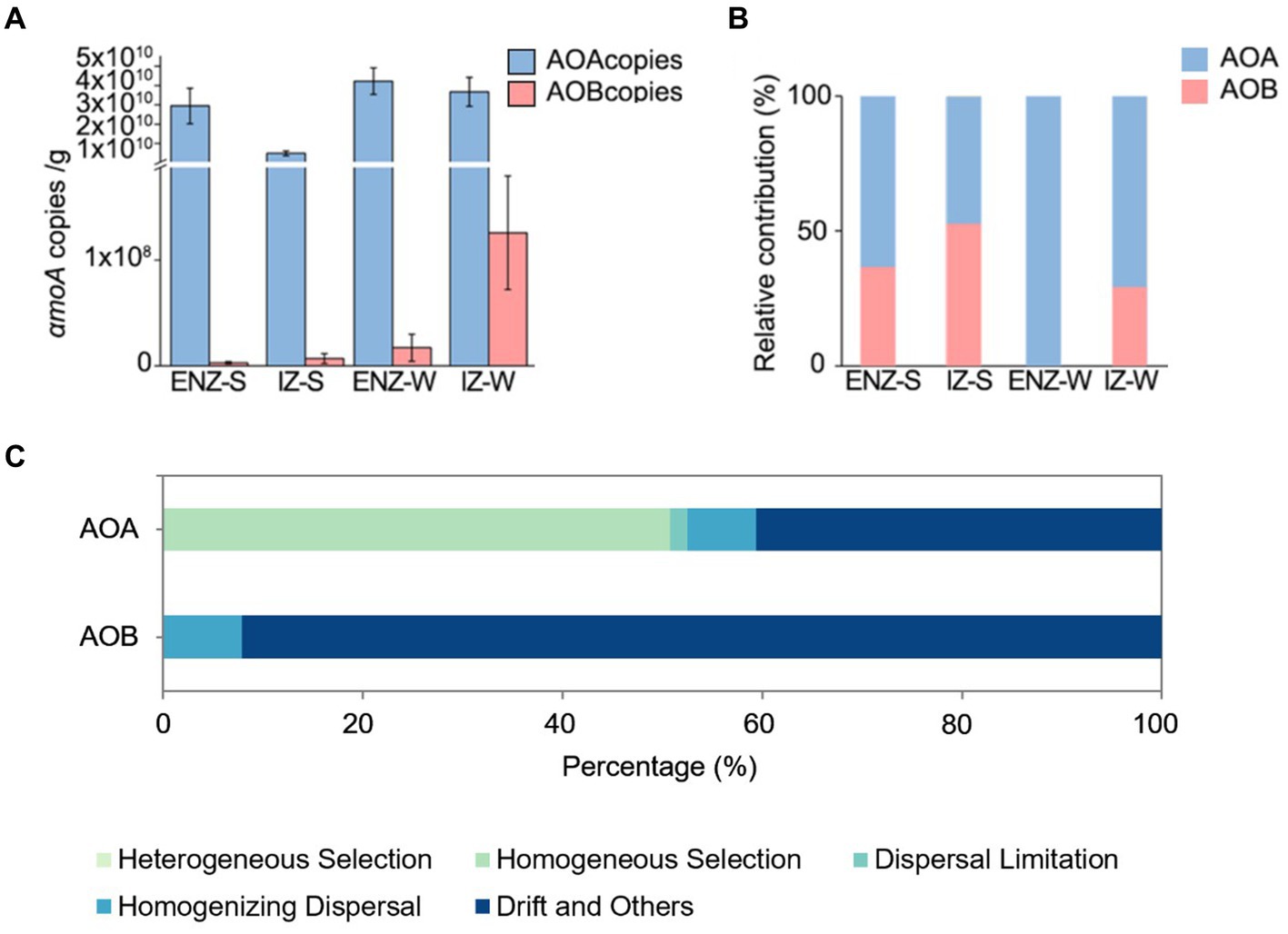
Figure 4. Absolute abundances of archaeal and bacterial amoA genes in the XCT Cave (A). Relative contribution to nitrification by AOA and AOB in the XCT Cave (B). The relative contribution of different ecological processes to ecological assembly processes of ammonia-oxidizing archaea and ammonia-oxidizing bacteria communities (C). Abbreviations of ENZ-S, ENZ-W, IZ-S ang IZ-W are the same as those in Figure 1.
Weathered rock samples showed higher PNR (0.37 ± 0.24 μM/g) than those in the sediment samples (0.22 ± 0.11 μM/g) (Supplementary Table S1) and AOA contribute more to PNR than AOB within the XCT Cave (Figure 4B). AOA exclusively contributed to PNR in rock samples close to the entrance and AOB contributed 52% to PNR in the sediments in IZ (Figure 4B).
The ENZ network of ammonia-oxidizers consisted of 134 nodes and 429 edges, and the IZ network consisted of 131 nodes and 546 edges, respectively (Table 2). Positive links dominated in all networks. Higher weighted degree, diameter, and modularity index were observed in the ENZ network compared with the IZ network, whereas the topology indices of density and mean clustering coefficient were higher in the IZ network (Table 2). The nodes of AOA predominated in all networks, accounted for 86.57% and 84.73% in the networks of ENZ (Figures 5A,B) and IZ (Figures 5D,E), respectively. More nodes were from weathered rocks in ENZ networks (56%) (Figure 5C), while more nodes were from sediments in the IZ networks (65%) (Figure 5F).
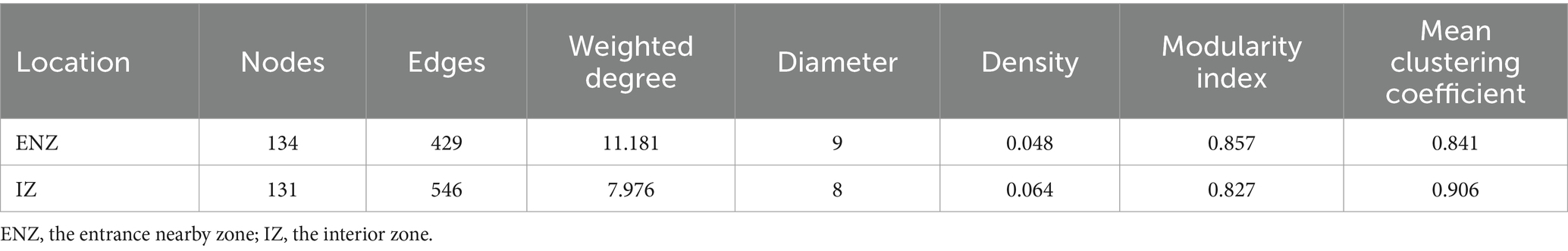
Table 2. Topology indices of ENZ and IZ co-occurrence network of ammonia oxidizing archaea and ammonia-oxidizing bacteria within the XCT cave, Guilin City, Guangxi Province.
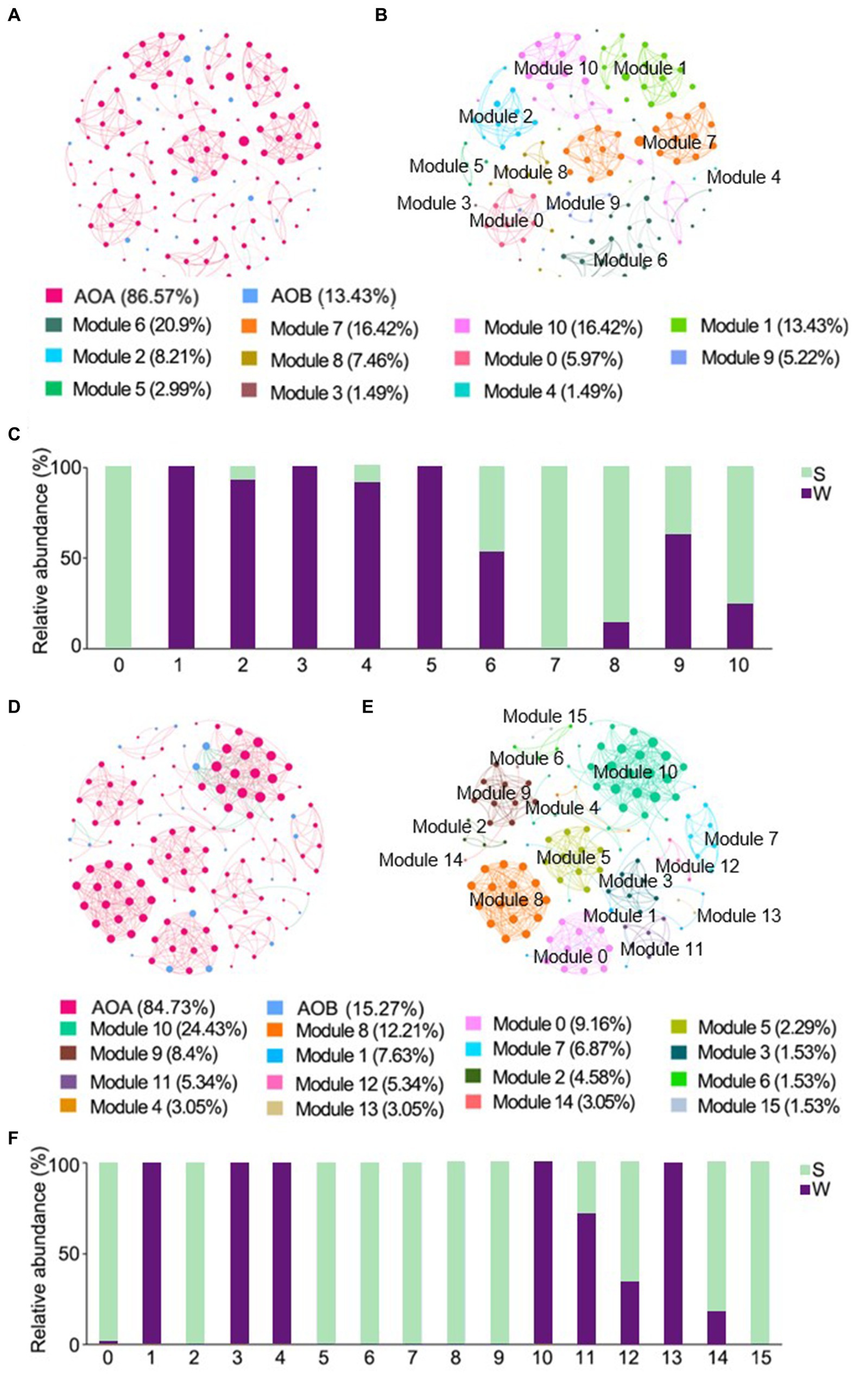
Figure 5. Cross-domain co-occurrence networks based on pairwise Spearman’s correlations between OTUs with a coefficient > |0.6| and a p-value <0.01. Cross-domain co-occurrence networks of ENZ with OTUs colored by ammonia-oxidizers (A) and modularity (B), and the histogram represented the relative abundance of nodes in each module in weathered rocks and sediments (C). Cross-domain co-occurrence networks of IZ with OTUs colored by ammonia-oxidizers (D) and modularity (E), and the histogram represented the relative abundance of nodes in each module in weathered rocks and sediments (F). The size of each node is proportional to the number of connections. Red lines represent positive correlations and green lines represent negative correlations. W, weathered rock; S, sediment.
The identification of keystone taxa showed that the most dominant taxon in the ENZ network was OTU320 (Nitrosopira) with the highest betweenness centrality value of 221, followed by OTU3 (Nitrosopumilus) with the highest betweenness centrality value of 194, in addition to Nitrososphaera, Nitrosomonas, Nitrosospira and Candidatus Nitrosocosmicus (Table 3). In contrast, the keystone taxa in the IZ network were Nitrosopira, Nitrososphaera, and Nitrosopumilus, with the highest betweenness centrality values 130 of OTU344 (Nitrosopira) (Table 3).
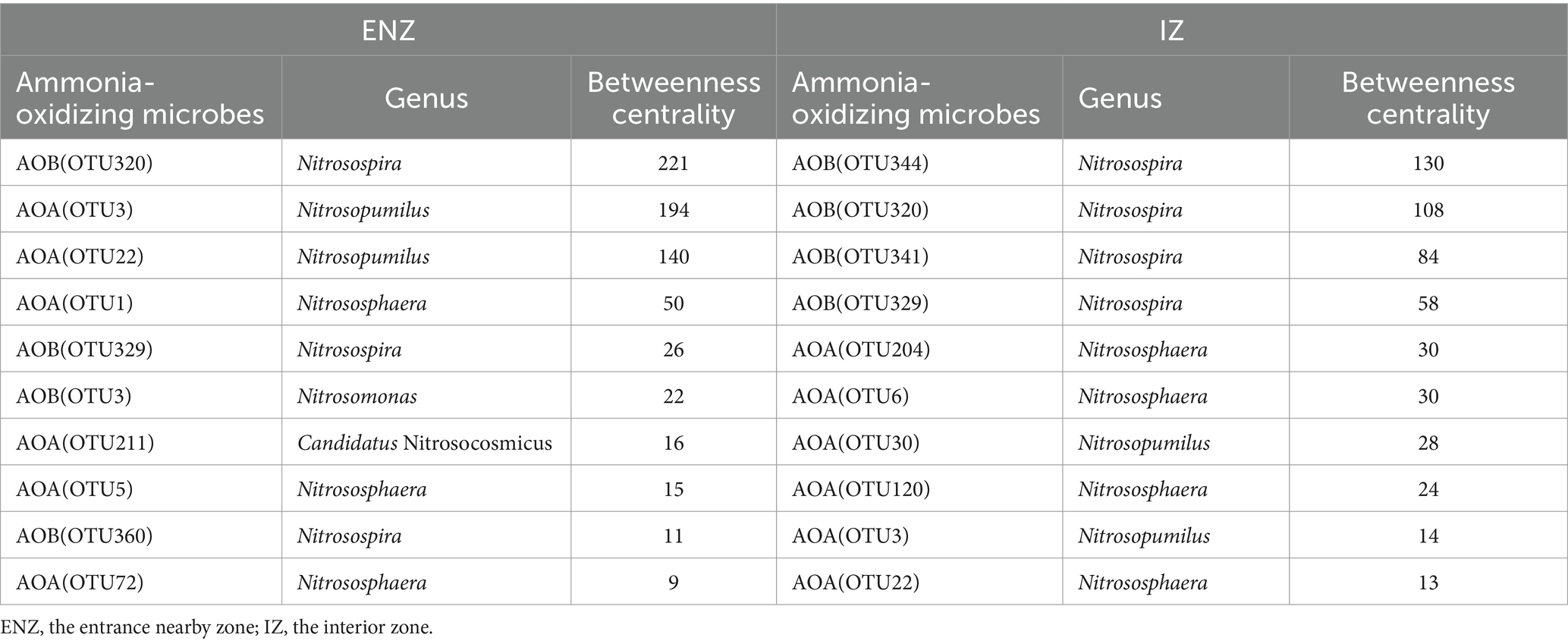
Table 3. Betweenness centrality of the top 10 nodes in the co-occurrence network of ammonia-oxidizing archaea and ammonia-oxidizing bacteria in ENZ and IZ of the XCT Cave, Guilin City, Guangxi Province.
Deterministic process dominated in AOA community assembly with a contribution of 50.71% (especially, homogeneous selection) (Figure 4C). NO3−, NH4+, TOC, and Cl− were the environmental factors that significantly affected the βNTI of AOA (Table 4). Contrasting with the ecological processes for AOA community assembly, stochastic processes contributed 100% to AOB community assembly, dominated by drift with a contribution of 92.09% (Figure 4C). None of the environmental factors investigated in this study had a significant correlation on the βNTI of AOB (Table 4).
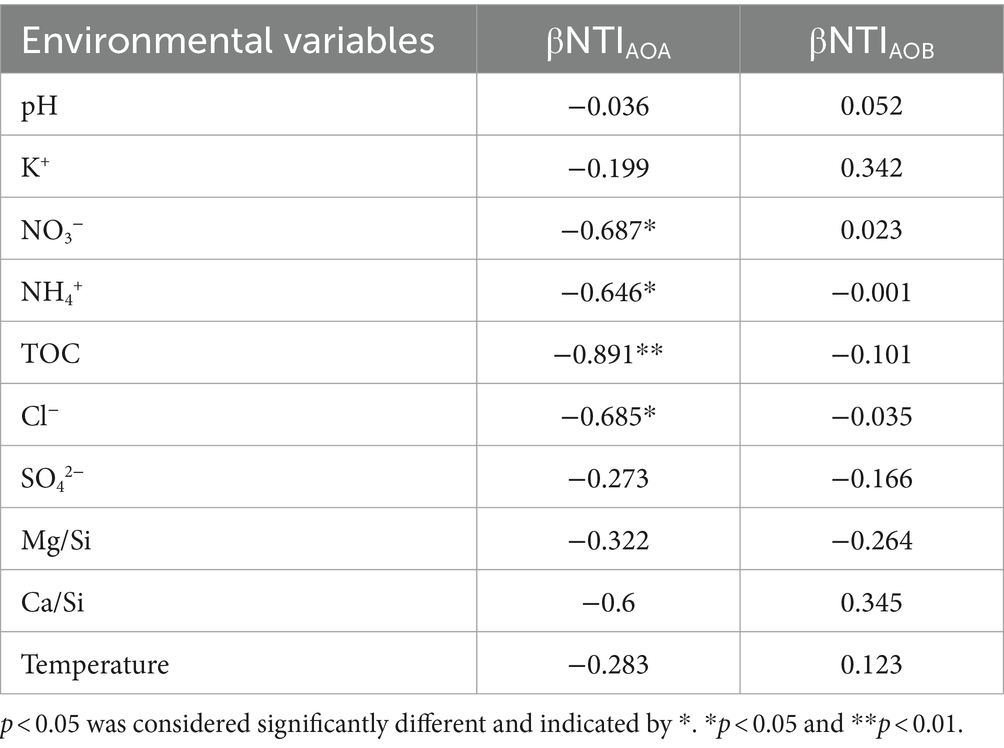
Table 4. Pearson and Spearman correlation analysis of environmental variables with βNTI of AOA and AOB in the XCT Cave, Guangxi Province.
The RDA analysis indicated that TOC, Cl− and NH4+ significantly impacted on ammonia-oxidizers (p < 0.05). RD1 and RD2 explained 67.60% and 21.00% of the variance, respectively (Figure 6A). Among them, TOC was positively correlated with AOTU3, AOTU11, AOTU2, and negatively correlated with most AOB OTUs and AOTU1, AOTU6 (Figure 6A). A linear fit of TOC to the AOTU1 showed that a negative correlation between the content of TOC and the relative abundance of AOTU1, R2 = 0.49 (Figure 6B). However, the linear fit effect of to the OTU of AOB was poor (Supplementary Figure S1).
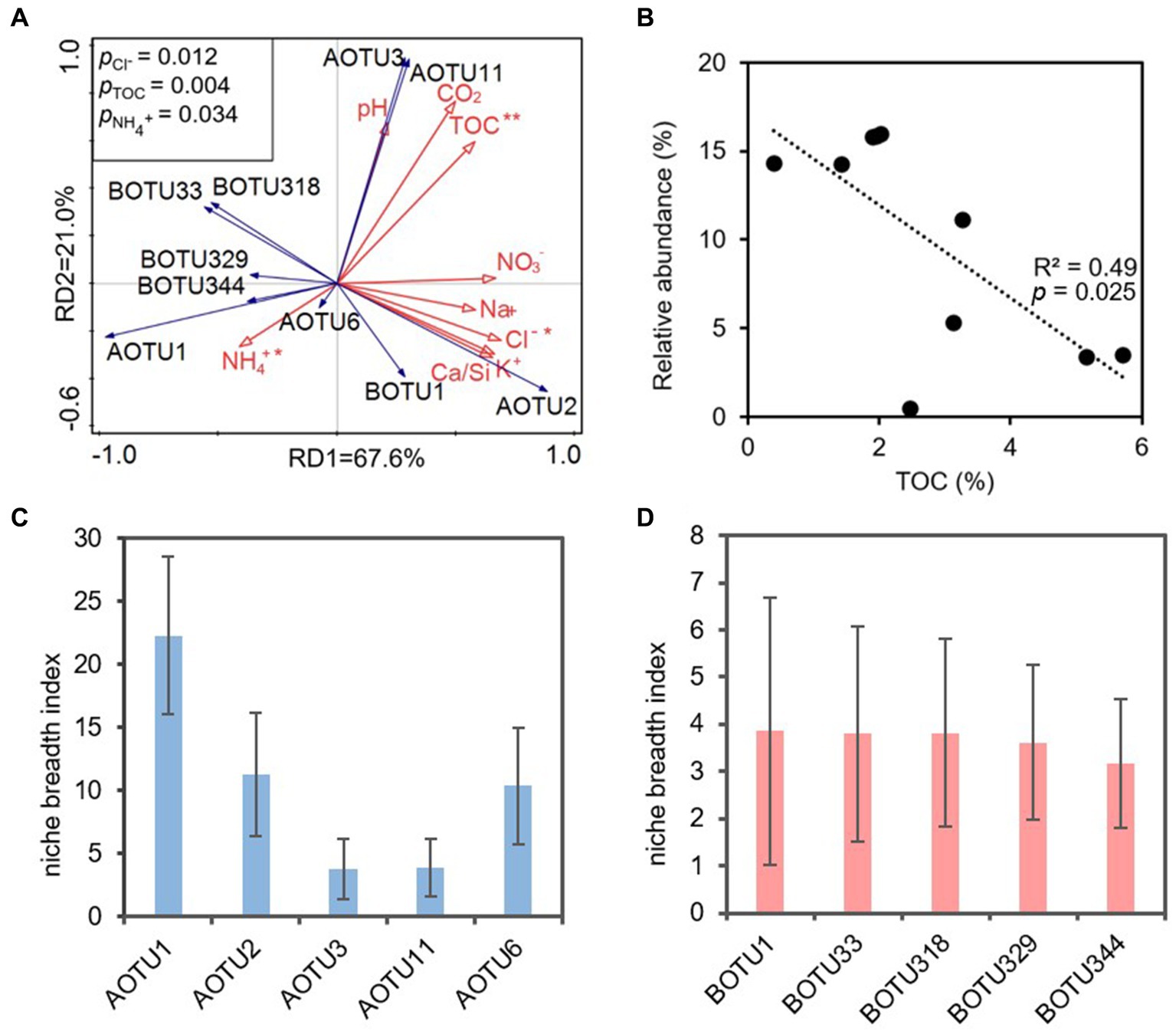
Figure 6. (A) Redundancy analysis (RDA) of physicochemical parameters (red solid arrows) and dominant OTUs (top 5) (blue solid arrows) based on ammonia-oxidizing archaeal and ammonia-oxidizing bacterial amoA genes. AOTU, archaeal OTU; BOTU, bacterial OTU. Asterisks indicate statistical significance (*p < 0.05, **p < 0.01, and ***p < 0.001), and the linear fit of TOC to the AOTU1 relative abundance (B). Panel (C,D) revealed the niche breath index of the dominant OTUs of AOA and AOB, respectively.
The niche breadth index showed that the dominant OTUs of AOA showed much wider niche breadth compared to that of AOB. The niche breadth index of AOA ranged from 3.8 (AOTU1) to 22.3 (AOTU3) (Figure 6C), whereas the BOTUs had niche breadth index ranging from 3.2 to 3.9 (Figure 6D).
The niche breadth index provided robust support for the superior adaption of AOA in oligotrophic karst cave compared to their bacterial counterparts. The elevated niche breadth index (Figures 6C,D) signified that AOA might effectively exploit a broader spectrum of resources in oligotrophic conditions when contrasted with AOB. On the contrary, the narrower niche breadth index indicated that AOB encountered environmental stress and heightened competition for resources (Vorobeichik, 1993; Martorell et al., 2015; Pannek et al., 2016).
The low concentration of ammonium in our cave strongly supports the dominance of AOA. AOA outnumbered their ammonia-oxidizing counterpart AOB by up to thousands of times in the copy numbers of amoA gene within the XCT Cave (Figure 4A), and contributed over 50% to PNR (Figure 4B), which matched well with the observation in other oligotrophic environments (Zhao et al., 2017; Marques et al., 2018; Cardarelli et al., 2020). Usually, AOA show high affinity to ammonium thus achieves a competitive advantage under oligotrophic conditions (Martens-Habbena et al., 2009). Experimental evidence with enriched culture of AOA and AOB from freshwater environments supported this, revealing AOA dominance at an initial ammonium concentration of 50 μM, while AOB dominate at 500 μM (French et al., 2021). A moderately thermophilic ammonia-oxidizing archaea is partially inhibited by a concentration of 3.08 mM ammonium, whereas active at 0.14 and 0.79 mM ammonium (Hatzenpichler et al., 2008). The ammonium concentrations in our cave varied from 10.40 ± 4.15 to 23.43 ± 4.54 mg/kg, equivalent to 0.58 ± 0.23 to 1.30 ± 0.25 mM (Table 1), favored for the establishment and thriving of AOA communities.
pH is another fundamental factor selecting on the nitrifying communities in natural environments (Li et al., 2018; Aigle et al., 2019). Thaumarchaeota Groups I.1a and I.1b are generally associated with neutrophilic or alkaline limestone systems (Chelius and Moore, 2004; Spear et al., 2007; Tetu et al., 2013), whereas Thaumarchaeota Group I.1c tend to dominate in sandstone caves with the pH ranging from 3 to 7 (Barton, 2013; Zhao et al., 2017; Marques et al., 2018). All of our dominant AOTUs affiliated with Group I.1b (Figure 3A), indicating a carbonate cave niche specificity. Compared with Group I.1a, the higher NH3 affinity [Km(app) ≈ 0.14–31.5 μM] (Jung et al., 2022) may help Group I.1b to dominate in karst caves. Due to the ionization of ammonia to ammonium, ammonia concentration decreases exponentially with the decreasing pH (Allison and Prosser, 1993; Wang et al., 2019). Therefore, AOA contributes more to nitrification under acidic pH conditions (Li et al., 2018, 2019; He et al., 2020). Different from the observation of the dominance of AOA in acidic soils, AOA has also been confirmed to dominate in alkaline and oligotrophic cave sediments (Zhao et al., 2017), which matched our results in this study (Figures 4A,B). At alkaline conditions with a pH > 7.3, a significant portion of the NH3/NH4+ pair exists as NH3, leading to NH3 concentrations exceeding 1 mM in copiotrophic systems. This creates an environment where AOB become more competitive. However, based on the pKa of ammonia, it’s worth noting that 1 mM NH3 is still 100-fold lower than the ammonia concentration typically used for AOB culturing (1.2 mM NH4+ equivalent to 100 mM NH3) (Martens-Habbena et al., 2009; Pester et al., 2011). The NH3 concentration in our cave, determined by the ionization constant of NH3 to NH4+, ranged from 5.18 ± 1.32 to 8.91 ± 2.78 mM. Although this range still favored the dominance of AOA, there was potential for competition from AOB for nitrogen resource to some extent.
The PCoA analysis indicated a pronounced segregation in AOA communities compared to AOB (Figures 1C,D), suggesting robust environmental selection on AOA (D'Amen et al., 2018; Shen et al., 2022). This aligned with the dominance of homogeneous selection in AOA community assembly (Figure 4C), revealing a strong influence of environmental factors driving AOA communities towards convergence (Wu et al., 2021). Pearson and Spearman tests furthered confirmed the significant impact of NO3−, NH4+, TOC and Cl− on AOA community assembly in our study (Table 4). In line with our findings, an increase in NH4+ supply has been reported to shift the ecological process from deterministic to stochastic process in AOA community assembly (Ma et al., 2023). This implied that AOA communities exhibited greater competitiveness and adaptability to oligotrophic conditions.
In contrast, AOB community assembly exhibited a predominance stochastic processes (Figure 4C), suggesting that the development of AOB communities was more self-regulated by intrinsic factors (Zhou and Ning, 2017). This observation was further supported by the Pearson and Spearman tests, revealing no significant correlations between environmental factors and βNTI of AOB (Table 4). Similar patterns have been demonstrated under other oligotrophic conditions, where AOB community establishment is also dominated by stochastic process (Yang et al., 2022; Fang J. et al., 2023). The dominance of genetic drift in the development of the AOB community in our study (Figure 4C) may suggest a relatively small AOB community (Ma et al., 2023; Ye et al., 2023; Zhang et al., 2023). This could be attributed to the challenges that AOB face in colonizing under lower ammonium concentrations (Keerio et al., 2020).
TOC, NH4+ and Cl− emerged as the primary environmental impact factors shaping ammonia oxidizers (Figure 6A). Nitrososphaera of AOA exhibited the dominance in the nearby zone of the cave entrance, declining in abundance inward to the cave, and was replaced by Nitrosopumilus in the IZ-W (Figure 2C). Similarly, IZ-W showed higher alpha diversity compared with other niches (Figure 1A). These findings suggest that environmental factors can influence the community composition and diversity of the AOA (Jin et al., 2011; Patil et al., 2021; Kou et al., 2023). The most abundant AOTU1 (Figures 2A) showed a negative correlation with TOC (Figure 6B), while the dominant OTU of AOB exhibited a positive correlation with TOC (Supplementary Figure S1). This aligned with findings in other environments, such as ponds, estuaries and upland soils (Dai et al., 2015, 2018; Wei et al., 2021; Zhu et al., 2023). Most AOA are autotrophs and well adapted to oligotrophic environments due to their highly efficient CO2-fixation pathway of hydroxypropionate/hydroxybutyrate (HP/HB) cycle, as indicated by genomic studies (Kobayashi et al., 2018; Zhao and Zhang, 2022). This may well explain the observation of the dominance of AOA in oligotrophic conditions and their higher contribution to nitrification in karst caves (Wang et al., 2021). However, lower TOC does not favor heterotrophic AOB in term of organic carbon sources. AOB typically show positive correlation with TOC (Zhang et al., 2010; Dai et al., 2018; Zhu et al., 2023).
Cl− showed a negative correlation with the dominant OTUs of both AOA and AOB, indicating the inhibition of Cl− on the functioning of amoA enzymes in ammonia-oxidizing microorganisms (Wang et al., 2014; Roy et al., 2020). Similar negative correlations have also been observed between Cl− and ammonia oxidizers in drinking water (Scott et al., 2015) and demonstrate inhibition on nitrification (Roy et al., 2020).
The AOA also dominated in the co-occurrence networks with AOB in caves. AOA occupied a greater nodes number in the cross-domain networks, forming closer links inward to the cave (Figures 5C,F), which suggested more collaboration for enhanced survival in subsurface caves. Interestingly, in the ENZ network, most nodes belonged to the weathered rock samples (Figure 5C), whereas the IZ network exhibited more sediment nodes (Figure 5F). In some oligotrophic environments, AOA have been found to dominate the nitrification network despite of the higher number of AOB (Zheng et al., 2022; Hu et al., 2023; Liu J. J. et al., 2023). These finding collectively highlighted the crucial role of AOA in the stabilizing the network (Jones and Hallin, 2019; He et al., 2021; Ma et al., 2023).
Nitrosospira and Nitrosopumilus were the keystone taxa in the ENZ network, while Nitrosospira and Nitrososphaera were the keystone taxa in IZ (Table 3). Nitrosospira are widely distributed in terrestrial and marine ecosystems (Lebedeva et al., 1997; Jiang and Bakken, 1999; Hollibaugh et al., 2002). AOA isolates such as Nitrosopumilus, Nitrososphaera, Nitrosopumilus, and Nitrososphaera are characterized by small size, and harbor flagellum-encoding genes (Tourna et al., 2011; Stieglmeier et al., 2014; Qin et al., 2017; Bayer et al., 2019). Although AOA and AOB both survive in oligotrophic environments (Ming et al., 2020; Guo et al., 2022), Nitrososphaera is more commonly found in areas with arsenic contamination and high salinity compared to Nitrosopumilus. This suggests that Nitrososphaera exhibits higher resistance to extreme conditions (Li et al., 2013, 2014; Liu et al., 2018; Zhu et al., 2022).
This study provides compelling evidence showcasing the superior adaption of ammonia-oxidizing archaea to oligotrophic subsurface karst caves. Dominant AOA OTUs exhibited broader ecological niche indexes compared to dominant AOB OTUs. AOA dominated ammonia-oxidizing communities with higher amoA gene copy numbers and significant contribution (48–100%) to potential nitrification rate. Deterministic processes dominate the ecological processes for the establishment of AOA communities, and TOC and NH4+ were identified as the primary environmental influence on AOA community assembly. In contrast, AOB is mainly governed by stochastic processes. Ammonia-oxidizing archaea contributed more nodes in the co-occurrence networks with ammonia-oxidizing bacteria and they collaborated more with AOB to survive the extreme conditions. These findings deepen our understanding of the ecology of ammonia-oxidizing microorganisms and nitrogen cycles in the subsurface biosphere.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.biosino.org/node/run/detail/OER444534, OER444534 for AOA and https://www.biosino.org/node/run/detail/OER445462, OER445462 for AOB.
QL: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Validation. XC: Data curation, Formal analysis, Investigation, Writing – review & editing. XL: Formal analysis, Investigation, Writing – review & editing. PG: Data curation, Validation, Writing – review & editing. HW: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing. CS: Investigation, Resources, Writing – review & editing. QH: Investigation, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (91951208).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1377721/full#supplementary-material
1. ^ https://github.com/torognes/vsearch/wiki/VSEARCH-pipeline
Aigle, A., Prosser, J. I., and Gubry-Rangin, C. (2019). The application of high-throughput sequencing technology to analysis of amoA phylogeny and environmental niche specialisation of terrestrial bacterial ammonia-oxidisers. Environ. Microb. 14:3. doi: 10.1186/s40793-019-0342-6
Allison, S. M., and Prosser, J. I. (1993). Ammonia oxidation at low ph by attached populations of nitrifying bacteria. Soil Biol. Biochem. 25, 935–941. doi: 10.1016/0038-0717(93)90096-t
Barton, H. A. (2013). “6.5 Biospeleogenesis” in Treatise on geomorphology. ed. J. F. Shroder (San Diego: Academic Press), 38–56.
Bastian, M., Heymann, S., and Jacomy, M. (2009). Gephi, An open source software for exploring and manipulating networks. Proc. Int. AAAI Confer. Web Soc. Media 3, 361–362. doi: 10.1609/icwsm.v3i1.13937
Bayer, B., Vojvoda, J., Reinthaler, T., Reyes, C., Pinto, M., and Herndl, G. J. (2019). Nitrosopumilus adriaticus sp. nov. and Nitrosopumilus piranensis sp. nov., two ammonia-oxidizing archaea from the Adriatic Sea and members of the class Nitrososphaeria. Int. J. Syst. Evol. Microbiol. 69, 1892–1902. doi: 10.1099/ijsem.0.003360
Beman, J. M., Popp, B. N., and Alford, S. E. (2012). Quantification of ammonia oxidation rates and ammonia-oxidizing archaea and bacteria at high resolution in the Gulf of California and eastern tropical North Pacific Ocean. Limnol. Oceanogr. 57, 711–726. doi: 10.4319/lo.2012.57.3.0711
Cao, J., Cheng, X. Y., Zeng, Z. L., Yang, Z. Q., Liu, X. Y., Wang, H. M., et al. (2021). Habitat specificity and co-occurrence network of bacterial communities in the Xincuntun cave, Guilin. Chin. Sci. Bull. 66, 4003–4016. doi: 10.1360/tb-2021-0021
Cardarelli, E. L., Bargar, J. R., and Francis, C. A. (2020). Diverse Thaumarchaeota dominate subsurface ammonia-oxidizing communities in semi-arid floodplains in the western United States. Microb. Ecol. 80, 778–792. doi: 10.1007/s00248-020-01534-5
Chelius, M. K., and Moore, J. C. (2004). Molecular phylogenetic analysis of archaea and bacteria in wind cave South Dakota. Geomicrobiol. J. 21, 123–134. doi: 10.1080/01490450490266389
Chen, C. H., Liang, H., and Gao, D. W. (2019). Community diversity and distribution of ammonia-oxidizing archaea in marsh wetlands in the black soil zone in north-East China. Front. Environ. Sci. Eng. 13:58. doi: 10.1007/s11783-019-1146-z
Cheng, X. Y., Liu, X. Y., Wang, H. M., Su, C. T., Zhao, R., Bodelier, P. L. E., et al. (2021). USCγ dominated community composition and co-occurrence network of methanotrophs and bacteria in subterranean karst caves. Microbiol. Spectr. 9:1. doi: 10.1128/Spectrum.00820-21
Cheng, Y., Wang, J., Chang, S. X., Cai, Z. C., Muller, C., and Zhang, J. B. (2019). Nitrogen deposition affects both net and gross soil nitrogen transformations in forest ecosystems. A Rev. Environ. Pollut. 244, 608–616. doi: 10.1016/j.envpol.2018.10.054
Cheng, X. Y., Wang, H. M., Zeng, Z. L., Li, L., Zhao, R., Bodelier, P. L. E., et al. (2022). Niche differentiation of atmospheric methane-oxidizing bacteria and their community assembly in subsurface karst caves. Environ. Microbiol. Rep. 14, 886–896. doi: 10.1111/1758-2229.13112
Cheng, X. Y., Xiang, X., Yun, Y., Wang, W. Q., Wang, H. M., and Bodelier, P. L. E. (2023). Archaea and their interactions with bacteria in a karst ecosystem. Front. Microbiol. 14:1068595. doi: 10.3389/fmicb.2023.1068595
Dai, L. L., Liu, C. Q., Yu, L. Q., Song, C. F., Peng, L., Li, X. L., et al. (2018). Organic matter regulates ammonia-oxidizing bacterial and archaeal communities in the surface sediments of Ctenopharyngodon idellus aquaculture ponds. Front. Microbiol. 9:2290. doi: 10.3389/fmicb.2018.02290
Dai, Y., Wu, Z., Zhou, Q. H., Zhao, Q., Li, N. N., Xie, S. G., et al. (2015). Activity, abundance and structure of ammonia-oxidizing microorganisms in plateau soils. Res. Microbiol. 166, 655–663. doi: 10.1016/j.resmic.2015.07.012
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
D'Amen, M., Mod, H. K., Gotelli, N. J., and Guisan, A. (2018). Disentangling biotic interactions, environmental filters, and dispersal limitation as drivers of species co-occurrence. Ecography 41, 1233–1244. doi: 10.1111/ecog.03148
Di, H. J., Cameron, K. C., Shen, J. P., Winefield, C. S., O'Callaghan, M., Bowatte, S., et al. (2009). Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat. Geosci. 2, 621–624. doi: 10.1038/ngeo613
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Dong, Y. Y., Gao, J., Wu, Q. S., Ai, Y. L., Huang, Y., Wei, W. Z., et al. (2020). Co-occurrence pattern and function prediction of bacterial community in karst cave. BMC Microbiol. 20:137. doi: 10.1186/s12866-020-01806-7
Emerson, K., Russo, R. C., Lund, R. E., and Thurston, R. V. (1975). Aqueous ammonia equilibrium calculations, effect of pH and temperature. J. Fish. Res. Board Can. 32, 2379–2383. doi: 10.1139/f75-274
Fang, W. K., Fan, T. Y., Wang, S., Yu, X. K., Lu, A. K., Wang, X. M., et al. (2023). Seasonal changes driving shifts in microbial community assembly and species coexistence in an urban river. Sci. Total Environ. 905:167027. doi: 10.1016/j.scitotenv.2023.167027
Fang, J., Lyu, T., Liu, J., He, S., Yang, X., Dou, H., et al. (2023). Response of nitrogen cycling and related microorganisms to brackish wetlands formed by evapotranspiration. Pedosphere 34, 252–266. doi: 10.1016/j.pedsph.2023.07.007
Ferrenberg, S., Martinez, A. S., and Faist, A. M. (2016). Aboveground and belowground arthropods experience different relative influences of stochastic versus deterministic community assembly processes following disturbance. Peerj 4:e2545. doi: 10.7717/peerj.2545
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 102, 14683–14688. doi: 10.1073/pnas.0506625102
French, E., Kozlowski, J. A., and Bollmann, A. (2021). Competition between Ammonia-oxidizing Archaea and Bacteria from freshwater environments. Appl. Environ. Microbiol. 87:e0103821. doi: 10.1128/aem.01038-21
Gao, J. F., Fan, X. Y., Wu, G. X., Li, T., and Pan, K. L. (2016). Changes of abundance and diversity of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in three nitrifying bioreactors with different ammonia concentrations. Desalin. Water Treat. 57, 21463–21475. doi: 10.1080/19443994.2015.1123196
Grobbelaar, J. U. (2000). Lithophytic algae, a major threat to the karst formation of show caves. J. Appl. Phycol. 12, 309–315. doi: 10.1023/A:1008172227611
Guo, J., Xuan, F. Q., Li, D. M., Wang, J. Q., and Zhang, B. C. A. (2022). Variations of soil chemical properties and microbial community around the acid reservoir in the mining area. Sustain. For. 14:10746. doi: 10.3390/su141710746
Hatzenpichler, R., Lebedeva, E. V., Spieck, E., Stoecker, K., Richter, A., Daims, H., et al. (2008). A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. USA 105, 2134–2139. doi: 10.1073/pnas.0708857105
He, X. J., Li, S., and Wu, F. Z. (2021). Responses of Ammonia-oxidizing microorganisms to intercropping Systems in Different Seasons. Agriculture 11:3. doi: 10.3390/agriculture11030195
He, Z. Y., Shen, J. P., Zhang, L. M., Tian, H. J., Han, B., Di, H. J., et al. (2020). DNA stable isotope probing revealed no incorporation of 13CO2 into comammox Nitrospira but ammonia-oxidizing archaea in a subtropical acid soil. J. Soils Sediments 20, 1297–1308. doi: 10.1007/s11368-019-02540-y
He, J., Shen, J., Zhang, L., Zhu, Y. G., Zheng, Y., Xu, M., et al. (2007). Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ. Microbiol. 9, 2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x
Heiss, E. M., Zawacki, V. W., Williams, A. A., Reed, M. H., Maguire, T. J., and Newell, S. E. (2022). Ammonia-oxidizing archaea and ammonium concentration as drivers of nitrification in a protected freshwater lake. Freshw. Sci. 41, 564–576. doi: 10.1086/721921
Hollibaugh, J. T., Bano, N., and Ducklow, H. W. (2002). Widespread distribution in polar oceans of a 16S rRNA gene sequence with affinity to Nitrosospira-like ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 68, 1478–1484. doi: 10.1128/aem.68.3.1478-1484.2002
Hu, X. Y., Huang, Y. J., Gu, G. K., Hu, H. J., Yan, H. Z., Zhang, H. J., et al. (2023). Distinct patterns of distribution, community assembly and cross-domain co-occurrence of planktonic archaea in four major estuaries of China. Environ. Microb. 18:75. doi: 10.1186/s40793-023-00530-9
Jiang, Q. Q., and Bakken, L. R. (1999). Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol. Ecol. 30, 171–186. doi: 10.1016/s0168-6496(99)00054-9
Jiao, S., Liu, Z. S., Lin, Y. B., Yang, J., Chen, W. M., and Wei, G. H. (2016). Bacterial communities in oil contaminated soils, biogeography and co-occurrence patterns. Soil Biol. Biochem. 98, 64–73. doi: 10.1016/j.soilbio.2016.04.005
Jin, T., Zhang, T., Ye, L., Lee, O. O., Wong, Y. H., and Qian, P. Y. (2011). Diversity and quantity of ammonia-oxidizing archaea and bacteria in sediment of the Pearl River estuary, China. Appl. Microbiol. Biotechnol. 90, 1137–1145. doi: 10.1007/s00253-011-3107-8
Jones, R. M., Goordial, J. M., and Orcutt, B. N. (2018). Low energy subsurface environments as extraterrestrial analogs. Front. Microbiol. 9:1605. doi: 10.3389/fmicb.2018.01605
Jones, C. M., and Hallin, S. (2019). Geospatial variation in co-occurrence networks of nitrifying microbial guilds. Mol. Ecol. 28, 293–306. doi: 10.1111/mec.14893
Jones, D. S., and Northup, D. E. (2021). Cave decorating with microbes, geomicrobiology of caves. Elements 17, 107–112. doi: 10.2138/gselements.17.2.107
Jung, M. Y., Sedlacek, C. J., Kits, K. D., Mueller, A. J., Rhee, S. K., Hink, L., et al. (2022). Ammonia-oxidizing archaea possess a wide range of cellular ammonia affinities. ISME J. 16, 272–283. doi: 10.1038/s41396-021-01064-z
Kandeler, E., and Gerber, H. (1988). Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fertil. Soils 6, 68–72. doi: 10.1007/BF00257924
Keerio, H. A., Bae, W., Park, J., and Kim, M. (2020). Substrate uptake, loss, and reserve in ammonia-oxidizing bacteria (AOB) under different substrate availabilities. Process Biochem. 91, 303–310. doi: 10.1016/j.procbio.2019.12.023
Khanom, A., Azad, M. A., Ali, M., Ali, M. Y., Biswas, S. K., and Rahman, M. M. (2021). Plants and microbes' responses to the net nitrification rates of chemical fertilizers in soils. Appl. Soil Ecol. 158:103783. doi: 10.1016/j.apsoil.2020.103783
Kobayashi, S., Hira, D., Yoshida, K., Toyofuku, M., Shida, Y., Ogasawara, W., et al. (2018). Nitric oxide production from nitrite reduction and hydroxylamine oxidation by copper-containing dissimilatory nitrite reductase (NirK) from the aerobic ammonia-oxidizing archaeon, Nitrososphaera viennensis. Microbes Environ. 33, 428–434. doi: 10.1264/jsme2.ME18058
Kou, Y. P., Li, C. N., Tu, B., Li, J. B., and Li, X. Z. (2023). The responses of ammonia-oxidizing microorganisms to different environmental factors determine their elevational distribution and assembly patterns. Microb. Ecol. 86, 485–496. doi: 10.1007/s00248-022-02076-8
Kozlowski, J. A., Stieglmeier, M., Schleper, C., Klotz, M. G., and Stein, L. Y. (2016). Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J. 10, 1836–1845. doi: 10.1038/ismej.2016.2
Lebedeva, E. V., Zherebyateva, T. V., Soina, V. S., Pommerening-Roser, A., Duske, D., Moller, U. C., et al. (1997). Taxonomic position of ammonia-oxidizing bacteria similar in their morphology to both Nitrosovibrio and Nitrosospira. Microbiology 66, 320–323. doi: 10.1007/0-387-30745-1_36
Li, Y. Y., Chapman, S. J., Nicol, G. W., and Yao, H. Y. (2018). Nitrification and nitrifiers in acidic soils. Soil Biol. Biochem. 116, 290–301. doi: 10.1016/j.soilbio.2017.10.023
Li, P., Jiang, D. W., Li, B., Dai, X. Y., Wang, Y. H., Jiang, Z., et al. (2014). Comparative survey of bacterial and archaeal communities in high arsenic shallow aquifers using 454 pyrosequencing and traditional methods. Ecotoxicology 23, 1878–1889. doi: 10.1007/s10646-014-1316-5
Li, P., Wang, Y. H., Jiang, Z., Jiang, H. C., Li, B., Dong, H. L., et al. (2013). Microbial diversity in high arsenic groundwater in hetao basin of Inner Mongolia, China. Geomicrobiol J. 30, 897–909. doi: 10.1080/01490451.2013.791354
Li, Y. Y., Xi, R. J., Wang, W. J., and Yao, H. Y. (2019). The relative contribution of nitrifiers to autotrophic nitrification across a pH-gradient in a vegetable cropped soil. J. Soils Sediments 19, 1416–1426. doi: 10.1007/s11368-018-2109-x
Liu, J. J., Li, C., Ma, W. D., Wu, Z. X., Liu, W., and Wu, W. X. (2023). Exploitation alters microbial community and its co-occurrence patterns in ionic rare earth mining sites. Sci. Total Environ. 898:165532. doi: 10.1016/j.scitotenv.2023.165532
Liu, Y. Y., Liu, J. W., Yao, P., Ge, T. T., Qiao, Y. L., Zhao, M. X., et al. (2018). Distribution patterns of ammonia-oxidizing archaea and bacteria in sediments of the eastern China marginal seas. Syst. Appl. Microbiol. 41, 658–668. doi: 10.1016/j.syapm.2018.08.008
Liu, H. Y., Qin, S. Y., Li, Y., Zhao, P., Nie, Z. J., and Liu, H. E. (2023). Comammox Nitrospira and AOB communities are more sensitive than AOA community to different fertilization strategies in a fluvo-aquic soil. Agric. Ecosyst. Environ. 342:108224. doi: 10.1016/j.agee.2022.108224
Liu, R., Suter, H., He, J. Z., Hayden, H., and Chen, D. (2015). Influence of temperature and moisture on the relative contributions of heterotrophic and autotrophic nitrification to gross nitrification in an acid cropping soil. J. Soils Sediments 15, 2304–2309. doi: 10.1007/s11368-015-1170-y
Liu, X. Y., Wang, H. M., Wang, W. Q., Cheng, X. Y., Wang, Y. H., Li, Q., et al. (2023). Nitrate determines the bacterial habitat specialization and impacts microbial functions in a subsurface karst cave. Front. Microbiol. 14:1115449. doi: 10.3389/fmicb.2023.1115449
Luvizott, D. M., Araujo, J. E., Silva, M. D. P., Dias, A. C. F., Kraft, B., Tegetmeye, H., et al. (2019). The rates and players of denitrification, dissimilatory nitrate reduction to ammonia (DNRA) and anaerobic ammonia oxidation (anammox) in mangrove soils. An Acad. Bras. Cienc. 91:e20180373. doi: 10.1590/0001-3765201820180373
Ma, M. C., Zhao, Y. B., Jiang, X., Guan, D. W., Yuan, M., Cao, F. M., et al. (2023). Fertilization altered co-occurrence patterns and microbial assembly process of ammonia-oxidizing microorganisms. Sci. Rep. 13:8234. doi: 10.1038/s41598-022-26293-w
Magnabosco, C., Lin, L. H., Dong, H., Bomberg, M., Ghiorse, W., Stan-Lotter, H., et al. (2018). The biomass and biodiversity of the continental subsurface. Nat. Geosci. 11, 707–717. doi: 10.1038/s41561-018-0221-6
Marques, E. L. S., Dias, J. C. T., Gross, E., Silva, A., de Moura, S. R., and Rezende, R. P. (2019). Purple sulfur bacteria dominate microbial community in Brazilian limestone cave. Microorganisms 7:29. doi: 10.3390/microorganisms7020029
Marques, E. D. S., Gross, E., Dias, J. C. T., Pirovani, C. P., and Rezende, R. P. (2018). Ammonia oxidation (amoA) and nitrogen fixation (nifH) genes along metasandstone and limestone caves of Brazil. Geomicrobiol J. 35, 869–878. doi: 10.1080/01490451.2018.1482386
Martens-Habbena, W., Berube, P. M., Urakawa, H., de la Torre, J. R., and Stahl, D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979. doi: 10.1038/nature08465
Martorell, C., Almanza-Celis, C. A. I., Pérez-García, E. A., and Sánchez-Ken, J. G. (2015). Co-existence in a species-rich grassland, competition, facilitation and niche structure over a soil depth gradient. J. Veg. Sci. 26, 674–685. doi: 10.1111/jvs.12283
Ming, H. X., Zhang, H. Z., Chen, Q. R., Wang, Y. T., Su, J., Zhao, X. H., et al. (2020). Abundance and community structure of ammonium monooxygenase (amoA) genes in the wet season of liaohe estuary sediments. Cont. Shelf Res. 209:104253. doi: 10.1016/j.csr.2020.104253
Mulec, J. (2008). Microorganisms in hypogeon, examples from Slovenian karst caves. Acta Carsologica 37, 153–160. doi: 10.3986/ac.v37i1.167
Ning, D. L., Yuan, M. T., Wu, L. W., Zhang, Y., Guo, X., Zhou, X. S., et al. (2020). A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 11:4717. doi: 10.1038/s41467-020-18560-z
Ouyang, Y., Norton, J. M., and Stark, J. M. (2017). Ammonium availability and temperature control contributions of ammonia oxidizing bacteria and archaea to nitrification in an agricultural soil. Soil Biol. Biochem. 113, 161–172. doi: 10.1016/j.soilbio.2017.06.010
Pannek, A., Manthey, M., and Diekmann, M. (2016). Comparing resource-based and co-occurrence-based methods for estimating species niche breadth. J. Veg. Sci. 27, 596–605. doi: 10.1111/jvs.12374
Park, S. J., Park, B. J., and Rhee, S. K. (2008). Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments. Extremophiles 12, 605–615. doi: 10.1007/s00792-008-0165-7
Patil, P. K., Baskaran, V., Vinay, T. N., Avunje, S., Leo-Antony, M., Shekhar, M. S., et al. (2021). Abundance, community structure and diversity of nitrifying bacterial enrichments from low and high saline brackishwater environments. Lett. Appl. Microbiol. 73, 96–106. doi: 10.1111/lam.13480
Pester, M., Rattei, T., Flechl, S., Grongroft, A., Richter, A., Overmann, J., et al. (2012). amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ. Microbiol. 14, 525–539. doi: 10.1111/j.1462-2920.2011.02666.x
Pester, M., Schleper, C., and Wagner, M. (2011). The Thaumarchaeota, an emerging view of their phylogeny and ecophysiology. Curr. Opin. Microbiol. 14, 300–306. doi: 10.1016/j.mib.2011.04.007
Phillips, L. A., Schefe, C. R., Fridman, M., O'Halloran, N., Armstrong, R. D., and Mele, P. M. (2015). Organic nitrogen cycling microbial communities are abundant in a dry Australian agricultural soil. Soil Biol. Biochem. 86, 201–211. doi: 10.1016/j.soilbio.2015.04.004
Qin, W., Heal, K. R., Ramdasi, R., Kobelt, J. N., Martens-Habbena, W., Bertagnolli, A. D., et al. (2017). Nitrosopumilus maritimus gen. Nov., sp. nov., Nitrosopumilus cobalaminigenes sp. nov., Nitrosopumilus oxyclinae sp. nov., and Nitrosopumilus ureiphilus sp. nov., four marine ammonia-oxidizing archaea of the phylum Thaumarchaeota. Int. J. Syst. Evol. Microbiol. 67, 5067–5079. doi: 10.1099/ijsem.0.002416
Ren, B. J., Shen, L. D., Liu, X., Jin, J. H., Huang, H. C., Yang, Y. L., et al. (2023). Effect of gradual increase of atmospheric CO2 concentration on nitrification potential and communities of ammonia oxidizers in paddy fields. J. Environ. Manag. 325:116597. doi: 10.1016/j.jenvman.2022.116597
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker, molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997
Roy, D., McEvoy, J., and Khan, E. (2020). Abundance and activity of ammonia oxidizing archaea and bacteria in bulk water and biofilm in water supply systems practicing chlorination and chloramination, full and laboratory scale investigations. Sci. Total Environ. 715:137043. doi: 10.1016/j.scitotenv.2020.137043
Rui, J., Li, J., Wang, S., An, J., Liu, W.-t., Lin, Q., et al. (2015). Responses of bacterial communities to simulated climate changes in alpine meadow soil of the Qinghai-Tibet plateau. Appl. Environ. Microbiol. 81, 6070–6077. doi: 10.1128/AEM.00557-15
Scott, D. B., Van Dyke, M. I., Anderson, W. B., and Huck, P. M. (2015). Influence of water quality on nitrifier regrowth in two full-scale drinking water distribution systems. Can. J. Microbiol. 61, 965–976. doi: 10.1139/cjm-2015-0375
Sebela, S., Miler, M., Skobe, S., Torkar, S., and Zupancic, N. (2015). Characterization of black deposits in karst caves, examples from Slovenia. Facies 61:6. doi: 10.1007/s10347-015-0430-z
Shen, D. J., Qian, H., Liu, Y. R., Zhao, S. M., and Luo, X. S. (2022). Nitrifier community assembly and species co-existence in forest and meadow soils across four sites in a temperate to tropical region. Appl. Soil Ecol. 171:104342. doi: 10.1016/j.apsoil.2021.104342
Spear, J. R., Barton, H. A., Robertson, C. E., Francis, C. A., and Pace, N. R. (2007). Microbial community biofabrics in a geothermal mine adit. Appl. Environ. Microbiol. 73, 6172–6180. doi: 10.1128/aem.00393-07
Stegen, J. C., Lin, X. J., Konopka, A. E., and Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664. doi: 10.1038/ismej.2012.22
Stieglmeier, M., Klingl, A., Alves, R. J. E., Rittmann, S., Melcher, M., Leisch, N., et al. (2014). Nitrososphaera viennensis gen. Nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int. J. Syst. Evol. Microbiol. 64, 2738–2752. doi: 10.1099/ijs.0.063172-0
Taylor, A. E., Zeglin, L. H., Dooley, S., Myrold, D. D., and Bottomley, P. J. (2010). Evidence for different contributions of archaea and bacteria to the ammonia-oxidizing potential of diverse Oregon soils. Appl. Environ. Microbiol. 76, 7691–7698. doi: 10.1128/aem.01324-10
Tetu, S. G., Breakwell, K., Elbourne, L. D. H., Holmes, A. J., Gillings, M. R., and Paulsen, I. T. (2013). Life in the dark, metagenomic evidence that a microbial slime community is driven by inorganic nitrogen metabolism. ISEM J. 7, 1227–1236. doi: 10.1038/ismej.2013.14
Tourna, M., Stieglmeier, M., Spang, A., Könneke, M., Schintlmeister, A., Urich, T., et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. USA 108, 8420–8425. doi: 10.1073/pnas.1013488108
Veresoglou, S. D., Chen, B. D., and Rillig, M. C. (2012). Arbuscular mycorrhiza and soil nitrogen cycling. Soil Biol. Biochem. 46, 53–62. doi: 10.1016/j.soilbio.2011.11.018
Vorobeichik, E. L. (1993). On some indexes of ecological niche breadth and overlap. Zhurnal Obshchei Biologii 54, 706–712.
Wang, Z. H., Meng, Y., Zhu-Barker, X., He, X. H., Horwath, W. R., Luo, H. Y., et al. (2019). Responses of nitrification and ammonia oxidizers to a range of background and adjusted pH in purple soils. Geoderma 334, 9–14. doi: 10.1016/j.geoderma.2018.07.038
Wang, H., Proctor, C. R., Edwards, M. A., Pryor, M., Domingo, J. W. S., Ryu, H., et al. (2014). Microbial community response to chlorine conversion in a chloraminated drinking water distribution system. Environ. Sci. Technol. 48, 10624–10633. doi: 10.1021/es502646d
Wang, D. D., Sheng, K., Zhao, W. D., Li, L. T., Zhang, Q., and Wang, Y. L. (2021). Dominance of archaeal ammonia-oxidizers in soil nitrification across different soil types and fertilities in North China plain. Eur. J. Soil Biol. 106:103354. doi: 10.1016/j.ejsobi.2021.103354
Wang, Y., Zhu, K., Chen, X., Wei, K. Y., Wu, R., and Wang, G. (2024). Stochastic and deterministic assembly processes of bacterial communities in different soil aggregates. Appl. Soil Ecol. 193:105153. doi: 10.1016/j.apsoil.2023.105153
Wei, J., Dong, X. Y., Liu, S. T., and Lin, X. B. (2023). Editorial, advances in estuarine and coastal nitrogen cycle. Front. Mar. Sci. 10:1130839. doi: 10.3389/fmars.2023.1130839
Wei, D., Zeng, S., Hou, D., Zhou, R., Xing, C., Deng, X., et al. (2021). Community diversity and abundance of ammonia-oxidizing archaea and bacteria in shrimp pond sediment at different culture stages. J. Appl. Microbiol. 130, 1442–1455. doi: 10.1111/jam.14846
Weiner, A. M., and Maizels, N. (1999). Evolution - a deadly double life. Science 284, 63–64. doi: 10.1126/science.284.5411.63
Wu, W. X., Xu, Z. M., Dai, M. H., Gan, J. P., and Liu, H. B. (2021). Homogeneous selection shapes free-living and particle-associated bacterial communities in subtropical coastal waters. Divers. Distrib. 27, 1904–1917. doi: 10.1111/ddi.13193
Xiang, X., Wang, R. C., Wang, H. M., Gong, L. F., Man, B. Y., and Xu, Y. (2017). Distribution of bathyarchaeota communities across different terrestrial settings and their potential ecological functions. Sci. Rep. 7:45028. doi: 10.1038/srep45028
Yang, X., You, L. C., Hu, H. W., and Chen, Y. L. (2022). Conversion of grassland to cropland altered soil nitrogen-related microbial communities at large scales. Sci. Total Environ. 816:151645. doi: 10.1016/j.scitotenv.2021.151645
Ye, F., Hong, Y. G., Yi, X. M., Sun, Z. H., Wu, J. P., and Wang, Y. (2023). Stochastic processes drive the soil fungal communities in a developing mid-channel bar. Front. Microbiol. 14:1104297. doi: 10.3389/fmicb.2023.1104297
Yu, Y. Y., Cui, M., Xiao, Y. T., Chang, M. Y., Wang, C., Zhao, L., et al. (2021). Quantitative estimation of stochastic and deterministic processes for soil prokaryotic community assembly in the Yellow River floodplain. Eur. J. Soil Sci. 72, 1462–1477. doi: 10.1111/ejss.13056
Yu, Y., Shi, W. J., and Yang, N. F. (2007). Study and spectrophotometric determination of nitrite with sulfanilamide and N-phenyl J-acid system. Spectrosc. Spectr. Anal. 27, 573–576.
Yuan, H. Y., Mei, R., Liao, J. H., and Liu, W. T. (2019). Nexus of stochastic and deterministic processes on microbial community assembly in biological systems. Front. Microbiol. 10:01536. doi: 10.3389/fmicb.2019.01536
Yun, Y., Wang, H. M., Man, B. Y., Xiang, X., Zhou, J. P., Qiu, X., et al. (2016). The relationship between pH and bacterial communities in a single karst ecosystem and its implication for soil acidification. Front. Microbiol. 7:01955. doi: 10.3389/fmicb.2016.01955
Zhang, L. M., Hu, H. W., Shen, J. P., and He, J. Z. (2012). Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 6, 1032–1045. doi: 10.1038/ismej.2011.168
Zhang, X., Wang, Y. L., Xu, Y., Babalola, B. J., Xiang, S. M., Ma, J. J., et al. (2023). Stochastic processes dominate community assembly of ectomycorrhizal fungi associated with Picea crassifolia in the Helan Mountains, China. Front. Microbiol. 13:1061819. doi: 10.3389/fmicb.2022.1061819
Zhang, S. J., Zeng, Y. H., Zhu, J. M., Cai, Z. H., and Zhou, J. (2022). The structure and assembly mechanisms of plastisphere microbial community in natural marine environment. J. Hazard. Mater. 421:126780. doi: 10.1016/j.jhazmat.2021.126780
Zhang, Z. F., Zhao, P., and Cai, L. (2018). Origin of cave Fungi. Front. Microbiol. 9:01407. doi: 10.3389/fmicb.2018.01407
Zhang, Y. J., Zhou, L. L., Zeng, G., Deng, H. P., and Li, G. B. (2010). Impact of total organic carbon and chlorine to ammonia ratio on nitrification in a bench-scale drinking water distribution system. Front. Environ. Sci. Eng. China. 4, 430–437. doi: 10.1007/s11783-010-0247-5
Zhao, P. Y., Bao, J. B., Wang, X., Liu, Y., Li, C., and Chai, B. F. (2019a). Deterministic processes dominate soil microbial community assembly in subalpine coniferous forests on the loess plateau. Peer J. 7:e6746. doi: 10.7717/peerj.6746
Zhao, R., Wang, H. M., Yang, H., Yun, Y., and Barton, H. A. (2017). Ammonia-oxidizing archaea dominate ammonia-oxidizing communities within alkaline cave sediments. Geomicrobiol J. 34, 511–523. doi: 10.1080/01490451.2016.1225861
Zhao, H. C., and Zhang, L. Q. (2022). Metagenome-assembled genomes of six ammonia-oxidizing archaea (AOA) from agricultural upland soil. Microbes Environ. 37:3. doi: 10.1264/jsme2.ME22035
Zhao, P. Y., Liu, J. X., Jia, T., Luo, Z. M., Li, C., and Chai, B. F. (2019b). Assembly mechanisms of soil bacterial communities in subalpine coniferous forests on the Loess Plateau, China. J. Microbiol. 57, 461–469. doi: 10.1007/s12275-019-8373-7
Zheng, J., Tao, L., Dini-Andreote, F., Luan, L., Kong, P. J., Xue, J. R., et al. (2022). Dynamic responses of ammonia-oxidizing archaea and bacteria populations to organic material amendments affect soil nitrification and nitrogen sse efficiency. Front. Microbiol. 13:911799. doi: 10.3389/fmicb.2022.911799
Zhou, J. Z., and Ning, D. L. (2017). Stochastic community assembly, does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81:4. doi: 10.1128/mmbr.00002-17
Zhu, X. F., Wang, C., Tang, S. Y., and Ji, G. D. (2023). Quantitative responses of active ammonia-oxidizing archaea and bacteria to the biological and abiotic factors across functional gene distribution in coastal wetlands. Wetlands 43:1. doi: 10.1007/s13157-022-01650-7
Keywords: ammonia-oxidizing community, nitrification, karst cave, niche breadth, subsurface biosphere
Citation: Li Q, Cheng X, Liu X, Gao P, Wang H, Su C and Huang Q (2024) Ammonia-oxidizing archaea adapted better to the dark, alkaline oligotrophic karst cave than their bacterial counterparts. Front. Microbiol. 15:1377721. doi: 10.3389/fmicb.2024.1377721
Received: 28 January 2024; Accepted: 28 March 2024;
Published: 10 April 2024.
Edited by:
Zhanfei He, Zhejiang University of Technology, ChinaReviewed by:
Diana Eleanor Northup, University of New Mexico, United StatesCopyright © 2024 Li, Cheng, Liu, Gao, Wang, Su and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongmei Wang, d2FuZ2htZWkwNEAxNjMuY29t; aG13YW5nQGN1Zy5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.