
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 08 April 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1376620
Staphylococcus species are the primary cause of mastitis in dairy cows across the world. Staphylococcus aureus has recently become a pathogen that is zoonotic and multidrug resistant. This study aimed to sequence whole genomes of 38 S. aureus isolates from 55 subclinical mastitis dairy cows of 7 small-scale farmers in the Free State Province, South Africa and document and their antimicrobial and virulence genes. The 38 isolates were grouped by the in silico multi-locus sequencing types (MLST) into seven sequence types (STs), that is (ST 97, 352, 152, 243) and three new STs (ST8495, ST8500, and ST8501). Thirty-three S. aureus isolates were divided into 7 core single-nucleotide polymorphism (SNP) clusters. Among the 9 distinct spa-types that were detected, Spa-types t2883 accounted for the majority of isolates at 12 (31.57%), followed by t416 with 11 (28.94%) and t2844 with 5 (13.15%). The data also revealed the identification of four (4) plasmids, with Rep_N (rep20) accounting for the majority of isolates with 17 (44.73%), followed by Inc18 (repUS5) with 2 (5.26%). These isolates included 11 distinct antimicrobial resistance genes and 23 genes linked to bacterial virulence. Surprisingly, no methicillin resistance associated genes were detected in these isolates. Genome data of the current study will contribute to understanding epidemiology S. aureus genotypes and ultimately aid in developing treatment and control plans to stop the spread of mastitis in the Free State province and South Africa as a whole.
Staphylococcus species are known to cause acute to chronic infections/diseases that are related to increased morbidity to infected hosts such as humans and animals (Pattabhiramaiah and Mallikarjunaiah, 2023). There are about 53 species with 28 sub-species within the Staphylococcus genus with S. aureus being the main cause of persistent clinical and subclinical intramammary infections (IMI; Vanderhaeghen et al., 2015). The pathogenesis of S. aureus starts with teat colonization, through the intramammary space by either progressive colonization or changes in intramammary pressure caused by the milking machines (Maity et al., 2020; Vargová et al., 2023). In the mammary alveolus, S. aureus adheres to and enters mammary epithelial cells, which serve as the site for multiplication, eventually resulting in a chronic IMI (Maity et al., 2020). The molecular mechanisms underlying S. aureus IMI still need to be fully deciphered. Generally, bacteria sense host signals and adapt gene expression to match environmental conditions to cause infection (Elhawy et al., 2021). Numerous virulence factors (VFs) involved in adhesion, invasion, and host defense evasion are known and well-studied in S. aureus. These VFs are either found in bacterial genomes or are within the transmissible genetic elements in a bacterium (Naushad et al., 2019). The emergence of drug resistance is a serious challenge for mastitis control due to their extensive use in the dairy industry, for example, through the dry cow therapy, contributes to the emergence of antimicrobial determinants in S. aureus, including development of multiple drug resistance (Klibi et al., 2018; Liu et al., 2020; Omwenga et al., 2021; Gelalcha et al., 2022). Molecular epidemiology of staphylococcal species involved in IMI of dairy cattle have focused more on the use of multi-locus enzyme electrophoresis (MLEE), pulsed-field gel electrophoresis (PFGE), sequence-based typing schemes, such as multiple-locus sequence typing (MLST), multiple-locus VNTR (variable number of tandem repeats) analysis (MLVA), random amplification of polymorphic DNA (RAPD) analysis and staphylococcal protein A (spa) typing (Li, 2008). Although these methods are helpful in genetic analysis, their resolution is often not strong enough to reveal genetic differences between strains. However, whole-genome sequencing (WGS) of bacterial genomes has become the preferred method to understand microevolution, phylogenies, and inter and intraspecies differences (Sivakumar et al., 2023). Thus, in the current study we utilized WGS to characterize and understand the virulence and antimicrobial resistance mechanisms in S. aureus isolates of subclinical mastitis (SCM) dairy cows from small-scale farmers of the Free State Province.
The S. aureus isolates used in the present study were all obtained from milk samples collected from seven (7) small scale farms in three local Municipalities (Maluti-A-Phofung, Mantsopa and Setsotso) in the Free State Province of South Africa between year 2021–2022. The sample collection was conducted according to the guidelines of National Mastitis Council (2004). A total of 166 composite milk samples from individual cows were randomly screened for intramammary infection by means of somatic cell count (SCC) assay using flow cytometry (Mérieux NutriSciences, South Africa). Thereafter, based on the SCC results, only 220 individual quarters from 55 of 166 cows were subjected to California mastitis test (CMT) according to manufacturer’s instructions (DeLaval, South Africa) on farm and subsequently only 160 quarter milk samples were collected for another round of SCC and microbiological analysis. The CMT results were scored and interpreted as recommended by Karzis et al. (2017). Isolates were defined as S. aureus on the basis of being gram-positive cocci and catalase positive. Thereafter, matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) and gene sequencing were employed for further identification of the isolates as reported in our previous study (Ozbey et al., 2022; Khasapane et al., 2024). Furthermore, we performed phenotypic and genomic antimicrobial resistance based on disk-diffusion and PCR techniques (Khasapane et al., 2024). To evaluate the susceptibility of Staphylococcus isolates to widely used antimicrobial drugs, the single disk diffusion technique was utilized. Antibiotic discs (ThermoFischer, South Africa) comprising of gentamicin (10 μg), ampicillin (10 μg), tetracycline (30 μg), penicillin (10 μg), erythromycin (15 μg), ciprofloxacin (5 μg), and cefoxitin (15 μg) were utilized according to the Clinical Laboratory Standards Institute (CLSI, 2023) guidelines which are interpreted as intermediate (I), sensitive (S), and resistant (R).
Whole genome sequencing of S. aureus isolates was conducted at the National Institute of Communicable Diseases (NICD) Sequencing Core Facility, South Africa. Briefly, multiplexed, paired-end libraries (2 × 150 bp) were prepared using the Illumina DNA Prep kit (Illumina, San Diego, United States), followed by sequencing on the Illumina NextSeq 2000 platform (Illumina, San Diego, United States) at 100× coverage.
Illumina paired-end reads were analyzed using the JEKESA bioinformatics pipeline v1.01 including quality control, species identification and de novo assembly as previously described in Souvorov et al. (2018). Multilocus sequence typing (MLST) was performed using mlst v2.19.0 (--legacy -scheme saureus; Gurevich et al., 2013), based on traditional PubMLST typing schemes.2
Detection of antimicrobial resistance determinants was performed using a combination of three popular tools, namely, AMRFinderPlus (Feldgarden et al., 2021), ABRicate3 and staramr,4 by scanning the assembled contigs against ResFinder (Bortolaia et al., 2020), PointFinder5 and AMRFinderPlus databases. The outputs from these tools were summarized using HAMRonization.6
Core genome SNPs (single nucleotide polymorphisms) were used to investigate the phylogeny and genetic relatedness of isolates. Briefly, whole genome alignments were performed using scapper7 and Staphylococcus aureus strain NCTC 8325 was used as a reference. Recombinant regions were removed using Gubbins v3.2.1(Croucher et al., 2015) and variable sites were obtained using snp-sites v2.5.1 (Page et al., 2016). Pairwise SNP distances were calculated using snp-dist v0.8.28 and a normalized pairwise SNP distance matrix was used as input for the cluster analysis using the R software environment v4.2.1. Assignment of SNP clusters was achieved by a combination of K-means clustering implemented in the eclust function (factoextra package; v1.0.7; Kassambara, 2016) and custom functions written in R using a silhouette score and SNP cut-off of 0.5 and 20, respectively. Visualization of cluster heat maps was performed using the ComplexHeatmap package v.14.0 (Gu, 2022). IQ-TREE v2.0.3 (Minh et al., 2020) was used to generate a maximum-likelihood phylogenetic tree using the GTR + F + ASC + R4 with 1,000 bootstrap approximations using UFBoot2 (Hoang et al., 2018). The phylogenetic tree was visualized and annotated using Microreact.9
Pairwise SNP distances were calculated using snp-dist (v0.8.2; https://github.com/tseemann/snp-dists) using the final alignment file based on variable sites only. A normalized pairwise SNP distance matrix was used as input for the cluster analysis performed using the R software environment (version 4.2.1). Briefly, K-means clustering was done using default parameters in the eclust function from the factoextra package (v1.0.7; Kassambara, 2016), but with bootstrapping set to 500, and only core SNP clusters with a silhouette score ≥ 0.5 and SNP cut-off ≤ 25 were considered. Visualization of cluster heat maps was performed using the ComplexHeatmap package (v.14.0; Gu, 2022). A simple dendrogram showing the SNP clusters was generated using the fviz_dend function from the factoextra package. The minimum spanning tree was generated using the ape package in R (Paradis et al., 2004) based on pairwise SNP distances. Visualization of the minimum spanning tree was done using the visNetwork package (v2.2.2; https://datastorm-open.github.io/visNetwork/).
As reported in our previous study (Khasapane et al., 2024) the isolates showed resistance to penicillin 43/50 (86%), ciprofloxacin 40/50 (80%), vancomycin 39/50 (76%), and cefoxitin 26/50 (52%). Observed resistance against gentamycin, ampicillin, tetracycline, and erythromycin was 18/50 (36%), 14/50 (28%), 9/50 (18%), and 9/50 (18%), respectively.
In the current study, whole genomes of 38 S. aureus isolates associated with bovine subclinical mastitis from Free State Province smallholder farms were sequenced. Table 1 provides an overview of genomic sequences.
The in silico MLST clustered the 38 isolates into 7 sequence types (STs; ST 97, 352, 152, 243) and 3 novel STs (ST8495, ST8500, and ST8501; Figure 1). Majority were assigned to the ST8500 n = 14 (36.8%) sequencing type, followed by ST97 n = 12 (31.5%), ST352 n = 7 (18.4%), ST8501 n = 2 (5.2%), while the latter STs [ST8495, ST152 and ST243] each had n = 1 (2.6%) isolates. Furthermore, whole genome sequencing analysis further revealed nine different spa-types of S. aureus and four types of plasmids from all isolates. Of all the spa-types, the t2883 accounted for most isolates with n = 12 (31.57%) followed by t416 with n = 11 (28.94%) and t2844 with n = 5 (13.15%). Interestingly, the study found 1 unknown spa-type t21 when the sequences were submitted to Ridom spa server,10 which we concluded to be novel spa type. While on the other hand plasmid Rep_N (rep20) was found in most of the isolates with n = 17 (44.73%), followed by Inc18 (repUS5) with n = 2 (5.26%) lastly one isolate (2.65%) had both plasmid rep16 and rep5a (Table 1; Figure 2).
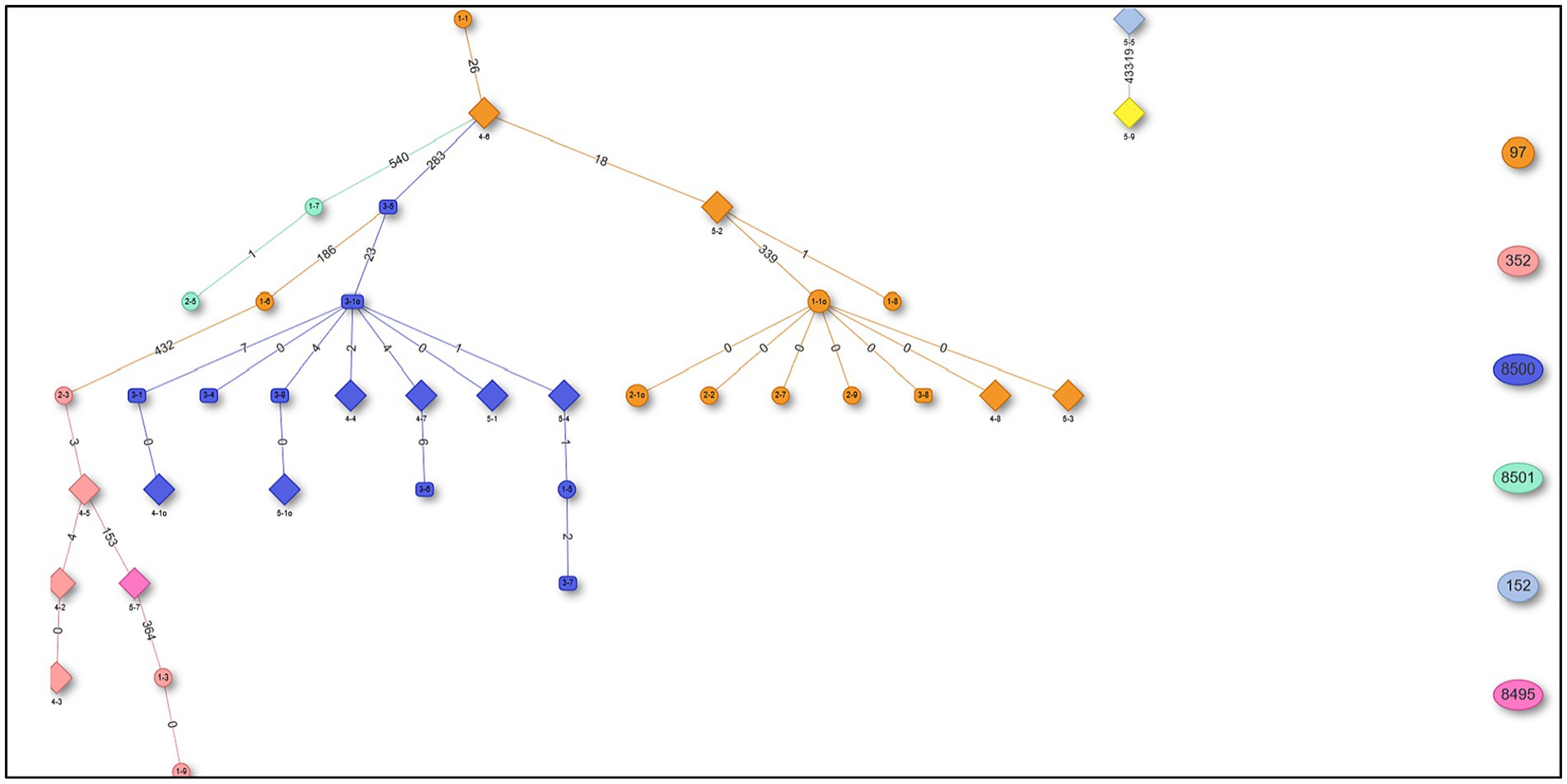
Figure 1. Minimum spanning tree (MST) showing sequencing types (STs) from 38 Staphylococcus aureus isolates.
All isolates belonging to ST8500, ST97, ST352, and ST8501 contained lmrS, mepA and tet (38) genes which indicates resistance to were resistant to macrolide/phenicol, efflux and tetracycline, respectively. On the other hand, 18 (47.36%) of the isolates belonging 204 to ST8500 (31.57%), ST97 (10.52%) and ST152 (2.63%) carried cadD gene encoding for resistance against cadmium. Finally, Fosfomycin (murA) encoding gene was carried by 2 (5.26%) isolates belonging to ST152 and ST243, respectively. Beta-lactam, ampicillin (blaI, blaR1, and blaZ), trimethoprim (dfrG and dfrG_1), and quinolone (parE) genes were carried by one isolate each (2.63%) belonging to ST152 (Figure 3).
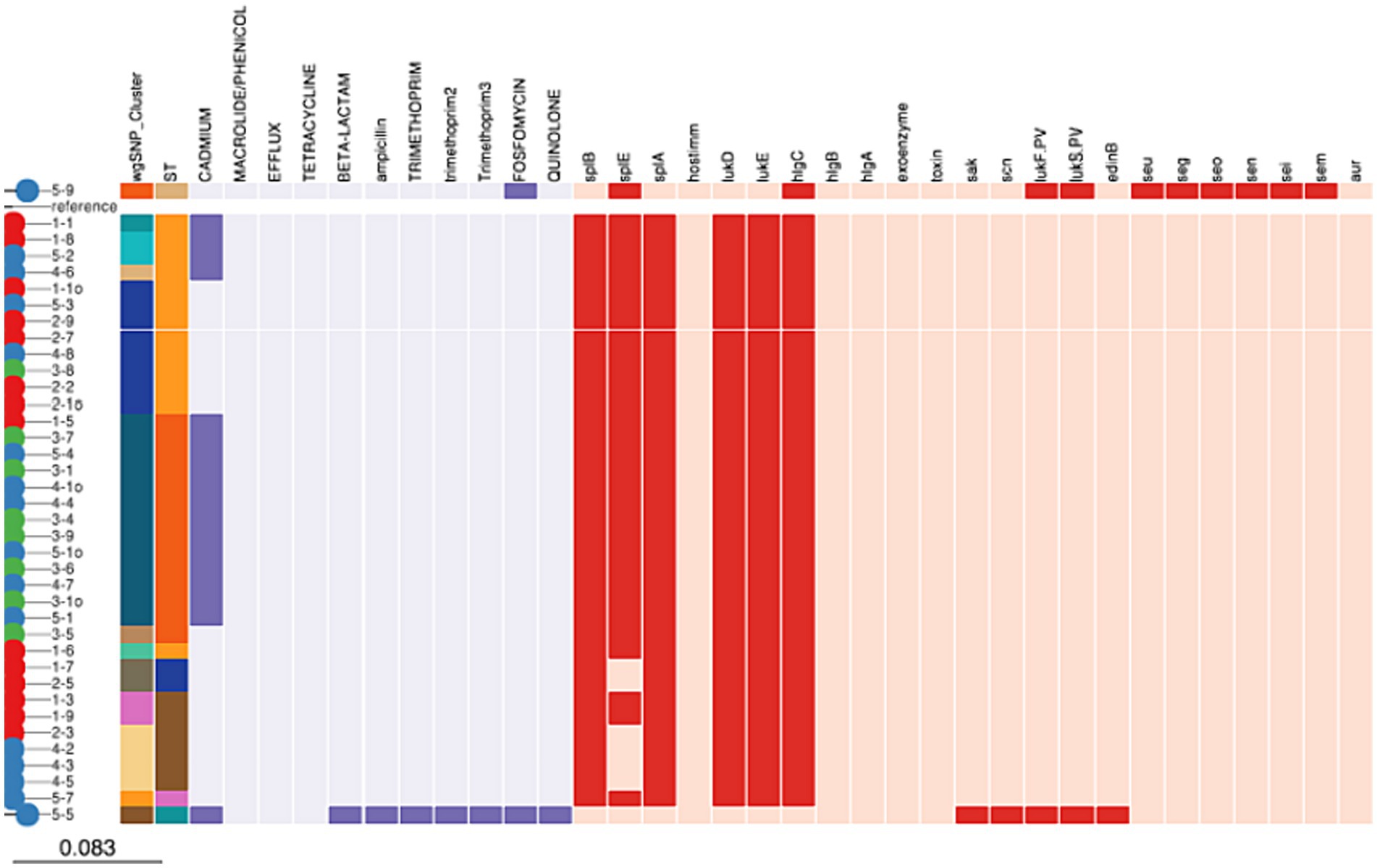
Figure 3. Distribution of antimicrobial resistance and virulence genes among Staphylococcus aureus isolates.
Our study further showed that all isolates (100%) belonging to ST8500, ST97, ST352, ST8501, ST243, ST152, and ST8495 carried aur, HlgA&B genes encoding for aureolysin factor, bi-component gamma hemolysin HlgAB subunit A and B, respectively. While 36 (94.7%) of the isolates belonging to ST8500, ST97, ST352, ST8500, and ST8495 carried LukD and E gene encoding for bi-component leukocidin LukE and D subunit D and E, respectively. Furthermore, splB, splE, and splA genes encoding for serine protease A, B, and E was carried by 36 (94.7%), 36 (94.7%) and 31 (81.5%) of all the isolates belonging to ST8500, ST97, ST152 and ST8495. Finally, 218 2 (5.26%) of the isolates contained LukF.PV and LukS.PV encoding for Panton-Valentine leucocidin only in ST152 and ST 243. While seu, seg, seo, sen, sei, sem, and edinB genes encoding for enterotoxins were detected in 1 (2.63%) of the isolates belonging to ST243 (Figure 3).
Numerous investigations have examined potential variations in virulence gene profiles between S. aureus isolates from clinical and subclinical mastitis; however, no discernible differences have been found (Rocha et al., 2019; Naushad et al., 2020; Åvall-Jääskeläinen et al., 2021). According to Wellnitz and Bruckmaier (2012), mastitis is a dynamic phenomenon in which the microbe enters the mammary gland, causing leukocytes in the milk and mammary-gland epithelial cells to react and start the immunologic defense process. This causes a large number of neutrophils to migrate to the mammary gland and attempt to kill the microbial cells. In this study, we examined the presence of S. aureus associated with subclinical mastitis in dairy cattle in addition to associated antimicrobial resistance and virulence factors in 38 isolates.
Our identified nine spa types from S. aureus isolates and majority of them were assigned to t2883 (31.57%), followed by t416 (28.94%). Prior research has revealed a high diversity of S. aureus spa types in samples collected along dairy chains, including t2883 (Ben Slama et al., 2011; Boero et al., 2022). Interestingly, a study by Ben Slama et al. (2011) found the t2883 spa type in human hands that had come into touch with animals in Tunisia, suggesting that there may be a chance of transmission from human to animal or vice versa. In the current analysis, the t416 spa type was also discovered, and it appears to be dominant in all 38 isolates of S. aureus. The high prevalence of t416 spa type discovery in our investigation is more than that reported by Mora-Hernández et al. (2021), who found t416 in 6.06% of the S. aureus isolates from dairy cows in Mexico that had mastitis. The widespread use of antibiotics to treat S. aureus-caused human infections and cow mastitis is endangering public health since AMR is emerging among dangerous bacteria (Zigo et al., 2019). Finding the ARGs is necessary to evaluate the pathogenic potential of S. aureus during mastitis. Scanning genome sequences facilitates the identification of genetic components linked to virulence and antibiotic resistance (Paramasivam et al., 2023).
The phylogenomic study aligned with the known mechanism of zoonotic transmission of S. aureus (Silva et al., 2023). This may be explained by data indicating that, although many S. aureus lineages are non-specific, others are suited to colonize and infect particular host species (Schmidt et al., 2017).
It has been observed that majority of isolates across all clusters and STs were carrying the multidrug resistance genes lmrS, mepA, and tet, which encode for resistance against macrolide/phenicol, multidrug efflux MATE transporter MepA and tetracycline. Pu et al. (2021) carried out an experiment to identify genes (cadD) that confer resistance to cadmium antibiotics from bacterial plasmids. Subsequent investigations discovered these genes in five strains of MRSA isolates from Spain. In addition, the cadD gene for cadmium resistance was found in ST97, ST152, and ST8500, which together account for 44.74% of the isolates. While this study is among the few that has detected this gene in raw milk, another study conducted in the UK found it in animal feed at a detection rate of only 23% (Smith et al., 2005). Sequence Type 243 of the isolates of S. aureus, carried the gene MurA, which codes for resistance against fosfomycin. At different levels, fosfomycin resistance genes and mutations were found, and they were unmistakably linked to specific clonal complexes. According to Michalopoulos et al. (2011), the antibiotic fosfomycin targets the UDP-N acetylglucosamine enolpyruvyl transferase, which is involved in cell wall construction and encoded by the murA gene. Interestingly, the only strain with all the antibiotic resistance genes in this investigation was ST152, with the exception of the cadmium resistance genes. Quinolone, beta-lactam, ampicillin (blaI, blaR1 and blaZ), and trimethoprim (dfrG and dfrG 1; parE) were also present in this ST152.
According to reports, beta-lactam antibiotics frequently cause staphylococci to exhibit resistance (Olsen et al., 2006; Zigo et al., 2022). The blaZ gene produces a penicillinase, also called a beta-lactamase, that imparts penicillin resistance by hydrolyzing the beta-lactam ring and rendering the drug inert (van den Borne et al., 2010; Zigo et al., 2022). Additionally, the results of this study support the conclusions that there is little to no Staphylococcus that is resistant to beta-lactam or penicillin (van den Borne et al., 2010). However, almost 45% of S. aureus isolates were found to be blaZ positive and phenotypically penicillin-resistant in a follow-up study conducted in New Zealand by Steele and McDougall (2014). This genotype/phenotype combination was associated with a very low cure following antibiotic treatment. The mecA and mecC genes were not present in any of the S. aureus isolates in the current investigation, which were all methicillin-sensitive S. aureus (MSSA). Furthermore, compared to a study by Haulisah et al. (2021) which revealed 80%–100% resistance to trimethropin, this study did not detect any of these genes from the isolates. Moreover, the production of toxins like hemolysins, leukotoxins, and enterotoxins as well as enzymes like serine proteases, cysteine proteases, and lipases that function as effectors during pathogenicity is what gives S. aureus its pathogenic potential (Sivakumar et al., 2023).
In addition to several enzyme-coding genes, the majority of the 38 S. aureus isolates included in this investigation carried multiple hemolysin genes. The genes encoding enterotoxins, enterotoxin-like proteins, and exfoliative toxins were found in just a small number of carefully selected genomes. Leukocidin D/E was found in all ST152 and ST342, while the Panton—Valentine leucocidin genes were only found in two sequence types (ST152 and ST243). Similar to this, all STs had higher frequencies of genes encoding adhesins and hemolysins, while isolates of S. aureus strains associated with bovine mastitis had lower frequencies of enterotoxins. Another crucial factor in determining virulence in staphylococci, particularly in S. aureus, is toxin synthesis. Inflammation and leukocyte cell death are promoted by these toxins, which include cytotoxins (hemolysins, leukotoxins, and leukocidins) and superantigens (enterotoxins, exfoliative toxins, and toxic shock syndrome toxins; TSST; Haag et al., 2019; Chen et al., 2022). In isolates from pus, skin infections, and abscesses, the lukD and lukE genes exhibited strong self-association. As their products are secreted prior to combining to create the PVL toxin, the genes lukF-PV and lukS-PV were associated in the current groups, which is consistent with the literature (Kaneko and Kamio, 2004; Rodrigues et al., 2022).
All S. aureus isolates in all clusters carried the cytotoxins hlgA, hlgB, and hlgC, which encode alpha, beta, and hemolysin, respectively. All S. aureus isolates used in the current investigation included leukocidin genes, such as ST97, ST352, lukS-PV, and lukF-PV (ST152 and ST243). This is consistent with research on bovine S. aureus isolates, which discovered leukocidin and leukotoxin genes in the majority of isolates (Wächter et al., 2021) from cows in India. This may explain similar observations from the current study because many of the lukS-PV and lukF-PV genes in our analysis shared the same percentage identity across different S. aureus isolates, suggesting that the gene sequences may be comparable to those of the genome even if they are absent. Many enterotoxin genes were also found in S. aureus isolates in this current study. The aur gene produces the protein aureolysin, which alters the adhesion factor CflB and triggers additional proteases to increase S. aureus pathogenicity (McAleese et al., 2001). The bi-component leukotoxins that are produced by the hglA, hglB, and hglC genes can create holes in cell membranes, which allows them to lyse cells (Staali and Colin, 2021). Since these gene products are linked to clinical mastitis, they could be valuable targets for the creation of vaccinations and therapeutic drugs (Ahmad-Mansour et al., 2021; Moawad et al., 2023).
The enterotoxins sec, sei, sen, sem, seo, and seu were also shown to be associated with similar frequencies in all analyzed groups; these findings have also been reported by other investigations (Indrawattana et al., 2013; Schwan, 2019; Ren et al., 2020). Additionally, prior research has demonstrated that see and sec, sel, are commonly found in MRSA strains (Hu et al., 2011); however, the current investigation did not find this association. It is crucial to prevent contamination of enterotoxin-producing S. aureus isolates throughout the food production chain since they can cause acute and severe food poisoning. Unpasteurized milk-based cheese and raw milk are well-known dietary sources of S. aureus food poisoning. It is believed that the chemotaxis inhibitory protein (chp product) and the scn gene product are highly specific for staphylococcal isolates of human origin (Pinchuk et al., 2010). The most typical cause of bovine mastitis is staphylococcal enterotoxin C. The primary enterotoxin gene of S. aureus isolated from cows with mastitis, according to other researchers, is called sea. Three enterotoxin-like genes have shown super-antigenic activity but no emetic qualities, two enterotoxin genes (seg and sei), and two enterotoxin genes make up the cluster (selo, selm, and seln). In the current investigation, 91.7% of the S. aureus isolates tested positive for the SEs coding genes classical expression (sea, seb, sec, or sed). The exfoliative toxins (eta and etb), toxic shock syndrome toxin-1 (tsst-1), staphylococcal enterotoxins (sea, seb, sec, sed, saw, seg, seh, sei, and sej), and pvl are all exoproteins that S. aureus is capable of producing (Ren et al., 2020). On the other hand, it is still unclear how staphylococcal enterotoxins affect mammary epithelial cells. Nineteen (19) serologically unique SEs have recently been found. Moreover, the genes most commonly found in S. aureus isolates from dairy cows with mastitis are sec, sed, seg, and sei and sea (Ren et al., 2020).
Understanding the epidemiology of S. aureus genotypes in dairy animals and herds may aid in developing treatment and control plans to stop the illness from spreading. Sequence Type 97, virulence genes like leucocidin, hemolysin, and aureolysin, and AMR genes like lmrS, mepA, and tet (38) were most frequently identified genes in this study. The capacity of S. aureus to colonize and penetrate the host may be influenced by the combination of these genes. Consequently, due to the global distribution of these genes, screening them in S. aureus isolates may be valuable for aiding in clinical outcome prediction and, specifically, for identifying hazardous strains. The current study also showed that all isolates had a nearly identical genotypic pattern and that certain isolates carried virulence factors such as PVL-encoding genes, suggesting that S. aureus isolates in animals should be closely monitored to prevent the spread of these genes. It is noteworthy that all isolates tested negative for mecA and mecC, Furthermore, the current study has also showed or revealed an association between ST97, spa type t2883 and t416 and the plasmid Rep_N. This study has shown that there is a wide range of S. aureus genotypes occurring in dairy cattle in the Free State province and that genetic variations are related to geographic origin of the isolates. This suggests that taking the region of interest and the strain virulence into consideration may help to formulate strategies directed to stop the spread of infection and to set up control measures in accordance with pathogen and host features. Hence, depending on the description of the circulating strain, the farmer would be able to choose whether to slaughter the sick animals or isolate positive cows using sanitary milking practices and an appropriate milking schedule.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA981445 and https://www.ncbi.nlm.nih.gov/, PRJNA1006054.
The animal studies were approved by Animal Research Ethics Committee of the University of Free State (UFS-AED2020/0060/21). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
NK: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Writing – original draft. JN: Supervision, Writing – review & editing. ZM: Supervision, Writing – review & editing. SK: Formal analysis, Methodology, Software, Visualization, Writing – review & editing. OT: Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Central University of Technology (UCDP M&D Grant) and the National Research Foundation (grant no: 134137).
The authors thank the state veterinarians of the three (3) study sites for their cooperation and patience. We are grateful to the farmers for their cooperation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. ^https://github.com/stanikae/jekesa
3. ^https://github.com/tseemann/abricate
4. ^https://github.com/phac-nml/staramr
5. ^http://www.genomicepidemiology.org/services/
6. ^https://github.com/pha4ge/hAMRonization
7. ^https://github.com/tseemann/scapper
Ahmad-Mansour, N., Loubet, P., Pouget, C., Dunyach-Remy, C., Sotto, A., Lavigne, J. P., et al. (2021). Staphylococcus aureus toxins: an update on their pathogenic properties and potential treatments. Toxins 13:677.
Åvall-Jääskeläinen, S., Koort, J., Simojoki, H., and Taponen, S. (2021). Genomic analysis of Staphylococcus aureus isolates associated with peracute non-gangrenous or gangrenous mastitis and comparison with other mastitis-associated Staphylococcus aureus isolates. Front. Microbiol. 12:688819. doi: 10.3389/fmicb.2021.688819
Ben Slama, K., Gharsa, H., Klibi, N., Jouini, A., Lozano, C., Gómez-Sanz, E., et al. (2011). Nasal carriage of Staphylococcus aureus in healthy humans with different levels of contact with animals in Tunisia: genetic lineages, methicillin resistance, and virulence factors. Eur. J. Clin. Microbiol. 30, 499–508.
Boero, E., Cruz, A. R., Pansegrau, W., Giovani, C., Rooijakkers, S. H., van Kessel, K. P., et al. (2022). Natural human immunity against staphylococcal protein a relies on effector functions triggered by IgG3. Front. Immunol. 13:834711. doi: 10.3389/fimmu.2022.834711
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). Res finder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Chen, H., Zhang, J., He, Y., Lv, Z., Liang, Z., Chen, J., et al. (2022). Exploring the role of Staphylococcus aureus in inflammatory diseases. Toxins 14:464. doi: 10.3390/toxins14070464
CLSI (2023). “Performance standards for antimicrobial disk and dilution susceptibility tests for Bacteria isolated from animals” in CLSI Document VET01-S. 6th ed. Eds. C. R. Burbick (Pittsburgh, PA, USA: CLSI)
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196
Elhawy, M. I., Huc-Brandt, S., Pätzold, L., Gannoun-Zaki, L., Abdrabou, A. M. M., Bischoff, M., et al. (2021). The Phosphoarginine phosphatase Ptp B from Staphylococcus aureus is involved in bacterial stress adaptation during infection. Cells 10:645. doi: 10.3390/cells10030645
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J. G., Haendiges, J., Haft, D. H., et al. (2021). AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11:12728. doi: 10.1038/s41598-021-91456-0
Gelalcha, B. D., Agga, G. E., and Dego, O. K. (2022). “Antimicrobial usage for the management of mastitis in the USA: impacts on antimicrobial resistance and potential alternative approaches” in Mastitis in dairy cattle, sheep and goats, Eds. O. K. Dego (Intech Open) 1–21.
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Haag, A. F., Fitzgerald, J. R., and Penadés, J. R. (2019). Staphylococcus aureus in animals. Microbiol Spectr 7, 10–1128. doi: 10.1128/microbiolspec.GPP3-0060-2019
Haulisah, N. A., Hassan, L., Bejo, S. K., Jajere, S. M., and Ahmad, N. I. (2021). High levels of antibiotic resistance in isolates from diseased livestock. Front Vet Sci 8:652351. doi: 10.3389/fvets.2021.652351
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hu, D.-L., Maina, E. K., Omoe, K., Inoue, F., Yasujima, M., and Nakane, A. (2011). Superantigenic toxin genes coexist with specific staphylococcal cassette chromosome mec genes in methicillin-resistant Staphylococcus aureus. Tohoku J. Exp. Med. 225, 161–169. doi: 10.1620/tjem.225.161
Indrawattana, N., Sungkhachat, O., Sookrung, N., Chongsa-nguan, M., Tungtrongchitr, A., Voravuthikunchai, S. P., et al. (2013). Staphylococcus aureus clinical isolates: antibiotic susceptibility, molecular characteristics, and ability to form biofilm. Biomed. Res. Int. 2013:654. doi: 10.1155/2013/314654
Kaneko, J., and Kamio, Y. (2004). Bacterial two-component and hetero-heptameric pore-forming cytolytic toxins: structures, pore-forming mechanism, and organization of the genes. Biosci. Biotechnol. Biochem. 68, 981–1003.
Karzis, J., Donkin, E. F., Webb, E. C., Etter, E. M., and Petzer, I. M. (2017). Somatic cell count thresholds in composite and quarter milk samples as indicator of bovine intramammary infection status. Onderstepoort J. Vet. Res. 84, 1–10. doi: 10.4102/ojvr.v84i1.1269
Kassambara, A. (2016). Factoextra: extract and visualize the results of multivariate data analyses. R package version, 1.
Khasapane, N. G., Koos, M., Nkhebenyane, S. J., Khumalo, Z. T., Ramatla, T., and Thekisoe, O. (2024). Detection of Staphylococcus isolates and their antimicrobial resistance profiles and virulence genes from subclinical mastitis cattle Milk using MALDI-TOF MS, PCR and sequencing in Free State Province, South Africa. Animals 14:154. doi: 10.3390/ani14010154
Klibi, A., Jouini, A., Gómez, P., Slimene, K., Ceballos, S., Torres, C., et al. (2018). Molecular characterization and clonal diversity of methicillin-resistant and-susceptible Staphylococcus aureus isolates of milk of cows with clinical mastitis in Tunisia. Microb. Drug Resist. 24, 1210–1216. doi: 10.1089/mdr.2017.0278
Li, L. (2008). Complete genome sequence and strain differentiation of Mycobacterium avium subspecies paratuberculosis. Minneapolis, United States: University of Minnesota.
Liu, K., Tao, L., Li, J., Fang, L., Cui, L., Li, J., et al. (2020). Characterization of Staphylococcus aureus isolates from cases of clinical bovine mastitis on large-scale Chinese dairy farms. Front Vet Sci 7:580129. doi: 10.3389/fvets.2020.580129
Maity, S., Das, D., and Ambatipudi, K. (2020). Quantitative alterations in bovine milk proteome from healthy, subclinical and clinical mastitis during S. aureus infection. J. Proteom. 223, p.103815.
McAleese, F. M., Walsh, E. J., Sieprawska, M., Potempa, J., and Foster, T. J. (2001). Loss of clumping factor B fibrinogen binding activity by staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease. J. Biol. Chem. 276, 29969–29978.
Michalopoulos, A. S., Livaditis, I. G., and Gougoutas, V. (2011). The revival of fosfomycin. Int. J. Infect. Dis. 15, e732–e739. doi: 10.1016/j.ijid.2011.07.007
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Moawad, A. A., El-Adawy, H., Linde, J., Jost, I., Tanja, G., Katja, H., et al. (2023). Whole genome sequence-based analysis of staphylococcus aureus isolated from bovine mastitis in Thuringia. Germany. Front. microbiol. 14:1216850.
Mora-Hernández, Y., Vera Murguía, E., Stinenbosch, J., Hernández Jauregui, P., van Dijl, J. M., and Buist, G. (2021). Molecular typing and antimicrobial resistance profiling of 33 mastitis-related Staphylococcus aureus isolates from cows in the Comarca Lagunera region of Mexico. Sci. Rep. 11:6912. doi: 10.1038/s41598-021-86453-2
National Mastitis Council. (2004). Microbiological procedures for the diagnosis of bovine udder infection and determination of Milk quality. Available at: http://www.nmconline.org/sampling.html (Accessed April 2021).
Naushad, S., Naqvi, S. A., Nobrega, D., Luby, C., Kastelic, J. P., Barkema, H. W., et al. (2019). Comprehensive virulence gene profiling of bovine non-aureus staphylococci based on whole-genome sequencing data. Msystems 4, 10–1128. doi: 10.1128/mSystems.00098-18
Naushad, S., Nobrega, D. B., Naqvi, S. A., Barkema, H. W., and De Buck, J. (2020). Genomic analysis of bovine staphylococcus aureus isolates from milk to elucidate diversity and determine the distributions of antimicrobial and virulence genes and their association with mastitis. Msystems 5, 10–1128.
Olsen, J. E., Christensen, H., and Aarestrup, F. M. (2006). Diversity and evolution of Bla Z from Staphylococcus aureus and coagulase-negative staphylococci. J. Antimicrob. Chemother. 57, 450–460. doi: 10.1093/jac/dki492
Omwenga, I., Aboge, G. O., Mitema, E. S., Obiero, G., Ngaywa, C., Ngwili, N., et al. (2021). Antimicrobial usage and detection of multidrug-resistant Staphylococcus aureus, including methicillin-resistant strains in raw milk of livestock from northern Kenya. Microb. Drug Resist. 27, 843–854. doi: 10.1089/mdr.2020.0252
Ozbey, G., Cambay, Z., Yilmaz, S., Aytekin, O., Zigo, F., Ozçelik, M., et al. (2022). Identification of bacterial species in milk by MALDI-TOF and assessment of some oxidant-antioxidant parameters in blood and milk from cows with different health status of the udder. Pol. J. Vet. Sci. 25, 269–277. doi: 10.24425/pjvs.2022.141811
Page, A. J., Taylor, B., Delaney, A. J., Soares, J., Seemann, T., Keane, J. A., et al. (2016). SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microbial Genom 2:e000056. doi: 10.1099/mgen.0.000056
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Paramasivam, R., Gopal, D. R., Dhandapani, R., Subbarayalu, R., Elangovan, M. P., Prabhu, B., et al. (2023). Is AMR in dairy products a threat to human health? An updated review on the origin, prevention, treatment, and economic impacts of subclinical mastitis. Infect Drug Resist 6, 155–178.
Pattabhiramaiah, M., and Mallikarjunaiah, S. (2023). “Foodborne Pathogens and food safety regulations” in Food microbial and molecular biology (Florida, USA: Apple Academic Press), 179–212.
Pinchuk, I. V., Beswick, E. J., and Reyes, V. E. (2010). Staphylococcal enterotoxins. Toxins 2, 2177–2197. doi: 10.3390/toxins2082177
Pu, Q., Fan, X. T., Sun, A. Q., Pan, T., Li, H., Lassen, S. B., et al. (2021). Co-effect of cadmium and iron oxide nanoparticles on plasmid-mediated conjugative transfer of antibiotic resistance genes. Environ. Int. 152:106453. doi: 10.1016/j.envint.2021.106453
Ren, Q., Liao, G., Wu, Z., Lv, J., and Chen, W. (2020). Prevalence and characterization of Staphylococcus aureus isolates from subclinical bovine mastitis in southern Xinjiang, China. J. Dairy Sci. 103, 3368–3380. doi: 10.3168/jds.2019-17420
Rocha, L. S., Silva, D. M., Silva, M. P., Vidigal, P. M. P., Silva, J. C. F., Guerra, S. T., et al. (2019). Comparative genomics of staphylococcus aureus associated with subclinical and clinical bovine mastitis. PLoS One 14:e0220804
Rodrigues, R. A., Pizauro, L. J. L., Varani, A. M., de Almeida, C. C., Silva, S. R., Cardozo, M. V., et al. (2022). Comparative genomics study of Staphylococcus aureus isolated from cattle and humans reveals virulence patterns exclusively associated with bovine clinical mastitis strains. Front. Microbiol. 13:1033675. doi: 10.3389/fmicb.2022.1033675
Schmidt, T., Kock, M. M., and Ehlers, M. M. (2017). Molecular characterization of staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: genetic diversity and inter-species host transmission. Front. microbiol. 8:248335.
Schwan, W. R. (2019). Staphylococcus aureus toxins: armaments for a significant pathogen. Toxins 11:457. doi: 10.3390/toxins11080457
Silva, V., Araújo, S., Monteiro, A., Eira, J., Pereira, J. E., Maltez, L., et al. (2023). Staphylococcus aureus and MRSA in livestock: antimicrobial resistance and genetic lineages. Microorganisms 11:124.
Sivakumar, R., Pranav, P. S., Annamanedi, M., Chandrapriya, S., Isloor, S., Rajendhran, J., et al. (2023). Genome sequencing and comparative genomic analysis of bovine mastitis-associated Staphylococcus aureus strains from India. BMC Genomics 24:44. doi: 10.1186/s12864-022-09090-7
Smith, E. M., Green, L. E., Medley, G. F., Bird, H. E., and Dowson, C. G. (2005). Multilocus sequence typing of Staphylococcus aureus isolated from high-somatic-cell-count cows and the environment of an organic dairy farm in the United Kingdom. J. Clin. Microbiol. 43, 4731–4736. doi: 10.1128/JCM.43.9.4731-4736.2005
Souvorov, A., Agarwala, R., and Lipman, D. J. (2018). SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 19:153. doi: 10.1186/s13059-018-1540-z
Staali, L., and Colin, D. A. (2021). Bi-component HlgC/HlgB and HlgA/HlgB γ-hemolysins from S. aureus: modulation of Ca2+ channels activity through a differential mechanism. Toxicon 201, 74–85.
Steele, N., and McDougall, S. (2014). Effect of prolonged duration therapy of subclinical mastitis in lactating dairy cows using penethamate hydriodide. N. Z. Vet. J. 62, 38–46. doi: 10.1080/00480169.2013.830350
van den Borne, B. H., van Schaik, G., Lam, T. J., and Nielen, M. (2010). Therapeutic effects of antimicrobial treatment during lactation of recently acquired bovine subclinical mastitis: two linked randomized field trials. J. Dairy Sci. 93, 218–233. doi: 10.3168/jds.2009-2567
Vanderhaeghen, W., Piepers, S., Leroy, F., Van Coillie, E., Haesebrouck, F., and De Vliegher, S. (2015). Identification, typing, ecology and epidemiology of coagulase negative staphylococci associated with ruminants. Vet. J. 203, 44–51. doi: 10.1016/j.tvjl.2014.11.001
Vargová, M., Zigo, F., Výrostková, J., Farkašová, Z., and Rehan, I. F. (2023). Biofilm-producing ability of Staphylococcus aureus obtained from surfaces and Milk of Mastitic cows. Vet Sci 10:386. doi: 10.3390/vetsci10060386
Wächter, H., Yörük, E., Becker, K., Görlich, D., and Kahl, B. C. (2021). Correlations of host and bacterial characteristics with clinical parameters and survival in Staphylococcus aureus bacteremia. J. Clin. Med. 10:1371. doi: 10.3390/jcm10071371
Wellnitz, O., and Bruckmaier, R. M. (2012). The innate immune response of the bovine mammary gland to bacterial infection. Vet. J. 192, 148–152.
Zigo, F., Elečko, J., Farkašová, Z., Zigová, M., Vasiľ, M., Ondrašovičová, S., et al. (2019). Preventive methods in reduction of mastitis PATHOGENS in dairy cows. J. Microbiol. Biotechnol. Food Sci. 9, 121–126. doi: 10.15414/jmbfs.2019.9.1.121-126
Keywords: subclinical mastitis, Staphylococcus aureus, virulence factors, antimicrobial resistance, whole genome sequencing
Citation: Khasapane NG, Nkhebenyane J, Mnisi Z, Kwenda S and Thekisoe O (2024) Comprehensive whole genome analysis of Staphylococcus aureus isolates from dairy cows with subclinical mastitis. Front. Microbiol. 15:1376620. doi: 10.3389/fmicb.2024.1376620
Received: 26 January 2024; Accepted: 19 March 2024;
Published: 08 April 2024.
Edited by:
Chang-Wei Lei, Sichuan University, ChinaReviewed by:
František Zigo, University of Veterinary Medicine and Pharmacy in Košice, SlovakiaCopyright © 2024 Khasapane, Nkhebenyane, Mnisi, Kwenda and Thekisoe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ntelekwane George Khasapane, bmtoYXNhcGFuZUBjdXQuYWMuemE=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.