- 1Guizhou University of Traditional Chinese Medicine, Department of Colorectal Medicine, Guiyang, Guizhou, China
- 2Guizhou University of Commerce, Computer and Information Engineering College, Guiyang, China
Background: Constipation is affected by a number of risk variables, including cardiovascular disease and growth factors. However, the impacts of gut flora on constipation incidence has not been shown. This work, Single-Variable Mendelian Randomization (SVMR) was utilized to estimate the causal relationship between the Eubacterium genus or Rumphococcus, and constipation.
Methods: Data for constipation, Eubacterium genus and Rumphococcus were taken from the Integrated Epidemiology Unit (IEU) open GWAS database. Including 218,792 constipation samples, and there were 16,380,466 Single Nucleotide Polymorphisms (SNPs) for constipation. The ids of Eubacterium genus and Rumphococcus were sourced from MiBioGen database. The sample count for the Eubacterium genus was 17,380, with 656 SNPs. In addition, the sample size for Rumphococcus was 15,339, with 545 SNPs. The SVMR was performed to assess the risk of Eubacterium genus and Rumphococcus in constipation using weighted median, MR Egger, simple mode, inverse variance weighted (IVW), and weighted mode. Finally, we did a sensitivity analysis that included a heterogeneity, horizontal pleiotropy, and Leave-One-Out (LOO) test to examine the viability of the MR data.
Results: The SVMR revealed that the Eubacterium genus and Rumphococcus were causally connected to constipation, with Rumphococcus (P = 0.042, OR = 1.074) as a hazardous factor and Eubacterium genus (P = 0.004, OR = 0.909) as a safety factor. Sensitivity tests then revealed the absence of variability between the constipation and the exposure factors (Eubacterium genus and Rumphococcus). Additionally, there were no other confounding factors and the examined SNPs could only influence constipation through the aforementioned exposure factors, respectively. As a result, the MR results were fairly robust.
Conclusion: Our investigation verified the causal links between the Eubacterium genus or Rumphococcus, and constipation, with greater Rumphococcus expression increasing the likelihood of constipation and the opposite being true for the Eubacterium genus.
1 Introduction
Constipation is a chronic gastrointestinal disease characterized by a high prevalence. Prolonged constipation symptoms have a detrimental impact on people's standard of life and place a significant financial and social burden on society (Dong et al., 2023). Constipation is also independently linked to poor clinical outcomes like cardiovascular disease (CV disease), end-stage renal disease (ESRD), and mortality, according to recent epidemiological investigations. This association may be mediated by changes in the gut microbiota and increased fecal metabolite production (Dong et al., 2023). Li et al. used Mendelian randomization to analyze the causal relationship between intestinal microorganisms and constipation. The results showed that the abundance of Fecal Coccus increased 1 and the risk of constipation decreased, while the abundance of Pseudomonas increased. Obviously, in the intestinal tract microorganism's fecal coccus genus 1, Pseudomonas and constipation related (Li et al., 2024). The Mancabelli et al. (2017) study found that the abundance of intestinal bacteria, including fecal cocci, decreased in patients with constipation. The Yarullina et al. (2020) study found decreased abundance of Fecal Coccus 1 in patients with constipation, but not in traditional probiotics such as Lactobacillus and Bifidobacterium. The Guo et al. (2020) study on the composition of intestinal microflora in the elderly patients with functional constipation showed that the spectrum of intestinal microflora in the elderly patients with functional constipation was significantly different from that in the healthy population, and the abundance of Bacteroides in the elderly patients with functional constipation was significantly higher than that in the healthy population. Bacteroides are gram-negative anaerobes and play an important role in the degradation and fermentation of colonic organic matter. Wu et al. (2011) confirmed that there was a negative correlation between Bacteroides in feces and dietary fiber intake. Low cellulose diet is the main cause of intestinal flora disorder in constipation patients. Lack of cellulose causes chyme to be difficult to promote intestinal peristalsis and constipation (Xu et al., 2021).
Constipation has also been reported more in Mendelian randomization (MR) studies on cardiovascular disease (CVD; Baumeister et al., 2021). Parkinson's disease (PD; Wang et al., 2023), Growth Factor (GF; Thursby and Juge, 2017) and other factors have been linked to constipation in a direct causative manner. The main treatment for functional defecatory diseases are biofeedback therapy combined with pelvic floor retraining (Bharucha, 2007). Moreover, In the US, constipation is the cause of 2.5 million doctor visits annually, with primary care physicians accounting for more than half of these encounters (Arce et al., 2002). Examining the patient's overall health, mental health, physical ailments, use of dietary fiber, and use of drugs that cause constipation are all crucial. The human gastrointestinal tract has a complex and dynamic microbial population, the intestinal flora, which has a significant impact on the host during homeostasis and disease. These bacteria are kept in a relatively stable state under normal conditions. They play a variety of roles in the body's physiological processes, secrete metabolites that contain enzymes to supplement the body's supply of digestive enzymes, aid in the digestion, absorption, and metabolism of substances, hasten the development of hyperproductive endo-epithelial cells and the regeneration of vascular tissue, maintain the immune system, and stop pathogenic bacteria from proliferating and differentiating (Arce et al., 2002; Cao et al., 2017; Chu et al., 2019). Changes in the composition of gut bacteria are associated with the pathogenesis of many inflammatory diseases and infections (Ohkusa et al., 2019). There is evidence that dysbacteriosis of the gut may lead to functional constipation and constipation-type irritable bowel syndrome. Constipation is made worse when the dynamic equilibrium of the intestinal flora is disrupted, which also affects the body's metabolism and causes a number of pathophysiological changes (Meng et al., 2020). However, studies of gut flora and constipation were not reported. Since there is currently little clinical understanding of constipation, this study looked into the regulation mechanism of intestinal flora in the pathogenesis of constipation as well as a novel approach to treating constipation through intestinal flora management.
Genetic variations are used by MR as instrumental variables (IVs) to determine if a exposure factor has a causal impact on an outcome. The MR offers an alternative way to probe the issue of causality in epidemiological research, by using additional genetic variants that are hypothesized to satisfy the IVs assumptions (Bowden and Holmes, 2019). Meanwhile, MR uses statistical methods from economics to let scientists to examine how human biology and disease are impacted by the environment, medications, and other factors. It is possible to use genetic variants known to influence factors like alcohol consumption or low-density lipoprotein (LDL) levels as IVs that can separate the effects of these factors on outcomes like pregnancy or cardiovascular disease, respectively, by taking advantage of the fact that genetic variation is randomized among children of the same parents. Although MR and similar techniques have limitations that researchers should be aware of, they are becoming more and more effective tools for addressing issues in human biology and epidemiology (Birney, 2022).
Imbalance of intestinal flora leads to a series of clinical symptoms. The disturbance of dynamic balance of intestinal flora leads to the proliferation of pathogenic bacteria, stimulates intestinal mucosal barrier, impairs immune system and leads to the decline of immune function. In addition, the release of a large number of pathogenic bacteria increased human intestinal endotoxin, disrupted the intestinal mucosal barrier, resulting in intestinal dysfunction. Eubacterium and Ruminococcus two kinds of intestinal microorganism, which can promote intestinal peristalsis, energy metabolism and maintain the balance of intestinal environment. Eubacterium is one of the first discovered metabolic microorganisms. It can degrade cellulose, also can ferment glucose, xylose and so on, and has certain digestive function to resistant starch. Important to Refractory Constipation, an analysis of the composition of the intestinal microflora of fresh feces and the accumulation of feces (old feces) in patients with refractory constipation has found a gap between the two. There are significant differences. The potential causal relationship between constipation and Eubacterium cocci and Ruminococcus coli was demonstrated.
In this study, based on single Nucleotide Polymorphisms (SNPs) data on constipation and exposure factors (Eubacterium genus and Rumphococcus) in GWAS database, this research performed Single-Variable MR (SVMR) methods to assess the causal links of Eubacterium genus and Rumphococcus on constipation separately, and a sensitivity analysis was conducted to assess the impact of hypotheses on the findings and ensure the rationality of findings.
2 Methods
2.1 Data source and pre-processing
The GWAS ids for constipation (finn-b-K11_CONSTIPATION) originated from the Integrative Epidemiology Unit (IEU) Open GWAS database (https://gwas.mrcieu.ac.uk/). The dataset on constipation contains 16,380,466 single nucleotide polymorphisms (SNPs) from 218,792 samples. The ids of Eubacterium genus and Rumphococcus were sourced from MiBioGen database (https://gwas.mrcieu.ac.uk/). The sample amount for Eubacterium genus (genus Eubacteriumcoprostanoligenesgroup.id.11375) was 17,380 with 656 SNPs. The sample size for Rumphococcus (genus.Ruminococcus2.id.11374) was 15339, and the amount of SNPs was 545. Thereafter, the “TwoSampleMR” R package (version 0.5.6; Hemani et al., 2018) was utilized to read and screen SNPs that were strongly linked to exposure factors. The linkage disequilibrium (LD) IVs were then eliminated using r2 = 0.001 and kb = 10,000 to prevent bias brought on by the chained IVs.
The three main assumptions that underpinned the MR investigation were that: (1) there is a strong and noteworthy association between IVs and exposure; (2) the IVs are independent of confounders; and (3) the IVs can only affect the outcome via exposure and not in any other way.
2.2 MR and sensitivity analysis
After the IVs were filtered, the input data for the SVMR analysis were collected. The most MR modes were Inverse Variance Weighted (IVW) regression (Burgess et al., 2015), weighted mode (Hartwig et al., 2017), weighted median (Bowden et al., 2016), simple mode, and MR-Egger (Bowden et al., 2015). Then, odds ratios (ORs) were computed, where a safety factor is represented by a value <1 and a risk factor by a value >1. Three types of plots were created to present the results: scatter, forest, and funnel. Sensitivity analysis, which mostly included the Leave-One-Out (LOO), heterogeneity, and horizontal pleiotropy, was conducted to assess the dependability of the MR data.
3 Results
3.1 Connection of Eubacterium genus and Rumphococcus to constipation
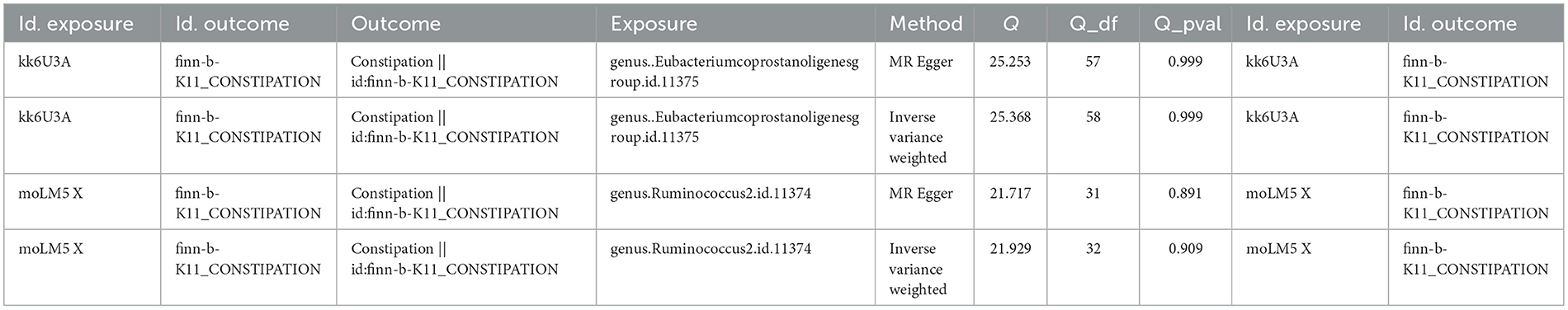
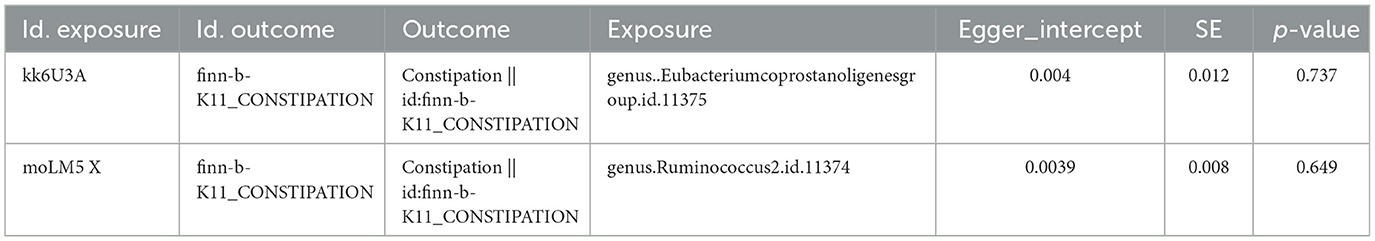
We identified 66 Eubacterium genus and 40 Rumphococcus individual SNPs as IVs after screening. The IVW findings suggested that Eubacterium genus (P = 0.004), Rumphococcus (P = 0.042) and constipation were linked by causality (Tables 1, 2). After in-depth analysis, OR values revealed that Rumphococcus (OR = 1.074) was a risk factor and Eubacterium genus (OR = 0.909) was a protective factor for constipation. The scatter plot showed that slope of the line for Rumphococcus was positive, indicating that increased amount of Rumphococcus led to increased risk of constipation, and the opposite result for Eubacterium genus (Figures 1A, B). The SNPs locations in the forest plot were protection on the left and risk on the right. Our findings supported that Rumphococcus and Eubacterium genus were a risk factor and a protective factor for constipation, respectively (Figures 1C, D). Also, Mendel's second law of random grouping was proven via the funnel plot (Figures 1E, F).
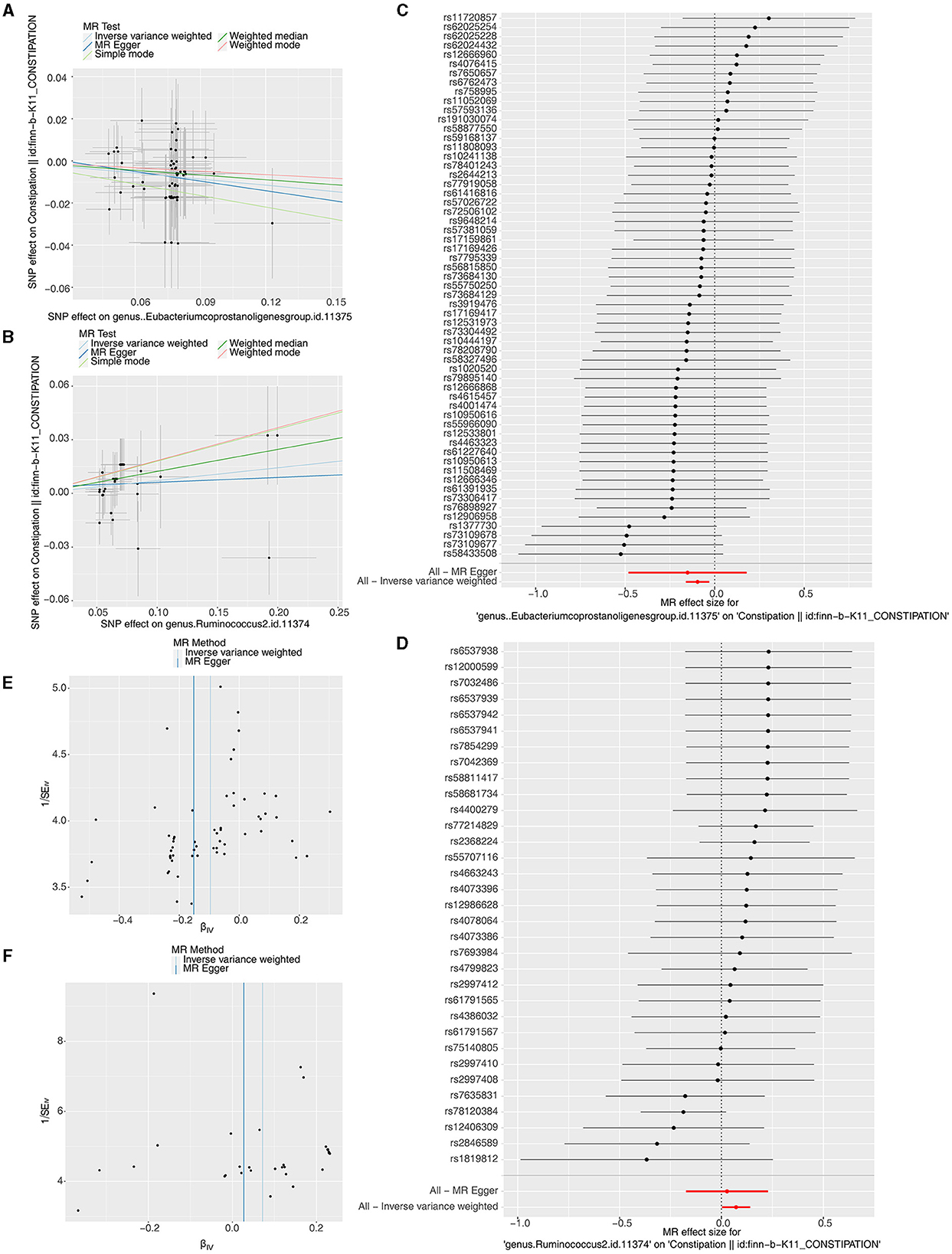
Figure 1. Scatter plots, forest plots, and funnel plots revealing the causative effects of the Eubacterium genus and Rumphococcus on constipation. (A, B) The effects of SNPs on microbiota and constipation. (A) Eubacterium genus; (B) Rumphococcus. (C, D) MR effect values of microbiota. (C) Eubacterium genus; (D) Rumphococcus. (E, F) Funnel plots of SNP distributions. (E) Eubacterium genus; (F) Rumphococcus.
3.2 Sensitivity analysis of MR
Several tests were run to assess the dependability of the analyses. The P-values of the heterogeneity tests were all larger than 0.05, indicating that there was no heterogeneity between exposure factors and outcomes (Table 1). The outcome of Horizontal pleiotropy test suggested that there was no confounding and SNPs can only influence constipation through two exposure factors independently (Eubacterium genus: P = 0.737, Rumphococcus: P = 0.648; Table 2). And, The LOO results reveal that there is no single SNP that has a major effect on the MR results, validating the correctness of the results (Figures 2A, B). In conclusion, Eubacterium genus and Rumphococcus were causally associated with development of constipation, with Rumphococcus as a risk factor and Eubacterium genus as a protective factor.
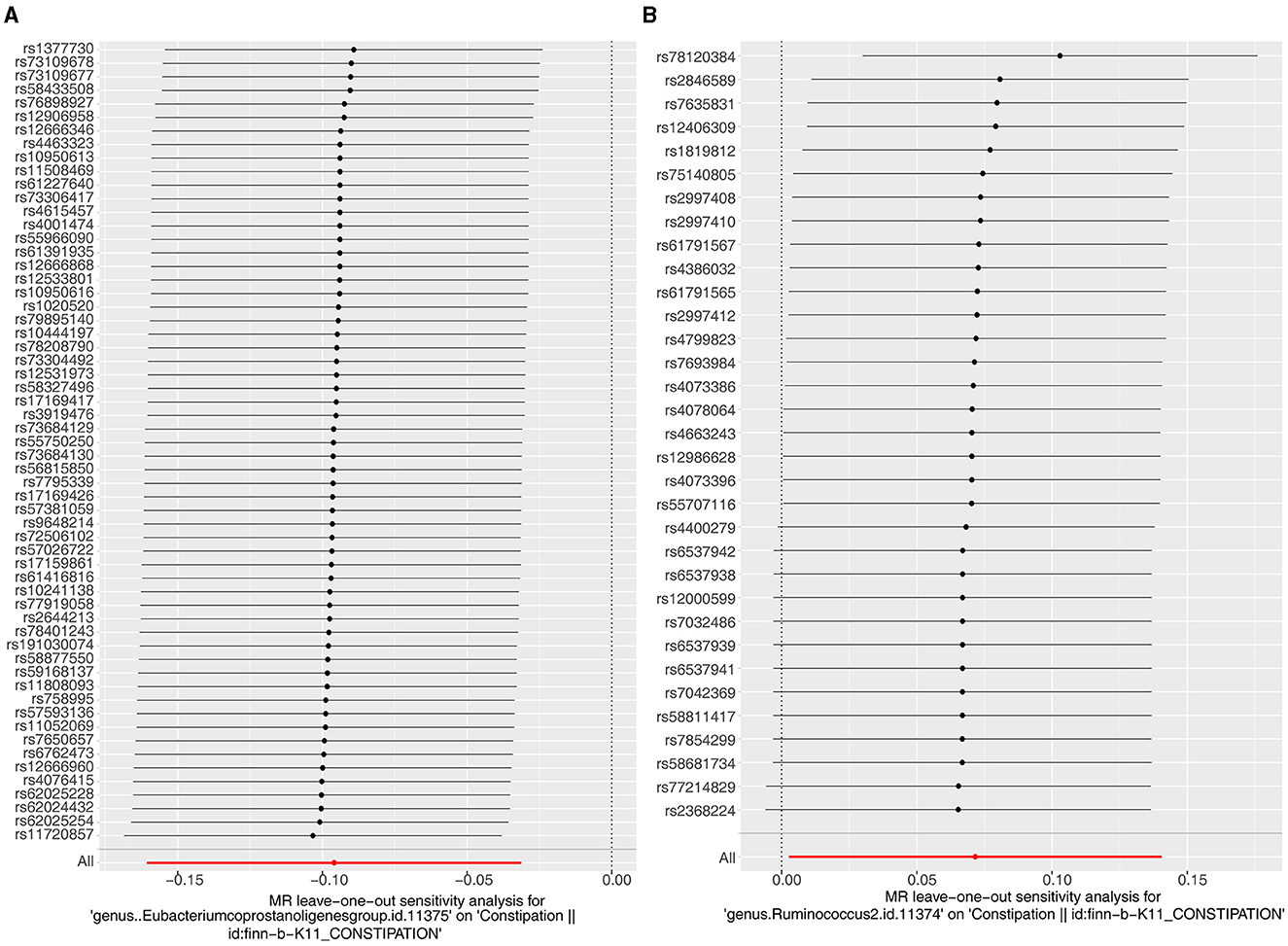
Figure 2. Leave one out of the sensitivity tests. Calculate the MR findings of the remaining IVs after deleting them one by one. (A) Eubacterium genus; (B) Rumphococcus.
4 Discussion
Constipation is one of the most frequent abnormalities of the gastrointestinal system that affects the patient's quality of life (Bisht et al., 2023). Based on SNPs data on constipation and exposure factors in public databases, this work performed a MR analysis using SVMR methods to investigate causality of Eubacterium genus and Rumphococcus on constipation. And a sensitivity analysis was conducted to assess the durability of the findings. Eubacterium genus and Rumphococcus were causally associated with constipation, Eubacterium genus was a protective factor of constipation, and Eubacterium genus was a risk factor for constipation.
It has been documented that each person has their own unique intestinal microbiota profile, which plays a role in nutrient metabolism, maintaining the integrity of the intestinal mucosal barrier, and preventing pathogen invasion. Qualitative and quantitative changes in the gut microbiota are related to the health status of our organism (Black and Ford, 2018). The overall composition of the colonic mucosal microbiota is associated with constipation, and the colonic mucosal microbiota in constipation patients is more abundant in Bacteroides. Probiotics can alter the altered intestinal microbiota of constipation patients, alter intestinal sensory and motor functions, and regulate the intraluminal environment (Di Domenico et al., 2022). Constipation is usually caused by dietary changes or inadequate fiber intake, where treatment with a multi-component probiotic formula consisting of Bifidobacterium bifidum and others has a positive effect on relieving constipation symptoms (Zhao and Yu, 2016).
Eubacterium genus is one of the first stomach bacteria to be discovered and plays a key role in metabolism. Eubacterium genus bacteria obtain nutrients by breaking down cellulose. It is also able to ferment glucose and xylose, which plays a significant role in the digestion of resistant starch. Refractory constipation is the most severe form of constipation, and its cause remains unknown. The symptoms of constipation appear repeatedly, causing great physical and psychological distress to the patient. A growing body of research shows that patients with constipation exhibit significant dysregulation of the gut microbiota compared to healthy individuals (Chen et al., 2021). Constipation is caused by dysbiosis. The study of the gut microbiota can identify the types of bacteria that contribute to the development of constipation (Ohkusa et al., 2019). Someone analyzed the composition of the gut microbiota of fresh feces and accumulated feces (old feces) in patients with refractory constipation and found significant differences between them. This study, we demonstrate a potential causative connection between Eubacterium genus and Rumphococcus and constipation. Rumphococcus is the direct cause of constipation, while Eubacterium genus is a protective factor for constipation. This implies that in order to prevent constipation, we should pay close attention to the content of Rumphococcus in patients and initiate targeted treatment as soon as feasible. Ruminococcus is a human gut symbiont that is found in the baby and adult gut microbiota and has been linked to a variety of intestinal and extra-intestinal illnesses. Meanwhile, Ruminococcus is becoming a major player in impacting health and illness outcomes in people ranging from infants to the elderly (Juge, 2023).
Since MR uses IVs analysis to simulate the randomization process of causal reasoning in randomized controlled trials (RCTs), the design is less susceptible to confounding and reverse causal bias, which improves the rigor and persuasiveness of the experiment and makes up for the shortcomings of the existing research design confounding and reverse causal bias, which is the advantage and innovation of this study. However, this study only targets two genera of bacteria, its coverage and generalization are limited, as we all know, constipation is the result of the interaction of a variety of factors, so it is undeniable that there are many other genera that affect the occurrence and development of constipation.
Current conjectures about the metabolic processes in which Eubacterium and Ruminococcus may be involved, the metabolites produced, and how these products affect the intestinal environment are as follows:
SCFAs are the main metabolites produced by the intestinal microbiota in the colon, such as dietary fiber, through fermentation in the colon (Yumiao et al., 2022). SCFAs are mainly involved in the regulation of intestinal function through the following two mechanisms: (1) regulate intestinal function by activating G protein-coupled receptors (GPRs) including GPR41, GPR43, and GPR109A (Li et al., 2022); (2) Regulate the transcription and expression of genes by inhibiting histone deacetylase (HDAC; Li et al., 2022), thereby exerting anti-inflammatory and intestinal barrier functions. SCFAs can maintain the homeostasis of intestinal flora. In a healthy state, the human intestinal microecosystem is always in a dynamic equilibrium. The imbalance of the intestinal microecosystem is closely related to the pathogenesis of the body (Jie et al., 2021), which can lead to low immunity and various inflammatory reactions (Bowen et al., 2020; Shufei et al., 2020). Studies have shown that when the concentration of SCFAs in the intestine increases, H+ can be released, thereby reducing the intestinal PH and causing the intestinal environment to form an acidic environment (Li et al., 2022), which can promote the reproduction of probiotics and maintain the homeostasis of the intestinal microbiota. Zichen et al. (2022) found that probiotic compound preparations could increase the content of SCFAs such as acetic acid in mice, promote the proliferation of intestinal probiotics, improve the intestinal microecology of mice, and promote the health of mice. Couto et al. (2020) found that SCFAs can promote the reproduction of probiotics such as Bifidobacteria, which in turn stimulate the synthesis of SCFAs, forming a virtuous cycle, thereby improving the body's intestinal microecology and promoting the balance and stability of the intestinal environment. SCFAs, FC and intestinal microbiota maintain intestinal health and improve constipation symptoms through the complex intestinal microecosystem. Aoki et al. (2016) demonstrated that Bifidobacterium gavage in mice could not only reduce loperamide-induced constipation, but also increase fecal SCFAs levels in mice. Matsumoto et al. (2010) also found that probiotic drinks were able to increase the abundance of bifidobacteria and the content of SCFAs, relieving symptoms in patients with constipation. This is consistent with the experimental findings of Aoki et al. (2016). SCFAs affect intestinal motility through the serotonin mechanism: 5-hydroxytryptamine (5-HT), also known as serotonin, produces 95% of 5-HT in the human body from the intestine (Li et al., 2019), which not only stimulates intestinal peristalsis, but is also essential for regulating visceral nerve sensation (Weiping and Bin, 2020). Renying (2022) found that SCFAs in the intestine of mice (B Chinese Journal of Anorectal Diseases, Vol. 44, No. 2, 2024, 17 acid, butyric acid, etc.) were significantly positively correlated with the level of 5-HT in the colon. Obata and Pachnis (2016) found that SCFAs can increase the level of 5-HT by stimulating ECs, thereby stimulating the vagus nerve and promoting gastrointestinal peristalsis. In addition, animal experiments have shown that Lactobacillus rhamnosus strains can specifically increase the concentration of 5-HT and fecal SCFAs in the colon of constipation model mice, thereby enhancing colonic smooth muscle contractility (Wang et al., 2020). SCFAs affect intestinal motility through the choline acetyltransferase mechanism: Choline acetyltransferase (ChAT) is an important biological enzyme required for acetylcholine synthesis, and studies have shown that SCFAs can promote the expression of ChAT to regulate neurogenes, thereby affecting intestinal tract movement (Soret et al., 2010). Suply et al. (2012) and Barichello et al. (2015) found that butyric acid in SCFAs, as an energy substrate in colon cells, can increase the proportion of cholinergic neurons and enhance the contraction function of colon muscles by restoring and increasing the content of glial cell-derived neurotrophic factors in mice. Soret et al. (2010) found that butyric acid can reduce the activity of deacetylase (HADC), thereby increasing the ChAT ratio to increase the proportion of cholinergic neurons and promote intestinal motility.
The enteric nervous system (ENS) is the core of the digestive and defense functions of the gastrointestinal tract, and is a complex system of neurons and glial cells in the intestinal wall, including reflex pathways related to normal intestinal peristalsis and sensory function. The gut microbiota is a vast ecosystem of symbiotic bacteria, fungi, viruses and other microorganisms. The gut microbiota not only regulates the motor program of the ENS, but is also essential for the normal structure and function of the ENS. The mechanism by which gut bacteria mediate movement through ENS is unknown. Traditionally, this interaction is thought to occur through immune mediators. However, there is growing evidence to support that the microbiota can interact directly with intestinal neurons. Intestinal neurons have been shown to express TLRs directly, and knockout models have shown a significant reduction in intestinal motility and thus exhibit a constipation phenotype. This suggests that bacteria or bacterial products may interact directly with intestinal neurons to promote ENS maturation, function, and intestinal motility. 5-HT is a key neurotransmitter in ENS that regulates intestinal motility and secretion responses, and studies have also confirmed that 5-HT can interact with the gut microbiota to initiate secretion and promote intestinal peristalsis by stimulating local intestinal nerve reflexes, and act on the vagus nerve to regulate intestinal contractility (Sikander et al., 2009). A mouse model of antibiotic-induced gut microbiota depletion found that the gut microbiota maintained the integrity of ENS by regulating the activity of intestinal neurons and promoting intestinal nerve development, in which LPS and short-chain fatty acids (SCFAs), the fermentation products of the microbiota, regulated the activity of intestinal neurons, and SCFA stimulated intestinal nerve development, so the gut microbiota is essential for the integrity of ENS (Vicentini et al., 2021). Under the influence of the natural and social environment, diet, brain, intestine and microbiota form a complex two-way network of interactions, which is called the “brain-gut-microbiota axis,” and a large amount of evidence confirms that the “brain-gut-microbiota axis” exists in both directions and plays a key role in the regulation of intestinal motility. Abnormal intestinal microbiota composition may lead to disruption of the brain-gut-microbiota axis signaling pathway, resulting in changes in intestinal motility. Impaired intestinal motility can be the result of dysfunction of control mechanisms at any level from the gut to the central nervous system, or it may be the result of a deficit in central nervous system regulation. The colonization process of intestinal flora initiates the signaling mechanism of the neural circuits that affect motor control and anxious behavior (Heijtz et al., 2011) and therefore plays an important role in the normal development of the central nervous system. It has been found that the gut microbiota is involved in controlling biological responses to stimuli such as digestion, immune system, and mood through the HPA axis (Walker et al., 2013). In addition, probiotics may reduce the expression of the inhibitory neurotransmitter GABA (γ-aminobutyric acid) receptor and the expression of cFos, a marker of neuronal activity, in the brain by modulating the gut-brain axis (Bercik et al., 2011; Bravo et al., 2011; Ait-Belgnaoui et al., 2014); It can also affect the activity of mi RNAs, which may be a mechanism by which intestinal microbes affect brain activity, thereby regulating the expression of important neuronal mRNAs (Laura De et al., 2021). In addition, the gut microbiota has the potential to directly or indirectly affect the nervous system through bacterial cellular components and microbiotic metabolites such as short-chain fatty acids, vitamins, and certain neurotransmitters (Badawy, 2017). Studies have shown that probiotics can stimulate intestinal endocrine cells to release gastrointestinal hormones, and immune cells can also stimulate the release of cytokines, which are then transported to the brain through vagus nerve afferents or blood, and the central nervous system changes the function of the gastrointestinal tract through return signals (Kennedy et al., 2017). A recent study found that the liver indirectly senses the intestinal microenvironment, activates hepatic vagus sensory afferents, and transmits signals to the brain, thereby regulating intestinal immune homeostasis through the vagus nerve, including the induction and maintenance of peripheral regulatory T cells (Teratani et al., 2020). Although ENS may be the main regulator of intestinal motility, both ENS and CNS are involved in intestinal motility, and both dysfunction or dysregulation can affect intestinal motility. Studies have shown that probiotics can mediate intestinal motility through the nervous system, providing evidence that probiotics may help regulate the intestinal tract or the central nervous system and normalize intestinal motility. The gastrointestinal microbiota plays a crucial role in intestinal motility, and studies conducted in germ-free mice have shown that both gastric emptying and intestinal motility are prolonged in the absence of gastrointestinal microbiota compared to wild-type mice (Waclawiková et al., 2022). One study showed that mice colonized with gastrointestinal microbiota had higher colonic contractility and significantly shorter intestinal peristalsis time (Kashyap et al., 2013). In rats colonized with specific pathogen-free microbiotas, colonization of Lactobacillus acidophilus, Bifidobacterium or Clostridium tobacco normalized the small intestine migratory motility complex and intestinal transit time, while colonization of E. coli inhibited intestinal electromyoelectric activity. Parthasarathy et al. (2016) found that fecal microbiota composition was correlated with colonic transit time, and the abundance of actinomycetes, bacteroides, lactococcus, and Rossiella was positively correlated with intestinal motility time, while Faecalibacterium was negatively correlated. Diet-induced changes in microbial composition may be partly mediated by changes in intestinal transit time, while dietary effects on intestinal transit time may be partly due to changes in gastrointestinal microbiota function caused by dietary changes. In addition, intestinal bacterial fermentation products can affect intestinal pH, permeability, and increase the production of gases (carbon dioxide, hydrogen, and methane) in the intestinal lumen, thereby reflexively causing intestinal smooth muscle contraction and promoting intestinal peristalsis (Bär et al., 2009).
We will continue to further explore in the future and continuously improve the relationship between microflora and constipation.
5 Conclusion
In conclusion, it can be inferred that there is a causal relationship between Eubacterium genus, Rumphococcus and constipation in Mendelian studies, respectively, with Eubacterium genus as a safety factor and Rumphococcus as a risk factor. In other words, an increase in the number of Rumphococcus leads to the direct occurrence of constipation, while the opposite is true for Eubacterium genus.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
XZ: Writing – original draft, Writing – review & editing. JC: Funding acquisition, Writing – review & editing. FH: Conceptualization, Writing – review & editing. WD: Project administration, Writing – review & editing. XY: Software, Visualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Guizhou Provincial Science and Technology Program, Contract No.: Qiankehe Foundation-ZK[2023] General 437.
Acknowledgments
XZ would like to thank Zhou Feng of Guizhou University for his support and the Big Data Lab for providing me with a place to obtain data. XZ would like to thank Wang Ziming of the First Affiliated Hospital of Guizhou University of Chinese Medicine for his valuable comments on the paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1376232/full#supplementary-material
References
Ait-Belgnaoui, A., Colom, A., Braniste, V., Ramalho, L., Marrot, A., Cartier, C., et al. (2014). Probiotic gut effectprevents the chronic psychological stress-induced brain activity abnormality in mice. Neurogastroenterol. Motil. 26, 510–520. doi: 10.1111/nmo.12295
Aoki, R., Tsuchida, S., Arai, Y., Ohno, K., Nishijima, T., Mawatari, T., et al. (2016). Effect of Bifidobacterium animalis subsp, lactis GCl.2505 on the physiological function of intestine in a rat model. Food Sci. Nutr. 4, 782–790. doi: 10.1002/fsn3.344
Arce, D. A., Ermocilla, C. A., and Costa, H. (2002). Evaluation of constipation. Am. Fam. Physician 65, 2283–2290.
Badawy, A. A. B. (2017). Tryptophan availability for kynurenine pathway metabolism across the life span: control mechanisms and focus on aging, exercise, diet and nutritional supplements. Neuropharmacology 112, 248–263. doi: 10.1016/j.neuropharm.2015.11.015
Bär, F., Von Koschitzky, H., Roblick, U., Bruch, H. P., Schulze, L., Sonnenborn, U., et al. (2009). Cell—free supernatants of Escherichia coli Nissle 1917 modulate human colonic motility: evidence from an in vitro organ bath study. Neurogastroenterol. Motil. 21, 559–566. doi: 10.1111/j.1365-2982.2008.01258.x
Barichello, T., Generoso, J. S., Simões, L. R., Faller, C. J., Ceretta, R. A., Petronilho, F., et al. (2015). Sodiunbutyrate prevents memory impairment by reestablishing BDNF and GDNFexpression in expenmental pneumococcal meningitis. Mol. Neurobiol. 52, 734–740. doi: 10.1007/s12035-014-8914-3
Baumeister, S., Meisinger, C., Leitzmann, M., Teumer, A., Bahls, M., Karch, A., et al. (2021). Physical activity and Parkinson's disease: a two-sample Mendelian randomisation study. J. Neurol. Neurosurg. Psychiatr. 92, 334–335. doi: 10.1136/jnnp-2020-324515
Bercik, P., Park, A. J., Sinclair, D., Khoshdel, A., Lu, J., Huang, X., et al. (2011). The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut—brain communication. Neurogastroenterol. Motil. 23, 1132–1139. doi: 10.1111/j.1365-2982.2011.01796.x
Bharucha, A. E. (2007). Constipation. Best. Pract. Res. Clin. Gastroenterol. 21, 709–731. doi: 10.1016/j.bpg.2007.07.001
Birney, E. (2022). Mendelian randomization. Cold Spring Harb. Perspect. Med. 12:a041302. doi: 10.1101/cshperspect.a041302
Bisht, P., Dagar, N., Kumar, N., Velayutham, R., and Arumugam, S. (2023). Potential targets in constipation research: a review. Curr. Drug Target. 24, 247–260. doi: 10.2174/1389450124666221209123541
Black, C. J., and Ford, A. C. (2018). Chronic idiopathic constipation in adults: epidemiology, pathophysiology, diagnosis and clinical management. Med. J. Aust. 209, 86–91. doi: 10.5694/mja18.00241
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525. doi: 10.1093/ije/dyv080
Bowden, J., Davey Smith, G., Haycock, P. C., and Burgess, S. (2016). Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314. doi: 10.1002/gepi.21965
Bowden, J., and Holmes, M. V. (2019). Meta-analysis and Mendelian randomization: a review. Res. Synth. Methods 10, 486–496. doi: 10.1002/jrsm.1346
Bowen, S., Mengxue, Y., Jun, L., Xue, Z., Fei, L., and Bo, Y. (2020). Analysis of intestinal microbiota diversity in patients with Graves' disease. J. Pract. Med. 36, 767–773.
Bravo, J. A., Forsythe, P., Chew, M. V., Escaravage, E., Savignac, H. M., Dinan, T. G., et al. (2011). Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U. S. A. 108, 16050–16055. doi: 10.1073/pnas.1102999108
Burgess, S., Scott, R. A., Timpson, N. J., Davey Smith, G., and Thompson, S. G. (2015). Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol. 30, 543–552. doi: 10.1007/s10654-015-0011-z
Cao, H., Liu, X., An, Y., Zhou, G., Liu, Y., Xu, M., et al. (2017). Dysbiosis contributes to chronic constipation development via regulation of serotonin transporter in the intestine. Sci. Rep. 7:10322. doi: 10.1038/s41598-017-10835-8
Chen, J., Chen, X., and Ho, C. L. (2021). Recent development of probiotic bifidobacteria for treating human diseases. Front. Bioeng. Biotechnol. 9:770248. doi: 10.3389/fbioe.2021.770248
Chu, J. R., Kang, S. Y., Kim, S. E., Lee, S. J., Lee, Y. C., and Sung, M. K. (2019). Prebiotic UG1601 mitigates constipation-related events in association with gut microbiota: a randomized placebo-controlled intervention study. World J. Gastroenterol. 25, 6129–6144. doi: 10.3748/wjg.v25.i40.6129
Couto, M. R., Gonçalves, P., Magro, F., and Martel, F. (2020). Microbiota-derived butyrate regulates intestinal infammation Focuson inflammatory bowel disease. Pharmacol. Res. 2020:104947.doi: 10.1016/j.phrs.2020.104947
Di Domenico, M., Ballini, A., Boccellino, M., Scacco, S., Lovero, R., Charitos, I. A., et al. (2022). The intestinal microbiota may be a potential theranostic tool for personalized medicine. J. Pers. Med. 12:40523. doi: 10.3390/jpm12040523
Dong, Q., Chen, D., Zhang, Y., Xu, Y., Yan, L., and Jiang, J. (2023). Constipation and cardiovascular disease: a two-sample Mendelian randomization analysis. Front. Cardiovasc. Med. 10:1080982. doi: 10.3389/fcvm.2023.1080982
Guo, M. Q., Yao, J. F., Yang, F., Liu, W., Bai, H., Ma, J., et al. (2020). The composition of intestinal microbiota and its association with functional constipation of the elderly patients. Fut. Microbiol. 15, 163–175. doi: 10.2217/fmb-2019-0283
Hartwig, F. P., Davey Smith, G., and Bowden, J. (2017). Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998. doi: 10.1093/ije/dyx102
Heijtz, R. D., Wang, S., Anuar, F., Qian, Y., Björkholm, B., Samuelsson, A., et al. (2011). Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U. S. A. 108, 3047–3052. doi: 10.1073/pnas.1010529108
Hemani, G., Zheng, J., Elsworth, B., Wade, K. H., Haberland, V., Baird, D., et al. (2018). The MR-Base platform supports systematic causal inference across the human phenome. Elife 7:12. doi: 10.7554/eLife.34408.012
Jie, P., Laihao, L., and Jianwei, M. (2021). Research progress on intestinal microbiota and human health. J. Shandong Norm. Univ. 36, 337–365.
Juge, N. (2023). Microbe profile: Ruminococcus gnavus: the yin and yang of human gut symbionts. Microbiology 169:1383. doi: 10.1099/mic.0.001383
Kashyap, P. C., Marcobal, A., Ursell, L. K., Larauche, M., Duboc, H., Earle, K. A., et al. (2013). Complex interactionsamong diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology 144, 967–977. doi: 10.1053/j.gastro.2013.01.047
Kennedy, P. J., Cryan, J. F., Dinan, T. G., and Clarke, G. (2017). Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology 112, 399–412. doi: 10.1016/j.neuropharm.2016.07.002
Laura De, V., Cara, H., Marie, F., and Clayton, C. (2021). Ingestion of Bifidobacterium longum changes mi RNA levels in the brains of mice. PLoS ONE 16:e0249817. doi: 10.1371/journal.pone.0249817
Li, C., Maijiao, P., and Zhoujin, T. (2022). Research progress on short-chain fatty acids associated with intestinal microbiota. World Chin. J. Digest. 30, 562–570. doi: 10.11569/wcjd.v30.i13.562
Li, H., Qian, X., Xiaoxiao, X., Chunqiang, L., and Yebao, H. (2024). Bidirectional Mendelian randomization of intestinal microorganisms and constipation. Prev. Med. 36, 198–202. doi: 10.19485/j.cnki.issn2096-5087.2024.03.004
Li, X., Shengfeng, L., Bingmei, Z., Xinyue, J., and Shuping, F. (2019). Research progress on 5-HT and intestinal microbiota and its role in gut-brain related diseases. Chin. J. Rehabil. Med. 34, 116–119.
Mancabelli, L., Milani, C., Lugli, G. A., Turroni, F., Mangifesta, M., Viappiani, A., et al. (2017). Unveiling the gut microbiota composition and functionality associated with constipation through metagenomic analyses. Sci. Rep. 7:10663. doi: 10.1038/s41598-017-10663-w
Matsumoto, K., Takada, T., Shimizu, K., Moriyama, K., Kawakami, K., Hirano, K., et al. (2010). Effects of aprobiotic fermented milk beverage containing Lactobacillus casei strain Shirota on defecation frequency, intestinal microbiota, and the intestinal environment of healthy individuals with soft stools. J. Biosci. Bioeng. 110, 547–552. doi: 10.1016/j.jbiosc.2010.05.016
Meng, X., Zhang, G., Cao, H., Yu, D., Fang, X., de Vos, W. M., et al. (2020). Gut dysbacteriosis and intestinal disease: mechanism and treatment. J. Appl. Microbiol. 129, 787–805. doi: 10.1111/jam.14661
Obata, Y., and Pachnis, V. (2016). The effect of microbiota and theimmune system on the development and organization ofthe enteric nervous system. Gastroenterology 151, 836–844. doi: 10.1053/j.gastro.2016.07.044
Ohkusa, T., Koido, S., Nishikawa, Y., and Sato, N. (2019). Gut microbiota and chronic constipation: a review and update. Front. Med. 6:19. doi: 10.3389/fmed.2019.00019
Parthasarathy, G., Chen, J., Chen, X., Chia, N., O'Connor, H. M., Wolf, P. G., et al. (2016). Relationship between microbiota of thecolonic mucosa vs. feces and symptoms, colonic transit, and methane production in female patients with chronic constipation. Gastroenterology 150, 367–379. doi: 10.1053/j.gastro.2015.10.005
Renying, Z. (2022). Evaluation of the Efficacy and Mechanism of Compound Probiotics in Alleviating Depression. Wuxi: Jiangnan University.
Shufei, Z., Lin, Z., and Ping, Z. (2020). Research progress on the correlation between chronic kidney disease and gut microbiota. J. Pract. Med. 36, 1684–1688.
Sikander, A., Rana, S. V., and Prasad, K. K. (2009). Role of serotonin in gastrointestinal motility and irritable bowel syndrome. Clin. Chim. Acta 403, 47–55. doi: 10.1016/j.cca.2009.01.028
Soret, R., Chevalier, J., De Coppet, P., Poupeau, G., Derkinderen, P., Segain, J. P., et al. (2010). Shorrchain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology 138, 1772–1782. doi: 10.1053/j.gastro.2010.01.053
Suply, E., de Vries, P., Soret, R., Cossais, F., and Neunlist, M. (2012). Butyrate enemas enhance both cholinergic and nitrergic phenotype of myenteric neurons and neuromuscular transmission irnewborn rat colon. Am. J. Physiol. Gastrointest. Liver Physiol. 302, G1373–G1380. doi: 10.1152/ajpgi.00338.2011
Teratani, T., Mikami, Y., Nakamoto, N., Suzuki, T., Harada, Y., Okabayashi, K., et al. (2020). The liver-brain -gutneural arc maintains the Treg cell niche in the gut. Nature 585, 591–596. doi: 10.1038/s41586-020-2425-3
Thursby, E., and Juge, N. (2017). Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836. doi: 10.1042/BCJ20160510
Vicentini, F. A., Keenan, C. M., Wallace, L. E., Woods, C., Cavin, J. B., Flockton, A. R., et al. (2021). Intestinal microbiota shapes gut physiology and regulates enteric neurons and glia. Microbiome 9:210. doi: 10.1186/s40168-021-01165-z
Waclawiková, B., Codutti, A., Alim, K., and Aidy, S. E. (2022). Gut microbiota-motility interregulation: insights from in vivo, ex vivo and in silico studies. Gut Microbes 14:1997296. doi: 10.1080/19490976.2021.1997296
Walker, E. F., Trotman, H. D., Pearce, B. D., Addington, J., Cadenhead, K. S., Cornblatt, B. A., et al. (2013). Cortisol levels and risk for psychosis: initial findings from the north american prodrome longitudinal study. Biol. Psychiatr. 74, 410–417. doi: 10.1016/j.biopsych.2013.02.016
Wang, G., Yang, S., Sun, S., Si, Q., Wang, L., Zhang, Q., et al. (2020). Lactobacillus rhamnosusstrains relieve loperamide induced constipation via different pathways independent of shortchain fatty acids. Front. Cell Infect. Microbiol. 2020:423. doi: 10.3389/fcimb.2020.00423
Wang, J., Yang, M., Xu, K., Wan, X., Xie, J., Yu, H., et al. (2023). The causal associations between growth factors and constipation: a two-sample Mendelian randomization study. Front. Physiol. 14:1204146. doi: 10.3389/fphys.2023.1204146
Weiping, Z., and Bin, J. (2020). Serotonin and chronic constipation. World Chin. J. Digest. 19, 2551–2554.
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y. Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. doi: 10.1126/science.1208344
Xu, H., Chen, X., Luge, W., Pengwei, Z., Yuwei, L., and Yanjun, Z. (2021). Analysis of intestinal flora in patients with slow transit constipation and defecation disorder. Gastroenterology 26, 724–731.
Yarullina, D. R., Shafigullin, M. U., Sakulin, K. A., Arzamastseva, A. A., Shaidullov, I. F., Markelova, M. I., et al. (2020). Characterization of gut contractility and microbiota in patients with severe chronic constipation. PLoS ONE 15:235985. doi: 10.1371/journal.pone.0235985
Yumiao, W., Yaowei, W., Wanqing, Z., Yayun, Y, and Weiwei, L. (2022). Research progress on the relationship between short-chain fatty acids and childhood asthma and their treatment. Chin. J. Modern Med. 32, 37–42.
Zhao, Y., and Yu, Y. B. (2016). Intestinal microbiota and chronic constipation. Springerplus 5:1130. doi: 10.1186/s40064-016-2821-1
Keywords: constipation, Mendelian randomization, causal relationship, GWAS, Rumphococcus, Eubacterium genus
Citation: Zhang X, Chen J, He F, Du W and Yu X (2024) Assessing the causal effects of Eubacterium and Rumphococcus on constipation: a Mendelian randomized study. Front. Microbiol. 15:1376232. doi: 10.3389/fmicb.2024.1376232
Received: 21 May 2024; Accepted: 17 July 2024;
Published: 31 July 2024.
Edited by:
Yu Pi, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Senhui Weng, Guangdong Provincial Hospital of Traditional Chinese Medicine, ChinaCosmin Mihai Vesa, University of Oradea, Romania
Copyright © 2024 Zhang, Chen, He, Du and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianhao Yu, MjIyNzA3MzRxcS5jb20=
 Xiao Zhang
Xiao Zhang Jiang Chen
Jiang Chen Feng He1
Feng He1