
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 12 April 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1370553
This article is part of the Research TopicOutbreak Investigations of Nosocomial InfectionsView all 14 articles
 Sandra Dos Santos1
Sandra Dos Santos1 Seydina M. Diene2,3,4
Seydina M. Diene2,3,4 Amina Benouda5
Amina Benouda5 Khalid Zerouali6
Khalid Zerouali6 Doaa M. Ghaith7
Doaa M. Ghaith7 Rasha H. El-Mahdy8
Rasha H. El-Mahdy8 Sawsan H. M. El Tayeb9
Sawsan H. M. El Tayeb9 Ilhem Boutiba10
Ilhem Boutiba10 Adnene Hammami11
Adnene Hammami11 Remie Chrabieh12
Remie Chrabieh12 Ziad Daoud13
Ziad Daoud13 Laurent Mereghetti14
Laurent Mereghetti14 Patrice Francois15
Patrice Francois15 Nathalie Van Der Mee-Marquet1*
Nathalie Van Der Mee-Marquet1*  on behalf of the consortium
on behalf of the consortiumIntroduction: The colonization of patients by carbapenemase-producing Enterobacterales (CPE) has been associated with heightened mortality, especially in vulnerable individuals within intensive care units (ICUs). Our study aimed to comprehensively assess CPE prevalence among ICU patients across the Mediterranean region pre-COVID-19, conducting a multicenter prevalence study in the first quarter of 2019.
Methods: We collected clinical data and rectal or fecal samples from 256 ICU patients for CPE testing. Additionally, we performed whole-genome sequencing on 40 representative CPE strains to document their molecular characteristics.
Results: Among the 256 patients, CPE was detected in 73 samples (28.5%), with prevalence varying from 3.3 to 69.0% across participating centers. We observed 13 colistin-resistant CPE strains, affecting three ICUs. Genetic analysis revealed highly diverse E. coli and K. pneumoniae strains, predominantly from international high-risk clones. Notably, blaOXA-48 and blaNDM-1 were the most prevalent carbapenemase genes. Molecular typing uncovered potential patient clusters in six centers. Significantly, longer hospital stays were associated with increased CPE carriage (p < 0.001). Nine centers across Morocco, Tunisia, Egypt, and Lebanon voluntarily participated.
Discussion: Our study provides CPE prevalence in Mediterranean ICUs and reaffirms established CPE presence in this setting but also provides updates on the molecular diversity of CPE strains. These findings highlight the imperative of reinforcing infection control measures in the participating ICUs to curtail escalated mortality rates, and of strictly applying isolation measures around patients originating from the Mediterranean region when transferred to other healthcare institutions.
The colonization of patients with carbapenemase-producing Enterobacterales (CPE) has been associated with increased mortality among vulnerable individuals, particularly those admitted to intensive care units (ICUs) (Dautzenberg et al., 2015; Tischendorf et al., 2016). Preventing the spread of these multidrug-resistant bacteria in ICUs involves a multifaceted approach, including the promotion and strict adherence to hand hygiene rules, proper disinfection of reusable medical equipment, implementation of strict cleaning and disinfection protocols to reduce environmental contamination, the appropriate use of broad-spectrum antibiotics, education and training of healthcare professionals, implementation of surveillance to quickly detect CPE-associated nosocomial infections, and the quick identification and isolation of CPE carriers to prevent transmission to other patients. Complementing the survey of CPE-associated infections and aiding in the implementation of preventive measures to curb the spread of CPE within ICUs, the epidemiological monitoring of patients carrying CPE is a key-tool to document the epidemiology of these bacteria in ICUs. The ongoing epidemiological data facilitates early detection of potential outbreaks, and assists in the better selection of probabilistic treatments to manage locally acquired infections.
Comprehensive epidemiological data on patient CPE-colonization within ICUs in Mediterranean countries are rather limited, in particular prevalence studies. Prior studies predominantly focused on outbreaks associated with K. pneumoniae carrying the blaOXA-48 gene, notably the 2011 Casablanca study (Barguigua et al., 2015), and those in 2014 involving patients from Algeria and Tunisia (Cuzon et al., 2015; Mathlouthi et al., 2016). Another significant source of clonal dissemination has been identified in K. pneumoniae carrying the blaNDM-1 gene, previously documented in Moroccan ICUs in 2011 (Barguigua et al., 2015) and in ICUs in Cairo in 2013 and 2016 (Wassef et al., 2016; Abdulall et al., 2018). A study conducted in 2016 by Hammami and colleagues highlighted that 34.9% of ICU patients at the University Hospital of Tunis harbored CPE with the blaOXA-48 and/or blaNDM-1 genes in their intestinal flora (Hammami et al., 2017). Additionally, to the best of our knowledge, the colonization of ICU patients in Mediterranean countries by colistin-resistant CPE has not been extensively reported, except for an outbreak of colistin-resistant blaOXA48 K. pneumoniae in 2015 among ICU patients at the University Hospital of Mahdia in Tunisia (Mansour et al., 2017). In this instance, the resistance to colistin was not attributed to the presence of mcr genes within the plasmids carried by this K. pneumoniae.
Our study aimed to assess the prevalence of CPE carriage among ICU patients in hospitals across diverse Mediterranean countries. This involved a multicenter prevalence study conducted just before the onset of the COVID-19 pandemic, in the first quarter of 2019 across a group of nine ICUs. Carbapenem-resistant strains, isolated from rectal or fecal samples, underwent molecular analyses to determine their susceptibility to antibiotics. Additionally, whole-genome sequencing was performed on 40 representative CPE strains to document the molecular characteristics of the predominant clones. Examination of the clinical data was also undertaken to investigate the risk factors associated with the intestinal carriage of CPE among patients in the participating ICUs.
A one-day prevalence study was conducted from January 1, 2019, to March 30, 2019, spanning nine centers in hospitals located in Morocco (2 centers), Tunisia (2), Egypt (3), and Lebanon (2). These centers—labeled C1 to C9—participated voluntarily, aiming to enroll 30 consecutive ICU patients in each. Prior consent from patients or their relatives was sought to access medical records and collect fecal or rectal samples for testing CPE.
The data were collected by the attending physicians. Collected data encompassed demographic and clinical information, including physical disabilities, significant underlying health conditions, and established risk factors associated with CPE carriage. These risk factors covered the duration of pre-screening hospitalization, prior hospitalization history, and recent antibiotic usage (on the study day and within the preceding 6 months).
Fecal or rectal swabs were collected using swabs with Amies-type preservation medium (Copan Italia SPA, Italy).
Clinical swabs were promptly screened for third-generation cephalosporin- and carbapenem- resistant Enterobacterales using CHROMagar ESBL and mSuperCARBA chromogenic agar plates (CHROMagar, Paris, France) at each participating center. After incubation of the chromogenic agar plates at 37°C for 24 h under aerobic conditions, each colony morphotype was taken into consideration. All Enterobacterales strains were then transferred to the central laboratory in Tours, France, following inoculation on a swab with Amies-type preservation medium. In Tours, we controlled the identification of each colony morphotype using Matrix-assisted laser desorption-ionization-time–of-flight mass spectrometry (microflex LT MALDI-TOF, MBT Compass software version 4.2.100.19; Bruker Daltonics, France). Subsequently, all Enterobacterales strains underwent antibiotic sensitivity testing using disc diffusion method (EUCAST, 2019), and determination of Ertapenem Minimum Inhibitory Concentration (MIC) (E-test strip; bioMérieux, Marcy-l’Étoile, France) and colistin MIC using a broth microdilution method as advised by EUCAST for Enterobacterales (EUCAST, 2019) (colistin UMIC plates; Bruker Daltonics, France). Carbapenemase production was investigated when Ertapenem MIC >0.5 mg/L using the immunochromatographic test RESIST-5 O.K.N.V.I (Coris BioConcept, Belgium) and confirmed using a PCR test for molecular detection of the genes encoding for the five carbapenemases of primary public health concern (blaOXA-48, blaNDM, blaKPC, blaVIM and blaIMP genes) (Doyle et al., 2012).
Among all CPE strains, 40 were thoughtfully chosen after applying the RAPD technique for epidemiological typing, using three primers as previously described (van der Mee-Marquet et al., 2010). To avoid redundancy, strains with similar RAPD profiles from the same center were excluded, retaining only one for sequencing. The chosen strains underwent whole-genome sequencing (WGS) via next-generation sequencers. Genomic DNA, purified using the DNeasy kit (Qiagen), was sequenced on the Illumina HiSeq, generating 100-base pairs (bp) paired-end reads with barcoding using the Nextera XT kit. Data Processing: Read quality assessment was performed using Trimmomatic v0.36, with specified parameters for paired-end reads. Genome assembly was conducted using the SPADES v3.12.0 assembler. Assembled genomes and plasmids1 were annotated using the RAST server.2 Multilocus sequence typing analysis (in silico) developed by Achtamn et al. 3was conducted using annotated genomes and submitted to the Center for Genomic Epidemiology database.4 The phylogenetic relationships of all isolates were investigated through genomic single-nucleotide polymorphism (SNP)–based analysis (CSIPhylogeny). Additional tools, available at the Center for Genomic Epidemiology,5 were used for detecting antibiotic resistance,6 mobile genetic elements, 7and virulence factors.8 The investigation of phylogenetic relationships among isolates involved genomic single-nucleotide polymorphism (SNP)–based analysis (CSIPhylogeny).
Statistical analysis was performed using univariable methods, employing chi2 and Fisher’s exact test as appropriate. All tests were 2-tailed, and a significant level of p < 0.05 was used.
The study was conducted in collaboration with hospital directors, attending physicians, and the infection control pilot team in Tours, France, in adherence to French Healthcare recommendations for infection prevention. Ethical approvals were obtained at eight hospital centers, ensuring anonymization of patient and sample data prior to analysis. Data from center 9 were not included in the study.
A total of 256 ICU patients were included in the study. The clinical data were available for 226 patients (Table 1). For all but one of the participating centers, the number of patients per center was approximately 30. Due to patient refusals to participate, center C8 enrolled only 16 patients. Out of the 256 patients, 100 were women (44.0%), and 126 were men (46.0%), with ages ranging from 6 days to 102 years (mean age: 57 years). Various medical conditions were observed among the patients, with 15.5% having experienced trauma, 31.4% having diabetes mellitus, and 26.1% having malignancies. Among the patient population, 44.5% had recent hospitalization history, and 48.0% had received antibiotic treatment in the last 6 months. The most common antibiotics used included third-generation cephalosporins (28.1%) and carbapenems (10.1%). On the day of the study, 74.2% of the patients were under antibiotic treatment, which primarily consisted of third-generation cephalosporins (37.2%), carbapenems (20.4%), amoxicillin-clavulanic acid (13.3%), or fluoroquinolones (7.5%).
A total of 110 strains of carbapenem-resistant Enterobacterales were cultured on SuperCarba plates of which 103 (93.6%) were confirmed as CPE strains. The remaining seven strains that did not produce carbapenemase were obtained from four different centers, consisting of two Proteus mirabilis strains, one Morganella morgannii, one K. pneumoniae, and three E. coli.
Out of the 103 CPE strains, 61 were K. pneumoniae (59.2%), while there were two K. aerogenes, 31 E. coli (30.1%), seven C. freundii, and two E. cloacae (Table 2). Among the 31 E. coli strains, 20 were resistant to ciprofloxacin (64.5%), 21 to trimethoprim-sulfamethoxazole (67.7%), 12 to gentamicin (38.7%), and 15 to tobramycin (48.4%). Two strains were resistant to all tested aminoglycosides and fluoroquinolones (6.4%) but remained susceptible to fosfomycin, tigecycline, and colistin. Multi-resistance to antibiotics was more prevalent among the 61 K. pneumoniae strains. Specifically, 59 were resistant to ciprofloxacin (93.8%), 56 to tobramycin (90.3%), 50 to fosfomycin (83.3%), 45 to gentamicin (75.0%), and 44 to trimethoprim-sulfamethoxazole (73.3%). Twenty-seven strains were resistant to all tested aminoglycosides and fluoroquinolones (43.5%) but remained susceptible to tigecycline (100.0%) and colistin (81.4%). The detected carbapenemase genes included blaOXA-48 (44.7%), blaOXA-181 (6.8%), blaOXA-244 (4.8%), blaNDM-1 (19.4%), blaNDM-5 (12.6%), blaNDM-7 (2.9%), and blaNDM-4 (1.0%). Ten out of the 103 CPE strains (9.7%) co-carried both blaOXA-48 and blaNDM carbapenemase genes (Table 2). The blaOXA-244 gene was found to be associated with E. coli (p = 0.002), and the blaNDM-1 gene showed an association with K. pneumoniae (p = 0.011).
CPE strains were detected in 73 out of the 256 clinical samples, accounting for 28.5% of the total (Supplementary Table 1). The number of CPE strains varied among carriers, with the majority of carriers exhibiting a single CPE strain (50 cases, 68.5%). The prevalence of CPE carriage ranged from 3.3 to 69.0% across the participating centers (Figure 1).
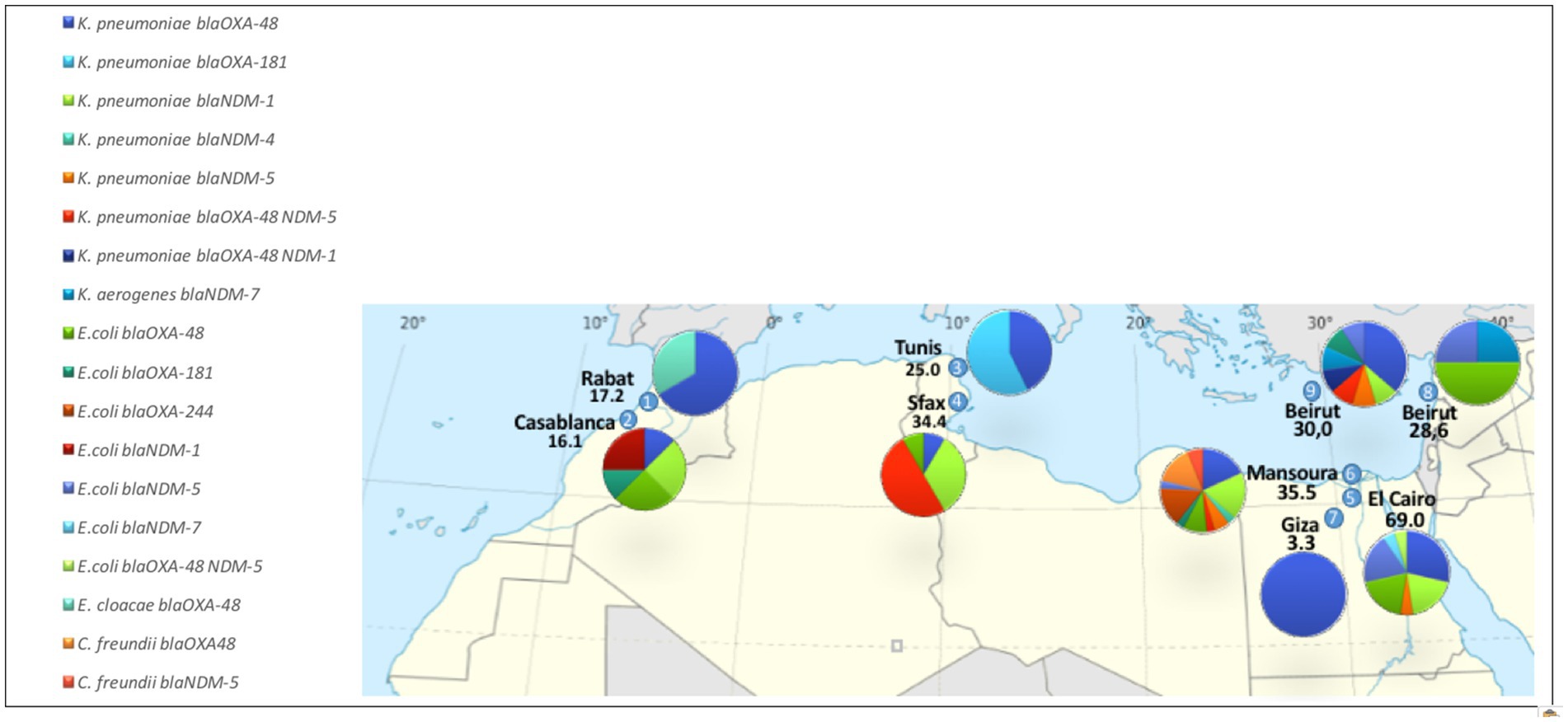
Figure 1. Prevalence of CPE carriage and distribution of the 103 CPE strains, stratified across participating centers.
CPE carriers experienced a prolonged duration of hospitalization before CPE screening. Specifically, 42.5% of CPE carriers had a hospitalization period exceeding 7 days, in contrast to 16.5% of non-carriers (p < 0.001). However, antibiotic therapy or hospitalization, both prevalent in the studied population (31.6 and 39.4%, respectively), did not show significant differences based on CPE carriage status.
Among the 110 Enterobacterales strains cultured on SuperCarba plates, 14 showed resistance to colistin, comprising four E. coli and ten K. pneumoniae. These 14 strains consisted of 13 CPE strains and one ESBL-producing E. coli. Notably, one individual from center C5 was found to carry two of these strains. This resulted in a colistin-resistant CPE carriage rate of 4.7%, affecting three of the eight ICUs (centers C3, C5, and C6). Among the 14 colistin-resistant strains identified, the four E. coli strains carried the mcr1 gene (28.6%)—three of which were CPE strains, alongside the ESBL-producing strain (Table 2). The three CPE strains carrying mcr1 were isolated from patients in center C6, co-harboring blaOXA-48, and remained susceptible to amikacin, fosfomycin, and tigecycline. Additionally, the ten colistin-resistant K. pneumoniae strains were isolated from eight patients in center C5 and two in center C3. These strains predominantly carried blaOXA-48 or blaNDM-5 and exhibited susceptibility solely to tigecycline (100.0%) and amikacin (30.0%).
Epidemiological typing of all strains was performed to select meticulously a representative set of CPE strains. RAPD typing revealed 15 clusters of strains, exhibiting similar RAPD-types in six of the nine centers (Supplementary Table 2). The number of clusters varied from zero (centers C2, C7, and C8) to six (center C6), with a median of one cluster. The clusters consisted of two to six strains each, predominantly represented by K. pneumoniae strains. There were ten K. pneumoniae clusters, encompassing strains belonging to eight different sequence-types (STs). Notably, the three E. coli strains carrying mcr1, isolated from patients in center C6, exhibited identical RAPD-types. There was no discernible correlation between a high prevalence of CPE carriers and the number of clusters. For instance, in center C6, where 35.5% of the patients were identified as CPE carriers, six clusters of strains with similar RAPD-types were identified. In contrast, in center C5, which had the highest rate of CPE carriage (69.0% of ICU patients), RAPD-typing revealed only two strains sharing the same RAPD-types among 26 CPE strains.
WGS was performed on a set of 40 CPE strains, along with the ESBL-producing E. coli carrying mcr1. The phylogenetic relationships among the 41 studied strains are depicted in Figure 2. Genetic diversity was notably observed among the eight studied E. coli strains, encompassing eight distinct sequence types (STs). Of these, the three CPE strains carrying mcr1 belonged to ST1196, distinct from the ESBL-producing strain carrying mcr1, which was identified as ST359. The 29 K. pneumoniae strains exhibited slightly less diversity, comprising 15 different sequence types (STs). The CPE strains identified in the nine ICUs predominantly belonged to various clones. Specifically, only four K. pneumoniae clones were found in more than one center: the ST101 clone, detected in centers C4, C5, C6, and C9; the ST383 clone, identified in centers C4, C5, and C9; and the ST147 and ST231 clones, each detected in two centers (centers C4 and C6, and centers C3 and C7, respectively). The five colistin-resistant K. pneumoniae strains that underwent WGS were associated with STs 383, 307, and 1626. A detailed analysis of amino acid sequences in these strains revealed a 663-bp sequence in the K. pneumoniae Sample_25 genome, replacing the mgrB gene with an insertion sequence from the IS1 family. Moreover, multiple mutations were identified in PmrA and PmrB proteins, including a common mutation (T246A) in PmrB shared among all isolates when compared to the colistin-susceptible MGH57578 isolate (Supplementary Table 3). Importantly, no mutations were found in PhoP and PhoQ proteins, and no significant hits to MCR-1 to MCR-10 were detected in the genomes of these colistin-resistant strains.
The resistome study confirmed that most isolates carried genes associated with resistance to fluoroquinolones (92.5%), aminoglycosides (97.5%), sulfonamides (90.0%), macrolides (50.0%), tetracyclines (70.0%), phenicoles (72.5%), and trimethoprim (90.0%) (Supplementary Tables 4, 5). In addition to the numerous genes associated with antimicrobials used in the human clinics, WGS analysis revealed the frequent carriage of genes associated with multidrug resistance, as well as resistance to bicyclomycin — an antimicrobial agent used in livestock environments —, to the antimalarial agent Fosmidomycin, and to numerous heavy metals (Supplementary Table 6).
The virulome study revealed that the majority of E. coli and K. pneumoniae CPE strains carried genes encoding adhesins, hemolysins, and siderophores (Supplementary Table 7). Additionnaly, E. coli strains exhibited the coiled surface structure curlin, which is involved in inert surface colonization and biofilm formation (csgA), along with numerous distinct toxin-antitoxin systems that are well-known for genetic element maintenance, virulence, stress resistance and phage inhibition (Qiu et al., 2022).
The plasmid study identified 219 plasmids within the genomes of the 40 CPE strains. The number of plasmid sequences per bacterial genome ranged from zero to 10, with a median value of six. Plasmid sequences were less prevalent among E. coli isolates, Citrobacter and E. cloacae (median value of one plasmid per genome) compared to the K. pneumoniae species (median value of seven plasmids per genome) (p = 0.005). Genomic comparison of the plasmidic sequences revealed eight plasmid groups (1–8), with plasmids belonging to groups 4, 6 and 7 carried by 65.5, 65.5 and 75.9% of the K. pneumoniae strains, respectively (Supplementary Figure 1).
Our comprehensive multicenter point-prevalence survey investigated CPE carriage among 226 ICU patients in nine hospitals across Morocco, Tunisia, Egypt, and Lebanon, conducted pre-COVID-19 pandemic. To our knowledge, this study is unique in providing comprehensive prevalence data on patient colonization in Mediterranean ICUs, distinguishing it as the most recent investigation of its kind.
Firstly, the study unveiled a noteworthy prevalence of CPE strains in eight of the participating ICU centers, demonstrating CPE carriage rates ranging from 16.1 to 69.0% among ICU patients. Additionally, we observed intestinal colonization by colistin-resistant CPE strains in patients from centers 3, 5, and 6. The heightened prevalence of CPE colonization across nearly all ICUs, with the exception of one, increases the risk of subsequent infections in these vulnerable patients harboring highly resistant isolates. This poses inherent challenges in treatment. While it’s highly probable that the prevalence of CPE and colistin-resistant CPE strains has increased during the pandemic (Pascale et al., 2021), our findings are particularly concerning due to the high mortality associated with the identified clones within intensive care settings. Strengthening or set up infection control measures in these ICUs is essential to mitigate the heightened mortality risk among patients. These findings also emphasize the importance of strictly implementing isolation measures for patients originating from the Mediterranean region when transferred to other healthcare institutions.
Secondly, molecular typing of the identified CPE strains unveiled distinct scenarios across centers. In the two ICUs with more than one CPE carrier and no identified clusters (centers C2 and C8), the predominant CPE strains were mostly E. coli strains (8/12; 66.6%). WGS analysis revealed carbapenemase-producing E. coli belonging to three international high-risk clones [ST410 (Roer et al., 2018), ST167 (Roer et al., 2018; Garcia-Fernandez et al., 2020), and ST69 (Shawa et al., 2022)], known for causing life-threatening infections, particularly affecting critically ill patients with severe underlying diseases and comorbidities, and five globally recognized clones, previously reported in livestock and food products (Stolle et al., 2013; Aizawa et al., 2014; Guo et al., 2015; Wang et al., 2018; Vasconcelos et al., 2020). It appears plausible that the acquisition of these genetically diverse and unrelated E. coli strains occurred in the community setting before patients’ hospitalization. In contrast, in the six remaining centers where clusters were identified, CPE strains were predominantly K. pneumoniae strains (32/49; 65.3%). The one-day prevalence study did not afford an investigation into the mechanisms of patient acquisition of CPE strains from these clusters. Longitudinal data tracking CPE colonization over time would offer insights into trends, persistence, and factors influencing acquisition and clearance of CPE strains. However, the WGS analysis showed that the identified K. pneumoniae strains were predominantly associated with international high-risk clones known for their propensity to thrive in the ICU bedside environment and initiate outbreaks (Weterings et al., 2015; Gordon et al., 2017; Liu et al., 2018; Naha et al., 2021; da Silva et al., 2022; De Koster et al., 2022; Edward et al., 2022; Shi et al., 2022). Thus, our data tentatively suggest nosocomial acquisition of CPE K. pneumoniae by patients in the ICUs where clusters were identified. Further evidence is needed for conclusive confirmation.
Concerning the genetic determinants responsible for carbapenem and colistin resistance, our study corroborates previous findings. In alignment with recent global epidemiology of carbapenemase genes (van Duin and Doi, 2017; Pitout et al., 2019), blaOXA-48 and blaNDM-1 were the most frequently detected genes among the identified CPE strains. Additionally, consistent with observations in Tunisian patients, the studied colistin-resistant K. pneumoniae strains did not show the presence of mcr-1 to mcr-10 genes (Jaidane et al., 2018).
When comparing carriers and non-carriers, our study did not identify prior hospitalization or antibiotic treatment as significant risk factors for CPE colonization. The limited number of ICU patients and the high rate of hospitalization and antibiotic treatment in our sample may have influenced these results. However, our findings confirmed that ICU stays of 7 days or more significantly increased the risk of CPE colonization [1;2]. Once again, the identification of this risk factor reinforces the hypothesis of a major acquisition of CPE strains in the ICU setting during patient care, potentially exacerbated by the failure to identify and isolate CPE carriers within the participating centers
Our research conclusively establishes the presence and widespread distribution of CPE strains within Mediterranean hosipitals. Significantly, our study adds value by furnishing data on the prevalence of patient colonization and the genetic characteristics of the detected CPE strains—information notoriously challenging to procure. However, the study including a relatively small sample size of ICU patients, with variable enrollment rates across centers, the generalizability of the findings to broader populations may be limited, and the variability in enrollment rates could introduce biases. Prioritizing the awareness of healthcare professionals regarding the escalating threat of antibiotic resistance is crucial, particularly within ICUs. Replicating this study in additional Mediterranean hospitals would promote the implementation of preventive measures and the adjustment of empirical antibiotic treatments for infected ICU patients, if necessary. Finally, our findings highlight the importance of vigilance concerning patients previously hospitalized in Mediterranean facilities and undergoing medical repatriations, to prevent the introduction of CPE into downstream healthcare facilities.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ebi.ac.uk/ena, PRJEB71867.
The study was conducted in collaboration with hospital directors, attending physicians, and the infection control pilot team in Tours, France, in adherence to French Healthcare recommendations for infection prevention. Ethical approvals were obtained at eight hospital centers, ensuring anonymization of patient and sample data prior to analysis. Data from center 9 were not included in the study. The study was conducted following a protocol in adherence to French Healthcare recommendations for infection prevention. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board also waived the requirement of written informed consent for participation from the participants or the participants' legal guardians/next of kin because prior verbal consent was obtained.
SS: Methodology, Writing – original draft, Writing – review & editing, Formal analysis, Investigation. SD: Formal analysis, Methodology, Validation, Writing – original draft, Writing – review & editing. AB: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. KZ: Data curation, Investigation, Writing – original draft, Writing – review & editing. DG: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. RE-M: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. SE: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. IB: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. AH: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. RC: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. ZD: Data curation, Investigation, Validation, Writing – original draft, Writing – review & editing. LM: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. PF: Formal analysis, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. NM-M: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
We acknowledge all the members of the group for CPE control (listed in alphabetic order): SH ABD EL AZIZ (Clinical and Chemical Pathology Department, National Research Centre, Giza, Egypt), AM ALAZAB (Critical Care Medicine, Faculty of Medicine, Cairo University, Cairo, Egypt), O ALMOAZZAMY (Microbiology Department, Faculty of Science, Zagazig University, Zagazig, Egypt), M EL DESOKY (Department of Surgery, Gastrointestinal Surgery Center, College of Medicine, Mansoura University, Mansoura, Egypt), JE MOKHBAT (Department of Medicine, Division of Infectious Diseases and Department of Clinical Microbiology, Lebanese American University G & RM Chagoury School of Medicine and Lebanese American University Medical Center Rizk Hospital, Beirut, Lebanon), R HUSNI (Division of Infectious Diseases and Department of Internal Medicine, Lebanese American University G & RM Chagoury School of Medicine and Lebanese American University Medical Center Rizk Hospital, Beirut, Lebanon), F ABILLAMA (Department of Medicine, Division of Critical Care and Respiratory Therapy, Lebanese American University G & RM Chagoury School of Medicine and Lebanese American University Medical Center Rizk Hospital, Beirut, Lebanon).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1370553/full#supplementary-material
1. ^Identified using mlplasmid available at https://sarredondo.shinyapps.io/mlplasmids/.
2. ^ https://rast.nmpdr.org/rast.cgi
3. ^ http://web.mpiib-berlin.mpg.de
4. ^Available at: https://cge.food.dtu.dk/services/MLST/.
5. ^ https://www.genomicepidemiology.org/
6. ^ https://cge.food.dtu.dk/services/ResFinder/
Abdulall, A. K., Tawfick, M. M., El Manakhly, A. R., and El Kholy, A. (2018). Carbapenem-resistant gram-negative bacteria associated with catheter-related bloodstream infections in three intensive care units in Egypt. Eur. J. Clin. Microbiol. Infect. Dis. 37, 1647–1652. doi: 10.1007/s10096-018-3294-7
Aizawa, J., Neuwirt, N., Barbato, L., Neves, P. R., Leigue, L., Padilha, J., et al. (2014). Identification of fluoroquinolone-resistant extended-spectrum β-lactamase (CTX-M-8)-producing Escherichia coli ST224, ST2179 and ST2308 in buffalo (Bubalus bubalis). J. Antimicrob. Chemother. 69, 2866–2869. doi: 10.1093/jac/dku218
Barguigua, A., Zerouali, K., Katfy, K., El Otmani, F., Timinouni, M., and Elmdaghri, N. (2015). Occurrence of OXA-48 and NDM-1 carbapenemase-producing Klebsiella pneumoniae in a Moroccan university hospital in Casablanca, Morocco. Infect. Genet. Evol. 31, 142–148. doi: 10.1016/j.meegid.2015.01.010
Cuzon, G., Bentchouala, C., Vogel, A., Héry, M., Lezzar, A., Smati, F., et al. (2015). First outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Constantine, Algeria. Int. J. Antimicrob. Agents 46, 725–727. doi: 10.1016/j.ijantimicag.2015.08.005
da Silva, L. C. B. A., Cardoso, B., Fontana, H., Esposito, F., Cortopassi, S. R. G., Sellera, F. P., et al. (2022). Human pandemic K27-ST392 CTX-M-15 extended-spectrum β-lactamase-positive Klebsiella pneumoniae: a one health clone threatening companion animals. One Health 15:100414. doi: 10.1016/j.onehlt.2022.100414
Dautzenberg, M. J. D., Wekesa, A. N., Gniadkowski, M., Antoniadou, A., Giamarellou, H., Petrikkos, G. L., et al. (2015). The association between colonization with carbapenemase-producing Enterobacterales and overall ICU mortality: an observational cohort study. Crit. Care Med. 43:11707. doi: 10.1097/CCM.0000000000001028
De Koster, S., Rodriguez Ruiz, J. P., Rajakani, S. G., Lammens, C., Glupczynski, Y., Goossens, H., et al. (2022). Diversity in the characteristics of Klebsiella pneumoniae ST101 of human, environmental, and animal origin. Front. Microbiol. 13:838207. doi: 10.3389/fmicb.2022.838207
Doyle, D., Peirano, G., Lascols, C., Lloyd, T., Church, D. L., and Pitout, J. D. (2012). Laboratory detection of Enterobacterales that produce carbapenemases. J. Clin. Microbiol. 50, 3877–3880. doi: 10.1128/JCM.02117-12
Edward, E. A., Mohamed, N. M., and Zakaria, A. S. (2022). Whole genome characterization of the high-risk clone ST383 Klebsiella pneumoniae with a simultaneous carriage of blaCTX-M-14 on IncL/M plasmid and blaCTX-M-15 on convergent IncHI1B/IncFIB plasmid from Egypt. Microorganisms. 10:1097. doi: 10.3390/microorganisms10061097
Garcia-Fernandez, A., Villa, L., Bibbolino, G., Bressan, A., Trancassini, M., Pietropaolo, V., et al. (2020). Novel insights and features of the NDM-5-producing Escherichia coli sequence type 167 high-risk clone. mSphere 5, e00269–e00220. doi: 10.1128/mSphere.00269-20
Gordon, A. E. K., Mathers, A. J., Cheong, E. Y. L., Gottlieb, T., Kotay, S., Walker, A. S., et al. (2017). The hospital water environment as a reservoir for Carbapenem-resistant organisms causing hospital-acquired infections-a systematic review of the literature. Clin. Infect. Dis. 64, 1435–1444. doi: 10.1093/cid/cix132
Guo, S., Wakeham, D., Brouwers, H. J., Cobbold, R. N., Abraham, S., Mollinger, J. L., et al. (2015). Human-associated fluoroquinolone-resistant Escherichia coli clonal lineages, including ST354, isolated from canine feces and extraintestinal infections in Australia. Microbes Infect. 17, 266–274. doi: 10.1016/j.micinf.2014.12.016
Hammami, S., Dahdeh, C., Mamlouk, K., Ferjeni, S., Maamar, E., Hamzaoui, Z., et al. (2017). Rectal carriage of extended-Spectrum Beta-lactamase and Carbapenemase producing gram-negative Bacilli in intensive care units in Tunisia. Microb. Drug Resist. 23, 695–702. doi: 10.1089/mdr.2016.0205
Jaidane, N., Bonnin, R. A., Mansour, W., Girlich, D., Creton, E., Cotellon, G., et al. (2018). Genomic insights into Colistin-resistant Klebsiella pneumoniae from a Tunisian teaching hospital. Antimicrob. Agents Chemother. 62:pii: e01601-17. doi: 10.1128/AAC.01601-17
Liu, L., Feng, Y., Long, H., McNally, A., and Zong, Z. (2018). Sequence type 273 Carbapenem-resistant Klebsiella pneumoniae carrying blaNDM-1 and blaIMP-4. Antimicrob. Agents Chemother. 62, e00160–e00118. doi: 10.1128/AAC.00160-18
Mansour, W., Haenni, M., Saras, E., Grami, R., Mani, Y., Ben Haj Khalifa, A., et al. (2017). Outbreak of colistin-resistant carbapenemase-producing Klebsiella pneumoniae in Tunisia. J Glob Antimicrob Resist. 10, 88–94. doi: 10.1016/j.jgar.2017.03.017
Mathlouthi, N., Al-Bayssari, C., El Salabi, A., Bakour, S., Ben Gwierif, S., Zorgani, A. A., et al. (2016). Carbapenemases and extended-spectrum β-lactamases producing Enterobacterales isolated from Tunisian and Libyan hospitals. J. Infect. Dev. Ctries. 10, 718–727. doi: 10.3855/jidc.7426
Naha, S., Sands, K., Mukherjee, S., Saha, B., Dutta, S., and Basu, S. (2021). OXA-181-like Carbapenemases in Klebsiella pneumoniae ST14, ST15, ST23, ST48, and ST231 from septicemic neonates: coexistence with NDM-5, Resistome, transmissibility, and genome diversity. mSphere 6, e01156–e01120. doi: 10.1128/mSphere.01156-20
Pascale, R., Bussini, L., Gaibani, P., Bovo, F., Fornaro, G., Lombardo, D., et al. (2021). Carbapenem-resistant bacteria in an intensive care unit during the COVID-19 pandemic: a multicenter before-and-after cross-sectional study. Infect. Control Hosp. Epidemiol. 16, 1–6. doi: 10.1017/ice.2021.144
Pitout, J. D. D., Peirano, G., Kock, M. M., Strydom, K. A., and Matsumura, Y. (2019). The global ascendency of OXA-48-type Carbapenemases. Clin. Microbiol. Rev. 33, e00102–e00119. doi: 10.1128/CMR.00102-19
Qiu, J., Zhai, Y., Wei, M., Zheng, C., and Jiao, X. (2022). Toxin-antitoxin systems: classification, biological roles, and applications. Microbiol. Res. 264:127159. doi: 10.1016/j.micres.2022.127159
Roer, L., Overballe-Petersen, S., Hansen, F., Schønning, K., Wang, M., Røder, B. L., et al. (2018). Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere 3:e00337-18. doi: 10.1128/mSphere.00337-18
Shawa, M., Furuta, Y., Paudel, A., Kabunda, O., Mulenga, E., Mubanga, M., et al. (2022). Clonal relationship between multidrug-resistant Escherichia coli ST69 from poultry and humans in Lusaka, Zambia. FEMS Microbiol. Lett. 368:fnac004. doi: 10.1093/femsle/fnac004
Shi, Q., Zhao, J., Wei, L., Zhu, F., Ji, J., Meng, Y., et al. (2022). J transmission of ST45 and ST2407 extended-spectrum β-lactamase-producing Klebsiella pneumoniae in neonatal intensive care units, associated with contaminated environments. Glob Antimicrob Resist. 31, 309–315. doi: 10.1016/j.jgar.2022.10.006
Stolle, I., Prenger-Berninghoff, E., Stamm, I., Scheufen, S., Hassdenteufel, E., Guenther, S., et al. (2013). Emergence of OXA-48 carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in dogs. J. Antimicrob. Chemother. 68, 2802–2808. doi: 10.1093/jac/dkt259
Tischendorf, J., Almeida de Avila, R., and Safdar, N. (2016). Risk of infection following colonization with carbapenem-resistant Enterobactericeae: a systematic review. Am. J. Infect. Control 44, 539–543. doi: 10.1016/j.ajic.2015.12.005
van der Mee-Marquet, N., Savoyen, P., Domelier-Valentin, A. S., Mourens, C., and Quentin, R. (2010). CTX-M-type fluoroquinolone-resistant E. coli: analysis of the colonization of residents and inanimate surfaces 1 year after a first case of urinary tract infection at a nursing home in France. Infect. Control Hosp. Epidemiol. 31, 968–970. doi: 10.1086/655835
van Duin, D., and Doi, Y. (2017). The global epidemiology of carbapenemase-producing Enterobacterales. Virulence 8, 460–469. doi: 10.1080/21505594.2016.1222343
Vasconcelos, P. C., Leite, E. L., Araújo, W. J., Silva, N. M. V., Saraiva, M. M. S., Santos Filho, L., et al. (2020). Draft genome sequence of mcr-1-mediated colistin-resistant Escherichia coli ST359 from chicken carcasses in northeastern Brazil. J Glob Antimicrob Resist. 23, 135–136. doi: 10.1016/j.jgar.2020.08.016
Wang, J., Huang, X. Y., Xia, Y. B., Guo, Z. W., Ma, Z. B., Yi, M. Y., et al. (2018). Clonal spread of Escherichia coli ST93 carrying mcr-1-harboring IncN1-IncHI2/ST3 plasmid among companion animals, China. Front. Microbiol. 9:2989. doi: 10.3389/fmicb.2018.02989
Wassef, M., Abdelhaleim, M., Ghaith, D., and El-Mahdy, Y. (2016). Emerging New Delhi metallo-β-lactamase-1-type-producing gram-negative bacteria isolated from Cairo University pediatric hospital, Cairo, Egypt. J Glob Antimicrob Resist. 7, 84–87. doi: 10.1016/j.jgar.2016.08.004
Weterings, V., Zhou, K., Rossen, J. W., van Stenis, D., Thewessen, E., Kluytmans, J., et al. (2015). An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1647–1655. doi: 10.1007/s10096-015-2401-2
Keywords: intensive care unit, carriage, carbapenem-resistant Enterobacterales, colistin-resistant Enterobacterales, Morocco, Tunisia, Lebanon, Egypt
Citation: Dos Santos S, Diene SM, Benouda A, Zerouali K, Ghaith DM, El-Mahdy RH, El Tayeb SHM, Boutiba I, Hammami A, Chrabieh R, Daoud Z, Mereghetti L, Francois P and Van Der Mee-Marquet N on behalf of the consortium (2024) Carbapenem- and colistin-resistant Enterobacterales in intensive care unit patients in Mediterranean countries, 2019. Front. Microbiol. 15:1370553. doi: 10.3389/fmicb.2024.1370553
Received: 14 January 2024; Accepted: 22 March 2024;
Published: 12 April 2024.
Edited by:
Miklos Fuzi, Independent Researcher, Budapest, HungaryReviewed by:
Salome N. Seiffert, Zentrum für Labormedizin (ZLM), SwitzerlandCopyright © 2024 Dos Santos, Diene, Benouda, Zerouali, Ghaith, El-Mahdy, El Tayeb, Boutiba, Hammami, Chrabieh, Daoud, Mereghetti, Francois, Van Der Mee-Marquet and Mediterranean infection control group. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nathalie Van Der Mee-Marquet, bi52YW5kZXJtZWVAY2h1LXRvdXJzLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.