- Department of Plant Pathology & Environmental Microbiology, The Pennsylvania State University, University Park, PA, United States
Trichoderma spp. are widely used to enhance crop growth and suppress diverse diseases. However, inconsistent field efficacy remains a major barrier to their use as a reliable alternative to synthetic pesticides. Various strategies have been investigated to enhance the robustness of their application. Here, we evaluated how T. virens application methods (pre-, at-, and post-transplant) affect the growth of two tomato varieties and their rhizosphere fungal and bacterial communities. Although the greatest rhizosphere abundance of T. virens was observed in the post-transplant application, the at-transplant application promoted tomato growth the most, indicating that greater rhizosphere abundance does not necessarily result in better tomato growth. None of the application methods significantly altered the global rhizosphere fungal and bacterial communities of the tested varieties. Changes in specific microbial genera and guilds may underpin the enhanced tomato growth. We also investigated whether the resulting microbiome changes affect the mycelial growth and conidial germination of Fusarium oxysporum f. sp. lycopersici and F. oxysporum f. sp. radicis-lycopersici, soilborne fungal pathogens of tomato, upon exposure to volatile compounds emitted by culturable rhizosphere microbes and metabolites extracted from the rhizosphere soils after Trichoderma treatments. Volatile compounds produced by cultured rhizosphere microbes after the at-transplant application suppressed the mycelial growth of both pathogens better than those after the other treatments. Similarly, water-soluble metabolites extracted from the rhizosphere soil samples after the at-transplant application most effectively suppressed the germination rate of F. oxysporum spores. Overall, our results suggest that the at-transplant application is most advantageous for promoting the growth of the tested tomato varieties and building soil suppressiveness against the tested fusaria. However, further studies are needed before applying this method to support tomato production. We discuss critical future questions.
Introduction
Biological control has been widely explored as a method for protecting crop health without disrupting environmental health and ecosystem services. Although many potential biological control agents (BCAs) have been evaluated, approximately 90% of the evaluated fungal BCAs belong to the genus Trichoderma (Hermosa et al., 2012; Woo et al., 2014, 2023), with >60% of the globally registered biofungicides containing Trichoderma (Verma et al., 2007). There are over 200 described Trichoderma species (Bissett et al., 2015; Dou et al., 2020), but only a few have been commercialized as BCAs. Trichoderma suppresses pathogens directly through mycoparasitism, competition, and antibiosis and indirectly by inducing plant defense responses (Benítez et al., 2004; Poveda, 2021; Guzmán-Guzmán et al., 2023). Many commercially available Trichoderma BCAs are effective against diverse pathogens (Guzmán-Guzmán et al., 2023; Woo et al., 2023). However, their inconsistent field efficacy in different production systems and under varying environmental conditions has restricted their utility as chemical pesticide alternatives. Limited knowledge about which factors cause biocontrol failures and how they work under complex and varied biotic and abiotic conditions has hampered efforts to enhance their effectiveness and design reliable Trichoderma deployment strategies (Mazzola and Freilich, 2017; Timmusk et al., 2017). Until reliability and effectiveness are assured, biocontrol will be perceived as a high-risk and high-cost practice.
Various strategies for improving the crop growth promotion and biocontrol efficacies of Trichoderma have been reviewed (Fraceto et al., 2018; Ferreira and Musumeci, 2021; Guzmán-Guzmán et al., 2023; Xiao et al., 2023). They include Trichoderma strain improvement (Szekeres et al., 2004; Montero-Barrientos et al., 2011; Hassan, 2014), formulation of Trichoderma with bioactive compounds (Yang et al., 2011; Nandini et al., 2017; Mbarga et al., 2020), co-application of Trichoderma with other beneficial microbes (Rudresh et al., 2005; Shanmugaiah et al., 2009; Mweetwa et al., 2016; Ghorchiani et al., 2018; Karuppiah et al., 2019; Szczałba et al., 2019; Konappa et al., 2020), and combining Trichoderma application with organic amendments (Haggag and Saber, 2000; Miranda et al., 2006; Joshi et al., 2009; Yang et al., 2011; Bhadauria et al., 2012; Hannan et al., 2012; Domínguez et al., 2014; Blaya et al., 2016; de Araujo et al., 2019; Sani et al., 2020; Da Silva et al., 2022; Amanullah and Khan, 2023). However, these strategies have their own limitations, calling for continued research to develop highly efficacious biocontrol products and application strategies that are economically viable, technically simple, regulatorily neutral, and easily adoptable.
Delivery methods likely influence the efficacy of BCAs. Generally, plant growth promotion and soilborne disease suppression by Trichoderma are achieved through its application to crop growth media (mostly soil) after seedling emergence. The application of Trichoderma as a seed treatment is also a promising strategy, especially for crops that are directly drilled into soils (e.g., field/row crops and some horticultural crops), because the soils likely harbor phytopathogenic fungi. The effectiveness of Trichoderma seed treatment in promoting crop growth under biotic and abiotic stress conditions has been demonstrated in field crops like pea (Harman et al., 1980), cotton (Hanson, 2000), wheat (Xue et al., 2017), cowpea (Adekunle et al., 2001), common bean (Carvalho et al., 2014), and corn (Estévez-Geffriaud et al., 2020). When directly planted in field soils, Trichoderma seed treatment alleviated biotic and abiotic stresses in tomatoes (Mastouri et al., 2010). Even when grown in autoclaved potting mix (e.g., sand and vermicompost) in the absence of any pathogens, T. asperellum seed treatment significantly enhanced the growth of tomato, brinjal, chili, okra, ridge gourd, and guar (Singh et al., 2016). However, in commercial production systems, most high-value brassicaceous (e.g., broccoli, Brussels sprouts) and solanaceous (e.g., tomato, pepper) vegetables are typically transplanted into the soil after raising them using autoclaved potting mixes in nursery trays. Therefore, priming the root system with Trichoderma during seedling growth could enhance seedling vigor and better protect the seedlings from soilborne diseases after transplantation.
Rapid advances in analyzing the composition and changes of plant-associated microbiomes under diverse conditions through massive sequencing of phylogenetically informative loci have uncovered how rhizosphere microbiomes affect crop yield, quality, and tolerance to biotic and abiotic stressors (Berendsen et al., 2012; Chaparro et al., 2012; Berg et al., 2017; Kwak et al., 2018; Zhou et al., 2019). Given their critical roles in plant growth and health, judicious manipulation of rhizosphere/bulk soil microbiomes is a promising approach for achieving greener agriculture. The effectiveness of Trichoderma-based biocontrol can be improved through a better understanding of how its introduction results in structural and functional changes in the soil microbiome and how the resulting changes affect crop growth and the suppressiveness of soilborne pathogens. We aimed to advance this understanding by initially assessing how the application method (collectively characterized by the time of application, the substrate to which the application is administered, and the duration of T. virens-tomato root system contact; see Materials and Methods for details) of T. virens affects the growth of two tomato varieties and subsequently analyzing how each application method affects the tomato rhizosphere fungal and bacterial community structure using amplicon metagenomic sequencing (metabarcoding). Trichoderma suppresses pathogens by secreting antifungal metabolites, including volatile compounds (Li et al., 2018; Woo et al., 2023). However, how antimicrobial compounds produced by soil microbes affect Trichoderma and pathogens remains largely unknown. We evaluated how volatile compounds emitted by culturable rhizosphere microbes of two tomato varieties after individual Trichoderma treatments affect the mycelial growth of T. virens and two soilborne fungal pathogens of tomatoes, F. oxysporum f. sp. lycopersici (Fusarium wilt) and F. oxysporum f. sp. radicis-lycopersici (Fusarium crown and root rot). They are among the most problematic Fusarium pathogens for tomato production (McGovern, 2015). We also assessed how water-soluble metabolites extracted from the rhizosphere soils of the two tomato varieties after different Trichoderma treatments affected the germination rate of T. virens and F. oxysporum conidia.
Materials and methods
Growing tomato plants, preparation and application of Trichoderma inoculum, measuring tomato growth, and collecting rhizosphere soils
Seeds of the tomato varieties Bonny Best (Fusarium wilt susceptible) and Red Deuce (Fusarium wilt resistant) were planted in seed starter trays filled with autoclaved (121°C and 20 psi for 30 min) Premier Horticulture 10380RG Pro-Mix Professional Grower Mix (Premier Horticulture Inc., Canada). Starter trays were kept on bottom watering trays and maintained in a growth chamber at 25°C, 75% RH, and 16 h light at 100 μmol/8 h dark.
The T. virens isolate used by Li et al. (2018) was cultured on PDA at 27°C for 1 week to harvest conidia using sterile distilled water. Conidial suspension was passed through four layers of sterile cheesecloth placed in sterile 50 mL Falcon tubes. After centrifuging the tubes at 4,000 rpm at 4°C for 10 min, the supernatant was discarded. The conidial concentration was determined using a hemocytometer after resuspension in sterile distilled water. The resulting conidial suspension was used for the following treatments: T1 (T. virens conidia in the bottom watering tray for a duration of 1 week prior to transplant, after introducing the conidia at 2 weeks after planting the seeds), T2 (T. virens conidia in the bottom watering tray for a duration of 30 min prior to transplantation, after introducing the conidia at 3 weeks after planting the seeds), T3 (T. virens conidia applied to the soil 1 week post-transplantation, after 3 weeks of pre-transplant growth upon planting the seeds in the bottom watering tray containing sterile distilled water), and CON (control treatment with sterile distilled water in the bottom watering tray for a duration of 3 weeks prior to transplant, after planting the seeds). Note that the T1, T2, and T3 treatments are hereafter referred to as pre-, at-, and post-transplant treatments/applications. Seedlings from all treatments were transplanted 3 weeks after planting seeds. The final conidial concentration was 106 conidia/mL of water for T1 and T2, and 106 conidia/cm3 of soil for T3. A batch of topsoil collected from the Penn State Russell E. Larson Agricultural Research Center at Rock Springs was used for transplantation. Forty-eight quart-sized Ziploc bags were filled with thoroughly homogenized soil. Seedlings were transplanted into bags placed in 3-inch plastic nursery pots. The pots were maintained in a growth chamber at 25°C, 75% RH, and 16 h light under a 100 μmol/8 h dark cycle. The amounts of water and fertilizers applied after transplantation were kept consistent across the treatments.
The plants were uprooted at 11 weeks after transplantation, and their rhizosphere soils (soil up to 2 mm surrounding the root) were collected in sterile 15 mL Falcon tubes. The tubes were stored at −80°C until genomic DNA extraction and other analyses. Fresh weights of the shoots and soil-free roots of each plant were measured. The root/shoot ratio of each plant was calculated. The treatment structure of the experiment was “two-factor factorial,” where the first factor was “Trichoderma treatment” with four levels (CON, T1, T2, and T3) and the second factor was “tomato variety” with two levels (Bonny Best and Red Deuce). Six plants/pots (replicates) were used for each Trichoderma treatment-tomato variety combination. The treated pots were randomly arranged in the growth chamber. Hence, the design structure of the experiment was completely randomized. The experiment was repeated one time.
Statistical analysis of the tomato growth traits
To test the main/simple effects of Trichoderma treatment and tomato variety on the tomato shoot and root weights, ANOVA was conducted using the PROC GLIMMIX procedure in SAS (version 9.4, SAS Institute, Cary, NC) at the 5% significance level (α = 0.05). Initially, the data from two repetitions were analyzed separately. As similar results were observed, the final analysis was performed using the average values of the two repetitions. As shoot and root weights are continuous measurements, statistical modeling was performed with the normal/Gaussian distribution with the “Identity” link function. The restricted maximum likelihood (REML) method was used to compute the variance components. Newton–Raphson with Ridging was used as the non-linear parameter optimization method. The assumptions of identical and independent (homogenous) distribution of residuals were tested using studentized residual plots and Brown and Forsythe’s test. The assumption of normal distribution of residuals was checked using Q–Q plots and the Shapiro–Wilk test. If residuals were found to be non-homogenously distributed (showing heteroskedasticity), appropriate heterogeneous variance models were fitted by specifying a “random residual/group = …” statement (where group = Trichoderma treatment or tomato variety). The computation of the degrees of freedom for the denominator of F-tests and the adjustment of the standard errors of fixed effects were performed using the Kenward-Roger option. Between the two heterogeneous variance models, the model with the lowest Bayesian Information Criterion (BIC) was selected as the best-fitting model. As the computed root/shoot ratios are proportions (between 0 and 1), the data were modeled with the beta distribution along with the “Logit” link function. The generalized linear mixed model (GLMM) estimation was based on integral approximations to the likelihood, where Laplace was used as the likelihood approximation method. The maximum likelihood (ML) method was used to compute variance components. The Newton–Raphson with Ridging method was used as the non-linear parameter optimization method. The degrees of freedom method was residual. The inverse link function was used to create the means and associated standard errors at the data scale. The mean separation (all pairwise comparisons of the levels of the factor “Trichoderma treatment”) was performed with adjustments for multiple comparisons using the Tukey–Kramer test at 5% significance level/experimental-wise error rate (αEER = 0.05).
DNA extraction, polymerase chain reaction, sequencing, sequence data processing, and analysis
DNA was extracted from 0.5 g of thoroughly homogenized rhizosphere soil using the FastDNA™ SPIN Kit for Soil (MP Biomedicals, Irvine, CA, United States). The concentration of genomic DNA was determined using the Qubit dsDNA BR Assay Kit and 1% agarose gel electrophoresis. PCR amplification of the rRNA ITS1 region was performed using ITS1F (5′-CTTGGTCATTTAGAG GAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′) primers. PCR amplification of the 16S rRNA V3–V4 region was performed using 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) primers. PCR, preparation of amplicon libraries, and sequencing were performed by the Beijing Genomic Institute (BGI). Sequencing was performed using the PE300 Illumina MiSeq platform.
Initial sequence data processing was performed in R (v.4.2.1) using the DADA2 (v.1.26) pipeline (Callahan et al., 2016). Primer sequences were removed from reads using cutadapt command-line version 2.8 (Martin, 2011), a plugin in DADA2. Fungal taxonomic assignment was performed using the UNITE General FASTA release (v.9) database (Kõljalg et al., 2005). Silva database (v.138.1) was used to assign taxonomy for bacteria (Quast et al., 2012). DADA2 outputs were converted into phyloseq objects using the phyloseq R package (v.1.42) (McMurdie and Holmes, 2013) and the Biostrings R package (v.2.66) (Pagès et al., 2022). These phyloseq objects were used as the source for downstream global and specific analyses. Following the standard procedure, all global analyses, including beta diversity, alpha diversity, and relative abundance (see below), were performed in R (v.4.2.1) using rarefied read counts (based on the minimum available reads per sample among all samples; fungi = 78,869, bacteria = 41,591). In addition, the rarefied read counts were converted into proportions to perform beta diversity and relative abundance analyses.
To analyze the β-diversity, the ordinate function of the phyloseq R package (v.1.42) (McMurdie and Holmes, 2013) was used with distance = bray and method = PCoA specifications. PCoA plots were visualized using the ggplot2 R package (v.3.3.3) (Wickham, 2016). A distance matrix was generated using the phyloseq::distance function of the phyloseq R package (v.1.42), with Bray–Curtis specification. The adonis2 function of the Vegan R package (v.2.6.4) (Oksanen et al., 2022) was applied to the Bray–Curtis distance matrix to perform a global scale permutational multivariate analysis of variance (PERMANOVA) to test the significance of the effect of Trichoderma treatment and tomato variety on the community composition dissimilarity between samples. A global scale multivariate homogeneity of group dispersions (variances) was performed using the betadisper and permutest functions of the Vegan R package (v.2.6.4) to test the null hypothesis of homogenous dispersions of samples belonging to different Trichoderma treatments and tomato varieties. The pairwise.adonis function of the pairwiseAdonis R package (v. 0.4) (Martinez Arbizu, 2020) was used as a post hoc test of the global PERMANOVA to assess which pairs of Trichoderma treatments have significantly different centroids. Similarly, the permutest function with pairwise = true specification was used as a post hoc test of the global beta dispersion test to assess which pairs of Trichoderma treatments have homogenous dispersions. The community composition dissimilarity between samples belonging to a given pair of Trichoderma treatment (and for two levels of tomato variety) levels was declared significant when the adjusted p-value obtained through pairwise.adonis test was smaller than 0.05, and the adjusted p-value obtained through the pairwise beta dispersion test was larger than 0.05. In both the global and pairwise analyses, significance was assessed using 999 permutations.
The alpha diversity in each sample was estimated using the microbiome R package (v.1.20.0) (Lahti and Shetty, 2023) with the following indicators: Chao1 richness, Shannon diversity, Coverage diversity (=number of species needed to cover 50% of the ecosystem), Pielou evenness, Core abundance (=relative proportion of the core species that exceed detection level 0.1% in over 50% of the samples), and Rare abundance (complement of the core abundance). Subsequently, ANOVA was performed using the GLIMMIX procedure in SAS (v.9.4) to test the main/simple effects (α = 0.05) of Trichoderma treatment and tomato variety on the estimated alpha diversity indicators. Because Pielou evenness, Core abundance, and Rare abundance values are proportions, data from those three indicators were modeled with the beta distribution. The Shannon diversity values of each sample were converted into proportions by dividing them by the largest Shannon diversity value in the dataset. Modified Shannon diversity values were also modeled using the beta distribution. As Caho1 richness and Coverage diversity values are counts, modeling was performed with a negative binomial distribution. In all cases, the remaining model specifications were identical to those used for the statistical analysis of root/shoot ratios.
The relative abundances of taxonomic groups at the phylum and genus levels were determined using the transform_sample_counts function in the R package phyloseq. The relative abundance of top fungal phyla/genera was depicted as stack bar plots using the ggplot() function in the R package ggplot2.
ANOVA was performed using the GLIMMIX procedure to test the main/simple effects (α = 0.05) of Trichoderma treatment and tomato variety on the proportional abundance (=ratio between “sum” of the “non-rarefied” read counts of a species/genus/guild of interest in a given rhizosphere sample and total read count of the same sample) of T. virens, beneficial fungi (Acremonium, Cadophora, Chaetomium, Clonostachys, Mortierella, Paraphaeosphaeria, Penicillium, Syncephalis, Humicola, Marquandomyces, Metacordyceps, Metarhizium, Linnemannia), Fusarium, other fungal phytopathogens (Albifimbria, Aspergillus, Cladosporium, Dactylonectria, Didymella, Gibellulopsis, Melanconiella, Microdochium, Mycoleptodiscus, Neocosmospora, Neonectria, Paecilomyces, Plectosphaerella, Pseudopithomyces, Rhizoctonia, Talaromyces), bacterial biocontrol agents (Arthrobacter, Bdellovibrio, Flavisolibacter, Lysobacter, Massilia), bacterial plant growth promoters (Devosia, Mucilaginibacter), nitrifiers (Chujaibacter, MND1, Nitrolancea, Nitrosospira, Nitrospira, Pseudarthrobacter, Pseudolabrys), symbiotic nitrogen fixers (Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Bradyrhizobium, Mesorhizobium), free-lining nitrogen fixers (Bacillus, Clostridium, Paenibacillus, Rhodopseudomonas), soil Polycyclic Aromatic Hydrocarbon/toxin degraders (Mycobacterium, Sphingomonas), phytopathogens (Burkholderia-Caballeronia-Paraburkholderia, Ralstonia, Streptomyces), and denitrifiers (Burkholderia-Caballeronia-Paraburkholderia, Conexibacter, Rhodanobacter). Modeling was performed using the beta distribution, and the model specifications were identical to those used for the statistical analysis of root/shoot ratios. We defined a guild as a group of genera that can potentially contribute to a specific ecological function/service (e.g., symbiotic nitrogen fixation). By using proportional abundance instead of absolute abundance, we standardized the effect of varying numbers of total read counts among samples (different library sizes) on the abundance of the species/genus/guild of interest in the respective samples. Furthermore, the use of “non-rarefied” read counts as the numerator is computationally and ecologically more realistic and sensical than computing the proportional abundance using “rarefied” read counts (=ratio between the sum of the “rarefied” read counts of a species/genus/guild in a given sample and the total number of read counts of the “smallest” sample), as rarefaction can artificially inflate or deflate the abundance of taxa as a result of the random resampling. Note that if proportional abundance is computed using “rarefied” read counts, the total read count number of the smallest sample should be used as the common denominator when rarefaction is performed based on the “minimum” read count sample among “all” samples, as rarefaction can be performed at any preferred number at the cost of removing samples from the data set or a given analysis.
Determination of whether volatiles emitted by cultured rhizosphere microbes after different Trichoderma treatments affect the in-vitro growth of T. virens, F. oxysporum f. sp. lycopersici, and F. oxysporum f. sp. radicis-lycopersici
We used a modified version of the sandwiched plate assay described in Li et al. (2018). First, X plates (plates with four sections) were prepared using four growth media, including R2A, tryptic soy agar (TSA), potato dextrose agar (PDA), and malt extract agar (MEA). Each plate contained all four media without any antibiotics or antifungal compounds. R2A/TSA and PDA/MEA were used to maximize the culturable fraction of bacteria and filamentous fungi/yeasts, respectively. Second, 2.000 g of each rhizosphere soil sample was homogenized in 10 mL of sterile distilled water, and 5 μL of each soil suspension was spread onto each section of the plate using sterile glass beads. The inoculated plates were incubated at 27.5°C for 2 days (dense colonies appeared at this time). After incubating PDA plates inoculated with mycelial plugs of T. virens, F. oxysporum f. sp. lycopersici, and F. oxysporum f. sp. radicis-lycopersici for 24 h, these plates were sandwiched with 2 days-old X plates containing rhizosphere microbial colonies and incubated at 27.5°C for 3 days before measuring the fungal colony diameters. This experiment was conducted two times using the soil samples collected after the two growth chamber experiments (see above). To test the main/simple effects of rhizomicrobial volatiles from Trichoderma treatment and tomato variety on the fungal colony diameters, ANOVA was performed using the PROC GLIMMIX procedure in SAS (v.9.4, SAS Institute, Cary, NC) at the 5% significance level (α = 0.05). Initially, the data from two repetitions were analyzed separately. However, the final analysis was performed using the average values of two repetitions because the results of the separate analyses were interpretationally identical. Model specifications and the method of mean separation were identical to those used for the statistical analysis of tomato shoot and root weights (see above).
Determination of whether water-soluble metabolites extracted from tomato rhizosphere soils after receiving different Trichoderma treatments affect the germination of T. virens, F. oxysporum f. sp. lycopersici, and F. oxysporum f. sp. radicis-lycopersici conidia
The original soil suspensions (n = 48) prepared for sandwich assays (see above) by suspending 2.000 g of soil in 10 mL of sterile distilled water were thoroughly vortexed and centrifuged for 10 min at 4,000 rpm. The supernatants were filtered using 0.2 μM filters to obtain sterile metabolite extracts. The ability of these metabolite extracts to modulate the conidial germination rate of T. virens and F. oxysporum was assayed under in vitro conditions using a redox reagent, Alamar Blue (AB: Thermo Fisher Scientific). As AB is an indicator of the cellular metabolic activity, it can be used for high-throughput determination of conidial viability and germination rate. AB, in its oxidized form, is blue and non-fluorescent. Germinating conidia cause a chemical reduction of AB from non-fluorescent blue to fluorescent red. Since AB reduction is directly proportional to the conidial germination rate, the effects of soil metabolites on the germination rate can be measured by monitoring the level of AB reduction indicated by the resulting absorbance changes.
For the assay, conidial suspensions of three fungi were prepared using pH-neutral potato dextrose broth (PDB). The assay was set up on a round bottom 96-well plate. One hundred μL of PDB containing 105 conidia/ml, 10 μL of AB, and 10 μL of metabolite extract were mixed in each well. For each fungus, 48 reaction wells (48 metabolite extracts = four treatments, two varieties, and six replicates) were used. The wells for negative controls contained the same volumes of PDB, AB, and metabolite extract. Plates were incubated at 25°C for 24 h before measuring the absorbance at 570 and 600 nm using a plate reader. AB reduction (proportional) in each well was computed using the following equation:
where.
O1 = molar extinction coefficient of oxidized AB at 570 nm (=80,586); O2 = molar extinction coefficient of oxidized AB at 600 nm (=117,216); R1 = molar extinction coefficient of reduced AB at 570 nm (=155,677); R2 = molar extinction coefficient of reduced AB at 600 nm (=14,652); A1 = absorbance value of test wells at 570 nm; A2 = absorbance value of test wells at 600 nm; N1 = absorbance value of negative control well at 570 nm; and N2 = absorbance value of negative control well at 600 nm.
The experiment was conducted two times using soil samples collected from the two growth chamber experiments (see above). To test the effects of soil metabolites resulting from Trichoderma treatments and tomato varieties on the germination rate of conidia (=AB reduction), ANOVA was conducted using the PROC GLIMMIX procedure in SAS (v.9.4) at the 5% significance level (α = 0.05). Initially, the data from two repetitions were analyzed separately. As the results of the separate analyses were interpretationally identical, the final analysis was performed using the average values of two repetitions. Model specifications and the method of mean separation were identical to those used for the statistical analysis of the tomato root/shoot weight ratio described above.
Results
Effect of Trichoderma treatment, tomato variety, and their interaction on tomato growth
ANOVA revealed a significant main effect of Trichoderma treatment on shoot weight and a significant Trichoderma treatment × tomato variety interaction effect on root weight and the root/shoot ratio (Supplementary Table S1). T2 (at-transplant application) significantly improved shoot weight compared to T1 (pre-transplant application) and CON (Figure 1). Both T2 and T3 (post-transplant application) increased the root weight and root/shoot ratio of Red Deuce compared to CON and T1 (Figure 1). However, none of the Trichoderma treatments did significantly better than others in enhancing the root weight or root/shoot ratio of Bonny Best (Figure 1).
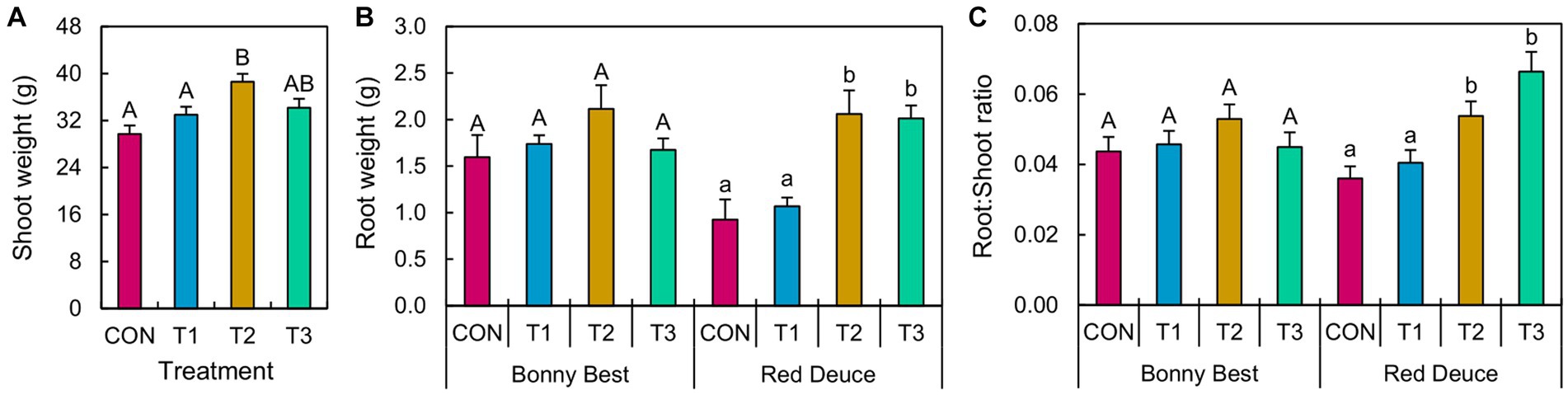
Figure 1. Effects of Trichoderma application methods on tomato growth. Mean (A) shoot weight, (B) root weight, and (C) root-to-shoot ratio of the tomato plants treated with CON (control treatment), T1 (pre-transplant treatment with T. virens), T2 (at-transplant treatment with T. virens), and T3 (post-transplant treatment with T. virens) are shown. Means followed by the same letters within each letter type (upper or lower case) are not significantly different at the 5% level (=5% experimental-wise error rate) of significance after adjusting the p-values for multiple comparisons using the Tukey test. Error bars represent standard errors. (A) shows the main effect of Trichoderma treatment, while (B) and (C) show its simple effects.
Effect of Trichoderma treatment, tomato variety, and their interaction on alpha diversity
The Trichoderma treatment × tomato variety interaction was significant (α = 0.05) for the fungal and bacterial Shannon diversity and fungal Pielou evenness, whereas the main effect of Trichoderma treatment was significant for the fungal core and rare abundances (Supplementary Table S2). The mean fungal and bacterial Shannon diversities were not significantly different among the four treatments in Red Duce (Supplementary Figures S1, S2). In Bonny Best, the fungal Shannon diversities of T1 and T2 were greater than those of CON and T3 (Supplementary Figure S1), whereas T1 increased the bacterial Shannon diversity compared to CON, T2, and T3 (Supplementary Figure S2). The mean fungal Pielou evenness among the treatments was not significantly different in Red Duce. However, in Bonny Best, T1 significantly increased the evenness compared to CON and T3 (Supplementary Figure S1). T3 decreased the fungal core abundance compared to the other Trichoderma treatments across both varieties, whereas T1 decreased the fungal core abundance compared to CON (Supplementary Figure S1). T3 increased the fungal rare abundance compared to the other treatments across both varieties, and T2 increased the abundance compared to CON (Supplementary Figure S1).
Effect of Trichoderma treatment and tomato variety on the rhizosphere fungal and bacterial community composition dissimilarity (beta diversity)
According to the principal coordinate analysis (PCoA), the first two principal coordinates explained 40.5 and 43.5% of the total variation for fungi and bacteria, respectively (Figure 2). The global PERMANOVA revealed a significant Trichoderma treatment effect on fungal and bacterial beta diversities, but the effects of tomato variety and its interaction with Trichoderma treatment were non-significant (Figure 2). Per pairwise PERMANOVA, both fungal and bacterial community compositions were significantly dissimilar between T2 and T3 (Supplementary Table S3). This finding was further supported by the non-significant beta dispersion between T2 and T3 (Supplementary Table S3). None of the Trichoderma treatments significantly altered the fungal or bacterial community composition compared to CON at the global scale (Supplementary Table S3).
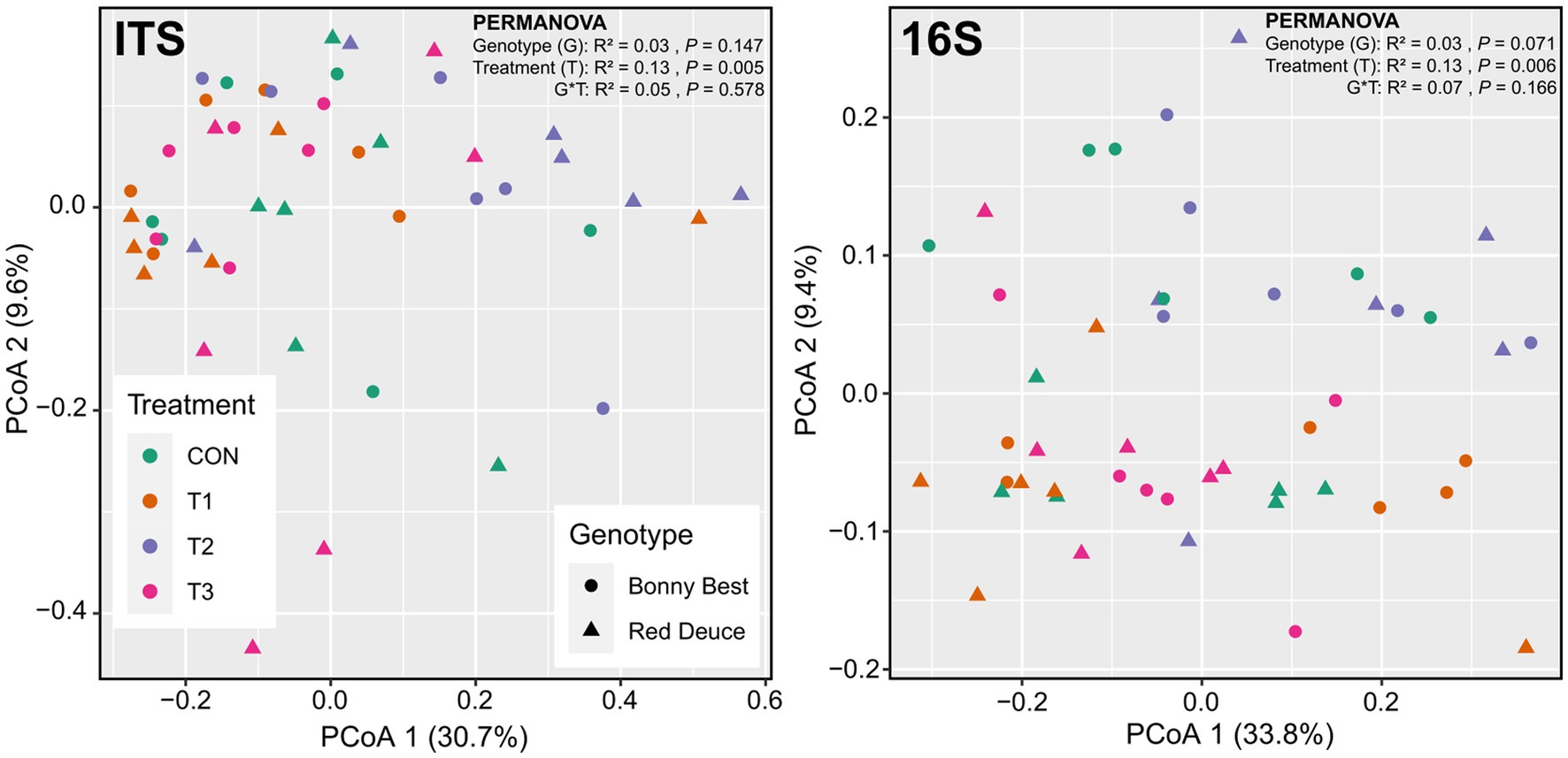
Figure 2. Principal coordinate analysis (PCoA) plots showing the Bray–Curtis dissimilarity-based distribution pattern of rhizosphere soil samples for fungal and bacterial communities. Samples belong to CON (control treatment), T1 (pre-transplant treatment with T. virens), T2 (at-transplant treatment with T. virens), and T3 (post-transplant treatment with T. virens), and two tomato varieties (Bonny Best and Red Deuce) are depicted. Marker color and shape correspond to the Trichoderma treatment and tomato variety, respectively.
Effects of Trichoderma treatments and tomato varieties on the relative abundance of major fungal and bacterial phyla and genera
All Trichoderma treatments altered the relative abundance of fungal phyla such as Ascomycota, Basidiomycota, Chytridiomycota, and Mortierellomycota, but the nature of alteration varied among the treatments (Figure 3). In both tomato varieties, the abundance of Ascomycota was highest in T3, followed by T1, CON, and T2. The opposite was observed for Mortierellomycota. T3 resulted in the highest abundance of Chytridiomycota in both varieties, whereas T2 caused the lowest. All Trichoderma treatments decreased the abundance of Basidiomycota compared to CON in Bonny Best, whereas the opposite was true in Red Deuce. For bacterial phyla, all Trichoderma treatments markedly decreased the abundance of Bacteroidota, Patescibacteria, and Proteobacteria compared to CON in Bonny Best, while increasing the abundance of Actinobacteriota, Chloroflexi, and Firmicutes (Figure 3). In Red Deuce, T1 resulted in the highest abundance of Acidobacteriota, Chloroflexi, Firmicutes, and Proteobacteria and the lowest abundance of Bacteroidota, Gemmatimonadota, and Patescibacteria.
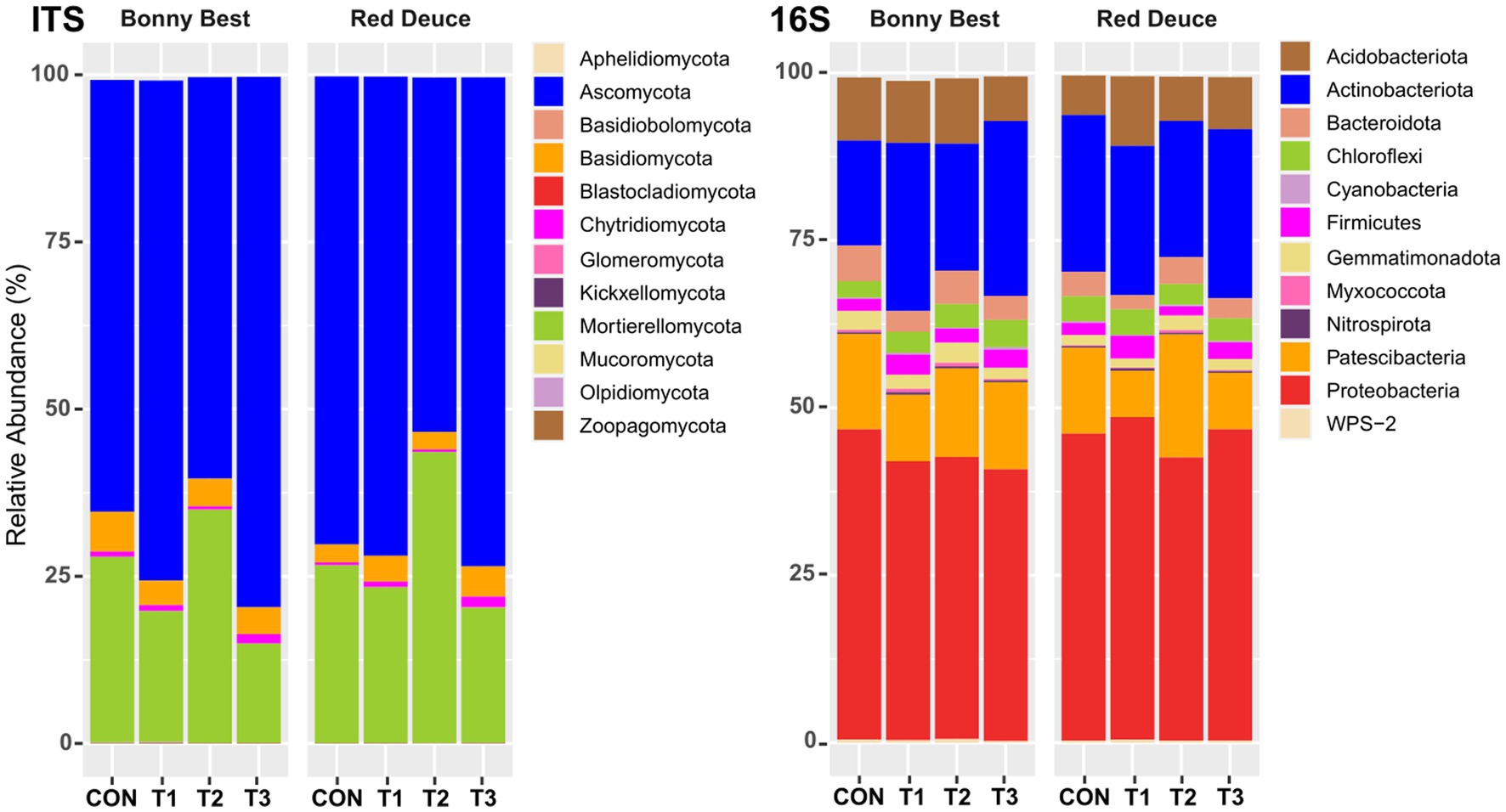
Figure 3. Effect of Trichoderma treatment on the mean relative abundance of the top 12 fungal and bacterial phyla. Treatments include CON (control treatment), T1 (pre-transplant treatment with T. virens), T2 (at-transplant treatment with T. virens), and T3 (post-transplant treatment with T. virens).
At the genus level, among all treatments, T1 resulted in the highest relative abundance of the fungal genera Albifimbria, Aspergillus, Chaetomium, Fusarium, Melanconiella, and Penicillium in the rhizosphere of both varieties (Figure 4). T2 resulted in the highest abundance of Linnemannia, Mortierella, and Tetracladium and the lowest abundance of Fusarium in both varieties. For bacterial genera, compared to the other treatments, T2 increased the abundance of Gemmatimonas, Pseudolabrys, Rhodanobacter, Sphingomonas, and TM7a but decreased the abundance of Chujaibacter, Devosia, Dyella, and Jatrophihabitans in both varieties (Figure 4).
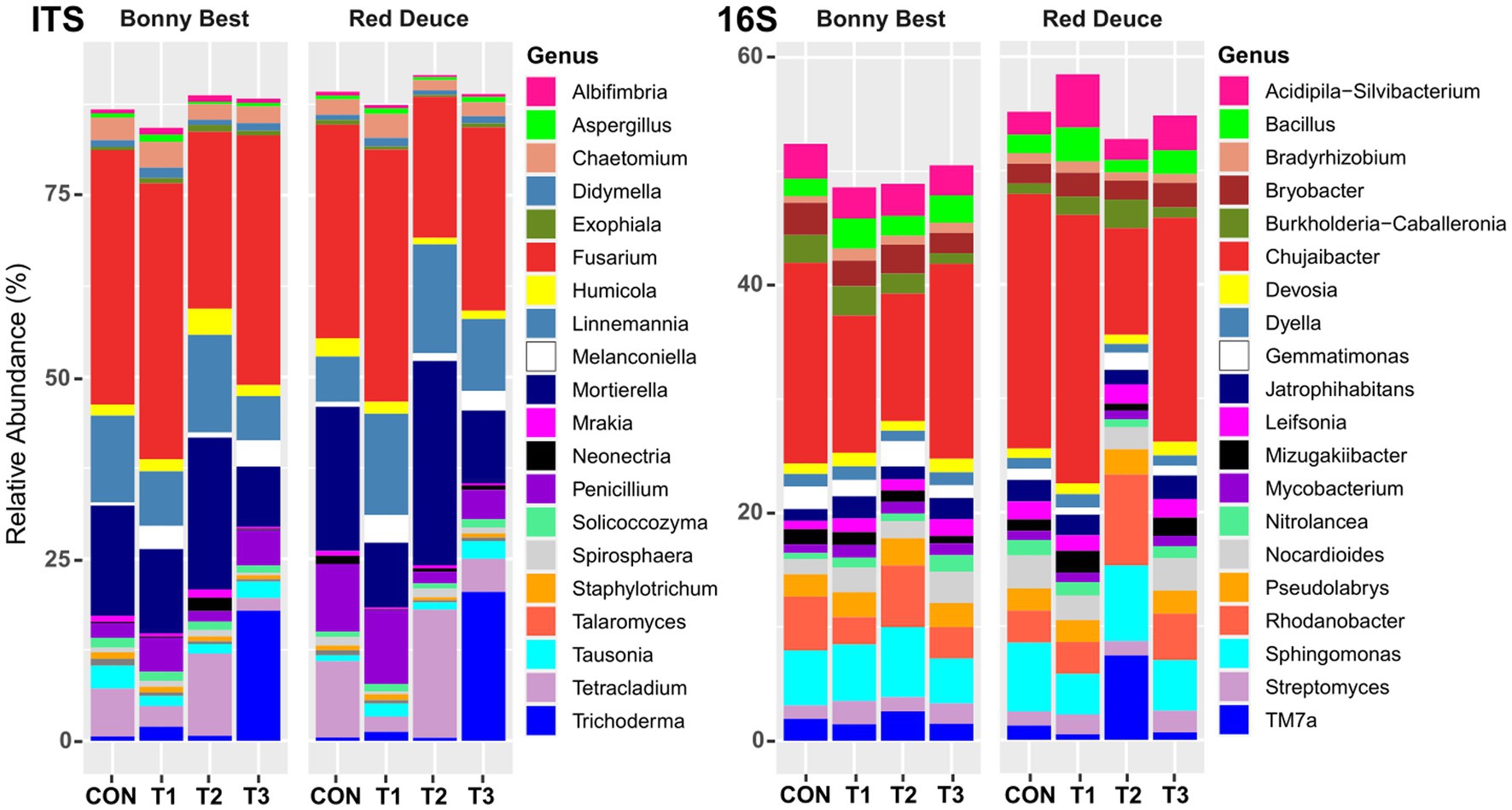
Figure 4. Effect of Trichoderma treatment on the mean relative abundance of the top 20 fungal and bacterial genera. Treatments include CON (control treatment), T1 (pre-transplant treatment with T. virens), T2 (at-transplant treatment with T. virens), and T3 (post-transplant treatment with T. virens).
Effect of Trichoderma treatment and tomato variety on the proportional abundance of specific species, genera, and guilds
ANOVA did not reveal a significant Trichoderma treatment × tomato variety interaction effect on the proportional abundance of any tested species, genera, or guilds (Supplementary Table S4). The main effect of Trichoderma treatment was significant in many cases, except for fungal pathogens other than Fusarium, bacterial biocontrol agents, and bacterial pathogens (Supplementary Table S4). The mean separation showed that the abundance of T. virens was higher in T3 than in the other treatments (Figure 5). T1 resulted in a significantly higher abundance of Fusarium compared to T2, bacterial plant growth promoters compared to CON, nitrifiers compared to CON and T2, and symbiotic nitrogen fixers compared to T2. T1 significantly decreased the abundance of free-living nitrogen fixers and denitrifiers compared to T2. T2 significantly increased the abundance of beneficial fungi compared to T3, free-living nitrogen fixers compared to T1 and T3, and denitrifiers compared to CON, T1, and T3. T2 significantly decreased the abundance of nitrifiers compared to T1 and T3 and soil chemicals/toxin detoxifiers compared to T3.
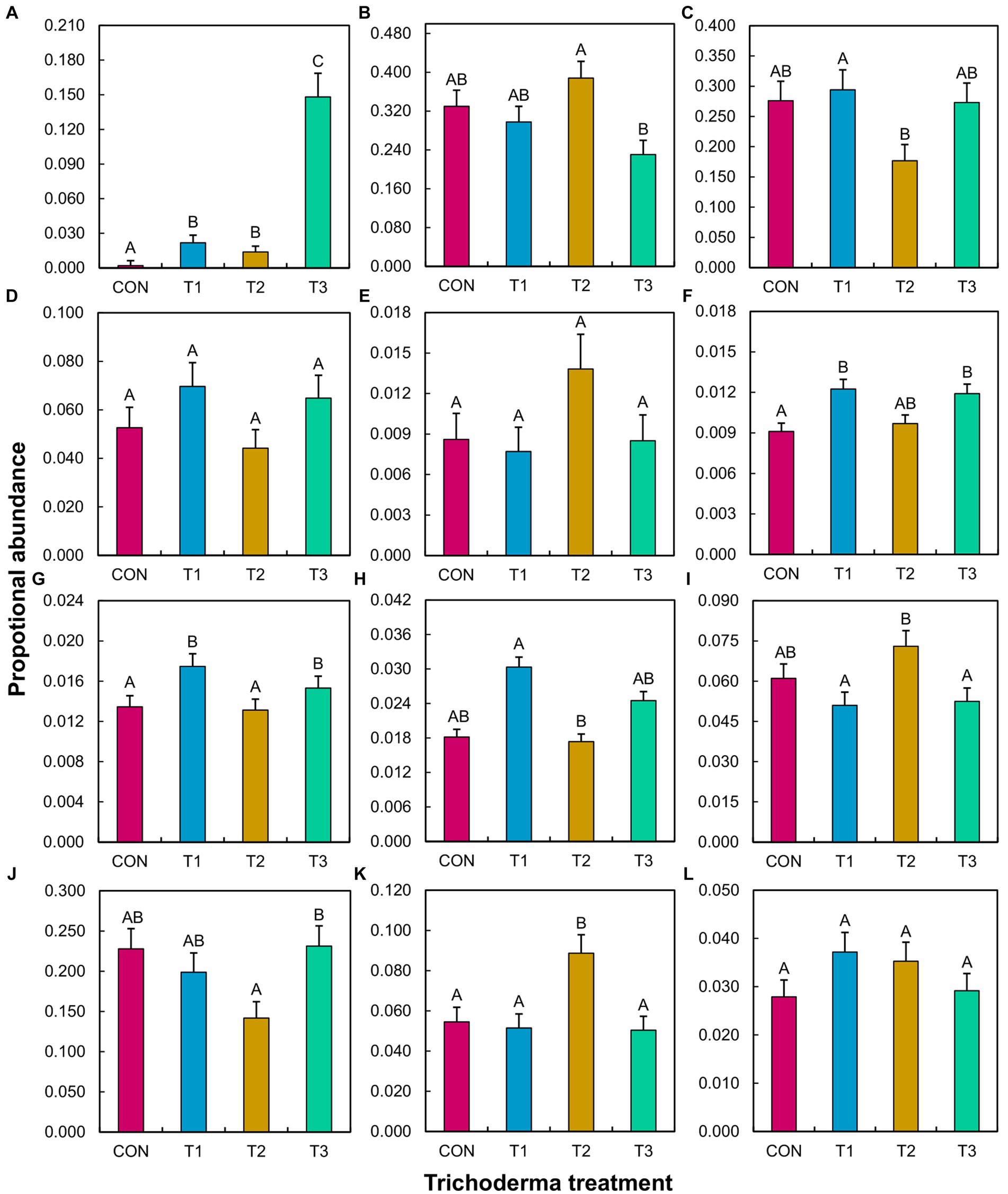
Figure 5. Effect of different Trichoderma treatments on the proportional abundance of a specific species, genus, or guild in the rhizosphere across tomato varieties (Bonny Best and Red Deuce). Treatments include CON (control treatment), T1 (pre-transplant treatment with T. virens), T2 (at-transplant treatment with T. virens), and T3 (post-transplant treatment with T. virens). Proportional abundance is the ratio between the read counts of a specific species/genus/guild in a sample and the total read count of the same sample. A guild is a combination of genera that perform a specific ecological role. (A) T. virens; (B) Beneficial fungi (Acremonium, Cadophora, Chaetomium, Clonostachys, Mortierella, Paraphaeosphaeria, Penicillium, Syncephalis, Humicola, Marquandomyces, Metacordyceps, Metarhizium, Linnemannia); (C) Fusarium; (D) Non-Fusarial fungal pathogens (Albifimbria, Aspergillus, Cladosporium, Dactylonectria, Didymella, Gibellulopsis, Melanconiella, Microdochium, Mycoleptodiscus, Neocosmospora, Neonectria, Paecilomyces, Plectosphaerella, Pseudopithomyces, Rhizoctonia, Talaromyces); (E) Biocontrol agents (Arthrobacter, Bdellovibrio, Flavisolibacter, Lysobacter, Massilia); (F) Plant growth promoters (Devosia, Mucilaginibacter); (G) Nitrifiers (Chujaibacter, MND1, Nitrolancea, Nitrosospira, Nitrospira, Pseudarthrobacter, Pseudolabrys); (H) Symbiotic Nitrogen fixers (Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Bradyrhizobium, Mesorhizobium); (I) Free-living Nitrogen fixers (Bacillus, Clostridium, Paenibacillus, Rhodopseudomonas); (J) soil chemicals/toxins detoxifiers (Mycobacterium, Sphingomonas); (K) Denitrifiers (Burkholderia-Caballeronia-Paraburkholderia, Conexibacter, Rhodanobacter); (L) Pathogens (Burkholderia-Caballeronia-Paraburkholderia, Ralstonia, Streptomyces). (E–L) represent bacteria. Per ANOVA (Supplementary Table S4), the main effect of Trichoderma treatment is depicted. Means followed by a common letter are not significantly different. The designation of the significant mean difference between the four levels of the factor “Trichoderma treatment” is based on the p-values that are adjusted for multiple comparisons using the Tukey–Kramer test at the 5% level of significance (=5% experiment-wise error rate). Error bars represent standard errors.
Effects of volatiles emitted by cultured rhizomicrobial communities from different Trichoderma treatments on the growth of T. virens and F. oxysporum
A significant Trichoderma treatment × tomato variety interaction effect on the colony diameter of T. virens and F. oxysporum f. sp. lycopersici was observed (Supplementary Table S5). The main effect of Trichoderma treatment was significant on the growth of F. oxysporum f. sp. radicis-lycopersici. According to the mean separation, volatiles released by the T2 rhizomicrobiome of Bonny Best increased the growth of T. virens compared to CON (Figure 6). In Red Deuce, T1 and T2 rhizomicrobiomes increased the growth of T. virens compared to CON. In Bonny Best, F. oxysporum f. sp. lycopersici growth was not significantly different among the four treatments (Figure 6). However, in Red Deuce, volatiles released by T1 and T2 rhizomicrobiomes significantly reduced the growth of F. oxysporum f. sp. lycopersici compared to CON and T3 (Figure 6). T1 and T2 rhizomicrobiomes significantly reduced the growth of F. oxysporum f. sp. radicis-lycopersici compared to CON and T3 across both varieties (Figure 6). Representative colonies of the tested fungi after each treatment are shown in Supplementary Figure S3.
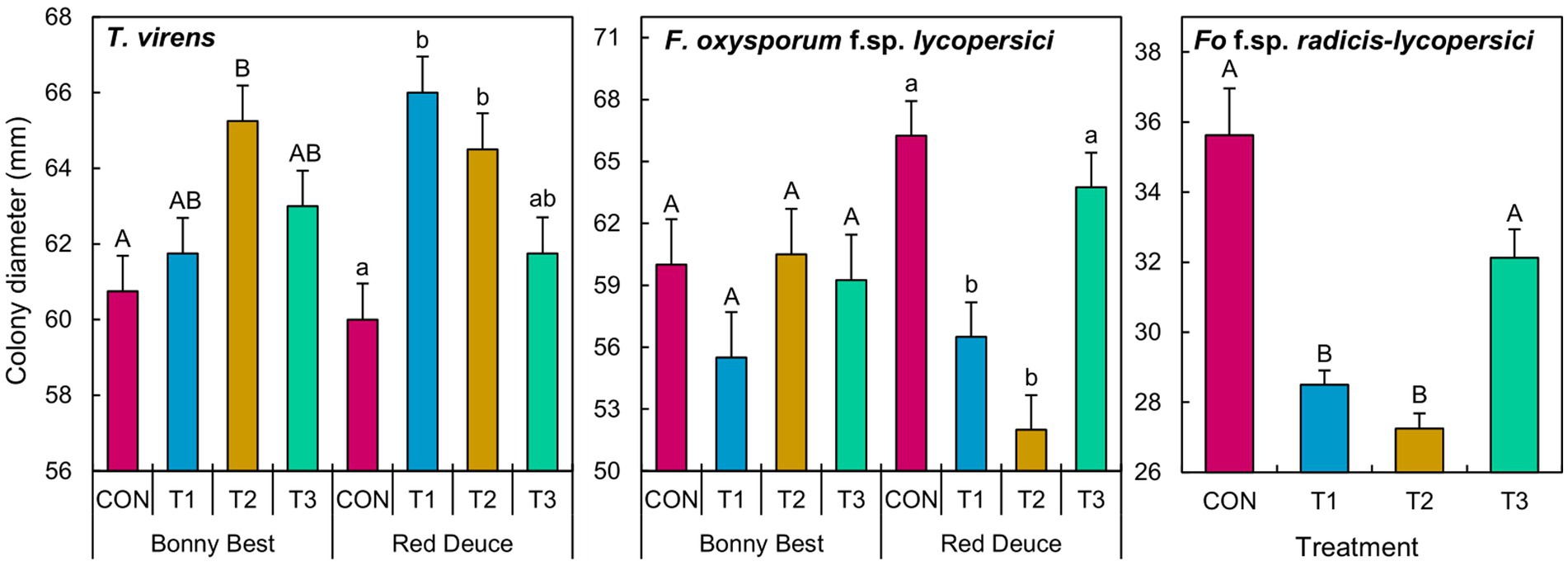
Figure 6. Effect of the volatiles emitted by the cultured fraction of the rhizomicrobiomes isolated from two tomato varieties (Bonny Best and Red Deuce) following Trichoderma treatments on the mycelial growth of T. virens, F. oxysporum f. sp. lycopersici, and F. oxysporum f. sp. radicis-lycopersici. Treatments include CON (control treatment), T1 (pre-transplant treatment with T. virens), T2 (at-transplant treatment with T. virens), and T3 (post-transplant treatment with T. virens). Means followed by the same letters within each letter type (upper or lower case) are not significantly different at the 5% level (=5% experimental-wise error rate) of significance after adjusting the p-values for multiple comparisons using the Tukey test. Error bars represent standard errors. In the case of F. oxysporum f. sp. radicis-lycopersici, the main effect of Trichoderma treatment is depicted across two tomato varieties.
Effects of water-soluble rhizosphere metabolites on the germination of T. virens and F. oxysporum conidia
The main effect of Trichoderma treatment was significant on the germination of T. virens conidia (Supplementary Table S6). A significant Trichoderma treatment × tomato variety interaction effect was observed for F. oxysporum f. sp. lycopersici and F. oxysporum f. sp. radicis-lycopersici (Supplementary Table S6). According to the mean separation, metabolites from T3 significantly reduced the conidial germination of T. virens compared to those of CON and T1 across tomato varieties (Figure 7). T2 and T3 metabolites from Bonny Best reduced the germination of both F. oxysporum strains compared to CON and T1 (Figure 7). T2 metabolites from Red Deuce reduced the germination of F. oxysporum f. sp. lycopersici compared to T1 (Figure 7). No significant germination difference in F. oxysporum f. sp. radicis-lycopersici was observed among the four treatments in Red Deuce (Figure 7).
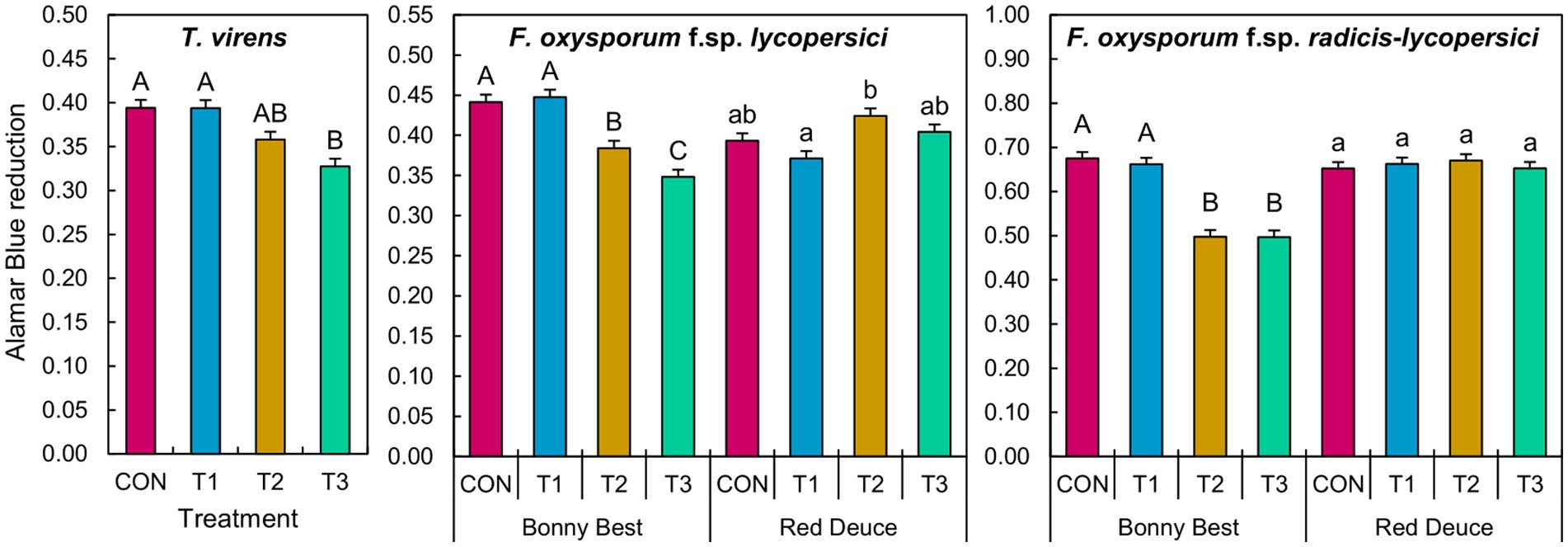
Figure 7. Effect of the water-soluble metabolites extracted from rhizosphere soils of two tomato varieties (Bonny Best and Red Deuce) following Trichoderma treatments on the germination of T. virens, F. oxysporum f. sp. lycopersici, and F. oxysporum f. sp. radicis-lycopersici conidia. The level of Almar Blue reduction is directly proportional to the conidial germination rate. Treatments include CON (control treatment), T1 (pre-transplant treatment with T. virens), T2 (at-transplant treatment with T. virens), and T3 (post-transplant treatment with T. virens). Means followed by the same letters within each letter type (upper or lower case) are not significantly different at the 5% level (=5% experimental-wise error rate) of significance after adjusting the p-values for multiple comparisons using the Tukey test. Error bars represent standard errors.
Discussion
The at-transplant application of T. virens enhances tomato growth the most
Our findings showed that the maximum shoot growth of both tomato varieties can be achieved by applying T. virens just before transplanting the seedlings. Although the at- and post-transplant applications were equally effective at enhancing tomato root growth and root/shoot ratio, this effect was only significant for Red Deuce. As the tomato variety influences the beneficial effect of Trichoderma (Tucci et al., 2011), the effect of its application method on tomato growth could also be influenced by the tomato variety. Evaluation of how the growth of diverse tomato varieties responds to the tested application strategies with more Trichoderma spp. and soil types is needed to determine which strategy works best for individual production systems and conditions. Tomato-growing soils likely harbor a variety of pathogens. Since we did not introduce any soilborne tomato pathogen(s) to the soil, an evaluation of how different application methods affect crop growth and health in the presence of known pathogens is also needed. Previous studies showed that Trichoderma did not enhance plant growth in the absence of F. oxysporum f. sp. lycopersici (Ghazalibiglar et al., 2016; Natsiopoulos et al., 2022).
Greater rhizosphere abundance of T. virens does not necessarily result in better tomato growth
The post-transplant application resulted in the highest rhizosphere abundance of T. virens in both tomato varieties. Although its abundance was substantially different between the at- and post-transplant applications, we did not observe a significant difference in the growth of both varieties between the two treatments. On the other hand, although the abundance following the at-transplant application was lower than that after the pre-transplant application, the former enhanced tomato growth better. These observations suggested that rhizosphere abundance may not be the primary factor determining the tomato growth-promoting ability of T. virens. In this experiment, we introduced T. virens inoculum in the form of conidia. Although the presence of conidia is important, how well they germinate and grow determines the success of root colonization and, in turn, the level of growth promotion. Previous studies have shown that Trichoderma must colonize plant roots to stimulate plant growth (Kleifeld and Chet, 1992; Olson and Benson, 2007), and that greater colonization results in a greater level of growth promotion (Eslahi et al., 2020). Therefore, although post-transplant application resulted in greater rhizosphere T. virens abundance (sequence read counts), it is possible that intact conidia in the rhizosphere, rather than mycelia, served as the main source of DNA contributing to this greater abundance/read count. In fact, the Alamar Blue assay showed that rhizosphere metabolites from post-transplant application significantly reduced the germination of T. virens conidia compared to those from control (see below). Additional experiments are needed to determine how different application strategies affect the rhizoplane and root endosphere abundance of T. virens and to unravel their possible association with tomato growth in the absence and presence of known soilborne pathogens.
Changes in the abundance of specific microbial genera and guilds caused by the at-transplant application may contribute to the enhancement of tomato growth
A previous study showed that both bacterial inoculants and inoculant-driven rhizosphere bacterial community shifts contribute to the early growth stimulation of tomato plants (Eltlbany et al., 2019). Soil application of T. guizhouense significantly increased cabbage plant biomass, and this effect was attributed to changes in the composition of the resident fungal community, including the increased abundance and number of resident plant growth-promoting fungal taxa (Wang et al., 2023). Our results showed that the at-transplant application of T. virens increased the abundance of potentially beneficial fungi (e.g., crop growth promoters, entomopathogenic and nematophagous fungi, mycoparasites) and bacterial biocontrol agents. Such changes may contribute to enhanced tomato growth. In addition, the at-transplant application decreased the abundance of rhizosphere fusaria, including F. fujikuroi, F. graminearum, F. neocosmosporiellum, F. nurragi, F. proliferatum, F. sporotrichioides, F. equiseti, and F. sacchari. Although they are not known to cause soilborne diseases in tomatoes, their opportunistic pathogenicity and asymptomatic growth hindrance cannot be ruled out. Therefore, the reduction in the abundance of potentially pathogenic fusaria may have helped enhance tomato growth. The increase in free-living nitrogen fixers (predominantly Bacillus species) could contribute to increased vegetative growth by enhancing nitrogen availability. A previous study showed that certain Bacillus species enhance tomato growth by improving nitrogen fixation and uptake (Masood et al., 2020). Of course, the at-transplant application-mediated increase in denitrifiers may reduce the available nitrogen for tomatoes. Further studies are needed to determine whether T. virens application-mediated changes in bacterial taxa related to the nitrogen cycle can significantly affect the overall nitrogen utilization efficiency of tomatoes.
T. virens treatments do not significantly alter the global rhizosphere fungal/bacterial community composition
Competitive displacement of non-target soil microbes and toxicity are key unintended impacts associated with using BCAs like Trichoderma (Cook et al., 1996; Brimner and Boland, 2003). Such potentially negative effects on resident soil microbiomes have become a concern (Brimner and Boland, 2003; Szczepaniak et al., 2015). Some microbiome studies provide evidence of Trichoderma-driven competitive displacement and significant soil microbial community shifts, whereas others do not. T. viride T23 induced significant shifts in the richness, structure, and composition of the rhizosphere fungal and bacterial communities of muskmelon (Zhang et al., 2023). T. koningiopsis application similarly shifted both communities in the Pinus massoniana rhizosphere (Yu and Luo, 2020). In contrast, Trichoderma did not affect the rhizosphere fungal community composition of lettuce (Bellini et al., 2023), the bacterial community composition of tomato (Eltlbany et al., 2019), or the fungal/bacterial community composition of zucchini (Cucu et al., 2020). Therefore, the degree to which Trichoderma application affects the non-target rhizosphere microbial community appears to be determined by multiple factors, such as Trichoderma species, plants, and soil sources. In the present study, none of the Trichoderma application methods significantly decreased the tomato rhizosphere’s fungal/bacterial richness, evenness, or diversity compared to CON. Furthermore, none of the application methods significantly altered the fungal and bacterial community composition of the field soil employed. Therefore, the pre-, at-, and post-transplant applications of T. virens did not appear to pose a threat to the rhizomicrobiome structure and function of the tested varieties. New experiments using different soils and tomato varieties are needed to further evaluate this neutral effect.
Application methods differentially affect mycelial growth and conidial germination of T. virens and F. oxysporum via rhizomicrobial volatiles and water-soluble soil metabolites
Several sequence data-based studies have suggested that Trichoderma-mediated changes in the rhizomicrobiome contribute to soilborne disease suppression. The control of damping-off caused by F. oxysporum in Pinus massoniana seedlings by T. koningiopsis was partly attributed to rhizosphere microbiome shifts (Yu and Luo, 2020). T. harzianum-mediated shifts in the maize rhizosphere microbiome contributed to suppressing Fusarium Stalk rot caused by F. graminearum (Saravanakumar et al., 2017). In these cases, disease suppression was attributed to an increase in beneficial microbial taxa such as resident rhizosphere biocontrol agents and plant growth promoters. Nonetheless, the mechanisms by which altered microbial communities confer better pathogen control are poorly understood. Microbial volatiles have been shown to play a key role in inter-microbial communication, and some microbes use them to suppress others. For example, Trichoderma was shown to use its volatile compounds to suppress F. oxysporum (Li et al., 2018). Therefore, extending the role of volatiles in microbe-microbe to microbiome–microbe interactions, we hypothesized that volatiles emitted by the culturable fraction of altered rhizosphere microbes upon different T. virens treatments differentially modulate the growth of T. virens and two F. oxysporum strains. We further hypothesized that water-soluble metabolites extracted from the rhizosphere soil of the two tomato varieties after four Trichoderma treatments differentially affect the germination of T. virens and F. oxysporum conidia. Although none of the Trichoderma treatments significantly altered the rhizosphere fungal and bacterial community composition at the global level, they altered the abundance of some major phyla and genera. These alterations can result in altered microbial volatile and rhizosphere metabolite profiles, resulting in differential effects on Trichoderma and F. oxysporum growth.
Our findings showed that the volatiles emitted by the rhizomicrobial community from both tomato varieties after the at-transplant application better promoted the in-vitro mycelial growth of T. virens. As discussed above, this may play a direct role in enhancing its root colonization, thus better promoting tomato growth. The same was observed with the pre-transplant application of Red Deuce. On the other hand, the post-transplant application did not cause volatile-mediated mycelial growth enhancement of T. virens in both varieties. These findings suggest that the effectiveness of T. virens application methods in enhancing its mycelial proliferation by altering rhizomicrobial volatiles is tomato variety-dependent. On the other hand, as revealed by the non-significant Trichoderma treatment × tomato variety interaction, the effect of treatment on the rhizosphere metabolite-mediated conidia germination of T. virens was not tomato variety-dependent. For instance, compared to the control, soil metabolites from the post-transplant application reduced conidial germination across varieties. Taken together, volatiles and water-soluble metabolites from a given application method do not appear to have similar effects (i.e., synergistic, antagonistic, or neutral) on the modulation of T. virens conidial germination and mycelial growth. A reciprocal experiment (effects of volatiles on conidial germination and water-soluble metabolites on mycelial growth) and simultaneous exposure (effects of volatiles and water-soluble metabolites on conidial germination and subsequent mycelial growth) could provide more mechanistic insights.
The volatiles emitted by culturable rhizomicrobial communities of both tomato varieties after the pre- and at-transplant applications significantly suppressed the mycelial growth of F. oxysporum f. sp. radicis-lycopersici. The same was true for F. oxysporum f. sp. lycopersici, but only for Red Duce. The post-transplant application had a non-significant effect on the growth of both Fusarium species. Metabolites from Bonny Best after the at- and post-transplant applications significantly suppressed the conidial germination of both F. oxysporum strains. Taken together, these results suggest that the application method matters for building suppressiveness against Fusarium pathogens. The importance of application methods was previously reported; for example, suppressiveness against pre-emergence damping-off of pea caused by Pythium spp. was only achieved by planting Trichoderma conidia-treated seeds but not by introducing them into the conducive soil (Harman et al., 1980). In contrast, suppressiveness against Rhizoctonia solani was induced by both seed and soil applications (Harman et al., 1980). Our results further suggested that Trichoderma application-mediated changes in both the rhizosphere metabolite and rhizomicrobial emitted volatile profiles may be important in suppressing F. oxysporum in susceptible tomato varieties such as Bonny Best, while volatiles are more important for resistant varieties such as Red Deuce.
The findings of our in-vitro assays, in general, suggest that some of the tested T. virens application methods could build rhizomicrobiome-mediated mycelial growth suppressiveness against some Fusarium pathogens in a tomato variety-independent manner, while the same for some other pathogens can be tomato variety-dependent. It would be vital to redo the experiment by introducing pathogens to the soil used in our experiments, growing the same tomato, and quantifying the actual pathogen levels in the rhizosphere to see if different T. virens application methods truly build suppressiveness against the pathogens. It is also important to investigate the durability of such microbial ecology-based pathogen suppressiveness by repeating the aforementioned experiment over multiple tomato growth cycles (with and without applying a new Trichoderma inoculant). Furthermore, as the influence of soil source on the outcomes described above cannot be neglected, expanded investigations are required to test whether different soils, Trichoderma species, application methods, and tomato varieties in combination similarly modulate the rhizosphere suppressiveness against soilborne pathogens via rhizomicrobial volatiles and water-soluble metabolites.
Conclusion
The at-transplant application of T. virens promoted tomato growth better than the pre- and post-transplant applications. In commercial tomato production, tomato seedlings are typically raised using a dual-tray system (starter and bottom trays). Therefore, the method we used to introduce Trichoderma can be readily incorporated into commercial systems to enhance the vigor and health of tomatoes. Our findings showed that T. virens (particularly through the at-transplant application) could potentially be used to build rhizomicrobiome-based suppressiveness against soilborne pathogens like F. oxysporum. However, further experiments with more explanatory variables are required before implementing this method for tomato production. Specific questions include whether different Trichoderma species work similarly, whether different tomato varieties and soil types result in similar tomato growth and rhizomicrobiome responses to Trichoderma treatments, whether rhizomicrobiome changes driven by Trichoderma treatments contribute to suppressing other soilborne pathogens, and whether the assays used to evaluate the effects of volatile and water-soluble soil metabolites are reliable indicators for predicting the (a) conduciveness/suppressiveness of diverse field soils for Trichoderma proliferation and (b) Trichoderma-mediated soil conduciveness/suppressiveness against diverse pathogens.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
AB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. SK: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We acknowledge support from the USDA Specialty Crop Multi-State Program (AM170200XXXXG006), USDA Specialty Crop Block Grant Program (C940000979), and USDA-NIFA and Hatch Appropriations (PEN4839).
Acknowledgments
The authors would like to thank Drs. Dilooshi Weerasooriya, Carl Schlagnhaufer, and Fabricio Rocha Vieira for their miscellaneous assistance. Authors also wish to thank the reviewers and the Pennsylvania State University One Health Microbiome Center.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1366690/full#supplementary-material
References
Adekunle, A. T., Cardwell, K. F., Florini, D. A., and Ikotun, T. (2001). Seed treatment with Trichoderma species for control of damping-off of cowpea caused by Macrophomina phaseolina. Biocontrol Sci. Tech. 11, 449–457. doi: 10.1080/09583150120067481
Amanullah, F., and Khan, W. U. D. (2023). Trichoderma asperellum L. coupled the effects of biochar to enhance the growth and physiology of contrasting maize cultivars under copper and nickel stresses. Plan. Theory 12:958. doi: 10.3390/plants12040958
Bellini, A., Gilardi, G., Idbella, M., Zotti, M., Pugliese, M., Bonanomi, G., et al. (2023). Trichoderma enriched compost, BCAs and potassium phosphite control fusarium wilt of lettuce without affecting soil microbiome at genus level. Appl. Soil Ecol. 182:104678. doi: 10.1016/j.apsoil.2022.104678
Benítez, T., Rincón, A. M., Limón, M. C., and Codón, A. C. (2004). Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 7, 249–260.
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., Köberl, M., Rybakova, D., Müller, H., Grosch, R., and Smalla, K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93:fix50. doi: 10.1093/femsec/fix050
Bhadauria, B. P., Singh, Y., Puri, S., and Singh, P. K. (2012). Ecofriendly management of fusarium wilt of Brinjal. Ecol Environ Conserv. 18, 1049–1052.
Bissett, J., Gams, W., Jaklitsch, W., and Samuels, G. J. (2015). Accepted Trichoderma names in the year 2015. IMA Fungus. 6, 263–295. doi: 10.5598/imafungus.2015.06.02.02
Blaya, J., Frutos, C., Marhuenda, J., Pascual, A., and Ros, M. (2016). Microbiota characterization of compost using omics approaches opens new perspectives for Phytophthora root rot control. PLoS One 11, 1–19. doi: 10.1371/journal.pone.0158048
Brimner, T. A., and Boland, G. J. (2003). A review of the non-target effects of fungi used to biologically control plant diseases. Agric. Ecosyst. Environ. 100, 3–16. doi: 10.1016/S0167-8809(03)00200-7
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Carvalho, D. D., Lobo Junior, M., Martins, I., Inglis, P. W., and Mello, S. (2014). Biological control of fusarium oxysporum f. sp. phaseoli by Trichoderma harzianum and its use for common bean seed treatment. Trop. Plant Pathol. 39, 384–391. doi: 10.1590/S1982-56762014000500005
Chaparro, J. M., Sheflin, A. M., Manter, D. K., and Vivanco, J. M. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 48, 489–499. doi: 10.1007/s00374-012-0691-4
Cook, R. J., Bruckart, W. L., Coulson, J. R., Goettel, M. S., Humber, R. A., Lumsden, R. D., et al. (1996). Safety of microorganisms intended for pest and plant disease control: a framework for scientific evaluation. Biol. Control 7, 335–351.
Cucu, M. A., Gilardi, G., Pugliese, M., Ferrocino, I., and Gullino, M. L. (2020). Effects of biocontrol agents and compost against the Phytophthora capsici of zucchini and their impact on the rhizosphere microbiota. Appl. Soil Ecol. 154:103659. doi: 10.1016/j.apsoil.2020.103659
Da Silva, J. S., de Medeiros, E. V., Da Costa, D. P., de Souza, C. A., De Oliveira, J. B., da França, R., et al. (2022). Biochar and Trichoderma aureoviride URM 5158 as alternatives for the management of cassava root rot. Appl. Soil Ecol. 172:104353. doi: 10.1016/j.apsoil.2021.104353
de Araujo, A. S., Blum, L. E. B., and de Figueiredo, C. C. (2019). The synergistic effect of sewage sludge biochar and Trichoderma harzianum on soybean yield. Aust. J. Crop. Sci. 13, 1183–1187. doi: 10.21475/ajcs.19.13.07.p1779
Domínguez, P., Miranda, L., Soria, C., de los Santos, B., Chamorro, M., Romero, F., et al. (2014). Soil biosolarization for sustainable strawberry production. Agron. Sustain. Dev. 34, 821–829. doi: 10.1007/s13593-014-0211-z
Dou, K., Lu, Z., Wu, Q., Ni, M., Yu, C., Wang, M., et al. (2020). MIST: a multilocus identification system for Trichoderma. Appl. Environ. Microbiol. 86, 1–13. doi: 10.1128/AEM.01532-20
Eltlbany, N., Baklawa, M., Ding, G. C., Nassal, D., Weber, N., Kandeler, E., et al. (2019). Enhanced tomato plant growth in soil under reduced P supply through microbial inoculants and microbiome shifts. FEMS Microbiol. Ecol. 95:fiz124. doi: 10.1093/femsec/fiz124
Eslahi, N., Kowsari, M., Motallebi, M., Zamani, M. R., and Moghadasi, Z. (2020). Influence of recombinant Trichoderma strains on growth of bean (Phaseolus vulgaris L) by increased root colonization and induction of root growth related genes. Sci. Hortic. 261:108932. doi: 10.1016/j.scienta.2019.108932
Estévez-Geffriaud, V., Vicente, R., Vergara-Díaz, O., Narváez Reinaldo, J. J., and Trillas, M. I. (2020). Application of Trichoderma asperellum T34 on maize (Zea mays) seeds protects against drought stress. Planta 252, 1–12. doi: 10.1007/s00425-020-03404-3
Ferreira, F. V., and Musumeci, M. A. (2021). Trichoderma as biological control agent: scope and prospects to improve efficacy. World J. Microbiol. Biotechnol. 37:90. doi: 10.1007/s11274-021-03058-7
Fraceto, L. F., Maruyama, C. R., Guilger, M., Mishra, S., Keswani, C., Singh, H. B., et al. (2018). Trichoderma harzianum-based novel formulations: potential applications for management of next-gen agricultural challenges. J. Chem. Technol. Biotechnol. 93, 2056–2063. doi: 10.1002/jctb.5613
Ghazalibiglar, H., Kandula, D. R. W., and Hampton, J. G. (2016). Biological control of fusarium wilt of tomato by Trichoderma isolates. N Z Plant Prot. 69, 57–63. doi: 10.30843/nzpp.2016.69.5915
Ghorchiani, M., Etesami, H., and Alikhani, H. A. (2018). Improvement of growth and yield of maize under water stress by co-inoculating an arbuscular mycorrhizal fungus and a plant growth promoting rhizobacterium together with phosphate fertilizers. Agric. Ecosyst. Environ. 258, 59–70. doi: 10.1016/j.agee.2018.02.016
Guzmán-Guzmán, P., Kumar, A., de Los Santos-Villalobos, S., Parra-Cota, F. I., Orozco-Mosqueda, M. D. C., Fadiji, A. E., et al. (2023). Trichoderma species: our best fungal allies in the biocontrol of plant diseases—a review. Plan. Theory 12:432. doi: 10.3390/plants12030432
Haggag, W. M., and Saber, M. S. (2000). Evaluation of three compost as multi-strain carriers for biofertilizer and biocontrol agents against fusarium wilt disease of some legume plants. Arab J. Biotechnol. 3, 133–144.
Hannan, M. A., Hasan, M. M., Hossain, I., Rahman, S. M. E., Ismail, A. M., and Oh, D. H. (2012). Integrated management of foot rot of lentil using biocontrol agents under field condition. J. Microbiol. Biotechnol. 22, 883–888. doi: 10.4014/jmb.1201.01008
Hanson, L. E. (2000). Reduction of Verticillium wilt symptoms in cotton following seed treatment with Trichoderma virens. J. Cotton Sci. 4, 224–231.
Harman, G. E., Chet, I., and Baker, R. (1980). Trichoderma hamatum effects on seed and seedling disease induced in radish and pea by Pythium spp. or Rhizoctonia solani. Phytopathology 70, 1167–1172. doi: 10.1094/Phyto-70-1167
Hassan, M. M. (2014). Influence of protoplast fusion between two Trichoderma spp. on extracellular enzymes production and antagonistic activity. Biotechnol. Biotechnol. Equip. 28, 1014–1023. doi: 10.1080/13102818.2014.978206
Hermosa, R., Viterbo, A., Chet, I., and Monte, E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158, 17–25. doi: 10.1099/mic.0.052274-0
Joshi, D., Hooda, K. S., and Bhatt, J. C. (2009). Integration of soil solarization with biofumigation and Trichoderma spp. for management of damping-off in tomato (Lycopersicon esculentum) in the mid altitude region of northwestern Himalayas. Indian J. Agric. Sci. 79, 754–757.
Karuppiah, V., Sun, J., Li, T., Vallikkannu, M., and Chen, J. (2019). Co-cultivation of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens 1841 causes differential gene expression and improvement in the wheat growth and biocontrol activity. Front. Microbiol. 10:1068. doi: 10.3389/fmicb.2019.01068
Kleifeld, O., and Chet, I. (1992). Trichoderma harzianum-interaction with plants and effect on growth response. Plant Soil 144, 267–272. doi: 10.1007/BF00012884
Kõljalg, U., Larsson, K. H., Abarenkov, K., Nilsson, R. H., Alexander, I. J., Eberhardt, U., et al. (2005). UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 166, 1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x
Konappa, N., Krishnamurthy, S., Arakere, U. C., Chowdappa, S., and Ramachandrappa, N. S. (2020). Efficacy of indigenous plant growth-promoting rhizobacteria and Trichoderma strains in eliciting resistance against bacterial wilt in a tomato. Egypt J. Biol. Pest. Control. 30, 1–13. doi: 10.1186/s41938-020-00303-3
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. doi: 10.1038/nbt.4232
Li, N., Alfiky, A., Wang, W., Islam, M., Nourollahi, K., Liu, X., et al. (2018). Volatile compound-mediated recognition and inhibition between Trichoderma biocontrol agents and fusarium oxysporum. Front. Microbiol. 9:2614. doi: 10.3389/fmicb.2018.02614
Martin, M. (2011). Cutadapt removes adapter sequences from high throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Martinez Arbizu, P. (2020). pairwiseAdonis: pairwise multilevel comparison using adonis. R package version 0.4.
Masood, S., Zhao, X. Q., and Shen, R. F. (2020). Bacillus pumilus promotes the growth and nitrogen uptake of tomato plants under nitrogen fertilization. Sci. Hortic. 272:109581. doi: 10.1016/j.scienta.2020.109581
Mastouri, F., Björkman, T., and Harman, G. E. (2010). Seed treatment with Trichoderma harzianum alleviates biotic, abiotic, and physiological stresses in germinating seeds and seedlings. Phytopathology 100, 1213–1221. doi: 10.1094/PHYTO-03-10-0091
Mazzola, M., and Freilich, S. (2017). Prospects for biological soilborne disease control: application of indigenous versus synthetic microbiomes. Phytopathology 107, 256–263. doi: 10.1094/PHYTO-09-16-0330-RVW
Mbarga, J., Begoude, B., Ambang, Z., Meboma, M., Kuaté, J., Ewbank, W., et al. (2020). Field testing an oil-based Trichoderma asperellum formulation for the biological control of cacao black pod disease, caused by Phytophthora megakarya. Crop Prot. 132:105134. doi: 10.1016/j.cropro.2020.105134
McGovern, R. J. (2015). Management of tomato diseases caused by fusarium oxysporum. Crop Prot. 73, 78–92. doi: 10.1016/j.cropro.2015.02.021
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Miranda, M. A., Estrella, A. H., and Cabriales, J. P. (2006). Colonization of the rhizosphere, rhizoplane and endorhiza of garlic (Allium sativum L.) by strains of Trichoderma harzianum and their capacity to control allium white-rot under field conditions. Soil Biol. Biochem. 38, 1823–1830. doi: 10.1016/j.soilbio.2005.11.036
Montero-Barrientos, M., Hermosa, R., Cardoza, R. E., Gutiérrez, S., and Monte, E. (2011). Functional analysis of the Trichoderma harzianum nox1 gene, encoding an NADPH oxidase, relates production of reactive oxygen species to specific biocontrol activity against Pythium ultimum. Appl. Environ. Microbiol. 77, 3009–3016. doi: 10.1128/AEM.02486-10
Mweetwa, A. M., Chilombo, G., and Gondwe, B. M. (2016). Nodulation, nutrient uptake and yield of common bean inoculated with rhizobia and Trichoderma in an acid soil. J. Agric. Sci. 8, 61–71. doi: 10.5539/jas.v8n12p61
Nandini, B., Hariprasad, P., Prakash, H. S., Shetty, H. S., and Geetha, N. (2017). Trichogenic-selenium nanoparticles enhance disease suppressive ability of Trichoderma against downy mildew disease caused by Sclerospora graminicola in pearl millet. Sci. Rep. 7:2612. doi: 10.1038/s41598-017-02737-6
Natsiopoulos, D., Tziolias, A., Lagogiannis, I., Mantzoukas, S., and Eliopoulos, P. A. (2022). Growth-promoting and protective effect of Trichoderma atrobrunneum and T. Simmonsii on tomato against soil-borne fungal pathogens. Crops 2, 202–217. doi: 10.3390/crops2030015
Oksanen, J., Kindt, R., Legendre, P., O’Hara, B., Stevens, M. H., Oksanen, M. J., et al. (2022). Vegan: community ecology package. R package version 2.6.4.
Olson, H. A., and Benson, D. M. (2007). Induced systemic resistance and the role of binucleate Rhizoctonia and Trichoderma hamatum 382 in biocontrol of Botrytis blight in geranium. Biol. Control 42, 233–241. doi: 10.1016/j.biocontrol.2007.05.009
Pagès, H., Aboyoun, P., Gentleman, R., and DebRoy, S., (2022). Biostrings: Efficient manipulation of biological strings. R package version 2.66.0.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res, 41, D590–D596.
Poveda, J. (2021). Trichoderma as biocontrol agent against pests: new uses for a mycoparasite. Biol. Control 159:104634. doi: 10.1016/j.biocontrol.2021.104634
Rudresh, D. L., Shivaprakash, M. K., and Prasad, R. D. (2005). Effect of combined application of rhizobium, phosphate solubilizing bacterium and Trichoderma spp. on growth, nutrient uptake and yield of chickpea (Cicer aritenium L.). Appl. Soil Ecol. 28, 139–146. doi: 10.1016/j.apsoil.2004.07.005
Sani, M. N. H., Hasan, M., Uddain, J., and Subramaniam, S. (2020). Impact of application of Trichoderma and biochar on growth, productivity and nutritional quality of tomato under reduced NPK fertilization. Ann. Agric. Sci. 65, 107–115. doi: 10.1016/j.aoas.2020.06.003
Saravanakumar, K., Li, Y., Yu, C., Wang, Q. Q., Wang, M., Sun, J., et al. (2017). Effect of Trichoderma harzianum on maize rhizosphere microbiome and biocontrol of fusarium stalk rot. Sci. Rep. 7:1771. doi: 10.1038/s41598-017-01680-w
Shanmugaiah, V., Balasubramanian, N., Gomathinayagam, S., Manoharan, P. T., and Rajendran, A. (2009). Effect of single application of Trichoderma viride and Pseudomonas fluorescens on growth promotion in cotton plants. Afr. J. Agric. Res. 4, 1220–1225.
Singh, V., Upadhyay, R. S., Sarma, B. K., and Singh, H. B. (2016). Trichoderma asperellum spore dose depended modulation of plant growth in vegetable crops. Microbiol. Res. 193, 74–86. doi: 10.1016/j.micres.2016.09.002
Szczałba, M., Kopta, T., Gąstoł, M., and Sękara, A. (2019). Comprehensive insight into arbuscular mycorrhizal fungi, Trichoderma spp. and plant multilevel interactions with emphasis on biostimulation of horticultural crops. J. Appl. Microbiol. 127, 630–647. doi: 10.1111/jam.14247
Szczepaniak, Z., Cyplik, P., Juzwa, W., Czarny, J., Staninska, J., and Piotrowska-Cyplik, A. (2015). Antibacterial effect of the Trichoderma viride fungi on soil microbiome during PAH’s biodegradation. Int. Biodeterior. Biodegrad. 104, 170–177. doi: 10.1016/j.ibiod.2015.06.002
Szekeres, A., Kredics, L., Antal, Z., Kevei, F., and Manczinger, L. (2004). Isolation and characterization of protease overproducing mutants of Trichoderma harzianum. FEMS Microbiol. Lett. 233, 215–222. doi: 10.1111/j.1574-6968.2004.tb09485.x
Timmusk, S., Behers, L., Muthoni, J., Muraya, A., and Aronsson, A. C. (2017). Perspectives and challenges of microbial application for crop improvement. Front. Plant Sci. 8:49. doi: 10.3389/fpls.2017.00049
Tucci, M., Ruocco, M., De Masi, L., De Palma, M., and Lorito, M. (2011). The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol. Plant Pathol. 12, 341–354. doi: 10.1111/j.1364-3703.2010.00674.x
Verma, M., Brar, S. K., Tyagi, R. D., Surampalli, R. Y., and Valero, J. R. (2007). Antagonistic fungi, Trichoderma spp.: panoply of biological control. Biochem. Eng. J. 37, 1–20. doi: 10.1016/j.bej.2007.05.012
Wang, Y., Liu, Z., Hao, X., Wang, Z., Wang, Z., Liu, S., et al. (2023). Biodiversity of the beneficial soil-borne fungi steered by Trichoderma-amended biofertilizers stimulates plant production. NPJ Biofilms Microbiomes. 9:46. doi: 10.1038/s41522-023-00416-1
Woo, S. L., Hermosa, R., Lorito, M., and Monte, E. (2023). Trichoderma: a multipurpose, plant-beneficial microorganism for eco-sustainable agriculture. Nat. Rev. Microbiol. 21, 312–326. doi: 10.1038/s41579-022-00819-5
Woo, S. L., Ruocco, M., Vinale, F., Nigro, M., Marra, R., Lombardi, N., et al. (2014). Trichoderma-based products and their widespread use in agriculture. Open Mycol. J. 8, 71–126. doi: 10.2174/1874437001408010071
Xiao, Z., Zhao, Q., Li, W., Gao, L., and Liu, G. (2023). Strain improvement of Trichoderma harzianum for enhanced biocontrol capacity: strategies and prospects. Front. Microbiol. 14:1146210. doi: 10.3389/fmicb.2023.1146210
Xue, A. G., Guo, W., Chen, Y., Siddiqui, I., Marchand, G., Liu, J., et al. (2017). Effect of seed treatment with novel strains of Trichoderma spp. on establishment and yield of spring wheat. Crop Prot. 96, 97–102. doi: 10.1016/j.cropro.2017.02.003
Yang, X., Chen, L., Yong, X., and Shen, Q. (2011). Formulations can affect rhizosphere colonization and biocontrol efficiency of Trichoderma harzianum SQR-T037 against fusarium wilt of cucumbers. Biol. Fertil. Soils 47, 239–248. doi: 10.1007/s00374-010-0527-z
Yu, C., and Luo, X. (2020). Trichoderma koningiopsis controls fusarium oxysporum, causing damping-off in Pinus massoniana seedlings by regulating active oxygen metabolism, osmotic potential, and the rhizosphere microbiome. Biol. Control 150:104352. doi: 10.1016/j.biocontrol.2020.104352
Zhang, Z., Tang, S., Liu, X., Ren, X., Wang, S., and Gao, Z. (2023). The effects of Trichoderma viride T23 on rhizosphere soil microbial communities and the metabolomics of muskmelon under continuous cropping. Agronomy 13:1092. doi: 10.3390/agronomy13041092
Keywords: biocontrol, crop growth promotion, Fusarium, amplicon metagenomic sequencing, soilborne diseases, suppressive soil, tomato, trichoderma
Citation: Bandara AY and Kang S (2024) Trichoderma application methods differentially affect the tomato growth, rhizomicrobiome, and rhizosphere soil suppressiveness against Fusarium oxysporum. Front. Microbiol. 15:1366690. doi: 10.3389/fmicb.2024.1366690
Edited by:
Shitou Xia, Hunan Agricultural University, ChinaCopyright © 2024 Bandara and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seogchan Kang, c3hrNTVAcHN1LmVkdQ==
†ORCID: Ananda Y. Bandara, https://orcid.org/0000-0003-0725-8226
Seogchan Kang, https://orcid.org/0000-0003-2291-5634
 Ananda Y. Bandara
Ananda Y. Bandara Seogchan Kang
Seogchan Kang