
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 July 2024
Sec. Microbial Physiology and Metabolism
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1366336
 Yunwen Bi1†
Yunwen Bi1† Hao An1†
Hao An1† Zhewei Chi1†
Zhewei Chi1† Zhongheng Xu1
Zhongheng Xu1 Yuan Deng1
Yuan Deng1 Yuxian Ren1
Yuxian Ren1 Rui Wang1
Rui Wang1 Xinyi Lu1
Xinyi Lu1 Jia Guo1*
Jia Guo1* Ren Hu1*
Ren Hu1* Marie-Joelle Virolle2*
Marie-Joelle Virolle2* Delin Xu1*
Delin Xu1*Streptomycetes are well-known antibiotic producers possessing in their genomes numerous silent biosynthetic pathways that might direct the biosynthesis of novel bio-active specialized metabolites. It is thus of great interest to find ways to enhance the expression of these pathways to discover most needed novel antibiotics. In this study, we demonstrated that the over-expression of acetyltransferase SCO0988 up-regulated the production of specialized metabolites and accelerated sporulation of the weak antibiotic producer, Streptomyces lividans and that the deletion of this gene had opposite effects in the strong antibiotic producer, Streptomyces coelicolor. The comparative analysis of the acetylome of a S. lividans strain over-expressing sco0988 with that of the original strain revealed that SCO0988 acetylates a broad range of proteins of various pathways including BldKB/SCO5113, the extracellular solute-binding protein of an ABC-transporter involved in the up-take of a signal oligopeptide of the quorum sensing pathway. The up-take of this oligopeptide triggers the “bald cascade” that regulates positively specialized metabolism, aerial mycelium formation and sporulation in S. coelicolor. Interestingly, BldKB/SCO5113 was over-acetylated on four Lysine residues, including Lys425, upon SCO0988 over-expression. The bald phenotype of a bldKB mutant could be complemented by native bldKB but not by variant of bldKB in which the Lys425 was replaced by arginine, an amino acid that could not be acetylated or by glutamine, an amino acid that is expected to mimic acetylated lysine. Our study demonstrated that Lys425 was a critical residue for BldKB function but was inconclusive concerning the impact of acetylation of Lys425 on BldKB function.
Streptomyces are widely distributed in terrestrial, marine and freshwater environments where they play important ecological roles (Chater, 2016). Streptomyces undergoes a complex morphological differentiation cycle starting with the germination of a spore that yields a substrate mycelium from which aerial hyphae arise. Subsequently, the tip ends of these aerial hyphae differentiate into spores. This morphological differentiation process is accompanied by the production of a variety of specialized bioactive metabolites of great interest in modern medicine (antibacterial, anticancer, and immuno-suppressive drugs) or agriculture (fungicides, herbicides, pesticides etc.…) (Niu et al., 2016). Indeed, more than 60% of clinically used antibiotics are original or chemically modified versions of metabolites produced by Streptomyces species (Lewis, 2020). A given Streptomyces specie is usually known to produce less than 5 characterized bio-active metabolites whereas the in silico analysis of the sequenced genome of numerous Streptomyces species revealed the presence of 5 to 10 fold more biosynthetic pathways that are likely to direct the biosynthesis of yet unknown specialized metabolites (Ikeda et al., 2014; Hannigan et al., 2019). Most of these pathways are unfortunately not expressed (cryptic) under normal laboratory conditions. In consequence, only a small fraction of the potential biosynthetic capacity of these bacteria is known and thus exploited. The implementation of novel strategies to activate the expression of these silent pathways is necessary to exploit the huge metabolic diversity of this genera and discover most needed novel antibiotics to face the worrying emergence and rapid spreading of antibiotic-resistant pathogens.
Several strategies were used to increase the expression of specialized metabolites biosynthetic pathways. These include genetic manipulations of genes encoding regulators linked or not to the pathways, use of elicitors or co-cultivation with other microorganisms (Ochi, 2017). However only few studies were conducted to determine the impact of post-translational modifications (PTM) on the function of proteins playing a role in the regulation of primary and specialized metabolism in Streptomyces species (Zhang and Xu, 2018). That is only in the last decade that extensive post-translational proteins modifications were discovered in S. coelicolor and in other Streptomyces species (Hesketh et al., 2002). These modifications include the phosphorylation of His and Asp residues of sensory kinases and response regulators of two component systems (Cruz-Bautista et al., 2023) or that of Ser or Thr residues of various proteins by eukaryotic-like Ser/Thr kinases (Wright and Ulijasz, 2014) but these modifications can also include succinylation (Yang et al., 2021), crotonylation (Sun et al., 2020b) as well as acetylation (Liao et al., 2014; Hamam et al., 2019).
Streptomyces coelicolor genome encodes 93 acetyltransferases (Bentley et al., 2002) but the biological function of most of them is unknown. Indeed, only a few studies demonstrated the involvement of acetyltransferases in the regulation of enzymes activities, protein–protein or DNA-transcriptional factors interactions in Streptomyces (Martín and Liras, 2020). In one of our previous published studies, we identified the acetyltransferase SCO0988 as a regulatory target of SCO3201, a regulator of the TetR-family whose over-expression caused a strong inhibition of the biosynthesis the blue polyketide antibiotic actinorhodin (ACT) and of sporulation in S. coelicolor (Zhang et al., 2020). Since the expression of sco0988 was repressed in condition of sco3201 over-expression (Xu et al., 2010), this suggested that SCO0988 might have a positive impact on ACT production and sporulation. In order to test this hypothesis, sco0988 was deleted in S. coelicolor (SC), and over-expressed in S. lividans (SL). SC and SL are phylogenetically closely related model strains that bear the same pathways directing the biosynthesis of the colored antibiotics, undecylprodigiosin (RED), a red-pigmented tripyrrole antibiotic (Feitelson et al., 1985), and actinorhodin (ACT), a blue pigmented polyketide antibiotic (Malpartida and Hopwood, 1986). However, these metabolites are abundantly and poorly produced by SC and SL, respectively. We demonstrated that the deletion of sco0988 in SC abolished antibiotic production and sporulation of SC whereas its over-expression in SL enhanced the weak antibiotic production of this strain as well as its sporulation. Our study thus confirmed that SCO0988 has a positive impact on both metabolic and morphological differentiation of these two model species. A comparative analysis of the acetylome of SL over-expressing SCO0988 with that of the native strain revealed the acetylation of lysine residues present in proteins belonging to diverse ontological classes. We decided to focus our study on BldKB/SCO5113, an extracellular solute-binding protein of an oligopeptide ABC transporter for two main reasons. Firstly, the intensity of acetylation of this protein was greatly enhanced upon SCO0988 over-expression and secondly because the BldK transporter is known to be involved in the bald signaling cascade of the quorum sensing pathway that plays a positive role in the control of antibiotic production and morphological differentiation in SC (Nodwell et al., 1996). Our results suggested that the acetylation of BldkB on the Lys 425 residue is necessary for its functionality and the way this acetylation could impact BldKB function is discussed. Our study thus contributes to a better understanding of the role of the acetyltransferase SCO0988 in the activation of antibiotic production and morphological differentiation in Streptomyces species.
The strains used in this study were S. coelicolor M145 and S. lividans TK24 (Bentley et al., 2002). These strains were grown at 28°C in MS (ACROS, Belgium) and YEME media (ACMEC, Shanghai) for spore generation and protoplast preparation, respectively, whereas R2YE medium (BD, United States) was used for protoplasts regeneration after transformation (Kieser et al., 2000). Apramycin, and thiostrepton (HARVEYBIO, Beijing) were added in solid R2YE medium at a final concentration of 50 μg/mL in cultures of SC or SL containing the plasmids pOJ260 and pSET152 and the plasmids pDH5 and pWHM3, respectively. The following E. coli strains were used, DH5α for routine cloning, ET12567 to prepare de-methylated plasmids to transform SC and C43 for efficient protein expression. These strains were grown in Luria broth (LB) medium. Ampicillin (50 μg/mL), chloramphenicol (25 μg/mL), and kanamycin (50 μg/mL) were added to growth media when required.
In order to disrupt the gene encoding the acetyltransferase SCO0988 in S. coelicolor M145 (SC), a 370 bp DNA fragment internal to the sco0988 coding region was amplified by PCR using the primer pair DisCsco0988-F/DisCsco0988-R (Table 1) and SC chromosomal DNA as template. The resulting PCR fragments were digested by BamHI and HindIII (Takara, Japan) and ligated into the plasmid pOJ260 that carries a gene conferring resistance to apramycin cut by the same enzymes (Kieser et al., 2000). The resulting plasmid, pOJ260-sco0988int, was transformed into E. coli ET12567 to prevent its methylation (Flett et al., 1997) and apramycin resistant (ApraR) transformants were selected. The non-methylated plasmids extracted from the ApraR transformants were transformed into SC and selected for ApraR. The sco0988 disrupted mutants (SC-∆sco0988) were confirmed by PCR using the primer pair DisCsco0988-F/DisCsco0988-R (Table 1).
In order to complement the SC-∆SCO0988 mutant and to over-express sco0988 in S. lividans TK24, the coding sequence of sco0988 was amplified by PCR using the primers OEsco0988-XbaI-F and OEsco0988-HindIII-R (Table 1) and SL genomic DNA as template. The resulting PCR products were digested by XbaI and HindIII and cloned into pWHM3-ermE cut by the same enzymes in order to put the expression of sco0988 under the control of the strong ermE promoter (Bibb et al., 1985). The resulting pWHM3-ermE-sco0988 plasmid was transformed in SL and in SC-∆sco0988 and protoplasts to generate the strains SL/pWHM3-ermE-sco0988 and SC-∆sco0988/pWHM3-ermE-sco0988. The SC and SL strains harboring pWHM3-ermE empty plasmid were used as a control (see Table 2).
In order to quantify biomass yield as well as RED (undecylprodigiosin) and ACT (actinorhodin) production of native and genetically modified derivatives of SC and SL constructed in this study, 107 spores of each strain were used to inoculate 200 mL of R2YE medium and grown for 72 h until stationary phase. Two hundred microliters of each culture was plated on cellophane discs deposited on the surface of 9 cm-diameter plates of R2YE solid medium in triplicates and the plates were incubated at 28°C. Half of the mycelium of each triplicate was collected every 12 h, from 24 h until 84 h of incubation, in order to establish a growth curve. Collected mycelial samples were dried at 55°C overnight and weighted. One quarter of the mycelium of each triplicate was used to assay intracellular RED and ACT concentrations and the agar medium below the last quarter was used to assay extracellular ACT concentrations as described previously but with slight modifications (Xu et al., 2010).
To assay intracellular RED, the mycelium was extracted by the addition of 1 mL of methanol then the mixture was acidified to pH 2–3 by addition of 1 mL HCl (1 M) whereas to assay intracellular ACT the mycelium was extracted by the addition of 1 mL of KOH (1 M). Samples were vortexed for 30 min at 4°C. To assay RED the optical density was measured at 530 nm against a blank constituted by methanol and HCl (1 M) (v/v:1/1) whereas to assay ACT the optical density was measured at 640 nm against a blank constituted by KOH (1 M).
To assay extracellular ACT, a quarter of each of the three R2YE agar plates was smashed and subjected to diffusion in 10 mL water at 4°C for at least 2 h. The mixture was then centrifuged and 10 mL of KOH (1N) was added to the collected supernatant. The solution was mixed by inversion, then 10 mL of HCL (3N) was added. The resulting mixture was incubated on ice for 10 min and the precipitated ACT was then collected by centrifugation. The supernatant was discarded and the ACT pellet was re-suspended in 1 mL KOH (1N). The optical density of the solution was measured at 640 nm against a blank constituted by KOH (1 M) in order to determine ACT concentration. In all cases a spectrophotometer (Molecular Devices, United States) was used to determine optical density.
In order to carry out comparative acetylome analysis of SL/pWHM3-ermE and SL/pWHM3-ermE-sco0988, 107 spores of these strains were plated on cellophane discs laid on the surface of plates of R2YE solid medium. Approximately 400 mg of dry mycelium of each strain was collected from 10 plates after 24 h and 36 h of incubation at 28°C. At 24 h the strains did not produce any colored antibiotics whereas at 36 h, the strains just started to produce colored antibiotics. Cells were lysed by sonication and total proteins were quantified using the Bradford reagent (ABP Biosciences, United States). To maintain reducing state and stabilize proteins, DTT was added at a final concentration of 10 mM to each extracted protein samples (20 μg) that were incubated for 1.5 h at 37°C, and a sulfhydryl alkylating reagent iodoacetamide (IAA) was the added to prevent disulfide bond formation at a final concentration 50 mM and the mixture was incubated in the dark for 30 min at room temperature.
The proteins were digested with trypsin (trypsin: protein = 1:50, weight ratio) at 37°C overnight. The pH of the mixture was adjusted to pH ≤3 with trifluoroacetic acid (TFA) 10% (final concentration 0.1%). Each digested peptides sample was desalted on C18 Cartridges (Cat. No. 66872-U, Sigma) and lyophilized. The digested peptides present in 1.4 mL of precooled IAP buffer (50 mM MOPS/NaOH pH 7.2, 10 mM Na2HPO4, 50 mM NaCl) (PTMScan®, #9993) were enriched by immunoaffinity purification. To do so they were incubated for 1.5 h at 4°C, with a PTMScan® Acetyl-Lysine Motif Antibody conjugated to protein A agarose beads (Cell Signaling Technology, United States). After centrifugation of the beads (2,000 × g, 30 s), the supernatant was discarded and the beads were washed thrice with 1 mL precooled IAP buffer then washed thrice again with precooled water. The washed beads were incubated with 40 μL TFA 10% (final concentration 0.15%) for 10 min at room temperature, then centrifuged (2,000 × g, 30 s). The supernatant containing the acetylated peptides was collected and this operation was repeated thrice. Desalted supernatants of the immunoprecipitated peptides utilizing C18 stage-tips (Cat. No. 22A00A001, Thermo) were prepared prior to LC-MS analysis (Nativio et al., 2020).
A Q-Exactive HF/HFX mass spectrometer (Thermo Scientific™, United States) coupled with an Easy nLC (Proxeon Biosystems) (Thermo Scientific™, United States) was used for LC-MS/MS analysis (Chen et al., 2019). Using IntelliFlow technology, the peptides in formic acid (0.1%) buffer were loaded onto the reverse phase trap column (Acclaim PepMap100, 100 pm*2 cm, nanoViper C18; Thermo Scientific™, United States) on the C18-reversed phase analytical column (Easy Column, 10 cm long, 75 pm inner diameter, 3 pm resin; Thermo Scientific™, United States) and separated with a linear gradient of acetonitrile (84%) and formic acid (0.1%) at a flow rate of 300 nL/min (Cox et al., 2020). MS data were acquired in a positive ion mode by a data-dependent top10 method (Barkovits et al., 2020).
The MS raw data for each sample were combined and searched using the MaxQuant software for protein identification and quantitation analysis. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium1 via the iProX partner repository with the dataset identifier PXD041540.
The protein sequences of the selected differentially acetylated proteins were locally searched using NCBI BLAST+ client software (ncbi-blast-2.2.28+-win32.exe). Homologous sequences were found using InterProScan (Quevillon et al., 2005) and the functional ontological category of the proteins was established using Blast2Go.2 Enrichment analysis, based on the Fisher’s exact test (Shan and Gerstenberger, 2017), was carried out the considering the whole quantified proteins as background dataset. Benjamini–Hochberg correction (Benjamini and Hochberg, 1995) for multiple testing was used to adjust derived p-values. p-values inferior to 0.05 were considered as significant. Sequences including the six amino acids upstream and downstream of the modified site were used to predict motifs with the MEME software.3
In order to delete bldKB/sco5113 in S. coelicolor M145 (SC), two 1.5-kb DNA fragments flanking the sco5113 coding region were amplified by PCR using primer pairs DelUpsco5113-HindIII-F/DelUpsco5113-XbaI-R and DelDownsco5113-XbaI-F/DelDownsco5113-KpnI-R (Table 1) and SC genomic DNA as template. The resulting two PCR fragments were individually digested with the restriction enzymes present in the primers (Takara, Japan) and cloned into the pDH5 plasmid that carries thiostrepton resistance determinant (Kieser et al., 2000), using a triple ligation strategy. The resulting plasmid pDH5-Delsco5113 suitable for sco5113 deletion was transformed into SC protoplasts. The transformants were grown for three generations on plates of R2YE medium in absence of any selective pressure. Subsequently, spores resulting from the transformants were diluted and spread to yield isolated colonies. The thiostrepton resistance or sensitivity of the resulting sporulated colonies was determined by their replicate plating on R2YE and on R2YE with thiosptrepton at 50 μg/mL. The chromosomal structure of the transformants, that had lost the delivery plasmid and were only able to grown on R2YE, was assessed by PCR using the primer pair VerifDelsco5113-F and VerifDelsco5113-R (Table 1) and genomic DNA originating from these transformants. The mutants in which the successful in-frame deletion of sco5113 was confirmed were called SC-ΔbldKB.
In order to generate a mutagenized version of BldKB/SCO5113, a plasmid containing BldKB/SCO5113 ORF together with its native promoter was first constructed. To do so, a PCR fragment encompassing this region was amplified using the primer pair CPsco5113-EcoRI-F/CPsco5113-XbaI-R (Table 1) and SC chromosomal DNA as template. The resulting PCR product cut by EcoRI and XbaI was ligated into pUC19 to generate pUC19-bldKB-wt (AAG).
Subsequently, in order to generate mutagenized BldKB, the primer pairs K425Rsco5113-F/K425Rsco5113-R and K425Qsco5113-F/K425Qsco5113-R (Table 1), in which the 425th Lys codon (AAG) was replaced by an Arg codon (AGG) or by a Gln codon (CAG), were used to amplify the mutated fragment by PCR using pUC19-bldKB-wt as a template. The resulting PCR products were treated with DpnI to digest the methylated parental DNA template, and subsequently transformed into E. coli DH5α competent cells. Plasmids isolated from various colonies were sequenced and the plasmids harboring the desired mutation (AAG/K to AGG/R) and (AAG/K to CAG/Q) were selected and named pUC19-bldKB-K425R and pUC19-bldKB-K425Q, respectively.
bldKB-wt (AAG), bldKB-K425R (AGG) and bldKB-K425Q (CAG) together with their native promoters were amplified by PCR using primer pair CPsco5113-EcoRI-F/CPsco5113-XbaI-R (Table 1) from pUC19-bldKB-wt (AAG), pUC19-bldKB-K425R (AGG) and pUC19-bldKB-K425Q (CAG), respectively. The resulting DNA fragments were ligated into the EcoRI and XbaI sites of pSET152 to generate pSET152-bldKB-wt, pSET152-bldKB-K425R and pSET152-bldKB-K425Q, respectively. These three plasmids were transformed into SC-ΔbldKB protoplasts to generate SC-ΔbldKB/pSET152-bldKB-wt, SC-ΔbldKB/pSET152-bldKB-K425R and SC-ΔbldKB/pSET152-bldKB-K425Q and the phenotype of the transformants was assessed for complementation.
In order to carry out in vitro acetylation of BldKB, recombinant His-tag fused BldKB was first expressed and purified in E. coli C to facilitate protein expression and purification (Miroux and Walker, 1996). To do so the whole bldKB coding region, except its termination codon, was amplified by PCR using primer pairs PEsco5113-EcoRI-F and PEsco5113-HindIII-R (Table 1). The resulting PCR fragments digested by EcoRI and HindIII (Takara, Japan) were cloned into the plasmid pET28a under the control of a LacI-like IPTG inducible promoter (Schifferli, 1995). In this plasmid, that carries a gene conferring resistance to kanamycin (Kanr), the cloned gene is fused to a DNA sequence translated as a 6-His-tag. The recombinant plasmid pET28a-bldKB was transformed into E. coli C43. E. coli C43/pET28a-bldKB was cultured in LB medium containing 25 μg/mL kanamycin at 37°C. When OD600 reached 0.4–0.5, IPTG was added at a final concentration of 0.5 mM and induction was allowed for 5–6 h. The cells were collected by centrifugation, washed in PBS (137 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH = 7.4) and sonicated to obtain homogenous cell crude extracts. His-tag fused BldKB was purified to near homogeneity using Ni-NTA column according to the manufacturer’s instructions (Sangon Biotech, Shanghai Co., Ltd.).
However, since the acetyltransferase SCO0988 is a membrane protein, that could not be purified in E. coli, crude extracts of SL/pWHM3-ermE and SL/pWHM3-ermE-sco0988 had to be prepared as described in You et al. (2019) in order to carry out the acetylation assays. To do so, these two strains were cultivated in R2YE liquid medium until stationary phase and their mycelium was collected by centrifugation, washed in PBS, re-suspended in sonication buffer (50 mM Tris-HCl pH 8.0, 10% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol) containing a deacetylase inhibitor cocktail (Beyotime, Shanghai) and sonicated in order to obtain a homogeneous solution that was centrifuged to remove cell debris. The protein concentration of each supernatant was determined using the Bradford reagent (Catalogue No. 500-0006; Bio-Rad) and adjusted to 500 μg/mL for further uses. In the acetylation assay, 5 μg of purified six-histidine-tagged BldKB (His6-BldKB) and 1 μg of the crude extract of either SL/pWHM3-ermE or SL/pWHM3-ermE-sco0988 were mixed together with 20 μM of acetyl-CoA in HAT buffer (50 mM Tris-HCl pH 8.0, 10% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol) and incubated at 30°C for 3 h. Control was carried out in parallel with the reaction mix containing BldKB alone in presence of AcetylCoA.
At the end of the incubation period, His6-BldKB present in the reaction mixture was purified using Ni-NTA column following the manufacturers’ instructions. The collected fractions were separated on 12% SDS-PAGE and transferred on a polyvinylidene fluoride (PVDF) membrane that was incubated with antibodies against acetylated lysine (Cat. No. ICP0380, Runzekang, Beijing). Visualization of the acetylated protein was achieved by the addition of secondary antibody (HRP Goat Anti-Rabbit IgG) and a solution containing 3,3′-Diaminobenzidine (DAB) (Cat. No. PK10005, Proteintech) the substrate of HRP as described in Kim et al. (2013).
SCO0988 belongs to the Gcn5-related N-acetyltransferase (GNAT) (Pfam00583) superfamily whose carboxyl terminal end catalyzes the transfer of the acetyl moiety of acetyl-CoA to the ε-amino group of a lysine residue (Vetting et al., 2005). The Kyte and Doolite hydropathic profile (Kyte and Doolittle, 1982) of this protein suggested that it is a membrane protein with multiple trans membrane segments (Supplementary Figure S1). The level of expression of sco0988 was shown to increase with time indicating that it might regulate late developmental processes such as morphological differentiation and secondary/specialized metabolism (Supplementary Figure S2) (Zhang et al., 2020).
In order to determine whether this acetyltransferase has an impact on these processes in the strong antibiotic producer, SC, a sco0988 disruptive mutant of SC, SC-Δsco0988 was constructed. The phenotypes of the mutant, and of the wt strain are shown in Figure 1. The sco0988 deletion mutant had a slightly slower growth rate than the original strain (Supplementary Figure S3) and was characterized by a strong inhibition of both antibiotic production and sporulation (Figure 1A). Quantitative analysis demonstrated a 2/3 reduction of RED production and a complete abolition of ACT biosynthesis in SC-Δsco0988 compared to the original strain (p < 0.05) (Figure 1B). The complementation of the SC-Δsco0988 by sco0998 carried by the high copy number plasmid pWHM3 (Bibb et al., 1985) restored growth (Supplementary Figure S3) as well as RED and ACT production to the level of the original strain whereas sporulation was enhanced compared to that of the wt strain (Figures 1A,B).
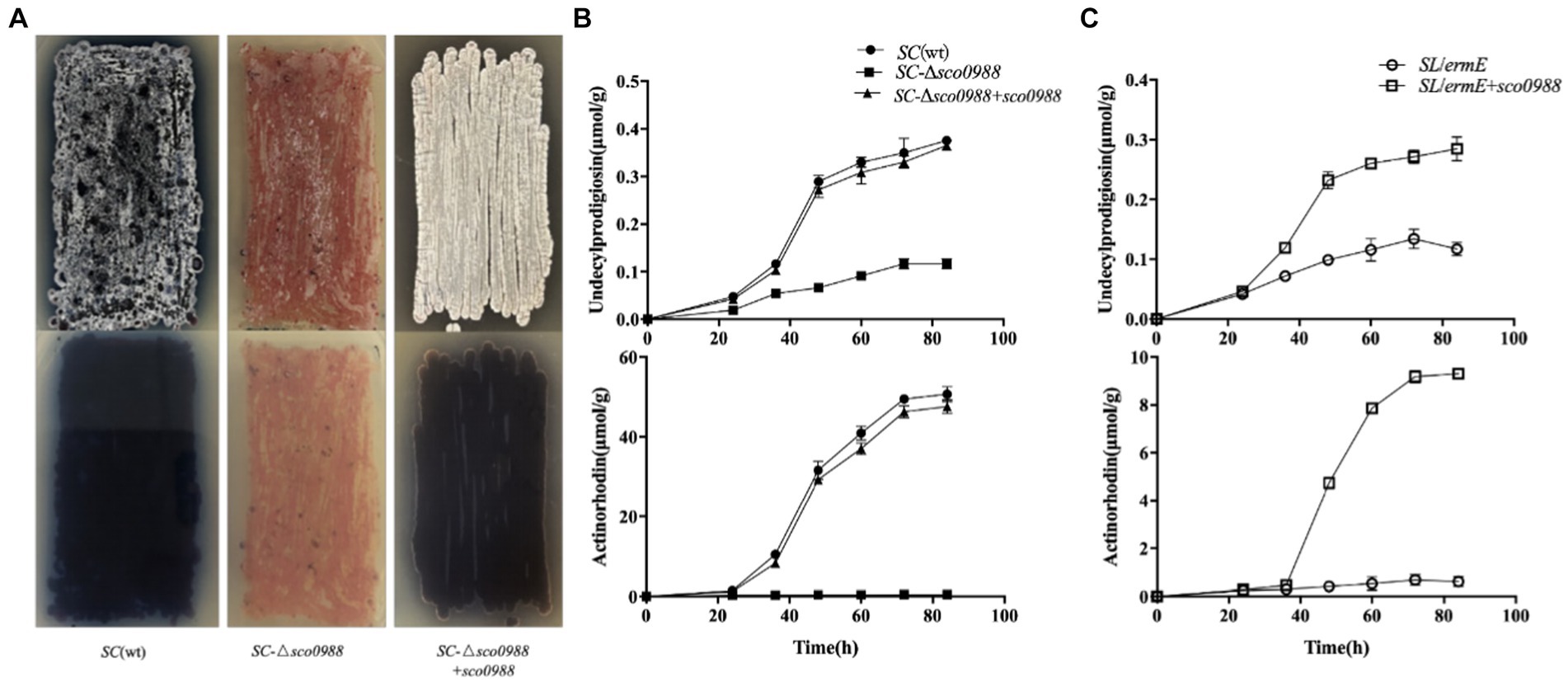
Figure 1. Phenotype resulting from the deletion of sco0988 in S. coelicolor M145 and from the over-expression of sco0988 in S. lividans. (A) Front/back pictures of lawns of the original strain S. coelicolor M145, of the mutant deleted for sco0988 and of this mutant complemented by sco0988 carried the plasmid pWHM3-ermE-sco0988. The strains were grown on solid R2YE medium for 48 h. (B) Quantitative analysis of RED and ACT production of the three S. coelicolor strains mentioned above. (C) Quantitative analysis of RED and ACT production of S. lividans TK24 and of this strain transformed by the plasmid pWHM3-ermE-sco0988. For (B,C) the strains were grown in liquid R2YE medium for 72 h at 28°C under constant shaking of 220 rpm.
In order to determine whether this acetyltransferase has an impact on these processes in the weak antibiotic producer, SL, sco0988 carried by pWHM3 was over-expressed in this strain. This over-expression led to a two-fold increase of RED production and the originally undetectable ACT production became now detectable (Figure 1C). The over-expression of sco0988 had no impact on the growth rate of the strain that was similar to that of the strain containing the empty plasmid (data not shown). These results clearly demonstrated that SCO0988 has a positive impact on both antibiotic production and morphological differentiation in these two model Streptomyces species.
In order to gain an insight into the biological processes affected by the over-expression of sco0988 in S. lividans TK24, a comparative analysis of the acetylome of the control strain (SL/pWHM3-ermE) and of the strain overexpressing sco0988 (SL/pWHM3-ermE-sco0988) was carried out. Acetylated lysine (acK) peptides were purified by immunoaffinity-based acetyl-lysine peptide enrichment, identified and quantified in both strains by high resolution mass spectrometry. An overview of the experimental procedures used is provided in Figure 2A.
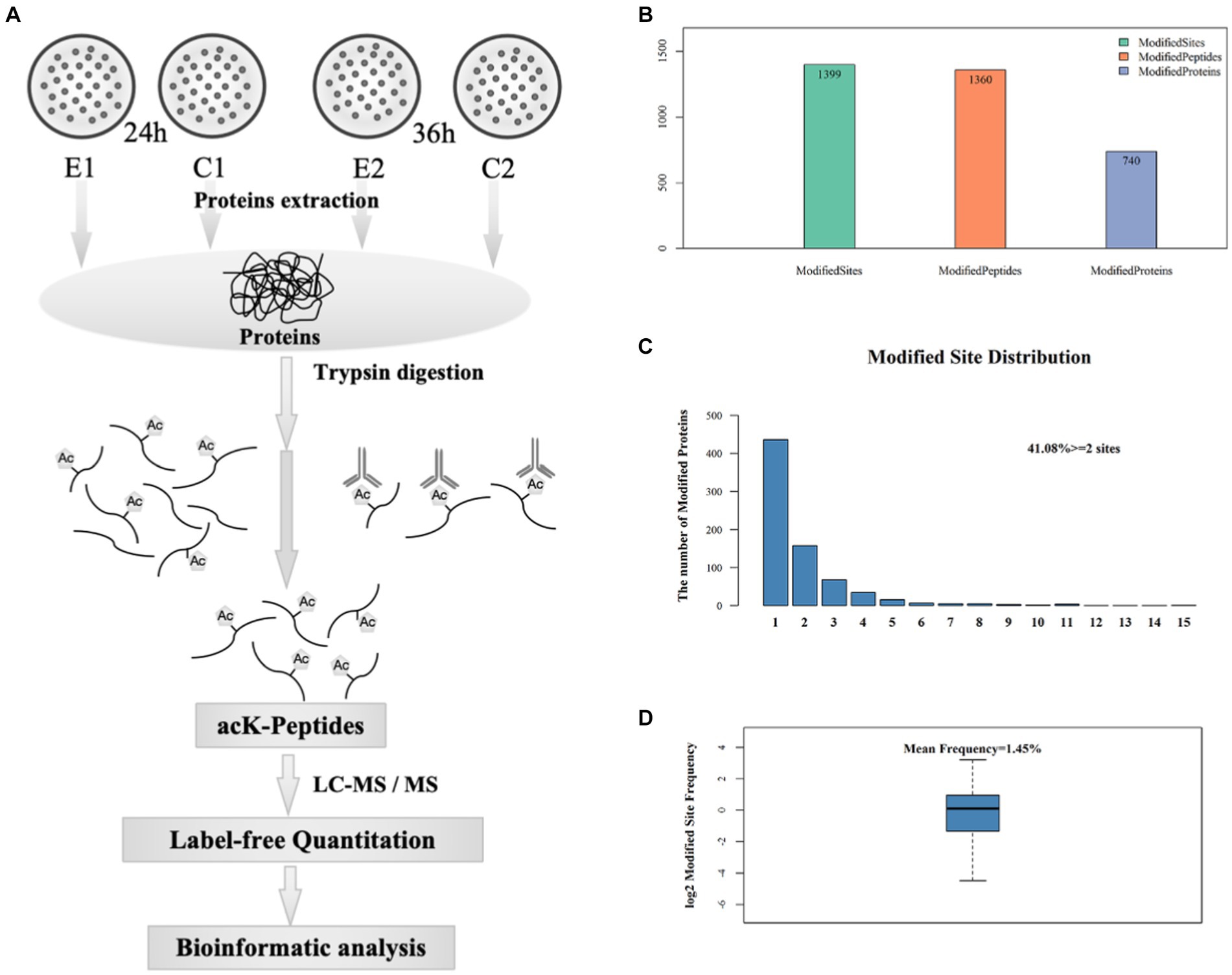
Figure 2. Preparation and analysis of acetylated peptides. (A) Workflow used to purify acetylated peptides. (B) Acetylome analysis: number of sites, peptides and proteins with acetyl modifications. (C) Distribution of the number of acetylated proteins bearing 1 to 15 acetylated sites. (D) Occurrence frequency of lysine acetylated sites.
A total of 1,399 acetylated lysine sites were discovered in 740 proteins (Figure 2B) that represented approximately 10% of S. lividans TK24 proteome (Rückert et al., 2015). 59% of these proteins, contained a single acetylated site, 21% had two acetylated sites and 9% contained three acetylated sites (Figure 2C). The average frequency of lysine acetylation was of 1.45 acetylated residue in 100 amino acids (Figure 2D). Detailed information concerning all acetylated peptides and the corresponding proteins is provided in Supplementary Table S1. A total of 643 statistically differentially acetylated peptides (p < 0.05) at 24 h and 36 h is shown in Supplementary Table S2. Considering that 54 of them were present at the two time points, the amount of the acetylated peptides is thus 589 (643-54). Among these 589 acetylated peptides, 177 (30.05%) were more acetylated while 358 (60.78%) were less acetylated in the strain overexpressing sco0988 than in the control strain. These observations suggested that in SL the pool of acetylCoA is limited and that the over-expression of sco0988 consumes acetylCoA and thus limits the acetylation of the specific targets of other acetyltransferases. Interestingly, 92 acetylated peptides were found exclusively present in the strain over-expressing sco0988 whereas 159 acetylated peptides were only present in the control strain (Supplementary Tables S1, S2). The proteins over-acetylated or exclusively acetylated in SL/pWHM3-ermE-sco0988 can thus be considered as potential specific SCO0988 targets.
Gene Ontology (GO) terms and KEGG pathway enrichment analysis of differentially acetylated proteins (DAPs) was performed with Applied Protein Technology Co., Ltd (Shanghai, China). The ontological classifications, determined by Blast2Go (see text footnote 2) and Interpro scan,4 are shown in Figure 3. A large proportion of DAPs (>60%) were classified as involved in nitrogen (19.6%) and carbon (14.93%) metabolisms, transcriptional and translational processes (18.97%) as well as transport (12.44%).
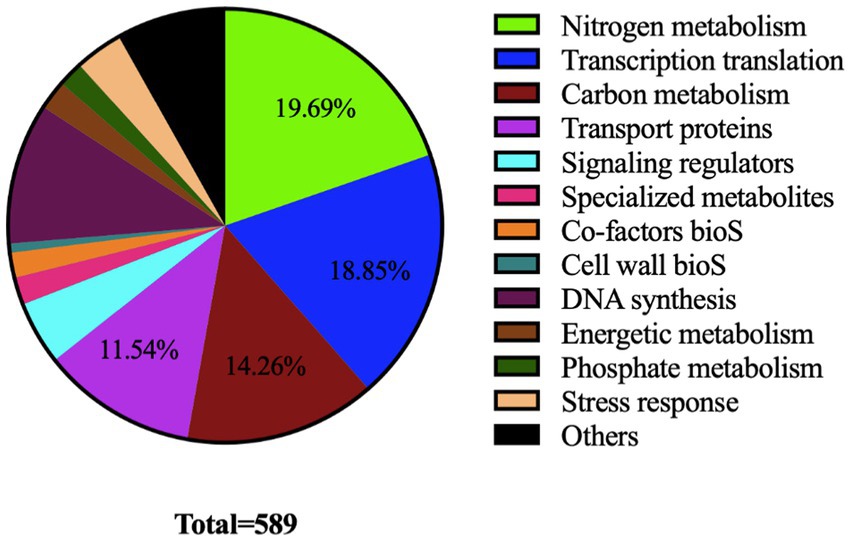
Figure 3. Pie chart showing ontological classification of proteins acetylated by the acetyltransferase SCO0988 in S. lividans.
In order to identify a possible consensus acetylation motif of SCO0988, the frequency of amino acids present from position −5 to +5 around the 92 lysines exclusively acetylated in the strain over-expressing SCO0988 were determined using the MEME program, and similarly, the frequency of amino acids presents from position −5 to +5 around 159 lysines exclusively acetylated in the control strain were determined using the MEME program. The results shown in Figure 4 demonstrated that, as expected, the consensus of the sites acetylated by SCO0988 was different and also far less diverse than the consensus of the other acetylated sites that are likely to be acetylated by multiple acetyltransferases other than SCO0988. In the latter the central pair KK is highly conserved but is replaced by KR in the SCO0988 consensus. K and R are both basic amino acids. In the SCO0988 consensus the positions 2, 3, 5 and 6 are highly conserved and are P, D, K and R, respectively and in the other positions only 2 alternative amino acids are present.
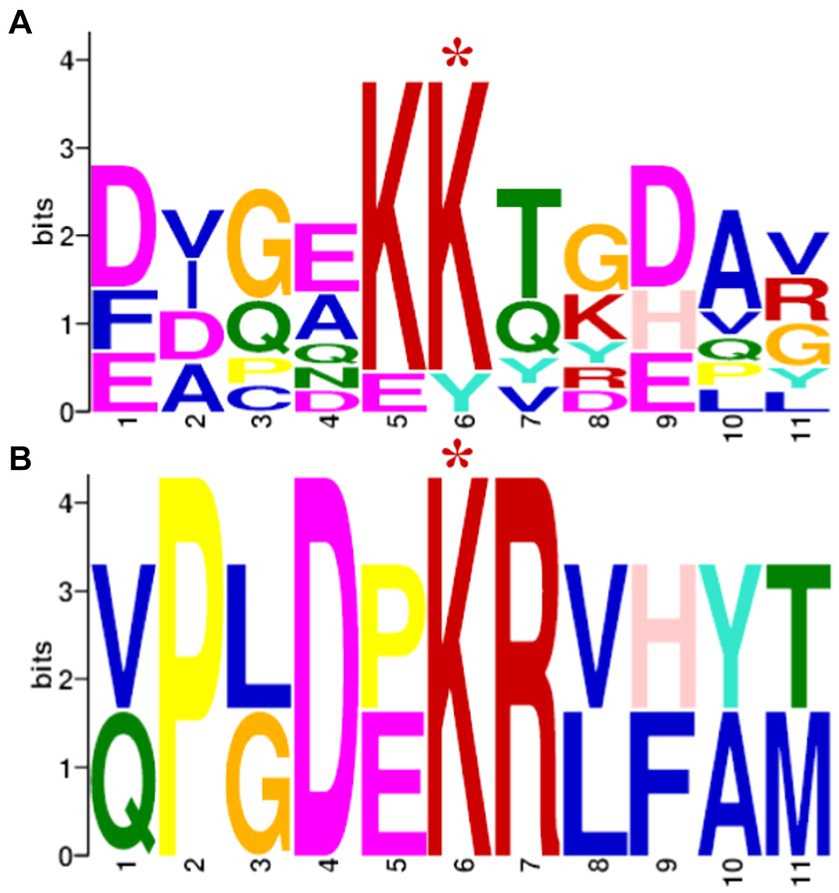
Figure 4. Probable sequence motifs of acetylation sites identified using the MEME software. These sites consist of 5 residues present on each side of the modified lysine residue signaled by a red asterisk. (A) Sequence motifs of the 159 acetylated lysine sites identified exclusively in the control strain. (B) Sequence motifs of the 92 acetylated lysine sites identified exclusively in the S. lividans strain over-expressing SCO0988.
BldKB/SCO5113 is the extracellular solute-binding protein of the ABC transporter BldKABCDE (SCO5112-SCO5116) also constituted of BldKA and BldKC (SCO5112 and SCO5114) two integral membrane proteins, BldKD/SCO5115/intracellular ATPase subunit and BldKE/SCO5116/ATP-binding protein. This ABC transporter is involved in the up-take of the BLD261 oligopeptide that is a signaling molecule of the quorum sensing pathway (Willey et al., 1993). The up-take of this signaling molecule by the BldK transporter triggers the expression of genes of the “bald” signaling cascade that controls positively antibiotic production as well as aerial mycelium formation and sporulation in SC (Nodwell et al., 1999). Since, 4 of the 7 acetylated lysine (acK) peptides detected in BldKB/SCO5113 showed a great increase in their acetylation level upon sco0988 overexpression, with the Lys425 showing the largest increase (Supplementary Table S2), BldKB was selected for further analysis.
In order to determine whether SCO0988 was able to acetylate BldKB in vitro, recombinant His6-BldKB was expressed in E. coli and purified (Figure 5A). Since the membrane protein SCO0988 formed inclusion bodies when E. coli/pET expression system was used (data not shown), cell crude extracts from S. lividans over-expressing sco0988 were prepared for the acetylation assay. Western blot probed with an antibody against acetylated lysine clearly demonstrated an intensified band corresponding to acetylated BldKB in the reaction carried out in the presence of crude extract of SL/pWHM3-ermE-sco0988 (Figure 5B) but not in that carried out in the presence of crude extract of SL/pHWM3-ermE (Figure 5B). No acetylation was observed in the control assays with BldKB alone in presence of AcetylCoA (data not shown). These data thus demonstrate the acetylation of BldKB by SCO0988.
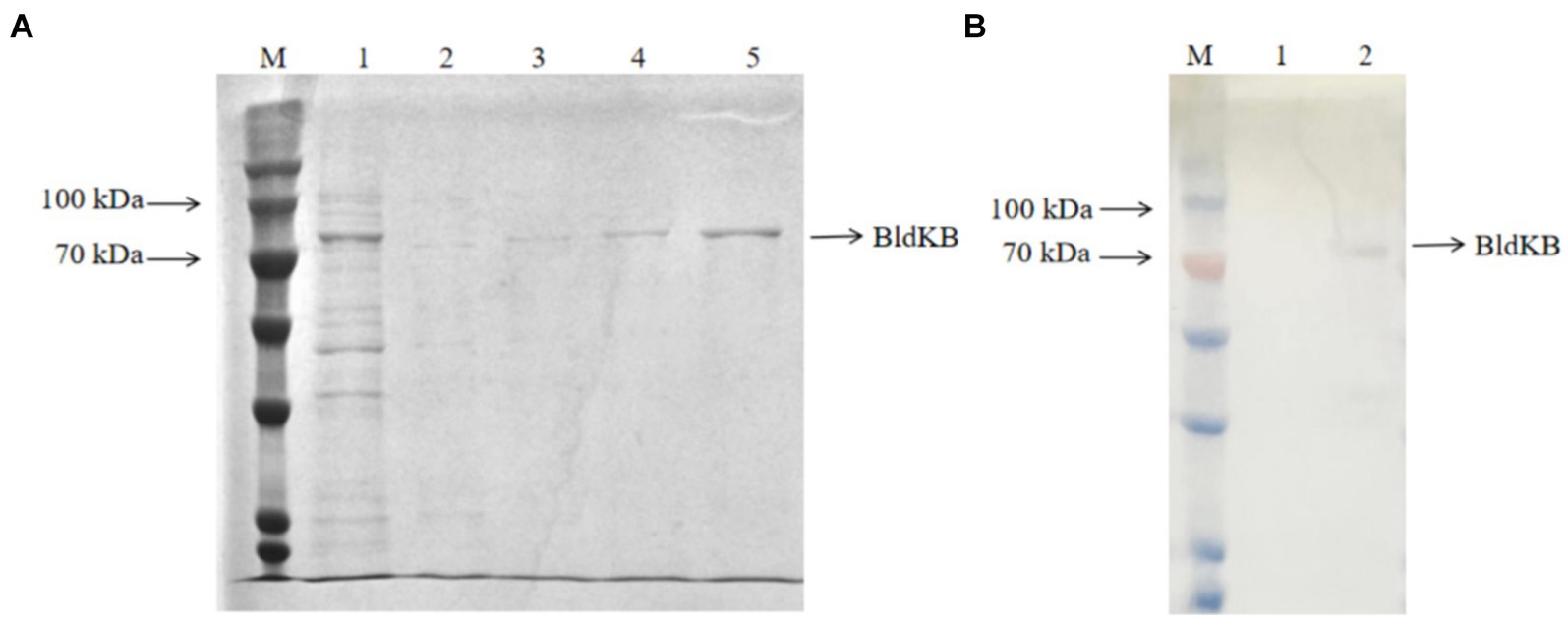
Figure 5. In vitro acetylation assay of BldKB. (A) SDS-PAGE analysis of His6-BldKB purified using E. coli/pET expression system. Lane M: marker (Cat. No. PMK06-01, Biocomma); lane 1: flow through; lanes 2–3: elution with 100 mM imidazone; lanes 4–5: elution with 250 mM imidazone. (B) Western blot analysis of BldKB acetylation in presence of cell crude extracts of S. lividans/pWHM3 (lane 1) and S. lividans/pWHM3-ermE-SCO0988 (lane 2) using antibodies against acetylated lysine.
According to NCBI’s Conserved Domain Database (CDD), four structural domains could be identified in BldKB: a periplasmic component DdpA-like (57–592), an oligopeptide binding domain OppA2-like (75–582), an extracellular solute-binding domain SBP_bac_5-like (120–516) and a substrate-binding domain PRK09755 (356–599) (Figure 6A).5 All four domains are critical for the BldKB function. Seven acetylated lysine sites were detected in BldKB and among them four (K/248, K/337, K/341 and K/425) were over-acetylated upon sco0988 over-expression (Supplementary Tables S1, S2 and Figure 6). Interestingly K425, the most sensitively acetylated Lys upon SCO0988 over-expression, is present in the four domains detected in BldKB (Figure 6). In order to determine whether K425 was important the function and/or the acetylation of BldKB, a SC strain disrupted for bldKB, SC-ΔbldKB, was first constructed. SC-ΔbldKB had a similar growth pattern as the original strain (Supplementary Figure S4) but its RED and ACT production levels were reduced, being 32.03 and 34.53% lower, respectively, than that of the original strain, at 72 h (Figures 7A,B). In contrast, morphological differentiation and sporulation seemed totally abolished in SC-ΔbldKB. These observations are consistent with those reported in previous studies (Nodwell et al., 1996; Nodwell and Losick, 1998; Park et al., 2005). Subsequently, variants of BldKB in which Lys425 was substituted by Arg/R or Gln/Q were constructed. Arg/R was chosen since it cannot be acetylated and Gln/Q was chosen since many reports in the literature mentioned that it mimics acetylated Lys (Kamieniarz and Schneider, 2009; Okada et al., 2021; Morse et al., 2023). When bldKB (K425R) and bldKB (K425Q) were introduced into SC-ΔbldKB, complementation did not occur since ACT and RED production as well as sporulation were not restored to wild type level (Figures 7A,B). In contrast, these features were restored to wild type level when the native bldKB gene was introduced into SC-ΔbldKB. These results indicated that Lys425 is crucial for BldKB function and that the replacement of Lys425 by Arg or Gln greatly alters BldKB function. Unfortunately, these results were inconclusive concerning the impact of acetylation on BldKB function (see discussion).
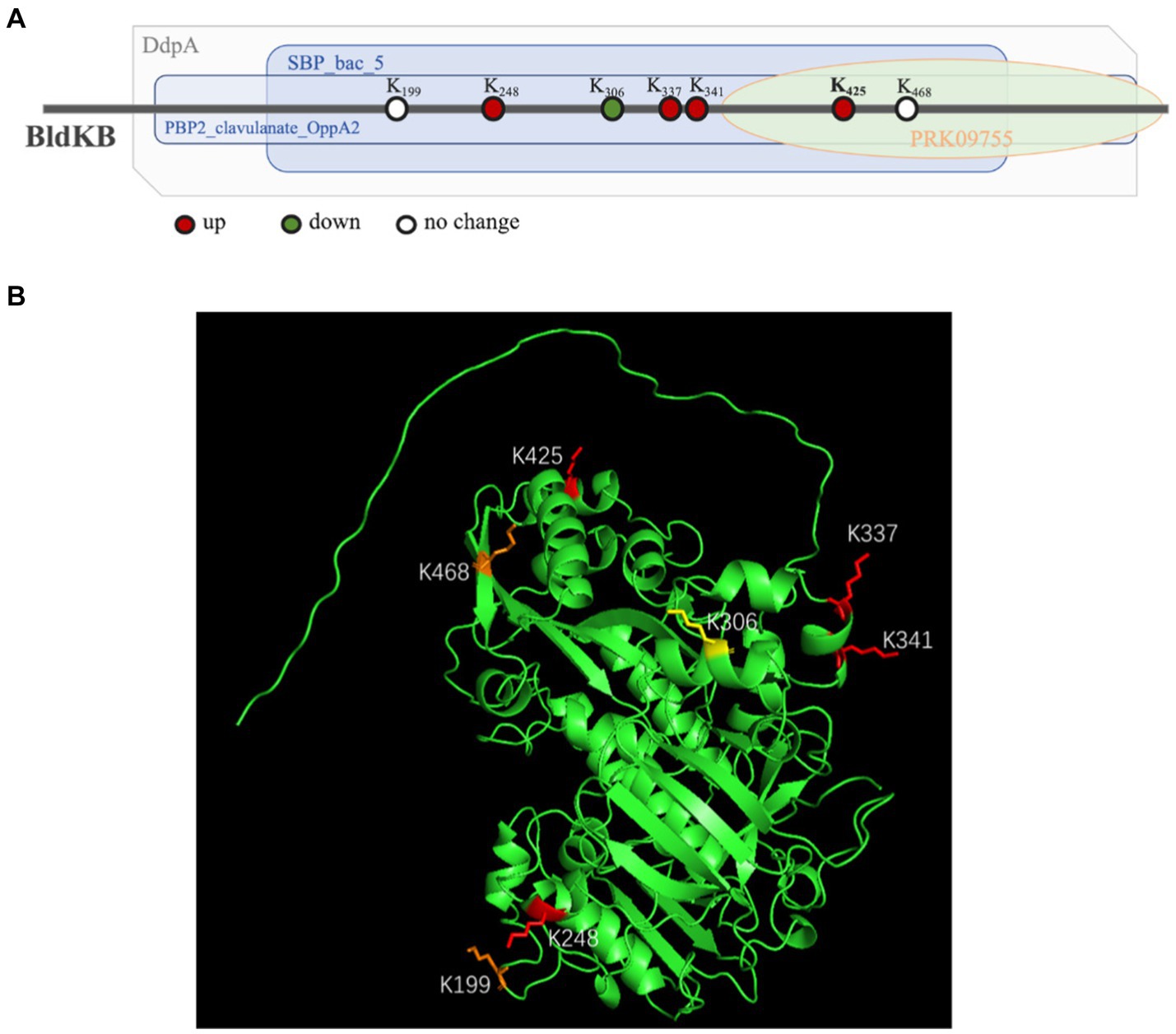
Figure 6. Structure of the BldKB protein. (A) Mapping of the four domains identified by InterProScan in the BldkB protein and positioning of the acetylated lysines (K). (B) Positions of the seven acetylated lysines of BldKB (K199, K248, K337, K341, K425, K468 and K306) on its three-dimensional structure predicted by AlphaFold and PyMol. The four Lys over-acetylated (K248, K337, K341 and K425), the two Lys with un-changed level of acetylation (K199 and K468) and the Lys under-acetylated (K306), in the strain SL/pWHM3-ermE-sco0988, are represented in red, orange and yellow, respectively.
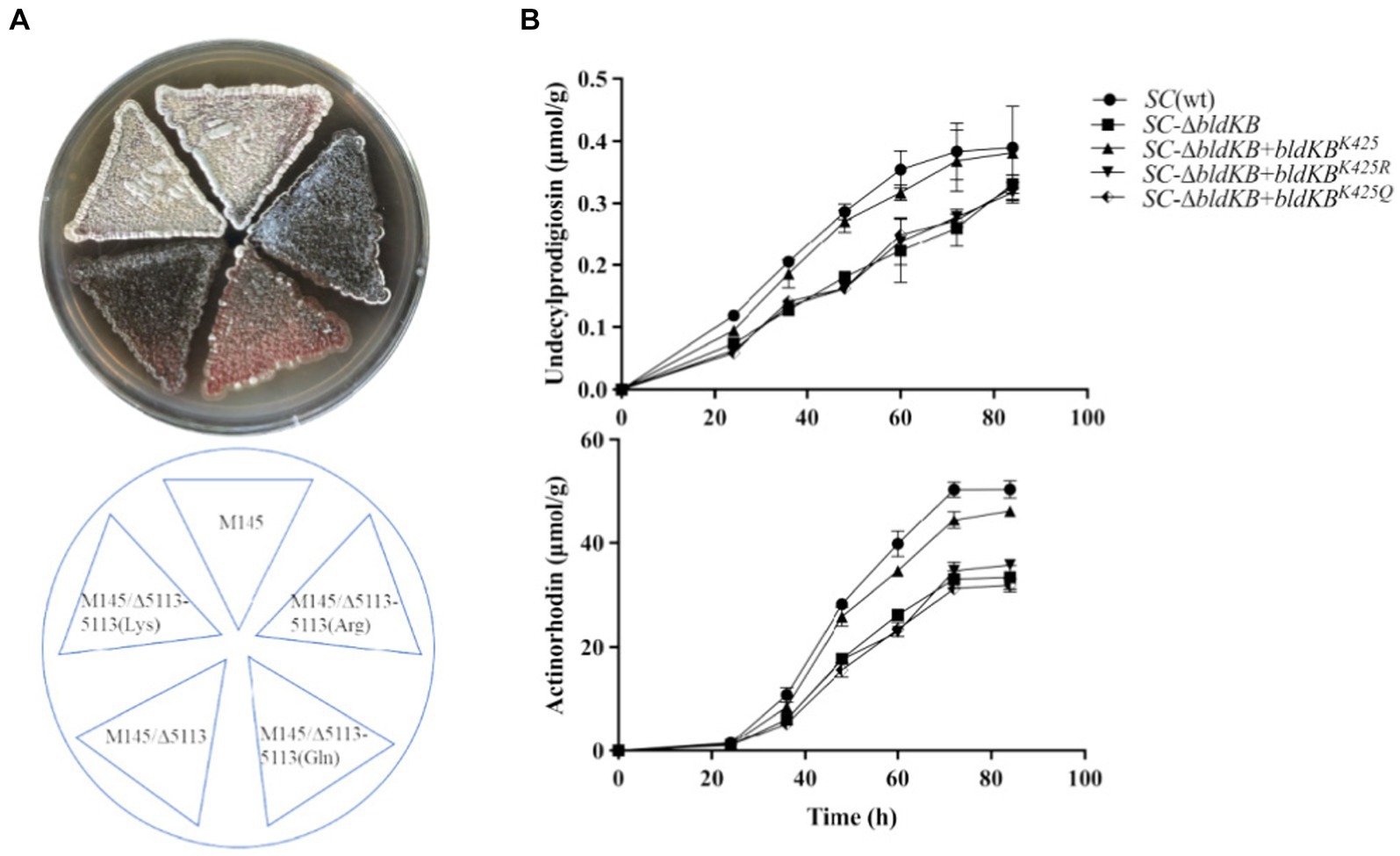
Figure 7. Phenotypes of the genetically modified strains under study. (A) Front and back pictures of lawns of S. coelicolor M145, of the bldKB deleted strain (SC-ΔbldKB) and of this mutant strain complemented by bldKBK425 (AAG/Lys), bldKBK425R (AGG/Arg) or bldKBK425Q (CAG/Gln). The strains were grown on solid R2YE medium for 72 h. (B) Quantitative analysis of RED and ACT productions by the various strains grown in liquid R2YE medium.
In bacteria Lys acetylation in proteins can result from two different processes: a chemical process using the high energy metabolites acetyl phosphate and acetylCoA as acetyl donor and an enzymatic process resulting from a catalytic reaction between an acetyltransferase, an acetyl donor (usually acetylCoA), and a lysine amino acid acceptor present in a protein substrate. Interestingly, Interestingly, non-enzymatic and acetyltransferase-dependent acetylation sites are usually different, suggesting that these two acetylation mechanisms play distinct roles in the post-translational modifications of bacterial proteins (Christensen et al., 2018, 2019). The “regulatory importance” of enzymatic protein acetylation has been demonstrated in many microbial processes including primary and specialized metabolisms, DNA replication, chemotaxis, virulence etc.… (Galdieri et al., 2014; Menzies et al., 2016) whereas a report suggested that AcP-driven acetylation have little functional consequences at least in E. coli (Schastnaya et al., 2023).
It is noteworthy that the impact of protein acetylation on the activity of target proteins remains largely unknown in Actinobacteria of the Streptomyces genus that are well known producers of bio-active specialized metabolites of great economic importance. Still, some proteins directly involved in the biosynthesis of bio-active specialized metabolites have been shown to be acetylated in Streptomyces roseosporus (Liao et al., 2014) and Streptomyces griseus (Ishigaki et al., 2017). The acetylation, on Lys70, of the deoxysugar epimerase StrM of the streptomycin biosynthetic pathway of S. griseus, lowers its activity and this results into a reduced streptomycin production (Ishigaki et al., 2017). Furthermore, GlnR, the central response regulator of nitrogen assimilation that governs the expression of numerous genes involved into N-assimilation in most Actinomycetes species (Amin et al., 2016), activates the expression of the Gcn5-type lysine acetyltransferase AcuA and of the NAD+ dependent deacetylase SrtN in Saccharopolyspora erythraea (You et al., 2017). These enzymes are involved in the acetylation of 3 AMP-forming acetyl-CoA synthetases and has a negative impact on their activity (You et al., 2016). The activity of an acetoacetyl-CoA synthetase (Martín et al., 2021) was also shown to be controlled negatively by O-serine and Nε-lysine acetylation in SL (VanDrisse and Escalante-Semerena, 2018). In SC, the function of the regulator GlnR/SCO4159, is modulated by post-translational modifications including phosphorylation of Ser/Thr residues and acetylation of Lys residues. GlnR was shown to be acetylated on 4 Lys residues when SC is grown in defined a media and this acetylation was independent of N-concentration. In contrast, GlnR was acetylated on a single Lys residue upon growth in a complex N-rich medium. However acetylation seems to have little influence on the formation of GlnR-DNA complex whereas phosphorylation inhibits the binding of GlnR to its targets genes (Sun et al., 2020a).
In our study, the comparative analysis of the acetylomes of the native strain of S. lividans and of the S. lividans strain overexpressing the acetyltransferase SCO0988 revealed a total of 1,399 acetylation sites in 740 proteins. Interestingly, among these 1,399 acetylated peptides detected, 589 (42.10%) are showing significant differences in their acetylation level between the two strains (ratio +/− 2 with p < 0.05) (Supplementary Table S2). Our study revealed that 92 proteins were exclusively acetylated and 118 were over-acetylated in the strain over-expressing SCO0988. These proteins can thus be considered as specific SCO0988 targets. We focused our study on one of the proteins extensively over-acetylated in the strain over-expressing SCO0988. This protein is the extracellular solute-binding protein BldKB of the oligopeptide transporter (BldKABCDE/SCO5112-SCO5116). This protein was chosen since it was shown to be acetylated on 7 different Lys residues. Four of these Lys (K248, K337, K341 and K425) were over-acetylated, in the strain over-expressing sco0988 whereas the acetylation level of K199 and K468 was unchanged and that of K306 was reduced (Figure 6). Lys are positively charged residues and their acetylation that neutralize these positive charges might be important for the BldKB function. Indeed, the BLD261 oligopeptide transported by the BldK ABC transporter is likely to be positively charged as its NH2+ groups could be ionized into NH3+. Since charges of the same sign repel each other, the similar electrostatic charges of these two “partners” might prevent their interaction whereas the neutralization of the positive charge of Lys by acetylation would allow this interaction. Considering that it was the acetylation level of K425 that showed the greatest increase in the SCO0988 over-expressing strain, we decided to replace K425 by an Arg/R or a Gln/Q residue, in order to determine whether these substitutions had an impact on the function and/or on the acetylation of BldKB. These residues were chosen since R cannot be acetylated whereas Q is expected to mimic acetylated K (Kamieniarz and Schneider, 2009; Okada et al., 2021; Morse et al., 2023). The native BldKB as well as the mutated versions of BldKB, BldKBK425R and BldKBK425Q, were used to complement the bldkB deletion mutant of SC. Our results revealed that only the native gene was able to restore the phenotype of the original strain indicating that Lys425 was critical for BldKB function and that its replacement by Arg or Gln deeply alters BldKB function. These replacements might alter the functionality, the stability or the proper folding of the protein. Alternatively, one cannot exclude that the replacement of K425 by Q does not effectively mimic acetylated K425, in our specific context.
In conclusion, we wish to stress that our study demonstrated that acetylation could constitute a valuable tool to enhance the expression of already known or of cryptic biosynthetic pathways of specialized metabolites present in the Streptomyces genomes in order to discover most needed novel antibiotics to face the worrying emergence and rapid spreading of antibiotic-resistant pathogens. However, considering the multiplicity of SCO0988 targets, including 10 proteins annotated as being involved in signalization and regulation, that are exclusively acetylated in the strain over-expressing SCO0988 (Supplementary Table S2), the lack of acetylation of these regulatory proteins and possibly that of other proteins, besides BldKB, might contribute to the reduction of antibiotic production and to the inhibition of morphological differentiation and sporulation of the sco0988 deletion mutant of SC.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
YB: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft. HA: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. ZC: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft. ZX: Data curation, Formal analysis, Investigation, Software, Writing – original draft. YD: Data curation, Investigation, Methodology, Writing – original draft. YR: Formal analysis, Investigation, Methodology, Writing – original draft. RW: Formal analysis, Investigation, Methodology, Writing – original draft. XL: Data curation, Investigation, Methodology, Writing – original draft. JG: Conceptualization, Data curation, Formal analysis, Methodology, Resources, Software, Validation, Writing – review & editing. RH: Conceptualization, Data curation, Formal analysis, Methodology, Software, Supervision, Validation, Writing – review & editing. M-JV: Conceptualization, Data curation, Formal analysis, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. DX: Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (No. 2022YFE0122100), the National Natural Science Foundation of China (Nos. 32271568 and 42030713), the Natural Science Foundation of Guangdong Province (No. 2023A1515012806), Key Program of Marine Economy Development (Six Marine Industries), Special Foundation of Department of Natural Resources of Guangdong Province (No. GDNRC [2021]37).
The authors thank the Shanghai Applied Protein Technology Co., Ltd. for technology support in quantitative acetylome analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1366336/full#supplementary-material
1. ^https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD041540
3. ^http://meme-suite.org/index.htm
4. ^http://www.ebi.ac.uk/InterProScan/
5. ^https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?INPUT_TYPE=precalc&SEQUENCE=1532202
Amin, R., Franz-Wachtel, M., Tiffert, Y., Heberer, M., Meky, M., Ahmed, Y., et al. (2016). Post-translational serine/threonine phosphorylation and lysine acetylation: a novel regulatory aspect of the global nitrogen response regulator GlnR in S. coelicolor M145. Front. Mol. Biosci. 3:38. doi: 10.3389/fmolb.2016.00038
Barkovits, K., Pacharra, S., Pfeiffer, K., Steinbach, S., Eisenacher, M., Marcus, K., et al. (2020). Reproducibility, specificity and accuracy of relative quantification using spectral library-based data-independent acquisition. Mol. Cell. Proteomics 19, 181–197. doi: 10.1074/mcp.RA119.001714
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. doi: 10.1038/417141a
Bibb, M. J., Janssen, G. R., and Ward, J. M. (1985). Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene 41, e357–e368. doi: 10.1016/0378-1119(86)90122-8
Chater, K. F. (2016). Recent advances in understanding Streptomyces. F1000Research 5:2795. doi: 10.12688/f1000research.9534.1
Chen, Z. L., Meng, J. M., Cao, Y., Yin, J. L., Fang, R. Q., Fan, S. B., et al. (2019). A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 10:3404. doi: 10.1038/s41467-019-11337-z
Christensen, D. G., Meyer, J. G., Baumgartner, J. T., D'Souza, A. K., Nelson, W. C., Payne, S. H., et al. (2018). Identification of novel protein lysine acetyltransferases in Escherichia coli. mBio 9:e01905. doi: 10.1128/mBio.01905-18
Christensen, D. G., Xie, X. S., Basisty, N., Byrnes, J., McSweeney, S., Schilling, B., et al. (2019). Post-translational protein acetylation: an elegant mechanism for bacteria to dynamically regulate metabolic functions. Front. Microbiol. 10:1604. doi: 10.3389/fmicb.2019.01604
Cox, R. M., Sourimant, J., Toots, M., Yoon, J. J., Ikegame, S., Govindarajan, M., et al. (2020). Orally efficacious broad-spectrum allosteric inhibitor of paramyxovirus polymerase. Nat. Microbiol. 5, 1232–1246. doi: 10.1038/s41564-020-0752-7
Cruz-Bautista, R., Ruíz-Villafán, B., Romero-Rodríguez, A., Rodríguez-Sanoja, R., and Sánchez, S. (2023). Trends in the two-component system’s role in the synthesis of antibiotics by Streptomyces. Appl. Microbiol. Biotechnol. 107, 4727–4743. doi: 10.1007/s00253-023-12623-z
Feitelson, J. S., Malpartida, F., and Hopwood, D. A. (1985). Genetic and biochemical-characterization of the red gene-cluster of Streptomyces coelicolor A3(2). J. Gen. Microbiol. 131, 2431–2441. doi: 10.1099/00221287-131-9-2431
Flett, F., Mersinias, V., and Smith, C. P. (1997). High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155, 223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x
Galdieri, L., Zhang, T. T., Rogerson, D., Lleshi, R., and Vancura, A. (2014). Protein acetylation and acetyl coenzyme a metabolism in budding yeast. Eukaryot. Cell 13, 1472–1483. doi: 10.1128/EC.00189-14
Hamam, H. J., Khan, M. A., and Palaniyar, N. (2019). Histone acetylation promotes neutrophil extracellular trap formation. Biomol. Ther. 9:32. doi: 10.3390/biom9010032
Hannigan, G. D., Prihoda, D., Palicka, A., Soukup, J., Klempir, O., Rampula, L., et al. (2019). A deep learning genome-mining strategy for biosynthetic gene cluster prediction. Nucleic Acids Res. 47:e110. doi: 10.1093/nar/gkz654
Hesketh, A. R., Chandra, G., Shaw, A. D., Rowland, J. J., Kell, D. B., Bibb, M. J., et al. (2002). Primary and secondary metabolism, and post-translational protein modifications, as portrayed by proteomic analysis of Streptomyces coelicolor. Mol. Microbiol. 46, 917–932. doi: 10.1046/j.1365-2958.2002.03219.x
Ikeda, H., Shin-ya, K., and Omura, S. (2014). Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J. Ind. Microbiol. Biotechnol. 41, 233–250. doi: 10.1007/s10295-013-1327-x
Ishigaki, Y., Akanuma, G., Yoshida, M., Horinouchi, S., Kosono, S., and Ohnishi, Y. (2017). Protein acetylation involved in streptomycin biosynthesis in Streptomyces griseus. J. Proteome 155, 63–72. doi: 10.1016/j.jprot.2016.12.006
Kamieniarz, K., and Schneider, R. (2009). Tools to tackle protein acetylation. Chem. Biol. 16, 1027–1029. doi: 10.1016/j.chembiol.2009.10.002
Kieser, T., Bibb, M. J., Chater, K., and Hopwood, D. A. (2000). Practical Streptomyces genetics. Norwich: John Innes Foundation.
Kim, H., Park, S., and Lee, H. K. (2013). Application of bispecific antibody against antigen and hapten for immunodetection and immunopurification. Exp. Mol. Med. 45, 1–7. doi: 10.1038/emm.2013.83
Kyte, J., and Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. doi: 10.1016/0022-2836(82)90515-0
Lewis, K. (2020). The science of antibiotic discovery. Cell 181, 29–45. doi: 10.1016/j.cell.2020.02.056
Liao, G. J., Xie, L. X., Li, X., Cheng, Z. Y., and Xie, J. P. (2014). Unexpected extensive lysine acetylation in the trump-card antibiotic producer Streptomyces roseosporus revealed by proteome-wide profiling. J. Proteome 106, 260–269. doi: 10.1016/j.jprot.2014.04.017
Malpartida, F., and Hopwood, D. A. (1986). Physical and genetic-characterization of the gene-cluster for the antibiotic actinorhodin in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 205, 66–73. doi: 10.1007/BF02428033
Martín, J. F., and Liras, P. (2020). The balance metabolism safety net: integration of stress signals by interacting transcriptional factors in Streptomyces and related Actinobacteria. Front. Microbiol. 10:3120. doi: 10.3389/fmicb.2019.03120
Martín, J. F., Liras, P., and Sánchez, S. (2021). Modulation of gene expression in Actinobacteria by translational modification of transcriptional factors and secondary metabolite biosynthetic enzymes. Front. Microbiol. 12:630694. doi: 10.3389/fmicb.2021.630694
Menzies, K. J., Zhang, H. B., Katsyuba, E., and Auwerx, J. (2016). Protein acetylation in metabolism—metabolites and cofactors. Nat. Rev. Endocrinol. 12, 43–60. doi: 10.1038/nrendo.2015.181
Miroux, B., and Walker, J. E. (1996). Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298. doi: 10.1006/jmbi.1996.0399
Morse, P. T., Pérez-Mejías, G., Wan, J. M., Turner, A. A., Márquez, I., Kalpage, H. A., et al. (2023). Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle. Nat. Commun. 14:4166. doi: 10.1038/s41467-023-39820-8
Nativio, R., Lan, Y. M., Donahue, G., Sidoli, S., Berson, A., Srinivasan, A. R., et al. (2020). An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat. Genet. 52, 1024–1035. doi: 10.1038/s41588-020-0696-0
Niu, G. Q., Chater, K. F., Tian, Y. Q., Zhang, J. H., and Tan, H. R. (2016). Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 40, 554–573. doi: 10.1093/femsre/fuw012
Nodwell, J. R., and Losick, R. (1998). Purification of an extracellular signaling molecule involved in production of aerial mycelium by Streptomyces coelicolor. J. Bacteriol. 180, 1334–1337. doi: 10.1128/JB.180.5.1334-1337.1998
Nodwell, J. R., McGovern, K., and Losick, R. (1996). An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22, 881–893. doi: 10.1046/j.1365-2958.1996.01540.x
Nodwell, J. R., Yang, R., Kuo, D., and Losick, R. (1999). Extracellular complementation and the identification of additional genes involved in aerial mycelium formation in Streptomyces coelicolor. Genetics 151, 569–584. doi: 10.1093/genetics/151.2.569
Ochi, K. (2017). Insights into microbial cryptic gene activation and strain improvement: principle, application and technical aspects. J. Antibiot. 70, 25–40. doi: 10.1038/ja.2016.82
Okada, A. K., Teranishi, K., Ambroso, M. R., Isas, J. M., Vazquez-Sarandeses, E., Lee, J. Y., et al. (2021). Lysine acetylation regulates the interaction between proteins and membranes. Nat. Commun. 12:6466. doi: 10.1038/s41467-021-26657-2
Park, H. S., Shin, S. K., Yang, Y. Y., Kwon, H. J., and Suh, J. W. (2005). Accumulation of S-adenosylmethionine induced oligopeptide transporters including BldK to regulate differentiation events in Streptomyces coelicolor M145. FEMS Microbiol. Lett. 249, 199–206. doi: 10.1016/j.femsle.2005.05.047
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120. doi: 10.1093/nar/gki442
Rückert, C., Albersmeier, A., Busche, T., Jaenicke, S., Winkler, A., Friojónsson, O. H., et al. (2015). Complete genome sequence of Streptomyces lividans TK24. J. Biotechnol. 199, 21–22. doi: 10.1016/j.jbiotec.2015.02.004
Schastnaya, E., Doubleday, P. F., Maurer, L., and Sauer, U. (2023). Non-enzymatic acetylation inhibits glycolytic enzymes in Escherichia coli. Cell Rep. 42:111950. doi: 10.1016/j.celrep.2022.111950
Schifferli, D. M. (1995). Use of TnphoA and T7 RNA polymerase to study fimbrial proteins. Methods Enzymol. 253, 242–258. doi: 10.1016/S0076-6879(95)53023-1
Shan, G. G., and Gerstenberger, S. (2017). Fisher’s exact approach for post hoc analysis of a chi-squared test. PLoS One 12:e0188709. doi: 10.1371/journal.pone.0188709
Sun, C. F., Li, Y. Q., and Mao, X. M. (2020a). Regulation of protein post-translational modifications on metabolism of actinomycetes. Biomol. Ther. 10:1122. doi: 10.3390/biom10081122
Sun, C. F., Xu, W. F., Zhao, Q. W., Luo, S., Chen, X. A., Li, Y. Q., et al. (2020b). Crotonylation of key metabolic enzymes regulates carbon catabolite repression in Streptomyces roseosporus. Commun. Biol. 3:192. doi: 10.1038/s42003-020-0924-2
VanDrisse, C. M., and Escalante-Semerena, J. C. (2018). In Streptomyces lividans, acetyl-CoA synthetase activity is controlled by O-serine and N-lysine acetylation. Mol. Microbiol. 107, 577–594. doi: 10.1111/mmi.13901
Vetting, M. W., de Carvalho, L. P. S., Yu, M., Hegde, S. S., Magnet, S., Roderick, S. L., et al. (2005). Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433, 212–226. doi: 10.1016/j.abb.2004.09.003
Willey, J., Schwedock, J., and Losick, R. (1993). Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7, 895–903. doi: 10.1101/gad.7.5.895
Wright, D. P., and Ulijasz, A. T. (2014). Regulation of transcription by eukaryotic-like serine-threonine kinases and phosphatases in Gram-positive bacterial pathogens. Virulence 5, 863–885. doi: 10.4161/21505594.2014.983404
Xu, D. L., Seghezzi, N., Esnault, C., and Virolle, M. J. (2010). Repression of antibiotic production and sporulation in Streptomyces coelicolor by overexpression of a TetR family transcriptional regulator. Appl. Environ. Microbiol. 76, 7741–7753. doi: 10.1128/AEM.00819-10
Yang, Y. J., Zhang, H., Guo, Z. Y., Zou, S. W., Long, F., Wu, J. C., et al. (2021). Global insights into lysine acylomes reveal crosstalk between lysine acetylation and succinylation in Streptomyces coelicolor metabolic pathways. Mol. Cell. Proteomics 20:100148. doi: 10.1016/j.mcpro.2021.100148
You, Z. Y., Jiang, W. X., Qin, L. Y., Gong, Z., Wan, W., Li, J., et al. (2019). Requirement for p62 acetylation in the aggregation of ubiquitylated proteins under nutrient stress. Nat. Commun. 10:5792. doi: 10.1038/s41467-019-13718-w
You, D., Wang, M. M., and Ye, B. C. (2017). Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and post-translational levels. Mol. Microbiol. 103, 845–859. doi: 10.1111/mmi.13595
You, D., Yin, B. C., Li, Z. H., Zhou, Y., Yu, W. B., Zuo, P., et al. (2016). Sirtuin-dependent reversible lysine acetylation of glutamine synthetases reveals an autofeedback loop in nitrogen metabolism. Proc. Natl. Acad. Sci. U.S.A. 113, 6653–6658. doi: 10.1073/pnas.1525654113
Zhang, J., Liang, Q. T., Xu, Z. H., Cui, M., Zhang, Q. Z., Abreu, S., et al. (2020). The inhibition of antibiotic production in Streptomyces coelicolor over-expressing the TetR regulator SCO3201 IS correlated with changes in the lipidome of the strain. Front. Microbiol. 11:1399. doi: 10.3389/fmicb.2020.01399
Keywords: acetyltransferase, SCO0988, Streptomyces, acetylome, BldKB
Citation: Bi Y, An H, Chi Z, Xu Z, Deng Y, Ren Y, Wang R, Lu X, Guo J, Hu R, Virolle M-J and Xu D (2024) The acetyltransferase SCO0988 controls positively specialized metabolism and morphological differentiation in the model strains Streptomyces coelicolor and Streptomyces lividans. Front. Microbiol. 15:1366336. doi: 10.3389/fmicb.2024.1366336
Received: 06 January 2024; Accepted: 12 July 2024;
Published: 24 July 2024.
Edited by:
Carsten Jers, Technical University of Denmark, DenmarkReviewed by:
Evgeniya Warmer, ETH Zürich, SwitzerlandCopyright © 2024 Bi, An, Chi, Xu, Deng, Ren, Wang, Lu, Guo, Hu, Virolle and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia Guo, Z2oyMDA5QGpudS5lZHUuY24=; Ren Hu, dGh1cmVuQGpudS5lZHUuY24=; Marie-Joelle Virolle, bWFyaWUtam9lbGxlLnZpcm9sbGVAaTJiYy5wYXJpcy1zYWNsYXkuZnI=; Delin Xu, eHVkZWxpbkBsaXZlLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.