
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 March 2024
Sec. Microbial Physiology and Metabolism
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1366111
 Delfina Ensinck1
Delfina Ensinck1 Edileusa C. M. Gerhardt2
Edileusa C. M. Gerhardt2 Lara Rollan1
Lara Rollan1 Luciano F. Huergo3,4
Luciano F. Huergo3,4 Hugo Gramajo1
Hugo Gramajo1 Lautaro Diacovich1*
Lautaro Diacovich1*PII proteins are signal transduction proteins that belong to a widely distributed family of proteins involved in the modulation of different metabolisms in bacteria. These proteins are homotrimers carrying a flexible loop, named T-loop, which changes its conformation due to the recognition of diverse key metabolites, ADP, ATP, and 2-oxoglutarate. PII proteins interact with different partners to primarily regulate a set of nitrogen pathways. In some organisms, PII proteins can also control carbon metabolism by interacting with the biotin carboxyl carrier protein (BCCP), a key component of the acetyl-CoA carboxylase (ACC) enzyme complex, inhibiting its activity with the consequent reduction of fatty acid biosynthesis. Most bacteria contain at least two PII proteins, named GlnB and GlnK, with different regulatory roles. In mycobacteria, only one PII protein was identified, and the three-dimensional structure was solved, however, its physiological role is unknown. In this study we purified the Mycobacterium tuberculosis (M. tb) PII protein, named GlnB, and showed that it weakly interacts with the AccA3 protein, the α subunit shared by the three different, and essential, Acyl-CoA carboxylase complexes (ACCase 4, 5, and 6) present in M. tb. A M. smegmatis deletion mutant, ∆MsPII, exhibited a growth deficiency on nitrate and nitrite as unique nitrogen sources, and accumulated nitrite in the culture supernatant. In addition, M. tb PII protein was able to interact with the C-terminal domain of the ammonium transporter Amt establishing the ancestral role for this PII protein as a GlnK functioning protein.
Nitrogen is a constituent of essential cellular metabolites, such as nucleotides and aminoacids, consequently, all organisms must be provided with this macro-element. Actinomycetes, including M. smegmatis, can metabolize different nitrogen sources, and may adapt their metabolism to a number of environmental conditions and situations of nitrogen starvation (Jeßberger et al., 2013). For instance, during the infection process within the eukaryotic host cells, Mycobacterium tuberculosis (M. tb) is exposed to nutrient starvation, hypoxia, low pH, and reactive species of oxygen and nitrogen; hence, the bacterium must adjust its global metabolism to establish an asymptomatic latent state. Mycobacteria are also capable of exploiting diverse nitrogen sources available within the host; consequently, the regulation of nitrogen metabolism is essential for the virulence and survival of M. tb (Gouzy et al., 2014).
In many bacteria and plants, the proteins belonging to the PII superfamily function as sensors and regulators of nitrogen status (Ninfa and Atkinson, 2000; Arcondéguy et al., 2001; Forchhammer et al., 2022). These proteins consist of 12–13 kDa homo-trimers with three exposed T-loops that can be modified allosterically by binding ATP, ADP and 2-oxoglutarate (2-OG) (Macpherson et al., 1998; Xu et al., 1998; Radchenko et al., 2010) or by reversible covalent modifications such as uridylation or adenylation of the conserve Tyrosine-51 residue (Son and Rhee, 1987; Hesketh et al., 2002; Strösser et al., 2004; Williams et al., 2013). These modifications may alter the structure of the T-loops, allowing or constraining the interaction of the PII proteins with different protein partners or targets (Huergo et al., 2013). The genome of Escherichia coli, and other bacteria, contains at least two genes that encode for two putative PII proteins, named as GlnB and GlnK, which share approximately 67% of sequence identity at the aminoacids levels. In E. coli, GlnB regulates the expression and activity of the glutamine synthetase (GlnA) either through its action on the two-component system (NtrB-NtrC) or by interacting with the adenylyl-transferase GlnE, depending on the levels of glutamine in the cells (Bueno et al., 1985). Recently, GlnB was also reported to be involved in the regulation of carbon metabolism, through its interaction with the biotin carboxyl carrier protein (BCCP) component of the acetyl-CoA carboxylase complex (Feria Bourrellier et al., 2010; Gerhardt et al., 2015; Hauf et al., 2016; Rodrigues et al., 2019) leading to the inhibition of the enzyme activity. On the other hand, glnK is arranged in an operon together with the gene of an ammonium transporter, amtB. In actinomycetes, this operon also contains the gene encoding for the adenylyl transferase, glnD (Thomas et al., 2000; Arcondéguy et al., 2001). The function of GlnK is to bind AmtB, under nitrogen excess conditions, preventing the entry of ammonium into the cell. When the concentration of nitrogen is limited, GlnK binds ATP and 2-OG and later is uridylated by GlnD; this modification impedes the interaction of this protein with the transporter AmtB allowing the influx of ammonium (Coutts and Thomas, 2002; Durand and Merrick, 2006).
Mycobacteria genomes contain a unique gene encoding for PII protein. The M. tb PII protein (encoded by Rv2919c) was annotated as GlnB due to the highest sequence similarity (61.6%) with E. coli GlnB. In contrast, the genetic environment of the GlnB encoding gene could suggest that this protein may have the role of GlnK, as it is part of the amt-glnB-glnD operon, where amt is an orthologue of amtB (Thomas et al., 2000). In addition, the structure of M. tb PII protein also shows that this polypeptide has features of GlnK-like proteins (Shetty et al., 2010). However, the resulting structure of the phylogenetic tree indicates that the PII proteins of actinobacteria would form a different lineage from the GlnK and GlnB of proteobacteria (Sant'Anna et al., 2009; Bandyopadhyay et al., 2010). Interestingly, the gene encoding M. tb PII is not essential for growth in rich media, but it is relevant for survival in primary murine macrophages (Rengarajan et al., 2005).
For mycobacteria, the preferred nitrogen sources are ammonium or aminoacids (Amon et al., 2009; Gouzy et al., 2014); however, they also have the capacity to metabolize other inorganics compounds like nitrate or nitrite. The utilization of different nitrogen sources is modulated at the transcriptional level by GlnR. This key regulator control the expression of a number of genes related to nitrogen metabolism in M. smegmatis, including ammonium transporters, and is required for M. tb to utilize nitrate and nitrite as unique nitrogen source (Amon et al., 2008; Malm et al., 2009; Jenkins et al., 2013; Williams et al., 2015). A second regulatory protein was identified in M. smegmatis, NnaR, which cooperates with GlnR to modulate the transcriptional activation of the nirBD operon (Antczak et al., 2018). However, several aspects of the control and connections of nitrogen pathways are still unknown in mycobacteria, including the role of PII protein (Gouzy et al., 2014). In this work, we evaluated the functions of the purified PII protein of M. tb at the biochemical level adding evidence for the role of this protein as a GlnK-like. Furthermore, we constructed a ∆MsPII deletion mutant strain in M. smegmatis, and its physiological characterization suggests a new role for the PII proteins in the assimilation of nitrogen in mycobacteria.
The bacterial strains and plasmids used in this work are listed in Table 1. The E. coli strains were used for cloning proposes and protein heterologous expression. They were transformed according to (Sambrook et al., 1989). Transformants were selected on Luria–Bertani (LB) media at 37°C supplemented with the appropriate antibiotics: 50 μg kanamycin (Km) ml−1 and/ or 20 μg chloramphenicol (Cm) ml−1. M. smegmatis mc2155, an electroporation proficient mutant of mc26 (Snapper et al., 1990), was routinely grown at 37°C in Middlebrook 7H9 (BD-Difco) medium supplemented with 0.03% tyloxapol or in solid medium Middlebrook 7H10 (BD-Difco) supplemented with the appropriate antibiotics if required 15 μg kanamycin ml−1, 20 μg gentamycin ml−1 or 50 μg hygromycin ml−1. For nitrogen analysis, bacteria were grown in modified Sauton’s minimal medium (Jenkins et al., 2013) supplemented with different nitrogen sources. Cells were cultivated in 15 mM ammonium sulfate (Sigma) to log phase and washed twice with nitrogen-free medium before inoculating fresh medium with the required nitrogen source.
Isolation of plasmid DNA, restriction enzyme digestion and agarose gel electrophoresis were carried out by conventional methods (Sambrook et al., 1989). The genomic DNA of M. smegmatis was obtained as described by Connell (1994).
The oligonucleotides oDE01 (5’-CGTATCATATGGAAAACCTG TACTTCCAGGGTATGAAGCTGAT CACTGCGA), used to introduce a NdeI site (underlined) and TEV site (bold) upstream the M. tuberculosis glnB gene, and oDE02 (5′- AGCACAGAAGCTTTA GTTTCATAACGCGTCGTGTC), which introduces a HindIII site (underlined) at the end of the open reading frame (ORF), were used to amplify the complete glnB gene. The PCR product was digested with NdeI and HindIII and cloned in NdeI/HindIII-cleaved pET24a(+), yielding pDE2.
For the construction of the M. smegmatis ∆MsPII mutant strain, the upstream region of MSMEG_2426 gene was amplified with the primers oDE09 (5’-ACTAGTCGGTCTGGGTGAGGG)- and oDE10 (5’-GCTAGCCGTCTTGACATCTTC) introducing SpeI and NheI sites at its extremes. The 1761 bp PCR product was cloned into pJET 1.2 Blunt (Thermo Fisher Scientific) yielding pDE8 plasmid. The downstream region of MSMEG_2426 gene was amplified with the primers oDE11 (5′- GCTAGCATCGGTGACGGCAAGGTG) and oDE12 (5’-ACTAGTGAGCACGTCGAGCAGTTCC) introducing NheI and SpeI sites at its extremes. The 1842 bp PCR product was cloned into pJET 1.2 Blunt (Thermo Fisher Scientific) yielding plasmid pDE9. Plasmids pDE8 and pDE9 were digested with SpeI and NheI, and the fragments obtained were cloned into SpeI-cleaved pPR27 plasmid, yielding pDE13.
The oligonucleotides oDE13 (5’-CATATGAAGCTGATTACTG CGAT), used to introduce a NdeI site (underlined) at the translational start codon of the M. smegmatis MSMEG_2426 gene, and oDE14 (5’-GAATTCTACAGGGCGTC GGCT), which introduces an EcoRI site (underlined) at the end of the open reading frame (ORF), were used to amplify the complete MSMEG_2426 gene. The PCR product was digested with NdeI and EcoRI and cloned in NdeI/EcoRI-cleaved pMVHG, yielding pDE35.
Using the vector pET32a(+) as a template and the primers 32AseIup (5′- GAGATATATTAATATGAGCGATAAAATTATTC), which introduces an AseI site (underlined), and 32TEVdn (5′- GGTG CATATGACCCTGGAAGTACAGGTTTTCGCCAGAACCAGAAC CGGCC) introducing NdeI (underlined) and TEV (bold) sites, a fragment of 384 pb was amplified. This PCR product was cloned into the pET28a(+) vector digested with NdeI, yielding pET2832. This vector allows the expression of proteins as His and TrxA tags fusions, with a TEV digestion site.
The oligonucleotides oDE27 (5′- CCTGTA CTTCCAGGGTCATA TGATCGGGCTCAGGC) and oDE28 (5′- GCTCGAGTGCGGC CGCAAGCT TTCATTTCGGCTC) were used to amplify the last 183 pb (61 codons) of the Rv2920c gene (amtCTR). The PCR product was cloned in NdeI/HindIII-cleaved pET2832 by AQUA Cloning (Beyer et al., 2015), yielding pNZn2.
Recombinant M. tuberculosis His-tagged ACCase subunits AccA3, AccD5, AccD6, and AccE5 were purified as described in Gago et al. (2006). The same protocol was applied for His-tagged M. tb PII and AmtCTR fused to TrxA. After dialysis, TrxA was removed through digestion with TEV protease. In brief, the AmtCTR-TrxA fraction was incubated with TEV protease (30:1) for 2 h at room temperature, followed by an additional 2 h at 4°C. The mixture was then loaded onto a Ni-NTA column, and cleaved AmtCTR was subsequently recovered. PII proteins from E. coli, A. brasilense and M. tb were obtained without tags, according to Gerhardt et al. (2015, 2017).
Proteins were analyzed by SDS-PAGE (Laemmli, 1970). Protein contents were determined by measuring A280 nm to the solutions and by the Bradford method, using BSA as standard.
For the production of polyclonal antibodies anti PII, a rabbit was immunized biweekly with purified M. tb PII, emulsified in Freund’s complete adjuvant (Sigma) at a 1:1 (v/v) ratio. Antisera against PII were elicited in rabbit following conventional procedures (Burnette, 1981, Anal. Biochem). The anti-M. tb PII was used to evaluate the presence of M. tb or M. smegmatis PII proteins (88.4% identity), and both proteins were recognized by the antibody (Supplementary Figure S1A).
PII from mycobacteria were detected by Western blot analysis. After electrophoretic separation, proteins were transferred onto nitrocellulose membranes (Bio-Rad). Anti-M. tb PII was used at a 1:300 dilution, and anti-RpoB at 1:10,000 dilution, as a control. Antigenic polypeptides were visualized using an alkaline phosphatase-tagged secondary antibody. For biotinylated proteins western blotting procedure was modified as described by Nikolau et al. (1985). Proteins were probed alkaline phosphatase-streptavidin conjugate (AP-streptavidin diluted 1:10,000) (Bio-Rad).
M. smegmatis mc2155 was grown in 0.5 mM (NH4)2SO4 to an OD 600 nm 0.6–0.8 and half of the culture was subjected to a shock of 15 mM of ammonium sulfate, for 15 min (Atkinson et al., 1994; Durand and Merrick, 2006; Moure et al., 2012; Rodrigues et al., 2014). The adenylation state of PII in cell-free extracts was determined by electrophoresis on SDS- PAGE or native PAGE and followed by Western blotting as described in (Bonatto et al., 2007).
Complex formation between M. tb His-AccA3 and PII without a tag were assessed by pull down as described by Rajendran et al. (2011), with modifications. Pull down assay was performed using 15 μL of magnetic beads (Promega) with interaction buffer (Tris–HCl 50 mM pH 8.0, NaCl 100 mM, glycerol 5%, LDAO 0,01%, imidazole 20 mM; MgCl2 5 mM) and saturating ATP (3.5 mM). Fifteen micrograms of purified His-AccA3 were mixed with 20 μg of PII protein from M. tb (Mt), A. brasilense (Ab) or E. coli (Ec). Proteins eluted from the Ni2+ were analyzed by SDS-PAGE and the gel was stained with Coomassie Blue.
The interaction between PII and AmtCTR was analyzed in vitro. 10 μg of cell extracts expressing His-tagged M. tb PII were mixed with 10 μg of purified AmtCTR without tag. The mixture was prepared in the presence of either 1 mM ATP or 1 mM ATP along with 1.5 mM 2-OG. After incubating the mixture for 1 h at 4°C, it was loaded into a Ni2+-NTA column that had been pre-equilibrated with PBS containing ATP, or ATP and 2-OG. The column was then washed with PBS, supplemented with the ATP and 2-OG, using 10 column volumes. Finally, the bound proteins were eluted from the column using PBS containing 250 mM imidazole. SDS-PAGE was performed to analyze the samples obtained from the column.
ACCase activities in cell-free extracts and in vitro reconstituted complexes were determined by following the incorporation of radioactive HCO3− into acid non-volatile material, as previously described (Diacovich et al., 2002). Substrate concentrations were 0.5 mM for acetyl-CoA or propionyl-CoA. One unit of enzyme activity catalyzed the incorporation of 1 mmol 14C into acid-stable products min−1.
For the construction of the M. smegmatis mutant ∆MsPII the plasmid pDE13 was used to transform M. smegmatis cells. The mutant strain generated through a double homologous recombination event was selected according to (Crotta Asis et al., 2021).
Nitrite concentration was determined using the Griess reaction (Peter, 1879). For the uptake assay, exponentially growing cells in modified Sauton’s minimal medium supplemented with 1 mM nitrite at an OD 600 nm of 0.6–0.8 were harvested by centrifugation at 3,000 × g for 10 min at 4° C. Afterward, they were washed twice using modified Sauton’s medium that lacked a nitrogen source. The cultures were then resuspended in fresh medium at pH 7.2 or 9 containing 0.2 mM sodium nitrite and 1 mM ammonium sulfate when indicated, and adjusting to an OD of 600 nm of 0.75. Cells were cultivated under constant shaking of 180 rpm at 37° C. At regular intervals of 15 min, aliquots of the cultures were sampled and centrifuged at 5,000 × g for 10 min to remove the cells previous to nitrite quantification.
Crude extracts were obtained from cultures grown in modified Sauton’s minimal medium with 1 mM nitrate. Appropriate amounts of extracts were incubated for 30 min at 30°C in a buffer composed of 0.1 M potassium phosphate (pH 7.5), 0.4 mM NADPH, 0.025 mM FAD, 0.025 mM FMN, and 1 mM NaNO3. To measure the concentration of accumulated nitrite, the Griess Reaction was performed (Peter, 1879).
Cell-free extracts excised from Coomassie-stained SDS-PAGE gels were subjected to digestion with trypsin. Peptide separations were carried out on a nanoHPLC Ultimate3000 (Thermo Scientific) using a nano column EASY-Spray ES903 (50 cm × 50 μm ID, PepMap RSLC C18). The mobile phase flow rate was 400 nl/min, using 0.1% formic acid in water (solvent A) and 0.1% formic acid and 100% acetonitrile (solvent B). The gradient profile was set as follows: 4–35% solvent B for 30 min, 35–90% solvent B for 1 min and 90% solvent B for 5 min. Three microliters of each sample were injected. MS analysis was performed by using a Q-Exactive HF mass spectrometer (Thermo Scientific). For ionization, 1.9 kV of liquid junction voltage and 250°C capillary temperature was used. The full scan method employed a m/z 375–1,600 mass selection, an Orbitrap resolution of 120,000 (at m/z 200), a target automatic gain control (AGC) value of 3e6, and maximum injection times of 100 ms. After the survey scan, the 7 most intense precursor ions were selected for MS/MS fragmentation. Fragmentation was performed with a normalized collision energy of 27 eV and MS/MS scans were acquired with a dynamic first mass. AGC target was 5e5, resolution of 30,000 (at m/z 200), intensity threshold of 4.0e4, isolation window of 1.4 m/z units and maximum IT was 200 ms. Charge state screening was enabled to reject unassigned, singly charged, and equal or more than seven protonated ions. A dynamic exclusion time of 15 s was used to discriminate against previously selected ions.
MS data were analyzed with MaxQuant (V: 2.1.4.00) using standardized workflows. Mass spectra *.raw files were searched against a database from Mycobacterium smegmatis mc2155 (reviewed and unreviewed proteins), entry UP00000075 from Uniprot. The “MSMEG_4206” sequence was added to the database. Precursor and fragment mass tolerance were set to 10 ppm and 0.02 Da, respectively, allowing 2 missed cleavages. Fixed modification: carbamidomethylation of cysteines. Variable modifications: protein N-terminal acetylation and methionine oxidation. The statistical analysis was performed with Perseus v1.6.15.0 software (MaxQuant).
The culture was grown in a modified Sautons’ medium supplemented with 0.5 mM (NH4)2SO4 until reaching an OD of 0.8 at 600 nm. The culture was then split into four fractions. In three of the cultures, (NH4)2SO4 was added to a final concentration of 15 mM, inducing an ammonium shock. After 15 min, the cultures were harvested by centrifugation at 3,000 x g for 10 min at 4°C. The cells were washed twice with PBS and resuspended in 100 μl of PBS containing 1 mM PMSF and either 0.6 mM ADP, or 4.5 mM ATP, and 1.5 mM 2-OG. Then, cultures were lysed using a water bath sonicator (Diagenode), and the lysates were clarified by centrifugation at 11,180 × g for 30 min at 4°C. The resulting cell-free extracts were further centrifuged at 16,000 x g for 1 h at 4°C, and the supernatant, representing the cytosolic fraction, was collected. The pellet, containing the membrane fraction, was washed once with PBS, containing the corresponding additives, and centrifuged again for 30 min at 16,000 g. The membranes were then resuspended in 50 μl of PBS. The samples were analyzed by SDS-PAGE followed by western blotting, performed to detect PII and RpoB, serving as a control.
The genomes of M. tb and M. smegmatis contain one type of PII protein-encoding gene. The PII protein expression in mycobacteria is induced in response to nitrogen limitation (Amon et al., 2008; Williams et al., 2013) and PII is rapidly adenylated by GlnD (Williams et al., 2013). To understand the dynamics of PII protein adenylation in response to the nutritional status, we analyzed the state of PII adenylation under different nitrogen regimen. M. smegmatis mc2155 was grown in modified Sauton’s minimal medium supplemented with 0.5 mM ammonium sulfate (Williams et al., 2013, 2015) to an OD 600 nm of 0.7; at this stage, PII expression can be detected by Western-blot (Supplementary Figure S1A). At this particular OD, the culture was divided into two fractions and one was subjected to a concentration of 30 mM of ammonium, which is considered an ammonium shock (Coutts and Thomas, 2002) After 15 min, cells were collected and protein extracts were analyzed by SDS-PAGE or native gels, and detected by Western blotting. As can be observed in Figures 1A,B, the migration pattern of PII changed after the ammonium shock, with a pattern consistent with the loss of the nucleoside (Atkinson et al., 1994; Durand and Merrick, 2006; Moure et al., 2012; Rodrigues et al., 2014). In native gels, this modification provokes PII to run faster than unmodified PII, due to PII-AMP’s higher negative charge imparted by the AMP residues. On the contrary, in SDS-PAGE, as the gel in Figure 1A, the adenylation of PII produces a delay in protein migration. The impact of adenylation on protein electrophoretic mobility, under denaturing conditions, was validated by LC-MS/MS (data not shown). These results suggest that the adenylation of PII is readily reversible by increasing the availability of ammonium.
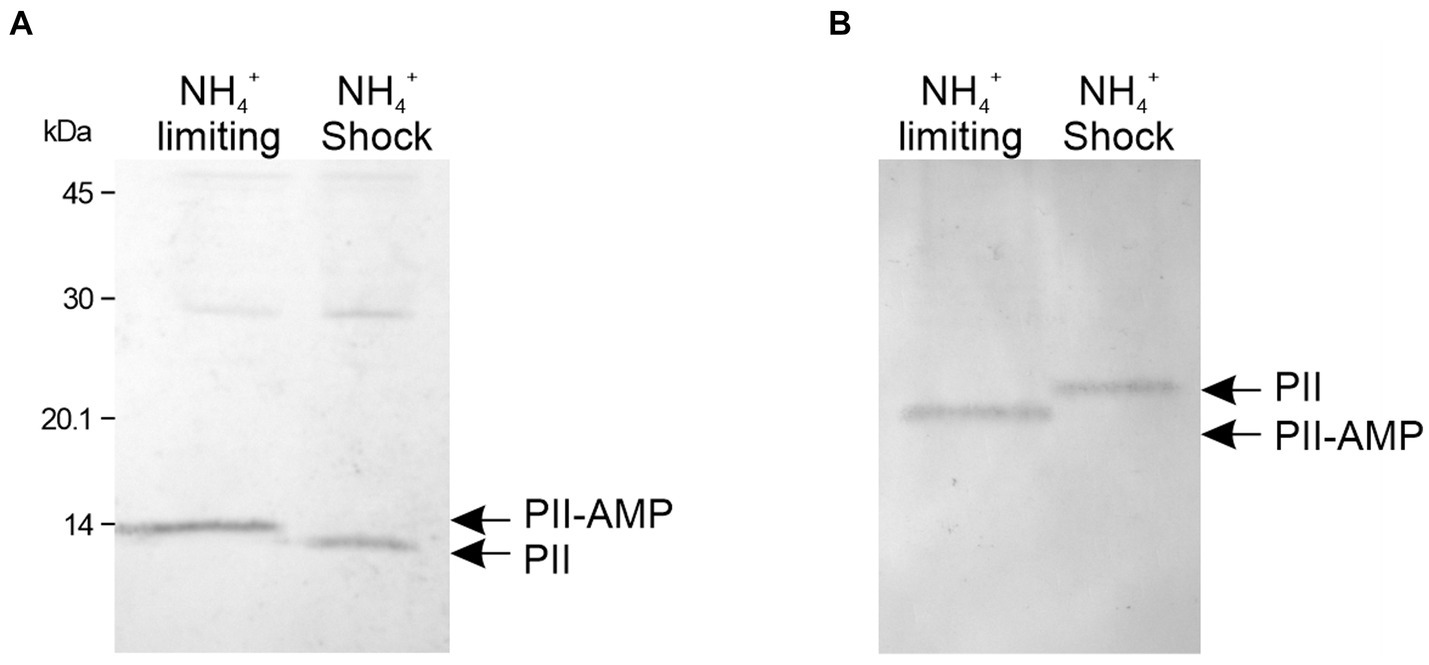
Figure 1. Determination of the adenylation state of PII from M. smegmatis. Bacteria were grown in nitrogen-limiting conditions with 0.5 mM of (NH4)2SO4. An aliquot of the culture was subject to an ammonium shock. The mobility of PII was analyzed before (NH4+ limiting) and after 15 min of ammonium shock (NH4+ shock). Cell-free extracts were separated by SDS-PAGE (A) or native PAGE (B) and analyzed by western blot, using a polyclonal anti M. tb PII antibody.
In E. coli two PII proteins were described, namely GlnB and GlnK. Both proteins can interact with the BCCP subunit of the acetyl-CoA carboxylase (ACC) complex, but only GlnB is able to interact with the complex inhibiting the ACC holoenzyme activity (Gerhardt et al., 2015). Given that the regulation of ACC by PII is conserved in distantly related organisms, ranging from plants to bacteria (Forchhammer et al., 2022), we hypothesized that such PII function could be also conserved in mycobacteria. In M. tb the acetyl-CoA carboxylase activity is present in two enzyme complexes, named acyl-CoA carboxylases (ACCases) based on their relaxed substrate specificity. These two complexes, ACCase 5 and ACCase 6, consist of two subunits: an α-subunit named AccA3, which is shared by both ACCases, containing the biotin carboxylase (BC) and the biotin carboxyl carrier protein (BCCP) domains, and a specific β-subunit carrying the carboxyltransferase (CT) domain, named AccD5 or AccD6, respectively (Gago et al., 2006; Daniel et al., 2007; Kurth et al., 2009; Pawelczyk et al., 2011; Bazet Lyonnet et al., 2014). A third subunit, named ε, is also required for maximal activity of the ACCase 5 complex. These two enzymes are able to carboxylate both acetyl-CoA (ACC activity) and propionyl-CoA (PCC activity), but with different substrate preferences (Gago et al., 2006; Daniel et al., 2007). On the other hand, in E. coli there is only one type of ACC enzyme of which the BC and BCCP components are encoded by separated polypeptides.
To investigate the interaction between PII and the biotinylated subunit of the ACCase, purified AccA3-His6 (α-subunit) was immobilized into nickel beads and used as bait which was challenged with purified M. tb PII without tags (Gerhardt et al., 2015). Orthologue PII proteins, GlnB from Azospirillum brasiliense (Ab) and GlnB from E. coli (Ec), which share 60.7 and 61.6% similarity with M. tb PII (Mt), respectively, were used as controls. The pull-down assay data revealed that AccA3 interacted strongly with Ab-GlnB but weakly with Ec-GlnB or Mt-PII (Figure 2A). We also observed that the specificity of the AccA3 PII proteins interaction is dependent on the PII effectors. The interaction between AccA3 and PII is only effective in the presence of ATP, but it does not occur in the presence of ADP (Figure 2A and Supplementary Figure S2A). On the other hand, we also used lysozyme as a non-specific protein and showed that this protein does not interact with AccA3, in the same conditions used to detect AccA3-PII interaction (Supplementary Figure S2B).
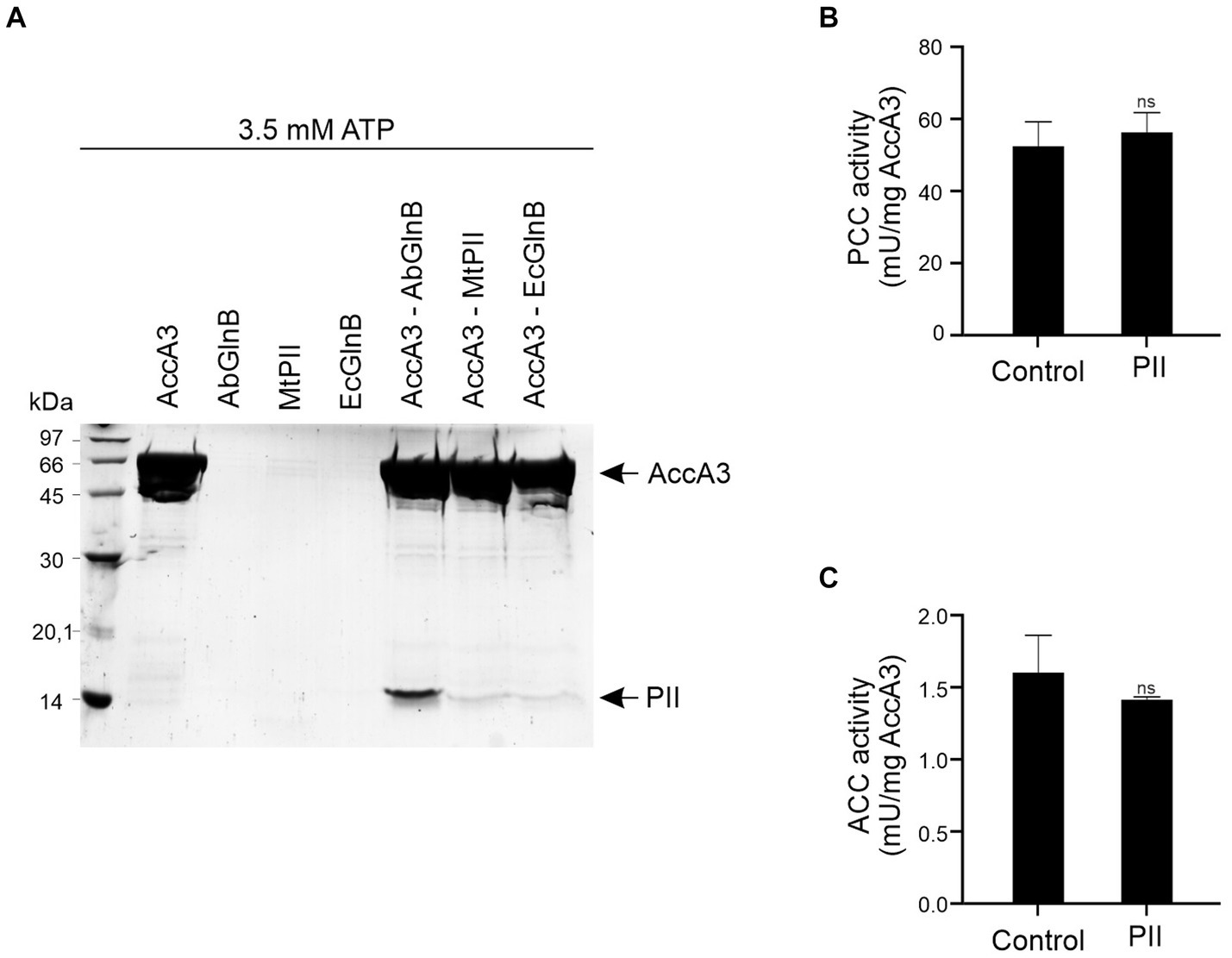
Figure 2. Interaction between PII and AccA3 and influence on ACCasa activities. (A) Complex formation between M. tb His-AccA3 and PII proteins was assessed by pull-down under saturating concentrations of ATP (3.5 mM), and MgCl2 (5 mM). Purified His-AccA3 was mixed with PII protein (without tags) from M. tb (Mt), A. brasilense (Ab), or E. coli (Ec). Proteins eluted from the Ni2+ resins were analyzed by SDS-PAGE and the gel was stained with Coomassie Blue. The effect of M. tb PII on PCC (B) and ACC (C) activities of the in vitro reconstituted complexes ACCase 5 and ACCase 6, respectively, was measured by determining the amount of radiolabeled HCO3− incorporated into malonyl-CoA or methylmalonyl-CoA, in presence or absence of M. tb PII. PII was added at a concentration equal to that of the AccA3 subunit (0.8 μM for ACCase 5 and 1.9 μM for ACCase 6). The specific activity is plotted as nmols of malonyl- or methylmalonyl-CoA produced per min, per mg of the AccA3 subunit. Data is reported as the average of quadruplicate and standard deviation. Statistical analysis was performed with a t-test at p < 0.05, ns, no significant.
In order to analyze if these interactions could affect M. tb ACCase activities, we assayed the propionyl-CoA carboxylase (PCC) activity in vitro using the reconstituted ACCase 5 complex which is composed of AccA3 (BC-BCCP α-subunit), AccD5 (CT), and AccE5 (ε subunit). When Ab-GlnB PII protein was incorporated into the reaction mix, an inhibition of the PCC activity was detected (Supplementary Figure S3), which is consistent with the strong interaction observed in the pull-down assay (Figure 2A). This data support that the interaction between ACCase and PII acts to reduce ACC/PCC activity as described previously in bacteria such as E. coli and Streptomyces hygroscopicus var. ascomyceticus (Wang et al., 2021). However, in the presence of M. tb PII no significant modulation of PCC activity was detected (Figure 2B), which is consistent with the weak interaction between M. tb PII and AccA3 (Figure 2A). Likewise, the ACC activity of the M. tb ACCase 6 complex, which shares the BC-BCCP subunit with ACCase 5, was not affected by the presence of PII either (Figure 2C).
In E. coli the formation of the ACC-GlnB complex is inhibited when GlnB is uridylated (Gerhardt et al., 2015). Hence, we analyzed ACCase activities in M. smegmatis cell-free extracts, grown in limiting nitrogen conditions (1 mM of ammonium) or after the ammonium shock (30 mM of ammonium), conditions were PII is adenylated or de-adenylated, respectively (Figure 1). Under these conditions, no changes were observed in the PCC or the ACC activities of the corresponding ACCases (Figure 3).
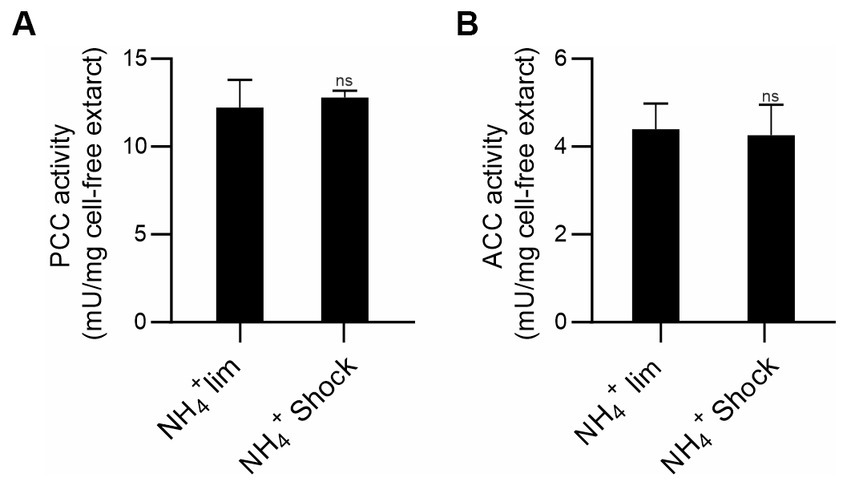
Figure 3. Determination of acyl-CoA carboxylase activities in M. smegmatis cell-free extracts. M. smegmatis cultures were grown under ammonium-limiting conditions and after ammonium shock, PCC (A) and ACC (B) activities were determined in cell-free extracts by incorporation of radiolabeled HCO3− into the propionyl-CoA or acetyl-CoA as substrates, respectively. The specific activity is plotted as nmols of the corresponding carboxy-acyl-CoA produced per min, per mg of total proteins. Data is reported as the average of quadruplicate and standard deviation. Statistical analysis was performed with Student’s t-test at p < 0.05, ns: no significant.
Previous high-throughput studies suggested that PII is not essential for the viability of M. tb (Sassetti et al., 2003). Assuming that this would also be applied to M. smegmatis PII coding gene (MSMEG_2426), we constructed a knockout (KO) mutant in M. smegmatis to gain information about the physiological relevance of PII. We disrupted MSMEG_2426 by using an in-frame deletion strategy, which would not have a polar effect on the expression of glnD. For this, we replaced MSMEG_2426, the PII coding gene, with a mutant allele, using a classic two-step homologous recombination approach (Crotta Asis et al., 2021; Figure 4A). The strain obtained was named ∆MsPII, confirming that the MSMEG_2426 gene is not essential for the viability of M. smegmatis. PII was not detected by Western blotting in cell-free extracts from two individual colonies of the putative mutants, while PII was perfectly distinguished in cell-free extracts deriving from wild-type strain, which confirmed that the expected allelic exchange at the chromosomal MSMEG_2426 locus had been successful (Figure 4B). The PII expression levels were partially complemented by the plasmid pDE35, expressing MSMEG_2426 under the control of the constitutive promoter Phsp60 (Figure 4B); this strain was named ∆MsPIIc. In all the cases, the anti-M. tb PII antibody was used to detect PII proteins from M. tb or M. smegmatis; this antibody was able to well recognize both proteins (Supplementary Figure S1).
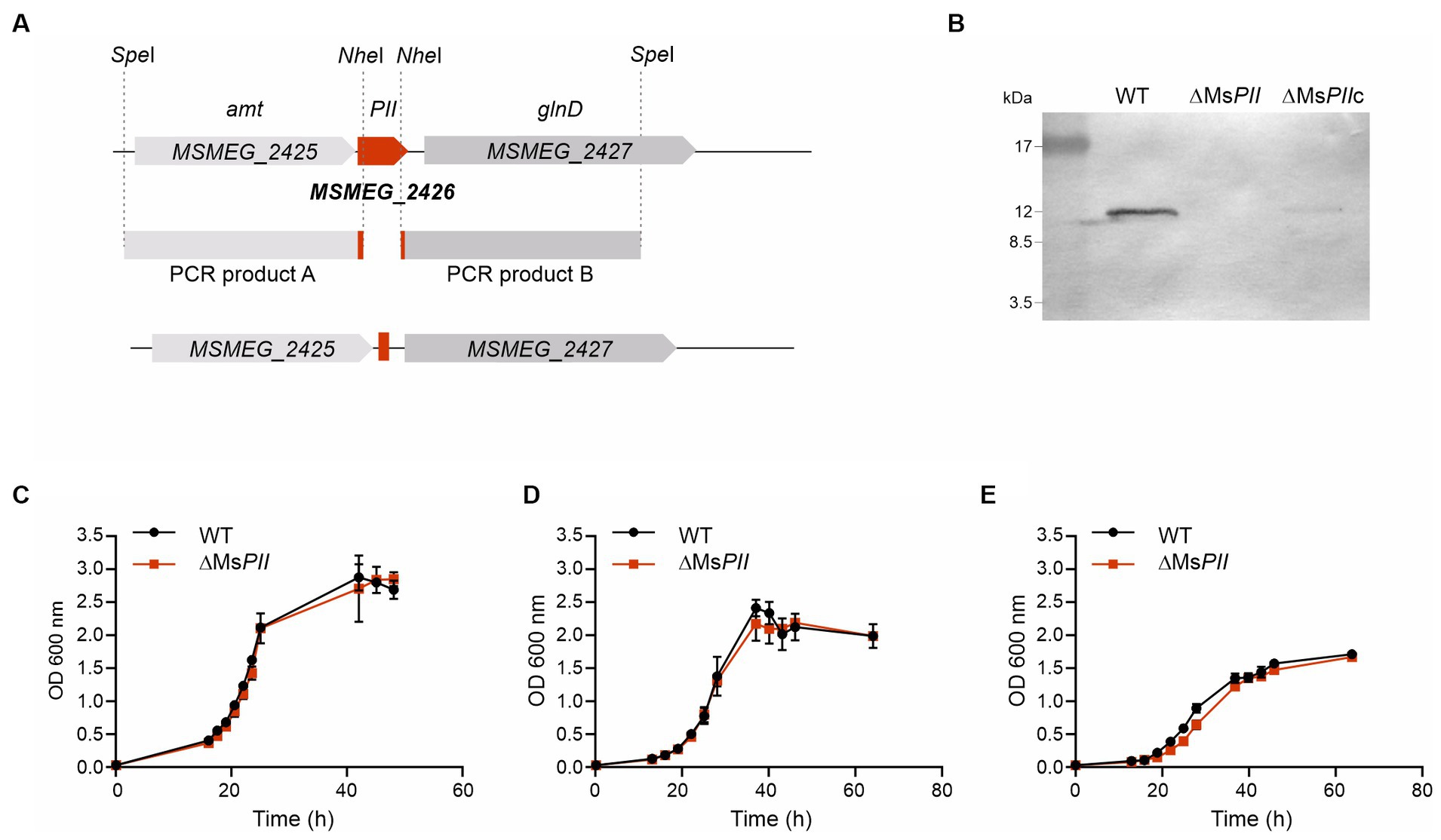
Figure 4. Construction of the M. smegmatis PII deletion mutant strain ΔMsPII. (A) An in-frame deletion of the PII gene (MSMEG_2426) was constructed by two-steps of homologous recombination with the thermo-sensitive plasmid pDE13. The schematic representation illustrates the genetic MSMEG_2426 chromosomal region of the wild-type strain mc2155, the PCR products A and B, that were amplified and cloned in the delivery vector, and the resulting genetic rearrangement in the mutant strain, obtained for recombination, generating the deletion. (B) The presence of the PII protein was analyzed by Western blotting in cell-free extract from the wild type, the ΔMsPII and in the ΔMsPIIc strains. Anti-M. tb antibody was used to detect endogenous PII protein from M. smegmatis. Growth curves of M. smegmatis mc2155 (WT) and ΔMsPII strains in rich medium 7H9, complemented with 0.2% glycerol and 0.03% tyloxapol (C) or in modified Sauton’s minimal media, supplemented with 15 mM (D) or 0.5 mM of (NH4)2SO4 (E).
To analyze the physiological consequences of altered PII protein levels, the wild-type, ∆MsPII, strains were grown in 7H9 liquid medium and modified Sauton’s minimal medium supplemented with 15 mM or 0.5 mM of (NH4)2SO4 (Williams et al., 2013, 2015). The growth dynamic of the ∆MsPII strain was very similar to that corresponding to the isogenic wild-type strain (Figures 4C–E). Hence, under these growth conditions, the absence of PII does not modify the cell growth rate.
It is well described that PII proteins are involved in the regulation of nitrogen metabolism. To further understand the metabolic role of mycobacterial PII, we grew the three strains, wild-type, ∆MsPII and ∆MsPIIc in Sauton’s minimal media where the unique source of nitrogen was nitrate or nitrite. When the medium was supplemented with 1 mM of nitrite (Akhtar et al., 2013) a clear growth delay was observed in the ∆MsPII strain, with a marked and prolonged lag phase, when compared to the wild-type; however, both strains reached similar final ODs. The complemented strain shows an intermediate behavior, suggesting that the expression of the M. smegmatis from the plasmid pDE35 partially compensates for the loss of M. smegmatis PII expression from the chromosome (Figure 5A). Simultaneously, the concentration of the nitrite remaining in the medium was measured through the Griess assay. The nitrite was not detectable after 33 h in the wild-type cultures, while it took over 41 and 52 h to become undetectable in the ∆MsPIIc and ∆MsPII strains, respectively (Figure 5B).
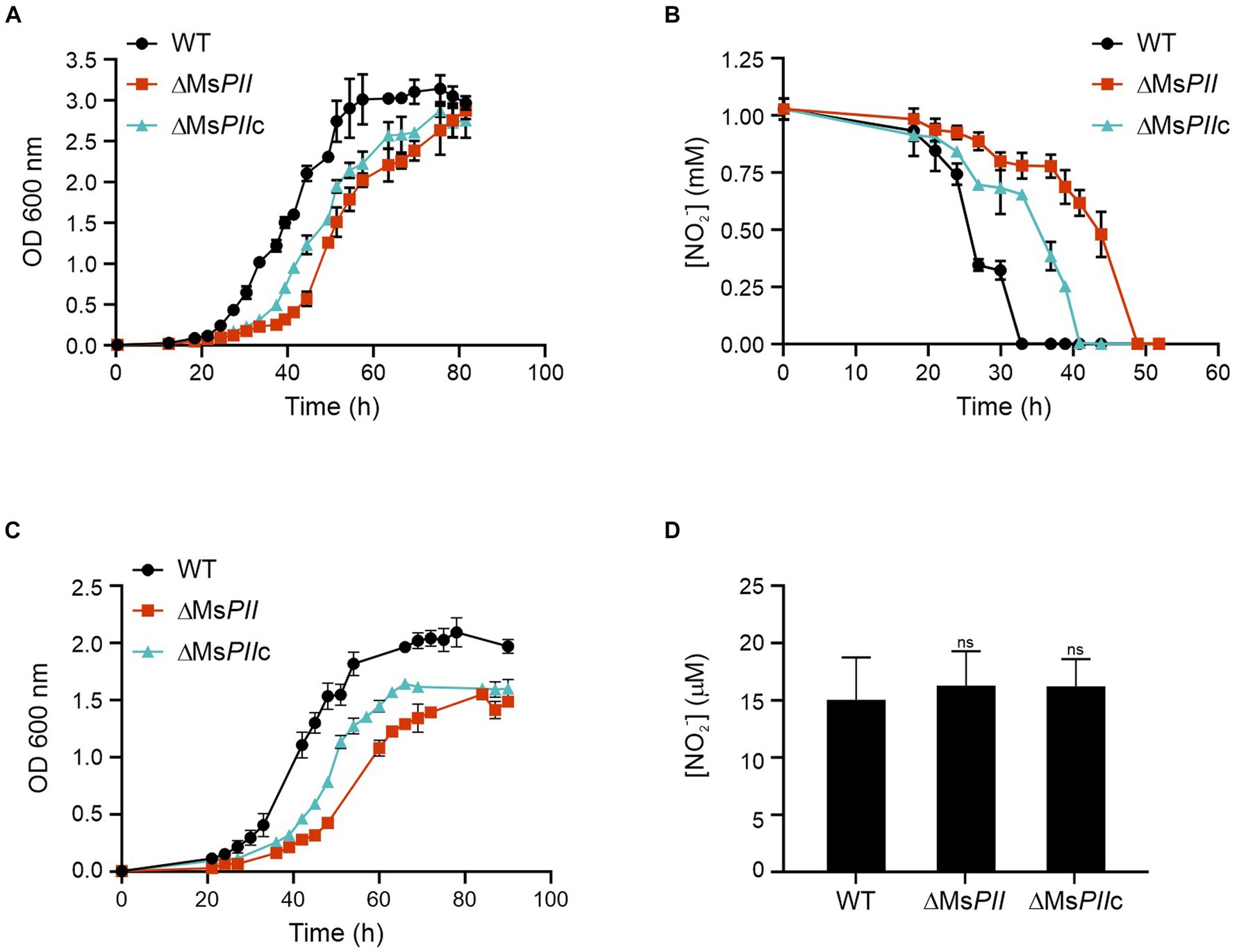
Figure 5. Utilization of nitrite and nitrate as the sole nitrogen source in the ΔMSPII strain. High ammonium-cultured cells from wild-type, ΔMSPII and ΔMSPIIc strains were inoculated in fresh modified Sauton’s minimal medium, supplemented with 1 mM of nitrite and growth at 37°C. Bacterial growth (A) and nitrite consumption (B) were followed. Cell growth was measured using 1 mM nitrate as a nitrogen source (C). The accumulation of nitrite in nitrate-cultures mycobacteria was determined in lysates of the wild-type, PII-null-mutant and ΔMSPIIc strains (D). Growth was measured as the OD 600 nm, while the nitrite concentration was measured through the Griess assay. Data is expressed as the mean ± S.D. of triplicates. Statistical analysis was performed using Student’s t-test at p < 0.05, ns, no significant.
Similar but strongest differences were observed when the strains were grown in Sauton’s media supplemented with 1 mM of nitrate (Figure 5C). The ∆MsPII strain had a reduced growth rate and lower final ODs when compared to the wild type. To determine whether the nitrite assimilation of ∆MsPII cells was the cause for the lower growth rate in nitrate, we measured the nitrite concentration in extracts of cells that were growing in 1 mM nitrate. In this assay, the three cultures resulted in comparable levels of nitrite, indicating the mutant cells are not accumulating or excreting nitrite (Figure 5D).
These experiments suggest that M. smegmatis PII protein could be involved in the modulation of nitrate and nitrite assimilation. We conclude that PII is not completely required for cell growth but it increases cell fitness when nitrate or nitrate are used as sole nitrogen sources.
The ability to take up nitrite at different pH was analyzed in wild-type, ΔMsPII and ΔMsPIIc cells, previously adapted to nitrite. The three strains presented a similar uptake activity when nitrite was the sole nitrogen source at pH 7.2 (Figure 6A). The addition of ammonium resulted in a diminution of the nitrite uptake in all the strains; however, this effect was retarded in the ΔMsPII strain (Figure 6B). At pH 9, the wild-type strain presented a similar ammonium-dependent inhibition, but this effect was not observed in the ΔMsPII strain. The ammonium inhibition resulted slower in ΔMsPIIc strain (Figures 6C,D). At pH 7.2, nitrite predominantly exists in the form of nitrous acid which could permeate the cell via diffusion. Conversely, at pH 9, the nitrite ions must be transported through the cell envelope by specific permeases (Lee et al., 1998). Therefore, the lack of ammonium-induced inhibition of nitrite uptake at pH 9 in the ΔMsPII strain would imply a potential role of M. smegmatis PII in orchestrating the hierarchical utilization of ammonium over nitrite as a nitrogen source.

Figure 6. Analysis of nitrite uptake in the wild-type, PII-null mutant and complemented strains. The nitrite uptake was measured in the M. smegmatis wild-type, PII-null mutant and complementing strains. The assay was performed on nitrite-grown cells, incubated in the presence of 0.2 mM of NaNO2 and in the absence (A,B) or presence (C,D) of 1 mM of (NH4)2SO4. The data is reported as the average of triplicates ± S.D.
To assimilate nitrate into the organic nitrogen pool, it undergoes two consecutive reductions. Initially, nitrate is mainly intracellularly reduced to nitrite by the action of a nitrate reductase activity, and subsequently, nitrite is further reduced to ammonia by the reaction catalyzed by the nitrite reductase (Morozkina and Zvyagilskaya, 2007; Gouzy et al., 2014). In mycobacteria, a unique nitrite reductase enzyme was identified (NirBD) (Malm et al., 2009), while several genes encode for proteins with nitrate reductase activity, including NarGHJI, and NarB. A novel type of nitrate reductase was recently described in M. smegmatis, which was named NasN. This NADPH-dependent diflavin enzyme is uniquely responsible for nitrate assimilation (Tan et al., 2020; Cardoso et al., 2021). We measured the cytoplasmic nitrate reductase activities in cell–free extracts of M. smegmatis wild-type and ∆MsPII strains and found that the protein extracts of the mutant strain had higher nitrate reductase activity than the wild-type (Figure 7). This variation could be the result of protein levels or a direct or indirect regulation of the nitrate reductase activity by PII. NasN is part of the GlnR regulon, the major nitrogen-regulator in M. smegmatis (Jeßberger et al., 2013). Even though there is no evidence that PII and GlnR may interact, PII could be modulating protein expression as seen in other systems (Huergo et al., 2013). Therefore, we compared the protein profiles of both strains when grown in 1 mM nitrate by LC–MS/MS. The proteins with significant differences were screened according to the criteria of │FC│ > 1.5 and p < 0.05 (Figure 8). Though, the amount of a few proteins involved in nitrogen metabolism was altered (Table 2), no differences were observed in the GlnR or NasN levels between these strains, suggesting that the modulation of nitrate reductase activity by PII occurs by posttranslational mechanism. The subunits of the NirBD complex, which is under the regulation of GlnR and NasR, were equally represented in both samples as well. The comprehensive roster of differentially expressed proteins is detailed in Supplementary Tables S1, S2.
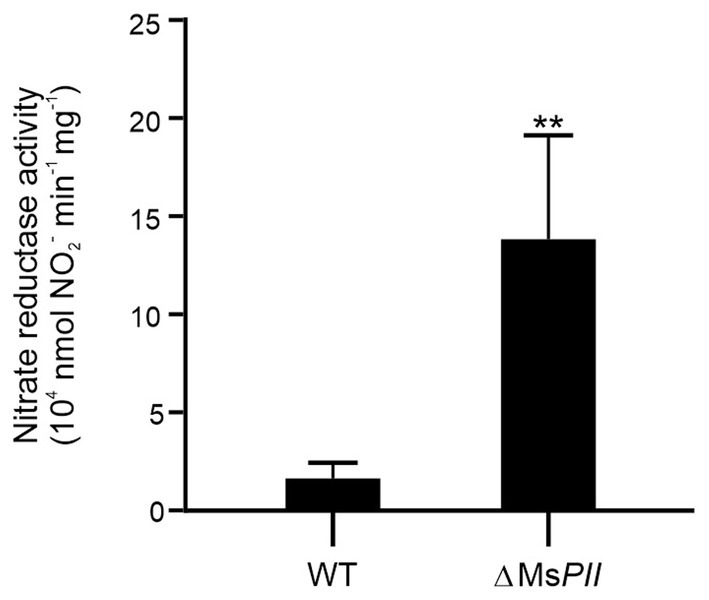
Figure 7. Effect of PII deletion in nitrate reductase activity. Specific nitrate reductase activity was quantified in the cytoplasmic fractions of the cell-free extracts from M. smegmatis wild-type and ΔMsPII strains. Bacteria were grown aerobically at 37°C in modified Sauton’s minimal medium, containing 1 mM of KNO3. Reactions were conducted in presence of the electron donors NADPH and the cofactors FMN and FAD+, at pH 7.5 and 30°C. Data are expressed as the mean ± SD of five biological and two technical replicates. Statistical analysis was performed using Student’s t-test. ***p < 0.001.
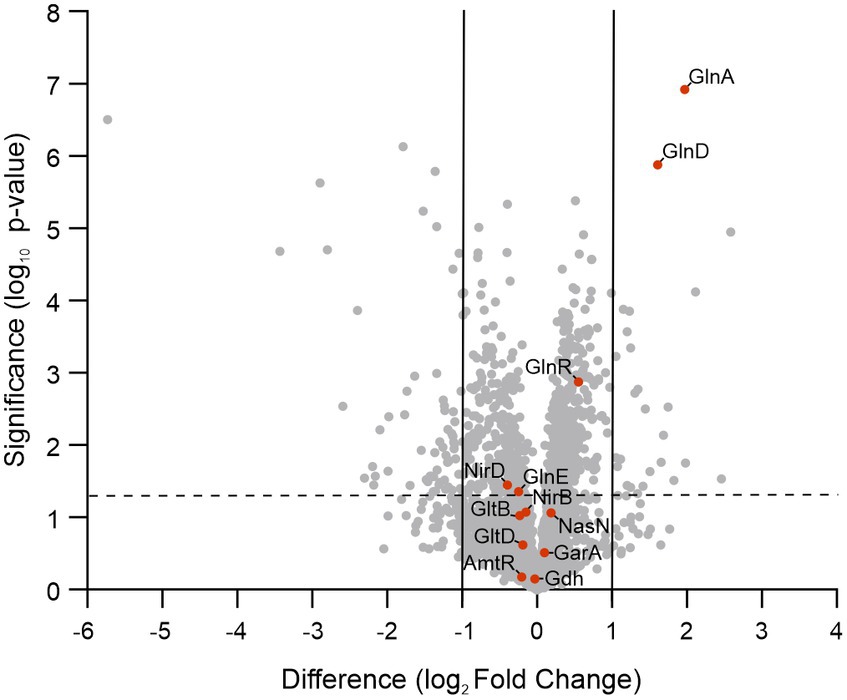
Figure 8. Volcano plot of log2 fold changes for ΔMsPII versus wild-type strains when grown in 1 mM of nitrate. Orange dots correspond to nitrogen assimilation and metabolism-related proteins. The vertical gray lines represent the log2 of the fold change of −2 and 2. The dotted line is the log10 of a p-value = 0.05.
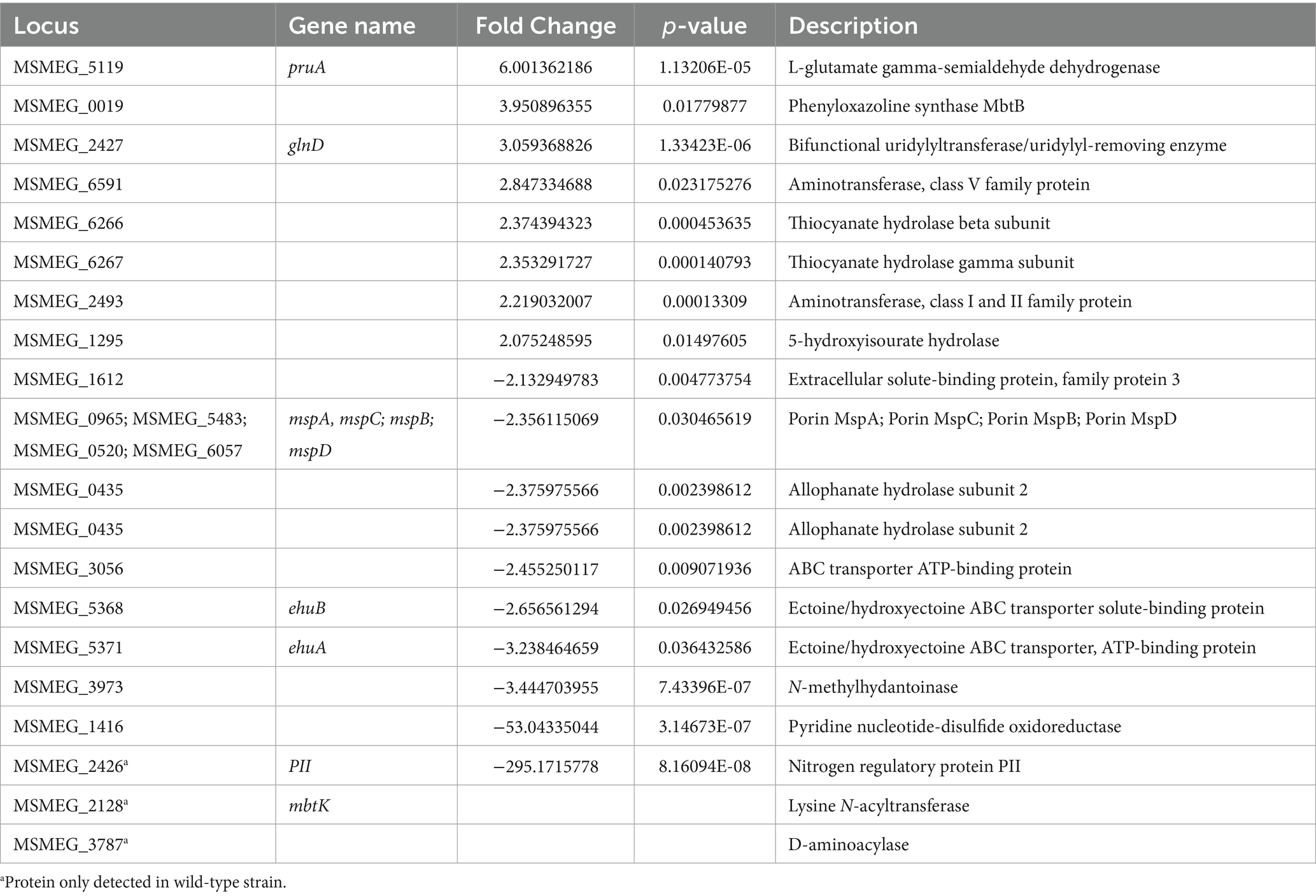
Table 2. Proteins involved in nitrogen metabolism exhibit differential expression during growth in 1 mM KNO3 in the M. smegmatis ΔMsPII strain in comparison with the wild-type.
The enrichment analysis using a Fisher’s exact test1 (Ge et al., 2018) of the significantly changed proteins screened from the LC–MS/MS showed that proteins involved in ammonium assimilation pathways were increased, including GlnA and GlnD, as well as proline and arginine metabolism (Supplementary Figure S4). On the other hand, many transmembrane transporters, including putative amino acid ABC transporters, were reduced. Surprisingly, one of these was the Mce4 transport system involved in cholesterol uptake (García-Fernández et al., 2017; Rank et al., 2021), and which is considered a virulence factor in M. tb. In addition, the expression of proteins related to the stress response was also altered.
In many bacteria and Archaea, the PII protein named GlnK, interacts with the ammonia channel AmtB to modulate its function (Thomas et al., 2000; Coutts and Thomas, 2002; Sant'Anna et al., 2009). When the cellular nitrogen status increases, the GlnK-AmtB binding promotes a block of ammonia transport into the cell (Coutts and Thomas, 2002; Zheng et al., 2004; Conroy et al., 2007). Regulation of AmtB activity seems to be the archetypical function of the GlnK protein, also these genes form a conserved glnK-amtB operon in a range of prokaryotes.
Based on both synteny and protein crystal structure analyses, it has been previously proposed that M. tb GlnB may in fact share an analogous function to E. coli GlnK and not GlnB (Shetty et al., 2010). However, the precise function of PII in mycobacteria has not been studied at a biochemical level.
To verify if M. tb PII function as GlnK interacting with Amt, we analyzed the presence of PII on the membrane and cytoplasmic fractions by SDS-PAGE and Western blotting. Cells were grown in ammonium–limiting conditions for 28 h. An aliquot of the culture was incubated in 30 mM of ammonium for 15 min. The membranes were isolated by centrifugation and washed in the presence of ATP, ADP, or ATP and 2-OG. PII was only present in the cytoplasmic fraction in the ammonium-limiting condition. In contrast, after the ammonium shock, PII was found in both the cytoplasmic and membrane fractions, indicating that PII associates with the membrane in an ammonium-dependent manner. This association was weakened by the presence of the PII protein allosteric effectors ATP and 2-OG (Figure 9A).
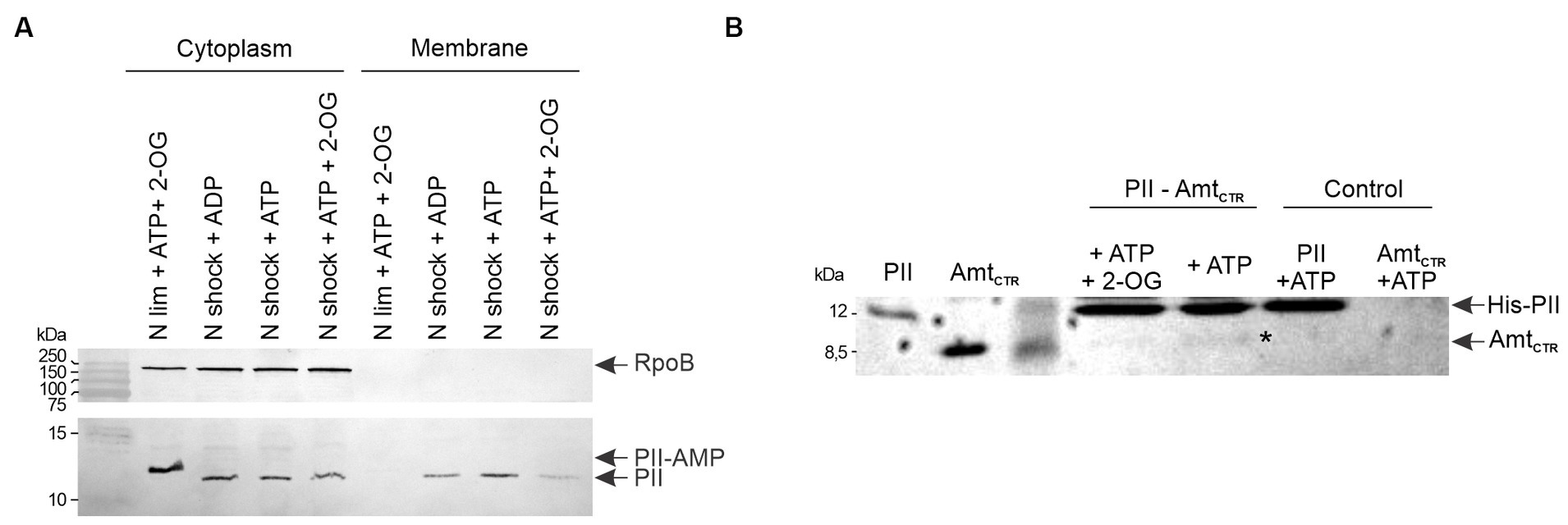
Figure 9. Interaction between PII and Amt, and effect of ammonium concentration on PII cellular localization. (A) The cellular localization profiles of PII in cell-free extracts from wild-type M. smegmatis were analyzed after an ammonium shock (NH4+), in the presence of the PII allosteric effectors, as indicated: 4.5 mM ATP, 0.6 mM ADP and/or 1.5 mM 2-OG. Cytoplasmic and membrane fractions were subjected to SDS-PAGE followed by Western blotting with anti-M. tb PII and RpoB antibodies. (B) The interaction of M. tb PII with the C-terminal cytosolic region of Amt (AmtCTR) was analyzed by a pull-down assay. Purified His-PII was mixed with purified Amt from M. tb. Proteins eluted from the Ni2+ resin were analyzed by SDS-PAGE and the gel was stained with Coomassie Blue.
To determine if the association with the membrane was through Amt transporter, we performed a pull-down assay in which purified M. tb His6-PII was mixed with a Ni-NTA resin (Novagen) and incubated with purified C-terminal Amt (AmtCTR), without tags, for 30 min. After three washes with buffer containing different combination of the PII protein allosteric effectors Mg-ATP and 2-OG, the elutions were analyzed by SDS-PAGE. Figure 9B shows that PII is able to interact with AmtCTR being negatively modulated by 2-OG, in agreement with the PII cellular localization analysis (Figure 9A). These data confirm that M. tb GlnB plays an ancestral role as a GlnK protein, in addition to its function as a modulator of nitrite assimilation.
Structural and biochemical investigations have been conducted on the PII proteins of mycobacteria M. tb and M. smegmatis; however, their specific functions remain elusive. A critical aspect in unraveling this role consists in elucidating their capacity to perceive nitrogen, carbon, and energy status. Like in other actinomycetes, it has been demonstrated that mycobacterial PII is adenylated upon induction by ammonium starvation (Williams et al., 2013). We verified that this modification is rapidly reversible in response to changes in ammonium levels in M. smegmatis (Figure 1). Moreover, we observed that PII can be localized at the membrane following an ammonium shock; with the PII membrane interaction being regulated by 2-OG (Figure 9A). This result is consistent with the PII: AmtB interaction that modulates the incorporation of ammonium through the AmtB channel. When ammonium is scarce, GlnK binds the allosteric effectors ATP and 2-OG and GlnK remains located in the cytosol allowing full AmtB activity. Conversely, when ammonium becomes available, GlnK binds the allosteric effector ADP and migrates to the cell membrane to obstruct the AmtB transporter via the interaction with the GlnK T-loop (Coutts and Thomas, 2002; Zheng et al., 2004; Conroy et al., 2007). The hypothesis the mycobacterial PII regulates Amt activity is reinforced by the genomic context of the PII gene (amt-PII-glnD), the conservation of amino acid residues involved in the interaction between GlnK and AmtB in E. coli, in both M. tb and M. smegmatis (Conroy et al., 2007) and the resolution of M. tb PII crystal structure (Shetty et al., 2010). Also, a homology model based on the crystal structure of the E. coli GlnK: AmtB complex showed that M. tb PII T-loop could engage in complex formation with AmtB (Shetty et al., 2010). Even though we were unable to heterologously express M. tb Amt in the soluble fraction of E. coli, we could observe an interaction between M. tb PII and the cytosolic C-terminal domain of Amt (Figure 9B). Previous studies have also reported that Ec-GlnK shows a reduced interaction with the C-terminal truncated version of AmtB (Coutts and Thomas, 2002). Taken together, these data provide strong evidence that the mycobacteria PII protein retained the archetypical GlnK-like function being able to regulate Amt activity by physical interaction with the Amt C-terminal domain, upon an ammonium shock.
Acetyl-CoA carboxylase complexes are also targets of PII regulation in many organisms (Feria Bourrellier et al., 2010; Gerhardt et al., 2015; Hauf et al., 2016; Rodrigues et al., 2019; Wang et al., 2021). We found that M. tb PII weakly interacts with the ACCase subunit AccA3, which contains the BCCP domain. However, this weak interaction did not affect ACC or PCC activities of the main complexes ACCases 5 and 6. As shown in Figure 2A, AccA3 from M. tb can bind GlnB protein from other species. This interaction seems stronger with GlnB from A. brasiliensis and it proved to inhibit its PCC activity (Supplementary Figure S3). In E. coli, both GlnB and GlnK, can interact with the BCCP subunit of the ACC complex, however, only GlnB regulates ACC activity in vitro (Gerhardt et al., 2015). These data support that the GlnB-like PII evolved to regulate fatty acid biosynthesis in agreement with a recent report from Streptomyces hygroscopicus var. ascomyceticus (Wang et al., 2021). This particular actinomycete encodes two PII proteins, named Sh-GlnK and Sh-GlnB. However, only Sh-GlnB has the ability to modulate ACCase activity in this microorganism. The fact that mycobacteria PII was not able to regulate ACC/PCC activities also suggests that it evolved to act as a GlnK-type of PII.
To gain deeper insights into the role of PII in mycobacteria, we decided to construct KO mutants. A high-throughput analysis carried out by transposon site hybridization (TraSH) indicated that M. tb glnB gene is not essential (Sassetti et al., 2003); however, after several attempts, we were unable to construct a deletion mutant in H37Rv strain. Similarly, Read et al. (2007) also failed to obtain a glnB mutant strain. However, we successfully obtained a PII mutant strain in M. smegmatis (ΔMsPII) by double homologous recombination, which presented normal growth in a media supplemented with low concentrations of ammonium. This result suggests that PII must not play an essential role in adaptation to low ammonium levels. In addition, the ΔMsPII strain did not present any growth difference in media with increased levels of nitrogen, when compared with the wild-type strain, indicating that the regulation of the Amt channel is not required for bacteria survival (Supplementary Figure S5).
The ΔMsPII mutant strain was also exposed to various organic and inorganic nitrogen sources, exhibiting a slight reduction of optical densities when glutamate or asparagine were used as sole nitrogen sources (Supplementary Figure S6). The most prominent defect of the ΔMsPII strain was the impairment to grow when nitrate or nitrite were used as the sole nitrogen source, primarily characterized by extended lag phases (Figure 5). Unlike the wild-type strain, nitrite uptake was not inhibited by ammonium in the ΔMsPII strain (Figure 6). This strain also presented higher nitrate reductase activity (Figure 7) and increased levels of glutamine synthetase when nitrate was the sole nitrogen source. These behaviors resemble those reported in cyanobacteria PII mutant strains (Forchhammer and De Marsac, 1995; Lee et al., 1998; Kobayashi et al., 2005). In photosynthetic bacteria, ammonium is the preferred nitrogen source and it negatively regulates the assimilation of alternative nitrogen sources, such as nitrate or nitrite, via the PII protein. A PII-null mutant strain of Synechococcus sp. PCC 7942 exhibited a slight decrease in growth rate when exposed to nitrate or glutamine. This was accompanied by an increase in nitrate reductase activity compared to the wild-type, along with elevated levels of glnA expression at both transcript and protein levels (Forchhammer and De Marsac, 1995). Glutamine synthetase accumulation was also observed in an E. coli mutant lacking GlnB (Bueno et al., 1985), regardless of the nitrogen status. In both, E. coli and cyanobacteria glutamine synthetase and E. coli’s glnD expression depends on transcriptional regulators, whose activity is indirectly modulated by GlnB (Jiang and Ninfa, 1999; Espinosa et al., 2006, 2007). In the case of M. smegmatis, glnD and glnA are under GlnR control (Jenkins et al., 2013; Jeßberger et al., 2013), but little is known about how GlnR is activated. Overexpression of GlnD and GlnA in the ΔMsPII strain could be the result of an attempt to compensate for the absence of PII. However, in the ΔMsPII mutant, only a subset of genes corresponding to the GlnR operon exhibited altered protein levels. In addition, in the M. tuberculosis’ glnD-null mutant, the expression of the amt-glnB-glnD operon is upregulated under nitrogen deficiency conditions (Read et al., 2007). Therefore, the possibility that PII is involved in the transcription regulation of these genes, through its interaction with as-yet-undetermined targets, cannot be ruled out.
In contrast to the M. smegmatis ΔMsPII, for Synechococcus sp. PCC 7942, nitrate-cultured mutant cells exhibited an exacerbated nitrite excretion. The PII-null mutant also displayed normal nitrite uptake activity at pH 7.2, but high assimilation of nitrate even in the presence of ammonium at high pH (9.6), bypassing the ammonium repression. Consequently, the modulation of the nitrate/nitrite uptake in M. smegmatis could potentially be attributed to an effect of PII on ABC transporters, as was observed for other systems (Watzer et al., 2019). However, the fact that the ammonium-inhibition takes longer to occur in the mutant strain at pH 7.2 suggests additional regulatory points in the process of nitrite assimilation that may also be under the influence of PII.
In M. smegmatis, NarK and NarK3 were designed to be involved in nitrate and nitrite transport (Amon et al., 2009). Nevertheless, there are numerous putative subunits for ABC transporters that are indicated to be involved in nitrate/nitrite assimilation. Further research should be carried out to thoroughly understand this mechanism. It may be interesting to analyze whether the role of PII in nitrate/nitrite assimilation is replicated in M. tuberculosis, considering that nitrate has a relevant role in virulence (Gouzy et al., 2013). Also, the PII-null mutant strain resulted defective in replication during the invasion of mouse macrophages (Rengarajan et al., 2005).
Altogether, the PII proteins of mycobacteria maintain their role as modulators of ammonium entry, although this function does not seem to be essential for the organism. However, their absence showed negative effects when cells are grown in nitrate or nitrite, indicating that PII is involved in the uptake or metabolization of these alternative nitrogen sources, as we indicate in a proposed model (Figure 10). It is worth noting the flexibility of PII proteins to regulate different groups of proteins depending on the specie, making in some cases the distinction between GlnK and GlnB types to be blurry.
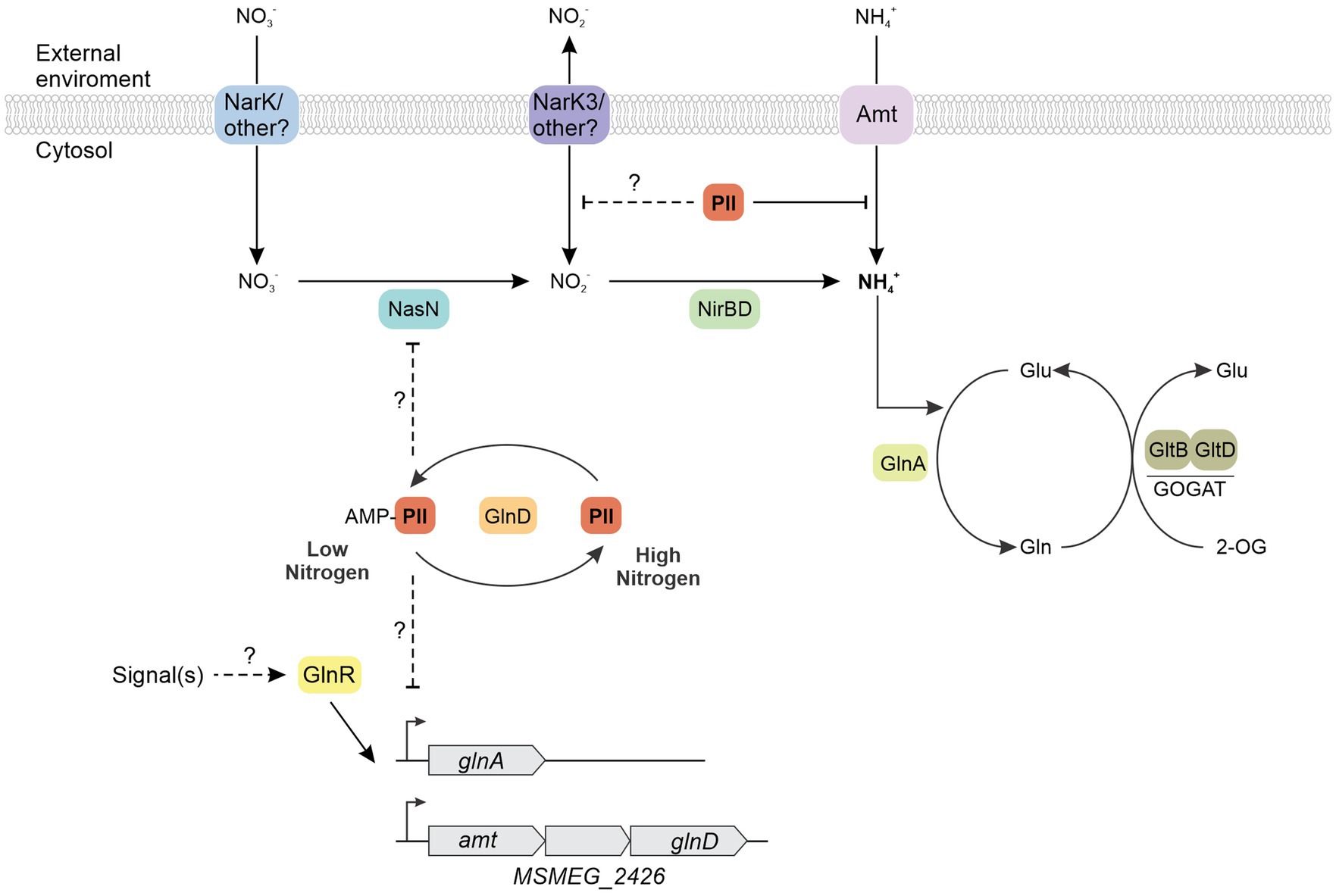
Figure 10. Proposed model about physiological roles of PII protein in mycobacteria. The putative ammonium transporter Amt mediates the ammonium (NH4+) enter to the bacterial cytosol in a low nitrogen regimen. The glutamine synthetase GlnA assimilates the ammonium to synthesize glutamine, which serves as the substrate for the glutamine oxoglutarate aminotransferase (GOGAT) in the synthesis of glutamate. The bacteria sense nitrogen status using the 2-OG/glutamine ratio. When ammonium is scarce, PII binds the allosteric effectors ATP and 2-OG and it is adenylated. PII-AMP remains located in the cytosol allowing full Amt activity. Conversely, when ammonium becomes available, PII is deadenylated and binds the allosteric effector ADP, migrating to the cell membrane and obstructing the Amt transporter via the interaction with the PII T-loop. The absence of PII, like in the ΔMsPII mutant strain, produces a growth defect when nitrate or nitrite were used as the sole nitrogen source. The ΔMsPII mutant strain presented higher nitrate reductase activity and increased levels of glutamine synthetase when nitrate was the sole nitrogen source.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifier PXD049128 and 10.6019/PXD049128.
DE: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft. EG: Formal analysis, Writing – review & editing, Investigation, Methodology. LR: Investigation, Formal analysis, Writing – review & editing, Methodology. LH: Conceptualization, Data curation, Formal analysis, Supervision, Writing – review & editing. HG: Conceptualization, Data curation, Formal analysis, Supervision, Writing – review & editing, Funding acquisition. LD: Conceptualization, Data curation, Formal analysis, Funding acquisition, Supervision, Writing – review & editing, Project administration, Resources, Validation, Visualization, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Support to LD from ANPCyT (grant P. BID - PICT 2019-03487) is acknowledged. Support to EG and LH from CNPq and CAPES are acknowledged.
We thank Germán Rosano and Lic. Alejo Cantoia from the Mass Spectrometry Unit (UEM) of the Institute of Molecular and Cellular Biology of Rosario (UEM-IBR), Argentina, for the LC–MS/MS analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1366111/full#supplementary-material
1. ^ShinyGO 0.77: http://bioinformatics.sdstate.edu/go/
Akhtar, S., Khan, A., Sohaskey, C. D., Jagannath, C., and Sarkar, D. (2013). Nitrite reductase NirBD is induced and plays an important role during in vitro dormancy of Mycobacterium tuberculosis. J. Bacteriol. 195, 4592–4599. doi: 10.1128/JB.00698-13
Amon, J., Bräu, T., Grimrath, A., Hänßler, E., Hasselt, K., Höller, M., et al. (2008). Nitrogen control in Mycobacterium smegmatis: nitrogen-dependent expression of ammonium transport and assimilation proteins depends on the OmpR-type regulator GlnR. J. Bacteriol. 190, 7108–7116. doi: 10.1128/JB.00855-08
Amon, J., Titgemeyer, F., and Burkovski, A. (2009). A genomic view on nitrogen metabolism and nitrogen control in mycobacteria. J. Mol. Microbiol. Biotechnol. 17, 20–29. doi: 10.1159/000159195
Antczak, M., Płocińska, R., Płociński, P., Rumijowska-Galewicz, A., Żaczek, A., Strapagiel, D., et al. (2018). The NnaR orphan response regulator is essential for the utilization of nitrate and nitrite as sole nitrogen sources in mycobacteria. Sci. Rep. 8, 17552–17515. doi: 10.1038/s41598-018-35844-z
Arcondéguy, T., Jack, R., and Merrick, M. (2001). P II signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65, 80–105. doi: 10.1128/mmbr.65.1.80-105.2001
Atkinson, M. R., Kamberov, E. S., Weiss, R. L., and Ninfa, A. J. (1994). Reversible uridylylation of the Escherichia coli PII signal transduction protein regulates its ability to stimulate the dephosphorylation of the transcription factor nitrogen regulator I (NRI or NtrC). J. Biol. Chem. 269, 28288–28293. doi: 10.1016/s0021-9258(18)46926-8
Bandyopadhyay, A., Arora, A., Jain, S., Laskar, A., Mandal, C., Ivanisenko, V. A., et al. (2010). Expression and molecular characterization of the Mycobacterium tuberculosis PII protein. J. Biochem. 147, 279–289. doi: 10.1093/jb/mvp174
Bazet Lyonnet, B., Diacovich, L., Cabruja, M., Bardou, F., Quémard, A., Gago, G., et al. (2014). Pleiotropic effect of AccD5 and AccE5 depletion in acyl-coenzyme a carboxylase activity and in lipid biosynthesis in mycobacteria. PLoS One 9:e99853. doi: 10.1371/journal.pone.0099853
Beyer, H. M., Gonschorek, P., Samodelov, S. L., Meier, M., Weber, W., and Zurbriggen, M. D. (2015). AQUA cloning: a versatile and simple enzyme-free cloning approach. PLoS One 10, 1–20. doi: 10.1371/journal.pone.0137652
Bonatto, A. C., Couto, G. H., Souza, E. M., Araújo, L. M., Pedrosa, F. O., Noindorf, L., et al. (2007). Purification and characterization of the bifunctional uridylyltransferase and the signal transducing proteins GlnB and GlnK from Herbaspirillum seropedicae. Protein Expr. Purif. 55, 293–299. doi: 10.1016/j.pep.2007.04.012
Bueno, R., Pahel, G., and Magasanik, B. (1985). Role of glnB and glnD gene products in regulation of the glnALG operon of Escherichia coli. J. Bacteriol. 164, 816–822. doi: 10.1128/jb.164.2.816-822.1985
Burnette, N. W. (1981). “Western blotting”: electrophoretic transfer of preoteins from sodium dodecyl sulfate-polyaclylamide gels to unmodified nitrocellulose and readiographic detection with antibody and radioiodinated protein. Rev. Sci. Instrum. 112, 195–203. doi: 10.1063/1.4897167
Cardoso, N. C., Papadopoulos, A. O., and Kana, B. D. (2021). Mycobacterium smegmatis does not display functional redundancy in nitrate reductase enzymes. PLoS One 16, 1–14. doi: 10.1371/journal.pone.0245745
Chapman-Smith, A., Turner, D. L., Cronan, J. E., Morris, T. W., and Wallace, J. C. (1994). Expression, biotinylation and purification of a biotin-domain peptide from the biotin carboxy carrier protein of Escherichia coli acetyl-CoA carboxylase. Biochem. J. 302, 881–887
Connell, N. D. (1994). Mycobacterium: transformation, isolation, and mutant selection. Methods Cell Biol. 45, 107–125. doi: 10.1016/s0091-679x(08)61848-8
Conroy, M. J., Durand, A., Lupo, D., Li, X. D., Bullough, P. A., Winkler, F. K., et al. (2007). The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. U. S. A. 104, 1213–1218. doi: 10.1073/pnas.0610348104
Coutts, G., and Thomas, G. (2002). Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 21, 536–545. doi: 10.1093/emboj/21.4.536
Crotta Asis, A., Savoretti, F., Cabruja, M., Gramajo, H., and Gago, G. (2021). Characterization of key enzymes involved in triacylglycerol biosynthesis in mycobacteria. Sci. Rep. 11, 13257–13213. doi: 10.1038/s41598-021-92721-y
Daniel, J., Oh, T. J., Lee, C. M., and Kolattukudy, P. E. (2007). AccD6, a member of the Fas II locus, is a functional carboxyltransferase subunit of the acyl-coenzyme a carboxylase in Mycobacterium tuberculosis. J. Bacteriol. 189, 911–917. doi: 10.1128/JB.01019-06
Diacovich, L., Peirú, S., Kurth, D., Rodríguez, E., Podestá, F., Khosla, C., et al. (2002). Kinetic and structural analysis of a new group of acyl-CoA carboxylases found in Streptomyces coelicolor A3(2). J. Biol. Chem. 277, 31228–31236. doi: 10.1074/jbc.M203263200
Durand, A., and Merrick, M. (2006). In vitro analysis of the Escherichia coli AmtB-GlnK complex reveals a stoichiometric interaction and sensitivity to ATP and 2-oxoglutarate. J. Biol. Chem. 281, 29558–29567. doi: 10.1074/jbc.M602477200
Espinosa, J., Forchhammer, K., Burillo, S., and Contreras, A. (2006). Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 61, 457–469. doi: 10.1111/j.1365-2958.2006.05231.x
Espinosa, J., Forchhammer, K., and Contreras, A. (2007). Role of the Synechococcus PCC 7942 nitrogen regulator protein PipX in NtcA-controlled processes. Microbiology 153, 711–718. doi: 10.1099/mic.0.2006/003574-0
Feria Bourrellier, A. B., Valot, B., Guillot, A., Ambard-Bretteville, F., Vidal, J., and Hodges, M. (2010). Chloroplast acetyl-CoA carboxylase activity is 2-oxoglutarate-regulated by interaction of PII with the biotin carboxyl carrier subunit. Proc. Natl. Acad. Sci. U. S. A. 107, 502–507. doi: 10.1073/pnas.0910097107
Forchhammer, K., and De Marsac, N. T. (1995). Functional analysis of the phosphoprotein P(II) (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 177, 2033–2040. doi: 10.1128/jb.177.8.2033-2040.1995
Forchhammer, K., Selim, K. A., and Huergo, L. F. (2022). New views on PII signaling: from nitrogen sensing to global metabolic control. Trends Microbiol. 30, 722–735. doi: 10.1016/j.tim.2021.12.014
Gago, G., Kurth, D., Diacovich, L., Tsai, S., and Gramajo, H. (2006). Biochemical and structural characterization of an essential acyl coenzyme a carboxylase from Mycobacterium tuberculosis. J. Bacteriol. 188, 477–486. doi: 10.1128/JB.188.2.477
García-Fernández, J., Papavinasasundaram, K., Galán, B., Sassetti, C. M., and García, J. L. (2017). Molecular and functional analysis of the mce4 operon in Mycobacterium smegmatis. Environ. Microbiol. 19, 3689–3699. doi: 10.1111/1462-2920.13869
Gerhardt, E. C. M., Moure, V. R., Souza, A. W., Pedrosa, F. O., Souza, E. M., Diacovich, L., et al. (2017). Expression and purification of untagged Glnk proteins from actinobacteria. EXCLI J. 16, 949–958. doi: 10.17179/excli2017-394
Gerhardt, E. C. M., Rodrigues, T. E., Müller-Santos, M., Pedrosa, F. O., Souza, E. M., Forchhammer, K., et al. (2015). The bacterial signal transduction protein GlnB regulates the committed step in fatty acid biosynthesis by acting as a dissociable regulatory subunit of acetyl-CoA carboxylase. Mol. Microbiol. 95, 1025–1035. doi: 10.1111/mmi.12912
Ge, S. X., Jung, D., Jung, D., and Yao, R. (2018). ShinyGO: a graphical enrichment tool for animals and plants. bioRxiv 36:315150. doi: 10.1101/082511.Lai
Gouzy, A., Larrouy-Maumus, G., Di Wu, T., Peixoto, A., Levillain, F., Lugo-Villarino, G., et al. (2013). Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate. Nat. Chem. Biol. 9, 674–676. doi: 10.1038/nchembio.1355
Gouzy, A., Poquet, Y., and Neyrolles, O. (2014). Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence. Nat. Rev. Microbiol. 12, 729–737. doi: 10.1038/nrmicro3349
Grzegorzewicz, A. E., Hess, T., Jones, V., Gruppo, V., Born, S. E. M., Chavadi, S. S., et al. (2012). Inhibition of Mycolic Acid Transport Across the Mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol. 8, 334–341. doi: 10.1038/nchembio.794.INHIBITION
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Hauf, W., Schmid, K., Gerhardt, E. C. M., Huergo, L. F., and Forchhammer, K. (2016). Interaction of the nitrogen regulatory protein GlnB (PII) with biotin carboxyl carrier protein (BCCP) controls acetyl-Coa levels in the Cyanobacterium synechocystis sp. PCC 6803. Front. Microbiol. 7, 1–14. doi: 10.3389/fmicb.2016.01700
Hesketh, A., Fink, D., Gust, B., Rexer, H. U., Scheel, B., Chater, K., et al. (2002). The GlnD and GlnK homologues of Streptomyces coelicolor A3(2) are functionally dissimilar to their nitrogen regulatory system counterparts from enteric bacteria. Mol. Microbiol. 46, 319–330. doi: 10.1046/j.1365-2958.2002.03149.x
Huergo, L. F., Chandra, G., and Merrick, M. (2013). PII signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol. Rev. 37, 251–283. doi: 10.1111/j.1574-6976.2012.00351.x
Huergo, L. F., Filipaki, A., Chubatsu, L. S., Yates, M. G., Steffens, M. B., Pedrosa, F. O., et al. (2005). Effect of the over-expression of PII and PZ proteins on the nitrogenase activity of Azospirillum brasilense. FEMS Microbiol. Lett. 253, 47–54. doi: 10.1016/j.femsle.2005.09.026
Jenkins, V. A., Barton, G. R., Robertson, B. D., and Williams, K. J. (2013). Genome wide analysis of the complete GlnR nitrogen-response regulon in Mycobacterium smegmatis. BMC Genomics 14:1. doi: 10.1186/1471-2164-14-301
Jeßberger, N., Lu, Y., Amon, J., Titgemeyer, F., Sonnewald, S., Reid, S., et al. (2013). Nitrogen starvation-induced transcriptome alterations and influence of transcription regulator mutants in Mycobacterium smegmatis. BMC. Res. Notes 6:482. doi: 10.1186/1756-0500-6-482
Jiang, P., and Ninfa, A. J. (1999). Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J. Bacteriol. 181, 1906–1911. doi: 10.1128/JB.181.6.1906-1911.1999
Kobayashi, M., Takatani, N., Tanigawa, M., and Omata, T. (2005). Posttranslational regulation of nitrate assimilation in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 187, 498–506. doi: 10.1128/JB.187.2.498-506.2005
Kurth, D. G., Gago, G. M., de la Iglesia, A., Lyonnet, B. B., Lin, T. W., Morbidoni, H. R., et al. (2009). ACCase 6 is the essential acetyl-CoA carboxylase involved in fatty acid and mycolic acid biosynthesis in mycobacteria. Microbiology 155, 2664–2675. doi: 10.1099/mic.0.027714-0
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Lee, H. M., Flores, E., Herrero, A., Houmard, J., and Tandeau De Marsac, N. (1998). A role for the signal transduction protein P(II) in the control of nitrate/nitrite uptake in a Cyanobacterium. FEBS Lett. 427, 291–295. doi: 10.1016/S0014-5793(98)00451-7
Macpherson, K. H. R., Xu, Y., Cheah, E., Carr, P. D., van Heeswijk, W. C., Westerhoff, H. V., et al. (1998). Crystallization and preliminary X-ray analysis of Escherichia coli GlnK. Acta Crystallogr. Sect. D Biol. Crystallogr. 54, 996–998. doi: 10.1107/S0907444998001887
Malm, S., Tiffert, Y., Micklinghoff, J., Schultze, S., Joost, I., Weber, I., et al. (2009). The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology 155, 1332–1339. doi: 10.1099/mic.0.023275-0
Morozkina, E. V., and Zvyagilskaya, R. A. (2007). Nitrate reductases: structure, functions, and effect of stress factors. Biochemist 72, 1151–1160. doi: 10.1134/S0006297907100124
Moure, V. R., Razzera, G., Araújo, L. M., Oliveira, M. A. S., Gerhardt, E. C. M., Müller-Santos, M., et al. (2012). Heat stability of Proteobacterial P II protein facilitate purification using a single chromatography step. Protein Expr. Purif. 81, 83–88. doi: 10.1016/j.pep.2011.09.008
Nikolau, B. J., Wurtele, E. S., and Stumpf, P. K. (1985). Use of streptavidin to detect biotin-containing proteins in plants. Anal. Biochem. 149, 448–453. doi: 10.1016/0003-2697(85)90596-2
Ninfa, A. J., and Atkinson, M. R. (2000). PII signal transduction proteins. Trends Microbiol. 8, 172–179. doi: 10.1016/S0966-842X(00)01709-1
Pawelczyk, J., Brzostek, A., Kremer, L., Dziadek, B., Rumijowska-Galewicz, A., Fiolka, M., et al. (2011). Accd6, a key carboxyltransferase essential for mycolic acid synthesis in Mycobacterium tuberculosis, is dispensable in a nonpathogenic strain. J. Bacteriol. 193, 6960–6972. doi: 10.1128/JB.05638-11
Pelicic, V., Jackson, M., Reyrat, J.-M., Jacobs, W. R. J., Gicquel, B., and Guilhot, C. (1997). Efficient allelic exchange and transposon mutagenesis in Mycobacterium avium by specialized transduction. Proc. Natl. Acad. Sci. USA 94, 10955–10960. doi: 10.1128/AEM.69.9.5039-5044.2003
Perez-Riverol, Y., Bai, J., Bandla, C., García-Seisdedos, D., Hewapathirana, S., Kamatchinathan, S., et al. (2022). The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552. doi: 10.1093/nar/gkab1038
Peter, G. (1879). Bemerkungen zu der abhandlung der H. H. Weselsky und Benedikt “Ueber einige azoverbindungen.”. Berichte Dtsch. Chem. Gesellschaft 12, 226–230. doi: 10.1002/cber.18790120165
Radchenko, M. V., Thornton, J., and Merrick, M. (2010). Control of AmtB-GlnK complex formation by intracellular levels of ATP, ADP, and 2-oxoglutarate. J. Biol. Chem. 285, 31037–31045. doi: 10.1074/jbc.M110.153908
Rajendran, C., Gerhardt, E. C. M., Bjelic, S., Gasperina, A., Scarduelli, M., Pedrosa, F. O., et al. (2011). Crystal structure of the GlnZ-DraG complex reveals a different form of P II-target interaction. Proc. Natl. Acad. Sci. U. S. A. 108, 18972–18976. doi: 10.1073/pnas.1108038108
Rank, L., Herring, L. E., and Braunstein, M. (2021). Evidence for the mycobacterial mce4 transporter being a multiprotein complex. J. Bacteriol. 203, 1–17. doi: 10.1128/JB.00685-20
Read, R., Pashley, C. A., Smith, D., and Parish, T. (2007). The role of GlnD in ammonia assimilation in Mycobacterium tuberculosis. Tuberculosis 87, 384–390. doi: 10.1016/j.tube.2006.12.003
Rengarajan, J., Bloom, B. R., and Rubin, E. J. (2005). Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 102, 8327–8332. doi: 10.1073/pnas.0503272102
Rodrigues, T. E., Gerhardt, E. C. M., Oliveira, M. A., Chubatsu, L. S., Pedrosa, F. O., Souza, E. M., et al. (2014). Search for novel targets of the PII signal transduction protein in Bacteria identifies the BCCP component of acetyl-CoA carboxylase as a PII binding partner. Mol. Microbiol. 91, 751–761. doi: 10.1111/mmi.12493
Rodrigues, T. E., Sassaki, G. L., Valdameri, G., Pedrosa, F. O., Souza, E. M., and Huergo, L. F. (2019). Fatty acid biosynthesis is enhanced in Escherichia coli strains with deletion in genes encoding the PII signaling proteins. Arch. Microbiol. 201, 209–214. doi: 10.1007/s00203-018-1603-2
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular cloning: a laboratory manual. 3rd. Nueva York: Cold Spring Harbor Laboratory Press.
Sant'Anna, F. H., Trentini, D. B., de Souto Weber, S., Cecagno, R., da Silva, S. C., and Schrank, I. S. (2009). The PII superfamily revised: a novel group and evolutionary insights. J. Mol. Evol. 68, 322–336. doi: 10.1007/s00239-009-9209-6
Sassetti, C. M., Boyd, D. H., and Rubin, E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84. doi: 10.1046/j.1365-2958.2003.03425.x
Shetty, N. D., Reddy, M. C. M., Palaninathan, S. K., Owen, J. L., and Sacchettini, J. C. (2010). Crystal structures of the apo and ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein. Protein Sci. 19, 1513–1524. doi: 10.1002/pro.430
Snapper, S. B., Melton, R. E., Mustafa, S., Kieser, T., and Jacobs, W. R. (1990). Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4, 1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x
Son, H. S., and Rhee, S. G. (1987). Cascade control of Escherichia coli glutamine synthetase. Purification and properties of PII protein and nucleotide sequence of its structural gene. J. Biol. Chem. 262, 8690–8695. doi: 10.1016/S0021-9258(18)47469-8
Strösser, J., Lüdke, A., Schaffer, S., Krämer, R., and Burkovski, A. (2004). Regulation of GlnK activity: modification, membrane sequestration and proteolysis as regulatory principles in the network of nitrogen control in Corynebacterium glutamicum. Mol. Microbiol. 54, 132–147. doi: 10.1111/j.1365-2958.2004.04247.x
Studier, F. W., and Moffatt, B. A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130. doi: 10.1016/0022-2836(86)90385-2
Tan, W., Liao, T. H., Wang, J., Ye, Y., Wei, Y. C., Zhou, H. K., et al. (2020). A recently evolved diflavin-containing monomeric nitrate reductase is responsible for highly efficient bacterial nitrate assimilation. J. Biol. Chem. 295, 5051–5066. doi: 10.1074/jbc.RA120.012859
Thomas, G., Coutts, G., and Merrick, M. (2000). The glnKamtB operon. A conserved gene pair in prokaryotes. Trends Genet. 16, 11–14. doi: 10.1016/S0168-9525(99)01887-9
Wang, P., Wang, X., Yin, Y., He, M., Tan, W., Gao, W., et al. (2021). Increasing the Ascomycin yield by relieving the inhibition of acetyl/Propionyl-CoA carboxylase by the signal transduction protein GlnB. Front. Microbiol. 12, 1–14. doi: 10.3389/fmicb.2021.684193
Watzer, B., Spät, P., Neumann, N., Koch, M., Sobotka, R., MacEk, B., et al. (2019). The signal transduction protein PII controls ammonium, nitrate and urea uptake in cyanobacteria. Front. Microbiol. 10, 1–20. doi: 10.3389/fmicb.2019.01428
Williams, K. J., Bennett, M. H., Barton, G. R., Jenkins, V. A., and Robertson, B. D. (2013). Adenylylation of mycobacterial Glnk (PII) protein is induced by nitrogen limitation. Tuberculosis 93, 198–206. doi: 10.1016/j.tube.2012.12.003
Williams, K. J., Jenkins, V. A., Barton, G. R., Bryant, W. A., Krishnan, N., and Robertson, B. D. (2015). Deciphering the metabolic response of Mycobacterium tuberculosis to nitrogen stress. Mol. Microbiol. 97, 1142–1157. doi: 10.1111/mmi.13091
Xu, Y., Cheah, E., Carr, P. D., van Heeswijk, W. C., Westerhoff, H. V., Vasudevan, S. G., et al. (1998). GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J. Mol. Biol. 282, 149–165. doi: 10.1006/jmbi.1998.1979
Keywords: mycobacteria, PII protein, nitrogen metabolism regulation, nitrate/nitrite assimilation, ammonium transport
Citation: Ensinck D, Gerhardt ECM, Rollan L, Huergo LF, Gramajo H and Diacovich L (2024) The PII protein interacts with the Amt ammonium transport and modulates nitrate/nitrite assimilation in mycobacteria. Front. Microbiol. 15:1366111. doi: 10.3389/fmicb.2024.1366111
Received: 05 January 2024; Accepted: 04 March 2024;
Published: 25 March 2024.
Edited by:
Robert Jansen, Radboud University, NetherlandsReviewed by:
Vineet Kumar, The University of Texas at Austin, United StatesCopyright © 2024 Ensinck, Gerhardt, Rollan, Huergo, Gramajo and Diacovich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lautaro Diacovich, ZGlhY292aWNoQGlici1jb25pY2V0Lmdvdi5hcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.