- 1College of Animal Science and Technology, Gansu Agricultural University, Lanzhou, China
- 2Gansu Key Laboratory of Animal Generational Physiology and Reproductive Regulation, Lanzhou, China
- 3Tianzhu County Animal Disease Prevention and Control Center, Wuwei, China
This study aimed to investigate the effect of prickly ash seeds (PAS) on the microbial community found in rumen microbes of Hu sheep by adding different percentages of prickly ash seeds and to carry out research on the relation between rumen flora and production performance. Twenty-seven male lambs of Hu sheep were classified into three groups based on the content of prickly ash seeds (PAS) fed for 90 days, i.e., 0%, 3%, and 6%. At the end of the feeding trial, rumen fluid samples were collected from six sheep in each group for 16S amplicon sequencing. The results showed that the addition of prickly ash seeds significantly increased both Chao1 and ACE indices (P < 0.05), and the differences between groups were greater than those within groups. The relative content of Bacteriodota decreased, and the relative content of Fusobacteriota, Proteobacteria, Acidobacteriota, and Euryarchaeota increased. The relative content of Papillibacter and Saccharofermentans was increased at the genus level, and the relative content of Bacteroides and Ruminococcus was decreased. The test group given 3% of prickly ash seeds was superior to the test group given 6% of prickly ash seeds. In addition, the addition of 3% of prickly ash seeds improved the metabolism or immunity of sheep. Fusobacteriota and Acidobacteriota were positively correlated with total weight, dressing percentage, and average daily gain (ADG) and negatively correlated with average daily feed intake (ADFI), feed-to-gain ratio (F/G), and lightness (L*). Methanobrevibacter and Saccharofermentans were positively correlated with ADG and negatively correlated with ADFI and L*. In conclusion, under the present experimental conditions, the addition of prickly ash seeds increased the abundance and diversity of rumen microorganisms in Hu sheep and changed the relative abundance of some genera. However, the addition of 6% prickly ash seeds may negatively affect the digestive and immune functions in sheep rumen.
1 Introduction
Complex and large microbial communities inhabit the rumen of ruminants and play a key role in ruminant health, growth performance, and immunity (Malmuthuge and Guan, 2017; Schären et al., 2018). Microorganisms in the rumen include four major groups: protozoa, fungi, archaea, and bacteria. Among the bacteria, Firmicutes and Bacteroidetes are the most abundant. Most of the species in Firmicutes are associated with carbohydrate metabolism (Fernando et al., 2010; Hart et al., 2018), whereas Bacteroidetes are mainly associated with starch digestion and metabolism (Jami and Mizrahi, 2012; Snelling and Wallace, 2017). In addition, there are other groups of bacteria such as Fibrobacterota, Gracilibacteria, Spirochaetota, and Euryarchaeota that work together to maintain and mutually regulate the stability and balance of the rumen's internal environment. It has been found that the composition of the microbial community in the rumen can be affected by various factors such as diet, environmental factors, species, sex, and age, of which diet composition, nutritional level, and feed intake are considered to be the key factors affecting the rumen microbial community. Henderson et al. measured rumen microbial samples from 35 countries and 32 animal species to assess whether they were influenced by diet, host, or geographic location and found that differences in microbial community composition were primarily attributable to diet, with a lesser influence of the host (Henderson et al., 2015). In addition, in long-term feeding trials, it was found that adding enzyme preparations, plant extracts, probiotics, herbal additives, and other substances to animal feed can improve the gastrointestinal health of livestock and poultry; enhance the body's immune system; promote the digestion and absorption of nutrients, which will have a positive impact on the growth of livestock and poultry; and reduce the cost of feeding to a certain extent (Mao et al., 2023; Wang et al., 2023a).
Prickly ash is a small perennial deciduous tree of the family Rutaceae. It has high economic value in terms of food and medicinal use and has become an important source of economy for farmers in some areas. Prickly ash seeds are the main byproduct of prickly ash production, accounting for approximately 60% of the total weight of prickly ash. Prickly ash seeds can be processed into prickly ash oil (an ideal edible vegetable oil) and can also be used as herbal medicine. Long-term studies have found that prickly ash seeds are rich in dietary fiber, melanin, polyphenols, flavonoids, quercetin, volatile oils, and other natural substances (Tang et al., 2014; Yang et al., 2014), which have better anti-inflammatory and antibacterial (Hou et al., 2021), antioxidant, antitumor, cholesterol-lowering, and immune-enhancing properties (Fortuoso et al., 2019; Zhong et al., 2022). In addition, the antimicrobial peptides of prickly ash seeds have antimicrobial activity, inhibiting Bacillus subtilis, Escherichia coli, Salmonella, and Staphylococcus aureus (Hou et al., 2019). Prickly ash seeds inhibit lung inflammation and tissue damage during asthma (Wang et al., 2016; Phuyal et al., 2020). Prickly ash seed oil extracted from prickly ash seeds inhibits osteoclastogenesis and anti-malignant melanoma (Pang et al., 2019; Zhang et al., 2022a). In recent years, a number of scholars have carried out studies using prickly ash seeds and their derivatives in animals. For example, the addition of prickly ash seeds to diets has the potential to enhance pork quality and growth performance (Song et al., 2020). Prickly ash essential oil can promote nutrient digestion and absorption as well as increase intestinal health in sheep, while a portion of the rumen microbiota was significantly associated with differential metabolite and enzyme activities (Zhang et al., 2022a,b).
With the development of modernization and large-scale farming, the concept of green and healthy farming has been proposed in animal husbandry. In addition, prickly ash seeds have the advantages of naturalness, non-resistance, and multifunctionality, making it an ideal green feed. As the raw materials are abundant, they do not need any processing, resulting in low economic cost. Previous experiments have shown that including prickly ash seeds in the diet can improve the beneficial bacteria in the intestinal tract of Hu sheep lambs, reduce harmful bacteria, reduce inflammation, and increase the immunity of sheep (Li et al., 2023a). Therefore, 16s rRNA sequencing technology was used in this study to compare the bacterial community structure and distribution in the rumen of sheep supplemented with 0%, 3%, and 6% prickly ash seeds and to analyze the correlation between rumen microorganisms and production traits. This study aimed to augment the limitation in studies on the effects of prickly ash seeds on the ruminal bacterial community and to find the key microorganisms affecting the production performance of sheep through a study on the correlation between rumen microbes and production traits so as to provide beneficial help for promoting the healthy growth of ruminants.
2 Materials and methods
2.1 Ethics statement
All experimental designs and procedures were approved by the Animal Care Committee for Animal Care and Experimental Procedures established by the Ministry of Science and Technology of the People's Republic of China (Approval No. 2006-398).
2.2 Animals and experimental design
Twenty-seven healthy 3-month-old Hu male sheep lambs (aged 90 ± 5 days; 25.66 ± 3.03 kg body weight) were selected from June to September 2022 at Dongwu Good Breeding Sheep Breeding Base, Dongxiang County (Linxia Prefecture, Gansu Province, China). They were randomly divided into three groups of nine lambs each. The three groups were supplemented with 0% (CK group), 3% (A group), and 6% (B group) of prickly ash seeds in the basal diet. At the end of the fattening test, six sheep in each group were randomly selected to determine production traits after slaughter, and rumen fluid was collected for 16S rRNA sequencing and analyzed for the correlation between the diversity of rumen flora and production traits.
2.3 Test diet
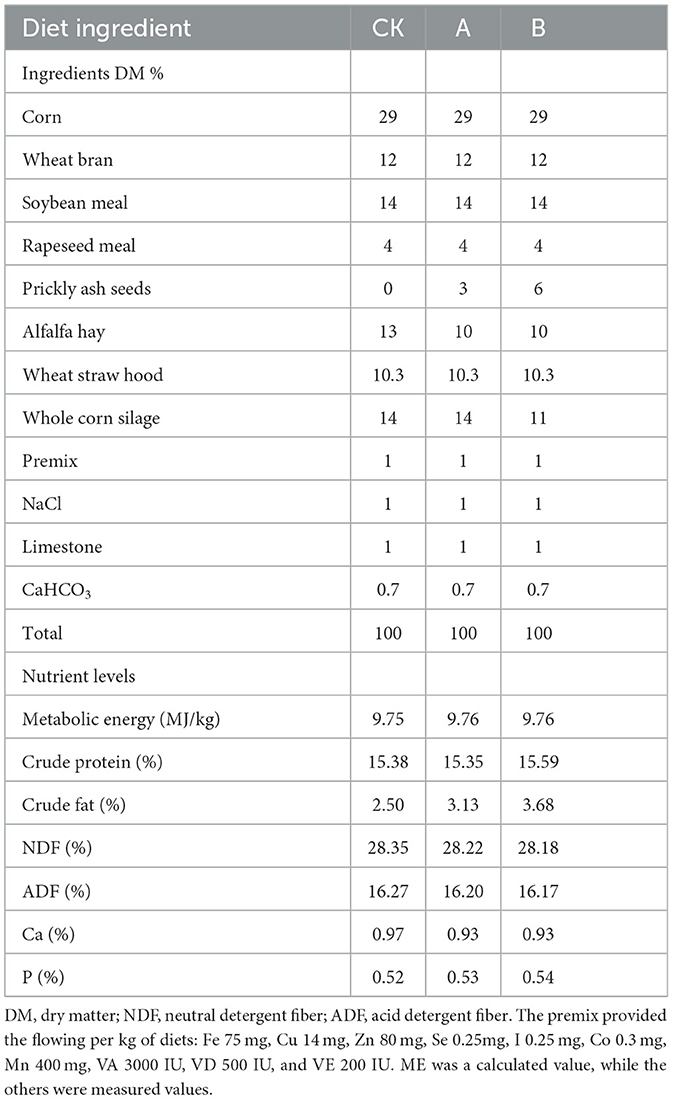
The basal diet was formulated with reference to the Nutritional Requirements of Chinese Meat Sheep and made into a total mixed ration (TMR). The composition and nutrient content of the experimental basal diet are shown in Table 1. Prickly ash seeds were purchased from the grain market in Guanghe County, and their conventional nutrient content (Supplementary Table S1) is given in Li et al. (2023a).
2.4 Feeding management
Before the start of the trial, the pen was thoroughly disinfected, the sheep were ear-tagged and vaccinated according to the procedure, and then each sheep was weighed and placed in pens. At the beginning of the trial, feeding was carried out at 07:00 am and 18:00 pm every day, during which the lambs had ad libitum access to food and drink, and the pens were disinfected regularly.
2.5 Sample collection and processing
At the end of the fattening test, six test sheep with close body condition were randomly selected from each group. The sheep were slaughtered, and the rumen fluid was collected after 12 h of fasting and finally filtered through a gauze into 50-mL centrifuge tubes. The centrifuge tubes were immediately placed in a liquid nitrogen tank and brought back to the laboratory, and there, they were transferred to a −80°C refrigerator for storage for the determination of rumen microbiota diversity.
2.6 16S rRNA amplicon sequencing
2.6.1 DNA extraction
Total microbial DNA was extracted from rumen samples with reference to the requirements of the TGuide S96 Magnetic Bead Method Soil/Fecal Genomic DNA Extraction Kit [Tiangen Biotechnology Co., Ltd. (Beijing, China)]. The concentration of the DNA samples was determined by an enzyme marker and then qualitatively detected by 2% agarose gel electrophoresis. The DNA samples that met the requirements for online library construction were sent to Wuhan Metavir Biotechnology Co. (Wuhan, China) for 16S amplicon sequencing.
2.6.2 Library construction and on-line sequencing
Using DNA diluted in sterile water as the template, specific primers were synthesized with barcode sequences (338F: ACTCCTACGGGGAGGCAGCAG; 806R: GGACTACHVGGGGTWTCTAAT) according to the sequence of primers in the V3–V4 region for PCR amplification. All PCR reactions were carried out with 15 μL of Phusion® High-Fidelity PCR Master Mix [New England Biolabs (Beijing, China) Ltd.], with 0.2 μM of forward and reverse primers, and approximately 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s, and final denaturation at 72°C for 5 min. Sequencing libraries were generated using the TruSeq® DNA PCR-Free Sample Preparation Kit [Illumina (Shanghai, China) Ltd.] following the manufacturer's instructions, and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer [Thermo Scientific (Shanghai, China) Ltd.] and Agilent Bioanalyzer 2,100 system (Beijing, China). Finally, the library was sequenced on an Illumina NovaSeq 6,000 platform (Shanghai, China), and 250-bp paired-end reads were generated.
2.6.3 Sequencing data processing
The downstream data (raw PE) from Illumina NovaSeq sequencing were spliced and quality-controlled to obtain clean tags, and then, chimera filtering was performed to obtain effective tags that could be used for subsequent analyses. The UPARSE algorithm (USEARCH v7, http://www.drive5.com/uparse/) was used to cluster the effective tags of all the samples into operational taxonomic units (OTUs) according to the 97% sequence similarity principle. The sequences of OTUs were then annotated with the SSUrRNA database of SILVA138.1 (http://www.arb-silva.de/) using the mothur method to obtain the microbial community composition of each sample or subgroup at each subgroup level (set the threshold to 0.8 ~ 1). Rapid multiple sequence comparisons were performed using MAFFT (v7.490, https://mafft.cbrc.jp/alignment/software/) software, and then, the data were homogenized across samples. The alpha-diversity and beta-diversity analyses were performed. Qiime software (Version 1.9.1) was used to calculate Observed_otus, Chao1, Shannon, Simpson, ace, Goods-coverage, and PD_whole_tree indices. Rarefaction curve, rank abundance curves, and species accumulation curves were plotted using R software (Version 4.1.2), and differences between groups were analyzed using R software for the alpha-diversity index. QIIME software (Version 1.9.1) was used to calculate the UniFrac distance and construct the UPGMA clustering tree. Principal component analysis (PCA), principal coordinates analysis (PCoA), and Non-Metric Multi-Dimensional Scaling (NMDS) plots were drawn using R software (Version 4.1.2). Analysis of similarity (ANOSIM) uses the anosim function of the R vegan package. Linear discriminant analysis effect size (LEfSe) analyses were performed using LEfSe software with the default filter value of LDA score set to 4. Finally, the 16S rRNA gene sequences were analyzed for functional prediction in Kyoto Encyclopedia of Genes and Genomes (KEGG) and Cluster of orthologous group (COG) databases using PICRUSt2. Other graphics used OmicStudio (https://www.omicstudio.cn/online).
2.7 Statistical analysis
The experimental data were preliminarily sorted out by Excel 2021 software, and then, the one-way ANOVA process of SPSS 26.0 statistical software was used to conduct the one-way ANOVA. Duncan's method was used to conduct multiple comparisons test, and the results were represented by mean ± standard deviation. A p-value of < 0.05 indicates statistically significant difference, and 0.05 ≤ P < 0.1 indicates that the difference tends to be significant. With reference to the previous results on production traits by our research group (Ma et al., 2024), we used Spearman's correlation test to analyze the correlation between rumen microorganisms and production traits. The greater the correlation coefficient, the stronger the correlation between the variables. Spearman's correlation coefficient greater than 0.5 means that there is a strong positive correlation between the two variables, and a correlation coefficient < 0.5 means that there is a strong negative correlation between the two variables.
3 Results
3.1 Sequencing results and bacterial diversity
A total of 1,379,355 reads pairs were obtained by 16S amplicon sequencing. A total of 1,340,281 clean tags were generated after paired-end reads were spliced and filtered, and 1,047,982 effective tags produced by chimeras were removed. At least 56,078 clean tags were generated for each sample, and 74,460 clean tags were generated on average. The average content of Q30 was 94.07%, and the average content of GC was 54.15%. On the principle of dividing different OTUs based on a sequence similarity >97%, there were 3,648, 3,930 and 4,260 OTUs in groups CK, A, and B, respectively (Figure 1A), with the lowest number of OTUs unique to group CK and the highest number of OTUs in group B. The number of OTUs common to each group was 2,042. The unique OTUs in each group were 990, 771, and 1,011, respectively. The sample sequencing rarefaction curve show that the curves flatten out as the number of samples sequenced increases, indicating an adequate amount of data (Figure 1B). Species accumulation box plots were used as a judgement of the adequacy of the sample size. The absence of a sharp increase in the position of the box plots indicated that the sampling was adequate and that data analysis could be carried out (Figure 1D). The rank clustering curve tends to flatten out, indicating that the amount of sequencing data is progressively reasonable (Figure 1C).
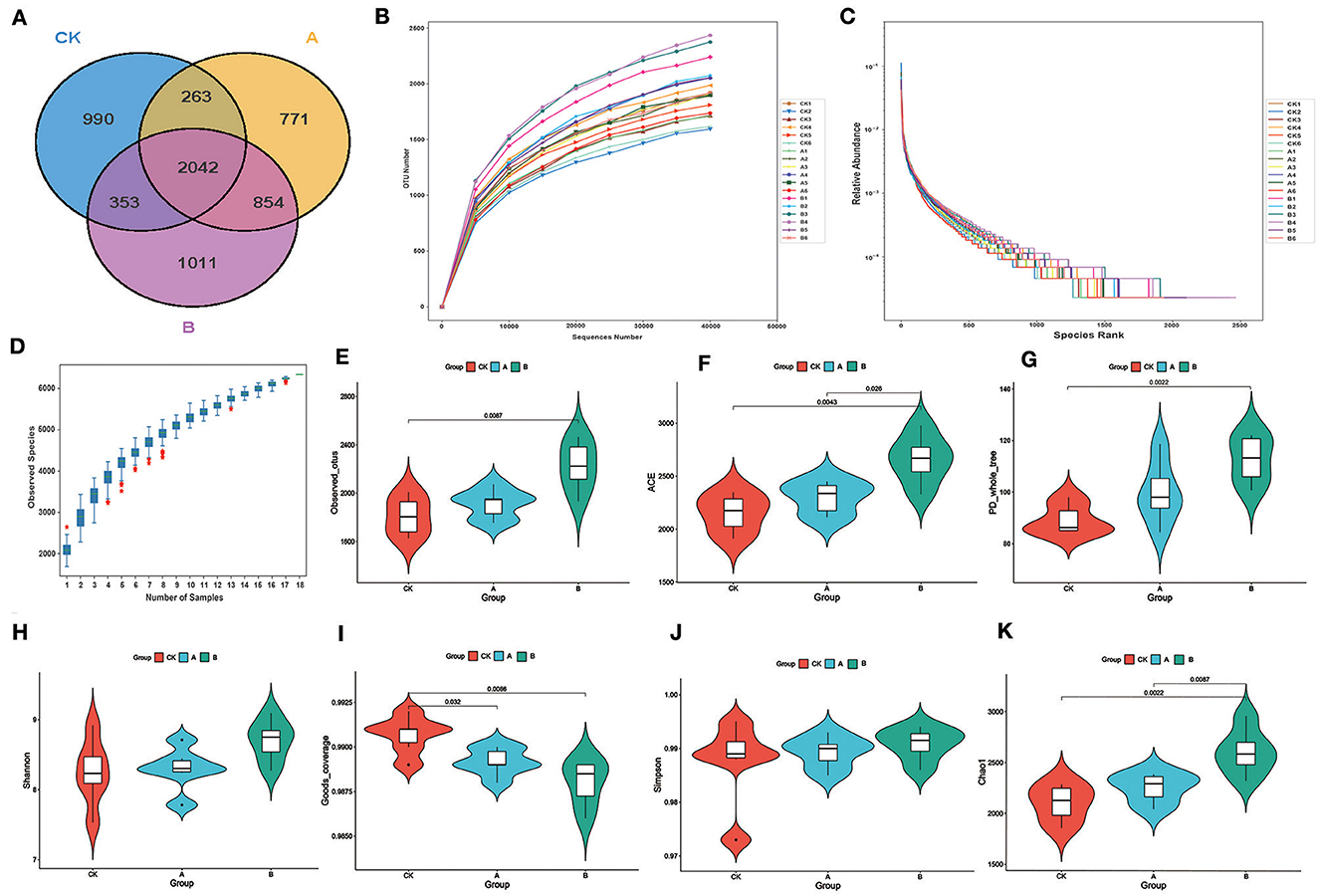
Figure 1. Sequencing results and statistical analysis of diversity. (A) The Venn diagram shows three groups of common or unique OTUs. (B) Rarefaction curve for all samples. (C) Rank abundance. (D) Species accumulation boxplot. (E) Violin plots of the number of observed_otus values between the three groups. (F) Violin plots of the number of ACE index values between the three groups. (G) Violin plots of the number of PD_whole_tree index values between the three groups. (H) Violin plots of the number of Shannon index values between the three groups. (I) Violin plots of the number of Goods_coverage between the three groups. (J) Violin plots of the number of Simpson index values between the three groups. (K) Violin plots of the number of Chao1 index values between the three groups.
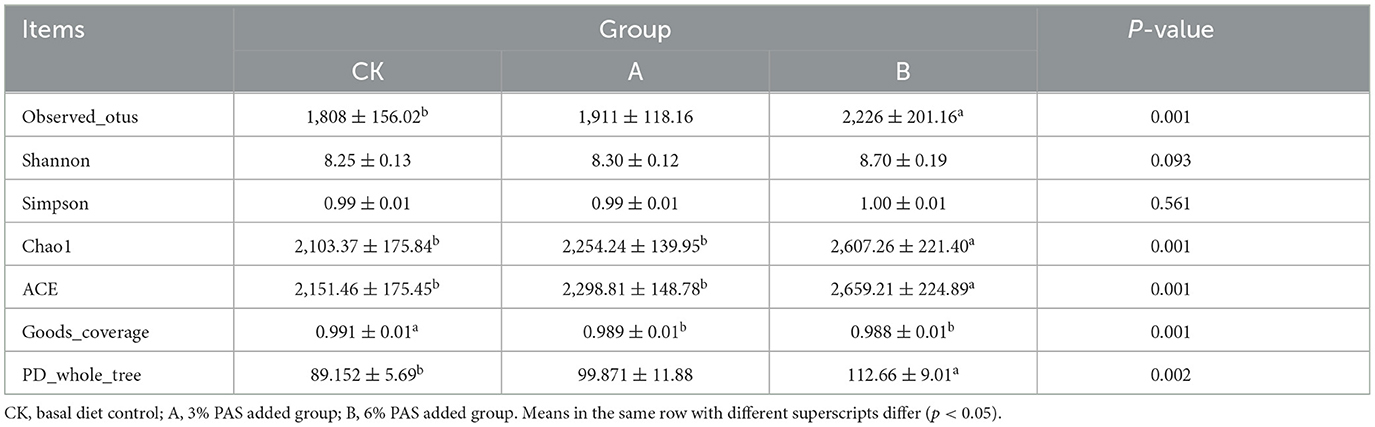
As can be seen from Table 2, the alpha diversity indices of groups A and B were mostly higher than those of group CK. The average number of visually observed OTUs, ACE index (estimation of the number of OTUs in the community), and PD_whole_tree index of each group showed CK < A < B (Figures 1E–G). The Shannon index of group A was higher than that of group CK (Figure 1H). The Simpson index did not differ significantly (P > 0.05) among groups (Figure 1J). In addition, ACE and Chao1 indices were significantly higher (P < 0.05) in group B than in groups CK and A (Figures 1F, K). The Goods_coverage index was significantly higher (P < 0.05) in group A than in groups CK and B (Figure 1I).
3.2 Cluster analysis and microbial composition
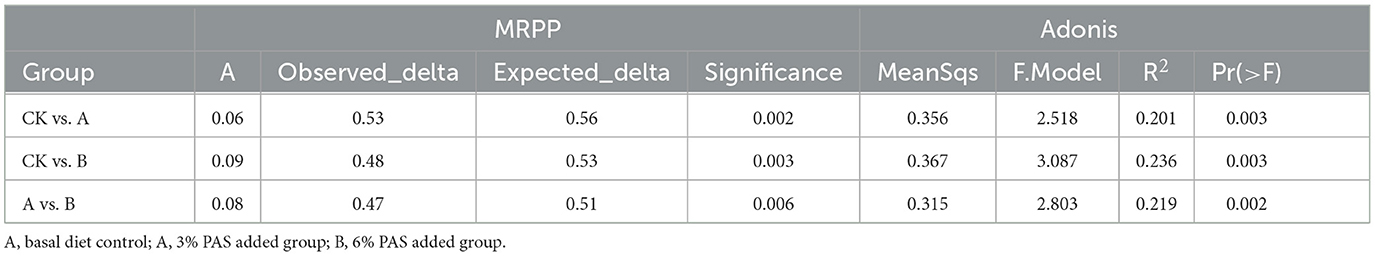
Principal coordinates analysis (PCoA) based on unweighted UniFrac showed that the rumen flora of lambs in group CK and groups A and B were significantly different, and the rumen flora of lambs in the two experimental groups were not significantly different (Figure 2A). PCA and NMDS analysis showed that the community composition was relatively similar within groups, but the community composition was more different between groups (Figures 2B, D). Moreover, the stress < 2 in NMDS analysis indicated that NMDS could accurately reflect the degree of differences between samples. The PCA plot showed that the contribution values of the two principal components were 11.58% and 8.26%, respectively, and group B and group A could be distinguished from the other two groups according to PC1 and PC2, respectively. ANOSIM showed that the range of the R-value between CK, A, and B groups was [−1.1], and the p-value was 0.001, which indicated that the difference between groups was significantly greater than the difference within groups (P < 0.05) (Figure 2C). In addition, the Multi-Response Permutation Procedure (MRPP) analysis and the Adonis analysis also indicated that the reliability of this test was high (Table 3). The UPGMA clustering tree based on the unweighted UniFrac distances of OTUs shows that half of group A is clustered with group CK and the other half is clustered with group B. The main rumen bacterial groups were Bacteroidota and Firmicutes, and the composition of the bacterial groups was basically similar among the groups. Interestingly, Acidobacteriota was present alone in all individuals of group B and in some individuals of group A (Figure 2E).
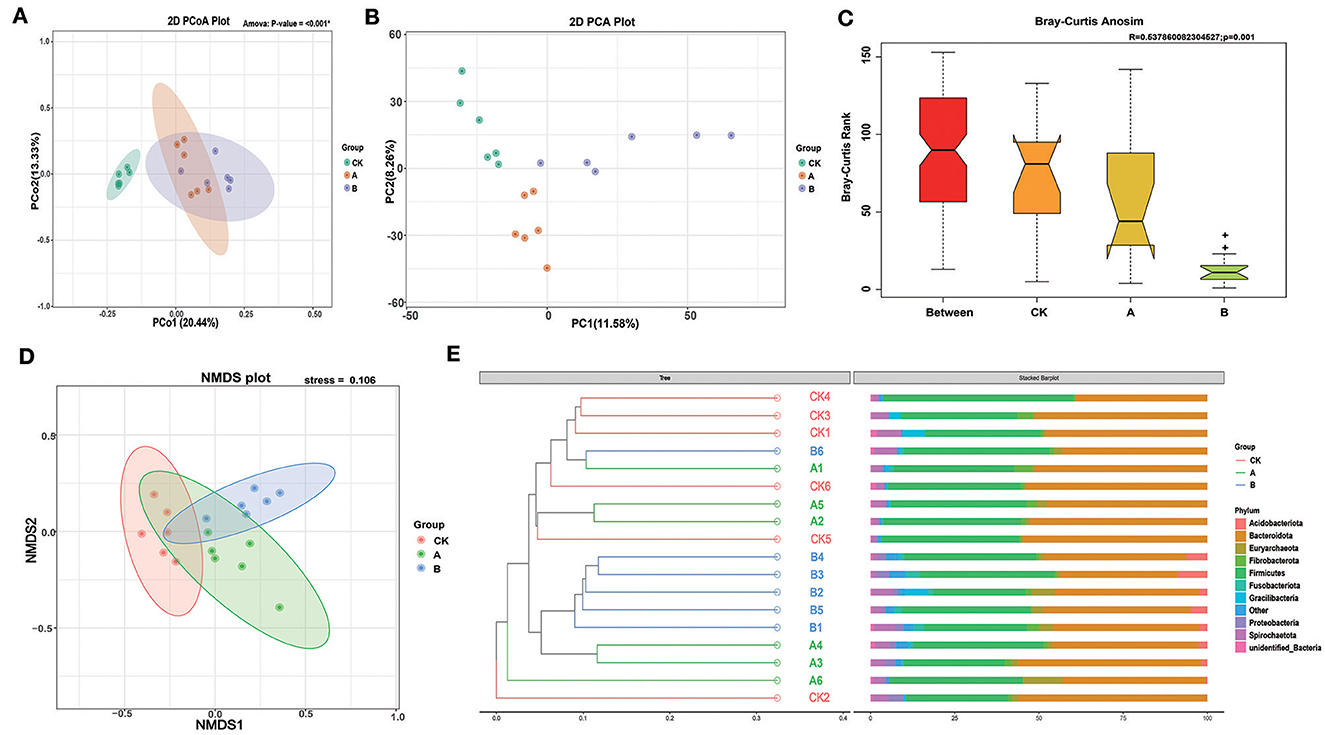
Figure 2. Cluster analysis and microbial composition. (A) Principal coordinate analysis (PCoA) based on all samples. (B) Principal component analysis (PCA) based on all samples. (C) OTU-based difference analysis of Anosim between groups. (D) Non-Metric Multi-Dimensional Scaling (NMDS) based on all samples. (E) Unweighted UniFrac distance UPGMA clustering tree based on OUT.
3.3 Relative abundance of rumen flora at the phylum level
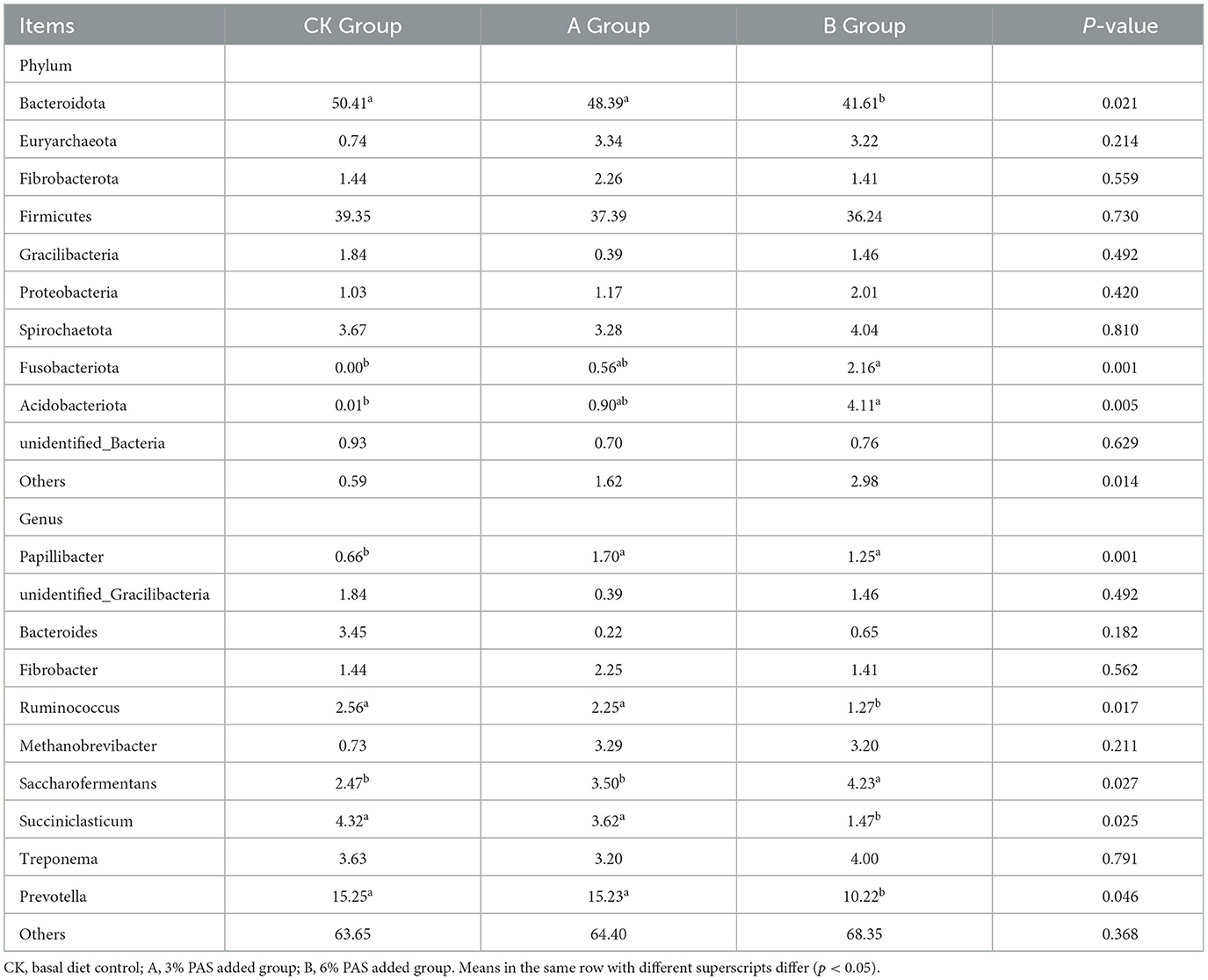
As shown in Table 4 and Figure 3A, Bacteroidota and Firmicutes were the main bacterial phyla comprising the rumen fluid flora of sheep lambs in the three groups, and their relative abundance accounted for more than 77.85% of the proportion of rumen flora. The abundance of Fusobacteriota, Proteobacteria, Acidobacteriota, and Euryarchaeota was lower in the CK group than in groups A and B. Gracilibacteria abundance was higher in the CK group than in groups A and B. The abundance of Fibrobacteriota and Euryarchaeota was higher in group A than in groups CK and B. The structure of the bacterial flora at the phylum level was basically similar for all samples within each group (Figure 3B). As can be seen from the heat map, most of the flora are higher in group B than in the other two groups in terms of content (Figure 3C). From the ternary plot, it can be seen that the abundance of Gracilibacteria was highest in group CK and Acidobacteriota, Proteobacteria, and Firmicutes in group B (Figure 3D). The final analysis of the species that differed between groups using SIMPLE revealed similar results to those of the previous graphs (Figures 3E, F).
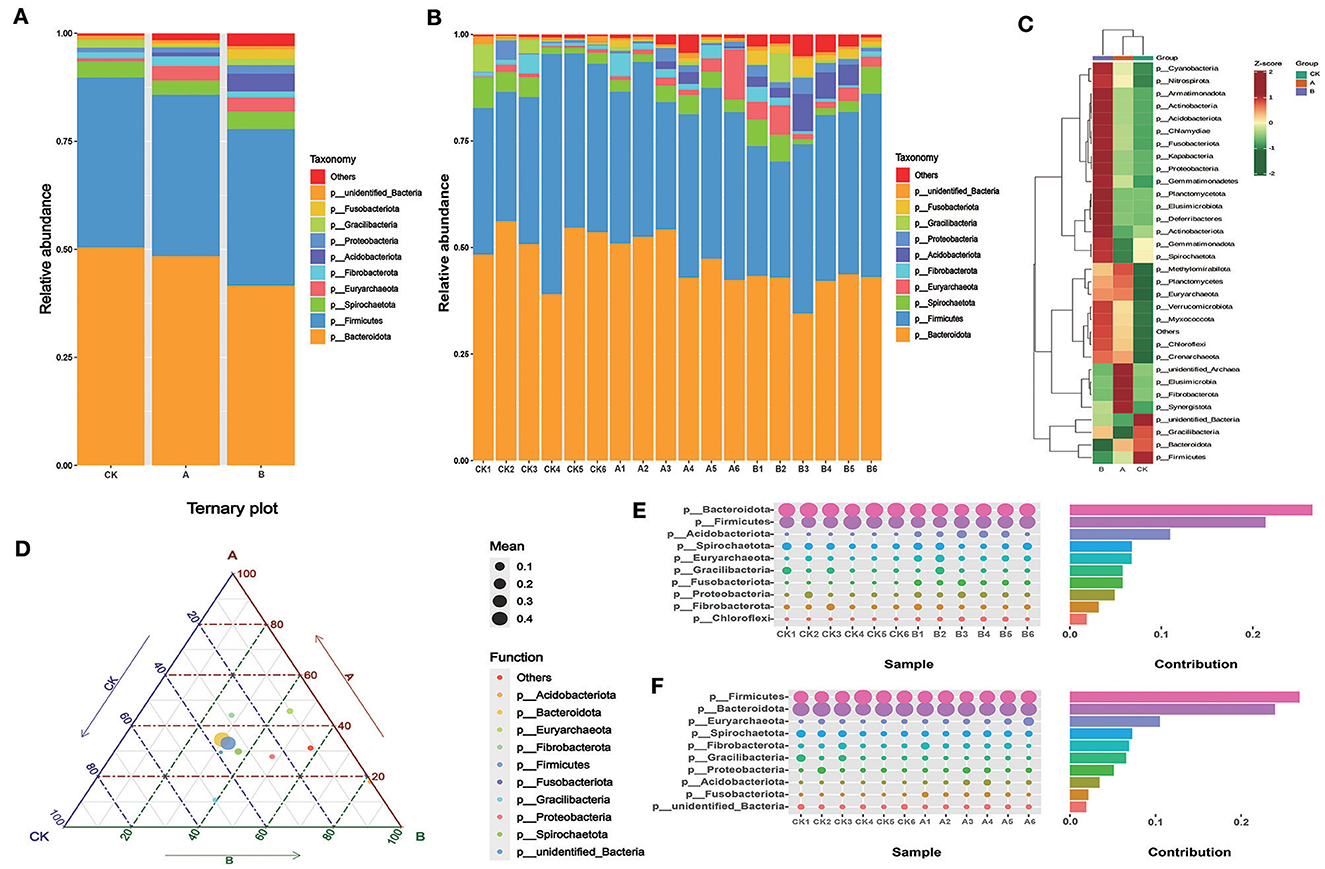
Figure 3. Relative abundance of rumen flora at the phylum level. (A) Microbial composition between the three groups at the phylum level. (B) Microbial composition of all samples at the phylum level between the three groups. (C) OTU-based species abundance heat maps for different groups at the phylum level. (D) Ternary-phase diagram based on the phylum level. (E) Contribution of Simper difference between group CK and group A based on the phylum level. (F) Contribution of Simper difference between group CK and group B based on the phylum level.
3.4 Relative abundance of rumen flora at genus level
According to Table 4 and Figure 4A, at the genus level, the abundance of Papillibacter in the CK group was significantly lower than that in groups A and B (P < 0.05). The abundances of Bacteroides, Ruminococcus, Succiniclasticum, and Prevotella in group CK were higher than those in groups A and B. The abundance of Saccharofermentans in group B was higher than that in groups CK and A (P < 0.05). There was no significant difference in the abundance of other bacteria between the control group and the experimental group. There was no significant difference in the microflora content among all groups (Figure 4B). According to the heat map, compared with other groups, the bacteria in group CK with different expression content primarily belong to Firmicutes (Figure 4C). The content of Bacteroides in group CK was higher than that in groups A and B (Figures 4D–F). The content of Methanobrevibacter in groups A and B was higher than that in group CK (Figures 4E, F).
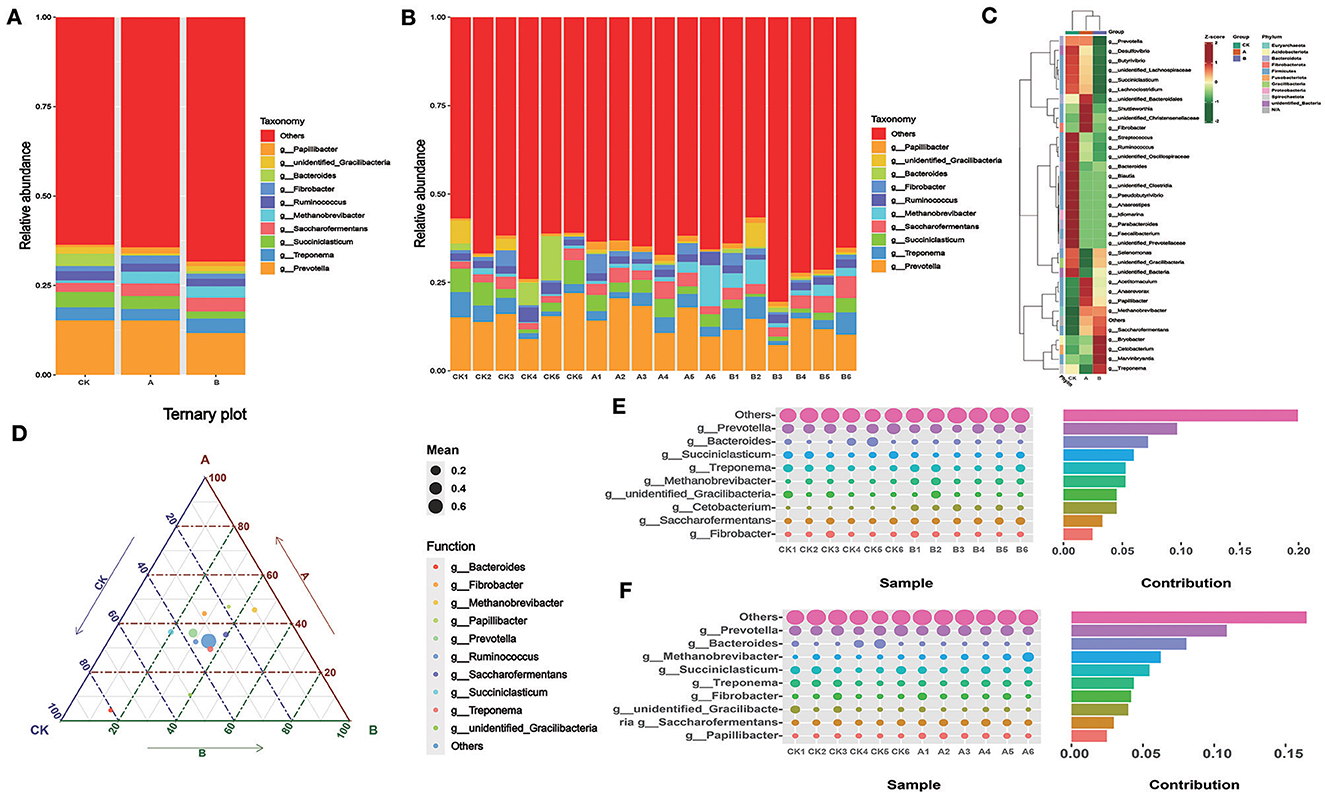
Figure 4. Relative abundance of rumen flora at the genus level. (A) Microbial composition between the three groups at the genus level. (B) Microbial composition of all samples at the genus level between the three groups (C) OTU-based species abundance heat maps for different groups at the genus level. (D) Ternary-phase diagram based on the genus level. (E) Contribution of Simper difference between group CK and group A based on the genus level. (F) Contribution of Simper difference between group CK and group B based on the genus level.
3.5 LEFse analysis and function prediction
According to LEFse analysis, significant differences occurred in the dominant flora of prickly ash seeds added in the rumen. A comparison of group CK and group A revealed that the dominant flora of the CK group was Faecalibacterium_prausnitzii. The dominant flora of group A was bacterium_P201, Papillibacteria, Oscillospiraceae, Methanobacteriales, and Lachnospiraceae_bacterium_CG2 (Figures 5B, F). A comparison of group CK and group B revealed that the dominant flora of group CK were Bactreoidota, unidentified_Prevotellaceae, and Acidaminococcales. The dominant flora of group B are then Fusobacteriota, Euryarchaeota, Bryobacter, Aeromonas_veronii, Fusobacteriaceae, Acidobacteriota, Saccharofermentans, Oscillospiraceae, Cetobacterium, and Bacteroides_paurosaccharolyticus (Figures 5C, D). The PICRUSt2 software package was used for functional prediction based on the KEGG database based on 16s sequencing data, and Stamp maps were drawn. The results showed that amino acid metabolism, carbohydrate metabolism, and the immune system in group A were significantly higher than those in group CK (P < 0.05), and antimicrobial activity and transport and catabolism in group CK were significantly higher than those in group A (P < 0.05) (Figure 5A). After comparison between group CK and group B, it was found that signal transduction, transcription, energy metabolism, cellular community-prokaryotes, cell motility and xenobiotics biodegradation, and metabolism in group B were significantly higher than those of group CK (P < 0.05). Glycan biosynthesis and metabolism, immune system, and antimicrobial activity in group CK were significantly higher than those in group B (Figure 5E).
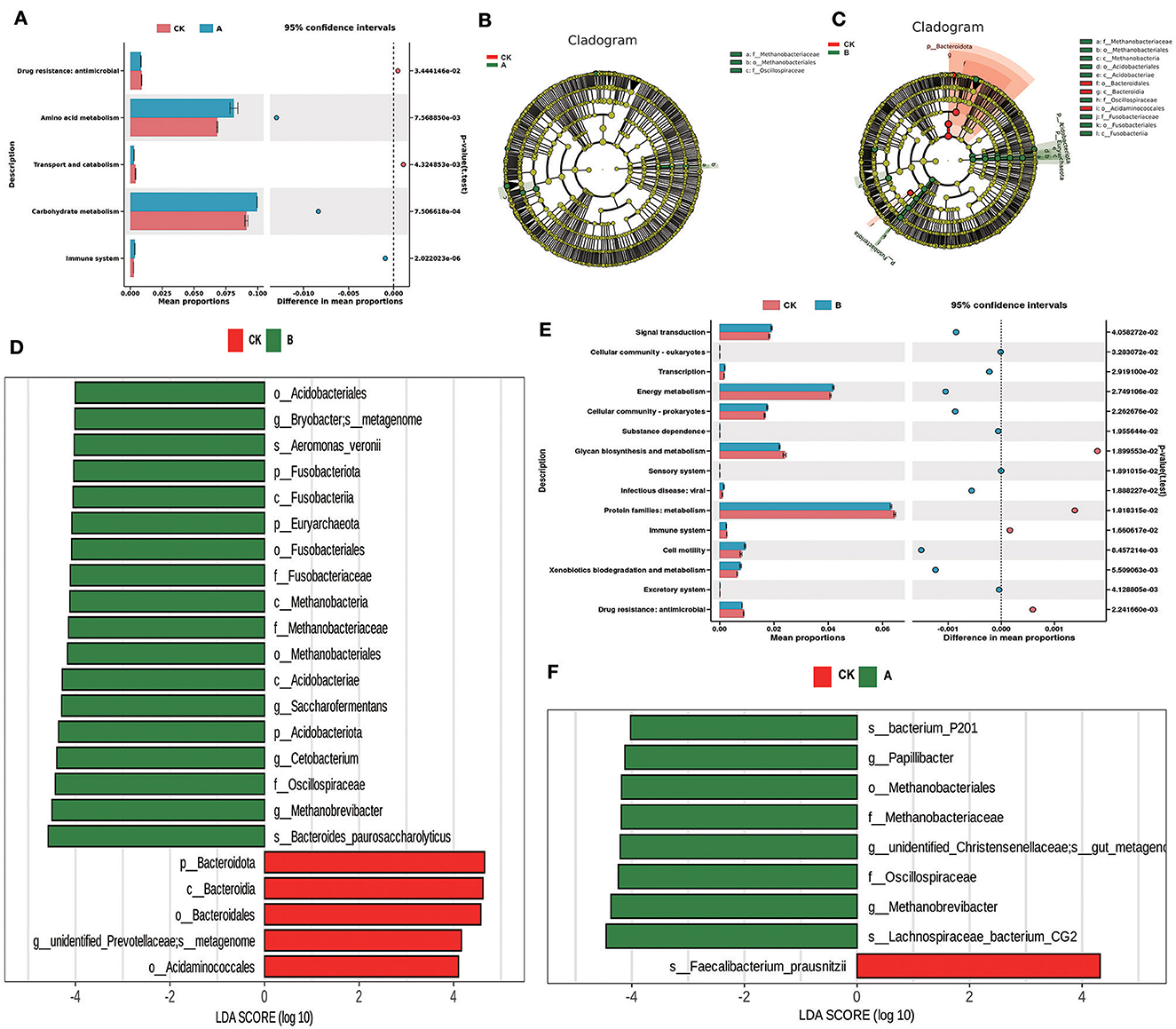
Figure 5. LEfSe analysis and PICRUST2 function prediction. (A) Functional differences between group CK and group A. (B) OTU-based evolutionary branching diagram between group CK and group A. (C) OTU-based evolutionary branching diagram between group CK and group B. (D) LDA value distribution histogram based on OTU between group CK and group B. (E) Functional differences between group CK and group B. (F) LDA value distribution histogram based on OTU between group CK and group A.
3.6 Correlation analysis of rumen microorganism and production traits
Through the analysis of the correlation between the phylum or genus level microorganisms and production traits, we found that many microorganisms had a significant correlation with production traits (Figures 6A, B). We have found many interesting phenomena. For example, b* and ADG(g/d) are positively correlated with Fusobacteriota, Acidobacteriota, Euryarchaeota, and Methanobrevibacter and negatively correlated with Bacteroidota. The total weight and dressing percentage were positively correlated with Fusobacteriota and Acidobacteriota. ADFI(g/d), F/G(g/d), and L* are negatively correlated with Fusobacteriota, Acidobacteriota, Methanobrevibacter, and Papillibacter. In addition, dressing percentage was negatively correlated with Bacteroidota. ADFI(g/d) was negatively correlated with Proteobacteria but positively correlated with Succiniclasticum. Saccharofermentans was positively correlated with ADG(g/d) but negatively correlated with ADFI(g/d) and L*. Shear force was negatively correlated with Papillibacter.
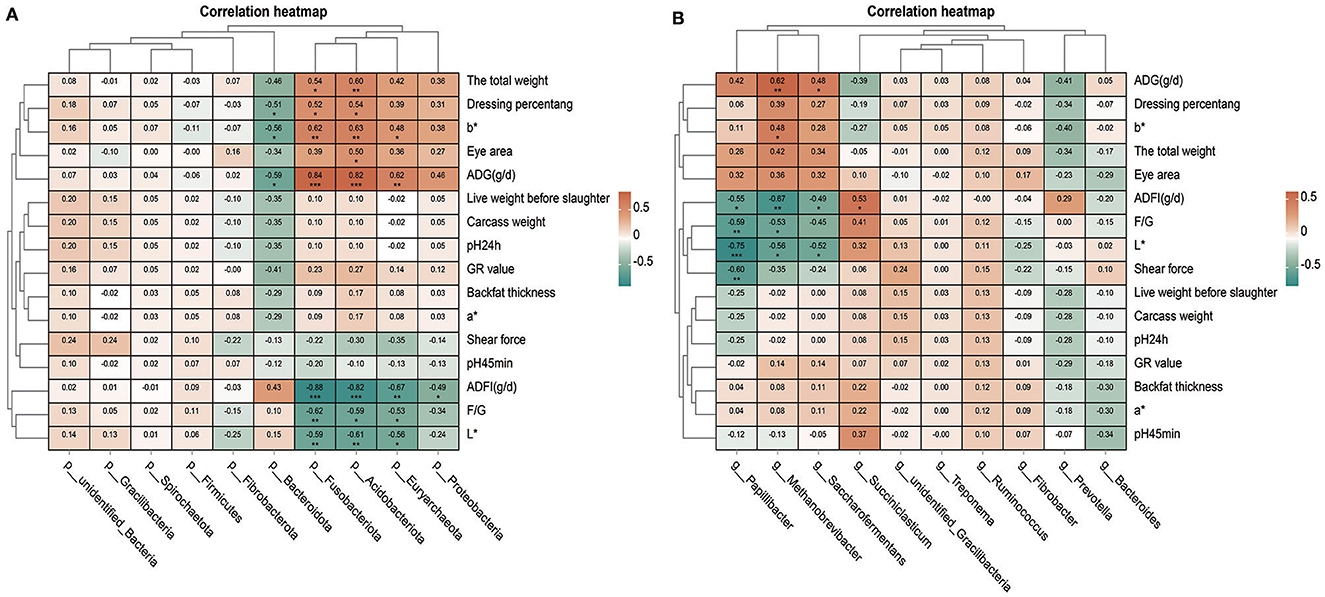
Figure 6. Correlation analysis between microorganisms and production traits. (A) Thermal map analysis of correlation between phylum level microorganisms and production traits. (B) Thermal map analysis of correlation between genus level microorganisms and production traits.
4 Discussion
As an important digestive organ of ruminants, the rumen microbes cannot function without the presence of various microorganisms in it, and the microbial diversity is closely related to species, age, diet, and feeding environment (Loh et al., 2020; Gresner et al., 2022). The digestion of more than 70% of carbohydrates and proteins in the feed is completed by rumen microorganisms (Wang et al., 2020; Zhang et al., 2021). Therefore, diet composition is an important factor affecting the structure and function of rumen microflora (Chai et al., 2021; Ge et al., 2023). Alpha diversity indexes can reflect the richness and diversity of species in the samples, of which the ACE index and Chao1 index were mainly related to the richness of microbial communities, and the larger the value, which indicates that there were more species in the samples, the higher the richness; whereas the Shannon index and the Simpson index were related to the diversity of microbial communities; the larger the Shannon index, the higher the diversity of microbial communities, and the smaller the Simpson index, the higher the diversity of microbial communities (Jiang et al., 2022; Li et al., 2022). Rumen microorganisms play an important role in the health and growth of sheep, and the higher the diversity of rumen microorganisms, the more stable the microbial ecosystem, which is more beneficial to the growth of sheep (Wang et al., 2023b). In this experiment, through alpha diversity index analysis, it was found that the Chao1 and Ace indices of the two experimental groups were significantly higher than those of the control group, indicating that prickly ash seeds had a significant impact on the abundance of rumen microorganisms, and rumen flora was more abundant in the experimental group. The beta diversity index is a comparative analysis of the microbial community composition of different samples. We choose four methods to carry out this analysis. All the methods showed that the degree of microbial community differences within groups was smaller than that between groups. PCoA showed that the CK group and two experimental groups were significantly separated. PCA and NMDS analysis showed that the three groups could be distinguished from each other. ANOSIM showed that there were significant differences among all groups. However, the three groups of sheep were not significantly separated in NMDS analysis, which was significantly different from PCA and PCoA, which may be due to differences in the clustering methods. At the same time, through the UPGMA tree diagram at the phylum level, we found that the dominant bacteria genera of all the sheep tested at the phylum level were Bacteroidota and Firmicutes, and the rumen flora structure in group CK and group B was similar, while the rumen flora structure in group A was different. Although the rumen microflora structure of sheep was affected by the amount of Prickly Ash Seeds (PAS) added, there were still individual differences in the same group of sheep (Sato et al., 2023).
There is a symbiotic relationship between ruminants and rumen microorganisms. The organism provides a suitable environment for microorganisms to grow, while the microorganisms decompose nutrients in the rumen for absorption and utilization by the organism (Malmuthuge and Guan, 2017). The composition and difference of rumen microorganisms in three groups of sheep were analyzed. We found that, after the addition of PAS, the upregulated microorganisms in sheep rumen included Methanobacteria, Papillibacter, Fusobacteriota, Acidobacteriota, Euryarchaeota, Saccharofermentans, Proteobacteria, and Oscillospiraceae, among others. Methanobacteria is a group of specialized methane-producing microorganisms that exist in the rumen of ruminants. They convert hydrogen and carbon dioxide or carbon monoxide into methane via the methanation pathway, a process that is critical for pH balance and energy metabolism in the rumen. In the gut of animals such as sheep (Drancourt et al., 2021), Euryarchaeota is also involved in methane production (Goffredi et al., 2008). Papillibacter helps the host break down food, promotes nutrient absorption, and is involved in maintaining microecological balance in the gut (Bonyadian et al., 2018). Saccharofermentans can produce acetic acid by fermenting carbohydrates, which contributes to the acidification of the rumen environment and the energy supply of the animals. Oscillospiraceae helps the host digest, absorb nutrients, and maintain intestinal barrier function (Aljumaah et al., 2022). In addition, it was found that the microorganisms that decreased the rumen content after PAS addition included Bacteroidota, Acidaminococcales, Faecalibacterium_Prausnitzii, Firmicutes, Gracilibacteria, Bacteroides, Prevotella, and Ruminococcus. Ruminococcus plays an important role in digesting resistant starch, and Prevotella is often considered a bacterium associated with a healthy plant-based diet, acting as a “probiotic” in the human body (Cheng et al., 2023). Prevotella plays a role in the diagnosis and treatment of inflammation-related diseases (Yang et al., 2018). Firmicutes can indirectly connect with other tissues and organs through metabolites and regulate hunger and satiety (Zhou et al., 2022a). Bacteroidetes and Firmicutes are often the “core microbiome” of rumen in ruminants, and this study was no exception in substantiating the abovementioned finding (Weimer, 2015). These two dominant phyla work together to promote the degradation of fibrous and non-fibrous materials in the rumen to produce acetic acid and propionic acid for absorption and utilization by the body (Ye et al., 2022; Zhou et al., 2022b). As a dominant phylum in the rumen, the main role of Bacteroidota is to promote the degradation of complex organic matter in ruminants, such as the degradation of carbohydrates, which can help the host to absorb and utilize polysaccharides efficiently (An et al., 2023; Wang et al., 2023c). Therefore, the experiment showed that the high addition of prickly ash seeds may have negative effects on the digestive function in the rumen. In conclusion, the richness and diversity of rumen flora of sheep can be enhanced by adding prickly ash seeds in the diet, and the diet supplemented with 3% prickly ash seeds may be more beneficial to the healthy development of sheep rumen. Some beneficial bacteria that have special effects in improving immunity and other aspects may work together to enhance sheep production traits.
Based on the above research results and the previous research results (Ma et al., 2024), the correlation analysis was carried out on the relationship between rumen microorganisms and production traits. It was found that Papillibacter were inversely correlated to ADFI(g/d), F/G, L*, and shear force, and Papillibacter may be associated with weight management and obesity (Zong et al., 2023). Saccharofermentans belongs to butyric acid-oxidizing bacteria, which can produce short-chain fatty acids such as butyric acid through fermentation (Li et al., 2018). With the addition of PAS, Saccharofermentans content and ADG(g/d) increased. It is likely that Saccharofermentans can reduce ADFI(g/d) while increasing ADG(g/d) in sheep. Methanobrevibacter is positively correlated with ADG(g/d) and may play a role in weight regulation by affecting the digestion of food and the absorption of calories. Euryarchaeota is able to produce a short-chain fatty acid called butyric acid, which is believed to help control appetite and fat storage (He et al., 2022; Li et al., 2023b). Fusobacteriota can also produce metabolites that are beneficial to the human body, such as pyruvate and butyric acid, which play an important role in intestinal health and the regulation of the immune system (Bleicher et al., 2022; Guo et al., 2022). It was found that Fusobacteriota, Acidobacteriota, and Euryarchaeota were positively correlated with ADG and negatively correlated with ADFI and F/G. The increase of these bacteria content may contribute to the increase of the average daily gain of sheep, the decrease of the average daily feed intake, and the decrease of the feed to gain ratio.
5 Conclusion
This study revealed the effects of adding different proportions of prickly ash seeds on the rumen flora of Hu sheep lambs and correlated the top ten bacteria in terms of relative abundance of species at the phylum and genus level with production traits. The results showed that the addition of different levels of prickly ash seeds could increase the diversity and abundance of sheep rumen microorganisms, influence the composition of sheep rumen community, and increase the content of rumen beneficial bacteria. It also promoted metabolic capacity and immune system in lambs. However, the addition of 6% prickly ash seeds resulted in greater changes in rumen flora, an increase in harmful bacteria, and a possible decrease in the excretory system and the immune system. Correlation analysis showed that Fusobacteriota, Acidobacteriota, Euryarchaeota, and Methanobrevibacter were positively correlated with ADG(g/d) and negatively correlated with ADFI(g/d) and F/G. Therefore, the present study provides new insights on improving rumen health of sheep through the addition of green additives and provides some help in understanding the correlation between rumen flora and production traits.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1058965.
Ethics statement
The animal studies were approved by the Animal Care Committee of Gansu Agricultural University (GSAU-AEW-2020-0057). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
DL: Methodology, Writing – original draft, Writing – review & editing. QL: Writing – review & editing. XM: Writing – review & editing. HuW: Writing – review & editing. CW: Writing – review & editing. HaW: Writing – review & editing. ZL: Writing – review & editing. TL: Writing – original draft, Writing – review & editing. YM: Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The study was supported by the Education Science and Technology Innovation project of Gansu Province (GSSYLXM-02), Excellent Doctoral Program of Gansu Province (23JRRA1450), Science and Technology Plan Project of Gansu Province (23JRRA1448), Research and Demonstration on Key Technology of Improving Quality and Efficiency of Meat Sheep Industry in Dongxiang County (GSAU-JSFW-2023-103), Linxia Cattle and Sheep Industry Major Scientific and Technological Support Project (KJJC-LX-2023-05), and Discipline Team Project of Gansu Agricultural University (GAU-XKTD-2022-20). The funders had no participation in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
Thanks to all participants for their advice and support of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1364517/full#supplementary-material
References
Aljumaah, M. R., Bhatia, U., Roach, J., Gunstad, J., and Azcarate, P. M. A. (2022). The gut microbiome, mild cognitive impairment, and probiotics: a randomized clinical trial in middle-aged and older adults. Clin. Nutr. 41, 2565–2576. doi: 10.1016/j.clnu.2022.09.012
An, Y. W., Wang, H. R., Zong, Z. C., Gao, Z. X., Shi, C. X., Li, S. F., et al. (2023). Sophora alopecuroidesEffects of adding to high concentrate diet on rumen fermentation parameters and microbial diversity of sheep. Front. Vet. Sci. 10:1200272. doi: 10.3389/fvets.2023.1200272
Bleicher, J., Ebner, E. E., and Bak, K. H. (2022). Formation and analysis of volatile and odor compounds in meat-a review. Molecules 27:6703. doi: 10.3390/molecules27196703
Bonyadian, M., Moshtaghi, H., and Nadi, H. (2018). PCR detection of Vibrio cholerae, Escherichia coli, and Salmonella sp. from bottled drinking water in Iran. J. Infect. Dev. Ctries 12, 700–705. doi: 10.3855/jidc.10160
Chai, J. M., Lv, X. K., Diao, Q. Y., Usdrowski, H., Zhuang, Y. M., Huang, W., et al. (2021). Solid diet manipulates rumen epithelial microbiota and its interactions with host transcriptomic in young ruminants. Environ. Microbiol. 23, 6557–6568. doi: 10.1111/1462-2920.15757
Cheng, S. S., Li, B. L., Ding, Y. X., Hou, B. C., Hung, W. L., He, J., et al. (2023). The probiotic fermented milk of Lacticaseibacillus paracasei JY062 and Lactobacillus gasseri JM1 alleviates constipation via improving gastrointestinal motility and gut microbiota. J. Dairy Sci. 107, 1857–1876. doi: 10.3168/jds.2023-24154
Drancourt, M., Djemai, K., Gouriet, F., Grine, G., Loukil, A., Bedotto, M., et al. (2021). Methanobrevibacter smithii archaemia in febrile patients with bacteremia, including those with endocarditis. Clin. Infect. Dis. 73, e2571–e2579. doi: 10.1093/cid/ciaa998
Fernando, S. C., Purvis, H. T., Najar, F. Z., Sukharnikov, L. O., Krehbiel, C. R., Nagaraja, T. G., et al. (2010). Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 76, 7482–7490. doi: 10.1128/AEM.00388-10
Fortuoso, B. F., Gebert, R. R., Griss, L. G., Glombovisky, P., Cazarotto, C. J., Rampazzo, L., et al. (2019). Reduction of stool bacterial counts and prevention of diarrhea using an oral homeopathic product in newborn lambs. Microb. Pathog. 127, 347–351. doi: 10.1016/j.micpath.2018.12.022
Ge, T., Yang, C., Li, B., Huang, X. Y., Zhao, L. Y., Zhang, X. Q., et al. (2023). High-energy diet modify rumen microbial composition and microbial energy metabolism pattern in fattening sheep. BMC Vet. Res. 19:32. doi: 10.1186/s12917-023-03592-6
Goffredi, S. K., Wilpiszeski, R., Lee, R., and Orphan, V. J. (2008). Temporal evolution of methane cycling and phylogenetic diversity of archaea in sediments from a deep-sea whale-fall in Monterey Canyon, California. ISME J. 2, 204–220. doi: 10.1038/ismej.2007.103
Gresner, N., Rodehutscord, M., and Südekum, K. H. (2022). Amino acid pattern of rumen microorganisms in cattle fed mixed diets-An update. J. Anim. Physiol. Anim. Nutr. 106, 752–771. doi: 10.1111/jpn.13676
Guo, H. R., Li, B. B., Gao, M. Q., Li, Q., Gao, Y. W., Dong, N., et al. (2022). Dietary nutritional level affects intestinal microbiota and health of goats. Microorganisms 10:2332. doi: 10.3390/microorganisms10122322
Hart, E. H., Creevey, C. J., Hitch, T., and Kingston-Smith, A. H. (2018). Meta-proteomics of rumen microbiota indicates niche compartmentalisation and functional dominance in a limited number of metabolic pathways between abundant bacteria. Sci. Rep. 8:10504. doi: 10.1038/s41598-018-28827-7
He, L. J., Guo, J. X., Wang, Y. B., Wang, L., Xu, D. D., Yan, E. F., et al. (2022). Effects of dietary yeast β-glucan supplementation on meat quality, antioxidant capacity and gut microbiota of finishing pigs. Antioxidants (Basel) 11:1340. doi: 10.3390/antiox11071340
Henderson, G., Cox, F., Ganesh, S., Jonker, A., Young, W., and Janssen, P. H. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5:14567. doi: 10.1038/srep14567
Hou, J., Wang, J., Meng, J., Zhang, X., Niu, Y., Gao, J., et al. (2021). Zanthoxylum bungeanum seed oil attenuates LPS-induced BEAS-2B cell activation and inflammation by inhibiting the TLR4/MyD88/NF-kappaB signaling pathway. Evid. Based Compl. Alternat. Med. 2021:2073296. doi: 10.1155/2021/2073296
Hou, X. Y., Li, S. S., Luo, Q. Y., Shen, G. H., Wu, H. J., Li, M. L., et al. (2019). Discovery and identification of antimicrobial peptides in Sichuan pepper (Zanthoxylum bungeanum Maxim) seeds by peptidomics and bioinformatics. Appl. Microbiol. Biotechnol. 103, 2217–2228. doi: 10.1007/s00253-018-09593-y
Jami, E., and Mizrahi, I. (2012). Composition and similarity of bovine rumen microbiota across individual animals. PLoS ONE 7:e33306. doi: 10.1371/journal.pone.0033306
Jiang, L. F., Wang, Y. Y., Peng, H., Li, R., Zhang, F., Wang, N., et al. (2022). Association between obesity with the diversity and genus of gut microbiota in school-aged children. Zhonghua Liu Xing Bing Xue Za Zhi 43, 260–268. doi: 10.3760/cma.j.cn112338-20210617-00478
Li, D. P., Yang, H., Li, Q., Ma, K. Y., Wang, H. H., Wang, C. H., et al. (2023a). Prickly Ash Seeds improve immunity of Hu sheep by changing the diversity and structure of gut microbiota. Front. Microbiol. 14:1273714. doi: 10.3389/fmicb.2023.1273714
Li, W. J., Cui, Z. Y., Jiang, Y. W., Aisikaer, A., Wu, Q. C., Zhang, F., et al. (2023b). Dietary guanidine acetic acid improves ruminal antioxidant capacity and alters rumen fermentation and microflora in rapid-growing lambs. Antioxidants 12:772. doi: 10.3390/antiox12030772
Li, W. W., Khalid, H., Zhu, Z., Zhang, R. H., Liu, G. Q., Chen, C., et al. (2018). Methane production through anaerobic digestion: participation and digestion characteristics of cellulose, hemicellulose and lignin. Appl. Energy 226, 1219–1228. doi: 10.1016/j.apenergy.2018.05.055
Li, Z. X., Zhou, J., Liang, H., Ye, L., Lan, L. Y., Lu, F., et al. (2022). Differences in alpha diversity of gut microbiota in neurological diseases. Front. Neurosci. 16:879318. doi: 10.3389/fnins.2022.879318
Loh, Z. H., Ouwerkerk, D., Klieve, A. V., Hungerford, N. L., and Fletcher, M. T. (2020). Toxin degradation by rumen microorganisms: a review. Toxins 12:664. doi: 10.3390/toxins12100664
Ma, X. Y., Li, Q., Qi, C. H., Sun, X. C., Su, M. C., Li, Y. Z., et al. (2024). Effects of Zanthoxylum bungeanum seeds on production performance, internal organ index and rumen tissue morphology of Hu sheep lambs. J. Gansu Agric. Univ. 1–12.
Malmuthuge, N., and Guan, L. L. (2017). Understanding host-microbial interactions in rumen: searching the best opportunity for microbiota manipulation. J. Anim. Sci. Biotechnol. 8:8. doi: 10.1186/s40104-016-0135-3
Mao, H. L., Ji, W. W., Yun, Y., Zhang, Y. F., Li, Z. f., et al. (2023). Influence of probiotic supplementation on the growth performance, plasma variables, and ruminal bacterial community of growth-retarded lamb. Front. Microbiol. 14:1216534. doi: 10.3389/fmicb.2023.1216534
Pang, W. W., Liu, S., He, F. T., Li, X. Y., Saira, B., Zheng, T. L., et al. (2019). Anticancer activities of Zanthoxylum bungeanum seed oil on malignant melanoma. J. Ethnopharmacol. 229, 180–189. doi: 10.1016/j.jep.2018.10.012
Phuyal, N., Jha, P. K., Raturi, P. P., and Rajbhandary, S. (2020). Total phenolic, flavonoid contents, and antioxidant activities of fruit, seed, and bark extracts of Zanthoxylum armatum DC. Sci. World J. 2020:8780704. doi: 10.1155/2020/8780704
Sato, Y., Takebe, H., Tominaga, K., Yasuda, J., Kumagai, H., Hirooka, H., et al. (2023). A rumen virosphere with implications of contribution to fermentation and methane production, and endemism in cattle breeds and individuals. Appl. Environ. Microbiol. 90:e0158123. doi: 10.1128/aem.01581-23
Schären, M., Frahm, J., Kersten, S., Meyer, U., Hummel, J., Breves, G., et al. (2018). Interrelations between the rumen microbiota and production, behavioral, rumen fermentation, metabolic, and immunological attributes of dairy cows. J. Dairy Sci. 101, 4615–4637. doi: 10.3168/jds.2017-13736
Snelling, T. J., and Wallace, R. J. (2017). The rumen microbial metaproteome as revealed by SDS-PAGE. BMC Microbiol. 17:9. doi: 10.1186/s12866-016-0917-y
Song, C. H., Oh, S. M., Lee, S., Choi, Y., Kim, J. D., Jang, A., et al. (2020). The ratio of dietary n-3 polyunsaturated fatty acids influences the fat composition and lipogenic enzyme activity in adipose tissue of growing pigs. Food Sci. Anim. Resour. 40, 242–253. doi: 10.5851/kosfa.2020.e8
Tang, W., Xie, Q., Guan, J., Jin, S., and Zhao, Y. (2014). Phytochemical profiles and biological activity evaluation of Zanthoxylum bungeanum maxim seed against asthma in murine models. J. Ethnopharmacol. 152, 444–450. doi: 10.1016/j.jep.2014.01.013
Wang, H. H., Su, M. C., Wang, C. H., Li, D. P., Li, Q., Liu, Z. L., et al. (2023a). Yeast culture repairs rumen epithelial injury by regulating microbial communities and metabolites in sheep. Front. Microbiol. 14:1305772. doi: 10.3389/fmicb.2023.1305772
Wang, H. N., Meng, L. B., and Mi, L. (2023b). Leymus chinensis effects of hay and alfalfa hay on growth performance, rumen microbiota, and untargeted metabolomics of meat in lambs. Front. Vet. Sci. 10:1256903. doi: 10.3389/fvets.2023.1256903
Wang, J. Q., Li, X. W., Liu, M., Wang, S. C., and Cao, Z. F. (2016). Inhibitory effect of Zanthoxylum bungeanum seed oil on ovalbumin-induced lung inflammation in a murine model of asthma. Mol. Med. Rep. 13, 4289–4302. doi: 10.3892/mmr.2016.5050
Wang, L. J., Li, Y., Zhang, Y. G., and Wang, L. H. (2020). The effects of different concentrate-to-forage ratio diets on rumen bacterial microbiota and the structures of holstein cows during the feeding cycle. Animals 10:957. doi: 10.3390/ani10060957
Wang, Y. J., Jiang, M. Y., Tang, Y. M., Qiu, S. N., Sun, Y. R., and Sun, H. X. (2023c). The effects of soil intake on the growth performance, rumen microbial community and tissue mineral deposition of German Mutton Merino sheep. Ecotoxicol. Environ. Saf. 263:115368. doi: 10.1016/j.ecoenv.2023.115368
Weimer, P. J. (2015). Redundancy, resilience, and host specificity of the ruminal microbiota: implications for engineering improved ruminal fermentations. Front. Microbiol. 6:296. doi: 10.3389/fmicb.2015.00296
Yang, H., Yang, M., Fang, S. M., Huang, X. C., He, M. Z., Ke, S. L., et al. (2018). Evaluating the profound effect of gut microbiome on host appetite in pigs. BMC Microbiol. 18:215. doi: 10.1186/s12866-018-1364-8
Yang, Q., Cao, W., Zhou, X., Cao, W., Xie, Y., and Wang, S. (2014). Anti-thrombotic effects of α-linolenic acid isolated from Zanthoxylum bungeanum maxim seeds. BMC Compl. Altern. Med. 14:348. doi: 10.1186/1472-6882-14-348
Ye, M. J., Hou, M., Peng, Q. M., Jia, S., Peng, B., Yin, F. F., et al. (2022). The microbiota and cytokines correlation between the jejunum and colon in altay sheep. Animals 12:1564. doi: 10.3390/ani12121564
Zhang, H., Lang, X., Li, X., Chen, G., and Wang, C. (2022a). Effect of Zanthoxylum bungeanum essential oil on rumen enzyme activity, microbiome, and metabolites in lambs. PLoS ONE 17:e0272310. doi: 10.1371/journal.pone.0272310
Zhang, H., Lang, X., Zhang, Y., and Wang, C. (2022b). Distribution of bacteria in different regions of the small intestine with Zanthoxylum bungeanum essential oil supplement in small-tailed Han sheep. Front. Microbiol. 13:1062077. doi: 10.3389/fmicb.2022.1062077
Zhang, Y., Wang, C., Peng, A., Zhang, H., and Wang, H. R. (2021). Metagenomic insight: dietary thiamine supplementation promoted the growth of carbohydrate-associated microorganisms and enzymes in the rumen of saanen goats fed high-concentrate diets. Microorganisms 9:632. doi: 10.3390/microorganisms9030632
Zhong, T., Wang, Y., Wang, X., Freitas-de-Melo, A., Li, H., Zhan, S., et al. (2022). Diarrhea in suckling lambs is associated with changes in gut microbiota, serum immunological and biochemical parameters in an intensive production system. Front. Microbiol. 13:1020657. doi: 10.3389/fmicb.2022.1020657
Zhou, A., Yuan, Y., Yang, M., Huang, Y. J., Li, X., Li, S. P., et al. (2022a). Crosstalk between the gut microbiota and epithelial cells under physiological and infectious conditions. Front. Cell Infect. Microbiol. 12:832672. doi: 10.3389/fcimb.2022.832672
Zhou, L. L., Li, H., Hou, G. Y., Hu, C. J., Ji, F. J., Peng, W. Q., et al. (2022b). Effects of blended microbial feed additives on performance, meat quality, gut microbiota and metabolism of broilers. Front. Nutr. 9:1026599. doi: 10.3389/fnut.2022.1026599
Keywords: prickly ash seeds, 16S rRNA, rumen microbial, sheep, production property
Citation: Li D, Li Q, Ma X, Wang H, Wang C, Wang H, Liu Z, Li T and Ma Y (2024) Prickly ash seeds can promote healthy production of sheep by regulating the rumen microbial community. Front. Microbiol. 15:1364517. doi: 10.3389/fmicb.2024.1364517
Received: 02 January 2024; Accepted: 06 May 2024;
Published: 20 May 2024.
Edited by:
Anusorn Cherdthong, Khon Kaen University, ThailandReviewed by:
Lihui Zhu, Shanghai Academy of Agricultural Sciences, ChinaChuanfa Liu, The Chinese University of Hong Kong, China
Copyright © 2024 Li, Li, Ma, Wang, Wang, Wang, Liu, Li and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youji Ma, eWptYUBnc2F1LmVkdS5jbg==
 Dengpan Li
Dengpan Li Qiao Li
Qiao Li Xueyi Ma1,2
Xueyi Ma1,2 Huihui Wang
Huihui Wang Chunhui Wang
Chunhui Wang Taotao Li
Taotao Li Youji Ma
Youji Ma