- 1Using Technology Development Department, Nakdonggang National Institute of Biological Resources (NNIBR), Sangju, Republic of Korea
- 2Industrial Insect and Sericulture Division, National Institute of Agricultural Sciences, Rural Development Administration (RDA), Wanju, Republic of Korea
Introduction: The white-spotted flower chafer (Protaetia brevitarsis seulensis), which is widely distributed in Asian countries, is traditionally used in oriental medicine. However, its larvae are prone to severe damage by green muscardine disease (caused by Metarhizium anisopliae) during breeding. The aim of this study was to characterize Bacillus velezensis TJS119, which has been isolated from freshwater, and investigate its potential as a biocontrol agent against M. anisopliae in insects.
Methods: TJS119 was obtained from freshwater samples in the Republic of Korea and was classified as B. velezensis. We evaluated its in vitro antifungal effect, sequenced the bacterial whole genome, mined genes responsible for the synthesis of secondary metabolites, performed secondary metabolite analysis Ultra performance liquid chromatography-mass spectrometry (UPLC–MS/MS), and conducted bioassays for determining green muscardine disease control ability.
Results: Bacillus velezensis TJS119 inhibited the mycelial growth of M. anisopliae in vitro. The size of the B. velezensis TJS119 genome was estimated to be 3,890,913 bp with a GC content of 46.67% and 3,750 coding sequences. Biosynthetic gene clusters for secondary metabolites with antifungal activity were identified in the genome. Lipopeptides, including fengycin secreted by TJS119 exhibit antifungal activity. Application of TJS119 for the biocontrol against green muscardine disease increased the viability of white-spotted flower chafer by 94.7% compared to the control.
Discussion: These results indicate that B. velezensis TJS119 is a potential biocontrol agent for insect pathogens.
1 Introduction
The white-spotted flower chafer Protaetia brevitarsis seulensis is traditionally used in oriental medicine. It is widely distributed in Asian countries including Korea, Southeast Asia, and China (Pan J. et al., 2017). Belonging to the family Scarabaeidae, which is globally distributed with nearly 6,200 species and 267 genera (Suh and Kang, 2012), white-spotted flower chafers play an important ecological role. The larvae of white-spotted flower chafers inhabit soil and feed on decaying organic matter. Thus, they are exposed to a wide range of pathogenic microbes that attack their immune systems, thereby experiencing considerable selective pressure (Li et al., 2019). In traditional Korean medicine, the larvae are used to treat microbial infections. Additionally, the white-spotted flower chafer is widely marketed as an edible insect, generating high revenue. In insect farms, the larvae are subjected to mass mortality owing to the occurrence of green muscardine disease.
Green muscardine infection is caused by a fungus belonging to the genus Metarhizium. The name of the disease is based on the observation that after the death of the host insect, the fungus covers its cuticle with a layer of green spores. Insect mycologists and microbial control specialists use the name “green muscardine” to specifically refer to infections caused by Metarhizium spp., with many of them infecting a wide range of insects, including the larvae of P. brevitarsis seulensis. Green muscardine reportedly affects over 200 known insect species (Calderone, 2002).
Microorganisms with antifungal activity are widely used in agriculture. Among these, Bacillus spp. have demonstrated high effectiveness as biocontrol agents. They produce spores and several bioactive compounds, making them resilient against challenging environmental conditions. Therefore, Bacillus spp. are among of the most extensively studied bacteria in the field of biocontrol (Li et al., 2021; Pang et al., 2021). Bacillus subtilis, B. amyloliquefaciens, B. cereus, B. megaterium, and B. velezensis are the most commonly used biocontrol agents, because they produce phytohormones that promote plant growth and a wide range of secondary metabolites that suppress competitive plant pathogens (Lugtenberg and Kamilova, 2009). Recently, B. velezensis has been reclassified as a heterotypic synonym of B. amyloliquefaciens subsp. plantarum, B. methylogrophicus, and B. oryzicola (Dunlap et al., 2015). Several strains of B. velezensis have been extensively used in agriculture and biotechnology due to their environmental safety, ease of industrial production, and biocontrol efficacy (Rabbee et al., 2023). These include B. velezensis BR-1 (Zhou et al., 2022), HNA3 (Zaid et al., 2022), GH1-13 (Kim et al., 2017b), S3-1 (Jin et al., 2017), M75 (Kim et al., 2017a), LS69 (Liu et al., 2017), 9912D (Pan H. Q. et al., 2017), and S499 (Molinatto et al., 2016). However, the relative efficacy of B. velezensis TJS119 against insect pathogens and its mechanism of biocontrol remain unknown.
The aim of this study was to investigate the mechanisms underlying the biocontrol ability of B. velezensis TJS119 and to determine its genome sequence. The results of bioassays indicated that B. velezensis TJS119 is a potential biocontrol agent that can be used against green muscardine (M. anisopliae). Genomic and secondary metabolite analysis were performed to determine the mechanism by which B. velezensis TJS119 controls diseases. Overall, the results of the present study support the application of B. velezensis for disease control in Protaetia brevitarsis seulensis larvae.
2 Materials and methods
2.1 Isolation of bacterial strains
One hundred and eighty-six bacterial strains were isolated from freshwater in the Nakdong River (128.26°E, 36.43°N, June 2021), Sangju, Republic of Korea. Freshwater samples were serially diluted in sterilized distilled water and inoculated on tryptic soy agar plates (TSA; BD Biosciences). The plates were incubated at 30°C for 5 days. Single colonies were selected and continuously sub-cultured in fresh TSA to obtain pure cultures.
2.2 Screening of biological control bacteria against green muscardine
Each bacterium was activated at 28°C for 24 h with agitation at 200 rpm and subsequently inoculated into tryptic soy broth (TSB) liquid medium with liquid loading of 10 mL·L−1 in a 200 mL Erlenmeyer flask under identical conditions for 2 days. Aseptic filtrate of the bacterium was obtained using an aseptically sterilized bacterial filter with a pore size of 0.22 μm. The targeted pathogen, M. anisopliae strain KACC 40969, was obtained from the Korean Agricultural Culture Collection (KACC). Metarhizium anisopliae was cultured on potato dextrose agar (PDA; BD Biosciences) medium at 28°C for 1 month, and then washed with sterile distilled water to obtain conidia. A hemocytometer was used to adjust the conidial suspension to 1 × 106 conidia/mL. Metarhizium anisopliae suspension was prepared by adding 1 × 105 conidia/g to PDA medium. Thereafter, a paper disk (8 mm) was placed on the PDA and loaded with 100 μL of the filtered culture supernatant of each bacterium. The plates were inverted and incubated at 28°C for 3 days. The inhibitory zone was determined in millimeters (mm) using the vernier calipers.
2.3 Genome sequencing and analysis of bacterial
For genome sequencing, TJS119 cells were cultured in TSA at 30°C for 48 h, and their genomic DNA (gDNA) was extracted using the Wizard Genomic DNA Kit (Promega, United States) according to the manufacturer’s instructions. The whole genome was sequenced using an Illumina NovaSeq platform at Macrogen (Seoul, Republic of Korea). gDNA libraries were generated using the TruSeq Nano DNA High Throughput Library Prep Kit (Illumina). The generated short and long reads were quality-filtered and adapter-trimmed using the Trimmomatic software (version 0.36) (Bolger et al., 2014). The quality of the Illumina reads was assessed using FastQC (v0.11.5; https://www.bioinformatics.babraham.ac.uk/projects/fastqc) for Illumina outputs. Library sequencing data were assembled using SPAdes version 3.15 (Bankevich et al., 2012). Genome sequence contamination was assessed using the contamination estimator in BUSCO (Simão et al., 2015). Protein-coding sequences, tRNAs, and insertion sequence elements were predicted using Prokka (version 1.14.6) (Seemann, 2014), tRNA-scan-SE (v2.0.9), and ISEScan (v1.7.2.3), respectively. Average nucleotide identity (ANI) values were calculated using OrthoANI1 (Lee et al., 2016). The default settings were used for all software analysis unless otherwise indicated. The draft genome sequence of TJS119 has been deposited at GenBank under the accession number JAXQPU000000000. Phylogenetic trees based on 92 bacterial core genes were constructed with an up-to-date bacterial core gene set (UBCG) (Na et al., 2018) using the maximum-likelihood method (Felsenstein, 1981). The whole-genome sequence of TJS119 was processed using the antibacterial version of the antibiotic and secondary metabolite analysis shell (antiSMASH, version 7.0.0) webserver.2 Biosynthetic gene clusters (BGCs) were identified by aligning the sequence against different genomes using the BLAST tool of the National Center for Biotechnology Information (NCBI) database.3 The identified BGCs were compared with those of other microbes compiled in the Minimum Information for Biosynthetic Gene Clusters (MIBiG) database4 to identify whether similar pathways exist in other organisms. Virulence factors were confirmed using the VFanalyzer online tool (Liu et al., 2022) (accessed on 4 March 2024). The default settings were used for all software analysis unless otherwise indicated. The PathogenFinder 1.1 online tool (Cosentino et al., 2013) was employed to conduct bacterial pathogenicity estimation. The assembled fasta files were uploaded to PathogenFinder, and Firmicutes was selected as the organism class (accessed on 4 March 2024).
2.4 Extraction of secondary metabolites from TJS119
To confirm the activity of the selected strain, it was inoculated in 1 L of TSB and incubated at 30°C for 3 days with constant shaking in a shaker incubator. Cell-free supernatant was then collected via centrifugation at 7,000 × g for 20 min at 4°C, solvent-extracted twice using an equal volume of n-butanol in a separating funnel with vigorous shaking for approximately 5 min, and allowed to stand for 30 min. The aqueous and organic phases were separated, and the organic phase was collected and evaporated using a rotary evaporator under vacuum at 35°C. After evaporation, the dried crude organic extract was dissolved in methanol. The crude extract was subjected to paper disk diffusion to confirm the presence of antifungal compounds. A hemocytometer was used to adjust the concentration conidial suspension to 1 × 106 conidia/mL. Metarhizium anisopliae plates ware prepared by adding 1 × 105 conidia/g to PDA medium. To these plates, 50, 100, 200, or 300 μL of the crude extract was added on the disk of each plate, with pure n-butanol as a control. The plates were then incubated at 28°C for 3 days and observed for antifungal activity.
2.5 Identification of antifungal compounds using UPLC–MS/MS analysis
The crude extract was passed through a syringe filter (0.22 μm). An Exion UPLC and a SCIRX QTRAP 4500 mass spectrometer equipped with an ESI interface were used (SCIEX, Framingham, MA, United States). The Luna Omega polar C18 column (150 mm × 2.1 mm, 1.6 μm; Phenomenex) was employed for chromatographic separation at 35°C. The mobile phase consisted of 0.1% formic acid (A) and acetonitrile (B), at a flow rate of 0.25 mL/min. Gradient elution was performed as follows: 0–1 min, 10% B; 1–16 min, 10–95% B; 16–19 min, 95% B. Nitrogen (N2) was used as the collision gas, the ion spray voltage was 5.5 kV, and the temperature was 600°C. The declustering potential (DP) voltage and collision energy were set at 30 and 10, respectively.
2.6 Bioassay
Protaetia brevitarsis (third instar) larvae were obtained from a local farm (Sangju Gapjangsan white grub farm, Sangju, Republic of Korea). They were reared at 25°C under 65% relative humidity in the dark. Plastic cages (L × W × H = 180.00 cm × 460.00 cm × 160.00 cm) were used for rearing. The experiment was conducted with 100 larvae placed in 2 kg of sterilized sawdust. Metarhizium anisopliae was cultured on PDA (BD Biosciences) medium at 28°C for 1 month, and then washed with sterile distilled water to obtain conidia. The conidia suspension was used at a concentration of 1 × 105 conidia/mL per 100 g of sawdust. In the experimental group, B. velezensis TJS119 strain was used at a concentration of 1 × 107 CFU/g; water was added to the control group. The final sawdust moisture content was 60%. After treating with the fungus, we confirmed the mortality of larvae due to green muscardine disease for 5 weeks. The presence of green muscardine disease was determined based on the formation of M. anisopliae conidia on the epidermis of dead larvae (Figure 1). We calculated the average number of individuals affected by the disease, aggregated through experiments conducted with three repetitions.
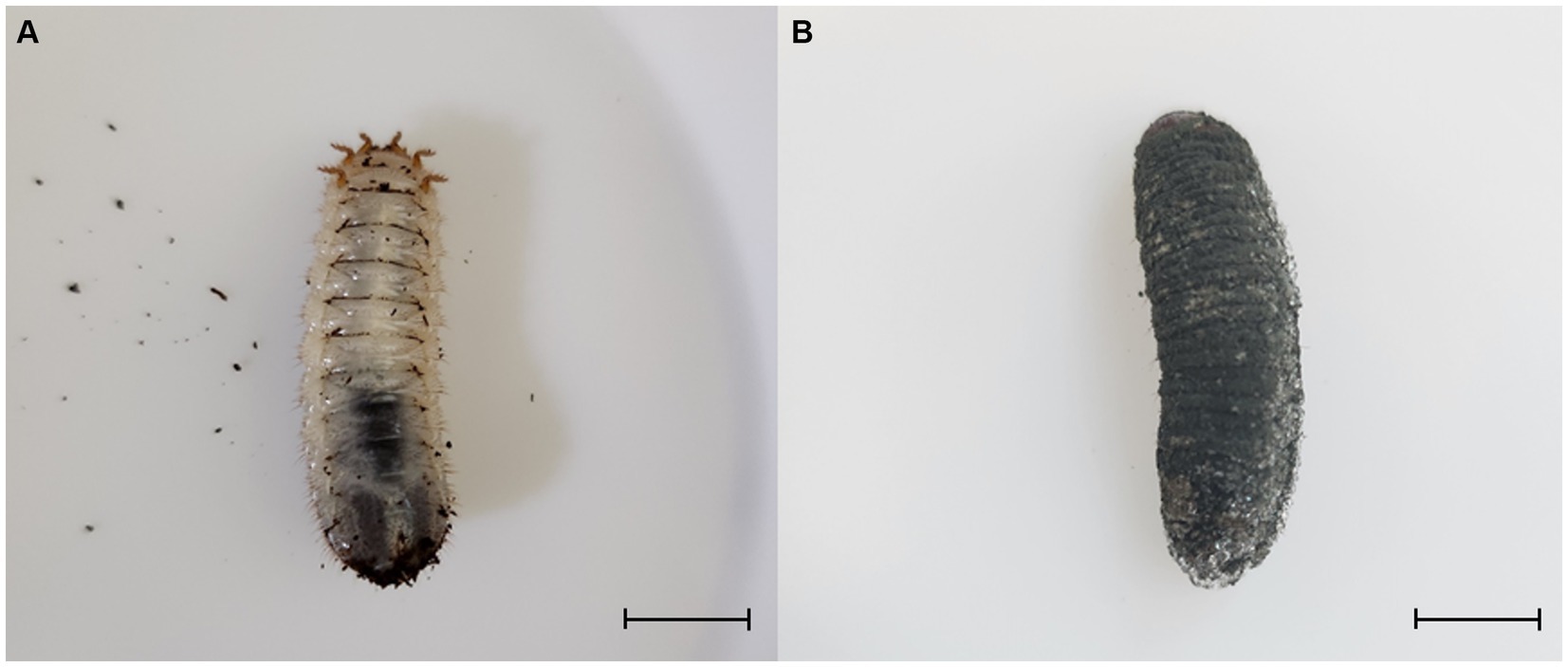
Figure 1. Third instar larvae of Protaetia brevitarsis were observed: (A) a non-infected larva, and (B) a larva infected with Metarhizium anisopliae. The infected larvae exhibited stiffness and displayed dark green spores on their epidermis. Scale bar: 10 mm.
3 Results
3.1 Isolation and identification of Bacillus velezensis TJS119
Out of 186 bacterial isolates obtained from the freshwater samples of the Nakdong River, isolate TJS119 exhibited the highest degree of fungal growth inhibition. Bacillus velezensis TJS119 was chosen for the maximum antagonistic activity against M. anisopliae in culture media, with an antagonistic zone width of 23.7 ± 1 mm (Figure 2). The strain has been maintained at the Nakdonggang National Institute of Biological Resources (NNIBR) and deposited as strain KACC 81272BP at the “Korean Agricultural Culture Collection (KACC)” of Wanju-gun, Korea.
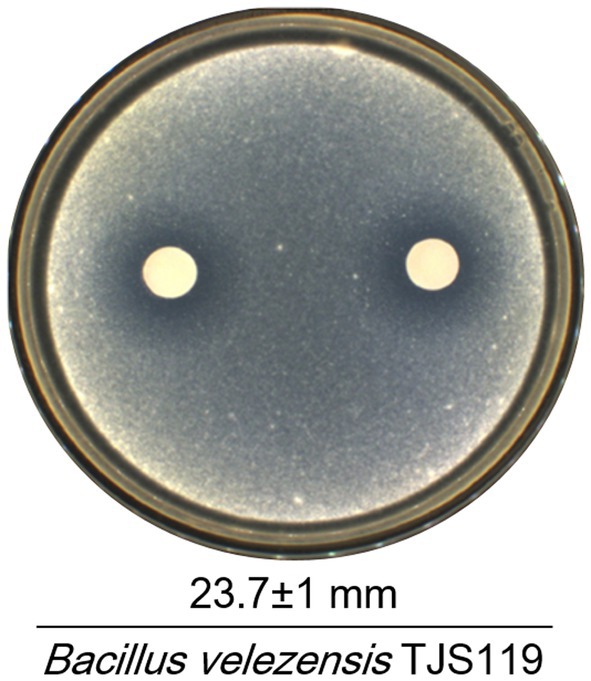
Figure 2. Antagonistic effect of Bacillus velezensis TJS119 on Metarhizium anisopliae. Values given are mean ± standard deviation of duplicate measurements.
3.2 Genome information of Bacillus velezensis TJS119
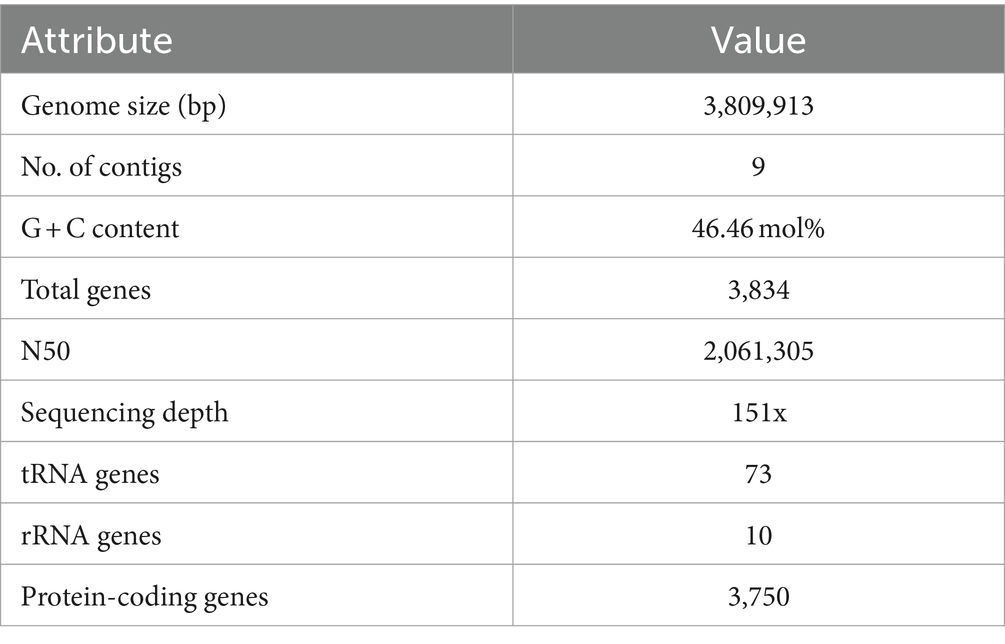
The final genome assembly of TJS119 contained nine contigs with a total length of 3,809,913 bp, an N50 of 2,061,305 bp, 151 × sequencing depth, and 3,750 protein-coding regions. Genome sequencing indicated that the DNA G + C content of TJS119 was 46.4 mol% (Table 1). The chromosome contains 73 tRNAs, 10 rRNAs, and 1 tmRNA. The ANI between TJS119 and B. velezensis was 98.26. The phylogenomic tree (Figure 3), based on 92 core bacterial gene sequences, also indicated that TJS119 is a B. velezensis strain. The genomic features of TJS119 were compared with those of other closely related Bacillus spp. as shown in Table 2. The genome size of the five B. velezensis strains ranged from 3.80 to 3.92 Mb; their G + C content ranged from 46.4 to 46.5% and predicted number of coding genes ranged from 3,684 to 3,750. TJS119 and five other strains possesses one circular chromosome without a plasmid.

Figure 3. Phylogenomic tree based on 92 bacterial core gene sequences, reconstructed with the maximum-likelihood method, showing the position of TJS119 among the related taxa within the genus Bacillus. Genome accession numbers are indicated in parentheses. The numbers at the nodes indicate the gene support index. Bar: 0.02 substitutions per position. The 92 bacterial core genes were alaS, argS, aspS, cgtA, coaE, cysS, dnaA, dnaG, dnaX, engA, ffh, fmt, frr, ftsY, gmk, hisS, ileS, infB, infC, ksgA, lepA, leuS, ligA, nusA, nusG, pgk, pheS, pheT, prfA, pyrG, recA, rbfA, rnc, rplA, rplB, rplC, rplD, rplE, rplF, rplI, rplJ, rplK, rplL, rplM, rplN, rplO, rplP, rplQ, rplR, rplS, rplT, rplU, rplV, rplW, rplX, rpmA, rpmC, rpmI, rpoA, rpoB, rpoC, rpsB, rpsC, rpsD, rpsE, rpsF, rpsG, rpsH, rpsI, rpsJ, rpsK, rpsL, rpsM, rpsO, rpsP, rpsQ, rpsR, rpsS, rpsT, secA, secG, secY, serS, smpB, tig, tilS, truB, tsaD, tsf, uvrB, ybeY, and ychF.
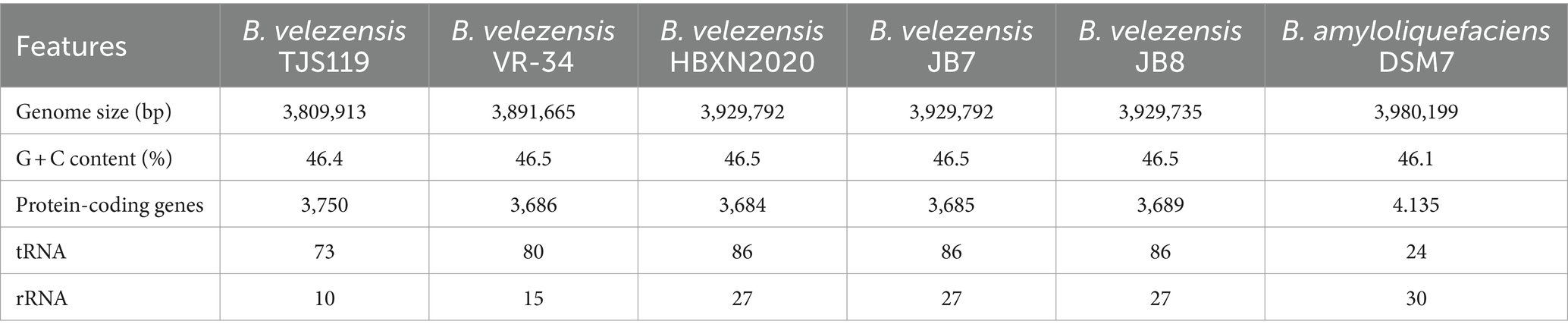
Table 2. Comparison of genomic features between Bacillus velezensis TJS119 and closely-related strains.
The ANI is a powerful approach for assessing evolutionary distances among bacterial species based on digital whole-genome comparisons, and the closer the value to 1 the higher the similarity. Based on the ANI values, the genome sequence of TJS119 is most similar to that of B. velezensis with ANI values > 97%. However, the ANI values between TJS119 and B. amyloliquefaciens were < 95%. Strains with ANI > 96% are typically considered to belong to the same species. Therefore, TJS119 does not belong to B. amyloliquefaciens.
The antiSMASH 7.0 webserver revealed 12 BGCs in the genome of B. velezensis TJS119, eight of which presented significant similarity with previously identified clusters in the Minimum Information about a Biosynthetic Gene cluster (MIBiG) repository that are involved in the synthesis of surfactin (82% similarity to known clusters revealed by antiSMASH), macrolactin H (100%), bacillaene (100%), fengycin (100%), difficidin (100%), bacillibactin (100%), bacilysin (100%), and butirosin A/butirosin B (7%). Four of the 12 clusters were not similar to any of the clusters in the antiSMASH database (Table 3). We compared the genome sequence of B. velezensis TJS119 with sequences on the VFDB database. Only a few virulence genes related to adherence, immune evasion, iron acquisition, regulation, secretion system, toxin, acid resistance, antiphagocytosis, cell surface components, copper uptake, invasion, iron uptake, stress adaptation, and surface protein anchoring factor were detected in the genome data of B. velezensis TJS119 (Supplementary Table S1). No pathogenic genes were detected in the genome.
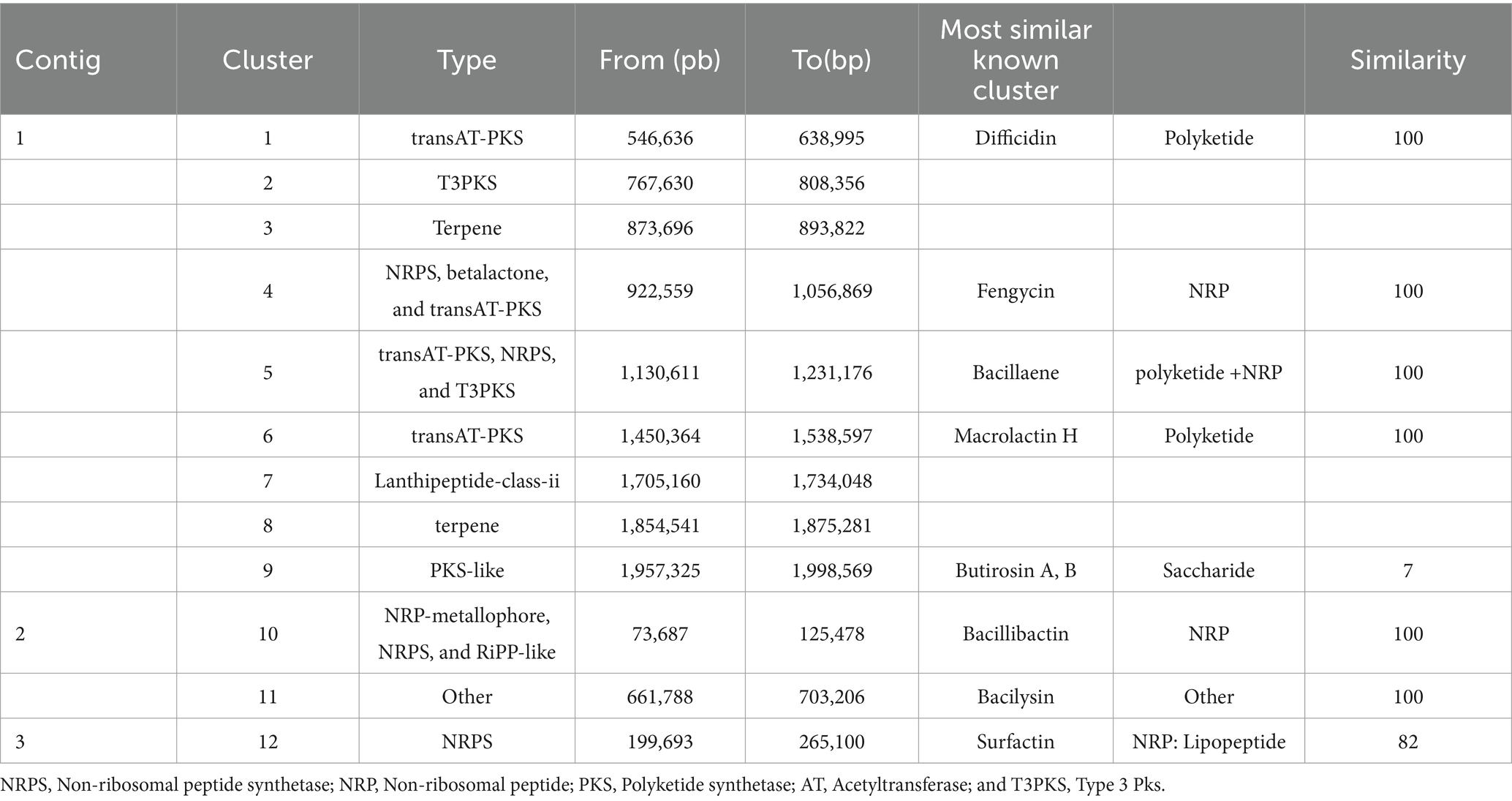
Table 3. Biosynthetic gene clusters (BGCs) found in the TJS119 genome using the webserver antiSMASH 7.0.0.
3.3 In vitro antifungal activity
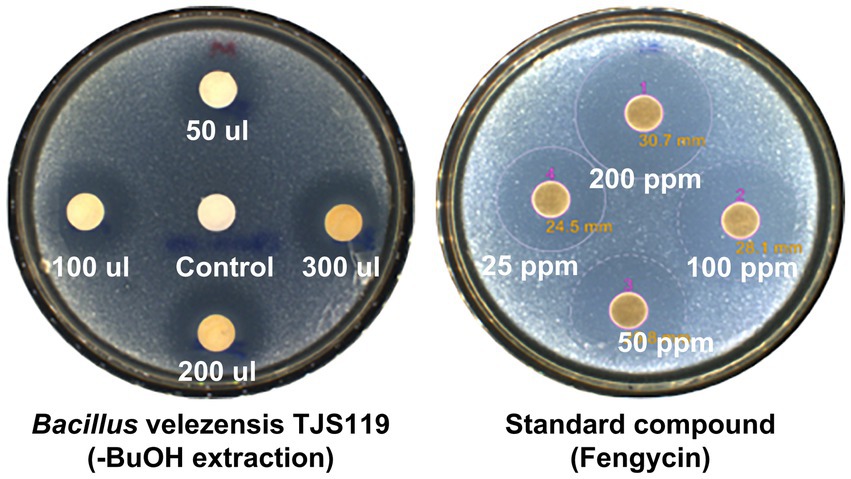
The butanol extract of TJS119 culture broth inhibited the growth of M. anisopliae in a concentration-dependent manner (Figure 4). This result confirmed the presence of antifungal compounds in the n-butanol extract. The fengycin standard (Sigma-Aldrich) inhibited M. anisopliae at all concentrations tested (25–200 ppm).
3.4 Identification of antifungal compounds
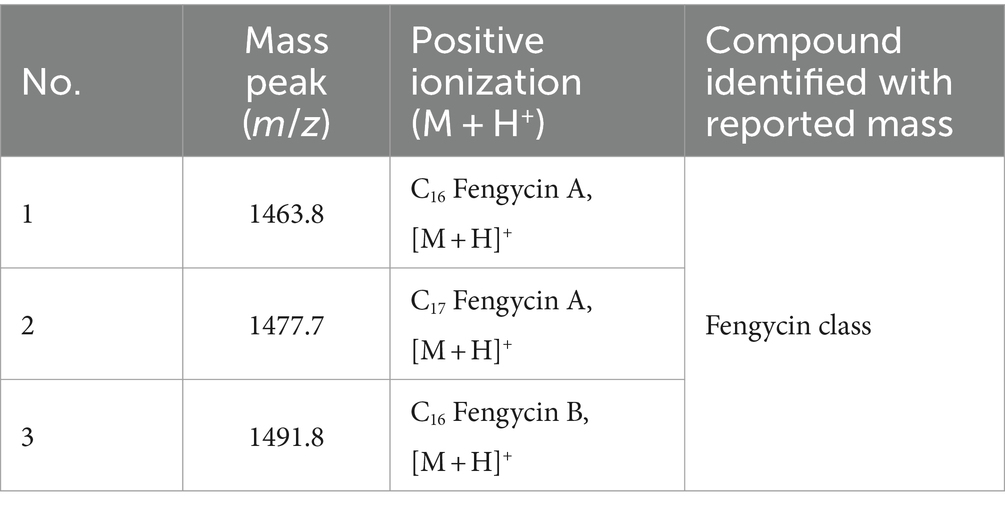
To identify the antifungal compounds produced by the bacterial isolate, the extract of TJS119 was subjected to UPLC–MS/MS analysis. The molecular ion masses of previously reported compounds were obtained from their LC–MS analysis. The UPLC–MS/MS data were analyzed for the presence of fingerprints of the compounds by matching them to those of previously reported structures. The results of the UPLC-MS/MS analysis suggested the presence of fengycin [m/z 1,063.8, 1,477.7, 1,491.8 (M + H+); Figure 5; Table 4].
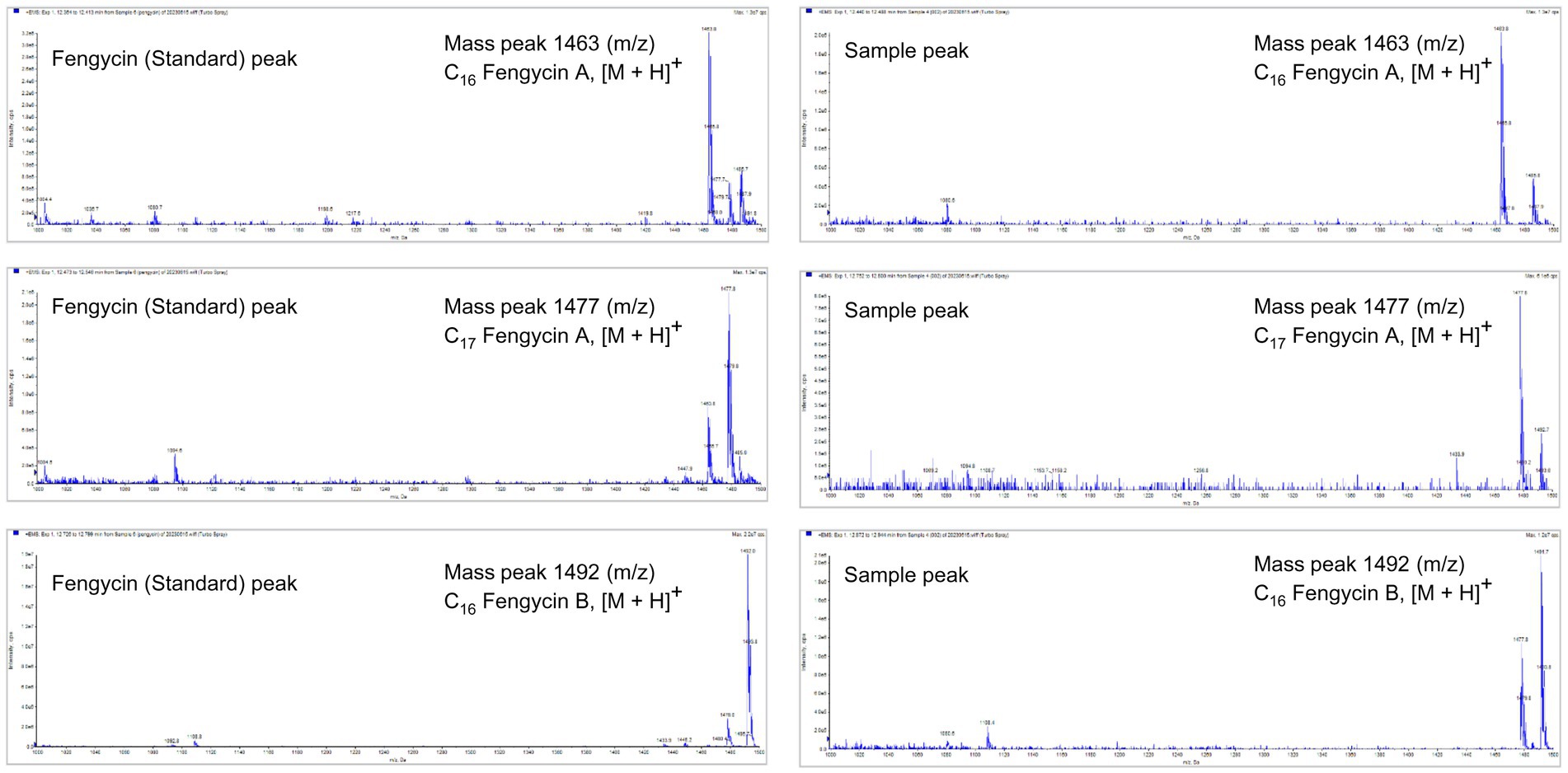
Figure 5. LC–MS/MS-based fragmentation analysis of fengycin (m/z 1463.8, 1477.7, and 1491.8) isolated from the n-butanol extract of Bacillus velezensis TJS119.
3.5 Biocontrol effect of TJS119 against Metarhizium anisopliae
One week 5, the incidence rate of fungal infection in the group was 60.83% ± 4.12%; whereas the incidence rate was 3.17% ± 4.08% when treated with B. velezensis TJS119 (1 × 107 CFU/g). The biocontrol effect of TJS119 against green muscardine disease (M. anisopliae) was 94.7%. The bioassay results indicated that TJS119 could control green muscardine disease (Table 5).
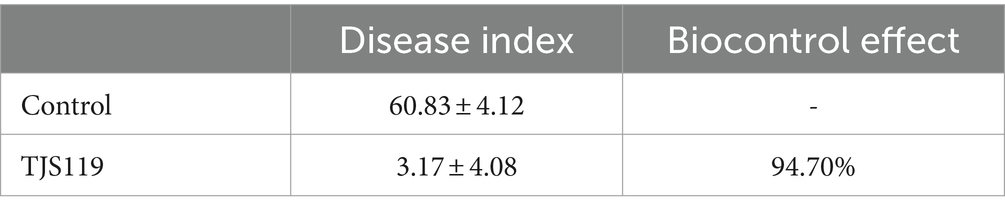
Table 5. Evaluation of biocontrol efficacy of Bacillus velezensis TJS119 against green muscardine disease.
4 Discussion
The genus Bacillus includes several species that are highly similar, and the taxonomic status of Bacillus strains cannot be easily determined using conventional classification methods (Hamdache et al., 2013). Several strains initially identified as B. amyloliquefaciens, including FZB42 (Schilling et al., 2018), vb7 (Saravanan et al., 2021), and SQR9 (Feng et al., 2018), are now classified as B. velezensis. To determine the relationship between TJS119 and other strains of Bacillus, a phylogenetic tree based on 92 core gene sequences was constructed in this study. The results indicate that TJS119 is closely related to B. velezensis NRRL B-41580T, B. amyloliquefaciens DSM 7 T, and B. siamensis KCTC 13613T.
Among biocontrol bacteria, Bacillus spp. are the most effective. The primary mechanisms employed by Bacillus spp. in disease control in plants involve antagonism, competition, and disease resistance induction. In addition, Bacillus spp. can enhance resistance by promoting plant growth in a synergistic manner (Backer et al., 2018; Rodriguez et al., 2019). However, no study has evaluated their ability to prevent diseases in industrial insect husbandry.
Recent studies have focused on secondary metabolites and enzymes produced by B. velezensis that confer resistance in plants against a broad-spectrum of pathogens (Wang et al., 2021). Antimicrobial cyclic lipopeptides synthesized by NRPS and polyketides synthesized via the polyketide pathway have broad-spectrum antimicrobial activity. Cyclic lipopeptides belong to three families, surfactin, iturin, and fengycin, all of which can induce systemic resistance in plants against pathogens (Wu et al., 2018). However, the gene cluster encoding iturin was not detected in TJS119. Polyketides, including macrolactins, bacillysins, and difficidins, can be used as antibiotics, antifungals, and natural pesticides. The most important Bacillus polyketides are those of B. subtilis and B. velezensis, with the former being particularly rich in these compounds (Rabbee and Baek, 2020). Using whole-genome sequencing, Zhang et al. (2022) found that B. velezensis GS-1 produces the lipopeptides surfactin, fengycin, and plantazolicin, which exhibit inhibitory effects against M. oryzae. Eight secondary metabolite-related gene clusters have been identified in B. velezensis VB7, which is protective against carnation infection by Sclerotinia sclerotiorum, cotton infection by tobacco streak virus, and tomato peanut shoot necrosis infection (Saravanan et al., 2021). Meanwhile, 12 secondary metabolite-related gene clusters have been identified in B. velezensis YC89, which effectively controls red rot disease in sugarcane (Xie et al., 2023). In the present study, 12 gene clusters were predicted in the TJS119 genome using antiSMASH, an online secondary metabolite gene cluster prediction tool, including four with unknown functions and seven highly similar antibiotic synthesis gene clusters (serpactin, macrolactin H, bacillin, fengycin, dipicidin, bacillibactin, and bacillicin). The 12 secondary metabolite gene clusters were identified using whole-genome sequencing. We further confirmed that fengycin was the main active secondary metabolite in the organic solvent extract.
The safety of bacteria used for biological control was confirmed. The analysis of genome of B. velezensis TJS119 against the VFDB database revealed the presence of some putative virulence genes, which cannot be considered harmful. Genes encoding hemolysin A (hlyA), cytolysin (cyl), enterotoxins hemolysin BL (Hbl), non-hemolytic enterotoxin (Nhe), and cytotoxin K (CytK), which are well-known potential virulence factors, are missing in B. velezensis TJS119 (Dietrich et al., 2021; Chen et al., 2023). In addition, no truly pathogenic coding genes were found in the genome of B. velezensis TJS119. Additionally, studies on B. velezensis using fish (Gao et al., 2017), pig (Bampidis et al., 2021), layer chicken (Ye et al., 2020), and mice (Chen et al., 2023) have suggested the safety of this strain controlling diseased in animals.
In the present study, we assessed the relative efficacy of B. velezensis TJS119 against green muscardine disease using laboratory tests. Bacillus velezensis TJS119 exhibited a relative efficacy of 94.7% against green muscardine disease. The successful introduction of biocontrol agents can be influenced by environmental various factors in the field, such as temperature, climate, and terrain. Therefore, further studies, particularly field trials with B. velezensis TJS119, are necessary.
5 Conclusion
In the field of industrial insect husbandry, the breeding of P. brevitarsis seulensis larvae is challenged by substantial mass mortality, primarily linked to the onset of diseases, specifically green muscardine disease. The use of chemical fertilizers and antifungal agents to increase production is unsustainable owing to their conspicuous environmental effects. Based on the findings of this study, B. velezensis TJS119 can be considered an alternative biocontrol agent to be formulated and extensively applied in bio-based industries. Our in vitro experiments and in vivo bioassay provide information on the genomic features of B. velezensis TJS119. The identified secondary metabolite clusters and the comparative analysis of the implicated genes indicate that the biological control properties of B. velezensis TJS119 are genetically associated, and thus inherently stable.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, JAXQPU000000000.
Author contributions
K-IH: Conceptualization, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. YN: Investigation, Resources, Writing – review & editing. BH: Formal Analysis, Investigation, Writing – review & editing. JK: Investigation, Writing – review & editing. JJ: Investigation, Resources, Writing – review & editing. EK: Funding acquisition, Writing – review & editing. M-HL: Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the Environmental Analysis of the Occurrence of Major Insect Diseases and Technology Development to Prevent Spread (RS-2021-RD009894) and the Nakdonggang National Institute of Biological Resources (NNIBR), funded by the Ministry of Environment (MOE) of Republic of Korea (NNIBR20243109).
Acknowledgments
We would like to thank Editage (www.editage.co.kr) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1361961/full#supplementary-material
Footnotes
1. ^ https://www.ezbiocloud.net/tools/orthoani
2. ^ https://antismash.secondarymetabolites.org/
References
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Bampidis, V., Azimonti, G., Bastos, M. D. L., Christensen, H., Dusemund, B., Durjava, M. F., et al. (2021). Safety and efficacy of a feed additive consisting of Bacillus velezensis DSM 15544 (Calsporin®) for piglets (suckling and weaned), pigs for fattening, sows in order to have benefit in piglets, ornamental fish, dogs, and all avian species (Asahi biocycle co.). EFSA J. 19:e06903. doi: 10.2903/j.efsa.2021.6903
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Calderone, R. (2002). Fungal Pathogenesis: Principles and Clinical Applications. Boca Raton: CRC Press.
Chen, B., Zhou, Y., Duan, L., Gong, X., Liu, X., Pan, K., et al. (2023). Complete genome analysis of Bacillus velezensis TS5 and its potential as a probiotic strain in mice. Front. Microbiol. 14:1322910. doi: 10.3389/fmicb.2023.1322910
Cosentino, S., Larsen, M. V., Aarestrup, F. M., and Lund, O. (2013). PathogenFinder--distinguishing friend from foe using bacterial whole genome sequence data. PLoS One 8:e77302. doi: 10.1371/journal.pone.0077302
Dietrich, R., Jessberger, N., Ehling-Schulz, M., Märtlbauer, E., and Granum, P. E. (2021). The food poisoning toxins of Bacillus cereus. Toxins 13:98. doi: 10.3390/toxins13020098
Dunlap, C. A., Kim, S. J., Kwon, S. W., and Rooney, A. P. (2015). Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and “Bacillus oryzicola” are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 66, 1212–1217. doi: 10.1099/ijsem.0.000858
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Feng, H., Zhang, N., Fu, R., Liu, Y., and Krell, T. (2018). Recognition of dominant attractants by key chemoreceptors mediates recruitment of plant growth-promoting rhizobacteria. Environ. Microbiol. 21, 402–415. doi: 10.1111/1462-2920.14472
Gao, X.-Y., Liu, Y., Miao, L., Li, E.-W., Sun, G.-X., Liu, Y., et al. (2017). Characterization and mechanism of anti-Aeromonas salmonicida activity of a marine probiotic strain, Bacillus velezensis V4. Appl. Microbiol. Biotechnol. 101, 3759–3768. doi: 10.1007/s00253-017-8095-x
Hamdache, A., Azarken, R., Lamarti, A., Aleu, J., and Collado, I. G. (2013). Comparative genome analysis of Bacillus spp. and its relationship with bioactive nonribosomal peptide production. Phytochem. Rev. 12, 685–716. doi: 10.1007/s11101-013-9278-4
Jin, Q., Jiang, Q., Zhao, L., Su, C., Li, S., Si, F., et al. (2017). Complete genome sequence of Bacillus velezensis S3-1, a potential biological pesticide with plant pathogen inhibiting and plant promoting capabilities. J. Biotechnol. 259, 199–203. doi: 10.1016/j.jbiotec.2017.07.011
Kim, S. Y., Lee, S. Y., Weon, H. Y., Sang, M. K., and Song, J. (2017a). Complete genome sequence of Bacillus velezensis M75, a biocontrol agent against fungal plant pathogens, isolated from cotton waste. J. Biotechnol. 241, 112–115. doi: 10.1016/j.jbiotec.2016.11.023
Kim, S. Y., Song, H., Sang, M. K., Weon, H. Y., and Song, J. (2017b). The complete genome sequence of bacillus velezensis strain GH1-13 reveals agriculturally beneficial properties and a unique plasmid. J. Biotechnol. 259, 221–227. doi: 10.1016/j.jbiotec.2017.06.1206
Lee, I., Kim, Y. O., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Li, C., Cheng, P., Zheng, L., Li, Y., Chen, Y., Wen, S., et al. (2021). Comparative genomics analysis of two banana fusarium wilt biocontrol endophytes Bacillus subtilis r31 and tr21 provides insights into their differences on phytobeneficial trait. Genomics 113, 900–909. doi: 10.1016/j.ygeno.2021.02.006
Li, Z., Meng, M., Li, S., and Deng, B. (2019). The transcriptome analysis of Protaetia brevitarsis Lewis larvae. PLoS One 14:e0214001. doi: 10.1371/journal.pone.0214001
Liu, G., Kong, Y., Fan, Y., Geng, C., Peng, D., and Sun, M. (2017). Whole-genome sequencing of Bacillus velezensis LS69, a strain with a broad inhibitory spectrum against pathogenic bacteria. J. Biotechnol. 249, 20–24. doi: 10.1016/j.jbiotec.2017.03.018
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Ann. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Molinatto, G., Puopolo, G., Sonego, P., Moretto, M., Engelen, K., Viti, C., et al. (2016). Complete genome sequence of Bacillus amyloliquefaciens subsp. plantarum S499, a rhizobacterium that triggers plant defences and inhibits fungal phytopathogens. J. Biotechnol. 238, 56–59. doi: 10.1016/j.jbiotec.2016.09.013
Na, S. I., Kim, Y. O., Yoon, S. H., Ha, S. M., Baek, I., and Chun, J. (2018). (UP: UBCG)-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285. doi: 10.1007/s12275-018-8014-6
Pan, H. Q., Li, Q. L., and Hu, J. C. (2017). The complete genome sequence of Bacillus velezensis 9912D reveals its biocontrol mechanism as a novel commercial biological fungicide agent. J. Biotechnol. 247, 25–28. doi: 10.1016/j.jbiotec.2017.02.022
Pan, J., Li, L., Liang, L., He, H., Su, Y., Wang, X., et al. (2017). Cytoprotective effects of cell-permeable bifunctional antioxidant enzyme, GST-TAT-SOD, against cisplatin-induced cell damage. Oxidative Med. Cell. Longev. 2017:9530791. doi: 10.1155/2017/9530791
Pang, Y., Yang, J., Chen, X., Jia, Y., Li, T., Jin, J., et al. (2021). An antifungal chitosanase from Bacillus subtilis SH21. Molecules 26:1863. doi: 10.3390/molecules26071863
Rabbee, M. F., and Baek, K. H. (2020). Antimicrobial activities of lipopeptides and polyketides of Bacillus velezensis for agricultural applications. Molecules 25:4973. doi: 10.3390/molecules25214973
Rabbee, M. F., Hwang, B., and Baek, K. (2023). Bacillus velezensis: a beneficial biocontrol agent or facultative phytopathogen for sustainable agriculture. Agronomy 13:840. doi: 10.3390/agronomy13030840
Rodriguez, P. A., Rothballer, M., Chowdhury, S. P., Nussbaumer, T., Gutjahr, C., and Falter-Braun, P. (2019). Systems biology of plant-microbiome interactions. Mol. Plant 12, 804–821. doi: 10.1016/j.molp.2019.05.006
Saravanan, R., Nakkeeran, S., Saranya, N., Senthilraja, C., Renukadevi, P., Krishnamoorthy, A. S., et al. (2021). Mining the genome of bacillus velezensis VB7 (CP047587) for MAMP genes and non-ribosomal peptide synthetase gene clusters conferring antiviral and antifungal activity. Microorganisms 9:2511. doi: 10.3390/microorganisms9122511
Schilling, T., Hoppert, M., Daniel, R., and Hertel, R. (2018). Complete genome sequence of vb_bvep-goe6, a virus infecting bacillus velezensis fzb42. Genome Announc. 6, e00008–e00018. doi: 10.1128/genomeA.00008-18
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Suh, H. J., and Kang, S. C. (2012). Antioxidant activity of aqueous methanol extracts of Protaetia brevitarsis Lewis (Coleoptera: Scarabaedia) at different growth stages. Nat. Prod. Res. 26, 510–517. doi: 10.1080/14786419.2010.530267
Wang, C., Ye, X., Ng, T. B., and Zhang, W. (2021). Study on the biocontrol potential of antifungal peptides produced by Bacillus velezensis against fusarium solani that infects the passion fruit Passiflora edulis. J. Agric. Food Chem. 69, 2051–2061. doi: 10.1021/acs.jafc.0c06106
Wu, G., Liu, Y., Xu, Y., Zhang, G., Shen, Q., and Zhang, R. (2018). Exploring elicitors of the beneficial Rhizobacterium Bacillus amyloliquefaciens SQR9 to induce plant systemic resistance and their interactions with plant signaling pathways. Mol. Plant-Microbe Interact. 31, 560–567. doi: 10.1094/MPMI-11-17-0273-R
Xie, L., Liu, L., Luo, Y., Rao, X., Di, Y., Liu, H., et al. (2023). Complete genome sequence of biocontrol strain Bacillus velezensis YC89 and its biocontrol potential against sugarcane red rot. Front. Microbiol. 14:1180474. doi: 10.3389/fmicb.2023.1180474
Ye, M., Wei, C., Khalid, A., Hu, Q., Yang, R., Dai, B., et al. (2020). Effect of Bacillus velezensis to substitute in-feed antibiotics on the production, blood biochemistry and egg quality indices of laying hens. BMC Vet. Res. 16:400. doi: 10.1186/s12917-020-02570-6
Zaid, D. S., Cai, S., Hu, C., Li, Z., and Li, Y. (2022). Comparative genome analysis reveals phylogenetic identity of Bacillus velezensis HNA3 and genomic insights into its plant growth promotion and biocontrol effects. Microbiol. Spectr. 10:e0216921. doi: 10.1128/spectrum.02169-21
Zhang, Y., Zhao, M., Chen, W., Yu, H., Jia, W., Pan, H., et al. (2022). Multi-omics techniques for analysis antifungal mechanisms of lipopeptides produced by Bacillus velezensis GS-1 against Magnaporthe oryzae in vitro. Int. J. Mol. Sci. 23:3762. doi: 10.3390/ijms23073762
Keywords: Bacillus velezensis , Protaetia brevitarsis seulensis larvae , biosynthetic genes, Metarhizium anisopliae , insect disease, green muscardine, biocontrol
Citation: Han K-I, Nam YH, Hwang BS, Kim JT, Jung JO, Kim E and Lee M-H (2024) Characterization of Bacillus velezensis TJS119 and its biocontrol potential against insect pathogens. Front. Microbiol. 15:1361961. doi: 10.3389/fmicb.2024.1361961
Edited by:
Oleksandr S. Yushchuk, University of Insubria, ItalyReviewed by:
Matteo Calcagnile, University of Salento, ItalyÇisem Bulut Albayrak, Adnan Menderes University, Türkiye
Copyright © 2024 Han, Nam, Hwang, Kim, Jung, Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kook-Il Han, aGtpNDYyOEBubmlici5yZS5rcg==
 Kook-Il Han
Kook-Il Han Young Ho Nam1
Young Ho Nam1 Mi-Hwa Lee
Mi-Hwa Lee