- 1Key Laboratory of Livestock Disease Prevention and Treatment of Guangdong Province, Institute of Animal Health, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2College of Veterinary Medicine, Inner Mongolia Agricultural University, Hohhot, China
Porcine epidemic diarrhea virus (PEDV) is responsible for causing fatal watery diarrhea in piglets, resulting in significant economic losses within the pig farming industry. Although vaccination is currently employed as a preventive measure, certain vaccines do not provide complete protection against PEDV field strains. Probiotics present a promising alternative due to their ability to regulate intestinal flora, enhance host immunity, and improve resistance against pathogenic microorganisms. We isolated six lactic acid bacteria (LAB) from the fecal microorganisms of Bama pigs, compared to Limosilactobacillus mucosae DSM13345 of the same genus in which Limosilactobacillus mucosae G01 (L. mucosae G01) proved to have a potent anti-PEDV effect. In a comprehensive manner, L. mucosae G01 significantly augmented the phosphorylation of IRF3 in IPEC-J2 cells, resulting in the induction of interferons (IFN α, IFN β, IFN λ1, and IFN λ3) and subsequent upregulation of interferon-stimulated genes (ISGs) (MX1, MX2, OAS1, and ZAP) in a dose-dependent fashion, consequently leading to the mitigation of PEDV replication. These findings underscore the promising prospects of L. mucosae G01 as a naturally derived substitute for combating PEDV and other enteric coronavirus infections.
1 Introduction
Piglet diarrhea has long been a significant challenge within the breeding industry, with the porcine epidemic diarrhea virus (PEDV) identified as the primary causative agent among various contributing factors. In particular, the infection of suckling piglets with PEDV often leads to severe diarrhea accompanied by a 100% mortality rate, resulting in significant economic losses (Jung et al., 2015; Kenney et al., 2021; Turlewicz-Podbielska and Pomorska-Mól, 2021). Although vaccination is commonly employed as a preventive measure, the effectiveness of certain vaccines may be compromised by genetic variations between the PEDV vaccine and the circulating strain in the field (Li et al., 2018; Luo et al., 2022). So, there is a need for developing complementary strategies that can prevent the spread of PEDV.
Probiotics have emerged as a promising alternative for restoring the ecological balance of the intestine and enhancing the intestinal microenvironment of the host (Kim et al., 2019). The beneficial effects of Lactobacillus on the gastrointestinal tract and overall health of the host are attributed to their ability to improve nutrient absorption, regulate harmful bacteria, and boost immune function (De Filippis et al., 2020; Di Giacomo et al., 2022). Additionally, the antiviral and antimicrobial properties of Lactobacillus further contribute to their potential as an alternative to conventional biological products (Arena et al., 2018). Lactobacillus has been found to disrupt viral adsorption and binding to epithelial surfaces, thereby inhibiting viral proliferation and stimulating immune responses against viruses (Wang et al., 2016; La Fata et al., 2018; Paone and Cani, 2020; Chen Y.-M. et al., 2022). Moreover, Lactobacillus has emerged as a leading microorganism in the colonization of the porcine intestinal tract, exerting a significant influence on the complex developmental trajectory of the intestinal immune system (Cebra, 1999). The strategic administration of Lactobacillus during episodes of gastrointestinal disturbance is crucial for maintaining the fragile balance of intestinal microflora (Tibary et al., 2006). This intervention provides a powerful method to protect the developing intestinal microbiota of nursing piglets during their critical early stages of growth and development (Siggers et al., 2008).
In comparison to other pig breeds in China, Bama sows exhibit enhanced adaptability and resistance to diseases, potentially due to the higher proportion of microbial species and greater abundance of gene copies of total bacteria in their gut microbiota (Yang et al., 2014). Previous studies have indicated the crucial involvement of intestinal microorganisms in the metabolic processes of Bama pigs, particularly during the pre-weaning stage (Yang et al., 2014; Ma et al., 2020; Wang K. et al., 2021). Chen C. et al. (2022) conducted an investigation into the intricate diversity of gut microbiota in Bama pigs, focusing on the effects of customized dietary interventions. The researchers discovered significant disparities in the composition of the gut microbiota in Bama pigs. By conducting thorough sequencing analysis of both gut flora and fecal microorganisms, they revealed a noteworthy abundance of probiotic species within the gastrointestinal ecosystem of these pigs. These findings serve as a relevant benchmark for studying probiotic populations present in the gut microbiota of Bama pigs (He et al., 2016; Chen C. et al., 2022). Consequently, these findings imply that the gut microbiota of Bama pigs might play a crucial role in their overall health and resistance to diseases.
In this study, our objective was to isolate Lactobacillus strains from Bama pig fecal and evaluate their capacity to inhibit PEDV replication in vitro. Through subsequent screening, we discovered a specific Limosilactobacillus mucosae G01 (L. mucosae G01) that exhibits antiviral properties. This L. mucosae G01 compound has the ability to enhance the phosphorylation of the crucial IRF3 protein in IPEC-J2 cells, leading to an upregulation of the transcription of type I and type III interferons, as well as interferon-stimulated genes (ISGs) such as MX1, MX2, OAS1, and ZAP. Consequently, this empowers IPEC-J2 cells to effectively combat PEDV infections.
2 Materials and methods
2.1 Cells and virus
The porcine intestinal epithelial cell line (IPEC-J2) were obtained from Dr. Li Wang at the Institute of Animal Science, Guangdong Academy of Agricultural Sciences, China. The cells were cultured in Roswell Park Memorial Institute (RPMI) Medium 1640 supplemented with 10% fetal bovine serum (FBS). Vero cells were stored in our laboratory at the Institute of Animal Health, Guangdong Academy of Agricultural Sciences, Guangzhou, China, were used for propagation of the GD/HZ/2016 (GenBank Accession: OP191700.1) strain. The vero cells were cultured in DMEM containing 15 μg/ml trypsin. The L. mucosae G01 strain uploaded with NCBI sequence number OR783178.
2.2 Isolation of Lactobacillus strains from fecal samples
The Lactobacillus strains utilized in this research were obtained from recently collected fecal samples of Guangxi Bama scented pigs. These samples were isolated from five sows in the early gestation stage and were collected over a span of three consecutive days. A total of 15 pig fecal samples were collected using sterile swabs. Subsequently, the samples were transferred into 1.5 mL centrifuge tubes containing PBS and promptly placed in ice boxes for transportation to the laboratory.
The procedures for isolating Lactobacillus strains from swine fecal samples were conducted following the methodology outlined by Halimi and Mirsalehian (2016). The collected fecal samples were introduced into de Man, Rogosa, and Sharpe (MRS) broth and incubated at a temperature of 37°C for a duration of 15 h. Subsequently, the samples were diluted and streaked onto MRS agar plates in triplicate. To amplify the 16S rRNA genes, the universal primers (27 F and 1492 R) were employed: 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′- GGTTACCTTGTTACGACTT-3′). The PCR products obtained were ligated into the pMD19T vector and subsequently subjected to sequencing analysis using the automated sequencing service offered by Sangon Bioengineering (Guangzhou) Co., Ltd. The 16S rRNA sequences obtained were subjected to comparison using the MUSCLE algorithm in Mega 11 v11.0.13 (Callaway and Singh-Cundy, 2019). Subsequently, a phylogenetic tree was constructed employing the maximum Neighbor Joining (NJ) algorithm to elucidate the evolutionary relationships among the isolated strains. The resulting files were subsequently uploaded to iTOL1 (Shi et al., 2018) for the purpose of visualizing the evolutionary tree, which included species information for each strain.
2.3 Cell viability test of Lactobacillus strains
The cell viability assay of Lactobacillus strains on IPEC-J2 cells was conducted following modifications to the method described by EL-Adawi (El-Adawi, 2012). IPEC-J2 cell suspensions were prepared at a concentration of 5×104 cells/ml and inoculated into 96-well plates, followed by incubation at 37°C for 12 h. Simultaneously, Lactobacillus strains were incubated overnight, washed three times with sterile PBS buffer, and subsequently centrifuged at 6,000 × g for 10 min. Subsequently, 10, 1, and 0.1 MOI of the Lactobacillus strains were added to each sample, which were then incubated for 24 h. The CCK-8 reagent was added and incubated at 37°C for 1 h. The absorbance at a wavelength of 450 nm was quantified utilizing the Multiskan Sky instrument manufactured by Thermo Scientific. The result of the calculation is: Cell viability (%) = (ODt / ODc) × 100%, where ODt and ODc are the absorbance of treated and control cells, respectively (Kurpe et al., 2020).
2.4 In vitro evaluation of PEDV with Lactobacillus strains
The experimental methodology employed to evaluate the therapeutic efficacy of Lactobacillus strains on IPEC-J2 cells infected with the PEDV strain GD/HZ/2016 was derived from the work of Chang-Liao et al. (2020). To assess the anti-PEDV effect, the IPEC-J2 cells were subjected to both prestimulation and poststimulation using Lactobacillus strains. For the prestimulation phase, the IPEC-J2 cells were cultured in 12-well plates and exposed to either 1 or 0.1 MOI of Lactobacillus strains. Following a 2 h incubation period, the cells were rinsed thrice with sterile PBS and subsequently infected with 0.1 MOI of PEDV. Post-stimulation, the IPEC-J2 cells were exposed to 0.1 MOI of PEDV, followed by three washes with sterile PBS. Subsequently, the cells were treated with either 1 or 0.1 MOI of Lactobacillus strains for a duration of 2 h. After another wash with sterile PBS, the cultures were incubated at 37°C in a 5% CO2 environment and harvested 24 h post-infection with PEDV. Cell samples were collected for qRT-PCR analyses to evaluate the impact of Lactobacillus strains on virus replication.
2.5 RNA extraction and reverse transcription quantitative polymerase chain reaction (RT-qPCR)
The TaKaRa MiniBEST Universal RNA Extraction Kit, manufactured in China by TaKaRa, was utilized to extract total RNA from IPEC-J2 cells subjected to various experimental conditions. For qRT-PCR analysis, the HiScript® II one Step qRT-PCR SYBR Green Kit, produced by Vazyme in China, was employed. The primer sequences employed for gene detection can be found in Table 1.
2.6 Western blot
The experimental procedure was performed according to the methodology outlined by Shen et al. (2016). In brief, IPEC-J2 cells were lysed using 150 μL of lysis buffer obtained from Thermo Fisher Scientific, China. The resulting proteins were then separated and transferred onto a polyvinylidene fluoride membrane. Subsequently, the membrane was incubated for 1 h with 5% non-fat milk, followed by a 2 h incubation with primary antibodies against PEDV N-protein (Medgene Labs, USA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ABclonal, China), synthetic phosphorylated peptide around interferon regulatory Factor 3 (pIRF3) (ABclonal, China), and interferon regulatory Factor 3 (IRF3) (ABclonal, China), followed by three washes with PBST. The PVDF membranes were subsequently subjected to incubation with horseradish peroxidase-conjugated goat anti-mouse and rabbit anti-lgG(H + L) secondary antibodies (Bioworld, China). The visualization of immunity protein bands was achieved by employing an ECL Kit (Millipore, China).
2.7 Immunofluorescence assay
Refer to Zhang W’s method (Zhang et al., 2023) as follows. The cells were fixed using a 4% paraformaldehyde solution (Beyotime, China) and permeabilized with a PBS solution containing 0.1% Triton X-100. Subsequently, the cells were blocked with a 1% bovine serum albumin solution (BBI Life Sciences, China) for 1 h. Following this, the cells were incubated overnight at 4°C with a mouse anti-PEDV N IgG antibody, and then with Alexa Fluor™ 594 goat anti-mouse IgG (Invitrogen, China) for 1 h at 37°C. After washing with PBS three times, the cells were incubated with 4,6-diamidino-2-phenylindole (DAPI, Beyotime) for 5 min. Finally, the cells were examined using a differential fluorescence microscope.
2.8 Virus titer detection
Refer to Zhang et al. (2023) method as follows. In order to ascertain the virus titers, Vero cells were cultivated in 96-well plates until reaching a confluence of 70–80%. Subsequently, the cells were infected with 10-fold serial dilutions of PEDV, with a total of four replicates. Following an incubation period of 72 h, the cells were evaluated using the immunofluorescence assay (IFA). The Reed-Muench method was employed to determine the virus titers, which were quantified as TCID50 per milliliter.
2.9 Data analysis
The data analysis was performed utilizing GraphPad Prism v8.0 software, employing an ANOVA to evaluate the statistical significance of differences in mean values. The mean values for each group were reported as the mean ± standard deviation (SD). Statistical analyses were conducted to ascertain the presence of significant differences between the treatment and control groups. One-way analysis of variance followed by the Tukey-Kramer post-test was used. A p < 0.05 was considered statistically significant.
3 Results
3.1 Molecular identification of isolated Lactobacillus strains from Bama pig fecal
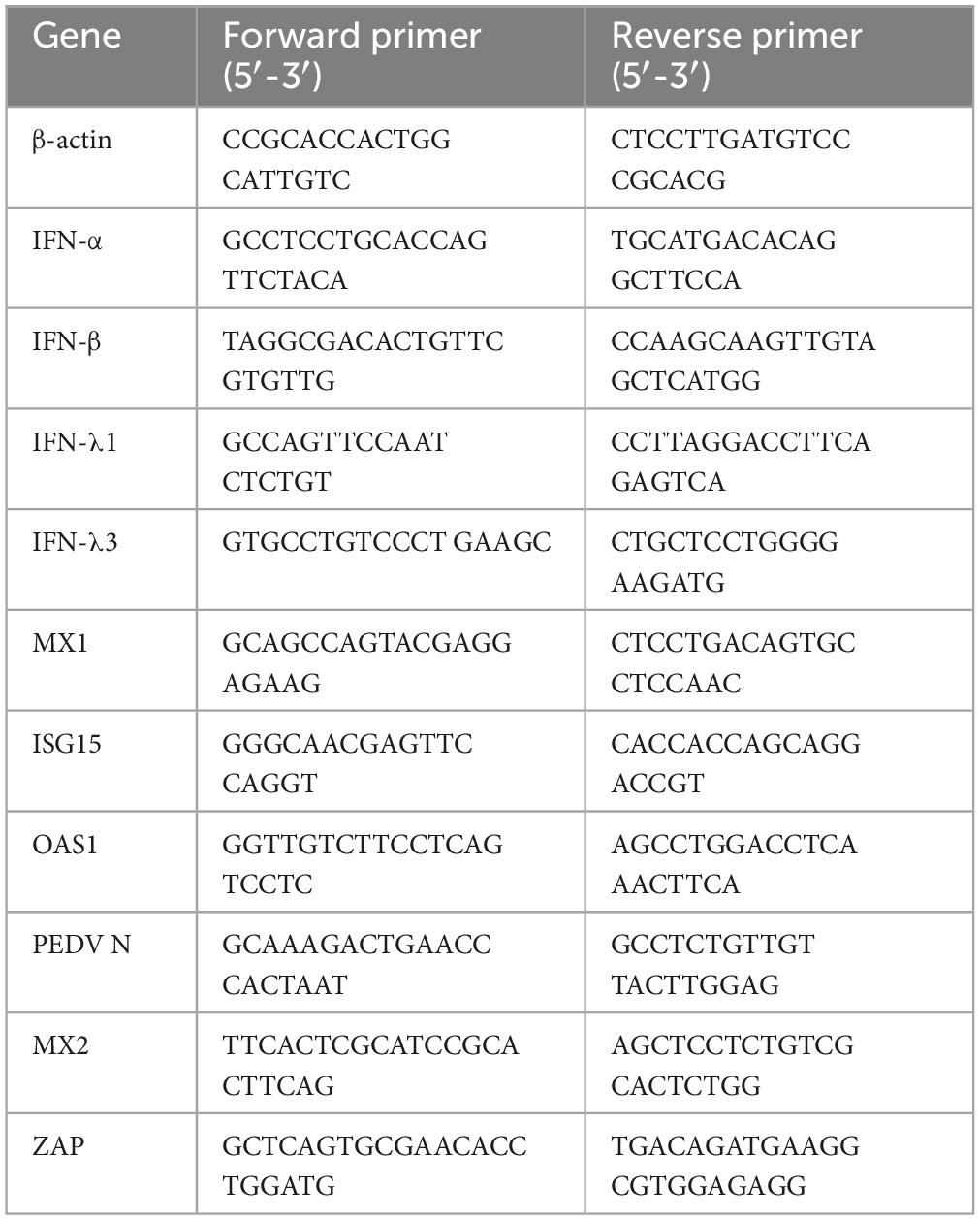
A total of 70 Lactobacillus strains were selected for polymerase chain reaction (PCR) using the 27F/1492R common primer specific for bacterial 16S rRNA. This PCR amplification resulted in a bright band at approximately 1500 base pairs, indicating successful amplification of the 16S rRNA product (Figure 1A). The PCR products were subsequently sequenced and compared to the National Center for Biotechnology Information (NCBI) database for further analysis of the isolated Lactobacillus species. The analysis revealed that 28 strains potentially belonged to Limosilactobacillus reuteri DSM 20016 (L. reuteri DSM20016), 3 strains to Weissella thailandensis FS61-1, and 4 strains to Limosilactobacillus vaginalis ATCC 49540 (L. vaginalis ATCC49540). Seventeen strains were identified as Limosilactobacillus mucosae G01 (L. mucosae G01), twelve as Limosilactobacillus johnsonii CIP103620 (L. johnsonii CIP103620), and 6 strains as Ligilactobacillus salivarius JCM1231 (L. salivarius JCM1231) (Figure 1B). The construction of a phylogenetic tree utilizing the 16S rRNA gene sequences revealed a range of Lactobacillus species that were obtainable from Bama pigs fecal (Figure 1C).
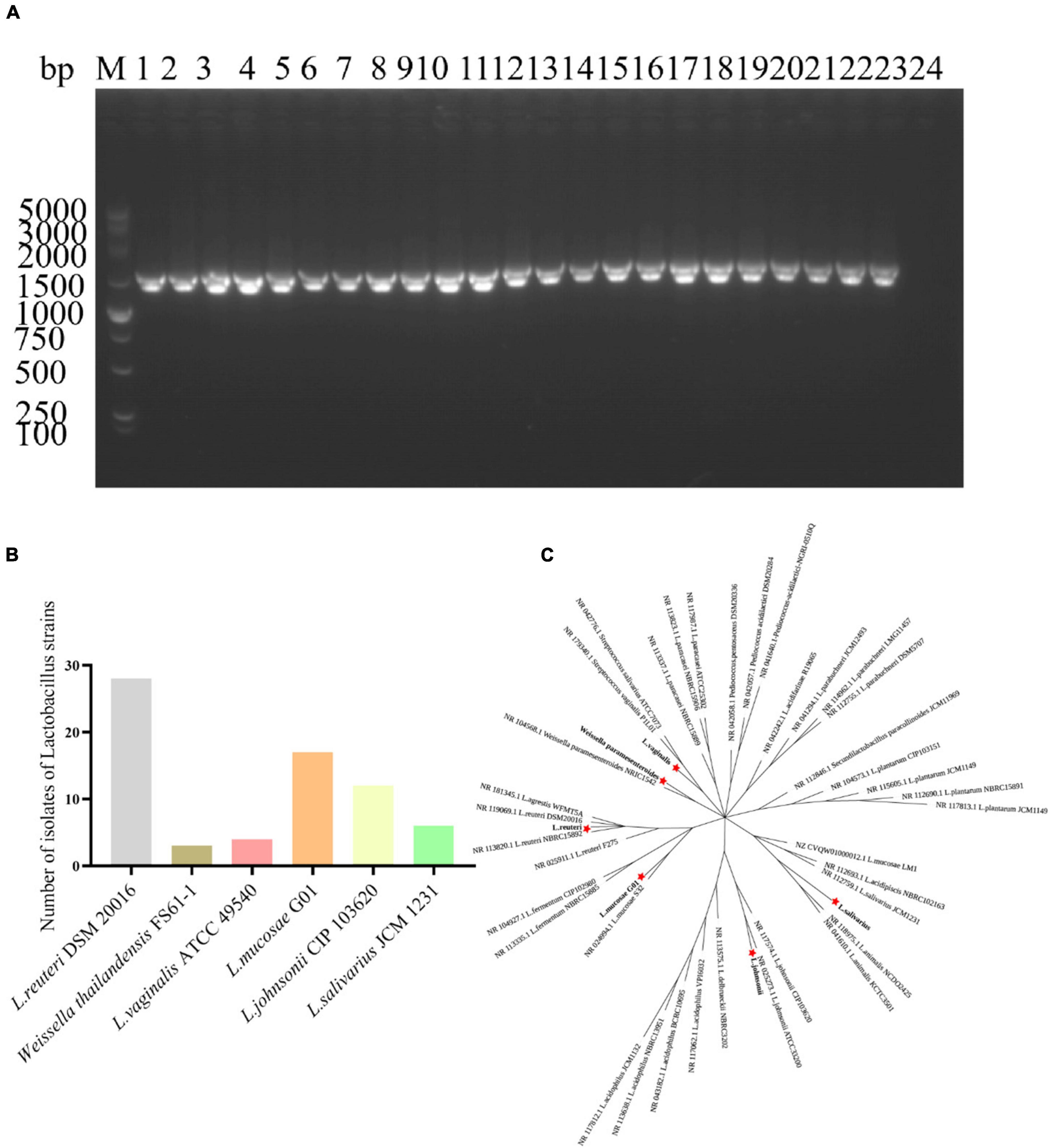
Figure 1. Isolation and characterization of Lactobacillus strains from fecal samples of Bama pigs. (A) Representative amplification of the PCR product using primers 27F/1492R. M denotes the DL5000 marker, and 1–23 correspond to the monoclonal Lactobacillus strains tested. The Line 24 was used as a negative control well. (B) Proportion of total isolates accounted for by Lactobacillus in fecal samples. Gray color is L. reuteri DSM20016 strain, brown color is Weissella thailandensis FS61-1 strain, light pink color is L. vaginalis ATCC49540 strain, orange color is L. mucosae G01, light yellow color is L. johnsonii CIP103620, and green color is L. salivarius JCM1231 strain. (C) An evolutionary tree was constructed employing the maximum likelihood method, utilizing 16S rRNA gene sequences of the isolated Lactobacillus strains. Mega 11 v11.0.13 was employed for construction, and the tree was enhanced using the iTOL online software. The red star pattern indicates the current isolate.
3.2 Cytotoxicity test of Lactobacillus strains
To assess the cytotoxic effects of various Lactobacillus strains on IPEC-J2 cells, a CCK-8 assay was conducted using three different bacterial concentrations (10, 1, and 0.1 MOI). As illustrated in Figure 2, the L. mucosae G01 strain was observed to promote the proliferation of IPEC-J2 cells, as evidenced by the optical density value (OD450) detected using the CCK-8 reagent. At a concentration of 0.1 MOI, L. johnsonii CIP103620, L. salivarius JCM1231, and the L. mucosae G01 strain exhibited no cytotoxicity toward IPEC-J2 cells. Conversely, Weissella thailandensis FS61-1 and L. vaginalis ATCC49540 demonstrated cytotoxic effects on IPEC-J2 cells.
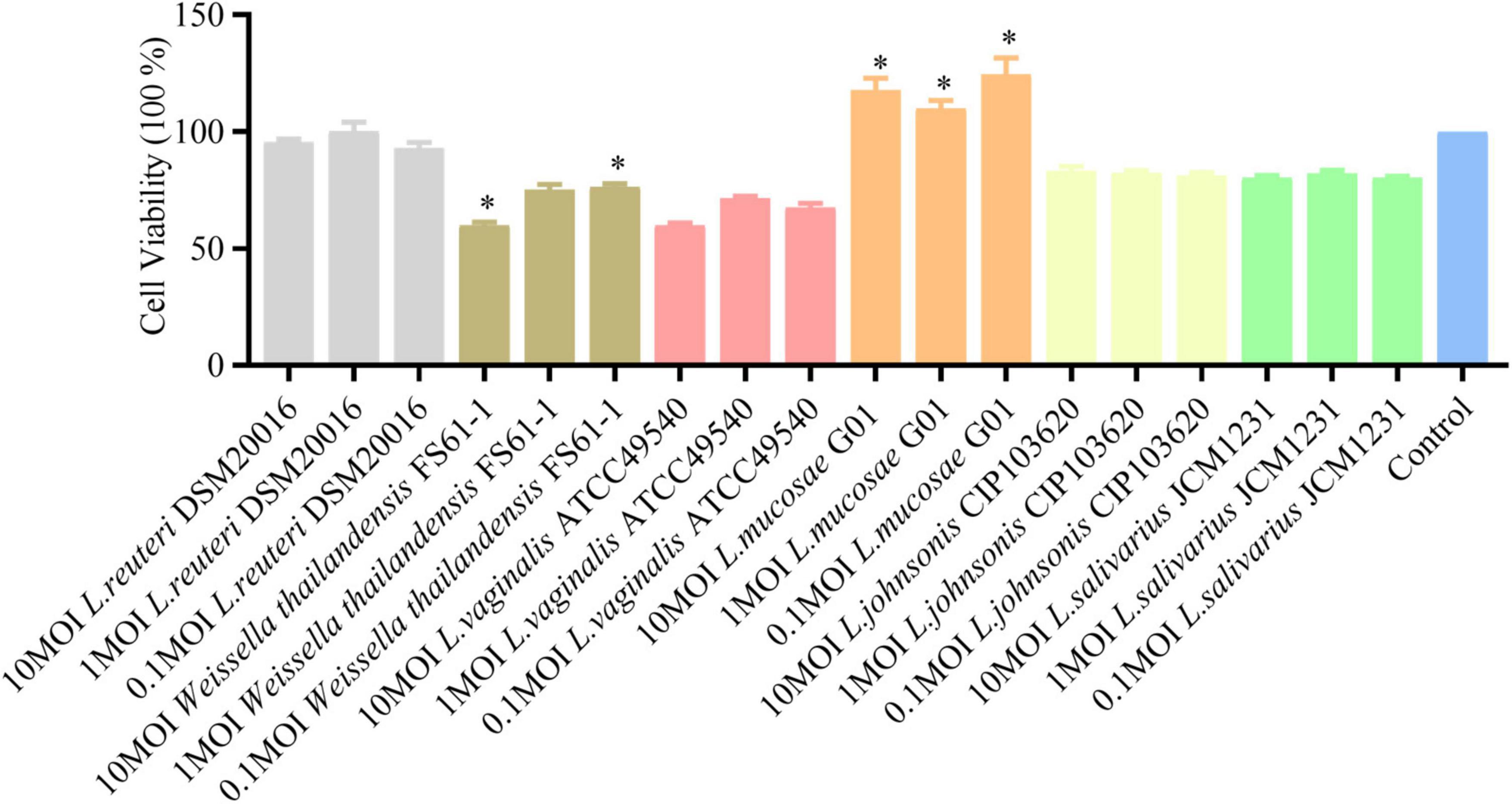
Figure 2. Effect of isolated lactic acid bacteria strains on IPEC-J2 cell viability. Lactobacillus strains, isolated from intestinal and fecal samples, were incubated with IPEC-J2 cells at concentrations of 10, 1, and 0.1 MOI for 24 h at 37°C, and cell viability was assessed using the CCK-8 (5 mg/ml, 10 μl/ well). OD450, a widely employed method for measuring cell viability, is presented. Gray color is L. reuteri DSM20016 strain, brown color is Weissella thailandensis FS61-1 strain, light pink color is L. vaginalis ATCC49540 strain, orange color is L. mucosae G01, light yellow color is L. johnsonii CIP103620, green color is L. salivarius JCM1231 strain and blue color is IPEC-J2 cells. All assays were performed in triplicates, with three replicates per experiment, and each bar is the mean ± SEM. The asterisk indicates significant differences compared to the control group (p < 0.05).
3.3 Screening of antiviral Lactobacillus strains
The inhibitory effects of L. reuteri DSM20016, L. mucosae G01, L. johnsonii CIP103620, and L. salivarius JCM1231 on PEDV infection in IPEC-J2 cells were assessed through prestimulation and poststimulation (Figure 3). Analysis using qRT-PCR revealed a significant reduction in PEDV N mRNA levels with L. mucosae G01 treatment. Furthermore, L. reuteri DSM20016 and L. mucosae G01 demonstrated inhibition of viral replication in the poststimulation mode, while the other strains did not exhibit a significant effect. Considering the antiviral properties observed, L. mucosae G01 was chosen for subsequent investigation.
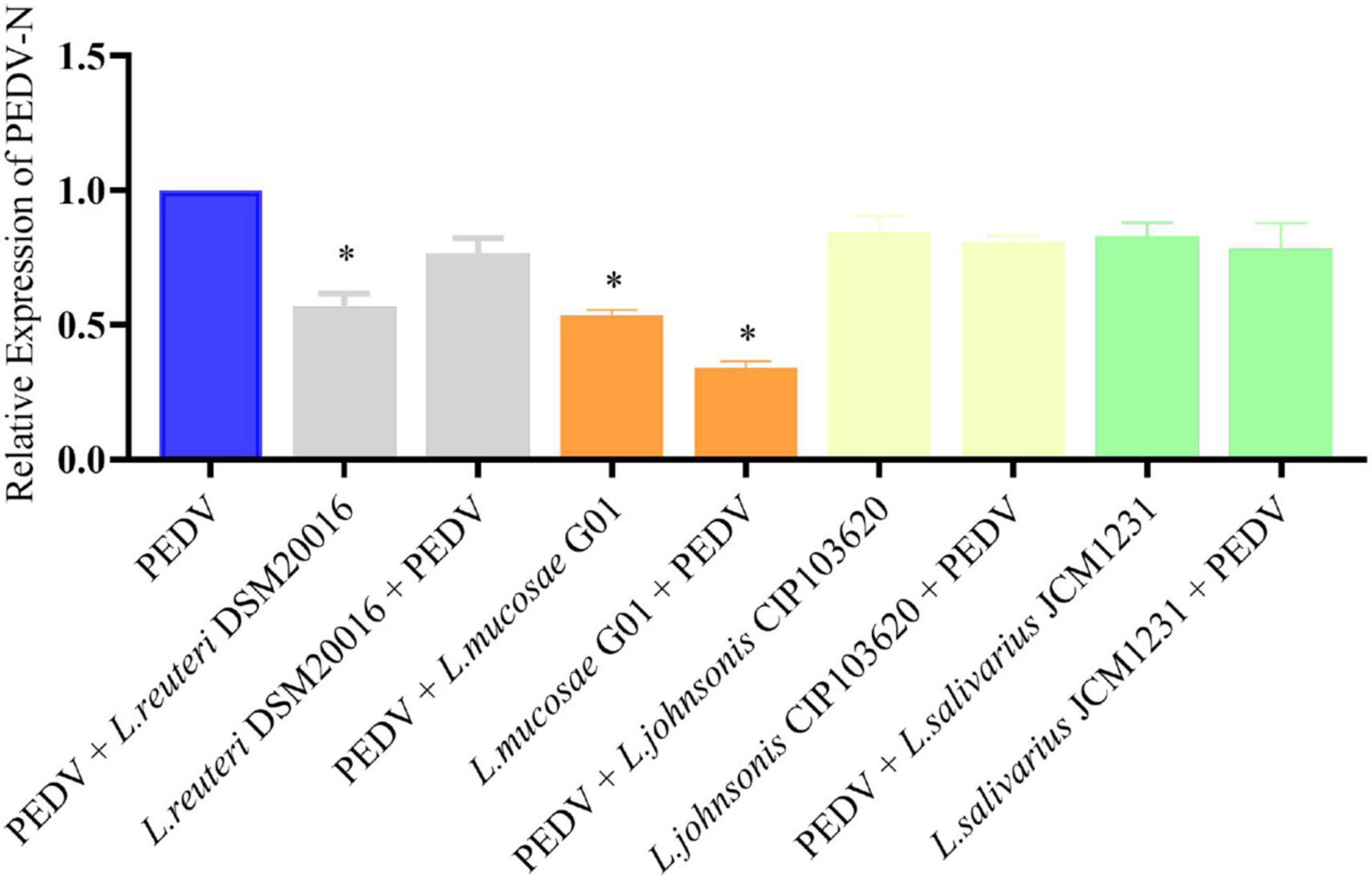
Figure 3. Lactobacillus strains screened for antiviral activity. IPEC-J2 cells were pre-stimulated or post-stimulated with Lactobacillus strains at 1 MOI for 2 h before or after PEDV infection. Following incubation at 37°C for 24 h, the copy number of the PEDV N gene was quantified using quantitative reverse transcription polymerase chain reaction (qRT-PCR). Gray color is L. reuteri DSM20016 strain, orange color is L. mucosae G01, light yellow color is L. johnsonii CIP103620, green color is L. salivarius JCM1231 strain and blue color is PEDV control group. All assays were performed in triplicates, with three replicates per experiment, and each bar is the mean ± SEM. The asterisk indicates significant differences compared to the control group (p < 0.05).
3.4 Comparison of the anti-PEDV effect of L. mucosae G01 with that of commercialized L. mucosae DSM13345
In order to assess the anti-PEDV efficacy of various L. mucosae G01 strains, the L. mucosae DSM13345 strain (obtained from Minebea) was utilized as a control. IPEC-J2 cells infected with PEDV were subjected to pre-stimulation with L. mucosae at a 1 MOI concentration, and the expression levels of PEDV N mRNA were determined using qPCR. The L. mucosae G01 strain isolated from the intestine demonstrated a significant inhibitory effect on PEDV replication in comparison to the L. mucosae DSM13345 strain (Figure 4). The L. mucosae G01 significantly reduced (p < 0.05) PEDV N mRNA expression compared to L. mucosae DSM13345 (Figure 4A), and supernatant viral titers were shown to be consistent with mRNA levels as well (Figure 4C). The findings from the figure strongly suggest that L. mucosae G01 possesses the potential to counteract PEDV infection in IPEC-J2 cells.
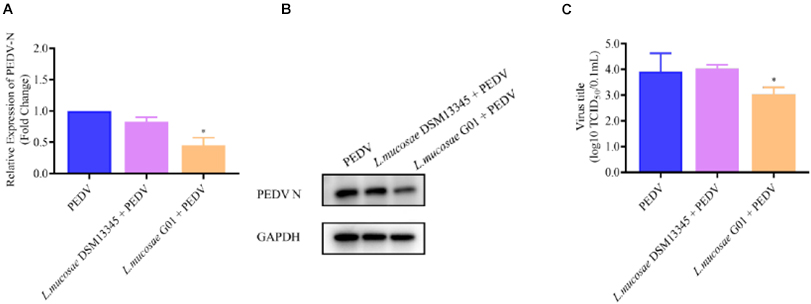
Figure 4. Depicts a comparison of the anti-PEDV effects of 1 MOI L. mucosae DSM13345 and L. mucosae G01 strains on IPEC-J2 cells. The method was utilized to evaluate the expression levels of the N protein in L. mucosae DSM13345 and L. mucosae G01 following 2 h interaction with IPEC-J2 cells, subsequent addition of PEDV-HZ, and after 24 h cell supernatants were harvested. Cell lysate containing PMSF was added for nucleic acid extraction and Western Blot assay. (A) RT-qPCR method was used to detect L. mucosae DSM13345 and L. mucosae G01 samples PEDV N mRNA expression level. (B) The quantification of N protein expression levels was performed through Western blot analysis. PEDV N is the viral content of the N nucleocapsid, and GAPDH is an internal reference for harvested cells. (C) TCID50 analysis cell supernatant titer. The purple color is L. mucosae DSM13345, orange color is L. mucosae G01, and blue color is PEDV control group. All assays were performed in triplicates, with three replicates per experiment, and each bar is the mean ± SEM. The asterisk indicates significant differences compared to the control group (p < 0.05).
To ascertain the potential correlation between the concentration of L. mucosae G01 and the potency of antiviral activity, our findings indicate that pre-stimulating IPEC-J2 cells with 0.1 and 1 MOI of L. mucosae G01 for a duration of 24 h resulted in a notable reduction in PEDV N mRNA levels (p < 0.05) (Figure 5A). Furthermore, a dose-dependent decline in PEDV N protein expression was observed (p < 0.05) (Figures 5B, E). These outcomes strongly imply that L. mucosae G01 effectively impedes PEDV production, with inhibition rates ranging from 21.90 to 68.10%. Furthermore, our study revealed a significant decrease in PEDV titers in a dose-dependent manner (p < 0.05), as evidenced by a 1.53-log reduction in progeny virus levels, indicating the inhibition of PEDV replication (Figures 5C, F). However, no statistically significant differences were observed in the expression levels of PEDV N mRNA in poststimulation IPEC-J2 cells following PEDV infection at 0.1 MOI L. mucosae G01 (Figure 5D). These findings suggest that L. mucosae G01 exhibits anti-PEDV activities, which hold potential for the development of effective interventions against this pathogen.
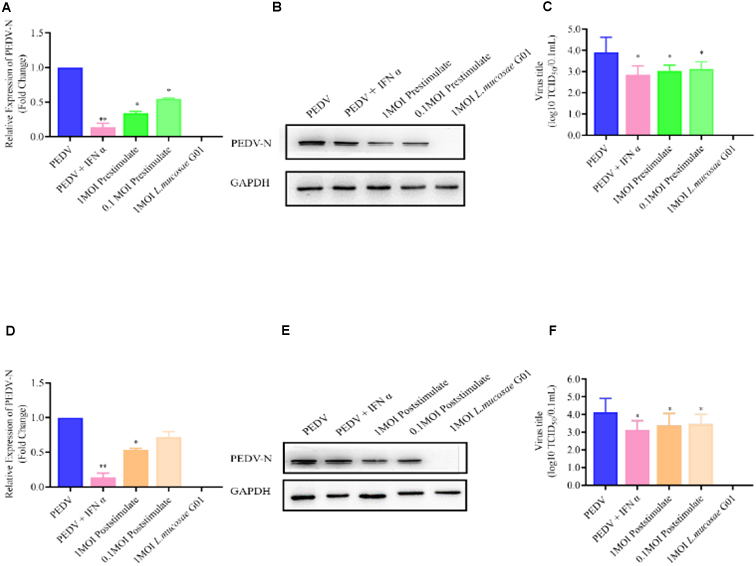
Figure 5. L. mucosae G01 inhibits PEDV replication with both prestimulate and post stimulate. Depicts the prestimulation and poststimulation effects of L. mucosae G01 strain were demonstrated in IPEC-J2 cells after 24 h with 1 and 0.1 MOI and expression level of PEDV N protein and treat vero cells with a 10-fold dilution of viral supernatant for 72 h. (A) Expression content of mRNA levels of PEDV N nucleocapsid protein in response to prestimulation at 1 and 0.1 MOI. (B) Expression levels of PEDV N protein in response to prestimulation with at 1 and 0.1 MOI. (C) TCID50 analysis cell supernatant titer were measured with prestimulation at 1 and 0.1 MOI. (D) Expression content of mRNA levels of PEDV N nucleocapsid protein in response to poststimulation at 1 and 0.1 MOI. (E) Expression levels of PEDV N protein in response to poststimulation with at 1 and 0.1 MOI. (F) TCID50 analysis cell supernatant titer were measured with prestimulation at 1 and 0.1 MOI. The experimental groups included PEDV-infected, virus infected after IFN treated cells, cells infected with L. mucosae G01 1 MOI and then treated with PEDV, cells infected with L. mucosae G01 0.1 MOI and then treated with PEDV, and cells infected with L. mucosae G01. Cells infected 0.1 MOI PEDV then treated with L. mucosae G01 at 1 and 0.1 MOI. Pink color is PEDV and IFN-α as positive control. Green is the prestimulation group: 1 MOI L. mucosae G01 interacted with IPEC-J2 cells followed by 0.1 MOI PEDV, light green is the prestimulation group: 0.1 MOI L. mucosae G01 interacted with IPEC-J2 cells followed by 0.1 MOI PEDV, and orange is the treatment group: 0.1 MOI PEDV interacted with IPEC-J2 cells before interacting with 1 MOI L. mucosae G01, light yellow is 0.1 MOI PEDV first interacting with IPEC-J2 cells before interacting with 0.1 MOI L. mucosae G01 and blue color is PEDV control group. All assays were performed in triplicates, with three replicates per experiment, and each bar is the mean ± SEM. Statistically significant differences between groups are indicated by *p < 0.05 or **p < 0.01.
3.5 The L. mucosae G01 strain induces interferon and interferon-stimulated genes (ISGs) production in IPEC-J2 cells
In the present study, the investigation focused on assessing the levels of interferons (IFN α, IFN β, IFN λ1, and IFN λ3) in IPEC-J2 cells before and after stimulation with the L. mucosae G01 strain. The obtained results revealed a significant elevation in the expression of IFN types of interferons in IPEC-J2 cells inoculated with L. mucosae G01 compared to the control group. Furthermore, both pre- and post-stimulation with L. mucosae G01 were found to significantly enhance the expression of IFN α, IFN β, IFN λ1, and IFN λ3 (Figures 6A–D). These findings suggest that L. mucosae G01 has the potential to augment the antiviral response by inducing the expression of interferons in IPEC-J2 cells.
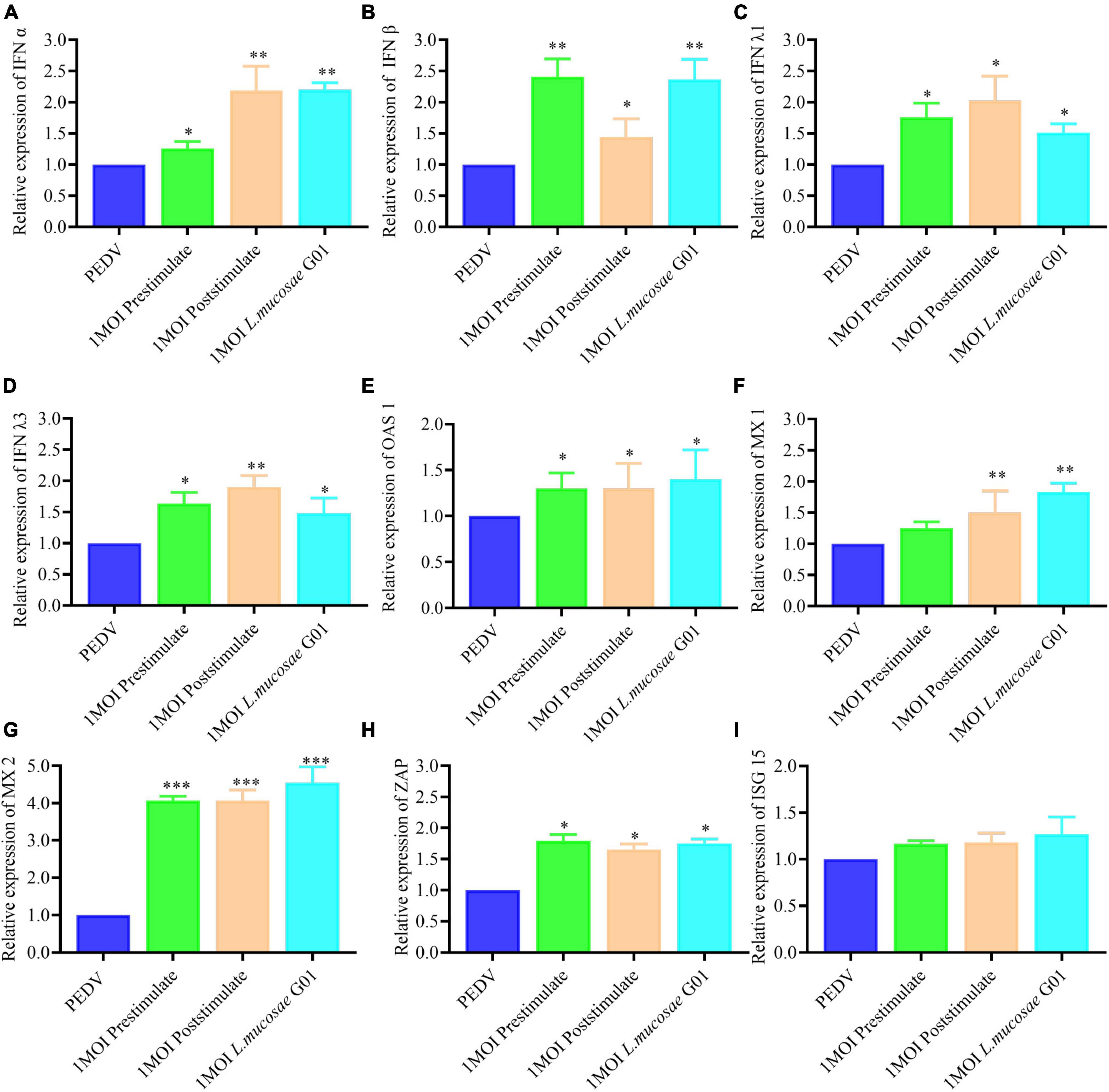
Figure 6. The L. mucosae G01 strain induces interferon and Interferon-stimulated genes (ISGs). The prestimulation and poststimulation effects of L. mucosae G01 strain were demonstrated in IPEC-J2 cells after 24 h with 1 MOI. Cells were harvested and cell lysate containing PMSF was added, followed by nucleic acid extraction. The mRNA levels of IFN α, IFN β, IFN λ1, IFN λ3 and effector protein were detected using qPCR. (A) Expression content of mRNA levels of IFN α in response to prestimulation or poststimulation at 1 MOI. (B) Expression content of mRNA levels of IFN β in response to prestimulation or poststimulation at 1 MOI. (C) Expression content of mRNA levels of IFN λ1 in response to prestimulation or poststimulation at 1 MOI. (D) Expression content of mRNA levels of IFN λ3 in response to prestimulation or poststimulation at 1 MOI. (E) Expression content of mRNA levels of OAS1 in response to prestimulation or poststimulation at 1 MOI. (F) Expression content of mRNA levels of MX1 in response to prestimulation or poststimulation at 1 MOI. (G) Expression content of mRNA levels of MX2 in response to prestimulation or poststimulation at 1 MOI. (H) Expression content of mRNA levels of ZAP in response to prestimulation or poststimulation at 1 MOI. (I) Expression content of mRNA levels of ISG15 in response to prestimulation or poststimulation at 1 MOI. The experimental groups included PEDV infected, cells infected with L. mucosae G01 1 MOI and then treated with PEDV, cells infected with L. mucosae G01 1 MOI and then treated with PEDV, and cells infected with L. mucosae G01. Green is the prestimulation group: 1 MOI L. mucosae G01 interacted with IPEC-J2 cells followed by 0.1 MOI PEDV, orange is the treatment group: 0.1 MOI PEDV interacted with IPEC-J2 cells before interacting with 1 MOI L. mucosae G01, azure is the L. mucosae G01 group, and blue color is PEDV control group. All assays were performed in triplicates, with three replicates per experiment, and each bar is the mean ± SEM. Statistically significant differences between groups are indicated by *p < 0.05, **p < 0.01 and ***p < 0.001.
When comparing the IPEC-J2 cells after stimulation with L. mucosae G01 to the PEDV cells, it was observed that both prestimulation and poststimulation resulted in a significant increase in MX2 expression (p < 0.001) (Figure 6G). Furthermore, the levels of OAS1, MX1, and ZAP expression in the prestimulated and post stimulated IPEC-J2 cells were found to be significantly different following L. mucosae G01 infection (p < 0.05) (Figures 6E, F, H). However, L. mucosae G01 did not cause changes in ISG15 (Figure 6I).
3.6 The L. mucosae G01 strain can stimulate IRF3 phosphorylation in IPEC-J2 cells
PEDV binds to the TLR3 receptor in IPEC-J2 cells, triggering an intracellular signaling cascade that results in the phosphorylation of interferon regulatory factor 3 (IRF3). Subsequently, the activated IRF3 translocates to the nucleus and stimulates the transcription of interferons (Stanifer et al., 2019).
To investigate the impact of L. mucosae G01 on IRF3 phosphorylation and IFN signaling, we conducted an assessment of the pIRF3/IRF3 ratio in IPEC-J2 cells subjected to L. mucosae G01 treatment. The outcomes demonstrate that exposure to 1 MOI and 0.1 MOI L. mucosae G01 strain resulted in a dose-dependent stimulation of IRF3 phosphorylation in IPEC-J2 cells (Figure 7). These findings imply that L. mucosae G01 possesses the ability to induce IRF3 phosphorylation in IPEC-J2 cells, potentially contributing to its anti-PEDV properties. The quantification of p-IRF3/IRF3 levels was accomplished through western blot analysis of lysed cellular proteins. The results of this study indicate that L. mucosae G01 potentially contributes to the activation of IFN signaling and the induction of IRF3 phosphorylation in IPEC-J2 cells.
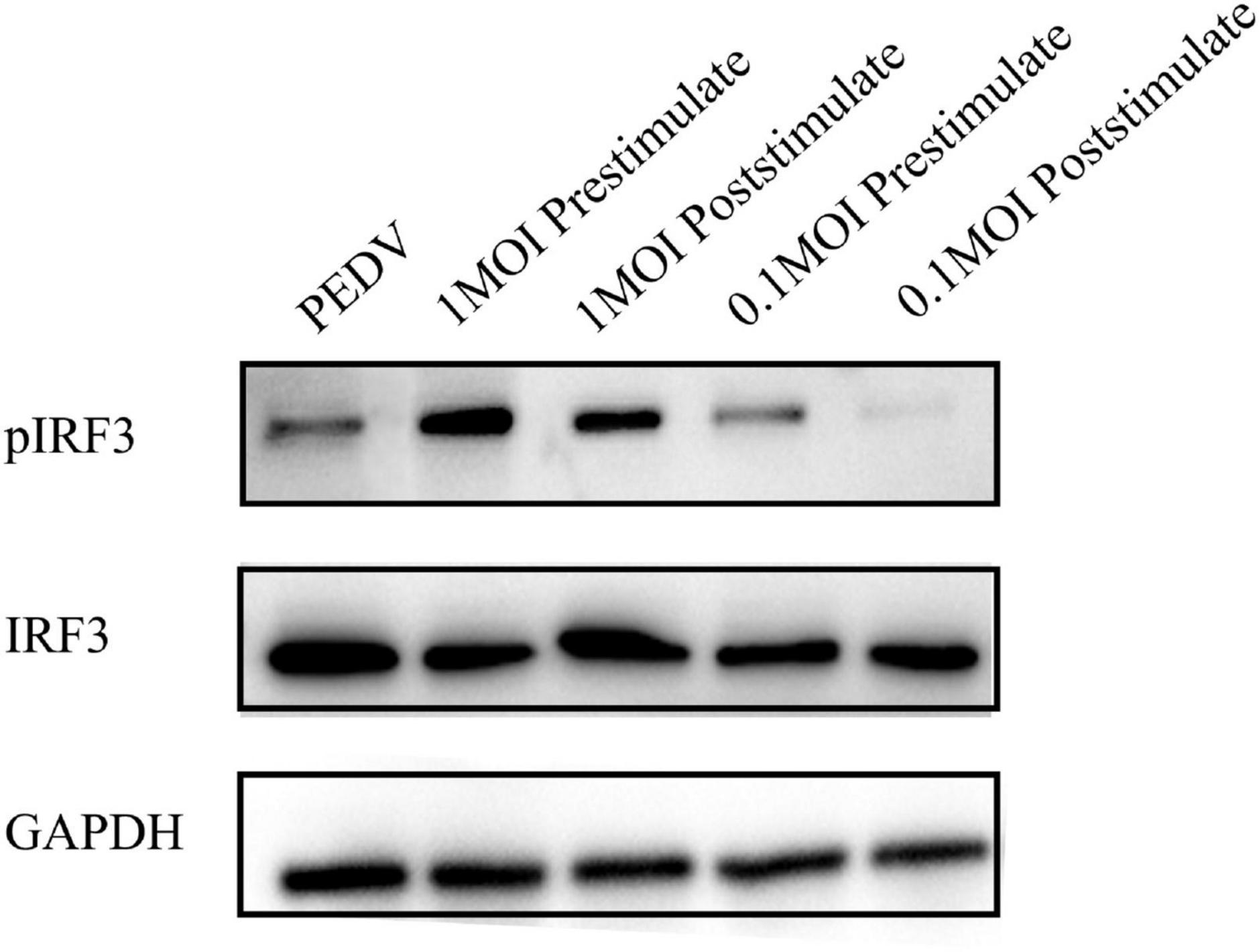
Figure 7. The L. mucosae G01 strain upregulates stimulate IRF3 phosphorylation in IPEC-J2 cells. The pIRF3 is phosphorylated IRF3, pIRF3/IRF3 ratio assesses whether the interferon pathway is activated, and GAPDH, an intracellular reference. The L. mucosae G01 strain at 1 and 0.1 MOI were demonstrated in IPEC-J2 cells after 30 min. IPEC-J2 cells were pre-treated with 1 and 0.1 MOI of L. mucosae G01 for 30 min, followed by the addition of 0.1 MOI PEDV, and harvested after 2 h. Western blot analysis was performed to quantify the levels of p-IRF3/IRF3 after prestimulation and poststimulation the cells with PEDV. The different treatment conditions included PEDVgroup, L. mucosae G01 1 MOI treated with PEDV, PEDV treated with L. mucosae G01 1 MOI, L. mucosae G01 0.1 MOI treated with PEDV, and 0.1 MOI L. mucosae G01 treated with PEDV.
4 Discussion
Since 2010, China has experienced a widespread outbreak of PEDV, which has had significant clinical implications for newborn piglets and resulted in substantial economic losses within the swine industry (Vlasova et al., 2014; Sujie et al., 2022). In recent years, there has been an increasing number of studies advocating for the use of probiotics to regulate the balance of intestinal flora and reduce piglet mortality (Inatomi et al., 2017; Wang et al., 2017; Hou et al., 2018). The Bama pig, a unique Chinese breed known for its strong disease resistance and diverse gut microbiota, has been subject to screening of Lactobacillus strains from piglet fecal samples, which has been reported to significantly improve the growth performance of weaned piglets (Wang W. et al., 2021). In this study, a total of six Lactobacillus strains were isolated from fecal samples obtained from Bama-scented pigs. The Lactobacillus species accounted for 61.95% of all isolates. The findings indicate that the relative abundance of Lactobacillus species in wild boar fecal was higher compared to that in domestic pigs, with L. mucosae G01 representing 54.54% of the total isolates (Zhong et al., 2022). The successful isolation and verification of L. mucosae’s widespread presence in Bama pigs provide a solid foundation for utilizing it as a probiotic food additive. Furthermore, probiotics play a crucial role in acting as a protective barrier against pathogenic microorganisms within the intestinal tract. Li et al. (2022) discovered that local specialty breeds of hogs exhibited a more diverse microflora population in the corresponding intestinal segments when compared to large white hogs. Notably, significant variations were observed in the abundance of L. mucosae G01 and Streptococcus equi. Furthermore, our isolated L. mucosae G01 demonstrated a remarkable 1.53-log reduction in PEDV activity, surpassing the effectiveness of commercially available L. mucosae DSM13345. The unique characteristics of this strain may be attributed to long-term dietary and environmental disparities among pigs. The study conducted by Huang et al. (2023) demonstrated that L. reuteri, obtained from the fecal of large white pigs, exhibited significant superiority in preventing, treating, and competing against PEDV, particularly the L. reuteri LRM strain. These findings align with our own assessment, conducted in a comparative manner.
Huang et al. (2023) conducted a study in which they employed L. reuteri, isolated from pig fecal, and observed a decrease in the mRNA expression of inflammatory cytokines. This methodology demonstrated efficacy in alleviating clinical symptoms and intestinal damage in piglets infected with PEDV. Additionally, Chang-Liao et al. (2020) performed an in vitro evaluation to assess the effectiveness of Ln. mesenteroides, derived from kefir grains. In contrast to the aforementioned approach, the Ln. mesenteroides strain exhibits direct interaction with PEDV in vitro, subsequently exerting an inhibitory effect on PEDV replication in Vero cells through neutralization of the viral form. Our dissimilarities stem from the bacteria’s capacity to occupy receptors on the cell surface in the simulated animal’s natural state, thereby impeding PEDV invasion into the organism, or the bacterial modulation of the cell itself to elicit a mechanistic response against PEDV subsequent to its invasion into the organism. It is noteworthy that the aforementioned favorable results were primarily observed in vero cells, yet there is a lack of lactobacilli evaluation regarding PEDV invasion in intestinal epithelial cells. The present study aimed to address this gap by conducting in vitro investigations, which revealed that L. mucosae G01 induced the activation and subsequent phosphorylation of the IRF3 protein. The subsequent activation resulted in an increase in the mRNA expression of IFN-I and IFN-III genes, alongside the induction of antiviral proteins including MX2, OAS1, and ZAP. These findings provide insights into the immunomodulatory function of L. mucosae G01 during PEDV infection.
During the initial phases of viral infections, interferons play a pivotal role in the host’s defense mechanism against pathogenic microorganisms. Both type I and type III interferons stimulate the expression of antiviral genes within cells, thereby enhancing their ability to withstand viral infections (Lazear et al., 2015; Stanifer et al., 2019). Despite their distinct receptors and secreted cell types, these interferons facilitate the natural immune response, activate immune cells, and provide protection against pathogenic infections (Mesev et al., 2019). Type I interferon is primarily secreted by infected and immune cells, whereas type III interferon is predominantly secreted by epithelial and specific immune cells, particularly on mucosal surfaces and within epithelial cells, thereby regulating viral infection (Li et al., 2017; Mesev et al., 2019). In the interferon signaling pathway, IRF3 plays a crucial role as it becomes phosphorylated and translocated to the nucleus upon PEDV infection, facilitating the transcription of both type I and type III interferons (Stanifer et al., 2019; Su et al., 2020). Lactobacillus strains possess the capacity to modulate the immune response within the intestines and combat pathogenic microorganisms by stimulating IRF3 and promoting the production of type I interferon (Gonzalez-Ochoa et al., 2017). Prior research has demonstrated that lactobacilli activate IRF3 and induce IFN-I expression via the Toll-like receptor pathway (Kanmani and Kim, 2019). Moreover, lactobacilli play a role in activating the innate immune system of intestinal epithelial cells to defend against viral intrusion. Our study provides evidence that L. mucosae G01 has the ability to induce the expression of IRF3 protein and enhance the secretion of both IFN-I and IFN-III, resulting in the upregulation of antiviral proteins MX1, MX2, OAS1, and ZAP. In the post-stimulation trial, the reduction in L. mucosae G01 virus can be attributed to the activation of the IFN signaling pathway, which facilitates the clearance of the virus by enhancing cellular antiviral responses. Several studies have provided support for our findings. In addition, the research conducted by Kanmani et al. has demonstrated that L. delbrueckii TUA4408L possesses the ability to impede rotavirus replication by activating IRF3 (Kanmani et al., 2018). Furthermore, administering probiotics to rats has been found to enhance the expression of interferon-induced MX1 mRNA (Yu et al., 2010). In the forthcoming stage of our research program, we will commence a comprehensive investigation into the in vivo verification of the antiviral attributes of L. mucosae G01. This crucial research endeavor aims to elucidate the degree of safeguarding that probiotics can bestow upon the gastrointestinal wellbeing of piglets, thereby augmenting our comprehension of their potential as an invaluable mechanism in mitigating PEDV-associated difficulties.
In summary, our research demonstrates the notable capacity of L. mucosae G01 to hinder PEDV replication through dual mechanisms, involving direct viral inhibition and the activation of the IFN signaling pathway in vitro. These findings have substantial implications for the development of strategies targeting the prevention and control of PEDV infections.
Data availability statement
The data presented in the study are deposited in the NCBI Nucleotide repository, accession number GenBank: OR783178.1.
Ethics statement
The approval for animal experiments was obtained from the Experimental Animal Ethics Committee of Institute of Animal Health, Guangdong Academy of Agricultural Sciences with the SYXK(Yue)X2021-0165. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
BZ: Writing – original draft. HS: Writing – review & editing. HG: Writing – review & editing. NW: Methodology, Writing – review & editing. CZ: Project administration, Writing – review & editing. ZL: Project administration, Writing – review & editing. HH: Investigation, Writing – review & editing. JN: Methodology, Writing – review & editing. YQ: Methodology, Writing – review & editing. LG: Data curation, Funding acquisition, Supervision, Writing – review & editing. JZ: Data curation, Funding acquisition, Supervision, Writing – review & editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Key Research and Development Program of China (grant number 2022YFD1800801-02), the open competition program of top ten critical priorities of Agricultural Science and Technology Innovation for the 14th Five-Year Plan of Guangdong Province (grant number 2022SDZG02), State Key Laboratory of Swine and Poultry Breeding Industry (grant numbers 2023QZ-NK13, ZQQZ-55), the Independent Research and Development Projects of Maoming Laboratory (grant number 2022KF010), the National Natural Science Foundation of China (grant number 31302101), the Scientific Observation and Experiment Station of Veterinary Drugs and Diagnostic Techniques of Guangdong Province, Ministry of Agriculture and Rural Affairs-Key Laboratory of Livestock Disease Prevention of Guangdong Province, P. R. China (grant number YDWS202107), and the Special Fund for Key Laboratory of Livestock Disease Prevention of Guangdong Province (grant number 2023B1212060040).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Arena, M. P., Capozzi, V., Russo, P., Drider, D., Spano, G., and Fiocco, D. (2018). Immunobiosis and probiosis: Antimicrobial activity of lactic acid bacteria with a focus on their antiviral and antifungal properties. Appl. Microbiol. Biotechnol. 102, 9949–9958. doi: 10.1007/s00253-018-9403-9
Callaway, T., and Singh-Cundy, A. (2019). HD-AGPs as speciation Genes: Positive selection on a proline-rich domain in non-hybridizing species of petunia, Solanum, and Nicotiana. Plants (Basel) 8:211. doi: 10.3390/plants8070211
Cebra, J. J. (1999). Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 69, 1046S–1051S. doi: 10.1093/ajcn/69.5.1046s
Chang-Liao, W.-P., Lee, A., Chiu, Y.-H., Chang, H.-W., and Liu, J.-R. (2020). Isolation of a Leuconostoc mesenteroides strain with anti-porcine epidemic diarrhea virus activities from kefir grains. Front. Microbiol. 11:1578. doi: 10.3389/fmicb.2020.01578
Chen, Y.-M., Limaye, A., Chang, H.-W., and Liu, J.-R. (2022). Screening of lactic acid bacterial strains with antiviral activity against porcine epidemic diarrhea. Probiotics Antimicrob. Proteins 14, 546–559. doi: 10.1007/s12602-021-09829-w
Chen, C., Zheng, J., Xiong, C., Zhou, H., Wei, C., Hu, X., et al. (2022). Metabolomics characterize the differential metabolic markers between Bama xiang pig and Debao pig to identify pork. Foods 12:5. doi: 10.3390/foods12010005
De Filippis, F., Pasolli, E., and Ercolini, D. (2020). The food-gut axis: Lactic acid bacteria and their link to food, the gut microbiome and human health. FEMS Microbiol. Rev. 44, 454–489. doi: 10.1093/femsre/fuaa015
Di Giacomo, S., Toussaint, F., Ledesma-García, L., Knoops, A., Vande Capelle, F., Fremaux, C., et al. (2022). Expanding natural transformation to improve beneficial lactic acid bacteria. FEMS Microbiol. Rev. 46, fuac014. doi: 10.1093/femsre/fuac014
El-Adawi, H. I. (2012). Cytotoxicity assay and antioxidant activities of the lactic acid bacterial strains. Afr. J. Microbiol. Res. 6:924. doi: 10.5897/AJMR11.924
Gonzalez-Ochoa, G., Flores-Mendoza, L. K., Icedo-Garcia, R., Gomez-Flores, R., and Tamez-Guerra, P. (2017). Modulation of rotavirus severe gastroenteritis by the combination of probiotics and prebiotics. Arch. Microbiol. 199, 953–961. doi: 10.1007/s00203-017-1400-3
Halimi, S., and Mirsalehian, A. (2016). Assessment and comparison of probiotic potential of four Lactobacillus species isolated from feces samples of Iranian infants: Probiotic potential of lactobacilli. Microbiol. Immunol. 60, 73–81. doi: 10.1111/1348-0421.12352
He, M., Fang, S., Huang, X., Zhao, Y., Ke, S., Yang, H., et al. (2016). Evaluating the contribution of gut microbiota to the variation of porcine fatness with the cecum and fecal samples. Front. Microbiol. 07:2108. doi: 10.3389/fmicb.2016.02108
Hou, X., Jiang, X., Jiang, Y., Tang, L., Xu, Y., Qiao, X., et al. (2018). Oral immunization against PEDV with recombinant Lactobacillus casei expressing dendritic cell-targeting peptide fusing COE protein of PEDV in piglets. Viruses 10:106. doi: 10.3390/v10030106
Huang, Z., Zhang, W., Su, L., Ma, G., Guo, J., et al. (2023). Isolation of Limosilactobacillus reuteri strain with anti-porcine epidemic diarrhea virus from swine feces. Probiotics Antimicrob Proteins doi: 10.1007/s12602-023-10138-7
Inatomi, T., Amatatsu, M., Romero-Pérez, G. A., Inoue, R., and Tsukahara, T. (2017). Dietary probiotic compound improves reproductive performance of porcine epidemic diarrhea virus-infected sows reared in a Japanese commercial swine farm under vaccine control condition. Front. Immunol. 8:1877. doi: 10.3389/fimmu.2017.01877
Jung, K., Annamalai, T., Lu, Z., and Saif, L. J. (2015). Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet. Microbiol. 178, 31–40. doi: 10.1016/j.vetmic.2015.04.022
Kanmani, P., Albarracin, L., Kobayashi, H., Hebert, E. M., Saavedra, L., Komatsu, R., et al. (2018). Genomic characterisation of Lactobacillus delbrueckii TUA4408L and evaluation of its extracellular polysaccharide for antiviral activity in porcine intestinal epithelial cells. Front. Immunol. 9:2178. doi: 10.3389/fimmu.2018.02178
Kanmani, P., and Kim, H. (2019). Immunobiotic strains modulate toll-like receptor 3 agonist induced innate antiviral immune response in human intestinal epithelial cells by modulating IFN regulatory factor 3 and NF-κB Signaling. Front. Immunol. 10:1536. doi: 10.3389/fimmu.2019.01536
Kenney, S. P., Wang, Q., Vlasova, A., Jung, K., and Saif, L. (2021). Naturally occurring animal coronaviruses as models for studying highly pathogenic human coronaviral disease. Vet. Pathol. 58, 438–452. doi: 10.1177/0300985820980842
Kim, S.-K., Guevarra, R. B., Kim, Y.-T., Kwon, J., Kim, H., Cho, J. H., et al. (2019). Role of probiotics in human gut microbiome-associated diseases. J. Microbiol. Biotechnol. 29, 1335–1340. doi: 10.4014/jmb.1906.06064
Kurpe, S., Grishin, S., Surin, A., Selivanova, O., Fadeev, R., Dzhus, U., et al. (2020). Antimicrobial and amyloidogenic activity of peptides synthesized on the basis of the ribosomal s1 protein from thermus thermophilus. Int. J. Mol. Sci. 21:6382. doi: 10.3390/ijms21176382
La Fata, G., Weber, P., and Mohajeri, M. H. (2018). Probiotics and the gut immune system: Indirect regulation. Probiotics Antimicrob. Proteins 10, 11–21. doi: 10.1007/s12602-017-9322-6
Lazear, H. M., Nice, T. J., and Diamond, M. S. (2015). Interferon-λ: Immune functions at barrier surfaces and beyond. Immunity 43, 15–28. doi: 10.1016/j.immuni.2015.07.001
Li, L., Fu, F., Xue, M., Chen, W., Liu, J., Shi, H., et al. (2017). IFN-lambda preferably inhibits PEDV infection of porcine intestinal epithelial cells compared with IFN-alpha. Antiviral Res. 140, 76–82. doi: 10.1016/j.antiviral.2017.01.012
Li, Y., Wu, Q., Huang, L., Yuan, C., Chen Yuan, Wang, J., et al. (2018). An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 9, 3811–3811. doi: 10.1038/s41467-018-06056-w
Li, Z., Zhang, W., Su, L., Huang, Z., Zhang, W., Ma, L., et al. (2022). Difference analysis of intestinal microbiota and metabolites in piglets of different breeds exposed to porcine epidemic diarrhea virus infection. Front. Microbiol. 13:990642. doi: 10.3389/fmicb.2022.990642
Luo, W.-R., Wu, X.-M., Wang, W., Yu, J.-L., Chen, Q.-Q., Zhou, X., et al. (2022). Novel coronavirus mutations: Vaccine development and challenges. Microb. Pathog. 173:105828. doi: 10.1016/j.micpath.2022.105828
Ma, C., Gao, Q., Zhang, W., Azad, A. K., and Kong, X. (2020). Alterations in the blood parameters and fecal microbiota and metabolites during pregnant and lactating stages in bama mini pigs as a model. Mediat. Inflamm. 2020:8829072. doi: 10.1155/2020/8829072
Mesev, E. V., LeDesma, R. A., and Ploss, A. (2019). Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 4, 914–924. doi: 10.1038/s41564-019-0421-x
Paone, P., and Cani, P. D. (2020). Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 69, 2232–2243. doi: 10.1136/gutjnl-2020-322260
Shen, H., Zhang, C., Guo, P., Liu, Z., Sun, M., Sun, J., et al. (2016). Short communication: Antiviral activity of porcine IFN-λ3 against porcine epidemic diarrhea virus in vitro. Virus Genes 52, 877–882. doi: 10.1007/s11262-016-1374-2
Shi, B., Zhan, X. M., Zheng, J. X., Qiu, H., Liang, D., Ye, Y. M., et al. (2018). Identifying key bird species and geographical hotspots of avian influenza A (H7N9) virus in China. Infect. Dis. Poverty. 7:97. doi: 10.1186/s40249-018-0480-x
Siggers, R. H., Siggers, J., Boye, M., Thymann, T., Mølbak, L., Leser, T., et al. (2008). Early administration of probiotics alters bacterial colonization and limits diet-induced gut dysfunction and severity of necrotizing Enterocolitis in Preterm Pigs3. J. Nutr. 138, 1437–1444. doi: 10.1093/jn/138.8.1437
Stanifer, M. L., Pervolaraki, K., and Boulant, S. (2019). Differential regulation of type I and type III interferon signaling. Int. J. Mol. Sci. 20:1445. doi: 10.3390/ijms20061445
Su, R., Shereen, M. A., Zeng, X., Liang, Y., Li, W., Ruan, Z., et al. (2020). The TLR3/IRF1/Type III IFN axis facilitates antiviral responses against enterovirus infections in the intestine. mBio 11, e2540–e2520. doi: 10.1128/mBio.02540-20
Sujie, D., Ning, K., Chunmei, W., Li, Y., and Dage, S. (2022). FUBP3 degrades the porcine epidemic diarrhea virus nucleocapsid protein and induces the production of type I interferon. J. Virol. 96:e0061822. doi: 10.1128/jvi.00618-22
Tibary, A., Fite, C., Anouassi, A., and Sghiri, A. (2006). Infectious causes of reproductive loss in camelids. Theriogenology 66, 633–647. doi: 10.1016/j.theriogenology.2006.04.008
Turlewicz-Podbielska, H., and Pomorska-Mól, M. (2021). Porcine coronaviruses: Overview of the state of the art. Virol. Sin. 36, 833–851. doi: 10.1007/s12250-021-00364-0
Vlasova, A. N., Marthaler, D., Wang, Q., Culhane, M. R., Rossow, K., Rovira, A., et al. (2014). Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 20, 1620–1628. doi: 10.3201/eid2010.140491
Wang, K., Kong, X., Azad, A. K., Zhu, Q., Xiong, L., Zheng, Y., et al. (2021). Maternal probiotic or synbiotic supplementation modulates jejunal and colonic antioxidant capacity, mitochondrial function, and microbial abundance in Bama mini-piglets. Oxid. Med. Cell. Longev. 2021, 1–14. doi: 10.1155/2021/6618874
Wang, W., Ma, H., Zhu, Y., Ni, K., Qin, G., Tan, Z., et al. (2021). Screening of lactic acid bacteria with inhibitory activity against ETEC K88 as feed additive and the effects on sows and piglets. Animals 11:1719. doi: 10.3390/ani11061719
Wang, M., Gao, Z., Zhang, Y., and Pan, L. (2016). Lactic acid bacteria as mucosal delivery vehicles: A realistic therapeutic option. Appl. Microbiol. Biotechnol. 100, 5691–5701. doi: 10.1007/s00253-016-7557-x
Wang, X., Wang, L., Huang, X., Ma, S., Yu, M., Shi, W., et al. (2017). Oral delivery of probiotics expressing dendritic cell-targeting peptide fused with porcine epidemic diarrhea virus COE antigen: A promising vaccine strategy against PEDV. Viruses 9:312. doi: 10.3390/v9110312
Yang, L., Bian, G., Su, Y., and Zhu, W. (2014). Comparison of faecal microbial community of lantang, bama, erhualian, meishan, xiaomeishan, duroc, landrace, and yorkshire sows. Asian Australas. J. Anim. Sci. 27, 898–906. doi: 10.5713/ajas.2013.13621
Yu, Z., Zeng, Z., Huang, Z., Lian, J., Yang, J., Deng, Q., et al. (2010). Increased mRNA expression of interferon-induced Mx1 and immunomodulation following oral administration of IFN-α2b-transformed B. longum to mice. Arch. Microbiol. 192, 633–638. doi: 10.1007/s00203-010-0589-1
Zhang, W., Shen, H., Wang, M., Fan, X., Wang, S., Wuri, N., et al. (2023). Fangchinoline inhibits the PEDV replication in intestinal epithelial cells via autophagic flux suppression. Front. Microbiol. 14:1164851. doi: 10.3389/fmicb.2023.1164851
Keywords: PEDV, antiviral activity, L. mucosae G01, IFN, Bama pig
Citation: Zhang B, Shen H, Gou H, Wuri N, Zhang C, Liu Z, He H, Nie J, Qu Y, Geri L and Zhang J (2024) Isolation of Limosilactobacillus mucosae G01 with inhibitory effects on porcine epidemic diarrhea virus in vitro from Bama pig gastroenteritis. Front. Microbiol. 15:1360098. doi: 10.3389/fmicb.2024.1360098
Received: 22 December 2023; Accepted: 25 June 2024;
Published: 07 August 2024.
Edited by:
Jose M. Jimenez-Guardeño, University of Malaga, SpainReviewed by:
Liu Sidang, Shandong Agricultural University, ChinaTongling Shan, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Zhang, Shen, Gou, Wuri, Zhang, Liu, He, Nie, Qu, Geri and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Letu Geri, Z2VyaWxldHVzeUBpbWF1LmVkdS5jbg==; Jianfeng Zhang, MTM2Njg5MzkyOThAMTM5LmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Bin Zhang
Bin Zhang Haiyan Shen
Haiyan Shen Hongchao Gou
Hongchao Gou Nile Wuri
Nile Wuri Chunhong Zhang
Chunhong Zhang Zhicheng Liu1,2
Zhicheng Liu1,2 Haiyan He
Haiyan He Letu Geri
Letu Geri Jianfeng Zhang
Jianfeng Zhang