- 1Engineering Research Center of Health Medicine Biotechnology of Guizhou Province & School of Biology and Engineering (Modern Industry College of Health Medicine) & School of Public Health, Guizhou Medical University, Guiyang, Guizhou, China
- 2Key Laboratory of Environmental Pollution Monitoring and Disease Control, China Ministry of Education (Guizhou Medical University), Guiyang, Guizhou, China
- 3School of Basic Medical Science, Guizhou Medical University, Guiyang, Guizhou, China
- 4Department of Laboratory Medicine, Guizhou Provincial People's Hospital, Affiliated Hospital of Guizhou University, Guiyang, Guizhou, China
Candida albicans (C. albicans), a microbe commonly isolated from Candida vaginitis patients with vaginal tract infections, transforms from yeast to hyphae and produces many toxins, adhesins, and invasins, as well as C. albicans biofilms resistant to antifungal antibiotic treatment. Effective agents against this pathogen are urgently needed. Antimicrobial peptides (AMPs) have been used to cure inflammation and infectious diseases. In this study, we isolated whole housefly larvae insect SVWC peptide 1 (WHIS1), a novel insect single von Willebrand factor C-domain protein (SVWC) peptide from whole housefly larvae. The expression pattern of WHIS1 showed a response to the stimulation of C. albicans. In contrast to other SVWC members, which function as antiviral peptides, interferon (IFN) analogs or pathogen recognition receptors (PRRs), which are the prokaryotically expressed MdWHIS1 protein, inhibit the growth of C. albicans. Eukaryotic heterologous expression of WHIS1 inhibited C. albicans invasion into A549 and HeLa cells. The heterologous expression of WHIS1 clearly inhibited hyphal formation both extracellularly and intracellularly. Furthermore, the mechanism of WHIS1 has demonstrated that it downregulates all key hyphal formation factors (ALS1, ALS3, ALS5, ECE1, HWP1, HGC1, EFG1, and ZAP1) both extracellularly and intracellularly. These data showed that heterologously expressed WHIS1 inhibits C. albicans invasion into epithelial cells by affecting hyphal formation and adhesion factor-related gene expression. These findings provide new potential drug candidates for treating C. albicans infection.
Introduction
Candidal vaginitis is a commonly observed gynecological disease in the clinic (Agrawal et al., 2023), and vulvovaginal candidiasis (VVC) affects women worldwide (Hong et al., 2022; Kan et al., 2022; Schwebke et al., 2022). The microbe Candida albicans (C. albicans) is commonly isolated from patients with vaginal tract infections (Roudbarmohammadi et al., 2016; Esfahani et al., 2022; Wagner et al., 2022) and is also normally present in the oral cavity and intestines (Mayer et al., 2013; Gao et al., 2019). Thus, this pathogen increases the occurrence of opportunistic mucosal infections, especially in immunocompromised individuals. According to many reports, C. albicans is becoming increasingly resistant to many antifungal agents, such as ketoconazole and amphotericin (Hong et al., 2022; Nyirjesy et al., 2022; Agrawal et al., 2023), possibly due to the potential formation of mycelia and biofilms that protect it against antibiotic treatment (Moyes et al., 2015; Gulati and Nobile, 2016; Kadosh, 2016).
C. albicans hyphae are key pathogenetic factors that differ between yeast cells and mycelia and are regulated by several networks (Fan et al., 2013; Jenull et al., 2017; Desai et al., 2018). In yeast form, cells can flow with the host digestive tract and migrate, exhibiting decreased pathogenicity in patients (Cleary et al., 2011; Mayer et al., 2013; Moyes et al., 2015). In hyphal form, C. albicans produces many mycelia and wraps together (Mayer et al., 2013; Moyes et al., 2015; Jenull et al., 2017). Mycelia readily adhere to host infection sites and form biofilms for long-term persistence in the host (da Silva Dantas et al., 2016; Gulati and Nobile, 2016). The genes encoding agglutinin-like sequences (ALS1, ALS3, and ALS5) promote the adhesion of C. albicans (Fan et al., 2013; Roudbarmohammadi et al., 2016; Chevalier et al., 2019). ALS3, a cell surface glycoprotein, promotes adhesion and biofilm formation and affects ALS1 and ALS5 (Cleary et al., 2011; Fan et al., 2013; Roudbarmohammadi et al., 2016). Secreted aspartyl proteinase 6 (SAP6) participates in maintaining the cell surface integrity of C. albicans (Buu and Chen, 2013). Moreover, the extent of cell elongation 1 (ECE1) and hyphal wall protein 1 (HWP1), which are hypha-specific genes and hyphal morphological switches, depend on the hyphal formation (Nobile et al., 2006; Fan et al., 2013; Richardson et al., 2018; Liu et al., 2021). In addition, hyphal G cyclin 1 (HGC1) and enhanced filamentous growth transcription factor (EFG1), which are major activators of hyphal development, regulate hypha-inducing signals (Fan et al., 2013; Panariello et al., 2017; Desai et al., 2018). Zinc-responsive activator protein (ZAP1) governs the balance between hyphal cells and yeast cells in the biofilms of C. albicans (Nobile et al., 2009; Ganguly et al., 2011).
Houseflies living in complex environments rely on their powerful innate immune system (Wang et al., 2020). Antimicrobial peptides (AMPs) play an important role in protecting insects against environmental pathogens via the regulation of the toll signaling pathway (Gao et al., 2015; Wang et al., 2020). Hence, insects represent a rich source for the discovery of new AMPs. Many studies have confirmed the biological functions of AMPs, such as their anticancer (Bao et al., 2021; Timmons and Hewage, 2022), antiviral (Liu R. et al., 2022; Nabeta et al., 2022; Nikyar et al., 2022), antibacterial (Maleki Dizaj et al., 2022; Wang et al., 2022b) and immunoregulatory activities (Liu et al., 2018; Liu H. et al., 2022). There are also some reports on the anti-C. albicans activity of AMPs (Hu et al., 2022; Nabeta et al., 2022; Peng-Wei et al., 2022).
The single von Willebrand factor C-domain protein (SVWC) family contains many members that are always found in arthropods (Wang et al., 2017). Macrobrachium nipponense SVWC reportedly functions as an extracellular pattern recognition receptor (PRR) in river prawns (Qin et al., 2020) and helps the host defend against white spot syndrome virus (WSSV) (Nan et al., 2023). Eriocheir sinensis SVWC is involved in antibacterial responses in the Chinese mitten crab as a PRR (Qin et al., 2022). Simultaneously, the SVWC peptide of Marsupenaeus japonicus Vago functions as an IFN analog with anti-WSSV activity via the JAK-STAT signaling pathway (Gao et al., 2021). Moreover, SVWC members also exist in insects. Drosophila melanogaster Vago (DmVago), as an insect SVWC, is reportedly an antiviral peptide (Gao et al., 2021), and Culex quinquefasciatus Vago (CqVago) functions as an IFN analog with anti-WNV activity (West Nile virus) (Paradkar et al., 2012). Most importantly, Zhu et al. identified 29 SVWC peptides in houseflies as AMPs using bioinformatic methods (Qi et al., 2021). Thus, the housefly SVWC peptide may have great potential as an AMP.
In this study, we identified an insect SVWC peptide, viz. WHIS1, from houseflies that inhibits the growth of C. albicans. Both heterologous prokaryotic and eukaryotic expressions of the WHIS1 protein could limit the growth of C. albicans. Furthermore, WHIS1 affected C. albicans invasion into A549 and HeLa cells. In addition, heterologous eukaryotic expression of WHIS1 inhibited hyphal formation and adhesion-related gene expression in A549 and HeLa cells both extracellularly and intracellularly. These findings also provide new candidates and a theoretical basis for the clinical treatment of C. albicans infection.
Materials and methods
Microbial strains and culture conditions
The bacterial strains (Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922) were cultured overnight in tryptic soy broth (TSB) or Mueller Hinton broth (MHB) at 37°C and 200 rpm. C. albicans ATCC10231 was precultured overnight at 37°C with shaking at 200 rpm in potato dextrose broth (PDB) and cultured in potato-dextrose agar (PDA) at 28 or 37°C according to the different experimental requirements.
Cell culture
Human lung carcinoma A549 cells were kindly provided by the School of Public Health, Guizhou Medical University. Human cervical cancer cells (HeLa cells) were kindly gifted by the School of Biology and Engineering (Modern Industry College of Health Medicine). The A549 cells and HeLa cells were maintained and grown to confluence in 25 cm2 cell culture flasks containing Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 100 μg/ml penicillin–streptomycin at 37°C in an atmosphere with 5% CO2. Once the cells had grown to 80% confluence in the culture flasks, they were detached with a solution containing 0.25% trypsin-0.53 mM EDTA (Gibco) and then seeded into six-well culture plates (5 × 105 cells/well) or 24-well culture plates (8 × 104 cells/well) in preparation for transfection and subsequent experiments without antibiotics (in DMEM).
cDNA cloning and sequence analysis and structural modeling of WHIS1
The WHIS1 gene and its homologous version WHISIM were amplified using PCR from the C. albicans-stimulated housefly 3rd larval cDNA template, and the sequences were subsequently submitted to NCBI GenBank under project SUB11062418 (GenBank accession number SUB11062418 Musca domestica WHIS1: OQ267400; M. domestica WHIS1 mutant: OQ267401). The open reading frames (ORFs) were analyzed by ORF Finder in DNAMAN. The ORFs without nucleotide sequences coding for a signal peptide of WHIS1 and its mutant were then cloned and inserted into the pET29a (+) vector for prokaryotic expression. The full-length ORF of WHIS1 was cloned and inserted into the PcDNA3.1(+) vector with the KpnI and NotI cleavage sites for eukaryotic expression. The signal peptide was analyzed using SignalP 5.0 online software (SignalP - 5.0 - Services - DTU Health Tech). Whole-ORF amino acid sequences were utilized for protein BLAST analysis. We set a target maximum of 1,000 sequences but a homologous sequence target of only 119 (Supplementary Excel 1 contains the BLAST details and accession number). The phylogenetic tree was constructed with NCBI Tree Viewer online software. The structural model of WHIS1 was generated with Phyre 2 online software. The amino acid sequence of WHIS1 was analyzed using the Helical Wheel Projection Program (http://rzlab.ucr.edu/scripts/wheel/wheel). The pI was predicted with ProtParam online software. Amino acid sequence alignment of MdWHIS1 with its homologs was performed using BioEdit software. The phylogenetic tree was subsequently constructed using MEGA software with the N-J method, and 1,000 bootstraps were generated.
Prokaryotic expression and protein purification of WHIS1
The WHIS1 ORF without the nucleotide sequence encoding the N-terminal 20-amino-acid signal peptide was cloned and inserted into the pET29a (+) prokaryotic expression vector between the NdeI and XhoI restriction endonuclease sites. The host cells, E. coli BL21(DE3), were cultured in LB liquid medium at 37°C and 200 rpm overnight. First, the liquid cultures were transferred into a new culture (1:1,000 volume) and incubated for 2–3 h until the OD600 value reached 0.4–0.6. Subsequently, the 0.2 mM IPTG was added, and the bacteria were cultured at 16 or 25°C for 24 h. IPTG was used to induce the prokaryotic expression of WHIS1. Then, the bacteria were collected from cultured liquid medium by centrifugation at 12,000 rpm. After washing with PBS 3 times, the bacteria cells were disrupted by hypothermic ultrasonication. Then, the lysed cells were centrifuged at 12,000 rpm at 4°C. Then, the cell lysate could be divided into supernatant and precipitate. These samples were analyzed by SDS–PAGE. The proteins in the supernatant samples were soluble proteins, and the proteins in precipitates consisted of inclusion bodies, which are usually not easily soluble in PBS or water. After a large number of WHIS1 inclusion bodies was obtained, 8M urea was utilized to make it soluble in 50 mM Tris buffer. Then, urea dialysis was performed with a dialysis bag. The urea concentration was reduced from 8 to 6, 5, 4, and 2 M, among other concentrations, in a urea buffer for gradient reduction. The His•Bind® Purification Kit (EMD Millipore Corp., Billerica, MA, USA) was subsequently used for protein purification according to the manufacturer's protocol. Subsequently, 30- and 15-kDa ultrafiltration devices (tube + membrane chips) from Millipore were used for further protein filtration. After centrifuging at 10,000 rpm at 4°C, the molecular weight of proteins less than the membrane pore size will be filtered into the bottom tube, and the molecular weight of the protein bigger than the membrane pore size will remain on the membrane chip of the ultrafiltration tube. The eluted protein mixture solution was first filtered through a 30-kDa ultrafiltration tube, and the filtrate was subsequently filtered through a 15-kDa ultrafiltration tube. The resulting filtrate was used for the minimum inhibitory concentration (MIC) test, and a time–growth curve was constructed. The homogeneity of the final preparations was analyzed by SDS–PAGE.
Western blot assay
As described above, WHIS1 without its signal peptide was fused with His Taq in pET29a(+). After inducing with IPTG for prokaryotic expression and purification of WHIS1 protein, different samples were treated by boiling with RIPA buffer. Then 25 μl of the sample was loaded into a gel well. After completing SDS–PAGE, the gel was transformed into a PVDF membrane. Then, the membrane was sealed with milk for 2 h. Then, the membranes were incubated with anti-His antibody at 4°C for 12 h. After incubation, the membranes were washed with TBST buffer for three times, for 10 min each time. The second antibody was incubated for 2 h. After washing with TBST another three times, ECL chemical kits were utilized for the development of chemiluminescent color in the BIORAD Scanner. Thus, we also detected the protein by Western blotting with the anti-His antibody of WHIS1.
Antimicrobial activity analysis and time–growth curve
The minimum inhibitory concentration (MIC) and minimum fungicidal concentration (MFC) were determined by the microbroth dilution method (Wang et al., 2021a,b) according to the conditions recommended by the Clinical and Laboratory Standards Institute (CLSI, a reference method for broth dilution antifungal susceptibility testing of yeasts, in Approved Standard M27-A3, third ed., Clinical and Laboratory Standards Institute, Wayne, PA, USA, 2008). A single colony of C. albicans was placed in a PDB medium and was cultured overnight (E. coli and S. aureus in TSB or MHB). WHIS1 and its mutant were obtained by prokaryotic expression at a concentration of 2–250 μg/ml in a sterile RPMI 1640 medium (Gibco). Then, 100 μl (5 × 107 CFU/ml) of working C. albicans solution was added to each well, and the final total volume in each well was 200 μl. The samples were incubated on a shaker at 150 rpm and 37°C for 12 h. The MIC and MFC were visually determined, and PDA plate diffusion verification was performed (Wang et al., 2021a,b). The kinetics of the anti-C. albicans activity of prokaryotically expressed WHIS1 and its mutant were studied. A 96-well plate containing a suspension (a final concentration of 5 × 107 CFU/ml) and PDB containing different concentrations of prokaryotically expressed WHIS1 and its mutant was incubated at 28°C, and equal amounts of medium were used as the solvent control. A microplate reader was subsequently used to measure the OD595nm value of the sample at different time points, and the OD595nm value was used as the ordinate to construct the time–growth curve.
Cell transfection procedures
Lipofectamine 2000 reagent was used for transfection according to the manufacturer's instructions. A549 and HeLa cells were divided into two groups: the WHIS1 group and the vector group. The culture medium was replaced with Opti-MEM before transfection. The samples for the six-well plates were prepared as follows: the plasmid (4 μg) was mixed in 200 μl of Opti-MEM and incubated for 5 min, and in a separate tube, the liposomes (10 μl) were suspended in 200 μl of Opti-MEM and incubated for 5 min. The samples for the 24-well plates were prepared as follows: the plasmid (1 μg) was mixed in 50 μl of Opti-MEM and incubated for 5 min, and in a separate tube, the liposomes (2.5 μl) were suspended in 50 μl of Opti-MEM and incubated for 5 min. Both the plasmid and liposomes in Opti-MEM were mixed gently by pipetting and incubated for 15 min at room temperature. The cells were seeded in 6- or 24-well culture plates, allowed to adhere for 24 h, and transfected with DNA-liposome complexes. The various mixtures were added carefully to each well of the cell culture plates, and the plates were then shaken gently. The plates were incubated at 37°C and 5% CO2 for 5 h, the medium was replaced with DMEM containing 10% FBS, and various experiments were subsequently conducted.
Toxic assay of amphotericin b to A549 cells
A549 cells were seeded with a density of 5,000 cells/well in 96-well plates containing the DMEM medium (10% FBS) overnight at 37°C in an atmosphere with 5% CO2. The cells were treated with different concentrations (30μg/ml, 35μg/ml, 40μg/ml, and 45μg/ml) of amphotericin B diluted in DMEM medium and were incubated for different times (20 min, 40 min, 1 h, and 24 h). Subsequently, 100 μl of 10% CCK8 solution was added to the cells and incubated for 1 h. Then the absorbance was read at a 450 nm test wavelength using a microplate reader.
Hyphal formation assay of C. albicans
Twenty-four hours after the transfection of A549 or HeLa cells with WHIS1 or vector, C. albicans suspensions were calibrated in RPMI 1640 with 10% FBS, and the transfected cells were infected at a multiplicity of infection (MOI) of 20 for 8 h at 37°C and 5% CO2. The C. albicans suspensions were used to calculate cell number (1.6 × 106 CFU/ml) according to the McFarland method (0.5). The MOI was calculated by the C. albicans cell number divided by the host cell number (A549/HeLa). After incubation, the vector and WHIS1 groups were observed using Cytation 5 at different magnifications under the same view field. This procedure was used for the total observation of all C. albicans specimens. After the supernatant was discarded and the cells were washed twice with PBS, Cytation 5 was used to capture images of C. albicans attached to the cell surface at different magnifications. Subsequently, RPMI 1640 containing 40 μg/ml of amphotericin B was added to kill the C. albicans cells outside the monolayer. The plate was incubated at 37°C in 5% CO2 for 20 min and washed twice with PBS. The monolayer cells were digested with 0.25% trypsin-0.53mM EDTA and subsequently lysed with 0.1% Triton X-100 for 10 min in a cell incubator. The two groups were subsequently photographed with Cytation 5 at different magnifications (Peng-Wei et al., 2022). This process was used for the analysis of C. albicans invasion into the cells.
Effect of WHIS1 on C. albicans invasion into A549 and HeLa cells: different MOIs
Before their use in the invasion assays, the cells were washed with phosphate-buffered saline (PBS). C. albicans suspensions were calibrated in DMEM supplemented with 10% FBS, and the transfected cells were infected at an MOI of approximately 15 or 20 to infect for 8 h at 37°C and 5% CO2. Following incubation, each well was washed twice with 500 μl of PBS, and DMEM containing 40 μg/ml of amphotericin B was added to kill C. albicans outside the monolayer cells. The plate was then incubated at 37 °C in 5% CO2 for 20 min and washed twice with 500 μl of PBS. The monolayer cells were digested with 100 μl of 0.25% trypsin-0.53mM EDTA and subsequently lysed with 500 μl of 0.1% Triton X-100 for 10 min in a cell incubator. The C. albicans was subsequently plated on PDA plates to enumerate the intracellular fungi (CFU). Three wells were used for each tested group.
Effect of WHIS1 on C. albicans invasion into A549 and HeLa cells: supernatant and intracellular microbes
Before their use in the invasion assays, the cells were washed with PBS. C. albicans suspensions were calibrated in DMEM supplemented with 10% FBS, and the transfected cells were incubated at an MOI of 20 for 8 h at 37°C and 5% CO2. After incubation, the supernatant of C. albicans was centrifuged at 3000 rpm for 3 min and resuspended in 600 μl of PBS. Serial dilution was then performed. Subsequently, 10 μl of each dilution was applied to a PDA plate and incubated at 37°C. For the collection of intracellular fungi, each well was washed twice with 500 μl of PBS, and DMEM containing 40 μg/ml of amphotericin B was added to kill C. albicans cells outside the monolayer. The plate was then incubated at 37 °C in 5% CO2 for 20 min and washed twice with 500 μl of PBS. The monolayer cells were digested with 100 μl of 0.25% trypsin-0.53 mM EDTA and subsequently lysed with 500 μl of 0.1% Triton X-100 for 10 min in a cell incubator. The C. albicans was then plated on PDA plates to enumerate the intracellular fungi (CFU). Three wells were used for each tested group.
Expression analysis of the WHIS1 gene in houseflies or A549 and HeLa cells
Houseflies at different developmental stages, different organs of 3rd-instar housefly larvae or 3rd-instar larval samples, after exposure to different stimuli were ground with TRIzol reagent. RNA was then extracted according to the manufacturer's protocol. Similarly, total RNA from A549 and HeLa cells was extracted using TRIzol reagent at 1.5, 12, and 24 h after transfection and 8 h and 12 h after invasion. The RNA was reverse transcribed to cDNA using a PrimeScript™ RT Reagent Kit with a gDNA Eraser. Quantitative real-time PCR (RT–PCR) was used for the detection of gene expression with the qPCR master mix obtained from Biogoethe Biotechnology Co., Ltd, Wuhan. Housefly β-actin or GAPDH was selected as the internal reference gene for the assessment of the WHIS1 expression pattern by real-time PCR. Human β-actin and HeLa or A549 cells, 1.5 h after transfection, were used as an internal reference and control, respectively. The reaction process was carried out as follows: 95°C for 30 s followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. The 2−ΔΔCt statistical method was used for data analysis (Wang et al., 2022a; Wang C. et al., 2022). The primers used are shown in Supplementary Table 2.
Total RNA extraction and real-time PCR analysis of gene expression in C. albicans
C. albicans suspension was added to the transfected cells, and the MOI was adjusted to 20. After being allowed to invade A549 or HeLa cells for 8 h at 37°C in 5% CO2, the cells were centrifuged at 12,000 rpm for 5 min for sample collection. DEPC-treated water was used for the initial washes. For the collection of intracellular C. albicans, the supernatant was discarded, each well was washed twice with 1 ml of PBS, and DMEM containing 40 μg/ml of amphotericin B was added to kill C. albicans cells outside the monolayer. The plate was then incubated at 37°C in 5% CO2 for 20 min and washed twice with PBS. The monolayer cells were digested with 0.25% trypsin-0.53 mM EDTA and subsequently lysed with 0.1% Triton X-100 for 10 min in a cell incubator. Intracellular C. albicans was centrifuged at 12,000 rpm for 5 min for sample collection. Total RNA was extracted using the Fungal RNA kit and then reverse transcribed to cDNA using the PrimeScript™ RT Reagent Kit with a gDNA Eraser. Real-time PCR was utilized for the detection of gene expression. 18S rRNA was selected as the internal reference gene. The reaction process and the statistical methods used were the same as those used for the abovementioned analyses. The sequences of the primers used are shown in Supplementary Table 3.
For the detection of target genes (WHIS1 and C. albicans genes) in different samples, including housefly larvae and organs and A549 and HeLa cells, different internal reference genes were selected. To detect the WHIS1 expression pattern in houseflies, GAPDH and β-actin were selected as the internal references for the tissue samples and the microbial stimulation samples (Figure 4), respectively, because these genes exhibit enhanced stability under the tested conditions, as described in previous reports (Gao et al., 2015). Human β-actin was selected as an internal reference for the detection of WHIS1 in A549 and HeLa cells (Figures 6E–H). 18S rRNA was selected as an internal reference for C. albicans gene detection in A549 and HeLa cells to determine host cell gene expression (Figures 8, 9). All the experimental procedures used in this study received ethical approval (no. 1702102) as described by Wang et al. (2021b). The statistical analyses were conducted using Microsoft Office Excel. Unpaired t-tests were used to compare two datasets. All the experiments were performed with three independent replicates.
Results
Characterization of WHIS1
As shown in Supplementary Figure 1, WHIS1 was significantly upregulated in the microbe-stimulated housefly 3rd larval transcriptome. A BLAST analysis of the protein sequence against the NCBI database showed that this protein belongs to the single von Willebrand factor C-domain protein (SVWC) superfamily (Figures 1, 2). This protein has a signal peptide (the first 20 amino acids) and 8 conserved cysteines, including those in the SVWC domain (Figure 1A). The theoretical molecular weight of WHIS1 is 13,353.79. The pI of this protein is 7.52. Moreover, as revealed by SDS–PAGE, the protein was close to 12 kDa. Furthermore, proteins with high homology to the identified protein have been predicted but remain uncharacterized, as detailed in the NCBI database (Supplementary Figure 2 and Supplementary Excel 1). Thus, we named this peptide whole-housefly larvae insect SVWC peptide 1 (WHIS1). The antiviral peptide Vago in D. melanogaster and Culex mosquitoes, as an SVWC member, serves as an IFN analog to help the host combat viruses (Paradkar et al., 2012; Gao et al., 2021). In addition, other members of the SVWC family function as PRRs in arthropods such as M. nipponense and E. sinensis (Qin et al., 2020, 2022). Therefore, gene homology was analyzed by sequence alignment and the construction of a phylogenetic tree. Amino acid sequence alignment revealed that housefly WHIS1 was more similar to uncharacterized Md39 (M. domestica39), Md68, and Md76 compared to others (Figure 2A). Moreover, with the exception of 8 cysteines conserved in SVWC members, MdWHIS1 was not similar to D. melanogaster Vago (DmVago). A phylogenetic tree showed that housefly WHIS1 was more homologous to insect SVWCs compared to arthropod SVWCs. Furthermore, MdWHIS1 clustered with the SVWCs of Stomoxys calcitrans and Lucilia cuprina (Figure 2B). These data indicate that MdWHIS1 may have different functions in the fruit fly than those of Vago and arthropod SVWCs.
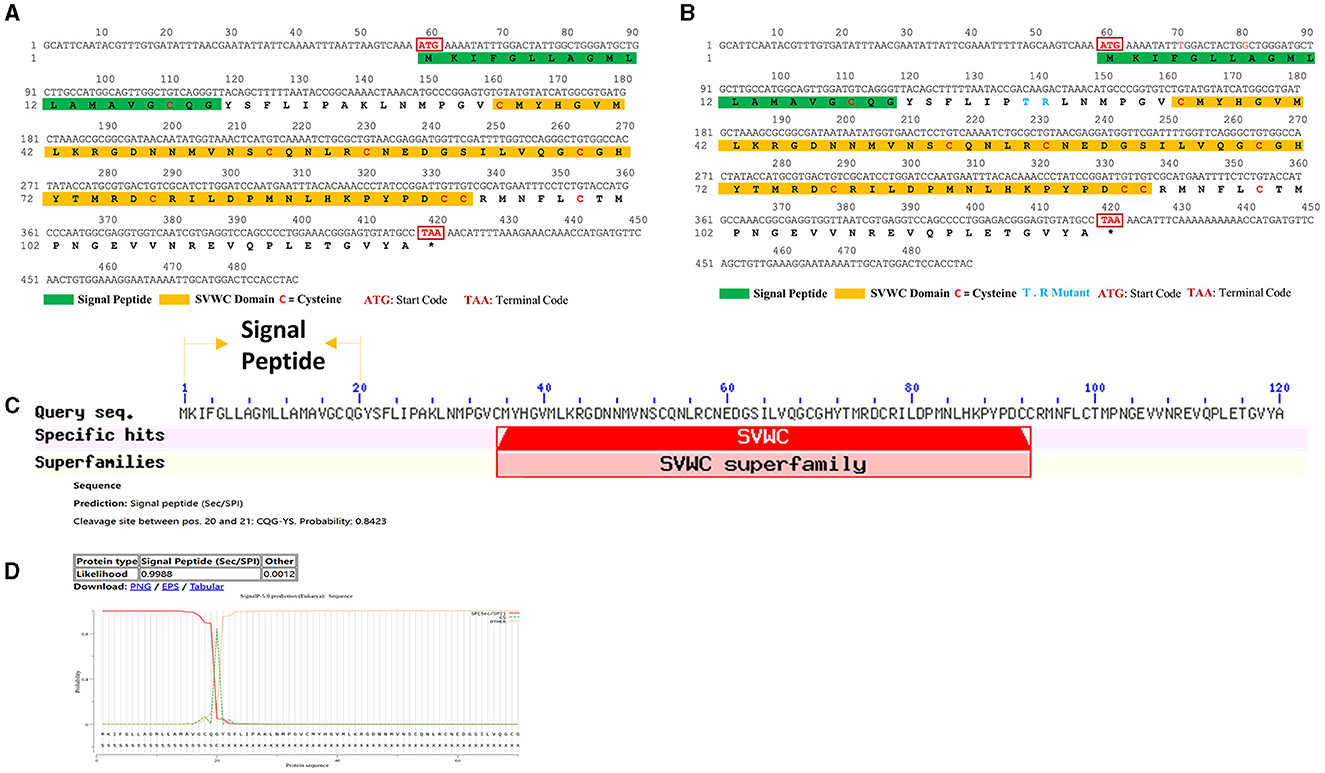
Figure 1. ORF and signal peptide of WHIS1. The ORFs of WHIS1 (A) or its mutant (B) were annotated with ORF Finder in DNAMAN. The signal peptide of WHIS1 was predicted by SignalP 5.0 (D), and its domains were subjected to BLAST searches against the NCBI database (C).
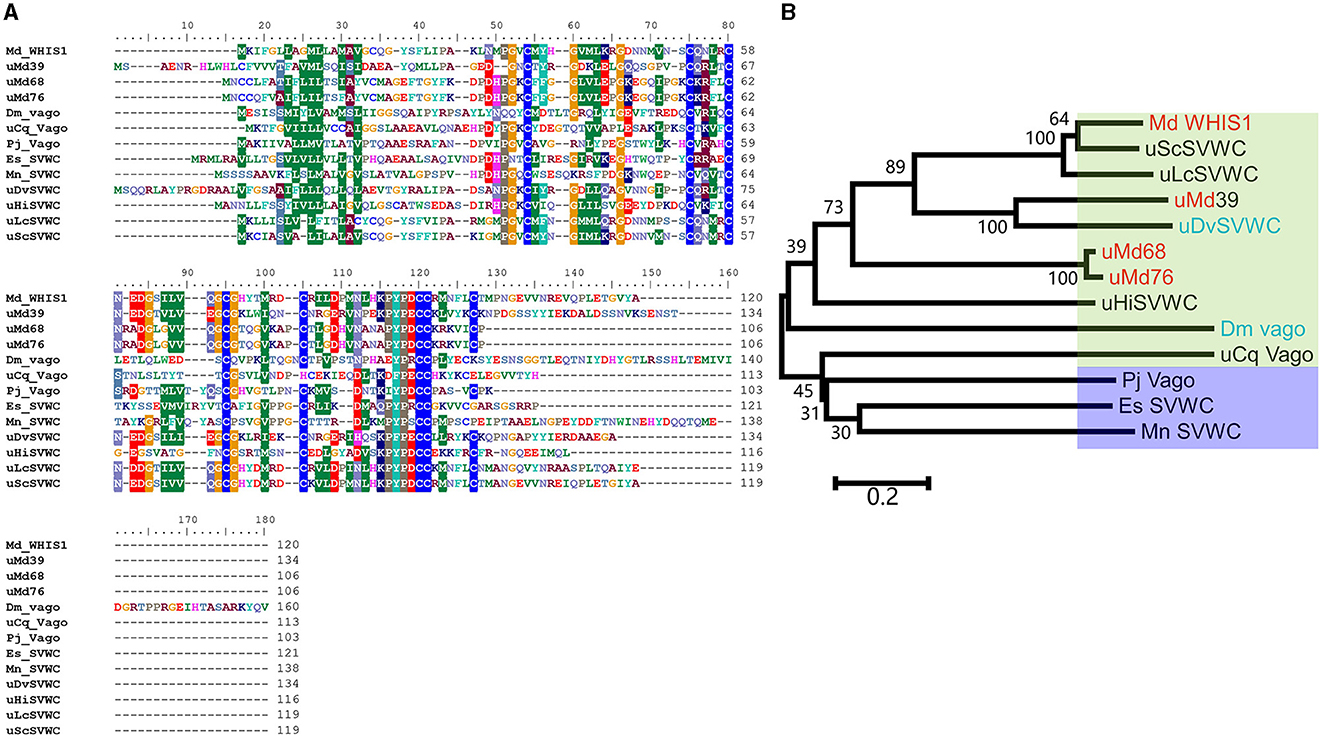
Figure 2. Sequence alignment and phylogenetic dendrogram of WHIS1 with other SVWC homologs. (A) Sequence alignment of Musca domestica WHIS1 with other SVWC homologs. (B) A phylogenetic tree was constructed using MEGA via the neighbor-joining tool with 100 bootstraps. The green background represents insects, the crystal represents arthropods, the red background represents Musca domestica, and the blue background represents Drosophila.
To further determine the function of the WHIS1, basic protein informatics analysis and structure mimicking were performed. As shown in Figure 3, the pI of WHIS1 is 7.52. Furthermore, structural modeling of WHIS1 showed that cysteine residues play an important role in the formation of internal S–S bonds to support the structure (Figures 3A, B). Moreover, we obtained a 2-amino-acid mutant or a variant of WHIS1 in houseflies (Figure 1B). Due to their high amino acid similarity, the proteins have similar properties.
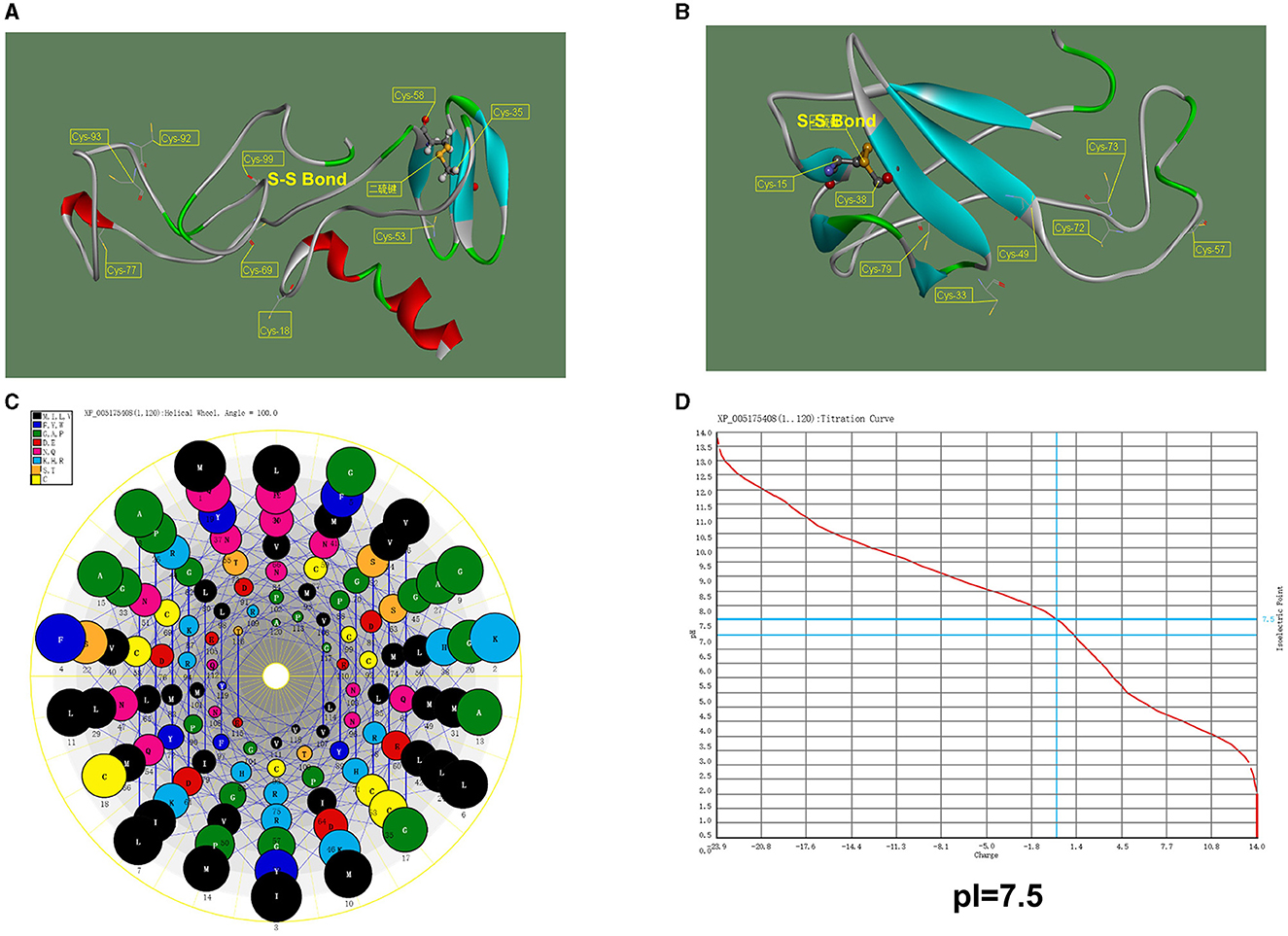
Figure 3. Structural model of WHIS1 and its mutant. Structural model of full-length WHIS1 with the signal peptide (A) and without the signal peptide (B), prepared using Phyre 2 online software. (C) Helix wheel of the WHIS1 amino acids. (D) pI of WHIS1.
Expression pattern of WHIS1 in houseflies
As shown in Figure 4A, WHIS1 was highly expressed from the 2nd larval stage to the adult stage and was expressed at high levels mainly in the fat body and salivary glands (Figure 4B). In addition, the 3rd-instar larvae showed upregulated expression of WHSI after stimulation with S. aureus, E. coli, and C. albicans for 6 h or 12 h (Figures 4C–E). These findings indicate that WHIS1 may participate in the antimicrobial infection process in houseflies.
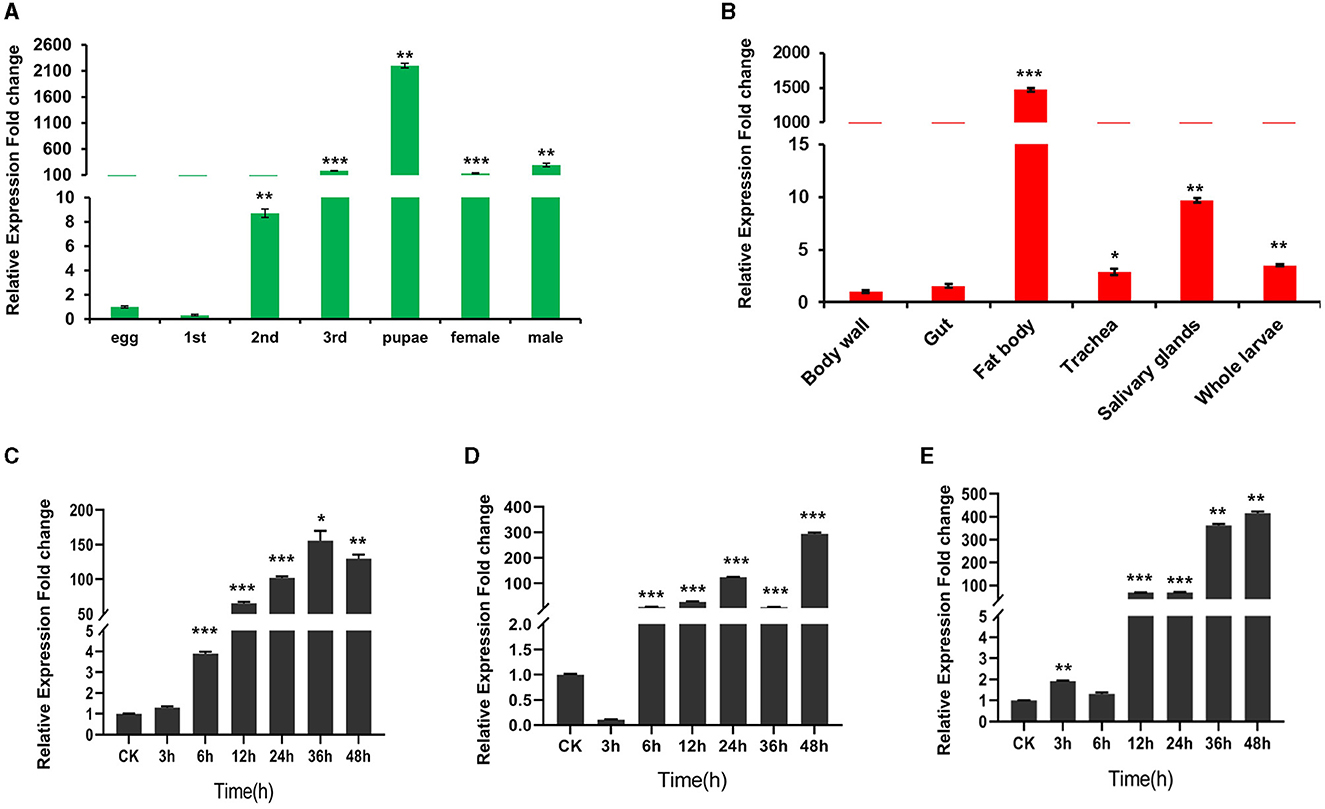
Figure 4. Expression pattern of WHIS1 in houseflies. (A) WHIS1 expression levels in houseflies at different developmental stages. (B) WHIS1 expression levels in different organs of 3rd-instar housefly larvae. WHIS1 expression levels in 3rd-instar housefly larvae after stimulation with Escherichia coli (C), Staphylococcus aureus (D), or Candida albicans (E). n = 3, *P < 0.05, **P < 0.01, and ***P < 0.001, vs. the control group (first column).
Prokaryotic expression of WHIS1 and antimicrobial activity analysis
To analyze the anti-C. albicans activity of WHIS1, the protein and its mutant was prokaryotically expressed. After purification of the protein, 30-kDa and 15-kDa ultrafiltration tubes were utilized to filter the elution mixture. The filtrate was used for the MIC test, and a time–growth curve was generated. All the samples were detected by SDS–PAGE (Figure 5). The MIC of WHIS1 was 0.011 mmol/L. The MIC and time–growth curves showed that WHIS1 and its mutant inhibited the growth of C. albicans (Supplementary Table 1 and Supplementary Figure 3).
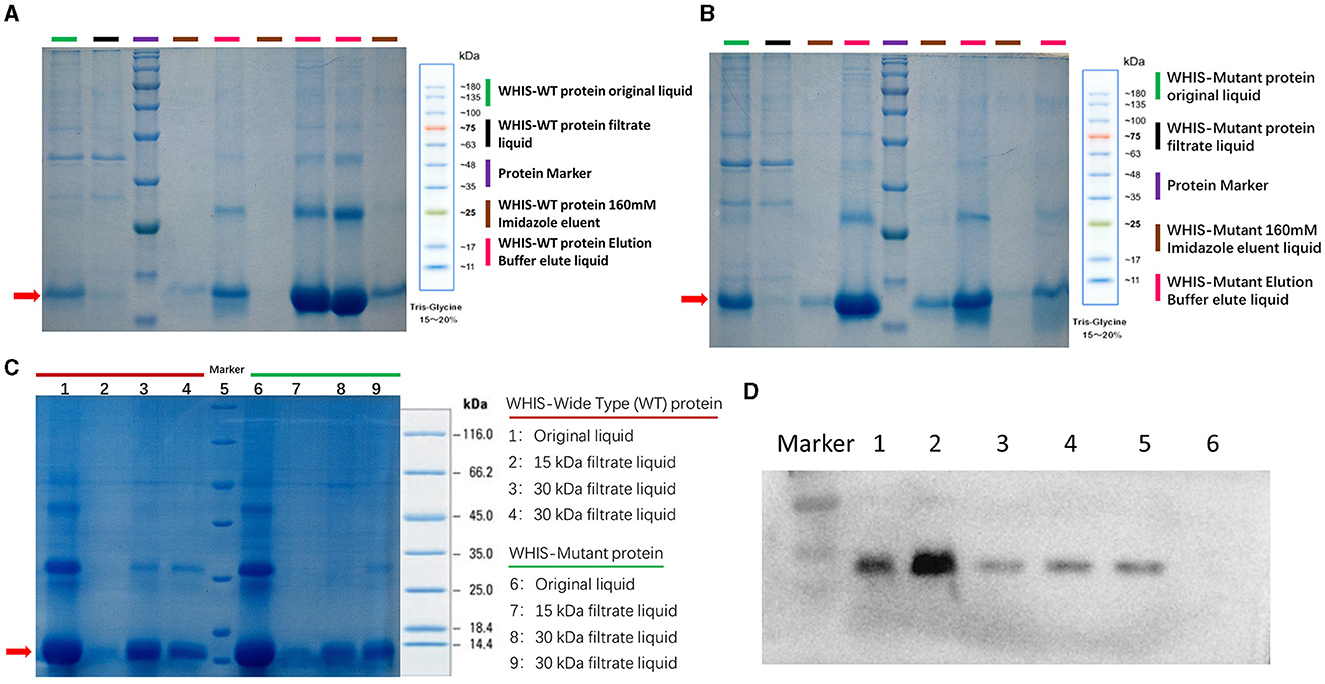
Figure 5. Prokaryotic expression of WHIS1 and its mutant. SDS–PAGE of purified wild-type (A) or mutant (B) WHIS1 protein or filtered protein (C). (D) Western blotting of His antibody to prokaryotic WHIS1 with His Taq. Channel 1: 16°C induced prokaryotic WHIS1 supernatant, 2: WHIS1 precipitate, 3: Eluting WHIS1 with a buffer of 200 mM imidazole, 4: Eluting WHIS1 with 250 mM of imidazole buffer, 5: Purified WHIS1, 6: Un-induction sample.
WHIS1 inhibits C. albicans invasion into A549 and HeLa cells
To further validate the anti-C. albicans activity of WHIS1, heterologous eukaryotic methods were used. WHIS1-PcDNA3.1(+) was transfected into A549 and HeLa cells for eukaryotic expression. Compared with the empty vector [PcDNA3.1(+)], WHIS1 decreased the invasion of C. albicans into A549 cells at an MOI of 15 or 20 (Figure 6A). Furthermore, both the supernatant and intracellular CFUs of C. albicans were reduced by transfection with WHIS1. Thus, WHIS1 can both inhibit cell invasion and limit the growth of C. albicans (Figure 6B). In addition, relatively high expression of WHIS1 was detected 24–36 h after transfection (Figure 6E). In addition, we detected the expression of WHIS1 after 8 h and 12 h of the same treatment similar to that used for the invasion experiment (Figure 6F). Both analyses showed that the WHIS1 expression level was high. Similar results were also observed in HeLa cells (Figures 6C, D, G, H). Overall, these data indicate that heterologous eukaryotic expression of WHIS1 inhibits C. albicans invasion and limits its growth.
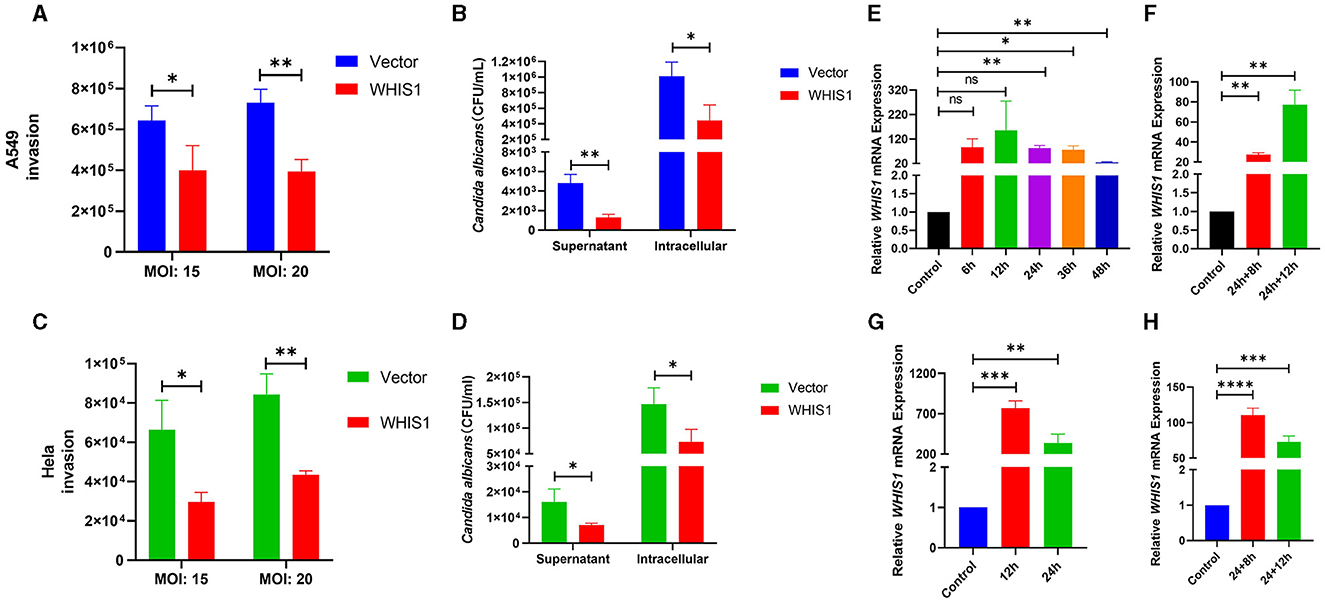
Figure 6. WHIS1 inhibits Candida albicans invasion into A549 and HeLa cells. Twenty-four hours after transfection with WHIS1, A549 (A) or HeLa (C) cells were infected with C. albicans at an MOI of 15 or 20, and the C. albicans CFUs were then calculated by the spread plate method. Similar to the abovementioned options, the supernatant or intracellular C. albicans CFUs in A549 (B) or HeLa (D) cells were counted. The WHIS1 expression level in A549 (E) or HeLa (G) cells at different time points post-transfection was measured. Using the same options as in (B), the WHIS1 expression level in A549 (F) or HeLa (H) cells 24 h after transfection and infected with C. albicans for different time points was measured. n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 vs. the control group.
WHIS1 affects C. albicans hyphal formation
Hyphal formation is a key factor in C. albicans invasion into host cells and its persistence at infection sites (Fan et al., 2013; Jenull et al., 2017; Nabeta et al., 2022). To further elucidate this phenomenon, a hyphal formation assay was performed using A549 and HeLa cells. The cell density and hyphal length of C. albicans were lower in the WHIS1 expression group compared with the vector group (Figures 7, 8). In addition, these phenomena were observed in the total supernatant (Figures 7A, D), on the cell surface (Figures 7B, E), and intracellularly (Figures 7C, F) in the A549. Similar data were observed in the total supernatant (Figures 8A, D), on the cell surface (Figures 8B, E), and intracellularly (Figures 8C, F) in the HeLa cells.
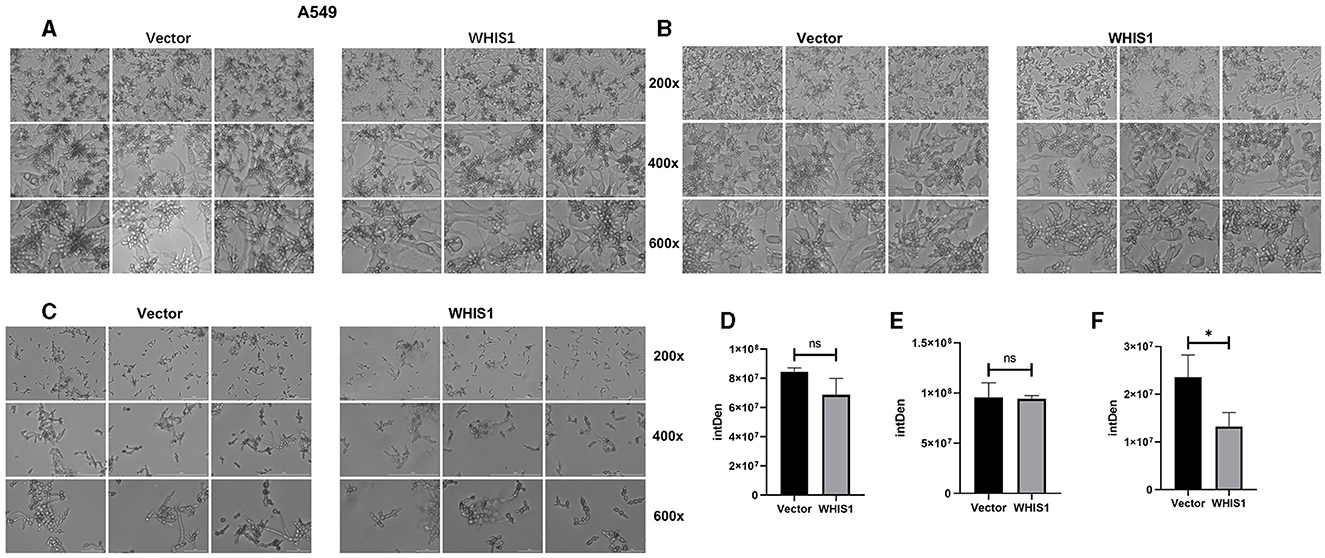
Figure 7. WHIS1 reduces the density and inhibits hyphal formation in Candida albicans-infected A549 cells. Morphological observation of C. albicans in medium (A), attached to the surface of A549 cells (B), after invading A549 cells (C). IMAGE J software was used for gray density analysis, total (D), on the surface (E), and after invading cells (F). Three replicate wells of each sample were observed with Cytation 5. n = 3, *P < 0.05, ns: not significant. vs. the control group.
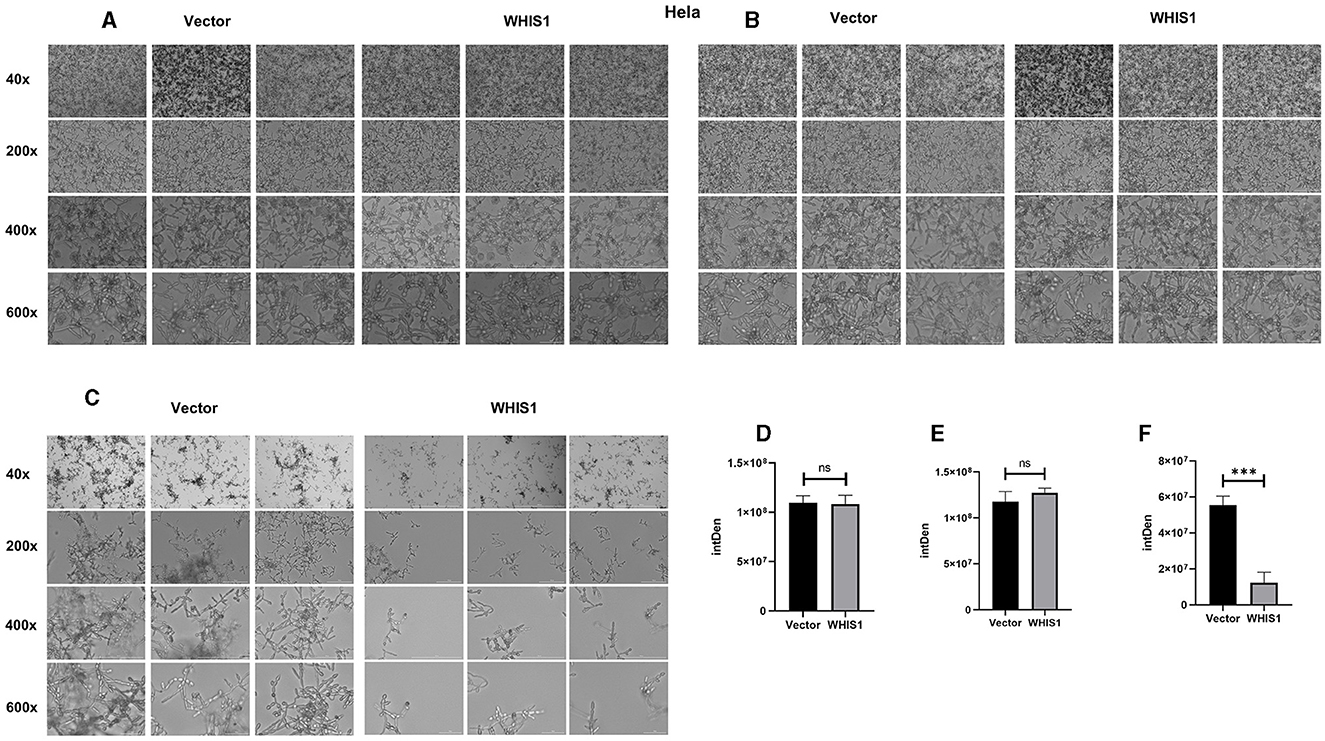
Figure 8. WHIS1 reduces the density and inhibits hyphal formation in Candida albicans-infected HeLa cells. Morphological observation of C. albicans in medium (A), attached to the surface of HeLa cells (B), after invading HeLa cells (C). IMAGE J software was utilized for gray density analysis, total (D), on the surface (E), and after invading cells (F). Three replicate wells of each sample were observed with Cytation 5. n = 3, ***P < 0.001, ns: not significant. vs. the control group.
WHIS1 affects C. albicans hyphae and biofilm-related gene expression and invasion into A549 and HeLa cells
Mycelial cells are a pathogenic form of C. albicans and are controlled by several genes (Fan et al., 2013; Kumar et al., 2015; Liu et al., 2021). To further elucidate the inhibitory mechanisms of WHIS1 against C. albicans, the key factors involved in the effects of WHIS1 treatment on A549 and HeLa cells were determined (Figure 9). According to the changes in gene expression, the expression of agglutinin-like sequence-encoding genes (ALS1, ALS3, and ALS5), which promote adhesion, and SAP6, a secreted aspartyl proteinase that participates in maintaining the cell surface integrity of C. albicans, were strongly downregulated in A549 cells (Figure 9A). Furthermore, several key mycelium formation-related factors (ECE1, HWP1, HGC1, EFG1, and ZAP1) of C. albicans were also significantly downregulated (Figure 9B). To further verify this finding, similar results were obtained with C. albicans-infected HeLa cells (Figures 9C, D).
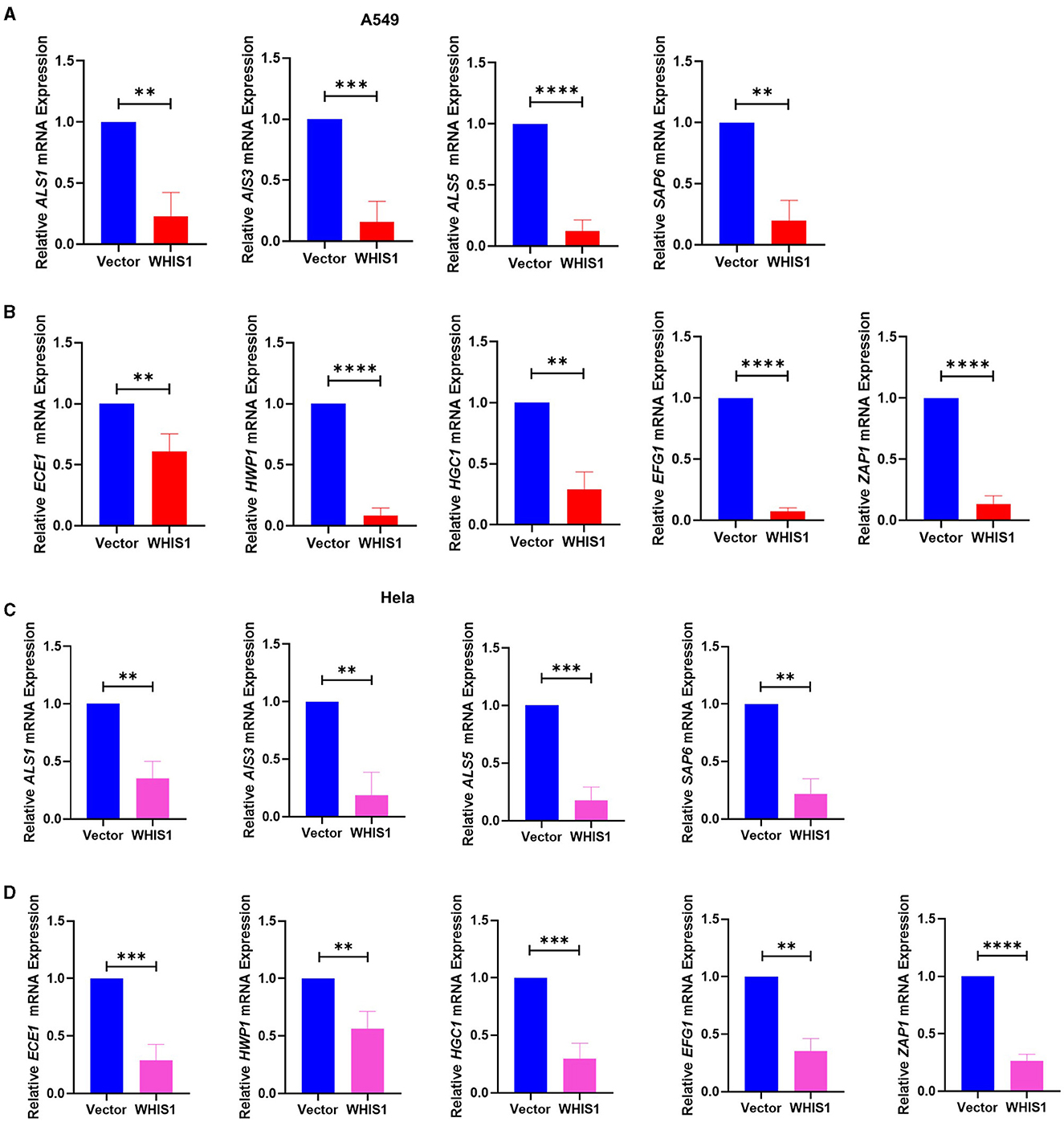
Figure 9. WHIS1 inhibits Candida albicans hyphae- and biofilm-related gene expression in A549 (A, B) and HeLa (C, D) cells. Twenty-four hours after transfection with WHIS1, A549 or HeLa cells were infected with C. albicans at an MOI of 20 for 8 h. Total samples (supernatant + attached cells) were subsequently collected for RNA extraction. The adhesion-related factors ALS1, ALS3, ALS5, and SAP6 (A, C) and key mycelium formation factors (ECE1, HWP1, HGC1, EFG1, and ZAP1) (B, D) were detected by real-time PCR. 18S rRNA was used as the internal reference gene. Statistical analyses were performed with an unpaired t-test, and the results are presented as the means ± standard deviations (n = 3). **P < 0.01, ***P < 0.001, and ****P < 0.0001 vs. the vector group.
To determine the depth to which C. albicans invade host cells, the intracellular conditions were analyzed. The same results were observed with intracellular C. albicans-infected A549 cells (Figures 10A, B), and the results were verified with C. albicans-infected HeLa cells (Figures 10C, D). These data suggest that WHIS1 inhibits C. albicans invasion into A549 and HeLa cells by affecting hyphal formation and adhesion-related gene expression both extracellularly and intracellularly.
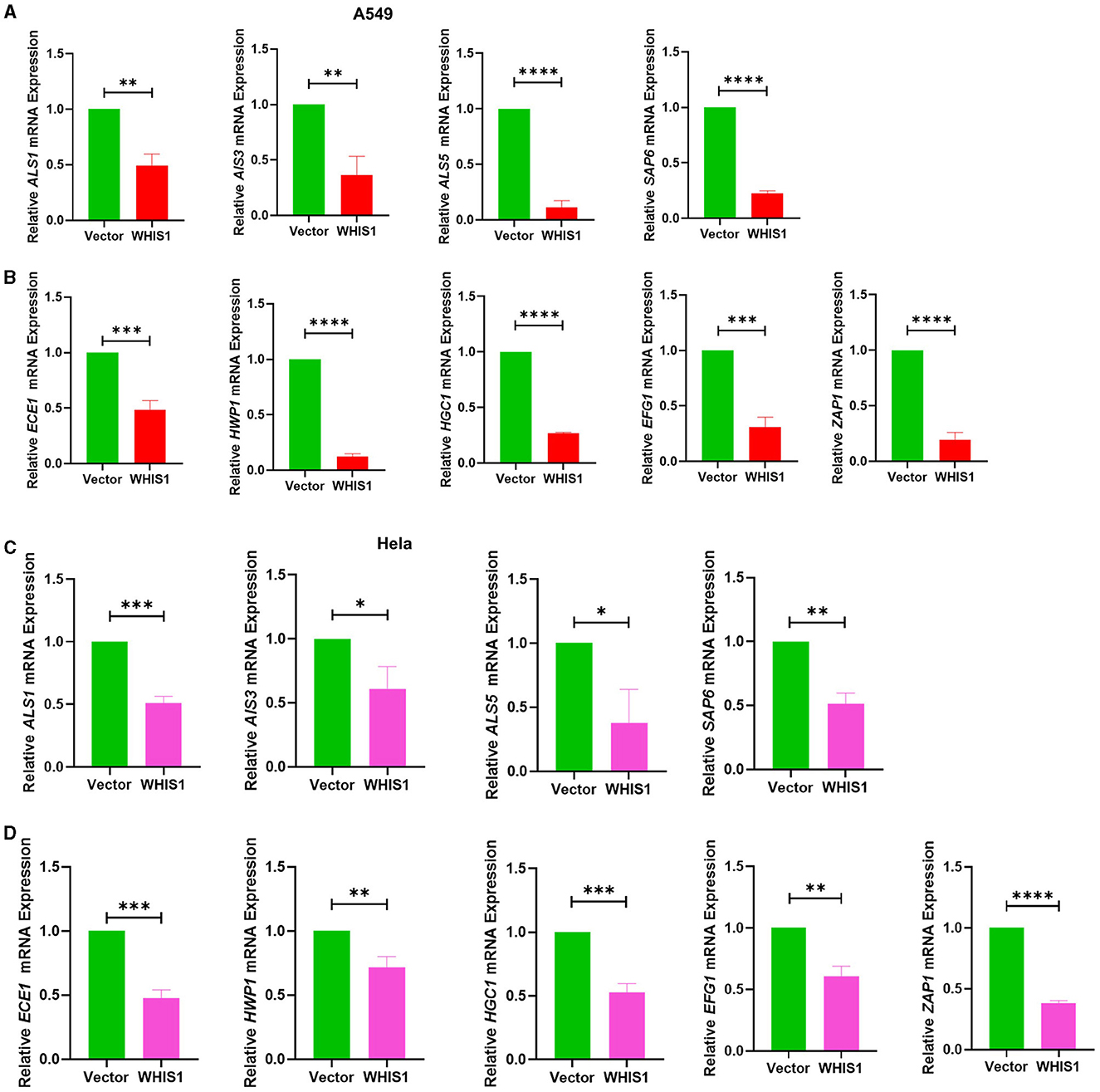
Figure 10. WHIS1 inhibits Candida albicans hyphae- and biofilm-related gene expression in A549 (A, B) and HeLa (C, D) cells. Twenty-four hours after transfection with WHIS1, HeLa cells were infected with C. albicans at an MOI of 20 for 8 h. The supernatant was discarded, and only the attached cells were collected for RNA extraction. The adhesion-related factors ALS1, ALS3, ALS5, and SAP6 (A, C) and key mycelium formation factors (ECE1, HWP1, HGC1, EFG1, and ZAP1) (B, D) were detected by real-time PCR. 18S rRNA was used as the internal reference gene. Statistical analyses were performed with an unpaired t-test, and the results are presented as the means ± standard deviations (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 vs. the vector group.
Discussion
Vaccination causes substantial pain in women (Hong et al., 2022; Rosati et al., 2022; Agrawal et al., 2023). C. albicans, as one of the main causative pathogens, can form hyphae and biofilms for long-term resistance and can also cause severe infection (Hameed and Fatima, 2013; Mayer et al., 2013; Naglik et al., 2019). Herein, we identified an insect-SVWC peptide, whole-housefly larvae insect SVWC peptide 1 (WHIS1), via a microbe-stimulated transcriptomic screen of housefly larvae. Here, we cloned WHIS1, modeled its structure, prokaryotically expressed the encoded protein, and tested its antimicrobial activity. Heterologous expressed MdWHIS1 clearly inhibited hyphal formation in C. albicans-infected A549 and HeLa cells, and an analysis of its expression pattern showed that WHIS1 was mainly expressed in the fat body and salivary glands of housefly larvae. Moreover, WHIS1 expression was rapidly upregulated by microbial stimulation in houseflies. Furthermore, infection experiments demonstrated that MdWHIS1 reduced the density and hyphae density of C. albicans in A549 and HeLa cells. To further elucidate the underlying mechanism involved, the gene expression patterns were assessed, and the results showed that WHIS1 downregulated all hyphal formation-related factors both extracellularly and intracellularly in A549 and HeLa cells. These data showed that WHIS1 inhibits C. albicans invasion into epithelial cells by affecting hyphal formation and adhesion factor-related gene expression. This study explored the potential mechanism of the insect SVWC peptide, WHIS1, against C. albicans and enhanced the understanding of the antifungal functions of insect SVWC members.
Houseflies live in complex environments due to their powerful innate immune system. AMPs play an important role as antimicrobial agents in the environment (Gao et al., 2015; Wang et al., 2021b, 2022b). WHIS1 expression was first found to be significantly upregulated through a screen of the microbe-stimulated 3rd-instar housefly larva transcriptome (Supplementary Figure 1). A protein BLAST search against the NCBI database showed that the targeted homologs of this protein belonged to insects (Supplementary Figure 2 and Supplementary Excel 1). All the homologs were putative or uncharacterized. Thus, housefly WHIS1 was cloned for the first time and submitted to the NCBI GenBank under project SUB11062418 (Figure 1). However, several members of the SVWC family have been found in arthropods (M. nipponense and E. sinensis) and function as PRRs (Qin et al., 2020, 2022). Simultaneously, the SVWC peptide M. japonicus Vago functions as an IFN analog with anti-WSSV activity via the JAK-STAT signaling pathway (Gao et al., 2021). Moreover, D. melanogaster Vago, an SVWC member, reportedly encodes an antiviral peptide (Gao et al., 2021), and C. quinquefasciatus Vago functions as an IFN analog with anti-WNV activity (Paradkar et al., 2012). This finding indicates that WHIS1 could function as a PRR or IFN analog. To address this question, sequence alignment was performed, and a phylogenetic tree was constructed. The amino acid sequence of MdWHIS1 is quite different from those of DmVago, CqVago, and arthropod SVWCs (Figure 2A). Furthermore, the genes also clustered in different branches of the phylogenetic tree (Figure 2B), which indicates that these genes show marked sequence differences, apart from the eight conserved cysteines in the SVWC domain (Figures 1–3). Intriguingly, Zhu et al. identified 29 SVWC members in houseflies as AMPs through bioinformatics methods (Qi et al., 2021). Thus, WHIS1 may have different functions although it belongs to the SVWC superfamily.
To further verify the transcriptome of WHIS1, its expression pattern was analyzed. WHIS1 could be upregulated by microbial stimulation and was expressed mainly in the fat body and salivary gland of 3rd-instar housefly larvae. This finding indicates that WHIS1 plays a pivotal role in antimicrobial defense in the host (Figure 4).
After determining the WHIS1 sequence, we used several methods to verify its function. First, we heterologously expressed 100 amino acids without the first 20 amino acids (the signal peptide). To analyze the anti-C. albicans activity of WHIS1, the WHIS protein was prepared using a prokaryotic expression method (Figure 5). Obtaining soluble WHIS1 protein posed a challenge due to the consistent formation of inclusion bodies in the initial experiments. Thus, we attempted to optimize the expression conditions, and after refolding the inclusion bodies by dialysis, we obtained some prokaryotically expressed WHIS1 protein. After purification with a His binding kit, an ultrafiltration tube was utilized to filter the elution mixture. MIC and time–growth curve tests were carried out with the purified prokaryotically expressed WHIS1 protein (Supplementary Table 1 and Supplementary Figure 3). The data showed that the prokaryotically expressed proteins inhibited the growth of C. albicans. The MIC was 0.011 mmol/L. As shown using ExPASy (https://web.expasy.org/cgi-bin/protparam/protparam), the molecular weight of WHIS1 (containing the signal peptide) was found to be 13,353.79, and the theoretical pI was calculated to be equal to 7.52. In the prokaryotic expression experiment, WHIS1 without its signal peptide was fused with His Taq. Thus, we also detected the protein by Western blotting with anti-His antibody (Figure 5D). The results showed that prokaryotically expressed WHIS1 hybridized with the band at 12.2 kDa. Moreover, WHIS1 comprised the key band. Furthermore, even some protein bands were found in the Coomassie-stained SDS–PAGE gel, and the purified sample protein was filtered by Millipore ultrafiltration devices (tube + membrane chips). Thus, this process prevents other impurity proteins from exerting further influence in subsequent experiments. The sample will be more pure with WIHS1 protein. However, this method used for WHIS1 protein expression is time-consuming and complex, especially due to the large number of inclusion bodies. Thus, a better protein production method is needed for WHIS1.
Subsequently, the eukaryotic expression method was used in this study. Moreover, heterologous expression of WHIS1 prevents the effects of the WHIS1-mediated signaling pathway in insects (if present) (Wang et al., 2017). Therefore, WHIS1 will function by directly affecting other cells. Furthermore, many insect defensins or AMPs are glycosylated. In some cases, glycosylation is necessary for full activity. Prokaryotic expression is unlikely to allow for glycosylation. To avoid this effect, we are interested in testing the eukaryotic expression method. The WHIS1 ORF with a signal peptide coding sequence was cloned and inserted into the eukaryotic overexpression vector PcDNA3.1(+). The construct was then transfected into A549 cells, and after 24 h, the cells were infected with C. albicans for 8 h prior to the invasion experiment (Figure 6). Transfection with the empty vector served as a control. The final CFUs of C. albicans in the supernatant and intracellular space were calculated. The WHIS1-transfected group showed decreased C. albicans counts after infection with different MOIs and in the supernatant or intracellular space of A549 cells (Figures 6A, B). To learn the cytotoxic effects of Amphotericin B to A549 cells, the CCK8 method of cell viability assay was performed. It was shown that 30–45 μg/ml of amphotericin B did not affect the viability of A549 cells (Supplementary Figure 4A). But as time extended, the toxic effect on the cell was constantly increasing (Supplementary Figures 4B–D).
The expression of WHIS1 in A549 cells post-transfection and post-C. albicans infection was also detected at different time points. These data showed that the WHIS1 expression level was relatively high at different time points (Figures 6E, F). To further demonstrate the function of WHIS1, all the assays were re-performed using HeLa cells, and the results were similar to those obtained with A549 cells (Figures 6C, D, G, H). PcDNA3.1 plasmid is a commercially available eukaryotic expression vector that has been used in various previous studies. In general, this plasmid can be transfected using Lipofectamine transfection regent into eukaryotic cells such as A549, HeLa, and 293T cells. Its transfection can induce over expression of the target gene. RNA was isolated without DNA and plasmid. We also detected the change in the WHIS1 expression level by using real-time quantitative PCR (Figures 6E–H). It also showed that WHIS1 was highly expressed for a long period. In addition, as the prokaryotically expressed WHIS1 protein directly adds to the A549 cells, it also shows an inhibition effect on C. albicans invasion of A549 (Supplementary Figure 5). These data demonstrated that heterologous eukaryotic expression of WHIS1 plays a pivotal role in helping host cells resist C. albicans invasion.
Hyphal formation is a key factor in C. albicans invasion into host cells (Kadosh, 2016; Jenull et al., 2017; Liu et al., 2021). C. albicans acquires infectivity during its transformation from yeast to hyphae stage (Lee et al., 2017; Kim et al., 2019; Liboro et al., 2021). In addition, the hyphae formation will promote the formation of biofilm of C.albicans (Kim et al., 2019, 2022; Liboro et al., 2021). Thus, several anti-C. albicans agents are targeting the hyphae formation to inhibit its biofilm formation (Lee et al., 2018; Kim et al., 2019, 2022). As shown in Figures 7, 8, WHIS1 reduced the cell density of C. albicans and inhibited hyphal formation in A549 and HeLa cells. The long and extensive hyphae of C. albicans formed a network in the vector group. However, the growth of C. albicans hyphae in A549 cells was inhibited in the WHIS1 group. WHIS1 shortened the length of hyphae and reduced the number of C. albicans aggregates. Furthermore, to determine the invasion conditions of C. albicans on host cells after WHIS1 treatment, the total supernatant (full view; Figures 7A, D), discarded supernatant (cell surface; Figures 7B, E), and cell digest obtained with ET and Triton X 100 (intracellular; Figures 7C, F) were collected. WHIS1 inhibited the hyphal growth of C. albicans invasion into A549 cells (Figures 7C, F). Furthermore, these results were verified in C. albicans-infected HeLa cells (Figures 8C, F). These results indicated that WHIS1 can inhibit the density and mycelial growth of C. albicans. To further investigate the underlying molecular mechanism, the gene expression changes were subsequently observed under these conditions.
C. albicans exhibits many virulence factors that cause disease, including the morphological transition between yeast and hyphal forms, the expression of adhesins and invasins on the cell surface, the formation of biofilms, phenotypic switching, and the secretion of hydrolytic enzymes (Nobile et al., 2009; Buu and Chen, 2013; Hameed and Fatima, 2013; Moyes et al., 2015; Gulati and Nobile, 2016). Some of these pathogeneses are associated with certain gene families, particularly the agglutinin-like sequence (ALS) family (Fan et al., 2013; Roudbarmohammadi et al., 2016). The formation of mycelium by C. albicans from yeast leads to increased pathogenicity and toxicity to host cells and plays an important role in the invasion ability of Candida species (Fan et al., 2013; Mayer et al., 2013; Moyes et al., 2015; Wongsuk et al., 2016; Wagner et al., 2022). In addition, C. albicans more easily adheres to substrates and forms biofilms, leading to long-term residence at the host infection site in the host and making its eradication difficult (Mayer et al., 2013; Moyes et al., 2015; Gulati and Nobile, 2016). ALS1, ALS3, and ALS5 promote the adhesion of C. albicans (Fan et al., 2013; Roudbarmohammadi et al., 2016; Chevalier et al., 2019). ALS3, a cell surface glycoprotein, promotes adhesion and biofilm formation and affects ALS1 and ALS5. ALS3 mediates binding to host ligands (such as E-cadherin on epithelial cells and N-cadherin on endothelial cells), thereby triggering engulfment of the fungal cell into the host cell (Cleary et al., 2011; Fan et al., 2013; Roudbarmohammadi et al., 2016).
SAP6, a secreted aspartyl proteinase, participates in maintaining the cell surface integrity of C. albicans. Following adhesion to host cell surfaces and hyphal growth, C. albicans hyphae can secrete hydrolases, such as Sap6, which have been proposed to facilitate active penetration into these cells. In addition, Sap6 is thought to enhance the efficiency of extracellular nutrient acquisition (Buu and Chen, 2013; Kumar et al., 2015; Jenull et al., 2017). Moreover, the levels of extent of cell elongation 1 (ECE1) and hyphal wall protein 1 (HWP1), which are hypha-specific genes and hyphal morphological switches, depend on hyphal formation, and the expression of both of these genes is regulated by EFG1 (Nobile et al., 2006; Fan et al., 2013; Richardson et al., 2018; Liu et al., 2021). In addition, HGC1 and EFG1, which are major activators of hyphal development, regulate hyphal-inducing signals. HGC1 encodes a protein involved in regulating mycelial growth. The expression of this protein is correlated with the growth and extension of hyphae. Hgc1 plays a role in polarized growth and represses cell separation from hyphae (Fan et al., 2013; Panariello et al., 2017; Desai et al., 2018). ZAP1 governs the balance between hyphal cells and yeast cells in the biofilms of C. albicans (Nobile et al., 2009; Ganguly et al., 2011). Together, the results indicate that transition to the hyphal form and adherence cause damage to the host mucosa via the combined action of secreted aspartyl proteases and phospholipases, facilitating the invasion of the epithelium by the organism (Fan et al., 2013; Desai et al., 2018).
To further elucidate the anti-C. albicans mechanism of WHIS1, these key hyphal formation factors of C. albicans were assessed. All C. albicans factors were significantly downregulated in A549 cells after WHIS1 treatment (Figures 9A, B). To further verify the underlying mechanism, HeLa cells, a cervical carcinoma cell line, were selected to mimic C. albicans infection. Intriguingly, WHIS1 also inhibited hyphal formation by limiting the expression of hyphal factors in infected cells (Figures 9C, D).
To investigate the role of gene expression of extracellular and intracellular C. albicans in host cell invasion, the expression of these factors was measured under different conditions, as described for the hyphal assay. Furthermore, in both the total samples (supernatant + intracellular), which represent all C. albicans cells, and the intracellular samples, which represent C. albicans cells inside A549 cells, the levels of all the factors were decreased in the WHIS1 group (Figures 10A, B). In addition, both hyphal formation and adhesion factors were downregulated in the intracellular and extracellular C. albicans-infected HeLa cell models (Figures 10C, D). Based on the results of this study, it is reasonable to speculate that WHIS1 disrupts the invasion ability of C. albicans by inhibiting the formation of hyphae and adhesion factor expression. In contrast to other SVWC members, WHIS1 has antifungal functions as an SVWC peptide in insects.
Conclusion
In general, our results demonstrated that WHIS1 inhibits C. albicans invasion into host cells by affecting hyphal formation and adhesion-related gene expression. These findings increase the understanding of the functions of SVWC members, especially in insects. This study identified a housefly peptide as a novel drug candidate and thus provides a theoretical basis for the clinical treatment of C. albicans infections.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov; OQ267401-OQ267401.
Ethics statement
No animal studies are presented in this manuscript.
Author contributions
MC: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing—original draft, Writing—review & editing. W-KH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing—review & editing. YY: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing—review & editing. S-MW: Conceptualization, Data curation, Investigation, Methodology, Writing—review & editing. Y-XY: Conceptualization, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing—review & editing, Investigation. W-XL: Conceptualization, Data curation, Investigation, Methodology, Writing—review & editing. GL: Conceptualization, Methodology, Resources, Supervision, Writing—review & editing, Investigation. S-FW: Conceptualization, Resources, Supervision, Validation, Writing—review & editing, Investigation. HZ: Methodology, Resources, Supervision, Validation, Writing—review & editing, Investigation. H-ML: Funding acquisition, Methodology, Resources, Supervision, Validation, Writing—review & editing, Investigation. BW: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing—original draft, Writing—review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (31760250, 32200157, 32160668), the Guizhou Provincial Natural Science Foundation (2020, No. 4Y237; 2019, No. 1273), the Guizhou Provincial Department of Education Young Talent Scientist Growth Program (Qian Jiao He KY No [2021]152), the China Scholarship Council (Grant No. 201908520005), and the Excellent Young Talents Plan of Guizhou Medical University (2023-104); funds from the central finance to support the development of local universities (Qian Jiao Ji No [2023]036 the Guizhou Provincial Department of Education); and the first class discipline construction project in Guizhou Province-Public Health and Preventive Medicine (No. 2017[85]).
Acknowledgments
We thank Prof. Jian-Wei Wu of Guizhou Medical University for providing the housefly larvae and microbe-stimulated cDNA for WHIS1 gene cloning. We thank Dr. Zu-Quan Hu of Guizhou Medical University for providing the HeLa cells.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1358752/full#supplementary-material
Supplementary Figure 1. Fold changes in the expression of WHIS1 in response to different stimuli identified from transcriptomic data.
Supplementary Figure 2. Phylogenetic dendrogram of WHIS1 homologs identified via a BLAST search of the NCBI protein database A phylogenetic tree was constructed using proteins identified from a BLAST search of the NCBI with the WHIS1 amino acid sequence. The top 119 homologous genes were visualized via the NCBI BLAST Tree View (A). WHIS1 is included in the yellow section. WHIS1 and the nine most similar homologs are shown in (B) and were visualized using NCBI BLAST Tree View with the neighbor-joining Kimura model (protein).
Supplementary Figure 3. The time–growth curve of C. albicans after treatment with prokaryotically expressed WHIS1 or its mutant. The strains were treated with different concentrations of WHIS1 or its mutant for 24 h, and the OD595 value was measured with a microplate reader at various time points was measured with a microplate reader. Time–growth curves of C. albicans (A) with WT WHIS1 or its mutant (B) were drawn, n = 3.
Supplementary Figure 4. Toxic Assay of Amphotericin B to A549 cells. Cell viability of different concentrations of amphotericin B on A549 cells for 20 min (A), 40 min (B), 1 h (C), and 24 h (D).
Supplementary Figure 5. Effect of prokaryotic WHIS1 inhibits C. albicans invasion of A549 cells. The A549 cells were seeded into 24-well culture plates (8 × 104 cells/well) overnight at 37°C in an atmosphere with 5% CO2. Then prokaryotically expressed recombinant WHIS1 was diluted to a final concentration of 0.011 mM (150 μg/ml) by a sterile RPMI 1640 medium. C. albicans suspensions and WHIS1 were added to each well. The A549 cells were infected for 8 h at 37°C and 5% CO2. Following incubation, each well was washed twice with 500 μl of PBS, and 1640 containing 40 μg/ml of amphotericin B was added to kill C. albicans outside the monolayer cells. The plate was then incubated at 37°C in 5% CO2 for 20 min and washed twice with 500 μl of PBS. The monolayer cells were digested with 100 μl of 0.25% trypsin-0.53 mM EDTA and subsequently lysed with 500 μl of 0.1% Triton X-100 for 10 min in a cell incubator, and the C. albicans was subsequently plated on PDA plates to enumerate the intracellular fungi (CFU). Three wells were used for each tested group, n = 3.
Supplementary Table 1. Anti-C. albicans activity of prokaryotically expressed WHIS1 protein.
Supplementary Table 2. List of primers used for WHIS1.
Supplementary Table 3. List of primers used for the real-time PCR assay of C. albicans biofilm-related genes.
Supplementary Excel 1. Alignment of proteins identified through a BLAST search of NCBI based on WHIS1.
Abbreviations
VVC, vulvovaginal candidiasis; MDR, multidrug resistant; C. albicans, Candida albicans; S. aureus, Staphylococcus aureus; E. coli, Escherichia coli; WHIS1, whole housefly larvae insect SVWC peptide 1; MIC, minimum inhibitory concentration; MFC, minimum fungicidal concentration; CK, control group; RT–PCR, real-time quantitative polymerase chain reaction; AMP, antimicrobial peptide; FBS, fetal bovine serum; CFU, colony-forming unit; ALS1, ALS3, and ALS5, agglutinin-like sequence 1, 3, and 5; ECE1, extent of cell elongation 1; HWP1, hyphal wall protein 1; HGC1, hyphal G cyclin 1; EFG1, enhanced filamentous growth transcription factor 1; ZAP1, zinc-responsive activator protein 1; SAP6, secreted aspartyl proteinase 6; PBS, phosphate-buffered saline; DMEM, Dulbecco's modified Eagle's medium; TSB, tryptic soy broth; PDA, potato-dextrose agar; PDB, potato dextrose broth; MHB, Mueller Hinton broth; SVWC, single von Willebrand factor C-domain protein; IFN, interferon; PRRs, pathogen recognition receptors.
References
Agrawal, P., Yazdy, G., Ghanem, K. G., Handa, V. L., Schumacher, C. M., Sobel, J. D., et al. (2023). Vaginal Candida albicans: high frequency of in vitro fluconazole resistance in a select population-a brief note. Sex. Transm. Dis. 50, 121–123. doi: 10.1097/OLQ.0000000000001730
Bao, Q., Ganbold, T., Qiburi, Q., Bao, M., Han, S., Baigude, H., et al. (2021). AMP functionalized curdlan nanoparticles as a siRNA carrier: synthesis, characterization and targeted delivery via adenosine A2B receptor. Int. J. Biol. Macromol. 193, 866–873. doi: 10.1016/j.ijbiomac.2021.10.138
Buu, L. M., and Chen, Y. C. (2013). Sap6, a secreted aspartyl proteinase, participates in maintenance the cell surface integrity of Candida albicans. J. Biomed. Sci. 20:101. doi: 10.1186/1423-0127-20-101
Chevalier, M., Doglio, A., Rajendran, R., Ramage, G., Precheur, I., Ranque, S., et al. (2019). Inhibition of adhesion-specific genes by Solidago virgaurea extract causes loss of Candida albicans biofilm integrity. J. Appl. Microbiol. 127, 68–77. doi: 10.1111/jam.14289
Cleary, I. A., Reinhard, S. M., Miller, C. L., Murdoch, C., Thornhill, M. H., Lazzell, A. L., et al. (2011). Candida albicans adhesin Als3p is dispensable for virulence in the mouse model of disseminated candidiasis. Microbiology 157, 1806–1815. doi: 10.1099/mic.0.046326-0
da Silva Dantas, A., Lee, K. K., Raziunaite, I., Schaefer, K., Wagener, J., Yadav, B., et al. (2016). Cell biology of Candida albicans-host interactions. Curr. Opin. Microbiol. 34, 111–118. doi: 10.1016/j.mib.2016.08.006
Desai, P. R., Lengeler, K., Kapitan, M., Janßen, S. M., Alepuz, P., Jacobsen, I. D., et al. (2018). The 5' untranslated region of the EFG1 transcript promotes its translation to regulate hyphal morphogenesis in Candida albicans. mSphere 3:e00280-18. doi: 10.1128/mSphere.00280-18
Esfahani, A., Omran, A. N., Salehi, Z., Shams-Ghahfarokhi, M., Ghane, M., Eybpoosh, S., et al. (2022). Molecular epidemiology, antifungal susceptibility, and ERG11 gene mutation of Candida species isolated from vulvovaginal candidiasis: comparison between recurrent and non-recurrent infections. Microb. Pathog. 170:105696. doi: 10.1016/j.micpath.2022.105696
Fan, Y., He, H., Dong, Y., and Pan, H. (2013). Hyphae-specific genes HGC1, ALS3, HWP1, and ECE1 and relevant signaling pathways in Candida albicans. Mycopathologia 176, 329–335. doi: 10.1007/s11046-013-9684-6
Ganguly, S., Bishop, A. C., Xu, W., Ghosh, S., Nickerson, K. W., Lanni, F., et al. (2011). Zap1 control of cell-cell signaling in Candida albicans biofilms. Eukaryotic Cell 10, 1448–1454. doi: 10.1128/EC.05196-11
Gao, J., Zhao, B. R., Zhang, H., You, Y. L., Li, F., Wang, X. W., et al. (2021). Interferon functional analog activates antiviral Jak/Stat signaling through integrin in an arthropod. Cell Rep. 36:109761. doi: 10.1016/j.celrep.2021.109761
Gao, Y., Liang, G., Wang, Q., She, X., Shi, D., Shen, Y., et al. (2019). Different host immunological response to C. albicans by human oral and vaginal epithelial cells. Mycopathologia 184, 1–12. doi: 10.1007/s11046-018-0301-6
Gao, Y., Tang, T., Gu, J., Sun, L., Gao, X., Ma, X., et al. (2015). Downregulation of the Musca domestica peptidoglycan recognition protein SC (PGRP-SC) leads to overexpression of antimicrobial peptides and tardy pupation. Mol. Immunol. 67, 465–474. doi: 10.1016/j.molimm.2015.08.007
Gulati, M., and Nobile, C. J. (2016). Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 18, 310–321. doi: 10.1016/j.micinf.2016.01.002
Hameed, S., and Fatima, Z. (2013). Novel regulatory mechanisms of pathogenicity and virulence to combat MDR in Candida albicans. Int. J. Microbiol. 2013:240209. doi: 10.1155/2013/240209
Hong, N., Lei, Y., Chen, H., Chen, X., Tsui, K. M., Hu, D., et al. (2022). Genotyping and drug resistance profile of clinical isolates of Candida albicans from vulvovaginal candidiasis in the Eastern China. Mycopathologia 187, 217–224. doi: 10.1007/s11046-022-00616-x
Hu, N., Mo, X. M., Xu, S. N., Tang, H. N., Zhou, Y. H., Li, L., et al. (2022). A novel antimicrobial peptide derived from human BPIFA1 protein protects against Candida albicans infection. Innate Immun. 28, 67–78. doi: 10.1177/17534259221080543
Jenull, S., Tscherner, M., Gulati, M., Nobile, C. J., Chauhan, N., Kuchler, K., et al. (2017). The Candida albicans HIR histone chaperone regulates the yeast-to-hyphae transition by controlling the sensitivity to morphogenesis signals. Sci. Rep. 7:8308. doi: 10.1038/s41598-017-08239-9
Kadosh, D. (2016). Control of Candida albicans morphology and pathogenicity by post-transcriptional mechanisms. Cell. Mol. Life Sci. 73, 4265–4278. doi: 10.1007/s00018-016-2294-y
Kan, S., Song, N., Pang, Q., Mei, H., Zheng, H., Li, D., et al. (2022). In vitro antifungal activity of azoles and other antifungal agents against pathogenic yeasts from vulvovaginal candidiasis in China. Mycopathologia 188, 99–109. doi: 10.1007/s11046-022-00687-w
Kim, S. W., Joo, Y. J., Chun, Y. J., Park, Y. K., and Kim, J. (2019). Cross-talk between Tor1 and Sch9 regulates hyphae-specific genes or ribosomal protein genes in a mutually exclusive manner in Candida albicans. Mol. Microbiol. 112, 1041–1057. doi: 10.1111/mmi.14346
Kim, Y. G., Lee, J. H., Park, S., Khadke, S. K., Shim, J. J., Lee, J., et al. (2022). Hydroquinones including tetrachlorohydroquinone inhibit Candida albicans biofilm formation by repressing hyphae-related genes. Microbiol. Spectr. 10:e0253622. doi: 10.1128/spectrum.02536-22
Kumar, R., Saraswat, D., Tati, S., and Edgerton, M. (2015). Novel aggregation properties of Candida albicans secreted aspartyl proteinase Sap6 mediate virulence in oral candidiasis. Infect. Immun. 83, 2614–2626. doi: 10.1128/IAI.00282-15
Lee, J. H., Kim, Y. G., Choi, P., Ham, J., Park, J. G., Lee, J., et al. (2018). Antibiofilm and antivirulence activities of 6-gingerol and 6-shogaol against Candida albicans due to hyphal inhibition. Front. Cell. Infect. Microbiol. 8:299. doi: 10.3389/fcimb.2018.00299
Lee, K. H., Park, S. J., Choi, S. J., and Park, J. Y. (2017). Proteus vulgaris and Proteus mirabilis decrease Candida albicans biofilm formation by suppressing morphological transition to its hyphal form. Yonsei Med. J. 58, 1135–1143. doi: 10.3349/ymj.2017.58.6.1135
Liboro, K., Yu, S. R., Lim, J., So, Y. S., Bahn, Y. S., Eoh, H., et al. (2021). Transcriptomic and metabolomic analysis revealed roles of Yck2 in carbon metabolism and morphogenesis of Candida albicans. Front. Cell. Infect. Microbiol. 11:636834. doi: 10.3389/fcimb.2021.636834
Liu, H., Wang, G., Hao, D., Wang, C., and Zhang, M. (2022). Antimicrobial and immunoregulatory activities of TS40, a derived peptide of a TFPI-2 homologue from black rockfish (Sebastes schlegelii). Mar. drugs 20:353. doi: 10.3390/md20060353
Liu, J., Willems, H. M. E., Sansevere, E. A., Allert, S., Barker, K. S., Lowes, D. J., et al. (2021). A variant ECE1 allele contributes to reduced pathogenicity of Candida albicans during vulvovaginal candidiasis. PLoS Pathog. 17:e1009884. doi: 10.1371/journal.ppat.1009884
Liu, R., Liu, Z., Peng, H., Lv, Y., Feng, Y., Kang, J., et al. (2022). Bomidin: an optimized antimicrobial peptide with broad antiviral activity against enveloped viruses. Front. Immunol. 13:851642. doi: 10.3389/fimmu.2022.851642
Liu, S. H., Li, H. F., Yang, Y., Wei, D., Jiang, H. B., Dou, W., et al. (2018). Antimicrobial peptide gene BdPho responds to peptidoglycan infection and mating stimulation in oriental fruit fly, Bactrocera dorsalis (Hendel). AMB Express 8:5. doi: 10.1186/s13568-017-0533-8
Maleki Dizaj, S., Salatin, S., Khezri, K., Lee, J. Y., and Lotfipour, F. (2022). Targeting multidrug resistance with antimicrobial peptide-decorated nanoparticles and polymers. Front. Microbiol. 13:831655. doi: 10.3389/fmicb.2022.831655
Mayer, F. L., Wilson, D., and Hube, B. (2013). Candida albicans pathogenicity mechanisms. Virulence 4, 119–128. doi: 10.4161/viru.22913
Moyes, D. L., Richardson, J. P., and Naglik, J. R. (2015). Candida albicans-epithelial interactions and pathogenicity mechanisms: scratching the surface. Virulence 6, 338–346. doi: 10.1080/21505594.2015.1012981
Nabeta, H. W., Lasnik, A. B., Fuqua, J. L., Wang, L., Rohan, L. C., Palmer, K. E., et al. (2022). Antiviral lectin Q-Griffithsin suppresses fungal infection in murine models of vaginal candidiasis. Front. Cell. Infect. Microbiol. 12:976033. doi: 10.3389/fcimb.2022.976033
Naglik, J. R., Gaffen, S. L., and Hube, B. (2019). Candidalysin: discovery and function in Candida albicans infections. Curr. Opin. Microbiol. 52, 100–109. doi: 10.1016/j.mib.2019.06.002
Nan, Q., Muyi, L., Han, Z., Feifei, L., Xinrui, G., Mengjia, W., et al. (2023). Single von Willebrand factor C-domain protein confers host defense against white spot syndrome virus by functioning as a pattern recognition receptor in Macrobrachium nipponense. Int. J. Biol. Macromol. 241:124520. doi: 10.1016/j.ijbiomac.2023.124520
Nikyar, A., Bolhassani, A., Rouhollah, F., and Heshmati, M. (2022). In vitro delivery of HIV-1 Nef-Vpr DNA construct using the human antimicrobial peptide LL-37. Curr. Drug Deliv. 19, 1083–1092. doi: 10.2174/1567201819666220217164055
Nobile, C. J., Nett, J. E., Andes, D. R., and Mitchell, A. P. (2006). Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryotic Cell 5, 1604–1610. doi: 10.1128/EC.00194-06
Nobile, C. J., Nett, J. E., Hernday, A. D., Homann, O. R., Deneault, J. S., Nantel, A., et al. (2009). Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 7:e1000133. doi: 10.1371/journal.pbio.1000133
Nyirjesy, P., Schwebke, J. R., Angulo, D. A., Harriott, I. A., Azie, N. E., Sobel, J. D., et al. (2022). Phase 2 randomized study of oral ibrexafungerp versus fluconazole in vulvovaginal candidiasis. Clin. Infect. Dis. 74, 2129–2135. doi: 10.1093/cid/ciab841
Panariello, B. H. D., Klein, M. I., Pavarina, A. C., and Duarte, S. (2017). Inactivation of genes TEC1 and EFG1 in Candida albicans influences extracellular matrix composition and biofilm morphology. J. Oral Microbiol. 9:1385372. doi: 10.1080/20002297.2017.1385372
Paradkar, P. N., Trinidad, L., Voysey, R., Duchemin, J. B., and Walker, P. J. (2012). Secreted vago restricts West Nile virus infection in culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. USA. 109, 18915–18920. doi: 10.1073/pnas.1205231109
Peng-Wei, W., Chao-Rong, S., Xu, W., Ming, C., Yong-Xin, Y., Cong, W., et al. (2022). A potential milk preservative—phormicin C-NS, sorbic acid-modified housefly antimicrobial peptide, inhibits Candida albicans hypha and biofilm formation. Lwt 168:113883. doi: 10.1016/j.lwt.2022.113883
Qi, S., Gao, B., and Zhu, S. (2021). Molecular diversity and evolution of antimicrobial peptides in Musca domestica. Diversity 13:107. doi: 10.3390/d13030107
Qin, N., Sun, H., Lu, M., Wang, J., Tang, T., Liu, F., et al. (2020). A single von Willebrand factor C-domain protein acts as an extracellular pattern-recognition receptor in the river prawn Macrobrachium nipponense. J. Biol. Chem. 295, 10468–10477. doi: 10.1074/jbc.RA120.013270
Qin, Y., Luo, Z., Zhao, K., Nan, X., Guo, Y., Li, W., et al. (2022). A new SVWC protein functions as a pattern recognition protein in antibacterial responses in Chinese mitten crab (Eriocheirsinensis). Fish Shellfish Immunol. 131, 1125–1135. doi: 10.1016/j.fsi.2022.11.004
Richardson, J. P., Mogavero, S., Moyes, D. L., Blagojevic, M., Krüger, T., Verma, A. H., et al. (2018). Processing of Candida albicans Ece1p is critical for candidalysin maturation and fungal virulence. MBio. 9:e02178-17. doi: 10.1128/mBio.02178-17
Rosati, D., Bruno, M., Jaeger, M., Kullberg, B. J., Van De Veerdonk, F., Netea, M. G., et al. (2022). An exaggerated monocyte-derived cytokine response to candida hyphae in patients with recurrent vulvovaginal candidiasis. J. Infect. Dis. 225, 1796–1806. doi: 10.1093/infdis/jiaa444
Roudbarmohammadi, S., Roudbary, M., Bakhshi, B., Katiraee, F., Mohammadi, R., Falahati, M., et al. (2016). ALS1 and ALS3 gene expression and biofilm formation in Candida albicans isolated from vulvovaginal candidiasis. Adv. Biomed. Res. 5:105. doi: 10.4103/2277-9175.183666
Schwebke, J. R., Sobel, R., Gersten, J. K., Sussman, S. A., Lederman, S. N., Jacobs, M. A., et al. (2022). Ibrexafungerp versus placebo for vulvovaginal candidiasis treatment: a phase 3, randomized, controlled superiority trial (VANISH 303). Clin. Infect. Dis. 74, 1979–1985. doi: 10.1093/cid/ciab750
Timmons, P. B., and Hewage, C. M. (2022). Biophysical study of the structure and dynamics of the antimicrobial peptide maximin 1. J. Pept. Sci. 28:e3370. doi: 10.1002/psc.3370
Wagner, A. S., Vogel, A. K., Lumsdaine, S. W., Phillips, E. K., Willems, H. M. E., Peters, B. M., et al. (2022). Mucosal infection with unmasked Candida albicans cells impacts disease progression in a host niche-specific manner. Infect. Immun. 90:e0034222. doi: 10.1128/iai.00342-22
Wang, B., Song, C. R., Zhang, Q. Y., Wei, P. W., Wang, X., Long, Y. H., et al. (2022a). The fusaric acid derivative qy17 Inhibits Staphylococcus haemolyticus by disrupting biofilm formation and the stress response via altered gene expression. Front. Microbiol. 13:822148. doi: 10.3389/fmicb.2022.822148
Wang, B., Wei, P. W., Wan, S., Yao, Y., Song, C. R., Song, P. P., et al. (2021a). Ginkgo biloba exocarp extracts inhibit S. aureus and MRSA by disrupting biofilms and affecting gene expression. J. Ethnopharmacol. 271:113895. doi: 10.1016/j.jep.2021.113895
Wang, B., Wei, P. W., Yao, Y., Song, C. R., Wang, X., Yang, Y. X., et al. (2022b). Functional and expression characteristics identification of Phormicins, novel AMPs from Musca domestica with anti-MRSA biofilm activity, in response to different stimuli. Int. J. Biol. Macromol. 209, 299–314. doi: 10.1016/j.ijbiomac.2022.03.204
Wang, B., Yao, Y., Wei, P., Song, C., Wan, S., Yang, S., et al. (2021b). Housefly Phormicin inhibits Staphylococcus aureus and MRSA by disrupting biofilm formation and altering gene expression in vitro and in vivo. Int. J. Biol. Macromol. 167, 1424–1434. doi: 10.1016/j.ijbiomac.2020.11.096
Wang, C., Wei, P. W., Song, C. R., Wang, X., Zhu, G. F., Yang, Y. X., et al. (2022). Evaluation of the antimicrobial function of Ginkgo biloba exocarp extract against clinical bacteria and its effect on Staphylococcus haemolyticus by disrupting biofilms. J. Ethnopharmacol. 298:115602. doi: 10.1016/j.jep.2022.115602
Wang, H., Smagghe, G., and Meeus, I. (2017). The role of a single gene encoding the Single von Willebrand factor C-domain protein (SVC) in bumblebee immunity extends beyond antiviral defense. Insect Biochem. Mol. Biol. 91, 10–20. doi: 10.1016/j.ibmb.2017.10.002
Wang, X., Gao, S., Hao, Z., Tang, T., and Liu, F. (2020). Involvement of TRAF6 in regulating immune defense and ovarian development in Musca domestica. Int. J. Biol. Macromol. 153, 1262–1271. doi: 10.1016/j.ijbiomac.2019.10.259
Keywords: AMP, C. albicans, antimicrobial activity, WHIS1, hypha
Citation: Chen M, Huang W-K, Yao Y, Wu S-M, Yang Y-X, Liu W-X, Luo G, Wei S-F, Zhang H, Liu H-M and Wang B (2024) Heterologous expression of the insect SVWC peptide WHIS1 inhibits Candida albicans invasion into A549 and HeLa epithelial cells. Front. Microbiol. 15:1358752. doi: 10.3389/fmicb.2024.1358752
Received: 20 December 2023; Accepted: 13 May 2024;
Published: 30 May 2024.
Edited by:
Pedro José Alcolea, Spanish National Research Council (CSIC), SpainReviewed by:
Slawomir Milewski, Gdansk University of Technology, PolandEdith Porter, California State University, Los Angeles, United States
Copyright © 2024 Chen, Huang, Yao, Wu, Yang, Liu, Luo, Wei, Zhang, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Wang, d2FuZ2JpbmdfZ211X2VkdUAxNjMuY29t; Shao-Feng Wei, c2hhb2ZlbmdneUAxNjMuY29t; Hua Zhang, NzgwODM3NDgyQHFxLmNvbQ==; Hong-Mei Liu, aG1saXVAZ21jLmVkdS5jbg==
†These authors have contributed equally to this work
 Ming Chen
Ming Chen Wei-Kang Huang1†
Wei-Kang Huang1† Gang Luo
Gang Luo Shao-Feng Wei
Shao-Feng Wei Hua Zhang
Hua Zhang Bing Wang
Bing Wang