
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 March 2024
Sec. Food Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1354696
Salmonella is one of the most important zoonotic pathogens and is mostly transmitted through food of animal origin. Application of bacteriophages is a promising tool to biocontrol Salmonella on both food and food contact surfaces. In this study, we evaluated the effectiveness of a six-phage cocktail for the reduction of Salmonella Enteritidis and a mixture of five major Salmonella serotypes (S. Enteritidis, Salmonella Typhimurium, Salmonella Infantis, Salmonella Paratyphi B, and Salmonella Indiana) on chicken skin and stainless steel. A phage cocktail with a final concentration of 107 PFU/cm2 was sprayed on these surfaces. After adding the phage cocktail, the samples were incubated at RT (~23°C) for different periods of time. The phage cocktail caused a significant reduction of S. Enteritidis and the mixed culture on chicken skin 30 min after phage addition, with 1.8 log10 and 1 log10 units, respectively. Reduction rates (1.2–1.7 log10 units) on stainless steel after 30 min were similar. Four hours after addition, the phage cocktail caused a significant reduction on both surfaces up to 3 log10 units on chicken skin and 2.4 log10 units on stainless steel. In a further experiment, bacteria added to stainless steel were not allowed to dry to simulate a fresh bacterial contamination. In this case, the bacterial count of S. Enteritidis was reduced below the detection limit after 2 h. The results demonstrate that this phage cocktail has potential to be used in post-harvest applications to control Salmonella contaminations.
Salmonella is a gram-negative, facultative anaerobic bacterium belonging to the Enterobacteriaceae family (Bahrani-Mougeot et al., 2009). It is classified into two species, Salmonella enterica and Salmonella bongori, and encompasses more than 2.600 serotypes (Issenhuth-Jeanjean et al., 2014). Serotypes of Salmonella enterica are of particular importance for human health as they are primarily associated with foodborne illnesses (Park and Andam, 2020). The illness caused by non-typhoidal Salmonella spp. (NTS) is called salmonellosis. The majority of outbreaks and sporadic cases of salmonellosis are attributed to contaminated food (Chanamé Pinedo et al., 2022; Teklemariam et al., 2023). Primary sources are poultry, raw and undercooked meat, egg and egg products and raw dairy products (Heredia and García, 2018).
In the European Union, 773 foodborne disease outbreaks occurred in 2021, with approximately 60,000 human cases (EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control), 2022). Up to 80% of illnesses are sporadic and not associated with outbreaks (Gut et al., 2018). The serotypes Salmonella Enteritidis and Salmonella Typhimurium are of significant importance, as they are mostly implicated in food-borne infections and outbreaks (Park and Andam, 2020). However, the prevalence of Salmonella in broiler farms is progressively linked to the S. Infantis serotype (Hauser et al., 2012; Mughini-Gras et al., 2021). After S. Enteritidis and S. Typhimurium, Salmonella Infantis is the third most frequently detected serotype in humans (Garcia-Soto et al., 2020). Furthermore, when compared to S. Enteritidis and S. Typhimurium, S. Infantis exhibited a significantly higher occurrence of persistent infections (Marzel et al., 2016). In addition, Salmonella Paratyphi B and Salmonella Indiana are epidemiologically important serotypes linked to chicken meat in Germany (Federal Office of Consumer Protection and Food Safety (BVL), 2020). Numerous positive cases of S. Paratyphi B and S. Indiana have been detected in poultry meat across various continents (Ferrari et al., 2019). Thus, Salmonella spp. are predominantly found in poultry products, with chicken being the primary reservoir (Ferrari et al., 2019; EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control), 2022). Although measures such as surveillance, biosecurity, and vaccination are adopted, salmonellosis remains a significant challenge for both public and animal health (Vandeplas et al., 2010; Ferrari et al., 2019). The safety of chicken meat products may be increased using chemical or physical antimicrobial measures (Wessels et al., 2021; Han et al., 2022). In this context, the use of antibiotics is of major concern, due to the global challenge of antibiotic resistance (Mehdi et al., 2018). Hence, it is of significant importance to enforce different control and preventive strategies to limit bacterial contamination, including Salmonella, in chicken-derived products.
The use of strictly lytic (virulent) bacteriophages, commonly designated as phages, represents an alternative approach, serving as a promising biocontrol tool to reduce agents either in living animals or during postharvest processing (Moye et al., 2018; Islam et al., 2021). Bacteriophages are viruses that infect exclusively bacteria by binding to specific cell wall receptors, followed by lysis of the bacterial cell. Phages are the most occurring biological entities in the biosphere (Clokie et al., 2011). There are temperate and virulent phages. Virulent phages undergo only a lytic cycle and are therefore utilized in therapy or for biocontrol (Sulakvelidze et al., 2001; Moye et al., 2018). Phages were discovered before antibiotics, but after the advent of antibiotics, phage research moved to the background (Clokie et al., 2011). Shortly after the discovery of antibiotics, the issue of antibiotic resistance emerged (Kutateladze and Adamia, 2010). Nowadays there is a global problem of antibiotic resistance leading to a revival of alternative approaches like harnessing phages (Willyard, 2017; Vikram et al., 2021). Phages, as antimicrobial agents, can be used for the therapy of both humans and animals, as well as for applications along the food chain (Hagens and Loessner, 2007; Abedon et al., 2014). Besides their specificity and the fact that they do not harm the normal microflora on food, one other advantage of using phages is that they do not change the organoleptic characteristics of food (Moye et al., 2018).
The use of phages in the poultry and chicken meat industry is currently receiving considerable attention. Several reviews have addressed this area of research in depth, particularly focusing on Salmonella spp. as target bacteria (Żbikowska et al., 2020; Islam et al., 2021; Wessels et al., 2021; Han et al., 2022; Khan and Rahman, 2022; Abd-El Wahab et al., 2023; Shahdadi et al., 2023). To date, there are a number of reports describing experimental studies on the reduction of Salmonella through post-harvest application (Goode et al., 2003; Hungaro et al., 2013; Duc et al., 2018; Atterbury et al., 2020; Esmael et al., 2021; Shakeri et al., 2021). In most studies, two Salmonella serotypes and a single phage or a phage cocktail were applied in a laboratory setting to chicken skin or to chicken meat. However, phage application is not confined to the research area; it is also effectively integrated into practical food processing. Among other commercially available phage products, the U.S. Food and Drug Administration (FDA) approved different Salmonella specific phage preparations for utilization within the poultry industry (Han et al., 2022). Several studies reported on the successful application of commercial Salmonella phages at the laboratory scale (Sukumaran et al., 2015, 2016; Grant et al., 2017; Yeh et al., 2017, 2018). In the USA and other developed countries, the use of phage cocktails with a Generally Recognized as Safe (GRAS) status is permitted. Conversely, within the European Union (EU), the application of phage preparations within the food industry is not allowed due to regulatory concerns (Abd-El Wahab et al., 2023). However, the use of phages to combat Listeria is regarded as safe within the EU (EFSA BIOHAZ Panel, 2016).
In this study, we evaluated the efficacy of a commercially available Salmonella phage product containing six phages on both chicken skin and stainless-steel surfaces. Some properties of the phages have already been described (Gvaladze et al., 2023). Four phages exhibited a myoviridal morphology, while the remaining two are siphoviruses. Five of them revealed a broad host range and a strong lytic activity on several Salmonella serotypes (S. Enteritidis, S. Typhimurium, S. Infantis, S. Paratyphi B, and S. Indiana) at low temperatures and at low MOIs in liquid cultures. Moreover, a mixture of ten strains belonging to five serotypes, was efficiently lysed down to 6°C. Here, the phages' ability to lyse these Salmonella serotypes on hard surfaces was studied. Stainless steel was selected as it is a common component of equipment used for food production. To our knowledge, only little data have been published yet on the application of phages on this surface. In addition, most studies used only S. Enteritidis and S. Typhimurium as the target serotypes.
The Salmonella strains used in this study were isolated from chicken products and stored at the National Reference Laboratory for Salmonella, German Federal Institute for Risk Assessment (BfR) Berlin, Germany (Table 1). Salmonella stock cultures were preserved at −80°C using glycerol as a cryoprotectant. Bacteria were grown overnight on lysogeny broth agar (LB; Carl Roth GmbH, Karlsruhe, Germany) at 37°C. Following this, individual colonies were selected and inoculated into LB broth, which was then incubated at 37°C overnight.
The six phages of the cocktail “Applied Phage Meat S2” were isolated from environmental sources (duck pond in Hamm, Germany and sewage treatment plant in Hamm, Germany) by the PTC Phage Technology Center (Bönen, Germany). The phages of this cocktail have been deposited in the German Collection of Microorganisms and Cell Cultures (DSMZ) and are listed in Table 2. “Applied Phage Meat S2” is a commercially available product (FinkTec GmbH, Hamm, Germany), holding an US FDA “Generally Recognized as Safe (GRAS)” status for its application to combat Salmonella during both meat and vegetable processing (FDA, 2023a,b). The cocktail covered over 400 Salmonella isolates belonging to 41 different serotypes (FDA, 2023a).
The phage cocktail was enumerated using a double agar overlay plaque assay (Kropinski et al., 2009). For each strain, the phage cocktail titer was determined (Table 1). Serial dilutions of phages were prepared in sodium magnesium buffer (SM, 50 mM Tris-HCl, 100 mM sodium chloride, 8 mM magnesium sulfate, pH 7.5). Overnight culture and each serial dilution of lysates (each 0.1 ml) were mixed and incubated for 5 min. Next, 5 ml of LB soft agar (0.6%) were added to the mixture and poured onto a LB plate. The LB plates were incubated at 37°C overnight.
To apply phages on chicken skin, 10 ml spray bottles (NeoLab Migge GmbH, Heidelberg, Germany) were used. The spray volume was initially determined at least 100 times to ensure the precise quantity of phages used for application. The average value of this measurement was 129 μl (±0.01 μl). We also determined the optimal distance between the spray bottle and skin surface. Sterile water mixed with bromophenol blue was used for this purpose and sprayed onto a filter paper. The ideal spraying distance was 18 cm, resulting in a circular coverage of ~5 cm diameter.
Fresh chicken was purchased on the day of the experiment from a local supermarket. The experiment was conducted with major modifications based on the description provided by Duc et al. (2020). The breast skin was carefully removed and cut into 4 × 4 cm2 pieces. First two different strains of S. Enteritidis (strain a and strain b, Table 1.) were used. For individual strains, a 1:100 dilution of the overnight culture was prepared. The cultures were incubated at 37°C with shaking at 200 rpm. After reaching an optical density at 588 nm (OD588) of 0.2 (corresponding to ~5 × 107 CFU/ml) each culture was diluted 10-fold and 0.1 ml of the dilution (10−1) was dropped onto the skin and spread with a Drigalski spatula to obtain ~5 × 104 CFU/cm2. The artificially contaminated skin was left to dry completely under a clean bench, which took 20–90 min depending on the skin structure. Once the bacterial culture completely dried on the skin, the phage cocktail was sprayed onto the sample. A 10 mL spray bottle was fixed on a stand at a distance of 18 cm, and the skin was sprayed two to three times. The phage cocktail titer was determined for each strain and the concentration of S. Enteritidis a and S. Enteritidis b on the skin reached a final concentration of 3 × 106 PFU/cm2 [multiplicity of infection (MOI) 50] and 2 × 107 PFU/cm2 (MOI 500), respectively. Controls were sprayed in the same way with PBS buffer (phosphate-buffered saline). After spraying the phage cocktail, the samples were incubated at room temperature (RT) for 2, 4, 6, and 24 h.
Following the experiment with the two S. Enteritidis strains, the stability of the phage cocktail was determined. For this, after each time point (2, 4, 6, 24 h), a 1 ml aliquot (after homogenization) of the treated sample of each strain was centrifuged at 11,000 × g, and the supernatant was filtered through a 0.22 μm pore-size filter (VWR International GmbH, Darmstadt, Germany). The filtered lysate was titrated using the double-agar overlay plaque assay. The resulting phage titer was then compared with the initial titer.
The experiment with a mixture of strains was conducted in the same way as described above for S. Enteritidis. However, here five different Salmonella serotypes were used: S. Enteritidis, S. Typhimurium, S. Infantis, S. Paratyphi B and S. Indiana (all strains except for S. Enteritidis a—see Table 1). LB broth was inoculated with 20 μl of an overnight culture (OD588 approx. 1.6) of each strain. The culture containing several strains was allowed to grow until it reached an OD588 of 0.2. The final concentrations of the bacteria and of the phage cocktail on skin was 5 × 104 CFU/cm2 and 3 × 106 PFU/cm2 (MOI 50), respectively. The experiment was performed in triplicate. After spraying the phage cocktail or PBS, the treated samples and controls were incubated at room temperature for 0.5, 1, 2, and 4 h.
Stainless steel plates 4 × 4 cm2 (N 1.4301) were produced by the Ottim Metall GmbH, Berlin, Germany. To mimic a contamination as applied in the chicken skin experiments, a single strain (S. Enteritidis b) was prepared. In the first experiment we simulated a new surface contamination prior to drying. In this case, the phage cocktail was sprayed onto liquid S. Enteritidis before being dried on stainless-steel. An amount of 0.1 ml S. Enteritidis b (5 × 105 CFU/cm2) was distributed on the stainless-steel surface in small drops and was directly treated with two–three sprays of the phage cocktail (2 × 107 PFU/cm2). In another experiment, we used a single strain (S. Enteritidis b was used as the positive control) and a mixture of the five serotypes. The experiment was carried out as described above. However, after contamination of the stainless-steel, Salmonella cells were allowed to dry completely under a clean bench. Upon drying (~20 min), the phage cocktail was sprayed onto the surface to achieve 2 × 107 PFU/cm2 (MOI 50). This experiment was carried out in triplicate and the control samples were sprayed with PBS.
After the treatment, the samples of chicken skin or stainless steel and their respective controls were placed in a cold storage room (4°C). In the cold room, the samples were placed in 15 × 80 cm stomacher bags containing 20 ml of PBS. The samples were gently massaged by hand and homogenized for 1 min at level 3 (MiniMix, INTERSCIENCE, Wiesbaden, Germany). Thereafter, the samples were taken to the laboratory in an ice-bath box and diluted to a 10−3 dilution level. All dilutions and the undiluted sample were plated on XLD plates (Xylose Lysine Deoxycholate agar, Oxoid Deutschland GmbH, Wesel, Germany).
The effect of exposure to the phage cocktail was tested using paired sample t-test as provided by SPSS (IBM SPSS statistics, Version 26). Pairs were defined by time points per trial. Comparisons were done per trial including all four–five time points. When three treated samples were compared to one control, three comparisons were carried out.
In a previous study performed under in vitro conditions, reduction of Salmonella strains by the phage cocktail investigated here has been demonstrated (Gvaladze et al., 2023). To examine the reduction on chicken skin, we initially tested two S. Enteritidis strains revealing the highest susceptibility to the phage cocktail. Reduction rates for both strains were similar, ~2 log10 units 2 h after addition of the phage cocktail at RT. Reduction of bacterial counts was also determined 4 and 6 h post treatment. The strongest decrease was observed after 4 h, which showed a 3 log10 unit reduction. Even after 24 h of treatment with the phage cocktail, a reduction of ~2.7 log10 units was observed for both strains (Figure 1). The results reached statistical significance with a p-value of ≤ 0.002. We also determined the bacterial count after-phage cocktail treatment relative to the initial count. The Salmonella count was 1.2 log10 units lower than the initial level after 6 h of treatment, whereas after 24 h similar numbers of bacteria were determined.
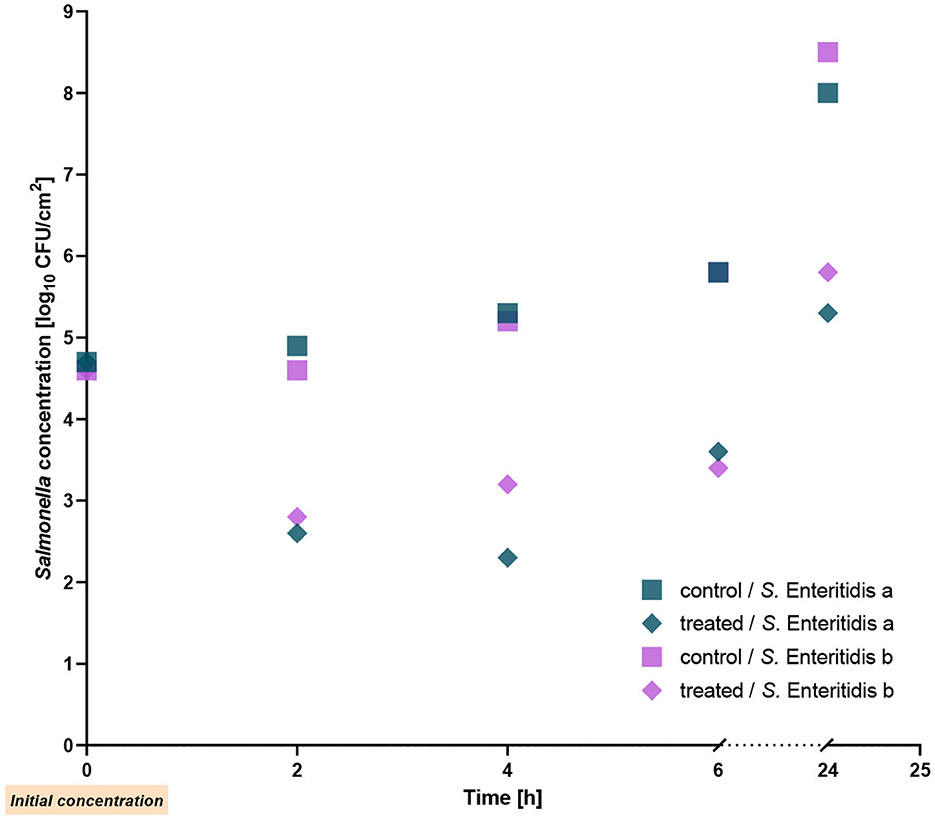
Figure 1. Reduction of two Salmonella Enteritidis strains on chicken skin by the phage cocktail at room temperature.
We also examined the stability of the phages on chicken skin. The initial titers of the phage cocktail for S. Enteritidis a and S. Enteritidis b were 7 × 107 PFU/ml and 5 × 108 PFU/ml, respectively. Throughout the entire experiment, the titers remained stable and showed only a slight decrease of ~0.5 log10 units (Figure 2).
In this experiment, the reduction of a mixture of five different Salmonella serotypes was studied. S. Enteritidis strain b was used as the positive control. The mixture showed a reduction of 1 log10 unit after 30 min of phage application, and the reduction remained stable throughout the entire experiment, resulting in a decrease of 1.2 log10 units (p ≤ 0.002) after 4 h incubation at RT. On the other hand, the single Enteritidis strain b was reduced by 1.5–1.8 log10 units during the 4 h (p < 0.001; Figure 3). Compared to the initial count, the bacterial counts of S. Enteritidis b and of the mixture were also lower (up to 1 log10 unit) after treatment for 4-h.
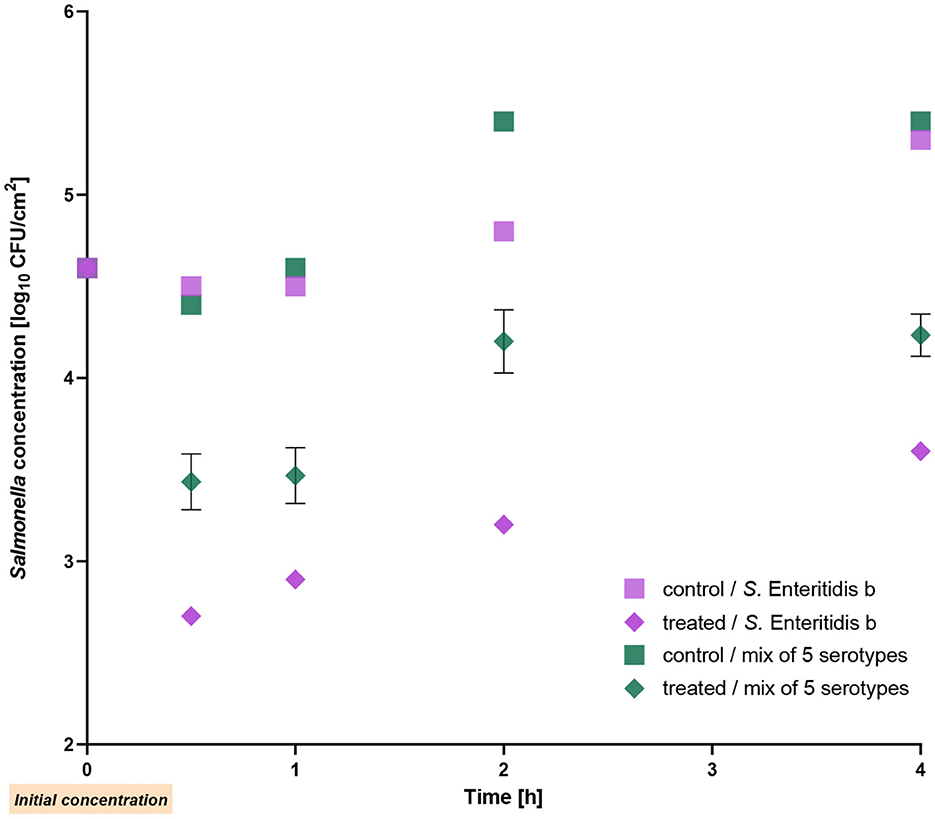
Figure 3. Reduction of a mixture of five Salmonella serotypes on chicken skin by the phage cocktail at room temperature. Salmonella Enteritidis strain b was used as a positive control. The error bars in the figure represent the mean and standard deviation (SD) for three phage-treated samples.
We now wanted to learn whether a significant reduction of Salmonella is also achievable on a stainless steel surface. The first thing we did was to simulate a new surface contamination prior to drying. In this case, the phage cocktail was used to infect liquid S. Enteritidis. The reduction on wet stainless steel was much higher than lysis after drying the bacteria (see below). Within the first hour of treatment, the single S. Enteritidis strain b exhibited a remarkable decrease of 3 log10 units (p = 0.012) and after 2 h, the bacterial count reached a level below the detection limit (Figure 4).
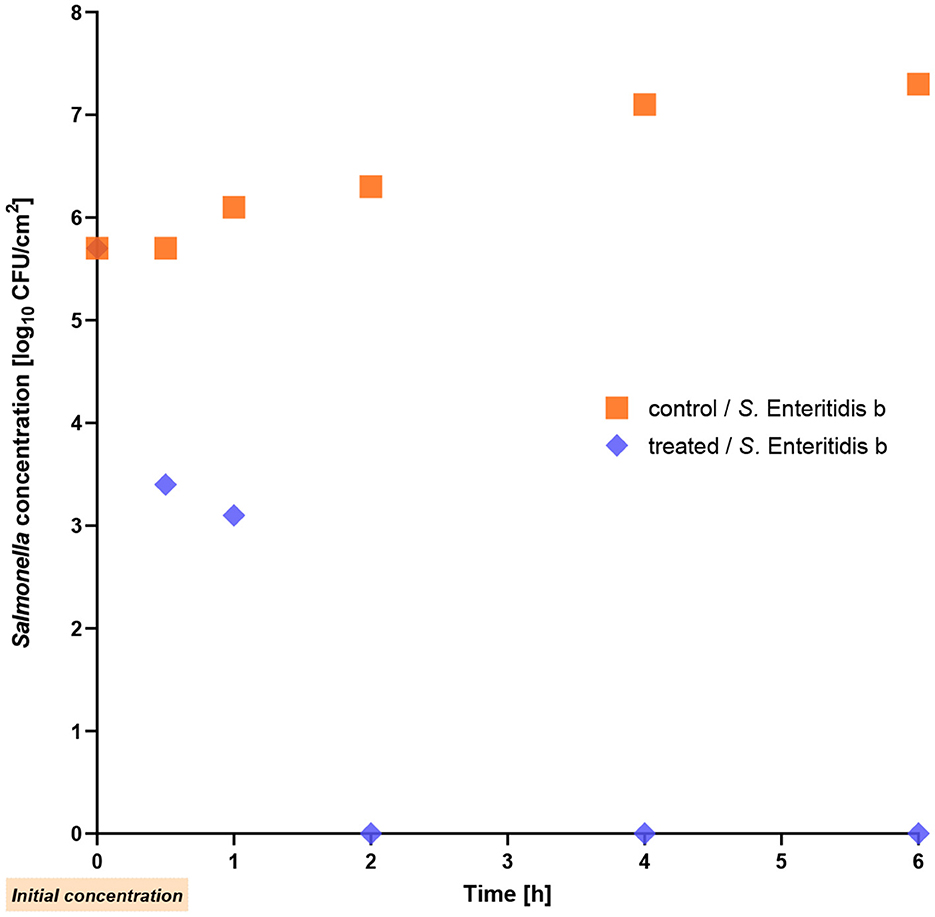
Figure 4. Reduction of Salmonella Enteritidis strain b on stainless-steel at room temperature. The phage cocktail was applied to the still wet contamination.
In the second experiment, the bacteria were allowed to dry completely on the stainless steel surface before adding the phages. The experiment was performed with S. Enteritidis strain b (positive control) and the mixture containing the five different serotypes. Figure 5 illustrates the obtained results indicating a similar reduction of both the single Salmonella strain and the mixture, with reductions of up to 2.4 log10 units (p ≤ 0.009) observed after 4 h of treatment. Whereas the mixture exhibited a reduction of 1.2 log10 units within the first 30 min, the single strain showed a reduction of 1.7 log10 units.
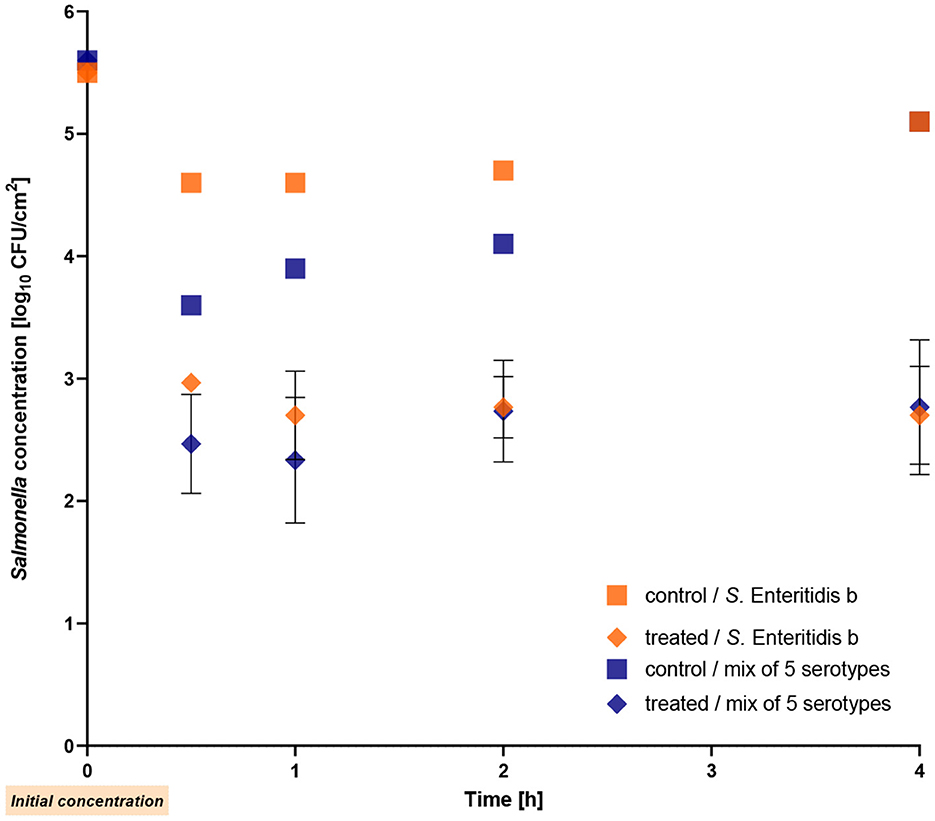
Figure 5. Reduction of the mixture of five Salmonella serotypes after drying on stainless-steel by the phage cocktail at room temperature. The Salmonella Enteritidis strain b was used as a positive control. The error bars in the figure represent the mean and standard deviation (SD) for three phage-treated samples.
In this study, we analyzed the effectiveness of a phage cocktail for the reduction of relevant Salmonella serotypes on both chicken skin and stainless-steel surfaces. Our findings clearly demonstrate that this phage cocktail could be a promising biocontrol tool, which could be employed for post-harvest applications during poultry production. Initially, the ability of the phage cocktail to reduce the predominant serotype S. Enteritidis was determined. Thereafter, its performance against a mixture of five important Salmonella serotypes (S. Enteritidis, S. Typhimurium, S. Infantis, S. Paratyphi B and S. Indiana) was examined. Chicken can contain multiple Salmonella serotypes simultaneously (Betancor et al., 2010). The coexistence of multiple Salmonella serotypes in chicken and poultry poses a significant challenge to food safety. A phage cocktail can effectively target multiple serotypes; therefore, it is common to use two or more phages for applications (Sevilla-Navarro et al., 2018). The cocktail comprising six-phages showed a significant reduction of the single Salmonella strain as well as of the mixture on both matrices. Slaughterhouses being potential sources of Salmonella cross-contamination of chicken meat play a pivotal role in controlling the spread of this pathogen throughout the food chain (Heyndrickx et al., 2007; Zeng et al., 2021). While in numerous studies the application of Salmonella phages has been explored on food samples spiked with a single serotype, there are few reports on the reduction of mixtures of different Salmonella serotypes. In addition, there is only a limited number of studies employing commercial phage cocktails against mixtures of bacteria. Yeh et al. (2017) applied two commercial phage cocktails at a concentration of up to 108 PFU/ml during the tumbling process to reduce a mixture of four Salmonella serotypes (S. Enteritidis, S. Infantis, S. Heidelberg, and S. Newport) in various ground meat types, including chicken, at 4°C. This application resulted in a reduction of ~1 log10 unit. Sukumaran et al. (2015) and Yeh et al. (2018) also compared the application of individual or combined commercial phage cocktails with chemical and/or physical methods to reduce mixed Salmonella serotypes. In both studies reductions of up to 1 log10 unit were achieved. In another study, Sukumaran et al. (2016) compared dip and surface applications of the commercial phage cocktail SalmoFresh™ on chicken breast using a three-serotype cocktail (S. Typhimurium, S. Enteritidis, and S. Heidelberg). They again achieved reductions of up to 1 log10 unit at 4°C and RT. According to this result, dipping of food in a phage solution does not achieve better reductions than spraying. Grant et al. (2017) used the Salmonelex™ phage cocktail to reduce a Salmonella contamination on two different bacterial mixtures: the first mixture contained S. Newport, S. Typhimurium, and S. Thomson while the second mixture comprised S. Enteritidis, S. Heidelberg, and S. Typhimurium. They achieved maximum reductions of 0.9 log10 units and 0.7 log10 units at 4°C after 8 h, respectively. The bacterial and phage doses used in these experiments were similar to those used in our study, with 104 CFU and 107 PFU per cm2 of chicken meat. Whereas almost all of these bacterial mixtures included S. Enteritidis or S. Typhimurium, Yeh et al.'s mixture was the only one that contained the relevant serotype S. Infantis. We went a step further by testing a combination of five different serotypes. As far as we are aware, no comparable results have been published as yet.
The use of phages on stainless steel remains an area of limited research. Stainless steel, a commonly used material in food processing equipment (Veluz et al., 2012), can act as a carrier of bacteria, thereby facilitating transmission of pathogens and cross-contamination (Arnold and Silvers, 2000). To the best of our knowledge, only a few studies have explored the reduction of Salmonella using phages on stainless-steel surfaces (Woolston et al., 2013; Gong and Jiang, 2017; Ge et al., 2022; Korzeniowski et al., 2022). Korzeniowski et al. (2022) investigated the impact of individual phages and a cocktail at various concentrations on S. Enteritidis biofilms, resulting in reductions of 60%−97% at 37°C. Ge et al. (2022) contaminated a metal surface with S. Enteritidis and applied a single phage, achieving a 1 log10 unit reduction. Woolston et al. (2013) employed a commercially available phage cocktail, similar to our approach. The cocktail, SalmoLyse™, reduced a surface contamination with Paratyphi B by 2.1–4.1 log10 units. Gong and Jiang (2017) studied a six-phage cocktail against S. Typhimurium and a mix of 10 S. Typhimurium isolates, with reductions of 3 log10 and 2 log10 units, respectively at 23°C. The last study is consistent with our results in the manner that it shows a greater reduction with a single strain. Though, even when mixed strains were used, significant reductions were observed.
In conclusion, our study demonstrated the effectiveness of a phage cocktail on both single S. Enteritidis strains and a mixture of five different Salmonella serotypes. Moreover, reductions persisted over an extended period. In a previous study, we assessed the impact of the phage cocktail at low temperatures ranging from 15 to 6°C, using individual and mixed Salmonella cultures. The significant reduction of a mixture of the five different Salmonella serotypes at 6°C suggests that the phage cocktail remains active within the cold chain. Resistance against the six phages was not observed, we identified at least one phage to which the bacteria exhibited sensitivity (Gvaladze et al., 2023). Thus, it is likely that some bacteria survived the infection not because of resistance, but rather because of the fact that they entered the late logarithmic or even stationary growth phase, in which propagation of phages is limited. It is of course also conceivable that they simply survived, because they did not encounter an infectious particle. The promising results of this study corroborate the potential of these phages for applications in post-harvest settings such as slaughterhouses under real conditions.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
TG: Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. HL: Conceptualization, Funding acquisition, Project administration, Resources, Validation, Writing – review & editing. SH: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research received financial backing from the Federal Ministry of Food and Agriculture (BMEL) through the Federal Office for Agriculture and Food (BLE) under the innovation support program (grant number: 281C104B18), in accordance with a resolution by the Parliament of the Federal Republic of Germany.
The authors express gratitude to Istvan Szabo for contributing Salmonella strains to the project. We are also highly indebted to Bernd-Alois Tenhagen for his expertise and support in conducting the statistical analysis. Furthermore, we appreciate the financial support from the Federal Ministry of Food and Agriculture (BMEL).
HL was employed by Phage Technology Center GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abd-El Wahab, A., Basiouni, S., El-Seedi, H. R., Ahmed, M. F. E., Bielke, L. R., Hargis, B., et al. (2023). An overview of the use of bacteriophages in the poultry industry: successes, challenges, and possibilities for overcoming breakdowns. Front. Microbiol. 14:1136638. doi: 10.3389/fmicb.2023.1136638
Abedon, S. T., Kuhl, S. J., Blasdel, B. G., and Kutter, E. M. (2014). Phage treatment of human infections. Bacteriophage 1, 66–85. doi: 10.4161/bact.1.2.15845
Arnold, J. W., and Silvers, S. (2000). Comparison of poultry processing equipment surfaces for susceptibility to bacterial attachment and biofilm formation. Poult. Sci. 79, 1215–1221. doi: 10.1093/ps/79.8.1215
Atterbury, R. J., Gigante, A. M., Rubio Lozano, M. S., Mendez Medina, R. D., Robinson, G., Alloush, H., et al. (2020). Reduction of Salmonella contamination on the surface of chicken skin using bacteriophage. Virol. J. 17:98. doi: 10.1186/s12985-020-01368-0
Bahrani-Mougeot, F. K., Scobey, M. W., and Sansonetti, P. J. (2009). “Enteropathogenic infections,” in Encyclopedia of Microbiology, 3rd ed., ed. M. Schaechter (Oxford: Academic Press), 329–343. doi: 10.1016/B978-012373944-5.00224-8
Betancor, L., Pereira, M., Martinez, A., Giossa, G., Fookes, M., Flores, K., et al. (2010). Prevalence of Salmonella enterica in poultry and eggs in Uruguay during an epidemic due to Salmonella enterica serovar Enteritidis. J. Clin. Microbiol. 48, 2413–2423. doi: 10.1128/JCM.02137-09
Chanamé Pinedo, L., Mughini-Gras, L., Franz, E., Hald, T., and Pires, S. M. (2022). Sources and trends of human salmonellosis in Europe, 2015–2019: an analysis of outbreak data. Int. J. Food Microbiol. 379:109850. doi: 10.1016/j.ijfoodmicro.2022.109850
Clokie, M. R., Millard, A. D., Letarov, A. V., and Heaphy, S. (2011). Phages in nature. Bacteriophage 1, 31–45. doi: 10.4161/bact.1.1.14942
Duc, H. M., Son, H. M., Honjoh, K. I., and Miyamoto, T. (2018). Isolation and application of bacteriophages to reduce Salmonella contamination in raw chicken meat. LWT 91, 353–360. doi: 10.1016/j.lwt.2018.01.072
Duc, H. M., Son, H. M., Yi, H. P. S., Sato, J., Ngan, P. H., Masuda, Y., et al. (2020). Isolation, characterization and application of a polyvalent phage capable of controlling Salmonella and Escherichia coli O157:H7 in different food matrices. Food Res. Int. 131:108977. doi: 10.1016/j.foodres.2020.108977
EFSA BIOHAZ Panel (2016). (EFSA Panel on Biological Hazards). Evaluation of the safety and efficacy of Listex™ P100 for reduction of pathogens on different ready-to-eat (RTE) food products. EFSA J. 14:e04565. doi: 10.2903/j.efsa.2016.4565
EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control) (2022). The European Union One Health 2021 Zoonoses Report. EFSAJournal 20:273. doi: 10.2903/j.efsa.2022.7666
Esmael, A., Azab, E., Gobouri, A. A., Nasr-Eldin, M. A., Moustafa, M. M. A., Mohamed, S. A., et al. (2021). Isolation and characterization of two lytic bacteriophages infecting a multi-drug resistant Salmonella Typhimurium and their efficacy to combat Salmonellosis in ready-to-use foods. Microorganisms 9:423. doi: 10.3390/microorganisms9020423
FDA (2023a). GRAS Notices GRN No. 1038 2023 [updated 10, 12.2023. Preparation containing six bacteriophages (phage) specific to Salmonella enterica spp. serovars]. Available online at: https://www.cfsanappsexternal.fda.gov/scripts/fdcc/index.cfm?set=GRASNoticesandid=1038 (accessed December 12, 2023).
FDA (2023b). GRAS Notices GRN No. 1070 2023 [updated 10, 12.2023. Preparations containing six bacteriophages (phage) specific to Salmonella enterica]. Available online at: https://www.cfsanappsexternal.fda.gov/scripts/fdcc/?set=GRASNoticesandid=1070 (accessed December 12, 2023).
Ferrari, R. G., Rosario, D. K. A., Cunha-Neto, A., Mano, S. B., Figueiredo, E. E. S., Conte-Junior, C. A., et al. (2019). Worldwide epidemiology of Salmonella serovars in animal-based foods: a meta-analysis. Appl. Environ. Microbiol. 85, e00591-19. doi: 10.1128/AEM.00591-19
Garcia-Soto, S., Abdel-Glil, M. Y., Tomaso, H., Linde, J., and Methner, U. (2020). Emergence of multidrug-resistant Salmonella enterica subspecies enterica serovar infantis of multilocus sequence type 2283 in German Broiler Farms. Front. Microbiol. 11:1741. doi: 10.3389/fmicb.2020.01741
Ge, H., Lin, C., Xu, Y., Hu, M., Xu, Z., Geng, S., et al. (2022). A phage for the controlling of Salmonella in poultry and reducing biofilms. Vet. Microbiol. 269:109432. doi: 10.1016/j.vetmic.2022.109432
Gong, C., and Jiang, X. (2017). Application of bacteriophages to reduce Salmonella attachment and biofilms on hard surfaces. Poult. Sci. 96, 1838–1848. doi: 10.3382/ps/pew463
Goode, D., Allen, V. M., and Barrow, P. A. (2003). Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl. Environ. Microbiol. 69, 5032–5036. doi: 10.1128/AEM.69.8.5032-5036.2003
Grant, A., Parveen, S., Schwarz, J., Hashem, F., and Vimini, B. (2017). Reduction of Salmonella in ground chicken using a bacteriophage. Poult. Sci. 96, 2845–2852. doi: 10.3382/ps/pex062
Gut, A. M., Vasiljevic, T., Yeager, T., and Donkor, O. N. (2018). Salmonella infection - prevention and treatment by antibiotics and probiotic yeasts: a review. Microbiology 164, 1327–1344. doi: 10.1099/mic.0.000709
Gvaladze, T., Lehnherr, H., Große-Kleimann, J., and Hertwig, S. A. (2023). Bacteriophage cocktail reduces five relevant Salmonella serotypes at low multiplicities of infection and low temperatures. Microorganisms 11:2298. doi: 10.3390/microorganisms11092298
Hagens, S., and Loessner, M. J. (2007). Application of bacteriophages for detection and control of foodborne pathogens. Appl. Microbiol. Biotechnol. 76, 513–519. doi: 10.1007/s00253-007-1031-8
Han, S., Byun, K.-H., Mizan, M. F. R., Kang, I., and Ha, S.-D. (2022). Bacteriophage and their lysins: a new era of biocontrol for inactivation of pathogenic bacteria in poultry processing and production—a review. Food Control. 137:108976. doi: 10.1016/j.foodcont.2022.108976
Hauser, E., Tietze, E., Helmuth, R., Junker, E., Prager, R., Schroeter, A., et al. (2012). Clonal dissemination of Salmonella enterica serovar infantis in Germany. Foodborne Pathog. Dis. 9, 352–360. doi: 10.1089/fpd.2011.1038
Heredia, N., and García, S. (2018). Animals as sources of food-borne pathogens: a review. Anim Nutr. 4, 250–255. doi: 10.1016/j.aninu.2018.04.006
Heyndrickx, M., Herman, L., Vlaes, L., Butzler, J. P., Wildemauwe, C., Godard, C., et al. (2007). L. Multiple typing for the epidemiological study of the contamination of broilers with Salmonella from the hatchery to the slaughterhouse. J. Food Prot. 70, 323–334. doi: 10.4315/0362-028X-70.2.323
Hungaro, H. M., Mendonça, R. C. S., Gouvêa, D. M., Vanetti, M. C. D., and Pinto, C. L. O. (2013). Use of bacteriophages to reduce Salmonella in chicken skin in comparison with chemical agents. Food Res. Int. 52, 75–81. doi: 10.1016/j.foodres.2013.02.032
Islam, M. R., Martinez-Soto, C. E., Lin, J. T., Khursigara, C. M., Barbut, S., Anany, H., et al. (2021). A systematic review from basics to omics on bacteriophage applications in poultry production and processing. Crit. Rev. Food Sci. Nutr. 63, 3097–3129. doi: 10.1080/10408398.2021.1984200
Issenhuth-Jeanjean, S., Roggentin, P., Mikoleit, M., Guibourdenche, M., de Pinna, E., Nair, S., et al. (2014). Supplement 2008-2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 165, 526–530. doi: 10.1016/j.resmic.2014.07.004
Khan, M. A. S., and Rahman, S. R. (2022). Use of phages to treat antimicrobial-resistant Salmonella infections in poultry. Vet Sci. 9:438. doi: 10.3390/vetsci9080438
Korzeniowski, P., Sliwka, P., Kuczkowski, M., Misic, D., Milcarz, A., Kuzminska-Bajor, M., et al. (2022). Bacteriophage cocktail can effectively control Salmonella biofilm in poultry housing. Front. Microbiol. 13:901770. doi: 10.3389/fmicb.2022.901770
Kropinski, A. M., Mazzocco, A., Waddell, T. E., Lingohr, E., and Johnson, R. P. (2009). “Enumeration of bacteriophages by double agar overlay plaque assay,” in Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions, eds M. R. J. Clokie, and A. M. Kropinski (Totowa, NJ: Humana Press), 69–76. doi: 10.1007/978-1-60327-164-6_7
Kutateladze, M., and Adamia, R. (2010). Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 28, 591–595. doi: 10.1016/j.tibtech.2010.08.001
Marzel, A., Desai, P. T., Goren, A., Schorr, Y. I., Nissan, I., Porwollik, S., et al. (2016). Persistent infections by nontyphoidal Salmonella in humans: epidemiology and genetics. Clin. Infect. Dis. 62, 879–886. doi: 10.1093/cid/civ1221
Mehdi, Y., Létourneau-Montminy, M. P., Gaucher, M. L., Chorfi, Y., Suresh, G., Rouissi, T., et al. (2018). Use of antibiotics in broiler production: global impacts and alternatives. Anim Nutr. 4, 170–178. doi: 10.1016/j.aninu.2018.03.002
Moye, Z. D., Woolston, J., and Sulakvelidze, A. (2018). Bacteriophage applications for food production and processing. Viruses 10:205. doi: 10.3390/v10040205
Mughini-Gras, L., van Hoek, A., Cuperus, T., Dam-Deisz, C., van Overbeek, W., van den Beld, M., et al. (2021). Prevalence, risk factors and genetic traits of Salmonella Infantis in Dutch broiler flocks. Vet. Microbiol. 258:109120. doi: 10.1016/j.vetmic.2021.109120
Park, C. J., and Andam, C. P. (2020). Distinct but intertwined evolutionary histories of multiple Salmonella enterica subspecies. mSystems 5:e00515-19. doi: 10.1128/mSystems.00515-19
Sevilla-Navarro, S., Marín, C., Cortés, V., García, C., Vega, S., Catalá-Gregori, P., et al. (2018). Autophage as a control measure for Salmonella in laying hens. Poult. Sci. 97, 4367–4373. doi: 10.3382/ps/pey294
Shahdadi, M., Safarirad, M., Berizi, E., Mazloomi, S. M., Hosseinzadeh, S., Zare, M., et al. (2023). A systematic review and modeling of the effect of bacteriophages on Salmonella spp. reduction in chicken meat. Heliyon 9:e14870. doi: 10.1016/j.heliyon.2023.e14870
Shakeri, G., Hammerl, J. A., Jamshidi, A., Ghazvini, K., Rohde, M., Szabo, I., et al. (2021). The lytic siphophage vB_StyS-LmqsSP1 reduces the number of Salmonella. Appl. Environ. Microbiol. 87:e0142421. doi: 10.1128/AEM.01424-21
Sukumaran, A. T., Nannapaneni, R., Kiess, A., and Sharma, C. S. (2015). Reduction of Salmonella on chicken meat and chicken skin by combined or sequential application of lytic bacteriophage with chemical antimicrobials. Int. J. Food Microbiol. 207, 8–15. doi: 10.1016/j.ijfoodmicro.2015.04.025
Sukumaran, A. T., Nannapaneni, R., Kiess, A., and Sharma, C. S. (2016). Reduction of Salmonella on chicken breast fillets stored under aerobic or modified atmosphere packaging by the application of lytic bacteriophage preparation SalmoFreshTM. Poult. Sci. 95, 668–675. doi: 10.3382/ps/pev332
Sulakvelidze, A., Alavidze, Z., and Morris, J. G. Jr. (2001). Bacteriophage therapy. Antimicrob. Agents Chemother. 45, 649–659. doi: 10.1128/AAC.45.3.649-659.2001
Teklemariam, A. D., Al-Hindi, R. R., Albiheyri, R. S., Alharbi, M. G., Alghamdi, M. A., Filimban, A. A. R., et al. (2023). Human salmonellosis: a continuous global threat in the farm-to-fork food safety continuum. Foods 12:1756. doi: 10.3390/foods12091756
Vandeplas, S., Dauphin, R. D., Beckers, Y., Thonart, P., and Théwis, A. (2010). Salmonella in chicken: current and developing strategies to reduce contamination at farm level. J. Food Prot. 73, 774–785. doi: 10.4315/0362-028X-73.4.774
Veluz, G. A., Pitchiah, S., and Alvarado, C. Z. (2012). Attachment of Salmonella serovars and Listeria monocytogenes to stainless steel and plastic conveyor belts. Poult. Sci. 91, 2004–2010. doi: 10.3382/ps.2011-01689
Vikram, A., Woolston, J., and Sulakvelidze, A. (2021). Phage biocontrol applications in food production and processing. Curr. Issues Mol. Biol. 40, 267–302. doi: 10.21775/cimb.040.267
Wessels, K., Rip, D., and Gouws, P. (2021). Salmonella in chicken meat: consumption, outbreaks, characteristics, current control methods and the potential of bacteriophage use. Foods 10:1742. doi: 10.3390/foods10081742
Willyard, C. (2017). The drug-resistant bacteria that pose the greatest health threats. Nature 543:15. doi: 10.1038/nature.2017.21550
Woolston, J., Parks, A. R., Abuladze, T., Anderson, B., Li, M., Carter, C., et al. (2013). Bacteriophages lytic for Salmonella rapidly reduce Salmonella contamination on glass and stainless steel surfaces. Bacteriophage 3:e25697. doi: 10.4161/bact.25697
Yeh, Y., de Moura, F. H., Van Den Broek, K., and de Mello, A. S. (2018). Effect of ultraviolet light, organic acids, and bacteriophage on Salmonella populations in ground beef. Meat Sci. 139, 44–48. doi: 10.1016/j.meatsci.2018.01.007
Yeh, Y., Purushothaman, P., Gupta, N., Ragnone, M., Verma, S. C., de Mello, A. S., et al. (2017). Bacteriophage application on red meats and poultry: effects on Salmonella population in final ground products. Meat Sci. 127, 30–34. doi: 10.1016/j.meatsci.2017.01.001
Żbikowska, K., Michalczuk, M., and Dolka, B. (2020). The use of bacteriophages in the poultry industry. Animals 10:872. doi: 10.3390/ani10050872
Keywords: Salmonella, phage, biocontrol, chicken, steel, post-harvest application
Citation: Gvaladze T, Lehnherr H and Hertwig S (2024) A bacteriophage cocktail can efficiently reduce five important Salmonella serotypes both on chicken skin and stainless steel. Front. Microbiol. 15:1354696. doi: 10.3389/fmicb.2024.1354696
Received: 12 December 2023; Accepted: 19 February 2024;
Published: 04 March 2024.
Edited by:
Alicja Wegrzyn, University of Gdansk, PolandReviewed by:
Sylwia Bloch, University of Gdansk, PolandCopyright © 2024 Gvaladze, Lehnherr and Hertwig. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefan Hertwig, c3RlZmFuLmhlcnR3aWdAYmZyLmJ1bmQuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.