- 1State Key Laboratory of Black Soils Conservation and Utilization, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Changchun, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Second Hospital of Jilin University, Changchun, China
- 4Key Lab of Groundwater Resources and Environment, Ministry of Education, Jilin University, Changchun, China
The over-utilizing of nitrogen fertilizers in paddy wetlands potentially threatens to the surrounding waterbody, and a deep understanding of the community and function of microorganisms is crucial for paddy non-point source pollution control. In this study, top soil samples (0–15 cm) of paddy wetlands under groundwater's irrigation at different depths (H1: 6.8 m, H2: 13.7 m, H3: 14.8 m, H4: 15.6 m, H5: 17.0 m, and H6: 17.8 m) were collected to investigate microbial community and function differences and their interrelation with soil properties. Results suggested some soil factor differences for groundwater's irrigation at different depths. Deep-groundwater's irrigation (H2-H6) was beneficial to the accumulation of various electron acceptors. Nitrifying-bacteria Ellin6067 had high abundance under deep groundwater irrigation, which was consistent with its diverse metabolic capacity. Meanwhile, denitrifying bacteria had diverse distribution patterns. Iron-reducing bacteria Geobacter was abundant in H1, and Anaeromyxobacter was abundant under deep groundwater irrigation; both species could participate in Fe-anammox. Furthermore, Geobacter could perform dissimilatory nitrate reduction to ammonia using divalent iron and provide substrate supply for anammox. Intrasporangium and norank_f_Gemmatimonadacea had good chromium- and vanadium-reducting potentials and could promote the occurrence of anammox. Low abundances of methanotrophs Methylocystis and norank_f_Methyloligellaceae were associated with the relatively anoxic environment of paddy wetlands, and the presence of aerobic methane oxidation was favorable for in-situ methane abatement. Moisture, pH, and TP had crucial effects on microbial community under phylum- and genus-levels. Microorganisms under shallow groundwater irrigation were highly sensitive to environmental changes, and Fe-anammox, nitrification, and methane oxidation were favorable under deep groundwater irrigation. This study highlights the importance of comprehensively revealing the microbial community and function of paddy wetlands under groundwater's irrigation and reveals the underlying function of indigenous microorganisms in agricultural non-point pollution control and greenhouse gas abatement.
Highlights
• Deep groundwater irrigation favors the accumulation of multi-electron acceptors
• Geobacter and Anaeromyxobacter could participate in Fe-anammox
• Methylocystis and norank_f_Methyloligellaceae involve in-situ abatement of methane
• Moisture, pH, and TP play important roles in shaping microbial community
• Deep groundwater irrigation promotes Fe-anammox, nitrification, and methane oxidation
1 Introduction
Paddy wetlands, in which the constructed wetlands were coupled with water and fertilizer, had typical wetland ecosystems and vegetation characteristics and could provide unique habitats for microorganisms to ensure global food security (Ding et al., 2021). However, the excessive nitrogen-fertilizers application under large-scale irrigation could cause serious soil degradation and surrounding-water pollution, posing a major risk to human's health (Sun et al., 2021). In addition, paddy wetlands were one of the major sources of methane emission (Sun et al., 2019). Soil microorganisms regulated wetland biogeochemical cycles (Torsvik and Øvreås, 2002). Microbial species had various functions, such as denitrification and methane oxidation (Li et al., 2014; Chi et al., 2021a). Some biotic or abiotic factors could control microbial community composition and function, and microorganisms were sensitive and could respond rapidly to environmental changes (Guo et al., 2019; Pérez Castro et al., 2019). Therefore, revealing the community and function of microorganisms in paddy wetlands was crucial for maintaining their ecological stability and material management.
As efficient agronomic measures, fertilization and irrigation could affect microbial community and diversity in paddy wetlands (Wang et al., 2021). Environmental changes obviously altered microbial communities and activities, thereby affecting biogeochemical processes. Long-term fertilization could alter soil physicochemical properties, thereby affecting microbial communities (Guo et al., 2020). Hou et al. (2022) found that the long-term application of pig manure could promote the enrichment of complex organic-degrading bacteria and improve soil enzyme activity. Inorganic fertilizers could increase microbial diversity and change microbial community composition in paddy wetlands (Huang et al., 2019). The type of fertilizer had minimal influence on microbial community under long-term fertilization (Liu et al., 2022). Furthermore, irrigation could ensure rice growth and affect microbial community. Jiang et al. (2021) showed that irrigation combined with biochar could alleviate greenhouse gas emissions and alter microbial community. Irrigation modes had some effects on microbial community of paddy wetlands (Jin et al., 2020). Reclaimed water irrigation could improve soil microbial activity and fertility to a certain extent (Wei et al., 2017). Although some studies investigated microbial community of paddy wetlands under fertilization and irrigation, the effects of groundwater's irrigation at different depths on microbial community and diversity in paddy wetlands remain unknown.
Recent studies were involved in the function and metabolic potential of microorganisms (Chi et al., 2021d). Li et al. (2019) revealed the responses of nitrogen-metabolized pathways and relevant function genes to coastal environmental variations. Exogenous pollution input could alter microbial community, thus changing their metabolic function (Chi et al., 2021b). The genes encoding exogenous metabolism and nitrate-reducting were significantly enriched on coastal sediments (Su et al., 2018). The Sanjiang Plain was regarded as a representative of large-scale paddy development, and paddy wetlands was mainly irrigated using groundwater with various electron acceptors (Cao et al., 2016; Chong et al., 2020; Meng et al., 2021; Chen et al., 2024). However, research on the community and function of microorganisms in paddy wetlands under groundwater's irrigation was still lacking.
Microorganisms in paddy wetlands had vital effects on the nitrogen cycle, but a large knowledge gap existed regarding the influence of groundwater irrigation. We hypothesized that groundwater irrigation had significant effects on microbial communities and functions of paddy wetlands, and that the input of exogenous electron acceptors could improve soil material transformation. To test this hypothesis, we analyzed soil genetic data from paddy wetlands under groundwater irrigation using high-throughput sequencing. The aims in this study were as follows: (1) to reveal the microbial community composition and difference in paddy wetlands under groundwater's irrigation at different depths, (2) to elucidate the responses in the metabolic-pathways and related-enzyme genes of microorganisms, and (3) to explore the dominant enablers involving in microbial community and functional variation in paddy wetlands. This study can offer valuable academic references for non-point source pollution control and greenhouse gas reduction of paddy wetlands.
2 Materials and methods
2.1 Sampling area description
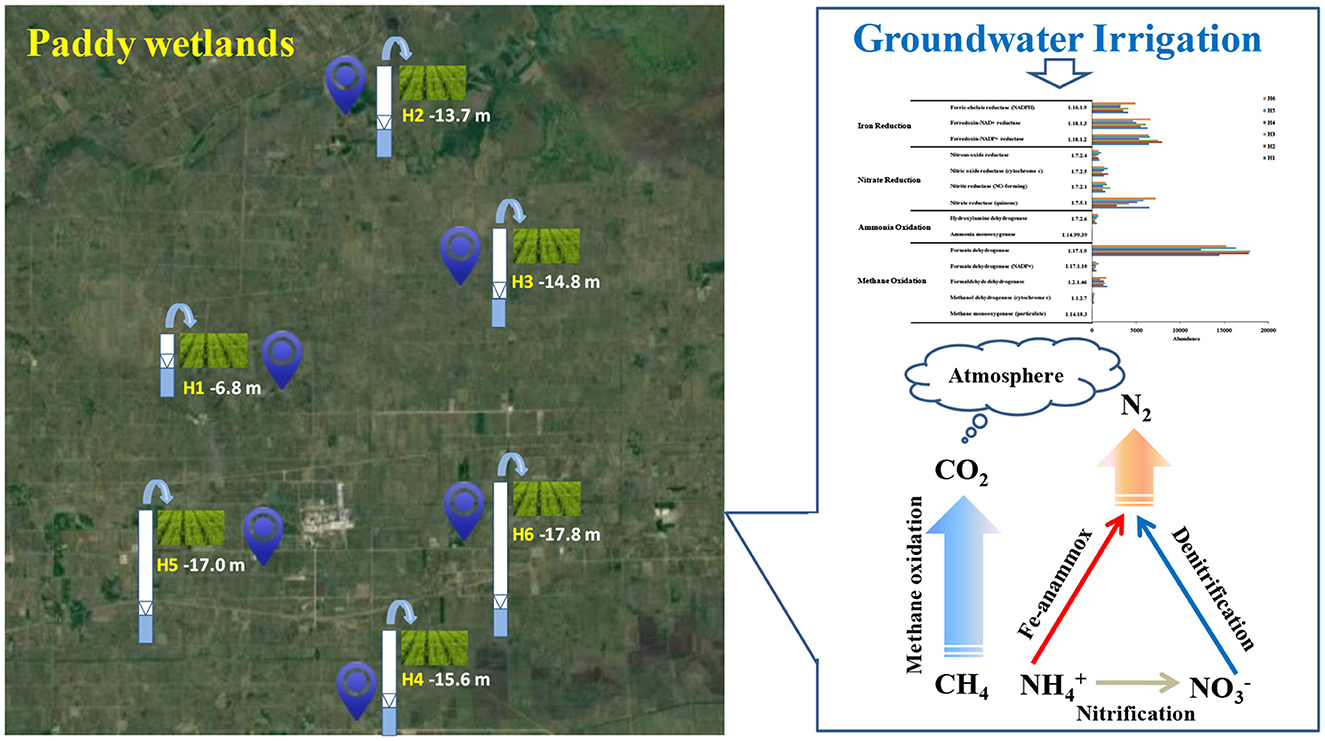
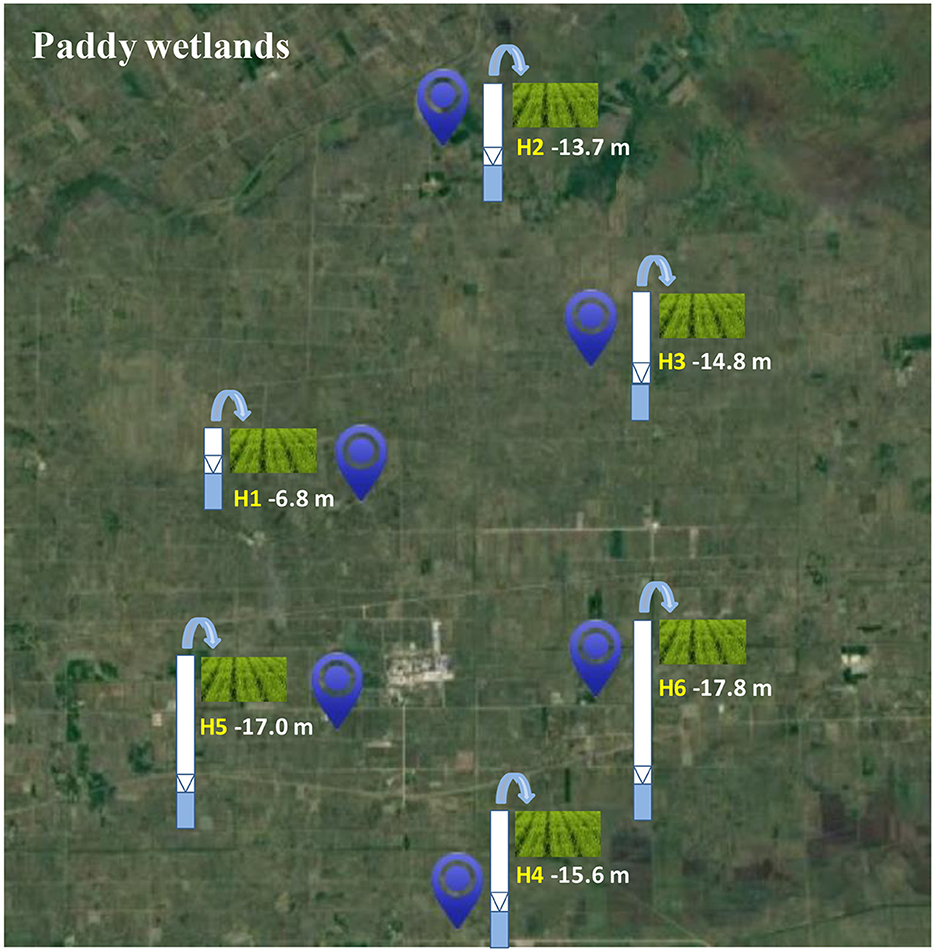
The research area was situated on Honghe Farm of Sanjiang Plain in the Heilongjiang Province, China, which belonged to the temperate- and continental-monsoon climate. The mean temperature and rainfall per year were 1.6°C−1.9°C and 565–600 mm, respectively. Precipitation was concentrated from June to August. Furthermore, the growing period of rice was from May to September, and the type of the soil was paddy soil. Urea was the main fertilizer with an average fertilization rate of 180 kg N/hm2. The paddy wetlands in this area were mainly assarted from swamps and dry land in the last few decades, which were under groundwater's irrigation. Therefore, paddy wetlands under groundwater's irrigation at different depths, such as H1 (−6.8 m), H2 (−13.7 m), H3 (−14.8 m), H4 (−15.6 m), H5 (−17.0 m), and H6 (−17.8 m), were selected to investigate the underlying influence of groundwater's irrigation on microbial community and function of paddy wetlands (Figure 1). Here, we defined shallow- and deep-groundwater as less and more than 10 m at depth.
2.2 Soil collecting and analysis
Top soil samples (0–15 cm) in paddy wetlands were collected in triplicgot and mixed into a composite sample at sampling sites H1 (47°37′39′′-N; 133°29′11′′-E), H2 (47°43′15′′-N; 133°31′6′′-E), H3 (47°39′47′′-N; 133°34′35′′-E), H4 (47°30′55′′-N; 133°31′27′′-E), H5 (47°34′4′′-N; 133°28′37′′-E), and H6 (47°34′35′′-N; 133°34′41′′-E) in May 2021. Six soil samples were obtained and named H1, H2, H3, H4, H5, and H6. All soil samples were placed in plastic bags and then sent to the nearby laboratory on ice packs. A part was air-dried for 3 weeks at room-temperature and then filtered using a 2-mm sifter in order to determinate soil properties. And the rest part was stored in a −80°C in order to analyse microbial community and function. Soil physicochemical factors were tested as previously described (Li et al., 2019).
2.3 Illumina sequence and statistical analyses
Total DNA from the six soil samples was extracted by means of PowerSoil-DNA-isolation kit (MoBio, Carlsbad, CA). Moreover, V3-V4 region in 16S rRNA gene was amplifed using primers 338F and 806R. PCR amplification was performed in a total volume of 20 μL (contained 4 μL Buffer, 0.4 μL FastPfu Polymerase, 2 μL dNTP, 5 μM of each primer and 10 ng genome DNA). Thermal cycling conditions were as follows: an initial denaturation at 95°C for 3 min, followed by 35 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 10 min. The PCR products were further performed based on Illumina-MiSeq-platform (Shanghai-Majorbio Bio-pharm Technology Co., Ltd., China). Processing of raw sequences obtained from Illumina sequencing was performed using QIIME software (version 1.9.1). We assembled paired-end reads using FLASH (version 1.2.11, https://ccb.jhu.edu/software/FLASH/index.shtml), where forward and reverse reads had overlapping base lengths ≥10 bp, and base mismatches were prevented. Reads with a quality score < 20, ambiguous bases, and improper primers were discarded before clustering. High-quality sequences of 252096 were acquired with 36934–48033 sequences for each soil sample. The obtained sequences were further clustered through USEARCH with 97% similarity as OTUs. And the taxonomy was verified by the Silva databases. To reduce the influence of sequencing depth on treatment effects, the samples were randomly resampled to the same sequence depth based on the least number of sequences. Correlation analysis (CA) was used to uncover the interrelation between crucial phyla, function gene, and soil property. Principal coordinate analysis (PCoA) was utilized to identify microbial community difference. Redundancy analysis was performed to investigate the key factors shaping microbial community and function. Shannon index, functional gene, and metabolic pathway were obtained using Quantitative Insights into Microbial Ecology (QIIME) and Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (Chen et al., 2016; Guo et al., 2022).
3 Results and discussion
3.1 Soil characteristics
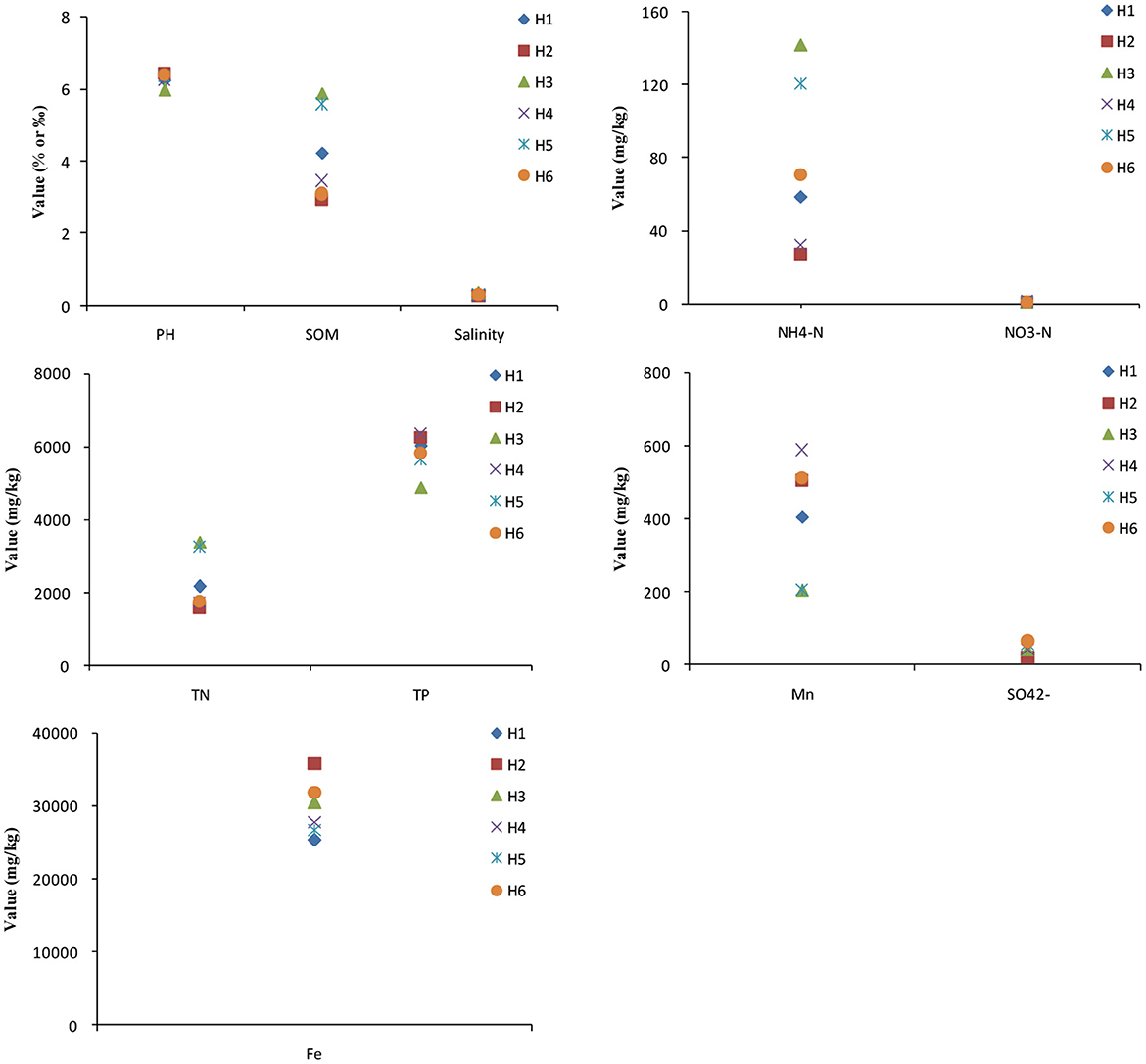
The soil of paddy wetlands was weakly acidic and had low salinity for groundwater's irrigation at various depths, whereas the contents of SOM, TN, and TP were relatively high in the ranges of 2.93–5.90%, 1571.46–3397.90 mg/kg, and 4892.93–6347.79 mg/kg, respectively (Figure 2). The level of -N was high and that of -N was low. Fe, Mn and were abundant in paddy soil. In particular, SOM, -N, and TN had high levels in H3 and H5, indicating their good nutrient condition that was conducive to paddy growth and material transformation. Meanwhile, the low levels in H2 and H4 may be related to their soil structure and properties. The low -N level was consistent with high SOM amount, suggesting that a high SOM content in paddy soil could promote denitrification (Li et al., 2019). Mn had high levels in H2, H4, and H6; Fe was significantly enriched in H2, H3, and H6; and was high in H6. These findings implied that deep-groundwater's irrigation was beneficial to the enrichment of multiple electron acceptors. CA showed that salinity had a positive correlation with -N, and TN levels (p < 0.05; Supplementary Table 1), indicating that high salinity had inhibitory effects on denitrification (Wang et al., 2018a). SOM had a positive correlation with TN (p < 0.01), suggesting that SOM accumulating was beneficial to TN enriching. Mn was negatively correlated with SOM, -N, and TN, indicating that Mn was mainly affected by groundwater irrigation. No significant correlation was found among Fe, , and other soil properties. In summary, SOM, TN, and TP were abundant in paddy soil; some differences were observed among the paddy soil samples for groundwater's irrigation at various depths; and deep-groundwater's irrigation could promote the accumulation of various electron acceptors.
3.2 Microbial community composition and difference
The community of microorganisms under phylum- and genus- levels was shown in Figure 3. Acidobacteriota, Desulfobacterota, Actinobacteriota, Myxococcota, Bacteroidota, Firmicutes, Chloroflexi, Nitrospirota, Planctomycetota, and Proteobacteria were the important phyla with over 97% bacterial abundance (Figure 3A). Among them, Actinobacteriota, Chloroflexi, and Firmicutes had a correlation with organics degradation (He et al., 2018; Chi et al., 2021b), and Acidobacteriota, Bacteroidota, Nitrospirota, Planctomycetota, and Proteobacteria were connected with nitrogen cycling (Chi et al., 2021e). Desulfobacterota and Myxococcota were involved in sulfur and iron cycle (Langwig et al., 2022). Bacteroidota and Firmicutes were the most abundant in H1, and Acidobacteriota and Proteobacteria had the highest abundance in H2. The abundance of Acidobacteriota was lower in H1 than in H2-H6; this species played key roles in denitrification (Zhang et al., 2020). In addition, low abundance of Nitrospiota was consistent with the relatively anoxic environment of paddy wetlands. High abundance of Proteobacteria in paddy wetlands was relating to its diverse substrate-utilizing and environmental-adaptability (Bernhard et al., 2005). High abundances of Actinobacteriota and Chloroflexi were consistent with high SOM amount in the soil. Therefore, the phyla related to carbon and nitrogen cycling were widespread and abundant in paddy wetlands, and some phyla had high abundance under deep groundwater irrigation.
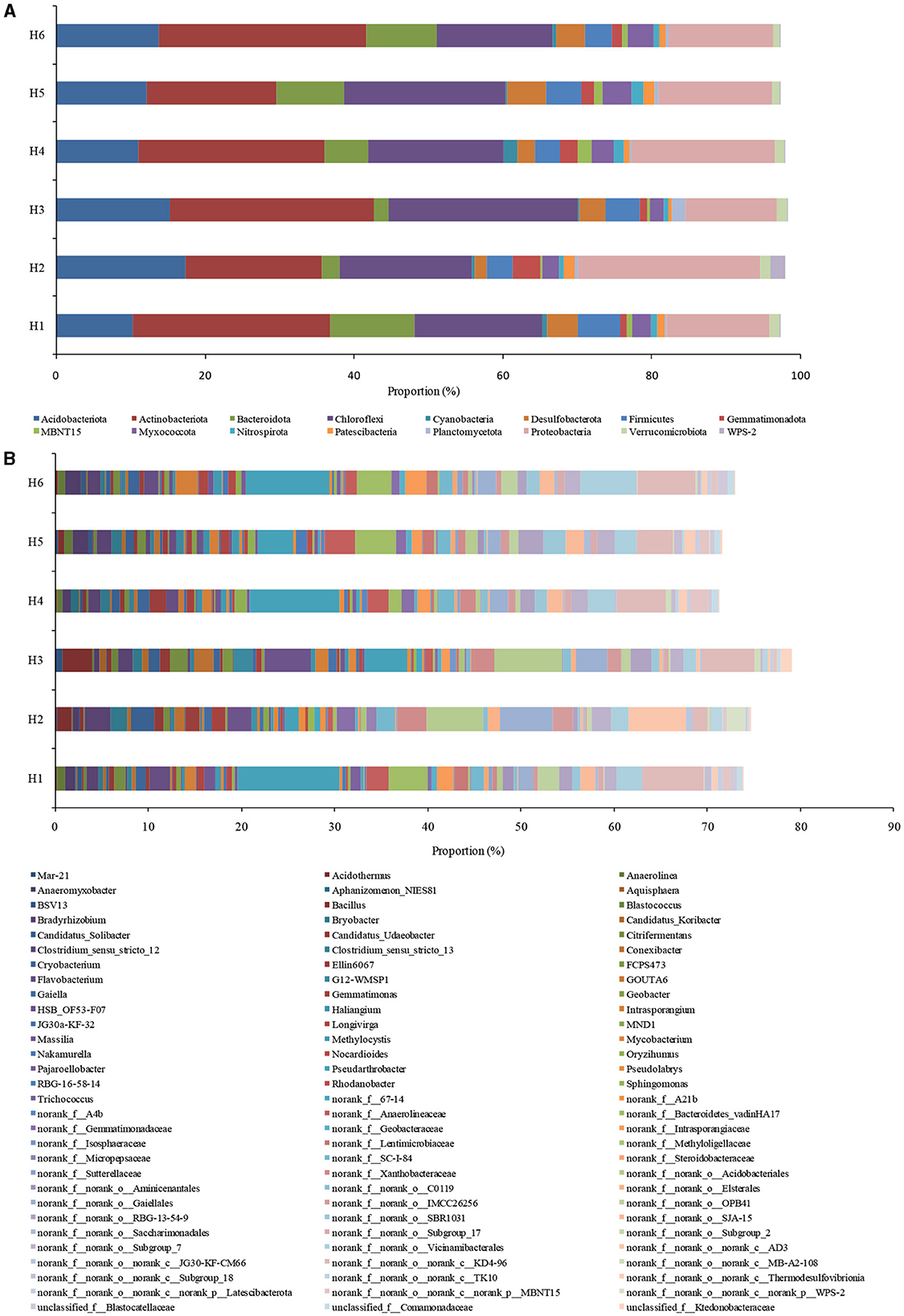
Figure 3. Taxonomic classification of bacterial community at (A) phylum and (B) genus levels. Less than 0.5% was not included.
The dominant genera were MND1, Sphingomonas, Ellin6067, Flavobacterium, Pseudolabrys, Rhodanobacter, Gemmatimonas, Anaeromyxobacter, Bacillus, Geobacter, Citrifermentans, Methylocystis, norank_f_Methyloligellaceae, Pseudarthrobacter, and Conexibacter (Figure 3B). The abundances of nitrifying bacteria MND1 and Sphingomonas were relatively low, although -N level was high in paddy soil; the discrepancy may be related to the relatively anoxic environment of paddy wetlands (Tolar et al., 2020; Gao et al., 2021). High abundance of nitrifying bacteria Ellin6067 was related to its diverse metabolic capabilities (such as nitrification and organic matter degradation) (Lezcano et al., 2017; Li et al., 2021b), and Ellin6067 was abundant in deep groundwater irrigation. In addition, aerobic Candidatus_Udaeobacter was widely distributed in paddy soil but had low abundance, further indicating the relatively anoxic environment of paddy wetlands (Li et al., 2021a). The denitrifying bacteria Flavobacterium was widely distributed in paddy wetlands, with the highest abundance found under shallow groundwater irrigation (Pishgar et al., 2020). Meanwhile, Pseudolabrys, Rhodanobacter, and Gemmatimonas were abundant under deep groundwater irrigation (Van Den Heuvel et al., 2010; Green et al., 2012; Yan et al., 2019), indicating the diversity of denitrifying bacteria. As an aerobic denitrifier, Gemmatimonas could reduce N2O to N2 and decrease greenhouse gas emissions (Park et al., 2017). The high presence of denitrifying bacteria was consistent with the high SOM amount in the soil. The organic-degrading bacteria Pseudarthrobacter was abundant in all the samples, with the highest abundance found under shallow groundwater irrigation. Conexibacter had high abundance under deep groundwater irrigation. Pseudarthrobacter showed the organic matter degradation potential at low temperature (Zhang et al., 2016), and Conexibacter could decompose refractory organic matter (Liang et al., 2021). Iron-reducing bacteria Geobacter was abundant under shallow groundwater irrigation, and Anaeromyxobacter, Bacillus, and Citrifermentans were dominant under deep groundwater irrigation; all these genera had Fe-anammox potential (Chi et al., 2021a; Ottoni et al., 2022). Studies have shown that Anaeromyxobacter and Geobacter could participate in Fe-anammox (Zhou et al., 2016). Furthermore, Geobacter had dissimilar nitrate reduction potential and could drive dissimilatory nitrate reduction to ammonia (DNRA) using divalent iron, thus providing substrate for anammox (Finneran et al., 2002). Excitedly, Intrasporangium had chromium-reducing potential and was widely distributed in paddy wetlands (Yang et al., 2009). norank_f_Gemmatimonadaceae was abundant under deep groundwater irrigation and had good nitrate- and vanadium- reduction ability (Jia et al., 2019; Fei et al., 2022). The existence of these species could promote anammox. The methanotrophs Methylocystis and norank_f_Methyloligellaceae were widely distributed with low abundance, possibly due to the relatively anoxic environment of paddy wetlands. The presence of aerobic methane oxidation was beneficial to in-situ reduction of methane in paddy wetlands. Moreover, HSB_OF53-F07 had nitrogen metabolism ability and diverse metabolic functions under aerobic/anoxic conditions (Wu et al., 2021). Bradyrhizobium with high abundance had nitrogen fixation function and could promote rice growth (Kaneko et al., 2002). Candidatus_Solibacter had high abundance under deep groundwater irrigation, which was consistent with the results in paddy soils under mine drainage irrigation and long-term fertilization (Wang et al., 2018b; Yu et al., 2019). Candidatus_Solibacter could degrade complex pollutants and outperform the competition in environmental filtration (Ward et al., 2009). Cryobacterium and Trichococcus had low temperature adaptations and may play important roles in material cycling (Pikuta et al., 2006; Teoh et al., 2021). Therefore, groundwater irrigation was beneficial to organics degradation, denitrification, and Fe-anammox. Meanwhile, nitrification and aerobic methane oxidation in paddy wetlands were relatively weak. Shallow groundwater irrigation increased organics degradation, but deep groundwater irrigation promoted the occurrence of Fe-anammox.
PCoA suggested that the microbial communities had minimal differences under shallow and deep groundwater irrigation, but significantly differed with H2 and H3, which was related to high the SOM, -N, TN, and Fe levels (Figure 4A). The abundance of Pseudarthrobacter under groundwater irrigation (above 15 m) was higher than that in other irrigation zones (below 15 m) (p > 0.05) (Figure 4B). Meanwhile Bradyrhizobium and HSB_OF53-F07 had high abundances in irrigation zones (below 15 m) (p > 0.05), which was consistent with high abundance of Bradyrhizobium and low count of HSB_OF53-F07 and Pseudarthrobacter in H2 and H3.
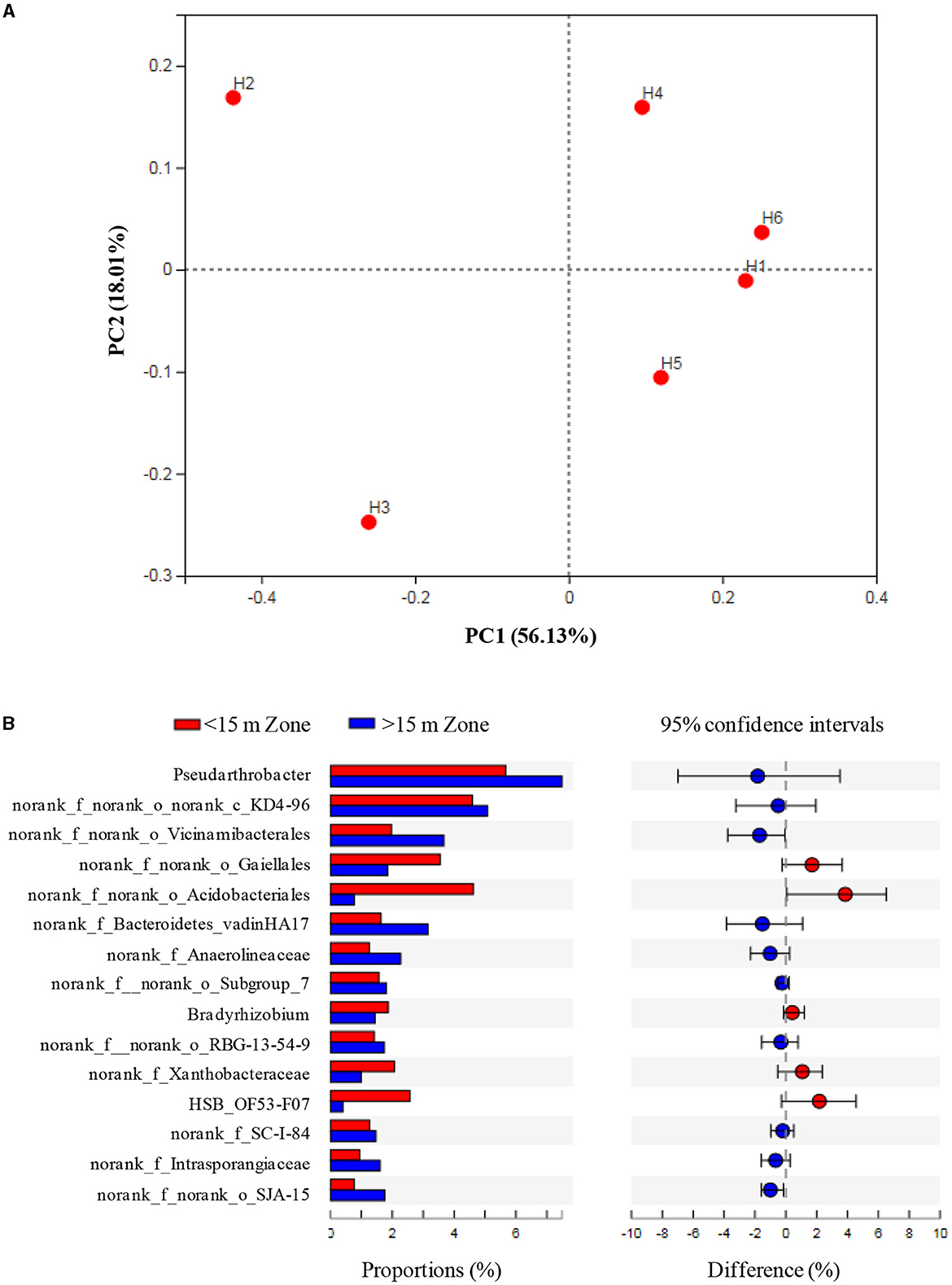
Figure 4. Principal coordinate analysis (PCoA) (A) and species differences (B) at different sampling zones.
3.3 Key variables influencing the community of microorganisms
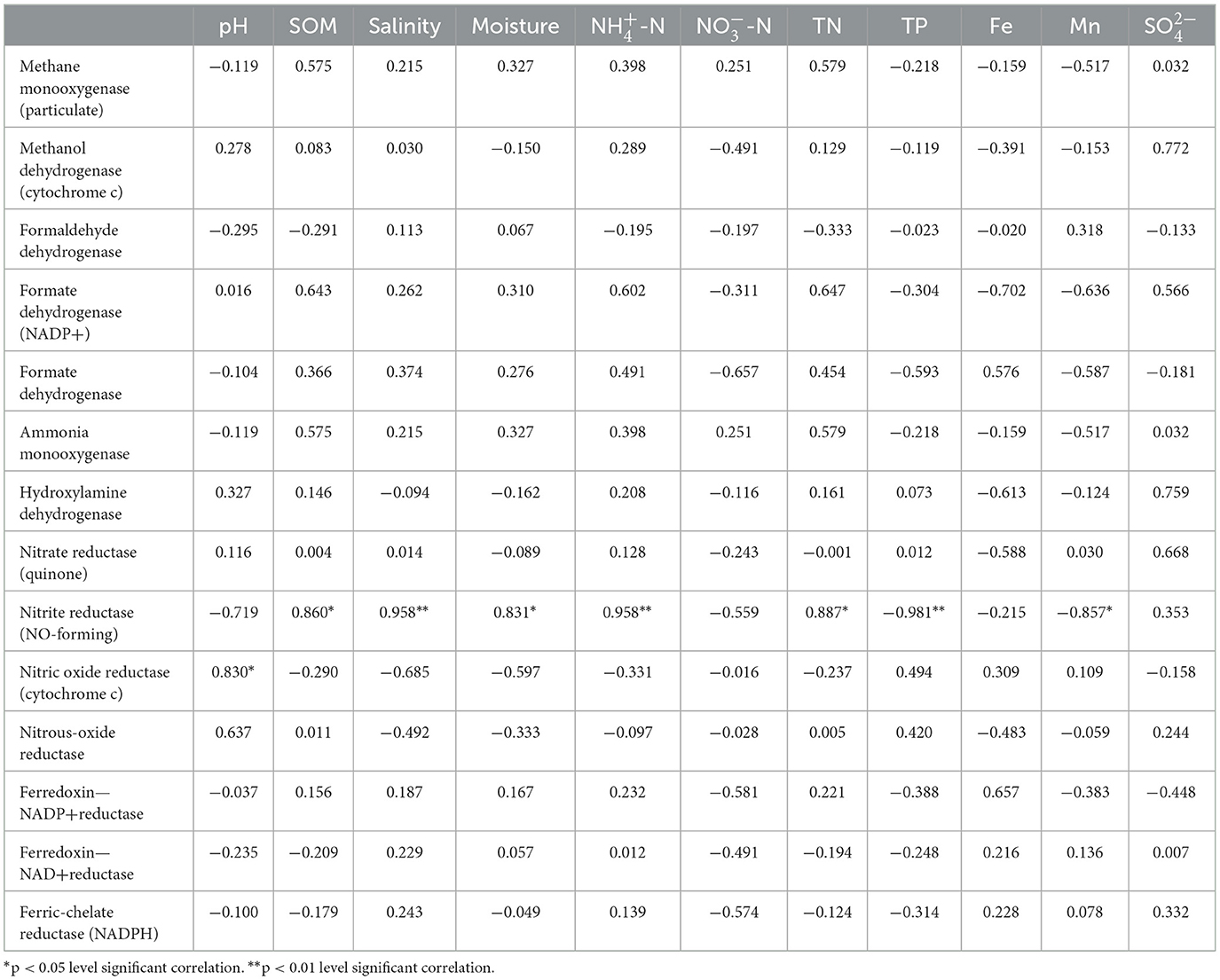
The Shannon's index was lower in shallow-groundwater's irrigation than in deep-groundwater's irrigation, indicating that deep-groundwater's irrigation with multiple electron acceptors was beneficial to the improvement of microbial diversity (Supplementary Figure 1). CA suggested that Acidobacteria was positively correlated with Fe (p < 0.01; Supplementary Table 2), expressing that Fe accumulation could promote Acidobacteria enrichment and facilitate denitrification. Chloroflexi had a positive correlation with SOM, salinity, moisture, -N, and TN, but negative correlation with TP, and Mn (p < 0.05), suggesting that SOM and TN accumulation was beneficial to Chloroflexi enrichment. Planctomycetes showed a significant positive correlation with salinity (p < 0.05). A previous study found that anammox bacteria Scalindua had favorable salinity adaptation (Zheng et al., 2016). RDA showed that moisture, pH, and TP were the major controlling factors at phylum level, and -N and multi-electron acceptors had significant effects at genus level (Figures 5A, B). Similarly, previous studies found that pH and moisture had crucial effects on the microbial community (Banning et al., 2011; Pan et al., 2021), and nutrients also could alter microbial composition (Francioli et al., 2018). Thus, moisture, pH, and TP had crucial effects in shaping microbial community under phylum- and genus-levels.
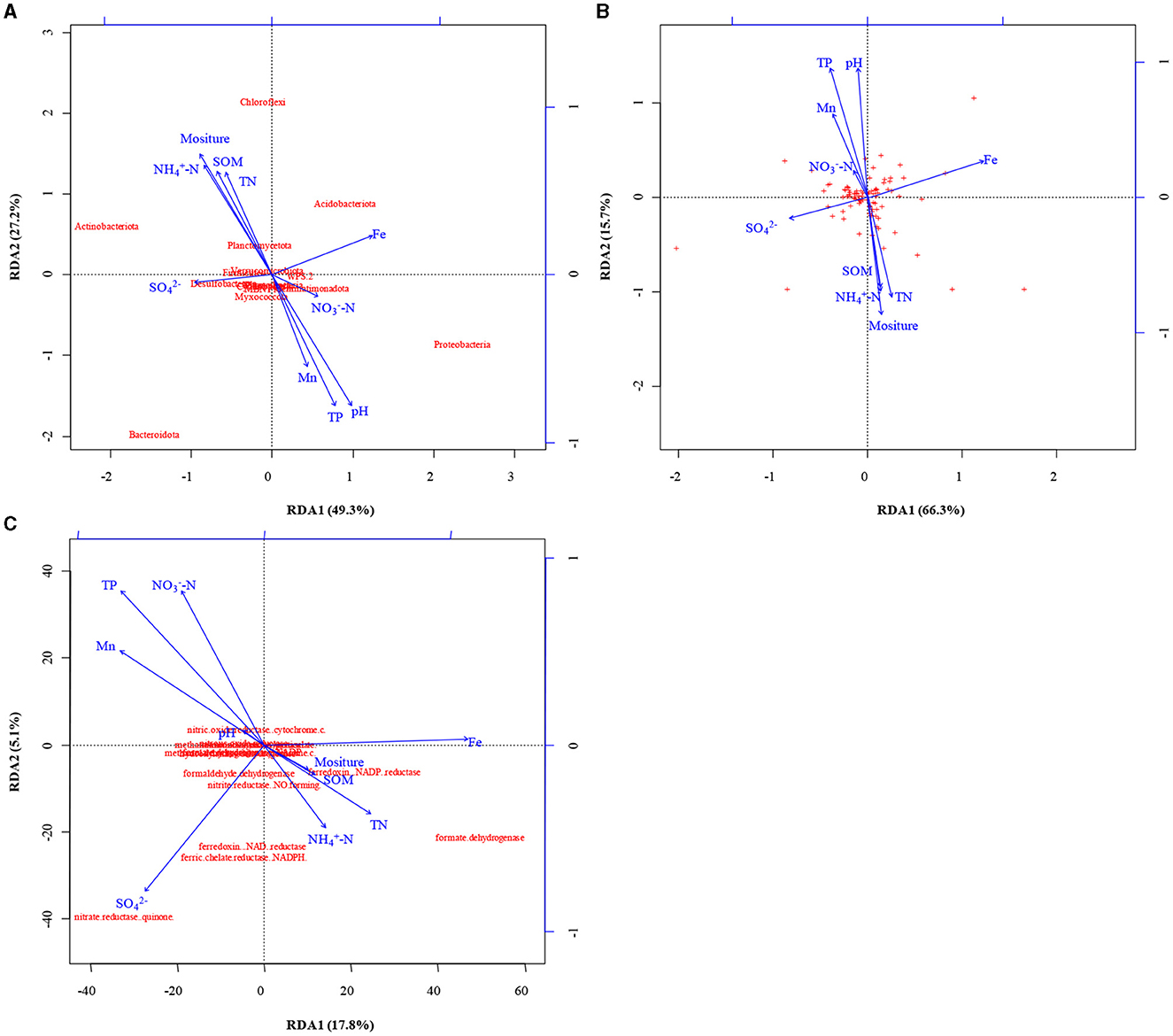
Figure 5. The results of redundancy analysis (RDA) between soil properties, microbial community at phyla (A) and genus (B) level, and functional genes (C).
3.4 Function prediction of microorganisms
Microbial functions were predicted by PICRUSt (Figure 6). Microorganisms were present in different metabolic pathways, such as metabolism, gene information's processing, environmental information's processing, and cellular processes (Li et al., 2018; Chi et al., 2021c); among which, their abundance in metabolism was the highest (Figure 6A). In particular, the genes involved in cell motility and growth/death were abundant, indicating the high microbial activity in paddy wetlands. High abundance of genes encoding carbohydrates, amino acids, and energy metabolism suggested the good material and energy metabolism of microorganisms. The gene encoding energy metabolism were abundant in H4-H6, and this finding was possibly related to the abundant supply of electron acceptors under deep groundwater irrigation. The expression of genes encoding xenobiotic biodegradation and terpenoids/polyketide metabolism was observed, indicating that microorganisms showed potential to metabolize exogenous substances. The genes related to signal transduction and membrane transport were highly abundant possibly due to the high nutrient levels in paddy wetlands. The repair-related gene was more expressed under shallow groundwater irrigation, indicating that microorganisms were alive to environmental variations. This findings were consistent with the low biodiversity in paddy wetlands.
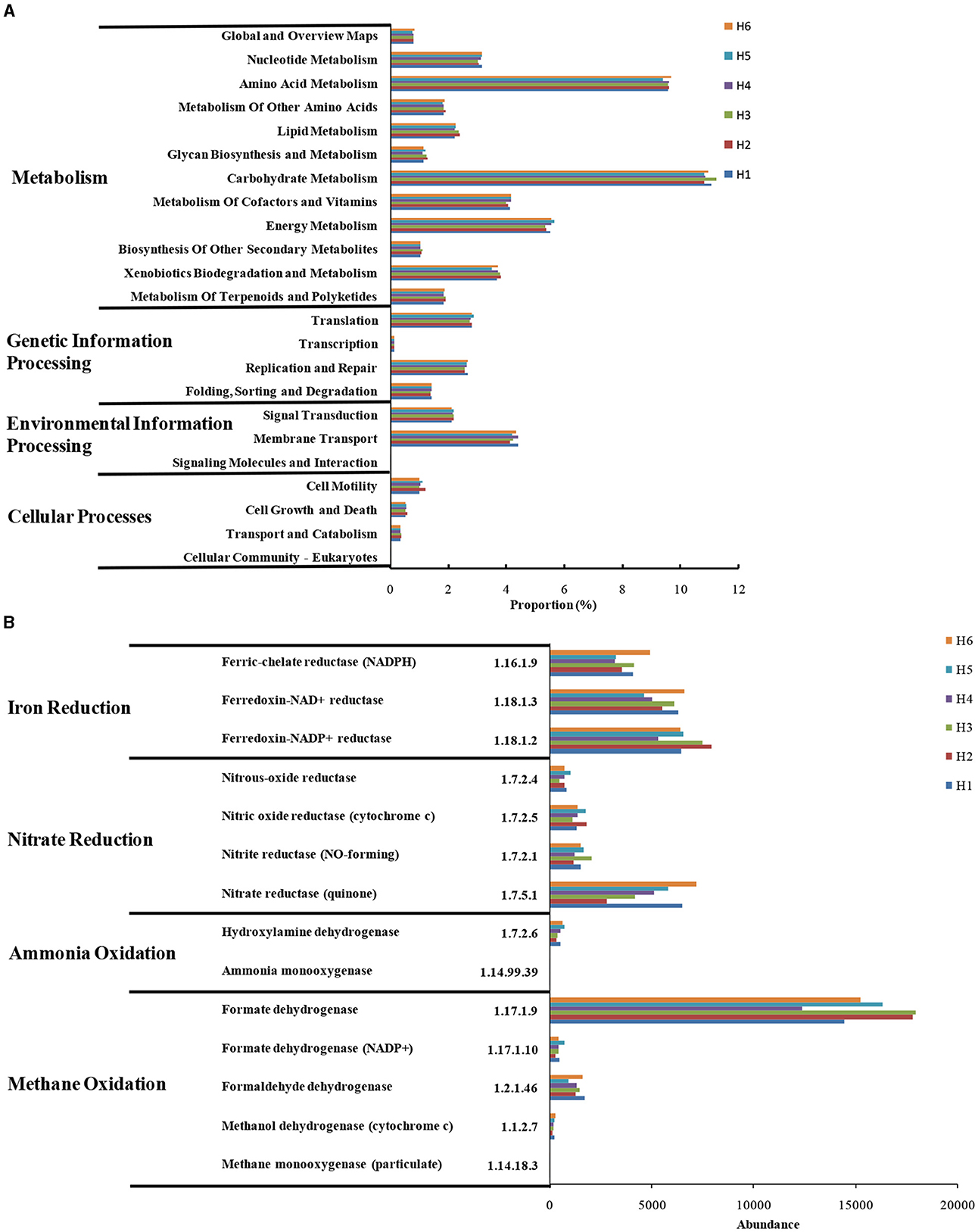
Figure 6. KEGG-predicted metabolic pathways (A) and functional genes (B) at different sampling sites.
These genes with regard to nitrogen and methane metabolism were analyzed as shown in Figure 6B. The nitrifying-related genes [EC:1.14.99.39 and EC:1.7.2.6] were abundant and highly expressed under deep groundwater irrigation, indicating that nitrification occurred in paddy wetlands. The nitrate-reductase gene [EC:1.7.5.1], which could promote denitrification and DNRA, was the most abundant (Chi et al., 2021f). High level of nitrite-reductase gene [EC:1.7.2.1] was beneficial to denitrification and anammox (Chi et al., 2021d). The abundance of denitrifying-related genes coincided with widespread denitrifying bacteria and low -N levels. Moreover, the genes related to iron reduction were highly expressed and highly abundant under deep groundwater irrigation. This finding indicated the existence of Fe-anammox in paddy wetlands, which was consistent with the enrichment of Fe-anammox bacteria. The genes encoding methane oxidation were abundant, especially formate dehydrogenase [EC:1.17.1.9], indicating the widespread occurrence of methane oxidation. In particular, the abundance of formate dehydrogenase [EC:1.17.1.9] was more abundant under deep groundwater's irrigation than that under shallow groundwater's irrigation, indicating that deep groundwater's irrigation favored its enrichment. The low level of methane-monooxygenase [EC:1.14.18.3] was consistent with the relatively anoxic environment of paddy wetlands. The complete methane oxidation-related genes was related to the widespread distribution of methanotrophs. Thus, deep groundwater irrigation could promote Fe-anammox, nitrification, and methane oxidation. Meanwhile, denitrification was ubiquitous under groundwater irrigation.
RDA indicated that electron acceptors were the main drivers of functional gene changes, and TN and TP had some effects (Figure 5C). Previous studies reported that nutrients expressed important effects on the functional genes (Chi et al., 2021b), and electron acceptors had a direct role (Li et al., 2022). CA showed that nitrite reductase (NO-forming) had a positive correlation with SOM, salinity, moisture, -N, and TN (p < 0.01 or p < 0.05) but negative correlation with TP, and Mn (p < 0.01 or p < 0.05; Table 1), indicating that SOM and TN accumulation was favorable for denitrification. Furthermore, nitrite reductase (NO-forming) showed a positive correlation with -N (p < 0.01), suggesting the possibility of anammox (Li et al., 2019). In addition, -N had a positive correlation with ammonia monooxygenase, and hydroxylamine dehydrogenase (p > 0.05), implying that -N accumulation could promote nitrification. The genes with respect to iron reduction [ferredoxin–NADP+ reductase, ferredoxin–NAD+ reductase, and ferric-chelate reductase (NADPH)] were positively correlated with -N, and Fe (p > 0.05), indicating the existence of Fe-anammox of paddy wetlands.
4 Conclusions
This study illustrated the potential effects of groundwater's irrigation on the community and function of microorganism in paddy wetlands. Deep-groundwater's irrigation favored the accumulation of multi-electron acceptors. Groundwater irrigation was beneficial to organics degradation, denitrification, and Fe-anammox, but nitrification and aerobic methane oxidation were relatively weak in paddy wetlands. Moisture, pH, and TP played vital roles for microbial communities shaping. Microorganisms were highly sensitive to environmental changes under shallow-groundwater's irrigation, and deep-groundwater's irrigation favored Fe-anammox, nitrification, and methane oxidation. Denitrification was widespread under groundwater's irrigation. The findings offer new ideas for non-point source pollution control and greenhouse gas reduction in paddy wetlands.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
HL: Writing—original draft. AS: Writing—review & editing. LQ: Writing—review & editing. SL: Writing—review & editing. ZC: Conceptualization, Funding acquisition, Supervision, Writing—review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Key R&D Program of China (Nos. 2023YFC3207704; 2022YFF1300901), Natural Science Foundation of Jilin Province (No. 20230101100JC), and National Natural Science Foundation of China (No. 42077353).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1354279/full#supplementary-material
References
Banning, N. C., Gleeson, D. B., Grigg, A. H., Grant, C. D., Andersen, G. L., Brodie, E. L., et al. (2011). Soil microbial community successional patterns during forest ecosystem restoration. Appl. Environ. Microbiol. 77, 6158–6164. doi: 10.1128/AEM.00764-11
Bernhard, A. E., Colbert, D., McManus, J., and Field, K. G. (2005). Microbial community dynamics based on 16S rRNA gene profiles in a Pacific Northwest estuary and its tributaries. FEMS Microbiol. Ecol. 52, 115–128. doi: 10.1016/j.femsec.2004.10.016
Cao, Y., Tang, C., Song, X., Liu, C., and Zhang, Y. (2016). Identifying the hydrochemical characteristics of rivers and groundwater by multivariate statistical analysis in the Sanjiang Plain, China. Appl. Water Sci. 6, 169–178. doi: 10.1007/s13201-014-0215-5
Chen, C., Sun, F., Zhang, H., Wang, J., Shen, Y., and Liang, X. (2016). Evaluation of COD effect on anammox process and microbial communities in the anaerobic baffled reactor (ABR). Bioresource Technol. 216, 571–578. doi: 10.1016/j.biortech.2016.05.115
Chen, X., Sheng, Y., and Wang, G. (2024). Spatiotemporal successions of N, S, C, Fe, and As cycling genes in groundwater of a wetland ecosystem: enhanced heterogeneity in wet season. Water Res. 251, 121105. doi: 10.1016/j.watres.2024.121105
Chi, Z., Hou, L., and Li, H. (2021a). Effects of pollution load and salinity shock on nitrogen removal and bacterial community in two-stage vertical flow constructed wetlands. Bioresource Technol. 342, 126031. doi: 10.1016/j.biortech.2021.126031
Chi, Z., Hou, L., Li, H., Li, J., Wu, H., and Yan, B. (2021c). Elucidating the archaeal community and functional potential in two typical coastal wetlands with different stress patterns. J. Clean. Prod. 285, 124894. doi: 10.1016/j.jclepro.2020.124894
Chi, Z., Hou, L., Li, H., Wu, H., and Yan, B. (2021b). Indigenous bacterial community and function in phenanthrene-polluted coastal wetlands: potential for phenanthrene degradation and relation with soil properties. Environ. Res. 199, 111357. doi: 10.1016/j.envres.2021.111357
Chi, Z., Ju, S., Li, H., Li, J., Wu, H., and Yan, B. (2021d). Deciphering edaphic bacterial community and function potential in a Chinese delta under exogenous nutrient input and salinity stress. Catena 201, 105212. doi: 10.1016/j.catena.2021.105212
Chi, Z., Wang, W., Li, H., Wu, H., and Yan, B. (2021e). Soil organic matter and salinity as critical factors affecting the bacterial community and function of Phragmites australis dominated riparian and coastal wetlands. Sci. Total Environ.762, 143156. doi: 10.1016/j.scitotenv.2020.143156
Chi, Z., Zhu, Y., Li, H., Wu, H., and Yan, B. (2021f). Unraveling bacterial community structure and function and their links with natural salinity gradient in the Yellow River Delta. Sci. Total Environ. 773, 145673. doi: 10.1016/j.scitotenv.2021.145673
Chong, L., Liu, H. J., Qiang, F. U., Guan, H. F., Ye, Q., Zhang, X., et al. (2020). Mapping the fallowed area of paddy fields on Sanjiang Plain of Northeast China to assist water security assessments. J. Integr. Agric. 19, 1885–1896. doi: 10.1016/S2095-3119(19)62871-6
Ding, B., Zhang, H., Luo, W., Sun, S., Cheng, F., and Li, Z. (2021). Nitrogen loss through denitrification, anammox and Feammox in a paddy soil. Sci. Total Environ. 773, 145601. doi: 10.1016/j.scitotenv.2021.145601
Fei, Y., Zhang, B., He, J., Chen, C., and Liu, H. (2022). Dynamics of vertical vanadium migration in soil and interactions with indigenous microorganisms adjacent to tailing reservoir. J. Hazard. Mater. 424, 127608. doi: 10.1016/j.jhazmat.2021.127608
Finneran, K. T., Housewright, M. E., and Lovley, D. R. (2002). Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ. Microbiol. 4, 510–516. doi: 10.1046/j.1462-2920.2002.00317.x
Francioli, D., Schulz, E., Buscot, F., and Reitz, T. (2018). Dynamics of soil bacterial communities over a vegetation season relate to both soil nutrient status and plant growth phenology. Microb. Ecol. 75, 216–227. doi: 10.1007/s00248-017-1012-0
Gao, F., Liu, G., She, Z., Ji, J., Gao, M., Zhao, Y., et al. (2021). Effects of salinity on pollutant removal and bacterial community in a partially saturated vertical flow constructed wetland. Bioresour. Technol. 329, 124890. doi: 10.1016/j.biortech.2021.124890
Green, S. J., Prakash, O., Jasrotia, P., Overholt, W. A., Cardenas, E., Hubbard, D., et al. (2012). Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Appl. Environ. Microb. 78, 1039–1047. doi: 10.1128/AEM.06435-11
Guo, M., Jiang, Y., Xie, J., Cao, Q., Zhang, Q., Mabruk, A., et al. (2022). Bamboo charcoal addition enhanced the nitrogen removal of anammox granular sludge with COD: performance, physicochemical characteristics and microbial community. J. Environ. Sci. 115, 55–64. doi: 10.1016/j.jes.2021.07.010
Guo, X., Zhou, X., Hale, L., Yuan, M., Ning, D, Feng, J., et al. (2019). Climate warming accelerates temporal scaling of grassland soil microbial biodiversity. Nat. Ecol. Evol. 3, 612–619. doi: 10.1038/s41559-019-0848-8
Guo, Z., Wan, S., Hua, K., Yin, Y., Chu, H., Wang, D., et al. (2020). Fertilization regime has a greater effect on soil microbial community structure than crop rotation and growth stage in an agroecosystem. Appl. Soil Ecol. 149, 103510. doi: 10.1016/j.apsoil.2020.103510
He, Z. W., Liu, W. Z., Gao, Q., Tang, C., Wang, L., Guo, Z., et al. (2018). Potassium ferrate addition as an alternative pre-treatment to enhance shortchain fatty acids production from waste activated sludge. Bioresource Technol. 247,174–181. doi: 10.1016/j.biortech.2017.09.073
Hou, Q., Lin, S., Ni, Y., Huang, S., Zuo, T., Wang, J., et al. (2022). Assembly of functional microbial communities in paddy soil with long-term application of pig manure under rice-rape cropping system. J. Environ. Manage. 305, 114374. doi: 10.1016/j.jenvman.2021.114374
Huang, Q., Wang, J., Wang, C., and Wang, Q. (2019). The 19-years inorganic fertilization increased bacterial diversity and altered bacterial community composition and potential functions in a paddy soil. Appl. Soil Ecol. 144, 60–67. doi: 10.1016/j.apsoil.2019.07.009
Jia, L., Jiang, B., Huang, F., and Hu, X. (2019). Nitrogen removal mechanism and microbial community changes of bioaugmentation subsurface wastewater infiltration system. Bioresource Technol. 294, 122140. doi: 10.1016/j.biortech.2019.122140
Jiang, Z., Yang, S., Pang, Q., Xu, Y., Chen, X., Sun, X., et al. (2021). Biochar improved soil health and mitigated greenhouse gas emission from controlled irrigation paddy field: insights into microbial diversity. J. Clean. Prod. 318, 128595. doi: 10.1016/j.jclepro.2021.128595
Jin, W., Cao, W., Liang, F., Wen, Y., Wang, F., Dong, Z., et al. (2020). Water management impact on denitrifier community and denitrification activity in a paddy soil at different growth stages of rice. Agric. Water Manage. 241, 106354. doi: 10.1016/j.agwat.2020.106354
Kaneko, T., Nakamura, Y., Sato, S., Minamisawa, K., Uchiumi, T., Sasamoto, S., et al. (2002). Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9, 189–197. doi: 10.1093/dnares/9.6.189
Langwig, M. V., De Anda, V., Dombrowski, N., Seitz, K. W., Rambo, I. A., Greening, C., et al. (2022). Large-scale protein level comparison of Deltaproteobacteria reveals cohesive metabolic groups. ISME J. 16, 307–320. doi: 10.1038/s41396-021-01057-y
Lezcano, M. Á., Velázquez, D., Quesada, A., and El-Shehawy, R. (2017). Diversity and temporal shifts of the bacterial community associated with a toxic cyanobacterial bloom: an interplay between microcystin producers and degraders. Water Res. 125, 52–61. doi: 10.1016/j.watres.2017.08.025
Li, H., Chi, Z., Li, J., Wu, H., and Yan, B. (2019). Bacterial community structure and function in soils from tidal freshwater wetlands in a Chinese delta: potential impacts of salinity and nutrient. Sci. Total Environ. 696, 134029. doi: 10.1016/j.scitotenv.2019.134029
Li, H., Chi, Z., Lu, W., and Wang, H. (2014). Sensitivity of methanotrophic community structure, abundance, and gene expression to CH4 and O2 in simulated landfill biocover soil. Environ. Pollut. 184, 347–353. doi: 10.1016/j.envpol.2013.09.002
Li, H., Chi, Z., and Yan, B. (2018). Insight into the impact of Fe3O4 nanoparticles on anammox process of subsurface-flow constructed wetlands under long-term exposure. Environ. Sci. Pollut. R 25, 29584–29592. doi: 10.1007/s11356-018-2975-1
Li, H., Liang, S., Chi, Z., Wu, H., and Yan, B. (2022). Unveiling microbial community and function involved in anammox in paddy vadose under groundwater irrigation. Sci. Total Environ. 849, 157876. doi: 10.1016/j.scitotenv.2022.157876
Li, Q., Song, A., Yang, H., and Muller, W. E. G. (2021a). Impact of rocky desertification control on soil bacterial community in Karst Graben Basin, southwestern China. Front. Microbiol. 12, 448. doi: 10.3389/fmicb.2021.636405
Li, X., Lu, Y., Chen, Y., Zhu, G., and Zeng, R. J. (2021b). Constraining nitrification by intermittent aeration to achieve methane-driven ammonia recovery of the mainstream anaerobic effluent. J. Environ. Manage. 295, 113103. doi: 10.1016/j.jenvman.2021.113103
Liang, J., Gao, S., Wu, Z., Rijnaarts, H. H. M., and Grotenhuis, T. (2021). DNA-SIP identification of phenanthrene-degrading bacteria undergoing bioaugmentation and natural attenuation in petroleum-contaminated soil. Chemosphere 266, 128984. doi: 10.1016/j.chemosphere.2020.128984
Liu, Q., Atere, C. T., Zhu, Z., Shahbaz, M., Wei, X., Pausch, J., et al. (2022). Vertical and horizontal shifts in the microbial community structure of paddy soil under long-term fertilization regimes. Appl. Soil Ecol. 169, 104248. doi: 10.1016/j.apsoil.2021.104248
Meng, L., Zuo, R., Wang, J. S., Li, Q., Du, C., Liu, X., et al. (2021). Response of the redox species and indigenous microbial community to seasonal groundwater fluctuation from a typical riverbank filtration site in Northeast China. Ecol. Eng. 159, 106099. doi: 10.1016/j.ecoleng.2020.106099
Ottoni, J. R., dos Santos Grignet, R., Barros, M. G. A., Bernal, S. P. F., Panatta, A. A. S., Lacerda-Júnior, G. V., et al. (2022). DNA metabarcoding from microbial communities recovered from stream and its potential for bioremediation processes. Curr. Microbiol. 79, 1–10. doi: 10.1007/s00284-021-02752-x
Pan, X., Li, H., Zhao, L., Yang, X., Su, J., Dai, S., et al. (2021). Response of syntrophic bacterial and methanogenic archaeal communities in paddy soil to soil type and phenological period of rice growth. J. Clean. Prod. 278, 123418. doi: 10.1016/j.jclepro.2020.123418
Park, D., Kim, H., and Yoon, S. (2017). Nitrous oxide reduction by an obligate aerobic bacterium, Gemmatimonas aurantiaca strain T-27. Appl. Environ. Microb. 83, e00502–e00517. doi: 10.1128/AEM.00502-17
Pérez Castro, S., Cleland, E. E., Wagner, R., Al Sawad, R., and Lipson, D. A. (2019). Soil microbial responses to drought and exotic plants shift carbon metabolism. ISME J. 13, 1776–1787. doi: 10.1038/s41396-019-0389-9
Pikuta, E. V., Hoover, R. B., Bej, A. K., Marsic, D., Whitman, W. B., Krader, P. E., et al. (2006). Trichococcus patagoniensis sp. nov., a facultative anaerobe that grows at−5°C, isolated from penguin guano in Chilean Patagonia. Int. J. Syst. Evol. Microbiol. 56, 2055–2062. doi: 10.1099/ijs.0.64225-0
Pishgar, R., Dominic, J. A., Tay, J. H., and Chu, A. (2020). Pilot-scale investigation on nutrient removal characteristics of mineral-rich aerobic granular sludge: identification of uncommon mechanisms. Water Res. 168, 115151. doi: 10.1016/j.watres.2019.115151
Su, Z., Dai, T., Tang, Y., Tao, Y., Huang, B., Mu, Q., et al. (2018). Sediment bacterial community structures and their predicted functions implied the impacts from natural processes and anthropogenic activities in coastal area. Mar. Pollut. Bull. 131, 481–495. doi: 10.1016/j.marpolbul.2018.04.052
Sun, R., Wang, F., Hu, C., and Liu, B. (2021). Metagenomics reveals taxon-specific responses of the nitrogen-cycling microbial community to long-term nitrogen fertilization. Soil Biol. Biochem. 156, 108214. doi: 10.1016/j.soilbio.2021.108214
Sun, X., Ding, J., Jiang, Z., Yang, S., Jiang, Z., Xu, J., et al. (2019). Biochar improved rice yield and mitigated CH4 and N2O emissions from paddy field under controlled irrigation in the Taihu Lake Region of China. Atmos. Environ. 200, 69–77. doi: 10.1016/j.atmosenv.2018.12.003
Teoh, C. P., Lavin, P., Lee, D. J. H., Gonz}'alez-Aravena, M., Najimudin, N., Lee, P. C., et al. (2021). Genomics and transcriptomics analyses provide insights into the cold adaptation strategies of an Antarctic bacterium, Cryobacterium sp. SO1. Polar Biol. 44, 1305–1319. doi: 10.1007/s00300-021-02883-8
Tolar, B. B., Boye, K., Bobb, C., Maher, K., Bargar, J. R., and Francis, C. A. (2020). Stability of floodplain subsurface microbial communities through seasonal hydrological and geochemical cycles. Front. Earth Sci. 8,338. doi: 10.3389/feart.2020.00338
Torsvik, V., and Øvreås, L. (2002). Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5, 240–245. doi: 10.1016/S1369-5274(02)00324-7
Van Den Heuvel, R. N., Van Der Biezen, E., Jetten, M. S. M., Hefting, M. M., and Kartal, B. (2010). Denitrification at pH 4 by a soil-derived Rhodanobacter-dominated community. Environ. Microbiol. 12, 3264–3271. doi: 10.1111/j.1462-2920.2010.02301.x
Wang, H., Gilbert, J. A., Zhu, Y., and Yang, X. (2018a). Salinity is a key factor driving the nitrogen cycling in the mangrove sediment. Sci. Total Environ. 631, 1342–1349. doi: 10.1016/j.scitotenv.2018.03.102
Wang, H., Zeng, Y., Guo, C., Bao, Y., Lu, G., Dang, Z., et al. (2018b). Bacterial, archaeal, and fungal community responses to acid mine drainage-laden pollution in a rice paddy soil ecosystem. Sci. Total Environ. 616, 107–116. doi: 10.1016/j.scitotenv.2017.10.224
Wang, J. L., Liu, K. L., Zhao, X. Q., Zhao, H. Q., Li, D., Li, J. J., et al. (2021). Balanced fertilization over four decades has sustained soil microbial communities and improved soil fertility and rice productivity in red paddy soil. Sci. Total Environ. 793, 148664. doi: 10.1016/j.scitotenv.2021.148664
Ward, N. L., Challacombe, J. F., Janssen, P. H., Henrissat, B., Coutinho, P. M., Wu, M., et al. (2009). Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microb. 75, 2046–2056. doi: 10.1128/AEM.02294-08
Wei, G., Andersen, M. N., Qi, X. B., Li, P., Li, Z., Fan, X., et al. (2017). Effects of reclaimed water irrigation and nitrogen fertilization on the chemical properties and microbial community of soil. J. Integr. Agric. 16, 679–690. doi: 10.1016/S2095-3119(16)61391-6
Wu, T., Liu, Y., Yang, K., Zhu, L., White, J. C., and Lin, D. (2021). Synergistic remediation of PCB-contaminated soil with nanoparticulate zero-valent iron and alfalfa: targeted changes in the root metabolite-dependent microbial community. Environ. Sci. Nano 8, 986–999. doi: 10.1039/D1EN00077B
Yan, Q., Ding, Y., Wen, H., Liu, F., Hu, J., and Cai, R. (2019). Effect of biologicalmatrix on denitrification and microorganisms of SWIS. Chin. J. Environ. Eng. 13, 1099–1105.
Yang, J., He, M., and Wang, G. (2009). Removal of toxic chromate using free and immobilized Cr (VI)-reducing bacterial cells of Intrasporangium sp. Q5-1. World J. Microb. Biot. 25, 1579–1587. doi: 10.1007/s11274-009-0047-x
Yu, Y., Wu, M., Petropoulos, E., Zhang, J., Nie, J., Liao, Y., et al. (2019). Responses of paddy soil bacterial community assembly to different long-term fertilizations in southeast China. Sci. Total Environ. 656, 625–633. doi: 10.1016/j.scitotenv.2018.11.359
Zhang, H., Sun, H., Yang, R., Li, S., Zhou, M., Gao, T., et al. (2016). Complete genome sequence of a psychotrophic Pseudarthrobacter sulfonivorans strain Ar51 (CGMCC 4.7316), a novel crude oil and multi benzene compounds degradation strain. J. Biotechnol. 231, 81–82. doi: 10.1016/j.jbiotec.2016.04.010
Zhang, X., Li, X., Zhao, X., Chen, X., Zhou, B., Weng, L., et al. (2020). Bioelectric field accelerates the conversion of carbon and nitrogen in soil bioelectrochemical systems. J. Hazard. Mater. 388, 121790. doi: 10.1016/j.jhazmat.2019.121790
Zheng, Y., Jiang, X., Hou, L., Liu, M., Lin, X., Gao, J., et al. (2016). Shifts in the community structure and activity of anaerobic ammonium oxidation bacteria along an estuarine salinity gradient. J. Geophys. Res. Biogeosci. 121, 1632–1645. doi: 10.1002/2015JG003300
Keywords: paddy wetlands, soil indigenous microorganisms, groundwater's irrigation, microbial community, microbial function
Citation: Li H, Song A, Qiu L, Liang S and Chi Z (2024) Deep groundwater irrigation altered microbial community and increased anammox and methane oxidation in paddy wetlands of Sanjiang Plain, China. Front. Microbiol. 15:1354279. doi: 10.3389/fmicb.2024.1354279
Received: 12 December 2023; Accepted: 05 February 2024;
Published: 21 February 2024.
Edited by:
Yizhi Sheng, China University of Geosciences, ChinaReviewed by:
Jia Meng, Harbin Institute of Technology, ChinaRen-Cun Jin, Hangzhou Normal University, China
Chongjun Chen, Suzhou University of Science and Technology, China
Copyright © 2024 Li, Song, Qiu, Liang and Chi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zifang Chi, Y2hpemlmYW5nQGpsdS5lZHUuY24=
 Huai Li
Huai Li Aiwen Song1,2
Aiwen Song1,2 Zifang Chi
Zifang Chi