
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 March 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1353849
 Fushan Zhang1*
Fushan Zhang1* Leyuan Li2,3
Leyuan Li2,3 Yuxin Zhao2,3
Yuxin Zhao2,3 Huiyue Dong2,3
Huiyue Dong2,3 Buhui Zhao2,3
Buhui Zhao2,3 Xiaoyu Zhao2,3
Xiaoyu Zhao2,3 Ziwei Xia2,3
Ziwei Xia2,3 Leizi Chi2,3
Leizi Chi2,3 Yan Wang2,3
Yan Wang2,3 Ruichao Li4
Ruichao Li4 Shangshang Qin2,3
Shangshang Qin2,3 Xiangjing Fu2,3*
Xiangjing Fu2,3*Introduction: Carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-HvKP) strains combining virulence and multidrug resistance (MDR) features pose a great public health concern. The aim of this study is to explore the evolutionary characteristics of virulence in CR-HvKP by investigating the genetic features of resistance and virulence hybrid plasmids.
Methods: The resistance and virulence phenotypes were determined by using antimicrobial susceptibility testing and the mouse bacteremia infection model, respectively. Plasmid profiles were investigated by S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and Southern blotting, conjugation assay, and whole genome sequencing (WGS). Bioinformatics tools were used to uncover the genetic features of the resistance and virulence hybrid plasmids.
Results: Two ST11-KL64 CRKP clinical isolates (KP18-3-8 and KP18-2079), which exhibited enhanced virulence compared with the classic CRKP, were detected positive for blaKPC−2 and rmpA2. The virulence level of the hypermucoviscous strain KP18-3-8 was higher than that of KP18-2079. S1-PFGE, Southern hybridization and WGS analysis identified two novel hybrid virulence plasmids in KP18-3-8 (pKP1838-KPC-vir, 228,158 bp) and KP18-2079 (pKP1838-KPC-vir, 182,326 bp), respectively. The IncHI1B/repB-type plasmid pKP1838-KPC-vir co-harboring blaKPC−2 and virulence genes (rmpA2, iucABCD and iutA) but lacking type IV secretion system could transfer into non-hypervirulent ST11 K. pneumoniae with the assistance of a helper plasmid in conjugation. The IncFII/IncR-type virulence plasmid pKP18-2079-vir may have been generated as a result of recombination between a typical pLVPK-like virulence plasmid and an MDR plasmid.
Conclusion: Our studies further highlight co-evolution of the virulence and resistance plasmids in ST11-CRKP isolates. Close surveillance of such hybrid virulence plasmids in clinical K. pneumoniae should be performed.
As the most predominant and prevalent species within carbapenem-resistant Enterobacterales (CRE), carbapenem-resistant Klebsiella pneumoniae (CRKP) is notable for its considerable resistance and the treatment challenges. Current studies revealed that multidrug-resistant (MDR) or extensive drug-resistant (XDR) CRKP isolates were only susceptible to tigecycline, polymyxin, and new β-lactamase inhibitor compound preparations such as ceftazidime/avibactam (Yang et al., 2022), and these agents were also prescribed for the treatment of CRKP infections in China (Yahav et al., 2020; Han et al., 2022).
Recently, the emergence and increasing reports of carbapenem-resistant hypervirulent K. pneumoniae (CR-HvKP), which are simultaneously hypervirulent and multidrug resistant, represent a new serious threat in China for clinical treatment and therefore be recognized as a real superbug (Gu et al., 2018). The occurrence of CR-HvKP could be through either horizontal transfer of resistance plasmids into HvKP strains or through acquisition of the virulence plasmids in CRKP strains (Yang et al., 2021; Han et al., 2022), and the latter pathway was more frequently reported to be responsible for the emergence of CR-HvKP isolates since the MDR K. pneumoniae isolates have demonstrated a greater tendency to acquire virulence genes than hypervirulent clones to acquire resistance genes (Wyres et al., 2019). A recent national surveillance concerning CRKP from China identified a high proportion of CRKP isolates (34.2%) co-harboring blaKPC−2 and virulence plasmids in China (Wang et al., 2018; Zhang et al., 2020). The co-existence of virulence and MDR plasmids in the same CR-HvKP strain provide the opportunity for recombination between the two types of plasmids. Our previous report described the fusion of a virulence plasmid p17-16-vir (MK191024) and an MDR plasmid p17-16-CTX (MK192097) during conjugation (Li et al., 2020), and recent studies also reported the fusion of the non-conjugative virulence plasmid with the blaKPC-positive plasmid through homologous recombination (Xie et al., 2021; Zhao et al., 2022). In addition, recent research studies revealed that the pLVPK-like virulence plasmid can be transferred from HvKP strains to ST11 CRKP and Escherichia coli strains via a self-transferable IncF plasmid in a variety of modes (Li et al., 2020; Xu et al., 2021). Furthermore, IS element-mediated insertion of blaKPC−2 into the virulence plasmid is another way to generate hybrid plasmids that simultaneously expressed the carbapenem resistance- and hypervirulence-associated phenotypes (Dong et al., 2018; Jin et al., 2021). The emergence of such plasmids, coupled with the co-transfer of resistance and virulence determinants, may directly facilitate the transformation of classical K. pneumoniae into CR-HvKP. This phenomenon further exacerbates the spread of CR-HvKP strains, posing a severe threat to public health and requiring continuous monitoring. In this study, we have reported two novel hybrid plasmids encoding MDR/carbapenem resistance and virulence phenotypes in KPC-2 producing K. pneumoniae belonging to ST11-KL64, which is regarded as the most commonly detected type responsible for the majority of CRKP infections in China (Zhang et al., 2020; Wang et al., 2022), indicating further evolution of the pLVPK-like virulence plasmids in CRKP.
Clinical K. pneumoniae strain 18-3-8 (KP18-3-8) was obtained from the urine culture of a 75-year-old man hospitalized in a respiratory unit of a local hospital in Pingdingshan city of Henan province on 14 July 2018, and KP18-2079 was recovered from the blood culture of a 42-year-old man, who was diagnosed with epilepsy and hospitalized at a neurosurgery unit of a teaching hospital of Zhengzhou University on 22 November 2018. The minimum inhibitory concentrations (MICs) of antimicrobial agents commonly used for the treatment of Enterobacterales infections were determined by the broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2018). The MIC results of the tested antibiotics were interpreted using the CLSI breakpoints, except tigecycline, which was interpreted in accordance with the EUCAST breakpoints (http://www.eucast.org/clinical_breakpoints/). An E. coli strain ATCC 25922 was used as the quality control. PCR and sequencing methods were employed to detect the presence of carbapenemase-encoding genes (blaKPC, blaNDM, blaIMP, and blaOXA−48−like) and virulence factors rmpA/A2 in CRKP isolates (Qin et al., 2014; Yu et al., 2018).
S1-PFGE and Southern blotting were performed according to the method described in our previous study (Li et al., 2020). In brief, the agarose gel plugs containing whole-cell DNA of the strain were treated with S1 nuclease (TaKaRa, Dalian, China) and then PFGE was performed under the following conditions: 1% agarose solution for 18 h at 6 V/cm and 14°C, with a pulse angle of 120° and the pulse time linearly ramped from 2.16 s to 63.8 s. Digoxigenin-labeled blaKPC−2 and rmpA2-specific probes were hybridized with linear plasmids separated by S1-PFGE on nylon membranes (Millipore, USA) and then detected using an NBT/BCIP color detection kit (Roche, Germany).
The transferability of hybrid plasmids was investigated by conducting conjugation experiments. KP18-3-8 and KP18-2079 served as the donors, with hygromycin-resistant ST11 K. pneumoniae HS11286YZ6 as the recipient strain (Xie et al., 2018). Donor and recipient strains, grown to logarithmic growth phase (OD600 of 0.6–0.8), were inoculated into Luria Bertani (LB) broth, were mixed at a ratio of 1:1 between donor and recipient strains, and were incubated for 24 h at 30°C. Transconjugants were selected on LB agar plates supplemented with 2 μg/mL meropenem or 2 μg/mL potassium tellurite (K2TeO3) plus 200 μg/mL hygromycin and identified by PCR targeting blaKPC−2 or rmpA2.
The hypermucoviscous phenotype was determined using the string test (Russo et al., 2018). The mouse bacteremia infection model was used to assess the virulence of various K. pneumoniae strains as described previously (Yang et al., 2019). The non-hypermucoviscous ST1-K19 K. pneumoniae strain, KP18-208, was used as the control for low virulence (Qin et al., 2020), while an ST268-K20 K. penumoniae strain, KP19-2065, served as the control for hypervirulence. Mice in each group were infected intravenously with 5.0 × 105 CFU and 5.0 × 106 CFU of each K. pneumoniae strain tested. The survival rate of the mice was recorded daily for 5 days. The survival curves were plotted using the Kaplan-Meier method, and the statistical analysis was performed using the Log-rank test in GraphPad 9.5.1.
Genome DNA of the two clinical strains was extracted using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI), quantified using Nanodrop and Qubit fluorometer, and sequenced via Illumina paired-end 2×150 bp platform. Nanopore sequencing long-read DNA libraries were constructed using a rapid sequencing kit (SQK-RAD004, ONT, Oxford, UK) and a rapid barcode kit (SQK-RAD004, ONT, Oxford, UK) according to the manufacturer's protocol. Sequencing was performed using MinION (ONT, Oxford, UK). Different bioinformatics tools including Unicycler and Flye were utilized to assemble the complete genome sequences (Wick et al., 2017; Kolmogorov et al., 2019). The RAST tool was used to annotate the genomes, and the sequences were further modified manually (https://rast.nmpdr.org/). Different databases and tools, including Resfinder and PlasmidFinder (https://cge.cbs.dtu.dk/services/) from the Center for Genomic Epidemiology (CGE) and ISFinder (https://www-is.biotoul.fr/), were used to perform genomic analysis and visualization. Easyfig (https://github.com/mjsull/Easyfig) and BRIG (http://sourceforge.net/projects/brig/) were used for displaying plasmid comparison maps (Alikhan et al., 2011; Sullivan et al., 2011).
Both isolates KP18-3-8 and KP18-2079 were resistant to carbapenem and cephalosporins but were susceptible to colistin (Table 1). PCR-based screening for carbapenemase and virulence-encoding genes identified both blaKPC−2 and rmpA2 genes in each isolate. Further analysis using S1-PFGE and Southern blotting techniques with blaKPC−2 and rmpA2-specific probes indicated that these two genes were localized on plasmids of the same size in both KP18-3-8 (approximately 220 kb) and KP18-2079 (approximately 180 kb; Figure 1) strains, and therefore, we speculated that the two genes were located on the same plasmid.
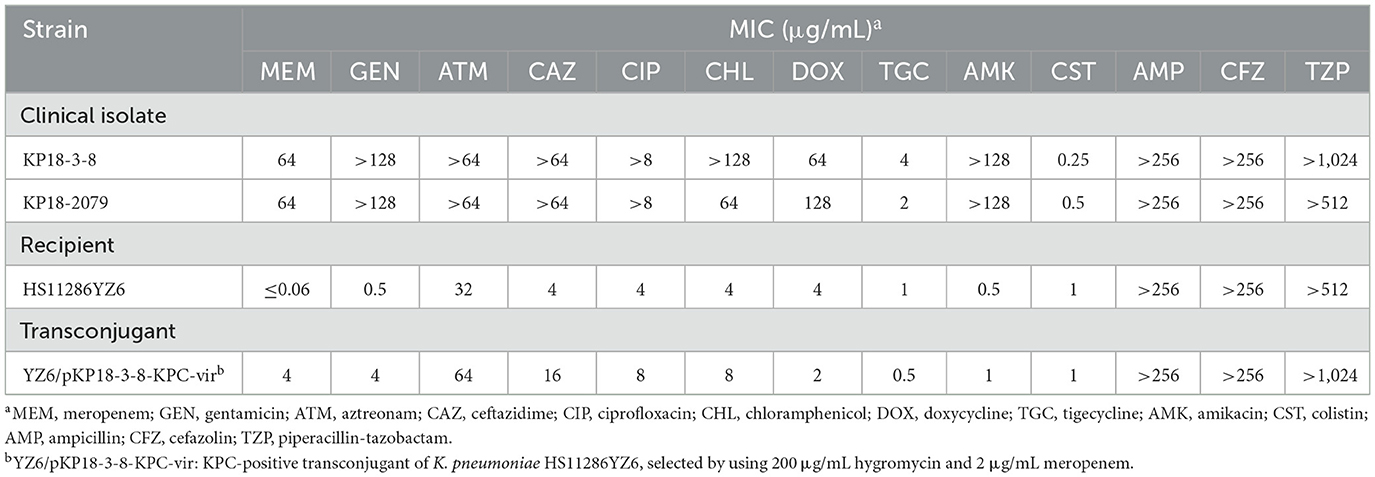
Table 1. Antimicrobial susceptibility testing of KP18-3-8, KP18-2079, and the transconjugant of KP18-3-8.
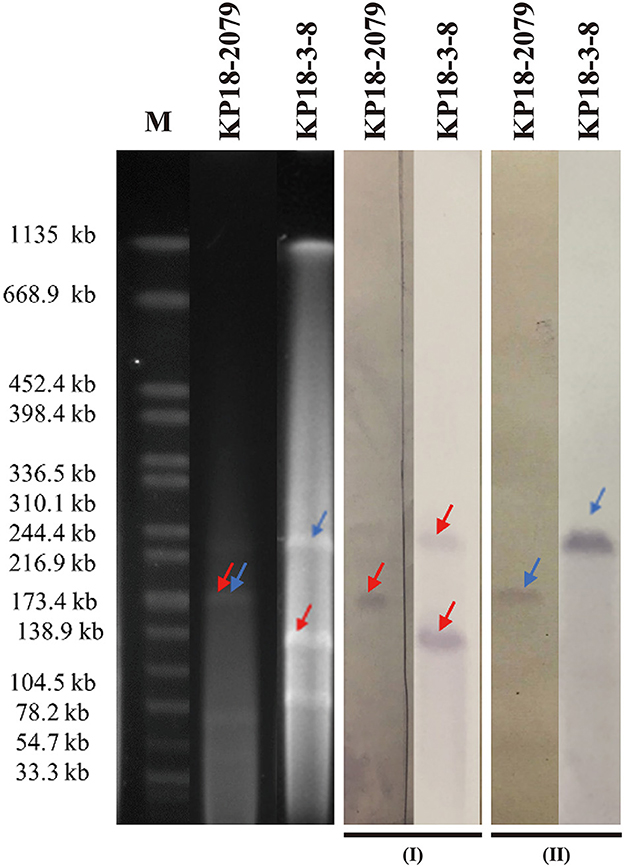
Figure 1. S1-PFGE and Southern hybridization analyses of KP18-2079 and KP18-3-8 using the blaKPC−2 (I) and rmpA2 probes (II). Lane M, reference standard strain H9812 restricted with XbaI (sizes are given in kilobases).
To uncover the genetic basis of the resistance and virulence in the two strains, whole-genome sequences of KP18-3-8 and KP18-2079 were obtained using a combination of the Illumina (short-read) and Nanopore MinION (long-read) sequencing platforms. KP18-3-8 belonged to ST11 and serotype KL64 and contained one chromosome of ~5.54 Mbp and five plasmids, namely pKP1838-KPC-vir (MT035874), pKP1838-KPC (MT232812), pKP1838-IncFII (MT035876), pKP1838-Col (MT035877), and pKP1838-5kb (MT035878) of 228,158, 130,717, 87,095, 10,060, and 5,596 bp in size, respectively. In addition, KP18-2079, which was also a ST11-KL64 strain, contained a chromosome of ~5.51 Mbp and harbored six plasmids, namely pKP18-2079-vir (MT090958), pKP18-2079-KPC (MT090959), pKP18-2079-tetA (MT090960), pKP18-2079-54kb (MT090961), pKP18-2079-11kb (MT090962), and pKP18-2079-5kb (MT090963), with sizes of 182,326, 186,564, 84,699, 54,874, 11,970, and 5,596 bp, respectively. Resistome and virulence genes of KP18-3-8 and KP18-2079 are summarized in Table 2. Interestingly, along with the results of S1-PFGE and Southern blotting, we found that KP18-3-8 carried two blaKPC−2-bearing plasmids, namely pKP1838-KPC-vir and p1838-KPC, and the co-occurrence of blaKPC−2 and rmpA2 was observed in pKP1838-KPC-vir. Meanwhile, the blaKPC−2 and rmpA2 genes in KP18-2079 were carried by separated plasmids (pKP18-2079-vir and pKP18-2079-KPC) with comparable size.
An IncHI1B/repB-type plasmid pKP1838-KPC-vir (MT035874) with a length of 228,158 bp was found to be the largest plasmid in the CRKP strain, KP18-3-8. It shared 99% of its identity and 93% of its coverage with a typical non-conjugative pLVPK-like virulence plasmid pvir-CR-HvKP267 (MG053312), which was carried by a tigecycline resistant KPC-2 producing hypervirulent K. pneumoniae KP267 isolated in 2015 in our previous study (Yao et al., 2018). Notably, compared with pvir-CR-HvKP267, the plasmid pKP1838-KPC-vir carried an additional 19 kb segment containing the carbapenemase-encoding blaKPC−2 gene, indicating that pKP1838-KPC-vir might have undergone further evolution by integrating exogenous drug resistance genes on the backbone of traditional virulence plasmids. In the KP18-3-8 strain, two blaKPC−2 genes were identified, each borne on distinct plasmids, pKP1838-KPC-vir and pKP1838-KPC. Detailed comparative analyses of the gene surrounding blaKPC−2 revealed a 45-kb segment harboring the gene within a non-Tn4401 element (NTEKPC−1b), along with gene clusters conferring resistance to heavy metals such as mercury, copper, and silver. Despite a high degree of sequence homology (99% query coverage and 100% nucleotide identity between the segments), notable variations were observed in their genomic arrangement, including an inversion of the entire NTEKPC−1b element associated with the IS26 element in pKP1838-KPC relative to pKP1838-KPC-vir (Figure 2). Based on the above observations, we supposed that the plasmid pKP1838-KPC-vir co-harboring blaKPC−2 and virulence factors possibly evolved from a typical pLVPK-like virulence plasmid, and the genetic environment surrounding blaKPC−2 on pKP1838-KPC-vir similar to that on the blaKPC−2-bearing plasmid pKP1838-KPC indicates that pKP1838-KPC might have been the source of the blaKPC−2-carrying fragment for the plasmid pKP1838-KPC-vir.
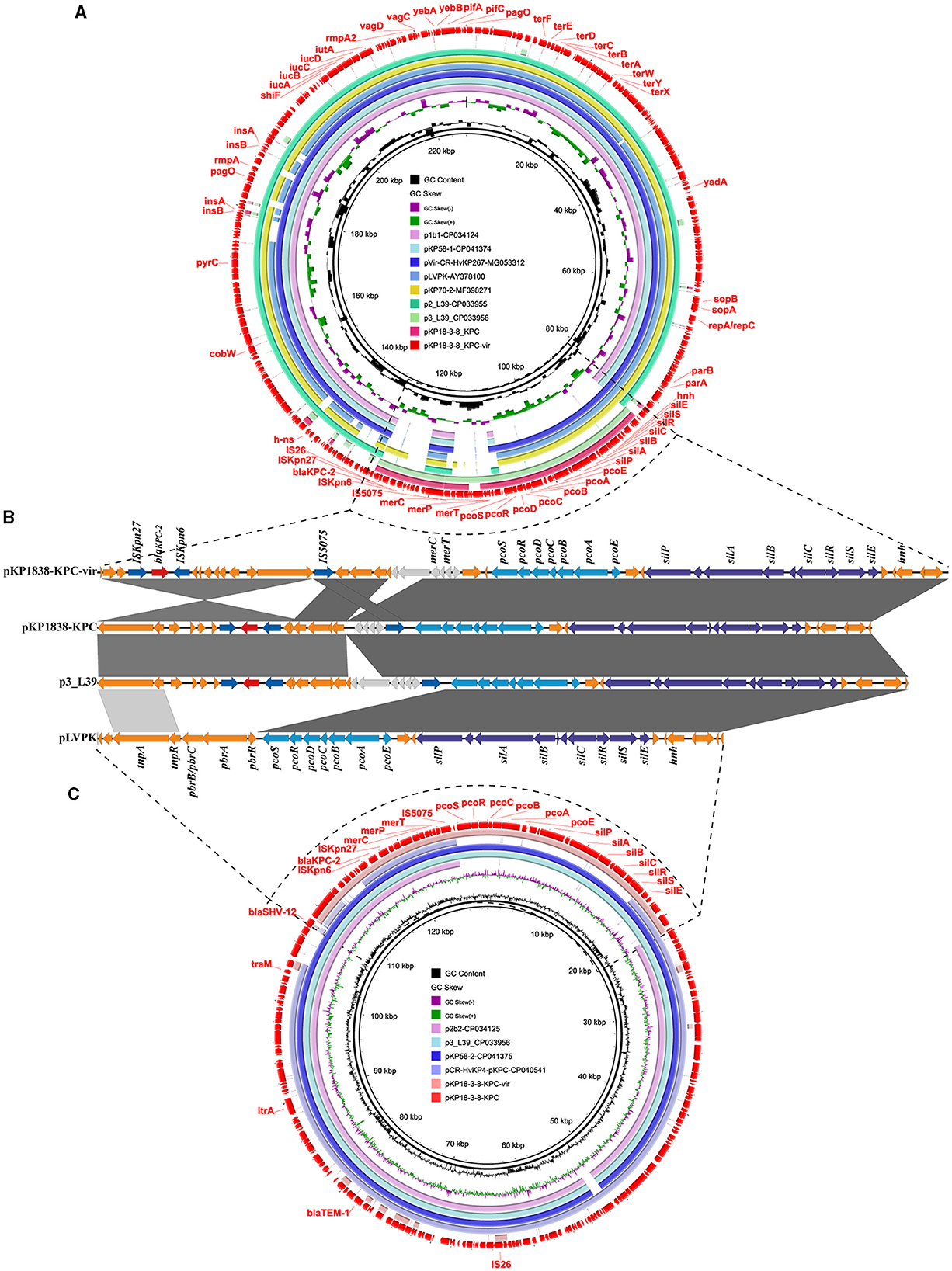
Figure 2. The rmpA2 and blaKPC−2 co-harboring plasmid pKP1838-KPC-vir and the blaKPC−2-bearing plasmid pKP1838-KPC in Klebsiella pneumoniae strain KP18-3-8. Circular and linear comparisons of pKP1838-KPC-vir, pKP1838-KPC, p3_L39, and pLVPK plasmids. The pKP1838-KPC-vir and pKP1838-KPC plasmids located at the outermost circle was used as the reference plasmid to perform sequence alignment with BLASTn by BRIG software.
The conjugative assay revealed that, despite lacking a type IV secretion system (T4SS), pKP1838-KPC-vir was successfully transferred to K. pneumoniae HS11286YZ6 at a frequency of 6.37 × 10−8 per donor cell. Our previous study and recent reports demonstrated that the non-conjugative virulence plasmids could be mobilized by the conjugative plasmids belonging to IncF (Li et al., 2020; Xu et al., 2021). Thus, we suggested that the plasmid pKP1838-IncFII, which contained four necessary components [oriT, relaxase gene, type IV coupling protein (T4CP) gene, and T4SS gene cluster] for self-transfer in the same strain, might have served as a helper plasmid by sharing its T4SS with pKP1838-KPC-vir to transfer.
A recent nationwide surveillance study on CRKP in China revealed that over 90% CRKP isolates were KPC-type carbapenemase producers, especially KPC-2, and over 30% KPC-2 producing CRKP isolates were positive for one or more plasmid-borne virulence genes such as rmpA, rmpA2, iucA, and iroN (Zhang et al., 2020). Typically, the blaKPC−2 and virulence genes were carried by distinct plasmids. However, two recent reports described the homologous recombination-mediated fusion of the KPC plasmid and the virulence plasmid during the conjugation process, resulting in the generation of conjugative hybrid plasmids (Xie et al., 2021; Zhao et al., 2022). In addition to this homologous recombination pathway to generate the hybrid blaKPC−2-bearing virulence plasmids, blaKPC−2 was rarely reported inserted into the virulence plasmid backbone. To date, only two plasmids, namely pKP70-2 (MF398271) and pCRHV-C2244 (MT644086), which were presumed to be generated as a consequence of an IS26-mediated blaKPC-bearing fragment inserted into the virulence plasmid, were identified in the ST23-K1 HvKP strain KP70-2 isolated from China and the ST11-K64 CRKP strain C2244 isolated from China, respectively (Dong et al., 2018; Jin et al., 2021). However, there was no additional blaKPC−2-bearing plasmid other than the hybrid virulence- and resistance-encoding plasmid in each strain. In this study, the co-occurrence of blaKPC−2 that was carried by the NTEKPC−1b element in both virulence and MDR plasmids in the same isolate provided a more direct evidence of recombination events that occurred between virulence and MDR plasmids.
An 182,326 bp IncFII/IncR-type virulence plasmid pKP18-2079-vir (MT090958) containing 237 open reading frames, with GC contents of 50.60%, was identified in K. pneumoniae KP18-2079 (Table 2). This hybrid plasmid harbored both virulence genes (iutA-iucABCD, rmpA, rmpA2, and peg344) and multiple antimicrobial resistance determinants, including beta-lactamase genes blaTEM−1B and blaCTX−M−65, aminoglycoside resistance gene rmtB1, and phosphonic acids resistance gene fosA3, simultaneously. The BLAST analysis showed that this type of plasmid has not been reported previously, and it shared 99% identity and 67% coverage with the virulence plasmid p1-L388 (CP029221), a typical pLVPK-like virulence plasmid carrying no resistance genes in a ST11 KPC-2-producing K. pneumoniae clinical isolate L388 (Figures 3A, C). The comparative genomic analysis further revealed that pKP18-2079-vir contained two genetically and physically distinct modules: an ~117 kb reversed p1-L388-derived module harboring virulence genes (iutA-iucABCD, rmpA, rmpA2, and peg344) and copper resistance genes (pcoEABCDRS) and an ~65 kb module carrying a multidrug resistance region that was essentially homologous to pKPC-L388 (CP029225), a common blaKPC−2-bearing IncFII plasmid other than p1-L388 in K. pneumoniae L388. The fosA3 gene encompassed by two IS26 elements was absent in pKPC-L388 (Figures 3A, C). Interestingly, we further found that the remaining parts of p1-L388 and pKPC-L388 were present in the plasmid pKP18-2079-KPC (MT090959) and formed a novel IncHI1B/IncFrepB-type blaKPC−2-bearing plasmid in K. pneumoniae KP18-2079 (Figures 3B, D). These observations indicated that pKP18-2079-vir (182,326 bp) and p18-2079-KPC (186,564 bp) harbored by KP18-2079 might be generated as a consequence of genetic rearrangement events between p1-L388 (217,870 bp) and pKPC-L388-like (145,851 bp) plasmids.
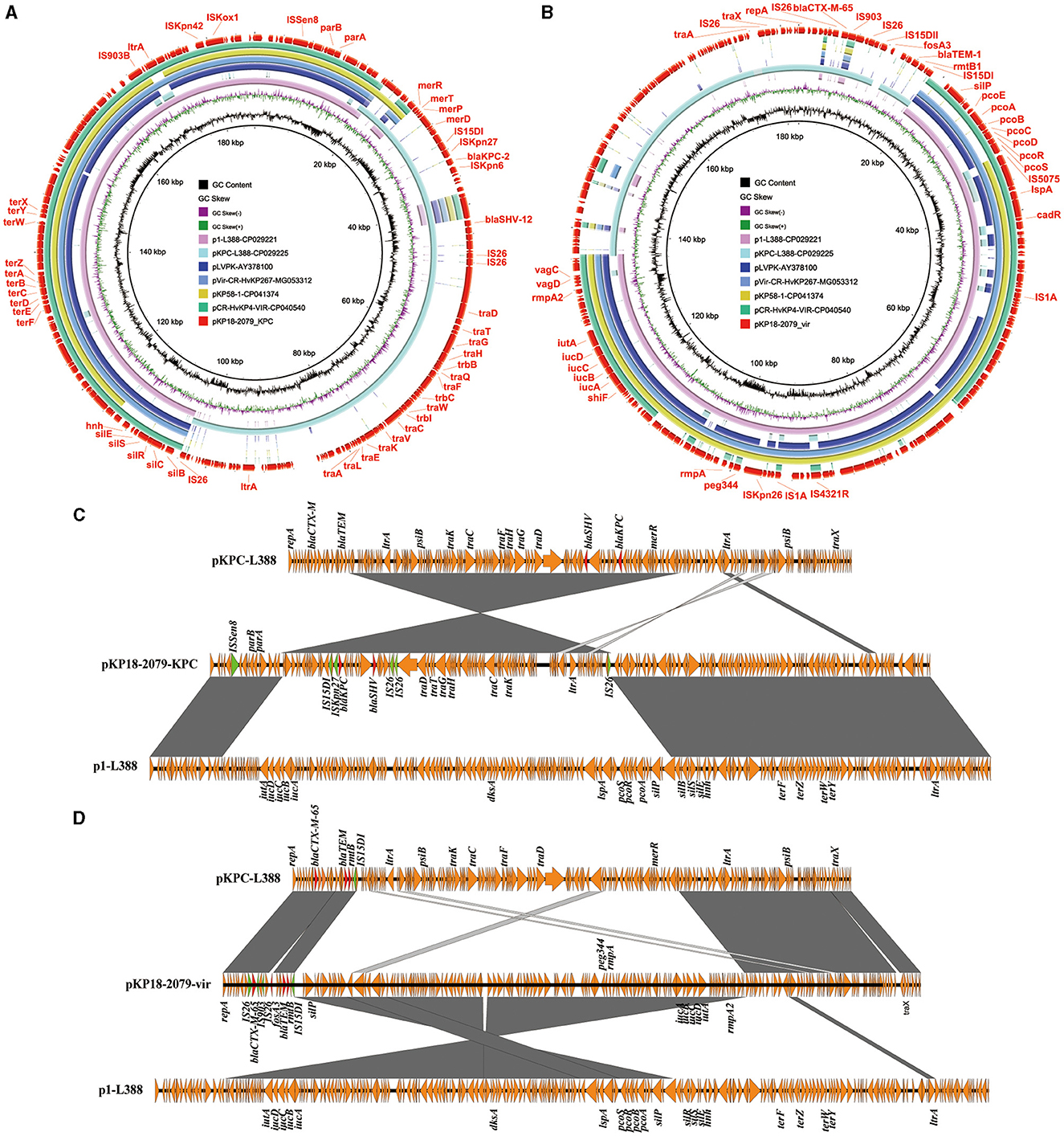
Figure 3. The MDR virulence plasmid p18-2079-vir and the blaKPC−2-bearing plasmid p18-2079-KPC in Klebsiella pneumoniae strain KP18-2079. (A, B) Circular maps of plasmid p18-2079-KPC and p18-2079-vir recovered from K. pneumoniae KP18-2079 with plasmids recorded in the NCBI database by BLAST Ring Image Generator (BRIG). (C, D) Linear alignment of plasmid p18-2079-KPC and p18-2079-vir recovered from K. pneumoniae KP18-2079 using Easyfig.
The mouse infection model was employed to assess the virulence of KP18-3-8 and KP18-2079. As shown in Figure 4, infections of mice with 5 × 106 CFU of strains KP19-2065, KP18-3-8, and KP18-2079 led to a 100% mortality rate at 12 h, whereas the mortality rate was recorded as 0% for the strain KP18-208, a classic CRKP control strain (Figure 4A). Consistently, when infected at the lower dose of 5 × 105 CFU of KP19-2065, KP18-3-8, and KP18-2079, it resulted in a 100% mortality rate at 12, 36, and 60 h, respectively, whereas a 0% mortality rate was observed for KP18-208 (Figure 4B). These data confirmed that KP18-3-8 and KP18-2079 exhibited a virulence level slightly lower than that of KP19-2065 but much higher than that of KP18-208, a classic CRKP strain without a virulence plasmid. In addition, the virulence level of KP18-2079 without the hypermucoviscosity phenotype was lower than that of KP18-3-8, a hypermucoviscous strain which was positive for the string test (Figure 5).
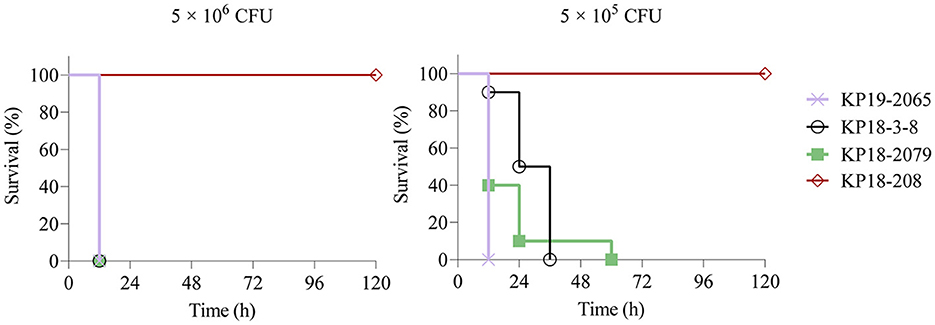
Figure 4. Virulence potential of Klebsiella pneumoniae KP18-3-8 and KP18-2079 in a mouse infection model with an inoculum of 5 × 106 CFU and 5 × 105 CFU. Survival of mice (n = 10) infected by the indicated inoculum of each K. pneumoniae strain at 120 h is shown. The test strains included K. pneumoniae strain KP18-3-8, K. pneumoniae strain KP18-2079, ST268-K20 hypervirulent K. pneumoniae strain KP19-2065 (hypervirulence control), and ST1-K19 classic K. pneumoniae strain KP18-208 (low-virulence control). A log-rank (Mantel-Cox) test was performed for the indicated curves. A significant difference (P < 0.0001) was observed between KP18-3-8/ KP18-2079 and KP18-208 in 5 × 106 CFU and 5 × 105 CFU.
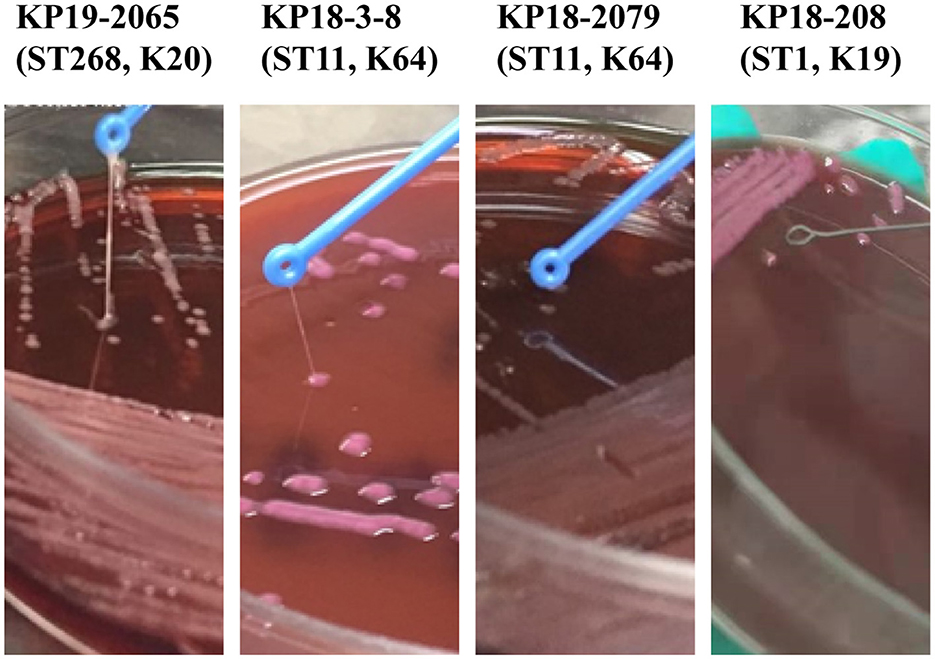
Figure 5. A comparison of Klebsiella pneumoniae KP18-2079 and KP18-3-8 with hvKp KP19-2065 and the non-mucoviscous KP18-208. The hypermucoviscosity phenotype of K. pneumoniae KP18-2079 and KP18-3-8 evidenced by the string test. After the overnight incubation at 37°C on blood agar, a mucoviscous string of at least 5 mm in length was observed when lifting the KP18-2079 and KP18-3-8 colony using a disposable inoculation loop. The ST268-K20 hvKP strain KP19-2065 was used as a positive control while the non-mucoviscous ST1-K19 strain KP18-208 was used as a negative control.
ST11-KL64 CR-HvKP, which was found to be derived from an ST11-KL47–like ancestor through recombination, has demonstrated enhanced virulence and higher mortality rates than that of ST11-KL47 CRKP (Zhou et al., 2020). Moreover, a recent surveillance study on CR-HvKP revealed that ST11-KL64 CR-HvKP became the most common detected CR-HvKP type in different regions of China, especially in Henan province (Zhang et al., 2020). In our study, we isolated two ST11-KL64 CRKP strains that exhibited an XDR profile, with susceptibility only to colistin. Notably, these strains demonstrated heightened virulence compared to their non-virulent plasmid-bearing CRKP counterparts. The presence of composite plasmids encoding both virulence and resistance factors significantly contributed to their carbapenem resistance/MDR profiles and virulent phenotypes.
Overall, our study identified two novel hybrid virulence plasmids in ST11-KL64 CRKP strains. The IncHI1B/repB-type plasmid pKP1838-KPC-vir co-harboring blaKPC−2 and virulence factors may have been generated as a consequence of integration of blaKPC−2-bearing fragment into a typical pLVPK-like virulence plasmid, and the IncFII/IncR-type virulence plasmid pKP18-2079-vir may have been generated as a result of recombination between a typical pLVPK-like virulence plasmid and an MDR plasmid. Our findings revealed further evolution of the pLVPK-like virulence plasmids which were found to be present in ST11 CRKP since 2015 (Gu et al., 2018). The acquisition of such a virulence- and resistance-encoding plasmid, especially the blaKPC−2-bearing virulence plasmid simultaneously conferring carbapenem resistance and enhanced virulence in classical K. pneumoniae isolates, may enable them to become CR-HvKP. The emergence of such strains, which may cause severe infections in individuals difficult to treat with current antibiotics, warrants worldwide attention. Close surveillance on the epidemiology of such hybrid plasmids is urgently warranted to control their further dissemination.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
The animal study was approved by the Institutional Animal Care and Use Committee of Zhengzhou University. The study was conducted in accordance with the local legislation and institutional requirements.
FZ: Writing – review & editing, Writing – original draft, Resources, Data curation. LL: Writing – original draft, Formal analysis, Data curation. YZ: Writing – original draft, Formal analysis, Data curation. HD: Writing – original draft, Software, Formal analysis, Data curation. BZ: Writing – original draft, Investigation. XZ: Writing – original draft, Methodology. ZX: Writing – original draft, Data curation. LC: Writing – original draft, Data curation. YW: Writing – original draft, Data curation. RL: Writing – review & editing, Funding acquisition. SQ: Writing – review & editing, Funding acquisition. XF: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Science and Technology Research Project of Henan Province (232102311002) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing, 28th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Dong, N., Lin, D., Zhang, R., Chan, E. W., and Chen, S. (2018). Carriage of blaKPC−2 by a virulence plasmid in hypervirulent Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 3317–3321. doi: 10.1093/jac/dky358
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Han, Y. L., Wen, X. H., Zhao, W., Cao, X. S., Wen, J. X., Wang, J. R., et al. (2022). Epidemiological characteristics and molecular evolution mechanisms of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 13:1003783. doi: 10.3389/fmicb.2022.1003783
Jin, L., Wang, R., Gao, H., Wang, Q., and Wang, H. (2021). Identification of a novel hybrid plasmid encoding KPC-2 and virulence factors in Klebsiella pneumoniae sequence type 11. Antimicrob. Agents Chemother. 65:e02435–20. doi: 10.1128/AAC.02435-20
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Li, R., Cheng, J., Dong, H., Li, L., Liu, W., Zhang, C., et al. (2020). Emergence of a novel conjugative hybrid virulence multidrug-resistant plasmid in extensively drug-resistant Klebsiella pneumoniae ST15. Int. J. Antimicrob. Agents 55:105952. doi: 10.1016/j.ijantimicag.2020.105952
Qin, S., Fu, Y., Zhang, Q., Qi, H., Wen, J. G., Xu, H., et al. (2014). High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob. Agents Chemother. 58, 4275–4282. doi: 10.1128/AAC.02813-13
Qin, S., Zhang, C., Schwarz, S., Li, L., Dong, H., Yao, H., et al. (2020). Identification of a novel conjugative mcr-8.2-bearing plasmid in an almost pan-resistant hypermucoviscous Klebsiella pneumoniae ST11 isolate with enhanced virulence. J. Antimicrob. Chemother. 75, 2696–2699. doi: 10.1093/jac/dkaa226
Russo, T. A., Olson, R., Fang, C. T., Stoesser, N., Miller, M., MacDonald, U., et al. (2018). Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J. Clin. Microbiol. 56:e00776–18. doi: 10.1128/JCM.00776-18
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Wang, M., Earley, M., Chen, L., Hanson, B. M., Yu, Y., Liu, Z., et al. (2022). Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect. Dis. 22, 401–412. doi: 10.1016/S1473-3099(21)00399-6
Wang, Q., Wang, X., Wang, J., Ouyang, P., Jin, C., Wang, R., et al. (2018). Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin. Infect. Dis. 67, S196–S205. doi: 10.1093/cid/ciy660
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wyres, K. L., Wick, R. R., Judd, L. M., Froumine, R., Tokolyi, A., Gorrie, C. L., et al. (2019). Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 15:e1008114. doi: 10.1371/journal.pgen.1008114
Xie, M., Yang, X., Xu, Q., Ye, L., Chen, K., Zheng, Z., et al. (2021). Clinical evolution of ST11 carbapenem resistant and hypervirulent Klebsiella pneumoniae. Commun. Biol. 4:650. doi: 10.1038/s42003-021-02148-4
Xie, Y., Tian, L., Li, G., Qu, H., Sun, J., Liang, W., et al. (2018). Emergence of the third-generation cephalosporin-resistant hypervirulent Klebsiella pneumoniae due to the acquisition of a self-transferable blaDHA−1-carrying plasmid by an ST23 strain. Virulence 9, 838–844. doi: 10.1080/21505594.2018.1456229
Xu, Y., Zhang, J., Wang, M., Liu, M., Liu, G., Qu, H., et al. (2021). Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 13:119. doi: 10.1186/s13073-021-00936-5
Yahav, D., Giske, C. G., Gramatniece, A., Abodakpi, H., Tam, V. H., Leibovici, L., et al. (2020). New beta-lactam-beta-lactamase inhibitor combinations. Clin. Microbiol. Rev. 34:e00115–20. doi: 10.1128/CMR.00115-20
Yang, X., Dong, N., Chan, E. W., Zhang, R., and Chen, S. (2021). Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 29, 65–83. doi: 10.1016/j.tim.2020.04.012
Yang, X., Sun, Q., Li, J., Jiang, Y., Li, Y., Lin, J., et al. (2022). Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg. Microbes Infect. 11, 841–849. doi: 10.1080/22221751.2022.2049458
Yang, X., Wai-Chi Chan, E., Zhang, R., and Chen, S. (2019). A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat. Microbiol. 4, 2039–2043. doi: 10.1038/s41564-019-0566-7
Yao, H., Qin, S., Chen, S., Shen, J., and Du, X. D. (2018). Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 18, 25. doi: 10.1016/S1473-3099(17)30628-X
Yu, F., Lv, J., Niu, S., Du, H., Tang, Y. W., Pitout, J. D. D., et al. (2018). Multiplex PCR analysis for rapid detection of Klebsiella pneumoniae carbapenem-resistant (sequence type 258 [ST258] and ST11) and hypervirulent (ST23, ST65. ST86, and ST375) strains. J. Clin. Microbiol. 56:e00731–18. doi: 10.1128/JCM.00731-18
Zhang, Y., Jin, L., Ouyang, P., Wang, Q., Wang, R., Wang, J., et al. (2020). Carbapenem-resistant Enterobacteriaceae, evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J. Antimicrob. Chemother. 75, 327–336. doi: 10.1093/jac/dkz446
Zhao, Q., Feng, Y., and Zong, Z. (2022). Conjugation of a hybrid plasmid encoding hypervirulence and carbapenem resistance in Klebsiella pneumoniae of sequence type 592. Front. Microbiol. 13:852596. doi: 10.3389/fmicb.2022.852596
Keywords: blaKPC−2-bearing virulence plasmid, MDR-virulence plasmid, hypervirulent, ST11-KL64, CRKP
Citation: Zhang F, Li L, Zhao Y, Dong H, Zhao B, Zhao X, Xia Z, Chi L, Wang Y, Li R, Qin S and Fu X (2024) Molecular characterization of hybrid virulence plasmids in ST11-KL64 KPC-2-producing multidrug-resistant hypervirulent Klebsiella pneumoniae from China. Front. Microbiol. 15:1353849. doi: 10.3389/fmicb.2024.1353849
Received: 11 December 2023; Accepted: 29 February 2024;
Published: 14 March 2024.
Edited by:
Goutam Chowdhury, National Institute of Cholera and Enteric Diseases, IndiaReviewed by:
Farah Al Marzooq, United Arab Emirates University, United Arab EmiratesCopyright © 2024 Zhang, Li, Zhao, Dong, Zhao, Zhao, Xia, Chi, Wang, Li, Qin and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fushan Zhang, emhmc2gwMUAxNjMuY29t; Xiangjing Fu, ZnV4aWFuZ2ppbmdAenp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.