- 1State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan University, Kunming, China
- 2Department of Biology, McMaster University, Hamilton, ON, Canada
- 3College of Agriculture and Life Sciences, Kunming University, Kunming, China
Purpureocillium lavendulum is an important biocontrol agent against plant-parasitic nematodes, primarily infecting them with conidia. However, research on the regulatory genes and pathways involved in its conidiation is still limited. In this study, we employed Agrobacterium tumefaciens-mediated genetic transformation to generate 4,870 random T-DNA insertion mutants of P. lavendulum. Among these mutants, 131 strains exhibited abnormal conidiation, and further in-depth investigations were conducted on two strains (designated as #5-197 and #5-119) that showed significantly reduced conidiation. Through whole-genome re-sequencing and genome walking, we identified the T-DNA insertion sites in these strains and determined the corresponding genes affected by the insertions, namely Plhffp and Plpif1. Both genes were knocked out through homologous recombination, and phenotypic analysis revealed a significant difference in conidiation between the knockout strains and the wild-type strain (ku80). Upon complementation of the ΔPlpif1 strain with the corresponding wildtype allele, conidiation was restored to a level comparable to ku80, providing further evidence of the involvement of this gene in conidiation regulation in P. lavendulum. The knockout of Plhffp or Plpif1 reduced the antioxidant capacity of P. lavendulum, and the absence of Plhffp also resulted in decreased resistance to SDS, suggesting that this gene may be involved in the integrity of the cell wall. RT-qPCR showed that knockout of Plhffp or Plpif1 altered expression levels of several known genes associated with conidiation. Additionally, the analysis of nematode infection assays with Caenorhabditis elegans indicated that the knockout of Plhffp and Plpif1 indirectly reduced the pathogenicity of P. lavendulum towards the nematodes. The results demonstrate that Agrobacterium tumefaciens - mediated T-DNA insertion mutagenesis, gene knockout, and complementation can be highly effective for identifying functionally important genes in P. lavendulum.
1 Introduction
Plant-Parasitic Nematodes (PPNs) have a wide range of hosts, ranging from lower plants like mosses, ferns, and algae, to higher plants such as gymnosperms and angiosperms (Shao et al., 2023). Almost all plants are affected by plant-parasitic nematodes to varying degrees. PPNs pose a serious threat to numerous economically important crops worldwide (Rehman et al., 2016) and are a significant limiting factor for global food security (Montarry et al., 2021). In economically underdeveloped regions, due to a lack of relevant control measures, the damage caused by nematodes to food crops is more severe than in developed regions (Coyne et al., 2018).
Currently, the control and management of PPNs mainly involve agricultural practices, physical control, chemical control, biological control, and the use of plant-based pesticides (Li et al., 2015). Biological control typically involves the use of nematode predators, such as Nematophagous microorganisms, to control nematode populations. Some bacterial pathogens of nematodes produce toxic substances that can kill nematodes, while certain fungi have evolved specific predatory structures to capture and feed on nematodes (Liang et al., 2019). Purpureocillium lavendulum is an important bio-control fungus against nematodes (Figure 1A). Compared to its sister species, P. lilacinum, its inability of growth at temperatures above 35°C makes it safe for humans and other mammals (Perdomo et al., 2013).
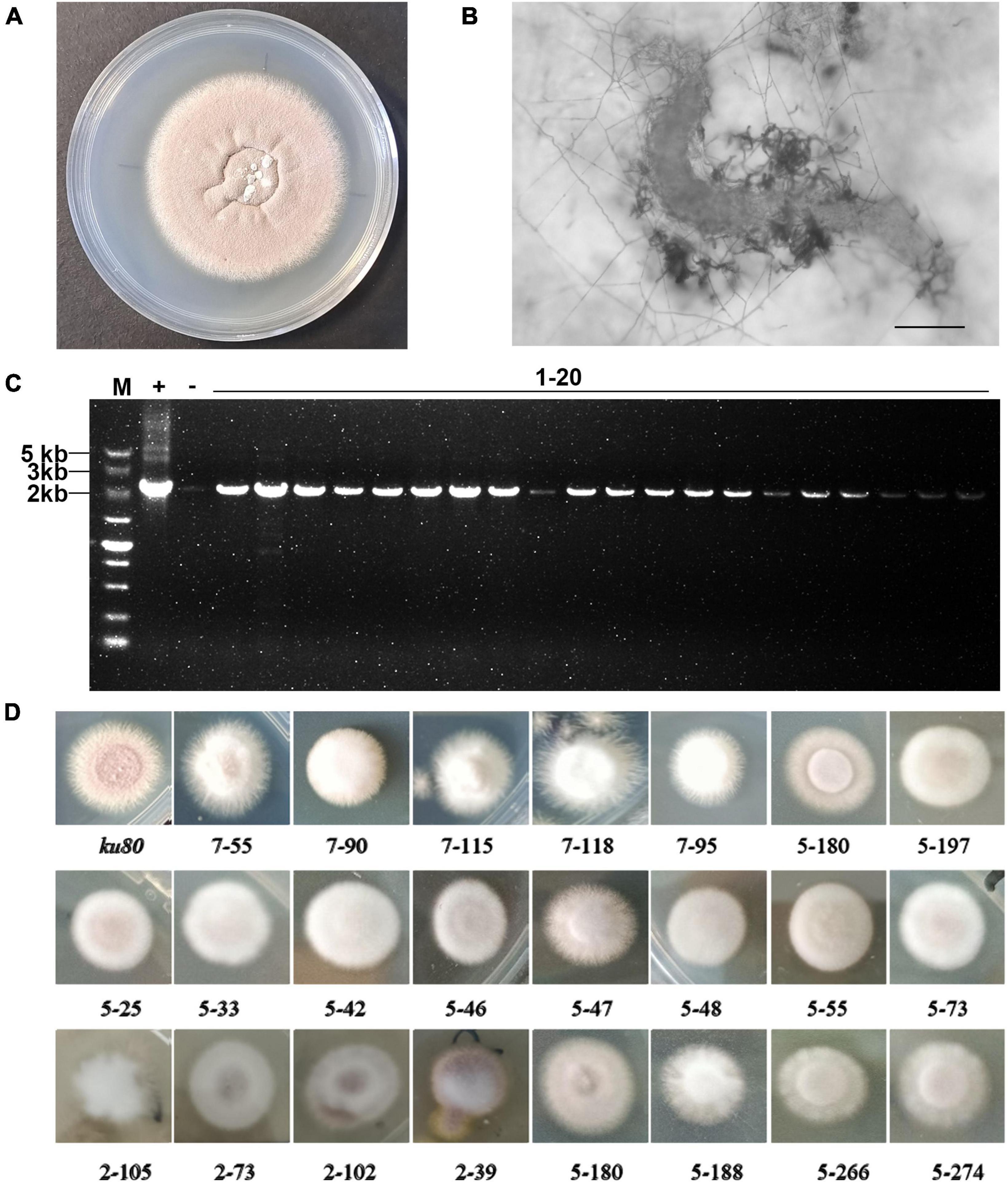
Figure 1. Construction and screening of the T-DNA random insertion library. (A) The purple-color colony of the P. lavendulum ku80 strain. (B) A nematode infected by P. lavendulum (scale bar, 50 μm). (C) T-DNA insertion in the transformants was confirmed by PCR amplification of the sur gene. M, DNA marker. + PCR with the plasmid pPK2-sur-GFP as template. –, PCR with the ku80 genomic DNA as template. (D) Colony morphology of ku80 and conidiation defect strains.
Conidiation, the formation of asexual conidia, is a crucial process employed by P. lavendulum for infecting nematodes, with conidia serving as the infectious propagule in this interaction (Figure 1B). However, research on conidiation regulatory genes and mechanisms in P. lavendulum is still limited. Our previous studies identified three genes involved in P. lavendulum conidiation: PlbrlA, PlabaA, and PlwetA (Chen et al., 2020). These genes respectively control the early, middle, and late stages of asexual development. Deletion of PlbrlA completely inhibits the formation of conidia, resulting in the production of conidiophores without conidial formation. PlabaA determines the differentiation of conidia on conidiophores. PlwetA affects various phenotypes related to conidial maturation, including conidial detachment from conidiophores, cell wall thickening, vacuole formation, trehalose production, and heat shock tolerance, among others (Chen et al., 2020). In addition, conidiation of P. lavendulum was also found be co-regulated by nitrogen sources and histone H3K14 acetylation (Tang et al., 2023). While the core regulatory genes and a few upstream genes have been defined, the genes that they interact with to impact conidiation remain largely unknown.
Mutant library construction and screening is an effective approach for studying unknown genes and their biological functions. In recent years, Agrobacterium-mediated genetic transformation to construct T-DNA random insertion mutant libraries has been widely used to study filamentous fungi, such as Aspergillus terreus, Colletotrichum higginsianum, and Trichoderma reesei (Yao Hua et al., 2007; Wang et al., 2014; Tanaka et al., 2023). In this study, we utilized Agrobacterium-mediated genetic transformation to construct a T-DNA random insertion mutant library of P. lavendulum. Screening of the library led to the identification of two conidiation mutants that we further characterized using targeted gene knockout and complementation.
2 Materials and methods
2.1 Strains and plasmids
Purpureocillium lavendulum ku80, a ku80 gene knockout strain with a high homologous recombination rate, was used as the parental strain for mutant library construction and gene knockout (Liu et al., 2019). E. coli DH5α competent cells were used to construct knockout plasmids. Agrobacterium tumefaciens AGL-1 was used for fungal transduction. The pPK2-sur-GFP plasmid was kindly provided by Professor Fang Weiguo from Zhejiang University, and it was used for constructing the mutant library of P. lavendulum (Xu et al., 2014).
2.2 Agrobacterium-mediated genetic transformation of P. lavendulum
Fungal transformation followed a previously described method (Zhuang et al., 2023). Briefly, the pPK2-sur-GFP plasmid was transferred into Agrobacterium tumefaciens AGL-1 competent cells using the liquid nitrogen freezing method described in the reference manual. The Agrobacterium cells harboring the pPK2-sur-GFP plasmid were inoculated into LB liquid medium and incubated on a shaker at 28°C until reaching an optical density (OD) of 0.45. At this point, the Agrobacterium culture was mixed with P. lavendulum conidia at a 1:1 ratio, avoiding light exposure. The mixture is evenly spread on IM medium supplemented with acetosyringone and covered with a microporous filter membrane. The plates were incubated inverted at 22°C for 48 hours. Afterward, the microporous filter membrane was transferred to M-100 medium (with 50 μg/ml chlorsulfuron) and incubated at 28°C for 4–6 days until transformants grow out. A 20 mutant strains were randomly selected for confirm of transformation analysis by PCR amplifying sulfonylurea resistance gene (sur) with the primers surF/surR (Supplementary Table 1).
2.3 Screening of the mutant library of P. lavendulum
The obtained transformants were subjected to screening by inoculating them on PDA agar plates and incubation at 28°C for 4 days and comparing their colony morphologies. The conidia of P. lavendulum are purple, and the wild-type strain (ku80) exhibits a high conidiation rate, resulting in colonies with a purple color. Therefore, to identify transformants with reduced conidiation, the ones showed pale purple or white colonies were selected.
2.4 Identification of the T-DNA insertion sites
Two strategies were utilized for identifying the T-DNA insertion sites, genome walking and genome re-sequencing, in the target strains with reduced conidiation. In the genome walking method, the Takara Genome Walking Kit (Takara, Catalog Number: 6108) was used for three rounds of PCR amplification using primers in the kit and designed according to the T-DNA sequence (Supplementary Table 1). The obtained bands were sequenced and compared using Local BLAST in the software BioEdit to identify fragments that match the P. lavendulum genome (Hall, 1999). By aligning these fragments with the T-DNA sequence, potential insertion sites of the T-DNA were identified.
Selected strains with reduced conidiation were subjected to whole-genome re-sequencing. The reads partially mapped to T-DNA were mapped to the reference genome and thus the potential insertion position was identified. After that, primers were designed for PCR reactions to validate the predicted T-DNA insertion site and determine the precise insertion point in each mutant (primers were listed in Supplementary Table 1).
2.5 Gene knockout and complementation
Gene knockouts were carried out by homologous recombination. The homologous arms were amplified with primers in Supplementary Table 1, and then inserted into the plasmid ppk2-sur-gfp by infusion method, at the XbaI/Bg1II and SpeI/EcoRV sites, respectively. To construct the gene complementation plasmid of Plpif1, the complete gene, including the open reading frame (ORF) region, along with 2kb upstream and 0.5 kb downstream regions, was amplified by PCR and then ligated to the ppk2-bml-GFP plasmid [reported previously(Liu et al., 2019)]. The plasmid was then transformed into the fungus using Agrobacterium-mediated transformation method. Transformants grew on screening plates were then confirmed by PCR (primers were listed in Supplementary Table 1).
2.6 Morphology observation and comparison
After obtaining the knockout strains as well as the complementation strain, they were cultured on regular solid media MM, TG, and CMA, as well as on stress-inducing media (MM with various concentrations of SDS, NaCl, or H2O2). The growth rates and conidia production were determined and compared with the ku80 strain.
2.7 Real-time quantitative PCR
The expression levels of conidiation regulation genes previously reported (Chen et al., 2020), such as PlfluG, PlflbA, PlbrlA, et.al., were selected to measure their expression levels with real-time quantitative PCR. RNA was extracted from mycelia of ku80 and the two mutant strains using RaPure Total RNA Micro Kit (Magen, Guangzhou, China). Real-time quantitative PCR were carried out by SYBR-Green method with KAPA SYBR FAST qPCR Kit (Roche) on the LightCycler-® 480 Instrument II (Roche). The actin gene was used as an internal reference. The primers used were listed in Supplementary Table 1.
3 Results
3.1 Establishment of a random T-DNA insertion mutant library in P. lavendulum using ATMT
Using the P. lavendulum ku80 strain as the parental strain, genetic transformation mediated by Agrobacterium tumefaciens was employed to introduce the T-DNA of the plasmid pPK2-sur-GFP into the ku80 genome, constructing a random T-DNA insertion mutant library of P. lavendulum. After multiple batches of genetic transformation, a total of 4,870 mutant strains were obtained. To evaluate the efficiency of T-DNA insertion into the P. lavendulum genome, 20 mutant strains were randomly selected for analysis. PCR results (The target band size amplified by the designed primers was 2145 bp) showed that all 20 randomly selected mutant strains had T-DNA insertions (Figure 1C). This indicated that the success rate of using Agrobacterium-mediated transformation in P. lavendulum was relatively high.
3.2 Identification, screening, and preservation of the random T-DNA insertion mutant library in P. lavendulum
A total of 2650 transformants were screened for defect in conidiation. Among them, 131 strains showed reduced conidiation and increased white mycelium production. 37 strains had colony diameters significantly larger or smaller than the parental ku80 strain when grown on the same culture medium. The colonies of some representative strains are shown in (Figure 1D). From the figure, we can see that the colony of ku80 was purple on PDA medium, with a powdery appearance due to abundant conidia production. Most of the screened mutants exhibited reduced conidiation, increased white or pale purple mycelium, irregular colony morphology with serrated edges or uniform extension of mycelium, while some colonies were round and full, fluffy, and exhibited a delicate mycelial appearance instead of a powdery appearance. The obtained mutant strains were classified based on phenotypes to establish a random mutant library of P. lavendulum.
Large-scale preservation of strains is crucial for the repeated utilization of the mutant library. The following preservation methods for mutant strains of P. lavendulum were selected and tested: (1) 25% glycerol at −80°C, (2) 50% glycerol at −80°C, and (3) deionized water at 4°C. To determine the best preservation method among the three mentioned above, 1 mL (1 × 108ml–1) conidia of the ku80 strain was preserved in each cryovial using the three preservation methods. One month later, three cryovials were taken out from each preservation method, and 100 μL of the thawed suspension was uniformly spread on MM medium. The results after 3 days of incubation is shown in Supplementary Figure 1A. It can be seen from the figure that the conidia preserved in 50% glycerol showed significantly slower germination and growth rates compared to the other two methods after one month of preservation. After six months, the same procedure was repeated with three cryovials from each preservation method, and the results are shown in Supplementary Figure 1B. It can be observed that conidia preserved using all three methods could germinate normally after six months, but the conidia preserved in 50% glycerol still exhibited slower germination and growth rates compared to the other two methods. Based on these results, we selected 25% glycerol at -80°C for preserving the mutant strains.
3.3 Identification of T-DNA insertion sites and affected genes
The T-DNA insertion site in strain #5-197 was identified in the gene contig1.1577 of P. lavendulum by whole-genome re-sequencing and was further confirmed through PCR. Through sequence BLAST in GenBank, its possible function was identified as an α, β hydrolase folds family protein (VFPFJ_05724) (Sequence ID: XM-018322804.1). This gene was named Plhffp.
The T-DNA insert site of strain #5-119 was identified by genome walking method. After sequencing and alignment, the PCR product was found to be 1388 bp in length. Among these, 834 bp matched the contig9 of the P. lavendulum genome, while 544 bp matched the T-DNA sequence. After PCR verification, the gene affected by the T-DNA insertion site was finally determined. Through Blast against the GenBank, its possible function was identified as ATP-dependent DNA helicase PIF1 (partial mRNA of PIF1) (VFPFJ_11645) (Sequence ID: XM-018328712.1). Therefore, this gene was named Plpif1.
3.4 The deletion of the Plhffp gene reduces conidiation and weakens cell wall synthesis and antioxidant capacity
To determine the function of the two genes, we performed targeted gene knockout and complementation experiments. Unfortunately, Plhffp could not be complemented due to its high GC content, which made PCR amplification impossible (Figure 2). The growth rates of ΔPlhffp on MM, TG, and CMA media were not significantly different from the ku80 strain (Supplementary Figure 2). However, the conidiation yield of ΔPlhffp was significantly decreased compared to ku80 on the MM medium (Figure 3A). The germination rate of the mutant was similar to the ku80 strain when cultured in MM medium (Figure 3B). This indicates that the knockout of the Plhffp gene affected conidiation without affecting the normal growth of the strain. On high osmolarity media, the growth rate of ΔPlhffp was not affected (Supplementary Figures 3A, C, E), indicating that the deletion of this gene does not impact the strain’s ability to resist high osmolarity. However, on media containing a high concentration of NaCl (0.3M), although the growth rate of ΔPlhffp was similar to ku80, the conidiation yield decreased significantly compared to ku80 (Supplementary Figure 3F). Both growth and conidiation of the ΔPlhffp mutant were lower than that of the ku80 strain when growing on MM media with 0.01–0.03% SDS, although not all of the differences were statistically significant (Supplementary Figure 4). High concentration of H2O2 also affected the growth and conidia of the ΔPlhffp mutant. The conidia production was significantly lower in the mutant than in ku80 on 2.5 and 5 mM H2O2 (Supplementary Figure 5). It is speculated that deletion of the Plhffp gene may affect the integrity of the strain’s cell wall and its antioxidant capacity.
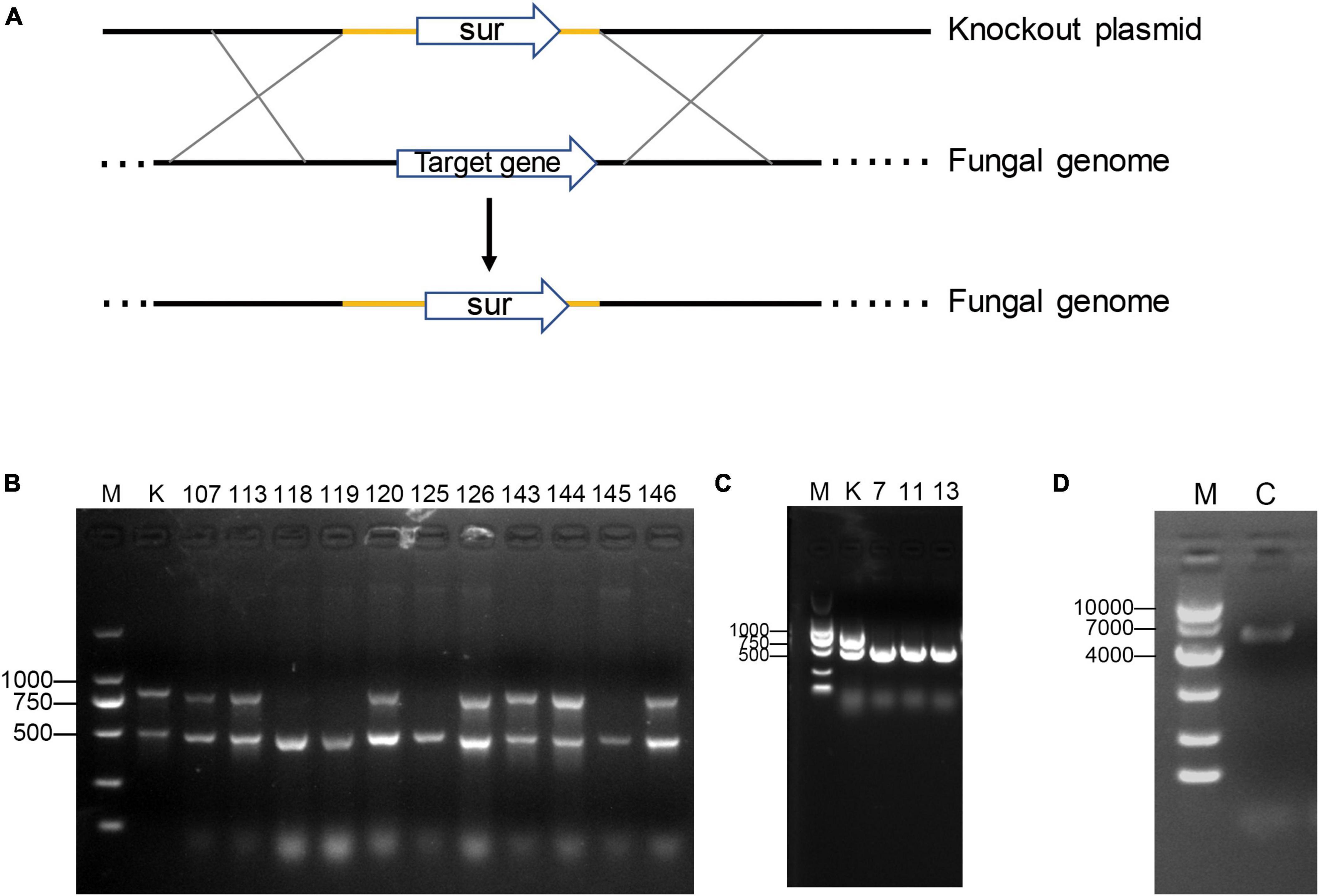
Figure 2. Gene knockout and complementation. (A) The Pattern diagram of homologous recombination knockout genes. (B) PCR screening of Plhffp mutants by amplifying the Plhffp gene (the upper band) and the beta-tubulin gene (the lower band). K, ku80. (C) PCR screening of Plpif1 mutants by amplifying the Plpif1 gene (the upper band) and the beta-tubulin gene (the lower band). K, ku80. (D) PCR confirmation of the complementation of Plpif1 gene.
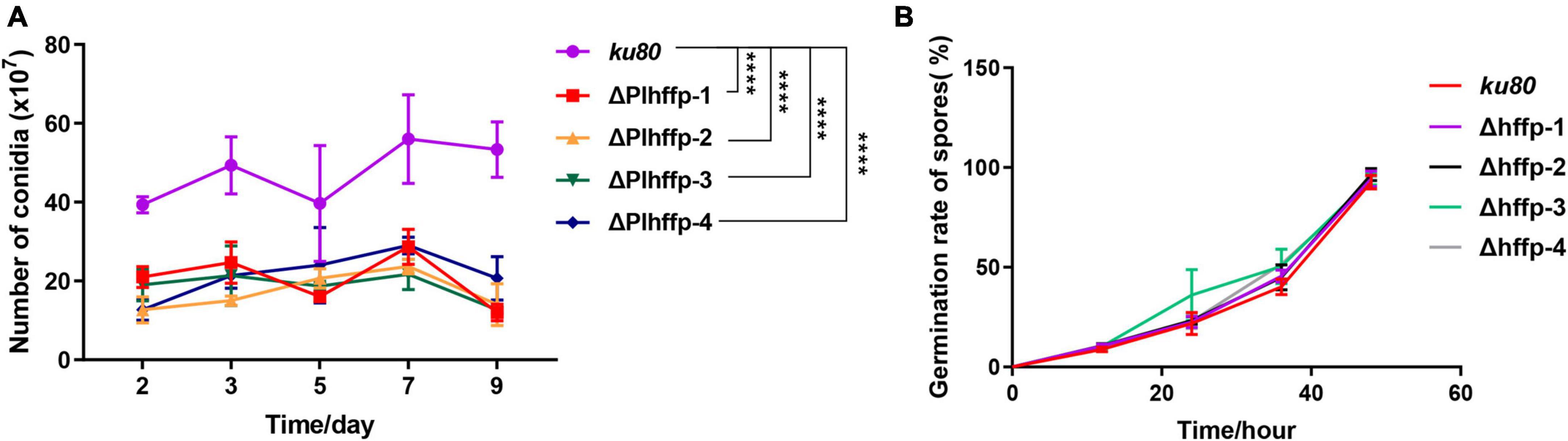
Figure 3. Comparison of conidiation (A) and conidia (B) germination of ku80 and the ΔPlhffp strains. ****, p < 0.0001.
3.5 The deletion of the Plpif1 gene reduces conidiation and weakens antioxidant capacity
The growth rates of ΔPlpif1 and the complementary strain ΔPlpif1-C on MM, TG, and CMA media were not significantly different from the ku80 strain (Figure 4). This indicates that the knockout of the Plpif1 gene does not affect the normal growth of the strain. However, it is worth noting that deletion of the Plpif1 gene significantly decreased conidiation on MM, TG, and CMA media compared to ku80. In addition, after complementation of the Plpif1 gene, the conidiation yield was restored and showed no significant difference compared to ku80 (Figure 4). This indicates that the knockout of the Plpif1 gene affects conidiation without impacting the normal growth of the strain. On high osmolarity media (with 0.1–0.3 M NaCl), the growth rates of ΔPlpif1 and ΔPlpif1-C were consistent with ku80, showing no significant differences. However, the conidiation yield of ΔPlpif1 was significantly lower than the control, while the conidiation yield of ΔPlpif1-C was restored to a level similar to ku80 (Figure 5). Similar to ΔPlhffp, on H2O2 media, when the H2O2 concentration were 1 and 2.5 mM, ΔPlpif1 strain grew similar to the ku80 and the complementation strain, while conidiation was significantly decreased. When grew on 5 mM H2O2 medium, the ΔPlpif1 mutant strain could hardly grow. However, after complementation of the Plpif1 gene, the growth rate no longer showed a significant difference compared to ku80 (Figure 6). This indicates that the deletion of the Plpif1 gene, like the deletion of the Plhffp gene, reduces the antioxidant capacity of the fungus.
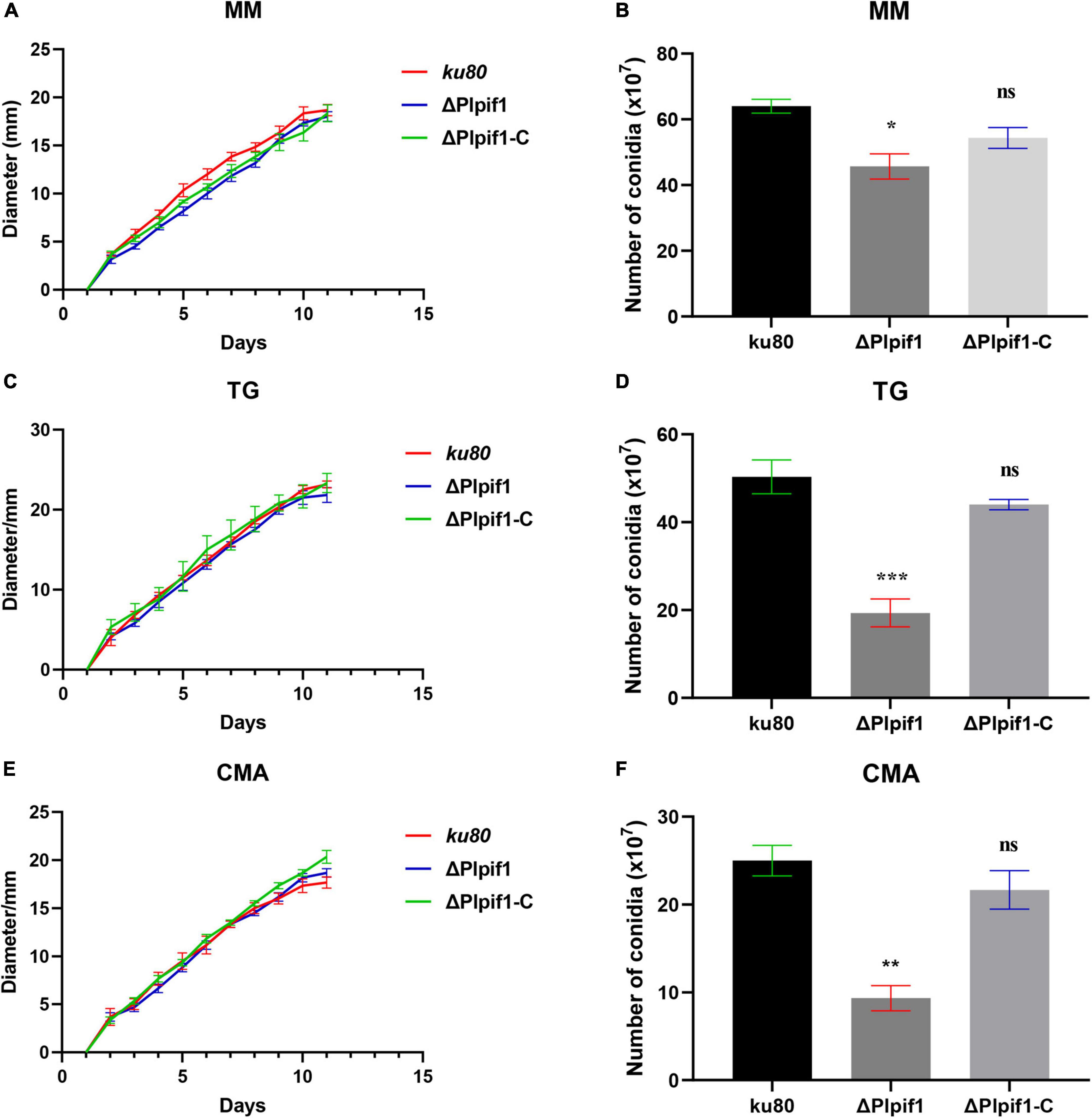
Figure 4. Comparison of growth and conidiation of ku80, the ΔPlpif1 and ΔPlpif1-C strains on different media. (A,B) MM media; (C,D) TG media; (E,F) CMA media. ΔPlpif1-C, the complementary strain of ΔPlpif1.*, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not statistically significant.
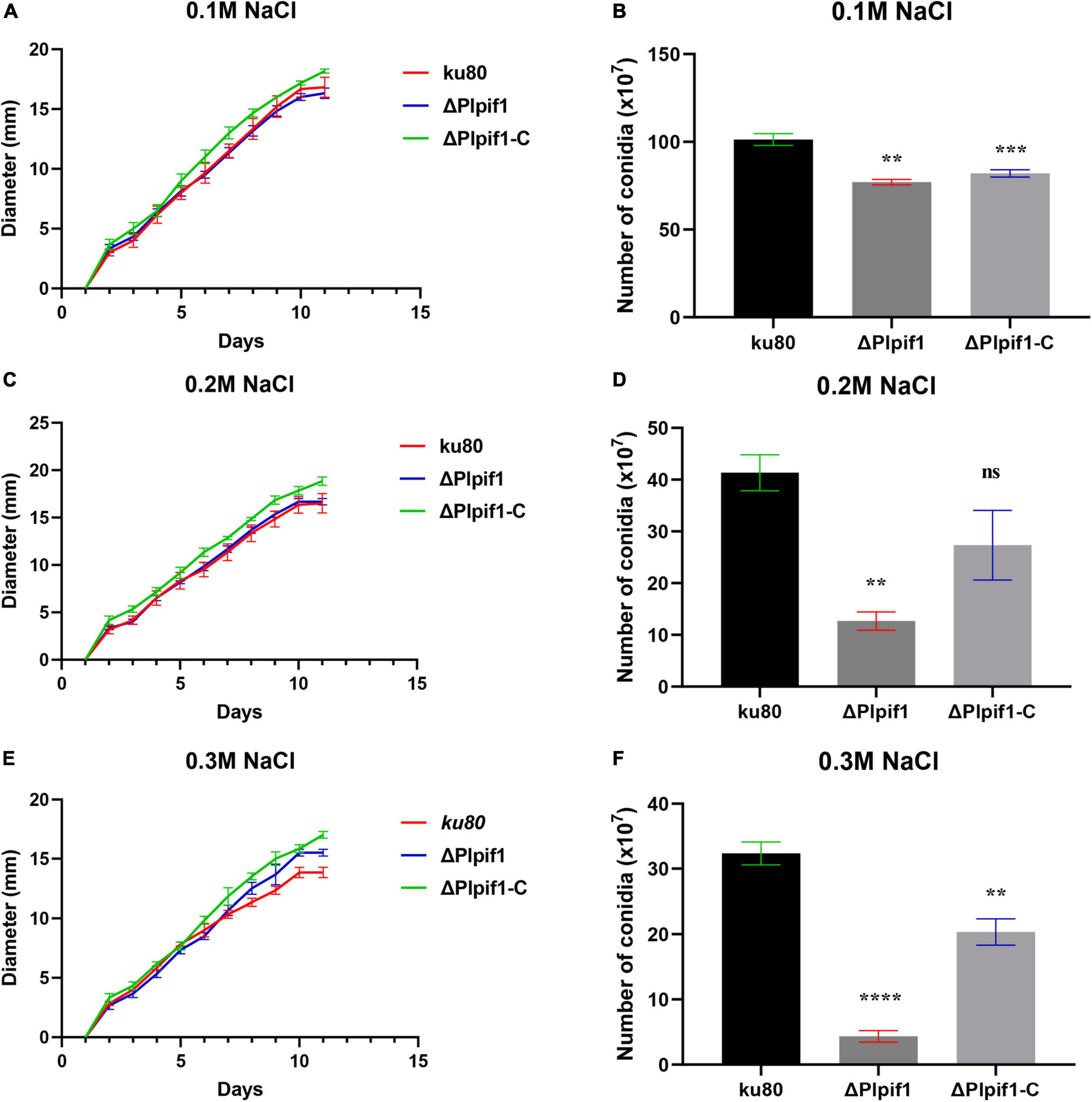
Figure 5. Comparison of growth and conidiation of ku80, the ΔPlpif1 and ΔPlpif1-C strains on high osmotic media. (A,B) media with 0.1 M NaCl; (C,D) media with 0.2 M NaCl; (E,F) media with 0.1 M NaCl. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, not statistically significant.
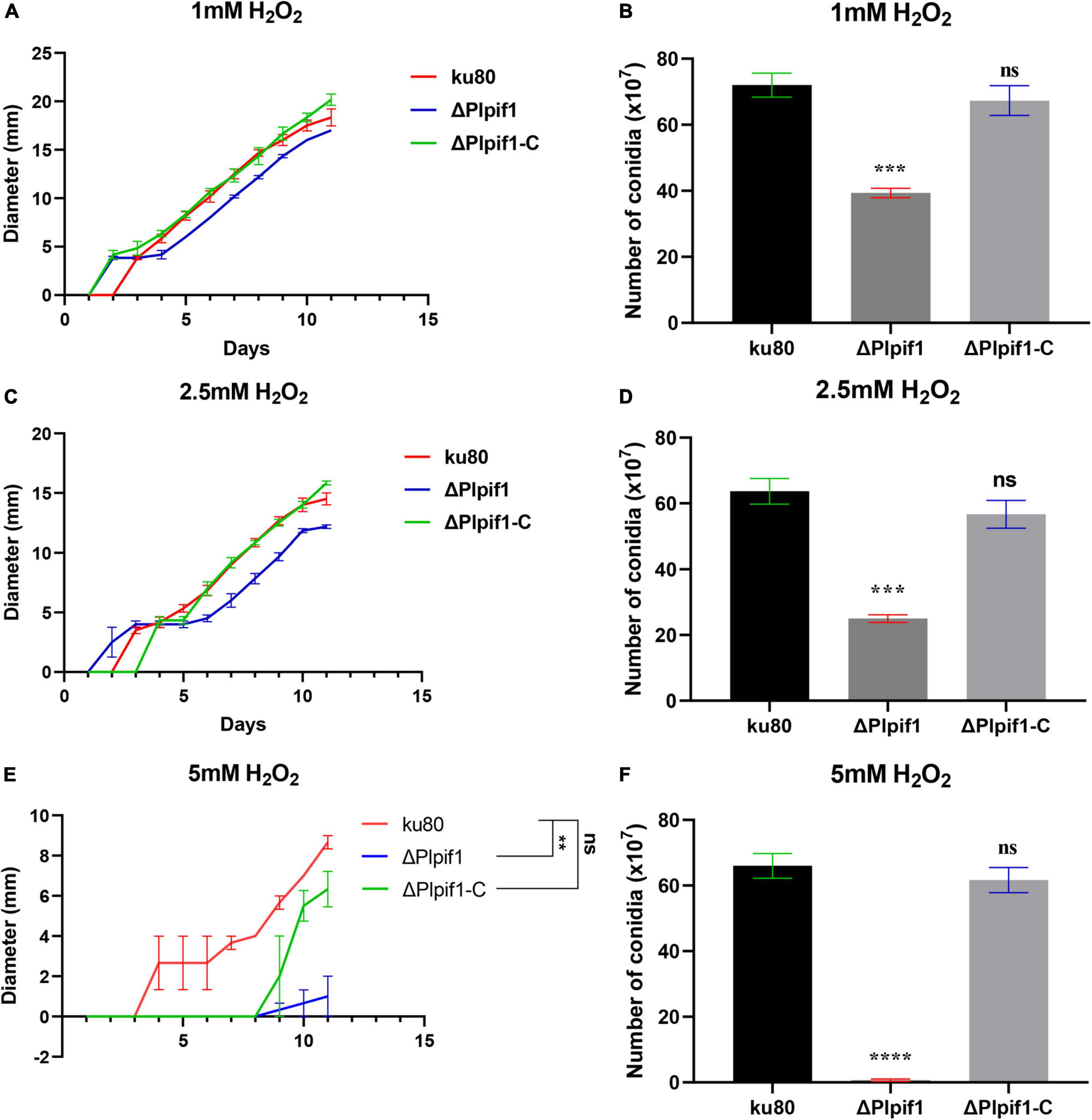
Figure 6. Comparison of growth and conidiation of ku80, the ΔPlpif1 and ΔPlpif1-C strains on H2O2 containing media. (A,B) media with 1 mM H2O2; (C,D) media with 2.5 mM H2O2; (E,F) media with 5 mM H2O2. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, not statistically significant.
3.6 Deletion of the Plhffp and Plpif1 genes affects the expression of key conidiation-related genes
Our previous study showed nine genes, PlabaA, PlbrlA, PlfadA, PlflbA, PlflbC, PlflbD, PlfluG, PlpkaA, and PlwetA to be associated with conidiation in P. lavendulum (Chen et al., 2020). Among them, three PlbrlA, PlabaA, PlwetA represented a central regulatory pathway for conidiation. To investigate the changes in the expression levels of these nine conidiation-related genes in the ΔPlhffp and ΔPlpif1 mutants, RT-qPCR was conducted to measure the expression levels of the nine genes, aiming to find the potential connection between the two deleted genes and these nine conidiation-related regulatory genes.
According to the experimental results (Figure 7), in ΔPlhffp mutant, the expression level of PlbrlA, which mainly regulates the production of conidia in P. lavendulum, was significantly decreased compared to the wildtype strain. In contrast, the expression level of PlflbD was significantly increased in ΔPlhffp mutant, consistent with its role of a negative regulator for conidiation in P. lavendulum. It is speculated that the Plhffp gene regulates conidiation in P. lavendulum through coordinated regulation of PlbrlA and PlflbD genes. Similarly, in the ΔPlpif1 mutant, the expression level of PlflbD also significantly increased, consistent with its inhibitory effect on conidiation in P. lavendulum.
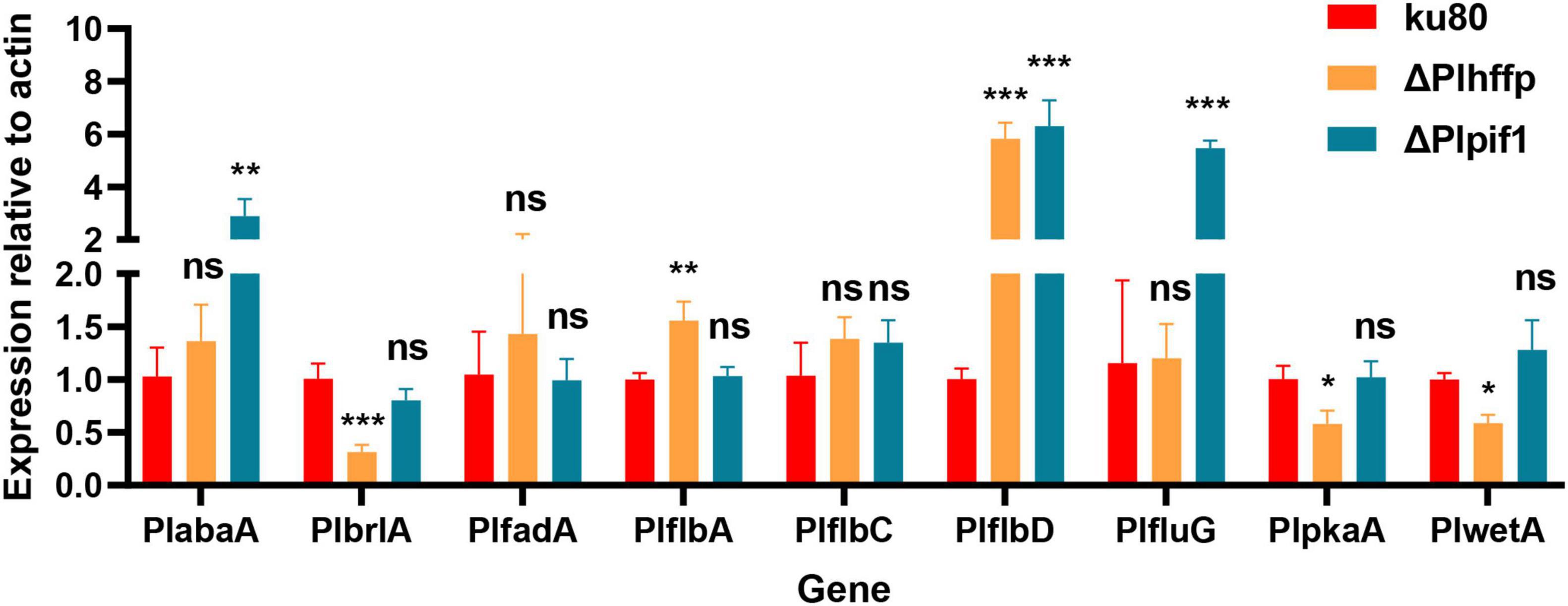
Figure 7. The expression levels of previously reported conidiation-regulating genes in ku80 and the two mutants. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not statistically significant.
3.7 The two conidiation-regulating genes do not directly affect the nematocidal activity of the fungus
The nematocidal activities of the ΔPlhffp and ΔPlpif1 mutants were measured and compared with the original strain ku80. Interestingly, there was no statistically significant difference in insecticidal activity between these two mutants and strain ku80 (Supplementary Figure 6). However, considering that the experiments were conducted with an equal number of conidia, while the actual conidiation yields of ΔPlhffp and ΔPlpif1 mutants were significantly lower than that of ku80, it can be concluded that the deletion of the Plhffp and Plpif1 genes indirectly reduces the nematocidal activity of P. lavendulum.
4 Discussion
Purpureocillium lavendulum is an important nematode biocontrol fungus with the potential to be developed into a biological nematicide. The production and regulation of conidia (asexual spores) are crucial for the nematocidal activity of P. lavendulum. In this study, to investigate the regulatory mechanisms associated with conidial production in P. lavendulum, we constructed a T-DNA mutant library. Through screening the mutant library, we successfully identified two genes involved in the regulation of conidial production in this fungus. Gene knockout and complementation experiments confirmed their involvement in conidial production.
To construct a large number of T-DNA random insertion mutants, the fungus P. lavendulum ku80 strain was transformed using ATMT. The initial screening of these mutants focused on their colony morphology to identify those with potentially reduced conidiation. Out of 2650 mutants screened initially, 131 exhibited abnormal conidiation, characterized by increased white mycelium production. Based on this result, we estimated that ∼5% of the genes in the entire genome of P. lavendulum may potentially affect conidia production. After the initial screening, two strategies, namely, second-generation sequencing and genome walking, were employed to identify the T-DNA insertion sites. These two approaches have been proven effective in other studies (Cottage et al., 2001; Chen et al., 2021; Edwards et al., 2022).
One mutant strain, #5-197, displayed white colonies, which remained unchanged even after several months of cultivation in test tubes, indicating low conidiation. The T-DNA insert site identified through whole-genome second-generation sequencing and confirmed by PCR, was predicted to disrupt an α, β-hydrolase-fold family protein, Plhffp. The α, β-hydrolase fold superfamily is an ancient and diverse group of major hydrolytic enzymes. The core structure of α, β-hydrolases consists of β-folds surrounded by α-helices (Wullich et al., 2020). These proteins are commonly found in bacteria and fungi. For example, 105 α, β-hydrolases have been identified in the human pathogenic bacterium Mycobacterium tuberculosis and many are involved in lipid metabolism, particularly in the biosynthesis and maintenance of its unique cell membrane. α, β-hydrolase fold proteins in M. tuberculosis also participate in immune evasion, immune regulation, detoxification, and metabolic adaptation. They play roles in growth, response to intracellular acidification, and dormancy (Johnson, 2017).
The growth of ΔPlhffp mutant on SDS-containing medium was retarded, suggesting that the deletion of the Plhffp gene may partially affect the integrity of the cell wall. Lipids are minor components of fungal cell walls and, in some cases, can participate in cell signaling (Beccaccioli et al., 2019). The homologous function of Plhffp is mainly associated with lipid metabolism (Lord et al., 2013), so it is speculated that the deletion of the Plhffp gene may affect the normal metabolism of some lipids in the cell wall, disrupting the integrity of the cell wall and reducing its ability to resist adverse environments. This is also evident in oxidative culture medium: when the concentration of H2O2 was 1 mM or 2.5 mM, the growth rate of ΔPlhffp showed no difference compared to the control, but when the H2O2 concentration increased to 5 mM, the growth of ΔPlhffp was significantly inhibited, and some plates did not produce any visible colonies. At H2O2 concentrations of 10 mM and 15 mM, ΔPlhffp failed to germinate and grow. In terms of conidiation, at concentrations of 1 mM, 2.5 mM, and 5 mM, the conidiation of ΔPlhffp was significantly lower than that of ku80. This indicates that the deletion of this gene severely affects the strain’s antioxidant capacity. Fungal cell walls are composed of complex components that may contain antioxidant substances such as glutathione, creating a suitable redox environment for the fungus. Therefore, it is hypothesized that the deletion of the Plhffp gene not only affects cell wall lipid metabolism but also reduces the content of antioxidant components in the cell wall, leading to a decreased ability to resist high oxidative conditions.
On a high-salt (NaCl) culture medium, the growth rate of ΔPlhffp was similar to ku80, indicating that the deletion of the Plhffp gene did not affect the strain’s ability to tolerate high osmolarity. In terms of conidiation, there was no significant difference in spore production between ΔPlhffp mutant and ku80 on 0.1 M and 0.2 M NaCl media. Considering the significantly lower conidiation of the ΔPlhffp mutant compared to ku80 on regular culture medium, it is speculated that the presence of NaCl creates a relatively unfavorable environment that triggers a response in ΔPlhffp mutant to counteract the adverse conditions. Conidiation is a fungal strategy to cope with unfavorable environments, and the increased conidiation in ΔPlhffp mutant may be a result of this response. As the NaCl concentration reached 0.3 M, there was no significant difference in colony size compared to ku80, but the final colony diameter of ΔPlhffp mutant was smaller, and the conidiation was significantly reduced compared to ku80. This suggests that the 0.3 M NaCl concentration has exceeded the threshold that the knockout strain can tolerate, thus affecting both colony growth and conidiation to some extent.
The results of RT-qPCR showed that in the ΔPlhffp mutant, the expression levels of three genes, PlbrlA, PlpkaA, and PlwetA, were significantly downregulated. The absence of PlbrlA inhibits conidial formation (Chen et al., 2020), PlpkaA is a suppressor gene for conidial production in P. lavendulum and other fungi (Baltussen et al., 2019), and PlwetA regulates conidial detachment, cell wall thickening, and other phenotypes related to conidial maturation (Etxebeste et al., 2010). The significant decrease in the expression levels of these three genes overall inhibits the production of conidia in P. lavendulum. On the other hand, the expression levels of two genes, PlflbA and PlflbD, were significantly upregulated. FlbA is an important upstream activating gene for conidial formation in Aspergillus nidulans, but its deletion in P. lavendulum does not affect conidiation (Chen et al., 2020). Further research is needed to understand its role in P. lavendulum. PlflbD suppresses conidial production in P. lavendulum. The significant upregulation of these two genes inhibits conidial production in P. lavendulum. The expression levels of PlfadA and PlflbC showed a slight increase but were not statistically significant. Both genes are suppressor genes for conidial production in P. lavendulum, and their partially increased expression levels have an inhibitory effect on conidial production.
After validating the results of genome walking, the T-DNA insertion site of mutant #5-119 was determined as an ATP-dependent DNA helicase. In wheat, knocking out the ATP-dependent DNA helicase gene (TaDHL-7B) using the CRISPR/Cas9 system resulted in reduced plant height and increased lodging resistance, while the yield remained unchanged (Guo et al., 2022). In the fission yeast, the ATP-dependent DNA helicase CHL1 can unfold DNA:RNA and DNA:DNA substrates in vitro. CHL1 plays an important role in maintaining genome stability by participating in DNA repair and sister chromatid cohesion (He et al., 2022). In eukaryotes, the Pif1 helicase family has multiple roles in maintaining nuclear DNA and mitochondrial DNA integrity. The ScPif1 (S. cerevisiae Pif1) helicase in the budding yeast Saccharomyces cerevisiae is involved in replication by participating in replication barriers such as G-quadruplexes and protein blocks, reducing genetic instability at these sites. ScPif1 also regulates telomerase, promotes Okazaki fragment processing, and works with polymerase δ in break-induced repair (Geronimo and Zakian, 2016). Another Pif1 helicase in the budding yeast, Rrm3, assists fork progression at replication fork barriers on rDNA loci and tRNA genes. (Byrd and Raney, 2017). In both budding and fission yeasts, the Pif1 helicase family affects both the semiconservative replication of telomeric DNA mediated by telomerase and the recombinational elongation of telomeres (Geronimo and Zakian, 2016).
The growth rate of ΔPlpif1 mutant was not significantly different from ku80 and the complementation strain on MM, TG, and CMA media, indicating that the deletion of the Plpif1 gene does not affect the normal growth of the strain. The deletion of the Plpif1 gene resulted in significantly decreased conidiation in all the three media. Complementation of the deletion mutant rescued that phenotype. On a culture medium containing NaCl, there were no significant differences in colony morphology, growth rate, or conidiation between the ΔPlpif1, ΔPlpif1-C and ku80 strain. This indicates that the knockout of the Plpif1 gene does not affect the strain’s ability to regulate osmotic pressure. On a medium containing SDS, the colony size and growth curve of the ΔPlpif1 strain were not significantly different from the ku80 strain, suggesting that the deletion of this gene does not affect the cell wall integrity of the strain.
On culture media with low concentration (1 mM) and median concentration (2.5 mM) of H2O2, the final colony diameter of the knockout strain was comparable to the control, but on a medium with a high concentration (5 mM) of H2O2, the colony diameter of the knockout strain was significantly smaller than the control. This indicates that the knockout strain has weaker antioxidant capacity. The conidiation of the knockout strain was similar to the control on 1 mM H2O2 medium but significantly lower than the control on 2.5 mM and 5m M H2O2 media. Overall, knockout of the Plpif1 gene mainly affects the strain’s conidiation but not growth and stress tolerance.
The results of RT-qPCR showed that the expression levels of PlbrlA were downregulated although the change was statistically not significant. The negative regulator PlflbD was found upregulated. Based on this, we hypothesize that Plpif1 may act on PlflbD, thereby inhibiting the expression of PlbrlA and suppressing conidiation. The specific interactions between these two genes require further investigations.
The above study identified two new genes involved in conidiation regulation in P. lavendulum, Plhffp and Plpif1. However, the regulatory mechanisms involving these two genes require further investigation. To comprehensively address conidiation regulation, more conidiation-deficient mutants in our library need to be analyzed.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI – PRJNA1066049.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
YW: Data curation, Investigation, Methodology, Writing—original draft. F-NQ: Investigation, Methodology, Writing—review and editing. Y-RX: Investigation, Methodology, Writing—review and editing. K-QZ: Funding acquisition, Supervision, Writing—review and editing. JX: Writing—review and editing. Y-RC: Funding acquisition, Methodology, Writing—review and editing. L-ML: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Writing—original draft, Writing—review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of the article. This work was supported in part by grants from the National Natural Science Foundation of China (32160045), the key Project of Yunnan Applied Basic Research (202201AS070052), the Applied Basic Research Foundation of Yunnan Province (202301AT070051), Yunnan Provincial Major Project for Basic Research (202201BC070004), Science and technology special project from southwest united graduate school of Yunnan province [202302CC4040021], Special fund of the Yunnan University “Double First-Class” construction, science and technology innovation base construction project (202307AB110011), and grants from the Young Academic and Technical Leader Raising Foundation of Yunnan Province (to L-ML) and the Ten-thousand Talents Plans for Young Top-notch Talents of Yunnan Province (YNWR-QNBJ-2018-135 and YNWR-QNBJ-2018-011).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1352989/full#supplementary-material
References
Baltussen, T., Zoll, J., Verweij, P. E., and Melchers, W. (2019). Molecular Mechanisms of Conidial Germination in Aspergillus spp. Microbiol. Mol. Biol. Rev. 84, e49–e19.
Beccaccioli, M., Reverberi, M., and Scala, V. (2019). Fungal lipids: biosynthesis and signalling during plant-pathogen interaction. Front. Biosci. 24:172–185. doi: 10.2741/4712
Byrd, A. K., and Raney, K. D. (2017). Structure and function of Pif1 helicase. Biochem. Soc. Trans. 45, 1159–1171.
Chen, M., Yang, H.-Y., Cao, Y.-R., Hui, Q.-Q., Fan, H.-F., Zhang, C.-C., et al. (2020). Functional characterization of core regulatory genes involved in sporulation of the nematophagous fungus Purpureocillium lavendulum. mSphere 5, e932–e920.
Chen, X., Dong, Y., Huang, Y., Fan, J., Yang, M., and Zhang, J. (2021). Whole-genome resequencing using next-generation and Nanopore sequencing for molecular characterization of T-DNA integration in transgenic poplar 741. BMC Genomics 22:329. doi: 10.1186/s12864-021-07625-y
Cottage, A., Yang, A., Maunders, H., de Lacy, R. C., and Ramsay, N. A. (2001). Identification of DNA sequences flanking T-DNA insertions by PCR-walking. Plant Mol. Biol. Rep. 19, 321–327.
Coyne, D. L., Cortada, L., Dalzell, J. J., Claudius-Cole, A. O., Haukeland, S., Luambano, N., et al. (2018). Plant-parasitic nematodes and food security in sub-Saharan Africa. Annu. Rev. Phytopathol. 56, 381–403.
Edwards, B., Hornstein, E. D., Wilson, N. J., and Sederoff, H. (2022). High-throughput detection of T-DNA insertion sites for multiple transgenes in complex genomes. BMC Genomics 23:685. doi: 10.1186/s12864-022-08918-6
Etxebeste, O., Garzia, A., Espeso, E. A., and Ugalde, U. (2010). Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol. 18, 569. doi: 10.1016/j.tim.2010.09.007
Geronimo, C. L., and Zakian, V. A. (2016). Getting it done at the ends: Pif1 family DNA helicases and telomeres. DNA Repair 44, 151–158. doi: 10.1016/j.dnarep.2016.05.021
Guo, B., Jin, X., Chen, J., Xu, H., Zhang, M., Lu, X., et al. (2022). DNA helicase (TaDHL), a Novel Reduced-Height (Rht) Gene in Wheat. Genes 13, 979.
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nuclc Acids Sympos. Ser.s 41, 95–98.
He, D., Du, Z., Xu, H., and Bao, X. (2022). Chl1, an ATP-Dependent DNA Helicase, Inhibits DNA: RNA Hybrids Formation at DSB sites to maintain genome stability in S. pombe. Int. J. Mol. Sci. 23, 6631. doi: 10.3390/ijms23126631
Johnson, G. (2017). The α/β Hydrolase Fold Proteins of Mycobacterium tuberculosis, with Reference to their Contribution to Virulence. Curr. Protein Pept. Sci. 18, 190–210.
Li, J., Zou, C. G., Xu, J. P., Ji, X. L., Niu, X. M., Yang, J. K., et al. (2015). Molecular Mechanisms of Nematode-Nematophagous Microbe Interactions: Basis for Biological Control of Plant-Parasitic Nematodes. Annu. Rev. Phytopathol. 53, 67–95. doi: 10.1146/annurev-phyto-080614-120336
Liang, L. M., Zou, C. G., Xu, J., and Zhang, K. Q. (2019). Signal pathways involved in microbe-nematode interactions provide new insights into the biocontrol of plant-parasitic nematodes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180317. doi: 10.1098/rstb.2018.0317
Liu, L., Cao, Y.-R., Zhang, C.-C., Fan, H.-F., Guo, Z.-Y., Yang, H.-Y., et al. (2019). An Efficient Gene Disruption System for the Nematophagous Fungus Purpureocillium lavendulum. Fungal Biol. 123, 274–282. doi: 10.1016/j.funbio.2018.10.009
Lord, C. C., Thomas, G., and Brown, J. M. (2013). Mammalian alpha beta hydrolase domain (ABHD) proteins: Lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochim. Biophys. Acta 1831, 792–802. doi: 10.1016/j.bbalip.2013.01.002
Montarry, J., Mimee, B., Danchin, E. G. J., Koutsovoulos, G. D., Ste-Croix, D. T., and Grenier, E. (2021). Recent Advances in Population Genomics of Plant-Parasitic Nematodes. Phytopathology 111, 40–48. doi: 10.1094/PHYTO-09-20-0418-RVW
Perdomo, H., Cano, J., Gene, J., Garcia, D., Hernandez, M., and Guarro, J. (2013). Polyphasic analysis of Purpureocillium lilacinum isolates from different origins and proposal of the new species Purpureocillium lavendulum. Mycologia 105, 151–161. doi: 10.3852/11-190
Rehman, S., Gupta, V. K., and Goyal, A. K. (2016). Identification and functional analysis of secreted effectors from phytoparasitic nematodes. BMC Microbiol. 16:48. doi: 10.1186/s12866-016-0632-8
Shao, H., Zhang, P., Peng, D., Huang, W., Kong, L. A., Li, C., et al. (2023). Current advances in the identification of plant nematode diseases: From lab assays to in-field diagnostics. Front. Plant Sci. 14:1106784. doi: 10.3389/fpls.2023.1106784
Tanaka, S., Yoshida, M., Natsume, H., Ohashi, Y., Takagi, H., Nishiyama, C., et al. (2023). Rapid detection of mutations and chromosomal rearrangements in T-DNA insertion mutants of Colletotrichum higginsianum using long-read whole-genome sequencing. J. Gener. Plant Pathol. 89, 206–210.
Tang, P., Han, J.-J., Zhang, C.-C., Tang, P.-P., Qi, F.-N., Zhang, K.-Q., et al. (2023). The growth and conidiation of Purpureocillium lavendulum are co-regulated by nitrogen sources and histone H3K14 Acetylation. J. Fungi 9, 325. doi: 10.3390/jof9030325
Wang, D., He, D., Li, G., Gao, S., Lv, H., Shan, Q., et al. (2014). An efficient tool for random insertional mutagenesis: Agrobacterium tumefaciens-mediated transformation of the filamentous fungus Aspergillus terreus. J. Microbiol. Methods 98, 114–118. doi: 10.1016/j.mimet.2014.01.007
Wullich, S. C., Martín, A. A. S., and Fetzner, S. (2020). An α/β -Hydrolase Fold Subfamily Comprising Pseudomonas Quinolone Signal-Cleaving Dioxygenases. Appl. Environ. Microbiol. 86, e279–e220.
Xu, C., Zhang, X., Qian, Y., Chen, X., Liu, R., Zeng, G., et al. (2014). A high-throughput gene disruption methodology for the entomopathogenic fungus Metarhizium robertsii. PLoS One 9:e107657. doi: 10.1371/journal.pone.0107657
Yao Hua, Z., Xiao Li, W., Tian Hong, W., and Qiao, J. (2007). Agrobacterium-mediated transformation (AMT) of Trichoderma reesei as an efficient tool for random insertional mutagenesis. Appl. Microbiol. Biotechnol. 73, 1348–1354. doi: 10.1007/s00253-006-0603-3
Keywords: Purpureocillium lavendulum, Agrobacterium tumefaciens – mediated transformation, T-DNA random insertion mutant library, conidiation genes, biocontrol
Citation: Wei Y, Qi F-N, Xu Y-R, Zhang K-Q, Xu J, Cao Y-R and Liang L-M (2024) Characterization of regulatory genes Plhffp and Plpif1 involved in conidiation regulation in Purpureocillium lavendulum. Front. Microbiol. 15:1352989. doi: 10.3389/fmicb.2024.1352989
Received: 09 December 2023; Accepted: 05 February 2024;
Published: 15 February 2024.
Edited by:
Abdelazeem Algammal, Suez Canal University, EgyptReviewed by:
Huan Peng, Chinese Academy of Agricultural Sciences, ChinaSuvendu Mondal, Bhabha Atomic Research Centre (BARC), India
Copyright © 2024 Wei, Qi, Xu, Zhang, Xu, Cao and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lian-Ming Liang, bGlhbmdsbUB5bnUuZWR1LmNu; Yan-Ru Cao, eWFucnVjYW8zQGFsaXl1bi5jb20=
 Yu Wei1
Yu Wei1 Jianping Xu
Jianping Xu Lian-Ming Liang
Lian-Ming Liang