
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 February 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1352851
This article is part of the Research Topic Opportunistic pathogens: pathogenesis and multi-drug resistance mechanisms View all 10 articles
 Saulo Henrique Rodrigues1†
Saulo Henrique Rodrigues1† Gustavo Dantas Nunes1†
Gustavo Dantas Nunes1† Gabriela Guerrera Soares1†
Gabriela Guerrera Soares1† Roumayne Lopes Ferreira1†
Roumayne Lopes Ferreira1† Marcelo Silva Folhas Damas1†
Marcelo Silva Folhas Damas1† Pedro Mendes Laprega1†
Pedro Mendes Laprega1† Rebecca Elizabeth Shilling1
Rebecca Elizabeth Shilling1 Leslie Camelo Campos2
Leslie Camelo Campos2 Andrea Soares da Costa1
Andrea Soares da Costa1 Iran Malavazi1
Iran Malavazi1 Anderson Ferreira da Cunha1
Anderson Ferreira da Cunha1 Maria-Cristina da Silva Pranchevicius1*
Maria-Cristina da Silva Pranchevicius1*Klebsiella aerogenes is an important opportunistic pathogen with the potential to develop resistance against last-line antibiotics, such as carbapenems, limiting the treatment options. Here, we investigated the antibiotic resistance profiles of 10 K. aerogenes strains isolated from patient samples in the intensive-care unit of a Brazilian tertiary hospital using conventional PCR and a comprehensive genomic characterization of a specific K. aerogenes strain (CRK317) carrying both the blaKPC-2 and blaNDM-1 genes simultaneously. All isolates were completely resistant to β-lactam antibiotics, including ertapenem, imipenem, and meropenem with differencing levels of resistance to aminoglycosides, quinolones, and tigecycline also observed. Half of the strains studied were classified as multidrug-resistant. The carbapenemase-producing isolates carried many genes of interest including: β-lactams (blaNDM-1, blaKPC-2, blaTEM-1, blaCTX-M-1 group, blaOXA-1 group and blaSHVvariants in 20-80% of the strains), aminoglycoside resistance genes [aac(6’)-Ib and aph(3’)-VI, 70 and 80%], a fluoroquinolone resistance gene (qnrS, 80%), a sulfonamide resistance gene (sul-2, 80%) and a multidrug efflux system transporter (mdtK, 70%) while all strains carried the efflux pumps Acr (subunit A) and tolC. Moreover, we performed a comprehensive genomic characterization of a specific K. aerogenes strain (CRK317) carrying both the blaKPC-2 and blaNDM-1 genes simultaneously. The draft genome assembly of the CRK317 had a total length of 5,462,831 bp and a GC content of 54.8%. The chromosome was found to contain many essential genes. In silico analysis identified many genes associated with resistance phenotypes, including β-lactamases (blaOXA-9, blaTEM-1, blaNDM-1, blaCTX-M-15, blaAmpC-1, blaAmpC-2), the bleomycin resistance gene (bleMBL), an erythromycin resistance methylase (ermC), aminoglycoside-modifying enzymes [aac(6’)-Ib, aadA/ant(3”)-Ia, aph(3’)-VI], a sulfonamide resistance enzyme (sul-2), a chloramphenicol acetyltransferase (catA-like), a plasmid-mediated quinolone resistance protein (qnrS1), a glutathione transferase (fosA), PEtN transferases (eptA, eptB) and a glycosyltransferase (arnT). We also detected 22 genomic islands, eight families of insertion sequences, two putative integrative and conjugative elements with a type IV secretion system, and eight prophage regions. This suggests the significant involvement of these genetic structures in the dissemination of antibiotic resistance. The results of our study show that the emergence of carbapenemase-producing K. aerogenes, co-harboring blaKPC-2 and blaNDM-1, is a worrying phenomenon which highlights the importance of developing strategies to detect, prevent, and control the spread of these microorganisms.
Klebsiella aerogenes, previously identified as Enterobacter aerogenes, is a Gram-negative bacterium belonging to the Enterobacteriaceae family. It can be found in various environments including water, soil, air, and the human digestive system as a commensal organism. However, this bacterium is also a significant opportunistic pathogen that has been associated with hospital-acquired diseases such as pneumonia, meningitis, skin, soft tissue and urinary tract infections (Chen et al., 2015).
β-lactam antibiotics are one of the most commonly prescribed drug classes with numerous clinical uses. These antibiotics are sub-classed as penicillins (these being the most commonly prescribed), cephalosporins, cephamycins, monobactams, and carbapenems (Sta Ana et al., 2021). Klebsiella aerogenes is intrinsically resistant to ampicillin, amoxicillin, first-generation cephalosporins, and cefoxitin due to the expression of a constitutive AmpC β-lactamase. AmpC type lactamases are also known as extended spectrum β-lactamases (ESBLs) and these provide resistance against the majority of β-lactam antibiotics, such as extended-spectrum cephalosporins and monobactams, with the exception of carbapenems and cephamycins (Davin-Regli and Pages, 2015; Castanheira et al., 2021).
The presence of ESBLs in K. aerogenes strains has been well-documented, leading to the use of carbapenems as a last-resort treatment for serious infections caused by these pathogens. However, previous studies have shown a high prevalence of antibiotic resistance to cephalosporins and carbapenems in clinically relevant K. aerogenes strains worldwide (Ma et al., 2020). These infections pose a significant public health challenge due to the limited treatment options available and their elevated mortality rates (Mulani et al., 2019; Denissen et al., 2022).
The main resistant mechanism of carbapenemase-producing K. aerogenes is the production of carbapenemases, although other mechanisms have been proposed, including overproduction of β-lactamases, efflux pumps, porin deficiency, and changes in penicillin-binding proteins (Pan et al., 2021). Carbapenem resistance in clinical isolates of Carbapenem Resistant Enterobacterales (CRE) is predominantly caused by the presence of these carbapenemases, especially Klebsiella pneumoniae Carbapenamase (KPC) and New Delhi Metallo-β-lactamase (NDM). blaKPC is commonly found on various plasmids like IncF-, IncI-, IncA/C-, IncX-, and IncR-type plasmids, while blaNDM is mostly associated with IncX3-type plasmids. These plasmids are easily transferable and can promote the dissemination of blaKPC and blaNDM through horizontal gene transfer among diverse bacterial populations spreading antibiotic resistance (Yuan et al., 2023).
Although carbapenemases such as KPC, NDM, and Imipenemase (IMP) have been detected in K. aerogenes, there are limited studies demonstrating the simultaneous presence of blaKPC and blaNDM genes. Here, we conducted an in-depth analysis of genes related to the antimicrobial resistance and mobile genetic elements in carbapenemase-producing K. aerogenes, found in Brazilian hospitals, to understand its genomic diversity. To the best of our knowledge, this paper is the first to report the simultaneous presence of both blaKPC and blaNDM in K. aerogenes isolated from clinical samples in Brazil.
A total of 10 K. aerogenes were isolated from clinical specimens and devices in the ICU and neonatal intensive care unit (NICU) at a tertiary care of a government hospital in Palmas, Tocantins, Brazil, between January 2017 and May 2020. The K. aerogenes strains were initially identified by the hospital’s clinical microbiology laboratory before being forwarded to the Central Public Health Laboratory of the State of Tocantins (LACEN/TO) for species confirmation and drug susceptibility testing. LACEN is a healthcare facility under the Brazilian Ministry of Health that receives samples for antimicrobial resistance surveillance.
Bacterial identification and determination of antibiotic susceptibility were carried out using the VITEK2 compact automated system (bioMerieux, Hazelwood, MO, USA). The susceptibility of the K. aerogenes isolates were tested against a panel of 16 antibiotics, which included ampicillin/sulbactam (SAM), piperacillin/tazobactam (TZP), cefuroxime sodium (CXM-S), cefuroxime axetil (CXM-AX), cefoxitin (FOX), ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), ertapenem (ETP), imipenem (IPM), meropenem (MEM), amikacin (AMK), gentamicin (GEN), ciprofloxacin (CIP), tigecycline (TGC), and colistin (CST). The findings were interpreted in accordance with the guidelines set forth by Clinical and Laboratory Standards Institute (CLSI, 2023). Phenotypic detection of carbapenemase production in K. aerogenes was carried out by modified Hodge test and ethylenediaminetetraacetic acid (EDTA) synergy tests under the CLSI guidelines (CLSI, 2023) as described elsewhere (Ferreira et al., 2019, 2020; Damas et al., 2022; Soares et al., 2023). K. aerogenes isolates were classified as multidrug-resistant (MDR) by non-susceptibility to at least one agent in three or more antimicrobial categories, as per the criteria established by Magiorakos et al. (2012). K. aerogenes are naturally resistant to ampicillin (AMP), amoxicillin/clavulanic acid (AMC), FOX, and cephalothin (CFL) due to the low production of the naturally induced cephalosporinase of Bush group 1 (class C) (Davin-Regli and Pages, 2015). Therefore, AMP and FOX were not included in the MDR classification (Magiorakos et al., 2012).
Klebsiella aerogenes strains were subcultured on Brain Heart Infusion (BHI) broth (Oxoid, United Kingdom) and incubated for 24 h at 37°C. Genomic DNA extraction was performed from an overnight culture using the Cellco Genomic DNA purification kit (Cellco Biotech., São Carlos, Brazil), according to the manufacturer’s instructions. The DNA was quantified using the NanoVue Plus instrument (GE Healthcare Life Sciences, Marlborough, MA, United States). The quality of the genomic DNA was examined through electrophoresis while the bacterial DNA concentration was determined using the Qubit® 3.0 fluorometer in combination with the Qubit® dsDNA Broad Range Assay Kit from Life Technologies (Carlsbad, CA, USA).
Polymerase chain reaction (PCR) was performed for the detection of resistance-related genes, such as ESBL-encoding genes (blaTEM, blaSHV variants, blaOXA-1, 4 and 30, blaCTX-M-1 group, blaGES, blaPER-1 and 3, blaVEB-1 to 6), carbapenemase genes (blaKPC, blaOXA-48, blaIMP-1, blaVIM-2, blaNDM, blaSPM-1, blaGIM-1, blaSIM-1), aminoglycosides [armA, rmtB, aph(3′)-VIa (aphA6)], tetracycline (tetB), sulfonamide (sul-1, sul-2), colistin resistance (mcr-1), plasmid mediated quinolone resistance (PMQR) gene [aac(6’)-Ib-cr, qnrS1 and qnrS2], efflux pump (acrAB, tolC, and mdtK) genes. Amplicons were analyzed by gel electrophoresis in 1.0% agarose and visualized under ultraviolet (UV) light. Supplementary Table S1 provides information on amplicons length and PCR conditions.
One amplicon of each studied gene was purified using the Gel Band Purification Kit (Cellco Biotech., São Carlos, Brazil) and sequenced using the Sanger DNA sequencing method. The sequences were edited using Bioedit v7.0.5 (Hall, 1999), then compared with GenBank and Refseq sequences using BlastX tools: ACT53230.1 (blaCTX-M-15), QXU68638.1 (blaTEM-1), EKZ5222878.1 (blaNDM), SCZ84112.1 (blaSHV-2), WEA84669.1 (blaKPC), WP_240093217.1 (blaOXA-1), WP_047046709.1 (tolC), EFZ4507594.1 (qnrS), MCL7674773.1 (acrA), HBS1035150.1 (aac-(6’)-Ib), HEC1006964.1 (aph(3’)-VIa), QDB65114.1 (sul-2), and PLP19006.1 (mdtK). Subsequently, the nucleotide sequences of the genes were submitted to the GenBank database and assigned accession numbers: SRX22793090 (sul-2), SRX22793089 (mdtK), SRX22793063 (qnrS), SRX22793062 (tolC), SRX22793031 (acrA), SRX22789929 (aph(3’)-VIa), SRX22789927 (aac-(6’)-Ib), SRX22789878 (blaSHV-2), SRX22789871 (blaOXA-1), SRX22789802 (blaNDM), SRX22789553 (blaKPC), SRX22789857 (blaTEM-1), and SRX22789858 (blaCTX-M-15).
The K. aerogenes CRKA317 was selected for whole genome sequencing (WGS). The Nextera XT DNA Library Prep Kit (Illumina, San Diego, California, United States) was utilized to conduct the library preparation using 1 ng of DNA as our material to sequence. A limited cycle polymerase chain reaction (PCR) program was employed to amplify the libraries introducing Index 1 (i7) adapters, Index 2 (i5) adapters, and the requisite sequences for generating sequencing clusters. The amplified library was purified using 0.6 x Agencourt AMPure XP beads (Beckman Coulter, Brea, California, USA). The quality of the library and the size of fragmented DNA was evaluated on a 1.5% electrophoresis agarose gel and quantified using a fluorometric method involving the Qubit® 3.0 instrument and the Qubit® dsDNA Broad Range Assay Kit (Life Technologies, Carlsbad, California, United States). The resulting library concentrations were subsequently normalized to 4 nM using a standard dilution method. The libraries were then combined, denatured with 0.2 N sodium hydroxide (NaOH), and diluted to attain a final concentration of 1.8 pM. To ensure the run’s accuracy and control, a PhiX control was added to achieve a final concentration of 1.5 pM. The sequencing run involved a paired-end run comprising 75 cycles for each read (2 × 75), plus up to eight cycles for two index reads.
Initially, FastQC v.0.12.01 was used to check the raw reads quality. The raw reads were filtered by quality, length, and adapter regions using Trim Galore! v.0.6.10.2 The genome assembly was made with SPAdes 3.2 (Bankevich et al., 2012) and SSPACE (Boetzer et al., 2011), using “careful” and “cov-cutoff auto” as settings. Contigs with less than 200 bp were discarded. PlasmidFinder 2.133 (Carattoli et al., 2014) and PlasmidSPAdes (Antipov et al., 2016) were used to plasmid detection and assembly attempts. QUAST v5.0.2 (Gurevich et al., 2013) were used to access the general statistics of assembled genome. The circular genome was built using Proksee4 (Grant et al., 2023). For the annotations of genome, both Prokka v.1.14.5 (Seemann, 2014) and Rapid Annotation using Subsytems Technology (RAST)5 (Aziz et al., 2008) servers were used. The completeness of the assembled genome was assessed using the BUSCO program (Simão et al., 2015).
The Clusters of Orthologous Group (COG) were annotated and distributed in categories using eggNOG-mapper v26 (Cantalapiedra et al., 2021). Kyoto Encyclopedia of Genes and Genomes (KEGG) was used to determinate Gene Ontology (GO)7 (Kanehisa et al., 2016).
Our 16S rRNA gene sequence from our genome annotation was used in the analysis with another 36 16S rRNA reference sequences of Klebsiella genus obtained from the GenBank database (Supplementary Table S2). The 16S rRNA analysis was performed using Escherichia coli as its outgroup. Nucleotide sequences were aligned using the online software MAFFT8 (Katoh and Standley, 2013). JModelTest v2.1.10 (Posada, 2008) was used to estimate the best-fitting nucleotide substitution model and PhyML v3.0 (Guindon et al., 2009) to construct a maximum likelihood (ML) phylogenetic tree. Branches was supported by bootstrap analysis of 1,000 replicates.
The OrthoANI v0.93.1 tool (Yoon et al., 2017) was used to calculate the Average Nucleotide Identity (ANI) between our genome and another 18 reference and uncharacterized complete Klebsiella genus genomes (Supplementary Table S3). For the in silico calculation of digital DNA–DNA Hybridization (dDDH), Genome to Genome Distance Calculator (GGDC 3.0)9 (Meier-Kolthoff et al., 2013) was used with the same genomes. A heatmap with the results from OrthoANI and dDDH was constructed using CIMminer.10 To reinforce our phylogenetic inference, the Type (Strain) Genome Server (TYGS)11 was performed using all strains from the server database (Meier-Kolthoff et al., 2022).
The online pipeline REALPHY12 (Bertels et al., 2014) was used to build a whole-genome sequence-based phylogenetic tree, using the 26 complete genomes of clinical strains of K. aerogenes available in NCBI (Supplementary Table S4). The result showed the closest Klebsiella species to our strain. These species were used in Orthovenn2 web server13 (Xu et al., 2019) to compare orthologous gene clusters using whole-genome sequence. Furthermore, Bacterial Pangenome Analysis Pipeline (BPGA) v.1.3 (Chaudhari et al., 2016) was performed against the Kyoto Encyclopedia Genomics and Genes Database (KEGG) to predict the core, accessory and unique genes as well as their functional distribution. REALPHY, Orthovenn2 and BPGA were used in default settings.
The annotation of antibiotic resistance genes, efflux pumps and porins was made by CARD online14 (Alcock et al., 2019), ResFinder 4.4.215 (Bortolaia et al., 2020), ABRicate16 (Afgan et al., 2018), BlastKOALA17 (Kanehisa et al., 2016) and CARD and ARG-ANNOT (Gupta et al., 2014) databases. The parameters used for databases were 1E-5 e-value, ≥ 70% of identity and ≥90% coverage cut-off.
Known mutations in gyrA, gyrB, and parC, that are responsible for quinolone resistance, were investigated using BLASTp comparison. Furthermore, even though K. aerogenes CRKA317 is not resistance to colistin, mutations in phoP and phoQ were also evaluated to investigate polymyxin resistance. For the alignment we used the following sequences: gyrA (Klebsiella [multispecies]: WP_004201688.1), gyrB (Klebsiella [multispecies]: WP_004173845.1), parC (Klebsiella [multispecies]: WP_004181324.1), phoP (Klebsiella [multispecies]: WP_025714403.1) and phoQ (Klebsiella [multispecies]: WP_045393745.1).
The presence of Genomic Islands (GIs) was investigated with IslandViewer 4 webserver18 (Bertelli et al., 2017), using K. aerogenes isolate 57 as the reference strain. Integrons, transposons and insertion sequences were evaluated using Integron Finder (Afgan et al., 2018), TnCentral19 (Ross et al., 2021) and ISfinder20 (Siguier, 2006), respectively. MGEfinder21 (Durrant et al., 2020) was used to understand the relation between resistance genes with mobile genetic elements. The webserver ICEfinder22 (Liu et al., 2019) was used to identify Integrative and Conjugative Elements (ICE). Sequences of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) were searched using CRISPRCas Finder23 (Couvin et al., 2018). To identify and annotate prophage sequences in genome, the PHASTER webserver24 (Arndt et al., 2016) was used. The Phigaro v.2.3.0 pipeline (Starikova et al., 2020) was used to indicate the possible phage family.
Raw reads were submitted to Sequence Reads Archives,25 with submission number JAXIVA000000000. The draft genome is available at GenBank BioProject accession PRJNA1047945.
A total of 10 non-repetitive clinical isolates of K. aerogenes were isolated from rectal swabs, (40%, n = 4), urine (30%, n = 3), tracheal aspirate (10%, n = 1), blood (10%, n = 1) and catheter tips (10%, n = 1) from adult patients admitted to the intensive care unit (ICU) of a tertiary hospital located in Brazil. All isolates were resistant to the β-lactam antibiotics tested, including SAM, TZP, CXM-S, CXM-AX, FOX, CAZ, CRO, FEP, ETP, IPM, and MEM. MDR was observed in 50% (n = 5) of the strains, and the most common MDR profiles were related to β-lactam-aminoglycosides-quinolone (20%, n = 2), β-lactam-quinolone-glicylcycline (20%, n = 2), and β-lactam-aminoglycosides-quinolone-glicylcycline (10%, n = 1). On the other hand, 80% of the isolates were susceptible to GEN, 80% to AMK, 70% to TGC, 50% to CIP, and 100% to CST. General data and susceptibility profiles of all clinical carbapenem-resistant K. aerogenes (CRKA) isolates are showed in Table 1 and Supplementary Figure S1.
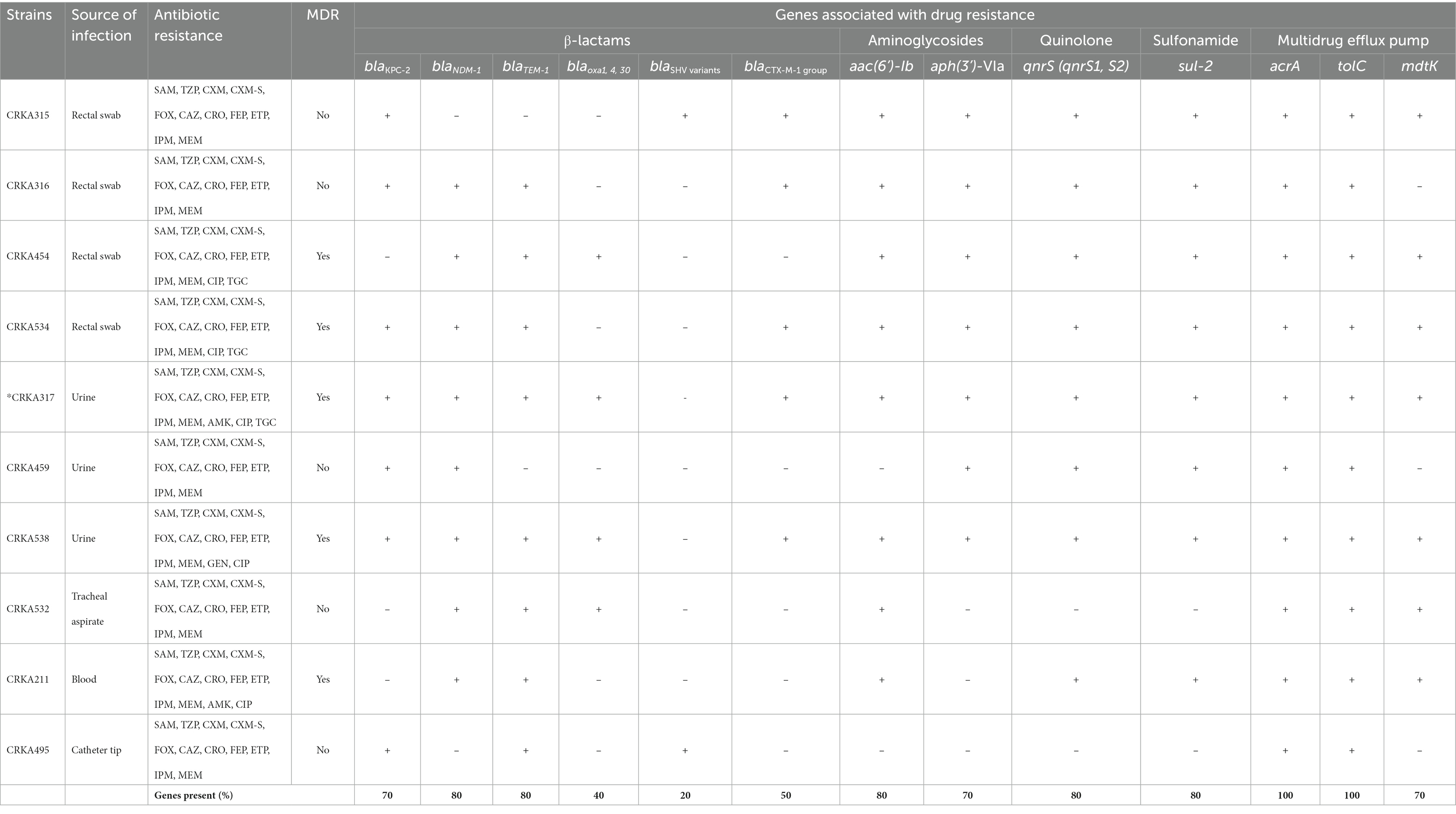
Table 1. Antimicrobial resistance of K. aerogenes isolates and presence of genes coding for resistance, and efflux pumps.
Of the 10 carbapenemase-producing K. aerogenes isolates, the blaNDM-1 gene was detected in 8 isolates (80%), followed by blaKPC-2 in 7 isolates (70%). Whereas the concomitant presence of blaKPC-2 with blaNDM-1 gene was detected in 5 isolates (50%). In addition, the blaTEM-1 (80%, n = 8) was the most common ESBL-encoding gene among K. aerogenes investigated, followed by blaCTX-M1-group (50%, n = 5), blaOXA-1,4, and 30 (40%, n = 4), and blaSHV variants (20%, n = 2) (Table 1 and Supplementary Figure S1).
Regarding the genes that provide resistance to aminoglycosides, 7 isolates (70%) carried the aph(3′)-VI (aphA6) and 8 strains (80%) carried the aac(6’)-Ib gene. Eight strains (80%) harbored qnrS (qnrS1 and/or qnrS2), capable of causing resistance fluoroquinolones antibiotics. The sulfonamide resistance gene (sul-2) gene was present in 8 isolates (80%). All the CRKA isolates we investigated had genes related to efflux pumps acrA and tolC. The mdtK gene, which is a multidrug efflux system transporter, was present in 7 strains (70%) (Table 1 and Supplementary Figure S1). The genes related to antibiotic resistance blaOXA-48; blaSPM-1; blaIMP-1; blaVIM-2; blaSIM-1; blaGIM-1, blaGES-1, 9,11; blaPER-1, 3; blaVEB-1 to 6, mcr-1, sul-1, aac(6′)-Ib-cr, armA, rmtB and tetB were not found in CRKA isolates.
β-lactams: SAM (ampicillin-sulbactam), TZP (piperacillin-tazobactam), CXM-S (cefuroxime sodium), CXM (cefuroxime axetil), FOX (cefoxitin), CAZ (ceftazidime), CRO (ceftriaxone), FEP (cefepime), ETP (ertapenem), IPM (imipenem), MEM (meropenem); aminoglycosides: GEN (gentamicin) and AMK (amikacin); quinolones: CIP. (ciprofloxacin); glycylcycline: TGC (tigecycline) and polymyxin: CST (colistin). MDR (multidrug-resistant) = resistance to at least one agent in three or more antibiotic categories. * Whole-genome sequencing was performed on CRKA317. +, the tested gene was detected by PCR and Sanger sequencing; –, the tested gene was not detected.
Given the existence of both the blaKPC-2 and blaNDM-1 genes in CRK317, as well as the strain’s resistance to a broad spectrum of antibiotics (except for gentamicin and colistin), whole genome sequencing (WGS) was employed to obtain comprehensive genomic data from the K. aerogenes CRKA317. The draft genome of CRKA317 comprised one circular chromosome, which is 5,462,831 bp in size, with an average GC content of 54.8%. The annotation of the bacterial genome predicted a total of 51 contigs, 5,403 coding sequences and 5,374 genes that covered 88.88% of genome. Of the 65 RNA genes predicted, 5 were rRNAs, 59 were tRNAs and one was a transfer-messenger RNA (tmRNA) (Supplementary Table S5 and Figure 1A). The assembly of plasmids was unsuccessful, but fragments of IncFIB (pQil), IncC, and IncFII (K) plasmids were detected.
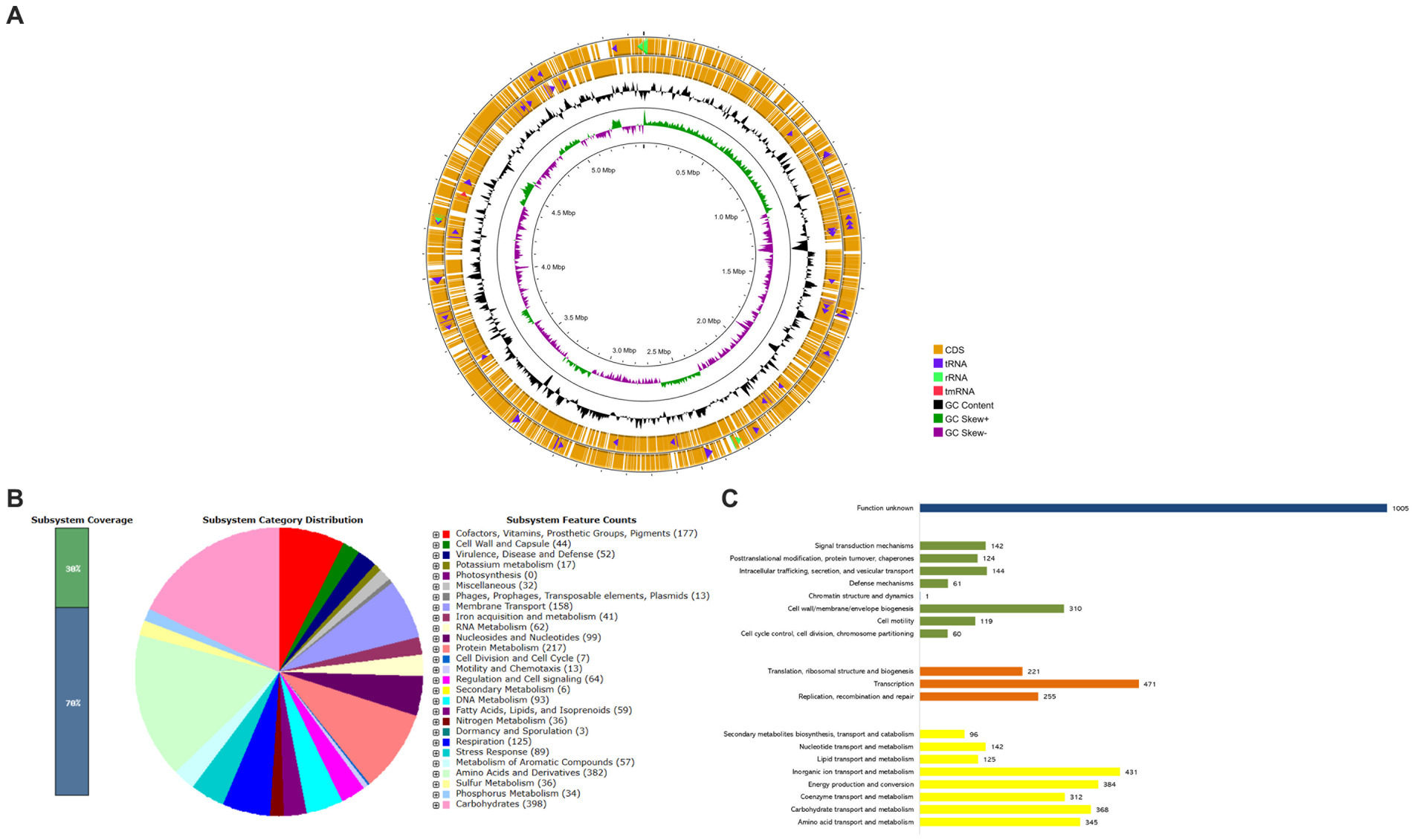
Figure 1. Whole-Genome Sequencing and functional annotations of K. aerogenes CRKA317. (A) The circular diagram consists of four circles, numbered from 1 (outer) to 4 (inner). The outer two orange circles represent the coding sequence (CDS), the transfer RNA (tRNA) is indicated by a purple arrowhead, the ribosomal RNA (rRNA) is shown with a green arrowhead, and the transfer-messenger RNA (tmRNA) is denoted by a red arrowhead. The third, black circle represents the GC content. The fourth circle displays the GC skew curve indicating a positive skew in green and negative skew in violet. (B) The bar indicates the percentage of subsystem coverage, with the green segment representing the proportion of proteins included. The pie chart presents an overview of the distribution of various categories within these subsystems. (C) COG classification in categories: metabolism (yellow bars), information processing and storage (orange bars), cellular processes and signaling (green bars), and unknown function (blue bar).
According to the RAST analysis, the genome of K. aerogenes CRKA317 is composed of 398 subsystems that can be categorized into 27 distinct categories (Figure 1B). The six most significant categories included “carbohydrates” with a total of 398 genes, followed by “amino acids and derivatives” (382 genes), “protein metabolism” (217 genes), “cofactors, vitamins, prosthetic groups, pigments” (177 genes), “membrane transport” (158 genes), and “respiration” (125 genes). In the specific category of “virulence, disease and defense,” (52 genes) there were 32 genes related to resistance against antibiotics and toxic compounds; such as β-lactamase enzymes (one gene), fluoroquinolone resistance (two genes), fosfomycin resistance (one gene), copper homeostasis (11 genes), copper homeostasis: cooper tolerance (ten genes), cobalt-zinc-cadmium resistance (four genes), zinc resistance (two genes), adaptation to d-cysteine (one gene); Furthermore, we found 14 genes associated with invasion and intracellular resistance, four genes linked to adhesion, and two genes related to bacteriocins, ribosomally synthetized antibacterial peptides.
The analysis of protein-coding genes resulted in a total of 5,116 genes distributed across different functional categories within the Cluster of Orthologous Groups. The largest proportion of known protein coding genes was related to “transcription” (471; 9.21%), followed by categories such as “inorganic ion transport and metabolism” (431; 8.42%), “energy production and conversion” (384; 7.50%), “carbohydrate transport and metabolism” (368; 7.19%), and “amino acid transport and metabolism” (345; 6.74%). There were also gene associations with defense mechanisms (61; 1.19%) and a significant portion classified as having unknown functions (1,005; 19.64%) (Figure 1C).
To gain insights into the evolutionary placement of K. aerogenes CRKA317, a phylogenetic tree was generated using 16S rRNA gene sequences from 36 reference sequences of Klebsiella species available at NCBI.26 Our findings indicated that CRKA317 was not closely related to K. aerogenes (Figure 2A). The use of 16S rRNA gene for species identification presents significant challenges in interpretation because of its hypervariable domains (Koroiva and Santana, 2022). Nevertheless, this approach allowed for an assessment of its relationship in the broader context of the genus.
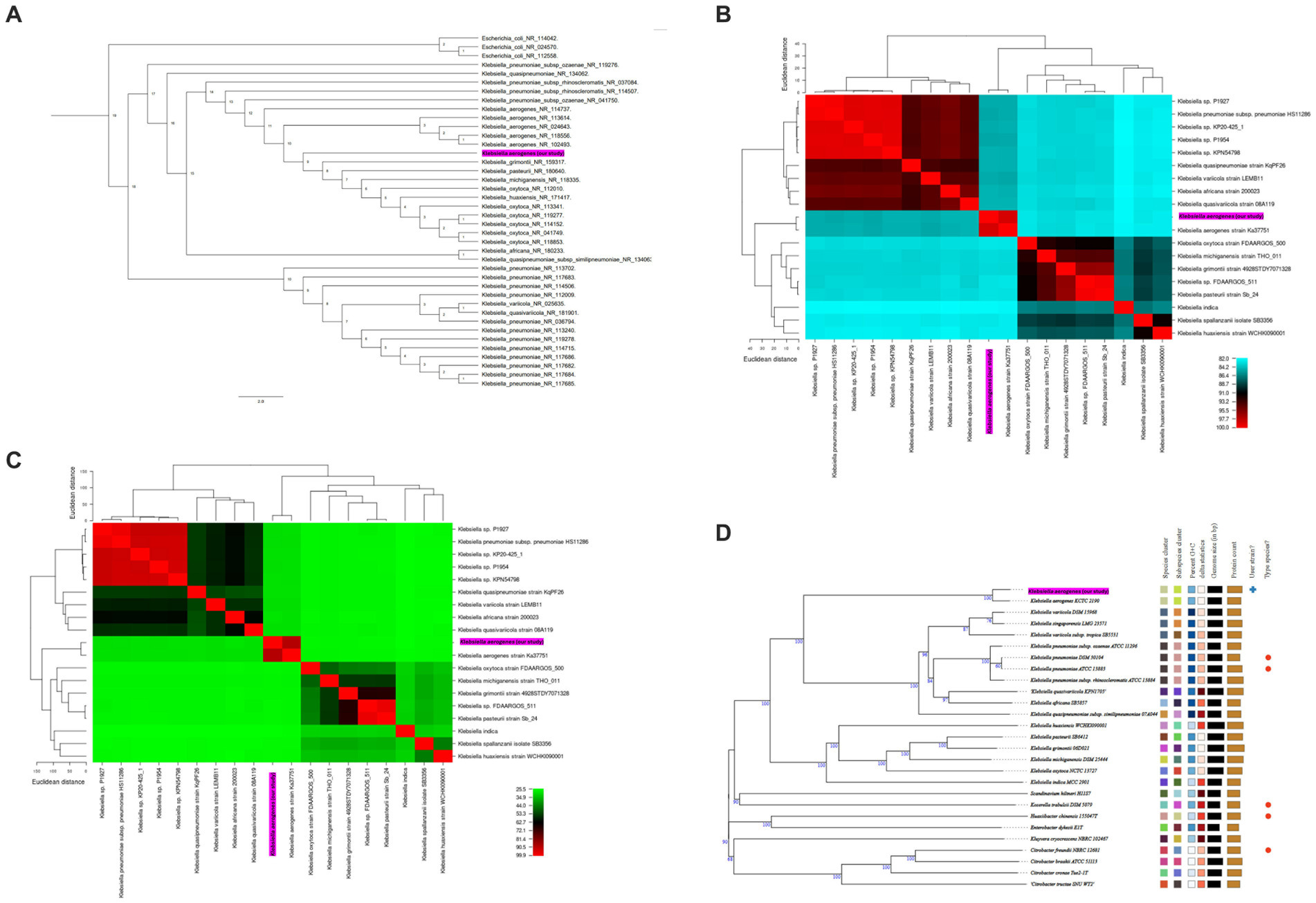
Figure 2. (A) Phylogenetic tree based on 16S rRNA gene sequences, which shows the relationship between K. aerogenes CRKA317 and other. The number next to the node represents the age value, and the scale bar indicates 2.0 substitutions per nucleotide position. (B,C) display heat maps of average nucleotide identity (ANI, B) and in silico DNA–DNA hybridization (DDH, C) respectively, which compare K. aerogenes CRKA317 to Klebsiella species. (D) Phylogenomic tree based on TYGS. The numbers above the branches represent GBDP pseudo-bootstrap support values >60% of 100 replications, with an average branch support of 86.2%. The tree was rooted at the midpoint. Our strain is highlighted in pink.
Next, species validation and genomic similarity were assessed through in silico ANI, DDH and TYGS analysis. Our next step was a comparative analysis of 13 reference sequences of Klebsiella species, including 5 clinical isolates of Klebsiella spp. and our K. aerogenes CRKA317. ANI analysis revealed high similarity between K. aerogenes CRKA317 and K. aerogenes (Ka37751; GCA_007632255.1), with a close match of approximately 98.52% (Figure 2B). The genetic relatedness between these two strains was also confirmed with a DDH value of 88.90% (Figure 2C). The TYGS-based results showed that K. aerogenes CRKA317 is most closely related to K. aerogenes KCTC 2190, with dDDH values of 89%, also positioning CKA317 as a K. aerogenes (Figure 2D).
Next, we determined the genetic similarity between K. aerogenes CRKA317 and 26 genomes of K. aerogenes obtained from the NCBI database. Our findings indicated that our K. aerogenes CRKA317 strain is more closely related to K. aerogenes 57, K. aerogenes CAVI1320, and K. aerogenes EA46506, which are forming a monophyletic clade (Figure 3A). Although the isolation source for the closest strain (K. aerogenes 57) was not specified in the NCBI website, the other two strains were isolated from clinical samples (Figure 3B).
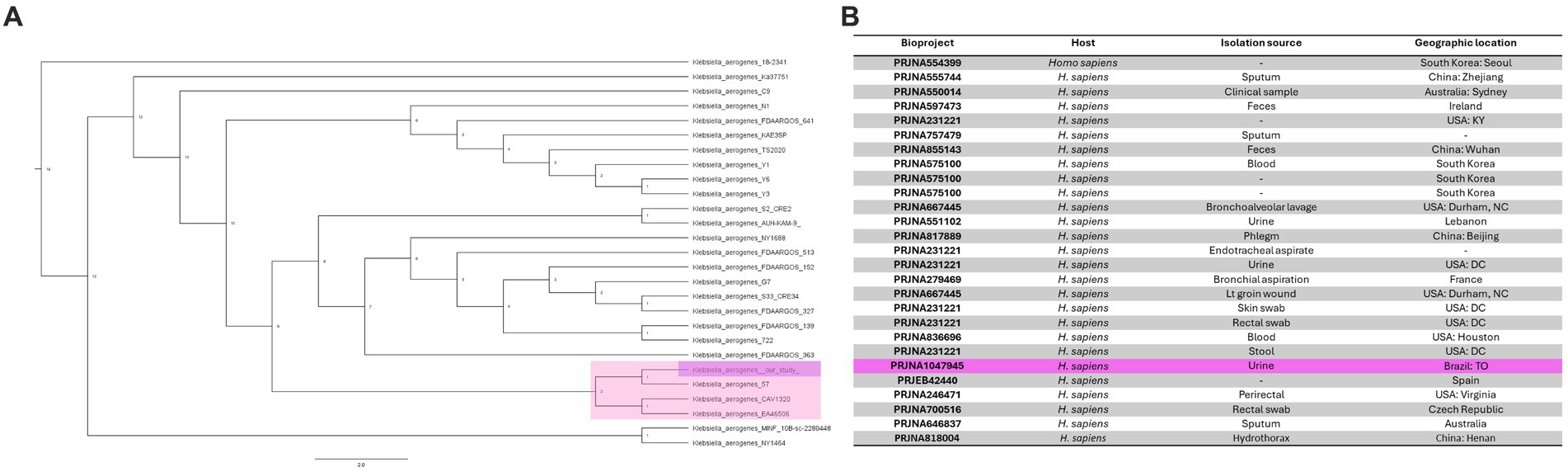
Figure 3. Phylogenomic analysis of K. aerogenes strains. (A) Phylogenetic tree showing the similarity between CRKA317 and other 26 selected strains of K. aerogenes. (B) Genome Assembly and Annotation report of K. aerogenes strains. Our strain is highlighted in pink.
We also conducted a comparative analysis of the predicted gene numbers in three closely related strains of K. aerogenes with our K. aerogenes CRKA317. This allowed us to identify both common genes shared across these strains, as well as those that were unique to each individual strain. Our data showed the four K. aerogenes strains shared 4,242 genes, and K. aerogenes CRKA317 was found to contain 16 strain-specific gene clusters which were associated to metal ion transport including response to cadmium ion, and copper ion transport (Figure 4A). The K. aerogenes CRKA317 showed a higher number of singleton genes (n = 574) compared to other K. aerogenes strains (Figure 4B), including genes associated with resistance such as blaNDM-1, aadA/ant(3”)-Ia, aph(3’)-VI, and qnrS1.
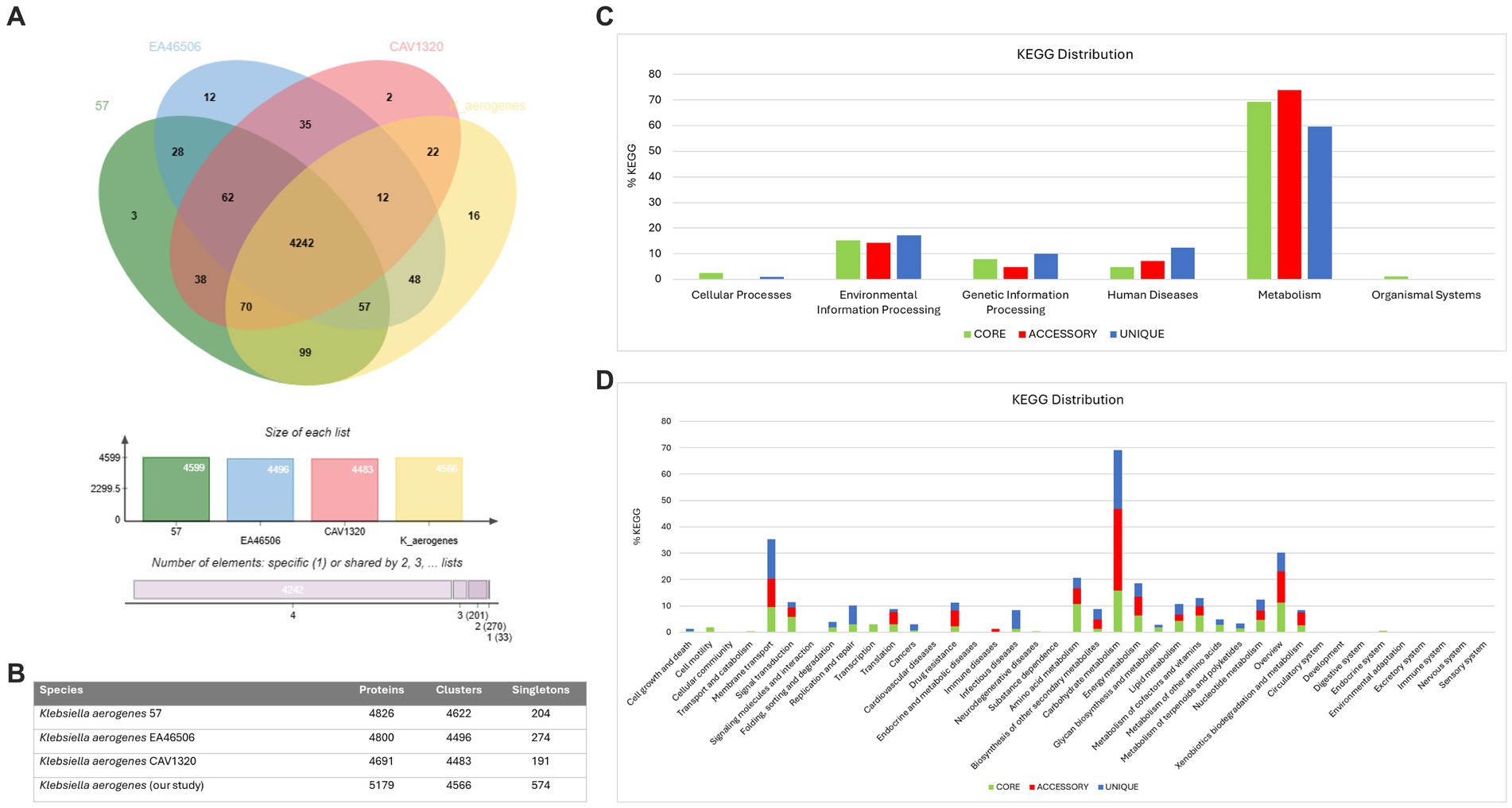
Figure 4. Comparative genomic analysis. (A) Venn diagram and bar chart showing the numbers of unique and shared orthologous genes present in most closely related strains of K. aerogenes. (B) Number of proteins, clusters and singletons. (C) KEGG pathway classification in core, accessory and unique genomes. (D) Distribution of KEGG pathway classification.
The analysis of KEGG functional distributions in the 4 strains (K. aerogenes 57, K. aerogenes CAVI1320, and K. aerogenes EA46506) showed that the majority of genes were associated with the core genomes (94.16%; n = 2,950), followed by unique genomes (3.16%; n = 99), and accessory genomes (2.7%; n = 84) (Figure 4C). The genes were associated mainly with “metabolism” and were highly abundant in the accessory genome (73.91%; n = 62), followed by the core (69.29%; n = 2047), and unique (59.59%, n = 59) genomes (Figure 4C). Out of the total number of genes linked to human diseases (n = 158), 12.3% were found in the unique gene clusters, while 7.14% were present in accessory gene clusters and 4.73% was assigned to core gene clusters (Figure 4C).
Analysis of the annotations for all core genes revealed that a majority were associated with “carbohydrate metabolism” (15.83%), “overview” (11.2%), and “amino acid metabolism” (10.68%). Within the unique genome, significant proportions of genes were identified as belonging to categories such as “carbohydrate metabolism” (22.22%), “membrane transport” (15.15%), and “overview,” “infectious disease” and “replication and repair” with similar values (7.07%). In the accessory genome, most genes were categorized as “carbohydrate metabolism” (30.95%), “overview” (11.9%) and “membrane transport” (10.7%) (Figure 4D). Notably, we observed the presence of genes related to β-lactam resistance (0.61%, n = 18), vancomycin resistance (0.27%, n = 8), and cationic antimicrobial peptide resistance (1.35%, n = 40) within the core gene clusters specifically linked to drug resistance (Figure 4D).
Resistome analysis using WGS revealed that K. aerogenes CRKA317 harbored dozens of antibiotic resistance-associated genes, including genes coding for β-lactamases (blaOXA-9, blaTEM-1, blaNDM-1, blaCTX-M-15, blaAmpC-1, blaAmpC-2); aminoglycoside-modifying enzymes [aac(6’)-Ib, aadA/ant(3”)-Ia, aph(3’)-VI], a chloramphenicol acetyltransferase (catA-like chloramphenicol resistance), an erythromycin resistance methylase (ermC), a plasmid-mediated quinolone resistance protein (qnrS1), a sulfonamide resistance enzyme (sul-2), a glutathione transferase (fosA: fosfomycin resistance), PEtN transferases (eptA and eptB: resistance to peptide antibiotic), a glycosyltransferase (arnT: resistance to peptide antibiotic) (Table 2). We also found the bleMBL gene that encodes a bleomycin resistance protein (BRP). Although the majority of resistance genes identified through sequencing were consistent with those detected in whole genome sequencing (Table 1), blaKPC-2 was only detected through Polymerase Chain Reaction-based amplification and sequencing, but not predicted from incomplete genomic sequence of K. aerogenes CRKA317.
A rich repertoire of genes related to efflux-mediated resistance was found in the genome of K. aerogenes CRKA317 (Table 3), including an ATP-binding cassette (ABC) antibiotic efflux pumps (tolC), resistance-nodulation-cell division (RND)-type efflux pumps (oqxA, oqxB, acrA, acrB, acrD, HAE1, EefA, EefB, mdtA, mdtB), major facilitator superfamily membrane transport proteins (mdtH, KdeA, MFS-MMR-like), a multidrug and toxic compound extrusion transporter (MATE) (mdtK) and an outer membrane efflux protein (oprM). In addition, we have identified multiple MDR efflux pump acrAB transcriptional activators/regulators (marR, ramA, soxS), as well as genes that code for porin-associated proteins such as oprD, ompA, ompX, and ompW. The ramA and soxS genes were specifically associated with mobile genetic elements (Figure 5).
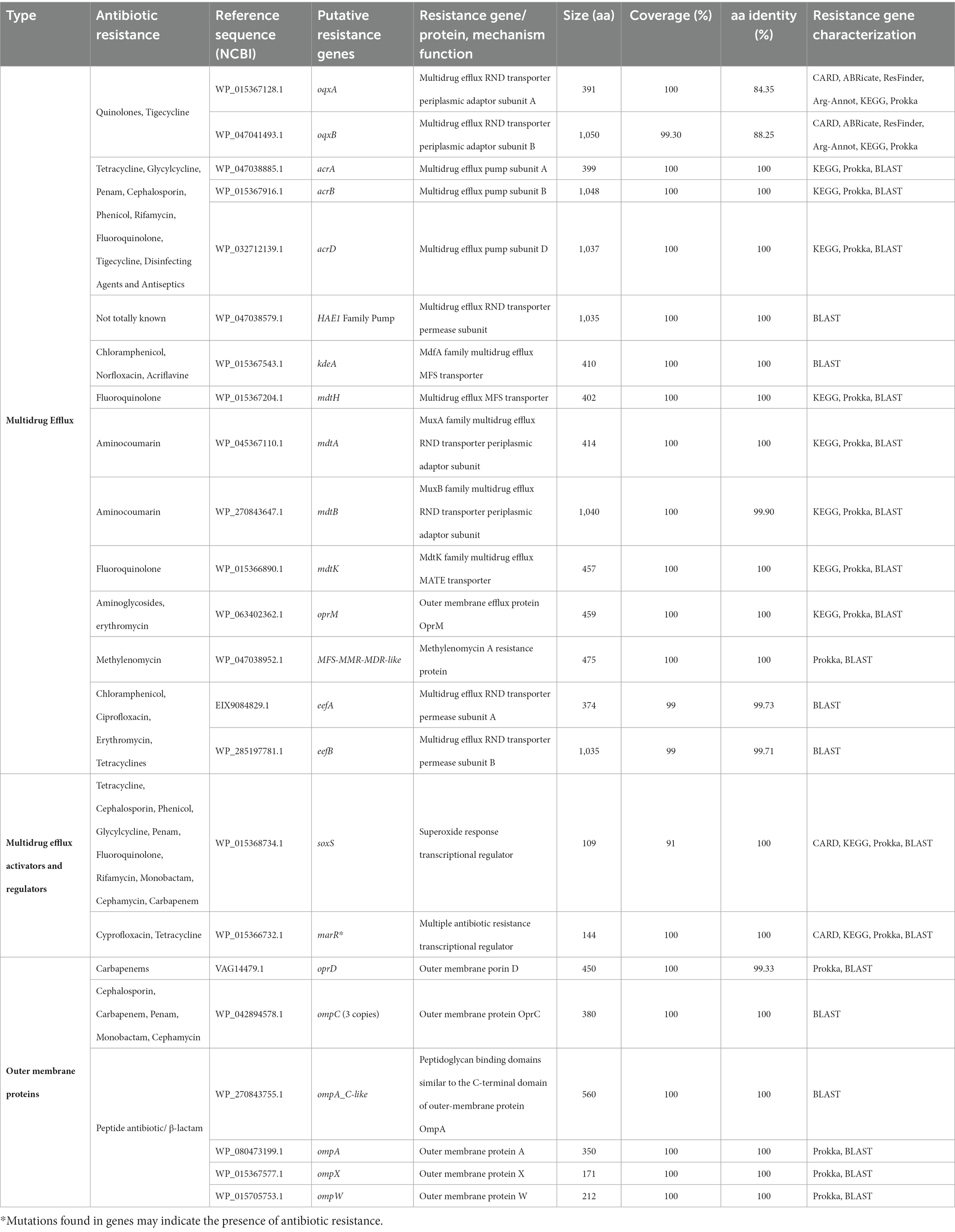
Table 3. Identification of the genes encoding multidrug efflux, activators/regulators and outer membrane proteins genes in the genome of K. aerogenes CRKA317.
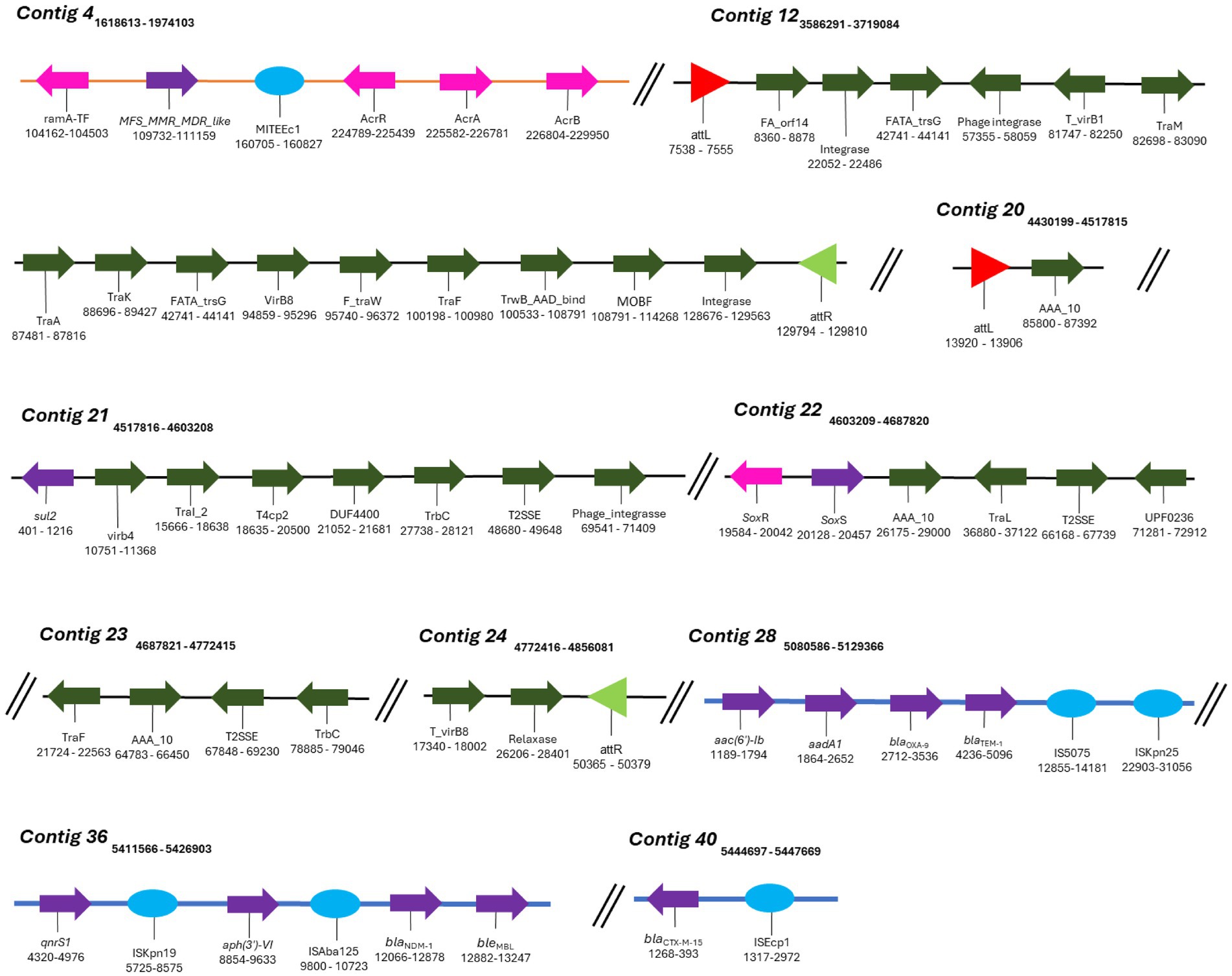
Figure 5. Mobile genetic elements (MGEs) identified in K. aerogenes CRKA317. Schematic representation of MGEs identified. The orange line represents the DNA strand (contig 4), blue lines represent GIs and black lines (contig 12 and contigs 20 to 24) represent the ICEs. The colored arrows represent the following genes: dark green - conjugation of the ICEs (e.g., contig 12), purple – resistance genes (e.g., contig 28), pink – efflux pumps and their respective activators/regulators (e.g., contig 4). The red (contig 20) and light green triangles (contig 24) represent attL and attR sequences, respectively. The light blue circles represent insertion sequences (e.g., contig 40).
Klebsiella aerogenes CRKA317 presented amino acid substitutions in marR (Ser3Asn), which may play a role in the development of quinolone resistance (Maneewannakul and Levy, 1996). Additionally, we found more two novel mutations in marR (Val96Ile and Gly103Glu), and one novel mutation (Ala12Glu) in the transcription factor of the regulon, soxS (Supplementary Figure S2).
We searched the K. aerogenes CRKA317 genome for the presence of GIs and Mobile Genetic Elements (MGEs). GIs are groups of genes within a bacterial genome that appear to have been obtained through horizontal gene transfer. We found 22 GIs in K. aerogenes CRKA317’s genome, which contained both resistance genes and insertion sequences. It is noteworthy that these antimicrobial resistance genes (blaOXA-9, blaTEM-1, blaNDM-1, bleMBL, blaCTX-M-15, aac(6’)-Ib, aadA/ant(3”)-Ia, aph(3’)-VI, and qnrS1) were located within an island in the contigs of the K. aerogenes CRKA317 genome that did not align with the reference strain K. aerogenes 57 isolate (Supplementary Figure S3). Furthermore, only one resistance gene (blaAmpC) was identified in islands located within the aligned contigs with reference strain K. aerogenes 57.
MGEs consist of a broad array of genomic sequences, such as plasmids, prophages, pathogenicity islands, restriction and modification systems, transposons, and Insertion Sequences (ISs). Our analyses showed that the K. aerogenes CRKA317 genome contained eight families of ISs, two copies of putative integrative and conjugative elements (ICE) with type IV secretion system (T4SS), and eight prophage regions. ISs are mobile repetitive DNA sequences that have the capacity to replicate and relocate within a host genome, playing a role in genetic diversity and regulation of gene expression in prokaryotes (Tempel et al., 2022). Our CRK317 strain contained two copies of MITEEc1 from the IS630 family, albeit only one copy carried genes such as MFS_MMR_MDR-like, acrR, acrA, and acrB (contig 4). The IS110 family presented one copy of IS4321/IS5075 and one copy of ISKpn25 harboring the aac(6’)-Ib, aadA1, blaOXA-9, and blaTEM-1 genes (contig 28). Contig 36 comprised the IS30 family harboring one copy of ISAba125, carrying blaNDM-1 and bleMBL genes and the ISKra4 family presenting one copy of ISKpn19, aph(3’)-VI and qnrS1 genes. The ISEcp1 member of the IS1380 family presented the blaCTX-M-15 gene (contig 40) (Figure 5). The ISRaq1 member of the IS3 family, the ISR1 member of the IS1 family and the IS26 member of the IS6 family did not present any resistance genes.
ICEs are found in the bacterial host chromosome and play a significant role in spreading resistance genes which significantly contribute to the evolution of discrete bacterial strains (Bean et al., 2022). Among the two putative ICEs containing a type IV secretion system (T4SS), contig 12 exhibited elements features of the ICE backbone, including a 16-base pair direct repeat attL sequence (5’-AAGAAGGGGAGTCCTG-3’); various integrases such as virB(s), rve, and phage integrases; T4SSs; T4CP; relaxases; and another 16-base pair direct repeat attrR sequence (5’-AAGAAGGGGAGTCCTG-3’). Additionally, contigs 20-24 contained similar components with a slight variation in the length of its direct repeats, 15 base pairs for both attL and attrR (Table 4). We did not detect CRISPR or Cas genes in K. aerogenes CRKA317 genome.
Among the eight prophage regions identified in CRK317, two were completely intact, four were incomplete, and two were classified as uncertain. Similarly, two other phages were observed in the fully intact regions: the first region (43.2 kb in size with 50.3% CG content and 63 CDS) showed similarities to Salmonella phage SEN34 (NC_028699.1), while the second region (54.5 kb in size with 52.32% CG content and 54 CDS) exhibited similarities to Escherichia phage vB_EcoM_ECO1230-10 (NC_027995.1) (Supplementary Figure S4).
Klebsiella aerogenes is known as an opportunistic pathogen of patients admitted to the intensive care unit and is frequently associated with multidrug resistance (MDR) (Azevedo et al., 2018). It has been detected in various hospital sources and is known for its ability to adapt to this setting. This study found that K. aerogenes was mainly detected in rectal swabs and urine samples, followed by tracheal aspirate, blood, and catheter tips. Previous studies have highlighted that K. aerogenes is commonly present in human specimens such as urinary, gastrointestinal, respiratory, blood, abscesses, and cutaneous samples (Davin-Regli et al., 2019). Additionally, rectal swabs are employed for active surveillance of asymptomatic carriers. These findings are also consistent with prior studies demonstrating the presence of E. aerogenes in surveillance rectal swabs (Vrioni et al., 2012). But the small number of K. aerogenes isolates is one of the major limitations of this study.
Our study found that all K. aerogenes isolates displayed some level of resistance to the β-lactams tested, including carbapenems, and 50% of K. aerogenes isolates exhibited a MDR profile (Table 1). Carbapenems have traditionally been a preferred treatment for infections caused by MDR Gram-negative strains (Bouza, 2021). Therefore, the emergence of carbapenem-resistant K. aerogenes strains present a new challenge in the treatment of these infections and pose a public health threat worldwide (Kamio and Espinoza, 2022).
The presence of resistance genes was verified using polymerase chain reactions. Out of the 10 isolates, we discovered that two only contained the blaKPC-2 gene and three only contained the blaNDM-1 gene while, interestingly, five strains carried both the blaKPC-2 and blaNDM-1 genes. Previous studies have reported the presence of blaKPC or blaNDM genes in clinical K. aerogenes isolates from various countries (Pulcrano et al., 2016; Franolić et al., 2019; Ma et al., 2020), including Brazil (Bispo Beltrão et al., 2020; Soares et al., 2021). However, our literature review indicated that there has been only one study of K. aerogenes co-harboring both blaKPC and blaNDM genes, which was observed in China (Zhang et al., 2017). Therefore, to the best of our knowledge, this is the first report describing clinical samples of K. aerogenes isolated from Brazil with simultaneous carriage of blaKPC and blaNDM genes. It is noteworthy that we found a significant number of K. aerogenes isolates exhibiting a blaKPC-2 or blaNDM-1 carbapenemase in addition to ESBLs genes, including members of the blaCTX-M-1; blaOXA1, 4, 30; blaTEM; and blaSHVvariants. Our findings are in line with studies that reported the concomitant presence of carbapenemase and ESBLs genes in K. aerogenes strains (Ma et al., 2020; Pan et al., 2021).
Although most of the CRKA strains were susceptible to aminoglycosides (gentamicin and/or amikacin), a high percentage of the isolates were found to contain the aph(3’)-VI gene, a plasmid-encoded aminoglycoside phosphotransferase that confers resistance to amikacin. Studies have shown that Enterobacter aerogenes can carry both aph(3’)-VI and blaKPC (Firmo et al., 2019), and also aph(3’)-VI with blaNDM (Chen et al., 2015). However, to the best of our knowledge, this is the first report of Klebsiella aerogenes concomitant harboring aph(3’)-VI, blaKPC-2, and blaNDM-1 genes, as can be seen in the CRKA315 strain. Most of the CRKA isolates also harbored the aac(6’)-Ib gene, responsible for encoding an aminoglycoside 6’-N-acetyltransferase type Ib, which provides resistance to amikacin (Machuca et al., 2016). Interestingly, amikacin susceptible isolates harboring the aac(6′)-Ib gene have been reported in K. pneumoniae strains (Almaghrabi et al., 2014; Haldorsen et al., 2014; Galani et al., 2019). Despite half of the CRKA strains being susceptible to ciprofloxacin, 80% of the isolates harbored the qnrS (qnrS1 and/or S2) gene. The qnrS genes have been detected in a variety of microorganisms and environments, where they can be located in both the chromosome and in plasmids (Xu et al., 2023). Studies have indicated an association between qnrS1 and a Tn3-like-blaTEM-1-containing transposon, leading to enhanced recombination and insertion effectiveness (Guan et al., 2013). The expression of the qnrS1 gene is enhanced by quinolones, in contrast to certain other qnr genes (Monárrez et al., 2018). QnrS2, associated with quinolone resistance which demonstrates a 92% similarity in amino acid composition with qnrS1, is frequently identified in IncQ, IncU, and ColE-type plasmids as part of a mobile insertion cassette element bracketing inverted repeats but lacking a transposase (Picão et al., 2008; Han et al., 2012; Dobiasova et al., 2016; Wen et al., 2016). Notably, a newly identified surrounding genetic structure of qnrS2 flanked by IS26 elements was observed in E. coli strains from China (Tao et al., 2020). This finding highlights the important role of IS26 in facilitating the horizontal spread of quinolone resistance genes. Therefore, our data suggests that the variance in results between phenotyping and genotyping may be linked to the presence of multiple concurrent resistance mechanisms (Galani et al., 2019). However, we should mention that the incomplete sequencing of K. aerogenes CRKA317 may undermine the confidence level of this speculation.
Efflux pumps are important membrane proteins that play a crucial role as defense mechanisms by actively exporting harmful substances, such as antibiotics, detergents, and heavy metals (Jang, 2023). In our study, all of the isolates studied harbored acrA, encoding a subunit that functions as an adapter protein linked to AcrB, and TolC outer membrane channel proteins. Although we did not identify acrB in our analysis, together these proteins form the AcrAB-TolC stable efflux complex, known to contribute to multidrug resistance in nosocomial pathogens (Chen et al., 2022). These genes have been identified in K. aerogenes clinical isolates and are responsible for expelling various compounds, including antimicrobial agents like quinolones, tetracyclines, and chloramphenicol (Pradel and Pagès, 2002; Masi et al., 2007; Chevalier et al., 2008). Although the expression levels of acrAB and tolC genes were not determined in our K. aerogenes isolates, it was observed that all strains carried both genes which might be involved in the development of MDR carbapenem-resistant profile of K. aerogenes. The mdtK gene encodes an efflux pump that has the ability to expel acriflavine, doxorubicin, norfloxacin, and dipeptides (Hayashi et al., 2010; Andersen et al., 2015). This gene was found in a majority of our CRKA strains. Previous studies have reported the presence of the mdtK gene in K. aerogenes isolated from river sediment (Iyer et al., 2017).
WGS is a valuable tool for identifying and characterizing disease-associated bacteria in clinical settings. Thus, we conducted a comprehensive analysis of the entire genome of K. aerogenes CRKA317 to gain deeper insights into its genomic diversity, and methods of resistance.
The draft genome of K. aerogenes CRKA317 comprised of a single circular chromosome with a length similar to most K. aerogenes genomes in NCBI GenBank and harbored various essential genes for bacterial cellular processes. Additionally, the results of the RAST and eggNOG analyses showed that our K. aerogenes CRKA317 carried genes linked to drug resistance.
Next, we explored the phylogenetic affiliation of K. aerogenes CRKA317. The16S rRNA gene sequence analysis showed that the K. aerogenes CRKA317 was not closely related to K. aerogenes as a specie. Although, 16S rRNA gene is used extensively in bacterial phylogenetics, the limitations of using 16S rRNA gene relatedness to classify bacteria have been extensively documented (Rossi-Tamisier et al., 2015; Thorell et al., 2019; Soares et al., 2023). Therefore, a combination of ANI, dDDH, and TYGS technologies were employed to determine the phylogenetic position of the K. aerogenes CRKA317, which predicted a close phylogenetic relationship with K. aerogenes.
In our investigation of the genetic relationship between K. aerogenes CRKA317 and another 26 K. aerogenes strains, we found three strains (K. aerogenes 57, K. aerogenes CAVI1320, and K. aerogenes EA46506) that were closely related to K. aerogenes CRKA317. When analyzing the distribution of shared gene families among four strains, we found that our K. aerogenes CRKA317 had the highest number of singleton genes compared to its closest three relatives. Some of these singleton genes were related to antibiotic resistance. Singleton genes are typically acquired through horizontal gene transfer (HGT) or mutations in pre-existing genes. These genes are often associated with specific metabolic pathways, virulence, antibiotic resistance mechanisms, or other environmental adaptations (Costa et al., 2020). Furthermore, the KEGG pathway analysis revealed that most of the genes were in the core genome and were related to metabolic pathways. As the analyses were based on small number of K. aerogenes, we cautiously speculated that K. aerogenes might have genomic plasticity, that may contribute to antibiotic resistance and environmental adaptation.
Our comprehensive analysis of the K. aerogenes CRKA317 using WGS confirmed the correlation between the genotype and the phenotype to its antimicrobial resistance. Furthermore, our analysis found a large number of antibiotic resistance-associated genes such as porin and efflux pump-encoding genes giving resistance to both previously tested and untested antibiotics, suggesting a wide-ranging antibiotic resistance profile (Tables 2, 3). AmpC β-lactamases are usually encoded within the chromosome or found as ampC genes on a plasmid. Our K. aerogenes CRKA317 harbored two copies of blaAmpC gene. Several Gram-negative organisms, including E. aerogenes, Enterobacter cloacae, Serratia marcescens, Providencia stuartii, Pseudomonas aeruginosa, Hafnia alvei, and Morganella morganii, have presented AmpC in their genomes (Jacoby, 2009; Ghanavati et al., 2018; Tamma et al., 2019). This enzyme provides resistance against aminopenicillins, cephalosporins, oxyimino-cephalosporins (e.g., ceftriaxone, cefotaxime, and ceftazidime), cephamycins (e.g., cefoxitin and cefotetan), and monobactams (aztreonam) (Jacoby, 2009). The gene sul-2 implicated in sulphonamide resistance due to inducing high levels of dihydropteroate synthase was found in our strain (Teichmann et al., 2014). This gene has been widely studied and its association with sulfamethoxazole resistance has been demonstrated in numerous studies worldwide (Teichmann et al., 2014; Shin et al., 2015), including in an E. aerogenes from Brazil (Grazziotin et al., 2016). The K. aerogenes CRKA317 contained the fosA gene, which is commonly found in the genomes of K. pneumoniae, K. oxytoca, E. cloacae, E. aerogenes, S. marcescens, M. morganii, P. stuartii, and P. aeruginosa. All these species haboring fosA gene presented intrinsic resistance or reduced susceptibility to fosfomycin (Ito et al., 2017). The ermC gene, known for its role in conferring erythromycin resistance in S. aureus and other Staphylococci (Jamrozy et al., 2017) was also detected in the K. aerogenes CRKA317. The K. aerogenes CRKA317 also contained the catA-like gene, responsible for producing a chloramphenicol acetyltransferase that catalyzes chloramphenicol (Huang et al., 2017). This gene is commonly present on transposons and plasmids, and it is widespread among a range of organisms such as Acinetobacter spp., Bacillus methylotrophicus and Chryseobacterium indologenes (Obayiuwana and Ibekwe, 2020; Damas et al., 2022).
We also detected three alterations in amino acids in marA in our K. aerogenes CRKA317 (Ser3Asn, Val96Ile, Gly103Glu) and one in soxS (Ala2Glu). These findings partially corroborate the results of Maneewannakul and Levy (1996), where they identified three mutations in marA (Ser3Asn, Val96Glu, Gly103Ser) leading to resistance to fluoroquinolones in E. coli. Moreover, Aly et al. (2015) demonstrated that a mutation in soxS (Ala12Ser) contributed to resistance against ciprofloxacin, enrofloxacin, chloramphenicol, and doxycycline in E. coli strains. We suggest these same mechanisms gave rise to antimicrobial resistance in our K. aerogenes CRKA317 due to the similar mutations noted.
Additionally, our K. aerogenes CRKA317 presented several MGEs harboring resistance genes (Table 4). MGEs are important tools for acquiring resistance genes through horizontal gene transfer. ISs, for example, are transposable DNA segments that have been previously linked to resistance genes and can be transferred horizontally by plasmids or by bacteriophages (Siguier et al., 2014). The blaOXA-9, blaTEM-1 genes encoding for resistance to β-lactams, and aac(6’)-Ib, aadA1 encoding resistance aminoglycosides were related to an IS110 family transposase (IS4321/IS5075 and ISKpn25 insertion elements). These findings are partially supported by previous studies that have shown ESBL-encoding genes (blaTEM-1B) and genes related to aminoglycoside resistance (aph-Id, aph-Ib) are located near to an IS5075 insert in K. pneumoniae (Pajand et al., 2023). The blaCTX-M-15 gene was in close proximity to ISEcp1, which is a member of the IS1380 family (Poirel et al., 2008), and has been identified as one of several elements responsible for facilitating the transfer of blaCTX-M genes across various species of Enterobacteriaceae (Shawa et al., 2021; Wang et al., 2023). The blaNDM-1 and bleMBL genes (a class B3 β-lactamases with carbapenemase activity) were close to the insertion sequence ISAba125 (IS30 family). Studies have proposed that the ISAba125–bleNDM combination occurred initially in Acinetobacter spp. and later transferred to other Gram-negative bacteria (Castanheira et al., 2023). Additionally, the ISAba125–blaNDM-1–bleMBL combination has been found in a structure referred to as NDM-GE-U.S, first observed in a K. pneumoniae strain from the United States and subsequently detected in various strains worldwide (Hudson et al., 2014; Peirano et al., 2018).
The qnrS1 the aph(3”)-VI genes, were found in close proximity to the ISKpn19 insertion sequence of the ISKra4 family in our isolate. Studies have shown that qnrS1 gene, which confers resistance to ciprofloxacin, is related to ISKpn19 in various bacteria, such as Leclercia adecarboxylata, Salmonella corvallis; K. pneumoniae, E. coli, and S. marcescens (Xu et al., 2020; Chen et al., 2023; Sano et al., 2023; Zheng et al., 2024). The aph(3”)-VI gene, responsible for amikacin modification, was not found to be flanked by the ISKpn19 insertion sequence in our literature review. It is worth noting that K. aerogenes CRKA317 presents a large number of singleton genes, including blaNDM-1, aadA/ant(3”)-Ia, aph(3’)-VI, and qnrS1 and its presence indicates the variability and diversity within the genome of our strain. It is possible that these genes were obtained from other lineages through horizontal gene transfer, leading to the development of new functions, such as resistance to antibiotics. This assumption can be reinforced by the presence of a genomic island that carries most of the resistance genes (blaOXA-9, blaTEM-1, blaNDM-1, bleMBL, blaCTX-M-15, aac(6’)-Ib, aadA/ant(3”)-Ia, aph(3’)-VI, and qnrS1) found in our genome. Genomic islands are distinct regions in the bacterial genome with genes related to each other and often associated with specific functions. These islands are often acquired through horizontal gene transfer events and are associated with the widespread distribution of antimicrobial resistance factors among bacteria (Juhas et al., 2009). The composition of genomic islands is conducive to the acquisition of new antibiotic resistance genes, as they include several MGEs, which facilitate the incorporation of new genes, but also their own transfer, for example, using tRNA genes as recombination sites into the chromosome (da Silva Filho et al., 2018).
Our K. aerogenes CRKA317 displayed many genes of interested linked to antibiotic resistance (Table 3). The ramA, acrR, acrA and acrB genes, and MFS_MMR_MDR-like gene were found to be associated with MITEEc. MITEEc belongs to the IS630 family and was found in an extensively drug-resistant (XDR) Escherichia coli isolate (Jain et al., 2021). RamA belongs to the AraC/XylS protein family and shows a close association with the marA and soxS proteins (Rosenblum et al., 2011). Elevated expression of ramA is associated with the activation of the acrAB efflux pump, which confers multidrug resistance in various bacterial species such as E. aerogenes, K. pneumoniae, and E. cloacae (Chollet et al., 2004; Keeney et al., 2007; Ruzin et al., 2008). The acrR gene regulates the multidrug efflux pump AcrAB-TolC (Subhadra et al., 2018). The MFS_MMR_MDR-like gene is linked to methylenomycin resistance, and the antibiotic Methylenomycin A is produced naturally by Streptomyces coelicolor A3, a model organism for streptomycetes (Bentley et al., 2002; Bowyer et al., 2017). The acrAB-tolC system where the acrAB fusion protein, members of the RND-type efflux family, function with another antibiotic efflux pump, tolC, to pump out various compounds such as SDS, novobiocin, deoxycholate, aminoglycosides, and dianionic β-lactams including carbenicillin, oxacillin, nafcillin, and aztreonam (Guérin et al., 2023). We also found many other RND antibiotic efflux pumps, the first pair being eefA and eefB which are part of the eefABC locus known to encode a tripartite efflux pump that gives rise to resistance to erythromycin and other antibiotics in E. aerogenes (Pradel and Pagès, 2002; Masi et al., 2007). The oqxA and oqxB operon has also been described to increase antibiotic resistance in E. aerogenes, this time to quinolones as they combine to make the oqxAB efflux pump (Wong et al., 2015; Moosavian et al., 2021). The HAE1 family, also contained in our isolate, contains large number of identified RND transporters (Nikaido, 2018). These pumps are commonly found in Gram-negative bacteria, typically existing as trimers, and are involved in the transportation of drugs and other hydrophobic substances (Nikaido, 2018). In addition, we found members of the acrB/acrD/acrF family, specifically acrD, mdtA and mdtB. The mdtABC operon is transcriptionally activated by baeR and leads to the formation of the mdtABC tripartite complex, which provides resistance to novobiocin and deoxycholate in E. coli (Nagakubo et al., 2002). Related to this complex, it was found mdtH and mdtK both of which give rise to proteins that function has multidrug efflux pumps that contribute to resistance against quinolone antibiotics such as norfloxacin and enoxacin in E. coli strains (Nishino and Yamaguchi, 2001; Yu et al., 2020). The final RND antibiotic efflux pump sequenced in our isolate is oprM which is part of MexX-MexY-OprM efflux systems that mediate intrinsic antibiotic resistance to aminoglycosides and erythromycin in bacteria such as P. aeruginosa (Wong et al., 2001), Brevundimonas brasiliensis sp. nov and Burkholderia vietnamiensis (Shinoy et al., 2013; Soares et al., 2023).
We also found putative ICE with T4SS just harboring sul-2 gene, encoding for resistance to sulfonamide, and sox gene, a key component of a central regulatory system present in all Enterobacteriaceae, which detects and reacts to internal chemical stressors like antibiotics (Chubiz, 2023). ICEs are mobile elements integrated into the chromosomes that can be excised and transferred horizontally to other bacteria and, therefore, have been associated with antimicrobial resistance genes (Johnson and Grossman, 2015). Despite this, due to the finding of only one resistance gene in the ICEs studied here (sul-2), we can assume that this structure is not the main source of resistance gene acquisition in K. aerogenes CRKA317.
Genes associated with porins were the second category identified in our isolate (Table 3). Porins belong to a category of transmembrane proteins called omps, which form small channels in the membrane and facilitate the passive movement of hydrophilic compounds. They regulate cellular permeability and can either enhance or reduce resistance to antibiotics. In our strain, we specifically found the outer membrane protein encoding genes oprD, ompC, ompA, ompX and ompW which have been found to have clinical significance. For instance, a reduction or absence of OmpC in clinical E. aerogenes isolates has been linked to a slight increase in imipenem MIC (Lavigne et al., 2012). Meanwhile, overexpressing ompX in E. aerogenes results in elevated resistance to β-lactam antibiotics, possibly due to significant reduction in the Omp36 porin (Hejair et al., 2017). OmpW expression in A. baumannii isolates was found to increase when exposed to ciprofloxacin and decrease when exposed to imipenem (Gurpinar et al., 2022). Conversely, A. baumannii strains with mutations in ompA exhibited reduced permeability for cephalothin/cephaloridine and lower minimum inhibitory concentrations for a range of antibiotics including imipenem, colistin, meropenem, chloramphenicol, aztreonam, and nalidixic acid (Smani et al., 2014; Tsai et al., 2020). Finally, in P. aeruginosa, the porin oprD plays a significant role in the uptake of basic amino acids and carbapenems (Wong et al., 2001).
Other genes of interest included: a superoxide response transcriptional regulator (soxS), a multiple antibiotic resistance transcriptional regulator (marA), and a major facilitator superfamily member (kdeA),
Prophages play a role in the survival mechanisms of their hosts and contribute to the enhancement of genetic diversity within the host genome (Kondo et al., 2021). In our study, we found two intact regions which were associated with the presence of a prophage highly similar to the Salmonella phage SEN34 (National Center for Biotechnology Information reference sequence NC_028699.1), and Escherichia phage vB_EcoM_ECO1230-10 (NC_027995.1). Prophage regions of Salmonella phage SEN34 (NC_028699.1) has been identified in Salmonella salamae (Hounmanou et al., 2022), and Salmonella enterica serovar Paratyphi B (Castellanos et al., 2020) and have been linked to drug resistance.
There are limitations to our study that need to be acknowledged. We encountered difficulties in assembling complete plasmid sequences, primarily due to the short reads generated by high-throughput sequencer. This can result in antimicrobial resistance genes being located on incomplete contigs, leading to uncertainty about whether they are situated on a plasmid or within the chromosome (Orlek et al., 2017; Berbers et al., 2020). Nonetheless, it is important to highlight those studies conducted in China have shown that clinical isolates of carbapenem-resistant K. aerogenes carried blaNDM-1 gene on plasmids of the IncFIIAs type. In another study, Shen et al. (2019) identified a plasmid (p1564) containing genes for plasmid replication (IncA/C repA), antibiotic resistance (blaNDM-1, rmtC, aacA4, bleMBL, blaCMY-6 and sul-1), and conjugation (tra clusters). In K. aerogenes, there is still no consensus on the location of the blaKPC-2 gene in the plasmid, the transposon variants capable of carrying this gene, and which incompatibility (Inc) groups carry the blaKPC gene (Bispo Beltrão et al., 2020). However, a study conducted by Bispo Beltrão et al. (2020) in Brazil has described a non-Tn4401 element (NTEKPC-IId) that carries the blaKPC-2 and aph(3’)-VII genes in IncQ1 plasmids in K. aerogenes. To date, the IncQ1 blaKPC-2-positive plasmids have been found in different strains such as E. coli, K. pneumoniae of CG258, Klebsiella quasipneumoniae, and P. aeruginosa (de Oliveira Santos et al., 2018).
Although K. aerogens has not been reported to carry the plasmids found in our sequencing study, it is important to note genes such as blaKPC and blaNDM are commonly associated with the plasmid fragments found in K. aerogenes CRKA317. Takei et al. (2022) found nine isolates of K. pneumoniae carrying blaNDM-1 and blaCTX-M-15 on the IncFIB (pQil) plasmid and another five isolates carrying blaNDM-1 on the IncC plasmid. Similar data were also observed in the results of Zeng et al. (2022), who identified the blaNDM-1 gene in IncC plasmids from 21 K. pneumoniae isolates. Finally, the fragmented INCFIIK plasmid observed in our genome has already been noted to carry genes such as blaKPC-2, blaCTX-M-15, blaTEM-1 and, less commonly, blaNDM-1 (Chen et al., 2013; Bi et al., 2018). This suggests an emerging mechanism, using Inc. groups, that plays a role in the dissemination of carbapenem resistance in clinically important bacteria.
In conclusion, our current research has uncovered a concerning scenario involving K. aerogenes demonstrating resistance to commonly utilized drugs for treating infections, including those considered as last-resort options for life-threatening infections in ICU patients. Moreover, the presence of mobile genetic elements highlights the alarming potential for the transmission of various resistance genes such as blaNDM-1 and blaKPC-2 within hospital settings to susceptible populations. This scenario poses significant challenges for managing infectious diseases and underscores the necessity of early detection of such genetic features or mutations.
Our study did not involve human genetic material or biological samples. The strains were obtained from the collection of the Central Laboratory of Public Health, a leading diagnostic center in Tocantins, Brazil. This was a retrospective study and epidemiological data were obtained from a database at LACEN-TO in accordance with Resolution 466/12 of the National Health Council (Conselho Nacional de Saúde/Ministério da Saúde, 2012). Informed consent was not required as per Resolution 466/12 regarding research involving humans by the National Health Council. The study received approval from the Committee of Ethics in Human Research at the Federal University of São Carlos (no. 1.088.936), and permissions to conduct it were obtained from the State Department of Health in Tocantins and LACEN/TO.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The studies involving humans were approved by Committee of Ethics in Human Research at the Federal University of São Carlos (no. 1.088.936). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
SR: Formal analysis, Investigation, Methodology, Writing – review & editing. GN: Formal analysis, Investigation, Methodology, Writing – review & editing. GS: Formal analysis, Investigation, Methodology, Writing – review & editing. MD: Formal analysis, Investigation, Methodology, Writing – review & editing. RF: Formal analysis, Investigation, Methodology, Writing – review & editing. PL: Formal analysis, Investigation, Methodology, Writing – review & editing. RS: Formal analysis, Methodology, Writing – review & editing. LC: Methodology, Writing – review & editing. AC: Formal analysis, Writing – review & editing. IM: Visualization, Writing – review & editing. AC: Visualization, Writing – review & editing. M-CP: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo-Brazil (FAPESP grant 2022/16872-6, 2020/11964-4, 2024/00886-3 to M-CP; and FAPESP grant 2022/01223-2, 2018/20697-0 to AC). This study was partially financed by the Fundação de Amparo à Pesquisa do Estado de São Paulo-Brazil (FAPESP) as a fellowship to SR. (FAPESP fellowship 2022/12429-0), GN (FAPESP fellowship 2023/08917-2), GS (FAPESP fellowship 2021/08423-4), MD (FAPESP fellowship 2018/24213-7), and PL (FAPESP fellowship 2021/00425-8).
The authors thank the Central Public Health Laboratory of the State of Tocantins (LACEN/TO) - Brazil for generously supplying the K. aerogenes strains and Tocantins State Health Department (SES/Tocantins) for enabling the progress of this project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1352851/full#supplementary-material
1. ^https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
2. ^https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
3. ^https://cge.food.dtu.dk/services/PlasmidFinder/
6. ^http://eggnog-mapper.embl.de/
7. ^https://www.kegg.jp/blastkoala/
8. ^https://www.ebi.ac.uk/Tools/msa/mafft/
9. ^https://ggdc.dsmz.de/ggdc.php
10. ^https://discover.nci.nih.gov/cimminer/oneMatrix.do
12. ^https://realphy.unibas.ch/realphy/
13. ^https://orthovenn2.bioinfotoolkits.net/task/create
14. ^https://card.mcmaster.ca/analyze/rgi
15. ^http://genepi.food.dtu.dk/resfinder
16. ^https://galaxy.pasteur.fr/
17. ^https://www.kegg.jp/blastkoala/
18. ^https://www.pathogenomics.sfu.ca/islandviewer/
19. ^https://tncentral.ncc.unesp.br/
20. ^https://www-is.biotoul.fr/index.php
21. ^https://cge.food.dtu.dk/services/MobileElementFinder/
22. ^https://bioinfo-mml.sjtu.edu.cn/ICEfinder/ICEfinder.html
23. ^https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index
Afgan, E., Baker, D., Batut, B., van den Beek, M., Bouvier, D., Čech, M., et al. (2018). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. doi: 10.1093/nar/gky379
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2019). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. doi: 10.1093/nar/gkz935
Almaghrabi, R., Clancy, C. J., Doi, Y., Hao, B., Chen, L., Shields, R. K., et al. (2014). Carbapenem-resistant Klebsiella pneumoniae strains exhibit diversity in aminoglycoside-modifying enzymes, which exert differing effects on Plazomicin and other agents. Antimicrob. Agents Chemother. 58, 4443–4451. doi: 10.1128/AAC.00099-14
Aly, S. A., Boothe, D. M., and Suh, S.-J. (2015). A novel alanine to serine substitution mutation in sox S induces overexpression of efflux pumps and contributes to multidrug resistance in clinical Escherichia coli isolates. J. Antimicrob. Chemother. 70, 2228–2233. doi: 10.1093/jac/dkv105
Andersen, J., He, G.-X., Kakarla, P. K. C. R., Kumar, S., Lakra, W., et al. (2015). Multidrug efflux pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus bacterial food pathogens. Int. J. Environ. Res. Public Health 12, 1487–1547. doi: 10.3390/ijerph120201487
Antipov, D., Hartwick, N., Shen, M., Raiko, M., Lapidus, A., and Pevzner, P. A. (2016). Plasmid SPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32, 3380–3387. doi: 10.1093/bioinformatics/btw493
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Azevedo, P. A. A., Furlan, J. P. R., Oliveira-Silva, M., Nakamura-Silva, R., Gomes, C. N., Costa, K. R. C., et al. (2018). Detection of virulence and β-lactamase encoding genes in Enterobacter aerogenes and Enterobacter cloacae clinical isolates from Brazil. Braz. J. Microbiol. 49, 224–228. doi: 10.1016/j.bjm.2018.04.009
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bean, E. L., Herman, C., Anderson, M. E., and Grossman, A. D. (2022). Biology and engineering of integrative and conjugative elements: construction and analyses of hybrid ICEs reveal element functions that affect species-specific efficiencies. PLoS Genet. 18:e1009998. doi: 10.1371/journal.pgen.1009998
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A.-M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. doi: 10.1038/417141a
Berbers, B., Ceyssens, P.-J., Bogaerts, P., Vanneste, K., Roosens, N. H. C., Marchal, K., et al. (2020). Development of an NGS-based workflow for improved monitoring of circulating plasmids in support of risk assessment of antimicrobial resistance gene dissemination. Antibiotics 9:503. doi: 10.3390/antibiotics9080503
Bertelli, C., Laird, M. R., Williams, K. P., Lau, B. Y., Hoad, G., Winsor, G. L., et al. (2017). Island viewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Bertels, F., Silander, O. K., Pachkov, M., Rainey, P. B., and van Nimwegen, E. (2014). Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 31, 1077–1088. doi: 10.1093/molbev/msu088
Bi, D., Zheng, J., Li, J.-J., Sheng, Z.-K., Zhu, X., Ou, H.-Y., et al. (2018). In silico typing and comparative genomic analysis of Inc FII K plasmids and insights into the evolution of replicons, plasmid backbones, and resistance determinant profiles. Antimicrob. Agents Chemother. 62. doi: 10.1128/AAC.00764-18
Bispo Beltrão, E. M., de Oliveira, É. M., dos Santos Vasconcelos, C. R., Cabral, A. B., Rezende, A. M., and Souza Lopes, A. C. (2020). Multidrug-resistant Klebsiella aerogenes clinical isolates from Brazil carrying Inc Q1 plasmids containing the Bla KPC-2 gene associated with non-Tn4401 elements (NTEKPC-IId). J. Glob. Antimicrob. Resist. 22, 43–44. doi: 10.1016/j.jgar.2020.05.001
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D., and Pirovano, W. (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579. doi: 10.1093/bioinformatics/btq683
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). Res finder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Bouza, E. (2021). The role of new carbapenem combinations in the treatment of multidrug-resistant gram-negative infections. J. Antimicrob. Chemother. 76, iv38–iv45. doi: 10.1093/jac/dkab353
Bowyer, J. E. L. C., De los Santos, E., Styles, K. M., Fullwood, A., Corre, C., and Bates, D. G. (2017). Modeling the architecture of the regulatory system controlling methylenomycin production in Streptomyces coelicolor. J. Biol. Eng. 11:30. doi: 10.1186/s13036-017-0071-6
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). Egg NOG-mapper v2: functional annotation, Orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). Detection and typing of plasmids using plasmid finder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Castanheira, M., Mendes, R. E., and Gales, A. C. (2023). Global epidemiology and mechanisms of resistance of Acinetobacter baumannii-calcoaceticus complex. Clin. Infect. Dis. 76, S166–S178. doi: 10.1093/cid/ciad109
Castanheira, M., Simner, P. J., and Bradford, P. A. (2021). Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC Antimicrob Resist 3. doi: 10.1093/jacamr/dlab092
Castellanos, L. R., van der Graaf-van Bloois, L., Donado-Godoy, P., Veldman, K., Duarte, F., Acuña, M. T., et al. (2020). Antimicrobial resistance in Salmonella enterica Serovar Paratyphi B variant Java in poultry from Europe and Latin America. Emerg. Infect. Dis. 26, 1164–1173. doi: 10.3201/eid2606.191121
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
Chen, L., Chavda, K. D., Melano, R. G., Jacobs, M. R., Levi, M. H., Bonomo, R. A., et al. (2013). Complete sequence of a Bla KPC-2-harboring Inc FII K1 plasmid from a Klebsiella pneumoniae sequence type 258 strain. Antimicrob. Agents Chemother. 57, 1542–1545. doi: 10.1128/AAC.02332-12
Chen, Z., Li, H., Feng, J., Li, Y., Chen, X., Guo, X., et al. (2015). NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front. Microbiol. 6:294. doi: 10.3389/fmicb.2015.00294
Chen, M., Shi, X., Yu, Z., Fan, G., Serysheva, I. I., Baker, M. L., et al. (2022). In situ structure of the Acr AB-Tol C efflux pump at subnanometer resolution. Structure 30, 107–113.e3. doi: 10.1016/j.str.2021.08.008
Chen, K., Xie, M., Wang, H., Chan, E. W.-C., and Chen, S. (2023). Intercontinental spread and clonal expansion of col RNA1 plasmid-bearing Salmonella Corvallis ST1541 strains: a genomic epidemiological study. One Health Advances 1:16. doi: 10.1186/s44280-023-00017-9
Chevalier, J., Mulfinger, C., Garnotel, E., Nicolas, P., Davin-Régli, A., and Pagès, J.-M. (2008). Identification and evolution of drug efflux pump in clinical Enterobacter aerogenes strains isolated in 1995 and 2003. PLoS One 3:e3203. doi: 10.1371/journal.pone.0003203
Chollet, R., Chevalier, J., Bollet, C., Pages, J.-M., and Davin-Regli, A. (2004). Ram a is an alternate activator of the multidrug resistance Cascade in Enterobacter aerogenes. Antimicrob. Agents Chemother. 48, 2518–2523. doi: 10.1128/AAC.48.7.2518-2523.2004
Chubiz, L. M. (2023). The mar, sox, and rob Systems. Eco Sal Plus 11. doi: 10.1128/ecosalplus.esp-0010-2022
CLSI. (2023). Performance Standards for Antimicrobial Susceptibility Testing. 33rd ed. Wayne PA: CLSI supplement M100.
Costa, S. S., Guimarães, L. C., Silva, A., Soares, S. C., and Baraúna, R. A. (2020). First steps in the analysis of prokaryotic Pan-genomes. Bioinform Biol Insights 14:117793222093806. doi: 10.1177/1177932220938064
Couvin, D., Bernheim, A., Toffano-Nioche, C., Touchon, M., Michalik, J., Néron, B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–W251. doi: 10.1093/nar/gky425
da Silva Filho, A. C., Raittz, R. T., Guizelini, D., De Pierri, C. R., Augusto, D. W., dos Santos-Weiss, I. C. R., et al. (2018). Comparative analysis of Genomic Island prediction tools. Front. Genet. 9:619. doi: 10.3389/fgene.2018.00619
Damas, M. S. F., Ferreira, R. L., Campanini, E. B., Soares, G. G., Campos, L. C., Laprega, P. M., et al. (2022). Whole genome sequencing of the multidrug-resistant Chryseobacterium indologenes isolated from a patient in Brazil. Front. Med (Lausanne) 9:931379. doi: 10.3389/fmed.2022.931379
Davin-Regli, A., Lavigne, J.-P., and Pagès, J.-M. (2019). Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin. Microbiol. Rev. 32. doi: 10.1128/CMR.00002-19
Davin-Regli, A., and Pages, J.-M. (2015). Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 6:392. doi: 10.3389/fmicb.2015.00392
de Oliveira Santos, I. C., Albano, R. M., Asensi, M. D., and D’Alincourt Carvalho-Assef, A. P. (2018). Draft genome sequence of KPC-2-producing Pseudomonas aeruginosa recovered from a bloodstream infection sample in Brazil. J. Glob. Antimicrob. Resist. 15, 99–100. doi: 10.1016/j.jgar.2018.08.021
Denissen, J., Reyneke, B., Waso-Reyneke, M., Havenga, B., Barnard, T., Khan, S., et al. (2022). Prevalence of ESKAPE pathogens in the environment: antibiotic resistance status, community-acquired infection and risk to human health. Int. J. Hyg. Environ. Health 244:114006. doi: 10.1016/j.ijheh.2022.114006
Dobiasova, H., Videnska, P., and Dolejska, M. (2016). Complete Sequences of IncU Plasmids Harboring Quinolone Resistance Genes qnrS2 and aac(6\u0027)-Ib-cr in Aeromonas spp. from Ornamental Fish. Antimicrob Agents Chemother 60, 653–657. doi: 10.1128/AAC.01773-15
Durrant, M. G., Li, M. M., Siranosian, B. A., Montgomery, S. B., and Bhatt, A. S. (2020). A Bioinformatic analysis of integrative Mobile genetic elements highlights their role in bacterial adaptation. Cell Host Microbe 27, 140–153.e9. doi: 10.1016/j.chom.2019.10.022
Ferreira, R. L., da Silva, B. C. M., Rezende, G. S., Nakamura-Silva, R., Pitondo-Silva, A., Campanini, E. B., et al. (2019). High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Front. Microbiol. 9:3198. doi: 10.3389/fmicb.2018.03198
Ferreira, R. L., Rezende, G. S., Damas, M. S. F., Oliveira-Silva, M., Pitondo-Silva, A., Brito, M. C. A., et al. (2020). Characterization of KPC-producing Serratia marcescens in an intensive care unit of a Brazilian tertiary hospital. Front. Microbiol. 11:956. doi: 10.3389/fmicb.2020.00956
Firmo, E. F., Cabral, A. B., De Oliveira, É. M., and Lopes, A. C. S. (2019). Emergence of aph(3’)-VI and accumulation of aminoglycoside modifying enzyme genes in KPC-2-possessing Enterobacter aerogenes isolates from infections and colonization in patients from Recife-PE, Brazil. Rev. Soc. Bras. Med. Trop. 52. doi: 10.1590/0037-8682-0460-2018
Franolić, I., Bedenić, B., Beader, N., Lukić-Grlić, A., Mihaljević, S., Bielen, L., et al. (2019). NDM-1-producing Enterobacter aerogenes isolated from a patient with a JJ ureteric stent in situ. CEN Case Rep. 8, 38–41. doi: 10.1007/s13730-018-0360-z
Galani, I., Nafplioti, K., Adamou, P., Karaiskos, I., Giamarellou, H., and Souli, M. (2019). Nationwide epidemiology of carbapenem resistant Klebsiella pneumoniae isolates from Greek hospitals, with regards to plazomicin and aminoglycoside resistance. BMC Infect. Dis. 19:167. doi: 10.1186/s12879-019-3801-1
Ghanavati, R., Emaneini, M., Kalantar-Neyestanaki, D., Maraji, A. S., Dalvand, M., Beigverdi, R., et al. (2018). Clonal relation and antimicrobial resistance pattern of extended-spectrum β-lactamase- and amp C β-lactamase-producing Enterobacter spp. isolated from different clinical samples in Tehran, Iran. Rev. Soc. Bras. Med. Trop. 51, 88–93. doi: 10.1590/0037-8682-0227-2017
Grant, J. R., Enns, E., Marinier, E., Mandal, A., Herman, E. K., Chen, C., et al. (2023). Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51, W484–W492. doi: 10.1093/nar/gkad326
Grazziotin, A. L., Vidal, N. M., Palmeiro, J. K., Dalla-Costa, L. M., and Venancio, T. M. (2016). Genome sequencing of four multidrug-resistant Enterobacter aerogenes isolates from hospitalized patients in Brazil. Front. Microbiol. 7:1649. doi: 10.3389/fmicb.2016.01649
Guan, X., Xue, X., Liu, Y., Wang, J., Wang, Y., Wang, J., et al. (2013). Plasmid-mediated quinolone resistance – current knowledge and future perspectives. J. Int. Med. Res. 41, 20–30. doi: 10.1177/0300060513475965
Guérin, F., Gravey, F., Reissier, S., Penven, M., Michaux, C., Le Hello, S., et al. (2023). Temocillin resistance in the Enterobacter cloacae Complex is conferred by a single point mutation in bae S, leading to overexpression of the Acr D efflux pump. Antimicrob. Agents Chemother. 67. doi: 10.1128/aac.00358-23
Guindon, S., Delsuc, F., Dufayard, J.-F., and Gascuel, O. (2009). Estimating maximum likelihood phylogenies with Phy ML, 113–137.
Gupta, S. K., Padmanabhan, B. R., Diene, S. M., Lopez-Rojas, R., Kempf, M., Landraud, L., et al. (2014). ARG-ANNOT, a new Bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220. doi: 10.1128/AAC.01310-13
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Gurpinar, S. S., Kart, D., and Eryilmaz, M. (2022). The effects of antidepressants fluoxetine, sertraline, and amitriptyline on the development of antibiotic resistance in Acinetobacter baumannii. Arch. Microbiol. 204:230. doi: 10.1007/s00203-022-02853-6
Haldorsen, B. C., Simonsen, G. S., Sundsfjord, A., and Samuelsen, Ø. (2014). Increased prevalence of aminoglycoside resistance in clinical isolates of Escherichia coli and Klebsiella spp. in Norway is associated with the acquisition of AAC (3)-II and AAC (6′)-Ib. Diagn. Microbiol. Infect. Dis. 78, 66–69. doi: 10.1016/j.diagmicrobio.2013.10.001
Hall, T. A. (1999). Bio edit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Han, J. E., Kim, J. H., Choresca, C. H., Shin, S. P., Jun, J. W., Chai, J. Y., et al. (2012). First description of ColE-type plasmid in Aeromonas spp. carrying quinolone resistance (qnrS2) gene. Lett Appl Microbiol 55, 290–294. doi: 10.1111/j.1472-765X.2012.03293.x
Hayashi, M., Tabata, K., Yagasaki, M., and Yonetani, Y. (2010). Effect of multidrug-efflux transporter genes on dipeptide resistance and overproduction in Escherichia coli. FEMS Microbiol. Lett. 304, 12–19. doi: 10.1111/j.1574-6968.2009.01879.x
Hejair, H. M. A., Zhu, Y., Ma, J., Zhang, Y., Pan, Z., Zhang, W., et al. (2017). Functional role of omp F and omp C porins in pathogenesis of avian pathogenic Escherichia coli. Microb. Pathog. 107, 29–37. doi: 10.1016/j.micpath.2017.02.033
Hounmanou, Y. M. G., Baniga, Z., García, V., and Dalsgaard, A. (2022). Salmonella Salamae and S. Waycross isolated from Nile perch in Lake Victoria show limited human pathogenic potential. Sci. Rep. 12:4229. doi: 10.1038/s41598-022-08200-5
Huang, L., Yuan, H., Liu, M.-F., Zhao, X.-X., Wang, M.-S., Jia, R.-Y., et al. (2017). Type B chloramphenicol acetyltransferases are responsible for chloramphenicol resistance in Riemerella anatipestifer, China. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00297
Hudson, C. M., Bent, Z. W., Meagher, R. J., and Williams, K. P. (2014). Resistance determinants and Mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS One 9:e99209. doi: 10.1371/journal.pone.0099209
Ito, R., Mustapha, M. M., Tomich, A. D., Callaghan, J. D., McElheny, C. L., Mettus, R. T., et al. (2017). Widespread Fosfomycin resistance in gram-negative Bacteria attributable to the chromosomal fos a gene. MBio :8. doi: 10.1128/mBio.00749-17
Iyer, R., Iken, B., and Damania, A. (2017). Whole genome of Klebsiella aerogenes PX01 isolated from San Jacinto River sediment west of Baytown, Texas reveals the presence of multiple antibiotic resistance determinants and mobile genetic elements. Genom Data 14, 7–9. doi: 10.1016/j.gdata.2017.07.012
Jacoby, G. A. (2009). Amp C β-Lactamases. Clin. Microbiol. Rev. 22, 161–182. doi: 10.1128/CMR.00036-08
Jain, P., Bepari, A. K., Sen, P. K., Rafe, T., Imtiaz, R., Hossain, M., et al. (2021). High prevalence of multiple antibiotic resistance in clinical E. coli isolates from Bangladesh and prediction of molecular resistance determinants using WGS of an XDR isolate. Sci. Rep. 11:22859. doi: 10.1038/s41598-021-02251-w
Jamrozy, D., Coll, F., Mather, A. E., Harris, S. R., Harrison, E. M., Mac Gowan, A., et al. (2017). Evolution of mobile genetic element composition in an epidemic methicillin-resistant Staphylococcus aureus: temporal changes correlated with frequent loss and gain events. BMC Genomics 18:684. doi: 10.1186/s12864-017-4065-z
Jang, S. (2023). Acr AB-Tol C, a major efflux pump in gram negative bacteria: toward understanding its operation mechanism. BMB Rep. 56, 326–334. doi: 10.5483/BMBRep.2023-0070
Johnson, C. M., and Grossman, A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 49, 577–601. doi: 10.1146/annurev-genet-112414-055018
Juhas, M., van der Meer, J. R., Gaillard, M., Harding, R. M., Hood, D. W., and Crook, D. W. (2009). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393. doi: 10.1111/j.1574-6976.2008.00136.x
Kamio, K., and Espinoza, J. L. (2022). The predominance of Klebsiella aerogenes among Carbapenem-resistant Enterobacteriaceae infections in Japan. Pathogens 11:722. doi: 10.3390/pathogens11070722
Kanehisa, M., Sato, Y., and Morishima, K. (2016). Blast KOALA and ghost KOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Keeney, D., Ruzin, A., and Bradford, P. A. (2007). Ram a, a transcriptional regulator, and Acr AB, an RND-type efflux pump, are associated with decreased susceptibility to Tigecycline in Enterobacter cloacae. Microb. Drug Resist. 13, 1–6. doi: 10.1089/mdr.2006.9990
Kondo, K., Kawano, M., and Sugai, M. (2021). Distribution of antimicrobial resistance and virulence genes within the prophage-associated regions in nosocomial pathogens. mSphere :6. doi: 10.1128/mSphere.00452-21
Koroiva, R., and Santana, D. J. (2022). Evaluation of partial 12S rRNA, 16S rRNA, COI and Cytb gene sequence datasets for potential single DNA barcode for hylids (Anura: Hylidae). An. Acad. Bras. Cienc. 94. doi: 10.1590/0001-3765202220200825
Lavigne, J.-P., Sotto, A., Nicolas-Chanoine, M.-H., Bouziges, N., Bourg, G., Davin-Regli, A., et al. (2012). Membrane permeability, a pivotal function involved in antibiotic resistance and virulence in Enterobacter aerogenes clinical isolates. Clin. Microbiol. Infect. 18, 539–545. doi: 10.1111/j.1469-0691.2011.03607.x
Liu, M., Li, X., Xie, Y., Bi, D., Sun, J., Li, J., et al. (2019). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47, D660–D665. doi: 10.1093/nar/gky1123
Ma, D.-Y., Huang, H.-Y., Zou, H., Wu, M.-L., Lin, Q.-X., Liu, B., et al. (2020). Carbapenem-resistant Klebsiella aerogenes clinical isolates from a teaching Hospital in Southwestern China: detailed molecular epidemiology, resistance determinants, risk factors and clinical outcomes. Infect. Drug Resist. 13, 577–585. doi: 10.2147/IDR.S235975
Machuca, J., Ortiz, M., Recacha, E., Díaz-De-Alba, P., Docobo-Perez, F., Rodríguez-Martínez, J.-M., et al. (2016). Impact of AAC (6′)-Ib-cr in combination with chromosomal-mediated mechanisms on clinical quinolone resistance in Escherichia coli. J. Antimicrob. Chemother. 71, 3066–3071. doi: 10.1093/jac/dkw258
Magiorakos, A.-P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Maneewannakul, K., and Levy, S. B. (1996). Identification for mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40, 1695–1698. doi: 10.1128/AAC.40.7.1695
Masi, M., Saint, N., Molle, G., and Pagès, J.-M. (2007). The Enterobacter aerogenes outer membrane efflux proteins Tol C and Eef C have different channel properties. Biochim. Biophys. Acta Biomembr. 1768, 2559–2567. doi: 10.1016/j.bbamem.2007.06.008
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L., and Göker, M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 50, D801–D807. doi: 10.1093/nar/gkab902
Monárrez, R., Wang, Y., Fu, Y., Liao, C.-H., Okumura, R., Braun, M. R., et al. (2018). Genes and proteins involved in qnr S1 induction. Antimicrob. Agents Chemother. 62. doi: 10.1128/AAC.00806-18
Moosavian, M., Khoshkholgh Sima, M., Ahmad Khosravi, N., and Abbasi Montazeri, E. (2021). Detection of Oqx AB efflux pumps, a multidrug-resistant agent in bacterial infection in patients referring to teaching hospitals in Ahvaz, southwest of Iran. Int. J. Microbiol. 2021, 1–5. doi: 10.1155/2021/2145176
Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S., and Pardesi, K. R. (2019). Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front. Microbiol. 10:539. doi: 10.3389/fmicb.2019.00539
Nagakubo, S., Nishino, K., Hirata, T., and Yamaguchi, A. (2002). The putative response regulator bae R stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, Mdt ABC. J. Bacteriol. 184, 4161–4167. doi: 10.1128/JB.184.15.4161-4167.2002
Nikaido, H. (2018). RND transporters in the living world. Res. Microbiol. 169, 363–371. doi: 10.1016/j.resmic.2018.03.001
Nishino, K., and Yamaguchi, A. (2001). Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183, 5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001
Obayiuwana, A., and Ibekwe, A. M. (2020). Antibiotic resistance genes occurrence in wastewaters from selected pharmaceutical facilities in Nigeria. Water (Basel) 12:1897. doi: 10.3390/w12071897
Orlek, A., Stoesser, N., Anjum, M. F., Doumith, M., Ellington, M. J., Peto, T., et al. (2017). Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front. Microbiol. 8:182. doi: 10.3389/fmicb.2017.00182
Pajand, O., Rahimi, H., Badmasti, F., Gholami, F., Alipour, T., Darabi, N., et al. (2023). Various arrangements of mobile genetic elements among CC147 subpopulations of Klebsiella pneumoniae harboring Bla NDM-1: a comparative genomic analysis of carbapenem resistant strains. J. Biomed. Sci. 30:73. doi: 10.1186/s12929-023-00960-0
Pan, F., Xu, Q., and Zhang, H. (2021). Emergence of NDM-5 producing Carbapenem-resistant Klebsiella aerogenes in a pediatric Hospital in Shanghai, China. Front. Public Health 9:621527. doi: 10.3389/fpubh.2021.621527
Peirano, G., Matsumura, Y., Adams, M. D., Bradford, P., Motyl, M., Chen, L., et al. (2018). Genomic epidemiology of global Carbapenemase-producing Enterobacter spp., 2008–2014. Emerg. Infect. Dis. 24, 1010–1019. doi: 10.3201/eid2406.171648
Picão, R. C., Poirel, L., Demarta, A., Silva, C. S. F., Corvaglia, A. R., Petrini, O., et al. (2008). Plasmid-mediated quinolone resistance in Aeromonas allosaccharophila recovered from a Swiss lake. J. Antimicrob. Chemother. 62, 948–950. doi: 10.1093/jac/dkn341
Poirel, L., Naas, T., and Nordmann, P. (2008). Genetic support of extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14, 75–81. doi: 10.1111/j.1469-0691.2007.01865.x
Posada, D. (2008). jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Pradel, E., and Pagès, J.-M. (2002). The Acr AB-Tol C efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob. Agents Chemother. 46, 2640–2643. doi: 10.1128/AAC.46.8.2640-2643.2002
Pulcrano, G., Pignanelli, S., Vollaro, A., Esposito, M., Iula, V. D., Roscetto, E., et al. (2016). Isolation of Enterobacter aerogenes carrying bla TEM-1 and Bla KPC-3 genes recovered from a hospital intensive care unit. APMIS 124, 516–521. doi: 10.1111/apm.12528
Rosenblum, R., Khan, E., Gonzalez, G., Hasan, R., and Schneiders, T. (2011). Genetic regulation of the ram a locus and its expression in clinical isolates of Klebsiella pneumoniae. Int. J. Antimicrob. Agents 38, 39–45. doi: 10.1016/j.ijantimicag.2011.02.012
Ross, K., Varani, A. M., Snesrud, E., Huang, H., Alvarenga, D. O., Zhang, J., et al. (2021). TnCentral: a prokaryotic transposable element database and web portal for transposon analysis. mBio :12. doi: 10.1128/mBio.02060-21
Rossi-Tamisier, M., Benamar, S., Raoult, D., and Fournier, P.-E. (2015). Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int. J. Syst. Evol. Microbiol. 65, 1929–1934. doi: 10.1099/ijs.0.000161
Ruzin, A., Immermann, F. W., and Bradford, P. A. (2008). Real-time PCR and statistical analyses of acr AB and ram a expression in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52, 3430–3432. doi: 10.1128/AAC.00591-08
Sano, E., Fontana, H., Esposito, F., Cardoso, B., Fuga, B., Costa, G. C. V., et al. (2023). Genomic analysis of fluoroquinolone-resistant Leclercia adecarboxylata carrying the ISKpn19-orf-qnr S1-ΔIS3-Bla LAP-2 module in a synanthropic pigeon, Brazil. J. Glob. Antimicrob. Resist. 33, 256–259. doi: 10.1016/j.jgar.2023.04.013
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shawa, M., Furuta, Y., Mulenga, G., Mubanga, M., Mulenga, E., Zorigt, T., et al. (2021). Novel chromosomal insertions of ISEcp1-Bla CTX-M-15 and diverse antimicrobial resistance genes in Zambian clinical isolates of Enterobacter cloacae and Escherichia coli. Antimicrob. Resist. Infect. Control 10:79. doi: 10.1186/s13756-021-00941-8
Shen, X., Liu, L., Yu, J., Cao, X., Zhan, Q., Guo, Y., et al. (2019). Coexistence of blaNDM-1 and rmt C on a transferrable plasmid of a novel ST192 Klebsiella aerogenes clinical isolate. Infect. Drug Resist. 12, 3883–3891. doi: 10.2147/IDR.S228130
Shin, H. W., Lim, J., Kim, S., Kim, J., Kwon, G. C., and Koo, S. H. (2015). Characterization of trimethoprim-sulfamethoxazole resistance genes and their relatedness to class 1 Integron and insertion sequence common region in gram-negative Bacilli. J. Microbiol. Biotechnol. 25, 137–142. doi: 10.4014/jmb.1409.09041
Shinoy, M., Dennehy, R., Coleman, L., Carberry, S., Schaffer, K., Callaghan, M., et al. (2013). Immunoproteomic analysis of proteins expressed by two related pathogens, Burkholderia multivorans and Burkholderia cenocepacia, during human infection. PLoS One 8:e80796. doi: 10.1371/journal.pone.0080796
Siguier, P. (2006). ISfinder: the reference Centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Siguier, P., Gourbeyre, E., and Chandler, M. (2014). Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol. Rev. 38, 865–891. doi: 10.1111/1574-6976.12067
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Smani, Y., Fàbrega, A., Roca, I., Sánchez-Encinales, V., Vila, J., and Pachón, J. (2014). Role of Omp a in the multidrug resistance phenotype of Acinetobacter baumannii. Antimicrob. Agents Chemother. 58, 1806–1808. doi: 10.1128/AAC.02101-13
Soares, G. G., Campanini, E. B., Ferreira, R. L., Damas, M. S. F., Rodrigues, S. H., Campos, L. C., et al. (2023). Brevundimonas brasiliensis sp. nov.: a new multidrug-resistant species isolated from a patient in Brazil. Microbiol. Spectr. 11. doi: 10.1128/spectrum.04415-22
Soares, C. R. P., Oliveira-Júnior, J. B., and Firmo, E. F. (2021). First report of a Bla NDM-resistant gene in a Klebsiella aerogenes clinical isolate from Brazil. Rev. Soc. Bras. Med. Trop. 54. doi: 10.1590/0037-8682-0262-2020
Sta Ana, K. M., Madriaga, J., and Espino, M. P. (2021). β-Lactam antibiotics and antibiotic resistance in Asian lakes and rivers: an overview of contamination, sources and detection methods. Environ. Pollut. 275:116624. doi: 10.1016/j.envpol.2021.116624
Starikova, E. V., Tikhonova, P. O., Prianichnikov, N. A., Rands, C. M., Zdobnov, E. M., Ilina, E. N., et al. (2020). Phigaro: high-throughput prophage sequence annotation. Bioinformatics 36, 3882–3884. doi: 10.1093/bioinformatics/btaa250
Subhadra, B., Kim, J., Kim, D. H., Woo, K., Oh, M. H., and Choi, C. H. (2018). Local repressor Acr R regulates Acr AB efflux pump required for biofilm formation and virulence in Acinetobacter nosocomialis. Front. Cell. Infect. Microbiol. 8:270. doi: 10.3389/fcimb.2018.00270
Takei, S., Lu, Y. J., Tohya, M., Watanabe, S., Misawa, S., Tabe, Y., et al. (2022). Spread of Carbapenem-resistant Klebsiella pneumoniae clinical isolates producing NDM-type Metallo-β-lactamase in Myanmar. Microbiol. Spectr. 10. doi: 10.1128/spectrum.00673-22
Tamma, P. D., Doi, Y., Bonomo, R. A., Johnson, J. K., Simner, P. J., Tamma, P. D., et al. (2019). A primer on amp C β-lactamases: necessary knowledge for an increasingly multidrug-resistant world. Clin. Infect. Dis. 69, 1446–1455. doi: 10.1093/cid/ciz173
Tao, Y., Zhou, K., Xie, L., Xu, Y., Han, L., Ni, Y., et al. (2020). Emerging coexistence of three PMQR genes on a multiple resistance plasmid with a new surrounding genetic structure of qnr S2 in E. coli in China. Antimicrob. Resist. Infect. Control 9:52. doi: 10.1186/s13756-020-00711-y
Teichmann, A., Agra, H. N. D. C., Nunes, L. D. S., Da Rocha, M. P., Renner, J. D. P., Possuelo, L. G., et al. (2014). Antibiotic resistance and detection of the sul 2 gene in urinary isolates of Escherichia coli in patients from Brazil. J. Infect. Develop. Countries 8, 039–043. doi: 10.3855/jidc.3380
Tempel, S., Bedo, J., and Talla, E. (2022). From a large-scale genomic analysis of insertion sequences to insights into their regulatory roles in prokaryotes. BMC Genomics 23:451. doi: 10.1186/s12864-022-08678-3
Thorell, K., Meier-Kolthoff, J. P., Sjöling, Å., and Martín-Rodríguez, A. J. (2019). Whole-genome sequencing redefines Shewanella taxonomy. Front. Microbiol. 10:1861. doi: 10.3389/fmicb.2019.01861
Tsai, Y.-K., Liou, C.-H., Lin, J.-C., Fung, C.-P., Chang, F.-Y., and Siu, L. K. (2020). Effects of different resistance mechanisms on antimicrobial resistance in Acinetobacter baumannii: a strategic system for screening and activity testing of new antibiotics. Int. J. Antimicrob. Agents 55:105918. doi: 10.1016/j.ijantimicag.2020.105918
Vrioni, G., Daniil, I., Voulgari, E., Ranellou, K., Koumaki, V., Ghirardi, S., et al. (2012). Comparative evaluation of a prototype chromogenic medium (Chrom ID CARBA) for detecting carbapenemase-producing enterobacteriaceae in surveillance rectal swabs. J. Clin. Microbiol. 50, 1841–1846. doi: 10.1128/JCM.06848-11
Wang, W., Wei, X., Arbab, S., Wu, L., Lu, N., Zhu, Q., et al. (2023). Multidrug-resistant Escherichia coli isolate of Chinese bovine origin carrying the Bla CTX-M-55 gene located in IS26-mediated composite Translocatable units. Microorganisms 11:2795. doi: 10.3390/microorganisms11112795
Wen, Y., Pu, X., Zheng, W., and Hu, G. (2016). High Prevalence of Plasmid-Mediated Quinolone Resistance and IncQ Plasmids Carrying qnrS2 Gene in Bacteria from Rivers near Hospitals and Aquaculture in China. PLoS One 11:e0159418. doi: 10.1371/journal.pone.0159418
Wong, K. K. Y., Brinkman, F. S. L., Benz, R. S., and Hancock, R. E. W. (2001). Evaluation of a structural model of Pseudomonas aeruginosa outer membrane protein Opr M, an efflux component involved in intrinsic antibiotic resistance. J. Bacteriol. 183, 367–374. doi: 10.1128/JB.183.1.367-374.2001
Wong, M. H. Y., Chan, E. W. C., and Chen, S. (2015). Evolution and dissemination of Oqx AB-like efflux pumps, an emerging quinolone resistance determinant among members of Enterobacteriaceae. Antimicrob. Agents Chemother. 59, 3290–3297. doi: 10.1128/AAC.00310-15
Xu, L., Dong, Z., Fang, L., Luo, Y., Wei, Z., Guo, H., et al. (2019). Ortho Venn 2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 47, W52–W58. doi: 10.1093/nar/gkz333
Xu, Q., Fu, Y., Zhao, F., Jiang, Y., and Yu, Y. (2020). Molecular characterization of Carbapenem-resistant Serratia marcescens clinical isolates in a tertiary Hospital in Hangzhou, China. Infect. Drug Resist. 13, 999–1008. doi: 10.2147/IDR.S243197
Xu, Y., Zheng, Z., Ye, L., Chan, E. W.-C., and Chen, S. (2023). Identification and genetic characterization of conjugative plasmids encoding Coresistance to ciprofloxacin and cephalosporin in foodborne Vibrio spp. Microbiol Spectr. 11. doi: 10.1128/spectrum.01032-23
Yoon, S.-H., Ha, S.-M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yu, L., Li, W., Li, Q., Chen, X., Ni, J., Shang, F., et al. (2020). Role of Lsr R in the regulation of antibiotic sensitivity in avian pathogenic Escherichia coli. Poult. Sci. 99, 3675–3687. doi: 10.1016/j.psj.2020.03.064
Yuan, Q., Xia, P., Xiong, L., Xie, L., Lv, S., Sun, F., et al. (2023). First report of coexistence of Bla KPC-2-, Bla NDM-1- and mcr-9-carrying plasmids in a clinical carbapenem-resistant Enterobacter hormaechei isolate. Front. Microbiol. 14:1153366. doi: 10.3389/fmicb.2023.1153366
Zeng, Z., Lei, L., Li, L., Hua, S., Li, W., Zhang, L., et al. (2022). In silico characterization of Bla NDM-harboring plasmids in Klebsiella pneumoniae. Front. Microbiol. 13:1008905. doi: 10.3389/fmicb.2022.1008905
Zhang, R., Liu, L., Zhou, H., Chan, E. W., Li, J., Fang, Y., et al. (2017). Nationwide surveillance of clinical Carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19, 98–106. doi: 10.1016/j.ebiom.2017.04.032
Keywords: Klebsiella aerogenes, whole-genome sequencing, resistance genes, mobile genetic elements, metabolic features, intensive care unit
Citation: Rodrigues SH, Nunes GD, Soares GG, Ferreira RL, Damas MSF, Laprega PM, Shilling RE, Campos LC, Costa AS, Malavazi I, Cunha AF and Pranchevicius M-CS (2024) First report of coexistence of blaKPC-2 and blaNDM-1 in carbapenem-resistant clinical isolates of Klebsiella aerogenes in Brazil. Front. Microbiol. 15:1352851. doi: 10.3389/fmicb.2024.1352851
Received: 09 December 2023; Accepted: 26 January 2024;
Published: 14 February 2024.
Edited by:
Mingxi Wang, Huaqiao University, ChinaReviewed by:
Tahir Hussain, Iowa State University, United StatesCopyright © 2024 Rodrigues, Nunes, Soares, Ferreira, Damas, Laprega, Shilling, Campos, Costa, Malavazi, Cunha and Pranchevicius. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria-Cristina da Silva Pranchevicius, bWNzcHJhbmNAZ21haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.