
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 07 February 2024
Sec. Microbe and Virus Interactions with Plants
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1352354
This article is part of the Research Topic Insights in Microbe and Virus Interactions with Plants: 2023 View all 12 articles
In eukaryotic cells, purine metabolism is the way to the production of deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) and plays key roles in various biological processes. Purine metabolism mainly consists of de novo, salvage, and catabolic pathways, and some components of these pathways have been characterized in some plant pathogenic fungi, such as the rice blast fungus Magnaporthe oryzae and wheat head blight fungus Fusarium graminearum. The enzymatic steps of the de novo pathway are well-conserved in plant pathogenic fungi and play crucial roles in fungal growth and development. Blocking this pathway inhibits the formation of penetration structures and invasive growth, making it essential for plant infection by pathogenic fungi. The salvage pathway is likely indispensable but requires exogenous purines, implying that purine transporters are functional in these fungi. The catabolic pathway balances purine nucleotides and may have a conserved stage-specific role in pathogenic fungi. The significant difference of the catabolic pathway in planta and in vitro lead us to further explore and identify the key genes specifically regulating pathogenicity in purine metabolic pathway. In this review, we summarized recent advances in the studies of purine metabolism, focusing on the regulation of pathogenesis and growth in plant pathogenic fungi.
Purines, one of the most abundant metabolites in eukaryotic cells, participate in a series of basic cellular processes. In addition to serving as building blocks of DNA and RNA, purine nucleotides play essential roles in cellular energy homeostasis and signal transduction. Purines can also be converted into other molecules to serve as cofactors, such as coenzyme A and NAD, to well regulate biochemical reactions.
Purine metabolism is achieved by three pathways: de novo pathway, salvage pathway, and catabolic pathway, all of which involve a series of well-regulated sequential biochemical reactions with metabolite conversion (Zrenner et al., 2006). New purine bases are obtained mainly by de novo and salvage pathways. Purine de novo pathway is well-conserved in the eukaryotic cells and consists of three branches of inosine monophosphate (IMP), adenosine monophosphate (AMP), and guanosine monophosphate (GMP) de novo synthesis. Purine salvage pathway recycles preformed nucleosides and nucleobases. To maintain the metabolic balance, purine metabolites need to be degraded by catabolic pathway, through which the nucleotides are dephosphorylated into nucleosides, and subsequently hydrolyzed into ribose and bases (Tozzi et al., 2006).
In eukaryotic organisms, purine metabolic pathways play crucial roles in promotion of growth, development and pathogenesis. In plant pathogenic fungi, various genes related to the de novo and salvage purine synthesis as well as purine catabolism have been functionally characterized. This review aims to summarize key observations from these studies and discuss the importance of purine metabolism in growth, development, and pathogenesis in necrotrophic, hemibiotrophic, and biotrophic pathogens.
Among eukaryotic organisms, purine metabolism is best characterized in the budding yeast Saccharomyces cerevisiae (Table 1), a model in which metabolic pathways are comprehensive described. In yeast, purine metabolism is orchestrated by purine de novo, salvage, and catabolic pathways, and each of them has different roles in regulating growth and development (Ljungdahl and Daignan-Fornier, 2012).
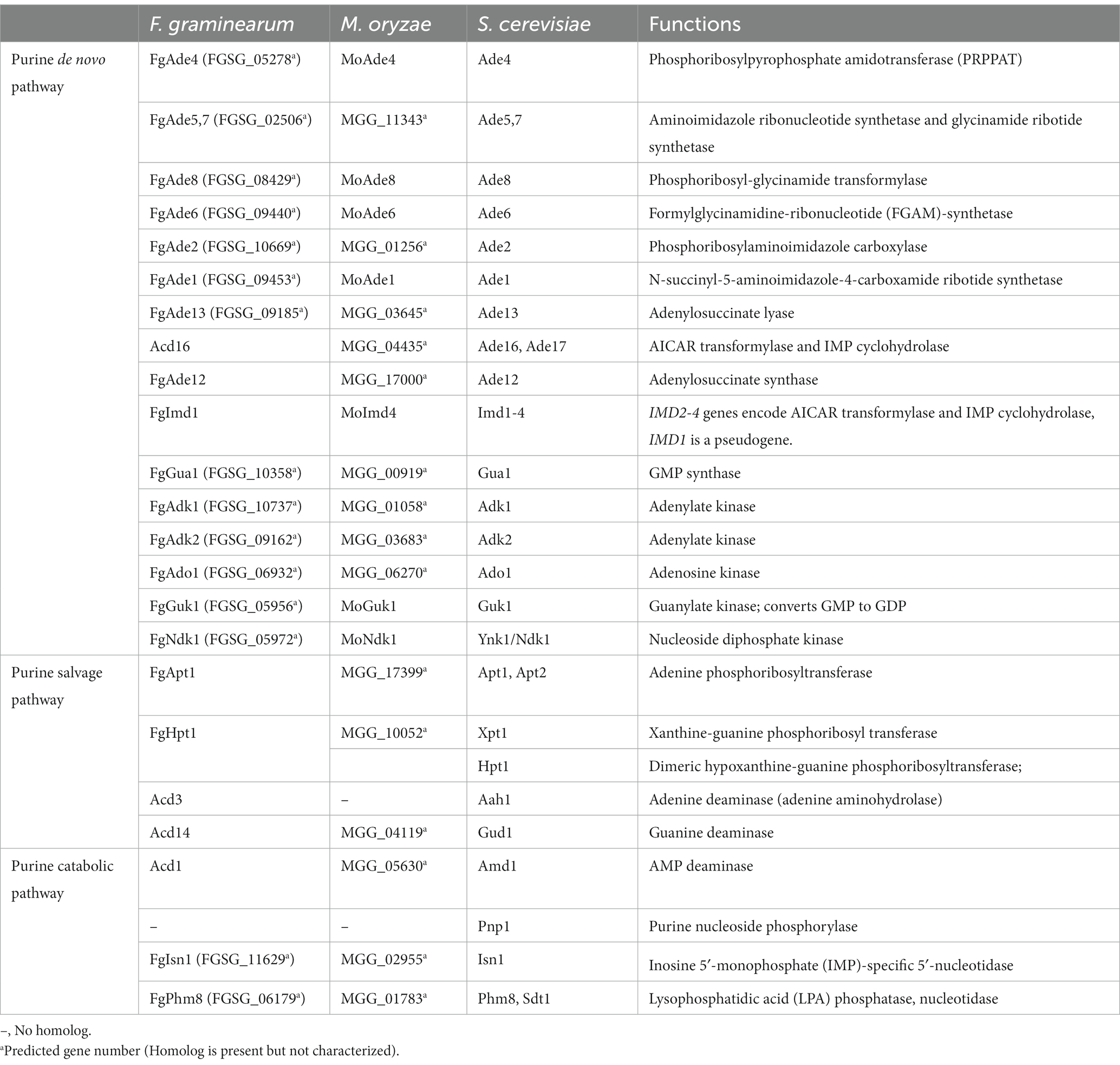
Table 1. Key components of purine metabolic pathways in F. graminearum, and their orthologs in M. oryzae and S. cerevisiae.
Purine de novo pathway consists of three branches including IMP, AMP and GMP de novo synthesis. In yeast, purine de novo pathway originates from 5-phosphoribosyl-1-pyrophosphate (PRPP) to produce the first purine nucleotide IMP by 10 enzymatic steps. IMP has an intermediate role as the precursor used for both AMP and GMP synthesis (Vavassori et al., 2005; Rolfes, 2006). Deletion of ADE16 and ADE17, two paralogous genes encoding the enzymes that catalyze the last two steps of IMP de novo synthesis from PRPP, resulted in adenine and histidine auxotrophy, and ADE16 may plays a role in respiration and sporulation in S. cerevisiae (Tibbetts and Appling, 2000). In addition, accumulation of 5-phosphoribosyl-5-amino-4-imidazole carboxamide ribonucleotide (AICAR) interferes methionine biosynthesis and other cellular processes when both ADE16 and ADE17 are deleted in the budding yeast (Rébora et al., 2005).
IMP formed by purine de novo synthesis is then converted to AMP and GMP, respectively (Rolfes, 2006). In this process, the rate-limiting reaction from IMP to XMP is catalyzed by IMP dehydrogenase (Imd) (Hyle et al., 2003). There are four IMD (IMD1-4) genes in S. cerevisiae, among them, IMD1 is a pseudogene, and Imd2 is intrinsically resistant to drug which could be significantly induced upon mycophenolic acid (MPA) treatment. Both IMD3 and IMD4 could not confer MPA resistance to cells lacking IMD2. Moreover, deletion of either of ScIMD family member confers yeast cells guanine auxotrophy (McPhillips et al., 2004).
S. cerevisiae could take up extracellular purine bases and re-utilize nucleosides and nucleotides through the salvage pathway (Kokina et al., 2019). In this process, xanthine-guanine phosphoribosyl transferase (Xpt1) is required for xanthine utilization and optimally utilization of guanine (Guetsova et al., 1999), hypoxanthine-guanine phosphoribosyltransferase (Hpt1) is used to convert hypoxanthine and guanine to IMP and GMP, respectively (Lecoq et al., 2000), and adenine phosphoribosyltransferase (Apt1) is recruited to convert adenine into AMP (Alfonzo et al., 1995).
In yeast, purine catabolic reactions ensure the balanced homeostasis of preformed nucleosides and nucleotides. High-energy demand leads to a high ATP availability by irreversibly degrading AMP using AMPD (AMP deaminase) (Sims et al., 1999). Conversion of AMP to IMP is the sole pathway of AMP catabolism in eukaryotic cells (Meyer et al., 1989; Saint-Marc et al., 2009). Yeast AMP deaminase Amd1 is responsible for AMP catabolism, and the deletion of Amd1 shows sporulation defect and exhibits hyper-accumulation of ATP and depletion of guanosine nucleotides (Walther et al., 2014).
Dysfunction of de novo pathway could result in the defects of vegetative growth and plant infection (Fernandez et al., 2013; Sun et al., 2021; Aron et al., 2022). Among plant pathogenic fungi, the purine de novo pathways are mostly characterized in two hemibiotrophic pathogens, rice blast fungus Magnaporthe oryzae and wheat head blight fungus Fusarium graminearum (Figure 1), both of which are ranked in the top 10 fungal pathogens in molecular plant pathology (Dean et al., 2012). Genes in the IMP de novo pathway play important roles in various developmental processes (Table 1), and are related to the pathogenicity in many cases.
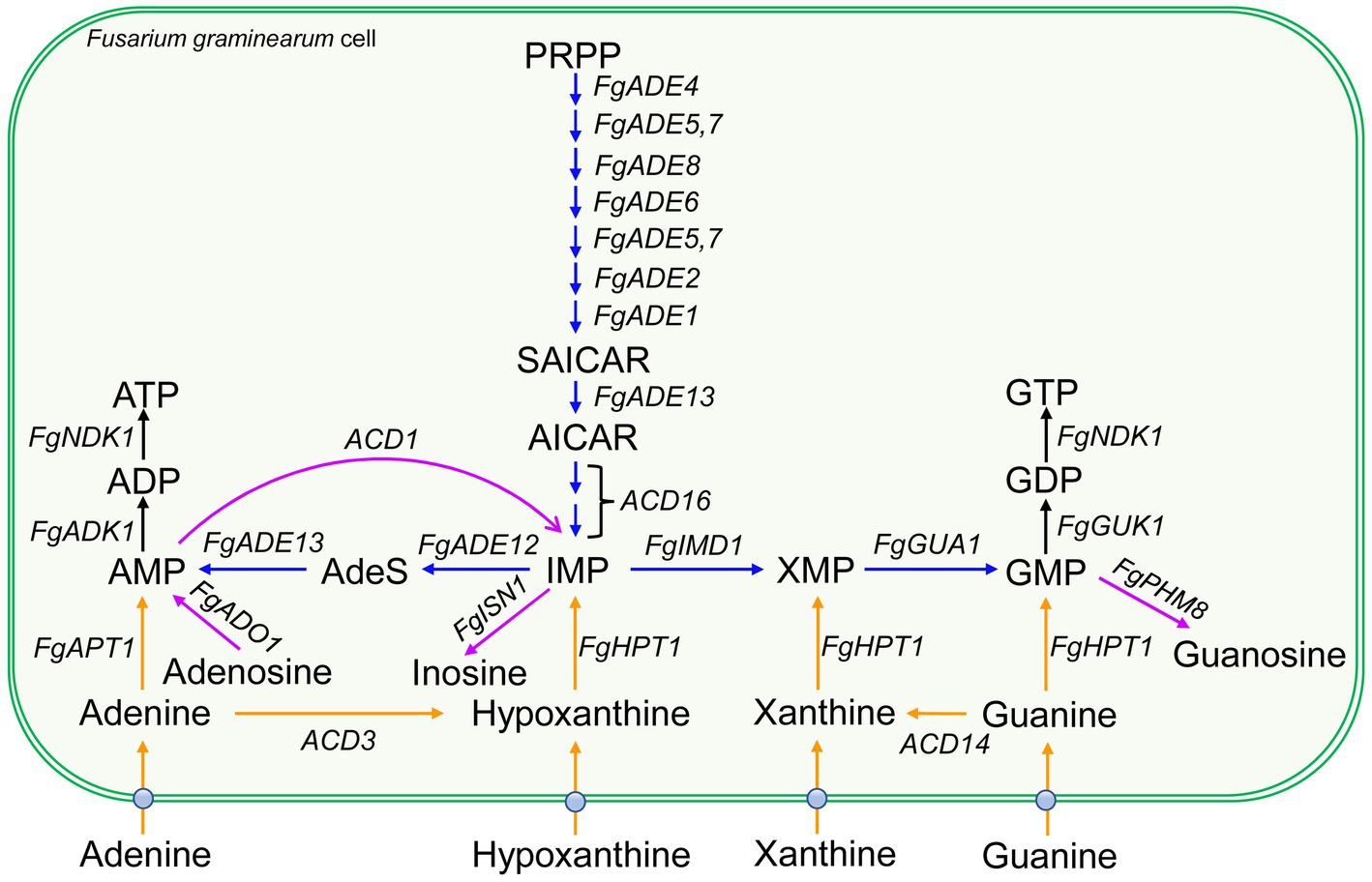
Figure 1. Purine de novo, salvage and catabolic pathways in Fusarium graminearum. Blue arrows represent the steps of purine de novo biosynthesis pathway. Orange arrows represent the steps of purine salvage biosynthesis pathway. The purple arrow indicates the degradation steps for purine catabolism. The ball on fungal plasma membrane indicates purine transporters. Gene names are italicized. AICAR, 5-phosphoribosyl-5-amino-4-imidazole carboxamide ribonucleotide; AdeS, adenylosuccinate; PRPP, 5-phosphoribosyl-1-pyrophosphate; SAICAR, 5-phosphoribosyl-4-(N-succinocarboxamide)-5-amino-imidazole ribonucleotide. The gene ID for each gene is listed in Table 1.
In Coniothyrium minitans, the first enzyme for purine de novo biosynthesis phosphoribosylpyrophosphate amidotransferase (PRPPAT) was firstly proved to be required for conidiation (Qin et al., 2011). In M. oryzae, MoADE4 encodes the corresponding enzyme catalyzing the first step for the conversion of PRPP into 5-phosphoribosyl-1-amine (PRA). The deletion of MoADE4 results in adenine, adenosine, and hypoxanthine auxotrophy on minimal medium. Interestingly, the conidia of Moade4 mutant could form appressoria but fails to penetrate the cuticle of rice and ultimately results in completely nonpathogenic on rice and barley leaves. Moreover, exogenous adenine could partially rescue conidiation, infectious growth, and pathogenicity defects (Aron et al., 2022). The hosphoribosylglycinamide formyltransferase Ade8 is functionally downstream of Ade4, catalyzing the third step of purine de novo pathway, and the deletion of ADE8 could cause defects in vegetative growth, conidiation, and pathogenicity (Liu et al., 2022). Because appressoria fail to penetrate host cells, Moade8 mutant shows obvious defects in virulence on rice and barley, and exogenous adenine could not rescue the appressoria defect (Liu et al., 2022). All these results indicate the key important roles of ADE8 in M. oryzae. However, ADE8 is not always indispensable, for example, in Candida albicans, a principal human fungal pathogen, ADE8 is conditionally essential and can be bypassed when exogenous adenine exists (Jiang et al., 2010). In addition, mutants of the other two genes (MoADE5,7 and MoADE6) involved in IMP de novo pathway show similar defects to Moade8 (Liu et al., 2022). Not only that, Ade6 catalyze the downstream reaction of Ade8 and was proved to be required for exogenous adenine in biotrophic pathogen Ustilago maydis, an ubiquitous smut fungus infecting maize (Heidenreich et al., 2008). As is reported, MoAde1, an ortholog of yeast Ade1 which is responsible for the seventh step for converting 4-carboxy-5-aminoimidazole ribonucleotide (CAIR) to 5-phosphoribosyl-4-(N-succinocarboxamide)-5-amino-imidazole ribonucleotide (SAICAR), is required for vegetative growth and pathogenicity. Different from the Moade4 mutant, the Moade1 mutant could form functional appressoria to penetrate host cells, but notably reduced for hyphae growth in planta (Fernandez et al., 2013). These studies in M. oryzae laid the foundation for research of IMP de novo pathway in plant pathogenic fungi.
The defects of ade16 ade17 double mutant in S. cerevisiae are consistent with disruption of the AICAR transformylase/IMP cyclohydrolase (ATIC) encoded by the gene ADE16 in Cryptococcus neoformans which is a fungal pathogen causing meningoencephalitis in the immunocompromised (Tibbetts and Appling, 2000; Wizrah et al., 2022). In C. neoformans, ADE16 deletion resulted in adenine and histidine auxotrophs and lead to the block of the establishment of infection to a murine model (Wizrah et al., 2022). Similar to C. neoformans, F. graminearum also has only one orthologous gene (ACD16) of yeast ADE16 and ADE17. The deletion mutant of ACD16 has severe defects in vegetative growth, conidiation, sexual reproduction, and plant infection. When supplied with exogenous adenine and histidine, the vegetative growth could be rescued, but the perithecia formation and plant infection could not. Because acd16 mutant fails to form an infection cushion to penetrate the host cell, it is nonpathogenic on wheat spikelets and corn silks (Sun et al., 2021). The two-step reaction from AICAR to IMP catalyzed by Ade16 and Ade17 in the budding yeast corresponds to the sole reaction catalyzed by Acd16 in F. graminearum and the enzyme encoded by MGG_04435 in M. oryzae (Table 1), respectively. Apart from this reaction, the purine de novo pathway is well conserved in these two plant pathogenic fungi. These findings suggest that IMP de novo pathway has a critical role in pathogenicity and growth of plant pathogenic fungi.
Collectively, these discoveries have shown that IMP de novo pathway is functionally well-conserved in plant pathogenic fungi, because all the genes involved in this pathway are crucial for vegetative growth and virulence with the common characteristic that the infection structures are almost defective in penetration and lead to nearly nonpathogenic to the host, even though the infectious processes vary among different fungi. The results in M. oryzae and F. graminearum shed light on that de novo synthesis is functionally conserved in growth, development and pathogenesis in plant pathogenic fungi.
Two key genes functioning in AMP and ADP de novo pathways, ADE12 and ADK1, have been shown to be tightly linked to the pathogenicity in phytopathogens. In M. oryzae, MoADE12 encoding an adenylosuccinate synthase is important for the conidiation, sexual reproduction, and pathogenicity. The Moade12 mutant does not affect appressoria function but has defects in infectious hyphae growth, indicating that MoADE12 is essential for invasive growth in M. oryzae (Zhang et al., 2022). In F. graminearum, the Fgade12 deletion mutant has pleiotropic defects in vegetative growth, conidiation, sexual reproduction, and pathogenicity (Sun et al., 2021). The functional characterization of ADK1 in M. oryzae is still lacking, however, Su et al. (2020) showed that the deletion mutant of ADK1 in the cotton Verticillium wilt fungus Verticillium dahlia, a hemibiotrophic pathogen, has reduced virulence on the host plants. More interestingly, host-induced gene silencing (HIGS) of the gene coding adenylate kinase improved plant resistance to V. dahlia (Su et al., 2020), inspiring us that purine metabolic genes perform great potential as targets for HIGS to enhance the resistance of transgenic plants. Taken together, these results suggest that the AMP de novo pathway is important for growth and essential for pathogenicity.
In C. albicans, GUA1 is conditionally essential and can be bypassed when exogenous adenine is enough and available (Jiang et al., 2010). In C. neoformans, loss of inosine monophosphate dehydrogenase (IMPDH) lead to the slow growth and virulence defects (Morrow et al., 2012). In plant pathogenic fungi, the M. oryzae Imd4 (MoImd4) is proved to partially recover the defect of S. cerevisiae imd4 mutant, and its deletion mutant has a significant growth difference compared to the wild-type strain. Moreover, the invasive hyphae of Moimd4 are defective in attenuated pathogenicity in M. oryzae (Yang et al., 2019). However, there is only one ortholog (FgImd1) of IMP dehydrogenase in F. graminearum, and the Fgimd1 mutant does not affect the formation of infection cushion, but shows a differentiation block of invasive hyphae. Furthermore, exogenous GMP and guanine could fully recover the vegetative growth of Fgimd1 mutant but do not rescue the defects of sexual reproduction, indicating that GMP de novo synthesis is distinct from AMP de novo pathway in the sexual reproduction stage (Sun et al., 2021). Intriguingly, IMPDH is found to be a DNA-binding transcriptional repressor in Drosophila by attenuating the expression of histone genes and E2f, a key driver of cell proliferation (Kozhevnikova et al., 2012). Considering that IMPDH binds a CT-rich ssDNA element in a sequence-specific manner in both Drosophila and Escherichia coli (Kozhevnikova et al., 2012), this ability is probably conserved from prokaryotes to eukaryotes and remains to be further investigated in fungi.
To date, Guk1 responsible for the conversion of GMP into GDP is only characterized in M. oryzae. There are two members of Guk1 in M. oryzae: MoGuk1 and MoGuk2. MoGuk1 is orthologous to the yeast Guk1 and essential for viability (Cai et al., 2017); MoGuk2 exhibited enzymatic activity, and its mutant is weakly reduced in vegetative growth, conidial germination, appressorial formation, but obviously reduced in sporulation, and pathogenicity (Cai et al., 2017). Moreover, Moguk2 mutant could not produce perithecia (Cai et al., 2017). These results indicate that MoGuk2 is functional for GTP de novo synthesis and is important for plant infection in the rice blast fungus.
Finally, Ndk1 is recruited to synthesize end-products ATP and GTP (Amutha and Pain, 2003). Intriguingly, there is evidence showed that Ndk1 is involved in suppressing host oxidative bursts that triggered immunity in rice cells. The Mondk1 mutant could form melanized appressoria and penetrate rice leaf sheath and onion epidermal cell surfaces successfully. However, invasive hyphae of Mondk1 mutant are confined in rice cells of the initial inoculated site and could not extend to neighboring cells (Rocha and Wilson, 2020).
Together, disruption phenotypes of GMP de novo pathway are less severe than that of AMP de novo genes, suggesting that AMP de novo pathway plays more important roles in growth and virulence of plant pathogenic fungi. Moreover, ATP and GTP synthesis processes are rewarding to be lucubrated in virulence and suppression of plant immunity in the host.
In C. neoformans, loss of the hypoxanthine-xanthine-guanine phosphoribosyltransferase Hpt1 in the salvage pathway had no phenotypes (Morrow et al., 2012). To date, functions of components catalyzing purine salvage pathway in plant pathogenic fungi are only characterized in F. graminearum. Unlike S. cerevisiae, F. graminearum has only a single gene (FgHPT1) orthologous to the XPT1. None of the deletion mutants of Fghpt1, Fgacd3 (named aah1 in S. cerevisiae), and Fgacd14 (named gud1 in S. cerevisiae) has detectable phenotypes (Sun et al., 2021), indicating that salvage synthesis of AMP/GMP is not important for growth, development and virulence. However, adding adenine could rescue the growth and sexual defects of the Fgade12 mutant. Also, addition of exogenous guanine rescues the vegetative growth of the Fgimd1 mutant (Sun et al., 2021), suggesting that purine salvage pathways are not defective in phenotypes but are indeed functional in F. graminearum.
Purine salvage pathway has dispensable role in growth, development and pathogenicity when the de novo pathway is functional in plant pathogenic fungi (Sun et al., 2021). Zrenner et al. (2006) suggested that due to its lower energy demand, the salvage pathway is likely used to maintain nucleotide pools in nongrowing cells, while de novo synthesis is predominantly utilized in proliferating cells (Zrenner et al., 2006). This may suggest that purine de novo synthesis is critical during stages of transition in pathogenic fungi, as new cells need to be rapidly proliferated, while the purine salvage pathway may supplement newly synthesized nucleotides to balance degradation processes in older cells.
Exogenous purines can be transported into intracellular space by purine transporters, which is a process required to balance nucleotide pools. Functional characterizations have been reported in several fungal organisms, for example, in Aspergillus nidulans, S. cerevisiae, and necrotrophic fungus Botrytis cinerea (Alguel et al., 2016; Daumann et al., 2016; Boswell-Casteel et al., 2018). Interestingly, the maize purine transporter LPE1 can complement the growth of uapA uapC azgA triple mutants on uric acid in A. nidulans (Argyrou et al., 2001), suggesting that the function of purine transporter is likely conserved among eukaryotes. In spite of these studies, purine transporters in plant pathogenic fungi are largely undefined, and further research is needed to unravel their roles in development and virulence.
Similar to its yeast orthologous gene AMD1, ACD1 is dispensable for vegetative growth but significantly affects conidiation and sexual reproduction in F. graminearum. By heterologous expression, yeast AMD1 partially recovers the defect of perithecia formation of acd1 mutant (Sun et al., 2021). This result indicates that the AMP catabolic pathways may have the conservative functions during sexual reproduction in fungi. Notably, the acd1 mutant is able to form abundant infection cushions similar to the wild-type strain during the stage of plant infection, but its invasive hyphae are blocked after penetration of host cells, suggesting that ACD1 is not essential for the initial penetration and colonization but is important for the differentiation and growth of invasive hyphae in the wheat rachis (Sun et al., 2021). Additionally, exogenous IMP or GMP (but not AMP) can rescue the defects in perithecia formation and development for acd1 mutant, but cannot recover the infectious defects. This significant difference in Acd1 during saprophytic and infectious growth suggests that purine catabolic metabolism maybe specifically regulated in plant infection. This is the first experimental evidence convincingly showing that purine metabolism has different roles during saprophytic and infectious growth in plant fungal pathogens. The stage-specific functions of the AMP deaminase gene maybe conserved in plant pathogenic fungi, and its regulation mechanism during the infectious stage needs to be disclosed and further elucidated.
In the hemibiotroph Colletotrichum graminicola, the maize anthracnose fungus, the purine degradation genes ALA1 and URE1 encoding ureases are required for appressorial penetration and full virulence. The urease inhibitor acetohydroxamic acid (AHA) significantly protects the host, and interferes the incasice process of hemibiotrophs and biotrophs fungi, and oomycetes (Benatto Perino et al., 2020). The inhibition of purine catabolic pathway could be a choice for fungicide exploration and application to control diseases caused by plant fungal pathogens and oomycetes. Other components responsible for enzymatic steps of the catabolic pathway remain to be characterized to elucidate their roles in growth and virulence in plant pathogenic fungi.
Overall, purine metabolic pathways play different roles in growth, conidiation, development, and infection in plant pathogenic fungi. Intriguingly, the genes encoding enzymes catalyzing the catabolic pathways showed stage-specific regulation of sexual and infectious growth in plant fungal pathogens. Notably, a few genes in salvage and catabolic pathways are absent or have a low identity of sequence similarity in M. oryzae in comparing with F. graminearum, indicating that the corresponding reactions are slightly different in these pathways between them, therefore, we recommend that more purine metabolic enzymes should be characterized in plant pathogenic fungi. Further investigation of metabolic enzymes, transporters, transcription factors, and crosstalk among purine metabolic pathways can lead to a better understanding of the purine metabolic network involved in the regulation of different development and infection processes.
MS: Writing – original draft, Writing – review & editing. PD: Writing – review & editing. ZC: Writing – review & editing. JD: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (32202239, 32302619), Natural Science Foundation of Hebei Province (C2023204188, C2022204186), S&T Program of Hebei (23567601H), State Key Laboratory of North China Crop Improvement and Regulation (NCCIR2023ZZ-15 and NCCIR2021RC-11), China Agriculture Research System (CARS-02), and the Basic Scientific Research Expenses of Hebei Provincial Universities (KY2023060).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alfonzo, J. D., Sahota, A., Deeley, M. C., Ranjekar, P., and Taylor, M. W. (1995). Cloning and characterization of the adenine phosphoribosyltransferase-encoding gene (APT1) from Saccharomyces cerevisiae. Gene 161, 81–85. doi: 10.1016/0378-1119(95)00239-3
Alguel, Y., Amillis, S., Leung, J., Lambrinidis, G., Capaldi, S., Scull, N. J., et al. (2016). Structure of eukaryotic purine/H(+) symporter UapA suggests a role for homodimerization in transport activity. Nat. Commun. 7:11336. doi: 10.1038/ncomms11336
Amutha, B., and Pain, D. (2003). Nucleoside diphosphate kinase of Saccharomyces cerevisiae, Ynk1p: localization to the mitochondrial intermembrane space. Biochem. J. 370, 805–815. doi: 10.1042/bj20021415
Argyrou, E., Sophianopoulou, V., Schultes, N., and Diallinas, G. (2001). Functional characterization of a maize purine transporter by expression in Aspergillus nidulans. Plant Cell 13, 953–964. doi: 10.1105/tpc.13.4.953
Aron, O., Otieno, F. J., Tijjani, I., Yang, Z., Xu, H., Weng, S., et al. (2022). De novo purine nucleotide biosynthesis mediated by MoAde4 is required for conidiation, host colonization and pathogenicity in Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 106, 5587–5602. doi: 10.1007/s00253-022-12100-z
Benatto Perino, E. H., Glienke, C., de Oliveira Silva, A., and Deising, H. B. (2020). Molecular characterization of the purine degradation pathway genes ALA1 and URE1 of the maize anthracnose fungus Colletotrichum graminicola identified urease as a novel target for plant disease control. Phytopathology 110, 1530–1540. doi: 10.1094/PHYTO-04-20-0114-R
Boswell-Casteel, R. C., Johnson, J. M., and Hays, F. A. (2018). Functional characterization of the Saccharomyces cerevisiae equilibrative nucleoside transporter 1 (ScENT1). Molecules 23:732. doi: 10.3390/molecules23040732
Cai, X., Zhang, X., Li, X., Liu, M., Liu, X., Wang, X., et al. (2017). The atypical guanylate kinase MoGuk2 plays important roles in asexual/sexual development, conidial septation, and pathogenicity in the rice blast fungus. Front. Microbiol. 8:2467. doi: 10.3389/fmicb.2017.02467
Daumann, M., Golfier, P., Knüppel, N., Hahn, M., and Möhlmann, T. (2016). Botrytis cinerea can import and utilize nucleosides in salvage and catabolism and BcENT functions as high affinity nucleoside transporter. Fungal Biol. 120, 904–916. doi: 10.1016/j.funbio.2016.05.012
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Fernandez, J., Yang, K. T., Cornwell, K. M., Wright, J. D., and Wilson, R. A. (2013). Growth in rice cells requires de novo purine biosynthesis by the blast fungus Magnaporthe oryzae. Sci. Rep. 3:2398. doi: 10.1038/srep02398
Guetsova, M. L., Crother, T. R., Taylor, M. W., and Daignan-Fornier, B. (1999). Isolation and characterization of the Saccharomyces cerevisiae XPT1 gene encoding xanthine phosphoribosyl transferase. J. Bacteriol. 181, 2984–2986. doi: 10.1128/JB.181.9.2984-2986.1999
Heidenreich, M., Budde, A., Zhiqiang, A., and Leong, S. (2008). Disruption of a yeast ADE6 gene homolog in Ustilago maydis. Fungal Genet. Rep. 55, 40–43. doi: 10.4148/1941-4765.1090
Hyle, J. W., Shaw, R. J., and Reines, D. (2003). Functional distinctions between IMP dehydrogenase genes in providing mycophenolate resistance and guanine prototrophy to yeast. J. Biol. Chem. 278, 28470–28478. doi: 10.1074/jbc.M303736200
Jiang, L., Zhao, J., Guo, R., Li, J., Yu, L., and Xu, D. (2010). Functional characterization and virulence study of ADE8 and GUA1 genes involved in the de novo purine biosynthesis in Candida albicans. FEMS Yeast Res. 10, 199–208. doi: 10.1111/j.1567-1364.2009.00600.x
Kokina, A., Ozolina, Z., and Liepins, J. (2019). Purine auxotrophy: possible applications beyond genetic marker. Yeast 36, 649–656. doi: 10.1002/yea.3434
Kozhevnikova, E. N., van der Knaap, J. A., Pindyurin, A. V., Ozgur, Z., van Ijcken, W. F., Moshkin, Y. M., et al. (2012). Metabolic enzyme IMPDH is also a transcription factor regulated by cellular state. Mol. Cell 47, 133–139. doi: 10.1016/j.molcel.2012.04.030
Lecoq, K., Konrad, M., and Daignan-Fornier, B. (2000). Yeast GMP kinase mutants constitutively express AMP biosynthesis genes by phenocopying a hypoxanthine-guanine phosphoribosyltransferase defect. Genetics 156, 953–961. doi: 10.1093/genetics/156.3.953
Liu, M. Y., Sun, L. X., Qian, H., Zhang, Y. R., Zhu, X. M., Li, L., et al. (2022). De novo purine nucleotide biosynthesis pathway is required for development and pathogenicity in Magnaporthe oryzae. J. Fungi 8:915. doi: 10.3390/jof8090915
Ljungdahl, P. O., and Daignan-Fornier, B. (2012). Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190, 885–929. doi: 10.1534/genetics.111.133306
McPhillips, C. C., Hyle, J. W., and Reines, D. (2004). Detection of the mycophenolate-inhibited form of IMP dehydrogenase in vivo. Proc. Natl. Acad. Sci. U. S. A. 101, 12171–12176. doi: 10.1073/pnas.0403341101
Meyer, S. L., Kvalnes-Krick, K. L., and Schramm, V. L. (1989). Characterization of AMD, the AMP deaminase gene in yeast. Production of AMD strain, cloning, nucleotide sequence, and properties of the protein. Biochemistry 28, 8734–8743. doi: 10.1021/bi00448a009
Morrow, C. A., Valkov, E., Stamp, A., Chow, E. W., Lee, I. R., Wronski, A., et al. (2012). De novo GTP biosynthesis is critical for virulence of the fungal pathogen Cryptococcus neoformans. PLoS Pathog. 8:e1002957. doi: 10.1371/journal.ppat.1002957
Qin, L., Gong, X., Xie, J., Jiang, D., Cheng, J., Li, G., et al. (2011). Phosphoribosylamidotransferase, the first enzyme for purine de novo synthesis, is required for conidiation in the sclerotial mycoparasite Coniothyrium minitans. Fungal Genet. Biol. 48, 956–965. doi: 10.1016/j.fgb.2011.06.007
Rébora, K., Laloo, B., and Daignan-Fornier, B. (2005). Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: a central role for a small molecule. Genetics 170, 61–70. doi: 10.1534/genetics.104.039396
Rocha, R. O., and Wilson, R. A. (2020). Magnaporthe oryzae nucleoside diphosphate kinase is required for metabolic homeostasis and redox-mediated host innate immunity suppression. Mol. Microbiol. 114, 789–807. doi: 10.1111/mmi.14580
Rolfes, R. (2006). Regulation of purine nucleotide biosynthesis: in yeast and beyond. Biochem. Soc. Trans. 34, 786–790. doi: 10.1042/BST0340786
Saint-Marc, C., Pinson, B., Coulpier, F., Jourdren, L., Lisova, O., and Daignan-Fornier, B. (2009). Phenotypic consequences of purine nucleotide imbalance in Saccharomyces cerevisiae. Genetics 183, 529–538. doi: 10.1534/genetics.109.105858
Sims, B., Mahnke-Zizelman, D. K., Profit, A. A., Prestwich, G. D., Sabina, R. L., and Theibert, A. B. (1999). Regulation of AMP deaminase by phosphoinositides. J. Biol. Chem. 274, 25701–25707. doi: 10.1074/jbc.274.36.25701
Su, X., Lu, G., Li, X., Rehman, L., Liu, W., Sun, G., et al. (2020). Host-induced gene silencing of an adenylate kinase gene involved in fungal energy metabolism improves plant resistance to Verticillium dahliae. Biomol. Ther. 10:127. doi: 10.3390/biom10010127
Sun, M., Bian, Z., Luan, Q., Chen, Y., Wang, W., Dong, Y., et al. (2021). Stage-specific regulation of purine metabolism during infectious growth and sexual reproduction in Fusarium graminearum. New Phytol. 230, 757–773. doi: 10.1111/nph.17170
Tibbetts, A. S., and Appling, D. R. (2000). Characterization of two 5-aminoimidazole-4-carboxamide ribonucleotide transformylase/inosine monophosphate cyclohydrolase isozymes from Saccharomyces cerevisiae. J. Biol. Chem. 275, 20920–20927. doi: 10.1074/jbc.M909851199
Tozzi, M. G., Camici, M., Mascia, L., Sgarrella, F., and Ipata, P. L. (2006). Pentose phosphates in nucleoside interconversion and catabolism. FEBS J. 273, 1089–1101. doi: 10.1111/j.1742-4658.2006.05155.x
Vavassori, S., Wang, K., Schweizer, L., and Schweizer, M. (2005). Ramifications of impaired PRPP synthesis in Saccharomyces cerevisiae. Biochem. Soc. Trans. 33, 1418–1420. doi: 10.1042/BST0331418
Walther, T., Létisse, F., Peyriga, L., Alkim, C., Liu, Y., Lardenois, A., et al. (2014). Developmental stage dependent metabolic regulation during meiotic differentiation in budding yeast. BMC Biol. 12:60. doi: 10.1186/s12915-014-0060-x
Wizrah, M. S. I., Chua, S. M. H., Luo, Z., Manik, M. K., Pan, M., Whyte, J. M. L., et al. (2022). AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans. J. Biol. Chem. 298:102453. doi: 10.1016/j.jbc.2022.102453
Yang, L., Ru, Y., Cai, X., Yin, Z., Liu, X., Xiao, Y., et al. (2019). MoImd4 mediates crosstalk between MoPdeH-cAMP signalling and purine metabolism to govern growth and pathogenicity in Magnaporthe oryzae. Mol. Plant Pathol. 20, 500–518. doi: 10.1111/mpp.12770
Zhang, Z., Hao, Z., Chai, R., Qiu, H., Wang, Y., Wang, J., et al. (2022). Adenylsuccinate synthetase MoADE12 plays important roles in the development and pathogenicity of the rice blast fungus. J. Fungi 8:780. doi: 10.3390/jof8080780
Keywords: purine de novo pathway, purine salvage pathway, purine catabolic pathway, plant pathogenic fungi, pathogenesis, purine transporters
Citation: Sun M, Dai P, Cao Z and Dong J (2024) Purine metabolism in plant pathogenic fungi. Front. Microbiol. 15:1352354. doi: 10.3389/fmicb.2024.1352354
Received: 08 December 2023; Accepted: 29 January 2024;
Published: 07 February 2024.
Edited by:
Trevor Carlos Charles, University of Waterloo, CanadaReviewed by:
Jaunius Urbonavičius, Vilnius Gediminas Technical University, LithuaniaCopyright © 2024 Sun, Dai, Cao and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manli Sun, c21sNjY2QHllYWgubmV0; Jingao Dong, ZG9uZ2ppbmdhb0AxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.