- 1Department of Pathogen Biology, School of Medicine, Qinghai University, Qinghai, China
- 2Department of Orthopaedic Trauma, The Affiliated Hospital of Qinghai University, Qinghai, China
- 3Department of Immunology, School of Medicine, Qinghai, China
- 4Department of Traditional Chinese Medicine, School of Medicine, Qinghai University, Qinghai, China
- 5Department of Public Health, School of Medicine, Qinghai University, Qinghai, China
- 6Department of Genetics, School of Medicine, Qinghai University, Qinghai, China
Enteroviruses (EVs) are the main cause of a number of neurological diseases. Growing evidence has revealed that successful infection with enteroviruses is highly dependent on the host machinery, therefore, host proteins play a pivotal role in viral infections. Both host and viral proteins can undergo post-translational modification (PTM) which can regulate protein activity, stability, solubility and interactions with other proteins; thereby influencing various biological processes, including cell metabolism, metabolic, signaling pathways, cell death, and cancer development. During viral infection, both host and viral proteins regulate the viral life cycle through various PTMs and different mechanisms, including the regulation of host cell entry, viral protein synthesis, genome replication, and the antiviral immune response. Therefore, protein PTMs play important roles in EV infections. Here, we review the role of various host- and virus-associated PTMs during enterovirus infection.
1 Introduction
Enterovirus infections caused by various pathogens, such as coxsackievirus B (CVB), poliovirus, and enterovirus 71 (EV71), can cause serious diseases (Bauer et al., 2019; Saguil et al., 2019). To achieve efficient infection, these viruses must co-opt the host cell machinery which is involved in the progression of every stage in the enterovirus life cycle (Figure 1) (Tabor-Godwin et al., 2012; Lyoo et al., 2017; Wu and Chu, 2017). Post-translational modification (PTM) of proteins diversify proteome function by covalently adding small proteins or other groups, potentially altering their cellular localization, interaction with partners, and activation state (Schweppe et al., 2003; Zolg et al., 2018). The disruption of PTMs can upset normal physiological processes (Gupta et al., 2021; He et al., 2021; Smith and Carregari, 2022). The rapid development of proteomics and mass spectrometry has identified an increasing number of PTMs, including glycosylation, ubiquitination, methylation, phosphorylation, acetylation, and lipidation (Aslebagh et al., 2019; Andres et al., 2020; Hermann et al., 2022). The progression of PTMs is dynamic, reversible, and catalyzed by specific enzymes; therefore, many compounds targeting these enzymes can effectively inhibit or promote PTM (Lopez et al., 2016; Sun et al., 2020; Zhu et al., 2022c). Different PTMs on different host and viral proteins can exert various effects on viral growth by targeting viral entry, replication, assembly, egress, and antiviral immune response (Kumar et al., 2020; Song et al., 2021; Cheng et al., 2022; Xue et al., 2022). Here, we review some essential PTMs, including ubiquitination, phosphorylation, acetylation, SUMOylation, tyrosine sulfation, neddylation, and ISGylation, associated with host-pathogen interactions to better understand the role and mechanism of PTMs during enterovirus infection.
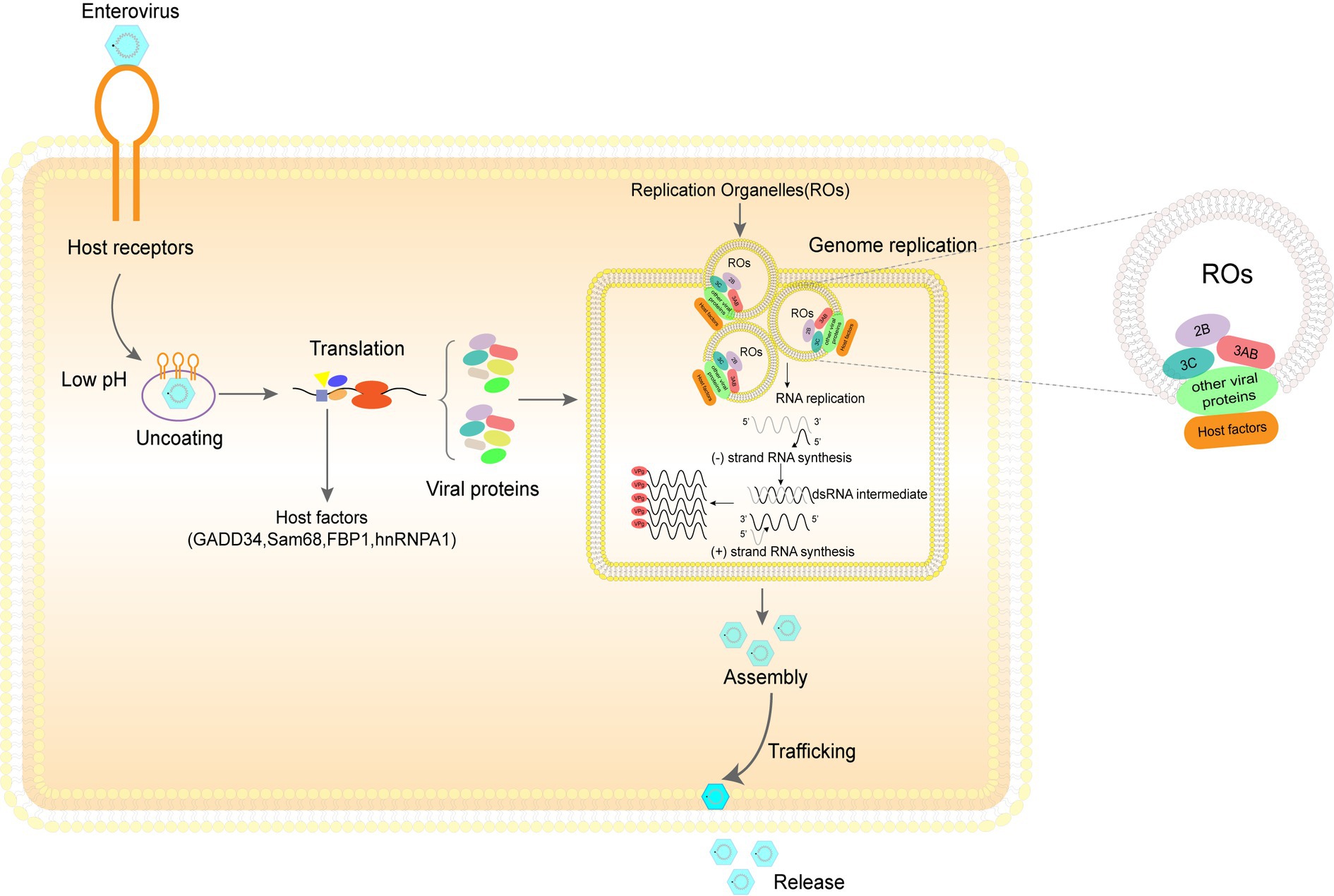
Figure 1. The enterovirus life cycle depends heavily on diverse host machinery. Step 1. Entry: The three main steps by which EV71 virus particles enter the host cell are adhesion onto the host cell surface, binding of viral particles to cell receptors, and entry of viral particles into cells through endocytic pathways. Step 2. Uncoating and genome release: under the influence of the pH environment of the host cell, the virus sheds its shell and releases its genome. Step 3. Translation and cleavage of viral polyprotein: under the influence of various host proteins, translation of viral RNA is initiated and the resulting polyprotein is cleaved to obtain the structural and non-structural proteins. Step 4. Replication of viral genomes: viral genome replication requires the participation of a large number of host factors and viral proteins to complete the formation of replication organelles (ROs) and genome replication. Step 5. Assembly and release of viral particles: the assembly of viral particles also requires coordination between various viral and host proteins. After the assembly, the virus particles are released into the extracellular space in various ways to initiate the next round of infection.
2 Ubiquitination
As a pivotal PTM, ubiquitination plays a role in various cellular processes in eukaryotes according to egiht differently-linked polyUb chains which established via conjugation to intrinsic ubiquitin residues, including seven Lys residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 or Lys63) or the N-terminal Met1 residue (Chau et al., 1989; Hoege et al., 2002; Lauwers et al., 2009). The differently-linked polyUb chains code for distinct signaling outcomes including cancer development, protein trafficking, cell death, immune response, signaling pathways, and viral infection, through its proteolytic and non-proteolytic functions (Komander, 2009; Dores and Trejo, 2014; van Wijk et al., 2019; Iwai, 2021; Pellegrino et al., 2022; Park et al., 2022a). This process is catalyzed by E1, E2, and E3 which enable ubiquitin (Desai et al.) and Ub-like modifiers, SUMO, NEDD8, and ISG15, to bind to substrate proteins, and is described as ubiquitination, SUMOylation, neddylation, and ISGylation, respectively (Kang and Yi, 2011; Zhang et al., 2017). These modifications play different roles in enterovirus infections.
2.1 The antiviral role of ubiquitination in enterovirus infection
Ubiquitination is widely involved in various physiological processes, and its dysregulation plays an important role in various diseases, including enterovirus infections (Swatek and Komander, 2016; Rape, 2018). Numerous studies have shown that protein ubiquitination can inhibit enterovirus infection through different mechanisms. An increasing number of studies have indicated that tripartite motif (TRIM) proteins, which have E3 Ub ligase activity, play pivotal roles in inhibiting viral infections including those caused by enteroviruses. A previous study revealed that TRIM7 inhibits enteroviral replication by ubiquitination and degradation of viral 2 BC (Change “2C” into “2 BC”) protein. Mechanistic studies have shown that TRIM7 recognizes the C-terminal region of 2C via the PRY-SPRY domain, thus mediating the degradation of the 2 BC (Change “2C” into “2 BC”) protein (Figure 2A) (Fan et al., 2021). Another study showed that the E3 Ub ligase TRIM38 disrupts EV71 infection by ubiquitinating cellular proteins that regulate immune signaling pathways or interact with viral proteins (Figure 2B) (Liu et al., 2011). In addition, TRIM proteins can suppress enterovirus infection by regulating the immune response through different mechanisms. TRIM21 can mediate the upregulation of type I interferon signaling by interacting with mitochondrial anti-viral signaling protein (MAVS), eventually catalyzing the K27-linked polyubiquitination of MAVS and inhibiting CVB3 infection (Figure 2C) (Liu et al., 2018). Enterovirus 3C proteins suppress retinoic acid-inducible gene I (RIG-I)-mediated type I interferon (IFN) responses via their cleavage activity, however, TRIM25 overexpression rescues this suppression. TRIM25 mediates the RIG-I ubiquitination and restores its expression and IFN-β production, indicating that TRIM25 can abolish enterovirus infection (Figure 2D) (Xiao et al., 2021). Arrestin domain containing 4 (ARRDC4) plays pivotal roles in G-protein-coupled receptor-associated physiological and pathological processes and glucose metabolism. A previous study reported that the expression of ARRDC4 was increased after EV71 infection, both in vitro and in vivo. Subsequently, ARRDC4 interacts with melanoma differentiation-associated protein 5 (MDA5) and recruits TRIM65 to increase K63-linked ubiquitination of MDA5 (Change “TRIM65 to increase MDA5 K63 ubiquitination” into “TRIM65 to increase K63-linked ubiquitination of MDA5”), eventually leading to the activation of the innate signaling pathway and inhibition of EV71 infection (Figure 2E) (Meng et al., 2017).
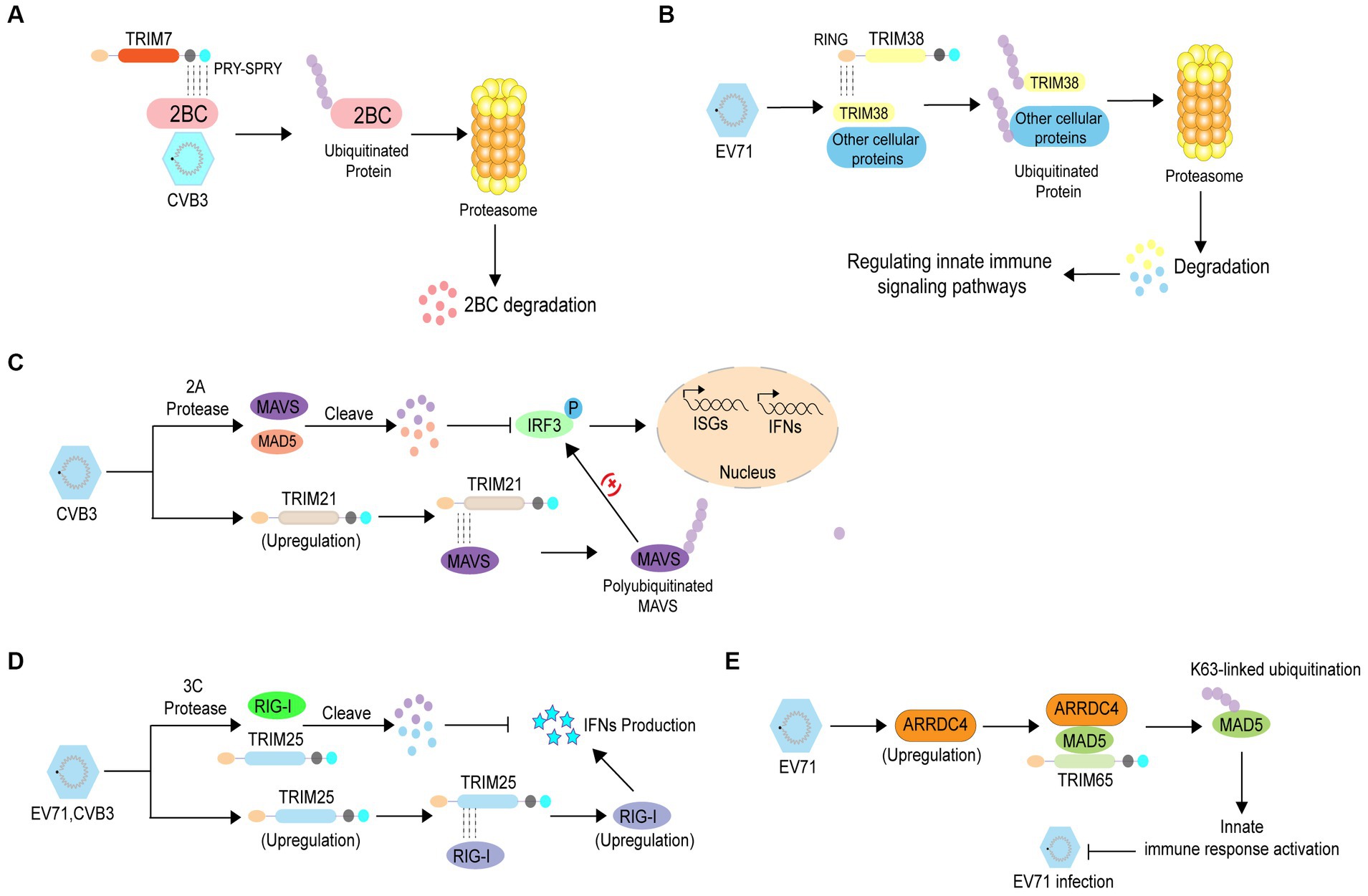
Figure 2. The antiviral role of ubiquitination in enterovirus infection. (A) TRIM7 recognizes the C-terminal region of 2C via the PRY-SPRY domain, thereby mediating degradation of the 2 BC (Change 2C into 2 BC) protein to inhibit enterovirus replication. (B) TRIM38 disrupts EV71 infection by ubiquitinating cellular proteins that regulate immune signaling pathways or their interactions with viral proteins. (C) TRIM21 mediates the enhancement of type I interferon signaling by interacting with mitochondrial anti-viral signaling protein to catalyze the K27-linked polyubiquitination of mitochondrial anti-viral signaling protein, which inhibits CVB3 infection. (D) TRIM25 mediates RIG-I ubiquitination and restores RIG-I expression and IFN-β production to prevent enterovirus infection. (E) EV71 infection induces ARRDC4 expression Subsequently, ARRDC4 interacts with MDA5 and recruits TRIM65 to increase MDA5 K63 ubiquitination, leading to the activation of the innate signaling pathway; thus, inhibiting EV71 infection.
The Ub-proteasome system (UPS) can also inhibit enterovirus infection through other mechanisms. The internal ribosome entry site (IRES) is an EV71 genomic functional element that is essential for the translation of viral proteins. Kung YA et al. reported that the negative IRES trans-acting factor KH-type splicing regulatory protein (KHSRP) downregulates IRES-mediated translation in enterovirus-infected cells by interacting with KLHL12, a substrate adaptor for the E3 ligase complex; leading to KHSRP ubiquitination (Table 1) (Kung et al., 2017). Many chemical compounds can also participate in the UPS to regulate enterovirus infections. Curcumin (diferuloylmethane) is a natural polyphenolic compound that dysregulates the UPS. Si X et al. found that curcumin treatment markedly decreases the replication of coxsackieviruses. Further investigation revealed that not only 20S proteasome proteolytic activities but also cellular deubiquitination activities were decreased after curcumin treatment, which led to the accumulation of ubiquitinated proteins and a reduction of free ubiquitin (Change “ubiquitinated protein” into “ubiquitin”) (Table 1) (Si et al., 2007). Pyrrolidine dithiocarbamate (PDTC) is an antioxidant that suppresses ubiquitin proteasome-mediated proteolysis. A previous study reported that the synthesis of CVB3 viral RNA, expression of viral protein VP1, and the release of viral progeny were significantly reduced after PDTC treatment. Further evidence indicated that the negative effect of PDTC on viral infection is probably related to the regulation of ubiquitination (Table 1) (Si et al., 2005b).
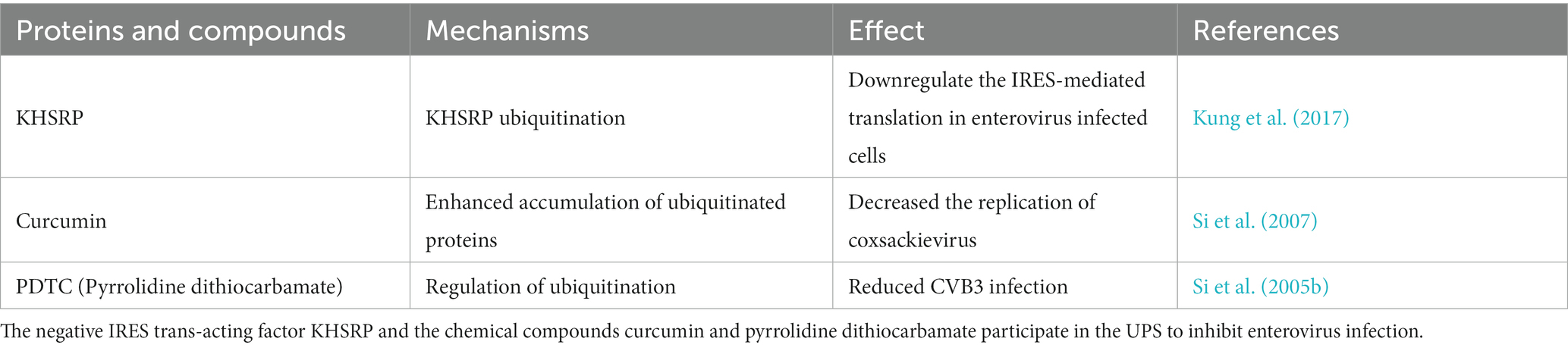
Table 1. The anti-enterovirus role of other proteins and chemical compounds involved in ubiquitin-proteasome system (UPS).
2.2 The proviral role of ubiquitination in enterovirus infection
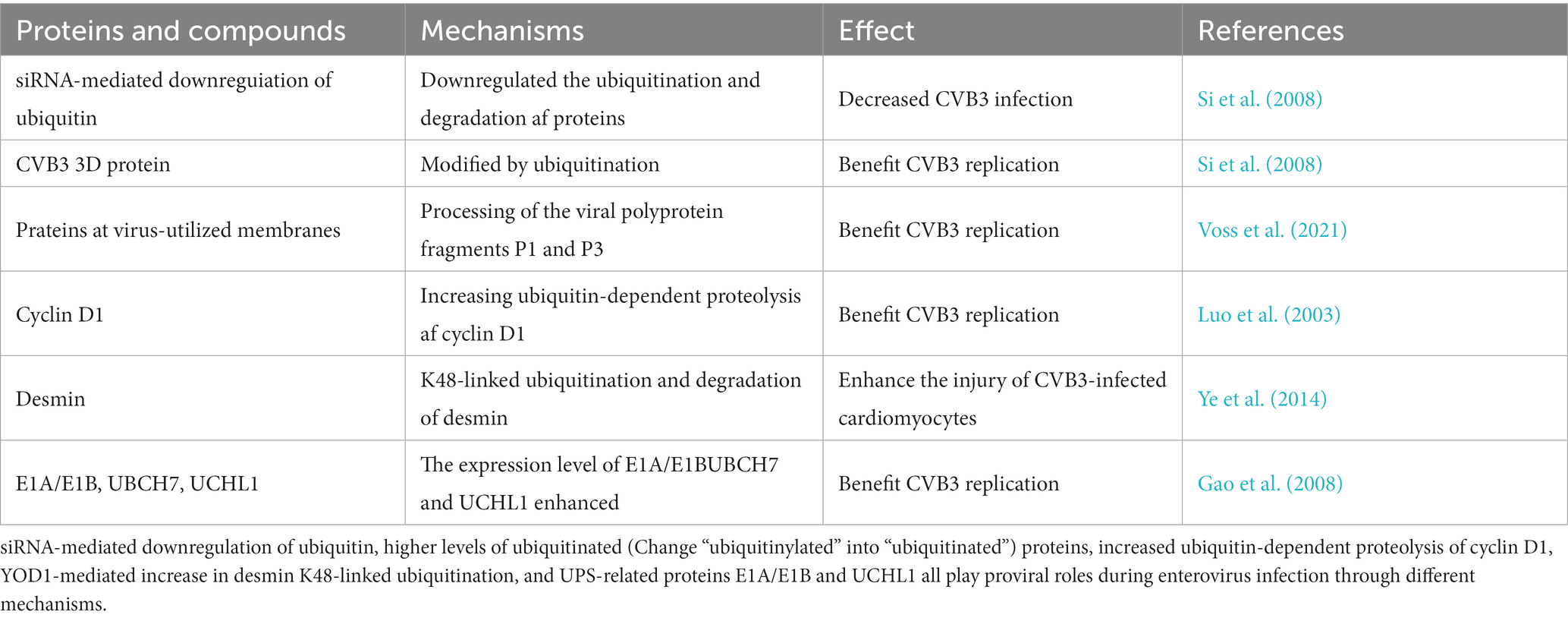
The ubiquitination of proteins can also play a positive role in enterovirus infection. siRNA-mediated downregulation of Ub can decrease CVB3 infection by downregulating the ubiquitination and degradation of proteins. Furthermore, the 3D protein of CVB3 is modified by ubiquitination, and this ubiquitination is required for efficient viral replication (Table 2) (Si et al., 2008). Additionally, Voss M et al. showed that during CVB3 infection, there is an increase of ubiquitinated (Change “ubiquitinylated” into “ubiquitinated”) proteins in virus-utilized membranes that direct proteins to proteasomal degradation. This process is exploited by CVB3 for the cleavage of viral polyprotein fragments P1 and P3 to ensure correct viral replication (Table 2) (Voss et al., 2021). The host cell cycle regulatory machinery can also be modified by CVB3 to promote viral replication. To create a favorable environment for CVB3 replication, CVB3 infection can disrupt the homeostasis of host cells by increasing the ubiquitin-dependent proteolysis of cyclin D1, which results in cell cycle arrest (Table 2) (Luo et al., 2003). Cardiac failure is a serious complication of CVB3 infections. Intercalated disks (ICDs) are connections that sustain cardiac structures and mediate signal communication among cardiomyocytes, and ICD deficiency can lead to heart dysfunction. Ye X et al. revealed that CVB3-induced miR-21 expression could target the deubiquitinating enzyme YOD1 to increase K48-linked ubiquitination and degradation of desmin, a component of ICDs; resulting in the destruction of desmosomes and enhanced cardiomyocyte injury (Table 2) (Ye et al., 2014). During CVB3 infection, the expression levels of the UPS-related proteins E1A/E1B、UBCH7 (Add“UBCH7”)and UCHL1 are significantly upregulated, indicating that the UPS plays a significant role in CVB3 infection (Table 2) (Gao et al., 2008).
Similarly, ubiquitination promotes enterovirus infection by regulating immune responses. SAM and HD Domain containing Deoxynucleoside Triphosphate Triphosphohydrolase 1 (SAMHD1) is an effector of innate immunity and has been confirmed as a restrictive factor for EV71. However, further studies have revealed that during EV71 infection, this inhibition can be overcome via ubiquitination of SAMHD1 by TRIM21 and its subsequently degradation through the proteasomal pathway (Figure 3A) (Li et al., 2020). Tumor necrosis factor receptor-associated factor 6 (TRAF6) is an important protein in the RIG-I-like receptor (RLR)-mediated antiviral signaling pathway that also regulates EV71 infection via a ubiquitination-associated pathway. RLR-induced NF-κB signaling is upregulated by ubiquitin-specific protease 4 (USP4)-mediated TRAF6 K48-linked deubiquitination, which ultimately suppresses EV71 replication (Figure 3B) (Xu et al., 2018). RNA interference (RNAi) is a conserved antiviral immune mechanism in various eukaryotes, and STIP1 homology and U-box containing protein 1 (STUB1) regulates the RNAi machinery in mammals. A previous study revealed that STUB1 interacts with Argonaute RISC Catalytic Component 2 (AGO2) and accelerates its K48-linked ubiquitination; thus facilitating its degradation, decreasing the RNAi response, and promoting EV71 replication in mammalian cells (Figure 3C) (Zhang et al., 2022).
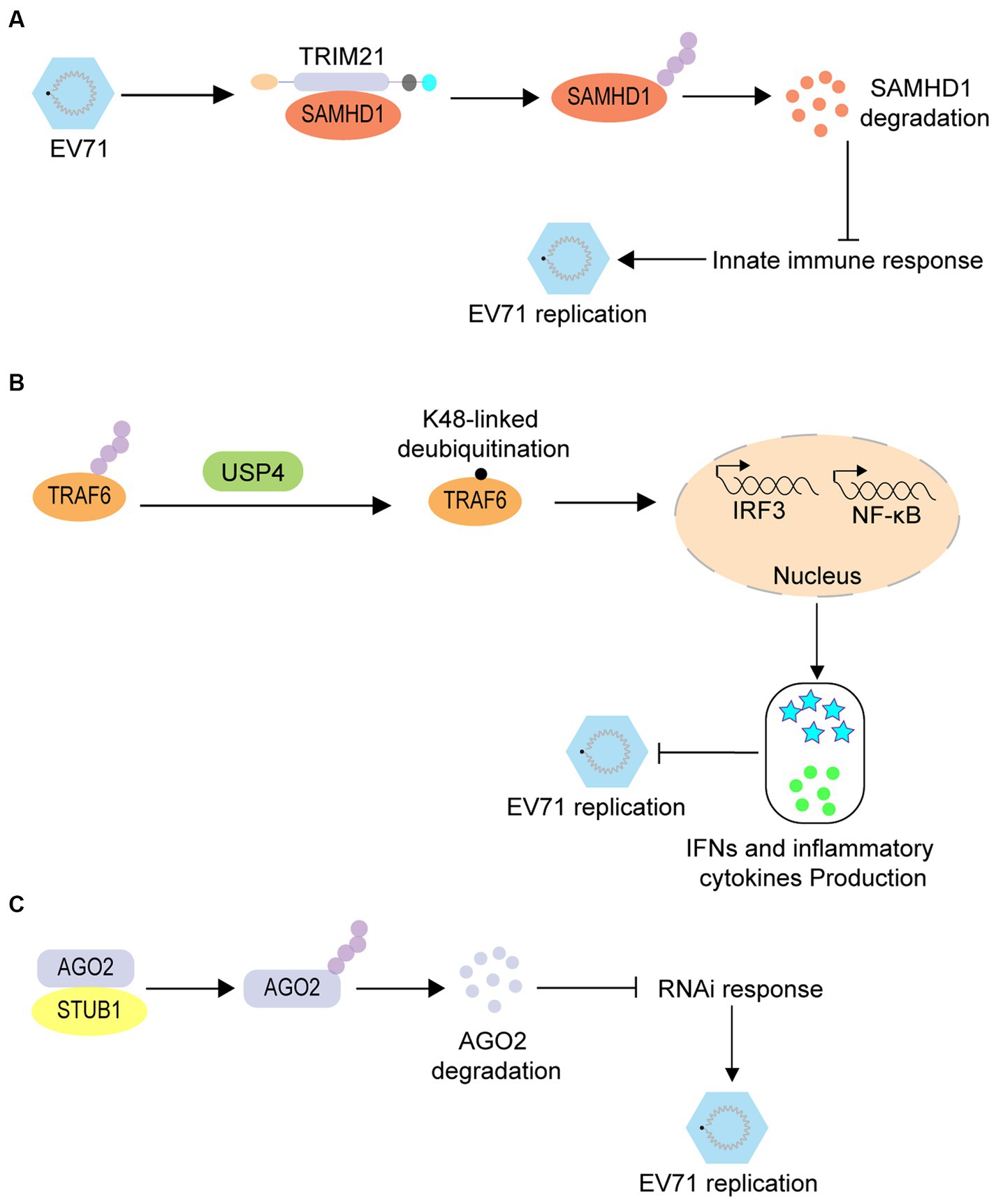
Figure 3. The proviral role of ubiquitination in enterovirus infection. (A) TRIM21-mediated ubiquitination of SAMHD1 leads to the degradation of SAMHD1, thus promoting EV71 infection. (B) USP4-mediated TRAF6 K48-linked deubiquitination induces the upregulation of RLR-induced NF-κB signaling to suppress the EV71 replication. (C) STUB1 interacts with AGO2 and accelerates the K48-linked ubiquitination of AGO2, thus promoting its degradation and decreasing the RNAi response; thereby promoting EV-71 replication.
2.3 The roles of Ub-like modification in enterovirus infection
Both of SUMO, NEDD8 and ISG15 are Ubiquitin-Like Proteins (Herrmann et al., 2007). The SUMO proteins share structural similarities with ubiquitin, and the conjugation of SUMO proteins to substrates happens via a serious of enzymatic cascade involving the E1,E2 and E3 protein ligase. When a SUMO peptide tagged on the lysine residue of the protein substrate it could regulated various cellular processes, including transcription, replication, chromosome segregation and DNA repair (Geiss-Friedlander and Melchior, 2007; Gareau and Lima, 2010). NEDD8 is 60% identical and 80% homologous to ubiquitin. When NEDD8 tagged to the substrate protein, it regulated protein metabolism and activity (Cappadocia and Lima, 2018). Interferon-Stimulated Gene 15 protein (ISG15) is expressed at low levels under physiological conditions (Desai et al., 2006). Compared with ubiquitin, ISG15 shows substantial sequence variation from species to species. Once ISG15 is tagged to targets within the cell upon interferon stimulation, these cellular targets are involved in every aspect of cellular function, including DNA replication/repair, cell metabolism, signal transduction, and cytoskeletal organization and others (Mirzalieva et al., 2022).
Viral proteins can be SUMOylated during the enterovirus life cycle. EV71 3C protein can be SUMO-modified by Ubc9, which decreases its protease activity and protein stability; thus decreasing EV71 replication (Figure 4A) (Chen et al., 2011). In addition, the 3D protein is modified by SUMO-1 during infection and combined with 3D ubiquitination, this can increase the stability of the 3D protein which promotes EV71 replication (Figure 4B) (Liu et al., 2016).

Figure 4. The roles of Ub-like modification in enterovirus infection. (A) EV71 3C protein can be SUMO-modified by Ubc9 which decreases the protease activity and protein stability of 3C, thereby decreasing EV71 replication. (B) SUMOylation of the EV71 3D protein cooperates with 3D ubiquitination to increase its stability, eventually promoting the replication of EV71. (C) SUMOylation of REGγ causes it to translocate from the nucleus to the cytoplasm, allowing the possible interaction of REG with viral or host proteins to play a proviral role during CVB3 infection. (D) The VP2 protein of EV71 is modified by NEDD8 at lysine 69, reducing its stability and decreasing viral replication. (E) Protein ISGylation blocks coxsackievirus pathology by increasing antiviral effectors, IFIT1/3 proteins, and metabolic reprogramming.
Host proteins can also be SUMOylated during viral infection. The SUMOylation of REGγ, a component of the 11S proteasome activator, causes it to translocate from the nucleus to the cytoplasm, offering possibilities for the interaction of REG with viral or host proteins to play a proviral role during CVB3 infection (Figure 4C) (Gao et al., 2010). Neddylation also affects enterovirus infection. The VP2 protein of EV71 can be modified by NEDD8 at lysine 69, which reduces VP2 stability and decreases viral replication (Figure 4D) (Wang et al., 2022). Several studies have shown that protein modification with ISG15, referred to as ISGylation, plays a pivotal role in type I IFN-induced antimicrobial systems. One study revealed that ISGylation blocks coxsackievirus pathology by increasing the levels of antiviral effector IFIT1/3 proteins and metabolic reprogramming (Figure 4E) (Kespohl et al., 2020).
3 Phosphorylation
Phosphorylation is a reversible process involving protein kinases and phosphatases which catalyze the phosphorylation of serine, threonine, and tyrosine in proteins. Protein phosphorylation plays a broad role in various biological events, including protein stability, protein interactions, transcription regulation, signal transduction, intracellular localization, and cell cycle progression (Zhao et al., 2010; Zhang and Pelech, 2012; Deutscher et al., 2014; Puertollano et al., 2018; Yamasaki et al., 2020; Bilbrough et al., 2022; Park et al., 2022b). In addition, phosphorylation of both viral and cellular proteins can significantly affect enterovirus pathogenesis.
3.1 The antiviral role of phosphorylation in the infection of enterovirus
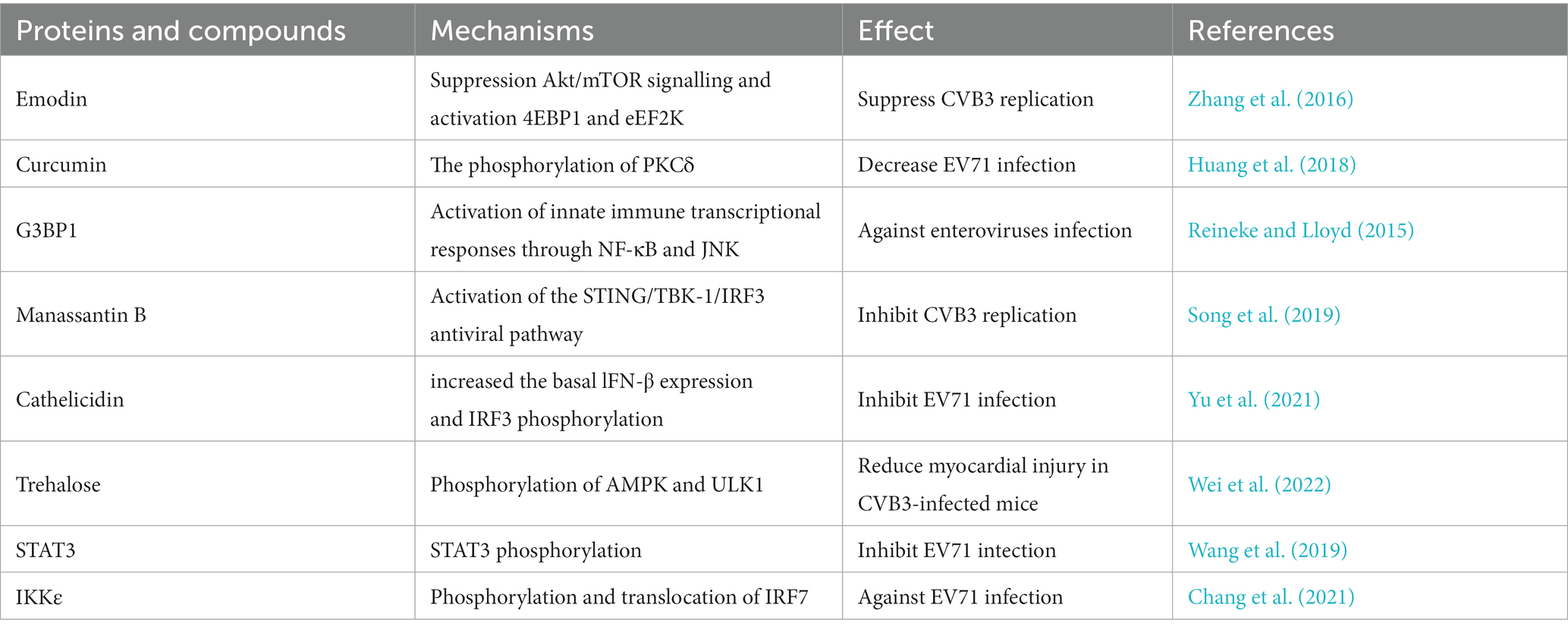
Currently, there are relatively few reports on inhibition of protein phosphorylation during enterovirus infection. One study revealed that the natural compound emodin, a natural compound derived from plant roots, can inhibit CVB3 replication by suppressing Akt/mTOR signaling and activating 4EBP1 and eEF2K (Table 3) (Zhang et al., 2016). Curcumin is a compound obtain anti-cancer properties that can decrease EV71 infection by inhibiting the phosphorylation of PKCδ (Table 3) (Huang et al., 2018). Additionally, protein phosphorylation plays an antiviral role in enterovirus infection via an immune-regulated pathway. A study revealed that G3BP1 plays an anti-enterovirus role by inducing stress granule formation, and is associated with the innate immune transcriptional response activation through NF-κB and JNK (Table 3) (Reineke and Lloyd, 2015). Manassantin B inhibits CVB3 replication by activating the STING/TKB-1/IRF3 antiviral pathway and increasing the production of mROS (Song et al., 2019). Cathelicidin antimicrobial peptides (human LL-37 and mouse CRAMP) (Add “human LL-37 and mouse CRAMP”) also play an antiviral role in EV71 infection and (Delete”and”) LL-37 and CRAMP were shown to markedly increase the basal IFN-β expression and IRF3 phosphorylation, effectively alleviating EV71 infection (Yu et al., 2021). Trehalose prevents cardiovascular diseases by regulating autophagy. In a viral myocarditis mouse model induced by CVB3, trehalose was shown to reduce myocardial injury and significantly enhance AMPK and ULK1 phosphorylation in B cells (Wei et al., 2022). During EV71 infection, STAT3 phosphorylation and the expression of downstream inflammatory regulators are increased, which activates the type I IFN-mediated antiviral response (Wang et al., 2019). In addition, the host IKKε gene leads to alterations in IFN production during EV71 infection through phosphorylation and translocation of IRF7 in the presence of ubiquitin, activating the expression of IFNβ and ISGs and attenuating viral propagation (Table 3) (Chang et al., 2021).
3.2 The proviral role of phosphorylation in enterovirus infection
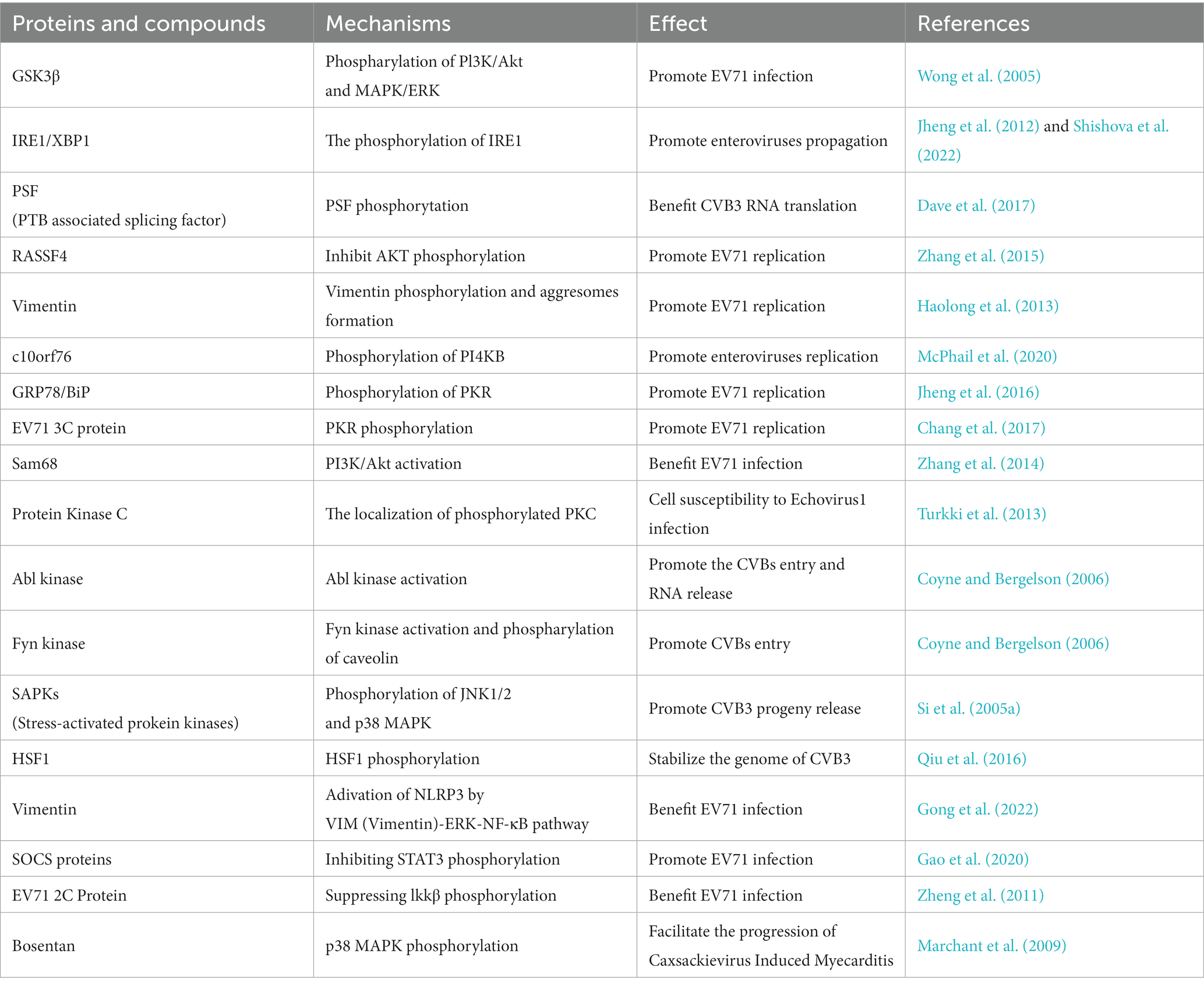
Phosphorylation also plays an important role in the promotion of enterovirus infection. The cellular phosphoproteome undergoes significant changes and approximately 85% of the quantified phosphoproteome is dynamically regulated during CVB3 infection (Giansanti et al., 2020). Once the EV71 virus binds to host receptors, the phosphorylation of PI3K/Akt and MAPK/ERK is initiated; immediately inactivating GSK3β, delaying host cell apoptosis, and promoting infection (Wong et al., 2005). The IRE1/XBP1 pathway plays a pivotal role in the endoplasmic reticulum (ER), or unfolded protein, stress response. One study revealed that XBP1 participates in EV71 replication by affecting viral entry. After EV71 infection, IRE1 undergoes phosphorylation and activation, whereas the downstream XBP1s (spliced XBP1) protein levels decreased (Jheng et al., 2012). During poliovirus and CVB3 infection, IRE1 undergoes the complicated dynamics of autophosphorylation and cleavage, indicating that enteroviruses utilize various mechanisms to regulate the Ire1-Xbp1 host defensive pathway in infected cells (Shishova et al., 2022) (Table 4).
The phosphorylation of many proteins can regulate stages in the life cycle of enteroviruses. PTB associated splicing factor (PSF) is an IRES trans-acting factor which is critical for CVB3 RNA translation. One study revealed that PSF protein levels in the cytoplasm are enhanced during CVB3 infection and its phosphorylation promotes CVB3 RNA translation (Dave et al., 2017). The tumor suppressor RASSF4, which participates in diverse biological processes including cell death, signal transduction, and tumor development, has been reported to promote EV71 infection and subsequently accelerate the inhibition of AKT phosphorylation in infected cells (Zhang et al., 2015). The type III intermediate filament vimentin has been reported to be a key factor in the process of viral infection through different mechanisms. In EV71 infection of human astrocytoma cells, vimentin undergoes rearrangement immediately following the formation of aggresome-like structures in the perinuclear region. Further research has indicated that vimentin is phosphorylated, and that both phosphorylation and aggresome formation are important for EV71 replication (Haolong et al., 2013) (Table 4).
Enterovirus replication is heavily reliant on PI4KB kinase activity, and the interaction between PI4KB and c10orf76 is a critical Golgi signaling complex and an important factor influencing the replication of several enteroviruses. The affinity between PI4KB and c10orf76 is determined by the phosphorylation status of PI4KB, and the phosphorylated PI4KB at Ser496 site shows a decrease in affinity for c10orf76 (McPhail et al., 2020). Enteroviruses can also use ER stress or the unfolded protein response to promote viral replication. Jheng JR et al. revealed that ER stress and the unfolded protein response, GRP78/BiP, were redistributed during EV71 infection, which contributed to the promotion of EV71 replication. Further studies showed that dsRNA, rather than a viral protein, induces the phosphorylation of PKR, which plays a role in GRP78/BiP redistribution (Jheng et al., 2016). However, another study showed that in addition to dsRNA, viral proteins can also cause PKR phosphorylation, and that the EV71 3C protein interacts with PKR and subsequently mediates its phosphorylation to promote viral replication (Chang et al., 2017) (Table 4).
Numerous studies have shown that nuclear proteins translocate into the cytoplasm to promote EV71 replication. During EV71 infection, Sam68 translocates to the cytoplasm and mediates the activation of PI3K/Akt to assist the infection (Zhang et al., 2014). The localization of phosphorylated host proteins is related to the susceptibility to viral infections. In a study of cell susceptibility to Echovirus1, phosphorylated PKC (Change “pPKC” into “phosphorylated PKC”) was mainly distributed in the cytoplasm and usually accumulated in the perinuclear compartment in non-permissive cells, whereas in permissive cells, it was evenly distributed and no perinuclear aggregation was observed (Turkki et al., 2013) (Table 4).
The activation of some kinases also plays a significant role in EV71 infection. The process of virus attachment to decay-accelerating factor on the surface of apical cells activates Abl kinase, which promotes the movement of the virus to tight junctions where it immediately interacts with coxsackievirus-adenovirus receptor, facilitating the alteration of conformation in the viral capsid. This process is critical for entry and RNA release. Additionally, interaction with decay-accelerating factor can activate Fyn kinase, which is necessary for caveolin phosphorylation and viral transportation into cells (Coyne and Bergelson, 2006). Another study reported that the phosphorylation of JNK1/2 and p38 MAPK was enhanced during CVB3 infection and that the stress-activated protein kinase pathway plays a critical role in the life cycle of CVB3, especially during viral progeny release (Si et al., 2005a). CVB3 infection induces phosphorylation of heat shock factor 1, leading to the upregulation of Hsp70-1 which contributes to the stabilization of the CVB3 genome; thus promoting viral infection (Qiu et al., 2016) (Table 4).
Phosphorylation also affects the host immune system, promoting EV infection, and the occurrence and development of enterovirus-related diseases. The inflammasome is an essential element of the natural immune system and is closely associated with EV71-induced central nervous system injury, which is regulated by signaling pathways such as VIM (Vimentin)-ERK-NF-κB pathway (Add “Vimentin”) (Gong et al., 2022). Viruses can evade the host immune response by increasing the expression of suppressor of cytokine signaling proteins. One study revealed that suppressor of cytokine signaling 3 promotes EV71 infection by inhibiting interferon-induced STAT3 phosphorylation and negatively regulates the JAK/STAT signaling pathway; thus, allowing EV71 to escape host immunity (Gao et al., 2020). Viral proteins also participate in immune responses related to phosphorylation. For example, EV71 2C protein inhibits TNF-α–mediated activation of NF-κB by suppressing Ikkβ phosphorylation (Zheng et al., 2011).
In addition to endogenous cellular phosphorylation, chemical drugs increase the phosphorylation of cellular proteins to regulate viral infections. The endothelin-1 receptor antagonist Bosentan can facilitate the progression of Coxsackievirus-induced myocarditis by inducing p38 MAPK phosphorylation, which increases the viral load in cells and tissues (Marchant et al., 2009) (Table 4).
4 Acetylation
Protein acetylation is the principal PTM catalyzed by acetyltransferases (Baeza et al., 2016). Protein acetylation affects numerous biological events, including gene replication, transcription, repair, and signal transduction pathways; thereby regulating diverse cellular processes (Arnesen, 2011; Verdin and Ott, 2015). Host and viral proteins undergo acetylation which can play pivotal roles in different phases of viral infection, including viral entry, genome replication, assembly and release of progeny viral particles, and host antiviral responses (Murray et al., 2018; Xue et al., 2022). With the rapid development and optimization of proteomics and mass spectrometry technologies, a new era of protein acetylation research during viral infection has recently emerged.
4.1 The antiviral role of acetylation in the infection of enterovirus
Sirtuin 1 (SIRT1) is a lysine deacetylase that regulates various processes including inflammation, metabolism, and aging. Han Y et al. revealed that reactive oxygen species generation and SIRT1 expression were downregulated in apoptotic cells infected with EV71, and treatment with a reactive oxygen species inhibitor decreased EV71 propagation and increased SIRT1 expression in EV71-infected cells. SIRT1 can inhibit the acetylation and RNA dependent RNA polymerase activity of 3D pol, thereby decreasing viral genome replication. Additionally, SIRT1 can interact with the 5′ UTR of EV71 RNA to disrupt viral RNA translation (Figure 5A) (Han et al., 2016). CVB3 infection induces HDAC2 activity and treatment with a HDAC inhibitor can inhibit CVB3 replication, indicating that the acetylation of proteins can suppress viral infection (Figure 5B) (Shim et al., 2013). There are relatively few studies on inhibition of enterovirus infection by acetylation and further research is required to fully understand this process.
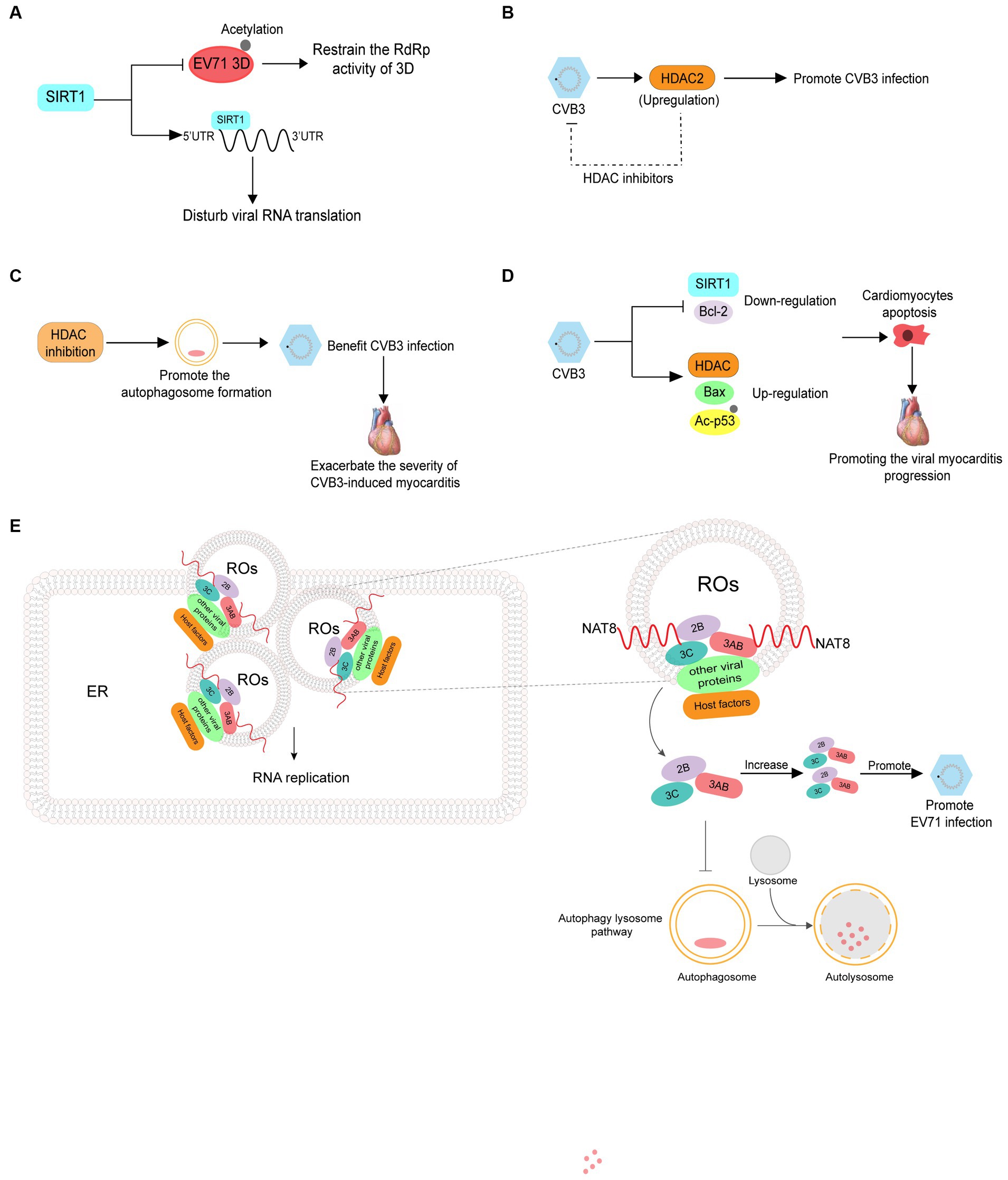
Figure 5. The roles of acetylation in the infection of enterovirus. (A) SIRT1 can inhibit the acetylation and RNA dependent RNA polymerase activity of 3D pol, thus reducing viral genome replication. SIRT1 also interacts with the 5′ UTR of EV71 RNA to disrupt viral RNA translation. (B) CVB3 infection induces histone deacetylase2 (HDAC2) activity, and treatment with HDAC inhibitors can inhibit CVB3 replication. (C) Inhibition of HDAC activity increases the formation of autophagosomes which promote CVB3 replication and ultimately exacerbate the severity of CVB3-induced myocarditis. (D) CVB3 infection induces the expression of HDAC1 and Bax while suppressing SIRT1 and Bcl-2, in addition to upregulating acetylated p53. (E) NAT8 promotes EV71 replication by increasing the stability of 2B, 3AB, and 3C proteins in an acetyltransferase-activity-dependent manner.
4.2 The proviral role of acetylation in enterovirus infection
Contrary to the results above, Zhou L et al. revealed that inhibition of HDAC can increase autophagosome formation, which benefits CVB3 replication and ultimately exacerbates the severity of CVB3-induced myocarditis (Figure 5C) (Zhou et al., 2015). In addition to promoting viral infections, acetylation is related to the pathogenesis of viral myocarditis. CVB3 infection has been reported to induce the expression of HDAC1 and Bax while suppressing SIRT1 and Bcl-2 and upregulating acetylated p53. These events lead to cardiomyocyte apoptosis, thus promoting viral myocarditis progression (Figure 5D) (Jiang et al., 2019). NAT8 is an ER-resident acetyltransferase and promotes EV71 replication by increasing the stability of 2B, 3AB, and 3C proteins in an acetyltransferase activity-dependent manner (Figure 5E) (Zhao et al., 2022).
5 Conclusion
PTM of proteins is involved in various biological events, and disruption of the modification process can lead to the development of diseases (Chaugule and Walden, 2016; Caruso Bavisotto et al., 2020; Morales-Tarre et al., 2021; Zhu et al., 2022b). PTM participates in diverse viral infections, including hepatitis B, influenza, SARS-CoV-2, and enterovirus (Karim et al., 2020; Hofmann et al., 2023; Liang et al., 2023; Park et al., 2023). Currently, over 200 distinct covalent modifications have been reported; with phosphorylation, ubiquitination, acetylation, glycosylation being among the most common (Tsikas, 2021). A growing body of work suggests that different PTMs play pro- or antiviral roles through different mechanisms, such as altering the abundance and biochemical properties of host and viral proteins, reducing proteins/protein or proteins/nucleic acid interaction, and regulating innate and adaptive immune responses. Although many studies have confirmed the roles of PTMs in viral infection, the understanding of PTMs has been limited by traditional approaches, owing to the difficulties in detection and complexity of these modifications (Dai et al., 2021; Zhu et al., 2022a). Traditionally, researchers utilize antibody-based assays to reveal specific PTMs; however, this detection approach fails to confirm the combinatorial patterns of diverse PTMs, and the antibodies have difficulty distinguishing specific modification sites (Hattori and Koide, 2018; Leutert et al., 2021). In recent years, increasing evidence of crosstalk between PTMs also has been reported (Cuijpers and Vertegaal, 2018; Vu et al., 2018; Kirsch et al., 2020; Leutert et al., 2021). Therefore, further in-depth investigation of the role and mechanisms of PTM crosstalk and other modifications in enterovirus infection is required.
Author contributions
XZ: Writing – original draft, Investigation, Software, Supervision, Writing – review & editing. YH: Writing – original draft. JZ: Writing – original draft. YL: Writing – original draft. XM: Writing – review & editing. HC: Writing – review & editing. YX: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andres, M., Garcia-Gomis, D., Ponte, I., Suau, P., and Roque, A. (2020). Histone H1 Post-translational modifications: update and future perspectives. Int. J. Mol. Sci. 21:5941. doi: 10.3390/ijms21165941
Arnesen, T. (2011). Towards a functional understanding of protein N-terminal acetylation. PLoS Biol. 9:e1001074. doi: 10.1371/journal.pbio.1001074
Aslebagh, R., Wormwood, K. L., Channaveerappa, D., Wetie, A. G. N., Woods, A. G., and Darie, C. C. (2019). Identification of posttranslational modifications (Ptms) of proteins by mass spectrometry. Adv. Exp. Med. Biol. 1140, 199–224. doi: 10.1007/978-3-030-15950-4_11
Baeza, J., Smallegan, M. J., and Denu, J. M. (2016). Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem. Sci. 41, 231–244. doi: 10.1016/j.tibs.2015.12.006
Bauer, L., Manganaro, R., Zonsics, B., Strating, J., El Kazzi, P., Lorenzo Lopez, M., et al. (2019). Fluoxetine inhibits enterovirus replication by targeting the viral 2C protein in a stereospecific manner. Acs Infect Dis 5, 1609–1623. doi: 10.1021/acsinfecdis.9b00179
Bilbrough, T., Piemontese, E., and Seitz, O. (2022). Dissecting the role of protein phosphorylation: a chemical biology toolbox. Chem. Soc. Rev. 51, 5691–5730. doi: 10.1039/D1CS00991E
Cappadocia, L., and Lima, C. D. (2018). Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 118, 889–918. doi: 10.1021/acs.chemrev.6b00737
Caruso Bavisotto, C., Alberti, G., Vitale, A. M., Paladino, L., Campanella, C., Rappa, F., et al. (2020). Hsp60 Post-translational modifications: functional and pathological consequences. Front. Mol. Biosci. 7:95. doi: 10.3389/fmolb.2020.00095
Chang, Y. H., Lau, K. S., Kuo, R. L., and Horng, J. T. (2017). Dsrna binding domain of Pkr is Proteolytically released by enterovirus A71 to facilitate viral replication. Front. Cell. Infect. Microbiol. 7:284. doi: 10.3389/fcimb.2017.00284
Chang, Y. L., Liao, Y. W., Chen, M. H., Chang, S. Y., Huang, Y. T., Ho, B. C., et al. (2021). Ikkepsilon isoform switching governs the immune response against Ev71 infection. Commun Biol 4:663. doi: 10.1038/s42003-021-02187-x
Chau, V., Tobias, J. W., Bachmair, A., Marriott, D., Ecker, D. J., Gonda, D. K., et al. (1989). A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583. doi: 10.1126/science.2538923
Chaugule, V. K., and Walden, H. (2016). Specificity and disease in the ubiquitin system. Biochem. Soc. Trans. 44, 212–227. doi: 10.1042/BST20150209
Chen, S. C., Chang, L. Y., Wang, Y. W., Chen, Y. C., Weng, K. F., Shih, S. R., et al. (2011). Sumoylation-promoted enterovirus 71 3C degradation correlates with a reduction in viral replication and cell apoptosis. J. Biol. Chem. 286, 31373–31384. doi: 10.1074/jbc.M111.254896
Cheng, N., Liu, M., Li, W., Sun, B., Liu, D., Wang, G., et al. (2022). Protein post-translational modification in Sars-CoV-2 and host interaction. Front. Immunol. 13:1068449. doi: 10.3389/fimmu.2022.1068449
Coyne, C. B., and Bergelson, J. M. (2006). Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124, 119–131. doi: 10.1016/j.cell.2005.10.035
Cuijpers, S. A. G., and Vertegaal, A. C. O. (2018). Guiding mitotic progression by crosstalk between Post-translational modifications. Trends Biochem. Sci. 43, 251–268. doi: 10.1016/j.tibs.2018.02.004
Dai, J., Yu, F., Zhou, C., and Yu, W. (2021). Understanding the limit of open search in the identification of peptides with Post-translational modifications - a simulation-based study. IEEE/ACM Trans. Comput. Biol. Bioinform. 18, 2884–2890. doi: 10.1109/TCBB.2020.2991207
Dave, P., George, B., Sharma, D. K., and Das, S. (2017). Polypyrimidine tract-binding protein (Ptb) and Ptb-associated splicing factor in Cvb3 infection: an Itaf for an Itaf. Nucleic Acids Res. 45, 9068–9084. doi: 10.1093/nar/gkx519
Desai, S. D., Haas, A. L., Wood, L. M., Tsai, Y. C., Pestka, S., Rubin, E. H., et al. (2006). Elevated expression of Isg15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res. 66, 921–928. doi: 10.1158/0008-5472.CAN-05-1123
Deutscher, J., Ake, F. M., Derkaoui, M., Zebre, A. C., Cao, T. N., Bouraoui, H., et al. (2014). The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78, 231–256. doi: 10.1128/MMBR.00001-14
Dores, M. R., and Trejo, J. (2014). Atypical regulation of G protein-coupled receptor intracellular trafficking by ubiquitination. Curr. Opin. Cell Biol. 27, 44–50. doi: 10.1016/j.ceb.2013.11.004
Fan, W., Mar, K. B., Sari, L., Gaszek, I. K., Cheng, Q., Evers, B. M., et al. (2021). Trim7 inhibits enterovirus replication and promotes emergence of a viral variant with increased pathogenicity. Cell 184:e17. doi: 10.1016/j.cell.2021.04.047
Gao, W., Hou, M., Liu, X., Li, Z., Yang, Y., and Zhang, W. (2020). Induction of Socs expression by Ev71 infection promotes Ev71 replication. Biomed. Res. Int. 2020:2430640. doi: 10.1155/2020/2430640
Gao, G., Wong, J., Zhang, J., Mao, I., Shravah, J., Wu, Y., et al. (2010). Proteasome activator Reggamma enhances coxsackieviral infection by facilitating p53 degradation. J. Virol. 84, 11056–11066. doi: 10.1128/JVI.00008-10
Gao, G., Zhang, J., Si, X., Wong, J., Cheung, C., Mcmanus, B., et al. (2008). Proteasome inhibition attenuates coxsackievirus-induced myocardial damage in mice. Am. J. Physiol. Heart Circ. Physiol. 295, H401–H408. doi: 10.1152/ajpheart.00292.2008
Gareau, J. R., and Lima, C. D. (2010). The Sumo pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871. doi: 10.1038/nrm3011
Geiss-Friedlander, R., and Melchior, F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956. doi: 10.1038/nrm2293
Giansanti, P., Strating, J., Defourny, K. A. Y., Cesonyte, I., Bottino, A. M. S., Post, H., et al. (2020). Dynamic remodelling of the human host cell proteome and phosphoproteome upon enterovirus infection. Nat. Commun. 11:4332. doi: 10.1038/s41467-020-18168-3
Gong, Z., Gao, X., Yang, Q., Lun, J., Xiao, H., Zhong, J., et al. (2022). Phosphorylation of Erk-dependent Nf-kappaB triggers Nlrp3 Inflammasome mediated by vimentin in Ev71-infected glioblastoma cells. Molecules 27:4190. doi: 10.3390/molecules27134190
Gupta, R., Sahu, M., Srivastava, D., Tiwari, S., Ambasta, R. K., and Kumar, P. (2021). Post-translational modifications: regulators of neurodegenerative proteinopathies. Ageing Res. Rev. 68:101336. doi: 10.1016/j.arr.2021.101336
Han, Y., Wang, L., Cui, J., Song, Y., Luo, Z., Chen, J., et al. (2016). Sirt1 inhibits Ev71 genome replication and Rna translation by interfering with the viral polymerase and 5'utr Rna. J. Cell Sci. 129, 4534–4547. doi: 10.1242/jcs.193698
Haolong, C., Du, N., Hongchao, T., Yang, Y., Wei, Z., Hua, Z., et al. (2013). Enterovirus 71 Vp1 activates calmodulin-dependent protein kinase ii and results in the rearrangement of vimentin in human astrocyte cells. PLoS One 8:e73900. doi: 10.1371/journal.pone.0073900
Hattori, T., and Koide, S. (2018). Next-generation antibodies for post-translational modifications. Curr. Opin. Struct. Biol. 51, 141–148. doi: 10.1016/j.sbi.2018.04.006
He, S., Wang, F., Yung, K. K. L., Zhang, S., and Qu, S. (2021). Effects of alpha-Synuclein-associated Post-translational modifications in Parkinson's disease. ACS Chem. Neurosci. 12, 1061–1071. doi: 10.1021/acschemneuro.1c00028
Hermann, J., Schurgers, L., and Jankowski, V. (2022). Identification and characterization of post-translational modifications: clinical implications. Mol. Asp. Med. 86:101066. doi: 10.1016/j.mam.2022.101066
Herrmann, J., Lerman, L. O., and Lerman, A. (2007). Ubiquitin and ubiquitin-like proteins in protein regulation. Circ. Res. 100, 1276–1291. doi: 10.1161/01.RES.0000264500.11888.f0
Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G., and Jentsch, S. (2002). Rad6-dependent Dna repair is linked to modification of Pcna by ubiquitin and Sumo. Nature 419, 135–141. doi: 10.1038/nature00991
Hofmann, S., Plank, V., Groitl, P., Skvorc, N., Hofmann, K., Luther, J., et al. (2023). Sumo modification of hepatitis B virus Core mediates nuclear entry, Promyelocytic leukemia nuclear body association, and efficient formation of covalently closed circular Dna. Microbiol Spectr 11:e0044623. doi: 10.1128/spectrum.00446-23
Huang, H. I., Chio, C. C., and Lin, J. Y. (2018). Inhibition of Ev71 by curcumin in intestinal epithelial cells. PLoS One 13:e0191617. doi: 10.1371/journal.pone.0191617
Iwai, K. (2021). Discovery of linear ubiquitination, a crucial regulator for immune signaling and cell death. FEBS J. 288, 1060–1069. doi: 10.1111/febs.15471
Jheng, J. R., Lin, C. Y., Horng, J. T., and Lau, K. S. (2012). Inhibition of enterovirus 71 entry by transcription factor Xbp1. Biochem. Biophys. Res. Commun. 420, 882–887. doi: 10.1016/j.bbrc.2012.03.094
Jheng, J. R., Wang, S. C., Jheng, C. R., and Horng, J. T. (2016). Enterovirus 71 induces dsrna/Pkr-dependent cytoplasmic redistribution of Grp78/BiP to promote viral replication. Emerg Microbes Infect 5:e23. doi: 10.1038/emi.2016.20
Jiang, D., Li, M., Yu, Y., Shi, H., and Chen, R. (2019). Microrna-34a aggravates coxsackievirus B3-induced apoptosis of cardiomyocytes through the Sirt1-p53 pathway. J. Med. Virol. 91, 1643–1651. doi: 10.1002/jmv.25482
Kang, C., and Yi, G. S. (2011). Identification of ubiquitin/ubiquitin-like protein modification from tandem mass spectra with various Ptms. BMC Bioinform 12:S8. doi: 10.1186/1471-2105-12-S14-S8
Karim, M., Biquand, E., Declercq, M., Jacob, Y., Van Der Werf, S., and Demeret, C. (2020). Nonproteolytic K29-linked ubiquitination of the Pb2 replication protein of influenza a viruses by Proviral Cullin 4-based E3 ligases. MBio 11. doi: 10.1128/mBio.00305-20
Kespohl, M., Bredow, C., Klingel, K., Voss, M., Paeschke, A., Zickler, M., et al. (2020). Protein modification with Isg15 blocks coxsackievirus pathology by antiviral and metabolic reprogramming. Sci. Adv. 6:eaay1109. doi: 10.1126/sciadv.aay1109
Kirsch, R., Jensen, O. N., and Schwammle, V. (2020). Visualization of the dynamics of histone modifications and their crosstalk using Ptm-CrossTalkMapper. Methods 184, 78–85. doi: 10.1016/j.ymeth.2020.01.012
Komander, D. (2009). The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 37, 937–953. doi: 10.1042/BST0370937
Kumar, R., Mehta, D., Mishra, N., Nayak, D., and Sunil, S. (2020). Role of host-mediated Post-translational modifications (Ptms) in Rna virus pathogenesis. Int. J. Mol. Sci. 22:323. doi: 10.3390/ijms22010323
Kung, Y. A., Hung, C. T., Chien, K. Y., and Shih, S. R. (2017). Control of the negative Ires trans-acting factor Khsrp by ubiquitination. Nucleic Acids Res. 45, 271–287. doi: 10.1093/nar/gkw1042
Lauwers, E., Jacob, C., and Andre, B. (2009). K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185, 493–502. doi: 10.1083/jcb.200810114
Leutert, M., Entwisle, S. W., and Villen, J. (2021). Decoding Post-translational modification crosstalk with proteomics. Mol. Cell. Proteomics 20:100129. doi: 10.1016/j.mcpro.2021.100129
Li, Z., Huan, C., Wang, H., Liu, Y., Liu, X., Su, X., et al. (2020). Trim21-mediated proteasomal degradation of Samhd1 regulates its antiviral activity. EMBO Rep. 21:e47528. doi: 10.15252/embr.201847528
Liang, B., Zhu, Y., Shi, W., Ni, C., Tan, B., and Tang, S. (2023). Sars-CoV-2 spike protein Post-translational modification landscape and its impact on protein structure and function via computational prediction. Research 6:0078. doi: 10.34133/research.0078
Liu, X., Lei, X., Zhou, Z., Sun, Z., Xue, Q., Wang, J., et al. (2011). Enterovirus 71 induces degradation of Trim38, a potential E3 ubiquitin ligase. Virol. J. 8:61. doi: 10.1186/1743-422X-8-61
Liu, H., Li, M., Song, Y., and Xu, W. (2018). Trim21 restricts Coxsackievirus B3 replication, cardiac and pancreatic injury via interacting with Mavs and positively regulating Irf3-mediated type-I interferon production. Front. Immunol. 9:2479. doi: 10.3389/fimmu.2018.02479
Liu, Y., Zheng, Z., Shu, B., Meng, J., Zhang, Y., Zheng, C., et al. (2016). Sumo modification stabilizes enterovirus 71 polymerase 3D to facilitate viral replication. J. Virol. 90, 10472–10485. doi: 10.1128/JVI.01756-16
Lopez, J. E., Sullivan, E. D., and Fierke, C. A. (2016). Metal-dependent deacetylases: Cancer and epigenetic regulators. ACS Chem. Biol. 11, 706–716. doi: 10.1021/acschembio.5b01067
Luo, H., Zhang, J., Dastvan, F., Yanagawa, B., Reidy, M. A., Zhang, H. M., et al. (2003). Ubiquitin-dependent proteolysis of cyclin D1 is associated with coxsackievirus-induced cell growth arrest. J. Virol. 77, 1–9. doi: 10.1128/JVI.77.1.1-9.2003
Lyoo, H., Dorobantu, C. M., Van Der Schaar, H. M., and Van Kuppeveld, F. J. M. (2017). Modulation of proteolytic polyprotein processing by coxsackievirus mutants resistant to inhibitors targeting phosphatidylinositol-4-kinase Iiibeta or oxysterol binding protein. Antivir. Res. 147, 86–90. doi: 10.1016/j.antiviral.2017.10.006
Marchant, D., Dou, Y., Luo, H., Garmaroudi, F. S., Mcdonough, J. E., Si, X., et al. (2009). Bosentan enhances viral load via endothelin-1 receptor type-A-mediated p38 mitogen-activated protein kinase activation while improving cardiac function during coxsackievirus-induced myocarditis. Circ. Res. 104, 813–821. doi: 10.1161/CIRCRESAHA.108.191171
Mcphail, J. A., Lyoo, H., Pemberton, J. G., Hoffmann, R. M., Van Elst, W., Strating, J., et al. (2020). Characterization of the c10orf76-Pi4kb complex and its necessity for Golgi Pi4P levels and enterovirus replication. EMBO Rep. 21:e48441. doi: 10.15252/embr.201948441
Meng, J., Yao, Z., He, Y., Zhang, R., Zhang, Y., Yao, X., et al. (2017). Arrdc4 regulates enterovirus 71-induced innate immune response by promoting K63 polyubiquitination of Mda5 through Trim65. Cell Death Dis. 8:e2866. doi: 10.1038/cddis.2017.257
Mirzalieva, O., Juncker, M., Schwartzenburg, J., and Desai, S. (2022). Isg15 and Isgylation in human diseases. Cell 11:538. doi: 10.3390/cells11030538
Morales-Tarre, O., Alonso-Bastida, R., Arcos-Encarnacion, B., Perez-Martinez, L., and Encarnacion-Guevara, S. (2021). Protein lysine acetylation and its role in different human pathologies: a proteomic approach. Expert Rev. Proteomics 18, 949–975. doi: 10.1080/14789450.2021.2007766
Murray, L. A., Sheng, X., and Cristea, I. M. (2018). Orchestration of protein acetylation as a toggle for cellular defense and virus replication. Nat. Commun. 9:4967. doi: 10.1038/s41467-018-07179-w
Park, E. S., Dezhbord, M., Lee, A. R., and Kim, K. H. (2022a). The roles of ubiquitination in pathogenesis of influenza virus infection. Int. J. Mol. Sci. 23, 4593. doi: 10.3390/ijms23094593
Park, J. W., Tyl, M. D., and Cristea, I. M. (2023). Orchestration of mitochondrial function and remodeling by Post-translational modifications provide insight into mechanisms of viral infection. Biomol. Ther. 13:869. doi: 10.3390/biom13050869
Park, K., Zhong, J., Jang, J. S., Kim, J., Kim, H. J., Lee, J. H., et al. (2022b). Zwc complex-mediated Spt5 phosphorylation suppresses divergent antisense Rna transcription at active gene promoters. Nucleic Acids Res. 50, 3835–3851. doi: 10.1093/nar/gkac193
Pellegrino, N. E., Guven, A., Gray, K., Shah, P., Kasture, G., Nastke, M. D., et al. (2022). The next frontier: translational development of ubiquitination, Sumoylation, and Neddylation in Cancer. Int. J. Mol. Sci. 23:3480. doi: 10.3390/ijms23073480
Puertollano, R., Ferguson, S. M., Brugarolas, J., and Ballabio, A. (2018). The complex relationship between Tfeb transcription factor phosphorylation and subcellular localization. EMBO J. 37. doi: 10.15252/embj.201798804
Qiu, Y., Ye, X., Hanson, P. J., Zhang, H. M., Zong, J., Cho, B., et al. (2016). Hsp70-1: upregulation via selective phosphorylation of heat shock factor 1 during coxsackieviral infection and promotion of viral replication via the au-rich element. Cell. Mol. Life Sci. 73, 1067–1084. doi: 10.1007/s00018-015-2036-6
Rape, M. (2018). Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 19, 59–70. doi: 10.1038/nrm.2017.83
Reineke, L. C., and Lloyd, R. E. (2015). The stress granule protein G3bp1 recruits protein kinase R to promote multiple innate immune antiviral responses. J. Virol. 89, 2575–2589. doi: 10.1128/JVI.02791-14
Saguil, A., Kane, S. F., Lauters, R., and Mercado, M. G. (2019). Hand-foot-and-mouth disease: rapid evidence review. Am. Fam. Physician 100, 408–414.
Schweppe, R. E., Haydon, C. E., Lewis, T. S., Resing, K. A., and Ahn, N. G. (2003). The characterization of protein post-translational modifications by mass spectrometry. Acc. Chem. Res. 36, 453–461. doi: 10.1021/ar020143l
Shim, S. H., Park, J. H., Ye, M. B., and Nam, J. H. (2013). Histone deacetylase inhibitors suppress coxsackievirus B3 growth in vitro and myocarditis induced in mice. Acta Virol. 57, 462–466. doi: 10.4149/av_2013_04_462
Shishova, A., Dyugay, I., Fominykh, K., Baryshnikova, V., Dereventsova, A., Turchenko, Y., et al. (2022). Enteroviruses manipulate the unfolded protein response through multifaceted deregulation of the Ire1-Xbp1 pathway. Viruses 14:2486. doi: 10.3390/v14112486
Si, X., Gao, G., Wong, J., Wang, Y., Zhang, J., and Luo, H. (2008). Ubiquitination is required for effective replication of coxsackievirus B3. PLoS One 3:e2585. doi: 10.1371/journal.pone.0002585
Si, X., Luo, H., Morgan, A., Zhang, J., Wong, J., Yuan, J., et al. (2005a). Stress-activated protein kinases are involved in coxsackievirus B3 viral progeny release. J. Virol. 79, 13875–13881. doi: 10.1128/JVI.79.22.13875-13881.2005
Si, X., Mcmanus, B. M., Zhang, J., Yuan, J., Cheung, C., Esfandiarei, M., et al. (2005b). Pyrrolidine dithiocarbamate reduces coxsackievirus B3 replication through inhibition of the ubiquitin-proteasome pathway. J. Virol. 79, 8014–8023. doi: 10.1128/JVI.79.13.8014-8023.2005
Si, X., Wang, Y., Wong, J., Zhang, J., Mcmanus, B. M., and Luo, H. (2007). Dysregulation of the ubiquitin-proteasome system by curcumin suppresses coxsackievirus B3 replication. J. Virol. 81, 3142–3150. doi: 10.1128/JVI.02028-06
Smith, B. J., and Carregari, V. C. (2022). Histone modifications in neurological disorders. Adv. Exp. Med. Biol. 1382, 95–107. doi: 10.1007/978-3-031-05460-0_7
Song, J. H., Ahn, J. H., Kim, S. R., Cho, S., Hong, E. H., Kwon, B. E., et al. (2019). Manassantin B shows antiviral activity against coxsackievirus B3 infection by activation of the Sting/Tbk-1/Irf3 signalling pathway. Sci. Rep. 9:9413. doi: 10.1038/s41598-019-45868-8
Song, B., Liu, D., Greco, T. M., and Cristea, I. M. (2021). Post-translational modification control of viral Dna sensors and innate immune signaling. Adv. Virus Res. 109, 163–199. doi: 10.1016/bs.aivir.2021.03.001
Sun, C. F., Li, Y. Q., and Mao, X. M. (2020). Regulation of protein Post-translational modifications on metabolism of Actinomycetes. Biomol. Ther. 10:1122. doi: 10.3390/biom10081122
Swatek, K. N., and Komander, D. (2016). Ubiquitin modifications. Cell Res. 26, 399–422. doi: 10.1038/cr.2016.39
Tabor-Godwin, J. M., Tsueng, G., Sayen, M. R., Gottlieb, R. A., and Feuer, R. (2012). The role of autophagy during coxsackievirus infection of neural progenitor and stem cells. Autophagy 8, 938–953. doi: 10.4161/auto.19781
Tsikas, D. (2021). Post-translational modifications (Ptm): analytical approaches, signaling, physiology and pathophysiology-part I. Amino Acids 53, 485–487. doi: 10.1007/s00726-021-02984-y
Turkki, P., Makkonen, K. E., Huttunen, M., Laakkonen, J. P., Yla-Herttuala, S., Airenne, K. J., et al. (2013). Cell susceptibility to baculovirus transduction and echovirus infection is modified by protein kinase C phosphorylation and vimentin organization. J. Virol. 87, 9822–9835. doi: 10.1128/JVI.01004-13
Van Wijk, S. J., Fulda, S., Dikic, I., and Heilemann, M. (2019). Visualizing ubiquitination in mammalian cells. EMBO Rep. 20. doi: 10.15252/embr.201846520
Verdin, E., and Ott, M. (2015). 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264. doi: 10.1038/nrm3931
Voss, M., Braun, V., Bredow, C., Kloetzel, P. M., and Beling, A. (2021). Coxsackievirus B3 exploits the ubiquitin-proteasome system to facilitate viral replication. Viruses 13:1360. doi: 10.3390/v13071360
Vu, L. D., Gevaert, K., and De Smet, I. (2018). Protein language: Post-translational modifications talking to each other. Trends Plant Sci. 23, 1068–1080. doi: 10.1016/j.tplants.2018.09.004
Wang, H., Yuan, M., Wang, S., Zhang, L., Zhang, R., Zou, X., et al. (2019). Stat3 regulates the type I Ifn-mediated antiviral response by interfering with the nuclear entry of Stat1. Int. J. Mol. Sci. 20:4870. doi: 10.3390/ijms20194870
Wang, H., Zhong, M., Cui, B., Yan, H., Wu, S., Wang, K., et al. (2022). Neddylation of enterovirus 71 Vp2 protein reduces its stability and restricts viral replication. J. Virol. 96:e0059822. doi: 10.1128/jvi.00598-22
Wei, B., Lu, F., Kong, Q., Huang, Y., Huang, K., and Wu, W. (2022). Trehalose induces B cell autophagy to alleviate myocardial injury via the Ampk/Ulk1 signalling pathway in acute viral myocarditis induced by Coxsackie virus B3. Int. J. Biochem. Cell Biol. 146:106208. doi: 10.1016/j.biocel.2022.106208
Wong, W. R., Chen, Y. Y., Yang, S. M., Chen, Y. L., and Horng, J. T. (2005). Phosphorylation of Pi3K/Akt and Mapk/Erk in an early entry step of enterovirus 71. Life Sci. 78, 82–90. doi: 10.1016/j.lfs.2005.04.076
Wu, K. X., and Chu, J. J. (2017). Antiviral screen identifies Ev71 inhibitors and reveals camptothecin-target, Dna topoisomerase 1 as a novel Ev71 host factor. Antivir. Res. 143, 122–133. doi: 10.1016/j.antiviral.2017.04.008
Xiao, H., Li, J., Yang, X., Li, Z., Wang, Y., Rui, Y., et al. (2021). Ectopic expression of Trim25 restores rig-I expression and Ifn production reduced by multiple enteroviruses 3C (pro). Virol. Sin. 36, 1363–1374. doi: 10.1007/s12250-021-00410-x
Xu, C., Peng, Y., Zhang, Q., Xu, X. P., Kong, X. M., and Shi, W. F. (2018). Usp4 positively regulates Rlr-induced Nf-kappaB activation by targeting Traf6 for K48-linked deubiquitination and inhibits enterovirus 71 replication. Sci. Rep. 8:13418. doi: 10.1038/s41598-018-31734-6
Xue, M., Feng, T., Chen, Z., Yan, Y., Chen, Z., and Dai, J. (2022). Protein acetylation going viral: implications in antiviral immunity and viral infection. Int. J. Mol. Sci. 23:11308. doi: 10.3390/ijms231911308
Yamasaki, A., Jin, Y., and Ohsumi, Y. (2020). Mitotic phosphorylation of the Ulk complex regulates cell cycle progression. PLoS Biol. 18:e3000718. doi: 10.1371/journal.pbio.3000718
Ye, X., Zhang, H. M., Qiu, Y., Hanson, P. J., Hemida, M. G., Wei, W., et al. (2014). Coxsackievirus-induced miR-21 disrupts cardiomyocyte interactions via the downregulation of intercalated disk components. PLoS Pathog. 10:e1004070. doi: 10.1371/journal.ppat.1004070
Yu, J., Dai, Y., Fu, Y., Wang, K., Yang, Y., Li, M., et al. (2021). Cathelicidin antimicrobial peptides suppress Ev71 infection via regulating antiviral response and inhibiting viral binding. Antivir. Res. 187:105021. doi: 10.1016/j.antiviral.2021.105021
Zhang, H., Cong, H., Song, L., and Tien, P. (2014). The nuclear protein Sam68 is redistributed to the cytoplasm and is involved in Pi3K/Akt activation during Ev71 infection. Virus Res. 180, 1–11. doi: 10.1016/j.virusres.2013.11.020
Zhang, Y., Li, Y., Tang, B., and Zhang, C. Y. (2017). The strategies for identification and quantification of Sumoylation. Chem. Commun. (Camb.) 53, 6989–6998. doi: 10.1039/C7CC00901A
Zhang, F., Liu, Y., Chen, X., Dong, L., Zhou, B., Cheng, Q., et al. (2015). Rassf4 promotes Ev71 replication to accelerate the inhibition of the phosphorylation of Akt. Biochem. Biophys. Res. Commun. 458, 810–815. doi: 10.1016/j.bbrc.2015.02.035
Zhang, H., and Pelech, S. (2012). Using protein microarrays to study phosphorylation-mediated signal transduction. Semin. Cell Dev. Biol. 23, 872–882. doi: 10.1016/j.semcdb.2012.05.009
Zhang, H. M., Wang, F., Qiu, Y., Ye, X., Hanson, P., Shen, H., et al. (2016). Emodin inhibits coxsackievirus B3 replication via multiple signalling cascades leading to suppression of translation. Biochem. J. 473, 473–485. doi: 10.1042/BJ20150419
Zhang, S., Zhang, X., Bie, Y., Kong, J., Wang, A., Qiu, Y., et al. (2022). Stub1 regulates antiviral Rnai through inducing ubiquitination and degradation of dicer and Ago2 in mammals. Virol. Sin. 37, 569–580. doi: 10.1016/j.virs.2022.05.001
Zhao, B., Li, L., Tumaneng, K., Wang, C. Y., and Guan, K. L. (2010). A coordinated phosphorylation by Lats and Ck1 regulates yap stability through Scf (beta-Trcp). Genes Dev. 24, 72–85. doi: 10.1101/gad.1843810
Zhao, X., Yuan, H., Yang, H., Liu, Y., Xun, M., Li, X., et al. (2022). N-acetyltransferase 8 promotes viral replication by increasing the stability of enterovirus 71 nonstructural proteins. J. Virol. 96:e0011922. doi: 10.1128/jvi.00119-22
Zheng, Z., Li, H., Zhang, Z., Meng, J., Mao, D., Bai, B., et al. (2011). Enterovirus 71 2C protein inhibits Tnf-alpha-mediated activation of Nf-kappaB by suppressing IkappaB kinase beta phosphorylation. J. Immunol. 187, 2202–2212. doi: 10.4049/jimmunol.1100285
Zhou, L., He, X., Gao, B., and Xiong, S. (2015). Inhibition of histone deacetylase activity aggravates Coxsackievirus B3-induced myocarditis by promoting viral replication and myocardial apoptosis. J. Virol. 89, 10512–10523. doi: 10.1128/JVI.01028-15
Zhu, G., Jin, L., Sun, W., Wang, S., and Liu, N. (2022b). Proteomics of post-translational modifications in colorectal cancer: discovery of new biomarkers. Biochim. Biophys. Acta Rev. Cancer 1877:188735. doi: 10.1016/j.bbcan.2022.188735
Zhu, Q., Liang, P., Chu, C., Zhang, A., and Zhou, W. (2022c). Protein sumoylation in normal and cancer stem cells. Front. Mol. Biosci. 9:1095142. doi: 10.3389/fmolb.2022.1095142
Zhu, F., Yang, S., Meng, F., Zheng, Y., Ku, X., Luo, C., et al. (2022a). Leveraging protein dynamics to identify functional phosphorylation sites using deep learning models. J. Chem. Inf. Model. 62, 3331–3345. doi: 10.1021/acs.jcim.2c00484
Zolg, D. P., Wilhelm, M., Schmidt, T., Medard, G., Zerweck, J., Knaute, T., et al. (2018). ProteomeTools: systematic characterization of 21 Post-translational protein modifications by liquid chromatography tandem mass spectrometry (Lc-Ms/Ms) using synthetic peptides. Mol. Cell. Proteomics 17, 1850–1863. doi: 10.1074/mcp.TIR118.000783
Keywords: enterovirus infection, post-translation modification, host factors, pathogenesis, enterovirus life cycle
Citation: Zhao X, Hu Y, Zhao J, Liu Y, Ma X, Chen H and Xing Y (2024) Role of protein Post-translational modifications in enterovirus infection. Front. Microbiol. 15:1341599. doi: 10.3389/fmicb.2024.1341599
Edited by:
Leiliang Zhang, Shandong First Medical University and Shandong Academy of Medical Sciences, ChinaReviewed by:
Girish Patil, Oklahoma State University, United StatesMir Mubashir Khalid, Gladstone Institutes, United States
Fuminori Tokunaga, Osaka Metropolitan University, Japan
Copyright © 2024 Zhao, Hu, Zhao, Liu, Ma, Chen and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohui Zhao, eGlhb2h1aXpoYW9AcWh1LmVkdS5jbg==
 Xiaohui Zhao
Xiaohui Zhao Yibo Hu
Yibo Hu Jun Zhao
Jun Zhao Yan Liu3
Yan Liu3 Xueman Ma
Xueman Ma