- 1College of Plant Protection and College of Bioscience and Biotechnology, Hunan Agricultural University, Changsha, China
- 2Hunan Tobacco Company, Changsha, China
- 3Institute of Plant Protection, Hunan Academy of Agricultural Sciences, Changsha, China
Background: Tobacco mosaic virus (TMV) is one famous plant virus responsible for substantial economic losses worldwide. However, the roles of bacterial communities in response to TMV in the tobacco rhizosphere remain unclear.
Methods: We explored the soil physicochemical properties and bacterial community succession of the healthy (YTH) and diseased (YTD) plants with TMV infection by 16S rRNA gene sequencing and bioinformatics analysis.
Results: We found that soil pH in the YTD group was significantly lower than in the YTH group, and the soil available nutrients were substantially higher. The bacterial community analysis found that the diversity and structure significantly differed post-TMV disease onset. With TMV inoculated, the alpha diversity of the bacterial community in the YTD was markedly higher than that in the YTH group at the early stage. However, the alpha diversity in the YTD group subsequently decreased to lower than in the YTH group. The early bacterial structure of healthy plants exhibited higher susceptibility to TMV infection, whereas, in the subsequent stages, there was an enrichment of beneficial bacterial (e.g., Ramlibacter, Sphingomonas, Streptomyces, and Niastella) and enhanced energy metabolism and nucleotide metabolism in bacteria.
Conclusion: The initial soil bacterial community exhibited susceptibility to TMV infection, which might contribute to strengthening resistance of Tobacco to TMV.
Introduction
Tobacco mosaic virus (TMV) is the first virus to be discovered and identified, a single-stranded RNA (Creager et al., 1999). The optical temperature for TMV proliferation is 25–30°C, and its reproduction is inhibited at above 37°C. TMV can infect more than 400 species of plants in 36 families, such as Cruciferae, Solanaceae, Compositae, and Chenopodium (Scholthof et al., 2011). It is also a common tobacco virus disease and can remain in the soil to become the primary infection source for the next year after the winter (Hossain et al., 2022). Field infection is mainly via sap transmission. The gentle friction between the sick and healthy leaves causes damage, and the virus can invade through micro-wounds. Secondly, human activities and chewing mouth insects (e.g., locusts and tobacco green worms) can spread the virus. During tobacco cultivation, the incidence rate of TMV ranges from 20 to 30%, with some instances reaching as high as 60–80%, resulting in significant economic losses for the industry (Islam et al., 2018).
The commonly used methods mainly include chemical pesticides and transgenic approaches, which cannot eliminate TMV load in infected tobacco plants and whose efficacy is still limited in the field. Microbes (e.g., Streptomyces, Pseudomonas, and Bacillus) have been considered potential alternatives for chemical pesticides to inhibit plant pathogens, including fungal, bacterial, and viral pathogens (Kim et al., 2022; Ren et al., 2022; Shi et al., 2022). Pseudomonas fluorescens could inhibit the activity of Cucumber mosaic virus, Tomato mottle virus, and Tobacco necrosis virus (Maurhofer et al., 1998; Murphy et al., 2000). Azotobacter vinelandii, and Bacillus subtilis could inhibit Potato virus, Potato leaf roll virus, and Potato virus Y in Solanum tuberosum (Beris et al., 2018). The study of Tollenaere et al. (2017) found that the bacterium Xanthomonas oryzae decreased the relative estimate of rice yellow mottle virus load by about 50%. As a result, it is essential to note that regulating bacterial communities and increasing beneficial bacteria could influence plant resistance to TMV.
Currently, numerous bacterial strains have been employed to prevent TMV infection. For instance, Damayanti and Katerina (2008) found that B. cereus (I–35) and Stenotrophomonas sp. (II-10) can reduce the infection of TMV in hot pepper. Lian et al. (2011) found that Rhodopseudomonas palustris GJ-22 could inhibit TMV in Nicotiana tabacum. In addition, P. chlororaphis O6N was resistant to TMV in N. tabacum cv Xanthi-nc (Park et al., 2012).
Many studies have focused on comparing the differences in soil microbial communities between diseased and healthy plants (Liang et al., 2021; Hossain et al., 2022; Tang et al., 2023). However, there is still limited research on the response mechanism of soil bacterial communities and plants to the pathogen attack (Lu et al., 2022; Kuang et al., 2023). In this study, we explored the succession of soil bacterial communities of the healthy and diseased plants after N. benthamiana inoculated with TMV to indicate the role of soil bacterial communities in controlling TMV and the potential beneficial microbes.
Materials and methods
Pot experiment and virus inoculation
The pot soils were obtained from the 0 to 20 cm depth layer at the Yunyuan test site of Hunan Agricultural University in Changsha, China. The soils were naturally air-dried and passed through a 5 mm sieve for the pot experiment. The properties of pot soil were as follows: the pH value was 5.32, and the contents of organic matter (OM), total nitrogen (TN), nitrate nitrogen (NO3–-N), ammonium nitrogen (NH4+-N), total phosphorus (TP), available phosphorus (AP), total potassium (TK), available potassium (AK) were 97.500, 3.790, 0.034, 0.108, 1.269, 0.046, 15.200, and 0.214 g/kg, respectively.
The healthy N. benthamiana seedlings (5–7 true leaves) were transplanted into the 40 pots (15.5 cm × 14 cm) containing 1.0 kg soil. One plant was transplanted in each pot. Fifteen days after transplantation, we rinsed 80 ml TMV suspensions into each plant root. TMV suspensions (wt: vol, 1:30) were prepared by abrasing infected tobacco leaves with carborundum abrasive powder and phosphate buffer (pH 7.0). No pest or disease controls were applied during the experiment. Soil samples (5.0 g) were collected from the vicinity of the roots (2–4 cm) on days 7, 14, 21, and 28 after viral exposure to the plants. The collected samples were then stored at −80°C. On day 28, the incidence TMV was assessed in accordance with the Chinese national standard (GB/T23222-2008) and molecular detection of tobacco leaves using a specific TMV primer (Kumar et al., 2011). After inoculating TMV, there were seven among 40 tobacco plants to be healthy, and 33 were diseased. The 40 plants were classified into healthy (YTH) and diseased (YTD) groups based on disease investigation results. Soil samples from seven healthy tobacco plants were pairwise mixed to form three replicates for the YTH group on days 7, 14, 21, and 28, respectively, and one plant soil sample was obsoleted. Similarly, soil samples from 33 diseased tobacco plants were equally divided into three parts, each of which was mixed to form three replicates for the YTD group. Finally, each group had three replicated soil samples on days 7, 14, 21, and 28, respectively, for the subsequent detection of the soil bacterial community succession and soil physicochemical properties.
Measurement of soil physicochemical properties
The soil physicochemical properties were measured as described in our previous study (Wang et al., 2024). The soil pH was determined by a pH meter. Soil organic matter content was determined by the potassium dichromate oxidation method. The soil AP content was determined by the molybdenum antimony resistance colorimetric method, the soil AK content was determined by flame photometer, the content of NH4+-N and NO3–-N were determined by ultraviolet spectrophotometry, and the soil AP content was determined by sulfuric acid-accelerator digestion. The TN content was determined by the Kjeller method, the TP content was determined by NaOH alkali melting and molybdenum antimony resistance spectrophotometry, and soil TK content was determined by NaOH alkali melting and flame photometer.
Soil DNA extraction, amplification, 16S rRNA gene sequencing, and data processing
DNA extraction, amplification, 16S rRNA gene amplicon sequencing, and data processing were described in detail in our previous study (Xiao et al., 2018). The PowerSoil DNA Isolation Kit (TIANGEN BIOTECH, Beijing, China) was used to isolate DNA from the soils. DNA extracts were purified by electrophoresis on a 1% agarose gel using a DNA gel extraction kit (OMEGA, USA) and the concentration was measured by a Nanodrop 2000 microspectrophotometer (NanoDrop Technologies, Wilmington, NC, USA). The 16S rRNA gene was amplified with primer pair 338F (5′-CTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). PCR products were purified, quantified, and homogenized to construct the library. The products were sequenced on an Illumina Novaseq machine (Illumina, San Diego, CA, USA). The raw data of 16S rRNA gene sequences was deposited in the Sequence Read Archive (SRA) of NCBI under the accession number PRJNA1021354.
Pair-end reads were processed using Trimmomatic v0.33 to detect and remove the sequences with a quality score below 20. Sequences were denoised and chimera filtered with the UCHIME v4.2 to obtain the effective reads, which were further clustered into operational taxonomic units (OTUs) using a 97% identity threshold. The taxonomic assignment used an RDP classifier with a minimal 50% confidence estimate.
Molecular ecological network construction and characterization
Random matrix theory (RMT)-based approaches were used for network construction (Xiao et al., 2018), hub and connector OTU identification and the topological property were determined with a similar threshold (0.96). OTUs, presented in 12 out of 12 replicates, were used for network analysis to ensure data reliability. Various network properties were characterized, such as average degree, average path distance, average clustering coefficient, and modularity index. The network modules were generated using rapid greedy modularity optimization. The experimental data used for constructing phylogenetic molecular ecological networks (pMENs) were based on 16S rRNA gene sequences, and Gephi 0.9.2 software was used to visualize network graphs. The pMENs were constructed separately based on sequencing data of two treatments to reveal the differences between the soil bacterial network interactions of healthy and diseased plants.
Functional profiling
Prior to functional gene prediction using PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states) described by Langille et al. (2013), the detected OTUs were reclassified using the GREENGENES reference database. Subsequently, PICRUSt uses 16S rRNA genes to infer metagenome gene functional content from phylogenetic information. The predictions are precalculated for genes in databases, including the Kyoto Encyclopedia of Genes and Genomes (KEGG). The input data were first normalized by copy number by dividing each OTU by the known 16S rRNA copy number abundance before metagenome predictions and subsequent collapse into functional pathways. The output of PICRUSt consists of a table of functional gene counts as KEGG orthologs (KOs). The Nearest Sequenced Taxon Index (NSTI) value was used to validate the reliability of predicted metagenomes and functional pathways.
Statistical analysis
The community diversity was assessed using the number of OTUs, Shannon diversity index (H’), Chao 1, and Simpson evenness index. Differences in diversity and relative abundances of bacterial composition based on Tukey’s test were conducted by a one-way analysis of variance (ANOVA) (Deng et al., 2012). Principal Coordinates Analysis (PCoA) was conducted to analyze the bacterial community structure. LDA Effect Size (LEfse) analysis was conducted to find the species with significant differences between groups (Barberán et al., 2014; Wagg et al., 2014; Cui et al., 2016). All analyses were performed using R v.3.6.3 and on Tutools.1
Results
TMV incidence and soil physicochemical properties
The incidence of N. benthamiana in the YTD and YTH groups at different stages is shown in Figure 1. Compared with healthy tobacco in the YTH group all the time, tobacco leaves curled on day 7 and then gradually became severe in the YTD group after TMV inoculation. According to the results of soil physicochemical properties on day 28 (Figure 2), we found the pH value (YTD: 5.01; YTH: 5.25) was significantly lower, and the contents of TN (YTD: 3.51 g/kg; YTH: 3.43 g/kg), NO3–-N (YTD: 0.045 g/kg; YTH: 0.023 g/kg), AP (YTD: 0.054 g/kg; YTH: 0.047 g/kg), and AK (YTD: 0.147 g/kg; YTH: 0.126 g/kg) were significantly higher in the YTD group than in the YTH group. These results suggested that tobacco plants grew better in the YTH group than in the YTD group.
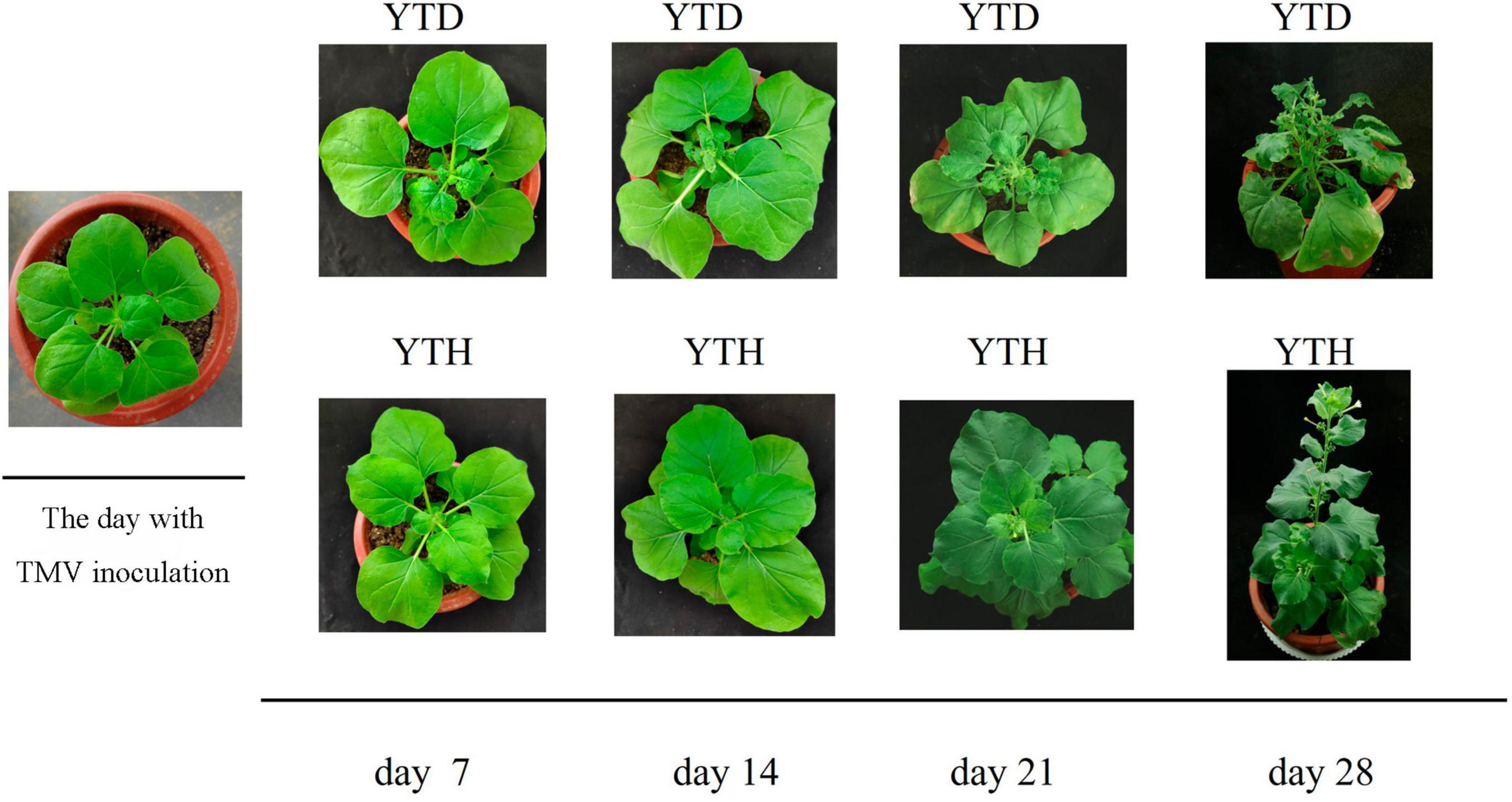
Figure 1. The incidence of N. benthamiana in YTD and YTH groups at different stages (day 7, 14, 21, and 28).
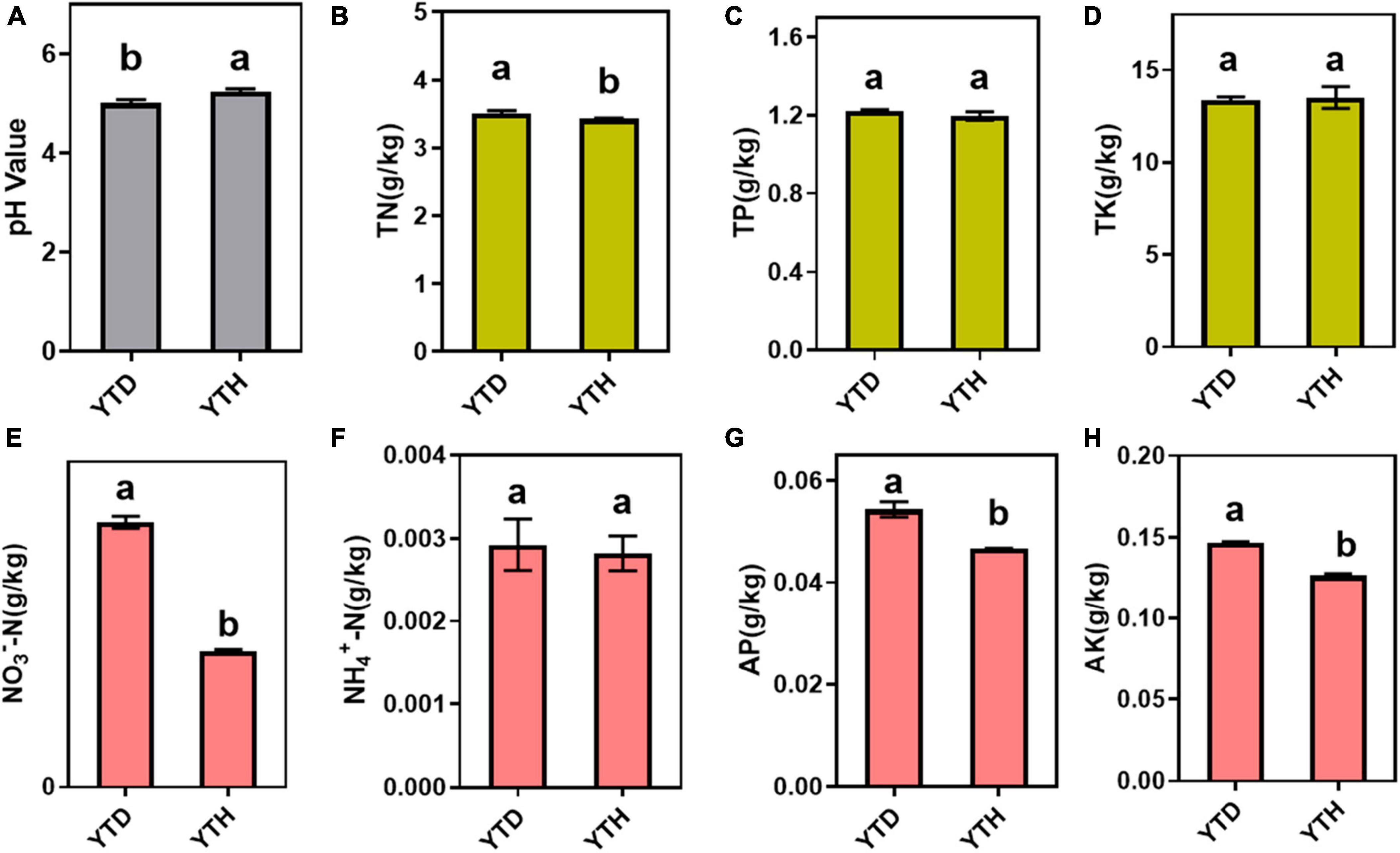
Figure 2. Soil physicochemical properties, including pH value (A) and the total contents of total nitrogen (B), total phosphorus (C), total potassium (D), NO3–-N (E), NH4+-N (F), available phosphorus (G), and available potassium (H). Differences in soil physicochemical properties based on Tukey’s test were conducted by ANOVA (n = 3). Different lowercase letters represent significant differences between groups at the 5% level.
Overview of soil bacterial communities
A total of 1,518,787 quality sequences were obtained, with between 60,756 and 65,938 sequences per sample. After clustering, 1473 OTUs were detected in all samples. Among them (Figure 3A), the number of core OTUs was the highest on day 21 (1382) and the lowest on day 7 (1335). The number of unique OTU in the YTD group was higher on day 7 and more down on days 14, 21, and 28 than that in the YTH group (Figure 3B). Heatmap analysis of the bacterial community showed that samples in YTH on day 7 were clustered with the samples in YTD on day 14, 21, and 28. In contrast, samples in YTD on day 7 were clustered with the samples in YTH on days 14, 21, and 28 (Figure 3C).
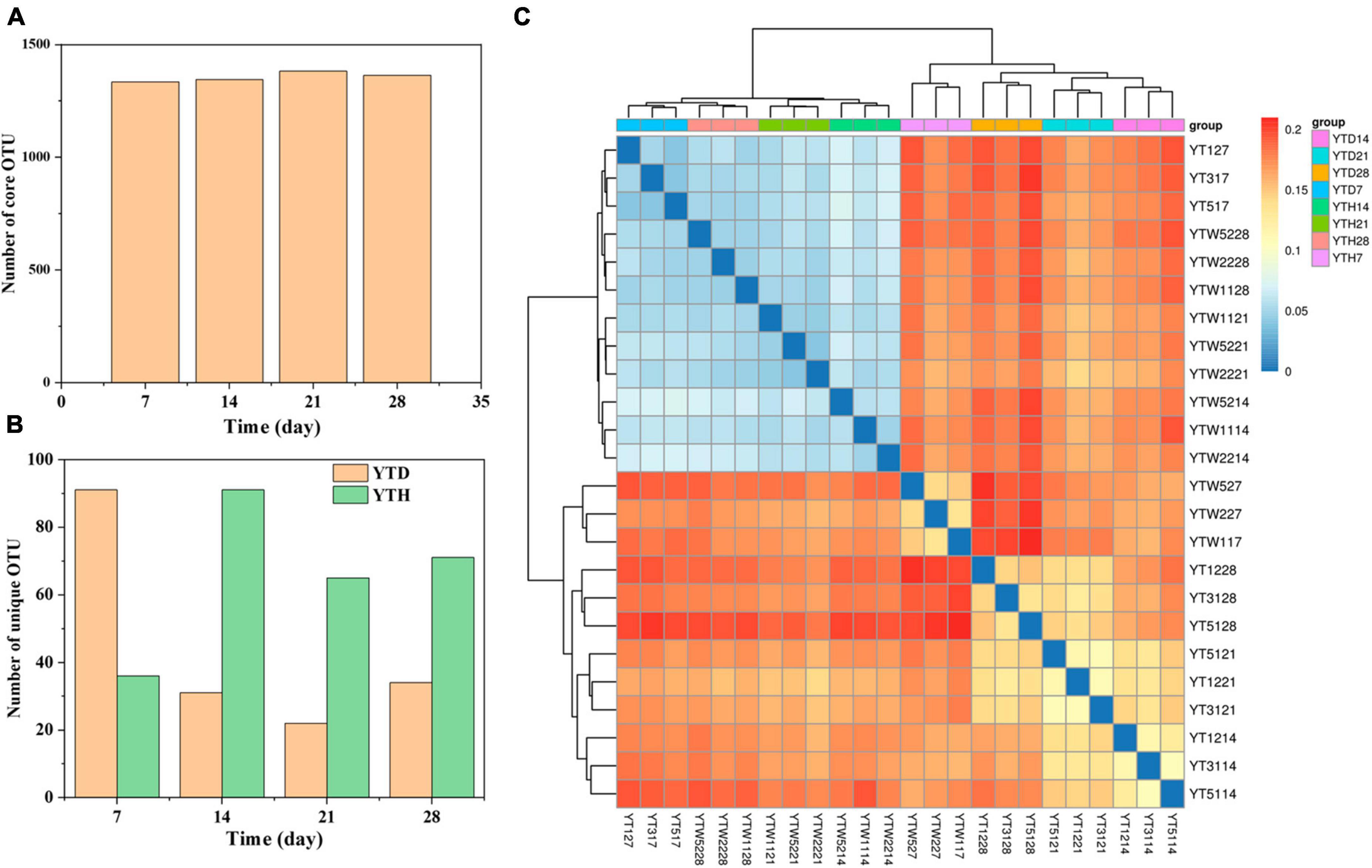
Figure 3. The number of core OTU (A) and unique OTU (B) in the YTD and YTH groups, and (C) heatmap analysis used Bray-Curtis dissimilarity metrics of bacterial community at OTU level. The number of core and unique OTU were statistic based on the 12 samples of each group.
The α-diversity indices, including the number of OTUs, Chao 1, Shannon diversity, and Simpson evenness, were shown in Figures 4A–D. After inoculating TMV, the α-diversity indices were significantly higher in the YTD group than in the YTH group on day 7. However, the diversity indices were increased constantly in the YTH group and decreased in the YTD group, resulting in the diversity indices being significantly higher in the YTH group than those in the YTD group on days 14, 21, and 28.
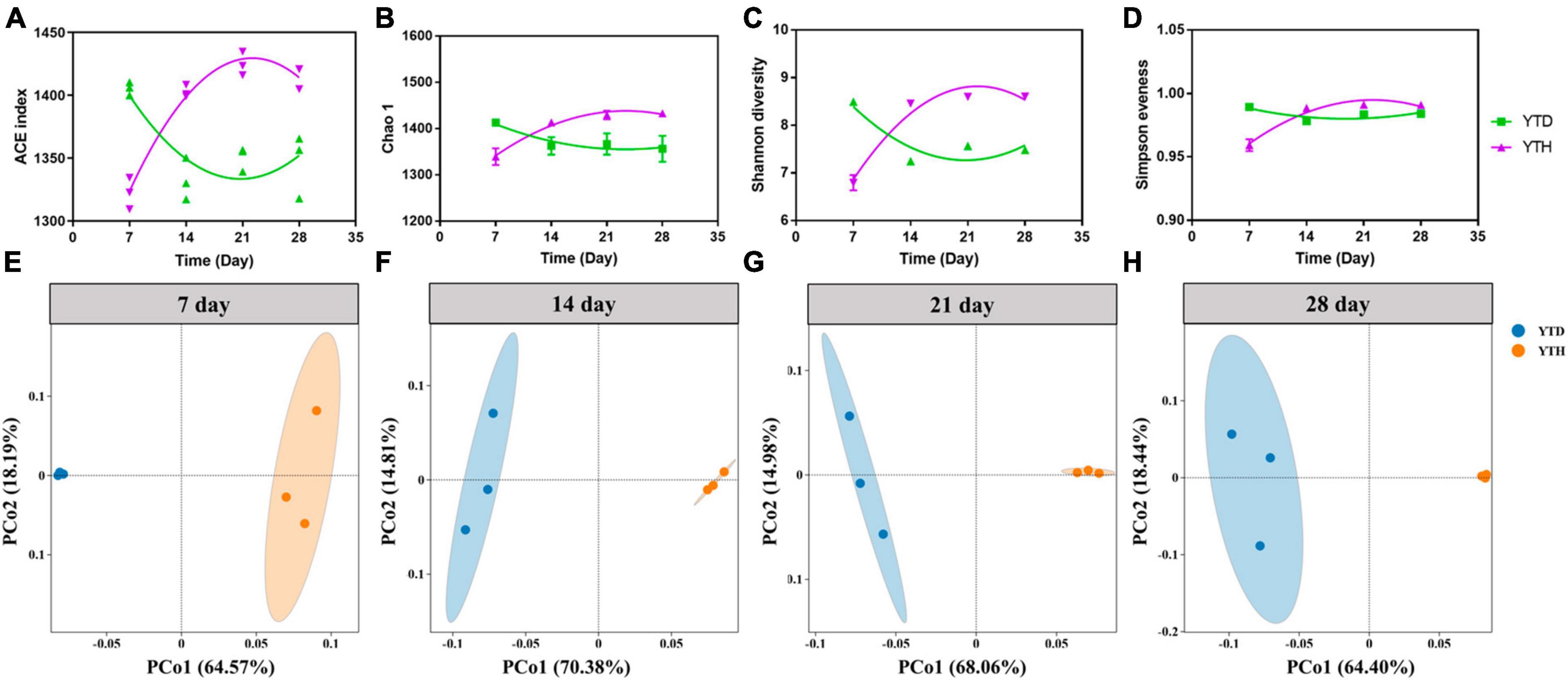
Figure 4. The change of diversity indices [(A) number of OTU; (B) Chao 1; (C) Shannon diversity; (D) Simpson evenness] and the ordination plots of all samples for the community structure analyzed by PCoA on day 5 (E), 14 (F), 21 (G), and 28 (H), respectively. Non-linear fitting was conducted in Graphpad software (version 8.0) to analysis the change trend of diversity indices.
Principal Coordinates Analysis results showed that the YTH samples were segregated from the YTD samples during the whole planting stage (Figures 4E–H). Additionally, samples within the YTD group were clustered more closely on day 7 than on other days, and it was interesting that samples within the YTH group were clustered more closely on days 14, 21, and 28 than on day 7.
The compositions of soil bacterial communities
After inoculating TMV, soil bacterial community compositions were explored and analyzed at four-time points. At the phylum level (Figure 5A), the bacterial composition consisted of 21 phyla. The dominant phyla (top 6) included Proteobacteria (50.34–70.99%), Acidobacteria (4.59–15.06%), Gemmatimonadetes (2.43–13.31%), Bacteroidetes (3.74–8.78%), Actinobacteria (2.41–10.67%) and Chloroflexi (2.59–5.98%), etc. At the family level (Figure 5B), 217 families were found among all soil samples, and the dominant families (top 6) were Burkholderiaceae (7.08–51.31%), Sphingomonadaceae (1.71–14.84%), Gemmatimonadaceae (2.26–13.10%), Caulobacteraceae (2.37–8.63%), Xanthobacteraceae (1.78–6.67%), Rhodanobacteraceae (1.21–5.67%), etc. In addition, all 1473 OTUs were affiliated to 345 genera, and the dominant genera (relative abundance >1%) included Burkholderia, Massilia, Sphingomonas, Bordetella, Mucilaginibacter, Gemmatimonas, Bacillus, Pseudolabrys, Flavisolibacter, Ramlibacter, Phenylobacterium, Dyella, Rhodanobacter, Gemmatirosa, Pandoraea, and Ellin6067.
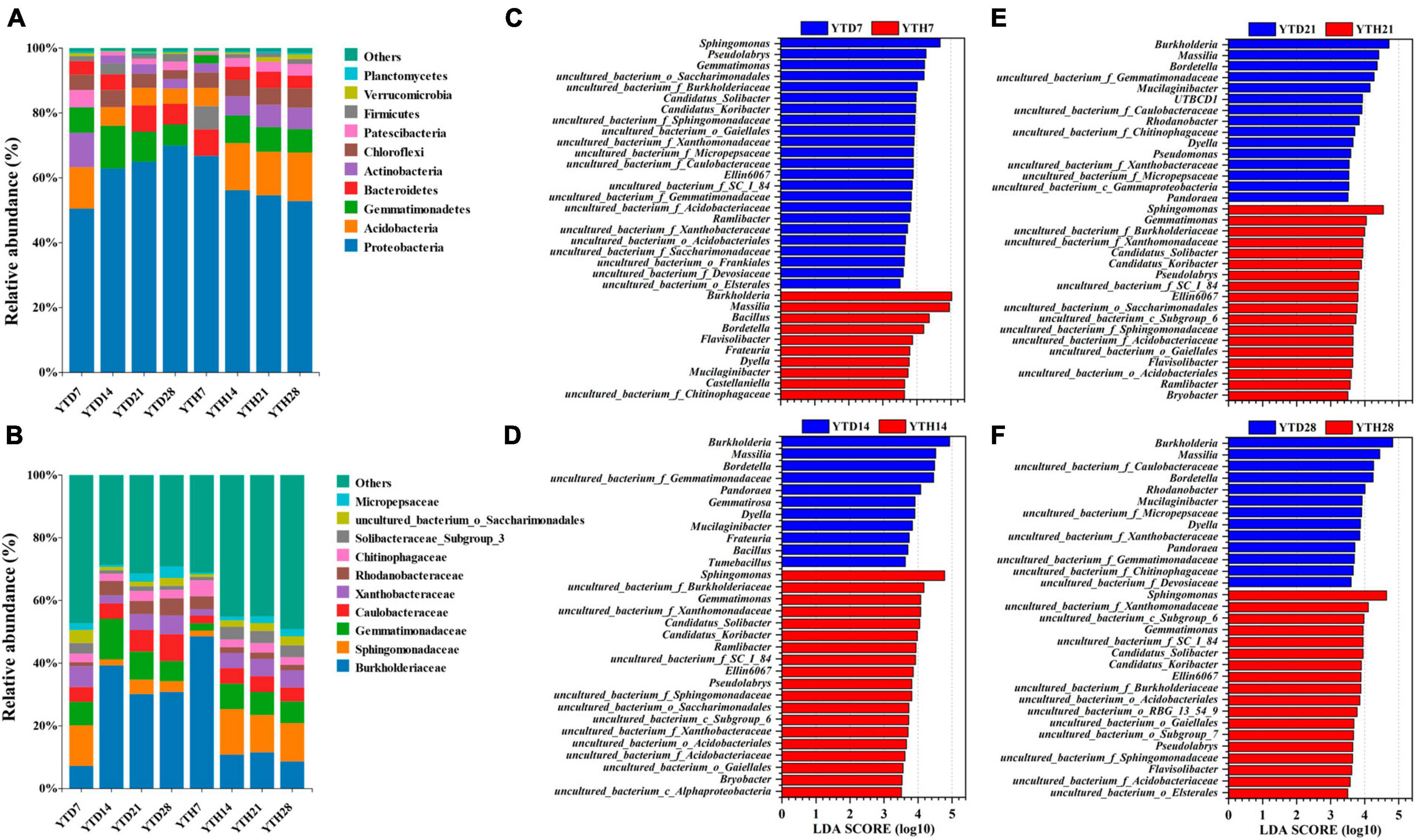
Figure 5. The compositions of bacterial communities in phylum (A), family level (B), and the significant genera between YTD and YTH groups according to LEfSe analysis on day 7 (C), 14 (D), 21 (E), and 28 (F), respectively.
To further explore the differences between the soil bacterial community compositions of healthy and diseased plants, LEfse analysis at the genus level was conducted, and LDA analysis showed the main genera with significant differences (Figures 5C–F). The findings were as follows: on day 7, the principal genera in YTD were Sphingomonas, Pseudolabrys, and Gemmatimonas, and in YTH were Burkholderia, Massilia, and Bacillus; on day 14, 21, and 28, it was contrary that the main genera in YTD were Burkholderia, Massilia, Bordetella and Dyella, and in YTH were Sphingomonas, Gemmatimonas, Ellin6067 and Pseudolabrys.
Network interactions of bacterial communities in healthy and diseased soil
We constructed bacterial networks to describe the symbiotic interactions at OTU level to better understand the interrelationships of soil bacterial communities in the healthy and diseased plants after inoculating TMV. The network nodes were mainly divided into Proteobacteria, Acidobacteria, Bacteroidetes, Actinobacteria, and Gemmatimonadetes (Figures 6A, B). The relative abundances of total nodes were about 63.44 and 69.38%, respectively, in the YTH and YTD groups. Further analysis found that the proportion of Sphingomonas (13.97%), Burkholderia (10.29%), and Massilia (9.11%) were the three genera with the enormous proportions in YTH group, which were different from that of Burkholderia (18.22%), Massilia (8.19%) and uncultured Gemmatimonadaceae (6.80%) in the YTD group.
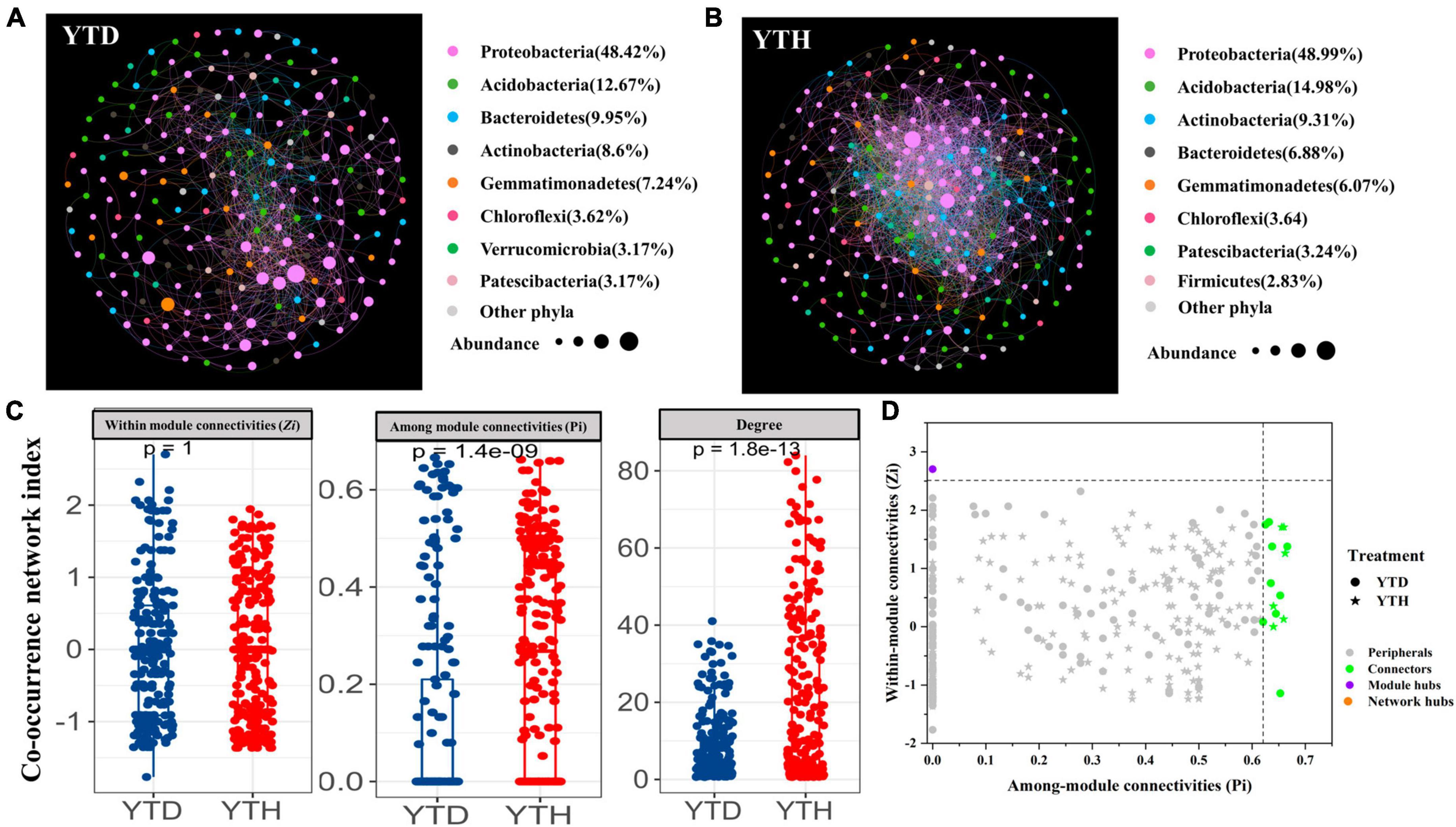
Figure 6. Molecular ecological networks in YTD (A), and YTH (B) groups, the co-occurrence network index (C), and the Z-P plot showing the distribution of OTUs based on their topological roles (D). The color of the circle represented the module in the network interaction. The topological role of each OTU was determined according to the scatter plot of within-module connectivity (Zi) and among-module connectivity (Pi).
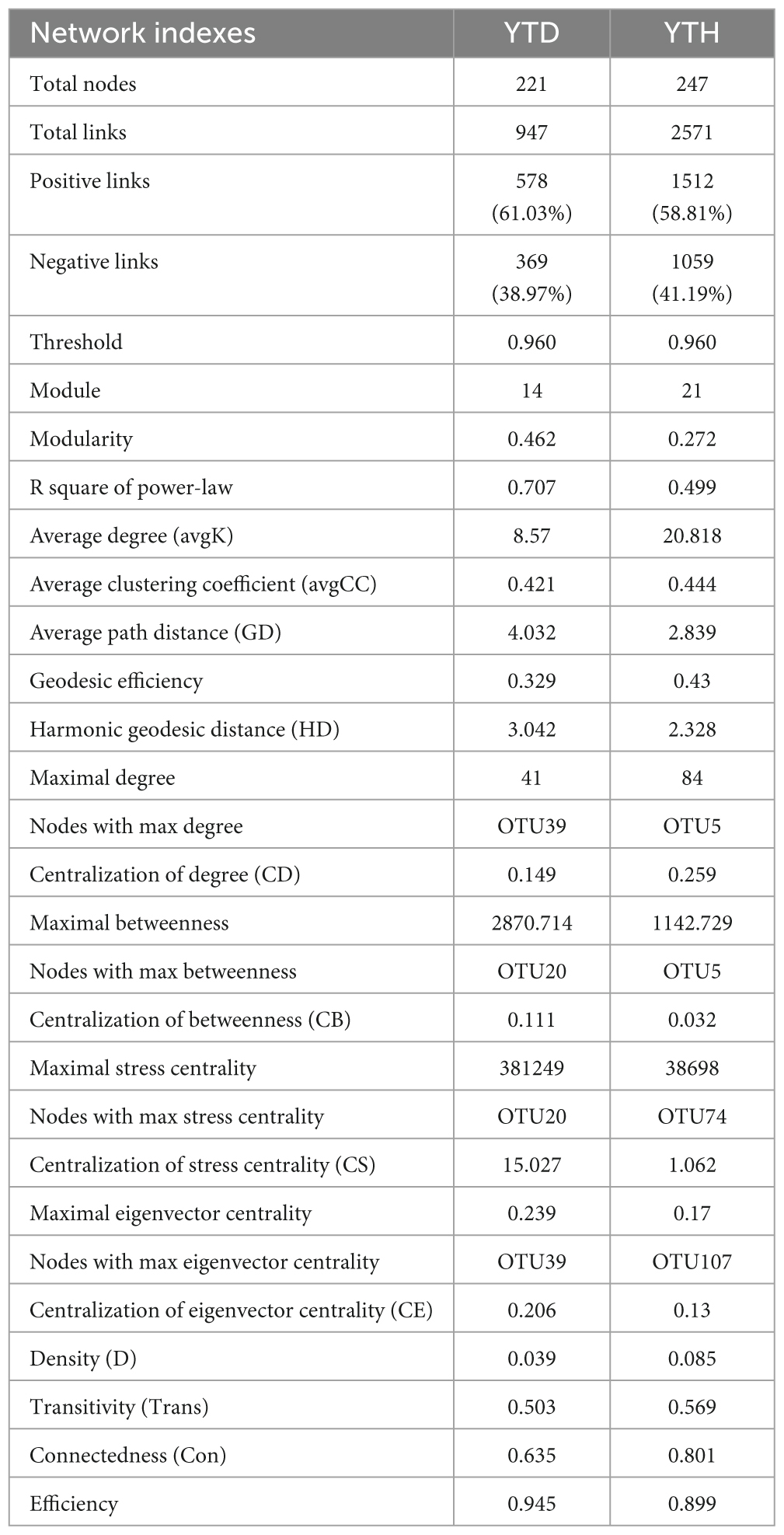
Major topological parameters of empirical MENs of bacterial communities in the two groups revealed that the soil bacterial community network of the YTD group was simpler than that of the YTH group (Table 1). With the same threshold (0.960), their correlations were more than 0.500, indicating that the degree distributions in both constructed molecular ecological networks fitted the power-law model well. There were more nodes and links in the YTH group (247 nodes and 2571 links) than in the YTD (221 and 947) group. The healthy network significantly increased the Zi, Pi, and degree, which indicated the creation of more intricate network patterns (Figure 6C). It showed that TMV might disrupt interactions of soil bacterial communities, and simpler eco-network showed weaker resistance to TMV, causing disease in the tobacco plant.
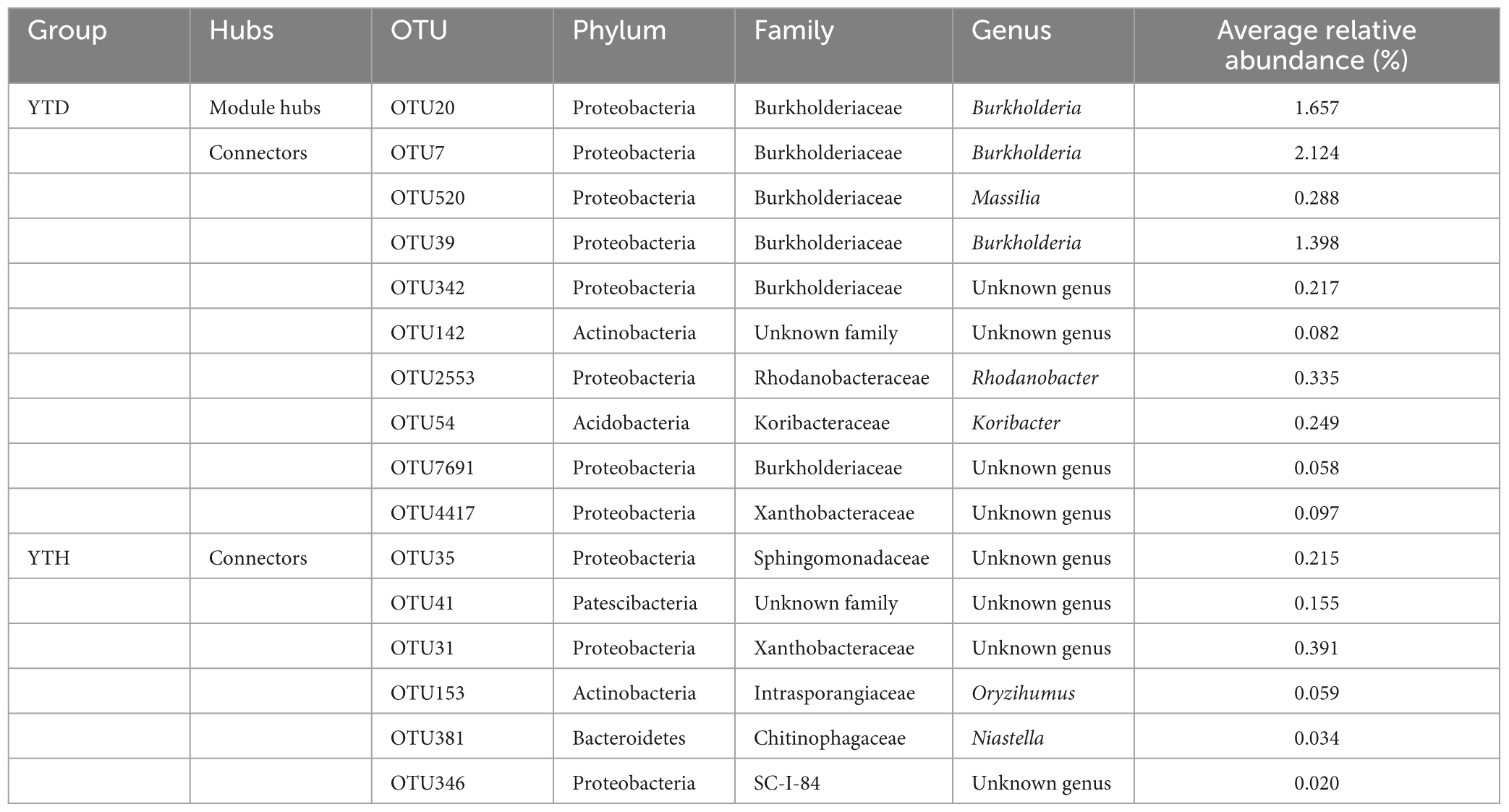
We classified nodes into four categories, including peripherals, connectors, module hubs, and network hubs. Connectors, module hubs, and network hubs are commonly considered to be the keystone in the co-occurrence network. The results found that there were also more module hubs (1 hub) and connectors (9 connectors) in the YTD group than those in the YTH group (Figure 6D). The keystone genera in the YTH group were classified as Oryzihumus, Niastella, and other unknown genera. The keystone families were classified as Sphingomonadaceae, Xanthobacteraceae, Intrasporangiaceae, Chitinophagaceae, and SC-I-84 (Table 2). However, with the TMV outburst, the keystone genera in the YTD group were replaced by Burkholderia, Massilia, Rhodanobacter, and Koribacter, as well as other unknown genera, and the keystone families were Burkholderiaceae, Xanthobacteraceae, Rhodanobacteraceae and Koribacteraceae (Table 2).
The succession of key genera or families with inoculating TMV
Based on the above analysis, we explored the succession of sixteen possible key genera (Figure 7). Among these genera, four including Bordetella, Burkholderia, Massilia, and Dyella, showed a similar succession tendency, that is, the relative abundances were higher in YTD than those in YTH group on day 7, while were decreased gradually in YTD and were progressively increased in YTH, resulting in that higher in YTH on the later time points. The succession of Nine genera, including Ramlibacter, Elin6067, Sphingomonas, Pseudolabrys, Gemmatimonas, Koribacter, Niastella, Oryzihumus, and Streptomyces, was an opposite tendency with the above four genera. The relative abundance of Rhodanobacter gradually increased in the YTH group and changed a little in YTD. The relative abundance of Bacillus decreased quickly on day 14 in the YTD group and changed slightly in YTH. The relative abundance of Flavisolibacter gradually reduced in both groups and was higher in the YTD group.
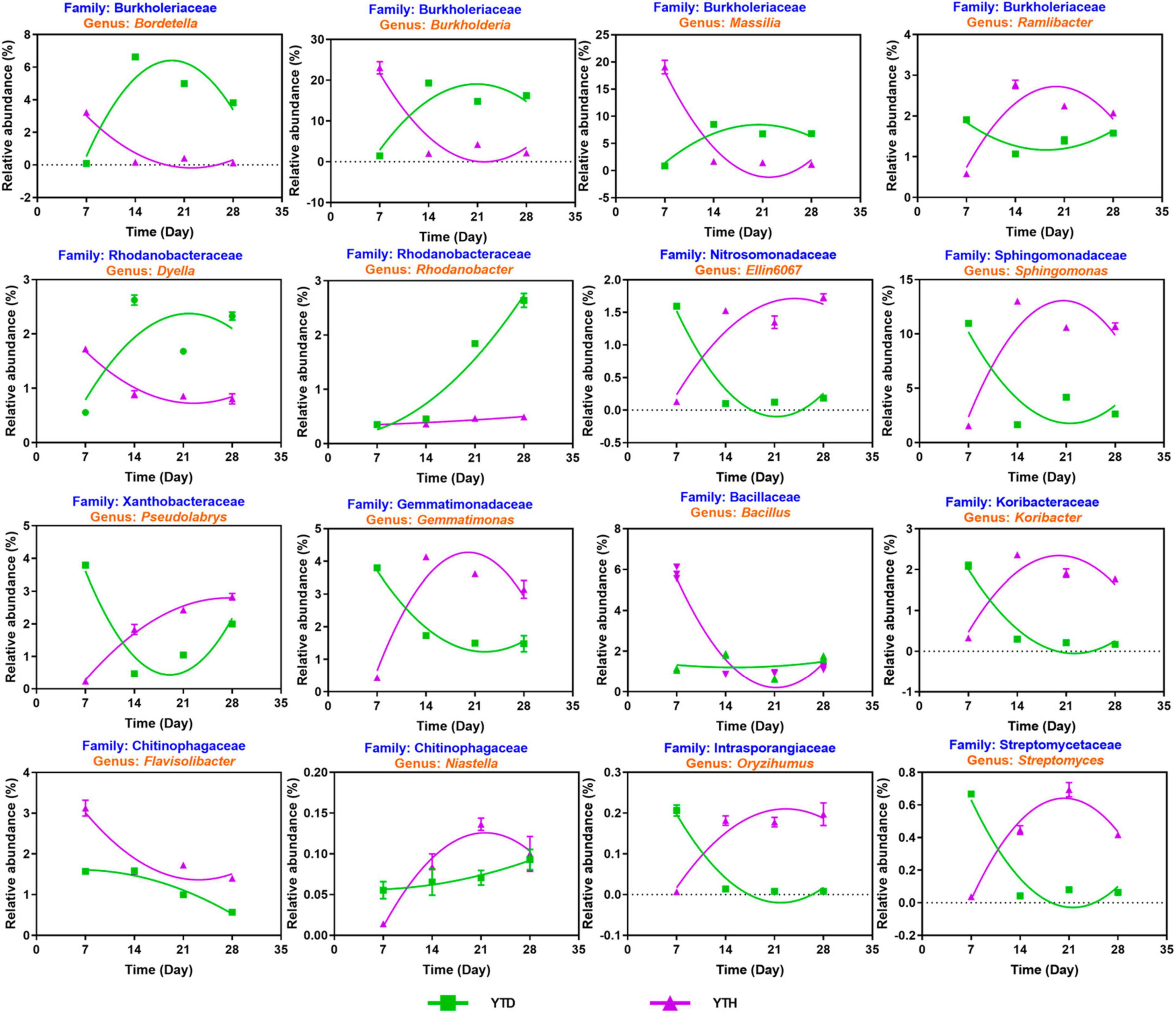
Figure 7. The succession of 16 key genera after inoculating TMV in the YTD and YTH groups by non-linear fitting in Graphpad software (version 8.0).
Metabolic diversity
Based on the 16S rRNA sequences, potential functions were predicted to explore the differences in soil bacterial functional genes between the YTH and YTD groups (Figure 8). It indicated that the relative abundance of genes changed considerably in the early stage (day 7 and 14) after TMV inoculated. The relative abundances of genes related to Transcription, Translation, environmental adaption, and metabolism (nucleotide, cofactors and vitamins, Glycan biosynthesis, other amino and other secondary metabolites) showed higher in the YTH group than that in the YTD group on day 7 while it was on the contrary on day 14. The relative abundances of cell mobility, transport and catabolism, membrane transport, and metabolism (amino acid and xenobiotics biodegradation) showed higher in the YTD group than in the YTH group on day 7, while it was on the contrary on day 14.
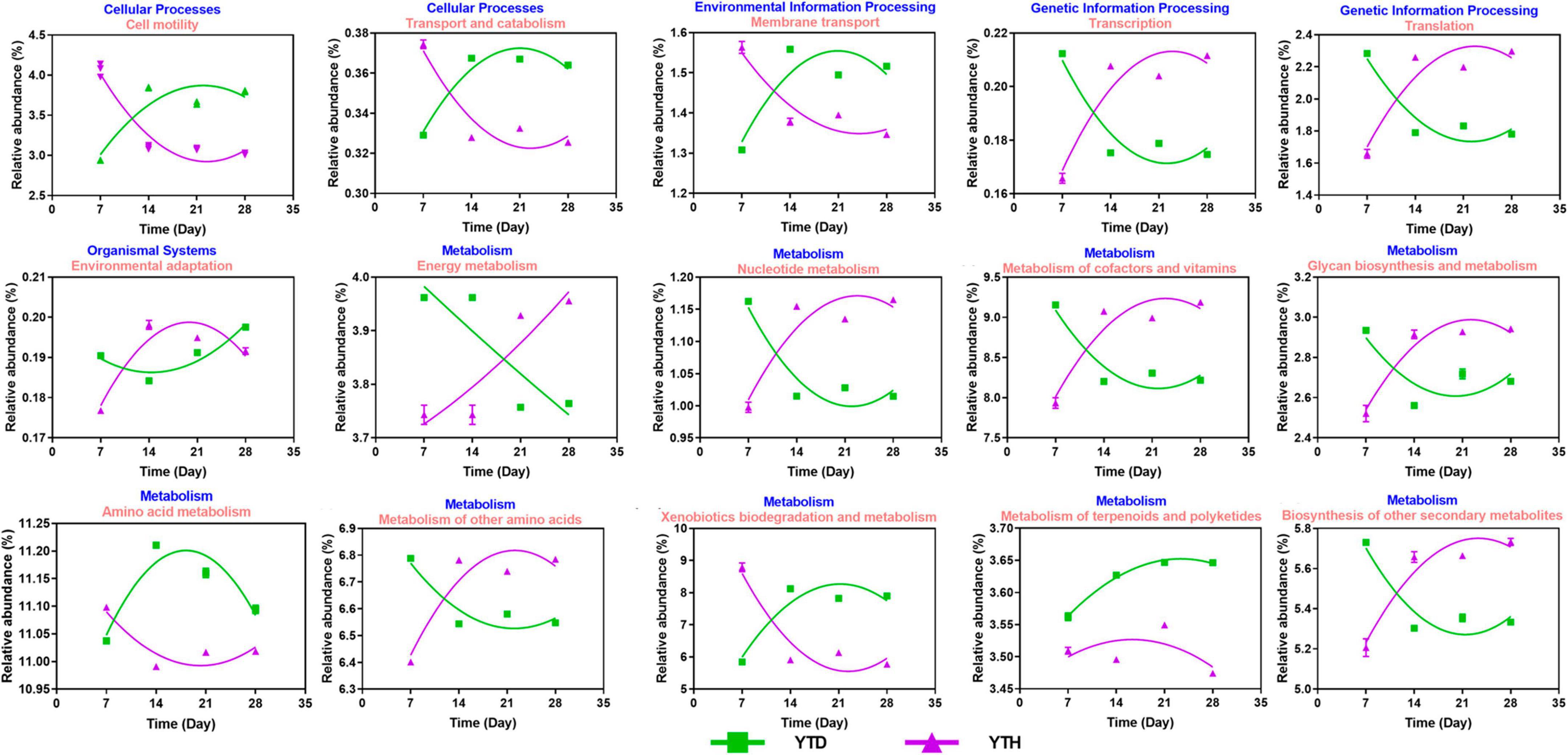
Figure 8. The change tendency of predicted metabolic pathways after inoculating TMV in the YTD and YTH groups by non-linear fitting in Graphpad software (version 8.0).
Discussion
Previous studies focused on isolating and applying beneficial microbes in controlling plant pathogens (Dunne et al., 1997; Liu et al., 2007; Shanks et al., 2012; Han et al., 2016). However, it is also essential to understand the response of plants and their soil microbial community when faced with pathogen infection. Research on plant-microbe interactions revealed that plants can shape their rhizosphere microbiome. Recent advances in pathogen or insect attacks show that plants can recruit protective microorganisms and enhance microbial activity to suppress pathogens (Berendsen et al., 2012; Stringlis et al., 2018). In this study, we studied which bacteria could be recruited by N. benthamiana and explored the possible mechanism of plant-bacteria interaction to respond to TMV attack.
This study found that there was significant variation in the bacteria community between the two groups of treatments. The network has become more complex in YTH, which is conducive to strengthening the stability and function of bacterial communities (Hernandez et al., 2021). At the same time, the proportion of beneficial bacteria (e.g., Ramlibacter, Sphingomonas, Streptomyces, and Niastella) in YTH shows an increasing trend compared to YTD. Ramlibacter and Sphingomonas can produce extracellular polysaccharides, which are beneficial for activating defense-related genes and enhancing the activity of defense-related enzymes to enhance plant disease resistance (Meneghine et al., 2017; Canwei et al., 2020; Jivkova et al., 2022). Streptomyces can improve the activity of plant antioxidant enzymes (SOD, CAT, POD, and MDA), assisting plants in resisting viral infections (Chen et al., 2022). Ara et al. (2012) found streptomyces derived from various bioactive compounds to effectively minimize the TMV local lesions on the leaves of weed plants. Niastella can promote plant growth and defense against pathogens (Cuartero et al., 2022). Overall, the beneficial microorganisms were consistent with the previous studies (Manjunatha et al., 2022).
In addition, most metabolic activities of bacteria in the two groups of treatments also showed an opposite variation trend. The enhancement of the genetic information process (Transcription and translation) and metabolism (nucleotide, glycan biosynthesis, and other amino acids) were crucial for bacterial growth, reproduction, and environment adaptability in YTH. Glycan and its compounds, e.g., mannosyl erythritol lipids (Cortés-Sánchez et al., 2013) and glycoprotein (Zhang et al., 2015), displayed promising adequate substitutes for biocontrol agents for many applications in agricultural production. Many studies found that amino acid metabolic pathways could regulate plant immunity (Zeier, 2013). Proline metabolism is involved in the oxidative burst and the hypersensitive response associated with pathogen recognition (Chen et al., 2011; Zeier, 2013). Asp-derived pyridine nucleotides influence invasion immunity by modulating the crosstalk of salicylic acid- and jasmonic acid-regulated defense pathways (Zhang and Mou, 2009).
Remarkably, we discovered that the bacteria community diversity, the proportion of beneficial bacteria, and bacteria metabolic levels of YTH were lower than YTD on day 7 in this study. However, on the 14th day, the opposite phenomenon was observed, YTH higher than YTD and remaining stable afterward. Based on the results of PCoA and bacterial succession, we propose a mechanism of soil bacterial community and N. benthamiana response to TMV infection. The reagency or sensibility of the soil bacterial community might influence the response of N. benthamiana to TMV attack. If the soil bacterial community is insensitive or sluggish, viral activity leads to bacterial lysis, facilitating the release of signal molecules that are subsequently transmitted to tobacco plants. The threatened tobacco plants activate their intrinsic defense mechanisms and excrete metabolites, leading to recruiting the beneficial bacteria. This restructuring includes increased helpful bacteria to combat the viral invasion, subsequently protecting the plant. However, suppose the soil bacterial community is sensitive or responds rapidly. In that case, some bacterial strains exhibit specific resistance to virus invasion, thereby impeding the prompt transmission of signal molecules to tobacco plants. Consequently, the activation of tobacco defense mechanisms is hindered, resulting in a rapid increase of TMV virosome. This abrupt rise in TMV levels disrupts the structure of the bacterial community and reduces the proportion of beneficial bacteria. Ultimately, these alterations in the microbial community significantly impact tobacco growth. It is also explained that it was similar between the soil bacterial communities in the YTH group disrupted by TMV on day 7 and in the YTD group disrupted by TMV on days 14, 21 and 28 (Figure 3C).
Further investigation is warranted to elucidate the precise response mechanisms of tobacco to soil TMV. Additionally, this study conducted a comparative analysis of the bacteria community present in various tobacco soils, identifying several potentially beneficial bacteria that hold promise for future research endeavors.
Conclusion
With TMV inoculated, although the alpha diversity of the bacterial community in the YTD was markedly higher at the early stage, it subsequently decreased to lower than in the YTH group. Ecological network was also more complexed in the YTH than in the YTD group. In addition, there was an increase of beneficial bacterial (e.g., Ramlibacter, Sphingomonas, Streptomyces, and Niastella) in the YTH group, which might contribute to strengthening resistance of Tobacco to TMV.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA1021354.
Author contributions
YZ: Data curation, Formal analysis, Investigation, Methodology, Writing – review and editing. TL: Data curation, Investigation, Project administration, Resources, Supervision, Writing – review and editing. SW: Data curation, Investigation, Writing – review and editing. DZ: Funding acquisition, Supervision, Writing – review and editing. ZX: Data curation, Writing – review and editing. ZR: Resources, Writing – review and editing. LL: Data curation, Writing – review and editing. SL: Data curation, Writing – review and editing. YX: Data curation, Formal analysis, Visualization, Writing – original draft. QT: Conceptualization, Formal analysis, Funding acquisition, Supervision, Visualization, Writing – review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Key Research Project of Hunan Province (2023NK2019), the Key Science and Technology Project of China National Tobacco Corporation [110202101050 (LS-10) and 110202201019 (LS-03)], the project of Science and Technology of Hunan Branch of China National Tobacco Corporation (HYYC2023KJ32 and XX2022-2024Aa01), Scientific Research Project of Education Department of Hunan Province (21A0142).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Ara, I., Bukhari, N. A., Aref, N. M., Shinwari, M., and Bakir, M. A. (2012). Antiviral activities of Streptomycetes against Tobacco mosaic virus (TMV) in Datura plant: Evaluation of different organic compounds in their metabolites. Afr. J. Biotechnol. 11, 2130–2138. doi: 10.5897/AJB11.3388
Barberán, A., Ramirez, K. S., Leff, J. W., Bradford, M. A., Wall, D. H., and Fierer, N. (2014). Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol. Lett. 17, 794–802. doi: 10.1111/ele.12282
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Beris, D., Theologidis, I., Skandalis, N., and Vassilakos, N. (2018). Bacillus amyloliquefaciens strain MBI600 induces salicylic acid dependent resistance in tomato plants against Tomato spotted wilt virus and Potato virus Y. Sci. Rep. 8:10320. doi: 10.1038/s41598-018-28677-3
Canwei, S., Xiaoyun, H., Ahmed, N., Shiqi, W., Erxun, Z., and Meide, L. (2020). Fructosan form Paenibacillus kribbensis PS04 enhance disease resistance against Rhizoctonia solani and Tobacco mosaic virus. Electron. J. Biotech. 47, 43–50. doi: 10.1016/j.ejbt.2020.07.002
Chen, D., Ali, M. N. H. A., Kamran, M., Magsi, M. A., Mora-Poblete, F., Maldonado, C., et al. (2022). The Streptomyces chromofuscus Strain RFS-23 induces systemic resistance and activates plant defense responses against Tomato yellow leaf curl virus infection. Agronomy 12:2419. doi: 10.3390/agronomy12102419
Chen, J., Zhang, Y., Wang, C., Weitao, L., Jin, J. B., and Hua, X. (2011). Proline induces calcium-mediated oxidative burst and salicylic acid signaling. Amino Acids 40, 1473–1484. doi: 10.1007/s00726-010-0757-2
Cortés-Sánchez, A. D. J., Hernández-Sánchez, H., and Jaramillo-Flores, M. E. (2013). Biological activity of glycolipids produced by microorganisms: New trends and possible therapeutic alternatives. Microbiol. Res. 168, 22–32. doi: 10.1016/j.micres.2012.07.002
Creager, A. N. H., Scholthof, K. G., Citovsky, V., and Scholthof, H. B. (1999). Tobacco mosaic virus: Pioneering research for a century. Plant Cell 11, 301–308. doi: 10.1105/tpc.11.3.301
Cuartero, J., Pascual, J. A., Vivo, J.-M., Özbolat, O., Sánchez-Navarro, V., Weiss, J., et al. (2022). Melon/cowpea intercropping pattern influenced the N and C soil cycling and the abundance of soil rare bacterial taxa. Front. Microbiol. 13:1004593. doi: 10.3389/fmicb.2022.1004593
Cui, P., Fan, F., Yin, C., Song, A., Huang, P., Tang, Y., et al. (2016). Long-term organic and inorganic fertilization alters temperature sensitivity of potential N2O emissions and associated microbes. Soil Biol. Biochem. 93, 131–141. doi: 10.1016/j.soilbio.2015.11.005
Damayanti, A. T., and Katerina, T. (2008). Protection of hot pepper against multiple infections of viruses by utilizing root colonizing bacteria. J. ISSAAS 14, 92–100.
Deng, Y., He, Z., Xu, M., Qin, Y., Van Nostrand, J. D., Wu, L., et al. (2012). Elevated carbon dioxide alters the structure of soil microbial communities. Appl. Environ. Microb. 78, 2991–2995.
Dunne, C., Crowley, J. J., Moënne-Loccoz, Y., Dowling, D. N., Bruijn, S., and O’Gara, F. (1997). Biological control of Pythium ultimum by Stenotrophomonas maltophilia W81 is mediated by an extracellular proteolytic activity. Microbiology 143, 3921–3931. doi: 10.1099/00221287-143-12-3921
Han, T., You, C., Zhang, L., Feng, C., Zhang, C., Wang, J., et al. (2016). Biocontrol potential of antagonist Bacillus subtilis Tpb55 against tobacco black shank. BioControl 61, 195–205. doi: 10.1007/s10526-015-9705-0
Hernandez, D. J., David, A. S., Menges, E. S., Searcy, C. A., and Afkhami, M. E. (2021). Environmental stress destabilizes microbial networks. ISME J. 15, 1722–1734. doi: 10.1038/s41396-020-00882-x
Hossain, C., Hernandez, V., McHugh, L., Tran, P., and Nash, B. (2022). Analyzing the impacts of Tobacco mosaic virus on the microbial diversity of Nicotiana benthamiana. J. Plant Pathol. 104, 959–967. doi: 10.1007/s42161-022-01103-4
Islam, W., Qasim, M., Noman, A., Tayyab, M., Chen, S., and Wang, L. (2018). Management of Tobacco mosaic virus through natural metabolites. Rec. Nat. Prod. 12, 403–415. doi: 10.25135/rnp.49.17.10.178
Jivkova, D., Sathiyanarayanan, G., Harir, M., Hertkorn, N., Schmitt-Kopplin, P., Sanhaji, G., et al. (2022). Production and characterization of a novel exopolysaccharide from Ramlibacter tataouinensis. Molecules 27:7172. doi: 10.3390/molecules27217172
Kim, B., Park, A. R., Song, C. W., Song, H., and Kim, J. (2022). Biological control efficacy and action mechanism of Klebsiella pneumoniae JCK-2201 producing meso-2,3-Butanediol against tomato bacterial wilt. Front. Microbiol. 13:914589. doi: 10.3389/fmicb.2022.914589
Kuang, L., Li, T., Wang, B., Peng, J., Li, J., Li, P., et al. (2023). Diseased-induced multifaceted variations in community assembly and functions of plant-associated microbiomes. Front. Microbiol. 14:1141585. doi: 10.3389/fmicb.2023.1141585
Kumar, S., Udaya, S. A., Nayaka, S. C., Lund, O. S., and Prakash, H. S. (2011). Detection of Tobacco mosaic virus and Tomato mosaic virus in pepper and tomato by multiplex RT-PCR. Lett. Appl. Microbiol. 53, 359–363. doi: 10.1111/j.1472-765X.2011.03117.x
Langille, M., Zaneveld, J., Caporaso, J., McDonald, D., Knights, D., Reyes, J., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lian, L., Xie, L., Zheng, L., and Lin, Q. (2011). Induction of systemic resistance in tobacco against Tobacco mosaic virus by Bacillus spp. Biocontrol Sci. Techn. 21, 281–292. doi: 10.1080/09583157.2010.543667
Liang, X., Wang, Y., Zhang, Y., Zhuang, J., and Radosevich, M. (2021). Viral abundance, community structure and correlation with bacterial community in soils of different cover plants. Appl. Soil Ecol. 168:104138. doi: 10.1016/j.apsoil.2021.104138
Liu, C. H., Chen, X., Liu, T. T., Lian, B., Gu, Y., Caer, V., et al. (2007). Study of the antifungal activity of Acinetobacter baumannii LCH001 in vitro and identification of its antifungal components. Appl. Microbiol. Biot. 76, 459–466. doi: 10.1007/s00253-007-1010-0
Lu, Q., Hu, C., Cai, L., Wu, C., Zhang, H., Wei, L., et al. (2022). Changes in soil fungal communities after onset of wheat yellow mosaic virus disease. Front. Bioeng. Biotech. 10:1033991. doi: 10.3389/fbioe.2022.1033991
Manjunatha, L., Rajashekara, H., Uppala, L. S., Ambika, D. S., Patil, B., Shankarappa, K. S., et al. (2022). Mechanisms of microbial plant protection and control of plant viruses. Plants 11:3449. doi: 10.3390/plants11243449
Maurhofer, M., Reimmann, C., Schmidli-Sacherer, P., Heeb, S., Haas, D., and Défago, G. (1998). Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain p3 improve the induction of systemic resistance in tobacco against Tobacco necrosis virus. Phytopathology 88, 678–684. doi: 10.1094/PHYTO.1998.88.7.678
Meneghine, A. K., Moretto, C., Castellane, T. C. L., and Carareto Alves, L. M. (2017). Production, characterization and bioemulsifying activity of an exopolysaccharide produced by Sphingomonas sp. Isolated from Freshwater. J. Polym. Environ. 25, 1080–1086. doi: 10.1007/s10924-016-0878-0
Murphy, J. F., Zehnder, G. W., Schuster, D. J., Sikora, E. J., Polston, J. E., and Kloepper, J. W. (2000). Plant growth-promoting rhizobacterial mediated protection in tomato against Tomato mottle virus. Plant Dis. 84, 779–784. doi: 10.1094/PDIS.2000.84.7.779
Park, J. Y., Yang, S. Y., Kim, Y. C., Kim, J., Dang, Q. L., Jeong, J. K., et al. (2012). Antiviral peptide from Pseudomonas chlororaphis O6 against Tobacco mosaic virus (TMV). J. Korean Soc. Appl. Bi. 55, 89–94. doi: 10.1007/s13765-012-0015-2
Ren, X., Wang, J., Zhu, F., Wang, Z., Mei, J., Xie, Y., et al. (2022). B -aminobutyric acid (BABA)-induced resistance to tobacco black shank in tobacco (Nicotiana tabacum L.). PLoS One 17:e267960. doi: 10.1371/journal.pone.0267960
Scholthof, K. G., Adkins, S., Czosnek, H., Palukaitis, P., Jacquot, E., Hohn, T., et al. (2011). Top 10 plant viruses in molecular plant pathology: Top 10 plant viruses. Mol. Plant Pathol. 12, 938–954. doi: 10.1111/j.1364-3703.2011.00752.x
Shanks, R. M. Q., Dashiff, A., Alster, J. S., and Kadouri, D. E. (2012). Isolation and identification of a bacteriocin with antibacterial and antibiofilm activity from Citrobacter freundii. Arch. Microbiol. 194, 575–587. doi: 10.1007/s00203-012-0793-2
Shi, H., Xu, P., Wu, S., Yu, W., Cheng, Y., Chen, Z., et al. (2022). Analysis of rhizosphere bacterial communities of tobacco resistant and non-resistant to bacterial wilt in different regions. Sci. Rep. 12:18309. doi: 10.1038/s41598-022-20293-6
Stringlis, I. A., Yu, K., Feussner, K., de Jonge, R., Van Bentum, S., Van Verk, M. C., et al. (2018). MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. U.S.A. 115, E5213–E5222. doi: 10.1073/pnas.1722335115
Tang, Q., Liu, T., Teng, K., Xiao, Z., Cai, H., Wang, Y., et al. (2023). Microbial interactions and metabolisms in response to bacterial wilt and black shank pathogens in the tobacco rhizosphere. Front. Plant Sci. 14:1200136. doi: 10.3389/fpls.2023.1200136
Tollenaere, C., Lacombe, S., Wonni, I., Barro, M., Ndougonna, C., Gnacko, F., et al. (2017). Virus-Bacteria rice Co-Infection in Africa: Field estimation, reciprocal effects, molecular mechanisms, and evolutionary implications. Front. Plant Sci. 8:645. doi: 10.3389/fpls.2017.00645
Wagg, C., Bender, S. F., Widmer, F., and van der Heijden, M. G. A. (2014). Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. 111, 5266–5270. doi: 10.1073/pnas.1320054111
Wang, Y., Chen, L., Li, Z., Duan, S., Zhang, X., Fang, J., et al. (2024). The role of iron-rich organic fertilizer in promoting the growth of Chinese cabbage and inhibiting the transformation of cadmium. Sci. Total Environ. 908:168430. doi: 10.1016/j.scitotenv.2023.168430
Xiao, Y., Liu, X., Meng, D., Tao, J., Gu, Y., Yin, H., et al. (2018). The role of soil bacterial community during winter fallow period in the incidence of tobacco bacterial wilt disease. Appl. Microbiol. Biot. 102, 2399–2412. doi: 10.1007/s00253-018-8757-3
Zeier, J. (2013). New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant, Cell Environ. 36, 2085–2103. doi: 10.1111/pce.12122
Zhang, G., Han, L., Zhang, G., Zhang, X., and Feng, J. (2015). Purification and characterization of a novel glycoprotein from Streptomyces sp. ZX01. Int. J. Biol. Macromol. 78, 195–201. doi: 10.1016/j.ijbiomac.2015.04.012
Keywords: Tobacco mosaic virus, bacterial interaction, sensitivity of bacterial community, beneficial bacteria, alpha diversity
Citation: Zhao Y, Liu T, Wu S, Zhang D, Xiao Z, Ren Z, Li L, Liu S, Xiao Y and Tang Q (2024) Insight into the soil bacterial community succession of Nicotiana benthamiana in response to Tobacco mosaic virus. Front. Microbiol. 15:1341296. doi: 10.3389/fmicb.2024.1341296
Received: 21 November 2023; Accepted: 15 January 2024;
Published: 31 January 2024.
Edited by:
Fei Yan, Ningbo University, ChinaReviewed by:
Yang Jian, Ningbo University, ChinaOrlando Borras-Hidalgo, Qilu University of Technology, China
Copyright © 2024 Zhao, Liu, Wu, Zhang, Xiao, Ren, Li, Liu, Xiao and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunhua Xiao, aHVhemlwaWFvbGluZy4xMjNAMTYzLmNvbQ==; Qianjun Tang, dGFuZ3FpYW5qdW43OEAxNjMuY29t
†These authors have contributed equally to this work
 Yuqiang Zhao1†
Yuqiang Zhao1† Deyong Zhang
Deyong Zhang Yunhua Xiao
Yunhua Xiao