
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 04 March 2024
Sec. Microbial Physiology and Metabolism
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1339576
Abscisic acid (ABA) is a conserved and important “sesquiterpene signaling molecule” widely distributed in different organisms with unique biological functions. ABA coordinates reciprocity and competition between microorganisms and their hosts. In addition, ABA also regulates immune and stress responses in plants and animals. Therefore, ABA has a wide range of applications in agriculture, medicine and related fields. The plant pathogenic ascomycete B. cinerea has been extensively studied as a model strain for ABA production. Nevertheless, there is a relative dearth of research regarding the regulatory mechanism governing ABA biosynthesis in B. cinerea. Here, we discovered that H3K9 methyltransferase BcDIM5 is physically associated with the H3K14 deacetylase BcHda1. Deletion of Bcdim5 and Bchda1 in the high ABA-producing B. cinerea TB-31 led to severe impairment of ABA synthesis. The combined analysis of RNA-seq and ChIP-seq has revealed that the absence of BcDIM5 and BcHda1 has resulted in significant global deficiencies in the normal distribution and level of H3K9me3 modification. In addition, we found that the cause of the decreased ABA production in the ΔBcdim5 and ΔBchda1 mutants was due to cluster gene repression caused by the emergence of hyper-H3K9me3 in the ABA gene cluster. We concluded that the ABA gene cluster is co-regulated by BcDIM5 and BcHda1, which are essential for the normal distribution of the B. cinerea TB-31 ABA gene cluster H3K9me3. This work expands our understanding of the complex regulatory network of ABA biosynthesis and provides a theoretical basis for genetic improvement of high-yielding ABA strains.
Abscisic acid (ABA) is a sesquiterpene signaling molecule produced in all kingdoms of life. Different organisms have developed different biosynthesis and signal transduction pathways related to ABA (Lievens et al., 2017). To date, the best-known functions of ABA are derived from its role as a major plant hormone in a variety of plant processes, including plant growth, seed dormancy and fruit ripening among others (Singh and Roychoudhury, 2023). Exogenous application has important effects on the promotion of plant growth and development, improvement of stress tolerance and increase in disease resistance (Zehra et al., 2021). In addition, ABA plays an important role in compatible mutualistic interactions such as mycorrhizal and rhizosphere bacteria in plants (Stec et al., 2016). In humans and animal models, studies have shown that the nutrient-derived ABA or ABA treatment is beneficial in the treatment of inflammatory diseases such as colitis and type 2 diabetes, which confer potential to ABA as an interesting nutraceutical or pharmacognostic drug. Meanwhile, the anti-inflammatory, anti-apoptotic, and anti-oxidative properties of ABA in humans and animal models have sparked an interest in this molecule and its signaling pathway as a novel pharmacological target (Sakthivel et al., 2016; Lievens et al., 2017; Shabani et al., 2023). All these points indicate that ABA has a broad application prospect in agriculture and medicine. At present, ABA has been industrialized through B. cinerea fermentation. B. cinerea is a typical phytopathogenic ascomycete that causes gray mold in over 1,000 host plants annually, but it is also a reservoir of secondary metabolites (Dean et al., 2012). Currently, the B. cinerea T4 and B05.10 genome sequences predicted 43 genes probably encoding key enzymes (KEs) for secondary metabolite (SM) biosynthesis (Amselem et al., 2011), including 21 polyketide synthases (PKS), 1 chalcone synthase, 6 sesquiterpene cyclases (STC), 9 non-ribosomal peptide synthases (NRPS), 2 dimethylallyl tryptophan synthases (DMATS), and 5 diterpene cyclases (DTC), suggesting that in addition to ABA, B. cinerea has the potential to produce the above SMs.
In our laboratory, the B. cinerea wild-type strain TBC-6 was isolated from wheat stems and leaves in southwest China. B. cinerea TBC-6 improvements were performed and generated a series of B. cinerea mutants with different yields of ABA (Gong et al., 2014), such as TB-31, TB-3-H8, and TBC-A, which were used for biosynthesis regulation research and industrial production of ABA (Ding et al., 2016). The ABA biosynthesis gene cluster and positive regulator in B. cinerea have been identified, including two hypothetical P450 monooxygenase coding genes Bcaba1 and Bcaba2, hypothetical FPP (Farnesyl Pyrophosphate) catalysis gene Bcaba3, presumptive short-chain dehydrogenase/reductase coding genes Bcaba4, and pathway-specific transcription factors BcabaR1 (Wang et al., 2018). Meanwhile, we found that the global regulator BcLAE1 is involved in the regulation of ABA biosynthesis. Interestingly, H3K9me3 was significantly altered in the BcLAE1 knockout mutant. We therefore suspect that ABA synthesis is regulated by higher-level epigenetic regulation in addition to pathway-specific transcription factors and global regulators.
Epigenetic regulation refers to heritable regulation independent of DNA sequence changes caused by RNA interference, DNA methylation, and Posttranslational modification of histones (Lai et al., 2022). Among them, histone posttranslational modification affects chromatin modification status (euchromatin and heterochromatin) through phosphorylation, methylation and acetylation of histone N-terminal tail, thus realizing the regulation of gene expression (Bind et al., 2022). Studies have shown that monomethylation, dimethylation, and trimethylation (me1/2/3) of histone lysine are catalyzed by histone lysine methyltransferase (KMT), mostly with a catalytic Set (Su(var)3–9, enhancer-of-zeste and trithorax) domain, which transfer a methyl group from S-Adenosyl-L-Methionine (SAM) to the lysine residues on the N-terminal of H3 or H4 (Brosch et al., 2008). Changes in chromatin structure achieved by histone methylation play a key role in regulation of fungal SM biosynthesis. For example, H3K4me2/3, H3K36me1/3 and H4K20me1 are commonly associated with transcriptional activation, whereas H3K9me2/3, H3K27me2/3 and H4K20me3 are associated with transcriptional repression (Lai et al., 2022). The H3K4me2/3 alteration in Fusarium Fujikuroi encouraged the expression of the Gibberellin (GA) gene cluster and increased GA production (Janevska et al., 2018). Deletion of FgKMT6 in Fusarium graminearum results in a genome-wide deficiency of H3K27me3, which activates the expression of mycotoxins, pigments, and some secondary metabolism-related genes in the ΔFgKMT6 (Tang et al., 2021). The methylation modification of H3K36 in Aspergillus flavus is caused by the AshA and SetB, which can directly control the synthesis of mycotoxins (Zhuang et al., 2022). However, the mechanism by which histone modifications regulate ABA biosynthesis in B. cinerea has not been reported.
In the current study, we discovered the histone H3K9 methyltransferase BcDIM5 is involved in the regulation of ABA synthesis. This regulatory role is achieved by synergizing with the H3K14 deacetylase BcHda1. Loss of BcDIM5 and BcHda1 led to an excessive accumulation of H3K9me3 occupancy in the ABA gene cluster, resulting in a severe impairment of ABA production. Furthermore, our observations implied that BcHda1 and BcDIM5 are required for normal distribution of constitutive heterochromatic modifications and the stable maintenance of ABA biosynthesis in B. cinerea TB-31.
In a previous study, we discovered that the global regulator BcLae1 knockdown significantly reduced the production of ABA and changed the global H3K9me3 modification (Wei et al., 2022). Therefore, we speculated whether changes in H3K9me3 modification would have an effect on ABA synthesis. Based on this, we knocked out the reported H3K9 methyltransferase BcDIM5 in B. cinerea TB-31 (Supplementary Figure S1). In comparison to TB-31, the ABA production of ΔBcdim5 was reduced by more than 97% during the 6–12 days culture period. ABA production was recovered following Bcdim5 complementation. At day 12, the randomly selected complement strains ΔBcdim5-C-X3, X6 and X7 restored ABA yield to more than 69% of that of TB-31 (Figure 1A), demonstrating that BcDIM5 is involved in the regulation of ABA synthesis.
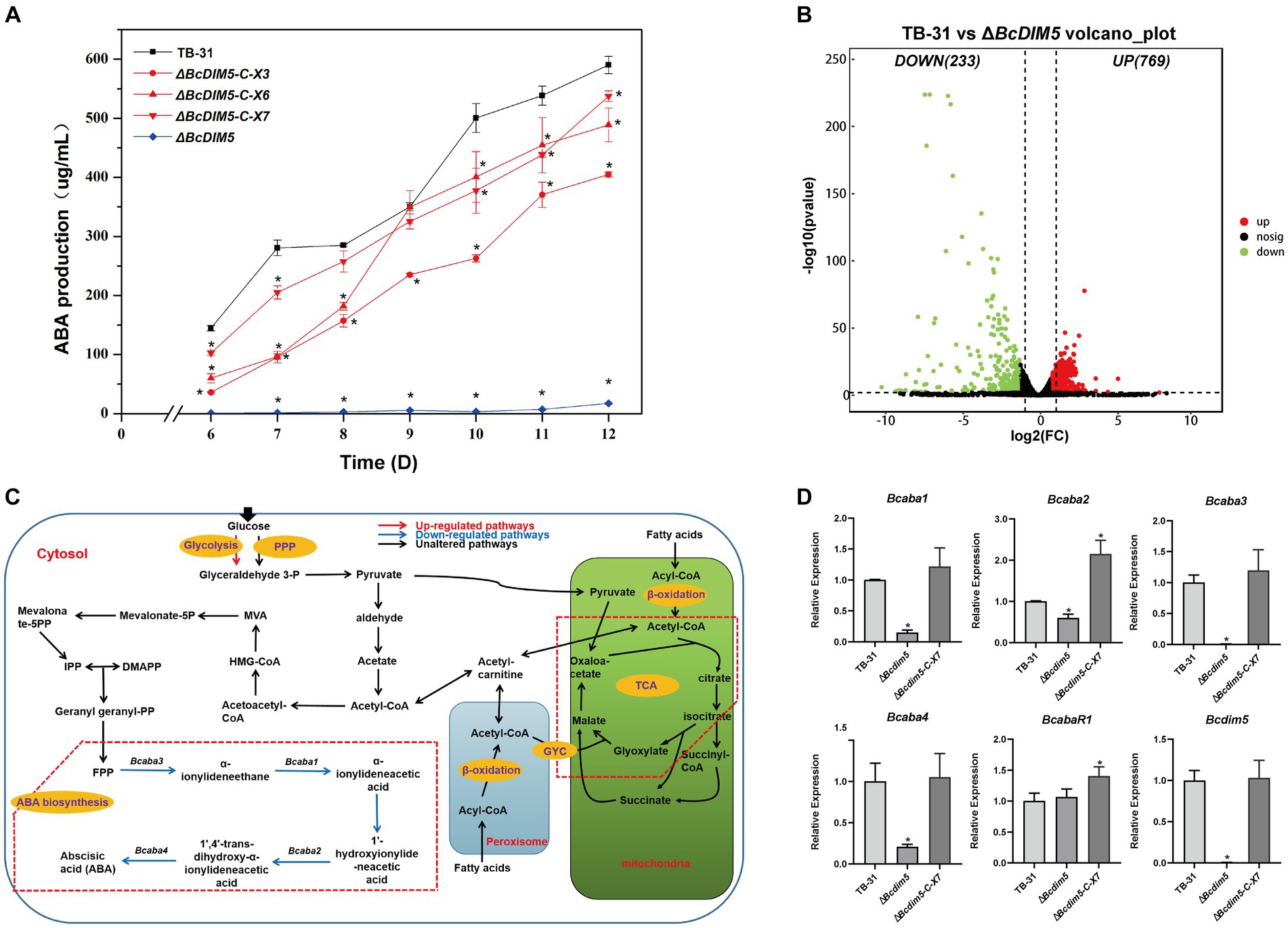
Figure 1. Bcdim5 is required for ABA production in B. cinerea TB-31. (A) ABA production of control strain TB-31, ΔBcdim5 and three randomly selected ΔBcdim5-C mutants at different culture time. Samples for the quantitative determination of ABA production were collected at 6–12 days. The error bars indicate the standard errors of the mean for three replicate cultures (n = 3). Asterisks indicate significant differences in ABA production between selected mutants and TB-31 (p < 0.05). (B) DEGs between TB-31 and ΔBcdim5 on day 6 of PDA culture. The criteria for the selection of DEGs were a log2 |fold change| ≥ 1, value of p < 0.05, and RPKM of at least one sample larger than 1. (C) Schematic of metabolic pathways associated with ABA production affected by Bcdim5 deletion. The red arrows indicate upregulated pathways. Blue arrows indicate downregulated pathways, and black arrows indicate pathways that were not observably regulated (log2 |fold change| ≥ 1, value of p < 0.05 and RPKM of at least one sample larger than 1). The red dashed box annotates the glyoxylic acid cycle (GYC) pathway. (D) RT-qPCR examining the transcriptional levels of ABA gene cluster, BcabaR1 and Bcdim5 in ΔBcdim5, ΔBcdim5-C-X7, and TB-31. The relative transcriptional levels of selected genes were obtained after normalization to the constitutive tubulin reference gene (BC1G_05600) at 6 days. The relative values for selected genes transcription at 6 days in TB-31 were assigned as 100%. Shown are means and SEM, n = 3 independent biological replicates. *p < 0.05 versus the same genes of the TB-31 group.
When Bcdim5 is knocked out, ABA synthesis is severely reduced from early to late stages. To understand the impact of Bcdim5 knockout on the related pathway of ABA synthesis, we selected the early stage of ABA synthesis for RNA-seq analysis. Based on the results of differential analysis, the differentially expressed genes (DEGs) were identified between the transformants using the threshold log2 |fold change| ≥ 1, p-value <0.05 and RPKM value of at least one sample larger than 1. And a total of 1,002 DEGs were obtained, of which 769 genes were up-regulated and 233 genes were down-regulated (Figure 1B).
Subsequently, we focused on analyzing changes in the expression of ABA synthesis-related pathways, including carbohydrate transport and utilization, acetyl-CoA synthesis and transport-related pathways, FPP synthesis and ABA synthesis gene cluster (Supplementary Dataset S1). It was found that the transcription levels of 1,4-Beta-mannopyranoside mannosidase manA (BCIN_02g07520), Arabinoglucosidase xylB (BCIN_08g00060) and Ferulic acid esterase faeb-2 (BCIN_09g05740) were reduced. The expression of the remaining enzymes involved in sugar transporters, degrading enzymes and permeases were not significantly altered. This result suggests that the carbon source utilization of ΔBcdim5 was not significantly changed compared with TB-31. The central metabolite acetyl-CoA is the link between the primary metabolic pathway and the secondary metabolic pathway, which is also the precursor of the synthesis of ABA in B. cinerea. Pyruvate is an important node in glucose synthesis of acetyl-coA. Both glycolysis and pentose phosphate pathway (PPP) convert glucose into intracellular pyruvate. However, there was no discernible variation in the transcription levels of the genes involved in the PPP. The expression levels of BCIN_15g04970 (glucose-6-phosphate isomerase) and BCIN_08g06300 (6-phosphofructose kinase subunit α) genes in glycolysis pathway were higher than that of TB-31, which ensured that the glucose catabolism of ΔBcdim5 was not inhibited. The expression levels of associated genes in the β-oxidation pathway, which generates acetyl-coA from fatty acids, did not differ significantly either. The transport of acetyl-coA involves the TCA cycle and carnitine shuttle system, but the alterations in gene expression pertaining to these transport systems were not significant. Consequently, the decrease in ABA production in ΔBcdim5 cannot be attributed to the synthesis and transport of acetyl-coA. Additionally, the transcription levels of enzymes involved in the MVA pathway remain unchanged, while the transcription level of the ABA synthesis gene cluster (Bcaba1, Bcaba2, Bcaba3 and Bcaba4) experienced a significant reduction in the ΔBcdim5 (Figure 1C). Therefore, we suggested that this is the direct cause of the reduced ABA production in ΔBcdim5, which was confirmed by RT-qPCR (Figure 1D).
In eukaryotes, histone modifying enzymes commonly bind to multiple subunits to affect chromatin homeostasis (Beisel and Paro, 2011; Song et al., 2015). The DIM-5 complex has been well characterized in fission yeast. In addition to forming a methyltransferase complex with RiK1, Raf1, Raf2, and Pcu4 subunits (Oya et al., 2019; Shan et al., 2021), DIM-5 exists in close association with the Argonaute complex to maintain heterochromatin assembly (Buker et al., 2007). In the B. cinerea genome, the Arb2 subunit of the Argonaute complex was found to be present in the putative H3K14 deacetylase BcHda1 by NCBI BLAST comparison (Figure 2A). We therefore speculated BcHda1 as a potential interacting partner of BcDIM5. Based on this, Y2H assays were performed on BcDIM5 and BcHda1, revealing that the yeast cells were capable of growing on the TDO/X/A plate and QDO/X/A plate following the interaction of BcHda1 and BcDIM5 (Figure 2B), suggesting a potential functional relationship between the two proteins. Additionally, the physical association of BcHda1 and BcDIM5 was confirmed in vitro through GST-pulldown assay after the purification of His-BcHda1 and GST-BcDIM5 (Figure 2C), suggesting a potential functional relationship between the two proteins.
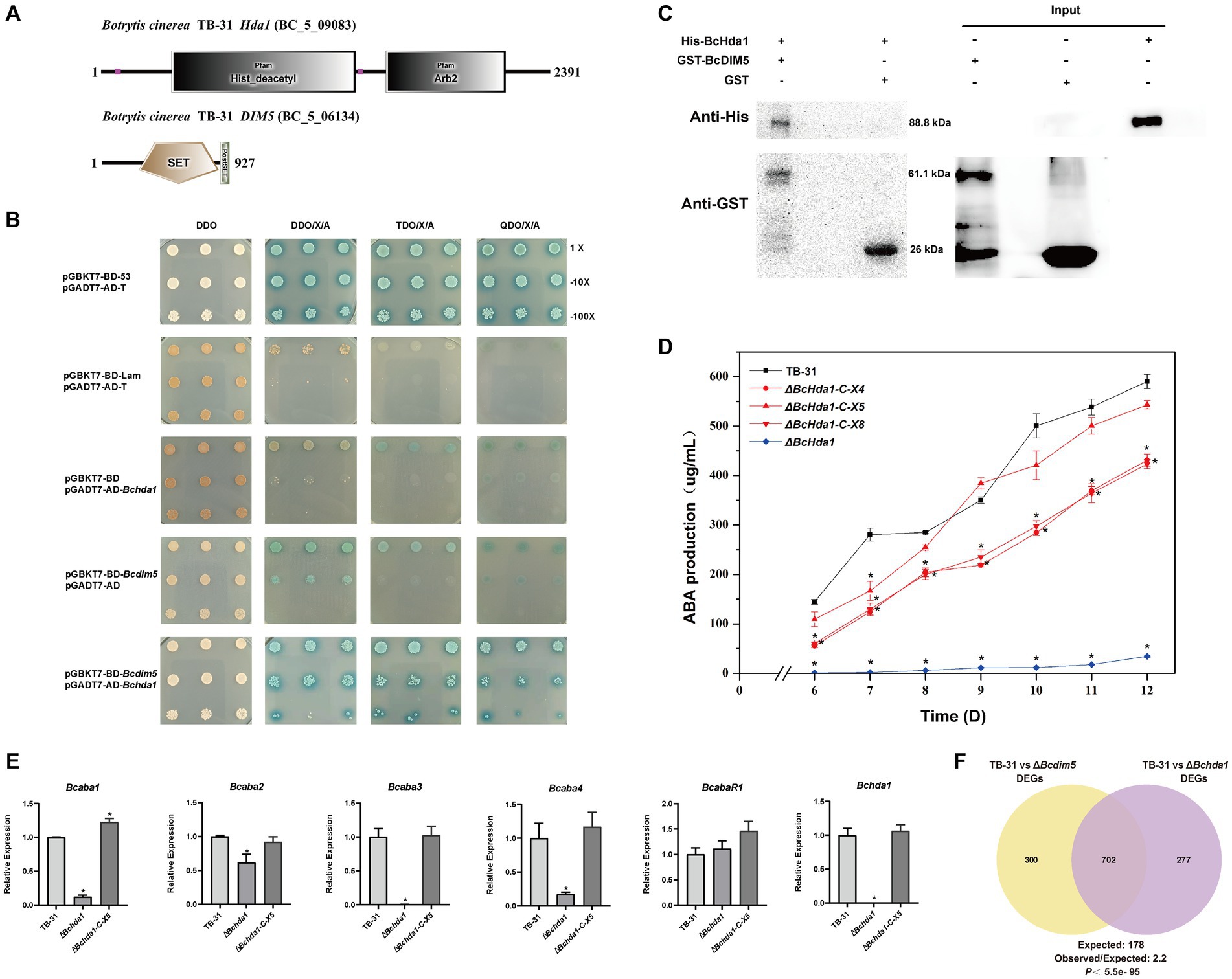
Figure 2. The interacting protein BcHda1 of BcDIM5 is also involved in ABA synthesis. (A) Structures and conserved domains of BcHda1 and BcDIM5. The indicated domains include: SET (IPR001214), Post-SET (IPR003616), His_deacetylse (IPR023801), Arb2 (IPR019154). (B) Yeast-two-hybrid assay between BcHda1 and BcDIM5. pGBKT7-BD-53/pGADT7-AD-T was the positive control, pGBKT7-BD-lam/pGADT7-AD-T, pGBKT7-BD/pGADT7-AD-Bchda1, and pGBKT7-BD- Bcdim5/pGADT7-AD were the negative controls. The experimental group pGBKT7-BD-Bcdim5/pGADT7-AD-Bchda1 could grow in TDO/X/A and QDO/X/A medium after hybridization. (C) GST-Pulldown validates the interaction of BcDIM5 with BcHda1. (D) ABA production of control strain TB-31, ΔBchda1 and three randomly selected ΔBchda1-C mutants at different culture time. Samples for the quantitative determination of ABA production were collected at 6–12 days. The error bars indicate the standard errors of the mean for three replicate cultures (n = 3). Asterisks indicate significant differences in ABA production between selected mutants and TB-31 (p < 0.05). (E) RT-qPCR examining the transcriptional levels of ABA gene cluster, BcabaR1 and Bchda1 in ΔBchda1, ΔBchda1-C-X5, and TB-31. The relative values for selected genes transcription at 6 days in TB-31 were assigned as 100%. Shown are means and SEM, n = 3 independent biological replicates. *p < 0.05 versus the same genes of the TB-31 group. (F) Venn diagram showing the significant overlap of DEGs in the ΔBcdim5 and ΔBchda1. p-values for the significance of the overlap between gene sets were calculated using Fisher’s exact test.
Subsequently, we also performed knockdown experiments on BcHda1. Knockdown of Bchda1 revealed that the ABA production of ΔBchda1 was similar to that of ΔBcdim5. During the 6–12 days culture period, ABA produced by ΔBchda1 was reduced by more than 94% compared to TB-31. The ABA yield was restored upon Bchda1 complementation, particularly in ΔBchda1-C-X5, which approached that of the control strain TB-31 (Figure 2D). These findings suggested that Bchda1 is also essential for the ABA biosynthesis in B. cinerea TB-31. The effect of Bchda1 knockdown on the ABA synthesis pathway was assessed by RNA-seq analysis (Supplementary Table S2). The results showed that except for a significant decrease in the transcription levels of the ABA synthesis gene cluster (Figure 2E), the expression changes of the other related pathways were not significant, implying that BcDIM5 and BcHda1 affect ABA synthesis by regulating the transcription levels of the ABA gene cluster. Comparison of RNA-seq data for ΔBchda1 and ΔBcdim5 revealed a large number of identical terms in the GO and KEGG enrichment results (Supplementary Figure S2). In particular, the expression of many DNA repair genes was upregulated, suggesting that knockdown of Bcdim5 and Bchda1 may have a significant impact on growth and development. From the phenotypic results we clearly observed that the growth of ΔBchda1 and ΔBcdim5 became retarded compared to the control strain TB-31 (Supplementary Figure S3). Meanwhile, the two sets of transcriptome data exhibit 702 identical DEGs, which constitute 71.7 and 70.1% of the total DEGs of TB-31 vs. ΔBchda1 and TB-31 vs. ΔBcdim5, respectively (Figure 2F). The overlap of the DEGs data demonstrates that the two proteins act synergistically to exert regulatory functions.
To further investigate whether the overexpression of Bchda1 and Bcdim5 can enhance ABA production, we generated overexpressed mutants and assessed ABA production in 9 randomly selected mutants after 7 days of incubation. The results showed that the ABA production of overexpressed strains was generally reduced (Figure 3A). In order to investigate the potential for ABA production to match or exceed that of the control strain TB-31 during later stages of culture, three Bchda1 and Bcdim5 overexpression mutants were randomly selected for 6–12 days to determine the ABA production (Supplementary Figure S4), but the ABA yield was still reduced (Figure 3B).
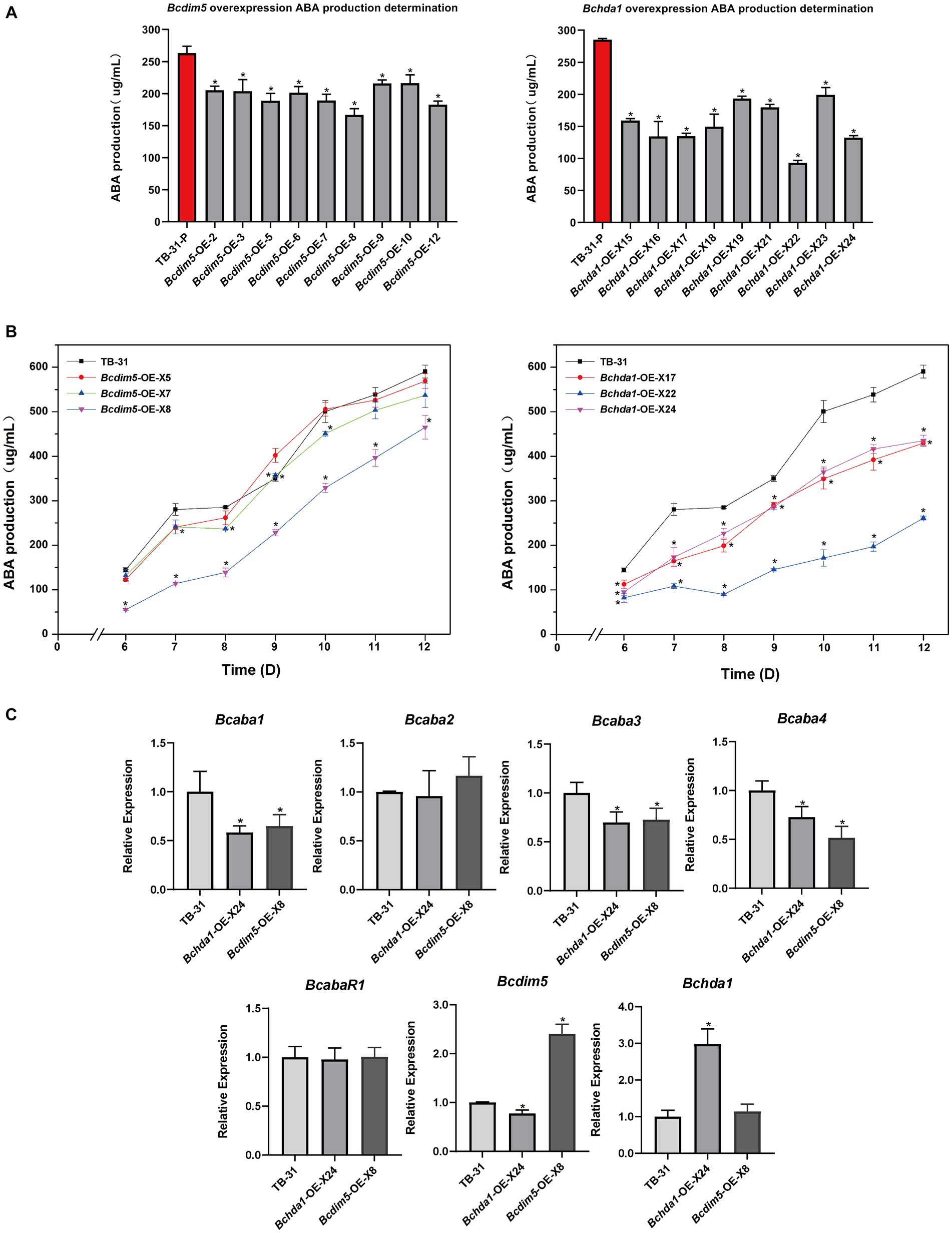
Figure 3. Overexpression of Bcdim5 and Bchda1 did not increase ABA production in B. cinerea TB-31. (A) ABA yield of Bcdim5 and Bchda1 overexpression strains cultured for 7 days. The error bars indicate the standard errors of the mean for three replicate cultures (n = 3). Asterisks indicate significant differences in ABA production between selected mutants and TB-31 (p < 0.05). (B) ABA production of control strain TB-31, three randomly selected Bcdim5-OE mutants and three randomly selected Bchda1-OE mutants at different culture time. Samples for the quantitative determination of ABA production were collected at 6–12 days. The error bars indicate the standard errors of the mean for three replicate cultures (n = 3). Asterisks indicate significant differences in ABA production between selected mutants and TB-31 (p < 0.05). (C) RT-qPCR examining the transcriptional levels of ABA gene cluster, BcabaR1, Bcdim5 and Bchda1 in Bcdim5-OE-X8, Bchda1-OE-X24, and TB-31. The relative values for selected genes transcription at 6 days in TB-31 were assigned as 100%. Shown are means and SEM, n = 3 independent biological replicates. *p < 0.05 versus the same genes of the TB-31 group.
RNA-seq analysis of ABA synthesis related pathways revealed that a large number of genes related to carbon metabolism was upregulated in Bchda1-OE-X24, which was beneficial for ABA synthesis. The increased transcription level of acetoacetyl-CoA synthetase (BCIN_07g02750) in the MVA pathway, enhancing the ability of acetyl-CoA to synthesize acetoacetyl-CoA. Otherwise, the expression changes of related genes in the remaining pathways were not significant in the Bchda1-OE-X24 (Supplementary Table S2). Additionally, no significant changes in expression were observed in any of the genes involved in ABA synthesis-related pathways in Bcdim5-OE-X8 strain. However, a significant decrease in the FPKM of the ABA cluster was noted in Bchda1-OE-X24 and Bcdim5-OE-X8 mutants, and further analysis via RT-qPCR revealed reduced expression levels of Bcaba1, Bcaba3 and Bcaba4 within the cluster (Figure 3C). Therefore, we suggested that the appropriate expression levels of Bcdim5 and Bchda1 are important factors for maintaining stable ABA synthesis.
Since BcDIM5 is a histone H3K9 methyltransferase and BcHda1 is a putative histone H3K14 deacetylase, their deletion and overexpression will inevitably affect the change of global chromatin modification status. The results showed that H3K9me and H3K9me2 did not change significantly in Bcdim5 and Bchda1 mutants. However, the levels of H3K9me3 modification were considerably reduced in ΔBchda1 and ΔBcdim5. The restoration of modification levels after gene complementation suggested that BcDIM5 and BcHda1 play a role in regulating the global H3K9me3 modification level. Consequently, it is hypothesized that the alteration of the global H3K9me3 modification state is the primary factor in regulating the DEGs shared by Bcdim5 and Bchda1 mutants. Meanwhile, we found that Bchda1 overexpression resulted in a decrease in the global H3K14ac modification level. The results suggested that BcHda1 functions as a histone H3K14 deacetylase, thereby influencing the global H3K14ac modification level (Figures 4A,B).
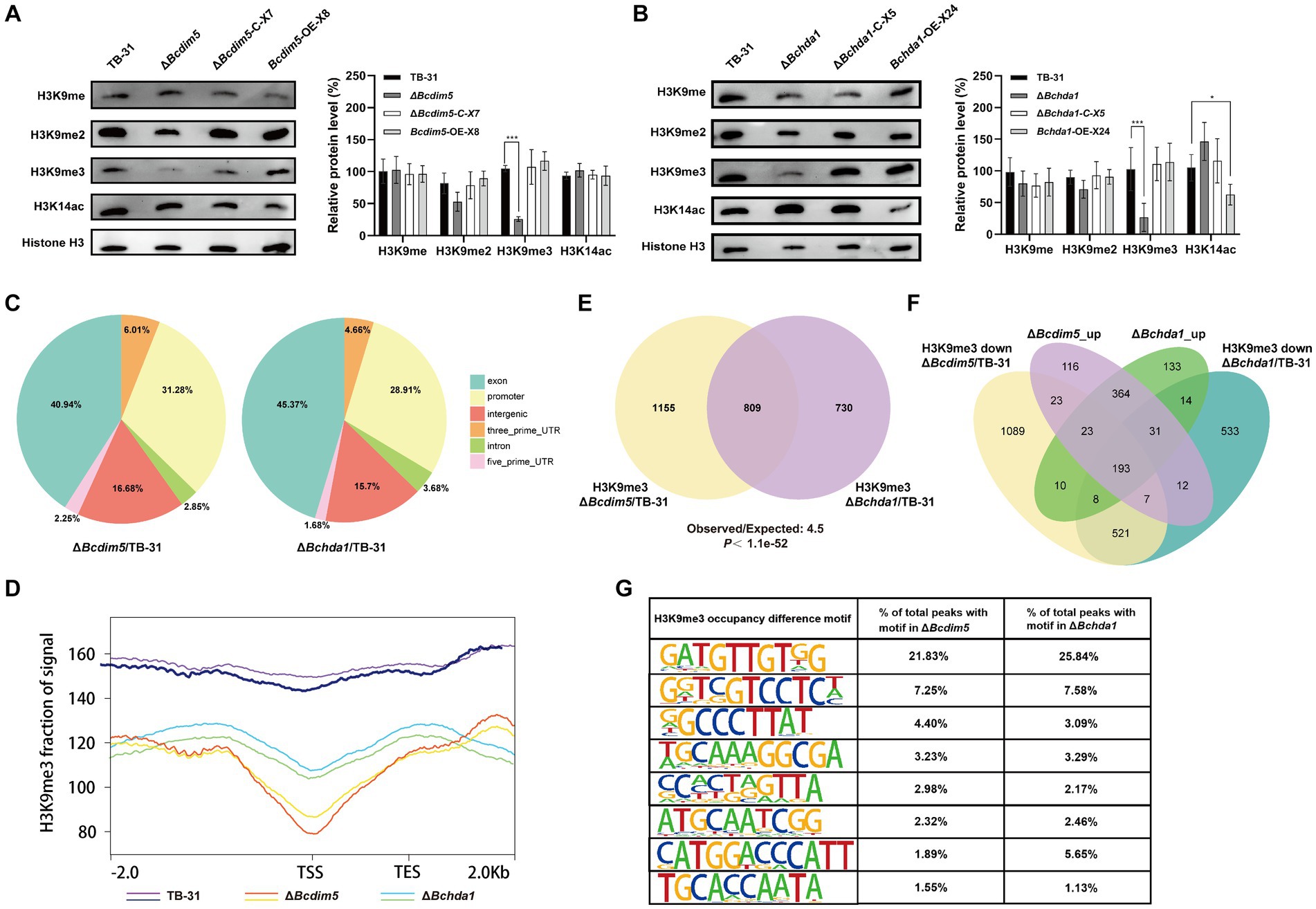
Figure 4. Bcdim5 and Bchda1 are essential for the normal distribution of genome-wide H3K9me3 occupancy in B. cinerea TB-31. (A,B) Global H3K9 methylation statues and H3K14 acetylation statues of the control strain TB-31, Bcdim5 gene mutants and Bchda1 gene mutants after dark cultivation for 6 days. Immunoblot signals relative to the TB-31 (set as 1) were quantified. Each value represents the mean ± SE of three biological replicates. The asterisks indicate significant differences between mutants and the TB-31 (*p < 0.05, ***p < 0.001). (C) Distribution of BcDIM5 and BcHda1 mediated H3K9me3 removal sites in different regions of annotated genes. The intergenic region refers to the region beyond 2000 bp of the transcription start site of the coding gene and 1,000 bp of the transcription stop site, including the untranscribed and post-transcriptional untranslated regions. (D) Metagene plot shows H3K9me3 ChIP-seq intensities within 2.0-kb genomic regions flanking the peak summits at the TSS in the TB-31, ΔBcdim5 and ΔBchda1 strains. (E) Venn diagram showing the significant overlap of H3K9me3-occupied genes in the ΔBcdim5 and ΔBchda1. p-values for the significance of the overlap between gene sets were calculated using Fisher’s exact test. (F) Venn diagram showing the significant overlap between upregulated genes and H3K9me3-occupied genes in the ΔBcdim5 and ΔBchda1. (G) H3K9me3 occupancy difference motif analysis in the ΔBcdim5 and ΔBchda1 strains. The 8 most abundant motifs are listed.
The H3K9me3 and H3K14ac play crucial roles in fungal development, virulence, and SMs (Gu et al., 2017; Chen et al., 2022; Zhang et al., 2023), with the former typically associated with gene repression (Gessaman and Selker, 2017) and the latter with gene activation (Wang et al., 2017). In order to gain a deeper understanding of the impact of Bchda1 and Bcdim5 loss on H3K9me3 and H3K14ac occupancy in strains, ChIP-seq was conducted to map the genomic distribution of these modifications in the ΔBcdim5 and ΔBchda1 mutants, with B. cinerea TB-31 serving as a background control (Supplementary Dataset S2). The results showed that the level of H3K14ac was almost unchanged in the ΔBcdim5 compared to TB-31. In contrast, the ΔBchda1 exhibited a higher occupancy of H3K14ac in comparison to TB-31. However, the identification of only 128 H3K14ac differential peaks corresponding to 105 genes indicated that the enriched regions accounted for a minute proportion of the entire genome. This suggested that the loss of Bchda1 had a relatively minor impact on global H3K14ac. Notably, both the ΔBchda1 and ΔBcdim5 mutants demonstrated a significant deficiency in the distribution of H3K9me3 occupancy when compared to TB-31. Specifically, the H3K9me3 levels of 998 genes in the ΔBchda1 and 1,191 genes in the ΔBcdim5 were reduced by at least one-fold. The BcHda1 and BcDIM5 co-mediated H3K9me3 removal sites were distributed in different regions of annotated genes, including exons, promoters, introns, and intergenic regions, with a larger proportion of exons and promoters (Figure 4C). Meta-gene analysis also showed decreased H3K9me3 levels in ΔBcdim5 and ΔBchda1 strains, especially between TSS and TES regions (Figure 4D). In addition, the additional regions with 254 genes and 80 genes gained H3K9me3 occupancy in the ΔBchda1 and ΔBcdim5 strains, respectively, but were absent in the TB-31 (Supplementary Dataset S2). Collectively, the loss of BcDIM5 and BcHda1 led to severe global defects in the normal distribution and level of H3K9me3 modification, including majority loss and reduction, and local hypermethylation of genomic sites in B. cinerea TB-31.
To further compare H3K9me3 distribution between the ΔBcdim5 and ΔBchda1 strains, we quantified the detailed numbers of H3K9me3-occupied genes with corresponding peaks. In total, we identified 2,846 H3K9me3-occupied peaks (ΔBcdim5 to TB-31) corresponding with 1964 genes in the ΔBcdim5 and 2069 peaks (ΔBchda1 to TB-31) corresponding with 1,539 genes in the ΔBchda1. Among them, 809 genes had overlapping H3K9me3-occupied peaks (Figure 4E), implying that the modification of H3K9me3-enriched regions of these genes may be co-regulated by BcHda1 and BcDIM5. Subsequently, we explored the biological connection between BcHda1 and BcDIM5 in gene repression. RNA-seq analysis indicated that DEGs of ΔBcdim5 and ΔBchda1 strains showed the same direction of dysregulation with significant overlap in upregulated genes (611 upregulated genes). Meanwhile, 193 genes in the upregulated cross set have H3K9me3 modifications missing or reduced in ΔBcdim5 and ΔBchda1 strains, indicating that BcDIM5 and BcHda1 are required for transcriptional silencing of these genes (Figure 4F). In addition, 158 genes in the upregulated cross set were adjacent to the H3K9me3 deletion peak domains, suggesting that the spread of H3K9me3 may be required for gene repression. These results indicated that the BcDIM5 and BcHda1 are critical for H3K9me3-mediated transcriptional silencing in B. cinerea TB-31. To gain more insights into the characterization of genome-wide H3K9me3 modification of B. cinerea TB-31 by BcDIM5 and BcHda1, the motifs of H3K9me3-occupied peaks were predicted and analyzed using HOMER software. The results showed that the predominant motif in abundance is GTTGTNG, followed by GTCCTC, GCCCTT and GGCGA, etc. (Figure 4G). The binding of H3K9me3 on these motifs may require the co-participation of BcDIM5 and BcHda1.
Histone modification changes induced by BcDIM5 and BcHda1 also affect the transcription levels of SM key enzyme genes of B. cinerea (Supplementary Table S2). The transcription levels of Bcstc1, Bcstc3, Bcpks11, Bcpks17, and Bcpks21 genes were decreased in ΔBcdim5, while the transcription levels of Bcstc4, Bcpks1, Bcpks10, and Bcpks14 genes were increased. In conjunction with ChIP-seq analysis, it was observed that the ΔBcdim5 exhibited an increase in H3K9me3 occupancy in the exon region of the Bcstc3, while a decrease in H3K9me3 occupancy in the exon region of the Bcpks1 and Bcpks10 was noted (Figures 5A–D). Additionally, the transcription levels of five polyketide synthases Bcpks6, Bcpks9, Bcpks11, Bcpks1, and Bcpks21 genes were all increased in Bchda1-OE-X24 strain. The ChIP-qPCR analysis revealed an increase in H3K14ac occupancy in the promoter region of the Bcpks6 and Bcpks21 genes (Supplementary Figure S5), which may account for the observed up-regulation of transcription. Histone modification reprogramming caused by BcDIM5 and BcHda1 facilitated the subsequent mining of novel secondary metabolites in B. cinerea.
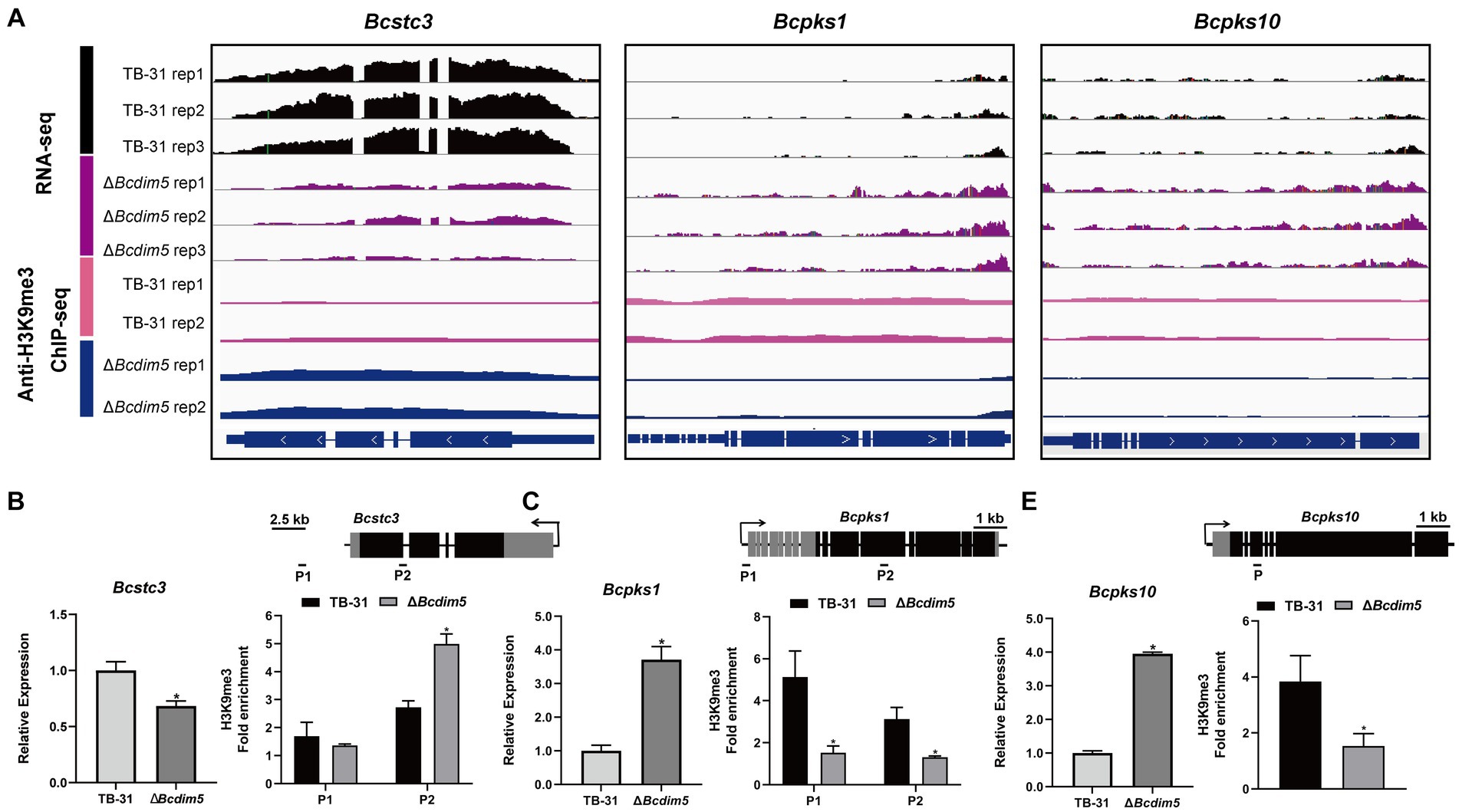
Figure 5. Differential H3K9me3 occupancy due to Bcdim5 deletion leads to changes in the expression of SM-related genes. (A) Genome tracks of RNA-seq, ChIP-seq with anti-H3K9me3 for Bcstc3, Bcpks1 and Bcpks10 loci in the TB-31 and ΔBcdim5 strains. Structures of Bcstc3, Bcpks1 and Bcpks10 are shown. (B) Bcstc3; (C) Bcpks1; (D) Bcpks10. [i in (B–D)] qRT-PCR analysis of Bcstc3, Bcpks1 and Bcpks10 expression levels. The B. cinerea tubulin gene (BC1G_05600) was used as an internal control. The relative values for selected genes transcription at 6 days in TB-31 were assigned as 100%. Shown are means and SEM, n = 3 independent biological replicates. *p < 0.05 versus the same genes of the TB-31 group. [ii in (B–D)] ChIP-qPCR analysis of H3K9me3 status at the Bcstc3, Bcpks1 and Bcpks10 loci. IgG was used as a negative control for comparison with IP to calculate the enrichment ploidy. Schematic genetic structures of Bcstc3, Bcpks1 and Bcpks10, represented as black boxes for exons, gray boxes for 5′ and 3′ untranslated regions and black lines for promoters and introns. Amplified regions are indicated below each locus (P). Data are presented as the mean ± SD (n = 3). *p < 0.05 versus the H3K9me3 occupancy of the TB-31 group.
Current studies suggest that H3K9me3 is commonly associated with the repression of biosynthetic gene clusters, and has been further identified as a mark of constitutive heterochromatin, principally associated with centromeric heterochromatin (Wiles and Selker, 2017; Torres-Garcia et al., 2020). Therefore, knockdown of DIM5 in pathogenic fungi leads to increased production of some secondary metabolites (Wang et al., 2022). However, DIM5 is often associated with the pathogenicity of the strain. The loss of DIM5 is accompanied by slow growth and development of strains, weakened toxicity, and reduced virulence factor synthesis (Gu et al., 2017), which also occurs in B. cinerea (Zhang et al., 2016). In the ΔBcdim5 and ΔBchda1 mutants, the production of the virulence factor ABA was severely impaired. IGV analysis of the H3K14ac and H3K9me3 enrichment of ABA gene clusters in TB-31, ΔBcdim5, and ΔBchda1 revealed that the H3K14ac modification did not exhibit a significant difference. Unexpectedly, the deletion of Bcdim5 and Bchda1 did not result in a decrease in H3K9me3 occupancy within the ABA gene cluster, but rather a hyper-H3K9me3 enrichment (Figure 6A). Subsequently, ChIP-qPCR was used to further validate that H3K9me3 occupancy in the promoter region and exon region of Bcaba1 and Bcaba3 genes increased significantly (Figure 6B), which in turn led to the down-regulation of the expression of the entire gene cluster. Whether the generation of this hyper-H3K9me3 modification is due to the deletion of Bcdim5 and Bchda1 resulting in the recruitment of other histone-modifying enzymes to the ABA gene cluster needs to be further investigated. However, there is no doubt that BcDIM5 and BcHda1 are required for ABA synthesis. Meanwhile, we also found that BcDIM5 and BcHda1 did not directly bind to the promoter region of the ABA gene cluster by electrophoretic mobility shift assays (data not shown). Therefore, we believe that other proteins assist BcDIM5 and BcHda1 in the H3K9me3 modification of ABA gene clusters (Figure 6C).
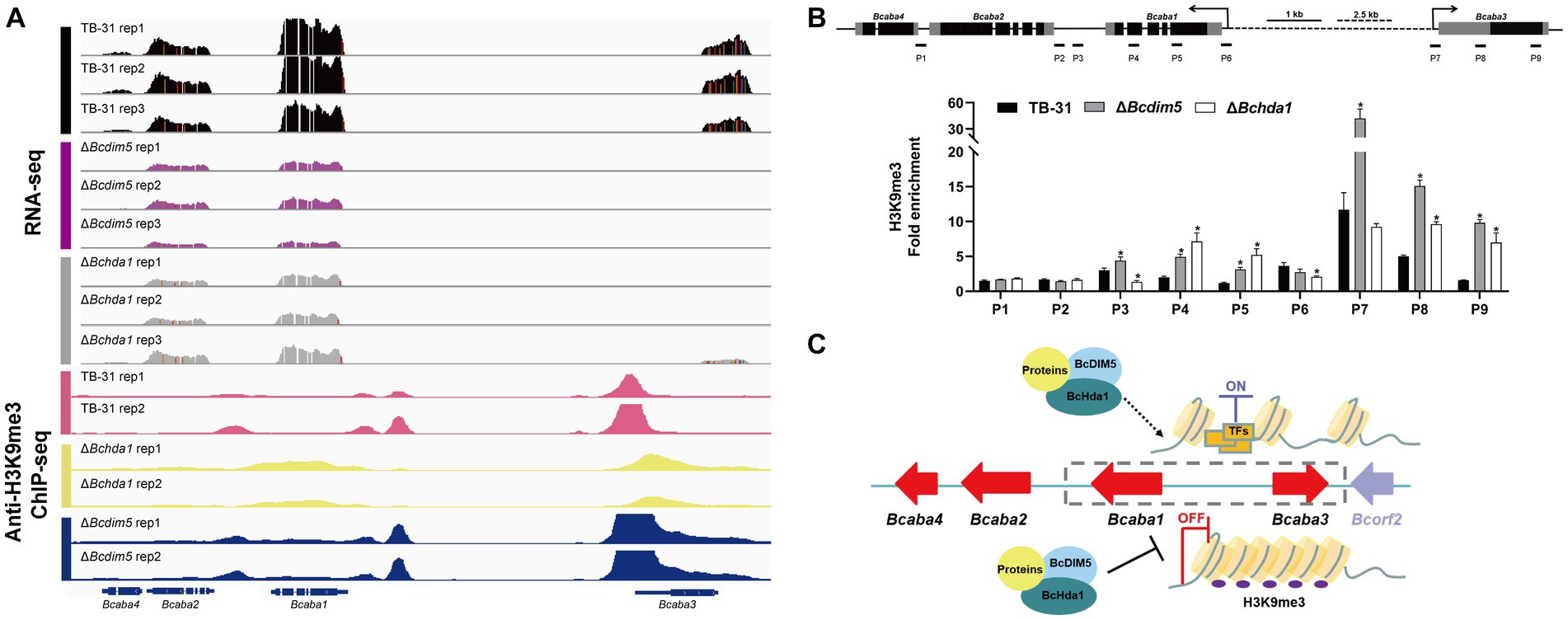
Figure 6. Reduced expression levels of the ABA gene cluster are associated with increased H3K9me3 occupancy. (A) Genome tracks of RNA-seq, chromatin immunoprecipitation followed by sequencing (ChIP-seq) with anti-H3K9me3 for ABA gene cluster (Bcaba1-4) loci in the TB-31, ΔBcdim5 and ΔBchda1 strains. Structures of Bcaba1-4 are shown. (B) ChIP-qPCR analysis of H3K9me3 methylation status at the ABA gene cluster loci. IgG was used as a negative control for comparison with IP to calculate the enrichment ploidy. Schematic genetic structures of Bcaba1, Bcaba2, Bcaba3 and Bcaba4, represented as black boxes for exons, gray boxes for 5′ and 3′ untranslated regions and black lines for promoters and introns. Amplified regions are indicated below each locus (P1-P9). Data are presented as the mean ± SD (n = 3). *p < 0.05 versus the H3K9me3 occupancy of the TB-31 group. (C) A proposed model to explain the role of BcDIM5 and BcHda1 in regulating ABA biosynthesis in B. cinerea TB-31.
Apart from the ABA gene cluster, we conducted an analysis of two other well-known virulence factors (Simon et al., 2013; Porquier et al., 2019), namely BOT (botrydial) and BOA (botcinic acid). RNA-seq data revealed that the expression levels of the BOA and BOT gene clusters remained unaltered in the ΔBchda1 strain. However, in the ΔBcdim5 strain, the expression levels of Bcboa3, Bcboa15, Bcboa16, Bcboa17 and the entire BOT gene cluster (Bcbot1-5) were downregulated. IGV showed increased H3K9me3 occupancy in the exon and promoter regions of the Bcboa3, Bcboa15, and Bcboa16 genes in ΔBcdim5 strain, which may be responsible for the down-regulation of expression (Figures 7A–D). Notably, the BOT gene cluster did not exhibit any significant change in H3K9me3 occupancy (Supplementary Figure S6), so we suggested that the knockdown of Bcdim5 indirectly led to the down-regulation of transcription levels in the whole cluster. Deletion of Bcdim5 and Bchda1 disrupted the normal post-translational modifications of some virulence factors (ABA, BOA), which also reflected the complexity of the regulatory network of virulence factors. Combined with the analysis of the above studies, we concluded that the ABA gene cluster is co-regulated by BcDIM5 and BcHda1, and they are essential for the normal distribution and maintenance of the B. cinerea TB-31 ABA gene cluster H3K9me3.
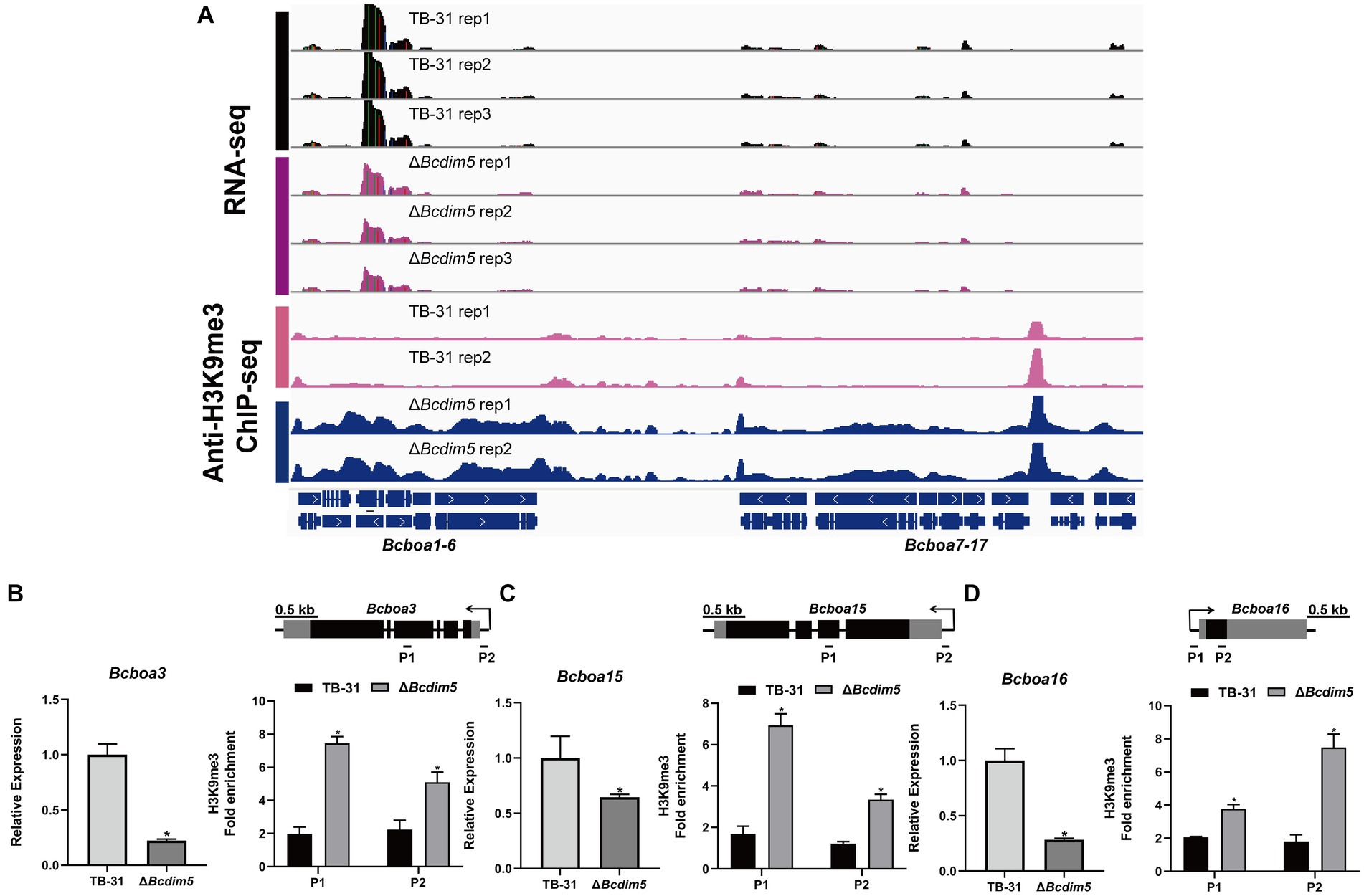
Figure 7. Reduced expression levels of the BOA gene cluster are associated with increased H3K9me3 occupancy. (A) Genome tracks of RNA-seq, ChIP-seq with anti-H3K9me3 for BOA gene cluster (Bcboa1-17) loci in the TB-31 and ΔBcdim5 strains. Structures of Bcboa1-17 are shown. (B) Bcboa3; (C) Bcboa15; (D) Bcboa16. [i in (B–D)] qRT-PCR analysis of Bcboa3, Bcboa15, Bcboa16 expression levels. The B. cinerea tubulin gene (BC1G_05600) was used as an internal control. The relative values for selected genes transcription at 6 days in TB-31 were assigned as 100%. Shown are means and SEM, n = 3 independent biological replicates. *p < 0.05 versus the same genes of the TB-31 group. [ii in (B–D)] ChIP-qPCR analysis of H3K9me3 status at the Bcboa3, Bcboa15, Bcboa16 loci. IgG was used as a negative control for comparison with IP to calculate the enrichment ploidy. Schematic genetic structures of Bcboa3, Bcboa15, Bcboa16, represented as black boxes for exons, gray boxes for 5′ and 3′ untranslated regions and black lines for promoters and introns. Amplified regions are indicated below each locus (P1, P2). Data are presented as the mean ± SD (n = 3). *p < 0.05 versus the H3K9me3 occupancy of the TB-31 group.
The effect of histone modification on the development and virulence of the common plant pathogenic fungus B. cinerea has been widely reported. However, the effect of this epigenetic modification on the synthesis of the virulence factor ABA in B. cinerea remains unclear. Here, we conducted an investigation on the H3K9 methyltransferase BcDIM5. Our findings indicate that knockdown of Bcdim5 led to a decrease in the overall expression level of the ABA gene cluster in B. cinerea TB-31, resulting in impaired ABA synthesis. Additionally, we identified a histone deacetylase BcHda1 that physically associates with BcDIM5, and both proteins play crucial roles in maintaining transcriptional silencing and normal distribution of H3K9me3 in B. cinerea TB-31. This work further extends our understanding of the complex regulatory network of ABA biosynthesis.
Deletion of the histone methyltransferase BcDIM5 has been reported to result in silencing of the pathogenic genes and to reduce the virulence of B. cinerea B05.10 (Zhang et al., 2016), which is consistent with the conclusions we reached. When BcDIM5 was knocked down, global H3K9me3 modification levels were reduced. As important virulence factors of B. cinerea, the reduced production of ABA and BOA in ΔBcdim5 will inevitably affect the virulence of the strain. In another study, Reyez-Dominguez and colleagues demonstrated that the loss of H3K9me3 caused a decrease in DON synthesis in F. graminearum, which hindered the transmission of the fungus from infected flowering wheat heads to neighboring spikelets (Etier et al., 2022). In the mango pathogen Fusarium mangiferae, the loss of H3K9 methyltransferase kmt1 almost completely inhibits the biosynthesis of the toxins fusapyrone and deoxyfusapyrone (Atanasoff-Kardjalieff et al., 2021). All these results indicate that H3K9 methyltransferase is closely related to virulence factors of strains. Therefore, we suggest that when H3K9 methyltransferase is deleted, rapid changes in the H3K9me3 modification pattern lead to a coordinated interaction between fungal development and virulence, affecting the normal post-translational modification (PTM) of some virulence factors.
Normal genome-wide H3K9me distribution is essential for both pathogens and symbionts in fungi–host interactions (Lai et al., 2022). In the present study we found that the genome-wide H3K9me3 occupancy distribution after Bchda1 and Bcdim5 loss showed significant defects, with 998 genes in the ΔBchda1 and 1,191 genes in the ΔBcdim5 showing at least a 1-fold decrease in H3K9me3 levels. Meanwhile RNA-seq and ChIP-seq data has revealed a significant intersection of DEGs and H3K9me3 differential peaks when Bchda1 and Bcdim5 were lost compared to TB-31. At least 193 genes were up-regulated in ΔBcdim5 and ΔBchda1 strains due to the loss or reduction of H3K9me3 modification, implying that BcHda1 and BcDIM5 would jointly maintain a normal distribution of H3K9me3 modifications.
Due to the versatile of the histone code, two different types of histone modification crosstalk have been proposed in yeast. The cis-effect is described as a “communication” of adjacent modifications within the same histone tails (Fischle et al., 2003). For example, in fission yeast, Clr4 has been shown to associate with the ubiquitin E3 ligase Cul4, forming the Clr4 methyltransferase complex (Oya et al., 2019). This complex preferentially ubiquitinates H3K14 and then promotes the methylation of H3K9. Trans-effects design cross-communications between different histone marks located within the same nucleosome (Fingerman et al., 2008). For example, H2BK123ub is essential for the deposition of H3K4me2/me3 and H3K79me2/me3, which in turn affects gene expression (van Welsem et al., 2018). In contrast, in the present study, H3K14ac did not change after Bcdim5 loss, indicating that Bcdim5 is not involved in the global regulation of H3K14ac modification. The enriched regions generated by H3K14ac modification after Bchda1 loss accounted for a very small proportion of the entire genome, with only 105 genes having altered H3K14ac occupancy. However, we found that global H3K14ac modification decreased when Bchda1 was overexpressed, suggesting that the function of its own deacetylase is only exercised when the normal distribution of H3K9me3 is steady. This may be a unique regulatory mechanism and is deserving of further research. In addition to ABA, BcDIM5 and BcHda1 also cause changes in transcription levels of other key enzymes of SM, most of which correspond to compounds not reported in the literature, which provides favorable support for the subsequent excavation of unknown SMs in B. cinerea.
In conclusion, our results identified a physical linkage between the H3K9 methyltransferase BcDIM5 and the H3K14 deacetylase BcHda1, revealing their regulatory roles in ABA biosynthesis. BcDIM5 and BcHda1 play a key role in the normal distribution of H3K9me3 occupancy and the stable synthesis of SMs in B. cinerea TB-31. To our knowledge, this is the first report on the involvement of histone modifications in B. cinerea ABA biosynthesis. This work provides a theoretical basis for further genetic improvement of high-yielding ABA strains and new inspiration for the discovery and production of unknown SMs in B. cinerea.
In our laboratory, the B. cinerea wild-type strain TBC-6 was isolated from wheat stems and leaves in southwest China (Tan et al., 1998). The ABA-hyper producing B. cinerea mutant TB-31 was generated from multiple rounds of mutagenesis and screening initiated from B. cinerea TBC-6 (Gong et al., 2014). Binary vectors pCBh1, pCBg1, and pCBsilent1 used in this study were constructed by our laboratory (Ding et al., 2015). After conidia maturation, Agrobacterium tumefaciens EHA105-mediated transformation (ATMT) was performed as previously described (Rolland et al., 2003). The detailed operation of ATMT, extracellular ABA quantification, quantitative RT-PCR, histone extraction analysis and DNA/RNA extraction were performed as previously described (Wei et al., 2022).
A list of all primers used to prepare transformants and RT-qPCR is shown in Supplementary Table S1.
Briefly, the knockout transformants were constructed using a double-combined PCR method (Yu et al., 2004). The upstream fragment and downstream fragment of Bcdim5 ORF were amplified with primer pairs Bcdim5-5-F1/−R1 and Bcdim5-3-F1/−R1, respectively. The hygromycin expression cassette fragment (PoliC::hph) was then amplified using the primer pair Hph-F1/Hph-R1. The knockout cassette was obtained by overlapping PCR with primer pair Bcdim5-5-F1/Bcdim5-3-R1, and then transformed into TB-31 protoplasts. The Bchda1 knockout mutant was constructed in the same way as above. The protoplasts were produced as previously described (Choquer et al., 2008). The construction of complement transformants and overexpression transformants (promoter: Aspergillus nidulans oliC) was carried out by ATMT transformation method to transfer target vectors into corresponding strains. Transformants were selected on PDA containing hygromycin (50 μg/mL) or glyphosate (100 μg/mL) by three rounds of subcultures with the same antibiotic selection.
Undifferentiated hyphae of B. cinerea TB-31, ΔBchda1, ΔBcdim5, Bchda1-OE-X24, Bcdim5-OE-X8 transformants were identified, and the hyphae were harvested for RNA extraction after dark cultivation on potato dextrose agar (PDA) plates at 25°C for 6 days. RNA quality was assessed on an Agilent 2,100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and checked using RNase free agarose gel electrophoresis. After total RNA was extracted, eukaryotic mRNA was enriched by Oligo (dT) beads. After total RNA was extracted, prokaryotic mRNA was enriched by removing rRNA by Ribo-ZeroTM Magnetic Kit (Epicentre, Madison, WI, USA). Then the enriched mRNA was fragmented into short fragments using fragmentation buffer and reversly transcribed into cDNA by using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB #7530, New England Biolabs, Ipswich, MA, USA). The purified double-stranded cDNA fragments were end repaired, A base added, and ligated to Illumina sequencing adapters. The ligation reaction was purified with the AMPure XP Beads (1.0X). Ligated fragments were subjected to size selection by agarose gel electrophoresis and poly merase chain reaction (PCR) amplified. The resulting cDNA library was sequenced using Illumina Novaseq 6000 by Gene Denovo Biotechnology Co. (Guangzhou, China).
Chromatin immunoprecipitation (ChIP) was performed as previously described with some necessary modification (Liu et al., 2019). B. cinerea strains were grown on potato dextrose agar (PDA) slants at 25°C for 6 days. Mycelia were collected in filtered and clarified 1xPDB liquid medium with 1% (v/v) formaldehyde. Crosslinking was performed at 25°C and 90 rpm for 15 min, then 125 mM glycine was added to terminate fixation, and incubated at room temperature for 5 min after mixing. Subsequently, the treated samples were rinsed with water, frozen and ground in liquid nitrogen for chromatin isolation. Anti-H3K9me3 (Active Motif) and anti-H3K14ac (Active Motif) were used for immunoprecipitation into the chromatin. Immunoprecipitated DNA was used for ChIP followed by sequencing (ChIP-seq) DNA library preparation or ChIP-qPCR analysis.
ChIP analysis was carried out following the previously provided protocol by Wuhan IGENEBOOK Biotechnology Co., Ltd.1 (Gendrel et al., 2005). ChIP-seq libraries were sequenced on the Illumina NovaSeq 6000 platform. Raw Data, processed using the software FastQC (version: 0.11.5) for quality control (de Sena Brandine and Smith, 2019). For the annotation of genomic distribution of peaks, we divided the genome into five fragments: promoter [the 2-kb region upstream of the transcription start site (TSS)], exon, intron, transcription termination site (TTS) [the 1-kb region downstream of the transcription end site (TES)] and Intergenic (outside of these regions).
ChIP-qPCR was analyzed by Fold Enrichment method. The CT value of IgG was used as a negative control for comparison with IP to calculate the fold enrichment (Zhuang et al., 2022). The abundance of immunoprecipitated chromatin was determined by qRT-PCR using the primers given in Supplementary Table S1. The data are presented as mean ± SD of three biological replicates.
The GST and GST-tag fusion protein were purified by pGEX-6P-1 vector. Use pET28a (+) vector to purify His-tag fusion protein. The constructed vectors were then transferred into E. coli Rosetta for expression. The expression of GST and GST-BcDIM5 proteins was induced at 16°C for 24 h (0.5 mM IPTG). The expression of BcHda1 protein was induced at 22°C for 16 h (0.5 mM IPTG). 500 μg purified GST and GST fusion protein were incubated with GST-Tag resin at 4°C for 4 h. Add the interaction His-tag fusion protein 500 μg and rotate overnight at 4°C. The protein was washed with elution buffer at least 7 times, and then use a small amount of eluting buffer to elute the protein for subsequent Western Blot.
To confirm interactions, yeast-two-hybrid analyses were applied using the Matchmaker® Gold Yeast-Two-Hybrid System (Takara Bio USA, Inc.) according to manufacturer’s instructions. Bcdim5 was inserted into the pGBKT7 bait vector, Bchda1 was inserted into the pGADT7 prey vector. Bait and prey constructs were transformed into Y2H Gold and Y187 strains as described in instruction for the Matchmaker system, respectively.
Data were expressed as the mean ± SD. Differences among different treatments were compared using SPSS v.16.0 (SPSS Inc., Chicago, IL, United States). p-values were calculated using Fisher’s exact test for overlapping using online tools to determine the significance of the overlap of two gene sets.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material. The data presented in the study are deposited in the “Science Data Bank” repository, accession number: 10.57760/sciencedb.09674.
ZW: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. DS: Funding acquisition, Writing – review & editing. XH: Data curation, Software, Writing – review & editing. TL: Data curation, Software, Writing – review & editing. ZL: Investigation, Project administration, Writing – review & editing. DL: Project administration, Writing – review & editing. JY: Validation, Writing – review & editing. HT: Funding acquisition, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research received financial support from the National Natural Science Foundation of China (32070059). We acknowledge funding from the Open Research Fund of Key Laboratory of Environmental and Applied Microbiology, Chengdu Institute of Biology, Chinese Academy of Sciences (KLEAMCAS201901).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1339576/full#supplementary-material
Amselem, J., Cuomo, C. A., van Kan, J. A., Viaud, M., Benito, E. P., Couloux, A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. doi: 10.1371/journal.pgen.1002230
Atanasoff-Kardjalieff, A. K., Lünne, F., Kalinina, S., Strauss, J., and Studt, L. (2021). Biosynthesis of Fusapyrone depends on the H3K9 methyltransferase, FmKmt1, in fusarium mangiferae. Front. Fungal Biol. 2:671796. doi: 10.3389/ffunb.2021.671796
Beisel, C., and Paro, R. (2011). Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 12, 123–135. doi: 10.1038/nrg2932
Bind, S., Bind, S., Sharma, A. K., and Chaturvedi, P. (2022). Epigenetic modification: a key tool for secondary metabolite production in microorganisms. Front. Microbiol. 13:784109. doi: 10.3389/fmicb.2022.784109
Brosch, G., Loidl, P., and Graessle, S. (2008). Histone modifications and chromatin dynamics: a focus on filamentous fungi. FEMS Microbiol. Rev. 32, 409–439. doi: 10.1111/j.1574-6976.2007.00100.x
Buker, S. M., Iida, T., Bühler, M., Villén, J., Gygi, S. P., Nakayama, J., et al. (2007). Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat. Struct. Mol. Biol. 14, 200–207. doi: 10.1038/nsmb1211
Chen, X., Wu, L., Lan, H., Sun, R., Wen, M., Ruan, D., et al. (2022). Histone acetyltransferases MystA and MystB contribute to morphogenesis and aflatoxin biosynthesis by regulating acetylation in fungus aspergillus flavus. Environ. Microbiol. 24, 1340–1361. doi: 10.1111/1462-2920.15856
Choquer, M., Robin, G., Pêcheur, P., Giraud, C., Levis, C., and Viaud, M. (2008). Ku70 or Ku80 deficiencies in the fungus Botrytis cinerea facilitate targeting of genes that are hard to knock out in a wild-type context. FEMS Microbiol. Lett. 289, 225–232. doi: 10.1111/j.1574-6968.2008.01388.x
de Sena Brandine, G., and Smith, A. D. (2019). Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res 8:1874. doi: 10.12688/f1000research.21142.2
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Ding, Z. T., Zhang, Z., Luo, D., Zhou, J. Y., Zhong, J., Yang, J., et al. (2015). Gene overexpression and RNA silencing tools for the genetic manipulation of the S-(+)-abscisic acid producing ascomycete Botrytis cinerea. Int. J. Mol. Sci. 16, 10301–10323. doi: 10.3390/ijms160510301
Ding, Z., Zhang, Z., Zhong, J., Luo, D., Zhou, J., Yang, J., et al. (2016). Comparative transcriptome analysis between an evolved abscisic acid-overproducing mutant Botrytis cinerea TBC-A and its ancestral strain Botrytis cinerea TBC-6. Sci. Rep. 6:37487. doi: 10.1038/srep37487
Etier, A., Dumetz, F., Chéreau, S., and Ponts, N. (2022). Post-translational modifications of histones are versatile regulators of fungal development and secondary metabolism. Toxins (Basel) 14:317. doi: 10.3390/toxins14050317
Fingerman, I. M., Du, H. N., and Briggs, S. D. (2008). Controlling histone methylation via trans-histone pathways. Epigenetics 3, 237–242. doi: 10.4161/epi.3.5.6869
Fischle, W., Wang, Y., and Allis, C. D. (2003). Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15, 172–183. doi: 10.1016/s0955-0674(03)00013-9
Gendrel, A. V., Lippman, Z., Martienssen, R., and Colot, V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213–218. doi: 10.1038/nmeth0305-213
Gessaman, J. D., and Selker, E. U. (2017). Induction of H3K9me3 and DNA methylation by tethered heterochromatin factors in Neurospora crassa. Proc. Natl. Acad. Sci. USA 114, E9598–e9607. doi: 10.1073/pnas.1715049114
Gong, T., Shu, D., Zhao, M., Zhong, J., Deng, H. Y., and Tan, H. (2014). Isolation of genes related to abscisic acid production in Botrytis cinerea TB-3-H8 by cDNA-AFLP. J. Basic Microbiol. 54, 204–214. doi: 10.1002/jobm.201200311
Gu, Q., Ji, T., Sun, X., Huang, H., Zhang, H., Lu, X., et al. (2017). Histone H3 lysine 9 methyltransferase FvDim5 regulates fungal development, pathogenicity and osmotic stress responses in fusarium verticillioides. FEMS Microbiol. Lett. 364:fnx184. doi: 10.1093/femsle/fnx184
Janevska, S., Güldener, U., Sulyok, M., Tudzynski, B., and Studt, L. (2018). Set1 and Kdm5 are antagonists for H3K4 methylation and regulators of the major conidiation-specific transcription factor gene ABA1 in fusarium fujikuroi. Environ. Microbiol. 20, 3343–3362. doi: 10.1111/1462-2920.14339
Lai, Y., Wang, L., Zheng, W., and Wang, S. (2022). Regulatory roles of histone modifications in filamentous fungal pathogens. J. Fungi (Basel) 8:565. doi: 10.3390/jof8060565
Lievens, L., Pollier, J., Goossens, A., Beyaert, R., and Staal, J. (2017). Abscisic acid as pathogen effector and immune regulator. Front. Plant Sci. 8:587. doi: 10.3389/fpls.2017.00587
Liu, Z., Jian, Y., Chen, Y., Kistler, H. C., He, P., Ma, Z., et al. (2019). A phosphorylated transcription factor regulates sterol biosynthesis in fusarium graminearum. Nat. Commun. 10:1228. doi: 10.1038/s41467-019-09145-6
Oya, E., Nakagawa, R., Yoshimura, Y., Tanaka, M., Nishibuchi, G., Machida, S., et al. (2019). H3K14 ubiquitylation promotes H3K9 methylation for heterochromatin assembly. EMBO Rep. 20:e48111. doi: 10.15252/embr.201948111
Porquier, A., Moraga, J., Morgant, G., Dalmais, B., Simon, A., Sghyer, H., et al. (2019). Botcinic acid biosynthesis in Botrytis cinerea relies on a subtelomeric gene cluster surrounded by relics of transposons and is regulated by the Zn(2)Cys(6) transcription factor BcBoa13. Curr. Genet. 65, 965–980. doi: 10.1007/s00294-019-00952-4
Rolland, S., Jobic, C., Fèvre, M., and Bruel, C. (2003). Agrobacterium-mediated transformation of Botrytis cinerea, simple purification of monokaryotic transformants and rapid conidia-based identification of the transfer-DNA host genomic DNA flanking sequences. Curr. Genet. 44, 164–171. doi: 10.1007/s00294-003-0438-8
Sakthivel, P., Sharma, N., Klahn, P., Gereke, M., and Bruder, D. (2016). Abscisic acid: a Phytohormone and mammalian cytokine as novel Pharmacon with potential for future development into clinical applications. Curr. Med. Chem. 23, 1549–1570. doi: 10.2174/0929867323666160405113129
Shabani, M., Soti, M., Ranjbar, H., and Naderi, R. (2023). Abscisic acid ameliorates motor disabilities in 6-OHDA-induced mice model of Parkinson's disease. Heliyon 9:e18473. doi: 10.1016/j.heliyon.2023.e18473
Shan, C. M., Kim, J. K., Wang, J., Bao, K., Sun, Y., Chen, H., et al. (2021). The histone H3K9M mutation synergizes with H3K14 ubiquitylation to selectively sequester histone H3K9 methyltransferase Clr4 at heterochromatin. Cell Rep. 35:109137. doi: 10.1016/j.celrep.2021.109137
Simon, A., Dalmaislenaers, B., Morgant, G., and Viaud, M. (2013). Screening of a Botrytis cinerea one-hybrid library reveals a Cys(2)his(2) transcription factor involved in the regulation of secondary metabolism gene clusters. Fungal Genet. Biol. 52, 9–19. doi: 10.1016/j.fgb.2013.01.006
Singh, A., and Roychoudhury, A. (2023). Abscisic acid in plants under abiotic stress: crosstalk with major phytohormones. Plant Cell Rep. 42, 961–974. doi: 10.1007/s00299-023-03013-w
Song, Z. T., Sun, L., Lu, S. J., Tian, Y., Ding, Y., and Liu, J. X. (2015). Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc. Natl. Acad. Sci. USA 112, 2900–2905. doi: 10.1073/pnas.1419703112
Stec, N., Banasiak, J., and Jasiński, M. (2016). Abscisic acid – an overlooked player in plant-microbe symbioses formation? Acta Biochim. Pol. 63, 53–58. doi: 10.18388/abp.2015_1210
Tan, H., Ge, G., Zhidong, L. I., Shulin, P., Baoliang, L., Amp, L. D., et al. (1998). High yield ABA producing strain obtained by UV irradiation of protoplasts. Chin. J. App. Environ. Biol. 3, 281–285.
Tang, G., Yuan, J., Wang, J., Zhang, Y. Z., Xie, S. S., Wang, H., et al. (2021). Fusarium BP1 is a reader of H3K27 methylation. Nucleic Acids Res. 49, 10448–10464. doi: 10.1093/nar/gkab844
Torres-Garcia, S., Yaseen, I., Shukla, M., Audergon, P., White, S. A., Pidoux, A. L., et al. (2020). Epigenetic gene silencing by heterochromatin primes fungal resistance. Nature 585, 453–458. doi: 10.1038/s41586-020-2706-x
van Welsem, T., Korthout, T., Ekkebus, R., Morais, D., Molenaar, T. M., van Harten, K., et al. (2018). Dot1 promotes H2B ubiquitination by a methyltransferase-independent mechanism. Nucleic Acids Res. 46, 11251–11261. doi: 10.1093/nar/gky801
Wang, L., Liu, J., Li, X., Lyu, X., Liu, Z., Zhao, H., et al. (2022). A histone H3K9 methyltransferase Dim5 mediates repression of sorbicillinoid biosynthesis in Trichoderma reesei. Microb. Biotechnol. 15, 2533–2546. doi: 10.1111/1751-7915.14103
Wang, L., Zhang, F., Rode, S., Chin, K. K., Ko, E. E., Kim, J., et al. (2017). Ethylene induces combinatorial effects of histone H3 acetylation in gene expression in Arabidopsis. BMC Genomics 18:538. doi: 10.1186/s12864-017-3929-6
Wang, Y., Zhou, J., Zhong, J., Luo, D., Li, Z., Yang, J., et al. (2018). Cys(2)his(2) zinc finger transcription factor BcabaR1 positively regulates abscisic acid production in Botrytis cinerea. Appl. Environ. Microbiol. 84:e00920. doi: 10.1128/aem.00920-18
Wei, Z., Shu, D., Sun, Q., Chen, D. B., Li, Z. M., Luo, D., et al. (2022). The BcLAE1 is involved in the regulation of ABA biosynthesis in Botrytis cinerea TB-31. Front. Microbiol. 13:969499. doi: 10.3389/fmicb.2022.969499
Wiles, E. T., and Selker, E. U. (2017). H3K27 methylation: a promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 43, 31–37. doi: 10.1016/j.gde.2016.11.001
Yu, J. H., Hamari, Z., Han, K. H., Seo, J. A., Reyes-Domínguez, Y., and Scazzocchio, C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. doi: 10.1016/j.fgb.2004.08.001
Zehra, A., Raytekar, N. A., Meena, M., and Swapnil, P. (2021). Efficiency of microbial bio-agents as elicitors in plant defense mechanism under biotic stress: a review. Curr. Res. Microb. Sci. 2:100054. doi: 10.1016/j.crmicr.2021.100054
Zhang, X., Liu, X., Zhao, Y., Cheng, J., Xie, J., Fu, Y., et al. (2016). Histone H3 lysine 9 methyltransferase DIM5 is required for the development and virulence of Botrytis cinerea. Front. Microbiol. 7:1289. doi: 10.3389/fmicb.2016.01289
Zhang, X., Noberini, R., Vai, A., Bonaldi, T., Seidl, M. F., and Collemare, J. (2023). Detection and quantification of the histone code in the fungal genus aspergillus. Fungal Genet. Biol. 167:103800. doi: 10.1016/j.fgb.2023.103800
Keywords: Botrytis cinerea, H3K9me3, abscisic acid, secondary metabolism, gene regulation
Citation: Wei Z, Shu D, Hou X, Li T, Li Z, Luo D, Yang J and Tan H (2024) Normal distribution of H3K9me3 occupancy co-mediated by histone methyltransferase BcDIM5 and histone deacetylase BcHda1 maintains stable ABA synthesis in Botrytis cinerea TB-31. Front. Microbiol. 15:1339576. doi: 10.3389/fmicb.2024.1339576
Received: 21 November 2023; Accepted: 02 February 2024;
Published: 04 March 2024.
Edited by:
XueHong Zhang, Shanghai Jiao Tong University, ChinaReviewed by:
Kunlong Yang, Jiangsu Normal University, ChinaCopyright © 2024 Wei, Shu, Hou, Li, Li, Luo, Yang and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Shu, c2h1ZGFuQGNpYi5hYy5jbg==; Hong Tan, YWJhdGhAY2liLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.