
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 29 January 2024
Sec. Extreme Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1338669
This article is part of the Research TopicAcidophile Microbiology: From Extreme Environments to Biotechnological ApplicationsView all 19 articles
Tetrathionate hydrolase (TTH) is a unique enzyme found in acidophilic sulfur-oxidizing microorganisms, such as bacteria and archaea. This enzyme catalyzes the hydrolysis of tetrathionate to thiosulfate, elemental sulfur, and sulfate. It is also involved in dissimilatory sulfur oxidation metabolism, the S4-intermediate pathway. TTHs have been purified and characterized from acidophilic autotrophic sulfur-oxidizing microorganisms. All purified TTHs show an optimum pH in the acidic range, suggesting that they are localized in the periplasmic space or outer membrane. In particular, the gene encoding TTH from Acidithiobacillus ferrooxidans (Af-tth) was identified and recombinantly expressed in Escherichia coli cells. TTH activity could be recovered from the recombinant inclusion bodies by acid refolding treatment for crystallization. The mechanism of tetrathionate hydrolysis was then elucidated by X-ray crystal structure analysis. Af-tth is highly expressed in tetrathionate-grown cells but not in iron-grown cells. These unique structural properties, reaction mechanisms, gene expression, and regulatory mechanisms are discussed in this review.
Tetrathionate (S4O62−) is a type of polythionate (SnO62−: n = 3–6) produced by the reaction of hydrogen sulfide with sulfite under acidic conditions, such as sulfur hot springs, acidic soil, and acid mine drainage (Karchmer, 1970; Williamson and Rimstidt, 1993). This compound is also produced by the oxidation of pyrite and other sulfide ores during abiotic and biological processes. Tetrathionate is soluble in water, in contrast to elemental sulfur, and is relatively more stable than thiosulfate at low pH, making it a common reduced inorganic sulfur compound (RISCs) in acidic environments. Therefore, tetrathionate is a suitable growth substrate for acidophilic sulfur-oxidizing microorganisms. Tetrathionate oxidation was first reported in the acidophilic iron- and sulfur-oxidizing bacterium Acidithiobacillus ferrooxidans (formerly Thiobacillus ferrooxidans) in the 1970s (Eccleston and Kelly, 1978).
Investigation of bacterial sulfur oxidation has found two major pathways for RISCs oxidation. In both pathways, thiosulfate is an important intermediate and a growth substrate for sulfur-oxidizing bacteria. The Sox system, one of the best-characterized pathways, has been investigated in the neutrophilic sulfur bacterium Paracoccus pantotrophus (Friedrich et al., 2001). Sox systems and their related genes are widespread among sulfur-oxidizing bacteria, including both photo- and chemoautotrophs (Hensen et al., 2006; Sato et al., 2012). The complete Sox system in P. pantotrophus contains four components: SoxXA, SoxYZ, SoxB, and Sox(CD)2. SoxXA catalyzes the oxidation of thiosulfate to generate a SoxY-cysteine-sulfhydryl group in the SoxYZ complex. SoxY acts as a thiosulfate carrier. SoxB hydrolyses the terminal sulphonate of the SoxYZ complex. Sox(CD)2 oxidizes the terminal sulfane sulfur of SoxY-cysteine persulfide into cysteine-S-sulfate. This system has been previously reviewed and summarized (Friedrich et al., 2005; Ghosh and Dam, 2009). Alternatively, the thiosulfate oxidation pathway (the S4I pathway) involves tetrathionate formation as an intermediate and is mainly observed in acidophilic sulfur-oxidizing bacteria and archaea (Figure 1). Whole-genome analysis indicates that only the S4I pathway is detected in the acidophilic sulfur-oxidizing bacterium A. ferrooxidans (Quatrini et al., 2009), even though both the Sox system and S4I pathway are detected in the genomes of other Acidithiobacillus spp., such as A. caldus (Mangold et al., 2011) and A. thiooxidans (Yin et al., 2014). This suggests that A. ferrooxidans mainly utilizes the S4I pathway for dissimilatory sulfur oxidation. In this pathway, two thiosulfate molecules are oxidized to tetrathionate by thiosulfate: quinone oxidoreductase (TQO) (Müller et al., 2004) or thiosulfate dehydrogenase (TSD) (Kikumoto et al., 2013). The membrane-bound TQO or periplasmic TSD take electrons from the oxidation of thiosulfate to the electron transport chain to produce energy or reducing power for CO2 fixation. Tetrathionate, a product of thiosulfate oxidation, is further metabolized by tetrathionate hydrolase (TTH). This enzyme has a unique primary structure (Buonfiglio et al., 1999) and its activity has only been detected in sulfur-oxidizing microorganisms using the S4I pathway for dissimilatory sulfur oxidation. TTH plays an important role in tetrathionate metabolism, not only in the S4I pathway, but also in the utilization of tetrathionate as a growth substrate. Here, the unique properties of TTHs are discussed to understand microbial sulfur metabolism and sulfur chemistry.
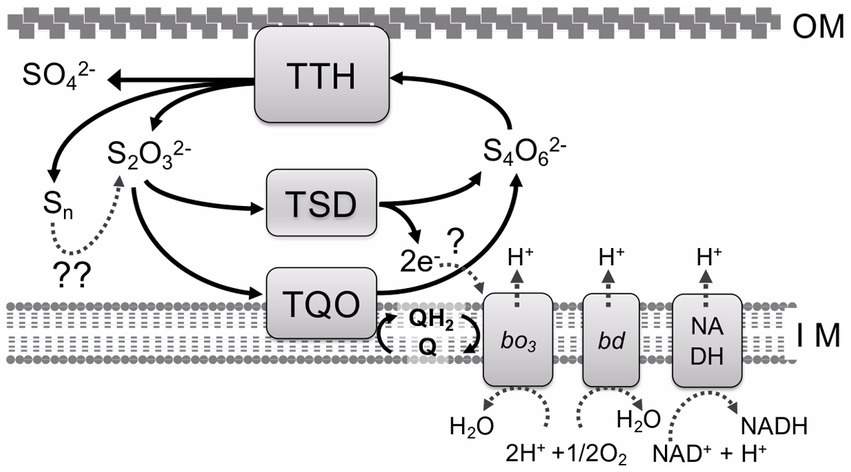
Figure 1. S4I pathway for tetrathionate and thiosulfate oxidation in A. ferrooxidans. TTH, tetrathionate hydrolase; TSD, thiosulfate dehydrogenase; TQO, thiosulfate:quinone oxidoreductase; Q, QH2, quinone pool; bd, bo3, terminal oxidases; NADH, NADH dehydrogenase complex I; OM, outer membrane; IM, inner membrane;?, electron acceptor;??, enzymatic and/or abiotic reactions.
It is well established that the acidophilic bacteria genus Thiobacillus can oxidize tetrathionate as an energy source. TTH was first purified and characterized in 1996 as a tetrathionate-decomposing enzyme from A. ferrooxidans Funis 2–1 (Sugio et al., 1996) and A. thiooxidans ON107 (Tano et al., 1996) (formerly Thiobacillus ferrooxidans and Thiobacillus thiooxidans, respectively). Although TTHs have been purified from Acidiphilium acidophilum (de Jong et al., 1997a) (formerly Thiobacillus acidophilus), A. ferrooxidans ATCC19859 (de Jong et al., 1997b), and A. caldus (Bugaytsova and Lindström, 2004), no genetic information on TTHs from sulfur-oxidizing bacteria was reported at that time. All TTHs described above show the maximum activity at a pH range of 2.5–4.0, which suggests that they are localized in the periplasm or on the outer membrane; different diffraction assays identified TTH from A. caldus as a periplasmic protein (Bugaytsova and Lindström, 2004). The properties of the TTHs from various species are summarized in Table 1. Kanao et al. (2007) purified TTH from the A. ferrooxidans type strain ATCC23270 and explored the N-terminal amino acid sequence of the enzyme using a BLAST search, finding that AFE_0029 encodes the N-terminal amino acid sequence. Although heterologous expression of the gene in Escherichia coli resulted in the formation of inclusion bodies in an inactive form, polyclonal antibodies against the recombinant protein clearly recognized the purified native TTH, indicating that the gene was immunologically identified as encoding TTH. Thus, AFE_0029 was named Af-tth (Kanao et al., 2007). The TTH gene from A. ferrooxidans is called tetH in several papers, however, tet is widely used for tetracycline-resistance genes, such as tetK, tetL, tetM, and tetO (Zilhao et al., 1988); to avoid confusion, the TTH gene will be termed Af-tth here. Interestingly, Af-tth has already been registered as a sulfur-regulated outer membrane protein from A. ferrooxidans MSR in the database but the function and physiological role of the gene product has not been elucidated (Buonfiglio et al., 1999). As the deduced amino acid sequence does not show similarity to any other protein in the database, this gene is reported as a novel gene encoding a sulfur-regulated outer membrane protein (Buonfiglio et al., 1999). Although Af-Tth is localized on the outer membrane (peripheral protein), it is detected in the culture medium supernatant, suggesting that Af-Tth acts in the extracellular space to generate hydrophilic sulfur globules (Beard et al., 2011). TTH genes from A. caldus (Ac-tetH) (Rzhepishevska et al., 2007) and the thermoacidophilic archaeon Acidianus ambivalens (Ad-tth1) (Protze et al., 2011) have been experimentally identified based on the amino acid sequences of purified TTHs and whole genome sequencing. The amino acid sequences of Af-tth, Ac-tetH, and Ad-tth1 are similar and encode proteins of the pyrroloquinoline quinone (PQQ)-containing protein family (Pfam family (El-Gebali et al., 2019): PQQ_2). Inactive recombinant Af-Tth can be obtained in inclusion bodies from E. coli host cells and successfully refolded in an active form under acidic conditions without PQQ or other cofactors (Kanao et al., 2010). This provides direct evidence that Af-tth encodes TTH in A. ferrooxidans and that the enzyme does not require any cofactors for the reaction. In contrast to Af-Tth, quinoid compounds have been detected in Ac-TetH (Rzhepishevska et al., 2007). Comparison of Ad-TTH1 with the quinoprotein alcohol dehydrogenase from Pseudomonas putida showed that the heme c domain and conserved PQQ-binding site residues are missing, including disulfide-forming cysteines and calcium-coordinating amino acids (Protze et al., 2011). Both Ad-TTH1 and Af-Tth do not require PQQ cofactor for the reaction. Although Ad-TTH1 activity is detected in the pseudo-periplasmic fraction, TTH is secreted extracellularly to form zipper-like particles in the closely related thermoacidophilic archaeon Acidianus hospitalis (Krupovic et al., 2012). Kanao et al. (2018) identified the TTH gene from the marine acidophilic Acidithiobacillus sp. strain SH (SH-tth) using partial purification and internal peptide sequences of the protein. Although recombinant SH-Tth can be synthesized in inclusion bodies during E. coli heterologous expression, TTH activity is successfully recovered by acidic refolding. Interestingly, SH-Tth is activated in the presence of 1 M NaCl. This indicates that the enzyme is from a marine bacterium.
Af-tth expression differs depending on the growth substrate used. Quantitative RT-PCR analysis of total RNA from A. ferrooxidans cells grown on ferrous iron (Fe2+), elemental sulfur (S0), and tetrathionate (S4) showed the Af-tth expression ratios of S0/Fe2+ and S4/Fe2+ are 68 ± 21 and 181 ± 5, respectively (Kanao et al., 2023). This result suggests that gene expression is upregulated in cells grown on elemental sulfur and tetrathionate, but not on ferrous iron. The Ac-tetH expression ratio of S4/S0 is 233.5 ± 134.0 (Rzhepishevska et al., 2007). Ac-tetH from A. caldus is also highly expressed in cells grown on tetrathionate compared to cells grown on elemental sulfur. TTH activity is detected in tetrathionate-grown cells, but not in elemental sulfur-grown cells of Ad. ambivalens. Northern blot hybridization analysis supports the enzyme activity results, which show a much stronger signal of Ad-tth1 in tetrathionate-grown cells and a faint signal in elemental sulfur-grown cells. This suggests that Ad-tth1 transcription is also upregulated depending on the growth substrate, similar to Ac-tetH expression in A. caldus.
The Ac-tetH gene cluster contains two component system-related genes (rsrS and rsrR) and a TQO-coding gene (doxD) (Rzhepishevska et al., 2007). Gene expression is regulated by a two-component system called the RsrS-RsrR system (Wang et al., 2016). The relative transcriptional levels of Ac-tetH in rsrR or rsrS knockout mutants are much lower than those in the wild-type when stimulated with potassium tetrathionate, indicating a positive effect of RsrS-RsrR on the transcriptional regulation of Ac-tetH. However, no rsrS and rerR disruption effects are observed during elemental sulfur growth. Furthermore, Ac-tetH deficient mutant strains cannot grow on tetrathionate (van Zyl et al., 2008). These results suggest that Ac-TetH is the sole enzyme that metabolizes tetrathionate in the S4I pathway and that the Sox system may function in the elemental sulfur-grown cells of this bacterium.
A stoichiometric model of RISCs oxidation has been proposed for A. thiooxidans (Bobadilla Fazzini et al., 2013). According to this model, although tetrathionate is generated by the TQO (DoxDA) reaction from thiosulfate, it is chemically reduced to elemental sulfur and thiosulfate in sulfur-grown cells. Thus, no metabolic flux by way of TTH is expected, and the Sox system mainly functions during elemental sulfur growth. A similar flux is observed, assuming the presence of elemental sulfur in the TTH reaction, even in cells grown on tetrathionate. When TTH without sulfur formation functions in tetrathionate-grown cells, sulfur oxidation occurs mainly via the S4I pathway, and TTH plays a central role in A. thiooxidans.
Two-component transcriptional regulation systems (RegB/RegA) have been previously investigated in A. ferrooxidans (Moinier et al., 2017). According to this study, the −12/24 element and predicted integration host factor (IHF) binding sites are also proposed. IHF is expected to bind to the region upstream of the −12/−24 element and regulate gene expression with a sigma-54 dependent transcriptional response regulator (AFE_0027). Yu et al. (2014) constructed Af-tth knockout and overexpression strains and showed that the Af-tth knockout strain could not grow on tetrathionate medium, suggesting that Af-Tth is the sole enzyme responsible for tetrathionate hydrolysis. However, cell growth of the knockout mutant was observed on elemental sulfur with a slightly lower cell yield than that of the wild-type. Genes related to Sox systems could not be assigned to the whole-genome sequence of A. ferrooxidans. Some sulfur oxidation systems unrelated to the S4I pathway or Sox system should exist in this bacterium. In contrast, the highest and lowest growth yields were observed in the Af-tth overexpression strain on tetrathionate and elemental sulfur, respectively. Thus, Af-Tth influences sulfur metabolism to some extent in A. ferrooxidans.
Zhang et al. (2014) identified the Ac-tetH promoter (PtetH) and constructed an expression vector utilizing PtetH and containing the eGFP gene to confirm the functionality of PtetH. Although recombinant E. coli harboring the expression vector emitted fluorescence regardless of the presence of tetrathionate, eGFP fluorescence was observed following tetrathionate induction in recombinant A. caldus. These results suggest that PtetH is constitutively active in E. coli host cells and is inducible in native host cells. For the Af-tth promoter (Ptth), Kanao et al. (2023) constructed the expression vector pMPJC with exogenous gfp under Ptth control using A. ferrooxidans host cells. Green fluorescence was observed in recombinant A. ferrooxidans grown on tetrathionate, but not in recombinant E. coli distinct from the PtetH expression vector. This suggests that Ptth does not function in E. coli and is suitable for the in vivo amplification of its expression vector, even if it contains genes encoding proteins that are toxic to E. coli host cells.
Heterologous recombinant gene expression using E. coli is a valid and profitable strategy to investigate the gene products of A. ferrooxidans; functional recombinant gene products can provide direct evidence of gene function. However, owing to differential growth conditions, particularly pH, it is frequently impossible to obtain a recombinant protein in its functional form. Recombinant Af-Tth is synthesized in inclusion bodies in an inactive form in E. coli cells. Kanao et al. (2023) examined homologous expression of Af-tth modifying His-tag codon using the endogenous promoter Ptth in A. ferrooxidans. The His-tag-modified Af-Tth was successfully obtained from native cells in its active form. The recombinant enzyme was easily purified by metal cation affinity column chromatography and the specific activity (2.2 ± 0.37 U·mg−1) was equivalent to the refolded Af-Tth (2.5 ± 0.18 U·mg−1).
A catalytic reaction for TTH has been proposed in experiments with intact A. ferrooxidans cells (Steudel et al., 1987; Hazeu et al., 1988). The first suggested reaction is:
This tetrathionate hydrolysis reaction has been proposed for an enzyme purified from A. ferrooxidans (Beard et al., 2011). One molecule of tetrathionate is hydrolyzed by Af-Tth to generate disulfane monosulfonic acid (DSMSA) and sulfuric acid (sulfate). DSMSA is a key intermediate in the biological oxidation of pyrite (Schippers et al., 1996). As DSMSA is highly reactive, several abiotic reactions may occur spontaneously. For example, four DSMSA molecules can elongate to form elemental sulfur (S8) and sulfurous acid (sulfite). Consequently, polythionates, S8, and sulfites are the final products (Pronk et al., 1990; Wentzien et al., 1994). The second reaction (Meulenberg et al., 1992) suggests that one tetrathionate molecule is hydrolyzed to thiosulfate, sulfur, and sulfate in equimolar amounts by TTH according to the following equation:
Reaction estimations using purified enzymes of A. thiooxidans (Tano et al., 1996) and the A. ferrooxidans strain Funis 2–1 (Sugio et al., 1996) are:
However, this equation is based on the ratio of tetrathionate consumption to thiosulfate production. Although the reaction mixture is turbid owing to the generation of elemental sulfur globules, elemental sulfur does not take into account the TTH enzymatic reaction product but was considered a secondary abiotic reaction product. HPLC analysis of Ac-TetH reaction products by Bugaytsova and Lindström (2004) demonstrated the presence of thiosulfate and pentathionate, suggesting the reaction:
Concerning the Af-Tth reaction, Kanao et al. (2007) detected the turbidity of the reaction mixture by elemental sulfur production, and the same level of thiosulfate production accompanying tetrathionate consumption was detected using the ion-pair HPLC method. Except for thiosulfate, no other polythionate peaks, such as tri-, penta-, or hexathionate peaks, were detected in the reaction mixture after the complete consumption of tetrathionate. This result suggests that the overall Af-Tth reaction is the same as Reaction (2). Because sulfur atoms (S0) are unstable, Reaction (2) should be repeated eight times to produce S8 as a stable end product, according to the following equation:
Determining the specific reaction of the enzyme is difficult because tetrathionate hydrolysis in Acidithiobacillus generates multiple products with chemical interactions between thiosulfate, sulfane sulfur chemicals, polythionate, sulfate, and possibly S8 at high sulfate concentrations and low pH. In addition, Af-Tth exhibits a unique primary structure, and the catalytic reaction is unprecedented. Therefore, the precise whole reaction and reaction mechanism of TTH remain unknown.
Cysteine residues in the catalytic center play important roles in many other enzymes involved in dissimilatory sulfur oxidation, including sulfide:quinone oxidoreductase (Cherney et al., 2010), sulfur oxygenase reductase (Veith et al., 2011), and thiosulfate dehydrogenase (Brito et al., 2015). Although cysteine residues are normally conserved in each enzyme among different sulfur-oxidizing microorganisms, only one cysteine residue (Cys301) can be detected in the primary structure of Af-Tth and is not conserved among TTHs from Acidithiobacillus spp. (Kanao et al., 2014). Moreover, the site-specific variant (Af-Tth C301A) exhibits similar activity and dimeric formation. These results suggest that the sole cysteine residue (Cys301) in Af-Tth is not involved in the tetrathionate hydrolysis reaction or subunit assembly, indicating a novel cysteine-independent reaction mechanism for this enzyme.
Recombinant Af-Tth crystals (hexagonal cylinder with dimensions of 0.2 mm × 0.05 mm × 0.05 mm) have been successfully obtained (Kanao et al., 2013) and the X-ray crystallographic results reported (Kanao et al., 2021). Af-Tth is a homodimer and its monomer structure exhibits an eight-bladed beta-propeller motif, as determined by X-ray crystallography at 1.95 Å resolution. Two insertion loops participate in dimerization, and one loop forms a cavity with the beta-propeller region (Figure 2). Analysis of the substrate-soaked structure of Af-Tth has shown the electron density of the polymerized sulfur atoms derived from the tetrathionate molecules and the catalytic center of the enzyme. The sole cysteine residue (Cys301) is located >25 Å from the active site. Asp325 of Af-Tth located within 4 Å of the electron density is conserved among TTHs from Acdithiobacillus spp. and Ad-TTH1 from Ad. ambivalens. TTH activity is completely abolished in a site-specific Af-Tth D325N variant, suggesting that Asp325 plays a crucial role in the first tetrathionate hydrolysis step. Considering that the Af-Tth reaction occurs only under acidic pH conditions, Asp325 acts as an acid for the tetrathionate hydrolysis reaction (Figure 3). If Asp325 acted as an acid, the Sα atom of the tetrathionate molecule could be protonated by Asp325. Owing to the protonated Sα atom, the terminal sulfur atom of the tetrathionate molecule is easily hit by the nucleophilic attack of a water molecule. The sulfur–sulfur bond between the terminal sulfur atom and protonated Sα atoms is broken in an SN2 reaction, thereby generating sulfate and DSMSA. This is consistent with the Reaction (1). The next step in the Af-Tth catalytic reaction is proton abstraction from DSMSA, followed by the release of a sulfur (S0) atom:
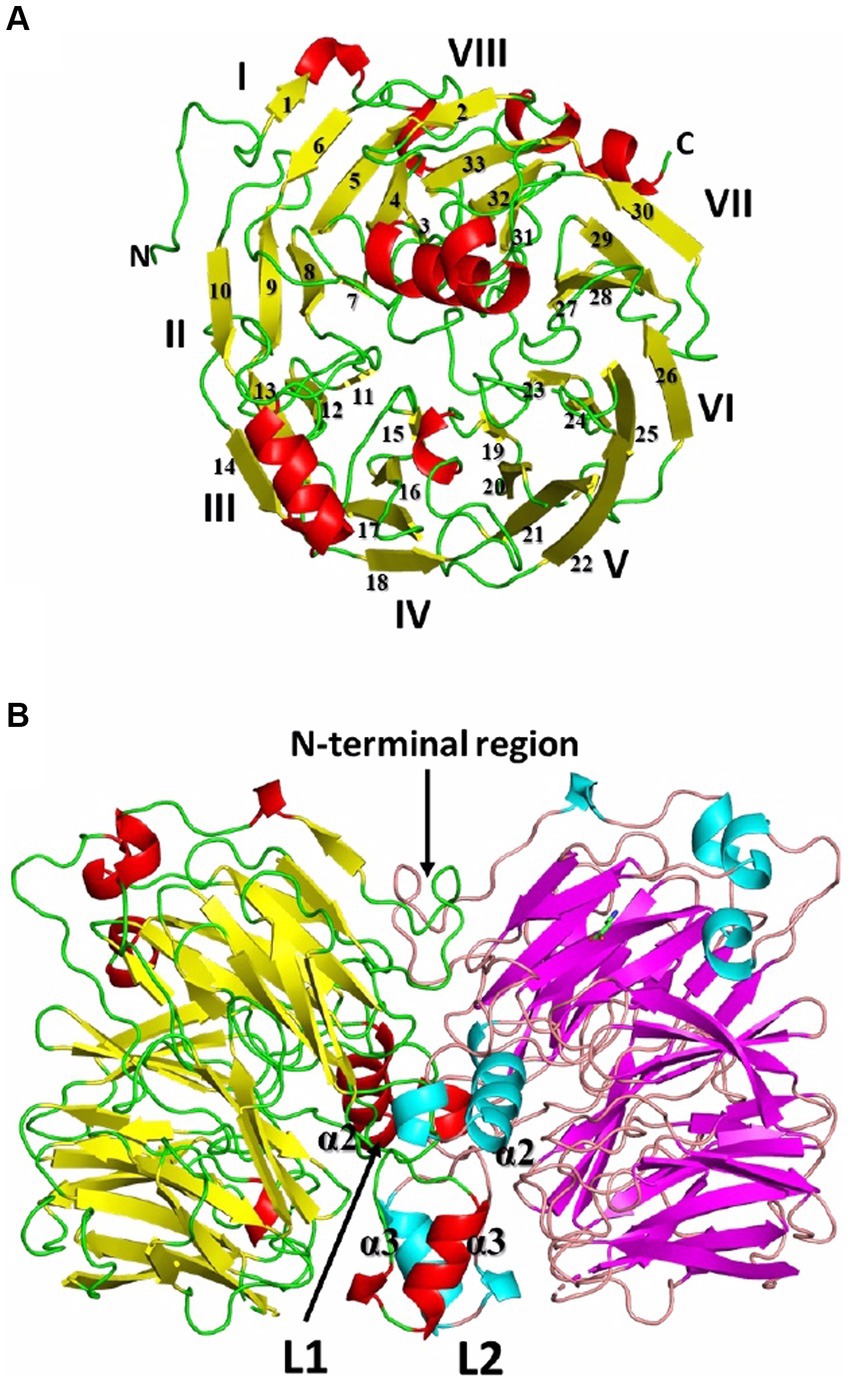
Figure 2. Overall structure of recombinant Af-Tth. (A) Monomer and (B) dimer structures. For each monomer of chains B and D in the dimer, beta-strands are colored yellow and magenta, alpha- and 310 helices are red and cyan, and loops are green and wheat, respectively. The monomer in (A) is the view from the right side of (B). The monomer structure exhibits an eight-bladed (I-VIII) beta-propeller motif. L1 and L2 are the insertion loops of the eight-bladed beta-propeller motif.
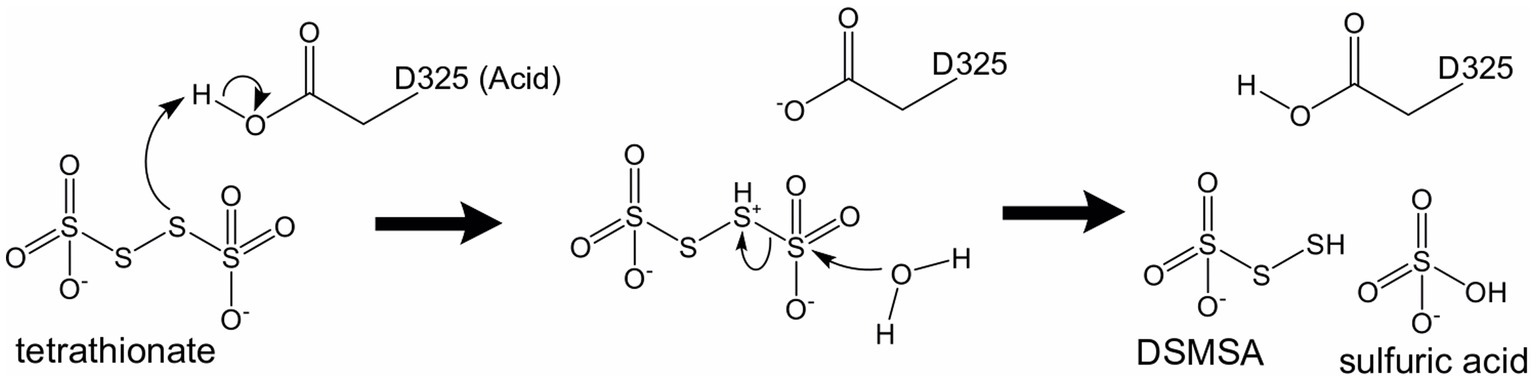
Figure 3. Proposed catalytic mechanism of tetrathionate hydrolysis by Af-Tth. The amino acid residue, Asp325, works as a general acid under neutral or acidic conditions. DSMSA, disulfane monosulfonic acid.
Reaction (2) is obtained by combining Reaction (1) and (A). Moreover, the Af-Tth reaction is multistep. The released sulfur (S0) atoms oligomerize and finally cyclize as S8. However, the underlying mechanisms remain unknown. Further studies are required to elucidate the complete reaction mechanism of this unique enzyme.
To date, various enzymes related to RISC oxidation metabolism have been investigated, but most are oxidoreductases, such as SoxXA, Sox(CD)2, TQO, TSD, sulfide:quinone oxidoreductase, sulfur dioxygenase, and sulfur oxygenase reductase, and a few hydrolases (only TTH and SoxB (Sauvé et al., 2009)). TTH, which catalyzes tetrathionate hydrolysis, is a unique enzyme that cooperates with TQO and/or TSD to function in the S4I pathway for RISC oxidation, particularly of thiosulfate (Figure 1). The S4I pathway is widely distributed in alpha-, beta-, and gamma-proteobacteria, in particular acidophilic and neutrophilic sulfur-oxidizing bacteria, including the genera Acidiphilium, Tetrathiobacter, Acidithiobacillus, and Thermithiobacillus and thermoacidophilic archaea belonging to Sulfolobaceae including the genus Acidianus. However, TTHs have only been purified and characterized from acidophilic autotrophic sulfur-oxidizing microorganisms (Wang et al., 2018). All purified TTHs show an optimum pH in the acidic range, suggesting that they are localized in the periplasmic space or outer membrane (Table 1). TTH catalyzes a unique cysteine-independent reaction. Elucidating the detailed reaction mechanism of how the enzyme cleaves the sulfur–sulfur bond in the DSMSA molecule is of interest from the viewpoint of both inorganic sulfur chemistry and enzymology. It is expected that detailed X-ray crystallographic analyses of both site-specific variants and wild-type enzymes will clarify the complete reaction mechanism of TTH.
TTH gene expression is regulated and enhanced by a two-component system in cells grown on tetrathionate in Acidithiobacillus spp. and Ad-tth1 expression is also regulated at the transcriptional level depending on the growth substrate. Although no homologous genes related to the Sox system were detected in the whole-genome sequence of A. ferrooxidans, Af-tth knockout variants showed good growth on elemental sulfur. This suggests that other RISC oxidation pathways may be distinct from the Sox and S4I pathways. For instance, TTH independent tetrathionate metabolism for thiosulfate oxidation was recently reported in the newly isolated bacterium, Erythrobacter flavus (Zhang et al., 2020). In addition, although the strain closely related to Acidicaldus sp. DX-1 can grow on tetrathionate overlay medium (Johnson et al., 2006), no candidate genes with significant similarity to tth and Sox relevant genes were found in the draft genome of the strain DX-1 (Liu et al., 2006). Investigation of microbial sulfur oxidation, especially in acidophilic sulfur-oxidizer, is essential and important not only for their applications but also for understanding the sulfur cycle in the environment.
TK: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the JSPS KAKENHI (grant number 21 K05876).
The author would like to thank Editage (www.editage.jp) for English language editing.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Beard, S., Paradela, A., Albar, J. P., and Jerez, C. A. (2011). Growth of Acidithiobacillus ferrooxidans ATCC 23270 in thiosulfate under oxygen-limiting conditions generates extracellular sulfur globules by means of a secreted tetrathionate hydrolase. Front. Microbiol. 2:79. doi: 10.3389/fmicb.2011.00079
Bobadilla Fazzini, R. A., Cortés, M. P., Padilla, L., Maturana, D., Budinich, M., Maass, A., et al. (2013). Stoichiometric modeling of oxidation of reduced inorganic sulfur compounds (Riscs) in Acidithiobacillus thiooxidans. Biotechnol. Bioeng. 110, 2242–2251. doi: 10.1002/bit.24875
Brito, J. A., Denkmann, K., Pereira, I. A., Archer, M., and Dahl, C. (2015). Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum: structural and functional insights into thiosulfate oxidation. J. Biol. Chem. 290, 9222–9238. doi: 10.1074/jbc.M114.623397
Bugaytsova, Z., and Lindström, E. B. (2004). Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus. Eur. J. Biochem. 271, 272–280. doi: 10.1046/j.1432-1033.2003.03926.x
Buonfiglio, V., Polidoro, M., Soyer, F., Valenti, P., and Shively, J. (1999). A novel gene encoding a sulfur-regulated outer membrane protein in Thiobacillus ferrooxidans. J. Biotechnol. 72, 85–93. doi: 10.1016/s0168-1656(99)00097-8
Cherney, M. M., Zhang, Y., Solomonson, M., Weiner, J. H., and James, M. N. (2010). Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification. J. Mol. Biol. 398, 292–305. doi: 10.1016/j.jmb.2010.03.018
de Jong, G. A. H., Hazeu, W., Bos, P., and Kuenen, J. G. (1997a). Isolation of the tetrathionate hydrolase from Thiobacillus acidophilus. Eur. J. Biochem. 243, 678–683. doi: 10.1111/j.1432-1033.1997.00678.x
de Jong, G. A. H., Hazeu, W., Bos, P., and Kuenen, J. G. (1997b). Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans. Microbiology 143, 499–504. doi: 10.1099/00221287-143-2-499
Eccleston, M., and Kelly, D. P. (1978). Oxidation kinetics and chemostat growth kinetics of Thiobacillus ferrooxidans on tetrathionate and thiosulfate. J. Bacteriol. 134, 718–727. doi: 10.1128/jb.134.3.718-727.1978
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Friedrich, C. G., Bardischewsky, F., Rother, D., Quentmeier, A., and Fischer, J. (2005). Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8, 253–259. doi: 10.1016/j.mib.2005.04.005
Friedrich, C. G., Rother, D., Bardischewsky, F., Quentmeier, A., and Fischer, J. (2001). Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67, 2873–2882. doi: 10.1128/AEM.67.7.2873-2882.2001
Ghosh, W., and Dam, B. (2009). Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 33, 999–1043. doi: 10.1111/j.1574-6976.2009.00187.x
Hazeu, W., Batenburg-van der Vegte, W. H., Bos, P., van der Pas, R. K., and Kuenen, J. G. (1988). The production and utilization of intermediary elemental sulfur during the oxidation of reduced sulfur compounds by Thiobacillus ferrooxidans. Arch. Microbiol. 150, 574–579. doi: 10.1007/BF00408252
Hensen, D., Sperling, D., Trüper, H. G., Brune, D. C., and Dahl, C. (2006). Thiosulphate oxidation in the phototrophic Sulphur bacterium Allochromatium vinosum. Mol. Microbiol. 62, 794–810. doi: 10.1111/j.1365-2958.2006.05408.x
Johnson, D. B., Stallwood, B., Kimura, S., and Hallberg, K. B. (2006). Isolation and characterization of Acidicaldus organivorus, gen. Nov., sp. nov.: a novel sulfur-oxidizing, ferric iron-reducing thermo-acidophilic heterotrophic Proteobacterium. Arch. Microbiol. 185, 212–221. doi: 10.1007/s00203-006-0087-7
Kanao, T., Hase, N., Nakayama, H., Yoshida, K., Nishiura, K., Kosaka, M., et al. (2021). Reaction mechanism of tetrathionate hydrolysis based on the crystal structure of tetrathionate hydrolase from Acidithiobacillus ferrooxidans. Protein Sci. 30, 328–338. doi: 10.1002/pro.3984
Kanao, T., Kamimura, K., and Sugio, T. (2007). Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans. J. Biotechnol. 132, 16–22. doi: 10.1016/j.jbiotec.2007.08.030
Kanao, T., Kosaka, M., Yoshida, K., Nakayama, H., Tamada, T., Kuroki, R., et al. (2013). Crystallization and preliminary X-ray diffraction analysis of tetrathionate hydrolase from Acidithiobacillus ferrooxidans. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69, 692–694. doi: 10.1107/S1744309113013419
Kanao, T., Kunihisa, T., Ohgimoto, S., Ito, M., Murakami, C., Nakayama, H., et al. (2023). Recombinant expression using the tetrathionate hydrolase promoter in Acidithiobacillus ferrooxidans. J. Biosci. Bioeng. 135, 176–181. doi: 10.1016/j.jbiosc.2022.12.005
Kanao, T., Matsumoto, C., Shiraga, K., Yoshida, K., Takada, J., and Kamimura, K. (2010). Recombinant tetrathionate hydrolase from Acidithiobacillus ferrooxidans requires exposure to acidic conditions for proper folding. FEMS Microbiol. Lett. 309, 43–47. doi: 10.1111/j.1574-6968.2010.02019.x
Kanao, T., Nakayama, H., Kato, M., and Kamimura, K. (2014). The sole cysteine residue (Cys301) of tetrathionate hydrolase from Acidithiobacillus ferrooxidans does not play a role in enzyme activity. Biosci. Biotechnol. Biochem. 78, 2030–2035. doi: 10.1080/09168451.2014.948374
Kanao, T., Onishi, M., Kajitani, Y., Hashimoto, Y., Toge, T., Kikukawa, H., et al. (2018). Characterization of tetrathionate hydrolase from the marine acidophilic sulfur-oxidizing bacterium, Acidithiobacillus thiooxidans strain SH. Biosci. Biotechnol. Biochem. 82, 152–160. doi: 10.1080/09168451.2017.1415128
Karchmer, J. H. (1970). The analytical chemistry of sulfur and its compounds. Wiley & Sans, New York
Kikumoto, M., Nogami, S., Kanao, T., Takada, J., and Kamimura, K. (2013). Tetrathionate-forming thiosulfate dehydrogenase from the acidophilic, chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 79, 113–120. doi: 10.1128/AEM.02251-12
Krupovic, M., Peixeiro, N., Bettstetter, M., Rachel, R., and Prangishvili, D. (2012). Archaeal tetrathionate hydrolase goes viral: secretion of a sulfur metabolism enzyme in the form of virus-like particles. Appl. Environ. Microbiol. 78, 5463–5465. doi: 10.1128/AEM.01186-12
Liu, Y., Yang, H., Zhang, X., Xiao, Y., Guo, X., and Liu, X. (2006). Genomic analysis unravels reduced inorganic sulfur compound oxidation of heterotrophic acidophilic Acidicaldus sp. strain DX-1. Biomed. Res. Int. 2016:8137012. doi: 10.1155/2016/8137012
Mangold, S., Valdés, J., Holmes, D. S., and Dopson, M. (2011). Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front. Microbiol. 2:17. doi: 10.3389/fmicb.2011.00017
Meulenberg, R., Pronk, J. T., Hazeu, W., Bos, P., and Kuenen, J. G. (1992). Oxidation of reduced sulfur compounds by intact cells of Thiobacillus acidophillus. Arch. Microbiol. 157, 161–168. doi: 10.1007/BF00245285
Moinier, D., Byrne, D., Amouric, A., and Bonnefoy, V. (2017). The global redox responding RegB/RegA signal transduction system regulates the genes involved in ferrous iron and inorganic sulfur compound oxidation of the acidophilic Acidithiobacillus ferrooxidans. Front. Microbiol. 8:1277. doi: 10.3389/fmicb.2017.01277
Müller, F. H., Bandeiras, T. M., Urich, T., Teixeira, M., Gomes, C. M., and Kletzin, A. (2004). Coupling of the pathway of Sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 53, 1147–1160. doi: 10.1111/j.1365-2958.2004.04193.x
Pronk, J. T., Meulenberg, R., Hazeu, W., Bos, P., and Kuenen, J. G. (1990). Oxidation of reduced inorganic Sulphur compounds by acidophilic thiobacilli. FEMS Microbiol. Rev. 6, 293–306. doi: 10.1111/j.1574-6968.1990.tb04103.x
Protze, J., Müller, F., Lauber, K., Naß, B., Mentele, R., Lottspeich, F., et al. (2011). An extracellular tetrathionate hydrolase from the thermoacidophilic archaeon Acidianus ambivalens with an activity optimum at pH 1. Front. Microbiol. 2:68. doi: 10.3389/fmicb.2011.00068
Quatrini, R., Appia-Ayme, C., Denis, Y., Jedlicki, E., Holmes, D. S., and Bonnefoy, V. (2009). Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394. doi: 10.1186/1471-2164-10-394
Rzhepishevska, O. I., Valdés, J., Marcinkeviciene, L., Gallardo, C. A., Meskys, R., Bonnefoy, V., et al. (2007). Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds. Appl. Environ. Microbiol. 73, 7367–7372. doi: 10.1128/AEM.01497-07
Sato, Y., Kanbe, H., Miyano, H., Sambongi, Y., Arai, H., Ishii, M., et al. (2012). Transcriptome analyses of metabolic enzymes in thiosulfate-and hydrogen-grown Hydrogenobacter thermophilus cells. Biosci. Biotechnol. Biochem. 76, 1677–1681. doi: 10.1271/bbb.120210
Sauvé, V., Roversi, P., Leath, K. J., Garman, E. F., Antrobus, R., Lea, S. M., et al. (2009). Mechanism for the hydrolysis of a sulfur-sulfur bond based on the crystal structure of the thiosulfohydrolase SoxB. J. Biol. Chem. 284, 21707–21718. doi: 10.1074/jbc.M109.002709
Schippers, A., Jozsa, P., and Sand, W. (1996). Sulfur chemistry in bacterial leaching of pyrite. Appl. Environ. Microbiol. 62, 3424–3431. doi: 10.1128/aem.62.9.3424-3431.1996
Steudel, R. H., Gobel, T., and Hazeu, W. (1987). Chromatographic separation of higher polythionates SnO62-(n=3… 22) and their detection in cultures of Thiobacillus ferrooxidans; molecular composition of bacterial sulfur secretion. Angew. Chem. Int. Ed. Engl. 26, 151–153. doi: 10.1002/anie.198701511
Sugio, T., Kanao, T., Furukawa, H., Nagasawa, T., and Blake, R. C. (1996). Isolation and identification of an iron-oxidizing bacterium which can grow on tetrathionate medium and the properties of a tetrathionate-decomposing enzyme isolated from the bacterium. J. Ferment. Bioeng. 82, 233–238. doi: 10.1016/0922-338X(96)88813-1
Tano, T., Kitaguchi, H., Harada, M., Nagasawa, T., and Sugio, T. (1996). Purification and some properties of a tetrathionate decomposing enzyme from Thiobacillus thiooxidans. Biosci. Biotechnol. Biochem. 60, 224–227. doi: 10.1271/bbb.60.224
van Zyl, L. J., van Munster, J. M., and Rawlings, D. E. (2008). Construction of arsB and tetH mutants of the sulfur-oxidizing bacterium Acidithiobacillus caldus by marker exchange. Appl. Environ. Microbiol. 74, 5686–5694. doi: 10.1128/AEM.01235-08
Veith, A., Urich, T., Seyfarth, K., Protze, J., Frazão, C., and Kletzin, A. (2011). Substrate pathways and mechanisms of inhibition in the sulfur oxygenase reductase of Acidianus ambivalens. Front. Microbiol. 2:37. doi: 10.3389/fmicb.2011.00037
Wang, Z. B., Li, Y. Q., Lin, J. Q., Pang, X., Liu, X. M., Liu, B. Q., et al. (2016). The two-component system RsrS-RsrR regulates the tetrathionate intermediate pathway for thiosulfate oxidation in Acidithiobacillus caldus. Front. Microbiol. 7:1755. doi: 10.3389/fmicb.2016.01755
Wang, R., Lin, J. Q., Liu, X. M., Pang, X., Zhang, C. J., Yang, C. L., et al. (2018). Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front. Microbiol. 9:3290. doi: 10.3389/fmicb.2018.03290
Wentzien, S. S., Sand, W., Albertsen, A., and Steudel, R. (1994). Thiosulfate and tetrathionate degradation as well as biofilm generation by Thiobacillus intermedius and Thiobacillus versutus studied by microcalorimetry, HPLC, and ion-pair chromatography. Arch. Microbiol. 161, 116–125. doi: 10.1007/BF00276471
Williamson, M. A., and Rimstidt, J. D. (1993). The rate of decomposition of the ferric-thiosulfate complex in acidic aqueous solutions. Geochim. Cosmochim. Acta 57, 3555–3561. doi: 10.1016/0016-7037(93)90138-M
Yin, H., Zhang, X., Li, X., He, Z., Liang, Y., Guo, X., et al. (2014). Whole-genome sequencing reveals novel insights into sulfur oxidation in the extremophile Acidithiobacillus thiooxidans. BMC Microbiol. 14:179. doi: 10.1186/1471-2180-14-179
Yu, Y., Liu, X., Wang, H., Li, X., and Lin, J. (2014). Construction and characterization of tetH overexpression and knockout strains of Acidithiobacillus ferrooxidans. J. Bacteriol. 196, 2255–2264. doi: 10.1128/JB.01472-13
Zhang, M. J., Jiang, C. Y., You, X. Y., and Liu, S. J. (2014). Construction and application of an expression vector from the new plasmid pLAtc1 of Acidithiobacillus caldus. Appl. Microbiol. Biotechnol. 98, 4083–4094. doi: 10.1007/s00253-014-5507-z
Zhang, J., Liu, R., Xi, S., Cai, R., Zhang, X., and Sun, C. (2020). A novel bacterial thiosulfate oxidation pathway provides a new clue about the formation of zero-valent sulfur in deep sea. ISME J. 14, 2261–2274. doi: 10.1038/s41396-020-0684-5
Keywords: tetrathionate hydrolase, reduced inorganic sulfur compounds, dissimilatory sulfur metabolism, S4-intermediate pathway, acidophiles, chemoautotroph
Citation: Kanao T (2024) Tetrathionate hydrolase from the acidophilic microorganisms. Front. Microbiol. 15:1338669. doi: 10.3389/fmicb.2024.1338669
Received: 15 November 2023; Accepted: 15 January 2024;
Published: 29 January 2024.
Edited by:
Elizabeth Watkin, Edith Cowan University, AustraliaReviewed by:
Ruiyong Zhang, Institute of Oceanology (CAS), ChinaCopyright © 2024 Kanao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tadayoshi Kanao, dGthbmFvQG9rYXlhbWEtdS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.