- Division of Geological and Planetary Sciences, California Institute of Technology, Pasadena, CA, United States
The hydrogen isotope ratios (δ2HAA values) of amino acids in all organisms are substantially fractionated relative to growth water. In addition, they exhibit large variations within microbial biomass, animals, and human tissues, hinting at rich biochemical information encoded in such signals. In lipids, such δ2H variations are thought to primarily reflect NADPH metabolism. Analogous biochemical controls for amino acids remain largely unknown, but must be elucidated to inform the interpretation of these measurements. Here, we measured the δ2H values of amino acids from five aerobic, heterotrophic microbes grown on different carbon substrates, as well as five Escherichia coli mutant organisms with perturbed NADPH metabolisms. We observed similar δ2HAA patterns across all organisms and growth conditions, which–consistent with previous hypotheses–suggests a first-order control by biosynthetic pathways. Moreover, δ2HAA values varied systematically with the catabolic pathways activated for substrate degradation, with variations explainable by the isotopic compositions of important cellular metabolites, including pyruvate and NADPH, during growth on each substrate. As such, amino acid δ2H values may be useful for interrogating organismal physiology and metabolism in the environment, provided we can further elucidate the mechanisms underpinning these signals.
1 Introduction
Stable hydrogen isotope analysis of amino acids (δ2HAA) is receiving growing attention due to its potential utility as a tracer of ecological and/or physiological processes, as well as the extreme fractionations recorded in laboratory-grown and natural organisms. In the first published study on terrestrial δ2HAA values, Fogel et al. (2016) discovered large (>100‰) variations in δ2HAA values in Escherichia coli cultured on glucose or tryptone (a complex protein source) in different growth waters, with two key insights emerging from their study: (1) patterns of δ2HAA values may be driven by ubiquitous biochemical mechanisms associated with amino acid synthesis in organisms, and (2) hydrogen can be directly routed from organic substrates, or incorporated from water via de novo amino acid synthesis, to variable extents depending on the protein content of the medium. Expanding this work to an animal model, Newsome et al. (2020) observed that hydrogen sources of amino acids in mouse muscle tissue are driven by similar metabolic factors as E. coli, but that carbohydrates and amino acids from both the diet and gut microbiome are particularly important hydrogen sources. Recently, Gharibi et al. (2022a) reported extreme 2H-enrichments in proline and hydroxyproline (δ2H values >1,000‰) from seal bone collagen, although the cause of these extreme δ2H values was not identified. Smith et al. (2022) ruled out growth rate as a primary control on δ2HAA values in E. coli and revealed that carbon and hydrogen isotope compositions of amino acids are governed by different biochemical factors. Drawing on the well-known spatial variations in precipitation isotope ratios (Craig, 1961; Dansgaard, 1964; Rozanski et al., 1993; Kendall and Coplen, 2001; Poage and Chamberlain, 2001), Mancuso et al. (2023) revealed the first systematic link between δ2HAA values in human tissue (scalp hair) and local water δ2H, supporting the utility of this compound-specific tool as a potential tracer of geographical origin (Rubenstein and Hobson, 2004; Bowen et al., 2005). Together, these results encourage a variety of potential exciting applications of δ2HAA analysis across diverse fields such as ecology, archaeology, microbiology, biogeochemistry, and forensics. However, these applications are limited by our lack of fundamental understanding of which biochemical controls set δ2HAA values in terrestrial organisms.
Here we seek to elucidate some of the mechanistic controls on biological δ2HAA values. We focus on microbes, which are simpler systems than animals because most microbes are unicellular, can synthesize all 20 amino acids (Price et al., 2018), and can be grown in defined media. Furthermore, microbes are the major drivers of biogeochemical processes such as energy and nutrient cycling in the environment (Falkowski et al., 2008), so understanding how their δ2HAA values relate to their metabolic activities may render δ2HAA analysis a useful tool for interrogating the critical microbial-driven changes to our planet's surface geochemistry. Amino acids are formed via biosynthetic pathways that are ubiquitous across most forms of life. Their carbon skeleton precursors are the intermediates of central metabolic pathways (Figure 1), and the hydrogen on each amino acid is derived from different combinations of sources, including the organic precursors, water, and NAD(P)H. As such, δ2HAA values are complicated to interpret, but may contain multiple layers of useful biochemical information.
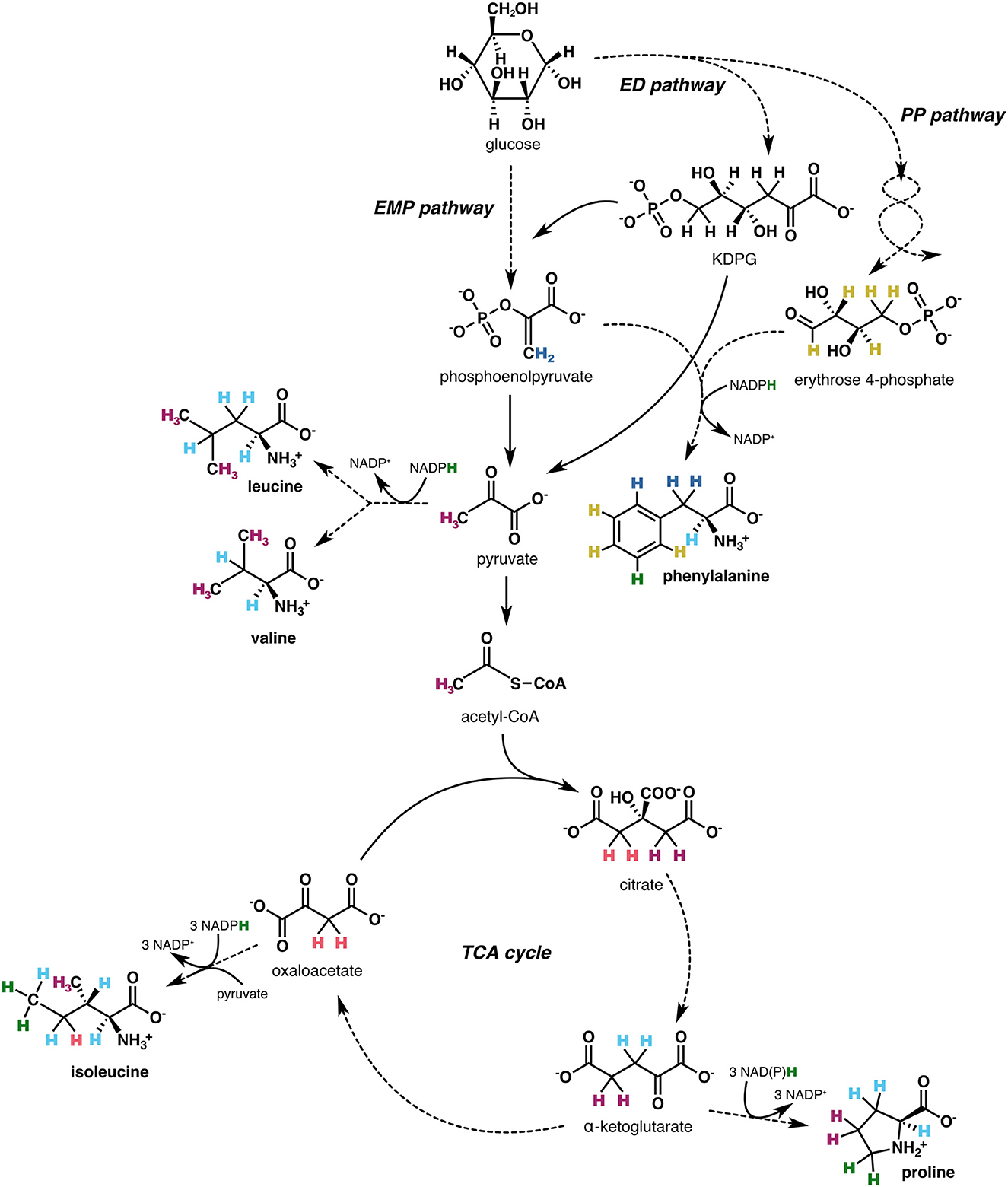
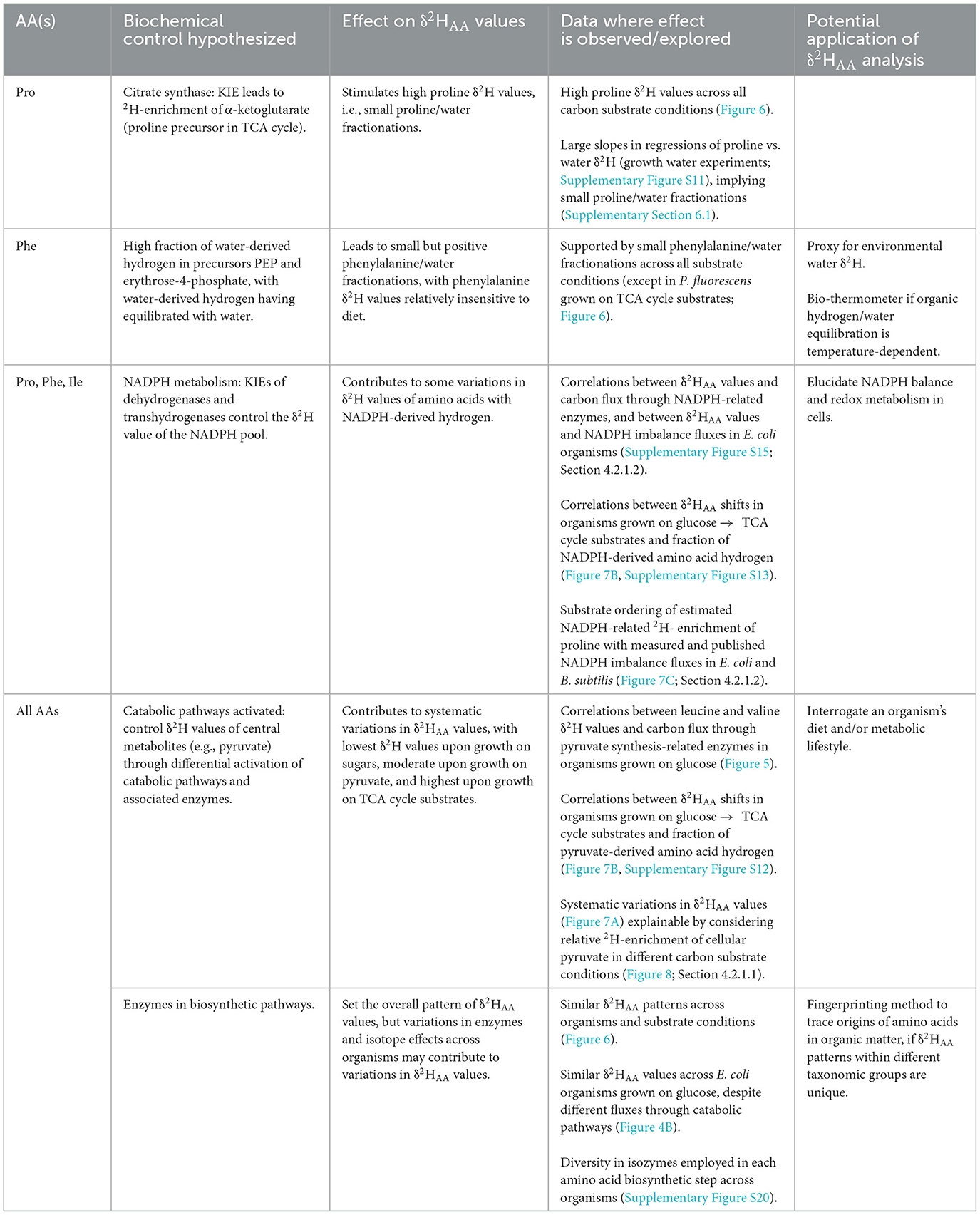
Figure 1. Simplified schematic of biosynthetic pathways (showing end-member compounds), central metabolic pathway precursors, and hydrogen sources for the five amino acids investigated in this study (proline, phenylalanine, leucine, valine, isoleucine). Hydrogen atoms are visually tracked from source to amino acid through colors: green indicates hydrogen from NAD(P)H, light blue from water, and the remaining colors correspond to hydrogen from organic precursors. Solid arrows denote single metabolic reactions; dashed arrows encompass multiple steps. Although NAD(P)H is used to reduce substrates in all amino acid biosynthetic pathways, some NAD(P)H-derived hydrogen is subsequently lost due to elimination or equilibration with water (see detailed biosynthetic pathways in Supplementary Figures S16–S19). EMP, Embden-Meyerhof-Parnas; ED, Entner-Doudoroff; PP, pentose phosphate; TCA, tricarboxylic acid.
In this study, we investigated the δ2H values of amino acids from the biomass of five aerobic, heterotrophic bacteria that was previously generated for lipid δ2H analysis (Wijker et al., 2019). The organisms included E. coli, Bacillus subtilis, Ensifer meliloti, Pseudomonas fluorescens, and Rhizobium radiobacter, as well as five mutant E. coli organisms lacking specific dehydrogenase or transhydrogenase enzymes. These organisms were grown on different carbon substrates, including on glucose for which their metabolic fluxes were characterized (Wijker et al., 2019), enabling investigation of the mechanistic link between δ2HAA values and microbial metabolism. This experimental system provides the opportunity to test a number of hypotheses about the mechanisms that govern amino acid δ2H values, including whether NADPH metabolism is a primary control (the case for lipids; Zhang et al., 2009; Wijker et al., 2019), and how varied fluxes through specific enzymes in central metabolism affect δ2HAA values. We targeted five amino acids–proline, phenylalanine, leucine, valine, and isoleucine–which were selected because (1) they span different parts of central metabolism (Figure 1), and (2) their hydrogen isotope compositions are among the most reliable to interpret, as these amino acids exhibit consistent baseline chromatographic separation (Supplementary Figure S1), have relatively high ionization efficiencies, and maintain stable hydrogen isotope compositions through hydrolysis and derivatization (Supplementary Figures S9, S10; Supplementary Table S4; Silverman et al., 2022; for further details, see Supplementary Section 1). We provide hypotheses for the observed δ2HAA patterns within and across organisms cultured under different conditions. As such, we aim to elucidate the underlying mechanisms that control 2H/1H fractionation in these five amino acids.
Additionally, δ2HAA analyses to-date have been hampered by the presence of “labile” organic hydrogen in the amine (–NH2) and carboxyl (–COOH) groups, which readily exchange with hydrogen in both water and ambient water vapor. Derivatization of amino acids removes the carboxyl- and one amine-bound hydrogen, but the remaining amine hydrogen cannot be excluded from the measured isotopic composition, and may dilute or obscure biological signals (i.e., those of non-exchangeable, C-bound hydrogen in the amino acids) and furthermore lead to incomparable results across laboratories. Previous studies (Fogel et al., 2016; Newsome et al., 2020; Smith et al., 2022; Mancuso et al., 2023) have attempted to correct for the contribution of derivative and exchangeable hydrogen to measured δ2HAA values through the use of amino acid standards, whereby the δ2H values of underivatized amino acid powders pre-equilibrated with ambient water vapor (following the comparative equilibration method; Wassenaar and Hobson, 2003) are measured via a high-temperature conversion elemental analyzer coupled to an isotope ratio mass spectrometer, then subtracted via mass balance from the δ2H values of corresponding derivatized amino acids. The central issue with this approach is that hydrogen in the derivative reagents, derivatized amino acids, and underivatized amino acids cannot be mass balanced, as (1) the isotopic fractionations between the exchangeable hydrogen and water (or ambient moisture) are unknown, thus the δ2H values of the carboxyl and amine hydrogen atoms removed during derivatization cannot be properly accounted for, and (2) the isotopic fractionation between the amine-bound hydrogen and water likely differs when amino acids are in derivatized (possessing a secondary amine) vs. underivatized (primary amine) form, so knowledge of the amine hydrogen δ2H value in the latter case may not help correct for exchangeable hydrogen in the former case. Independent measurements of the derivative reagent δ2H values are possible in some cases, but without accompanying correction for the amine-bound hydrogen in derivatized amino acids, errors in the reported δ2H values of amino acid carbon-bound hydrogen may be significant (on the order of 10 to 100‰; Supplementary Figure S7; Supplementary Section 4). Here we have developed a new, simple procedure for controlling this exchangeable amine-bound hydrogen based on separate oxidation and derivatization of a diamine compound to obtain the combined δ2H value of our amine group derivative and the exchangeable hydrogen. By subtracting the isotopic contribution of both the derivative hydrogen and exchangeable amine-bound hydrogen, we are able to accurately calculate the δ2H value of pure carbon-bound hydrogen in amino acids.
2 Materials and methods
2.1 Strain and culture conditions
The microbial biomass measured here was generated in a prior study targeting lipid δ2H analysis (Wijker et al., 2019); all relevant culturing details are recapitulated here. Five wildtype aerobic heterotrophic microbes (Escherichia coli MG1655, Bacillus subtilis PY79, Ensifer meliloti Young 2003, Pseudomonas fluorescens 2-79, and Rhizobium radiobacter C58) and five mutant E. coli organisms carrying specific deletions of dehydrogenase or transhydrogenase genes (glucose 6-phosphate dehydrogenase deleted in JW1841, phosphoglucose isomerase deleted in JW3985, membrane-bound transhydrogenase deleted in PntAB, soluble transhydrogenase deleted in UdhA, and both transhydrogenases deleted in UdhA-PntAB) were cultured on unlabeled glucose for hydrogen isotope analysis, and on 13C-labeled glucose (100% 1-13C-glucose and a mixture of 20% (wt/wt) U-13C6-glucose + 80% (wt/wt) unlabeled glucose) for metabolic flux analysis. The relative metabolic fluxes were calculated based on the 13C-labeling pattern of proteinogenic amino acids—see Wijker et al. (2019) for more details. Wildtype organisms were additionally cultured on acetate, citrate, fructose, pyruvate, and/or succinate in an isotopically constant growth water; as well as on glucose in growth waters with different isotopic compositions, which were manipulated by adding specific volumes of 99.9% purity D2O to distilled, deionized water. Each organism was grown with 4 g/L of carbon source in M9 minimal medium (prepared as described in Fuhrer et al., 2005) in batch culture on a rotary shaker at 200 rpm, thereby ensuring aerobic conditions were maintained and fermentation was avoided throughout the course of the experiments. The carbon source served as the limiting nutrient in each culture, causing cells to transition to stationary growth phase upon depletion. B. subtilis and R. radiobacter cultures were supplemented with a vitamin mixture, while strains JW1841 and JW3985 were given 50 μg/mL of kanamycin. B. subtilis and E. coli cultures were incubated at 37°C; all other organisms were incubated at 30°C. All wildtype cultures were prepared in duplicate except for organisms grown in D2O-spiked media (for growth water experiments) and for E. coli, which was grown on pyruvate and acetate in single cultures, and on glucose in two non-replicate cultures: culture #1 was grown along with the rest of the wildtype organisms, E. coli mutants, and growth water experiments for non-E. coli organisms; culture #2 was grown at a later date as one of four cultures in E. coli growth water experiments. Although culturing conditions were identical between E. coli cultures #1 and #2 grown on glucose, the different timing of culturing, and different methods used to process the biomass (see Section 2.2), renders culture #2 a repeat experiment, but not true biological replicate, to culture #1. Culture growth was monitored by measuring optical density at 600 nm (OD600), and cells were harvested in late-exponential phase, lyophilized, and stored at -80°C until further processing for lipid and amino acid δ2H analysis (Wijker et al., 2019 and this study, respectively).
2.2 Amino acid hydrolysis, derivatization, extraction, and quantification
A 10–20 mg of dry biomass from each sample was hydrolyzed anoxically in 6N HCl at 110°C for 24 h in tightly capped VOA vials. Following hydrolysis, samples were uncapped and left on the hot plates until the 6N HCl was completely evaporated, then samples were resuspended in 0.5 ml of 0.1N HCl. Amino acids in all samples except E. coli culture #2 were derivatized with 7:6:3 (v/v/v) anhydrous methanol (MeOH), pyridine, and methyl chloroformate (MCF); reagents were added at room temperature, then samples were immediately capped and sonicated for ~5 min (procedure adapted from Husek, 1991a,b and Zampolli et al., 2007). E. coli culture #2 was derivatized with the same reagent bottles and reaction procedure as the other samples, but was placed on dry ice while derivative reagents were added to slow the derivatization reaction. Note that in contrast to most published δ2HAA studies (Fogel et al., 2016; Newsome et al., 2020; Smith et al., 2022; Mancuso et al., 2023), we avoid using fluorinated derivative reagents, as hydrofluoric acid can form during pyrolysis in the gas chromatograph/isotope ratio mass spectrometer (GC/IRMS), leading to potential hydrogen isotope fractionation (Sauer et al., 2001; Renpenning et al., 2017; Silverman et al., 2022).
The resulting methoxycarbonyl (MOC) esters (Figure 2A) were extracted twice with methyl tert-butyl ether (MTBE) and filtered through a sodium sulfate column to adsorb any water present. Samples were concentrated to ~0.25–0.5 ml under N2. MOC ester peaks were identified via gas chromatography/mass spectrometry (GC/MS) on a Thermo-Scientific Trace ISQ equipped with a Zebron ZB-5 ms column (30-m × 0.25-mm i.d., 0.25 μm film thickness) and programmable temperature vaporizing (PTV) injector operated in splitless mode, using He as a carrier gas (flow rate = 1.4 ml/min). The GC oven was held at 80°C for 1 min, ramped at 5°C/min to 280°C with no hold, then ramped at 20°C/min to 310°C with a final 5 min temperature hold. Peaks were identified by comparing the relative retention times and mass spectra to those of known MOC ester standards, as well as to mass spectra in the NIST MS Library database.
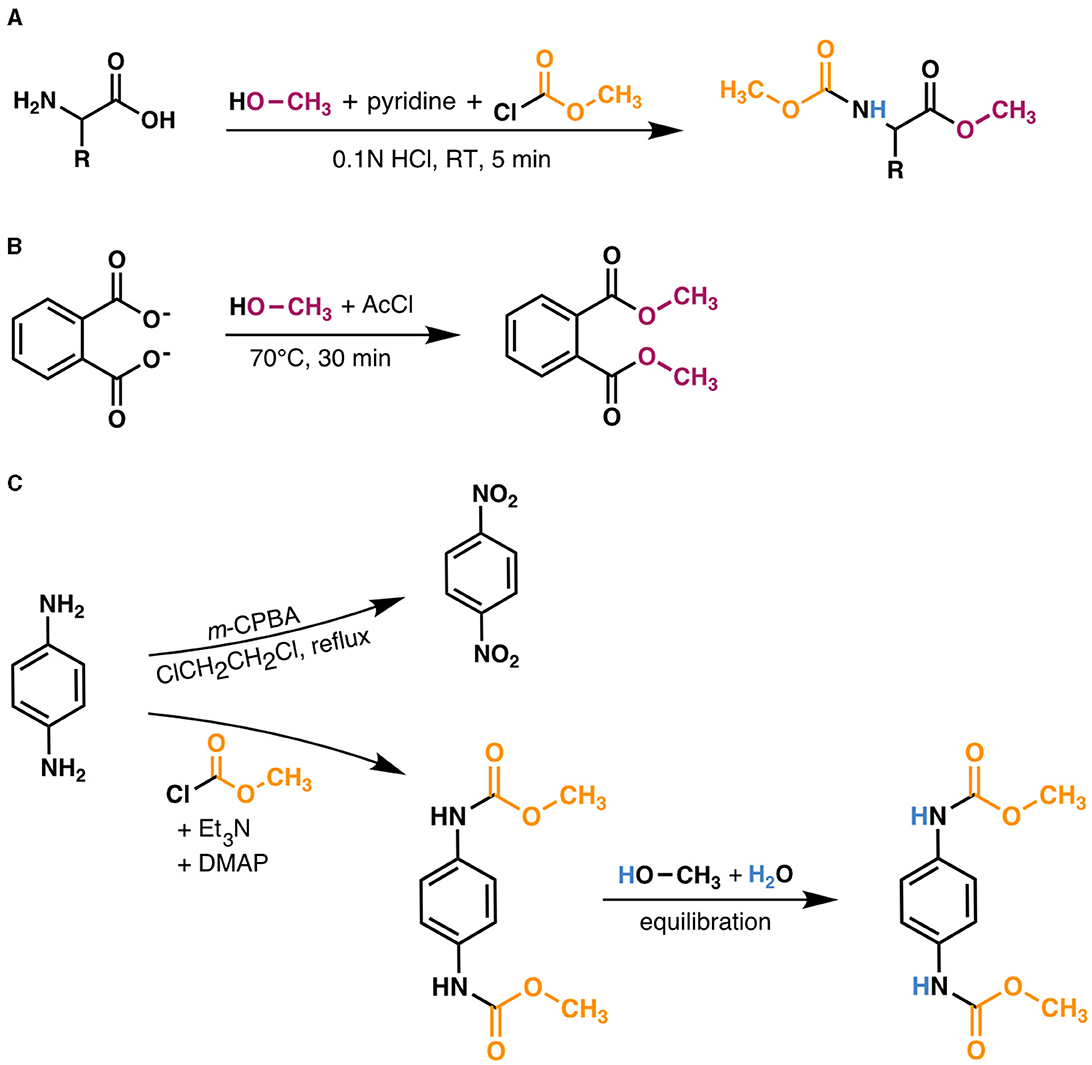
Figure 2. Derivatization scheme for amino acids and methods used to measure δ2H values of derivative reagents. (A) Amino acids were derivatized with anhydrous methanol, pyridine, and methyl chloroformate in 0.1N HCl; see Section 2.2 for details. The exchangeable amine hydrogen atom (blue in derivatized product) was equilibrated in the solvent (0.1N HCl), which was prepared using the same water supply as that used to equilibrate the N-bound hydrogen in the dimethyl 1,4-phenylenedicarbamate product [DCP; depicted in (C)]. (B) The δ2H value of anhydrous methanol was measured by derivatizing disodium phthalate with known isotopic composition (Sessions et al., 2002) with anhydrous methanol in acetyl chloride (AcCl); see Section 2.5 for details. (C) The δ2H value of methyl chloroformate was measured by first oxidizing p-phenylenediamine (PPD) to dinitrobenzene (DNB) to obtain the aromatic hydrogen δ2H value (top reaction), then separately derivatizing PPD with methyl chloroformate to produce the DCP (bottom reaction). DCP was purified, then dissolved in a 1:1 (v/v) mixture of anhydrous methanol:water to equilibrate the N-bound hydrogen atoms before extraction and measurement via GC/P/IRMS. See Section 2.5 for details.
2.3 Isotope analysis
The δ2H values of MOC esters were measured by a gas chromatograph coupled to an isotope ratio mass spectrometer (Thermo Finnigan Delta+XP) using a pyrolysis interface (i.e., GC/P/IRMS). Chromatographic separation was achieved on a thick-film Zebron ZB-5ms column (30-m × 0.25-mm i.d., 1.00 μm film thickness) with a nearly identical chromatographic method as used in GC/MS analysis [exceptions included a higher carrier gas flow rate (1.7 ml/min) and slight modifications to the temperature program to optimize MOC ester separation] so peaks could be identified by retention order and relative height. Measured isotope ratios were calibrated using hydrogen gas of known isotopic composition and are reported in δ notation (in units of ‰, or parts per thousand; Urey, 1948; McKinney et al., 1950) relative to the Vienna Standard Mean Ocean Water (VSMOW) international standard (δ2H = RAA/RVSMOW − 1), where R = 2H/1H. Additionally, an eight-compound fatty acid methyl ester standard mixture was analyzed between every 5–6 samples to verify instrument accuracy and precision. Samples were analyzed in triplicate, and the MOC ester δ2H values were corrected for the addition of methyl hydrogen from the derivative reagents, as well as for the remaining exchangeable amine hydrogen (see Section 2.5). The standard deviation of triplicate analyses for individual amino acids was typically ≤6‰. The average root-mean-square error of the external FAME standard was 3.2‰ across all analyses. δ2H values of the culture media (δ2Hw) were measured previously using a Los Gatos Research DLT-100 liquid water isotope analyzer and calibrated against up to four working standards with δ2H values ranging from –73 to +458‰ (Wijker et al., 2019). Data are reported as apparent fractionations between amino acids (AA) and culture medium water (w) according to the equation = (δ2HAA + 1)/(δ2Hw + 1) − 1, with uncertainty propagated as Equation 1
2.4 Hydrolysis and derivatization tests for isotopic alteration
Potential changes in amino acid isotopic compositions during acid hydrolysis were investigated by varying the hydrolysis conditions used (temperature, duration, and O2 presence). For a control treatment, standard bovine serum albumin (BSA) was hydrolyzed in 6N HCl for 24 h at 110°C under anoxic conditions (achieved by sparging samples with N2 for 2 min with vigorous shaking). Variations on these conditions were achieved by either hydrolyzing BSA (1) without sparging with N2 (oxic hydrolysis), (2) at 105°C, or (3) for 20 or 48 h. All conditions were prepared in duplicate. Amino acids were derivatized to MOC esters, extracted, and analyzed via GC/P/IRMS using methods described in Sections 2.2, 2.3.
Additionally, to test for hydrogen isotope exchange with aqueous medium during hydrolysis and derivatization (Hill and Leach, 1964; Fogel et al., 2016; Silverman et al., 2022), BSA and a mixture of pure amino acid standards were separately hydrolyzed (6N HCl, 24 h, 110°C, oxic) and derivatized (0.1N HCl, 7:2:3 v/v/v anhydrous MeOH, pyridine, MCF) in aqueous solvent with different hydrogen isotope compositions.
2.5 Correction for derivative and exchangeable amine hydrogen
To determine the δ2H value of MeOH, 100 μg of disodium phthalate with known isotopic composition (Sessions et al., 2002) was derivatized in 2 ml of 20:1 (v/v) anhydrous MeOH:acetyl chloride (70°C, 30 min; Figure 2B). The derivatized product was extracted with 4 ml of 1:1 water:hexane, then measured by GC/P/IRMS, and contribution of disodium phthalate hydrogen was subtracted by mass balance.
The combined δ2H value of MCF and the remaining exchangeable amine hydrogen was characterized via derivatization of p-phenylenediamine (PPD)—a compound with two primary amine groups—with MCF and separate oxidation of PPD to dinitrobenzene (DNB), which has no nitrogen-bound hydrogen. One hundred mM of PPD was dissolved in 5 ml of anoxic dichloromethane with 3 eq. triethylamine (pre-distilled with CaH2 to remove any HCl generated in the reaction) and 0.1 eq. 4-dimethylamino pyridine, and was subsequently derivatized via an overnight reaction with 2.5 eq. MCF to yield dimethyl 1,4-phenylenedicarbamate (hereafter, “dicarbamate product”, or DCP in Equation 2; Figure 2C). The DCP was purified via flash column chromatography with silica gel. Five mg of DCP was dissolved in 2 ml of anhydrous MeOH, then 2 ml of distilled, deionized water was slowly added. The solution was mixed on a shaker for 2 h to ensure complete equilibration of the two amine hydrogen atoms with water, then the DCP was extracted once with 4 ml MTBE, filtered through a sodium sulfate column, and analyzed by GC/P/IRMS. In a separate reaction, PPD was oxidized to DNB using 8 eq. of m-chloroperbenzoic acid added under refluxing 1,2-dichloroethane in a procedure adapted from Liu et al. (2014) (Figure 2C). DNB was purified via flash column chromatography with silica gel, then 3 mg of DNB was dissolved in MTBE and analyzed via GC/P/IRMS. The δ2H value of MCF and the exchangeable nitrogen-bound hydrogen (MCF+NH) was obtained by solving for FMCF+NH in the mass balance equation
where F is the fractional abundance (i.e., mole fraction) of 2H in each compound. PPD was chosen for these reactions (1) because of the favorable 8/4 ratio of (MCF + amine)/aromatic hydrogen in the dicarbamate product, and (2) because the exchangeable amine hydrogen atoms on the dicarbamate product and on MOC esters should have similar hydrogen isotope compositions when equilibrated in the same water supply, as the amine hydrogen in the dicarbamate product and in MOC esters share similar intramolecular bonding environments so should be controlled by similar equilibrium 2H/1H fractionation factors at a constant temperature (Wang et al., 2009).
3 Results
3.1 Derivative correction
Hydrogen atoms on amine and carboxyl groups rapidly exchange with water so do not contribute information about native δ2HAA values. Derivatization of the carboxyl group removes the exchangeable hydrogen, but derivatization of the amine group removes only one of the two exchangeable hydrogen atoms (Figure 2A). In order to determine the isotope compositions of the native, non-exchangeable (i.e., carbon-bound) hydrogen on the amino acids, it is necessary to correct δ2HAA values not only for the added derivative (MeOH and MCF) hydrogen, but also for the exchangeable amine hydrogen atom remaining after derivatization. A suitable method for this latter correction has eluded prior studies of δ2HAA thus far, yet is imperative, as errors in reported δ2HAA values can be on the order of 10 to 100‰ when the amine-bound hydrogen is improperly accounted for (Supplementary Figure S7; Supplementary Section 4). Here we developed a method to characterize the combined δ2H values of MCF and the exchangeable amine hydrogen by derivatizing PPD with MCF, separately oxidizing PPD to DNB (which removes all four of the nitrogen-bound hydrogens; Figure 2C), and analyzing both resulting products using GC/P/IRMS. Importantly, this approach requires derivatizing samples with the same reagents and water as those used to measure PPD.
The δ2H value of the MCF + amine hydrogen was –111.43 ± 1.96‰ when equilibrated with water of δ2H = –86.50 ± 0.36‰. The δ2H value of MeOH was –67.27 ± 1.78‰. Correction for these non-biological contributions generally shifted δ2HAA values lower (Figure 3). For the relatively 2H-depleted amino acids (leucine, valine, and isoleucine), changes in δ2H values were substantial, reaching up to 123‰ for wildtype organisms grown on glucose. For the relatively 2H-enriched amino acids (proline and phenylalanine), this correction resulted in small or negligible changes, but in some cases resulted in higher δ2H values. However, note that correction of proline δ2H values using this approach may introduce small (<20‰) errors, as proline does not contain amine-bound hydrogen after derivatization, but the isotopic composition of MCF cannot be isolated from our measured δ2H value for MCF + amine hydrogen (see Supplementary Section 4). For other researchers to adopt our correction method, they will need to re-analyze PPD of known δ2H (available by request) with their own reagents and water.
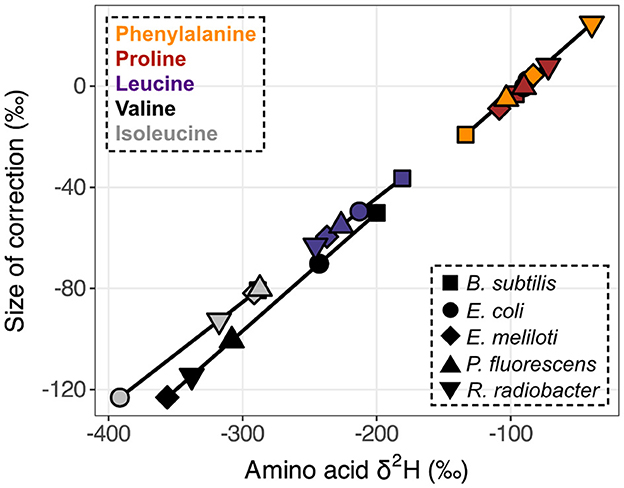
Figure 3. Size of derivative δ2H corrections vs. native amino acid isotopic compositions. Measured δ2HAA values were corrected for derivative and exchangeable amine hydrogen contributions (δ2HMeOH = –67.27 ± 1.78‰, δ2HMCF+NH = –111.43 ± 1.96‰). Data displayed are from one replicate of wildtype organisms grown on glucose (shapes denote organisms as defined in the lower right legend). Amino acids are denoted by colors (upper left legend) and corresponding symbols are connected to highlight compound-specific magnitudes of correction effects. Horizontal error bars (indicating the propagated uncertainties (±1σ) from the amino acid and derivative measurements) are smaller than symbols.
3.2 Tests for isotopic alteration during sample preparation
Certain preparatory steps can alter the isotopic compositions of amino acids (reviewed in Silverman et al., 2022). In particular, degradation or non-quantitative recovery of amino acids during acid hydrolysis can lead to isotopic fractionation (e.g., Bada et al., 1989; Phillips et al., 2021). To assess the isotopic consequences of different hydrolysis conditions on δ2HAA values, standard BSA protein was hydrolyzed at different temperatures (105 or 110°C), for different durations (20, 24, or 48 h), anoxically or with O2 present. Compared to conventional hydrolysis conditions (6N HCl, 110°C, 20-24 h, anoxic; Silverman et al., 2022), no treatment significantly altered the hydrogen isotope composition of amino acids (Supplementary Table S4).
To investigate whether carbon-bound hydrogen in amino acids exchanges with aqueous medium during hydrolysis or derivatization, BSA and a mixture of amino acid standards were separately hydrolyzed (6N HCl, 110°C, 24 h) and derivatized to MOC esters in solvents with different isotopic compositions. Slopes of regressions of amino acid vs. water δ2H values (Supplementary Figures S9, S10) represent the equilibrium fractionation factor (αeq) between organic hydrogen and water, multiplied by the fraction of hydrogen exchanged in the amino acid. To estimate the maximum percent of carbon-bound hydrogen exchanged, each slope was divided by αeq = 0.9, an estimate based on fractionation factors for a variety of hydrogen positions in linear and cyclic organic molecules (Wang et al., 2009, 2013). These calculations indicate that ten amino acids experienced negligible (<2%) hydrogen exchange with aqueous medium during hydrolysis, while tryptophan experienced significant exchange (~27%; Supplementary Figure S9). Asparagine + aspartic acid (Asx), glutamine + glutamic acid (Glx), and tyrosine experienced moderate exchange (4%–10%); this effect has been previously demonstrated through deuterated and tritiated hydrolysis experiments (Hill and Leach, 1964; Fogel et al., 2016) and is likely due to the increased lability of hydrogen adjacent to the polar—R groups. Hydrogen exchange in tryptophan may have occurred through a reversible reaction with sulfur-containing amino acids in the presence of oxygen (common tryptophan degradation mechanisms summarized in Silverman et al., 2022). All amino acids experienced low (<2%) hydrogen exchange during derivatization (Supplementary Figure S10).
3.3 2H/1H fractionations and carbon fluxes across wildtype organisms grown on glucose
The substantial variations in values within wildtype organisms grown on glucose are summarized in Figure 4A and Supplementary Table S2 for the five amino acids analyzed, and in Supplementary Figure S6 and Supplementary Table S3 for the other amino acids measured in this study. All organisms produced similar patterns, where phenylalanine and proline were the most 2H-enriched, while isoleucine and valine were the most 2H-depleted. This pattern mirrors that previously observed for E. coli (Fogel et al., 2016), with the exception of phenylalanine, which in our study was significantly more 2H-enriched relative to the average. Within a single organism, the five values spanned large ranges (220‰–352‰). Valine exhibited the largest variation across organisms (190‰) while proline and leucine values varied the least (86‰–93‰). values from biological replicates of wildtype organisms grown on glucose were generally reproducible (within 30‰) except for phenylalanine and isoleucine from P. fluorescens cultures (Supplementary Figure S5; Supplementary Table S2), which differed between replicates for unknown reasons. values in E. coli cultures grown on glucose also differed by >30‰ for four of the amino acids, but these cultures are not considered biological replicates due to differences in sample preparation (see Sections 2.1, 2.2).
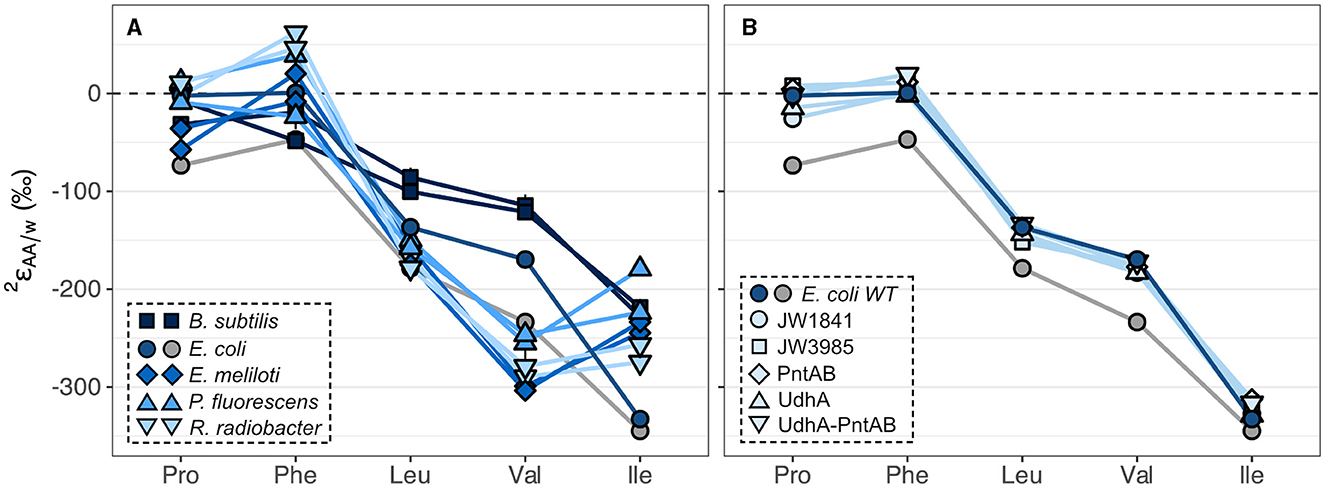
Figure 4. Summary of 2H/1H fractionations between amino acids and water in wildtype organisms (A) and in E. coli wildtype (WT) and mutant organisms (B) grown on glucose. Wildtype cultures were grown in biological duplicate, except for E. coli, which was grown in two non-replicate cultures (see Sections 2.1, 2.2) distinguished by blue and gray symbols for cultures #1 and #2, respectively. Error bars indicate the propagated uncertainties (±1σ) from the amino acid, derivative, and water measurements and are smaller than symbols. Amino acids are proline (Pro), phenylalanine (Phe), leucine (Leu), valine (Val), and isoleucine (Ile).
Metabolic flux analysis carried out in a previous study (Wijker et al., 2019) revealed substantial differences in pathways used for glucose breakdown and in carbon fluxes through central metabolic enzymes (Supplementary Figure S3; Supplementary Table S1; Wijker et al., 2019). E. coli and B. subtilis primarily used the EMP pathway for glucose catabolism and excreted high fluxes of acetate. In contrast, E. meliloti and R. radiobacter mainly relied on the ED pathway to metabolize glucose and exhibited moderate fluxes through the TCA cycle. P. fluorescens exhibited high fluxes through the ED pathway and TCA cycle, as well as periplasmic conversion of glucose to gluconate and 2-ketogluconate, and cyclic flux through the EDEMP pathway (Nikel et al., 2015; Wijker et al., 2019). values for leucine and valine correlated with carbon fluxes through enzymes related to pyruvate synthesis: KDPG aldolase (ED pathway), PEP carboxykinase (anaplerotic pathway), phosphoglucose isomerase (EMP pathway), pyruvate kinase (EMP + ED pathways), and transketolase (PP pathway; Figure 5). Directions of correlations between values and carbon flux through EMP and ED pathways opposed those observed for lipid/water fractionations (, presented in Wijker et al., 2019), although these relationships may not be directly comparable, as values are primarily controlled by NADPH metabolism (Zhang et al., 2009; Wijker et al., 2019) while leucine and valine do not inherit hydrogen from NADPH. values for other amino acids did not correlate with carbon flux through any central metabolic enzyme in wildtype organisms.
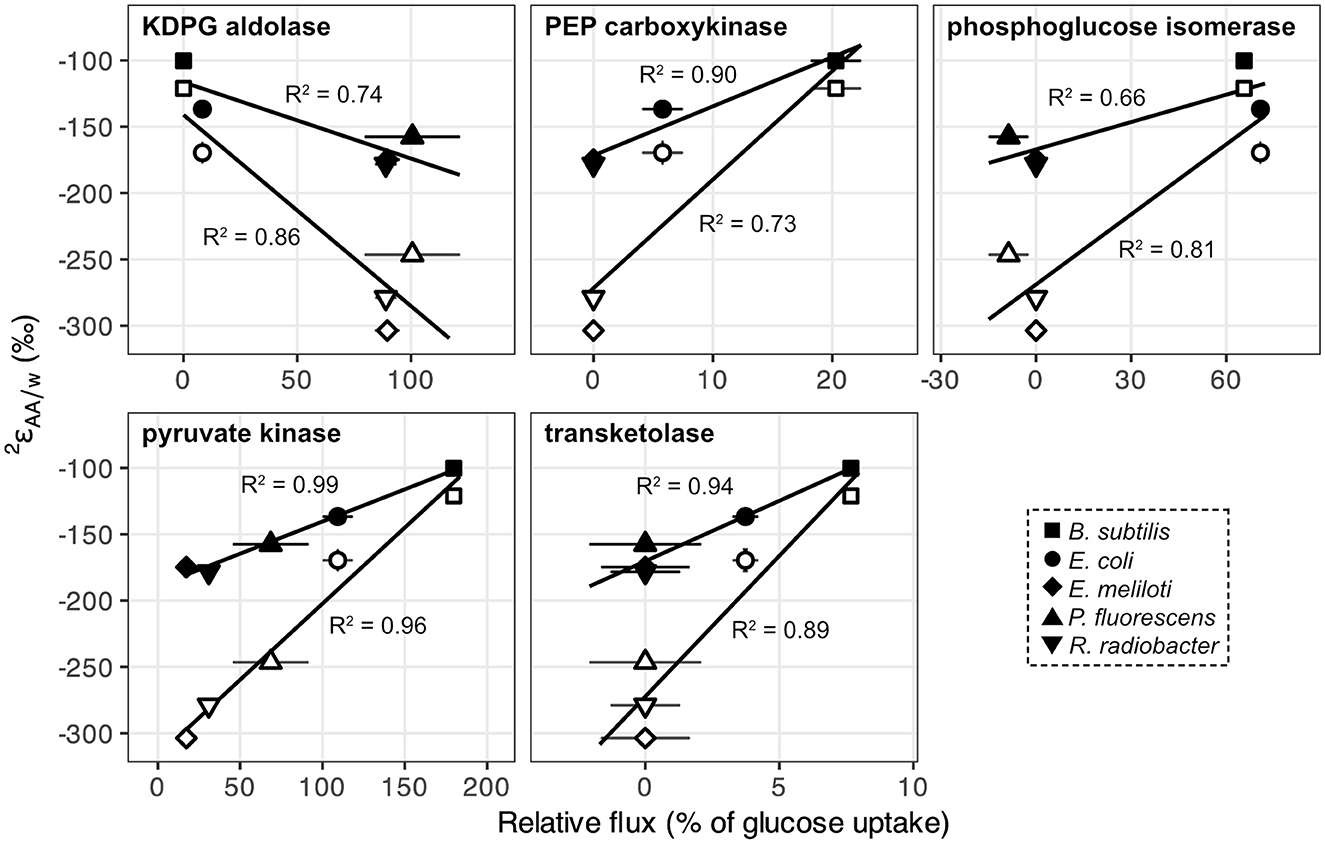
Figure 5. values for leucine (black) and valine (white) in wildtype organisms grown on glucose vs. relative carbon flux (i.e., normalized to glucose uptake rates) through pyruvate synthesis-related enzymes in central metabolism: KDPG aldolase (ED pathway), PEP carboxykinase (anaplerotic pathway), phosphoglucose isomerase (EMP pathway), pyruvate kinase (EMP + ED pathways), and fructose 6-phosphate-forming transketolase (PP pathway). Error bars represent ±1σ. Regression analyses were performed using values from the first of each biological replicate condition (see Supplementary Table S2).
3.4 2H/1H fractionations and carbon fluxes in E. coli knockout mutants grown on glucose
Specific dehydrogenase and transhydrogenase genes were deleted in E. coli organisms in a previous study (Wijker et al., 2019) to interrogate the influence of NADPH on lipid δ2H values. Deletion of these genes forced carbon flux through alternative central metabolic enzymes to accomplish glucose catabolism and NADPH balance (Supplementary Figure S4; Table S1). Despite drastic differences in the magnitudes of carbon fluxes, E. coli mutant organisms produced similar values compared to wildtype E. coli culture #1, differing by <40‰ for any given amino acid (Figure 4B; Supplementary Table S2; see note about E. coli culture #2 in Section 3.3). Lipid δ2H values showed a similar response in these organisms (Wijker et al., 2019), as did δ2HAA values in E. coli mutants with inhibited glycolysis or oxidative pentose phosphate pathways in a previous study (Smith et al., 2022). Nevertheless, variations in isotopic compositions hint at some control by NADPH, as δ2HAA values correlated with carbon fluxes through all NADPH-related enzymes (Supplementary Figure S15), with proline exhibiting the strongest correlations (R2 = 0.70–0.86), followed by phenylalanine (R2 = 0.58–0.71), then isoleucine (R2 = 0.36–0.49). The PGI knockout mutant, JW3985, had a severely perturbed metabolism and fell off the regressions in most cases so was excluded from the regression analyses.
3.5 2H/1H fractionations across wildtype organisms grown on different substrates
In addition to glucose, wildtype organisms were cultured on acetate, citrate, fructose, pyruvate, and/or succinate, which enter central metabolism at different nodes and activate different catabolic pathways for substrate breakdown. values from biological replicates of wildtype organisms grown on each substrate were generally reproducible (within 30‰) except for proline and phenylalanine in B. subtilis grown on succinate, and proline in R. radiobacter grown on succinate, which differed between replicates for unclear reasons (Figure 6, Supplementary Figure S5, Supplementary Table S2). Fructose led to similar values as glucose-based growth, while pyruvate and TCA cycle substrates (acetate, citrate, and succinate) led to 2H-enrichment of all amino acids (Figure 6), mirroring phenomena observed for lipids (Zhang et al., 2009; Osburn et al., 2016; Wijker et al., 2019). The amount of 2H-enrichment varied widely for each amino acid. Proline in B. subtilis grown on succinate, and isoleucine and phenylalanine in P. fluorescens grown on acetate, exhibited the largest singular 2H-enrichments (286‰–360‰ higher values relative to those during glucose-based growth). However, phenylalanine was generally 2H-enriched by the least amount (<100‰ difference between values upon growth on TCA cycle substrates relative to on glucose for all organisms except P. fluorescens), while valine was generally 2H-enriched by the greatest amount (142–245‰). These differences were significantly greater than those between growth water (<15‰) or substrate δ2H values (<85‰; Supplementary Table S2).
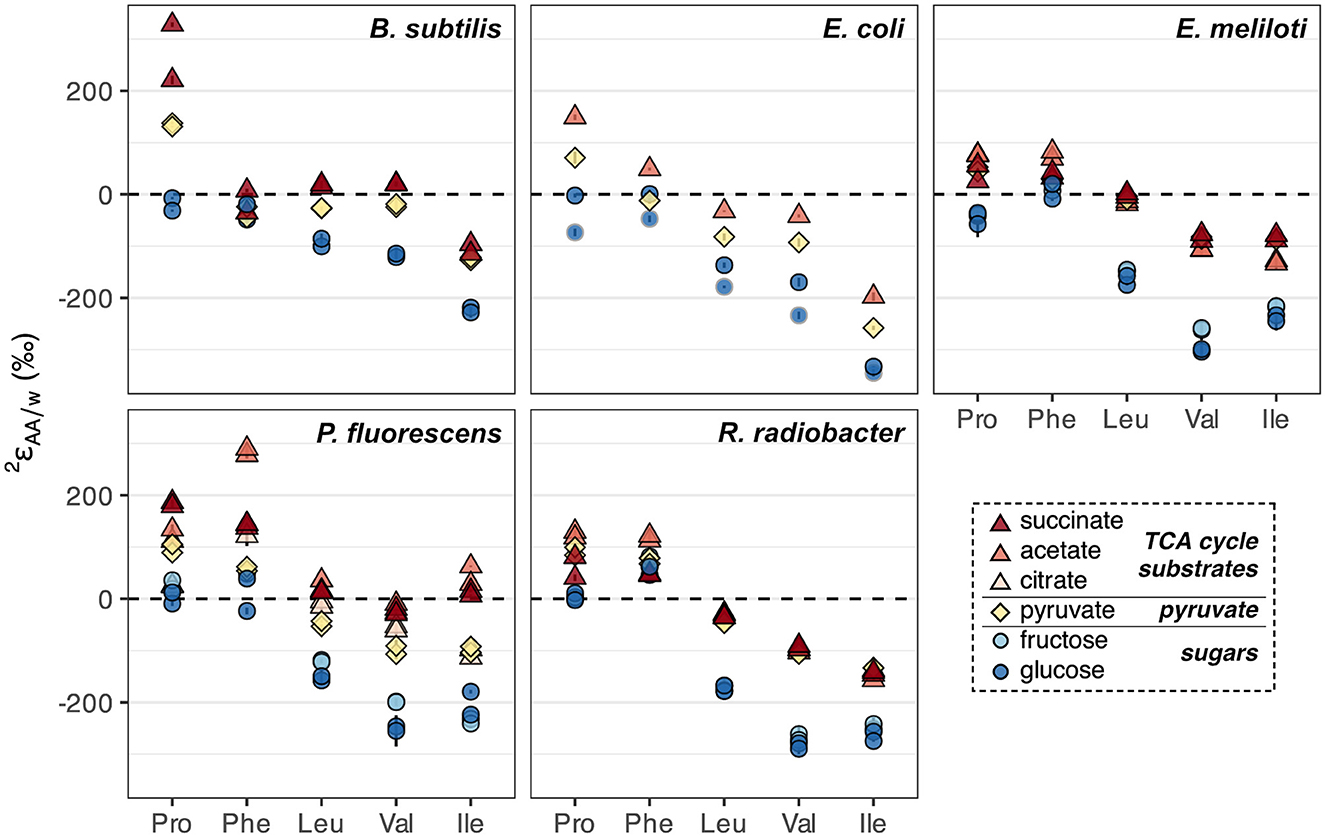
Figure 6. Summary of 2H/1H fractionations between amino acids and water in wildtype organisms grown on different substrates (denoted by colors) spanning metabolically distinct classes (denoted by shapes). Error bars indicate the propagated uncertainties (±1σ) from the amino acid, derivative, and water measurements, and are smaller than symbols. Duplicate cultures were set up for all organisms and conditions except E. coli, which was grown on acetate and pyruvate in a single replicate, and on glucose in two different (non-replicate) experiments (with cultures #1 and #2 distinguished by dark blue circles with black and gray borders, respectively; see Sections 2.1, 2.2).
4 Discussion
Consistencies in patterns (i.e., the relative ordering of values) within each growth condition, coupled with the substantial shifts in values across growth conditions, underscore the existence of systematic controls on δ2HAA values. Initial investigations (Fogel et al., 2016; Newsome et al., 2020; Gharibi et al., 2022a; Smith et al., 2022; Mancuso et al., 2023) have begun to explore the complicated factors driving δ2HAA signals in heterotrophic microbes, mammals, and humans, but we are still far from mechanistic understanding of these controls. In the following sections we interrogate several biochemical controls on the patterns and variations in microbial δ2HAA values in an attempt to elucidate how these signals can be used as tracers for microbial or ecological studies in the environment (summarized in Table 1). Our data allow us to provide a mechanistic explanation for some, though not all, of the observed patterns. As numerous enzymes in central metabolism are referenced throughout the discussion, a schematic of the central metabolic pathways with all enzymes annotated is provided for reference (Supplementary Figure S2).
4.1 Controls on the pattern during glucose metabolism
patterns were strikingly similar across wildtype organisms and E. coli mutants grown on all carbon substrates (Figures 4, 6), indicating that biosynthetic pathways (as opposed to catabolic pathways) serve as first-order controls on δ2HAA values. Our interpretation is consistent with Fogel et al. (2016), who observed similar δ2HAA patterns in E. coli cultured on glucose or tryptone in different growth waters. In this section, we investigate the biochemical factors that set the general pattern in glucose-grown cultures, focusing our interpretation on hydrogen sources and mechanisms of hydrogen exchange, as well as relevant isotope effects associated with enzymes in central metabolic and biosynthetic pathways. Based on these interpretations, we speculate on the most prominent types of biological information that can be obtained from δ2HAA measurements. Amino acids are discussed in order from most to least 2H-enriched. As these microbes share the same biosynthetic pathways, variations in 2ε values of a given amino acid across organisms (e.g., Figure 4A) hint at the importance of additional, second-order controls, which are examined in Section 4.2.
4.1.1 Proline
The high δ2H values of proline are likely due in large part to the 2H-enriching kinetic isotope effect (KIE) of citrate synthase in the TCA cycle. Proline is mainly synthesized from the TCA cycle intermediate α-ketoglutarate and inherits four hydrogen atoms from α-ketoglutarate, two from NAD(P)H, and one from water (Figure 1, Supplementary Figure S16). In the first step of the TCA cycle, citrate synthase combines acetyl-CoA with oxaloacetate to form citrate, abstracting a proton from acetyl-CoA's methyl group with a large KIE (1.94 measured in vitro; Lenz et al., 1971). The resulting 2H-enriched hydrogen in citrate is retained through formation of α-ketoglutarate (and ultimately, synthesis of proline), as aconitase and isocitrate dehydrogenase stereospecifically remove the oxaloacetate-derived hydrogen from citrate and isocitrate, respectively (Supplementary Figure S16; Lowenstein, 1967; Smith and York, 1970; Csonka and Fraenkel, 1977; Ochs and Talele, 2020). As proline inherits ~30% of its hydrogen from NAD(P)H through this synthesis pathway, and its δ2H value appears to be controlled to some extent by NADPH metabolism (see Section 4.2.1.2), proline may be a sensitive indicator of redox balance in cells. Some microbial species within the families Rhizobiaceae and Pseudomonadaceae can additionally synthesize proline from ornithine, which in turn is synthesized from arginine (Stalon et al., 1987; Schindler et al., 1989). The prevalence of this pathway in the P. fluorescens and R. radiobacter strains examined in this study is unclear, but its operation would presumably reduce the fraction of NAD(P)H-derived hydrogen in proline.
4.1.2 Phenylalanine
The source of phenylalanine's high δ2H values is unclear, but may be due to relatively large fractions of water-derived hydrogen in phenylalanine's organic precursors (phosphoenolpyruvate and erythrose-4-phosphate; Figure 1, Supplementary Figure S17). During glucose metabolism, phosphoenolpyruvate (PEP) is primarily synthesized through the EMP or ED pathway, and its hydrogen can be directly routed from glucose or partially exchanged with water (e.g., at the triose phosphate level; Rose and O'Connell, 1961; Saur et al., 1968; Reynolds et al., 1971; Russell and Young, 1990). Erythrose-4-phosphate is synthesized through the PP pathway, which includes numerous isomerizations and reversible reactions that exchange organic hydrogen with water (Russell and Young, 1990). As these equilibrations are presumably controlled by equilibrium rather than kinetic isotope effects, the resulting fractionations are unlikely to be strongly negative (in contrast to the potentially large normal KIEs expressed during water incorporation into pyruvate; Section 4.1.3). Indeed, theoretical calculations predict slightly negative to relatively positive equilibrium fractionations for the hydrogen sites susceptible to equilibration with water in the organic intermediates (Wang et al., 2009). The relatively small isotopic fractionation of phenylalanine across carbon substrate conditions (Figure 6) further supports a large fraction of water-derived hydrogen, which may render phenylalanine a useful proxy for environmental water δ2H, and potentially a bio-thermometer if equilibration with water is temperature-dependent.
4.1.3 Leucine and valine
The low δ2H values of leucine and valine, as well as the consistent 2H-depletion of valine relative to leucine in glucose-grown organisms, may be attributed in part to low pyruvate δ2H values. Leucine and valine are formed through overlapping biosynthetic pathways, initiated by condensation of two pyruvate molecules to form 2-acetolactate. Both pyruvate methyl groups remain intact through this and subsequent steps, ultimately becoming part of the isopropyl groups of these amino acids (Figure 1, Supplementary Figure S18). During glucose metabolism, pyruvate's methyl hydrogen is likely to become 2H-depleted relative to the methylene hydrogen in its central metabolite precursors (PEP, KDPG, and malate), as all pyruvate synthesis reactions incorporate solvent hydrogen with potentially large isotope effects, ranging from –141‰ for equilibrium-controlled incorporation (Wang et al., 2009) to presumably larger fractionations for kinetically-controlled transfers. For example, solvent isotope effects measured in vitro for pyruvate kinase and KDPG aldolase were 1,700 and 3,250‰, respectively (Meloche, 1975; Bollenbach et al., 1999), although note that the applicability of such measurements to in vivo studies is untested, and enzyme reversibility (such as with KDPG aldolase; Jacobson et al., 2019) as well as keto-enol tautomerization of pyruvate (Chiang et al., 1992) would drive pyruvate δ2H values toward equilibrium. As valine inherits a larger proportion of pyruvate hydrogen (Figure 1, Supplementary Figure S18), variations in the isotopic composition of pyruvate would result in more pronounced changes in the δ2H value of valine compared to leucine, as observed here (Figure 5). All additional carbon-bound hydrogen in leucine and valine are likely transferred from water, and the isotope compositions of these hydrogen atoms may reflect equilibrium and/or kinetic control. Pyruvate occupies a crucial node at the intersection of many branches of central metabolism. The isotope composition of its methyl hydrogen should therefore be sensitive to the central metabolic pathways activated (see Section 4.2.1.1), and consequently to the types of carbon substrates consumed by an organism. As leucine and valine inherit unaltered pyruvate methyl hydrogen (in addition to isotopically invariant water hydrogen), these amino acids should also be sensitive to organisms' diet if they are synthesized de novo.
4.1.4 Isoleucine
As with leucine and valine, low isoleucine δ2H values in organisms grown on glucose may be the result of relatively 2H-depleted hydrogen sources (pyruvate and NADPH) and large normal KIEs associated with hydrogen transfer from water. Isoleucine is formed from oxaloacetate, which in turn is synthesized from malate in the TCA cycle, or from PEP or pyruvate through anaplerotic pathways (Figure 1, Supplementary Figure S2, S19). The pro-S position of oxaloacetate's methylene group is retained through biosynthesis of isoleucine (Supplementary Figure S19); depending on the extent to which oxaloacetate undergoes keto-enol tautomerization (i.e., intercoverts between a ketone and enol(ate) structure; Kosicki, 1962; Bruice and Bruice, 1978) and/or is synthesized by malate dehydrogenase (thereby incorporating solvent hydrogen into oxaloacetate's pro-S methylene position; Gawron and Fondy, 1959; Omi et al., 2003), the hydrogen atom retained may originate from water (see details in Supplementary Figure S19 caption). Together, this hydrogen, along with the water-derived fraction in pyruvate's methyl group (Section 4.1.3) and those transferred from or equilibrated with water during isoleucine biosynthesis, contribute to a potentially large (≥60%) fraction of carbon-bound hydrogen in isoleucine sourced from water (Supplementary Figure S19)—an estimate consistent with model predictions by Fogel et al. (2016). In contrast to phenylalanine, whose water-derived hydrogen may be predominantly acquired through equilibrium exchange reactions, isoleucine's water-derived hydrogen may be primarily transferred from water by enzymatic reactions with large KIEs, contributing to very low isoleucine δ2H values. Despite its high water fraction, the δ2H value of isoleucine remains sensitive to the metabolic programming of cells (i.e., both redox balance and central metabolic pathways activated) through its pyruvate and NADPH hydrogen, as evident through the large δ2H variations across growth conditions (Figure 6). This sensitivity may be enhanced by the different origins of oxaloacetate-sourced hydrogen in isoleucine (water vs. organic hydrogen), which vary depending on which carbon substrates are being catabolized.
4.2 Controls on variations in values
While the similar patterns of values across glucose-grown organisms reveals remarkable consistencies in net isotopic fractionations of central metabolic and biosynthetic pathways, the variations in values hint at subtle physiological differences in the organisms. Culturing experiments in which the δ2H value of an organism's growth water is manipulated can provide some constraints on the biochemical causes of these differences (presented in Supplementary Section 6.1). The substantial systematic shifts in values upon growth on different carbon substrates provide further insight into biochemical controls on these isotopic signals, including the differential activation of enzymes across catabolic pathways (Section 4.2.1). Finally, potential differences in values due to enzymatic variations in biosynthetic pathways are considered (Section 4.2.2).
4.2.1 Metabolite pools in activated catabolic pathways
Growth of organisms on carbon substrates that activated different catabolic pathways led to substantial shifts in δ2HAA values (Figures 6, 7A). In general, growth on sugars (glucose and fructose) led to the most 2H-depleted amino acids, followed by growth on pyruvate, then TCA cycle substrates (acetate, citrate, and succinate). The overall pattern is similar to that observed in lipids (Zhang et al., 2009; Wijker et al., 2019). Moreover, patterns of 2H-enrichment relative to δ2HAA values in glucose-grown cells were similar within pairs of metabolically-related organisms (B. subtilis + E. coli and E. meliloti + R. radiobacter), highlighting organismal physiology as an important control on δ2HAA values. The altered metabolic programming in organisms grown on different substrates undoubtedly alters the hydrogen isotope composition of central metabolites that feed into amino acid biosynthesis. In particular, pyruvate occupies a crucial node in central metabolism and contributes hydrogen to all amino acids either directly (the case for leucine, valine, and isoleucine), or indirectly (for proline and phenylalanine; see Supplementary Section 6.2), so likely controls some of the variation in the δ2HAA values. Furthermore, some amino acids inherit hydrogen from NADPH (29% in proline, 20% in isoleucine, and 13% in phenylalanine; Figure 1, Supplementary Figures S16–S19). NADPH serves as a hydride carrier in organisms, both providing reducing power for anabolic reactions and transmitting isotopic information to products. As lipid δ2H variations are thought to be primarily controlled by the hydrogen isotope composition of the NADPH pool (Zhang et al., 2009; Wijker et al., 2019), amino acid δ2H values may be similarly influenced by NADPH metabolism. Together, changes in the isotopic composition of the pyruvate-related and NADPH fractions of hydrogen in the amino acids explain 29%–70% of the shifts in δ2HAA values between pairs of substrate conditions (e.g., when E. coli is grown on acetate vs. glucose; Figure 7B; Supplementary Sections 6.2, 6.3). Stated a different way, δ2HAA values may vary with the inverse of the fraction of water-derived hydrogen in amino acids, however further work is required to quantify these fractions under different metabolic conditions. In the following two sections we separately explore the influences of pyruvate and NADPH on δ2HAA variations, with mechanisms affecting pyruvate δ2H summarized in Figure 8.
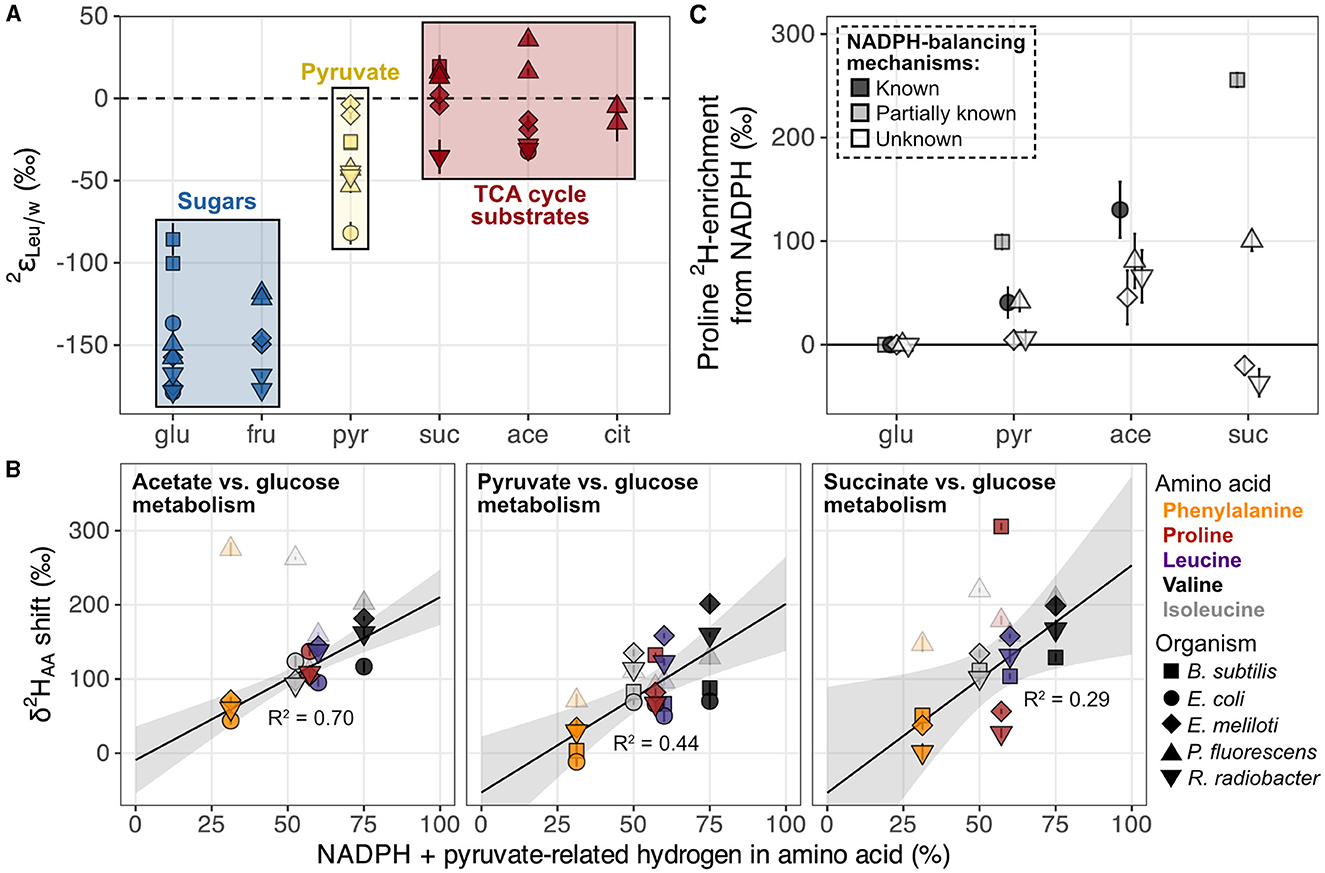
Figure 7. Hypotheses explored for pyruvate and NADPH controls on δ2HAA variations in organisms grown on different substrates. (A) Example of systematic shifts in values for one amino acid (leucine) in wildtype organisms grown on metabolically distinct classes of carbon substrates (denoted by colors). All values from Figure 6 are plotted. Error bars indicate the propagated uncertainties (±1σ) from the amino acid, derivative, and water measurements, and are smaller than symbols. (B) Estimated influence of NADPH- and pyruvate-derived hydrogen on shifts in δ2HAA values across growth conditions. See Section 4.2.1 and Supplementary Sections 6.2, 6.3 for details on accounting estimates. δ2HAA shifts were calculated as the difference in δ2H value of a given amino acid between the two substrate conditions compared within each panel (using data from replicate culture #1 for each condition). Error bars on individual data points are the propagated uncertainties (±1σ) from each pair of δ2HAA values measured and are smaller than symbols. The shaded gray region indicates the 95% confidence interval of the coefficients from the linear regression fit; R2 values are adjusted for the number of predictors in the model. P. fluorescens data are shown as transparent symbols but excluded from the regressions due to multiple instances as extreme outliers. When P. fluorescens data are included in the regressions, adjusted R2 values are 0.02, 0.43, and 0.21 for shifts from glucose to acetate, pyruvate, and succinate metabolism, respectively. Colors denote amino acids as shown in the legend to the right of the plot. (C) NADPH-driven 2H-enrichment of proline (i.e., that beyond 2H-enrichment due to acetyl-CoA alone) estimated in wildtype organisms cultured on the indicated substrate compared to during growth on glucose (data from replicate #1 cultures used). See Section 4.2.1.2 and Supplementary Section 6.3 for calculation details. Error bars indicate combined uncertainties in measured proline δ2H values (Supplementary Table S2), estimated shifts in pyruvate δ2H values (Supplementary Table S6), and/or measured acetate and pyruvate δ2H values (reported in Zhang et al., 2009). Data are slightly spread about the x-axis to increase ease of visualization. In all three figures, shapes denote the organism as shown in the bottom right legend. Growth substrates are glu, glucose; fru, fructose; pyr, pyruvate; suc, succinate; ace, acetate; and cit, citrate.
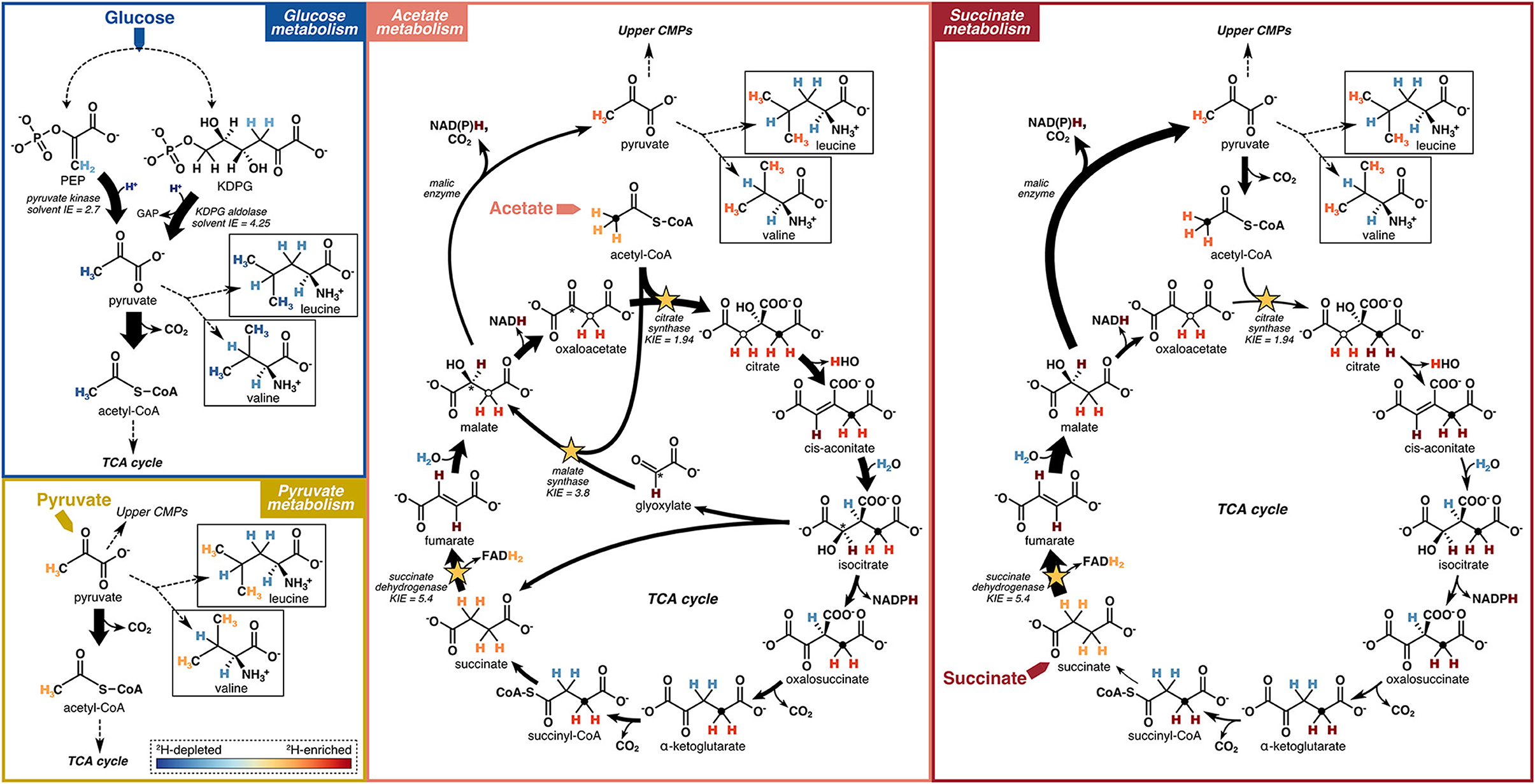
Figure 8. Schematic models of hydrogen flow through relevant central metabolic pathways (CMPs), corresponding isotopic fractionations, and relative 2H-enrichment of leucine and valine upon growth of microbes on different carbon substrates. Reactions potentially having a strong influence on the hydrogen isotope compositions of pyruvate and acetyl-CoA, and thus on the amino acids investigated in this study, are highlighted. Arrow widths indicate flux magnitudes estimated based on 13C-based metabolic flux maps reported in Gerosa et al. (2015). Dashed lines summarize a series of reactions not shown. Filled circles in the TCA cycle metabolites trace the methyl group of acetyl-CoA through succinyl-CoA. Open circles trace the methyl group of acetyl-CoA into malate and through citrate via the glyoxylate shunt (acetate metabolism) and the methylene group of oxaloacetate into citrate (succinate metabolism). The asterisk traces the alcohol carbon position in isocitrate to oxaloacetate via the glyoxylate shunt (acetate metabolism). The color gradient representing the relative 2H-enrichments or 2H-depletions of hydrogen within biomolecules is shown as an inset in the “Pyruvate metabolism” box. Isotope effects (KIEs and solvent IEs) shown are reported from Rétey et al. (1970), Lenz et al. (1971), Meloche (1975), O'Leary (1989), and Bollenbach et al. (1999).
4.2.1.1 Pyruvate δ2H
The substrate-driven ordering of amino acid 2H-enrichment (TCA cycle substrates > pyruvate > sugars) can be explained by the flow of hydrogen and activated enzymes in the different conditions. Growth on sugars that activate upper EMP/ED pathways should fuel an overall 2H-depleted pyruvate pool, as pyruvate is predominantly synthesized by enzymes that transfer hydrogen from water with potentially large normal isotope effects (e.g., pyruvate kinase and KDPG aldolase; Figure 8; Meloche, 1975; Bollenbach et al., 1999; Wang et al., 2009; although note that the expression of such isotope effects may be diluted by any enzyme reversibility). The influence of pyruvate on values in glucose-grown organisms is visible through the correlations between leucine or valine values (which should only reflect pyruvate and water hydrogen) and carbon flux through enzymes related to pyruvate synthesis (Figure 5). In particular, the ED pathway appears to have a more 2H-depleting effect on pyruvate δ2H than does the EMP pathway, as increased flux through KDPG aldolase leads to net 2H-depletion of leucine and valine, while increased flux through both phosphoglucose isomerase and pyruvate kinase stimulate 2H-enrichment of the amino acids (Figure 5). These trends are consistent with the magnitudes of solvent isotope effects observed for enzymes in the respective pathways (1,700‰ for pyruvate kinase and 3,250‰ for KDPG aldolase; Meloche, 1975; Bollenbach et al., 1999). 2H-enrichment by PEP carboxykinase (anaplerotic pathway) may be due to the 2H-enriched hydrogen transferred from the TCA cycle into pyruvate through PEP. Transketolase influences the hydrogen isotope composition of pyruvate by combining PP pathway intermediates to produce fructose 6-phosphate and glyceraldehyde 3-phosphate, which feed into glycolysis (Supplementary Figure S2). In contrast to the wildtype microbes, mutant E. coli organisms mainly synthesized pyruvate via glycolysis (Supplementary Figure S4, Supplementary Table S1), resulting in similar δ2HAA values as wildtype E. coli. This particular result highlights how variations in amino acid hydrogen isotope compositions are driven by differences in hydrogen routing through central metabolism rather than merely by changes in the magnitude of flux through a given enzyme.
Compared to glucose metabolism, the cellular pyruvate pool should be more 2H-enriched when organisms are grown on pyruvate (Figure 8), as pyruvate is directly assimilated with presumably minimal isotopic alteration, so the resulting cellular pyruvate δ2H value should be close to that of the starting substrate (–12‰ as measured by Zhang et al., 2009; Supplementary Table S2). Even higher cellular pyruvate δ2H values are likely produced in organisms grown on TCA cycle substrates (succinate, citrate, and acetate) due to the large KIEs of three differentially activated enzymes: succinate dehydrogenase, citrate synthase, and malate synthase. These enzymes abstract hydrogen from their respective substrates, leading to strong 2H-enrichment of residual hydrogen in the organic products (Figure 8). Succinate dehydrogenase removes one hydrogen from each methylene group in succinate with a large isotope effect (in vitro KIE = 5.4; Rétey et al., 1970), producing highly 2H-enriched fumarate. Succinate metabolism stimulates high flux through malic enzyme (Gerosa et al., 2015), which carries the 2H-enriched fumarate hydrogen through malate into pyruvate (although with isotopic dilution via solvent hydrogen incorporation by fumarase and malic enzyme; Figure 8), contributing to among the highest δ2HAA values across growth conditions, including a 228‰–360‰ 2H-enrichment of proline in B. subtilis grown on succinate vs. glucose (Figure 6). This contrasts with low δ2HAA values produced during sugar metabolism, whereby malate is predominantly routed around the TCA cycle through oxaloacetate, so the 2H-enriched hydrogen does not end up in pyruvate, nor in α-ketoglutarate due to stereospecific abstraction by aconitase and isocitrate dehydrogenase (Figure 8; Smith and York, 1970; Gerosa et al., 2015; Ochs and Talele, 2020). During acetate metabolism, acetyl-CoA is either routed around the TCA cycle through citrate synthase, or through the glyoxylate shunt by malate synthase, which conserves carbon by bypassing the decarboxylating steps of the TCA cycle (Zhao and Shimizu, 2003; Gerosa et al., 2015). Both citrate synthase and malate synthase abstract a hydrogen from the methyl group of acetyl-CoA with large KIEs (in vitro KIEs = 1.94 and 3.8, respectively; Lenz et al., 1971; O'Leary, 1989), leading to 2H-enriched methylene hydrogen in citrate and malate that are routed into pyruvate through malic enzyme (Figure 8). The enzymatic reactions comprising the glyoxylate shunt do not introduce water hydrogen into organic intermediates, thus 2H-enrichment of malate when the glyoxylate shunt is activated is likely more significant than when malate is synthesized by fumarase. Phenylalanine in P. fluorescens experienced the most 2H-enrichment from glucose to acetate metabolism (239‰–313‰), potentially in part due to transfer of 2H-enriched TCA cycle hydrogen into PEP through PEP carboxykinase (Gerosa et al., 2015; Dolan et al., 2020). However, in other organisms, the δ2H value of phenylalanine shifted by much smaller amounts. Citrate metabolism bypasses the 2H-enriching citrate synthase step, leading to significantly lower proline δ2H values in P. fluorescens compared to pyruvate, acetate, or succinate metabolism (Figure 6). However, flux through succinate dehydrogenase and malic enzyme still carries 2H-enriched hydrogen into pyruvate, likely contributing to the strong 2H-enrichment of the other four amino acids during citrate metabolism compared to glucose metabolism. Note that reversibility of any central metabolic enzymatic reaction contributing to equilibration of organic hydrogen with water could dilute any of the aforementioned signals.
Overall, pyruvate appears to exert an important influence on the δ2H values of all amino acids. The δ2H value of pyruvate is likely sensitive to the catabolic pathways activated for substrate degradation, which contributes to some of the variations in δ2HAA values across substrate conditions. Thus, the hydrogen isotope compositions of amino acids—particularly leucine and valine—may be useful for identifying the types of substrates consumed, and thus metabolic pathways used, by an organism. In other words, δ2HAA values may provide insight into how organisms process carbon from their environment. Future work should explore the mechanistic link between pyruvate (and other important central metabolites) and amino acid δ2H values under different metabolic conditions.
4.2.1.2 NADPH δ2H
Proline, phenylalanine, and isoleucine derive hydrogen from NADPH (Figure 1, Supplementary Figures S16–S19). The isotopic composition of NADPH is thought to be primarily driven by the relative fluxes through dehydrogenases (which produce NADPH) in central metabolism, and through transhydrogenases that interconvert NADH and NADPH to maintain NADPH balance between catabolic and anabolic processes (Zhang et al., 2009; Wijker et al., 2019). The dehydrogenase and transhydrogenase enzymes have different KIEs (O'Leary, 1989; Bizouarn et al., 1995; Venning et al., 1998; Fjellström et al., 1999), and their fluxes vary across organisms and substrate conditions, leading to large variations in the isotopic composition of NADPH (Wijker et al., 2019). In wildtype organisms grown on glucose, these effects are visible through correlations between lipid δ2H values and fluxes through dehydrogenases (6PGDH, ICDH) or enzymes that directly compete with dehydrogenases (PGI, KDPG aldolase; Wijker et al., 2019). However, amino acid δ2H values across the same organisms grown on glucose showed no correlation with carbon flux through any NADPH-related enzyme, nor with overall NADPH imbalance flux (calculated as the difference between all NADPH-producing and -consuming fluxes; Supplementary Figure S14). In fact, despite nearly identical carbon fluxes in E. meliloti and R. radiobacter grown on glucose (Supplementary Figure S3; Supplementary Table S1), these organisms exhibited the largest differences in δ2H values for proline (Figure 6; Supplementary Table S2), whose hydrogen is derived from the same sources as lipids: NAD(P)H, acetyl-CoA, and water. Lack of clear control by NADPH on δ2HAA values during glucose metabolism may be due to the fact that amino acids inherit only a small fraction of hydrogen from NADPH (13–30%) relative to other sources (e.g., ~75% of hydrogen in phenylalanine derived from PEP + erythrose 4-phosphate), so any control by NADPH may be obscured by variations in isotope compositions of the other sources. Alternatively, these results could be due to different cofactor specificities (i.e., use of NADH vs. NADPH; Fuhrer and Sauer, 2009) of biosynthetic enzymes, different isotope effects within amino acid biosynthetic pathways (Section 4.2.2.), downstream processing of amino acids after synthesis, or an outsized influence of other unknown factors on δ2HAA values in wildtype organisms.
While the extent of NADPH influence on δ2HAA values in wildtype organisms is unclear when comparing different organisms grown under the same condition, it is more apparent when physiological variability is controlled for—i.e., by comparing single organisms grown under different conditions. In E. coli wildtype and mutant organisms with perturbed NADPH metabolisms, δ2H values for proline, phenylalanine, and isoleucine were weakly to moderately correlated with carbon flux through NADPH-related enzymes, and δ2H values for isoleucine and phenylalanine positively correlated with NADPH imbalance fluxes in the cells (Supplementary Figure S15). This latter result is presumably due to increased activity of soluble transhydrogenase UdhA, which corrects NADPH overproduction by converting NADPH to NADH with an accompanying normal KIE, leading to 2H-enrichment of the residual NADPH pool. Surprisingly, NADPH imbalance was not correlated with proline δ2H values in E. coli organisms, possibly because these modest effects were overprinted by changes in the δ2H value of acetyl-CoA—the other major hydrogen source of proline. We attempted to disentangle these influences by isolating the contribution of NADPH to proline δ2H variations in wildtype organisms. We subtracted the relative contribution of acetyl-CoA δ2H variations () from total shifts in proline δ2H values () between pairs of glucose and non-glucose substrate conditions:
where is the NADPH-driven variation in proline δ2H. In turn, can be estimated based on assumptions about how hydrogen is routed through the catabolic pathways. During glucose, pyruvate, and succinate metabolism, the majority of acetyl-CoA is produced from pyruvate via pyruvate dehydrogenase (Gerosa et al., 2015), and as the methyl group remains intact, no hydrogen isotope alteration is presumed to occur (Figure 8). Thus, from glucose to pyruvate or succinate metabolism can be approximated as equal to shifts in cellular pyruvate δ2H (; Supplementary Equation S6), which in turn can be estimated through shifts in leucine or valine δ2H (see Supplementary Section 6.3 for details). During acetate metabolism, pyruvate hydrogen does not route into proline (Figure 8; Gerosa et al., 2015; Dolan et al., 2020), so individual cellular acetyl-CoA δ2H values upon growth on acetate and glucose were estimated based on measured substrate δ2H values and assumptions about hydrogen routing from substrates into acetyl-CoA (see Supplementary Section 6.3 for details). The magnitudes of estimated NADPH-driven 2H-enrichment of proline varied widely across organisms (Figure 7C). Interestingly, for E. coli and B. subtilis in particular, the magnitudes of NADPH imbalances in our glucose-grown cultures (–32 and -11%, respectively, measured in Wijker et al., 2019), as well as those from published work on E. coli grown on pyruvate (13%), acetate (50%), and succinate (72%; Gerosa et al., 2015; Haverkorn van Rijsewijk et al., 2016), appear to scale with the substrate ordering of NADPH-driven 2H-enrichment of proline (Figure 7C). In E. coli and B. subtilis, significant soluble transhydrogenase activity has been demonstrated in association with NADPH overproduction (Sauer et al., 2004; Fuhrer and Sauer, 2009; Haverkorn van Rijsewijk et al., 2016), so the potentially progressive increase in 2H-enrichment of the NADPH pool across growth on glucose, pyruvate, acetate, and succinate, may be contributing to the associated increase in proline (and other amino acid) δ2H values. Control on δ2HAA values by NADPH metabolism may also be important in E. meliloti, P. fluorescens, and R. radiobacter, but the extent of this control is unclear due to uncertainty in NADPH balancing mechanisms in these organisms.
Overall, the isotope composition of the NADPH pool appears to exert some control on the δ2H values of amino acids that inherit NADPH hydrogen. Unlike for lipids, the influence of NADPH may be difficult to observe across different organisms, but more readily apparent when considering a given organism grown under different physiological conditions. In this way, primary controls on amino acid δ2H values are different from those for lipids, yet amino acids may offer the unique advantage of isolating the isotopic influence of NADPH—i.e., by comparing the δ2H values of amino acids containing vs. lacking NADPH-derived hydrogen—which may enable researchers to probe NADPH-related metabolic phenomena such as redox balance in cells.
4.2.2 Enzymatic variations in biosynthetic pathways
Variations in δ2HAA values across organisms grown on the same substrate (e.g., Figure 4) may be driven not only by differences in the organisms' catabolic fluxes and pathways employed for substrate degradation, but also by species-specific differences in the amino acid biosynthetic enzymes and their isotope effects. Throughout the evolution of amino acid biosynthetic pathways, events such as gene duplication, functional convergence, and emergence of alternative pathways have contributed to a diversity in the enzymes and mechanisms of amino acid synthesis employed across different clades and species (Hernández-Montes et al., 2008). These enzymatic variations may contribute to different isotope effects expressed at each biosynthetic step, and consequently, different net fractionations expressed for the overall biosynthetic pathways. The five microbes investigated in this study largely employ the same enzymatic reactions to synthesize their amino acids, but exhibit large variations in isozymes expressed for each step (Supplementary Figure S20). For example, B. subtilis expresses three isozymes of pyrroline-5-carboxylate reductase (ProG, ProH, and ProI; EC 1.5.1.2)—which transfers hydrogen from NAD(P)H to catalyze the final step of proline biosynthesis (Supplementary Figure S16)—while the other four organisms express the same single enzyme (ProC). These differences may contribute to some of the variations in δ2HAA values across the five organisms; however, a general lack of data on isotope effects in these pathways challenges interrogation of this hypothesis in our study. Future studies that elucidate the net hydrogen isotope fractionations in each amino acid biosynthetic pathway will be invaluable in facilitating a more comprehensive, mechanistic understanding of δ2HAA controls and variations.
4.3 Conclusions and potential applications
Here we have explored several hypotheses regarding biochemical controls on δ2HAA values in aerobic heterotrophic microbes. Our results demonstrate that the overall pattern of amino acid/water fractionations is highly correlated with the individual biosynthetic pathways in organisms, while magnitudes of fractionations are likely controlled by the organic precursor and NADPH isotope compositions. In turn, the δ2H values of organic precursors and the NADPH pool are driven by the relative fluxes through different central metabolic pathways, which vary depending on the catabolic pathways activated for substrate degradation. Together, these results suggest that δ2HAA values may be useful tracers for carbon processing within organisms and the environment. As the 20 biological amino acids are ubiquitous across the tree of life, and organisms that share similar biosynthetic pathways should produce similar patterns of amino acid/water fractionations, we may expect the δ2H values of amino acids synthesized de novo across microbial and metazoan taxa to be governed by the same controls. Quantitative interrogations of the hypotheses presented in this study will likely require modeling work (e.g., Mueller et al., 2022), but are needed in order to fully understand the information encoded in these signals.
While controls on δ2HAA values are clearly nuanced and it may not be possible to uniquely relate all δ2HAA values to simple biological or environmental properties, the different combinations of hydrogen sources in amino acids leads to rich variability in δ2HAA signals, thus numerous potential applications of δ2HAA analysis. For example, δ2HAA values may provide insight into the metabolic strategies that microbes employ for carbon and energy acquisition. Microbes are the major drivers of nutrient and energy cycling in the environment, thereby playing substantial roles in shaping the geochemistry of our planet. While we observed large systematic variations in amino acid/water fractionations across heterotrophic microbes grown on different carbon substrates, we predict that even larger variations should exist between organisms of different metabolic classes (e.g., heterotrophs vs. autotrophs), as is the case for lipids (Zhang et al., 2009; Osburn et al., 2016). If true, δ2HAA values and patterns may be used to decipher the metabolisms of unculturable organisms, to distinguish contributions by metabolically distinct organisms to geochemical processes in nature (e.g., in largely inaccessible environments such as deep subsurface biospheres), and/or to quantify contributions by different metabolisms to bulk organic matter in the environment (including by mixotrophic organisms that operate on a continuum of carbon and energy acquisition strategies, or across diverse taxa such as plants, algae, bacteria, and fungi). While lipid δ2H values have been suggested as a potential tool for this latter application (Cormier et al., 2022), amino acids may offer distinct advantages, as their δ2H values can be linked to host organisms through the isolation and sequencing of proteins (Gharibi et al., 2022b), and their different combinations of hydrogen sources capture a more diverse suite of information than is encoded in lipids (which inherit hydrogen from essentially the same three sources: acetyl-CoA, NADPH, and water). However, we view amino acid and lipid δ2H analyses as complementary, as each can help inform interpretations from the other. Leucine and valine δ2H may provide the most direct information about the metabolic ‘state' or lifestyle of an organism, as the positioning of pyruvate as a central node in metabolism makes it relatively sensitive to the molecular wiring of central metabolic pathways in cells. In turn, accounting for these metabolic signals may help isolate the NADPH-driven signals in proline and lipid δ2H values, thereby increasing their sensitivity as redox indicators in cells. Moreover, if the majority of phenylalanine hydrogen is indeed equilibrated with water at the level of metabolic intermediates, the δ2H value of phenylalanine may provide insight into water δ2H in places where water sources are unclear (e.g., in environments with intermittent wet/dry cycles), and if such equilibrium is temperature-dependent, phenylalanine could additionally serve as a potential bio-thermometer.
In addition to geomicrobiology-based applications, δ2HAA analyses may provide useful information about eukaryotic organisms, including their stressors, diets, and migration patterns. The application of δ2HAA values to human and wildlife forensics is in the early stages of exploration, with links between mammal diet, drinking water, and δ2HAA values beginning to emerge (Newsome et al., 2020; Mancuso et al., 2023). The biochemical controls discussed here may only be relevant for interpreting δ2H values of non-essential amino acids (e.g., proline) as well as those with relatively high contributions from the gut microbiome (e.g., phenylalanine; Newsome et al., 2020). However, the differentiated tissues in animals, variable residence times of proteins in cells, and integration of numerous dietary hydrogen sources significantly increase the complexity of information encoded in mammalian δ2HAA signals, which will require detailed investigations to disentangle. δ2HAA signals in plants likely carry important physiological information as well. Like microbes, plants can synthesize all 20 proteinogenic amino acids, yet their hydrogen metabolism may be simpler to interpret, as plants derive their organic hydrogen exclusively from water. Amino acids are involved in numerous mechanisms of stress alleviation in plants (e.g., Batista-Silva et al., 2019), so their δ2HAA values may encode information about their physiological status. For example, synthesis and degradation of proline helps maintain redox balance (i.e., the NADP/NADPH ratio) in plants and appears to facilitate drought tolerance (Sharma et al., 2011; Bhaskara et al., 2015; Batista-Silva et al., 2019). In the first step of proline catabolism, proline dehydrogenase removes a carbon-bound hydrogen atom, which should lead to 2H-enrichment of the residual proline pool. Consequently, proline δ2H values may serve as a sensitive indicator of oxidative and drought stress in plants, which are highly important aspects of crop health. This mechanism may additionally play a role in the extreme 2H-enrichment of proline observed in gray seals (Gharibi et al., 2022a), which endure significant periods of oxidative stress while diving. Overall, δ2HAA analysis has potential to become a highly useful isotopic tool for a variety of diverse applications, which will undoubtedly emerge as we continue to unravel the biochemical mechanisms underpinning δ2HAA signals in organisms.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SS: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Writing—original draft, Writing—review & editing. RW: Conceptualization, Methodology, Writing—review & editing. AS: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing—review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by an NSF GRFP and CEMI (Center for Environmental Microbial Interactions) award to SS and an NSF award OCE-2023687 and NSF Geobiology and Low-Temperature Geochemistry Program award (#1921330) to AS.
Acknowledgments
The authors would like to thank Tyler Fulton and Dr. Brian Stoltz for assistance with the chemical reactions used to measure the isotope composition of methyl chloroformate, as well as Fenfang Wu, Patrick Almhjell, and Elliott Mueller for informative discussions. We would also like to thank both reviewers for their constructive comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1338486/full#supplementary-material
References
Bada, J. L., Schoeninger, M. J., and Schimmelmann, A. (1989). Isotopic fractionation during peptide bond hydrolysis. Geochimica et Cosmochimica Acta 53, 3337–3341. doi: 10.1016/0016-7037(89)90114-2
Batista-Silva, W., Heinemann, B., Rugen, N., Nunes-Nesi, A., Araújo, W. L., Braun, H., et al. (2019). The role of amino acid metabolism during abiotic stress release. Plant, Cell Environ. 42, 1630–1644. doi: 10.1111/pce.13518
Bhaskara, G. B., Yang, T.-H., and Verslues, P. E. (2015). Dynamic proline metabolism: importance and regulation in water limited environments. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00484
Bizouarn, T., Grimley, R. L., Cotton, N. P., Stilwell, S. N., Hutton, M., and Jackson, J. B. (1995). The involvement of NADP(H) binding and release in energy transduction by proton-translocating nicotinamide nucleotide transhydrogenase from Escherichia coli. Biochim. Biophys. Acta 1229, 49–58. doi: 10.1016/0005-2728(94)00186-9
Bollenbach, T. J., Mesecar, A. D., and Nowak, T. (1999). Role of lysine 240 in the mechanism of yeast pyruvate kinase catalysis. Biochemistry 38, 9137–9145. doi: 10.1021/bi990690n
Bowen, G. J., Wassenaar, L. I., and Hobson, K. A. (2005). Global application of stable hydrogen and oxygen isotopes to wildlife forensics. Oecologia 143, 337–348. doi: 10.1007/s00442-004-1813-y
Bruice, P. Y., and Bruice, T. C. (1978). Lack of concertedness in the catalysis of the enolization of oxaloacetic acid by general acids and bases. Formation of a carbinolamine intermediate in the tertiary amine catalyzed enolization reaction. J. Am. Chem. Soc. 100, 4793–4801. doi: 10.1021/ja00483a027
Chiang, Y., Kresge, A. J., and Pruszynski, P. (1992). Keto-enol equilibria in the pyruvic acid system: determination of the keto-enol equilibrium constants of pyruvic acid and pyruvate anion and the acidity constant of pyruvate enol in aqueous solution. J. Am. Chem. Soc. 114, 3103–3107. doi: 10.1021/ja00034a053
Cormier, M.-A., Berard, J.-B., Bougaran, G., Trueman, C. N., Mayor, D. J., Lampitt, R. S., et al. (2022). Deuterium in marine organic biomarkers: toward a new tool for quantifying aquatic mixotrophy. New Phytol. 234, 776–782. doi: 10.1111/nph.18023
Craig, H. (1961). Isotopic variations in meteoric waters. Science 133, 1702–1703. doi: 10.1126/science.133.3465.1702
Csonka, L. N., and Fraenkel, D. G. (1977). Pathways of NADPH formation in Escherichia coli. J. Biol. Chem. 252, 3382–3391. doi: 10.1016/S0021-9258(17)40402-9
Dansgaard, W. (1964). Stable isotopes in precipitation. Tellus 16, 436–468. doi: 10.1111/j.2153-3490.1964.tb00181.x
Dolan, S. K., Kohlstedt, M., Trigg, S., Ramirez, P. V., Kaminski, C. F., Wittmann, C., et al. (2020). Contextual flexibility in Pseudomonas aeruginosa central carbon metabolism during growth in single carbon sources. Am. Soc. Microbiol. 11, e02684–e02619. doi: 10.1128/mBio.02684-19
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Fjellström, O., Bizouarn, T., Zhang, J.-W., Rydström, J., Venning, J. D., and Jackson, J. B. (1999). Catalytic properties of hybrid complexes of the NAD(H)-binding and NADP(H)-binding domains of the proton-translocating transhydrogenases from Escherichia coli and Rhodospirillum rubrum. Biochemistry 38, 415–422. doi: 10.1021/bi9817111
Fogel, M. L., Griffin, P. L., and Newsome, S. D. (2016). Hydrogen isotopes in individual amino acids reflect differentiated pools of hydrogen from food and water in Escherichia coli. Proc. Natl. Acad. Sci. 113, E4648–E4653. doi: 10.1073/pnas.1525703113
Fuhrer, T., Fischer, E., and Sauer, U. (2005). Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187, 1581–1590. doi: 10.1128/JB.187.5.1581-1590.2005
Fuhrer, T., and Sauer, U. (2009). Different biochemical mechanisms ensure network-wide balancing of reducing equivalents in microbial metabolism. J. Bacteriol. 191, 2112–2121. doi: 10.1128/JB.01523-08
Gawron, O., and Fondy, T. P. (1959). Stereochemistry of the fumarase and aspartase catalyzed reactions and of the krebs cycle from fumaric acid to d-isocitric acid. J. Am. Chem. Soc. 81, 6333–6334. doi: 10.1021/ja01532a059
Gerosa, L., Haverkorn van Rijsewijk, B., Christodoulou, D., Kochanowski, K., Schmidt, T., Noor, E., et al. (2015). Pseudo-transition analysis identifies the key regulators of dynamic metabolic adaptations from steady-state data. Cell Syst. 1, 270–282. doi: 10.1016/j.cels.2015.09.008
Gharibi, H., Chernobrovkin, A. L., Eriksson, G., Saei, A. A., Timmons, Z., Kitchener, A., et al. (2022a). Abnormal (hydroxy)proline deuterium content redefines hydrogen chemical mass. J. Am. Chem. Soc. 144, 2484–2487. doi: 10.1021/jacs.1c12512
Gharibi, H., Chernobrovkin, A. L., Saei, A. A., Zhang, X., Gaetani, M., Makarov, A. A., et al. (2022b). Proteomics-compatible Fourier transform isotopic ratio mass spectrometry of polypeptides. Analyt. Chem. 94, 15048–15056. doi: 10.1021/acs.analchem.2c03119
Haverkorn van Rijsewijk, B. R. B., Kochanowski, K., Heinemann, M., and Sauer, U. (2016). Distinct transcriptional regulation of the two Escherichia coli transhydrogenases PntAB and UdhA. Microbiology 162, 1672–1679. doi: 10.1099/mic.0.000346
Hernández-Montes, G., Díaz-Meja, J. J., Pérez-Rueda, E., and Segovia, L. (2008). The hidden universal distribution of amino acids biosynthetic networks: a genomic perspective on its origins and evolution. Genome Biol. 9:R95. doi: 10.1186/gb-2008-9-6-r95
Hill, J., and Leach, S. J. (1964). Hydrogen exchange at carbon-hydrogen sites during acid or alkaline treatment of proteins. Biochemistry 3, 1814–1818. doi: 10.1021/bi00900a003
Husek, P. (1991a). Amino acid derivatization and analysis in five minutes. Federat. Eur. Biochem. Soc. 280, 354–356. doi: 10.1016/0014-5793(91)80330-6
Husek, P. (1991b). Rapid derivatization and gas chromatographic determination of amino acids. J. Chromatogr. A 552, 289–299. doi: 10.1016/S0021-9673(01)95945-X
Jacobson, T. B., Adamczyk, P. A., Stevenson, D. M., Regner, M., Ralph, J., Reed, J. L., et al. (2019). 2H and 13C metabolic flux analysis elucidates in vivo thermodynamics of the ED pathway in Zymomonas mobilis. Metab. Eng. 54, 301–316. doi: 10.1016/j.ymben.2019.05.006
Kendall, C., and Coplen, T. B. (2001). Distribution of oxygen-18 and deuterium in river waters across the United States. Hydrol. Proc. 15, 1363–1393. doi: 10.1002/hyp.217
Kosicki, G. W. (1962). Isotope rate effects in the enolization of oxalacetic acid. Canad. J. Chem. 40, 1280–1284. doi: 10.1139/v62-196
Lenz, H., Buckel, W., Wunderwald, P., Biedermann, G., Buschmeier, V., Eggerer, H., et al. (1971). Stereochemistry of si-citrate synthase and ATP-citrate-lyase reactions. Eur. J. Biochem. 24, 207–215. doi: 10.1111/j.1432-1033.1971.tb19672.x
Liu, J., Li, J., Ren, J., and Zeng, B.-B. (2014). Oxidation of aromatic amines into nitroarenes with m-CPBA. Tetrahedr. Lett. 55, 1581–1584. doi: 10.1016/j.tetlet.2014.01.073
Lowenstein, J. M. (1967). “The tricarboxylic acid cycle,” in Metabolic Pathways, ed. D. M. Greenberg (New York: Academic Press), 146–270. doi: 10.1016/B978-0-12-299251-3.50010-0
Mancuso, C. J., Ehleringer, J. R., and Newsome, S. D. (2023). Examination of amino acid hydrogen isotope measurements of scalp hair for region-of-origin studies. Rapid Commun. Mass Spectr. 37:e9442. doi: 10.1002/rcm.9442
McKinney, C. R., McRea, I. M., Epstein, S., Allen, H. A., and Urey, H. C. (1950). Improvements in mass spectrometers for measurement of small differences in isotope abundance ratios. Rev. Sci. Instr. 21, 724–730. doi: 10.1063/1.1745698
Meloche, H. P. (1975). An enzymic synthesis yielding crystalline sodium pyruvate labeled with isotopic hydrogen. Methods Enzymol. 41, 106–110. doi: 10.1016/S0076-6879(75)41027-8
Mueller, E. P., Wu, F., and Sessions, A. L. (2022). Quantifying Isotopologue Reaction Networks (QIRN): a modelling tool for predicting stable isotope fractionations in complex networks. Chem. Geol. 610:121098. doi: 10.1016/j.chemgeo.2022.121098
Newsome, S. D., Nakamoto, B. J., Curras, M. R., and Fogel, M. L. (2020). Compound-specific δ2H analysis highlights the relationship between direct assimilation and de novo synthesis of amino acids from food and water in a terrestrial mammalian omnivore. Oecologia 193, 827–842. doi: 10.1007/s00442-020-04730-9
Nikel, P. I., Chavarría, M., Fuhrer, T., Sauer, U., and de Lorenzo, V. (2015). Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and pentose phosphate pathways. J. Biol. Chem. 290, 25920–25932. doi: 10.1074/jbc.M115.687749
Ochs, R. S., and Talele, T. T. (2020). Revisiting prochirality. Biochimie 170, 65–72. doi: 10.1016/j.biochi.2019.12.009
O'Leary, M. H. (1989). Multiple isotope effects on enzyme-catalyzed reactions. Ann. Rev. Biochem. 58, 377–401. doi: 10.1146/annurev.bi.58.070189.002113
Omi, R., Goto, M., Miyahara, I., Mizuguchi, H., Hayashi, H., Kagamiyama, H., et al. (2003). Crystal structures of threonine synthase from Thermus thermophilus HB8. J. Biol. Chem. 278, 46035–46045. doi: 10.1074/jbc.M308065200
Osburn, M. R., Dawson, K. S., Fogel, M. L., and Sessions, A. L. (2016). Fractionation of hydrogen isotopes by sulfate- and nitrate-reducing bacteria. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01166
Phillips, A. A., Wu, F., and Sessions, A. L. (2021). Sulfur isotope analysis of cysteine and methionine via preparatory liquid chromatography and elemental analyzer isotope ratio mass spectrometry. Rapid Commun. Mass Spectr. 35:e9007. doi: 10.1002/rcm.9007
Poage, M. A., and Chamberlain, C. P. (2001). Empirical relationships between elevation and the stable isotope composition of precipitation and surface waters: considerations for studies of paleoelevation change. Am. J. Sci. 301, 1–15. doi: 10.2475/ajs.301.1.1
Price, M. N., Zane, G. M., Kuehl, J. V., Melnyk, R. A., Wall, J. D., Deutschbauer, A. M., et al. (2018). Filling gaps in bacterial amino acid biosynthesis pathways with high-throughput genetics. PLoS Genet. 14:e1007147. doi: 10.1371/journal.pgen.1007147
Renpenning, J., Schimmelmann, A., and Gehre, M. (2017). Compound-specific hydrogen isotope analysis of fluorine-, chlorine-, bromine- and iodine-bearing organics using gas chromatography-chromium-based high-temperature conversion (Cr/HTC) isotope ratio mass spectrometry. Rapid Commun. Mass Spectrom. 31, 1095–1102. doi: 10.1002/rcm.7872
Rétey, J., Seibl, J., Arigoni, D., Cornforth, J. W., Ryback, G., Zeylemaker, W. P., et al. (1970). Stereochemical studies of the exchange and abstraction of succinate hydrogen on succinate dehydrogenase. Eur. J. Biochem. 14, 232–242. doi: 10.1111/j.1432-1033.1970.tb00282.x
Reynolds, S. J., Yates, D. W., and Pogson, C. I. (1971). Dihydroxyacetone Phosphate: its structure and reactivity with α-glycerophosphate dehydrogenase, aldolase and triose phosphate isomerase and some possible metabolic implications. Biochem. J. 122, 285–297. doi: 10.1042/bj1220285
Rose, I. A., and O'Connell, E. L. (1961). Intramolecular hydrogen transfer in the phosphoglucose isomerase reaction. J. Biol. Chem. 236, 3086–3092. doi: 10.1016/S0021-9258(18)93975-X
Rozanski, K., Araguás-Araguás, L., and Gonfiantini, R. (1993). “Isotopic patterns in modern global precipitation,” in Climate Change in Continental Isotopic Records, eds. P. K. Swart, K. C. Lohmann, J. Mckenzie, and S. Savin (Washington, DC: American Geophysical Union), 1–36. doi: 10.1029/GM078p0001
Rubenstein, D. R., and Hobson, K. A. (2004). From birds to butterflies: animal movement patterns and stable isotopes. Trends Ecol. Evolut. 19, 256–263. doi: 10.1016/j.tree.2004.03.017
Russell, R. W., and Young, J. W. (1990). A review of metabolism of labeled glucoses for use in measuring glucose recycling. J. Dairy Sci. 73, 1005–1016. doi: 10.3168/jds.S0022-0302(90)78759-0
Sauer, P. E., Eglinton, T. I., Hayes, J. M., Schimmelmann, A., and Sessions, A. L. (2001). Compound-specific D/H ratios of lipid biomarkers from sediments as a proxy for environmental and climatic conditions. Geochim. Cosmochim. Acta 65, 213–222. doi: 10.1016/S0016-7037(00)00520-2
Sauer, U., Canonaco, F., Heri, S., Perrenoud, A., and Fischer, E. (2004). The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279, 6613–6619. doi: 10.1074/jbc.M311657200
Saur, W. K., Crespi, H. L., Halevi, E. A., and Katz, J. J. (1968). Deuterium isotope effects in the fermentation of hexoses to ethanol by Saccharomyces cerevisiae. I. Hydrogen exchange in the glycolytic pathway. Biochemistry 7, 3529–3536. doi: 10.1021/bi00850a030
Schindler, U., Sans, N., and Schröder, J. (1989). Ornithine cyclodeaminase from octopine Ti plasmid Ach5: identification, DNA sequence, enzyme properties, and comparison with gene and enzyme from nopaline Ti plasmid C58. J. Bacteriol. 171, 847–854. doi: 10.1128/jb.171.2.847-854.1989
Sessions, A. L., Jahnke, L. L., Schimmelmann, A., and Hayes, J. M. (2002). Hydrogen isotope fractionation in lipids of the methane-oxidizing bacterium Methylococcus capsulatus. Geochim. Cosmochim. Acta 66, 3955–3969. doi: 10.1016/S0016-7037(02)00981-X
Sharma, S., Villamor, J. G., and Verslues, P. E. (2011). Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiol. 157, 292–304. doi: 10.1104/pp.111.183210
Silverman, S. N., Phillips, A. A., Weiss, G. M., Wilkes, E. B., Eiler, J. M., and Sessions, A. L. (2022). Practical considerations for amino acid isotope analysis. Organic Geochem. 164:104345. doi: 10.1016/j.orggeochem.2021.104345
Smith, D. A., Nakamoto, B. J., Suess, M. K., and Fogel, M. L. (2022). Central metabolism and growth rate impacts on hydrogen and carbon isotope fractionation during amino acid synthesis in E. coli. Front. Microbiol. 13:840167. doi: 10.3389/fmicb.2022.840167
Smith, W. G., and York, J. L. (1970). Stereochemistry of the citric acid cycle. J. Chem. Educ. 47:588. doi: 10.1021/ed047p588
Stalon, V., Vander Wauven, C., Momin, P., and Legrain, C. (1987). Catabolism of arginine, citrulline and ornithine by Pseudomonas and related bacteria. J. General Microbiol. 133, 2487–2495. doi: 10.1099/00221287-133-9-2487
Urey, H. C. (1948). Oxygen isotopes in nature and in the laboratory. Science 108, 489–496. doi: 10.1126/science.108.2810.489
Venning, J. D., Bizouarn, T., Cotton, N. P. J., Quirk, P. G., and Jackson, J. B. (1998). Stopped-flow kinetics of hydride transfer between nucleotides by recombinant domains of proton-translocating transhydrogenase. Eur. J. Biochem. 257, 202–209. doi: 10.1046/j.1432-1327.1998.2570202.x
Wang, Y., Sessions, A. L., Nielsen, R. J., and Goddard, W. A. (2009). Equilibrium 2H/1H fractionations in organic molecules. II: Linear alkanes, alkenes, ketones, carboxylic acids, esters, alcohols and ethers. Geochim. Cosmochim. Acta 73, 7076–7086. doi: 10.1016/j.gca.2009.08.018
Wang, Y., Sessions, A. L., Nielsen, R. J., and Goddard, W. A. (2013). Equilibrium 2H/1H fractionation in organic molecules: III. Cyclic ketones and hydrocarbons. Geochim. Cosmochim. Acta 73107, 82–95. doi: 10.1016/j.gca.2013.01.001
Wassenaar, L. I., and Hobson, K. A. (2003). Comparative equilibration and online technique for determination of non-exchangeable hydrogen of keratins for use in animal migration studies. Isotopes Environ. Health Stud. 39, 211–217. doi: 10.1080/1025601031000096781
Wijker, R. S., Sessions, A. L., Fuhrer, T., and Phan, M. (2019). 2H/1H variation in microbial lipids is controlled by NADPH metabolism. Proc. Nat. Acad. Sci. 116, 12173–12182. doi: 10.1073/pnas.1818372116
Zampolli, M., Basaglia, G., Dondi, F., Sternberg, R., Szopa, C., and Pietrogrande, M. (2007). Gas chromatography-mass spectrometry analysis of amino acid enantiomers as methyl chloroformate derivatives: application to space analysis. J. Chromatogr. A 1150, 162–172. doi: 10.1016/j.chroma.2006.12.033
Zhang, X., Gillespie, A. L., and Sessions, A. L. (2009). Large D/H variations in bacterial lipids reflect central metabolic pathways. Proc. Nat. Acad. Sci. 106, 12580–12586. doi: 10.1073/pnas.0903030106
Keywords: amino acids, hydrogen isotopes, isotope fractionation, aerobic metabolism, heterotrophic bacteria, pyruvate, NADPH
Citation: Silverman SN, Wijker RS and Sessions AL (2024) Biosynthetic and catabolic pathways control amino acid δ2H values in aerobic heterotrophs. Front. Microbiol. 15:1338486. doi: 10.3389/fmicb.2024.1338486
Received: 14 November 2023; Accepted: 20 February 2024;
Published: 05 April 2024.
Edited by:
Shuhei Ono, Massachusetts Institute of Technology, United StatesReviewed by:
Naohiko Ohkouchi, Japan Agency for Marine-Earth Science and Technology (JAMSTEC), JapanJeemin H. Rhim, University of California, Santa Barbara, United States
Copyright © 2024 Silverman, Wijker and Sessions. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaelyn N. Silverman, c3NpbHZlcm1hbkBjYWx0ZWNoLmVkdQ==
†Present address: Reto S. Wijker, Geological Institute, Department of Earth Sciences, ETH Zürich, Zürich, Switzerland
 Shaelyn N. Silverman
Shaelyn N. Silverman Reto S. Wijker†
Reto S. Wijker† Alex L. Sessions
Alex L. Sessions