
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 February 2024
Sec. Evolutionary and Genomic Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1338231
Wickerhamomyces is a well-known genus of the family Wickerhamomycetaceae in the class Ascomycetes. These fungi can survive in a variety of substrates and environments and perform many valuable roles in both industrial processes and the natural ecosystems. During our investigation of yeast diversity associated with plant materials, 53 Wickerhamomyces isolates were obtained from rotting wood and plant leaves collected in Fujian, Guizhou, Henan, and Yunnan Provinces of China. Isolates were identified as 14 Wickerhamomyces species, including 1 species known previously to occur in China (W. anomalus), 9 new record species in China (W. arborarius, W. ciferrii, W. edaphicus, W. lynferdii, W. pijperi, W. subpelliculosa, W. xylosica, W. strasburgensis, and W. sydowiorum), and 4 novel species (W. guiyangensis sp. nov., W. paramyanmarensis sp. nov., W. quanzhouensis sp. nov., and W. phyllophilus sp. nov.). This study presents a detailed account of these new species, illustrating their morphology and analyzing their phylogenetic relationships with other Wickerhamomyces species. Our study is the first comprehensive study on Wickerhamomyces species associated with plant materials from tropical and subtropical China. The results of this study update our understanding of the phylogenetic relationships, systematics, and ecology of Wickerhamomyces.
The genus Wickerhamomyces was first established by Kurtzman et al. (2008) with the type species Wickerhamomyces canadensis, which formerly belonged to the genus Pichia (Kurtzman, 2011a). Based on the phylogenetic analysis of concatenated sequences of LSU, SSU, and EF-1α, this genus was placed in the family Wickerhamomycetaceae of the order Saccharomycetales (Kurtzman et al., 2008), which was further confirmed by using a genome-scale phylogeny (Groenewald et al., 2023). With the progress of yeast isolation technology and molecular biology, a number of Wickerhamomyces species have been discovered in different regions of the world. Currently, Wickerhamomyces consists of 38 defined species, including the most recent additions of W. lannaensis, W. nanensis, and W. sinyiensis (Nundaeng et al., 2021; Chang et al., 2022). Among the 38 valid species, 12 are asexual morphs and 26 have known for their ascosporic states (Kurtzman, 2011b; Shimizu et al., 2020; Nundaeng et al., 2021; Chang et al., 2022). In addition, 10 anamorphic Candida species, including C. mycetangii, C. namnaoensis, C. nitrativorans, C. odintsovae, C. peoriensis, C. ponderosae, C. quercuum, C. silvicultrix, C. solani, and C. ulmi, belong to the Wickerhamomyces clade based on phylogenetic analysis (Kurtzman, 2011b; Lachance et al., 2011; Shimizu et al., 2020), which will be transferred to the genus Wickerhamomyces as new combinations in the future. Phenotypically, the asexual morphs of Wickerhamomyces reproduced by multilateral budding on a narrow base, and some species produce pseudohyphae and/or true hyphae (Shimizu et al., 2020; Nundaeng et al., 2021; Chang et al., 2022). The sexual morphs of Wickerhamomyces are characterized by the production of hat-shaped or spherical ascospores with equatorial ledges (Kurtzman et al., 2008; Kurtzman, 2011b). Members of the genus Wickerhamomyces ferment glucose and possess Q-7 as a predominant ubiquinone. They can utilize various carbon sources but not methanol or hexadecane (Kurtzman et al., 2008; Kurtzman, 2011b). Due to the similarity in phenotypic characteristics within the genus, a combination of phenotypical characteristics and phylogenetic analysis has been adopted as the standard method for concretely identifying Wickerhamomyces species (Kurtzman et al., 2008; Nundaeng et al., 2021).
Members of the genus Wickerhamomyces have been studied for a variety of applications, and most previous studies have focused on the most widely distributed species W. anomalus (Nundaeng et al., 2021). The strains of this species offer a diversity of desirable characteristics; BS91 and DMKU-RP04 demonstrated resistance to plant pathogenic fungi (Czarnecka et al., 2019; Limtong et al., 2020); SDBR-CMU-S1-06 and Wa-32 produced the plant growth promoter indole-3-acitic acid (IAA) (Kumla et al., 2018); CBS261, HN006, and HN010 produced ethylacetate, which is used to enhance the aroma and quality of Chinese liquor (Ravasio et al., 2018; Yan et al., 2019; Li et al., 2021). Notably, most strains of W. anomalus are known to produce killer toxins that possess antimicrobial and larvicidal activities (Walker, 2011; Nundaeng et al., 2021). In addition, several other species, including W. bovis, W. silvicola, W. edaphicus, W. siamensis, W. rabaulensis, W. psychrolipolyticus, and W. chambardii, can be produced mycocin, biosurfactants, saturated fatty acids, xylitol, cellulase, and lipases that could be applied in the bioremediation, biotechnology, food, and cosmetic industries (Shimizu et al., 2020; Nundaeng et al., 2021). On the other hand, some Wickerhamomyces species, such as W. anomalus and W. lynferdii, have been responsible for the spoilage of beer and bakery products (Lanciotti et al., 1998; Timke et al., 2005). W. anomalus, W. myanmarensis, and W. onychis, which have been reported to exist clinical specimens, appear to be opportunistic human pathogens (Kurtzman, 2011b; Arastehfar et al., 2019).
Species of Wickerhamomyces are widely distributed in tropical, subtropical, and temperate regions (Kurtzman, 2011b; Nundaeng et al., 2021). Presently, 20 Wickerhamomyces species have been reported in Asia, with 10 species (W. anomalus, W. ciferrii, W. edaphicus, W. lannaensis, W. nanensis, W. rabaulensis, W. siamensis, W. sydowiorum, W. tratensis, and W. xylosicus) found in Thailand (Limtong et al., 2012; Nakase et al., 2012; Kaewwichian et al., 2013), two (W. psychrolipolyticus and W. scolytoplatypi) in Japan (Ninomiya et al., 2013; Shimizu et al., 2020), two (W. silvicola and W. ochangensis) in South Korea (Shin et al., 2011), one (W. xylosivorus) in Indonesia (Kobayashi et al., 2017), and four (W. anomalus, W. kurtzmanii, W. menglaensis, and W. mori) in China (Hui et al., 2013; You et al., 2016; Chai et al., 2019; Zhou et al., 2019).
The presence of Wickerhamomyces in China has not been extensively documented. Current members of the genus have been reported only in Sichuan Province (W. anomalus) (You et al., 2016), the Inner Mongolia Autonomous Region (W. kurtzmanii) (Zhou et al., 2019), Yunnan Province (W. menglaensis) (Chai et al., 2019), and Henan Province (W. mori) (Hui et al., 2013). China is located in the northern hemisphere, providing a temperate environment suitable for hosting Wickerhamomyces species. To investigate the diversity of Wickerhamomyces in China, we isolated yeast strains from plant materials collected in Fujian, Guizhou, Henan, and Yunnan Provinces using a culture-based approach. A total of 14 species, including 4 new species, were identified from the isolated yeast strains. This study aims to characterize the diversity of Wickerhamomyces species inhabiting these substrates and describe four new species based on morphological characteristics and phylogenetic analyses of ITS and LSU.
Thirty-six strains used in this study were isolated from rotting wood by an enrichment method described by Shi et al. (2020). An additional 17 strains were isolated from plant leaves by the ballistospore-fall method as previously described (Nakase and Takashima, 1993; Hu et al., 2022). Vaseline was employed to adhere semi-withered leaf samples inside the lids of Petri plates containing yeast malt (YM) agar (0.3% yeast extract, 0.3% malt extract, 0.5% peptone, 1% glucose, and 2% agar). The plates were then incubated at 25°C for 7 days until colonies gradually formed. Different yeast morphotypes were picked and purified by streaking onto fresh YM agar. All purified yeast strains were then suspended in YM broth supplemented with 20% (v/v) glycerol and stored at −80°C. All isolates used in this study and their origins are presented in Table 1.
The morphological and physiological characteristics of each yeast strain were examined according to the methods established by Kurtzman et al. (2011). Colony characteristics were observed on YM agar after 2 weeks of incubation at 25°C. Mycelium formation was investigated by cultivation on corn meal (CM) agar (2% cornmeal infusion and 2% agar) in slide culture at 25°C for 2 weeks. Sexual processes of all strains were studied individually and in strain pairs on YM agar, CM agar, 5% malt extract (ME) agar, and V8 agar at 20°C and 25°C for 2 months with weekly observation (Kurtzman, 2011b). Fermentation of glucose was carried out in liquid medium using Durham fermentation tubes. Carbon source and nitrogen source assimilation tests were conducted in liquid medium, and starved inoculum was used in nitrogen assimilation tests (Kurtzman et al., 2011). Cycloheximide resistance was performed in liquid medium, and urea hydrolysis was conducted on agar slants. Acid production and the diazonium blue B (DBB) reaction tests were performed in Petri dish with a solid medium. Growth potential at various temperatures (15°C, 20°C, 25°C, 30°C, 35°C, and 37°C) was determined by cultivation on YM agar. Cell morphology was examined through a Leica DM 2500 microscope (Leica Microsystems GmbH, Wetzlar, Germany) with a Leica DFC295 digital microscope color camera employing bright field, phase contrast, and differential interference contrast (DIC) optics. Novel taxonomic descriptions and proposed names were deposited in MycoBank (http://www.mycobank.org; 8 November 2023).
Genomic DNA was extracted from each yeast isolate using the Ezup Column Yeast Genomic DNA Purification Kit, according to the manufacturer’s protocol (Sangon Biotech, Shanghai, China). Sequenced data were generated from the internal transcribed spacer (ITS) region and the D1/D2 domain of the large subunit (LSU) rRNA gene using primer pairs ITS1/ITS4 (White et al., 1990) and NL1/NL4 (Kurtzman and Robnett, 1998). PCR amplification was performed in a 25 μL reaction volume containing 9.5 μL of ddH2O, 12.5 μL of 2 × Taq PCR Master Mix with blue dye (Sangon Biotech, Shanghai, China), 1 μL of DNA template, and 1 μL of each primer. PCR reactions were carried out according to the following conditions: initial denaturation step at 95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 56°C for 30 s, 72°C for 40 s, and a final extension at 72°C for 10 min. PCR products were checked and purified in 1% agarose gels before being sequenced by Sangon Biotech (Shanghai) Co., Ltd. The identity and accuracy of each sequence were determined by GenBank sequences and assembled using BioEdit 7.1.3.0 (Hall, 1999). Newly obtained sequences were then submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/; Table 2).
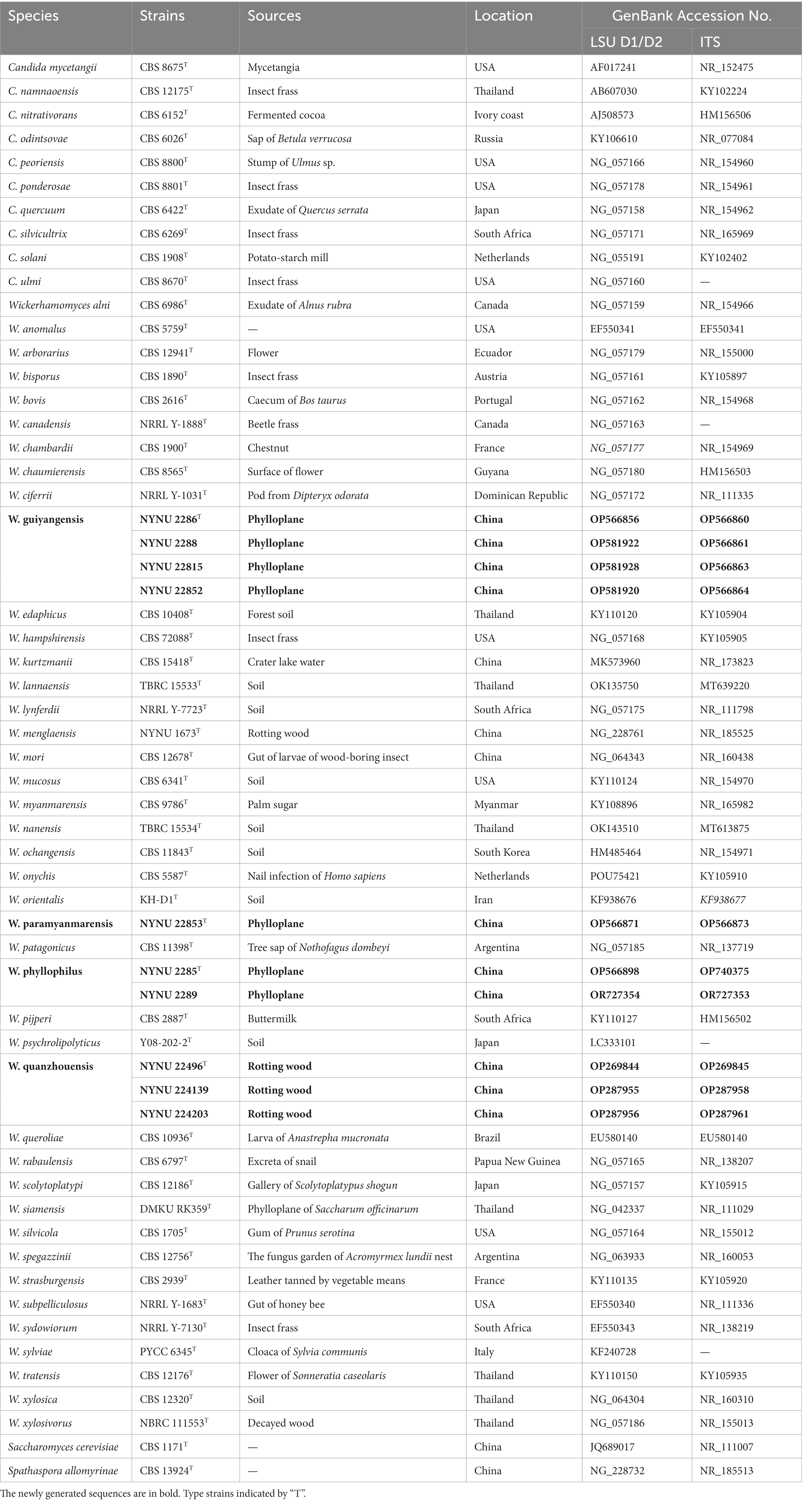
Table 2. Taxa, strains, sources, locations, and corresponding GenBank numbers of the taxa used in this study.
A total of 116 nucleotide sequences that belonged to 54 taxa were included in the phylogenetic analyses. Except for 20 sequences recognized in this study, the other sequences were obtained from previous studies (Shimizu et al., 2020; Nundaeng et al., 2021; Chang et al., 2022) and GenBank (Table 2). Spathaspora allomyrinae CBS 13924T and Saccharomyces cerevisiae CBS 1171T were used as the outgroup. The phylogenetic relationships between Wickerhamomyces species and related species were determined through the analysis of combined ITS and LSU datasets. Single gene sequence alignments were generated using CLUSTAL_X v.1.83 (Thompson et al., 1997) or MAFFT 7.110 (Katoh and Standley, 2013) before being manually adjusted with BioEdit v. 7.1.3.0 (Hall, 1999). Multiple genes were concatenated using SequenceMatrix v. 1.7.8 (Vaidya et al., 2011). Gblocks 0.91b was used to detect and remove ambiguously aligned regions from each sequence before phylogenetic analysis (Castresana, 2000). The best-fit evolutionary model was estimated by using Modelfinder (Kalyaanamoorthy et al., 2017).
Phylogenetic analyses were performed using both maximum likelihood (ML) and Bayesian (BI) analyses. ML analysis was performed using RAxML v. 8.2.10 (Stamatakis, 2006) with PHY files generated by CLUSTAL_X v.1.83 (Thompson et al., 1997). The best scoring tree was selected among suboptimal trees from each run by comparing likelihood scores under the GTRGAMMA substitution model. The resulting replicates were plotted on the best scoring tree, which was obtained previously. Statistical support values (BS) were obtained using non-parametric bootstrapping with 1,000 replicates. BI analysis was conducted by MrBayes v. 3.2.7a (Ronquist et al., 2012) in the CIPRES Science Gateway version 3.3. GTR + I + G was selected as the best-fit model for the concatenated dataset. Posterior probabilities (PP) were determined by Markov chain Monte Carlo sampling (MCMC) (Rannala and Yang, 1996). Six simultaneous Markov chains were run for 50 million generations, and the trees were sampled every 1000th generation; thus, 50,000 trees were obtained. The first 25% of the saved trees, representing the burn-in phase of the analysis, were discarded. The remaining trees were used for calculating posterior probabilities (PP) in the majority rule consensus tree.
The phylogenetic trees were viewed with FigTree v. 1.4.3 (Andrew, 2016) and processed with Adobe Illustrator CS5. ML bootstrap support (BS) ≥50% and Bayesian posterior probabilities (PP) ≥0.95 are displayed on the edited phylogenetic tree in the first and second positions, respectively.
During this investigation, we isolated 36 and 17 yeast strains from rotting wood and plant leaf samples, respectively. Each strain represents the morphology in an individual sample (Table 1). All isolates were identified at the species level based on the threshold of >99% sequence identity with the type strain of described species in the D1/D2 domain or ITS region (Kurtzman and Robnett, 1998; Fell et al., 2000; Vu et al., 2016). In total, we identified 14 distinct species belonging to Wickerhamomyces. Among the wood samples, eight known species were represented (W. anomalus, W. arborarius, W. ciferrii, W. edaphicus, W. lynferdii, W. pijperi, W. subpelliculosa, and W. xylosica) as well as one potentially novel (Wickerhamomyces sp. 1). W. anomalus and W. pijperi were the most dominant, occurring in seven to 10 samples collected across the collection locations. W. arborarius, W. ciferrii, and W. subpelliculosa were the most scarce, occurring only in one sample or location (Table 1). The 17 Wickerhamomyces strains collected from plant leaf samples were identified as belong to two known species (W. strasburgensis and W. sydowiorum) and three potentially new species (Wickerhamomyces sp. 2, Wickerhamomyces sp. 3, and Wickerhamomyces sp. 4). W. sydowiorum was the most abundant yeast species among the leaf samples, followed by Wickerhamomyces sp. 3, which occurred in four to seven samples (Table 1).
Among the 53 yeast strains isolated, 10 strains represented 4 new species in the genus Wickerhamomyces. To reveal the phylogenetic position of the new species, phylogenetic analyses were performed with a combination of ITS and LSU datasets. A total of 1,276 characteristics, including gaps (ITS: 1–666 and LSU: 667–1,276), were included in the sequence dataset for the phylogenetic analysis. Of these characteristics, 517 were constant, 156 were parsimony-uninformative, and 603 were parsimony-informative. The topology of the ML and Bayesian trees was consistent, therefore only the ML analysis is displayed with BS (≥50%) and PP (≥0.95) at the nodes (Figure 1). In our phylogenetic tree, 10 newly isolated strains were formed into four separate groups within the Wickerhamomyces clade, all of which were well supported (100% BS/1 PP) and distinct from other known species.
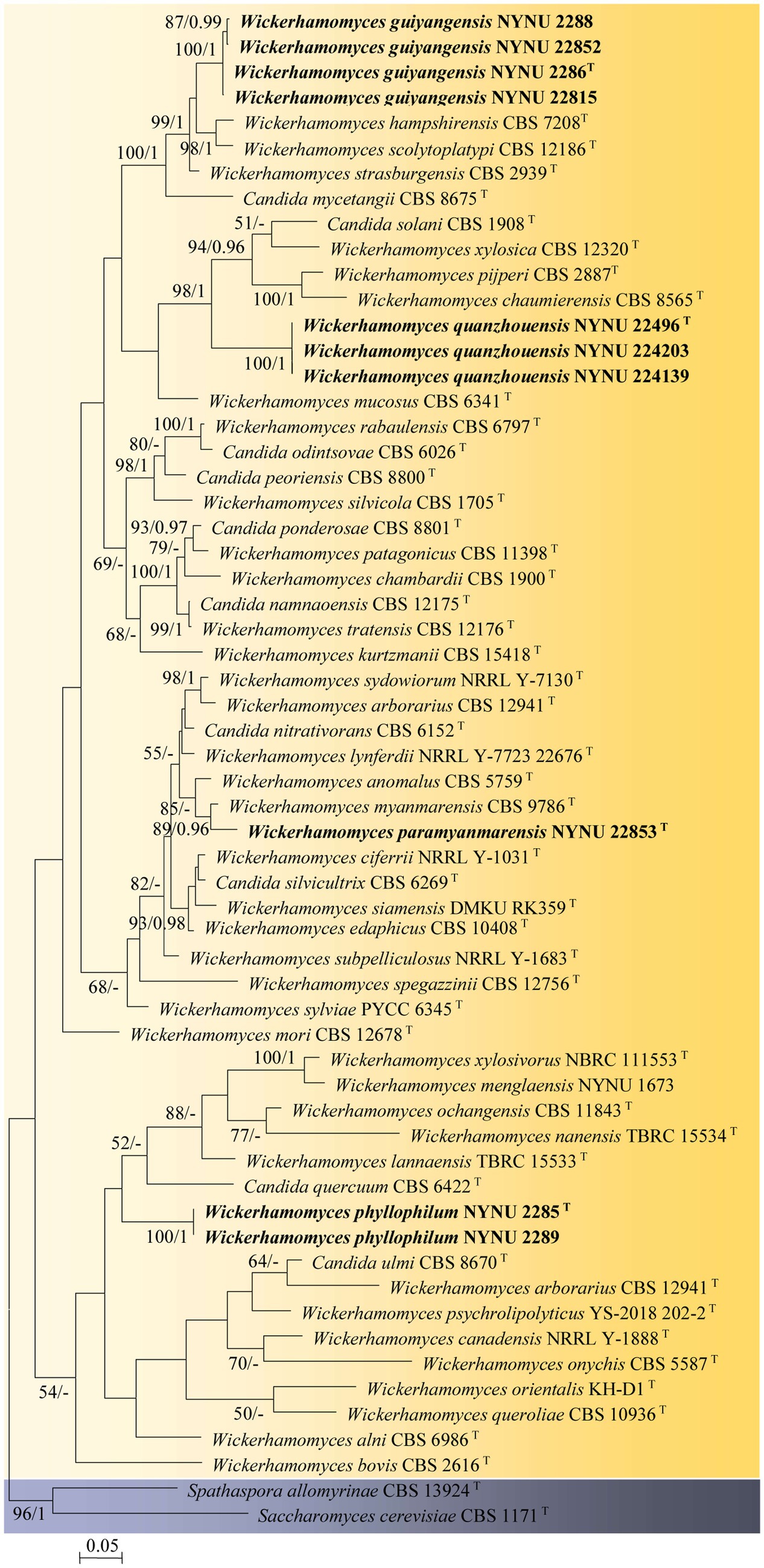
Figure 1. Maximum likelihood phylogram of Wickerhamomyces based on the combined ITS and LSU sequence data. Spathaspora allomyrinae CBS 13924T and Saccharomyces cerevisiae CBS 1171T were used as outgroups. Branches are labeled with BS >50% and PP >0.95, respectively. Strains obtained in the present study are shown in bold.
Four strains isolated from the phylloplane, NYNU 2286T, NYNU 2288, NYNU 22815, and NYNU 22852, formed a well-supported clade that clustered with W. hampshirensis, W. scolytoplatypi, and W. strasburgensis with high statistical support (99% BS/1 PP; Figure 1). The four strains of the NYNU 2286T group possess similar sequences with zero to six nt substitutions in the D1/D2 domain and ITS region, indicating that they belong to the same species. BLASTn searches of the D1/D2 and ITS sequences indicate that the NYNU 2286T group is most closely related to type strain of W. strasburgensis, differing by 19 nt (~3.3%) substitutions in the D1D2 domain and by 21–29 nt (~3.4%–4.7%) mismatches in the ITS region, respectively. Therefore, the NYNU 2286T group represents a novel Wickerhamomyces species, for which the name W. guiyangensis sp. nov. is proposed.
Strain NYUN 22853T, isolated from the phylloplane, was found to be closely related to W. myanmarensis CBS 9786T with strong statistical support (89% BS/0.96 PP; Figure 1). The two strains differed by 7 nt (~1.2%) substitutions in the D1/D2 domain and 19 nt (~3%) mismatches in the ITS region, respectively. These findings suggest that NYUN 22853T represents a novel species in the genus Wickerhamomyces, for which the name W. paramyanmarensis sp. nov. is proposed.
Three isolates collected from rotting wood, NYNU 22496T, NYNU 224139, and NYNU 224203, formed a separate branch, clustering with C. solani, W. xylosica, W. pijperi, and W. chaumierensis with good statistical support (98% BS/1 PP; Figure 1). Three isolates of the NYNU 22496T group shared 100% nucleotide identity based on D1/D2 and ITS sequences, indicating that they are conspecific. This group differed from these four known species by 44–53 nt (~7.7–9.3%) substitutions in the D1/D2 domain and more than 26–74 nt (~7.9–11.7%) mismatches in the ITS region. Hence, the NYNU 22496T group represents a novel Wickerhamomyces species, for which the name W. quanzhouensis sp. nov. is proposed.
Two strains isolated from the phylloplane, NYNU 2285T and NYNU 2289, were located at a basal branch related to C. quercuum CBS 6422T but without support (Figure 1). The two strains of the NYNU 2285T group had identical sequences in both the D1/D2 domain and ITS region. This group differed from the closest relative C. quercuum CBS 6422T by 20 nt (~3.4%) substitutions in the D1/D2 domain and 18 nt (~5.9%) mismatches in the ITS region, respectively, suggesting that the NYNU 2285T group represents a new species in Wickerhamomyces, for which the name W. phyllophilus sp. nov. is proposed.
Wickerhamomyces guiyangensis C. Y. Chai and F. L. Hui, sp. nov., Figure 2.
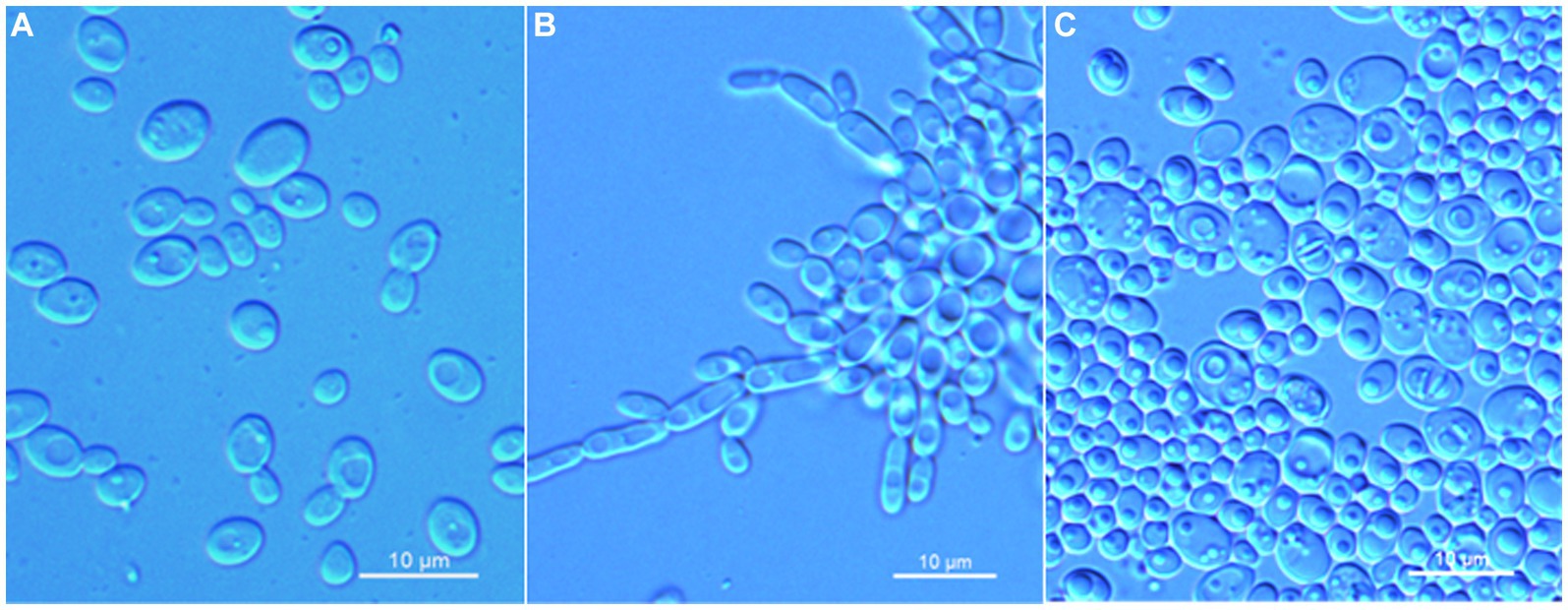
Figure 2. Morphological characteristics of Wickerhamomyces guiyangensis sp. nov. (GDMCC 2.320T, holotype). (A) Budding yeast cells, 3 days, YM broth, 25°C. (B) Pseudohyphae, 7 days, CM agar, 25°C. (C) Ascospores, 7 days, 5% ME agar, 25°C. Scale bars = 10 μm.
MycoBank: MB 850882.
Etymology: the specific epithet “guiyangensis” refers to the geographic origin of the type strain, Guiyang city, Guizhou.
Typus: China, Guizhou Province, Guiyang City, Guiyang Botanical Garden, in the phylloplane from undetermined leaf, August 2022, L. Zhang and F. L. Hui, NYUN 2286 (holotype GDMCC 2.320T preserved as a metabolically inactive state, culture ex-type PYCC 9922).
Description: On YM agar, after 7 days at 25°C, colonies are white to cream, raised and butyrous with a smooth surface and an entire margin. In YM broth, after 3 days at 25°C, cells are ovoid or ellipsoidal (3.1–5.9 × 3.5–7.3 μm), occur singly or budding pairs. Budding is multilateral. After 1 month at 25°C, a ring and sediment are present. In Dalmau plate culture on corn meal agar, pseudohyphae are formed, but not true hyphae. Ascospores are observed on 5% ME agar after 7 days of culture at 25°C. D-Glucose and sucrose are fermented, but not D-galactose, maltose, lactose, raffinose, trehalose, or D-xylose. Glucose, inulin, sucrose, raffinose, galactose, trehalose, maltose, melezitose, methyl-α-D-glucoside, cellobiose, salicin, L-rhamnose, D-xylose, L-arabinose, D-arabinose, 5-keto-D-gluconate, glycerol, D-mannitol, D-glucitol, DL-lactate, succinate, citrate, D-gluconate, and D-glucono-1,5-lactone are assimilated as sole carbon sources. Melibiose, lactose, L-sorbose, D-ribose, methanol, ethanol, erythritol, ribitol, galactitol, myo-inositol, D-glucosamine, 2-keto-D-gluconate, and D-glucuronate are not assimilated. Ethylamine and L-lysine are assimilated as sole nitrogen sources. Nitrite, nitrate, and cadaverine are not assimilated. Maximum growth temperature is 35°C. Growth in vitamin-free medium is positive. Growth with 0.01% cylcloheximide, 10% NaCl/5% glucose, and 1% acetic acid is negative. Starch-like compounds are not produced. Diazonium blue B color and urease reaction are negative.
Additional strain examined: China, Guizhou Province, Guiyang City, Guiyang Botanical Garden, in the phylloplane from undetermined leaf, August 2022, L. Zhang and F. L. Hui, NYUN 2288, NYUN 22815 and NYUN 22852.
GenBank accession numbers: holotype GDMCC 2.320T (ITS: OP566860, D1/D2: OP566856); additional strains NYUN 2288 (ITS: OP566861, D1/D2: OP581922), NYUN 22815 (ITS: OP566863, D1/D2: OP581928), and NYUN 22852 (ITS: OP566864, D1/D2: OP581920).
Note: Physiologically, W. guiyangensis sp. nov. differed from its close relative W. strasburgensis (Kurtzman, 2011b) by its ability to assimilate inulin, D-arabinose, and 5-keto-D-gluconate and its inability to grow in 10% NaCl/5% glucose. Moreover, W. strasburgensis weakly ferments galactose and raffinose, while the new species does not.
Wickerhamomyces paramyanmarensis C. Y. Chai and F. L. Hui, sp. nov., Figure 3.
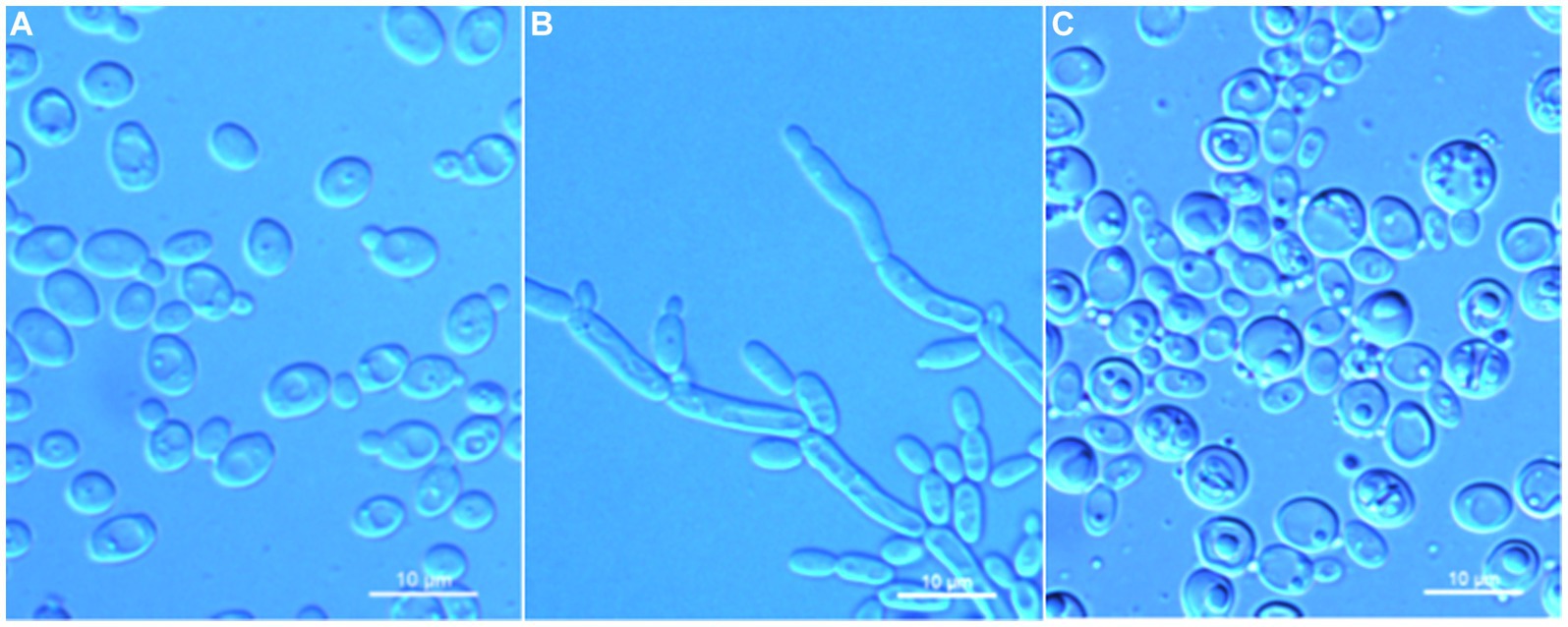
Figure 3. Morphological characteristics of Wickerhamomyces paramyanmarensis sp. nov. (GDMCC 2.304T, holotype). (A) Budding yeast cells, 3 days, YM broth, 25°C. (B) Pseudohyphae, 7 days, CM agar, 25°C. (C) Ascospores, 7 days, 5% ME agar, 25°C. Scale bars = 10 μm.
MycoBank: MB 850883.
Etymology: the specific epithet “paramyanmarensis” refers to its phylogenetic closeness to W. myanmarensis.
Typus: China, Guizhou Province, Guiyang City, Guiyang Botanical Garden, in the phylloplane from undetermined leaf, August 2022, L. Zhang and F. L. Hui, NYNU 22853 (holotype GDMCC 2.304T preserved as a metabolically inactive state, culture ex-type PYCC 9924).
Description: On YM agar, after 7 days at 25°C, colonies are white to cream, raised and butyrous with a smooth surface and an entire margin. In YM broth, after 3 days at 25°C, cells are ovoid or ellipsoidal (3.5–4.9 × 4.6–7.2 μm), occur singly or budding pairs. Budding is multilateral. After 1 month at 25°C, a ring and sediment are present. In Dalmau plate culture on corn meal agar, pseudohyphae are formed but not true hyphae. Ascospores are observed on 5% ME agar after 7 days of culture at 25°C. D-Glucose and sucrose are fermented but not D-galactose, maltose, lactose, raffinose, trehalose, or D-xylose. Glucose, inulin, sucrose, raffinose, melibiose, galactose, lactose, trehalose, maltose, melezitose, methyl-α-D-glucoside, cellobiose, L-rhamnose, D-xylose, L-arabinose, D-ribose, glycerol, erythritol, D-mannitol, D-glucitol, DL-lactate, D-gluconate, 2-keto-D-gluconate, and D-glucuronate are assimilated as sole carbon sources. Salicin, L-sorbose, D-arabinose, 5-keto-D-gluconate, methanol, ethanol, ribitol, galactitol, myo-inositol, succinate, citrate, D-glucosamine, and glucono-1,5-lactone are not assimilated. Nitrite, nitrate, ethylamine, L-lysine, and cadaverine are not assimilated. Maximum growth temperature is 35°C. Growth in vitamin-free medium is negative. Growth with 0.01% cylcloheximide, 10% NaCl/5% glucose, and 1% acetic acid is negative. Starch-like compounds are not produced. Diazonium blue B color and urease reaction are negative.
GenBank accession numbers: holotype GDMCC 2.304T (ITS: OP566873, D1/D2: OP566871).
Note: Physiologically, W. paramyanmarensis sp. nov. differed from its close relative W. myanmarensis (Kurtzman, 2011a) in the ability to assimilate inulin, melibiose, lactose, and L-rhamnose and inability to assimilate salicin, D-arabinose, ethanol, ribitol, succinate, and citrate and growth at 37°C. Moreover, W. myanmarensis latently ferments galactose, maltose, and raffinose, while the new species does not.
Wickerhamomyces quanzhouensis C. Y. Chai and F. L. Hui, sp. nov., Figure 4.
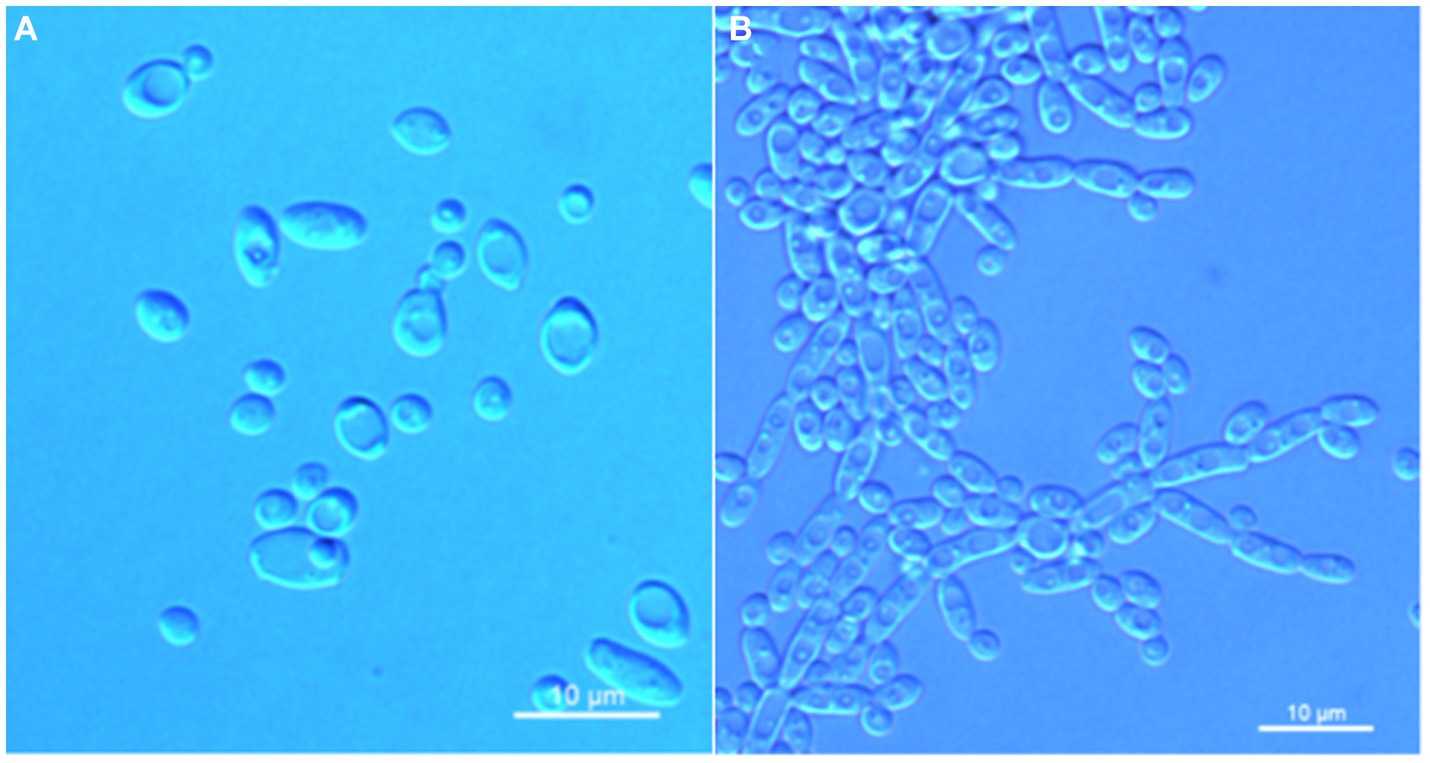
Figure 4. Morphological characteristics of Wickerhamomyces quanzhouensis sp. nov. (GDMCC 2.308T, holotype). (A) Budding yeast cells, 3 days, YM broth, 25°C. (B) Pseudohyphae, 7 days, CM agar, 25°C. Scale bars = 10 μm.
MycoBank: MB 850884.
Etymology: the specific epithet “quanzhouensis” refers to the geographic origin of the type strain, Quanzhou city, Fujian.
Typus: China, Fujian Province, Quanzhou City, Qingyuan Mountain, in rotting wood, April 2022, W. T. Hu and S. B. Chu, NYNU 22496 (holotype GDMCC 2.308T preserved as a metabolically inactive state, culture ex-type PYCC 9923).
Description: On YM agar, after 7 days at 25°C, colonies are white to cream color, raised and butyrous with a smooth surface and an entire margin. In YM broth, after 3 days at 25°C, cells are ovoid to ellipsoid (2.0–3.7 × 2.6–7.5 μm), occur singly or budding pairs. Budding is multilateral. After 1 month at 25°C, a ring and sediment are present. In Dalmau plate culture on corn meal agar, pseudohyphae are formed but not true hyphae. Ascospores were not obtained for individual strains and strain pairs on YM agar, CM agar, 5% ME agar, and V8 agar at 20°C and 25°C for 2 months. D-Glucose is fermented but not D-galactose, sucrose, maltose, lactose, raffinose, trehalose, or D-xylose. Glucose, inulin, sucrose, raffinose, trehalose, maltose, cellobiose, salicin, D-xylose, D-arabinose, ethanol, glycerol, ribitol, galactitol, D-mannitol, D-glucitol, DL-lactate, succinate, citrate, and glucono-1,5-lactone are assimilated as sole carbon sources. Melibiose, galactose, lactose, melezitose, methyl-α-D-glucoside, L-sorbose, L-rhamnose, L-arabinose, D-ribose, methanol, erythritol, myo-inositol, D-gluconate, D-glucosamine, 2-keto-D-gluconate, and D-glucuronate are not assimilated. Ethylamine and L-lysine are assimilated as sole nitrogen sources. Nitrite, nitrate, and cadaverine are not assimilated. Maximum growth temperature is 30°C. Growth in vitamin-free medium is positive. Growth with 0.01% cylcloheximide, 10% NaCl/5% glucose, and 1% acetic acid is negative. Starch-like compounds are not produced. Diazonium blue B color and urease reaction are negative.
Additional strain examined: China, Fujian Province, Quanzhou City, Qingyuan Mountain, in rotting wood, April 2022, W. T. Hu and S. B. Chu, NYUN 224139 and NYUN 224203.
GenBank accession numbers: holotype GDMCC 2.308T (ITS: OP269845, D1/D2: OP269844); additional strains NYUN 224139 (ITS: OP287958, D1/D2: OP287955) and NYUN 224203 (ITS: OP287961, D1/D2: OP287956).
Note: Physiologically, W. quanzhouensis sp. nov. differed from its close relative W. xylosica (Limtong et al., 2012) in the ability to assimilate inulin, raffinose, D-arabinose, ribitol, galactitol, D-glucitol, and citrate and inability to assimilate melezitose, L-sorbose, and 2-keto-D-gluconate.
Wickerhamomyces phyllophilus C. Y. Chai and F. L. Hui, sp. nov., Figure 5.
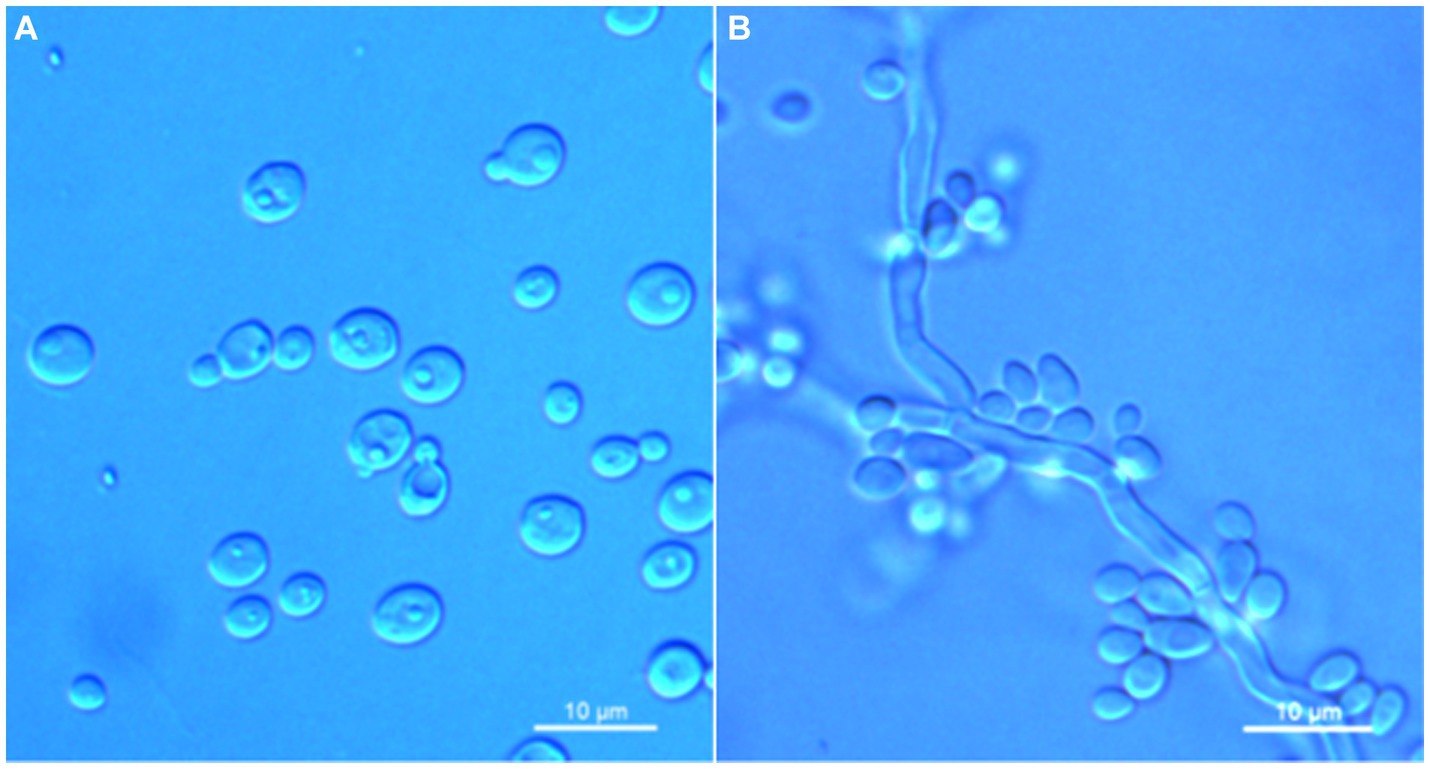
Figure 5. Morphological characteristics of Wickerhamomyces phyllophilus sp. nov. (GDMCC 2.301T, holotype). (A) Budding yeast cells, 3 days, YM broth, 25°C. (B) Pseudohyphae, 7 days, CM agar, 25°C. Scale bars = 10 μm.
MycoBank: MB 850885.
Etymology: the specific epithet “phyllophilus” refers to leaves, the substrate origin of the type strain.
Typus: China, Guizhou Province, Guiyang City, Guiyang Botanical Garden, in the phylloplane from leaf of Celtis sp., August 2022, L. Zhang and F. L. Hui, NYNU 2285 (holotype GDMCC 2.301T preserved as a metabolically inactive state, culture ex-type PYCC 9921).
Description: On YM agar, after 7 days at 25°C, colonies are white to cream color, raised, and butyrous with a smooth surface and an entire margin. In YM broth, after 3 days at 25°C, cells are ovoid to ellipsoid (3.4–5.7 × 3.6–6.7 μm), occur singly or budding pairs. Budding is multilateral. After 1 month at 25°C, a ring and sediment are present. In Dalmau plate culture on corn meal agar, pseudohyphae are formed but not true hyphae. Ascospores were not obtained for individual strains and strain pairs on YM agar, CM agar, 5% ME agar, and V8 agar at 20°C and 25°C for 2 months. D-Glucose is fermented but not D-galactose, sucrose, maltose, lactose, raffinose, trehalose, or D-xylose. Glucose, inulin, sucrose, galactose, trehalose, maltose, melezitose, methyl-α-D-glucoside, cellobiose, D-xylose, glycerol, D-mannitol, D-glucitol, DL-lactate, and D-gluconate (weak) are assimilated as sole carbon sources. Raffinose, melibiose, lactose, salicin, L-sorbose, L-rhamnose, L-arabinose, D-arabinose, 5-keto-D-gluconate, D-ribose, methanol, ethanol, erythritol, ribitol, galactitol, myo-inositol, succinate, citrate, D-glucosamine, 2-keto-D-gluconate, D-glucuronate, and glucono-1,5-lactone are not assimilated. Nitrite, nitrate, ethylamine, L-lysine, and cadaverine are not assimilated. Maximum growth temperature is 35°C. Growth in vitamin-free medium is negative. Growth with 0.01% cylcloheximide, 10% NaCl/5% glucose, and 1% acetic acid is negative. Starch-like compounds are not produced. Diazonium blue B color and urease reaction are negative.
Additional strain examined: China, Guizhou Province, Guiyang City, Guiyang Botanical Garden, in the phylloplane from leaf of Celtis sp., August 2022, L. Zhang and F. L. Hui, NYUN 2289.
GenBank accession numbers: holotype GDMCC 2.301T (ITS: OP740375, D1/D2: OP566898); additional strains NYUN 2289 (ITS: OR727354, D1/D2: OR727353).
Note: Physiologically, W. phyllophilus sp. nov. differed from its close relative C. quercuum (Lachance et al., 2011) in the ability to assimilate inulin, galactose, and trehalose and inability to assimilate salicin, ethanol, succinate, and citrate.
Before advances in gene sequencing, the identification of Wickerhamomyces species was primarily based on phenotypic characteristics. However, due to the presence of many shared polymorphic characteristics and similar appearances across different species, this method is often inaccurate (Nundaeng et al., 2021). Now, ribosomal DNA gene sequence analysis has become the dominant form of identification as it has proven to be very effective (Vu et al., 2016). Molecular taxonomic studies have deeply improved our understanding of the phylogenetic relationships, systematics, and ecology of yeasts. As a result, a combination of phenotypic and phylogenetic data is used to concretely identify the Wickerhamomyces species (Kurtzman and Robnett, 1998; Nundaeng et al., 2021). In this study, we introduced four new Wickerhamomyces species, consisting of W. guiyangensis sp. nov., W. paramyanmarensis sp. nov., W. quanzhouensis sp. nov., and W. phyllophilus sp. nov., and describe them in terms of both phenotype and phylogeny. These new species formed four well-separated clades in the resultant phylogram, which indicated the distinct phylogenetic positions of each species within the genus Wickerhamomyces. Pairwise sequence comparisons of the D1/D2 domain and the ITS region between these four species and closely related species showed that they had lower similarity values than the common threshold for species demarcation in ascomycetous yeasts (Fell et al., 2000; Kurtzman et al., 2008; Vu et al., 2016). However, they were found to be highly similar in cell shape, colony morphology, and color, but they differed from the closest related species in terms of their physiological and biochemical characteristics. Therefore, a combination of phenotypic characteristics and molecular phylogenetic analyses conducted in our study confirmed the existence of these new species in China.
Before our study, only four Chinese species, W. anomalus, W. kurtzmanii, W. menglaensis, and W. mori, were known from China (Hui et al., 2013; You et al., 2016; Chai et al., 2019; Zhou et al., 2019). This study provides 14 additional species, including 1 previously known species (W. anomalus), 9 new recorded species (W. arborarius, W. ciferrii, W. edaphicus, W. lynferdii, W. pijperi, W. subpelliculosa, W. xylosica, W. strasburgensis, and W. sydowiorum), and 4 novel species (W. guiyangensis sp. nov., W. paramyanmarensis sp. nov., W. quanzhouensis sp. nov., and W. phyllophilus sp. nov.), increasing the number of Wickerhamomyces species from 4 to 17. In China, there are likely species that need to be identified, such as GenBank accession JQ901898. These studies suggest that there are likely even more Wickerhamomyces species waiting to be discovered. So far, almost no Wickerhamomyces species have been reported from eastern China, which also have abundant forest resources, so it is necessary to investigate yeast resources in these regions in the future studies.
Members of the Wickerhamomyces clade are widely distributed and are found in different habitats, as shown in Table 2. They can be successfully isolated from soil (Limtong et al., 2012; Shimizu et al., 2020), phylloplane (Kaewwichian et al., 2013; Into et al., 2020), tree exudates (Kurtzman, 2011b), flowers (Nakase et al., 2012), rotting wood (Kobayashi et al., 2017; Chai et al., 2019), insect (Hui et al., 2013), insect frass (Nakase et al., 2012), birds (Kurtzman, 2011b), patients (Kurtzman, 2011b; Arastehfar et al., 2019), fermented food (Ravasio et al., 2018; Yan et al., 2019; Li et al., 2021), brined vegetables (Kurtzman, 2011b), and crater lake water (Zhou et al., 2019), but most known species are found mostly in association with plant materials. However, only a few species, such as W. edaphicus, W. siamensis, W. menglaensis, and W. xylosivorus, have been isolated from rotting wood and plant leaf samples, respectively. In this study, we isolated nine species (W. anomalus, W. arborarius, W. ciferrii, W. edaphicus, W. lynferdii, W. pijperi, W. quanzhouensis sp. nov., W. subpelliculosa, and W. xylosica) from rotting wood and an additional five species (W. guiyangensis sp. nov., W. paramyanmarensis sp. nov., W. phyllophilus sp. nov., W. strasburgensis, and W. sydowiorum) from plant leaves. Previous studies showed that phylloplane yeast strains of W. anomalus have a variety of biological functions, such as resistance to plant pathogenic fungi and biosynthesis of the plant growth promoter indole-3-acitic acid (IAA) (Kumla et al., 2018; Czarnecka et al., 2019; Limtong et al., 2020). In this study, we also isolated 17 strains of Wickerhamomyces from the phylloplane, which may own similar ecological functions as W. anomalus. In addition, D-xylose assimilation was observed for W. quanzhouensis sp. nov., which was similar to those found for W. menglaensis and W. xylosivorus (Kobayashi et al., 2017; Chai et al., 2019). D-xylose assimilation may be an important physiological trait for these yeasts in the colonization of rotting wood. These findings expanded our knowledge of where Wickerhamomyces species can survive and demonstrates the complicated ecological function of this genus. Future research will focus on Wickerhamomyces diversity from a broader range of substrates and environments. Ultimately, these findings will help researchers gain a better understanding of the diversity, distribution, and ecology of Wickerhamomyces.
In this study, the species diversity of the genus Wickerhamomyces in China was studied. A total of 14 species were obtained and circumscribed as four novel species, namely, W. guiyangensis sp. nov., W. paramyanmarensis sp. nov., W. quanzhouensis sp. nov., and W. phyllophilus sp. nov., nine newly recorded species W. arborarius, W. ciferrii, W. edaphicus, W. lynferdii, W. pijperi, W. subpelliculosa, W. xylosica, W. strasburgensis, and W. sydowiorum in China, and a known species W. anomalus. All the studied species were identified by morphological characteristics and phylogenetic analysis of combined ITS and LSU sequences. Four new species are described based on their asexual and/or sexual states, and their differences with the close relatives were compared and discussed.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
C-YC: Investigation, Methodology, Writing – original draft. TK: Investigation, Methodology, Resources, Software, Writing – original draft. Q-HN: Funding acquisition, Resources, Software, Validation, Writing – review & editing. F-LH: Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (Project No. 31570021), Agricultural Biomass Green Conversion Technology University Scientific Innovation Team in Henan Province, China (Project No. 24IRTSTHN036), Key Scientific Research Project of Colleges and Universities in Henan Province, China (Project No. 23A210018), NSFC (Project No. 3217010010) and program for the Outstanding Youth Science Fund Project of Henan province (Project No. 222300420014).
The authors are very grateful to their colleagues at the School of Life Science and Agricultural Engineering, Nanyang Normal University, including Dr. Lin Zhang for providing specimens; Dr. Ting Lei for help with phylogenetic analysis; and Ya-Zhuo Qiao and Wen-Ting Hu for help with morphological observations.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Andrew, R. (2016). FigTree: Tree figure drawing tool version 1.4.3. University of Edinburgh Edinburgh, UK.
Arastehfar, A., Bakhtiari, M., Daneshnia, F., Fang, W., Sadati, S. K., Al-Hatmi, A. M., et al. (2019). First fungemia case due to environmental yeast Wickerhamomyces myanmarensis: detection by multiplex qPCR and antifungal susceptibility. Future Microbiol. 14, 267–274. doi: 10.2217/fmb-2018-0253
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chai, C. Y., Huang, L. N., Cheng, H., Liu, W. J., and Hui, F. L. (2019). Wickerhamomyces menglaensis f. a., sp. nov., a yeast species isolated from rotten wood. Int. J. Syst. Evol. Microbiol. 69, 1509–1514. doi: 10.1099/ijsem.0.003350
Chang, C. F., Hsu, I. M., Liu, C. H., and Lee, C. F. (2022). Wickerhamomyces sinyiensis f. a., sp. nov., a new ascomycetous yeast species in the Wickerhamomyces clade isolated in Taiwan. Int. J. Syst. Evol. Microbiol. 72:005466. doi: 10.1099/ijsem.0.005466
Czarnecka, M., Zarowska, B., Połomska, X., Restuccia, C., and Cirvilleri, G. (2019). Role of biocontrol yeasts Debaryomyces hansenii and Wickerhamomyces anomalus in plants’ defence mechanisms against Monilinia fructicola in apple fruits. Food Microbiol. 83, 1–8. doi: 10.1016/j.fm.2019.04.004
Fell, J. W., Boekhout, T., Fonseca, A., Scorzetti, G., and Statzell-Tallman, A. (2000). Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50, 1351–1371. doi: 10.1099/00207713-50-3-1351
Groenewald, M., Hittinger, C. T., Bensch, K., Opulente, D. A., Shen, X. X., Li, Y., et al. (2023). A genome-informed higher rank classification of the biotechnologically important fungal subphylum Saccharomycotina. Stud. Mycol. 105, 1–22. doi: 10.3114/sim.2023.105.01
Hall, T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hu, W. T., Chu, S. B., Li, Y., and Hui, F. L. (2022). Hyphopichia xiaguanensis f. a., sp. nov., an ascomycetous yeast species isolated from plant leaves. Int. J. Syst. Evol. Microbiol. 72:5398. doi: 10.1099/ijsem.0.005398
Hui, F. L., Chen, L., Chu, X. Y., Niu, Q. H., and Ke, T. (2013). Wickerhamomyces mori sp. nov., an anamorphic yeast species found in the guts of wood-boring insect larvae. Int. J. Syst. Evol. Microbiol. 63, 1174–1178. doi: 10.1099/ijs.0.048637-0
Into, P., Pontes, A., Sampaio, J. P., and Limtong, S. (2020). Yeast diversity associated with the phylloplane of corn plants cultivated in Thailand. Microorganisms 8:80. doi: 10.3390/microorganisms8010080
Kaewwichian, R., Kawasaki, H., and Limtong, S. (2013). Wickerhamomyces siamensis sp. nov., a novel yeast species isolated from the phylloplane in Thailand. Int. J. Syst. Evol. Microbiol. 63, 1568–1573. doi: 10.1099/ijs.0.050013-0
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). Modelfinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kobayashi, R., Kanti, A., and Kawasaki, H. (2017). Three novel species of D-xylose-assimilating yeasts, Barnettozyma xylosiphila sp. nov., Barnettozyma xylosica sp. nov. and Wickerhamomyces xylosivorus f. a., sp. nov. Int. J. Syst. Evol. Microbiol. 67, 3971–3976. doi: 10.1099/ijsem.0.002233
Kumla, J., Nundaeng, S., Suwannarach, N., and Lumyong, S. (2018). Evaluation of multifarious plant growth promoting trials of yeast isolated from the soil of Assam tea (Camellia sinensis var. assamica) plantations in northern Thailand. Microorganisms 8:1168. doi: 10.3390/microorganisms8081168
Kurtzman, C. P. (2011a). “Pichia E. C. Hansen (1904)” in The yeasts, a taxonomic study. eds. C. P. Kurtzman, J. W. Fell, and T. Boekhout. 5th ed (Amsterdam: Elsevier), 685–707.
Kurtzman, C. P. (2011b). “Wickerhamomyces Kurtzman, Robnett & Basehoar-Powers (2008)” in The yeasts, a taxonomic study. eds. C. P. Kurtzman, J. W. Fell, and T. Boekhout. 5th ed (Amsterdam: Elsevier), 899–917.
Kurtzman, C. P., Fell, J. W., and Boekhout, T. (2011). “Methods for isolation, phenotypic characterization and maintenance of yeasts” in The yeasts, a taxonomic study. eds. C. P. Kurtzman, J. W. Fell, and T. Boekhout. 5th ed (Amsterdam: Elsevier), 87–110.
Kurtzman, C. P., and Robnett, C. J. (1998). Identification and phylogeny ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73, 331–371. doi: 10.1023/a:1001761008817
Kurtzman, C. P., Robnett, C. J., and Basehoar-Powers, E. (2008). Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Res. 8, 939–954. doi: 10.1111/j.1567-1364.2008.00419.x
Lachance, M. A., Boekhout, T., Scorzetti, G., Fell, J. W., and Kurtzman, C. P. (2011). “Candida Berkhout (1923)” in The yeasts, a taxonomic study. eds. C. P. Kurtzman, J. W. Fell, and T. Boekhout. 5th ed (Amsterdam: Elsevier), 987–1278.
Lanciotti, R., Sinigaglia, M., Gardini, F., and Guerzoni, M. E. (1998). Hansenula anomala as spoilage agent of cream-filled cakes. Microbiol. Res. 153, 145–148. doi: 10.1016/s0944-5013(98)80032-3
Li, W., Shi, C., Guang, J., Ge, F., and Yan, S. (2021). Development of Chinese chestnut whiskey: yeast strains isolation, fermentation system optimization, and scale-up fermentation. AMB Express 11:17. doi: 10.1186/s13568-020-01175-4
Limtong, S., Into, P., and Attarat, P. (2020). Biocontrol of rice seedling rot disease caused by Curvularia lunata and Helminthosporium oryzae by epiphytic yeasts from plant leaves. Microorganisms 8:647. doi: 10.3390/microorganisms8050647
Limtong, S., Nitiyon, S., Kaewwichian, R., Jindamorakot, S., Am-In, S., and Yongmanitchai, W. (2012). Wickerhamomyces xylosica sp. nov. and Candida phayaonensis sp. nov., two xylose-assimilating yeast species from soil. Int. J. Syst. Evol. Microbiol. 62, 2786–2792. doi: 10.1099/ijs.0.039818-0
Nakase, T., Jindamorakot, S., Am-In, S., Ninomiya, S., and Kawasaki, H. (2012). Wickerhamomyces tratensis sp. nov. and Candida namnaoensis sp. nov., two novel ascomycetous yeast species in the Wickerhamomyces clade found in Thailand. J. Gen. Appl. Microbiol. 58, 145–152. doi: 10.2323/jgam.58.145
Nakase, T., and Takashima, M. A. (1993). Simple procedure for the high frequency isolation of new taxa of ballistosporous yeasts living on the surfaces of plants. RIKEN Rev. 3, 33–34.
Ninomiya, S., Mikata, K., Kajimura, H., and Kawasaki, H. (2013). Two novel ascomycetous yeast species, Wickerhamomyces scolytoplatypi sp. nov. and Cyberlindnera xylebori sp. nov., isolated from ambrosia beetle galleries. Int. J. Syst. Evol. Microbiol. 63, 2706–2711. doi: 10.1099/ijs.0.050195-0
Nundaeng, S., Suwannarach, N., Limtong, S., Khuna, S., Kumla, J., and Lumyong, S. (2021). An updated global species diversity and phylogeny in the genus Wickerhamomyces with addition of two new species from Thailand. J. Fungi 7:957. doi: 10.3390/jof7110957
Rannala, B., and Yang, Z. (1996). Probability distribution of molecular evolutionary trees a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Ravasio, D., Carlin, S., Boekhout, T., Groenewald, M., Vrhovsek, U., Walther, A., et al. (2018). Adding flavor to beverages with non-conventional yeasts. Fermentation 4:15. doi: 10.3390/fermentation4010015
Ronquist, F., Teslenko, M., Mark, P., Avres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes3.2: efficient Bayesian phylogenetic inference and model choice, across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Shi, C. F., Zhang, K. H., Chai, C. Y., Yan, Z. L., and Hui, F. L. (2020). Diversity of the genus Sugiyamaella and description of two new species from rotting wood in China. MycoKeys 77, 27–39. doi: 10.3897/mycokeys.77.60077
Shimizu, Y., Konno, Y., and Tomita, Y. (2020). Wickerhamomyces psychrolipolyticus f. a., sp. nov., a novel yeast species producing two kinds of lipases with activity at different temperatures. Int. J. Syst. Evol. Microbiol. 70, 1158–1165. doi: 10.1099/ijsem.0.003894
Shin, K. S., Bae, K. S., Lee, K. H., Park, D. S., Kwon, G. S., and Lee, J. B. (2011). Wickerhamomyces ochangensis sp. nov., an ascomycetous yeast isolated from the soil of a potato field. Int. J. Syst. Evol. Microbiol. 61, 2543–2546. doi: 10.1099/ijs.0.026682-0
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Timke, M., Wang-Lieu, N. Q., Altendorf, K., and Lipski, A. (2005). Fatty acid analysis and spoilage potential of biofilms from two breweries. J. Appl. Microbiol. 99, 1108–1122. doi: 10.1111/j.1365-2672.2005.02714.x
Vaidya, G., Lohman, D. J., and Meier, R. (2011). Sequence matrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vu, D., Groenewald, M., Szöke, S., Cardinali, G., Eberhardt, U., Stielow, B., et al. (2016). DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 85, 91–105. doi: 10.1016/j.simyco.2016.11.007
Walker, G. M. (2011). Pichia anomala: cell physiology and biotechnology relative to other yeasts. Antonie Van Leeuwenhoek 99, 25–34. doi: 10.1007/s10482-010-9491-8
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR protocols: a guide to methods and applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322.
Yan, S., Xiangsong, C., and Xiang, X. (2019). Improvement of the aroma of lily rice wine by using aroma-producing yeast strain Wickerhamomyces anomalus HN006. AMB Express 9:89. doi: 10.1186/s13568-019-0811-8
You, L., Wang, S., Zhou, R., Hu, X., Chu, Y., and Wang, T. (2016). Characteristics of yeast flora in Chinese strong-flavoured liquor fermentation in the Yibin region of China. J. Inst. Brew. 122, 517–523. doi: 10.1002/jib.352
Keywords: Ascomycetes, morphology, phylogenetic analyses, taxonomy, plants, tropical and subtropical regions
Citation: Chai C-Y, Ke T, Niu Q-H and Hui F-L (2024) Diversity of Wickerhamomyces (Wickerhamomycetaceae, Saccharomycetales) in China with the description of four new species. Front. Microbiol. 15:1338231. doi: 10.3389/fmicb.2024.1338231
Received: 14 November 2023; Accepted: 15 January 2024;
Published: 08 February 2024.
Edited by:
Xiong Zhang, Nanjing Agricultural University, ChinaReviewed by:
Mengmeng Wang, Ministry of Ecology and Environment, ChinaCopyright © 2024 Chai, Ke, Niu and Hui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiu-Hong Niu, cWl1aG9uZ25pdTcyM0AxNjMuY29t; Feng-Li Hui, ZmVuZ2xpaHVpQHllYWgubmV0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.