
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 March 2024
Sec. Microbial Symbioses
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1334918
 Yayun Zhao1,2†
Yayun Zhao1,2† Tao Sun1,3†
Tao Sun1,3† Yang Li1,4
Yang Li1,4 Zhibo Yang1
Zhibo Yang1 Jun Chen1
Jun Chen1 Jing Wang1
Jing Wang1 Xinlong Yu1
Xinlong Yu1 Xuexi Tang1,2*
Xuexi Tang1,2* Hui Xiao1,2*
Hui Xiao1,2*Endophytic bacteria have a complex coevolutionary relationship with their host macroalgae. Dioecious macroalgae are important producers in marine ecosystems, but there is still a lack of research on how sex influences their endophytic bacteria. In this study, the endophytic bacterial communities in male and female S. thunbergii and their reproductive tissues (receptacles) were compared using culture methods and high-throughput sequencing. The endophytic bacterial communities detected by the two methods were different. Among the 78 isolated strains, the dominant phylum, genus, and species were Bacillota, Alkalihalobacillus, and Alkalihalobacillus algicola, respectively, in the algal bodies, while in the receptacles, they were Bacillota, Vibrio, and Vibrio alginolyticus. However, 24 phyla and 349 genera of endophytic bacteria were identified by high-throughput sequencing, and the dominant phylum and genus were Pseudomonadota and Sva0996_ Marine_ Group, respectively, in both the algal body and the receptacles. The two methods showed similar compositions of endophytic bacterial communities between the samples of different sexes, but the relative abundances of dominant and specific taxa were different. The high-throughput sequencing results showed more clearly that the sex of the host alga had an effect on its endophyte community assembly and a greater effect on the endophytic bacterial community in the receptacles. Moreover, most specific bacteria and predicted functional genes that differed between the samples from the males and females were related to metabolism, suggesting that metabolic differences are the main causes of sex differences in the endophytic bacterial community. Our research is the first to show that host sex contributes to the composition of endophytic bacterial communities in dioecious marine macroalgae. The results enrich the database of endophytic bacteria of dioecious marine macroalgae and pave the way for better understanding the assembly mechanism of the endophytic bacterial community of algae.
Endophytic bacteria are commonly present in plants and coevolve with their hosts, playing an important role in host growth, development and disease resistance. Studying the community structure of endophytic bacteria in plants is necessary for analyzing the functions of these bacteria and elucidating endophyte-host interactions.
Macroalgae are an important component of marine ecosystems and play a crucial role in material cycling and energy flow. At present, research on the community structure of endophytic bacteria in plants mainly focuses on higher terrestrial plants, and much work has been done on isolating, identifying, and describing the physiological and biochemical characteristics of endophytic bacteria, as well as uncovering their interactions with hosts (Afzal et al., 2019; Ling et al., 2022; Yarte et al., 2022). In recent years, high-throughput sequencing technology has also been widely used to obtain a large amount of information on the endophytic bacterial community in terrestrial plants, includingthe crops Oryza sativa (Liu Y. et al., 2019), Zea mays (Liu Y. et al., 2020) and the medicinal plants Panax notoginseng (Zhang C. et al., 2020) and Gastrodia elata f. glauca (Zheng et al., 2022), as well as some aquatic plants, such as Lemnaceae, Pontederia crassipes, Pistia (Pramanic et al., 2023), and Glehnia littoralis (Huo et al., 2020). There have also been some studies on isolating, identifying, and describing the physiological activity and function of algal endophytic bacteria (Song et al., 2015; Zhang, 2017; Feng et al., 2020; Amri et al., 2023) based on culture methods. However, Hollants et al. (2011) used the denaturing gradient gel electrophoresis (DGGE) method to study the endophytic bacterial community of Bryopsidales (Chlorophyta), and Mei (2019) applied high-throughput sequencing to investigate the endophytic bacterial community in Sargassum horneri and Ulva prolifera, but there have been no studies on the differences in endophytic bacteria between the different sexes of algae.
There are many factors that can affect the composition of endophytic bacterial communities, but relevant studies have mainly been carried out on land plants. Previous studies have shown that both the external environment and internal factors influence plant endophytic bacterial composition (Afzal et al., 2019). The number and species of endophytic bacteria have different distributions in plants of different habitats (Zhang A. et al., 2020). The reason why the external environment can influence bacterial composition is that bacterial metabolic function changes depending on soil nutrients, contaminants and temperature, which lead to a strong selection effect on the bacterial population (Zhang et al., 2015). On the other hand, internal factors such as host plant species, tissue location and life history stage all have effects on the endophytic bacterial community (Dastogeer et al., 2018; Mei et al., 2021). The vertical transmission of endophytic bacteria enables these bacteria to be transferred between generations of plants and form a stable symbiotic relationship with plants. Endophytic bacteria differ among different host plants (Liaqat and Eltem, 2016), different varieties of the same plant species (Munir et al., 2020) and even among different seed genotypes (Liu Y. et al., 2020). There are also differences in endophytic bacterial communities among different tissues of the same plant, as confirmed by high-throughput sequencing in Arachis hypogaea Linn (Li et al., 2021), Zea mays L. (Marag and Suman, 2018), and Hippophae tibetana (Zhang A. et al., 2021). Additionally, at different growth stages of the plant, the endophytic bacterial community will undergo corresponding changes due to alterations in the internal and external environments (Mahmood et al., 2019; Song et al., 2020).
However, host sex is rarely mentioned among the factors affecting the community structure of endophytic bacteria. In fact, sex can lead to differences in the morphology, structure and function of dioecious plants (Tang, 2020; Lu et al., 2021; Zeng et al., 2022), and there are also some differences in enzyme activity (Chen et al., 2020), secondary metabolites (Li et al., 2022) and endogenous hormone levels (Ge et al., 2021), but there have been few studies on sex differences in endophytic bacteria in plants. Some studies have revealed differences in epiphytic bacterial communities in plants of different sexes, such as in Populus cathayana (Liu et al., 2021), Sargassum thunbergii (Wang et al., 2022b), and Porphyra haitanensis (Yang et al., 2022). However, for marine macroalgae, whether sex has an impact on the assembly of endophytic bacterial communities is still unknown.
Sargassum thunbergii is a common intertidal macroalgae in coastal China. As the feed of valuable aquaculture organisms and the preferred plants for marine pastures, there is an urgent need for artificial cultivation of S. thunbergii. Endophytic bacteria are closely related to the growth and development of S. thunbergii, and their study will facilitate its cultivation. S. thunbergii is a typical dioecious alga with receptacles appearing in the reproduction stage (Wang, 2007). There are obvious differences in the appearance and internal structure of female and male receptacles, and they perform different reproductive functions (Wang et al., 2007). Whether these differences will lead to differences in the endophytic bacterial communities between male and female S. thunbergii and their receptacles and whether host algal sex will affect the endophytic bacterial community have not yet been explored.
In this study, the endophytic bacterial community was compared between males and females of the intertidal macroalga S. thunbergii and their receptacles from Shandong Peninsula by culture-dependent and high-throughput sequencing technologies, aiming at enriching the basic information on the endophytic bacterial community in marine macroalgae and elucidating the role of host sex in the assembly of the endophytic bacterial community in S. thunbergii.
The algal samples were collected from a 5 m × 100 m sampling square in the continuous intertidal sea area at Taipingjiao (120°21′34.2″E, 36°14′58.3″N) along the coast of Qingdao (China) on July 21, 2021, during the reproductive period of S. thunbergii, and then placed in sterile sample bags and brought back to the laboratory for further processing within 30 min. The sex of the algae was confirmed in the laboratory by observing the internal structure of the receptacles using a microscope (Nikon H600L, Tokyo, Japan).
For the culture dependent test, six strains of S. thunbergii (3 males and 3 females) were used and these samples were from 3 different sampling sties, respectively. The surface of the algae was disinfected on a clean bench by a method established in the pre-experiment (75% alcohol for 5 min + 2.5% sodium hypochlorite for 10 min) and finally washed with sterile water 7 times. Then, 0.1 mL of the final sterile water rinse was collected and spread on Zobell 2216E medium plates. When cultured in a biochemistry incubator (SPX-150, Wanfeng Instrument Co. Ltd., China) at 25°C for 48 h, if no colonies had grown, the surface disinfection of the algae was considered successful.
On the clean bench, 2 g of male and female body tissues and their receptacles (picked with sterile tweezers were taken from disinfected S. thunbergii individuals and transferred to a sterile mortar). Eight milliliters of sterile seawater were added to grind the samples into a homogenized suspension. The dilution 100, 10–1, and 10–2 were prepared and 0.1 mL of the suspension was spread on Zobell 2216E medium plate with three replicates. Then, the plates were placed in a biochemical incubator at 25°C for 48 h. Since the number of colonies of dilution 100 group was less than thirty while that of dilution 10–1 group was less five and that of dilution 10–2 group was zero, all colonies grown on the plates of dilution 100 group were isolated and purified by the continuous streaking method, and the obtained strains were stored in stroke-physiological saline solution containing 15% glycerol at −80°C.
Bacterial DNA was extracted from the isolated strains using a TIANamp Bacterial DNA Kit [Tiangen Biotech (Beijing) Co., Ltd.]. The 16S rDNA sequence was then amplified using the forward primer 27F and reverse primer 1492R, after that sequencing was conducted by Sangon Biotech (Shanghai) Co., Ltd. The sequencing results were spliced and compared with the EzBioCloud database.1 The top 10 strains with the closest similarity among the strains with more than 98% similarity were selected. A phylogenetic tree was constructed with the sequences through three methods with MEGA 11.0 software, namely, neighbor-joining (NJ), maximum likelihood (ML) and minimum-evolution (ME), to determine the species of the strains.
On the clean bench, 2 g of male and female body tissues and receptacles (picked with sterile tweezers in an ice bath) was taken from the disinfected S. thunbergii individuals, transferred to a sterile sample tube and stored at −80°C. DNA extraction and sequencing were performed by Guangzhou Kidio Biotechnology Co., Ltd. The samples were labeled Male-ENDO (endophytic bacteria in the male algal body), Female-ENDO (endophytic bacteria in the female algal body), M-ENDO-Receptacles (endophytic bacteria in male receptacles) and F-ENDO-Receptacles (endophytic bacteria in female receptacles). Each group had 8 replicates and each sample came from one alga.
After genomic DNA was extracted from the samples, the V3 + V4 region of 16S rDNA was amplified with a specific primer with a barcode. The primer sequences were 341F: CCTACGGGNGGCWGCAG and 806R: GGACTACHVGGG TATCTAAT. Then, the PCR amplification products were cut and recovered, the purified amplification products were mixed in equal amounts, the sequencing joints were connected, the sequencing library was constructed, and high-throughput sequencing was performed on the Illumina NovaSeq 6000 PE250 platform (Illumina, San Diego, CA, USA).
UPARSE software (version 9.2.64_i86linux32) was used to concatenate and deduplicate sequences, sequences with more than 97% similarity were clustered into an operational taxonomic unit (OUT), and the SILVA (version 132) database was used to classify the OTUs. The Chao1, Ace, Shannon and Simpson α-diversity indices were calculated by QIIME (version 1.9.1.) The significance of intergroup index comparisons was determined by the Kruskal-Wallis (KW) test and Welch’s t-test. β-diversity was analyzed using principal coordinate analysis (PCoA) and the unweighted pair-group method using arithmetic averages (UPGMA) approach based on Bray-Curtis distances. According to the OTU classification, the high-throughput sequencing data were used to cluster the bacteria at the phylum and genus levels, the species not clearly classified or with relative abundance less than 1% were classified as “others,” and a histogram was drawn. Linear discriminant analysis effect size (LEfSe) software was used to analyze the differences between groups. The KW rank sum test was performed among samples from all groups first, and then the Wilcoxon rank sum test was used to compare the selected species between the two groups. Linear discriminant analysis (LDA) was used to sort the selection results and generate the LDA difference analysis diagram, and then an evolutionary branching diagram was obtained by mapping the differences onto the classification tree with a known hierarchical structure. Finally, PICRUSt (version 2.1.4) was used to predict the function of endophytic bacteria, and the KW test was used to analyze the significance of functional differences.
The bacterial sequences obtained in this study have been saved to the National Center for Biotechnology Information (NCBI) with BioProject IDs: PRJNA830829 and PRJNA830307.
In this study, a total of 78 bacterial strains were isolated from male and female S. thunbergii and their receptacles and their phylogenetic tree is shown in Supplementary Figure 1. Additionally, the functions of isolated strains reported in previous studies were listed in Supplementary Table 1.
Thirty-one bacterial strains were isolated from algal bodies (19 from males and 12 from females), which belonged to 2 phyla, 7 genera, 14 species, and 1 suspected new species (Supplementary Table 1A). The dominant phylum, genus, and species were Bacillota, Alkalihalobacillus and Alkalihalobacillus algicola, respectively. In addition, 1 phylum (Bacillota), 3 genera (e.g., Alkalihalobacillus), and 2 species (A. algicola and A. berkeleyi) were detected in the algal bodies of both sexes. However, 1 phylum (Pseudomonadota), 3 genera (e.g., Rossellomorea), and 8 species (e.g., A. hwajinpoensis) were isolated only from the male algal body, while 1 genus (Mesobacillus) and five species (e.g., A. caeni) were isolated only from the female algal body (Figure 1).
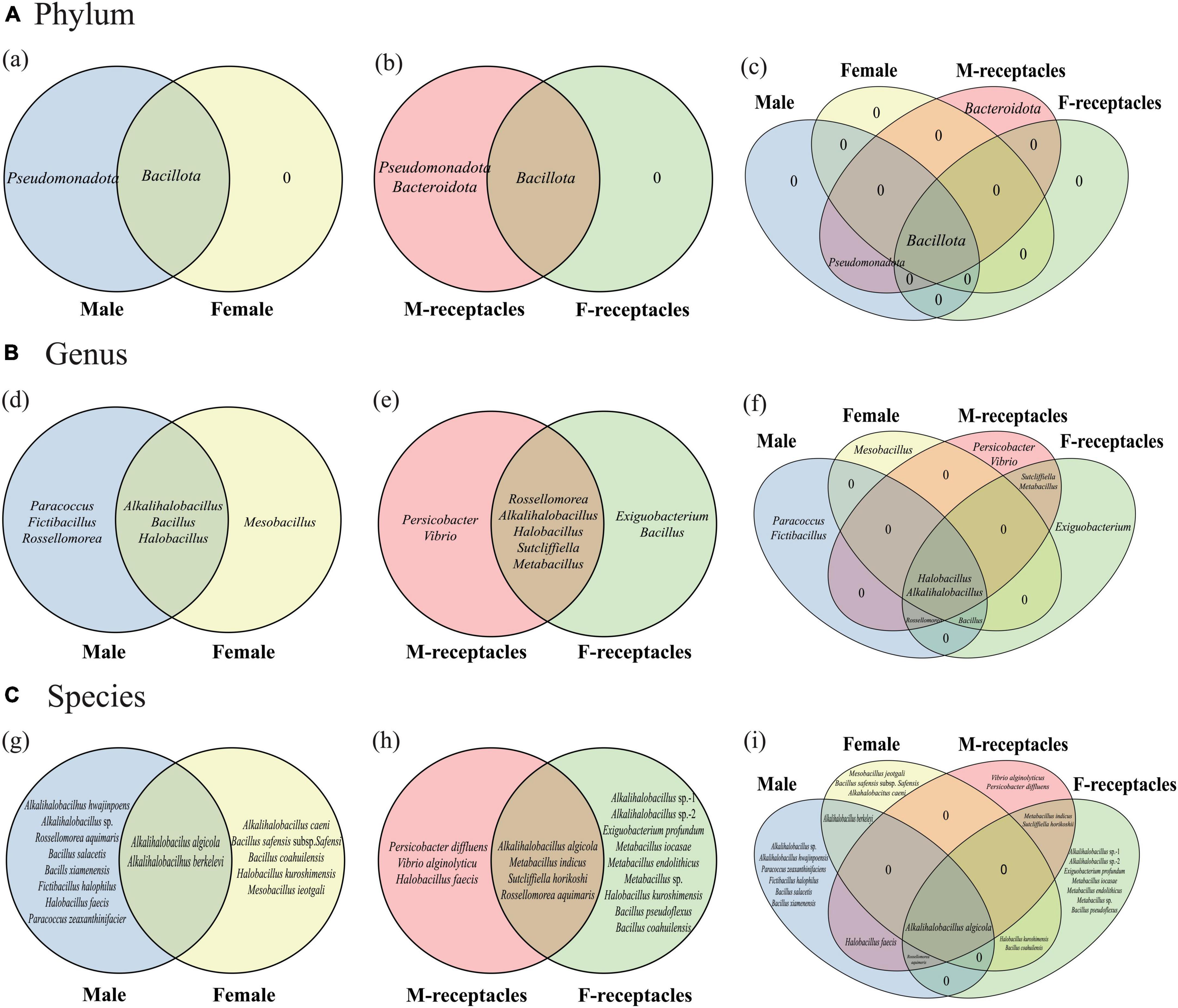
Figure 1. Venn diagrams of culturable heterotrophic bacteria of male and female S. thunbergii and their receptacles. (A) Phylum level. (B) Genus level. (C) Species level.
The culturable endophytic bacteria in receptacles were more diverse than those in algal bodies. Forty-seven strains (21 from males and 26 from females) belonging to 3 phyla, 9 genera, 13 species (Supplementary Table 1B), and 3 suspected new species were also isolated. One phylum (Bacteroidota) and five genera (e.g., Exiguobacterium) were isolated only from the receptacles (Figure 1), and the dominant phylum was the same as in the algal body, but the dominant genus and species changed to Vibrio and V. alginolyticus, respectively. In addition, various samples shared 1 phylum (Bacillota), 2 genera (Alkalihalobacillus and Halobacillus) and 1 species (A. algicola), but each kind of sample had specific bacteria (Figure 1).
Moreover, the culturable endophytic bacteria isolated from the receptacles were very different between the sexes. The dominant phylum, genus and species in the male receptacles were Pseudomonadota, Vibrio, and V. alginolyticus, while in the female receptacles, they were Bacillota, Alkalihalobacillus, Metabacillus, and E. profundum, respectively. One phylum (Bacillota), 5 genera (e.g., Alkalihalobacillus), and 4 species (e.g., A. algicola) were shared by male and female receptacles. However, many bacterial taxa could be isolated only from the receptacles of one sex. For example, Pseudomonadota and Bacteroidota were specific to male receptacles, while there were no specific phyla in female receptacles. Additionally, there were two genera (Vibrio and Persicobacter) and three species (e.g., V. alginolyticus) specific to male receptacles, while two genera (Exiguobacterium and Bacillus) and nine species (such as E. profundum) were specific to female receptacles.
Because the DNA extracted from one Male-ENDO sample was unqualified, the results of 31 samples were ultimately analyzed, and a total of 3,377,830 sequences were obtained. After mass filtering and removal of chimeric, chloroplast and mitochondrial sequences, 3,195,870 optimized sequences were obtained. The coverage of all samples was above 99% (Supplementary Figure 1), indicating that the sequencing depth covered most of the bacteria in the samples and the sequencing data were reliable and effective.
The results of α-diversity analysis (Figure 2) revealed significant differences in the four indices among the four groups of samples (KW test, P < 0.05). Both the Chao1 and Ace index results showed that the abundance of bacteria in female algae was the highest, followed by that in female receptacles, that in male receptacles and that in male algal bodies. The Shannon and Simpson indices showed that the bacterial diversity in the algal body was higher than that in the receptacle. Interestingly, the four indices were higher in females than in males, indicating that the abundance and diversity of endophytic bacteria in the female samples were higher than those in the male samples.
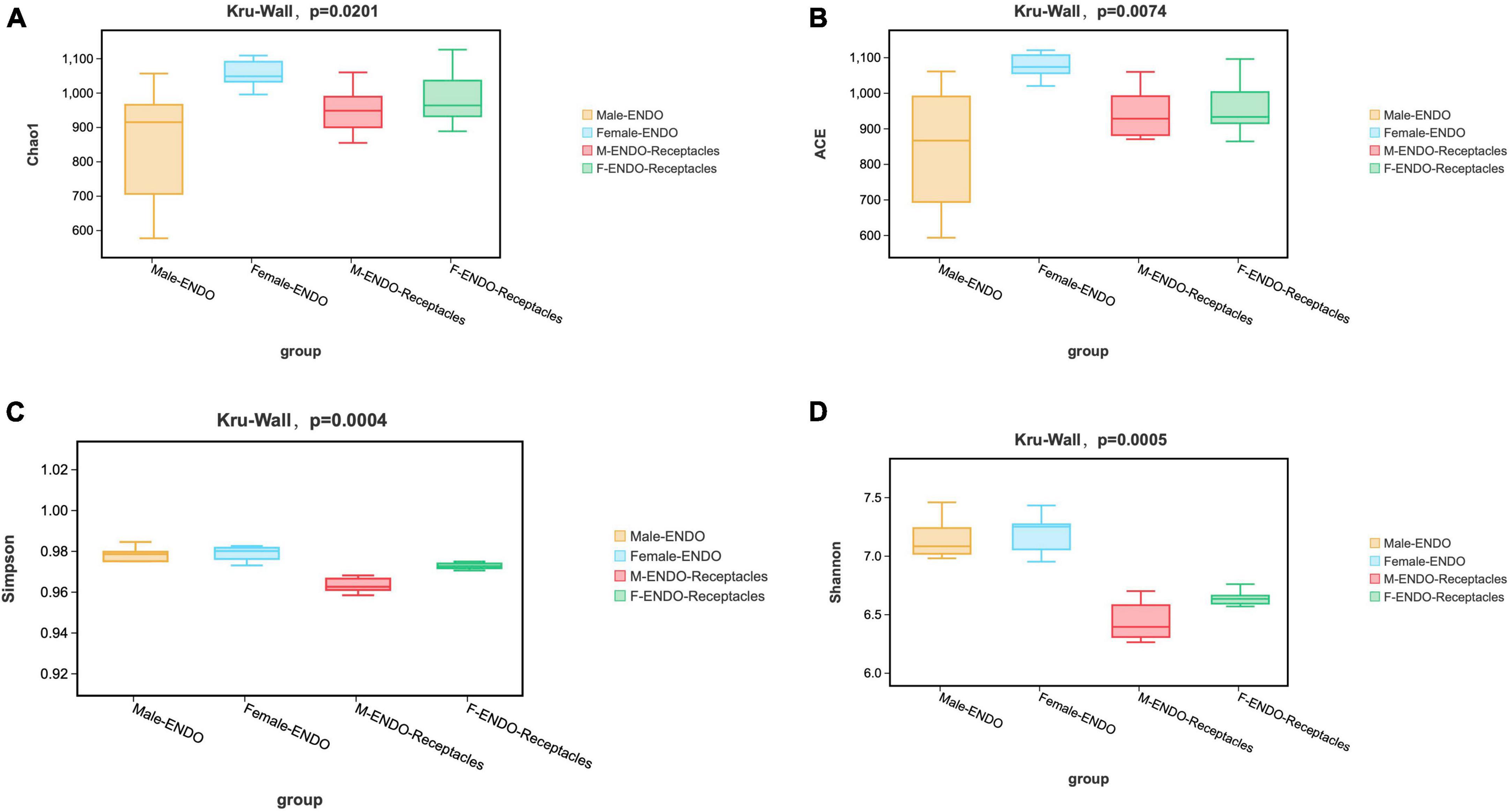
Figure 2. The α-diversity of endophytic bacterial communities in male and female S. thunbergii and their receptacles. (A) Chao1 index. (B) ACE index. (C) Simpson index. (D) Shannon index.
The results of UPGMA and PCoA based on Bray-Curtis distances (Figure 3) clustered the bacteria from male and female S. thunbergii and their receptacles, indicating that the samples in each group were similar, but the differences between groups were significant (P < 0.01). Notably, the clustering of endophytic bacteria in male and female receptacles was more obvious than that in algal bodies, indicating that the differences between male and female receptacles were larger than those between male and female algal bodies.
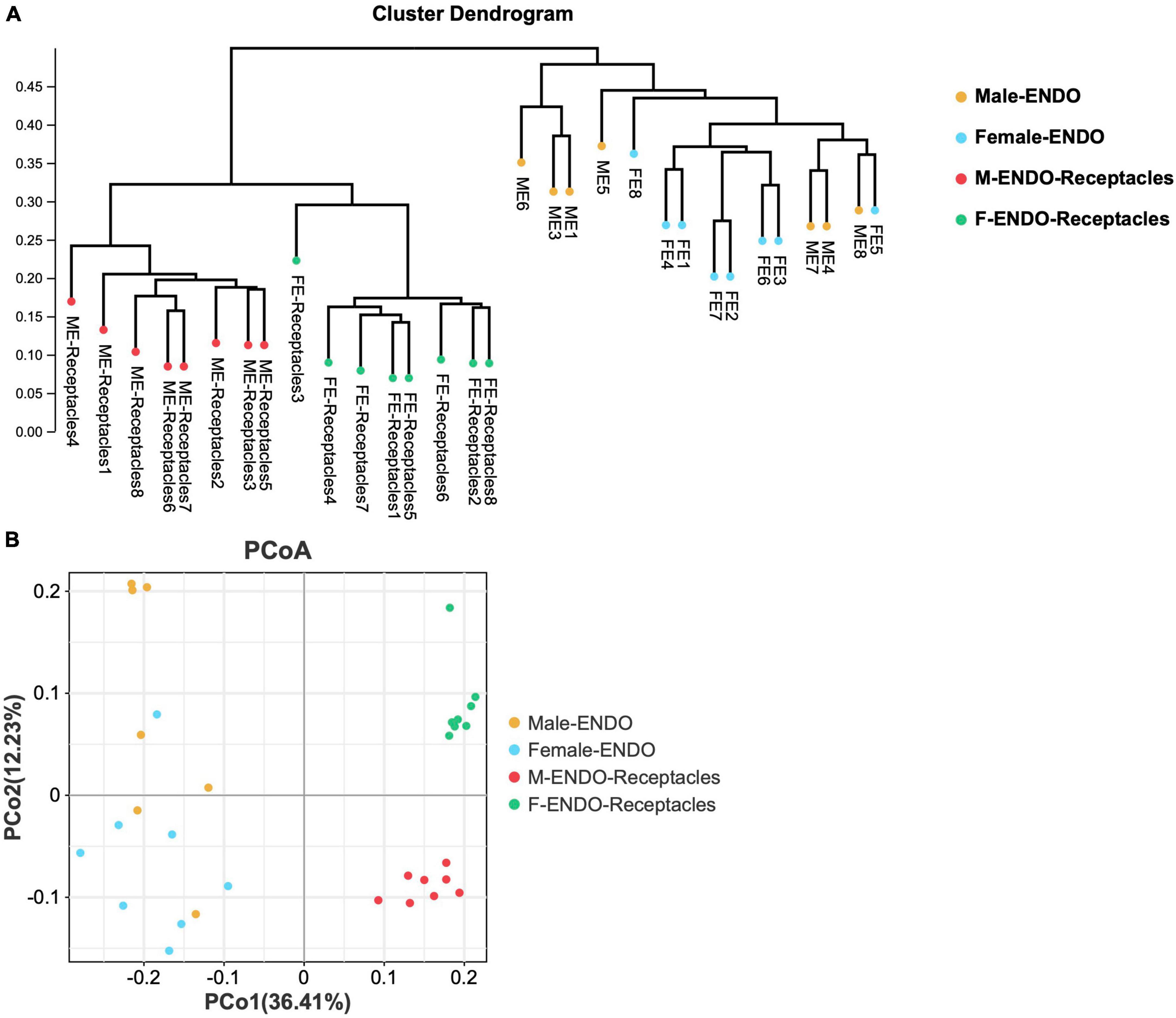
Figure 3. The β-diversity of endophytic bacterial communities in male and female S. thunbergii and receptacles. (A) Results of the unweighted pair-group method with arithmetic mean (UPGMA) approach. (B) Results of principal coordinate analysis (PCoA).
The Venn diagram (Figure 4) showed that the shared bacterial taxa accounted for the majority in the four kinds of samples. There were 16 shared endophytic bacteria at the phylum level (Figure 4A), and the core phyla (with relative abundances higher than 1%) included Pseudomonadota (39.89–56.03%), Bacteroidota (11.73–16.70%), Actinomycetota (11.37–30.31%), Planctomycetota (1.13–3.75%), Verrucomicrobiota (2.68–5.97%), and Cyanobacteria (2.57–10.52%). At the genus level (Figure 4B), there were 129 shared genera, and the core genera (with relative abundances higher than 1%) included Sva0996_ Marine_ Group (6.27–13.01%), Loktanella (4.44–7.46%), Burkholderia Caballeronia Paraburkholderia (1.39–7.51%), Acinetobacter (1.35–3.66%), Mariactor (2.12–3.23%), Pseudoruegeria (3.19–4.70%), Granulosicoccus (1.81–3.62%), and Phormidesmis_ ANTLACV51 (1.23–4.50%).
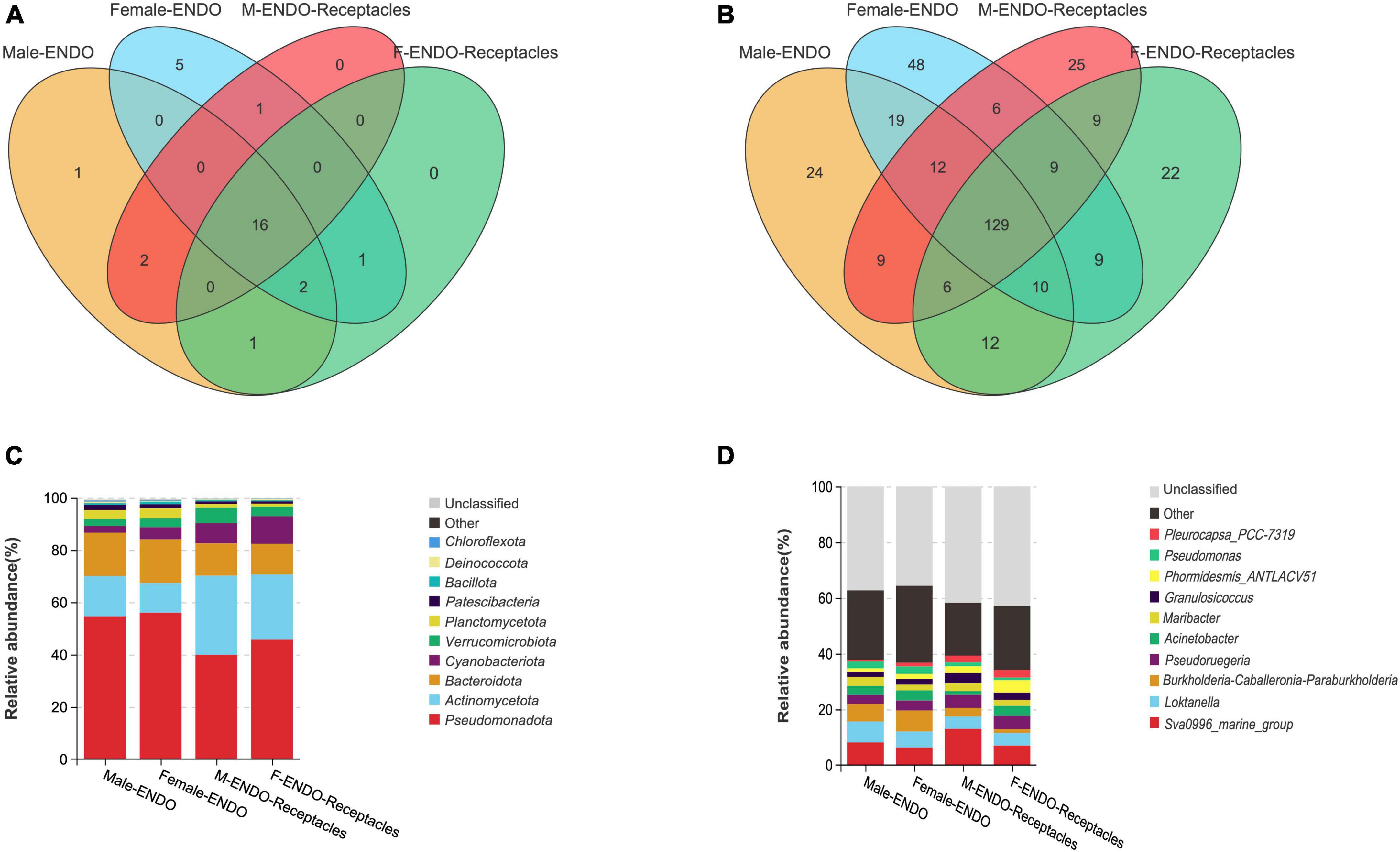
Figure 4. Venn diagram and relative abundance of endophytic bacteria in algal bodies and receptacles of male and female S. thunbergii. (A,C) Phylum level. (B,D) Genus level.
Each kind of sample also had specific endophytic bacterial taxa. For instance, there were 5 specific phyla (e.g., Entotheonellaeota and Halanaerobiaeota) in female algal bodies and only one specific phylum (Elusimicrobiota) in male algal bodies, but there were no specific phyla in the receptacles. There were 24 genera specific to male algal bodies, including Brachybacterium, Oikopleura_dioica, and Magnetospira, while 48 genera were specific to female algal bodies, including Macrococcus, Candidatus_Actinomarina, and Marinobacterium. For the receptacles, there were 25 specific genera in males (e.g., Prevotella_1, Muricauda, and Erysipelotrichaceae_UCG-007), while 22 genera were specific to female receptacles (e.g., Shewanella, Minicystis, and Steroidobacter).
The community composition and relative abundance of the endophytic bacteria in male and female S. thunbergii and their receptacles are shown in Figure 4. The compositions were similar among the four kinds of samples, but the abundance of some bacterial taxa differed obviously. At the phylum level, the top three phyla in algal bodies were Pseudomonadota, Bacteroidota, and Actinomycetota, with relative abundances of 54.62, 16.58, and 15.42% in male algal bodies and 56.03, 16.70, and 11.37% in female bodies, respectively. In receptacles, the top three phyla were the same, but the order changed to Pseudomonadota, Actinomycetota, and Bacteroidota, and the abundance of the top three phyla was 39.89, 30.31, and 12.35% in males and 45.72, 24.93, and 11.73% in females, respectively. Notably, Cyanobacteria was ranked fourth in female algal bodies and the receptacles of both sexes but was ranked sixth in male algal bodies.
Additionally, the dominant genera of endophytic bacteria in the four kinds of samples were basically the same but differed in relative abundance. The top three dominant genera in male algal bodies were Sva0996_marine_group (8.16%). Loktanella (7.46%) and Burkholderia-Caballeronia-Paraburkholderia (6.37%), while they were Burkholderia-Caballeronia-Paraburkholderia (7.51%), Sva0996_marine_group (6.27%), and Loktanella (5.82%) in female algal bodies. The top three dominant genera in male and female receptacles were the same, namely, Sva0996_marine_group (males: 13.01%; females: 7.00%), Pseudoruegeria (males: 4.70%; females: 4.70%), and Loktanella (males: 4.44%; females: 4.53%).
The network analysis (Figure 5) based on the relationship pairs with the top-50 correlation coefficient showed the specific network of the endophytic bacterial community in male and female algal bodies and receptacles of S. thunbergia. The results indicated that the correlation between the endophytic bacteria in four kinds of samples was significantly different (P < 0.05). The node number of endophytic bacteria at genus level in female algal bodies were larger than in male algal bodies, while the results in receptacles was opposite.
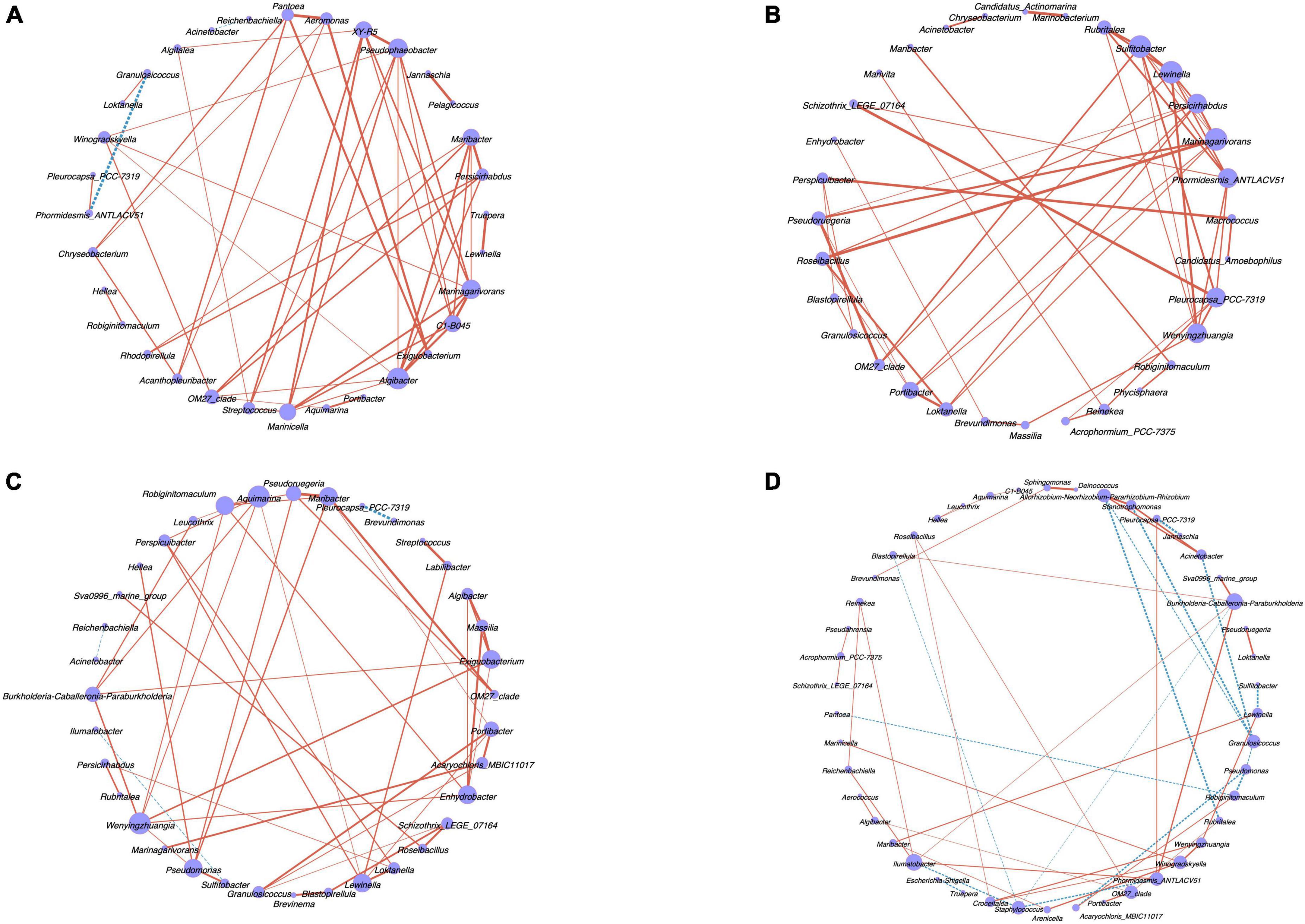
Figure 5. The network analysis of endophytic bacteria in algal bodies and receptacles of male and female S. thunbergia at genus level (P < 0.05). (A) Male algal bodies. (B) Female algal bodies. (C) Male receptacles. (D) Female receptacles. Node color corresponds to genus taxonomic classification. Edge color represents positive (red) and negative (blue) correlations.
Compared to the interaction between the endophytes in different sex algal bodies through the network analysis, 50 nodes and 1225 edges in the microbial network from S. thunbergia were found. Also, the endophytes in female S. thunbergia had 56 nodes and 1540 edges. Algibacter, Marinagarivorans, Pseudophaeobacter, and Maribacter showed high betweenness in male algal bodies, which indicated that these microbes were the key nodes of this sample group. The key nodes of the endophytes in female algal bodies were Sulfitobacter, Marinagarivorans, Lewinella, and Phormidesmis_ANTLACV51 with a high level of betweenness.
The interaction between the endophytes in male and female receptacles of S. thunbergia was also different. The microbial network from female receptacles had 40 nodes and 780 edges, while that from male receptacles had 54 nodes and 1431 edges. In the microbiome of male receptacles from S. thunbergia, Aquimarina, Wenyingzhuangia, Maribacter, and Pseudomonas showed high betweenness centrality in the community network, while Burkholderia-Caballeronia-Paraburkholderia, Ilumatobacter, Phormidesmis_ANTLACV51, and Granulosicoccus were the key nodes of the endophytic microbial community in female receptacles.
The LEfSe analysis (Figure 6) revealed the endophytic bacterial taxa with significant differences between groups (LDA > 4), namely, biomarkers, and the results indicated that there were many biomarkers between the four kinds of samples. Alphaproteobacteria (class), Flavobacteriales (order), and Flavobacteriaceae (family) were enriched in male algal bodies, while Pseudomonadota (phylum), Gammaproteobacteria (class), and Pseudomonadales (order) were enriched in female algal bodies; Microtrichales (order), Acidimicrobiia (class), and Actinomycetota (phylum) were abundant in male receptacles, while Cyanobacteria (phylum), Oxyphotobacteria (class), and Arenicellaceae (family) were abundant in female receptacles.
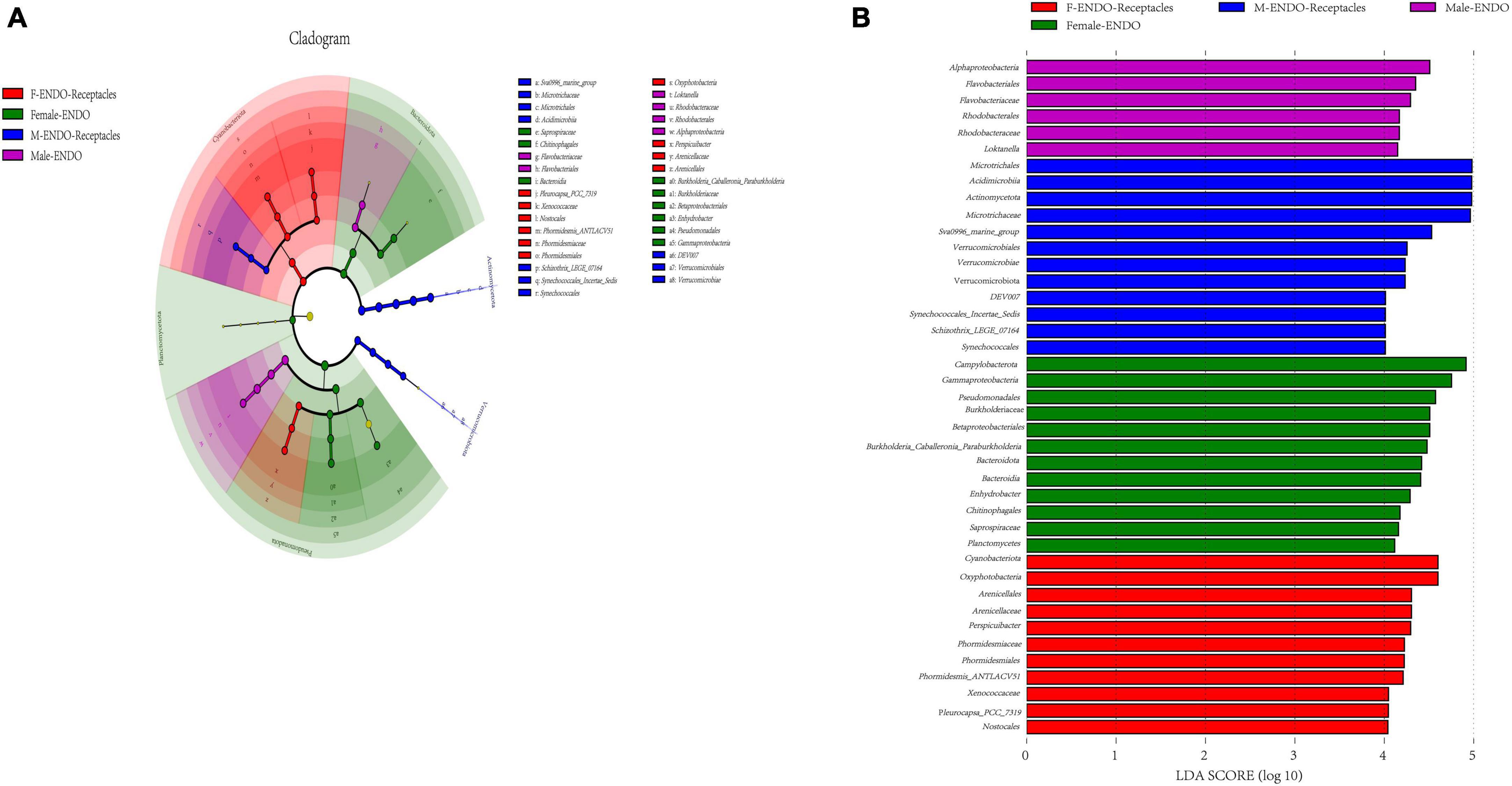
Figure 6. Biomarker analysis of male and female S. thunbergii and their receptacles. The cladogram shows the phylogenetic structure of the microbiota. The circles that radiate from inside to outside represent taxonomic levels from phylum to genus, and each small circle represents an individual taxon. The diameter of the circles is proportional to the relative abundance. The linear discriminant analysis (LDA) scores indicate significant differences in bacterial taxa (LDA score >4.0). (A) Cladogram. (B) LDA score chart.
In addition, Figures 7A, B. showed the indicator values of the groups with significant differences (P < 0.01) in abundance between male and female S. thunbergii and their receptacles at the phylum and genus levels. The groups showed large differences in indicator values. Among the indicator species at the genus level, Planctomicrobium, Anderseniella, and Algitalea were abundant in the male algal bodies; HIMB11, Lactobacillus, and Marinobacterium were enriched in the female algal bodies; Rhodococcus and Delftia were abundant in the male receptacles; and Perspicuibacter, Aquimarina, and Coraliomargarita were enriched in the female receptacles. This suggests that the highest frequency or abundance of endophytic bacteria was not the same between the algal body and the receptacles or between samples of different sexes.
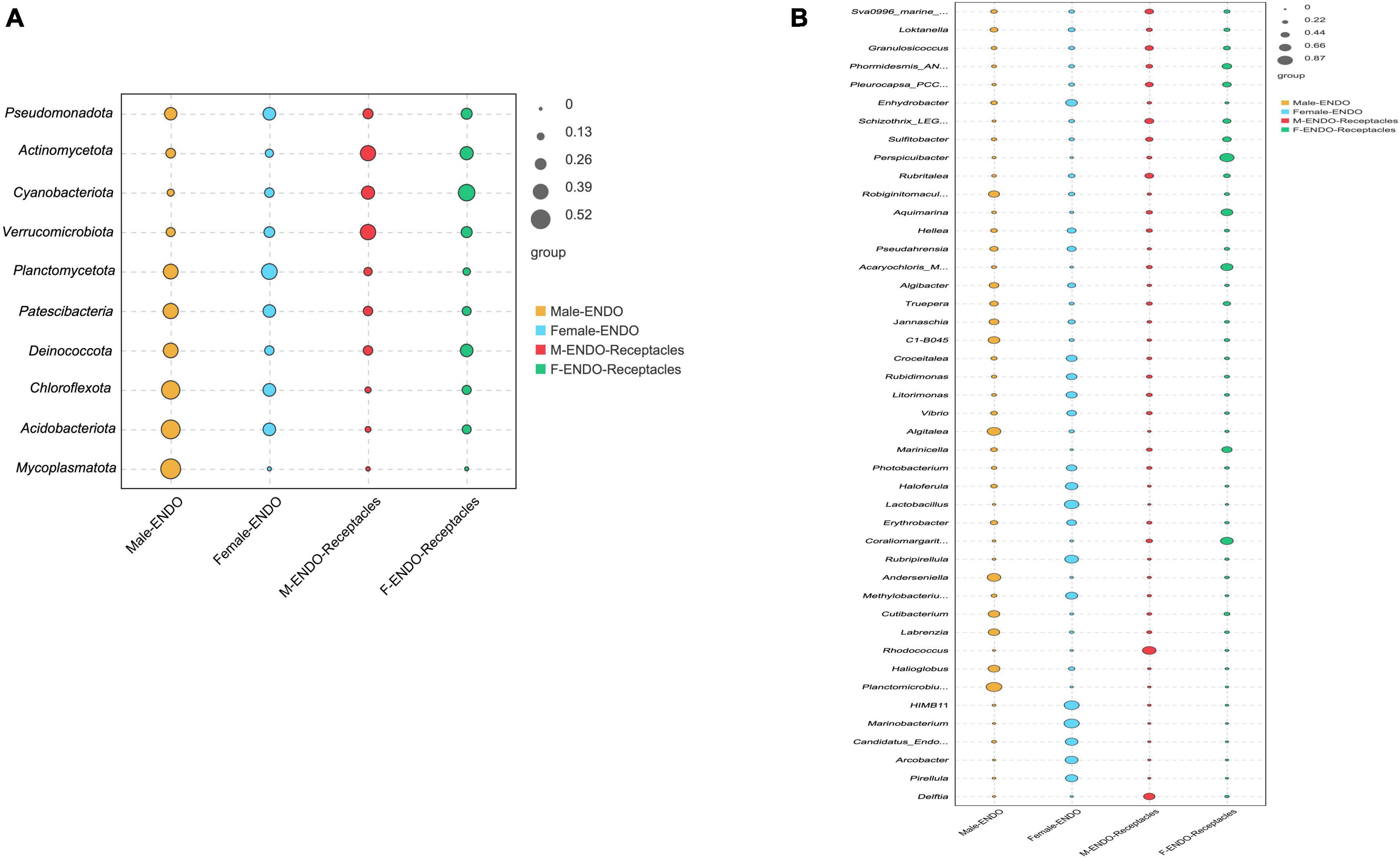
Figure 7. Indicator value analysis of endophytic bacteria in male and female S. thunbergii and their receptacles at the phylum and genus levels (P < 0.01). (A) Phylum level. (B) Genus level.
Based on PICRUSt2, the gene functional abundance of the endophytic bacteria in male and female S. thunbergii and their receptacles was predicted. The results showed significant differences among the 4 groups of samples (Figure 8). At the secondary level, there was a significant difference (P < 0.01) in the function of 27 out of 33 genes among the 4 kinds of samples. These functions included five categories: metabolism, genetic information processing, cellular processes, organismal systems, and environmental information processing. At the third level, there was a significant difference (P < 0.05) among the four kinds of samples in 151 out of 169 gene functions. Among them, 109 out of 151 gene functions were related to metabolism, such as xenobiotic biodegradation and metabolism (18 types), carbohydrate metabolism (15 types), amino acid metabolism (13 types), and lipid metabolism (12 types), and others included environmental adaptability, infectious diseases, and cell motility. Another interesting finding was that the abundance of predicted genes with significant differences in female receptacles was higher than that in males, but in the algal bodies of the two sexes, the abundances of predicted genes with significant differences were basically the same, and both were lower than those in the receptacles.
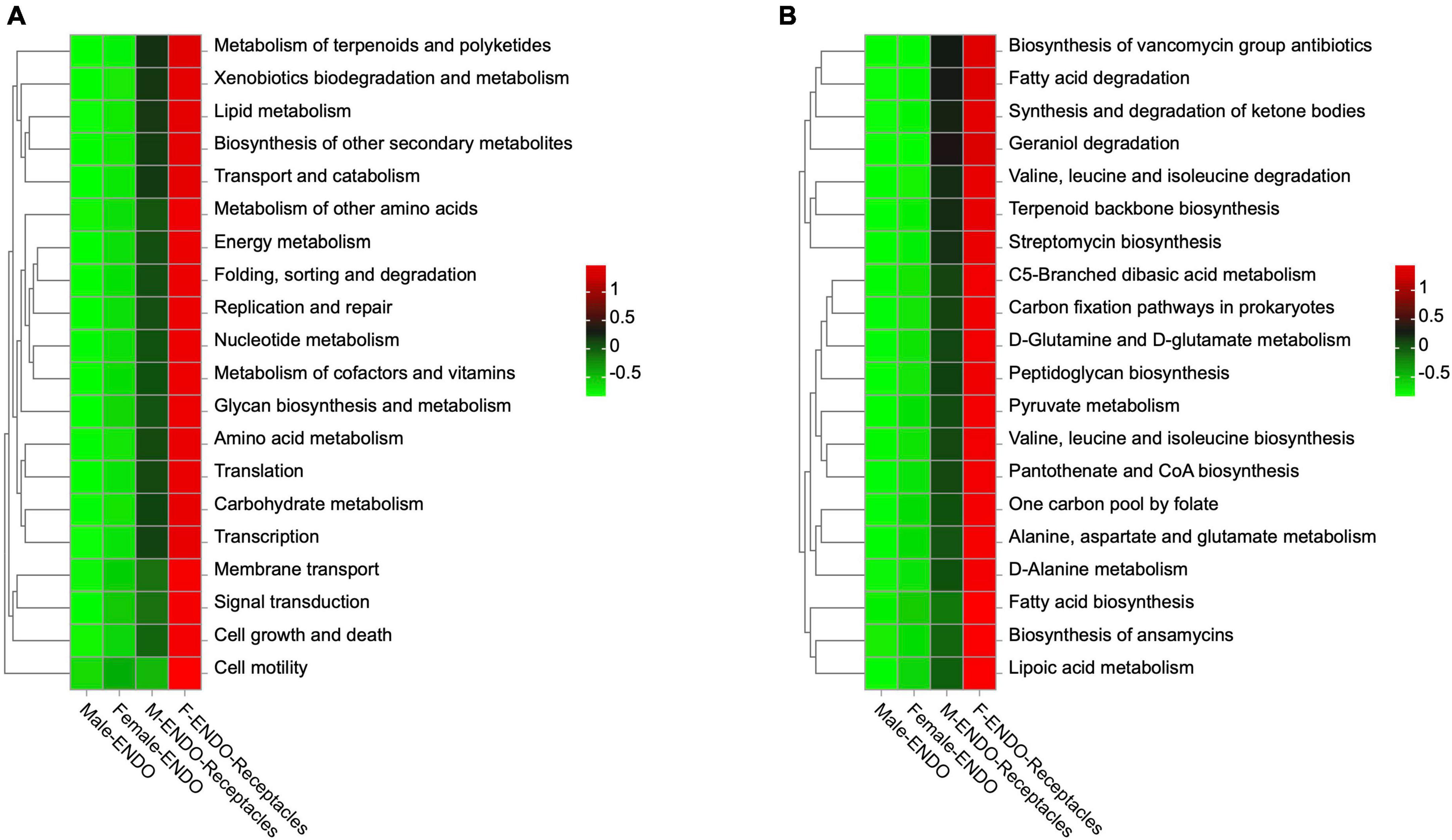
Figure 8. Functional prediction of genes of endophytic bacteria in male and female S. thunbergii and their receptacles. (A) Level 2. P < 0.01. (B) Level 3. P < 0.05 (KW rank sum test).
The results of both culture and high-throughput sequencing in this study showed that the endophytic bacterial species were very rich in S. thunbergii and their receptacles. Among the strains isolated from algal bodies, Bacillota dominated at the phylum level, which was consistent with the results of studies on terrestrial plants (Webster et al., 2020). However, the results at the genus level and species were different from those in terrestrial plants, with most of the detected bacteria being saline-alkali tolerant or salt tolerant bacteria (Supplementary Table 1), such as A. algicola (Ivanova et al., 2004), A. berkeleyi (Patel and Gupta, 2020), A. caeni (Patel and Gupta, 2020), R. aquimaris (Wang et al., 2023), F. halophilus (Sharma et al., 2016), H. faecis (Zhang et al., 2019), and H. kuroshimensis (Shi et al., 2020), indicating the adaptation of algal endophytic bacteria to the marine environment. It was interesting that the dominant species was A. algicola, which was an algae-dweller with alginolytic abilities and a lot of other isolated endophytic bacteria such as H. kuroshimensis (Shi et al., 2020), V. alginolyticus (de Souza Valente and Wan, 2021), and P. diffluens (Nikolaeva et al., 1999) were also alginic acid-dissolving bacteria. In addition, there were many bacterial species involved in material metabolism, included P. caeni (Baker et al., 1998) with multiple metabolic functions, M. jeotgali (Green-Ruiz et al., 2008) and S. horikoshii (Gupta et al., 2020) related to heavy metal absorption, etc. These results indicated that the metabolic relationship between endophytes and hosts, especially the metabolism of algal derived substances by endophytes, which was very important in the relationship between endophytes and hosts. Moreover, there are also some beneficial endophytic bacteria which were beneficial to the host, including antibacterial B. Safensis (Zhang Z. et al., 2020) and B. xiamenensis (Amna et al., 2020), as well as M. indicus (Falkenberg et al., 2023), which induced host metamorphosis, indicating that there was a very complex relationship between endophytic bacteria and host algae.
In addition, our study revealed that endophytic bacteria were more abundant in the receptacles than in the algal bodies, with five genera isolated only in the receptacles. These genera were rich in functions; for example, Exiguobacterium, which is adapted to extreme environmental conditions such as salt and alkali (He et al., 2012; Zhang et al., 2013), the biodegrading bacterium Persicobacter (Han et al., 2012), and Vibrio (de Souza Valente and Wan, 2021), which is a common pathogen in marine organisms.
The high-throughput sequencing results showed that the core phyla of endophytic bacteria in S. thunbergii were Pseudomonadota, Bacteroidota, and Actinomycetota, which were similar to those in terrestrial plants (Fadiji et al., 2020) and rice seeds (Feng et al., 2023). However, there was a significant difference in the core genera of endophytic bacteria in S. thunbergii compared with terrestrial plants and the other macroalgae S. horneri and U. prolifera (Mei, 2019). This indicated that the species specificity of host algae is an important factor determining the composition of endophytic bacterial communities. The core endophytic bacteria in S. thunbergii included the genus Sva0996_marine_group, which is related to the metabolism of alga-derived substances (Orsi et al., 2016), the dimethylsulfoniopropionate (DMSP) degrader Loktanella (Sun et al., 2020), and pollutant-degrading bacteria, such as the phenanthrene (PHE)-degrading bacterium Acinetobacter (Li et al., 2017), the polyaromatic hydrocarbon (PAH)-degrading bacterium Pseudoruegeria (Yuan, 2008), and the hydrocarbon-degrading bacteria Granulosicoccus (Rizzo et al., 2019) and Robiginitomaculum (Verhoeven et al., 2017). In addition, there were some bacteria that can promote and inhibit the growth of algae, such as Burkholderia-Caballeronia-Paraburkholderia (Wu et al., 2022), Enhydrobacter (Dartora et al., 2016), and Maribacter (Ghaderiardakani et al., 2019). This indicated that the composition of endophytic bacteria was closely related to the external environment and host alga. Interestingly, these endophytic bacteria were also abundant in S. thunbergii (Wang et al., 2022a), which is consistent with the findings of studies on S. horneri and U. prolifera (Mei, 2019), indicating that the endophytic bacteria and epiphytic bacteria in macroalgae have a common source and close connection.
Previous studies paid little attention to the differences in endophytic bacterial communities between hosts of different sexes, and similar studies have not been carried out in macroalgae. However, the results of both the culture method and high-throughput sequencing in this study revealed differences between endophytic bacteria in algal bodies and receptacles of hosts of different sexes.
The results of both methods showed a difference in the endophytic bacterial community between male and female S. thunbergii and their receptacles. Although the dominant bacteria of female and male algal bodies were basically the same, some species could be isolated from only a single sex. Interestingly, some endophytes isolated from algae of different sexes, although not the same species, performed similar functions. For example, H. trueperi with antibacterial activity was isolated from male algae (Chen et al., 2010), while B. safensis (Zhang Z. et al., 2020), which were isolated from female algal bodies, also have antibacterial properties. There have been many relevant studies on endophytic bacteria isolated from different plant organs (Wang et al., 2021; Zhang A. et al., 2021). However, the differences in culturable bacteria among different reproductive tissues of dioecious plants have not been reported. This study revealed that the dominant endophytic bacteria differed between the male and female receptacles of S. thunbergia. For example, V. neocaledonicus, the dominant bacterium isolated only from the male receptacles, has been reported as a pathogen and has the ability to prevent oxygen from entering rusted biofilm (Moradi et al., 2018). The dominant genus Alkalihalobacillus in female receptacles has a degradation function (Song et al., 2023).
The results of high-throughput sequencing of the specific taxa more clearly showed the differences in endophytic bacterial communities between sexes. The endophytic bacteria of samples from different sexes included specific phyla and genera, and most of them were metabolism-related bacteria. For example, among the specific bacterial phyla in the female algal bodies, Entotheonellaeota has the ability to synthesize bioactive substances (Ho et al., 2021), Armatimonadota and Planctomycetota can decompose and utilize complex polysaccharides (Wang et al., 2015), Nitrospirota is associated with N metabolism (Daims et al., 2015), and Marinimicrobia_SAR406_clade is associated with the dark fixation of dissolved inorganic carbon (DIC) (Guerrero-Feijóo et al., 2018). The only specific phylum in the male algal bodies was Elusimicrobiota, which consists of nitrogen-fixing bacteria (Zheng et al., 2016; Méheust et al., 2020). Interestingly, there were no specific phyla in either male or female receptacles. At the genus level, the results were more striking. For example, the specific genera in male and female algal bodies were still related to metabolism. The specific genera in male algal bodies included the alginate-degrading bacterium Brachybacterium (Wang M. et al., 2018), phosphorus-solubilizing bacterium Psychrobacillus (Chiba et al., 2022), carotenoid-producing bacterium Flavicella (Teramoto and Nishijima, 2015), and Rubricoccus (Nakajima et al., 2017), which contains rhodopsin-producing genes and plays an important role in energy conversion. Interestingly, there were also some gut metabolic bacteria present, such as Lentisphaera (related to pentose metabolism) and Dialister (Louis et al., 2014) (related to propionic acid production). In addition, the group also included the autotrophic bacterium Magnetospira involved in CO2 fixation (He, 2014). The number of specific genera in the female algal bodies was much greater than that in the male algal bodies, and their functions were more diverse. However, most of them were involved in nutrient cycling and metabolism, such as Macrococcus (Mazhar et al., 2018), with the highest abundance, which can promote carbohydrate and amino acid metabolism; bacteria involved in sulfur metabolism, including Marinobacterium (Fuse et al., 2000), Desulfatitalea (Pang et al., 2021), Halanaerobium (Ravot et al., 2005), and Sva0081_sediment_group (Fan et al., 2018); bacteria involved in nitrogen metabolism, including Pirellula (Han et al., 2019), Costertonia (Kwon et al., 2006) and Gemmatimonas (Park et al., 2017); and complex compound-degrading bacteria, including SM1A02 (Zheng et al., 2020), and Chryseolinea (Milkereit et al., 2021). Interestingly, many bacteria have been reported in human and animal intestines, such as Cloacibacterium (Nouha et al., 2016), Prevotella_9, Prevotella_2 (Liu Z. et al., 2020), and Ruminococcaceae_UCG-005 (Gebeyew et al., 2021), which showed the various metabolic capacities of endophytic bacteria specific to female algal bodies. In addition, Fusobacterium is an opportunistic anaerobic pathogen (Castellarin et al., 2012), and Geodermatophilus has significant antioxidant capacity and can resist multiple environmental stresses (Hongmin et al., 2015). Additionally, there were phototrophic/heterotrophic bacteria of Candidatus_Actinomarina that absorb and assimilate dissolved organic matter (DOM) (Xie Z., 2018) and RB41 bacteria important for controlling carbon circulation (Stone et al., 2021).
The differences in specific genera of endophytic bacteria were more apparent in the receptacles. The specific genera with the highest abundance in male receptacles were almost all intestinal bacteria, including Prevotella_ 1 (Liu Z. et al., 2020), Erysipelotrichaceae_ UCG-007 (Guo et al., 2020), Shuttleworthia (Jin-Shun et al., 2017), Defluviitaleaceae_U (Yang et al., 2020), Faecalibacterium (Heinken et al., 2014), Rubellimicrobium (Xie D., 2018), Ruminococcus_2 (Ma et al., 2022), Candidatus_Saccharimonas (Cao et al., 2022), and Terrimicrobium (Qiu et al., 2014). They also included the specific pathogenic bacteria Peptococcus (Bourgault et al., 1980) and Roseomonas (Rihs et al., 1993). However, in the female receptacles, there were many pathogenic bacteria, such as Legionella (Machner and Isberg, 2006), Mycobacterium (Palomino, 2009), and Kocuria (Basaglia et al., 2002). The bacterial genus Promicromonospora specific to the female receptacles also has antagonistic effects against Fusarium oxysporum and can produce antioxidants. It was particularly interesting that Steroidobactor (Fahrbach et al., 2008) was the first known bacteria to grow on estradiol (C-18) and testosterone (C-19), while Minicystis (Garcia et al., 2014) could produce steroids. In addition, there were few degrading bacteria in the receptacles of both sexes, and the degrading bacteria in the male receptacles were the phenol-degrading anaerobic bacterium Thermicanus (Qi, 2021) and the sulfate-degrading bacterium Desulfobulbus (Samain et al., 1986). The female receptacles included the polyethylene degrader Brevibacillus (Stone et al., 2021), biopolymer degrader Tepidisphaera (Jiang et al., 2020) and various macromolecular degraders such as Luteimonas (Lin et al., 2020; Zhou et al., 2021). Moreover, there were alga-soluble bacteria (Muricauda) in the male receptacles (Shi et al., 2012), while there were algicidal bacteria [Kordia (Sohn et al., 2004) and Saprospira (Furusawa et al., 2003)] in the female receptacles.
The results of LEfSe analysis also showed that the genera with high indicator values were different between the sexes and were mainly related to metabolism. For example, Planctomicrobium, the bacterium with the highest indicator value in male algal bodies, can participate in the degradation of biopolymers in plant and fungal cell walls (Kulichevskaya et al., 2019), and the bacteria with high indicator values in male receptacles included the degrading bacterium Rhodococcus (Larkin et al., 2005) and endophytic nitrogen-fixing bacterium Delftia (Han, 2004). Marinobacterium, with a high indicator value in female algal bodies, participates in the sulfur cycle (Fuse et al., 2000), and Coraliomargarita, with a high indicator value in female receptacles, has been reported as a specific microfloral member in the gut of Apostichopus japonicus (Quan et al., 2019).
In summary, there were significant differences in the composition of endophytic bacteria between males and females, mainly associated with material metabolism, as well as the degradation of pollutants, pathogen resistance and antioxidant activity. Studies on higher plants have shown significant differences in enzyme activity, secondary metabolites and endogenous hormone levels between male and female individuals and reported that estrogen is a unique secreted hormone in various brown algae (Li et al., 2006) and that the content of bromophenols in the reproductive tissues of the red alga Neorhodomela larix varies between the sexes (Carlson et al., 1989). This indicates that the differences in endophytic bacterial communities mentioned above may mainly be due to differences in material metabolism within the host algae and reproductive tissues of different sexes. However, further confirmation based on combining the metabolomic and transcriptome differences between the sexes of S. thunbergii and their receptacles is needed.
The results of the two methods in this study revealed differences in the structure and function of endophytic bacterial communities between male and female S. thunbergii and their receptacles. The β-diversity analysis revealed that endophytic bacterial communities in samples from different sexes could be clustered separately. Moreover, the clustering between male and female receptacles was more obvious, indicating that the sex differences in endophytic bacterial communities from receptacles were greater than those from algal bodies. This can be explained by the fact that the differences in the male and female algal bodies of S. thunbergii were smaller than those in their receptacles. The smaller difference in endophytic bacterial communities between male and female algal bodies means that the bacterial community in algal bodies of different sexes was more stable than that in the receptacles, but the significant differences in β-diversity between groups indicated that the sex of S. thunbergii has a certain impact on the structure of the endophytic bacterial communities in S. thunbergii and that the impact was stronger on the endophytic bacterial communities in the receptacles.
The α-diversity results also showed that the abundance and evenness of endophytic bacteria in S. thunbergii were higher in females than in males. Previous studies have found that the reproductive tissues of brown macroalgae can secrete unique estrogenic hormones, which may correspond to their metabolic bacteria. Additionally, this study showed the bacterial genera Steroidobactors (Fahrbach et al., 2008) that can degrade sex hormones and Minicystis (Garcia et al., 2014) that can produce steroids in female receptacles, indicating that the sex of the host can directly affect the composition of endophytic bacteria in the receptacles of S. thunbergii.
Moreover, the results of predicted gene function indicated that the difference in the abundance of functional genes between male and female algal bodies was less obvious than that between male and female receptacles. This further confirmed that the differences in bacterial communities of endophytic bacteria between different sexes were mainly due to differences in the receptacles. These differences were mainly related to metabolic genes, which suggested that metabolic differences were the main reason for the differences in endophytic bacteria between male and female algal bodies and receptacles. In addition, the abundance of predicted genes with significant differences was basically the same in both male and female algal bodies but lower than that in the receptacles, and the abundance of predicted genes in the female receptacles was much higher than that in the male receptacles. Many studies have shown significant differences in the content of various chemicals in male and female flowers in plants (Quan et al., 2019; Sowndhararajan et al., 2020). Although algae do not have floral organs, studies have shown that the content of bromophenols in the reproductive structures of the two sexes varies in the red alga Neorhodomela larix (Carlson et al., 1989). In this study, it was also found that both Sva0996_marine_group, which is involved in the utilization of organic matter (Wang Y. et al., 2018), and Phormidesmis, which is related to organophosphorus decomposition, were more abundant in female receptacles. It can be speculated that the differences in the chemicals between male and female receptacles lead to differences in endophytic bacterial communities closely related to host metabolism in the receptacles of different sexes. However, few studies have focused on the differences between male and female algae and their receptacles, and further studies will need to analyze the metabolic differences between algae and receptacles of different sexes and their correlation with endophytic bacterial communities to reveal the mechanism by which the sex of dioecious macroalgae affects the assembly of the endophytic bacterial community.
In this study, the endophytic bacterial community structures of male and female S. thunbergii and their receptacles were compared based on a culture method and high-throughput sequencing. The results of both methods showed that the majority of endophytic bacteria in the two sexes of S. thunbergii and their receptacles were the same, but the diversity, abundance of dominant taxa, specific bacterial taxa, and biomarkers differed between the sexes, especially in the samples from receptacles. There was a significant difference in predicted functional abundance between male and female samples, and most of the functions were related to metabolism. It was found for the first time that the sex of the host alga contributes to the community assembly of endophytic bacteria in S. thunbergii, and the impact on the endophytic bacterial community was greater in the receptacles. Moreover, many endophytic bacterial strains were obtained, providing experimental materials for the effective utilization and development of algal microbial resources. The results of this study help elucidate the mechanism of endophytic bacterial community assembly in dioecious marine macroalgae and further the understanding of the interaction between endophytic bacteria and macroalgae.
The original contributions presented in the study are publicly available. The bacterial sequences obtained in this study have been deposited to National Center for Biotechnology Information (NCBI) under BioProject accession numbers PRJNA830307 and PRJNA830329.
YZ: Data curation, Formal Analysis, Investigation, Writing – original draft. TS: Data curation, Formal Analysis, Investigation, Writing – original draft. YL: Data curation, Formal Analysis, Investigation, Writing – review and editing. ZY: Data curation, Formal Analysis, Investigation, Writing – review and editing. JC: Writing – review and editing. JW: Writing – review and editing. XY: Writing – review and editing. XT: Funding acquisition, Project administration, Supervision, Writing – review and editing. HX: Supervision, Writing – review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (42176154).
TS was employed by Qingdao Branch CCCC Water Transportation Consultants Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1334918/full#supplementary-material
Afzal, I., Shinwari, Z. K., Sikandar, S., and Shahzad, S. (2019). Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 221, 36–49.
Amini Hajiabadi, A., Mosleh Arani, A., Ghasemi, S., Rad, M. H., Etesami, H., Shabazi Manshadi, S., et al. (2021). Mining the rhizosphere of halophytic rangeland plants for halotolerant bacteria to improve growth and yield of salinity-stressed wheat. Plant Physiol. Biochem. 163, 139–153.
Ammar, E. M., Ellogheshtawy, H. S., Elloghatoury, E. H., and Amer, S. K. (2021). Green synthesis of polyhydroxyalkanoates polymer by bacillus iocasae. Poly. Int. 70, 1478–1485.
Amna Xia, Y., Farooq, M. A., Javed, M. T., Kamran, M. A., Mukhtar, T., et al. (2020). Multi-stress tolerant PGPR Bacillus xiamenensis PM14 activating sugarcane (Saccharum officinarum L.) red rot disease resistance. Plant Physiol. Biochem. 151, 640–649. doi: 10.1016/j.plaphy.2020.04.016
Amri, M., Rjeibi, M. R., Gatrouni, M., Mateus, D. M. R., Asses, N., Pinho, H. J. O., et al. (2023). Isolation, identification, and characterization of phosphate-solubilizing bacteria from tunisian soils. Microorganisms 11:783. doi: 10.3390/microorganisms11030783
Baker, S. C., Ferguson, S. J., Ludwig, B., Page, M. D., Richter, O. M., and van Spanning, R. J. (1998). Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol. Mol. Biol. Rev. 62, 1046–1078. doi: 10.1128/MMBR.62.4.1046-1078.1998
Basaglia, G., Carretto, E., Barbarini, D., Moras, L., Scalone, S., Marone, P., et al. (2002). Catheter-related bacteremia due to Kocuria kristinae in a patient with ovarian cancer. J. Clin. Microbiol. 40, 311–313. doi: 10.1128/JCM.40.1.311-313.2002
Bourgault, A. M., Rosenblatt, J. E., and Fitzgerald, R. H. (1980). Peptococcus magnus: a significant human pathogen. Ann. Intern. Med. 93, 244–248. doi: 10.7326/0003-4819-93-2-244
Cao, C., Wang, L., Ai, C., Gong, G., Wang, Z., Huang, L., et al. (2022). Impact of Lycium barbarum arabinogalactan on the fecal metabolome in a DSS-induced chronic colitis mouse model. Food Funct. 13, 8703–8716. doi: 10.1039/d2fo01283a
Carlson, D. J., Lubchenco, J., Sparrow, M. A., and Trowbridge, C. D. (1989). Fine-scale variability of lanosol and its disulfate ester in the temperate red alga Neorhodomela larix. J. Chem. Ecol. 15, 1321–1333. doi: 10.1007/BF01014833
Castellarin, M., Warren, R. L., Freeman, J. D., Dreolini, L., Krzywinski, M., Strauss, J., et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306. doi: 10.1101/gr.126516.111
Cerritos, R., Vinuesa, P., Eguiarte, L. E., Herrera-Estrella, L., Alcaraz-Peraza, L. D., Arvizu-Gómez, J. L., et al. (2008). Bacillus coahuilensis sp. nov., a moderately halophilic species from a desiccation lagoon in the Cuatro Ciénegas Valley in Coahuila. Mexico. Int. J. Syst. Evol. Microbiol. 58(Pt 4), 919–923. doi: 10.1099/ijs.0.64959-0
Chandna, P., Mayilraj, S., and Kuhad, R. C. (2016). Bacillus pseudoflexus sp. nov., a moderately halophilic bacterium isolated from compost. Ann. Microbiol. 66, 895–905.
Chen, H., Zhao, L., Huang, X., He, J., Huang, Y., and Zhu, L. (2020). Comparison on leaf physiological properties of male and female Podocarpus macrophyllus. GuangXi For. Sci. 49, 125–129.
Chen, L., Wang, G., Bu, T., Zhang, Y., Liu, M., Zhang, J., et al. (2010). Identification of a moderately halophilic bacterium whb45 and screening of its antimicrobial and antitumor activity. Microbiol. Chin. 37, 85–90.
Chiba, A., Peine, M., Kublik, S., Baum, C., Schloter, M., and Schulz, S. (2022). Complete genome sequence of Psychrobacillus sp. Strain INOP01, a phosphate-solubilizing bacterium isolated from an agricultural soil in Germany. Microbiol. Resour. Announc. 11:e0020722. doi: 10.1128/mra.00207-22
Crapart, S., Fardeau, M. L., Cayol, J., Thomas, P., Sery, C., Ollivier, B., et al. (2007). Exiguobacterium profundum sp. nov., a moderately thermophilic, lactic acid-producing bacterium isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 57, 287–292. doi: 10.1099/ijs.0.64639-0
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509.
Daroonpunt, R., Yiamsombut, S., Sitdhipol, J., and Tanasupawat, S. (2019). Bacillus salacetis sp. nov., a slightly halophilic bacterium from Thai shrimp paste (Ka-pi). Int. J. Syst. Evol. Microbiol. 69, 1162–1168. doi: 10.1099/ijsem.0.003286
Dartora, J., Guimarães, V. F., Menezes, C. R. J., Freiberger, M. B., Castoldi, G., and Gonçalves, D. V. (2016). Maize response to inoculation with strains of plant growth-promoting bactéria. Rev. Bras. Eng. Agríc. Ambient. 20, 606–611.
Dastogeer, K. M. G., Li, H., Sivasithamparam, K., Jones, M. G. K., and Wylie, S. J. (2018). Host specificity of endophytic mycobiota of wild nicotiana plants from arid regions of northern Australia. Microbiol. Ecol. 75, 74–87. doi: 10.1007/s00248-017-1020-0
de Souza Valente, C., and Wan, A. H. L. (2021). Vibrio and major commercially important vibriosis diseases in decapod crustaceans. J. Invertebr. Pathol. 181:107527.
Fadiji, A. E., Ayangbenro, A. S., and Babalola, O. O. (2020). Metagenomic profiling of the community structure, diversity, and nutrient pathways of bacterial endophytes in maize plant. Antonie Van Leeuwenhoek 113, 1559–1571. doi: 10.1007/s10482-020-01463-w
Fahrbach, M., Kuever, J., Remesch, M., Huber, B. E., Kämpfer, P., Dott, W., et al. (2008). Steroidobacter denitrificans gen. nov., sp. nov., a steroidal hormone-degrading gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 58, 2215–2223. doi: 10.1099/ijs.0.65342-0
Falkenberg, F., Voß, L., Bott, M., Bongaerts, J., and Siegert, P. (2023). New robust subtilisins from halotolerant and halophilic Bacillaceae. Appl. Microbiol. Biotechnol. 107, 3939–3954. doi: 10.1007/s00253-023-12553-w
Fan, X., Ding, S., Gong, M., Chen, M., Gao, S., Jin, Z., et al. (2018). Different influences of bacterial communities on Fe (III) reduction and phosphorus availability in sediments of the Cyanobacteria- and macrophyte-dominated zones. Front. Microbiol. 9:2636. doi: 10.3389/fmicb.2018.02636
Feng, S., Xie, G., Liu, N., Xu, C., Lu, F., and Feng, X. (2020). Isolation and antioxidative activities of algal endophytes. J. Food Biotechnol. 39, 99–105.
Feng, X., Wang, Z., Li, X., Wang, W., Gu, A., and Liu, Y. (2023). Analysis of endophytic bacterial diversity in rice seeds with regional characteristics in Yunnan Province, China, based on high-throughput sequencing technology. Curr. Microbiol. 80:287. doi: 10.1007/s00284-023-03399-6
Furusawa, G., Yoshikawa, T., Yasuda, A., and Sakata, T. (2003). Algicidal activity and gliding motility of Saprospira sp. SS98-5. Can. J. Microbiol. 49, 92–100. doi: 10.1139/w03-017
Fuse, H., Takimura, O., Murakami, K., Yamaoka, Y., and Omori, T. (2000). Utilization of dimethyl sulfide as a sulfur source with the aid of light by Marinobacterium sp.strain DMS-S1. Appl. Environ. Microbiol. 66, 5527–5532. doi: 10.1128/AEM.66.12.5527-5532.2000
Garcia, R., Gemperlein, K., and Müller, R. (2014). Minicystis rosea gen. nov., sp. nov., a polyunsaturated fatty acid-rich and steroid-producing soil myxobacterium. Int. J. Syst. Evol. Microbiol. 64, 3733–3742. doi: 10.1099/ijs.0.068270-0
Ge, L., Huang, Y., Xue, J., Qi, Q., Zhang, Q., Xu, X., et al. (2021). Dynamic changes of endogenous hormones in female and male flower of Taxus cuspidata during flower development. J. Beihua Univer. 22, 456–461.
Gebeyew, K., Chen, K., Wassie, T., Azad, M. A. K., He, J., Jiang, W., et al. (2021). Dietary amylose/amylopectin ratio modulates cecal microbiota and metabolites in weaned goats. Front. Nutr. 8:774766. doi: 10.3389/fnut.2021.774766
Ghaderiardakani, F., Califano, G., Mohr, J., Abreu, M., Coates, J., and Wichard, T. (2019). Analysis of algal growth- and morphogenesis-promoting bacterial factors (AGPFs) in an integrated multi-trophic aquaculture system for farming the green seaweed Ulva. Aquacult. Environ. Interactions 11, 375–391.
Green-Ruiz, C., Rodriguez-Tirado, V., and Gomez-Gil, B. (2008). Cadmium and zinc removal from aqueous solutions by Bacillus jeotgali: pH, salinity and temperature effects. Bioresour. Technol. 99, 3864–3870. doi: 10.1016/j.biortech.2007.06.047
Guerrero-Feijóo, E., Sintes, E., Herndl, G. J., and Varela, M. M. (2018). High dark inorganic carbon fixation rates by specific microbial groups in the Atlantic off the Galician coast (NW Iberian margin). Environ. Microbiol. 20, 602–611. doi: 10.1111/1462-2920.13984
Guo, J., Zhang, X., Saiganesh, A., Peacock, C., Chen, S., Dykes, G. A., et al. (2020). Linking the westernised oropharyngeal microbiome to the immune response in Chinese immigrants. Allergy Asthma Clin. Immunol. 16:67. doi: 10.1186/s13223-020-00465-7
Gupta, R. S., Patel, S., Saini, N., and Chen, S. (2020). Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the subtilis and cereus clades of species. Int. J. Syst. Evol. Microbiol. 70, 5753–5798. doi: 10.1099/ijsem.0.004475
Han, J. (2004). The Studies on the Identification and Biological Characteristics of a Novel rice Endophytic Bacteria Strain Delftia tsuruhatensis HR4 with N2-Fixing Activity. Doctor’s Thesis. China: Capital Normal University.
Han, W., Zhao, S., Liu, H., Wu, Z., Gu, Q., and Li, Y. (2012). Isolation, identification and agarose degradation of a polysaccharide-degrading marine bacterium Persicobacter sp. JZB09. Microbiol. Chin. 52, 776–783.
Han, X., Feng, J., Zhang, L., Cui, B., Zhang, J., Pan, L., et al. (2019). Micro-polluted water treatment by biological contact oxidation process: aeration mode and bacteria community analysis. Environ. Eng. Sci. 36, 1491–1502.
He, P., Xu, S., Huang, X., Seswita-Zilda, D., and Zhang, Q. (2012). Isolation and identification of microorganisms from high temperature seaweed beds and study on the characteristics of thermo and salt tolerances. Microbiol. China 39, 1769–1777.
He, Q. (2014). Autotrophic Bacteria and Their Community in the Pearl River. Master’s Thesis. Guangzhou: South China University of Technology.
Heinken, A., Khan, M. T., Paglia, G., Rodionov, D. A., Harmsen, H. J. M., and Thiele, I. (2014). Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J. Bacteriol. 196, 3289–3302. doi: 10.1128/JB.01780-14
Ho, X. Y., Katermeran, N. P., Deignan, L. K., Phyo, M. Y., Ong, J. F. M., Goh, J. X., et al. (2021). Assessing the diversity and biomedical potential of microbes associated with the neptune’s cup sponge Cliona patera. Front. Microbiol. 12:631445. doi: 10.3389/fmicb.2021.631445
Hollants, J., Leroux, O., Leliaert, F., Decleyre, H., De Clerck, O., and Willems, A. (2011). Who is in there? exploration of endophytic bacteria within the siphonous green seaweed Bryopsis (Bryopsidales, Chlorophyta). PLoS One 6:e26458. doi: 10.1371/journal.pone.0026458
Huo, X., Wang, Y., Zhang, D., Gao, T., and Liu, M. (2020). Characteristics and diversity of endophytic bacteria in endangered Chinese herb Glehnia littoralis based on illumina sequencing. Pol. J. Microbiol. 69, 283–291. doi: 10.33073/pjm-2020-031
Ivanova, E. P., Alexeeva, Y. A., Zhukova, N. V., Gorshkova, N. M., Buljan, V., Nicolau, D. V., et al. (2004). Bacillus algicola sp. nov., a novel filamentous organism isolated from brown alga Fucus evanescens. Syst. Appl. Microbiol. 27, 301–307. doi: 10.1078/0723-2020-00269
Jiang, B., Zeng, Q., Liu, J., Hou, Y., Xu, J., Li, H., et al. (2020). Enhanced treatment performance of phenol wastewater and membrane antifouling by biochar-assisted EMBR. Bioresour. Technol. 306:123147. doi: 10.1016/j.biortech.2020.123147
Jin-Shun, Z., Cai-Xia, W. U., Ming-Mei, L. I. U., Xiao-Shuang, S. U., Kang, Z., and Guo-Qi, Z. (2017). Effects of alfalfa flavonoids as dietary additives on bacterial flora in the rumen of dairy cows. Acta Pratacult. Sinica 26:82.
Kang, C. L., Li, Q. F., Zhang, Y., Chen, S. B., and Wang, Y. (2018). Purifying effect of three heterotrophic nitrification-aerobic denitrification bacteria strains on the farming water of verasper variegates. Prog. Fishery Sci. 39, 42–48.
Kulichevskaya, I. S., Naumoff, D. G., Ivanova, A. A., Rakitin, A. L., and Dedysh, S. N. (2019). Detection of chitinolytic capabilities in the freshwater planctomycete planctomicrobium piriforme. Microbiology 88, 423–432.
Larkin, M. J., Kulakov, L. A., and Allen, C. C. R. (2005). Biodegradation and rhodococcus–masters of catabolic versatility. Curr. Opin. Biotechnol. 16, 282–290. doi: 10.1016/j.copbio.2005.04.007
Li, J., Luo, C., Song, M., Dai, Q., Jiang, L., Zhang, D., et al. (2017). Biodegradation of phenanthrene in polycyclic aromatic hydrocarbon-contaminated wastewater revealed by coupling cultivation-dependent and -independent approaches. Environ. Sci. Technol. 51, 3391–3401. doi: 10.1021/acs.est.6b04366
Li, L., Lei, G., Li, L., Du, Z., Zhen, J., Wang, J., et al. (2021). Diversity analysis on endophytic bacterial community in different organs of Arachis hypogaea Linn, based on high-throughput sequencing. J. Peanut. Sci. 50, 1–20.
Li, W., Li, D., and Du, X. (2006). Advance in male-attractants from Phaeophyta. Mar. Sci. 30, 79–84.
Li, Z., Li, J., Wu, C., Xu, B., Song, P., Liu, Z., et al. (2022). Total flavonoid content and antioxidant activity of female and male flower buds of Populus tomentosa. Central S. Pharmacy 20, 1034–1038.
Liaqat, F., and Eltem, R. (2016). Identification and characterization of endophytic bacteria isolated from in vitro cultures of peach and pear rootstocks. 3 Biotech 6:120. doi: 10.1007/s13205-016-0442-6
Lin, P., Yan, Z.-F., and Li, C.-T. (2020). Luteimonas cellulosilyticus sp. nov., cellulose-degrading bacterium isolated from soil in changguangxi national wetland park. China. Curr. Microbiol. 77, 1341–1347. doi: 10.1007/s00284-020-01934-3
Ling, L., Yang, C., Li, Z., Luo, H., Feng, S., Zhao, Y., et al. (2022). Plant endophytic bacteria: a potential resource pool of electroactive micro-organisms. J. Appl. Microbiol. 132, 2054–2066. doi: 10.1111/jam.15368
Liu, L., Lu, L., Li, H., Meng, Z., Dong, T., Peng, C., et al. (2021). Divergence of phyllosphere microbial communities between females and males of the dioecious Populus cathayana. Mol. Plant Microbe Interact. 34, 351–361. doi: 10.1094/MPMI-07-20-0178-R
Liu, Y., Xu, P., Yang, F., Li, M., Yan, H., Li, N., et al. (2019). Composition and diversity of endophytic bacterial community in seeds of super hybrid rice ‘Shenliangyou 5814’ (Oryza sativa L.) and its parental lines. Plant Growth Regul. 87, 257–266.
Liu, Y., Yan, H., Zhang, X., Zhang, R., Li, M., Xu, T., et al. (2020). Investigating the endophytic bacterial diversity and community structures in seeds of genetically related maize (Zea mays L.) genotypes. 3 Biotech 10:27. doi: 10.1007/s13205-019-2034-8
Liu, Z., Guo, F., Zhang, B., Xaing, D., Zhao, Z., Zhao, S., et al. (2020). Effects of complex probiotics derived from different variety on enteric duct microbes in duroc×diannian smallear pig. mLife 40, 21–29.
Louis, P., Hold, G. L., and Flint, H. J. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672.
Lu, J., Chen, Y., and Yin, T. (2021). Research progress on sex determination genes of woody plants. Chin. Bull. Bot. 56:90.
Ma, J., Zhang, Y., Dong, C., Chai, Y., and Guo, Q. (2022). Comparison of the chemical constituents and in vitro hypoglycemic effect of Polygonatum from different origins. Modern Food Sci. Technol. 38, 116–126.
Machner, M. P., and Isberg, R. R. (2006). Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 11, 47–56. doi: 10.1016/j.devcel.2006.05.013
Mahmood, A., Takagi, K., Ito, K., and Kataoka, R. (2019). Changes in endophytic bacterial communities during different growth stages of cucumber (Cucumis sativus L.). World J. Microbiol. Biotechnol. 35:104. doi: 10.1007/s11274-019-2676-z
Marag, P. S., and Suman, A. (2018). Growth stage and tissue specific colonization of endophytic bacteria having plant growth promoting traits in hybrid and composite maize (Zea mays L.). Microbiol. Res. 214, 101–113. doi: 10.1016/j.micres.2018.05.016
Mazhar, S., Hill, C., and McAuliffe, O. (2018). The genus Macrococcus: an insight into its biology, evolution, and relationship with Staphylococcus. Adv. Appl. Microbiol. 105, 1–50. doi: 10.1016/bs.aambs.2018.05.002
Méheust, R., Castelle, C. J., Matheus Carnevali, P. B., Farag, I. F., He, C., Chen, L.-X., et al. (2020). Groundwater Elusimicrobia are metabolically diverse compared to gut microbiome Elusimicrobia and some have a novel nitrogenase paralog. ISME J. 14, 2907–2922. doi: 10.1038/s41396-020-0716-1
Mei, X. (2019). Community Structures and Functions of Bacteria Associated with Blooming Seaweeds in the Yellow Sea. Master’s Thesis. China: University of Chinese Academy of Sciences.
Mei, Y., Shayimu, G., Zhi, D., Jing, Z., Xiao, J., Qi, Y., et al. (2021). Diversity and function analysis of endophytic bacterial community in different tissues of Lycium ruthenicum Murr. Acta Microbiol. Sin. 61, 152–166.
Milkereit, J., Geisseler, D., Lazicki, P., Settles, M. L., Durbin-Johnson, B. P., and Hodson, A. (2021). Interactions between nitrogen availability, bacterial communities, and nematode indicators of soil food web function in response to organic amendments. Appl. Soil Ecol. 157:103767.
Moradi, M., Song, Z., and Xiao, T. (2018). Exopolysaccharide produced by Vibrio neocaledonicus sp. as a green corrosion inhibitor: production and structural characterization. J. Mater. Sci. Technol. 34, 2447–2457.
Munir, S., Li, Y., He, P., Huang, M., He, P., He, P., et al. (2020). Core endophyte communities of different citrus varieties from citrus growing regions in China. Sci. Rep. 10:3648. doi: 10.1038/s41598-020-60350-6
Nakajima, Y., Yoshizawa, S., Park, S., Kumagai, Y., Wong, S.-K., Ogura, Y., et al. (2017). Draft genome sequence of Rubricoccus marinus SG-29T, a marine bacterium within the family Rhodothermaceae, which contains two different Rhodopsin genes. Genome Announc. 5:e00990-17. doi: 10.1128/genomeA.00990-17
Nedashkovskaya, O. I., Van Trappen, S., Frolova, G. M., and De Vos, P. (2012). Bacillus berkeleyi sp. nov., isolated from the sea urchin Strongylocentrotus intermedius. Arch. Microbiol. 194, 215–221. doi: 10.1007/s00203-011-0771-0
Nikolaeva, E. V., Usov, A. I., Sinitsyn, A. P., and Tambiev, A. H. (1999). Degradation of agarophytic red algal cell wall components by new crude enzyme preparations. J. Appl. Phycol. 11, 385–389.
Nouha, K., Kumar, R. S., and Tyagi, R. D. (2016). Heavy metals removal from wastewater using extracellular polymeric substances produced by Cloacibacterium normanense in wastewater sludge supplemented with crude glycerol and study of extracellular polymeric substances extraction by different methods. Bioresour. Technol. 212, 120–129. doi: 10.1016/j.biortech.2016.04.021
Orsi, W. D., Smith, J. M., Liu, S., Liu, Z., Sakamoto, C. M., Wilken, S., et al. (2016). Diverse, uncultivated bacteria and archaea underlying the cycling of dissolved protein in the ocean. ISME J. 10, 2158–2173. doi: 10.1038/ismej.2016.20
Palomino, J. C. (2009). Molecular detection, identification and drug resistance detection in Mycobacterium tuberculosis. FEMS Immunol. Med. Microbiol. 56, 103–111. doi: 10.1111/j.1574-695X.2009.00555.x
Pang, Y., Wang, J., Li, S., and Ji, G. (2021). Long-term sulfide input enhances chemoautotrophic denitrification rather than DNRA in freshwater lake sediments. Environ. Pollut. 270:116201. doi: 10.1016/j.envpol.2020.116201
Park, D., Kim, H., and Yoon, S. (2017). Nitrous oxide reduction by an obligate aerobic bacterium, Gemmatimonas aurantiaca Strain T-27. Appl. Environ. Microbiol. 83:e00502-17. doi: 10.1128/AEM.00502-17
Patel, S., and Gupta, R. (2020). A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 70, 406–438. doi: 10.1099/ijsem.0.003775
Pramanic, A., Sharma, S., Dhanorkar, M., Prakash, O., and Singh, P. (2023). Endophytic microbiota of floating aquatic plants: recent developments and environmental prospects. World J. Microbiol. Biotechnol. 39:96. doi: 10.1007/s11274-023-03543-1
Qi, Y. (2021). Study on the Degradation of Phenolic Pollutants in Coal Chemical Wastewater by Fenton-Like and Anaerobic Digestion Mythod. Master’s Thesis. Qingdao: Qingdao University of Science and Technology.
Qiu, Y.-L., Kuang, X.-Z., Shi, X.-S., Yuan, X.-Z., and Guo, R.-B. (2014). Terrimicrobium sacchariphilum gen. nov., sp. nov., an anaerobic bacterium of the class “Spartobacteria” in the phylum Verrucomicrobia, isolated from a rice paddy field. Int. J. Syst. Evol. Microbiol. 64, 1718–1723. doi: 10.1099/ijs.0.060244-0
Quan, Z., Zhang, P., Zhang, Y., Wang, L., Ding, J., and Chang, Y. (2019). Bacterial community and function in the intestinal tracts of sea cucumber (Apostichopus japonicus) at different temperatures. Chinese J. Ecol. 38, 2756–2764.
Ravot, G., Casalot, L., Ollivier, B., Loison, G., and Magot, M. (2005). rdlA, a new gene encoding a rhodanese-like protein in Halanaerobium congolense and other thiosulfate-reducing anaerobes. Res. Microbiol. 156, 1031–1038. doi: 10.1016/j.resmic.2005.05.009
Rihs, J. D., Brenner, D. J., Weaver, R. E., Steigerwalt, A. G., Hollis, D. G., and Yu, V. L. (1993). Roseomonas, a new genus associated with bacteremia and other human infections. J. Clin. Microbiol. 31, 3275–3283. doi: 10.1128/jcm.31.12.3275-3283.1993
Rizzo, C., Malavenda, R., Gerçe, B., Papale, M., Syldatk, C., Hausmann, R., et al. (2019). Effects of a simulated acute oil spillage on bacterial communities from arctic and antarctic marine sediments. Microorganisms 7:632. doi: 10.3390/microorganisms7120632
Samain, E., Albagnac, G., and Legall, J. (1986). Redox studies of the tetraheme cytochrome c3 isolated from the propionate-oxidizing, sulfate-reducing bacterium Desulfobulbus elongatus. FEBS Lett. 204, 247–250.
Sharma, A., Kohli, P., Singh, Y., Schumann, P., and Lal, R. (2016). Fictibacillus halophilus sp. nov., from a microbial mat of a hot spring atop the Himalayan Range. Int J. Syst. Evol. Microbiol. 66, 2409–2416. doi: 10.1099/ijsem.0.001051
Shi, R., Huang, H., Qi, Z., Hu, W., Tian, Z., and Dai, M. (2012). Algicidal activity against Prorocentrum micans by a marine bacterium isolated from a HABs area. South China. Acta Ecol. Sin. 32, 4993–5001.
Shi, X. G., Li, Y., Zheng, W. H., Xiao, Y. C., Liu, L. M., and Chen, J. F. (2020). Isolation and algicidal characteristics of a specific algicidal bacterium on Skeletonema costatum. Microbiol. China 47, 3527–3538.
Sohn, J. H., Lee, J.-H., Yi, H., Chun, J., Bae, K. S., Ahn, T.-Y., et al. (2004). Kordia algicida gen. nov., sp. nov., an algicidal bacterium isolated from red tide. Int. J. Syst. Evol. Microbiol. 54, 675–680. doi: 10.1099/ijs.0.02689-0
Son, H., and Lee, K. (2023). Genome sequences of Metabacillus sp. Strain B2-18, isolated from human skin, and Metabacillus endolithicus KCTC 33579T. Microbiol. Resour. Announc. 12:e0134822. doi: 10.1128/mra.01348-22
Song, F., Li, C., Zhang, N., He, X., Yang, H., Yan, Z., et al. (2023). Alkalihalobacillus clausii PA21 transcriptome profiling and functional analysis revealed the metabolic pathway involved in glycoalkaloids degradation. Int. J. Biol. Macromol. 242:124682. doi: 10.1016/j.ijbiomac.2023.124682
Song, T., Zhang, W., Wei, C., Jiang, T., Xu, H., Cao, Y., et al. (2015). Isolation and characterization of agar-degrading endophytic bacteria from plants. Curr. Microbiol. 70, 275–281. doi: 10.1007/s00284-014-0713-6
Song, X., Li, L., Cui, G., and Ding, G. (2020). Population structure analysis of endophytic bacteria in roots of Platycodon grandiflorum (Jacq)A. DC with different growth years. Guangxi Agric. Sci. 51, 2358–2366.
Sowndhararajan, K., Kim, J.-H., Song, J. E., Kim, M., and Kim, S. (2020). Chemical components of male and female flowers of Schisandra chinensis. Biochem. Syst. Ecol. 92:104121.
Stone, B. W., Li, J., Koch, B. J., Blazewicz, S. J., Dijkstra, P., Hayer, M., et al. (2021). Nutrients cause consolidation of soil carbon flux to small proportion of bacterial community. Nat. Commun. 12, 3381.
Sun, H., Tan, S., Liang, J., Yang, G., Xin, Y., and Zhang, X. (2020). Horizontal and vertical distribution of dimethyl sulfoniopropionate (DMSP) producing and catabolizing bacteria in the East China Sea. Microbiol. Chin. 60, 1865–1881.
Tang, X. (2020). Characteristics and research progress of sex-specific responses to environmental stresses of dioecious plants. Periodical Ocean Univer. China 50, 74–81.
Teramoto, M., and Nishijima, M. (2015). Flavicella marina gen. nov., sp. nov., a carotenoid-producing bacterium from surface seawater. Int. J. Syst. Evol. Microbiol. 65, 799–804. doi: 10.1099/ijs.0.000018
Verhoeven, J. T. P., Kavanagh, A. N., and Dufour, S. C. (2017). Microbiome analysis shows enrichment for specific bacteria in separate anatomical regions of the deep-sea carnivorous sponge Chondrocladia grandis. FEMS Microbiol. Ecol. 93:fiw214. doi: 10.1093/femsec/fiw214
Wang, C., Sun, Q., Ji, Y. N., Liu, L., Zhang, K., Li, J. K., et al. (2023). A high saline-alkali tolerance strain Rossellomorea aquimaris S-2 and its application. Shandong Agricultural University, CN116948867A.
Wang, F., Sun, X., and Li, F. (2007). Studies on sexual reproduction and seedling-rearing of Sargassum thunbergii. Mar. Fish. Res. 27, 1–6.
Wang, J., Li, Y., Yang, Z., Sun, T., Yu, X., Zhao, Y., et al. (2022a). Sex plays a role in the construction of epiphytic bacterial communities on the algal bodies and receptacles of Sargassum thunbergii. Front. Microbiol. 13:935222. doi: 10.3389/fmicb.2022.935222
Wang, J., Yang, Z., Wang, G., Shang, S., Tang, X., and Xiao, H. (2022b). Diversity of epiphytic bacterial communities on male and female Sargassum thunbergii. AMB Express 12:97.
Wang, M., Wang, X., and Chen, L. (2018). In situ screening of cultivable alginate-degrading microorganism on surface of brown seaweed. Microbiol. China 45, 1853–1860.
Wang, S., Liu, J., Sun, J., Jia, N., Jin, N., Liu, J., et al. (2021). Diversity and antibacterial activity of endophytic bacteria in roots and stems of Dendrobium officinale with different cultivation patterns. Microbiol. Chin. 61, 4006–4025.
Wang, X., Sharp, C. E., Jones, G. M., Grasby, S. E., Brady, A. L., and Dunfield, P. F. (2015). Stable-isotope probing identifies uncultured planctomycetes as primary degraders of a complex heteropolysaccharide in soil. Appl. Environ. Microbiol. 81, 4607–4615. doi: 10.1128/AEM.00055-15
Wang, Y., Wang, B., Dann, L. M., Mitchell, J. G., Hu, X., Tang, H., et al. (2018). Bacterial community structure in the Bohai Strait provides insights into organic matter niche partitioning. Continental Shelf Res. 169, 46–54.
Wang, Z. (2007). The Physiological Ecology and Reproduction Biology of Sargassum thunbergii. Master’s Thesis. China: University of Chinese Academy of Sciences.
Webster, G., Mullins, A. J., Cunningham-Oakes, E., Renganathan, A., Aswathanarayan, J. B., Mahenthiralingam, E., et al. (2020). Culturable diversity of bacterial endophytes associated with medicinal plants of the Western Ghats, India. FEMS Microbiol. Ecol. 96:fiaa147. doi: 10.1093/femsec/fiaa147
Wu, Q., Chen, D., Zhou, W., Zhang, X., and Ao, J. (2022). Long-term fertilization has different impacts on bacterial communities and phosphorus forms in sugarcane rhizosphere and bulk soils under low-P stress. Front. Plant Sci. 13:1019042. doi: 10.3389/fpls.2022.1019042
Xie, D. (2018). Research of Synergistic Reaction Between Intestinal Microflora Quorum Sensing Inhibitor and Jiedu Quyu ziyin Recipe on Treating MRL/lpr Mice. Master’s Thesis. Hangzhou: Zhejiang Chinese Medical University.
Xie, Z. (2018). The Molecular Biogeochemistry of Marine Organic Matter in the Oligotrophic South China Sea. Doctor’s Thesis. China: XiaMen University.
Yang, Q., Liang, Q., Balakrishnan, B., Belobrajdic, D. P., Feng, Q. J., and Zhang, W. (2020). Role of dietary nutrients in the modulation of gut microbiota: a narrative review. Nutrients 12:381.
Yang, Z., Chen, J., Shang, S., Wang, J., Xue, S., Tang, X., et al. (2022). Diversity of epiphytic bacterial communities on male and female Porphyra haitanensis. Ann. Microbiol. 72, 1–8.
Yarte, M. E., Gismondi, M. I., Llorente, B. E., and Larraburu, E. E. (2022). Isolation of endophytic bacteria from the medicinal, forestal and ornamental tree Handroanthus impetiginosus. Environ. Technol. 43, 1129–1139. doi: 10.1080/09593330.2020.1818833
Yuan, J. (2008). Diversity of PAH-Degrading Bacteria in Deep Water of Indian Ocean, Classification and Degradation Pathway Research of Some Novel Bacteria. Doctor’s Thesis. China: Xiamen University.
Zeng, S., Zeng, S., Liang, T., Li, L., Xing, X., Chen, H., et al. (2022). Pollination system and reproductive allocation strategies of dioecious tree Salix dunnii. J. Trop. Subtrop. Bot. 30, 357–366.
Zhang, A., Guo, B., Han, X., and Li, X. (2020). Diversity of endophytic bacteria in seeds of Hippophae rhamnoides subsp. sinensis in two different habitats. Acta Ecol. Sin. 40, 5247–5257.
Zhang, A., Yin, Y., Kong, W., Zhu, X., and Yang, Y. (2021). Diversity of endophytic bacteria in five types of tissues of Hippophae tibetana. Biodiv. Sci. 29, 1236–1244.
Zhang, C., Ma, X., Zhu, R., Liu, Z., Gu, M., Zhang, J., et al. (2020). Analysis of the endophytic bacteria community structure and function of Panax notoginseng based on high-throughput sequencing. Curr. Microbiol. 77, 2745–2750. doi: 10.1007/s00284-020-02068-2
Zhang, K. (2017). Manganese Tolerance and Enhanced Phytoremediation Potential of Endophytic Bacteria in Foxtail algae. Master’s Thesis. China: Guangxi University.
Zhang, Q., Zhang, Q., Huang, X., and Guo, X. (2015). Advance in endophytic bacterial diversity of wetland plants. Wetland Sci. 13, 233–243.
Zhang, Y., Shi, P., and Ma, J. (2013). Exiguobacterium spp. and their applications in environmental remediation. Chin. J. Appl. Environ. Biol. 19, 898–904.
Zhang, Y., Yang, Q., Ling, J., Tang, X., Zhang, W., and Dong, J. (2021). Complete genome sequence of Metabacillus sp. cB07, a bacterium inducing settlement and metamorphosis of coral larvae. Mar. Genomics 60:100877. doi: 10.1016/j.margen.2021.100877
Zhang, Z., Hu, L., Liu, L., and Ji, M. (2020). Identification and antibacterial properties of an antagonistic bacterium Bacillus safensis against walnut fungal disease. J. Henan Agric. Sci. 49, 97–104.
Zhang, Z. Y., Zhou, J., Yuan, L., and Gao, R. C. (2019). Effects of mixed starter cultures and exogenous L-Lys on fermentation quality of fish paste. Schl. Food Biol. Eng. 40, 108–114.
Zheng, H., Dietrich, C., Radek, R., and Brune, A. (2016). Endomicrobium proavitum, the first isolate of Endomicrobia class. nov. (phylum Elusimicrobia)–an ultramicrobacterium with an unusual cell cycle that fixes nitrogen with a Group IV nitrogenase. Environ. Microbiol. 18, 191–204. doi: 10.1111/1462-2920.12960
Zheng, H., Zhang, P., Qin, J., Guo, J., and Deng, J. (2022). High-throughput sequencing-based analysis of the composition and diversity of endophytic bacteria community in tubers of Gastrodia elata f.glauca. Front. Microbiol. 13:1092552. doi: 10.3389/fmicb.2022.1092552
Zheng, Q., Wang, Y., Lu, J., Lin, W., Chen, F., and Jiao, N. (2020). Metagenomic and metaproteomic insights into photoautotrophic and heterotrophic interactions in a Synechococcus culture. mBio 11:e03261-19. doi: 10.1128/mBio.03261-19
Keywords: endophytic bacteria, host sex, macroalgae, Sargassum thunbergii, receptacles
Citation: Zhao Y, Sun T, Li Y, Yang Z, Chen J, Wang J, Yu X, Tang X and Xiao H (2024) The host sex contributes to the endophytic bacterial community in Sargassum thunbergii and their receptacles. Front. Microbiol. 15:1334918. doi: 10.3389/fmicb.2024.1334918
Received: 08 November 2023; Accepted: 14 February 2024;
Published: 15 March 2024.
Edited by:
Decai Jin, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Bao-zhu Fang, Chinese Academy of Sciences (CAS), ChinaCopyright © 2024 Zhao, Sun, Li, Yang, Chen, Wang, Yu, Tang and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuexi Tang, dGFuZ3h4QG91Yy5lZHUuY24=; Hui Xiao, eGlhb2h1aUBvdWMuZWR1LmNu
†These authors have contributed equally to this work and share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.