- 1Department of Outpatient Clinical Research Development, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named After Academician V.I. Kulakov, Ministry of Health of the Russian Federation, Moscow, Russia
- 2Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Pirogov Medical University, Moscow, Russia
Introduction: Human papilloma virus (HPV) is the most common sexually transmitted infection worldwide. Cervicovaginal microbiota plays an important role in HPV infection and is associated with the development of squamous intraepithelial lesions (SIL). The natural history of cervical cancer involves reversible changes in the cervical tissue from a normal state, in which no neoplastic changes are detected in the squamous epithelium, to varying states of cellular abnormalities that ultimately lead to cervical cancer. Low-grade SIL (LSIL), like another cytological category - atypical squamous cells of undetermined significance (ASCUS), may progress to high-grade SIL (HSIL) and invasive cervical cancer or may regress to a normal state.
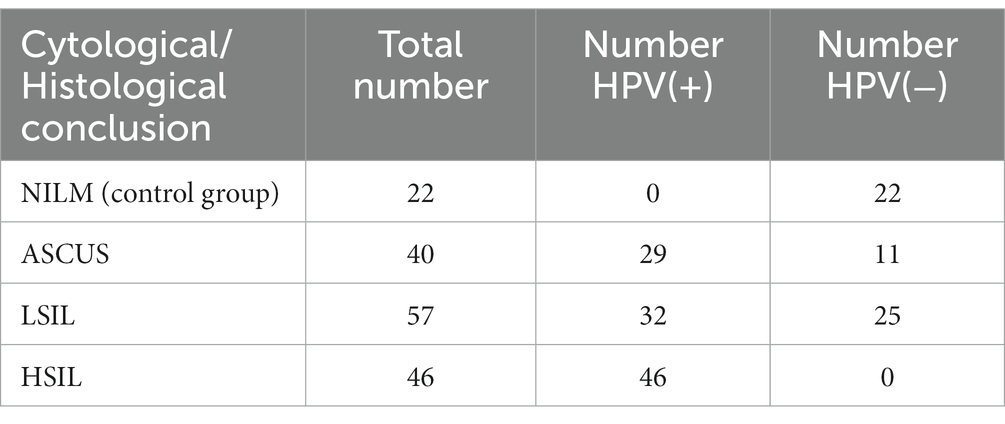
Methods: In this work, we studied cervical canal microbiome in 165 HPV-positive and HPV-negative women of a reproductive age with ASCUS [HPV(+) n = 29; HPV(−) n = 11], LSIL [HPV(+) n = 32; HPV(−) n = 25], HSIL [HPV(+) n = 46], and the control group with negative for intraepithelial lesion malignancy (NILM) [HPV(−) n = 22].
Results and Discussion: HPV16 is the most prevalent HPV type. We have not found any differences between diversity in studied groups, but several genus [like Prevotella (p-value = 0.026), Gardnerella (p-value = 0.003), Fannyhessea (p-value = 0.024)] more often occurred in HSIL group compared by NILM or LSIL regardless of HPV. We have found statistically significant difference in occurrence or proportion of bacterial genus in studied groups. We also identified that increasing of the ratio of Lactobacillus iners or age of patient lead to higher chance to HSIL, while increasing of the ratio of Lactobacillus crispatus lead to higher chance to LSIL. Patients with a moderate dysbiosis equally often had either of three types of vaginal microbial communities (CST, Community State Type) with the prevalence of Lactobacillus crispatus (CST I), Lactobacillus gasseri (CST II), and Lactobacillus iners (CST III); whereas severe dysbiosis is linked with CST IV involving the microorganisms genera associated with bacterial vaginosis and aerobic vaginitis: Gardnerella, Fannyhessea, Dialister, Sneathia, Anaerococcus, Megasphaera, Prevotella, Finegoldia, Peptoniphilus, Porphyromonas, Parvimonas, and Streptococcus.
1 Introduction
Studies on the occurrence and mortality in 36 cancer types conducted in 185 countries show that cervical cancer is the 4th cancer among all cancer types in women (Bray et al., 2018). In developing countries, cervical cancer occurrence and mortality occupies the second place (Qingqing et al., 2021).
The natural history of cervical cancer includes reversible changes in the cervical tissue from a normal state with no neoplastic changes, to varying states of cellular abnormalities in the squamous epithelium that ultimately lead to cervical cancer. The Bethesda “Epithelial cell abnormality: Squamous” category encompasses a spectrum of squamous cell lesions starting from the precancerous lesions of low-grade dysplasia associated with transient human papilloma virus (HPV) infection to higher grade lesions. The two-tiered system of low-grade SIL (LSIL) and high-grade SIL (HSIL) matches the HPV carcinogenic potential. LSIL is now recommended to be used as a diagnostic category to describe HPV transient infection-related changes, while HSIL is used to categorize true precancerous lesion. However, depending on qualitative and quantitative factors, some equivocal morphological features may fall under the category “Atypical Squamous Cells” (ASCs), which are subdivided into two categories; “Atypical Squamous Cells of Undetermined Significance” (ASC-US) or “Atypical Squamous Cells-HSIL cannot be excluded” (ASC-H), based on the suspected underlying lesion LSIL versus HSIL, respectively (Alrajjal et al., 2021).
Development of squamous intraepithelial lesions (SIL) and cervical cancer is mostly caused by the persistence of the human papillomavirus (HPV) (Qingqing et al., 2021; Hu et al., 2022; Zuñiga Martinez et al., 2022). According to the pertinent literature, the group of viruses related to high oncogenic risk includes 13 genotypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66), two of them (16, 18) being associated with 70% of all cases of cervical cancer (Burd, 2003; Cogliano et al., 2005). The World Health Organization (WHO) estimates that HPV infections worldwide are increasing by 9 to 13% each year (Santella et al., 2022).
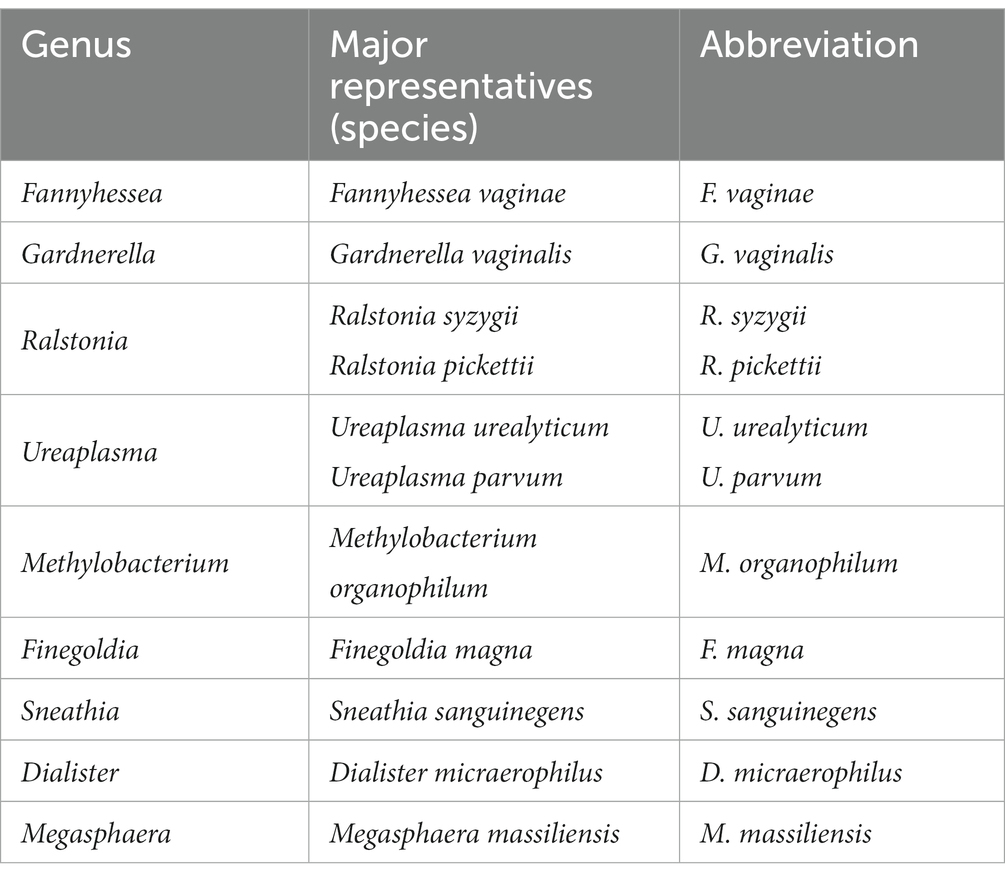
The microbial communities of the female reproductive system have been extensively studied over the past decade (Al-Nasiry et al., 2020; Andralojc et al., 2021). According to the published data, one of the risk factors promoting persistent HPV infection is the cervicovaginal microbiota disruption (Ritu et al., 2019; Nieves-Ramírez et al., 2021). Studying the cervicovaginal microbiota in the ‘Human Microbiome’ project revealed that the bacteria colonizing the female genital tract make up 9% of the entire human microbiome (Proctor et al., 2019). In one work, five types of vaginal microbial communities have been outlined (CST – Community State Type) (McClymont et al., 2022; Molina et al., 2022). Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus iners, and Lactobacillus jensenii prevail in four CST (I, II, III, and V), respectively, while no Lactobacillus species is predominant in CST IV (‘diverse’ group) which is further divided into the following subtypes: CST IV-A comprising the Anaerococus, Peptoniphilus, Corynebacterium, Prevotella, Finegoldia, and Streptococcus genera; CST IV-В including the Atopobium, Fannyhessea, Gardnerella, Sneathia, Mobiluncus, Megaspheragenera.
Some authors consider microbial diversity as well as changes in the qualitative and quantitative composition of the cervicovaginal microbiota to be crucial for triggering the proliferation of the stratified squamous epithelial cells and to accompany HPV persistence. Conversely, the viral infection can alter bacterial composition irrespective of the SIL severity looping a ‘vicious circle’ in the development and progression of the disease (Kang et al., 2021; Naidoo et al., 2022; Ntuli et al., 2022). Recently, special attention has been paid to the influence of individual microorganisms on SIL development (Norenhag et al., 2020; Gardella et al., 2022; Lin et al., 2022).
In our study, we aimed to estimate the characteristics of the microbiome by NGS sequencing of 16S ribosomal genes in the HPV-positive [HPV(+)] and HPV-negative [HPV(−)] women of a reproductive age with cervical pathology.
2 Methods
2.1 Clinical material
From January 2022 to May 2022, 165 women of reproductive age who sought consultation at the Department of Outpatient Clinical Research Development of Kulakov National Medical Research Center for Obstetrics, Gynecology, and Perinatology for diagnosis and treatment of cervical pathology. The patients visited the department because of a wide list of reasons, for example, pregnancy management, absence of menstruation, examination and treatment of female infertility and etc. The primary diagnosis was established based on the results of liquid-based cytology. The cytological material was obtained in accordance with the laws and regulations of the Russian Federation. Air-dried cervical epithelial swabs were prepared by routine Papanicolaou staining. The specimens were collected from patients undergoing cytological and histological examination Departments of Kulakov National Medical Research Center for Obstetrics, Gynecology, and Perinatology (Moscow, Russia). The samples were classified according to the Bethesda system: normal cytology (negative for intraepithelial lesion or malignancy; NILM), low- and high-grade squamous intraepithelial lesions (LSILs and HSILs), or atypical squamous cells of undetermined significance (ASCUS). The final diagnosis was confirmed by histological examination carried out for 76 (46%) women. If the diagnosis had not been verified by histology, patients were placed under dynamic monitoring for cervix condition due to the lack of indications for treatment, and their diagnosis was interpreted by cytological methods. The criteria for inclusion in the study were as follows:
1. The age of women ranging from 19 to 45 years old;
2. The cytological conclusion: ASCUS, low-grade squamous intraepithelial lesion (LSIL), high-grade squamous intraepithelial lesion (HSIL), and negative for intraepithelial lesion or malignancy (NILM);
3. The provided written informed consent for enrollment in the study.
The criteria for exclusion were as follows:
1. Pregnancy;
2. Lactation;
3. Treatment with antibiotics in the previous 14 days;
4. Neuropsychiatric diseases;
5. Acute inflammatory diseases;
6. Renal, liver, or lung dysfunction at the decompensation stage.
This study conformed to the principles of the Declaration of Helsinki. The involved human participants were reviewed and approved by the Local Ethics Committee at the FSBI “National Medical Research Center For Obstetrics, Gynecology And Perinatology Named After Academician V.I.Kulakov” Ministry of Health of the Russian Federation (protocol no. 3 of 26 March 2020).
2.2 HPV testing
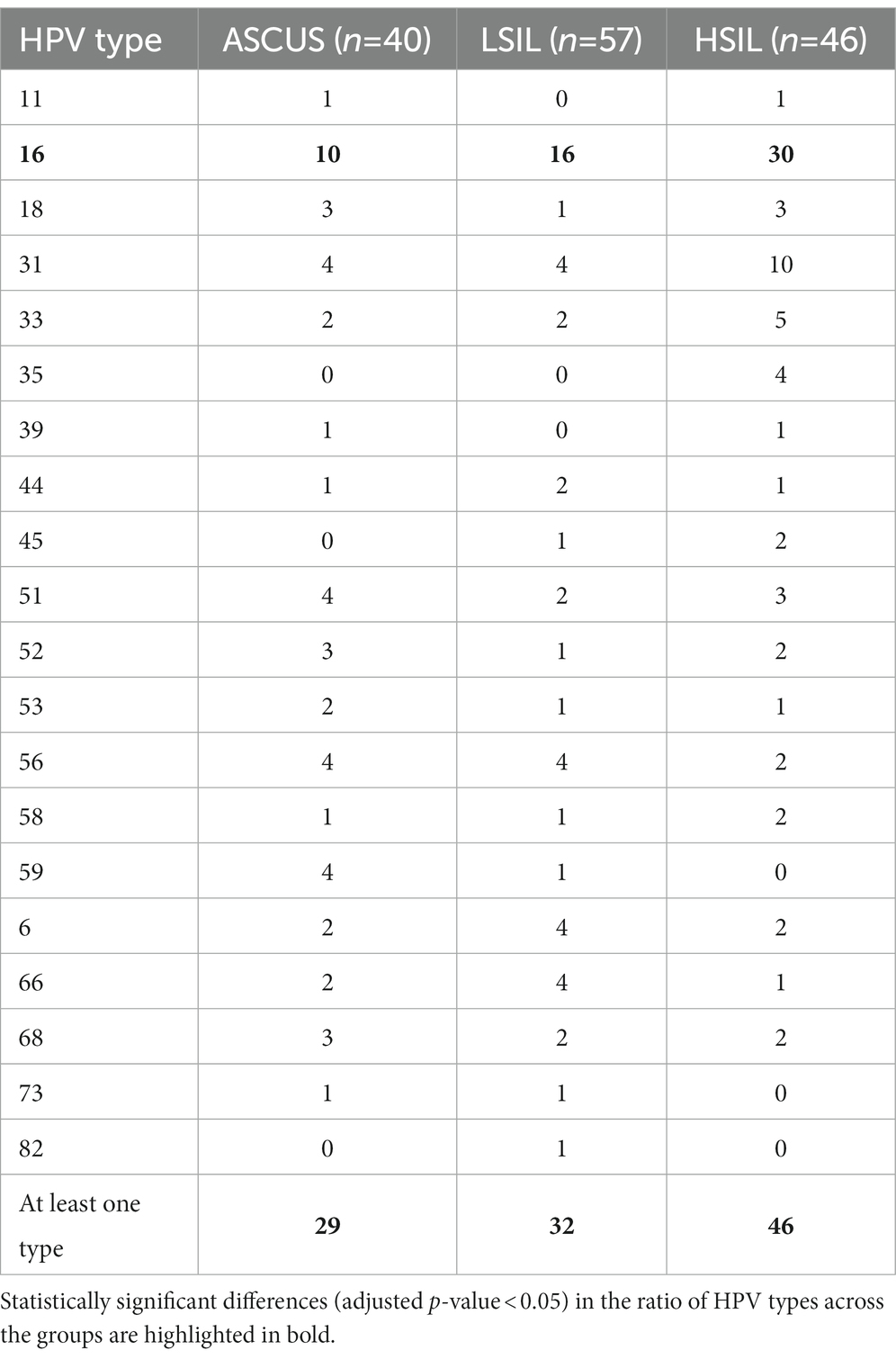
HPV testing was performed by qPCR using the reagent kit for detecting, genotyping, and identifying of the HPV viral load by PCR “HPVquant-21” (DNA technology, Russia) and involved 21 HPV types (6, 11, 16, 18, 26, 31, 33, 35, 39, 44(55), 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, 82).
2.3 Collecting the biomaterial for metagenomic study
The swab was collected into the 1.5 tubes containing 300 microliters of 20 mM Tris–HCl buffer (pH = 7.2).
2.4 DNA isolation and library preparation for sequencing
DNA was isolated from the biomaterial using the DNeasy Blood and Tissue kit (Qiagen, United States) following the manufacturer’s instructions. Quality control of the isolated prokaryotic DNA was performed by qPCR using the primers recognizing the V4 DNA region encoding 16S rRNA (515F and 806R) (Parada et al., 2016).
The libraries were prepared for sequencing in two steps. At the first step, the V4 region of 16S rRNA was amplified using the primers to the prokaryotic V4 DNA region encoding 16S rRNA (515F and 806R) containing technical sequences for the MGI adapters. At the second step, amplification was performed using primers containing unique barcodes and primer technical sequences. The concentrations of prepared libraries were measured by Qubit Flex (Life Techonologies, United States) using dsDNA HS Assay Kit (Life Technologies, United States) following the manufacturer’s protocol. The quality of the prepared libraries was assessed using Bioanalyzer 2,100 with the High Sensitivity DNA kit (Agilent Technologies) according to the manufacturer’s instructions. Next, the libraries were circularized and sequenced in the paired-end mode using the DNBSEQG-400 platform with the DNBSEQ-G400RS High-throughput Sequencing Set PE150 kit according to manufacturer’s protocol (MGI Tech). FastQ files were generated using the zebracallV2 software by the manufacturer (MGI Tech).
2.5 Sequencing data processing
The obtained overlapping paired reads were merged into unified nucleotide sequences and grouped based on the sequence identity and possible polymerase errors using Qiime2 v.2022.8. Each sequence group was assigned with the taxonomic class (family and genus) using the RDP classificator. Furthermore, each sequence group was aligned using blast v2.13.0 with default settings against the 16S rRNA database followed by determining the species in a read group. Sequence groups with the content of <0.01% in the sample were excluded from the analysis.
2.6 Statistical analysis
For statistical analysis, we used compositional data without any transformation to avoid complexity with clinical interpretation. The differences in age between the groups were assessed using One-way ANOVA, followed by multiple pairwise comparisons using the t-test with Bonferroni correction. The statistical significance of differences in the ratio of categorical variables (including the qualitative assessment of microorganisms) between the groups was assessed using the Chi-square test, provided that there were no cells in the contingency table with the values below 5. The exact Fisher’s test was used if at least one cell in the contingency table contained a value of <5. Only those microorganisms with the content of >1% in at least two patients were subjected to further qualitative estimation (the frequency of occurrence of a microorganism in the sample). The microorganisms with the content >5% for at least two patients were subjected to further quantitative estimation. The statistical significance of differences upon quantitative assessment between groups was evaluated using the Kruskal–Wallis test. Alpha diversity was assessed using the Shannon and the Simpson index with using amount of reads per taxon. Beta diversity between all groups was assessed by adonis algorithm (Bray-Curtis distance) from “vegan” package (R language). Statistical significance was assessed using the Kruskal–Wallis test for all groups, followed by pairwise comparisons using the Mann–Whitney test with Bonferroni correction. The differences were considered statistically significant at p < 0.05. The charts were created using the R programming language (ver 4.2.0). Logistic regression was created using the ‘glm’ and ‘step’ functions from the ‘stats’ package in the R programming language (ver 4.2.0).
3 Results
3.1 Demographic and clinical indicators did not revealed any significant differences
The study included 165 female participants with the age ranging from 19 to 45 years (mean age: 31 ± 6.3 years). The patients were stratified into 4 groups (Table 1).
The mean ages of the patients from the ASCUS, HSIL, LSIL, and NILM groups were 30.7 ± 6.2, 33 ± 5.8, 30 ± 6.3, and 30.4 ± 5.8 years (Figure 1). There were no statistically significant differences between the groups (p = 0.093).

Figure 1. The age distribution in the studied groups. Boxplot shows standard sample metrics (median, Q25–Q75 and Q25 − 1.5 * IQR; Q75 + 1.5 * IQR by whiskers). No statistically significant difference were found between groups.
The median ages of sexual initiation in the ASCUS, LSIL, HSIL, and NILM groups were 18 years (IQR (Q25–Q75) is 16–18 years), 18 years (16–18 years), 17 years (16–18 years), and 18 years (18–19 years), respectively (Figure 2). We detected statistically significant differences in the age of sexual initiation between the LSIL and NILM groups (adjusted p-value = 0.018) and between the HSIL and NILM groups (adjusted p-value = 0.03). On average, patients from the HSIL and LSIL groups have a lower age of sexual initiation (1 year less) compared to the control group.
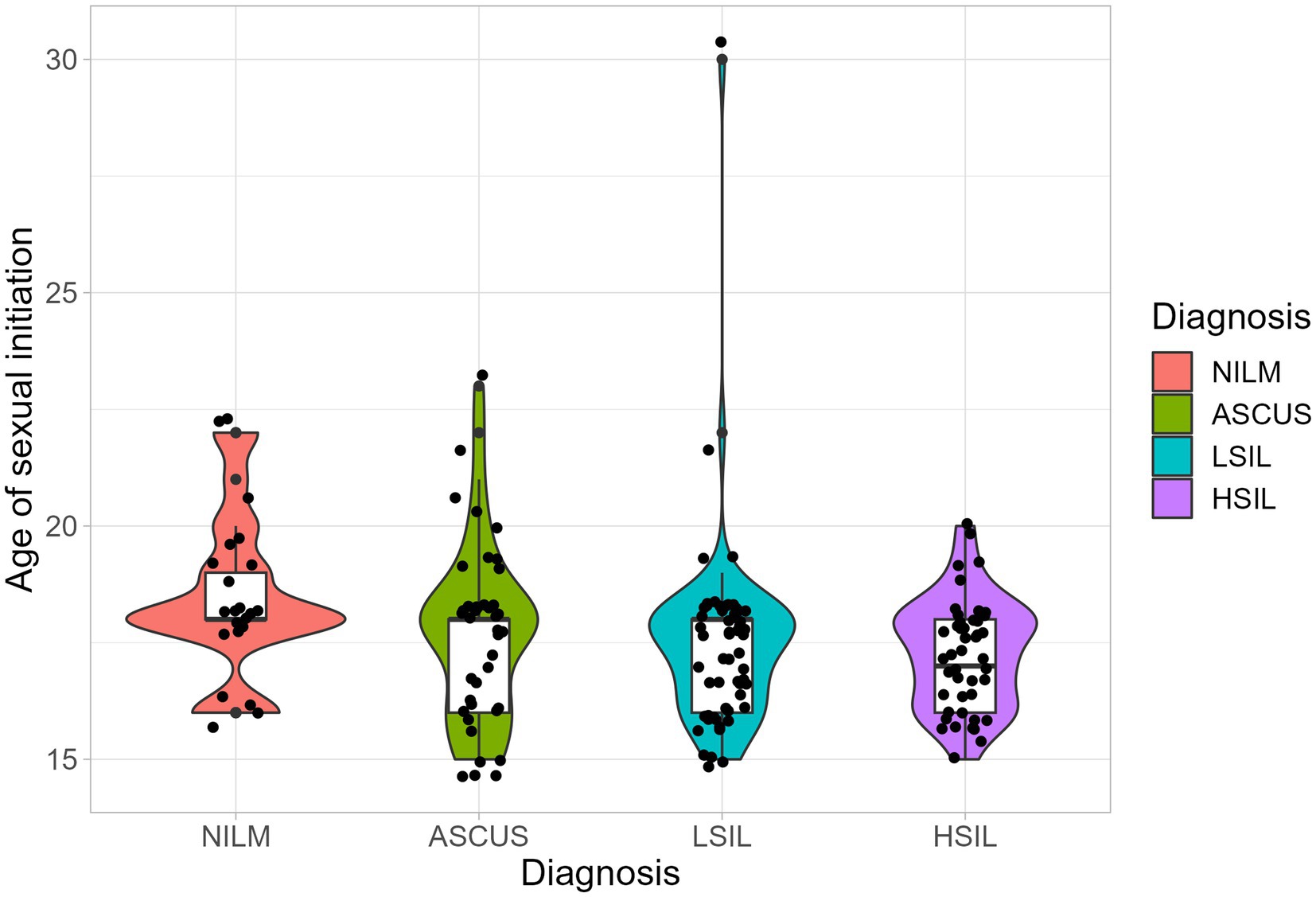
Figure 2. The distribution of the age of sexual initiation. Boxplot shows standard sample metrics (median, Q25−Q75 and Q25 − 1.5 * IQR; Q75 + 1.5 * IQR by whiskers). No statistically significant difference were found between groups.
Statistical analysis of the patients based on anamnestic and clinical parameters (somatic morbidity, pregnancies, childbirths, contraception methods, surgical interventions, gynecological diseases, and past infectious diseases) did not reveal any significant differences between the groups (the data are shown in Supplementary Table S1).
3.2 HPV16 type and overall amount of infected patient was higher in HSIL group
The number of patients with at least one HPV type was higher in the HSIL group (100%) than in the LSIL (72.5%) and ASCUS (56.1%) groups (adjusted p-value = 0.0086). The most common HPV type in the LSIL, HSIL, NILM was HPV16. After applying the correction for multiple comparisons, only HPV16 exhibited statistically significant differences (adjusted p-value = 0.013) (Tables 2, 3; Figure 3). The Spearman’s correlation coefficient between the age and the Lactobacillus representation across all studied groups was 0.095 being not statistically significant (p-value = 0.22), similarly to the correlation coefficient between the age and HPV carriage (rho = −0.059, p-value = 0.46). Estimating the correlation between the Lactobacillus representation among the studied groups did not provide any statistically significant results (Table 3). The groups under scrutiny showed the reverse correlation between the age and Lactobacillus representation in the sample. In particular, the correlation coefficient raised from -0.28 in the HSIL group to 0.38 in the control NILM group (Figure 4, Table 3). Estimating the linear correlation between the HPV carriage and age in the HSIL and NILM groups was impossible since they comprised only one HPV carriage group. The ASCUS group showed the negative statistically significant correlation (p-value = 0.047) between the age and HPV carriage (Table 3).

Table 3. The Spearman’s correlation coefficient and statistical significance between the age and Lactobacillus representation.
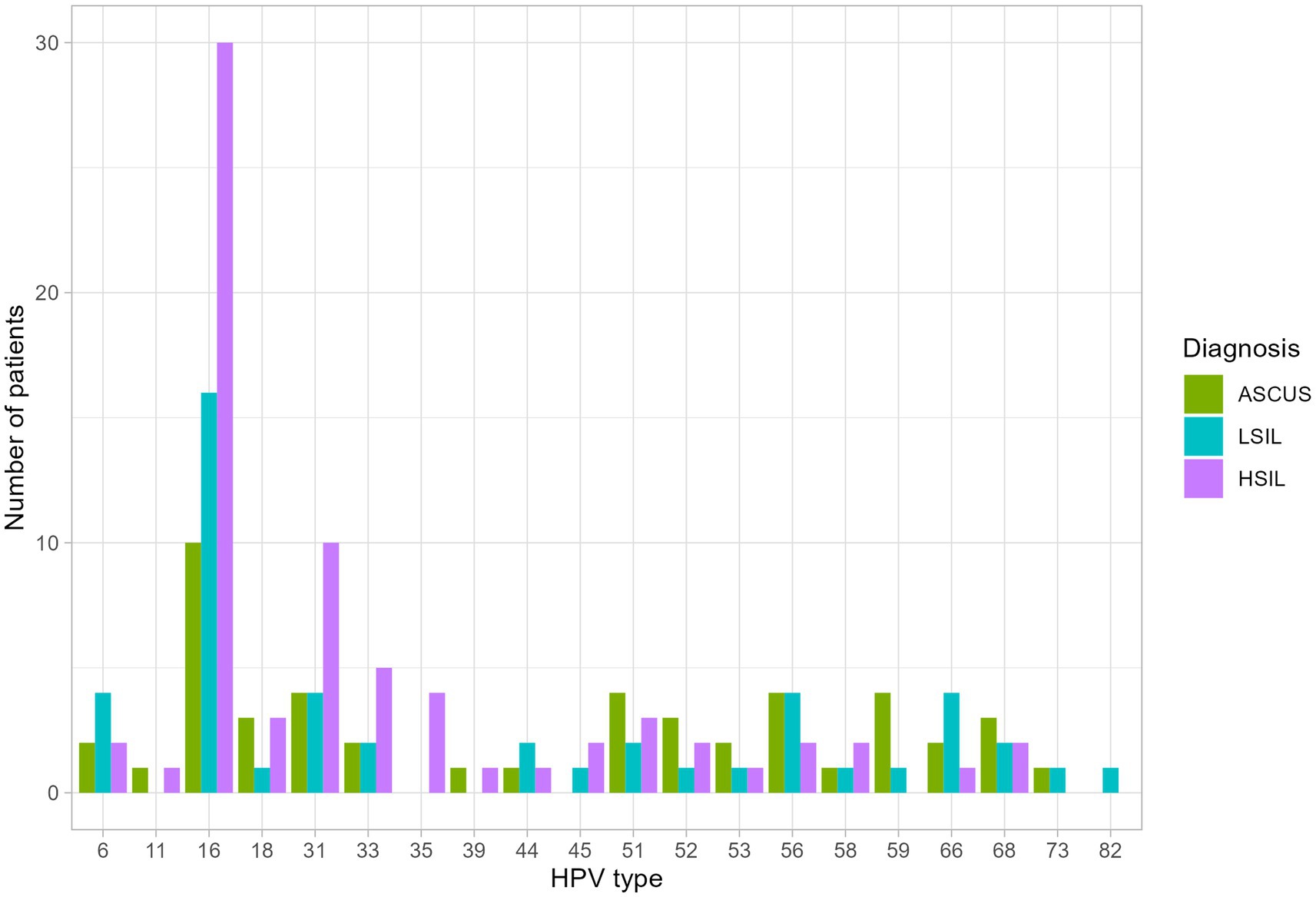
Figure 3. The distribution of HPV types among the studied groups. Statistically significant difference were identified for HPV 16 type in HSIL group and at least one HPV type was higher in the HSIL group.
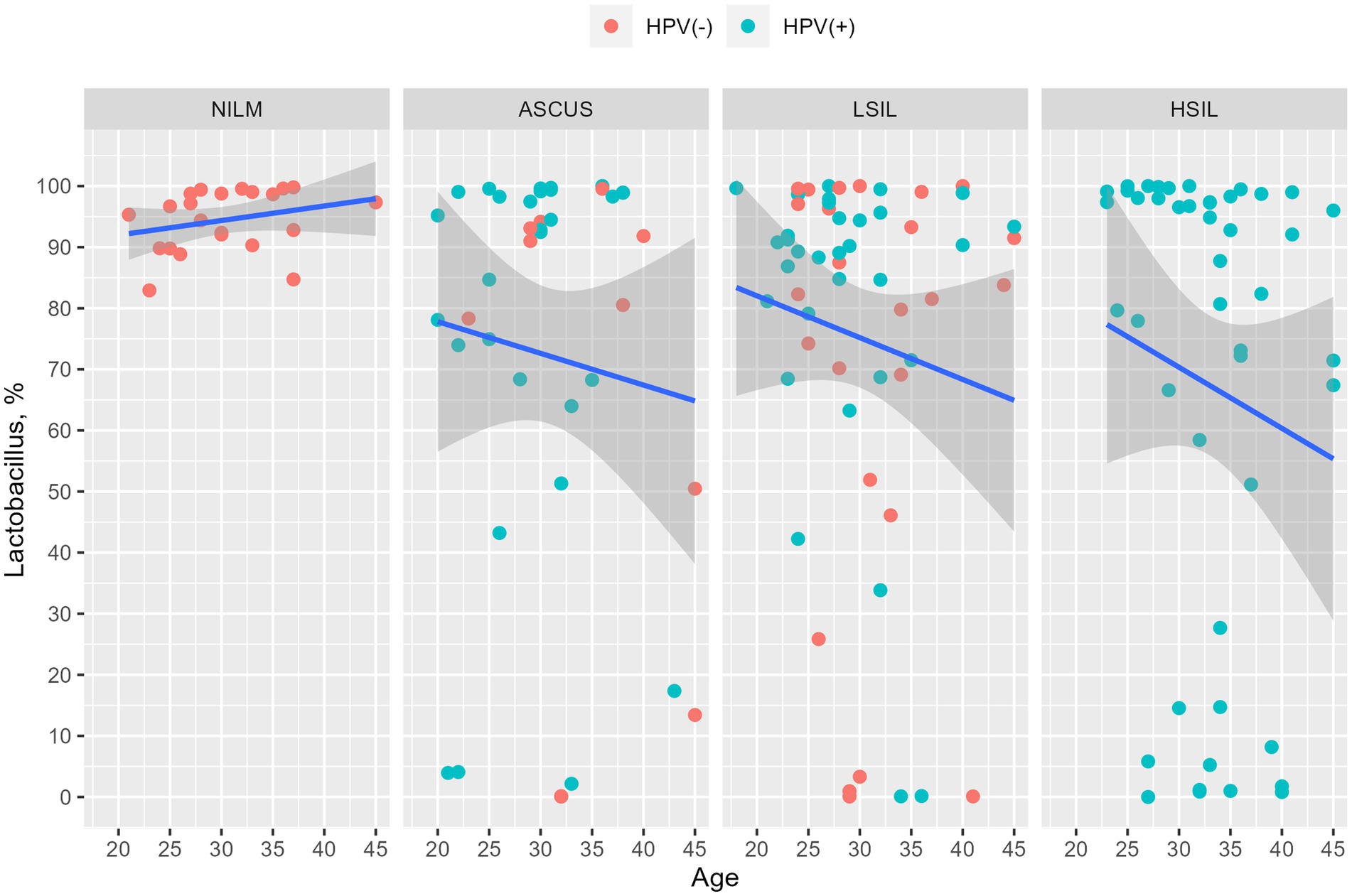
Figure 4. Dependency of the Lactobacillus genus representation from age. The HPV-negative and HPV-positive patients are indicated in red and blue, respectively. It was observed that the percentage of Lactobacillus is decreased in all groups except NILM. The changes were not statistically significant.
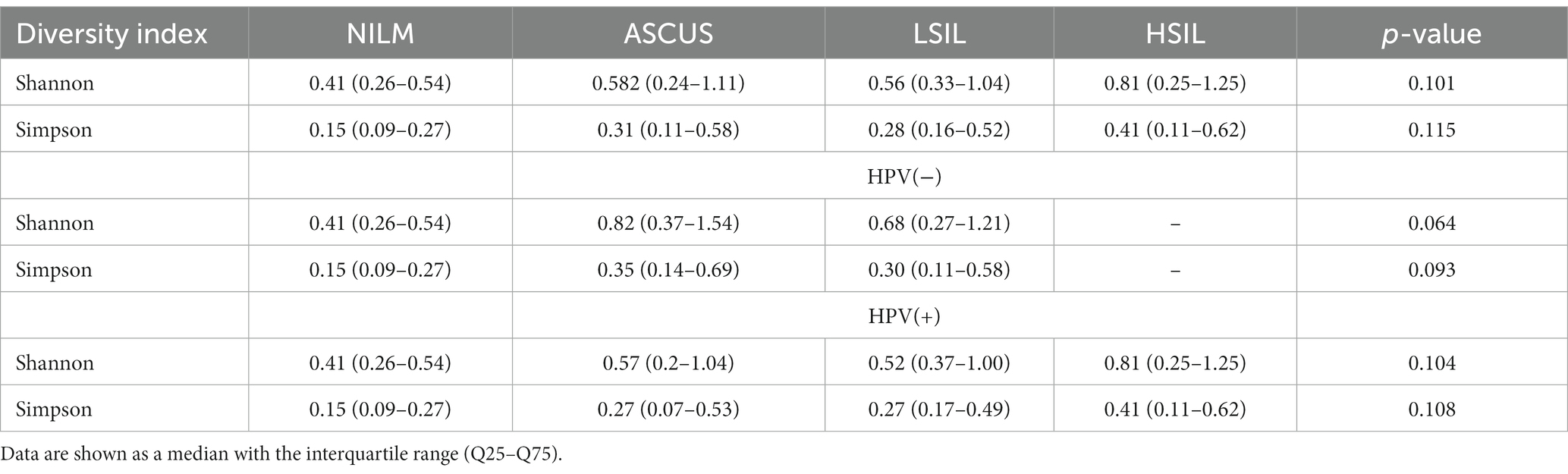
3.3 Evaluation of the alpha and beta microorganism diversity
Statistically significant differences in the alpha diversity between the groups were not detected (Table 4). The control group (NILM) displayed the lowest diversity level. Analyzing all patients as well as only HPV(+) patients revealed the greatest level of diversity in the HSIL group. In the HPV(−) patients, the highest diversity level was observed for the ASCUS group.
The Beta diversity analysis did not revealed any statistically significant difference between groups (p-value = 0.15). Pairwise comparison taking into account HPV status also did not show difference (Supplementary Table S8). Pairwise comparison without HPV revealed only one statistically significant difference (p-value = 0.036) between HSIL and LSIL group, but after applying multiple comparisons correction it was gone (p-value = 0.22).
3.4 The qualitative and quantitative estimations of the Lactobacillus species did not reveal any significant difference
The qualitative and quantitative estimations of the Lactobacillus content in the HPV(+) or HPV(−) patients, and the control group are shown in Tables 5, 6. There was no statistically significant difference in the Lactobacillus content between the groups (p > 0.05). Below is the list of the species sorted based on their content: Lactobacillus crispatus, L. iners, L. jensenii, L. gasseri, L. delbrueckii. It is worth noting that among the Lactobacillaceae family, we also detected the species of Limosilactobacillus genus (including L. antri, L. coleohominis, L. reuteri, L. mucosae) in the HPV(+) patients with ASCUS (2 cases, 5%), LSIL (4 cases, 12.5%), HSIL (3 cases, 6.52%), and in the control group (2 cases, 9.09%). No statistically significant difference was observed between studied groups by genus.
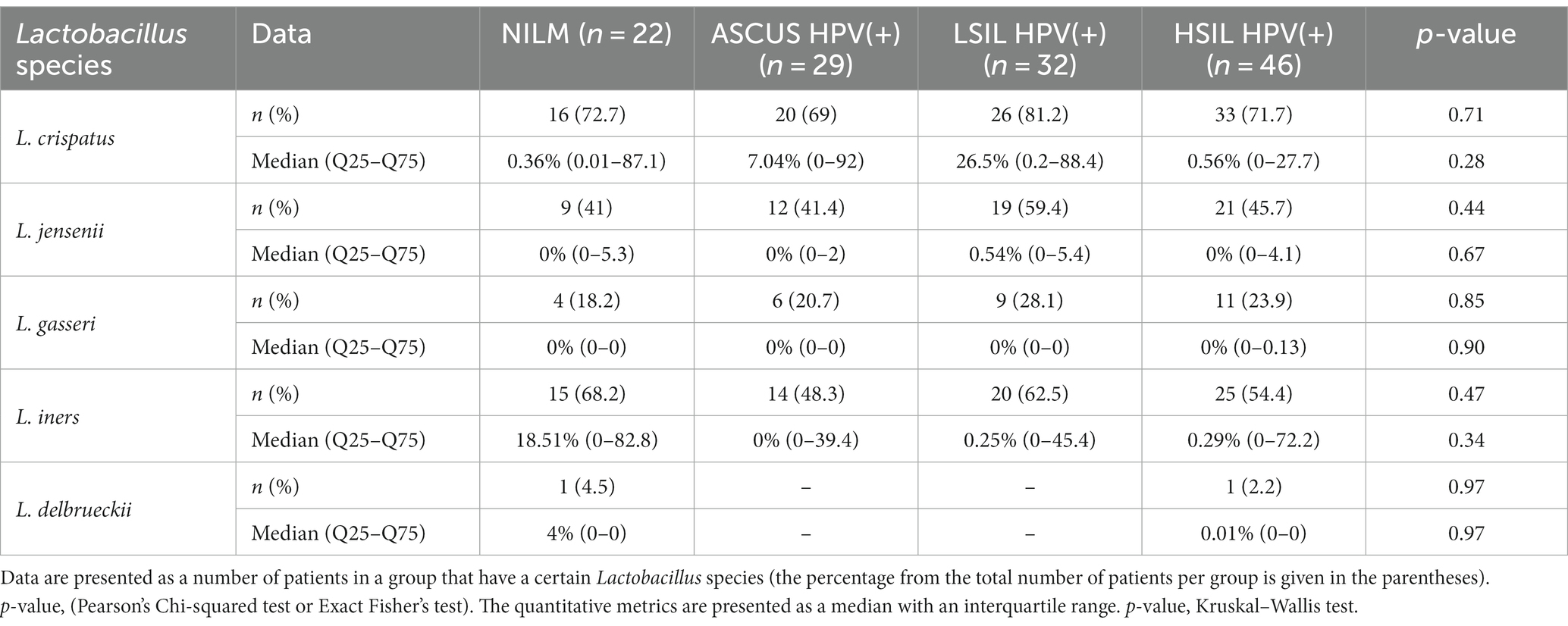
Table 5. The qualitative and quantitative characteristics of the Lactobacillus species content in the HPV(+) patients and control group.
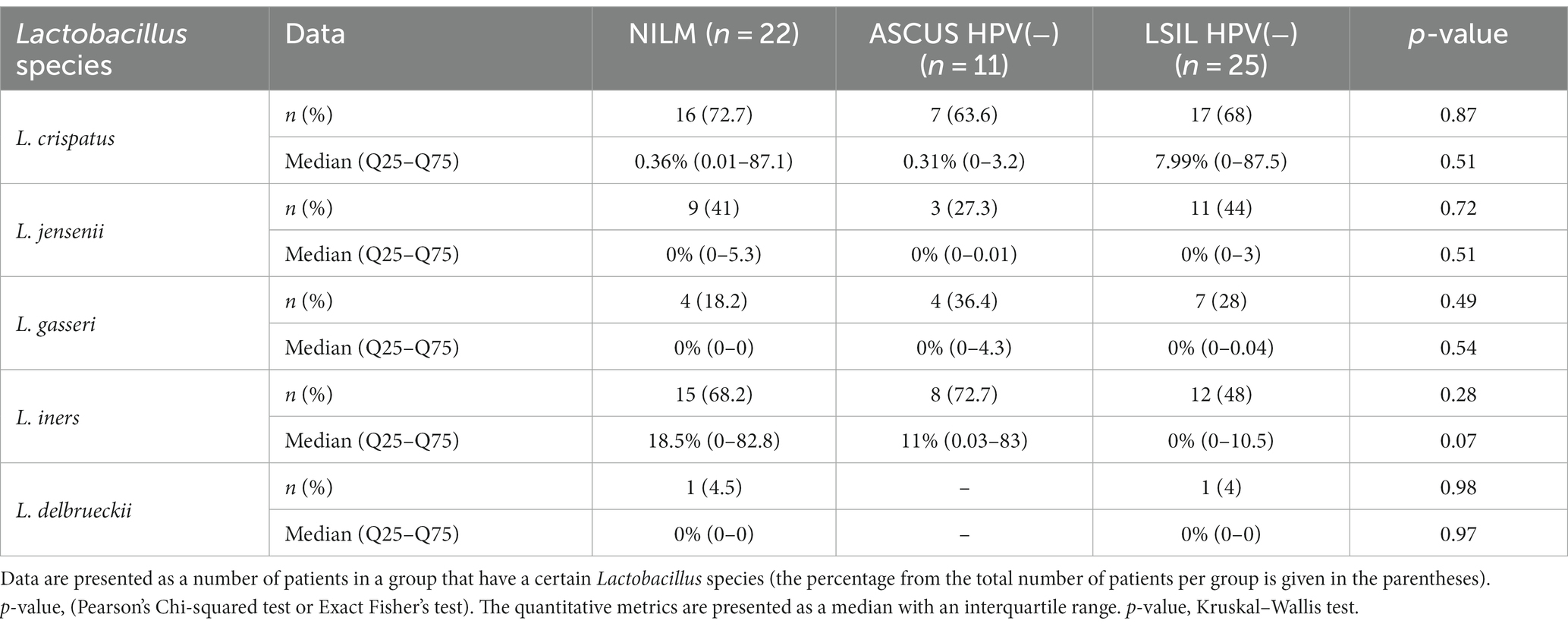
Table 6. The qualitative and quantitative characteristics of the content of various Lactobacillus species in the HPV(−) patients and the control group.
3.5 The qualitative and quantitative estimations of the content of microorganisms (except Lactobacillus species) in the HPV(+) and HPV(−) patients
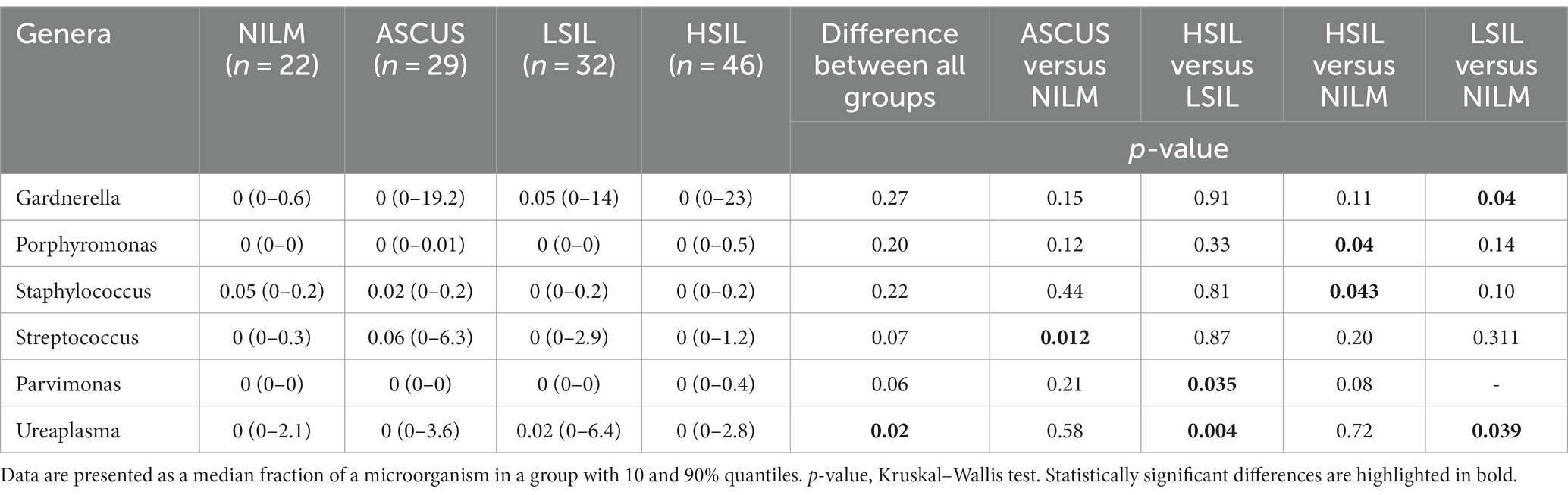
In this work, the cervical canal of the women of reproductive age comprised more than 100 various bacterial genera, 44 being the most common (not <1% of reads in two or more samples). Estimating the frequencies of detecting taxons across all the groups studied revealed statistically significant differences in the occurrence frequencies of Fannyhessea and Gardnerella among the ASCUS, HSIL, LSIL, and NILM groups (Table 7). Pairwise group comparisons showed the differences in the occurrence frequencies between the control group and the groups under scrutiny (ASCUS, HSIL, LSIL). For the Prevotella genus, significant differences were found only between the HSIL and NILM groups. In all detected statistically significant differences, these microorganism genera were more common in the groups under investigation (ASCUS, HSIL, LSIL) compared to the NILM group. Other comparisons did not provide any statistically significant results (Supplementary Table S2).
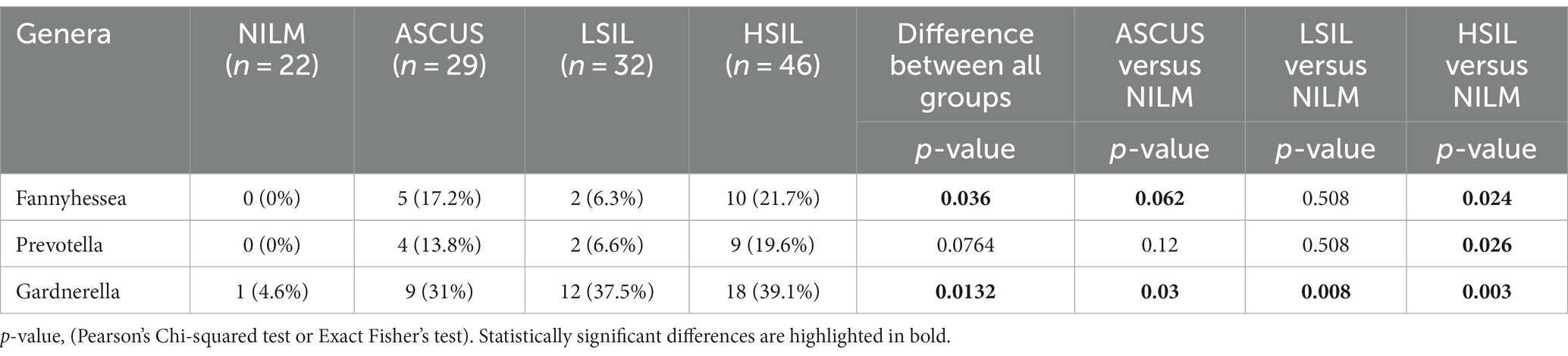
Table 7. The qualitative estimation of the microorganism occurrence frequencies in the HPV(+) patients.
The qualitative estimation of all groups of HPV(−) patients revealed statistically significant differences for 4 microorganism genera: Prevotella, Gardnerella, Porphyromonas, Dialister (Table 8). Pairwise comparisons demonstrated the significant differences only between the ASUS and control groups for all 4 genera. They occurred significantly more frequently in the ASCUS group compared to the control group. Other comparisons did not provide any statistically significant results (Supplementary Table S3).
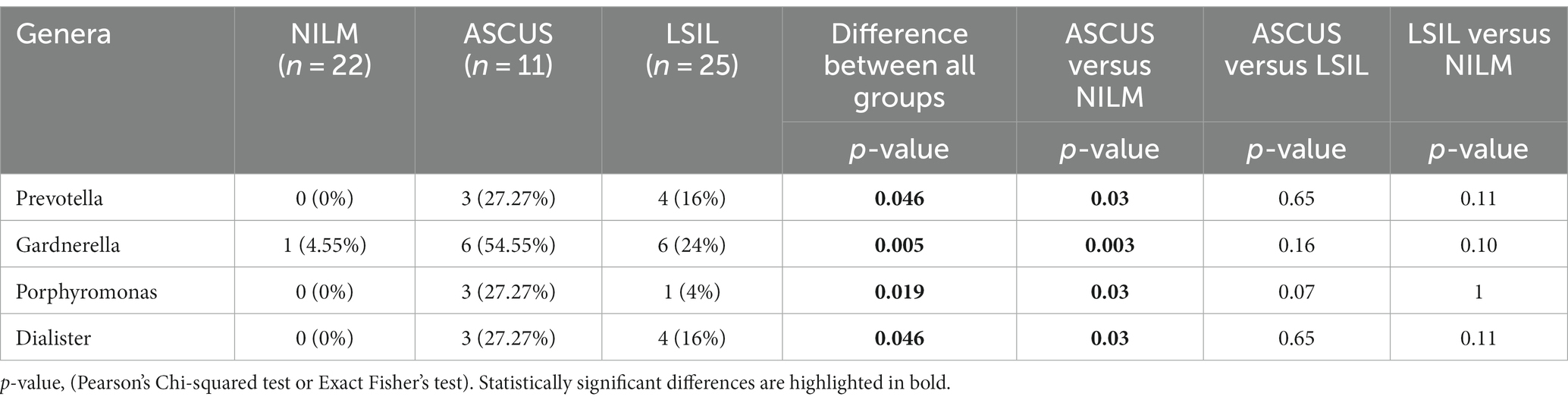
Table 8. The qualitative estimation of the microorganism occurrence frequencies in the HPV(−) patients.
The quantitative estimation of the microorganism content across all studied groups revealed statistically significant differences in case of the Ureaplasma genus (Table 9). On average, Ureaplasma is more common in the LSIL group than in the other groups. Pairwise comparisons confirmed the statistically significant differences of Ureaplasma occurrence between the HSIL and LSIL as well as LSIL and NILM groups. Furthermore, the Parvimonas species was shown to occur more frequently in the HSIL group compared to the LSIL group. Significant differences were observed also for the Parvimonas, Streptococcus, Staphylococcus, Porphyromonas, Gardnerella genera. On average, the representation percentages of Streptococcus, Staphylococcus, Porphyromonas, Gardnerella genera were higher in the ASCUS, HSIL, and LSIL groups compared to the NILM group. Other comparisons did not provide any statistically significant results (Supplementary Table S4).
The quantitative estimation of the microorganism representation among all studied groups in HPV(−) patients revealed statistically significant differences in the mean fraction for 4 microorganism genera Gardnerella, Porphyromonas, Dialister, Campylobacter (Table 10). Pairwise comparisons showed most differences in the mean microorganism fractions between the ASCUS and NILM groups. On average, the fractions of six microorganism genera (Gardnerella, Porphyromonas, Dialister, Campylobacter, Prevotella, Fusobacterium) and two family (Ruminococcaceae, Porphyromonadaceae) were higher fraction in the ASCUS group than in the NILM group. Other comparisons did not provide any statistically significant results (Supplementary Table S5).
The microorganisms detected in the ASCUS HPV(+) group (along with the microorganisms listed in Table 11) included F. vaginae (with median of 0 and 10% quantile – 90% quantile of 0–8.3) as well as small fractions (90% quantile not exceeding 3%) of Prevotella spp., M. organophilum, Anaerococcus spp., F. magna, Peptoniphilus spp., R. syzygii, and R. pickettii.
In the LSIL HPV(+), U. urealyticum and U. parvum occurred more frequently and had a higher mean content compared to the other HPV(+) groups. The microorganisms detected in the LSIL group (apart from the microorganisms listed in Table 9): R. syzygii and R. pickettii (0%, 0–3.5) as well as small fractions (90% quantile not exceeding 3%) of F. magna, M. organophilum, Sphingomonas spp., Acinetobacter spp., Streptococcus spp., R. syzygii, and R. pickettii.
In the HSIL HPV(+) groups, the fractions of F. vaginae, G. vaginalis, Prevotella spp. were 0% (0–15.52), 0% (0–23), 0% (0–10.3), respectively. They had the highest occurrence frequencies (Table 7) among all HPV(+) groups. The microbiome of the cervical canal in the HSIL HPV(+) group comprised the following microorganisms (along with the microorganisms listed in Table 9): Prevotella spp. (0%, 0–10.3) and S. sanguinegens (0%, 0–6.35) as well as small fractions (90% quantile not exceeding 3%) of Streptococcus spp., Anaerococcus spp., F. magna, D. micraerophilus, Parvimonas spp., Peptoniphilus spp., R. syzygii, R. pickettii, U. urealyticum, U. parvum, Gemella spp., M. massiliensis, and Sphingomonas spp.
Along with the microorganisms listed in Table 10, the microorganisms detected in the LSIL HPV(−) group included Bifidobacterium (0, 0–7.79); U. urealyticum and U. parvum (0, 0–6.26). They also comprised small fractions (90% quantile not exceeding 3%) of Sphingomonas spp., Peptoniphilus spp., F. magna.
Comparing the quantitative contents of anaerobic microorganisms (Anaerococcus spp. - (2.3%, 0–0.1), D. micraerophilus (1.8%, 0–1.5), S. sanguinegens (6.6%, 0–0.03), G. vaginalis (8.3%, 0.2–8.7), Prevotella (5.6%, 0–8.7), Porphyromonas (1.6%, 0–1.3)) showed that their average percentage was the highest in the ASCUS HPV(−) group compared to the other groups (Table 11).
3.6 Influence of age and proportion of Lactobacillus iners and Lactobacillus crispatus on SIL status
The logistic regression based on the age and age of sexual initiation as interacting independent variables and on the SIL status (HSIL/LSIL) as a dependent variable revealed no significant difference for either feature. The HPV status was not included in the model since the HSIL group comprised no HPV(−) patients. The stepwise selection of the best model based on the AIC parameter revealed the statistically significant relationship between the age and an elevated probability of being sorted into the HSIL group rather than LSIL (Table 12; age only model).
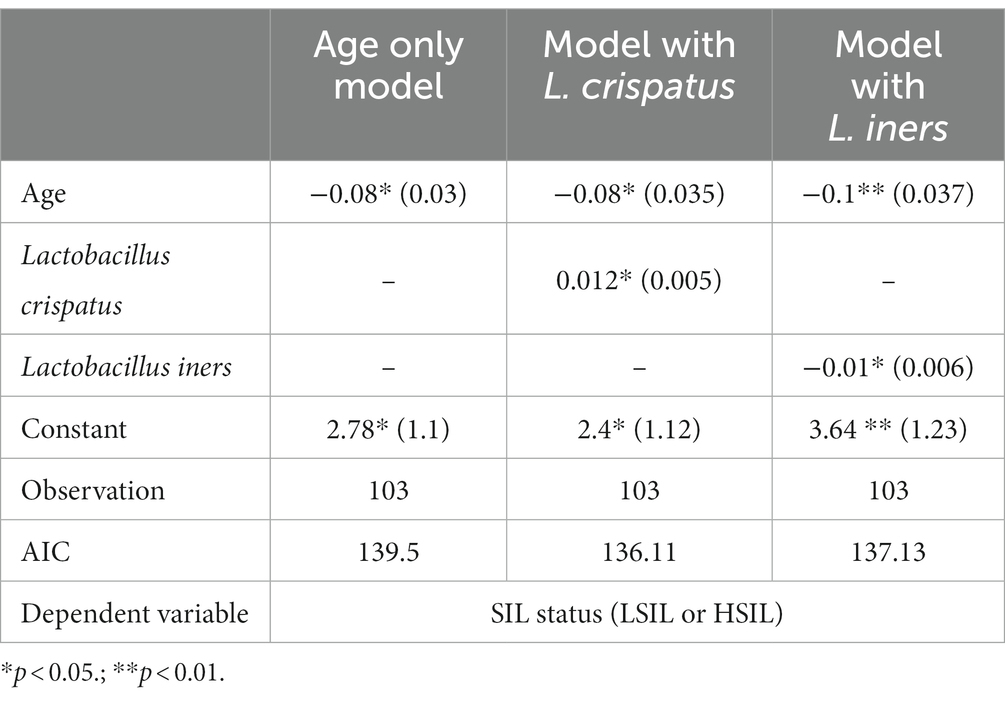
Table 12. The coefficients (standard error) of the logistic regression models including different type of predictors.
Adding the data on L. jensenii, L. gasseri as well as the most represented dominants from CST IV such as Prevotella, Streptococcus, Gardnerella, Sneathia, Fannyhessea did not exert significant effects on the coefficients for the other features included in the model. The statistical significance remained unaltered, while the absolute value changed only within 2 decimal places. The coefficient for the feature indicating the representation of the taxon based on an absolute value did not exceed 0.05 and was not statistically significant.
Creating the model that includes the values of representation of L. crispatus, L. iners did not affect the features previously included in the model as well (Table 12, model with L. crispatus and L. iners, respectively). The coefficients for these features were statistically significant (p-value < 0.05) yet opposite in sign.
In particular, the positive coefficient for L. crispatus shows that an elevated percentage leads to an increase of the probability of detecting LSIL rather than HSIL. At the same time, elevated content of L. iners results in a higher probability of detecting HSIL rather than LSIL given the equal age of a patient.
3.7 The predominant Lactobacillus species patients with normocenosis, moderate and severe dysbiosis (anaerobic, aerobic, and mixed)
In this study, we detected all previously isolated CST I-V including IV-A and IV-B (Figure 5). However, in two patients (one was from the NILM group, the other one - from the LSIL group), L. delbrueckii prevailed with the contents of 90 and 74%, respectively. CST was termed СST Ldelb in these patients.
Among all groups under investigation, the predominant microorganisms were CST I and II, with L. crispatus and L. iners prevailing in the cervical canal. The highest fraction of the CST IV group which correlates with pathologic microflora state was observed in the HSIL group (Figure 5).
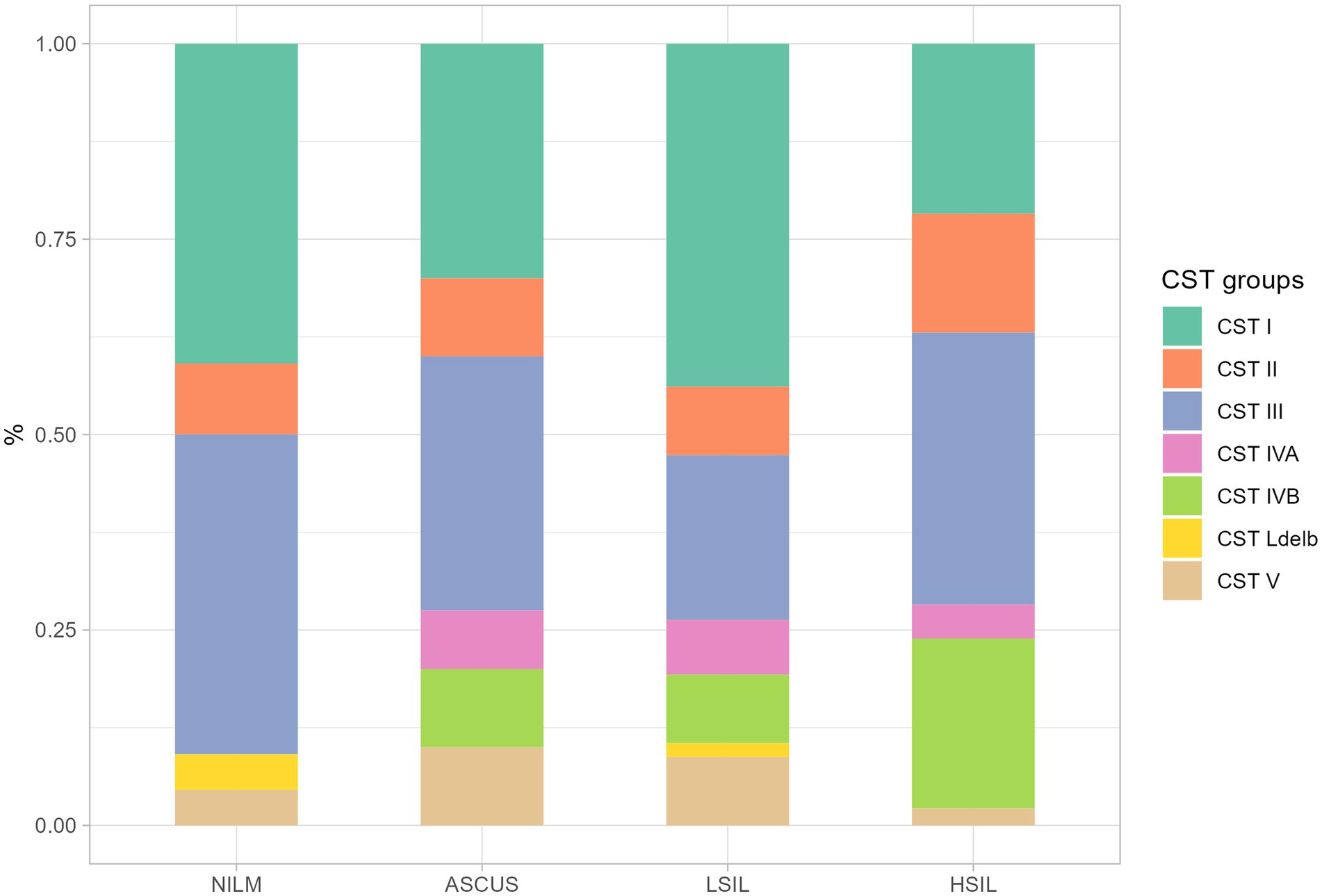
Figure 5. The ratio of CST fraction in the groups under scrutiny. The plot shows the proportion of samples from different. All CST groups was determined by predominant Lactobacillus (CST I – CST III, CST V, CST Ldelb) or other non-Lactobacillus (CST IVA, CST IVB) genus.
In the HSIL group, the most severe dysbiosis was found in 24% of patients being the most common dysbiosis among all studied groups. In the patients with the established normocenosis, L. delbrueckii was absent in all the groups (ASCUS, LSIL, HSIL).
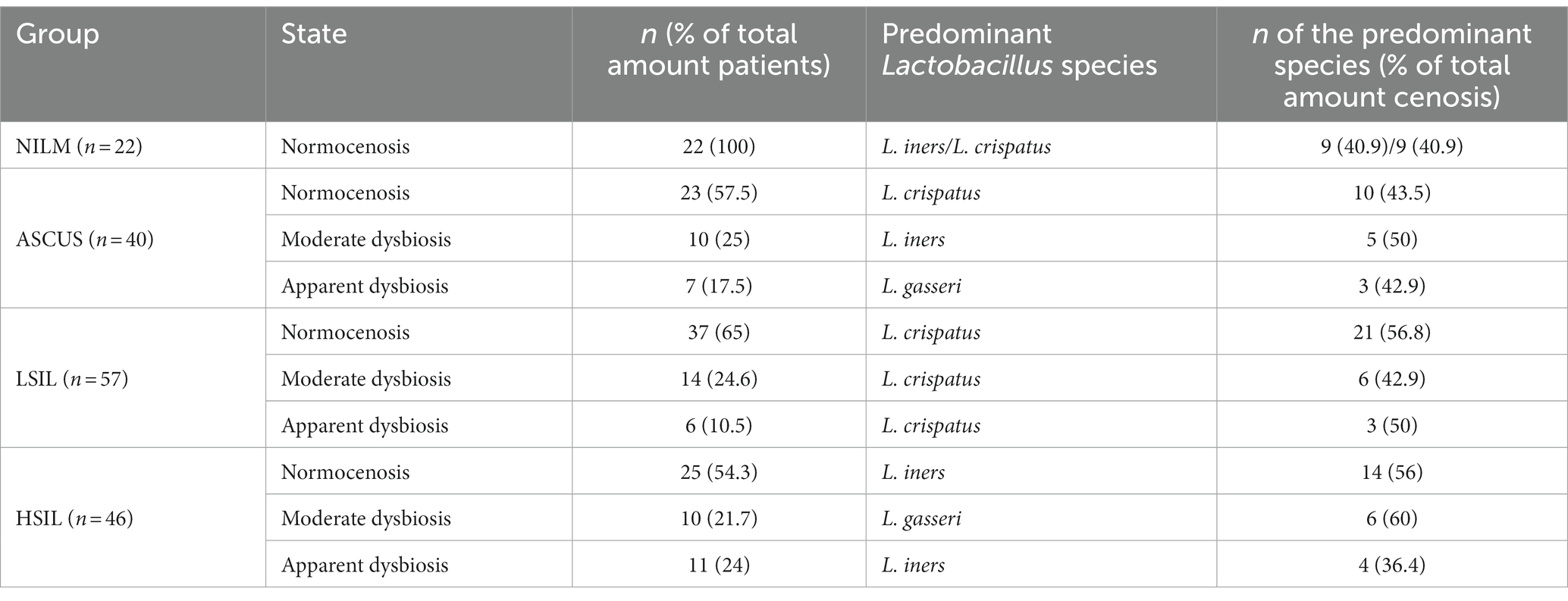
The microorganisms most frequently prevailing in the NILM, ASCUS, LSIL, HSIL groups were L. crispatus, L. iners, and L. gasseri (Figures 6, 7; Table 13). The highest L. gasseri content was observed in the HSIL patients. The L. jensenii fraction was relatively small. The fraction of the samples containing L. crispatus was the smallest in the HSIL group. Furthermore, in normocenosis, L. gasseri content was the smallest in all groups.
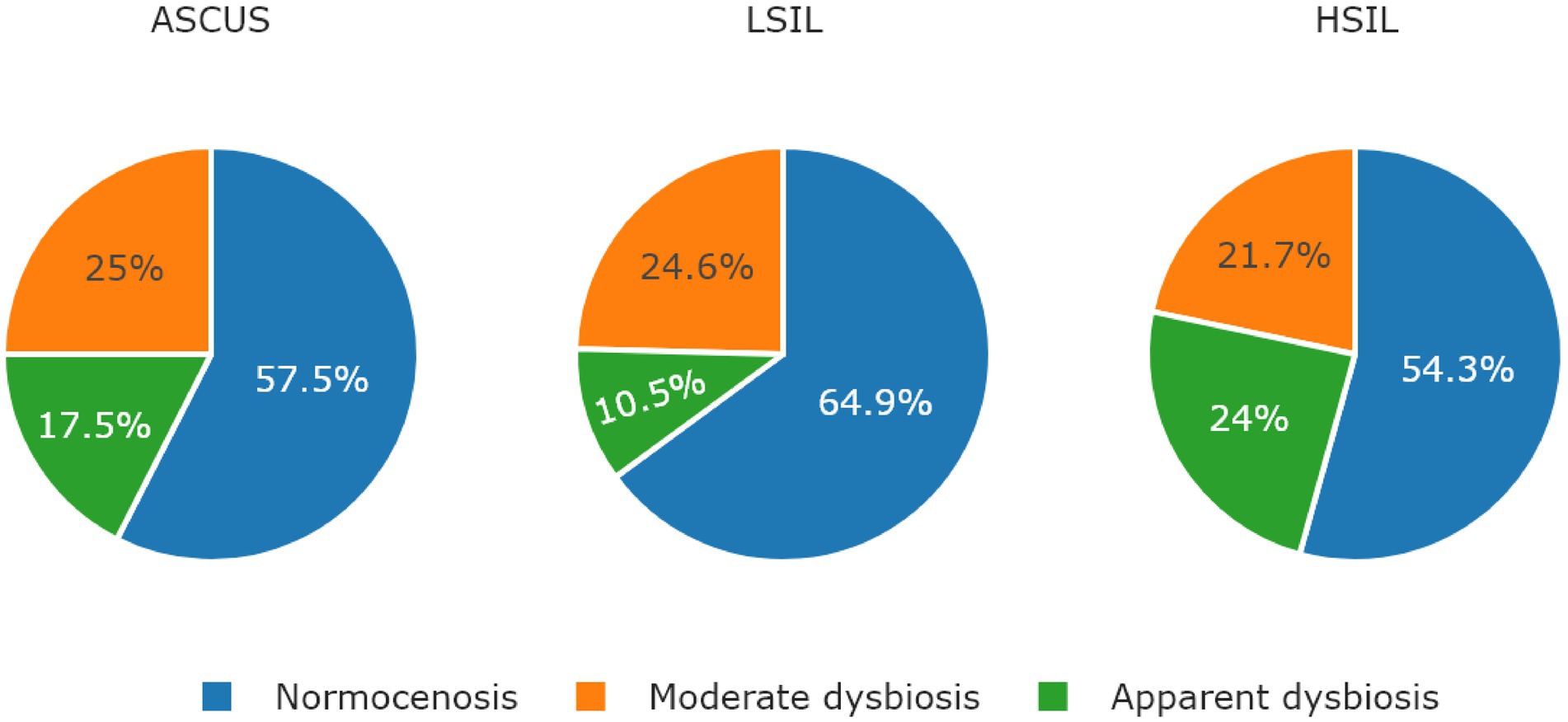
Figure 6. The ratio of microbiological cenosises were observed in the studied groups. The type of cenosis was determined by the percentage of Lactobacillus genus in the sample. Normocenosis – more than or equal 80%; moderate dysbiosis – from 20 to 80%; apparent dysbiosis – <20%.
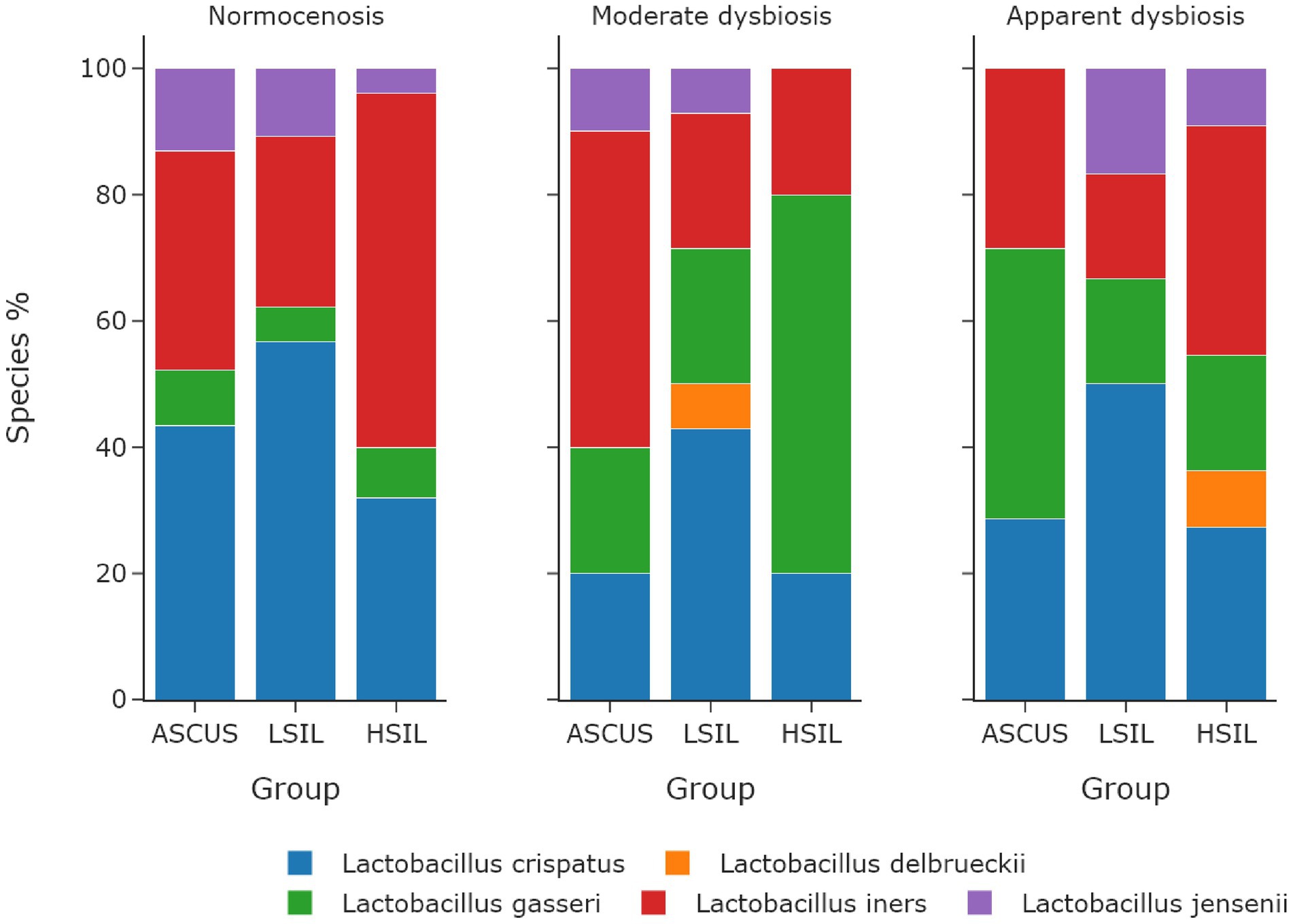
Figure 7. The dependence of the fraction of predominant Lactobacillus species on microbiome cenosises. The type of cenosis was determined by the percentage of Lactobacillus genus in the sample. Normocenosis – more than or equal 80%; moderate dysbiosis – from 20 to 80%; apparent dysbiosis – <20%.
In each microbiota state (normocenosis, moderate dysbiosis, pronounced dysbiosis), HSIL samples exhibit a certain decrease of the representation of L. crispatus with the predominance of other species (Table 13; Supplementary Table S6).
4 Discussion
Among women with cervical dysfunction confirmed by cytological and/or histological examination, cervical HPV colonization was observed in 100, 72.5, and 56.1% for HSIL, ASCUS, and LSIL, respectively. The most common HPV type was HPV16 which occurred twice as often in the patients with severe cervical dysfunction (HSIL, 58.8%) compared to LSIL (28%) and ASCUS (25%) patients. Alpha and beta diversity did not show any statistically significant difference between studied group. We suggest that it may be due to lack of influence of HPV to cervical canal microbiota. Nevertheless, some CST (for example CST IV or CST III) could be more vulnerable to entry and spread of HPV. In healthy women of a reproductive age, equally often prevailing types of Lactobacillus are L. crispatus and L. iners. For example, L. crispatus turned out to be the predominant Lactobacillus species in women with normocenosis for the ASCUS and LSIL groups, while L. iners prevailed in the HSIL group. There are contradictions in the works of other authors concerning L. iners: some studies show that it accompanies HPV infection and SIL (Wang et al., 2019; Sasivimolrattana et al., 2022; Xu et al., 2022), whereas other studies demonstrate no association of the kind (McKee et al., 2020; Gomez Cherey et al., 2023; Ivanov et al., 2023). L. crispatus is considered to protect from SIL progressing in HPV (Cheng et al., 2020), since it produces a lot of the D-isomer of lactic acid which increases the viscosity of the vaginal secrete and elevates its ability to sequester virions, in contrast to L. iners which produces only the L-isomer (Kalia et al., 2020; Nicolò et al., 2023). Moreover, L. iners produces inerolysin that creates pores in the vaginal epithelium potentially promoting HPV infection (Xu et al., 2022; Sharifian et al., 2023). This theory agrees to our results. All taxons with statistically significant difference are usually found in cervical microbiota, but they have higher occurrence and ratio in HSIL group compared with NILM and LSIL groups. The logistic regression allowed for individually evaluating the impact of the age and the role of certain microorganisms in HSIL and LSIL development. The older was the patient as well as the higher was L. iners fraction in the sample from the cervical canal, the higher was the probability of detecting HSIL rather than LSIL. Conversely, the younger age and high L. crispatus content increased the chance of being sorted into the LSIL group. In our work, the ASCUS patients with moderate dysbiosis more often contained L. iners, whereas LSIL patients contained L. crispatus, and HSIL patients had L. gasseri. Severe dysbiosis correlated with L. gasseri, L. crispatus, and with both L. crispatus and L. iners in the ASCUS, LSIL, and HSIL groups, respectively. L. gasseri was the least common Lactobacillus in the women of reproductive age. L. delbrueckii was the predominant Lactobacillus being present in 1.8% of the enrolled women, although it did not belong to any type of vaginal communities. In the ASCUS group, LSIL, and HSIL dysbiosis were observed in 42.5, 35, and 45.6% of patients, respectively. Anaerobic dysbiosis or bacterial vaginosis was the most prevalent in all groups. In bacterial vaginosis, the following anaerobic microorganisms prevailed: G. vaginalis, F. vaginae, D. micraerophilus, S. sanguinegens, Anaerococcus spp., M. massiliensis, Prevotella spp., F. magna, Peptoniphilus spp., Porphyromonas spp., and Parvimonas spp. Aerobic vaginitis was more rare, and in all cases under scrutiny, the predominant microorganism to cause it was Streptococcus spp. The bacteria associated with bacterial vaginitis produce carcinogenic nitrosamines, which trigger the release of cytokines such as interleukine-1b involved in SIL development. During HPV infection, the cytokines weaken the immune system, while carcinogenic nitrosamines facilitate DNA damage (Muzny et al., 2020; Tidbury et al., 2021). The predominance of certain groups of microorganisms such as Sneathia, Prevotella, Megasphaera, Dialister, Gardnerella, Streptococcus, and Fannyhessea is associated with SIL development (Mitra et al., 2020; Dong et al., 2022) which is in line with our data. However, it is believed that the predominance of anaerobic microorganisms is associated with LSIL (Lin et al., 2021), whereas aerobic vaginitis is associated with HSIL (Plisko et al., 2021). Colonization of the cervical canal and vagina by U. urealyticum and U. parvum lead to SIL development (Condic et al., 2023) which is supported by our results, since Ureaplasma spp. prevailed both qualitatively and quantitatively in the LSIL HPV(+) patients. Other works suggest that microbial diversity is elevated upon HPV infection (Chen et al., 2020; Usyk et al., 2020) which, however, was not observed in this work (Figure 7).
The qualitative and quantitative comparisons of microbiomes between the groups allowed us to detect statistically significant differences for several genera: Prevotella, Gardnerella, Porphyromonas, Dialister, Staphylococcus, Ureaplasma. Estimating the possible relationship of various Lactobacillus species with dysbiotic impairments of the microbiome and HPV-associated cervical diseases did not reveal any statistical significant difference in the Lactobacillus species composition and its relationship with HPV-associated cervical disorders. Of note, that L. iners, L. gasseri, L. crispatus were often detected upon dysbiosis, whereas L. gasseri was the least common in all groups. Evaluating dysbiotic impairments of the microbiome in HPV(+) patients based on 16S sequencing demonstrated that dysbiosis was most common in the patients with ASCUS (42.5%), LSIL (35%), and HSIL (45.6%). Bacterial vaginosis was associated with G. vaginalis, F. vaginae, D. micraerophilus, S. sanguinegens, Anaerococcus spp., M. massiliensis, Prevotella spp., F. magna, Peptoniphilus spp., Porphyromonas spp., and Parvimonas spp., while aerobic vaginitis was related to Streptococcus spp. Detecting HPV along with the impaired microbiome, such as bacterial vaginosis and aerobic vaginitis, may allow for identifying the groups of women with the high risk of SIL development and progression.
Data availability statement
The data presented in the study are deposited in the NCBI BioProject repository, accession number PRJNA1041987.
Ethics statement
The studies involving humans were approved by Local Ethics Committee at the FSBI “National Medical Research Center for Obstetrics, Gynecology and Perinatology Named After Academician V.I.Kulakov” Ministry of Health of the Russian Federation (protocol no. 3 of 26 March 2020). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
AP: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. VC: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. AK: Conceptualization, Investigation, Methodology, Software, Writing – original draft. AAn: Conceptualization, Investigation, Methodology, Writing – original draft. ZR: Conceptualization, Data curation, Formal analysis, Software, Visualization, Writing – original draft. AAs: Conceptualization, Investigation, Writing – original draft. DK: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. DR: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. GB: Conceptualization, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by grant no. 075-15-2019-1789 from the Ministry of Science and Higher Education of the Russian Federation allocated to the Center for Precise Genome Editing and Genetic Technologies for Biomedicine.
Acknowledgments
This is a short text to acknowledge the contributions of specific colleagues, institutions, or agencies that aided the efforts of the authors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1334502/full#supplementary-material
References
Al-Nasiry, S., Ambrosino, E., Schlaepfer, M., Morré, S. A., Wieten, L., Voncken, J. W., et al. (2020). The interplay between reproductive tract microbiota and immunological system in human reproduction. Front. Immunol. 11, 1–20. doi: 10.3389/fimmu.2020.00378
Alrajjal, A., Pansare, V., Choudhury, M. S. R., Khan, M. Y. A., and Shidham, V. B. (2021). Squamous intraepithelial lesions (SIL: LSIL, HSIL, ASCUS, ASC-H, LSIL-H) of uterine cervix and Bethesda system. Cytojournal 18:16. doi: 10.25259/Cytojournal_24_2021
Andralojc, K. M., Molina, M. A., Qiu, M., Spruijtenburg, B., Rasing, M., Pater, B., et al. (2021). Novel high-resolution targeted sequencing of the cervicovaginal microbiome. BMC Biol. 19, 267–218. doi: 10.1186/s12915-021-01204-z
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Burd, E. M. (2003). Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 16, 1–17. doi: 10.1128/CMR.16.1.1-17.2003
Chen, Y., Qiu, X., Wang, W., Li, D., Wu, A., Hong, Z., et al. (2020). Human papillomavirus infection and cervical intraepithelial neoplasia progression are associated with increased vaginal microbiome diversity in a Chinese cohort. BMC Infect. Dis. 20, 629–612. doi: 10.1186/s12879-020-05324-9
Cheng, L., Norenhag, J., Hu, Y. O. O., Brusselaers, N., Fransson, E., Ährlund-Richter, A., et al. (2020). Vaginal microbiota and human papillomavirus infection among young Swedish women. NPJ Biofilms Microbiomes 6, 39–10. doi: 10.1038/s41522-020-00146-8
Cogliano, V., Baan, R., Straif, K., Grosse, Y., Secretan, B., El Ghissassi, F., et al. (2005). Carcinogenicity of human papillomaviruses. Lancet Oncol. 6:204. doi: 10.1016/s1470-2045(05)70086-3
Condic, M., Neidhöfer, C., Ralser, D. J., Wetzig, N., Thiele, R., Sieber, M., et al. (2023). Analysis of the cervical microbiome in women from the German national cervical cancer screening program. J. Cancer Res. Clin. Oncol. 149, 6489–6500. doi: 10.1007/s00432-023-04599-0
Dong, B., Huang, Y., Cai, H., Chen, Y., Li, Y., Zou, H., et al. (2022). Prevotella as the hub of the cervicovaginal microbiota affects the occurrence of persistent human papillomavirus infection and cervical lesions in women of childbearing age via host NF-κB/C-myc. J. Med. Virol. 94, 5519–5534. doi: 10.1002/jmv.28001
Gardella, B., Pasquali, M. F., La Verde, M., Cianci, S., Torella, M., and Dominoni, M. (2022). The complex interplay between vaginal microbiota, HPV infection, and immunological microenvironment in cervical intraepithelial neoplasia: a literature review. Int. J. Mol. Sci. 23:7174. doi: 10.3390/ijms23137174
Gomez Cherey, J. F., Payalef, S. N., Fleider, L., Reyes, A. P., Maldonado, V. A., Losada, M. O., et al. (2023). Microbiota unbalance in relation to high-risk human papillomavirus cervical infection. Int. J. Gynecol. Cancer 33, 482–488. doi: 10.1136/ijgc-2022-003760
Hu, C., Liu, T., Han, C., Xuan, Y., Jiang, D., Sun, Y., et al. (2022). HPV E6/E7 promotes aerobic glycolysis in cervical cancer by regulating IGF2BP2 to stabilize m6A-MYC expression. Int. J. Biol. Sci. 18, 507–521. doi: 10.7150/ijbs.67770
Ivanov, M. K., Brenner, E. V., Hodkevich, A. A., Dzyubenko, V. V., Krasilnikov, S. E., Mansurova, A. S., et al. (2023). Cervicovaginal-microbiome analysis by 16S sequencing and real-time PCR in patients from Novosibirsk (Russia) with cervical lesions and several years after Cancer treatment. Diagnostics 13:140. doi: 10.3390/diagnostics13010140
Kalia, N., Singh, J., and Kaur, M. (2020). Microbiota in vaginal health and pathogenesis of recurrent vulvovaginal infections: a critical review. Ann. Clin. Microbiol. Antimicrob. 19:5. doi: 10.1186/s12941-020-0347-4
Kang, G.-U., Jung, D.-R., Lee, Y. H., Jeon, S. Y., Han, H. S., Chong, G. O., et al. (2021). Potential association between vaginal microbiota and cervical carcinogenesis in Korean women: a cohort study. Microorganisms 9:294. doi: 10.3390/microorganisms9020294
Lin, W., Zhang, Q., Chen, Y., Chen, L., Dong, B., and Sun, P. (2021). The prevalence of human papillomavirus and bacterial vaginosis among young women in China: a cross-sectional study. BMC Womens Health 21, 409–410. doi: 10.1186/s12905-021-01504-0
Lin, S., Zhang, B., Lin, Y., Lin, Y., and Zuo, X. (2022). Dysbiosis of cervical and vaginal microbiota associated with cervical intraepithelial neoplasia. Front. Cell. Infect. Microbiol. 12:767693. doi: 10.3389/fcimb.2022.767693
McClymont, E., Albert, A. Y., Wang, C., Dos Santos, S. J., Coutlée, F., Lee, M., et al. (2022). Vaginal microbiota associated with oncogenic HPV in a cohort of HPV-vaccinated women living with HIV. Int. J. STD AIDS 33, 847–855. doi: 10.1177/09564624221109686
McKee, K. S., Carter, K. A., Bassis, C., Young, V. B., Reed, B., Harper, D. M., et al. (2020). The vaginal microbiota, high-risk human papillomavirus infection, and cervical cytology: results from a population-based study. Gynecol. pelvic Med. 3:18. doi: 10.21037/gpm-20-10
Mitra, A., MacIntyre, D. A., Ntritsos, G., Smith, A., Tsilidis, K. K., Marchesi, J. R., et al. (2020). The vaginal microbiota associates with the regression of untreated cervical intraepithelial neoplasia 2 lesions. Nat. Commun. 11:1999. doi: 10.1038/s41467-020-15856-y
Molina, M. A., Andralojc, K. M., Huynen, M. A., Leenders, W. P. J., and Melchers, W. J. G. (2022). In-depth insights into cervicovaginal microbial communities and hrHPV infections using high-resolution microbiome profiling. npj Biofilms Microbiomes 8:8. doi: 10.1038/s41522-022-00336-6
Muzny, C. A., Łaniewski, P., Schwebke, J. R., and Herbst-Kralovetz, M. M. (2020). Host-vaginal microbiota interactions in the pathogenesis of bacterial vaginosis. Curr. Opin. Infect. Dis. 33, 59–65. doi: 10.1097/QCO.0000000000000620
Naidoo, K., Abbai, N., Tinarwo, P., and Sebitloane, M. (2022). BV associated bacteria specifically BVAB 1 and BVAB 3 as biomarkers for HPV risk and progression of cervical neoplasia. Infect. Dis. Obstet. Gynecol. 2022, 1–11. doi: 10.1155/2022/9562937
Nicolò, S., Antonelli, A., Tanturli, M., Baccani, I., Bonaiuto, C., Castronovo, G., et al. (2023). Bacterial species from vaginal microbiota differently affect the production of the E6 and E7 Oncoproteins and of p53 and p-Rb Oncosuppressors in HPV16-infected cells. Int. J. Mol. Sci. 24:7173. doi: 10.3390/ijms24087173
Nieves-Ramírez, M. E., Partida-Rodríguez, O., Moran, P., Serrano-Vázquez, A., Pérez-Juárez, H., Pérez-Rodríguez, M. E., et al. (2021). Cervical squamous intraepithelial lesions are associated with differences in the vaginal microbiota of Mexican women. Microbiol. Spectr. 9:e0014321. doi: 10.1128/Spectrum.00143-21
Norenhag, J., Du, J., Olovsson, M., Verstraelen, H., Engstrand, L., and Brusselaers, N. (2020). The vaginal microbiota, human papillomavirus and cervical dysplasia: a systematic review and network meta-analysis. BJOG An Int. J. Obstet. Gynaecol. 127, 171–180. doi: 10.1111/1471-0528.15854
Ntuli, L., Mtshali, A., Mzobe, G., Liebenberg, L. J. P., and Ngcapu, S. (2022). Role of immunity and vaginal microbiome in clearance and persistence of human papillomavirus infection. Front. Cell. Infect. Microbiol. 12, 1–10. doi: 10.3389/fcimb.2022.927131
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Plisko, O., Zodzika, J., Jermakova, I., Pcolkina, K., Prusakevica, A., Liepniece-Karele, I., et al. (2021). Aerobic vaginitis—underestimated risk factor for cervical intraepithelial neoplasia. Diagnostics 11, 1–9. doi: 10.3390/diagnostics11010097
Proctor, L. M., Creasy, H. H., Fettweis, J. M., Lloyd-Price, J., Mahurkar, A., Zhou, W., et al. (2019). The integrative human microbiome project. Nature 569, 641–648. doi: 10.1038/s41586-019-1238-8
Qingqing, B., Jie, Z., Songben, Q., Juan, C., Lei, Z., and Mu, X. (2021). Cervicovaginal microbiota dysbiosis correlates with HPV persistent infection. Microb. Pathog. 152:104617. doi: 10.1016/j.micpath.2020.104617
Ritu, W., Enqi, W., Zheng, S., Wang, J., Ling, Y., and Wang, Y. (2019). Evaluation of the associations between cervical microbiota and HPV infection, clearance, and persistence in Cytologically Normal women. Cancer Prev Res (Phila) 12, 43–56. doi: 10.1158/1940-6207.CAPR-18-0233
Santella, B., Schettino, M. T., Franci, G., De Franciscis, P., Colacurci, N., Schiattarella, A., et al. (2022). Microbiota and HPV: the role of viral infection on vaginal microbiota. J. Med. Virol. 94, 4478–4484. doi: 10.1002/jmv.27837
Sasivimolrattana, T., Chantratita, W., Sensorn, I., Chaiwongkot, A., Oranratanaphan, S., Bhattarakosol, P., et al. (2022). Cervical microbiome in women infected with HPV16 and high-risk HPVs. Int. J. Environ. Res. Public Health 19:14716. doi: 10.3390/ijerph192214716
Sharifian, K., Shoja, Z., and Jalilvand, S. (2023). The interplay between human papillomavirus and vaginal microbiota in cervical cancer development. Virol. J. 20, 73–79. doi: 10.1186/s12985-023-02037-8
Tidbury, F. D., Langhart, A., Weidlinger, S., and Stute, P. (2021). Non-antibiotic treatment of bacterial vaginosis—a systematic review. Arch. Gynecol. Obstet. 303, 37–45. doi: 10.1007/s00404-020-05821-x
Usyk, M., Zolnik, C. P., Castle, P. E., Porras, C., Herrero, R., Gradissimo, A., et al. (2020). Cervicovaginal microbiome and natural history of HPV in a longitudinal study. PLoS Pathog. 16, e1008376–e1008320. doi: 10.1371/journal.ppat.1008376
Wang, H., Ma, Y., Li, R., Chen, X., Wan, L., and Zhao, W. (2019). Associations of cervicovaginal lactobacilli with high-risk human papillomavirus infection, cervical intraepithelial neoplasia, and cancer: a systematic review and meta-analysis. J. Infect. Dis. 220, 1243–1254. doi: 10.1093/infdis/jiz325
Xu, X., Zhang, Y., Yu, L., Shi, X., Min, M., Xiong, L., et al. (2022). A cross-sectional analysis about bacterial vaginosis, high-risk human papillomavirus infection, and cervical intraepithelial neoplasia in Chinese women. Sci. Rep. 12, 6609–6612. doi: 10.1038/s41598-022-10532-1
Keywords: squamous intraepithelial lesion, 16S rRNA gene sequencing, HPV, cervicovaginal microbiota, SIL prediction
Citation: Peremykina A, Cheranev V, Krivoy A, Andreev AO, Repinskaia Z, Asaturova AV, Korostin D, Rebrikov D and Bayramova GR (2024) Microbiome markers in HPV-positive and HPV-negative women of reproductive age with ASCUS and SIL determined by V4 region of 16S rRNA gene sequencing. Front. Microbiol. 15:1334502. doi: 10.3389/fmicb.2024.1334502
Edited by:
Jean-Philippe Lavigne, Centre Hospitalier Universitaire de Nîmes, FranceReviewed by:
Tarik Gheit, International Agency for Research on Cancer (IARC), FranceNi Zhao, Johns Hopkins University, United States
Copyright © 2024 Peremykina, Cheranev, Krivoy, Andreev, Repinskaia, Asaturova, Korostin, Rebrikov and Bayramova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Valery Cheranev, ZmVyb3ZhbEB5YW5kZXgucnU=
†These authors have contributed equally to this work and share first authorship
 Anastasiya Peremykina
Anastasiya Peremykina Valery Cheranev
Valery Cheranev Andrey Krivoy2
Andrey Krivoy2 Aleksandra V. Asaturova
Aleksandra V. Asaturova Denis Rebrikov
Denis Rebrikov