In spite of a clear molecular explanation presented more than four and half decades ago (Kandler and König, 1978), a misconception that archaea lack peptidoglycan and some of these organisms carry pseudomurein or pseudopeptidoglycan has proliferated both in peer-reviewed publications and textbooks and online educational materials (Albers and Meyer, 2011; Visweswaran et al., 2011; Rodrigues-Oliveira et al., 2017; Bruslind, 2019; Baker et al., 2020; Madigan et al., 2021; Medvedeva et al., 2023; Salas et al., 2024) (bio.libretexts.org). This article is intended to describe how this misconception arose and when and with what rationale a clear attempt was made to remove it.
In 1977, based on a 16S rRNA-based phylogenetic analysis of several methanogens, Woese and Fox identified archaebacteria as a prokaryotic group that is distinct from bacteria (Woese and Fox, 1977); later, archaebacteria were renamed as archaea (Woese et al., 1990). The specialized cell walls of methanogens that were assumed to lack peptidoglycan (Kandler and Hippe, 1977) were a key feature supporting the declaration (Fox et al., 1977). In 1978, Kandler and König reported that the rigid components of the cell walls of six species of Methanobacterium, which are currently known as Methanobacterium formicicum, Methanothermobacter thermautotrophicus, Methanobrevibacter ruminantium, Methanobrevibacter arboriphilus, and Methanobacterium bryantii, and members of the order Methanobacteriales within the class of Methanobacteria (Rinke et al., 2021), lacked muramic acid or D-amino acids, which are typical constituents of bacterial peptidoglycan. Instead, they consisted of L-amino acids (Lys, Glu, and Ala) and N-acetylglucosamine or N-acetylgalactosamine (Kandler and König, 1978). For apparent similarities to peptidoglycan, which is also called “murein” for one of its key components, muramic acid, Kandler proposed that this structure of Methanobacterium should be termed “pseudomurein” (Kandler, 1979). König and Kandler showed that, in Methanobacterium thermoautotrophicum (currently, Methanothermobacter thermautotrophicus), the peptide moiety of this polymer contains an unusually high number of ε- and γ-bonds, and it is coupled to the glycan strand via a glutamyl residue (Konig and Kandler, 1979), and N-acetyltalosaminuronic acid is a component of the glycan strand (König and Kandler, 1979; König et al., 1983). These observations solidified their proposal that pseudomurein is distinct from peptidoglycan and is a result of convergent “invention” (König and Kandler, 1979). They showed that the glycan strands of pseudomurein are composed of alternating N-acetylglucosamine or N-acetylgalactosamine and N-acetyltalosaminuronic acid residues linked through β-1,3 glycosidic bonds, whereas, in the strands of bacterial peptidoglycan, one finds β-1,4-linked N-acetylglucosamine and N-acetylmuramic acid units (König et al., 1982) (Figure 1).
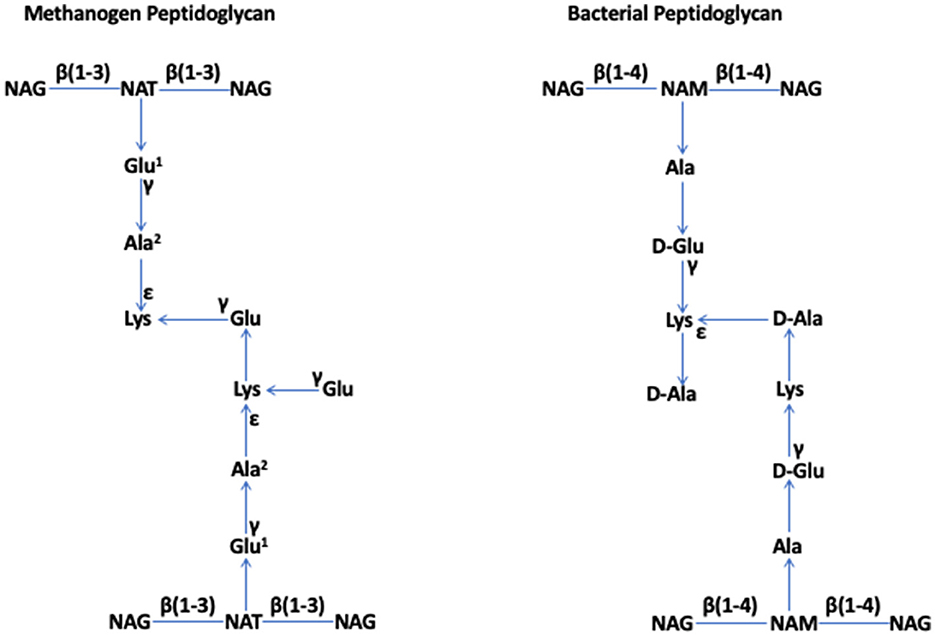
Figure 1. Methanogen and bacterial peptidoglycan (Kandler, 1993; Kandler and Konig, 1993; Albers and Meyer, 2011; Kandler and König, 2014; Tashiro et al., 2017; Subedi et al., 2021). NAG, N-acetylglucosamine; NAT, N-acetyltalosaminuronic acid; NAM, N-acetylmuramic acid. Special features. Methanogen peptidoglycan: β (1–3) bonds in the glycan strand and L-amino acids in the peptide moiety, where Glu1 and Ala2 can be replaced by Asp and Thr or Ser, respectively, in some members. Bacterial peptidoglycan: β (1–4) bonds in the glycan strand and both L- and D-amino acids in the peptide moiety. The figure has been adapted and modified from Subedi et al. (2021).
The aforementioned effort to define the rigid component of the methanobacterial cell wall as a novel entity came to face reality when conformational energy calculations suggested that murein and pseudomurein share a similar three-dimensional (3D) architecture (Leps et al., 1984a,b). Kandler and König recognized that their concept that the cell walls of methanogens lacked peptidoglycan (Kandler and König, 1978) was not correct and they issued a correction: “Such a polymer must also be considered as a peptidoglycan, a very general chemical term” (Kandler and König, 1985); a similar clarification has appeared more recently as well (Claus and König, 2010). A peptidoglycan cell wall has also been found in Methanopyrus kandleri, the only characterized representative of Methanopyrus (of the Methanopyri class and Methanopyrales order) (Kurr et al., 1991).
The above-described findings need to be considered in the context that, even within the bacterial domain, the peptidoglycan structure varies (Vollmer, 2008). Thus, the versions of this polymer that one finds in the methanogens of the orders of Methanobacteriales and Methanopyrales are not pseudomurein as it does not contain muramic acid but simply another type of peptidoglycan, where glycan strands are connected by peptide linkages. The name archaeal peptidoglycan that has been used recently (Khairunisa et al., 2023) is appropriate in this context.
To summarize, in most methanogens and almost all other archaea, a crystalline surface layer (S-layer) composed of one or two glycoproteins encases the cytoplasmic membrane formed by either a diether lipid bilayer or a tetraether lipid monolayer (Albers and Meyer, 2011; Klingl et al., 2019; Meyer et al., 2022; van Wolferen et al., 2022). In some cases, a protein sheath and a layer of methanochondroitin (a proteoglycan made of N-acetylgalactosamine and glucuronic acid) are found to occur over the S-layer (Albers and Meyer, 2011; Klingl et al., 2019; Meyer et al., 2022). In contrast, in a limited number of methanogens that belong to the genera of Methanobacterium, Methanothermobacter, Methanosphaera, and Methanobrevibacter (of the class of Methanobacteria and the order of Methanobacteriales) and Methanopyrus (of the Methanopyri class and the Methanopyrales order) of the Methanobacteriota phylum (Rinke et al., 2021), a rigid sacculus made of glycan strands connected by peptide linkages or peptidoglycan takes the place of the S-layer as described above (Albers and Meyer, 2011; Klingl et al., 2019; Meyer et al., 2022; van Wolferen et al., 2022); in Methanothermus, which also belongs to the Methanobacteriales order, an S-layer is placed over the peptidoglycan.
The recognition that certain archaea contain peptidoglycan brings the studies on the cell walls of Methanobacteria and Methanopyri to a larger arena, and consequently, this recognition will accelerate the discovery process that will provide products of applied value, such as improved methods for environmental detection, creation of archaeal-specific antibiotics, and improvements in genetics and cell biology research. These advances would facilitate the development of better methods for mitigating methane emission from livestock, manipulating human gut metabolism/microbiome toward better health, and improving processes for methane production and biofuel from renewable resources. Methanobacteriales constitute a great portion of the bovine rumen and human gut methanogen population (Eckburg et al., 2005; Borrel et al., 2020; Khairunisa et al., 2023) and are attractive for industrial bioproduction of methane, as well as amino acids (Pappenreiter et al., 2019; Pfeifer et al., 2021; Taubner et al., 2023).
Author contributions
BM: Writing – original draft, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The relevant research on methanogens in BM's laboratory has been supported in part by the National Aeronautics and Space Administration Astrobiology: Exobiology and Evolutionary Biology grant NNX13AI05G and the Virginia Tech Agricultural Experiment Station Hatch Program (CRIS project VA-160021). The funders have no role in the design and writing this opinion article.
Acknowledgments
The idea of writing this opinion article originated when the author (BM) brought up the issue addressed here to the attention of Dr. Nika Pende of the Archaea Physiology & Biotechnology Group at the University of Vienna at the 2023 Archaea: Ecology, Metabolism and Molecular Biology Gordon Conference. They were introduced by Dr. Simon K.-M.R. Rittmann, a senior colleague of Dr. Pende, which led to a fruitful discussion, resulting in an earlier version of this manuscript. BM remains grateful for the conversations that he had with Dr. Pende. We thank the reviewers and the editor for the valuable suggestions, which have been incorporated into this article.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The reviewer BW declared a past collaboration with the author BM and the handling editor.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Albers, S.-V., and Meyer, B. H. (2011). The archaeal cell envelope. Nat. Rev. Microbiol. 9, 414–426. doi: 10.1038/nrmicro2576
Baker, B. J., De Anda, V., Seitz, K. W., Dombrowski, N., Santoro, A. E., and Lloyd, K. G. (2020). Diversity, ecology and evolution of Archaea. Nat. Microbiol. 5, 887–900. doi: 10.1038/s41564-020-0715-z
Borrel, G., Brugere, J. F., Gribaldo, S., Schmitz, R. A., and Moissl-Eichinger, C. (2020). The host-associated archaeome. Nat. Rev. Microbiol. 18, 622–636. doi: 10.1038/s41579-020-0407-y
Claus, H., and König, H. (2010). “Cell envelopes of methanogens,” in Prokaryotic Cell Wall Compounds, 231–251. doi: 10.1007/978-3-642-05062-6_7
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
Fox, G. E., Magrum, L. J., Balch, W. E., Wolfe, R. S., and Woese, C. R. (1977). Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc. Natl. Acad. Sci. U S A. 74, 4537–4541. doi: 10.1073/pnas.74.10.4537
Kandler, O. (1979). Zellwandstrukturen bei Methan-Bakterien. Naturwissenschaften 66, 95–105. doi: 10.1007/BF00373500
Kandler, O. (1993). Cell Wall biochemistry and three-domain concept of life. System. Appl. Microbiol. 16, 501–509. doi: 10.1016/S0723-2020(11)80319-X
Kandler, O., and Hippe, H. (1977). Lack of peptidoglycan in the cell walls of Methanosarcina barkeri. Arch. Microbiol. 113, 57–60. doi: 10.1007/BF00428580
Kandler, O., and König, H. (1978). Chemical composition of the peptidoglycan-free cell walls of methanogenic bacteria. Arch. Microbiol. 118, 141–152. doi: 10.1007/BF00415722
Kandler, O., and König, H. (1985). “Cell envelopes of archaebacteria,” in Archaebacteria, eds. C. R. Woese, and R. S. Wolfe (New York, London: Academic Press). doi: 10.1016/B978-0-12-307208-5.50015-5
Kandler, O., and Konig, H. (1993). “Chapter 8 Cell envelopes of archaea: Structure and chemistry,” in The Biochemistry of Archaea (Archaebacteria), eds. C. R. Woese, and R. S. Wolfe (New York, London: Academic Press). doi: 10.1016/S0167-7306(08)60257-4
Kandler, O., and König, H. (2014). Cell wall polymers in archaea (Archaebacteria). Cell. Molec. Life Sci. 54, 305–308. doi: 10.1007/s000180050156
Khairunisa, B. H., Heryakusuma, C., Ike, K., Mukhopadhyay, B., and Susanti, D. (2023). Evolving understanding of rumen methanogen ecophysiology. Front. Microbiol. 14:1296008. doi: 10.3389/fmicb.2023.1296008
Klingl, A., Pickl, C., and Flechsler, J. (2019). “Archaeal cell walls,” in Bacterial Cell Walls and Membranes. doi: 10.1007/978-3-030-18768-2_14
Konig, H., and Kandler, O. (1979). The amino acid sequence of the peptide moiety of the pseudomurein from Methanobacterium thermoautotrophicum. Arch. Microbiol. 121, 271–275. doi: 10.1007/BF00425067
König, H., and Kandler, O. (1979). N-Acetyltalosaminuronic acid a constituent of the pseudomurein of the genus Methanobacterium. Arch. Microbiol. 123, 295–299. doi: 10.1007/BF00406664
König, H., Kandler, O., Jensen, M., and Rietschel, E. T. (1983). The primary structure of the glycan moiety of pseudomurein frommethanobacterium thermoautotrophicum. Hoppe-Seyler's Zeitschr. Physiol. Chem. 364, 627–636. doi: 10.1515/bchm2.1983.364.1.627
König, H., Kralik, R., and Kandler, O. (1982). Structure and modifications of pseudomurein in methano-bacleriales. Zentralblatt Bakteriol. Mikrobiol. Hygien 3, 179–191. doi: 10.1016/S0721-9571(82)80031-8
Kurr, M., Huber, R., König, H., Jannasch, H. W., Fricke, H., Trincone, A., et al. (1991). Methanopyrus kandleri, gen. and sp. nov. represents a novel group of hyperthermophilic methanogens, growing at 110°C. Arch. Microbiol. 156, 239–247. doi: 10.1007/BF00262992
Leps, B., Barnickel, G., and Bradaczek, H. (1984a). Structural studies on the bacterial cell wall peptidoglycan pseudomurein. I. Conformational energy calculations on the glycan strands in C1 conformation and comparison with murein. J. Theor. Biol. 107, 85–114. doi: 10.1016/S0022-5193(84)80123-X
Leps, B., Labischinski, H., Barnickel, G., Bradaczek, H., and Giesbrecht, P. (1984b). A new proposal for the primary and secondary structure of the glycan moiety of pseudomurein. Conformational energy calculations on the glycan strands with talosaminuronic acid in 1C conformation and comparison with murein. Eur. J. Biochem. 144, 279–286. doi: 10.1111/j.1432-1033.1984.tb08461.x
Madigan, M. T., Bender, K. S., Buckley, D. H., Sattley, W. M., and Stahl, D. A. (2021). Brock Biology of Microorganisms, Hoboken, NJ: Pearson Education.
Medvedeva, S., Borrel, G., Krupovic, M., and Gribaldo, S. (2023). A compendium of viruses from methanogenic archaea reveals their diversity and adaptations to the gut environment. Nat. Microbiol. 8, 2170–2182. doi: 10.1038/s41564-023-01485-w
Meyer, B. H., Albers, S. V., Eichler, J., and Aebi, M. (2022). “Archaea,” in Essentials of Glycobiology, eds. A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, et al. (New York, NY: Cold Spring Harbor).
Pappenreiter, P. A., Zwirtmayr, S., Mauerhofer, L. M., Rittmann, S. K. R., and Paulik, C. (2019). Development of a simultaneous bioreactor system for characterization of gas production kinetics of methanogenic archaea at high pressure. Eng. Life Sci. 19, 537–544. doi: 10.1002/elsc.201900035
Pfeifer, K., Ergal, I., Koller, M., Basen, M., Schuster, B., and Rittmann, S. K. M. R. (2021). Archaea biotechnology. Biotechnol. Adv. 47:107668. doi: 10.1016/j.biotechadv.2020.107668
Rinke, C., Chuvochina, M., Mussig, A. J., Chaumeil, P.-A., Davín, A. A., Waite, D. W., et al. (2021). A standardized archaeal taxonomy for the Genome Taxonomy Database. Nat. Microbiol. 6, 946–959. doi: 10.1038/s41564-021-00918-8
Rodrigues-Oliveira, T., Belmok, A., Vasconcellos, D., Schuster, B., and Kyaw, C. M. (2017). Archaeal S-layers: overview and current state of the art. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02597
Salas, E., Gorfer, M., Bandian, D., Eichorst, S. A., Schmidt, H., Horak, J., et al. (2024). Reevaluation and novel insights into amino sugar and neutral sugar necromass biomarkers in archaea, bacteria, fungi, and plants. Sci. Total Environ. 906:167463. doi: 10.1016/j.scitotenv.2023.167463
Subedi, B. P., Martin, W. F., Carbone, V., Duin, E. C., Cronin, B., Sauter, J., et al. (2021). Archaeal pseudomurein and bacterial murein cell wall biosynthesis share a common evolutionary ancestry. FEMS Microbes 2:xtab012. doi: 10.1093/femsmc/xtab012
Tashiro, T., Ishida, A., Hori, M., Igisu, M., Koike, M., Méjean, P., et al. (2017). Early trace of life from 3.95 Ga sedimentary rocks in Labrador, Canada. Nature 549, 516–518. doi: 10.1038/nature24019
Taubner, R.-S., Baumann, L. M. F., Steiner, M., Pfeifer, K., Reischl, B., Korynt, K., et al. (2023). Lipidomics and Comparative Metabolite Excretion Analysis of Methanogenic Archaea Reveal Organism-Specific Adaptations to Varying Temperatures and Substrate Concentrations. mSystems 8, e01159–22. doi: 10.1128/msystems.01159-22
van Wolferen, M., Pulschen, A. A., Baum, B., Gribaldo, S., and Albers, S. V. (2022). The cell biology of archaea. Nat. Microbiol. 7, 1744–1755. doi: 10.1038/s41564-022-01215-8
Visweswaran, G. R. R., Dijkstra, B. W., and Kok, J. (2011). Murein and pseudomurein cell wall binding domains of bacteria and archaea—a comparative view. Appl. Microbiol. Biotechnol. 92, 921–928. doi: 10.1007/s00253-011-3637-0
Vollmer, W. (2008). Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32, 287–306. doi: 10.1111/j.1574-6976.2007.00088.x
Woese, C. R., and Fox, G. E. (1977). Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U S A. 74, 5088–5090. doi: 10.1073/pnas.74.11.5088
Keywords: archaea, methanogen, cell wall, peptidoglycan, misconception, reminder
Citation: Mukhopadhyay B (2024) A reminder—peptidoglycan cell walls indeed occur in the archaeal domain, specifically in the members of Methanobacteria and Methanopyri classes. Front. Microbiol. 15:1329047. doi: 10.3389/fmicb.2024.1329047
Received: 27 October 2023; Accepted: 27 March 2024;
Published: 09 May 2024.
Edited by:
Zhe Lyu, North Carolina State University, United StatesReviewed by:
Barny Whitman, University of Georgia, United StatesNicole Buan, University of Nebraska-Lincoln, United States
Copyright © 2024 Mukhopadhyay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Biswarup Mukhopadhyay, Ymlzd2FydXBAdnQuZWR1
 Biswarup Mukhopadhyay
Biswarup Mukhopadhyay