
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 April 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1328766
Aeromonas species cause a wide spectrum of human diseases, primarily gastroenteritis, septicemia, and wound infections. Several studies have shown that about 40% of these cases involve mixed or polymicrobial infections between Aeromonas spp. and bacteria from other genera. However, the immune response of macrophages in front of the bacteria present in the mixed infections, as well as their impact on antimicrobial therapy, have not been investigated. This study evaluated the cell damage and immune response of the mouse macrophage BALB/c cell line (J774A.1) after performing a single and a mixed infection with a strain of Aeromonas caviae and Yersinia enterocolitica, both recovered from the same fecal sample from a patient with diarrhea. Macrophage cell damage was measured by the release of lactate dehydrogenase (LDH) while the immune response was evaluated studying the expression by RT-qPCR of six relevant immune-related genes. Additionally, the antimicrobial susceptibility pattern of the single and mixed strains in front of seventeen antibiotics was evaluated to determine the potential impact on the infection treatment. Macrophages infected with the mixture of the two strains showed a higher cell damage in comparison with the single infections and the immune-related genes, i.e., cytokines and chemokines genes (TNF-α, CCL20), and apoptotic and pyroptotic genes (TP53 and IL-1β) were overexpressed. After infection with the mixed cultures, an increase in the antimicrobial resistance was observed for ciprofloxacin, trimethoprim, chloramphenicol, gentamicin and ertapenem. This study increased the knowledge about the synergetic effect of the bacteria involved in mixed infection and on their potential impact on the treatment and evolution of the infection.
Some species of the genus Aeromonas are considered human opportunistic emerging pathogens because they cause a wide spectrum of diseases, mainly gastroenteritis, wound infections, and bacteremia (Janda and Abbott, 2010; Figueras and Beaz-Hidalgo, 2015; Fernández-Bravo and Figueras, 2020). Infections caused by Aeromonas may involve more than one type of bacteria within the same clinical sample either of the same genera of other genera (Lamy et al., 2009; Mosser et al., 2015; Fernández-Bravo and Figueras, 2020). For instance, a co-infection involving two strains of Aeromonas hydrophila which were isolated before and after amputation and surgical debridement from the wound site of a severe case of necrotizing fasciitis (NF) occurred in a previously healthy young girl (Grim et al., 2014; Ponnusamy et al., 2016). The interaction of the two involved A. hydrophila strains in relation to the outcome of the infection was investigated using different murine animal models and results demonstrated that the enhanced virulence of the mixed infection was related to the existence of specific virulence genes in both bacteria (Grim et al., 2014; Ponnusamy et al., 2016; Fernández-Bravo et al., 2019). Experimental mixed infections with different Aeromonas spp. and/or strains using other animal models like Caenorhabditis elegans (Mosser et al., 2015) and Drosophila melanogaster (Barraud et al., 2020; Robert et al., 2023) showed, in general, an increase of the virulence as determined by the survival experiment after infection with the two Aeromonas strains. However, Robert et al. (2023) showed that the enhanced virulence was strain dependent.
According to Lamy et al. (2009) polymicrobial infections with Aeromonas represents 42.3% of the clinical cases, while in a study included in the Ph.D. of Latif (2015) were only 20%. In cases of Aeromonas diarrhea, the most frequent associated bacteria corresponded to Campylobacter and Salmonella, while in cases of wound infections predominate other bacteria like Klebsiella, Enterococcus, Escherichia coli, Staphylococcus aureus, and Yersinia enterocolitica (Lamy et al., 2009; Chao et al., 2013; Zhou et al., 2019; Fernández-Bravo and Figueras, 2020; Yuwono et al., 2021; Robert et al., 2023). However, the interactions between the isolated strains of the different genera with Aeromonas have not yet been investigated. Previous studies demonstrated that the synergism between bacteria from different genera such as Pseudomonas aeruginosa, S. aureus or Salmonella increases the pathogenicity and the complexity of the disease (Nguyen and Oglesby-Sherrouse, 2016; Xu et al., 2019; Wilson et al., 2022). In this sense, polymicrobial infections tend to be associated with treatment failure and worse patient prognoses (Adamowicz et al., 2018; Little et al., 2021). Vega et al. (2013) demonstrated that the mixed culture of E. coli and Salmonella Typhimurium increases the tolerance to ciprofloxacin and in a study performed by Adamowicz et al. (2018) these mixed culture increased the tolerance to tetracycline.
Although Y. enterocolitica is a worldwide distributed agent of gastrointestinal infections, primarily affecting young individuals and commonly causing acute diarrhea (Hering et al., 2016; Nesbakken, 2021) like Aeromonas (Fernández-Bravo and Figueras, 2020). However, co-infections between Aeromonas spp. and Yersinia spp. in fecal samples are rarely reported (Latif, 2015; Hering et al., 2016; Nesbakken, 2021; Grave et al., 2022). In a study conducted in Australia, Aeromonas was the only enteropathogen in 1755 (82.3%) of the patients and out of the 377 patients that showed a mixed infection with another enterobacteria only 6 patients (1.4%) showed a coinfection with Yersinia (Yuwono et al., 2021). In another study performed, in Latvia, by Grave et al. (2022) about the prevalence of Aeromonas in a hospitalized pediatric population with gastroenteritis, 3 patients (6%) were affected by a co-infection of Aeromonas and Y. enterocolitica. In addition, from the unpublished data of the Ph.D. thesis of Latif (2015) only in one of the 409 patients studied a co-infection of Aeromonas caviae and Y. enterocolitica was observed. Recently, a new case of a patient that presented a gastrointestinal infection from which the same species, i.e., A. caviae and Y. enterocolitica were identified from the same feces culture was found. Considering the rarity of these cases and the lack of studies investigating the potential interaction between the bacteria of these genera, the present study aimed to evaluate the cell damage and immune response of macrophages after performing the single and dual infection with those strains. Furthermore, the antimicrobial susceptibility pattern of the single and mixed cultures was determined as well as the in vitro growth dynamics of the mixed strains.
A stool sample from a patient with diarrhea was screened for bacterial pathogens, i.e., Salmonella spp., Shigella spp., Campylobacter jejuni, Yersinia spp., and diarrheagenic E. coli using specific culture media such as Cefsulodin-Irgasan-Novobiocin (CIN) agar, Salmonella-Shigella (SS), Campylosel agar, and MacConkey agar with sorbitol (SMAC) (bioMérieux®, France). Eight colonies were isolated from the CIN plates for preliminary identification. Subsequently, the Enterobacterial Repetitive Intergenic Consensus PCR (ERIC-PCR) was performed to identify specific fingerprint patterns among the isolates with the conditions previously described by Versalovic et al. (1991), using the primers referred to in Table 1. After the genotyping, two different isolates were primarily identified by Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) with V3.1 of the Biotyper database (Bruker®). The cut-offs recommended were as follows: highly probable species level (scores ≥2.300), probable species level (scores 2.000–2.299), probable genus level (scores 1.700–1.999), and no reliable identification (scores ≤1.699). Both isolates were preserved in Tryptone Soya Broth (TSB) (Becton Dickinson GmbH, Germany) plus glycerol (20%) at −80°C. After growing the bacteria in Tryptone Soya Agar (TSA, Becton Dickinson GmbH, Germany) at 37°C for 24 h, the genomic DNA was extracted using the InstaGene™ DNA Purification Matrix (Bio-Rad Hercules CA, USA) following the manufacturer’s instructions (Fernández-Bravo et al., 2020). Their identity was determined after sequencing the rpoD and 16S rRNA genes using primers and conditions previously described (Soler et al., 2004; Savin et al., 2014). A BLASTn analysis was conducted to find similarities between sequences. Subsequently, the sequences were aligned with 36 type strains of Aeromonas species and 12 strains of Yersinia species using the ClustalW algorithm, and the phylogenetic analysis was performed with the Neighbor-Joining (NJ) and maximum-likelihood algorithms (ML) in MEGA v6.0 (Fernández-Bravo and Figueras, 2020).
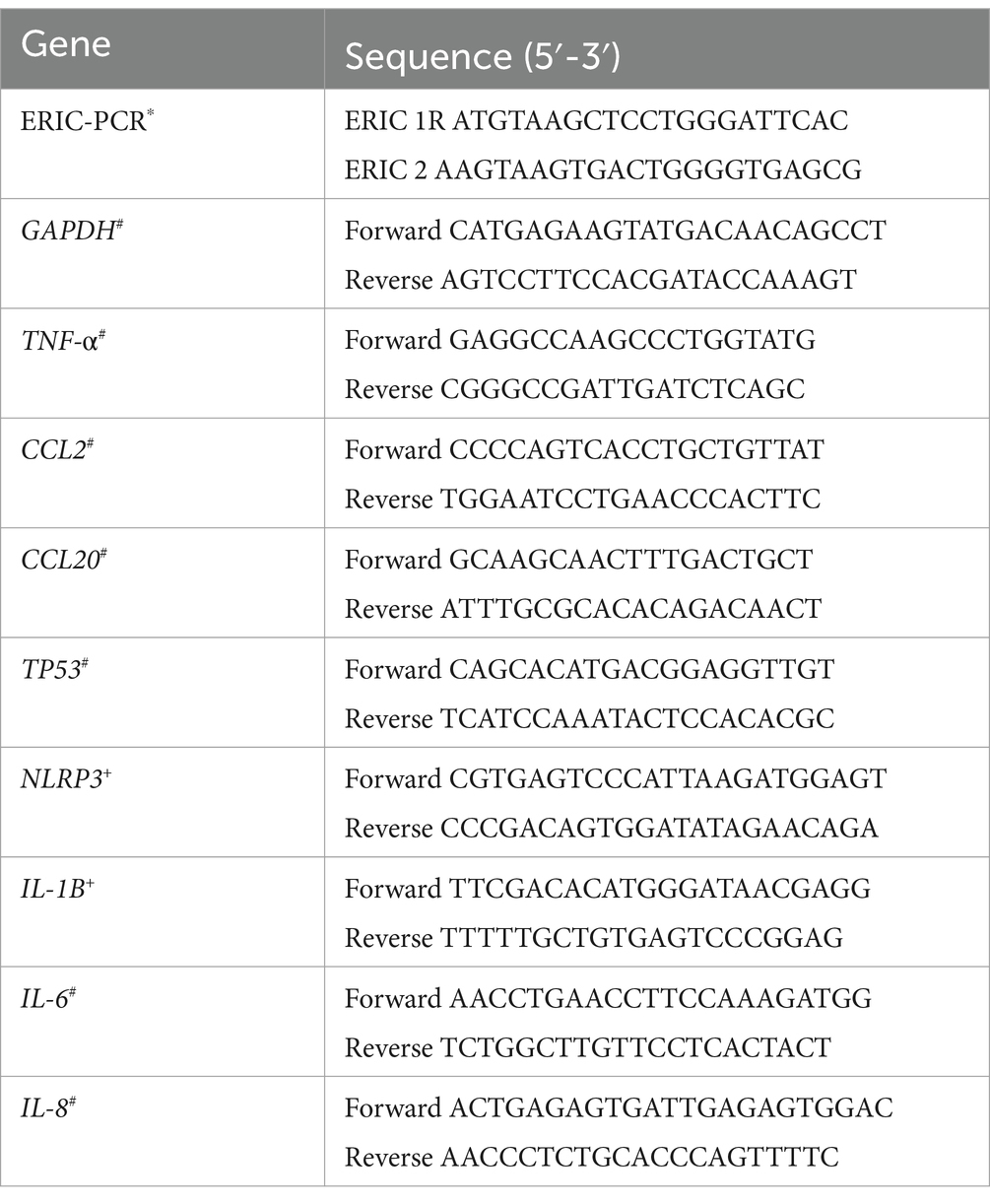
Table 1. Primers used in the study for ERIC-PCR and for gene expression analysis (Versalovic et al., 1991*; Murciano et al., 2015#; Zhao et al., 2017+).
The mouse macrophage BALB/c cell line J774A.1 (Tsuchiya et al., 1980) was selected for the infection experiments due to its significance as a model for studying the innate immune response against bacteria. This cell line was maintained as a cell adhesion in Dulbecco’s Modified Eagle’s Medium (DMEM, PAA Laboratories GmbH, Germany) supplemented with 10% FBS (fetal bovine serum, PAA Laboratories) plus 1% P/S solution (penicillin–streptomycin, PAA Laboratories) at 37°C and 5% CO2. Prior to infection experiments, cells were seeded in tissue culture plates containing DMEM without FBS and P/S (serum and antibiotic-free conditions) at a concentration of 0.5 × 106 cells/mL to obtain 1 × 106 cells/mL after 24 h (Murciano et al., 2017).
Before the infection, bacteria were grown at 37°C in serum and antibiotic-free DMEM under shaking conditions (100 rpm) for 24 h. Cell line J774A.1 was infected with the individual strains of A. caviae and Y. enterocolitica using overnight cultures at multiplicity of infection (MOI) of 10, i.e., the ratio between the number of bacteria and the number of targeted cells (Fernández-Bravo and Figueras, 2022). For the polymicrobial studies, the bacterial strains were mixed in a ratio 1:1 and the cells were infected at MOI 10. The infected cell lines were then incubated at 37°C and 5% CO2 and sampled at specific times depending on the experiment.
After infection with the single and the mixed strains at MOI 10, supernatants were collected at 4 h and 6 h post-infection (Fernández-Bravo and Figueras, 2022). Cell damage was determined by measuring the lactate dehydrogenase (LDH) enzyme released into the supernatants, indicating plasma membrane damage. The Cytox 96 Non-Radioactive Cytotoxicity Assay (Promega, Madison, WI, USA) was performed according to the manufacturer’s instructions and read with the SPECTRA spectrophotometer (Biotek, Spain). A standard curve was generated using bovine recombinant LDH (Sigma-Aldrich, St Louis, MO, USA). LDH levels in the samples were determined by extrapolation from the curve (Murciano et al., 2015; Fernández-Bravo and Figueras, 2022).
The MTT Cell Proliferation Assay 30-1010K™ (ATCC) was used as a measure of macrophage viability after infection with the mixed ant the single cultures. Macrophages were seeded in 96-well microtiter plates. Then, macrophages were infected at MOI 10 and subsequently incubated with gentamicin or streptomycin (50 μg/mL) for 45 min (Fernández-Bravo and Figueras, 2022). Later, the medium was replaced with fresh DMEM for 6 h at 37°C and 5% CO2. After incubation, the MTT reagent was added to the wells (10 μL/well), and microplates were incubated at 37°C and 5% CO2 for 2 h. At that point, 100 μL of the detergent reagent was added following incubation in darkness at room temperature for 2 h (Andersson et al., 2016). Finally, absorbance was measured at 570 nm in a Spectramax M5e microplate reader (Molecular Devices, Sunnyvale, CA, USA). Viability percentages were calculated with respect to non-infected macrophages incubated with DMEM, for which a value of 100% of viability was established.
Monocytes infected at MOI 10 were incubated at 37°C with 5% CO2 for 1 h. Subsequently, gentamicin or streptomycin treatment (50 μg/mL) for 1 h was administered to eliminate extracellular bacteria (time 0), after which the bacterial count within the monocytes was assessed (Fernández-Bravo and Figueras, 2022). The infected monocytes were then incubated further with fresh DMEM and a maintenance dose of gentamicin or streptomycin (2 μg/mL) for an additional 4 h. Following this, the bacterial count within J7741.A cells was determined. Percent survival was calculated based on the bacterial count after 4 h of incubation and after gentamicin or streptomycin treatment (50 μg/mL) (Fernández-Bravo and Figueras, 2022).
Six different genes implicated in the immune response against pathogens were selected to quantify their transcription levels in macrophages (J774A.1) in response to the infections produced with the single and mixed strains in relation to the non-infected cells. The primers used to evaluate the expression of the selected genes were those from Murciano et al. (2017) and Zhao et al. (2017) and are listed in Table 1. The selected genes encoded for: cytokines and chemokines (TNF- α, IL-6, IL-8, CCL2 and CCL20), apoptosis (TP53) and pyroptosis (NLRP3 and IL-1β). After 4 h of infection at MOI 10, J774A.1 cells were washed twice with PBS, and the RNA was isolated from the samples by using the GenElute™ Mammalian Total RNA Miniprep Kit (Sigma-Aldrich). The cDNA was transcribed from total RNA by using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Inc. Hercules, CA, USA). A Real-time PCR was performed with cDNA for quantification by using the Power SYBR® green PCR Master Mix (Applied Biosystems®, Life Technologies, Glasgow, UK) on a StepOnePlus™ Real-Time PCR System (Applied Biosystems®). The thermal cycling conditions were: 94°C for 5 min, followed by 40 cycles of 30s at 94°C, 30s at 61°C, 30s at 72°C, and finally 20s at 80°C. Threshold cycle (CT) values were obtained to establish the relative RNA levels of the tested genes, using the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene as a housekeeping gene of reference. The relative gene expression was determined by using the delta–delta Ct (2–∆∆Ct) method that relays on the signal from the real-time polymerase chain reaction, as done in previous studies (Murciano et al., 2015; Fernández-Bravo and Figueras, 2022).
Overnight cultures of both strains were mixed in a ratio 1:1, spotted on nonselective LB agar plates, and incubated for 4 h at 37°C. The number of each strain before and after incubation for 4 h in mixtures was determined by serial dilution and plating on bacterial selective medium ampicillin dextrin agar (ADA) and CIN for Aeromonas and Yersinia, respectively, (Fernández-Bravo et al., 2019). Percent survival was obtained based on the number of bacteria after 4 h of incubation in relation to the number before incubation (Murciano et al., 2017).
In vitro antimicrobial susceptibility testing of both single strains and mixed cultures was determined by BD BBL™ Sensi-Disc™ (Oxoid, London, UK). The commercial antibiotics used for Aeromonas and Yersinia infections and other included as controls were ciprofloxacin (30 μg), trimethoprim (25 μg), piperacillin-tazobactam (100/10 μg), chloramphenicol (10 μg), penicillin (10 μg), cefepime (30 μg), amikacin (30 μg), amoxicillin-clavulanic (20/10 μg), imipenem (10 μg), ampicillin (10 μg), gentamicin (10 μg), aztreonam (30 μg), cefotaxime (30 μg), cephalotin (30 μg), ceftazidime (30 μg), tobramycin (10 μg), and ertapenem (10 μg). First, a fresh culture with a turbidity of 0.5 McFarland was swabbed onto the surface of Mueller-Hinton agar (MHA) (HiMedia, Mumbai, India). For the mixed culture of Aeromonas and Yersinia the concentration was calculated 1:1 with a final turbidity of 0.5 McFarland. The antibiotic disks were placed on the MHA surface using sterile forceps, and the agar plates were incubated at 37°C for 24 h. The inhibitory zones were interpreted according to the measurements provided by both the Clinical and Laboratory Standards Institute (CLSI, 2020) and the European Committee on Antimicrobial Susceptibility Testing Guidelines (EUCAST, 2021). Finally, as a control, the amount of each strain in the co-culture with the antibiotic gentamicin, after the antimicrobial test, was determined by collecting all the bacteria, performing a serial dilution and plating on selective bacterial medium ampicillin dextrin agar (ADA) and CIN for Aeromonas and Yersinia, respectively, (Fernández-Bravo et al., 2019).
All the experiments were performed in triplicate and the statistical significance was determined by using the Student’s two-tailed t-test or two-way ANOVA at p < 0.05 using the GraphPad Prism 6.0 (GraphPad Software, CA, USA). Analysis was chosen based on the type of experiment.
The preliminary identification of the strain 1185C with the MALDI-TOF Biotyper, considering exclusively the first match in the identification and applying manufacturer recommended cut-off, resulted in a score lower than 2.0 for A. hydrophila (1.893) and higher for A. caviae (2.051). The strain 1186C showed a higher score than 2.0 for Y. enterocolitica (2.121). A BLASTn analysis using the obtained rpoD and 16S rRNA sequences revealed 99% similarity with a strain of A. caviae (accession number PP404053) and Y. enterocolitica (accession number PP280108), respectively. Furthermore, the phylogenetic trees constructed with the rpoD and 16S rRNA gene of these strains and the sequences of the type strains of Aeromonas spp. and Yersinia spp. revealed that the sequences of the isolated strains clustered with the sequences of the type strains of A. caviae and Y. enterocolitica (Supplementary Figures 1, 2).
The capacity of the single and mixed infections to induce cell damage in macrophages (J774A.1) was evaluated through the release of LDH to the cell culture supernatant is shown in Figure 1. All infections, regardless of mixed or single at MOI 10, resulted in a significant degree of cell damage at 4 h (p < 0.05) when compared to the non-infected cells. Furthermore, it was found that the degree of cell damage increased at 6 h (p < 0.05) (Figure 1). The strain of Y. enterocolitica (1186C) was able to induce a higher degree (p < 0.05) of cell damage than the strain of A. caviae (1185C) at 4 h (Figure 1A). However, no significant difference in cell damage between Y. enterocolitica and A. caviae was observed at 6 h (Figure 1B). In addition, co-infection with both strains 1185C and 1186C caused significantly higher cell damage than the single infection, independently of the exposure time (p < 0.05) (Figure 1).
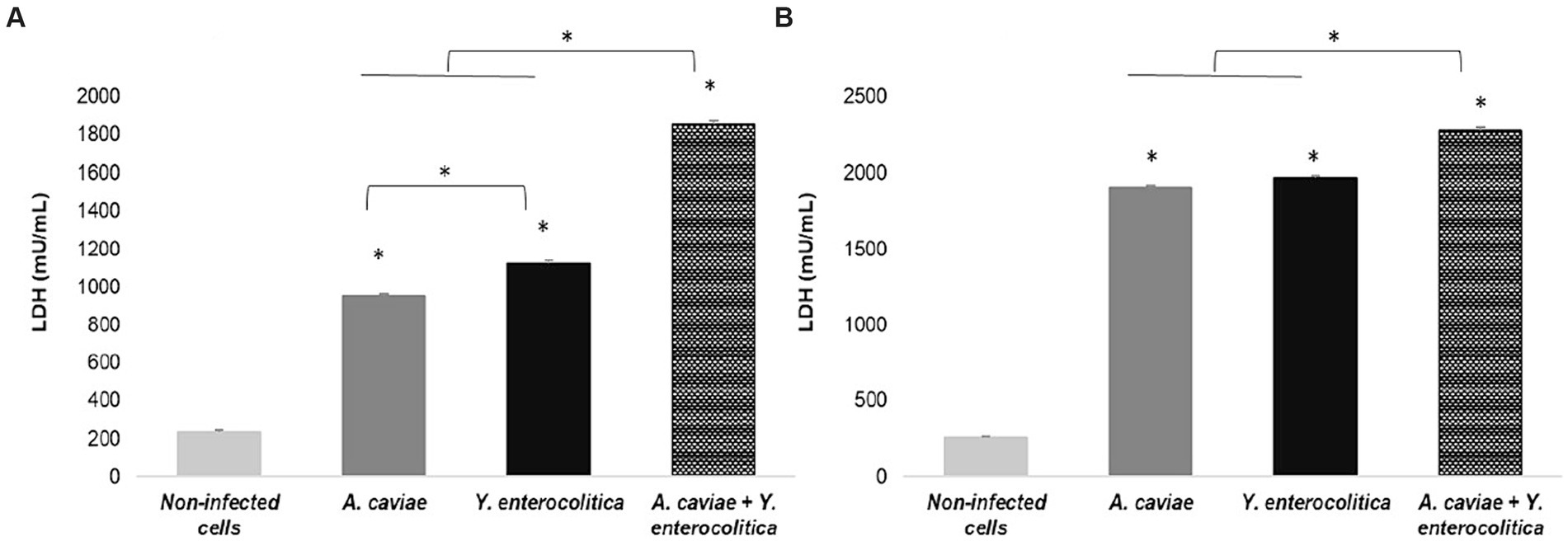
Figure 1. Cell damage in J774A.1 macrophages induced at 4 h (A) and 6 h (B) by the single and mixed infection at MOI 10 in relation to the non-infected cells, measured by the release of lactate dehydrogenase (LDH) enzyme. Asterisks indicate statistically significant difference *p < 0.05.
As shown in Figure 2, the mixed infection at 4 h with Y. enterocolitica 1186C and A. caviae 1185C at MOI 10 showed a significant higher intracellular survival (p < 0.05) with a 53.21% of survival percentage than the single infections. The viability in the mixed culture of Yersinia was 32.11% being higher than that of Aeromonas (21.10%). The percentage of survival with the single infections was higher (p < 0.05) after Y. enterocolitica (41.54%) infection in comparison with A. caviae (34.32%).
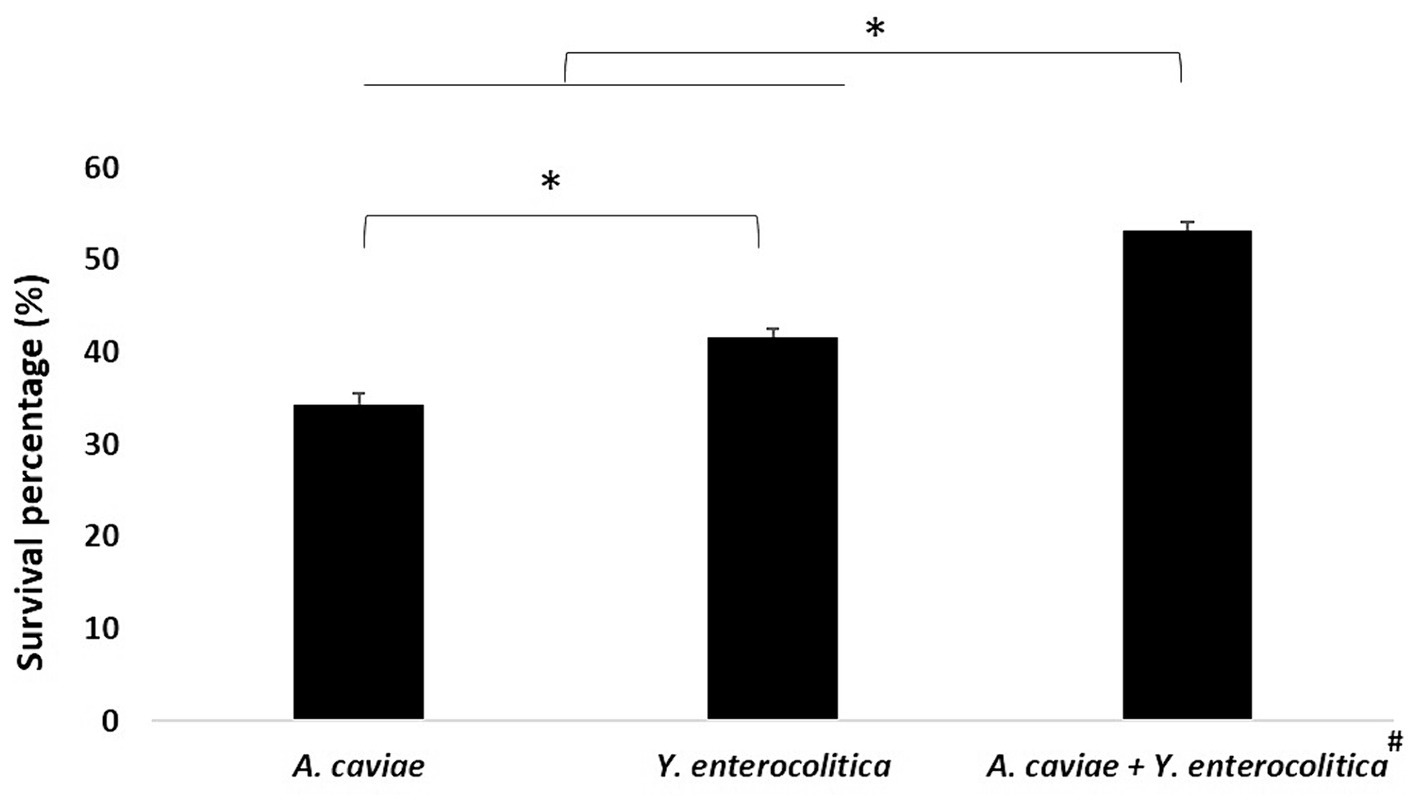
Figure 2. Intracellular survival at 4 h in J774A.1 macrophages after the single and mixed infections at MOI 10. Asterisks indicate statistically significant difference *p < 0.05. The viability of Yersinia (32.11%) was higher than that Aeromonas (21.10%)#.
The results on viability of infected macrophages incubated with different single and mixed cultures, measured as the reduction of MTT, are shown in Figure 3. Infected macrophages with Y. enterocolitica (1186C), showed a significant reduction in their viability (p < 0.05), 100 to 53.87% in relation to A. caviae (1185C), 100 to 65.78%. The highest level of viability was observed for macrophages incubated with the mixed culture (34.89%).
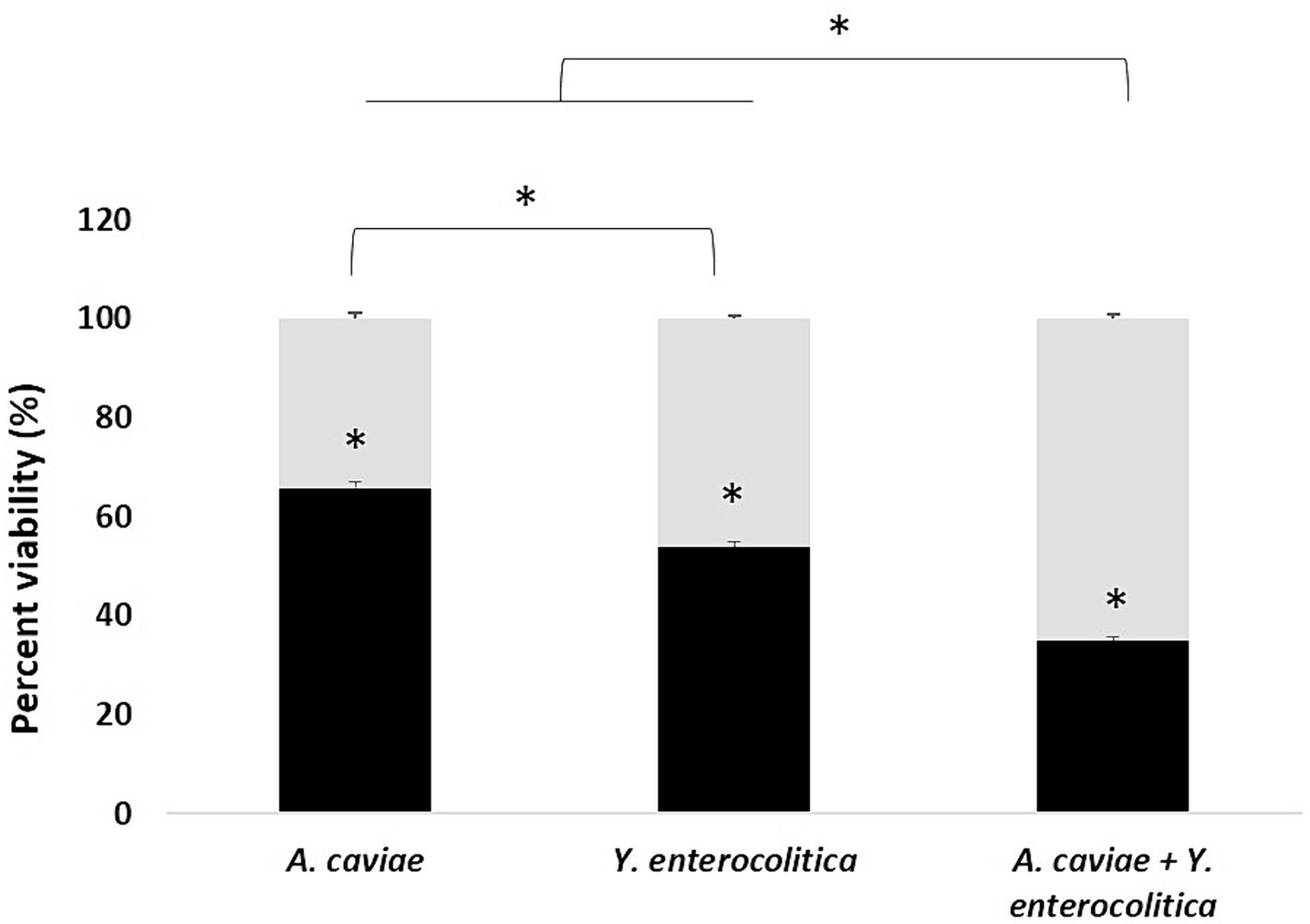
Figure 3. Viability percentage of J774A.1 macrophages at 5 h after infection with the single and mixed infection at MOI 10. Asterisks indicate statistically significant difference p < 0.05*.
All macrophage infections induced the expression of the genes encoding cytokine TNF-α and chemokines (CCL2, CCL20), with a significant increase (p < 0.05) in the transcript expression levels in relation to the non-infected cells (Figure 4). In addition, the TNF-α and CCL2 gene expression levels were significantly higher after infection with the Y. enterocolitica strain 1186C in comparison with the A. caviae strain 1185C. However, the expression of the CCL20 gene was significantly higher after infection with the latter strain (1185C) than with the Y. enterecolitica (1186C) infection (p < 0.05) (Figure 4). The transcriptional level after the macrophage infections with the mix of both strains was higher for the three genes TNF-α, CCL2 and CCL20 in comparison to the single infections (p < 0.05). However, the expression level of the proinflammatory cytokine genes IL-6 and IL-8 was close to the limit of detection for all single and mixed infections and no significant differences were found compared to non-infected cells (Supplementary Figure 3).
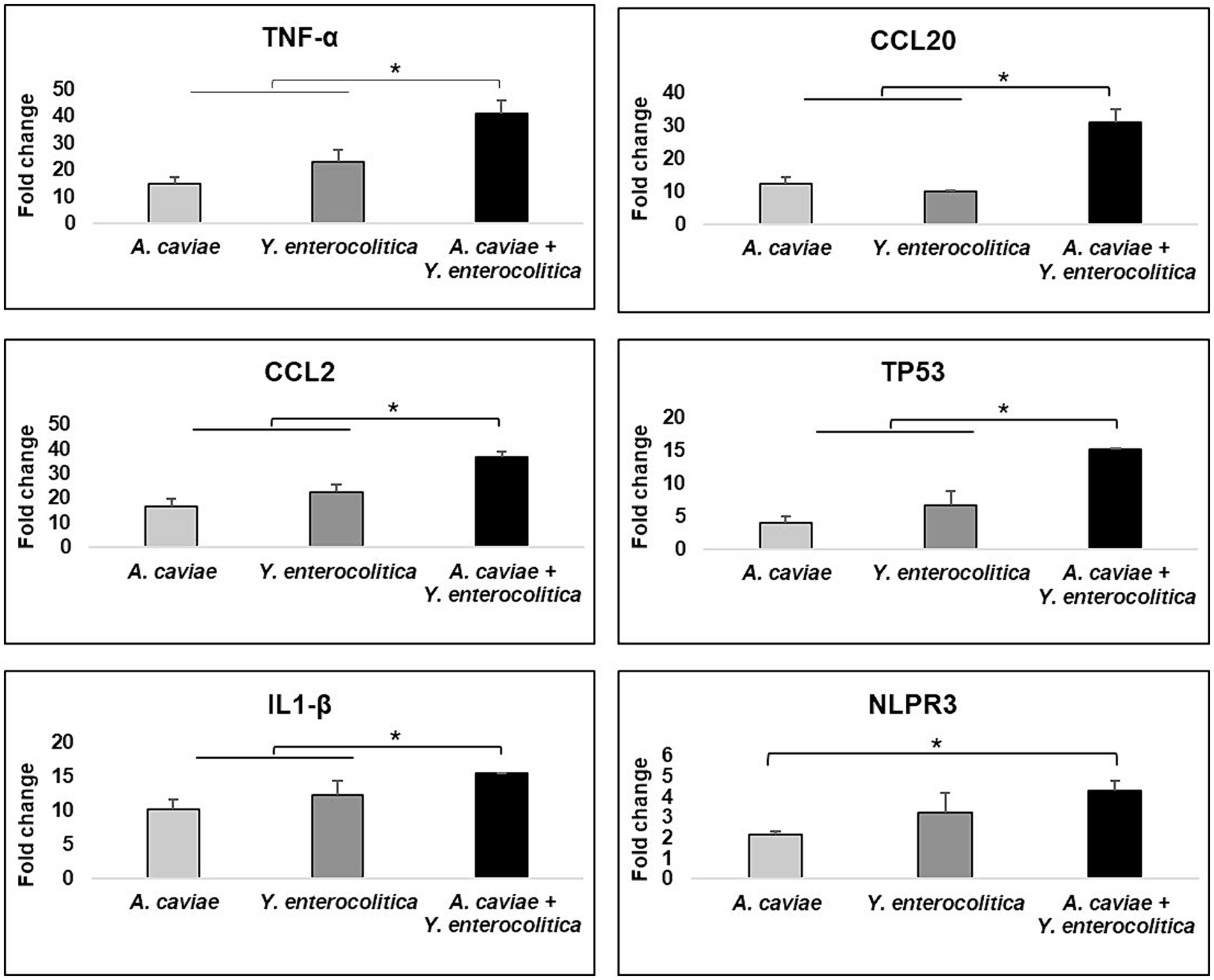
Figure 4. Gene expression profile of J774A.1 macrophages in relation to the non-infected cells induced by the single and mixed infections at MOI 20 d1termined by RT-qPCR. Transcript levels of the genes were normalized to the expression of the GAPDH gene. Expression fold change respect to the non-infected cells was calculated using the comparative ∆∆Ct method. Asterisks indicate statistically significant difference *p < 0.05.
The transcriptional level of TP53 apoptosis gene increased after infection with all single and mixed cultures (p < 0.05) (Figure 4). The expression of the TP53 gene showed no significant differences between the single infections with Y. enterocolitica 1186C and A. caviae 1185C (Figure 4). However, the expression levels of TP53 were significantly higher after the infection was produced with the mix of both strains in comparison with the single infections (p < 0.05) (Figure 4).
The NLRP3 and IL1-β genes which are associated with pyroptosis, i.e., cell death mediated by the formation of the inflammasome, were overexpressed in J774A.1 cells in response to all infections when compared with the non-infected cells (p < 0.05) (Figure 4). The expression levels of both genes were significantly higher after the single infection with Y. enterocolitica strain 1186C in comparison with A. caviae strain 1185C. The upregulation of IL-1β showed an increase (p < 0.05) when J774A.1 cells were infected with a mixed culture of both strains. No significant differences in the level of the NLRP3 expression were observed when comparing the mixed infection with the Y. enterocolitica single infection. However, significant differences in the expression of the NLRP3 were detected (p < 0.05) for the mixed infection in comparison with the A. caviae single infection (Figure 4).
When the A. caviae strain 1185C was mixed 1:1 with the Y. enterocolitica strain 1186C during 4 h and plated on ADA and CIN agar, the colony count for A. caviae strain 1185C decreased from 100 to 73% (Figure 5). However, the number of colonies for the strain Y. enterocolitica 1186C increased from 100 to 127% in mixed culture after 4 h (Figure 5).
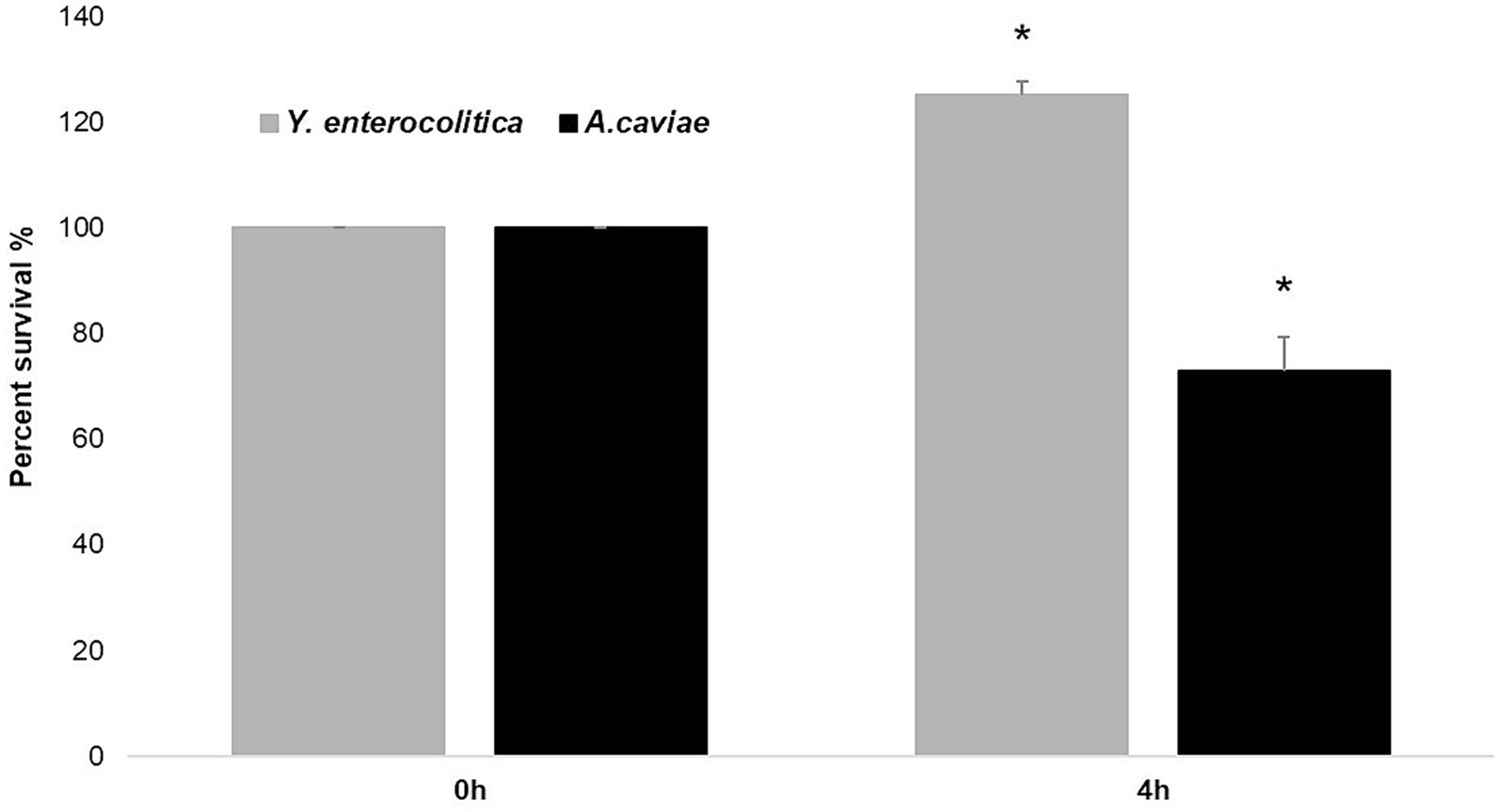
Figure 5. Killing effect of Y. enterocolitica to A. caviae after mixed culture during 4 h in a ratio of 1: 1. Asterisks indicate statistically significant difference *p < 0.05.
The antimicrobial susceptibility results showed that individually, the A. caviae 1185C and the Y. enterocolitica 1186C strains were resistant to penicillin, amoxicillin/clavulanic, ampicillin and cephalothin and with an intermediate resistance to trimethoprim. For the other antibiotics, these strains were sensible (Table 2). However, the co-culture of both strains showed a different antimicrobial pattern in comparison with the single strains for ciprofloxacin, trimethoprim, chloramphenicol, gentamicin and ertapenem, increasing the resistance to these antibiotics. The results of co-cultures after gentamicin treatment showed that the inhibition halos overlapped between Aeromonas and Yersinia and the amount of bacteria in relation to the total colonies was 37.15% and 62.85%, respectively.
The infections caused by Aeromonas can involve more than one strain or strains belonging to Aeromonas or other genera (Lamy et al., 2009; Chao et al., 2013; Barraud et al., 2020; Lin et al., 2020; Vithiya and Raja, 2022). However, to the best of our knowledge, very few studies have investigated the potential interactions of the isolated bacteria. Mosser et al. (2015), Fernández-Bravo et al. (2019), and Robert et al. (2023) showed that the presence of two Aeromonas strains can sometimes increase the virulence in the host. Therefore, the interest of this study resides in evaluating whether a mixed infection between A. caviae and Y. enterocolitica isolated from the same patient can increase virulence and what are the differences in terms of antimicrobial susceptibility.
Our results showed that Y. enterocolitica induces higher cell damage on macrophages than A. caviae after 4 h of infection. However, the cell damage was higher after co-infecting the macrophages with a mixed culture of both bacteria. Therefore, the dual infection with A. caviae and Y. enterocolitica increases the pathogenicity. Consistent results were obtained in the MTT assay being the highest survival percentage after single infection in relation to the mixed infection with both bacteria. Furthermore, intracellular survival was higher after infection with Y. enterocolitica compared to A. caviae, and the survival rate was higher after mixed infection. These results agree with those obtained by Mosser et al. (2015) that demonstrated using the Caenorhabditis elegans animal model an increase of the virulence after infection with two Aeromonas strains in relation to a single strain, indicating a synergism between both strains. In a recent study performed by Barraud et al. (2020) in which they investigate the behavior of two strains of Aeromonas veronii, recovered from an infected flap after leech therapy, all the analyses showed that the virulence of both recovered strains did not differ when tested individually but virulence was enhanced in the mixed infection. The authors showed that their results suggest that the outcome of the infection could have been worsened due to the synergies produced by the two isolates. However, Robert et al. (2023) using Drosophila melanogaster as a model, detected various effects depending on the strains used for co-cultures, with virulence sometimes more significant and others less or the same. Xu et al. (2019) demonstrated that the presence of polymicrobial infections between Pseudomonas aeruginosa and Staphylococcus aureus induces biofilm formation, increasing the level of pathogenicity. The recent literature review has shown an increase in virulence factor production by P. aeruginosa in a polymicrobial environment with S. aureus (Little et al., 2021). Some Yersinia and Aeromonas species have type three secretion systems (T3SS), and both are capable of inflicting gastroenteritis via the fecal-oral route of infection (Frey and Origgi, 2016; Romero et al., 2016; Mandal et al., 2017). This T3SS acts as a vehicle that can inject or translocate different toxins in the host immune cells and increase virulence (Rosenzweig and Chopra, 2013; Rangel et al., 2019). Therefore, considering the relevance of this finding, further studies must be carried out to elucidate the responsible mechanism of such behavior.
In relation to the expression of immune-related genes, higher transcriptional levels were observed after mixed infection in comparison with the single infections, except for the NLRP3 pyroptosis gene that recognizes pathogen-associated molecular patterns (PAMPs) to induce the activation of the immune response (Kumar et al., 2011). In a previous study using the monocytic human cell line THP-1 as in vitro infection model, Fernández-Bravo and Figueras (2022) investigated the innate human immune response against six commonly found clinical Aeromonas species, evaluating cell damage, intracellular survival and the expression of 11 immune-related genes and results demonstrated that A. caviae induce a cytokine storm that resulted in an increase in virulence. In the present study, results suggest that after the mixed infection, the virulence would increase due to the overexpression of the immune-related genes. In addition, this previous study suggested that different pathways of cell death were activated depending on the Aeromonas species. For example, in A. caviae both routes (apoptosis and pyroptosis) were induced, as it was shown to occur in the present study by the overexpression of TP53 and NLRP3 genes. These findings on the immune response are very interesting since they reinforce the hypothesis that the innate immune response is species-specific for Aeromonas, since the results of this study with A. caviae are similar to those obtained in the previous study by Fernández-Bravo and Figueras (2022).
The antimicrobial patterns of the bacteria varied when they were analyzed in pure cultures or in mixed cultures. Both single bacteria were susceptible to ciprofloxacin and ertapenem, however, thesusceptibility was intermediate in the mixed culture. In addition, the single cultures were susceptible to trimethoprim, chloramphenicol, and gentamicin, while the mixed cultures were resistant to these antibiotics. This is relevant because some of these antibiotics, i.e., ertapenem, trimethoprim, or ciprofloxacin are used empirically for the treatment of Aeromonas and Yersinia and this may hamper the effectivity of the treatment (González-Torralba et al., 2018; Dhanapala et al., 2021). These results are in agreement with previous studies of polymicrobial infections between other bacterial genera. In fact, Vega et al. (2013) demonstrated that the mixed culture of E. coli and S. typhimurium increases the tolerance to ciprofloxacin as occurred in our study. Our hypothesis about this increase in tolerance is that in polymicrobial infections Quorum-Sensing would have a role in inducing the expression of different genes, as well as increasing the production of enzymes and even the formation of a barrier that reduces the permeability of drugs.
The growth dynamics in our study showed that when the strains were mixed for the contact-killing assay in LB, colony counts of Y. enterocolitica increased (and the A. caviae decreased at 4 h). This result could be associated with the antagonism of both bacteria as occurred in a previous study with two strains of A. hydrophila in which one eliminated the other (Fernández-Bravo et al. 2019). In the latter study, the type six secretion system (T6SS) of one strain produced the TseC effector toxin that directly killed the other in vitro and in vivo. Our results in the present study could be associated with the presence and the overexpression of the T6SS in Y. enterocolitica. Ongoing studies (unpublished) have demonstrated the presence of genes that encode different proteins related to the T6SS by PCR in both strains. However, further studies to clarify the functionality of this T6SS by transcriptomic analysis are necessary to prove this suspicion.
To our knowledge, this is the first study that demonstrates the interactions between co-isolated bacteria of Aeromonas with other different genera associated with a diarrheal process and their potential impacts on the evolution of the disease and its treatment. This study shows the need for performing further studies to better understand the complexity of occurring mixed infections and pretends to alert clinicians of the importance to take into account the potential interactions among these bacteria in relation to the treatment efficacy and outcome of the disease.
Mixed infection with A. caviae and Y. enterocolitica induces differences in the activation of the immune response in macrophages compared with single infections, as well as an increase in the cell damage that may enhance the infection disease process. Apparently, both synergistic and antagonistic effects may coexist as well as an increase in the antimicrobial resistance of the antimicrobials used to empirically treat infections produced by these bacteria. The latter may potentially hamper antimicrobial treatment. This study indicates for the first time the importance of performing further studies on Aeromonas mixed infections to clarify the specific mechanisms of those interactions and their impact on the evolution of infectious diseases.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
AF-B: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. GR: Data curation, Methodology, Writing – review & editing. MF: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was performed with the ESCMID Research Grant 2024 awarded to AF-B.
The authors thank the staff in the Microbiology Unit at the University Rovira i Virgili (URV) and to Carme Sanmartí Solé for technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1328766/full#supplementary-material
Adamowicz, E. M., Flynn, J., Hunter, R. C., and Harcombe, W. R. (2018). Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J. 12, 2723–2735. doi: 10.1038/s41396-018-0212-z
Andersson, J. A., Fitts, E. C., Kirtley, M. L., Ponnusamy, D., Peniche, A. G., Dann, S. M., et al. (2016). New role for FDA-approved drugs in combating antibiotic-resistant bacteria. Antimicrob. Agents Chemother. 60, 3717–3729. doi: 10.1128/AAC.00326-16
Barraud, O., Robert, A., Laval, L., Ruimy, R., Morquin, D., Boyer, L., et al. (2020). It takes two to tango: two Aeromonas isolates combine virulence and multidrug resistance in flap infection following leech therapy. Clin. Microbiol. Infect. 26, 793–794. doi: 10.1016/j.cmi.2019.12.021
Chao, C. M., Lai, C. C., Tang, H. J., Ko, W. C., and Hsueh, P. R. (2013). Skin and soft-tissue infections caused by Aeromonas species. Eur. J. Clin. Microbiol. Infect. Dis. 32, 543–547. doi: 10.1007/s10096-012-1771-y
CLSI. Clinical and laboratory standards institute (CLSI). Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI Suppl M100. Wayne, PA: CLSI. (2020)
Dhanapala, P. M., Kalupahana, R. S., Kalupahana, A. W., Wijesekera, D. P. H., Kottawatta, S. A., Jayasekera, N. K., et al. (2021). Characterization and antimicrobial resistance of environmental and clinical Aeromonas species isolated from fresh water ornamental fish and associated farming environment in Sri Lanka. Microorganisms 9:2106. doi: 10.3390/MICROORGANISMS9102106
EUCAST (2021). EUCAST. Available at: https://www.eucast.org/ (Accessed December 21, 2021)
Fernández-Bravo, A., and Figueras, M. J. (2020). An update on the genus Aeromonas: taxonomy, epidemiology, and pathogenicity. Microorganisms 8:129. doi: 10.3390/MICROORGANISMS8010129
Fernández-Bravo, A., and Figueras, M. J. (2022). Immune response of the monocytic cell line THP-1 against six Aeromonas spp. Front. Immunol. 13:875689. doi: 10.3389/FIMMU.2022.875689
Fernández-Bravo, A., Fort-Gallifa, I., Ballester, F., Pujol, I., Gomez-bertomeu, F., Domínguez, M., et al. (2020). A case of Aeromonas trota in an immunocompromised patient with diarrhea. Microorganisms 8:399. doi: 10.3390/microorganisms8030399
Fernández-Bravo, A., Kilgore, P. B., Andersson, J. A., Blears, E., Figueras, M. J., Hasan, N. A., et al. (2019). T6SS and ExoA of flesh-eating Aeromonas hydrophila in peritonitis and necrotizing fasciitis during mono- and polymicrobial infections. Proc. Natl. Acad. Sci. U. S. A. 116, 24084–24092. doi: 10.1073/pnas.1914395116
Figueras, M. J., and Beaz-Hidalgo, R. (2015). “Aeromonas infections in humans” in Aeromonas. ed. J. Graf (Norfolk: Academic Press), 65–108.
Frey, J., and Origgi, F. C. (2016). Type III secretion system of Aeromonas salmonicida undermining the host’s immune response. Front. Mar. Sci. 3:130. doi: 10.3389/FMARS.2016.00130/BIBTEX
González-Torralba, A., García-Esteban, C., and Alós, J. I. (2018). Enteropathogens and antibiotics. Enferm. Infecc. Microbiol. Clin. 36, 47–54. doi: 10.1016/j.eimc.2015.06.015
Grave, I., Rudzate, A., Nagle, A., Miklasevics, E., and Gardovska, D. (2022). Prevalence of Aeromonas spp. infection in pediatric patients hospitalized with gastroenteritis in Latvia between 2020 and 2021. Children 9:1684. doi: 10.3390/children9111684
Grim, C. J., Kozlova, E. V., Ponnusamy, D., Fitts, E. C., Sha, J., Kirtley, M. L., et al. (2014). Functional genomic characterization of virulence factors from necrotizing fasciitis-causing strains of Aeromonas hydrophila. Appl. Environ. Microbiol. 80, 4162–4183. doi: 10.1128/AEM.00486-14
Hering, N. A., Fromm, A., Kikhney, J., Lee, I. F. M., Moter, A., Schulzke, J. D., et al. (2016). Yersinia enterocolitica affects intestinal barrier function in the colon. J. Infect. Dis. 213, 1157–1162. doi: 10.1093/infdis/jiv571
Janda, J. M., and Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
Kumar, H., Kawai, T., and Akira, S. (2011). Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30, 16–34. doi: 10.3109/08830185.2010.529976
Lamy, B., Kodjo, A., and Laurent, F. (2009). Prospective nationwide study of Aeromonas infections in France. J. Clin. Microbiol. 47, 1234–1237. doi: 10.1128/JCM.00155-09
Latif, F. L. (2015). Aeromonas, un microorganismo ambiental de importancia en salud humana y animal. PhD thesis. Available at: http://www.tdx.cat/handle/10803/334686
Lin, C. C., Chen, Y. C., Goh, Z. N. L., Seak, C. K., Seak, J. C. Y., Shi-Ying, G., et al. (2020). Wound infections of snakebites from the venomous protobothrops mucrosquamatus and viridovipera stejnegeri in Taiwan: bacteriology, antibiotic susceptibility, and predicting the need for antibiotics—a BITE study. Toxins 12:575. doi: 10.3390/toxins12090575
Little, W., Black, C., and Smith, A. C. (2021). Clinical implications of polymicrobial synergism effects on antimicrobial susceptibility. Pathogens 10:144. doi: 10.3390/pathogens10020144
Mandal, D., Basu, A., and Biswas, M. (2017). Structural analysis of major translocator - chaperone interaction from Ysa-Ysp T3SS of Yersinia enterocolitica. Can J Biotechnol 1., 1–2. doi: 10.24870/cjb.2017-a214
Mosser, T., Talagrand-Reboul, E., Colston, S. M., Graf, J., Figueras, M. J., Jumas-Bilak, E., et al. (2015). Exposure to pairs of Aeromonas strains enhances virulence in the Caenorhabditis elegans infection model. Front. Microbiol. 6:1218. doi: 10.3389/fmicb.2015.01218
Murciano, C., Hor, L. I., and Amaro, C. (2015). Host-pathogen interactions in Vibrio vulnificus: responses of monocytes and vascular endothelial cells to live bacteria. Future Microbiol. 10, 471–487. doi: 10.2217/fmb.14.136
Murciano, C., Te, L. C., Fernández-Bravo, A., Hsieh, T. H., Fouz, B., Hor, L. I., et al. (2017). MARTX toxin in the zoonotic serovar of vibrio vulnificus triggers an early cytokine storm in mice. Front. Cell. Infect. Microbiol. 7:332. doi: 10.3389/fcimb.2017.00332
Nesbakken, T. (2021). “Yersinia” in Foodborne infections and intoxications. J. Glenn Morris, Jr. and Duc J. Vugia Eds. (Norfolk: Academic Press), 187,199.
Nguyen, A. T., and Oglesby-Sherrouse, A. G. (2016). Interactions between Pseudomonas aeruginosa and Staphylococcus aureus during co-cultivations and polymicrobial infections. Appl. Microbiol. Biotechnol. 100, 6141–6148. doi: 10.1007/s00253-016-7596-3
Ponnusamy, D., Kozlova, E. V., Sha, J., Erova, T. E., Azar, S. R., Fitts, E. C., et al. (2016). Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc. Natl. Acad. Sci. USA 113, 722–727. doi: 10.1073/pnas.1523817113
Rangel, L. T., Marden, J., Colston, S., Setubal, J. C., Graf, J., and Gogarten, J. P. (2019). Identification and characterization of putative Aeromonas spp. T3SS effectors. PLoS One 14:e0214035. doi: 10.1371/JOURNAL.PONE.0214035
Robert, A., Talagrand-Reboul, E., Figueras, M.-J., Ruimy, R., Boyer, L., and Lamy, B. (2023). Drosophila melanogaster systemic infection model to study altered virulence during polymicrobial infection by Aeromonas. Pathogens 12:405. doi: 10.3390/pathogens12030405
Romero, A., Saraceni, P. R., Merino, S., Figueras, A., Tomás, J. M., and Novoa, B. (2016). The animal model determines the results of Aeromonas virulence factors. Front. Microbiol. 7:1574. doi: 10.3389/fmicb.2016.01574
Rosenzweig, J. A., and Chopra, A. K. (2013). Modulation of host immune defenses by aeromonas and yersinia species: convergence on toxins secreted by various secretion systems. Front. Cell. Infect. Microbiol. 3:70. doi: 10.3389/fcimb.2013.00070
Savin, C., Martin, L., Bouchier, C., Filali, S., Chenau, J. Ô., Zhou, Z., et al. (2014). The Yersinia pseudotuberculosis complex: characterization and delineation of a new species, Yersinia wautersii. Int. J. Med. Microbiol. 304, 452–463. doi: 10.1016/j.ijmm.2014.02.002
Soler, L., Yáñez, M. A., Chacon, M. R., Aguilera-Arreola, M. G., Catalán, V., Figueras, M. J., et al. (2004). Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. Int. J. Syst. Evol. Microbiol. 54, 1511–1519. doi: 10.1099/IJS.0.03048-0
Tsuchiya, S., Yamabe, M., Yamaguchi, Y., Kobayashi, Y., Konno, T., and Tada, K. (1980). Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26, 171–176. doi: 10.1002/ijc.2910260208
Vega, N. M., Allison, K. R., Samuels, A. N., Klempner, M. S., and Collins, J. J. (2013). Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. U. S. A. 110, 14420–14425. doi: 10.1073/pnas.1308085110
Versalovic, J., Koeuth, T., and Lupski, R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19, 6823–6831. doi: 10.1093/NAR/19.24.6823
Vithiya, G., and Raja, S. (2022). Clinical significance and outcome of Aeromonas infection among 19 patients-a descriptive study from South India. Indian J. Med. Microbiol. 40, 299–302. doi: 10.1016/j.ijmmb.2022.03.001
Wilson, A., Fegan, N., and Turner, M. S. (2022). Co-culture with Acinetobacter johnsonii enhances benzalkonium chloride resistance in Salmonella enterica via triggering lipid a modifications. Int. J. Food Microbiol. 381:109905. doi: 10.1016/j.ijfoodmicro.2022.109905
Xu, Z., Xie, J., Soteyome, T., Peters, B. M., Shirtliff, M. E., Liu, J., et al. (2019). Polymicrobial interaction and biofilms between Staphylococcus aureus and Pseudomonas aeruginosa: an underestimated concern in food safety. Curr. Opin. Food Sci. 26, 57–64. doi: 10.1016/j.cofs.2019.03.006
Yuwono, C., Wehrhahn, M. C., Liu, F., Riordan, S. M., and Zhang, L. (2021). The isolation of Aeromonas species and other common enteric bacterial pathogens from patients with gastroenteritis in an Australian population. Microorganisms 9:1440. doi: 10.3390/microorganisms9071440
Zhao, X., Zhang, C., Hua, M., Wang, R., Zhong, C., Yu, J., et al. (2017). NLRP3 inflammasome activation plays a carcinogenic role through effector cytokine IL-18 in lymphoma. Oncotarget 8:108571, –108583. doi: 10.18632/oncotarget.21010
Keywords: Aeromonas caviae, Yersinia enterocolitica, mixed infections, macrophages, immune response, antimicrobial susceptibility
Citation: Fernández-Bravo A, Recio G and Figueras MJ (2024) Interactions between Aeromonas caviae and Yersinia enterocolitica isolated from a case of diarrhea: evaluation of antimicrobial susceptibility and immune response of infected macrophages. Front. Microbiol. 15:1328766. doi: 10.3389/fmicb.2024.1328766
Received: 19 January 2024; Accepted: 09 April 2024;
Published: 24 April 2024.
Edited by:
Juan A. Ayala, Autonomous University of Madrid, SpainReviewed by:
Jose Ramos-Vivas, Universidad Europea del Atlántico, SpainCopyright © 2024 Fernández-Bravo, Recio and Figueras. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria José Figueras, bWFyaWFqb3NlLmZpZ3VlcmFzQHVydi5jYXQ=; Ana Fernández-Bravo, YW5hLmZlcm5hbmRlekB1cnYuY2F0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.