- 1China Animal Health and Epidemiology Center, Qingdao, Shandong, China
- 2College of Life Sciences and Food Engineering, Hebei University of Engineering, Handan, Hebei, China
Avian astrovirus can infect a variety of poultry species and cause viral diarrhea, with a wide epidemic range strong pathogenicity and a high incidence. Among them, Duck astrovirus 3(DAstV-3), Duck astrovirus 4(DAstV-4), Goose astrovirus 1(GoAstV-1) and Goose astrovirus 2(GoAstV-2) are four types of astroviruses newly discovered in waterfowl in recent years. In order to realize the rapid detection of these four kinds of waterfowl stellate viruses, specific primers and probes were engineered to target a highly conserved region of ORF1b gene of DAstV-3, GoAstV-1 and GoAstV-2 and the ORF2 gene of DAstV-4, and a quadruple fluorescence quantitative RT-PCR method was developed. The results indicate that the method established in this study has good specificity and no cross reactivity with other pathogens. This method can detect viruses with a minimum concentration of 1 × 101 copies/μL for DAstV-4, GoAstV-1 and GoAstV-2, respectively, while the minimum concentration for DAstV-3 is 1 × 102 copies/μL. Compared with the routinely used RT-PCR method, the limit of detection by the multiplex RT-PCR lower. Both intra- and inter-assay variability tests revealed excellent reproducibility. This method was then used to analyze 269 field samples, and the results were verified by genome sequencing. In conclusion, this study presents a sensitive, accurate, and specific method for detecting DAstV-3, DAstV-4, GoAstV-1, and GoAstV-2 in a single reaction, enabling the monitoring and differential diagnosis of these four types of waterfowl astroviruses.
1 Introduction
Astroviruses are a class of small, non-enveloped virus with a single-stranded RNA that belong to the family Astroviridae. This family contains two genera: Mamastroviruses (MAstVs) and Avastroviruses (AAstVs) according to the different types of infected hosts (Koci and Schultz-Cherry, 2002). The genus Mamastrovirus includes astrovirus species that have been isolated from humans and a variety of mammals, such as porcine astrovirus, mink astrovirus, bovine astrovirus, and canine astrovirus. Avastrovirus includes astrovirus species that have been isolated from poultry, such as Turkey astrovirus, Chicken astrovirus, Avian nephritis virus, Duck astrovirus, and Goose astrovirus, etc. (De Benedictis et al., 2011; Chen et al., 2021). In addition, some astroviruses have not been classified. The genetic evolution of astroviruses shows that astroviruses have the potential to cross species barriers and adapt to new hosts (Roach and Langlois, 2021). The genome length of the virus is 6.8~7.9 kb, consisting of a 5′ untranslated region (UTR), three open reading frames (ORFs)-ORF1a, ORF1b, and ORF2, a 3′ UTR, and a poly-A tail. ORF1a and ORF1b are highly conserved regions that code for nonstructural proteins. ORF1b encodes RNA dependent RNA polymerase (RDRP) while ORF2 encodes the viral capsid protein. ORF2 is a variable region that interacts specifically with virus-specific antibodies and helps in determining phylogenetic relationships between viruses (Strain et al., 2008). The research of the Astrovirus Research Group of the International Committee of Taxonomy of Viruses (ICTV) shows that the classification of viruses is generally based on the genetic distance between the host and the whole amino acid sequence of the structural protein, but the viruses with more than 75% homology in the whole gene sequence of ORF2 protein should be considered as members of the same genotype (Tan et al., 2023).
Avian astrovirus can infect a variety of avian species and cause intestinal and other internal organ diseases, such as broiler dysplasia syndrome, gosling gout infection, duck viral hepatitis, viral diarrhea (Chu et al., 2012; Donato and Vijaykrishna, 2017; Niu et al., 2018). At present, the reported waterfowl astrovirus mainly includes six types, Duck astrovirus 1 (DAstV-1), DAstV-2, DAstV-3, DAstV-4, Goose astrovirus 1 (GoAstV-1) and GoAstV-2. DAstV-1 and DAstV-2 have been officially classified by the ICTV as astroviruses and the main symptom of infection being duck hepatitis, which has a rapid onset (Yugo et al., 2016; Wei et al., 2022). DAstV-3 was first reported in Peking ducks in 2013. In recent years, it has been widely spread with high infection rate and morbidity, which is harmful to ducklings and has a certain effect on duck embryo hatching, leading to serious damage to the chorioallantoic membrane, growth retardation and embryo death (Liu et al., 2014; Li et al., 2021). There were few reports about DAstV-4, and its pathogenicity needs further investigation (Liao et al., 2015). GoAstV-1 causes urate deposition in the liver, ureters, kidneys, joints, and other internal organs, leading to swelling of the kidneys and liver (Zhang et al., 2022). GoAstV-2 is a serious infectious disease found in 2016–2018, which is characterized by gout, hemorrhage and swelling of the kidneys, accompanied by urate deposits, white feces and paralysis (Chen et al., 2020; Zhu et al., 2022). Among them, waterfowl-derived astroviruses, such as DAstV-3, DAstV-4, GoAstV-1, and GoAstV-2, were newly discovered over recent years, which bring great harm to poultry industry and there were no effective biological agents to prevent and control. Therefore, accurate detection method is very important for the diagnosis, prevention and control of this disease.
At present, the detection methods for waterfowl astrovirus include virus isolation, RT-PCR (Reverse Transcription-Polymerase Chain Reaction), fluorescence quantitative RT-PCR, Loop-mediated isothermal amplification (LAMP), and so on (Liu et al., 2016; Yu et al., 2020; Wang et al., 2022). Virus isolation, RT-RT-PCR take a long time and are not suitable for rapid clinical detection. The LAMP technology takes a short time and can realize macroscopic observation, but is prone to false positive results. On the other hand, fluorescent quantitative RT-PCR has the advantages of simple operation, fewer steps, no need for agarose gel electrophoresis, and the lab capability of avoiding contamination to a certain extent; and simultaneously, the multiple fluorescent quantitative RT-PCR method simultaneously detects a plurality of viruses and realizes the detection of a plurality of pathogens.
This study intends to develop a rapid, sensitive and specific quadruple fluorescence quantitative RT-PCR method simultaneous detection of DAstV-3, DAstV-4, GoAstV-1 and GoAstV-2, and provides technical support for epidemiological investigation and prevention and control of waterfowl-derived astrovirus.
2 Materials methods
2.1 Virus and clinical samples
The DAstV-3, DAstV-4, GoAstV-1, GoAstV-2, Newcastle disease virus (NDV), H9 subtype avian influenza Virus (H9-AIV), Avian reovirus (ARV) were isolated and stored in the China Animal Health and Epidemiology Center, Qingdao, China. There were 269 clinical samples of throat and anal swabs from ducks and geese collected from 11 regions including Anhui, Fujian, Guangdong, Guangxi, Henan, Hubei, Hunan, Jiangsu, Jiangxi, Shandong and Sichuan.
2.2 Primer and probe design
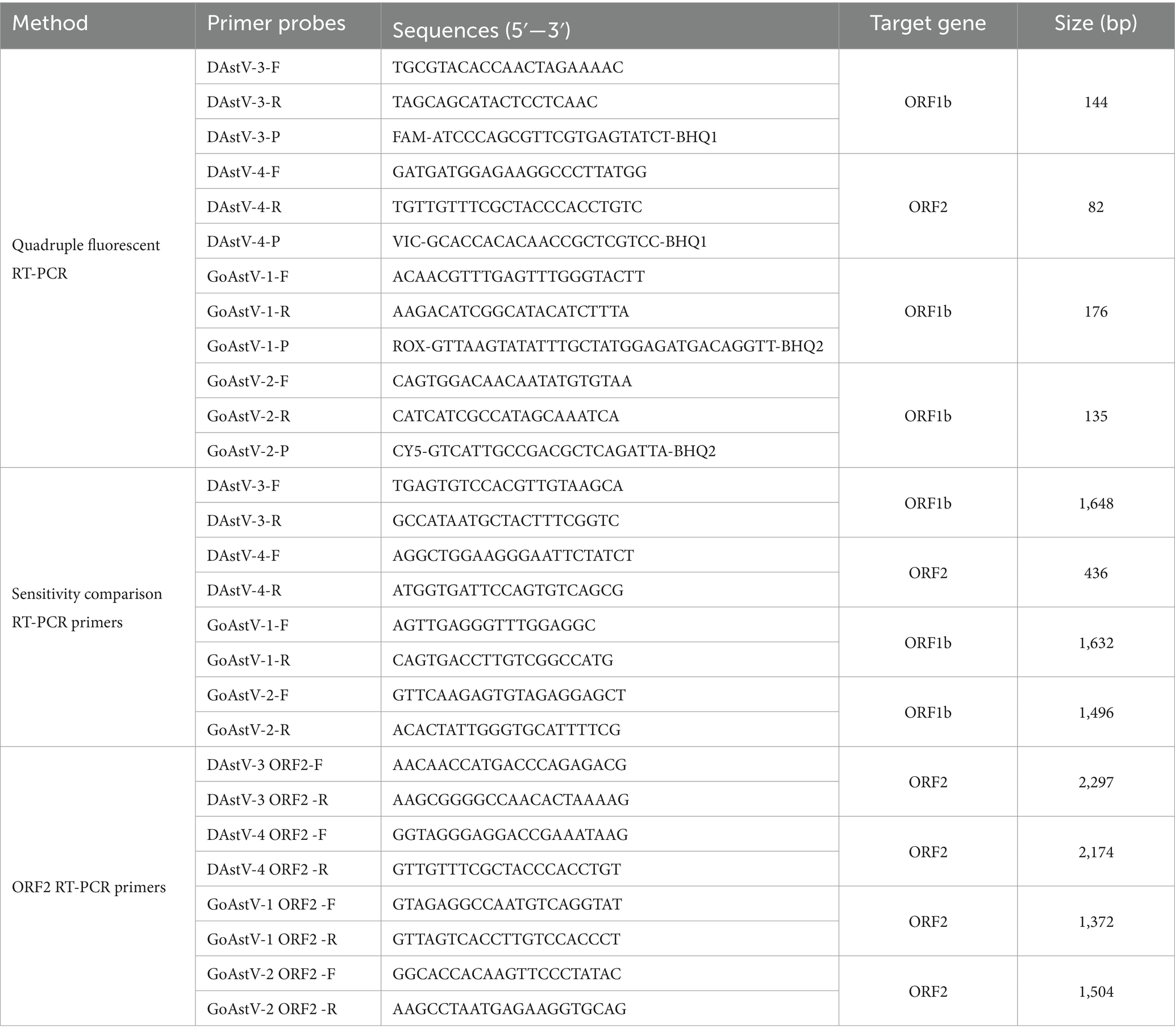
The Mega 7 software was used to compare the genomes of DAstV-3, DAstV-4, GoAstV-1 and GoAstV-2, respectively, to identify the conserved regions of the virus. The primer and probe were designed with reference to the sequence of DAstV-3 (accession number: KJ020899), DAstV-4 (accession number: JX624774), GoAstV-1 (accession number: MH410610) and GoAstV-2 (accession number: ON745304). Primers and probes used for fluorescent RT-PCR were designed based on conserved sequence of gene using Oligo 7, and RT-PCR primers were designed to construct standard templates. To avoid the mutual interference between the emission spectra of fluorescent groups affecting the accuracy and specificity of the detection results of the established method, and considering the brand and model of the quantitative fluorescence PCR instrument, FAM, VIC, ROX and CY5 were used to label the fluorescent probes, respectively, in this study. Details of the final primers and exo-probes are shown in Table 1. All primers and exo-probes were synthesized by Sangon Biotech (Shanghai) Co., Ltd.
2.3 Preparation of standard templates
The target gene fragment was amplified by RT-PCR using DAstV-3 ORF1bF/R, DAstV-4 ORF2F/R, GoAstV-1 ORF1bF/R, GoAstV-2 ORF1bF/R respective by HiScript High Fidelity One step RT-PCR Kit (Vazyme Biotech, China). The RT-PCR product was purified using a gel Recovery Kit (Thermo Fisher Scientific, China), then linked to the TA vector (Clone Smarter technologies, the United States of America) and transferred into Escherichia coli DH5α competent cells (Ruiboxingke biotechnology, China). Positive colonies were chosen for DNA sequencing identification after selection with ampicillincon LB plates. A plasmid Extraction Kit (Tiangen Biotech, China) was used to extract plasmids. Using Spe I incision enzyme to single enzyme of plasmids, and then the digested plasmid was purified by gel recovery kit (Takara Biotechnology, China). Using T7 (Yeasen Biotechnology, China) was transcribed kits for in vitro transcription, then using RNeasy Mini purification kits (QIAGEN Enterprise Management, China) to purification of transcription products to remove the impurity protein and various kinds of ions. The concentration of the recombinant plasmid was quantified using a NanoDrop 2000 spectrophotometer, and the copy number was calculated according to the formula: copies/μL = (6.02 × 1023) × ng/μL × 10−9/ (DNA length×660). Each plasmid was diluted from 1 × 107 to 1 × 100 copies/μL for use.
2.4 Condition optimization
The reaction condition of the quadruple fluorescence RT-PCR was optimized refer to requirements the Evo M-MLV One Step RT-qRT-PCR Kit II (Accurate Biotechnology, China), including the concentration of primers and probes, temperature of annealing.
2.5 Standard curve preparation
A quadruple fluorescence quantitative RT-PCR reaction system was configured, and the standard curves were generated and analyzed by using five gradients dilution standards of 1.0 × 106~1.0 × 102 copies/μL of four viruses as templates. RNase-free water was used as the negative control. All reactions were conducted in triplicates.
2.6 Specificity, sensitivity and reproducibility of quadruple fluorescence quantitative RT-PCR
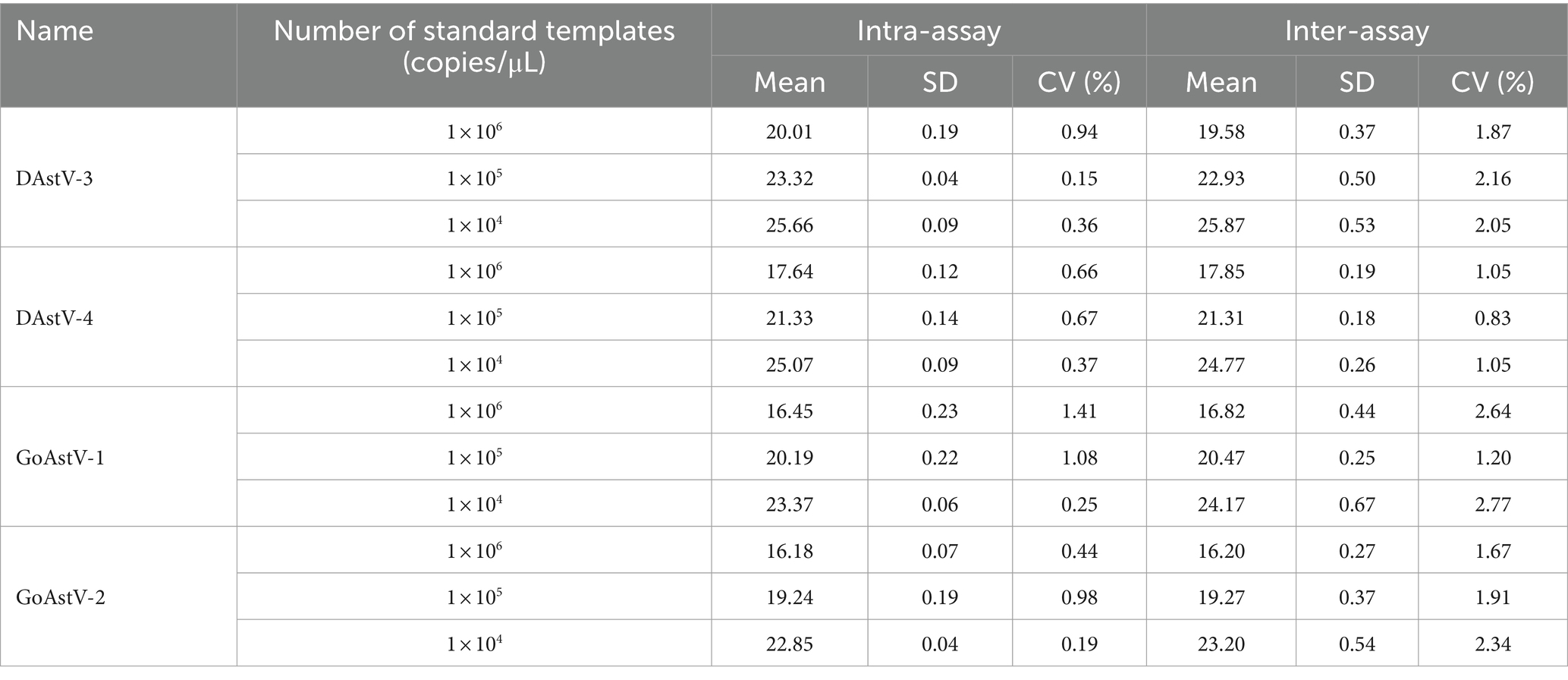
The specificity of the quadruple fluorescence quantitative RT-PCR assay was evaluated based on cross-reactivity with other goose and duck viruses, includingDAstV-1 H9-AIV, NDV, ARV. In order to test the sensitivity of this method, four virus standards 1.0 × 107~1.0 × 100 diluted 10 times were used as templates, and the amplification was carried out according to the established quadruple fluorescence quantitative RT-PCR method, and compared with the standard RT-PCR method (Table 2). The repeatability experiment was carried in triplicate under optimized reaction conditions using three serial 10-fold diluted from 1.0 × 106 copies/μL to 1.0 × 104 copies/μL of standard, and the intra- and inter-group coefficient of variation (CV) was calculated for each assay.
2.7 Clinical sample detection
To further verify the clinical potential of this quadruple fluorescence quantitative RT-PCR method assay, we tested 269 clinical samples from waterfowl. ORF2 sequence amplification was performed on the positive samples detected by multiplex fluorescence quantitative RT-PCR, and the primers for ORF2 amplification were established by our laboratory (Table 1).
3 Results
3.1 Quadruple fluorescence quantitative RT-PCR
To develop a quadruple fluorescence quantitative RT-PCR assay, different fluorescent-labeled target probes (DAstV-3 gene: FAM, DAstV-4 gene: VIC, GoAstV-1 gene: ROX, and GoAstV-2 gene: CY5) were first used. The concentration of recombinant plasmids was measured by NanoDrop 2000 spectrophotometer and the copy number of each virus plasmid was calculated. The initial concentration of virus was: 2.9 × 109 for DAstV-3, 4 × 1010 for DAstV-4, 2.6 × 1010 for GoAstV-1, 4.6 × 1010 for GoAstV-2. Based on the single fluorescence quantitative RT-PCR method, the quadruple fluorescence quantitative RT-PCR method was established, and the reaction system and annealing temperature were optimized by the quadruple fluorescence quantitative RT-PCR method. The results of the optimal reaction system was determined by optimization as follows: 10 μL 2× One step RT-PCR Buffer II, 0.5 μL Pro Taq HS DNA Polymerase, 0.5 μL Evo M-MLV RTase Enzyme Mix II, the primers (10 μmol/L) of DAstV-3, DAstV-4, GoAstV-1 and GoAstV-2 are 0.8 μL, 0.8 μL, 0.6 μL and 0.6 μL respectively, and the probes (10 μmol/L) are 0.8, 0.6, 0.8, and 0.6 μL, respectively. Finally, 2.0 μL of RNA template and RNase-free water were added to a final volume of 25 μL. All reactions were performed on a QuantStudio™ 5 Real-time fluorescence quantitative PCR instrument (Thermo Fisher Technologies, Shanghai, China). The detection program was as follows: 42°C for 5 min and 95°C 30s, followed by 40 cycles of 95°C for 5 s and 60°C 30s.
3.2 Standard curve for the quadruple fluorescence quantitative RT-PCR assay
Quadruple fluorescence quantitative RT-PCR was performed using 1.0 × 106 ~ 1.0 × 102 copies/μL of standards. The standard curves for DAstV-3, DAstV-4, GoAstV-1 and GoAstV-2 were drawn by plotting threshold cycle (Ct) on the x-axis and the log of the starting quantity on the y-axis. The standard equation are as follows (Figure 1): Y = −3.327Χ + 39,218, R2 = 0.998 (DAstV-3); Y = −3.587Χ + 39.685, R2 = 0.998 (DAstV-4); Y = −3.21Χ + 36.328, R2 = 0.998 (GoAstV-1); Y = −3.25Χ + 37.145, R2 = 0.997 (GoAstV-2).
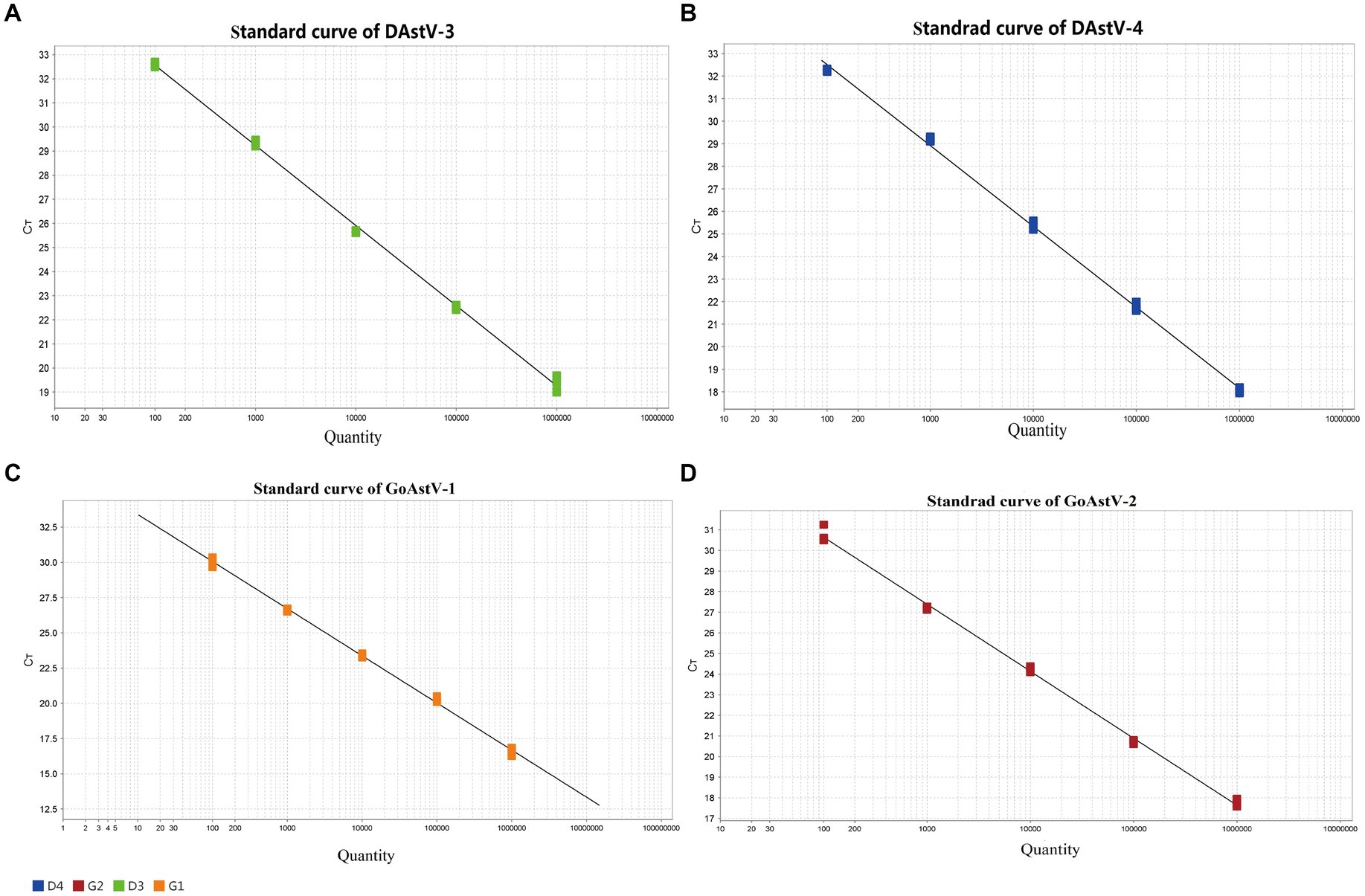
Figure 1. Establishment of a standard curve for the quadruple fluorescent quantitative RT-PCR assay. (A) Standard curve for DAstV-3; (B) Standard curve for DAstV-4; (C) Standard curve for GoAstV-1; (D) Standard curve for GoAstV-2.
3.3 Specificity of the quadruple fluorescence quantitative RT-PCR assay
The results showed that positive for the nucleic acids of DAstV-3, DAstV-4, GoAstV-1 and GoAstV-2, and negative for the DAstV-1, H9-AIV, NDV, ARV and Negative controls. The developed multiplex RT-PCR assay demonstrated high specificity and did not cross-react with other viruses tested (Figure 2).
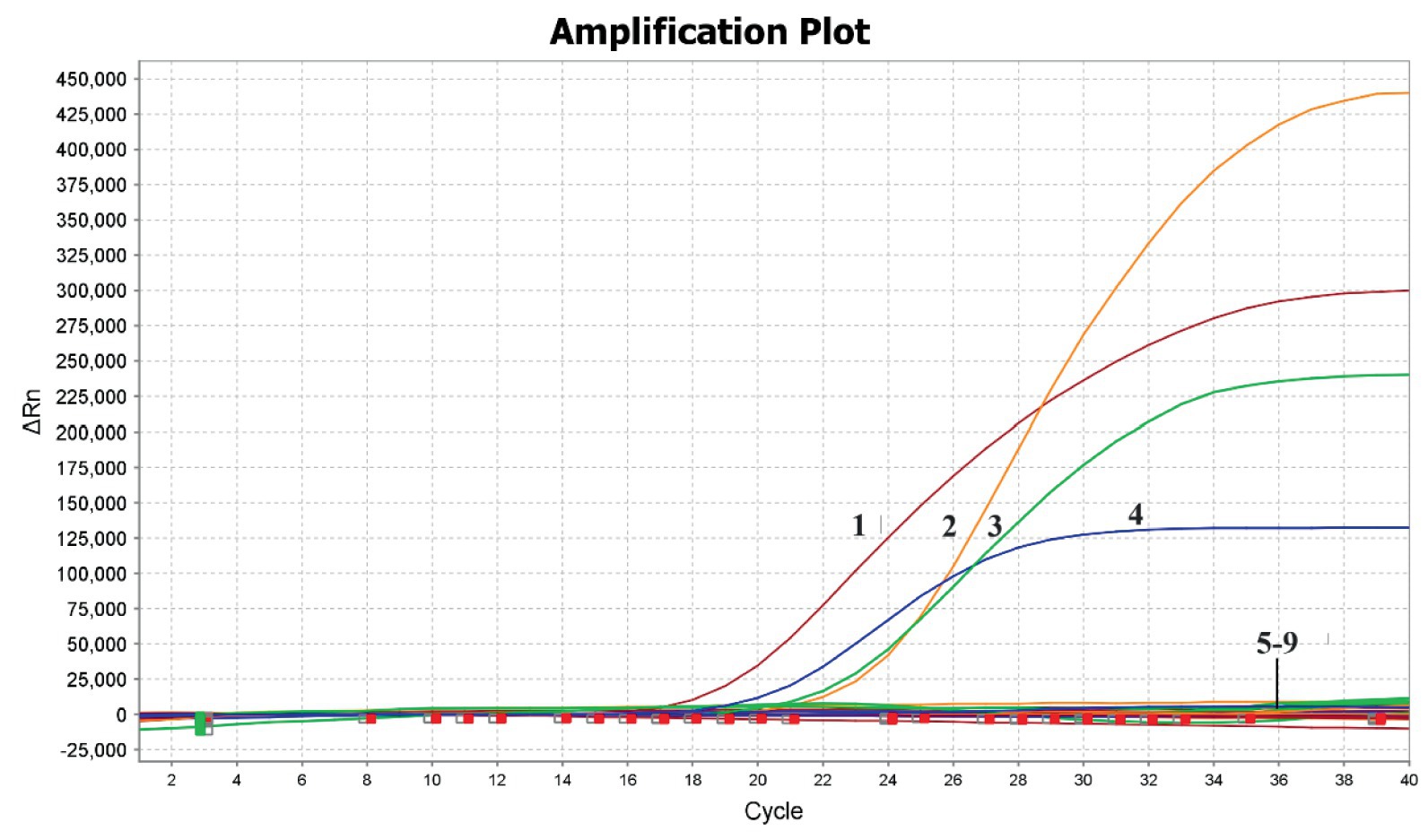
Figure 2. Specificity of the quadruple fluorescent quantitative RT-PCR assay. 1, GoAstV-2; 2, GoAstV-1; 3, DAstV-3; 4, DAstV-4; 5–9, DAstV-1, H9-AIV, NDV, ARV, Negative controls.
3.4 Sensitivity of the quadruple fluorescence quantitative RT-PCR assay
The sensitivity of the quadruple fluorescence quantitative RT-PCR assay was tested using serial 10-fold dilutions of the constructed four standard samples of waterfowl astrovirus. The results showed that this method has good sensitivity, while the sensitivity of DAstV-4, GoAstV-1 and GoAstV-2 was 101 copies/μL, respectively, and the sensitivity of DAstV-3 was 102 copies/μL (Figure 3). Whereas the sensitivity of the standard RT-PCR assay for DAstV-3 is 1 × 103 copies/μL and that of DAstV-4, GoAstV-1, GoAstV-2 is 1 × 102 copies/μL, respectively (Figure 4). Therefore, the sensitivity of the new method was 10 times higher than those of the conventional assay, respectively.
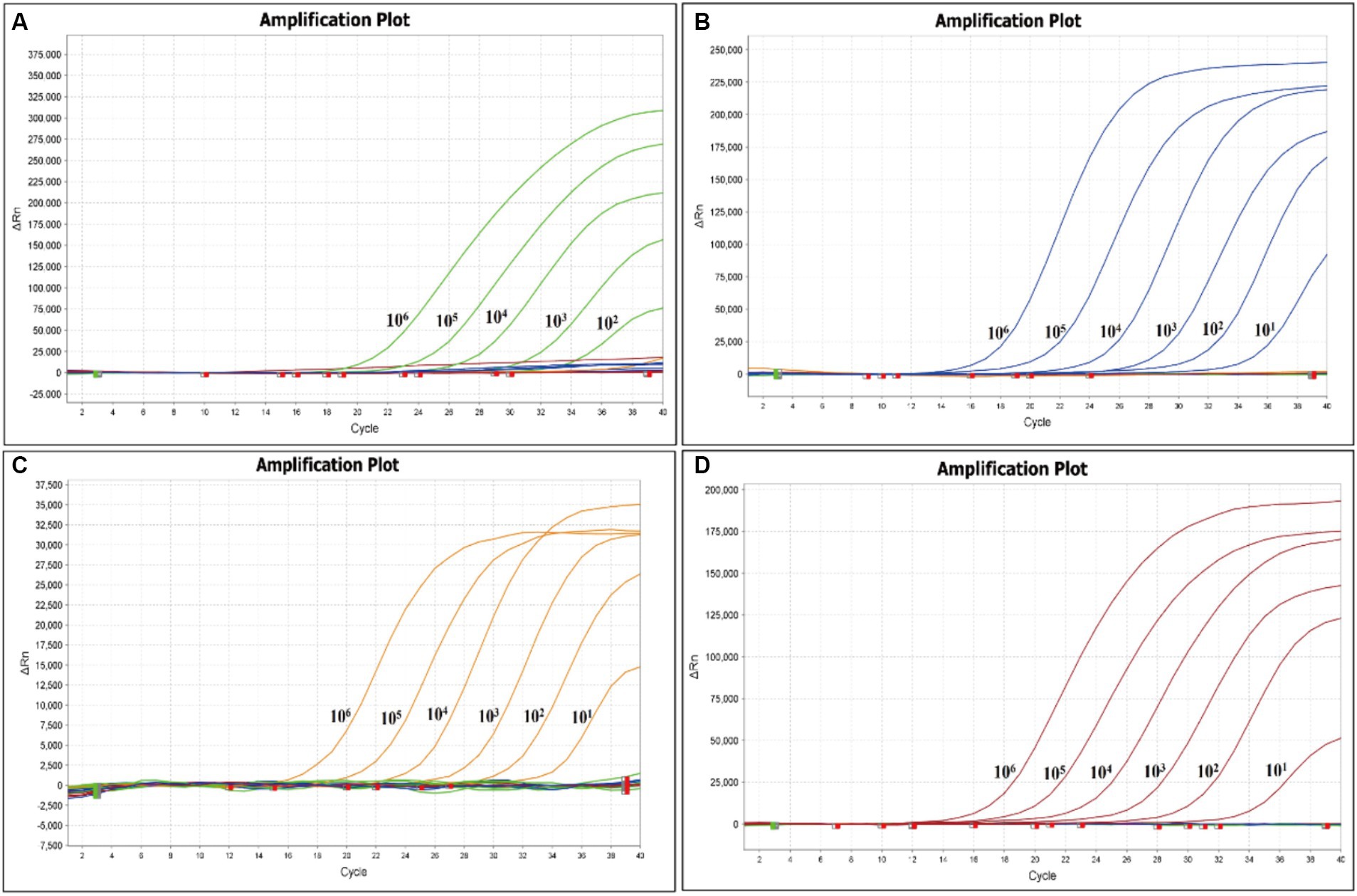
Figure 3. Sensitivity of the quadruple fluorescent quantitative RT-PCR assay. (A) Amplification plot for the DAstV-3; (B) Amplification plot for the DAstV-4; (C) Amplification plot for the GoAstV-1; (D) Amplification plot for the GoAstV-2.
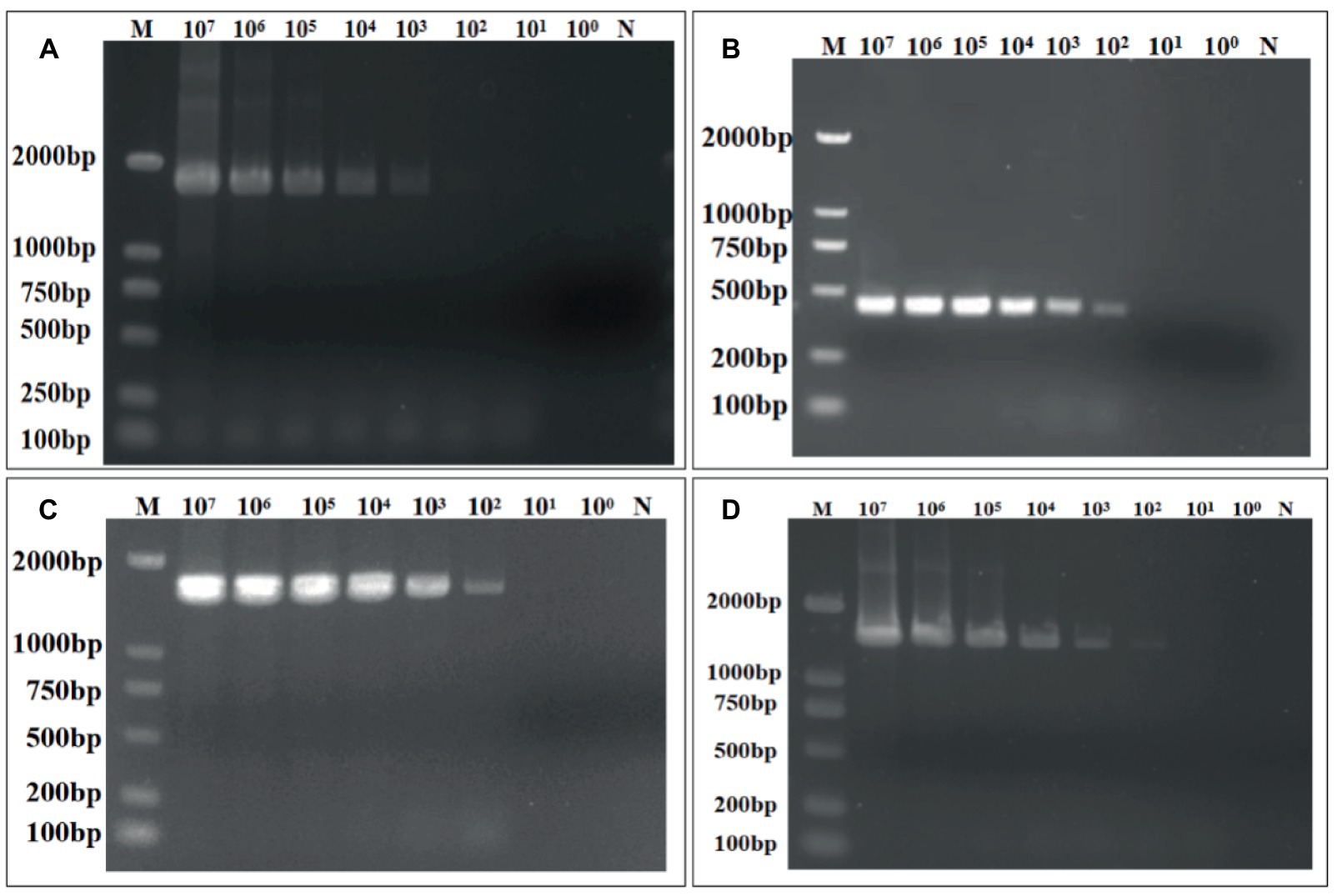
Figure 4. Sensitivity of the conventional RT-RT-PCR assay for the detection of DAstV-3, DAstV-4, GoAstV-1 and GoAstV-2. (A) DAstV-3 ORF1b; (B) DAstV-4 ORF2; (C) GoAstV-1 ORF1b; (D) GoAstV-2 ORF1b.
3.5 Repeatability and reproducibility of the quadruple fluorescence quantitative RT-PCR assay
The intra- and inter-assay CVs were evaluated at concentrations of 1.0 × 106, 1.0 × 105, 1.0 × 104 copies/μL of standard, respectively. The intra-assay CV. was <1.5% and the inter-assay CV was <3%, indicating that the quadruple fluorescence quantitative RT-PCR assay is repeatable and reproducible (Table 2).
3.6 Clinical sample detection
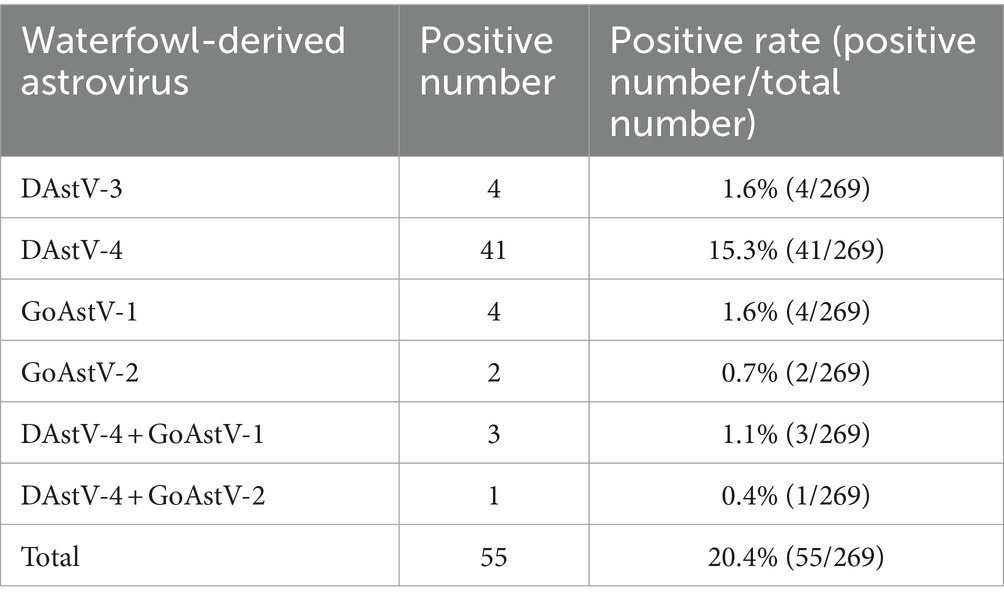
Based on the above results of sensitivity, we next sought to verify the capability of this quadruple fluorescence quantitative RT-PCR assay to detect clinical samples. As shown in Table 3, we tested to detect these viruses in clinical samples, 15.3% were found to be positive for DAstV-4, 1.6% for DAstV-3, 1.6% for GoAstV-1 and 0.7% for GoAstV-2. Besides, we also identified the presence of co-infection of DAstV-4 with GoAstV-1 and DAstV-4 with GoAstV-2, respectively. Sequencing was performed on RT-PCR products of the ORF2 gene, which verified the results of the quadruple fluorescence quantitative RT-PCR assay.
4 Discussion
DAstV-3, DAstV-4, GoAstV-1 and GoAstV-2 as newly discovered waterfowl-derived astrovirus that have caused reduced production performance and even death of ducks and geese in recent years, there is no clear classification. To control the disease, it is important to establish a reliable, it is important to establish a reliable, rapid, accurate, and sensitive method for the detection of DAstV-3, 4 and GoAstV-1, 2. At present, the main laboratory detection methods for avian astrovirus are molecular biological diagnostic techniques. Among them, fluorescence quantitative RT-PCR detection method is widely used in laboratory detection because of its short time consumption and high sensitivity. In previous studies, a TaqMan-based quantitative RT-PCR method was established for the detection of GoAstV-2 alone and a duplex RT-PCR method to simultaneously detect and differentiate between GoAstV-1 and GAstV-2 (Yi et al., 2022). However, it is more practical to develop a quadruplex quantitative RT-PCR method to simultaneously detect and discriminate between DAstV-3,4 and GoAstV-1,2 in field clinical samples.
The genome structure of AAstV consists of three open reading frames, among which ORF1b encodes RDRP, which is a conserved sequence of the virus. ORF2 encodes virus coat protein, which can react specifically with virus-specific antibodies and help to determine the phylogenetic relationship between viruses. In this study, in order to achieve rapid detection of DAstV-3, DAstV-4, GoAstV-1 and GoAstV-2, initially selected four conserved genes of waterfowl astroviruses, ORF1b, and designed specific primers and probes. However, when the primers and probes of DAstV-4 were in the ORF1b gene region, The specificity and sensitivity effects were not ideal, so attempts were made to design primer probes in the ORF2 region that determines the typing of avian astrovirus, which could cooperate with the other three waterfowl-derived astrovirus primer probes without cross-reaction. Therefore, in this study, a quadruple fluorescent quantitative RT-PCR method was established for the highly conserved ORF1b region of DAstV-3, GoAstV-1 and GoAstV-2 and the ORF2 region of DAstV-4. The method had good specificity and had no cross-reaction with other avian viruses. The sensitivity of the quadruple fluorescence quantitative RT-PCR for DAstV-3, 4 and GoAstV-1, 2 were 10 times higher than those of the conventional assay, respectively. The intra-assay and inter-assay coefficients of variation were less than 3%, suggesting good reproducibility.
The results of clinical samples showed that the positive rate of DAstV-4 was the highest, with 15.3%, that of DAstV-3 and GoAstV-1 was 1.6%, and that of GoAstV-2 was 0.7%. There are co-infections between DAstV-4 and GoAstV-1 and between DAstV-4 and GoAstV-2, but the specific infection and transmission mechanism need to be studied. In order to further verify the method, ORF2 amplification was carried out on the samples which were positive by quadruple fluorescence quantitative detection, and the amplified products were sent to sequencing. The RT-PCR products were sequenced and the results confirmed that quadruple fluorescence quantitative RT-PCR assay was reliable.
In summary, a quadruple fluorescent quantitative RT-PCR method was successfully established in this study. The assay proved to be specific, sensitive, reliable, and feasible for testing field clinical samples. This newly developed assay represents a useful tool for the diagnosis and detection of DAstV-3, 4 and GoAstV-1, 2, and their co-infection, and should facilitate molecular epidemiological investigations of these viruses.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal studies were approved by Animal Welfare Committee of the China Animal Health and Epidemiology Center; It is attributed to China Animal Health and Epidemiology Center. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
YL: Funding acquisition, Writing – original draft, Writing – review & editing. JL: Methodology, Writing – original draft, Writing – review & editing. FZ: Data curation, Writing – review & editing. JS: Investigation, Writing – review & editing. CD: Conceptualization, Investigation, Writing – review & editing. YF: Investigation, Writing – review & editing. GM: Validation, Writing – review & editing. WJ: Methodology, Writing – review & editing. XY: Data curation, Writing – review & editing. GL: Data curation, Writing – review & editing. HL: Funding acquisition, Resources, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program (2022YFC2305101).
Acknowledgments
We are grateful to all the authors who have contributed to this manuscript, and we would like to thank you for giving your journal valuable time to review this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Chen, Q., Yu, Z., Xu, X., Ji, J., Yao, L., Kan, Y., et al. (2021). First report of a novel goose astrovirus outbreak in Muscovy ducklings in China. Poult. Sci. 100:101407. doi: 10.1016/j.psj.2021.101407
Chen, H., Zhang, B., Yan, M., Diao, Y., and Tang, Y. (2020). First report of a novel goose astrovirus outbreak in Cherry Valley ducklings in China. Transbound. Emerg. Dis. 67, 1019–1024. doi: 10.1111/tbed.13418
Chu, D. K., Leung, C. Y., Perera, H. K., Ng, E. M., Gilbert, M., Joyner, P. H., et al. (2012). A novel group of avian astroviruses in wild aquatic birds. J. Virol. 86, 13772–13778. doi: 10.1128/jvi.02105-12
De Benedictis, P., Schultz-Cherry, S., Burnham, A., and Cattoli, G. (2011). Astrovirus infections in humans and animals—molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 11, 1529–1544. doi: 10.1016/j.meegid.2011.07.024
Donato, C., and Vijaykrishna, D. (2017). The broad host range and genetic diversity of mammalian and avian Astroviruses. Viruses 9:102. doi: 10.3390/v9050102
Koci, M. D., and Schultz-Cherry, S. (2002). Avian astroviruses. Avian Pathol. 31, 213–227. doi: 10.1080/03079450220136521
Li, Y., Cui, Y., Liu, H., Wang, J., Li, Y., and Wang, Y. (2021). Establishment of duplex SYBR green I-based real-time RT-PCR assay for simultaneous detection of duck hepatitis a Virus-1 and duck Astrovirus-3. Avian Dis. 65, 281–286. doi: 10.1637/aviandiseases-D-20-00115
Liao, Q., Liu, N., Wang, X., Wang, F., and Zhang, D. (2015). Genetic characterization of a novel astrovirus in Pekin ducks. Infect. Genet. Evol. 32, 60–67. doi: 10.1016/j.meegid.2015.02.025
Liu, N., Jiang, M., Wang, M., Wang, F., Zhang, B., and Zhang, D. (2016). Isolation and detection of duck astrovirus CPH: implications for epidemiology and pathogenicity. Avian Pathol. 45, 221–227. doi: 10.1080/03079457.2016.1143549
Liu, N., Wang, F., and Zhang, D. (2014). Complete sequence of a novel duck astrovirus. Arch. Virol. 159, 2823–2827. doi: 10.1007/s00705-014-2141-0
Niu, X., Tian, J., Yang, J., Jiang, X., Wang, H., Chen, H., et al. (2018). Novel goose Astrovirus associated gout in Gosling, China. Vet. Microbiol. 220, 53–56. doi: 10.1016/j.vetmic.2018.05.006
Roach, S. N., and Langlois, R. A. (2021). Intra- and cross-species transmission of Astroviruses. Viruses 13:1127. doi: 10.3390/v13061127
Strain, E., Kelley, L. A., Schultz-Cherry, S., Muse, S. V., and Koci, M. D. (2008). Genomic analysis of closely related astroviruses. J. Virol. 82, 5099–5103. doi: 10.1128/jvi.01993-07
Tan, Z., Zhai, H., Sun, R., Sun, Z., Xie, R., Zhang, L., et al. (2023). Complete genome sequence and phylogenetic analysis of a goose astrovirus isolate in China. Braz. J. Microbiol. 54, 427–434. doi: 10.1007/s42770-022-00854-7
Wang, A., Liu, L., Zhang, S., Ye, W., Zheng, T., Xie, J., et al. (2022). Development of a duplex real-time reverse transcription-polymerase chain reaction assay for the simultaneous detection of goose astrovirus genotypes 1 and 2. J. Virol. Methods 310:114612. doi: 10.1016/j.jviromet.2022.114612
Wei, F., Wang, Y., Wang, Q., Yang, J., Jiang, X., He, D., et al. (2022). The isolation and characterization of duck astrovirus type-1 remerging in China. Transbound. Emerg. Dis. 69, 2890–2897. doi: 10.1111/tbed.14444
Yi, Z., Ding, R., Cao, R., Sun, W., Sun, M., Dong, Y., et al. (2022). Development of a duplex Taq man real-time RT-PCR assay for simultaneous detection of goose astrovirus genotypes 1 and 2. J. Virol. Methods 306:114542. doi: 10.1016/j.jviromet.2022.114542
Yu, Z., Zhang, D., Yang, K., Bai, C., Li, Y., Li, J., et al. (2020). A simple and rapid diagnostic method to detect new goose astrovirus using reverse-transcription loop-mediated isothermal amplification. 3 Biotech 10:20. doi: 10.1007/s13205-019-2006-z
Yugo, D. M., Hauck, R., Shivaprasad, H. L., and Meng, X. J. (2016). Hepatitis virus infections in poultry. Avian Dis. 60, 576–588. doi: 10.1637/11229-070515-Review.1
Zhang, F., Li, H., Wei, Q., Xie, Q., Zeng, Y., Wu, C., et al. (2022). Isolation and phylogenetic analysis of goose astrovirus type 1 from goslings with gout in Jiangxi province, China. Poult. Sci. 101:101800. doi: 10.1016/j.psj.2022.101800
Keywords: DAstV-3, DAstV-4, GoAstV-1, GoAstV-2, quadruple fluorescence quantitative RT-PCR
Citation: Li Y, Luo J, Zhang F, Shang J, Deng C, Feng Y, Meng G, Jiang W, Yu X, Liu G and Liu H (2024) Establishment and application of quadruple fluorescence quantitative RT-PCR method for the identification of waterfowl astrovirus. Front. Microbiol. 15:1328243. doi: 10.3389/fmicb.2024.1328243
Edited by:
Torsten Seuberlich, University of Bern, SwitzerlandReviewed by:
Qing Pan, Qingdao Agricultural University, ChinaJianqiang Ye, Yangzhou University, China
Liu Sidang, Shandong Agricultural University, China
Copyright © 2024 Li, Luo, Zhang, Shang, Deng, Feng, Meng, Jiang, Yu, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohui Yu, eXV4aWFvaHVpQGNhaGVjLmNu; Guanhui Liu, bGl1Z3Vhbmh1aS4xOTg2QDE2My5jb20=; Hualei Liu, bGl1aHVhbGVpQGNhaGVjLmNu
†These authors have contributed equally to this work and share first authorship
 Yang Li
Yang Li Juan Luo
Juan Luo Fuyou Zhang
Fuyou Zhang Jiajing Shang
Jiajing Shang Chunran Deng1,2
Chunran Deng1,2 Guanhui Liu
Guanhui Liu Hualei Liu
Hualei Liu