
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 02 February 2024
Sec. Systems Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1327010
This article is part of the Research Topic Emerging Technologies for Viability Enumeration of Live Microorganisms View all 12 articles
 Thulani Sibanda1,2,3†
Thulani Sibanda1,2,3† Tlaleo Azael Marole1†
Tlaleo Azael Marole1† Ursula Louise Thomashoff1
Ursula Louise Thomashoff1 Mapitsi S. Thantsha4†
Mapitsi S. Thantsha4† Elna M. Buys1*†
Elna M. Buys1*†Bifidobacterium species are essential members of a healthy human gut microbiota. Their presence in the gut is associated with numerous health outcomes such as protection against gastrointestinal tract infections, inflammation, and metabolic diseases. Regular intake of Bifidobacterium in foods is a sustainable way of maintaining the health benefits associated with its use as a probiotic. Owing to their global acceptance, fermented dairy products (particularly yogurt) are considered the ideal probiotic carrier foods. As envisioned in the definition of probiotics as “live organisms,” the therapeutic functionalities of Bifidobacterium spp. depend on maintaining their viability in the foods up to the point of consumption. However, sustaining Bifidobacterium spp. viability during the manufacture and shelf-life of fermented dairy products remains challenging. Hence, this paper discusses the significance of viability as a prerequisite for Bifidobacterium spp. probiotic functionality. The paper focuses on the stress factors that influence Bifidobacterium spp. viability during the manufacture and shelf life of yogurt as an archetypical fermented dairy product that is widely accepted as a delivery vehicle for probiotics. It further expounds the Bifidobacterium spp. physiological and genetic stress response mechanisms as well as the methods for viability retention in yogurt, such as microencapsulation, use of oxygen scavenging lactic acid bacterial strains, and stress-protective agents. The report also explores the topic of viability determination as a critical factor in probiotic quality assurance, wherein, the limitations of culture-based enumeration methods, the challenges of species and strain resolution in the presence of lactic acid bacterial starter and probiotic species are discussed. Finally, new developments and potential applications of next-generation viability determination methods such as flow cytometry, propidium monoazide–quantitative polymerase chain reaction (PMA-qPCR), next-generation sequencing, and single-cell Raman spectroscopy (SCRS) methods are examined.
The genus Bifidobacterium consists of Gram-positive bacteria belonging to the family Bifidobacteriaceae (Biavati and Mattarelli, 2015). Members of this genus are anaerobic or sometimes aerotolerant, non-spore-forming pleomorphic bacteria (Eckel et al., 2020). The taxonomy of the genus has been steadily changing over time due to advances in genomic characterization techniques, with the discovery of new species and subspecies in recent years (Lugli et al., 2018, 2021; Neuzil-Bunesova et al., 2020; Chen et al., 2021). As of the end of 2021, there were 98 documented species of the Bifidobacterium genus (Turroni et al., 2021). The primary ecological niche of Bifidobacterium species is the gastrointestinal tract (GIT) of mammals, birds, and insects (Alessandri et al., 2021). Within this diverse genus, some Bifidobacterium species were historically assumed to colonize only the GIT of specific animal host species (Turroni et al., 2011). However, advanced metagenomic approaches, such as bifidobacterial internally transcribed spacer (ITS) profiling, revealed that several of the Bifidobacterium species are ubiquitous within the GIT of different host animal species (Alessandri et al., 2021). Based on the core genome analysis, the genus is divided into 10 phylogenetic groups (B. adolescentis, B. boum, B. pullorum, B. asteroides, B. longum, B. psychraerophilum, B. bifidum, B. pseudolongum, B. bombi and B. tissieri) that partially correlate with animal host ecological niche (Alessandri et al., 2021; Duranti et al., 2021). Four of these phylogenetic groups are typical colonizers of the human GIT (Duranti et al., 2021). These include, the B. adolescentis group (B. adolescentis and B. catenulatum strains), the B. longum group (B. breve and B. longum strains), the B. pseudolongum group (B. animalis strains) and the B. bifidum group (B. bifidum strains) (Alessandri et al., 2021; Duranti et al., 2021).
Bifidobacteria are an integral component of the human gut microbiota, and their presence is associated with several health benefits (Sharma et al., 2021; Li et al., 2023). Due to their strong association with breast milk, Bifidobacterium species are among the earliest and most dominant colonizers of the GIT of neonates, making up to 90% of the microbiota of infants (Wong et al., 2020; Yang et al., 2021). Their relative abundance decreases in adult humans but remains stable at about 10–40% of the microbiota (Arboleya et al., 2016). In old age, the proportion decreases to about 5% (Arboleya et al., 2016). In addition to the decline in the relative abundance, there is also a change in the species diversity with age. The species, B. breve, B. longum subsp. infantis, and B. bifidum are the most dominant in infants, while B. longum, B. catenulatum and B. adolescentis dominate in adults (Derrien et al., 2022). Moreover, variations have also been reported among the elderly populations (Wang et al., 2015; Kato et al., 2017). For instance, the microbiota of Chinese centenarians was found to comprise of unique species, such as B. minimum, B. gallinarum/B. pullorum/B. saecularmay, and B. mongoliense which were absent in younger elderlies of 80–90 years (Wang et al., 2015). Due to dietary and stress factors such as antibiotic use, bifidobacterial levels in the GIT may be depleted, resulting in a dysbiosis of gut microbiota (Derrien et al., 2022). Evidence has shown that their reduction in the GIT is correlated with adverse health outcomes such as an increased risk of obesity, type 2 diabetes, allergic pathologies, irritable bowel syndrome, colorectal cancer and infections due to enteric viruses and bacterial pathogens (Xu et al., 2012; Akay et al., 2014; Taverniti and Guglielmetti, 2014; Gao et al., 2015; Kosumi et al., 2018; Wei et al., 2018; Li L. et al., 2021; Colston et al., 2022).
A sustainable approach to mitigate dysbiosis of the human gut microbiota and the attendant adverse health effects is the supplementation of bifidobacteria in foods as probiotics (He et al., 2023). According to the Food and Agriculture Organization of the United Nations [FAO], and World Health Organization [WHO] (2001) definition, probiotics are “live micro-organisms, which when consumed in adequate amounts confer a health benefit on the host.” Among the foods used as potential probiotic carriers, fermented dairy products, especially yogurt, are the most consumed. However, the sustenance of probiotic viability during processing and shelf-life of foods like yogurt is challenging. Viability is a prerequisite for probiotic functionality and therapeutic benefits (Terpou et al., 2019). The significance of viability has been demonstrated by the fact that probiotic functionalities such as antimicrobial effects, lactose intolerance relief and immune stimulation depend on cell viability (Terpou et al., 2019). Thus, for any therapeutic effects associated with probiotic intake, it is recommended that the levels of viable cells must be at least 106 CFU/g of food product at the time of consumption (Nyanzi et al., 2021). Several process factors such as homogenization, mixing, heating, fermentation, and cooling in the manufacturing of fermented dairy products potentially influence Bifidobacterium viability.
Physicochemical stresses such as dissolved oxygen, acidic pH, homogenization pressure and storage temperature constitute the main inhibitory factors (Meybodi et al., 2020). Moreover, antagonistic effects of the starter cultures, such as the production of H2O2, could also negatively impact Bifidobacterium viability (Meybodi et al., 2020). Due to the poor technological robustness, many potentially beneficial Bifidobacterium species with superior probiotic functionalities in the human GIT cannot be effectively incorporated into fermented dairy foods. Although most of the identified Bifidobacterium species are from the GIT of animals, some species and strains have recently been shown to be endogenously present in fermented foods (Laureys et al., 2016; Eckel et al., 2020). So far, a few species and strains, such as the moderately aerotolerant B. animalis subsp. lactis are used in the commercial production of probiotic foods (He et al., 2023). The non-aerotolerant strains cannot be used due to their susceptibility to oxygen exposure.
Accurate enumeration of viable Bifidobacterium spp. in dairy products is a critical factor in probiotic quality assurance. Specific enumeration of Bifidobacterium viability in yogurt is complicated by the co-occurrence of lactic acid bacteria (LAB) starter cultures. In mixed species products with Lacticaseibacillus rhamnosus and Lactobacillus acidophilus strains, the widely used medium for enumeration of Bifidobacterium spp. [De Man, Rogosa and Sharpe (MRS) agar supplemented with neomycin, nalidixic acid, lithium chloride and paromomycin (MRS-NNLP agar)] could not select for Bifidobacterium spp. (Van de Casteele et al., 2006; Ashraf and Smith, 2015). Moreover, probiotic functionality is strain and species-specific, yet culture-based methods are unable to selectively differentiate between individual species and strains of bifidobacteria (Hagen and Skelley, 2019; Yoon et al., 2021). This paper reviews the subject of Bifidobacterium spp. viability and its significance in probiotic functionality. Given that yogurt is a widely consumed dairy product that is globally accepted as a delivery vehicle for probiotics, the paper focuses on the factors that influence Bifidobacterium spp. viability during the manufacturing as well as the strategies for viability retention. Furthermore, the article explores the next-generation methods for Bifidobacterium spp. viability determination and their applications in industrial probiotic viability quality assurance. The review was primarily based on literature published in the past 15 years, However, some earlier key studies with an enduring relevance and impact on the subject were selectively incorporated.
A significant quantity of in vitro and in vivo evidence has demonstrated the probiotic functionalities of Bifidobacterium spp. (Konieczna I. et al., 2012; Groeger et al., 2013; Turroni et al., 2014; Din et al., 2020; Shang et al., 2020; Zhang et al., 2020; van der Hee and Wells, 2021; Schiweck et al., 2022; Álvarez-Mercado et al., 2022; He et al., 2023). These diverse probiotic functionalities include the enhancement of the host immune system, protection against communicable and non-communicable diseases, as well as improvement of nutritional metabolism (He et al., 2023). The immunomodulatory properties of Bifidobacterium species include the stimulation of both innate and adaptive immune defense systems (He et al., 2023). Some of the compelling empirical evidence for the immunostimulatory effects was demonstrated in immunosuppressed mice gavaged with B. bifidum strains, in which the oral administration of the probiotic resulted in increased secretion of immunoglobulin A and enhanced production and activity of lymphocytes, natural killer cells and macrophages (Turroni et al., 2014; Shang et al., 2020). In addition, experimental evidence in mice models, epithelial cell lines and human trial studies has shown that species such as B. longum subsp. infantis, B. animalis subsp. lactis and B. infantis can offer protection against chronic gastrointestinal inflammatory diseases such as inflammatory bowel disease and other non-enteric inflammatory diseases like autoimmune hepatitis (Konieczna I. et al., 2012; Groeger et al., 2013; Din et al., 2020; Zhang et al., 2020; Álvarez-Mercado et al., 2022). The protective mechanism is attributed to the ability of Bifidobacterium species to inhibit the release of proinflammatory cytokines while stimulating the release of anti-inflammatory cytokines (Konieczna I. et al., 2012; Zhang et al., 2020; Álvarez-Mercado et al., 2022; He et al., 2023). A further beneficial function of Bifidobacterium species relates to their role in the metabolism of dietary and human-derived heteroglycans (Li et al., 2023). Dietary heteroglycans such as arabinoxylans, pectin, and inulin are plant-derived components of dietary fiber that are not metabolized by human digestive enzymes (Kelly et al., 2021). Human-derived heteroglycans include mucin and human milk oligosaccharides (HMOs) (Luo et al., 2021). Except for a few other genera, Bifidobacterium species are the most prominent part of the gut microbiota capable of metabolizing heteroglycans (Li et al., 2023). The fermentation of heteroglycans has profound implications for health-promoting functions (Li et al., 2023). Among the fermentation products, short-chain fatty acids (SCFAs) such as acetate, propionate, butyrate and valerate that are linked to numerous beneficial effects (Parada Venegas et al., 2019; Schiweck et al., 2022). Using a murine model, Zhang et al. (2020) showed that oral intake of B. animalis subsp. lactis increased the concentration of fecal butyric acid in mice with experimentally induced autoimmune hepatitis. An abundance of evidence has shown that SCFAs are central to the regulation and induction of the immune system (van der Hee and Wells, 2021; Schiweck et al., 2022). They function as signaling molecules through cell surface G-protein coupled receptors (GPCRs) to control immune and metabolic functions (van der Hee and Wells, 2021; Li et al., 2023).
As enunciated in the original definition, viability is a primary criterion for describing an organism as a probiotic (Food and Agriculture Organization of the United Nations [FAO], and World Health Organization [WHO], 2002). However, there have been many scientific reports and reviews indicating that many of the known health benefits previously ascribed to live probiotics can also be exhibited by their metabolites and/or non-viable cells (Geraldo et al., 2020; Vallejo-Cordoba et al., 2020; Martorell et al., 2021). Previous experiments comparing the physiological functionalities of live and heat-killed B. breve in murine models concluded that while both heat-killed and live cells exhibited similar activities in the suppression of pro-inflammatory cytokine secretion, live cells had a more significant effect on the regulation of intestinal metabolism (Sugahara et al., 2017). Similar comparisons of protective properties of heat-inactivated and live B. longum subsp. longum and B. animalis subsp. lactis on ovalbumin-sensitized mice and cultured Caco-2 cells, respectively, showed that live cells exhibited a stronger inflammation-suppressing effect and increased barrier-integrity of epithelial cells (Castro-Herrera et al., 2020; Pyclik et al., 2021). Despite this recent evidence, it is undeniable that the requirement for viability continues to be the standard for the incorporation of probiotics into functional foods. While some physiological effects of probiotics, such as immunomodulatory properties, can be elicited by bacterial cell components such as lipoteichoic acids and peptidoglycan, some functions are dependent on metabolic activity and thus are a product of viable cells (Castro-Herrera et al., 2020). Several aspects of Bifidobacterium spp. probiotic functionality depend on their metabolic activities and secretion of enzymes and bioactive metabolites. A quintessential illustration of the viability-dependent function of Bifidobacterium spp. in gut health is their fermentation of dietary heteroglycans and human-derived oligosaccharides (Luo et al., 2021; Li et al., 2023). Apart from the direct benefits of such metabolism of complex carbohydrates, the unique carbohydrate-active enzymes of bifidobacteria enable them to support the growth and survival of other members of the gut microbiota through cross-feeding (An et al., 2014). In vitro experiments with co-cultures of Bifidobacterium and other genera of the human gut microbiota, such as Faecalibacterium, Eubacterium and Anaerostipes on fructooligosaccharide substrates, showed an enhanced production of butyrate, a bioactive SCFA (Belenguer et al., 2006; Rios-Covian et al., 2015; Kim H. et al., 2020). Thus, through synergistic metabolic interactions with other members of the gut microbiota, viable Bifidobacterium spp. can result in the amplification of biological signals that lead to enhanced probiotic functionality.
Fermented dairy products such as yogurts, cheeses, acidified milks and kefir are the most known category of food-based probiotic carrier systems for the human intake of Bifidobacterium spp. (Terpou et al., 2019; González-Orozco et al., 2022). Above all, the ability to survive the fermentation and associated processes during the production of these foods ultimately determines the number of viable Bifidobacterium spp. that reach the GIT, and the therapeutic benefit derived therefrom (Meybodi et al., 2020). This paper focuses on the viability of Bifidobacterium spp. in yogurt as an archetypal dairy-based probiotic carrier. Several physical and chemical stress factors associated with the yogurt manufacturing process and the subsequent storage period of the shelf-life impose adverse effects on Bifidobacterium viability (Meybodi et al., 2020). The sources of the physical and chemical stress factors associated with the yogurt manufacturing process are summarized in Figure 1. The ensuing subsections of the paper explore the physiological and genetic responses of Bifidobacterium spp. to the main stress factors associated with yogurt production (acid, osmotic, heat, oxidative and cold stress). A summary of the elucidated and postulated mechanisms is illustrated in Figure 2.
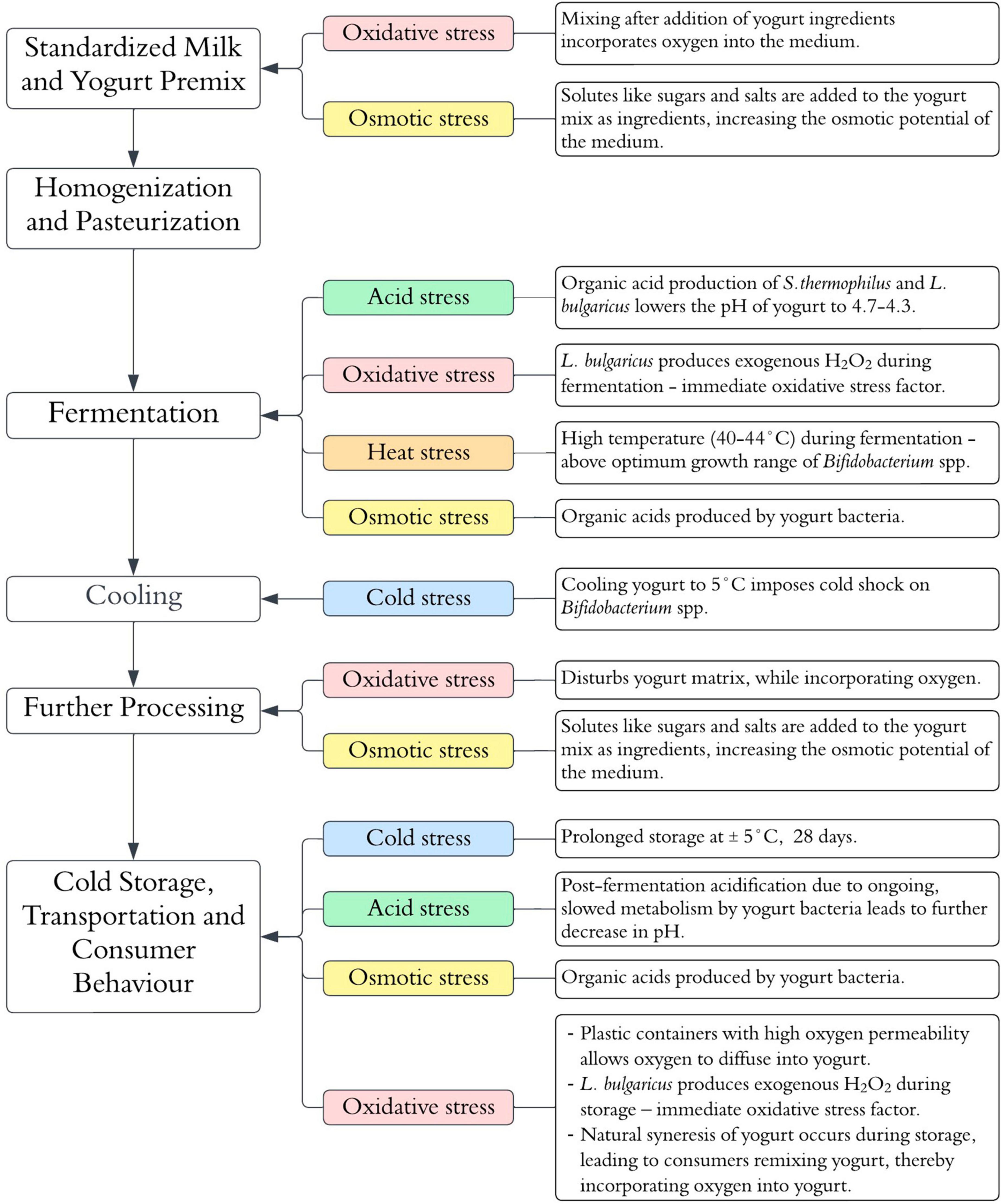
Figure 1. Stress factors potentially influencing Bifidobacterium spp. viability during yogurt manufacturing and shelf-life.
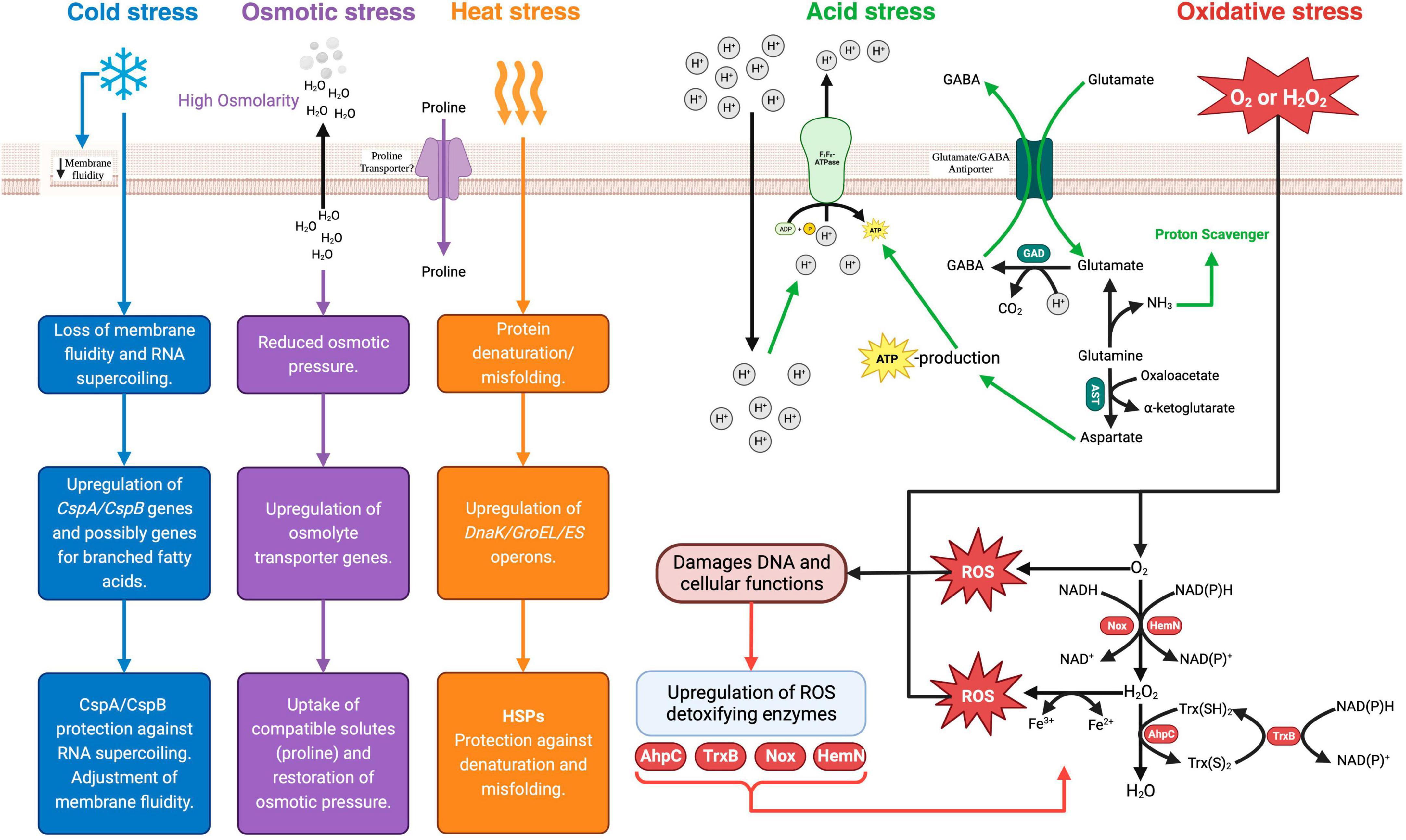
Figure 2. Bifidobacterium spp. stress response mechanisms. Csp, cold shock protein; Hsp, heat shock protein; GAD, glutamate decarboxylase; AST, aspartate transaminase; GABA, γ-amino butyric acid; EPS, exopolysaccharide; Nox, NADH oxidase; HemN, oxygen-independent coproporphyrinogen III oxidase; AhpC, Alkyl hydroperoxide reductase C-subunit; Trx(S)2, oxidized thioredoxin; Trx(SH)2, reduced thioredoxin; TrxB, thioreductase-like protein. Created with BioRender.com.
In most cases, probiotic Bifidobacterium spp. are incorporated into yogurt together with the starter culture at the onset of fermentation. As a neutrophile, the typical optimum pH for Bifidobacterium spp. growth is 6.5–7.0 (Biavati and Mattarelli, 2015). Given the low pH conditions in yogurt, the survival of Bifidobacterium spp. depend on the ability to activate an acid tolerance response needed to maintain intracellular pH homoeostasis. The mechanisms of acid stress response have been well elucidated and involve the increased expression of the proton-translocating F1F0-ATPase (Sánchez et al., 2007; Waddington et al., 2010; Jin et al., 2012). The F1F0-ATPase is an active proton pump whose activity results in the exclusion of protons using energy derived from the hydrolysis of ATP (Fiocco et al., 2020). In addition to the F1F0-ATPase-dependent acid tolerance response, other mechanisms include the alkalization of the cytoplasm through processes that consume intracellular H+ protons (Fiocco et al., 2020). These include the glutamate decarboxylase (GAD) and branched-chain amino acid metabolism pathways, that generate ammonia (Schöpping et al., 2022a). The GAD system comprises of the enzyme glutamate decarboxylase (GadB) and an antiporter (GadC) encoded by the GadB and GadC loci, respectively (Yunes et al., 2016). In this pathway, glutamate is converted by the activity of GadB to γ-aminobutyrate (GABA), with the consumption of H+ protons and the release of CO2 (Duranti et al., 2020). Ammonia production is thought to play a crucial role in maintaining intracellular pH equilibrium by functioning as a proton scavenger (Wei et al., 2019). When exposed to acidity, acid-sensitive strains such as B. longum exhibit increased production of enzymes responsible for branched-chain amino acid (BCAA) biosynthesis (Wei et al., 2019). Furthermore, sulfur-containing amino acids, such as cysteine and methionine, are hypothesized to play a role in the acid stress response of Bifidobacterium spp. (Sánchez et al., 2007; Schöpping et al., 2022b). An analysis of the proteome of low pH adapted mutants of B. longum identified a higher constitutive presence of methionine synthase, cystathionine gamma-lyase and cystathionine gamma-synthase compared to the unadapted wild type (Sánchez et al., 2007). All these enzymes are involved in additional pathways of NH3 generation and alkalization of the cytoplasm (Schöpping et al., 2022b). Furthermore, the acid tolerance response in B. longum strains has also been linked to an overall increase in cell envelope components under sub-lethal and lethal acid environments (Jin et al., 2015). Higher transcription rates were reported for genes associated with the synthesis of peptidoglycan, exopolysaccharides, and undecaprenyl-PP (UND-PP) in B. breve adapted at pH 3.2 (Jin et al., 2015). The high synthesis of peptidoglycan under acid stress conditions is postulated to strengthen the cell wall structure and provide protection against acid stress-induced cell damage (Jin et al., 2015). Moreover, acid stress induces changes in the fatty acid profiles of the cell membrane with a shift toward long-chain fatty acids (C16:0 and C18:0) (Wei et al., 2019).
Processes that allow oxygen diffusion into the milk or yogurt during manufacturing include stirring, homogenization, mixing and agitation (Figure 1). In addition, the packaging of yogurt in oxygen-permeable plastic containers can lead to an increased dissolved oxygen content over the storage period of the shelf life (Cruz et al., 2013). As anaerobes, Bifidobacterium spp. are intrinsically sensitive to oxygen and its derived reactive oxygen species (ROS). However, the sensitivities differ within the genus. Species such as B. animalis subspp. lactis, B. asteroides, B. minimum and B. indicum, are considered aerotolerant, while species such as B. boum and B. thermophilum are considered hyper-aerotolerant (Kawasaki et al., 2018). The primary response mechanisms to oxidative stress are based on the production of enzymes that can detoxify ROS. Except for a few aerotolerant species like B. asteroides and B. indicum, Bifidobacterium spp. generally lack the genes for the primary antioxidant enzymes like catalase and superoxide dismutase (Zuo et al., 2018). However, some Bifidobacterium spp. contain some inducible enzymes that can detoxify ROS (Schöpping et al., 2022b). One such enzyme is alkyl hydroperoxide reductase catalytic subunit C (AhpC) (Zuo et al., 2014). AhpC is a peroxidase component of alkyl hydroperoxide reductase enzymes systems that are found in many prokaryotes (Zuo et al., 2014). It functions together with the flavoprotein disulfide reductase (AhpF) to covert H2O2 to alcohol and water (Zuo et al., 2014). In some facultative anaerobes such as E. coli, AhpC is the main ROS detoxifying enzyme instead of catalase (Ma and Payne, 2012). In support of the hypothesis of inducible oxidative response systems in bifidobacteria, Xiao et al. (2011), observed that in aerobically grown B. longum, AhpC was one of the upregulated proteins together with DNA oxidative damage-protective proteins such as pyridine nucleotide-disulfide reductase (PNDR) and ribonucleotide reductase (NrdA). Similarly, Satoh et al. (2019), identified a thioredoxin reductase (TrxR) whose expression was induced by oxygen exposure in B. bifidum. Subsequently, TrxR was identified to be a reductase homologue of AhpF that interacted with AhpC in vitro to achieve H2O2 reduction. Hence, in B. bifidum, the oxidative stress response possibly involves an increased expression of thioredoxin reductase and the AhpF-AhpC H2O2 degradation system (Satoh et al., 2019).
To date, only a few studies have tried to elucidate the bifidobacterial osmoregulatory system (Cui et al., 2022; Zhang et al., 2022). Cui et al. (2016) experimentally determined the osmolarity limits for the growth of Bifidobacterium spp. to range from 850 to 1,300 mosM kg–1. The presence of solutes in yogurt (Figure 1) results in a hyperosmotic extracellular environment that can lead to the loss of water from bacterial cells, and consequently a loss of turgor pressure and plasmolysis (Brauer et al., 2023). Based on the established osmoregulatory systems in other probiotic bacterial genera that involve the intracellular accumulation of compatible solutes such as trehalose, glycine betaine and amino acids such as proline, Zhang et al. (2022) and Cui et al. (2022) sought to characterize the osmoregulatory mechanisms in B. bifidum and B. longum, respectively. Using a genome and metabolome analysis, hyper-osmotolerant mutants of B. bifidum were observed to have an increased accumulation of amino acids, especially proline, compared to non-osmotically adapted wild types (Zhang et al., 2022). As no potential proline transporter proteins were identified in the organism, the proposed osmotic protection mechanism was that proline accumulation occurred through an endogenous synthesis from glutamic acid (Zhang et al., 2022). In contrast, Cui et al. (2022) observed that when the osmotic pressure of a culture environment was increased, the addition of proline substantially improved the survival of B. longum, suggesting the presence of proline transporter proteins in this organism.
Except for a few thermophilic species like B. thermacidophilum, bifidobacteria are generally mesophilic organisms with an optimum growth temperature range of 37–41°C (Biavati and Mattarelli, 2015). Hence, a yogurt fermentation, which typically occurs at 40–44°C, can impose a mild heat stress on probiotic Bifidobacterium species. In general, the physiological effects of mild heat challenges on bacterial cells include the destabilization of non-covalent molecular bonds, ribosome dysfunctionality, and protein denaturation (Fiocco et al., 2020). Notably, apart from the earlier work by Ventura et al. (2004)2005 and Rezzonico et al. (2007), there has not been any studies on the bifidobacterial heat stress response systems in recent years. Like other prokaryotes, the heat stress response of Bifidobacterium spp. involves the increased production of heat shock proteins (HSPs) (Ventura et al., 2011). HSPs are ubiquitous and conserved proteins across the prokaryotic kingdom. They are encoded in two operons (dnaK and groEL-groES) and function as chaperones that protect physiological proteins against misfolding under conditions of heat stress (Hu et al., 2022). A transcriptomic analysis of B. longum exposed to heat shock treatment at 50°C found that the dnaK operon-encoded molecular chaperones (DnaK, GrpE, DnaJ) were the main HSPs produced (Rezzonico et al., 2007). The second class of molecular chaperones (GroEL and GroES) encoded by the groEL-groES operon were less expressed in B. longum in response to mild heat stress (Rezzonico et al., 2007). Studies on B. breve heat stress response showed that the GroEL and GroES chaperones are required for mild heat shock survival while dnaK, GrpE, DnaJ chaperones are necessary for survival under extreme heat stress (Ventura et al., 2004, 2005).
At the end of fermentation, yogurt is typically cooled to approximately 5°C, to allow for gel setting and inhibit the growth and metabolic activities of the yogurt bacteria (Sfakianakis and Tzia, 2014). In addition to a drastic drop in temperature after fermentation, the yogurt is further stored at cold temperatures for the duration of the shelf-life, typically 28 days. Except for a few species, such as B. mongoliense and B. psychraerophilum, which can grow under cold conditions, most Bifidobacterium spp., minimum growth temperature is 25–28°C (Biavati and Mattarelli, 2015). The physiological effects of cold stress in mesophilic bacteria include the changes in the cell membrane from an elastic liquid crystalline state to a rigid gel-phase state that impairs nutrient uptake, and the stabilization of nucleic acid secondary structures that impede DNA replication and protein synthesis (Phadtare, 2004). So far, the molecular mechanisms behind the cold stress response of Bifidobacterium spp. have not specifically been elucidated. However, based on knowledge from other Gram-positive organisms, the adjustment of membrane fluidity through the incorporation of unsaturated anteiso-branched-chain fatty acids (BCFA) is one of the mechanisms of bacterial cold stress adaptation (Yoon et al., 2015). A second mechanism of bacterial cold stress adaptation involves the increased production of cold shock proteins (CSPs) that act as RNA chaperones to prevent supercoiling and facilitate effective translation under cold conditions (Phadtare, 2004). CPSs belong to a family of small, highly conserved, structurally related proteins widely distributed in the prokaryotic kingdom (Phadtare, 2004). So far, a few studies have observed the presence of CSP gene homologs (CspA and CspB) in bifidobacterial genomes (Ventura et al., 2004; Rezzonico et al., 2007; Schöpping et al., 2022b). Interestingly, CspA expression in B. breve was activated by heat stress exposure together with the groEL-groES operon, suggesting a heat-induced co-transcription of both genes (Ventura et al., 2004).
The dissolved oxygen content of milk is a crucial factor influencing Bifidobacterium viability in yogurt. Hence, processes targeted at reducing the dissolved oxygen content of milk before fermentation may present some rational chances of viability retention. A comparative summary of these methods is given in Table 1. Previous studies have investigated the use of gasses like nitrogen to achieve milk deaeration (Bolduc et al., 2006; Ebel et al., 2011). The bubbling of pasteurized milk with a gas mixture of N2 and 4% (v/v) H2 (N2-H2) for 4 h at a flow rate of 20 mL/min decreased the redox potential of milk from +440 mV to +350 mV and −300 mV, respectively (Ebel et al., 2011). When the de-aerated milk was fermented by yogurt starter cultures together with B. bifidum, the fermented products made from milk treated with N2H2 had higher survival of B. bifidum during storage, and the treatment had no adverse effects on the fermentation kinetics and starter cultures (Ebel et al., 2011). In a similar deaeration treatment with N2, the dissolved oxygen concentration of milk was reduced from an average of 6.7 ppm to 0.3 ppm (Bolduc et al., 2006).
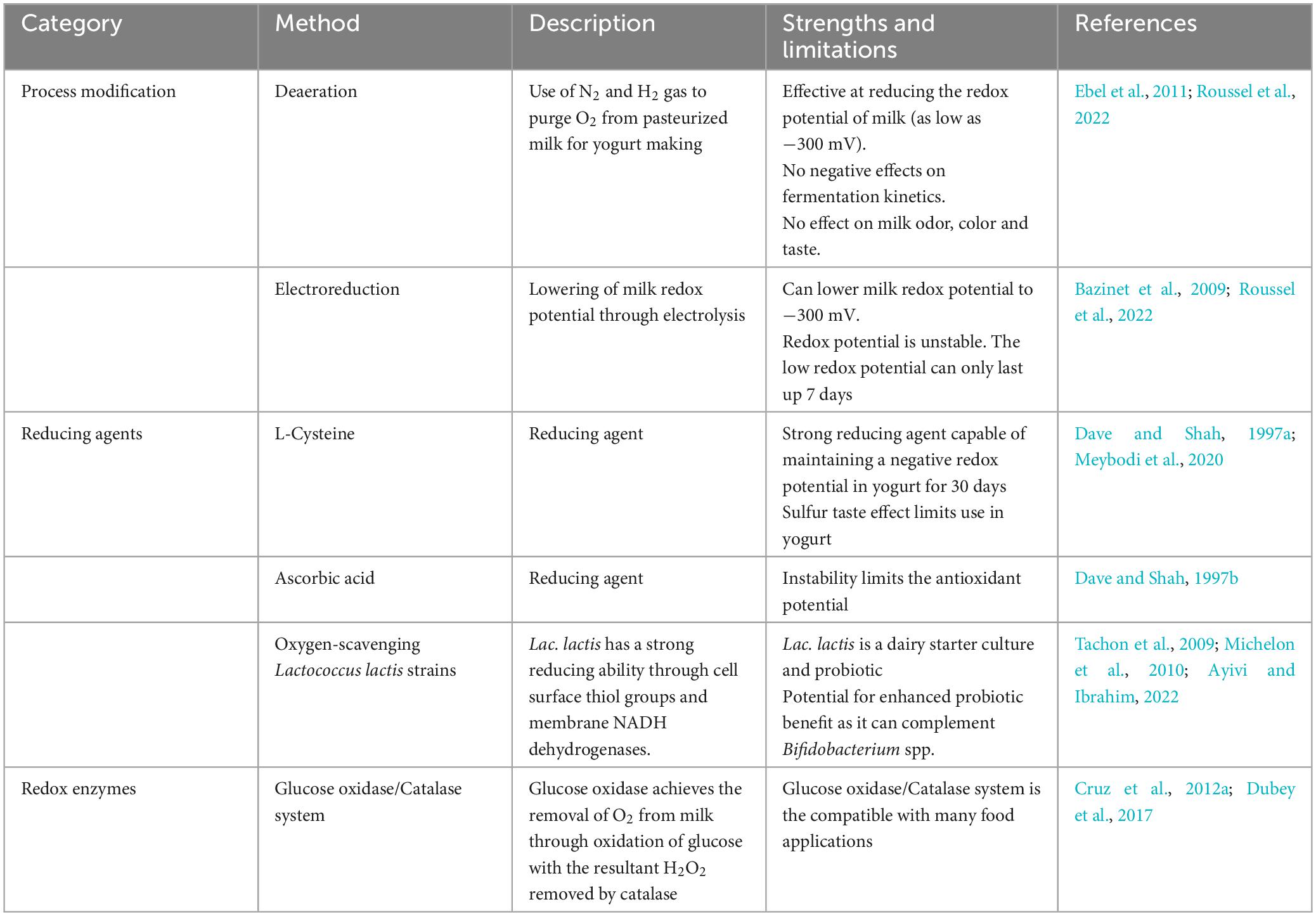
Table 1. Methods for reducing the redox potential of milk and yogurt for the enhancement of Bifidobacterium spp. viability.
An alternative method of lowering redox potential is electroreduction. This is a physical treatment involving voltage application to reduce the redox potential by electrolysis (Roussel et al., 2022). It is an efficient process of decreasing the redox potential of milk. Through the application of a voltage of −1.55 V through a milk sample for 40 minutes, Bolduc et al. (2006) reported a reduction in redox potential of milk from > + 200 mV to < −300 mV and a decrease in dissolved oxygen concentrations to between 2–3 ppm from 6.7 ppm in untreated milk. When treated by electroreduction, it is possible to maintain a negative redox potential in milk for up to 7 days (Bazinet et al., 2009).
Another technological process modification considered a potential method for viability improvement is high-pressure homogenization (HPH). HPH is a non-thermal milk preservation method in which the milk is exposed to pressure above 100 MPa (Massoud et al., 2016). Experiments with B. lactis have shown that an increase in homogenization pressure from 100 to 200 MPa, combined with increasing temperature from 50 to 70°C, led to a significant improvement in the viability of the probiotic in the resultant yogurt (Massoud et al., 2015). The improvement in viability has been attributed to the increase in free amino acids needed for probiotic nutrition (Massoud et al., 2016). Most significantly, the release of cysteine has a positive effect on Bifidobacterium viability.
An additional approach to reducing the redox potential of milk and yogurt is using Lactococcus lactis as an oxygen scavenger (Tachon et al., 2009). Lac. lactis is known for its strong ability to decrease the redox potential of milk to as low as −220 mV (Tachon et al., 2009). Its co-inoculation with probiotics in milk remarkably improved the viability of Bifidobacterium spp. (Yonezawa et al., 2010; Odamaki et al., 2011). Interestingly, Lac. lactis is a dairy starter culture used extensively in cheese fermentations and as a probiotic (Ruggirello et al., 2016; Jaskulski et al., 2020). The incorporation non-starter, adjunct lactic acid bacterial cultures in yogurt is a worthwhile proposition in the production of yogurt with enhanced health benefits (Ayivi and Ibrahim, 2022). However, the potential benefits must be balanced against any potential negative effects on yogurt fermentation kinetics, flavor and texture (Ayivi and Ibrahim, 2022). Generally, the combination of Bifidobacterium spp. with other probiotic lactic acid bacterial species have shown positive outcomes in terms of bioactive metabolites and therapeutic benefits (Pápai et al., 2021; Peng et al., 2022).
In addition, processes targeting the control of post-fermentation acidification in yogurt have been proposed for managing Bifidobacterium viability in yogurt. The H+-ATPase defective mutants of L. delbrueckii subsp. bulgaricus cannot exclude H+ protons and are sensitive to acidity due to their inability to maintain cytoplasmic pH homeostasis (Wang et al., 2013). As the primary organism responsible for post-fermentation acidification, the use of such mutants in yogurt fermentation can reduce the accumulation of acidity post-fermentation (Wang et al., 2013). Ongol et al. (2007) reported an enhanced viability of B. breve during storage in yogurt fermented with H+-ATPase defective mutants.
Stress adaptation involves the pre-exposure of an organism to sub-lethal stress conditions that induce the development of tolerance to subsequent lethal stress exposure (Fiocco et al., 2020). The process involves several repetitive generations of exposure to mild stress, punctuated by incremental stress intensification (Jiang et al., 2016). A typical stress adaptation process can involve up to 50 generations of repeated stress exposures in which stress-resistant variants are isolated every few generations (Jiang et al., 2016). Variants that exhibit a stable stress resistance phenotype are subsequently preserved as genetically adapted mutants (Berger et al., 2010). Some successful adaptation experiments have been reported for different Bifidobacterium species. An acid-resistant mutant strain of B. longum subsp. longum was isolated after a 50-generation successive subculturing in MRS broth adjusted to pH 2.5 (Jiang et al., 2016). Similarly, thermal stress- and oxidative stress-adapted bifidobacteria cells were isolated after successive exposures to heat and hydrogen peroxide, respectively (Berger et al., 2010; Mozzetti et al., 2010). The genetic basis of sustained stress adaptation is the evolutionary development of mutants that overexpress stress response genes. Using transcriptomic profiling of induced oxidative stress adaptation in B. longum subsp. longum, Xiao et al. (2011) observed an upregulation of genes encoding essential proteins involved in the protection or repair mechanisms of damaged cell components, such as alkyl hydroperoxide reductase C22 (AhpC), DNA-binding ferritin-like protein (Dps), ribonucleotide reductase (NrdA), and enolase. After successive heat shock treatments, Berger et al. (2010) also observed that heat-adapted mutants overexpressed the dnaK operon and the clpB gene in B. longum. Apart from the adaptive tolerance to the same stress factor used to induce adaptation (homologous adaptation), cross-protection to different stress factors (heterologous adaptation) can also occur (Chen et al., 2017). Central to the cross-tolerance is the role of the transcriptional general stress response regulators (Averina et al., 2012). In Bifidobacterium species, the WhiB-like proteins encoded by the whiB gene are the universal transcriptional regulators of stress response genes (Averina et al., 2012). As pleiotropic regulators, the effect of any induced stress adaptation can likely overlap among different stress factors (Averina et al., 2012).
Microencapsulation involves the use of biopolymers that entrap bacterial cells in a polymer matrix prepared into microgel spheres (Frakolaki et al., 2021). The entrapment within the microgel particles protects probiotics against environmental stress and aids their survival during food processing and storage (Yeung et al., 2016). While several biopolymers can be used in the preparation of probiotic microcapsules, alginate is by far the most widely used food-grade biopolymer (Liu et al., 2019; Abbas et al., 2022). A naturally occurring polysaccharide extracted from brown algae, alginate consists of a linear polymer of α-L-guluronic acid (G) and β-D-mannuronic acids (M) units in repetitive blocks (G-blocks and M-blocks) (Alba and Kontogiorgos, 2019). Solutions of alginates readily form gels in the presence of divalent cations such as Ca2+ through their interaction with the G-blocks in an egg-box model (Alba and Kontogiorgos, 2019). The microgel preparation process usually involves a solution of alginate mixed with a bacterial suspension that is subjected to extrusion to form droplets that are instantaneously hardened by treatment with CaCl2 solution into three-dimensional gel spheres entrapping the probiotic bacteria (Liu et al., 2019; Abbas et al., 2022). Another commonly used method in the preparation of alginate gel droplets is emulsion. This method involves the alginate-bacterial mixture suspended in an oil bath with a surfactant to produce a water-in-oil emulsion (Frakolaki et al., 2021). The emulsion is subsequently treated with a CaCl2 solution, and the formed beads are harvested by centrifugation (Liu et al., 2019). The gel spheres prepared by either of these methods are subsequently freeze-dried (Liu et al., 2019). Several studies have shown that encapsulation enhances the survival ability under environmental and simulated GIT stress conditions (Yeung et al., 2016; Ji et al., 2019; Cedran et al., 2021; Zhang et al., 2021). For instance, alginate- and chitosan-coated/alginate-encapsulated B. longum showed a 0.20–1.72 log10 CFU/g viability loss at 55–65°C compared to unencapsulated cells that had a viability loss of 3.0–5.0 log10 CFU/g loss (Ji et al., 2019). When subjected to simulated gastric conditions (pH 2.5), the viability of chitosan-coated/alginate-encapsulated B. longum decreased by 1.27 log10 CFU after 120 min compared to the viability of unencapsulated cells that had declined to undetected levels after the same time period (Ji et al., 2019). Besides the observed enhancement of survival under simulated stress conditions, the benefit of encapsulation has also been demonstrated in alginate-encapsulated Bifidobacterium spp. incorporated into yogurt and other foods. Cedran et al. (2021) observed a two-fold loss of viability of unencapsulated B. lactis compared to the alginate-encapsulated cells in jam after 6 days of storage. Similarly, Mousa et al. (2023) reported a higher viability of B. bifidum encapsulated in a double layer of whey protein and alginate in set yogurt during a 14-day storage at 4°C than free cells. Pradeep Prasanna and Charalampopoulos (2019) observed a > 3.0 log10 CFU/g viability decline for unencapsulated B. animalis subsp. lactis in goat milk yogurt over 28 days while the viability of alginate-encapsulated cells remained stable. However, it is worth noting that while the benefits of encapsulation indicate a better survival compared to unencapsulated cells, a general decline in viability especially over the duration of shelf life still occurs (Yeung et al., 2016; Mousa et al., 2023). Moreover, a disproportionately large number of encapsulation studies have been based on B. animalis subspp. lactis, an intrinsically acid and oxidative stress tolerant strain. Hence, there is still need for viability retention methods for the more stress-sensitive species like B. bifidum. Besides the viability retention benefits, the effects on microencapsulation on yogurt texture are as important. Mousa et al. (2023), observed an increase in viscosity, gumminess, chewiness, and adhesiveness in yogurt with alginate-encapsulated B. bifidum. Similarly, Li H. et al. (2021) observed an increase in water holding capacity and cohesiveness of yogurt with microcapsules of L. paracasei. Importantly most of the changes in texture are beneficial in yogurt quality and the microcapsule incorporated yogurt have been reported to be acceptable by sensory panels (Dimitrellou et al., 2019; Mousa et al., 2023).
Among the physicochemical stress factors associated with yogurt processing and storage, oxidative stress has the most significant effect on viability of Bifidobacterium spp. (Bolduc et al., 2006). Hence, the ability to control the dissolved oxygen content and the redox potential of yogurt can provide a sustainable approach to preserve viability. The incorporation of oxygen-scavenging compounds, such as ascorbic acid and cysteine, has been shown to improve Bifidobacterium spp. survival in yogurt (Norouzbeigi et al., 2021). At 500 mg/l, cysteine can maintain a negative redox potential in yogurt for 30 days (Dave and Shah, 1997a). However, despite its strong reducing capacity, cysteine is considered unsuitable for use in yogurt, as it impacts a sulfur taste (Meybodi et al., 2020). Regarding sensory effects, ascorbic acid is more compatible with dairy products (van Aardt et al., 2005). However, the antioxidant capacity of ascorbic acid is hindered by the gradual loss of its stability over the storage shelf life of yogurt (Dave and Shah, 1997b). From the highest concentration of 250 mg/Kg, only 15–20% of the ascorbic acid was retained in yogurt after 35 days of storage at 4°C (Dave and Shah, 1997b). An additional approach to protect Bifidobacterium spp. from the effects of oxidative stress is the addition of glucose oxidase and glucose in yogurt (Cruz et al., 2012a). Glucose oxidase utilizes oxygen as it oxidizes D-glucose to gluconic acid and hydrogen peroxide, thus causing a reduction in the dissolved oxygen content of yogurt (Afjeh et al., 2019). Cruz et al. (2012a) reported a significant increase in the viable population of B. longum in yogurts added with glucose oxidase and glucose. While the activity of glucose oxidase reduces the oxygen content of the yogurt, the release of hydrogen peroxide as a by-product has a negative impact on Bifidobacterium spp. Hence, the protective effect of the glucose oxidase + glucose system requires the addition of catalase as an accessory enzyme to eliminate the toxic effects of hydrogen peroxide (Cruz et al., 2012b). The use of protective agents in preserving probiotic viability is a technique that is already extensively used in spray-drying and freeze-drying processes for probiotic microencapsulation (Fiocco et al., 2020). A diverse range of substances have shown to be effective as protective agents against the osmotic, heat and cold stresses associated with spray- and freeze-drying processes (Fiocco et al., 2020). Some of these substances, such as skim milk powder, are milk by-products that are readily acceptable as ingredients in yogurt making. When used as a protectant, skim milk solids stabilize the bacterial cell membrane by forming a protective coating on cell wall proteins (Terpou et al., 2019). This prevents cell damage due to thermal and osmotic stress associated with spray drying (Terpou et al., 2019). Other substances, such as sugars, sugar alcohols and complex carbohydrates, have proven to be effective cryoprotectants during freeze-drying (Rockinger et al., 2021). Although they are used as protective agents against freezing, during freeze drying, cryoprotective agents can be valuable in preserving Bifidobacterium viability during the long period of cold stress during yogurt shelf-life. Among the cryoprotective agents, trehalose and glycerol are the most favorable for use in yogurt. These compounds lower the phase transition temperature of the cell membrane under cold conditions, thus maintaining it in a flexible liquid crystalline state while also keeping the cell membrane hydrated through their hydrogen bond interactions with phospholipid heads (Rockinger et al., 2021). Furthermore, due to their water-binding abilities, the compounds can suppress ice nucleation and prevent the damaging effect of ice crystal formation (Rockinger et al., 2021).
In the last 20 years, a broad range of culture media have been proposed for the enumeration of Bifidobacterium spp. in dairy products (Van de Casteele et al., 2006; Lima et al., 2009; Ashraf and Smith, 2015). Among them, is MRS-NNLP agar, a widely used selective media (Dave and Shah, 1996; Ashraf and Shah, 2011; Karimi et al., 2012). In such media, the use of selective supplements may lead to an underestimation of viability as some live Bifidobacterium spp. cells may be sensitive to the selective agents (Dave and Shah, 1997a). For example, Van de Casteele et al. (2006), reported a lower recovery of Bifidobacterium spp. on MRS-NNLP agar than on MRS agar. Secondly, the selectivity of the medium depends on the type of non-target species present in the product (Van de Casteele et al., 2006; Ashraf and Smith, 2015). In mixed species products with L. rhamnosus and L. acidophilus strains, MRS-NNLP agar could not select for Bifidobacterium spp. (Van de Casteele et al., 2006; Ashraf and Smith, 2015). Currently, the International Organization of Standardization (ISO) and International Diary Federation (IDF) recommended culture-based method for the enumeration of Bifidobacterium spp. in dairy products (ISO 29981:2010/IDF 220:2010) is based on trans-galactosylated oligosaccharides (TOS) propionate agar containing lithium mupirocin as a selective agent (TOS-Mup media) (International Organization for Standardization [ISO], and International Dairy Federation [IDF], 2010). Like MRS-NNLP agar, TOS-Mup media has a low recovery for some Bifidobacterium spp. (Bunesova et al., 2015). Table 2 summarizes the recent applications of the recommended media for selective enumeration of Bifidobacterium spp. in yogurt and other dairy-based products. It is evident that despite all the intensive work, there is still a need for a medium that could be used as a standard for the quantification of all Bifidobacterium spp. Moreover, species and strain-specific physiological requirements affect quantification efficiency (Van de Casteele et al., 2006; Bunesova et al., 2015). In addition to the drawbacks mentioned above, culture-based methods are laborious and have long results turnaround time of up to 72 h as agar plates need to be incubated under specific growth conditions (Davis, 2014; Geng et al., 2014). Since these methods are based on the cultivability of the cells, they cannot quantify cells that are in a viable but non-culturable (VBNC) state (Jackson et al., 2019; Vinderola et al., 2019). Hence, culture-based methods may underestimate viable counts of beneficial probiotic bacteria (Davis, 2014).

Table 2. Summary of the recent applications of the recommended media used for the selective enumeration of Bifidobacterium spp. in yogurt, lyophilized cultures, and other dairy-based products.
Flow cytometry is a single-cell analysis technique that is used to explore the physical and physiological characteristics of microbial cells as they pass through a beam of light (usually blue laser, 488 nm) (Davis, 2014). When used with fluorescent staining, the technique can distinguish between live and dead cells based on viability markers such as membrane integrity and intracellular enzyme activity (Wendel, 2022). Cell integrity is often determined by double staining with the DNA binding dyes such as diamidinophenylindole (DAPI), acridine orange and the SYTO dye series, which emits green fluorescence after excitation with 488 nm laser and propidium iodide (PI), which emits red fluorescence after excitation at the same wavelength (Veal et al., 2000). The exclusion of PI by cells with intact membranes gives viable cells a green fluorescence, while non-viable cells with damaged membranes fluoresce red (Veal et al., 2000). Intracellular enzyme activity is often determined using membrane-permeant fluorogenic substrates such as 5,6-carboxyfluorescein diacetate (5,6-cFDA), which, upon enzymatic hydrolysis by intracellular esterases from live cells, release a green, fluorescent carboxyfluorescein (Hoefel et al., 2003). Following a gating for live- (green fluorescent) and dead-cell (red fluorescent) subpopulations, viability determination is then based on the enumeration of cells from an appropriately diluted sample that falls within the live-cell region (Foglia et al., 2020). Although FCM has been available for a long time as a high throughput method of studying bacterial cell viability, its use in the enumeration of probiotic viability in foods has been limited. So far, one protocol by the ISO and IDF (ISO 19344 – IDF 232: 2015) is available for the flow cytometric enumeration of starter cultures and probiotics in fermented products (International Organization for Standardization [ISO], and International Dairy Federation [IDF], 2015). However, the non-specificity of the method due to its inability to selectively enumerate viable probiotics in the presence of the starter cultures, is a significant limitation. Some studies have attempted to improve the species selectivity of flow cytometry by incorporating antibody labeling in conjunction with membrane integrity and enzyme activity fluorescent probes (Geng et al., 2014; Chiron et al., 2018). The immuno-flow cytometry assay utilizes the specific binding of a primary polyclonal antibody to ligands on the bacterial cell, which is subsequently bound to a secondary antibody conjugated to a fluorescent tag and further stained with a viability probe (Wilkinson, 2018). Using this concept of dual labeling with polyclonal antibodies and 5,6-cFDA, only viable B. lactis were enumerated from mixed cultures and fermented products containing L. bulgaricus, S. thermophilus and Lac. lactis (Geng et al., 2014). Similarly, polyclonal antibodies specific for B. bifidum, B. longum subsp. infantis, B. longum subsp. longum, L. helveticus and L. rhamnosus in combination with SYTO®24 and PI staining were used to selectively enumerate the viable individual strains in multi-strain probiotic products (Chiron et al., 2018). Another immuno-flow cytometry concept with a potential application in probiotic enumeration is the immunomagnetic separation of specific probiotic strains from a mixed species using antibody-coated magnetic beads (Wilkinson, 2018). This technique, commonly used for the recovery and enrichment of pathogens, was recently used to isolate L. paracasei from human feces (Takada et al., 2023). When applied for probiotic viability enumeration, recovered cells from immunomagnetic separation can be analyzed by flow cytometry after staining with viability probes (Wilkinson, 2018). Apart from its value in viability determination, flow cytometry offers other benefits in studying some of the physical and physiological characteristics of viable cells that could relate to their stress responses and probiotic functionalities (Wendel, 2022). The additional element of flow cytometry, fluorescence-activated cell sorting (FACS), allows for the isolation and recovery of different subpopulations from a flow cytometry assay (viable, dead, and injured cells) for further analysis of metabolic, physiological, and genetic characteristics relating to probiotic functionality (Wendel, 2022).
Real-time quantitative polymerase chain reaction (qPCR)-based methods are premised on the detection and amplification of DNA of target organisms using fluorescent DNA intercalating dyes (e.g., SYBR green) or sequence-specific fluorogenic probes (e.g., TaqMan probes) (Agrimonti et al., 2019; Ruijter et al., 2021). Sequence-specific oligonucleotide primers are used to flank specific fragments of a target gene to be amplified. As the amount of PCR amplicon increases during PCR, the fluorescent signal accumulates. The quantification cycle (Cq) is then measured in the exponential phase of qPCR when the fluorescence signal has accumulated above the background fluorescence (Davis, 2014). During exponential phase, the amount of PCR amplicon is directly proportional to the DNA template (Davis, 2014). Hence, using standard curves established by plotting the Cq values against DNA copies, the number of copies of target species in the food sample can be determined (Davis, 2014). While the 16S rRNA gene has frequently been used in experimental qPCR-based methods (Zhang and Fang, 2006; Kim H. B. et al., 2020), its use poses challenges for quantification as it may exist as more than one copy in some bacterial genomes and it has a high sequence similarity between the Bifidobacterium spp. (Kim H. B. et al., 2020; Fan et al., 2021; Shi et al., 2022). For reliable quantification, the target gene should be a single copy within the bacterial genome (Shi et al., 2022). Recently, protein-encoding housekeeping genes such as the translation elongation factor EF-TU (tuf) and phenylalanine tRNA ligase subunit alpha (pheS) genes have been successfully used in qPCR methods for probiotic quantification in dairy products (Scariot et al., 2018; Fan et al., 2021; Shi et al., 2022). Alternatively, comparative genomics can be used to find unique and specific genetic markers for primer design and selective detection of closely related species and subspecies, especially of Bifidobacterium, where the use of housekeeping genes may be limited (Kim H. B. et al., 2020). A recent study by Kim H. B. et al. (2020) successfully designed species and subspecies-specific primers for 22 Bifidobacterium species and subspecies based on the genetic markers identified using comparative genomics. The main challenge of qPCR-based methods, however, is their inability to differentiate between DNA from live and dead cells (Scariot et al., 2018; Shehata and Newmaster, 2021; Guo et al., 2022). This implies that the use of qPCR methods may overestimate counts. Therefore, its application in probiotic viability determination is limited unless coupled with another technique that allows selective quantification of viable counts.
The challenge of distinguishing between live and dead cells encountered with general qPCR methods can be circumvented with the inclusion of viability dyes (Scariot et al., 2018; Shi et al., 2022; Shehata et al., 2023). At present, there are three types of viability dyes used for the selective quantification of viable bacterial cells, namely ethidium monoazide (EMA), propidium monoazide (PMA), and PMAxx, an improved version of PMA (Lv et al., 2021; Shehata and Newmaster, 2021; Mu et al., 2022). PMA is a next-generation viability dye developed in 2006 to overcome the challenges of EMA, which was found to penetrate cell membranes of live cells of some bacterial species (Nocker et al., 2006).
Propidium monoazide was produced through the chemical modification of propidium iodide by replacing the amino group on the phenanthridine ring with the azide group that can form a covalent crosslink with the DNA (Nocker et al., 2006; Cangelosi and Meschke, 2014). Recently, an improved and more effective version of PMA with the same spectral properties, PMAxx, was developed by Biotium Inc. PMAxx is a new generation DNA intercalating and membrane impermeant dye that can only penetrate the cell membranes of dead cells (Guo et al., 2022; Kallastu et al., 2023). The procedure for the PMAxx-based method involves an initial stage of incubation of the food sample with about 25–150 μM of PMAxx in the dark to allow the dye to penetrate compromised cell membranes and intercalate with the DNA (Nocker et al., 2006; Mu et al., 2022). Upon exposure to bright light, the azide group of PMAxx produces nitrene, a highly photo-reactive molecule, that forms a covalent crosslink with DNA or reacts with water to form hydroxylamine (an inactivated form of PMAxx) (Nocker et al., 2006; Fittipaldi et al., 2012). The PMAxx-DNA conjugate is insoluble and, therefore, is removed with cell debris during DNA extraction, while the remaining conjugates are not amplified during PCR (Nocker et al., 2006). Hence, only DNA from live cells is amplified during qPCR.
A few studies have reported the application of PMA-qPCR methods for quantification of Bifidobacterium spp. in fermented dairy products (Table 3). PMA-qPCR methods can provide insight into the physiological state of probiotics due to their ability to detect cells in the VBNC state (Dong et al., 2020). Kibbee and Örmeci (2017) reported the application of PMA-qPCR for quantification of VBNC cells of E. coli in wastewater effluents, while Guo et al. (2021) used the same method for VBNC cell enumeration in drinking water. Bifidobacterium spp. can exhibit the VBNC state as a protective mechanism when under stressful environments (Lahtinen et al., 2008). For example, B. longum and B. animalis subsp. lactis were found to exhibit the VBNC state under acidic conditions in fermented products (Lahtinen et al., 2008). The presence of VBNC Bifidobacterium spp. in fermented dairy products, can lead to an underestimation of viability, a key parameter in probiotic quality assurance. Several factors must be considered for the effective application of PMA-qPCR methods in yogurt. A review by Fittipaldi et al. (2012) identified factors that can affect the efficiency of PMA-qPCR methods. These factors include probiotic species type, dye concentration, pH and turbidity of the sample. The authors recommended a pH adjustment and dilution for highly acidic (pH ≤ 4) and turbid (≥10 Nephelometric Turbidity Units, NTU) samples, respectively. Several PMA-qPCR studies have included the pretreatment step of fermented dairy products before PMA treatment to disperse the casein micelles and to adjust the pH to 6.5 (Scariot et al., 2018; Yang et al., 2021). In addition, PMA treatment in these studies was carried out in clear media such as water or phosphate-buffered saline (PBS). However, the inclusion of this pretreatment step is not consistent with the PMA-qPCR methods for the quantification of probiotics in dairy-fermented products. For example, Shi et al. (2022) added PMA directly to the fermented milk sample (1 mL) without including a pretreatment step. In all these studies, PMA-qPCR methods selectively quantified live probiotic cells in fermented dairy products (Scariot et al., 2018; Yang et al., 2021; Shi et al., 2022). This shows that PMA may be applied directly to fermented dairy milk. However, additional PMA-qPCR studies on fermented dairy milk are necessary to support this conclusion and to comprehend the impact of fermented milk product pH, such as yogurt, on PMA efficacy.
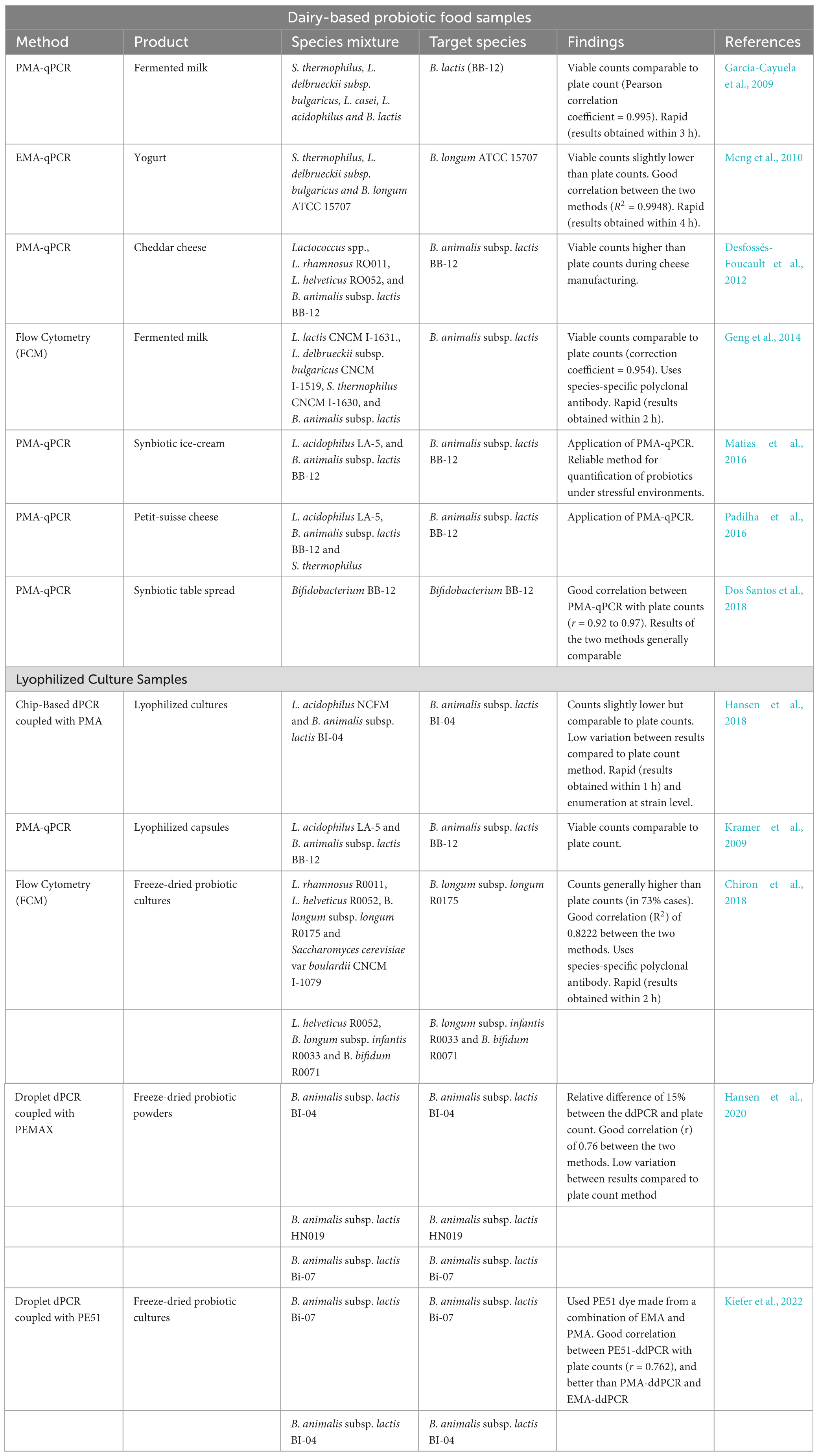
Table 3. Summary of culture-independent techniques that have been used for the determination of Bifidobacterium spp. viability in yogurt and other dairy-based products.
Digital PCR (dPCR) is a third-generation PCR and an emerging technology for microbial quantification (Agrimonti et al., 2019; Kiefer et al., 2022). Unlike qPCR, dPCR does not require standard curves for absolute quantification, and its sample preparation and amplification confirmation methods are different (Salipante and Jerome, 2020). In dPCR, a PCR reaction mixture containing the target sequence is partitioned randomly into thousands of small individual microreactors, such as oil droplets and chip wells (Hansen et al., 2020; Salipante and Jerome, 2020; Lv et al., 2021). A signal fluorescence from partitions containing a single copy of the target sequence is measured at the end of a PCR run (Hansen et al., 2018, 2020; Salipante and Jerome, 2020). Poisson statistics based on comparing the number of positive (with signal fluorescence) and negative reactions is used to determine the absolute quantity of the target sequence (Hansen et al., 2020; Salipante and Jerome, 2020). Like normal qPCR, dPCR cannot distinguish between live and dead cells; hence, it is coupled with viability dyes for selective quantification of live cells (Kiefer et al., 2022). Studies have reported the application of dPCR methods for quantification of Bifidobacterium spp. and other lactic acid bacterial species, mainly in freeze-dried products, indicating a good correlation with plate counts (Summarized in Table 3). In addition, dPCR methods hold several advantages over culture-based methods. These include short results turnaround time, low variation between results and the ability to quantify probiotics at strain levels (Hansen et al., 2020). However, high DNA concentrations can affect probiotic quantification by dPCR (Kim et al., 2023). For example, a recent study quantifying probiotic L. casei in milk showed that quantification at high DNA concentrations was not possible as dPCR was saturated and resulted in a narrow linear dynamic range (Kim et al., 2023). However, this can be solved by diluting the sample (Kim et al., 2023).
Although the application of next generation sequencing (NGS) methods in the study of food microbiomes has been widely reported (Cao et al., 2017; Jagadeesan et al., 2019), their use as quality assurance tools for probiotic viability in foods is still limited. NGS applications in foods have mainly been limited to metagenomic analysis that provides data on relative abundance of different taxa (Cao et al., 2017). Despite the high-resolution ability of NGS methods, the inability to provide information on the absolute quantities and viability status of the organisms in the food has limited its use in viability enumeration. Interestingly, some recent studies have shown the potential application of NGS in viability determination when the technology is coupled with a viability dye (Kallastu et al., 2023). Using the 16S rRNA gene amplicon sequencing coupled with PMAxx reagent, Kallastu et al. (2023) elaborated a workflow for the determination of absolute numbers of viable organisms in kimchi and sauerkraut. The developed workflow involved the addition of a spike-in control (standard) into the sample following PMAxx treatment. After the 16S rRNA gene amplicon sequencing, the number of viable bacteria was determined from the relative abundance and the ratios of the spike-in reads (Kallastu et al., 2023).
Culture-independent, label-free, non-invasive, rapid single-cell Raman spectroscopy (SCRS)-based techniques that give a collective insight into the organism’s phenome and genome are emerging as potential methods of microbial characterization (Jayan et al., 2022; Zhang et al., 2023). Recently, a novel automated SCRS-based technique that combines single-cell identification, viability, vitality and sequencing (SCIVVS) for the characterization of probiotics including Bifidobacterium spp., was described by Zhang et al. (2023). In the SCIVVS technique, probiotic characterization is a stepwise process where cells are first harvested from the probiotic sample, treated with a stable isotope probing (SIP), namely, deuterium oxide (D2O), which can only be taken up by live cells and subjected to Raman spectroscopy at single-cell resolution (Zhang et al., 2023). Other isotopes, such as 13C and 15N, respectively, can also be used for SIP in SCRS-based techniques (Jayan et al., 2022). SIP is based on the principle that the Raman spectra shift when an atom is substituted with its heavier isotope (Jayan et al., 2022). Hence, in the case of H2O and D2O (heavy water), the uptake of H2O in live and metabolically active cells is in the silent region (2040–2300 cm–1) of SCRS (He Y. et al., 2019). The uptake of D2O by the cells results in the partial replacement of the hydrogen (H) atom with the deuterium atom (D) (He Y. et al., 2019; Jayan et al., 2022). This results in the production of C – D bands, which can be modeled to deduce the metabolic state and viability of the cell (He Y. et al., 2019; Zhang et al., 2023). Hence, the SCRS of D2O-treated cells can be used to determine probiotic viability, vitality, and species-level identification (based on a compiled SCRS database of reference species) (Zhang et al., 2023). SCIVVS is also coupled with a single-cell assembly genome sequencing and thus gives comprehensive analysis from genome to phenome (Zhang et al., 2023). However, more studies are needed on applying and assessing the suitability of SCRS-based techniques for probiotic quantification.
Bifidobacterium species are one of the most important members of the GIT of healthy humans, with a substantial body of evidence showing their beneficial probiotic functionalities in experimental models and human trials. Notably, while evidence has shown that some physiological effects of probiotics, such as immunomodulatory properties, can be elicited by bacterial cell components like lipoteichoic acids and peptidoglycan, which can be constituents of both live and dead cells, some probiotic functionalities are dependent on viability and are species and strain specific. Therefore, as envisioned in the definition, viability is a critical quality assurance parameter for probiotics and probiotic foods. Unlike other common probiotic organisms such as lactic acid bacteria, the incorporation and sustained survival of bifidobacteria in probiotic carrier foods like yogurt is a significant challenge. Yogurt process stress factors such as acidity, oxygen, heat, osmotically active solutes, and cold storage impede Bifidobacterium spp. survival and result in viability decline over the product shelf life. Although several studies have investigated the phenotypic responses of different Bifidobacterium spp. to these stress factors, molecular stress response mechanisms still need to be fully elucidated. Response mechanisms to osmotic and cold stress particularly, are yet to be deciphered. Understanding the bifidobacterial stress response mechanisms is critical to the development of strategies to preserve cell viability. Hence, approaches such as stress adaptation, process modifications, microencapsulation and the use of stress protective agents have been investigated, with varying levels of success, as viability retention methods. In addition to viability retention approaches, viability measurement is the ultimate quality assurance requirement for probiotic foods. However, the available standard culture-based methods have proved inadequate for accurately determining Bifidobacterium viability in probiotic yogurt.
To a limited extent, flow cytometry has been considered an alternative method for the viability determination of yogurt cultures. However, the method lacks specificity and has limited application for Bifidobacterium viability. New innovations in immuno- flow cytometry, where fluorescent viability staining is linked to monoclonal antibodies, are expected to improve the applicability of the technology. Moving forward, molecular-based methods such as PMA-qPCR, digital PCR and sequencing, represent the future generation of methods for viability determination. Already, metagenomic sequencing, is used in microbiome analysis and relative quantification in foods. Innovative approaches to adapt the next generation sequencing for absolute microbial and viability quantification by coupling the sequencing with viability dyes and spike-in controls represents novel methods for the future. The adaptation of a recently described novel next-generation method called single-cell identification, viability and vitality tests, and source-tracking (SCIVVS) utilizes a D2O-probed single-cell raman spectrum (SCRS) that can accurately quantify cell viability at the species level based on the C–D band. Crucially, these alternative methods with the accompanying techno-economic assessments, should be developed into standardized and validated protocols for industrial probiotic viability quality assurance applications.
TS: Conceptualization, Formal analysis, Investigation, Methodology, Writing—original draft, Writing—review and editing. TM: Conceptualization, Formal analysis, Investigation, Methodology, Writing—original draft, Writing—review and editing. UT: Conceptualization, Formal analysis, Investigation, Methodology, Writing—original draft, Writing—review and editing. MT: Conceptualization, Funding acquisition, Project administration, Supervision, Writing—review and editing. EB: Conceptualization, Funding acquisition, Project administration, Supervision, Writing—review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors acknowledge Milk South Africa (Milk SA) (Project Number: PRJ-0344) for providing financial support for project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbas, M. S., Saeed, F., Afzaal, M., Jianfeng, L., Hussain, M., Ikram, A., et al. (2022). Recent trends in encapsulation of probiotics in dairy and beverage: a review. J. Food Processing Preserv. 46:e16689.
Afjeh, M. E. A., Pourahmad, R., Akbari-adergani, B., and Azin, M. (2019). Use of glucose oxidase immobilized on magnetic chitosan nanoparticles in probiotic drinking yogurt. Food Sci. Anim. Resour. 39, 73–83. doi: 10.5851/kosfa.2019.e5
Agrimonti, C., Bottari, B., Sardaro, M. L. S., and Marmiroli, N. (2019). Application of real-time PCR (qPCR) for characterization of microbial populations and type of milk in dairy food products. Crit. Rev. Food Sci. Nutrition 59, 423–442. doi: 10.1080/10408398.2017.1375893
Akalin, A., Kesenkas, H., Dinkci, N., Unal, G., Ozer, E., and Kınık, O. (2018). Enrichment of probiotic ice cream with different dietary fibers: structural characteristics and culture viability. J. Dairy Sci. 101, 37–46. doi: 10.3168/jds.2017-13468
Akalin, A. S., Unal, G., and Dinkci, N. (2018). Angiotensin-converting enzyme inhibitory and starter culture activities in probiotic yoghurt: effect of sodium–calcium caseinate and whey protein concentrate. Int. J. Dairy Technol. 71, 185–194.
Akay, H. K., Bahar Tokman, H., Hatipoglu, N., Hatipoglu, H., Siraneci, R., Demirci, M., et al. (2014). The relationship between bifidobacteria and allergic asthma and/or allergic dermatitis: a prospective study of 0–3 years-old children in Turkey. Anaerobe 28, 98–103.
Alba, K., and Kontogiorgos, V. (2019). “Seaweed polysaccharides (agar, alginate carrageenan,” in Encyclopedia of Food Chemistry, eds L. Melton, F. Shahidi, and P. Varelis (Oxford: Academic Press), 240–250.
Alessandri, G., van Sinderen, D., and Ventura, M. (2021). The genus Bifidobacterium: from genomics to functionality of an important component of the mammalian gut microbiota. Comput. Struct. Biotechnol. J. 19, 1472–1487. doi: 10.1016/j.csbj.2021.03.006
Álvarez-Mercado, A. I., Plaza-Díaz, J., de Almagro, M. C., Gil, Á, Moreno-Muñoz, J. A., and Fontana, L. (2022). Bifidobacterium longum subsp. infantis CECT 7210 reduces inflammatory cytokine secretion in Caco-2 cells cultured in the presence of Escherichia coli CECT 515. Int. J. Mol. Sci. 23:10813. doi: 10.3390/ijms231810813
An, H., Douillard, F. P., Wang, G., Zhai, Z., Yang, J., Song, S., et al. (2014). Integrated transcriptomic and proteomic analysis of the bile stress response in a centenarian-originated probiotic Bifidobacterium longum BBMN68. Mol. Cell Proteomics 13, 2558–2572. doi: 10.1074/mcp.M114.039156
Arboleya, S., Watkins, C., Stanton, C., and Ross, R. P. (2016). Gut bifidobacteria populations in human health and aging. Front. Microbiol. 7:1204. doi: 10.3389/fmicb.2016.01204
Ashraf, R., and Shah, N. P. (2011). Selective and differential enumerations of Lactobacillus delbrueckii subsp. bulgaricus, Streptococcus thermophilus, Lactobacillus acidophilus, Lactobacillus casei and Bifidobacterium spp. in yoghurt - a review. Int. J. Food Microbiol. 149, 194–208. doi: 10.1016/j.ijfoodmicro.2011.07.008
Ashraf, R., and Smith, S. C. (2015). Selective enumeration of dairy based strains of probiotic and lactic acid bacteria. Int. Food Res. J. 22, 2576–2586.
Averina, O. V., Zakharevich, N. V., and Danilenko, V. N. (2012). Identification and characterization of WhiB-like family proteins of the Bifidobacterium genus. Anaerobe 18, 421–429. doi: 10.1016/j.anaerobe.2012.04.011
Ayivi, R. D., and Ibrahim, S. A. (2022). Lactic acid bacteria: an essential probiotic and starter culture for the production of yoghurt. Int. J. Food Sci. Technol. 57, 7008–7025. doi: 10.3390/nu14245357
Bazinet, L., Péricou, J., and Araya-Farias, M. (2009). Effect of flow rate and acid molarity on redox potential modulation during electroreduction of milk and simulated milk aqueous mineral phase. Food Chem. 114, 919–926.
Belenguer, A., Duncan, S. H., Calder, A. G., Holtrop, G., Louis, P., Lobley, G. E., et al. (2006). Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 72, 3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006
Berger, B., Moine, D., Mansourian, R., and Arigoni, F. (2010). HspR mutations are naturally selected in Bifidobacterium longum when successive heat shock treatments are applied. J. Bacteriol. 192, 256–263. doi: 10.1128/JB.01147-09
Biavati, B., and Mattarelli, P. (2015). “Bifidobacterium,” in Bergey’s Manual of Systematics of Archaea and Bacteria, eds D. R. Boone, R. W. Castenholz, and G. M. Garrity (Berlin: Springer).
Bolduc, M.-P., Raymond, Y., Fustier, P., Champagne, C. P., and Vuillemard, J.-C. (2006). Sensitivity of bifidobacteria to oxygen and redox potential in non-fermented pasteurized milk. Int. Dairy J. 16, 1038–1048.
Brauer, A. M., Shi, H., Levin, P. A., and Huang, K. C. (2023). Physiological and regulatory convergence between osmotic and nutrient stress responses in microbes. Curr. Opin. Cell Biol. 81:102170. doi: 10.1016/j.ceb.2023.102170
Bunesova, V., Musilova, S., Geigerova, M., Pechar, R., and Rada, V. (2015). Comparison of mupirocin-based media for selective enumeration of bifidobacteria in probiotic supplements. J. Microbiol. Methods 109, 106–109. doi: 10.1016/j.mimet.2014.12.016
Buran, İ, Akal, C., Ozturkoglu-Budak, S., and Yetisemiyen, A. (2021). Rheological, sensorial and volatile profiles of synbiotic kefirs produced from cow and goat milk containing varied probiotics in combination with fructooligosaccharide. LWT 148:111591.
Cangelosi, G. A., and Meschke, J. S. (2014). Dead or alive: molecular assessment of microbial viability. Appl. Environ. Microbiol. 80, 5884–5891.
Cao, Y., Fanning, S., Proos, S., Jordan, K., and Srikumar, S. (2017). A review on the applications of next generation sequencing technologies as applied to food-related microbiome studies. Front. Microbiol. 8:1829. doi: 10.3389/fmicb.2017.01829
Castro-Herrera, V. M., Rasmussen, C., Wellejus, A., Miles, E. A., and Calder, P. C. (2020). In vitro effects of live and heat-inactivated Bifidobacterium animalis subsp. lactis, BB-12 and Lactobacillus rhamnosus GG on Caco-2 Cells. Nutrients 12:1719. doi: 10.3390/nu12061719
Cedran, M. F., Rodrigues, F. J., and Bicas, J. L. (2021). Encapsulation of Bifidobacterium BB12§in alginate-jaboticaba peel blend increases encapsulation efficiency and bacterial survival under adverse conditions. Appl. Microbiol. Biotechnol. 105, 119–127. doi: 10.1007/s00253-020-11025-9
Chen, J., Wang, J., and Zheng, H. (2021). Characterization of Bifidobacterium apousia sp. nov., Bifidobacterium choladohabitans sp. nov., and Bifidobacterium polysaccharolyticum sp. nov., three novel species of the genus Bifidobacterium from honey bee gut. Syst. Appl. Microbiol. 44:126247. doi: 10.1016/j.syapm.2021.126247
Chen, M.-J., Tang, H.-Y., and Chiang, M.-L. (2017). Effects of heat, cold, acid and bile salt adaptations on the stress tolerance and protein expression of kefir-isolated probiotic Lactobacillus kefiranofaciens M1. Food Microbiol. 66, 20–27. doi: 10.1016/j.fm.2017.03.020
Chiron, C., Tompkins, T. A., and Burguičre, P. (2018). Flow cytometry: a versatile technology for specific quantification and viability assessment of micro-organisms in multistrain probiotic products. J. Appl. Microbiol. 124, 572–584. doi: 10.1111/jam.13666
Colston, J. M., Taniuchi, M., Ahmed, T., Ferdousi, T., Kabir, F., Mduma, E., et al. (2022). Intestinal colonization with Bifidobacterium longum subspecies is associated with length at birth, exclusive breastfeeding, and decreased risk of enteric virus infections, but not with histo-blood group antigens, oral vaccine response or later growth in three birth cohorts. Front. Pediatrics 10:804798. doi: 10.3389/fped.2022.804798
Cruz, A. G., Castro, W. F., Faria, J. A. F., Bogusz, S., Granato, D., Celeguini, R. M. S., et al. (2012a). Glucose oxidase: a potential option to decrease the oxidative stress in stirred probiotic yogurt. LWT 47, 512–515.
Cruz, A. G., Castro, W. F., Faria, J. A. F., Lollo, P. C. B., Amaya-Farfán, J., Freitas, M. Q., et al. (2012b). Probiotic yogurts manufactured with increased glucose oxidase levels: Postacidification, proteolytic patterns, survival of probiotic microorganisms, production of organic acid and aroma compounds. J. Dairy Sci. 95, 2261–2269. doi: 10.3168/jds.2011-4582
Cruz, A. G., Castro, W. F., Faria, J. A. F., Bolini, H. M. A., Celeghini, R. M. S., Raices, R. S. L., et al. (2013). Stability of probiotic yogurt added with glucose oxidase in plastic materials with different permeability oxygen rates during the refrigerated storage. Food Res. Int. 51, 723–728.
Cui, S., Zhou, W., Tang, X., Zhang, Q., Yang, B., Zhao, J., et al. (2022). The effect of proline on the freeze-drying survival rate of Bifidobacterium longum CCFM 1029 and its inherent mechanism. Int. J. Mol. Sci. 23:13500. doi: 10.3390/ijms232113500
Cui, S. M., Zhao, J. X., Liu, X. M., Chen, Y. Q., Zhang, H., and Chen, W. (2016). Maximum-biomass concentration prediction for Bifidobacteria in the pH-controlled fed-batch culture. Lett. Appl. Microbiol. 62, 256–263. doi: 10.1111/lam.12540
Dave, R. I., and Shah, N. P. (1996). Evaluation of media for selective enumeration of Streptococcus thermophilus, Lactobacillus delbrueckii ssp. bulgaricus, Lactobacillus acidophilus, and Bifidobacteria. J. Dairy Sci. 79, 1529–1536. doi: 10.3168/jds.S0022-0302(96)76513-X
Dave, R. I., and Shah, N. P. (1997a). Effect of cysteine on the viability of yoghurt and probiotic bacteria in yoghurts made with commercial starter cultures. Int. Dairy J. 7, 537–545. doi: 10.3168/jds.S0022-0302(98)75839-4
Dave, R. I., and Shah, N. P. (1997b). Effectiveness of ascorbic acid as an oxygen scavenger in improving viability of probiotic bacteria in yoghurts made with commercial starter cultures. Int. Dairy J. 7, 435–443.
Davis, C. (2014). Enumeration of probiotic strains: review of culture-dependent and alternative techniques to quantify viable bacteria. J. Microbiol. Methods 103, 9–17. doi: 10.1016/j.mimet.2014.04.012
Derrien, M., Turroni, F., Ventura, M., and Sinderen, D. V. (2022). Insights into endogenous Bifidobacterium species in the human gut microbiota during adulthood. Trends Microbiol. 30, 940–947. doi: 10.1016/j.tim.2022.04.004
Desfossés-Foucault, É, Dussault-Lepage, V., Le Boucher, C., Savard, P., LaPointe, G., and Roy, D. (2012). Assessment of probiotic viability during Cheddar cheese manufacture and ripening using propidium monoazide-PCR quantification. Front. Microbiol. 3:350. doi: 10.3389/fmicb.2012.00350
Dimitrellou, D., Kandylis, P., and Kourkoutas, Y. (2019). Assessment of freeze-dried immobilized Lactobacillus casei as probiotic adjunct culture in yogurts. Foods 8:374. doi: 10.3390/foods8090374
Din, A. U., Hassan, A., Zhu, Y., Zhang, K., Wang, Y., Li, T., et al. (2020). Inhibitory effect of Bifidobacterium bifidum ATCC 29521 on colitis and its mechanism. J. Nutritional Biochem. 79:108353. doi: 10.1016/j.jnutbio.2020.108353
Dong, K., Pan, H., Yang, D., Rao, L., Zhao, L., Wang, Y., et al. (2020). Induction, detection, formation, and resuscitation of viable but non-culturable state microorganisms. Comprehensive Rev. Food Sci. Food Safety 19, 149–183.
Dos Santos, C. S., de Souza, C. H. B., Padilha, M., Gioielli, L. A., Ract, J. N. R., and Saad, S. M. I. (2018). Milk fat protects Bifidobacterium animalis subsp. lactis Bb-12 from in vitro gastrointestinal stress in potentially synbiotic table spreads. Food Function 9, 4274–4281. doi: 10.1039/c8fo00506k
Dubey, M., Zehra, A., Aamir, M., Meena, M., Ahirwal, L., Singh, S., et al. (2017). Improvement strategies, cost effective production, and potential applications of fungal glucose oxidase (GOD): current updates. Front. Microbiol. 13:1032. doi: 10.3389/fmicb.2017.01032
Duranti, S., Longhi, G., Ventura, M., van Sinderen, D., and Turroni, F. (2021). Exploring the ecology of bifidobacteria and their genetic adaptation to the mammalian gut. Microorganisms 9:8. doi: 10.3390/microorganisms9010008
Duranti, S., Ruiz, L., Lugli, G. A., Tames, H., Milani, C., Mancabelli, L., et al. (2020). Bifidobacterium adolescentis as a key member of the human gut microbiota in the production of GABA. Sci. Rep. 10:14112. doi: 10.1038/s41598-020-70986-z
Ebel, B., Martin, F., Le, L. D. T., Gervais, P., and Cachon, R. (2011). Use of gases to improve survival of Bifidobacterium bifidum by modifying redox potential in fermented milk. J. Dairy Sci. 94, 2185–2191. doi: 10.3168/jds.2010-3850
Eckel, V. P. L., Ziegler, L.-M., Vogel, R. F., and Ehrmann, M. (2020). Bifidobacterium tibiigranuli sp. nov. isolated from homemade water kefir. Int. J. Syst. Evol. Microbiol. 70, 1562–1570. doi: 10.1099/ijsem.0.003936
Erkaya-Kotan, T. (2020). In vitro angiotensin converting enzyme (ACE)-inhibitory and antioxidant activity of probiotic yogurt incorporated with orange fibre during storage. J. Food Sci. Technol. 57, 2343–2353. doi: 10.1007/s13197-020-04272-1
Fan, X., Li, X., Zhang, T., Xu, J., Shi, Z., Wu, Z., et al. (2021). A novel qPCR method for the detection of lactic acid bacteria in fermented milk. Foods 10:3066.
Fiocco, D., Longo, A., Arena, M. P., Russo, P., Spano, G., and Capozzi, V. (2020). How probiotics face food stress: they get by with a little help. Crit. Rev. Food Sci. Nutrition 60, 1552–1580. doi: 10.1080/10408398.2019.1580673
Fittipaldi, M., Nocker, A., and Codony, F. (2012). Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 91, 276–289. doi: 10.1016/j.mimet.2012.08.007
Foglia, C., Allesina, S., Amoruso, A., De Prisco, A., and Pane, M. (2020). New insights in enumeration methodologies of probiotic cells in finished products. J. Microbiol. Methods 175:105993. doi: 10.1016/j.mimet.2020.105993
Food and Agriculture Organization of the United Nations [FAO], and World Health Organization [WHO] (2001). Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria. Report from FAO/WHO expert consultation. Rome: Food and Agriculture Organization.
Food and Agriculture Organization of the United Nations [FAO], and World Health Organization [WHO] (2002). Guidelines for the evaluation of probiotics in food. Report of a Joint FAO/WHO Working group on drafting guidelines for the evaluation of probiotics in food. Rome: Food and Agriculture Organization.
Frakolaki, G., Giannou, V., Kekos, D., and Tzia, C. (2021). A review of the microencapsulation techniques for the incorporation of probiotic bacteria in functional foods. Crit. Rev. Food Sci. Nutrition 61, 1515–1536.
Frakolaki, G., Kekes, T., Lympaki, F., Giannou, V., and Tzia, C. (2022). Use of encapsulated Bifidobacterium animalis subsp. lactis through extrusion or emulsification for the production of probiotic yogurt. J. Food Process Eng. 45:e13792.
Gao, X., Jia, R., Xie, L., Kuang, L., Feng, L., and Wan, C. (2015). Obesity in school-aged children and its correlation with gut E. coli and Bifidobacteria: a case–control study. BMC Pediatrics 15:64. doi: 10.1186/s12887-015-0384-x
García-Cayuela, T., Tabasco, R., Peláez, C., and Requena, T. (2009). Simultaneous detection and enumeration of viable lactic acid bacteria and bifidobacteria in fermented milk by using propidium monoazide and real-time PCR. Int. Dairy J. 19, 405–409.
Geng, J., Chiron, C., and Combrisson, J. (2014). Rapid and specific enumeration of viable Bifidobacteria in dairy products based on flow cytometry technology: a proof of concept study. Int. Dairy J. 37, 1–4.
Geraldo, B. M. C., Batalha, M. N., Milhan, N. V. M., Rossoni, R. D., Scorzoni, L., and Anbinder, A. L. (2020). Heat-killed Lactobacillus reuteri and cell-free culture supernatant have similar effects to viable probiotics during interaction with Porphyromonas gingivalis. J. Periodontal Res. 55, 215–220. doi: 10.1111/jre.12704
Ghaderi-Ghahfarokhi, M., Yousefvand, A., Ahmadi Gavlighi, H., and Zarei, M. (2021). The effect of hydrolysed tragacanth gum and inulin on the probiotic viability and quality characteristics of low-fat yoghurt. Int. J. Dairy Technol. 74, 161–169.
González-Orozco, B. D., García-Cano, I., Jiménez-Flores, R., and Alvárez, V. B. (2022). Invited review: milk kefir microbiota—direct and indirect antimicrobial effects. J. Dairy Sci. 105, 3703–3715. doi: 10.3168/jds.2021-21382
Groeger, D., O’Mahony, L., Murphy, E. F., Bourke, J. F., Dinan, T. G., Kiely, B., et al. (2013). Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut Microbes 4, 325–339. doi: 10.4161/gmic.25487
Guo, J., Wang, W., Zhao, H., Luo, Y., Wan, M., and Li, Y. (2022). A new PMA-qPCR method for rapid and accurate detection of viable bacteria and spores of marine-derived Bacillus velezensis B-9987. J. Microbiol. Methods 199:106537. doi: 10.1016/j.mimet.2022.106537
Guo, L., Wan, K., Zhu, J., Ye, C., Chabi, K., and Yu, X. (2021). Detection and distribution of VBNC/viable pathogenic bacteria in full-scale drinking water treatment plants. J. Hazardous Materials 406:124335. doi: 10.1016/j.jhazmat.2020.124335
Hagen, P. C., and Skelley, J. W. (2019). Efficacy of Bifidobacterium species in prevention of necrotizing enterocolitis in very-low birth weight infants. a systematic review. J. Pediatric Pharmacol. Therapeutics 24, 10–15. doi: 10.5863/1551-6776-24.1.10
Hansen, S. J. Z., Morovic, W., DeMeules, M., Stahl, B., and Sindelar, C. W. (2018). Absolute enumeration of probiotic strains Lactobacillus acidophilus NCFM§and Bifidobacterium animalis subsp. lactis Bl-04§via Chip-Based Digital PCR. Front. Microbiol. 9:704. doi: 10.3389/fmicb.2018.00704
Hansen, S. J. Z., Tang, P., Kiefer, A., Galles, K., Wong, C., and Morovic, W. (2020). Droplet digital PCR is an improved alternative method for high-quality enumeration of viable probiotic strains. Front. Microbiol. 10:3025. doi: 10.3389/fmicb.2019.03025
Hao, Y., Wu, T., Guo, S., Kwok, L.-Y., Zhang, H., and Wang, J. (2023). Metabolic dynamics of fermented milk beverages co-fermented with Bifidobacterium animalis subsp. lactis Probio-M8 and Lacticaseibacillus paracasei PC-01 during storage. LWT 185:115196.
He, B.-L., Xiong, Y., Hu, T.-G., Zong, M.-H., and Wu, H. (2023). Bifidobacterium spp. as functional foods: a review of current status, challenges, and strategies. Crit. Rev. Food Sci. Nutrition 63, 8048–8065. doi: 10.1080/10408398.2022.2054934
He, J., Han, Y., Liu, M., Wang, Y., Yang, Y., and Yang, X. (2019). Effect of 2 types of resistant starches on the quality of yogurt. J. Dairy Sci. 102, 3956–3964.
He, Y., Wang, X., Ma, B., and Xu, J. (2019). Ramanome technology platform for label-free screening and sorting of microbial cell factories at single-cell resolution. Biotechnol. Adv. 37:107388. doi: 10.1016/j.biotechadv.2019.04.010
Hoefel, D., Grooby, W. L., Monis, P. T., Andrews, S., and Saint, C. P. (2003). A comparative study of carboxyfluorescein diacetate and carboxyfluorescein diacetate succinimidyl ester as indicators of bacterial activity. J. Microbiol. Methods 52, 379–388. doi: 10.1016/s0167-7012(02)00207-5
Hu, C., Yang, J., Qi, Z., Wu, H., Wang, B., Zou, F., et al. (2022). Heat shock proteins: biological functions, pathological roles, and therapeutic opportunities. MedComm 3:e161.
International Organization for Standardization [ISO], and International Dairy Federation [IDF] (2010). ISO 29981:2010 — IDF 220:2010. Milk products - Enumeration of presumptive bifidobacteria - Colony count technique at 37 degrees C. Geneva: International Organization for Standardization.
International Organization for Standardization [ISO], and International Dairy Federation [IDF] (2015). ISO 19344: IDF 232 - Milk and milk products - Starter cultures, probiotics and fermented products–Quantification of lactic acid bacteria by flow cytometry. Geneva: International Organization for Standardization.
Jackson, S. A., Schoeni, J. L., Vegge, C., Pane, M., Stahl, B., Bradley, M., et al. (2019). Improving end-user trust in the quality of commercial probiotic products. Front. Microbiol. 10:739. doi: 10.3389/fmicb.2019.00739
Jagadeesan, B., Gerner-Smidt, P., Allard, M. W., Leuillet, S., Winkler, A., Xiao, Y., et al. (2019). The use of next generation sequencing for improving food safety: translation into practice. Food Microbiol. 79, 96–115. doi: 10.1016/j.fm.2018.11.005
Jaskulski, I. B., Uecker, J., Bordini, F., Moura, F., Gonçalves, T., Chaves, N. G., et al. (2020). In vivo action of Lactococcus lactis subsp. lactis isolate (R7) with probiotic potential in the stabilization of cancer cells in the colorectal epithelium. Process Biochem. 91, 165–171.
Jayan, H., Pu, H., and Sun, D.-W. (2022). Recent developments in Raman spectral analysis of microbial single cells: techniques and applications. Crit. Rev. Food Sci. Nutrition 62, 4294–4308. doi: 10.1080/10408398.2021.1945534
Ji, R., Wu, J., Zhang, J., Wang, T., Zhang, X., Shao, L., et al. (2019). Extending viability of Bifidobacterium longum in chitosan-coated alginate microcapsules using emulsification and internal gelation encapsulation technology. Front. Microbiol. 10:1389. doi: 10.3389/fmicb.2019.01389
Jiang, Y., Ren, F., Liu, S., Zhao, L., Guo, H., and Hou, C. (2016). Enhanced acid tolerance in Bifidobacterium longum by adaptive evolution: comparison of the genes between the acid-resistant variant and wild-type strain. J. Microbiol. Biotechnol. 26, 452–460. doi: 10.4014/jmb.1508.08030
Jin, J., Qin, Q., Guo, H., Liu, S., Ge, S., Zhang, H., et al. (2015). Effect of pre-stressing on the acid-stress response in Bifidobacterium revealed using proteomic and physiological approaches. PLoS One 10:e0117702. doi: 10.1371/journal.pone.0117702
Jin, J., Zhang, B., Guo, H., Cui, J., Jiang, L., Song, S., et al. (2012). Mechanism analysis of acid tolerance response of Bifidobacterium longum subsp. longum BBMN 68 by gene expression profile using RNA-sequencing. PLoS One 7:e50777. doi: 10.1371/journal.pone.0050777
Kallastu, A., Malv, E., Aro, V., Meikas, A., Vendelin, M., Kattel, A., et al. (2023). Absolute quantification of viable bacteria abundances in food by next-generation sequencing: quantitative NGS of viable microbes. Curr. Res. Food Sci. 6:100443. doi: 10.1016/j.crfs.2023.100443
Karimi, R., Mortazavian, A. M., and Amiri-Rigi, A. (2012). Selective enumeration of probiotic microorganisms in cheese. Food Microbiol. 29, 1–9. doi: 10.1016/j.fm.2011.08.008
Kato, K., Odamaki, T., Mitsuyama, E., Sugahara, H., Xiao, J. Z., and Osawa, R. (2017). Age-related changes in the composition of gut Bifidobacterium species. Curr. Microbiol. 74, 987–995. doi: 10.1007/s00284-017-1272-4
Kawasaki, S., Watanabe, M., Fukiya, S., and Yokota, A. (2018). “Stress responses of bifidobacteria: oxygen and bile acid as the stressors,” in The Bifidobacteria and Related Organisms, eds P. Mattarelli, B. Biavati, W. H. Holzapfel, and B. J. B. Wood (Cambridge, CA: Academic Press), 131–143. doi: 10.1128/mmbr.00170-21
Kelly, S. M., Munoz-Munoz, J., and Van Sinderen, D. (2021). Plant glycan metabolism by bifidobacteria. Front. Microbiol. 12:609418. doi: 10.3389/fmicb.2021.609418
Kibbee, R. J., and Örmeci, B. (2017). Development of a sensitive and false-positive free PMA-qPCR viability assay to quantify VBNC Escherichia coli and evaluate disinfection performance in wastewater effluent. J. Microbiol. Methods 132, 139–147. doi: 10.1016/j.mimet.2016.12.004
Kiefer, A., Byrd, P. M., Tang, P., Jones, G., Galles, K., Fallico, V., et al. (2022). Viability droplet digital polymerase chain reaction accurately enumerates probiotics and provides insight into damage experienced during storage. Front. Microbiol. 13:966264. doi: 10.3389/fmicb.2022.966264
Kim, E., Yang, S.-M., Choi, C. H., Shin, M.-K., and Kim, H.-Y. (2023). Droplet digital PCR method for the absolute quantitative detection and monitoring of Lacticaseibacillus casei. Food Microbiol. 113:104265. doi: 10.1016/j.fm.2023.104265
Kim, H., Jeong, Y., Kang, S., You, H. J., and Ji, G. E. (2020). Co-Culture with Bifidobacterium catenulatum improves the growth, gut colonization, and butyrate production of Faecalibacterium prausnitzii: in vitro and in vivo studies. Microorganisms 8:788. doi: 10.3390/microorganisms8050788
Kim, H. B., Kim, E., Yang, S. M., Lee, S., Kim, M. J., and Kim, H. Y. (2020). Development of real-time PCR assay to specifically detect 22 Bifidobacterium species and subspecies using comparative genomics. Front. Microbiol. 11:2087. doi: 10.3389/fmicb.2020.02087
Konieczna, I., Zarnowiec, P., Kwinkowski, M., Kolesinska, B., Fraczyk, J., Kaminski, Z., et al. (2012). Bacterial urease and its role in long-lasting human diseases. Curr. Protein Peptide Sci. 13, 789–806. doi: 10.2174/138920312804871094
Konieczna, P., Akdis, C. A., Quigley, E. M. M., Shanahan, F., and O’Mahony, L. (2012). Portrait of an immunoregulatory Bifidobacterium. Gut Microbes 3, 261–266.
Kosumi, K., Hamada, T., Koh, H., Borowsky, J., Bullman, S., Twombly, T. S., et al. (2018). The amount of Bifidobacterium genus in colorectal carcinoma tissue in relation to tumor characteristics and clinical outcome. Am. J. Pathol. 188, 2839–2852. doi: 10.1016/j.ajpath.2018.08.015
Kramer, M., Obermajer, N., Bogovič Matijašić, B., Rogelj, I., and Kmetec, V. (2009). Quantification of live and dead probiotic bacteria in lyophilised product by real-time PCR and by flow cytometry. Appl. Microbiol. Biotechnol. 84, 1137–1147. doi: 10.1007/s00253-009-2068-7
Lahtinen, S. J., Ahokoski, H., Reinikainen, J. P., Gueimonde, M., Nurmi, J., Ouwehand, A. C., et al. (2008). Degradation of 16S rRNA and attributes of viability of viable but nonculturable probiotic bacteria. Lett. Appl. Microbiol. 46, 693–698. doi: 10.1111/j.1472-765X.2008.02374.x
Laureys, D., Cnockaert, M., De Vuyst, L., and Vandamme, P. (2016). Bifidobacterium aquikefiri sp. nov., isolated from water kefir. Int. J. Syst. Evol. Microbiol. 66, 1281–1286. doi: 10.1099/ijsem.0.000877
Li, H., Liu, T., Yang, J., Wang, R., Li, Y., Feng, Y., et al. (2021). Effect of a microencapsulated synbiotic product on microbiology, microstructure, textural and rheological properties of stirred yogurt. LWT 152:112302.
Li, L., Yan, Q., Ma, N., Chen, X., Li, G., and Liu, M. (2021). Analysis of intestinal flora and inflammatory cytokine levels in children with non-infectious diarrhea. Translational Pediatrics 10, 1340–1345. doi: 10.21037/tp-21-168
Li, L.-Q., Chen, X., Zhu, J., Zhang, S., Chen, S.-Q., Liu, X., et al. (2023). Advances and challenges in interaction between heteroglycans and Bifidobacterium: utilization strategies, intestinal health and future perspectives. Trends Food Sci. Technol. 134, 112–122.
Lima, K. G. D. C., Kruger, M. F., Behrens, J., Destro, M. T., Landgraf, M., Gombossy, et al. (2009). Evaluation of culture media for enumeration of Lactobacillus acidophilus, Lactobacillus casei and Bifidobacterium animalis in the presence of Lactobacillus delbrueckii subsp bulgaricus and Streptococcus thermophilus. LWT Food Sci. Technol. 42, 491–495.
Liu, H., Cui, S. W., Chen, M., Li, Y., Liang, R., Xu, F., et al. (2019). Protective approaches and mechanisms of microencapsulation to the survival of probiotic bacteria during processing, storage and gastrointestinal digestion: a review. Crit. Rev. Food Sci. Nutrition 59, 2863–2878. doi: 10.1080/10408398.2017.1377684
Lugli, G. A., Calvete-Torre, I., Alessandri, G., Milani, C., Turroni, F., Laiolo, P., et al. (2021). Phylogenetic classification of ten novel species belonging to the genus Bifidobacterium comprising B. phasiani sp. nov., B. pongonis sp. nov., B. saguinibicoloris sp. nov., B. colobi sp. nov., B. simiiventris sp. nov., B. santillanense sp. nov., B. miconis sp. nov., B. amazonense sp. nov., B. pluvialisilvae sp. nov., and B. miconisargentati sp. nov. Syst. Appl. Microbiol. 44:126273.
Lugli, G. A., Mangifesta, M., Duranti, S., Anzalone, R., Milani, C., Mancabelli, L., et al. (2018). Phylogenetic classification of six novel species belonging to the genus Bifidobacterium comprising Bifidobacterium anseris sp. nov., Bifidobacterium criceti sp. nov., Bifidobacterium imperatoris sp. nov., Bifidobacterium italicum sp. nov., Bifidobacterium margollesii sp. nov. and Bifidobacterium parmae sp. nov. Syst. Appl. Microbiol. 41, 173–183. doi: 10.1016/j.syapm.2018.01.002
Luo, Y., Xiao, Y., Zhao, J., Zhang, H., Chen, W., and Zhai, Q. (2021). The role of mucin and oligosaccharides via cross-feeding activities by Bifidobacterium: a review. Int. J. Biol. Macromol. 167, 1329–1337. doi: 10.1016/j.ijbiomac.2020.11.087
Lv, X., Gu, X., Wang, L., He, X., He, C., Zhang, J., et al. (2021). Rapid and absolute quantification of VBNC Cronobacter sakazakii by PMAxx combined with single intact cell droplet digital PCR in infant foods. LWT 145:111388.
Ma, L., and Payne, S. M. (2012). AhpC is required for optimal production of enterobactin by Escherichia coli. J. Bacteriol. 194, 6748–6757. doi: 10.1128/JB.01574-12
Martorell, P., Alvarez, B., Llopis, S., Navarro, V., Ortiz, P., Gonzalez, N., et al. (2021). Heat-treated Bifidobacterium longum CECT-7347: a whole-cell postbiotic with antioxidant, anti-inflammatory, and gut-barrier protection properties. Antioxidants 10:536. doi: 10.3390/antiox10040536
Massoud, R., Belgheisi, S., and Massoud, A. (2016). Effect of high pressure homogenization on improving the quality of milk and sensory properties of yogurt: a review. Int. J. Chem. Eng. Appl. 7, 66–70.
Massoud, R., Fadaei, V., Khosravi-Darani, K., and Nikbakht, H. R. (2015). Improving the viability of probiotic bacteria in yoghurt by homogenization. J. Food Processing Preserv. 39, 2984–2990. doi: 10.1111/jfpp.12551
Matias, N. S., Padilha, M., Bedani, R., and Saad, S. M. I. (2016). In vitro gastrointestinal resistance of Lactobacillus acidophilus La-5 and Bifidobacterium animalis Bb-12 in soy and/or milk-based synbiotic apple ice creams. Int. J. Food Microbiol. 234, 83–93. doi: 10.1016/j.ijfoodmicro.2016.06.037
Meng, X. C., Pang, R., Wang, C., and Wang, L. Q. (2010). Rapid and direct quantitative detection of viable bifidobacteria in probiotic yogurt by combination of ethidium monoazide and real-time PCR using a molecular beacon approach. J. Dairy Res. 77, 498–504. doi: 10.1017/S0022029910000658
Meybodi, N. M., Mortazavian, A. M., Arab, M., and Nematollahi, A. (2020). Probiotic viability in yoghurt: a review of influential factors. Int. Dairy J. 109, 104793.
Michelon, D., Abraham, S., Ebel, B., De Coninck, J., Husson, F., Feron, G., et al. (2010). Contribution of exofacial thiol groups in the reducing activity of Lactococcus lactis. FEBS J. 277, 2282–2290. doi: 10.1111/j.1742-4658.2010.07644.x
Mousa, A. H., Korma, S. A., Ali, A. H., Abdeldaiem, A. M., Bakry, I. A., Liu, X.-M., et al. (2023). Microencapsulation of Bifidobacterium bifidum F-35 via modulation of emulsifying technique and its mechanical effects on the rheological stability of set-yogurt. J. Food Sci. Technol. 60, 2968–2977. doi: 10.1007/s13197-023-05812-1
Mozzetti, V., Grattepanche, F., Moine, D., Berger, B., Rezzonico, E., Meile, L., et al. (2010). New method for selection of hydrogen peroxide adapted bifidobacteria cells using continuous culture and immobilized cell technology. Microbial Cell Factories 9:60. doi: 10.1186/1475-2859-9-60
Mu, D., Zhou, D., Xie, G., Liu, J., Wang, Z., Xiong, Q., et al. (2022). Real-time recombinase-aided amplification with improved propidium monoazide for the rapid detection of viable Escherichia coli O157:H7 in milk. J. Dairy Sci. 105, 1028–1038. doi: 10.3168/jds.2021-21074
Najgebauer-Lejko, D., Liszka, K., Tabaszewska, M., and Domagała, J. (2021). Probiotic yoghurts with sea buckthorn, elderberry, and sloe fruit purees. Molecules 26:2345. doi: 10.3390/molecules26082345
Neuzil-Bunesova, V., Lugli, G. A., Modrackova, N., Vlkova, E., Bolechova, P., Burtscher, J., et al. (2020). Five novel bifidobacterial species isolated from faeces of primates in two Czech zoos: Bifidobacterium erythrocebi sp. nov., Bifidobacterium moraviense sp. nov., Bifidobacterium oedipodis sp. nov., Bifidobacterium olomucense sp. nov. and Bifidobacterium panos sp. nov. Int. J. Syst. Evol. Microbiol. 71:4573. doi: 10.1099/ijsem.0.004573
Nocker, A., Cheung, C.-Y., and Camper, A. K. (2006). Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67, 310–320. doi: 10.1016/j.mimet.2006.04.015
Norouzbeigi, S., Vahid-Dastjerdi, L., Yekta, R., Farhoodi, M., and Mortazavian, A. M. (2021). Effects of using different O2 scavengers on the qualitative attributes of bifidus yogurt during refrigerated storage. Food Res. Int. 140:109953. doi: 10.1016/j.foodres.2020.109953
Nyanzi, R., Jooste, P. J., and Buys, E. M. (2021). Invited review: probiotic yogurt quality criteria, regulatory framework, clinical evidence, and analytical aspects. J. Dairy Sci. 104, 1–19. doi: 10.3168/jds.2020-19116
Odamaki, T., Xiao, J. Z., Yonezawa, S., Yaeshima, T., and Iwatsuki, K. (2011). Improved viability of bifidobacteria in fermented milk by cocultivation with Lactococcus lactis subspecies lactis. J. Dairy Sci. 94, 1112–1121. doi: 10.3168/jds.2010-3286
Ongol, M. P., Sawatari, Y., Ebina, Y., Sone, T., Tanaka, M., Tomita, F., et al. (2007). Yoghurt fermented by Lactobacillus delbrueckii subsp. bulgaricus H+-ATPase-defective mutants exhibits enhanced viability of Bifidobacterium breve during storage. Int. J. Food Microbiol. 116, 358–366. doi: 10.1016/j.ijfoodmicro.2007.02.019
Padilha, M., Morales, M. L. V., Vieira, A. D. S., Costa, M. G. M., and Saad, S. M. I. (2016). A prebiotic mixture improved Lactobacillus acidophilus and Bifidobacterium animalis gastrointestinal in vitro resistance in petit-suisse. Food Function 7, 2312–2319. doi: 10.1039/c5fo01592h
Pápai, G., Torres-Maravilla, E., Chain, F., Varga-Visi, É, Antal, O., Naár, Z., et al. (2021). The administration matrix modifies the beneficial properties of a probiotic mix of Bifidobacterium animalis subsp. lactis BB-12 and Lactobacillus acidophilus LA-5. Probiotics Antimicrob. Proteins 13, 484–494.
Parada Venegas, D., De la Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277.
Peng, C., Yao, G., Sun, Y., Guo, S., Wang, J., Mu, X., et al. (2022). Comparative effects of the single and binary probiotics of Lacticaseibacillus casei Zhang and Bifidobacterium lactis V9 on the growth and metabolomic profiles in yogurts. Food Res. Int. 152:110603. doi: 10.1016/j.foodres.2021.110603
Phadtare, S. (2004). Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6, 125–136.
Pradeep Prasanna, P. H., and Charalampopoulos, D. (2019). Encapsulation in an alginate–goats’ milk–inulin matrix improves survival of probiotic Bifidobacterium in simulated gastrointestinal conditions and goats’ milk yoghurt. Int. J. Dairy Technol. 72, 132–141.
Pyclik, M. J., Srutkova, D., Razim, A., Hermanova, P., Svabova, T., Pacyga, K., et al. (2021). Viability status-dependent effect of Bifidobacterium longum ssp. longum CCM 7952 on prevention of allergic inflammation in mouse model. Front. Immunol. 12:707728. doi: 10.3389/fimmu.2021.707728
Rezzonico, E., Lariani, S., Barretto, C., Cuanoud, G., Giliberti, G., Delley, M., et al. (2007). Global transcriptome analysis of the heat shock response of Bifidobacterium longum. FEMS Microbiol. Lett. 271, 136–145. doi: 10.1111/j.1574-6968.2007.00704.x
Rios-Covian, D., Gueimonde, M., Duncan, S. H., Flint, H. J., and de los Reyes-Gavilan, C. G. (2015). Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol. Lett. 362:fnv176. doi: 10.1093/femsle/fnv176
Rockinger, U., Funk, M., and Winter, G. (2021). Current approaches of preservation of cells during (freeze-) drying. J. Pharmaceutical Sci. 110, 2873–2893.
Roussel, C., Ebel, B., Munier, E., Michelon, D., Martin-Dejardin, F., Beuvier, E., et al. (2022). Green strategies to control redox potential in the fermented food industry. Food Res. Int. 156:111154. doi: 10.1016/j.foodres.2022.111154
Ruggirello, M., Cocolin, L., and Dolci, P. (2016). Fate of Lactococcus lactis starter cultures during late ripening in cheese models. Food Microbiol. 59, 112–118. doi: 10.1016/j.fm.2016.05.001
Ruijter, J. M., Barnewall, R. J., Marsh, I. B., Szentirmay, A. N., Quinn, J. C., van Houdt, R., et al. (2021). Efficiency correction is required for accurate quantitative PCR analysis and reporting. Clin. Chem. 67, 829–842.
Salipante, S. J., and Jerome, K. R. (2020). Digital PCR—an emerging technology with broad applications in microbiology. Clin. Chem. 66, 117–123. doi: 10.1373/clinchem.2019.304048
Sánchez, B., Champomier-Vergčs, M.-C., Collado, M. D. C., Anglade, P., Baraige, F., Sanz, Y., et al. (2007). Low-pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum. Appl. Environ. Microbiol. 73, 6450–6459. doi: 10.1128/AEM.00886-07
Satoh, T., Todoroki, M., Kobayashi, K., Niimura, Y., and Kawasaki, S. (2019). Purified thioredoxin reductase from O2-sensitive Bifidobacterium bifidum degrades H2O2 by interacting with alkyl hydroperoxide reductase. Anaerobe 57, 45–54. doi: 10.1016/j.anaerobe.2019.03.012
Scariot, M. C., Venturelli, G. L., Prudêncio, E. S., and Arisi, A. C. M. (2018). Quantification of Lactobacillus paracasei viable cells in probiotic yoghurt by propidium monoazide combined with quantitative PCR. Int. J. Food Microbiol. 264, 1–7. doi: 10.1016/j.ijfoodmicro.2017.10.021
Schiweck, C., Edwin Thanarajah, S., Aichholzer, M., Matura, S., Reif, A., Vrieze, E., et al. (2022). Regulation of CD4+ and CD8+ T cell biology by short-chain fatty acids and its relevance for autoimmune pathology. Int. J. Mol. Sci. 23:8272. doi: 10.3390/ijms23158272
Schöpping, M., Vesth, T., Jensen, K., Franzén, C. J., and Zeidan, A. A. (2022a). Genome-wide assessment of stress-associated genes in Bifidobacteria. Appl. Environ. Microbiol. 88:e0225121. doi: 10.1128/aem.02251-21
Schöpping, M., Zeidan, A. A., and Franzén, C. J. (2022b). Stress response in Bifidobacteria. Microbiol. Mol. Biol. Rev. 86:e0017021.
Sfakianakis, P., and Tzia, C. (2014). Conventional and innovative processing of milk for yogurt manufacture; development of texture and flavor: a review. Foods 3, 176–193. doi: 10.3390/foods3010176
Shang, J., Wan, F., Zhao, L., Meng, X., and Li, B. (2020). Potential immunomodulatory activity of a selected strain Bifidobacterium bifidum H3-R2 as evidenced in vitro and in immunosuppressed mice. Front. Microbiol. 11:2089. doi: 10.3389/fmicb.2020.02089
Sharma, M., Wasan, A., and Sharma, R. K. (2021). Recent developments in probiotics: an emphasis on Bifidobacterium. Food Biosci. 41:100993.
Shehata, H. R., Hassane, B., and Newmaster, S. G. (2023). Real-time polymerase chain reaction methods for strain specific identification and enumeration of strain Lacticaseibacillus paracasei 8700:2. Front. Microbiol. 13:1076631. doi: 10.3389/fmicb.2022.1076631
Shehata, H. R., and Newmaster, S. G. (2021). Enumeration of probiotic strain Lacticaseibacillus rhamnosus GG (ATCC 53103) using viability real-time PCR. Probiotics Antimicrob. Proteins 13, 1611–1620. doi: 10.1007/s12602-021-09849-6
Shi, Z., Li, X., Fan, X., Xu, J., Liu, Q., Wu, Z., et al. (2022). PMA-qPCR method for the selective quantitation of viable lactic acid bacteria in fermented milk. Front. Microbiol. 13:984506. doi: 10.3389/fmicb.2022.984506
Sugahara, H., Yao, R., Odamaki, T., and Xiao, J. Z. (2017). Differences between live and heat-killed bifidobacteria in the regulation of immune function and the intestinal environment. Beneficial Microbes 8, 463–472. doi: 10.3920/BM2016.0158
Tachon, S., Michelon, D., Chambellon, E., Cantonnet, M., Mezange, C., Henno, L., et al. (2009). Experimental conditions affect the site of tetrazolium violet reduction in the electron transport chain of Lactococcus lactis. Microbiology 155, 2941–2948. doi: 10.1099/mic.0.029678-0
Takada, T., Makino, H., Katto, M., Oishi, K., Hori, T., and Kurakawa, T. (2023). Immunomagnetic separation for isolation and enrichment of Lacticaseibacillus paracasei strain Shirota from human feces. Microbe 1:100007.
Taverniti, V., and Guglielmetti, S. (2014). Methodological issues in the study of intestinal microbiota in irritable bowel syndrome. World J. Gastroenterol. 20, 8821–8836. doi: 10.3748/wjg.v20.i27.8821
Terpou, A., Papadaki, A., Lappa, I., Kachrimanidou, V., Bosnea, L., and Kopsahelis, N. (2019). Probiotics in food systems: significance and emerging strategies towards improved viability and delivery of enhanced beneficial value. Nutrients 11:1591. doi: 10.3390/nu11071591
Tsevdou, M., Ouli-Rousi, M., Soukoulis, C., and Taoukis, P. (2020). Impact of high-pressure process on probiotics: viability kinetics and evaluation of the quality characteristics of probiotic yoghurt. Foods 9:360. doi: 10.3390/foods9030360
Turgut, T., and Cakmakci, S. L. (2018). Probiotic strawberry yogurts: microbiological, chemical and sensory properties. Probiotics Antimicrob. Proteins 10, 64–70. doi: 10.1007/s12602-017-9278-6
Turroni, F., Taverniti, V., Ruas-Madiedo, P., Duranti, S., Guglielmetti, S., Lugli, G. A., et al. (2014). Bifidobacterium bifidum PRL2010 modulates the host innate immune response. Appl. Environ. Microbiol. 80, 730–740. doi: 10.1128/AEM.03313-13
Turroni, F., van Sinderen, D., and Ventura, M. (2011). Genomics and ecological overview of the genus Bifidobacterium. Int. J. Food Microbiol. 149, 37–44. doi: 10.1016/j.ijfoodmicro.2010.12.010
Turroni, F., van Sinderen, D., and Ventura, M. (2021). Bifidobacteria: insights into the biology of a key microbial group of early life gut microbiota. Microbiome Res. Rep. 1:2. doi: 10.20517/mrr.2021.02
Vallejo-Cordoba, B., Castro-López, C., García, H. S., González-Córdova, A. F., and Hernández-Mendoza, A. (2020). “Chapter one - postbiotics and paraprobiotics: a review of current evidence and emerging trends,” in Advances in Food and Nutrition Research, eds A. G. da Cruz, E. S. Prudencio, E. A. Esmerino, and M. C. da Silva (Cambridge, CA: Academic Press), 1–34. doi: 10.1016/bs.afnr.2020.06.001
van Aardt, M., Duncan, S. E., Marcy, J. E., Long, T. E., O’Keefe, S. F., and Nielsen-Sims, S. R. (2005). Effect of antioxidant (α-tocopherol and ascorbic acid) fortification on light-induced flavor of milk. J. Dairy Sci. 88, 872–880. doi: 10.3168/jds.S0022-0302(05)72753-3
Van de Casteele, S., Vanheuverzwijn, T., Ruyssen, T., Van Assche, P., Swings, J., and Huys, G. (2006). Evaluation of culture media for selective enumeration of probiotic strains of lactobacilli and bifidobacteria in combination with yoghurt or cheese starters. Int. Dairy J. 16, 1470–1476.
van der Hee, B., and Wells, J. M. (2021). Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol. 29, 700–712.
Veal, D. A., Deere, D., Ferrari, B., Piper, J., and Attfield, P. V. (2000). Fluorescence staining and flow cytometry for monitoring microbial cells. J. Immunol. Methods 243, 191–210.
Ventura, M., Canchaya, C., Zink, R., Fitzgerald, G. F., and van Sinderen, D. (2004). Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analyses. Appl. Environ. Microbiol. 70, 6197–6209. doi: 10.1128/AEM.70.10.6197-6209.2004
Ventura, M., Margolles, A., Turroni, F., Zomer, A., de los Reyes-Gavilán, C. G., and van Sinderen, D. (2011). “Stress responses of Bifidobacteria,” in Stress Responses of Lactic Acid Bacteria, eds E. Tsakalidou and K. Papadimitriou (Boston, MA: Springer).
Ventura, M., Zink, R., Fitzgerald, G. F., and van Sinderen, D. (2005). Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing. Appl. Environ. Microbiol. 71, 487–500. doi: 10.1128/AEM.71.1.487-500.2005
Vinderola, G., Reinheimer, J., and Salminen, S. (2019). The enumeration of probiotic issues: from unavailable standardised culture media to a recommended procedure? Int. Dairy J. 96, 58–65.
Waddington, L., Cyr, T., Hefford, M., Hansen, L. T., and Kalmokoff, M. (2010). Understanding the acid tolerance response of bifidobacteria. J. Appl. Microbiol. 108, 1408–1420.
Wang, F., Huang, G., Cai, D., Li, D., Liang, X., Yu, T., et al. (2015). Qualitative and semiquantitative analysis of fecal Bifidobacterium species in centenarians living in Bama, Guangxi, China. Curr. Microbiol. 71, 143–149. doi: 10.1007/s00284-015-0804-z
Wang, X., Ren, H., Liu, D., Wang, B., Zhu, W., and Wang, W. (2013). H+-ATPase-defective variants of Lactobacillus delbrueckii subsp. bulgaricus contribute to inhibition of postacidification of yogurt during chilled storage. J. Food Sci. 78, M297–M302. doi: 10.1111/1750-3841.12038
Wei, Y., Gao, J., Liu, D., Li, Y., and Liu, W. (2019). Adaptational changes in physiological and transcriptional responses of Bifidobacterium longum involved in acid stress resistance after successive batch cultures. Microb. Cell Factories 18:156. doi: 10.1186/s12934-019-1206-x
Wei, Y., Yang, F., Wu, Q., Gao, J., Liu, W., Liu, C., et al. (2018). Protective effects of bifidobacterial strains against toxigenic Clostridium difficile. Front. Microbiol. 9:888. doi: 10.3389/fmicb.2018.00888
Wendel, U. (2022). Assessing viability and stress tolerance of probiotics—a review. Front. Microbiol. 12:818468. doi: 10.3389/fmicb.2021.818468
Wilkinson, M. G. (2018). Flow cytometry as a potential method of measuring bacterial viability in probiotic products: a review. Trends Food Sci. Technol. 78, 1–10. doi: 10.3389/fmicb.2023.1304621
Wong, C. B., Odamaki, T., and Xiao, J.-Z. (2020). Insights into the reason of human-residential bifidobacteria (HRB) being the natural inhabitants of the human gut and their potential health-promoting benefits. FEMS Microbiol. Rev. 44, 369–385. doi: 10.1093/femsre/fuaa010
Xiao, M., Xu, P., Zhao, J., Wang, Z., Zuo, F., Zhang, J., et al. (2011). Oxidative stress-related responses of Bifidobacterium longum subsp. longum BBMN68 at the proteomic level after exposure to oxygen. Microbiology 157, 1573–1588. doi: 10.1099/mic.0.044297-0
Xu, X., Hui, H., and Cai, D. (2012). Differences in fecal Bifidobacterium species between patients with type 2 diabetes and healthy individuals. J. Southern Med. Univer. 32, 531–533.
Yang, B., Ding, M., Chen, Y., Han, F., Yang, C., Zhao, J., et al. (2021). Development of gut microbiota and bifidobacterial communities of neonates in the first 6 weeks and their inheritance from mother. Gut Microbes 13, 1–13. doi: 10.1080/19490976.2021.1908100
Yerlikaya, O., Akpinar, A., Saygili, D., and Karagozlu, N. (2020). Incorporation of Propionibacterium shermanii subsp. freudenreichii in probiotic dairy drink production: physicochemical, rheological, microbiological and sensorial properties. Int. J. Dairy Technol. 73, 392–402.
Yeung, T. W., Üçok, E. F., Tiani, K. A., McClements, D. J., and Sela, D. A. (2016). Microencapsulation in alginate and chitosan microgels to enhance viability of Bifidobacterium longum for oral delivery. Front. Microbiol. 7:494. doi: 10.3389/fmicb.2016.00494
Yonezawa, S., Xiao, J. Z., Odamaki, T., Ishida, T., Miyaji, K., Yamada, A., et al. (2010). Improved growth of bifidobacteria by cocultivation with Lactococcus lactis subspecies lactis. J. Dairy Sci. 93, 1815–1823.
Yoon, Y., Kim, G., Jeon, B. N., Fang, S., and Park, H. (2021). Bifidobacterium Strain-Specific enhances the efficacy of cancer therapeutics in tumor-bearing mice. Cancers 13:957. doi: 10.3390/cancers13050957
Yoon, Y., Lee, H., Lee, S., Kim, S., and Choi, K.-H. (2015). Membrane fluidity-related adaptive response mechanisms of foodborne bacterial pathogens under environmental stresses. Food Res. Int. 72, 25–36. doi: 10.1128/JB.00148-18
Yunes, R. A., Poluektova, E. U., Dyachkova, M. S., Klimina, K. M., Kovtun, A. S., Averina, O. V., et al. (2016). GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe 42, 197–204. doi: 10.1016/j.anaerobe.2016.10.011
Zhang, H., Liu, M., Liu, X., Zhong, W., Li, Y., Ran, Y., et al. (2020). Bifidobacterium animalis ssp. lactis 420 mitigates autoimmune hepatitis through regulating intestinal barrier and liver immune cells. Front. Immunol. 11:569104. doi: 10.3389/fimmu.2020.569104
Zhang, J., Ren, L., Zhang, L., Gong, Y., Xu, T., Wang, X., et al. (2023). Single-cell rapid identification, in situ viability and vitality profiling, and genome-based source-tracking for probiotics products. iMeta 2:e117.
Zhang, T., and Fang, H. H. P. (2006). Applications of real-time polymerase chain reaction for quantification of microorganisms in environmental samples. Appl. Microbiol. Biotechnol. 70, 281–289.
Zhang, T., Jeong, C. H., Cheng, W. N., Bae, H., Seo, H. G., Petriello, M. C., et al. (2019). Moringa extract enhances the fermentative, textural, and bioactive properties of yogurt. LWT Food Sci. Technol. 101, 276–284.
Zhang, Y., Mao, B., Tang, X., Liu, X., Zhao, J., Zhang, H., et al. (2022). Integrative genome and metabolome analysis reveal the potential mechanism of osmotic stress tolerance in Bifidobacterium bifidum. LWT 159:113199.
Zhang, Z., Gu, M., You, X., Sela, D. A., Xiao, H., and McClements, D. J. (2021). Encapsulation of Bifidobacterium in alginate microgels improves viability and targeted gut release. Food Hydrocolloids 116:106634.
Zuo, F., Yu, R., Khaskheli, G. B., Ma, H., Chen, L., Zeng, Z., et al. (2014). Homologous overexpression of alkyl hydroperoxide reductase subunit C (AhpC) protects Bifidobacterium longum strain NCC2705 from oxidative stress. Res. Microbiol. 165, 581–589. doi: 10.1016/j.resmic.2014.05.040
Keywords: Bifidobacterium, viability, yogurt, probiotic, gut microbiota, stress response, viability improvement, next-generation methods
Citation: Sibanda T, Marole TA, Thomashoff UL, Thantsha MS and Buys EM (2024) Bifidobacterium species viability in dairy-based probiotic foods: challenges and innovative approaches for accurate viability determination and monitoring of probiotic functionality. Front. Microbiol. 15:1327010. doi: 10.3389/fmicb.2024.1327010
Received: 24 October 2023; Accepted: 15 January 2024;
Published: 02 February 2024.
Edited by:
José David Flores Félix, University of Salamanca, SpainReviewed by:
Diego Armando Esquivel Hernandez, Metropolitan Autonomous University, MexicoCopyright © 2024 Sibanda, Marole, Thomashoff, Thantsha and Buys. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elna M. Buys, ZWxuYS5idXlzQHVwLmFjLnph
†ORCID: Thulani Sibanda, orcid.org/0000-0002-6422-2871; Tlaleo Azael Marole, orcid.org/0000-0003-0794-6977; Mapitsi S. Thantsha, orcid.org/0000-0002-6212-2770; Elna M. Buys, orcid.org/0000-0001-7836-9295
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.