
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 January 2024
Sec. Food Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1324999
 Ryusuke Ohgi1†
Ryusuke Ohgi1† Sudeb Saha1,2†‡
Sudeb Saha1,2†‡ Binghui Zhou1,3
Binghui Zhou1,3 Taiga Sakuma1
Taiga Sakuma1 Mitsuki Sakurai1
Mitsuki Sakurai1 Yuhka Nakano1
Yuhka Nakano1 Fu Namai1,3
Fu Namai1,3 Wakako Ikeda-Ohtsubo1,3
Wakako Ikeda-Ohtsubo1,3 Yoshihito Suda4
Yoshihito Suda4 Keita Nishiyama1,3
Keita Nishiyama1,3 Julio Villena1,5*
Julio Villena1,5* Haruki Kitazawa1,3*
Haruki Kitazawa1,3*The emergence and spread of antibiotic resistance threat forced to explore alternative strategies for improving the resistance to pathogens in livestock production. Probiotic lactic acid bacteria represent an alternative for this objective. In this study, seven Lactiplantibacillus plantarum strains from porcine colostrum and milk were isolated, identified and characterized in terms of their abilities to modulate immunity in porcine intestinal epithelial (PIE) cells. Then, two potential immunoregulatory strains were studied in terms of their ability to utilize and grow in wakame (Undaria pinnafida). Isolates were identified by 16S rRNA gene and evaluated by studying their interaction with PIE cells. The expressions of peptidoglycan recognition proteins (PGRPs), nucleotide-binding oligomerization domain (NODs), host defense peptides (pBD), and type I interferons (IFNs) were evaluated by RT-qPCR. The strain 4M4417 showed a remarkable capacity to differentially regulate the expression of PGRP1, PGRP3, NOD1, NOD2, and pBD1 in PIE cells. On the other hand, the strain 4M4326 was the most efficient to improve the expression of IFN-α and IFN-β in PIE cells challenged with poly (I:C). Both L. plantarum 4M4326 and 4M4417 were characterized in terms of their ability to utilize wakame. Results demonstrated that both strains efficiently grew in wakame-based broth. Our results suggest that L. planatrum 4M4326 and 4M4417 are interesting candidates to develop immunomodulatory feeds based on wakame utilization. These new immunosynbiotic feeds could help to reduce severity of intestinal infections and improve immune health status in pigs.
Milk is considered as the main source of nutrients for neonates, providing optimal energy and bioactive compounds in their early life (Saha and Ara, 2012; Wang et al., 2022). In addition, it was reported that commensal bacteria from milk are key actors during the early stage of neonatal gut colonization, being major regulators of the host’s immune responses (Stewart et al., 2018). Newborn piglets suckle the sow and rely on the nutrients ingested with milk until weaning (Vodolazska et al., 2023). In general, weaning is performed in piglets around 3–4 weeks when the feeding changes from milk to solid feed. This practice induces a stressful condition to the pig’s life leading to the disruption the gut microbial compositions. This stress can induce alterations of the intestinal morphology and physiological functions and increase the susceptibility to enteric pathogens infections leading to diarrhea (Gresse et al., 2017; Ding et al., 2021). Post weaning diarrhea has been documented as a severe problem after weaning in swine industry globally, and it is one of the leading causes of the mortality observed during this stage. This infectious disease is usually caused by enterotoxigenic Escherichia coli, Salmonella spp., and rotavirus infection (Cremonesi et al., 2022). To curb post weaning diarrhea there has been a widespread and irrational use of antibiotics as feed additives (Pan et al., 2017) causing the increase of antibiotic resistance in the animals and in the consumers of their products. Thus, the use of antibiotics has been banned in many regions of the world, such as the European Union in 2006 (Chen et al., 2005) or its use has been limited in many countries such as USA, Japan, and China. In order to reduce the use of antimicrobials in livestock, several alternatives are being explored, including the use of immunomodulatory probiotics (immunobiotics) (Villena et al., 2018; Saha et al., 2023).
Recent research has shown that human breast milk is an excellent source of beneficial lactic acid bacteria (LAB) that are vertically transferred to the infant gut (Martín et al., 2007; Qi et al., 2022). Remarkably, some of the LAB strains isolated from breast milk can reduce the severity of infections and inhibit the growth of pathogenic bacteria by competitive exclusions and/or the production of antimicrobial compounds such as bacteriocins or organic acids (Martín et al., 2009). Recent studies have demonstrated that such bacteria could be transferred from the maternal gut to the mammary gland during late pregnancy and lactation through an immunologically based internal route that involves dendritic cells and macrophages: the entero-mammary pathway (Rodríguez, 2014; Selvamani et al., 2021; Qi et al., 2022). The beneficial effects of probiotic LAB in the gut are associated with several mechanisms, including the increase of mucus production, enhancement of epithelial barrier, the competition with harmful microbes for attachment and nutrients in the mucosa, as well as the modulation of the local and systemic immune systems (Elean et al., 2021; Kober et al., 2022). In terms of immunomodulation, it has been demonstrated that the oral administration of probiotic strains can prime the host immune system allowing a faster and more effective response to microbial infections, suggesting that these strains could be used as dietary supplements to enhance innate and adaptive immune system response (Sandes et al., 2017; Abramov et al., 2020).
In general, human breast milk contains approximately 103 to 104 colony forming units/mL of microbes such as Lactobacilli, Lactococci, Enterococci, and Leuconostoc spp. (Keddar et al., 2023). Among bacteria present in breast milk, Lactiplantibacillus plantarum strains have been identified as potential probiotics (Mollova et al., 2023). L. plantarum can survive in the gastrointestinal tract of humans and other mammals including pigs (de Vries et al., 2006; Masumizu et al., 2019; Mollova et al., 2023). In addition, several L. plantarum strains have shown to possess beneficial properties for the host, including their ability to beneficially modulate the immune system (Villena et al., 2018). In this regard, we found that different strains of L. plantarum such as CRL1506, CRL681, and MPL16 have a remarkable ability to modulate antibacterial and antiviral immune responses (Albarracin et al., 2020; Zhou et al., 2020; Baillo et al., 2022). Considering these facts, we hypothesized that, sow milk contains probiotic Lactobacillus strains which may have the capacity to beneficially modulate the immune system of piglets. Few studies have explored the potential probiotic properties of bacterial strains from sow milk (Martín et al., 2009; Gyawali et al., 2015).
In the present work, we aimed to isolate and characterize the immunomodulatory properties of L. plantarum from sow milk, particularly focused on their ability to modulate the expression of antimicrobial factors in the context of poly (I:C) challenge in porcine intestinal epithelial (PIE) cells. Considering that research has demonstrated that some prebiotic seaweed can enhance the beneficial effects of probiotic lactobacilli (Makkar et al., 2016; Masumizu et al., 2019), we also evaluated the ability of the strains to grow in wakame (Undaria pinnafida), with the aim of developing an immunosynbiotic feed to improve immune health of pigs in the future.
All pregnant sows (Landrace × Yorkshire × Duroc) were kept at Tsubonuma Farm, Faculty of Food Industry, Miyagi University, Japan. Animals were reared according to the guidelines of the Japanese Association of Laboratory Animal Science (JALAS) for care and use of animals in research. The protocol was approved by the laboratory health and safety committee of Miyagi University (Japan) with an approved protocol no. 2016–2023. The milk sample was collected from healthy pig. Corn-soybean based diet fed to the animals.
Milk samples were collected in a sterile tube. During milk sampling, sterile gloves were worn, nipple and the surrounding area were soaked with 75% ethyl alcohol to avoid the contamination by skin bacteria. The milk samples were immediately refrigerated at 4°C and transferred to the laboratory of Animal Food Function, Tohoku University. After transportation, milk samples were dispensed into serum tubes, mixed with equal amount of 60% (v/v) glycerol (final concentration was 30% glycerol sample stock) and stored at −80°C for the cultivation of bacteria.
For DNeasy Blood & Tissue Kit (Qiagen, United States), 0.2 mL milk sample were centrifuged at 6000 × g for 3 min, total DNA was isolated from its pellet using according to the manufacturer’s instructions for gram positive bacteria and then sent for Bioengineering Lab. Co., Ltd. (Kanagawa, Japan). Total DNA was subjected to 16S rRNA analysis, and read counts were calculated. DNA was amplified using the 2-step tailed PCR to target the V3–V4 regions of bacterial 16S rRNA. 1st PCR was performed with the 1st-341f_MIX (5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCT-NNNNN-CCTACGGGNGGCWGCAG-3′) and the 1st-805r_MIX (5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-NNNNN-GACTACHVGGGTATCTAATCC-3′) primers. Subsequently, 2nd PCR was performed with the 2ndF (5′-AATGATACGGCGACCACCGAGATCTACAC-Index2-ACACTCTTTCCCTACACGACGC-3′) and the 2ndR (5′-CAAGCAGAAGACGGCATACGAGAT-Index1-GTGACTGGAGTTCAGACGTGTG-primers). A 16S rRNA metagenomic sequencing library was prepared according to manufacturer’s instructions (Illumina, San Diego, CA, United States). The PCR products were pooled to construct the sequencing library and the quality of the library was confirmed using Fragment Analyzer and dsDNA915 Reagent Kit (Advanced Analytical Technologies). Sequencing was performed using the MiSeq Reagent Kit v3 (Illumina, United States) under the condition of 2 × 300 bp. The paired raw fastq data were merged and quality filtered. The FASTQ data from four breasts underwent analysis within the Quantitative Insights into Microbial Ecology version 2 (Qiime2) platform.1 Initially, the dada2 plugin in Qiime2 was employed to filter and denoise the raw FASTQ data. The passed forward and reverse sequences were then merged. Then the chimera sequences were eliminated from the merged sequences to construct the amplicon sequence variant (ASV) table. The ASV tables from four breasts were combined to create one ASV table. Finally, taxonomy analysis was performed by classifying each ASV using the SILVA 138 database (Quast et al., 2012).
To isolate Lactobacillus strains, primarily, collected milk samples were thawed and 50 μL of samples were 10-fold serially diluted with sterile phosphate buffered saline (PBS) solution. Later, an aliquot (50 μL) of mixtures was plated onto de Man, Rogosa and Sharpe (MRS, Becton Dickinson Company, United States) and incubated at 37oC for 48 h. Single colonies were collected, and pre-cultured onto MRS broth for up to 24 h and stored at −80°C with equal 60% glycerol solution for further experiments. The morphology of isolates was examined by Gram staining, and Gram-positive bacteria were used for 16S rRNA gene sequencing.
Genomic DNA was extracted from selected bacterial isolates using DNeasy Blood & Tissue Kit (Qiagen, United States) by following the manufacturer’s protocol for gram positive bacteria. The DNA was measured with a Nanodrop ND-2000 spectrophotometer (NanoDrop Technologies Wilimington, DE). 16S rDNA fragment was amplified by PCR using the bacterial universal primers 27f (5´-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5´-GGTTACCTTGTTACGACTT-3′) (Ref). After amplification, PCR products were purified by gel electrophoresis and the purified PCR products were sequenced by BigDye Terminator v1.1 Cycle Sequencing Kit (Thermo, Co., Ltd., Foster City, CA, United States). The obtained sequences were examined by basic Local Alignment Search Tool (BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi) through the alignment of 16 s rRNA gene sequences of isolates with the 16 s rRNA gene sequences of known bacteria in GeneBank database. More than 95% of sequence with a type of strain were used to identify the isolates.
The porcine intestinal epithelial cell lines (PIE) were used as an in vitro cellular model. PIE cells were initially derived from the intestinal epithelium of neonatal unsuckled piglets, previously developed by our research group (Moue et al., 2008), and was maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with fetal calf serum (10%), penicillin (100 U/mL), and streptomycin (100 μg/mL). The PIE cells cultures were grown in a flask. 75 cm2 flask at 37°C with 5% CO2. The cultures were passaged routinely to reach the confluent 80%–90% and used for experiments between 25th and 35th passages. PIE cells were counted and later seeded at 3.0 × 104 cells in Type I collagen coated 12-well plates (SUMILON, Tokyo, Japan) and incubate at 37°C with 5% CO2 for 3 days. On the day 3, L. plantarum isolates 5 × 107 cells/well (2.5 × 108 cells/mL) were added and stimulation was continued for 48 h at 37°C with 5% CO2.
For TLR3 activation experiments, PIE cells were seeded at 3.0 × 104 cells per well in 12-well type I collagen-coated plates and cultured for 3 days. After changing medium, lactobacilli (2.5 × 108 cells/mL) were added, and 48 h later, each well was washed vigorously with medium at least three times to eliminate all stimulants. Then, cells were stimulated with 5 μL of poly (I:C) (100 ng/mL) (Sigma-Aldrich, St. Louis, MI, United States) for 12 h.
After the treatment with lactobacilli (basal expression) and 12 h after poly (I:C) stimulation, PIE cells were washed twice PBS, treated with 500 μL of TRIzol reagent (Invitrogen, CA, United States) per well, and transferred to a 1.5 mL microtube for RNA extraction according to the manufacturer’s protocol.
Total RNA was extracted from treated and control PIE cells using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) and reverse transcribed into cDNA using the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara Bio Inc., Shiga, Japan) by following the manufacturer’s protocol. The synthesized cDNA was then used for quantitative PCR analysis using Platinum SYBR Green qPCR Super Mix UDG with ROX (Invitrogen, Carlsbad, CA, United States) on a 7,300 real-time PCR system (Applied Biosystems, Warrington, United Kingdom). In brief, cDNA 2.5 μL, Syber MIX 7.5 μL (reverse primer 1.25 μL (1 pM), Platinum SYBR Green qPCR SuperMIX-UDG with POX 5 μL) into each well of the dedicated 96 well plates. Primers used in this study are described in Supplementary Table S1. Amplification was carried out at the following conditions: 50°C for 2 min and 95°C for 5 min, followed by 40 cycles consisting of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. β-actin was used as an internal standard to normalize cDNA levels for differences in total cDNA levels in the samples. The samples were run thrice for each experimental condition and the average values were used to statistical analysis.
Bacterial growth assay was performed according to our previous publication (Masumizu et al., 2019). In brief, 0.1% (w/v) of wakame powder was suspended in water and the suspension was autoclaved at 121°C for 15 min. The wakame solution adjusted to pH 5 with 1 mol/L HCl and incubated with the two enzymes: 0.25% (w/v) cellulase and 0.25% (w/v) hemicellulose (Mitsubishi-Chemical Foods Corporation, Tokyo, Japan) at 50°C for 24 h. The hydrolyzed wakame was again autoclaved at 121°C for 15 min, and the supernatant was separated by centrifugation at 6,000 rpm for 20 min. The supernatant containing the wakame extract was further supplemented with 0.6% (w/v) Trypticase soy broth and 0.5% (w/v) NaCl. The pH was adjusted to 6.8 and the broth was autoclaved at 121°C for 15 min. This wakame broth obtained at the end of these procedures was considered as ready for use.
The lactobacilli strains used for the study of wakame assimilation were first grown in MRS agar at 37°C for 24 h. A single colony was picked for the pre-culture. After growth, 2% (v/v) of bacteria culture broth was added 5 mL of wakame broth or non-sugar MRS and incubated at 37°C for 48 h. Optical density was monitored for 48 h (measurement interval: 30 min, penetration rate: 60 rpm, total operation time: 48 h) using spectrometer system (TVS026CA, Advantec, Tokyo, Japan).
Wakame solution with or without enzyme treatment, the supernatant of the wakame broth fermented with L. plantarum strains, and 1% (w/v) of standard saccharides solutions (glucose, galactose, fructose, lactose, mannose, raffinose, alginate, and cellobiose) were dropped on TLC Silica gel 60 (Merck kGaA, Darmstadt, Germany). Butyl alcohol, isopropyl alcohol, and water (3:12:4) mixture was used as the solvent. After 2 times developing, 5% (v/v) sulfuric acid in methanol was sprayed on the plate and it was heated at 150°C for 10 min until spot visualizing.
The statistical analyses were performed by using Bell Curve for Excel (Social Survey Research Information Co., Ltd., Tokyo, Japan). Data were presented as the mean ± SD. Differences between the control were assessed using one-way ANOVA followed by the independent two-tailed t-test. Differences were considered as significant when p < 0.05.
The analysis of the microbial composition of porcine colostrum revealed the presence of a complex bacterial community (Figure 1A). Chryseobacterium and Staphylococcus were abundant genera, with Lactobacillus comprising approximately 2% of the colostrum microbiome (Figure 1B). Our results are consistent with previous studies that have identified porcine milk as a rich source of Lactobacillus (Martín et al., 2009; Wang et al., 2022). In a study by Chen et al. (2018) demonstrated that the microbial composition and diversity of the porcine colostrum changed significantly but was relatively stable in transitional and mature milk. The work described that the genera Corynebacterium and Streptococcus were significantly higher in colostrum while the bacterial groups Lactobacillus, Ruminococcaceae, unclassified Lachnospiraceae, and unclassified Clostridiales were more abundant in milk. In addition, L. reuteri was the most dominant species in milk (Chen et al., 2018). However, it should be noted that the milk microbiota of sows has not been systematically investigated, therefore there is not enough information to compare similarities and differences between studies and draw robust conclusions.
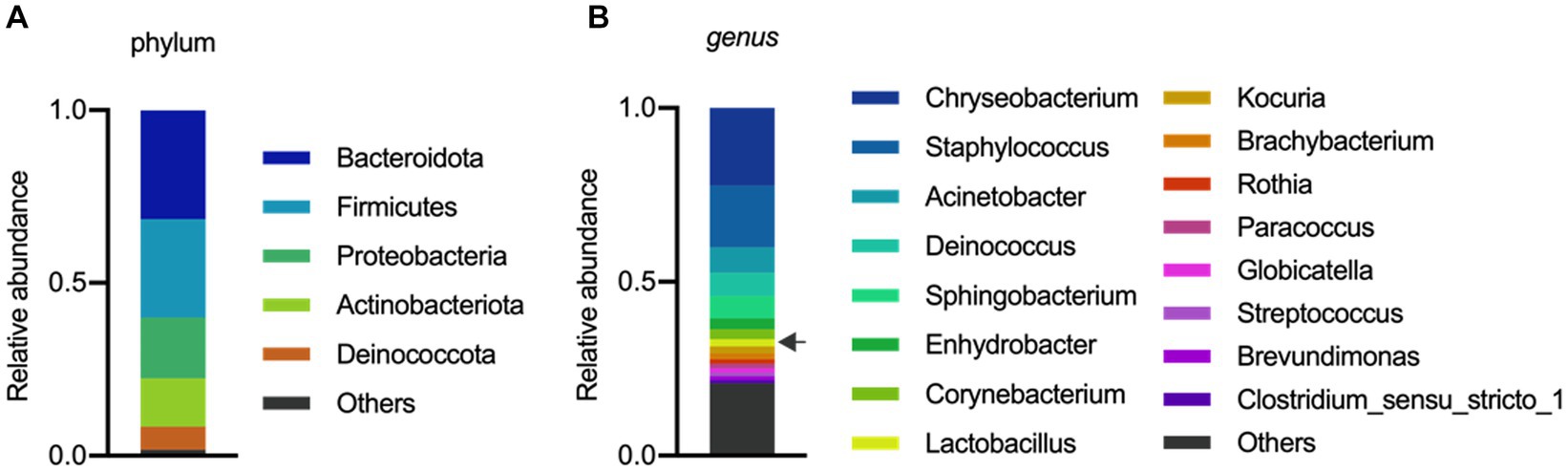
Figure 1. Microbial composition of porcine colostrum (day0) at Phylum level (A) and Genus level (B). Microbiome data from four breasts were combined in a single bar plot. The arrow indicates Lactobacillus.
Next, we aimed to isolate LAB strains for samples using MRS plates. Most of the isolated strains belonged to the species W. thailandensis, W. cibaria, W. paramesenteroides, and L. lactis (Supplementary Table S2). The identification by 16S rDNA PCR of isolated colonies revealed that among lactobacilli all the strains belonged to the species L. plantarum (Supplementary Table S2). Thus, our experimental procedures did not allow the isolation of other lactobacilli species present in the porcine milk. Similarly, the study of Wang et al. (2022) showed that from a total of 1,240 isolates from porcine milk, 922 belonged to the group of Lactobacillales, which was dominated by L. lactis. In addition, L. reuteri, L. salivarius, L. plantarum, L. paraplantarum, L. brevis, and W. paramesenteroides were the most abundant species isolated from sow milk (Martín et al., 2009). Considering that we were interested in L. plantarum strains because of their potential higher capacity to grow in wakame (Masumizu et al., 2019), we selected seven strains belonging to this species of lactobacilli (4cs321, 4cs331, 4M4326, 4M4338, 4M4346, 4M4347, and 4M4417) for the evaluation of the immunomodulatory properties in PIE cells.
We demonstrated previously that PIE cells are a useful in vitro tool to evaluate the ability of beneficial bacteria to modulate the innate immune responses, which has a high correlation with posterior in vivo studies (Shimazu et al., 2012; Suda et al., 2014; Albarracin et al., 2020). Then, in a first set of experiments we assessed whether the pre-stimulation of PIE cells with lactobacilli differentially modulate the expression of immune genes in PIE cells stimulated with the isolates of L. plantarum (Figure 2). A clear strain-dependent effect was observed when the expressions of PGRPs, NODs, and pBDs genes were analyzed, which is in line with our previous reports evaluating LAB strains of different origins (Villena et al., 2016; Masumizu et al., 2019; Mizuno et al., 2020; Baillo et al., 2022).
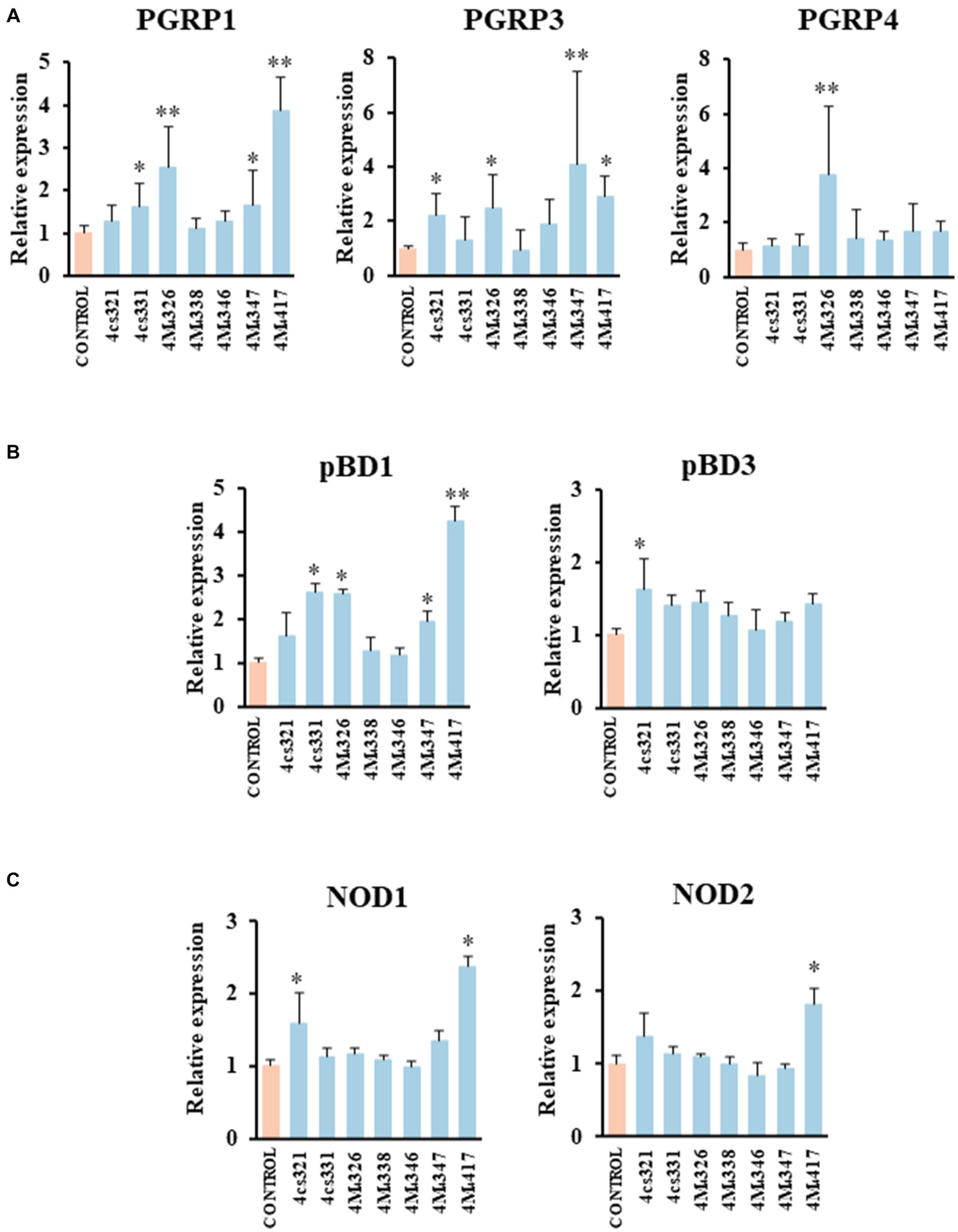
Figure 2. Effect of porcine lactobacilli on the expression of peptidoglycan recognition proteins (PGRP) (A) PGRP1, PGRP3, PGRP4, (B) host defense peptides pBD1, pBD3, and nucleotide-binding oligomerization domain (NOD), (C) NOD1 and NOD2 in PIE cells. The cells were stimulated with different Lactiplantibacillus plantarum strains (4cs321, 4cs331, 4M4326, 4M4338, 4M4346, 4M4347, and 4M4417) and the expression of immune factors was evaluated. PIE cells without lactobacilli stimulation were used as control. Values are presented as mean ± SD of three independent experiments (n = 3 per experiment). *p < 0.05 and **p < 0.01.
L. plantarum 4M4326 was the only strain able to increase PGRP4 expression. This strain also upregulated PGRP1 and PGRP3. L. plantarum 4cs331 increased PGRP1 while the 4M4321 strain upregulated PGRP3 (Figure 2A). The most remarkable effect was found for L. plantarum 4M4417 that enhanced the expression of PGRP1, and PGRP3 in PIE cells. PGRPs possess antibacterial and immunomodulatory properties that allow them to actively participate in the interaction of host’s cells with the intestinal microbiota. It was reported that mice lacking any of the four genes coding for PGRPs are more susceptible to dextran sulfate sodium (DSS)-induced colitis (Saha et al., 2010). Furthermore, stools from mice deficient in PGRPs when transferred to wild type germ-free mice can increase the severity of colitis when compared to the stools from normal mice (Jing et al., 2014). Although significant advances have been made in the biology of PGRPs in human and mice, their functions in pigs have been less explored. Our group demonstrated that the four PGRPs are expressed in PIE cells and that their expression can be modulated by probiotic microorganisms (Iida et al., 2019). Our data provided evidence of the potential capacity of the modulation of PGRPs by beneficial microbes to enhance resistance and reduce inflammatory damage severity during intestinal infections.
The results showed that L. plantarum 4cs321 significantly upregulated the expression of pBD3 while the 4cs331 strain increased pBD1 in PIE cells stimulated with the isolates of L. plantarum when compared to controls (Figure 2B). The expression of pBD1 was also enhanced by the strains 4M4326 and 4M4347. L. plantarum 4M4417 significantly increased the expression of pBD1 (Figure 2B). The secretion of host defence peptides by the intestinal epithelium is of importance since these factors can exert both antimicrobial and immunomodulatory activities, providing an early response to bacterial infections in the gut (Veldhuizen et al., 2008). Interestingly, the treatment of weaned piglets with synthetic host defence peptides was shown to improve their growth performance, nutrient digestion, and intestinal health (Yoon et al., 2013). An alternative to the administration of synthetic peptides is the induction of their expression using beneficial microorganisms. Some studies have shown the ability of L. plantarum strains to differentially modulate the expression of host defence peptides in porcine intestinal epithelial cells. The probiotic strain L. plantarum ZLP001 was shown to upregulate the expression of pBD2 (a β-defensin) and PG1-5 (a cathelicidin) in IPEC-J2 cells (Wang et al., 2018) in a TLR2/MAPK/AP-1 signaling dependent pathway (Wang et al., 2019). This effect has been described as one of the mechanisms involved in the augmented growth performance in post-weaning piglets upon the treatment with the ZLP001 strain (Wang et al., 2012). Similar studies have demonstrated the ability of lactobacilli to modulate host defence peptides in porcine intestinal epithelial cells including L. reuteri I5007 (Liu et al., 2017), L. salivarius B1 (Zhang et al., 2011), and L. plantarum Lac16 (Zhou et al., 2021).
We also investigated the effect of L. plantarum isolates on the expression of NOD1 and NOD2 in PIE cells (Figure 2C). The expression level of NOD1 was increased in PIE cells treated with the 4cs331 strain while L. plantarum 4M4417 was the only one with the capacity to improve the expression of both NOD1 and NOD2. The modulation of the expression of NOD1 and NOD2 in the intestinal epithelium has been connected to the ability of the intestinal microbiota and probiotics to beneficially modulate the inflammatory response. Studies in specific pathogen-free Nod1−/− and Nod2−/− mice showed that both receptors are necessary for the intestinal microbiota or probiotics to reduce the severity of colitis (Natividad et al., 2012).
On the other hand, we evaluated the ability of the seven L. plantarum strains from porcine milk to modulate TLR3-mediated immune responses in PIE cells. For this purpose, cells were stimulated with lactobacilli, challenged with the TLR3 agonist poly (I:C), and then the expression of IFN-α and IFN-β was evaluated (Figure 3A). Like the experiments with L. plantarum stimulation (Figure 2), a clear strain-dependent effect was observed when the expressions of type I IFNs genes were analyzed, which is in line with our previous reports evaluating LAB strains of different origins (Albarracin et al., 2020; Zhou et al., 2020). Most of the strains had no effect on the expression of IFN-α and IFN-β in PIE cells stimulated with poly (I:C) (Figure 3A). The strain 4cs331 increased the expression of IFN-α while the most remarkable effect was detected for L. plantarum 4M4326 that significantly enhanced both IFN-α and IFN-β. We previously reported that L. plantarum MPL16 and L. plantarum CRL1506 had the capacity to enhance the expression of type I IFNs and antiviral factors in poly (I:C)-challenged PIE cells, an effect that is not sheared by strains of the same species (Albarracin et al., 2017, 2020). Of note, both CRL1506 and MPL16 strains efficiently regulate the production of IFN-α and IFN-β in the gut of mice challenged with poly (I:C), which indicates that results obtained in PIE cells could be extrapolated to in vivo situations (Albarracin et al., 2020).
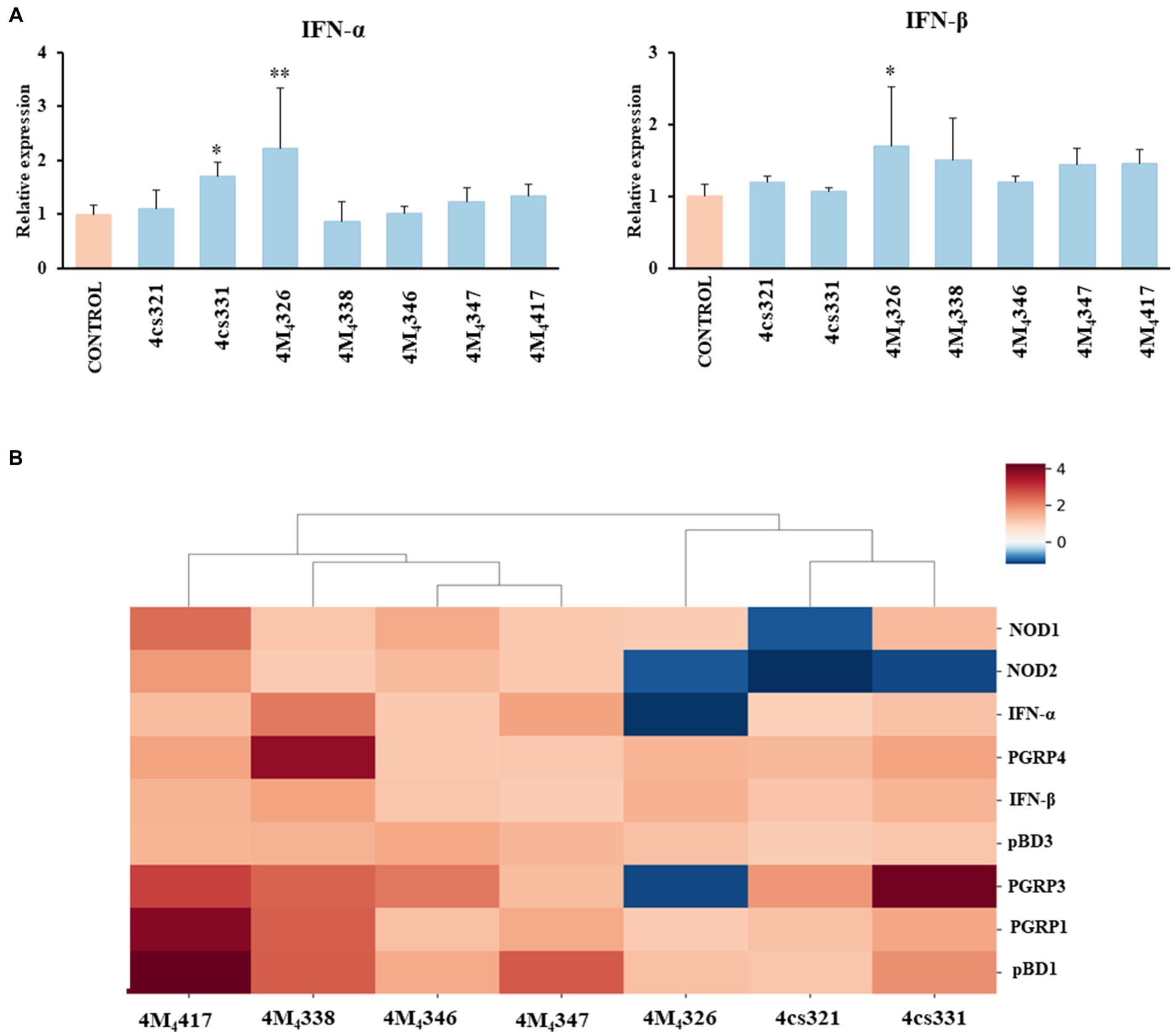
Figure 3. Effect of porcine lactobacilli on the expression of type I interferons (IFNs) (A) IFN-α and IFN-β in PIE cells challenged with poly (I:C). PIE cells were stimulated with different Lactiplantibacillus plantarum strains (4cs321, 4cs331, 4M4326, 4M4338, 4M4346, 4M4347, and 4M4417) and then challenged with poly (I:C). PIE cells without lactobacilli treatment and challenged with poly (I:C) were used as controls. Values are presented as mean ± SD of three independent experiments (n = 3 per experiment). *p < 0.05 and **p < 0.01. Heat map analysis (B) of the differentially regulated genes in PIE cells treated with L. plantarum strains and challenged with poly (I:C).
Taken together, our results show that some of the L. plantarum strains isolated from porcine milk have an excellent potential to be used in the formulation of immunomodulatory feeds and to be tested in vivo in pigs. To establish the best strains for further studies, we jointly analyzed gene expression changes in the two experiments (L. plantarum strains and poly (I:C) stimulation) using a heat map (Figure 3B). All the strains were clearly differentiated from each other, observing the most notorious effect for L. plantarum 4M4417. This strain was selected for the following studies. In addition, the strain 4M4326 was selected because was the only one with a noticeable effect on the modulation of antiviral immunity.
Wakame, an edible brown alga, contains various biologically active components that regulate the immune function (Jimenez-Escrig et al., 2011). Therefore, wakame has been proposed as a potential prebiotic feed supplement, however, only some microorganisms have the capacity to utilize its polysaccharides to grow (Tang et al., 2007). Our previous in vivo studies demonstrated that the administration of wakame to pigs as a feed significantly elevated the abundance of Lactobacillus spp. in the gut (Masumizu et al., 2019). Indeed, some L. plantarum strains isolated from the feces of wakame fed pig showed well growth in the presence of wakame in vitro (Masumizu et al., 2019). Therefore, wakame is expected to function as a natural prebiotic for selected L. plantarum strains. Accordingly, we conducted growth assay of L. plantarum 4M4326 and 4M4417 isolated in this work, on a non-sugar MRS medium supplemented with wakame as a sole carbon source (Figure 4A). In the early growth phase, both strains showed fast growth in the non-sugar MRS (control) medium, but the OD600 values decreased within 24 h. By contrast, in the wakame-supplemented medium, both strains exhibited a continuous growth pattern even after 40 h cultivation. TLC analysis indicated that the spots corresponding to monosaccharide were decreased in the culture medium of strains 4M4326 and 4M4417, compared to enzyme-treated wakame (Figure 4B). Of note, the Rf of the glucose standard did not match with the one from enzyme-treated wakame. Polysaccharide present in wakame seaweed is not completely degraded to pure monosaccharide like the standard glucose, and the glucose in the enzyme-treated wakame would be connected with some residues such as sulfates which are considered abundant in wakame. These results suggest that the wakame can enhance the growth of L. plantarum 4M4326 and 4M4417, although more studies are needed to determine the sugars present in the enzyme-treated wakame that favor the growth of lactobacilli. Remarkably there were other sugars in the TLC analysis that were not to be consumed by lactobacilli. The enzyme-treated wakame also contain disaccharides (cellobiose) and trisaccharide that cannot be assimilated by L. plantarum 4M4326 and 4M4417 but could be fermented by other intestinal bacteria in the porcine host when orally administered. Investigating the ability of the wakame fermented with 4M4326 or 4M4417 strains to modulate the composition of the porcine intestinal microbiota would be of value to completely characterize the potential beneficial effects of the symbiotic wakame-lactobacilli.
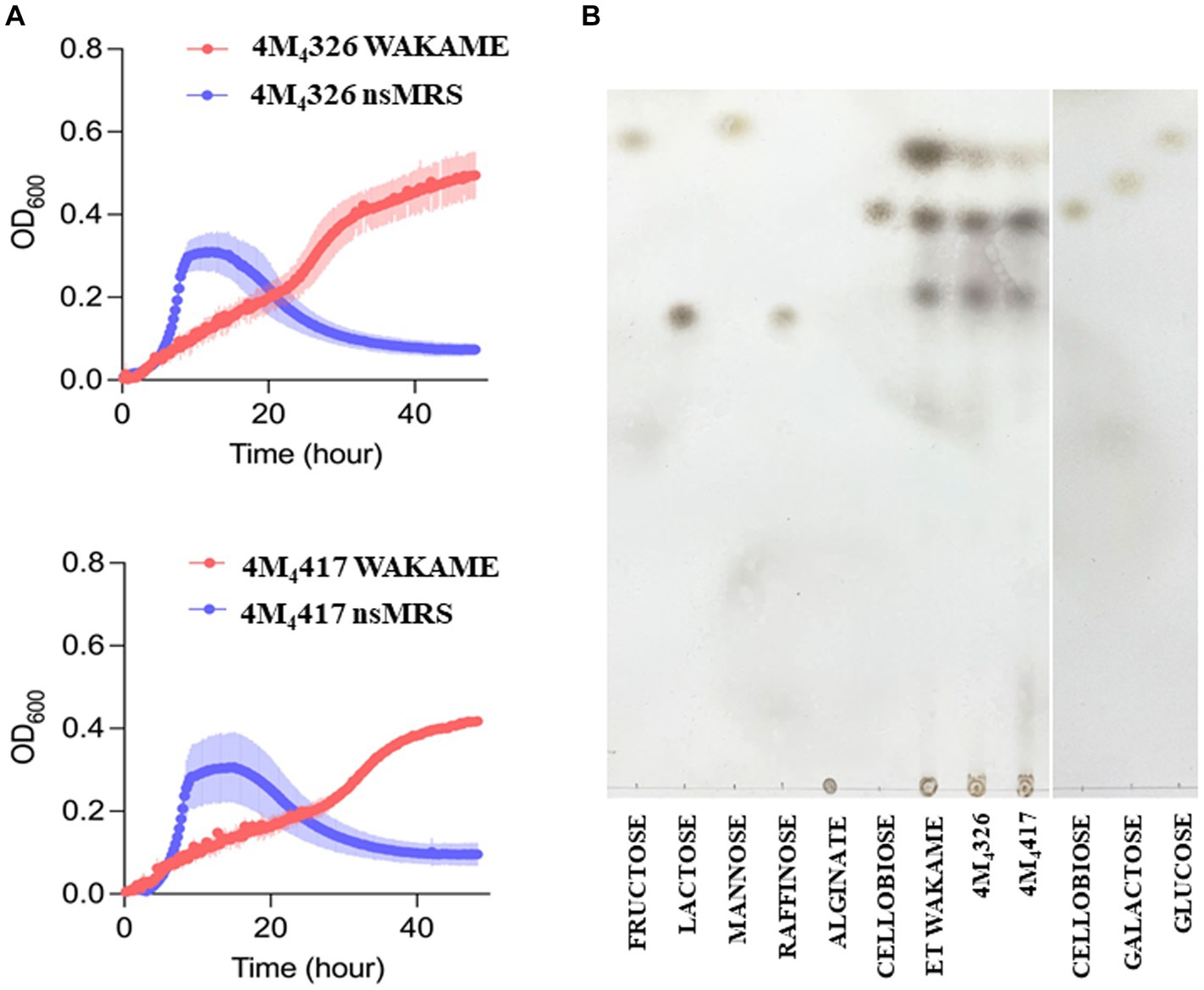
Figure 4. Growth of Lactiplantibacillus plantarum strains on wakame based-medium. The ability of 4M4326, and 4M4417 strains to utilize wakame to grow was evaluated estimating the optical density (OD) and compared with the growth in non-sugar MRS medium (A). Thin layer chromatography of different sugar and wakame fermentation in nutrient broth (B).
In the present work, we demonstrated that sow milk can be an excellent source for the isolation of potential probiotic (immunobiotic) LAB strains. Our experiments indicated that two L. plantarum strains derived from pig milk have immunomodulatory activities in PIE cells since they are able to modulate the expressions of genes involved in innate immune responses. L. plantarum 4M4417 efficiently regulated the expression of PGRPs, pBD, and NODs indicating its potential to beneficially influence the innate defence mechanism of the porcine intestinal mucosa. On the other hand, L. plantarum 4M4326 improved the expression of type I IFNs in PIE cells indicating its potential to be used as a probiotic enhancer of antiviral immunity in the porcine gut. Further in vivo investigations are needed to document the immunomodulatory activities of the 4M4326 and 4M4417 strains in pigs. In addition, future in vivo research on the synergistic combination of the immunomodulatory effects of wakame and the L. plantarum strains selected here are necessary to advance in the development of a functional immunosymbiotic feed with the capacity to efficiently improve immune health status and reduce the severity of intestinal infections in weaned piglets.
All raw data from the microbiome analysis have been deposited in the following public database: DDBJ (https://www.ddbj.nig.ac.jp/index-e.html), accession no. PRJDB16792.
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
RO: Formal analysis, Investigation, Methodology, Writing – original draft. SS: Formal analysis, Investigation, Methodology, Writing – original draft. BZ: Formal analysis, Investigation, Methodology, Writing – review & editing. TS: Formal analysis, Investigation, Methodology, Writing – review & editing. MS: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. YN: Formal analysis, Investigation, Methodology, Writing – review & editing. FN: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. WI-O: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. YS: Data curation, Formal analysis, Investigation, Project administration, Resources, Writing – review & editing. KN: Data curation, Formal analysis, Investigation, Methodology, Resources, Writing – review & editing. JV: Funding acquisition, Investigation, Visualization, Writing – original draft. HK: Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Grants-in-Aid for Scientific Research (A) (19H00965, 23H00354 HK), Grant-in-Aid for Scientific Research (B) (23H02358, KN), Grant-in-Aid for Early-Career Scientists (22K14982, BZ), Grant-in-Aid for Research Activity Start-up (23K19327, FN), and a Challenging Research (Exploratory, 23K18072, HK) and JSPS Fellows (22F22080, HK) from the Japan Society for the Promotion of Science (JSPS). This research was also supported by the Research Program on Development of Innovative Technology Grants (JPJ007097) from the Project of the Bio-Oriented Technology Research Advancement Institution (BRAIN) and by the Japan Racing Association to HK. This work was also supported by the JSPS Core-to-Core Program, A. Advanced Research Networks, entitled Establishment of International Agricultural Immunology Research Core for Quantum Improvement in Food Safety.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1324999/full#supplementary-material
Abramov, V. M., Kosarev, I. V., Priputnevich, T. V., Machulin, A. V., Khlebnikov, V. S., and Pchelintsev, S. Y. (2020). S-layer protein 2 of Lactobacillus crispatus 2029, its structural and immunomodulatory characteristics and roles in protective potential of the whole bacteria against foodborne pathogens. Int. J. Biol. Macromol. 150, 400–412. doi: 10.1016/j.ijbiomac.2020.02.065
Albarracin, L., García-Castillo, V., Masumizu, Y., Indo, Y., Islam, M. A., Suda, Y., et al. (2020). Efficient selection of new immunobiotic strains with antiviral effects in local and distal mucosal sites by using porcine intestinal epitheliocytes. Front. Immunol. 11:543. doi: 10.3389/fimmu.2020.00543
Albarracin, L., Kobayashi, H., Iida, H., Sato, N., Nochi, T., and Aso, H. (2017). Transcriptomic analysis of the innate antiviral immune response in porcine intestinal epithelial cells: influence of immunobiotic lactobacilli. Front. Immunol. 8:57. doi: 10.3389/fimmu.2017.00057
Baillo, A., Villena, J., Albarracín, L., Tomokiyo, M., Elean, M., and Fukuyama, K. (2022). Lactiplantibacillus plantarum strains modulate intestinal innate immune response and increase resistance to Enterotoxigenic Escherichia coli infection. Microorganisms 11:63. doi: 10.3390/microorganisms11010063
Chen, Y. J., Kwon, O. S., Min, B. J., Son, K. S., Cho, J. H., and Hong, J. W. (2005). The effects of dietary biotite V supplementation as an alternative substance to antibiotics in growing pigs. Asian Australas. J. Anim. Sci. 18, 1642–1645. doi: 10.5713/ajas.2005.1642
Chen, W., Mi, J., Lv, N., Gao, J., Cheng, J., and Wu, R. (2018). Lactation stage-dependency of the sow milk microbiota. Front. Microbiol. 9:945. doi: 10.3389/fmicb.2018.00945
Cremonesi, P., Biscarini, F., Castiglioni, B., Sgoifo, C. A., and Compiani, R. (2022). Gut microbiome modifications over time when removing in-feed antibiotics from the prophylaxis of post-weaning diarrhea in piglets. PLoS One 17:e0262199. doi: 10.1371/journal.pone.0262199
de Vries, M. C., Vaughan, E. E., Kleerebezem, M., and de Vos, W. M. (2006). Lactobacillus plantarum-survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 16, 1018–1028. doi: 10.1016/j.idairyj.2005.09.003
Ding, S., Yan, W., Fang, J., Jiang, H., and Liu, G. (2021). Potential role of Lactobacillus plantarum in colitis induced by dextran sulfate sodium through altering gut microbiota and host metabolism in murine model. Sci. China Life Sci. 64, 1906–1916. doi: 10.1007/s11427-020-1835-4
Elean, M., Albarracin, L., Fukuyama, K., Zhou, B., Tomokiyo, M., and Kitahara, S. (2021). Lactobacillus delbrueckii CRL 581 differentially modulates TLR3-triggered antiviral innate immune response in intestinal epithelial cells and macrophages. Microorganisms 9:2449. doi: 10.3390/microorganisms9122449
Gresse, R., Chaucheyras-Durand, F., Fleury, M. A., Van de Wiele, T., Forano, E., and Blanquet-Diot, S. (2017). Gut microbiota dysbiosis in postweaning piglets: understanding the keys to health. Trends Microbiol. 25, 851–873. doi: 10.1016/j.tim.2017.05.004
Gyawali, R., Minor, R. C., Donovan, B., and Ibrahim, S. A. (2015). Inclusion of oat in feeding can increase the potential probiotic bifidobacteria in sow milk. Animals 5, 610–623. doi: 10.3390/ani5030375
Iida, H., Tohno, M., Islam, M. A., Sato, N., Kobayashi, H., and Albarracin, L. (2019). Paraimmunobiotic bifidobacteria modulate the expression patterns of peptidoglycan recognition proteins in porcine intestinal epitheliocytes and antigen presenting cells. Cell 8:891. doi: 10.3390/cells8080891
Jimenez-Escrig, A., Gomez-Ordonez, E., and Ruperez, P. (2011). Seaweed as a source of novel nutraceuticals: sulfated polysaccharides and peptides. Adv. Food Nutr. Res. 64, 325–337. doi: 10.1016/B978-0-12-387669-0.00026-0
Jing, X., Zulfiqar, F., Park, S. Y., Núñez, G., Dziarski, R., and Gupta, D. (2014). Peptidoglycan recognition protein 3 and nod 2 synergistically protect mice from dextran sodium sulfate-induced colitis. J. Immunol. 193, 3055–3069. doi: 10.4049/jimmunol.1301548
Keddar, K., Ziar, H., Belmadani, N., Monnoye, M., Gérard, P., and Riazi, A. (2023). Probiotic bacteria from human milk can alleviate oral bovine casein sensitization in juvenile Wistar rats. Microorganisms 11:1030. doi: 10.3390/microorganisms11041030
Kober, A. K. M. H., Riaz Rajoka, M. S., Mehwish, H. M., Villena, J., and Kitazawa, H. (2022). Immunomodulation potential of probiotics: a novel strategy for improving livestock health, immunity, and productivity. Microorganisms 10:388. doi: 10.3390/microorganisms10020388
Liu, H., Hou, C., Wang, G., Jia, H., Yu, H., and Zeng, X. (2017). Lactobacillus reuteri I5007 modulates intestinal host defense peptide expression in the model of IPEC-J2 cells and neonatal piglets. Nutrients 9:559. doi: 10.3390/nu9060559
Makkar, H. P., Tran, G., Heuzé, V., Giger-Reverdin, S., and Lessire, M. (2016). Seaweeds for livestock diets: a review. Anim. Feed Sci. Technol. 212, 1–17. doi: 10.1016/j.anifeedsci.2015.09.018
Martín, R., Delgado, S., Maldonado, A., Jiménez, E., Olivares, M., and Fernández, L. (2009). Isolation of lactobacilli from sow milk and evaluation of their probiotic potential. J. Dairy Res. 76, 418–425. doi: 10.1017/S0022029909990124
Martín, R., Heilig, G. H. J., Zoetendal, E. G., Smidt, H., and Rodríguez, J. M. (2007). Diversity of the lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J. Appl. Microbiol. 103, 2638–2644. doi: 10.1111/j.1365-2672.2007.03497.x
Masumizu, Y., Zhou, B., Kober, A. H., Islam, M. A., Iida, H., and Ikeda-Ohtsubo, W. (2019). Isolation and immunocharacterization of Lactobacillus salivarius from the intestine of wakame-fed pigs to develop novel immunosynbiotics. Microorganisms 7:167. doi: 10.3390/microorganisms7060167
Mizuno, H., Arce, L., Tomotsune, K., Albarracin, L., Funabashi, R., and Vera, D. (2020). Lipoteichoic acid is involved in the ability of the immunobiotic strain Lactobacillus plantarum CRL1506 to modulate the intestinal antiviral innate immunity triggered by TLR3 activation. Front. Immunol. 11:571. doi: 10.3389/fimmu.2020.00571
Mollova, D., Gozmanova, M., Apostolova, E., Yahubyan, G., Iliev, I., and Baev, V. (2023). Lluminating the genomic landscape of Lactiplantibacillus plantarum PU3 – a novel probiotic strain isolated from human breast Milk, explored through Nanopore sequencing. Microorganisms 11:2440. doi: 10.3390/microorganisms11102440
Moue, M., Tohno, M., Shimazu, T., Kido, T., Aso, H., and Saito, T. (2008). Toll-like receptor 4 and cytokine expression involved in functional immune response in an originally established porcine intestinal epitheliocyte cell line. Biochim. Biophys. Acta 1780, 134–144. doi: 10.1016/j.bbagen.2007.11.006
Natividad, J. M., Petit, V., Huang, X., de Palma, G., and Jury, J. (2012). Commensal and probiotic bacteria influence intestinal barrier function and susceptibility to colitis in nod1−/−; nod2−/− mice. Inflamm. Bowel Dis. 18, 1434–1446. doi: 10.1002/ibd.22848
Pan, L., Zhao, P. F., Ma, X. K., Shang, Q. H., Xu, Y. T., and Long, S. F. (2017). Probiotic supplementation protects weaned pigs against enterotoxigenic Escherichia coli K88 challenge and improves performance similar to antibiotics. J. Anim. Sci. 95, 2627–2639. doi: 10.2527/jas.2016.1243
Qi, C., Zhou, J., Tu, H., Tu, R., Chang, H., and Chen, J. (2022). Lactation-dependent vertical transmission of natural probiotics from the mother to the infant gut through breast milk. Food Funct. 13, 304–315. doi: 10.1039/D1FO03131G
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., and Schweer, T. Y. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rodríguez, J. M. (2014). The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv. Nutr. 5, 779–784. doi: 10.3945/an.114.007229
Saha, S., and Ara, A. (2012). Chemical and microbiological evaluation of pasteurized milk available in Sylhet city of Bangladesh. Agriculturists 10, 104–108. doi: 10.3329/agric.v10i2.13147
Saha, S., Fukuyama, K., Debnath, M., Namai, F., Nishiyama, K., and Kitazawa, H. (2023). Recent advances in the use of probiotics to improve meat quality of small ruminants: a review. Microorganisms 11:1652. doi: 10.3390/microorganisms11071652
Saha, S., Jing, X., Park, S. Y., Wang, S., Li, X., Gupta, D., et al. (2010). Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-γ. Cell Host Microbe 8, 147–162. doi: 10.1016/j.chom.2010.07.005
Sandes, S., Alvim, L., Silva, B., Acurcio, L., Santos, C., and Campos, M. (2017). Selection of new lactic acid bacteria strains bearing probiotic features from mucosal microbiota of healthy calves: looking for immunobiotics through in vitro and in vivo approaches for immunoprophylaxis applications. Microbiol. Res. 200, 1–13. doi: 10.1016/j.micres.2017.03.008
Selvamani, S., Dailin, D. J., Gupta, V. K., Wahid, M., and Keat, H. C. (2021). An insight into probiotics bio-route: translocation from the mother’s gut to the mammary gland. Appl. Sci. 11:7247. doi: 10.3390/app11167247
Shimazu, T., Villena, J., Tohno, M., Fujie, H., Hosoya, S., and Shimosato, T. (2012). Immunobiotic Lactobacillus jensenii elicits anti-inflammatory activity in porcine intestinal epithelial cells by modulating negative regulators of the toll-like receptor signaling pathway. Infect. Immun. 80, 276–288. doi: 10.1128/IAI.05729-11
Stewart, C. J., Ajami, N. J., O’Brien, J. L., Hutchinson, D. S., Smith, D. P., and Wong, M. C. (2018). Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588. doi: 10.1038/s41586-018-0617-x
Suda, Y., Villena, J., Takahashi, Y., Hosoya, S., Tomosada, Y., and Tsukida, K. (2014). Immunobiotic Lactobacillus jensenii as immune-health promoting factor to improve growth performance and productivity in post-weaning pigs. BMC Immunol. 15:24. doi: 10.1186/1471-2172-15-24
Tang, J. C., Wei, J. H., Maeda, K., Kawai, H., Zhou, Q. X., and Hosoi-Tanabe, S. (2007). Degradation of seaweed wakame (Undaria pinnatifida) by composting process with inoculation of Bacillus sp. HR6. Biocontrol Sci. 12, 47–54. doi: 10.4265/bio.12.47
Veldhuizen, E. J., Rijnders, M., Claassen, E. A., van Dijk, A., and Haagsman, H. P. (2008). Porcine β-defensin 2 displays broad antimicrobial activity against pathogenic intestinal bacteria. Mol. Immunol. 45, 386–394. doi: 10.1016/j.molimm.2007.06.001
Villena, J., Aso, H., Rutten, V. P., Takahashi, H., Van Eden, W., and Kitazawa, H. (2018). Immunobiotics for the bovine host: their interaction with intestinal epithelial cells and their effect on antiviral immunity. Front. Immunol. 9:326. doi: 10.3389/fimmu.2018.00326
Villena, J., Vizoso-Pinto, M. G., and Kitazawa, H. (2016). Intestinal innate antiviral immunity and immunobiotics: beneficial effects against rotavirus infection. Front. Immunol. 7:563. doi: 10.3389/fimmu.2016.00563
Vodolazska, D., Feyera, T., and Lauridsen, C. (2023). The impact of birth weight, birth order, birth asphyxia, and colostrum intake per se on growth and immunity of the suckling piglets. Sci. Rep. 13:8057. doi: 10.1038/s41598-023-35277-3
Wang, J., Ji, H., Wang, S., Liu, H., Zhang, W., and Zhang, D. (2018). Probiotic Lactobacillus plantarum promotes intestinal barrier function by strengthening the epithelium and modulating gut microbiota. Front. Microbiol. 9:1953. doi: 10.3389/fmicb.2018.01953
Wang, J., Ji, H., Wang, S., Zhang, D., Liu, H., and Shan, D. (2012). Lactobacillus plantarum ZLP001: in vitro assessment of antioxidant capacity and effect on growth performance and antioxidant status in weaning piglets. Asian Austral. J. Anim. Sci. 25, 1153–1158. doi: 10.5713/ajas.2012.12079
Wang, L., Liu, Q., Chen, Y., Zheng, X., Wang, C., and Qi, Y. (2022). Antioxidant potential of Pediococcus pentosaceus strains from the sow milk bacterial collection in weaned piglets. Microbiome 10:83. doi: 10.1186/s40168-022-01278-z
Wang, Q., Sun, Q., Qi, R., Wang, J., Qiu, X., and Liu, Z. (2019). Effects of Lactobacillus plantarum on the intestinal morphology, intestinal barrier function and microbiota composition of suckling piglets. J. Anim. Physiol. Anim. Nutr. 103, 1908–1918. doi: 10.1111/jpn.13198
Wang, J., Zhang, W., Wang, S., Liu, H., Zhang, D., and Wang, Y. (2019). Swine-derived probiotic Lactobacillus plantarum modulates porcine intestinal endogenous host defense peptide synthesis through TLR2/MAPK/AP-1 signaling pathway. Front. Immunol. 10:2691. doi: 10.3389/fimmu.2019.02691
Yoon, J. H., Ingale, S. L., Kim, J. S., Kim, K. H., Lohakare, J., and Park, Y. K. (2013). Effects of dietary supplementation with antimicrobial peptide-P5 on growth performance, apparent total tract digestibility, faecal and intestinal microflora and intestinal morphology of weanling pigs. J. Sci. Food Agric. 93, 587–592. doi: 10.1002/jsfa.5840
Zhang, J., Deng, J., Li, Y., and Yang, Q. (2011). The effect of Lactobacillus on the expression of porcine β-defensin-2 in the digestive tract of piglets. Livest. Sci. 138, 259–265. doi: 10.1016/j.livsci.2011.01.001
Zhou, B., Albarracin, L., Indo, Y., Arce, L., Masumizu, Y., and Tomokiyo, M. (2020). Selection of immunobiotic Ligilactobacillus salivarius strains from the intestinal tract of wakame-fed pigs: functional and genomic studies. Microorganisms 8:1659. doi: 10.3390/microorganisms8111659
Keywords: porcine intestinal epithelial cells, pig milk, Lactobacillus plantarum, immunomodulatory potential, wakame
Citation: Ohgi R, Saha S, Zhou B, Sakuma T, Sakurai M, Nakano Y, Namai F, Ikeda-Ohtsubo W, Suda Y, Nishiyama K, Villena J and Kitazawa H (2024) In vitro evaluation of the immunomodulatory and wakame assimilation properties of Lactiplantibacillus plantarum strains from swine milk. Front. Microbiol. 15:1324999. doi: 10.3389/fmicb.2024.1324999
Received: 27 October 2023; Accepted: 05 January 2024;
Published: 26 January 2024.
Edited by:
Analia Graciela Abraham, National University of La Plata, ArgentinaReviewed by:
Harpreet Kaur, Vanderbilt University Medical Center, United StatesCopyright © 2024 Ohgi, Saha, Zhou, Sakuma, Sakurai, Nakano, Namai, Ikeda-Ohtsubo, Suda, Nishiyama, Villena and Kitazawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julio Villena, amN2aWxsZW5hQGNlcmVsYS5vcmcuYXI=; Haruki Kitazawa, aGFydWtpLmtpdGF6YXdhLmM3QHRvaG9rdS5hYy5qcA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.