
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 January 2024
Sec. Systems Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1302822
Background: Accumulating evidence suggests that alterations in gut microbiota composition are associated with the hidradenitis suppurativa (HS). However, the causal association between gut microbiota and HS remain undetermined.
Methods: We performed a bidirectional two-sample Mendelian randomization (MR) analysis using genome-wide association study summary data of gut microbiota and hidradenitis suppurativa from the MiBioGen consortium which concluded 18,340 individuals analyzed by the MiBioGen Consortium, comprising 211 gut microbiota. HS data were acquired from strictly defined HS data collected by FinnGenbiobank analysis, which included 211,548 European ancestors (409 HS patients, 211,139 controls). The inverse variance weighted method (IVW), weighted median (WME), simple model, weighted model, weighted median, and MR-Egger were used to determine the changes of HS pathogenic bacterial taxa, followed by sensitivity analysis including horizontal pleiotropy analysis. The MR Steiger test evaluated the strength of a causal association and the leave-one-out method assessed the reliability of the results. Additionally, a reverse MR analysis was carried out to seek for possible reverse causality.
Results: By combining the findings of all the MR steps, we identified four causal bacterial taxa, namely, Family XI, Porphyromonadaceae, Clostridium innocuum group and Lachnospira. The risk of HS might be positively associated with a high relative abundance of Clostridium innocuum group (Odds ratio, OR 2.17, p = 0.00038) and Lachnospira (OR 2.45, p = 0.017) but negatively associated with Family XI (OR 0.67, p = 0.049) and Porphyromonadaceae (OR 0.29, p = 0.014). There were no noticeable outliers, horizontal pleiotropy, or heterogeneity. Furthermore, there was no proof of reverse causation found in the reverse MR study.
Conclusion: This study indicates that Clostridium innocuum group and Lachnospira might have anti-protective effect on HS, whereas Family XI and Porphyromonadaceae might have a protective effect on HS. Our study reveals that there exists a beneficial or detrimental causal effect of gut microbiota composition on HS and offers potentially beneficial methods for therapy and avoidance of HS.
Hidradenitis suppurativa (HS), also known as acne inversa, is a chronic, persistent, inflammatory skin condition brought on by obstruction of the infundibular portion of the pilosebaceous unit (Rathod et al., 2023). It often misinterprets as an infection, with the high impact on the patient’s quality of life among all the assessed dermatological diseases. The global epidemiological survey indicates that HS prevalence varies from 0.03 to 4% (Calao et al., 2018). The pathophysiology and etiology of HS are yet undetermined. However, several variables are thought to be linked to the onset and worsening of HS, including as inflammation, genetics, microbiome, environmental components, age, BMI, smoking status, work status, and income (Nguyen et al., 2021).
Recent research has also revealed an underlying gut-skin axis, where the gut and immune system are in close communication and collaborate to guard against external antigens (De Pessemier et al., 2021; Sinha et al., 2021; Mahmud et al., 2022). Utilizing antibiotics with extra anti-inflammatory properties is one of the treatment strategies for HS, while it is yet unknown how their antimicrobial action directly affects the microbiota (Frew et al., 2019). Multiple studies have shown a connection between changes in intestinal flora abundance and the onset and progression of HS, albeit the nature of this connection is still unidentified (Ellis et al., 2019). For instance, in HS patients, research found that Bilophila species were more prevalent while Lachnobacterium species were less prevalent (Kam et al., 2021). Further research found that the increased inflammatory markers, such as TNF-α, IL-6, IL-8, and C-reactive protein, were linked to lower microbial diversity and increased sluggishness and deteriorating wellness metrics which might be related to the HS (Öğüt et al., 2022). Yet, there was occasionally disagreement in the epidemiological data supporting these relationships, which might be caused by different measuring techniques, selective bias and confounding factors. As an instance, it had been found that Firmicutes levels were either greater (Haskin et al., 2016) or lower (Kam et al., 2021) in HS patients compared to healthy controls.
Although clinical data validates this link, common cofounders, selective bias (such as age, gender, and area), and variability in investigations tend to render it tough to comprehend. So as to better understand the processes behind HS and supply evidence-based strategies for clinical counseling, it may be desirable to investigate the relationship between the gut microbiota and HS as well as to further examine a causative correlation.
Based on a genetic standpoint, Mendelian randomization (MR) examines potential causal relationships between exposures and outcomes through applying the genetic law of random distribution of gamete alleles (Richmond and Davey, 2022; Sanderson et al., 2022). The genetic variations are limited to influencing the exposure components in order to impact the outcome indicators, which can lower the interference of confounding and reverse causal relationships. MR requires a genetic variant that is strongly related with exposure as an instrumental variable (Lousdal, 2018). Using the abundance of the gut microbiota as the exposure factor and the occurrence of HS as the outcome, the two-sample Mendelian randomization method was used in this study to analyze the potential causal relationship between the gut microbiota and HS while examining their genetic relationship (Sekula et al., 2016; Bowden and Holmes, 2019).
In the present research, we used a Mendelian randomization methodology to assess a deeper connection between the gut responsible microbiota and HS. In addition to providing possible targets for therapy for the medical treatment of psoriasis, it was projected that our results might assist in the study of the inflammatory conditions underlying HS.
Our analysis used the summary statistics of publicly available GWASs. No new data was collected, and no new ethical approval was conducted. The whole process that we studied was presented in the flow chat in Figure 1. Concisely, we conducted a two-sample MR study to evaluate the causal effect between gut microbiota and HS (Jia et al., 2019). The validity of MR is based on three main assumptions (Bowden et al., 2015): (1) relevance--the relationship between genetic variants and exposure was robust; (2) independence the genetic variants were independent of confounding factors affecting exposure and outcome; and (3) exclusion restriction-the genetic variants influenced the risk of the outcome through exposure rather than other potential pathways. We used a two-sample MR computational model to investigate if there were directional causal relationships between gut microbiota and HS (the random-effects inverse variance weighted (IVW), MR-Egger regression, weighted median, reverse MR, and MR Steiger). Finally, several sensitivity analyses (the heterogeneity test, the pleiotropy test, and leave-one-out) were conducted sequentially.
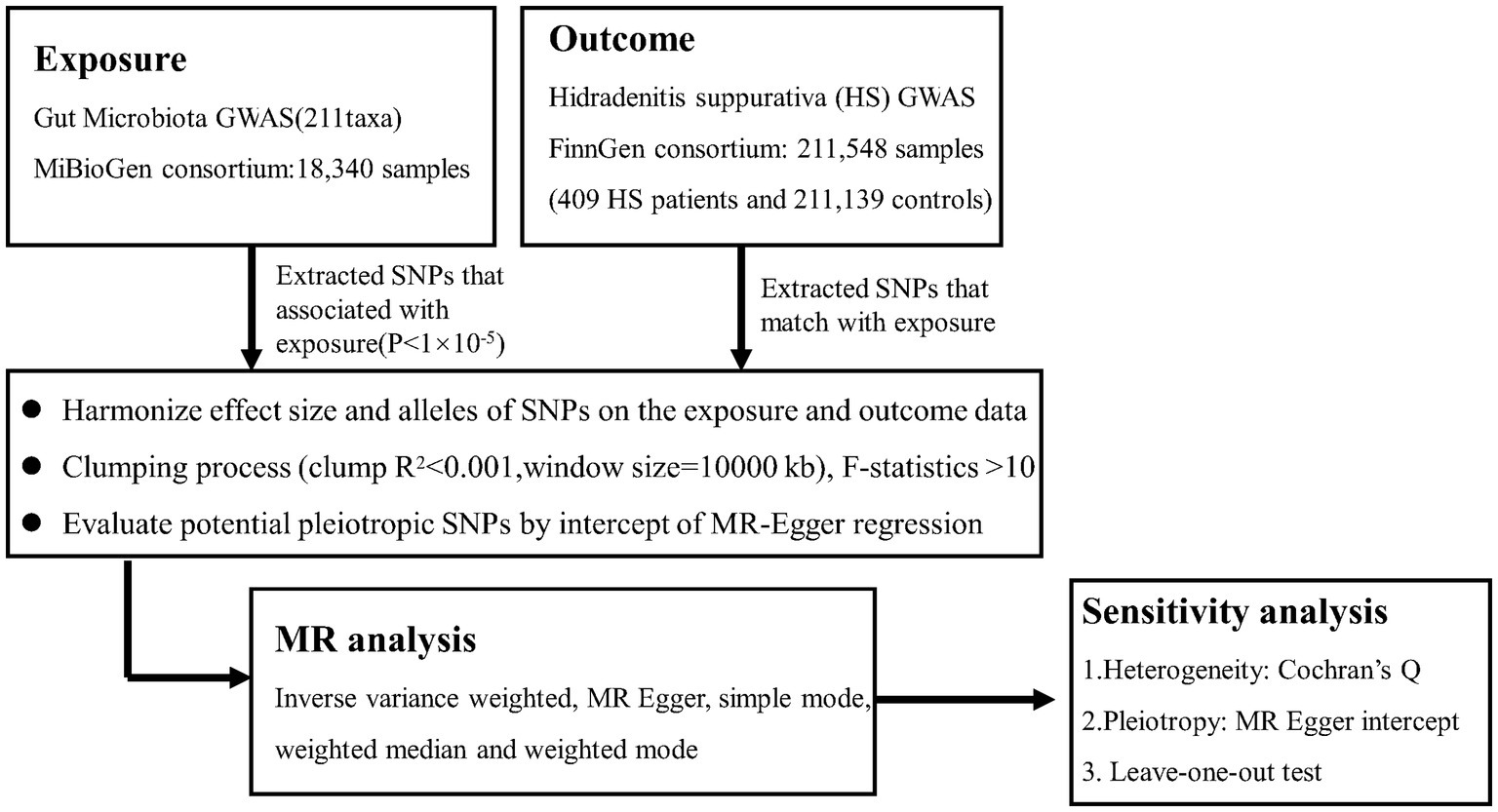
Figure 1. Flowchart of this Mendelian randomization study. MR. Mendelian randomization; HS. Hidradenitis suppurativa.
The GWAS summary statistics used in our study were presented in Table 1. The individuals from the data sources were from European ancestry mostly.
GWAS data for intestinal bacteria were extracted from the MiBioGen consortium1, which gathered whole genome genotyping data from 18,340 participants as long as the 16S rRNA genes from participant’s fecal microbiomes (Kurilshikov et al., 2021). The 20 cohort studies included single ancestry samples from various regions, such as European (n = 13,266), Middle Eastern (n = 481), Latin American (n = 1,097), East Asian (n = 811), African American (n = 114). The data collected from four cohorts included multiple ancestries (n = 2,571). Three different variable regions (V1-V2, V3-V4, and V4) of the 16S rRNA gene were targeted in order to profile the microbial composition. A total of 211 taxa (131 genera, 35 families, 20 orders, 16 classes, and 9 phyla) were included. In our study, we finally included 194 taxa (119 genera30 families, 20 orders, 16 classes, and 9 phyla) after excluding unknown gut microbes.
Summary statistics data for HS in individuals of European ancestry were obtained from the publicly available GWAS analyses. The study investigated HS cases (409) and controls (211,139), which included over 16 million genetic variants (Bao et al., 2023).2
We selected single-nucleotide polymorphisms (SNPs) associated with gut microbiota with a relatively tolerant significance level (p < 1.0 × 10−5) (ensuring sufficient variables for screening) (Din et al., 2019). Then, we applied chain disequilibrium r2 < 0.001 within the distance of 10,000 kb, as a cutoff of linkage disequilibrium, for respective independence before being used as primary genetic instruments. After coordinating with responsive results, every pair of combinations was retrieved for additional analysis. To detect bias from weak instrumental factors, the F statistic of IVs was determined (F = R2(n-k-1)/k(1-R2)) (n is the sample size, k is the number of included instrumental variables (IVs), and R2 is the exposure variance explained by the selected SNPs). An F statistic of more than 10 is suggestive of a strong instrument. Finally, a reverse MR study was performed to investigate the reverse causal relationship, and a nominal causal effect was defined as one with a p-value between 0.05.
The inverse variance weighted (IVW fixed model) method was employed as the main analysis, to obtain an unbiased estimate of the causal relationship between gut microbiota and HS. To evaluate the effect sizes of causation, odds ratios (ORs) of the exponential type and related confidence intervals (CIs) were mostly used and p value <0.05 was considered statically significant. Furthermore, the weighted median, MR Egger, simple mode, and weighted mode methods were applied as additional methods to estimate causal effects under different conditions. If at least half of the weight was generated from accurate IVs, the weighted median technique might incorporate data on many genetic variants into a single causal estimate and produce a consistent estimate (Bowden et al., 2016). The MR-Egger technique and MR-PRESSO (MR pleiotropy residual sum and outlie) might quantify the causal influence and determine whether genetic variations exhibit directional pleiotropy (Li and Li, 2023). In order to evaluate horizontal pleiotropy, the intercept of MR-Egger regression and MR-PRESSO were computed. The p value of >0.05 suggested that there was a low likelihood that pleiotropy would have an impact on the causal analysis. The IVW estimation method was used to produce Cochran’s Q test, which was utilized to find heterogeneity among instrumental variables. In order to determine horizontal pleiotropy and eliminate probable outliers, we also used the Mendelian randomization pleiotropy residual sum approach. To assess the level of influence of a single SNP’s causal association effect, the leave-one-out strategy, which systematically deleted one of the SNPs and utilized the remaining SNPs as instrumental variables for two-sample MR analysis, was applied (Dong et al., 2023). Moreover, MR Steiger directionality test was conducted to comprehensively assess the association between exposure and outcomes. The MR Steiger process made the assumption that a suitable genetic variation should explain more variance during exposure than during outcome. This strategy assured that the genetic instruments meet the requirements for a legitimate MR inquiry and aid in the identification of potential bidirectional effects (Hemani et al., 2017). Finally, we carried out reverse MR analysis to examine the effect of hidradenitis suppurativa on the identified gut microbiota. SNPs related to hidradenitis suppurativa were used as IVs.
If the following conditions were satisfied, we thought there was a significant causal link between systemic gut microbiota and HS: (1) The IVW method showed a significant difference (p < 0.05); (2) the five methods yielded consistent estimates; (3) the Cochran’s Q test, MR-Egger and MR-PRESSO were not significant (p > 0.05); and (4) the value of MR Steiger directionality <0.05. The ‘TwoSampleMR’ package in R software (version 4.2.2) were used for all MR analyses.
After screening at a relatively loose threshold (p < 1 × 10−5) and LD clumping, SNPs of gut microbiota ranging from phylum to genus levels were applied (Supplementary Table S1). The detailed information of the final SNPs for each bacterial trait were shown in (Supplementary Table S2). All the F statistics of the instrumental variables were over 10, implying that there was no weak instrument bias.
An overview of the causal effect of 211 gut microbiota taxa on HS is shown in Figure 2. Of all the genera, four significant bacterial genera were selected for further MR analyses. Furthermore, eight independent SNPs were associated with Family XI, nine independent SNPs were associated with Porphyromonadaceae and Clostridium innocuum group, respectively. Fifteen independent SNPs were associated with Lachnospira. SNP detailed message (Position, SD, R2, F) of significant genera in MR analyses were shown in Table 2.
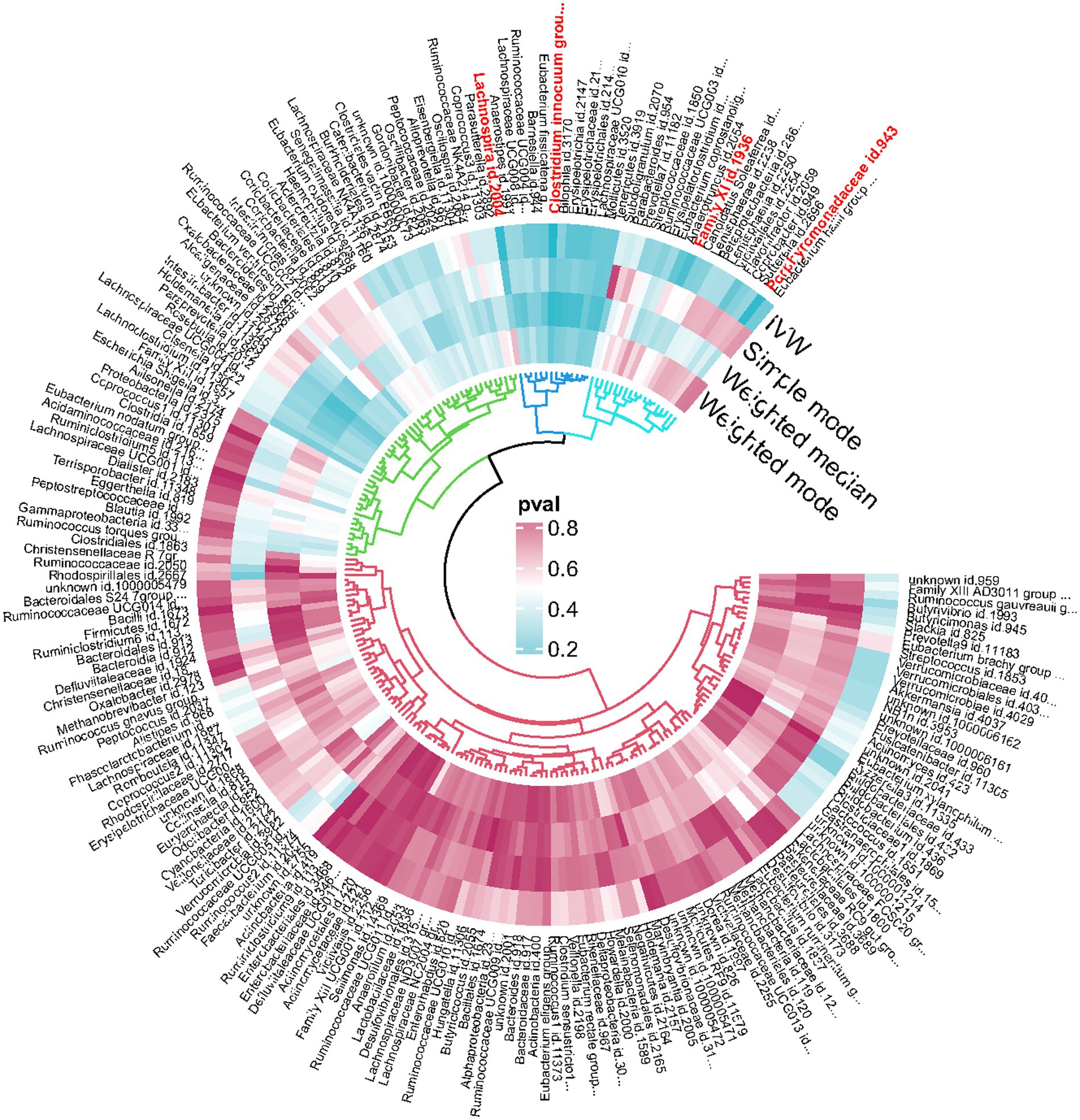
Figure 2. Overview of the causal role of gut microbiota in hidradenitis suppurativa. The Cyan Process color indicates statistical significance (p < 0.05). IVW inverse variance weighted method.
At the family level, we found that Family XI (OR 0.67, 95% CI 0.45–1.00, p = 0.049) and Porphyromonadaceae (OR 0.29, 95% CI 0.11–0.78, p = 0.014) had a nominally protective role in HS by the primary IVW method (Table 3). For another, at genus level Clostridium innocuum group (OR 2.17, 95% CI 1.42–3.34, p = 0.00038) and Lachnospira (OR 2.45, 95% CI 1.17–5.14, p = 0.017) had an increased risk of developing HS (Figure 3). All the MR Steiger directionality tests indicated a consistent trend from gut microbiota to HS for all outcomes.
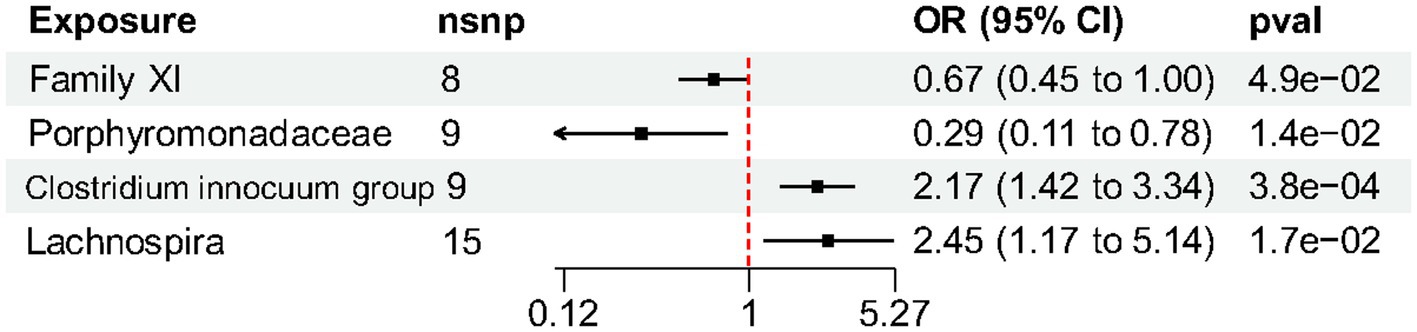
Figure 3. Forest plot of gut microbiota taxa associated with hidradenitis suppurativa identified by the inverse variance weighted method.
No heterogeneity was found within the IVs of all the four genera by Cochrane’s Q test (Table 4). The MR-Egger regression intercepts and the MR-PRESSO indicated no horizontal pleiotropy and outlier values (p > 0.05). The scatter plots illustrated that Family XI and Porphyromonadaceae might have protective effect on HS, meanwhile, Clostridium innocuum and Lachnospira might have anti-protective effect on HS. The IVW method, MR-Egger, weighted median, weighted mode, and simple mode are the methods of MR analysis that have weights and were described in the scatter plots. Positive markers of the association between the genus and HS were discovered to be the lines sloping upward from left to right, whereas protective genera were found to be those sliding downward from left to right (Figure 4). There were no potential outliers of the IVs of all four genera for HS in “leave-one-out” analysis (Figure 5), implying that all the identified causal associations were not influenced by single IV. Furthermore, the funnel plots showed no observable horizontal pleiotropy for any outcome (Figure 6). Analysis of the reverse MR data showed that HS had no causal effect on the screened gut microbiota (Table 5).
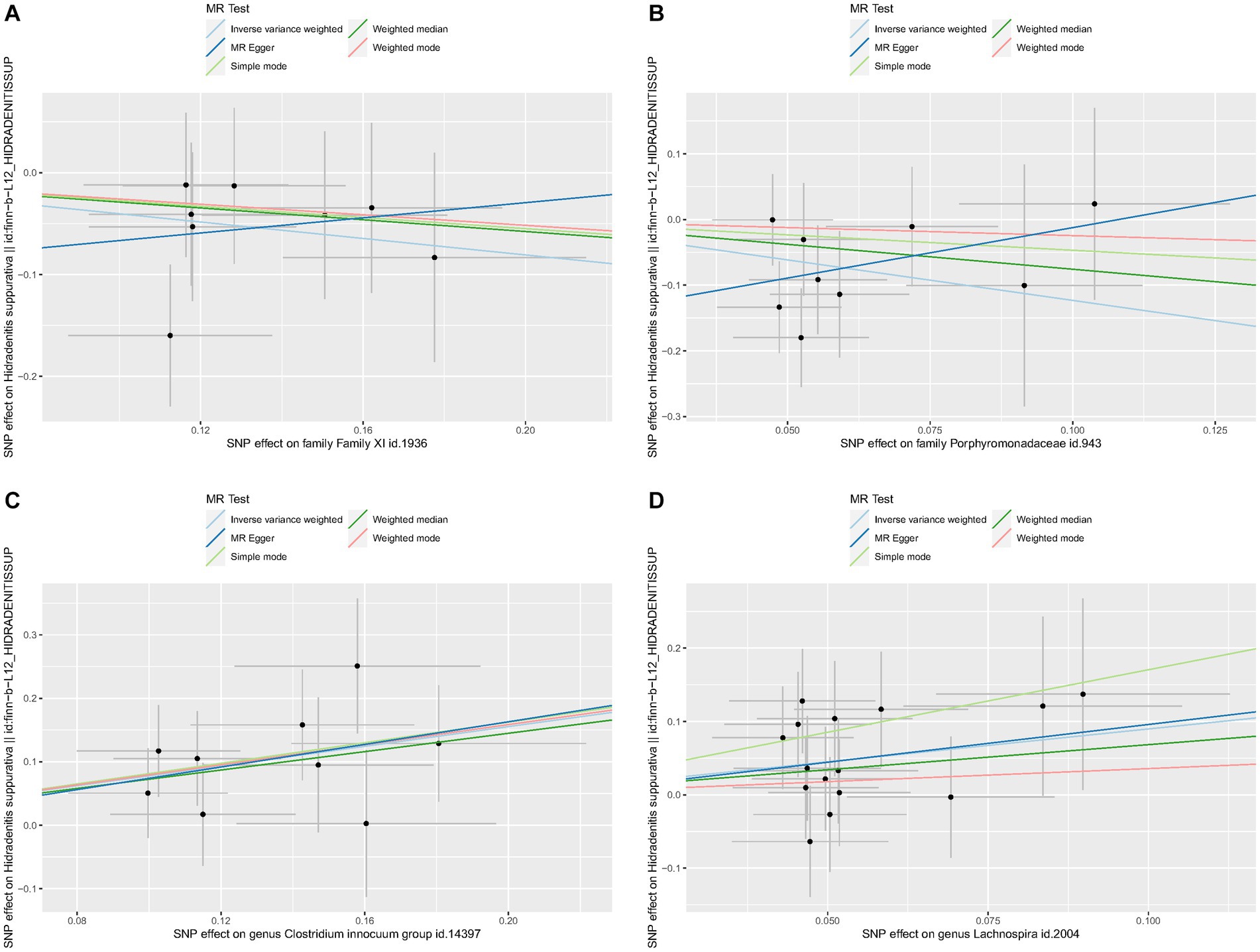
Figure 4. Scatter plots of each genus associated with the risk of hidradenitis suppurativa. (A) Family XI, (B) Porphyromonadaceae, (C) Clostridium innocuum group, (D) Lachnospira.
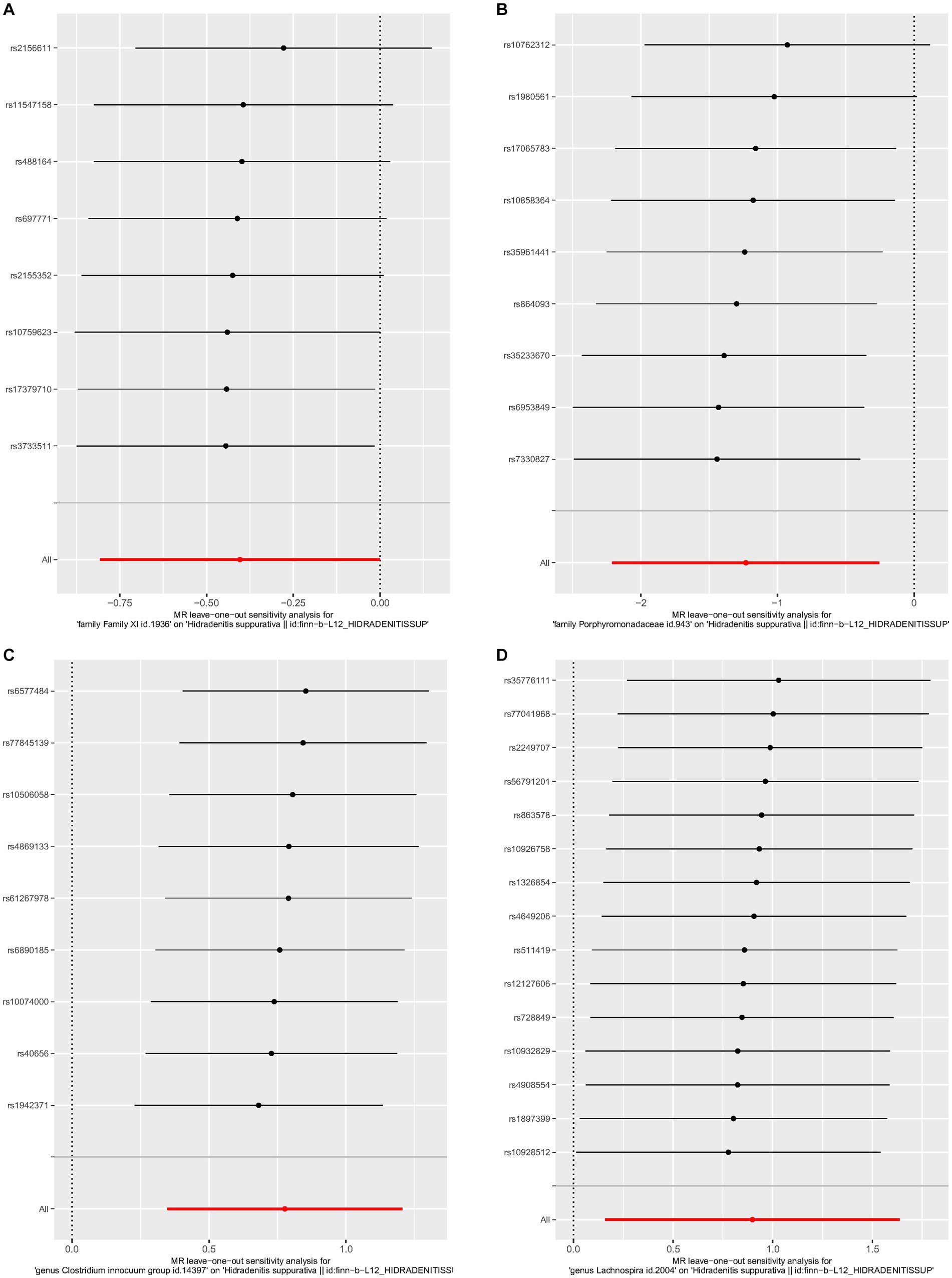
Figure 5. Leave-one-out analysis of each genus associated with hidradenitis suppurativa. (A) Family XI, (B) Porphyromonadaceae, (C) Clostridium innocuum group, (D) Lachnospira.
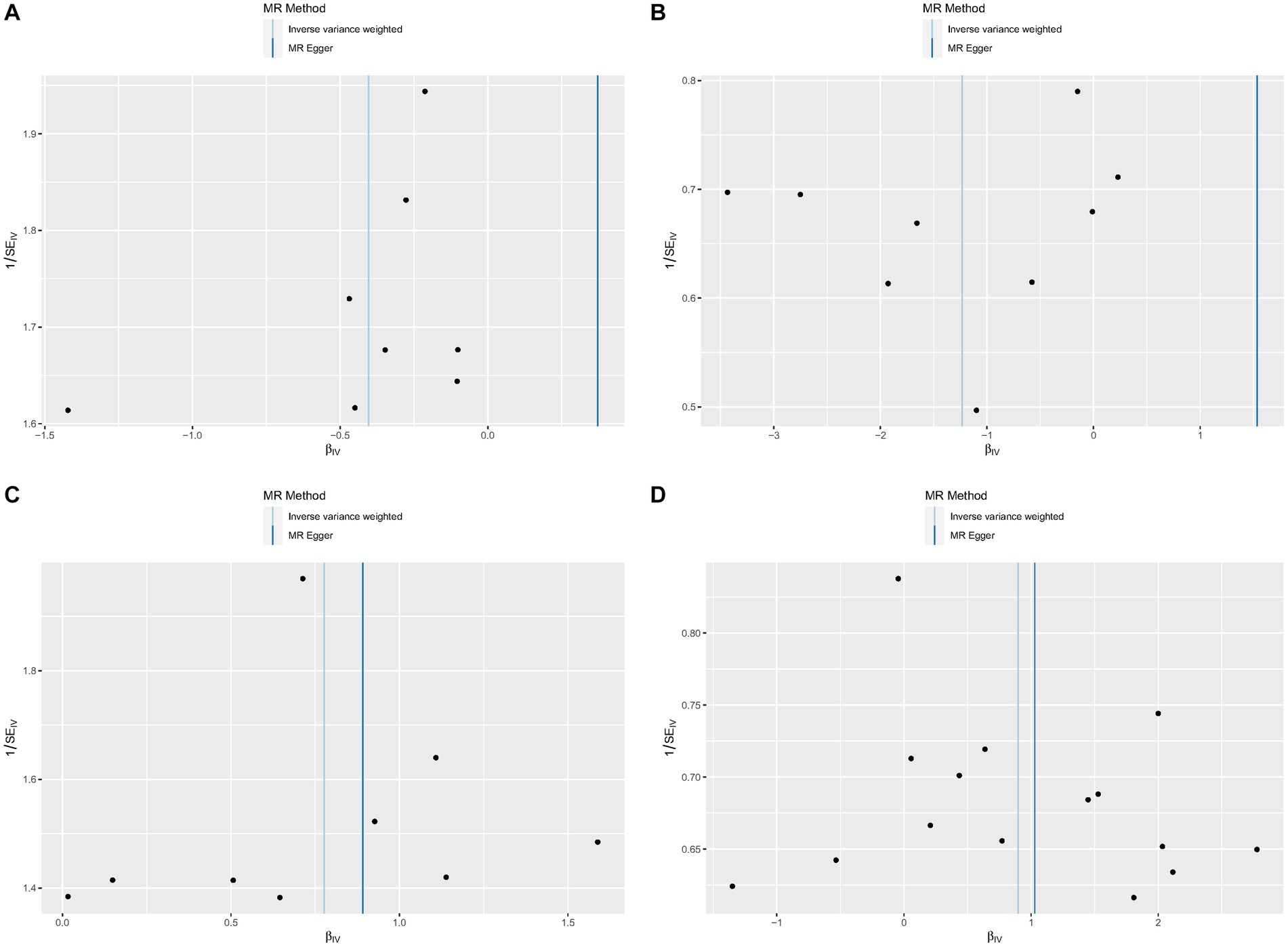
Figure 6. Funnel plots of each genus associated with hidradenitis suppurativa. (A) Family XI, (B) Porphyromonadaceae, (C) Clostridium innocuum group, (D) Lachnospira.
Firmicutes, Bacterioidetes, Actinobacteria, and Proteobacteria constitute the majority of normal gut microbiota (Liu et al., 2023). These organisms are largely steady and unchanging at the phylum level but vary greatly at the species level (Christovich and Luo, 2022). In conjunction with immunological modulation, the gut microbiota serves as essential underlying metabolism and intestinal permeability equilibrium (Qiu et al., 2022; McCallum and Tropini, 2023). According to epidemiological data, there was a link between HS disease development and gut microbiota. Our current study comprehensively examined the link of causality between gut microbiota and HS using pooled GWAS data. To the best of our knowledge, this study is the first to use MR method to investigate the bidirectional causative relationship between gut microbiota and HS. With the help of extensive GWAS summary statistics, we thoroughly assessed the causal impact of 211 GM taxa (from phylum to genus level) on HS in this investigation. Finally, we revealed several types of microbes in the gut that are formally linked to HS. Family XI (a family in Clostridiales also known as Clostridium cluster XI) and Porphyromonadaceae were identified to possess a role of prevention in HS, however the Clostridium innocuum group and Lachnospira were ostensibly linked to higher risk for HS. By using pleiotropy analysis, no significant pleiotropic variant among the chosen genetic instrumental variants was discovered in the datasets. Notably, results from five MR analysis techniques showed that the specific microbiome instrumental variations strongly affected the risk of HS not via other mechanisms. These findings suggest a causal relationship between HS and gut microbiome.
It is assumed that HS has a complex pathophysiology, with factors including microbiome, environment, lifestyle, and genetics all playing a role. There is a growing mount of knowledge on HS that suggests pathogenic bacteria may play a part and mounting evidence that the development and maintenance of host homeostasis depend on gut microbiota having constant communication with one another (Alves et al., 2022). Unfortunately, a complete understanding of the precise mechanism underpinning gut-skin microbial interactions is still lacking. McCarthy and Kam found that HS patients exhibited lower abundances of beneficial Lachnobacterium and Veillonella and higher levels of Ruminococcus gnavus, Clostridium ramosum, Bilophila, and Holdemania when compared to the healthy control group (Kam et al., 2021; McCarthy et al., 2022). Increased amounts of the bacterial metabolite Trimethylamine oxide (TMAO) in the bloodstream were discovered by Barrea in HS patients, and these levels were found to be correlated with higher HS Sartorius scores (Barrea et al., 2021). Luck et al. (2022) speculated that in addition to microbial dysbiosis, bacterial processes producing toxic compounds might also be linked to or involved in the development of HS.
In our study, one of the guarded microbiota (Porphyromonadaceae and Family XI) found come from the phylum Bacteroidetes and Clostridiales, which were capable of producing short-chain fatty acids (SCFAs) and were thought to preserve intestinal barrier function by preventing the passage of proinflammatory molecules into the systemic circulation and preventing the occurrence of metabolic endotoxemia (Brahe et al., 2013). There was evidence linking higher levels of acetate, n-Butyrate, and propionate to a higher Porphyromonadaceae abundance (Kelder et al., 2014). Through the production of short-chain fatty acids, Porphyromonadaceae taxa might likely play a role as adiposity modulators (Peng et al., 2009). To fulfill its function of safeguarding the gut and the human body, Jennings et al. discovered that individuals with higher relative abundances of Porphyromonadaceae had lower levels of adipose tissue and systemic inflammation (Jennings et al., 2023). Moreover, Valkonen et al. observed that Family XI could provide protection against allergic illness by contrasting the intestinal flora of the healthy control group with that of allergic disease patients (Valkonen et al., 2015). Previous research investigations had also noted that patients with HS showed a reduction in SCFAs and bacteria that produced SCFAs, such as Veillonella and Prevotella, which was consistent with our findings (Kam et al., 2021; Jiminez and Yusuf, 2023). At the same time, Tatian et al. found that HS individuals who received the adalimumab treatment showed a shift in the composition and function of the gut microbiota with significantly increased SCFA acetate (Tatian et al., 2022). The primary byproducts of the gut microbial fermentation of dietary fiber were SCFAs, which also included butyrate and propionate (Kam et al., 2021). By lowering the synthesis of pro-inflammatory cytokines including IL-1β, IL-6, IL-17, TNF-α and Th17 as well as immune cell proliferation, SCFAs and butyrate exhibited anti-inflammatory properties (Vinolo et al., 2011). Additionally, the intestinal dysbiosis and this increased cytokine outputs were what caused the HS to emerge (Molnar et al., 2020). More precisely, butyrate could stimulate Foxp3 enhancer and suppress histone deacetylase (HDAC). The host was thus protected against inflammation by peripherally derived Treg cells, which were stimulated to develop into naive CD4 + T cells when Foxp3 became expressed (Scheinfeld, 2013; Singh et al., 2014; Li et al., 2021). It suggested that the loss of butyrate-producing bacteria might be a major factor in the pathophysiology of HS by promoting a local inflammatory response, which weakened the gut epithelial barrier’s ability to regulate the presentation of gastrointestinal antigens to immune cells and systemic circulation (Bischoff et al., 2014).
Clostridium innocuum was a gram-positive, spore-forming, anaerobic bacterium (Cherny et al., 2021). According to recent research, it had been linked to diarrhea caused by antibiotics that resembled Clostridium difficile and extraintestinal infections (Chia et al., 2018). Despite the fact that no direct research had explored at the pathophysiology of Clostridium innocuum in HS, there was evidence that the IBD (inflammatory bowel disease) group had an excessive number of Clostridium innocuum colonization (Yu et al., 2023). Clostridium innocuum group had been postulated as a potential causative relation with Crohn’s disease (CD) by leading to creeping fat, stimulating tissue remodeling and causing inflamed and fibrotic intestine via M2 macrophages (Ha et al., 2020) which could partly explain the anti-protective mechanism the HS.
However, given the intricacy of the gut microbiota and the significant intra- and inter-species variation that might have an effect on host health, there was in fact a discrepancy between our findings and the available data. Conclusive evidence also needed to confirm how Lachnospira group increased the risk of IDB because Lachnospira group as one of butyrate-producing fora could benefit to certain inflammatory disorder (Wright et al., 2017; Iavarone et al., 2023; Ordoñez-Rodriguez et al., 2023). On the other hand, Lachnospira eligens, previously were known as Eubacterium eligens (Oren and Garrity, 2020). There was documentation of the evidence against the Eubacterium group which suppressed CD83 to preserve mice in systemic inflammation, corresponding to research by Islam et al. (2021). The Eubacterium group could contribute a proinflammatory function in colon carcinoma which could be similarly relevant to the role of nucleotide-binding oligomerization domain 2 linking HS with IBD, as demonstrated by Wang et al. (2021), Mintoff et al. (2023), and Ring et al. (2023). We thus concluded that the Lachnospira group might worsen HS by triggering systemic inflammation in the dysbacteriosis environment. Meanwhile, additional investigation was required done to determine this precise process. Research on the gut microbiota’s function in hidradenitis suppurativa is still under progress. The fundamental connection between the gut microbiota and hidradenitis suppurativa was based on the casual relation between gut microbiota and IBD (Chen and Chi, 2019). Moreover, anti-TNFα medications were effective in treating both HS and IBD, suggesting comparable inflammatory pathomechanisms. A potential new field of study involved employing bacteria to influence the immune system in therapeutics, since our awareness of the gut microbiome’s function in disease is expanding.
Our research revealed a link between four gut bacteria genera and HS. But HS is a multifactorial illness that may be impacted by the environment, gender, lifestyle, food, age, epigenetics, and genetics (Nomura, 2020). A previous study found that people with HS had considerably lower microbial makeup (α-diversity), and that Ruminococcus gnavus and Clostridium ramosum levels were higher in their microbiota than in those of healthy controls (McCarthy et al., 2022). Additionally, in 2020, Kam et al. analyzed fecal samples from three individuals with Hurley stage II or III in a case series. According to the study, gut microbiota species diversity might have diminished, there might be a rise in the number of Bilophila and Holdemania, a drop in the number of protective Lachnobacterium and Veillonella, and HS might be related to these changes. Additionally, it was observed that the phylum Firmicutes was decreased in the HS group compared to controls. But the authors pointed out that studies had shown that smoking can lower the relative abundance of Firmicutes in the gut (Kam et al., 2021). Lam and colleagues discovered that none of the healthy controls and the majority of HS patients had Robinsoniella in their feces, which might be properly quantified in the further inquiry (Lam et al., 2021). The occurrence of HS could not be explained by a change in a single bacterium, although a particular species might have a different sort of effect in a particular biological setting which demonstrated that a single bacterium would not have an impact on HS susceptibility. Hence, the discrepancy between our results and certain other studies might have the explanation.
As far as we are aware, this is the first MR research to examine the genetic relationship between HS and the gut flora. This study’s primary strength was its use of MR analysis to reduce confounding-related bias, which increased its trustworthiness when compared to traditional observational research. Through the targeting of certain gut flora, these discoveries provided new insights into the prevention, development, and therapy of HS. To investigate the precise processes behind the relationships between the gut microbiota and HS, further clinical trials and mechanism studies are required in the future.
However, our study has a number of drawbacks. First off, the majority of individuals in the two GWASs were of European heritage, so further research is needed to determine whether our findings are applicable. Second, the present microbiome GWAS research methodologies constrained the depth of our investigation. The generalizability of our results and the precision of our study may both be enhanced by adopting an advanced analytical approach to boost the specificity and accuracy of the existing results. What’s more, in this investigation, gender was not restricted. Hence, it was imperative to examine if there was a distinction between the male and female populations exclusively. By merging the information from cohort studies, clinical trials, and functional investigations, further work must be done to find connections between HS and gut microbiota which will be helpful for examining the pathophysiology of HS.
With final analysis, our bi-directional MR analysis revealed evidence of a putative causal link between certain gut microbiota and HS. Our findings would strengthen the case for gut microecological therapy of HS and establish a strong framework for more research into the pathophysiology of the gut microbiota that causes HS.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
CL: Writing – original draft, Writing – review & editing. XCL: Data curation, Writing – review & editing. XL: Funding acquisition, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors declare that the research received the following funding: Guangxi Zhuang Autonomous Region Health Commission Scientific Research Project (Z-C20231103).
It is highly appreciated that two sizable GWAS databases, MBioGen and FinnGen collaboration, share data on study findings. We also would like to express our sincere gratitude to all of the participants and researchers in the MBioGen and FinnGen studies.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1302822/full#supplementary-material
1. ^https://mibiogen.gcc.rug.nl/, Accessed on March 12, 2023
2. ^https://finngen.gitbook.io/documentation/, Accessed on March 12, 2023
Alves, E., Gregório, J., Rijo, P., Rosado, C., and Monteiro Rodrigues, L. (2022). Kefir and the gut-skin Axis. Int. J. Environ. Res. Public Health 19:13791. doi: 10.3390/ijerph192113791
Bao, B., Zhu, C., Shi, J., and Lu, C. (2023). Causal association between inflammatory bowel disease and hidradenitis suppurativa: a two-sample bidirectional Mendelian randomization study. Front. Immunol. 14:1071616. doi: 10.3389/fimmu.2023.1071616
Barrea, L., Muscogiuri, G., Pugliese, G., de Alteriis, G., Maisto, M., Donnarumma, M., et al. (2021). Association of Trimethylamine N-oxide (TMAO) with the clinical severity of hidradenitis Suppurativa (acne Inversa). Nutrients 13:1997. doi: 10.3390/nu13061997
Bischoff, S. C., Barbara, G., Buurman, W., Ockhuizen, T., Schulzke, J. D., Serino, M., et al. (2014). Intestinal permeability--a new target for disease prevention and therapy. BMC Gastroenterol. 14:189. doi: 10.1186/s12876-014-0189-7
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int. J. Epidemiol. 44, 512–525. doi: 10.1093/ije/dyv080
Bowden, J., Davey Smith, G., Haycock, P. C., and Burgess, S. (2016). Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314. doi: 10.1002/gepi.21965
Bowden, J., and Holmes, M. V. (2019). Meta-analysis and Mendelian randomization: a review. Res. Synth. Methods 10, 486–496. doi: 10.1002/jrsm.1346
Brahe, L. K., Astrup, A., and Larsen, L. H. (2013). Is butyrate the link between diet, intestinal microbiota and obesity-related metabolic diseases. Obes. Rev. 14, 950–959. doi: 10.1111/obr.12068
Calao, M., Wilson, J. L., Spelman, L., Billot, L., Rubel, D., Watts, A. D., et al. (2018). Hidradenitis Suppurativa (HS) prevalence, demographics and management pathways in Australia: a population-based cross-sectional study. PLoS One 13:e0200683. doi: 10.1371/journal.pone.0200683
Chen, W. T., and Chi, C. C. (2019). Association of Hidradenitis Suppurativa with Inflammatory Bowel Disease: a systematic review and meta-analysis. JAMA Dermatol. 155, 1022–1027. doi: 10.1001/jamadermatol.2019.0891
Cherny, K. E., Muscat, E. B., Reyna, M. E., and Kociolek, L. K. (2021). Clostridium innocuum: microbiological and clinical characteristics of a potential emerging pathogen. Anaerobe 71:102418. doi: 10.1016/j.anaerobe.2021.102418
Chia, J. H., Wu, T. S., Wu, T. L., Chen, C. L., Chuang, C. H., Su, L. H., et al. (2018). Clostridium innocuum is a vancomycin-resistant pathogen that may cause antibiotic-associated diarrhoea. Clin. Microbiol. Infect. 24, 1195–1199. doi: 10.1016/j.cmi.2018.02.015
Christovich, A., and Luo, X. M. (2022). Gut microbiota, leaky gut, and autoimmune diseases. Front. Immunol. 13:946248. doi: 10.3389/fimmu.2022.946248
De Pessemier, B., Grine, L., Debaere, M., Maes, A., Paetzold, B., and Callewaert, C. (2021). Gut-skin Axis: current knowledge of the interrelationship between microbial Dysbiosis and skin conditions. Microorganisms 9:353. doi: 10.3390/microorganisms9020353
Din, A. U., Hassan, A., Zhu, Y., Yin, T., Gregersen, H., and Wang, G. (2019). Amelioration of TMAO through probiotics and its potential role in atherosclerosis. Appl. Microbiol. Biotechnol. 103, 9217–9228. doi: 10.1007/s00253-019-10142-4
Dong, Y., Zou, Z., Deng, P., Fan, X., and Li, C. (2023). Circulating metabolites and depression: a bidirectional Mendelian randomization. Front. Neurosci. 17:1146613. doi: 10.3389/fnins.2023.1146613
Ellis, S. R., Nguyen, M., Vaughn, A. R., Notay, M., Burney, W. A., Sandhu, S., et al. (2019). The skin and gut microbiome and its role in common dermatologic conditions. Microorganisms 7:550. doi: 10.3390/microorganisms7110550
Frew, J. W., Hawkes, J. E., and Krueger, J. G. (2019). Topical, systemic and biologic therapies in hidradenitis suppurativa: pathogenic insights by examining therapeutic mechanisms. Ther. Adv. Chronic Dis. 10:204062231983064. doi: 10.1177/2040622319830646
Ha, C., Martin, A., Sepich-Poore, G. D., Shi, B., Wang, Y., and Gouin, K. (2020). Translocation of viable gut microbiota to mesenteric adipose drives formation of creeping fat in humans. Cell 183, 666–683.e17. doi: 10.1016/j.cell.2020.09.009
Haskin, A., Fischer, A. H., and Okoye, G. A. (2016). Prevalence of Firmicutes in lesions of hidradenitis Suppurativa in obese patients. JAMA Dermatol. 152, 1276–1278. doi: 10.1001/jamadermatol.2016.2337
Hemani, G., Tilling, K., and Davey, S. G. (2017). Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 13:e1007081. doi: 10.1371/journal.pgen.1007081
Iavarone, I., Greco, P. F., la Verde, M., Morlando, M., Torella, M., de Franciscis, P., et al. (2023). Correlations between gut microbial composition, pathophysiological and surgical aspects in endometriosis: a review of the literature. Medicina (Kaunas) 59:347. doi: 10.3390/medicina59020347
Islam, S., Ryu, H. M., Sayeed, H. M., Byun, H. O., Jung, J. Y., Kim, H. A., et al. (2021). Eubacterium rectale attenuates HSV-1 induced systemic inflammation in mice by inhibiting CD83. Front. Immunol. 12:712312. doi: 10.3389/fimmu.2021.712312
Jennings, A., Kühn, T., Bondonno, N. P., Waniek, S., Bang, C., Franke, A., et al. (2023). The gut microbiome modulates associations between adherence to a Mediterranean-style diet, abdominal adiposity, and C-reactive protein in population-level analysis. Am. J. Clin. Nutr. 119, 136–144. doi: 10.1016/j.ajcnut.2023.11.001
Jia, J., Dou, P., Gao, M., Kong, X., Li, C., Liu, Z., et al. (2019). Assessment of causal direction between gut microbiota-dependent metabolites and Cardiometabolic health: a bidirectional Mendelian randomization analysis. Diabetes 68, 1747–1755. doi: 10.2337/db19-0153
Jiminez, V., and Yusuf, N. (2023). Bacterial metabolites and inflammatory skin diseases. Meta 13:952. doi: 10.3390/metabo13080952
Kam, S., Collard, M., Lam, J., and Alani, R. M. (2021). Gut microbiome perturbations in patients with hidradenitis Suppurativa: a case series. J. Invest. Dermatol. 141, 225–228.e2. doi: 10.1016/j.jid.2020.04.017
Kelder, T., Stroeve, J. H., Bijlsma, S., Radonjic, M., and Roeselers, G. (2014). Correlation network analysis reveals relationships between diet-induced changes in human gut microbiota and metabolic health. Nutr. Diabetes 4:e122. doi: 10.1038/nutd.2014.18
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165. doi: 10.1038/s41588-020-00763-1
Lam, S. Y., Radjabzadeh, D., Eppinga, H., Nossent, Y. R. A., van der Zee, H. H., Kraaij, R., et al. (2021). A microbiome study to explore the gut-skin axis in hidradenitis suppurativa. J. Dermatol. Sci. 101, 218–220. doi: 10.1016/j.jdermsci.2020.12.008
Li, H., and Li, C. (2023). Causal relationship between gut microbiota and type 2 diabetes: a two-sample Mendelian randomization study. Front. Microbiol. 14:1184734. doi: 10.3389/fmicb.2023.1184734
Li, G., Lin, J., Zhang, C., Gao, H., Lu, H., Gao, X., et al. (2021). Microbiota metabolite butyrate constrains neutrophil functions and ameliorates mucosal inflammation in inflammatory bowel disease. Gut Microbes 13:1968257. doi: 10.1080/19490976.2021.1968257
Liu, L., Wang, H., Chen, X., Zhang, Y., Zhang, H., and Xie, P. (2023). Gut microbiota and its metabolites in depression: from pathogenesis to treatment. EBioMedicine 90:104527. doi: 10.1016/j.ebiom.2023.104527
Lousdal, M. L. (2018). An introduction to instrumental variable assumptions, validation and estimation. Emerg. Themes Epidemiol. 15:1. doi: 10.1186/s12982-018-0069-7
Luck, M. E., Tao, J., and Lake, E. P. (2022). The skin and gut microbiome in hidradenitis Suppurativa: current understanding and future considerations for research and treatment. Am. J. Clin. Dermatol. 23, 841–852. doi: 10.1007/s40257-022-00724-w
Mahmud, M. R., Akter, S., Tamanna, S. K., Mazumder, L., Esti, I. Z., Banerjee, S., et al. (2022). Impact of gut microbiome on skin health: gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes 14:2096995. doi: 10.1080/19490976.2022.2096995
McCallum, G., and Tropini, C. (2023). The gut microbiota and its biogeography. Nat. Rev. Microbiol. 22:969. doi: 10.1038/s41579-023-00969-0
McCarthy, S., Barrett, M., Kirthi, S., Pellanda, P., Vlckova, K., Tobin, A. M., et al. (2022). Altered skin and gut microbiome in hidradenitis Suppurativa. J. Invest. Dermatol. 142, 459–468.e15. doi: 10.1016/j.jid.2021.05.036
Mintoff, D., Borg, I., and Pace, N. P. (2023). NOD2 at the interface of hidradenitis suppurativa and inflammatory bowel disease-an in silico analysis. Exp. Dermatol. 11:14928. doi: 10.1111/exd.14928
Molnar, J., Mallonee, C. J., Stanisic, D., Homme, R. P., George, A. K., Singh, M., et al. (2020). Hidradenitis Suppurativa and 1-carbon metabolism: role of gut microbiome, matrix metalloproteinases, and Hyperhomocysteinemia. Front. Immunol. 11:1730. doi: 10.3389/fimmu.2020.01730
Nguyen, T. V., Damiani, G., Orenstein, L., Hamzavi, I., and Jemec, G. B. (2021). Hidradenitis suppurativa: an update on epidemiology, phenotypes, diagnosis, pathogenesis, comorbidities and quality of life. J. Eur. Acad. Dermatol. Venereol. 35, 50–61. doi: 10.1111/jdv.16677
Nomura, T. (2020). Hidradenitis Suppurativa as a potential subtype of autoinflammatory keratinization disease. Front. Immunol. 11:847. doi: 10.3389/fimmu.2020.00847
Öğüt, N. D., Hasçelik, G., and Atakan, N. (2022). Alterations of the human gut microbiome in patients with hidradenitis Suppurativa: a case-control study and review of the literature. Dermatol. Pract. Concept. 12:e2022191. doi: 10.5826/dpc.1204a191
Ordoñez-Rodriguez, A., Roman, P., Rueda-Ruzafa, L., Campos-Rios, A., and Cardona, D. (2023). Changes in gut microbiota and multiple sclerosis: a systematic review. Int. J. Environ. Res. Public Health 20:4624. doi: 10.3390/ijerph20054624
Oren, A., and Garrity, G. (2020). List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 70, 4043–4049. doi: 10.1099/ijsem.0.004244
Peng, L., Li, Z. R., Green, R. S., Holzman, I. R., and Lin, J. (2009). Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 139, 1619–1625. doi: 10.3945/jn.109.104638
Qiu, P., Ishimoto, T., Fu, L., Zhang, J., Zhang, Z., and Liu, Y. (2022). The gut microbiota in inflammatory bowel disease. Front. Cell. Infect. Microbiol. 12:733992. doi: 10.3389/fcimb.2022.733992
Rathod, U., Prasad, P. N., Patel, B. M., Patel, B., Patel, C., Gandhi, S. K., et al. (2023). Hidradenitis Suppurativa: a literature review comparing current therapeutic modalities. Cureus 15:e43695. doi: 10.7759/cureus.43695
Richmond, R. C., and Davey, S. G. (2022). Mendelian randomization: concepts and scope. Cold Spring Harb. Perspect. Med. 12:a040501. doi: 10.1101/cshperspect.a040501
Ring, H. C., Thorsen, J., Lilje, B., Bay, L., Bjarnsholt, T., Fuursted, K., et al. (2023). Predictive metagenomic analysis identifies specific bacterial metabolic pathways in hidradenitis suppurativa tunnels. J. Eur. Acad. Dermatol. Venereol. 38, e63–e65. doi: 10.1111/jdv.19433
Sanderson, E., Glymour, M. M., Holmes, M. V., Kang, H., Morrison, J., Munafò, M. R., et al. (2022). Mendelian randomization. Nat. Rev. Methods Primers 2:6. doi: 10.1038/s43586-021-00092-5
Scheinfeld, N. (2013). Treatment of hidradenitis supprurativa associated pain with nonsteroidal anti-inflammatory drugs, acetaminophen, celecoxib, gabapentin, pegabalin, duloxetine, and venlafaxine. Dermatol. Online J. 19:20616. doi: 10.5070/D31911020616
Sekula, P., Del Greco, M. F., Pattaro, C., and Köttgen, A. (2016). Mendelian randomization as an approach to assess causality using observational data. J. Am. Soc. Nephrol. 27, 3253–3265. doi: 10.1681/ASN.2016010098
Singh, N., Gurav, A., Sivaprakasam, S., Brady, E., Padia, R., Shi, H., et al. (2014). Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139. doi: 10.1016/j.immuni.2013.12.007
Sinha, S., Lin, G., and Ferenczi, K. (2021). The skin microbiome and the gut-skin axis. Clin. Dermatol. 39, 829–839. doi: 10.1016/j.clindermatol.2021.08.021
Tatian, A., Bordbar, S., Sarkissian, S., Woods, J. A., Cains, G. D., and Chong, C. W. (2022). Adalimumab therapy is associated with increased faecal short chain fatty acids in hidradenitis suppurativa. Exp. Dermatol. 31, 1872–1880. doi: 10.1111/exd.14665
Valkonen, M., Wouters, I. M., Täubel, M., Rintala, H., Lenters, V., Vasara, R., et al. (2015). Bacterial exposures and associations with atopy and asthma in children. PLoS One 10:e0131594. doi: 10.1371/journal.pone.0131594
Vinolo, M. A., Rodrigues, H. G., Nachbar, R. T., and Curi, R. (2011). Regulation of inflammation by short chain fatty acids. Nutrients 3, 858–876. doi: 10.3390/nu3100858
Wang, Y., Wan, X., Wu, X., Zhang, C., Liu, J., and Hou, S. (2021). Eubacterium rectale contributes to colorectal cancer initiation via promoting colitis. Gut Pathog. 13:2. doi: 10.1186/s13099-020-00396-z
Wright, E. K., Kamm, M. A., Wagner, J., Teo, S. M., Cruz, P. D., Hamilton, A. L., et al. (2017). Microbial factors associated with postoperative Crohn's disease recurrence. J. Crohns Colitis 11, 191–203. doi: 10.1093/ecco-jcc/jjw136
Keywords: hidradenitis suppurativa, gut microbiota, Mendelian randomization, gut-skin axis, genome-wide association study
Citation: Liu C, Liu X and Li X (2024) Causal relationship between gut microbiota and hidradenitis suppurativa: a two-sample Mendelian randomization study. Front. Microbiol. 15:1302822. doi: 10.3389/fmicb.2024.1302822
Received: 27 September 2023; Accepted: 08 January 2024;
Published: 29 January 2024.
Edited by:
Mariusz Sikora, National Institute of Geriatrics, Rheumatology and Rehabilitation, PolandCopyright © 2024 Liu, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Li, bHh4eGlhbmdAcXEuY29t
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.