- 1Department of General Surgery, Ningbo No.2 Hospital, Ningbo, China
- 2Department of Emergency, Ningbo Medical Center Lihuili Hospital, The Lihuili Affiliated Hospital of Ningbo University, Ningbo, China
- 3Department of Radiotherapy and Chemotherapy, Ningbo No.2 Hospital, Ningbo, China
Background: Over the years, numerous studies have explored the relationship between gut microbiota and lower extremity deep vein thrombosis (LEDVT) and pulmonary embolism (PE). The present study utilized Mendelian randomization (MR) to assess the causal link between gut microbiota and LEDVT combined with PE.
Methods: Human gut microbiota genome-wide association study (GWAS) summary data from the MiBioGen consortium (n = 18,340) were utilized. Summary-level data on LEDVT (2,116 cases and 359,078 controls) and LEDVT combined with PE (4,319 cases and 356,875 controls) were obtained from the IEU Open GWAS project. MR analysis was conducted using the inverse variance weighted (IVW) method as the primary analysis. Additionally, MR-Egger, weighted median, weighted mode, and simple mode were employed as supplementary methods. Sensitivity analyses, including tests for heterogeneity and horizontal pleiotropy, were performed. Lastly, reverse MR analysis was performed.
Results: The IVW analyses revealed seven causal relationships between genetic liability in the gut microbiota and LEDVT and five causal relationships between genetic liability in the gut microbiota and LEDVT combined with PE. The intersection of these outcomes identified that the genus Butyricicoccus reduced the risk of both LEDVT and LEDVT combined with PE, while the genus Clostridium innocuum increased the risk for both conditions.
Conclusion: This study demonstrates that the gut microbiota is causally associated with LEDVT and LEDVT combined with PE. Our findings provide valuable insights into the underlying mechanisms and suggest potential avenues for further clinical investigations of these conditions.
1 Introduction
Venous thromboembolism (VTE), including deep vein thrombosis (DVT) and PE (Konstantinides et al., 2020), is the third most common acute cardiovascular disease in the world (Raskob et al., 2014). According to literature reports, the estimated incidence rates of PE range from 39 to 115 cases per 100,000 (Cheuk et al., 2004; Centers for Disease Control and Prevention (CDC), 2012), while the estimated incidence rates of DVT range from 53.1 to 162 per 100,000 (Jang et al., 2011; Nordström et al., 1992). LEDVT is the most common form of DVT (Stubbs et al., 2018), and if left untreated, 30–50% of patients with LEDVT eventually develop PE (Anderson et al., 1991). It is now understood that 90% of pulmonary embolisms are caused by LEDVT (Kruit et al., 1991), when both LEDVT and PE are present, PE is almost always caused by LEDVT. Current evidence suggests that the mortality rate of PE is high, with an estimated 126,145 (34%) sudden fatal cases in six European countries with a total population of 454.4 million (Cohen et al., 2007). VTE also has a significant economic burden, with annual costs ranging from 2 to 10 billion in the United States (Park et al., 2009). VTE is a preventable condition associated with genetic and acquired risk factors. Genetic risk factors, such as the factor V Leiden variant (Simone et al., 2013) and prothrombin F2 gene variant G20210A (Morange et al., 2015), play a crucial role in the development of VTE. Therefore, a comprehensive understanding of the genetic factors associated with DVT and PE is essential for advancing future research in this area.
The gut microbiota is a complex community of microorganisms residing in the digestive system, consisting primarily of bacteria but also viruses and fungi (Turnbaugh et al., 2007). It is vital in regulating human health and diseases (Heintz-Buschart and Wilmes, 2018). The composition of the gut microbiota is influenced by various factors, including dietary choices (David et al., 2014) and genetic elements within the host (Bonder et al., 2016). Several studies have found that the gut microbiota composition can modulate the risk of VTE. For example, antibiotics have been shown to modulate the gut microbiome and inhibit pulmonary thrombosis in mice (Zhang et al., 2023). Gut microbiota metabolites, such as trimethylamine N-oxides (TMAO), may also contribute to VTE risk and recurrence (Fraser et al., 2020). Gut dysbiosis can lead to the translocation of lipopolysaccharides (LPS) into the systemic circulation, causing inflammation and affecting coagulation and platelet activation (Violi et al., 2023). The Gram-negative transition of gut microbes has been associated with systemic inflammation and increased FVIII activity, contributing to VTE development (Grimnes et al., 2022). A recent study revealed that intestinal microbiota colonization restored the production of von Willebrand factor in hepatic endothelial cells and prevented platelet aggregation in Toll-like receptor 2 (TLR2) deficient mice, thereby correcting plasma vWF levels and thrombus growth in TLR2 deficient mice (Jäckel et al., 2017). In another study, fecal microbiota transplant (FMT) from healthy donors altered the microbiome composition in patients with metabolic syndrome and the coagulation system (Emdin et al., 2017). Although previous studies have suggested a link between gut microbiota, LEDVT, and PE, the genetic association between gut microbiota and LEDVT combined with PE has been largely understudied.
MR is a method that combines data from GWAS to infer causality by minimizing the influence of confounding factors such as the environment (Emdin et al., 2017). By utilizing MR methods, the present study sought to clarify the relationship between the gut microbiota and LEDVT, as well as LEDVT combined with PE, providing new evidence for further research in this field and building upon the reliability of previous MR studies in assessing causality.
2 Materials and methods
2.1 Exposure data
Genetic variants for human gut microbiota were taken from the latest GWAS summary data from the MiBioGen Consortium (2022) (Kurilshikov et al., 2021). This comprehensive investigation encompassed 18,340 individuals from 24 cohorts from the USA, Canada, Israel, South Korea, Germany, Denmark, the Netherlands, Belgium, Sweden, Finland, and the UK. The primary objective was to examine the relationship between autosomal human genetic variants and the gut microbiome, with a particular emphasis on characterizing the gut microbial composition at the genus level. Advanced taxonomic classification methods and Microbiota quantitative trait loci (mbQTL) mapping analysis were employed to identify host genetic variants associated with the abundance levels of bacterial taxa within the gut microbiota. Ultimately, a total of 211 taxa were identified, comprising 131 genera, 35 families, 20 orders, 16 classes, and 9 phyla (Kurilshikov et al., 2021).
2.2 Outcome data
Summary-level data of LEDVT and LEDVT combined with PE were retrieved from the IEU Open GWAS project: https://gwas.mrcieu.ac.uk/ (v7.5.3-2023-07-19, N = 42,350). Data for LEDVT (2,116 cases and 359,078 controls), LEDVT combined with PE (4,319 cases and 356,875 controls) were acquired from the UK Biobank (UKB) of European ancestry (Sudlow et al., 2015). The details are presented in Table 1.
2.3 Instrumental variables selection
The study flowchart is illustrated in Figure 1. The exposure variable was the gut microbiota, while the outcome variable was LEDVT and LEDVT combined with PE. To enhance the validity and reliability of our findings, we used specific selection criteria for the identification of IVs: (1) single nucleotide polymorphisms (SNPs) associated with gut microbes were obtained with a significance threshold of (p < 1 × 10−5) (Sanna et al., 2019); (2) to account for the effect of linkage disequilibrium (LD), SNPs were clumped using an r2 threshold of 0.001 and a distance of 10,000 base pairs (Walker et al., 2019); (3) palindromic A/T or G/C alleles were excluded; (4) PhenoScanner1 was utilized to identify potential confounders (MR hypothesis II) by examining all SNPs with positive results (Kamat et al., 2019). The risk factors and confounders considered included smoking, obesity, hypercholesterolemia, hypertension, diabetes mellitus, Factor V Leiden, cancer, and, for women only, the use of oral contraceptives, hormone-replacement therapy, and menopausal status (Konstantinides et al., 2020); (5) the strength of the IVs was assessed using the F statistic (F = β2/se2). IVs with an F statistic >10 were considered to have no significant weak instrumental bias (Burgess et al., 2011).
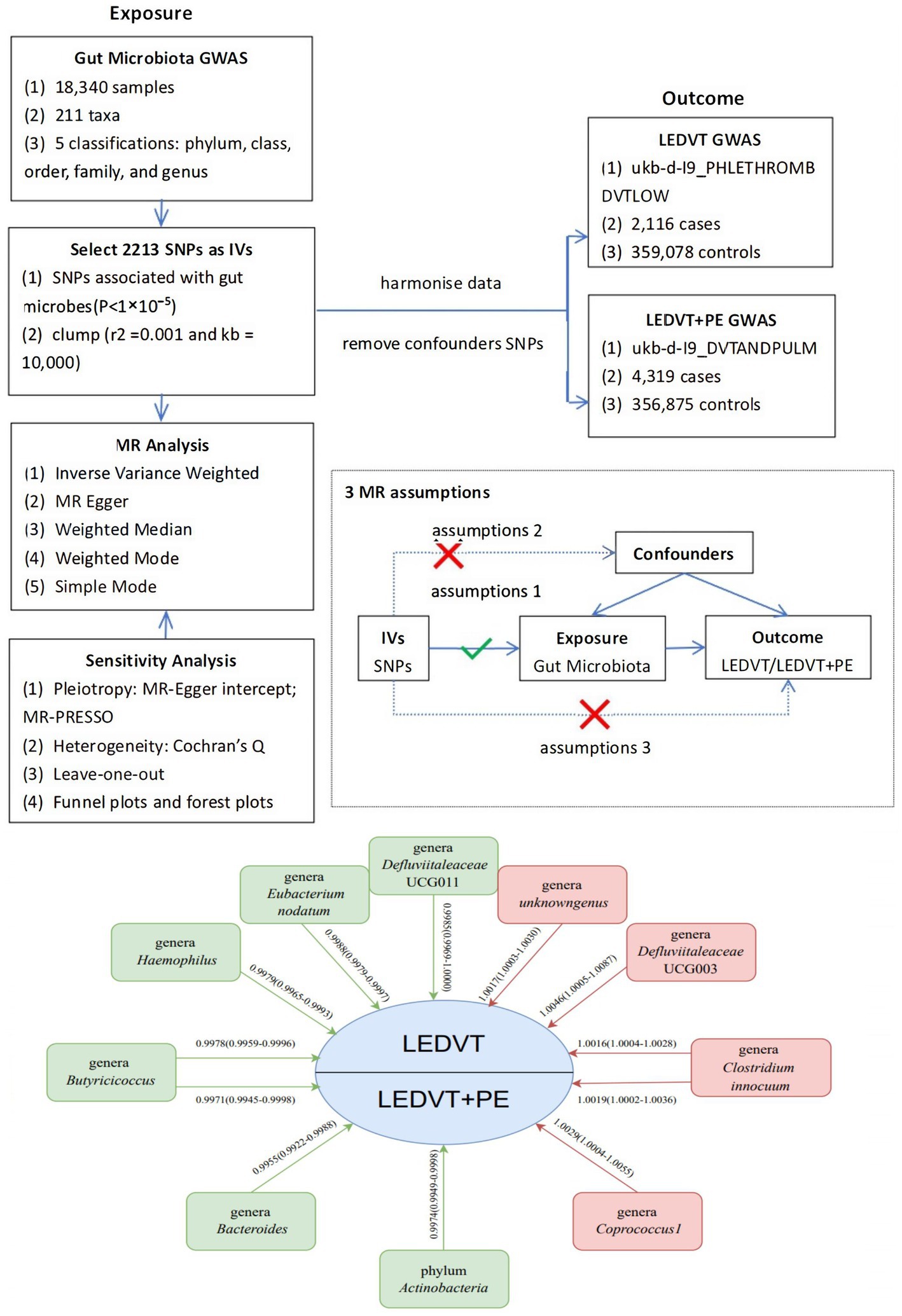
Figure 1. Overview of MR analyses process, major assumptions and major outcomes. GWAS, genome wide association study; SNPs, single nucleotide polymorphisms; IVs, instrumental variables; MR, Mendelian randomization; LEDVT, lower extremity deep vein thrombosis; PE, pulmonary embolism.
2.4 Statistical analysis
All statistical analyses were performed using R software (Version 4.2.2). MR analyses were performed using the TwosampleMR (version 0.5.6) (Hemani et al., 2017) and MR-PRESSO (version 1.0) (Verbanck et al., 2018) R packages. A p < 0.05 was statistically significant.
2.5 Mendelian randomization analysis
We performed a MR analysis to investigate the causal relationship between the gut microbiome and LEDVT or LEDVT combined with PE. To ensure reliable results, the MR analysis satisfied three assumptions, as illustrated in Figure 1: (1) The IVs included in the analysis were strongly correlated with the exposure (gut microbiome). (2) The IVs used were not associated with the selected confounding factors related to both the exposure and the outcome. (3) The IVs affected the outcome solely through the exposure, without any direct effect (Davey Smith and Hemani, 2014).
In the MR analysis, we utilized several methods to estimate the causal effects. The IVW method assumes that all SNPs are valid instruments and estimates the overall effect by calculating the inverse variance-weighted mean of the individual instrument estimates. It has been reported that the IVW method has greater statistical efficiency when there is no horizontal pleiotropy bias (Burgess et al., 2013). In contrast, the MR-Egger method allows for some degree of pleiotropy, meaning that the resulting effect remains even when the intercept (effect of the instrument on the outcome when the exposure is zero) is zero (Bowden et al., 2015). The weighted median approach can yield reliable estimates even when it accommodates invalid instruments, such as in cases where 50% of the IVs are considered invalid (Bowden et al., 2016). Although the IVW method is commonly more effective than other methods in most cases, we also employed the MR-Egger, weighted median, weighted mode (Hartwig et al., 2017), and simple mode methods as supplementary analyses.
2.6 Sensitivity analysis
We conducted several sensitivity analyses to assess the robustness of our results. Cochran’s Q statistics were employed to assess the heterogeneity of the IVs. A p-value exceeding 0.05 suggested a lack of significant heterogeneity (Bowden and Holmes, 2019). We utilized the MR-Egger method to quantify the presence of pleiotropy in the IVs. A p > 0.05 for the MR-Egger intercept indicated the absence of horizontal pleiotropy (Burgess and Thompson, 2017). Acknowledging that MR-Egger regression may exhibit reduced accuracy and statistical power, we also implemented the MR pleiotropy residual sum and outlier (MR-PRESSO) method to identify any outliers that could suggest pleiotropic biases. A global test p > 0.05 in MR-PRESSO indicated no evidence of horizontal pleiotropy (Verbanck et al., 2018). The leave-one-out sensitivity analyses were conducted to assess the stability of results by excluding a single SNP each time.
3 Results
3.1 Instrumental variables selection
Initially, a total of 14,587 SNPs were identified as IVs from a GWAS focusing on the human gut microbiota, encompassing 211 bacterial traits categorized into five biological classifications: phylum, class, order, family, and genus (p < 1 × 10−5). To ensure the independence of the selected SNPs from the outcomes of interest (LEDVT and LEDVT combined with PE), we removed those SNPs with LD effects, yielding a final set of 2,213 SNPs as IVs. Comprehensive data regarding these SNPs, including the effect allele, alternative allele, beta coefficient, standard error (SE), and p-value, were systematically compiled for subsequent analysis.
3.2 Two-sample MR analysis
3.2.1 Lower extremity deep vein thrombosis
The results of IVW analyses suggested that the genera Butyricicoccus [odds ratio (OR) = 0.9978, 95% confidence interval (CI), 0.9959–0.9996, p = 0.0193], Haemophilus (OR = 0.9979, 95% CI, 0.9965–0.9993, p = 0.0039), Eubacterium nodatum (OR = 0.9988, 95% CI, 0.9979–0.9997, p = 0.0094), and Defluviitaleaceae UCG011 (OR = 0.9985, 95% CI, 0.9969–1.0000, p = 0.0468) were protective factors for LEDVT, while the genera Clostridium innocuum (OR = 1.0016, 95% CI, 1.0004–1.0028, p = 0.0110), unknowngenus (OR = 1.0017, 95% CI, 1.0003–1.0030, p = 0.0170), and Erysipelotrichaceae UCG003 (OR = 1.0046, 95% CI, 1.0005–1.0087, p = 0.0221) were risk factors for LEDVT (Figure 2).
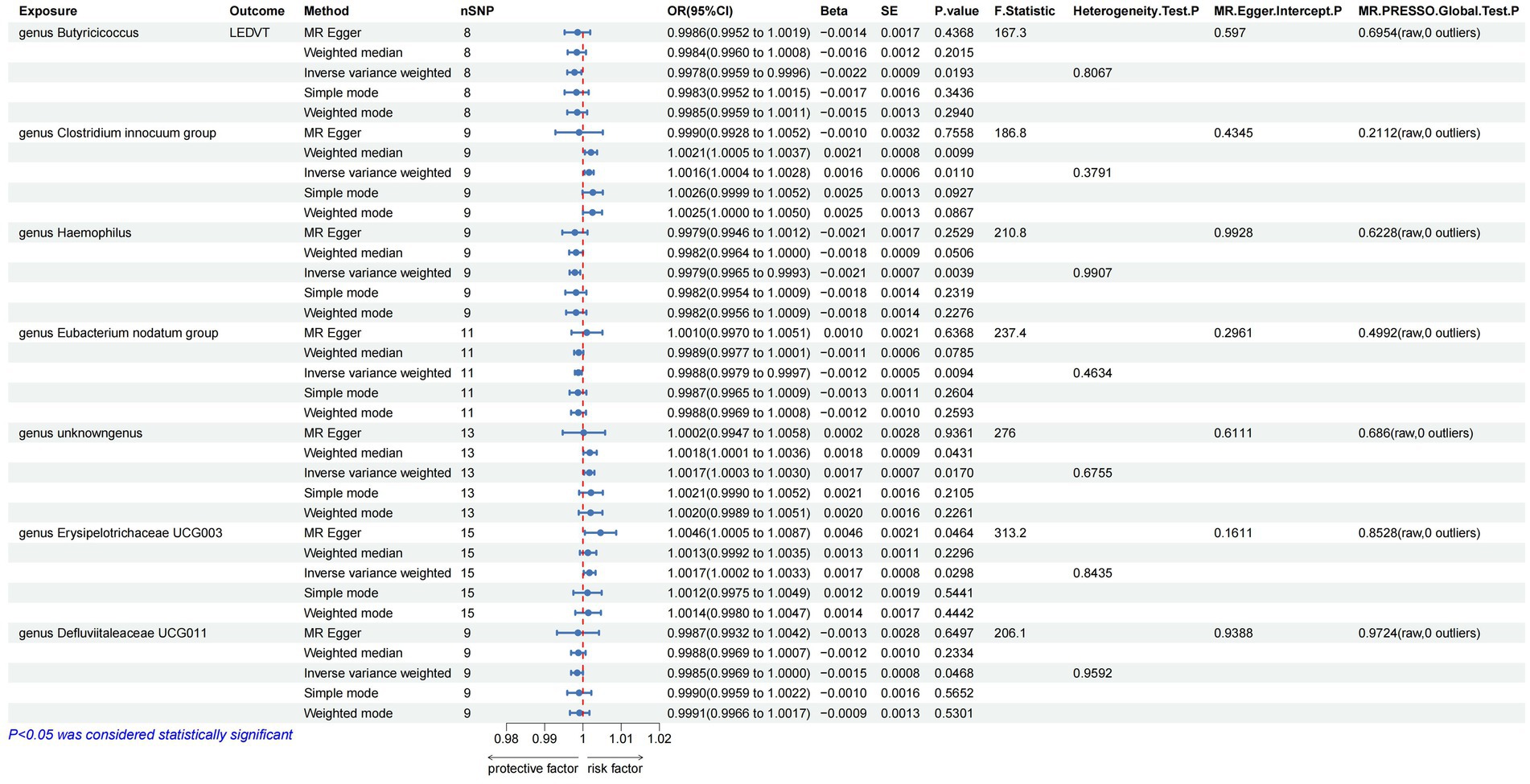
Figure 2. Mendelian randomization results of causal effects between gut microbiota and LEDVT (P < 1 × 10–5). LEDVT, lower extremity deep vein thrombosis; nSNPs, number of single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; SE, standard error.
3.2.2 Lower extremity deep vein thrombosis combined with pulmonary embolism
The results of IVW analyses suggested that the genera Butyricicoccus (OR = 0.9971, 95% CI, 0.9945–0.9998, p = 0.0340) and Bacteroides (OR = 0.9955, 95% CI, 0.9922–0.9988, p = 0.0077), and the phylum Actinobacteria (OR = 0.9974, 95% CI, 0.9949–0.9998, p = 0.0369) were negatively correlated with LEDVT combined with PE risk, while the genera Clostridium innocuum (OR = 1.0019, 95% CI, 1.0002–1.0036, p = 0.0250) and Coprococcus1 (OR = 1.0029, 95% CI, 1.0004–1.0055, p = 0.0216) were positively correlated with LEDVT combined with PE risk (Figure 3).
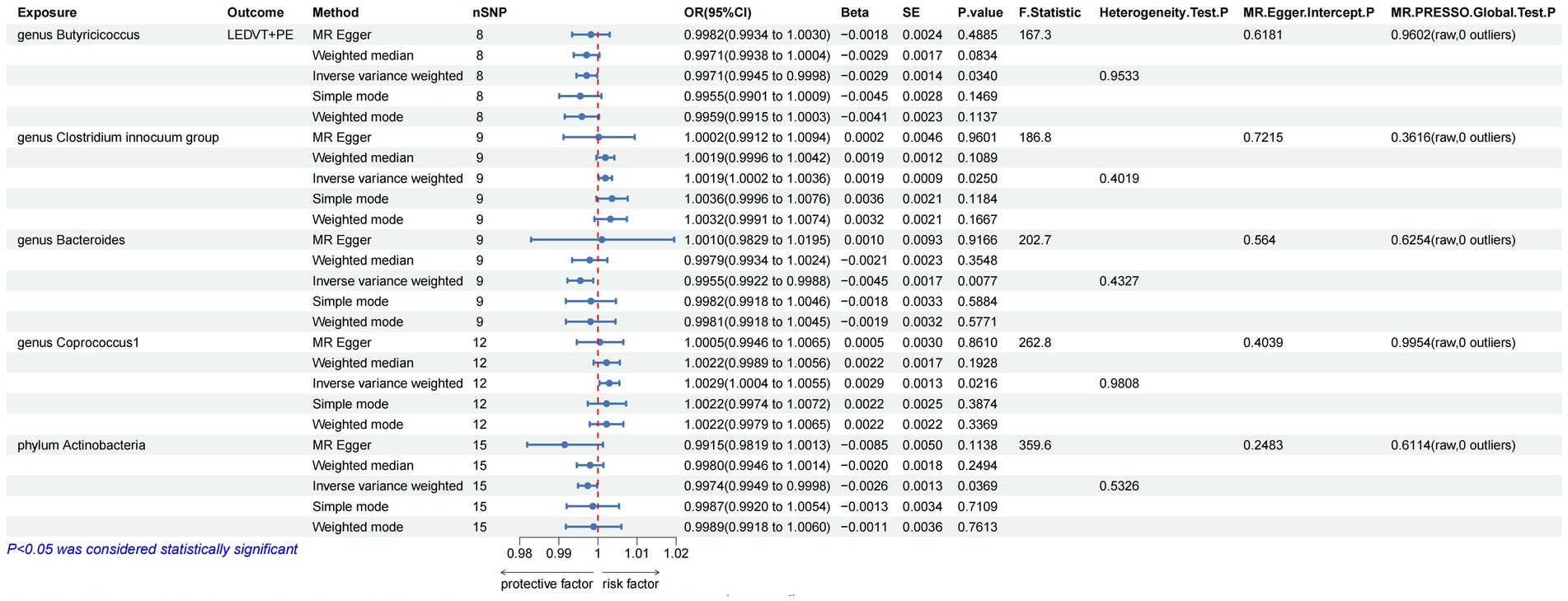
Figure 3. Mendelian randomization results of causal effects between gut microbiota and LEDVT + PE (P < 1 × 10−5). LEDVT, lower extremity deep vein thrombosis; PE, pulmonary embolism; nSNPs, number of single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; SE, standard error.
After the two sets of results were intersected using a Venn plot (Figure 4), it was found that the genus Butyricicoccus reduced the risk of both LEDVT and LEDVT combined with PE (Figure 5), while the genus Clostridium innocuum increased the risk of both conditions (Figure 6).
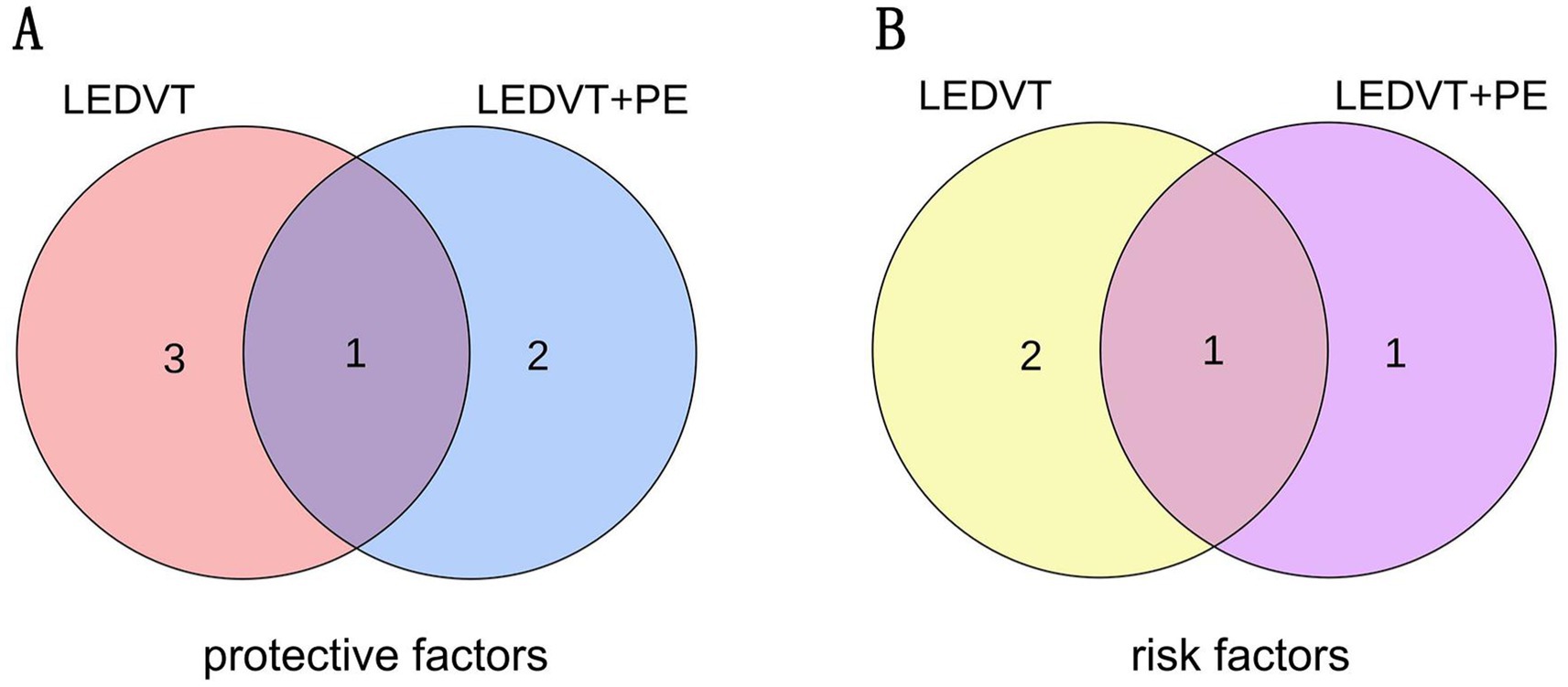
Figure 4. (A) The intersection of protective gut microbiota in LEDVT and LEDVT + PE GWAS. (B) The intersection of risk gut microbiota in LEDVT and LEDVT + PE GWAS. LEDVT, lower extremity deep vein thrombosis; PE, pulmonary embolism; GWAS, genome wide association study.
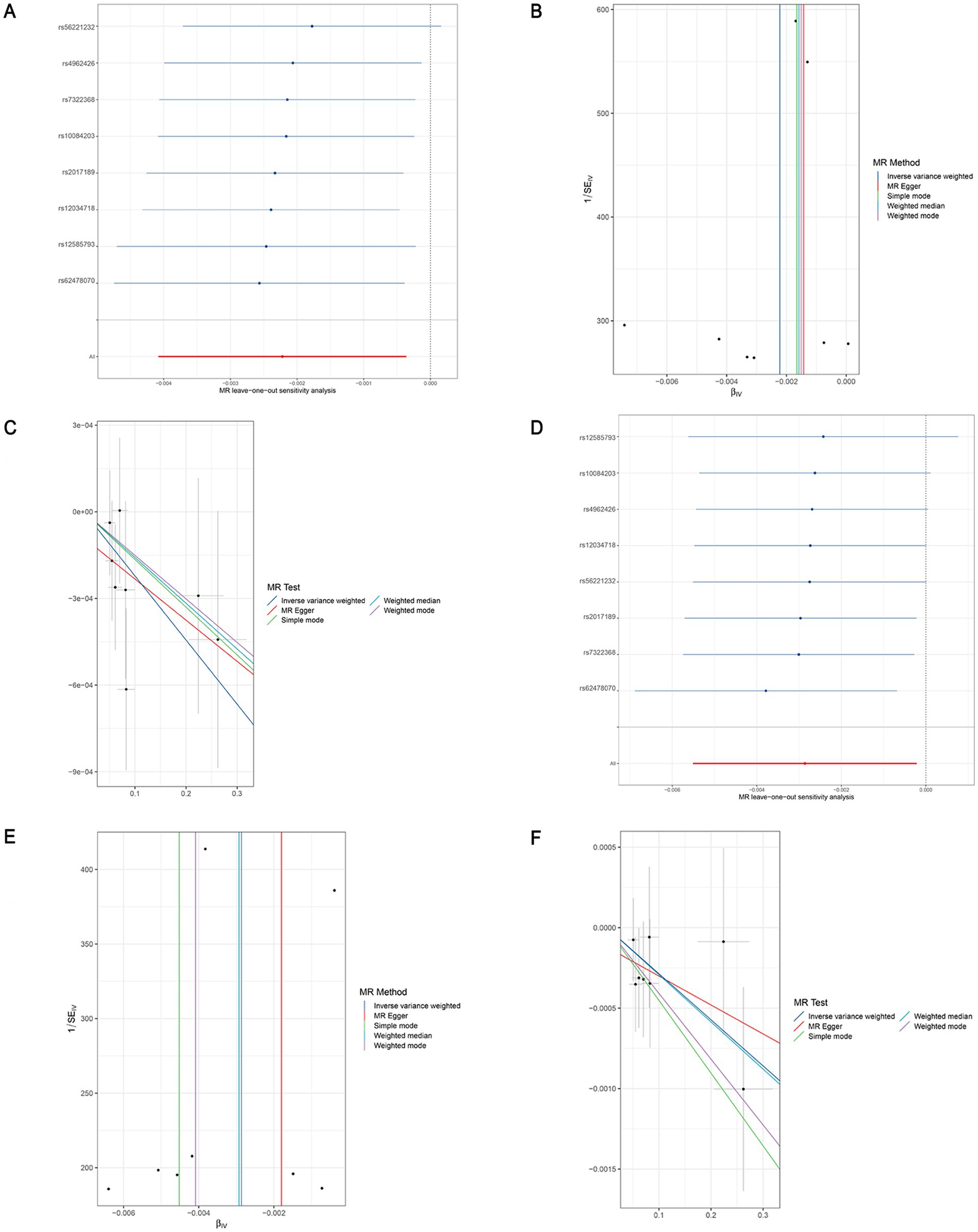
Figure 5. Leave-one-out sensitivity analyses (A), funnel plot (B), and scatter plot (C) of the causal effect of genus Butyricicoccus on LEDVT risk. Leave-one-out sensitivity analyses (D), funnel plot (E), and scatter plot (F) of the causal effect of genus Butyricicoccus on LEDVT + PE risk. LEDVT, lower extremity deep vein thrombosis; PE, pulmonary embolism.
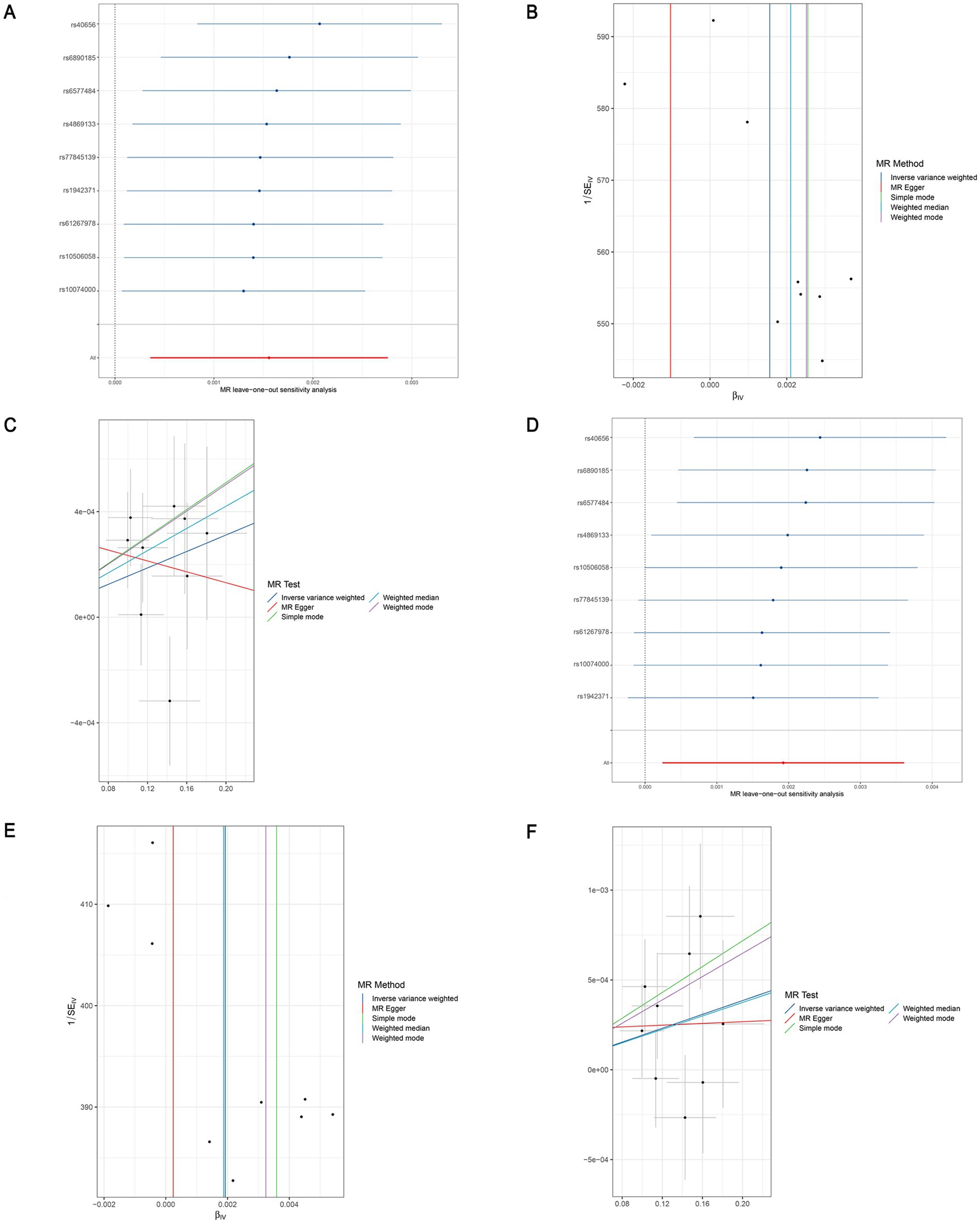
Figure 6. Leave-one-out sensitivity analyses (A), funnel plot (B), and scatter plot (C) of the causal effect of genus Clostridium innocuum group on LEDVT risk. Leave-one-out sensitivity analyses (D), funnel plot (E), and scatter plot (F) of the causal effect of genus Clostridium innocuum group on LEDVT + PE risk. LEDVT, lower extremity deep vein thrombosis; PE, pulmonary embolism.
3.3 Sensitivity analysis
In the analysis of causal associations, the F-statistics for the IVs exhibited a range from 167.331 to 359.589, indicating that weak IV bias was effectively mitigated (Figures 2, 3). The outcomes of Cochran’s IVW Q test indicated no significant heterogeneity among these IVs (p > 0.05) (Figures 2, 3). MR-Egger intercept test yielded a p > 0.05, suggesting the absence of horizontal pleiotropy (Figures 2, 3). Additionally, the MR-PRESSO global test confirmed the validity of the findings (Figures 2, 3). The leave-one-out analysis revealed that no individual SNP significantly influenced the MR estimation results (Supplementary Figure S1). Furthermore, the funnel plots (Supplementary Figure S2) demonstrated that all estimates adhered to the underlying assumptions, thereby reinforcing the credibility of the results.
3.4 Reverse MR
We performed MR Analysis to determine whether LEDVT or LEDVT combined with PE affects specific gut microbiota. We selected SNPs that were strongly associated with LEDVT or LEDVT combined with PE (p < 1 × 10−5, r2 = 0.001, kb = 10,000). The results of the Egger regression showed that there was no evidence of horizontal pleiotropy. Furthermore, the Cochran’s Q test indicated that there was no heterogeneity in the replication cohort, and all selected SNPs acted as strong IVs with F-statistics >10. The findings from the reverse MR analysis did not support causal relationships between the LEDVT or LEDVT combined with PE traits and the gut microbiota discussed earlier (Supplementary Table S1).
4 Discussion
In this study, we performed a two-sample MR analysis to evaluate the causal association between gut microbiota and LEDVT, as well as LEDVT combined with PE. Our findings revealed that certain genera, such as Butyricicoccus, Haemophilus, Eubacterium nodatum, and Defluviitaleaceae UCG011, were associated with a decreased risk of LEDVT. Conversely, genera like Clostridium innocuum, unknowngenus, and Erysipelotrichaceae UCG003 were identified as risk factors for LEDVT. Furthermore, genera including Butyricicoccus, Bacteroides, and phylum Actinobacteria showed a negative correlation with the risk of LEDVT combined with PE, while Clostridium innocuum and Coprococcus1 were positively correlated with the risk of LEDVT combined with PE. To enhance the accuracy of the results, we intersected both sets of findings. Ultimately, our analysis indicated that the genus Butyricicoccus reduced the risk of both LEDVT and LEDVT combined with PE, whereas the genus Clostridium innocuum increased the risk of both conditions.
Among the gut microbiota that reduce the risk of LEDVT, the genus Haemophilus has been reported to be negatively associated with Multiple Sclerosis (Schepici et al., 2019) and decreased in patients with rheumatoid arthritis (Zhang et al., 2015), whereas a negative association with type 2 diabetes was found in a Systematic Review (Letchumanan et al., 2022). In an animal experiment, Heat-treated adzuki bean protein hydrolysates (APH) increased the genus Eubacterium nodatum to reduce cholesterol in mice (Zhao et al., 2022). In another report, the genus Defluviitaleaceae UCG011 was negatively associated with obesity in rats fed a high-fat diet (Zhang et al., 2021). Inflammation, diabetes, and hyperlipidemia are widely acknowledged as risk factors for LEDVT. Accordingly, we hypothesized that the reduction of LEDVT by these bacteria may be related to them. Considering that the genus Erysipelotrichaceae UCG003 has been linked to a heightened risk of inflammatory bowel disease in a previous study, it is plausible to suggest that it may also play a role in elevating the risk of LEDVT through the promotion of inflammatory processes.
Research reports indicate that in cases of weight loss due to obesity, the relative abundance of the genus Bacteroides tends to increase. Furthermore, when transferring the microbial communities of obese mice into germ-free mice, mice that receive the microbiota from obese mice gain more fat than those that receive microbiota from lean mice (Turnbaugh et al., 2006). It has been observed that Bacteroides can inhibit atherosclerosis by reducing the levels of LPS (Yoshida et al., 2018). The reduction of LPS, in turn, inhibits inflammation, lowers blood coagulation, and reduces platelet activity (Violi et al., 2023). A decrease in the phylum Actinobacteria has been associated with increased intestinal permeability, which allows LPS to enter the bloodstream. This leads to the activation of the immune system and the maintenance of chronic inflammatory states such as insulin resistance, diabetes, and liver disease (Binda et al., 2018). Moreover, the genus Coprococcus1 can increase the risk of inflammatory bowel disease (Liu et al., 2022), and is related to high cholesterol (Morales et al., 2023), which increases the risk of obesity, which may account for the increased risk of LEDVT and PE (Wang et al., 2021).
In our study, the genus Butyricicoccus, mainly refers to the Butyricicoccus pullicaecorum, it is famous for producing butyric acid. It has been established that the genus Butyricicoccus is a gram-positive bacterium that is anaerobic and produces butyrate, found in the feces of healthy individuals. While no direct causal relationship has been established between Butyricicoccus and thrombosis, there are several reports indicating a negative correlation between Butyricicoccus and obesity, blood lipids (Zeng et al., 2019), and diabetes (Han et al., 2022). These factors suggest that Butyricicoccus may reduce the occurrence of coronary heart disease (Sun et al., 2022) and decrease liver injury by reducing the levels of LPS and inflammatory response (Li et al., 2023), which are high-risk factors for thrombosis patients, leading to the hypothesis that Butyricicoccus may reduce the likelihood of thrombus formation. The genus Clostridium innocuum is an anaerobic, Gram-positive, spore-forming bacterium first identified as a new species of the genus Clostridium by Smith and King in 1962 from a patient with an appendiceal abscess (Smith and King, 1962). Interestingly, the genus Clostridium innocuum has been positively associated with inflammatory bowel disease and increases the risk and prognosis of this patient population (Lunken et al., 2021). Additionally, it has been demonstrated to lead to severe infections, including endocarditis, osteomyelitis, peritonitis, and bacteremia (Cobo et al., 2023), and may promote adipogenesis, which may cause thrombosis (Ha et al., 2020). Nevertheless, further randomized controlled trials are needed to confirm these findings. In this study, the genera Butyricicoccus and Clostridium innocuum appeared in the two sets of GWAS data results, highlighting the accuracy and validity of this method, which is worthy of further verification and research.
The sample of our study was drawn from a large-scale multicenter database covering individuals of different ages, genders, and health status, which provides some support for the generality of the findings. However, it is important to note that the sample mainly comes from the specific geographical area and population, and other areas people may have different characteristics of the microbial community. Therefore, although our study had high internal validity, it should be taken with caution when generalized to other populations, and further validation in different contexts is needed.
In future studies, we will plan to conduct similar studies in different geographic regions and populations to verify the generalizability and applicability of our findings in other contexts. In addition, it is necessary to further study the biological mechanism between microbial genera and LEDVT and LEDVT combined with PE, and to use molecular biology and metabolomics technology to explore how microbial metabolites affect host physiological functions and cause thrombosis. Further, intervention studies can explore the potential of microbial genera in the prevention and treatment of VTE, such as evaluating the effect of dietary modification or probiotic intervention, and ultimately provide new strategies for clinical practice.
Nevertheless, some limitations of this study should be considered. First, in our GWAS data, all study populations were of European ancestry. Nonetheless, it should be borne in mind that disease patterns may differ between different ancestors, highlighting the need to interpret our conclusions with caution when extending our findings to other populations. Secondly, the analyses exhibited some heterogeneity, which can be attributed to the utilization of GWAS data for investigating potential nonlinear relationships and variations in age, health status, sex, and different layering effects. This heterogeneity could impact the results. Thirdly, it is important to note that potential pleiotropy cannot be completely ruled out, which may introduce biased estimates of causal effects. Fourthly, the low power in this study indicates that this may limit the statistical power of the study and suggests that future studies need larger sample sizes or stronger effect sizes to validate your study findings. Lastly, the analysis of bacterial taxa was limited to the genus level and did not consider more specialized levels, such as species or strain. By utilizing more advanced shotgun metagenomic sequencing in microbiota GWAS, more specific and accurate results can be obtained.
5 Conclusion
To summarize, our study revealed four positive causal directions and three negative causal directions with LEDVT, as well as three positive causal directions and two negative causal directions with LEDVT combined with PE. Notably, both the genera Butyricicoccus and Clostridium innocuum were present in these results, suggesting a closer association with the disease. These findings offer a new avenue for future research on the disease, which could have implications for its prediction, prevention, and treatment.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found at: https://mibiogen.gcc.rug.nl/; https://gwas.mrcieu.ac.uk/datasets/ukb-d-I9_DVTANDPULM/; https://gwas.mrcieu.ac.uk/datasets/ukb-d-I9_PHLETHROMBDVTLOW/.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
QX: Writing – original draft, Investigation, Formal analysis, Data curation, Conceptualization. JF: Writing – review & editing, Investigation, Formal analysis. YW: Writing – review & editing, Investigation, Formal analysis. DL: Writing – review & editing, Supervision. BX: Writing – original draft, Investigation, Formal analysis, Data curation, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This project was supported by the Project of NINGBO Leading Medical and Health Discipline (Project number: 2022-F21), Ningbo Natural Science Foundation (Project number: 2022J249), and Medical Scientific Research Foundation of Zhejiang Province (Project number: 2023KY1078).
Acknowledgments
We thank the studies or consortiums referenced and included in the present analysis for providing public datasets and managing summary statistics. And we thank Home for Researchers editorial team (www.home-for-researchers.com) for language editing service.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1301737/full#supplementary-material
Footnotes
References
Anderson, F. A., Wheeler, H. B., Goldberg, R. J., Hosmer, D. W., Patwardhan, N. A., Jovanovic, B., et al. (1991). A population-based perspective of the hospital incidence and case-fatality rates of deep vein thrombosis and pulmonary embolism. The Worcester DVT study. Arch. Intern. Med. 151, 933–938. doi: 10.1001/archinte.1991.00400050081016
Binda, C., Lopetuso, L. R., Rizzatti, G., Gibiino, G., Cennamo, V., and Gasbarrini, A. (2018). Actinobacteria: a relevant minority for the maintenance of gut homeostasis. Dig. Liver Dis. 50, 421–428. doi: 10.1016/j.dld.2018.02.012
Bonder, M. J., Kurilshikov, A., Tigchelaar, E. F., Mujagic, Z., Imhann, F., Vila, A. V., et al. (2016). The effect of host genetics on the gut microbiome. Nat. Genet. 48, 1407–1412. doi: 10.1038/ng.3663
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int. J. Epidemiol. 44, 512–525. doi: 10.1093/ije/dyv080
Bowden, J., Davey Smith, G., Haycock, P. C., and Burgess, S. (2016). Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314. doi: 10.1002/gepi.21965
Bowden, J., and Holmes, M. V. (2019). Meta-analysis and Mendelian randomization: a review. Res. Synth. Methods 10, 486–496. doi: 10.1002/jrsm.1346
Burgess, S., Butterworth, A., and Thompson, S. G. (2013). Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665. doi: 10.1002/gepi.21758
Burgess, S., and Thompson, S. G. (2017). Interpreting findings from Mendelian randomization using the MR-egger method. Eur. J. Epidemiol. 32, 377–389. doi: 10.1007/s10654-017-0255-x
Burgess, S., and Thompson, S. G.CRP CHD Genetics Collaboration (2011). Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 40, 755–764. doi: 10.1093/ije/dyr036
Centers for Disease Control and Prevention (CDC) (2012). Venous thromboembolism in adult hospitalizations - United States, 2007-2009. MMWR 61, 401–404.
Cheuk, B. L., Cheung, G. C., and Cheng, S. W. (2004). Epidemiology of venous thromboembolism in a Chinese population. Br. J. Surg. 91, 424–428. doi: 10.1002/bjs.4454
Cobo, F., Pérez-Carrasco, V., Tarriño-León, M., Aguilera-Franco, M., García-Salcedo, J. A., and Navarro-Marí, J. M. (2023). Bacteremia due to Clostridium innocuum: analysis of four cases and literature review. Anaerobe 83:102771. doi: 10.1016/j.anaerobe.2023.102771
Cohen, A. T., Agnelli, G., Anderson, F. A., Arcelus, J. I., Bergqvist, D., Brecht, J. G., et al. (2007). Venous thromboembolism (VTE) in Europe. The number of VTE events and associated morbidity and mortality. Thromb. Haemost. 98, 756–764
Davey Smith, G., and Hemani, G. (2014). Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23, R89–R98. doi: 10.1093/hmg/ddu328
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
Emdin, C. A., Khera, A. V., and Kathiresan, S. (2017). Mendelian randomization. JAMA 318, 1925–1926. doi: 10.1001/jama.2017.17219
Fraser, K., Roy, N. C., Goumidi, L., Verdu, A., Suchon, P., Leal-Valentim, F., et al. (2020). Plasma biomarkers and identification of resilient metabolic disruptions in patients with venous thromboembolism using a metabolic systems approach. Arterioscler. Thromb. Vasc. Biol. 40, 2527–2538. doi: 10.1161/ATVBAHA.120.314480
Grimnes, G., Bhoelan, S., Hindberg, K., Davids, M., Nieuwdorp, M., Mollnes, T. E., et al. (2022). Impact of a vancomycin-induced shift of the gut microbiome in a gram-negative direction on plasma factor VIII: C levels: results from a randomized controlled trial. Thromb. Haemost. 122, 540–551. doi: 10.1055/s-0041-1733906
Ha, C. W. Y., Martin, A., Sepich-Poore, G. D., Shi, B., Wang, Y., Gouin, K., et al. (2020). Translocation of viable gut microbiota to mesenteric adipose drives formation of creeping fat in humans. Cell 183, 666–683.e17. doi: 10.1016/j.cell.2020.09.009
Han, S., Chen, M., Cheng, P., Zhang, Z., Lu, Y., Xu, Y., et al. (2022). A systematic review and meta-analysis of gut microbiota in diabetic kidney disease: comparisons with diabetes mellitus, non-diabetic kidney disease, and healthy individuals. Front. Endocrinol. 13:1018093. doi: 10.3389/fendo.2022.1018093
Hartwig, F. P., Davey Smith, G., and Bowden, J. (2017). Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998. doi: 10.1093/ije/dyx102
Heintz-Buschart, A., and Wilmes, P. (2018). Human gut microbiome: function matters. Trends Microbiol. 26, 563–574. doi: 10.1016/j.tim.2017.11.002
Hemani, G., Tilling, K., and Davey Smith, G. (2017). Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 13:e1007081. doi: 10.1371/journal.pgen.1007081
Jäckel, S., Kiouptsi, K., Lillich, M., Hendrikx, T., Khandagale, A., Kollar, B., et al. (2017). Gut microbiota regulate hepatic von Willebrand factor synthesis and arterial thrombus formation via toll-like receptor-2. Blood 130, 542–553. doi: 10.1182/blood-2016-11-754416
Jang, M. J., Bang, S. M., and Oh, D. (2011). Incidence of venous thromboembolism in Korea: from the health insurance review and assessment service database. J. Thromb. Haemost. 9, 85–91. doi: 10.1111/j.1538-7836.2010.04108.x
Kamat, M. A., Blackshaw, J. A., Young, R., Surendran, P., Burgess, S., Danesh, J., et al. (2019). PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 35, 4851–4853. doi: 10.1093/bioinformatics/btz469
Konstantinides, S. V., Meyer, G., Becattini, C., Bueno, H., Geersing, G. J., Harjola, V. P., et al. (2020). 2019 ESC guidelines for the diagnosis and management of acute pulmonary embolism developed in collaboration with the European Respiratory Society (ERS). Eur. Heart J. 41, 543–603. doi: 10.1093/eurheartj/ehz405
Kruit, W. H. J., De Boer, A. C., Sing, A. K., and Van Roon, F. (1991). The significance of venography in the management of patients with clinically suspected pulmonary embolism. J. Intern. Med. 230, 333–339. doi: 10.1111/j.1365-2796.1991.tb00453.x
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165. doi: 10.1038/s41588-020-00763-1
Letchumanan, G., Abdullah, N., Marlini, M., Baharom, N., Lawley, B., Omar, M. R., et al. (2022). Gut microbiota composition in prediabetes and newly diagnosed type 2 diabetes: a systematic review of observational studies. Front. Cell. Infect. Microbiol. 12:943427. doi: 10.3389/fcimb.2022.943427
Li, Q., Liu, W., Zhang, H., Liu, R., Hou, H., Luo, Q., et al. (2023). ɑ-D-1,3-glucan from Radix Puerariae thomsonii improves NAFLD by regulating the intestinal flora and metabolites. Carbohydr. Polym. 299:120197. doi: 10.1016/j.carbpol.2022.120197
Liu, B., Ye, D., Yang, H., Song, J., Sun, X., Mao, Y., et al. (2022). Two-sample Mendelian randomization analysis investigates causal associations between gut microbial genera and inflammatory bowel disease, and specificity causal associations in ulcerative colitis or Crohn's disease. Front. Immunol. 13:921546. doi: 10.3389/fimmu.2022.921546
Lunken, G. R., Tsai, K., Schick, A., Lisko, D. J., Cook, L., Vallance, B. A., et al. (2021). Prebiotic enriched exclusive enteral nutrition suppresses colitis via gut microbiome modulation and expansion of anti-inflammatory T cells in a mouse model of colitis. Cell. Mol. Gastroenterol. Hepatol. 12, 1251–1266. doi: 10.1016/j.jcmgh.2021.06.011
MiBioGen Consortium. (2022). MiBioGen. Available at: https://mibiogen.gcc.rug.nl/ (Accessed September 16, 2022).
Morales, C., Arias-Carrasco, R., Maracaja-Coutinho, V., Seron, P., Lanas, F., Salazar, L. A., et al. (2023). Differences in bacterial small RNAs in stool samples from hypercholesterolemic and normocholesterolemic subjects. Int. J. Mol. Sci. 24:7213. doi: 10.3390/ijms24087213
Morange, P. E., Suchon, P., and Trégouët, D. A. (2015). Genetics of venous thrombosis: update in 2015. Thromb. Haemost. 114, 910–919. doi: 10.1160/TH15-05-0410
Nordström, M., Lindblad, B., Bergqvist, D., and Kjellström, T. (1992). A prospective study of the incidence of deep-vein thrombosis within a defined urban population. J. Intern. Med. 232, 155–160. doi: 10.1111/j.1365-2796.1992.tb00565.x
Park, B., Messina, L., Dargon, P., Huang, W., Ciocca, R., and Anderson, F. A. (2009). Recent trends in clinical outcomes and resource utilization for pulmonary embolism in the United States findings from the nationwide inpatient sample. Chest 136, 983–990. doi: 10.1378/chest.08-2258
Raskob, G. E., Angchaisuksiri, P., Blanco, A. N., Gallus, A., Hunt, B. J., Hylek, E. M., et al. (2014). Thrombosis. Arterioscler. Thromb. Vasc. Biol. 34, 2363–2371. doi: 10.1161/ATVBAHA.114.304488
Sanna, S., van Zuydam, N. R., Mahajan, A., Kurilshikov, A., Vich Vila, A., Võsa, U., et al. (2019). Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 51, 600–605. doi: 10.1038/s41588-019-0350-x
Schepici, G., Silvestro, S., Bramanti, P., and Mazzon, E. (2019). The gut microbiota in multiple sclerosis: an overview of clinical trials. Cell Transplant. 28, 1507–1527. doi: 10.1177/0963689719873890
Simone, B., de Stefano, V., Leoncini, E., Zacho, J., Martinelli, I., Emmerich, J., et al. (2013). Risk of venous thromboembolism associated with single and combined effects of factor V Leiden, prothrombin 20210A and methylenetethraydrofolate reductase C677T: a metaanalysis involving over 11,000 cases and 21,000 controls. Eur. J. Epidemiol. 28, 621–647. doi: 10.1007/s10654-013-9825-8
Smith, L. D., and King, E. (1962). Clostridium innocuum, SP. N., a spore-forming anaerobe isolated from human infections. J. Bacteriol. 83, 938–939. doi: 10.1128/jb.83.4.938-939.1962
Stubbs, M. J., Mouyis, M., and Thomas, M. (2018). Deep vein thrombosis. BMJ 360:k351. doi: 10.1136/bmj.k351
Sudlow, C., Gallacher, J., Allen, N., Beral, V., Burton, P., Danesh, J., et al. (2015). UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12:e1001779. doi: 10.1371/journal.pmed.1001779
Sun, B., Ma, T., Li, Y., Yang, N., Li, B., Zhou, X., et al. (2022). Bifidobacterium lactis Probio-M8 adjuvant treatment confers added benefits to patients with coronary artery disease via target modulation of the gut-heart/-brain axes. mSystems. 7:e0010022. doi: 10.1128/msystems.00100-22
Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., Knight, R., and Gordon, J. I. (2007). The human microbiome project. Nature 449, 804–810. doi: 10.1038/nature06244
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Verbanck, M., Chen, C. Y., Neale, B., and do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698. doi: 10.1038/s41588-018-0099-7
Violi, F., Pignatelli, P., Castellani, V., Carnevale, R., and Cammisotto, V. (2023). Gut dysbiosis, endotoxemia and clotting activation: a dangerous trio for portal vein thrombosis in cirrhosis. Blood Rev. 57:100998. doi: 10.1016/j.blre.2022.100998
Walker, V. M., Davies, N. M., Hemani, G., Zheng, J., Haycock, P. C., Gaunt, T. R., et al. (2019). Using the MR-base platform to investigate risk factors and drug targets for thousands of phenotypes. Wellcome Open Res. 4:113. doi: 10.12688/wellcomeopenres.15334.2
Wang, Z., Usyk, M., Vázquez-Baeza, Y., Chen, G. C., Isasi, C. R., Williams-Nguyen, J. S., et al. (2021). Microbial co-occurrence complicates associations of gut microbiome with US immigration, dietary intake and obesity. Genome Biol. 22:336. doi: 10.1186/s13059-021-02559-w
Yoshida, N., Emoto, T., Yamashita, T., Watanabe, H., Hayashi, T., Tabata, T., et al. (2018). Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation 138, 2486–2498. doi: 10.1161/CIRCULATIONAHA.118.033714
Zeng, Q., Li, D., He, Y., Li, Y., Yang, Z., Zhao, X., et al. (2019). Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci. Rep. 9:13424. doi: 10.1038/s41598-019-49462-w
Zhang, Z., Chen, H., Huang, J., Li, Z., Kong, C., Mao, Y., et al. (2023). Early administration of vancomycin inhibits pulmonary embolism by remodeling gut microbiota. J. Pers. Med. 13:537. doi: 10.3390/jpm13030537
Zhang, Q., Fan, X. Y., Cao, Y. J., Zheng, T. T., Cheng, W. J., Chen, L. J., et al. (2021). The beneficial effects of Lactobacillus brevis FZU0713-fermented Laminaria japonica on lipid metabolism and intestinal microbiota in hyperlipidemic rats fed with a high-fat diet. Food Funct. 12, 7145–7160. doi: 10.1039/d1fo00218j
Zhang, X., Zhang, D., and Jia, H. (2015). The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21, 895–905. doi: 10.1038/nm.3914
Keywords: gut microbiota, deep vein thrombosis, pulmonary embolism, Mendelian randomization, genetic
Citation: Xu Q, Fang J, Wang Y, Lang D and Xu B (2024) The causal relationship between gut microbiota and lower extremity deep vein thrombosis combined with pulmonary embolism. Front. Microbiol. 15:1301737. doi: 10.3389/fmicb.2024.1301737
Edited by:
Georgia Damoraki, National and Kapodistrian University of Athens, GreeceReviewed by:
Akihiko Oka, Shimane University, JapanJoseph Atia Ayariga, Alabama State University, United States
Copyright © 2024 Xu, Fang, Wang, Lang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Xu, SW51eWFzaGFAMTI2LmNvbQ==; aW51eWFzaGFAenVhYS56anUuZWR1LmNu
†ORCID: Bin Xu, https://orcid.org/0000-0001-8258-4414
 Qiyang Xu1
Qiyang Xu1 Yi Wang
Yi Wang Bin Xu
Bin Xu