
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 January 2024
Sec. Food Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1297220
This article is part of the Research Topic Fermentation of Silage in Cold and Tropical Regions: Effects of Phyllosphere Microbiota View all 6 articles
Jerusalem Artichoke (Helianthus tuberosus L.), an emerging “food and fodder” economic crop on the Qinghai-Tibet Plateau. To tackle problems such as incomplete fermentation and nutrient loss occurring during the low-temperature ensilage of Jerusalem Artichokes in the plateau’s winter, this study inoculated two strains of low-temperature resistant lactic acid bacteria, Lactobacillus plantarum (GN02) and Lactobacillus brevis (XN25), along with their mixed components, into Jerusalem Artichoke silage material. We investigated how low-temperature resistant lactic acid bacteria enhance the quality of low-temperature silage fermentation for Jerusalem Artichokes and clarify its mutual feedback effect with microorganisms. Results indicated that inoculating low-temperature resistant lactic acid bacteria significantly reduces the potential of hydrogen and water-soluble carbohydrates content of silage, while increasing lactic acid and acetic acid levels, reducing propionic acid, and preserving additional dry matter. Inoculating the L. plantarum group during fermentation lowers pH and propionic acid levels, increases lactic acid content, and maintains a dry matter content similar to the original material. Bacterial community diversity exhibited more pronounced changes than fungal diversity, with inoculation having a minor effect on fungal community diversity. Within the bacteria, Lactobacillus remains consistently abundant (>85%) in the inoculated L. plantarum group. At the fungal phylum and genus levels, no significant changes were observed following fermentation, and dominant fungal genera in all groups did not differ significantly from those in the raw material. L. plantarum exhibited a positive correlation with lactic acid and negative correlations with pH and propionic acid. In summary, the inoculation of L. plantarum GN02 facilitated the fermentation process, preserved an acidic silage environment, and ensured high fermentation quality; it is a suitable inoculant for low-temperature silage in the Qinghai-Tibet Plateau.
Qinghai Province is situated in the northeastern region of the Qinghai-Tibet Plateau. The growth of animal husbandry has been significantly impeded by insufficient grass during the cold season and other factors. Planting ‘food and fodder’ cash crops to augment the forage supply is considered an effective approach to address this issue (Xin, 2013; Qi, 2021). Jerusalem Artichoke (JA, Helianthus tuberosus L.), a ‘food and fodder’ cash crop cultivated for its tubers. It exhibits characteristics such as cold tolerance (Zhao et al., 2020), drought resistance (Zhou et al., 2022), salinity resistance (Li et al., 2016; Teng, 2019), disease resistance, and robust stress resilience (Qiu et al., 2018). It thrives in adverse growth conditions, including wasteland, saline-alkali soil, and gravel. The crop boasts a high yield of aboveground stems and leaves, enriched nutritional value, and favorable palatability. The feed value of its stems and leaves surpasses that of Solanum tuberosum and Helianthus annuus, rendering it a superior-quality roughage featured in the China Feed Database1 (Wang et al., 2020a,b). Research has demonstrated that JA tubers, stems, and leaves are abundant in fructans. When used as dietary additives, they can significantly enhance the intestinal microorganisms of ruminants, bolster lactation performance in dairy cows, and lower the occurrence of mastitis in dairy cows (Wang et al., 2021a,b,c).
Silage involves the storage of feed through lactic acid bacteria (LAB) fermentation (Ren et al., 2021; Okoye et al., 2023). The post-fermentation lactic acid, dry matter, and ammonia nitrogen contents of JA all align with the benchmarks for high-quality silage (Monllor et al., 2020). Ambient temperature is an important factor affecting the quality of silage (Zhou et al., 2016). Ordinarily, JA silage undergoes fermentation at room temperature for 40–45 days (Wu et al., 2022). However, the average daily temperature during JA harvesting in Qinghai Province hovers around 10°C, occasionally plummeting to below 0°C at night. The fermentation process is inhibited at low temperatures (Li et al., 2021, 2022). In colder temperatures (<5°C), inoculating with low-temperature resistant LAB can expedite the low-temperature fermentation process, inhibit the proliferation of detrimental microorganisms, and substantially enhance silage quality. Li (2016) employed corn (Zea mays) and alfalfa (Lotuscorniculatus) as silage substrates and inoculated three low-temperature resistant LAB at 15°C. This intervention resulted in reduced pH and ammonia nitrogen content and a noteworthy elevation in silage lactic acid content, and enhanced fermentation. Consequently, the inoculation of low-temperature resistant LAB can enhance the low-temperature ensiling of Jerusalem Artichokes on the Qinghai-Tibet Plateau. Research indicates that particular LAB strains, isolated from silage within distinct environmental contexts, could be optimal for enhancing silage fermentation quality (Ju, 2020; Lin et al., 2022). Initially in this study, we isolated high acid-producing, fast-growing, and low-temperature resistant strains—Lactobacillus plantarum GN02 and Lactobacillus brevis XN25—from JA silage on the Qinghai-Tibet Plateau, utilized for both homotypic and heterotypic LAB fermentation (Wei et al., 2023). Homotypic fermentation LAB rapidly generates lactic acid throughout fermentation, thereby lowering the silage pH and creating an acidic milieu. In contrast, heterotypic fermentation LAB generates both lactic acid and acetic acid during silage fermentation, enhancing silage’s aerobic stability (Xu et al., 2021; Liu et al., 2022). Nonetheless, the current understanding of how the mechanisms and microbial progression of homotypic and heterotypic fermentation by low-temperature resistant LAB influence the low-temperature silage fermentation of JA remains incomplete. Furthermore, effective assessment criteria for JA silage are lacking. The quality of low-temperature silage fermentation is influenced by microbial community structure and abundance. The elevated presence of harmful bacterial species and the inadequate representation of beneficial bacteria during silage, thereby contributing to fermentation failure (Cheng et al., 2021). Previous studies have highlighted the contribution of bacterial community to silage fermentation. However, Li et al. (2021) reported that Saccharomyces and spoilage fungi can impact silage fermentation quality by depleting water-soluble carbohydrates and lactic acid, alongside mycotoxin generation and pH elevation. It is unclear how the response of microbial community of JA silage to the inoculation of low-temperature resistant LAB.
This study utilized JA stems and leaves as experimental materials. They were inoculated with homotypic fermentation and heterotypic fermentation low-temperature resistant LAB during silage preparation in a natural low-temperature environment. The impact of low-temperature resistant LAB on the fermentation quality of JA low-temperature silage was assessed by tracking fermentation parameter changes. Additionally, high-throughput sequencing was employed to unveil the microbial succession throughout the fermentation process, elucidating the dominant species within JA silage and their correlation with fermentation products. This exploration aims to elucidate the regulatory mechanism of low-temperature resistant LAB on JA silage. The findings of this study offer a theoretical foundation for developing JA silage on the Qinghai-Tibet Plateau, providing substantial guidance for the sustainable and stable advancement of animal husbandry in this region.
Test materials were sourced from the Academy of Agriculture and Forestry Sciences at Qinghai University. The JA variety ‘QingYu No. 2’ was selected and cultivated in the experimental field of the Academy of Agriculture and Forestry Sciences of Qinghai University, Xining, Qinghai Province, China. Planting took place on early April 20, 2022, at geographical coordinates E101 °44 ′57.7572 ″, N36 °43 ′8.346 ″, and an elevation of 2266.6 m. On October 21, 2022, the above-ground stems and leaves were crushed to a length of 2 ± 1 cm and utilized as raw materials for JA silage production. The low-temperature resistant LAB employed in the experiment was derived from L. plantarum GN02 and L. brevis XN25, which were initially selected in the laboratory (Wei et al., 2023). The NCBI registration numbers for these strains were OP740787 and OP740791, respectively. Viable bacterial counts were determined using OD values and centrifugation (4,000 rpm, 5 min). The resulting bacterial pellet was resuspended in sterile water to achieve the number of viable bacteria 1 × 108 cfu/mL, creating a bacterial solution for backup. A 500 g sample of crushed JA raw materials was selected at random. The prepared LAB inoculant was manually and uniformly sprayed onto the raw materials using a sterile watering can (Inoculation dose is 1.0 × 106 cfu/g of FM.). After thorough mixing, the mixture was placed in a vacuum-sealed polyethylene plastic bag (280 mm × 350 mm, Zhejiang, China). The bag was vacuum-sealed using a vacuum machine (model 14,891, Deli, Shandong, China), and the mixture was then stored in a natural environment for fermentation. The experiment comprised 4 distinct treatments: CK (sterile water, 5 mL), FD (L. brevis XN25, 1.0 × 106 cfu/g of FM), FZ (L. plantarum GN02, 1.0 × 106 cfu/g of FM), and FZD (L. brevis XN25 and L. plantarum GN02 were mixed, 1.0 × 106 cfu/g of FM). Each treatment was replicated three times and bags were opened for analyses after 7 and 60 days of ensiling. In total, 27 bags were sampled (2 storage periods × 4 treatments × 3 replicates +3 raw materials). Silage samples were collected to assess fermentation parameters and microbial communities.
Raw materials and silage samples are analyzed for pH, dry matter, crude protein, water-soluble carbohydrates, neutral detergent fiber, and acid detergent fiber content. Silage samples additionally necessitate assessing lactic acid, acetic acid, and propionic acid levels. Homogenize 20 g of the sample with 180 mL of distilled water in a high-speed juicer for 30 s. Filter the mixture through 4 layers of medical gauze to collect the filtrate. Centrifuge the filtrate at 5,000 rpm for 10 min. Measure the pH of the filtrate using a glass electrode pH meter (pH S-2F, LEICI, Shanghai, China). Store the filtrate at −20°C for subsequent analysis of lactic acid (LA), acetic acid (AA), propionic acid (PA), and ammonia nitrogen (NH3-N) content. LA, AA, and PA contents were quantified using high-performance liquid chromatography (Li et al., 2018). NH3-N content was extracted using the Plant Ammonia Nitrogen Content Assay Kit (Beijing Boxbio Science & Technology Co., Ltd.) and measured at 570 nm using a microplate reader (EPOCH2, BioTek, Vermont, USA). Another 300 g sample was dried at 65°C in an oven for 48 h until a constant weight was achieved. The dry matter (DM) content was determined using an electronic balance (PTX-FA210S, HuaZhi, Connecticut, USA). The dried sample was ground using a grinder and sieved through a 1 mm screen. The resulting sample was utilized for quantifying crude protein (CP), water-soluble carbohydrates (WSC), neutral detergent fiber (NDF), and acid detergent fiber (ADF) content. CP content was quantified using a Kjeldahl nitrogen analyzer (K9860, Hainon, Shandong, China) following the analytical method outlined by the Society of Chemists (AOAC, 1990). WSC content analysis was conducted using the anthrone reagent colorimetric method (Thomas, 1977). NDF and ADF contents were measured using a fiber analyzer (F800, Hainon, Shandong, China) following the methods described by Van Soest et al. (1991).
Immediately after collection, silage samples were rapidly frozen and stored at −80°C. Bacterial and fungal DNA were extracted from the silage using a MagPure Soil DNA LQ Kit (Yesen, Shanghai, China), in accordance with the manufacturer’s instructions. DNA concentration was quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), while integrity was assessed through agarose gel electrophoresis. PCR amplification targeting the V3-V4 hypervariable region of bacterial 16S rRNA and the ITS1 region of fungal ITS gene was conducted in 25 mL reaction volumes, employing universal primer pairs. For bacteria, the primers were 343F (5’-TACGGRAGGCAGCAG-3′) and 798R (5’-AGGGTATCTAATCCT-3′). For fungi, the primers were ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5’-GCTGCGTTCTTCATCGATGC-3′). The reverse primer included a sample-specific barcode, and both primers were linked to an Illumina sequencing adapter.
Amplicon quality was assessed through gel electrophoresis. PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter Co., USA) and quantified with the Qubit dsDNA assay kit. Subsequently, the concentrations were adjusted for sequencing purposes. Sequencing was conducted on an Illumina NovaSeq6000 using two paired-end read cycles of 250 bases each (Illumina Inc., San Diego, CA; OE Biotech Company, Shanghai, China). The raw sequencing data were in FASTQ format. Subsequently, paired-end reads underwent preprocessing using cutadapt software to identify and remove adapters. Following trimming, the paired-end reads were filtered for low-quality sequences, denoised, merged, and subjected to chimera detection and removal using DADA2 (Callahan et al., 2016) with QIIME2’s default parameters (Bolyen et al., 2019). Finally, the software generated representative reads and an abundance table for ASVs. The representative read for each ASV was chosen utilizing the QIIME 2 package. These representative reads were then annotated and compared to the Silva database Version 138 (16 s rDNA) and Unite (ITS) using q2-feature-classifier with the default parameters. Microbial diversity within the silage samples was assessed using alpha diversity metrics, including the Chao1 index (Chao and Bunge, 2002) and Shannon index (Hill et al., 2003). The Unifrac distance matrix, computed using QIIME software, was employed for unweighted UniFrac principal coordinates analysis (PCoA). Bacterial differential markers were identified using LEfSe (LDA) analysis, employing an LDA Score screening threshold of 4 (Weiss, 2017).
We utilized Excel 2019 for fundamental statistical analysis. Additionally, we employed IBM SPSS 27.0 for two-way analysis of variance, including Duncan’s multiple comparisons, to assess fermentation attributes and chemical parameters. A value of p < 0.05 was considered statistically significant, and a value of p < 0.01 was considered very significant. We analyzed and plotted microbial sequencing data on the Oebiotech Cloud Platform.2
As shown in Table 1, throughout the ensiling fermentation period, pH levels in all groups decreased relative to F0. Significant pH differences were observed among groups during ensiling. After 60 days of ensiling, only FZ silage had pH values lower than 5.0, whereas the pH values of other silages remained at high levels (values for CK and FD silages). LA content exhibited an upward trajectory over time in the CK and FD groups, in contrast to the decreasing trend observed in the FZ and FZD groups, aligning with the pH pattern. Substantial variations in LA content were evident among groups on 7, 60 days. Notably, the FZ group exhibited the highest LA content on both 7, 60 days (26.93 g/kg and 24.09 g/kg FM). In contrast, the CK group had the lowest LA content (0.57 g/kg and 4.08 g/kg FM) on the respective days. Gradual AA content increments were observed across all groups over time. Specifically, on 7, 60 days, the FD group demonstrated notably elevated AA content in comparison to the remaining groups (11.66 g/kg and 16.37 g/kg FM). Conversely, PA content exhibited a gradual decline across all groups over time. On 7, 60 days, the PA content of the CK group exhibited significant elevation in comparison to the other groups, whereas the FZ group demonstrated considerable reduction (2.15 g/kg and 2.17 g/kg FM). The FZD group exhibited significantly higher NH3-N content compared to the others. Interestingly, on 60 days, the FD group demonstrated a marked rise in NH3-N content (17.61 g/kg FM), significantly surpassing the other groups. Two-way analysis of variance indicates noteworthy interactions (p < 0.01) among the fermentation attributes (pH, LA, AA, PA, NH3-N) concerning fermentation period and microbial strains.
As shown in Table 2, throughout the ensiling fermentation period, the DM contents of each group exhibited a progressive decrease. On 7 days, no noteworthy disparities in DM contents were observed among the groups. The CK group displayed a swifter decline during fermentation and exhibited a notably lower level than the other groups by 60 days. The DM content of the FZ group at the 60 days mark (333.17 g/kg) was markedly greater than that of the CK and FZD groups. The WSC content of all groups exhibited a notable decrease over time, and it was significantly lower than that of the initial state (F0). Throughout fermentation, the FD group displayed elevated WSC content (24.83 g/kg, 12.85 g/kg DM) in comparison to the other groups, yet no significant difference was observed between FD and FZ on 60 days. The CP content of all groups experienced a slight increase over time, and the FZD group exhibited significant elevation over the others on 7 days; no significant differences were detected among the groups on 60 days. Compared to the F0 group, the NH3-N content decreased in all groups except for the FZD group on 7 days. The NDF content of all groups saw an increase relative to the F0 state, and the FZ group exhibited both the highest content and significant elevation on 7 days. No significant differences were observed among the groups on 60 days. Analogous to the NDF pattern, the ADF content of all groups showed an increase in relation to the F0 state, with the FZ and FZD groups displaying higher content on 60 days. Intriguingly, on 7, 60 days, no significant differences were found between the CK group and the FZ or FZD groups; all three showed higher values than the FD group. Two-way analysis of variance shows notable disparities (p < 0.01) in DM and WSC chemical parameters among fermentation periods and microbial strains. However, the interaction between them is not statistically significant (p > 0.05), suggesting a lack of interaction between fermentation periods and microbial strains in DM and WSC. On the contrary, a significant interaction (p < 0.01) is observed among the chemical parameters of CP, NDF, and ADF across fermentation periods and microbial strains.
We use the Chao1 and Shannon indices of α-diversity to describe the abundance and diversity of bacterial and fungal populations within groups. The Chao1 index estimates the number of ASVs in the community, while the Shannon index estimates intragroup community diversity. Higher values for both indices suggest greater richness and diversity within intragroup communities. In bacteria (Figures 1A,B), the Chao1 (771.73) and Shannon (6.23) indices of the F0 group were significantly higher than those of the other groups. On 7 days, the Chao1 index of the FZ group (301.24) was significantly higher than that of the FZD group (227.77), while the Chao1 indices of the other groups showed no significant differences. Moreover, the Shannon indices of the CK and FD groups (2.45, 2.64) were significantly higher than that of the FZ group (1.54), and there were no significant differences from the FZD group. These findings indicate that the FZD group exhibited the lowest community richness on 7 days, whereas the FZ group had the lowest community diversity. On 60 days, the Chao1 indices of the FZ (237.31) and FZD (233.11) groups were significantly higher than those of the CK (159.61) and FD (144.87) groups and no significant differences were observed within the CK, FD, and FZ, FZD groups. Additionally, the Shannon index of the FZ group (1.44) was significantly lower than that of the other groups, with no significant differences observed among the other groups. These findings suggest that the CK and FD groups exhibited the lowest community richness on 60 days, while the FZ group had the lowest community diversity. With the progression of fermentation, variations in the Chao1 index were evident within the identical groups on 7, 60 days. Specifically, the Chao1 indices of the CK groups exhibited a substantial decrease over time, whereas those of the FZD groups displayed a significant increase. No noteworthy effects were observed in the other groups. Similarly, the Shannon index indicated no significant distinctions among the identical groups on 7, 60 days. Within fungi (Figures 1C,D), the Chao1 indices of all groups exhibited no noteworthy variances on fermentation 7, 60 days, suggesting minimal discrepancies in fungal community richness during the fermentation phase. Additionally, the Shannon index indicated no significant distinctions among the groups on 7 days, with no notable differences observed between the groups and the F0 group. However, on 60 days, the CK (6.26) and FZD (6.09) groups demonstrated a substantial elevation compared to the FD (5.31) group. Although the FD group did not display significant differences when compared to the groups on 0 and 7 days, this suggests alterations in fungal community diversity during the later stages of fermentation, particularly evident in the CK and FZD groups.
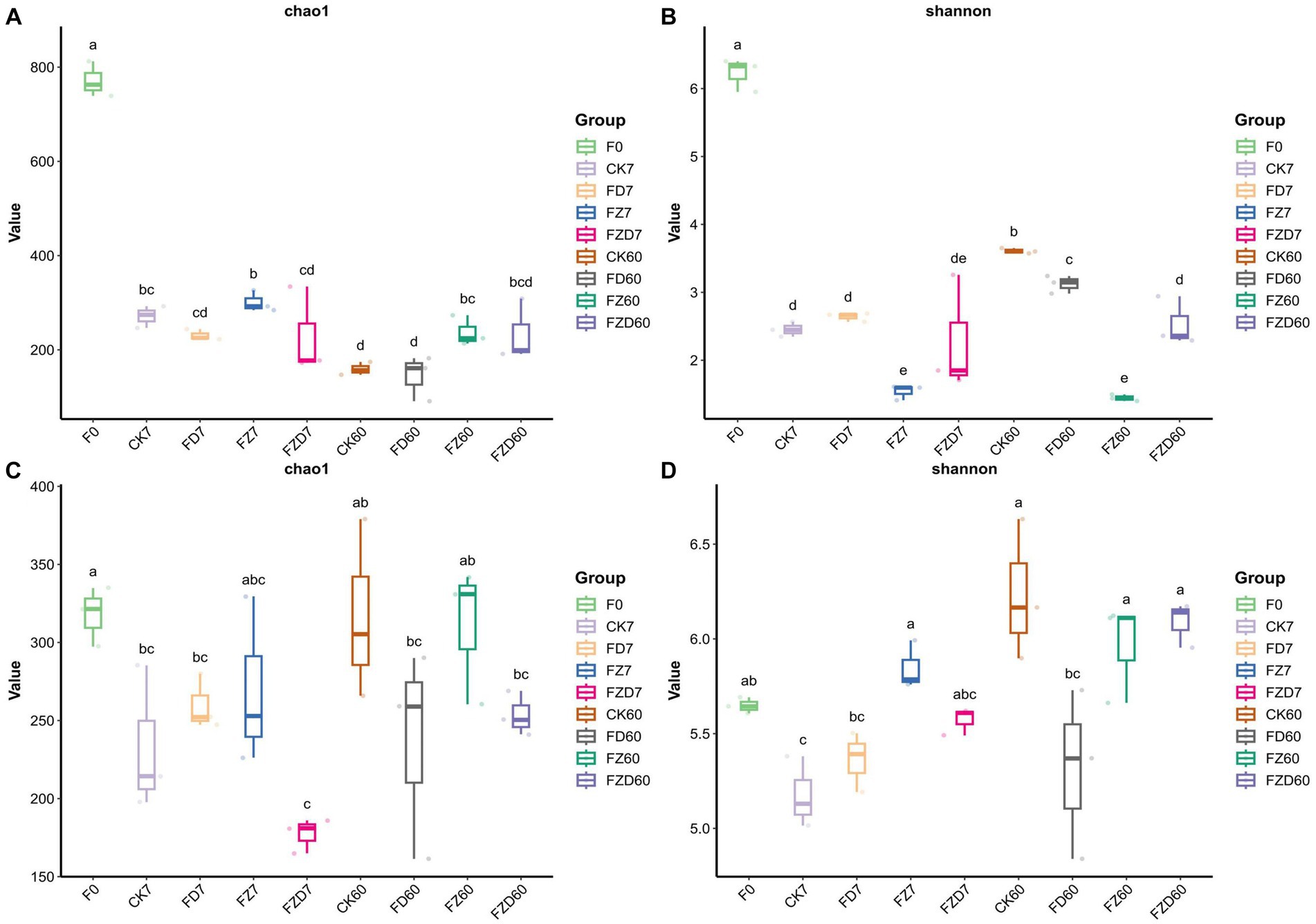
Figure 1. The Chao1 and Shannon indices represent the α-diversity of bacteria (A,B) and fungi (C,D). F0, JA raw material; CK, sterile water; FD, Lactobacillus brevis; FZ, Lactobacillus plantarum; FZD, the combination of L. brevis and L. plantarum; The numbers 7 and 60 correspond to fermentation duration; The use of different lowercase letters signifies statistically significant differences (p < 0.05) among various treatments on either the same or different ensiling days, while treatments sharing the same lowercase letters indicate no statistically significant differences (p > 0.05) between them.
PCoA was performed to analyze the microbial community changes in JA silage during fermentation. Within bacteria (Figure 2A), the F0 group showed substantial separation along the PC2 axis from other groups, while the remaining groups demonstrated dispersed distribution, signifying noteworthy differences between the groups. On 7, 60 days, the CK and FD groups cluster together along the PC1 axis, similar to the clustering of the FZ and FZD groups, emphasizing how the presence of added microbial species significantly contributes to distinct inter-group bacterial variations. Importantly, data for the FZ group at 7, 60 days exhibited substantial overlap. Among fungi (Figure 2B), all groups were segregated along the PC2 axis, and a more pronounced separation was noticeable among the 7 days groups than the 60 days groups. ANOSIM tests utilizing Euclidean distance revealed that differences in bacterial and fungal community structures between groups during fermentation were markedly more substantial than differences within groups (R2 = 0.98, p = 0.001; R2 = 0.99, p = 0.001), affirming the robustness of PCoA clustering. Therefore, it is clear that the introduction of microbial species impacts alterations in the microbial community throughout the fermentation of JA silage.
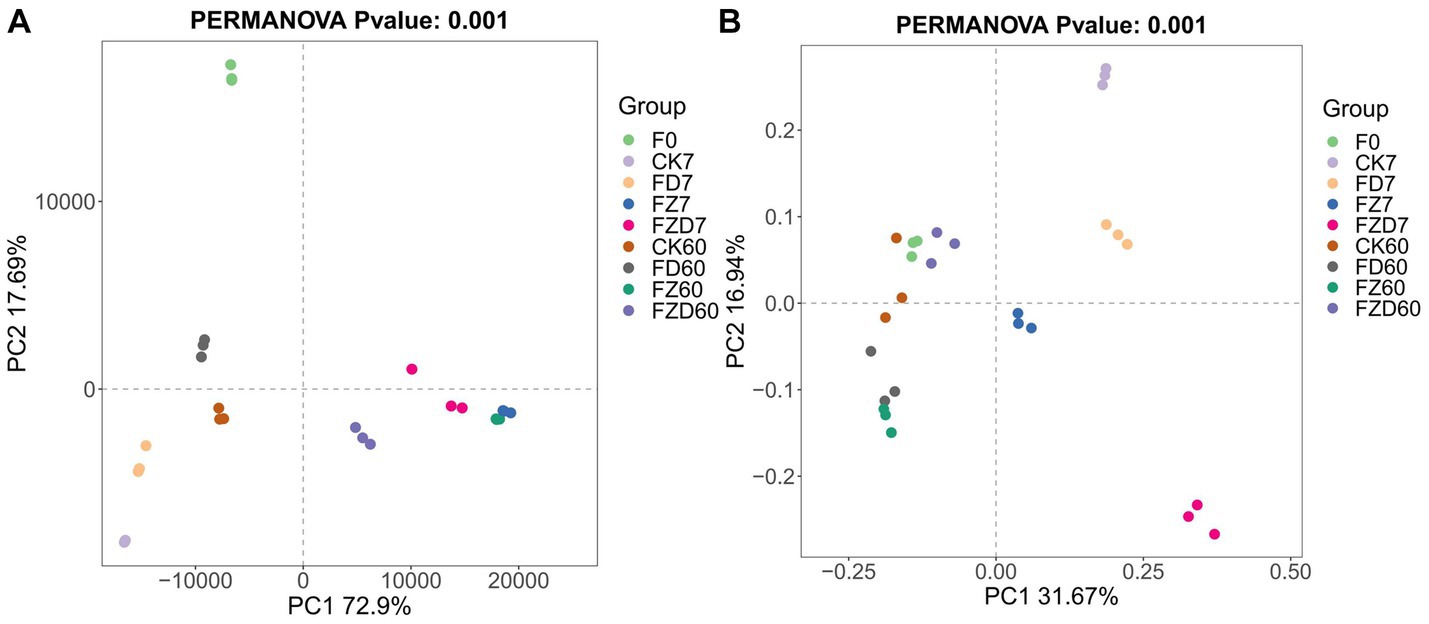
Figure 2. PCoA examines the community structure of bacteria [(A) Euclidean method] and fungi [(B) Bray curtis method]. F0, JA raw material; CK, sterile water; FD, L. brevis; FZ, L. plantarum; FZD, the combination of L. brevis and L. plantarum; The numbers 7 and 60 correspond to fermentation duration; ANOSIM test (p < 0.05).
We conducted high-throughput sequencing on 27 samples utilizing the Illumina MiSeq sequencing platform. Following sequencing, the distribution of raw bacterial reads ranged from 78,045 to 81,711, and after quality control, the distribution of clean tags ranged from 58,290 to 75,142. After eliminating chimeras from the clean tags, valid tags (the data used for analysis) spanned from 44,672 to 70,842. ASV counts per sample varied between 91 and 813. Using 100% sequence similarity, all sequences were classified, yielding a total of 25 phyla, 48 classes, 128 orders, 215 families, 477 genera, and 883 species. Regarding fungi, the distribution of raw reads data spanned from 78,201 to 81,996, clean tags data distribution ranged from 67,322 to 78,183, and valid tags data distribution ranged from 67,322 to 78,064. After classification, ASVs ranged from 152 to 378, resulting in a total of 13 phyla, 33 classes, 72 orders, 162 families, 362 genera, and 508 species.
In bacterial classification at the phylum level (Figure 3A), the species composition of the F0 group distinguishes it from the other groups. The dominant phyla within it are Proteobacteria, Firmicutes, Bacteroidota, and Actinobacteriota, comprising relative abundances of 54.26, 16.59, 12.48, and 8.74%, respectively, contributing to a total community composition of 92.07%. On the 7 days of fermentation, Firmicutes (88.48, 91.37, 91.61, 93.07%) were the predominant phylum in all groups, constituting over 85% of the total. By the 60 days of fermentation, the CK and FD groups were characterized by dominant Firmicutes (64.71, 74.91%) and Proteobacteria (34.24, 23.92%), while the FZ and FZD groups exhibited Firmicutes (95.22, 94.38%) as the prevailing phylum. The evolution of ensiling fermentation reveals shifts in the species composition at the phylum level within each group compared to 0 day. On 7 days, Firmicutes’ relative abundance markedly surged in all groups, establishing it as the prevailing phylum, while the relative abundances of Proteobacteria, Bacteroidota, and Actinobacteriota conspicuously declined, leading to an overall relative abundance falling below 3%. On 60 days, the CK and FD groups exhibited a notable increase in the relative abundance of Proteobacteria, whereas the relative abundance of Firmicutes decreased by 23.76 and 16.46%, respectively. In the FZ and FZD groups, Firmicutes showed a slight increase, maintaining its dominant status at the phylum level.
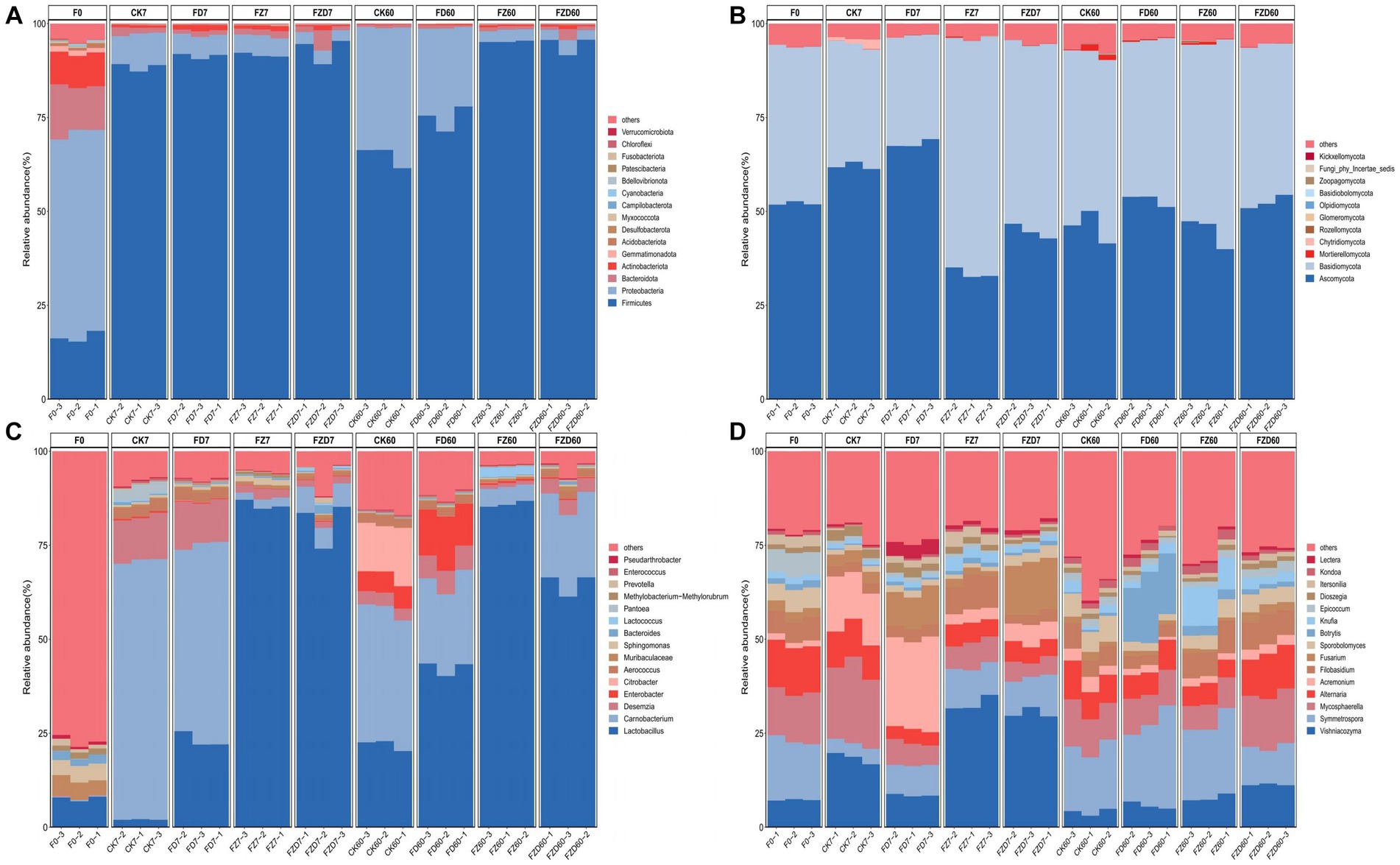
Figure 3. During silage fermentation, the relative abundance of bacteria (A,C) and fungi (B,D) is assessed at both the phylum and genus levels. F0, JA raw material; CK, sterile water; FD, L. brevis; FZ, L. plantarum; FZD, the combination of L. brevis and L. plantarum; The numbers 7 and 60 correspond to fermentation duration.
At the fungal phylum level (Figure 3B), in contrast to bacteria, the dominant phyla were Ascomycota and Basidiomycota across all groups throughout each period, comprising over 90% of the total community. As fermentation progressed, the relative abundances of the two phyla experienced alterations. In comparison to the F0 group, the CK and FD groups showed an initial increase followed by a decrease in the relative abundance of Ascomycota at both 7, 60 days, whereas the relative abundance of Basidiomycota exhibited an initial decrease followed by an increase. Conversely, the FZ and FZD groups demonstrated trends opposite to those of the CK and FD groups in terms of the relative abundance changes of Ascomycota and Basidiomycota. In both time periods, the FD group exhibited the highest relative abundance of Ascomycota (68.03, 53.07%), whereas the FZ group showed the highest relative abundance of Basidiomycota (62.59, 50.22%).
Significant differences in bacterial and fungal genus composition exist among the groups at the genus level. In bacteria (Figure 3C), the relatively abundant genera are Lactobacillus (7.66%), Muribaculaceae (4.68%), and Sphingomonas (4.28%). By the 7 days of fermentation, dominant genera had begun to emerge in all groups. In the CK and FD groups, the dominant genera included Carnobacterium (68.82, 51.96%), Desemzia (11.63, 11.52%), and Lactobacillus (2.05, 23.18%), while Lactobacillus was comparatively less abundant in the CK group. Conversely, in the FZ and FZD groups, Lactobacillus emerged as the dominant genus (85.70, 81.03%). On 60 days, alterations in dominant genera were evident across all groups. In the CK group, the relative abundance of Carnobacterium (35.78%) and Desemzia (3.41%) decreased, while Lactobacillus (21.90%) increased. This period also saw the emergence of Citrobacter (13.52%) and Enterobacter (5.61%). In the FD group, the relative abundance of Lactobacillus (42.40%) increased, while Carnobacterium (23.17%) and Desemzia (6.24%) declined. Enterobacter (12.65%) began to appear. The FZ group maintained Lactobacillus (85.94%) as the dominant genus, experiencing minimal changes in relative abundance. Conversely, the FZD group witnessed a decrease in Lactobacillus (64.84%) alongside an increase in Carnobacterium (22.26%) and a slight presence of Desemzia (3.89%).
At the genus level of fungi (Figure 3D), in the F0 group, the relatively abundant genera are Symmetrospora (15.77%), Mycosphaerella (12.96%), Alternaria (12.51%), and Vishniacozyma (7.28%). On 7 days, the CK group experienced an increase in the relative abundance of Vishniacozyma (18.4%), Mycosphaerella (20.07%), and Acremonium (13.61%), which became dominant genera. Similarly, the FD group saw Acremonium (24.04%) emerge as a dominant genus, while in the FZ and FZD groups, the dominant genus is Vishniacozyma (32.90, 30.32%). On 60 days, shifts in genus abundance occurred, with Symmetrospora, Mycosphaerella, Alternaria, and Vishniacozyma emerging as dominant genera in each group. Notably, these genera showed relatively uniform distribution among groups.
LEfSe is a frequently employed tool for analyzing taxa that exhibit significant differences across various taxonomic levels. The preset LDA score threshold for filtering is set at 4. Analysis reveals the enrichment of diverse bacterial species among groups during the fermentation process. On 7 days (Figure 4A), the CK group exhibits notable species enrichment, including Carnobacterium, Desemzia, and Pantoea; the FD group shows enrichment of Duganella and Aerococcus; the FZ group demonstrates Lactobacillus enrichment; the FZD group displays enrichment of Exiguobacterium. On 60 days (Figure 4B), the CK group shows significant enrichment of Carnobacterium and Aerococcus; the FD group displays enrichment of Enterobacter and Desemzia; the FZ group presents enrichment of Lactobacillus, Burkholderia-Caballeronia-Paraburkholderia, Lactococcus, and Weissella; the FZD group does not exhibit distinct enriched taxa.
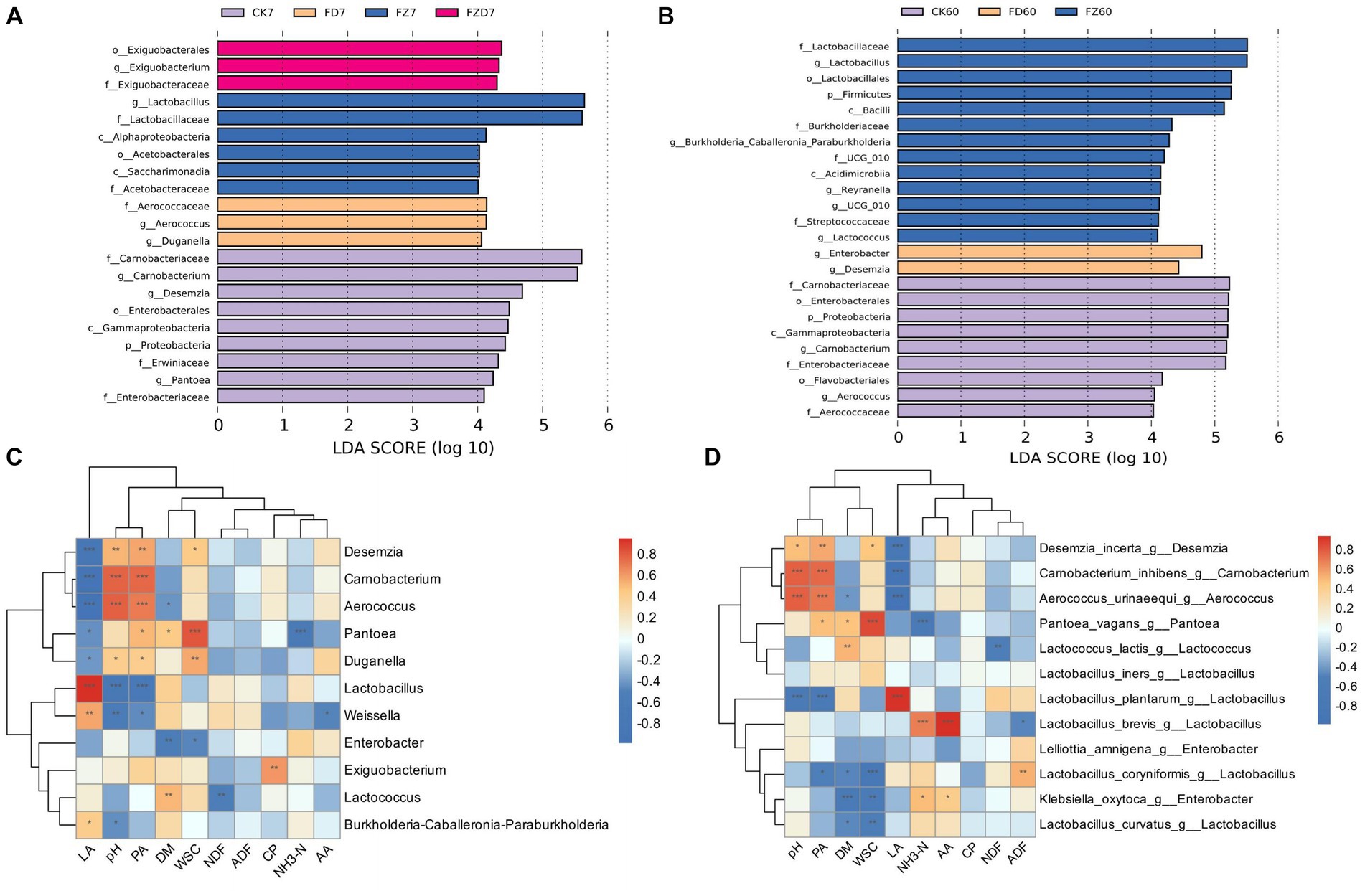
Figure 4. LEfSe analysis results for bacterial genus-level differential biomarkers on both the 7 d (A) and the 60 d (B) of ensiling fermentation (LDA score = 4). CK, sterile water; FD, L. brevis; FZ, L. plantarum; FZD, the combination of L. brevis and L. plantarum; The numbers 7 and 60 correspond to fermentation duration. The Spearman correlation heatmap (C) depicts the correlation between bacterial genus-level differential biomarkers and ensiling fermentation parameters; The analysis in Spearman correlation heatmap (D) included the selection of the top 30 species-level differential biomarkers from the bacterial genus-level differential biomarkers and their correlation analysis with fermentation parameters; Significance levels are denoted as *** (p < 0.001), ** (p < 0.01), and * (p < 0.05); red signifies a positive correlation, blue represents a negative correlation, while white indicates no correlation; Decimal values indicate the correlation coefficient R.
Correlation heatmaps illustrate the relationship between microbial communities and silage fermentation indicators. Spearman correlation analysis was performed on fermentation indicators, leveraging LEfSe analysis on taxa with notable differences at 7, 60 days, resulting in correlation coefficients (R = ±0.6) and significance indexes (p < 0.001). Figure 4C reveals that LA exhibits a positive correlation with Lactobacillus and a negative correlation with Desemzia, Carnobacterium, and Aerococcus; pH and PA display a positive correlation with Carnobacterium and Aerococcus while having a negative correlation with Lactobacillus; WSC indicates positive correlation with Pantoea; NH3-N exhibits negative correlation with Pantoea; DM, NDF, ADF, CP, and AA show limited correlations with distinct taxa.
To better understand the pivotal taxonomic species affecting fermentation, species that exhibited differential abundances within the top 30 species were chosen for Spearman correlation analysis in relation to fermentation indicators (R = ±0.6, p < 0.001). As depicted in Figure 4D, LA exhibits a positive correlation with Lactobacillus plantarum and negative correlations with Desemzia incerta, Carnobacterium inhibens, and Aerococcus urinaeequi; pH and PA show positive correlations with A. urinaeequi and C. inhibens, but negative correlations with L. plantarum; WSC displays a positive correlation with Pantoea vagans, but a negative correlation with L. coryniformis; NH3-N presents a positive correlation with L. brevis, while exhibiting a negative correlation with P. vagans; interestingly, at the species level, AA exhibits a positive correlation with L. brevis, whereas DM displays a negative correlation with Klebsiella oxytoca.
DM and WSC in raw materials are critical for ensuring fermentation quality (Wang et al., 2022). Typically, a DM content between 25–35% and a WSC content exceeding 50 g/kg ensure an ample ensiling fermentation substrate (Hu et al., 2009; Wang et al., 2018). JA raw material has a DM content of 35.02%, surpassing that of corn and oats, and a WSC content of 61.21 g/kg, meeting the criteria for high-quality ensiling material (Chen et al., 2020; Li et al., 2022). The storage temperature significantly influences ensiling success (Zhou et al., 2016). The Tibetan Plateau’s low temperatures make it prone to incomplete ensiling fermentation (Li et al., 2022), but inoculating low-temperature resistant LAB offers a potential solution for winter low-temperature ensiling fermentation (McDonald et al., 1991; Zhou et al., 2016). A pH below 4.6 during ensiling inhibits the activity of undesirable microorganisms and protein hydrolyzing enzymes (Ogunade et al., 2018a,b). In line with prior studies (Chen et al., 2020), inoculating low-temperature resistant LAB results in a gradual pH decrease, correlated with reduced DM and WSC content; microbial metabolism transforms WSC and available nutrients in DM into easily preservable organic acids and other substances (Xia et al., 2022).
Organic acid content is a vital indicator to evaluate fermentation success. Inoculating homotypic fermentation L. plantarum increased LA concentration and accelerated feed acidification, aligning with Chen et al.'s (2020) results. Interestingly, in the later fermentation stages, the LA content decreases, possibly due to high raw material WSC content. Homotypic fermentation LAB can produce a substantial amount of LA. As fermentation progresses, excessive WSC consumption shifts the fermentation from homotypic to heterotypic. LA is then converted into CO2, ethanol, or AA and volatile fatty acids. This clarifies why the WSC content in L. plantarum-inoculated silage is lower than in L. brevis-inoculated silage (Li et al., 2018; Bai et al., 2022; Tahir et al., 2023). These gases and volatile components contribute to a decline in ensiled DM and an increase in the relative content of NDF and ADF.
The NH3-N content indicates the extent of CP and amino acid decomposition in ensiled feed. Higher values suggest excessive consumption of CP and amino acids, resulting in a decline in silage quality (Hassanat et al., 2007). According to Pahlow et al. (2003) and Wilkinson (2005), proteinase hydrolysis of CP generates NH3-N, leading to an elevation in NH3-N content and a reduction in CP content in ensiled feed (Guan et al., 2018). At the conclusion of fermentation, there is no significant difference in CP content between groups and the raw material. However, inoculation with L. brevis leads to higher NH3-N content, possibly due to the elevated pH facilitating the normal activity of protein hydrolyzing enzymes. Moreover, Enterobacteria exhibit increased activity at pH 5–6, generating ammonia and biogenic amines through deamination and decarboxylation, resulting in an elevated NH3-N content (Queiroz et al., 2018; Wang et al., 2019). Additionally, microorganisms synthesize microbial protein during fermentation, offsetting the decline in CP content (He et al., 2020). However, the specific mechanism requires further investigation.
The quality of ensilage fermentation is intricately linked to microbial community changes, with diverse microbial interactions occurring throughout the process (Ni et al., 2017; Wang et al., 2022). This study utilizes high-throughput sequencing for the first time to unveil the distribution of bacteria and fungi in low-temperature ensiled JA. Inoculating with L. plantarum results in the lowest bacterial diversity index, signifying a higher proportion of beneficial microorganisms (Du et al., 2021). The fungal Chao1 index shows no significant difference, indicating that inoculation does not affect the richness of the fungal community. At the fermentation’s end, L. brevis inoculation yields a lower Shannon index, diminishing fungal community diversity. Factors like low temperature and anaerobiosis inhibit fungal activity, resulting in a gradual succession of fungal communities (Keshri et al., 2018; Cheng et al., 2022). The competition between LAB and undesirable microorganisms decides the success or failure of ensiling, and alterations in the microbial community elucidate the quality disparities among silages (Ni et al., 2017; Bai et al., 2021). Bacterial PCoA illustrates alterations in the bacterial community throughout ensiling, leading to variations in fermentation products (Ni et al., 2017). Inoculating with L. brevis results in sluggish bacterial activity, hindering dominant species formation, possibly determined by the inoculant. Conversely, L. plantarum inoculation dominates the early ensiling stages, promptly establishing an acidic environment and maintaining stability (Tahir et al., 2023). Fungal PCoA reveals a complete separation of the fungal community on 7 days of ensiling, but no apparent separation on 60 days. After fermentation, raw materials and silage samples intriguingly cluster together. This phenomenon may be because, in the early ensiling stages, aerobic bacteria heavily rely on residual air for reproduction. However, as fermentation advances, low-temperature anaerobic and acidic conditions suppress the activity of aerobic and neutral bacteria, inducing dormancy and eventual stabilization at the same level. The community’s species composition remains unaltered (Bao, 2019). The impact of inoculation on the fungal community composition may not be significant (Duniere et al., 2017; Wang et al., 2022), but specific reasons require further investigation.
Proteobacteria and Firmicutes prevail as bacterial phyla in silages, aligning with our study’s results (Chen et al., 2020; Fu et al., 2023). L. plantarum inoculation leads to Lactobacillus dominance throughout fermentation, improving low-temperature silage fermentation (Muck et al., 2018; Yan et al., 2019). In contrast to prior studies, Carnobacterium and Desemzia are notably abundant in both non-inoculated and L. brevis-inoculated low-temperature silage of JA. Carnobacterium, a Gram-positive bacterium, thrives in frozen soil or decaying plants, displaying facultative anaerobic, cold-tolerant, heterofermentative traits (Leisner, 1992; Meisel et al., 1994; Leisner et al., 2007). Desemzia, a thermophilic bacterium, degrades fats, proteins, and simple carbohydrates under aerobic conditions, commonly found in compost fermentation (Zainudin et al., 2017; Qiao et al., 2019). Both bacteria, saprophytes, likely originate from the soil at the base of JA stems and decaying leaves, resulting in silage fermentation failure. The primary fungal phyla in JA silage are Ascomycota and Basidiomycota. Inoculation with L. brevis enhances Ascomycota abundance, whereas L. plantarum inoculation elevates Basidiomycota abundance. This aligns with Cheng et al.’s (2022) research, highlighting varied LAB effects on fungal communities across distinct feed materials. Regarding genus-level composition, prevalent fungal genera in JA raw materials consist of Symmetrospora, Mycosphaerella, Alternaria, and Vishniacozyma. This contrasts with prior research (Gao et al., 2019; Chen et al., 2020; Li et al., 2021), with the disparity linked to distinct raw material types. In the initial fermentation phases, non-inoculated silage samples exhibit dominance of Vishniacozyma, Mycosphaerella, and Acremonium as genera. Vishniacozyma, an antagonistic yeast, possesses inhibitory effects against fungal diseases like gray mold (Nian et al., 2023), whereas Mycosphaerella and Acremonium can induce fungal diseases in plants (Bowness et al., 2016; Hou et al., 2019). This suggests that pathogenic fungi exploit residual air for extensive reproduction in early fermentation stages, prompting allergic reactions and a substantial rise in corresponding antagonistic fungi. L. brevis inoculation leads to an increased abundance of Acremonium, inhibiting the proliferation and metabolism of Vishniacozyma. On the other hand, L. plantarum inoculation efficiently inhibits the proliferation of pathogenic fungi but does not suppress the proliferation of Vishniacozyma (Santos et al., 2016). The fungal genera mentioned above are infrequently documented in silage. This study reveals interspecies interactions among them in JA silage for the first time. This phenomenon may be linked to the distinctive geographical and climatic features of the Qinghai-Tibet Plateau, along with the rich fructooligosaccharide content in JA stems and leaves. While offering a reference for future investigations into aerobic exposure in JA silage, the specific mechanisms require additional exploration.
LEfSe analysis suggests a positive correlation between biomarker function and the role of community abundance (Barberán et al., 2012; Weiss, 2017). Correlation heatmaps can assess how microbial species affect fermentation parameters, visually illustrating their interaction with species-level biomarkers. L. plantarum is positively correlated with LA, while pH and PA show negative correlations, consistent with previous research results (Fu et al., 2023). L. brevis is positively correlated with NH3-N and AA, while NH3-N is negatively or not correlated with Lactobacillus (Wang et al., 2022; Tahir et al., 2023). L. brevis is less likely to dominate in silage; its increased abundance accompanies changes in various heterofermentative bacteria, leading to AA synthesis and concurrent NH3-N production. The correlation analysis is considered positive, despite changes in heterofermentative bacteria types and continuous NH3-N synthesis. Inoculating L. brevis did not yield the anticipated results; instead, a significant proliferation of spoilage microorganisms occurred, causing nutrient loss, a considerable increase in NH3-N, and possible toxin production, detrimentally affecting livestock consumption. Spoilage microorganisms, including Carnobacterium inhibens, Desemzia incerta, and Aerococcus urinaeequi, show a negative correlation with LA and a positive correlation with pH and PA. As mentioned before, these spoilage bacteria are predominantly found in both non-inoculated and L. brevis-inoculated samples, providing additional evidence that L. brevis inoculation does not enhance low-temperature fermentation.
Inoculating low-temperature resistant LAB alleviates the negative impact of low temperatures in JA silage. Inoculating Lactobacillus plantarum GN02, which engages in homolactic fermentation, enhances LA levels, lowers pH and PA, sustains an acidic fermentation environment, preserves higher levels of DM and WSC, prevents nutrient degradation, and enhances the quality of low-temperature silage feed in JA. Additionally, it enhances the abundance of Lactobacillus, thereby inhibiting the proliferation of detrimental microorganisms like Carnobacterium, Desemzia, and Enterobacter. Inoculating Lactobacillus brevis XN25, which undergoes heterolactic fermentation, raises AA levels, but results in a gradual pH decrease. Moreover, it leads to a higher abundance of spoilage microorganisms in silage, consequently compromising fermentation efficacy. In conclusion, Lactobacillus plantarum GN02, utilizing homolactic fermentation, exhibits enhanced capabilities for low-temperature fermentation. Therefore, it is the recommended inoculant for winter silage of JA on the Qinghai-Tibet Plateau. This study addresses the deficiency in winter low-temperature silage techniques for JA on the Qinghai-Tibet Plateau. It not only establishes a theoretical foundation for winter silage but also contributes to advancing the development of a green, organic agricultural, and livestock product output center on the Qinghai-Tibet Plateau.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1013828.
XW: Writing – original draft. XS: Writing – review & editing. HZ: Writing – review & editing. QZ: Writing – review & editing. GL: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Financial support was provided by the Special Project for the Fundamental Research Program of Qinghai (2021-ZJ-928), the Transformation of Scientific and Technological Achievements in Qinghai Province (2022-NK-122) and National Natural Science Foundation of China (32260764).
We appreciate the Agriculture and Forestry Sciences Institute of Qinghai University for providing the cultivar. This work received support from the Laboratory for Research and Utilization of Germplasm Resources in the Qinghai-Tibet Plateau.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer HL declared a shared affiliation with the authors XW and GL to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AOAC . (1990). Official methods of analysis, Association of Official Analytical Chemists. 15th Edn. Artington, VA: AOAC.
Bai, J., Ding, Z. T., Ke, W. C., Xu, D. M., Wang, M. S., Huang, W. K., et al. (2021). Different lactic acid bacteria and their combinations regulated the fermentation process of ensiled alfalfa: ensiling characteristics, dynamics of bacterial community and their functional shifts. Microb. Biotechnol. 14, 1171–1182. doi: 10.1111/1751-7915.13785
Bai, J., Ding, Z., Su, R., Wang, M., Cheng, M., Xie, D., et al. (2022). Storage temperature is more effective than lactic acid bacteria inoculations in manipulating fermentation and bacterial community diversity, co-occurrence and functionality of the whole-plant corn silage. Microbiol. Spectr. 10:e0010122. doi: 10.1128/spectrum.00101-22
Bao, W. (2019). Dynamics variation of microbial diversity and functional geneomics during alfalfa silage fermentation process. Hohhot: Inner Mongolia Agricultural University.
Barberán, A., Bates, S. T., Casamayor, E. O., and Fierer, N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351. doi: 10.1038/ismej.2011.119
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bowness, R., Gossen, B. D., Chang, K., Goswani, R., Willenborg, C. J., Holtz, M., et al. (2016). Sensitivity of Mycosphaerella pinodes to Pyraclostrobin fungicide. Plant Dis. 100, 192–199. doi: 10.1094/PDIS-03-15-0350-RE
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chao, J., and Bunge, J. (2002). Estimatin the number of species in a stochastic abundance model. Biometrics. Biometrics. 58, 531–539. doi: 10.1111/j.0006-341x.2002.00531.x
Chen, L., Bai, S., You, M., Xiao, B., Li, P., and Cai, Y. (2020). Effect of a low temperature tolerant lactic acid bacteria inoculant on the fermentation quality and bacterial community of oat round bale silage. Anim. Feed Sci. Technol. 269:114669. doi: 10.1016/j.anifeedsci.2020.114669
Chen, S., Chang, Y., Huang, H., Kuo, S., and Wang, H. (2020). Application of condensed molasses fermentation solubles and lactic acid bacteria in corn silage production. J. Sci. Food Agric. 100, 2722–2731. doi: 10.1002/jsfa.10304
Chen, P., Hu, Y., Tang, F., Zhao, M., Peng, X., and Shen, S. (2020). Cooperation between Broussonetia papyrifera and its symbiotic fungal community to improve local adaptation of the host. Appl. Environ. Microb. 86, e00464–e00420. doi: 10.1128/AEM.00464-20
Cheng, Q., Chen, Y., Bai, S., Chen, L., You, M., Zhang, K., et al. (2021). Study on the bacterial community structure and fermentation characteristics of fresh and ensiled paper mulberry. Anim. Sci. J. 92:e13656. doi: 10.1111/asj.13656
Cheng, Q., Li, M., Fan, X., Chen, Y., Sun, H., Xie, Y., et al. (2022). Effects of epiphytic and exogenous lactic acid bacteria on fermentation quality and microbial community compositions of paper mulberry silage. Front. Microbiol. 13:973500. doi: 10.3389/fmicb.2022.973500
Du, Z., Sun, L., Lin, Y., Yang, F., and Cai, Y. (2021). The use of PacBio SMRT technology to explore the microbial network and fermentation characteristics of woody silage prepared with exogenous carbohydrate additives. J. Appl. Microbiol. 131, 2193–2211. doi: 10.1111/jam.15124
Duniere, L., Xu, S., Long, J., Elekwachi, C., Wang, Y., Turkington, K., et al. (2017). Bacterial and fungal core microbiomes associated with small grain silages during ensiling and aerobic spoilage. BMC Microbiol. 17:50. doi: 10.1186/s12866-017-0947-0
Fu, Z., Sun, L., Wang, Z., Liu, Y., Hao, J., Gao, C., et al. (2023). Effect of different regions on fermentation profiles, microbial communities, and their metabolomic pathways and properties in Italian ryegrass silage. Front. Microbiol. 13:1076499. doi: 10.3389/fmicb.2022.1076499
Gao, F., Chen, J., Xiao, J., Cheng, W., Zheng, X., Wang, B., et al. (2019). Microbial community composition on grape surface controlled by geographical factors of different wine regions in Xinjiang, China. Food Res. Int. 122, 348–360. doi: 10.1016/j.foodres.2019.04.029
Guan, H., Yan, Y., Li, X., Li, X., Shuai, Y., Feng, G., et al. (2018). Microbial communities and natural fermentation of corn silages prepared with farm bunker-silo in Southwest China. Bioresour. Technol. 265, 282–290. doi: 10.1016/j.biortech.2018.06.018
Hassanat, F., Mustafa, A. F., and Seguin, P. S. (2007). Effects of inoculation on ensiling characteristics, chemical composition and aerobic stability of regular and brown midrib millet silages. Anim. Feed Sci. Technol. 139, 125–140. doi: 10.1016/j.anifeedsci.2007.01.005
He, L., Wang, C., Xing, Y., Zhou, W., Pian, R., Chen, X., et al. (2020). Ensiling characteristics, proteolysis and bacterial community of high-moisture corn stalk and stylo silage prepared with Bauhinia variegate flower. Bioresour. Technol. 296:122336. doi: 10.1016/j.biortech.2019.122336
Hill, T. C., Walsh, K. A., Harris, J. A., and Moffett, B. F. (2003). Using ecological diversity measures with bacterial communities. FEMS Micro. Ecol. 43, 1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x
Hou, Y., Zhang, X., Zhang, N., Naklumpa, W., Zhao, W., Liang, X., et al. (2019). Genera Acremonium and Sarocladium cause Brown spot on bagged apple fruit in China. Plant Dis. 103, 1889–1901. doi: 10.1094/PDIS-10-18-1794-RE
Hu, W., Schmidt, R. J., Mcdonell, E. E., Klingerman, C. M., and Kung, L. (2009). The effect of Lactobacillus buchneri 40788 or Lactobacillus plantarum MTD-1 on the fermentation and aerobic stability of corn silages ensiled at two dry matter contents. J. Dairy Sci. 92, 3907–3914. doi: 10.3168/jds.2008-1788
Ju, Z. (2020). Screening of low temperature tolerant lactobacillus from oat epiphytes on Qinghai-Tibet plateau and study on their silage fermentation performance. Lanzhou: Gansu Agricultural University.
Keshri, J., Chen, Y., Pinto, R., Kroupitski, Y., Weinberg, Z. G., and Sela, S. S. (2018). Microbiome dynamics during ensiling of corn with and without Lactobacillus plantarum inoculant. Appl. Micro. Biot. 102, 4025–4037. doi: 10.1007/s00253-018-8903-y
Leisner, J. (1992). Characterisation of lactic acid bacteria isolated from lightly preserved fish products and their ability to metabolise various carbohydrates and amino acids [Ph. D. thesis]. Royal Veterinary and Agricultural University: Copenhagen, Denmark.
Leisner, J. J., Laursen, B. G., Prévost, H., Drider, D., and Dalgaard, P. (2007). Carnobacterium: positive and negative effects in the environment and in foods. FEMS Microbiol. Rev. 31, 592–613. doi: 10.1111/j.1574-6976.2007.00080.x
Li, P. (2016). Study on improvement of ensiing characteristics of siberian wildrye and oat in alpine pastoral area of northwest. Nanjing Agricultural University, Nanjing.
Li, N., Chen, M., Gao, X., Long, X., Shao, H., Liu, Z., et al. (2016). Carbon sequestration and Jerusalem artichoke biomass under nitrogen applications in coastal saline zone in the northern region of Jiangsu, China. Sci. Total Environ. 568, 885–890. doi: 10.1016/j.scitotenv.2016.06.074
Li, X., Chen, F., Wang, X., Sun, L., Guo, L., Xiong, Y., et al. (2021). Impacts of low temperature and ensiling period on the bacterial community of oat silage by SMRT. Microorganisms 9:274. doi: 10.3390/microorganisms9020274
Li, Y., Du, S., Sun, L., Cheng, Q., Hao, J., Lu, Q., et al. (2022). Effects of lactic acid bacteria and molasses additives on dynamic fermentation quality and microbial community of native grass silage. Front. Microbiol. 13:830121. doi: 10.3389/fmicb.2022.830121
Li, H., Guan, H., Jia, Z., Liu, W., Ma, X., Liu, Y., et al. (2022). Freeze-thaw condition limits the fermentation process and accelerates the aerobic deterioration of oat (Avena sativa) silage in the Qinghai-Tibet plateau. Front. Microbiol. 13:944945. doi: 10.3389/fmicb.2022.944945
Li, P., Lu, Y., Zhao, M., Chen, L., Zhang, C., Cheng, Q., et al. (2021). Effects of phenyllactic acid, lactic acid bacteria, and their mixture on fermentation characteristics and microbial community composition of timothy silage. Front. Microbiol. 12:743433. doi: 10.3389/fmicb.2021.743433
Li, P., Tang, H., Feng, T., Zhang, Y., Qu, H., Bai, S., et al. (2018). Effects of UV radiation on the process of wilting and application of additives at ensiling on the fermentation quality of Siberian wildrye silage on the Qinghai-Tibetan plateau. Grassl. Sci. 64, 61–68. doi: 10.1111/grs.12174
Lin, D., Ju, Z., Chai, J., and Zhao, G. (2022). Screening and identification of low temperature tolerant lactic acid bacterial epiphytes from oats on the Qinghai-Tibetan plateau. Acta Pratacul. Sin. 31, 103–114. doi: 10.11686/cyxb2021358
Liu, Y., Li, Y., Lu, Q., Sun, L., Du, S., Liu, T., et al. (2022). Effects of lactic acid bacteria additives on the quality, volatile chemicals and microbial community of leymus chinensis silage during aerobic exposure. Front. Microbiol. 13:938153. doi: 10.3389/fmicb.2022.938153
Mcdonald, P., Henderson, A. R., and Heron, S. J. E. (1991). The biochemistry of silage. Marlow: Chalcombe Publications.
Meisel, J., Wolf, G., and Hammes, W. P. (1994). Heme-dependent cytochrome formation in Lactobacillus maltaromicus. Syst. Appl. Microbiol. 17, 20–23. doi: 10.1016/S0723-2020(11)80026-3
Monllor, P., Muelas, R., Roca, A., Atzori, A. S., Díaz, J. R., Sendra, E., et al. (2020). Long-term feeding of dairy goats with broccoli by-product and artichoke plant silages: milk yield, quality and composition. Animals (Basel) 10:1670. doi: 10.3390/ani10091670
Muck, R., Nadeau, E., Mcallister, T., Contreras, G. F., Santos, M., and Kung, L. J. (2018). Silage review: recent advances and future uses of silage additives. J. Dairy Sci. 101, 3980–4000. doi: 10.3168/jds.2017-13839
Ni, K., Wang, F., Zhu, B., Yang, J., Zhou, G., Pan, Y., et al. (2017). Effects of lactic acid bacteria and molasses additives on the microbial community and fermentation quality of soybean silage. Bioresour. Technol. 238, 706–715. doi: 10.1016/j.biortech.2017.04.055
Nian, L., Xie, Y., Zhang, H., Wang, M., Yuan, B., Cheng, S., et al. (2023). Vishniacozyma victoriae: an endophytic antagonist yeast of kiwifruit with biocontrol effect to Botrytis cinerea. Food Chem. 411:135442. doi: 10.1016/j.foodchem.2023.135442
Ogunade, I. M., Jiang, Y., Cervantes, A. A., Kim, D. H., Oliveira, A. S., Vyas, D., et al. (2018b). Bacterial diversity and composition of alfalfa silage as analyzed by Illumina MiSeq sequencing: effects of Escherichia coli O157:H7 and silage additives. J. Dairy Sci. 101, 2048–2059. doi: 10.3168/jds.2017-12876
Ogunade, I. M., Jiang, Y., Kim, D. H., Cervantes, A. A., Arriola, K. G., Vyas, D., et al. (2018a). Fate of Escherichia coli O157:H7 and bacterial diversity in corn silage contaminated with the pathogen and treated with chemical or microbial additives. J. Dairy Sci. 100, 1780–1794. doi: 10.3168/jds.2016-11745
Okoye, C. O., Wang, Y., Gao, L., Wu, Y., Li, X., Sun, J., et al. (2023). The performance of lactic acid bacteria in silage production: a review of modern biotechnology for silage improvement. Microbiol. Res. 266:127212. doi: 10.1016/j.micres.2022.127212
Pahlow, G., Muck, R. E., Driehuis, F., Elferink, S. J. W. H., and Spoelstra, S. F. (2003). Microbiology of ensiling. Agron. Monogr. 42, 31–94. doi: 10.2134/agronmonogr42.c2
Qi, X. (2021). Compacting responsibility, go all out to tackle tough problems, a summary of the third land survey work in Qinghai Province. Qinghai Land Econ. 2021, 55–57.
Qiao, C., Ryan, P. C., Liu, C., Shen, Z., Ou, Y., Liu, Z., et al. (2019). Key extracellular enzymes triggered high-efficiency composting associated with bacterial community succession. Bioresour. Technol. 288:121576. doi: 10.1016/j.biortech.2019.121576
Qiu, P., Du, Y., Yu, Y., Song, M., Kong, G., and Ni, M. (2018). Research progress on saline-alkali and drought rolerance of Helianthus tuberous. Northern Horticult. 2018, 168–171. doi: 10.11937/bfyy.20174027
Queiroz, O. C. M., Ogunade, I. M., Weinberg, Z., and Adesogan, A. T. (2018). Silage review: foodborne pathogens in silage and their mitigation by silage additives. J. Dairy Sci. 101, 4132–4142. doi: 10.3168/jds.2017-13901
Ren, H., Sun, W., Yan, Z., Zhang, Y., Wang, Z., Song, B., et al. (2021). Bioaugmentation of sweet sorghum ensiling with rumen fluid: fermentation characteristics, chemical composition, microbial community, and enzymatic digestibility of silages. J. Clean. Prod. 294:126308. doi: 10.1016/j.jclepro.2021.126308
Santos, A. O., Ávila, C., Pinto, J. C., Carvalho, B. F., Dias, D. R., and Schwan, R. F. (2016). Fermentative profile and bacterial diversity of corn silages inoculated with new tropical lactic acid bacteria. J. Appl. Microbiol. 120, 266–279. doi: 10.1111/jam.12980
Tahir, M., Li, J., Xin, Y., Wang, T., Chen, C., Zhong, Y., et al. (2023). Response of fermentation quality and microbial community of oat silage to homofermentative lactic acid bacteria inoculation. Front. Microbiol. 13:1091394. doi: 10.3389/fmicb.2022.1091394
Teng, M. (2019). Responses of Jerusalem artichoke growth to saline-sodic stress and soil improvement. Changchun: Jilin Agricultural University.
Thomas, T. A. (1977). An automated procedure for the determination of soluble carbohydrates in herbage. J. Sci. Food Agric. 28, 639–642. doi: 10.1002/jsfa.2740280711
Van Soest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Wang, Y., He, L., Xing, Y., Zhou, W., Pian, R., Yang, F., et al. (2019). Bacterial diversity and fermentation quality of Moringa oleifera leaves silage prepared with lactic acid bacteria inoculants and stored at different temperatures. Bioresour. Technol. 284, 349–358. doi: 10.1016/j.biortech.2019.03.139
Wang, C., He, L., Xing, Y., Zhou, W., Yang, F., Chen, X., et al. (2019). Fermentation quality and microbial community of alfalfa and stylo silage mixed with Moringa oleifera leaves. Bioresour. Technol. 284, 240–247. doi: 10.1016/j.biortech.2019.03.129
Wang, Y., Liu, J., Yan, G., and Xiong, B. (2020a). The research progress on variety breeding, nutritional value and feeding effect of Jerusalem artichoke as feedstuff. China Feed. 19, 106–114. doi: 10.15906/j.cnki.cn11-2975/s.20201923
Wang, Y., Nan, X., Zhao, Y., Jiang, L., Wang, H., Hua, D., et al. (2021b). Dietary supplementation with inulin improves lactation performance and serum lipids by regulating the rumen microbiome and metabolome in dairy cows. Anim. Nutr. 7, 1189–1204. doi: 10.1016/j.aninu.2021.09.007
Wang, Y., Nan, X., Zhao, Y., Jiang, L., Wang, M., Wang, H., et al. (2021a). Rumen microbiome structure and metabolites activity in dairy cows with clinical and subclinical mastitis. J. Anim. Sci. Biotechnol. 12:36. doi: 10.1186/s40104-020-00543-1
Wang, Y., Nan, X., Zhao, Y., Jiang, L., Wang, H., Zhang, F., et al. (2021c). Dietary supplementation of inulin ameliorates subclinical mastitis via regulation of rumen microbial community and metabolites in dairy cows. Microbiol. Spectr. 9:e0010521. doi: 10.1128/Spectrum.00105-21
Wang, Z., Tan, Z., Wu, G., Wang, L., Qin, G., Wang, Y., et al. (2022). Microbial community and fermentation characteristic of whole-crop wheat silage treated by lactic acid bacteria and Artemisia argyi during ensiling and aerobic exposure. Front. Microbiol. 13:1004495. doi: 10.3389/fmicb.2022.1004495
Wang, Y., Wang, C., Zhou, W., Yang, F. Y., Chen, X. Y., and Zhang, Q. (2018). Effects of wilting and Lactobacillus plantarum addition on the fermentation quality and microbial community of Moringa oleifera leaf silage. Front. Microbiol. 9:1817. doi: 10.3389/fmicb.2018.01817
Wang, Y., Zhao, Y., Xue, F., Nan, X., Wang, H., Hua, D., et al. (2020b). Nutritional value, bioactivity, and application potential of Jerusalem artichoke (Helianthus tuberosus L.) as a neotype feed resource. Anim. Nutr. 6, 429–437. doi: 10.1016/j.aninu.2020.09.001
Wei, X., Sun, X., Zhang, H., Wang, L., Yang, S., Si, C., et al. (2023). Identification of low-temperature-tolerant lactic acid bacteria in Helianthus tuberosus silage in alpine regions. Pratacult. Sci. 41, 1–11. doi: 10.11829/j.issn.1001-0629
Weiss, K. (2017). Volatile organic compounds in silages-efects of management factors on their formation: a review. Slovak. J. Anim. Sci. 50, 55–67.
Wu, T., Liu, Y., and Zhao, F. (2022). Effects of different additives on the quality and nutritional value of the stem and leaf silage of Helianthus tuberosus. Pratacult. Sci. 39, 770–777. doi: 10.11829/j.issn.1001-0629.2021-0140
Xia, T., Wang, T., Sun, J., Shi, W., Liu, Y., Huang, F., et al. (2022). Modulation of fermentation quality and metabolome in co-ensiling of Sesbania cannabina and sweet sorghum by lactic acid bacterial inoculants. Front. Microbiol. 13:851271. doi: 10.3389/fmicb.2022.851271
Xin, Y. (2013). Grassland management and the sustainable development of Qinghai Province. Qinghai Pratacult. 22, 15–19.
Xu, D., Wang, N., Rinne, M., Ke, W., Weinberg, Z. G., Da, M., et al. (2021). The bacterial community and metabolome dynamics and their interactions modulate fermentation process of whole crop corn silage prepared with or without inoculants. Microb. Biotechnol. 14, 561–576. doi: 10.1111/1751-7915.13623
Yan, Y., Li, X., Guan, H., Huang, L., Ma, X., Peng, Y., et al. (2019). Microbial community and fermentation characteristic of Italian ryegrass silage prepared with corn Stover and lactic acid bacteria. Bioresour. Technol. 279, 166–173. doi: 10.1016/j.biortech.2019.01.107
Zainudin, M. H. M., Ramli, N., Hassan, M. A., Shirai, Y., Tashiro, K., Sakai, K., et al. (2017). Bacterial community shift for monitoring the co-composting of oil palm empty fruit bunch and palm oil mill effluent anaerobic sludge. J. Ind. Microbiol. Biotechnol. 44, 869–877. doi: 10.1007/s10295-017-1916-1
Zhao, M., Guo, Y., Sun, S., and Ren, Y. (2020). Evaluation of cold resistance of four Jerusalem artichoke (Helianthus tuberosus) under low temperature stress. Plant Physiol. J. 56, 1419–1431. doi: 10.13592/j.cnki.ppj.2020.0131
Zhou, Y., Drouin, P., and Lafreniere, C. (2016). Effect of temperature (5-25 degrees°C) on epiphytic lactic acid bacteria populations and fermentation of whole-plant corn silage. J. Appl. Microbiol. 121, 657–671. doi: 10.1111/jam.13198
Keywords: Qinghai-Tibet Plateau, Jerusalem Artichoke silage, low-temperature resistant lactic acid bacteria, winter fermentation, microbial community structure
Citation: Wei X, Sun X, Zhang H, Zhong Q and Lu G (2024) The influence of low-temperature resistant lactic acid bacteria on the enhancement of quality and the microbial community in winter Jerusalem Artichoke (Helianthus tuberosus L.) silage on the Qinghai-Tibet Plateau. Front. Microbiol. 15:1297220. doi: 10.3389/fmicb.2024.1297220
Received: 04 October 2023; Accepted: 11 January 2024;
Published: 29 January 2024.
Edited by:
Ping Li, Guizhou University, ChinaReviewed by:
Zhihao Dong, Nanjing Agricultural University, ChinaCopyright © 2024 Wei, Sun, Zhang, Zhong and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiwen Zhong, MjkwODk3NDRAcXEuY29t; Guangxin Lu, bHVneDc0QHFxLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.