- 1Shunyi District Center for Disease Control and Prevention, Beijing, China
- 2Workstation for Microbial Infectious Disease, Shunyi District Center for Disease Control and Prevention, Beijing, China
- 3National Key Laboratory of Intelligent Tracking and Forecasting for Infectious Diseases, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention (China CDC), Beijing, China
- 4Hangzhou Center for Disease Control and Prevention, Hangzhou, Zhejiang, China
Introduction: Shewanella is an important opportunistic pathogen distributed in marine environments that has caused an increasing number of clinical infections. However, there are few reports on the distribution and characteristics of Shewanella in the diarrheal pathogen spectrum. In this study, we have systematically described the prevalence of Shewanella infections in diarrhea patients in Beijing, China 2017–2019, and genome characteristics and antimicrobial susceptibility of Shewanella isolates.
Methods: Stool samples were collected from diarrhea patients in a surveillance project from 2017 to 2019. Shewanella strains were isolated, and identified using VITEKR 2 COMPACT and MALDI-TOF MS. Average nucleotide identity (ANI) analysis, multi-locus sequence typing (MLST), phylogenetic analysis, virulence-associated genes and antimicrobial resistance genes analysis were used for genome characteristics description. The antibiotic susceptibility test was performed with microbroth dilution method.
Results: 1104 fecal samples were collected, and the Shewanella detection rate was 2.36% (26/1104). The main manifestations of infection caused by Shewanella spp. were diarrhea (100%, 26/26), abdominal pain (65.38%, 17/26), and vomiting (38.46%, 10/26). The 26 isolates were classified into 3 species (S. algae (n = 18), S. indica (n = 5), and S. chilikensis (n = 3)) and 22 sequence types. Core genome single nucleotide polymorphism-based evolutionary tree identified three clone groups corresponding to three infection events in the same months in 2017 and 2019. The putative virulence-associated gene pool consisted of 56 potential virulence genes, including 19 virulence gene factors. The resistance rates of the 26 isolates to 17 antibiotics from high to low were as follows: polymyxin E (76.92%), cefotaxime (57.69%), ampicillin (50%), ampicillin-sulbactam (34.62%), nalidixic acid (15.38%), ciprofloxacin (11.54%), selectrin (3.846%,1/26), and tetracycline (3.846%, 1/26). The rate of multidrug resistance was 38.46% (10/26).
Discussion: Monitoring for Shewanella spp. should be added to the routine surveillance of infectious diarrhea during the epidemic season.
1 Introduction
Shewanella spp. are a group of gram-negative, oxidase-positive, facultatively anaerobic bacteria distributed in marine environments and the digestive tracts of marine animals. They can also be found in extreme environments with low temperature, high pressure, and high salinity (Kouzuma et al., 2015). Clinical cases related to Shewanella, which are opportunistic pathogens, have been frequently reported (Yousfi et al., 2017a). Some Shewanella species, such as S. algae (Chia-Wei et al., 2022), S. putrefaciens (Holt et al., 2005), and S. xiamenensis (Zong, 2011; Antonelli et al., 2015), are directly associated with clinical infections. Furthermore, Shewanella often causes skin and soft tissue infections, ear, nose, and throat-related diseases, chest and abdominal cavity infections, blood infections, and even cardiovascular and central nervous system diseases (Yu et al., 2022), causing widespread concern both domestically and internationally. Such bacteria have been isolated from clinical samples of patients with diarrhea in recent years (Yiallouros et al., 2013; Dey et al., 2015; Fang et al., 2017). Yonglu et al. (2009) isolated them for the first time from patients with food poisoning. Therefore, diarrheal diseases caused by Shewanella spp. require more attention.
However, systematic taxonomic studies of the Shewanella genus are not yet complete. Traditional classification and identification methods based on bacterial morphology, cultural characteristics, and biochemical results are difficult to apply to the genus Shewanella, because the phenotypic characteristics of its species are very similar (Liu et al., 2013). In addition, it is difficult to distinguish genetically closely related Shewanella species through homology analysis of the 16S rRNA gene sequence (Yarza et al., 2014). Molecular typing techniques, such as multilocus sequence typing (MLST), have been applied for surveillance of the molecular epidemiology of Shewanella (Thorell et al., 2019). Whole-genome sequencing can be used to quickly determine the genetic and evolutionary characteristics of Shewanella. Moreover, whole-genome sequencing has good sensitivity and specificity for predicting drug resistance and virulence-related genes (Lee et al., 2016).
However, relevant studies on the distribution and characteristics of Shewanella in the diarrheal pathogen spectrum in China are still lacking. In the present study, we used whole-genome sequencing and bioinformatics tools for an in-depth analysis of epidemic and pathogenic characteristics based on monitoring the diarrheal pathogen in Beijing, China, from 2017 to 2019.
2 Materials and methods
2.1 Sample collection
According to the guidelines of the local foodborne disease surveillance project in Beijing, patients with diarrhea enrolled in this study were outpatients who presented with acute diarrhea. This was defined as ≥3 passages of watery, loose, mucus, or bloody stools during a 24 h period in two clinics in Shunyi district, Beijing from January 1st, 2017 to December 30th, 2019. Age, sex, occupation, clinical symptoms, and other information of the patients with diarrhea were collected, and 5 g of fresh stool samples were collected from each patient. The samples were stocked in Cary-Blair medium at 4°C and transported to the laboratory for bacterial isolation within 24 h to isolate and culture Shewanella.
2.2 Shewanella isolation
Fecal samples from the patients were isolated and cultured immediately after delivery to the laboratory. Briefly, a 200 mg stool sample was inoculated into 3% NaCl alkaline peptone water and enriched at 37°C for 24 h. After selective enrichment, a loop of the culture was streaked on TCBS and CHROMagar Vibrio color red medium and incubated at 37°C for 24 h. Five presumptive Shewanella colonies on the selective agar plate (medium-sized, smooth, raised, rounded colonies with colorless edges and black centers on TCBS agar plates and round, translucent, smooth-surfaced pink-purple colonies on CHROMagar Vibrio color red medium) were inoculated in 3% NaCl tryptic soy agar and incubated at 37°C for 24 h. The pure culture of each colony was identified using VITEKR 2 COMPACT (BioMerieux) and MALDI-TOF MS (Bruker).
2.3 Whole-genome sequencing and bioinformatics analysis
Genomic DNA was extracted using a Wizard Genomic DNA Extraction Kit (Promega, Madison, WI, USA) following the manufacturer’s instructions. DNA samples were sent to the Beijing Genomics Institution for next-generation sequencing, requiring a total amount > 20 μg. The concentration reached 50 ng/μL and OD260/280 of 1.8–2.0 for individual samples. Low-quality reads were discarded, and clean data were assembled using SOAP de novo (version 2.04). After removing contigs with less than 500 bp, QUAST (version 5.0.1) was applied to evaluate the quality of assembled genomes (Gurevich et al., 2013). Prokka (version 1.12) (Seemann, 2014), Prodigal (version 2.6.3) (Hyatt et al., 2010), and RAST were used to annotate genomes.1
The genome sequences of 26 laboratory isolates and four type strains (S. algae JCM 21037T, S. chilikensis KCTC 22540T, S. indica KCTC 23171T, and S. carassii 08MAS2251T) were included in this study. Average nucleotide identity (ANI) analysis was used to evaluate the evolutionary distance between bacteria at the genomic level based on a Perl script as previously described (Huang et al., 2022). ANI values >95% were considered indicative of the same species. MLST was performed using seven housekeeping genes (16S rRNA, gyrA, gyrB, infB, recN, rpoA, and topA) (Huang et al., 2023). Housekeeping gene sequences were extracted from the genomes to obtain the allele numbers and sequence type (ST). BioNumerics 7.1 was used to analyze allele profiles and construct a minimum spanning tree. Prokka (version 1.12) and Roary pan-genome pipeline with an identity cut-off of 95% were used for gene annotation and pan genome analysis, respectively. Snippy was used to refer to core genome single nucleotide polymorphisms, with the genome sequence of S. algae JCM 21037T used as a reference. Gubbins was used to remove recombinant sequence sites. The phylogenetic evolutionary tree was constructed using IQ-TREE based on the decombined core genome single nucleotide polymorphisms (maximum likelihood estimation, bootstraps 1,000). In the Virulence Factor Database (VFDB) database, owing to the lack of a virulence factor library related to the Shewanella genus, we used Vibrio species as a reference for virulence gene analysis because of their close phylogenetic relationship. Potential antimicrobial resistance genes were predicted by Comprehensive Antibiotic Research Database (CARD) (http://arpcard.mcmaster.ca).
2.4 Antimicrobial susceptibility testing (AST) for Shewanella
An AST panel for aerobic Gram-negative bacilli (Shanghai Fosun Long March Medical Science Co., Ltd., China) was performed using the microbroth dilution method. As there were no breakpoints for Shewanella, the results for susceptibility (S), intermediate (I), and drug resistance (R) were interpreted according to the Enterobacteriaceae standards of the American Committee for Clinical Laboratory Standardization (Clinical and Laboratory Standards Institute, 2019). Escherichia coli ATCC 25922 was used as a control for ampicillin, ampicillin-salbactam, tetracycline, meropenem, polymyxin E, ertapenem, ceftazidime/avibactam, tigecycline, cefotaxime, ceftazidime-avibactam, ciprofloxacin, azithromycin, chloramphenicol, nalidixic acid, streptomycin, selectrin, and amikacin.
2.5 Statistical analysis
All statistical analyses were performed using the Stata software version 12.0. The chi-squared test was used to compare the isolation ratios of different pathogens in different months. The Shewanella infection ratios between different sexes, among different age groups, and contamination in different suspected food groups were also analyzed using the chi-squared test. Statistical significance was set at p < 0.05.
2.6 Ethics statement
All aspects of the study were performed in accordance with the national ethics regulations and were approved by the Ethics Committee of the Shunyi District CDC, China. Participants received information on the purpose of the study and their right to keep their information confidential. Written informed consent was obtained from each participant or their parents/guardians.
3 Results
3.1 Prevalence and clinical characteristics of Shewanella infection
In this surveillance study, 1,104 fecal samples were collected. The detection rate of Shewanella was 2.36% (26/1104). The detection rates in 2017, 2018, and 2019 were 2.43% (9/371), 2.67% (10/374), and 1.95% (7/359), respectively (Table 1). The major clinical symptoms reported in infectious cases caused by Shewanella spp. were diarrhea (100%, 26/26), abdominal pain (65.38%, 17/26), and vomiting (38.46%, 10/26) (Table 1). Each of the 26 patients had diarrhea more than three times per day; among them, six had loose stools (23.08%, 6/26), and the remaining had liquid stools (76.92%, 20/26). The Shewanella infection rates in male and female patients were 2.17% (14/644) and 2.61% (12/460), respectively, with no significant difference between the infection rate and sex (p = 0.639). The infection rate of Shewanella in the 61–80-year-old group ranked first at 2.99% (4/134), but there was no significant difference in Shewanella infection among the different age groups (p = 0.745, Fisher’s exact test) (Table 2). The three occupations with the highest Shewanella infection ratios were commercial service personnel, workers, and household workers, with infection ratios of 4.11% (3/73), 2.82% (6/213), and 2.71% (6/221), respectively. Again, there was no significant difference in Shewanella infection among the different occupational groups (p = 0.950) (Table 2).
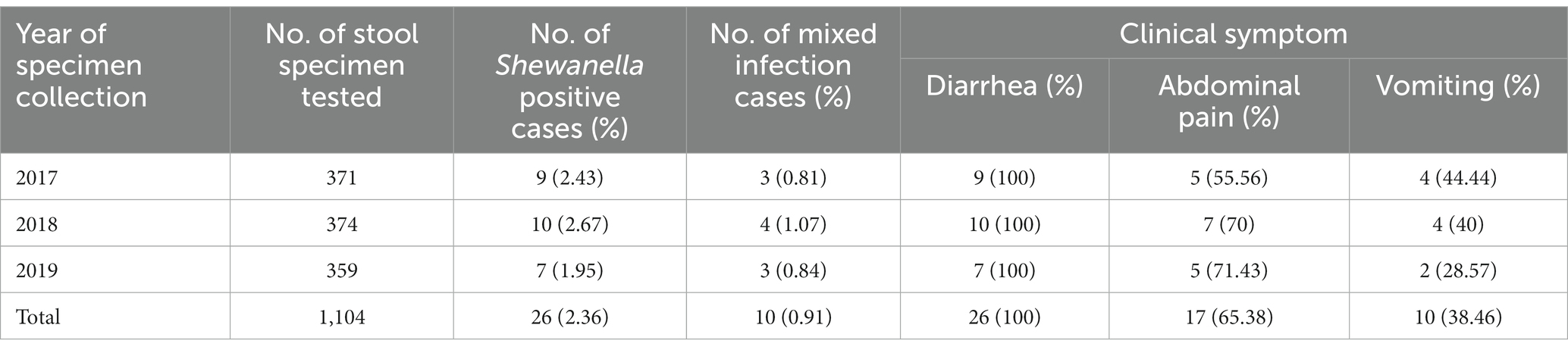
Table 1. Prevalence of Shewanella infections in diarrhea patients in S District, Beijing, 2017–2019.
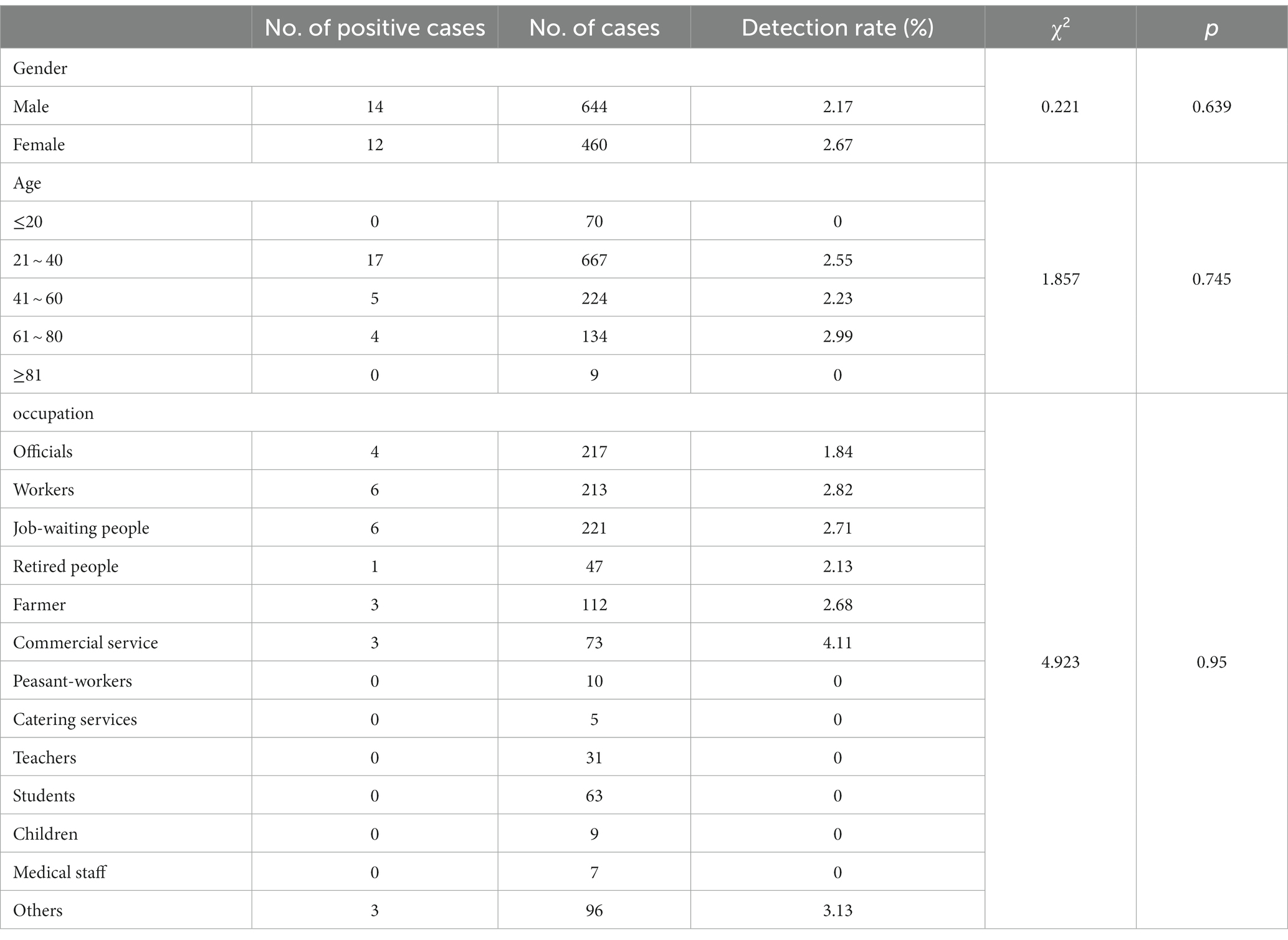
Table 2. The detection rate of Shewanella in different populations in S District, Beijing, 2017–2019.
3.2 Identification of Shewanella species
The quality and complpeteness of the genomes was shown in the Supplementary Table S1. The ANI heatmap revealed identified 26 Shewanella strains (Figure 1). These strains were classified into three distinct species, namely S. algae (n = 18, 69.23%), S. indica (n = 5, 19.23%), and S. chilikensis (n = 3, 11.54%). Both S. algae and S. chilikensis were detected in 2017–2019, whereas S. indica was detected only in 2018–2019.
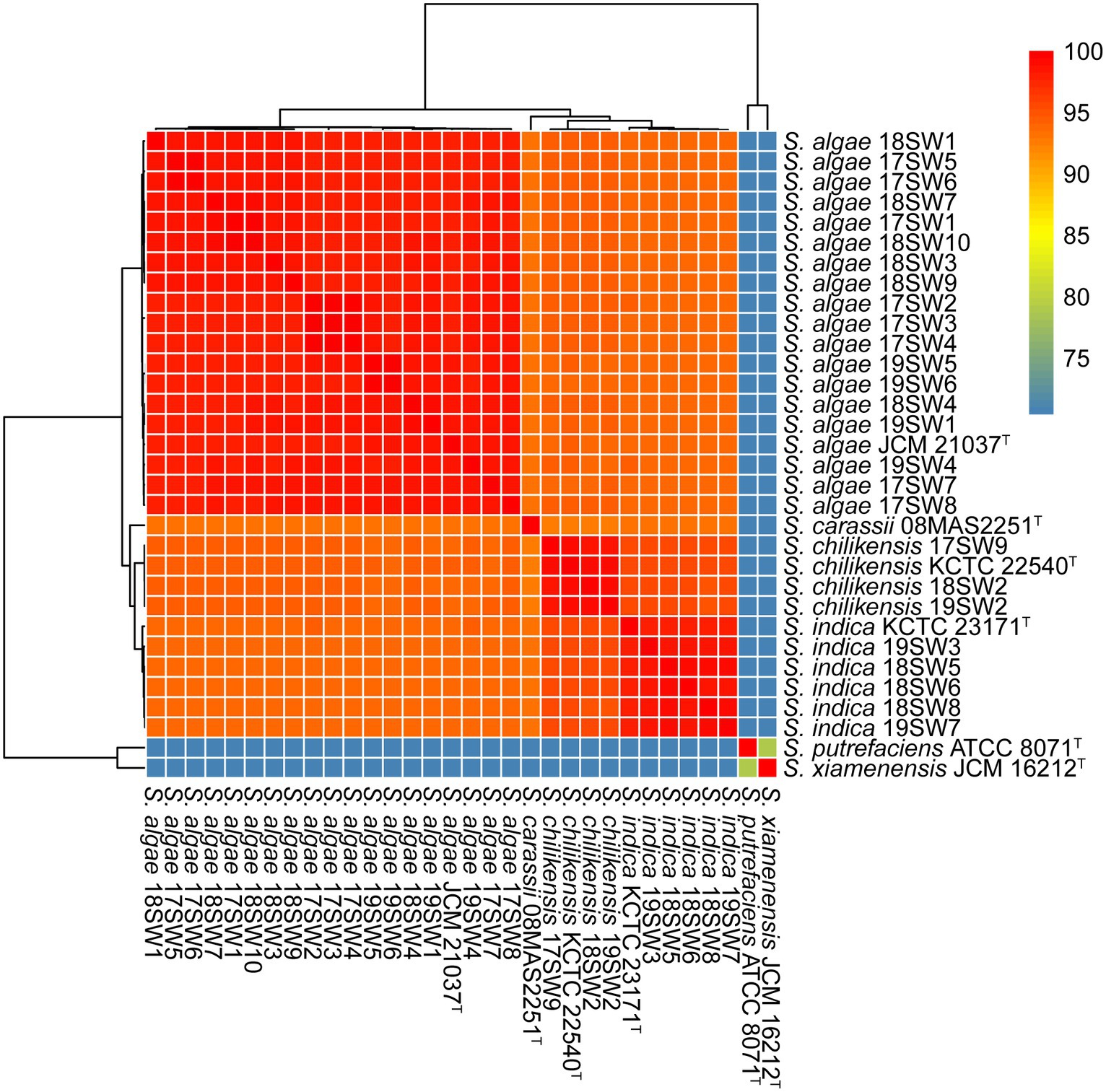
Figure 1. Average nucleotide identity (ANI) values for the species identification for the 26 Shewanella strains. Six type strains (S. algae JCM 21037T S. carassii 08MAS2251T S. chilikensis KCTC 22540T S. indica KCTC 23171T S. putrefaciens ATCC 8071T S. xiamenensis JCM 16212T) were used as reference genome. The color from blue to red represents the value of ANI from <75 to 100.
3.3 MLST for Shewanella
The allelic profiles of Shewanella isolates were classified into 22 STs. Among these, S. algae were classified into 16 STs, accounting for 66.7%. S. indica and S. chilikensis were classified into 5 and 3 STs, accounting for 8.8 and 7.4%, respectively. The minimum spanning tree of different Shewanella isolates classified by species, isolation region, and isolation source is shown in Figure 2. No significant regional or isolation source clustering was observed, suggesting high genetic diversity among these strains.
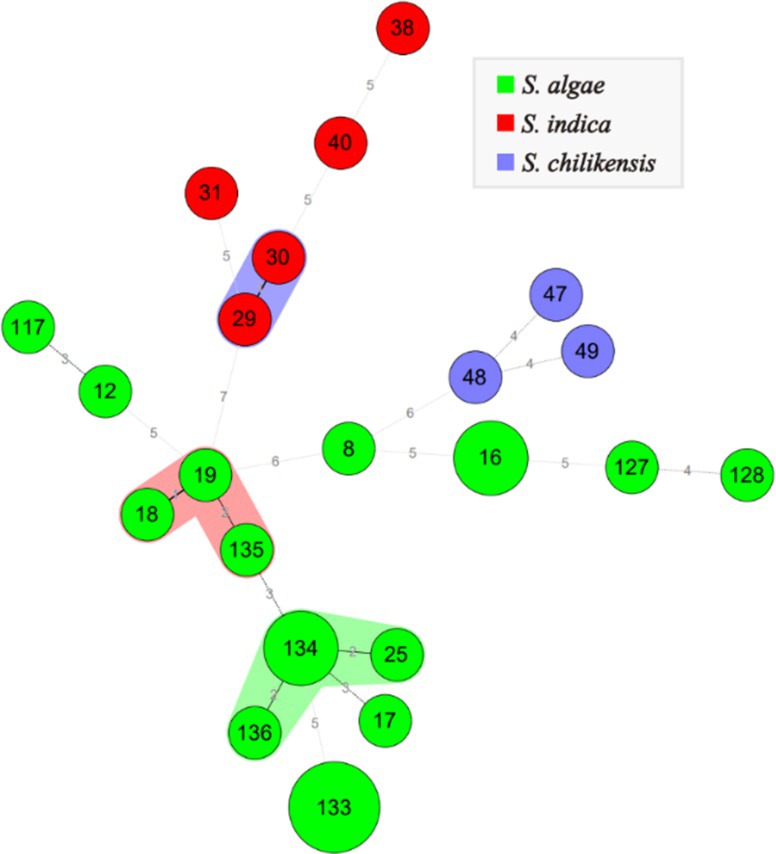
Figure 2. MLST for 26 Shewanella strains. Green, red and blue color represent S. algae, S. indica and S. chilikensis, respectively. The size of the circle represents the number of strains.
3.4 Genomic evolutionary characteristics of Shewanella
A total of 301,356 cg of single nucleotide polymorphisms were identified. A phylogenetic tree was constructed using maximum likelihood (Figure 3). Three clone groups were identified among them, namely strains 17SW2/3/4, 17SW5/6, and 19SW5/6, corresponding to three infection events in the same months in 2017 and 2019. However, different clones were detected even in the same month, such as strains 17SW2/3/4, 17SW5/6, 17SW1, and 17SW7, which were isolated in July 2017. Different Shewanella species were also detected during the same period: S. algae 17SW8 and S. chilikensis 17SW9 were detected in August 2017, and S. algae 19SW1, S. chilikensis 19SW2, and S. indica 19SW3 were detected in June 2019. According to statistical analysis, the positive ratio between S. algae and other Shewanella spp. showed no significant difference in terms of sex, age, or mixed infections.
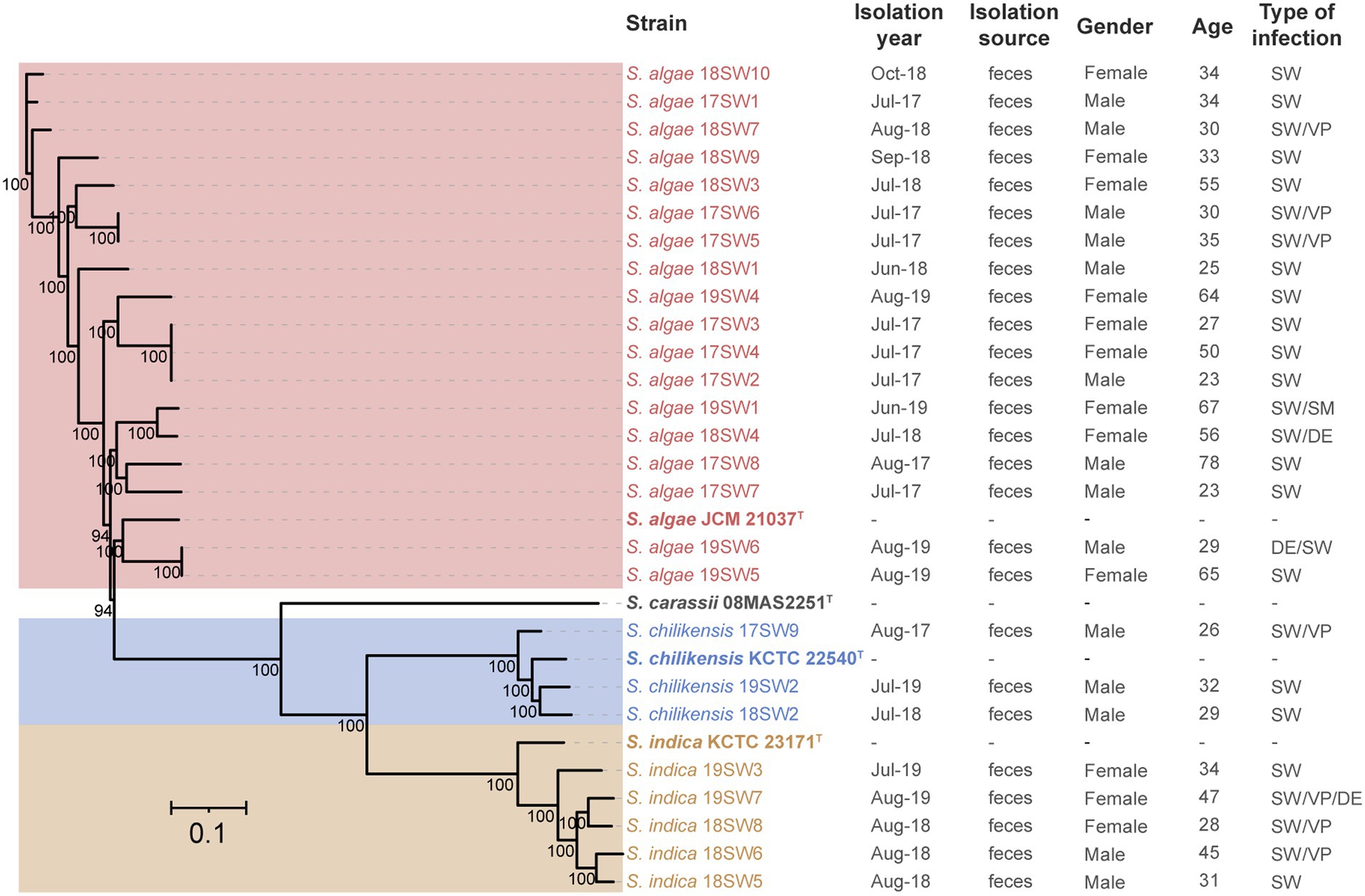
Figure 3. Phylogenetic tree of 26 Shewanella isolates and four type strains based on cgSNPs by the maximum-likelihood method.
3.5 Distribution of virulence-associated genes
The putative virulence-associated gene pool comprised 56 potential virulence genes, including 19 virulence gene factors (Figure 4). Different Shewanella species exhibited specific virulence gene patterns. Most strains carried potential virulence genes related to flagella and chemotactic proteins, but rarely carried genes of the III and I secretion systems. Most S. algae carried potential virulence genes related to the VI secretion system, including the hcp gene related to the inner tube and the effector protein vgrG, suggesting that T6SS may be an important virulence factor in S. algae. S. indica lacked the potential virulence genes related to the VI secretion system, but most strains carried genes related to the flagella. Most S. chilikensis strains also harbored potential virulence genes related to the flagella.
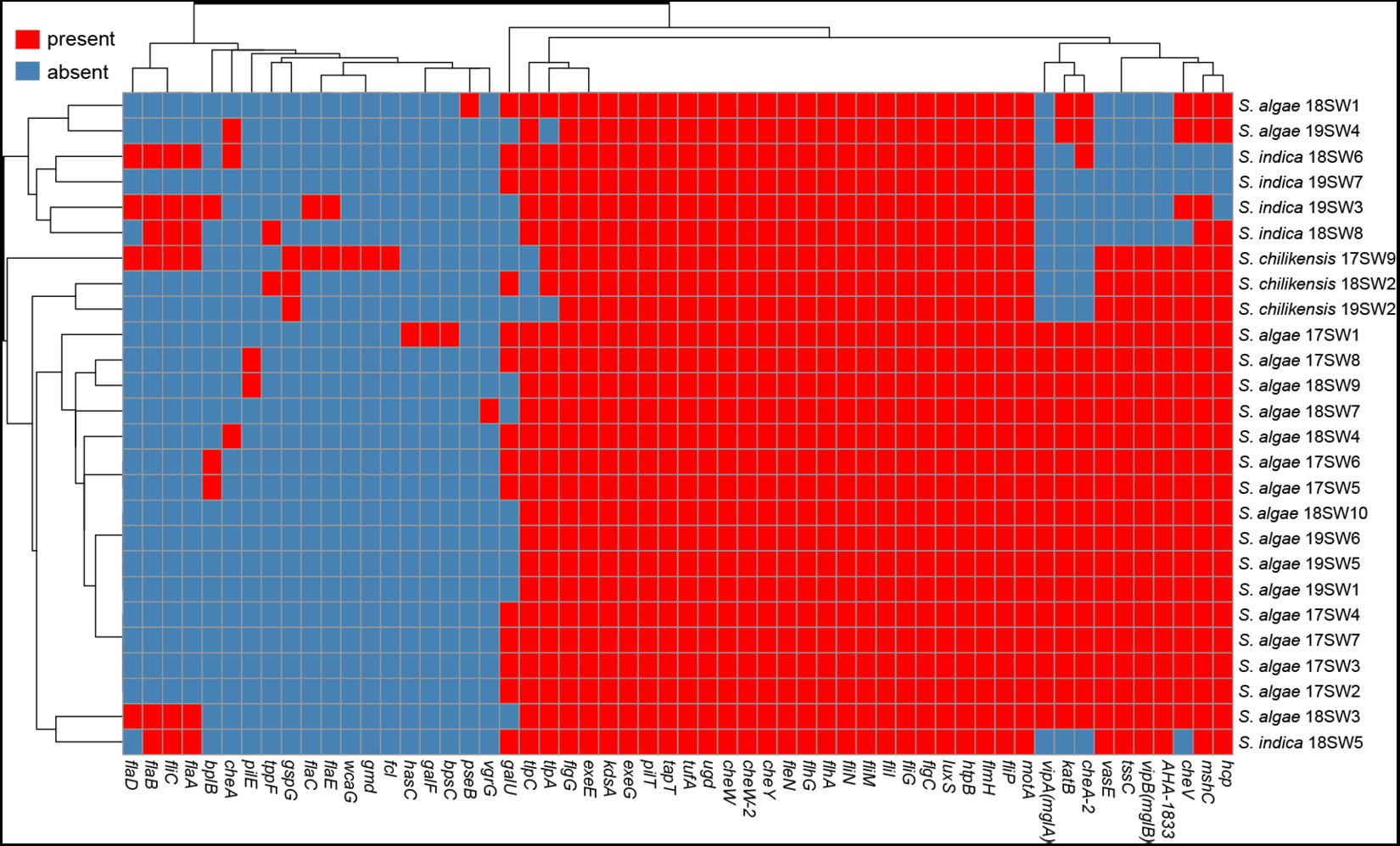
Figure 4. The heatmap of the virulence genes in 26 Shewanella strains. Red and blue colors represent the presence and absence of virulence genes, respectively.
3.6 Antimicrobial resistance genes analysis
A total of nine ARGs were identified by searching against the CARD (Figure 5). Only The strain 17SW1 was a multidrug-resistant stain, which carries the highest number of antimicrobial resistance genes (aph(3″)-Ib, aph(6)-Id, blaOXA-SHE, floR, qnrA7, sul1, sul2), indicating a capacity to resist to aminoglycosides, β-lactam, amide alcohols, quinolones, sulfonamides. The blaOXA gene was located in all strains with the major genotype of blaOXA-SHE (80.77%). Nine strains carried qnr with the most common genotype of qnrA3.
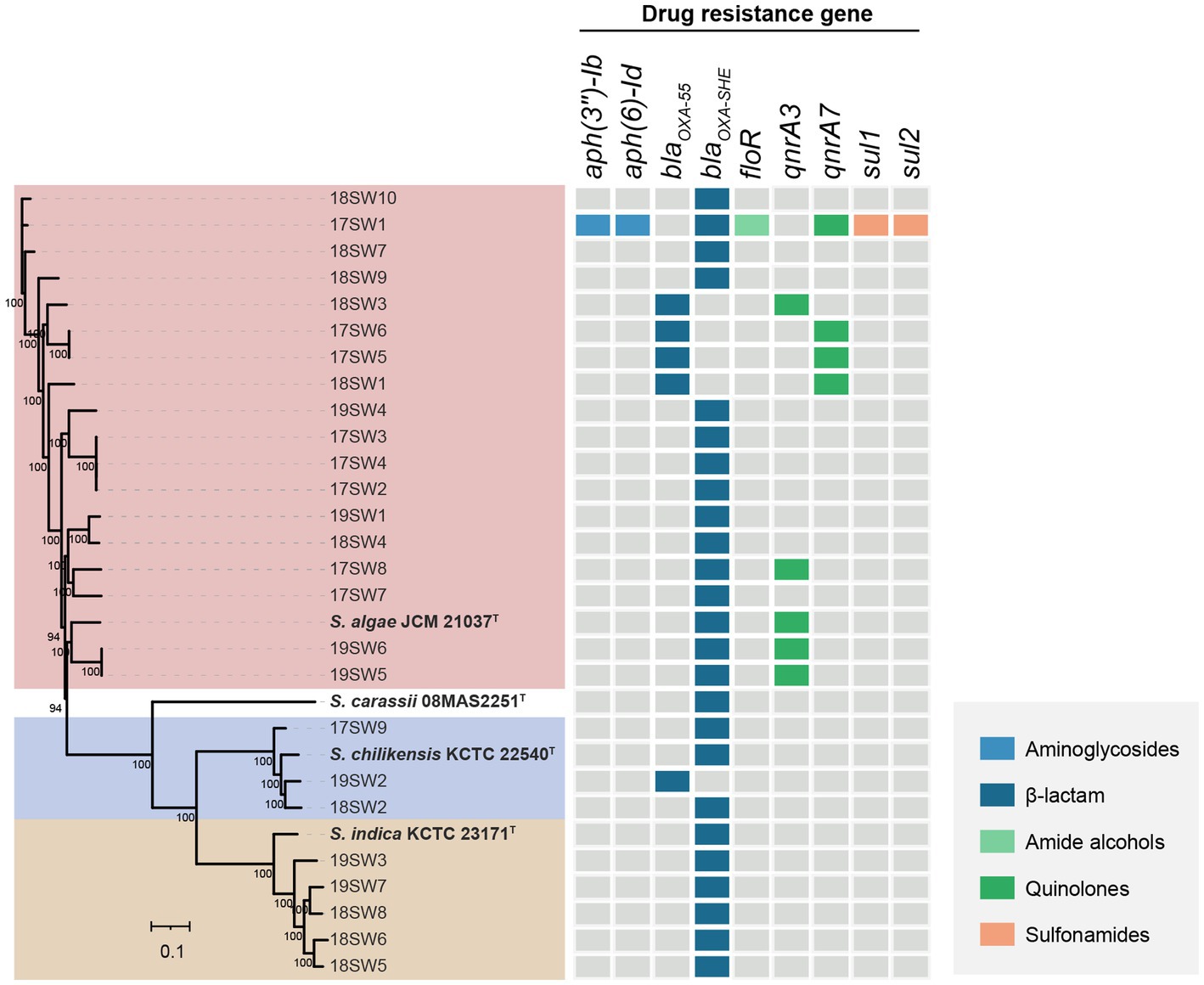
Figure 5. The heatmap of the antimicrobial resistance genes in 26 Shewanella strains. Gray color represents the presence of antimicrobial resistance gene and other colors represent the presence of different antimicrobial resistance genes.
3.7 Antibiotic susceptibility of Shewanella spp.
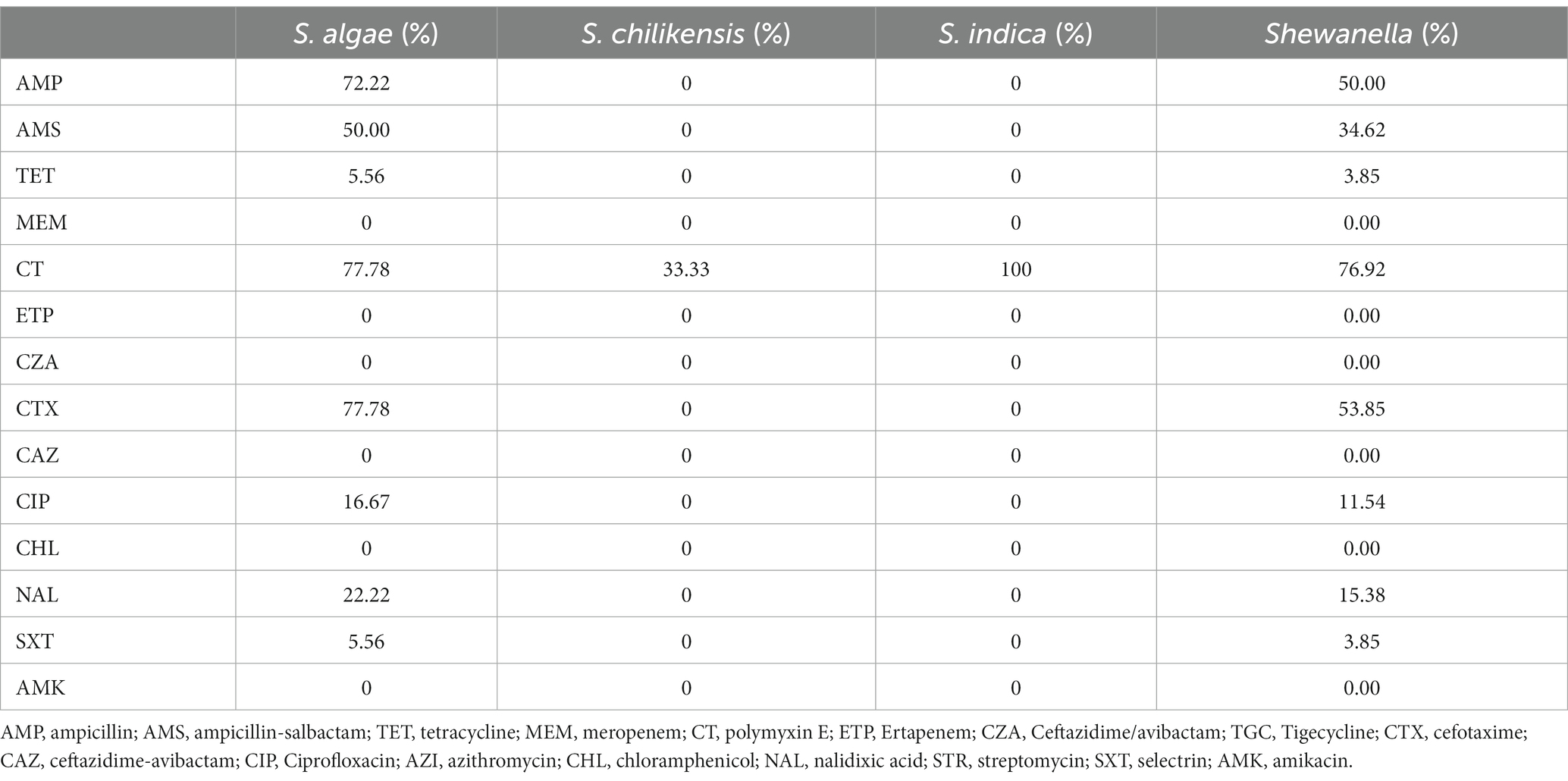
The resistance rates of 26 strains to 17 antibiotics from high to low were polymyxin E (76.92%, 20/26), cefotaxime (53.85%, 14/26), ampicillin (50%, 13/26), ampicillin-salbactam (34.62%, 9/26), nalidixic acid (15.38%, 4/26), ciprofloxacin (11.54%, 3/26), selectrin (3.85%, 1/26), and tetracycline (3.85%, 1/26). None of the strains were resistant to ceftazidime/avibactam, ceftazidime-avibactam, tigecycline, meropenem, chloramphenicol, streptomycin, ertapenem, azithromycin, or amikacin. The rate of multidrug resistance was 38.46% (10/26). There were 12 types of drug resistance spectra (Figure 6), with the dominant spectrum being polymyxin E. The antimicrobial susceptibilities of the different strains are shown in Table 3.
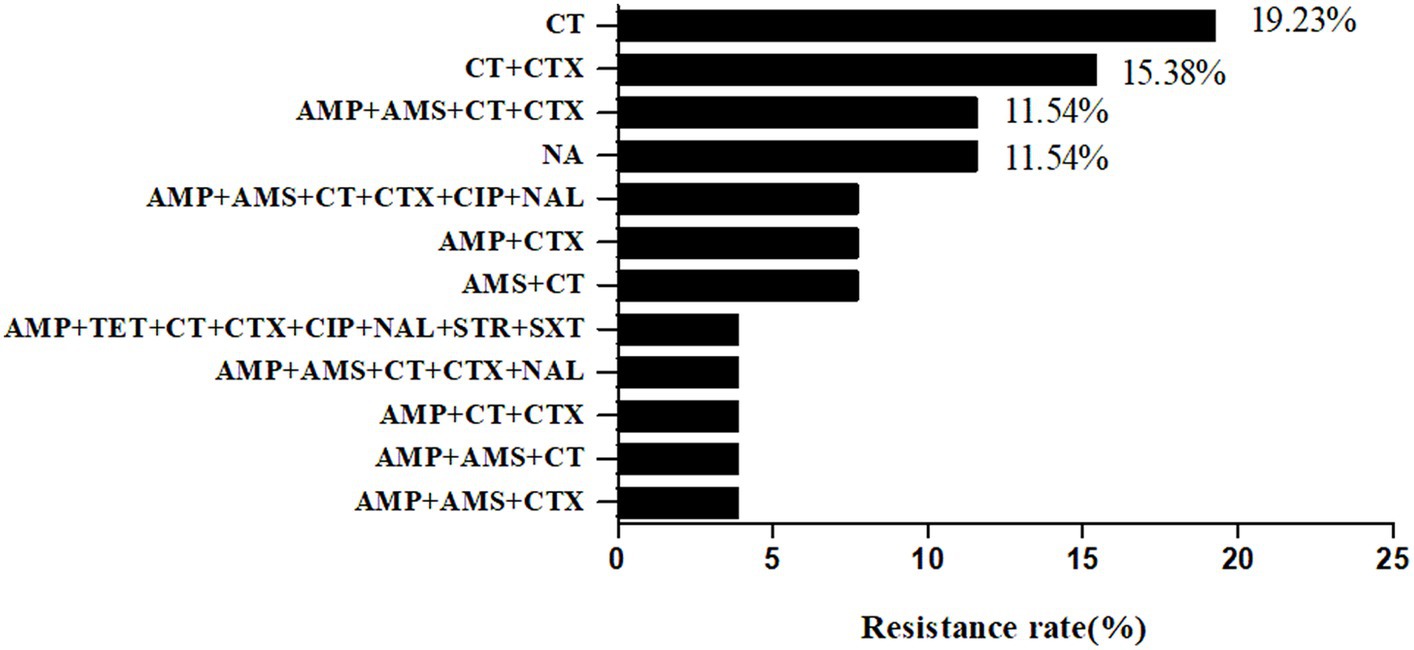
Figure 6. Antibiotic Susceptibility spectrum of 26 Shewanella strains. AMP, ampicillin; AMS, ampicillin-salbactam; TET, tetracycline; MEM, meropenem; CT, polymyxin E; ETP, Ertapenem; CZA, Ceftazidime/avibactam; TGC, Tigecycline; CTX, cefotaxime; CAZ, ceftazidime-avibactam; CIP, Ciprofloxacin; AZI, azithromycin; CHL, chloramphenicol; NAL, nalidixic acid; STR, streptomycin; SXT, selectrin; AMK, amikacin. NA, no resistance to all antibiotics.
4 Discussion
To date, more than 70 species of Shewanella have been identified.2 Several Shewanella species are opportunistic pathogens that cause infectious diseases in aquatic animals and humans. Shewanella has aroused widespread concern owing to the rapid increase in the number of identified species and the increasing reports of relevant clinical cases (Yu et al., 2022).
In this study, a long-term diarrheal disease-based surveillance program showed a detection rate of 2.36% for Shewanella. The epidemiological and clinical symptoms of Shewanella infection are similar to those of halophilic marine Vibrio species (Holt et al., 2005). Shewanella showed a clear seasonal distribution. Specifically, peak detection was concentrated in July and August (summer), whereas no detection occurred from December to April (the following year, winter), consistent with previous monitoring surveys. This may be because seafood circulation is frequent in the summer, with temperature and humidity facilitating Shewanella growth and foods being prone to cross-contamination, which leads to food-borne infections. A report from Anhui Province, China, demonstrated the isolation of S. algae from patients with food poisoning (Wang et al., 2013).
The Shewanella samples collected in this study included three main species: S. algae (69.23%), S. indica (19.23%), and S. chilikensis (11.54%). The allelic profiles of Shewanella isolates were classified into 22 STs. Among these, S. algae were classified into 16 STs, accounting for 66.7%. S. indica and S. chilikensis were classified into five and three STs, accounting for 8.8% and 7.4%, respectively. The dominant species isolated from clinical settings, such as blood, sputum, urine, and abdominal cavity effusion, were S. algae and S. putrefaciens (Yu et al., 2022). Moreover, S. xiamenensis has also been isolated from anal swab samples (Antonelli et al., 2015). S. indica and S. chilikensis are often isolated from seafood and cooked food (Wang et al., 2013), leading to an increased risk of foodborne diseases. In our study, S. algae was the dominant species detected in the genus Shewanella, which is highly associated with clinical diseases. Fang et al. (2019) showed that S. algae, S. indica, and S. chilikensis are clinically relevant with highly similar 16S rRNA sequences. Therefore, S. indica and S. chilikensis isolates may have been misidentified as S. algae in the molecular diagnosis. We sequenced the whole genome of 26 Shewanella isolates and performed higher-resolution MLST and ANI for the accurate identification of Shewanella species.
MLST is a common molecular typing method with high discriminatory power that has developed rapidly in recent years. MLST is suitable for both molecular epidemiology and evolution studies, as well as for international reference laboratories to establish a typing system for comparing strains worldwide (Perez-Losada et al., 2013; Floridia-Yapur et al., 2021). This study utilized seven previously described housekeeping genes to explore the genetic correlation of Shewanella isolates, and provide a scientific basis for studying the overall evolutionary structure and phylogenetic relationships of Shewanella. MLST classified the isolated strains of Shewanella into 22 STs, and the pathogen spectrum of Shewanella was rich and diverse, indicating that these strains have high genetic diversity. MLST is simple to use and can provide quick and easy comparisons between different laboratories. It has become a routine typing method for various bacteria, and it can be used for epidemiological monitoring and evolutionary research on Shewanella. MLST can be used as a molecular typing method for routine Shewanella surveillance, which should be increased to provide a scientific basis for the prevention and control of clinical infections.
Pan-genome analysis revealed approximately 3,000 core genomes from 18 S. algae strains. Core genome function annotation showed that the main functions were bacterial basal metabolism and biosynthesis, such as metabolism and energy production of various substances. This result is consistent with that of a core genome analysis of S. algae (Huang et al., 2022). The functions of auxiliary genomes, especially those of specific genes, are primarily related to DNA replication and repair, biofilm formation, and cell movement. Auxiliary genomes, represented by virulence islands and prophages, are one of the main driving forces of the genetic evolution and environmental adaptation of S. algae (Huang et al., 2022). S. algae has high intra-species diversity in population structure and genetic evolution, which contributes to the evolution of its pathogenic mechanism and environmental adaptability (Janda and Abbott, 2014). By exploring the relationship between the sequence diversity of virulence factors and different Shewanella strains, this study revealed differences in the composition of virulence genes among S. algae, S. indica, and S. chilikensis, indicating that the virulence spectra of the different Shewanellae species were different. However, all three Shewanella species were isolated from clinical samples, indicating that Shewanella pathogenesis may be influenced by multiple factors and may be the result of complex interactions between the host and the environment. This suggests that different Shewanella strains are initiation factors, rather than determining factors, for human diseases, and that the pathogenic process may result from the synergistic effect of multiple factors (Yu et al., 2022).
Cerbino et al. (2023) performed virulome analysis on Shewanella spp. They put forward that there is a correlation between the VAS T6SS and the S. algae lineage. T6SS may be a key virulence system that contributes to S. algae virulence. Furthermore, irgA (iron-regulated adhesin), lasB (elastase), and zmp1 (Zn-metalloprotease) homologs were detected mostly in S. algae and S. xiamenensis. Chemosensory and c-di-GMP signal transduction systems integrate environmental stimuli to modulate gene expression, including the switch from a planktonic to sessile lifestyle and pathogenicity. Alberto et al. provide an inventory of the c-di-GMP turnover proteome and chemosensory networks of S. algae. S. algae strains encoded 61–67 c-di-GMP turnover proteins and 28–31 chemoreceptors, placing S.algae near the top in terms of these signaling capacities per Mbp of genome (Cerbino et al., 2023).
Differences in antibiotic susceptibilities of clinical Shewanella isolates have been reported. However, they are usually susceptible to third- and fourth-generation cephalosporins, β-lactamase inhibitor combinations, and quinolones (Vignier et al., 2013; Yousfi et al., 2017a). Shewanella isolates in this study showed resistance to ampicillin-sulbactam (β-lactamase inhibitor combination), cefotaxime, ciprofloxacin (quinolones), and nalidixic acid (quinolones). This suggests that an increased risk of drug resistance in Shewanella may contribute to clinical treatment failure. The high resistance rate of S. algae to ampicillin in this study (72.22%, 13/18), despite its large variations in practice (Holt et al., 2005), suggests that S. algae drug resistance is more serious in this region and should be continuously monitored. Moreover, 76.92% of Shewanella isolates were resistant to polymyxin E in this study. Related studies confirmed that Shewanella spp. isolated from neonatal patients with sepsis (Charles et al., 2015) and wounds of patients bitten by cobras (Liu et al., 2014) were resistant to polymyxin E, which may be related to the different sources and regions of strain isolation. Surprisingly, the strains exhibited a polymyxin resistance phenotype yet no determinants were detected. The molecular mechanisms of polymyxin resistance has been characterized, including specific modification of outer membrane porins, reductions in the overall negative charge of the LPS, overexpression of efflux pump systems, and overproduction of capsule polysaccharide (Bialvaei and Samadi Kafil, 2015). Polymyxin resistance in Gram-negative bacteria is commonly due to decreased binding to the bacterial outer membrane because of lipopolysaccharide remodeling that is caused by changes in PhoPQ and PmrAB, both two-component regulatory systems (Zavascki et al., 2007; Lopez-Rojas et al., 2011). Acquired polymyxin resistance most often mediated by replacement of lipid A by addition of 4-amino-4-deoxy-L-arabinose (L-Ara4N) and/or phosphoethanolamine (PEtn) (Moffatt et al., 2010). Because of this effect, which is mediated by pmrC and necessitates the products of the ugd and pbg loci and ethanolamine, these alterations eliminate negative charges, decreasing the affinity of LPS and boosting resistance to polymyxins (Beceiro et al., 2011). In addition, a non-specific mechanism for the tolerance of polymyxin was shown to be up-regulation of the MexAB-OprM efflux pump (Beceiro et al., 2011). The genetic mechanisms underlying colistin resistance in Shewanella are not well understood. Huang et al. elaborated a combination of three mutations (PmrB 451, PmrE168, PmrH292) that were strongly associated with colistin resistance in S. algae (Beceiro et al., 2011). Therefore, the mechanism of polymycin resistance in Shewanella needs to be deeply explored in combination with molecular experiments and genomic information.
Compared to Vibrio parahaemolyticus isolated from patients with diarrhea in Beijing, China, which is widely distributed in the marine environment and is halophilic, Shewanella spp. showed a high level of resistance to various antibiotics. In addition, one S. algae isolate in this study also showed combined resistance to ampicillin + tetracycline + polymyxin E + cefotaxime + ciprofloxacin + nalidixic acid + streptomycin + cotrimoxazole, suggesting that S. algae may be prone to carrying resistance elements that can become carriers of resistance genes or even undergo horizontal transfer. In 2017, S. xiamenensis isolated from hospital wastewater had several drug resistance genes that were resistant to trimethoprim, aminoglycosides, quaternary ammonium compounds, β-lactams, chloramphenicol, sulfonamides, and tetracyclines (Yousfi et al., 2017b). Due to the widespread use of antibiotics, Shewanella is at risk of becoming a superbacterium.
Data availability statement
The data presented in this study are deposited in the NCBI database under accession number PRJNA1015006.
Ethics statement
The studies involving humans were approved by The Ethical committee of Beijing Center for Disease Prevention and Control. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from primarily isolated as part of our previous study for which ethical approval was obtained. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
YK: Writing – original draft, Data curation, Formal analysis, Visualization, Validation. KY: Data curation, Writing – original draft, Software. ZH: Data curation, Writing – original draft, Software. BP: Methodology, Writing – review & editing. SL: Writing – original draft, Investigation, Validation. TP: Writing – original draft, Investigation, Validation. YL: Writing – review & editing, Conceptualization, Methodology, Supervision. DW: Writing – review & editing, Conceptualization, Funding acquisition, Resources.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Science and Technology Fundamental Resources Investigation Program of China (2021FY100904).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1293577/full#supplementary-material
Footnotes
References
Antonelli, A., Di Palo, D. M., Galano, A., Becciani, S., Montagnani, C., Pecile, P., et al. (2015). Intestinal carriage of Shewanella xiamenensis simulating carriage of OXA-48-producing Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 82, 1–3. doi: 10.1016/j.diagmicrobio.2015.02.008
Beceiro, A., Llobet, E., Aranda, J., Bengoechea, J. A., Doumith, M., Hornsey, M., et al. (2011). Phosphoethanolamine modification of lipid a in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55, 3370–3379. doi: 10.1128/AAC.00079-11
Bialvaei, A. Z., and Samadi Kafil, H. (2015). Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 31, 707–721. doi: 10.1185/03007995.2015.1018989
Cerbino, G. N., Traglia, G. M., Ayala Nunez, T., Parmeciano Di Noto, G., Ramirez, M. S., Centron, D., et al. (2023). Comparative genome analysis of the genus Shewanella unravels the association of key genetic traits with known and potential pathogenic lineages. Front. Microbiol. 14:1124225. doi: 10.3389/fmicb.2023.1124225
Charles, M. V., Srirangaraj, S., and Kali, A. (2015). Neonatal sepsis caused by Shewanella algae: a case report. Australas. Med. J. 8, 64–66. doi: 10.4066/AMJ.2015.2292
Chia-Wei, L., Cheng, J. F., Tung, K. C., Hong, Y. K., Lin, J. H., Lin, Y. H., et al. (2022). Evolution of trimethoprim/sulfamethoxazole resistance in Shewanella algae from the perspective of comparative genomics and global phylogenic analysis. J. Microbiol. Immunol. Infect. 55, 1195–1202. doi: 10.1016/j.jmii.2021.09.014
Dey, S., Bhattacharya, D., Roy, S., Nadgir, S. D., Patil, A., and Kholkute, S. D. (2015). Shewanella algae in acute gastroenteritis. Indian J. Med. Microbiol. 33, 172–175. doi: 10.4103/0255-0857.148442
Fang, Y., Wang, Y., Liu, Z., Dai, H., Cai, H., Li, Z., et al. (2019). Multilocus sequence analysis, a rapid and accurate tool for taxonomic classification, evolutionary relationship determination, and population biology studies of the genus Shewanella. Appl. Environ. Microbiol. 85, e03126–e03118. doi: 10.1128/AEM.03126-18
Fang, Y., Wang, Y., Liu, Z., Lu, B., Dai, H., Kan, B., et al. (2017). Shewanella carassii sp. nov., isolated from surface swabs of crucian carp and faeces of a diarrhoea patient. Int. J. Syst. Evol. Microbiol. 67, 5284–5289. doi: 10.1099/ijsem.0.002511
Floridia-Yapur, N., Rusman, F., Diosque, P., and Tomasini, N. (2021). Genome data vs MLST for exploring intraspecific evolutionary history in bacteria: much is not always better. Infect. Genet. Evol. 93:104990. doi: 10.1016/j.meegid.2021.104990
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Holt, H. M., Gahrn-Hansen, B., and Bruun, B. (2005). Shewanella algae and Shewanella putrefaciens: clinical and microbiological characteristics. Clin. Microbiol. Infect. 11, 347–352. doi: 10.1111/j.1469-0691.2005.01108.x
Huang, Z., Yu, K., Fu, S., Xiao, Y., Wei, Q., and Wang, D. (2022). Genomic analysis reveals high intra-species diversity of Shewanella algae. Microb. Genom. 8:000786. doi: 10.1099/mgen.0.000786
Huang, Z., Yu, K., Wang, Y., Li, Y., Gao, H., Bai, X., et al. (2023). Distribution and molecular characterization of Shewanella in China. Disease Surve. 38:384. doi: 10.3784/jbjc.202212300556
Hyatt, D., Chen, G. L., Locascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Janda, J. M., and Abbott, S. L. (2014). The genus Shewanella: from the briny depths below to human pathogen. Crit. Rev. Microbiol. 40, 293–312. doi: 10.3109/1040841X.2012.726209
Kouzuma, A., Kasai, T., Hirose, A., and Watanabe, K. (2015). Catabolic and regulatory systems in Shewanella oneidensis MR-1 involved in electricity generation in microbial fuel cells. Front. Microbiol. 6:609. doi: 10.3389/fmicb.2015.00609
Lee, I., Ouk Kim, Y., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Liu, P. Y., Lin, C. F., Tung, K. C., Shyu, C. L., Wu, M. J., Liu, J. W., et al. (2013). Clinical and microbiological features of shewanella bacteremia in patients with hepatobiliary disease. Intern. Med. 52, 431–438. doi: 10.2169/internalmedicine.52.8152
Liu, P. Y., Shi, Z. Y., Shyu, C. L., Wu, Z. Y., Lai, K. L., Chang, C. Y., et al. (2014). Cobra bite wound infection caused by Shewanella algae. Int. J. Infect. Dis. 20, 11–12. doi: 10.1016/j.ijid.2013.08.014
Lopez-Rojas, R., Dominguez-Herrera, J., McConnell, M. J., Docobo-Perez, F., Smani, Y., Fernandez-Reyes, M., et al. (2011). Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J. Infect. Dis. 203, 545–548. doi: 10.1093/infdis/jiq086
Martin-Rodriguez, A. J., Higdon, S. M., Thorell, K., Tellgren-Roth, C., Sjoling, A., Galperin, M. Y., et al. (2022). Comparative genomics of cyclic di-GMP metabolism and chemosensory pathways in Shewanella algae strains: novel bacterial sensory domains and functional insights into lifestyle. Regulation 7:e0151821. doi: 10.1128/msystems.01518-21
Moffatt, J. H., Harper, M., Harrison, P., Hale, J. D., Vinogradov, E., Seemann, T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977. doi: 10.1128/AAC.00834-10
Perez-Losada, M., Cabezas, P., Castro-Nallar, E., and Crandall, K. A. (2013). Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infect. Genet. Evol. 16, 38–53. doi: 10.1016/j.meegid.2013.01.009
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Thorell, K., Meier-Kolthoff, J. P., Sjöling, Å., and Martín-Rodríguez, A. J. (2019). Whole-genome sequencing redefines Shewanella taxonomy. Front. Microbiol. 10:1861. doi: 10.3389/fmicb.2019.01861
Vignier, N., Barreau, M., Olive, C., Baubion, E., Theodose, R., Hochedez, P., et al. (2013). Human infection with Shewanella putrefaciens and S. algae: report of 16 cases in Martinique and review of the literature. Am. J. Trop. Med. Hyg. 89, 151–156. doi: 10.4269/ajtmh.13-0055
Wang, D., Wang, Y., Huang, H., Lin, J., Xiao, D., and Kan, B. (2013). Identification of tetrodotoxin-producing Shewanella spp. from feces of food poisoning patients and food samples. Gut Pathog. 5:15. doi: 10.1186/1757-4749-5-15
Yarza, P., Yilmaz, P., Pruesse, E., Glockner, F. O., Ludwig, W., Schleifer, K. H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. doi: 10.1038/nrmicro3330
Yiallouros, P., Mavri, A., Attilakos, A., Moustaki, M., Leontsini, F., and Karpathios, T. (2013). Shewanella putrefaciens bacteraemia associated with terminal ileitis. Paediatr. Int. Child. Health 33, 193–195. doi: 10.1179/2046905512Y.0000000045
Yonglu, W., Duochun, W., Shengwei, Z., Jinxiu, Z., Yan, L., Yong, T., et al. (2009). Isolation and characterization of Shewanella spp.from patients of food poisoning. Chin. J. Epidemiol. 8, 836–840. doi: 10.3760/cma.j.issn.0254-6450.2009.08.018
Yousfi, K., Bekal, S., Usongo, V., and Touati, A. (2017a). Current trends of human infections and antibiotic resistance of the genus Shewanella. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1353–1362. doi: 10.1007/s10096-017-2962-3
Yousfi, K., Touati, A., Lefebvre, B., Fournier, E., Cote, J. C., Soualhine, H., et al. (2017b). A novel plasmid, pSx1, harboring a new Tn1696 derivative from extensively drug-resistant Shewanella xiamenensis encoding OXA-416. Microb. Drug Resist. 23, 429–436. doi: 10.1089/mdr.2016.0025
Yu, K., Huang, Z., Xiao, Y., and Wang, D. (2022). Shewanella infection in humans: epidemiology, clinical features and pathogenicity. Virulence 13, 1515–1532. doi: 10.1080/21505594.2022.2117831
Zavascki, A. P., Goldani, L. Z., Li, J., and Nation, R. L. (2007). Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60, 1206–1215. doi: 10.1093/jac/dkm357
Keywords: Shewanella, prevalence, molecular characteristics, diarrhea, antibiotic susceptibility
Citation: Kang Y, Yu K, Huang Z, Pang B, Liu S, Peng T, Li Y and Wang D (2024) Prevalence and molecular characteristics of Shewanella infection in diarrhea patients in Beijing, China 2017–2019. Front. Microbiol. 15:1293577. doi: 10.3389/fmicb.2024.1293577
Edited by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyReviewed by:
Miklos Fuzi, Independent Researcher, Budapest, HungaryYadong Zhang, Harvard Medical School, United States
Copyright © 2024 Kang, Yu, Huang, Pang, Liu, Peng, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Li, liying19830805@126.com; Duochun Wang, wangduochun@icdc.cn
†These authors have contributed equally to this work
 Ying Kang
Ying Kang Keyi Yu3†
Keyi Yu3† Bo Pang
Bo Pang Duochun Wang
Duochun Wang