- 1Department of Industrial Biotechnology, Institute of Biotechnology, University of Gondar, Gondar, Ethiopia
- 2Department of Biotechnology, University of Gondar, Gondar, Ethiopia
- 3Department of Industrial Biotechnology, Institute of Biotechnology, University of Gondar, Gondar, Ethiopia
Amino acid depletion therapy is a promising approach for cancer treatment. It exploits the differences in the metabolic processes between healthy and cancerous cells. Certain microbial enzymes induce cancer cell apoptosis by removing essential amino acids. L-asparaginase is an enzyme approved by the FDA for the treatment of acute lymphoblastic leukemia. The enzymes currently employed in clinics come from two different sources: Escherichia coli and Erwinia chrysanthemi. Nevertheless, the search for improved enzymes and other sources continues because of several factors, including immunogenicity, in vivo instability, and protease degradation. Before determining whether L-asparaginase is clinically useful, research should consider the Michaelis constant, turnover number, and maximal velocity. The identification of L-asparaginase from microbial sources has been the subject of various studies. The primary goals of this review are to explore the most current approaches used in the search for therapeutically useful L-asparaginases and to establish whether these investigations identified the crucial characteristics of L-asparaginases before declaring their therapeutic potential.
Introduction
Amino acid deprivation is an approach that shows promise for the development of novel cancer therapies. This natural remedy relies on the difference in metabolic processes between healthy and cancerous cells (Dhankhar et al., 2020). Because of their rapid growth, cancer cells produce fewer enzymes overall, which leads to auxotrophy for a subset of amino acids and makes them a target for enzymes that deplete amino acids. Therapeutic enzymes can only suppress tumor cells by cutting off amino acids, since normal cells can make their own. Amino acid deprivation therapy requires the use of certain enzymes, such as L-asparaginase, arginine deiminase, methionine, lysine oxidase, glutaminase, and phenylalanine ammonia-lyase (Dhankhar et al., 2020). L-asparaginases were first discovered as anticancer enzymes in 1922 (El-fakharany et al., 2020). The 22nd World Health Organization List of Essential Medicines now includes L-asparaginase as a cytotoxic therapy for acute lymphoblastic leukemia (WHO, 2021).
Leukemia is usually a cancer that causes an overabundance of aberrant white blood cells (blasts) in the bone marrow. Lymphoblastic leukemia affects lymphocytes that make up the lymph tissues 4. Acute lymphoblastic leukemia (ALL) is characterized by unchecked lymphocyte immature growth. L-asparaginase is one of the most widely used medications for treating ALL (Díaz-Barriga et al., 2021).
Antineoplastic drug successfully treated acute lymphoblastic leukemia (Radadiya et al., 2020; Sobat et al., 2020). L-asparaginase converts L-asparagine into aspartic acid and ammonia. When administered systemically, L-asparaginase (L-ASNase) reduces the availability of L-asparagine and prevents cancer cells from multiplying quickly and with a prime need for exogenous asparagine (van Trimpont et al., 2022). Lymphoblasts do not express asparagine synthetase (ASNS); hence, these cells must take extracellular asparagine to survive.
The L-asparaginases from Escherichia coli and Erwinia chrysanthemi have been used extensively in medicine, but growing complications like hypersensitivity, antigenicity, short half-life, temporary blood clearance, and unfavorable L-glutaminase-dependent neurotoxicity require ongoing research to find more suitable alternatives (Nguyen et al., 2018; Radadiya et al., 2020; Sobat et al., 2020). Problems with commercial L-ASNases have prompted research into better sources of the enzyme. This article summarizes the bacterial and fungal based L-asparaginases that are medically significant.
L-asparaginases in the modern clinics
Today’s clinics use five distinct L-ASNase preparations (van Trimpont et al., 2022). The two ansB gene products from E. chrysanthemi are in the market under the trade names Erwinase and Rylaze. The ansB gene from E. coli was the source of the remaining three. Two of the three type II E. coli ASNases are stabilized formulations produced by the covalent conjugation of monomethoxy polyethylene glycol (PEG) to lysine on the enzyme via a succinimidyl succinate linker (Oncaspar) or succinimidyl carbonate linker (Asparlas). One of the three type II E. coli ASNases is a native formulation (Elspar and Kidrol). The addition of a PEG tag to L-ASNases prolongs their half-life and delays their removal from the body (van Trimpont et al., 2022).
The primary treatment for most ALL patients is PEG-native E. coli L-ASNase (EcA); unfortunately, many patients experience hypersensitivity reactions. This hypersensitivity may result from either a negative immunological response to the enzyme or the formation of neutralizing antibodies (often known as silent inactivation; Modi and Gervais, 2022). Native E. chrysanthemi L-ASNase (ErA) differs immunologically from PEG-native EcA and does not react with PEG-nEcA-derived antibodies. Therefore, patients who acquired an allergy to PEG-native EcA were administered native ErA (or, starting in July 2021, recombinant ErA) as second-line treatment (Modi and Gervais, 2022).
L-asparaginases important in clinics
Type-I L-asparaginase, which has a low affinity for asparagine, is encoded by ansA in E. coli, whereas type-II L-asparaginase, which has a higher affinity for asparagine, is encoded by ansB. Acute lymphoblastic leukemia treatment employs type-II enzymes (Vimal and Kumar, 2017). The activity of an enzyme is often determined by adding a substrate to L-asparaginase and quantifying the amount of the output produced. While this measurement is a useful indicator for comparing activity between multiple enzymes within the same study, it is not always possible to compare this type of measurement to those from other studies. It is challenging to compare results from different research organizations because experiment-to-experiment variations in temperature, pH, and substrate quantity exist. In order to truly understand the therapeutic potential of a novel L-asparaginase, the kinetic parameters offer better insights (Beckett and Gervais, 2019).
The kinetic parameters provide insight into substrate affinity and turnover, which are unconditional values that can be consistently compared between study groups, and are essential for fully appreciating the therapeutic potential of a novel L-asparaginase. Before making claims regarding the therapeutic potential of a novel enzyme, Beckett and Gervais (2019) state that, at the very least, Michaelis constant (Km), turnover number (Kcat), and maximum velocity (Vmax) should be established. However, many of the articles published in recent years lack this information (Abdelrazek et al., 2019; Kumar et al., 2019; Moguel et al., 2020; Prakash et al., 2020; Chakraborty and Shivakumar, 2021). In this review, few L-asparaginases from bacterial and fungal origin are reviewed, along with their corresponding kinetic properties and other characteristics.
Michaelis constant (Km value)
One requirement for the therapeutic effectiveness of L-asparaginases is the Km value. Michaelis constant (Km) is the amount of a substrate at which the reaction rate is half of the maximum reaction rate. A lower Km value shows that the enzyme is more active toward that substrate and can perform at half its maximum rate at lower substrate concentrations (Baral et al., 2020). For L-asparagine to completely disappear from circulation, a micromolar Km is needed.
The Km value is a quantifiable measure of the enzyme’s affinity for a substrate; the lower the Km value, the greater the affinity of the enzyme for that substrate, and, in a therapeutic setting, the more effective the enzyme (Beckett and Gervais, 2019; Baral et al., 2020). Because intravenous L-asparaginase must be able to remove circulating pools of L-asparagine, which are not high (approximately 50 M), a low Km (at least micromolar) of L-asparaginase is necessary (Loch and Jaskolski, 2021).
Turnover number (Kcat)
Turnover number, often known as Kcat, is another helpful indicator for assessing enzyme activity. It provides the number of substrates that a single enzyme molecule converts into a product per unit of time (Baral et al., 2020). The substrate fully occupied the catalytic sites of the enzyme at the maximum reaction speed (Vmax). Vmax is the amount of substrate that an enzyme can convert into a product in a second. The turnover number (Kcat) is therefore the Vmax divided by the enzyme concentration. The rate of transforming substrates into products increases as Kcat increases. A good therapeutic L-asparaginase must have a low Km value and a high Kcat value to lower endogenous asparagine levels from 40 to 80 M to 0.02 M (Beckett and Gervais, 2019). Oncaspar and Erwinase, the two L-asparaginases that are currently on the market, have asparagine Km and Kcat values of roughly 0.05 mM and 200–560 s−1, respectively (Beckett and Gervais, 2019; Baral et al., 2020).
Glutaminase coactivity
The literature contains conflicting information regarding the secondary glutaminase activity of L-ASNases. From the FDA (United States)-approved E. chrysanthemi L-asparaginase, Nguyen et al. (2018) created a novel low glutaminase enzyme that was highly effective against T and B cell ALL (Nguyen et al., 2018). However, Chan et al. (2019) examined both the wild-type ASNase and its glutaminase-deficient mutant. They discovered that L-ASNase glutaminase activity, rather than ASNase activity alone, contributed to substantial anticancer efficacy against xenografts of the ASNS-negative leukemia cell line in NSG mice (Chan et al., 2019). This finding implies that, even against ASNS-negative cancer types, long-lasting, single-agent anticancer efficacy in vivo requires ASNase glutaminase activity. Horvath et al. (2019) investigated the pharmacodynamics of ASNase in a mouse model of cancer. They showed that asparagine was detected after L-ASNase treatment, in contrast to previously published methods that produced post-ASNase asparagine levels below the detection threshold (Horvath et al., 2019). They ascribed these differences to the use of whole blood, which collects the target analyte in both the red blood cell and plasma, as opposed to only serum or plasma.
A different study by Nguyen et al. (2017) suggested that a single-point mutation controls the substrate selection of L-asparaginases. They claimed that whereas the L-glutaminase activities of Wolinella succinogenes L-asparaginase (WoA) with proline at position 121 (WoA-P121) and WoA with serine at position 121 (WoA-S121) were noticeably different, their kinetic characteristics were identical. L-glutaminase activity is present in the WoA variant with a proline at position 121 but not in WoA-S121 (Nguyen et al., 2017). Nguyen et al. (2017) concluded that the residue at position 121 controls Asn vs. Gln selectivity via a mechanism other than substrate binding, which calls for further research.
Toxicity
L-asparaginase’s toxicity is caused by either a secondary L-glutaminase activity or a hypersensitivity to the foreign protein (Baral et al., 2020; Fonseca et al., 2021; van Trimpont et al., 2022). When the body perceives an enzyme as foreign, it triggers an immunological reaction that results in a minor allergic reaction to anaphylactic shock (Baral et al., 2020). Glutaminase coactivity results in the hydroxylation of glutamine to glutamate and ammonia. Excessive ammonia generation may cause neurotoxicity and liver malfunction (van Trimpont et al., 2022). Ashok et al. (2019) eliminated toxicity caused by E. coli and E. chrysanthemi L-ASNases by purifying the urease (Ashok et al., 2019). Urease catalyzes the hydrolysis of urea, which results in the production of ammonia and carbon dioxide. As a result, ammonia is excreted from the bloodstream as urea is converted back to ammonia, resulting in hyperammonemia, a condition that damages the brain (Ashok et al., 2019). Chakraborty and Shivakumar (2021) also described a glutaminase and urease-free L-asparaginase from Agaricomycete sp. Ganoderma australe GPC191 (Chakraborty and Shivakumar, 2021). The urease activity of L-ASNases was not, however, reported in many of the studies.
The need for exogenous sources of L-asparaginases
Humans possess a type III L-asparaginase that needs a threonine residue at the N-terminus to catalyze reactions. Unfortunately, because of its millimolar Km value for L-asparagine, natural human L-asparaginase is not suitable for therapeutic uses. L-asparaginases must have a micromolar Km for asparagine in order to remove it from the blood (Rigouin et al., 2017). To get rid of the circulating L-asparagine in ALL patients, type II L-asparaginases from E. coli or E. chrysanthemi are utilized (Radadiya et al., 2020; Andrade et al., 2021). Wild type II L-asparaginases exhibit high selectivity for L-asparagine and low glutaminase side activity (2%–10%).
Kinetic parameters of microbial L-asparaginases in recent studies
L-asparaginase is a therapeutic enzyme used to treat cancer. The major obstacles to the therapeutic potential of this enzyme are immunogenicity, short plasma half-life, and glutaminase activity (Vidya et al., 2017). Several research and development projects are underway to address these problems. To identify a novel therapeutic L-asparaginase, studies must consider Vmax, Km, and Kcat values comparable to or superior to those of current therapeutic therapies (Beckett and Gervais, 2019). Plants, animals, and microbes naturally produce L-asparaginases. Compared to plant and animal sources, microbes are more cost-effective and easier to modify, optimize, extract, and purify for enzyme synthesis (Fazeli et al., 2021). In addition, genetically altering microbes to boost yields is simple (Costa-Silva et al., 2019). Because bacterial and fungal L-asparaginases are thought to be important in both medicine and industry, this review focuses on their kinetic properties.
Kinetic parameters of bacterial L-asparaginases
Although other bacterial species can produce L-asparaginases, only type-II L-asparaginases from E. coli and E. chrysanthemi have been produced on an industrial scale (Vimal and Kumar, 2017). There are two forms of bacterial L-asparaginases: intracellularly produced type I, or cytosolic, and extracellularly created type II, or periplasmic (El-fakharany et al., 2020). Most bacteria produce type I, or cytosolic, L-asparaginases within their cells, but a few secrets type II, or periplasmic, L-asparaginases outside. Since gram-positive bacteria do not have a periplasmic gap, they secrete much more enzymes into the environment than gram-negative bacteria (Vimal and Kumar, 2017).
The production of extracellularly secreted enzymes is more beneficial for industrial processes because these can be made in large quantities in a culture of ideal conditions, and it takes less money and effort to purify them (Vimal and Kumar, 2017; El-fakharany et al., 2020). As a result, it would be more profitable to test for extracellular enzymes in gram-positive bacteria. The type II bacterial L-asparaginases are the focus of this review since they are crucial for clinical outcomes. Table 1 shows bacterial strains that produce L-ASNase, along with their characteristics. The review includes studies from 2017 to present and reported the kinetic property of L-ASNases. According to the data in Table 1, Bacillus velezensis, Streptomyces brollosae NEAE-115, and Bacillus halotolerans OHEM18 have low Km values as compared to commercial L-ASNases, signifying that they might be potential candidates. L-ASNase from Bacillus halotolerans OHEM18 also showed an antioxidant activity against 2, 2′-azino-bis [3-ethylbenzothiazoline-6-sulfonic acid (ABTS) and 2, 2′-diphenyl-1-picrylhydrazyl (DPPH) radicals (Table 1)].
Kinetic parameters of fungi-derived L-asparaginases
Bacterial sources of L-asparaginase have been linked to a variety of immunological reactions, including hypersensitivity, irregular clotting, and allergic reactions. To reduce these immunological issues, a different supply is required. Fungi have evolved more closely with humans than bacteria, so it is likely that the enzymes they produce will have lower immunogenicity than bacteria (Ashok et al., 2019; Costa-Silva et al., 2019).
It has been claimed that a different source of L-asparaginases with fewer negative effects comes from fungi. Fungal species can be employed more successfully than other microbes to cure cancer because of their eukaryotic origin and their ability to reproduce the actions of human cells (Ashok et al., 2019). There may be fewer negative effects because yeast ASNase is more similar to human congeners (Karla et al., 2020). A variety of yeast genera, including Candida, Pichia, Rhodosporidium, Saccharomyces, and Yarrowia, have been reported to generate ASNase (Karla et al., 2020). Owing to appropriate post-translational protein modification and glycosylation, yeasts are good eukaryotic candidates for the production of asparaginase (Darvishi et al., 2018). The kinetic characteristics of L-asparaginases from fungi, as reported in recent literature, are shown in Table 2. According to Table 2, Aspergillus terreus, a marine sediment isolate, had a significantly high Km compared to E. coli and E. chrysanthemi. Due to its kinetic characteristics, Km = 9.37 M, Vmax = 127 M/ML/min, and specific activity of 468.03 U/mg, the plant parasitic fungus Lasiodiplodia theobromae appears to be a suitable candidate for L-ASNase synthesis (Table 3).
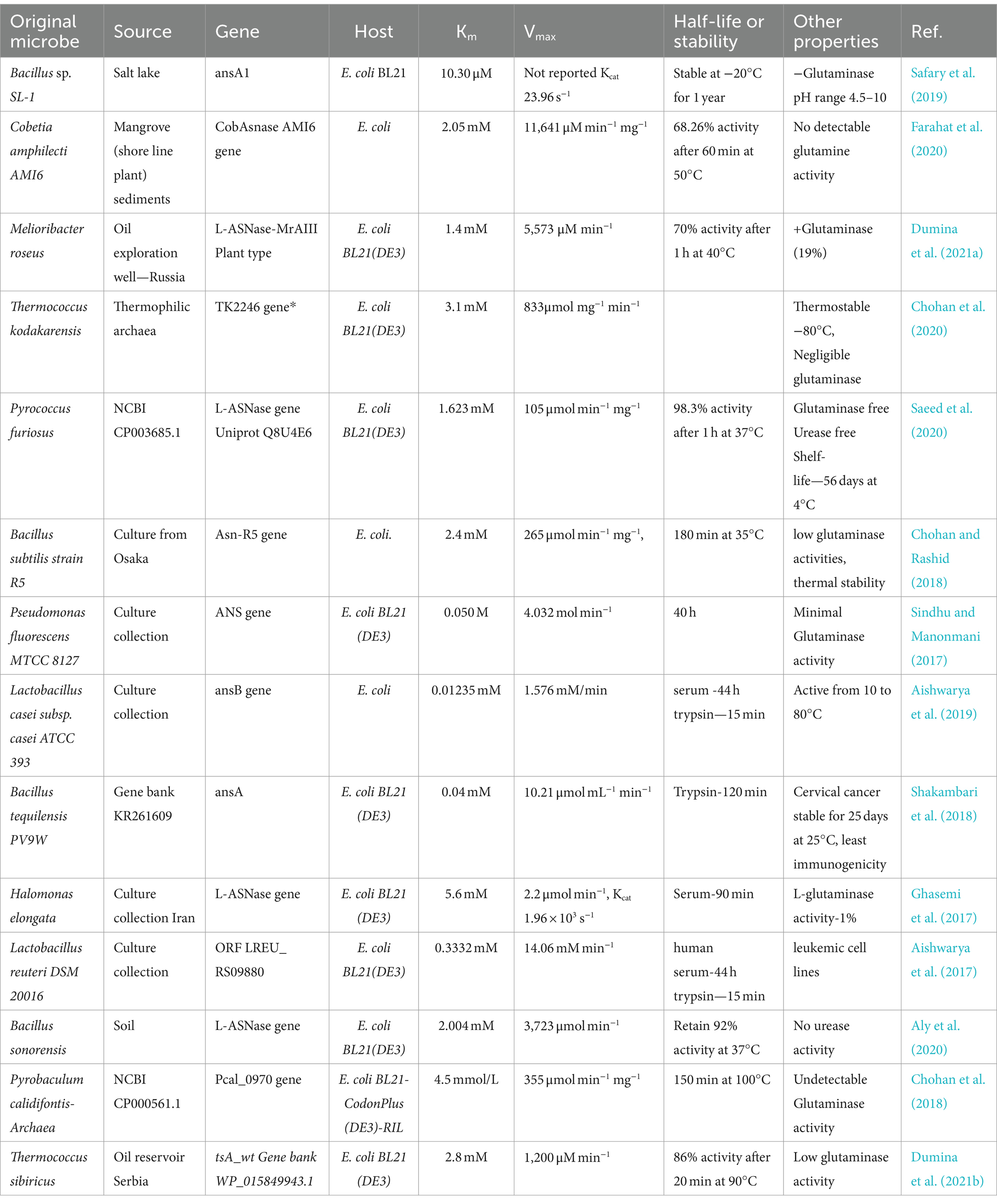
Table 2. The table lists recent studies that include recombinant microbial L-asparaginases and their accompanying characteristics.
The search for L-asparaginase is still ongoing
The E. chrysanthemi (ErAII) enzyme is only utilized when there are immunological reactions to E. coli-derived ASNases, and E. coli (EcAII) type II ASNase, whether in its native or PEGylated form, is currently the medication of choice. However, these L-ASNases have a high degree of in vivo instability, a short half-life, and require many administrations to be therapeutically effective (Maggi et al., 2017). Additionally, the immunological response, proteolytic degradation, and toxic side effects of type II bacterial L-ASNases limit their clinical usage (Radadiya et al., 2020). Numerous trials have been conducted worldwide in an attempt to reduce these problems.
Different strategies have been used to produce L-asparaginases with the best qualities for medicinal use. Recombinant ASNase synthesis, enzyme encapsulation, structural alteration of the enzyme, and the hunt for new ASNase-producing microbes are some of these methods (Sobat et al., 2020; Belén et al., 2021; Díaz-Barriga et al., 2021). L-asparaginase is found throughout all spheres of life; thus, a variety of sources, including bacteria, fungi, plants, and mammals, have been screened to identify enzymes with superior characteristics (Sobat et al., 2020).
Finding new L-asparaginases from new sources
Extreme conditions, such as salinity or high temperature, which cause the inactivation of enzymes isolated from terrestrial microorganisms, are widely used to produce enzymatic operations on an industrial scale. The marine environment attracts special interest in the search for new sources for economically significant products due to its extraordinary diversity and harsh circumstances (Alrumman et al., 2019; Mostafa et al., 2019; Sharma et al., 2019; Sobat et al., 2020; Bhargavi and Madhuri, 2021). Bacteria that are halotolerant or halophilic often express and synthesize enzymes that can withstand extreme conditions, such as high temperatures, high salt concentrations, the presence of organic solvents, pH, and non-physiological values (Dumina et al., 2020; El-fakharany et al., 2020). Given that these enzymes are naturally exposed to large levels of osmolytes, such as NaCl or other suitable solutes, it would make sense to anticipate enhanced biological activity and/or osmolarity tolerance in physiological circumstances of blood serum (Ghasemi et al., 2017).
A new L-asparaginase from the hyperthermophilic archaeon Thermococcus sibiricus has been described by Dumina et al. (2021a). They cloned the L-ASNase gene of Thermococcus sibiricus into E. coli and studied its kinetics. The findings indicated a good contender, with Km and Vmax for the enzyme being 2.8 mM and 1,200 M/min, respectively (Dumina et al., 2021b). El-fakharany et al. (2020) discovered Bacillus halotolerans OHEM18, which can produce L-ASNases with Km = 0.0047 M and Vmax = 92.74 2. This strain makes an essential extracellular L-ASNase, which is crucial for industry. In addition, Chakravarty et al. (2021) found Bacillus australimaris NJB19 in oceanic sediments and cloned the ASNase genes into E. coli to produce enzyme (Chakravarty et al., 2021). They discovered that the enzyme was effective against leukemic cells and had high affinity for asparagine. Tables 2, 3 show the results of several other investigations into the search for novel L-ASNases.
Reduced or absent co-activity in L-ASNase variants
The glutaminase co-activity of L-ASNase therapy has been implicated in the majority of its non-immune-related severe side effects. Thus, a decrease in glutaminase coactivity might significantly enhance the toxicity profile of L-ASNase. Alrumman et al. (2019) isolated Bacillus licheniformis from the Red Sea, which produces a glutaminase-free ASNase and may be a future candidate for pharmaceutical use as an anticancer medication 34. Ashok et al. (2019) screened fungal species from Antarctic soil and moss that generate L-asparaginase devoid of glutaminase and urease (Ashok et al., 2019). Glutaminase-free L-ASNase has been tested in numerous investigations from various sources (El-naggar et al., 2018; Ashok et al., 2019; Mostafa et al., 2019; Safary et al., 2019; Karla et al., 2020; Prakash et al., 2020; Saeed et al., 2020; Prihanto et al., 2022). Further studies are necessary because there is still controversy regarding the coactivity of L-glutaminase asparaginase.
Encapsulation
Encapsulation is a promising and innovative strategy to improve the in vivo performance of ASNase because it prevents the enzyme from coming into direct contact with the environment, shields it from protease degradation, extends the enzyme’s catalytic half-life, and, in some cases, lowers immunogenicity (Villanueva-flores et al., 2021; Guimarães et al., 2022). Shakambari et al. (2018) encapsulated L-ASNase from Bacillus tequilensis PV9W in solid lipid particles made from palmitic acid (Shakambari et al., 2018). In comparison to the natural L-asparaginase, the lipid particle-encapsulated version had increased Vmax (7.790.34 to 10.211.43), a Km that was cut in half (0.0700.01 to 0.040.001), a better half-life (50.19 to 120.56 min), and was stable for 25 days when stored at 25°C (Shakambari et al., 2018).
Encapsidation in a scaffold or substrate is another highly effective method to enhance the pharmacodynamics and pharmacokinetics of ASNase. ASNase II from E. coli was genetically engineered by Dáz-Barriga and colleagues into virus-like proteins of the bacteriophage P22. According to their findings, the encapsulated ASNase had a 15-fold and a 2-fold increase in Km and Kcat compared to the unbound enzyme (Díaz-Barriga et al., 2021). These virus-like particles are made up of numerous copies of self-assembling proteins, but because they lack genetic material, they are unable to spread diseases. Their self-assembly abilities enable the development of a nanoparticle that is identical to the original viral capsid, except that it does not contain genetic material and instead includes an interesting chemical, in this example L-ASNase (Díaz-Barriga et al., 2021). The key findings, limitations, and existing knowledge gaps related to ASNase nano- and micro-encapsulation are well-reviewed in Villanueva-flores et al. (2021).
Site-directed mutagenesis
Site-directed mutagenesis can be used to generate EcAII variants with improved characteristics (Maggi et al., 2017). The stability, immunogenicity, and substrate specificity of L-ASNases can be significantly enhanced by using this method. According to a study by Maggi et al. (2017), Asn24 plays a critical role in maintaining EcAII active sites. For proteases that break down, asn24 serves as a cleavage point. Maggi et al. created the EcAII N24S variant through site-directed mutagenesis, which maintains asparaginase and glutaminase functions while exhibiting long-term storage stability and protease resistance (Maggi et al., 2017).
Structure-based rational design
Structure-based rational design is a potent protein engineering strategy to enhance the enzymatic capabilities of ASNases. Using bioinformatics servers (HotSpot Wizard and EVcoupling), rational design approach first screens active center residues using structural analysis, compares sequences, and identifies distal residues, allowing the construction of smart libraries for performing mutations at specific sites with less screening work (Zhou et al., 2022). Zhou et al. (2022) increased the type II ASNase from Bacillus licheniformis’s catalytic activity by using semi-rational design (Zhou et al., 2022). According to their findings, the mutant’s Km value was 1.45 mM instead of 2.33 mM, and its Kcat value was 778.87 min-1 as opposed to 197.95 min−1 in the wild type. According to Aghaeepoor et al. (2018), the N248S mutation is linked to an appropriate L-ASNase function, while disrupting glutaminase activity due to hampered interactions. They stated that in silico analysis methods would provide useful information for creating mutant enzymes (Aghaeepoor et al., 2018). Yari et al. (2019) also described the significance of computational techniques for preliminary screening of appropriate mutations.
Using in silico screening
Traditional approaches to drug discovery are mainly based on in vitro drug screening and in vivo animal experiments; however, these methods are usually expensive and laborious. In recent years, omics data have provided an opportunity for the computational prediction of anti-cancer drugs, improving the efficiency of drug discovery (Baral et al., 2020; Li et al., 2020). Over time, there has been significant improvement in bioinformatics databases and methods used for drug screening, structural design, and immunogenicity prediction. This offers an opportunity to find anticancer medications that are less expensive and time consuming. Li et al. (2020) reviewed the databases and computational tools available for creating new cancer treatment approaches.
The basis of enzyme catalysis is the binding energy, which decreases the activation energy and circumvents the adverse entropic conditions necessary for the correct alignment of the enzyme and its substrates. Free energy was used to measure the binding energy, which is the energy generated when a substrate forms a weak connection with an enzyme’s active site (Delta G). It is challenging to calculate this energy experimentally; therefore, an in silico method utilizing docking software would be more effective (Baral et al., 2020).
According to research by Baral et al. (2020), asnB genes from Streptomyces griseus, Streptomyces venezuelae, and Streptomyces collinus have higher binding energies than those from E. coli and E. chrysanthemi and were predicted to have the lowest Kms because binding energy and Km are inversely correlate 20. As-Suhbani and Bhosale (2020) demonstrated in a different investigation that the interactions between the ligand L-asparagine and Fusarium solani CLR-36 L-asparaginase occurred at the active site with good binding energy (−6.85 kcal/mol; As-Suhbani and Bhosale, 2020).
Numerous studies have addressed the use of bioinformatics tools to analyze immune responses using in silico methods. Using the prediction tool EMBOSS antigenic explorer, Abdelrazek et al. (2019) identified possible antigenic areas of the L-asparaginase sequence of Bacillus licheniformis and compared them to those of E. coli and E. chrysanthemi. For the L-asparaginases of E. coli, E. chrysanthemi, and Bacillus licheniformis, 18, 16, and 17 antigenic areas, respectively, were discovered (Abdelrazek et al., 2019). Using a bioinformatics-based methodology, Belén et al. (2021) also predicted the immunogenicity of L-ASNases from nine filamentous fungi and compared the outcomes with those of E. coli L-ASNase (Belén et al., 2021).
Using recombinant DNA technology
Although hypersensitivity reactions frequently prevent their use, bacterial L-asparaginase has long been a crucial component of ALL treatment. For individuals with hypersensitivity, alternative asparaginase preparations are required to guarantee asparaginase availability. Recombinant technology has the potential to fill this unmet need by engineering cells to produce recombinant asparaginase with higher recovery, lower immunogenicity, and cheaper production costs (Vimal and Kumar, 2017), as well as by enhancing the characteristics of numerous commercially significant enzymes (Shakambari et al., 2018). Maese et al. (2021) described a recombinant Erwinia asparaginase (JZP-458) with no immunological cross-reactivity with E. coli-derived asparaginases in vitro. It was derived from a new Pseudomonas fluorescens production platform. The US Food and Drug Administration approved JZP-458 (Rylaze) in June 2021 for the treatment of ALL in patients younger than 1 month old who exhibited hypersensitivity to asparaginase produced by E. coli (Maese et al., 2021). The process of cloning the L-asparaginase gene from an organism into E. coli has been attempted numerous times with success. Table 2 presents the results of a few of these attempts.
Author contributions
KT: Conceptualization, Writing – original draft, Writing – review & editing, Methodology, Validation, Investigation. BT: Validation, Writing – review & editing, Conceptualization, Supervision. BG: Validation, Writing – review & editing, Methodology.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1269282/full#supplementary-material
References
Abdelrazek, N. A., Elkhatib, W. F., Raafat, M. M., and Aboulwafa, M. M. (2019). Experimental and bioinformatics study for production of L-asparaginase from Bacillus licheniformis: a promising enzyme for medical application. AMB Exp 9, 1–16. doi: 10.1186/s13568-019-0751-3
Aghaeepoor, M., Akbarzadeh, A., Mirzaie, S., Hadian, A., Jamshidi Aval, S., and Dehnavi, E. (2018). Selective reduction in glutaminase activity of l'Asparaginase by asparagine 248 to serine mutation: a combined computational and experimental effort in blood cancer treatment. Int. J. Biol. Macromol. 120, 2448–2457. doi: 10.1016/j.ijbiomac.2018.09.015
Aishwarya, S. S., Iyappan, S., Lakshmi, K. V., and Rajnish, K. N. (2017). In silico analysis, molecular cloning, expression and characterization of l-asparaginase gene from Lactobacillus reuteri DSM 20016. 3 Biotech 7:348. doi: 10.1007/s13205-017-0974-4
Aishwarya, S. S., Selvarajan, E., Iyappan, S., and Rajnish, K. (2019). Recombinant L-asparaginase II from Lactobacillus casei subsp.casei ATCC 393 and its anticancer activity. Indian J. Microbiol. 59, 313–320. doi: 10.1007/s12088-019-00806-0
Alrumman, S. A., Mostafa, Y. S., Al-izran, K. A., Alfai, M. Y., and Taha, T. H. (2019). Production and anticancer activity of an L-asparaginase from Bacillus licheniformis isolated from the Red Sea, Saudi Arabia. Sci. Rep. 9:3756. doi: 10.1038/s41598-019-40512-x
Aly, N., El-Ahwany, A., Ataya, F. A., and Saeed, H. (2020). Bacillus sonorensis L. Asparaginase: cloning, expression in E. Coli and characterization. Protein J. 39, 717–729. doi: 10.1007/s10930-020-09932-x
Andrade, K. C. R., Fernandes, R. A., Pinho, D. B., de Freitas, M. M., Filho, E. X. F., Pessoa, A., et al. (2021). Sequencing and characterization of an L-asparaginase gene from a new species of penicillium section Citrina isolated from Cerrado. Sci. Rep. 11:17861. doi: 10.1038/s41598-021-97316-1
Ashok, A., Doriya, K., Rao, J. V., Qureshi, A., and Tiwari, A. K. (2019). Microbes producing L-asparaginase free of glutaminase and urease isolated from extreme locations of Antarctic soil and Moss. Sci. Rep. 9:38094. doi: 10.1038/s41598-018-38094-1
As-Suhbani, A. E., and Bhosale, H. (2020). Homology MODELING of glutaminase free l-asparaginase from FUSARIUM SOLANI CLR-36. J Adv. Sci. Res. 11, 48–54.
Baral, A., Gorkhali, R., Basnet, A., Koirala, S., and Kumar, H. (2020). Selection of the optimal L-asparaginase II against acute lymphoblastic Leukemia: an in silico approach. JMIRx Med 2, 1–22. doi: 10.2196/29844
Beckett, A., and Gervais, D. (2019). What makes a good new therapeutic L-asparaginase? World J. Microbiol. Biotechnol. 9, 1–13. doi: 10.1007/s11274-019-2731-9
Belén, L. H., Lissabet, J. F. B., Rangel-Ygaui, C. O., Monteriro, G., Pessoa, A., and Farias, J. G. (2021). Immunogenicity assessment of fungal L-asparaginases: an in silico approach. SN Appl. Sci. 2, 1–10. doi: 10.1007/s42452-020-20
Bhargavi, M., and Madhuri, R. J. (2021). Optimization, Production, purification and characterization of anti-cancer enzyme L-asparaginase from marine Sarocladium kiliense. Biosc.Biotech.res.Comm. Spec. Iss 13, 93–102. doi: 10.21786/bbrc/13.15/17
Chakraborty, M., and Shivakumar, S. (2021). Bioprospecting of the agaricomycete Ganoderma australe GPC191 as novel source for l-asparaginase production. Sci. Rep. 11, 1–8. doi: 10.1038/s41598-021-84949-5
Chakravarty, N., Singh, J., and Singh, R. P. (2021). A potential type-II L-asparaginase from marine isolate bacillus australimaris NJB19: statistical optimization, in silico analysis and structural modeling. Int. J. Biol. Macromol. 174, 527–539. doi: 10.1016/j.ijbiomac.2021.01.130
Chan, W. K., Horvath, T. D., Tan, L., Link, T., Harutyunyan, K. G., Pontikos, M. A., et al. (2019). Glutaminase activity of L-asparaginase contributes to durable preclinical activity against acute lymphoblastic leukemia. Mol. Cancer Ther. 18, 1587–1592. doi: 10.1158/1535-7163.MCT-18-1329
Chohan, S. M., and Rashid, N. (2018). Gene cloning and characterization of recombinant L-asparaginase from Bacillus subtilis strain R5. Biologia 73, 537–543. doi: 10.2478/s11756-018-0054-1
Chohan, S. M., Rashid, N., Sajed, M., and Imanaka, T. (2018). Pcal _ 0970: an extremely thermostable L-asparaginase from Pyrobaculum calidifontis with no detectable glutaminase activity. Folia Microbiol. 64, 313–320. doi: 10.1007/s12223-018-0656-6
Chohan, S. M., Sajed, M., Naeem, S., and Rashid, N. (2020). Heterologous gene expression and characterization of TK2246, a highly active and thermostable plant type l-asparaginase from Thermococcus kodakarensis. Int. J. Biol. Macromol. 147, 131–137. doi: 10.1016/j.ijbiomac.2020.01.012
Costa-Silva, T. A., Camacho-Córdova, D. I., Agamez-Montalvo, G. S., Parizotto, L. A., Sánchez-Moguel, I., and Pessoa-Jr, A. (2019). Optimization of culture conditions and bench-scale production of anticancer enzyme L-asparaginase by submerged fermentation from aspergillus terreus CCT 7693. Prep. Biochem. Biotechnol. 49, 95–104. doi: 10.1080/10826068.2018.1536990
Da Cunha, M. C., Aguilar, J. G., Lindo, M., Castro, R. J. S., and Sato, H. H. (2021). L-asparaginase from Aspergillus oryzae spp.: effects of production process and biochemical parameters. Prep. Biochem. Biotechnol. 52, 1–11. doi: 10.1080/10826068.2021.1931881
Darvishi, F., Faraji, N., and Shamsi, F. (2018). Production and structural modeling of a novel asparaginase in Yarrowia lipolytica. Int. J. Biol. Macromol. 125, 955–961. doi: 10.1016/j.ijbiomac.2018.12.162
Desai, S. S., and Hungund, B. S. (2018). Submerged fermentation, purification, and characterization of L-asparaginase from streptomyces sp. isolated from soil. J. Appl. Biol. Biotechnol. 6, 17–23. doi: 10.7324/JABB.2018.60503
Dhankhar, R., Gupta, V., Kumar, S., Kapoor, R. K., and Gulati, P. (2020). Microbial enzymes for deprivation of amino acid metabolism in malignant cells: biological strategy for cancer treatment. Appl. Microbiol. Biotechnol. 104, 2857–2869. doi: 10.1007/s00253-020-10432-2
Dias, F. F. G., Gonçalves, J., Helia, A., and Sato, H. (2019). L-asparaginase from aspergillus spp.: production based on kinetics. 3 Biotech 9, 1–10. doi: 10.1007/s13205-019-1814-5
Díaz-Barriga, C., Villanueva-Flores, F., Quester, K., Zárate-Romero, A., Cadena-Nava, R. D., and Huerta-Saquero, A. (2021). Asparaginase-phage P22 nanoreactors: toward a biobetter development for acute lymphoblastic Leukemia treatment. Pharmaceutics 13, 1–16. doi: 10.3390/pharmaceutics13050604
Dumina, M. V., Eldarov, M. A., Zdanov, D. D., and Sokolov, N. N. (2020). L-asparaginases of extremophilic microorganisms in biomedicine. Biochem. Chem. 14, 277–296. doi: 10.1134/S1990750820040046
Dumina, M., Zhgun, A., Pokrovskaya, M., Aleksandrova, S., Zhdanov, D., Sokolov, N., et al. (2021b). Highly active thermophilic L-asparaginase from Melioribacter roseus represents a novel large Group of Type II bacterial L-asparaginases from Chlorobi-Ignavibacteriae-Bacteroidetes clade. Int. J. Mol. Sci. 22:632. doi: 10.3390/ijms222413632
Dumina, M., Zhgun, A., Pokrovskaya, M., Aleksandrova, S., Zhdanov, D., Sokolov, N., et al. (2021a). A novel l-asparaginase from hyperthermophilic archaeon thermococcus sibiricus: heterologous expression and characterization for biotechnology application. Int. J. Mol. Sci. 22, 1–17. doi: 10.3390/ijms22189894
El-fakharany, E., Orabi, H., Abdelkhalek, E., and Sidkey, N. (2020). Purification and biotechnological applications of L-asparaginase from newly isolated bacillus halotolerans OHEM18 as antitumor and antioxidant agent. J. Biomol. Struct. Dyn. 40, 1–13. doi: 10.1080/07391102.2020.1851300
El-Gendy, M. M. A. A., Awad, M. F., El-Shenawy, F. S., El-Bondkly, A. M. A., and El-bondkly, A. M. A. (2021). Production, purification, characterization, antioxidant and antiproliferative activities of extracellular L-asparaginase produced by Fusarium equiseti AHMF4. Saudi J. Biol. Sci. 28, 2540–2548. doi: 10.1016/j.sjbs.2021.01.058
El-naggar, N. E., Deraz, S. F., El-ewasy, S. M., and Suddek, G. M. (2018). Purification, characterization and immunogenicity assessment of glutaminase free L-asparaginase from streptomyces brollosae NEAE-115. BMC Pharmacol. Toxicol. 19, 1–15. doi: 10.1186/s40360-018-0242-1
Farahat, M. G., Amr, D., and Galal, A. (2020). Molecular cloning, structural modeling and characterization of a novel glutaminase-free L-asparaginase from Cobetia amphilecti AMI6. Int. J. Biol. Macromol. 143, 685–695. doi: 10.1016/j.ijbiomac.2019.10.258
Fazeli, N., Alimadadi, N., and Nasr, S. (2021). Screening and optimization of process parameters for the production of L-asparaginase by indigenous fungal-type strains. Iran. J. Sci. Technol. Trans. A Sci. 45, 409–416. doi: 10.1007/s40995-020-01056-2
Fonseca, M. H. G., Fiúza, T. D. S., De Morais, S. B., De Souza, T. D. A. C. B., and Trevizani, R. (2021). Circumventing the side effects of L-asparaginase. Biomed. Pharmacother. 139:111616. doi: 10.1016/j.biopha.2021.111616
Ghasemi, A., Asad, S., Kabiri, M., and Dabirmanesh, B. (2017). Cloning and characterization of Halomonas elongata L-asparaginase, a promising chemotherapeutic agent. Appl. Microbiol. Biotechnol. 101, 7227–7238. doi: 10.1007/s00253-017-8456-5
Golbabaie, A., Nouri, H., Moghimi, H., and Khaleghian, A. (2020). L-asparaginase production and enhancement by Sarocladium strictum: in vitro evaluation of anti-cancerous properties. J. Appl. Microbiol. 129, 356–366. doi: 10.1111/jam.14623
Guimarães, M., JJM, C., Bueno, C. Z., Torres-Obreque, K. M., GVR, L., Monteiro, G., et al. (2022). Peg-grafted liposomes for L-asparaginase encapsulation. Pharmaceutics 14, 1–18. doi: 10.3390/pharmaceutics14091819
Hassan, S. W. M., Farag, A. M., and Beltagy, E. A. (2018). Purification, characterization and anticancer activity of L-asparaginase produced by marine aspergillus terreus. J Pure Appl Microbiol 12, 1845–1854. doi: 10.22207/JPAM.12.4.19
Horvath, T. D., Chan, W. K., Pontikos, M. A., Martin, L. A., du, D., Tan, L., et al. (2019). Assessment of L-asparaginase pharmacodynamics in mouse models of cancer. Metabolites 9:10. doi: 10.3390/metabo9010010
Karla, R., CMN, M., Ferraro, R. B., Moguel, I. S., Tonso, A., Lourenço, F. R., et al. (2020). Glutaminase-free L-asparaginase production by Leucosporidium muscorum isolated from Antarctic glutaminase-free L-asparaginase production by Leucosporidium muscorum. Prep. Biochem. Biotechnol. 51, 1–12. doi: 10.1080/10826068.2020.1815053
Kumar, V., Kumar, S., Darnal, S., Patial, V., Singh, A., Thakur, V., et al. (2019). Optimized chromogenic dyes—based identification and quantitative evaluation of bacterial L-asparaginase with low/no glutaminase activity bioprospected from pristine niches in Indian trans—Himalaya. 3 Biotech 9, 1–9. doi: 10.1007/s13205-019-1810-9
Li, K., Du, Y., Li, L., and Wei, D. (2020). Bioinformatics approaches for anti-cancer drug discovery. Curr. Drug Targets 21, 3–17. doi: 10.2174/1389450120666190923162203
Loch, J. I., and Jaskolski, M. (2021). Structural and biophysical aspects of l-asparaginases: a growing family with amazing diversity. IUCrJ 8, 514–531. doi: 10.1107/S2052252521006011
Maese, L., Rizzari, C., Coleman, R., Power, A., van der Sluis, I., and Rau, R. E. (2021). Can recombinant technology address asparaginase Erwinia chrysanthemi shortages? Pediatr. Blood Cancer 68, 1–7. doi: 10.1002/pbc.29169
Maggi, M., Mittelman, S. D., Parmentier, J. H., Colombo, G., Meli, M., Whitmire, J. M., et al. (2017). A protease-resistant Escherichia coli asparaginase with outstanding stability and enhanced anti-leukaemic activity in vitro. Sci. Rep. 7:4. doi: 10.1038/s41598-017-15075-4
Modi, T., and Gervais, D. (2022). Improved pharmacokinetic and pharmacodynamic profile of a novel PEGylated native Erwinia chrysanthemi L-asparaginase. Invest. New Drugs 40, 21–29. doi: 10.1007/s10637-021-01173-8
Moguel, I. S., Yamakawa, C. K., Pessoa, A., and Mussatto, S. I. (2020). L-asparaginase production by Leucosporidium scottii in a bench-scale bioreactor with co-production of lipids. Front. Bioeng. Biotechnol. 8, 1–11. doi: 10.3389/fbioe.2020.576511
Mostafa, Y., Alrumman, S., Alamri, S., Hashem, M., al-izran, K., Alfaifi, M., et al. (2019). Enhanced production of glutaminase-free L-asparaginase by marine bacillus velezensis and cytotoxic activity against breast cancer cell lines. Electron. J. Biotechnol. 42, 6–15. doi: 10.1016/j.ejbt.2019.10.001
Moubasher, H. A., Balbool, B. A., Helmy, Y. A., Alsuhaibani, A. M., Atta, A. A., Sheir, D. H., et al. (2022). Insights into asparaginase from endophytic fungus Lasiodiplodia theobromae: purification, characterization and antileukemic activity. Int. J. Environ. Res. Public Health 19, 1–14. doi: 10.3390/ijerph19020680
Nguyen, H. A., Durden, D. L., and Lavie, A. (2017). The differential ability of asparagine and glutamine in promoting the closed/active enzyme conformation rationalizes the Wolinella succinogenes L-asparaginase substrate specificity. Sci. Rep. 7, 1–14. doi: 10.1038/srep41643
Nguyen, H. A., Su, Y., Zhang, J. Y., Antanasijevic, A., Caffrey, M., Schalk, A. M., et al. (2018). A novel L-asparaginase with low L-glutaminase coactivity is highly Ef fi cacious against both T- and B-cell acute lymphoblastic Leukemias in vivo. Cancer Res. 78, 1549–1560. doi: 10.1158/0008-5472.CAN-17-2106
Prakash, P., Singh, H. R., and Jha, S. K. (2019). Production, purification and kinetic characterization of glutaminase free anti-leukemic L-asparaginase with low endotoxin level from novel soil isolate. Prep. Biochem. Biotechnol. 50, 1–12. doi: 10.1080/10826068.2019.1692221
Prakash, P., Singh, H. R., and Jha, S. K. (2020). Indicator dye based screening of glutaminase free L-asparaginase producer and kinetic evaluation of enzyme production process. Prep. Biochem. Biotechnol. 50, 803–813. doi: 10.1080/10826068.2020.1737942
Prihanto, A. A., Jatmiko, Y. D., Yanti, I., and Murtazam, M. A. (2022). Optimization of glutaminase-free L-asparaginase production using mangrove endophytic Lysinibacillus fusiformis B27 [version 2; peer review: 2 approved]. F1000Res 8, 1–18. doi: 10.12688/f1000research.21178.2
Radadiya, A., Zhu, W., Coricello, A., Alcaro, S., and Richards, N. G. J. (2020). Improving the treatment of acute lymphoblastic Leukemia. Biochemistry 59, 3193–3200. doi: 10.1021/acs.biochem.0c00354
Rigouin, C., Nguyen, H. A., Schalk, A. M., and Lavie, A. (2017). Discovery of human-like L-asparaginases with potential clinical use by directed evolution. Sci. Rep. 7:10224. doi: 10.1038/s41598-017-10758-4
Saeed, H., Hemida, A., El-Nikhely, N., Abdel-Fattah, M., Shalaby, M., Hussein, A., et al. (2020). Highly efficient Pyrococcus furiosus recombinant L-asparaginase with no glutaminase activity: expression, purification, functional characterization, and cytotoxicity on THP-1, A549 and Caco-2 cell lines. Int. J. Biol. Macromol. 156, 812–828. doi: 10.1016/j.ijbiomac.2020.04.080
Safary, A., Moniri, R., Hamzeh-mivehroud, M., and Dastmalchi, S. (2019). Highly efficient novel recombinant L-asparaginase with no glutaminase activity from a new halo-thermotolerant bacillus strain. Bioimpacts 9, 15–23. doi: 10.15171/bi.2019.03
Shakambari, G., Sameer Kumar, R., Ashokkumar, B., Ganesh, V., Vasantha, V. S., and Varalakshmi, P. (2018). Cloning and expression of L-asparaginase from Bacillus tequilensis PV9W and therapeutic efficacy of solid lipid particle formulations against cancer. Sci. Rep. 8:18013. doi: 10.1038/s41598-018-36161-1
Sharma, S., Fulke, A. B., and Chaubey, A. (2019). Bioprospection of marine actinomycetes: recent advances, challenges and future perspectives. Acta Ocean. Sin 38, 1–17. doi: 10.1007/s13131-018-1340-z
Sindhu, R., and Manonmani, H. K. (2017). Expression and characterization of recombinant L-asparaginase from Pseudomonas fluorescens. Protein Expr. Purif. 143, 83–91. doi: 10.1016/j.pep.2017.09.009
Sobat, M., Asad, S., Kabiri, M., and Mehrshad, M. (2020). Metagenomic discovery and functional validation of L-asparaginases with anti-leukemic effect from the Caspian Sea. iScience 24:101973. doi: 10.1016/j.isci.2020.101973
van Trimpont, M., Peeters, E., de Visser, Y., Schalk, A. M., Mondelaers, V., de Moerloose, B., et al. (2022). Novel insights on the use of L-asparaginase as an efficient and safe anti-cancer therapy. Cancers 14:902. doi: 10.3390/cancers14040902
Vidya, J., Sajitha, S., Ushasree, M. V., Sindhu, R., Binod, P., Madhavan, A., et al. (2017). Genetic and metabolic engineering approaches for the production and delivery of L-asparaginases: an overview. Bioresour. Technol. 245, 1775–1781. doi: 10.1016/j.biortech.2017.05.057
Villanueva-flores, F., Zárate-Romero, A., Torres, A. G., and Huerta-Saquero, A. (2021). Encapsulation of asparaginase as a promising strategy to improve in vivo drug performance. Pharmaceutics 13:1965. doi: 10.3390/pharmaceutics13111965
Vimal, A., and Kumar, A. (2017). Biotechnological production and practical application of L-asparaginase enzyme. Biotechnol. Genet. Eng. Rev. 33, 40–61. doi: 10.1080/02648725.2017.1357294
WHO (2021). World Health Organization model list of essential medicines 22nd list Geneva: World Health Organization, 1–66.
Yari, M., Eslami, M., Ghoshoon, M. B., Nezafat, N., and Ghasemi, Y. (2019). Decreasing the immunogenicity of Erwinia chrysanthemi asparaginase via protein engineering: computational approach. Mol. Biol. Rep. 46, 4751–4761. doi: 10.1007/s11033-019-04921-5
Keywords: acute lympho blastic leukemia, cancer, kinetic properties, L-asparaginase, Michaelis constant
Citation: Tsegaye K, Tsehai BA and Getie B (2024) Desirable L-asparaginases for treating cancer and current research trends. Front. Microbiol. 15:1269282. doi: 10.3389/fmicb.2024.1269282
Edited by:
Monika Prakash Rai, Motilal Nehru National Institute of Technology Allahabad, IndiaReviewed by:
Arun Karnwal, Lovely Professional University, IndiaLeonardo Teixeira Dall'Agnol, Federal University of Maranhão, Brazil
Nigel Richards, Cardiff University, United Kingdom
Copyright © 2024 Tsegaye, Tsehai and Getie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kindu Tsegaye, a2luZHUubmlicmV0QHVvZy5lZHUuZXQ=
 Kindu Tsegaye
Kindu Tsegaye Berhanu Andualem Tsehai2
Berhanu Andualem Tsehai2 Birhan Getie
Birhan Getie